Abstract
Colorectal cancer (CRC) represents one of the most prevalent and lethal malignant neoplasms and every individual of age 50 and above should undergo regular CRC screening. Currently, the most effective preventive screening procedure to detect adenomatous polyps, the precursors to CRC, is colonoscopy. Since every colorectal cancer starts as a polyp, detecting all polyps and removing them is crucial. By exactly doing that, colonoscopy reduces CRC incidence by 80%, however it is an invasive procedure that might have unpleasant and, in rare occasions, dangerous side effects. Despite numerous efforts over the past two decades, a non-invasive screening method for the general population with detection rates for adenomas and CRC similar to that of colonoscopy has not yet been established. Recent advances in next generation sequencing technologies have yet to be successfully applied to this problem, because the detection of rare mutations has been hindered by the systematic biases due to sequencing context and the base calling quality of NGS.
We present the first study that applies the high read accuracy and depth of single molecule, real time, circular consensus sequencing (SMRT-CCS) to the detection of mutations in stool DNA in order to provide a non-invasive, sensitive and accurate test for CRC. In stool DNA isolated from patients diagnosed with adenocarcinoma, we are able to detect mutations at frequencies below 0.5% with no false positives. This approach establishes a foundation for a non-invasive, highly sensitive assay to screen the population for CRC and the early stage adenomas that lead to CRC.
Keywords: Colorectal cancer, Single molecule sequencing, Third generation sequencing, High sensitivity, Stool DNA, APC gene
1. Introduction
Although the rate of colorectal cancer (CRC) has been declining at 3.0% per year over the past decade, in 2014 there were about 140,000 new cases in the United States making it the third most common cancer after lung-bronchus and prostate (Cancer Facts and Figures, American Cancer Society, 2014) and this figure in North America essentially mirrors the latest available worldwide survey (www.globocan.iarc.fr).
A substantial amount of research has been conducted in the past thirty years to demonstrate that the molecular genetic landscape of CRC is extremely complex. Single Nucleotide Polymorphisms (SNPs), Insertion–Deletions (Indels), Microsatellite Instabilities (MSI), and alteration in methylation patterns can all occur at different loci depending on the site of tumor and its stage (Fearon, 2011, The Cancer Genome Atlas Network, 2012, Tomlinson et al., 2010, Houlston, 2012).
The molecular mechanisms behind the formation of adenomas and their progression into CRC were first presented 25 years ago in a model proposed by Fearon and Vogelstein (Fearon and Vogelstein, 1990).
Briefly, early adenomas emerge on the normal epithelium and this event is associated with mutations in the Adenomatous Polyposis Coli (APC) or β-catenin (CTNNB1) genes (Morin et al., 1997, Sparks et al., 1998). Most early adenomas suffer additional mutations in either the Kirsten Rat Sarcoma (KRAS) or v-raf murine sarcoma viral oncogene homolog B (BRAF) genes (or other genes of the RTK-RAS pathway), which lead to the formation of intermediate and larger adenomas (Chan, 2003). It is at this point that chromosomal instabilities (CIN) or deficiency in the mismatch repair (MMR) system begin to occur, which leads to an increased mutation rate in the neoplastic cells. CIN in colorectal cancer is often observed on the long arm of chromosome 18 and it is associated with mutations in genes SMAD2 and SMAD4 (Takagi et al., 1996, Miyaki et al., 1999). These genes represent human homologs of mothers against decapentaplegic (MAD) in drosophila and of the SMA protein in Caenorhabditis elegans. On the other hand, MMR is usually caused by a decrease in the expression of the MutL-homolog 1 (MLH1) gene on chromosome 3 through the hyper methylation of the promoter region (Vilar and Gruber, 2010). MMR induces alteration of microsatellite sequences in the tumor cells and increases the overall mutation rate for all genes, including oncogenes and tumor suppressor genes (Parsons et al., 1993). Finally, the progression to carcinoma is often accompanied by the malfunctioning of the cellular tumor antigen TP53, a protein with tumor suppressor activity.
The progression of CRC shows a significant acceleration between the second and the third stage, i.e., upon the onset of CIN or MMR, with survival rates dropping from 80% to less than 40%, whereas early detection (stage I) is associated with survival rates above 90% (Sameer, 2013). It is therefore critical to base the screening of the general population on approaches capable of confidently identifying the first, local instances of adenomas and CRC neoplastic formation.
The methods currently used to test for CRC can be divided into two general categories: invasive methods such as colonoscopy, and non-invasive methods that detect biomarkers in stool. Colonoscopy, i.e., the direct inspection of the colon, is certainly the most accurate approach as the physician is able to visualize the epithelium and eventually ascertain the presence of abnormalities on its surface. However it presents a number of drawbacks. It can still miss a significant percentage of adenomas, even upon repeated examination and it tends to have unpleasant, and in rare occasions, dangerous side effects, which are mostly connected to the preparation that the bowel requires prior to the procedure (Senore et al., 2011, Lebwohl et al., 2011). Moreover, this preparation represents an additional step in this screening process which seems can be sub-optimally performed in a significant number of cases (Lebwohl et al., 2011). Not the least, its invasiveness is the source of reluctance for a considerable number of individuals, particularly among certain populations (Ramos et al., 2011, Talaat and Harb, 2013). Despite its lower level of discomfort, since only the rectum and the lower colon are inspected, similar arguments can be put forward for flexible sigmoidoscopy (FSG) and computed tomography colonography (Senore et al., 2011).
Non-invasive tests based on fecal occult blood (FOBTs) are a widely used screening method. A small sample of stool is self-collected, placed on a card, and sent to the physician. There are several methods for testing for occult blood in the feces: for instance, fecal immunochemical testing (FIT) utilizes antibodies to detect human hemoglobin whereas in guaiac tests (gFOBT) the stool is smeared on a chemically treated paper which, if blood is present, will change color when absorbing hydrogen peroxide. FIT has been shown to perform more effectively than gFOBT (Sharp et al., 2012), however FOBTs in general are much less effective in reducing CRC incidence compared to colonoscopy (Moayyedi and Achkar, 2006), mostly due to their inherent difficulties in detecting early stages polyps, which do not bleed.
Finally, non-invasive testing of stool can be done by interrogating the genetic material present in a stool sample. The use of DNA testing for screening is certainly extremely promising, however so far it has been hindered by its limited sensitivity since, when the presence of mutated DNA is to be inferred from body fluids, the concentration of mutated cells becomes extremely low. Being continuously available and easy to collect and containing mutant DNA from exfoliating adenomas at the earliest stages of CRC development, stool is the obvious choice for inspecting potential CRC mutations and it is not surprising that results based on stool samples are superior to those based on cell free DNA in blood plasma (Diehl et al., 2008). Early studies using multi-target panels, based on methylation, mutation and hemoglobin assays reported sensitivities between 70% and 90% for CRC, where the sensitivity would typically increase with the number of markers included and the size of the neoplasm (Dong et al., 2001, Ahlquist et al., 2000, Ahlquist et al., 2012, Traverso et al., 2002a). Recently, a large study of 9989 patients showed that a multi-panel assay of stool DNA markers could detect colorectal cancer with a sensitivity of 92.3%, while 42.4% of patients with polyps were detected. The false positive rate of the DNA based test in this study was 13.4% (Imperiale et al., 2014), which would generate 134 false positives, and consequently 134 unnecessary follow-ups in the form of colonoscopy, for each 1000 patients screened.
Massive parallel sequencing has been proven successful in identifying mutations from cancerous tissues (Gerecke et al., 2013, Kinde et al., 2011) and specific cancer panels, such as the Ion AmpliSeq™Cancer Hotspot Panel v2 and the TruSeq Amplicon — Cancer Panel (TSACP) are now available for second generation platforms. However, despite covering a broad spectrum of genes, their sensitivity is bound at 5% (Frampton et al., 2013, Fang et al., 2013; http://www.edgebio.com/ion-ampliseq-fixed-panels-hct-15-colon-carcinoma-cell-line; Singh et al., 2013), a constraint which makes them ineffective when screening for CRC using stool DNA.
A recent survey of 224 CRC tumors by whole exome DNA sequencing (The Cancer Genome Atlas Network, 2012) shows that 93% of all tumors have mutations in the wnt signaling pathway, 62% have mutations in the RTK-RAS pathway and 61% have mutations in TP53 signaling. The fraction of CRC tumors that will not have a mutation in any of these pathways is then (1–0.93) × (1–0.62) × (1–0.61) = 0.6%, thus the theoretical false negative rate due to not looking for the driver mutations present in a tumor would be 0.6% if all possible mutations were screened in all of the genes involved in these pathways. The 15 amplicon assays described in this paper will not detect all mutations in these 3 key pathways, but is designed to detect the maximum number with a self-imposed limit of 15 amplicons. The number of amplicons in such an assay, i.e., the genomic spectrum that could be interrogated by deep sequencing, could certainly be enlarged, however, at the time of writing, a region of about 5000 nucleotide seemed to be the most appropriate projected trade-off between the size of the region to inspect and a cost-effective clinical setting, whereby sufficient sequencing data for a sample could be obtained by employing only on one SMRT cell.
The rationale behind such an assay is closely reinforced by the fact that, apart from the aforementioned genes, no equally informative biomarkers for CRC have been so far reported, as the cases of microRNA and gut microbiotas witness (Hrašovec and Glavač, 2012, Mazeh et al., 2013, Zhu et al., 2013, Dejea et al., 2013), where no definite conclusion can be drawn regarding an association between biomarkers of that type and the development of CRC.
In the current study, we present the first application of single molecule, real time, circular consensus sequencing (SMRT-CCS) (Travers et al., 2010) to the detection of mutations associated with CRC using stool DNA as analyte. The high quality of the raw sequence data produced by SMRT-CCS allows for a sensitivity of detection in the range of 0.5–2%, which is required to detect polyps using stool DNA as analyte, as most of the human DNA in a stool sample from such patients will be wild type (Traverso et al., 2002b).
2. Materials and methods
2.1. Experimental design
The main goal of this study is to test the specificity and sensitivity of third generation, single molecule sequencing in detecting mutated DNA at concentrations comparable to those observed in stool from exfoliated cells derived from early stage adenomas.
We performed a series of experiments to detect low frequency CRC mutations using an assay consisting of fifteen amplicons covering key regions of the genes most frequently mutated in CRC. The test sequence includes 8 overlapping amplicons covering codons 840–1581 of the APC gene, which is twice the size of the Mutation Cluster Region (MCR) covering codons 1210–1581 that has been used previously to detect mutations in stool DNA from patients with polyps (Traverso et al., 2002b). About 83% of APC mutations in sporadic CRC are found in the MCR, while closer to 90% are found in the region covering codons 840–1581 (Laurent-Puig et al., 1998). Additional test sequences include exon 3 of Beta Catenin (CTNNB1) which contains serine and threonine residues that are mutated in 48% of all CR tumors lacking APC mutations (Sparks et al., 1998), exon 2 of KRAS containing codons 12 and 13 of this gene, exon 15 of BRAF which contains the critical codon 600, often mutated in a number of cancers, and exons 5–8 of the TP53 gene. The amplicons are generated using genomic DNA as template, pooled at equal molarities and sequenced using the third generation, single molecule sequencing Pacific Biosciences RS II platform. The amplicons were optimized to minimize length variability, resulting in a range of 327–344 bp.
2.2. Simulated stool sample (DLD1/wild type titration)
We simulated a stool sample having 3 mutations in CRC genes present at the 1.5% level by mixing 97% wild type DNA with 3% DNA extracted from the DLD1 CRC cell line, which contains heterozygous mutations in APC (codon 1416, deletion 1c), KRAS (G13D caused by a g > a transversion), and TP53 (Ser241Pro caused by a c > t transition). To compare the performances of SMRT-CCS against second generation sequencing technologies, we sequenced the mixed sample, as well as the wild type, on three platforms: Pacific Biosciences RS II, Illumina MiSeq and IonTorrent PGM.
For the sequencing on the Pacbio RS II, the SMRT bell libraries were produced using the DNA Template Prep Kit 2.0 (250 bp — 3 Kb) according to the supplied protocol for blunt ended ligations (Pacific Biosciences p/n 001-540-726).
Paired end, 2 × 250 bp libraries prepared with the The TruSeq DNA Sample Prep Kit v2 (Illumina, Inc., California, USA) were processed on the Illumina MiSeq.
Finally, template preparation with the Ion PGM™ Template OT2 400 Kit (Ion Torrent, Life Technologies, USA) preceded sequencing on an Ion 316 (100 Mb.p.) micro-chip using the Ion Torrent Personal Genome Machine (Ion Torrent, Life Technologies, Carlsbad, USA) with the Ion PGM™ Sequencing 400 Kit for 850 flows.
2.3. CRC tumor tissue
After establishing the workflow on DNA derived from cell lines with known mutations, we proceeded to analyze DNA from CRC tumor samples. Genomic DNA extracted from tumor tissue and matched, adjacent normal tissue (colon) from a 37 y/o male patient diagnosed with adenocarcinoma. The 15 pooled amplicons from the 2 DNA samples were used to make 2 PacBio libraries and the libraries were sequenced with one SMRT cell being used per sample.
2.4. Stool samples from CRC patients
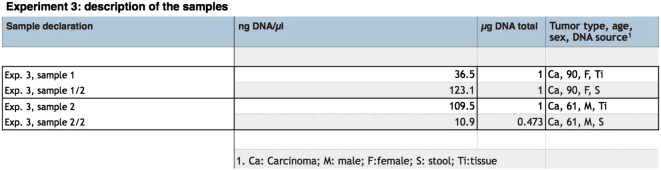
Finally, clinical samples from CRC patients were analyzed. DNA was first isolated from the stool of two sporadic CRC cases, a 90 y/o female and a 61 y/o male, taken after they were diagnosed with CRC by colonoscopy and prior to surgical removal of the tumor (Supplementary Table 3). After the excision of the cancerous neoplasm, DNA was extracted from the tissue itself in order to act as a control sample against which to confirm any mutations found in the DNA isolated from the stool samples. Only the MCR of the APC gene (codons 1210–1581) was analyzed for these experiments using 4 overlapping segments of the APC gene and one SMRT cell was used per library.
Supplementary Table 3.
Experiment 3 : description of the samples.
2.5. Data analysis
In all cases, CCS reads were generated using the SMRT Analysis 2.1 software. The resulting consensus sequences were aligned to the reference pools of either 4 or 15 amplicons using the PacBio read-alignment software Blasr. with the following settings: minMatch = 8, sdpTupleSize = 8, minPctIdentity = 75, bestn = 1, nCandidates = 10, and maxScore = 500. Following the alignments, consensus calls were generated by counting at each locus the occurrences of the reference allele and those of the alternative allele in the wild type and the tumor sample. By doing so, at each position a contingency table was then produced, leading to a Fisher's exact test to test the hypothesis that the alternative allele frequency in the non-WT sample is significantly different from that of the WT. Based on the assumption that at most one mutation per codon results in a functional disruption, the resulting P-values were then Bonferroni-corrected by the number of codons in each amplicon. In the case of the simulated stool sample sequenced on the Ion Torrent PGM, data analysis was also streamlined by means of the variant caller pipeline available on the Torrent Suite, the software specifically designed by Life Technologies for the downstream analysis of sequences generated by Ion Torrent.
3. Results
3.1. Simulated stool sample from wild-type/DLD1 mixture
The purpose of sequencing the mixture of wild-type and 3% DLD1 DNA was to compare 3 different massively parallel sequencing technologies with respect to their performances in detecting different types of mutations at an expected frequency of 1.5%.
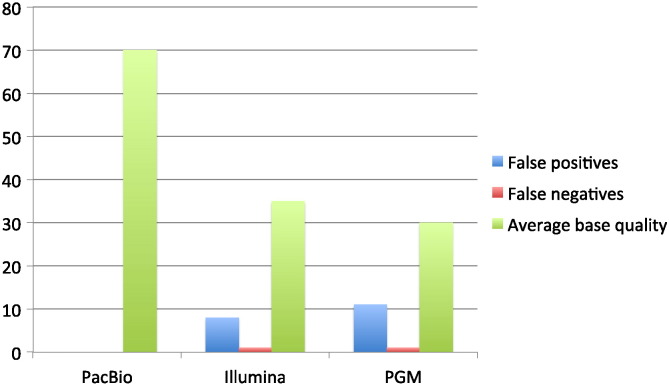
When looking at the results, we have obtained the difference between the Illumina and Ion Torrent platforms and PacBio RS II is evident. Pacbio RS shows 100% sensitivity and specificity (Table 1). The three bona fide mutations known to be present in the DLD1 cell line mutations give strong signals well above background, as the first locus outside the three known polymorphisms has a P-value thirty times higher than any of the true mutations.
Table 1.
Experiment 1: mutations reported based on the sequencing run on the PacBio RS II.
| Segment | Relative position | RA | AA | Cov. WT | AA Freq. WT (%) | Cov. DLD1-M | AA Freq. DLD1-M (%) | P | Adj. P |
|---|---|---|---|---|---|---|---|---|---|
| KRAS (*) | 214 | G | A | 2095 | 0 | 3335 | 1.29 | 0 | 0 |
| APC7 (*) | 94 | G | − C | 3346 | 0.04 | 4012 | 0.51 | 2.30E-05 | 0.0078 |
| TP53_exon7 (*) | 161 | C | T | 2485 | 0.04 | 2569 | 0.70 | 7.80E-05 | 0.0284 |
| APC2 | 26 | A | + T | 3380 | 0 | 3753 | 0.2 | 0.0021 | 0.750 |
Note: (*) True positive. RA: Reference allele. AA: Alternative allele. WT: wild type. DLD1-M: DLD1 — mixed sample.
On the contrary, both the Illumina MiSeq and IonTorrent PGM report a number of false positives and fail to identify one of the bona fide mutations (Fig. 1). Moreover, when the PGM data are analyzed directly on the IonTorrent server through the proprietary aligner, i.e., T-map (http://mendel.iontorrent.com/ion-docs/Technical-Note-TMAP-Alignment_9012907.html), and variant caller, no mutations are found either with stringent or with default settings. This is a further confirmation that, in the workflow of this platform, a mutation with a frequency of 1.5% is treated as an error and therefore discarded.
Fig. 1.
Simulated stool sample: comparison of three different sequencing platforms. With extremely high CCS-reads quality, the results obtained using PacBio are 100% specific and sensitive. On the contrary, both IonTorrent PGM and Illumina MiSeq report several false positive and miss to identify one variant (Supplementary Table 1, Supplementary Table 2).
Simulated stool sample: comparison of three different sequencing platforms. With extremely high CCS-reads quality, the results obtained using PacBio are 100% specific and sensitive. On the contrary, both IonTorrent PGM and Illumina MiSeq report several false positive and miss to identify one variant (Supplementary Tables 1 and 2).
3.2. DNA from a CRC tumor
The analysis of the tumor sample from a young patient revealed a somewhat expected scenario. Two codons were identified as mutated (Table 2): a 5-base, germline deletion was detected at codon 1309 of the APC gene, indicating that this individual was a carrier for the APC mutation and had familial adenomatous polyposis coli (FAP). The increased frequency of the mutation (above the expected 50% level) in the tumor tissue is expected due to loss of the wild type allele in that fraction of the tissue sample that consists of tumor cells. Furthermore a somatic, missense substitution was identified at codon 237 of TP53. Both mutations are known hotspots associated with CRC and were confirmed by Sanger sequencing (Fig. 2, Fig. 3). The frequency of the TP53 mutation indicates that the cellularity of the tumor sample was approximately 16%, which is in the normal range for CRC biopsies.
Table 2.
Experiment 2: mutations found in the DNA samples extracted from 37 y/o patient diagnosed with adenocarcinoma.
| Germline mutation | Somatic mutation | |
|---|---|---|
| Gene | APC | TP53 |
| Codon | 1309 | 237 |
| Reference allele | T | G |
| Alternative allele (AA) | -AAAAG | A |
| Coverage in the wild type (WT) | 2022 | 1728 |
| AA frequency in the WT | 48.22% | 0.00% |
| Coverage in the patient sample | 1271 | 872 |
| AA frequency in the patient sample | 68.61% | 12.96% |
| P-value | 0 | 1.45E-56 |
| Adjusted P-value | 0 | 4.98E-54 |
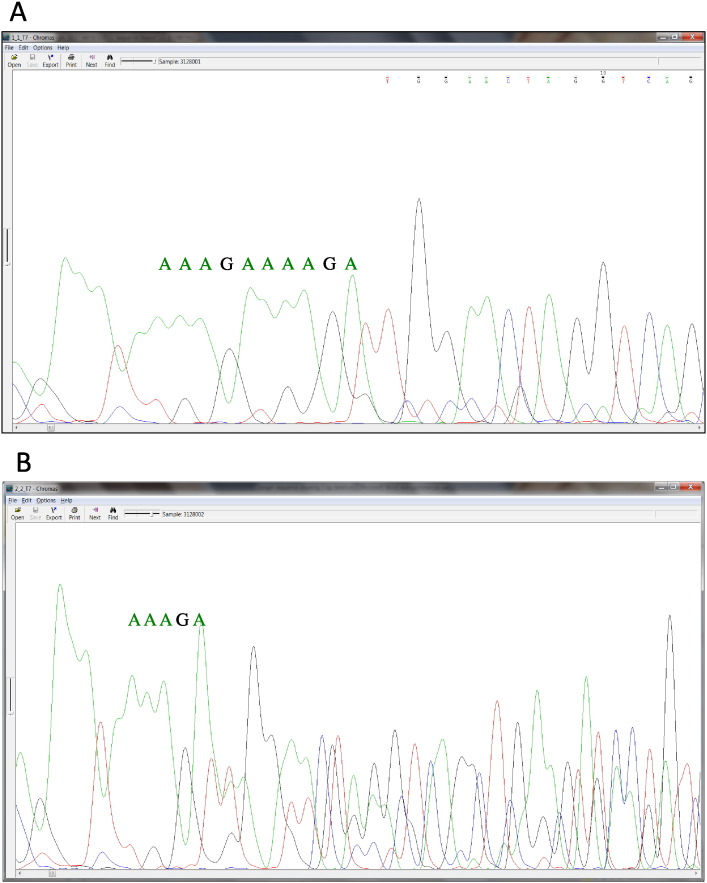
Fig. 2.
Confirmation by Sanger sequencing of the germline, 5 bp deletion in the tumor sample from a 37 y/o patient diagnosed with adenocarcinoma. This mutation represents the most frequently mutated codon in the APC gene. The DNA samples extracted form both the tumor and the healthy, adjacent tissues were sequenced on one SMRT cell. By means of third generation sequencing, the deletion was identified with frequencies of 48.22% and 68.61% in the normal (A) and the tumor (B) tissues, respectively (P < 1e-256 in both cases, Fisher's exact test).
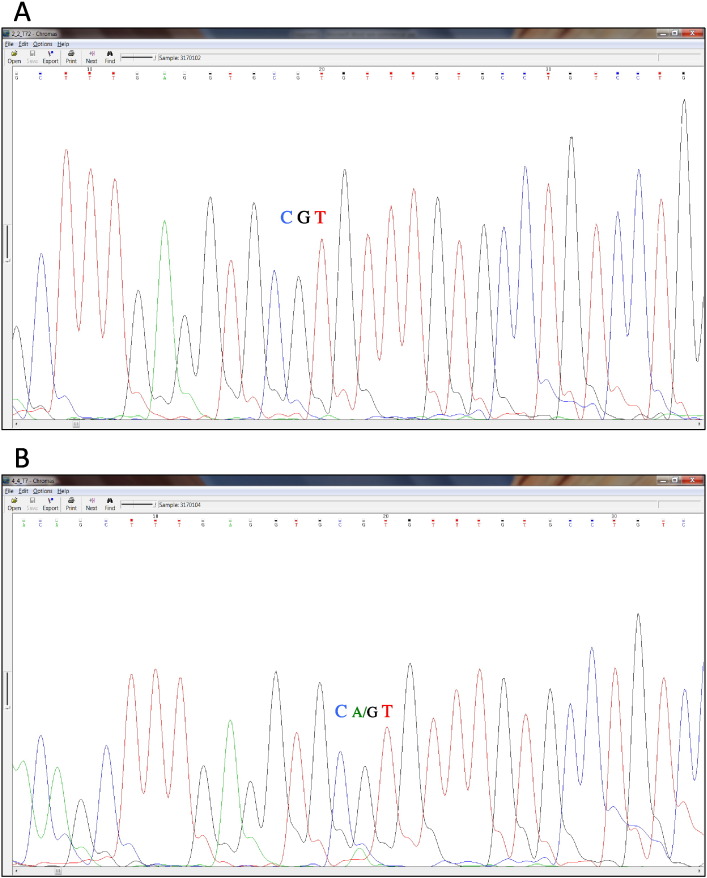
Fig. 3.
Confirmation by Sanger sequencing of the somatic point substitution in the tumor sample from a 37 y/o patient diagnosed with adenocarcinoma. This mutation represents the third most frequently mutated codon in the TP53 gene. The mutation in the tumor sample (B) is a heterozygous substitution which occurs at a frequency of 15.96% (highlighted in purple). Despite this rather low cellularity, the total absence of background noise allows for an easy detection of the green A signal in the Sanger sequencing track, which is instead clearly absent in the healthy tissue (A).
This experiment shows that PacBio RS sequencing is capable of identifying not only mutations in DNA derived from cells cultivated in vitro, but also from DNA extracted from an in vivo tumor sample where the nature of the mutations was not known beforehand.
3.3. Stool samples from CRC patients
In order to test the assay on clinically relevant DNA samples, we analyzed paired stool and tumor DNA samples extracted from each of the two CRC patients. DNA was isolated from stool prior to the surgical resection of the tumor as well as from the cancerous tissues. Only the MCR of the APC gene was analyzed in these experiments. In one patient, the 5 bp deletion located in the tandem repeat at codon 1309 on the APC gene was again encountered. This is not surprising, as this deletion is the most common APC mutation found in sporadic CRC. The mutation was detected at a frequency in the stool sample (FS) of 0.57% and confirmed at a frequency in the cancer sample (FC) of 82.22%. No other mutations were reported in this patient (Table 3). In the second patient, we identified two APC polymorphisms in the DNA isolated from stool: a rare, but known mutation (a deletion in a homopolymer stretch) at codon 1491 of the APC gene (FS = 0.37%) and an additional homopolymer insertion at codon 1556 (FS = 1.32%). In this case, we could confirm the presence of the deletion at codon 1491 in the cancer sample (FC = 19.05%) but no alterations were detected at codon 1556 (FC = 0.05%) (Table 4).
Table 3.
Experiment 3: mutations found in the DNA samples extracted from patient 1.
| Tumor | Stool | |
|---|---|---|
| Gene | APC6 | APC6 |
| Codon | 1309 | 1309 |
| Reference allele | T | T |
| Alternative allele (AA) | -AAAAG | -AAAAG |
| Coverage in the wild type (WT) | 4658 | 4658 |
| AA frequency in the WT (%) | 0.00 | 0.00 |
| Coverage in the patient sample | 3897 | 4550 |
| AA frequency in the patient sample (%) | 82.22 | 0.57 |
| P-value | 0 | 1.22E-08 |
| Adjusted P-value | 0 | 3.66E-06 |
Table 4.
Experiment 3: mutations found in the DNA samples extracted from patient 2.
| Tumor | Stool | ||
|---|---|---|---|
| Gene | APC | APC | APC |
| Codon | 1491 | 1491 | 1556 |
| Reference allele | A | A | G |
| Alternative allele (AA) | − T | − T | + A |
| Coverage in the wild type (WT) | 7372 | 7372 | 7359 |
| AA frequency in the WT (%) | 0.00 | 0.00 | 0.04 |
| Coverage in the patient sample | 8005 | 8005 | 7984 |
| AA frequency in the patient sample (%) | 19.05 | 0.37 | 1.32 |
| P-value | 0 | 3.04E-09 | 2.72E-26 |
| Adjusted P-value | 0 | 3.43E-07 | 8.16E-24 |
4. Discussion
The study presented here is the first attempt at determining the feasibility of using single molecule, third generation sequencing for the detection of CRC driving mutations in stool samples. The first step was to show that mutations present at low levels (1.5%) could be detected above background using a sequencing-based approach capable of detecting any mutation present in the amplicon. The raw error rate of the sequence data is critical in this respect because for genes such as APC, the cancer causing mutations can be found at any position in the amplicon and often consist of micro insertions and deletions, so that an error rate as low as 0.7% (the estimated error rate for the Illumina platform) will produce far too many false positives when scanning a test sequence of 1116 bp, which is the size of the MCR of the APC gene. Circular Consensus Sequencing of small amplicons, on the other hand, generates a highly accurate consensus sequence where the background is low enough to confidently call mutations at the 1.5% level.
When a stool DNA based screening test is performed for CRC, the mutations present in a patient sample, if any are indeed present, are unknown and can be found at many locations in the genes in the assay, thus the need of deep sequencing for unbiased interrogation of all the genomic loci. In this respect, the major problem associated with second generation sequencing technologies is the sensitivity of detection. In particular, the error in calling the nucleotides has systematic bias due to sequence context, often associated with the PCR amplification process required for such assays. As a consequence, within a sequencing run, the error will appear more frequently at the same loci, making vain any attempt of correction by enlarging the sampling space, i.e. by increasing the coverage. The error rate of the sequencing method thereby sets an upper limit to the quality of the raw sequences and thus the inherent sensitivity of the assay.
This type of issue has been already addressed in a couple of recent works:Lou et al.(2013) generated “circular sequencing” of short reads prior to the preparation of standard Illumina libraries, with the outcome of reducing the error rates by two to three orders of magnitude. Chen-Harris et al.(2013) applied the concept of overlapping read pairs (ORP) to screen for rare mutants in a viral ecosystem, detecting strains as rare as 0.1%. When compared to our approach, the former method has certainly the disadvantage of an extra wet-lab step prior to the start of the standard protocol, which translates into additional time consumption and cost. In particular, the error rates are yes improved, but still, at a reported 7.6 × 10− 6, they remain at most comparable to those reached by the CCS algorithm (Table 1). As for the latter, the results are very promising, but in principle ORP only take advantage of two copies of the same molecule (the forward and the reverse read). With PacBio RS II, sequencing continues as long as the polymerase is active and generates raw reads with an average length of 8000 nucleotides, scanning the same molecules many times over. Moreover, the distribution of base calling error is random (Carneiro et al., 2012). Consequently targeted amplicons, whose length is usually in the range of hundreds of bases, are each sequenced many times and any random sequencing error will be canceled out when consensus sequence is generated (Carneiro et al., 2012).
In the analysis of the simulated stool sample, the differences in the results between PacBio RS and the two 2nd generation platforms, Illumina MiSeq and IonTorrent PGM, are striking. PacBio RS shows 100% specificity and sensitivity. Remarkably, the fraction of the mutant allele in the APC deletion and in the TP53 substitution is smaller than targeted (0.5% and 0.75%, respectively) but the total absence of error in the wild type ensures that the mutations are still significant, even after Bonferroni correction (Table 1).
On the contrary, both the MiSeq and PGM are unable to identify one mutation. In the case of the MiSeq, despite a sequencing run beyond the manufacturer's specs in terms of quality (Supplementary Fig. S1), a PCR-induced, sequence context systematic bias has likely occurred. This is augmented by the observation that the alternative nucleotide at relative position 161 of TP53_exon7 is observed as being adenine instead of thymine in both the wild type and the mutant samples (Supplementary Table 1).
The lower overall quality of the PGM (Supplementary Fig. S2) instead generates too much background noise in the wild-type which masks the correct alternative observation of adenine as mutated allele in the KRAS gene. Furthermore, the average sequencing quality of the second generation technologies, which is two to three orders of magnitude lower than the PacBio CCS, generates a large amount of false positives when a direct case/control Fisher's test is adopted (Supplementary Table 1, Supplementary Table 2). When such false positives are present in a test for CRC they would result in a substantial number of unnecessary follow-up colonoscopies which would increase costs and cause emotional stress for the patients. The relatively high raw error rate for second generation sequencing in its present form allows it to be used for detecting mutations in tumor tissue where the mutant allele frequency is often above 5%, but makes them inadequate to identify low frequency mutations in DNA isolated from stool.
As expected, PacBio RS II was able to detect driver mutations in DNA derived from a CRC tumor in the case of the 37 year old patient. However, in order to test the feasibility of applying our approach for an unmet clinical need, we have isolated DNA from the stool of two patients diagnosed with adenocarcinoma and from the tumor tissues after surgery. In both patients, we identified a known CRC associated mutation which was then confirmed in the tumor DNA. At 0.37% and 0.57%, the frequencies of mutated DNA in the stool sample were low; however, the absence of sequencing errors at this level in the wild type allowed those observations to be confidently reported as significantly different from the wild type even after a stringent Bonferroni correction.
In one stool sample, we also identified a second mutation at codon 1556 of the APC gene. This codon is the eighth most frequently mutated codon in CRC and this mutation is extremely significant in our stool sample (P = 2.72e-26, PBonf = 9.35e-24). Therefore, despite the fact that this polymorphism was not observed in the DNA from the tumor tissue, it is unlikely that represents a false positive, as there are no other positives reported, not even at lower frequencies. We believe that this mutation might either come from a secondary neoplastic formation, which perhaps was not fully developed yet at the time of the colonoscopy and therefore was not resected; alternatively, and most probably, is the result of tumor cellular polyclonality, whereby the isolated DNA from the resected tumor sample captured only one cellular sub-population.
In conclusion, the use of third generation, single molecule sequencing with PacBio RS technology allows for the identification of mutations at 0.5% frequency, and as this is a sequencing method, any mutation within the amplicon can be detected. As most APC mutations detected in stool DNA from patients with polyps present in at least in this amount (Traverso et al., 2002b), the potential exists to screen non-symptomatic patients for polyps using this approach. In this respect, this type of screening could result in a much earlier identification of neoplastic formation than currently non-invasive tests such as FOBTs. Although within FOBTs the antibodies-based detection (FIT) has proven to be superior to other tests (Sharp et al., 2012), the fact that polyps and early adenomas do not bleed de facto limits the scope of FOBTs to already cancerous lesions, which often appear only in stage III of the development of the malignancy, when survival rated have already dropped significantly (Sameer, 2013).
Despite these encouraging results, we clearly need to underline that the current study only represents a pilot experiments, and a large scale, cohort study will be certainly required to eventually validate our finding in a definitive manner, as well as to leverage sufficient statistical power to infer meaningful false positive and miss rates. Moreover, a larger sample should ideally include CRC lesions at different stages, so to produce an even finer granularity in the results.
With respect to cost, PacBio RS II is currently generating enough throughput to sequence at least one sample on one SMRT cell. At the time of writing, the full cost for sequencing one SMRT cell, i.e., from sample delivery to data generation, is around $700 to $1000, depending on the provider and whether the customer is a research or a for-profit institution. This figure does not include the cost to isolate DNA from stool and amplify the genes of interest, an expense that gravitates (e.g., QIAmp mini stool kit and Haloplex custom) around $100 per samples. Therefore, even assuming that further advances in research will not reduce the current cost per sample for preparing and sequencing an assay like the one we described, the current price is already competitive with that of a colonoscopy, at least in the USA, where it already exceeds $1000 for Medicare patients (Pyenson et al., 2014). Moreover, the total cost of a colonoscopy can vary enormously, reaching peaks of $9000 in certain States for commercial payers, because a lot of factors can play a role in rising the price. For example, outpatients in hospitals will pay more than patients in doctor's offices, and general anesthesia represents another significant expense (International Federation of Health Plans, 2012). Instead, a genetic screening test is immune to all this, as its cost can be easily broken down into simple, stable components.
With respect to the cost of other sequencing technology, Illumine MiSeq might have an edge at this regard, given the larger throughput generated, however the key aspect of sensitivity discussed previously would still hold. Certainly a deep, large scale cost analysis, expanding also outside the US health system (which is notably expensive), will be required to provide a more comprehensive picture.
Finally, a few words about the perspective of introducing such a screening procedure in the public health sector: given the investment required to purchase and operate at capacity a Pacific Biosciences instrument, a centralized approached seems favorable and a strategic planning would be needed to ensure a sufficiently capillary network of centers able to offer such a sequencing service. As far as turnaround time is concerned, an optimized, streamlined version of our current workflow could be reasonably completed in 36 h for a full run, which would scan at least 16 samples. This is, per sample, faster than a colonoscopy (which involves also a preparatory day), however, even by increasing the instrument capacity by means of pooling multiple samples in one SMRT cell, the number of instrument required to keep up with the 14 million procedures performed yearly in the USA alone would be too large to envisage this sequencing approach to quickly become mainstream.
The following are the supplementary material related to this article.
Quality control report of the sequencing run with Illumina MiSeq in experiment 1.
Quality control report of the sequencing run with IonTorrent PGM in experiment 1.
Quality control report of the sequencing run with Pacific Bioscience in experiment 1 for the DLD1-mixed sample.
Quality control report of the sequencing run with Pacific Bioscience in experiment 1 for the wild type sample.
Quality control report of the sequencing run with Pacific Bioscience in experiment 2 for the tumor sample.
Quality control report of the sequencing run with Pacific Bioscience in experiment 2 for the adjacent healthy tissue sample.
Quality control report of the sequencing run with Pacific Bioscience in experiment 3 for sample Exp. 3, sample 1, as described in Supplementary Table 3.
Quality control report of the sequencing run with Pacific Bioscience in experiment 3 for sample Exp. 3, sample 1/2, as described in Supplementary Table 3.
Quality control report of the sequencing run with Pacific Bioscience in experiment 3 for sample Exp. 3, sample 2, as described in Supplementary Table 3.
Quality control report of the sequencing run with Pacific Bioscience in experiment 3 for sample Exp. 3, sample 2/2, as described in Supplementary Table 3.
Quality control report of the sequencing run with Pacific Bioscience in experiment 3 for the control APC sequence.
Experiment 1 : Putative mutations reported based on the sequencing run on the Illumina MiSeq.
Experiment 1 : Putative mutations reported based on the sequencing run on the IonTorrent PGM.
Authors' contributions
Study design, conception and coordination: AG, GR.
DNA Template and sequencing libraries preparation, sequencing runs: AG, LP, AP.
Data analysis: GR.
Writing and revising the manuscript: GR, AG, BS, LP and AP.
Aid in study coordination and administrative, technical and material support: RS, FH.
All authors read and approved the final manuscript.
References
- Ahlquist D.A1., Skoletsky J.E., Boynton K.A., Harrington J.J., Mahoney D.W., Pierceall W.E. Colorectal cancer screening by detection of altered human DNA in stool: feasibility of a multitarget assay panel. Gastroenterology. 2000;119(5):1219–1227. doi: 10.1053/gast.2000.19580. [DOI] [PubMed] [Google Scholar]
- Ahlquist D.A1., Zou H., Domanico M., Mahoney D.W., Yab T.C., Taylor W.R. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology. 2012;142(2):248–256. doi: 10.1053/j.gastro.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Facts and Figures, American Cancer Society 2014. www.cancer.org
- Carneiro M.O., Russ C., Ross M.G., Gabriel S.B., Nusbaum C., MA D.P. Pacific biosciences sequencing technology for genotyping and variation discovery in human data. BMC Genomics. 2012;13:375. doi: 10.1186/1471-2164-13-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan T.L., Zhao W., Leung S.Y., Yuen S.T. BRAF and KRAS mutations in colorectal hyperplastic polyps and serrated adenomas. Cancer Res. 2003;63:4878–4881. [PubMed] [Google Scholar]
- Chen-Harris H., Borucki M.K., Torres C., Slezak T.R., Allen J.E. Ultra-deep mutant spectrum profiling: improving sequencing accuracy using overlapping read pairs. BMC Genomics. 2013;14:96. doi: 10.1186/1471-2164-14-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejea C., Wick E., Sears C.L. Bacterial oncogenesis in the colon. Future Microbiol. 2013;8(4):445–460. doi: 10.2217/fmb.13.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl F., Schmidt K., Durkee K.H., Moore K.J., Goodman S.N., Shuber A.P. Analysis of mutations in DNA isolated from plasma and stool of colorectal cancer patients. Gastroenterology. 2008;135(2):489–498. doi: 10.1053/j.gastro.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S.M1., Traverso G., Johnson C., Geng L., Favis R., Boynton K. Detecting colorectal cancer in stool with the use of multiple genetic targets. J. Natl. Cancer Inst. 2001;93(11):858–865. doi: 10.1093/jnci/93.11.858. (Jun 6) [DOI] [PubMed] [Google Scholar]
- Fang P., Yan Z., Liu W., Darwanto A., Pelak K., Anoe K. Validation of Illumina TruSeq Amplicon Cancer Panel with concordance testing using Ion AmpliSeq Cancer Panel and other methods. Cancer Res. 2013;73(8) Supplement 1. [Google Scholar]
- Fearon E.R. Molecular genetics of colorectal cancer. Annu. Rev. Pathol.: Mech. Dis. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- Fearon E.R., Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Frampton G.M., Fichtenholtz A., Otto G.A., Wang K., Downing S.R., He J. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 2013;31(11):1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerecke C., Mascher C., Gottschalk U., Kleuser B., Scholtka B. Ultrasensitive detection of unknown colon cancer-initiating mutations using the example of the adenomatous polyposis coli gene. Cancer Prev. Res. 2013;6(9):898–907. doi: 10.1158/1940-6207.CAPR-13-0145. [DOI] [PubMed] [Google Scholar]
- Houlston R.S. and members of COGENT, COGENT (COlorectal cancer GENeTics) revisited. Mutagenesis. 2012;27:143–151. doi: 10.1093/mutage/ger059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrašovec S., Glavač D. MicroRNAs as novel biomarkers in colorectal cancer. Front. Genet. 2012;3:1–9. doi: 10.3389/fgene.2012.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://mendel.iontorrent.com/ion-docs/Technical-Note-TMAP-Alignment_9012907.html
- http://www.edgebio.com/ion-ampliseq-fixed-panels-hct-15-colon-carcinoma-cell-line
- Imperiale T.F1., Ransohoff D.F., Itzkowitz S.H., Levin T.R., Lavin P., Lidgard G.P. Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 2014;370(14):1287–1297. doi: 10.1056/NEJMoa1311194. Apr 3. [DOI] [PubMed] [Google Scholar]
- International Federation of Health Plans . 2012. Comparative Price Report. [Google Scholar]
- Kinde I., Wu J., Papadopoulos N., Kinzler K.W., Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. U. S. A. 2011;108(23):9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent-Puig P., Béroud C., Soussi T. APC gene: database of germline and somatic mutations in human tumors and cell lines. Nucleic Acids Res. 1998;26(1):269–270. doi: 10.1093/nar/26.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebwohl B., Kastrinos F., Glick M., Rosenbaum A.J., Wang T., Neugut A.I. The impact of suboptimal bowel preparation on adenoma miss rates and the factors associated with early repeat colonoscopy. Gastrointest. Endosc. 2011;73:1207–1214. doi: 10.1016/j.gie.2011.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou D.I., Hussmann J.A., McBee R.M., Acevedo A., Andino R., Press W.H. High-throughput DNA sequencing errors are reduced by orders of magnitude using circle sequencing. Proc. Natl. Acad. Sci. U. S. A. 2013;110(49):19872–19877. doi: 10.1073/pnas.1319590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazeh H., Mizrahi I., Ilyayev N., Halle D., Brücher B.L.D.M., Bilchik A. The diagnostic and prognostic role of microRNA in colorectal cancer — a comprehensive review. J. Cancer. 2013;4:281–295. doi: 10.7150/jca.5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaki M., Iijima T., Konishi M., Sakai K., Ishii A., Yasuno M. Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene. 1999;20:3098–3103. doi: 10.1038/sj.onc.1202642. [DOI] [PubMed] [Google Scholar]
- Moayyedi P., Achkar E. Does fecal occult blood testing really reduce mortality? A reanalysis of systematic review data. Am. J. Gastroenterol. 2006;101(2):380–384. doi: 10.1111/j.1572-0241.2006.00537.x. [DOI] [PubMed] [Google Scholar]
- Morin P.J., Sparks A.B., Korinek V., Barker N., Clevers H., Vogelstein B. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275(5307):1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Parsons R., Li G.M., Longley M.J., Fang W.H., Papadopoulos N. Hypermutability and mismatch repair deficiency in RER + tumor cells. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- Pyenson B., Scammell C., Broulette J. Costs and repeat rates associated with colonoscopy observed in medical claims for commercial and Medicare populations. BMC Health Serv. Res. 2014;14:92. doi: 10.1186/1472-6963-14-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos M., Llagostera M., Esteva M., Cabeza E., Cantero X., Segarra M. Knowledge and attitudes of primary healthcare patients regarding population-based screening for colorectal cancer. BMC Cancer. 2011;11(1):408. doi: 10.1186/1471-2407-11-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sameer A.S. Colorectal cancer: molecular mutations and polymorphisms. Front Oncol. 2013;3:114. doi: 10.3389/fonc.2013.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senore C., Ederle A., Fantin A., Andreoni B., Bisanti L., Grazzini G. Acceptability and side-effects of colonoscopy and sigmoidoscopy in a screening setting. J. Med. Screen. 2011;18(3):128–134. doi: 10.1258/jms.2011.010135. [DOI] [PubMed] [Google Scholar]
- Sharp L., Tilson L., Whyte S., O'Ceilleachair A., Walsh C., Usher C. Cost-effectiveness of population-based screening for colorectal cancer: a comparison of guaiac-based faecal occult blood testing, faecal immunochemical testing and flexible sigmoidoscopy. Br. J. Cancer. 2012;106:805–816. doi: 10.1038/bjc.2011.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R.R., Patel K.P., Routbort M.J., Reddy N.G., Barkoh B.A., Handal B. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J. Mol. Diagn. 2013;15(5):607–622. doi: 10.1016/j.jmoldx.2013.05.003. [DOI] [PubMed] [Google Scholar]
- Sparks A.B., Morin P.J., Vogelstein B., Kinzler K.W. Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer. Cancer Res. 1998;58(6):1130–1134. [PubMed] [Google Scholar]
- Takagi Y., Kohmura H., Futamura M., Kida H., Tanemura H., Shimokawa K. Somatic alterations of the DPC4 gene in human colorectal cancers in vivo. Gastroenterology. 1996;111:1369–1372. doi: 10.1053/gast.1996.v111.pm8898652. [DOI] [PubMed] [Google Scholar]
- Talaat N., Harb W. Reluctance to screening colonoscopy in Arab Americans: a community based observational study. J. Community Health. 2013;38(4):619–625. doi: 10.1007/s10900-013-9688-7. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson I.P.M., Dunlop M., Campbell H., Zanke B., Gallinger S., Hudson T. COGENT (COlorectal cancer GENeTics): an international consortium to study the role of polymorphic variation on the risk of colorectal cancer. Br. J. Cancer. 2010;102:447–454. doi: 10.1038/sj.bjc.6605338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers K.J., Chin C.S., Rank D.R., Eid J.R., Turner S.W. Flexible and efficient template format for circular consensus sequencing and SNP detection. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverso G., Shuber A., Levin B., Johnson C., Olsson L., Schoetz D.J. Detection of APC mutations in fecal DNA from patients with colorectal tumors. N. Engl. J. Med. 2002;346(5):311–320. doi: 10.1056/NEJMoa012294. [DOI] [PubMed] [Google Scholar]
- Traverso G., Shuber A., Olsson L., Levin B., Johnson C., Hamilton S.R. Detection of proximal colorectal cancers through analysis of faecal DNA. Lancet. 2002;359(9304):403–404. doi: 10.1016/S0140-6736(02)07591-8. [DOI] [PubMed] [Google Scholar]
- Vilar E., Gruber S.B. Microsatellite instability in colorectal cancer—the stable evidence. Nat. Rev. Clin. Oncol. 2010;7:153–162. doi: 10.1038/nrclinonc.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- www.globocan.iarc.fr
- Zhu Q., Gao R., Wu W., Qin H. The role of gut microbiota in the pathogenesis of colorectal cancer. Tumour Biol. 2013;34(3):1285–2300. doi: 10.1007/s13277-013-0684-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quality control report of the sequencing run with Illumina MiSeq in experiment 1.
Quality control report of the sequencing run with IonTorrent PGM in experiment 1.
Quality control report of the sequencing run with Pacific Bioscience in experiment 1 for the DLD1-mixed sample.
Quality control report of the sequencing run with Pacific Bioscience in experiment 1 for the wild type sample.
Quality control report of the sequencing run with Pacific Bioscience in experiment 2 for the tumor sample.
Quality control report of the sequencing run with Pacific Bioscience in experiment 2 for the adjacent healthy tissue sample.
Quality control report of the sequencing run with Pacific Bioscience in experiment 3 for sample Exp. 3, sample 1, as described in Supplementary Table 3.
Quality control report of the sequencing run with Pacific Bioscience in experiment 3 for sample Exp. 3, sample 1/2, as described in Supplementary Table 3.
Quality control report of the sequencing run with Pacific Bioscience in experiment 3 for sample Exp. 3, sample 2, as described in Supplementary Table 3.
Quality control report of the sequencing run with Pacific Bioscience in experiment 3 for sample Exp. 3, sample 2/2, as described in Supplementary Table 3.
Quality control report of the sequencing run with Pacific Bioscience in experiment 3 for the control APC sequence.
Experiment 1 : Putative mutations reported based on the sequencing run on the Illumina MiSeq.
Experiment 1 : Putative mutations reported based on the sequencing run on the IonTorrent PGM.