Abstract
The proteasome is a giant protease responsible for degradation of the majority of cytosolic proteins. Competitive inhibitors of the proteasome are used against aggressive blood cancers. However, broadening the use of proteasome-targeting drugs requires new mechanistic approaches to the enzyme’s inhibition. In our previous studies we described Tat1 peptide, an allosteric inhibitor of the proteasome derived from a fragment of the basic domain of HIV-Tat1 protein. Here, we attempted to dissect the structural determinants of the proteasome inhibition by Tat1. Single- and multiple- alanine walking scans were performed. Tat1 analogs with stabilized beta-turn conformation at positions 4–5 and 8–9, pointed out by the molecular dynamics modeling and the alanine scan, were synthesized. Structure of Tat1 analogs were analyzed by circular dichroism, Fourier transform infrared and nuclear magnetic resonance spectroscopy studies, supplemented by molecular dynamics simulations. Biological activity tests and structural studies revealed that high flexibility and exposed positive charge are hallmarks of Tat1 peptide. Interestingly, stabilization of a beta-turn at the 8–9 position was necessary to significantly improve the inhibitory potency.
Introduction
The 26S proteasome, a main component of the ubiquitin-proteasome proteolytic system, consists of the 20S catalytic core and two 19S regulatory particles (RP) attached to both sides of the core. The 19S RP is responsible for recognition and binding of polyubiquitinylated protein substrates, their deubiquitinylation, unfolding, and translocation to the catalytic chamber [1]. Two other proteasome activators - 11S (PA28/REG) and PA200 do not process polyubiquitinylated proteins. The PA28 αβ/REG/11S activator enhances proteolysis of peptides and unstructured proteins and its physiological role is to activate production of antigenic peptides [2]. PA200 is predominantly found in PA200-20S-19S complexes, which are involved mainly in DNA repair and maintaining mitochondrial function [3].
The 20S core is built with four stacked heptameric rings arranged in the αββα order [4]. The inner β-rings harbor six active centers with three distinct activities: post-glutamyl peptide hydrolyzing (PGPH), trypsin-like (T-L) and chymotrypsin-like (ChT-L), located in β1, β2 and β5 subunits, respectively. An access of protein substrates to the catalytic chamber is restricted by the tightly packed N-terminal tails of the α subunits, forming the regulated gate [5], which can be opened by binding the 11S, 19S or PA200 activators. It is postulated that all the activating particles display a common mechanism of action, involving anchoring through their C-terminal residues to the proteasome surface, and opening the gate by repositioning the Pro17 reverse turn in one or multiple α-subunits [6]. The activators do not only force gate opening, but also influence the proteasome catalytic performance by allosteric signalling [7]. Importantly, allosteric signalling plays a critical yet not fully understood role in the functioning of the 20S core and its interactions with ligands [8–10].
The ubiquitin-proteasome system is involved in degradation of a variety of proteins, regulating either directly or indirectly many cellular processes, such as signal transduction, metabolism, cell cycle, and apoptosis [11]. This broad influence on physiological processes makes the proteasome an important target for drugs [12], especially anti-cancer [13,14]. Many competitive active sites-directed (orthosteric) inhibitors of the proteasome have been developed. Two of them, bortezomib and carfilzomib, are already used in the treatment of blood cancers, and a few others are in clinical trials [15,16]. Unfortunately, a significant number of patients do not respond to these drugs or develop the drug resistance [17]. Allosteric modulators provide a promising alternative to the orthosteric competitive inhibitors. Allosteric regulation of an enzyme or receptor activity involves binding of an effector to a site distinct from the active site, modifying its affinity toward natural substrates through long-distance conformational changes [18]. Allosteric modulators may serve as exceptionally precise tools, since they bind to very unique sites that are generally independent from the main function of the protein. If allosteric binding sites are not already involved in regulation of the target protein they are likely under lower evolutionary pressure than catalytic sites [19,20]. Often allosteric modulators may distinguish between protein targets belonging to the same family and possessing similar active sites, what in turn allows for avoiding many side effects typical for the therapy with orthosteric modulators. It is thus not surprising there is an increasing interest in utilizing the allosteric modulation phenomena in drug design [21–23].
We postulate that allosteric modulators of the proteasome may improve efficacy of treatment, limit off-target effects, and help to overcome resistance to competitive drugs. Recently, several allosteric inhibitors of the proteasome have been identified, including proline and arginine rich peptides [24], compounds with imidazoline scaffolds [25,26], chloroquine [27], clioquinol [28], rapamycin [29] and 5-amino-8-hydroxyquinoline, some of them overcoming certain forms of bortezomib resistance in vitro [26,30]. In our search for allosteric modulators of the proteasome proteolytic activity, we focused on fragments of Tat (transactivator) protein from human immunodeficiency virus type-1. This poorly structured 86-residue, RNA-binding protein regulates expression of viral and host genes, including genes encoding immunoproteasome subunits, in virus-infected human cells [31]. It was observed that, besides its canonical function, the HIV-1 Tat protein competes with the 11S activator and also inhibits the core 20S proteasome [32]. These actions are a part of the broad assault of the virus on cellular immune response [31]. The protein was found to bind to several single, separately expressed α and β subunits [33], however the binding was likely encouraged by the lack of whole-assembly context. To the contrary, the competition with PA28/REG activator is likely a result of much more specific interactions of HIV-1 Tat with the α-face of the core proteasome. Interestingly, the 11S activator shares the RTP (REG/Tat proteasome binding) motif with the viral protein [34]. The RTP site is located near the C-terminal region and activation loop of the PA28/REG and in the essential basic domain of HIV-1 Tat [35]. In our previous studies we concentrated on these specific interactions and designed short, twelve-residue peptides–fragments of the basic domain containing the RTP site [36,37] We found that the peptide Tat1 (G48RKKRRQRRRPS59), inhibits the ChT-L peptidase of the activated 20S proteasome in a noncompetitive manner [36]. The result, together with competition of the peptide with 11S activator [36], strongly implies that Tat1 does not bind to the enzyme active sites, but instead it interferes with binding of the 11S activator. This suggests that the binding site for the peptide resides on the α face, within the proteasome α-subunits. Moreover, the Tat1 peptide is most likely an allosteric inhibitor: it affects the three proteasome peptidase activities in a very distinct manner [36]. This excludes the notion that the Tat1 actions stem from a simple blocking of the proteasome gate and attenuating the flow of substrates. Interestingly, in preliminary experiments we found that treatment with Tat-derived peptides, including Tat1, does affect cell viability and activities of proteasome assemblies in cultured cells (data not shown). To optimize the structure of Tat1-based modulators, we attempted to identify residues that are crucial for the peptide activity. To define the pharmacophores we performed single and multiple alanine scans and tested activity of the 20S proteasome treated with the modified peptides. We also characterized the structure of the peptides with Fourier transform infrared (FTIR) and circular dichroism (CD) spectroscopy. As a result, we identified two “hot spots” in the sequence of Tat1, comprising the residues 4–6 and 8–10. According to the molecular modeling (MD) these regions are prone to the formation of a structural turn [36]. Therefore, we incorporated a potential β-turn-inducer, the Tic-Oic moiety, to these regions to obtain peptidomimetics with a stabilized secondary structure. Next, we characterized these peptidomimetics with CD, FTIR and nuclear magnetic resonance (NMR) spectroscopy, and with molecular modeling. Propensity of certain analogs to bind the proteasome was studied with microscale thermophoresis. Biological and structural properties of the peptidomimetics indicate that although a strong positive charge is important for their inhibitory potency, a proper spatial orientation of the charged amino acid side chains is no less significant.
Results
Ala substitutions decreased Tat1 inhibitory potency of Tat1
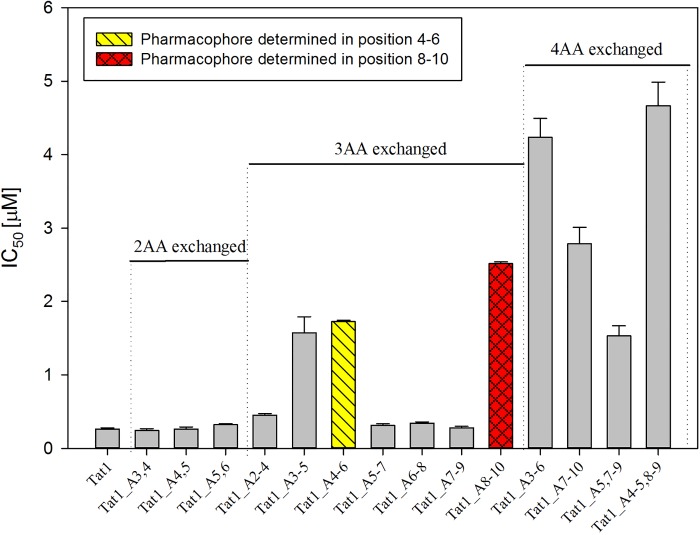
To identify a pharmacophore responsible for the biological activity of Tat1 we systematically replaced selected residues in the Tat1 sequence to alanine (Table 1, S1 Table).
Table 1. The Ala-scan analogs of Tat1 diversely affect the ChT-L peptidase of the 20S proteasome.
| Compound | Sequence | IC50 [μM] |
|---|---|---|
| Tat1 | H-GRKKRRQRRRPS-OH | 0.26 (±0.00) |
| Tat1_A3-4 | H-GRAARRQRRRPS-OH | 0.24 (±0.02) |
| Tat1_A4-5 | H-GRKAARQRRRPS-OH | 0.26 (±0.03) |
| Tat1_A5-6 | H-GRKKAAQRRRPS-OH | 0.32 (±0.02) |
| Tat1_A2-4 | H-GAAARRQRRRPS-OH | 0.45 (±0.02) |
| Tat1_A3-5 | H-GRAAARQRRRPS-OH | 1.57 (±0.22) |
| Tat1_A4-6 | H-GRKAAAQRRRPS-OH | 1.72 (±0.02) |
| Tat1_A5-7 | H-GRKKAAARRRPS-OH | 0.31 (± 0.00) |
| Tat1_A5,7–9 | H-GRKKARAAARPS-OH | 1.53 (±0.14) |
| Tat1_A6-8 | H-GRKKRAAARRPS-OH | 0.34 (±0.01) |
| Tat1_A7-9 | H-GRKKRRAAARPS-OH | 0.28 (±0.01) |
| Tat1_A8-10 | H-GRKKRRQAAAPS-OH | 2.52 (±0.03) |
| Tat1_A7-10 | H-GRKKRRAAAAPS-OH | 2.79 (±0.22) |
| Tat1_A3-6 | H-GRAAAAQRRRPS-OH | 4.24 (±0.26) |
| Tat1_A4-5,8–9 | H-GRKAARQAARPS-OH | 4.66 (±0.32) |
| Tat1_4-5TO | H-GRK-d-TicOicRQRRRPS-OH | 0.24 (±0.02) |
| Tat1_8-9TO | H-GRKKRRQ-d-TicOicRPS-OH | 0.17 (±0.03) |
| Tat1_8-9TOD | H-GRKKRRQTic-d-OicRPS-OH | 0.12 (±0.01) |
| Tat1_4-5TO,8-9TOD | H-GRK-d-TicOicRQTic-d-OicRPS-OH | 0.24 (±0.05) |
An initial single-Ala walking scan did not identify residues that influenced the ability of Tat1 to inhibit the ChT-L peptidase of the 20S core proteasome (CP). Indeed, the activity of CP treated with 1 μM of these peptides varied by no more than 4% comparing to Tat1 (S1 Fig). Therefore, in the next step we proceeded with multiple Ala walking scans by exchanging blocks of 2–4 adjacent amino acid residues into alanines.
Values of half maximal inhibitory concentration (IC50) calculated for these analogs are presented in Table 1. Substitution of two adjacent residues (Tat1_A3-4, Tat1_A4-5, and Tat1_A5-6) had a weak (Tat1_A5-6) or insignificant influence on the inhibitory potency of the peptides. Similarly, the peptides in which three consecutive residues were exchanged but only two of them were basic (Tat1_A5-7, Tat1_A6-8 and Tat1_A7-9), displayed activity comparable to Tat1. However, the importance of charge became apparent when three basic residues were replaced with hydrophobic Ala. Such peptides (Tat1_A2-4, Tat1_A3-5, Tat1_A4-6, Tat1_A8-10) inhibited the 20S proteasome less efficiently than Tat1. Furthermore, the effect depended on the position of the exchanged residues. The most pronounced response to the basic residues substitution was observed for Tat1_A8-10, for which IC50 increased almost 10 times. In contrast, substitutions of residues closer to the N-terminus showed a limited influence on the ChT-L activity. The substitution of residues had the least significant effect in the case of Tat1_A2-4, for which IC50 was only about two times higher than for Tat1. The essential role of the C-terminal basic residues for the peptides inhibitory potency was further demonstrated with Tat1_A5,7–9 and Tat1_A7-10. In both peptides Ala replaced four residues where three were basic. Despite the identical number of the exchanged residues, the peptide which lost three positively charged residues in the 8–10 positions was a weaker inhibitor of ChT-L peptidase (IC50 = 2.79 μM) than the compound, in which two basic residues from the C-terminus and one in the position 5 were replaced by Ala (IC50 = 1.53 μM). Analysis of biological activity of Tat1 analogs with Ala-substitutions in the central part of the sequence suggested importance of yet another region in the efficient inhibition of the 20S proteasome. Among the peptides in which three Arg/Lys residues between position 3 and 8 were replaced by Ala, the Tat1_A4-6 was the poorest inhibitor. Furthermore, removal of four basic residues from the position 3 to 6 resulted in the loss of inhibitory potency of the Tat1_A3-6, further confirming importance of this region (Table 1).
Incorporation of a Tic-Oic moiety into the pharmacophore sites specifically improves inhibitory potential of Tat1
Interestingly, the central and close to the C-terminus regions identified by the multiple-alanine walking scan were also recognized in our previous study [36]. Namely, results of molecular dynamics modeling suggested that the regions comprising residues 4–5 and 8–9 have a high propensity to form structural turns. To test whether the turns influence Tat1 activity, we introduced a dipeptide mimetic Tic-Oic (Tic: 1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid, Oic: (2S,3aS,7aS)-octahydro-1H-indole-2-carboxylic acid) into the two postulated pharmacophore sites (Table 1), aiming to induce a stabilized β-turn conformation [38]. To check how stereochemical properties influence biological activity, we utilized both d- and l- Tic and Oic units in two combinations: d-Tic,l-Oic, incorporated to the peptides Tat1_4-5TO and Tat1_8-9TO, and l-Tic,d-Oic utilized in the peptides Tat1_8-9TOD and Tat1_4-5TO,8-9TOD. We assumed that the turn structures induced by d-Tic,l-Oic and l-Tic,d-Oic would trigger a distinct spatial arrangement of the side chains, what might impact biological activity of the peptides.
Due to the Tic-Oic modification the overall positive charge of the peptides decreased because Tic-Oic moiety substituted either two (Tat1_4-5TO, Tat1_8-9TO, Tat1_8-9TOD) or four Arg/Lys residues (Tat1_4-5TO,8-9TOD). As a control we prepared Tat1_A4-5,8–9 peptide, in which four Ala residues were introduced in the relevant regions. Comparison of biological effectiveness of the obtained compounds indicated that their inhibitory capacity is not purely dependent on charge. While the peptide with four basic residues replaced by Ala had drastically reduced potency to inhibit the 20S proteasome (Table 1), Tat1_4-5TO, Tat1_8-9TO/TOD and Tat1_4-5TO,8-9TOD inhibited the 20S core with at least comparable effectiveness as Tat1 (Fig 1). Importantly, Tat1_8-9TO and especially Tat1_8-9TOD inhibited the 20S proteasome significantly better than Tat1, with IC50 reduced by half.
Fig 1. Inhibitory activity of the peptides with “pharmacophore” regions modified by the Tic-Oic moiety.
It thus seems that incorporation of the Tic-Oic moiety as a β-turn inducer leads to a locally more rigid peptide structure that in the certain context supports a better fit to proteasome binding domain(s) and consequently stimulates more efficient propagation of allosteric signals. Moreover, l-Tic,d-Oic at the position 8–9 provides a better fitting of the ligand since IC50 for Tat1_8-9TOD is about 30% lower than for its counterpart with d-Tic,l-Oic unit.
The peptide with Ala substitutions in both pharmacophore regions forms the least stable complex with the proteasome
Microscale thermophoresis detects changes in the mobility of protein molecules in temperature gradients generated by IR laser radiation. The thermophoretic effect is very sensitive to induced by a ligand binding alterations in the charge, size and/or the hydration shell of a studied protein [39]. Quantification of these changes is based on either intrinsic fluorophores such as tryptophan or covalently bound dyes. Titration of the selected peptides/peptidomimetics to a constant concentration of the fluorescently labeled human 20S proteasome allowed us to determine ligand dissociation constants (Kd), and systematically compare their binding affinity. The calculated Kd values (S2 Table) were substantially higher than those expected based on the corresponding IC50, which most probably were the result of the introduced labeling, likely decreasing the affinity of the peptides to the 20S proteasome. Moreover, the labelling also decreased the peptidase activity of the proteasome. Despite these limitations, the binding tendency of the studied Tat1 analogs followed their inhibitory capacity (S2 Fig). For example, Tat1_A4-5,8–9, the worst 20S inhibitor in this set of modulators, was found as forming the least stable complex with the proteasome (Kd = 21.40 μM). The equivalent peptidomimetic with Tic-Oic units simultaneously incorporated into the 4–5 and 8–9 positions was characterized by a much lower dissociation constant (Kd = 8.38 μM) indicating stronger interaction. Importantly, a comparison of the Kd values for Tat1_8-9TO and Tat1_8-9TOD confirmed a better fitting of the l-Tic,d-Oic than d-Tic,l-Oic moiety to the proteasome surface, as was suggested on the basis of the biological activity results.
FTIR determined conformation of Tat1 analogs depends on their structural flexibility
The amide I band (1600–1700 cm-1), that represents predominantly the stretching vibration of the amide C = O group, usually serves as a structural probe in FTIR of peptides and proteins [40]. Since the strength of C = O bond depends greatly on its hydrogen bonds network, various elements of secondary structure give rise to different but strongly overlapping signature bands that form an amide I envelope. These bands can be extracted from the envelope by carrying out the second derivative transformation or deconvolution of the spectra [41,42].
FTIR spectra of the parental Tat1 peptide were recorded either in a form of an aqueous solution or as a film prepared by solvent evaporation from the samples spotted on a FTIR window. The latter approach allowed elimination of subtracting a water background though not interfering with the band deconvolution. We applied this method since it was reported (and confirmed by us [43]) that evaporation of a solvent in most cases preserves the conformation [44]. Apparently, short linear peptides such as Tat1 and its analogs did not follow this rule, since the spectra of the peptides in solution and in the form of a film were distinct.
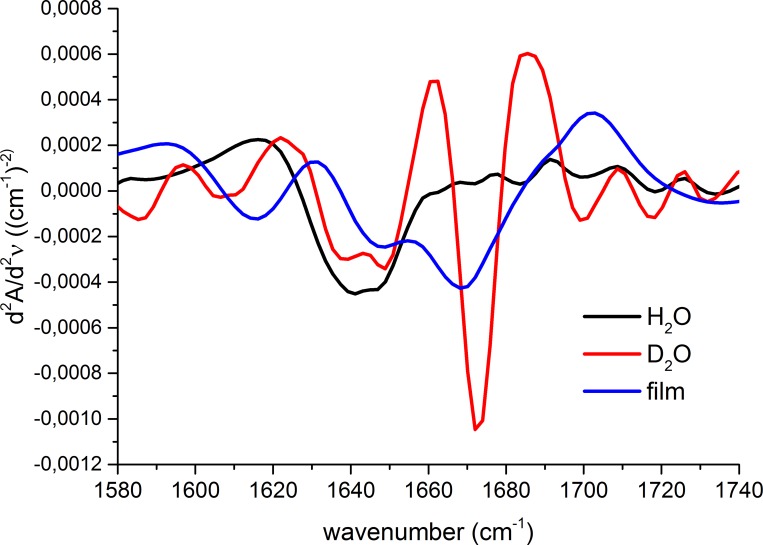
For Tat1 peptide in water, a single broad minimum extending between 1630 and 1655 cm-1 was detected (Fig 2). This minimum was probably formed by a combination of at least two bands centered at about 1638 and 1645 cm-1, which can be assigned to a β-sheet and a random coil conformation, respectively [45]. The β-sheet conformation was further confirmed by presence of another band typical for an antiparallel β-sheet, above 1690 cm-1. The other, very weak, minima localized at about 1670 and above 1680 cm-1 can be ascribed to turns.
Fig 2. The second derivative of FTIR spectra of Tat1, recorded in H2O, D2O and in a film mode.
In contrast, Tat1 in the form of a film produced three separate minima–at about 1615, 1645, and 1670 cm-1, which were attributed to the polyproline II (PPII), random coil, and turn conformations, respectively [36,46] (Fig 2). Among them, the minimum located at 1670 cm-1 was the most pronounced suggesting that the structural constrains imposed by the film increased preferences for the turn conformation. Surprisingly, this minimum also appeared as the main feature of Tat1 spectrum recorded in D2O (Fig 2). Preservation of this minimum implicates that although the bent structure of Tat1 was not stabilized by O…H and/or N…H hydrogen bonds strongly enough to be a preferred conformation in aqueous solution, the O…D and/or N…D bonds were sufficiently stronger to support the turn.
All the Ala-scan peptides in an aqueous solution consistently produced a broad minimum with the center between 1640 and 1650 cm-1 (S3 Fig), representing an unordered conformation. Spectra of these peptides in the form of a film displayed two minima. The first broad minimum extended between 1625 and 1650 cm-1 indicating an unordered conformation with some preferences for a β-sheet. The second and dominating minimum at about 1680 cm-1 was assigned to turns. For the selected peptides we also collected spectra in D2O. Similarly to the response of Tat1 to heavy water, these peptides adopted a more ordered conformation reflected by predominance of the minimum at 1670 cm-1 that is a hallmark of a turn structure (S4 Fig).
CD confirms propensity of the Ala-scan peptides for adopting an extended conformation
A CD spectrum of Tat1 displayed a weak maximum around 220 nm and a deep minimum at 195–198 nm [36], which together constitute a signature of an unordered or polyproline II (PPII) conformation [47,48]. The previously observed temperature-dependent changes in the spectrum of Tat1 strongly support the latter assignment [36]. The recorded CD spectra for Tat1 analogs, with both the single and multiple Ala substitutions, in general follow the pattern observed for the parental Tat1, indicating the Ala scan peptides also adopt an extended PPII conformation (S5 Fig).
A β-turn is present in the peptides with the TOD unit in the position 8–9
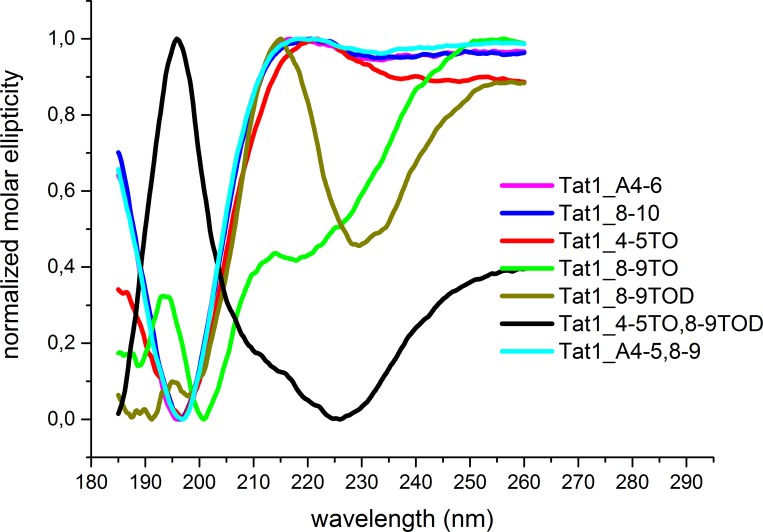
More variations appeared in CD spectra of Tat1 analogs with the Tic-Oic moiety. Tat1_4-5TO peptide displayed a CD spectrum similar to Tat1 and its Ala-scan analogs, the most probably assuming the PPII conformation (Fig 3). In contrast, CD spectra of Tat1-8-9TOD and Tat1_4-5TO,8-9TOD peptides indicated that the incorporation of the l-Tic,d-Oic moiety in the position 8–9 induced a stable turn, likely of the β-type (Fig 3).
Fig 3. Normalized CD spectra of Tat1 analogs in which the pharmacophore residues were substituted by either Ala or Tic-Oic moiety.
Turns are generally aperiodic secondary structures that are defined by four successive backbone torsional angles. Since the average dihedral angle values may vary at least by ±10° due to the flexibility of the structure, the turn character can be manifested in many different substructures. Using a semi-empirical approach, Woody distinguished four patterns of CD spectra characteristic for the β-turn motifs [49]. Tat1_4-5TO,8-9TOD peptide displayed a typical class C spectrum, since it showed an intensive maximum at about 195 nm and a broad α-helix-like minimum between 210 and 230 nm. This type of a spectrum was indicative of the βI conformation [50] (Fig 3). In turn, a CD spectrum of the Tat1_8-9TOD peptide, showing a minimum around 190 nm and a maximum at 215 nm, belonged to a class B spectrum, diagnostic for the βII conformation [50]. Finally, the CD spectrum of Tat1_8-9TO suggested a random coil conformation. It thus seems that l-Tic,d-Oic incorporated in the 8–9 region stabilizes the conformation better than its d-Tic,l-Oic counterpart.
Changes in chemical shifts indicate structural rearrangements imposed by the Tic-Oic moiety
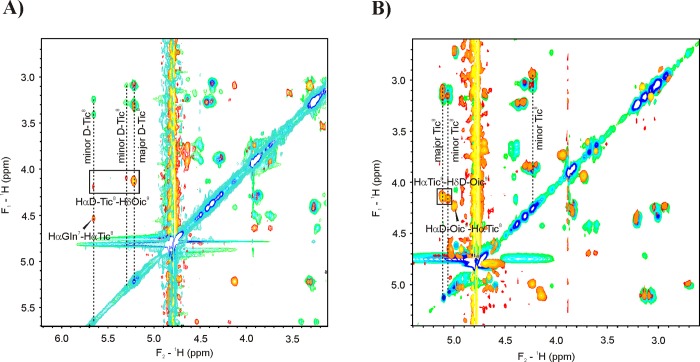
A cis-trans interconversion of the amide bond leads in some cases to two or more sets of resonances observed in NMR spectra [51]. Indeed, a proline residue present in the sequence of Tat1 induced the NMR detectable cis/trans isomerization [52–55]. Due to additional tertiary amide groups present in the TO/TOD analogs of Tat1, their spectra show distinct sets of proton resonances connected with cis/trans isomerization of not only Arg-Pro but also Xaa-Tic and Tic-Oic bonds (S3–S7 Tables). For each Tic-Oic modified peptide the abundance of major isomer was estimated based on the intensity of the non-overlapping signals of HN-Hα of the Gln7 residue of each isomer observed in the TOCSY spectrum. The content of the major conformation was usually in the range of 50–80%, the highest content was observed for the analog with Tic-Oic moiety in the positions 4–5. A poor separation of signals of the major and minor conformers, or low intensities of the latter, made a complete signal assignment for the minor isomers unreliable. Thus, the full assignment was done only for the major conformer of each peptide. The sequential connectivities dαδ(i,i+1) found in the Tic-Oic and Arg-Pro fragments indicated presence of trans peptide bonds. Additionally, the chemical shift dispersion of 13Cβ and 13Cγ (ΔδCβ-Cγ) in Pro residue, which falls in the range 4.7–5.1 ppm, confirmed the presence of the trans Arg-Pro peptide bond in the major conformations of all the peptides studied. The dαδ(d-Tic,Oic) cross peak was also found for the minor conformers of Tat1_4-5TO and Tat1_8-9TO analogs, indicating trans geometry of the Tic-Oic bond also in these isomers. In contrast, for Tat1_8-9TOD and Tat1_4-5TO,8-9TOD, the dαα(Tic,d-Oic) connectivity observed for one of the minor resonances evinces cis Tic8-d-Oic9 peptide bond (Fig 4). The absence of the sequential connectivities in the X-Tic dipeptide unit, i.e. dαδ(i,i+1) or dαα(i,i+1), precluded unambiguous determination of the peptide bond geometry even for the major species. However, in the ROESY spectrum of Tat1_8-9TO, the dαα(7,8) cross peak found for one of the minor isomers indicated a cis Gln7-Tic8 peptide bond (Fig 4).
Fig 4. Overlaid fragments of TOCSY (green-blue) and ROESY (red-yellow) spectra of Tat1_8-9TO (A) and Tat1_8-9TOD (B).
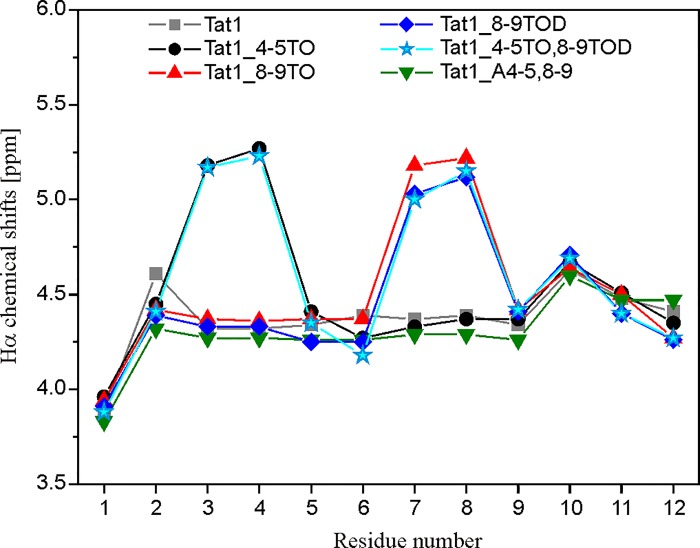
Incorporation of Ala residue in positions 4, 5, 8 and 9 of Tat1 did not affect the Hα chemical shifts comparing to Tat1 [36]. In turn, in the NMR spectra of the peptides modified with Tic-Oic unit a large downfield shift of the Hα of the amino acid preceding Tic was observed. The chemical shift variation was more than 0.8 ppm for both Lys3 (Tat1_4-5TO and Tat1_4-5TO,8-9TOD) and Gln7 (Tat1_8-9TO, Tat1_8-9TOD and Tat1_4-5TO,8-9TOD) (Fig 5). These variations are potential indicators of conformational changes imposed by the TO/TOD replacement. On the other hand, by analogy to Pro [56,57], Tic and Oic are expected to have extensive effects on Hα chemical shift of the preceding residues due to the reduced conformational degrees of freedom associated with the cyclic nature of this amino acids.
Fig 5. Chemical shift variation plot between Hα resonances of major conformations of Tat1, Tat1_4-5TO, Tat1_8-9TO, Tat1_8-9TOD, Tat1_4,5TO,8-9TOD and Tat1_A4-5,8–9.
The narrow HN chemical shift range (8.7–7.8 pm) found for the major conformation of each peptide was consistent with a solvent-exposed NH group that was not involved in hydrogen bonding [58]. Besides, no measurable ROEs were observed between any pair of the successive backbone amide groups (S6 Fig), which rather excluded a folded conformation. Nevertheless, the strong dαN(i,i+1) and dαδ(i,i+1) ROE cross-peaks indicated a short distance between these protons, which were expected in the ψ region of ~120±30° [59].
All the vicinal coupling constants, 3JHNHα, fell in the range of 6.5–9.3 Hz (S3–S7 Tables). Values in the range of 6.5–8 Hz are usually considered to be consistent with conformational averaging, whereas those greater than 8 Hz correspond to an extended state [51,60]. However, when the X-Pro and Pro-X pairs are in the β conformation, proline restricts conformational freedom of both the preceding and following it residues. Besides, there is a significant tendency of the residues which flank Pro to adopt the βP conformation, like the proline itself [59,61]. The same behavior should be expected for the proline-like Tic and Oic residues. Thus, in the case of TO/TOD analogs of Tat1, the 3JHNHα coupling constant values may suggest a backbone φ angle values of about -80°±20°, typical for a polyproline conformation [59].
Conformational ensemble obtained by MD with Time-Averaged Distance Restraints
Because our attempts to determine a secondary structure of the selected Tat1 analogs using ROE patterns and 3JHNHα scalar coupling constants did not give conclusive results, especially in regard to minor conformations, molecular dynamics simulations were performed. Due to overlapping signals in the NMR spectra, the structures of Tat1_4-5TO, Tat1_8-9TO, Tat1_8-9TOD, Tat1_4,5TO,8-9TOD, and Tat1_A4-5,8–9 were solved on the basis of only small number of NMR distance restraints (Table 2). The structures obtained in the last 800 ps of MD simulations with the time-averaged distance and dihedral angle restraints are displayed in Fig 6.
Table 2. Statistics of distance and dihedral angle restraints used for 3D structure calculations and quality ensemble of 200 NMR-derived structures of the peptides on the last steps of MD simulations with the time-averaged distance and dihedral angle restraints.
| Peptide | ||||||
|---|---|---|---|---|---|---|
| Tat1_4-5TO | Tat1_8-9TO | Tat1_8-9TOD | Tat1_4-5TO,8-9TOD | Tat1_A4-5,8–9 | ||
| Interproton distances: | total number | 57 | 36 | 48 | 48 | 71 |
| intra-residue | 46 | 27 | 39 | 37 | 59 | |
| i, i+1 | 11 | 9 | 9 | 11 | 11 | |
| i, i+2 | 0 | 0 | 0 | 0 | 1 | |
| Dihedral angle restraints a | 18 | 22 | 23 | 17 | 25 | |
| Structure b | 8,9 β I or IV (38%) | - | - | 6 γ(63%) | 5,6 β I or IV (81%) | |
| Hydrogen bonds c | s.c.6-CO5 | - | Hε10-CO8 | HN7-CO5 | s.c10-CO12 | |
| RMSD 1-9bb [Å] | 1.008 | 1.357 | 1.187 | 0.865 | 1.140 | |
| Rg heavy [Å] | 8.4 | 9.7 | 7.7 | 8.3 | 9.5 | |
aThe distance constraints and the 3JHNHα vicinal coupling constants were used in the HABAS algorithm of the DYANA package to generate φ, ψ and χ1 dihedral angle restraints.
b The content of the conformers with indicated turn structure is given in parentheses.
c Hydrogen bonds appearing in more than 50% of the structures in the final ensemble.
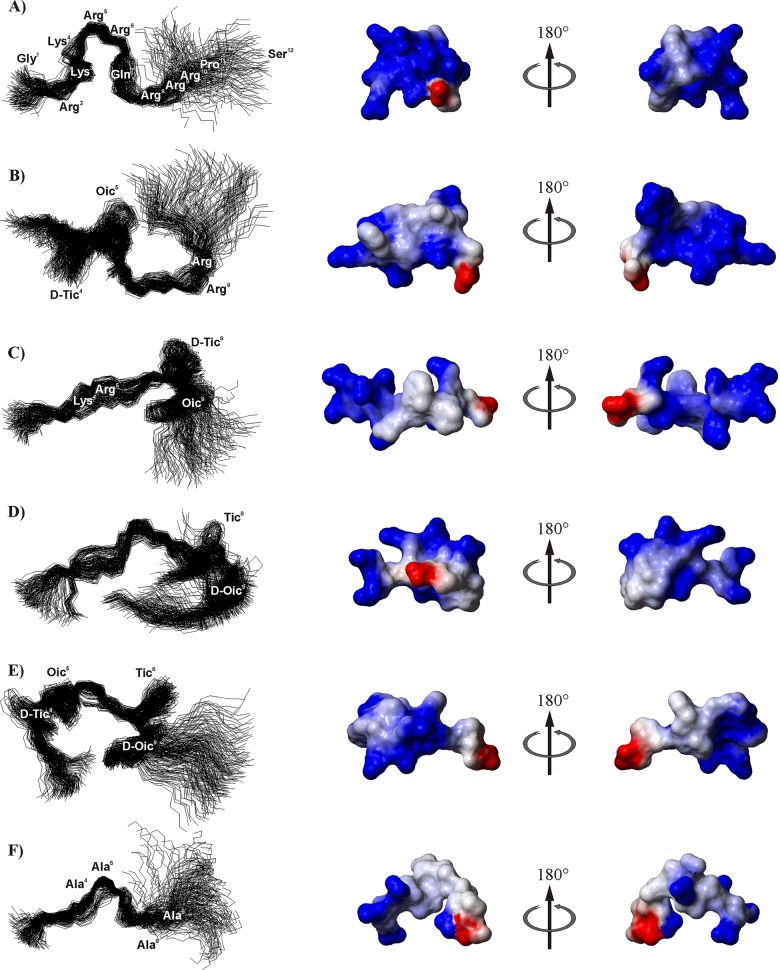
Fig 6. Superposed conformations obtained in the last 800 ps of MD simulations and surface electrostatic potential maps.
The maps indicate regions of negative potential in red, positive potential in blue, and neutral potential in white. The figures were generated by the MOLMOL). (A) Tat1 [36], (B) Tat1_4-5TO, (C) Tat1_8-9TO, (D) Tat1_8-9TOD, (E) Tat1_4-5TO,8-9TOD and (F) Tat1_A4-5,8–9. RMSD values are 0.800, 1.008, 1.357, 1.187, 0.865 and 1.140 Å for the backbone atoms in the fragment 1–9 of Tat1, Tat1_4-5TO, Tat1_8-9TO, Tat1_8-9TOD, Tat1_4-5TO,8-9TOD and Tat1_A4-5,8–9, respectively.
Collective scatter plots of the conformational states of each amino acid residue (S7 Fig) showed a high propensity of the peptides to adopt an extended (β-strand and/or PPII) conformation. The presence of Oic residue stabilizes locally the peptide conformation by fixing its φ and ψ dihedral angles to about -60° and 150°, respectively, similarly as proline [60,61]. In Tat1_4-5TO,8-9TOD, the change of Oic9 configuration from L into D resulted in the “mirror image” (φ,ψ) angles, i.e. 60° and -150°, respectively. In turn, in the case of Tat1_8-9TOD, the ψ dihedral angle of d-Oic9 was able to populate the left-handed α-helical region (ψ ≈ 40°) of the Ramachandran plot. The available backbone φ and ψ dihedral angles of l-Tic residue in Tat1_8-9TOD and Tat1_4-5TO,8-9TOD were limited to a small range around -120° and 60°, respectively, whereas the presence of d-Tic enantiomer gave rise to slight increase in population of distinct local conformations. Thus, based on the values of φ and ψ dihedral angles of the d-Tic residue, the conformations of Tat1_4-5TO and Tat1_8-9TO were grouped into two families, either with φ and ψ fixed at ca. 95° and -150° or at 130° and -70°. In turn, the d-Tic4 in Tat1_4-5TO,8-9TOD adopted preferably a 120° φ angle, whereas ψ angle was changing in the range of ca. -120 to -40°.. Nevertheless, according to the MD simulations, the Tic-Oic unit did not induce the turn structure in the place of its incorporation. Tat1_8-9TO analog adopted an extended conformation without any tendency for bent structures. In Tat1_4-5TO peptide, a single β-turn appeared exactly on the opposite terminus of the molecule than the Tic-Oic template, i.e. in the fragment Gln7-Arg8-Arg9-Arg10. This proposed β-turn was not stabilized by hydrogen bonds and was found in only ca. 40% of the structures (Table 2).
There were no β-turn structures detected in the peptides containing the Tic-d-Oic unit, i.e. Tat1_8-9TOD and Tat1_4-5TO,8-9TOD. Only a single inverse γ-turn with Arg6 at its apex was observed for the latter. In the case of Tat1_A4-5,8–9, a single non-hydrogen-bonded β-turn of type I or IV occupied a fragment Ala4-Ala5-Arg6-Gln7 of the peptide.
The average radii of gyration (Rg, the root mean square distance of the heavy atoms from their common center of gravity [62,63]) were 8.4, 9.7, 7.7, 8.3 and 9.5 Å, for Tat1_4-5TO, Tat1_8-9TO, Tat1_8-9TOD, Tat1_4-5TO,8-9TOD, and Tat1_A4-5,8–9, respectively. A lower value of Rg calculated for Tat1_8-9TOD was comparable with that obtained for Tat1 (7.5 Å) [36] and suggests a more compact structure than in other analogs.
The modification with neutral residues changed significantly the electrostatic potential of the peptides in comparison to Tat1 (Fig 6). Among the peptides studied, the Tat1_4-5TO,8-9TOD and Tat1_A4-5,8–9, due to replacement of as many as four positively charged residues with neutral residues, displayed the lowest total positive charge (+4). However, for the former the positively charged centers are rather concentrated on one side of the molecule, as for the other Tic-Oic modified peptides, while for the latter they seem to be distributed randomly over the whole peptide. Importantly, due to the extended conformation, all the residues were strongly water-exposed. The solvent-accessible surface area of each residue exceeded 30% with only two exceptions, i.e. Tic8 in Tat1_8-9TOD and Gln7 in Tat1_4-5TO,8-9TOD (S8 Fig).
Discussion
Since the proteasome is an attractive drug target there have been multiple attempts to find the best way to inhibit its proteolytic activity. The preference in the search for suitable compounds at first was directed toward short peptides equipped with a reactive electrophilic group that rendered the N-terminal threonine inactive. This approach resulted in a discovery of several peptide inhibitors that have been enormously useful in further deciphering of physiological role of the proteasome. The crown achievement of this strategy was a fast track introduction of bortezomib, a dipeptide equipped with a boronate group, to fight against advanced myeloma. The role of a peptide moiety in this design was to precisely bring the war head to its designated target. Since the reactive group chemically modifies the catalytic residue of the essential enzyme, there is a strong possibility of irreversible poisoning of healthy cells and off target biomolecules. To limit such effects, a line of inhibitors has been introduced that still compete with substrates for the active centers but they achieve their goal without modification of the catalytic residues. They represent a broad range of usually short peptides build with unnatural or modified amino acids that render the compounds indigestible by the proteases [64]. Finally, the third group of inhibitors takes advantage of the allosteric regulation of the 20S core activity. Currently, two main modes of allosteric regulation are explored. The more popular way employs the binding sites of the proteasome activators located on the α face to interfere with a signaling mostly from the gate to active centers. The second group focuses on hampering interactions between the α and β rings.
Tat1 peptide is derived from the binding domain of HIV-1 Tat protein that competes with the 11S activator, therefore it likely binds to the α face of the core. However, its exact binding site(s) remain elusive. Our previous studies established that Tat1 is a non-competitive inhibitor of 20S proteasome. Although in molecular modeling it predictably shows a high level of structural flexibility, it also exhibits a marked propensity to form two β-like turns. Here, we set out to rigorously test the hypothesis that the turns are at the core of the inhibitory properties of Tat1. The sequence of Tat1 is quite monotonous with two blocks of basic residues separated by a single glutamine residue and flanked with a single glycine on its amino terminus and Pro-Ser sequence on its carboxyl terminus. Long side chains of arginine and lysine residues somewhat limit spatial flexibility of the peptide. Not surprisingly, we had to apply alanine scans involving multiple consecutive residues to disable the quite impressive inhibitory potency of Tat1 and to identify two potential pharmacophores. This way, we successfully confirmed the previously suggested with the MD studies positioning of hot spots between residues 4–6 and 8–10. Indeed, substitution of residues within these regions with alanines led to at least 5 fold increase of IC50 measured for ChT-L activity of SDS activated 20S proteasome. All the above differences in IC50, although respectable, may not seem very impressive. However, these peptides are inherently flexible and sequentially monotonous therefore their residues likely substitute each other in interactions with the proteasome. Stiffening of Tat1 structure would restrict such flexibility. If the stiffening is accompanied by the proper exposure of the basic side chains, it will allow for not only retaining but even enhancement of the inhibitory capacity. Following this lead, we introduced a set of peptidomimetic residues Tic and Oic in the identified hot spots. The role of the Tic-Oic moiety was to induce or stabilize β-type turns strongly suggested by the MD. Unexpectedly, simultaneous incorporation of the beta turn inducer in the two hot spots did not bring substantial improvement to IC50. Moreover, the TO moiety at the position 4–5 helped with only a modest gain in IC50. We observed a significant decrease of IC50 only when the TO group was placed in the position 8–9. Importantly, this site was sensitive to stereochemistry of the moiety since incorporation of the TOD improved IC50 about 30% when compared with the TO group. Based on these results we propose that the positioning of the stable β-turn at the C-terminus is beneficial for inhibitory potency of the peptide. Although suggested by the MD simulations of Tat1, the simultaneous presence of stabilized rigid turns in both pharmacophore positions is not favorable. This indicates the improved design of Tat1 derivatives that includes a flexible longer N-terminal arm with a single β-turn placed close to the C-terminus capped with the Pro-Ser dipeptide. We predict that for correct binding either this arm should stay flexible or its correct structure is induced only when the β-turn docks.
The 12-mer of Tat1 is built with 8 basic residues grouped in sets of 5 and 3 members. Therefore, we expect that such accumulation of the positive charge has to play an important role in both maintaining preferred structure of the peptide and in its interactions with the target protein. Indeed, replacement of the charged residues with a much shorter and hydrophobic alanine partially removes steric hindrance of a long aliphatic chain decorated with a bulky guanidine group. Apparently, a high mobility decreases chances for the peptide to productively fit into the binding pocket(s). Additionally, the peptide may lose its specificity and probe alternative binding sites that have no or even an opposite effect on the peptidase activity. We are currently working toward solving a molecular structure of the proteasome with the bound Tat1 peptide to determine location of the binding pocket/s specific for allosteric inhibitors.
Intriguingly, we first identified the importance of structural dynamics of this peptide, when we compared the FTIR spectra collected in liquid and films. Contrary to our predictions, the Tat1 derivatives immobilized in a film or dissolved in D2O showed a clear propensity to form β-turns. The differences between the structures of Tat1 were surprising since we did not observe such dependence in a Tat2 structure. This linear peptide of identical length as Tat1 is also a non-competitive inhibitor of 20S proteasome, albeit with twice as high IC50 [37]. However, Tat2 contains an acidic Asp residue in its C-terminal part, which may influence the structural stability by formation of a salt bridge with one of several basic Arg/Lys residues present in the N-terminal part of the peptide. Since in the case of Tat1 there are no residues that could stabilize the structure through charge pairing, the peptide is in a random coil conformation when dissolved in water. On the other hand, however, Tat1 and its analogs seem to exist at the boundary of a more stable structure since limiting their flexibility by solvent evaporation or tightening the bonds due to the H/D exchange, enabled the peptides to achieve more ordered architecture with evident preferences for a turn conformation.
We recorded also CD spectra that reflect the averaged conformations of the peptides. The spectra were surprisingly sensitive to the position and stereochemistry of the introduced Tic-Oic substitution. We noticed that contrary to the CD spectrum of Tat1_8-9TO, representing random coil structure (Fig 4), Tat1_8-9TOD peptide preferentially adopted the βII turn conformation. This observation suggests that the change of stereochemistry around the Tic-Oic moiety in position 8–9 profoundly influenced flexibility of the peptide making it more rigid. Unexpectedly, the d-Tic,l-Oic unit, which stabilized an extended PPII conformation while present alone in the region 4–5, in the presence of 8-9TOD altered the bias from the βII toward the βI turn [44]. We concluded that it is necessary to take into account the complete structural context including both pharmacophore regions, to define the conformational preferences of Tat1-based modulators.
In our attempt to better structurally characterize the peptides we showed using NMR spectroscopy that the trans geometry of a peptide bond was preferred around the proline-like Tic and Oic residues. Since no measurable ROE cross-peaks were observed between the successive backbone amide groups (S6 Fig), we could not ascribe folded conformation to the major conformers of any TO/TOD peptides. Based on the strong dαN(i,i+1) and dαδ(i,i+1) ROE cross-peaks and the values of 3JHNHα coupling constant we suggested an extended, the most probably PPII conformation, for the major conformers of Tat1_4-5TO, Tat1_8-9TO, Tat1_8-9TOD and Tat1_4-5TO,8-9TOD.
We performed the MD simulations using NMR-derived time-averaged distance and dihedral angle restraints. In general, they confirmed a high propensity of the studied compounds to adopt an extended (β-strand and/or PPII) conformation. MD proposed also turn structures, such as a β-turn involving the fragment Gln7-Arg8-Arg9-Arg10 in Tat1_4-5TO; an inverse γ-turn with Arg6 at its apex in Tat1_4-5TO,8-9TOD; and a β-turn of type I or IV occupying a fragment Ala4-Ala5-Arg6-Gln7 in Tat1_A4-5,8–9, but none of them was hydrogen bonded. Flexibility of the peptides due to the lack of structure stabilizing intramolecular hydrogen interactions, as well as lack of clear relationship between the structural preferences and affinity toward the proteasome seems to advocate for the induced-fit binding of the TO/TOD Tat1 analogs to the 20S proteasome. On the other hand, while NMR and FTIR spectroscopy did not reveal any turns for Tat1_8-9TOD nor Tat1_4,5TO,8-9TOD (Table 2, S9 Fig), CD spectra of these two peptidomimetics displayed distinct spectral features of the βII and βI turn, respectively (Fig 3). One of possible explanations for this discrepancy is concentration-induced conformational changes. NMR and FTIR experiments required concentration of the sample about 25 to 100 times higher than CD spectroscopy. Likely the concentration of the analyte for the very polar and basic Tat1 analogs is crucial to stabilize the extended structure with the extensive solvent-mediated interactions. Significantly lower peptide concentrations used in the CD experiments were closer to the concentrations of the modulators employed in the biological tests what implies that the CD derived structural data may be more relevant when establishing the structure-activity correlation for the case of Tat1 analogs.
Interestingly, Tat1 peptide is not the sole example of a basic, Arg-rich proteasome modulator. The most notable examples are Pro and Arg-rich peptides (PR peptides), which inhibit the activated 20S proteasome in vitro at high nanomolar/low-micromolar concentrations, and preferentially inhibit degradation of selected protein substrates in vivo. The best studied and shortest 11-residue PR11 may contain a structural turn and is hypothesized to bind at the outer edge of the proteasomal α ring [24]. Another example is octa-arginine, inhibiting the proteasome peptidase activity in vitro with high-nanomolar IC50, and leading to accumulation of proteasome substrates in cultured cells [65]. The binding site(s) and mode(s) of this mixed-action inhibitor are unknown.
It remains to be established whether the introduction of β-turns via the Tic-Oic moiety has a unique influence on the inhibitory properties of Tat1 peptide or it could be recapitulated with other turn inducers. Our preliminary exploration of the chemical space indicates that other turn inducers can to some extent substitute for Tic-Oic [66]. The structure of the region 4–6 is important for inhibition of the 20S proteasome. However, it is not clear if the region would remain flexible after docking to the proteasome or if it strikes a unique pose. Apparently, stiffening of the 4–6 region before the peptide docking is not deleterious for the inhibitory potency but it does not improve it either. Finally, although we are confident that our peptides are not digested by proteasome at a MS-detectable rate, they may still influence other than ChT-L active centers and the digest of protein substrates in a hard to predict way. We currently work fervently toward tackling these questions.
Conclusions
In this work we attempted to identify and improve pharmacophores of Tat1 peptide which constitutes an important example of a non-competitive, likely allosteric inhibitor of the 20S proteasome. We found a single β-turn promoting region specifically positioned close to the C-terminus in the sequence of Tat1, which when stiffened with the β-turn inducing Tic-Oic moiety produced a substantially improved inhibitor. The accumulated spectroscopic and biological data for Ala and Tic-Oic substituted analogs of Tat1 suggests a complex interplay of the structural flexibility and charge in regulation of the peptides inhibitory potency. The current data also support the possibility that due to the structural flexibility of these peptides they can follow an induced fit pathway when they probe the 20S proteasome as a potential receptor. It appears that the Tat1 constitutes a unique structural scaffold on which we plan to build further allosteric inhibitors of 20S proteasome that could be expanded into specific regulators of this essential protease.
Materials and Methods
Synthesis of Ta1 analogs
The alanine analogs of Tat1 peptide were synthesized on a pre-loaded Wang resin (100–200 mesh, loading capacity 0.6 mmol/g) purchased from Advanced ChemTech (USA). The synthesis was performed on an automated peptide synthesizer 443A (Applied Biosystems, USA) utilizing a standard Fmoc/tBu strategy. Peptides Tat1_4-5TO, Tat1_8-9TO, Tat1_8-9TOD and Tat1_4-5TO,8-9TOD were obtained using a microwave-assisted manual Fmoc/tBu methodology in a microwave reactor Plazmatronika RM800 (Ertec Inc., Poland) equipped with a reaction vessel with nitrogen agitation.
Crude peptides were purified by reversed-phase high-performance liquid using a C8 semi-preparative Luna column (21.2 x 250 mm, 5 μm; Phenomenex). A linear gradient of acetonitrile in 0.1% aqueous trifluoroacetic acid or acetonitrile in 0.1 M triethylamine phosphate buffer, pH 3.0, was used as a mobile phase. The molecular weights of the peptides were ascertained by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry or electrospray ionization ion trap time-of-flight liquid chromatography mass spectrometry with C12 Jupiter Proteo column (150 x 2 mm, 4μ, 90Å; Phenomenex). The purity of the synthesized compounds was confirmed by analytical reversed phase chromatography using a C8 Kromasil column (4.6 x 250 mm, 5 μm) and a 30 min linear gradient of 5–80% acetonitrile in 0.1% aqueous trifluoroacetic acid. UV absorption was observed at λ = 223 nm.
Activity assays
Influence of the designed peptides on catalytic properties of the 20S proteasome was tested using the proteasome at a final concentration of 1.5 nM, isolated from human erythrocytes (Enzo Life Sciences) and activated with 0.01% sodium dodecyl sulfate. Succinyl-Leu-Leu-Val-Tyr-4-methylcoumarin-7-amide (SucLLVY-MCA) was employed as a model fluorogenic substrate to determine the activity of ChT-L peptidase. All assays were performed in the 96-well plate format using a reaction volume of 100 uL. Tests were carried out in 50 mM Tris/HCl, pH 8.0 containing 0.5 mM ethylenediamine tetraacetic acid. Stock solutions of the substrates and the tested peptides were prepared by dissolution of the lyophilized compounds in dimethyl sulfoxide (DMSO). The content of DMSO never exceeded 3% of the final reaction volume. Compounds were tested in the concentration range of 0.05–10 μM. The substrate was added at 100 μM final concentration (1% of the volume). The release of an aminomethyl coumarin (AMC) group was followed by measuring fluorescence emission at 460 nm in 2 min. intervals for up to 60 min. at 37°C (Fluoroskan Ascent). The peptidolytic activity was calculated as nanomoles of the released AMC product per mg of CP per second. The half maximal inhibitory concentration expressed in μM concentration was calculated with SigmaPlot 12.3 where exponential equation estimates based on fitting of two or three logistic parameters were employed.
Fourier Transform Infrared Spectroscopy
The peptides were dissolved in H2O to a final concentration of 20 mg/ml. Measurements were performed in a demountable cell equipped with CaF2 windows and 6 μm spacer. For the film mode 15 μl of the peptide solution was spotted on a CaF2 window and the solvent evaporated under a stream of gaseous nitrogen. FTIR spectra were collected with a Bruker IFS66 spectrophotometer (Bruker Inc., Germany) using a deuterated triglycine sulfate detector. During data collection the spectrometer was continuously purged with dry air free of CO2. Typically, 240 scans at 4 cm-1 resolution were averaged for each spectrum. At least three independent measurements were performed for each sample. Data processing was executed with GRAMS 5.0 (Galactic Enterprises). The spectra of water and pure TFA were subtracted from the peptides spectra, as it was described [37]. The spectral 2nd derivative was calculated with the application of a 9-point cubic Sawitzky-Golay smoothing function.
Circular dichroism spectroscopy
For CD measurements, 1 mm path length quartz cuvettes were used. Protein samples were dissolved in H2O or in 50 mM Tris/HCl, pH 8.0 at a concentration 0.2 mg/ml. Spectra were acquired at 25°C on a Jasco J-815 spectrometer in a range 185–260 nm for aqueous solutions and 195–260 nm for solutions of the peptides in the buffer. Each spectrum is a result of three independent measurements.
Nuclear Magnetic Resonance
The NMR samples were dissolved in H2O:D2O (9:1, v/v) (Tat1_A4-5,8–9) or partially deuterated phosphate buffer at pH 7.4 (Tat1_4-5TO, Tat1_8-9TO, Tat1_8-9TOD and Tat1_4-5TO,8-9TOD). Due to low quality of NMR spectra at pH 7.4 for the two latter, the pH was decreased to 4.5 by addition of the diluted acetic acid. The concentration of the samples was 5 mM in the case of peptides modified with Tic-Oic moiety and 10 mM in the case of Tat1_A4-5,8–9. For all peptides with Tic-Oic unit, NMR data sets were acquired at 301 K using a Varian Unity Plus 500 MHz operated at 11.7 T (1H resonance frequency 499.83 MHz). With Tat1_A4-5,8–9, due to a poor amide proton resonance dispersion, the 2D NMR spectra were recorded at 283 K and 298 K on a Bruker AVANCE III 700 MHz spectrometer (16.4 T, 1H resonance frequency 700.62 MHz). Proton resonance assignments were achieved by analysis of the 2D total chemical shift correlation spectroscopy (TOCSY) [67], the double-quantum filtered correlation spectroscopy (DQF-COSY) [68], the nuclear Overhauser effect spectroscopy (NOESY) [69], the rotating-frame Overhauser enhancement spectroscopy (ROESY) [70,71] as well as the gradient heteronuclear single quantum coherence experiment (1H-13C gHSQC) [72–74]. The volumes of cross peaks were picked up for the ROESY spectra with a mixing time of 300 ms. In the case of Tat1_A4-5,8–9, the ROESY spectrum recorded at 283 K was considered due to better signal dispersion. Vicinal coupling constants, 3JHNHa, were assigned using DQF-COSY. For Tat1_A4-5,8–9, the coupling constants were also measured from 1D NMR spectrum and the final values were obtained by averaging values from both DQF-COSY and 1D NMR spectra.
All chemical shifts were referenced with respect to external sodium 2,2-dimethyl-2-silapentane-5-sulfonate using a Ξ = 0.251449530 ratio for indirectly referenced 13C resonances [75]. The data were processed using VNMR (Varian Inc., Palo Alto, CA) and NMRPipe [76] and analyzed with the SPARKY software [77].
Molecular Dynamics Simulations
Molecular dynamics simulations were carried out with the parm99 force field in the AMBER11.0 package [78]. The valence geometry of the residues not specified in the standard AMBER database was parameterized as recommended by the AMBER 11.0 manual. Specifically, these residues were modeled using bond lengths, the valence and torsion angles of appropriate residues and compatible molecular segments taken from the CSDS [79] database. The point charges were optimized by fitting them to the ab initio molecular electrostatic potential (6-31G* basis set, GAMESS’04 [80]—ab initio molecular electronic structure program) for three different conformations of every nonstandard residue, followed by consecutive averaging the charges over all conformations, as recommended by the RESP protocol [81].
The structures of the major species of the peptides were computed using molecular dynamics simulations with explicit water and NMR-derived restraints. All MD simulations were run with the SHAKE algorithm (with a tolerance of 0.0005 Å) to constraint covalent bonds involving hydrogens with a 2-fs time step using periodic boundary conditions, and a temperature of 303 K (Berendsen temperature regulation). The electrostatic interactions were evaluated using the PME summation method with cutoff of 9 Å. All models for the MD simulations were constructed using LEAP (AMBER 11 package). The initially random structure of a peptide was embedded into a TIP3P water box enlarged by 10 Å in each direction from the solute. Each system was neutralized by a minimum amount of chloride ions. To equilibrate the solution density, the initial simulations were carried out in NTP ensemble at 1 atm (1 ns). The final density was close to 1.0 g/mL. In the next step, the NTV MD simulations (4 ns) with time-averaged distance restraints and dihedral angle restraints derived from the NMR spectroscopy were employed. The interproton distances were restrained with the force constants f = 50 kcal/(mol x Å2), and the dihedral angles with f = 2 kcal/(mol x rad2). The geometry of the peptide groups was kept fixed according to the NMR data (f = 50 kcal/(mol x rad2)). The inter-proton distances were calculated on the basis of ROEs intensities by the CALIBA algorithm of the DYANA [82] program. The upper limit distance restraints and the 3JHNHα vicinal coupling constants were used in the HABAS algorithm of the DYANA package to generate φ, ψ and χ 1 dihedral angle restraints and stereospecific assignments.
The coordinates were collected every 2000th step. The results were analyzed using Ptraj program from the AMBER 11 package. The conformations obtained in the last 800 ps of simulations were considered in further analysis. The final structures were visualized by MOLMOL [83].
Microscale thermophoresis
The human 20S proteasome was labeled using monolith NTTM Protein Labeling Kit RED-NHS (NanoTemper Technologies), according to the protocol provided by the supplier. During the measurements concentration of the labeled protein was kept constant at 3 nM. The peptides used as unlabeled binding partners were titrated in 3:1 dilutions in the concentration range of 0.2 to 120 μM (dependently on Kd values). Concentrations below 0.2 μM were tested as well but thermophoretic effects were not observed. The binding reactions were carried out in 50 mM Tris/HCl buffer containing 0.5 mM EDTA, pH 8.0 with an addition of sodium dodecyl sulfate at the final concentration of 0.005%. Measurements were performed in standard treated capillaries on a Monolith NT.115 system (NanoTemper Technologies). Capillaries were illuminated at 25°C with 80% LED power for 5 s. Thermophoresis was tracked for 30 s in three laser intensity sets: 80, 60 and 40%, back-diffusion was observed for 5 s. Fluorescence intensities were measured at 6–8 s and 34–36 s, and fluorescence intensity ratios of hot/cold were used as the assay readout. Kd values were determined using the internal evaluation software.
Peptide degradation tests
Susceptibility of the designed modulators to degradation by the 20S proteasome was examined by reversed-phase high-performance liquid chromatography (S10 Fig). The compounds were dissolved in water or DMSO to 10 mM concentration and then diluted with 25 mM Tris buffer containing 0.25 mM EDTA (pH 8.0) to the final concentration of 1 mM. 2.5 μg of human 20S proteasome (Enzo Life Sciences) was added to the mixture, which then, after adjustment of its final volume to 100 μl, was incubated at 37°C for 3 h. The reaction was stopped by adding 5 μl of 10% aqueous trifluoroacetic acid. Next, 40 μl of the mixture, along with the control not containing the 20S, were injected onto C8 Kromasil column (4,6 x 250 mm, 100 Å, 5 μm). The linear gradient of acetonitrile in 0.1% aqueous trifluoroacetic acid was performed with SIL-20AHT UFLC system equipped with SPD-M20A PDA detector (SHIMADZU), and the elution was observed at the wavelength of 223 nm.
Supporting Information
(XLSX)
(XLSX)
The peptides concentration was either 0.1 or 1 μM.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
The conformations obtained in the last 800 ps of MD simulations with time-averaged distance restraints and dihedral angle restraints.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
The equipment used for microscale thermophoresis was sponsored in part by the Centre for Preclinical Research and Technology (CePT), a project co-sponsored by European Regional Development Fund and Innovative Economy, The National Cohesion Strategy of Poland. The project was sponsored in part by The William and Ella Owens Foundation for Medical Research award (MEG and PAO).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The equipment used for microscale thermophoresis was sponsored in part by the Centre for Preclinical Research and Technology (CePT), a project co-sponsored by European Regional Development Fund and Innovative Economy, The National Cohesion Strategy of Poland. The project was sponsored in part by The William and Ella Owens Foundation for Medical Research award (MEG and PAO), 2011/01/B/ST5/06616 (EJ), National Science Center (https://www.ncn.gov.pl) and DS/530-8725-D496-15, Ministry of Science and Higher Education (http://www.nauka.gov.pl). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Stadtmueller BM, Hill CP. Proteasome activators. Mol Cell. 2011; 41: 8–19. 10.1016/j.molcel.2010.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stohwasser R, Salzmann U, Giesebrecht J, Kloetzel PM, Holzhütter, HG. Kinetic evidences for facilitation of peptide channeling by the proteasome activator PA28. Eur J Biochem. 2000; 267: 6221–6230. [DOI] [PubMed] [Google Scholar]
- 3. Ustrell V, Hoffman L, Pratt G, Rechsteiner M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002; 21: 3516–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik HD, et al. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997; 386: 463–471. [DOI] [PubMed] [Google Scholar]
- 5. Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, et al. A gated channel into the proteasome core particle. Nat Struct Biol. 2000; 7: 1062–1067. [DOI] [PubMed] [Google Scholar]
- 6. Sandre-Bazzaz K, Whitby FG, Robinson H, Formosa T, Hill CP. Structure of a Blm10 complex reveals common mechanism for proteasome binding and gate opening. Mol Cell. 2010; 37: 728–735. 10.1016/j.molcel.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Realini C, Jensen CC, Zhang Z, Johnston SC, Knowlton JR, Hill CP, et al. Characterization of recombinant REGalpha, REGbeta, and REGgamma proteasome activators. J Biol Chem. 1997; 272: 25483–25492. [DOI] [PubMed] [Google Scholar]
- 8. Gaczynska M, Osmulski PA. Harnessing proteasome dynamics and allostery in drug design. Antiox Redox Sign. 2014; 21: 2286–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Osmulski PA, Hochstrasser M, Gaczynska M. A tetrahedral transition state at the active sites of the 20S proteasome is coupled to opening of the α-ring channel. Structure. 2009; 17: 1137–1147. 10.1016/j.str.2009.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ruschak AM, Kay LE. Proteasome allostery as a population shift between interchanging conformers. Proc Natl Acad Sci USA. 2012; E3454–3462. 10.1073/pnas.1213640109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jung T, Catalgol B, Grune T. The proteasomal system. Mol Asp Med. 2009; 30: 191–296. [DOI] [PubMed] [Google Scholar]
- 12. Jankowska E, Stoj J, Karpowicz P, Osmulski PA, Gaczynska M. The proteasome in health and disease. Curr Pharm Des. 2013; 19: 1010–1028. [PubMed] [Google Scholar]
- 13. Pajonk F, McBride WH. (2004). The proteasome in cancer biology and therapy, In: Adams J, editor. Proteasome inhibitors in cancer therapy, Humana Press; 2004. pp. 111–121. [Google Scholar]
- 14. Frankland-Searby S, Bhaumik SR. The 26S proteasome complex: an attractive target for cancer therapy. Biochim Biophys Acta. 2012; 1825: 64–76. 10.1016/j.bbcan.2011.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen D, Frezza M, Schmitt S, Kanwar J, Dou QP. Bortezomib as the first proteasome inhibitor anticancer dryug: current status and future perspectives. Curr Cancer Drug Targets. 2011; 11: 239–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kortuem KM, Stewart AK. Carfilzomib. Blood. 2013; 121: 893–897. 10.1182/blood-2012-10-459883 [DOI] [PubMed] [Google Scholar]
- 17. Ruschak AM, Slassi M, Kay LE, Schimmer AD. Novel proteasome inhibitors to overcome bortezomib resistance. J Natl Cancer Inst. 2011; 103: 1–11. [DOI] [PubMed] [Google Scholar]
- 18. Laskowski RA, Gerick F, Thornton JM. The structural basis of allosteric regulation in proteins. FEBS Lett. 2009; 583: 1692–1698. 10.1016/j.febslet.2009.03.019 [DOI] [PubMed] [Google Scholar]
- 19. Merdanovic M, Mönig T, Ehrmann M, Kaiser M. Diversity of allosteric regulation in proteases. ACS Chem Biol. 2013; 8: 19–26. 10.1021/cb3005935 [DOI] [PubMed] [Google Scholar]
- 20. Hauske P, Ottmann C, Meltzer M, Ehrmann M, Kaiser M. Allosteric regulation of proteases. Chem BioChem. 2008; 9: 2929–2928. [DOI] [PubMed] [Google Scholar]
- 21. Melancon BJ, Hopkins CR, Wood MR, Emmitte KA, Niswender CM, Christopoulos A, et al. Allosteric modulation of seven transmembrane spanning receptors: theory, practice, and opportunities for central nervous system drug discovery. J Med Chem. 2012; 55: 1445–1464. 10.1021/jm201139r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kar G, Keskin O, Gursoy A, Nussinov R. Allostery and population shift in drug discovery. Curr Opin Pharm. 2010; 10: 715–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nussinov R, Tsai CJ. Allostery in disease and in drug discovery. Cell. 2013; 153: 293–305. 10.1016/j.cell.2013.03.034 [DOI] [PubMed] [Google Scholar]
- 24. Gaczynska M, Osmulski PA, Gao Y, Post MJ, Simons M. Proline- and arginine-rich peptides constitute a novel class of allosteric inhibitors of proteasome activity. Biochemistry. 2003; 42: 8663–8670. [DOI] [PubMed] [Google Scholar]
- 25. Azevedo LM, Landsdell TA, Ludwig JR, Mosey RA, Woloch DK, Cognan DP, et al. Inhibition of the human proteasome by imidazoline scaffolds. J Med Chem. 2013; 56: 5974–5978. 10.1021/jm400235r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lansdell TA, Hurchla MA, Xiang J, Hovde S, Weilbaechter KN, Henry RW, Tepe JJ. Non-competitive modulation of proteasome by imidazoline scaffolds overcome bortezomib resistant and delay MM tumor growth in vivo. ACS Chem Biol. 2013; 8: 578–587. 10.1021/cb300568r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sprangers R, Li X, Mao X, Rubinstein JL, Schimmer AD, Kay LE. TROSY-based NMR evidence for a novel class of 20S proteasome inhibitors. Biochemistry. 2008; 47: 6727–6734. 10.1021/bi8005913 [DOI] [PubMed] [Google Scholar]
- 28. Mao X, Li X, Sprangers R, Wang X, Venugopal A, Wood T, et al. Clioquinol inhibits the proteasome and displays preclinical activity in leukemia and myeloma. Leukemia. 2009; 23: 585–590. 10.1038/leu.2008.232 [DOI] [PubMed] [Google Scholar]
- 29. Gaczynska M, Osmulski PA. Rapamycin allosterically inhibits the proteasome. Mol Pharmacol. 2013; 84: 104–113. 10.1124/mol.112.083873 [DOI] [PubMed] [Google Scholar]
- 30. Li X, Wood TE, Sprangers R, Jansen G, Franke NE, Mao X, et al. Effect of noncompetitive proteasome inhibition on bortezomib resistance. J Natl Cancer Inst. 2010; 102: 1069–1082. 10.1093/jnci/djq198 [DOI] [PubMed] [Google Scholar]
- 31. Remoli AL, Marsili G, Perrotti E, Gallerani E, Ilari R, Nappi F, Cafaro A, et al. Intracellular HIV-1 Tat protein represses constitutive LMP2 transcription increasing proteasome activity by interfering with the binding of IRF-1 to STAT1. Biochem J. 2006; 396: 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seeger M, Ferre K, Frank R, Dubiel W. HIV-1 Tat inhibits the 20S proteasome and its 11S regulator-mediated activation. J Biol Chem. 1997; 272: 8145–8148. [DOI] [PubMed] [Google Scholar]
- 33. Apcher GS, Heink S, Zantopf D, Kloetzel PM, Schmid HP, Mayer RJ, Kruger E. Human immunodeficiency virus-1 Tat protein interacts with distinct proteasomal α and β subunits. FEBS Lett. 2003; 553: 200–204. [DOI] [PubMed] [Google Scholar]
- 34. Huang X, Seifert U, Salzmann U, Henklein P, Preissner R, Henke W, et al. The RTP site shared by the HIV-1 Tat protein and the 11S regulator subunit α is crucial for their effects on proteasome function including antigen processing. J Mol Biol. 2002; 323: 771–782. [DOI] [PubMed] [Google Scholar]
- 35. Zhang Z, Clawson A, Realini C, Jensen CC, Knowlton JR, Hill CP, Rechsteiner M. Identification of an activation region in the proteasome activator REGα. Proc Natl Acad Sci USA. 1998; 95: 2807–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jankowska E, Gaczynska M, Osmulski P, Sikorska E, Rostankowski R, Madabhushi S, et al. Potential allosteric inhibitors of the proteasome activity. Biopolymers. 2010; 93: 481–495. 10.1002/bip.21381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Witkowska J, Karpowicz P, Gaczynska M, Osmulski PA, Jankowska E. Dissecting a role of a charge and conformation of Tat2 peptide in allosteric regulation of 20S proteasome. J Pept Sci. 2014; 20: 649–656. 10.1002/psc.2642 [DOI] [PubMed] [Google Scholar]
- 38. Sawutz DG, Salvino JM, Seoane PR, Douty BD, Houck WT, Bobko MA, et al. Synthesis, characterization, and conformational analysis of the d/l-Tic7 stereoisomers of the bradykinin receptor antagonist D-Arg0[Hyp3,Thi5,d-Tic7,Oic8]bradykinin. Biochemistry. 1994; 33: 2373–2379. [DOI] [PubMed] [Google Scholar]
- 39. Duhr S, Brown D. Why molecules move along a temperature gradient. Proc Natl Acad Sci USA. 2006; 103: 19678–19682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Twardowski J, Anzenbacher P. Raman and IR spectroscopy in biology and biochemistry 1st ed. New York: Ellis Horwood; 1994. [Google Scholar]
- 41. Krimm S, Bandekar J. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv Protein Chem. 1986; 38: 181–364. [DOI] [PubMed] [Google Scholar]
- 42. Surewicz WK, Mantsch HH, Chapman D. Determination of protein secondary structure by Fourier transform infrared spectroscopy: a critical assessment. Biochemistry. 1993; 32: 389–394. [DOI] [PubMed] [Google Scholar]
- 43. Jankowska E, Wiczk W, Grzonka Z. Thermal and guanidine hydrochloride-induced denaturation of human cystatin C. Eur J Biophys. 2004; 33: 454–461. [DOI] [PubMed] [Google Scholar]
- 44. Oberg KA, Fink AL. A new attenuated total reflectance Fourier transform infrared spectroscopy method for the study of proteins in solution. Anal Biochem. 1998; 256: 92–106. [DOI] [PubMed] [Google Scholar]
- 45. Kong J, Yu S. Fourier transform infrared spectroscopy of protein secondary structure. Acta Biochim Biophys Sin. 2007; 39: 549–559. [DOI] [PubMed] [Google Scholar]
- 46. Malin EL, Alaimo MH, Brown EM, Aramini JM, Germann MW, Farrel HM, et al. Solution structures of casein peptides: NMR. FTIR, CD, and molecular modeling studies of αs1-casein, 1–23. J Protein Chem. 2001; 20: 391–404. [DOI] [PubMed] [Google Scholar]
- 47. Bhatnagar RS, Gough CA. Circular dichroism of collagen and related polypeptides In: Fasman GD, editor. Circular dichroism and the conformational analysis of biomolecules. Plenum Press; 1996. pp. 183–199. [Google Scholar]
- 48. Eker F, Griebenov K, Cao X, Nafie LA, Schweitzer-Stenner R. Tripetides with ionizable side chains adopt a perturbed polyproline II structure in water. Biochemistry 2004; 43: 613–621. [DOI] [PubMed] [Google Scholar]
- 49. Woody RW. Circular dichroism of peptides In: Hruby J, editor. The Peptides. Academic Press; 1985. pp. 15–144. [Google Scholar]
- 50. Perczel A, Hollosi M. Turns In: Fasman GD, editor. Circular dichroism and the conformational analysis of biomolecules. Plenum Press; 1996. pp. 306–318 [Google Scholar]
- 51. Lee YC, Jackson PL, Jablonsky MJ, Muccio DD, Pfister RR, Haddox JL, et al. NMR conformational analysis of cis and trans proline isomers in the neutrophil chemoattractant, N-acetyl-proline-glycine-proline. Biopolymers 2001; 58: 548–561. [DOI] [PubMed] [Google Scholar]
- 52. Guba W, Haessner R, Breipohl G, Henke S, Knolle J, Santagada V, et al. Combined approach of NMR and molecular dynamics within a biphasic membrane mimetic: conformation and orientation of the bradykinin antagonist Hoe140. J Am Chem Soc 1994; 116: 7532–7540. [Google Scholar]
- 53. Williamson MP, Waltho JP. Peptide structure from NMR. Chem Soc Rev. 1992; 21: 227–236. [Google Scholar]
- 54. Sikorska E, Sobolewski D, Kwiatkowska A. Conformational Preferences of Proline Derivatives Incorporated into Vasopressin Analogues: NMR and Molecular Modelling Studies. Chem Biol Drug Des. 2012; 79: 535–547. 10.1111/j.1747-0285.2012.01318.x [DOI] [PubMed] [Google Scholar]
- 55. Dyson HJ, Rance M, Houghten RA, Lerner RA, Wright PE. Folding of immunogenic peptide-fragments of proteins in water solution. I. Sequence requirements for the formation of a reverse turn. J Mol Biol. 1988; 201: 161–200. [DOI] [PubMed] [Google Scholar]
- 56. Schwarzinger S, Kroon GJ, Foss TR, Chung J, Wright PE, Dyson HJ. Sequence-dependent correction of random coil NMR chemical shifts. J Am Chem Soc. 2001; 123: 2970–2978. [DOI] [PubMed] [Google Scholar]
- 57. Carlisle EA, Holder JL, Maranda AM, de Alwis AR, Selkie EL, McKay SL. Effect of pH, urea, peptide length, and neighboring amino acids on alanine α proton random coil chemical shifts. Biopolymers. 2007; 85: 72–80. [DOI] [PubMed] [Google Scholar]
- 58. Misicka A, Verheyden PMF, Van Binst G. Equilibrium of the cis-trans isomeriation of the peptide bond with N-alkyl amino acids measured by 2D NMR. Lett Pept Sci. 1998; 5: 375–377. [Google Scholar]
- 59. Ma K, Kan LS, Wang K. Polyproline II helix is a key structural motif of the elastic PEVK segment of titin. Biochemistry. 2001; 40: 3427–3438. [DOI] [PubMed] [Google Scholar]
- 60. Eberstadt M, Gemmecker G, Mierke DF, Kessler H. Scalar coupling-constants—their analysis and their application for the elucidation of structures. Angew Chemie Int Ed. 1995; 34: 1671–1695. [Google Scholar]
- 61. Macarthur MW, Thornton JM. Influence of proline residues on protein conformation. J Mol Biol. 1991; 218: 397–412. [DOI] [PubMed] [Google Scholar]
- 62. Lobanov MY, Bogatyreva NS, Galzitskaya OV. Radius of gyration as an indicator of protein structure compactness. Mol Biol. 2008; 42: 623–628. [PubMed] [Google Scholar]
- 63. Narang P, Bhushan K, Bose S, Jayaram B. A computational pathway for bracketing native-like structures for small alpha helical globular proteins. Phys Chem Chem Phys. 2005; 7: 2364–2375. [DOI] [PubMed] [Google Scholar]
- 64. Formicola L, Maréchal X, Basse N, Bouvier-Durand M, Bonnet-Delpon D,Milcent T, et al. Novel fluorinated pseudopeptides as proteasome inhibitors. Bioorg Med Chem Lett. 2009; 19: 83–86. 10.1016/j.bmcl.2008.11.012 [DOI] [PubMed] [Google Scholar]
- 65. Kloß A, Henklein P, Siele D, Schmolke M, Apcher S, Kuehn L, et al. The cell-penetrating peptide octa-arginine is a potent inhibitor of proteasome activities. Eur J Pharm Biopharm. 2009; 72: 219–225. 10.1016/j.ejpb.2008.10.016 [DOI] [PubMed] [Google Scholar]
- 66. Sikorska E, Tuwalska D, Karpowicz P, Jankowska E, Nowacki A, Liberek B. Oligomers and peptidomimetics consisting of methyl 3-amino-2,3-dideoxyhexopyranosiduronic acids. Tetrahedron. 2015; 71: 2013–2024. [Google Scholar]
- 67. Bax A.; Davis D. G. Mlev-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Res. 1985, 65, 355–360. [Google Scholar]
- 68. Rance M, Sorensen OW, Bodenhausen G, Wagner G, Ernst RR, Wuthrich K. Improved spectral resolution in cosy h-1-nmr spectra of proteins via double quantum filtering. Biochem Biophys Res Commun. 1983; 117: 479–485. [DOI] [PubMed] [Google Scholar]
- 69. Kumar A, Ernst RR, Wuthrich K. A two-dimensional nuclear overhauser enhancement (2d noe) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biophys Res Commun. 1980; 95: 1–6. [DOI] [PubMed] [Google Scholar]
- 70. Bothnerby AA, Stephens RL, Lee JM, Warren CD, Jeanloz RW. Structure determination of a tetrasaccharide—transient nuclear overhauser effects in the rotating frame. J Am Chem Soc. 1984; 106: 811–813. [Google Scholar]
- 71. Bax A, Davis DG. Practical aspects of two-dimensional transverse noe spectroscopy. J Magn Res. 1985; 63: 207–213. [Google Scholar]
- 72. Palmer AG, Cavanagh J, Wright PE, Rance M. Sensitivity improvement in proton-detected 2-dimensional heteronuclear correlation nmr-spectroscopy. J Magn Res 1991; 93: 151–170. [Google Scholar]
- 73. Kay LE, Keifer P, Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc. 1992; 114: 10663–10665. [Google Scholar]
- 74. Schleucher J, Schwendinger M, Sattler M, Schmidt P, Schedletzky O, Glaser SJ, et al. A general enhancement scheme in heteronuclear multidimensional nmr employing pulsed-field gradients. J Biomol NMR. 1994; 4: 301–306. [DOI] [PubMed] [Google Scholar]
- 75. Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. H-1, C-13 and N-15 random coil NMR chemical-shifts of the common amino-acids. I. Investigations of nearest-neighbor effects. J Biomol NMR. 1995; 5: 67–81. [DOI] [PubMed] [Google Scholar]
- 76. Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMR Pipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995; 6: 277–293. [DOI] [PubMed] [Google Scholar]
- 77. Goddard TD, Kneller DG. SPARKY 3. University of California: San Francisco; 2001. [Google Scholar]
- 78. Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, et al. AMBER 11. University of California: San Francisco; 2010. [Google Scholar]
- 79. Allen FH, Motherwell WDS. Applications of the Cambridge Structural Database in organic chemistry and crystal chemistry. Acta Cryst B. 2002; 58: 407–422. [DOI] [PubMed] [Google Scholar]
- 80. Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS., Jensen JH, et al. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges—the resp model. J Phys Chem. 1993; 97: 10269–10280. [Google Scholar]
- 81. Bayly CI, Cieplak P, Cornell WD, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges—the resp model. J Phys Chem 1993; 97: 10269–10280. [Google Scholar]
- 82. Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997; 273: 283–298. [DOI] [PubMed] [Google Scholar]
- 83. Koradi R, Billeter M, Wuthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graph. 1996; 14: 51–55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
The peptides concentration was either 0.1 or 1 μM.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
The conformations obtained in the last 800 ps of MD simulations with time-averaged distance restraints and dihedral angle restraints.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.