 Improving enzymes by directed evolution requires the navigation of very large search spaces; we survey how to do this intelligently.
Improving enzymes by directed evolution requires the navigation of very large search spaces; we survey how to do this intelligently.
Abstract
The amino acid sequence of a protein affects both its structure and its function. Thus, the ability to modify the sequence, and hence the structure and activity, of individual proteins in a systematic way, opens up many opportunities, both scientifically and (as we focus on here) for exploitation in biocatalysis. Modern methods of synthetic biology, whereby increasingly large sequences of DNA can be synthesised de novo, allow an unprecedented ability to engineer proteins with novel functions. However, the number of possible proteins is far too large to test individually, so we need means for navigating the ‘search space’ of possible protein sequences efficiently and reliably in order to find desirable activities and other properties. Enzymologists distinguish binding (K d) and catalytic (k cat) steps. In a similar way, judicious strategies have blended design (for binding, specificity and active site modelling) with the more empirical methods of classical directed evolution (DE) for improving k cat (where natural evolution rarely seeks the highest values), especially with regard to residues distant from the active site and where the functional linkages underpinning enzyme dynamics are both unknown and hard to predict. Epistasis (where the ‘best’ amino acid at one site depends on that or those at others) is a notable feature of directed evolution. The aim of this review is to highlight some of the approaches that are being developed to allow us to use directed evolution to improve enzyme properties, often dramatically. We note that directed evolution differs in a number of ways from natural evolution, including in particular the available mechanisms and the likely selection pressures. Thus, we stress the opportunities afforded by techniques that enable one to map sequence to (structure and) activity in silico, as an effective means of modelling and exploring protein landscapes. Because known landscapes may be assessed and reasoned about as a whole, simultaneously, this offers opportunities for protein improvement not readily available to natural evolution on rapid timescales. Intelligent landscape navigation, informed by sequence-activity relationships and coupled to the emerging methods of synthetic biology, offers scope for the development of novel biocatalysts that are both highly active and robust.
Introduction
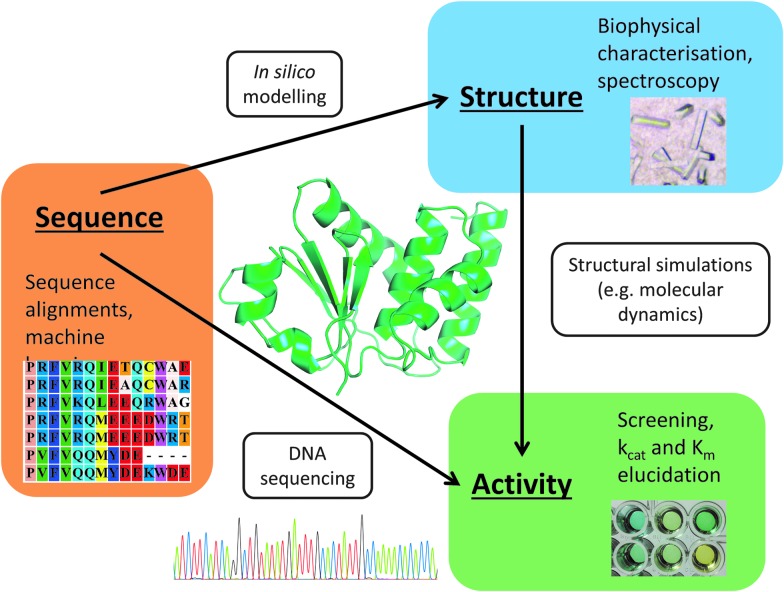
Much of science and technology consists of the search for desirable solutions, whether theoretical or realised, from an enormously larger set of possible candidates. The design, selection and/or improvement of biomacromolecules such as proteins represents a particularly clear example. 1 This is because natural molecular evolution is caused by changes in protein primary sequence that (leaving aside other factors such as chaperones and post-translational modifications) can then fold to form higher-order structures with altered function or activity; the protein then undergoes selection (positive or negative) based on its new function (Fig. 1). Bioinformatic analyses can trace the path of protein evolution at the sequence level 2–4 and match this to the corresponding change in function.
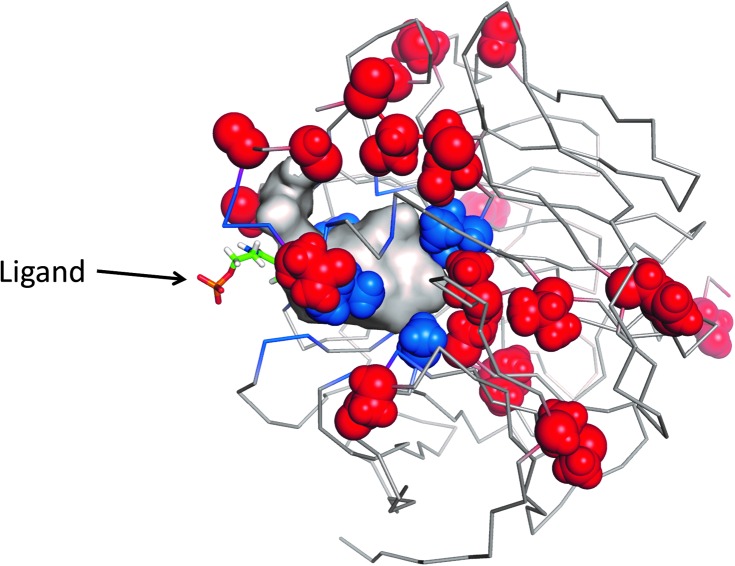
Fig. 1. Relationship between amino acid sequence, 3D structure (and dynamics) and biocatalytic activity. Implicitly, there is a host in which these manipulations take place (or they may be done entirely in vitro). This is not a major focus of the review. Typically, a directed evolution study concentrates on the relationships between protein sequence, structure and activity, and the usual means for assessing these are outlined (within the boxes). Many methods are available to connect and rationalise these relationships and some examples are shown (grey boxes). Thorough directed evolution studies require understanding of each of these parameters so that the changes in protein function can be rationalised, thereby to allow effective search of the sequence space. The key is to use emerging knowledge from multiple sources to navigate the search spaces that these represent. Although the same principles apply to multi-subunit proteins and protein complexes, most of what is written focuses on single-domain proteins that, like ribonuclease, 1342,1343 can fold spontaneously into their tertiary structures without the involvement of other proteins, chaperones, etc.
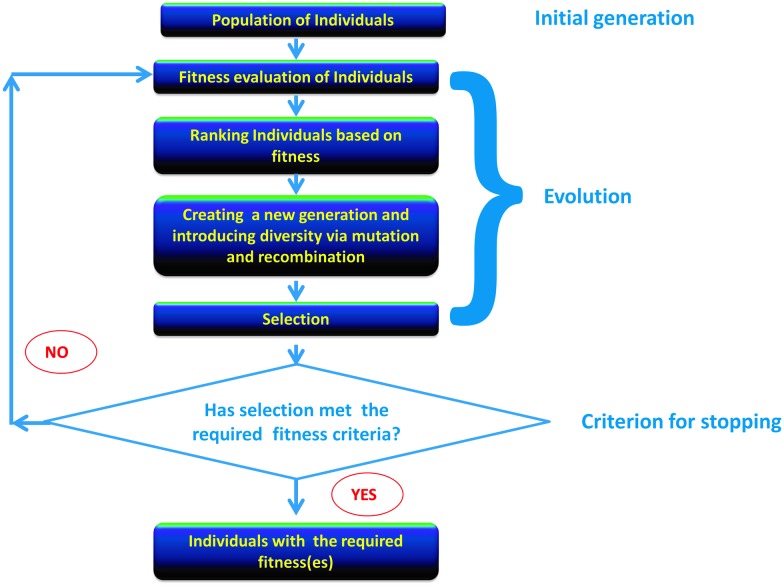
Proteins are nature's primary catalysts, and as the unsustainability of the present-day hydrocarbon-based petrochemicals industry becomes ever more apparent, there is a move towards carbohydrate feedstocks and a parallel and burgeoning interest in the use of proteins to catalyse reactions of non-natural as well as of natural chemicals. Thus, as well as observing the products of natural evolution we can now also initiate changes, whether in vivo or in vitro, for any target sequence. When the experimenter has some level of control over what sequence is made, variations can be introduced, screened and selected over several iterative cycles (‘generations’), in the hope that improved variants can be created for a particular target molecule, in a process usually referred to as directed evolution (Fig. 2) or DE. Classically this is achieved in a more or less random manner or by making a small number of specific changes to an existing sequence (see below); however, with the emergence of ‘synthetic biology’ a greater diversity of sequences can be created by assembling the desired sequence de novo (without a starting template to amplify from). Hence, almost any bespoke DNA sequence can be created, thus permitting the engineering of biological molecules and systems with novel functions. This is possible largely due to the reducing cost of DNA oligonucleotide synthesis and improvements in the methods that assemble these into larger fragments and even genomes. 5,6 Therefore, the question arises as to what sequences one should make for a particular purpose, and on what basis one might decide these sequences.
Fig. 2. The essential components of an evolutionary system. At the outset, a starting individual or population is selected, and one or more fitness criteria that reflect the objective of the process are determined. Next, the ability to rank these fitnesses and to select for diversity is created (by breeding individuals with variant sequences, introduced typically by mutation and/or recombination) in a way that tends to favour fitter individuals, this is repeated iteratively until a desired criterion is met.
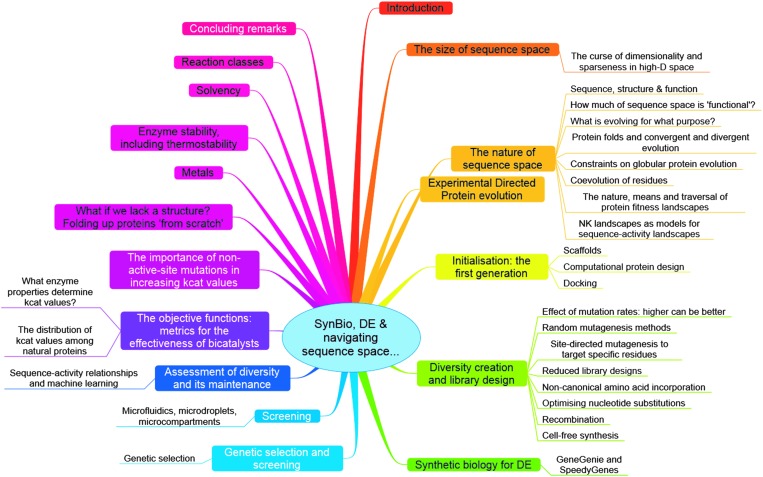
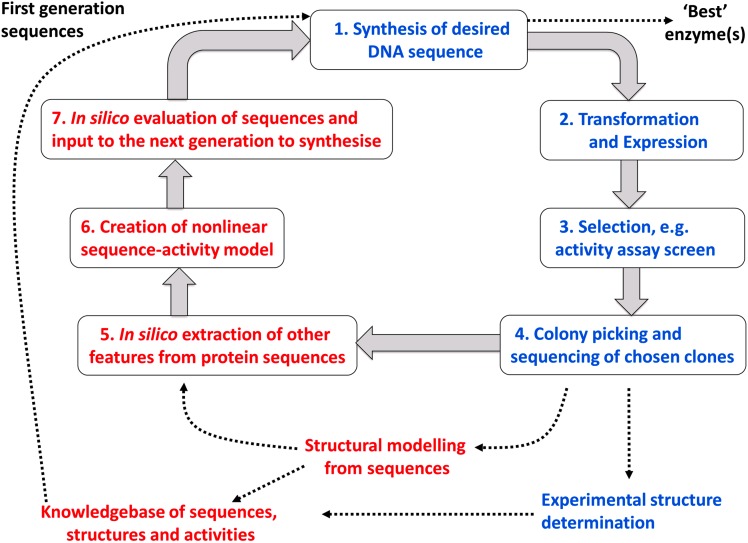
In this intentionally wide-ranging review, we introduce the basis of protein evolution (sequence spaces, constraints and conservation), discuss the methodologies and strategies that can be utilised for the directed evolution of individual biocatalysts, and reflect on their applications in the recent literature. To restrict our scope somewhat, we largely discount questions of the directed evolution of pathways (i.e. series of reactions) or gene clusters (e.g. ref. 7 and 8) and of the choice 9 or optimization of the host organism or expression conditions in which such directed evolution might be performed or its protein products expressed, nor the process aspects of any fermentation or biotransformation. We also focus on catalytic rate constants, albeit we recognize the importance of enzyme stability as well. Most of the strategies we describe can equally well be applied to proteins whose function is not directly catalytic, such as vaccines, binding agents, and the like. Consequently we intend this review to be a broadly useful resource or portal for the entire community that has an interest in the directed evolution of protein function. A broad summary is given as a mind map in Fig. 3, while the various general elements of a modern directed evolution program, on which we base our development of the main ideas, appears as Fig. 4.
Fig. 3. A ‘mind map’ 1344 of the contents of this paper; to read this start at “twelve o'clock” and read clockwise.
Fig. 4. An example of the basic elements of a mixed computational and experimental programme in directed evolution. Implicit are the choice of objective function (e.g. a particular catalytic activity with a certain turnover number) and the starting sequences that might be used with an initial or ‘wild type’ activity from which one can evolve improved variants. The core experimental (blue) and computational (red) aspects are shown as seven steps of an iterative cycle involving the creation and analysis of appropriate protein sequences and their attendant activities. Additional facets that can contribute to the programme are also shown (connected using dotted lines).
The size of sequence space
An important concept when considering a protein's amino acid sequence is that of (its) sequence space, i.e. the number of variations of that sequence that can possibly exist. Straightforwardly, for a protein that contains just the 20 main natural amino acids, a sequence length of N residues has a total number of possible sequences of 20 N . For N = 100 (a rather small protein) the number 20 100 (∼1.3 × 10 130 ) is already far greater than the number of atoms in the known universe. Even a library with the mass of the Earth itself – 5.98 × 1027 g – would comprise at most 3.3 × 1047 different sequences, or a miniscule fraction of such diversity. 10 Extra complexity, even for single-subunit proteins, also comes with incorporation of additional structural features beyond the primary sequence, like disulphide linkages, metal ions, 11 cofactors and post-translational modifications, and the use of non-standard amino acids (outwith the main 20). Beyond this, there may be ‘moonlighting’ activities 12 by which function is modified via interaction with other binding partners.
Considering sequence variation, using only the 20 ‘common’ amino acids, the number of sequence variants for M substitutions in a given protein of N amino acids is 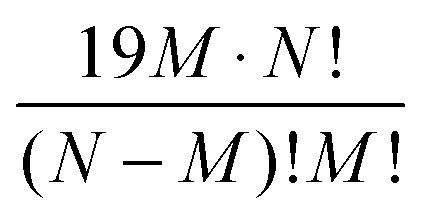 .
13
For a protein of 300 amino acids with random changes in just 1, 2 or 3 amino acids in the whole protein this is 5700, ca. 16 million and ca. 30 billion, while even for a comparatively small protein of N = 100 amino acids, the number of variants exceeds 1015 when M = 10. Insertions can be considered as simply increasing the length of N and the number of variants to 21 (a ‘gap’ being coded as a 21st amino acid), respectively.
.
13
For a protein of 300 amino acids with random changes in just 1, 2 or 3 amino acids in the whole protein this is 5700, ca. 16 million and ca. 30 billion, while even for a comparatively small protein of N = 100 amino acids, the number of variants exceeds 1015 when M = 10. Insertions can be considered as simply increasing the length of N and the number of variants to 21 (a ‘gap’ being coded as a 21st amino acid), respectively.
Consequently, the search for variants with improved function in these large sequence spaces is best treated as a combinatorial optimization problem, 1 in which a number of parameters must be optimised simultaneously to achieve a successful outcome. To do this, heuristic strategies (that find good but not provably optimal solutions) are appropriate; these include algorithms based on evolutionary principles.
The ‘curse of dimensionality’ and the sparseness or ‘closeness’ of strings in sequence space
One way to consider protein sequences (or any other strings of this type) is to treat each position in the string as a dimension in a discrete and finite space. In an elementary way, an amino acid X has one of 20 positions in 1-dimensional space, an individual dimer X k Y l has a specified position or represents a point (from 400 discrete possibilities) in 2D space, a trimer X k Y l Z m a specified location (from 8000) in 3D space, and so on. Various difficulties arise, however (‘the curse of dimensionality’ 14,15 ) as the number of dimensions increases, even for quite small numbers of dimensions or string length, since the dimensionality increases exponentially with the number of residues being changed. One in particular is the potential ‘closeness’ to each other of various randomly selected sequences, and how this effectively diverges extremely rapidly as their length is increased.
Imagine (as in ref. 16) that we have examples uniformly distributed in a p-dimensional hypercube, and wish to surround a target point with a hypercubical ‘neighbourhood’ to capture a fraction r of all the samples. The edge length of the (hyper)cube will be e p (r) = r (1/p). In just 10 dimensions e 10(0.01) = 0.63 and e 10(0.1) = 0.79 while the range (of a unit hypercube) for each dimension is just 1. Thus to capture even just 1% or 10% of the observations we need to cover 63% or 80% of the range (i.e. values) of each individual dimension. Two consequences for any significant dimensionality are that even large numbers of samples cover the space only very sparsely indeed, and that most samples are actually close to the edge of the n-dimensional hypercube. We shall return later to the question of metrics for the effective distance between protein strings and for the effectiveness of protein catalysts; for the latter we shall assume (and discuss below) that the enzyme catalytic rate constant or turnover number (with units of s–1, or in less favourable cases min–1, h–1, or d–1) is a reasonable surrogate for most functional purposes.
Overall, it is genuinely difficult to grasp or to visualise the vastness of these search spaces, 17 and the manner in which even very large numbers of examples populate them only extremely sparsely. One way to visualise them 18–22 is to project them into two dimensions. Thus, if we consider just 30mers of nucleic acid sequences, and in which each position can be A, T, G or C, the number of possible variants is 430, which is ∼1018, and even if arrayed as 5 μm spots the array would occupy 29 km2! 23 The equivalent array for proteins would contain only 14mers, in that there are more than 1018 possible proteins containing the 20 natural amino acids when their length is just 14 amino acids.
The nature of sequence space
Sequence, structure and function
One of the fundamental issues in the biosciences is the elucidation of the relationship between a protein's primary sequence, its structure and its function. Difficulties arise because the relationship between a protein's sequence and structure is highly complex, as is the relationship between structure and function. Even single mutations at an individual residue can change a protein's activity completely – hence the discovery of ‘inborn errors of metabolism’. 24,25 (The same is true in pharmaceutical drug discovery, with quite small changes in small molecule structure often leading to a dramatic change in activity – so-called ‘activity cliffs’ 26–33 – and with similar metaphors of structure–activity relationships, rather than those of sequence-activity, being equally explicit. 34–37 ) Annotation of putative function from unknown sequences is largely based upon sequence homology (similarity) to proteins of known characterised function and particularly the presence of specific sequence/structure motifs (such as the Rossmann fold 38 or the P-loop motif 39 ). While there have been great advances in predicting protein structure from primary sequence (see later), the prediction of function from structure (let alone sequence) remains an important (if largely unattained) aim. 40–54
How much of sequence space is ‘functional’?
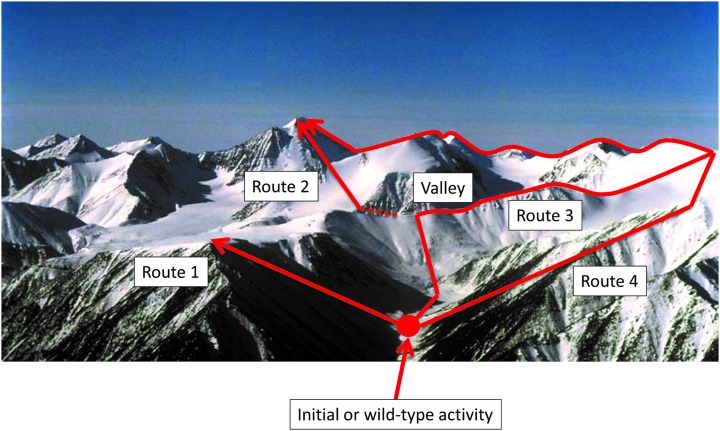
The relationship between sequence and function is often considered in terms of a metaphor in which their evolution is seen as akin to traversing a ‘landscape’, 55 that may be visualised in the same way as one considers the topology of a natural landscape, 56,57 with the ‘position’ reflecting the sequence and the desirable function(s) or fitness reflected in the ‘height’ at that position in the landscape (Fig. 5).
Fig. 5. A fitness landscape and its navigation. The initial or wild-type activity denotes the starting point (initialisation) for a directed evolution study (red circle). Accumulation of mutations that increase activity is represented by four routes to different positions in the landscape. Route 1 successfully increases activity through a series of additive mutations, but becomes stuck in a local optimum. Due to the nature of rugged fitness landscapes some of the shorter paths to a maximum possible (global optimum) fitness (activity) can require movement into troughs before navigating a new higher peak (route 2). Alternatively, one can arrive at the global optimum using longer but typically less steep routes without deep valleys (equivalent over flat ground to neutral mutations – routes 3 and 4).
Given the enormous numbers for populating sequence space, and the present impossibility of computing or sampling function from sequence alone, it is clear that natural evolution cannot possibly have sampled all possible sequences that might have biological function. 58 Hence, the strategy of a DE project faces the same questions as those faced in nature: how to navigate sequence space effectively while maintaining at least some function, but introducing sufficient variation that is required to improve that function. For DE there are also the practical considerations: how many variants can be screened (and/or selected for) and analysed with our current capabilities?
The first general point to be made is that most completely random proteins are practically non-functional. 10,56,59–66 Indeed, many are not even soluble, 67,68 although they may be evolved to become so. 69 Keefe and Szostak noted that ca. 1 in 1011 of random sequences have measurable ATP-binding affinity. 70 Consistent with this relative sparseness of functional protein space is the fact that even if one does have a starting structure(/function), one typically need not go ‘far’ from such a structure to lose structure quite badly, 71 albeit that with a ‘density’ of only 1 in 1011 proteins being functional this implies that all such functional sequences are connected by trajectories involving changes in only a single amino acid 72 (and see ref. 58). This is also consistent with the fact that sequence space is vast, and only a tiny fraction of possible sequences tend to be useful and hence selected for by natural evolution. One may note 70,73 that at least some degree of randomness will be accompanied by some structure, 74,75 functionality or activity. For proteins, secondary structure is understood to be a strong evolutionary driver, 76 particularly through the binary-patterning (arrangement of hydrophilic/hydrophobic residues), 64,77–84 and so is the (somewhat related) packing density. 85–89 In a certain sense, proteins must at some point have begun their evolution as more or less random sequences. 90 Indeed “Folded proteins occur frequently in libraries of random amino acid sequences”, 91 but quite small changes can have significantly negative effects. 92 Harms and Thornton give a very thoughtful account of evolutionary biochemistry, 4 recognizing that the “physical architecture {of proteins both} facilitates and constrains their evolution”. This means that it will be hard (but not impossible), especially without plenty of empirical data, 93 to make predictions about the best trajectories. Fortunately, such data are now beginning to appear. 57,94 Indeed, the leitmotiv of this review is that understanding such (sequence-structure–activity) landscapes better will assist us considerably in navigating them.
What is evolving and for what purpose?
In a simplistic way, it is easy to assume that protein sequences are being selected for on the basis of their contribution to the host organism's fitness, without normally having any real knowledge of what is in fact being implied or selected for. However, a profound and interesting point has been made by Keiser et al. 95 to the effect that once a metabolite has been ‘chosen’ (selected) to be part of a metabolic or biochemical network, proteins are somewhat constrained to evolve as ‘slaves’, to learn to bind and react with the metabolites that exist. Thus, in evolution, the proteins follow the metabolites as much as vice versa, making knowledge of ligand binding 96,97 and affinity 98 to protein binding sites a matter of primary interest, especially if (as in the DE of biocatalysts) we wish to bind or evolve catalysts for novel (and xenobiotic) small molecule substrates. In DE we largely assume that the experimenter has determined what should be the objective function(s) or fitness(es), and we shall indicate the nature of some of the choices later; notwithstanding, several aspects of DE do tend to differ from those selected by natural evolution (Table 1). Thus, most mutations are pleiotropic in vivo, 99,100 for instance. As DNA sequencing becomes increasingly economical and of higher throughput 101,102 a greater provenance of sequence data enables a more thorough knowledge of the entire evolutionary landscape to be obtained. In the case of short sequences most 103 or all 104 of the entire genotype-fitness landscape may be measured experimentally. We note too (and see later) that there are equivalent issues in the optimization and algorithms of evolutionary computing (e.g. ref. 105–107), where strategies such as uniform cross-over, 108 with no real counterpart in natural or experimental evolution, have been shown to be very effective.
Table 1. Some features by which natural evolution, classical DE of biocatalysts, and directed evolution of biocatalysts using synthetic biology differ from each other. Population structures also differ in natural evolution vs. DE, but in the various strategies for DE they follow from the imposed selection in ways that are difficult to generalize.
| Feature | Natural evolution | Classical DE | DE with synthetic biology |
| Objective function and selection pressure | Unclear; there is only a weak relation of a protein's function with organismal fitness; 117 k cat is not strongly selected for. Although presumably multi-objective, actual selection and fitness are ‘composites’. If there is no redundancy, organisms must retain function during evolution. 58,118 | Typically strong selection weak mutation (rarely was sequencing done so selection was based on fitness only). Can select explicitly for multiple outputs (e.g. k cat, thermostability). | Much as with classical DE, but diversity maintenance can be much enhanced via high-throughput methods of DNA synthesis and sequencing. |
| Mutation rates | Varies with genome size over orders of magnitude, 119 but typically (for organisms from bacteria to humans) <10–8 per base per generation. 120,121 Can itself be selected for. 122 | Mutation rates are controlled but often limited to only a few residues per generation, e.g. to 1/L where L is the aa length of the protein; much more can lead to too many stop codons. | Library design schemes that permit stop codons only where required mean that mutation rates can be almost arbitrarily high. |
| Recombination rates | Very low in most organisms (though must have occurred in cases of ‘horizontal gene transfer’); in some cases almost non-existent. 123 | Could be extremely high in the various schemes of DNA shuffling, including the creation of chimaeras from different parents. | Again it can be as high or low as desired; the experimenter has (statistically) full control. |
| Randomness of mutation | Although there are ‘hot spots’, mutations in natural evolution are considered to be random and not ‘directed’. 124 | In error-prone PCR, mutations are seen as essentially random. Site-directed methods offer control over mutations at a small number of specified positions. | As much or as little randomness may be introduced as the experimenter desires by using defined mixtures of bases for each codon, e.g. NNN or NNK as alternatives to specific subsets such as polar or apolar. |
| Evolutionary ‘memory’ | For individuals (cf. populations 125 ) there is no ‘memory’ as such, although the sequence reflects the evolutionary ‘trace’ (but not normally the pathway – cf. ref. 126 and 127). | Again, there is no real ‘memory’ in the absence of large-scale sequencing, but there is potential for it. 56 | With higher-throughput sequencing we can create an entire map of the landscape as sampled to date, to help guide the informed assessment of which sequences to try next. |
| Degree of epistasis | It exists, but only when there is a more or less neutral pathway joining the epistatic sites. | It is comparatively hard to detect at low mutation rates. | Potentially epistasis is much more obvious as sites can be mutated pairwise or in more complex designed patterns. |
| Maintenance of individuals of lower or similar fitness in population | They are soon selected out in a ‘strong selection, weak mutation’ regime; this limits jumps via lower fitness, and enforces at least neutral mutations. | It is in the hands of the experimenter, and usually not done when only fitnesses are measured. | Again it is entirely up to the experimenter; diversity may be maintained to trade exploration against exploitation. |
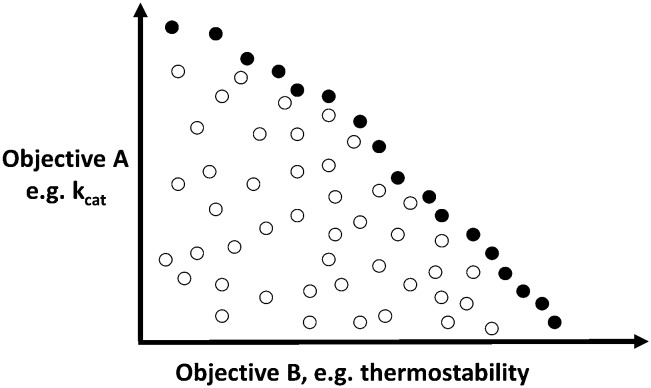
However, in the case of multi-objective optimisation (e.g. seeking to optimise two objectives such as both k cat and thermostability, or activity vs. immunogenicity 109 ), there is normally no individual preferred solution that is optimal for all objectives, 110 but a set of them, known as the Pareto front (Fig. 6), whose members are optimal in at least one objective while not being bettered (not ‘dominated’) in any other property by any other individual. The Pareto front is thus also known as the non-dominated front or ‘set’ of solutions. A variety of algorithms in multi-objective evolutionary optimisation (e.g. ref. 111–116) use members of the Pareto front as the choice of which ‘parents’ to use for mutation and recombination in subsequent rounds.
Fig. 6. A two-objective optimisation problem, illustrating the non-dominated or Pareto front. In this case we wish to maximise both objectives. Each individual symbol is a candidate solution (i.e. protein sequence), with the filled ones denoting an approximation to the Pareto front.
Protein folds and convergent and divergent evolution
What is certain, given that form follows function, is that natural evolution has selected repeatedly for particular kinds of secondary and tertiary structure ‘domains’ and ‘folds’. 128,129 It is uncertain as to how many more are ‘common’ and are to be found via the methods of structural genomics, 130 but many have been expertly classified, 131 e.g. in the CATH, 132–134 SCOP 135–137 or InterPro 138,139 databases, and do occur repeatedly.
Given that structural conservation of protein folds can occur for sequences that differ markedly from each other, it is desirable that these analyses are done at the structural (rather than sequence) level (although there is a certain arbitrariness about where one fold ends and another begins 140,141 ). Some folds have occurred and been selected via divergent evolution (similar sequences with different functions) 142 and some via convergent evolution (different sequences with similar functions). 143,144 This latter in particular makes the nonlinear mapping of sequence to function extremely difficult, and there are roughly two unrelated sequences for each E.C. (Enzyme Commission classification) number. 145 As phrased by Ferrada and colleagues, 146 “two proteins with the same structure and/or function in our data…{have} a median amino acid divergence of no less than 55 percent”. However, normally information is available only for extant molecules but not their history and precise evolutionary path (in contrast to DE). One conclusion might be that conventional means of phylogenetic analysis are not necessarily best placed to assist the processes of directed evolution, and we argue later (because a protein has no real ‘memory’ of its full evolutionary pathway) that modern methods of machine learning that can take into account ensembles of sequences and activities may prove more suitable. However, we shall first look at natural evolution.
Constraints on globular protein evolution structure in natural evolution
In gross terms, a major constraint on protein evolution is provided by thermodynamics, in that proteins will have a tendency to fold up to a state of minimum free energy. 147–149 Consequently, the composition of the amino acids has a major influence over protein folding because this means satisfying, so far as is possible, the preference of hydrophilic or polar amino acids to bind to each other and the equivalent tendency of hydrophobic residues to do so. 150–152 Alteration of residues, especially non-conservatively, often leads to a lowering of thermodynamic folding stability, 153 which may of course be compensated by changes in other locations. Naturally, at one level proteins need to have a certain stability to function, but they also need to be flexible to effect catalysis. This is coupled to the idea that proteins are marginally stable objects in the face of evolution. 154–159 Overall, this is equivalent to ‘evolution to the edge of chaos’, 160,161 a phenomenon recognizing the importance of trading off robustness with evolvability that can also be applied 162,163 to biochemical networks. 164–170 Thermostability (see later) may also sometimes (but not always 171–173 ) correlate with evolvability. 174,175
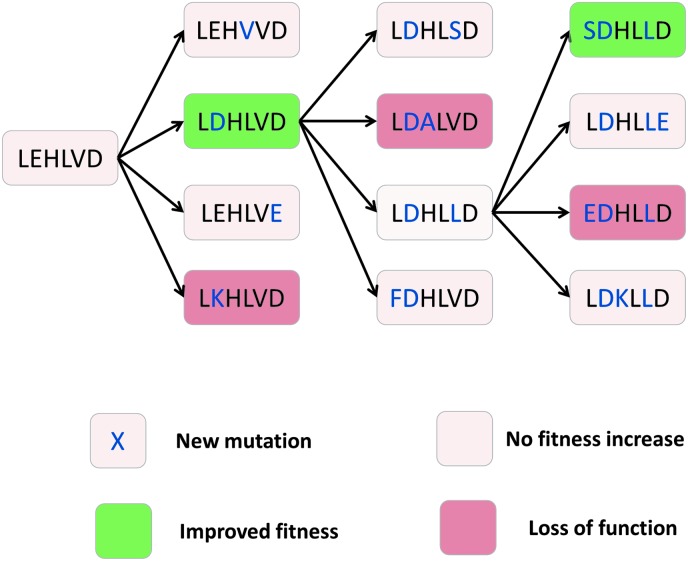
Given the thermodynamic and biophysical 157,176,177 constraints, that are related to structural contacts, various models (e.g. ref. 147 and 178) have been used to predict the distribution of amino acids in known proteins. As regards to specific mechanisms, it has been stated that “solvent accessibility is the primary structural constraint on amino acid substitutions and mutation rates during protein evolution.”, 148 while “satisfaction of hydrogen bonding potential influences the conservation of polar sidechains”. 179 Overall, given the tendency in natural evolution for strong selection, it is recognized that a major role is played by neutral mutations 180–182 or neutral evolution 183–188 (see Fig. 5 and 7). Gene duplication provides another strategy, allowing redundancy followed by evolution to new functions. 189
Fig. 7. Some evolutionary trajectories of a peptide sequence undergoing mutation. Mutations in the peptide sequence can cause an increase in fitness (e.g. enzyme activity, green), loss of fitness (salmon pink) or no change in fitness (grey). Typically, improved fitness mutations are selected for and subjected to further modification and selection. Neutral mutations keep sequences ‘alive’ in the series, and these can often be required for further improvements in fitness, as shown in steps 2 and 3 of this trajectory.
Coevolution of residues
Thus far, we have possibly implied that residues evolve (i.e. are selected for) independently, but that is not the case at all. 190–192 There can be a variety of reasons for the conservation of sequence (including correlations between ‘distant’ regions 193 ), but the importance to structure and function, and functional linkage between them, underlie such correlations. 194–209 Covariation in natural evolution reflects the fact that, although not close in primary sequence, distal residues can be adjacent in the tertiary structure and may represent an interaction favourable to protein function. Covariation also provides an important computational approach to protein folding more generally (see below).
The nature, means of analysis and traversal of protein fitness landscapes
Since John Holland's brilliant and pioneering work in the 1970s (reprinted as ref. 210), it has been recognized that one can search large search spaces very effectively using algorithms that have a more or less close analogy to that of natural evolution. Such algorithms are typically known as genetic or evolutionary algorithms (e.g. ref. 106 and 211–213, and their implementation is referred to as evolutionary computing. 106,214–216 The algorithms can be classified according to whether one knows only the fitnesses (phenotypes) of the population or also the genotypes (sequences). 107
Since we cannot review the very large literature, essentially amounting to that of the whole of molecular protein evolution, on the nature of (natural) protein landscapes, we shall therefore seek to concentrate on a few areas where an improved understanding of the nature of the landscape may reasonably be expected to help us traverse it. Importantly, even for single objectives or fitnesses, a number of important concepts of ruggedness, additivity, promiscuity and epistasis are inextricably intertwined; they become more so where multiple and often incommensurate objectives are considered.
Additivity
Additivity implies simple continuing fixing of improved mutations, 217–220 and follows from a model in which selection in natural evolution quite badly disfavours lower fitnesses, 221 a circumstance known from Gillespie 222,223 as ‘strong selection, weak mutation’ (SSWM, see also ref. 224–229). For small changes (close to neutral in a fitness or free energy sense), additivity may indeed be observed, 230,231 and has been exploited extensively in DE. 232–236 If additivity alone were true, however (and thus there is no epistasis for a given protein at all) then a rapid strategy for DE would be to synthesise all 20L amino acid variants at each position (of a starting protein of length L) and pick the best amino acid at each position. However, the very existence of convergent and divergent evolution implies that landscapes are rugged 237 (and hence epistatic), so at the very least additivity and epistasis must coexist. 236,238
Epistasis
The term ‘epistasis’ in DE covers a concept in which the ‘best’ amino acid at a given position depends on the amino acid at one or more other positions. In fact, we believe that one should start with an assumption of rather strong epistasis, 238–248 as did Wright. 55 Indeed the rugged fitness landscape is itself a necessary reflection of epistasis and vice versa. Thus, epistasis may be both cryptic and pervasive, 249 the demonstrable coevolution goes hand in hand with epistasis, and “to understand evolution and selection in proteins, knowledge of coevolution and structural change must be integrated”. 250
Promiscuity
The concept of enzyme promiscuity mainly implies that some enzymes may bind, or catalyse reactions with, more than one substrate, and this is inextricably linked to how one can traverse evolutionary landscapes. 251–270 It clearly bears strongly on how we might seek to effect the directed evolution of biocatalysts.
NK landscapes as models for sequence-activity landscapes
A very important class of conceptual (and tunable) landscapes are the so-called NK landscapes devised by Kauffman 161,271 and developed by many other workers (e.g. ref. 220, 221, 237 and 272–278). The ‘ruggedness’ of a given landscape is a slightly elusive concept, 279 but can be conceptualized 56,220 in a manner that implies that for a smooth landscape (like Mt Fuji 280,281 ) fitness and distance tend to be correlated, while for a very ‘rugged’ landscape the correlation is much weaker (since as one moves away from a starting sequence one may pass through many peaks and troughs of fitness). In NK landscapes, K is the parameter that tunes the extent of ruggedness, and it is possible to seek landscapes whose ruggedness can be approximated by a particular value of K, since one of the attractions of NK is that they can reproduce (in a statistical sense) any kind of landscape. 282 Indeed, we can use the comparatively sparse data presently available to determine that experimental sequence-fitness landscapes reflect NK landscapes that are fairly considerably (but not pathologically) rugged, 23,57,104,241,251,274,276,283 and that there is likely to be one or more optimal mutation rates that themselves depend on the ruggedness (see later). Note too that the landscapes for individual proteins, as discussed here, are necessarily more rugged than are those of pathways or organisms, due to the more profound structural constraints in the former. 57,157 (Parenthetically, NK-type landscapes and the evolutionary metaphor have also proved useful in a variety of other ‘complex’ spheres, such as business, innovation and economics (e.g. ref. 278 and 284–295, though a disattraction of NK landscapes in evolutionary biology itself is that they do not obey evolutionary rules. 224 )
Experimental directed protein evolution
A number of excellent books and review articles have been devoted to DE, and a sampling with a focus on biocatalysis includes. 296–334 As indicated above, DE begins with a population that we hope contains at least one member that displays some kind of activity of interest, and progresses through multiple rounds of mutation, selection and analysis (as per the steps in Fig. 4).
Initialisation; the first generation
During the preliminary design of a DE project the main objective and required fitness criteria must be defined and these criteria influence the experimental design and screening strategy.
We consider in this review that a typical scenario is that one has a particular substrate or substrate class in mind, as well as the chemical reaction type (oxidation, hydroxylation, amination and so on) that one wishes to catalyse. If any activity at all can be detected then this can be a starting point. In some cases one does not know where to start at all because there are no proteins known either to catalyse a relevant reaction or to bind the substrate of interest. For pharmaceutical intermediates, it can still be useful to look for reactions involving metabolites, as most drugs do bear significant structural similarities to known metabolites, 335,336 and it is possible to look for reactions involving the latter. A very useful starting point may be the structure-function linkage database ; http://sfld.rbvi.ucsf.edu/django/. 337 There are also ‘hub’ sequences that can provide useful starting points, 338 while Verma, 330 Nov 339 and Zaugg 340 list various computational approaches. If one has a structure in the form of a PDB file one can try HotSpotWizard ; http://loschmidt.chemi.muni.cz/hotspotwizard/. 341 Analysing the diversity of known enzyme sequences is also a very sensible strategy. 342,343 Nowadays, an increasing trend is to seek relevant diversity, aligned using tools such as Clustal Omega, 344,345 MUSCLE, 346 PROMALS, 347,348 or other methods based on polypharmacology, 141,349,350 that one may hope contains enzymes capable of effecting the desired reaction. Another strategy is to select DNA from environments that have been exposed to the substrate of interest, using the methods of functional metagenomics. 351,352 More commonly, however, one does have a very poor protein (clone) with at least some measurable activity, and the aim is to evolve this into a much more active variant.
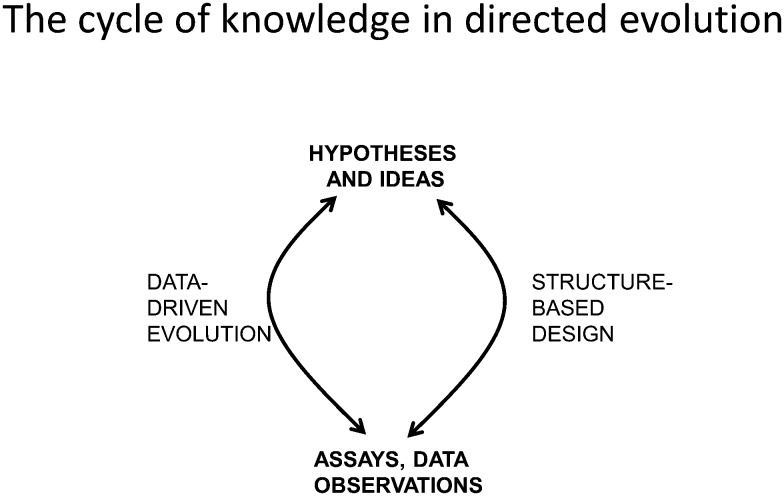
In general, scientific advance is seen in a Popperian view (see e.g. ref. 353–357) as an iterative series of ‘conjectures’ and ‘refutations’ by which the search for scientific truth is ‘narrowed’ by finding what is not true (may be falsified) via predictions based on hypothetico-deductive reasoning and their anticipated and experimental outcomes. However, Popper was purposely coy about where hypotheses actually came from, and we prefer a variant 358–362 (see also ref. 363 and 364) that recognises the equal contribution of a more empirical ‘data-driven’ arc to the ‘cycle of knowledge’ (Fig. 8).
Fig. 8. The ‘cycle of knowledge’ in modern directed evolution. Both structure-based design and a more empirical data-driven approach can contribute to the evolution of a protein with improved properties, in a series of iterative cycles.
In a similar vein, many commentators (e.g. ref. 365–368) consider the best strategy for both the starting population and the subsequent steps to be a judicious blend between the more empirical approaches of (semi-)directed evolution and strategies more formally based on attempts to design 369 (somewhat in the absence of fully established principles) sequences or structures based on what is known of molecular interactions. We concur with this, since at the present time it is simply not possible to design enzymes with high activities de novo (from scratch, or from sequence alone), despite progress in simple 4-helix-bundle and related ‘maquettes’. 370–373 David Baker, probably the leading expert in protein design, considers that design is still incapable of predicting active enzymes even when the chemistry and active sites appear good. 374,375 Several reviews attest to this, 329,376–379 but crowdsourcing approaches have been shown to help, 380 and computational design (and see below) certainly beats random sequences. 381 Overall, the fairest comment is probably that we can benefit from design for binding, specificity and active site modelling, but that for improving k cat we need the more empirical methods of DE, especially (see below) of residues distant from the active site.
Scaffolds
Because natural evolution has selected for a variety of motifs that have been shown in general terms to admit a wide range of possible enzyme activities, a number of approaches have exploited these motifs or ‘scaffolds’. 382 Triose phosphate isomerase (TIM) has proved a popular enzyme since the pioneering work of Albery and Knowles 383 and more recent work on TIM energetics, 384 and TIM (βα)8 barrels can be found in 5 of 6 EC classes. 146 TIM and many (but not all) such natural enzymes are most active as dimers, 385,386 caused by a tight interaction of 32 residues of each subunit in the wild type, though functional monomers can be created. 387,388
Thus, (βα)8 barrel enzymes 389–402 have proven particularly attractive as scaffolds for DE. 403–407 Some use or need cofactors like PLP, FMN, etc., 392,408 and their folding mechanisms are to some degree known. 385,409–411 We note, however, that virtual screening of substrates against these 412 has shown a relative lack of effectiveness of consensus design because of the importance of correlations (i.e. epistasis). 386
α/β and (α/β)2 barrels have also been favoured as scaffolds, 395,413–417 while attempts at automated scaffold selection can also be found. 374,418,419 A very interesting suggestion 420 is that the polarity of a fold may determine its evolvability.
Although not focused on biocatalysis, other scaffolds such as lipocalins 421–426 and affibodies 427–437 have proved useful for combinatorial biosynthesis and directed evolution.
Computational protein design
While computational protein design completely from scratch (in silico) is not presently seen as reasonable, probably (as we stress later) because we cannot yet use it to predict dynamics effectively, significant progress continues to be made in a number of areas, 373,438–456 including ‘fold to function’, 457 combinatorial design, 458 and a maximum likelihood framework for protein design. 459 Notable examples include a metalloenzyme for organophosphate hydrolysis, 460,461 aldolase 462,463 and others. 464–468 Theozymes 469–472 (theoretical catalysts, constructed by computing the optimal geometry for transition state stabilization by model functional) groups represent another approach.
Arguably the most advanced strategies for protein design and manipulation in silico are Rosetta 374,473–483 and RosettaBackrub, 484,485 while more ‘bottom-up’ approaches, based on some of the ideas of synthetic biology, are beginning to appear. 486–492 It is an easy prediction that developments in synthetic biology will have highly beneficial effects on de novo design, and vice versa.
Docking
If one is to find an enzyme that catalyses a reaction, one might hope to be able to predict that it can at least bind that substrate using the methods of in silico docking. 493 To date, methods based on Autodock, 494–499 APoC, 500 Glide 501–503 or other programs 504–511 have been proposed, but this strategy is not yet considered mainstream for the DE of a first generation of biocatalysts (and indeed is subject to considerable uncertainty 512 ). Our experience is that one must have considerable knowledge of the approximate answer (the binding site or pocket) before one tries these methods for DE of a biocatalyst.
Having chosen a member (or a population) as a starting point, the next step in any DE program is the important one of diversity creation. Indeed, the means of creating and exploiting suitable libraries that focus on appropriate parts of the protein landscape lies at the heart of any intelligent search method. 513
Diversity creation and library design
A diversity of sequences can be created in many ways, 514 but mutation or recombination methods are most commonly used in DE. Some are purely empirical and statistical (e.g. N mutations per sequence), while others are more focused to a specific part of the sequence (Fig. 9). Strategies may also be discriminated in terms of the degree of randomness of the changes and their extensiveness (Fig. 10). Two useful reviews include 515 and, 516 while others 334,517–519 cover computational approaches. A DE library creation bibliography is maintained at ; http://openwetware.org/wiki/Reviews:Directed_evolution/Library_construction/bibliography.
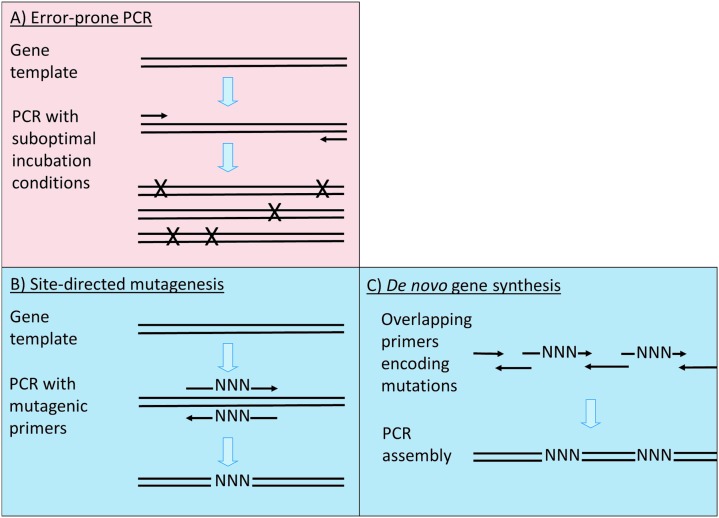
Fig. 9. Overview of the different mutagenesis strategies commonly employed to create variant protein libraries. Random methods (pink background) can create the greatest diversity of sequences in an uncontrolled manner. Mutations during error-prone PCR (A) are typically introduced by a polymerase amplifying sequences imperfectly (by being used under non-optimal conditions). In contrast, directed mutagenesis methods (blue background) introduce mutations at defined positions and with a controlled outcome. Site-directed mutagenesis (B) introduces a mutation, encoded by oligonucleotides, onto a template gene sequence in a plasmid. However, gene synthesis (C) can encode mutations on the oligonucleotides used to synthesise the sequence de novo, hence multiple mutations can be introduced simultaneously. X = random mutation, N = controlled mutation. →= PCR primer.
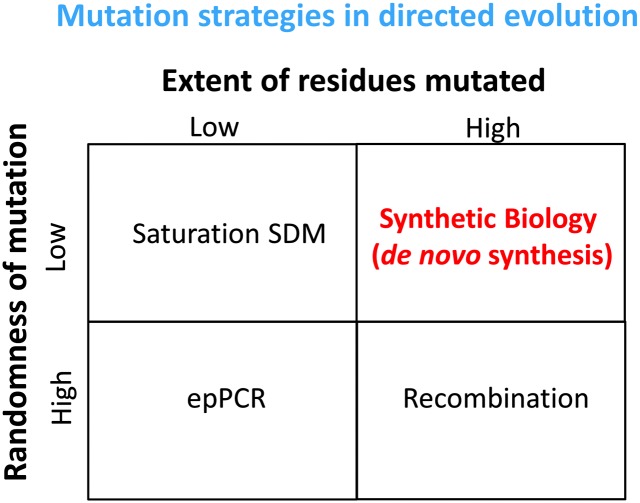
Fig. 10. A Boston matrix of the different strategies for variant libraries. Methods are identified in terms of the randomness of the mutations they create and the number of residues that can be targeted.
Effect of mutation rates, implying that higher can be better
In classical evolutionary computing, the recognition that most mutations were or are deleterious meant that mutation rates were kept low. If only one in 103 sequences is an improvement when the mutation rate is 1/L per position (L being the length of the string), then (in the absence of epistasis) only 1 in 106 is at 2/L. (Of course 1/L is far greater than the mutation rates common in natural evolution, which scales inversely with genome size, 119 may depend on cell–cell interactions, 520 and is normally below 10–8 per base per generation for organisms from bacteria to humans. 119–121 ) This logic is persuasive but limited, since it takes into account only the frequency but not the quality of the improvement (and as mentioned essentially does not consider epistasis). Indeed there is evidence that higher mutation rates are favoured both in silico 220,521–524 and experimentally. 525–528 This is especially the case for directed mutagenesis methods (especially those of synthetic biology), where stop codons can be avoided completely. We first discuss the more classical methods.
Random mutagenesis methods
Error-prone PCR (epPCR) is probably the most commonly used method for introducing random mutations. PCR amplification using Taq polymerase is performed under suboptimal conditions by altering the components of the reaction (in particular polymerase concentration, MgCl2 and dNTP concentration, or supplementation with MnCl2 (ref. 529)) or cycling conditions (increased extension times). 530 Although epPCR is the simplest to implement and most commonly used method for library creation, it is limited by its failure to access all possible amino acid changes with just one mutation, 339,531–533 a strong bias towards transition mutations (AT to GC mutations), 531 and an aversion to consecutive nucleotide mutations. 532,534
Refinement of these methods has allowed greater control over the mutation bias, rate of mutations 530,535–537 and the development of alternative methodologies like Mutagenic Plasmid Amplification, 538 replication, 539 error-prone rolling circle 540 and indel 541–543 mutagenesis. Typically, for reasons indicated above, the epPCR mutation rate is tuned to produce a small number of mutations per gene copy (although orthogonal replication in vivo may improve this 544 ), since entirely random epPCR produces multiple stop codons (3 in every 64 mutations) and a large proportion of non-functional, truncated or insoluble proteins. 545 The library size also dictates that a large number of mutants must be screened to test for all possibilities, which may also be impractical depending on the screening strategy available. While random methods for library design can be successful, intelligent searching of the sequence space, as per the title of this review, does not include purely random methods. 546 In particular, these methods do not allow information about which parts of the sequence have been mutated or whether all possible mutations for a particular region of interest have been screened.
Site-directed mutagenesis to target specific residues
Since the combinatorial explosion means that one cannot try every amino acid at every residue, one obvious approach is to restrict the number of target residues (in the following sections we will discuss why we do not think this is the best strategy for making faster biocatalysts). Indeed, mutagenesis directed at specific residues, usually referred to as site-directed mutagenesis, 547,548 dates from the origins of modern protein engineering itself. 549
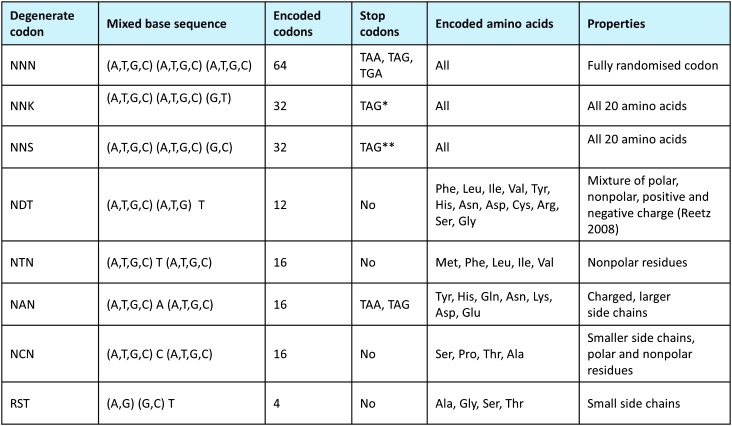
In site-directed mutagenesis, an oligonucleotide encoding the desired mutation is designed with flanking sequences either side that are complementary to the target sequence and these direct its binding to the desired sequence on a template. This oligomer is used as a PCR primer to amplify the template sequence, hence all amplicons encode the desired mutation. This control over the mutation enables particular types of mutation to be made by using mixed base codons, i.e. codons that contain a mixture of bases at a specified position (e.g. N denotes an equal mixture of A, T, G or C at a single position). Fig. 11 shows a compilation of the more common types of mixed codons used. These range from those capable of encoding all 20 amino acids (e.g. NNK) to a small subset of residues with a particular physicochemical property (e.g. NTN for nonpolar residues only).
Fig. 11. Examples of some of the common degenerate codons used in DE studies. A codon containing specific mixed bases is used to encode a particular set of amino acids, ranging from all twenty amino acids (NNN or NNK) to those with particular properties. Hence, choice of degenerate codons to use depends on the design and objective of the study. In the IUPAC terminology 590 K = G/T, M = A/C, R = A/G, S = C/G, W = A/T, Y = C/T, B = C/G/T, D = A/G/T, H = A/C/T, V = A/C/G, N = A/C/G/T. (*Typically with low codon usage; suppressor mutation may be used to block it. **Typically with low codon usage, especially in yeast; suppressor mutation may be used to block it).
The most common method (QuikChange and derivatives thereof) uses mutagenic oligonucleotides complementary to both strands of a target sequence, which are used as primers for a PCR amplification of the plasmid encoding the gene. Following DpnI digestion of the template, the PCR product is transformed into E. coli and the nicked plasmid is repaired in vivo. 550,551 Despite its popularity, QuikChange is somewhat limited by aspects like primer design and efficiency, and a variety of derivatives have been published that improve upon the original method. 552,553
Given that site-directed mutagenesis provides a way of mutating a small number of residues with high levels of accuracy, several approaches have been developed to identify possible positions to target to increase the hit rate and success. Combinatorial alanine scanning 554,555 is well known, while other flavours include the Mutagenesis Assistant Program, 531,556 and the semi-rational CASTing and B-FIT approaches 323,557 that employ a Mutagenic Plasmid Amplification method. 558
In addition to these more conventional methods, new approaches are continually being developed to improve efficiency and to reduce the number of steps in the workflow, for example Mutagenic Oligonucleotide-Directed PCR Amplification (MOD-PCR), 559 Overlap Extension PCR (OE-PCR), 560–564 Sequence Saturation Mutagenesis, 565–571 Iterative Saturation Mutagenesis, 557,572–579 and a variety of transposon-based methods. 580–583 However, a common issue with site-directed mutagenesis methods is the large number of steps involved and the limited number of positions that can be efficiently targeted at a time. The ability to mutate residues in multiple positions in a sequence is of particular interest as this can be used to address the question of combinatorial mutations simultaneously. Hence, methods like those by Liu et al., 584 Seyfang et al., 585 Fushan et al. 586 and Kegler-Ebo et al. 587 are important developments in mutagenesis strategies. Rational approaches have been reviewed, 588 including from the perspective of the necessary library size. 589 As a result, there is significant interest in the development of novel methodologies that can address these issues to produce accurate variant libraries, with larger numbers of simultaneous mutations in an economical workflow.
Optimising nucleotide substitutions
Following the selection of residues to target for mutation an important choice is the type of mutation to create. This choice is not obvious but determines the type of mutations that are made and the level of screening required. The experimenter needs to consider the nature of the mutations that they want to introduce for each position and this relates to the objective of the study. Using the common mixed base IUPAC terminology 590 (Fig. 11) there are a large number of codons that can be chosen, ranging from those encoding all 20 amino acids (the NNK or NNS codons), to a particular characteristic (e.g. NTN encodes just nonpolar residues 64 ) and a limited number of defined residues (GAN encoding just aspartate or glutamate). Importantly, choosing to use these specified mixed base codons in mutagenesis can reduce the possibility of premature stop codons and increase the chance of creating functional variants. For example, if a wild-type sequence encodes a nonpolar residue at a particular position then the number of functional variants is likely to be higher if the nonpolar codon NTN is used, encoding what are conserved substitutions, compared to encoding all possible residues with the NNK codon. 591,592
Indeed, it is known to be better to search a large library sparsely than a small library thoroughly. 593 Thus, a general strategy that seeks to move the trade-off between numbers of changes and numbers of clones to be assessed recognizes that one can design libraries that cover different general amino acid properties (such as charged, hydrophobic) while not encoding all 20 amino acids, thereby reducing (somewhat) the size of the search space. These are known as reduced library designs (see Fig. 11).
Reduced library designs
One limitation with the use of single degenerate codons is that for some sequences not all amino acids are equally represented and sometimes rare codons or stop codons are encoded. To circumvent this issue “small-intelligent” or “smart” libraries have been developed to provide equal frequency of each amino acid without bias. 594 Using a mixture of oligonucleotides, Kille et al. 595 created a restricted library with three codons NDT, VHG and TGG that encode 12, 9 and 1 codon, respectively. Together these encode 22 codons for all 20 amino acids in equal frequency, which provides good coverage of possible mutations but reduces the screening effort required to cover the sequence space. Alternative methods with the same objective include the MAX randomisation strategy 596 and using ratios of different degenerate codons designed by software (DC-Analyzer 597 ). Alternatively, the use of a reduced amino acid alphabet can also search a relevant sequence space whilst reducing the screening effort further. For example, the NDT codon encodes 12 amino acids of different physicochemical properties without encoding stop codons and has been shown to increase the number of positive hits (versus full randomization) in directed evolution studies. 324 Overall, a considerable number of such strategies have been used (e.g. ref. 64, 67, 68, 81, 82, 324, 513, 556, 592 and 596–603).
The opposite strategy to reduced library designs is to increase them by modifying the genetic code. While one may think that there is enough potential in the very large search spaces using just 20 amino acids, such approaches have led to some exceptionally elegant work that bears description.
Non-canonical amino acid incorporation
If the existing protein synthetic machinery of the host cell is able to recognise a novel amino acid, it is possible to take an auxotroph and add the non-canonical amino acid (NCAA) 604 that is thereby incorporated non-selectively. If one wishes to have site specificity, there are two main ways to increase the number of amino acids that can be incorporated into proteins. 605 First, the specificity of a tRNA molecule (e.g. one encoding a stop codon) can be modified to accommodate non-canonical amino acids; in this way, the use of the relevant codon can introduce an NCAA at the specified position. 606,607 Using this method, eight NCAAs were incorporated into the active site of nitroreductase (NTR, at Phe124) and screened for activity. One Phe analogue, p-nitrophenylalanine (pNF), exhibited more than a two-fold increase in activity over the best mutant containing a natural amino acid substitution (P124K), showing that NCAAs can produce higher enzyme activity than is possible with natural amino acids. 608
The other, considerably more radical and potentially ground-breaking, is effectively to evolve the genetic code and other apparatus such that instead of recognising triplets a subset of mRNAs and the relevant translational machinery can recognise and decode quadruplets. 609–619 To date, some 100 such NCAAs have been incorporated. However, the incorporation of NCAAs can often impact negatively on protein folding and thermostability, an issue that can be addressed through further rounds of directed evolution. 620
Recombination
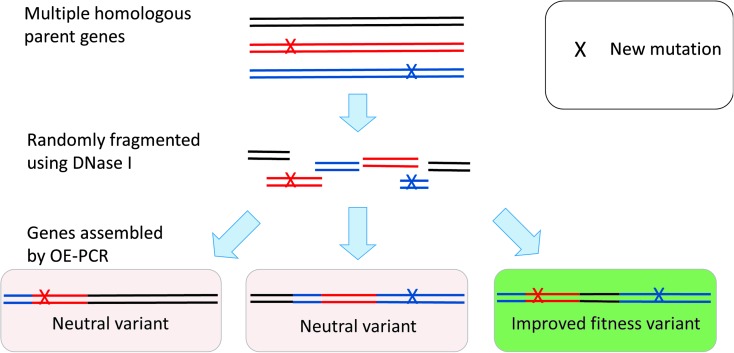
In contrast to the mutagenesis methods of library creation outlined above, but entirely consistent with our knowledge from strategies used in evolutionary computing (e.g. ref. 106), recombination is an alternative (or complementary) and effective strategy for DE (Fig. 12). Recombination techniques offer several advantages that reflect aspects of natural evolution that differ from random mutagenesis methods, not least because such changes can be combinatorial and hence able to search more areas of the sequence space in a given experiment. Recombination for the purposes of DE was popularized by Stemmer and his colleagues under the term ‘DNA shuffling’. 621–625 This used a pool of parental genes with point mutations that were randomly fragmented by DNAseI and then reassembled using OE-PCR. Since then, a variety of further methods have been developed using different fragmentation and assembly protocols. 626–629 Parental genes for DNA shuffling can be generated by random mutagenesis (epPCR) or from homologous gene families; such chimaeras may be particularly effective. 630–633
Fig. 12. The traditional recombination method for diversity creation. Recombination requires a sample of different variants of a gene (parents), which can be derived from a family of homologous genes or generated by random mutagenesis methods. The random fragmentation of these genes (using DNase I or other method) cleaves them into small constituent parts. Importantly, as the parental genes are all homologous, the fragments overlap in sequence thus allowing them to be reassembled by overlap extension PCR (OE-PCR) producing products that encode a random mixture of the parental genes. A key advantage of recombination methods is the improved ability to create combinatorial mutations. This is illustrated using two mutations (present in two different parental sequences) that when recombined separately produce no fitness improvement, but when combined together produce a variant with improved fitness.
Despite its advantages for searching wider sequence space, however, such recombination does not yield chimaeric proteins with balanced mutation distribution. Bias occurs in crossover regions of high sequence identity because the assembly of these sequences is more favourable during OE-PCR. 634,635 As a result, this reduces the diversity of sequences in the variant library. Alternative methods like SCRATCHY 636,637 generate chimaeras from genes of low sequence homology and so may help to reduce the extent of bias at the crossover points.
In addition to these more traditional methods of DNA shuffling, a number of variations have been developed (often with a penchant for a quirky acronym), such as Iterative Truncation for the Creation of HYbrid enzymes (ITCHY 638,639 ), RAndom CHImeragenesis on Transient Templates (RACHITT), 640 Recombined Extension on Truncated Templates (RETT), 641 One-pot Simple methodology for CAssette Randomization and Recombination) OSCARR, 642,643 DNA shuffling Frame shuffling, 644 Synthetic shuffling, 645 Degenerate Oligonucleotide Gene Shuffling (DOGS), 646 USERec, 647,648 SCOPE 649–651 and Incorporation of Synthetic Oligos duRing gene shuffling (ISOR). 652,653 Other methods of recombination that have been used for the improvement of proteins include the Protamax approach, 654 DNA assembler, 655,656 homologous recombination in vitro 657 and Recombineering (e.g. ref. 658 and 659). Circular permutation, in which the beginning and end of a protein are effectively recombined in different places, provides a (perhaps surprisingly) effective strategy. 17,660–667
There has long been a recognition that the better kind of chimaeragenesis strategies are those that maintain major structural elements, 668,669 by ensuring that crossover occurs mainly or solely in what are seen as structurally the most ‘suitable’ locations. This is the basis of the OPTCOMB, 670,671 RASPP, 672 SCHEMA (e.g. ref. 673–683) and other types of approach. 109,684–691
Thus, in the directed evolution of a cytochrome P450, Otey et al. 674 utilized the SCHEMA algorithm to approximate the effect of recombination with different parent P450s on the protein structure. SCHEMA provided a prediction of preferred positions for crossovers, which enabled the creation of a mutant with a 40-fold higher peroxidase activity. 673,678 Similarly, the recombination of stabilizing fragments was also able to increase the thermostability of P450s using the same approach. 692
Cell-free synthesis
Although the majority of the mutations and recombinations described above have been performed in vitro, the actual expression of the proteins themselves, and the analysis of their functionalities, is usually done in vivo. However, we should mention a series of purely in vitro strategies that have also been used to identify good sequences when coupled to suitable in vitro translation systems with functional assays. 693–700
Synthetic biology for directed evolution
With the recent improvements in DNA synthesis technology and reducing costs it is becoming increasingly feasible to synthesise sequences on a large scale. The most widely used methods for DNA synthesis continue to be short single-stranded oligodeoxyribonucleotides (typically 10–100 nt in length, often abbreviated to oligonucleotides or oligos) using phosphoramidite chemistry, 701,702 although syntheses from microarrays have particular promise. 546,703–708 Following synthesis, these oligonucleotides are assembled into larger constructs using enzymatic methods.
Hence, the foundation of synthetic biology is based on the ability to design and assemble novel biological systems ‘from the ground up’, i.e. synthetically at the DNA level. 709–713 As a result, gene synthesis and genome assembly methods have been developed to create novel sequences of several kilobases in length. 714 In particular, Gibson et al. recently assembled sections of the Mycoplasma genitalium genome (each 136 to 166 kb) using overlapping synthesised oligonucleotides. 5,6
These developments in DNA synthesis technology (and lowered cost) can greatly benefit directed evolution studies. In particular, gene synthesis using overlapping oligonucleotides presents a particularly promising method for introducing controlled mutations into a gene sequence. As these methods assemble the gene de novo, multiple mutations at different positions in the gene can be introduced simultaneously in a single workflow, decreasing the need for iterative rounds of mutagenesis.
In this process, oligonucleotide sequences are designed to be overlapping and span the length of the gene of interest, following synthesis they are assembled by either PCR-based 715,716 or ligation-based 717–720 methods. Variant libraries can be created using this process by encoding mixed base codons on the oligonucleotides and at multiple positions if required. 721 However, a limitation of the conventional gene synthesis procedure is the inherent error rate (primarily single base inserts or deletions), 722,723 which arises from errors in the phosphoramidite synthesis of the oligonucleotides. As a result, clones encoding the desired sequence must be verified by DNA sequencing and an error-correction procedure is often required. Several error-correction methods are used, including site-directed mutagenesis, 724 mismatch binding proteins 725 and mismatch cleaving endonucleases. 726,727 Of these, mismatch endonucleases are the most commonly used, and they are amenable to high throughput and automation.
SpeedyGenes and GeneGenie: tools for synthetic biology applied to the directed evolution of biocatalysts
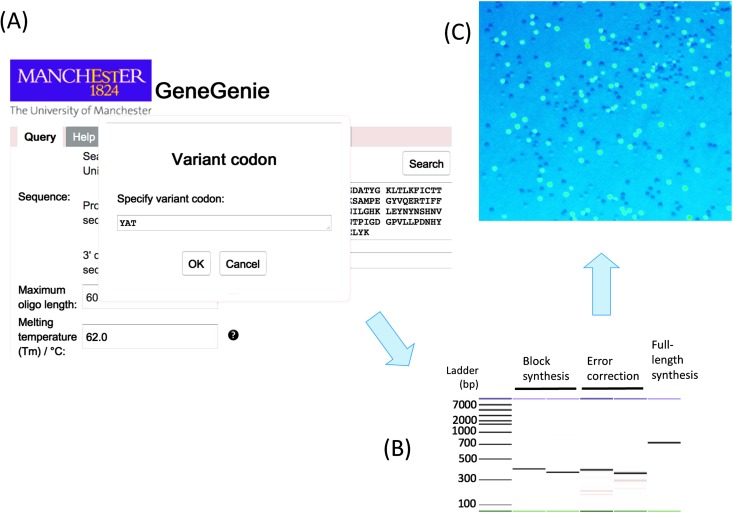
Mismatch endonucleases recognise and cleave heteroduplexes in a DNA sequence. Consequently, they can be used as an effective method for the removal of errors during gene synthesis. However, when using mixed-base codons in directed evolution this is problematic, as these mixed sequences will form heteroduplexes and so will be heavily cleaved, thus preventing assembly of the required full-length sequence. Hence, we have developed an improved gene synthesis method, SpeedyGenes, which both improves the accurate synthesis of larger genes and can also accommodate mixed-base codon sequences. 728 SpeedyGenes integrates a mismatch endonuclease step to cleave mismatched bases and, anticipating complete digestion of the mixed-base sequences, then restores these mixed base sequences by reintroducing the oligonucleotides encoding the mutation back into the PCR (“spiking in”) to allow the full length, error corrected gene to be synthesised. Importantly, multiple variant codons can be encoded at different positions of the gene simultaneously, enabling greater search of the sequence space through combinatorial mutations. This was illustrated 728 by the synthesis of a monoamine oxidase (MAO-N) with three contiguous mixed-base codons mutated at two different positions in the gene. The known structure of MAO-N showed that the side chains of these residues were known to interact, hence these libraries could be screened for combinatorial coevolutionary mutations.
As with most synthetic biology methods, the use of sequence design in silico is crucial to the successful synthesis in vitro. In the case of SpeedyGenes, a parallel, online software design tool, GeneGenie, was developed to automate the design of DNA sequences and the desired variant library. 729 By calculating the melting temperature (T m) of the overlapping sequences, and minimising the potential mis-annealing of oligomers, GeneGenie greatly improves the success rate of assembly by PCR in vitro. In addition, codons are selected according to the codon usage of the expression host organism, and cloning sequences can be encoded ab initio to facilitate downstream cloning. Importantly, any mixed base codon can be added to incorporate into the designed sequence, hence automating the design of the variant library. As an example, a limited library of enhanced green fluorescent protein (EGFP) were designed to encode two variant codons (YAT at Y66 and TWT at Y145), the product of which would encode a limited variant library of green and blue variants of EGFP 728 (Fig. 13).
Fig. 13. GeneGenie and SpeedyGenes: synthetic biology tools for the purposes of directed evolution. The integration of computational design and accurate gene synthesis methodology provide a strong platform that can be utilised for directed evolution. As an example, the design, synthesis and screening of a small library of EGFP variants is shown. Mixed base codons are used to encode the green and blue variants of EGFP in a single library. (A) GeneGenie (www.gene-genie.org/) designs overlapping oligonucleotides for a given protein together with any specific mixed base codon (here YAT denoting C/T,A,T). (B) SpeedyGenes assembles the gene sequence using these oligonucleotides, accurately (using error correction) producing variant libraries with the desired mutations. (C) Direct expression (no pre-selection) of the library in E. coli yielded colonies with the desired mutations (green or blue fluorescence).
Genetic selection and screening
An important aspect of any experiment exploiting directed evolution for the development of improved biocatalysts is how one determines which of the many millions (or more) of the different clones that are created is worth testing further and/or retaining for subsequent generations. If it is possible to include a (genetic) selection step prior to any screening, this is always likely to prove valuable. 303,730–732
Genetic selection
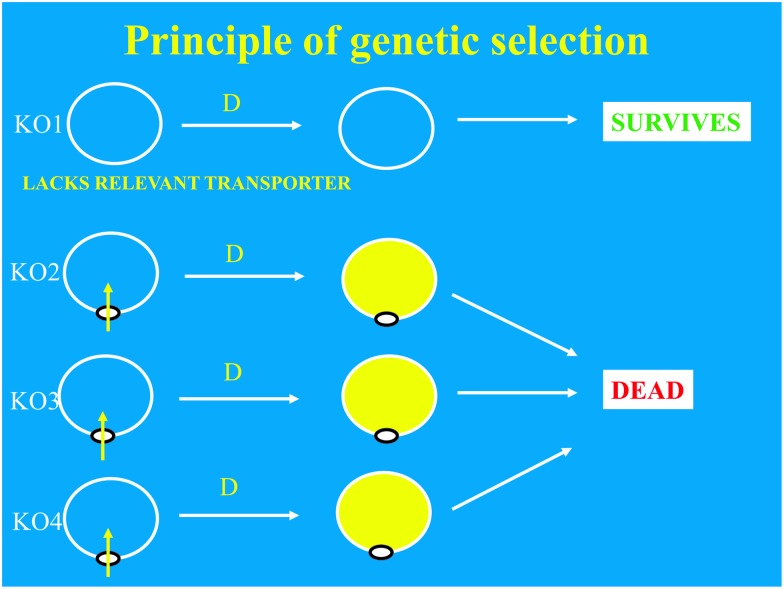
Most strategies for selection are unique to the protein of interest, and hence need to be designed empirically. Generally, this entails selection of a clone containing a desirable protein because it leads the cell to have a higher fitness. 599,733 Examples including those based on enantioselectivity, 734,735 substrate utilisation, 736 chemical complementation, 737,738 riboswitches, 739–743 and counter-selection 744 can be given. An ideal is when the selection rescues cells from a toxic insult that would otherwise kill them 745 (see Fig. 14) or repairs a growth defect 746–748 ). Two such examples 749,750 of genetic selection are based on transporter engineering. However, most of the time it is quite difficult to develop such a genetic selection assay, so one must resort to screening.
Fig. 14. The principle of genetic selection, here illustrated with a transporter gene knockout mutant in competition with others 749 that does not take up toxic levels of an otherwise cytotoxic drug D.
Screening
Microtitre plates are the standard in biomolecular screening, and this is no different in DE. 751 Herein, clones are seeded such that one clone per well is cultured, the substrates added, and the activity or products screened, primarily using chromogenic or fluorogenic substrates. This said, flow cytometry and fluorescence-activated cell sorting (FACS) have the benefit of much higher throughputs and have been widely applied (e.g. ref. 415 and 752–790) (and see below for microchannels and picodroplets). 2D arrays using immobilized proteins may also be used. 791,792 However, not all products of interest are fluorescent, and these therefore need alternative methods of detection.
Thus, other techniques have included Raman spectroscopy for the chemical imaging of productive clones, 793,794 while IR spectroscopy has been used to assess secondary structure (i.e. folding). 795 Various constructs have been used to make non-fluorescent substrates or product produce a fluorescence signal. 796 These include substrate-induced gene expression screening 797–799 and product-induced gene expression, 800 fluorescent RNAs, 801 reporter bacteria, 773,802 the detection of metabolites by fluorogenic RNA aptamers, 803–811 colourimetric aptamers and Au particles, 812 or appropriate transcription factors. 787 Riboswitches that respond to product formation, 742,743 chemical tags, 813,814 and chemical proteomics 815 have also been used as reporters for the production of small molecules.
Solid-phase screening with digital imaging is another alternative used for the engineering of biocatalysts. These methods generally use microbial colonies expressing the protein of interest to screen for activity directly in situ. 816–818 Advantages to this include the ability to use enzyme-coupled assays (like HRP) 819,820 or substrates of poor solubility or viscosity. 821
Microfluidics, microdroplets and microcompartments
Sometimes the ‘host’ and the screen are virtually synonymous, as this kind of miniaturisation can also offer considerable speeds. 822–825 Thus, there are trends towards the analysis of directed evolution experiments in microcompartments, 766,826–831 using suitable microfluidics 777,832–838 or picodroplets. 831,839–843 Agresti et al. 844 have shown that microfluidics using picolitre-volume droplets can screen a library of 108 HRP mutants in 10 hours. Although further refinement of microfluidics-based screening is required before its use becomes commonplace, it is clear that it has the capability to process the larger and more diverse libraries that one wishes to investigate.
Assessment of diversity and its maintenance
By now we have acquired a population of clones that are ‘better’ in some sense(s) than those of their parents. If we measure only fitnesses, however, as we have implicitly done thus far, we have only half the story, and we now return to the question of using knowledge of where we are or have been in a search space to optimize how we navigate it. There is of course a considerable literature on the role of ‘genetic’ and related searches in all kinds of single and multi-objective optimisation (see e.g. ref. 106, 107, 110, 113, 116, 210–213 and 845–858), all of which recognises that there is a trade-off between ‘exploration’ (looking for productive parts of the landscape) and ‘exploitation’ (performing more local searches in those parts). Methods or algorithms such as ‘efficient global optimisation’ 859 calculate these explicitly. Of course ‘where’ we are in the search space is simply encoded by the protein's sequence.
There is thus an increasing recognition that for the assessment 860–863 and maintenance 864 of diversity under selection one needs to study sequence-activity relationships. When DNA sequencing was much more expensive, methods were focused on assessing functionally important residues (e.g. ref. 865–868). As sequences became more readily available, methods such as PROSAR 219,232,233,869 were used to fix favourable amino acids, a strategy that proved rather effective (albeit that it does not consider epistasis). Now (although sequence-free methods are also possible 340,870–872 ), as large-scale DNA (including ‘next-generation’) sequencing becomes commonplace in DE, 873–876 we may hope to see large and rich datasets becoming openly available to those who care to analyse them.
Sequence-activity relationships and machine learning
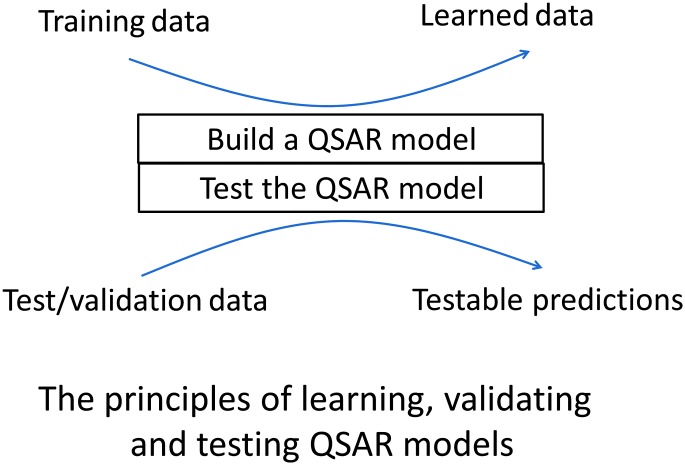
A historically important development in what is nowadays usually known as machine learning (ML) 877–879 was the recognition that it is possible to learn relationships (in the form of mathematical models) between paired inputs and outputs – in the classical case between mass spectra and the structures of the molecules that had generated them 880–884 – and more importantly that one could apply such models successfully in a predictive manner to molecules and spectra not used in the generation of the model. Such models are thus said to ‘learn’, or to ‘generalise’ to unseen samples (Fig. 15).
Fig. 15. The principles of building and testing a machine learning model, illustrated here with a QSAR model. We start with paired inputs and outputs (here sequences and activities) and learn a nonlinear mapping between the two. Methods for doing this that we have found effective include genetic programming 1345 and random forests. 23 In a second phase, the learned model is used to make predictions on an unseen validation and/or test set 1346 to establish that the model has generalized well.
In a similar vein, the first implementation of the idea that one could learn a mathematical model that captured the (normally rather nonlinear) relationships between a macromolecule's sequence and its activity in an assay of some kind, and thereby use that model to predict (in silico) the activities of untested sequences, seems to be that of Jonsson et al. 885 These authors 885 used partial least squares regression (a statistical model rather than ML – for the key differences see ref. 886) to establish a ‘quantitative sequence-activity model’ (QSAM) between (a numerical description of) 68-base-pair fragments of 25 E. coli promoters and their corresponding promoter strengths. The QSAM was then used to predict two 68 bp fragments that it was hoped would be more potent promoters than any in the training set. While extrapolation, to ‘fitnesses’ beyond what had been seen thus far, was probably a little optimistic, this work showed that such kinds of mappings were indeed possible (e.g. ref. 887–891). We have used such methods for a variety of protein-related problems, including predicting the nature and visibility of protein mass spectra. 892–894
As a separate example, we used another ML method known as ‘random forests’ 895 to learn the relationship between features of some 40 000 macromolecular (DNA aptamer) sequences and their activities, 23 and could use this to predict (from a search space some 14 orders of magnitude greater) the activities of previously untested sequences. While considerable work is going on in structural biology, we are always going to have very many more (indeed increasingly more) sequences than we have structures; thus we consider that approaches such as this are going to be very important in speeding up DE in biocatalysis and improving the functional annotation of proteins. In particular, those performing directed evolution can have simultaneous access to all sequences and activities for a given protein. 896,897 In contrast, an individual protein undergoing natural evolution cannot in any sense have a detailed ‘memory’ of its evolutionary past or pathway and in any event cannot (so far as is known, but cf. ref. 122 and 898) itself determine where to make mutations (only what to select on the basis of a poorly specified fitness). Machine learning methods seem extremely well suited for searching landscapes of this type. 23,56,107,677,899 Overall, this is a very important difference between natural evolution and (Experimenter-) Directed Evolution.
The objective function(s): metrics for the effectiveness of biocatalysts
This is not a review of enzyme kinetics and kinetic mechanisms, 549,900–902 and for our purposes we shall mainly assume that we are dealing with enzymes that catalyse thermodynamically favourable reactions, operating via a Michaelis–Menten type of reaction whose kinetic properties can largely be characterized via binding or Michaelis constants plus a (slower) catalytic rate constant k cat that is equivalent to the enzyme's turnover number (with units of reciprocal time). Much literature (e.g. ref. 549, 902 and 903) summarises the view that an appropriate measure of the effectiveness of an enzyme is a high value of k cat/K m, effected via the transduction of the initial energy of substrate/cofactor binding. 903–905 Certainly the lowering of K m alone is a very poor target for most purposes in directed evolution where initial substrate concentrations are large. Better (as an objective function) than enantiomeric excess for chiral reactions producing a preferred R form (preferred over the S form) is a P factor or E factor (k cat,R /K m,R )/(k cat,S /K m,S ) 906 of a product. For industrial purposes, we are normally much more interested in the overall conversion in a reactor, rather than any specific enzyme kinetic parameter. Hence, the space-time yield (STY) or volume–time output (VTO) over a specified period, whose units are expressed in amount × (volume × time)–1 (e.g. ref. 907–911) has also been preferred as an objective function. This is clearly more logical from the engineering point of view, but for understanding how best to drive directed evolution at the molecular level, it is arguably best to concentrate on k cat, i.e. the turnover number, which is what we do here.
The distribution of k cat values among natural proteins
Not least because of the classic and beautiful work on triose phosphate isomerase, an enzyme that is operating almost at the diffusion-controlled limit, 383,912 there is a quite pervasive view that natural evolution has taken enzymes ‘as far as they can go’ to make ‘proficient’ enzymes (e.g. ref. 913–915). Were this to be the case, there would be little point in developing directed evolution save for artificial substrates. However, it is not; most enzymes operate in vivo (and in vitro) at rates much lower than diffusion-controlled limits 916,917 (online databases of enzyme kinetic parameters include BRENDA 918 and SABIO-RK 919 ). One assumes that this is largely because evolution simply had no need (i.e. faster enzymes did not confer sufficient evolutionary advantage) 920 to select for them to increase their rates beyond that necessary to lose substantial flux control (a systems property 921–925 ). It is this in particular that makes it both desirable and possible to improve k cat or k cat/K m values over what Nature thus far has commonly achieved.
In biotransformations studies, most papers appear to report processes in terms of g product × (g enzyme × day)–1; while process parameters are important, 907 this serves (and is probably designed) to hide the very poor molecular kinetic parameters that actually pertain. K m is largely irrelevant because the concentrations in use are huge; thus our focus is on k cat. While DE has been shown to be capable of improving enzyme turnover numbers significantly, calculations show that even the ‘poster child’ examples (prositagliptin ketone transaminase, 926 ∼0.03 s–1; halohydrin dehydrogenase, 219 ∼2 s–1; isopropylmalate dehydrogenase, 927 ∼5 s–1; lovD, 368 ∼2 s–1) have turnover numbers that are very poor compared to those typical of primary metabolism, let alone the diffusion-controlled rates (depending on K m) of nearer 106–107 s–1. 916,917
What enzyme properties determine k cat values?
Almost since the beginning of molecular enzymology, scientists have come to wonder what particular features of enzymes are ‘responsible’ for enzyme catalytic power (i.e. can be used to explain it, from a mechanistic point of view). 928–930 It is implausible that there will be a unitary answer, as different sources will contribute differently in different cases. Scientifically, one may assume from the many successes of protein engineering that comparing various related sequences (and structures) by homology will be productive for our understanding of enzymology. Directed evolution studies increase the opportunities massively.
In general terms (e.g. ref. 930–933), preferred contributions to mechanisms have their different proponents, with such contributions being ascribed variously to a ‘Circe effect’, 904 strain or distortion, 934 electrostatic pre-organisation, 933,935–939 hydrogen tunneling, 940–947 reorganization energy, 948 and in particular various kinds of fluctuations and enzyme dynamics. 254,379,930,942,944–946,949–978 Less well-known flavours of dynamics include the idea that solitons may be involved. 979,980 Overall, we consider that the ‘dynamics’ view of enzyme action is especially attractive for those seeking to increase the turnover number of an enzyme.
This is because what is not in doubt is that following substrate binding at the active site (that is dominantly responsible for substrate affinity and the degree of specificity), the binding energy has been ‘used up’ (Fig. 16) and is not thereby available to drive the catalytic step in a thermodynamic sense. This means that the protein must explore its energy landscape via conformational fluctuations that are essentially isoenergetic, 951,966,981 before finding a configuration that places the active site residues into positions appropriate to effect the chemical catalysis itself, that happens as a ‘protein quake’ 981,982 in picoseconds or less. 983 The source of these motions, whether normal mode or otherwise, 984,985 can only be the protein and solvent fluctuations in the heat bath, and this means that their origins can lie in any parts of the protein, not just the few amino acids at the active site. Two exceedingly important corollaries of this follow. The first is that one may hope to predict or reflect this through the methods of molecular dynamics even when the active site is essentially unchanged, and this has recently been shown 368 ). The second corollary is that one should expect it to be found that successful directed evolution programs that increase k cat lead to many mutations that are very distant from the active site. This can also serve, at least in part, to account for why surface post-translational modifications such as glycosylation can have significant effects on turnover (e.g. ref. 986).
Fig. 16. A standard representation of an energy diagram for enzyme catalysis. Substrate binding is thermodynamically favourable, but to effect the catalytic reaction thermal energy is used to take the reaction to the right, often shown as a barrier represented by one or more ‘transition states’. Changes in the K m and K d (affecting substrate affinity) can be influenced most directly by mutagenesis of the residues at the active site whilst changes in the k cat occur primarily from mutagenesis of residues away from the active site (which can affect the fluctuations in enzyme structure required either for crossing the transition state ES‡ or by tunnelling under the barrier). At all points there are multiple roughly iso-energetic conformational (sub)states. Figure based on elements of those in ref. 930 and 966.
The importance of non-active-site mutations in increasing k cat values
In general terms, it is known that there is a considerable amount of long-range allostery in proteins, 987 such that distant mutations couple to the active site. 988 Indeed, most mutations (and amino acid residues) are necessarily distant from the active site, and there is a lack of correlation between a mutation's influence on k cat and its proximity to the active site 989 (by contrast, specificity is determined much more by the closeness to the active site 990 ). We still do not have that much data, since we require 3D structural information on many related variants; however, a number of excellent examples (Table 2) are indeed consistent with this recognition that effective DE strategies that raise k cat require that we spread our attention throughout the protein, and do not simply concentrate on the active site (Fig. 17). Indeed mutants with major improvements in k cat may display only minor changes in active site structure. 368,938,974,991
Table 2. Some examples of improvements in biocatalytic activities that have been achieved using directed evolution, focusing on examples where most relevant mutations are in amino acids that are distal to the active site.
| Target | Fold improvement over starting point | Ref. | Other notes |
| Cytochrome P450 | 9000 | 56 and 992 | 20 from generation 5, more than 15 away from active site |
| Diels–Alderase | k cat 108-fold; catalytic power 9000-fold | 993 | 21 aa, 16 outside active site |
| Glycerol dehydratase | 336 | 994 | 2 aa, both very distant from active site |
| Glyphosate acyltransferase | 200 in k cat | 995 | 21 mutations, only 4 at active site |
| Halohydrin dehalogenase | 4000 in volumetric productivity | 219 | 35 mutations, only 8 at active site |
| 3-Isopropylmalate dehydrogenase | 65 | 927 | 8 mutations, 6 distant from active site |
| LovD | >1000 | 368 | 29 mutations, 18 on enzyme surface |
| Phosphotriesterase | 25 | 996 | 7, only 1 at active site |
| Prositagliptin ketone transaminase | ∞ (no starting activity) | 926 | 27 mutations, 17 binding substrate. 200 g L–1, >99.5 ee |
| Triose phosphate isomerase | >10 000 | 386 | 36 mutations, only 1 at active site (NB effects on dimerisation, also implying distant effects) |
| Valine aminotransferase | 21 000 000 | 997 | 17 mutations, only 1 at active site |
Fig. 17. The residues that influence k cat tend to be distributed throughout an enzyme. The amino acid side chains of each of the 24 mutations obtained 993 by the directed evolution of a Dielsalderase (PDB: ; 4O5T) are highlighted. The active site pocket is shown in grey, while all mutated residues within 5 Å of the ligand (blue) are differentiated from those more than 5 Å away (red). This illustrates that the majority of mutations influencing k cat are not in close proximity to the active (substrate-binding) site. The figure was prepared using PyMol.
What if we lack the structure? Folding up proteins ‘from scratch’
A very particular kind of dynamics is that which leads a protein to fold into its tertiary structure in the first place, and the purpose of this brief section is to draw attention to some recent advances that might allow us to do this computationally. Advances in specialist computer hardware (albeit not yet widely available) can now make a prediction of how a given (smallish) protein sequence will fold up ‘ab initio’. 998–1002 However, there are many more protein sequences than there are structures, and this gap is destined to become considerably wider 1003 as sequencing methods continue to increase in speed. 102 The need for methods that can fold up proteins accurately ‘de novo’ (from their sequences) is thus acute. 1004 However, despite a number of advances (e.g. ref. 1000, 1005 and 1006) this is not yet routine. The problem is, of course, that the search space of possible structures is enormous, 1 and largely unconstrained. As well as using more powerful hardware, the real key is finding suitable constraints. An important recognition is that the covariance of residues in a series of homologous functional proteins provides a massive constraint on the inter-residue contacts and thus what structures they might adopt, and substantial advances have recently been made by a number of groups 197,199,1007–1011 in this regard. Directed evolution supplies an obvious means of creating and assessing suitable sequences.
Metals
As mentioned above, many proteins use metals and cofactors to aid the chemistry that they can catalyse, and while we shall not discuss cofactors, a short section on metalloenzymes is warranted, not least since nearly half of natural enzymes contain metals, 1012 albeit that free metals can be quite toxic. 1013–1015
To this end, if one wishes to keep open the possibility of incorporating metals into proteins undergoing DE (sometimes referred to as hybrid enzymes 1016–1019 ), it is necessary to understand the common mechanisms, residues and structures involved. 460,461,1020–1042
Some specific and unusual examples include high-valent metal catalysis, 1043 multi-metal designs as in a di-iron hydrylation reaction, 1044 a protein whose fluorescence is metal-dependent 1045 and various chelators, quantum dots and so on 1046–1050 and metallo-enzymes based on (strept)avidin–biotin technology. 1051–1053
A particular attraction of DE is that it becomes possible to incorporate metal ions that are rarely (or never) used in living organisms, to provide novel functions. Examples include iridium, 1054 rhodium 1055 and uranium (uranyl). 1056,1057
Enzyme stability, including thermostability
In general, the rates of chemical reactions increase with temperature, and if we evolve k cat to high levels we may create processes in which temperature may rise naturally anyway (and some processes may simply require it 1058 ). In a similar vein, protein stability tends to decrease with increasing temperature, and there is commonly 1059–1061 (though not always 1062 ) a trade-off between k cat and thermostability, including at the cellular level. 1063 This relationship depends effectively on the evolutionary pathway followed. 1062 As discussed above, thermostability may also sometimes (but not always 171–173 ) correlate with evolvability, 175 and is the result of multiple mutations each contributing a small amount. 1064–1069
Of course the ‘first law’ of directed evolution is that you get what you select for (even if you did not mean to). Thus if thermostability is important one must incorporate it into one's selection regime, typically by screening for it. 1070,1071 Of course if one uses a thermophile such as T. thermophilus then in vivo selection is possible, too. 1072
As rehearsed above, protein flexibility (a somewhat ill-defined concept 87,1073 ) is related to k cat, and most residues involved in improving k cat are away from the active site, at the protein surface (where they are bombarded by solvent thermal fluctuations). The connection between flexibility and thermostability is not well understood, and it does not always follow that less flexibility provides greater stability. 1074,1075 However, one might suppose that some residues that contribute flexibility are most important for (i.e. contribute significantly to) thermostability too. This is indeed the case. 989,1076,1077 Indeed, the same blend of design and focused (thus semi-empirical) DE that has proved valuable for improving k cat values seems to be the best strategy for enhancing thermostability too. 1078–1080
Some aspects of thermostability 1081–1087 can be related to individual amino acids (e.g. an ionic or H-bond formed by an arginine is of greater strength than that formed by a corresponding lysine, or thermophilic enzymes have more charged and hydrophobic but fewer polar residues 1088,1089 ). However, some aspects are best based on analyses of the 3D structure, 456,1090,1091 e.g. intra-helix ion pairs 1092,1093 and packing density. 1068,1094 Thus, Greaves and Warwicker 1095 conclude that “charge number relates to solubility, whereas protein stability is determined by charge location”. The choice of which residues to focus on can be assisted (if a structure is available) by looking at the local flexibility via methods such as mutability 1096,1097 or via B-factors, 1062,1098,1099 or via certain kinds of mass spectrometry. 1100–1108 Constraint Network Analysis 1109 provides a useful strategy for choosing which residues might be most important for thermostability. Unnatural amino acids may be beneficial too; thus fluoro-aminoacids can increase stability. 1110–1112
To disentangle the various contributions to k cat and thermostability, what we need are detailed studies of sequences and structures as they relate to both of these, and published ones remain largely lacking. However, the goal of finding sequence changes that improve both k cat and thermostability is exceptionally desirable. It should also be attainable, on the grounds that protein structural constraints that increase the rate of desirable conformational fluctuations while minimizing those that do not help the enzyme to its catalytically active confirmations must exist and will tend to have this precise effect.
Finally, thermal stresses are not the only stresses that may pertain during a biocatalytic process, albeit sometimes the same mutations can be beneficial in both (e.g. in permitting resistance to oxidation 1113,1114 or catalysis in organic solvents 1115 ).
Solvency
While our focus is on evolving proteins, those that are catalyzing reactions are always immersed in a solvent, and we cannot ignore this completely. Although ‘bulk’ measurements of solvent properties are typically unsuitable for molecular analyses of transport across membranes, 269,335,356,362,1116–1119 it is the case that some of the binding energy used in enzyme catalysis is effectively used in transferring a substrate from a usually hydrophilic aqueous phase to a usually more ‘hydrophobic’ protein phase. In general, the increased mass/hydrophobicity is also accompanied by a changed value for K m. 916 This can lead to some interesting effects of organic solvents, and solvent mixtures, 1120 on the specificity, 1121–1126 equilibria 1127 and catalytic rate constants 1128,1129 of enzymes, for reasons that are still not entirely understood. However, because the intention of many DE programs is the production of enzymes for use in industrial processes, the ability to function in organic solvents is often another important objective function, and can be solved via the above strategies. 577 One recent trend of note is the exploitation of ionic liquids 1130,1131 and ‘deep eutectic solvents’ 1132–1135 in biocatalysis.
Reaction classes
Apart from circumstances involving extremely reactive substrates and products, there is no known reason of principle why one might not be able to evolve a biocatalyst for any more-or-less simple (i.e. one-step, mono- or bi-molecular) chemical reaction. Thus, one's imagination is limited only by the reactions chosen (nowadays, for a more complex pathway, via retrosynthetic and related strategies (ref. 1136–1148)). Given that these are practically limitless (even if one might wish to start with ‘core’ molecules 1149,1150 ), we choose to be illustrative, and thereby provide a table of some of the kinds of reaction, reaction class or products for which the methods of DE have been used, with a slight focus on non-mainstream reactions. (Curiously, a convenient online database for these is presently lacking.) Our main point is that there seems no obvious limitation on reactions, beyond the case of very highly reactive substrates, intermediates or products, for which an enzymatic reaction cannot be evolved. Since the search space of possible enzymes can never be tested exhaustively, it is a safe prediction that we should expect this to hold for many more, and more complex, chemistries than have been tried to date, provided that the thermodynamics are favourable.
While the focus of this review is about how best to navigate the very large search spaces that pertain in directed enzyme evolution, we recognize that a number of processes including enzymes evolved by DE are now operated industrially. 327,328 Examples include sitagliptin, 926 generic chiral amine APIs, 1151 bio-isoprene, 1152 and atorvastatin. 1153
Concluding remarks and future prospects
In our review above, we have developed the idea that the most appropriate strategy for improving biocatalysts involves a judicious interplay between design and empiricism, the former more focused at the active site that determines binding and specificity, while the latter might usefully be focussed more on other surface and non-active-site residues to improve k cat and (in part) (thermo)stability. As our knowledge improves, design may begin to play a larger role in optimising k cat, but we consider that this will still require a considerable improvement in our understanding of the relationships between enzyme sequence, structure and dynamics. Thus, protein improvement is likely to involve the creation of increasingly ‘smart’ variant libraries over larger parts of the protein.
Another such interplay relates to the combination of experimental (‘wet’) and computational (‘dry’) approaches. We detect a significant trend towards more of the latter, 519 for instance in the use 368,1235,1296,1297 of molecular dynamics to calculate properties that suggest which residues might be creating internal friction 1298,1299 and hence lowering k cat. These examples help to illustrate that predictions and simulations in silico are likely to play an increasingly important role in predicting strategies for mutagenesis in vitro.
The increasing availability of genomic and metagenomic data, coupled to improvements in the design and prediction of protein structures (and maybe activities) will certainly contribute to improving the initialisation steps of DE. The availability of large sets of protein homologues and analogues will lead to a greater understanding of the relationships (Fig. 1) between protein sequence, structure, dynamics and catalytic activities, all of which can contribute to the design of DE experiments. Together with the development of improved synthetic biology methodology for DNA synthesis and variation, the tools for designing and initialising DE experiments are increasing greatly.
Specifically, the availability of large numbers of sequence-activity pairs may be used to learn to predict where mutations might best be tested. This decreases the empiricism of entirely random mutations in favour of synthetic biology strategies in which one has (at least statistically) more or less complete control over which sequences to make and test. Thus we see a considerable role for modern versions of sequence-activity mapping based on appropriate machine learning methods as a means of predicting where searches might optimally be conducted; this can be done in silico before creating the sequences themselves. 23 No doubt many useful datasets of this type exist in the databases of commercial organisations, but they need to become public as the likelihood is that crowdsourcing analyses would add value for their originators 1300 as well as for the common good. 1301
In terms of optimisation algorithms, we have already pointed out that very few of the modern algorithms of evolutionary optimisation have been applied to the DE problem, 107 and the advent of synthetic biology now makes their development and comparison (given that no one size will fit all 1302–1306 ) a worthwhile and timely endeavour. Complex DE algorithms that have no real counterpart in natural evolution can also now be carried out using the methods of synthetic biology.
Searching our empirical knowledge of reactions is becoming increasingly straightforward as it becomes digitised. As implied above, we expect to see an increasing cross-fertilisation between the fields of bioinformatics and cheminformatics 1307,1308 and text mining; 1309–1311 a very interesting development in this direction is that of Cadeddu et al. 1136
Conspicuous by their absence in Table 3 are the members of one important set of reactions that are widely ignored (because they do not always involve actual chemical transformations). These are the transmembrane transporters, and they make up fully one third of the reactions in the reconstructed yeast 1312 and human 25,1313 metabolic networks. Despite a widespread and longstanding assumption (e.g. ref. 1314) that xenobiotics simply tend to ‘float’ across biological membranes according to their lipophilicity, it is here worth highlighting the considerable literature (that we have reviewed elsewhere, e.g. ref. 269, 335, 356, 357, 362, 1116–1119 and 1315), including a couple of experimental examples (ref. 749 and 750), that implies that the diffusion of xenobiotics through phospholipid bilayers in intact cells is normally negligible. It is now clear that transporters enhance (and are probably required for) the transmembrane transport even of hydrophobic molecules such as alkanes, 1316–1321 terpenoids, 1319,1322,1323 long-chain, 1324–1328 and short-chain 1329–1332 fatty acids, and even CO2. 1333,1334 This may imply a significantly enhanced role for transporter engineering in whole cell biocatalysis.
Table 3. Some reactions, reaction classes or product types for which DE has proved successful. We largely exclude the very well-established programmes such as ketone and other stereoselective reductases, which along with various other reactions aimed at pharmaceutical intermediates have recently been reviewed in e.g. ref. 326, 328 and 1154–1162. Chiralities are implicit.
| Reaction (class) or substrate/product | Illustrative ref. |
| Aldolases e.g. R1CHO + R2C( O)R3 ⇌ R1C( O)CH2C(O)R3 | 462, 1163 and 1164 |
| Alkenyl and arylmalonate decarboxylases e.g. HOOCC(R1R2)COOH → HC(R1R2)COOH | 1165 |
| Amines | 1166–1169 |
| Amine dehydrogenase RC( O)Me + NH3 + NADH + H+ → RCHNH2Me + H2O + NAD+ | 1167 |
| Antifreeze proteins | 1170 |
| Azidation RH → RN3 | 1171 and 1172 |
Baeyer–Villiger monooxygenases 
|
1173 and 1174 |
| Beta-keto adipate HOOCCH2CH2C( O)CH2COOH | 1175 |
| Carotenoid biosynthesis | 1176 |
| Chlorinase Ar–H → Ar–Cl | 1177–1180 |
| Chloroperoxidase RH + Cl– + H2O2 → RCl + H2O + OH– | 1181–1187 |
| CO groups | 1157 |
| Cytochromes P450 e.g. R–H → R–OH | 56, 366, 632, 633, 674, 692, 992 and 1188–1208 |
| Diels–Alderases e.g. CH2 CHCH CH2 + CH2 CH2 → cyclohexene | 378, 380, 993, 1209 and 1210 |
| DNA polymerase | 1211 and 1212 |
| Endopeptidases | 769 |
| Esterase enantioselectivity | 1213 |
Epoxide hydrolase 
|
947 |
Flavanones 
|
1214–1221 |
| Fluorinase | 1178 and 1222 (and see ref. 1223) |
| Fatty acids | 1224–1226 |
| Glyphosate acyltransferase HOOCCH2NHCH2PO3 2– + AcCoA → HOOCCH2N(CH3C O)CH2PO3 2– + CoA | 995 and 1227–1229 |
| Glycine (glyphosate) oxidase HOOCCH2NHCH2PO3 2– + O2 → OHC–COOH + H2NCH2 PO3 2– + H2O2 | 1229–1231 |
| Haloalkane dehalogenase R1C(HBr)R2 + H2O → R1C(HOH)R2 + H+ + Br– | 1232–1235 |
| Halogenase Ar–H → Ar–Hal | 1236–1239 |
| Hydroxytyrosol 2–NO2–Ph–CH2CH2OH + O2 → 2–OH, 3–OH–Ph–CH2CH2OH + NO2 | 1240 |
| Kemp eliminase | 377 and 1241–1247 |
| Ketone reductions R1–C( O)–R2 → R1–CH(OH)–R2 | 1248 and 1249 |
| Laccase | 1250–1252 |
| Michael addition R–CH2CHO + Ph–CH CHNO2 → OHC–CH(R)–CH(Ph)CH2NO2 | 1253 |
| Monoamine oxidase R1R2CHCH2NR3R4 + 1/2O2 →R1R2C NR3 (R4 = H) or R1R2CH N+R3R4 (R4 = alkyl) + H2O | 728, 1151 and 1254–1256 |
| Nitrogenase N2 + 3H2 → 2NH3 | 1257 |
| Nucleases | 1258 |
| Old yellow enzyme (activated alkene reductions) | 664, 1259 and 1260 |
| Paraoxonase R1(R2O)(R3O)–P O → R1(R2O)(HO)P O + R3OH | 1261–1263 |
| Peroxidase | 1264 and 1265 |
| Phospho(mono/di/tri)esterases | 255, 831 and 1266–1271 |
| Polyketides | 1272–1275 |
| Polylactate | 1276 |
| Redox enzymes | 1277–1279 |
| Reductive cyclisation | 1280 |
| Restriction protease | 1281 |
| Retro-aldolase e.g. R1C( O)CH2C(O)R3 ⇌ R1CHO + R2C( O)R3 | 463 and 1282 |
| Sesquiterpene synthases | 1283 |
| Tautomerases Ar–CH C(OH)COOH ⇌ Ar–CH2COCOOH | 1284 |
| Terpene synthase/cyclase | 1285–1287 |
| Transaldolase erythrose-4-phosphate + fructose-6-phosphate → glyceraldehyde-3-phosphate + sedoheptulose-7-phosphate | 1288 and 1289 |
| Transketolase RCHO + HOCH2COCOOH → RCH(OH)COCH2OH | 1290–1293 |
| Zinc finger proteins | 1294 and 1295 |
The recent introduction of the community standard Synthetic Biology Open Language (SBOL) will certainly facilitate the sharing and re-use of synthetic biology designed sequences and modules. Beginning in 2008, the development of SBOL has been driven by an international community of computational synthetic biologists, and has led to the introduction of an initial standard for the sharing of synthetic DNA sequences 1335 and also for their visualisation. A recent proposal has introduced a more complete extension to the language, covering interactions between synthetic sequences, the design of modules and specification of their overall function. 1336 Just as with the Systems Biology Markup Language, 1337 the Systems Biology Graphical Notation, 1338 and related controlled vocabularies, metadata and ontologies for knowledge exchange in systems biology 1339,1340 and metabolomics, 1341 the availability of these kinds of standards will help move the field forward considerably.
Overall, we conclude that existing and emerging knowledge-based methods exploiting the strategies and capabilities of synthetic biology and the power of e-science will be a huge driver for the improvement of biocatalysts by directed evolution. We have only just begun.
Acknowledgments
We thank Chris Knight, Rainer Breitling, Nigel Scrutton and Nick Turner for very useful comments on the manuscript, Colin Levy for some material for the figures, and the Biotechnology and Biological Sciences Research Council for financial support (grant BB/M017702/1). This is a contribution from the Manchester Centre for Synthetic Biology of Fine and Speciality Chemicals (SYNBIOCHEM).
Biographies

Andrew Currin
Andrew Currin is a research associate at the Manchester Institute of Biotechnology, University of Manchester. He received his undergraduate degree (First Class Honours) in Biomedical Science in 2008 from the University of Birmingham, followed by a PhD in Biochemistry in 2012 at the University of Manchester. His work now focuses on the engineering of biocatalysts and developing DNA technology as a tool for synthetic biology. His interests lie in protein engineering, molecular biology, synthetic biology and drug discovery, with a particular focus on protein structure–function relationships and developing improved methodologies to investigate them.

Neil Swainston
Neil Swainston is a Research Fellow at the Manchester Institute of Biotechnology, University of Manchester. Following several years of industrial experience in proteomics bioinformatics software development with the Waters Corporation, he began his research career 8 years ago in the Manchester Centre for Integrative Systems Biology. His research interests span 'omics data analysis and management, genome-scale metabolic modelling, and enzyme optimisation through synthetic biology, and he has published over 25 papers covering these subjects. Driving all of these interests is a continued commitment to software development, data standardisation and reusability, and the development of novel informatics approaches.

Philip J. Day
Philip Day is Reader in Quantitative Analytical Genomics and Synthetic Biology, Manchester University, Philip leads interdisciplinary research for developing innovative tools in genomics and for pediatric cancer studies. Current research focuses on closed loop strategies for directed evolution gene synthesis and aptamer developments, and the development of active drug uptake using membrane transporters. Philip applies miniaturization for single cell analyses to decipher molecules per cell activities across heterogeneous cell populations. His research aims to providing exquisite quantitative data for systems biology applications and pathway analysis as a central theme for enabling personalised healthcare.

Douglas B. Kell
Douglas Kell is Research Professor in Bioanalytical Science at the University of Manchester, UK. His interests lie in systems biology, iron metabolism and dysregulation, cellular drug transporters, synthetic biology, e-science, chemometrics and cheminformatics. He was Director of the Manchester Centre for Integrative Systems Biology prior to a 5 year secondment (2008–2013) as Chief Executive of the UK Biotechnology and Biological Sciences Research Council. He is a Fellow of the Learned Society of Wales and of the American Association for the Advancement of Science, and was awarded a CBE for services to Science and Research in the New Year 2014 Honours list.
References
- Kell D. B. BioEssays. 2012;34:236–244. doi: 10.1002/bies.201100144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phylogenetic analysis of DNA sequences, ed. M. M. Miyamoto and J. Cracraft, Oxford University Press, Oxford, 1991. [Google Scholar]
- Page R. D. M. and Holmes E. C., Molecular evolution: a phylogenetic approach, Blackwell Science, Oxford, 1998. [Google Scholar]
- Harms M. J., Thornton J. W. Nat. Rev. Genet. 2013;14:559–571. doi: 10.1038/nrg3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Benders G. A., Axelrod K. C., Zaveri J., Algire M. A., Moodie M., Montague M. G., Venter J. C. Proc. Natl. Acad. Sci. U. S. A. 2008;105:20404–20409. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Benders G. A., Andrews-Pfannkoch C., Denisova E. A., Baden-Tillson H., Zaveri J., Stockwell T. B., Brownley A., Thomas D. W., Algire M. A., Merryman C., Young L., Noskov V. N., Glass J. I., Venter J. C., Hutchison 3rd C. A., Smith H. O. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- Frasch H. J., Medema M. H., Takano E., Breitling R. Curr. Opin. Biotechnol. 2013;24:1144–1150. doi: 10.1016/j.copbio.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Ongley S. E., Bian X., Neilan B. A., Muller R. Nat. Prod. Rep. 2013;30:1121–1138. doi: 10.1039/c3np70034h. [DOI] [PubMed] [Google Scholar]
- Pourmir A., Johannes T. W. Comput. Struct. Biotechnol. J. 2012;2:e201209012. doi: 10.5936/csbj.201209012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. V., Walter K. U., Kast P., Hilvert D. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10596–10601. doi: 10.1073/pnas.191159298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron K. J., Rutherford J. C., Ford D., Robinson N. J. Nature. 2009;460:823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- Jeffery C. J. Mol. BioSyst. 2009;5:345–350. doi: 10.1039/b900658n. [DOI] [PubMed] [Google Scholar]
- Moore J. C., Jin H. M., Kuchner O., Arnold F. H. J. Mol. Biol. 1997;272:336–347. doi: 10.1006/jmbi.1997.1252. [DOI] [PubMed] [Google Scholar]
- Bellman R., Adaptive control processes: a guided tour, Princeton University Press, Princeton, NJ, 1961. [Google Scholar]
- Manning C. D., Raghavan P. and Schütze H., Introduction to information retrieval, CUP, Cambridge, 2009. [Google Scholar]
- Hastie T., Tibshirani R. and Friedman J., The elements of statistical learning: data mining, inference and prediction, Springer-Verlag, Berlin, 2001. [Google Scholar]
- Arnold F. H. Nat. Biotechnol. 2006;24:328–330. doi: 10.1038/nbt0306-328. [DOI] [PubMed] [Google Scholar]
- Cleveland W. S., Visualizing data, Hobart Press, Summit, NJ, 1993. [Google Scholar]
- Beautiful visualization: looking at data through the eyes of experts, ed. J. Steele and N. Iliinsky, O'Reilly, Sebastopol, CA, 2010. [Google Scholar]
- Ware C., Information visualization, Morgan Kaufmann, San Francisco, 2000. [Google Scholar]
- McCandlish D. M. Evolution. 2011;65:1544–1558. doi: 10.1111/j.1558-5646.2011.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau N., Visualize this: the FlowingData guide to design, visualization and statistics, Wiley, Indianapolis, IN, 2011. [Google Scholar]
- Knight C. G., Platt M., Rowe W., Wedge D. C., Khan F., Day P., McShea A., Knowles J., Kell D. B. Nucleic Acids Res. 2009;37:e6. doi: 10.1093/nar/gkn899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo S., Franzson L., Jonsson J. J., Thiele I. Mol. BioSyst. 2012;8:2545–2558. doi: 10.1039/c2mb25075f. [DOI] [PubMed] [Google Scholar]
- Thiele I., Swainston N., Fleming R. M. T., Hoppe A., Sahoo S., Aurich M. K., Haraldsdottír H., Mo M. L., Rolfsson O., Stobbe M. D., Thorleifsson S. G., Agren R., Bölling C., Bordel S., Chavali A. K., Dobson P., Dunn W. B., Endler L., Goryanin I., Hala D., Hucka M., Hull D., Jameson D., Jamshidi N., Jones J., Jonsson J. J., Juty N., Keating S., Nookaew I., Le Novère N., Malys N., Mazein A., Papin J. A., Patel Y., Price N. D., Selkov Sr E., Sigurdsson M. I., Simeonidis E., Sonnenschein N., Smallbone K., Sorokin A., Beek H. V., Weichart D., Nielsen J. B., Westerhoff H. V., Kell D. B., Mendes P., Palsson B. Ø. Nat. Biotechnol. 2013;31:419–425. doi: 10.1038/nbt.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova D., Wawer M., Wassermann A. M., Bajorath J. J. Chem. Inf. Model. 2011;51:258–266. doi: 10.1021/ci100477m. [DOI] [PubMed] [Google Scholar]
- Maggiora G. M. J. Chem. Inf. Model. 2006;46:1535. doi: 10.1021/ci060117s. [DOI] [PubMed] [Google Scholar]
- Guha R., Van Drie J. H. J. Chem. Inf. Model. 2008;48:646–658. doi: 10.1021/ci7004093. [DOI] [PubMed] [Google Scholar]
- Medina-Franco J. L. Chem. Biol. Drug Des. 2013;81:553–556. doi: 10.1111/cbdd.12115. [DOI] [PubMed] [Google Scholar]
- Stumpfe D., Bajorath J. J. Chem. Inf. Model. 2012;52:2348–2353. doi: 10.1021/ci300288f. [DOI] [PubMed] [Google Scholar]
- Stumpfe D., Bajorath J. J. Med. Chem. 2012;55:2932–2942. doi: 10.1021/jm201706b. [DOI] [PubMed] [Google Scholar]
- Stumpfe D., Hu Y., Dimova D., Bajorath J. J. Med. Chem. 2014;57:18–28. doi: 10.1021/jm401120g. [DOI] [PubMed] [Google Scholar]
- Guha R., Medina-Franco J. L. J. Cheminf. 2014;6:11. doi: 10.1186/1758-2946-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha R. Abstracts of Papers of the American Chemical Society. 2010;240 [Google Scholar]
- Guha R. Methods Mol. Biol. 2011;672:101–117. doi: 10.1007/978-1-60761-839-3_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha R. J. Chem. Inf. Model. 2012;52:2181–2191. doi: 10.1021/ci300047k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha R. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2012;2:829–841. doi: 10.1002/wcms.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S. T., Rossmann M. G. J. Mol. Biol. 1973;76:241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- Saraste M., Sibbald P. R., Wittinghofer A. Trends Biochem. Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- Dobson P. D., Doig A. J. J. Mol. Biol. 2003;330:771–783. doi: 10.1016/s0022-2836(03)00628-4. [DOI] [PubMed] [Google Scholar]
- Dobson P. D., Doig A. J. J. Mol. Biol. 2005;345:187–199. doi: 10.1016/j.jmb.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Minshull J., Ness J. E., Gustafsson C., Govindarajan S. Curr. Opin. Chem. Biol. 2005;9:202–209. doi: 10.1016/j.cbpa.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Han L., Cui J., Lin H., Ji Z., Cao Z., Li Y., Chen Y. Proteomics. 2006;6:4023–4037. doi: 10.1002/pmic.200500938. [DOI] [PubMed] [Google Scholar]
- Tang Z. Q., Lin H. H., Zhang H. L., Han L. Y., Chen X., Chen Y. Z. Bioinf. Biol. Insights. 2007;1:19–47. doi: 10.4137/bbi.s315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulon J. L., Misra M., Martin S., Sale K., Sapra R. Bioinformatics. 2008;24:225–233. doi: 10.1093/bioinformatics/btm580. [DOI] [PubMed] [Google Scholar]
- Strömbergsson H., Daniluk P., Kryshtafovych A., Fidelis K., Wikberg J. E., Kleywegt G. J., Hvidsten T. R. J. Chem. Inf. Model. 2008;48:2278–2288. doi: 10.1021/ci800200e. [DOI] [PubMed] [Google Scholar]
- Bray T., Chan P., Bougouffa S., Greaves R., Doig A. J., Warwicker J. BMC Bioinf. 2009;10:379. doi: 10.1186/1471-2105-10-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvidsten T. R., Lægreid A., Kryshtafovych A., Andersson G., Fidelis K., Komorowski J. PLoS One. 2009;4:e6266. doi: 10.1371/journal.pone.0006266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler D. M., Araya C. L., Fleishman S. J., Kellogg E. H., Stephany J. J., Baker D., Fields S. Nat. Methods. 2010;7:741–746. doi: 10.1038/nmeth.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Min H., Kim S. J., Yoon S. Biochem. Biophys. Res. Commun. 2010;400:219–224. doi: 10.1016/j.bbrc.2010.08.042. [DOI] [PubMed] [Google Scholar]
- Shyu C. R., Pang B., Chi P. H., Zhao N., Korkin D., Xu D. Nucleic Acids Res. 2010;38:W53–W58. doi: 10.1093/nar/gkq522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. D., Babbitt P. C. J. Biol. Chem. 2012;287:35–42. doi: 10.1074/jbc.R111.283408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ferrari L., Aitken S., van Hemert J., Goryanin I. BMC Bioinf. 2012;13:61. doi: 10.1186/1471-2105-13-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Oja M., Weston J., Noble W. S. PLoS One. 2012;7:e32235. doi: 10.1371/journal.pone.0032235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., The roles of mutation, inbreeding, crossbreeding and selection in evolution, in Proc. Sixth Int. Conf. Genetics, ed. D. F. Jones, Genetics Society of America, Austin TX, Ithaca, NY, 1932, pp. 356–366. [Google Scholar]
- Romero P. A., Arnold F. H. Nat. Rev. Mol. Cell Biol. 2009;10:866–876. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser J. A. G. M., Krug J. Nat. Rev. Genet. 2014;15:480–490. doi: 10.1038/nrg3744. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J. Nature. 1970;225:563–564. doi: 10.1038/225563a0. [DOI] [PubMed] [Google Scholar]
- Davidson A. R., Lumb K. J., Sauer R. T. Nat. Struct. Biol. 1995;2:856–864. doi: 10.1038/nsb1095-856. [DOI] [PubMed] [Google Scholar]
- Beasley J. R., Hecht M. H. J. Biol. Chem. 1997;272:2031–2034. doi: 10.1074/jbc.272.4.2031. [DOI] [PubMed] [Google Scholar]
- Matsuura T., Ernst A., Plückthun A. Protein Sci. 2002;11:2631–2643. doi: 10.1110/ps.0215102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T., Plückthun A. Origins Life Evol. Biospheres. 2004;34:151–157. doi: 10.1023/b:orig.0000009836.86519.eb. [DOI] [PubMed] [Google Scholar]
- Axe D. D. J. Mol. Biol. 2004;341:1295–1315. doi: 10.1016/j.jmb.2004.06.058. [DOI] [PubMed] [Google Scholar]
- Bradley L. H., Thumfort P. P., Hecht M. H. Methods Mol. Biol. 2006;340:53–69. doi: 10.1385/1-59745-116-9:53. [DOI] [PubMed] [Google Scholar]
- Graziano J. J., Liu W., Perera R., Geierstanger B. H., Lesley S. A., Schultz P. G. J. Am. Chem. Soc. 2008;130:176–185. doi: 10.1021/ja074405w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Goenner T., Guerler A., Kolbeck B., Knapp E. W. Proteins. 2010;78:1618–1630. doi: 10.1002/prot.22678. [DOI] [PubMed] [Google Scholar]
- Tanaka J., Doi N., Takashima H., Yanagawa H. Protein Sci. 2010;19:786–795. doi: 10.1002/pro.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht M. H., West M. W., Patterson J., Mancias J. D., Beasley J. R., Broome B. M., Wang W. Self-Assem. Pept. Syst. Biol., Med. Eng., [Workshop] 2001:127–138. [Google Scholar]
- Ito Y., Kawama T., Urabe I., Yomo T. J. Mol. Evol. 2004;58:196–202. doi: 10.1007/s00239-003-2542-2. [DOI] [PubMed] [Google Scholar]
- Keefe A. D., Szostak J. W. Nature. 2001;410:715–718. doi: 10.1038/35070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman B., Baker D. Proc. Natl. Acad. Sci. U. S. A. 2000;97:10383–10388. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yockey H. P., Information Theory and Molecular Biology, Cambridge University Press, Cambridge, 1992. [Google Scholar]
- Wei Y., Hecht M. H. Protein Eng., Des. Sel. 2004;17:67–75. doi: 10.1093/protein/gzh007. [DOI] [PubMed] [Google Scholar]
- Minervini G., Evangelista G., Villanova L., Slanzi D., De Lucrezia D., Poli I., Luisi P. L., Polticelli F. BMC Bioinf. 2009;10(suppl 6):S22. doi: 10.1186/1471-2105-10-S6-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prymula K., Piwowar M., Kochanczyk M., Flis L., Malawski M., Szepieniec T., Evangelista G., Minervini G., Polticelli F., Wisniowski Z., Salapa K., Matczynska E., Roterman I. Chem. Biodiversity. 2009;6:2311–2336. doi: 10.1002/cbdv.200800338. [DOI] [PubMed] [Google Scholar]
- Scalley-Kim M., Baker D. J. Mol. Biol. 2004;338:573–583. doi: 10.1016/j.jmb.2004.02.055. [DOI] [PubMed] [Google Scholar]
- Kamtekar S., Schiffer J. M., Xiong H., Babik J. M., Hecht M. H. Science. 1993;262:1680–1685. doi: 10.1126/science.8259512. [DOI] [PubMed] [Google Scholar]
- West M. W., Hecht M. H. Protein Sci. 1995;4:2032–2039. doi: 10.1002/pro.5560041008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H., Buckwalter B. L., Shieh H. M., Hecht M. H. Proc. Natl. Acad. Sci. U. S. A. 1995;92:6349–6353. doi: 10.1073/pnas.92.14.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffet D. A., Foley J., Hecht M. H. Biophys. Chem. 2003;105:231–239. doi: 10.1016/s0301-4622(03)00072-3. [DOI] [PubMed] [Google Scholar]
- Hecht M. H., Das A., Go A., Bradley L. H., Wei Y. Protein Sci. 2004;13:1711–1723. doi: 10.1110/ps.04690804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley L. H., Wei Y., Thumfort P., Wurth C., Hecht M. H. Methods Mol. Biol. 2007;352:155–166. doi: 10.1385/1-59745-187-8:155. [DOI] [PubMed] [Google Scholar]
- Patel S. C., Bradley L. H., Jinadasa S. P., Hecht M. H. Protein Sci. 2009;18:1388–1400. doi: 10.1002/pro.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher M. A., McKinley K. L., Bradley L. H., Viola S. R., Hecht M. H. PLoS One. 2011;6:e15364. doi: 10.1371/journal.pone.0015364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih C. H., Chang C. M., Lin Y. S., Lo W. C., Hwang J. K. Proteins. 2012;80:1647–1657. doi: 10.1002/prot.24058. [DOI] [PubMed] [Google Scholar]
- Chang C. M., Huang Y. W., Shih C. H., Hwang J. K. Proteins. 2013;81:1192–1199. doi: 10.1002/prot.24268. [DOI] [PubMed] [Google Scholar]
- Huang T. T., Marcos M. L. D., Hwang J. K., Echave J. BMC Evol. Biol. 2014;14:78. doi: 10.1186/1471-2148-14-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh S. W., Huang T. T., Liu J. W., Yu S. H., Shih C. H., Hwang J. K., Echave J. BioMed Res. Int. 2014:572409. doi: 10.1155/2014/572409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh S. W., Liu J. W., Yu S. H., Shih C. H., Hwang J. K., Echave J. Mol. Biol. Evol. 2014;31:135–139. doi: 10.1093/molbev/mst178. [DOI] [PubMed] [Google Scholar]
- Yamauchi A., Nakashima T., Tokuriki N., Hosokawa M., Nogami H., Arioka S., Urabe I., Yomo T. Protein Eng. 2002;15:619–626. doi: 10.1093/protein/15.7.619. [DOI] [PubMed] [Google Scholar]
- Davidson A. R., Sauer R. T. Proc. Natl. Acad. Sci. U. S. A. 1994;91:2146–2150. doi: 10.1073/pnas.91.6.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H. H., Choe J., Loeb L. A. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9205–9210. doi: 10.1073/pnas.0403255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver N., The signal and the noise: the art and science of prediction, Penguin, New York, 2012. [Google Scholar]
- Poelwijk F. J., Kiviet D. J., Weinreich D. M., Tans S. J. Nature. 2007;445:383–386. doi: 10.1038/nature05451. [DOI] [PubMed] [Google Scholar]
- Keiser M. J., Roth B. L., Armbruster B. N., Ernsberger P., Irwin J. J., Shoichet B. K. Nat. Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- Bashton M., Nobeli I., Thornton J. M. Nucleic Acids Res. 2008;36:D618–D622. doi: 10.1093/nar/gkm611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams R., Worth C. L., Guenther S., Dunkel M., Lehmann R., Preissner R. Eur. J. Cell Biol. 2012;91:326–339. doi: 10.1016/j.ejcb.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Wassermann A. M., Bajorath J. Expert Opin. Drug Discovery. 2011;6:683–687. doi: 10.1517/17460441.2011.579100. [DOI] [PubMed] [Google Scholar]
- Featherstone D. E., Broadie K. BioEssays. 2002;24:267–274. doi: 10.1002/bies.10054. [DOI] [PubMed] [Google Scholar]
- Soskine M., Tawfik D. S. Nat. Rev. Genet. 2010;11:572–582. doi: 10.1038/nrg2808. [DOI] [PubMed] [Google Scholar]
- Shendure J., Ji H. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Shendure J., Aiden E. L. Nat. Biotechnol. 2012;30:1084–1094. doi: 10.1038/nbt.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez J. I., Xulvi-Brunet R., Campbell G. W., Turk-MacLeod R., Chen I. A. Proc. Natl. Acad. Sci. U. S. A. 2013;110:14984–14989. doi: 10.1073/pnas.1307604110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe W., Platt M., Wedge D., Day P. J., Kell D. B., Knowles J. J. R. Soc., Interface. 2010;7:397–408. doi: 10.1098/rsif.2009.0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiben Á. E., Hinterding R., Michalewicz Z. IEEE Trans. Evol. Comput. 1999;3:124–141. [Google Scholar]
- Handbook of evolutionary computation., ed. T. Bäck, D. B. Fogel and Z. Michalewicz, IOP Publishing/Oxford University Press, Oxford, 1997. [Google Scholar]
- O'Hagan S., Knowles J., Kell D. B. PLoS One. 2012;7:e48862. doi: 10.1371/journal.pone.0048862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syswerda G., Uniform crossover in genetic algorithms, in Proc 3rd Int Conf on Genetic Algorithms, ed. J. Schaffer, Morgan Kaufmann, 1989, pp. 2–9. [Google Scholar]
- He L., Friedman A. M., Bailey-Kellogg C. Proteins. 2012;80:790–806. doi: 10.1002/prot.23237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handl J., Kell D. B., Knowles J. IEEE/ACM Trans. Comput. Biol. Bioinf. 2007;4:279–292. doi: 10.1109/TCBB.2007.070203. [DOI] [PubMed] [Google Scholar]
- Knowles J. D., Corne D. W. Evol. Comput. 2000;8:149–172. doi: 10.1162/106365600568167. [DOI] [PubMed] [Google Scholar]
- Knowles J. D. and Corne D. W., M-PAES: a memetic algorithm for multiobjective optimization, Proc. 2000 Congr. Evol. Computation, 2000, vol. 1 and 2, pp. 325–332. [Google Scholar]
- Deb K., Multi-objective optimization using evolutionary algorithms, Wiley, New York, 2001. [Google Scholar]
- Zitzler E., Thiele L., Laumanns M., Fonseca C. M., da Fonseca V. G. IEEE Trans. Evol. Comput. 2003;7:117–132. [Google Scholar]
- Knowles J. IEEE Trans. Evol. Comput. 2006;10:50–66. [Google Scholar]
- Multiobjective Problem Solving from Nature, ed. J. Knowles, D. Corne and K. Deb, Springer, Berlin, 2008. [Google Scholar]
- Maher B. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- Weinreich D. M., Delaney N. F., Depristo M. A., Hartl D. L. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- Sung W., Ackerman M. S., Miller S. F., Doak T. G., Lynch M. Proc. Natl. Acad. Sci. U. S. A. 2012;109:18488–18492. doi: 10.1073/pnas.1216223109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman M. W., Crowell S. L. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D. Genetics. 2012;190:295–304. doi: 10.1534/genetics.111.134668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski P. D., Gerrish P. J., Lenski R. E. Nature. 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- Rockman M. V., Kruglyak L. PLoS Genet. 2009;5:e1000419. doi: 10.1371/journal.pgen.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski P. D., Lenski R. E. Annu. Rev. Ecol. Evol. Syst. 1995;26:553–578. [Google Scholar]
- Peck J. R., Waxman D. Evolution. 2010;64:3300–3309. doi: 10.1111/j.1558-5646.2010.01074.x. [DOI] [PubMed] [Google Scholar]
- Franke J., Klözer A., de Visser J. A. G. M., Krug J. PLoS Comput. Biol. 2011;7:e1002134. doi: 10.1371/journal.pcbi.1002134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B., Notebaart R. A., Pál C. Nat. Rev. Genet. 2011;12:591–602. doi: 10.1038/nrg3033. [DOI] [PubMed] [Google Scholar]
- Orengo C. A., Thornton J. M. Annu. Rev. Biochem. 2005;74:867–900. doi: 10.1146/annurev.biochem.74.082803.133029. [DOI] [PubMed] [Google Scholar]
- Moore A. D., Grath S., Schuler A., Huylmans A. K., Bornberg-Bauer E. Biochim. Biophys. Acta. 2013;1834:898–907. doi: 10.1016/j.bbapap.2013.01.007. [DOI] [PubMed] [Google Scholar]
- Lewis T. E., Sillitoe I., Andreeva A., Blundell T. L., Buchan D. W., Chothia C., Cuff A., Dana J. M., Filippis I., Gough J., Hunter S., Jones D. T., Kelley L. A., Kleywegt G. J., Minneci F., Mitchell A., Murzin A. G., Ochoa-Montano B., Rackham O. J., Smith J., Sternberg M. J., Velankar S., Yeats C., Orengo C. Nucleic Acids Res. 2013;41:D499–D507. doi: 10.1093/nar/gks1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Protein databases on the internet, Curr. Protoc. Mol. Biol., 2012, ch. 19, Unit 19.14. [DOI] [PubMed] [Google Scholar]
- Greene L. H., Lewis T. E., Addou S., Cuff A., Dallman T., Dibley M., Redfern O., Pearl F., Nambudiry R., Reid A., Sillitoe I., Yeats C., Thornton J. M., Orengo C. A. Nucleic Acids Res. 2007;35:D291–D297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff A. L., Sillitoe I., Lewis T., Redfern O. C., Garratt R., Thornton J., Orengo C. A. Nucleic Acids Res. 2009;37:D310–D314. doi: 10.1093/nar/gkn877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff A. L., Sillitoe I., Lewis T., Clegg A. B., Rentzsch R., Furnham N., Pellegrini-Calace M., Jones D., Thornton J., Orengo C. A. Nucleic Acids Res. 2011;39:D420–D426. doi: 10.1093/nar/gkq1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin A. G., Brenner S. E., Hubbard T., Chothia C. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Andreeva A., Howorth D., Chandonia J. M., Brenner S. E., Hubbard T. J., Chothia C., Murzin A. G. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox N. K., Brenner S. E., Chandonia J. M. Nucleic Acids Res. 2014;42:D304–D309. doi: 10.1093/nar/gkt1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S., Jones P., Mitchell A., Apweiler R., Attwood T. K., Bateman A., Bernard T., Binns D., Bork P., Burge S., de Castro E., Coggill P., Corbett M., Das U., Daugherty L., Duquenne L., Finn R. D., Fraser M., Gough J., Haft D., Hulo N., Kahn D., Kelly E., Letunic I., Lonsdale D., Lopez R., Madera M., Maslen J., McAnulla C., McDowall J., McMenamin C., Mi H., Mutowo-Muellenet P., Mulder N., Natale D., Orengo C., Pesseat S., Punta M., Quinn A. F., Rivoire C., Sangrador-Vegas A., Selengut J. D., Sigrist C. J., Scheremetjew M., Tate J., Thimmajanarthanan M., Thomas P. D., Wu C. H., Yeats C., Yong S. Y. Nucleic Acids Res. 2011;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge S., Kelly E., Lonsdale D., Mutowo-Muellenet P., McAnulla C., Mitchell A., Sangrador-Vegas A., Yong S. Y., Mulder N., Hunter S. Database. 2012;2012:bar068. doi: 10.1093/database/bar068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff A., Redfern O. C., Greene L., Sillitoe I., Lewis T., Dibley M., Reid A., Pearl F., Dallman T., Todd A., Garratt R., Thornton J., Orengo C. Structure. 2009;17:1051–1062. doi: 10.1016/j.str.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Bourne P. E. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5441–5446. doi: 10.1073/pnas.0704422105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnham N., Sillitoe I., Holliday G. L., Cuff A. L., Laskowski R. A., Orengo C. A., Thornton J. M. PLoS Comput. Biol. 2012;8:e1002403. doi: 10.1371/journal.pcbi.1002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett G. J., Borkakoti N., Thornton J. M. J. Mol. Biol. 2003;331:829–860. doi: 10.1016/s0022-2836(03)00734-4. [DOI] [PubMed] [Google Scholar]
- Gherardini P. F., Wass M. N., Helmer-Citterich M., Sternberg M. J. J. Mol. Biol. 2007;372:817–845. doi: 10.1016/j.jmb.2007.06.017. [DOI] [PubMed] [Google Scholar]
- Holliday G. L., Andreini C., Fischer J. D., Rahman S. A., Almonacid D. E., Williams S. T., Pearson W. R. Nucleic Acids Res. 2011;40:D783–D789. doi: 10.1093/nar/gkr799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrada E., Wagner A. PLoS One. 2010;5:e14172. doi: 10.1371/journal.pone.0014172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastolla U., Porto M., Roman H. E., Vendruscolo M. BMC Evol. Biol. 2006;6:43. doi: 10.1186/1471-2148-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worth C. L., Gong S., Blundell T. L. Nat. Rev. Mol. Cell Biol. 2009;10:709–720. doi: 10.1038/nrm2762. [DOI] [PubMed] [Google Scholar]
- Worth C. L., Preissner R., Blundell T. L. Nucleic Acids Res. 2011;39:W215–W222. doi: 10.1093/nar/gkr363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overington J., Donnelly D., Johnson M. S., Šali A., Blundell T. L. Protein Sci. 1992;1:216–226. doi: 10.1002/pro.5560010203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šali A., Blundell T. L. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Peterson M. E., Chen F., Saven J. G., Roos D. S., Babbitt P. C., Sali A. Protein Sci. 2009;18:1306–1315. doi: 10.1002/pro.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R., Fritsche M., Porto M., Bastolla U. PLoS Comput. Biol. 2010;6:e1000767. doi: 10.1371/journal.pcbi.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna D. M., Goldstein R. A. Biopolymers. 2000;53:1–8. doi: 10.1002/(SICI)1097-0282(200001)53:1<1::AID-BIP1>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Taverna D. M., Goldstein R. A. J. Mol. Biol. 2002;315:479–484. doi: 10.1006/jmbi.2001.5226. [DOI] [PubMed] [Google Scholar]
- Taverna D. M., Goldstein R. A. Proteins. 2002;46:105–109. doi: 10.1002/prot.10016. [DOI] [PubMed] [Google Scholar]
- DePristo M. A., Weinreich D. M., Hartl D. L. Nat. Rev. Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- Williams P. D., Pollock D. D., Goldstein R. A. Evol. Bioinf. Online. 2006;2:91–101. [PMC free article] [PubMed] [Google Scholar]
- Goldstein R. A. Proteins. 2011;79:1396–1407. doi: 10.1002/prot.22964. [DOI] [PubMed] [Google Scholar]
- Langton C. G. SFI S Sci. C. 1992;10:41–91. [Google Scholar]
- Kauffman S. A., The origins of order, Oxford University Press, Oxford, 1993. [Google Scholar]
- Csermely P., Sandhu K. S., Hazai E., Hoksza Z., Kiss H. J., Miozzo F., Veres D. V., Piazza F., Nussinov R. Curr. Protein Pept. Sci. 2012;13:19–33. doi: 10.2174/138920312799277992. [DOI] [PubMed] [Google Scholar]
- Csermely P., Korcsmáros T., Kiss H. J. M., London G., Nussinov R. Pharmacol. Therapeut. 2013;138:333–408. doi: 10.1016/j.pharmthera.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson J. M., Doyle J. Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top. 1999;60:1412–1427. doi: 10.1103/physreve.60.1412. [DOI] [PubMed] [Google Scholar]
- Carlson J. M., Doyle J. Proc. Natl. Acad. Sci. U. S. A. 2002;99(suppl 1):2538–2545. doi: 10.1073/pnas.012582499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csete M. E., Doyle J. C. Science. 2002;295:1664–1669. doi: 10.1126/science.1069981. [DOI] [PubMed] [Google Scholar]
- Csete M., Doyle J. Trends Biotechnol. 2004;22:446–450. doi: 10.1016/j.tibtech.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Zhou T., Carlson J. M., Doyle J. J. Theor. Biol. 2005;236:438–447. doi: 10.1016/j.jtbi.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Doyle 3rd F. J., Stelling J. J. R. Soc., Interface. 2006;3:603–616. doi: 10.1098/rsif.2006.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao P. J. Genet. Genomics. 2009;36:63–73. doi: 10.1016/S1673-8527(08)60093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giver L., Gershenson A., Freskgard P. O., Arnold F. H. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12809–12813. doi: 10.1073/pnas.95.22.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold F. H., Wintrode P. L., Miyazaki K., Gershenson A. Trends Biochem. Sci. 2001;26:100–106. doi: 10.1016/s0968-0004(00)01755-2. [DOI] [PubMed] [Google Scholar]
- Tokuriki N., Stricher F., Serrano L., Tawfik D. S. PLoS Comput. Biol. 2008;4:e1000002. doi: 10.1371/journal.pcbi.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J. D., Drummond D. A., Arnold F. H., Wilke C. O. Mol. Biol. Evol. 2006;23:1751–1761. doi: 10.1093/molbev/msl040. [DOI] [PubMed] [Google Scholar]
- Bloom J. D., Labthavikul S. T., Otey C. R., Arnold F. H. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke C. O., Bloom J. D., Drummond D. A., Raval A. Biophys. J. 2005;89:3714–3720. doi: 10.1529/biophysj.105.062125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke C. O., Drummond D. A. Curr. Opin. Struct. Biol. 2010;20:385–389. doi: 10.1016/j.sbi.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastolla U., Porto M., Roman H. E., Vendruscolo M. Gene. 2005;347:219–230. doi: 10.1016/j.gene.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Worth C. L., Blundell T. L. Proteins. 2009;75:413–429. doi: 10.1002/prot.22248. [DOI] [PubMed] [Google Scholar]
- Bloom J. D., Raval A., Wilke C. O. Genetics. 2007;175:255–266. doi: 10.1534/genetics.106.061754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J. D., Romero P. A., Lu Z., Arnold F. H. Biol. Direct. 2007;2:17. doi: 10.1186/1745-6150-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. D., Tawfik D. S. Nat. Methods. 2008;5:939–942. doi: 10.1038/nmeth.1262. [DOI] [PubMed] [Google Scholar]
- Hartl D. L., Dykhuizen D. E., Dean A. M. Genetics. 1985;111:655–674. doi: 10.1093/genetics/111.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen M. A., Stadler P. F., Fontana W. Proc. Natl. Acad. Sci. U. S. A. 1996;93:397–401. doi: 10.1073/pnas.93.1.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidys C. M., Stadler P. F. Appl. Math. Comput. 2001;117:321–350. [Google Scholar]
- Amitai G., Gupta R. D., Tawfik D. S. HFSP J. 2007;1:67–78. doi: 10.2976/1.2739115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noirel J., Simonson T. J. Chem. Phys. 2008;129:185104. doi: 10.1063/1.2992853. [DOI] [PubMed] [Google Scholar]
- Wagner A. Nat. Rev. Genet. 2008;9:965–974. doi: 10.1038/nrg2473. [DOI] [PubMed] [Google Scholar]
- Peisajovich S. G., Rockah L., Tawfik D. S. Nat. Genet. 2006;38:168–174. doi: 10.1038/ng1717. [DOI] [PubMed] [Google Scholar]
- Pritchard L., Bladon P., Mitchell J. M. O., Dufton M. J. Protein Eng. 2001;14:549–555. doi: 10.1093/protein/14.8.549. [DOI] [PubMed] [Google Scholar]
- Govindarajan S., Ness J. E., Kim S., Mundorff E. C., Minshull J., Gustafsson C. J. Mol. Biol. 2003;328:1061–1069. doi: 10.1016/s0022-2836(03)00357-7. [DOI] [PubMed] [Google Scholar]
- Neuwald A. F. Stat. Appl. Genet. Mol. Biol. 2011;10:36. doi: 10.2202/1544-6115.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard L., Dufton M. J. J. Theor. Biol. 2000;202:77–86. doi: 10.1006/jtbi.1999.1043. [DOI] [PubMed] [Google Scholar]
- Chelliah V., Chen L., Blundell T. L., Lovell S. C. J. Mol. Biol. 2004;342:1487–1504. doi: 10.1016/j.jmb.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Chelliah V., Blundell T. L. Biochemistry. 2005;70:835–840. doi: 10.1007/s10541-005-0192-2. [DOI] [PubMed] [Google Scholar]
- Chelliah V., Blundell T., Mizuguchi K. Proteins. 2005;61:722–731. doi: 10.1002/prot.20617. [DOI] [PubMed] [Google Scholar]
- Marks D. S., Colwell L. J., Sheridan R., Hopf T. A., Pagnani A., Zecchina R., Sander C. PLoS One. 2011;6:e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf T. A., Colwell L. J., Sheridan R., Rost B., Sander C., Marks D. S. Cell. 2012;149:1607–1621. doi: 10.1016/j.cell.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcos F., Pagnani A., Lunt B., Bertolino A., Marks D. S., Sander C., Zecchina R., Onuchic J. N., Hwa T., Weigt M. Proc. Natl. Acad. Sci. U. S. A. 2011;108:E1293–E1301. doi: 10.1073/pnas.1111471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin Jr R. N., Poelwijk F. J., Raman A., Gosal W. S., Ranganathan R. Nature. 2012;491:138–142. doi: 10.1038/nature11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomatis P. E., Fabiane S. M., Simona F., Carloni P., Sutton B. J., Vila A. J. Proc. Natl. Acad. Sci. U. S. A. 2008;105:20605–20610. doi: 10.1073/pnas.0807989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski M. I., Jones D. T. BMC Struct. Biol. 2009;9:10. doi: 10.1186/1472-6807-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abriata L. A., ML M. S., Tomatis P. E. FEBS Lett. 2012;586:3330–3335. doi: 10.1016/j.febslet.2012.07.010. [DOI] [PubMed] [Google Scholar]
- Burger L., van Nimwegen E. Mol. Syst. Biol. 2008;4:165. doi: 10.1038/msb4100203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigt M., White R. A., Szurmant H., Hoch J. A., Hwa T. Proc. Natl. Acad. Sci. U. S. A. 2009;106:67–72. doi: 10.1073/pnas.0805923106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger L., van Nimwegen E. PLoS Comput. Biol. 2010;6:e1000633. doi: 10.1371/journal.pcbi.1000633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausell A., Juan D., Pazos F., Valencia A. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1995–2000. doi: 10.1073/pnas.0908044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strafford J., Payongsri P., Hibbert E. G., Morris P., Batth S. S., Steadman D., Smith M. E. B., Ward J. M., Hailes H. C., Dalby P. A. J. Biotechnol. 2012;157:237–245. doi: 10.1016/j.jbiotec.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Juan D., Pazos F., Valencia A. Nat. Rev. Genet. 2013;14:249–261. doi: 10.1038/nrg3414. [DOI] [PubMed] [Google Scholar]
- Holland J. H., Adaption in natural and artificial systems: an introductory analysis with applications to biology, control, and artificial intelligence, MIT Press, 1992. [Google Scholar]
- Goldberg D. E., Genetic algorithms in search, optimization and machine learning, Addison-Wesley, 1989. [Google Scholar]
- Goldberg D. E., The design of innovation: lessons from and for competent genetic algorithms, Kluwer, Boston, 2002. [Google Scholar]
- Knowles J. IEEE Computational Intelligence Magazine. 2009;4:77–91. [Google Scholar]
- Fogel D. B., Evolutionary computation: toward a new philosophy of machine intelligence, IEEE Press, Piscataway, 1995. [Google Scholar]
- Evolutionary computation in bioinformatics, ed. G. B. Fogel and D. W. Corne, Morgan Kaufmann, Amsterdam, 2003. [Google Scholar]
- Ashlock D., Evolutionary computation for modeling and optimization, Springer, New York, 2006. [Google Scholar]
- Hamamatsu N., Aita T., Nomiya Y., Uchiyama H., Nakajima M., Husimi Y., Shibanaka Y. Protein Eng., Des. Sel. 2005;18:265–271. doi: 10.1093/protein/gzi028. [DOI] [PubMed] [Google Scholar]
- Hamamatsu N., Nomiya Y., Aita T., Nakajima M., Husimi Y., Shibanaka Y. Protein Eng., Des. Sel. 2006;19:483–489. doi: 10.1093/protein/gzl034. [DOI] [PubMed] [Google Scholar]
- Fox R. J., Davis S. C., Mundorff E. C., Newman L. M., Gavrilovic V., Ma S. K., Chung L. M., Ching C., Tam S., Muley S., Grate J., Gruber J., Whitman J. C., Sheldon R. A., Huisman G. W. Nat. Biotechnol. 2007;25:338–344. doi: 10.1038/nbt1286. [DOI] [PubMed] [Google Scholar]
- Wedge D., Rowe W., Kell D. B., Knowles J. J. Theor. Biol. 2009;257:131–141. doi: 10.1016/j.jtbi.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Carneiro M., Hartl D. L. Proc. Natl. Acad. Sci. U. S. A. 2011;107(suppl 1):1747–1751. doi: 10.1073/pnas.0906192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J. H. Theor. Popul. Biol. 1983;23:202–215. doi: 10.1016/0040-5809(83)90014-x. [DOI] [PubMed] [Google Scholar]
- Gillespie J. H. Evolution. 1984;38:1116–1129. doi: 10.1111/j.1558-5646.1984.tb00380.x. [DOI] [PubMed] [Google Scholar]
- Orr H. A. Nat. Rev. Genet. 2005;6:119–127. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- Orr H. A. Evolution. 2006;60:1113–1124. [PubMed] [Google Scholar]
- Orr H. A. J. Theor. Biol. 2006;238:279–285. doi: 10.1016/j.jtbi.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Orr H. A. Nat. Rev. Genet. 2009;10:531–539. doi: 10.1038/nrg2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unckless R. L., Orr H. A. Genetics. 2009;183:1079–1086. doi: 10.1534/genetics.109.106757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szendro I. G., Franke J., de Visser J. A. G. M., Krug J. Proc. Natl. Acad. Sci. U. S. A. 2013;110:571–576. doi: 10.1073/pnas.1213613110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J. A. Biochemistry. 1990;29:8509–8517. doi: 10.1021/bi00489a001. [DOI] [PubMed] [Google Scholar]
- Lunzer M., Miller S. P., Felsheim R., Dean A. M. Science. 2005;310:499–501. doi: 10.1126/science.1115649. [DOI] [PubMed] [Google Scholar]
- Fox R., Roy A., Govindarajan S., Minshull J., Gustafsson C., Jones J. T., Emig R. Protein Eng. 2003;16:589–597. doi: 10.1093/protein/gzg077. [DOI] [PubMed] [Google Scholar]
- Fox R. J. Theor. Biol. 2005;234:187–199. doi: 10.1016/j.jtbi.2004.11.031. [DOI] [PubMed] [Google Scholar]
- Iwakura M., Maki K., Takahashi H., Takenawa T., Yokota A., Katayanagi K., Kamiyama T., Gekko K. J. Biol. Chem. 2006;281:13234–13246. doi: 10.1074/jbc.M508823200. [DOI] [PubMed] [Google Scholar]
- Tracewell C. A., Arnold F. H. Curr. Opin. Chem. Biol. 2009;13:3–9. doi: 10.1016/j.cbpa.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reetz M. T. Angew. Chem., Int. Ed. 2013;52:2658–2666. doi: 10.1002/anie.201207842. [DOI] [PubMed] [Google Scholar]
- Hayashi Y., Aita T., Toyota H., Husimi Y., Urabe I., Yomo T. PLoS One. 2006;1:e96. doi: 10.1371/journal.pone.0000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J. D., Arnold F. H., Wilke C. O. Mol. Syst. Biol. 2007;3:76. doi: 10.1038/msb4100119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain K., Seetharaman S. Genetics. 2011;189:1029–1043. doi: 10.1534/genetics.111.134163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J. D., Gong L. I., Baltimore D. Science. 2010;328:1272–1275. doi: 10.1126/science.1187816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aita T., Hamamatsu N., Nomiya Y., Uchiyama H., Shibanaka Y., Husimi Y. Biopolymers. 2002;64:95–105. doi: 10.1002/bip.10126. [DOI] [PubMed] [Google Scholar]
- Østman B., Hintze A., Adami C. Proc. R. Soc. B. 2011;279:247–256. doi: 10.1098/rspb.2011.0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen M. S., Kemena C., Vlasov P. K., Notredame C., Kondrashov F. A. Nature. 2012;490:535–538. doi: 10.1038/nature11510. [DOI] [PubMed] [Google Scholar]
- Natarajan C., Inoguchi N., Weber R. E., Fago A., Moriyama H., Storz J. F. Science. 2013;340:1324–1327. doi: 10.1126/science.1236862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rummer J. L., McKenzie D. J., Innocenti A., Supuran C. T., Brauner C. J. Science. 2013;340:1327–1329. doi: 10.1126/science.1233692. [DOI] [PubMed] [Google Scholar]
- Mirceta S., Signore A. V., Burns J. M., Cossins A. R., Campbell K. L., Berenbrink M. Science. 2013;340:1234192. doi: 10.1126/science.1234192. [DOI] [PubMed] [Google Scholar]
- Alexander P. A., He Y., Chen Y., Orban J., Bryan P. N. Proc. Natl. Acad. Sci. U. S. A. 2009;106:21149–21154. doi: 10.1073/pnas.0906408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L. I., Bloom J. D. PLoS Genet. 2014;10:e1004328. doi: 10.1371/journal.pgen.1004328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunzer M., Golding G. B., Dean A. M. PLoS Genet. 2010;6:e1001162. doi: 10.1371/journal.pgen.1001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. G., Lovell S. C. Mol. Biol. Evol. 2009;26:1055–1065. doi: 10.1093/molbev/msp020. [DOI] [PubMed] [Google Scholar]
- Yoshikuni Y., Ferrin T. E., Keasling J. D. Nature. 2006;440:1078–1082. doi: 10.1038/nature04607. [DOI] [PubMed] [Google Scholar]
- Hult K., Berglund P. Trends Biotechnol. 2007;25:231–238. doi: 10.1016/j.tibtech.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Nobeli I., Favia A. D., Thornton J. M. Nat. Biotechnol. 2009;27:157–167. doi: 10.1038/nbt1519. [DOI] [PubMed] [Google Scholar]
- Tokuriki N., Tawfik D. S. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- Babtie A., Tokuriki N., Hollfelder F. Curr. Opin. Chem. Biol. 2010;14:200–207. doi: 10.1016/j.cbpa.2009.11.028. [DOI] [PubMed] [Google Scholar]
- Khersonsky O., Tawfik D. S. Annu. Rev. Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- Khersonsky O. and Tawfik D. S., Enzyme promiscuity: evolutionary and mechanistic aspects, in Comprehensive Natural Products II Chemistry and Biology, ed. L. Mander and H. -W. Lui, Elsevier, Oxford, 2010, pp. 48–90. [Google Scholar]
- Aharoni A., Gaidukov L., Khersonsky O., Mc Q. G. S., Roodveldt C., Tawfik D. S. Nat. Genet. 2005;37:73–76. doi: 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
- Humble M. S., Berglund P. Eur. J. Org. Chem. 2011:3391–3401. [Google Scholar]
- Huang R., Hippauf F., Rohrbeck D., Haustein M., Wenke K., Feike J., Sorrelle N., Piechulla B., Barkman T. J. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2966–2971. doi: 10.1073/pnas.1019605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D. R., Poignard P., Stanfield R. L., Wilson I. A. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Seisdedos H., Ibarra-Molero B., Sanchez-Ruiz J. M. PLoS Comput. Biol. 2012;8:e1002558. doi: 10.1371/journal.pcbi.1002558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam H., Lewis N. E., Lerman J. A., Lee D. H., Chang R. L., Kim D., Palsson B. Ø. Science. 2012;337:1101–1104. doi: 10.1126/science.1216861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J. H., van Loo B., Kamerlin S. C. L. FEBS Lett. 2012;586:1622–1630. doi: 10.1016/j.febslet.2012.04.012. [DOI] [PubMed] [Google Scholar]
- Chakraborty S. PLoS One. 2012;7:e40408. doi: 10.1371/journal.pone.0040408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti-Lafranconi P., Hollfelder F. ChemBioChem. 2013;14:285–292. doi: 10.1002/cbic.201200628. [DOI] [PubMed] [Google Scholar]
- Carbonell P., Lecointre G., Faulon J. L. J. Biol. Chem. 2011;286:43994–44004. doi: 10.1074/jbc.M111.274050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell P., Faulon J. L. Bioinformatics. 2010;26:2012–2019. doi: 10.1093/bioinformatics/btq317. [DOI] [PubMed] [Google Scholar]
- Kell D. B., Dobson P. D., Bilsland E., Oliver S. G. Drug Discovery Today. 2013;18:218–239. doi: 10.1016/j.drudis.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Chakraborty S., Minda R., Salaye L., Dandekar A. M., Bhattacharjee S. K., Rao B. J. Methods Mol. Biol. 2013;978:205–216. doi: 10.1007/978-1-62703-293-3_15. [DOI] [PubMed] [Google Scholar]
- Kauffman S. A., Weinberger E. D. J. Theor. Biol. 1989;141:211–245. doi: 10.1016/s0022-5193(89)80019-0. [DOI] [PubMed] [Google Scholar]
- Barnett L., Ruggedness and neutrality: the NKp family of fitness landscapes, Proc. 6th Int'l Conf. on Artificial Life, MIT Press, 1998, pp. 17–27. [Google Scholar]
- Aita T. J. Theor. Biol. 2008;254:252–263. doi: 10.1016/j.jtbi.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Aita T., Husimi Y. J. Theor. Biol. 2008;253:151–161. doi: 10.1016/j.jtbi.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Østman B., Hintze A., Adami C. Proc XII Conf Alife. 2010:126–132. [Google Scholar]
- Rowe W., Wedge D. C., Platt M., Kell D. B., Knowles J. Bioinformatics. 2010;26:2125–2142. doi: 10.1093/bioinformatics/btq353. [DOI] [PubMed] [Google Scholar]
- Rosenberg N. A. J. Theor. Biol. 2005;237:17–22. doi: 10.1016/j.jtbi.2005.03.026. [DOI] [PubMed] [Google Scholar]
- Kell D. B., Lurie-Luke E., J. R. Soc., Interface, 2015. , , in the press . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves C. R. and Rowe J. E., Genetic algorithms – principles and perspectives: a guide to GA theory, Kluwer Academic Publishers, Dordrecht, 2002. [Google Scholar]
- Aita T., Husimi Y. J. Math. Biol. 2000;41:207–231. doi: 10.1007/s002850000046. [DOI] [PubMed] [Google Scholar]
- Aita T., Uchiyama H., Inaoka T., Nakajima M., Kokubo T., Husimi Y. Biopolymers. 2000;54:64–79. doi: 10.1002/(SICI)1097-0282(200007)54:1<64::AID-BIP70>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Weinberger E. D., NP completeness of Kauffman's NK model: a tuneably rugged fitness landscape, Santa Fe Institute Technical Report, 1996, 96-02-003. [Google Scholar]
- Tokuriki N., Jackson C. J., Afriat-Jurnou L., Wyganowski K. T., Tang R., Tawfik D. S. Nat. Commun. 2012;3:1257. doi: 10.1038/ncomms2246. [DOI] [PubMed] [Google Scholar]
- Campbell D. T. Psychol. Rev. 1960;67:380–400. doi: 10.1037/h0040373. [DOI] [PubMed] [Google Scholar]
- Rivkin J. W., Siggelkow N. Complexity. 2002;7:31–43. [Google Scholar]
- Frenken K. Econ. Innov. New Technol. 2006;15:137–155. [Google Scholar]
- Geisendorf S. Comput. Econ. 2010;35:395–406. [Google Scholar]
- Johnson S., Where good ideas come from: the seven patterns of innovation, Penguin, London, 2011. [Google Scholar]
- Auerswald P., Kauffman S., Lobo J., Shell K. J. Econ. Dyn. Control. 2000;24:389–450. [Google Scholar]
- Kauffman S., Lobo J., Macready W. G. J. Econ. Behav. Organ. 2000;43:141–166. [Google Scholar]
- Ganco M., Hoetker G. Res Methodol Strat Mangement. 2009;5:237–268. [Google Scholar]
- Hodgson G. M., Knudsen T. J. Econ. Issues. 2006;40:287–295. [Google Scholar]
- Gabora L. Phys. Life Rev. 2013;10:117–145. doi: 10.1016/j.plrev.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Solé R. V., Valverde S., Casals M. R., Kauffman S. A., Farmer D., Eldredge N. Complexity. 2013;18:15–27. [Google Scholar]
- Wagner A., Rosen W. J. R. Soc., Interface. 2014;11:20131190. doi: 10.1098/rsif.2013.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Directed evolution library creation: methods and protocols, ed. F. H. Arnold and G. Georgiou, Springer, Berlin, 1996. [Google Scholar]
- Directed molecular evolution of proteins, ed. S. Brakmann and K. Johnsoon, Wiley-VCH, Weinheim, 2002. [Google Scholar]
- Voigt C. A., Kauffman S. and Wang Z. G., Rational evolutionary design: The theory of in vitro protein evolution, in Adv. Protein Chem., ed. F. M. Arnold, 2001, vol. 55, pp. 79–160. [DOI] [PubMed] [Google Scholar]
- Arnold F. H., Volkov A. A. Curr. Opin. Biotechnol. 1999;3:54–59. doi: 10.1016/s1367-5931(99)80010-6. [DOI] [PubMed] [Google Scholar]
- Evolutionary protein design, ed. F. H. Arnold, Academic Press, San Diego, 2001. [Google Scholar]
- Powell K. A., Ramer S. W., Del Cardayré S. B., Stemmer W. P. C., Tobin M. B., Longchamp P. F., Huisman G. W. Angew. Chem., Int. Ed. 2001;40:3948–3959. doi: 10.1002/1521-3773(20011105)40:21<3948::aid-anie3948>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Schmidt-Dannert C. Biochemistry. 2001;40:13125–13136. doi: 10.1021/bi011310c. [DOI] [PubMed] [Google Scholar]
- Taylor S. V., Kast P., Hilvert D. Angew. Chem., Int. Ed. 2001;40:3311–3335. doi: 10.1002/1521-3773(20010917)40:18<3310::aid-anie3310>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Tao H., Cornish V. W. Curr. Opin. Chem. Biol. 2002;6:858–864. doi: 10.1016/s1367-5931(02)00396-4. [DOI] [PubMed] [Google Scholar]
- Dalby P. A. Curr. Opin. Struct. Biol. 2003;13:500–505. doi: 10.1016/s0959-440x(03)00101-5. [DOI] [PubMed] [Google Scholar]
- Turner N. J. Trends Biotechnol. 2003;21:474–478. doi: 10.1016/j.tibtech.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Neylon C. Nucleic Acids Res. 2004;32:1448–1459. doi: 10.1093/nar/gkh315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leemhuis H., Stein V., Griffiths A. D., Hollfelder F. Curr. Opin. Struct. Biol. 2005;15:472–478. doi: 10.1016/j.sbi.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Yuan L., Kurek I., English J., Keenan R. Microbiol. Mol. Biol. Rev. 2005;69:373–392. doi: 10.1128/MMBR.69.3.373-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T., Yomo T. J. Biosci. Bioeng. 2006;101:449–456. doi: 10.1263/jbb.101.449. [DOI] [PubMed] [Google Scholar]
- Dalby P. A. Recent Pat. Biotechnol. 2007;1:1–9. doi: 10.2174/187220807779813929. [DOI] [PubMed] [Google Scholar]
- Sen S., Venkata Dasu V., Mandal B. Appl. Biochem. Biotechnol. 2007;143:212–223. doi: 10.1007/s12010-007-8003-4. [DOI] [PubMed] [Google Scholar]
- Akoh C. C., Chang S. W., Lee G. C., Shaw J. F. J. Agric. Food Chem. 2008;56:10445–10451. doi: 10.1021/jf801928e. [DOI] [PubMed] [Google Scholar]
- Chaput J. C., Woodbury N. W., Stearns L. A., Williams B. A. Expert Opin. Biol. Ther. 2008;8:1087–1098. doi: 10.1517/14712598.8.8.1087. [DOI] [PubMed] [Google Scholar]
- Patel R. N. Coord. Chem. Rev. 2008;252:659–701. [Google Scholar]
- Jäckel C., Kast P., Hilvert D. Annu. Rev. Biophys. 2008;37:153–173. doi: 10.1146/annurev.biophys.37.032807.125832. [DOI] [PubMed] [Google Scholar]
- Arnold F. H. Cold Spring Harbor Symp. Quant. Biol. 2009;74:41–46. doi: 10.1101/sqb.2009.74.046. [DOI] [PubMed] [Google Scholar]
- Stebel S. C., Gaida A., Arndt K. M. and Müller K. M., Directed Protein Evolution, in Molecular Biomethods Handbook, ed. J. M. Walker and R. Rapley, Humana Press, Totowa, NJ, 2nd edn, 2008, pp. 631–656. [Google Scholar]
- Turner N. J. Nat. Chem. Biol. 2009;5:567–573. doi: 10.1038/nchembio.203. [DOI] [PubMed] [Google Scholar]
- Jäckel C., Hilvert D. Curr. Opin. Biotechnol. 2010;21:753–759. doi: 10.1016/j.copbio.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Dalby P. A. Curr. Opin. Struct. Biol. 2011;21:473–480. doi: 10.1016/j.sbi.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Turner N. J., Truppo M. D. Curr. Opin. Chem. Biol. 2013;17:212–214. doi: 10.1016/j.cbpa.2013.02.026. [DOI] [PubMed] [Google Scholar]
- Reetz M. T., Carballeira J. D., Peyralans J., Hobenreich H., Maichele A., Vogel A. Chemistry. 2006;12:6031–6038. doi: 10.1002/chem.200600459. [DOI] [PubMed] [Google Scholar]
- Reetz M. T., Kahakeaw D., Lohmer R. ChemBioChem. 2008;9:1797–1804. doi: 10.1002/cbic.200800298. [DOI] [PubMed] [Google Scholar]
- Behrens G. A., Hummel A., Padhi S. K., Schätzle S., Bornscheuer U. T. Adv. Synth. Catal. 2011;353:2191–2215. [Google Scholar]
- Reetz M. T. Angew. Chem., Int. Ed. 2011;50:138–174. doi: 10.1002/anie.201000826. [DOI] [PubMed] [Google Scholar]
- Strohmeier G. A., Pichler H., May O., Gruber-Khadjawi M. Chem. Rev. 2011;111:4141–4164. doi: 10.1021/cr100386u. [DOI] [PubMed] [Google Scholar]
- Bornscheuer U. T., Huisman G. W., Kazlauskas R. J., Lutz S., Moore J. C., Robins K. Nature. 2012;485:185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- Goldsmith M., Tawfik D. S. Curr. Opin. Struct. Biol. 2012;22:406–412. doi: 10.1016/j.sbi.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Verma R., Schwaneberg U., Roccatano D. Comput. Struct. Biotechnol. J. 2012;2:e201209008. doi: 10.5936/csbj.201209008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davids T., Schmidt M., Bottcher D., Bornscheuer U. T. Curr. Opin. Chem. Biol. 2013;17:215–220. doi: 10.1016/j.cbpa.2013.02.022. [DOI] [PubMed] [Google Scholar]
- Kumar A., Singh S. Crit. Rev. Biotechnol. 2013;33:365–378. doi: 10.3109/07388551.2012.716810. [DOI] [PubMed] [Google Scholar]
- Reetz M. T. J. Am. Chem. Soc. 2013;135:12480–12496. doi: 10.1021/ja405051f. [DOI] [PubMed] [Google Scholar]
- Damborsky J., Brezovsky J. Curr. Opin. Chem. Biol. 2014;19:8–16. doi: 10.1016/j.cbpa.2013.12.003. [DOI] [PubMed] [Google Scholar]
- Dobson P. D., Patel Y., Kell D. B. Drug Discovery Today. 2009;14:31–40. doi: 10.1016/j.drudis.2008.10.011. [DOI] [PubMed] [Google Scholar]
- O'Hagan S., Swainston N., Handl J., Kell D. B. Metabolomics. 2015 doi: 10.1007/s11306-11014-10733-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiva E., Brown S., Almonacid D. E., Barber, 2nd A. E., Custer A. F., Hicks M. A., Huang C. C., Lauck F., Mashiyama S. T., Meng E. C., Mischel D., Morris J. H., Ojha S., Schnoes A. M., Stryke D., Yunes J. M., Ferrin T. E., Holliday G. L., Babbitt P. C. Nucleic Acids Res. 2014;42:D521–D530. doi: 10.1093/nar/gkt1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong K. A., Tidor B. Biotechnol. Prog. 2008;24:62–73. doi: 10.1021/bp070134h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nov Y. Methods Mol. Biol. 2014;1179:261–278. doi: 10.1007/978-1-4939-1053-3_18. [DOI] [PubMed] [Google Scholar]
- Zaugg J., Gumulya Y., Gillam E. M., Boden M. Methods Mol. Biol. 2014;1179:315–333. doi: 10.1007/978-1-4939-1053-3_21. [DOI] [PubMed] [Google Scholar]
- Pavelka A., Chovancova E., Damborsky J. Nucleic Acids Res. 2009;37:W376–W383. doi: 10.1093/nar/gkp410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhne M., Schätzle S., Jochens H., Robins K., Bornscheuer U. T. Nat. Chem. Biol. 2010;6:807–813. doi: 10.1038/nchembio.447. [DOI] [PubMed] [Google Scholar]
- Jochens H., Bornscheuer U. T. ChemBioChem. 2010;11:1861–1866. doi: 10.1002/cbic.201000284. [DOI] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Soding J., Thompson J. D., Higgins D. G. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Higgins D. G. Methods Mol. Biol. 2014;1079:105–116. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- Edgar R. C. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J., Kim B. H., Tang M., Grishin N. V. Nucleic Acids Res. 2007;35:W649–W652. doi: 10.1093/nar/gkm227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J., Grishin N. V. Methods Mol. Biol. 2014;1079:263–271. doi: 10.1007/978-1-62703-646-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Xie L., Bourne P. E. Bioinformatics. 2009;25:i305–312. doi: 10.1093/bioinformatics/btp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Xie L., Bourne P. E. Curr. Opin. Struct. Biol. 2011;21:189–199. doi: 10.1016/j.sbi.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama T., Miyazaki K. Curr. Opin. Biotechnol. 2009;20:616–622. doi: 10.1016/j.copbio.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Schofield M. M., Sherman D. H. Curr. Opin. Biotechnol. 2013;24:1151–1158. doi: 10.1016/j.copbio.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medawar P., Pluto's republic, Oxford University Press, Oxford, 1982. [Google Scholar]
- Popper K. R., Conjectures and refutations: the growth of scientific knowledge, Routledge & Kegan Paul, London, 5th edn, 1992. [Google Scholar]
- Chalmers A. F., What is this thing called Science? An assessment of the nature and status of science and its methods, Open University Press, Maidenhead, 1999. [Google Scholar]
- Kell D. B., Oliver S. G. Front. Pharmacol. 2014;5:231. doi: 10.3389/fphar.2014.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kell D. B. Trends Pharmacol. Sci. 2015 doi: 10.1016/j.tips.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Kell D. B., Oliver S. G. BioEssays. 2004;26:99–105. doi: 10.1002/bies.10385. [DOI] [PubMed] [Google Scholar]
- Kell D. B. Curr. Opin. Microbiol. 2004;7:296–307. doi: 10.1016/j.mib.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Kell D. B. and Knowles J. D., The role of modeling in systems biology, in System modeling in cellular biology: from concepts to nuts and bolts, ed. Z. Szallasi, J. Stelling and V. Periwal, MIT Press, Cambridge, 2006, pp. 3–18. [Google Scholar]
- Kell D. B. FEBS J. 2006;273:873–894. doi: 10.1111/j.1742-4658.2006.05136.x. [DOI] [PubMed] [Google Scholar]
- Kell D. B. FEBS J. 2013;280:5957–5980. doi: 10.1111/febs.12268. [DOI] [PubMed] [Google Scholar]
- Franklin L. R. Philos. Sci. 2005;72:888–899. [Google Scholar]
- Elliott K. C. Stud. Hist. Philos Sci. 2012;43:376–382. [Google Scholar]
- Doyle S. A., Fung S. Y., Koshland, Jr. D. E. Biochemistry. 2000;39:14348–14355. doi: 10.1021/bi001458g. [DOI] [PubMed] [Google Scholar]
- Li Q. S., Schwaneberg U., Fischer M., Schmitt J., Pleiss J., Lutz-Wahl S., Schmid R. D. Biochim. Biophys. Acta. 2001;1545:114–121. doi: 10.1016/s0167-4838(00)00268-5. [DOI] [PubMed] [Google Scholar]
- Gustafsson C., Govindarajan S., Minshull J. Curr. Opin. Biotechnol. 2003;14:366–370. doi: 10.1016/s0958-1669(03)00101-0. [DOI] [PubMed] [Google Scholar]
- Jiménez-Osés G., Osuna S., Gao X., Sawaya M. R., Gilson L., Collier S. J., Huisman G. W., Yeates T. O., Tang Y., Houk K. N. Nat. Chem. Biol. 2014;10:431–436. doi: 10.1038/nchembio.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian P. Chem. Soc. Rev. 2010;39:2071–2082. doi: 10.1039/b810924a. [DOI] [PubMed] [Google Scholar]
- Lichtenstein B. R., Farid T. A., Kodali G., Solomon L. A., Anderson J. L., Sheehan M. M., Ennist N. M., Fry B. A., Chobot S. E., Bialas C., Mancini J. A., Armstrong C. T., Zhao Z., Esipova T. V., Snell D., Vinogradov S. A., Discher B. M., Moser C. C., Dutton P. L. Biochem. Soc. Trans. 2012;40:561–566. doi: 10.1042/BST20120067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farid T. A., Kodali G., Solomon L. A., Lichtenstein B. R., Sheehan M. M., Fry B. A., Bialas C., Ennist N. M., Siedlecki J. A., Zhao Z., Stetz M. A., Valentine K. G., Anderson J. L., Wand A. J., Discher B. M., Moser C. C., Dutton P. L. Nat. Chem. Biol. 2013;9:826–833. doi: 10.1038/nchembio.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood C. W., Bruning M., Ibarra A. A., Bartlett G. J., Thomson A. R., Sessions R. B., Brady R. L., Woolfson D. N. Bioinformatics. 2014;30:3029–3035. doi: 10.1093/bioinformatics/btu502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson A. R., Wood C. W., Burton A. J., Bartlett G. J., Sessions R. B., Brady R. L., Woolfson D. N. Science. 2014;346:485–488. doi: 10.1126/science.1257452. [DOI] [PubMed] [Google Scholar]
- Richter F., Leaver-Fay A., Khare S. D., Bjelic S., Baker D. PLoS One. 2011;6:e19230. doi: 10.1371/journal.pone.0019230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Althoff E. A., Bolduc J., Jiang L., Moody J., Lassila J. K., Giger L., Hilvert D., Stoddard B., Baker D. J. Mol. Biol. 2012;415:615–625. doi: 10.1016/j.jmb.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Althoff E. A., Clemente F. R., Doyle L., Rothlisberger D., Zanghellini A., Gallaher J. L., Betker J. L., Tanaka F., Barbas, 3rd C. F., Hilvert D., Houk K. N., Stoddard B. L., Baker D. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röthlisberger D., Khersonsky O., Wollacott A. M., Jiang L., DeChancie J., Betker J., Gallaher J. L., Althoff E. A., Zanghellini A., Dym O., Albeck S., Houk K. N., Tawfik D. S., Baker D. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- Siegel J. B., Zanghellini A., Lovick H. M., Kiss G., Lambert A. R., St Clair J. L., Gallaher J. L., Hilvert D., Gelb M. H., Stoddard B. L., Houk K. N., Michael F. E., Baker D. Science. 2010;329:309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykora J., Brezovsky J., Koudelakova T., Lahoda M., Fortova A., Chernovets T., Chaloupkova R., Stepankova V., Prokop Z., Smatanova I. K., Hof M., Damborsky J. Nat. Chem. Biol. 2014;10:428–430. doi: 10.1038/nchembio.1502. [DOI] [PubMed] [Google Scholar]
- Eiben C. B., Siegel J. B., Bale J. B., Cooper S., Khatib F., Shen B. W., Players F., Stoddard B. L., Popovic Z., Baker D. Nat. Biotechnol. 2012;30:190–192. doi: 10.1038/nbt.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipnis Y., Baker D. Protein Sci. 2012;21:1388–1395. doi: 10.1002/pro.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersma Y. L., Pluckthun A. Curr. Opin. Biotechnol. 2011;22:849–857. doi: 10.1016/j.copbio.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Albery W. J., Knowles J. R. Biochemistry. 1976;15:5631–5640. doi: 10.1021/bi00670a032. [DOI] [PubMed] [Google Scholar]
- Nickbarg E. B., Knowles J. R. Biochemistry. 1988;27:5939–5947. doi: 10.1021/bi00416a018. [DOI] [PubMed] [Google Scholar]
- Zárate-Pérez F., Chánez-Cárdenas M. E., Vázquez-Contreras E. Prog. Mol. Biol. Transl. Sci. 2008;84:251–267. doi: 10.1016/S0079-6603(08)00407-8. [DOI] [PubMed] [Google Scholar]
- Sullivan B. J., Durani V., Magliery T. J. J. Mol. Biol. 2011;413:195–208. doi: 10.1016/j.jmb.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Alahuhta M., Salin M., Casteleijn M. G., Kemmer C., El-Sayed I., Augustyns K., Neubauer P., Wierenga R. K. Protein Eng., Des. Sel. 2008;21:257–266. doi: 10.1093/protein/gzn002. [DOI] [PubMed] [Google Scholar]
- Borchert T. V., Abagyan R., Jaenicke R., Wierenga R. K. Proc. Natl. Acad. Sci. U. S. A. 1994;91:1515–1518. doi: 10.1073/pnas.91.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmquist M. Curr. Protein Pept. Sci. 2000;1:209–235. doi: 10.2174/1389203003381405. [DOI] [PubMed] [Google Scholar]
- Henn-Sax M., Hocker B., Wilmanns M., Sterner R. Biol. Chem. 2001;382:1315–1320. doi: 10.1515/BC.2001.163. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K. FEBS Lett. 2001;492:193–198. doi: 10.1016/s0014-5793(01)02236-0. [DOI] [PubMed] [Google Scholar]
- Nagano N., Orengo C. A., Thornton J. M. J. Mol. Biol. 2002;321:741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- Schmidt D. M. Z., Mundorff E. C., Dojka M., Bermudez E., Ness J. E., Govindarajan S., Babbitt P. C., Minshull J., Gerlt J. A. Biochemistry. 2003;42:8387–8393. doi: 10.1021/bi034769a. [DOI] [PubMed] [Google Scholar]
- Vick J. E., Schmidt D. M., Gerlt J. A. Biochemistry. 2005;44:11722–11729. doi: 10.1021/bi050963g. [DOI] [PubMed] [Google Scholar]
- Park H. S., Nam S. H., Lee J. K., Yoon C. N., Mannervik B., Benkovic S. J., Kim H. S. Science. 2006;311:535–538. doi: 10.1126/science.1118953. [DOI] [PubMed] [Google Scholar]
- Vick J. E., Gerlt J. A. Biochemistry. 2007;46:14589–14597. doi: 10.1021/bi7019063. [DOI] [PubMed] [Google Scholar]
- Leopoldseder S., Claren J., Jurgens C., Sterner R. J. Mol. Biol. 2004;337:871–879. doi: 10.1016/j.jmb.2004.01.062. [DOI] [PubMed] [Google Scholar]
- Sterner R., Hocker B. Chem. Rev. 2005;105:4038–4055. doi: 10.1021/cr030191z. [DOI] [PubMed] [Google Scholar]
- Seitz T., Bocola M., Claren J., Sterner R. J. Mol. Biol. 2007;372:114–129. doi: 10.1016/j.jmb.2007.06.036. [DOI] [PubMed] [Google Scholar]
- Claren J., Malisi C., Höcker B., Sterner R. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3704–3709. doi: 10.1073/pnas.0810342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höcker B., Lochner A., Seitz T., Claren J., Sterner R. Biochemistry. 2009;48:1145–1147. doi: 10.1021/bi802125b. [DOI] [PubMed] [Google Scholar]
- Yang X., Kathuria S. V., Vadrevu R., Matthews C. R. PLoS One. 2009;4:e7179. doi: 10.1371/journal.pone.0007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlt J. A. Nat. Struct. Biol. 2000;7:171–173. doi: 10.1038/73249. [DOI] [PubMed] [Google Scholar]
- Bharat T. A., Eisenbeis S., Zeth K., Höcker B. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9942–9947. doi: 10.1073/pnas.0802202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höcker B. Biomol. Eng. 2005;22:31–38. doi: 10.1016/j.bioeng.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Fischer A., Seitz T., Lochner A., Sterner R., Merkl R., Bocola M. ChemBioChem. 2011;12:1544–1550. doi: 10.1002/cbic.201100051. [DOI] [PubMed] [Google Scholar]
- Eisenbeis S., Proffitt W., Coles M., Truffault V., Shanmugaratnam S., Meiler J., Höcker B. J. Am. Chem. Soc. 2012;134:4019–4022. doi: 10.1021/ja211657k. [DOI] [PubMed] [Google Scholar]
- Deng X., Lee J., Michael A. J., Tomchick D. R., Goldsmith E. J., Phillips M. A. J. Biol. Chem. 2010;285:25708–25719. doi: 10.1074/jbc.M110.121137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deechongkit S., Nguyen H., Jager M., Powers E. T., Gruebele M., Kelly J. W. Curr. Opin. Struct. Biol. 2006;16:94–101. doi: 10.1016/j.sbi.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Forsyth W. R., Bilsel O., Gu Z., Matthews C. R. J. Mol. Biol. 2007;372:236–253. doi: 10.1016/j.jmb.2007.06.018. [DOI] [PubMed] [Google Scholar]
- Gu Z., Rao M. K., Forsyth W. R., Finke J. M., Matthews C. R. J. Mol. Biol. 2007;374:528–546. doi: 10.1016/j.jmb.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyanaraman C., Bernacki K., Jacobson M. P. Biochemistry. 2005;44:2059–2071. doi: 10.1021/bi0481186. [DOI] [PubMed] [Google Scholar]
- Sakai A., Fedorov A. A., Fedorov E. V., Schnoes A. M., Glasner M. E., Brown S., Rutter M. E., Bain K., Chang S., Gheyi T., Sauder J. M., Burley S. K., Babbitt P. C., Almo S. C., Gerlt J. A. Biochemistry. 2009;48:1445–1453. doi: 10.1021/bi802277h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourist R., Jochens H., Bartsch S., Kuipers R., Padhi S. K., Gall M., Böttcher D., Joosten H. J., Bornscheuer U. T. ChemBioChem. 2010;11:1635–1643. doi: 10.1002/cbic.201000213. [DOI] [PubMed] [Google Scholar]
- Jochens H., Hesseler M., Stiba K., Padhi S. K., Kazlauskas R. J., Bornscheuer U. T. ChemBioChem. 2011;12:1508–1517. doi: 10.1002/cbic.201000771. [DOI] [PubMed] [Google Scholar]
- Gerlt J. A., Babbitt P. C., Jacobson M. P., Almo S. C. J. Biol. Chem. 2012;287:29–34. doi: 10.1074/jbc.R111.240945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukk T., Sakai A., Kalyanaraman C., Brown S. D., Imker H. J., Song L., Fedorov A. A., Fedorov E. V., Toro R., Hillerich B., Seidel R., Patskovsky Y., Vetting M. W., Nair S. K., Babbitt P. C., Almo S. C., Gerlt J. A., Jacobson M. P. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4122–4127. doi: 10.1073/pnas.1112081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanghellini A., Jiang L., Wollacott A. M., Cheng G., Meiler J., Althoff E. A., Rothlisberger D., Baker D. Protein Sci. 2006;15:2785–2794. doi: 10.1110/ps.062353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malisi C., Kohlbacher O., Höcker B. Proteins. 2009;77:74–83. doi: 10.1002/prot.22418. [DOI] [PubMed] [Google Scholar]
- Dellus-Gur E., Toth-Petroczy A., Elias M., Tawfik D. S. J. Mol. Biol. 2013;425:2609–2621. doi: 10.1016/j.jmb.2013.03.033. [DOI] [PubMed] [Google Scholar]
- Gebauer M., Skerra A. Methods Enzymol. 2012;503:157–188. doi: 10.1016/B978-0-12-396962-0.00007-0. [DOI] [PubMed] [Google Scholar]
- Gebauer M., Schiefner A., Matschiner G., Skerra A. J. Mol. Biol. 2012;425:780–802. doi: 10.1016/j.jmb.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Hohlbaum A. M., Skerra A. Expert Rev. Clin. Immunol. 2007;3:491–501. doi: 10.1586/1744666X.3.4.491. [DOI] [PubMed] [Google Scholar]
- Schlehuber S., Skerra A. Expert Opin. Biol. Ther. 2005;5:1453–1462. doi: 10.1517/14712598.5.11.1453. [DOI] [PubMed] [Google Scholar]
- Skerra A. Curr. Opin. Mol. Ther. 2007;9:336–344. [PubMed] [Google Scholar]
- Skerra A. FEBS J. 2008;275:2677–2683. doi: 10.1111/j.1742-4658.2008.06439.x. [DOI] [PubMed] [Google Scholar]
- Gunneriusson E., Nord K., Uhlen M., Nygren P. A. Environ. Prot. Eng. 1999;12:873–878. doi: 10.1093/protein/12.10.873. [DOI] [PubMed] [Google Scholar]
- Gunneriusson E., Samuelson P., Ringdahl J., Gronlund H., Nygren P. A., Stahl S. Appl. Environ. Microbiol. 1999;65:4134–4140. doi: 10.1128/aem.65.9.4134-4140.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronvall G., Jonsson K. J. Mol. Recognit. 1999;12:38–44. doi: 10.1002/(SICI)1099-1352(199901/02)12:1<38::AID-JMR378>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Nord K., Gunneriusson E., Ringdahl J., Stahl S., Uhlen M., Nygren P. A. Nat. Biotechnol. 1997;15:772–777. doi: 10.1038/nbt0897-772. [DOI] [PubMed] [Google Scholar]
- Nord K., Nord O., Uhlen M., Kelley B., Ljungqvist C., Nygren P. A. Eur. J. Biochem. 2001;268:4269–4277. doi: 10.1046/j.1432-1327.2001.02344.x. [DOI] [PubMed] [Google Scholar]
- Feldwisch J., Tolmachev V. Methods Mol. Biol. 2012;899:103–126. doi: 10.1007/978-1-61779-921-1_7. [DOI] [PubMed] [Google Scholar]
- Löfblom J., Feldwisch J., Tolmachev V., Carlsson J., Ståhl S., Frejd F. Y. FEBS Lett. 2010;584:2670–2680. doi: 10.1016/j.febslet.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Nygren P.-Å. FEBS J. 2008;275:2668–2676. doi: 10.1111/j.1742-4658.2008.06438.x. [DOI] [PubMed] [Google Scholar]
- Orlova A., Feldwisch J., Abrahmsén L., Tolmachev V. Cancer Biother. Radiopharm. 2007;22:573–584. doi: 10.1089/cbr.2006.004-U. [DOI] [PubMed] [Google Scholar]
- Tolmachev V., Orlova A., Nilsson F. Y., Feldwisch J., Wennborg A., Abrahmsén L. Expert Opin. Biol. Ther. 2007;7:555–568. doi: 10.1517/14712598.7.4.555. [DOI] [PubMed] [Google Scholar]
- Fernández L. A. Curr. Opin. Biotechnol. 2004;15:364–373. doi: 10.1016/j.copbio.2004.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner R., Schmid F. X. Science. 2004;304:1916–1917. doi: 10.1126/science.1100482. [DOI] [PubMed] [Google Scholar]
- Cheng G., Qian B., Samudrala R., Baker D. Nucleic Acids Res. 2005;33:5861–5867. doi: 10.1093/nar/gki894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chica R. A., Doucet N., Pelletier J. N. Curr. Opin. Biotechnol. 2005;16:378–384. doi: 10.1016/j.copbio.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Saunders C. T., Baker D. J. Mol. Biol. 2005;346:631–644. doi: 10.1016/j.jmb.2004.11.062. [DOI] [PubMed] [Google Scholar]
- Floudas C. A., Fung H. K., McAllister S. R., Monnigmann M., Rajgaria R. Chem. Eng. Sci. 2006;61:966–988. [Google Scholar]
- Alvizo O., Allen B. D., Mayo S. L., BioTechniques, 2007, 42 , 31 , 33, 35 passim . [DOI] [PubMed] [Google Scholar]
- Anderson J. L., Koder R. L., Moser C. C., Dutton P. L. Biochem. Soc. Trans. 2008;36:1106–1111. doi: 10.1042/BST0361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward T. R. Angew. Chem., Int. Ed. 2008;47:7802–7803. doi: 10.1002/anie.200802865. [DOI] [PubMed] [Google Scholar]
- Bellows M. L., Floudas C. A. Curr. Drug Targets. 2010;11:264–278. doi: 10.2174/138945010790711914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry H. C., Lehmann A., Saven J. G., DeGrado W. F., Therien M. J. J. Am. Chem. Soc. 2010;132:3997–4005. doi: 10.1021/ja907407m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz S. Curr. Opin. Biotechnol. 2010;21:734–743. doi: 10.1016/j.copbio.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazes R. J., Grisewood M. J., Maranas C. D. Curr. Opin. Struct. Biol. 2011;21:467–472. doi: 10.1016/j.sbi.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Pleiss J. Curr. Opin. Biotechnol. 2011;22:611–617. doi: 10.1016/j.copbio.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Samish I., MacDermaid C. M., Perez-Aguilar J. M., Saven J. G. Annu. Rev. Phys. Chem. 2011;62:129–149. doi: 10.1146/annurev-physchem-032210-103509. [DOI] [PubMed] [Google Scholar]
- Hilvert D. Annu. Rev. Biochem. 2013;82:447–470. doi: 10.1146/annurev-biochem-072611-101825. [DOI] [PubMed] [Google Scholar]
- Kries H., Blomberg R., Hilvert D. Curr. Opin. Chem. Biol. 2013;17:221–228. doi: 10.1016/j.cbpa.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Privett H. K., Kiss G., Lee T. M., Blomberg R., Chica R. A., Thomas L. M., Hilvert D., Houk K. N., Mayo S. L. Proc. Natl. Acad. Sci. U. S. A. 2012;109:3790–3795. doi: 10.1073/pnas.1118082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saven J. G. Curr. Opin. Chem. Biol. 2011;15:452–457. doi: 10.1016/j.cbpa.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P. S., Oberdorfer G., Xu C., Pei X. Y., Nannenga B. L., Rogers J. M., DiMaio F., Gonen T., Luisi B., Baker D. Science. 2014;346:481–485. doi: 10.1126/science.1257481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B. A., Hecht M. H. Curr. Opin. Chem. Biol. 2011;15:421–426. doi: 10.1016/j.cbpa.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Bhattacherjee A., Biswas P. J. Chem. Phys. 2009;131:125101. doi: 10.1063/1.3236519. [DOI] [PubMed] [Google Scholar]
- Kleinman C. L., Rodrigue N., Bonnard C., Philippe H., Lartillot N. BMC Bioinf. 2006;7:326. doi: 10.1186/1471-2105-7-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S. D., Kipnis Y., Greisen Jr P., Takeuchi R., Ashani Y., Goldsmith M., Song Y., Gallaher J. L., Silman I., Leader H., Sussman J. L., Stoddard B. L., Tawfik D. S., Baker D. Nat. Chem. Biol. 2012;8:294–300. doi: 10.1038/nchembio.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höcker B. Nat. Chem. Biol. 2012;8:224–225. doi: 10.1038/nchembio.800. [DOI] [PubMed] [Google Scholar]
- Althoff E. A., Wang L., Jiang L., Giger L., Lassila J. K., Wang Z., Smith M., Hari S., Kast P., Herschlag D., Hilvert D., Baker D. Protein Sci. 2012;21:717–726. doi: 10.1002/pro.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giger L., Caner S., Obexer R., Kast P., Baker D., Ban N., Hilvert D. Nat. Chem. Biol. 2013;9:494–498. doi: 10.1038/nchembio.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmeier K., Höcker B. Curr. Opin. Chem. Biol. 2013;17:929–933. doi: 10.1016/j.cbpa.2013.10.002. [DOI] [PubMed] [Google Scholar]
- Božič S., Doles T., Gradišar H., Jerala R. Curr. Opin. Chem. Biol. 2013;17:940–945. doi: 10.1016/j.cbpa.2013.10.014. [DOI] [PubMed] [Google Scholar]
- Simms J., Booth P. J. Curr. Opin. Chem. Biol. 2013;17:976–981. doi: 10.1016/j.cbpa.2013.10.005. [DOI] [PubMed] [Google Scholar]
- Hallen M. A., Keedy D. A., Donald B. R. Proteins. 2013;81:18–39. doi: 10.1002/prot.24150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss G., Çelebi-ölçüm N., Moretti R., Baker D., Houk K. N. Angew. Chem., Int. Ed. 2013;52:5700–5725. doi: 10.1002/anie.201204077. [DOI] [PubMed] [Google Scholar]
- Tantillo D. J. Org. Lett. 2010;12:1164–1167. doi: 10.1021/ol9028435. [DOI] [PubMed] [Google Scholar]
- Zhang X., DeChancie J., Gunaydin H., Chowdry A. B., Clemente F. R., Smith A. J., Handel T. M., Houk K. N. J. Org. Chem. 2008;73:889–899. doi: 10.1021/jo701974n. [DOI] [PubMed] [Google Scholar]
- Dechancie J., Clemente F. R., Smith A. J., Gunaydin H., Zhao Y. L., Zhang X., Houk K. N. Protein Sci. 2007;16:1851–1866. doi: 10.1110/ps.072963707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantillo D. J., Chen J., Houk K. N. Curr. Opin. Chem. Biol. 1998;2:743–750. doi: 10.1016/s1367-5931(98)80112-9. [DOI] [PubMed] [Google Scholar]
- Bouvignies G., Vallurupalli P., Hansen D. F., Correia B. E., Lange O., Bah A., Vernon R. M., Dahlquist F. W., Baker D., Kay L. E. Nature. 2011;477:111–114. doi: 10.1038/nature10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S., Khatib F., Treuille A., Barbero J., Lee J., Beenen M., Leaver-Fay A., Baker D., Popović Z., Players F. Nature. 2010;466:756–760. doi: 10.1038/nature09304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio F., Leaver-Fay A., Bradley P., Baker D., Andre I. PLoS One. 2011;6:e20450. doi: 10.1371/journal.pone.0020450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleishman S. J., Leaver-Fay A., Corn J. E., Strauch E. M., Khare S. D., Koga N., Ashworth J., Murphy P., Richter F., Lemmon G., Meiler J., Baker D. PLoS One. 2011;6:e20161. doi: 10.1371/journal.pone.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handl J., Knowles J., Vernon R., Baker D., Lovell S. C. Proteins. 2012;80:490–504. doi: 10.1002/prot.23215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A., Tyka M., Lewis S. M., Lange O. F., Thompson J., Jacak R., Kaufman K., Renfrew P. D., Smith C. A., Sheffler W., Davis I. W., Cooper S., Treuille A., Mandell D. J., Richter F., Ban Y. E., Fleishman S. J., Corn J. E., Kim D. E., Lyskov S., Berrondo M., Mentzer S., Popovic Z., Havranek J. J., Karanicolas J., Das R., Meiler J., Kortemme T., Gray J. J., Kuhlman B., Baker D., Bradley P. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange O. F., Rossi P., Sgourakis N. G., Song Y., Lee H. W., Aramini J. M., Ertekin A., Xiao R., Acton T. B., Montelione G. T., Baker D. Proc. Natl. Acad. Sci. U. S. A. 2012;109:10873–10878. doi: 10.1073/pnas.1203013109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T. C., Dimaio F., Read R. J., Baker D., Bunkóczi G., Adams P. D., Grosse-Kunstleve R. W., Afonine P. V., Echols N. J. Struct. Funct. Genomics. 2012;13:81–90. doi: 10.1007/s10969-012-9129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz C., Vernon R., Otting G., Baker D., Huber T. J. Mol. Biol. 2012;416:668–677. doi: 10.1016/j.jmb.2011.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gront D., Kulp D. W., Vernon R. M., Strauss C. E., Baker D. PLoS One. 2011;6:e23294. doi: 10.1371/journal.pone.0023294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J., Baker D. Proteins. 2011;79:2380–2388. doi: 10.1002/prot.23046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauck F., Smith C. A., Friedland G. F., Humphris E. L., Kortemme T. Nucleic Acids Res. 2010;38:W569–W575. doi: 10.1093/nar/gkq369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A., Kortemme T. PLoS One. 2011;6:e20451. doi: 10.1371/journal.pone.0020451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromley E. H. C., Channon K., Moutevelis E., Woolfson D. N. ACS Chem. Biol. 2008;3:38–50. doi: 10.1021/cb700249v. [DOI] [PubMed] [Google Scholar]
- Armstrong C. T., Boyle A. L., Bromley E. H., Mahmoud Z. N., Smith L., Thomson A. R., Woolfson D. N., Faraday Discuss., 2009, 143 , 305 –317 , , discussion 359–372 . [DOI] [PubMed] [Google Scholar]
- Bromley E. H. C., Sessions R. B., Thomson A. R., Woolfson D. N. J. Am. Chem. Soc. 2009;131:928–930. doi: 10.1021/ja804231a. [DOI] [PubMed] [Google Scholar]
- Gordon S. R., Stanley E. J., Wolf S., Toland A., Wu S. J., Hadidi D., Mills J. H., Baker D., Pultz I. S., Siegel J. B. J. Am. Chem. Soc. 2012;134:20513–20520. doi: 10.1021/ja3094795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D., Cheng J. Proteins. 2013;81:119–131. doi: 10.1002/prot.24167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey J. A., Chica R. A. Protein Sci. 2012;21:1241–1252. doi: 10.1002/pro.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanf K. J. M. Methods Mol. Biol. 2012;899:127–144. doi: 10.1007/978-1-61779-921-1_8. [DOI] [PubMed] [Google Scholar]
- Kim R., Skolnick J. J. Comput. Chem., Jpn. 2008;29:1316–1331. doi: 10.1002/jcc.20893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham P., Afzal-Ahmed I., Naftalin R. J. J. Biol. Chem. 2006;281:5797–5803. doi: 10.1074/jbc.M509422200. [DOI] [PubMed] [Google Scholar]
- Hetényi C., Maran U., García-Sosa A. T., Karelson M. Bioinformatics. 2007;23:2678–2685. doi: 10.1093/bioinformatics/btm431. [DOI] [PubMed] [Google Scholar]
- Cosconati S., Forli S., Perryman A. L., Harris R., Goodsell D. S., Olson A. J. Expert Opin. Drug Discovery. 2010;5:597–607. doi: 10.1517/17460441.2010.484460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A. J. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandeep G., Nagasree K. P., Hanisha M., Kumar M. M. BMC Res. Notes. 2011;4:445. doi: 10.1186/1756-0500-4-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoko S. D., Ouyang X., Su C. T., Kwoh C. K., Ong Y. S. IEEE/ACM Trans. Comput. Biol. Bioinf. 2012;9:1266–1272. doi: 10.1109/TCBB.2012.82. [DOI] [PubMed] [Google Scholar]
- Gao M., Skolnick J. Bioinformatics. 2013;29:597–604. doi: 10.1093/bioinformatics/btt024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repasky M. P., Murphy R. B., Banks J. L., Greenwood J. R., Tubert-Brohman I., Bhat S., Friesner R. A. J. Comput.-Aided Mol. Des. 2012;26:787–799. doi: 10.1007/s10822-012-9575-9. [DOI] [PubMed] [Google Scholar]
- Halgren T. A., Murphy R. B., Friesner R. A., Beard H. S., Frye L. L., Pollard W. T., Banks J. L. J. Med. Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- Friesner R. A., Banks J. L., Murphy R. B., Halgren T. A., Klicic J. J., Mainz D. T., Repasky M. P., Knoll E. H., Shelley M., Perry J. K., Shaw D. E., Francis P., Shenkin P. S. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Schomburg K. T., Ardao I., Gotz K., Rieckenberg F., Liese A., Zeng A. P., Rarey M. J. Biotechnol. 2012;161:391–401. doi: 10.1016/j.jbiotec.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Vass M., Tarcsay A., Keserü G. M. J. Comput.-Aided Mol. Des. 2012;26:821–834. doi: 10.1007/s10822-012-9578-6. [DOI] [PubMed] [Google Scholar]
- Greenidge P. A., Kramer C., Mozziconacci J. C., Wolf R. M. J. Chem. Inf. Model. 2012;53:201–209. doi: 10.1021/ci300425v. [DOI] [PubMed] [Google Scholar]
- Plewczynski D., Laźniewski M., Augustyniak R., Ginalski K. J. Comput. Chem., Jpn. 2011;32:742–755. doi: 10.1002/jcc.21643. [DOI] [PubMed] [Google Scholar]
- Wang R., Fang X., Lu Y., Yang C. Y., Wang S. J. Med. Chem. 2005;48:4111–4119. doi: 10.1021/jm048957q. [DOI] [PubMed] [Google Scholar]
- Yuan Y. X., Pei J. F., Lai L. H. J. Chem. Inf. Model. 2011;51:1083–1091. doi: 10.1021/ci100350u. [DOI] [PubMed] [Google Scholar]
- Sirin S., Kumar R., Martinez C., Karmilowicz M. J., Ghosh P., Abramov Y. A., Martin V., Sherman W. J. Chem. Inf. Model. 2014;54:2334–2346. doi: 10.1021/ci5002185. [DOI] [PubMed] [Google Scholar]
- Zhou H. Y., Skolnick J. J. Chem. Inf. Model. 2013;53:230–240. doi: 10.1021/ci300510n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faver J. C., Benson M. L., He X. A., Roberts B. P., Wang B., Marshall M. S., Kennedy M. R., Sherrill C. D., Merz K. M. J. Chem. Theory Comput. 2011;7:790–797. doi: 10.1021/ct100563b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith M., Tawfik D. S. Methods Enzymol. 2013;523:257–283. doi: 10.1016/B978-0-12-394292-0.00012-6. [DOI] [PubMed] [Google Scholar]
- Ruff A. J., Dennig A., Schwaneberg U. FEBS J. 2013;280:2961–2978. doi: 10.1111/febs.12325. [DOI] [PubMed] [Google Scholar]
- Zhang T., Wu Z. F., Chen H., Wu Q., Tang Z. Z., Gou J. B., Wang L. H., Hao W. W., Wang C. M., Li C. M. Afr. J. Biotechnol. 2010;9:9277–9285. [Google Scholar]
- Directed Evolution Library Creation: Methods and Protocols, ed. E. M. J. Gillam, J. N. Copp and D. Ackerley, Springer, Berlin, 2014. [Google Scholar]
- Damborsky J., Brezovsky J. Curr. Opin. Chem. Biol. 2009;13:26–34. doi: 10.1016/j.cbpa.2009.02.021. [DOI] [PubMed] [Google Scholar]
- Brezovsky J., Chovancova E., Gora A., Pavelka A., Biedermannova L., Damborsky J. Biotechnol. Adv. 2013;31:38–49. doi: 10.1016/j.biotechadv.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Sebestova E., Bendl J., Brezovsky J., Damborsky J. Methods Mol. Biol. 2014;1179:291–314. doi: 10.1007/978-1-4939-1053-3_20. [DOI] [PubMed] [Google Scholar]
- Krašovec R., Belavkin R. V., Aston J. A. D., Channon A., Aston E., Rash B. M., Kadirvel M., Forbes S., Knight C. G. Nat. Commun. 2014;5:3742. doi: 10.1038/ncomms4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates M., Corne D. and Loader R., Investigation of a characteristic bimodal convergence-time/mutation-rate feature in evolutionary search, Proc. Congr. Evol. Comput., IEEE, 1999, pp. 2175–2182. [Google Scholar]
- Oates M. and Corne D. W., Overcoming fitness barriers in multi-modal search spaces, in Foundation of Genetic Algorithms 6, ed. W. N. Martin and W. M. Spears, Academic Press, London, 2001, pp. 5–26. [Google Scholar]
- Oates M. J., Corne D. and Loader R., Tri-phase performance profile of evolutionary search on uni- and multi-modal search spaces, in Proc. IEEE Congr. Evol. Computation, IEEE Neural Networks Council, San Diego, 2000, pp. 357–364. [Google Scholar]
- Oates M. J., Corne D. W. and Kell D. B., The bimodal feature at large population sizes and high selection pressure: implications for directed evolution, in Recent advances in simulated evolution and learning, ed. K. C. Tan, M. H. Lim, X. Yao and L. Wang, World Scientific, Singapore, 2003, pp. 215–240. [Google Scholar]
- Zaccolo M., Gherardi E. J. Mol. Biol. 1999;285:775–783. doi: 10.1006/jmbi.1998.2262. [DOI] [PubMed] [Google Scholar]
- Daugherty P. S., Chen G., Iverson B. L., Georgiou G. Proc. Natl. Acad. Sci. U. S. A. 2000;97:2029–2034. doi: 10.1073/pnas.030527597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond D. A., Iverson B. L., Georgiou G., Arnold F. H. J. Mol. Biol. 2005;350:806–816. doi: 10.1016/j.jmb.2005.05.023. [DOI] [PubMed] [Google Scholar]
- Leconte A. M., Dickinson B. C., Yang D. D., Chen I. A., Allen B., Liu D. R. Biochemistry. 2013;52:1490–1499. doi: 10.1021/bi3016185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D. W., Chen E., Goeddel D. V. Technique. 1989;1:11–15. [Google Scholar]
- McCullum E. O., Williams B. A., Zhang J., Chaput J. C. Methods Mol. Biol. 2010;634:103–109. doi: 10.1007/978-1-60761-652-8_7. [DOI] [PubMed] [Google Scholar]
- Wong T. S., Roccatano D., Zacharias M., Schwaneberg U. J. Mol. Biol. 2006;355:858–871. doi: 10.1016/j.jmb.2005.10.082. [DOI] [PubMed] [Google Scholar]
- Wong T. S., Roccatano D., Schwaneberg U. Biocatal. Biotransform. 2007;25:229–241. [Google Scholar]
- Rasila T. S., Pajunen M. I., Savilahti H. Anal. Biochem. 2009;388:71–80. doi: 10.1016/j.ab.2009.02.008. [DOI] [PubMed] [Google Scholar]
- Zhao J., Kardashliev T., Joelle Ruff A., Bocola M., Schwaneberg U. Biotechnol. Bioeng. 2014;111:2380–2389. doi: 10.1002/bit.25302. [DOI] [PubMed] [Google Scholar]
- Cadwell R. C., Joyce G. F. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- Camps M., Naukkarinen J., Johnson B. P., Loeb L. A. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9727–9732. doi: 10.1073/pnas.1333928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp J. N., Hanson-Manful P., Ackerley D. F., Patrick W. M. Methods Mol. Biol. 2014;1179:3–22. doi: 10.1007/978-1-4939-1053-3_1. [DOI] [PubMed] [Google Scholar]
- Matsumura I., Rowe L. A. Biomol. Eng. 2005;22:73–79. doi: 10.1016/j.bioeng.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Alexander D. L., Lilly J., Hernandez J., Romsdahl J., Troll C. J., Camps M. Methods Mol. Biol. 2014;1179:31–44. doi: 10.1007/978-1-4939-1053-3_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii R., Kitaoka M., Hayashi K. Nucleic Acids Res. 2004;32:e145. doi: 10.1093/nar/gnh147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii R., Kitaoka M., Hayashi K. Nucleic Acids Res. 2006;34:e30. doi: 10.1093/nar/gnj032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipnis Y., Dellus-Gur E., Tawfik D. S. Protein Eng., Des. Sel. 2012;25:437–444. doi: 10.1093/protein/gzs023. [DOI] [PubMed] [Google Scholar]
- Fujii R., Kitaoka M., Hayashi K. Methods Mol. Biol. 2014;1179:151–158. doi: 10.1007/978-1-4939-1053-3_10. [DOI] [PubMed] [Google Scholar]
- Ravikumar A., Arrieta A., Liu C. C. Nat. Chem. Biol. 2014;10:175–177. doi: 10.1038/nchembio.1439. [DOI] [PubMed] [Google Scholar]
- Pritchard L., Corne D. W., Kell D. B., Rowland J. J., Winson M. K. J. Theor. Biol. 2004;234:497–509. doi: 10.1016/j.jtbi.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Hoebenreich S., Zilly F. E., Acevedo-Rocha C. G., Zilly M., Reetz M. T. ACS Synth. Biol. 2014 doi: 10.1021/sb5002399. [DOI] [PubMed] [Google Scholar]
- Wei D., Li M., Zhang X., Xing L. Anal. Biochem. 2004;331:401–403. doi: 10.1016/j.ab.2004.04.019. [DOI] [PubMed] [Google Scholar]
- Steffens D. L., Williams J. G. J. Biomol. Tech. 2007;18:147–149. [PMC free article] [PubMed] [Google Scholar]
- Fersht A., Enzyme structure and mechanism, W.H. Freeman, San Francisco, 2nd edn, 1977. [Google Scholar]
- Braman J., Papworth C., Greener A. Methods Mol. Biol. 1996;57:31–44. doi: 10.1385/0-89603-332-5:31. [DOI] [PubMed] [Google Scholar]
- Papworth C., Bauer J. C., Braman J., Wright D. A. Strategies. 1996;9:3–4. [Google Scholar]
- Zheng L., Baumann U., Reymond J. L. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchis J., Fernandez L., Carballeira J. D., Drone J., Gumulya Y., Hobenreich H., Kahakeaw D., Kille S., Lohmer R., Peyralans J. J., Podtetenieff J., Prasad S., Soni P., Taglieber A., Wu S., Zilly F. E., Reetz M. T. Appl. Microbiol. Biotechnol. 2008;81:387–397. doi: 10.1007/s00253-008-1678-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison K. L., Weiss G. A. Curr. Opin. Chem. Biol. 2001;5:302–307. doi: 10.1016/s1367-5931(00)00206-4. [DOI] [PubMed] [Google Scholar]
- Weiss G. A., Watanabe C. K., Zhong A., Goddard A., Sidhu S. S. Proc. Natl. Acad. Sci. U. S. A. 2000;97:8950–8954. doi: 10.1073/pnas.160252097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong T. S., Roccatano D., Schwaneberg U. Environ. Microbiol. 2007;9:2645–2659. doi: 10.1111/j.1462-2920.2007.01411.x. [DOI] [PubMed] [Google Scholar]
- Reetz M. T., Wang L. W., Bocola M. Angew. Chem., Int. Ed. 2006;45:1236–1241. doi: 10.1002/anie.200502746. [DOI] [PubMed] [Google Scholar]
- Kirsch R. D., Joly E. Nucleic Acids Res. 1998;26:1848–1850. doi: 10.1093/nar/26.7.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang L. W., Kovari I., Howe M. M. PCR Methods Appl. 1993;2:210–217. doi: 10.1101/gr.2.3.210. [DOI] [PubMed] [Google Scholar]
- Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Horton R. M., Hunt H. D., Ho S. N., Pullen J. K., Pease L. R. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Peng R. H., Xiong A. S., Yao Q. H. Appl. Microbiol. Biotechnol. 2006;73:234–240. doi: 10.1007/s00253-006-0583-3. [DOI] [PubMed] [Google Scholar]
- Heckman K. L., Pease L. R. Nat. Protoc. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- Williams E. M., Copp J. N., Ackerley D. F. Methods Mol. Biol. 2014;1179:83–101. doi: 10.1007/978-1-4939-1053-3_6. [DOI] [PubMed] [Google Scholar]
- Wong T. S., Tee K. L., Hauer B., Schwaneberg U. Nucleic Acids Res. 2004;32:e26. doi: 10.1093/nar/gnh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong T. S., Roccatano D., Loakes D., Tee K. L., Schenk A., Hauer B., Schwaneberg U. Biotechnol. J. 2008;3:74–82. doi: 10.1002/biot.200700193. [DOI] [PubMed] [Google Scholar]
- Mundhada H., Marienhagen J., Scacioc A., Schenk A., Roccatano D., Schwaneberg U. ChemBioChem. 2011;12:1595–1601. doi: 10.1002/cbic.201100010. [DOI] [PubMed] [Google Scholar]
- Ruff A. J., Kardashliev T., Dennig A., Schwaneberg U. Methods Mol. Biol. 2014;1179:45–68. doi: 10.1007/978-1-4939-1053-3_4. [DOI] [PubMed] [Google Scholar]
- Dennig A., Shivange A. V., Marienhagen J., Schwaneberg U. PLoS One. 2011;6:e26222. doi: 10.1371/journal.pone.0026222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennig A., Marienhagen J., Ruff A. J., Schwaneberg U. Methods Mol. Biol. 2014;1179:139–149. doi: 10.1007/978-1-4939-1053-3_9. [DOI] [PubMed] [Google Scholar]
- Shivange A. V., Dennig A., Schwaneberg U. J. Biotechnol. 2014;170:68–72. doi: 10.1016/j.jbiotec.2013.11.014. [DOI] [PubMed] [Google Scholar]
- Acevedo-Rocha C. G., Hoebenreich S., Reetz M. T. Methods Mol. Biol. 2014;1179:103–128. doi: 10.1007/978-1-4939-1053-3_7. [DOI] [PubMed] [Google Scholar]
- Parra L. P., Agudo R., Reetz M. T. ChemBioChem. 2013;14:2301–2309. doi: 10.1002/cbic.201300486. [DOI] [PubMed] [Google Scholar]
- Reetz M. T., Carballeira J. D., Vogel A. Angew. Chem., Int. Ed. 2006;45:7745–7751. doi: 10.1002/anie.200602795. [DOI] [PubMed] [Google Scholar]
- Reetz M. T., Carballeira J. D. Nat. Protoc. 2007;2:891–903. doi: 10.1038/nprot.2007.72. [DOI] [PubMed] [Google Scholar]
- Reetz M. T., Kahakeaw D., Sanchis J. Mol. BioSyst. 2009;5:115–122. doi: 10.1039/b814862g. [DOI] [PubMed] [Google Scholar]
- Reetz M. T., Soni P., Fernandez L., Gumulya Y., Carballeira J. D. Chem. Commun. 2010;46:8657–8658. doi: 10.1039/c0cc02657c. [DOI] [PubMed] [Google Scholar]
- Reetz M. T., Prasad S., Carballeira J. D., Gumulya Y., Bocola M. J. Am. Chem. Soc. 2010;132:9144–9152. doi: 10.1021/ja1030479. [DOI] [PubMed] [Google Scholar]
- Zheng H., Reetz M. T. J. Am. Chem. Soc. 2010;132:15744–15751. doi: 10.1021/ja1067542. [DOI] [PubMed] [Google Scholar]
- Baldwin A. J., Busse K., Simm A. M., Jones D. D. Nucleic Acids Res. 2008;36:e77. doi: 10.1093/nar/gkn358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin A. J., Arpino J. A., Edwards W. R., Tippmann E. M., Jones D. D. Mol. BioSyst. 2009;5:764–766. doi: 10.1039/b904031e. [DOI] [PubMed] [Google Scholar]
- Edwards W. R., Busse K., Allemann R. K., Jones D. D. Nucleic Acids Res. 2008;36:e78. doi: 10.1093/nar/gkn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. D., Arpino J. A. J., Baldwin A. J., Edmundson M. C. Methods Mol. Biol. 2014;1179:159–172. doi: 10.1007/978-1-4939-1053-3_11. [DOI] [PubMed] [Google Scholar]
- Liu H., Naismith J. H. BMC Biotechnol. 2008;8:91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfang A., Jin J. H. Anal. Biochem. 2004;324:285–291. doi: 10.1016/j.ab.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Fushan A. A., Drayna D. T. BMC Biotechnol. 2009;9:83. doi: 10.1186/1472-6750-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegler-Ebo D. M., Docktor C. M., DiMaio D. Nucleic Acids Res. 1994;22:1593–1599. doi: 10.1093/nar/22.9.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner K., Schwab H. Comput. Struct. Biotechnol. J. 2012;2:e201209010. doi: 10.5936/csbj.201209010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nov Y. PLoS One. 2013;8:e68069. doi: 10.1371/journal.pone.0068069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomenclature Committee of the International Union of Biochemistry (NC-IUB) Eur. J. Biochem. 1985;150:1–5. doi: 10.1111/j.1432-1033.1985.tb08977.x. [DOI] [PubMed] [Google Scholar]
- Mena M. A., Daugherty P. S. Protein Eng., Des. Sel. 2005;18:559–561. doi: 10.1093/protein/gzi061. [DOI] [PubMed] [Google Scholar]
- Tang L., Wang X., Ru B., Sun H., Huang J., Gao H. BioTechniques. 2014;56:301–310. doi: 10.2144/000114177. [DOI] [PubMed] [Google Scholar]
- Muñoz E., Deem M. W. Protein Eng., Des. Sel. 2008;21:311–317. doi: 10.1093/protein/gzn007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee K. L., Wong T. S. Biotechnol. Adv. 2013;31:1707–1721. doi: 10.1016/j.biotechadv.2013.08.021. [DOI] [PubMed] [Google Scholar]
- Kille S., Acevedo-Rocha C. G., Parra L. P., Zhang Z. G., Opperman D. J., Reetz M. T., Acevedo J. P. ACS Synth. Biol. 2013;2:83–92. doi: 10.1021/sb300037w. [DOI] [PubMed] [Google Scholar]
- Hughes M. D., Nagel D. A., Santos A. F., Sutherland A. J., Hine A. V. J. Mol. Biol. 2003;331:973–979. doi: 10.1016/s0022-2836(03)00833-7. [DOI] [PubMed] [Google Scholar]
- Tang L. X., Gao H., Zhu X. C., Wang X., Zhou M., Jiang R. X. BioTechniques. 2012;52:149–157. doi: 10.2144/000113820. [DOI] [PubMed] [Google Scholar]
- Akanuma S., Kigawa T., Yokoyama S. Proc. Natl. Acad. Sci. U. S. A. 2002;99:13549–13553. doi: 10.1073/pnas.222243999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley L. H., Kleiner R. E., Wang A. F., Hecht M. H., Wood D. W. Protein Eng., Des. Sel. 2005;18:201–207. doi: 10.1093/protein/gzi020. [DOI] [PubMed] [Google Scholar]
- Chaparro-Riggers J. F., Polizzi K. M., Bommarius A. S. Biotechnol. J. 2007;2:180–191. doi: 10.1002/biot.200600170. [DOI] [PubMed] [Google Scholar]
- Tanaka J., Yanagawa H., Doi N. PLoS One. 2011;6:e18034. doi: 10.1371/journal.pone.0018034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandström A. G., Wikmark Y., Engström K., Nyhlén J., Bäckvall J. E. Proc. Natl. Acad. Sci. U. S. A. 2012;109:78–83. doi: 10.1073/pnas.1111537108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacardit J., Stout M., Hirst J. D., Valencia A., Smith R. E., Krasnogor N. BMC Bioinf. 2009;10:6. doi: 10.1186/1471-2105-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Kwon I. Biotechnol. J. 2012;7:47–60. doi: 10.1002/biot.201100267. [DOI] [PubMed] [Google Scholar]
- Hoesl M. G., Budisa N. Angew. Chem., Int. Ed. 2011;50:2896–2902. doi: 10.1002/anie.201005680. [DOI] [PubMed] [Google Scholar]
- Wang L., Brock A., Herberich B., Schultz P. G. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- Wang Q., Parrish A. R., Wang L. Chem. Biol. 2009;16:323–336. doi: 10.1016/j.chembiol.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J. C., Duffy S. P., Hess K. R., Mehl R. A. J. Am. Chem. Soc. 2006;128:11124–11127. doi: 10.1021/ja061099y. [DOI] [PubMed] [Google Scholar]
- Chin J. W., Cropp T. A., Anderson J. C., Mukherji M., Zhang Z., Schultz P. G. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- Anderson J. C., Wu N., Santoro S. W., Lakshman V., King D. S., Schultz P. G. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7566–7571. doi: 10.1073/pnas.0401517101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H., Wang K., Davis L., Garcia-Alai M., Chin J. W. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- Chin J. W. Science. 2012;336:428–429. doi: 10.1126/science.1221761. [DOI] [PubMed] [Google Scholar]
- Davis L., Chin J. W. Nat. Rev. Mol. Cell Biol. 2012;13:168–182. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- Wang K., Schmied W. H., Chin J. W. Angew. Chem., Int. Ed. 2012;51:2288–2297. doi: 10.1002/anie.201105016. [DOI] [PubMed] [Google Scholar]
- Liu C. C., Schultz P. G. Annu. Rev. Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- Liu C. C., Mack A. V., Tsao M. L., Mills J. H., Lee H. S., Choe H., Farzan M., Schultz P. G., Smider V. V. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17688–17693. doi: 10.1073/pnas.0809543105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Schultz P. G. Nat. Rev. Mol. Cell Biol. 2006;7:775–782. doi: 10.1038/nrm2005. [DOI] [PubMed] [Google Scholar]
- Wang L., Xie J., Schultz P. G. Annu. Rev. Biophys. Biomol. Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- Bradley K. M., Benner S. A. Beilstein J. Org. Chem. 2014;10:1826–1833. doi: 10.3762/bjoc.10.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Schultz P. G. Curr. Opin. Chem. Biol. 2005;9:548–554. doi: 10.1016/j.cbpa.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Stemmer W. P. C. Nature. 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- Stemmer W. P. C. Proc. Natl. Acad. Sci. U. S. A. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Arnold F. H. Nucleic Acids Res. 1997;25:1307–1308. doi: 10.1093/nar/25.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coco W. M., Levinson W. E., Crist M. J., Hektor H. J., Darzins A., Pienkos P. T., Squires C. H., Monticello D. J. Nat. Biotechnol. 2001;19:354–359. doi: 10.1038/86744. [DOI] [PubMed] [Google Scholar]
- Coco W. M., Encell L. P., Levinson W. E., Crist M. J., Loomis A. K., Licato L. L., Arensdorf J. J., Sica N., Pienkos P. T., Monticello D. J. Nat. Biotechnol. 2002;20:1246–1250. doi: 10.1038/nbt757. [DOI] [PubMed] [Google Scholar]
- Kikuchi M., Ohnishi K., Harayama S. Gene. 1999;236:159–167. doi: 10.1016/s0378-1119(99)00240-1. [DOI] [PubMed] [Google Scholar]
- Kikuchi M., Ohnishi K., Harayama S. Gene. 2000;243:133–137. doi: 10.1016/s0378-1119(99)00547-8. [DOI] [PubMed] [Google Scholar]
- Miyazaki K. Nucleic Acids Res. 2002;30:e139. doi: 10.1093/nar/gnf139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Y., Wu W., Lv A. Biochimie. 2010;92:1081–1084. doi: 10.1016/j.biochi.2010.03.016. [DOI] [PubMed] [Google Scholar]
- Crameri A., Raillard S. A., Bermudez E., Stemmer W. P. C. Nature. 1998;391:288–291. doi: 10.1038/34663. [DOI] [PubMed] [Google Scholar]
- Behrendorff J. B. Y. H., Johnston W. A., Gillam E. M. J. Methods Mol. Biol. 2014;1179:175–187. doi: 10.1007/978-1-4939-1053-3_12. [DOI] [PubMed] [Google Scholar]
- Behrendorff J. B. Y. H., Johnston W. A., Gillam E. M. J. Methods Mol. Biol. 2013;987:177–188. doi: 10.1007/978-1-62703-321-3_16. [DOI] [PubMed] [Google Scholar]
- Rosic N. N., Huang W., Johnston W. A., DeVoss J. J., Gillam E. M. J. Gene. 2007;395:40–48. doi: 10.1016/j.gene.2007.01.031. [DOI] [PubMed] [Google Scholar]
- Joern J. M., Meinhold P., Arnold F. H. J. Mol. Biol. 2002;316:643–656. doi: 10.1006/jmbi.2001.5349. [DOI] [PubMed] [Google Scholar]
- Chaparro-Riggers J. F., Loo B. L., Polizzi K. M., Gibbs P. R., Tang X. S., Nelson M. J., Bommarius A. S. BMC Biotechnol. 2007;7:77. doi: 10.1186/1472-6750-7-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawarasaki Y., Griswold K. E., Stevenson J. D., Selzer T., Benkovic S. J., Iverson B. L., Georgiou G. Nucleic Acids Res. 2003;31:e126. doi: 10.1093/nar/gng126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz S., Ostermeier M. Methods Mol. Biol. 2003;231:143–151. doi: 10.1385/1-59259-395-X:143. [DOI] [PubMed] [Google Scholar]
- Ostermeier M., Lutz S. Methods Mol. Biol. 2003;231:129–141. doi: 10.1385/1-59259-395-X:129. [DOI] [PubMed] [Google Scholar]
- Patrick W. M., Gerth M. L. Methods Mol. Biol. 2014;1179:225–244. doi: 10.1007/978-1-4939-1053-3_16. [DOI] [PubMed] [Google Scholar]
- Coco W. M. Methods Mol. Biol. 2003;231:111–127. doi: 10.1385/1-59259-395-X:111. [DOI] [PubMed] [Google Scholar]
- Lee S. H., Ryu E. J., Kang M. J., Wang E. S., Piao Z., Choi Y. J., Jung K. H., Jeon J. Y. J., Shin Y. C. J. Mol. Catal. 2003;26:119–129. [Google Scholar]
- Hidalgo A., Schliessmann A., Molina R., Hermoso J., Bornscheuer U. T. Protein Eng., Des. Sel. 2008;21:567–576. doi: 10.1093/protein/gzn034. [DOI] [PubMed] [Google Scholar]
- Hidalgo A., Schliessmann A., Bornscheuer U. T. Methods Mol. Biol. 2014;1179:207–212. doi: 10.1007/978-1-4939-1053-3_14. [DOI] [PubMed] [Google Scholar]
- Kashiwagi K., Isogai Y., Nishiguchi K., Shiba K. Protein Eng., Des. Sel. 2006;19:135–140. doi: 10.1093/protein/gzj008. [DOI] [PubMed] [Google Scholar]
- Ness J. E., Kim S., Gottman A., Pak R., Krebber A., Borchert T. V., Govindarajan S., Mundorff E. C., Minshull J. Nat. Biotechnol. 2002;20:1251–1255. doi: 10.1038/nbt754. [DOI] [PubMed] [Google Scholar]
- Bergquist P. L., Reeves R. A., Gibbs M. D. Biomol. Eng. 2005;22:63–72. doi: 10.1016/j.bioeng.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Villiers B. R., Stein V., Hollfelder F. Protein Eng., Des. Sel. 2010;23:1–8. doi: 10.1093/protein/gzp063. [DOI] [PubMed] [Google Scholar]
- Villiers B., Hollfelder F. Methods Mol. Biol. 2014;1179:213–224. doi: 10.1007/978-1-4939-1053-3_15. [DOI] [PubMed] [Google Scholar]
- O'Maille P. E., Bakhtina M., Tsai M. D. J. Mol. Biol. 2002;321:677–691. doi: 10.1016/s0022-2836(02)00675-7. [DOI] [PubMed] [Google Scholar]
- O'Maille P. E., Tsai M. D., Greenhagen B. T., Chappell J., Noel J. P. Methods Enzymol. 2004;388:75–91. doi: 10.1016/S0076-6879(04)88008-X. [DOI] [PubMed] [Google Scholar]
- Dokarry M., Laurendon C., O'Maille P. E. Methods Enzymol. 2012;515:21–42. doi: 10.1016/B978-0-12-394290-6.00002-1. [DOI] [PubMed] [Google Scholar]
- Herman A., Tawfik D. S. Protein Eng., Des. Sel. 2007;20:219–226. doi: 10.1093/protein/gzm014. [DOI] [PubMed] [Google Scholar]
- Rockah-Shmuel L., Tawfik D. S., Goldsmith M. Methods Mol. Biol. 2014;1179:129–137. doi: 10.1007/978-1-4939-1053-3_8. [DOI] [PubMed] [Google Scholar]
- Hughes M. D., Zhang Z. R., Sutherland A. J., Santos A. F., Hine A. V. Nucleic Acids Res. 2005;33:e32. doi: 10.1093/nar/gni031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z., Zhao H., Zhao H. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z., Luo Y., Zhao H. Mol. BioSyst. 2011;7:1056–1059. doi: 10.1039/c0mb00338g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. Z., Elledge S. J. Nat. Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- Mosberg J. A., Gregg C. J., Lajoie M. J., Wang H. H., Church G. M. PLoS One. 2012;7:e44638. doi: 10.1371/journal.pone.0044638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordwald E. M., Garst A., Gill R. T., Kaar J. L. Curr. Opin. Biotechnol. 2013;24:1017–1022. doi: 10.1016/j.copbio.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Qian Z., Lutz S. J. Am. Chem. Soc. 2005;127:13466–13467. doi: 10.1021/ja053932h. [DOI] [PubMed] [Google Scholar]
- Qian Z., Fields C. J., Lutz S. ChemBioChem. 2007;8:1989–1996. doi: 10.1002/cbic.200700373. [DOI] [PubMed] [Google Scholar]
- Lutz S., Daugherty A. B., Yu Y., Qian Z. Methods Mol. Biol. 2014;1179:245–258. doi: 10.1007/978-1-4939-1053-3_17. [DOI] [PubMed] [Google Scholar]
- Fischereder E., Pressnitz D., Kroutil W., Lutz S. Bioorg. Med. Chem. 2014;22:5633–5637. doi: 10.1016/j.bmc.2014.06.023. [DOI] [PubMed] [Google Scholar]
- Daugherty A. B., Govindarajan S., Lutz S. J. Am. Chem. Soc. 2013;135:14425–14432. doi: 10.1021/ja4074886. [DOI] [PubMed] [Google Scholar]
- Yu Y., Lutz S. Trends Biotechnol. 2011;29:18–25. doi: 10.1016/j.tibtech.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Haglund E., Lindberg M. O., Oliveberg M. J. Biol. Chem. 2008;283:27904–27915. doi: 10.1074/jbc.M801776200. [DOI] [PubMed] [Google Scholar]
- Lindberg M., Tangrot J., Oliveberg M. Nat. Struct. Biol. 2002;9:818–822. doi: 10.1038/nsb847. [DOI] [PubMed] [Google Scholar]
- Smith M. A., Romero P. A., Wu T., Brustad E. M., Arnold F. H. Protein Sci. 2013;22:231–238. doi: 10.1002/pro.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudeau D. L., Smith M. A., Arnold F. H. Curr. Opin. Chem. Biol. 2013;17:902–909. doi: 10.1016/j.cbpa.2013.10.007. [DOI] [PubMed] [Google Scholar]
- Saraf M. C., Gupta A., Maranas C. D. Proteins. 2005;60:769–777. doi: 10.1002/prot.20490. [DOI] [PubMed] [Google Scholar]
- Pantazes R. J., Saraf M. C., Maranas C. D. Protein Eng., Des. Sel. 2007;20:361–373. doi: 10.1093/protein/gzm030. [DOI] [PubMed] [Google Scholar]
- Endelman J. B., Silberg J. J., Wang Z. G., Arnold F. H. Protein Eng., Des. Sel. 2004;17:589–594. doi: 10.1093/protein/gzh067. [DOI] [PubMed] [Google Scholar]
- Voigt C. A., Martinez C., Wang Z. G., Mayo S. L., Arnold F. H. Nat. Struct. Biol. 2002;9:553–558. doi: 10.1038/nsb805. [DOI] [PubMed] [Google Scholar]
- Otey C. R., Silberg J. J., Voigt C. A., Endelman J. B., Bandara G., Arnold F. H. Chem. Biol. 2004;11:309–318. doi: 10.1016/j.chembiol.2004.02.018. [DOI] [PubMed] [Google Scholar]
- Heinzelman P., Snow C. D., Smith M. A., Yu X., Kannan A., Boulware K., Villalobos A., Govindarajan S., Minshull J., Arnold F. H. J. Biol. Chem. 2009;284:26229–26233. doi: 10.1074/jbc.C109.034058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzelman P., Komor R., Kanaan A., Romero P., Yu X., Mohler S., Snow C., Arnold F. Protein Eng., Des. Sel. 2010;23:871–880. doi: 10.1093/protein/gzq063. [DOI] [PubMed] [Google Scholar]
- Heinzelman P., Romero P. A., Arnold F. H. Methods Enzymol. 2013;523:351–368. doi: 10.1016/B978-0-12-394292-0.00016-3. [DOI] [PubMed] [Google Scholar]
- Meyer M. M., Silberg J. J., Voigt C. A., Endelman J. B., Mayo S. L., Wang Z. G., Arnold F. H. Protein Sci. 2003;12:1686–1693. doi: 10.1110/ps.0306603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M. M., Hochrein L., Arnold F. H. Protein Eng., Des. Sel. 2006;19:563–570. doi: 10.1093/protein/gzl045. [DOI] [PubMed] [Google Scholar]
- Romero P. A., Stone E., Lamb C., Chantranupong L., Krause A., Miklos A. E., Hughes R. A., Fechtel B., Ellington A. D., Arnold F. H., Georgiou G. ACS Synth. Biol. 2012;1:221–228. doi: 10.1021/sb300014t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberg J. J., Endelman J. B., Arnold F. H. Methods Enzymol. 2004;388:35–42. doi: 10.1016/S0076-6879(04)88004-2. [DOI] [PubMed] [Google Scholar]
- Smith M. A., Arnold F. H. Methods Mol. Biol. 2014;1179:335–343. doi: 10.1007/978-1-4939-1053-3_22. [DOI] [PubMed] [Google Scholar]
- Smith M. A., Arnold F. H. Methods Mol. Biol. 2014;1179:345–352. doi: 10.1007/978-1-4939-1053-3_23. [DOI] [PubMed] [Google Scholar]
- Parker A. S., Griswold K. E., Bailey-Kellogg C., Research in Computational Molecular Biology, 2011, 6577 , 321 –335 , , 580 . [Google Scholar]
- Zhao H., Blazanovic K., Choi Y., Bailey-Kellogg C., Griswold K. E. Appl. Environ. Microbiol. 2014;80:2746–2753. doi: 10.1128/AEM.03914-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Friedman A. M., Bailey-Kellogg C. BMC Bioinf. 2012;13(suppl 3):S3. doi: 10.1186/1471-2105-13-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Friedman A. M., Bailey-Kellogg C. J. Comput. Biol. 2009;16:1151–1168. doi: 10.1089/cmb.2009.0090. [DOI] [PubMed] [Google Scholar]
- Ye X., Friedman A. M., Bailey-Kellogg C. Comput. Syst. Bioinf., CSB2007 Conf. Proc., 6th. 2008;7:99–108. [PubMed] [Google Scholar]
- Zheng W., Ye X., Friedman A. M., Bailey-Kellogg C. Comput. Syst. Bioinf., CSB2007 Conf. Proc., 6th. 2007;6:31–40. [PubMed] [Google Scholar]
- Ye X., Friedman A. M., Bailey-Kellogg C. J. Comput. Biol. 2007;14:777–790. doi: 10.1089/cmb.2007.R016. [DOI] [PubMed] [Google Scholar]
- Saftalov L., Smith P. A., Friedman A. M., Bailey-Kellogg C. Proteins. 2006;64:629–642. doi: 10.1002/prot.20984. [DOI] [PubMed] [Google Scholar]
- Li Y., Drummond D. A., Sawayama A. M., Snow C. D., Bloom J. D., Arnold F. H. Nat. Biotechnol. 2007;25:1051–1056. doi: 10.1038/nbt1333. [DOI] [PubMed] [Google Scholar]
- Lipovsek D., Plückthun A. J. Immunol. Methods. 2004;290:51–67. doi: 10.1016/j.jim.2004.04.008. [DOI] [PubMed] [Google Scholar]
- He M. Y. New Biotechnol. 2008;25:126–132. doi: 10.1016/j.nbt.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Okano T., Matsuura T., Suzuki H., Yomo T. ACS Synth. Biol. 2014;3:347–352. doi: 10.1021/sb400087e. [DOI] [PubMed] [Google Scholar]
- Nishikawa T., Sunami T., Matsuura T., Yomo T. J. Nucleic Acids. 2012;2012:923214. doi: 10.1155/2012/923214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano T., Matsuura T., Kazuta Y., Suzuki H., Yomo T. Lab Chip. 2012;12:2704–2711. doi: 10.1039/c2lc40098g. [DOI] [PubMed] [Google Scholar]
- Nishimura K., Matsuura T., Sunami T., Suzuki H., Yomo T. Langmuir. 2012;28:8426–8432. doi: 10.1021/la3001703. [DOI] [PubMed] [Google Scholar]
- Yanagida H., Matsuura T., Yomo T. Methods Mol. Biol. 2010;634:257–267. doi: 10.1007/978-1-60761-652-8_19. [DOI] [PubMed] [Google Scholar]
- Smith M. T., Wilding K. M., Hunt J. M., Bennett A. M., Bundy B. C. FEBS Lett. 2014;588:2755–2761. doi: 10.1016/j.febslet.2014.05.062. [DOI] [PubMed] [Google Scholar]
- Caruthers M. H., Barone A. D., Beaucage S. L., Dodds D. R., Fisher E. F., McBride L. J., Matteucci M., Stabinsky Z., Tang J. Y. Methods Enzymol. 1987;154:287–313. doi: 10.1016/0076-6879(87)54081-2. [DOI] [PubMed] [Google Scholar]
- Tian J. D., Ma K. S., Saaem I. Mol. BioSyst. 2009;5:714–722. doi: 10.1039/b822268c. [DOI] [PubMed] [Google Scholar]
- LeProust E. M., Peck B. J., Spirin K., McCuen H. B., Moore B., Namsaraev E., Caruthers M. H. Nucleic Acids Res. 2010;38:2522–2540. doi: 10.1093/nar/gkq163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond K. E., Li M. H., Rodesch M. J., Patel M., Lowe A. M., Kim C., Chu L. L., Venkataramaian N., Flickinger S. F., Kaysen J., Belshaw P. J., Sussman M. R., Cerrina F. Nucleic Acids Res. 2004;32:5011–5018. doi: 10.1093/nar/gkh793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovkov A. Y., Loskutov A. V., Robida M. D., Day K. M., Cano J. A., Le Olson T., Patel H., Brown K., Hunter P. D., Sykes K. F. Nucleic Acids Res. 2010;38:e180. doi: 10.1093/nar/gkq677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzas M., Stähler P. F., Kefer N., Siebelt N., Boisguérin V., Leonard J. T., Keller A., Stähler C. F., Häberle P., Gharizadeh B., Babrzadeh F., Church G. M. Nat. Biotechnol. 2010;28:1291–1294. doi: 10.1038/nbt.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Han H., Ahn J., Lee J., Cho N., Jang H., Kim H., Kwon S., Bang D. Nucleic Acids Res. 2012;40:e140. doi: 10.1093/nar/gks546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Elowitz M. B., Smolke C. D., Voigt C. A., Weiss R. Nat. Rev. Mol. Cell Biol. 2014;15:289–294. doi: 10.1038/nrm3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi I., Soramoto S., Ueno S., Husimi Y. BMC Biotechnol. 2004;4:19. doi: 10.1186/1472-6750-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Luo Y., Zhao H. Wiley Interdiscip. Rev.: Syst. Biol. Med. 2011;3:7–20. doi: 10.1002/wsbm.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch S. A., Gill R. T. Metab. Eng. 2011;14:205–211. doi: 10.1016/j.ymben.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Foo J. L., Ching C. B., Chang M. W., Leong S. S. Biotechnol. Adv. 2012;30:541–549. doi: 10.1016/j.biotechadv.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Watkins D. W., Armstrong C. T., Anderson J. L. Curr. Opin. Chem. Biol. 2014;19:90–98. doi: 10.1016/j.cbpa.2014.01.016. [DOI] [PubMed] [Google Scholar]
- Ma S., Tang N., Tian J. Curr. Opin. Chem. Biol. 2012;16:260–267. doi: 10.1016/j.cbpa.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer W. P. C., Crameri A., Ha K. D., Brennan T. M., Heyneker H. L. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- Xiong A. S., Yao Q. H., Peng R. H., Li X., Fan H. Q., Cheng Z. M., Li Y. Nucleic Acids Res. 2004;32:e98. doi: 10.1093/nar/gnh094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Hutchison, 3rd C. A., Pfannkoch C., Venter J. C. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15440–15445. doi: 10.1073/pnas.2237126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G. H., Wang S. Q., Wei H. L., Ping J., Liu J., Xu L. M., Zhang W. W. Biotechnol. Lett. 2012;34:721–728. doi: 10.1007/s10529-011-0832-0. [DOI] [PubMed] [Google Scholar]
- de Kok S., Stanton L. H., Slaby T., Durot M., Holmes V. F., Patel K. G., Platt D., Shapland E. B., Serber Z., Dean J., Newman J. D., Chandran S. S. ACS Synth. Biol. 2014;3:97–106. doi: 10.1021/sb4001992. [DOI] [PubMed] [Google Scholar]
- Roth T. L., Milenkovic L., Scott M. P. PLoS One. 2014;9:e107329. doi: 10.1371/journal.pone.0107329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J., Nieuwenhuijsen B. W., Kaftan E. J., Kennedy J. D., Chanda P. K. J. Biochem. Biophys. Methods. 2008;70:820–822. doi: 10.1016/j.jprot.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Xiong A. S., Peng R. H., Zhuang J., Gao F., Li Y., Cheng Z. M., Yao Q. H. FEMS Microbiol. Rev. 2008;32:522–540. doi: 10.1111/j.1574-6976.2008.00109.x. [DOI] [PubMed] [Google Scholar]
- Binkowski B. F., Richmond K. E., Kaysen J., Sussman M. R., Belshaw P. J. Nucleic Acids Res. 2005;33:e55. doi: 10.1093/nar/gni053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong A. S., Yao Q. H., Peng R. H., Duan H., Li X., Fan H. Q., Cheng Z. M., Li Y. Nat. Protoc. 2006;1:791–797. doi: 10.1038/nprot.2006.103. [DOI] [PubMed] [Google Scholar]
- Carr P. A., Park J. S., Lee Y. J., Yu T., Zhang S., Jacobson J. M. Nucleic Acids Res. 2004;32:e162. doi: 10.1093/nar/gnh160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann M., Oertel W., Berthold P., Hegemann P. Nucleic Acids Res. 2005;33:e58. doi: 10.1093/nar/gni058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saaem I., Ma S., Quan J., Tian J. Nucleic Acids Res. 2012;40:e23. doi: 10.1093/nar/gkr887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currin A., Swainston N., Day P. J., Kell D. B. Protein Evol Design Sel. 2014;27:273–280. doi: 10.1093/protein/gzu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swainston N., Currin A., Day P. J., Kell D. B. Nucleic Acids Res. 2014;12:W395–W400. doi: 10.1093/nar/gku336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Cornish V. W. Angew. Chem., Int. Ed. 2002;41:4402–4425. doi: 10.1002/1521-3773(20021202)41:23<4402::AID-ANIE4402>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Boersma Y. L., Dröge M. J., Quax W. J. FEBS J. 2007;274:2181–2195. doi: 10.1111/j.1742-4658.2007.05782.x. [DOI] [PubMed] [Google Scholar]
- Troll C., Alexander D., Allen J., Marquette J., Camps M. J. Visualized Exp. 2011:49. doi: 10.3791/2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh M. R., Greene D. N., Woods K. K., Matsumura I. Protein Eng., Des. Sel. 2006;19:113–119. doi: 10.1093/protein/gzj010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reetz M. T., Hobenreich H., Soni P., Fernandez L. Chem. Commun. 2008:5502–5504. doi: 10.1039/b814538e. [DOI] [PubMed] [Google Scholar]
- Acevedo-Rocha C. G., Agudo R., Reetz M. T. J. Biotechnol. 2014;191C:3–10. doi: 10.1016/j.jbiotec.2014.1004.1009. [DOI] [PubMed] [Google Scholar]
- Boersma Y. L., Dröge M. J., van der Sloot A. M., Pijning T., Cool R. H., Dijkstra B. W., Quax W. J. ChemBioChem. 2008;9:1110–1115. doi: 10.1002/cbic.200700754. [DOI] [PubMed] [Google Scholar]
- Peralta-Yahya P., Carter B. T., Lin H., Tao H., Cornish V. W. J. Am. Chem. Soc. 2008;130:17446–17452. doi: 10.1021/ja8055744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H., Peralta-Yahya P., Lin H., Cornish V. W. Bioorg. Med. Chem. 2006;14:6940–6953. doi: 10.1016/j.bmc.2006.06.034. [DOI] [PubMed] [Google Scholar]
- Desai S. K., Gallivan J. P. J. Am. Chem. Soc. 2004;126:13247–13254. doi: 10.1021/ja048634j. [DOI] [PubMed] [Google Scholar]
- Winkler W. C., Breaker R. R. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- Nomura Y., Yokobayashi Y. J. Am. Chem. Soc. 2007;129:13814–13815. doi: 10.1021/ja076298b. [DOI] [PubMed] [Google Scholar]
- Dixon N., Duncan J. N., Geerlings T., Dunstan M. S., McCarthy J. E., Leys D., Micklefield J. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2830–2835. doi: 10.1073/pnas.0911209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon N., Robinson C. J., Geerlings T., Duncan J. N., Drummond S. P., Micklefield J. Angew. Chem., Int. Ed. 2012;51:3620–3624. doi: 10.1002/anie.201109106. [DOI] [PubMed] [Google Scholar]
- Wingler L. M., Cornish V. W. ChemBioChem. 2011;12:715–717. doi: 10.1002/cbic.201000543. [DOI] [PubMed] [Google Scholar]
- Yokobayashi Y., Arnold F. H. Nat. Comput. 2005;4:245–254. [Google Scholar]
- Besenmatter W., Kast P., Hilvert D. Methods Enzymol. 2004;388:91–102. doi: 10.1016/S0076-6879(04)88009-1. [DOI] [PubMed] [Google Scholar]
- Hibbert E. G., Dalby P. A. Microb. Cell Fact. 2005;4:29. doi: 10.1186/1475-2859-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M. M., Kries H., Csuhai E., Kast P., Hilvert D. Protein Sci. 2010;19:1000–1010. doi: 10.1002/pro.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanthaler K., Bilsland E., Dobson P., Moss H. J., Pir P., Kell D. B., Oliver S. G. BMC Biol. 2011;9:70. doi: 10.1186/1741-7007-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G. E., Radic B., Mayor-Ruiz C., Blomen V. A., Trefzer C., Kandasamy R. K., Huber K. V. M., Gridling M., Chen D., Klampfl T., Kralovics R., Kubicek S., Fernandez-Capetillo O., Brummelkamp T. R., Superti-Furga G. Nat. Chem. Biol. 2014;10:768–773. doi: 10.1038/nchembio.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geddie M. L., Rowe L. A., Alexander O. B., Matsumura I. Methods Enzymol. 2004;388:134–145. doi: 10.1016/S0076-6879(04)88012-1. [DOI] [PubMed] [Google Scholar]
- An G., Bielich J., Auerbach R., Johnson E. A. Bio/Technology. 1991;9:69–73. doi: 10.1038/nbt0191-70. [DOI] [PubMed] [Google Scholar]
- Azuma T., Harrison G. I., Demain A. L. Appl. Microbiol. Biotechnol. 1992;38:173–178. doi: 10.1007/BF00174463. [DOI] [PubMed] [Google Scholar]
- Cormack B. P., Valdivia R. H., Falkow S. Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Reckermann M. Sci. Mar. 2000;64:235–246. [Google Scholar]
- Hewitt C. J., Nebe-Von-Caron G. Cytometry. 2001;44:179–187. doi: 10.1002/1097-0320(20010701)44:3<179::aid-cyto1110>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Rieseberg M., Kasper C., Reardon K. F., Scheper T. Appl. Microbiol. Biotechnol. 2001;56:350–360. doi: 10.1007/s002530100673. [DOI] [PubMed] [Google Scholar]
- Vidal-Mas J., Resina P., Haba E., Comas J., Manresa A., Vives-Rego J. Antonie van Leeuwenhoek. 2001;80:57–63. doi: 10.1023/a:1012208225286. [DOI] [PubMed] [Google Scholar]
- Santoro S. W., Schultz P. G. Proc. Natl. Acad. Sci. U. S. A. 2002;99:4185–4190. doi: 10.1073/pnas.022039799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernath K., Hai M., Mastrobattista E., Griffiths A. D., Magdassi S., Tawfik D. S. Anal. Biochem. 2004;325:151–157. doi: 10.1016/j.ab.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Buskirk A. R., Ong Y. C., Gartner Z. J., Liu D. R. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10505–10510. doi: 10.1073/pnas.0402762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt C. J., Nebe-Von-Caron G. Adv. Biochem. Eng./Biotechnol. 2004;89:197–223. doi: 10.1007/b93997. [DOI] [PubMed] [Google Scholar]
- Aharoni A., Thieme K., Chiu C. P., Buchini S., Lairson L. L., Chen H., Strynadka N. C., Wakarchuk W. W., Withers S. G. Nat. Methods. 2006;3:609–614. doi: 10.1038/nmeth899. [DOI] [PubMed] [Google Scholar]
- Davey H. M., Kell D. B. Microbiol. Rev. 1996;60:641–696. doi: 10.1128/mr.60.4.641-696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattanovich D., Borth N. Microb. Cell Fact. 2006;5:12. doi: 10.1186/1475-2859-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller O. J., Bernath K., Agresti J. J., Amitai G., Kelly B. T., Mastrobattista E., Taly V., Magdassi S., Tawfik D. S., Griffiths A. D. Nat. Methods. 2006;3:561–570. doi: 10.1038/nmeth897. [DOI] [PubMed] [Google Scholar]
- Valli M., Sauer M., Branduardi P., Borth N., Porro D., Mattanovich D. Appl. Environ. Microbiol. 2006;72:5492–5499. doi: 10.1128/AEM.00683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S., Hobenreich H., Vogel A., Knorr J., Wilhelm S., Rosenau F., Jaeger K. E., Reetz M. T., Kolmar H. Angew. Chem., Int. Ed. 2008;47:5085–5088. doi: 10.1002/anie.200705236. [DOI] [PubMed] [Google Scholar]
- Varadarajan N., Rodriguez S., Hwang B. Y., Georgiou G., Iverson B. L. Nat. Chem. Biol. 2008;4:290–294. doi: 10.1038/nchembio.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Li Y., Liotta D., Lutz S. Nucleic Acids Res. 2009;37:4472–4481. doi: 10.1093/nar/gkp400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Withers S. G. ChemBioChem. 2009;10:2704–2715. doi: 10.1002/cbic.200900384. [DOI] [PubMed] [Google Scholar]
- Díaz M., Herrero M., García L. A., Quirós C. Biochem. Eng. J. 2010;48:385–407. [Google Scholar]
- Dietrich J. A., McKee A. E., Keasling J. D. Annu. Rev. Biochem. 2010;79:563–590. doi: 10.1146/annurev-biochem-062608-095938. [DOI] [PubMed] [Google Scholar]
- Iijima S., Shimomura Y., Haba Y., Kawai F., Tani A., Kimbara K. J. Biosci. Bioeng. 2010;109:645–651. doi: 10.1016/j.jbiosc.2009.11.023. [DOI] [PubMed] [Google Scholar]
- Ishii S., Tago K., Senoo K. Appl. Microbiol. Biotechnol. 2010;86:1281–1292. doi: 10.1007/s00253-010-2524-4. [DOI] [PubMed] [Google Scholar]
- Lidstrom M. E., Konopka M. C. Nat. Chem. Biol. 2010;6:705–712. doi: 10.1038/nchembio.436. [DOI] [PubMed] [Google Scholar]
- Lim S. W., Abate A. R. Lab Chip. 2013;13:4563–4572. doi: 10.1039/c3lc50736j. [DOI] [PubMed] [Google Scholar]
- Müller S., Nebe-von-Caron G. FEMS Microbiol. Rev. 2010;34:554–587. doi: 10.1111/j.1574-6976.2010.00214.x. [DOI] [PubMed] [Google Scholar]
- Rhee S. G., Chang T. S., Jeong W., Kang D. Mol. Cells. 2010;29:539–549. doi: 10.1007/s10059-010-0082-3. [DOI] [PubMed] [Google Scholar]
- Stadlmayr G., Benakovitsch K., Gasser B., Mattanovich D., Sauer M. Biotechnol. Bioeng. 2010;105:543–555. doi: 10.1002/bit.22573. [DOI] [PubMed] [Google Scholar]
- Throndset W., Kim S., Bower B., Lantz S., Kelemen B., Pepsin M., Chow N., Mitchinson C., Ward M. Enzyme Microb. Technol. 2010;47:335–341. [Google Scholar]
- Throndset W., Bower B., Caguiat R., Baldwin T., Ward M. Enzyme Microb. Technol. 2010;47:342–347. [Google Scholar]
- Tracy B. P., Gaida S. M., Papoutsakis E. T. Curr. Opin. Biotechnol. 2010;21:85–99. doi: 10.1016/j.copbio.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Eun Y. J., Utada A. S., Copeland M. F., Takeuchi S., Weibel D. B. ACS Chem. Biol. 2011;6:260–266. doi: 10.1021/cb100336p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Álvaro E., Snajdrova R., Jochens H., Davids T., Böttcher D., Bornscheuer U. T. Angew. Chem., Int. Ed. 2011;50:8584–8587. doi: 10.1002/anie.201102360. [DOI] [PubMed] [Google Scholar]
- Doan T. T. Y., Sivaloganathan B., Obbard J. P. Biomass Bioenergy. 2011;35:2534–2544. [Google Scholar]
- Binder S., Schendzielorz G., Stäbler N., Krumbach K., Hoffmann K., Bott M., Eggeling L. Genome Biol. 2012;13:R40. doi: 10.1186/gb-2012-13-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lönneborg R., Varga E., Brzezinski P. PLoS One. 2012;7:e29994. doi: 10.1371/journal.pone.0029994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo T. H., Pogson M., Iverson B. L., Georgiou G. ChemBioChem. 2012;13:649–653. doi: 10.1002/cbic.201100718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes da Silva T., Roseiro J. C., Reis A. Trends Biotechnol. 2012;30:225–232. doi: 10.1016/j.tibtech.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Uttamchandani M., Huang X., Chen G. Y. J., Yao S. Q. Bioorg. Med. Chem. Lett. 2005;15:2135–2139. doi: 10.1016/j.bmcl.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Gordeev A. A., Samatov T. R., Chetverina H. V., Chetverin A. B. Biotechnol. Bioeng. 2011;108:2682–2690. doi: 10.1002/bit.23226. [DOI] [PubMed] [Google Scholar]
- Huang W. E., Li M., Jarvis R. M., Goodacre R., Banwart S. A. Adv. Appl. Microbiol. 2010;70:153–186. doi: 10.1016/S0065-2164(10)70005-8. [DOI] [PubMed] [Google Scholar]
- Peng L., Wang G., Liao W., Yao H., Huang S., Li Y. Q. Lett. Appl. Microbiol. 2010;51:632–638. doi: 10.1111/j.1472-765X.2010.02941.x. [DOI] [PubMed] [Google Scholar]
- Lees J. G., Janes R. W. BMC Bioinf. 2008;9:24. doi: 10.1186/1471-2105-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rossum T., Kengen S. W., van der Oost J. FEBS J. 2013;280:2979–2996. doi: 10.1111/febs.12281. [DOI] [PubMed] [Google Scholar]
- Uchiyama T., Watanabe K. Biotechnol. Genet. Eng. Rev. 2007;24:107–116. doi: 10.1080/02648725.2007.10648094. [DOI] [PubMed] [Google Scholar]
- Uchiyama T., Watanabe K. Nat. Protoc. 2008;3:1202–1212. doi: 10.1038/nprot.2008.96. [DOI] [PubMed] [Google Scholar]
- Uchiyama T., Miyazaki K. Methods Mol. Biol. 2010;668:153–168. doi: 10.1007/978-1-60761-823-2_10. [DOI] [PubMed] [Google Scholar]
- Uchiyama T., Miyazaki K. Appl. Environ. Microbiol. 2010;76:7029–7035. doi: 10.1128/AEM.00464-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictenberg J. Trends Biotechnol. 2012;30:621–626. doi: 10.1016/j.tibtech.2012.09.004. [DOI] [PubMed] [Google Scholar]
- van der Meer J. R., Belkin S. Nat. Rev. Microbiol. 2010;8:511–522. doi: 10.1038/nrmicro2392. [DOI] [PubMed] [Google Scholar]
- Stojanovic M. N., de Prada P., Landry D. W. J. Am. Chem. Soc. 2001;123:4928–4931. doi: 10.1021/ja0038171. [DOI] [PubMed] [Google Scholar]
- Stojanovic M. N., Kolpashchikov D. M. J. Am. Chem. Soc. 2004;126:9266–9270. doi: 10.1021/ja032013t. [DOI] [PubMed] [Google Scholar]
- Kikuchi K. Chem. Soc. Rev. 2010;39:2048–2053. doi: 10.1039/b819316a. [DOI] [PubMed] [Google Scholar]
- Kikuchi K. Adv. Biochem. Eng./Biotechnol. 2010;119:63–78. doi: 10.1007/10_2008_42. [DOI] [PubMed] [Google Scholar]
- Okumoto S., Jones A., Frommer W. B. Annu. Rev. Plant Biol. 2012;63:663–706. doi: 10.1146/annurev-arplant-042110-103745. [DOI] [PubMed] [Google Scholar]
- Okumoto S. Plant J. 2012;70:108–117. doi: 10.1111/j.1365-313X.2012.04910.x. [DOI] [PubMed] [Google Scholar]
- Paige J. S., Wu K. Y., Jaffrey S. R. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige J. S., Nguyen-Duc T., Song W., Jaffrey S. R. Science. 2012;335:1194. doi: 10.1126/science.1218298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack R. L., Jaffrey S. R. Curr. Opin. Chem. Biol. 2013;17:651–655. doi: 10.1016/j.cbpa.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhang J., Yang F., Yang X. Chem. Commun. 2011;47:9435–9437. doi: 10.1039/c1cc13459k. [DOI] [PubMed] [Google Scholar]
- Wombacher R., Cornish V. W. J. Biophotonics. 2011;4:391–402. doi: 10.1002/jbio.201100018. [DOI] [PubMed] [Google Scholar]
- Jing C., Cornish V. W. Acc. Chem. Res. 2011;44:784–792. doi: 10.1021/ar200099f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemical proteomics, ed. G. Drewes and M. Bantscheff, Springer, Berlin, 2012. [Google Scholar]
- Arkin A. P. and Youvan D. C., Digital imaging spectroscopy, in The photosynthetic reaction center, ed. J. Deisenhofer and J. R. Norris, Academic Press, New York, 1993, vol. 1, pp. 133–155. [Google Scholar]
- Goldman E. R., Youvan D. C. Bio/Technology. 1992;10:1557–1561. doi: 10.1038/nbt1292-1557. [DOI] [PubMed] [Google Scholar]
- Joo H., Arisawa A., Lin Z. L., Arnold F. H. Chem. Biol. 1999;6:699–706. doi: 10.1016/s1074-5521(00)80017-4. [DOI] [PubMed] [Google Scholar]
- Alexeeva M., Enright A., Dawson M. J., Mahmoudian M., Turner N. J. Angew. Chem., Int. Ed. 2002;41:3177–3180. doi: 10.1002/1521-3773(20020902)41:17<3177::AID-ANIE3177>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Alexeeva M., Carr R., Turner N. J. Org. Biomol. Chem. 2003;1:4133–4137. doi: 10.1039/b311055a. [DOI] [PubMed] [Google Scholar]
- Delagrave S., Murphy D. J., Pruss J. L., Maffia, 3rd A. M., Marrs B. L., Bylina E. J., Coleman W. J., Grek C. L., Dilworth M. R., Yang M. M., Youvan D. C. Protein Eng. 2001;14:261–267. doi: 10.1093/protein/14.4.261. [DOI] [PubMed] [Google Scholar]
- Weaver J. C. Biotechnol. Bioeng. Symp. 1987;17:185–195. [Google Scholar]
- Weaver J. C., Williams G. B., Klibanov A., Demain A. L. Bio/Technology. 1988;6:1084–1089. [Google Scholar]
- Weaver J. C., Bliss J. G., Powell K. T., Harrison G. I., Williams G. B. Bio/Technology. 1991;9:873–876. doi: 10.1038/nbt0991-873. [DOI] [PubMed] [Google Scholar]
- Weaver J. C., Bliss J. G., Harrison G. I., Powell K. T., Williams G. B. Methods. 1991;2:234–247. [Google Scholar]
- Tawfik D. S., Griffiths A. D. Nat. Biotechnol. 1998;16:652–656. doi: 10.1038/nbt0798-652. [DOI] [PubMed] [Google Scholar]
- Griffiths A. D., Tawfik D. S. Curr. Opin. Biotechnol. 2000;11:338–353. doi: 10.1016/s0958-1669(00)00109-9. [DOI] [PubMed] [Google Scholar]
- Courtois F., Olguin L. F., Whyte G., Theberge A. B., Huck W. T., Hollfelder F., Abell C. Anal. Chem. 2009;81:3008–3016. doi: 10.1021/ac802658n. [DOI] [PubMed] [Google Scholar]
- Devenish S. R. A., Kaltenbach M., Fischlechner M., Hollfelder F. Methods Mol. Biol. 2013;996:269–286. doi: 10.1007/978-1-62703-354-1_16. [DOI] [PubMed] [Google Scholar]
- Lu W. C., Ellington A. D. Methods. 2013;60:75–80. doi: 10.1016/j.ymeth.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Fischlechner M., Schaerli Y., Mohamed M. F., Patil S., Abell C., Hollfelder F. Nat. Chem. 2014;6:791–796. doi: 10.1038/nchem.1996. [DOI] [PubMed] [Google Scholar]
- Fallah-Araghi A., Baret J. C., Ryckelynck M., Griffiths A. D. Lab Chip. 2012;12:882–891. doi: 10.1039/c2lc21035e. [DOI] [PubMed] [Google Scholar]
- Grünberger A., Paczia N., Probst C., Schendzielorz G., Eggeling L., Noack S., Wiechert W., Kohlheyer D. Lab Chip. 2012;12:2060–2068. doi: 10.1039/c2lc40156h. [DOI] [PubMed] [Google Scholar]
- Levin I., Aharoni A. Chem. Biol. 2012;19:929–931. doi: 10.1016/j.chembiol.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Bai Y. P., Weibull E., Joensson H. N., Andersson-Svahn H. Sens. Actuators, B. 2014;194:249–254. [Google Scholar]
- Ma F., Xie Y., Huang C., Feng Y., Yang G. PLoS One. 2014;9:e89785. doi: 10.1371/journal.pone.0089785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld L., Lin T., Derda R., Tang S. K. Y. Microfluid. Nanofluid. 2014;16:921–939. [Google Scholar]
- Sjostrom S. L., Bai Y. P., Huang M. T., Liu Z. H., Nielsen J., Joensson H. N., Svahn H. A. Lab Chip. 2014;14:806–813. doi: 10.1039/c3lc51202a. [DOI] [PubMed] [Google Scholar]
- Kintses B., van Vliet L. D., Devenish S. R. A., Hollfelder F. Curr. Opin. Chem. Biol. 2010;14:548–555. doi: 10.1016/j.cbpa.2010.08.013. [DOI] [PubMed] [Google Scholar]
- Kintses B., Hein C., Mohamed M. F., Fischlechner M., Courtois F., Laine C., Hollfelder F. Chem. Biol. 2012;19:1001–1009. doi: 10.1016/j.chembiol.2012.06.009. [DOI] [PubMed] [Google Scholar]
- Shim J. U., Ranasinghe R. T., Smith C. A., Ibrahim S. M., Hollfelder F., Huck W. T., Klenerman D., Abell C. ACS Nano. 2013;7:5955–5964. doi: 10.1021/nn401661d. [DOI] [PubMed] [Google Scholar]
- Smith C. A., Li X., Mize T. H., Sharpe T. D., Graziani E. I., Abell C., Huck W. T. S. Anal. Chem. 2013;85:3812–3816. doi: 10.1021/ac400453t. [DOI] [PubMed] [Google Scholar]
- Zinchenko A., Devenish S. R., Kintses B., Colin P. Y., Fischlechner M., Hollfelder F. Anal. Chem. 2014;86:2526–2533. doi: 10.1021/ac403585p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agresti J. J., Antipov E., Abate A. R., Ahn K., Rowat A. C., Baret J. C., Marquez M., Klibanov A. M., Griffiths A. D., Weitz D. A. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4004–4009. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks J., Welch W., Mitchell T., Wynn H. Statist Sci. 1989;4:409–435. [Google Scholar]
- Koza J. R., Genetic programming: on the programming of computers by means of natural selection, MIT Press, Cambridge, Mass, 1992. [Google Scholar]
- Koza J. R., Genetic programming II: automatic discovery of reusable programs, MIT Press, Cambridge, Mass, 1994. [Google Scholar]
- Bäck T., Evolutionary algorithms in theory and practice, Oxford University Press, Oxford, 1996. [Google Scholar]
- Langdon W. B., Genetic programming and data structures: genetic programming+data structures=automatic programming!, Kluwer, Boston, 1998. [Google Scholar]
- New ideas in optimization, ed. D. Corne, M. Dorigo and F. Glover, McGraw Hill, London, 1999. [Google Scholar]
- Koza J. R., Bennett F. H., Keane M. A. and Andre D., Genetic Programming III: Darwinian Invention and Problem Solving, Morgan Kaufmann, San Francisco, 1999. [Google Scholar]
- Evolutionary Computation 1: basic algorithms and operators, ed. T. Bäck, D. B. Fogel and Z. Michalewicz, IOP Publishing, Bristol, 2000. [Google Scholar]
- Evolutionary Computation 2: advanced algorithms and operators, ed. T. Bäck, D. B. Fogel and Z. Michalewicz, IOP Publishing, Bristol, 2000. [Google Scholar]
- Langdon W. B. and Poli R., Foundations of genetic programming, Springer-Verlag, Berlin, 2002. [Google Scholar]
- Koza J. R., Keane M. A., Streeter M. J. Sci Am. 2003;288:52–59. doi: 10.1038/scientificamerican0203-52. [DOI] [PubMed] [Google Scholar]
- Koza J. R., Keane M. A., Streeter M. J., Mydlowec W., Yu J. and Lanza G., Genetic programming: routine human-competitive machine intelligence, Kluwer, New York, 2003. [Google Scholar]
- Handl J., Knowles J. IEEE Trans. Evol. Comput. 2007;11:56–76. [Google Scholar]
- Poli R., Langdon W. B. and McPhee N. F., A Field Guide to Genetic Programming, http://www.lulu.com/product/file-download/a-field-guide-to-genetic-programming/2502914, 2009.
- Jones D. R., Schonlau M., Welch W. J. J. Global. Opt. 1998;13:455–492. [Google Scholar]
- Patrick W. M., Firth A. E., Blackburn J. M. Protein Eng. 2003;16:451–457. doi: 10.1093/protein/gzg057. [DOI] [PubMed] [Google Scholar]
- Firth A. E., Patrick W. M. Bioinformatics. 2005;21:3314–3315. doi: 10.1093/bioinformatics/bti516. [DOI] [PubMed] [Google Scholar]
- Patrick W. M., Firth A. E. Biomol. Eng. 2005;22:105–112. doi: 10.1016/j.bioeng.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Firth A. E., Patrick W. M. Nucleic Acids Res. 2008;36:W281–W285. doi: 10.1093/nar/gkn226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starrfelt J., Kokko H. Biol. Rev. Cambridge Philos. Soc. 2012;87:742–755. doi: 10.1111/j.1469-185X.2012.00225.x. [DOI] [PubMed] [Google Scholar]
- del Sol Mesa A., Pazos F., Valencia A. J. Mol. Biol. 2003;326:1289–1302. doi: 10.1016/s0022-2836(02)01451-1. [DOI] [PubMed] [Google Scholar]
- Herrgard S., Cammer S. A., Hoffman B. T., Knutson S., Gallina M., Speir J. A., Fetrow J. S., Baxter S. M. Proteins. 2003;53:806–816. doi: 10.1002/prot.10458. [DOI] [PubMed] [Google Scholar]
- Lopez G., Maietta P., Rodriguez J. M., Valencia A., Tress M. L. J. Chem. Inf. Model. 2011;39:W235–W241. doi: 10.1093/nar/gkr437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addington T. A., Mertz R. W., Siegel J. B., Thompson J. M., Fisher A. J., Filkov V., Fleischman N. M., Suen A. A., Zhang C. S., Toney M. D. J. Mol. Biol. 2013;425:1378–1389. doi: 10.1016/j.jmb.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox R. J., Huisman G. W. Trends Biotechnol. 2008;26:132–138. doi: 10.1016/j.tibtech.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Liang F., Feng X. J., Lowry M., Rabitz H. J. Phys. Chem. B. 2005;109:5842–5854. doi: 10.1021/jp045926y. [DOI] [PubMed] [Google Scholar]
- McAllister S. R., Feng X. J., DiMaggio, Jr. P. A., Floudas C. A., Rabinowitz J. D., Rabitz H. Bioorg. Med. Chem. Lett. 2008;18:5967–5970. doi: 10.1016/j.bmcl.2008.09.068. [DOI] [PubMed] [Google Scholar]
- Feng X., Sanchis J., Reetz M. T., Rabitz H. Chemistry. 2012;18:5646–5654. doi: 10.1002/chem.201103811. [DOI] [PubMed] [Google Scholar]
- Araya C. L., Fowler D. M. Trends Biotechnol. 2011;29:435–442. doi: 10.1016/j.tibtech.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N. MAbs. 2011;3:17–20. doi: 10.4161/mabs.3.1.14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciani F., Bull R. A., Lloyd A. R. Trends Biotechnol. 2012;30:443–452. doi: 10.1016/j.tibtech.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead T. A., Chevalier A., Song Y., Dreyfus C., Fleishman S. J., De Mattos C., Myers C. A., Kamisetty H., Blair P., Wilson I. A., Baker D. Nat. Biotechnol. 2012;30:543–548. doi: 10.1038/nbt.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi P. and Brunak S., Bioinformatics: the machine learning approach, MIT Press, Cambridge, MA, 1998. [Google Scholar]
- Machine learning and data mining. Methods and applications, ed. R. S. Michalski, I. Bratko and M. Kubat, Wiley, Chichester, 1998. [Google Scholar]
- Mitchell T. M., Machine learning, McGraw Hill, New York, 1997. [Google Scholar]
- Buchanan B. G., Feigenbaum E. A. Artif. Intell. Mater. Process., Proc. Int. Symp. 1978;11:5–24. [Google Scholar]
- Buchanan B. G., Feigenbaum E. A., Lederberg J. Chemom. Intell. Lab. Syst. 1988;5:33–35. [Google Scholar]
- Feigenbaum E. A., Buchanan B. G. Artif. Intell. 1993;59:223–240. [Google Scholar]
- J. Lederberg, How DENDRAL was conceived and born, ACM Symp Hist Med Informatics, 1987, http://profiles.nlm.nih.gov/ps/access/BBALYP.pdf
- Lindsay R. K., Buchanan B. G., Feigenbaum E. A., Lederberg J. Artif. Intell. Mater. Process., Proc. Int. Symp. 1993;61:209–261. [Google Scholar]
- Jonsson J., Norberg T., Carlsson L., Gustafsson C., Wold S. J. Chem. Inf. Model. 1993;21:733–739. [Google Scholar]
- Breiman L. Stat Sci. 2001;16:199–215. [Google Scholar]
- Liao J., Warmuth M. K., Govindarajan S., Ness J. E., Wang R. P., Gustafsson C., Minshull J. BMC Biotechnol. 2007;7:16. doi: 10.1186/1472-6750-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Li Z. J. Mol. Graphics Modell. 2007;26:269–281. doi: 10.1016/j.jmgm.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Petersen B., Petersen T. N., Andersen P., Nielsen M., Lundegaard C. BMC Struct. Biol. 2009;9:51. doi: 10.1186/1472-6807-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Chen X., Wu Y., Shang Z. Amino Acids. 2010;38:199–212. doi: 10.1007/s00726-008-0228-1. [DOI] [PubMed] [Google Scholar]
- van den Berg B. A., Reinders M. J. T., Hulsman M., Wu L., Pel H. J., Roubos J. A., de Ridder D. PLoS One. 2012;7:e45869. doi: 10.1371/journal.pone.0045869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan S., Broadhurst D. I., Kell D. B., Goodacre R. Anal. Chem. 2003;75:6679–6686. doi: 10.1021/ac034669a. [DOI] [PubMed] [Google Scholar]
- Vaidyanathan S., Kell D. B., Goodacre R. Anal. Chem. 2004;76:5024–5032. doi: 10.1021/ac049684+. [DOI] [PubMed] [Google Scholar]
- Wedge D., Gaskell S. J., Hubbard S., Kell D. B., Lau K. W. and Eyers C., Peptide detectability following ESI mass spectrometry: prediction using genetic programming, in GECCO 2007, ed. D. Thierens et al., ACM, New York, 2007, pp. 2219–2225. [Google Scholar]
- Breiman L. Machine Learning. 2001;45:5–32. [Google Scholar]
- Fowler D. M., Stephany J. J., Fields S. Nat. Protoc. 2014;9:2267–2284. doi: 10.1038/nprot.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler D. M., Fields S. Nat. Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J., Overbaugh J., Miller S. Nature. 1988;335:142–145. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- Romero P. A., Krause A., Arnold F. H. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E193–E201. doi: 10.1073/pnas.1215251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleti T., Basic enzyme kinetics, Akadémiai Kiadó, Budapest, 1986. [Google Scholar]
- Cornish-Bowden A., Fundamentals of enzyme kinetics, Portland Press, London, 2nd edn, 1995. [Google Scholar]
- Fersht A., Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding, W.H. Freeman, San Francisco, 1999. [Google Scholar]
- Fersht A. R. Proc. R. Soc. London, Ser. B. 1974;187:397–407. doi: 10.1098/rspb.1974.0084. [DOI] [PubMed] [Google Scholar]
- Jencks W. P. Adv. Enzymol. Relat. Areas Mol. Biol. 1975;43:219–410. doi: 10.1002/9780470122884.ch4. [DOI] [PubMed] [Google Scholar]
- Whitty A., Fierke C. A., Jencks W. P. Biochemistry. 1995;34:11678–11689. doi: 10.1021/bi00037a005. [DOI] [PubMed] [Google Scholar]
- Liebeton K., Zonta A., Schimossek K., Nardini M., Lang D., Dijkstra B. W., Reetz M. T., Jaeger K. E. Chem. Biol. 2000;7:709–718. doi: 10.1016/s1074-5521(00)00015-6. [DOI] [PubMed] [Google Scholar]
- Greiner L., Laue S., Liese A., Wandrey C. Chemistry. 2006;12:1818–1823. doi: 10.1002/chem.200500675. [DOI] [PubMed] [Google Scholar]
- Fox R. J., Clay M. D. Trends Biotechnol. 2009;27:137–140. doi: 10.1016/j.tibtech.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Porro D., Gasser B., Fossati T., Maurer M., Branduardi P., Sauer M., Mattanovich D. Appl. Microbiol. Biotechnol. 2011;89:939–948. doi: 10.1007/s00253-010-3019-z. [DOI] [PubMed] [Google Scholar]
- Becker J., Wittmann C. Curr. Opin. Biotechnol. 2012;23:718–726. doi: 10.1016/j.copbio.2011.12.025. [DOI] [PubMed] [Google Scholar]
- Dach R., Song J. H. J., Roschangar F., Samstag W., Senanayake C. H. Org. Process Res. Dev. 2012;16:1697–1706. [Google Scholar]
- Knowles J. R., Albery W. J. Acc. Chem. Res. 1977;10:105–111. [Google Scholar]
- Miller B. G., Wolfenden R. Annu. Rev. Biochem. 2002;71:847–885. doi: 10.1146/annurev.biochem.71.110601.135446. [DOI] [PubMed] [Google Scholar]
- Radzicka A., Wolfenden R. Science. 1995;267:90–93. doi: 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- Miller B. G., Hassell A. M., Wolfenden R., Milburn M. V., Short S. A. Proc. Natl. Acad. Sci. U. S. A. 2000;97:2011–2016. doi: 10.1073/pnas.030409797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Even A., Noor E., Savir Y., Liebermeister W., Davidi D., Tawfik D. S., Milo R. Biochemistry. 2011;50:4402–4410. doi: 10.1021/bi2002289. [DOI] [PubMed] [Google Scholar]
- Milo R., Last R. L. Science. 2012;336:1663–1667. doi: 10.1126/science.1217665. [DOI] [PubMed] [Google Scholar]
- Schomburg I., Chang A., Placzek S., Söhngen C., Rother M., Lang M., Munaretto C., Ulas S., Stelzer M., Grote A., Scheer M., Schomburg D. Nucleic Acids Res. 2013;41:D764–D772. doi: 10.1093/nar/gks1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig U., Kania R., Golebiewski M., Rey M., Shi L., Jong L., Algaa E., Weidemann A., Sauer-Danzwith H., Mir S., Krebs O., Bittkowski M., Wetsch E., Rojas I., Müller W. J. Chem. Inf. Model. 2012;40:D790–D796. doi: 10.1093/nar/gkr1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacser H., Burns J. A. Genetics. 1981;97:639–666. doi: 10.1093/genetics/97.3-4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacser H. and Burns J. A., The control of flux, in Rate Control of Biological Processes. Symposium of the Society for Experimental Biology, ed. D. D. Davies, Cambridge University Press, Cambridge, 1973, vol. 27, pp. 65–104. [PubMed] [Google Scholar]
- Kell D. B., Westerhoff H. V. FEMS Microbiol. Rev. 1986;39:305–320. [Google Scholar]
- Kell D. B., Westerhoff H. V. Trends Biotechnol. 1986;4:137–142. [Google Scholar]
- Fell D. A., Understanding the control of metabolism, Portland Press, London, 1996. [Google Scholar]
- Heinrich R. and Schuster S., The regulation of cellular systems., Chapman & Hall, New York, 1996. [Google Scholar]
- Savile C. K., Janey J. M., Mundorff E. C., Moore J. C., Tam S., Jarvis W. R., Colbeck J. C., Krebber A., Fleitz F. J., Brands J., Devine P. N., Huisman G. W., Hughes G. J. Science. 2010;329:305–309. doi: 10.1126/science.1188934. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Asada K., Miyazaki J., Tomita T., Kuzuyama T., Nishiyama M. Biochem. J. 2010;431:401–410. doi: 10.1042/BJ20101246. [DOI] [PubMed] [Google Scholar]
- Benkovic S. J., Hammes-Schiffer S. Science. 2003;301:1196–1202. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- Garcia-Viloca M., Gao J., Karplus M., Truhlar D. G. Science. 2004;303:186–195. doi: 10.1126/science.1088172. [DOI] [PubMed] [Google Scholar]
- Hammes G. G., Benkovic S. J., Hammes-Schiffer S. Biochemistry. 2011;50:10422–10430. doi: 10.1021/bi201486f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruice T. C. Chem. Rev. 2006;106:3119–3139. doi: 10.1021/cr050283j. [DOI] [PubMed] [Google Scholar]
- Gao J. L., Ma S. H., Major D. T., Nam K., Pu J. Z., Truhlar D. G. Chem. Rev. 2006;106:3188–3209. doi: 10.1021/cr050293k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M. H., Parson W. W., Warshel A. Chem. Rev. 2006;106:1737–1756. doi: 10.1021/cr040427e. [DOI] [PubMed] [Google Scholar]
- Carey P. R. Chem. Rev. 2006;106:3043–3054. doi: 10.1021/cr0502854. [DOI] [PubMed] [Google Scholar]
- Warshel A., Sharma P. K., Kato M., Xiang Y., Liu H. B., Olsson M. H. M. Chem. Rev. 2006;106:3210–3235. doi: 10.1021/cr0503106. [DOI] [PubMed] [Google Scholar]
- Pisliakov A. V., Cao J., Kamerlin S. C., Warshel A. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17359–17364. doi: 10.1073/pnas.0909150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerlin S. C., Warshel A. Proteins. 2010;78:1339–1375. doi: 10.1002/prot.22654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamczyk A. J., Cao J., Kamerlin S. C., Warshel A. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14115–14120. doi: 10.1073/pnas.1111252108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frushicheva M. P., Mills M. J., Schopf P., Singh M. K., Prasad R. B., Warshel A. Curr. Opin. Chem. Biol. 2014;21C:56–62. doi: 10.1016/j.cbpa.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel Z. D., Klinman J. P. Chem. Rev. 2006;106:3095–3118. doi: 10.1021/cr050301x. [DOI] [PubMed] [Google Scholar]
- Pu J. Z., Gao J. L., Truhlar D. G. Chem. Rev. 2006;106:3140–3169. doi: 10.1021/cr050308e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay S., Scrutton N. S. Biochemistry. 2008;47:9880–9887. doi: 10.1021/bi8005972. [DOI] [PubMed] [Google Scholar]
- Hay S., Johannissen L. O., Sutcliffe M. J., Scrutton N. S. Biophys. J. 2010;98:121–128. doi: 10.1016/j.bpj.2009.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudney C. R., Johannissen L. O., Sutcliffe M. J., Hay S., Scrutton N. S. J. Am. Chem. Soc. 2010;132:11329–11335. doi: 10.1021/ja1048048. [DOI] [PubMed] [Google Scholar]
- Hay S., Scrutton N. S. Nat. Chem. 2012;4:161–168. doi: 10.1038/nchem.1223. [DOI] [PubMed] [Google Scholar]
- Hay S., Johannissen L. O., Hothi P., Sutcliffe M. J., Scrutton N. S. J. Am. Chem. Soc. 2012;134:9749–9754. doi: 10.1021/ja3024115. [DOI] [PubMed] [Google Scholar]
- Widersten M. Curr. Opin. Chem. Biol. 2014;21C:42–47. doi: 10.1016/j.cbpa.2014.03.015. [DOI] [PubMed] [Google Scholar]
- Fuxreiter M., Mones L. Curr. Opin. Chem. Biol. 2014;21C:34–41. doi: 10.1016/j.cbpa.2014.03.011. [DOI] [PubMed] [Google Scholar]
- Gavish B., Werber M. M. Biochemistry. 1979;18:1269–1275. doi: 10.1021/bi00574a023. [DOI] [PubMed] [Google Scholar]
- Gavish B. Phys. Rev. Lett. 1980;44:1160–1163. [Google Scholar]
- Beece D., Eisenstein L., Frauenfelder H., Good D., Marden M. C., Reinisch L., Reynolds A. H., Sorensen L. B., Yue K. T. Biochemistry. 1980;19:5147–5157. doi: 10.1021/bi00564a001. [DOI] [PubMed] [Google Scholar]
- Kell D. B. Trends Biochem. Sci. 1982;7:349. [Google Scholar]
- Welch G. R., Somogyi B., Damjanovich S. Prog. Biophys. Mol. Biol. 1982;39:109–146. doi: 10.1016/0079-6107(83)90015-9. [DOI] [PubMed] [Google Scholar]
- Somogyi B., Welch G. R., Damjanovich S. Biochim. Biophys. Acta. 1984;768:81–112. doi: 10.1016/0304-4173(84)90001-6. [DOI] [PubMed] [Google Scholar]
- Vitkup D., Ringe D., Petsko G. A., Karplus M. Nat. Struct. Biol. 2000;7:34–38. doi: 10.1038/71231. [DOI] [PubMed] [Google Scholar]
- Daniel R. M., Dunn R. V., Finney J. L., Smith J. C. Annu. Rev. Biophys. Biomol. Struct. 2003;32:69–92. doi: 10.1146/annurev.biophys.32.110601.142445. [DOI] [PubMed] [Google Scholar]
- Basner J. E., Schwartz S. D. J. Am. Chem. Soc. 2005;127:13822–13831. doi: 10.1021/ja043320h. [DOI] [PubMed] [Google Scholar]
- Antoniou D., Basner J., Nunez S., Schwartz S. D. Chem. Rev. 2006;106:3170–3187. doi: 10.1021/cr0503052. [DOI] [PubMed] [Google Scholar]
- Callender R., Dyer R. B. Chem. Rev. 2006;106:3031–3042. doi: 10.1021/cr050284b. [DOI] [PubMed] [Google Scholar]
- Finkelstein I. J., Massari A. M., Fayer M. D. Biophys. J. 2007;92:3652–3662. doi: 10.1529/biophysj.106.093708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauenfelder H., Fenimore P. W., Young R. D. IUBMB Life. 2007;59:506–512. doi: 10.1080/15216540701194113. [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman K., Kern D. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman K. A., Lei M., Thai V., Kerns S. J., Karplus M., Kern D. Nature. 2007;450:913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman K. A., Thai V., Lei M., Ott M., Wolf-Watz M., Fenn T., Pozharski E., Wilson M. A., Petsko G. A., Karplus M., Hubner C. G., Kern D. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- Frauenfelder H., Chen G., Berendzen J., Fenimore P. W., Jansson H., McMahon B. H., Stroe I. R., Swenson J., Young R. D. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5129–5134. doi: 10.1073/pnas.0900336106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkovic S. J., Hammes G. G., Hammes-Schiffer S. Biochemistry. 2008;47:3317–3321. doi: 10.1021/bi800049z. [DOI] [PubMed] [Google Scholar]
- Eppler R. K., Hudson E. P., Chase S. D., Dordick J. S., Reimer J. A., Clark D. S. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15672–15677. doi: 10.1073/pnas.0804566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehr D. D., Nussinov R., Wright P. E. Nat. Chem. Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S. D., Schramm V. L. Nat. Chem. Biol. 2009;5:551–558. doi: 10.1038/nchembio.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar I., Lezon T. R., Yang L. W., Eyal E. Annu. Rev. Biophys. 2010;39:23–42. doi: 10.1146/annurev.biophys.093008.131258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csermely P., Palotai R., Nussinov R. Trends Biochem. Sci. 2010;35:539–546. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitnitsky A. E. Phys. A. 2008;387:5483–5497. [Google Scholar]
- Sitnitsky A. E. Chem. Phys. 2010;369:37–42. [Google Scholar]
- Bhabha G., Lee J., Ekiert D. C., Gam J., Wilson I. A., Dyson H. J., Benkovic S. J., Wright P. E. Science. 2011;332:234–238. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shushanyan M., Khoshtariya D. E., Tretyakova T., Makharadze M., van Eldik R. Biopolymers. 2011;95:852–870. doi: 10.1002/bip.21688. [DOI] [PubMed] [Google Scholar]
- Jones A. R., Levy C., Hay S., Scrutton N. S. FEBS J. 2013;280:2997–3008. doi: 10.1111/febs.12223. [DOI] [PubMed] [Google Scholar]
- Dynamics in enzyme catalysis, ed. J. P. Klinman and S. Hammes-Schiffer, Springer, Berlin, 2013. [Google Scholar]
- Świderek K., Javier Ruiz-Pernía J., Moliner V., Tuñon I. Curr. Opin. Chem. Biol. 2014;21C:11–18. doi: 10.1016/j.cbpa.2014.03.005. [DOI] [PubMed] [Google Scholar]
- Davydov A. S. J. Theor. Biol. 1977;66:379–387. doi: 10.1016/0022-5193(77)90178-3. [DOI] [PubMed] [Google Scholar]
- Davydov A. S. Int. Rev. Cytol. 1987;106:183–225. doi: 10.1016/s0074-7696(08)61713-1. [DOI] [PubMed] [Google Scholar]
- Ansari A., Berendzen J., Bowne S. F., Frauenfelder H., Iben I. E., Sauke T. B., Shyamsunder E., Young R. D. Proc. Natl. Acad. Sci. U. S. A. 1985;82:5000–5004. doi: 10.1073/pnas.82.15.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadusc G., Ogilvie J. P., Schulenberg P., Marvet U., Miller R. J. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6110–6115. doi: 10.1073/pnas.101130298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnlund D., Johansson L. C., Wickstrand C., Barty A., Williams G. J., Malmerberg E., Davidsson J., Milathianaki D., DePonte D. P., Shoeman R. L., Wang D., James D., Katona G., Westenhoff S., White T. A., Aquila A., Bari S., Berntsen P., Bogan M., van Driel T. B., Doak R. B., Kjaer K. S., Frank M., Fromme R., Grotjohann I., Henning R., Hunter M. S., Kirian R. A., Kosheleva I., Kupitz C., Liang M., Martin A. V., Nielsen M. M., Messerschmidt M., Seibert M. M., Sjohamn J., Stellato F., Weierstall U., Zatsepin N. A., Spence J. C., Fromme P., Schlichting I., Boutet S., Groenhof G., Chapman H. N., Neutze R. Nat. Methods. 2014;11:923–926. doi: 10.1038/nmeth.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglebakk E., Echave J., Reuter N. Bioinformatics. 2012;28:2431–2440. doi: 10.1093/bioinformatics/bts445. [DOI] [PubMed] [Google Scholar]
- Jimenez-Roldan J. E., Freedman R. B., Romer R. A., Wells S. A. Phys. Biol. 2012;9:016008. doi: 10.1088/1478-3975/9/1/016008. [DOI] [PubMed] [Google Scholar]
- Pelis R. M., Suhre W. M., Wright S. H. Am. J. Physiol.: Renal, Fluid Electrolyte Physiol. 2006;290:F1118–F1126. doi: 10.1152/ajprenal.00462.2005. [DOI] [PubMed] [Google Scholar]
- Süel G. M., Lockless S. W., Wall M. A., Ranganathan R. Nat. Struct. Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- Wong K. F., Selzer T., Benkovic S. J., Hammes-Schiffer S. Proc. Natl. Acad. Sci. U. S. A. 2005;102:6807–6812. doi: 10.1073/pnas.0408343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley K. L., Kazlauskas R. J. Trends Biotechnol. 2005;23:231–237. doi: 10.1016/j.tibtech.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Paramesvaran J., Hibbert E. G., Russell A. J., Dalby P. A. Protein Eng., Des. Sel. 2009;22:401–411. doi: 10.1093/protein/gzp020. [DOI] [PubMed] [Google Scholar]
- Leferink N. G. H., Antonyuk S. V., Houwman J. A., Scrutton N. S., Eady R. R., Hasnain S. S. Nat. Commun. 2014;5:4395. doi: 10.1038/ncomms5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasan R., Meharenna Y. T., Snow C. D., Poulos T. L., Arnold F. H. J. Mol. Biol. 2008;383:1069–1080. doi: 10.1016/j.jmb.2008.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiswerk N., Beck T., Schulz J. D., Milovnik P., Mayer C., Siegel J. B., Baker D., Hilvert D. Proc. Natl. Acad. Sci. U. S. A. 2014;111:8013–8018. doi: 10.1073/pnas.1401073111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Chen Y., Jiang K., Zuo W., Luo Z., Wei Y., Du L., Wei H., Huang R., Du Q. J. Biotechnol. 2009;144:43–50. doi: 10.1016/j.jbiotec.2009.06.015. [DOI] [PubMed] [Google Scholar]
- Siehl D. L., Castle L. A., Gorton R., Keenan R. J. J. Biol. Chem. 2007;282:11446–11455. doi: 10.1074/jbc.M610267200. [DOI] [PubMed] [Google Scholar]
- Cho C. M., Mulchandani A., Chen W. Appl. Environ. Microbiol. 2002;68:2026–2030. doi: 10.1128/AEM.68.4.2026-2030.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oue S., Okamoto A., Yano T., Kagamiyama H. J. Biol. Chem. 1999;274:2344–2349. doi: 10.1074/jbc.274.4.2344. [DOI] [PubMed] [Google Scholar]
- Lindorff-Larsen K., Piana S., Dror R. O., Shaw D. E. Science. 2011;334:517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- Piana S., Sarkar K., Lindorff-Larsen K., Guo M., Gruebele M., Shaw D. E. J. Mol. Biol. 2011;405:43–48. doi: 10.1016/j.jmb.2010.10.023. [DOI] [PubMed] [Google Scholar]
- Piana S., Lindorff-Larsen K., Shaw D. E. Proc. Natl. Acad. Sci. U. S. A. 2013;110:5915–5920. doi: 10.1073/pnas.1218321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raval A., Piana S., Eastwood M. P., Dror R. O., Shaw D. E. Proteins. 2012;80:2071–2079. doi: 10.1002/prot.24098. [DOI] [PubMed] [Google Scholar]
- Shaw D. E., Deneroff M. M., Dror R. O., Kuskin J. S., Larson R. H., Salmon J. K., Young C., Batson B., Bowers K. J., Chao J. C., Eastwood M. P., Gagliardo J., Grossman J. P., Ho C. R., Ierardi D. J., Kolossvary I., Klepeis J. L., Layman T., Mcleavey C., Moraes M. A., Mueller R., Priest E. C., Shan Y. B., Spengler J., Theobald M., Towles B., Wang S. C. Commun. ACM. 2008;51:91–97. [Google Scholar]
- Schwede T. Structure. 2013;21:1531–1540. doi: 10.1016/j.str.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill K. A., MacCallum J. L. Science. 2012;338:1042–1046. doi: 10.1126/science.1219021. [DOI] [PubMed] [Google Scholar]
- Khatib F., Dimaio F., Cooper S., Kazmierczyk M., Gilski M., Krzywda S., Zabranska H., Pichova I., Thompson J., Popovic Z., Jaskolski M., Baker D. Nat. Struct. Mol. Biol. 2011;18:1175–1177. doi: 10.1038/nsmb.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg E. H., Lange O. F., Baker D. J. Phys. Chem. B. 2012;116:11405–11413. doi: 10.1021/jp3044303. [DOI] [PubMed] [Google Scholar]
- Marks D. S., Hopf T. A., Sander C. Nat. Biotechnol. 2012;30:1072–1080. doi: 10.1038/nbt.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor W. R., Jones D. T., Sadowski M. I. Protein Sci. 2012;21:299–305. doi: 10.1002/pro.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. T., Buchan D. W., Cozzetto D., Pontil M. Bioinformatics. 2012;28:184–190. doi: 10.1093/bioinformatics/btr638. [DOI] [PubMed] [Google Scholar]
- Nugent T., Jones D. T. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E1540–E1547. doi: 10.1073/pnas.1120036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosciolek T., Jones D. T. PLoS One. 2014;9:e92197. doi: 10.1371/journal.pone.0092197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreini C., Bertini I., Cavallaro G., Holliday G. L., Thornton J. M. JBIC, J. Biol. Inorg. Chem. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- Kell D. B. BMC Med. Genomics. 2009;2:2. doi: 10.1186/1755-8794-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kell D. B. Arch. Toxicol. 2010;577:825–889. doi: 10.1007/s00204-010-0577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kell D. B., Pretorius E. Metallomics. 2014;6:748–773. doi: 10.1039/c3mt00347g. [DOI] [PubMed] [Google Scholar]
- Reetz M. T., Peyralans J. J., Maichele A., Fu Y., Maywald M. Chem. Commun. 2006:4318–4320. doi: 10.1039/b610461d. [DOI] [PubMed] [Google Scholar]
- Reetz M. T., Rentzsch M., Pletsch A., Maywald M., Maiwald P., Peyralans J. J. P., Maichele A., Fu Y., Jiao N., Hollmann F., Mondière R., Taglieber A. Tetrahedron. 2007;63:6404–6414. [Google Scholar]
- Reetz M. T. Tetrahedron. 2002;58:6595–6602. [Google Scholar]
- Reetz M. T., Rentzsch M., Pletsch A., Maywald M. Chimia. 2002;56:721–723. [Google Scholar]
- Benson D. E., Wisz M. S., Hellinga H. W. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6292–6297. doi: 10.1073/pnas.97.12.6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgewater J. D., Lim J., Vachet R. W. Anal. Chem. 2006;78:2432–2438. doi: 10.1021/ac051983r. [DOI] [PubMed] [Google Scholar]
- Petros A. K., Reddi A. R., Kennedy M. L., Hyslop A. G., Gibney B. R. Inorg. Chem. 2006;45:9941–9958. doi: 10.1021/ic052190q. [DOI] [PubMed] [Google Scholar]
- Mantion A., Massuger L., Rabu P., Palivan C., McCusker L. B., Taubert A. J. Am. Chem. Soc. 2008;130:2517–2526. doi: 10.1021/ja0762588. [DOI] [PubMed] [Google Scholar]
- Pirngruber G. D., Frunz L., Lüchinger M. Phys. Chem. Chem. Phys. 2009;11:2928–2938. doi: 10.1039/b819678h. [DOI] [PubMed] [Google Scholar]
- Pedersen J. T., Teilum K., Heegaard N. H., Ostergaard J., Adolph H. W., Hemmingsen L. Angew. Chem., Int. Ed. 2011;50:2532–2535. doi: 10.1002/anie.201006335. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Mizuno T., Fukui S., Hiroaki H., Oku J., Kanaori K., Tajima K., Shirakawa M. J. Am. Chem. Soc. 2004;126:14023–14028. doi: 10.1021/ja047945r. [DOI] [PubMed] [Google Scholar]
- Tamerler C., Khatayevich D., Gungormus M., Kacar T., Oren E. E., Hnilova M., Sarikaya M. Biopolymers. 2010;94:78–94. doi: 10.1002/bip.21368. [DOI] [PubMed] [Google Scholar]
- Koder R. L., Anderson J. L., Solomon L. A., Reddy K. S., Moser C. C., Dutton P. L. Nature. 2009;458:305–309. doi: 10.1038/nature07841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock A. F., Iranzo O., Pecoraro V. L. Dalton Trans. 2009:2271–2280. doi: 10.1039/b818306f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranzo O., Chakraborty S., Hemmingsen L., Pecoraro V. L. J. Am. Chem. Soc. 2011;133:239–251. doi: 10.1021/ja104433n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun P., Goldberg E., Negron C., von Jan M., Xu F., Nanda V., Koder R. L., Noy D. Proteins. 2010;79:463–476. doi: 10.1002/prot.22895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran F. V., Wu S. P., Wang W., Nanda V., Saven J. G., Therien M. J., DeGrado W. F. J. Am. Chem. Soc. 2005;127:1346–1347. doi: 10.1021/ja044129a. [DOI] [PubMed] [Google Scholar]
- Kuroda K., Ueda M. Curr. Opin. Biotechnol. 2011;22:427–433. doi: 10.1016/j.copbio.2010.12.006. [DOI] [PubMed] [Google Scholar]
- González J. M., Meini M. R., Tomatis P. E., Medrano Martín F. J., Cricco J. A., Vila A. J. Nat. Chem. Biol. 2012;8:698–700. doi: 10.1038/nchembio.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sóvágó I., Kállay C., Várnagy K. Coord. Chem. Rev. 2012;256:2225–2233. [Google Scholar]
- Lu Y., Yeung N., Sieracki N., Marshall N. M. Nature. 2009;460:855–862. doi: 10.1038/nature08304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K. L., Lim S., Franklin S. J. Inorg. Chem. 2006;45:10002–10012. doi: 10.1021/ic060877k. [DOI] [PubMed] [Google Scholar]
- Sapsford K. E., Algar W. R., Berti L., Gemmill K. B., Casey B. J., Oh E., Stewart M. H., Medintz I. L. Chem. Rev. 2013;113:1904–2074. doi: 10.1021/cr300143v. [DOI] [PubMed] [Google Scholar]
- Happe T., Hemschemeier A. Trends Biotechnol. 2014;32:170–176. doi: 10.1016/j.tibtech.2014.02.004. [DOI] [PubMed] [Google Scholar]
- Dürrenberger M., Ward T. R. Curr. Opin. Chem. Biol. 2014;19:99–106. doi: 10.1016/j.cbpa.2014.01.018. [DOI] [PubMed] [Google Scholar]
- Bos J., Roelfes G. Curr. Opin. Chem. Biol. 2014;19:135–143. doi: 10.1016/j.cbpa.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Petrik I. D., Liu J., Lu Y. Curr. Opin. Chem. Biol. 2014;19:67–75. doi: 10.1016/j.cbpa.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman A. J., Sanford M. S. Nature. 2012;484:177–185. doi: 10.1038/nature11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reig A. J., Pires M. M., Snyder R. A., Wu Y., Jo H., Kulp D. W., Butch S. E., Calhoun J. R., Szyperski T., Solomon E. I., DeGrado W. F. Nat. Chem. 2012;4:900–906. doi: 10.1038/nchem.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Murao K., Tanabe Y., Oda M., Tanaka T. J. Am. Chem. Soc. 2007;129:11378–11383. doi: 10.1021/ja0685102. [DOI] [PubMed] [Google Scholar]
- Bae W., Mulchandani A., Chen W. J. Inorg. Biochem. 2002;88:223–227. doi: 10.1016/s0162-0134(01)00392-0. [DOI] [PubMed] [Google Scholar]
- Cutler C. S., Hennkens H. M., Sisay N., Huclier-Markai S., Jurisson S. S. Chem. Rev. 2012;113:858–883. doi: 10.1021/cr3003104. [DOI] [PubMed] [Google Scholar]
- Ferreirós-Martínez R., Esteban-Gómez D., Platas-Iglesias C., de Blas A., Rodríguez-Blas T. Dalton Trans. 2008:5754–5765. doi: 10.1039/b808631a. [DOI] [PubMed] [Google Scholar]
- Boros E., Ferreira C. L., Cawthray J. F., Price E. W., Patrick B. O., Wester D. W., Adam M. J., Orvig C. J. Am. Chem. Soc. 2010;132:15726–15733. doi: 10.1021/ja106399h. [DOI] [PubMed] [Google Scholar]
- Stürzenbaum S. R., Hockner M., Panneerselvam A., Levitt J., Bouillard J. S., Taniguchi S., Dailey L. A., Ahmad Khanbeigi R., Rosca E. V., Thanou M., Suhling K., Zayats A. V., Green M. Nat. Nanotechnol. 2013;8:57–60. doi: 10.1038/nnano.2012.232. [DOI] [PubMed] [Google Scholar]
- Lo C., Ringenberg M. R., Gnandt D., Wilson Y., Ward T. R. Chem. Commun. 2011;47:12065–12067. doi: 10.1039/c1cc15004a. [DOI] [PubMed] [Google Scholar]
- Ward T. R. Acc. Chem. Res. 2011;44:47–57. doi: 10.1021/ar100099u. [DOI] [PubMed] [Google Scholar]
- Heinisch T., Ward T. R. Curr. Opin. Chem. Biol. 2010;14:184–199. doi: 10.1016/j.cbpa.2009.11.026. [DOI] [PubMed] [Google Scholar]
- You Y. Curr. Opin. Chem. Biol. 2013;17:699–707. doi: 10.1016/j.cbpa.2013.05.023. [DOI] [PubMed] [Google Scholar]
- Hyster T. K., Knorr L., Ward T. R., Rovis T. Science. 2012;338:500–503. doi: 10.1126/science.1226132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner S. V., Boyaci H., Chen H., Jensen M. P., He C. Angew. Chem., Int. Ed. 2009;48:2339–2341. doi: 10.1002/anie.200805262. [DOI] [PubMed] [Google Scholar]
- Zhou L., Bosscher M., Zhang C., özçubukçu S., Zhang L., Zhang W., Li C. J., Liu J., Jensen M. P., Lai L., He C. Nat. Chem. 2014;6:236–241. doi: 10.1038/nchem.1856. [DOI] [PubMed] [Google Scholar]
- Turner P., Mamo G., Karlsson E. N. Microb. Cell Fact. 2007;6:9. doi: 10.1186/1475-2859-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low P. S., Somero G. N. Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol. 1974;49:307–312. doi: 10.1016/0305-0491(74)90165-5. [DOI] [PubMed] [Google Scholar]
- Wintrode P. L., Arnold F. H. Adv. Protein Chem. 2000;55:161–225. doi: 10.1016/s0065-3233(01)55004-4. [DOI] [PubMed] [Google Scholar]
- Socha R. D., Tokuriki N. FEBS J. 2013;280:5582–5595. doi: 10.1111/febs.12354. [DOI] [PubMed] [Google Scholar]
- Gumulya Y., Reetz M. T. ChemBioChem. 2011;12:2502–2510. doi: 10.1002/cbic.201100412. [DOI] [PubMed] [Google Scholar]
- Chang R. L., Andrews K., Kim D., Li Z., Godzik A., Palsson B. Ø. Science. 2013;340:1220–1223. doi: 10.1126/science.1234012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M., Kostrewa D., Wyss M., Brugger R., D'Arcy A., Pasamontes L., van Loon A. Protein Eng. 2000;13:49–57. doi: 10.1093/protein/13.1.49. [DOI] [PubMed] [Google Scholar]
- Lehmann M., Pasamontes L., Lassen S. F., Wyss M. Biochim. Biophys. Acta. 2000;1543:408–415. doi: 10.1016/s0167-4838(00)00238-7. [DOI] [PubMed] [Google Scholar]
- Lehmann M., Wyss M. Curr. Opin. Biotechnol. 2001;12:371–375. doi: 10.1016/s0958-1669(00)00229-9. [DOI] [PubMed] [Google Scholar]
- Lehmann M., Loch C., Middendorf A., Studer D., Lassen S. F., Pasamontes L., van Loon A. P. G. M., Wyss M. Protein Eng. 2002;15:403–411. doi: 10.1093/protein/15.5.403. [DOI] [PubMed] [Google Scholar]
- Coleman R. G., Sharp K. A. Proteins. 2010;78:420–433. doi: 10.1002/prot.22558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokanson C. A., Cappuccilli G., Odineca T., Bozic M., Behnke C. A., Mendez M., Coleman W. J., Crea R. Protein Eng., Des. Sel. 2011;24:597–605. doi: 10.1093/protein/gzr028. [DOI] [PubMed] [Google Scholar]
- Aucamp J. P., Cosme A. M., Lye G. J., Dalby P. A. Biotechnol. Bioeng. 2005;89:599–607. doi: 10.1002/bit.20397. [DOI] [PubMed] [Google Scholar]
- Aucamp J. P., Martinez-Torres R. J., Hibbert E. G., Dalby P. A. Biotechnol. Bioeng. 2008;99:1303–1310. doi: 10.1002/bit.21705. [DOI] [PubMed] [Google Scholar]
- Schwab T., Sterner R. ChemBioChem. 2011;12:1581–1588. doi: 10.1002/cbic.201000770. [DOI] [PubMed] [Google Scholar]
- Pfleger C., Radestock S., Schmidt E., Gohlke H. J. Comput. Chem., Jpn. 2013;34:220–233. doi: 10.1002/jcc.23122. [DOI] [PubMed] [Google Scholar]
- Wintrode P. L., Zhang D., Vaidehi N., Arnold F. H., Goddard 3rd W. A. J. Mol. Biol. 2003;327:745–757. doi: 10.1016/s0022-2836(03)00147-5. [DOI] [PubMed] [Google Scholar]
- Kamerzell T. J., Middaugh C. R. J. Pharm. Sci. 2008;97:3494–3517. doi: 10.1002/jps.21269. [DOI] [PubMed] [Google Scholar]
- Bae E., Bannen R. M., Phillips G. N. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9594–9597. doi: 10.1073/pnas.0800938105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid F. X. ChemBioChem. 2011;12:1501–1507. doi: 10.1002/cbic.201100018. [DOI] [PubMed] [Google Scholar]
- Vázquez-Figueroa E., Chaparro-Riggers J., Bommarius A. S. ChemBioChem. 2007;8:2295–2301. doi: 10.1002/cbic.200700500. [DOI] [PubMed] [Google Scholar]
- Polizzi K. M., Chaparro-Riggers J. F., Vazquez-Figueroa E., Bommarius A. S. Biotechnol. J. 2006;1:531–536. doi: 10.1002/biot.200600029. [DOI] [PubMed] [Google Scholar]
- Wijma H. J., Floor R. J., Janssen D. B. Curr. Opin. Struct. Biol. 2013;23:588–594. doi: 10.1016/j.sbi.2013.04.008. [DOI] [PubMed] [Google Scholar]
- Vieille C., Burdette D. S., Zeikus J. G. Biotechnol. Annu. Rev. 1996;2:1–83. doi: 10.1016/s1387-2656(08)70006-1. [DOI] [PubMed] [Google Scholar]
- Zeikus J. G., Vieille C., Savchenko A. Extremophiles. 1998;2:179–183. doi: 10.1007/s007920050058. [DOI] [PubMed] [Google Scholar]
- Maheshwari R., Bharadwaj G., Bhat M. K. Microbiol. Mol. Biol. Rev. 2000;64:461–488. doi: 10.1128/mmbr.64.3.461-488.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriprapundh D., Vieille C., Zeikus J. G. Protein Eng. 2000;13:259–265. doi: 10.1093/protein/13.4.259. [DOI] [PubMed] [Google Scholar]
- Vieille C., Zeikus G. J. Microbiol. Mol. Biol. Rev. 2001;65:1–43. doi: 10.1128/MMBR.65.1.1-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruins M. E., Janssen A. E., Boom R. M. Appl. Biochem. Biotechnol. 2001;90:155–186. doi: 10.1385/abab:90:2:155. [DOI] [PubMed] [Google Scholar]
- Li W. F., Zhou X. X., Lu P. Biotechnol. Adv. 2005;23:271–281. doi: 10.1016/j.biotechadv.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Wu L. C., Lee J. X., Huang H. D., Liu B. J., Horng J. T. Expert Syst. Appl. 2009;36:668–674. [Google Scholar]
- Berezovsky I. N. Phys. Biol. 2011;8:035002. doi: 10.1088/1478-3975/8/3/035002. [DOI] [PubMed] [Google Scholar]
- Imanaka T. Proc. Jpn. Acad., Ser. B. 2011;87:587–602. doi: 10.2183/pjab.87.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsworth L. D., van der Oost J., Koutsopoulos S. FEBS J. 2007;274:4044–4056. doi: 10.1111/j.1742-4658.2007.05954.x. [DOI] [PubMed] [Google Scholar]
- Hashimoto H., Inoue T., Nishioka M., Fujiwara S., Takagi M., Imanaka T., Kai Y. J. Mol. Biol. 1999;292:707–716. doi: 10.1006/jmbi.1999.3100. [DOI] [PubMed] [Google Scholar]
- Matsui I., Harata K. FEBS J. 2007;274:4012–4022. doi: 10.1111/j.1742-4658.2007.05956.x. [DOI] [PubMed] [Google Scholar]
- Chan C. H., Liang H. K., Hsiao N. W., Ko M. T., Lyu P. C., Hwang J. K. Proteins. 2004;57:684–691. doi: 10.1002/prot.20263. [DOI] [PubMed] [Google Scholar]
- Greaves R. B., Warwicker J. Biochem. Biophys. Res. Commun. 2009;380:581–585. doi: 10.1016/j.bbrc.2009.01.145. [DOI] [PubMed] [Google Scholar]
- Rathi P. C., Radestock S., Gohlke H. J. Biotechnol. 2012;159:135–144. doi: 10.1016/j.jbiotec.2012.01.027. [DOI] [PubMed] [Google Scholar]
- Radestock S., Gohlke H. Proteins. 2011;79:1089–1108. doi: 10.1002/prot.22946. [DOI] [PubMed] [Google Scholar]
- Lin C. P., Huang S. W., Lai Y. L., Yen S. C., Shih C. H., Lu C. H., Huang C. C., Hwang J. K. Proteins: Struct., Funct., Bioinf. 2008;72:929–935. doi: 10.1002/prot.21983. [DOI] [PubMed] [Google Scholar]
- Blum J. K., Ricketts M. D., Bommarius A. S. J. Biotechnol. 2012;160:214–221. doi: 10.1016/j.jbiotec.2012.02.014. [DOI] [PubMed] [Google Scholar]
- Engen J. R. Anal. Chem. 2009;81:7870–7875. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltashov I. A., Bobst C. E., Abzalimov R. R. Anal. Chem. 2009;81:7892–7899. doi: 10.1021/ac901366n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esswein S. T., Florance H. V., Baillie L., Lippens J., Barran P. E. J. Chromatogr. A. 2010;1217:6709–6717. doi: 10.1016/j.chroma.2010.05.028. [DOI] [PubMed] [Google Scholar]
- Konermann L., Pan J., Liu Y. H. Chem. Soc. Rev. 2011;40:1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- Jurneczko E., Cruickshank F., Porrini M., Nikolova P., Campuzano I. D., Morris M., Barran P. E. Biochem. Soc. Trans. 2012;40:1021–1026. doi: 10.1042/BST20120125. [DOI] [PubMed] [Google Scholar]
- Nakazawa S., Ahn J., Hashii N., Hirose K., Kawasaki N. Biochim. Biophys. Acta. 2013;1834:1210–1214. doi: 10.1016/j.bbapap.2012.11.012. [DOI] [PubMed] [Google Scholar]
- Resetca D., Wilson D. J. FEBS J. 2013;280:5616–5625. doi: 10.1111/febs.12332. [DOI] [PubMed] [Google Scholar]
- Goswami D., Callaway C., Pascal B. D., Kumar R., Edwards D. P., Griffin P. R. Structure. 2014;22:961–973. doi: 10.1016/j.str.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciano D. P., Dharmarajan V., Griffin P. R. Curr. Opin. Struct. Biol. 2014;28C:105–111. doi: 10.1016/j.sbi.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger C., Rathi P. C., Klein D. L., Radestock S., Gohlke H. J. Chem. Inf. Model. 2013;53:1007–1015. doi: 10.1021/ci400044m. [DOI] [PubMed] [Google Scholar]
- Buer B. C., Levin B. J., Marsh E. N. G. J. Am. Chem. Soc. 2012;134:13027–13034. doi: 10.1021/ja303521h. [DOI] [PubMed] [Google Scholar]
- Buer B. C., Marsh E. N. G. Protein Sci. 2012;21:453–462. doi: 10.1002/pro.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer B. C., Meagher J. L., Stuckey J. A., Marsh E. N. G. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4810–4815. doi: 10.1073/pnas.1120112109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh K. H., Nam S. H., Kim H. S. Protein Eng. 2002;15:689–695. doi: 10.1093/protein/15.8.689. [DOI] [PubMed] [Google Scholar]
- Oh K. H., Nam S. H., Kim H. S. Biotechnol. Prog. 2002;18:413–417. doi: 10.1021/bp0101942. [DOI] [PubMed] [Google Scholar]
- Vázquez-Figueroa E., Yeh V., Broering J. M., Chaparro-Riggers J. F., Bommarius A. S. Protein Eng., Des. Sel. 2008;21:673–680. doi: 10.1093/protein/gzn048. [DOI] [PubMed] [Google Scholar]
- Dobson P. D., Kell D. B. Nat. Rev. Drug Discovery. 2008;7:205–220. doi: 10.1038/nrd2438. [DOI] [PubMed] [Google Scholar]
- Dobson P., Lanthaler K., Oliver S. G., Kell D. B. Curr. Top. Med. Chem. 2009;9:163–184. doi: 10.2174/156802609787521616. [DOI] [PubMed] [Google Scholar]
- Kell D. B., Dobson P. D., Oliver S. G. Drug Discovery Today. 2011;16:704–714. doi: 10.1016/j.drudis.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Kell D. B., Goodacre R. Drug Discovery Today. 2014;19:171–182. doi: 10.1016/j.drudis.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter G. J., Kell D. B. CRC Crit. Rev. Biotechnol. 1995;15:139–177. doi: 10.3109/07388559509147404. [DOI] [PubMed] [Google Scholar]
- Wescott C. R., Klibanov A. M. J. Am. Chem. Soc. 1993;115:10362–10363. [Google Scholar]
- Wescott C. R., Klibanov A. M. Biochim. Biophys. Acta. 1994;1206:1–9. doi: 10.1016/0167-4838(94)90065-5. [DOI] [PubMed] [Google Scholar]
- Carrea G., Ottolina G., Riva S. Trends Biotechnol. 1995;13:63–70. [Google Scholar]
- Sardessai Y., Bhosle S. Res. Microbiol. 2002;153:263–268. doi: 10.1016/s0923-2508(02)01319-0. [DOI] [PubMed] [Google Scholar]
- Sardessai Y. N., Bhosle S. Biotechnol. Prog. 2004;20:655–660. doi: 10.1021/bp0200595. [DOI] [PubMed] [Google Scholar]
- Liu C., Yang G., Wu L., Tian G., Zhang Z., Feng Y. Protein Cell. 2011;2:497–506. doi: 10.1007/s13238-011-1057-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halling P. J. Biotechnol. Bioeng. 1990;35:691–701. doi: 10.1002/bit.260350706. [DOI] [PubMed] [Google Scholar]
- Xu K., Griebenow K., Klibanov A. M. Biotechnol. Bioeng. 1997;56:485–491. doi: 10.1002/(SICI)1097-0290(19971205)56:5<485::AID-BIT2>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Gupta M. N., Roy I. Eur. J. Biochem. 2004;271:2575–2583. doi: 10.1111/j.1432-1033.2004.04163.x. [DOI] [PubMed] [Google Scholar]
- Nordwald E. M., Kaar J. L. Biotechnol. Bioeng. 2013;110:2352–2360. doi: 10.1002/bit.24910. [DOI] [PubMed] [Google Scholar]
- Nordwald E. M., Kaar J. L. J. Phys. Chem. B. 2013;117:8977–8986. doi: 10.1021/jp404760w. [DOI] [PubMed] [Google Scholar]
- Maugeri Z., Leitner W., de Maria P. D. Tetrahedron Lett. 2012;53:6968–6971. [Google Scholar]
- Maugeri Z., de Maria P. D. RSC Adv. 2012;2:421–425. [Google Scholar]
- Domínguez de María P., Maugeri Z. Curr. Opin. Chem. Biol. 2011;15:220–225. doi: 10.1016/j.cbpa.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Zhang Q. H., Vigier K. D., Royer S., Jérôme F. Chem. Soc. Rev. 2012;41:7108–7146. doi: 10.1039/c2cs35178a. [DOI] [PubMed] [Google Scholar]
- Cadeddu A., Wylie E. K., Jurczak J., Wampler-Doty M., Grzybowski B. A. Angew. Chem., Int. Ed. 2014;53:8108–8112. doi: 10.1002/anie.201403708. [DOI] [PubMed] [Google Scholar]
- Carbonell P., Planson A. G., Fichera D., Faulon J. L. BMC Syst. Biol. 2011;5:122. doi: 10.1186/1752-0509-5-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell P., Planson A. G., Faulon J. L. Methods Mol. Biol. 2013;985:149–173. doi: 10.1007/978-1-62703-299-5_9. [DOI] [PubMed] [Google Scholar]
- Huang Q., Li L. L., Yang S. Y. J. Chem. Inf. Model. 2011;51:2768–2777. doi: 10.1021/ci100216g. [DOI] [PubMed] [Google Scholar]
- Law J., Zsoldos Z., Simon A., Reid D., Liu Y., Khew S. Y., Johnson A. P., Major S., Wade R. A., Ando H. Y. J. Chem. Inf. Model. 2009;49:593–602. doi: 10.1021/ci800228y. [DOI] [PubMed] [Google Scholar]
- Lewell X. Q., Judd D. B., Watson S. P., Hann M. M. J. Chem. Inf. Comput. Sci. 1998;38:511–522. doi: 10.1021/ci970429i. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Lergier J., Broadbelt L. J., Hatzimanikatis V. J. Am. Chem. Soc. 2005;127:9930–9938. doi: 10.1021/ja051586y. [DOI] [PubMed] [Google Scholar]
- Hatzimanikatis V., Li C., Ionita J. A., Henry C. S., Jankowski M. D., Broadbelt L. J. Bioinformatics. 2005;21:1603–1609. doi: 10.1093/bioinformatics/bti213. [DOI] [PubMed] [Google Scholar]
- Henry C. S., Broadbelt L. J., Hatzimanikatis V. Biotechnol. Bioeng. 2010;106:462–473. doi: 10.1002/bit.22673. [DOI] [PubMed] [Google Scholar]
- Soh K. C., Hatzimanikatis V. Trends Biotechnol. 2010;28:501–508. doi: 10.1016/j.tibtech.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Planson A. G., Carbonell P., Grigoras I., Faulon J. L. Curr. Opin. Biotechnol. 2012;23:948–956. doi: 10.1016/j.copbio.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Turner N. J., O'Reilly E. Nat. Chem. Biol. 2013;9:285–288. doi: 10.1038/nchembio.1235. [DOI] [PubMed] [Google Scholar]
- Campodonico M. A., Andrews B. A., Asenjo J. A., Palsson B. O., Feist A. M. GEM-Path, Metab Eng. 2014;25:140–158. doi: 10.1016/j.ymben.2014.07.009. [DOI] [PubMed] [Google Scholar]
- Bishop K. J., Klajn R., Grzybowski B. A. Angew. Chem., Int. Ed. 2006;45:5348–5354. doi: 10.1002/anie.200600881. [DOI] [PubMed] [Google Scholar]
- Fialkowski M., Bishop K. J., Chubukov V. A., Campbell C. J., Grzybowski B. A. Angew. Chem., Int. Ed. 2005;44:7263–7269. doi: 10.1002/anie.200502272. [DOI] [PubMed] [Google Scholar]
- Ghislieri D., Green A. P., Pontini M., Willies S. C., Rowles I., Frank A., Grogan G., Turner N. J. J. Am. Chem. Soc. 2013;135:10863–10869. doi: 10.1021/ja4051235. [DOI] [PubMed] [Google Scholar]
- Whited G. M., Feher F. J., Benko D. A., Cervin M. A., Chotani G. K., McAuliffe J. C., LaDuca R. J., Ben-Shoshan E. A., Sanford K. J. Ind. Biotechnol. 2010;6:152–163. [Google Scholar]
- DeSantis G., Wong K., Farwell B., Chatman K., Zhu Z., Tomlinson G., Huang H., Tan X., Bibbs L., Chen P., Kretz K., Burk M. J. J. Am. Chem. Soc. 2003;125:11476–11477. doi: 10.1021/ja035742h. [DOI] [PubMed] [Google Scholar]
- Bommarius A. S., Blum J. K., Abrahamson M. J. Curr. Opin. Chem. Biol. 2011;15:194–200. doi: 10.1016/j.cbpa.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Faber K., Biotransformations in organic chemistry. A textbook., Springer, Berlin, 2011. [Google Scholar]
- Patel R. N. ACS Catal. 2011;1:1056–1074. [Google Scholar]
- Schrewe M., Julsing M. K., Buhler B., Schmid A. Chem. Soc. Rev. 2013;42:6346–6377. doi: 10.1039/c3cs60011d. [DOI] [PubMed] [Google Scholar]
- Simon R. C., Mutti F. G., Kroutil W. Drug Discovery Today: Technol. 2013;10:e37–44. doi: 10.1016/j.ddtec.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Wang M., Si T., Zhao H. Bioresour. Technol. 2012;115:117–125. doi: 10.1016/j.biortech.2012.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhan N., Xu P., Koffas M. A. G. Curr. Opin. Biotechnol. 2013;24:1137–1143. doi: 10.1016/j.copbio.2013.02.019. [DOI] [PubMed] [Google Scholar]
- Huisman G. W., Collier S. J. Curr. Opin. Chem. Biol. 2013;17:284–292. doi: 10.1016/j.cbpa.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Nestl B. M., Hammer S. C., Nebel B. A., Hauer B. Angew. Chem., Int. Ed. 2014;53:3070–3095. doi: 10.1002/anie.201302195. [DOI] [PubMed] [Google Scholar]
- Bolt A., Berry A., Nelson A. Arch. Biochem. Biophys. 2008;474:318–330. doi: 10.1016/j.abb.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windle C. L., Muller M., Nelson A., Berry A. Curr. Opin. Chem. Biol. 2014;19:25–33. doi: 10.1016/j.cbpa.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okrasa K., Levy C., Wilding M., Goodall M., Baudendistel N., Hauer B., Leys D., Micklefield J. Angew. Chem., Int. Ed. 2009;48:7691–7694. doi: 10.1002/anie.200904112. [DOI] [PubMed] [Google Scholar]
- Abrahamson M. J., Vazquez-Figueroa E., Woodall N. B., Moore J. C., Bommarius A. S. Angew. Chem., Int. Ed. 2012;51:3969–3972. doi: 10.1002/anie.201107813. [DOI] [PubMed] [Google Scholar]
- Abrahamson M. J., Wong J. W., Bommarius A. S. Adv. Synth. Catal. 2013;355:1780–1786. [Google Scholar]
- Sattler J. H., Fuchs M., Tauber K., Mutti F. G., Faber K., Pfeffer J., Haas T., Kroutil W. Angew. Chem., Int. Ed. 2012;51:9156–9159. doi: 10.1002/anie.201204683. [DOI] [PubMed] [Google Scholar]
- Kohls H., Steffen-Munsberg F., Höhne M. Curr. Opin. Chem. Biol. 2014;19:180–192. doi: 10.1016/j.cbpa.2014.02.021. [DOI] [PubMed] [Google Scholar]
- Meister K., Ebbinghaus S., Xu Y., Duman J. G., Devries A., Gruebele M., Leitner D. M., Havenith M. Proc. Natl. Acad. Sci. U. S. A. 2013;110:1617–1622. doi: 10.1073/pnas.1214911110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews M. L., Chang W. C., Layne A. P., Miles L. A., Krebs C., Bollinger, Jr. J. M. Nat. Chem. Biol. 2014;10:209–215. doi: 10.1038/nchembio.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brustad E. M. Nat. Chem. Biol. 2014;10:170–171. doi: 10.1038/nchembio.1457. [DOI] [PubMed] [Google Scholar]
- Zhang Z. G., Parra L. P., Reetz M. T. Chemistry. 2012;18:10160–10172. doi: 10.1002/chem.201202163. [DOI] [PubMed] [Google Scholar]
- Polyak I., Reetz M. T., Thiel W. J. Phys. Chem. B. 2013;117:4993–5001. doi: 10.1021/jp4018019. [DOI] [PubMed] [Google Scholar]
- Wells, Jr. T., Ragauskas A. J. Trends Biotechnol. 2012;30:627–637. doi: 10.1016/j.tibtech.2012.09.008. [DOI] [PubMed] [Google Scholar]
- Schmidt-Dannert C., Umeno D., Arnold F. H. Nat. Biotechnol. 2000;18:750–753. doi: 10.1038/77319. [DOI] [PubMed] [Google Scholar]
- Butler A., Sandy M. Nature. 2009;460:848–854. doi: 10.1038/nature08303. [DOI] [PubMed] [Google Scholar]
- Deng H., O'Hagan D. Curr. Opin. Chem. Biol. 2008;12:582–592. doi: 10.1016/j.cbpa.2008.06.036. [DOI] [PubMed] [Google Scholar]
- Gribble G. W. Prog. Chem. Org. Nat. Prod. 2010;91:349–365. doi: 10.1007/978-3-211-99323-1_4. [DOI] [PubMed] [Google Scholar]
- Runguphan W., Qu X., O'Connor S. E. Nature. 2010;468:461–464. doi: 10.1038/nature09524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Duhalt R., Ayala M., Márquez-Rocha F. J. Phytochemistry. 2001;58:929–933. doi: 10.1016/s0031-9422(01)00326-0. [DOI] [PubMed] [Google Scholar]
- De Mot R., De Schrijver A., Schoofs G., Parret A. H. FEMS Microbiol. Lett. 2003;224:197–203. doi: 10.1016/S0378-1097(03)00452-X. [DOI] [PubMed] [Google Scholar]
- Hasan Z., Renirie R., Kerkman R., Ruijssenaars H. J., Hartog A. F., Wever R. J. Biol. Chem. 2006;281:9738–9744. doi: 10.1074/jbc.M512166200. [DOI] [PubMed] [Google Scholar]
- Hofrichter M., Ullrich R. Appl. Microbiol. Biotechnol. 2006;71:276–288. doi: 10.1007/s00253-006-0417-3. [DOI] [PubMed] [Google Scholar]
- Winter J. M., Moore B. S. J. Biol. Chem. 2009;284:18577–18581. doi: 10.1074/jbc.R109.001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofrichter M., Ullrich R., Pecyna M. J., Liers C., Lundell T. Appl. Microbiol. Biotechnol. 2010;87:871–897. doi: 10.1007/s00253-010-2633-0. [DOI] [PubMed] [Google Scholar]
- Bernhardt P., Okino T., Winter J. M., Miyanaga A., Moore B. S. J. Am. Chem. Soc. 2011;133:4268–4270. doi: 10.1021/ja201088k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otey C. R., Landwehr M., Endelman J. B., Hiraga K., Bloom J. D., Arnold F. H. PLoS Biol. 2006;4:e112. doi: 10.1371/journal.pbio.0040112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S. L., Kelly D. E. Philos. Trans. R. Soc. London, Ser. B. 2013;368:20120476. doi: 10.1098/rstb.2012.0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavira C., Höfer R., Lesot A., Lambert F., Zucca J., Werck-Reichhart D. Metab. Eng. 2013;18:25–35. doi: 10.1016/j.ymben.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Lamb D. C., Waterman M. R. Philos. Trans. R. Soc. London, Ser. B. 2013;368:20120434. doi: 10.1098/rstb.2012.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell J. M., O'Neill M., Taylor S. J. C., Moody T. S. Curr. Opin. Chem. Biol. 2013;17:271–275. doi: 10.1016/j.cbpa.2013.01.028. [DOI] [PubMed] [Google Scholar]
- Roberts G. A., Celik A., Hunter D. J. B., Ost T. W. B., White J. H., Chapman S. K., Turner N. J., Flitsch S. L. J. Biol. Chem. 2003;278:48914–48920. doi: 10.1074/jbc.M309630200. [DOI] [PubMed] [Google Scholar]
- Hunter D. J. B., Roberts G. A., Ost T. W. B., White J. H., Muller S., Turner N. J., Flitsch S. L., Chapman S. K. FEBS Lett. 2005;579:2215–2220. doi: 10.1016/j.febslet.2005.03.016. [DOI] [PubMed] [Google Scholar]
- O'Reilly E., Corbett M., Hussain S., Kelly P. P., Richardson D., Flitsch S. L., Turner N. J. Catal. Sci. Technol. 2013;3:1490–1492. [Google Scholar]
- O'Reilly E., Aitken S. J., Grogan G., Kelly P. P., Turner N. J., Flitsch S. L. Beilstein J. Org. Chem. 2012;8:496–500. doi: 10.3762/bjoc.8.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin A., Kohler V., Jones A., Ali A., Kelly P. P., O'Reilly E., Turner N. J., Flitsch S. L. Beilstein J. Org. Chem. 2011;7:1494–1498. doi: 10.3762/bjoc.7.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin A., Roberts G. A., Kisch J., Sabbadin F., Grogan G., Bruce N., Turner N. J., Flitsch S. L. Chem. Commun. 2009:2478–2480. doi: 10.1039/b901716j. [DOI] [PubMed] [Google Scholar]
- Fasan R., Chen M. M., Crook N. C., Arnold F. H. Angew. Chem., Int. Ed. 2007;46:8414–8418. doi: 10.1002/anie.200702616. [DOI] [PubMed] [Google Scholar]
- Trefzer A., Jungmann V., Molnar I., Botejue A., Buckel D., Frey G., Hill D. S., Jorg M., Ligon J. M., Mason D., Moore D., Pachlatko J. P., Richardson T. H., Spangenberg P., Wall M. A., Zirkle R., Stege J. T. Appl. Environ. Microbiol. 2007;73:4317–4325. doi: 10.1128/AEM.02676-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. C., Mantovani S. M., Fu Y., Snow C. D., Komor R. S., Wong C. H., Arnold F. H. ChemBioChem. 2010;11:2502–2505. doi: 10.1002/cbic.201000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlacher V. B., Girhard M. Trends Biotechnol. 2012;30:26–36. doi: 10.1016/j.tibtech.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Seifert A., Antonovici M., Hauer B., Pleiss J. ChemBioChem. 2011;12:1346–1351. doi: 10.1002/cbic.201100067. [DOI] [PubMed] [Google Scholar]
- Zilly F. E., Acevedo J. P., Augustyniak W., Deege A., Hausig U. W., Reetz M. T. Angew. Chem., Int. Ed. 2011;50:2720–2724. doi: 10.1002/anie.201006587. [DOI] [PubMed] [Google Scholar]
- Jung S. T., Lauchli R., Arnold F. H. Curr. Opin. Biotechnol. 2011;22:809–817. doi: 10.1016/j.copbio.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roiban G. D., Reetz M. T. Angew. Chem., Int. Ed. 2013;52:5439–5440. doi: 10.1002/anie.201301083. [DOI] [PubMed] [Google Scholar]
- Jiang D., Tu R., Bai P., Wang Q. Biotechnol. Lett. 2013;35:1663–1668. doi: 10.1007/s10529-013-1254-y. [DOI] [PubMed] [Google Scholar]
- Roiban G. D., Agudo R., Ilie A., Lonsdale R., Reetz M. T. Chem. Commun. 2014;50:14310–14313. doi: 10.1039/c4cc04925j. [DOI] [PubMed] [Google Scholar]
- Kim H. J., Ruszczycky M. W., Choi S. H., Liu Y. N., Liu H. W. Nature. 2011;473:109–112. doi: 10.1038/nature09981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend C. A. ChemBioChem. 2011;12:2267–2269. doi: 10.1002/cbic.201100431. [DOI] [PubMed] [Google Scholar]
- Obeid S., Schnur A., Gloeckner C., Blatter N., Welte W., Diederichs K., Marx A. ChemBioChem. 2011;12:1574–1580. doi: 10.1002/cbic.201000783. [DOI] [PubMed] [Google Scholar]
- Loakes D., Gallego J., Pinheiro V. B., Kool E. T., Holliger P. J. Am. Chem. Soc. 2009;131:14827–14837. doi: 10.1021/ja9039696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Morley K. L., Horsman G. P., Holmquist M., Hult K., Kazlauskas R. J. Chem. Biol. 2005;12:45–54. doi: 10.1016/j.chembiol.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Fowler Z. L., Koffas M. A. G. Appl. Microbiol. Biotechnol. 2009;83:799–808. doi: 10.1007/s00253-009-2039-z. [DOI] [PubMed] [Google Scholar]
- Fowler Z. L., Shah K., Panepinto J. C., Jacobs A., Koffas M. A. G. PLoS One. 2011;6:e25681. doi: 10.1371/journal.pone.0025681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler Z. L., Gikandi W. W., Koffas M. A. G. Appl. Environ. Microbiol. 2009;75:5831–5839. doi: 10.1128/AEM.00270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard E., Yan Y., Fowler Z. L., Li Z., Lim C. G., Lim K. H., Koffas M. A. G. Mol. Pharmaceutics. 2008;5:257–265. doi: 10.1021/mp7001472. [DOI] [PubMed] [Google Scholar]
- Xu P., Ranganathan S., Fowler Z. L., Maranas C. D., Koffas M. A. Metab. Eng. 2011;13:578–587. doi: 10.1016/j.ymben.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Mora-Pale M., Sanchez-Rodriguez S. P., Linhardt R. J., Dordick J. S., Koffas M. A. G. Plant Sci. 2013;210:10–24. doi: 10.1016/j.plantsci.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Wu C. X., Liu R., Gao M., Zhao G., Wu S., Wu C. F., Du G. H. Neurosci. Lett. 2013;546:57–62. doi: 10.1016/j.neulet.2013.04.060. [DOI] [PubMed] [Google Scholar]
- Wu J., Du G., Zhou J., Chen J. Metab. Eng. 2013;16:48–55. doi: 10.1016/j.ymben.2012.11.009. [DOI] [PubMed] [Google Scholar]
- Deng H., Cross S. M., McGlinchey R. P., Hamilton J. T., O'Hagan D. Chem. Biol. 2008;15:1268–1276. doi: 10.1016/j.chembiol.2008.10.012. [DOI] [PubMed] [Google Scholar]
- Liu W., Huang X., Cheng M. J., Nielsen R. J., Goddard, 3rd W. A., Groves J. T. Science. 2012;337:1322–1325. doi: 10.1126/science.1222327. [DOI] [PubMed] [Google Scholar]
- Lennen R. M., Politz M. G., Kruziki M. A., Pfleger B. F. J. Bacteriol. 2013;195:135–144. doi: 10.1128/JB.01477-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennen R. M., Pfleger B. F. Trends Biotechnol. 2012;30:659–667. doi: 10.1016/j.tibtech.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngquist J. T., Lennen R. M., Ranatunga D. R., Bothfeld W. H., Marner 2nd W. D., Pfleger B. F. Biotechnol. Bioeng. 2012;109:1518–1527. doi: 10.1002/bit.24420. [DOI] [PubMed] [Google Scholar]
- Castle L. A., Siehl D. L., Gorton R., Patten P. A., Chen Y. H., Bertain S., Cho H. J., Duck N., Wong J., Liu D., Lassner M. W. Science. 2004;304:1151–1154. doi: 10.1126/science.1096770. [DOI] [PubMed] [Google Scholar]
- Siehl D. L., Castle L. A., Gorton R., Chen Y. H., Bertain S., Cho H. J., Keenan R., Liu D., Lassner M. W. Pest Manage. Sci. 2005;61:235–240. doi: 10.1002/ps.1014. [DOI] [PubMed] [Google Scholar]
- Pollegioni L., Schonbrunn E., Siehl D. FEBS J. 2011;278:2753–2766. doi: 10.1111/j.1742-4658.2011.08214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedotti M., Rosini E., Molla G., Moschetti T., Savino C., Vallone B., Pollegioni L. J. Biol. Chem. 2009;284:36415–36423. doi: 10.1074/jbc.M109.051631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan T., Zhang K., Chen Y., Lin Y., Wu G., Zhang L., Yao P., Shao Z., Liu Z. PLoS One. 2013;8:e79175. doi: 10.1371/journal.pone.0079175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokop Z., Sato Y., Brezovsky J., Mozga T., Chaloupkova R., Koudelakova T., Jerabek P., Stepankova V., Natsume R., van Leeuwen J. G., Janssen D. B., Florian J., Nagata Y., Senda T., Damborsky J. Angew. Chem., Int. Ed. 2010;49:6111–6115. doi: 10.1002/anie.201001753. [DOI] [PubMed] [Google Scholar]
- Koudelakova T., Chovancova E., Brezovsky J., Monincova M., Fortova A., Jarkovsky J., Damborsky J. Biochem. J. 2011;435:345–354. doi: 10.1042/BJ20101405. [DOI] [PubMed] [Google Scholar]
- van Leeuwen J. G. E., Wijma H. J., Floor R. J., van der Laan J. M., Janssen D. B. ChemBioChem. 2012;13:137–148. doi: 10.1002/cbic.201100579. [DOI] [PubMed] [Google Scholar]
- Floor R. J., Wijma H. J., Colpa D. I., Ramos-Silva A., Jekel P. A., Szymanski W., Feringa B. L., Marrink S. J., Janssen D. B. ChemBioChem. 2014;15:1660–1672. doi: 10.1002/cbic.201402128. [DOI] [PubMed] [Google Scholar]
- Glenn W. S., Nims E., O'Connor S. E. J. Am. Chem. Soc. 2011;133:19346–19349. doi: 10.1021/ja2089348. [DOI] [PubMed] [Google Scholar]
- Herrera-Rodriguez L. N., Meyer H. P., Robins K. T., Khan F. Chim. Oggi. 2011;29:47–49. [Google Scholar]
- Herrera-Rodriguez L. N., Khan F., Robins K. T., Meyer H. P. Chim Oggi. 2011;29:31–33. [Google Scholar]
- Wong S. D., Srnec M., Matthews M. L., Liu L. V., Kwak Y., Park K., Bell 3rd C. B., Alp E. E., Zhao J., Yoda Y., Kitao S., Seto M., Krebs C., Bollinger, Jr. J. M., Solomon E. I. Nature. 2013;499:320–323. doi: 10.1038/nature12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernath-Levin K., Shainsky J., Sigawi L., Fishman A. Appl. Microbiol. Biotechnol. 2014;98:4975–4985. doi: 10.1007/s00253-013-5505-6. [DOI] [PubMed] [Google Scholar]
- Khersonsky O., Röthlisberger D., Dym O., Albeck S., Jackson C. J., Baker D., Tawfik D. S. J. Mol. Biol. 2010;396:1025–1042. doi: 10.1016/j.jmb.2009.12.031. [DOI] [PubMed] [Google Scholar]
- Khersonsky O., Röthlisberger D., Wollacott A. M., Murphy P., Dym O., Albeck S., Kiss G., Houk K. N., Baker D., Tawfik D. S. J. Mol. Biol. 2010;407:391–412. doi: 10.1016/j.jmb.2011.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg R., Kries H., Pinkas D. M., Mittl P. R., Grutter M. G., Privett H. K., Mayo S. L., Hilvert D. Nature. 2013;503:418–421. doi: 10.1038/nature12623. [DOI] [PubMed] [Google Scholar]
- Labas A., Szabo E., Mones L., Fuxreiter M. Biochim. Biophys. Acta. 2013;1834:908–917. doi: 10.1016/j.bbapap.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Khersonsky O., Kiss G., Rothlisberger D., Dym O., Albeck S., Houk K. N., Baker D., Tawfik D. S. Proc. Natl. Acad. Sci. U. S. A. 2012;109:10358–10363. doi: 10.1073/pnas.1121063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frushicheva M. P., Cao J., Warshel A. Biochemistry. 2011;50:3849–3858. doi: 10.1021/bi200063a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frushicheva M. P., Cao J., Chu Z. T., Warshel A. Proc. Natl. Acad. Sci. U. S. A. 2010;107:16869–16874. doi: 10.1073/pnas.1010381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. C., Pollard D. J., Kosjek B., Devine P. N. Acc. Chem. Res. 2007;40:1412–1419. doi: 10.1021/ar700167a. [DOI] [PubMed] [Google Scholar]
- Agudo R., Roiban G. D., Reetz M. T. J. Am. Chem. Soc. 2013;135:1665–1668. doi: 10.1021/ja3092517. [DOI] [PubMed] [Google Scholar]
- Camarero S., Pardo I., Cañas A. I., Molina P., Record E., Martínez A. T., Martínez M. J., Alcalde M. Appl. Environ. Microbiol. 2012;78:1370–1384. doi: 10.1128/AEM.07530-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J. R., Chang Y. S. Trends Biotechnol. 2013;31:335–341. doi: 10.1016/j.tibtech.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Mate D. M., Gonzalez-Perez D., Falk M., Kittl R., Pita M., De Lacey A. L., Ludwig R., Shleev S., Alcalde M. Chem. Biol. 2013;20:223–231. doi: 10.1016/j.chembiol.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Miao Y., Geertsema E. M., Tepper P. G., Zandvoort E., Poelarends G. J. ChemBioChem. 2013;14:191–194. doi: 10.1002/cbic.201200676. [DOI] [PubMed] [Google Scholar]
- Atkin K. E., Reiss R., Koehler V., Bailey K. R., Hart S., Turkenburg J. P., Turner N. J., Brzozowski A. M., Grogan G. J. Mol. Biol. 2008;384:1218–1231. doi: 10.1016/j.jmb.2008.09.090. [DOI] [PubMed] [Google Scholar]
- Bailey K. R., Ellis A. J., Reiss R., Snape T. J., Turner N. J. Chem. Commun. 2007:3640–3642. doi: 10.1039/b710456a. [DOI] [PubMed] [Google Scholar]
- Rowles I., Malone K. J., Etchells L. L., Willies S. C., Turner N. J. ChemCatChem. 2012;4:1259–1261. [Google Scholar]
- Anderson J. S., Rittle J., Peters J. C. Nature. 2013;501:84–87. doi: 10.1038/nature12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingoud A., Wende W. ChemBioChem. 2011;12:1495–1500. doi: 10.1002/cbic.201100055. [DOI] [PubMed] [Google Scholar]
- Toogood H. S., Scrutton N. S. Catal. Sci. Technol. 2013;3:2182–2194. doi: 10.1039/c5cy01642h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toogood H. S., Scrutton N. S. Curr. Opin. Chem. Biol. 2014;19:107–115. doi: 10.1016/j.cbpa.2014.01.019. [DOI] [PubMed] [Google Scholar]
- Ashani Y., Gupta R. D., Goldsmith M., Silman I., Sussman J. L., Tawfik D. S., Leader H. Chem.-Biol. Interact. 2010;187:362–369. doi: 10.1016/j.cbi.2010.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcolombri U., Elias M., Tawfik D. S. J. Mol. Biol. 2011;411:837–853. doi: 10.1016/j.jmb.2011.06.037. [DOI] [PubMed] [Google Scholar]
- Gupta R. D., Goldsmith M., Ashani Y., Simo Y., Mullokandov G., Bar H., Ben-David M., Leader H., Margalit R., Silman I., Sussman J. L., Tawfik D. S. Nat. Chem. Biol. 2011;7:120–125. doi: 10.1038/nchembio.510. [DOI] [PubMed] [Google Scholar]
- Garcia-Ruiz E., Gonzalez-Perez D., Ruiz-Dueñas F. J., Martínez A. T., Alcalde M. Biochem. J. 2012;441:487–498. doi: 10.1042/BJ20111199. [DOI] [PubMed] [Google Scholar]
- Patel S. C., Hecht M. H. Protein Eng., Des. Sel. 2012;25:445–452. doi: 10.1093/protein/gzs025. [DOI] [PubMed] [Google Scholar]
- Olguin L. F., Askew S. E., O'Donoghue A. C., Hollfelder F. J. Am. Chem. Soc. 2008;130:16547–16555. doi: 10.1021/ja8047943. [DOI] [PubMed] [Google Scholar]
- Babtie A. C., Bandyopadhyay S., Olguin L. F., Hollfelder F. Angew. Chem., Int. Ed. 2009;48:3692–3694. doi: 10.1002/anie.200805843. [DOI] [PubMed] [Google Scholar]
- van Loo B., Jonas S., Babtie A. C., Benjdia A., Berteau O., Hyvonen M., Hollfelder F. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2740–2745. doi: 10.1073/pnas.0903951107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afriat-Jurnou L., Jackson C. J., Tawfik D. S. Biochemistry. 2012;51:6047–6055. doi: 10.1021/bi300694t. [DOI] [PubMed] [Google Scholar]
- Mohamed M. F., Hollfelder F. Biochim. Biophys. Acta. 2013;1834:417–424. doi: 10.1016/j.bbapap.2012.07.015. [DOI] [PubMed] [Google Scholar]
- Wiersma-Koch H., Sunden F., Herschlag D. Biochemistry. 2013;52:9167–9176. doi: 10.1021/bi4010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Khosla C. Mol. Diversity. 1996;1:121–124. doi: 10.1007/BF01721327. [DOI] [PubMed] [Google Scholar]
- Ma S. M., Li J. W., Choi J. W., Zhou H., Lee K. K., Moorthie V. A., Xie X., Kealey J. T., Da Silva N. A., Vederas J. C., Tang Y. Science. 2009;326:589–592. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha W., Rubin-Pitel S. B., Zhao H. Mol. BioSyst. 2008;4:246–248. doi: 10.1039/b717705d. [DOI] [PubMed] [Google Scholar]
- Lee H. Y., Harvey C. J., Cane D. E., Khosla C. J. Antibiot. 2011;64:59–64. doi: 10.1038/ja.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T. H., Kim T. W., Kang H. O., Lee S. H., Lee E. J., Lim S. C., Oh S. O., Song A. J., Park S. J., Lee S. Y. Biotechnol. Bioeng. 2010;105:150–160. doi: 10.1002/bit.22547. [DOI] [PubMed] [Google Scholar]
- Hollmann F., Arends I. W. C. E., Buehler K. ChemCatChem. 2010;2:762–782. [Google Scholar]
- Hollmann F., Arends I. W. C. E., Buehler K., Schallmey A., Buhler B. Green Chem. 2011;13:226–265. [Google Scholar]
- Hollmann F., Arends I. W. C. E., Holtmann D. Green Chem. 2011;13:2285–2314. [Google Scholar]
- Geu-Flores F., Sherden N. H., Courdavault V., Burlat V., Glenn W. S., Wu C., Nims E., Cui Y., O'Connor S. E. Nature. 2012;492:138–142. doi: 10.1038/nature11692. [DOI] [PubMed] [Google Scholar]
- Schöpfel M., Tziridis A., Arnold U., Stubbs M. T. ChemBioChem. 2011;12:1523–1527. doi: 10.1002/cbic.201000787. [DOI] [PubMed] [Google Scholar]
- Obexer R., Studer S., Giger L., Pinkas D. M., Grütter M. G., Baker D., Hilvert D. ChemCatChem. 2014;6:1043–1050. [Google Scholar]
- Wymore T., Chen B. Y., Nicholas H. B., Ropelewski A. J., Brooks C. L. Mol. Inf. 2011;30:896–906. doi: 10.1002/minf.201100087. [DOI] [PubMed] [Google Scholar]
- Baas B. J., Zandvoort E., Geertsema E. M., Poelarends G. J. ChemBioChem. 2013;14:917–926. doi: 10.1002/cbic.201300098. [DOI] [PubMed] [Google Scholar]
- Diaz J. E., Lin C. S., Kunishiro K., Feld B. K., Avrantinis S. K., Bronson J., Greaves J., Saven J. G., Weiss G. A. Protein Sci. 2011;20:1597–1606. doi: 10.1002/pro.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauchli R., Rabe K. S., Kalbarczyk K. Z., Tata A., Heel T., Kitto R. Z., Arnold F. H. Angew. Chem., Int. Ed. 2013;52:5571–5574. doi: 10.1002/anie.201301362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major D. T., Freud Y., Weitman M. Curr. Opin. Chem. Biol. 2014;21C:25–33. doi: 10.1016/j.cbpa.2014.03.010. [DOI] [PubMed] [Google Scholar]
- Chen S. H., Hwang D. R., Chen G. H., Hsu N. S., Wu Y. T., Li T. L., Wong C. H. ACS Chem. Biol. 2012;7:481–486. doi: 10.1021/cb200396b. [DOI] [PubMed] [Google Scholar]
- Samland A. K., Rale M., Sprenger G. A., Fessner W. D. ChemBioChem. 2011;12:1454–1474. doi: 10.1002/cbic.201100072. [DOI] [PubMed] [Google Scholar]
- Hibbert E. G., Senussi T., Costelloe S. J., Lei W., Smith M. E. B., Ward J. M., Hailes H. C., Dalby P. A. J. Biotechnol. 2007;131:425–432. doi: 10.1016/j.jbiotec.2007.07.949. [DOI] [PubMed] [Google Scholar]
- Hibbert E. G., Senussi T., Smith M. E. B., Costelloe S. J., Ward J. M., Hailes H. C., Dalby P. A. J. Biotechnol. 2008;134:240–245. doi: 10.1016/j.jbiotec.2008.01.018. [DOI] [PubMed] [Google Scholar]
- Cázares A., Galman J. L., Crago L. G., Smith M. E. B., Strafford J., Ríos-Solís L., Lye G. J., Dalby P. A., Hailes H. C. Org. Biomol. Chem. 2010;8:1301–1309. doi: 10.1039/b924144b. [DOI] [PubMed] [Google Scholar]
- Payongsri P., Steadman D., Strafford J., MacMurray A., Hailes H. C., Dalby P. A. Org. Biomol. Chem. 2012;10:9021–9029. doi: 10.1039/c2ob25751c. [DOI] [PubMed] [Google Scholar]
- Minczuk M., Kolasinska-Zwierz P., Murphy M. P., Papworth M. A. Nat. Protoc. 2010;5:342–356. doi: 10.1038/nprot.2009.245. [DOI] [PubMed] [Google Scholar]
- Papworth M., Kolasinska P., Minczuk M. Gene. 2006;366:27–38. doi: 10.1016/j.gene.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Wijma H. J., Marrink S. J., Janssen D. B. J. Chem. Inf. Model. 2014;54:2079–2092. doi: 10.1021/ci500126x. [DOI] [PubMed] [Google Scholar]
- Wijma H. J., Janssen D. B. FEBS J. 2013;280:2948–2960. doi: 10.1111/febs.12324. [DOI] [PubMed] [Google Scholar]
- Rauscher A. Á., Simon Z., Szöllösi G. J., Gráf L., Derényi I., Málnási-Csizmadia A. FASEB J. 2011;25:2804–2813. doi: 10.1096/fj.11-180794. [DOI] [PubMed] [Google Scholar]
- Rauscher A., Derényi I., Gráf L., Málnási-Csizmadia A. IUBMB Life. 2013;65:35–42. doi: 10.1002/iub.1101. [DOI] [PubMed] [Google Scholar]
- Tapscott D. and Williams A., Wikinomics: how mass collaboration changes everything, New Paradigm, 2007. [Google Scholar]
- Rinaldi A. EMBO Rep. 2009;10:439–443. doi: 10.1038/embor.2009.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corne D. and Knowles J., No free lunch and free leftovers theorems for multiobjecitve optimisation problems., in Evolutionary Multi-criterion Optimization (EMO 2003), LNCS, ed. C. Fonseca et al., Springer, Berlin, 2003, vol. 2632, pp. 327–341. [Google Scholar]
- Culberson J. C. Evol. Comput. 1998;6:109–127. doi: 10.1162/evco.1998.6.2.109. [DOI] [PubMed] [Google Scholar]
- Radcliffe N. J., Surry P. D. Computer Science Today. 1995;1995:275–291. [Google Scholar]
- Wolpert D. H., Macready W. G. IEEE Trans. Evol. Comput. 1997;1:67–82. [Google Scholar]
- Rowe J. E., Vose M. D., Wright A. H. Evol. Comput. 2009;17:117–129. doi: 10.1162/evco.2009.17.1.117. [DOI] [PubMed] [Google Scholar]
- Zalatan J. G., Herschlag D. Nat. Chem. Biol. 2009;5:516–520. doi: 10.1038/nchembio0809-516. [DOI] [PubMed] [Google Scholar]
- Computational approaches in cheminformatics and bioinformatics, ed. R. Guha and A. Bender, Wiley, Hoboken, NJ, 2012. [Google Scholar]
- Ananiadou S., Kell D. B., Tsujii J.-i. Trends Biotechnol. 2006;24:571–579. doi: 10.1016/j.tibtech.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Ananiadou S., Pyysalo S., Tsujii J. i., Kell D. B. Trends Biotechnol. 2010;28:381–390. doi: 10.1016/j.tibtech.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Ananiadou S., Thompson P., Nawaz R., McNaught J., Kell D. B. Briefings Funct. Genomics. 2014 doi: 10.1093/bfgp/elu1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrgård M. J., Swainston N., Dobson P., Dunn W. B., Arga K. Y., Arvas M., Blüthgen N., Borger S., Costenoble R., Heinemann M., Hucka M., Le Novère N., Li P., Liebermeister W., Mo M. L., Oliveira A. P., Petranovic D., Pettifer S., Simeonidis E., Smallbone K., Spasić I., Weichart D., Brent R., Broomhead D. S., Westerhoff H. V., Kırdar B., Penttilä M., Klipp E., Palsson B. Ø., Sauer U., Oliver S. G., Mendes P., Nielsen J., Kell D. B. Nat. Biotechnol. 2008;26:1155–1160. doi: 10.1038/nbt1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swainston N., Mendes P., Kell D. B. Metabolomics. 2013;9:757–764. doi: 10.1007/s11306-013-0564-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman P. Pharmacol. Rev. 1972;24:583–655. [PubMed] [Google Scholar]
- Kell D. B. and Dobson P. D., The cellular uptake of pharmaceutical drugs is mainly carrier-mediated and is thus an issue not so much of biophysics but of systems biology, in Proc. Int. Beilstein Symposium on Systems Chemistry, ed. M. G. Hicks and C. Kettner, Logos Verlag, Berlin, 2009, pp. 149–168, http://www.beilstein-institut.de/Bozen2008/Proceedings/Kell/Kell.pdf. [Google Scholar]
- Doshi R., Nguyen T., Chang G. Proc. Natl. Acad. Sci. U. S. A. 2013;110:7642–7647. doi: 10.1073/pnas.1301358110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H., Chen B., Kang A., Lee J. M., Chang M. W. Biotechnol. Biofuels. 2013;6:95. doi: 10.1186/1754-6834-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Ling H., Chang M. W. Biotechnol. Biofuels. 2013;6:21. doi: 10.1186/1754-6834-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo J. L., Leong S. S. J. Biotechnol. Biofuels. 2013;6:81. doi: 10.1186/1754-6834-6-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida N., Ozato N., Matsui K., Kuroda K., Ueda M. J. Biotechnol. 2013;165:145–152. doi: 10.1016/j.jbiotec.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Grant C., Deszcz D., Wei Y. C., Martinez-Torres R. J., Morris P., Folliard T., Sreenivasan R., Ward J., Dalby P., Woodley J. M., Baganz F. Sci. Rep. 2014;4:5844. doi: 10.1038/srep05844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasiński M., Stukkens Y., Degand H., Purnelle B., Marchand-Brynaert J., Boutry M. Plant Cell. 2001;13:1095–1107. [PMC free article] [PubMed] [Google Scholar]
- Yazaki K. FEBS Lett. 2006;580:1183–1191. doi: 10.1016/j.febslet.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Wu Q., Kazantzis M., Doege H., Ortegon A. M., Tsang B., Falcon A., Stahl A. Diabetes. 2006;55:3229–3237. doi: 10.2337/db06-0749. [DOI] [PubMed] [Google Scholar]
- Wu Q., Ortegon A. M., Tsang B., Doege H., Feingold K. R., Stahl A. J. Mol. Cell Biol. 2006;26:3455–3467. doi: 10.1128/MCB.26.9.3455-3467.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khnykin D., Miner J. H., Jahnsen F. Dermatoendocrinol. 2011;3:53–61. doi: 10.4161/derm.3.2.14816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M. H., Khnykin D. Biochim. Biophys. Acta. 2014;1841:362–368. doi: 10.1016/j.bbalip.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba M. S., Alvarez H. M. Microbiology. 2014;160:1523–1532. doi: 10.1099/mic.0.078477-0. [DOI] [PubMed] [Google Scholar]
- Gimenez R., Nuñez M. F., Badia J., Aguilar J., Baldoma L. J. Bacteriol. 2003;185:6448–6455. doi: 10.1128/JB.185.21.6448-6455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam R., Anzai N., Ahmed N., Ellapan B., Jin C. J., Srivastava S., Miura D., Fukutomi T., Kanai Y., Endou H. J. Pharmacol. Sci. 2008;106:525–528. doi: 10.1254/jphs.sc0070291. [DOI] [PubMed] [Google Scholar]
- Moschen I., Bröer A., Galić S., Lang F., Bröer S. Neurochem. Res. 2012;37:2562–2568. doi: 10.1007/s11064-012-0857-3. [DOI] [PubMed] [Google Scholar]
- Sá-Pessoa J., Paiva S., Ribas D., Silva I. J., Viegas S. C., Arraiano C. M., Casal M. Biochem. J. 2013;454:585–595. doi: 10.1042/BJ20130412. [DOI] [PubMed] [Google Scholar]
- Kaldenhoff R., Kai L., Uehlein N. Biochim. Biophys. Acta. 2014;1840:1592–1595. doi: 10.1016/j.bbagen.2013.09.037. [DOI] [PubMed] [Google Scholar]
- Kai L., Kaldenhoff R. Sci. Rep. 2014:6665. doi: 10.1038/srep06665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdzicki M., Clancy K. P., Oberortner E., Pocock M., Quinn J. Y., Rodriguez C. A., Roehner N., Wilson M. L., Adam L., Anderson J. C., Bartley B. A., Beal J., Chandran D., Chen J., Densmore D., Endy D., Grunberg R., Hallinan J., Hillson N. J., Johnson J. D., Kuchinsky A., Lux M., Misirli G., Peccoud J., Plahar H. A., Sirin E., Stan G. B., Villalobos A., Wipat A., Gennari J. H., Myers C. J., Sauro H. M. Nat. Biotechnol. 2014;32:545–550. doi: 10.1038/nbt.2891. [DOI] [PubMed] [Google Scholar]
- Roehner N., Oberortner E., Pocock M., Beal J., Clancy K., Madsen C., Misirli G., Wipat A., Sauro H., Myers C. J. ACS Synth. Biol. 2014 doi: 10.1021/sb500176h. [DOI] [PubMed] [Google Scholar]
- Hucka M., Finney A., Sauro H. M., Bolouri H., Doyle J. C., Kitano H., Arkin A. P., Bornstein B. J., Bray D., Cornish-Bowden A., Cuellar A. A., Dronov S., Gilles E. D., Ginkel M., Gor V., Goryanin I. I., Hedley W. J., Hodgman T. C., Hofmeyr J. H., Hunter P. J., Juty N. S., Kasberger J. L., Kremling A., Kummer U., Le Novere N., Loew L. M., Lucio D., Mendes P., Minch E., Mjolsness E. D., Nakayama Y., Nelson M. R., Nielsen P. F., Sakurada T., Schaff J. C., Shapiro B. E., Shimizu T. S., Spence H. D., Stelling J., Takahashi K., Tomita M., Wagner J., Wang J. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Le Novère N., Hucka M., Mi H., Moodie S., Schreiber F., Sorokin A., Demir E., Wegner K., Aladjem M., Wimalaratne S. M., Bergman F. T., Gauges R., Ghazal P., Hideya K., Li L., Matsuoka Y., Villéger A., Boyd S. E., Calzone L., Courtot M., Dogrusoz U., Freeman T., Funahashi A., Ghosh S., Jouraku A., Kim S., Kolpakov F., Luna A., Sahle S., Schmidt E., Watterson S., Wu G., Goryanin I., Kell D. B., Sander C., Sauro H., Snoep J. L., Kohn K., Kitano H. Nat. Biotechnol. 2009;27:735–741. doi: 10.1038/nbt.1558. [DOI] [PubMed] [Google Scholar]
- Le Novère N., Finney A., Hucka M., Bhalla U. S., Campagne F., Collado-Vides J., Crampin E. J., Halstead M., Klipp E., Mendes P., Nielsen P., Sauro H., Shapiro B., Snoep J. L., Spence H. D., Wanner B. L. Nat. Biotechnol. 2005;23:1509–1515. doi: 10.1038/nbt1156. [DOI] [PubMed] [Google Scholar]
- Courtot M., Juty N., Knüpfer C., Waltemath D., Zhukova A., Dräger A., Dumontier M., Finney A., Golebiewski M., Hastings J., Hoops S., Keating S., Kell D. B., Kerrien S., Lawson J., Lister A., Lu J., Machne R., Mendes P., Pocock M., Rodriguez N., Villéger A., Wilkinson D. J., Wimalaratne S., Laibe C., Hucka M., Le Novère N. Mol. Syst. Biol. 2011;7:543. doi: 10.1038/msb.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodacre R., Broadhurst D., Smilde A., Kristal B. S., Baker J. D., Beger R., Bessant C., Connor S., Capuani G., Craig A., Ebbels T., Kell D. B., Manetti C., Newton J., Paternostro G., Somorjai R., Sjöström M., Trygg J., Wulfert F. Metabolomics. 2007;3:231–241. [Google Scholar]
- Anfinsen C. B., Haber E., Sela M., White F. H. Proc. Natl. Acad. Sci. U. S. A. 1961;47:1309–1314. doi: 10.1073/pnas.47.9.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfinsen C. B. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Buzan T., How to mind map, Thorsons, London, 2002. [Google Scholar]
- Kell D. B. Trends Genet. 2002;18:555–559. doi: 10.1016/s0168-9525(02)02765-8. [DOI] [PubMed] [Google Scholar]
- Broadhurst D., Kell D. B. Metabolomics. 2006;2:171–196. [Google Scholar]