Abstract
Mass spectrometry is a powerful alternative to antibody-based methods for the analysis of histone post-translational modifications (marks). A key development in this approach was the deliberate propionylation of histones to improve sequence coverage across the lysine-rich and hydrophilic tails that bear most modifications. Several marks continue to be problematic however, particularly di- and tri-methylated lysine 4 of histone H3 which we found to be subject to substantial and selective losses during sample preparation and liquid chromatography-mass spectrometry. We developed a new method employing a “one-pot” hybrid chemical derivatization of histones, whereby an initial conversion of free lysines to their propionylated forms under mild aqueous conditions is followed by trypsin digestion and labeling of new peptide N termini with phenyl isocyanate. High resolution mass spectrometry was used to collect qualitative and quantitative data, and a novel web-based software application (Fishtones) was developed for viewing and quantifying histone marks in the resulting data sets. Recoveries of 53 methyl, acetyl, and phosphoryl marks on histone H3.1 were improved by an average of threefold overall, and over 50-fold for H3K4 di- and tri-methyl marks. The power of this workflow for epigenetic research and drug discovery was demonstrated by measuring quantitative changes in H3K4 trimethylation induced by small molecule inhibitors of lysine demethylases and siRNA knockdown of epigenetic modifiers ASH2L and WDR5.
The field of Epigenetics has become important in drug discovery as many diseases have been linked to aberrations in chromatin and changes of histone post-translational modifications (PTMs)1 (1, 2). The core histones (H2A, H2B, H3, and H4 and their variants) undergo a multitude of PTMs. Some, like lysine acetylation, lysine mono-, di-, and trimethlyation, and serine/threonine phosphorylation are well documented, with over 100 distinct, albeit generally low abundance, modifications reported for H3 alone (3). Mass spectrometry provides an alternative to antibody-based methods for detecting and quantifying histone PTMs, as the latter are prone to problems of specificity and epitope occlusion (4, 5). The most commonly applied approach to date is known as “bottom-up” mass spectrometry and involves an initial processing of the histones into smaller peptides (6). A key development in histone PTM analysis was the deliberate chemical modification of histone tail lysines by propionic anhydride, preventing digestion of these Lys- and Arg-rich domains into peptides too short or hydrophilic to be detected in reverse-phase liquid chromatography-mass spectrometry experiments (7–9).
Despite this advance, some marks like H3K4 di- and tri-methylation remain problematic; in several examples from the recent literature the H3K4me3 mark is detected either only by means of specifically targeted methods (5), with larger quantitative variation than other marks (10), or not reported among detected marks at all (3, 11–13). Alternative approaches include top-down or middle-down mass spectrometry, in which entire histones, or large segments thereof are analyzed directly (14–16), but these techniques still suffer from relatively poor sensitivity in comparison to bottom-up workflows, and must contend with the full combinatorial complexity of histone PTMs (17).
The H3K4me3 mark is of low natural abundance, having a very restricted genomic localization strongly associated with active gene promotors and enhancers (18, 19), and aberrant activities of writers and erasers of that mark are associated with a variety of diseases (1, 2). Difficulties in its quantitation thus hinder the investigation of both fundamental biology and the discovery of lifesaving drugs. We therefore undertook a re-evaluation of the bottom-up histone PTM workflow, streamlining sample preparation and investigating sources of bias or sample loss. Alternatives to the standard propionylation technique were also explored, resulting in a new hybrid chemical modification workflow yielding across-the-board improvements in recovery of peptides from the N-terminal tail of histone H3, and dramatically improved detection of hydrophilic peptides with marks like H3K4me2/me3.
EXPERIMENTAL PROCEDURES
Materials
Chemical regents used in this study purchased from Sigma-Aldrich: Propionic anhydride and phenyl isocyanate (Fluka brand), perchloric acid, hydroxylamine (50% wt%), phenyl 13C6 isocyanate (Aldrich brand), 1 m Triethylammonium bicarbonate buffer solution (Sigma brand). Phenyl isothiocyanate was purchased from Acros Organics (Geel, Belgium). Agilent Technologies (Santa Clara, CA) supplied orthopthalaldehyde and FMOC-Cl. Synthetic peptides were purchased from JPT Peptide Technologies (Berlin, Germany). Lysine demethylase inhibitors were synthesized at WuXi AppTec (Shanghai, China).
Cell Culture and siRNA-Transfection
HEK293T cells were grown in Dulbecco's modified Eagle's medium containing antibiotics (100 units/L Pen/Strep, Gibco) and l-Glutamine (1× Glutamax, Gibco; Grand Island, NY) and were harvested at around 90% confluency.
PC9 cells were grown in RPMI 1640 medium under similar conditions as described above. 3 × 106 cells were transfected using Dharmafect 1 according to the manufacturer's protocol with the following siRNAs (Ambion, Grand Island, NY): s17302 (siAsh2L), s225470 (siWdr5), s17302 and s225470 (siAsh-Wdr). Nontarget-control (NTC): siGenome nontargeting siRNA #4.
HeLa cells were grown in Dulbecco's modified Eagle's medium as above. 2.5 × 106 cells were transfected with the same siRNA's targeting ASH2, WDR5, β-actin, or a nontargeting control at a final concentration of 10 nm using Hiperfect transfection reagent as per the manufacturer's protocol (Qiagen; Venlo, The Netherlands). Seventy-two hour post-transfection, cells were trypsinized and reseeded at 1 × 106 cells per plate. The remaining cells were pelleted by centrifugation at 500 × g for 3 min, washed once in PBS, pelleted again, and stored for processing. Twenty-four hours after replating, cells were transfected a second time as described above. Seventy-two hours following the second transfection, cells were harvested as above.
Western Blot Analysis
All cell pellets were lysed in a hypotonic lysis buffer (10 mm Tris, pH 7.9, 1.5 mm MgCl2, 10 mm KCl, 0.5 mm dithiotreitol, supplemented with protease inhibitors from Roche) on ice for 15 min, followed by centrifugation at 2500 × g at 4 °C. The supernatant was saved as the cytoplasmic fraction, and the pellet resuspended in 20 mm HEPES pH 7.5, 1 m NaCl, 20% glycerol, 1.5 mm MgCl2, 0.1 mm EDTA, 0.5 mm dithiotreitol, and protease inhibitors, then sonicated with a Branson sonicator for 3 s at setting 3. Lysates were spun again at 16,000 × g for 20 min at 4 °C, and the supernatant containing nuclear and chromatin fractions was pooled with the cytoplasmic fraction. Protein concentration was determined via Bradford assay, and equal total protein loads were run on 4–12% gradient Bis-Tris gel (Life Technologies; Grand Island, NY) using MES running buffer, before transferring to 0.2 micron nitrocellulose (Bio-Rad; Hercules, CA). Blots were blocked in 5% milk in PBS, probed with indicated primary antibodies overnight at 4 °C, followed by incubation with DyLight coupled secondary antibodies (Thermo) for 1 h at room temperature, before detection on the LiCor Odyssey imaging system.
Histone Purification
Core histones were extracted from cultured cells and column-purified using the “Histone Purification Mini Kit” (Active Motif, #40026) according to the manufacturer's instructions. Briefly, histones were acid-extracted, enriched on ion-exchange spin-columns and desalted by perchloric acid precipitation, yielding a mixture of the core histones H2A, H2B, H3, and H4 in high purity. The purified histones were resuspended in HPLC-grade dH2O (ddH2O) to a final protein concentration of between 0.5 and 1.0 μg/μl. Aliquots of 10 μg core histones were flash-frozen and stored at −80 °C.
Chemical Labeling of Histone Peptides
Standard Propionylation Method (Prop-x2)
Aliquots of 1 to 5 μg of purified core histones were prepared as described (9, 10) except that isopropanol replaced methanol in the propionylation reaction to minimize methyl ester side products (20). Briefly, samples were diluted in 100 mm ammonium bicarbonate to a total volume of 10 μl and 0.5 μl ammonium hydroxide was added to bring the pH between 8.0 and 9.0. Propionic anhydride was mixed 1:3 with reagent isopropanol and added at amounts of half the sample's starting volume (5 μl). Ammonia was added (3 μl immediately, and subsequently as needed) to maintain a pH of ∼8 during the 15 min reaction at 37 °C. Samples were dried by vacuum centrifugation and the propionylation reaction repeated. Dried samples were resuspended in 50 μl ammonium bicarbonate (100 mm, pH 8) and digested at 37 °C overnight with trypsin (1:20 enzyme-to-substrate by weight). The resulting peptides were dried by vacuum centrifugation and two more rounds of propionylation performed on the free peptide N termini as described above before desalting the sample (below).
Hybrid Propionylation-PIC Method (Prop-PIC)
For each sample reaction, 1 to 5 μg aliquots of purified core histones were diluted with ddH2O to a total volume of 9 μl and buffered to pH 8.5 by addition of 1 μl of 1 m Triethylammonium bicarbonate buffer. Propionic anhydride was mixed with ddH2O in a ratio of 1:100 and 1 μl of the anhydride-mixture was added immediately to the histone sample, with vortexing, and incubation for 2 min at room temperature. The reaction was quenched with 1 μl of 80 mm hydroxylamine (20 min at room temperature). Tryptic digestion was performed for 4 h or overnight with 0.1 μg trypsin (Promega Sequencing Grade; Madison, WI) per sample. A 1% v/v solution of phenyl isocyanate (PIC) in acetonitrile was freshly prepared and 3 μl added to each sample (17 mm final concentration) and incubated for 60 min at 37 °C. Efficiency of PIC-labeling was checked by MALDI-TOF (4800 Plus; Applied Biosystems, Grand Island, NY) and samples were acidified by adding 8 μl of 1% trifluoroacetic acid (TFA) to each sample prior to C18 stage-tip purification.
Stage-Tip Purification of Labeled Histone Peptides
Labeled histone samples were desalted using C18-stage-tips (Thermo Scientific; #SP201), which were wetted with 60% Acetonitrile/0.1% TFA and then equilibrated with 20 μl of 0.1% TFA. Acidified peptide samples were diluted in 60 μl ddH2O, loaded onto the stage-tip in and washed twice with 20 μl of 0.1% TFA. Peptides were eluted with 3 μl of 60% Acetonitrile/0.1% TFA and used immediately or dried in a vacuum centrifuge and stored at −20 °C.
QTRAP Flow-injection Analysis of Synthetic Peptides
Synthetic peptides with base sequences from Histone H3, residues 1–17 (ARTKQTARKSTGGKAPR) were procured as unmodified, or bearing monomethyl, dimethyl, trimethyl, or acetyl modifications on K4, or with phosphorylation on T6, alone or in combination with monomethyl or dimethyl K4. A 200 pmol aliquot of each peptide in 10 μl of 100 mm Triethylammonium bicarbonate buffer was propionylated as described in the hybrid method above, and digested with 0.1 μg trypsin (Promega) for 1 h at 37 °C, or at 1, 2, 5 10, 20, and 30 min for the time course of digestion. The resulting products of digestion were either used without further labeling in the time-course experiment, or derivatized with another round of propionylation or PIC on their new N termini. Fully derivatized peptides were analyzed before and after stage-tip desalting.
Five-μl aliquots containing nominally 1 pmol/μl of each synthetic peptide sample were injected into a 200 μl per minute solvent stream (0.1% v/v formic acid, 20% v/v acetonitrile in water) directed to the ion source (Turbo Ion Spray) of an Applied Biosystems 4000 QTRAP mass spectrometer. This instrument was operated in selected ion monitoring mode, with 125 ms dwell times at each peptide's [M+H]+ and [M+2H]2+ mass to charge ratio at unit resolution. Peak areas were extracted using the vendor's MultiQuant software, and areas for each charge state summed to obtain a total area under the curve for each peptide.
Orbitrap LC-MS/MS of Endogenous Histone Peptides
Stage-tip desalted histone peptides were resuspended with HPLC solvent A (0.1% v/v formic acid, 2% v/v acetonitrile in HPLC grade water) to final concentrations of 200 to 500 ng/μl total histones in order that ∼500 ng was analyzed by capillary LC-MS/MS using a hybrid linear ion trap/Orbitrap mass spectrometer (Orbitrap-Elite; Thermo Fisher Scientific, Waltham, MA). Peptides were loaded onto a C18 column (BEH-C18; 100 μm i.d. x 10 cm; 1.7 μm particles, 130Å pores; Waters Corp., Milford, MA) for 10 min at 1.5 μl per minute in 2% solvent B (0.1% v/v formic acid, 98% v/v acetonitrile) and separated at 1 μl per minute by a linear gradient from 2% solvent B to 25% solvent B over 60 min followed by a ramp to 40% B in 15 min, then to 90% B and re-equilibration at 2% B for a 90-min total run time. Full mass range spectra were collected at 60,000 resolution (M/Δ M at m/z 400), and product ions were collected in a “top 15” data dependent scan cycle at unit resolution in the ion trap mass analyzer (resonance collision-induced dissociation) or at 15,000 resolution in the Orbitrap (collision cell CID; i.e. higher energy collisional dissociation, (HCD)), respectively. A mass inclusion list for histone H3 peptides (as shown in supplemental Table S1) was specified to ensure that tandem mass spectra were acquired for as many histone peptides as possible. Certain co-eluting, isobaric peptides were the subject of scheduled, full mass range tandem mass spectra so that they might be distinguished at the MS/MS level. These included K18ac and K23ac; K36me3 and K27me2K36me1; K27me3K36me2 and K27me2K36me3; K36me2S28pr, and were quantified as detailed below.
Histone PTM Identification and Quantitation
Area-under-the-curve values for peptide peaks were calculated using the “Fishtones” program, a web application based on fishTones.js, a JavaScript library to visualize liquid chromatography-tandem mass spectrometry experiments. It offers a variety of features and graphical widgets including the ability to extract ion chromatograms for multiple experiments simultaneously, given peptide sequences and defined modifications and derivatization/isotopic labeling conditions. Tandem mass spectra associated with the extracted peaks can be displayed with annotated product ions, and viewed interactively to test the influence of hypothetical modifications. The user-selected peaks are then integrated and extracted for further processing. Designed to meet the challenges of histone PTM analysis, the library is of general applicability to LC-MS data analysis and is publicly available at http://research-pub.gene.com/fishtones-js/howto.
For those combinations of histone marks on two co-eluting peptides with identical masses, the relative abundance of each peptide was derived from PTM-specific ions in the product ion spectra across the chromatographic peak, and the full-mass range scan's integrated peak area apportioned accordingly in a manner similar to previous reports (10, 21, 22). The following marks subject to this procedure were: K18ac and K23ac; K36me3 and K27me2K36me1; K27me3K36me2 and K27me2K36me3; K36me2S28pr and K27me3K36me2T32pr; K27me2K36me3S28pr and K27me2K36me3T32pr. Integrated peak areas from the raw data were converted to relative abundances by dividing each measured peptide by the sum of all peak areas covering the same base sequence, for example, if the peaks covering H3 residues 3–8 had areas: K4unmod, 1 × 107counts; K4me1, 1 × 106counts; K4me2 2 × 105counts, and K4me3, 1 × 105counts, the relative abundance of K4me2 would be 2 × 105/1.13 × 107 = 0.0177. In cases where two or more marks are present on a single peptide, the peak's area contributes to both marks taken individually. For example, if the peptide covering H3 residues 18–26 yielded peaks: K18unK23un, K18me1K23un, K18acK23un, K18unK23ac, and K18acK23ac, then the relative abundance of K18ac would be (K18acK23un + K18acK23ac)/(sum of all). Once peaks' relative abundances for each mark are calculated, these are used to measure mark-by-mark changes, for examplein a two-sample comparison (chemical labeled or SILAC), among samples with a common internal standard (chemical labeled or super-SILAC), or among samples without internal standard (label free).
RESULTS
Peptides Modified at H3K4 are Not Equally Represented in LC-MS/MS Experiments
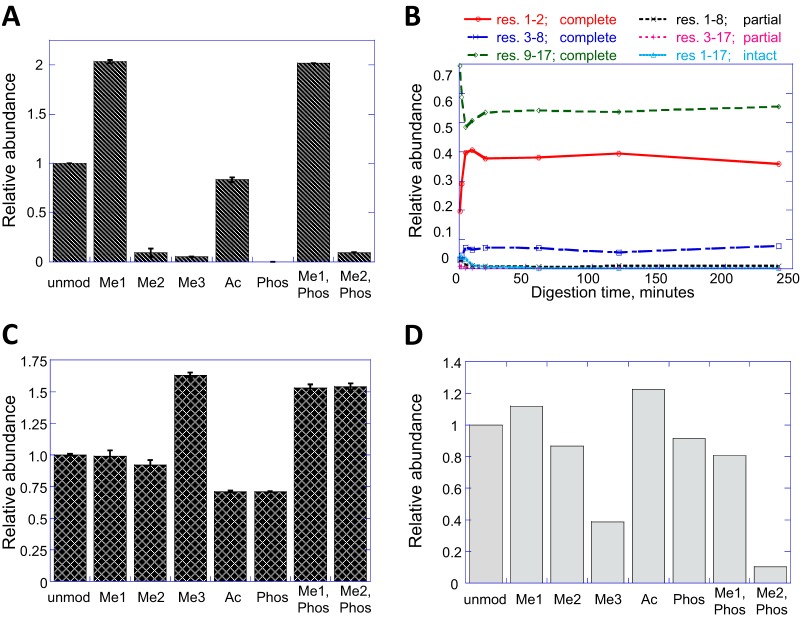
Eight synthetic peptides corresponding to histone H3 residues 1–17 (ARTKQTARKSTGGKAPR) were synthesized, identical except for their modification states at lysine-4 (unmodified, acetylated, mono-/di- and tri-methylated), and/or phosphorylated at threonine-6. Each of these peptides was treated according to published protocols (10) with isotopically labeled (d10) propionic anhydride to block the peptide N terminus and the unmodified lysines. Monomethyl K4 was also thereby converted to monomethyl, propionyl K4. Trypsin digestion was followed by a second round of propionylation to block the new peptide N termini. These peptides, in nominally equimolar ratios, were mixed with endogenous histones from 293T cells that had been prepared identically except without stable isotope labeling, and analyzed by capillary LC-MS/MS on an Orbitrap mass spectrometer. When the peak areas of the synthetic peptides - distinguishable by their stable isotope labels - were extracted, it was evident that the “recoveries” of differently modified forms were quite different (Fig. 1A). Whereas most versions of the peptide were detected with abundance equal to or greater than the unmodified peptide, the dimethyl, trimethyl, phosphoryl, and dimethyl/phosphoryl forms were all below 10% abundance relative to their unmodified counterpart. Although different peptides are not expected to have identical responses in the mass spectrometer, such large and selective discrimination against specific modified forms could confound quantitative experiments, particularly where the modified form is of low natural abundance to begin with, as is the cases of H3K4me2 and -me3.
Fig. 1.
Sources of discrepancy in H3K4 quantitation. A, Equimolar mixture of isotope-labeled, propionylated H3 T3-R8 peptide in eight modification states, injected in the milieu of endogenous histones from 293T cells. B, Time course of digestion releasing propionylated H3 T3-R8 from propionylated H3 A1-R17, where K4 is trimethylated. C, Relative ionization efficiencies of the propionylated H3 T3-R8 peptide in its various modified forms. D, Recovery of H3 T3-R8 peptides following StageTip C18 cleanup. In panels A, C, D, all abundances are expressed relative to the unmodified peptide.
Modifications at H3 K4 Do Not Impair Trypsin Digestion
The same synthetic peptides employed above were used to determine whether the variable recovery of modified peptides was because of some resistance to digestion, or instability of the digested peptides. The H3 A1-R17 peptides were propionylated and digested with trypsin, with aliquots taken at 1, 2, 5, 10, 20, and 30-minute time points. With all lysines blocked by modification, tryptic cleavage should occur at the two arginines, yielding three fully processed peptides and potentially two additional peptides resulting from partial digestion. In the time course for digestion of the H3 K4me3 peptide (Fig. 1B), all of these potential products can be observed at early time points. Within 5 mins however, the starting peptide is much attenuated and the partial digestion products have already peaked and are declining in abundance, to be replaced with the products of complete digestion. By 20 min only the fully processed peptides remain and their abundances are constant thereafter, indicating complete digestion to stable end products. Such time course experiments were performed for each of the modification states at K4 with similar results (data not shown). There is no apparent bias against production of any H3 T3-R8 tryptic peptide by modification at lysine-4.
Ionization Efficiency and Modification at H3K4
The abundance of peaks in a mass spectrum depends on multiple factors including the chemical properties of the analytes (23, 24), so it seemed plausible that differently modified forms of one peptide might have different intrinsic responses in the mass spectrometer. Complete digestion of the H3.1 A1-R17 synthetic peptides necessarily produces the modified H3 T3-R8 peptides in exactly 1:1 molar ratios with the unmodified H3 K9-R17 peptide. The H3 K9-R17 peptide can therefore be used to relate the ionization efficiencies of all modified forms of the H3 T3-R8 peptide to each other. The results, obtained by loop injection into the electrospray source of a mass spectrometer are shown in Fig. 1C. Direct injection was performed rather than LC-MS in order to isolate intrinsic mass spectrometric response from chromatographic behavior. It is evident that the relative detection efficiencies of the modified forms of H3 T3-R8 are on the same order of magnitude. Indeed, the peptide containing H3 K4me3 has a greater response than the unmodified peptide. This is likely because of the fixed positive charge conferred by the quaternary amine (25), consistent with published reports (5). The intrinsic responses of these peptides in electrospray ionization are not biased against di- and tri-methylation of K4, and only modestly against K4 acetylation and T6 phosphorylation.
Selective Losses During Sample Preparation
It is a striking feature of the H3T3-R8 peptide that the problematic di- and tri-methyl forms are more hydrophilic than the readily detected unmodified and monomethyl forms, the latter two being subject to K4-propionylation whereas the former two are not. This is reflected in their chromatographic behavior, with H3K4me2 and -me3 forms eluting very early, indeed within the loading volume of our HPLC column during LC-MS. Published protocols for histone PTM analysis often include a small trapping column or solid phase extraction step between the second propionylation and LC-MS in order to remove interfering salts and reaction byproducts that might foul the column or mass spectrometer inlet (11, 12, 26). It therefore seemed possible that these most hydrophilic peptides could be selectively lost during sample preparation. This suspicion was confirmed by measuring the recoveries of propionylated H3 A1-R17 tryptic peptides upon desalting with a C18 (StageTip) trapping column (Fig. 1D). Whereas most of the modified forms of the residue H3 T3-R8 peptide were similar to the unmodified form, approximately two thirds of the K4-trimethyl peptide, and even more of the dimethyl, phosphorylated peptide were lost. These losses, combined with the poor peak shape and such ion suppression as may occur because of salts present early in the LC-MS chromatography could account for the observed discrimination against H3K4me2 and -me3 marks seen in Fig. 1A.
Alternatives to Propionylation
If the H3K4 dimethyl and trimethyl peptides are problematic because they are too hydrophilic, then modifying their N termini with a reagent more hydrophobic than propionic anhydride might be a remedy. A series of alternative labels and chemistries were considered, including acetylation, trifluoroacetylation, and derivatization with ortho pthalaldehyde, fluorenylmethoxy chloroformate, tandem mass tags (activated esters of a substituted piperidine compound (27)), phenyl isothiocyanate (Edman's reagent), and phenyl isocyanate. Desired behavior included ease and efficiency of labeling and increased reverse-phase retention times relative to propionylation, without being so hydrophobic as to lose the later eluting histone peptides on-column. Commercial availability of a stable-isotope labeled version was also desirable.
Of all the reagents tested, PIC proved most advantageous, reacting quantitatively in less than an hour in mild aqueous conditions and conferring a retention time shift of 5 to 8 mins for a variety of hydrophilic peptides (data not shown). This reagent has been previously used in quantitative proteomic experiments (28, 29) and is available in both deuterated (d5) and carbon-13 (13C6) labeled forms. When PIC labeling was applied to the synthetic H3 A1-R17 peptide with trimethyl-K4, the effect on recovery was striking. The residue H3 T3-R8 tryptic peptide shifted in retention time from within the loading volume of the column to over 12 min into the gradient, and increased in abundance 10-fold, both in absolute terms and relative to the “adjacent” tryptic peptide spanning residues K9-R17 (supplemental Fig. S1).
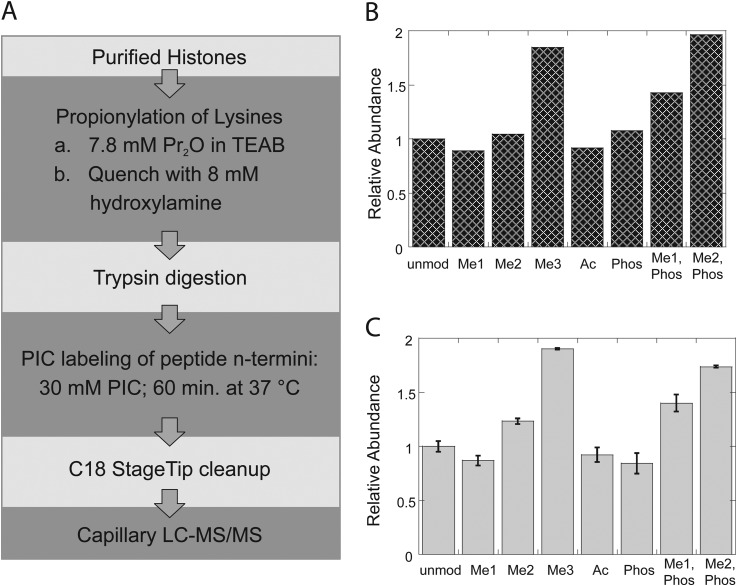
A new protocol was therefore implemented (Fig. 2A and supplemental Fig. S2), similar to the “standard” protocol in that predigestion propionylation was used to block unmodified and mono-methyl lysine, but differing in that postdigestion labeling is done with PIC, resulting in a single PIC label at the N terminus of each tryptic peptide. Another difference is that propionylation is now performed in aqueous solution at pH 8.5, buffered by triethylammonium bicarbonate, with excess propionic anhydride quenched stoichiometrically in situ, allowing the procedure to be performed in a single tube without vacuum drying to remove reagents between steps. Neither is it necessary to repeatedly check and adjust pH, as is necessary during the multiple propionylation reactions of the standard protocol, resulting in a significant reduction in the number of sample handling steps at which losses might be accrued. The entire protocol can be performed in less than a day (four hours of which are incubation with trypsin), so that samples can be prepared and loaded onto an autosampler for overnight LC-MS/MS. Ionization efficiencies and reverse-phase trapping column recoveries were determined for the propionylated/PIC labeled peptides as before (Figs. 2B and 2C). Relative ionization efficiencies among the various H3 K4 modifications were not significantly changed, but the recoveries of K4me3 and K4me2/T6 phosphoryl peptides were dramatically improved. Indeed, if the peptide abundances were corrected for ionization efficiency, the recoveries of the H3 T3-R8 peptide in all modified forms would be approximately equal. N-terminal PIC labeling does not strongly direct peptide fragmentation patterns under collision-induced dissociation, but a subtle decrease in b-ion abundances can be discerned, particularly in the first ions of the series. Interestingly, b1 ions are not normally observed in collision-induced dissociation of peptides with free N termini, but are prominent in the product ion spectra of peptides N-terminally modified by either propionylation or PIC, reflecting the presumed mechanism of low energy fragmentation (25). Indeed, the combination of Lys-propionylation and N-terminal PIC labeling produces other low mass ions in HCD that are diagnostic of several modifications including propionyl-Lys (m/z 140.107), [monomethyl, propionyl]-Lys (m/z 171.14), and N-terminal lysine with propionyl, [monomethyl, propionyl], dimethyl, and acetyl modifications at the epsilon amine (m/z 276.17, 318.18, 290.15, respectively). The slight decrease in b-ion abundances with PIC labeling seems correlated with higher abundances of these diagnostic ions, although the matter has not been explored systematically.
Fig. 2.
Histone preparation with Lys-propionylation, trypsin digestion, and N-terminal PIC-labeling. A, Sample preparation workflow. B, Relative ionization efficiencies H3 T3-R8 peptide in all modified forms. C, Recovery of H3 T3-R8 peptides following StageTip C18 cleanup. All abundances are expressed relative to the unmodified peptide.
Comparison of Prop-x2 to Prop-PIC Labeling Methods on Endogenous Histones
Identical 5 μg aliquots of purified histones from HEK293T cells were processed according to either the standard method (propionylation before and after digestion; “Prop-x2”) or the new method (propionylation before digestion, PIC after; “Prop-PIC”) combining them immediately prior to C18 cleanup and orbitrap LC-MS/MS analysis. Data were acquired in a data-dependent fashion typical of proteomic experiments. An in-house software application (Fishtones; supplemental Fig. S3) was used to extract ion chromatograms for all known combinations of lysine methylation, acetylation, and serine and threonine phosphorylation on the N-terminal tail on histone H3 as modified by either sample preparation method. Peptide identities were confirmed in the Fishtones application by reference to their collision-induced dissociation tandem mass spectra (HCD, see “Methods”). After selection by the user, individual peak areas for valid identifications were computed by the program. In the cases of co-eluting peptides of identical mass, e.g. residues K18-R26 acetylated on either K18 or K23, the abundances of product ions specific to each form were used to apportion the MS1 peak area between the two forms(10, 21, 22).
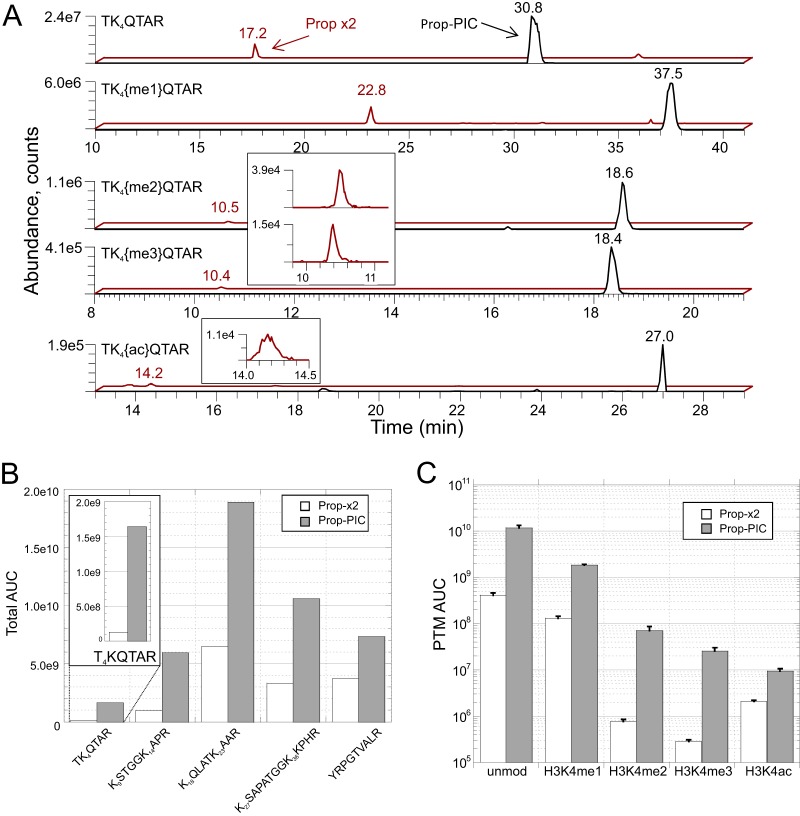
Several features of the new method are immediately apparent from the extracted ion chromatograms for various H3 T3-R8 peptides (Fig. 3A). First, as expected, all detected forms of the peptide are shifted to later retention times than their Prop-x2 counterparts. Additionally, use of the Prop-PIC labeling resulted in significant increases in peak areas for all forms of the peptide, but most dramatically in the cases of H3K4me2 and -me3, which gained approximately two orders of magnitude in peak areas. The peak shape of the peptide with H3K4ac was also improved. Three technical replicates on the full procedure confirmed the consistency and superiority of the Prop-PIC protocol for H3K4 modifications (Fig. 3B). Interestingly, although not quantified in this experiment (a peak with correct mass/retention time is present but MS/MS was not recorded), H3T3 phosphorylation has been observed in the course of our COMPASS complex studies, (below; supplemental Fig. S5). This mark is associated with mitosis and has been previously identified by mass spectrometry when expedients such as phosphopeptide enrichment and/or alternative enzymatic digestions were employed.
Fig. 3.
Direct comparison of standard propionylation (Prop-x2) labeling and the hybrid (Prop-PIC) labeling method via LC-MS using a 1:1 mixture of labeled histone samples. A, Extracted ion chromatograms of the hydrophilic H3T3-R8 peptide (TK4QTAR) and its modified versions on C18 reverse-phase LC-MS. Retention times and relative abundances are increased by the hybrid labeling method. B, Quantitative comparison of the recoveries of each H3.1 histone tail peptide, where integrated peak areas for all modification states are combined. C, Quantitative comparison of the recoveries of the H3 T3-R8 peptide in its various modification states for the Prop-PIC versus Propx2 (integrated peak areas; n = 3, error bars are standard deviations).
The new method would be of limited value if gains in quantifying H3K4me3 were offset by losses elsewhere. A full workup of histone H3.1 tryptic peptides from the N-terminal tail (residues T3-R49) using the Fishtones program revealed a total of 53 peptides in various combinations of lysine acetylation and methylation, and serine phosphorylation. These included acetylation plus mono-, di-, and trimethylation on K4, K9, K27, and K36, acetylation and monomethylation on K18 and K23, and acetylation on K14. Phosphorylation on serines 10 and 28 was also observed. Peptides derived from histone H4, isoforms of H2A, H2B, and the K27-R40 peptide of histone H3.3 (the only N-terminal peptide distinguishable from the other H3 variants) are also identifiable but were not quantified for the purposes of this comparison (not shown). All quantified peptides and their relative abundances in both Prop-x2 and Prop-PIC states are detailed in supplemental Fig. S4, supplemental Table S1, and their annotated spectra in supplemental Annotated Spectra files. Every peptide detected with the Prop-x2 labeling was also detected with Prop-PIC labeling, along with several peptides (K23me1, K9me1S10phosK14ac, S28phos, K27ac, K36ac) that were not detected with Prop-x2 (marked with asterisks in supplemental Fig. S4).
Quantitative analysis showed the recovery of peptides from the N-terminal tail of histone H3 to be markedly higher with Prop-PIC than with Prop-x2 labeling, whether taken peptide-by-peptide (supplemental Fig. S4) or summing peak areas for all modifications states of each tryptic peptide (Fig. 3C). On average, the new method yielded integrated peak areas four times those produced with the standard method for the same input of purified histones, and even more for certain marks such as the previously detailed H3K4me2 and -me3. Phosphopeptides were beneficiaries of the new method, with multiple peaks containing S10phos and S28phos elevated from just above detection limits with Prop-x2 to readily quantifiable by Prop-PIC (supplemental Figs. S4D and S4E). Low-abundance acetylation events such as K27ac and K36ac were likewise transformed from barely (or not) detectable to peaks with respectable signal-to-noise (supplemental Fig. S4E).
Unintended modification of serine and threonine residues is known to accompany the propionylation reaction (30). Despite generally higher peptide recovery with PIC-labeling, such artifactual peaks were lower in abundance than when Prop-x2 was used to prepare the samples, presumably because only one round of propionylation was performed, rather than the repeated exposures to propionic anhydride in the original method (9). Others have addressed this problem by replacing propionic anhydride with a less reactive ester of propionic acid (12, 30), but even without taking that measure the relative abundances of these side-products can be kept below one percent of their correctly labeled forms (supplemental Fig. S4).
Quantifying Increases in H3K4-methylation Induced by Lysine Demethylase Inhibitors
One of the many important applications of histone PTM analysis is the screening and characterization of small molecule drugs that inhibit histone tail-modifying enzymes; the “writers” and “erasers” of epigenetic marks. Mass spectrometry can establish whether, and to what extent, inhibition of a histone modifying enzyme alters modification states at defined sites on histone tails, and look for “off-target” effects at other histone loci. Tri- and di-methylation at H3K4 is associated with active gene expression, and demethylases that decrease H3K4me are generally presumed to be transcriptional repressors. These include members of the KDM1 and KDM5 families, both of which are overexpressed in multiple types of cancer and have drawn the attention of pharmaceutical companies as potential drug targets (2).
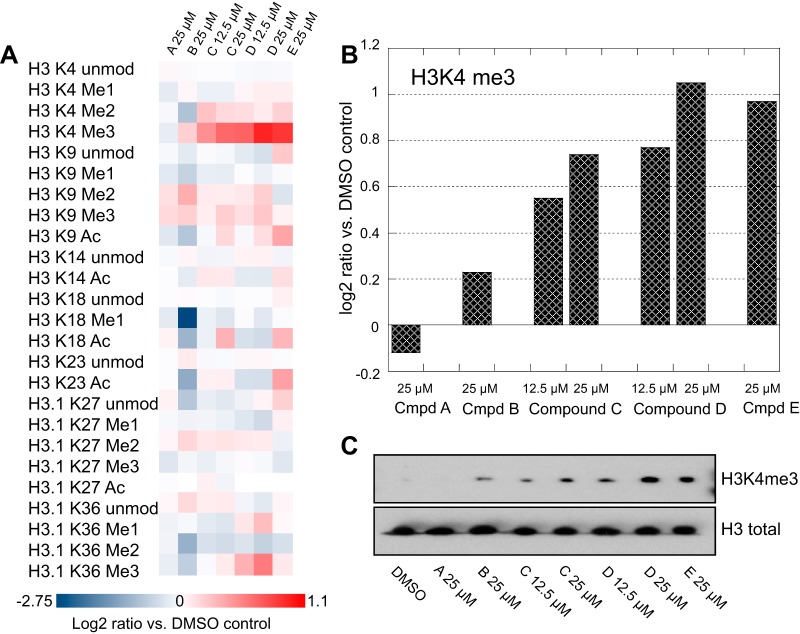
Cells derived from non small cell lung cancer (PC9) were treated in order to assess the activity and specificity of several putative KDM-inhibitors, and subjected to mass spectrometric analysis. Fig. 4 illustrates the results for an inactive control compound (compound A) and four active compounds (B, C, D, and E) to assess their potency and selectivity. A heat-map representation shows how each of 25 marks changes upon drug treatment relative to cells treated with DMSO diluent alone (Fig. 4A). All of the active compounds increased H3 K4-trimethylation to some degree, and in a dose dependent manner for compounds C and D, which were each tested at two concentrations (Fig. 4B). Each sample also contained stable isotope-labeled histones from untreated PC9 cells as internal standards. These constitute technical replicates and should in principle have identical abundances in all samples. Their variability therefore sets a lower bar for the reliability of quantifying each observed histone mark. In this case the 95% confidence interval (n = 10) for H3K4me3 is ± 0.2 log units; smaller changes are unlikely to be significant. Western blots of the same samples for H3K4me3 (Fig. 4C) are consistent with the mass spectrometric results, but are much less quantitative. Compound B showed the least change in H3K4-trimethylation, and was also the least selective in terms of changing modification levels at individual histone lysine residues. Indeed, the most dramatic effects were decreases in monomethylation at H3K18 as well as dimethylation at both H3K4 and H3K36, making this compound unattractive for further investigation. The other three compounds had much cleaner profiles, with small, across-the board increases in H3K4-dimethylation, and more variable, but relatively small H3K9 di- and trimethylation, and H3K36 trimethylation. Interestingly, increases of acetylation at H3K9 were observed for compounds C, D, and E, and were dose-dependent for compounds C and D. More subtle increases in acetylation at H3K14 and H3K18 were found for compounds C and E. These might be off-target effects of the drugs, or secondary effects of chromatin remodeling that follow from changes in H3K4-methylation.
Fig. 4.
Quantitative changes in histone H3 marks in PC9 cells upon treatment with DMSO vehicle, an inactive control (compound A) or histone lysine- demethylase inhibitors (compounds B, C, D, and E). A, Heat map displaying changes in each mark relative to the DMSO control. B, Effect of each compound on the H3K4 trimethyl mark, as log2 ratios versus DMSO control. C, Western blots of the same samples, probed for H3K4me3 and total H3.
Quantitative Decreases in H3K4 Methylation After Knockdown of Core Components of the COMPASS Complex
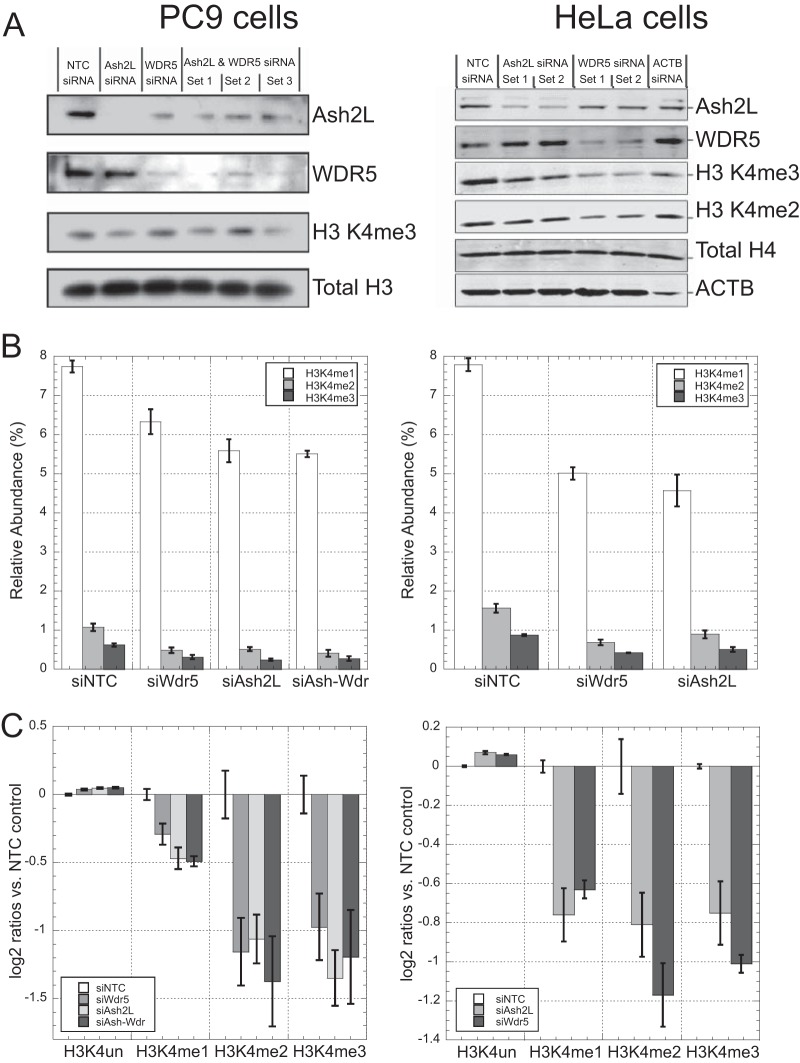
Given the low baseline abundance of H3K4me3, demonstrating a quantitative decrease in the mark is particularly challenging. We evaluated the Prop-PIC method in this scenario in two different cell lines by depleting the protein levels of ASH2L, WDR5, or both. These are core-subunits of the COMPASS-complex, and play an essential role in the methylation of H3K4 (31). Western blots (Fig. 5A) indicated that the ASH2L and WDR5 were robustly knocked down in each cell line, although not depleted entirely. It can be seen that the H3K4me3 mark was decreased in the knock-downs, but the extent of its change is difficult to quantify. The best conditions—set 3 for the combination knock-down in PC9 cells and set 1 in HeLa cells—were chosen for mass spectrometric analysis.
Fig. 5.
Changes in H3K4-methylation upon siRNA-knockdown of members of the COMPASS complex in PC9 cells (left-hand panels) and HeLa cells (right hand panels). A, Western blots showing efficiency of knock-downs, H3K4me3, and loading controls. B, Relative abundance of H3K4 methyl marks for each siRNA transfection. C, Mark-by-mark changes in H3K4 methylation-levels for each siRNA transfection, expressed as log2 ratios versus siNTC control. In B and C, NTC = nontarget control, and error bars are standard deviations with n = 3.
PC9 cells treated with siRNA against ASH2L, WDR5, or both showed a significant decrease in H3K4 methylation levels when compared with cells treated with control-siRNA (Fig. 5B, left hand graph). HeLa cells treated with siRNA against either gene individually followed a similar pattern (Fig. 5B, right). When expressed as log2-ratios versus the nontarget control siRNA, H3K4me1 levels declined modestly upon siRNA treatment (by 30% in PC9 cells; 40% in HeLa), but H3K4me2 and H3K4me3 levels each decreased by more than 50% in each cell line (Fig. 5C). A combinatorial siRNA-knockdown of both ASH2L and WDR5 in PC9 cells did not result in further reduction of H3K4me levels indicating that knockdown of either COMPASS core subunit is sufficient in reducing methylation via COMPASS HMTs in this cell line. In comparison to the Western blots, mass spectrometry is clearly more suitable for quantitation, and avoids certain technical challenges affecting antibody affinity, such as low antibody specificity for di- and trimethylated lysines, or epitope occlusion because of neighboring marks (e.g. H3T3-phosphorylation).
DISCUSSION
The most common approach to bottom-up mass spectrometric analysis of histone post-translational modifications is based on reaction of histones with propionic anhydride in organic solvents before and after trypsin digestion. Although generally effective, this protocol has several limitations, the most significant being poor recovery of the peptides containing H3K4 di- and trimethyl marks. We systematically evaluated sample preparation methodology and found that the problem lies not with enzymatic digestion or an instrumental bias against these modified peptides, but with sample recovery and chromatographic behavior. A modified protocol was developed, incorporating predigestion aqueous phase lysine-propionylation and postdigestion labeling of peptide N termini with phenylisocyanate in a one-pot workflow. The method is simple, efficient, and enables detection and quantitation of H3K4 in all of its methylation states, along with all of the major methyl and acetyl marks for the other lysines on the N-terminal tail of histone H3. It is compatible with experimental designs employing label free quantitation as well as SILAC or stable isotope incorporation via phenyl isocyanate or propionylation. It is likewise compatible with all typical approaches to mass spectrometric detection and quantitation, including selected ion monitoring, selected reaction monitoring, parallel reaction monitoring, and data-independent MS/MS.
The new method increased the detected abundance of histone peptides by an average of over fourfold relative to the standard Prop-x2 workflow, and the most hydrophilic peptides such as H3K4me2 and me3 by almost two orders of magnitude. Better recovery of peptides is advantageous for quantitative comparisons, particularly for low abundance marks that would otherwise fall near their limits of detection. This is evident in experiments such as the siRNA knock-down of Wdr5 and Ash2, where H3K4 trimethyl marks are quantitatively decreased from an already low abundance. Other low abundance marks such as H3K4me2, H3K27me and H3K36me should likewise benefit from this technique. Although discrepancies in peptide recoveries can be compensated for with internal standards of defined concentration(32, 33), increased signal also affords better quality tandem mass spectra, leading to higher confidence assignment of spectra to peptide sequences and localization of the post-translational modifications.
Importantly, we show that phosphorylated proteins and their various combinations with other marks are also more readily detected. Among these was a tryptic peptide bearing the H3T3-phospho mark in the two RNAi-treated PC9 and HeLa cell lines (supplemental Fig. S5). Phosphorylation of T3 has been previously reported in studies that used IMAC-enrichment for phosphosites and chymotrypsin digestion (26), or limited endoproteinase Arg-C digestion to produce peptides amenable to LC-MS analysis (21). It is an axiom of proteomics that any protein or modification can be detected given sufficient material and degrees of fractionation, but our workflow allows histone marks varying in abundance by over three orders of magnitude to be detected and quantified in a single experiment. The simplicity and convenience of this one-pot/one shot workflow combined with bioinformatic tools such as the Fishtones application allows mass spectrometry to contribute powerfully to epigenetic research programs, through both fundamental biochemical studies as well as in the context of drug discovery and characterization.
Supplementary Material
Acknowledgments
We thank Benjamin Garcia for technical advice and invaluable discussions on histone PTM analysis, and to Tommy Lai, Liao Jiangpeng and Zheng Xiaoping of WuXi AppTec for synthesis of KDM small molecule inhibitors.
Footnotes
Author contributions: T.M., P.T., M.C., and D.A. designed research; T.M., A.I., T.C., G.G., C.T., F.Z., M.C., and D.A. performed research; A.M., J.L., and D.A. contributed new reagents or analytic tools; T.M., A.I., T.C., A.M., and D.A. analyzed data; T.M. and D.A. wrote the paper.
 This article contains supplemental Figs. S1 to S5, Table S1, and Annotated Spectra files.
This article contains supplemental Figs. S1 to S5, Table S1, and Annotated Spectra files.
1 The abbreviations used are:
- PTM
- post-translational modification
- H3
- histone H3
- me1
- mono-methylation
- me2
- di-methylation
- me3
- tri-methylation
- LC-MS/MS
- Tandem liquid chromatography mass spectrometry
- PIC
- Phenyl isocyanate
- HCD
- higher energy collisional dissociation
- C18
- Carbon 18 column
- ac
- acetylation
- phos
- phosphorylation
- pr
- propionylation
- HMT
- histone methyl transferase
- FMOC-Cl
- Fluorenylmethyloxycarbonyl chloride
- SILAC
- Stable isotope labeling by amino acids in cell culture.
REFERENCES
- 1. Helin K., Dhanak D. (2013) Chromatin proteins and modifications as drug targets. Nature 502, 480–488 [DOI] [PubMed] [Google Scholar]
- 2. Rotili D., Mai A. (2011) Targeting Histone Demethylases: A New Avenue for the Fight against Cancer. Genes Cancer 2, 663–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tan M., Luo H., Lee S., Jin F., Yang J. S., Montellier E., Buchou T., Cheng Z., Rousseaux S., Rajagopal N., Lu Z., Ye Z., Zhu Q., Wysocka J., Ye Y., Khochbin S., Ren B., Zhao Y. (2011) Identification of 67 Histone Marks and Histone Lysine Crotonylation as a New Type of Histone Modification. Cell 146, 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Egelhofer T. A., Minoda A., Klugman S., Lee K., Kolasinska-Zwierz P., Alekseyenko A. A., Cheung M. S., Day D. S., Gadel S., Gorchakov A. A., Gu T., Kharchenko P. V., Kuan S., Latorre I., Linder-Basso D., Luu Y., Ngo Q., Perry M., Rechtsteiner A., Riddle N. C., Schwartz Y. B., Shanower G. A., Vielle A., Ahringer J., Elgin S. C., Kuroda M. I., Pirrotta V., Ren B., Strome S., Park P. J., Karpen G. H., Hawkins R. D., Lieb J. D. (2011) An assessment of histone-modification antibody quality. Nat. Struct. Mol. Biol. 18, 91–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peach S. E., Rudomin E. L., Udeshi N. D., Carr S. A., Jaffe J. D. (2012) Quantitative assessment of chromatin immunoprecipitation grade antibodies directed against histone modifications reveals patterns of co-occurring marks on histone protein molecules. Mol. Cell. Proteomics 11, 128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Britton L. M., Gonzales-Cope M., Zee B. M., Garcia B. A. (2011) Breaking the histone code with quantitative mass spectrometry. Expert Rev. Proteomics 8, 631–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smith C. M., Gafken P. R., Zhang Z., Gottschling D. E., Smith J. B., Smith D. L. (2003) Mass spectrometric quantification of acetylation at specific lysines within the amino-terminal tail of histone H4. Anal. Biochem. 316, 23–33 [DOI] [PubMed] [Google Scholar]
- 8. Syka J. E., Marto J. A., Bai D. L., Horning S., Senko M. W., Schwartz J. C., Ueberheide B., Garcia B., Busby S., Muratore T., Shabanowitz J., Hunt D. F. (2004) Novel linear quadrupole ion trap/FT mass spectrometer: performance characterization and use in the comparative analysis of histone H3 post-translational modifications. J. Proteome Res. 3, 621–626 [DOI] [PubMed] [Google Scholar]
- 9. Garcia B. A., Mollah S., Ueberheide B. M., Busby S. A., Muratore T. L., Shabanowitz J., Hunt D. F. (2007) Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat. Protocols 2, 933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Plazas-Mayorca M. D., Zee B. M., Young N. L., Fingerman I. M., LeRoy G., Briggs S. D., Garcia B. A. (2009) One-pot shotgun quantitative mass spectrometry characterization of histones. J. Proteome Res. 8, 5367–5374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Drogaris P., Wurtele H., Masumoto H., Verreault A., Thibault P. (2008) Comprehensive Profiling of Histone Modifications Using a Label-Free Approach and Its Applications in Determining Structure-Function Relationships. Anal. Chem. 80, 6698–6707 [DOI] [PubMed] [Google Scholar]
- 12. Jaffe J. D., Wang Y., Chan H. M., Zhang J., Huether R., Kryukov G. V., Bhang H. E., Taylor J. E., Hu M., Englund N. P., Yan F., Wang Z., Robert McDonald E., Wei L., Ma J., Easton J., Yu Z., deBeaumount R., Gibaja V., Venkatesan K., Schlegel R., Sellers W. R., Keen N., Liu J., Caponigro G., Barretina J., Cooke V. G., Mullighan C., Carr S. A., Downing J. R., Garraway L. A., Stegmeier F. (2013) Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat. Genet. 45, 1386–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garcia B. A., Hake S. B., Diaz R. L., Kauer M., Morris S. A., Recht J., Shabanowitz J., Mishra N., Strahl B. D., Allis C. D., Hunt D. F. (2007) Organismal differences in post-translational modifications in histones H3 and H4. J. Biol. Chem. 282, 7641–7655 [DOI] [PubMed] [Google Scholar]
- 14. Pesavento J. J., Kim Y. B., Taylor G. K., Kelleher N. L. (2004) Shotgun annotation of histone modifications: A new approach for streamlined characterization of proteins by top down mass spectrometry. J. Am. Chem. Soc. 126, 3386–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Young N. L., DiMaggio P. A., Plazas-Mayorca M. D., Baliban R. C., Floudas C. A., Garcia B. A. (2009) High throughput characterization of combinatorial histone codes. Mol. Cell. Proteomics 8, 2266–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tang H., Fang H., Yin E., Brasier A. R., Sowers L. C., Zhang K. (2014) Multiplexed Parallel Reaction Monitoring Targeting Histone Modifications on the QExactive Mass Spectrometer. Anal. Chem. 86, 5526–5534 [DOI] [PubMed] [Google Scholar]
- 17. Soldi M., Cuomo A., Bremang M., Bonaldi T. (2013) Mass spectrometry-based proteomics for the analysis of chromatin structure and dynamics. Int. J. Mol. Sci. 14, 5402–5431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., Wang W., Weng Z., Green R. D., Crawford G. E., Ren B. (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 [DOI] [PubMed] [Google Scholar]
- 19. Barski A., Cuddapah S., Cui K., Roh T.-Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 20. Zee B. M., Levin R. S., Dimaggio P. A., Garcia B. A. (2010) Global turnover of histone post-translational modifications and variants in human cells. Epigenetics Chromatin 3, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jung H. R., Pasini D., Helin K., Jensen O. N. (2010) Quantitative Mass Spectrometry of Histones H3.2 and H3.3 in Suz12-deficient Mouse Embryonic Stem Cells Reveals Distinct, Dynamic Post-translational Modifications at Lys-27 and Lys-36. Mol. Cell. Proteomics 9, 838–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Phanstiel D., Brumbaugh J., Berggren W. T., Conard K., Feng X., Levenstein M. E., McAlister G. C., Thomson J. A., Coon J. J. (2008) Mass spectrometry identifies and quantifies 74 unique histone H4 isoforms in differentiating human embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 105, 4093–4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao Y., Wang Y. (2007) A method to determine the ionization efficiency change of peptides caused by phosphorylation. J. Am. Soc. Mass Spectrom. 18, 1973–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Janek K., Wenschuh H., Bienert M., Krause E. (2001) Phosphopeptide analysis by positive and negative ion matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 15, 1593–1599 [DOI] [PubMed] [Google Scholar]
- 25. Arnott D., Liu P., Molina P., Phu L., Sandoval W. (2011) Manipulating the mass spectrometric properties of peptides through selective chemical modification. In: Ivanov A. R., Lazarev A. V., eds. Sample preparation in biological mass spectrometry, pp. 19–40, Springer; Netherlands [Google Scholar]
- 26. Garcia B. A., Barber C. M., Hake S. B., Ptak C., Turner F. B., Busby S. A., Shabanowitz J., Moran R. G., Allis C. D., Hunt D. F. (2005) Modifications of human histone H3 variants during mitosis. Biochemistry 44, 13202–13213 [DOI] [PubMed] [Google Scholar]
- 27. Dayon L., Hainard A., Licker V., Turck N., Kuhn K., Hochstrasser D. F., Burkhard P. R., Sanchez J. C. (2008) Relative quantification of proteins in human cerebrospinal fluids by MS/MS Using 6-Plex Isobaric Tags. Anal. Chem. 80, 2921–2931 [DOI] [PubMed] [Google Scholar]
- 28. Lyons C. E., Victor K. G., Moshnikov S. A., Bachmann L. M., Baras A. S., Dettmann K. M., Cross J. V., Templeton D. J. (2011) PICquant: a quantitative platform to measure differential peptide abundance using dual-isotopic labeling with 12C6- and 13C6-phenyl isocyanate. Anal. Chem. 83, 856–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mason D. E., Liebler D. C. (2003) Quantitative analysis of modified proteins by LC-MS/MS of peptides labeled with phenyl isocyanate. J. Proteome Res. 2, 265–272 [DOI] [PubMed] [Google Scholar]
- 30. Liao R., Wu H., Deng H., Yu Y., Hu M., Zhai H., Yang P., Zhou S., Yi W. (2013) Specific and efficient N-propionylation of histones with propionic acid N-hydroxysuccinimide ester for histone marks characterization by LC-MS. Anal. Chem. 85, 2253–2259 [DOI] [PubMed] [Google Scholar]
- 31. Shilatifard A. (2012) The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Ann. Rev. Biochem. 81, 65–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Creech A. L., Taylor J. E., Maier V. K., Wu X., Feeney C. M., Udeshi N. D., Peach S. E., Boehm J. S., Lee J. T., Carr S. A., Jaffe J. D. (2015) Building the Connectivity Map of epigenetics: Chromatin profiling by quantitative targeted mass spectrometry. Methods 72, 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin S., Wein S., Gonzales-Cope M., Otte G. L., Yuan Z. F., Afjehi-sadat L., Maile T., Berger S. L., Rush J., Lill J. R., Arnott D., Garcia B. A. (2014) Stable Isotope labeled histone peptide library for histone post-translational modification and variant quantification by mass spectrometry. Mol. Cell. Proteomics Mol. Cell. Proteomics 13, 2450–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.