Abstract
Defining the populations of tumor-initating cells that are present in tumors is a first step in developing therapeutics to target these cells. We show here that both CD44posCD24neg and CD44posCD24pos cell populations in estrogen receptor (ER) α–negative breast tumors are tumorigenic in murine xenograft models. We also describe a third population of xenograft-initiating cells (XIC) enriched in CD44posCD49fhiCD133/2hi cells that display heightened tumorigenicity, self-renewal in vivo, and the capacity to give rise to functional and molecular heterogeneity. Consistent with their capacity for self-renewal, these cells express elevated levels of Sox2, Bmi-1, and/or Nanog and their CpG islands are hypermethylated relative to nontumorigenic cells. These differences in methylome regulation may be responsible for the dramatic functional differences between the two populations. The identification of CD44posCD49fhiCD133/2hi XIC in ER-negative tumors may lead to expanded understanding of these tumors and ultimately the development of therapeutics designed to specifically target the cells.
Introduction
The cancer stem cell hypothesis states that tumors consist of a cellular hierarchy. Cancer stem cells reside at the peak of this hierarchy, and the root of the cancer, and give rise to nontumorigenic, differentiated progeny that make up the tumor bulk (1). Initially identified in leukemia (2), the identification of solid tumor cancer stem cells was first described in 2003 by Al-Hajj and coworkers in breast cancer (3). In the ensuing years, cancer stem cells have also been identified in a wide range of solid tumors, including brain, colon, lung, skin, ovary, pancreas, and head and neck (4). Herein, we refer to these cells as xenograft-initiating cells (XIC) in a deliberate effort to emphasize their primary functional characteristic, the ability to yield xenografts in immunocompromised mice.
CD49f (α6 integrin) homodimerizes with CD29 (β1 integrin) or CD104 (β4 integrin) and binds laminin, facilitating attachment of epithelial cells to the extracellular matrix. In addition to this mechanical role, CD49f cooperates with receptor tyrosine kinases to communicate, bidirectionally, between the cell and the extracellular matrix. In the normal mouse (5) and human (6) mammary gland, only CD49f-positive cells possess in vivo repopulating ability. In breast cancer, elevated CD49f expression is associated with reduced survival (7) and knockdown of its partner CD104 retards in vivo tumorigenicity (8). Although a functional role of CD133 in the stem cell phenotype seems lacking (9), this protein has been shown to mark normal stem cells of the hematopoietic system, kidney, neural tissue, skin, prostate, and pancreas (10). Furthermore, CD133 is expressed by XIC in the prostate, colon, brain, and kidney (10). In the mammary gland, however, CD133 localizes to estrogen receptor (ER)–positive luminal cells that lack in vivo repopulating capabilities (6, 11).
Herein, we show that CD44posCD49fhiCD133/2hi cells, isolated from four independent ER-negative patient tumors and xenografts, are enriched for XIC capable of giving rise to triple-negative tumors [those lacking ER, progesterone receptor (PR), and HER2] and ER-negative/HER2-positive tumors. These cells have elevated expression of genes known to play key roles in both stem cell self-renewal and cancer cell tumorigenicity. Furthermore, the methylome of CD44posCD49fhiCD133/2hi cells is divergent from that of the tumor bulk, providing a possible means by which their functional heterogeneity exists. Whereas targeted therapies for ER- and HER2-positive breast tumors have been successful (12, 13), nontargeted, cytotoxic therapies are currently the primary means of treating triple-negative tumors. The identification of triple-negative and ER-negative/HER2-positive tumor CD44posCD49fhiCD133/2hi XIC may lead to the improved understanding of these tumors and ultimately the development of targeted therapies.
Materials and Methods
Patient samples
Primary breast specimens and pleural effusion samples were collected from breast cancer patients in accordance with the National Cancer Institute (NCI) Review Board. Cells from pleural effusions were collected by centrifugation, washed twice with HBSS (Invitrogen), frozen viably in DMSO freeze media (Millipore), and frozen in liquid nitrogen until used. Tumor fragments were cut into 1-mm pieces and placed immediately into the intact abdominal fat pads of nonobese diabetic–severe combined immunodeficient (NOD-SCID) mice. Sorted or unsorted cells from pleural effusions were similarly injected.
In vivo tumorigenicity and processing of xenografts
In vivo tumorigenicity was assessed by both frequency and latency of tumor formation in the abdominal mammary gland fat pad of 4- to 8-week-old NOD-SCID mice obtained from the NCI colony (APA, Frederick, MD). All animal experiments were conducted in accord with accepted standards of humane animal care and approved by the Animal Care and Use Committee at the NIH. Five days before injection of cells, mice were administered etoposide i.p. (30 mg/kg body weight; Calbiochem) and estrogen via a subcutaneous pellet (0.72 mg β-estradiol, 90-d release; Innovative Research of America). Mice were anesthetized by an i.p. injection of ketamine/xylazine (750 and 50 mg/kg body weight, respectively) in 200 µL HBSS before surgically exposing the gland for injection. Cells were suspended in an F12 (Invitrogen)/Matrigel (high concentration; BD Biosciences) mixture (4:1) and injected into the mammary fat pad in a 50 µL volume. Tumor sizes were monitored weekly for 90 days and were not permitted to exceed 1 cm3. Xenografts were minced into <1-mm pieces and dissociated in F12 media containing collagenase type 3 (100 units/mL; Worthington Biochemical Corp.), dispase (0.8 units/mL; Invitrogen), and penicillin-streptomycin (100 units/mL; Invitrogen); incubated at 37°C with gentle agitation (250 rpm) for 90 to 120 minutes; and collected. Single cells were generated by an additional incubation in 0.05% trysin-EDTA (Invitrogen) for 5 minutes at 37°C. H&E-stained sections of mammary glands devoid of frank tumors were examined for the presence of macroscopic lesions.
Flow cytometric analysis and sorting
Antibodies were purchased from BD Pharmingen unless otherwise noted. Anti-human CD44 conjugated to allophycocyanin or phycoerythrin-cyanine 7 (PE-Cy7; eBioscience), anti-human CD24 conjugated to PE, anti-human CD49f–PE-Cy5, and anti-human CD133/2-PE (Miltenyi Biotech) were used for both analysis and live cell sorting. 7-Aminoactinomycin D (1 µg/mL final concentration; BD Pharmingen) or 4′,6-diamidino-2-phenylindole (DAPI; 0.1 µg/mL final concentration; Invitrogen) was used for live/dead cell distinction. Antibodies used for depletion of nontumor cells in pleural effusions (FITC-conjugated anti-human CD2, CD3, CD10, CD16, CD18, CD31, CD64, and CD140b) were similar to those described by Al Hajj and colleagues (3). For analysis of xenograft tumor cells, FITC-conjugated anti-mouse Ter119, CD31, CD45, and H-2Kd were used to exclude cells of mouse origin.
For flow cytometric analysis, cells were stained with antibodies in a 1× PBS solution containing 0.1% bovine serum albumin (BSA) and 0.1% sodium azide (Sigma) for 25 minutes at 4°C. Analyses were performed on either a BD FACSCalibur or BD LSR II. For live sorting, cells were stained with antibodies in a 1× PBS solution containing 0.1% fetal bovine serum and 100 units/mL penicillin-streptomycin for 25 minutes at 4°C. For dissociated xenografts, gates were established after compensation with lineageneg cells (Ter119neg, CD31neg, CD45neg, and H-2Kdneg cells) that were not exposed to anti-CD44, anti-CD49f, or anti-CD133/2 antibodies. This was done because lineagepos cells were found to have different background autofluorescence than lineageneg cells (Supplementary Fig. S1). Cell sorting was performed on a BD FACSAria operating at low pressure (20 p.s.i.) using a 100-µm nozzle. Cell clusters and doublets were electronically gated out. Post-sort analysis routinely indicated purities of >95%. Fluorescence-activated cell sorting data were analyzed using FlowJo v8.7.3 (Tree Star).
Tumor sphere formation assay
Xenografts were sorted as described above directly into 96-well, ultralow attachment plates (Corning Life Sciences) containing 50 µL of DMEM/F12 (Invitrogen) supplemented with 20 ng/mL human epidermal growth factor (PeproTech), 20 ng/mL basic fibroblast growth factor (PeproTech), 2% B27 (Invitrogen), 100 units/mL penicillin-streptomycin, and 1 µg/mL fungizone (Invitrogen). Cells were sorted in the following dilutions: 50, 250, and 500 cells per well. Cells were cultured at 37°C for 14 days. Growth factors were replenished 7 days after initiation of experiments. Tumor sphere growth was quantified microscopically under ×20 magnification.
Cytosine extension assay
Alterations in DNA methylation were evaluated via a modified cytosine extension assay (14). Briefly, duplicate tubes containing 50 ng of genomic DNA [isolated via Trizol (Invitrogen) from live sorted xenografts] were digested overnight with a 10-fold excess of HpaII or BssHII endonuclease (New England Biolabs); in addition, one tube was incubated without restriction enzyme addition and served as a nonspecific background control. The 35-µL nucleotide extension reaction [containing 50 ng of DNA, 1× PCR buffer (Invitrogen), 1.0 mmol/L MgCl2, 0.35 unit of Taq DNA polymerase (Invitrogen), and 8.0 µCi [32P]dCTP (Perkin-Elmer)] was incubated at 72°C for 2 hours. Triplicate 10-µL aliquots from each reaction were applied to DE-81 ion-exchange filters (Whatman). Unincorporated [32P]dCTP was removed with three 10-minute washes of 1.0 mol/L sodium phosphate buffer (pH 7.0). Incorporated [32P]dCTP was quantified with a Beckman LS600IC liquid scintillation counter (Beckman Coulter). [32P]dCTP that bound nonspecifically to uncut DNA was subtracted from enzyme-treated samples, and the results were expressed as relative [32P]dCTP incorporation. With this methodology, reduced [32P]dCTP incorporation is associated with hypermethylation.
Real-time reverse transcription-PCR
Total RNA was isolated from cells using the Trizol reagent and treated with amplification-grade DNase I (Invitrogen) to remove genomic contamination. RNA was then reverse transcribed using SuperScript III reverse transcriptase (Invitrogen) primed with oligo(dT) and random hexamers. cDNA was subjected to real-time PCR amplification using gene-specific primers and 2× Brilliant II SYBR Green QPCR Mastermix (Roche Applied Science). Primer sequences and PCR conditions are given in Supplementary Table S1. Data are presented using the mean ΔΔCt method.
Immunohistochemistry
Cross-sections of xenografts were formalin fixed and paraffin embedded. Five-micrometer-thick sections were deparaffinized with xylene and rehydrated with decreasing concentrations of ethanol. Antigen retrieval was achieved with citrate buffer (Dako). Endogenous peroxidase activity was blocked by a 10-minute incubation in 3% hydrogen peroxide followed by a 10-minute wash in 1× PBS. Immunostaining was performed using Vectastain ABC kit (Vector) according to the manufacturer’s instruction. Color was developed with diaminobenzidine peroxidase substrate kit (Vector), and sections were counterstained with hematoxylin (Sigma). Antibodies were obtained from the following sources: cytokeratin 5 (CK5), CK8, ER, PR (Novacastra, Leica Microsystems), p63 (Abcam), and HER2 (Dako). A breast tissue sample that had been previously determined to express high levels of the protein of interest was used as a positive control. A serial section of each sample that received all staining steps, with the exception of the primary antibody, was used as a negative control.
Immunofluorescence and confocal microscopy
Two human breast tumor tissue microarrays (U.S. Biomax, Inc.) were used for analysis; each contained 2 invasive lobular and 35 invasive ductal carcinoma specimens and 3 normal breast samples. One array was stained with the antibodies of interest, whereas the other was processed appropriately as a control. Arrays were briefly fixed in 100% methanol, washed, and then blocked in 1× PBS containing 10% goat serum and 2% BSA (blocking buffer) for 30 minutes at room temperature. Arrays were then incubated with a 1:50 dilution of the primary antibodies in 2% BSA and 2% goat serum sequentially as follows: overnight at 4°C with anti-CD44 (rabbit polyclonal; Abcam), 1 hour at room temperature with anti-CD49f (rat monoclonal; Affinity BioReagents, Thermo Fisher Scientific), and 1 hour at room temperature with anti-CD133/2 (mouse monoclonal; Miltenyi Biotech). Each slide was subsequently washed with 1× PBS and then incubated with appropriate secondary antibody (anti-rabbit Alexa Fluor 594, anti-rat Alexa Fluor 647, or anti-mouse Alexa Fluor 488; Invitrogen) at a 1:1,000 dilution in blocking buffer for 30 minutes at room temperature. Coverslips were mounted using Prolong Gold antifade reagent with DAPI (Invitrogen). Imaging was performed using the Carl Zeiss LSM510 confocal imaging system (Carl Zeiss MicroImaging) at ×63 magnification.
Statistics
ANOVA was performed using StatView 5.0.1 (SAS Institute).
Results
Tumorigenicity of CD44posCD24pos and CD44posCD24neg/low cells from ER-negative tumors
Breast XICs were previously identified as CD44posCD24neg/low (3). We set out to expand on this seminal work by identifying novel markers capable of further enriching XIC from the CD44posCD24neg/low population of ER-negative breast tumors. Xenografts were established with pleural effusions by injecting sorted or unsorted cells into the fat pad of NOD-SCID mice. Ten of 14 pleural effusions with unknown ER status failed to engraft (data not shown). Xenografts were characterized for the expression of ER, PR, HER2, CK5, CK8, and p63 by immunohistochemistry. All xenografts possessed basal/myoepithelial markers (CK5 and p63) and were ER/PR negative. Two expressed elevated levels of the HER2 oncoprotein (Supplementary Table S2).
Whereas CD44neg cells were largely nontumorigenic, experiments evaluating the tumorigenicity of CD44posCD24pos and CD44posCD24neg/low cells sorted from ER-negative tumors indicated that both populations were tumorigenic (Table 1). Specifically, 250 cells (both CD44posCD24pos and CD44posCD24neg/low) were frequently able to form tumors. Furthermore, CD44posCD24pos cells readily gave rise to CD44posCD24neg/low progeny and vice versa (Supplementary Fig. S2A). Pleural effusions SH1 and SH12 were the notable deviations from this observation. In the case of pleural effusion SH1, 1,000 CD44posCD24pos pleural cells were tumorigenic, whereas an equal number of CD44posCD24neg/low cells were not (Table 1; Supplementary Fig. S2B). SH12 CD44posCD24neg/low pleural cells were tumorigenic, whereas all other cells failed to yield a xenograft, consistent with observations made by Al-Hajj and colleagues (3). However, after one passage in vivo, SH12 XIC acquired CD24 expression. Specifically, both CD44posCD24neg/low and CD44posCD24pos cells from the passage 1 SH12 xenograft were tumorigenic (Table 1; Supplementary Fig. S2C). The purity of post-sorted cells routinely approached or exceeded 95% (Supplementary Fig. S3). Taken together, these data strongly suggest that in ER/PR-negative tumors, XICs are not always restricted to the CD44posCD24neg/low population.
Table 1.
Summary of in vivo tumorigenicity of six pleural effusions and xenografts sorted into CD44posCD24neg/dim and CD44posCD24pos populations
| Tumor | Tumors/injection |
||
|---|---|---|---|
| 250 | 500 | 1,000 | |
| SH5 xenograft | |||
| CD44posCD24pos | 2/4 | 2/2 | |
| CD44posCD24neg/dim | 3/4 | 4/4 | |
| CD44neg | 0/2 | 0/3 | |
| SH12 pleural effusion | |||
| CD44posCD24pos | 0/4 | 0/6 | |
| CD44posCD24neg/dim | 2/6 | 2/4 | |
| SH12 xenograft | |||
| CD44posCD24pos | 1/4 | 4/4 | |
| CD44posCD24neg/dim | 3/5 | 1/2 | |
| SH1 pleural effusion | |||
| CD44posCD24pos | 4/4 | ||
| CD44posCD24neg/dim | 0/3 | ||
| CD44neg | 0/5 | ||
| SH1 xenograft | |||
| CD44posCD24pos | 1/5 | ||
| CD44neg | 1/5 | ||
| 012 xenograft | |||
| CD44posCD24pos | 1/5 | ||
| CD44 pos CD24 neg/dim | 1/5 | ||
| CD44neg | 0/5 | ||
CD44posCD49fhiCD133/2hi cells are enriched for tumor-forming ability
After confirming that CD44neg cells are largely nontumorigenic, CD44pos cells were evaluated to further segregate this population into a tumorigenic and nontumorigenic fraction. With the tumors described above, CD44pos cells were evaluated for the expression of CD29, SSEA4, CD49f, CD133/2, TDGF1, CD200, CD117, CD135, or CD205. All markers, with the exception of CD205, were initially selected because they had previously been associated with normal stem cells and/or XIC (6, 10, 15–19). CD205 was chosen based on unpublished data suggesting it may decorate XIC. These populations were then sorted and injected into the abdominal mammary fat pad of NOD-SCID mice to evaluate the ability of each marker to segregate out a tumorigenic and nontumorigenic fraction from the CD44pos population. The expression of CD29, SSEA4, and CD205 failed to distinguish XIC from non-XIC (Supplementary Table S3). Likewise, we failed to detect cells positive for other candidate markers, including TDGF1, CD200, CD117, or CD135 (data not shown). Ultimately, CD44posCD49fhi cells were more tumorigenic than CD44posCD49fneg/low cells and CD44posCD133/2hi cells were more tumorigenic than CD44posCD133/2neg/low cells (Table 2). However, in both cases, the CD44posCD49fneg/low and CD44posCD133/2neg/low fractions retained considerable capacity for xenograft formation. Using the sorting scheme in Fig. 1, CD44posCD49fhiCD133/2hi and CD44posCD49fneg/lowCD133/2neg/low populations were sorted and injected into the mammary fat pad. Adding a third marker had mixed results in further enriching XIC activity, but CD44posCD49fneg/lowCD133/2neg/low cells were consistently less tumorigenic than CD44posCD49fneg/low or CD44posCD133/2neg/low cells (Table 2). Across all tumors evaluated, CD44posCD49fhiCD133/2hi cells were enriched for XIC relative to CD44posCD49fneg/lowCD133/2neg/low cells, which were largely depleted of this population. Despite this commonality, variation in the tumorigenicity of CD44posCD49fhiCD133/2hi cells exists across samples tested, with SH12 being most tumorigenic. Likewise, SH12 CD44posCD49fneg/lowCD133/2neg/low cells retained some capacity for forming xenografts, whereas this population sorted from other tumors was nontumorigenic.
Table 2.
Tumor-initiating ability of CD44pos-CD49fhiCD133/2hi and CD44posCD49fneg/low-CD133/2neg/low cells
| Tumor, population | Tumor/injectison |
||
|---|---|---|---|
| 50 | 250 | 2,500 | |
| 012 xenograft | |||
| CD44posCD49fhiCD133/2hi | 0/5 | 2/5 | |
| CD44posCD49fneg/lowCD133/2neg/low | 0/5 | 0/5 | |
| CD44posCD49fhi | 1/3 | ||
| CD44posCD49fneg/low | 0/5 | ||
| CD44posCD133/2hi | 0/5 | ||
| CD44posCD133/2neg/low | 1/5 | ||
| SH5 xenograft | |||
| CD44posCD49fhiCD133/2hi | 0/5 | 7/10 | 3/4 |
| CD44posCD49fneg/lowCD133/2neg/low | 0/5 | 0/10 | 0/5 |
| CD44posCD49fhi | 9/9 | ||
| CD44posCD49fneg/low | 1/8 | ||
| CD44posCD133/2hi | 4/10 | ||
| CD44posCD133/2neg/low | 2/10 | ||
| SH1 xenograft | |||
| CD44posCD49fhiCD133/2hi | 3/5 | 4/5 | |
| CD44posCD49fneg/lowCD133/2neg/low | 0/5 | 0/5 | |
| SH12 xenograft | |||
| CD44posCD49fhiCD133/2hi | 3/6 | 4/4 | |
| CD44posCD49fneg/lowCD133/2neg/low | 0/6 | 1/4 | |
| CD44posCD49fhi | 2/5 | ||
| CD44posCD49fneg/low | 4/5 | ||
| CD44posCD133/2hi | 9/10 | ||
| CD44posCD133/2neg/low | 4/9 | ||
| SH12 pleural effusion | |||
| CD44posCD49fhiCD133/2hi | 3/6 | 4/4 | |
| CD44posCD49fneg/lowCD133/2neg/low | 0/6 | 1/5 | |
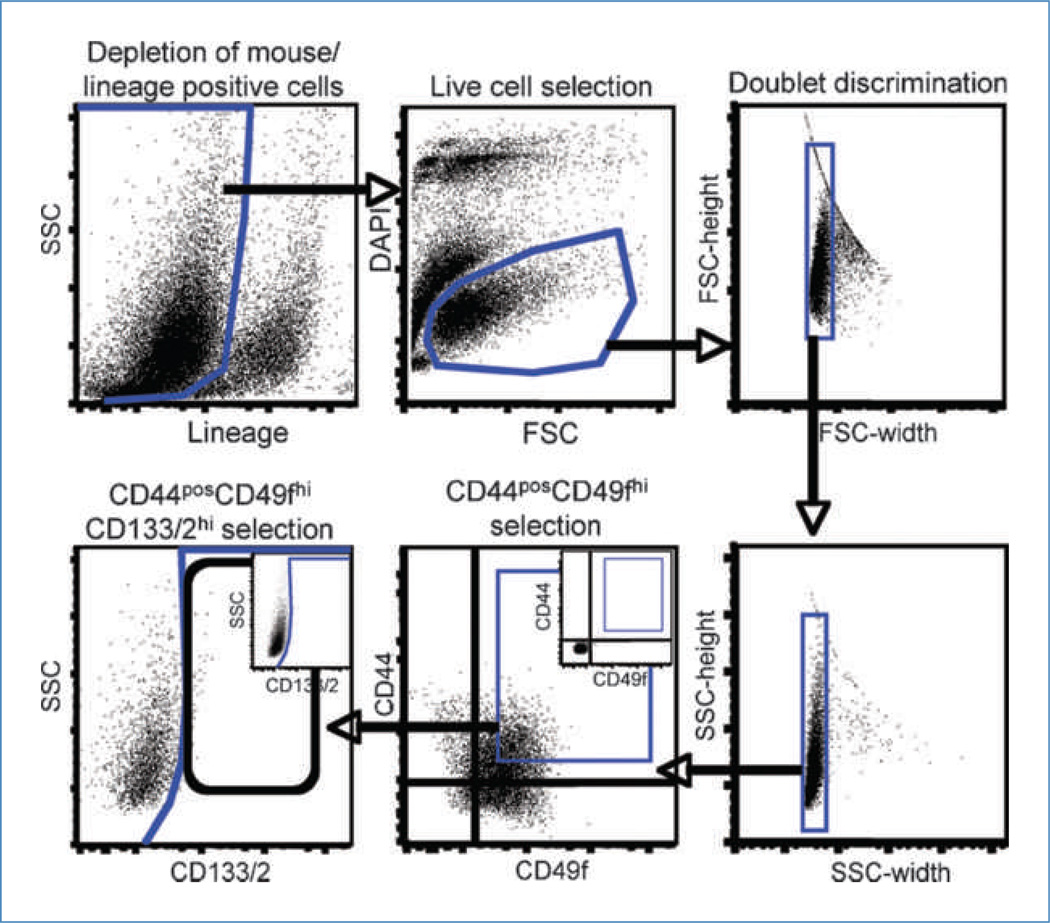
Figure 1.
Sorting strategy used for the isolation of CD44posCD49fhiCD133/2hi cells. Cells were depleted of nontumor cells from pleural effusions or mouse cells from xenografts by negative selection as described in Materials and Methods. Live cells were identified by their ability to exclude DAPI and discriminated from debris by their forward scatter (FSC) versus DAPI profile. Following depletion of doublets, CD44posCD49fhi cells were plotted by their side scatter (SSC) versus CD133/2 profile permitting identification of CD133/2hi cells. Gates were established (insets of CD44 versus CD49f and CD133/2 versus SSC) after compensation with live, lineage-negative cells not exposed to CD44, CD49f, or CD133/2 antibodies.
CD44posCD49fhiCD133/2hi cells give rise to immunophenotypic and functional heterogeneity
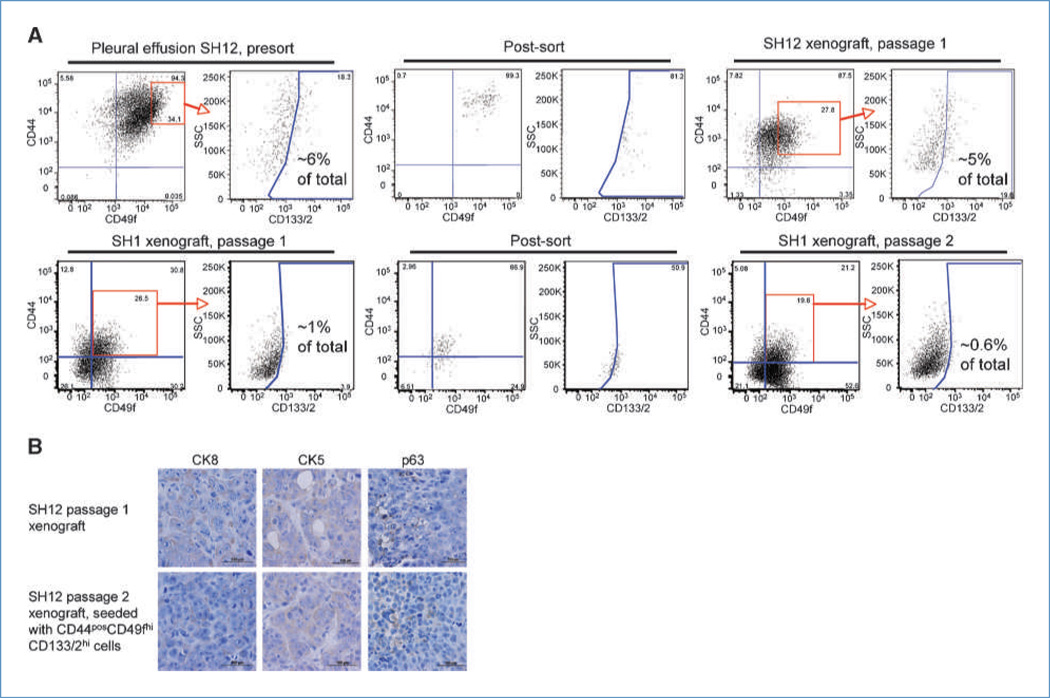
Xenografts initiated with CD44posCD49fhiCD133/2hi cells were dissociated and evaluated for their expression of CD44, CD49f, and CD133/2. In all cases, the immunophenotype of the xenograft was similar to that of the parental tumor (Fig. 2A; Supplementary Figs. S4 and S5; unstained gating controls in Supplementary Fig. S6). This was observed in both passage 1 xenografts initiated with sorted pleural cells and in xenografts passaged as many as six times. Xenografts resulting from CD44posCD49fhiCD133/2hi cells were also the same molecular subtype as the parental tumor. Specifically, the expression level and distribution of CK8, CK5, and p63 (Fig. 2B) as well as ER, PR, and HER2 (data not shown) were similar between the parental tumor from which CD44posCD49fhiCD133/2hi cells were derived and the resulting xenografts.
Figure 2.
CD44posCD49fhiCD133/2hi cells recapitulate the immunohistochemical heterogeneity of the parental tumor. A, expression profile of CD44, CD49f, and CD133/2 in a pleural effusion (top) and xenograft (bottom) and their xenografts resulting from sorted CD44posCD49fhiCD133/2hi cells. B, tumor tissues from passage 1 SH12 xenograft and passage 2 SH12 xenograft initiated with CD44posCD49fhiCD133/2hi cells were subjected to immunohistochemical analysis with the specific antibodies indicated.
CD44posCD49fhiCD133/2hi cells also expanded in vivo, giving rise to functional heterogeneity. Specifically, these XICs expanded via self-renewal and gave rise to nontumorigenic progeny. Furthermore, this ability was maintained over multiple in vivo passages. In the case of passage 5 xenograft SH5, CD44posCD49fneg/lowCD133/2neg/low cells were nontumorigenic, whereas CD44posCD49fhiCD133/2hi cells gave rise to passage 6 xenografts. Likewise, passage 6 CD44posCD49fhiCD133/2hi cells formed passage 7 xenografts that contained nontumorigenic CD44posCD49f neg/lowCD133/2neg/low cells (Supplementary Table S4).
In addition to giving rise to immunophenotypic and functional heterogeneity, CD44posCD49fhiCD133/2hi cells were enriched for the ability to grow in suspension culture. With all three xenografts evaluated, CD44posCD49fhiCD133/2hi cells were more efficient at forming tumor spheres than CD44posCD49fneg/lowCD133/2neg/low cells (P < 0.05; Supplementary Fig. S7). The number of cells sorted into each well did not affect the frequency of tumor sphere formation (data not shown). Despite differences between the populations within a xenograft, there were marked differences among xenografts in the efficiency with which CD44posCD49fhiCD133/2hi cells formed tumor spheres. Whereas ~2% of SH5 CD44posCD49fhiCD133/2hi cells had the capacity to grow in suspension, <0.5% ofSH1 and SH12 CD44posCD49fhiCD133/2hi cells had this ability. Whereas some CD44posCD49fneg/lowCD133/2neg/low cells grew in suspension, the resulting spheres were typically smaller than those formed by CD44posCD49f hi-CD133/2hi cells (Supplementary Fig. S7).
Molecular heterogeneity in ER-negative tumors
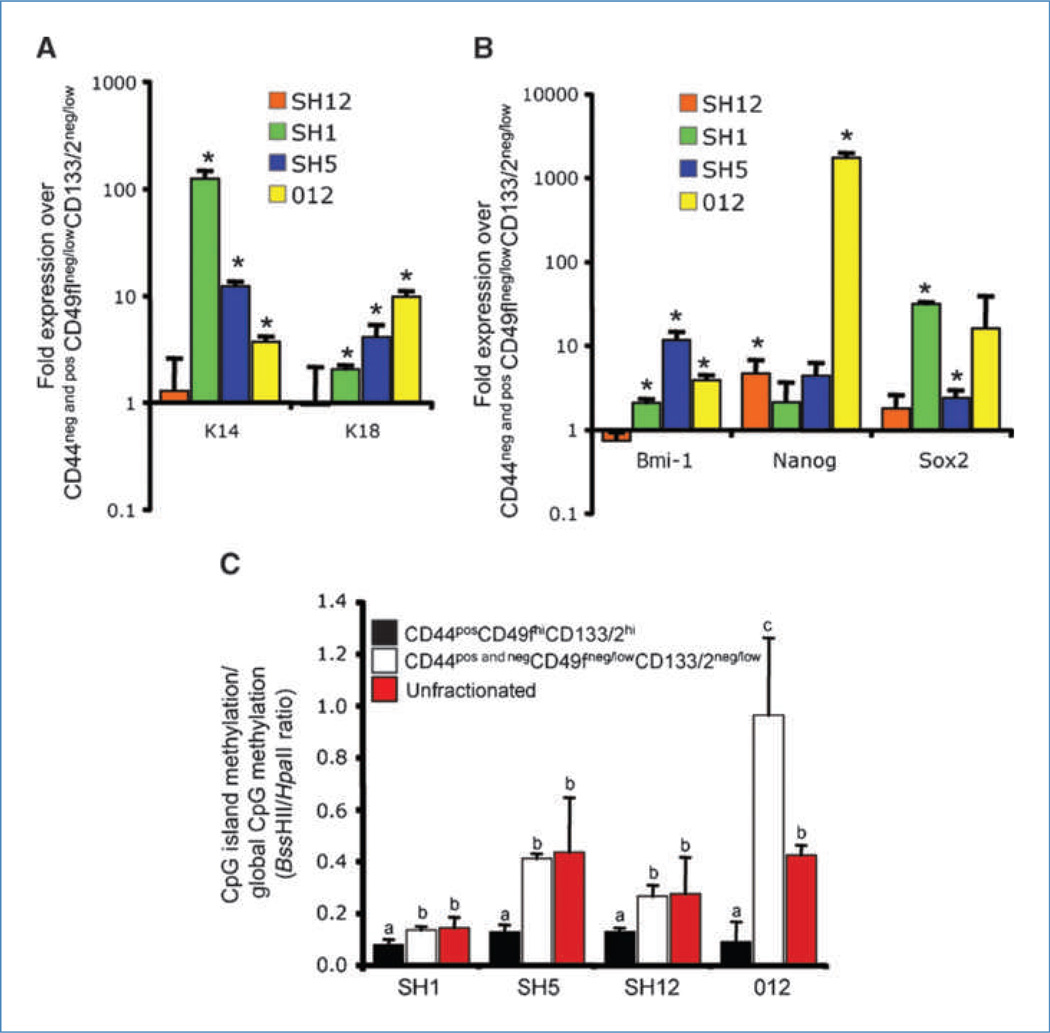
Data presented above show that CD44posCD49fhiCD133/2hi cells represent a functionally unique population. Quantitative real-time reverse transcription-PCR was used to determine if these cells also possessed divergent transcript abundance of specific genes. Relative to CD44negCD49fneg/lowCD133/2neg/low cells and CD44posCD49fneg/lowCD133/2neg/low cells (hereon referred to as CD44pos and negCD49fneg/lowCD133/2neg/low cells), CD44posCD49f hiCD133/2hi cells had elevated levels of the luminal marker K18 and the basal marker K14 (Fig. 3A). Additionally, these cells expressed elevated levels of genes critical for self-renewal (Bmi-1, Nanog, and Sox2; Fig. 3B). These data show that key transcripts associated with a primitive cell phenotype were upregulated in the CD44posCD49fhiCD133/2hi population. Differences in cell function and transcript abundance between CD44posCD49fhiCD133/2hi and CD44pos and neg-CD49fneg/lowCD133/2neg/low cells suggest that differential transcriptional regulation may exist between these two populations. A means by which these differences could arise is through divergent CpG methylation patterns. The cytosine extension assay (14) was used to evaluate the methylation status of CpG dinucleotides of sorted cells and unfractionated xenografts. This assay takes advantage of the methylation-sensitive restriction enzymes HpaII and BssHII and the incorporation of [32P]dCTP at the guanine overhang resulting from restriction enzyme activity. With this methodology, reduced [32P]dCTP incorporation is associated with hypermethylation. Whereas HpaII cleaves unmethylated CpG sites scattered randomly throughout the genome, BssHII sites are enriched in CpG islands (14).
Figure 3.
Molecular heterogeneity within tumors. A, transcript abundance of K14 and K18. Within each tumor, mRNA abundance in CD44pos CD49fhiCD133/2hi cells is expressed as a fold change relative to CD44pos and negCD49fneg/low CD133/2neg/low cells. *, P < 0.05. B, transcript abundance of Bmi-1, Nanog, and Sox2. Within each tumor, mRNA abundance in CD44posCD49fhiCD133/2hi cells is expressed as a fold change relative to CD44pos and negCD49fneg/low CD133/2neg/low cells. *, P < 0.05. C, ratio of promoter CpG methylation to global CpG methylation in sorted and unfractionated tumors. Histogram bar height corresponds to the ratio of cutting by methylation-sensitive restriction enzymes BssHII and HpaII. Lower restriction enzymatic activity is associated with hypomethylation. a, b, and c, within tumor, values with dissimilar letters are different at P < 0.05.
Among all four xenografts, the ratio of CpG island methylation to global CpG methylation (BssHII to HpaII ratio; hereon referred to as island to global CpG methylation index) was lower for CD44posCD49fhiCD133/2hi cells than both CD44pos and negCD49fneg/lowCD133/2neg/low cells and the unfractionated xenograft (P < 0.05; Fig. 3C). The lower island to global CpG methylation index was attributed largely to hypermethylation (P = 0.054) of CpG islands of CD44posCD49fhiCD133/2hi cells relative to CD44pos and negCD49fneg/lowCD133/2neg/low cells (data not shown). These data indicate that the CD44posCD49fhiCD133/2hi DNA methylome is unique from that of nontumorigenic cancer cells.
Localization of cells positive for CD44, CD49f, and CD133/2 in normal and cancerous breast tissue
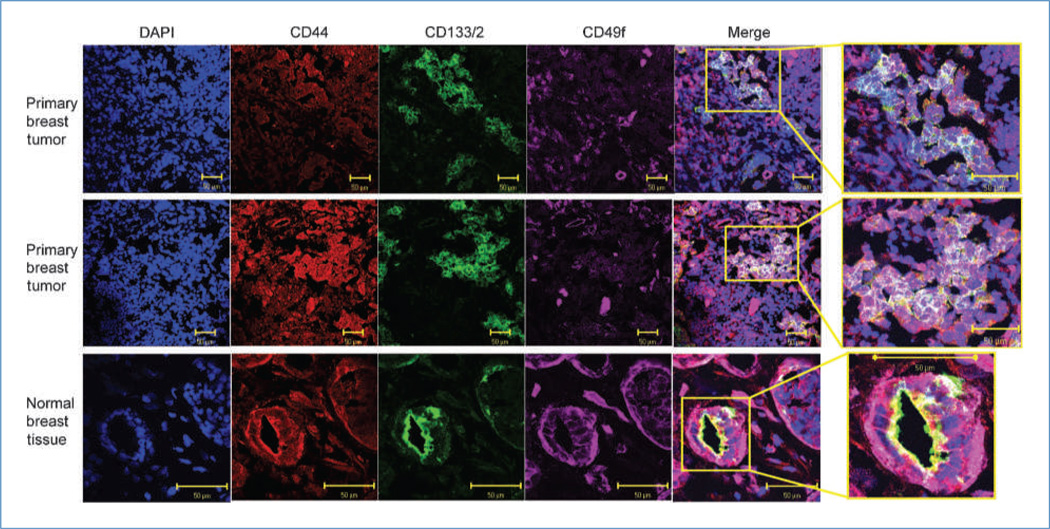
We showed above that CD44posCD49fhiCD133/2hi cells represent a unique, highly tumorigenic population of cells. Next, we set out to evaluate the localization of these cells in primary, ER-negative breast tumors and normal breast tissue using confocal microscopy. A high percentage of tumor cells expressed CD44 (red) and had some degree of reactivity to CD49f (Fig. 4, purple). A smaller fraction of primary tumor cells stained positive for CD133 (green). Merging of confocal images permitted visualization of cells positive for all three antigens (white). In tumors, CD44posCD49fhiCD133/2hi cells were primarily observed in unorganized clusters and only rarely as a single cell. In the normal breast, CD44posCD49fhiCD133/2hi cells were very abundant and were frequently found to be the polarized cells that make up the lobular/alveolar subunit, with CD133/2 marking the apical side and CD49f marking the basolateral side of the cells (Fig. 4; Supplementary Fig. S8A). This highly organized localization pattern is in stark contrast to that observed in primary ER-negative breast tumor tissue. In an effort to confirm whether the structures evaluated in Fig. 4 were indeed mammary epithelium, reduction mammoplasties were queried for the expression of CK8 and CK5. As seen in Supplementary Fig. S8B, normal epithelial structures reminiscent of those seen in Fig. 4 and Supplementary Fig. S8A stained positive for the mammary epithelial keratins CK8 and CK5. Negative controls for confocal images are provided in Supplementary Fig. S9.
Figure 4.
Localization of CD44, CD49f, and CD133/2 in primary ER-negative breast tissue and normal breast tissue. Confocal microscopy images of representative patient tumors and a normal breast sample illustrating localization of DNA (DAPI; blue), CD44 (red), CD133/2 (green), and CD49f (purple). In the merged images, cells simultaneously positive for all three markers appear white.
Discussion
The cancer stem cell hypothesis suggests that cancers are made up of a cellular hierarchy, with the tumor bulk consisting of differentiated cells that lack self-renewal mechanisms. Instead, the ability to self-renew (and conceivably seed distant metastases) resides exclusively in a population of cancer stem cells. The first characterization of solid tumor XIC was described by Al-Hajj and colleagues (3). Subsequently, the prospective identification of XIC has been described in a broad range of solid tumors (reviewed in ref. 4).
Here, we provide the first evidence that the XIC immunophenotype of ER-negative breast cancer is rarely CD44posCD24neg. Specifically, using four triple-negative and ER-negative/HER2-positive tumors, we found that XICs were equally dispersed between the CD44posCD24neg and CD44posCD24pos populations (Table 1). Similarly, we recently reported that CD44posCD24neg and CD44posCD24pos populations sorted from breast cancer cell lines are equally tumorigenic (20).
Whereas XICs reside in both CD44posCD24neg and CD44posCD24pos populations in ER-negative tumors, we found that CD44neg cells are largely nontumorigenic (Table 1), a finding consistent with that reported by Al Hajj and colleagues (3). CD44 expression is known to support survival, invasiveness, and tumorigenicity of cancer cells, and depletion of the protein ablates these phenotypes (21–25). Given the dominant phenotype associated with CD44 negativity, we chose to identify markers that could segregate CD44pos cells into a tumorigenic and nontumorigenic population. This work led to our observation that the tumorigenic capacity of ER-negative breast cancer is enriched in the CD44posCD49fhiCD133/2hi population relative to CD44posCD49fneg/lowCD133/2neg/low cells, which were largely nontumorigenic. In addition to enriching for tumorigenic cells, this population also possesses other hallmarks of XIC, including the capacity to form tumor spheres in vivo and give rise to functional and molecular heterogeneity. Consistent with their ability to self-renew in vivo, CD44posCD49fhiCD133/2hi cells expressed elevated levels of Sox2, Bmi-1, and Nanog, transcription factors known to play key roles in the self-renewal process. The relationships between these factors and transforming growth factor β (TGFβ) signaling (the latter is upregulated in CD44posCD24neg/dim breast cancer cells; ref. 26) are inconsistent and seem to be cell type dependent (27–29).
The CD44posCD49fhiCD133/2hi population obtained from two pleural effusions and four xenografts consistently enriched for cells capable of initiating xenografts in mouse mammary fat pads. Across pleural effusions and xenografts, however, there were marked differences in the tumorigenicity of the CD44posCD49fhiCD133/2hi population. Specifically, SH12 CD44posCD49fhiCD133/2hi cells were between 5- and 6-fold more tumorigenic than those cells sorted from SH5, SH1, and 012 xenografts. Similar differences were observed in the frequency of tumor sphere–forming efficiency. These data show that CD44posCD49fhiCD133/2hi cells remain a highly heterogeneous population. Similar heterogeneity in tumorigenicity across xenografts was observed in the CD44posCD49fneg/lowCD133/2neg/low population. Specifically, these cells from SH5, SH1, and 012 xenografts were nontumorigenic, whereas those from SH12 pleural effusion and xenograft possessed a very limited capacity to form xenografts.
Identification of the cell population targeted for transforming mutations and ultimately becoming a XIC is of great interest. Given their shared capacity for unlimited self-renewal, fewer accumulated mutations would presumably be required to transform a normal stem cell into a cancer stem cell or XIC than would be required for a differentiated cell. Indeed, in the case of most acute myeloid leukemias, hematopoietic progenitors, as opposed to more differentiated cells, are the target of mutations leading to the disease (2). CD49f positivity has been associated with primitive cells uniquely possessing in vivo mammary gland repopulating ability in both mice (5) and humans (6). Stem cells of multiple mouse and human organs express CD133 (10). In the breasts of both mice (11) and humans (6), however, its CD133 expression is limited to ER-positive, differentiated luminal cells devoid of in vivo repopulating capability (6, 11). Indeed, in the normal human mammary gland, we report that CD133-positive cells are frequently polarized and reside in the luminal/alveolar structure (Fig. 4). Given its localization in the normal breast and the fact that the ER-negative XICs described herein express CD133 support the hypothesis that they may originate from luminal-committed cells (6).
The observation that a differentiated cell can be the target of mutations yielding a XIC phenotype is not unprecedented. Specifically, differentiated populations are targets of transformation in both the hematopoietic system (30) and prostate (31). More recently, Lim and colleagues (6) showed that basal-like breast tumors likely originate from luminal progenitors as opposed to the more primitive population enriched for in vivo repopulating ability. These findings support the hypothesis that normal CD44posCD49fhiCD133/2hi cells, once transformed, become enriched with a XIC population. In addition to the acquisition of unlimited self-renewal capabilities, the normal, luminal CD44posCD49fhiCD133/2hi cell must lose ER expression on its journey to becoming a XIC capable of giving rise to ER-negative tumors. Indeed, the fact that the ER promoter becomes hypermethylated in ER-negative breast cancer is well established (32). An alternative hypothesis is that normal cells acquire the CD44posCD49fhiCD133/2hi immunophenotype on transformation, independent of their previous marker profile.
It is understood that cancer has both a genetic and epigenetic underpinning. Relative to normal tissue, CpG islands of tumor cells tend to be hypermethylated (33). We show that a similar relationship exists between nontumorigenic cells and XIC isolated from the same tumor. Specifically, relative to the tumor bulk and CD44pos and negCD49fneg/lowCD133/2neg/low cells, the island to global CpG methylation ratio was lower in CD44posCD49fhiCD133/2hi cells due to hypermethylation in the CpG islands of XIC. It is plausible that these differences in the epigenetic landscape between XIC and nontumorigenic cells lie at the heart of their functional differences. This is supported by the role DNA methylation plays in human embryonic stem cell differentiation (34). Furthermore, TGFβ signaling in ER-positive breast XIC is under epigenetic control (35) and tumor suppressor expression is frequently repressed in cancer by CpG island methylation (33, 36). We are currently comparing the methylomes of CD44pos and neg CD49fneg/low CD133/2neg/low and CD44POS-CD49fhiCD133/2hi cells to identify specific genes with divergent methylation patterns.
Consistent with their in vivo self-renewal capability, CD44posCD49fhiCD133/2hi cells have elevated levels of Nanog, Sox2, and/or Bmi-1. In addition to their well-described role in stem cell self-renewal, these genes play key roles in tumorigenicity and the XIC phenotype. The polycomb group gene Bmi-1 maintains self-renewal capabilities by suppression of Ink4a/ARF cell cycle inhibitory proteins in neural stem cells (37) and similarly regulates cell cycle inhibitory proteins in cancer (38). Nanog and Sox2 collaborate to maintain self-renewal in embryonic stem cells (39), and the depletion of either gene results in the loss of cancer cell tumorigenicity (40, 41). Together, these data support our observations that CD44posCD49f hiCD133/2hi cells are XIC with unlimited self-renewal capabilities. Additionally, we observed that CD44posCD49fhiCD133/2hi cells expressed elevated levels of the basal marker K14 and the luminal marker K18. This could be attributed to high levels of heterogeneous K14 and K18 expression by CD44posCD49fhiCD133/2hi cells or simultaneous expression of both markers. The simultaneous expression of two or more markers of divergent lineages has been observed in hematopoietic stem cells (42) and has been postulated to occur in mammary stem cells (43). Therefore, the coexpression of both luminal and basal lineage markers would be consistent with the primitive nature of transformed CD44posCD49fhiCD133/2hi cells.
The cancer stem cell hypothesis has garnered considerable research interest; however, it remains controversial. Among the concerns is the profound host/cancer cell interaction associated with melanoma xenograft models (44). Additionally, Shipitsin and colleagues (26) showed in primarily ER-positive breast cancer that, although CD44negCD24pos cells were related to CD44posCD24neg cells, they were not clones. The latter findings do not disprove the cancer stem cell hypothesis but instead suggest that tumors experience continued clonal evolution beyond the appearance and expansion of the first cancer stem cells/XIC. Although definitive evidence confirming the cancer stem cell hypothesis in its purest incarnation is lacking, the concept of functional and molecular heterogeneity in solid tumors is irrefutable. Identifying therapeutic approaches that take advantage of this heterogeneity is an expanding area of research (45–47).
Targeted therapies for ER-positive breast tumors have been highly successful (12). To a lesser degree, the humanized monoclonal antibody trastuzumab has been useful in the treatment of tumors overexpressing the oncoprotein HER2 (13). Although targeted therapies including poly (ADP-ribose) polymerase inhibitors show promise (48), to date, nontargeted, cytotoxic therapies are the primary option for triple-negative tumors. The true test of these markers awaits a larger cohort of ER-negative tumors with full clinical data and follow-up. However, the CD44posCD49fhiCD133/2hi marker profile may lead to the improved understanding of these tumors and ultimately the development of drugs designed to specifically target them.
Supplementary Material
Acknowledgments
We thank Barbara Taylor and Subhadra Banerjee of the Center for Cancer Research Flow Cytometry Core for their expert advice and patience, Max Bush and Mustapha Ali for their assistance with the xenograft experiments, and Sarah Lieber and Stefanie Alexander for their assistance with immunohistochemistry.
Grant Support
Center for Cancer Research, an Intramural Research Program of the NCI, and Breast Cancer Research Stamp proceeds awarded through competitive peer review.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
M.J. Meyer developed ideas, conceived the experiments, and wrote the manuscript. M.J. Meyer, J.M. Fleming, A.F. Lin, S.A. Hussnain, and E. Ginsburg conducted the experiments. J.M. Fleming and E. Ginsburg edited the manuscript. B.K. Vonderhaar developed ideas, conceived the experiments, and edited the manuscript. All authors contributed to the analysis of data.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 2.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–7. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 3.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–68. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 5.Stingl J, Eirew P, Ricketson I, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–7. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 6.Lim E, Vaillant F, Wu D, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–13. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 7.Friedrichs K, Ruiz P, Franke F, Gille I, Terpe HJ, Imhof BA. High expression level of α6 integrin in human breast carcinoma is correlated with reduced survival. Cancer Res. 1995;55:901–6. [PubMed] [Google Scholar]
- 8.Lipscomb EA, Simpson KJ, Lyle SR, Ring JE, Dugan AS, Mercurio AM. The α6β4 integrin maintains the survival of human breast carcinoma cells in vivo . Cancer Res. 2005;65:10970–6. doi: 10.1158/0008-5472.CAN-05-2327. [DOI] [PubMed] [Google Scholar]
- 9.Zhu L, Gibson P, Currle DS, et al. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature. 2009;457:603–7. doi: 10.1038/nature07589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizrak D, Brittan M, Alison MR. CD133: molecule of the moment. J Pathol. 2008;214:3–9. doi: 10.1002/path.2283. [DOI] [PubMed] [Google Scholar]
- 11.Sleeman KE, Kendrick H, Robertson D, Isacke CM, Ashworth A, Smalley MJ. Dissociation of estrogen receptor expression and in vivo stem cell activity in the mammary gland. J Cell Biol. 2007;176:19–26. doi: 10.1083/jcb.200604065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Early Breast Cancer Trialists' Collaborative Group. Tamoxifen for early breast cancer: an overview of the randomised trials. Lancet. 1998;351:1451–67. [PubMed] [Google Scholar]
- 13.Smith I, Procter M, Gelber RD, et al. 2-Year follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer: a randomised controlled trial. Lancet. 2007;369:29–36. doi: 10.1016/S0140-6736(07)60028-2. [DOI] [PubMed] [Google Scholar]
- 14.Pogribny I, Yi P, James SJ. A sensitive new method for rapid detection of abnormal methylation patterns in global DNA and within CpG islands. Biochem Biophys Res Commun. 1999;262:624–8. doi: 10.1006/bbrc.1999.1187. [DOI] [PubMed] [Google Scholar]
- 15.King FW, Ritner C, Liszewski W, et al. Subpopulations of human embryonic stem cells with distinct tissue-specific fates can be selected from pluripotent cultures. Stem Cells Dev. 2009;18:1441–50. doi: 10.1089/scd.2009.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bearzi C, Rota M, Hosoda T, et al. Human cardiac stem cells. Proc Natl Acad Sci U S A. 2007;104:14068–73. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shackleton M, Vaillant F, Simpson KJ, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–8. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 18.Ohyama M, Terunuma A, Tock CL, et al. Characterization and isolation of stem cell-enriched human hair follicle bulge cells. J Clin Invest. 2006;116:249–60. doi: 10.1172/JCI26043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adewumi O, Aflatoonian B, Ahrlund-Richter L, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–16. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- 20.Meyer MJ, Fleming JM, Ali MA, Pesesky M, Ginsburg E, Vonderhaar BK. Dynamic regulation of CD24 and the invasive, CD44posCD24-neg phenotype in breast cancer cell lines. Breast Cancer Res. 2009;11:R82. doi: 10.1186/bcr2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godar S, Ince TA, Bell GW, et al. Growth-inhibitory and tumor-suppressive functions of p53 depend on its repression of CD44 expression. Cell. 2008;134:62–73. doi: 10.1016/j.cell.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bourguignon LY, Peyrollier K, Xia W, Gilad E. Hyaluronan-CD44 interaction activates stem cell marker Nanog, Stat-3-mediated MDR1 gene expression, and ankyrin-regulated multidrug efflux in breast and ovarian tumor cells. J Biol Chem. 2008;283:17635–51. doi: 10.1074/jbc.M800109200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du L, Wang H, He L, et al. CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res. 2008;14:6751–60. doi: 10.1158/1078-0432.CCR-08-1034. [DOI] [PubMed] [Google Scholar]
- 24.Subramaniam V, Vincent IR, Gardner H, Chan E, Dhamko H, Jothy S. CD44 regulates cell migration in human colon cancer cells via Lyn kinase and AKT phosphorylation. Exp Mol Pathol. 2007;83:207–15. doi: 10.1016/j.yexmp.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 25.Subramaniam V, Vincent IR, Gilakjan M, Jothy S. Suppression of human colon cancer tumors in nude mice by siRNA CD44 gene therapy. Exp Mol Pathol. 2007;83:332–40. doi: 10.1016/j.yexmp.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 26.Shipitsin M, Campbell LL, Argani P, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–73. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 27.Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF-β signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–14. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 28.Ichida JK, Blanchard J, Lam K, et al. A small-molecule inhibitor of Tgf-β signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greber B, Lehrach H, Adjaye J. Control of early fate decisions in human ES cells by distinct states of TGFβ pathway activity. Stem Cells Dev. 2008;17:1065–77. doi: 10.1089/scd.2008.0035. [DOI] [PubMed] [Google Scholar]
- 30.Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–35. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Kruithof-de Julio M, Economides KD, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ottaviano YL, Issa JP, Parl FF, Smith HS, Baylin SB, Davidson NE. Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res. 1994;54:2552–5. [PubMed] [Google Scholar]
- 33.Bibikova M, Chudin E, Wu B, et al. Human embryonic stem cells have a unique epigenetic signature. Genome Res. 2006;16:1075–83. doi: 10.1101/gr.5319906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bloushtain-Qimron N, Yao J, Snyder EL, et al. Cell type-specific DNA methylation patterns in the human breast. Proc Natl Acad Sci U S A. 2008;105:14076–81. doi: 10.1073/pnas.0805206105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–16. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 37.Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–7. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Godlewski J, Nowicki MO, Bronisz A, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–30. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 39.Boyer LA, Lee TI, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gangemi RM, Griffero F, Marubbi D, et al. SOX2 silencing in glioblas-toma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells. 2009;27:40–8. doi: 10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- 41.You JS, Kang JK, Seo DW, et al. Depletion of embryonic stem cell signature by histone deacetylase inhibitor in NCCIT cells: involvement of Nanog suppression. Cancer Res. 2009;69:5716–25. doi: 10.1158/0008-5472.CAN-08-4953. [DOI] [PubMed] [Google Scholar]
- 42.Hu M, Krause D, Greaves M, et al. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997;11:774–85. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- 43.Villadsen R. In search of a stem cell hierarchy in the human breast and its relevance to breast cancer evolution. APMIS. 2005;113:903–21. doi: 10.1111/j.1600-0463.2005.apm_344.x. [DOI] [PubMed] [Google Scholar]
- 44.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–8. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoey T, Yen WC, Axelrod F, et al. DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell Stem Cell. 2009;5:168–77. doi: 10.1016/j.stem.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 46.Gupta PB, Onder TT, Jiang G, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–59. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan KS, Espinosa I, Chao M, et al. Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor-initiating cells. Proc Natl Acad Sci U S A. 2009;106:14016–21. doi: 10.1073/pnas.0906549106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rottenberg S, Jaspers JE, Kersbergen A, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci U S A. 2008;105:17079–84. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.