Abstract
Background and Aims
MADS-box genes comprise a gene family coding for transcription factors. This gene family expanded greatly during land plant evolution such that the number of MADS-box genes ranges from one or two in green algae to around 100 in angiosperms. Given the crucial functions of MADS-box genes for nearly all aspects of plant development, the expansion of this gene family probably contributed to the increasing complexity of plants. However, the expansion of MADS-box genes during one important step of land plant evolution, namely the origin of seed plants, remains poorly understood due to the previous lack of whole-genome data for gymnosperms.
Methods
The newly available genome sequences of Picea abies, Picea glauca and Pinus taeda were used to identify the complete set of MADS-box genes in these conifers. In addition, MADS-box genes were identified in the growing number of transcriptomes available for gymnosperms. With these datasets, phylogenies were constructed to determine the ancestral set of MADS-box genes of seed plants and to infer the ancestral functions of these genes.
Key Results
Type I MADS-box genes are under-represented in gymnosperms and only a minimum of two Type I MADS-box genes have been present in the most recent common ancestor (MRCA) of seed plants. In contrast, a large number of Type II MADS-box genes were found in gymnosperms. The MRCA of extant seed plants probably possessed at least 11–14 Type II MADS-box genes. In gymnosperms two duplications of Type II MADS-box genes were found, such that the MRCA of extant gymnosperms had at least 14–16 Type II MADS-box genes.
Conclusions
The implied ancestral set of MADS-box genes for seed plants shows simplicity for Type I MADS-box genes and remarkable complexity for Type II MADS-box genes in terms of phylogeny and putative functions. The analysis of transcriptome data reveals that gymnosperm MADS-box genes are expressed in a great variety of tissues, indicating diverse roles of MADS-box genes for the development of gymnosperms. This study is the first that provides a comprehensive overview of MADS-box genes in conifers and thus will provide a framework for future work on MADS-box genes in seed plants.
Keywords: MADS-box gene, flower development, gymnosperm, conifer, seed plant, spruce, pine, Picea, Pinus, ancestral gene set, most recent common ancestor, MRCA
INTRODUCTION
MADS-box genes comprise a large gene family coding for transcriptions factors (Gramzow et al., 2010). They are characterized by the presence of a MADS-box that encodes the DNA-binding domain of the corresponding MADS-domain proteins. Two types of MADS-box genes are distinguished, Type I or SRF-like and Type II or MEF2-like, which had probably already been established in the most recent common ancestor (MRCA) of extant eukaryotes (Alvarez-Buylla et al., 2000; Gramzow et al., 2010). In line with this, both types of MADS-box genes have been identified in most eukaryotes studied so far, even though taxa exist in which the one or other type of MADS-box genes has been lost (Gramzow et al., 2010). In plants, the proteins encoded by Type II MADS-box genes have a conserved, characteristic domain structure, with the MADS (M) domain followed by an Intervening (I) domain, a Keratin-like (K) domain and a C-terminal domain. The genes encoding these types of proteins have therefore been termed MIKC-type genes (Ma et al., 1991). In plants, the total number of MADS-box genes increased greatly to about 100 in flowering plants (angiosperms), while it remained low in all other eukaryotic groups (Alvarez-Buylla et al., 2000; Gramzow et al., 2010). MADS-box genes in land plants have been further subdivided into groups and clades based on their phylogeny and structural features (Gramzow and Theissen, 2010). The Type I genes have been subdivided into the three groups, Mα, Mβ and Mγ based solely on phylogenetic criteria, while, in the case of Type II genes, MIKCC- and MIKC*-group genes are distinguished by different lengths of their encoded K-domains and also on phylogenetic criteria (Henschel et al., 2002; Parenicova et al., 2003; Kwantes et al., 2012). Finally, about a dozen ancient clades of MIKCC-group genes have been recognized in angiosperms (Becker and Theissen, 2003), and a few other clades in mosses and ferns (Münster et al., 2002).
Our knowledge of the functional importance of the different types of MADS-box genes differs greatly in plants. Only a few Type I genes have been functionally characterized and it has been shown that they are mainly involved in female gametophyte, embryo and seed development (Yoo et al., 2006; Bemer et al., 2008; Kang et al., 2008; Steffen et al., 2008; Walia et al., 2009). Large fractions of their function may be hidden by redundancy, and many of these genes may have a function that is only weak, at best (Bemer et al., 2010). In contrast, the crucial functions of MIKC-type genes have long been recognized based on informative mutant phenotypes and are well studied (Schwarz-Sommer et al., 1990; Yanofsky et al., 1990; Trobner et al., 1992; Mandel et al., 1992; Pelaz et al., 2001). These genes are involved in controlling nearly all aspects of sporophyte and male gametophyte development (for recent reviews see Gramzow and Theissen, 2010; Smaczniak et al., 2012). Most prominent are their roles in flower and fruit development of angiosperms.
Land plants evolved from unicellular green algae (Cronk, 2001). The transition to land was accompanied by the evolution of structures that allow the regulation of water loss, such as cuticles and stomata (Peterson et al., 2010). Land plants comprise liverworts, mosses, hornworts (collectively called bryophytes) and tracheophytes. The tracheophytes evolved roots and vascular tissue for the transport of water and nutrients, with lycophytes being the most basal group that has these structures. The next clade that branches off from the tracheophyte tree comprises ferns and their allies such as horsetails. These ‘euphyllophytes’ represent the most basal group of land plants having true leaves. Thereafter, seeds evolved facilitating the dispersal of the corresponding plants. Seed plants comprise the ancestral gymnosperms and angiosperms. According to most molecular analyses, extant gymnosperms, comprising conifers, gnetophytes, cycads and Ginkgo, are monophyletic (Bowe and Coat, 2000; Chaw et al., 2000; Xi et al., 2013). However, the phylogenetic relationships between the different gymnosperm groups remain equivocal; nevertheless, the most recent comprehensive analyses suggest that cycads plus Ginkgo form a clade that is sister to all remaining extant gymnosperms (Wu et al., 2011; Xi et al., 2013). Angiosperms evolved having flowers that develop ovules enclosed in carpels, and seeds protected and distributed by fruits as new structural features. Hence, the evolution of some land plant lineages is characterized by the addition of new structures leading to more complex body plans.
Whole genome sequences are available for several green algae species, the moss Physcomitrella patens, the spikemoss (lycophyte) Selaginella moellendorffii and a number of angiosperms (The Arabidopsis Genome Initiative, 2000; Goff et al., 2002; Derelle et al., 2006; Tuskan et al., 2006; Jaillon et al., 2007; Merchant et al., 2007; Rensing et al., 2008; Banks et al., 2011). Hence, the complete set of MADS-box genes in the genomes of green algae, moss, spikemoss and angiosperms can be evaluated. While only one or two MADS-box genes have been identified in green algae, moss and spikemoss genomes encode around 20 MADS-box genes (Gramzow et al., 2012; Barker and Ashton, 2013). This number further increases in angiosperms, which have roughly about 100 MADS-box genes (Parenicova et al., 2003; Leseberg et al., 2006; Arora et al., 2007).
With these data, it is also possible to infer the minimal ancestral sets of MADS-box genes in the MRCA of plants, land plants, vascular plants and angiosperms. The MRCA of plants probably encoded only few MADS-box genes, but at least one Type I and one Type II gene (Gramzow and Theissen, 2010). In contrast, the MRCA of mosses and vascular plants and the MRCA of vascular plants both probably encoded at least two Type I, one MIKCC- and one MIKC*-group gene. The MRCA of extant angiosperms probably already possessed at least three Type I, 11 MIKCC- and two MIKC*-group genes, a great increase as compared with the ancestor of vascular plants. Looking at these numbers it appears likely that the increase in the number of MADS-box genes is correlated with the increasing complexity of land plants and hence, given the function of these genes in developmental control of extant organisms, that MADS-box genes are probably involved in the phenotypic evolution of plants (Theissen et al., 2000).
Due to the previous lack of whole genome data for ferns and allies and gymnosperms, the set of MADS-box genes in the MRCA of euphyllophytes and seed plants, respectively, could not be reliably inferred. From studies aiming at the isolation of MADS-box genes from gymnosperm transcriptomes, it is known that gymnosperms have several MIKCC-group genes that are orthologous to those known from angiosperms (Becker et al., 2000; Futamura et al., 2008; Carlsbecker et al., 2013). Hence, the MRCA of seed plants also probably contained at least ten MIKCC-group genes. However, whether the MRCA of extant seed plants contained even more MIKCC-group genes and what the ancestral number of Type I and MIKC*-group genes is in seed plants is not known.
Recently, the genomes of Picea abies, Picea glauca and Pinus taeda have been sequenced (Birol et al., 2013; Nystedt et al., 2013). Together with the increasing amount of transcriptome data (Wegrzyn et al., 2008; Lorenz et al., 2012), it is now possible to get a much more detailed picture about the ancestral set of MADS-box genes in seed plants and correlate MADS-box gene evolution with the evolution of seeds and flowers. Here we use this treasure trove of data to get a more detailed picture of the dynamics of MADS-box gene evolution during the origin of seed plants and the diversification of conifers.
MATERIALS AND METHODS
Identification of MADS-box genes
The whole genome assembly of Picea abies (Nystedt et al., 2013) was downloaded from Umea University in July 2012 (now available at http://congenie.org/). The whole genome assembly of Picea glauca (Birol et al., 2013) was downloaded from SMarTForests (http://www.smartforests.ca/) in January 2013 and the 0.8 version of the assembly of Pinus taeda was downloaded from PINEREFSEQ (http://pinegenome.org/pinerefseq/). Similarly, transcriptome data were downloaded from Dendrome (http://dendrome.ucdavis.edu/; Wegrzyn et al., 2008). Further-more, transcriptome data described by Lorenz et al. (2012) were downloaded from the NCBI Short Read Archive (Sayers et al., 2012).
The scaffolds of the whole genome assemblies as well as the assembled transcriptomes were translated in all six possible reading frames to create amino acid sequences using a customized perl script. These amino acid sequences were searched for MADS domains using hmmsearch of the HMMer package (Eddy, 1996) with a customized Hidden Markov Model for plant MADS domains (Gramzow and Theißen, 2013). For the whole genome data, all results with a length of at least 30 amino acids were kept and used for phylogeny reconstruction. For transcriptome data, the complete transcript sequences were obtained for each of the HMM results with a length of at least 30 amino acids. The identified transcript sequences were assembled separately for each gymnosperm species using Sequencher v 5.1 (Gene Codes Corp., Ann Arbor, MI, USA) with a minimum match percentage of 95 and a minimum overlap of 20. For the assembled transcript sequences, the open reading frames (ORFs) were then determined using Batch ORF Finder at greengene.uml.edu or ORF Finder at NCBI (Sayers et al., 2012). The ORFs were translated into protein sequences.
To determine which transcript sequences of P. abies, P. glauca and P. taeda correspond to which MADS-domain sequences identified from these genomes, we conducted local BLAST searches (Altschul et al., 1990) using the transcript sequences as query sequences and the whole genome assemblies of the P. abies, P. glauca and P. taeda genomes as database. If a transcript sequence had a match with more than 97 % sequence identity over a length of approx. 180 nucleotides to a genomic region that was identified to have a MADS-domain by hmmsearch, only the transcript sequence was kept and the MADS-domain sequence identified from the genome was removed from the dataset. As the number of transcript sequences for which no genomic region was identified was quite high using these stringent criteria, we later also considered shorter BLAST results.
The remaining MADS-domain-containing sequences identified from gymnosperm genomes and transcriptomes were combined and identical sequences were kept only once using the function ‘Remove Redundancy’ with a threshold of 100 of the program Jalview (Waterhouse et al., 2009). The MADS-domain-containing sequences remaining after this step were named using a two- or three-letter code for the species from which the sequences were identified followed by the keyword ‘MADS’ and incrementing numbers.
Phylogeny reconstruction
The reduced dataset of MADS-domain sequences identified from gymnosperm genomes and transcriptomes were aligned with all MADS-domain proteins from Arabidopsis thaliana, Oryza sativa, Populus trichocarpa, Vitis vinifera, Amborella trichopoda, Selaginella moellendorffii and Physcomitrella patens and with MADS-domain proteins annotated from gymnosperms and ferns as far as known using hmmalign (Eddy, 1996) with a hidden Markov model for plant MADS domains (Gramzow and Theißen, 2013) and the –trim option to remove non-homologous residues from the alignment. Furthermore, amino acids corresponding to insert states were removed from the alignment. The alignment was used to reconstruct a phylogeny using the RAxML program (Stamatakis, 2006) with the -f option to conduct a rapid bootstrap analysis with 1000 replicates and search for the best-scoring maximum-likelihood (ML) tree in one program run at the CIPRES Science gateway (Miller et al., 2010). Based on this ML phylogeny, MADS-domain sequences from gymnosperms were classified into Type I and Type II MADS domains depending on their grouping relative to the Type I and Type II MADS-domain sequences of the other species which had been classified previously (Parenicova et al., 2003; Leseberg et al., 2006; Arora et al., 2007; Diaz-Riquelme et al., 2009; Gramzow et al., 2012; Barker and Ashton, 2013).
We then aligned Type I and Type II MADS-domain sequences separately using sequences from the same species as described above and the program Probalign (Roshan and Livesay, 2006). For Type I MADS-domain proteins, the alignment was cropped to contain only the MADS domain, and for Type II proteins, the alignment was cropped to contain only the MADS and K domains. Phylogenies for these two datasets were reconstructed using RAxML as described above.
Based on the ML phylogeny for Type II proteins, these proteins were separated into 11 clades. To test the stability of these clades, separate datasets were compiled containing all the MADS sequences belonging to a specific clade according to the Type II ML phylogeny plus MADS sequences from other clades of A. thaliana, V. vinifera and O. sativa. Separate phylogenies for each of the 12 clades were constructed as described above for Type I and Type II phylogenies. Finally, datasets for each clade were compiled containing the MADS proteins belonging to the corresponding clade in the Type II as well as in the separate phylogeny. SEP3 from A. thaliana was used as a representative of the outgroup for all clades except the clade containing AGL2-, AGL6-, FLC- and SQUA-like genes, in which AGL12 of A. thaliana was used as an outgroup representative, and the clade containing AG-like genes in which AGL15 of A. thaliana was used. Alignments and phylogenies were constructed as described above. For each clade, two phylogenies were reconstructed in this way, one containing all MADS proteins belonging to this clade and another only containing MADS proteins with transcript support. To test the stability of the phylogenies for the different clades of Type II proteins, we also reconstructed phylogenies with MrBayes (Ronquist and Huelsenbeck, 2003) where we only included MADS proteins with transcript support. We used the WAG model of amino acid substitutions (Whelan and Goldman, 2001), generated 6 million generations, sampled every 1000th phylogeny and excluded the first 25 % from further analysis.
Gene expression data
Information about gene expression as revealed by transcriptome data was obtained from the NCBI expressed sequence tag database and the NCBI short read archive (Sayers et al., 2012).
RESULTS
Number of MADS sequences in gymnosperms
Our search of genome and transcriptome data for Gnetum gnemon and 18 conifer species (Fig. 1) identified 1064 MADS-domain sequences (Table 1, Supplementary Data Table S1). In species for which whole-genome information is available, Picea abies, Picea glauca and Pinus taeda, 261, 121 and 367 MADS sequences were found, respectively. However, there is evidence of transcription only for 49, 23 and 60 of these MADS sequences, respectively. We found a total of 43 sequences in transcriptome data of P. abies, P. glauca and P. taeda for which we did not find a corresponding sequence in the genome. This number decreases to 19 transcriptome sequences that were not found in the genomes when less stringent criteria were used. The fact that we did not find some transcript sequences in the genomes may be due to the incomplete knowledge of gymnosperm genomes. For P. abies and P. glauca, only about 61 % of the whole genome has been sequenced (Birol et al., 2013; Michael and Jackson, 2013; Nystedt et al., 2013). While most of the missing sequence is supposed to represent repetitive elements, some genes may have also escaped sequencing so far. We believe, however, with our combination of genome and transcriptome data, that we have made a major leap towards a complete overview of the MADS-box genes in gymnosperm genomes.
Fig. 1.
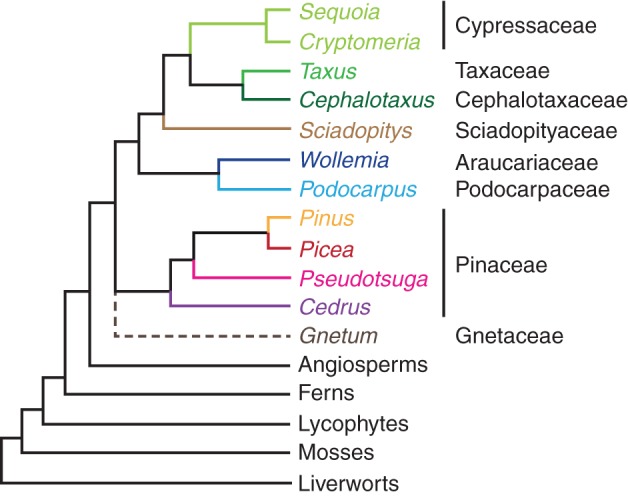
Relationships of conifer genera considered here, modified after Chaw et al. (1997) and Gugerli et al. (2001). Gnetum, angiosperms, ferns, lycophytes, mosses and liverworts are shown as outgroup representatives; Gnetum is shown on a dashed line due to the unresolved position. The different colours of the branches are used in the following figures to indicate the host taxa of the corresponding genes.
Table 1.
Number of MADS sequences identified from gymnosperm genome and transcriptome data
| Order | Family | Abbreviation | Total | Type I | Type II | |
|---|---|---|---|---|---|---|
| Gnetales | Gnetaceae | Gnetum gnemon | GgMADS | 41 | 0 | 41 |
| Coniferales | Pinaceae | Cedrus atlantica | CaMADS | 13 | 0 | 13 |
| Picea abies | PaMADS | 253 (41) + 8 | 12 | 249 | ||
| Picea glauca | PgMADS | 107 (9) + 14 | 3 | 118 | ||
| Picea sitchensis | PsMADS | 17 | 1 | 16 | ||
| Pinus banksiana | PbMADS | 2 | 0 | 2 | ||
| Pinus contorta | PcMADS | 14 | 0 | 14 | ||
| Pinus lambertiana | PlMADS | 41 | 0 | 41 | ||
| Pinus palustris | PpaMADS | 21 | 0 | 21 | ||
| Pinus pinaster | PpiMADS | 10 | 0 | 10 | ||
| Pinus taeda | PtaMADS | 346 (39) + 21 | 17 | 350 | ||
| Pseudotsuga menziesii | PmeMADS | 40 | 0 | 40 | ||
| Podocarpaceae | Podocarpus macrophyllus | PmaMADS | 16 | 1 | 15 | |
| Araucariaceae | Wollemia nobilis | WnMADS | 11 | 0 | 11 | |
| Sciadopityaceae | Sciadopitys verticillata | SvMADS | 22 | 1 | 21 | |
| Taxaceae | Taxus baccata | TbMADS | 3 | 0 | 3 | |
| Cephalotaxaceae | Cephalotaxus harringtonia | ChMADS | 35 | 0 | 35 | |
| Cryptomerica japonica | CjMADS | 10 | 0 | 10 | ||
| Cupressaceae | Sequoia sempervirens | SsMADS | 19 | 0 | 19 |
For species for which whole-genome information is available the numbers are given as follows: number of MADS sequences identified from genome data (number of MADS sequences identified from genome data and supported by transcriptome data) + number of MADS sequences identified from transcriptome data for which the genomic locus could not be identified.
Based on our RAxML phylogeny (Supplementary Data Fig. S1), we classified the identified MADS sequences into Type I and Type II. Bootstrap values are quite low. However, the grouping of the known MADS-box genes is correct and thus the classification of the gymnosperm sequences into Type I and Type II should also be accurate in most cases. Only 35 gymnosperm MADS sequences were classified as Type I. Even for the species for which whole-genome information is available, the percentage of Type I sequences is always less than 5 % of all MADS sequences. In contrast, 1029 gymnosperm MADS sequences were classified as Type II. In the species for which whole genome information is available, P. abies, P. glauca and P. taeda, the number of Type II sequences is 249, 118 and 350, respectively.
Type I genes
In our phylogenies of Type I genes there are two major branches that may represent large clades (Fig. 2, Supplementary Data Fig. S2). One putative clade contains genes classified as Mα genes in A. thaliana, P. trichocarpa and O. sativa (Parenicova et al., 2003; Leseberg et al., 2006; Arora et al., 2007) and MADS-box genes from V. vinifera, gymnosperms, S. moellendorffii and P. patens. The other clade comprises genes classified as Mβ and Mγ genes in A. thaliana, P. trichocarpa and O. sativa and MADS-box genes from V. vinifera, gymnosperms, S. moellendorffii and P. patens. This suggests that there are two ancient clades of Type I MADS-box genes in land plants. Again, the bootstrap values supporting the two groups are quite low but our results concur with previous studies (Gramzow et al., 2012).
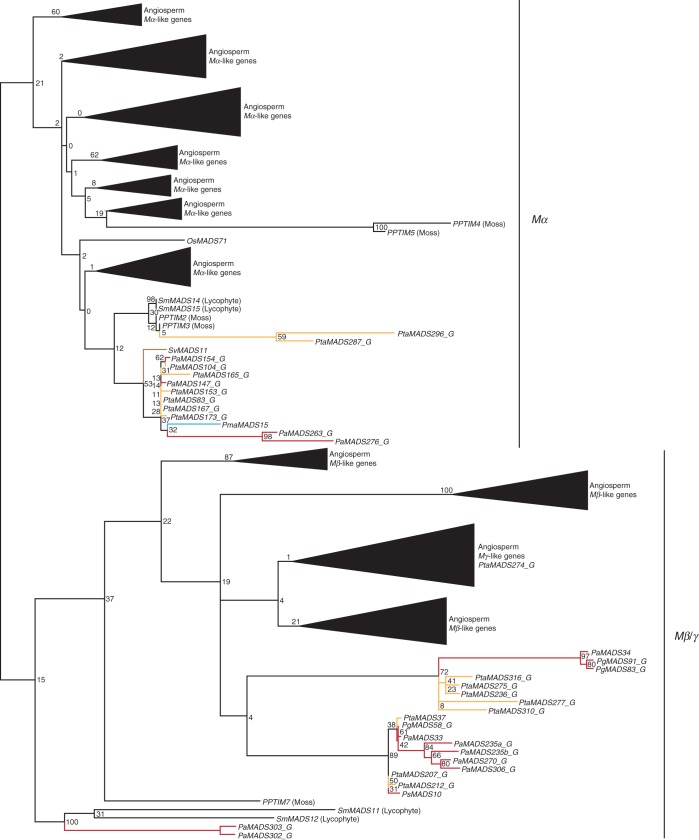
Fig. 2.
Phylogeny of Type I MADS-box genes. The two major putative clades of Type I genes, Mα and Mβ/γ (Gramzow et al., 2012), are marked by labelled lines on the right. The colours of the branches correspond to the colours of the gymnosperm genera in Fig. 1. Abbreviations of gymnosperm gene names are described in Table 1. Lycophyte sequences are from Selaginella moellendorffii and moss sequences are from Physcomitrella patens. Clades of angiosperm genes are collapsed into triangles and their classification according to (Parenicova et al., 2003; Leseberg et al., 2006; Arora et al., 2007) is shown on the right. Numbers at nodes denote bootstrap values. The fully resolved phylogeny is available as Supplementary Data Fig. S2.
As mentioned above, the number of Type I genes identified in gymnosperms is very low. The 35 Type I genes of gymnosperms are distributed approximately evenly between the two clades with 14 genes belonging to putative clade I (Mα) and 21 genes belonging to putative clade II (Mβ/Mγ). Evidence of transcription exists only for few of these genes. Our analysis of transcriptome data revealed that two genes of clade I are expressed in mixed shoot tissues. For clade II, four genes were found in transcriptome data derived from bud, male cone and embryo tissues.
Clades of Type II genes
Based on our phylogeny of Type II sequences (Supplementary Data Fig. S3), we defined 11 branches (putative clades, hereafter termed ‘clades’ for simplicity) for separate analyses (MIKC*, AGL2/AGL6/FLC/SQUA, DEF/GLO/OsMADS32/GGM13, AGAMOUS, AGL12, AGL15, AGL17, GpMADS4, StMADS11, TM3 and TM8). All of these clades contain gymnosperm sequences (Table 2).
Table 2.
Number of gymnosperm genes in different clades of Type II MADS-box genes
| Species | MIKC* | AGL2/AGL6/FLC/SQUA | DEF/GLO/OsMADS32/GGM13 | AGAMOUS | AGL12 | AGL15 | AGL17 | GpMADS4 | StMADS11 | TM3 | TM8 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| S. sempervirens | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 4 | 4 |
| C. japonica | 0 | 1 | 6 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| T. baccata | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| C. harringtonia | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 10 | 13 |
| S. verticillata | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 14 |
| W. nobilis | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 6 |
| P. macrophyllus | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 6 | 1 | 6 |
| P. taeda | 1 (41) | 11 (38) | 2 (64) | 3 (24) | 4 (15) | 0 (0) | 0 (4) | 0 (2) | 7 (20) | 25 (77) | 6 (58) |
| P. pinaster | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 4 | 3 |
| P. palustris | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 5 |
| P. lambertiana | 0 | 6 | 1 | 0 | 1 | 0 | 0 | 2 | 6 | 19 | 6 |
| P. contorta | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 8 | 2 |
| P. banksiana | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| P. abies | 4 (5) | 2 (15) | 8 (20) | 2 (15) | 0 (5) | 0 (1) | 0 (13) | 2 (9) | 14 (53) | 14 (85) | 4 (27) |
| P. glauca | 0 (1) | 3 (10) | 1 (8) | 2 (5) | 1 (6) | 0 (0) | 0 (6) | 0 (4) | 7 (26) | 8 (32) | 1 (17) |
| P. sitchensis | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 5 | 6 | 3 |
| P. menziesii | 0 | 1 | 2 | 2 | 0 | 0 | 0 | 0 | 1 | 31 | 3 |
| C. atlantica | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 8 | 0 |
| G. gnemon | 2 | 4 | 3 | 4 | 0 | 0 | 0 | 0 | 4 | 2 | 18 |
| Sum | 8 (50) | 39 (86) | 24 (105) | 15 (52) | 6 (27) | 0 (1) | 0 (23) | 5 (18) | 76 (147) | 163 (310) | 97 (188) |
Generally, the number of expressed genes is given. For P. taeda, P. abies, P. glauca and sum, the total number of genes including those solely identified from genome data without evidence of transcription is given in parentheses.
MIKC*-group genes
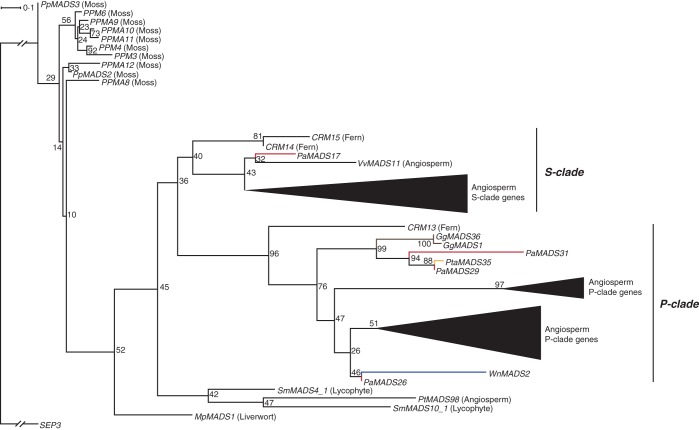
In our type II phylogeny, 50 gymnosperm sequences grouped together with MIKC*-group genes from mosses, ferns and angiosperms (Supplementary Data Fig. S3). Only eight of these are supported by expression data. The bryophyte sequences branch off before the differentiation of the S- and P-clade, each containing sequences from ferns, gymnosperms and angiosperms (Supplementary Data Fig. S4). In the phylogeny containing only sequences with expression data (Fig. 3), PaMADS17 is the only gymnosperm member of the S-clade. Interestingly, also 39 genomic sequences from P. taeda without evidence of transcription belong to the S-clade in our phylogeny. Ten gymnosperm sequences (seven with evidence of transcription) form the P-clade together with sequences from ferns and angiosperms. The association of the gymosperm sequences to the P- and S-clades in our MrBayes phylogeny is consistent with that in our RAxML phylogenies (Supplementary Data Fig. S4). The identified MIKC*-group genes of the P-clade, PaMADS29, PaMADS31 and PaMADS26 from P. abies, show expression in stem, wood, vegetative shoots and female cones. PaMADS26 was also identified from male cones. cDNA of GgMADS1 from G. gnemon was isolated from female cones. In contrast, the expression of the single S-clade gene PaMADS17 was detected only in male cones.
Fig. 3.
Phylogeny of MIKC*-group genes. Colours of the branches, gene names, triangles and numbers at nodes are as described in Fig. 2. The subclades S and P (Nam et al., 2003) are marked by labelled lines on the right. The liverwort sequence is from Marchantia polymorpha, fern sequences are from Ceratopteris richardii, lycophyte sequences are from Selaginella moellendorffii, moss sequences are from Physcomitrella patens and the separate angiosperm sequences are from Vitis vinifera and from Populus trichocarpa. The fully resolved phylogeny is available as Supplementary Data Fig. S4b.
AGL2/AGL6/FLC/SQUA-like genes
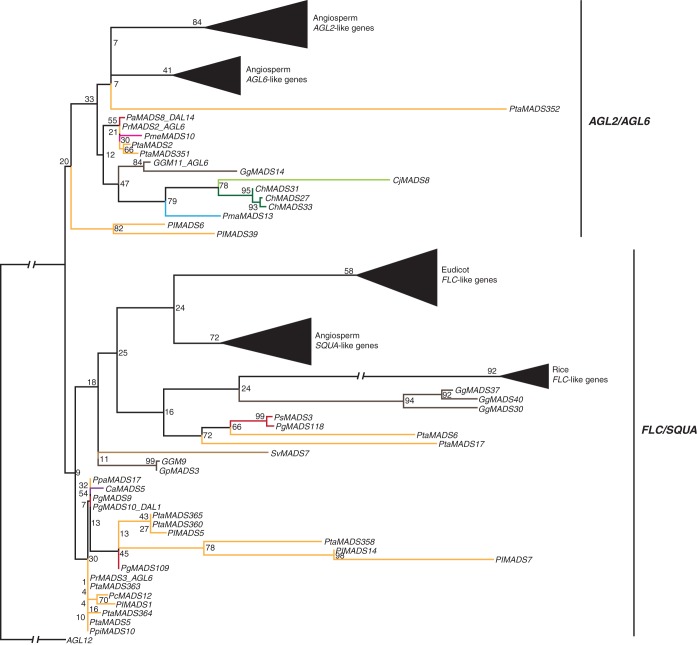
Recently, it was established that FLC-like genes belong to the superclade of AGL2-, AGL6- and SQUA-like genes (Ruelens et al., 2013). Hence, we here analysed these genes together. In our phylogenies, two large subclades can be defined, one consisting of AGL2- and AGL6-like genes and the other consisting of FLC- and SQUA-like genes (Supplementary Data Fig. S5a). Of the gymnosperm sequences identified here, 19 belong to the clade of AGL2/AGL6-like genes and 67 belong to the clade of FLC/SQUA-like genes. When we restrict our phylogeny to sequences for which we have evidence of transcription, the AGL2- and the AGL6-like genes of angiosperms form sister clades as well as the FLC- and SQUA-like genes of angiosperms, with the exception of FLC-like genes from rice, which cluster within a clade of gymnosperm sequences (Fig. 4). This suggests that the duplications giving rise to AGL2- and AGL6-like genes and to FLC- and SQUA-like genes occurred in angiosperms and that the MRCA of extant seed plants possessed at least one AGL2/AGL6-like gene and at least one FLC/SQUA-like gene. As the bootstrap values for our RAxML phylogenies were quite low we also reconstructed a phylogeny using MrBayes (Supplementary Data Fig. 5c). The sister-group relationships of AGL2- and AGL6-like genes and of FLC- and SQUA-like genes of angiosperms was confirmed in our MrBayes phylogeny with stronger support (posterior probability of 0·51 and 0·90, respectively). However, all gymnosperm sequences appear more closely related to AGL2/AGL6-like genes than to FLC/SQUA-like genes in our MrBayes phylogeny.
Fig. 4.
Phylogeny of AGL2/AGL6/FLC/SQUA-like genes. Colours of the branches, gene names, triangles and numbers at nodes are as described in Fig. 2. The subclades AGL2/AGL6 and FLC/SQUA are marked by labelled lines on the right. The fully resolved phylogeny is available as Supplementary Data Fig. S5b.
Analysing transcriptome data, we found expression of gymnosperm genes belonging to the AGL2/AGL6 clade in shoots, needles and reproductive tissues. FLC/SQUA-like genes from gymnosperms were identified in transcriptome data from a wide variety of tissues (roots, shoots, stems, bark and female cones).
DEF/GLO/OsMADS32/GGM13-like genes
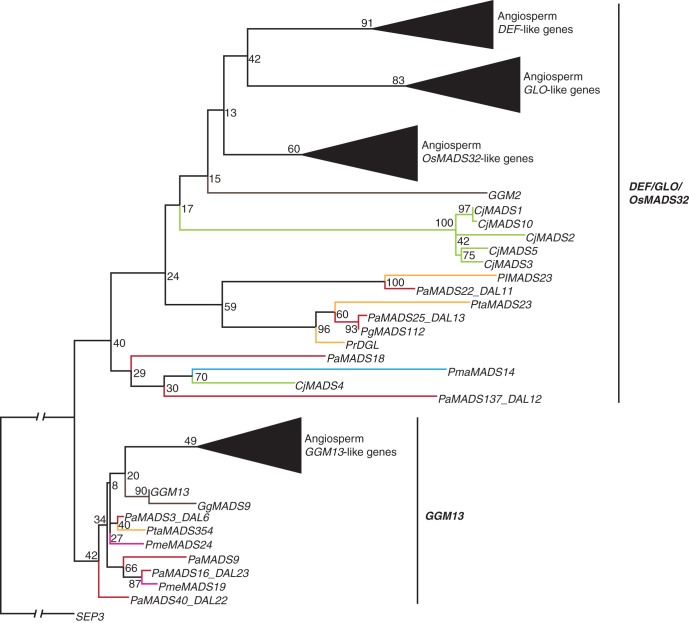
In our phylogenies, OsMADS32-like genes cluster with DEF-like, GLO-like and GGM13-like genes (Supplementary Data Fig. S3). Conse-quently, we analysed these clades together. Based on our Type II phylogeny including MADS sequences identified from genome and transcriptome projects, 105 sequences belong to this clade (Supplementary Data Fig. S3a). In our separate phylogenies for this superclade, DEF- and GLO-like genes from angiosperms form sister clades that in turn are sister clades to OsMADS32-like genes of angiosperms (Supplementary Data Fig. S6a). This suggests that there have been two duplications near the base of extant angiosperms giving rise first to OsMADS32- and DEF/GLO-like genes and then to DEF- and GLO-like genes. A clade of 60 MADS sequences from gymnosperms is sister to the DEF/GLO/OsMADS32-superclade. GGM13-like genes from angiosperms form a clade in our phylogeny to which 45 MADS sequences from gymnosperms are related.
When we exclude gymnosperm MADS sequences solely predicted on genomic sequences (Fig. 5), 24 MADS sequences from eight gymnosperm species cluster with DEF/GLO/OsMADS32-like genes from angiosperms and nine MADS sequences from five gymnosperm species cluster with GGM13-like genes from angiosperms. The phylogeny and distribution of the sequences suggests that there has been at least one DEF/GLO/OsMADS32-like gene and one GGM13-like gene in the MRCA of extant seed plants. In our MrBayes phylogeny, some gymnosperm sequences are sister to the OsMADS32- and to the DEF-like genes from angiosperms, respectively, rather than being ancestral to a superclade of DEF-, GLO- and OsMADS32-like genes from angiosperms (Supplementary Data Fig. 6c).
Fig. 5.
Phylogeny of DEF/GLO/OsMADS32/GGM13-like genes. Colours of the branches, gene names, triangles and numbers at nodes are as described in Fig. 2. The subclades DEF/GLO/OsMADS32 and GGM13 are marked by labelled lines on the right. The fully resolved phylogeny is available as Supplementary Data Fig. S6b.
Gymnosperm DEF/GLO/OsMADS32-like sequences are mainly derived from transcriptome data of male reproductive tissues. GGM13-like genes were mainly identified from mixed tissues. Only for GGM13-like genes of P. abies are specific tissues, namely female reproductive organs, given.
AGAMOUS- and AGL12-like genes
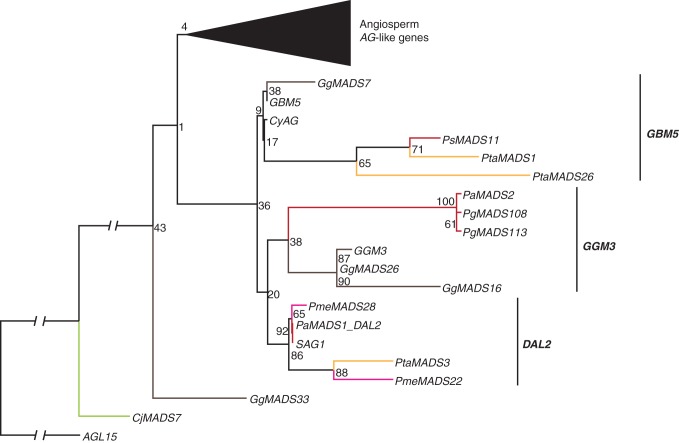
In our phylogenies, 52 of all identified gymnosperm MADS sequences belong to the AGAMOUS clade (Supplementary Data Fig. S7a). When we exclude sequences for which no evidence of transcription exists, 15 sequences remain. In this reduced phylogeny, all AG-like genes of gymnosperms are sister to the AG-like genes of angiosperms, suggesting that the MRCA of extant seed plants had at least one but may not have had more than one AG-like gene (Fig. 6). Furthermore, the AG-like genes of gymnosperms form three subclades in our RAxML phylogeny and two subclades in our MrBayes phylogeny (Supplementary Data Fig. 7c), indicating that the MRCA of extant gymnosperms may have possessed at least two to three AG-like genes. Interestingly, AG-like sequences derived from transcripts were identified from a variety of tissues, including roots, shoots, stems, bark, leaves and reproductive organs.
Fig. 6.
Phylogeny of AG-like genes. Colours of the branches, gene names, triangles and numbers at nodes are as described in Fig. 2. Three putative subclades of AG-like genes in gymnosperms, GBM5, GGM3 and DAL2, are marked by labelled lines on the right. The fully resolved phylogeny is available as Supplementary Data Fig. S7b.
The sister group of AG-like genes are the AGL12-like genes (Becker and Theissen, 2003). We identified 27 AGL12-like sequences from gymnosperms, where there is evidence of transcription for six of them (Fig. 7, Supplementary Data Fig. S8). Expression was mostly found in mixed tissues, twice in root and once in shoot and/or needles. Our phylogeny suggests that one AGL12-like gene was present in the MRCA of extant seed plants.
Fig. 7.
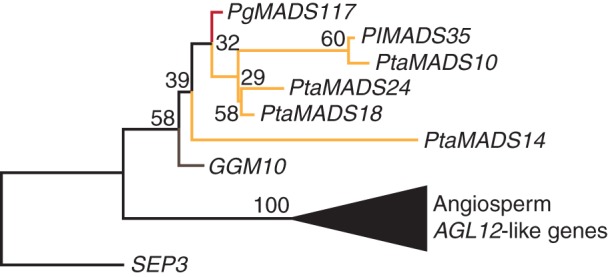
Phylogeny of AGL12-like genes. Colours of the branches, gene names, triangles and numbers at nodes are as described in Fig. 2. The fully resolved phylogeny is available as Supplementary Data Fig. S8b.
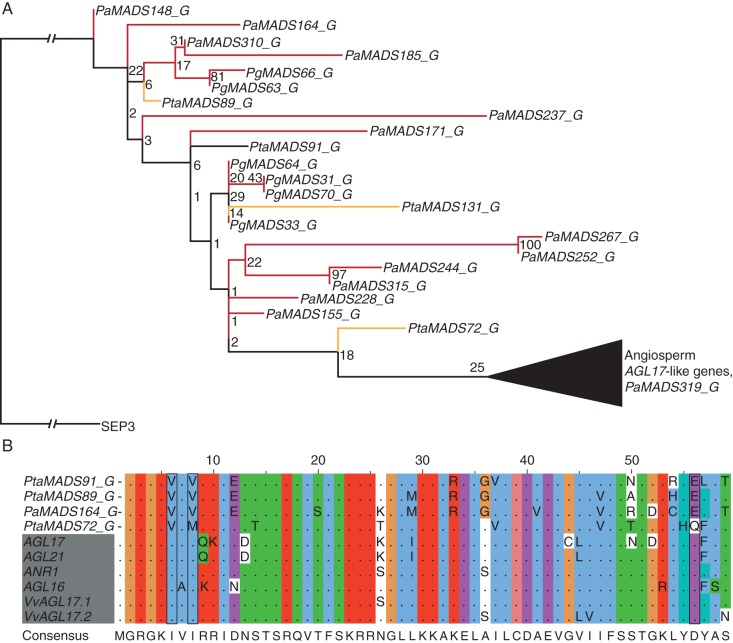
AGL15- and AGL17-like genes
In our Type II phylogeny, one sequence of P. abies clusters with AGL15-like genes from angiosperms and 23 sequences from P. taeda, P. abies and P. glauca cluster with AGL17-like genes of angiosperms (Supplementary Data Figs S3, S9 and S10). However, all of these gymnosperm sequences have been identified solely from genomic sequences and no evidence of transcription exists for these genes. Alignments reveal strong similarity of at least some of these gymnosperm sequences to the AGL15- and AGL17-like MADS sequences of angiosperms, respectively (Figs 8 and 9). Nevertheless, the alignments also reveal three positions each where the gymnosperm sequences are different from all of the angiosperm MADS sequences of the respective clades. Hence, further analyses are needed to confirm or deny the association of the gymnosperm sequences to the clades of AGL15- and AGL17-like genes.
Fig. 8.
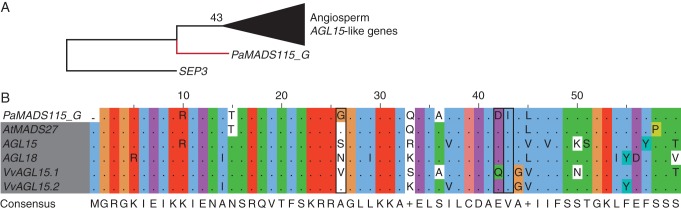
Phylogeny of AGL15-like genes (A) and alignment of the MADS domains of AGL15-like proteins (B). For the phylogeny, colours of the branches, gene names, triangle and number at nodes are as described in Fig. 2. The alignment is coloured according to conservation and biochemical properties of amino acids (Thompson et al., 1997) and shows only non-conserved residues. Names of angiosperm genes are shaded grey. Positions at which the gymnosperm sequence differs from all angiosperm sequences are boxed. The consensus sequence is given below the alignment.
Fig. 9.
Phylogeny of AGL17-like genes (A) and alignment of the MADS domains of AGL17-like proteins (B). For the phylogeny, colours of the branches, gene names, triangle and numbers at nodes are as described in Fig. 2. Colouring and labelling of the alignment is described in Fig. 8. Positions at which the gymnosperm sequences differ from all angiosperm sequences are boxed. The consensus sequence is given below the alignment.
GpMADS4-like genes
We found 18 sequences belonging to the gymnosperm-specific clade of GpMADS4-like genes (Supplementary Data Fig. S3). The known genes DAL10 of P. abies and GpMADS4 of G. parvifolium constitute the GpMADS4-subclade together with 13 other gymnosperm sequences (Supplementary Data Fig. S11). DAL21 of P. abies forms another subclade together with three other gymnosperm sequences, termed the DAL21-subclade here. These clades are also stable in our MrBayes phylogeny (Supplementary Data Fig. S11b). For both subclades, there are three genes supported by expression data (Fig. 10), where expression was found in bark, shoots, stems, needles, buds and cones.
Fig. 10.
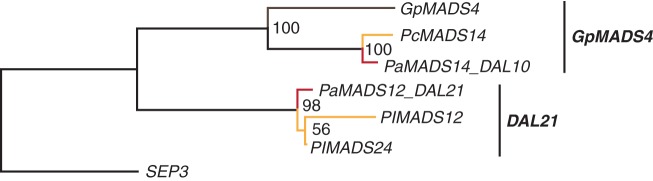
Phylogeny of GpMADS4-like genes. Colours of the branches, gene names and numbers at nodes are as described in Fig. 2. The two putative subclades GpMADS4 and DAL21 are marked by labelled lines on the right.
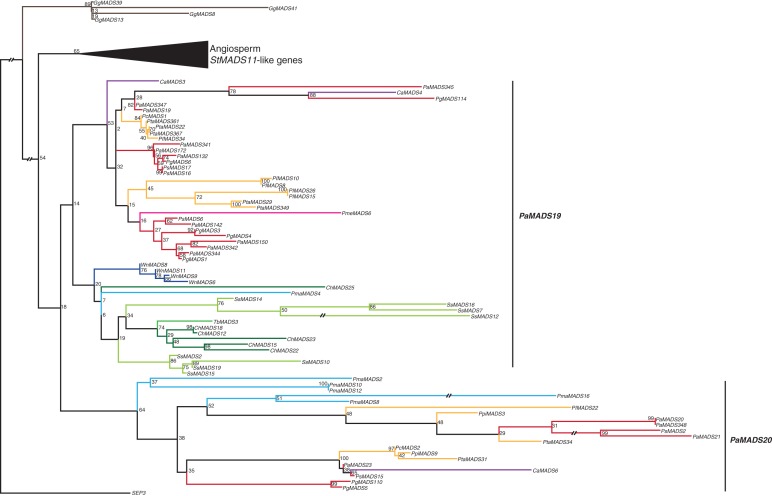
StMADS11-like genes
In our phylogenies, 147 gymnosperm sequences cluster together with StMADS11-like genes from angiosperms, of which 76 gymnosperm sequences have expression data support (Supplementary Data Fig. S12a). There are two subclades of StMADS11-like genes from conifers in our phylogeny (Fig. 11), suggesting that there was a duplication of an ancestral StMADS11-like gene near the base of extant conifers. We termed the two resulting subclades PaMADS19-like and PaMADS20-like genes after the P. abies gene with the lowest number in the corresponding clade. These two subclades also appear in our MrBayes phylogeny (Supplementary Data Fig. S12c). For the PaMADS19 subclade, sequences of all the conifer species studied here were found with the exception of those of S. verticillata. The subclade PaMADS20 contains only sequences of P. macrophyllus and the family Pinaceae. In both subclades several species-specific duplications occurred, for example in S. sempervirens, W. nobilis, C. harringtonia, P. macrophyllus and species of the family Pinaceae. Expression of gymnosperm StMADS11-like genes was found mainly in shoots, but also in other tissues such as roots, stems, wood, bark and needles.
Fig. 11.
Phylogeny of StMADS11-like genes. Colours of the branches, gene names, triangle and numbers at nodes are as described in Fig. 2. The two putative subclades of StMADS11-like genes in gymnosperms, PaMADS19 and PaMADS20, are as marked by labelled lines on the right. The fully resolved phylogeny is available as Supplementary Data Fig. S12b.
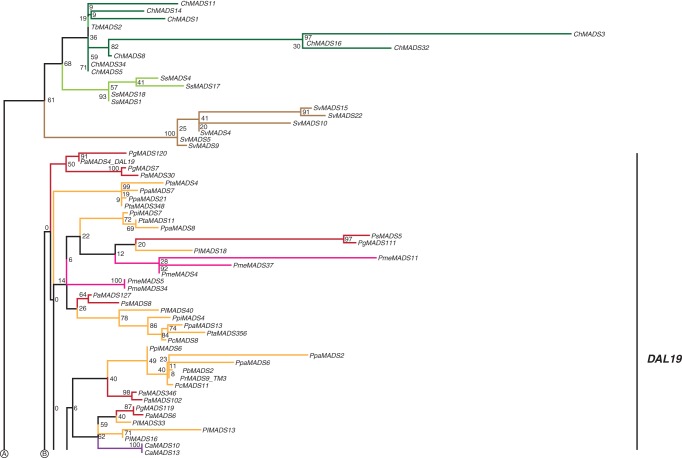
TM3-like genes
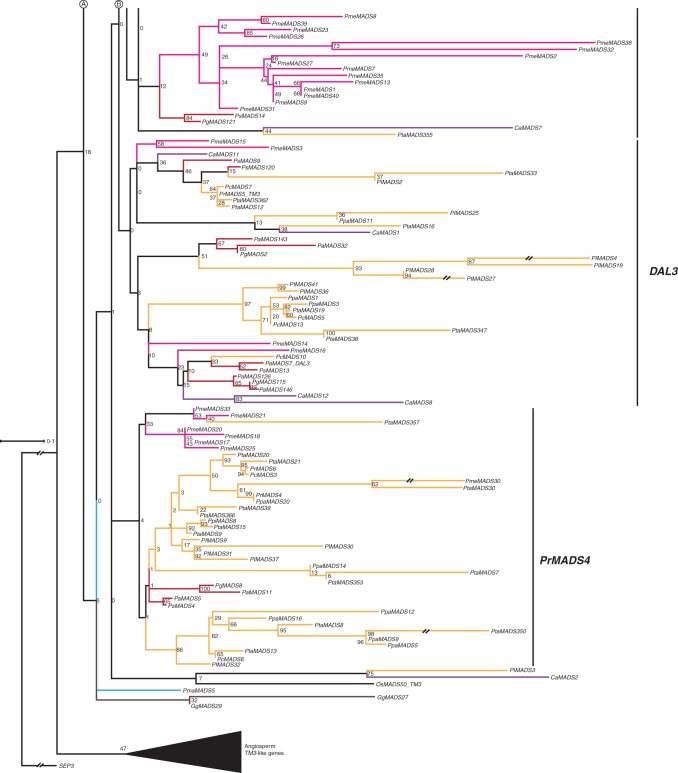
The clade of TM3-like genes includes 310 gymnosperm sequences from the species studied here, 163 of which have expression support, and four sequences from Pinus radiata (Supplementary Data Fig. S13). In our reduced phylogeny there are two main clades of gymnosperm sequences, one comprising genes from the conifer species S. verticillata, S. sempervirens, T. baccata and C. harringtonia (Fig. 12), and the other containing sequences of G. gnemon, P. macrophyllus and Pinaceae species. The clade of Pinaceae TM3-like genes can be further subdivided into three subclades, termed DAL19-, DAL3- and PrMADS4-like genes here. There are a number of species-specific expansions of TM3-like genes, for example in Pinaceae, S. verticillata, S. sempervirens and C. harringtonia. The MrBayes phylogeny shows different subclades to those observed in our RAxML phylogenies (Supplementary Data Fig. S13c). Hence, even though the sheer number of TM3-like genes in conifers indicates a number of duplications, the timing and extent of duplications cannot be determined based on our data. Transcripts of TM3-like genes were isolated from various tissues, such as shoots, stems, needles, buds, and female and male cones.
Fig. 12.
Phylogeny of TM3-like genes. Colours of the branches, gene names, triangle and numbers at nodes are as described in Fig. 2. Three putative subclades of TM3-like genes in Pinaceae, DAL19, DAL3 and PrMADS4, are marked by labelled lines on the right. The fully resolved phylogeny is available as Supplementary Data Fig. S13b.
TM8-like genes
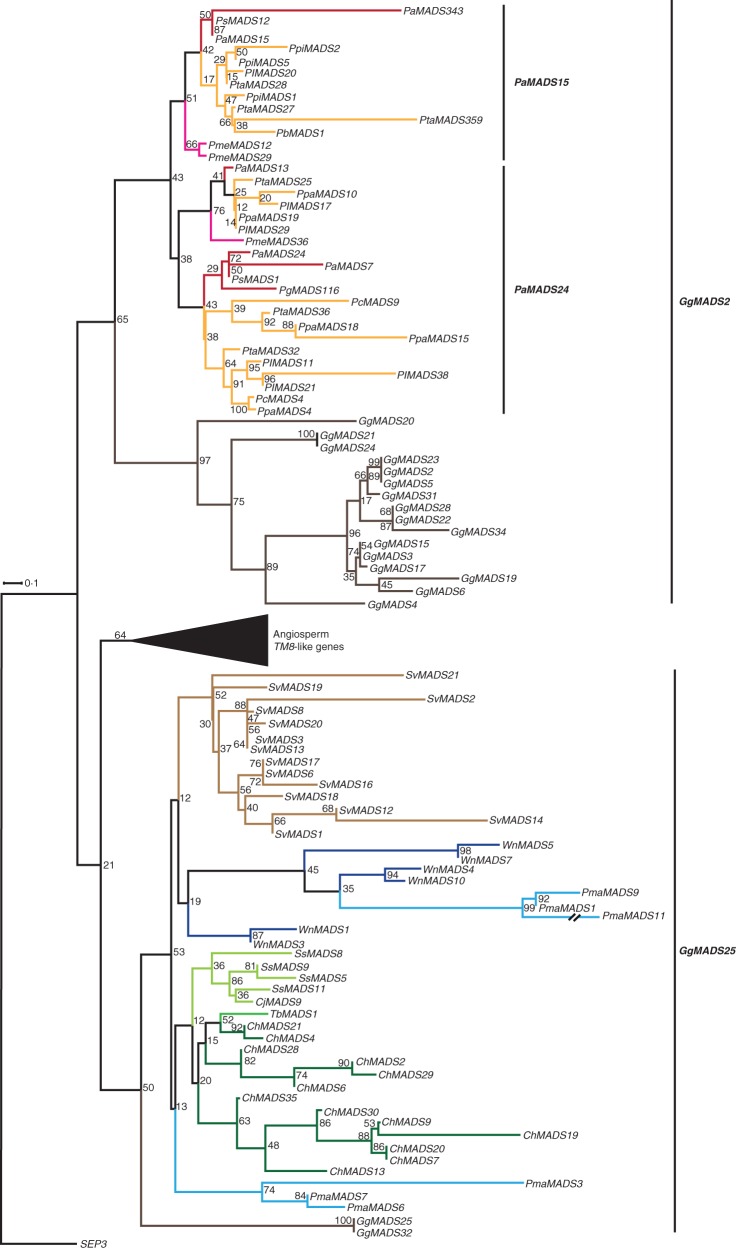
The angiosperm TM8-like genes group with 188 MADS-sequences from gymnosperms (Supplementary Data Fig. S14a). For 97 sequences expression data are available. In the reduced phylogeny including only genes with expression data, two subclades are evident, termed as GgMADS2-like and GgMADS25-like genes (Fig. 13). The clade of GgMADS2 contains numerous sequences of the family Pinaceae and 16 sequences of G. gnemon. The Pinaceae sequences can be further subdivided into the subclades PaMADS15 and PaMADS24. Transcript-based sequences belonging to these two subclades were isolated from buds, shoots, needles and stems and only one sequence of P. taeda PtaMADS27, belonging to the PaMADS15 subclade, was found to be expressed in roots. The clade of GgMADS25-like genes contains sequences of non-Pinaceae conifers and two sequences from G. gnemon. The clades of GgMADS25-like genes also appears in our MrBayes phylogeny (Supplementary Data Fig. S14c). However, the genes belonging to the GgMADS2-like clade in our RAxML phylogeny are split into two clades that are not sister clades in our MrBayes phylogeny. Expression of genes from this clade was also found in a variety of tissues, such as shoots, needles and male strobili.
Fig. 13.
Phylogeny of TM8-like genes. Colours of the branches, gene names, triangle and numbers at nodes are as described in Fig. 2. Two putative subclades of TM8-like genes in gymnosperms, GgMADS2 and GgMADS25, are marked by labelled lines on the right. Two putative subclades of the GgMADS2-subclade in Pinaceae, PaMADS15 and PaMADS24, are likewise marked by labelled lines. The fully resolved phylogeny is available as Supplementary Data Fig. S14b.
DISCUSSION
Large total number of MADS-box genes but low percentage of Type I genes in conifer genomes
The total number of MADS-box sequences identified from the three sequenced conifer genomes ranges from 121 to 367 with an average of 249. In sequenced angiosperm genomes, this number ranges from 60 to 265 where the average number is 116 (Gramzow and Theißen, 2013). Hence, the number of putative MADS-box genes (including pseudogenes) in conifers is on average much higher than in angiosperms. This is consistent with the large genome size of gymnosperms. The genomes studied here of Picea abies, Picea glauca and Pinus taeda have genome sizes of more than 20 Gbp (Birol et al., 2013; Nystedt et al., 2013). Despite the large genome sizes, there is no evidence for genome duplications in the gymnosperm lineage (Nystedt et al., 2013). The total number of genes also seems to be higher in gymnosperms than in most angiosperms. In P. abies, 70 968 protein-coding loci were predicted, of which 28 354 were considered to be of high confidence (Nystedt et al., 2013). Hence, the large number of MADS-box genes we found in gymnosperm genomes correlates with the large genome size and gene number in gymnosperms. However, many of the MADS-box genes we found are probably pseudogenes. We identified premature stop codons in the MADS boxes of 17 sequences from P. abies, eight sequences from P. glauca and 22 sequences from P. taeda. A complete analysis of the number of pseudogenes in the MADS-box gene family is not possible yet due to the short length of some scaffolds on which MADS-boxes have been identified. A large fraction of pseudogenes has also been identified for other gene families in gymnosperms, such as phytochrome, cdc2 and WUSCHEL-type homeobox genes (Kinlaw and Neale, 1997; Kvarnheden et al., 1998; Garcia-Gil, 2008; Hedman et al., 2013). How many of the identified MADS-box genes in conifers are functional remains to be determined. The number of genes supported by transcripts identified for P. abies, P. glauca and P. taeda ranges only from 23 to 60. This low number of MADS-box genes for which transcriptome data were found may have several reasons. First, the number of different tissues sampled was low to moderate. For P. abies, 22 transcriptome samples of different tissues were determined (Nystedt et al., 2013) while for P. taeda and P. glauca six and eight transcriptome datasets, respectively, were available (Wegrzyn et al., 2008; Lorenz et al., 2012). Furthermore, many MADS-box genes are known to be expressed at low level or only at specific stages of development (Becker and Theissen, 2003; De Bodt et al., 2003; Nam et al., 2004; Bemer et al., 2010). Hence, more transcriptome data may help to identify more MADS-box genes as expressed genes. However, as mentioned above, a number of the MADS-box genes we identified may also be pseudogenes.
In contrast to the large overall number of identified MADS-box genes in conifer genomes, the number of Type I MADS-box genes ranges from only three to 17 in the sequenced conifer genomes. Only two angiosperm genomes examined so far have fewer than 17 Type I MADS-box genes, namely Cucumis sativus (11) and Zea mays (14) (Gramzow and Theißen, 2013). Percentage-wise, the difference between the amount of Type I genes in conifer and angiosperm genomes is even clearer: in all angiosperm genomes that have been examined, the percentage of Type I genes is greater than 20 % of all MADS-box genes, while in examined conifers, the percentage is always lower than 5 %. For angiosperms, it has been shown that Type I genes have higher birth-and-death rates than Type II genes (Nam et al., 2004). Our phylogeny of Type I genes shows lineage-specific expansions of Type I genes for conifers, similar to what is observed for Type I genes in angiosperms (Fig. 2). However, there are fewer clades of conifer Type I genes, which generally also have fewer genes than the clades of angiosperm Type I genes (Supplementary Data Fig. S2). Hence, either the birth rate of Type I genes is lower or the death rate of Type I genes is higher in conifers than in angiosperms. Further studies are needed to clarify the evolutionary patterns of Type I genes in conifers.
Strengths and weaknesses of MADS-box gene phylogenies
In general, the support values in our phylogenies are quite low. This may be due to large datasets including quite diverse sequences and is common for MADS-box gene phylogenies in plants (Gramzow and Theißen, 2013). Therefore, we used two independent phylogenetic reconstructions using ML and Bayesian methods to test the stability of clades. Furthermore, we often observe paraphyletic relationships between the different gymnosperm sequences (Figs 2–6, 9, 11 and 13). Long branch attraction may, at least partially, explain the apparent paraphyletic relationship even though the methods we used here are less prone to long branch attraction than other phylogenetic methods (Bergsten, 2005). Also, some of the gymnosperm sequences are very short and may not provide enough information for the phylogenetic reconstruction algorithms to correctly place them in the phylogeny. Hence, the paraphyletic pattern of gymnosperm genes may often be an artefact and we largely ignored paraphyletic relationships and rather assumed monophyly of gymnosperm genes when estimating the number of MADS-box genes in the MRCAs of seed plants and gymnosperms.
Number of MADS-box genes in the MRCA of extant seed plants
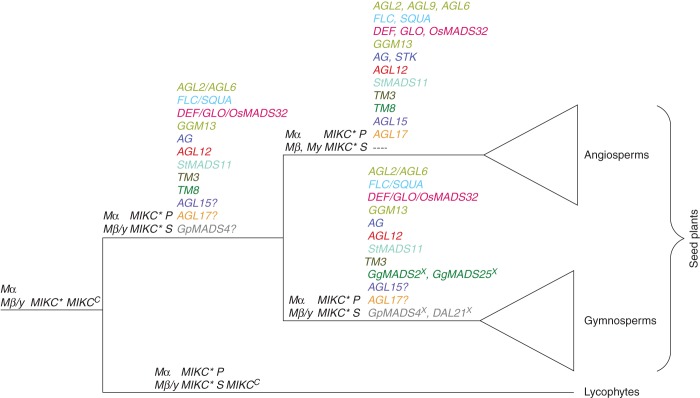
According to our phylogenies, the MRCA of extant seed plants contained at least 13–16 different MADS-box genes (Fig. 14). Of these, at least two were Type I genes [one Type I gene of clade I (Mα) and one Type I of clade II (Mβ/Mγ)], at least two were MIKC*-group genes [one MIKC*-group gene of the S clade and one MIKC*-group gene of the P clade (Nam et al., 2004)] and at least 9–12 were MIKCC-group genes (one each of the following clades: AGL2/AGL6, FLC/SQUA, DEF/GLO/OsMADS32, GGM13, AG, AGL12, StMADS11, TM3 and TM8; and possibly AGL15, AGL17 and GpMADS4). For MIKCC-group genes, the presence of AGL15- and AGL17-like genes needs to be verified as thus far gymnosperm genes with high similarity to the angiosperm AGL15- and AGL17-like genes have been identified solely based on genomic data and no evidence of transcription exists. Furthermore, support values for the corresponding clades are quite low and alignments of the MADS domains do not give a clear signal of whether the gymnosperm sequences belong to the AGL15- and AGL17-clades or not. The clade of GpMADS4-like genes is specific to gymnosperms. The deep branching of this clade in relation to the other ancient clades of Type II genes was not clear in our phylogenies. As it does not form a clear clade with another ancient clade of Type II genes, we hypothesize that this clade was present in the MRCA of seed plants and was subsequently lost in angiosperms. Given that phylogenies from previous studies also did not reveal any clear association of this clade with another major clade of MADS-box genes (Melzer et al., 2010; Carlsbecker et al., 2013), this hypothesis seems reasonable. However, it remains possible that phylogenic reconstructions have failed to place GpMADS4-like genes correctly thus far. Hence, more transcript sequences from gymnosperms may help to clarify the origin of AGL15-, AGL17- and GpMADS4-like genes.
Fig. 14.
Clades of MADS-box genes plotted on a simplified phylogeny of vascular plants. The crown groups of gymnosperms and angiosperms are shown as open triangles. At each node, the clades of MADS-box genes that had been established at this node according to our phylogenies are listed. The clades are sorted into three columns at each node where the first column lists Type I genes, the second column lists MIKC*-group genes and the third column lists MIKCC-group genes. Clade names generally refer to the first member that had been published, except for Mα and Mβ/γ, which denote different groups of Type I MADS-box genes, MIKCc and MIKC*, which refer to the two groups of Type II MIKC-type genes, and MIKC* S and MIKC* P, which are subclades of MIKC*-group genes. Clade names composed of clade names separated by slashes (‘/’) refer to clades that have not undergone the corresponding split. For example, the AGL2/AGL6-like genes in the stem group of seed plants gave rise to the clades of AGL2-like, AGL6-like and AGL9-like genes in the MRCA of extant angiosperms. MIKCc-group clades belonging to the same ancestral seed plant clade are shown in the same colour. A question mark behind a clade name indicates that the presence of this clade in the corresponding MRCA is less strongly supported than that of the other clades because putative clade members were only found in genome but not transcriptome data (AGL15 and AGL17) or because of ambiguities in tree topology (GpMADS4). The putative loss or absence of GpMADS4-like genes early in the evolution of angiosperms is indicated by minus signs. For lycophytes, the clades given at the terminal branch indicate that at least one clade member has been identified in at least one extant species of lycophytes. For angiosperms and gymnosperms, the clades given have been established in the stem group except for the two clades of TM8-like genes and the two clades of GpMADS4-like genes in gymnosperms (indicated by x). In these cases, our taxon sampling indicates presence of these clades in the MRCA of gnetophytes and conifers but as data from cycads and Ginkgo are missing we cannot infer presence in the MRCA of all extant gymnosperms.
Our work extends previous studies and increases the minimum number of MADS-box genes identified in the MRCA of extant seed plants by one to four. Earlier studies recognized the presence of two ancient clades of Type I genes that trace back close to the MRCA of land plants (Gramzow et al., 2012). At this stage, however, no gymnosperm Type I genes of clade I (Mα) could be identified but only a few gymnosperm Type I genes of clade II (Mβ/γ). Here we demonstrate that Type I genes of clade I are also present in gymnosperms. Similarly, the presence of two ancient clades of MIKC*-group genes that originated in the MRCA of euphyllophytes, the P- and the S-clade, has been described (Kwantes et al., 2012; Gramzow et al., 2012). In these previous studies, however, only gymnosperm members of the P-clade had been found whereas here we identified gymnosperm genes of both clades. At least eight MIKCC-group genes were predicted to be present in the MRCA of seed plants (Becker et al., 2000; Melzer et al., 2010). Our analyses reveal that this number should be increased to 9–12. In contrast to previous studies (Becker et al., 2000; Melzer et al., 2010; Kim et al., 2013), our phylogenies indicate that the duplication giving rise to AGL2- and AGL6-like genes may be specific to angiosperms and that the genes previously thought to be orthologues of AGL6-like genes in gymnosperms are actually orthologues to an ancestral AGL2/AGL6-like gene (Fig. 4). According to our scenario, only one AGL2/AGL6-like gene, instead of one AGL2- and one AGL6-like gene, was present in the MRCA of extant seed plants. Consequently, we do not have to assume a loss of AGL2-like genes in gymnosperms but two consecutive duplications at the base of angiosperms giving rise first to AGL2- and AGL6-like genes and then to SEP1- (AGL2-) and SEP3- (AGL9-) like genes. In line with a recent study (Ruelens et al., 2013), FLC- and SQUA-like genes of angiosperms form a clade in our phylogenies with the exception of FLC-like genes from rice. Neighbouring this clade are a number of MADS-box genes from gymnosperms. This suggests that FLC/SQUA-like genes are present in gymnosperms and that the MRCA of extant seed plants possessed at least one FLC/SQUA-like gene. Previous studies could not identify SQUA-like genes in gymnosperms (Becker et al., 2000; Melzer et al., 2010). Furthermore, our study indicates that AGL15-, AGL17- and GpMADS4-like genes were also present in the MRCA of extant seed plants. However, this finding needs to be tested with additional data, as described above. Finally, our study clarifies the origin of OsMADS32-like genes, which had previously been thought to be specific to monocots (Sang et al., 2012). In our phylogeny, we identify an OsMADS32-like gene in the basal angiosperm species Amborella trichopoda, Am.tr.OsM32 (Supplementary Data Fig. S6) as confirmed by other studies (The Amborella Genome Project, 2013). The clade of OsMADS32-like genes is sister to a clade of DEF- and GLO-like genes (Fig. 5), suggesting that OsMADS32-like genes originated by a duplication of an ancestral DEF/GLO/OsMADS32-like gene near the base of extant angiosperms.
Number of MADS-box genes in ancestral gymnosperms
From our phylogenies, we can infer some duplications in the lineage leading to the MRCA of gnetophytes (represented by Gnetum) and conifers after the lineage that led to angiosperms split off (Fig. 14). One duplication each happened in the TM8-clade, leading to GgMADS2- and GgMADS25-like genes, and in the GpMADS4-clade, generating GpMADS4- and DAL21-like genes. Hence, the number of MIKCC-group MADS-box genes is nearly as high in the MRCA of gnetophytes and conifers (14) as in the MRCA of angiosperms (17). If recent phylogenetic reconstructions are correct (Wu et al., 2011; Xi et al., 2013), data from Ginkgo and cycads will be required to determine whether the two gene duplications discussed above pre-date even earlier diversifications of extant gymnosperms than the split between gnetophytes and conifers.
Inferred functions of MADS-box genes in the MRCA of seed plants
Type I MADS-box genes
Only a few studies of Type I MADS-box gene functions have been published so far (Portereiko et al., 2006; Yoo et al., 2006; Bemer et al., 2008; Colombo et al., 2008; Kang et al., 2008; Steffen et al., 2008). These studies suggest as commonality a role for Type I MADS-box genes in female gametophyte, embryo and seed development in angiosperms (Gramzow and Theissen, 2010). Expression of one gymnosperm Type I gene was also found in embryo. Hence, a role in embryo development may be an ancestral function of Type I genes in seed plants. The expression of other gymnosperm Type I genes in a wide range of tissues (e.g. shoots, needles and cones) may represent transcriptional noise, indicate additional functions of gymnosperm Type I genes or restriction of Type I gene functions in angiosperms. Unfortunately, there are currently no data available, such as detailed gene expression pattern or mutant phenotypes, that would clarify the function of Type I genes in gymnosperms.
MIKC*-group genes
In ferns, these genes are expressed in male and hermaphroditic gametophytic tissue and in sporophytic tissue such as roots and stipes (Kwantes et al., 2012). In contrast, their expression is usually restricted to male gametophytic tissue (pollen) in basal and in derived angiosperms (Kwantes et al., 2012; Liu et al., 2013). The identified gymnosperm MIKC*-group genes are expressed in gametophytic and sporophytic tissues as well, indicating that no restriction to male gametophytic tissue occurred before the split of angiosperms and gymnosperms. Our analyses of transcriptome data reveal that P-clade genes are expressed in vegetative shoots, wood, and female and male reproductive organs, while the S-clade gene was found to be expressed in male reproductive organs. The broad expression pattern of genes of the P-clade suggests diverse functions during plant development, whereas S-clade genes seem to have a more restricted function in specifying male organs in gymnosperms. However, further investigations are needed to clarify the functions of MIKC*-group genes of extant gymnosperms. The data suggest that MIKC*-group genes may have had a role in the development of both male and female reproductive organs and in the development of vegetative tissues in the MRCA of extant seed plants. After the divergence of the lineages that led to extant gymnosperms and angiosperms, MIKC*-group genes became functionally restricted to the male gametophyte in angiosperms while they may have kept a broader role in gymnosperms.
AGL2/AGL6- and FLC/SQUA-like genes
The gymnosperm AGL2/AGL6-like genes identified here are expressed in shoots, needles and reproductive tissues. DAL1 and DAL14 from P. abies, which have been described as AGL6-like genes previously, are expressed in male and female cones (Carlsbecker et al., 2004, 2013). DAL1 is additionally expressed in vegetative tissues and has therefore been proposed to be involved in vegetative development and to regulate phase change from juvenile to adult (Carlsbecker et al., 2004). Angiosperm AGL2- and AGL6-like genes have roles in the transition to flowering and in lateral organ and flower development (Pelaz et al., 2000; Koo et al., 2010; Yoo et al., 2011). It has also been shown that some AGL6-like genes can act redundantly with AGL2-like genes in flower organ formation (Rijpkema et al., 2009). Combining this information suggests that ancestral AGL2/AGL6-like genes may have had roles in the transition to reproductive development and in the development of vegetative and reproductive organs. Angiosperm FLC-like genes have roles in developmental phase changes (Michaels and Amasino, 2001; Deng et al., 2011). The expression of gymnosperm FLC/SQUA-like genes identified here in a number of vegetative tissues may point to a similar role for these genes. Furthermore, gymnosperm FLC/SQUA-like genes are expressed in female cones. The function of SQUA-like genes in angiosperms in the development of the sterile organs of the flower and in meristem identity specification (Mandel et al., 1992; Ferrándiz et al., 2000) may represent a new function for these genes in angiosperms.
DEF/GLO/OsMADS32-like and GGM13-like genes
The gymnosperm genes that are sister to the clades of DEF-, GLO- and OsMADS32-like genes of angiosperms were found to be expressed in male reproductive tissues. This is consistent with previous reports about the expression of GGM2 of G. gnemon in male cones, of DAL11, DAL12 and DAL13 of P. abies in male bud meristems, and of PrDGL of P. radiata as well as of CjMADS1 and CjMADS2 of C. japonica in male strobili (Mouradov et al., 1999; Winter et al., 1999; Fukui et al., 2001; Sundstrom and Engstrom, 2002). Similarly, DEF- and GLO-like genes in angiosperms are expressed in male reproductive organs and in petals and specify stamen and petal identity (Schwarz-Sommer et al., 1990; Goto and Meyerowitz, 1994). These expression patterns point to an ancestral function of DEF/GLO/OsMADS32-like genes in the development of male reproductive organs. In contrast, gymnosperm GGM13-like genes are mainly expressed in female reproductive organs (Becker et al., 2002; Carlsbecker et al., 2013; Lovisetto et al., 2013), which is again consistent with what is known about the expression of GGM13-like genes in angiosperms (Becker et al., 2002; Yang et al., 2012). Hence, these genes may have an ancestral function in the development of female reproductive organs.
AG-like and AGL12-like genes
In previous publications, expression of gymnosperm AG-like genes has been observed mainly in reproductive organs, such as DAL2 of P. abies in female cones, JcMADS2 of Juniperus communis, TdMADS3 of Thujopsis dolabrata and CjMADS4 of C. japonica in seed cones and pollen cones, and SAG1 of P. mariana and GGM3 of G. gnemon in male and female cones (Rutledge et al., 1998; Tandre et al., 1998; Winter et al., 1999; Englund et al., 2011; Groth et al., 2011). However, GBM5 of G. biloba was reported to be expressed not only in reproductive organs, but also in vegetative leaves (Jager et al., 2003). Interestingly, our analysis of transcriptome data reveals expression of gymnosperm AG-like genes in many different tissues ranging from roots via shoots and leaves to reproductive organs. The possibly wide expression pattern of gymnosperm AG-like genes as observed in G. biloba and in transcriptome data does not completely comply with the function of AG-like genes in angiosperms in the development of reproductive organs and roots (Yanofsky et al., 1990; Liljegren et al., 2000; Pinyopich et al., 2003; Moreno-Risueno et al., 2010). The expression of gymnosperm AG-like genes in other tissues may represent just transcriptional noise, or the ancestral function of AG-like genes in the MRCA of seed plants might have been broader, and AG-like genes in angiosperms lost functions outside of roots and reproductive organs.
The expression of gymnosperm AGL12-like genes in roots and shoots/needles was found in trancriptome data here and had previously been described for DAL5 of P. abies (Carlsbecker et al., 2013). This fits with the expression of AGL12-like genes from angiosperms in roots, leaves and floral meristems and their function in root development and transition to reproductive development (Tapia-López et al., 2008). Hence, the ancestral function of AGL12-like genes in the MRCA of extant seed plants probably involved root development and phase change to reproductive development.
AGL12-like genes form the sister clade of AG-like genes (Becker and Theissen, 2003). The probably broad functions of the ancestral AG-like gene as described above and the probable involvement of the ancestral AGL12-like gene in root development and transition to reproductive development suggest that the ancestral AG/AGL12-like gene had broad functions as well, and that the functions of AGL12-like genes were restricted to root development and phase change to reproductive development early after the duplication leading to AG- and AGL12-like genes before the divergence of angiosperms and gymnosperms.
AGL15- and AGL17-like genes
The presence of AGL15- and AGL17-like genes in the MRCA of extant seed plants is not clear but is suggested by our phylogeny. For possible gymnosperm AGL15- and AGL17-like genes, no expression data are available. From angiosperms, some functions of AGL15- and AGL17-like genes are known, such as root and leaf development and transition to reproductive development (Zhang and Forde, 1998; Adamczyk et al., 2007; Kutter et al., 2007; Han et al., 2008). It is possible that MADS-box genes are involved in these processes in gymnosperms as well and that putative AGL15- and AGL17-like genes carry out these functions also in gymnosperms.
GpMADS4-like genes
Expression of GpMADS4 was detected in female reproductive organs (Shindo et al., 1999). However, this was the only tissue studied. The GpMADS4 orthologues GGM7 and DAL10 were found to be expressed during the development of female and male cones (Becker and Theissen, 2003; Carlsbecker et al., 2003, 2013). DAL10 has been hypothesized to be involved in meristem determination of reproductive buds establishing reproductive identity (Carlsbecker et al., 2013). In contrast to genes of the GpMADS4-subclade, DAL21 of the DAL21-subclade is specifically expressed in female cones (Carlsbecker et al., 2013). Hence, the ancestral GpMADS4-like gene in gymnosperms may have had a function in determining both types of reproductive structures. After the duplication event during gymnosperm evolution, the function of the DAL21-subclade may have been restricted to the development of female cones. According to our phylogeny, GpMADS4-like genes were present in the MRCA of seed plants but lost in angiosperms. Speculation about the function of GpMADS4-like genes in the MRCA of seed plants is difficult, but their expression patterns in extant gymnosperms suggest a role in reproductive organ development also in the MRCA of extant seed plants. This role may have been taken over by other genes or may have become dispensable in angiosperms leading to a loss of GpMADS4-like genes in angiosperms.
Expansions of the clades of StMADS11-, TM3- and TM8-like genes in gymnosperms
By far the highest numbers of MADS-box genes in gymnosperms were found for the StMADS11, TM3 and TM8 clades. MADS-box genes belonging to these clades were identified in nearly all gymnosperm species studied.
We identified two large subclades, PaMADS19- and PaMADS20-like genes of conifer StMADS11-like genes. Similarly, four subclades, SVP-, ZMM17-, MPF1- and MPF2-like genes, have been defined for angiosperm StMADS11-like genes (Khan and Ali, 2013). Angiosperm StMADS11-like genes are mainly expressed in vegetative tissues such as roots, leaves and shoots (Borner et al., 2000; Lee et al., 2000; Michaels et al., 2003; Fornara et al., 2008; Wingen et al., 2012). Different functions have been described for these genes. AGL24 and SVP of A. thaliana act as promoter and repressor of floral transition, respectively (Hartmann et al., 2000; Michaels et al., 2003). The three StMADS11-like genes of rice, OsMADS22, OsMADS47 and OsMADS55, are involved in the negative regulation of brassinosteroid responses (Fornara et al., 2008; Lee et al., 2008) and MPF2 of Physalis floridana is important for male fertility and the development of the ‘inflated calyx syndrome’ (He and Saedler, 2005). The identification of conifer StMADS11-like genes from transcriptome data of mixed shoots, developing buds and roots indicates that these genes may have diverse functions in conifers as well.
We observed two basal duplications of TM3-like genes from Pinaceae resulting in the three subclades DAL19-, DAL3- and PrMADS4-like genes. In P. radiata an expansion of the clade of TM3-like genes has been previously suggested (Walden et al., 1999). The four sequences studied by Walden et al. (1999) are distributed over the three subclades of our phylogeny. All PrMADS genes are expressed in male cones, roots, needles and shoots with the exception of PrMADS6, which is not expressed in roots. Furthermore, four TM3-like genes have been identified from P. abies previously (Tandre et al., 1995; Carlsbecker et al., 2013; Uddenberg et al., 2013) of which two – DAL3 and DAL19 – are expressed in seedlings, cambium, vegetative shoots, and female and male cones at different developmental stages. This indicates that conifer TM3-like genes may be involved in the development of different tissues. DAL19 is hypothesized to be involved in phase change from vegetative to reproductive development (Uddenberg et al., 2013). The angiosperm genes of the TM3-clade AGL20/SOC1 from A. thaliana, its paralogues AGL19, AGL42, AGL71 and AGL72, and the rice genes OsMADS50 and OsMADS56 act as key regulators of flowering time (Lee et al., 2000; Tadege et al., 2003; Schonrock et al., 2006; Ryu et al., 2009; Dorca-Fornell et al., 2011). Hence, a function in the transition to reproductive growth may have already been performed by the ancestral TM3-like gene in the MRCA of extant seed plants. Subsequently, some TM3-like genes evolved other functions, such as AGL14/XAL2 from A. thaliana, which is involved in root development (Garay-Arroyo et al., 2013) and AGL42/FYF, which is involved in flower senescence/abscission (Chen et al., 2011). Similarly, diversification of the TM3-like genes of conifers could have led to different functions similar to the evolution of TM3-like genes in angiosperms.
TM8 was first described in tomato where its transcripts were detected in gynoecia, stamens and petals (Pnueli et al., 1991). The TM8-like gene ERAF17 of cucumber is expressed in sepals, petals and ovaries of female flowers but not in male flowers (Ando et al., 2001). TM8-like genes have been lost in a number of angiosperm lineages such as Brassicaceae and Poaceae (Parenicova et al., 2003; Arora et al., 2007). In gymnosperms three TM8-like genes from G. biloba and one TM8-like gene from T. baccata were described and their expression patterns were examined recently (Lovisetto et al., 2012). GbMADS11 and GbMADS6 from G. biloba are expressed in leaves, male strobili and ovules, whereas GbMADS7 is expressed only weakly in these tissues. The TbTM8 gene from T. baccata is expressed strongly in ovules and developing arils. Two transcript sequences from C. japonica were identified from male strobili. The sequences from P. abies identified here were detected only in wood tissues and buds. Other conifer sequences were isolated from needles, shoot tips and wood tissues as well. Given the diverse expression patterns of TM8-like genes in gymnosperms and angiosperms, it is difficult to infer a function for the ancestral TM8-like gene in the MRCA of seed plants. Judged by the number of gene duplications and gene losses observed for this clade, it seems that TM8-like genes are a clade of fast evolving genes, which have acquired different functions in different species of conifers as well as of angiosperms.
CONCLUSIONS
We show that the minimal number of Type I genes was much lower than the minimal number of Type II MADS-box genes in the MRCA of seed plants. Our analysis of transcriptome data reveals that gymnosperm MADS-box genes are expressed in a great variety of tissues, indicating diverse roles of MADS-box genes for the development of gymnosperms. Our study is the first that provides a comprehensive overview about MADS-box genes in conifers and thus will provide a framework for future work on MADS-box genes in seed plants.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We are grateful to Ove Nilsson (Umea Plant Science Center, Sweden) for his invitation to participate in the Picea abies genome project, and to Charlie Scutt (ENS Lyon, France) for his invitation to contribute to this special issue of Annals of Botany. Part of this work has been funded by a grant (Th 417/8–1) from the Deutsche Forschungsgemeinschaft (DFG) to G.T.
LITERATURE CITED
- Adamczyk BJ, Lehti-Shiu MD, Fernandez DE. The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. The Plant Journal. 2007;50:1007–1019. doi: 10.1111/j.1365-313X.2007.03105.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Pelaz S, Liljegren SJ, et al. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5328–5333. doi: 10.1073/pnas.97.10.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando S, Sato Y, Kamachi S, Sakai S. Isolation of a MADS-box gene (ERAF17) and correlation of its expression with the induction of formation of female flowers by ethylene in cucumber plants (Cucumis sativus L.) Planta. 2001;213:943–52. doi: 10.1007/s004250100571. [DOI] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, et al. MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics. 2007;8:242. doi: 10.1186/1471-2164-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks JA, Nishiyama T, Hasebe, et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 2011;332:960–3. doi: 10.1126/science.1203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker EI, Ashton NW. A parsimonious model of lineage-specific expansion of MADS-box genes in Physcomitrella patens. Plant Cell Reports. 2013;32:1161–1177. doi: 10.1007/s00299-013-1411-8. [DOI] [PubMed] [Google Scholar]
- Becker A, Theissen G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Molecular Phylogenetics and Evolution. 2003;29:464–489. doi: 10.1016/s1055-7903(03)00207-0. [DOI] [PubMed] [Google Scholar]
- Becker A, Winter KU, Meyer B, Saedler H, Theissen G. MADS-Box gene diversity in seed plants 300 million years ago. Molecular Biology and Evolution. 2000;17:1425–34. doi: 10.1093/oxfordjournals.molbev.a026243. [DOI] [PubMed] [Google Scholar]
- Becker A, Kaufmann K, Freialdenhoven A, et al. A novel MADS-box gene subfamily with a sister-group relationship to class B floral homeotic genes. Molecular Genetics and Genomics. 2002;266:942–50. doi: 10.1007/s00438-001-0615-8. [DOI] [PubMed] [Google Scholar]
- Bemer M, Wolters-Arts M, Grossniklaus U, Angenent GC. The MADS domain protein DIANA acts together with AGAMOUS-LIKE80 to specify the central cell in Arabidopsis ovules. The Plant Cell. 2008;20:2088–101. doi: 10.1105/tpc.108.058958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemer M, Gordon J, Weterings K, Angenent GC. Divergence of recently duplicated Mγ-type MADS-box genes in Petunia. Molecular Biology and Evolution. 2010;27:481–495. doi: 10.1093/molbev/msp279. [DOI] [PubMed] [Google Scholar]
- Bergsten J. A review of long-branch attraction. Cladistics. 2005;21:163–193. doi: 10.1111/j.1096-0031.2005.00059.x. [DOI] [PubMed] [Google Scholar]
- Birol I, Raymond A, Jackman SD, et al. Assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data. Bioinformatics. 2013;29:1492–1497. doi: 10.1093/bioinformatics/btt178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner R, Kampmann G, Chandler J, et al. A MADS domain gene involved in the transition to flowering in Arabidopsis. The Plant Journal. 2000;24:591–599. doi: 10.1046/j.1365-313x.2000.00906.x. [DOI] [PubMed] [Google Scholar]
- Bowe LM, Coat G. Phylogeny of seed plants based on all three genomic compartments: extant gymnosperms are monophyletic and Gnetales' closest relatives are conifers. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:4092–4097. doi: 10.1073/pnas.97.8.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A, Sundstrom J, Tandre K, et al. The DAL10 gene from Norway spruce (Picea abies) belongs to a potentially gymnosperm-specific subclass of MADS-box genes and is specifically active in seed cones and pollen cones. Evolution and Development. 2003;5:551–61. doi: 10.1046/j.1525-142x.2003.03060.x. [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Tandre K, Johanson U, Englund M, Engstrom P. The MADS-box gene DAL1 is a potential mediator of the juvenile-to-adult transition in Norway spruce (Picea abies) The Plant Journal. 2004;40:546–57. doi: 10.1111/j.1365-313X.2004.02226.x. [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Sundstrom JF, Englund M, et al. Molecular control of normal and acrocona mutant seed cone development in Norway spruce (Picea abies) and the evolution of conifer ovule-bearing organs. New Phytologist. 2013;200:261–75. doi: 10.1111/nph.12360. [DOI] [PubMed] [Google Scholar]
- Chaw S-M, Zharkikh A, Sung HM, Lau TC, Li WH. Molecular phylogeny of extant gymnosperms and seed plant evolution: analysis of nuclear 18S rRNA sequences. Molecular Biology and Evolution. 1997;14:56–68. doi: 10.1093/oxfordjournals.molbev.a025702. [DOI] [PubMed] [Google Scholar]
- Chaw S-M, Parkinson CL, Cheng Y, Vincent TM, Palmer JD. Seed plant phylogeny inferred from all three plant genomes: monophyly of extant gymnosperms and origin of Gnetales from conifers. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:4086–4091. doi: 10.1073/pnas.97.8.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MK, Hsu WH, Lee PF, Thiruvengadam M, Chen HI, Yang CH. The MADS box gene, FOREVER YOUNG FLOWER, acts as a repressor controlling floral organ senescence and abscission in Arabidopsis. The Plant Journal. 2011;68:168–185. doi: 10.1111/j.1365-313X.2011.04677.x. [DOI] [PubMed] [Google Scholar]
- Colombo M, Masiero S, Vanzulli S, Lardelli P, Kater MM, Colombo L. AGL23, a type I MADS-box gene that controls female gametophyte and embryo development in Arabidopsis. The Plant Journal. 2008;54:1037–48. doi: 10.1111/j.1365-313X.2008.03485.x. [DOI] [PubMed] [Google Scholar]
- Cronk QCB. Plant evolution and development in a post-genomic context. Nature Reviews Genetics. 2001;2:607–619. doi: 10.1038/35084556. [DOI] [PubMed] [Google Scholar]
- De Bodt S, Raes J, Florquin K, et al. Genomewide structural annotation and evolutionary analysis of the type I MADS-box genes in plants. Journal of Molecular Evolution. 2003;56:573–86. doi: 10.1007/s00239-002-2426-x. [DOI] [PubMed] [Google Scholar]
- Deng W, Ying H, Helliwell CA, Taylor JM, Peacock WJ, Dennis ES. FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6680–6685. doi: 10.1073/pnas.1103175108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle E, Ferraz C, Rombauts S, et al. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11647–52. doi: 10.1073/pnas.0604795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Riquelme J, Lijavetzky D, Martinez-Zapater JM, Carmona MJ. Genome-wide analysis of MIKCC-type MADS box genes in grapevine. Plant Physiology. 2009;149:354–369. doi: 10.1104/pp.108.131052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorca-Fornell C, Gregis V, Grandi V, Coupland G, Colombo L, Kater MM. The Arabidopsis SOC1-like genes AGL42, AGL71 and AGL72 promote flowering in the shoot apical and axillary meristems. The Plant Journal. 2011;67:1006–17. doi: 10.1111/j.1365-313X.2011.04653.x. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Hidden Markov models. Current Opinion in Structural Biology. 1996;6:361–365. doi: 10.1016/s0959-440x(96)80056-x. [DOI] [PubMed] [Google Scholar]
- Englund M, Carlsbecker A, Engstrom P, Vergara-Silva F. Morphological ‘primary homology’ and expression of AG -subfamily MADS-box genes in pines, podocarps, and yews. Evolution & Development. 2011;13:171–181. doi: 10.1111/j.1525-142X.2011.00467.x. [DOI] [PubMed] [Google Scholar]
- Ferrándiz C, Gu Q, Martienssen R, Yanofsky MF. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development. 2000;127:725–734. doi: 10.1242/dev.127.4.725. [DOI] [PubMed] [Google Scholar]
- Fornara F, Gregis V, Pelucchi N, Colombo L, Kater M. The rice StMADS11-like genes OsMADS22 and OsMADS47 cause floral reversions in Arabidopsis without complementing the svp and agl24 mutants. Journal of Experimental Botany. 2008;59:2181–2190. doi: 10.1093/jxb/ern083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui M, Futamura N, Mukai Y, Wang Y, Nagao A, Shinohara K. Ancestral MADS box genes in Sugi, Cryptomeria japonica D. Don (Taxodiaceae), homologous to the B function genes in angiosperms. Plant and Cell Physiology. 2001;42:566–75. doi: 10.1093/pcp/pce069. [DOI] [PubMed] [Google Scholar]
- Futamura N, Totoki Y, Toyoda A, et al. Characterization of expressed sequence tags from a full-length enriched cDNA library of Cryptomeria japonica male strobili. BMC Genomics. 2008;9:383. doi: 10.1186/1471-2164-9-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garay-Arroyo A, Ortiz-Moreno E, de la Paz Sanchez M, et al. The MADS transcription factor XAL2/AGL14 modulates auxin transport during Arabidopsis root development by regulating PIN expression. EMBO Journal. 2013;32:2884–95. doi: 10.1038/emboj.2013.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gil MR. Evolutionary aspects of functional and pseudogene members of the phytochrome gene family in Scots pine. Journal of Molecular Evolution. 2008;67:222–232. doi: 10.1007/s00239-008-9135-z. [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, et al. A draft sequence of the rice genome (Oryza sativa L. ssp japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes & Development. 1994;8:1548–1560. doi: 10.1101/gad.8.13.1548. [DOI] [PubMed] [Google Scholar]
- Gramzow L, Theissen G. A hitchhiker's guide to the MADS world of plants. Genome Biology. 2010;11:214. doi: 10.1186/gb-2010-11-6-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramzow L, Theißen G. Phylogenomics of MADS-box genes in plants—two opposing life styles in one gene family. Biology. 2013;2:1150–1164. doi: 10.3390/biology2031150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramzow L, Ritz MS, Theissen G. On the origin of MADS-domain transcription factors. Trends in Genetics. 2010;26:149–153. doi: 10.1016/j.tig.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Gramzow L, Barker E, Schulz C, et al. Selaginella genome analysis – entering the ‘homoplasy heaven’ of the MADS world. Frontiers in Plant Science. 2012;3:214. doi: 10.3389/fpls.2012.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth E, Tandre K, Engstrom P, Vergara-Silva F. AGAMOUS subfamily MADS-box genes and the evolution of seed cone morphology in Cupressaceae and Taxodiaceae. Evolution and Development. 2011;13:159–70. doi: 10.1111/j.1525-142X.2011.00466.x. [DOI] [PubMed] [Google Scholar]
- Gugerli F, Sperisen C, Büchler U, Brunner I, Brodbeck S, Palmer JD, Qiu YL. The evolutionary split of Pinaceae from other conifers: evidence from an intron loss and a multigene phylogeny. Molecular Phylogenetics and Evolution. 2001;21:167–175. doi: 10.1006/mpev.2001.1004. [DOI] [PubMed] [Google Scholar]
- Han P, García-Ponce B, Fonseca-Salazar G, Alvarez-Buylla ER, Yu H. AGAMOUS-LIKE 17, a novel flowering promoter, acts in a FT-independent photoperiod pathway. The Plant Journal. 2008;55:253–265. doi: 10.1111/j.1365-313X.2008.03499.x. [DOI] [PubMed] [Google Scholar]
- Hartmann U, Höhmann S, Nettesheim K, Wisman E, Saedler H, Huijser P. Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. The Plant Journal. 2000;21:351–360. doi: 10.1046/j.1365-313x.2000.00682.x. [DOI] [PubMed] [Google Scholar]
- He C, Saedler H. Heterotopic expression of MPF2 is the key to the evolution of the Chinese lantern of Physalis, a morphological novelty in Solanaceae. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5779–5784. doi: 10.1073/pnas.0501877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedman H, Zhu TQ, von Arnold S, Sohlberg JJ. Analysis of the WUSCHEL-RELATED HOMEOBOX gene family in the conifer picea abies reveals extensive conservation as well as dynamic patterns. BMC Plant Biology. 2013;13 doi: 10.1186/1471-2229-13-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henschel K, Kofuji R, Hasebe M, Saedler H, Munster T, Theissen G. Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Molecular Biology and Evolution. 2002;19:801–814. doi: 10.1093/oxfordjournals.molbev.a004137. [DOI] [PubMed] [Google Scholar]
- Jager M, Hassanin A, Manuel M, Le Guyader H, Deutsch J. MADS-box genes in Ginkgo biloba and the evolution of the AGAMOUS family. Molecular Biology and Evolution. 2003;20:842–54. doi: 10.1093/molbev/msg089. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Kang IH, Steffen JG, Portereiko MF, Lloyd A, Drews GN. The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. The Plant Cell. 2008;20:635–647. doi: 10.1105/tpc.107.055137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MR, Ali GM. Functional evolution of cis-regulatory modules of STMADS11 superclade MADS-box genes. Plant Molecular Biology. 2013;83:489–506. doi: 10.1007/s11103-013-0105-5. [DOI] [PubMed] [Google Scholar]
- Kim S, Soltis PS, Soltis DE. AGL6-like MADS-box genes are sister to AGL2-like MADS-box genes. Journal of Plant Biology. 2013;56:315–325. [Google Scholar]
- Kinlaw CS, Neale DB. Complex gene families in pine genomes. Trends in Plant Science. 1997;2:356–359. [Google Scholar]
- Koo SC, Bracko O, Park MS, et al. Control of lateral organ development and flowering time by the Arabidopsis thaliana MADS-box gene AGAMOUS-LIKE6. The Plant Journal. 2010;62:807–816. doi: 10.1111/j.1365-313X.2010.04192.x. [DOI] [PubMed] [Google Scholar]
- Kutter C, Schob H, Stadler M, Meins F, Jr, Si-Ammour A. MicroRNA-mediated regulation of stomatal development in Arabidopsis. The Plant Cell. 2007;19:2417–2429. doi: 10.1105/tpc.107.050377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvarnheden A, Albert VA, Engstrom P. Molecular evolution of cdc2 pseudogenes in spruce (Picea) Plant Molecular Biology. 1998;36:767–774. doi: 10.1023/a:1005901413475. [DOI] [PubMed] [Google Scholar]
- Kwantes M, Liebsch D, Verelst W. How MIKC* MADS-box genes originated and evidence for their conserved function throughout the evolution of vascular plant gametophytes. Molecular Biology and Evolution. 2012;29:293–303. doi: 10.1093/molbev/msr200. [DOI] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, et al. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes and Development. 2000;14:2366–76. doi: 10.1101/gad.813600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Choi SC, An G. Rice SVP-group MADS-box proteins, OsMADS22 and OsMADS55, are negative regulators of brassinosteroid responses. The Plant Journal. 2008;54:93–105. doi: 10.1111/j.1365-313X.2008.03406.x. [DOI] [PubMed] [Google Scholar]
- Leseberg CH, Li A, Kang H, Duvall M, Mao L. Genome-wide analysis of the MADS-box gene family in Populus trichocarpa. Gene. 2006;378:84–94. doi: 10.1016/j.gene.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature. 2000;404:766–770. doi: 10.1038/35008089. [DOI] [PubMed] [Google Scholar]
- Liu Y, Cui S, Wu F, et al. Functional conservation of MIKC*-Type MADS box genes in Arabidopsis and rice pollen maturation. The Plant Cell. 2013;25:1288–303. doi: 10.1105/tpc.113.110049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz WW, Ayyampalayam S, Bordeaux JM, et al. Conifer DBMagic: a database housing multiple de novo transcriptome assemblies for 12 diverse conifer species. Tree Genetics & Genomes. 2012;8:1477–1485. [Google Scholar]
- Lovisetto A, Guzzo F, Tadiello A, Toffali K, Favretto A, Casadoro G. Molecular analyses of MADS-box genes trace back to Gymnosperms the invention of fleshy fruits. Molecular Biology and Evolution. 2012;29:409–419. doi: 10.1093/molbev/msr244. [DOI] [PubMed] [Google Scholar]
- Lovisetto A, Guzzo F, Busatto N, Casadoro G. Gymnosperm B-sister genes may be involved in ovule/seed development and, in some species, in the growth of fleshy fruit-like structures. Annals of Botany. 2013;112:535–544. doi: 10.1093/aob/mct124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Yanofsky MF, Meyerowitz EM. AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes and Development. 1991;5:484–95. doi: 10.1101/gad.5.3.484. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- Melzer R, Wang YQ, Theissen G. The naked and the dead: the ABCs of gymnosperm reproduction and the origin of the angiosperm flower. Seminars in Cell and Developmental Biology. 2010;21:118–128. doi: 10.1016/j.semcdb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–251. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Jackson S. The first 50 plant genomes. Plant Genome. 2013;6 [Google Scholar]
- Michaels SD, Amasino RM. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. The Plant Cell Online. 2001;13:935–941. doi: 10.1105/tpc.13.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Ditta G, Gustafson-Brown C, Pelaz S, Yanofsky M, Amasino RM. AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. The Plant Journal. 2003;33:867–874. doi: 10.1046/j.1365-313x.2003.01671.x. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. Gateway Computing Environments Workshop (GCE), 2010. Piscataway, NJ: IEEE; 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. [Google Scholar]
- Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science. 2010;329:1306–1311. doi: 10.1126/science.1191937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov A, Hamdorf B, Teasdale RD, Kim JT, Winter KU, Theissen G. A DEF/GLO-like MADS-box gene from a gymnosperm: Pinus radiata contains an ortholog of angiosperm B class floral homeotic genes. Developmental Genetics. 1999;25:245–252. doi: 10.1002/(SICI)1520-6408(1999)25:3<245::AID-DVG7>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Münster T, Faigl W, Saedler H, Theißen G. Evolutionary aspects of MADS-box genes in the eusporangiate fern Ophioglossum. Plant Biology. 2002;4:474–483. [Google Scholar]
- Nam J, Kim J, Lee S, An G, Ma H, Nei M. Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1910–5. doi: 10.1073/pnas.0308430100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, et al. The Norway spruce genome sequence and conifer genome evolution. Nature. 2013;497:579–84. doi: 10.1038/nature12211. [DOI] [PubMed] [Google Scholar]
- Parenicova L, de Folter S, Kieffer M, et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. The Plant Cell. 2003;15:1538–51. doi: 10.1105/tpc.011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405:200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- Pelaz S, Tapia-Lopez R, Alvarez-Buylla ER, Yanofsky MF. Conversion of leaves into petals in Arabidopsis. Current Biology. 2001;11:182–184. doi: 10.1016/s0960-9822(01)00024-0. [DOI] [PubMed] [Google Scholar]
- Peterson KM, Rychel AL, Torii KU. Out of the mouths of plants: the molecular basis of the evolution and diversity of stomatal development. The Plant Cell. 2010;22:296–306. doi: 10.1105/tpc.109.072777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyopich A, Ditta GS, Savidge B, et al. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature. 2003;424:85–88. doi: 10.1038/nature01741. [DOI] [PubMed] [Google Scholar]
- Pnueli L, Abu-Abeid M, Zamir D, Nacken W, Schwarz-Sommer Z, Lifschitz E. The MADS box gene family in tomato: temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. The Plant Journal. 1991;1:255–266. [PubMed] [Google Scholar]
- Portereiko MF, Lloyd A, Steffen JG, Punwani JA, Otsuga D, Drews GN. AGL80 is required for central cell and endosperm development in Arabidopsis. The Plant Cell. 2006;18:1862–1872. doi: 10.1105/tpc.106.040824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Rijpkema AS, Zethof J, Gerats T, Vandenbussche M. The petunia AGL6 gene has a SEPALLATA-like function in floral patterning. The Plant Journal. 2009;60:1–9. doi: 10.1111/j.1365-313X.2009.03917.x. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Roshan U, Livesay DR. Probalign: multiple sequence alignment using partition function posterior probabilities. Bioinformatics. 2006;22:2715–2721. doi: 10.1093/bioinformatics/btl472. [DOI] [PubMed] [Google Scholar]
- Ruelens P, de Maagd RA, Proost S, Theißen G, Geuten K, Kaufmann K. FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nature Communications. 2013;4:2280. doi: 10.1038/ncomms3280. [DOI] [PubMed] [Google Scholar]
- Rutledge R, Regan S, Nicolas O, et al. Characterization of an AGAMOUS homologue from the conifer black spruce (Picea mariana) that produces floral homeotic conversions when expressed in Arabidopsis. The Plant Journal. 1998;15:625–634. doi: 10.1046/j.1365-313x.1998.00250.x. [DOI] [PubMed] [Google Scholar]
- Ryu CH, Lee S, Cho LH, et al. OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant, Cell & Environment. 2009;32:1412–27. doi: 10.1111/j.1365-3040.2009.02008.x. [DOI] [PubMed] [Google Scholar]
- Sang X, Li Y, Luo Z, et al. CHIMERIC FLORAL ORGANS1, encoding a monocot-specific MADS box protein, regulates floral organ identity in rice. Plant Physiology. 2012;160:788–807. doi: 10.1104/pp.112.200980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research. 2012;40:D13–25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonrock N, Bouveret R, Leroy O, et al. Polycomb-group proteins repress the floral activator AGL19 in the FLC-independent vernalization pathway. Genes Development. 2006;20:1667–78. doi: 10.1101/gad.377206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Huijser P, Nacken W, Saedler H, Sommer H. Genetic control of flower development by homeotic genes in Antirrhinum majus. Science. 1990;250:931–936. doi: 10.1126/science.250.4983.931. [DOI] [PubMed] [Google Scholar]
- Shindo S, Ito M, Ueda K, Kato M, Hasebe M. Characterization of MADS genes in the gymnosperm Gnetum parvifolium and its implication on the evolution of reproductive organs in seed plants. Evolution and Development. 1999;1:180–190. doi: 10.1046/j.1525-142x.1999.99024.x. [DOI] [PubMed] [Google Scholar]
- Smaczniak C, Immink RG, Angenent GC, Kaufmann K. Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development. 2012;139:3081–3098. doi: 10.1242/dev.074674. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Steffen JG, Kang IH, Portereiko MF, Lloyd A, Drews GN. AGL61 interacts with AGL80 and is required for central cell development in Arabidopsis. Plant Physiology. 2008;148:259–268. doi: 10.1104/pp.108.119404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundstrom J, Engstrom P. Conifer reproductive development involves B-type MADS-box genes with distinct and different activities in male organ primordia. The Plant Journal. 2002;31:161–169. doi: 10.1046/j.1365-313x.2002.01343.x. [DOI] [PubMed] [Google Scholar]
- Tadege M, Sheldon CC, Helliwell CA, Upadhyaya NM, Dennis ES, Peacock WJ. Reciprocal control of flowering time by OsSOC1 in transgenic Arabidopsis and by FLC in transgenic rice. Plant Biotechnology Journal. 2003;1:361–369. doi: 10.1046/j.1467-7652.2003.00034.x. [DOI] [PubMed] [Google Scholar]
- Tandre K, Albert VA, Sundas A, Engstrom P. Conifer homologs to genes that control floral development in angiosperms. Plant Molecular Biology. 1995;27:69–78. doi: 10.1007/BF00019179. [DOI] [PubMed] [Google Scholar]
- Tandre K, Svenson M, Svensson ME, Engstrom P. Conservation of gene structure and activity in the regulation of reproductive organ development of conifers and angiosperms. The Plant Journal. 1998;15:615–623. doi: 10.1046/j.1365-313x.1998.00236.x. [DOI] [PubMed] [Google Scholar]
- Tapia-López R, García-Ponce B, Dubrovsky JG, et al. An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiology. 2008;146:1182–1192. doi: 10.1104/pp.107.108647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Amborella Genome Project. The Amborella genome and the evolution of flowering plants. Science. 2013;342:1241089. doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Theissen G, Becker A, Di Rosa A, et al. A short history of MADS-box genes in plants. Plant Molecular Biology. 2000;42:115–149. [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobner W, Ramirez L, Motte P, et al. GLOBOSA – a homeotic gene which interacts with DEFICIENS in the control of antirrhinum floral organogenesis. EMBO Journal. 1992;11:4693–4704. doi: 10.1002/j.1460-2075.1992.tb05574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Uddenberg D, Reimegard J, Clapham D, et al. Early cone setting in Picea abies acrocona is associated with increased transcriptional activity of a MADS box transcription factor. Plant Physiology. 2013;161:813–823. doi: 10.1104/pp.112.207746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden AR, Walter C, Gardner RC. Genes expressed in Pinus radiata male cones include homologs to anther-specific and pathogenesis response genes. Plant Physiology. 1999;121:1103–1116. doi: 10.1104/pp.121.4.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia H, Josefsson C, Dilkes B, Kirkbride R, Harada J, Comai L. Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility. Current Biology. 2009;19:1128–1132. doi: 10.1016/j.cub.2009.05.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn JL, Lee JM, Tearse BR, Neale DB. TreeGenes: a forest tree genome database. International Journal of Plant Genomics. 2008;2008:412875. doi: 10.1155/2008/412875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Molecular Biology and Evolution. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Wingen LU, Münster T, Faigl W, et al. Molecular genetic basis of pod corn (Tunicate maize) Proceedings of the National Academy of Sciences of the United States of America. 2012;109:7115–7120. doi: 10.1073/pnas.1111670109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter KU, Becker A, Munster T, Kim JT, Saedler H, Theissen G. MADS-box genes reveal that gnetophytes are more closely related to conifers than to flowering plants. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:7342–7347. doi: 10.1073/pnas.96.13.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Wang YN, Hsu CY, Lin CP, Chaw SM. Loss of different inverted repeat copies from the chloroplast genomes of Pinaceae and cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biology and Evolution. 2011;3:1284–1295. doi: 10.1093/gbe/evr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Rest JS, Davis CC. Phylogenomics and coalescent analyses resolve extant seed plant relationships. PLoS One. 2013;8:e80870. doi: 10.1371/journal.pone.0080870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wu F, Lin X, et al. Live and let die-the Bsister MADS-box gene OsMADS29 controls the degeneration of cells in maternal tissues during seed development of rice (Oryza sativa) PLoS One. 2012;7:e51435. doi: 10.1371/journal.pone.0051435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature. 1990;346:35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
- Yoo SK, Lee JS, Ahn JH. Overexpression of AGAMOUS-LIKE 28 (AGL28) promotes flowering by upregulating expression of floral promoters within the autonomous pathway. Biochemistry and Biophysics Research Communications. 2006;348:929–936. doi: 10.1016/j.bbrc.2006.07.121. [DOI] [PubMed] [Google Scholar]
- Yoo SK, Wu X, Lee JS, Ahn JH. AGAMOUS-LIKE 6 is a floral promoter that negatively regulates the FLC/MAF clade genes and positively regulates FT in Arabidopsis. The Plant Journal. 2011;65:62–76. doi: 10.1111/j.1365-313X.2010.04402.x. [DOI] [PubMed] [Google Scholar]
- Zhang H, Forde BG. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science. 1998;279:407–409. doi: 10.1126/science.279.5349.407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.