Abstract
The transcription factor CLOCK (CLK) is essential for the development and maintenance of circadian rhythms in Drosophila. However, little is known about how CLK levels are controlled. Here, we show that Clk mRNA is strongly regulated post-transcriptionally through its 3’UTR. Flies expressing Clk transgenes missing their normal 3’UTR, exhibited variable CLK-driven transcription and circadian behavior, as well as ectopic expression of CLK-target genes in the brain. Surprisingly, in these flies, the numbers of the key circadian neurons differs stochastically between individuals and within the two hemispheres of the same brain. In addition, flies carrying Clk transgenes with deletions in the binding sites for the miRNA bantam have stochastic number of pacemaker neurons, suggesting that this miRNA mediates the deterministic expression of CLK. Overall our results demonstrate a key role of Clk post-transcriptional control in stabilizing circadian transcription, which is essential for proper development and maintenance of circadian rhythms in Drosophila.
Keywords: circadian, clock, Clk, miRNAs, noise, Drosophila
Introduction
Most organisms use circadian clocks to keep temporal order and anticipate daily environmental changes. Circadian clocks appear to function on a cell-autonomous basis and are generated by interconnected complex transcriptional and post-translational feedback loops1. In Drosophila, the master genes Clock (Clk) and cycle (cyc) activate the circadian system by promoting rhythmic transcription of a number of key genes2–4. Three of these target gene products, PERIOD (PER)5, TIMELESS (TIM)6, and CLOCKWORK ORANGE (CWO)7–9 repress CLK-CYC-mediated transcription on a daily basis. CLK activates transcription only while associated with CYC. The latter is constitutively expressed whereas CLK is present in limiting amounts4,10. The CLK-CYC heterodimer also activates the expression of VRILLE (VRI) and PAR-DOMAIN-PROTEIN 1 (PDP1), which have been postulated to be responsible for the oscillation of Clk mRNA11,12. Although the role of VRI in repressing Clk transcription is well established, recent reports suggest that PDP1 might not directly regulate Clk13. In addition to transcriptional control, post-transcriptional and post-translational regulations play also a central role in circadian timekeeping (for reviews see 14–16).
Circadian clocks are widespread through the fly body 17. However, a group of approximately 150 neurons in the brain drive circadian rhythms in locomotor activity18. These neurons have been divided into several subgroups based on their anatomical location and expression of the core clock genes19,20. These groups are: small and large ventral lateral (sLNvs and lLNvs), dorsal lateral (LNds), and dorsal (DN1s, DN2s, and DN3s) neurons. The neuropeptide PIGMENT DISPERSING FACTOR (PDF), which is expressed exclusively in the LNvs, is essential for normal circadian patterns of activity in light and dark and for persistent circadian rhythms in constant darkness (DD)18.
Despite the central role of Clk in the circadian system, it is still not well understood how CLK levels and activity are controlled. The current model states that most (if not all) of this control is through post-translational regulation such as phosphorylation, ubiquitination and other types of modifications21–25. Indeed, although Clk mRNA levels display strong transcriptional oscillations11,12, CLK protein levels are nearly constant though the day26. Moreover, expression of CLK under the tim or per promoter in ClkARK fly strains (in which transcription has the opposite daily phase relative to that under control of the Clk promoter21) does not disrupt circadian behavior, indicating that flies can adapt or compensate for high levels of CLK21. As these experiments were performed in a wild-type background, whether constant or shifted Clk transcription alone can drive circadian behavioral rhythms was not determined.
In addition, it is well established that the levels or activity of CLK dramatically alter the circadian period and amplitude27–29. Moreover, uncontrolled expression of CLK leads to death. For example, expression of CLK using the circadian driver tim-gal4 leads to embryonic lethality30, and induction of CLK-driven transcription using the CLKGR system results in adult death (Ron Weiss, personal communication). Some of the toxic effects of CLK overexpression are likely due to a central role of Clk in cell determination. Indeed, CLK expression in non-circadian cells is enough to generate ectopic circadian clocks in the fly brain30. In addition, CLK-driven transcription may have a role in the development of the circadian system as Clk and cyc fly mutants display abnormalities in the morphology of circadian neurons29,31. These data demonstrate that post-translational control alone may not be enough to buffer big changes in CLK levels. Indeed, post-transcriptional regulation might also be important, as we demonstrated in the past that Clk is strongly regulated by miRNAs32. Briefly, Clk is strongly bound to AGO1 and the miRNA bantam can regulate Clk levels in Drosophila S2 cells32. Moreover, we showed that the putative bantam binding sites on Clk 3’ UTR are essential for normal circadian rhythms32. However, how this regulation impacts CLK levels and activity in time and space is still unknown.
Circadian clocks are extraordinarily robust systems; they are able to keep time accurately without any timing cues. This robustness is likely the result of multiple layers of regulation that assure accurate timekeeping by buffering stochastic changes of clock-relevant activities. For example, it has been proposed that the main function of the redundancy and interlocked transcriptional feedback loops of the circadian system is to provide robustness to circadian transcription33–36. These layers of regulation extend even beyond the single-cell level. Circadian neurons in the brain are organized in a network that synchronizes and likely amplifies the individual neuronal oscillators thereby contributing to a coherent and robust behavioral output2,37–39. Circadian clocks must also control or buffer transcriptional noise and its consequences, especially for genes that are expressed at limiting levels (i.e. Clk), as it is well known that transcriptional noise is inversely correlated with transcriptional rate40.
Here we demonstrate that the Clk 3’ UTR sets a threshold for meaningful circadian gene expression. Flies in which Clk is expressed without post-transcriptional control (ClkSV40 flies), have ectopic circadian cells in the brain. Surprisingly the levels of CLK per cell are normal in these flies, indicating that Clk post-transcriptional regulation does not control the overall CLK levels. Interestingly, ClkSV40 flies have aberrant circadian transcription and behavior, as well as widespread expression of CLK-target gene products in the brain. These behavioural deficits are accompanied by the stochastic development of pdf-expressing LNvs cells that varied in number between individuals and even between the two lobes of the same brain. Similarly, flies carrying Clk transgenes with deletions in the binding sites for the miRNA bantam display stochastic number of pacemaker neurons, strongly suggesting that this miRNA is the key factor mediating the deterministic expression of CLK. We backed up this role for post-transcriptional control by a mathematical model that predicts the central role of Clk mRNA turnover in minimizing noise of the circadian feedback loops. All together, our results demonstrate that post-transcriptional control of a master transcriptional regulator is an efficient way of assuring deterministic transcriptional responses.
Results
Clk mRNA is under strong post-transcriptional control
We recently showed that Clk mRNA levels are regulated by microRNAs (miRNAs)32. To address the importance of Clk post-transcriptional regulation, we compared Clk mRNA levels with those of other circadian mRNAs using previously published microarray and RNA-seq data from fly heads and brains7,41,42. In these datasets Clk mRNA levels were the lowest of the core circadian components (Figure 1a, Supplementary Figures 1a and 1b).
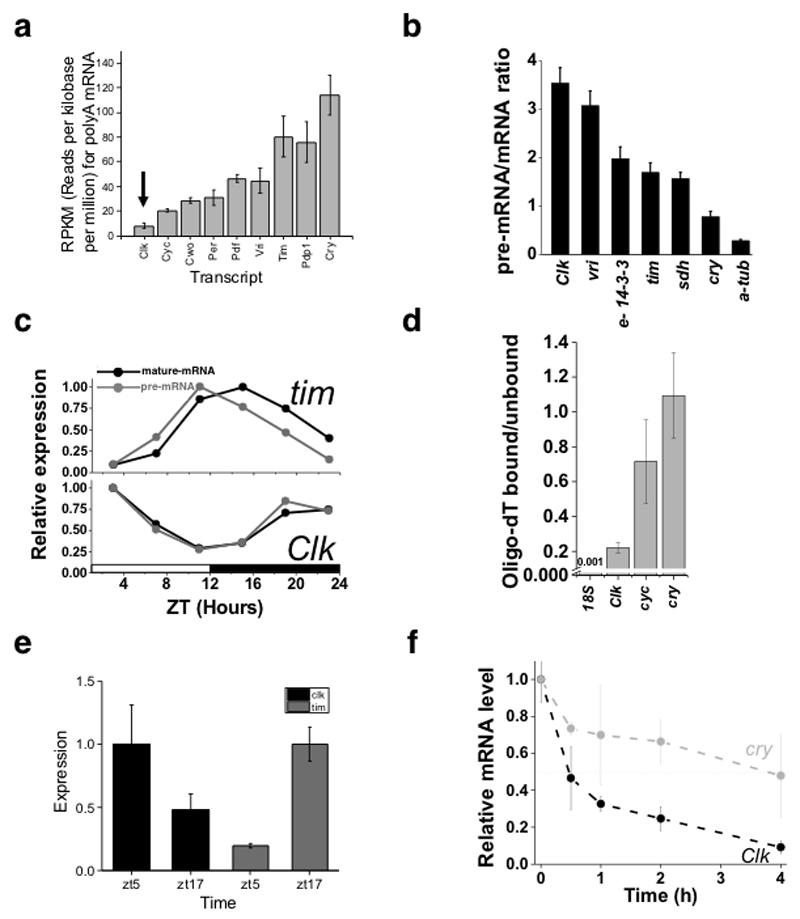
Figure 1. Clk is under strong post-transcriptional regulation.
a. Comparison of clock transcript levels. RNA-seq data 41 was used to compare expression levels of core circadian components in fly heads; an average of six timepoints ± SE in LD conditions (ZT 2,6,10,14,18,22) were analyzed. b. The levels of pre-mRNA and mRNA of core clock components were measured by real-time PCR (RT-PCR) from a mixture of six circadian time points and scaled using an equimolar mixture of the PCR products to obtain absolute measurements between the different mRNAs or pre-mRNAs in each sample (mean ± SE; three biological replicas). As the values in each sample (pre-mRNA and mRNA) are normalized, the pre-mRNA/mRNA ratio is relative, not absolute (see methods for details). c. RT-PCR measurements of transcriptional (gray) and steady state (black) levels of Clk and tim. Transcriptional/pre mRNA transcripts were detected using primers for intronic sequences of the genes, whereas steady state/ mature mRNAs were detected using primers flanking two exons. Expression was normalized to RP49 and RpS18. Representative experiment of three repeats is shown. d. RT-PCR results presenting the ratio of mRNA bound to oligo-dT beads vs. unbound mRNA. Data pooled from a mixture of six time points (mean ± SE; three biological replicates). e. A Drosophila fly wing system was developed to monitor circadian gene mRNA expression levels (normalized to RP49 at ZT5, 17). Clk and tim levels cycle with the expected phase. The measurements were performed in triplicated for each time point using the Canton-S strain. f. RT-PCR mRNA half-life measurements for Clk and cry mRNAs from fly wings treated with actinomycin D in five different time time-points (0-4h after actinomycin D exposure, 12 h light/12 hour dark conditions,mean ± SE; three biological replicates). See also Supplementary Figure 1
To determine whether the low mRNA levels are due to low transcription and/or high post-transcriptional control, we estimated the extent of Clk post-transcriptional regulation by comparing the ratio of pre-mRNA to steady-state mRNA using quantitative scaled RT-PCR. This comparison revealed that Clk as well as vri are under very strong post-transcriptional regulation (Figure 1b). This finding is in agreement with calculations made using previously published data comparing the signal obtained from nascent mRNA with steady-state mRNA41 (Supplementary Figure 1c). Additional lines of evidence also support strong Clk post-transcriptional regulation. For example, we observed no difference between the temporal profiles of mature and pre-mRNAs encoding CLK, suggesting that Clk is a very short-lived mRNA (Figure 1c). A delay, such as that observed in the profiles for tim mature and pre-mRNAs (Figure 1c), indicates a longer mRNA half-life. Clk mRNA binds weakly to oligo-dT beads (Figure 1d), likely reflecting a short polyA-tail43 and strongly binds to the miRNA-effector protein AGO1 demonstrating miRNA-mediated regulation of this mRNA32.
To determine whether Clk post-transcriptional regulation is at least partially due to rapid mRNA turnover, we estimated the Clk mRNA half-life in an oscillating system. Because Drosophila cells in culture do not exhibit circadian oscillations and mRNA half-lives cannot be easily measured in living flies, we first examined whether fly-wing cultures could be used for mRNA measurements and estimation of mRNA half-lives. These cultures are widely utilized to monitor circadian rhythms with luciferase reporters17,44 but have never been validated for mRNA measurements. We measured the amounts of Clk and tim mRNAs at two different time points in cultured wings. As observed in fly heads and brains, Clk and tim mRNAs displayed oscillations with opposite phase (high levels in the morning and night respectively; Figure 1e). We then determined the half-life of Clk and cry mRNAs by treating wings with the transcriptional inhibitor actinomycin D. The Clk mRNA half-life was very short (~0.5 h), considerably shorter than the half-life of cry mRNA (half-life ~4 h, Figure 1f), another circadian mRNA that oscillates with a similar phase.
Clk 3’ UTR sets a threshold for CLK-driven transcription
To study the functional significance of the short Clk mRNA half-life, we utilized Drosophila S2 cells, which do not express endogenous Clk mRNA or protein45. We co-transfected these cells with two different fluorescent reporters, both under the control of the copper-inducible metallothionein (MT) promoter, harboring either a control or a Clk 3’ UTR (Figure 2a). These reporters do not respond to transcriptional feedback (by VRI) and hence only evaluate the effect of the 3’ UTR. Expression from the reporter with the Clk 3’ UTR was significantly lower than that from control reporters, demonstrating that this 3’ UTR is strongly regulated also in S2 cells (Figure 2b). Importantly, the Clk 3’ UTR rendered the reporter insensitive to increasing levels of transcription at low concentrations of inducer (Figure 2b, inset), suggesting a suppression/threshold mechanism to prevent leaky Clk expression from stochastic transcription events.
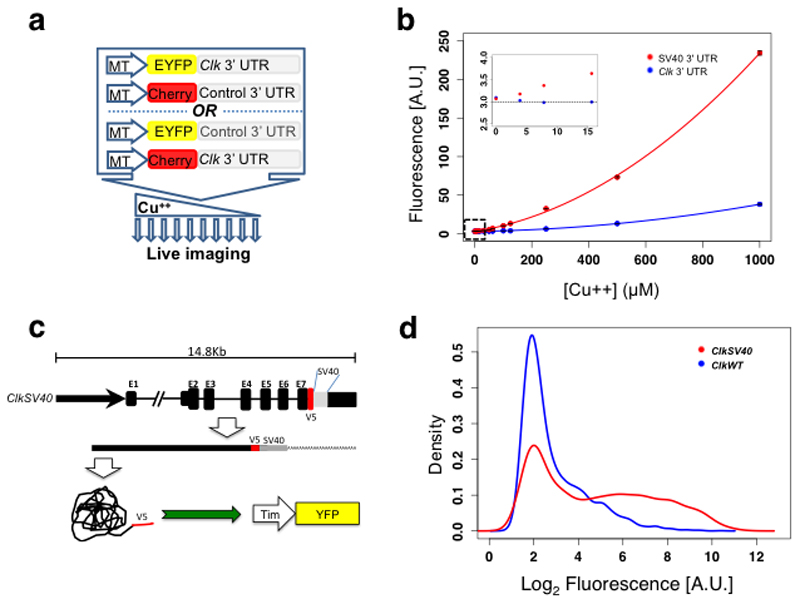
Figure 2. Clk 3’ UTR establishes a threshold for CLK-driven transcription.
a. Schematics of the system for monitoring post-transcriptional regulation in single Drosophila S2 cells. b. Mean fluorescent levels of S2 cell population expressing the Clk and SV40 3’ UTR fluorescent reporters at different copper levels. The inset in the left represents a closer view of the signal at low copper concentrations (0-15 µM). Cyan fluorescent protein (CFP) expressed from a pAc-CFP plasmid, was utilized as transfection control. c. Schematics of the ClkSV40 and tim-YFP constructs. d. Histogram representing the intensity of signal due to the tim-YFP reporter upon transfection with ClkSV40 or ClkWT plasmids. See also Supplementary Figure 2. Red fluorescent protein (RFP) expressed from a pAc-RFP plasmid, was utilized as transfection control.
To study the functional significance of the rapid Clk mRNA turnover in the context of the normal gene regulation, we generated a 14.5-kilobase (kb) Clk construct that includes all known Clk coding and regulatory sequences but also carries a SV40 3’ UTR inserted immediately downstream the open reading frame (ClkSV40; Figure 2c). We transfected ClkSV40 or ClkWT (same transgene but carrying the wild-type Clk 3’ UTR32) into Drosophila S2 cells along with a newly generated tim-YFP reporter (tim promoter driving the expression of YFP; tim is a direct CLK-CYC target). The ClkWT construct did not detectably activate expression of the tim-YPF reporter, indicating very low CLK expression (most cells show only background fluorescence; Figure 2d, blue line). In contrast, the presence of the ClkSV40 construct significantly increased tim-YFP levels compared to YFP levels in the presence of the ClkWT construct (Figure 2d; compare meanClkWT=15.9 with meanClkSV40=127). Importantly, tim-YFP reporter levels were highly variable among ClkSV40-transfected cells, suggesting that the Clk 3’ UTR not only decreases Clk mRNA levels but makes them more uniform.
Clk 3’ UTR prevents leaking CLK activity in the brain
To extend these observations, we generated transgenic flies carrying the ClkSV40 transgenes. These flies express a V5 tag fused to the CLK C-terminus to allow the identification of the encoded mRNAs and proteins (Figure 2c). We examined flies with the ClkWT or ClkSV40 transgene in a wild-type Clk genetic background (ClkWT or ClkSV40 flies, respectively). Like the endogenous Clk gene, the ClkWT transgene was under tight post-transcriptional control: the mRNAs expressed from the transgene were strongly associated with AGO1 and bound poorly to oligo-dT beads (Figures 3a, b). In contrast, ClkSV40 mRNAs did not associate with AGO1 and bound strongly to oligo-dT beads (Figures 3a, b). This indicates that Clk post-transcriptional regulation is directed predominantly if not exclusively by the 3’ UTR.
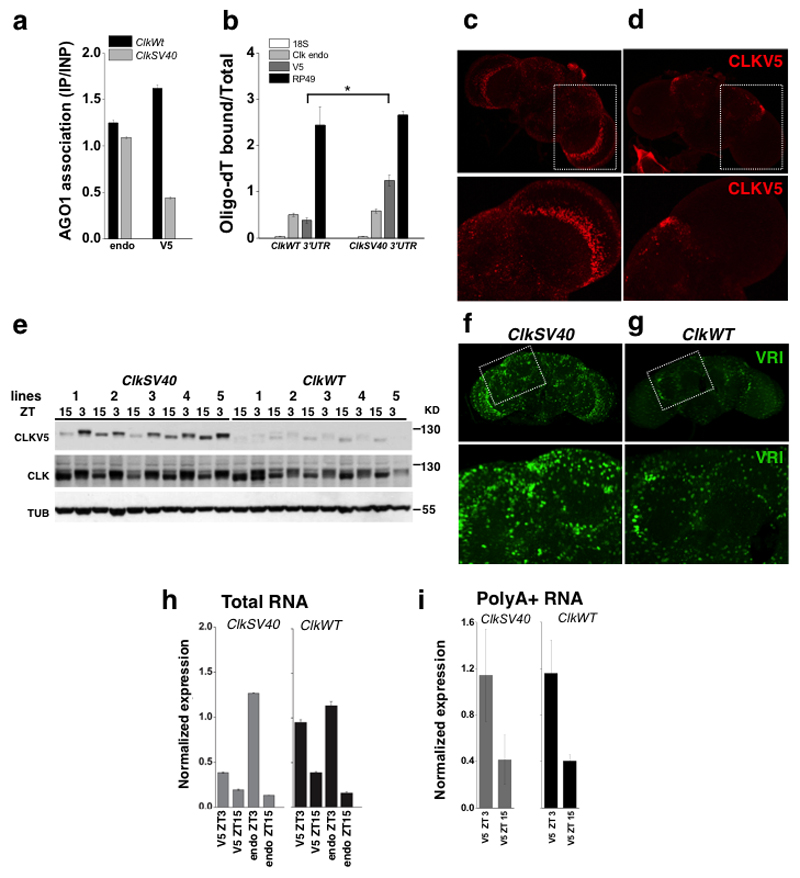
Figure 3. ClkSV40 flies express a stable mRNA that generates ectopic clock cells.
a. AGO1 binding to endogenous genomic Clk mRNA or ClkV5 from ClkWT or ClkSV40 transgenic fly heads (measured by RT-PCR, ratio of IP vs input is shown, one representative experiment out of three is presented). b. RT-PCR results showing the ratio of Clk mRNA bound vs. unbound- to oligo-dt beads in ClkWT and ClkSV40 fly heads (* p<0.036, student t-test; mean ± SE, n=6 for each genotype, average of five independent insertions of these transgenes were plotted). c, d. Immunofluorescence (IF) analysis of (c) ClkSV40 or (d) ClkWT Drosophila brains using an anti-V5 antibody (lower panel represents magnification of rectangle area). e. Expression levels of CLKV5, total CLK, and tubulin proteins determined by western blot analysis of head lysates from ClkWT or ClkSV40 flies (five independent insertions for each transgene). The showed experiment is one out of four representative experiments. f, g. Immunofluorescence analysis of (f) ClkSV40 or (g) ClkWT Drosophila brains using an anti-VRI antibody. h, i. ClkV5 and endogenous Clk mRNAs levels from ClkSV40 and ClkWT fly heads determined by RT-PCR from (h) total RNA and (i) polyA+-selected RNA (for each time point, the average of five independent insertions for each transgene is plotted). See also Supplementary Movie 1 and Supplementary Movie 2.
To address the ectopic and variable expression issue suggested by the S2 cells experiments, we assayed the expression of CLKV5 protein by immunohistochemistry. In ClkWT flies, expression of CLKV5 was restricted to the normal pattern (approx. 150 circadian neurons2). In ClkSV40 flies, however, there was widespread expression of CLKV5 throughout the brain (Figures 3c, d). Importantly, this ectopic expression was observed in five independent insertions of the ClkSV40 transgene and in none of the four ClkWT insertions that we tested (Supplementary Figure 2a). Moreover, ClkSV40 fly heads contained significantly higher levels of V5-tagged CLK than ClkWT heads as determined by western blot (Figure 3e). We assume that this increase is due to the presence of the ectopic circadian cells in the brain.
To determine whether this ectopic expression also generated ectopic CLK-driven transcription, we analyzed levels of two CLK-transcriptional targets: VRI and TIM. Immunocytochemistry using anti-VRI and anti-TIM antibodies revealed a massive increase in the number of VRI- and TIM-positive cells in ClkSV40 compared to ClkWT fly brains (Figures 3f,g and Supplementary Figure 2a and b; also see Supplementary Movies 1 and 2 for 3D visualization of VRI-positive cells in brains of ClkWT and ClkSV40 flies).
To rule out the possibility that CLK ectopic expression is due to enhanced transcription mediated by the introduction of the SV40 sequence, we compared ClkV5 mRNA levels between ClkWT and ClkSV40 fly heads at two different times of the day. Surprisingly mRNA levels from the ClkSV40 transgene were even lower than those from the ClkWT transgene, demonstrating that the higher levels of CLK in ClkSV40 flies are not due to an increase in Clk transcription (Figure 3h, compare levels of V5-tagged Clk mRNAs). The substantial differences in the levels of CLKV5 protein (Figure 3e) indicate that the Clk 3’ UTR also contributes to Clk translational regulation. This lack of a transcriptional effect was confirmed by examining the levels of V5-tagged Clk polyA+ mRNAs, which were similar in flies that expressed the ClkWT and the ClkSV40 transgenes (Figure 3i).
ClkSV40 flies have abnormal and variable circadian behavior
To test the importance of the Clk 3’ UTR on circadian behavior, we assessed locomotor activity rhythms in ClkWT and ClkSV40 flies. In flies with one copy of ClkWT, we observed a slightly shortened circadian period and little effect on the overall rhythm strength (Figure 4a and Supplementary Figure 3a) as previously described28. However, in flies with one copy of the ClkSV40 transgene, we observed a slightly more pronounced shortening of the circadian period, which was accompanied by a higher proportion of arrhythmic flies (Figure 4a and Supplementary Figure 3a). Interestingly, there was also high variability among the nine tested ClkSV40 fly insertion lines (Figure 4a and Supplementary Figure 3a). Moreover, a subpopulation of ClkSV40 rhythmic flies displayed atypical locomotor activity rhythms after a few days in constant darkness (e.g., variable and in some cases split behavior; Figures 4b, c and Supplementary Figure 3a).
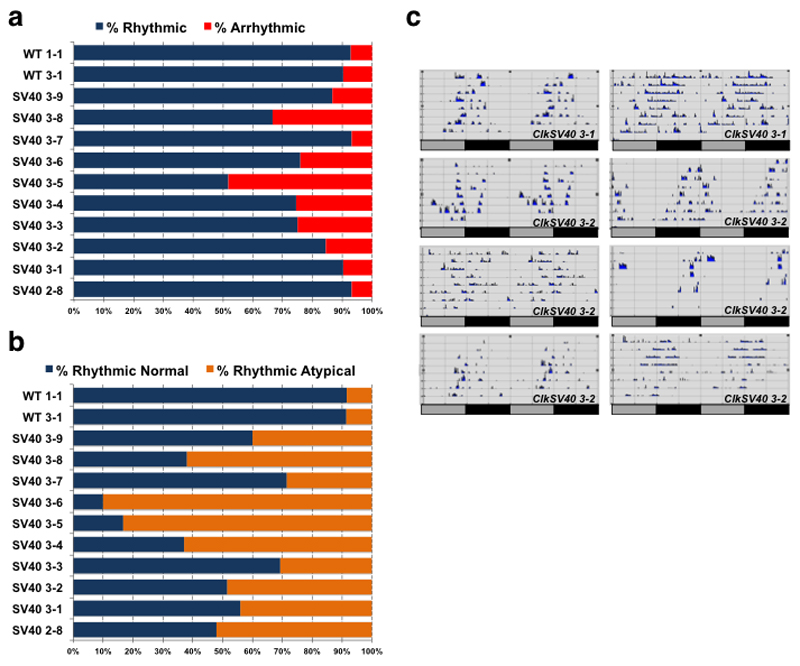
Figure 4. ClkSV40 flies display variable circadian behavior.
a. Bar graph summarizing the behavioral analyses of ClkWT and ClkSV40 flies. Each row and set of numbers (e.g. 1-1) represents a strain with an independent insertion of the wild type (ClkWT) or SV40 (ClkSV40) transgene. In all cases flies carry one extra copy of the Clk gene (either ClkWT or ClkSV40) in a wild-type background. Flies were classified as rhythmic or arrhythmic in the first 5 days in constant darkness using the Behavioral toolbox 44. b. Classification of rhythmic flies as normal and atypical. The classification was done manually after 10 days in constant darkness. c. Examples of individual ClkSV40 flies displaying atypical circadian behavior. See also Supplementary Figure 3 and 4.
The variable behavior observed in ClkSV40 flies could be due to higher CLK levels, higher CLK-driven transcription in circadian cells or ectopic circadian gene expression. Importantly, we found no correlation between the number of ectopic VRI-expressing cells and the behavioral defects (Supplementary Figure 3b). We then carefully quantified CLKV5 levels in flies carrying ClkWT and ClkSV40 transgenes using immunostaining. Surprisingly, and despite the overall difference in total protein levels and the larger number of CLKV5-positive cells (Supplementary Figure 4a), there were no significant differences in the CLKV5 protein level per cell between ClkWT and ClkSV40 flies (Supplementary Figure 4b). In contrast, we observed a strong effect on the levels of VRI, which was expressed in more cells and was also significantly upregulated on a per cell basis in ClkSV40 compared to ClkWT flies (Supplementary Figure 4c, d). VRI and CLK form a well-characterized transcriptional feedback loop: CLK is a strong transcriptional activator of vri, and VRI is the only known repressor of Clk transcription. Therefore, we surmised that the similar CLK protein levels per cell arise from very different levels of Clk transcription (high in ClkWT and low in ClkSV40) due to feedback transcriptional regulation by VRI. We assume that in steady state, in ClkSV40 flies CLK-activity per cell is slightly higher than that in ClkWT flies, which could explain both the increased levels of VRI as well as the slightly shorter period observed in these flies.
A mathematical model of the CLK-VRI feedback loop
It has been previously suggested that post-transcriptional control (e.g., by miRNAs) could be used to diminish variability in biological systems46–51. In this context, we speculate that the high-transcriptional/high-turnover Clk profile stabilizes the circadian system by minimizing the consequences of transcriptional noise and/or by allowing rapid transcriptional feedback control. To evaluate this hypothesis, we formulated a mathematical model for uncovering factors that could modulate noise in the levels of Clk (see Supplementary Methods). Interestingly, our model predicts that for a given target level of CLK protein, increasing the rate of Clk transcription while increasing the rate of mRNA degradation is an effective way of decreasing noise in the system (Supplementary Figure 5). As stated above, the levels of CLK per cell are similar in ClkWT and ClkSV40 flies; however, due to the strong post-transcriptional regulation provided by the Clk 3’UTR the same levels of protein are achieved by much lower transcriptional activity of the Clk promoter in ClkSV40 flies. In agreement with the established inverse relationship between transcriptional levels and noise52–55, our model predicted that in ClkSV40 flies CLK-driven activity would be more sensitive to transcriptional noise and hence more variable. Clk is involved in the development of the circadian neurons as well as in the control of accurate circadian transcription. Therefore, our model predicts that ClkSV40 flies might display stochastic development of the circadian neurons, which should be accompanied by random/variable circadian transcription.
Decanalized development of pacemaker cells in ClkSV40 flies
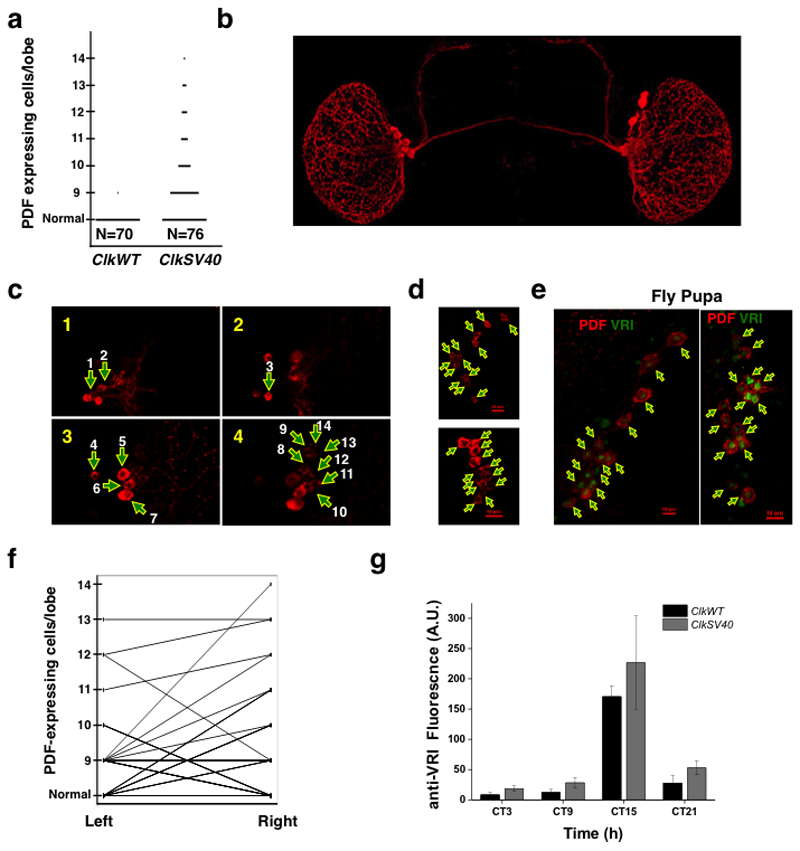
To test these possibilities, we first determined whether ClkSV40 flies show developmental abnormalities in the main pacemaker cells, the pdf-expressing neurons or LNvs. Pigment dispersing factor (PDF) is expressed in eight neurons in each brain lobe18. Most (69 out of 70) examined ClkWT brain hemispheres had eight canonical LNvs per brain lobe (Figure 5a). However, flies from three independent insertions of the ClkSV40 transgene had brains with more than eight pdf-expressing cells (Figures 5a-d). Interestingly, the number of pdf-expressing cells varied between brains in these three ClkSV40 insertions. In particular, one of the lines had between eight and fourteen pdf-expressing cells per brain hemisphere (Figures 5a-d ; see Supplementary Movie 3 and 4 for 3D visualization of pdf-positive cells in brains of ClkWT and ClkSV40 flies). Although these cells resemble sLNvs due to their smaller size and lower intensity (Supplementary Figure 6a,b), we did not detect them in larvae brains, suggesting that they develop later than canonical sLNvs. On the other hand, we found that most brains of ClkSV40 pupae had many additional pdf-expressing cells (Figure 5e and Supplementary Figure 7a,b). Interestingly, we found that most ClkSV40 pupae brains hemispheres have larger number of pdf-expressing cells than adults from the same strain (see Supplementary Figure 7b), suggesting that some of the additional pdf-expressing cells might die or lose pdf expression later in development (or after eclosion).
Figure 5. Stochastic development of the circadian system in ClkSV40 flies.
a. Number of PDF-positive cell bodies in ClkWT and ClkSV40 brains. Data is from the ClkSV40 (2-8) and ClkWT (1-1) fly strains. b. Representative example of a ClkSV40 fly brain in which the position of extra pdf-expressing cells can be visualized. c. Representative example of a ClkSV40 fly brain in which 14 pdf-expressing cells were observed in one of the brain hemispheres. Z stack of 3 images (each taken every 1 µm). Numbers in the upper left corner of each picture indicate the relative position of the Z stack. Arrows indicate the position of the pdf-expressing cells. d. Z stack (maximal projection) of 2 brain hemispheres of ClkSV40 flies immunostained for PDF showing the number of pdf-expressing cells (13 and 12 for the top and bottom brain hemispheres respectively). Arrows indicate the position of the pdf-expressing cells. Red scale bar indicates 10μm. e. Z stack (maximal projection) of 2 brains of ClkSV40 pupae immunostained for PDF (in red) and VRI (in green) showing the number of pdf-expressing cells (14 and 15 for the left and right images respectively). Arrows indicate the position of the pdf-expressing cells. Red scale bar indicates 10μm. f. Comparison of left and right hemispheres in ClkWT and ClkSV40 flies based on PDF immunofluorescent staining. g. Brain to brain variation in the levels of VRI (measured by IF) protein in the sLNvs at 4 time points (day 10 in DD), in ClkSV40 (line 2-8, red) and ClkWT (line 1-1, black) flies. See also Supplementary Figure 5, 6, 7 and 8 Supplementary Movies 3 and 4.
Interestingly, we observed that the two lobes of individual flies carrying ClkSV40 transgenes flies did not necessarily have the same number of pdf-expressing neurons, demonstrating that the number of these neurons is variable in the absence of Clk post-transcriptional regulation (Figures 5b and f). We observed the same phenomenon in pupae of ClkSV40 flies (Supplementary Figure 7c) demonstrating that this asymmetry is established during development.
To determine whether Clk 3’UTR also diminishes variability in daily CLK-driven transcription within adult flies; we determined VRI levels throughout the day in flies carrying ClkWT or ClkSV40 transgenes. We focused on one ClkWT (1-1) and one ClkSV40 (2-8) insertions of the transgenes. In order to minimize the effects of an endogenous WT Clk gene, we performed the experiment in the background of the Clk hypomorphic mutant ClkAR 29. In these transgenic lines, the ClkWT transgene rescued the arrhytmicity of the ClkAR mutation much more effectively (>75%) than the ClkSV40 transgene (<40%). Therefore we stained ClkWT; ClkAR and ClkSV40; CLKAR flies for VRI and PDF at four different circadian time points after eight days in constant darkness (DD8). We focused on the small lateral-ventral neurons (sLNvs), which display strong protein oscillations in constant darkness37. Whereas ClkWT; ClkAR flies displayed robust VRI protein oscillations and little brain-to-brain variability, ClkSV40; ClkAR flies expressed higher and more variable VRI levels with a significant number of flies showing inappropriate VRI staining at the early circadian time points (CT3 and CT7; Figure 5g and Supplementary Figure 8).
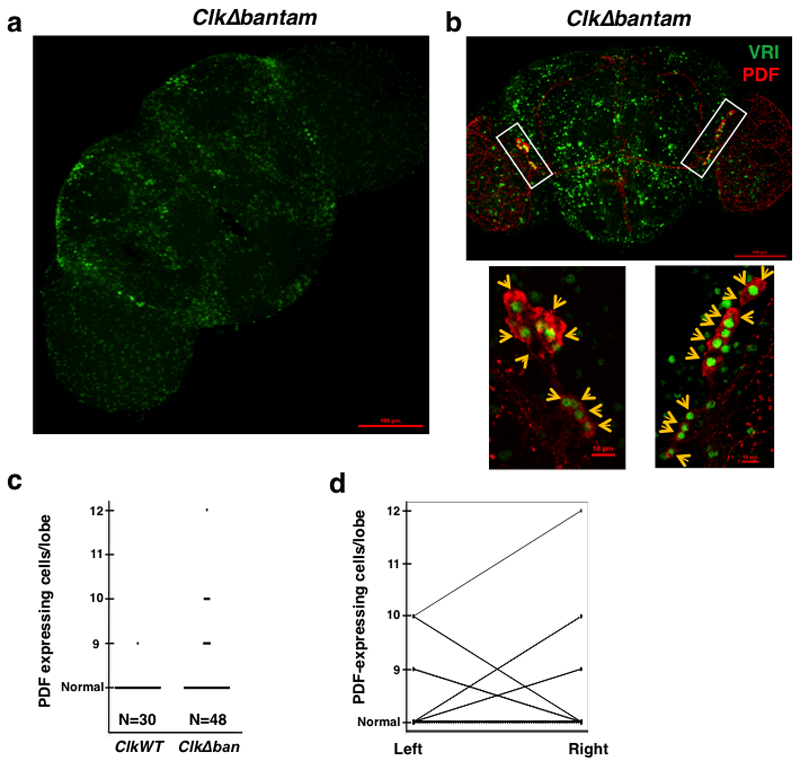
A Role of bantam in the development of the circadian neurons
We recently showed that the miRNA bantam regulates the Clk 3’ UTR, at least in Drosophila S2 cells32. Hence, we decided to define whether this regulation is key to the deterministic development and maintenance of the circadian neuronal network. For doing so, we examined by immunocytochemistry the number and intensity of circadian neurons (VRI and PDF positive cells) in flies carrying a Clk transgene with deletion of the putative bantam binding sites [ClkΔban32]. Indeed, and similarly to ClkSV40 flies, ClkΔban displayed ectopic expression of VRI across the fly brain (Figure 6a). In addition, ClkΔban showed variable and increased number of pdf-expressing cells (Figure 6b,c). Importantly, and as in ClkSV40 flies, we did not observe a correlation between the numbers of pdf-expressing cells in each hemisphere of individual fly brains (Figure 6d). These results demonstrate the importance of the bantam binding sites in the Clk 3’ UTR for normal development of the pdf-expressing cells. Interestingly, we found that some of these brains also have heterogeneous VRI expression in the PDF positive cells. For example we found brains in which VRI expression differs or is even absent in some of the PDF positive cells (i.e. see Figure 6b). We observed a similar phenomenon in ClkSV40 pupae (see Figure 5e and Supplementary Figure 6a). This strongly suggests that CLK activity is variable across circadian neurons within the same brain.
Figure 6. Deletion of the bantam binding sites on the Clk 3’UTR lead to stochastic development of the pdf-expressing cells.
a. Immunofluorescence (IF) analysis of a representative ClkΔban (3-7) Drosophila brain using an anti-VRI antibody. Flies were collected and dissected at ZT15. b. Representative example of a ClkΔban fly brain in which 21 LNvs cells were observed, 9 in one hemisphere and 12 in the other (top panel). Lower panel represents magnification of rectangle area. Of the 12 PDF positive cell bodies in the right hemisphere: 9 cells are VRI positive, one cell display low intensity of VRI immunostaining and 2 cells are VRI negative. In the left hemisphere of the brain, 9 PDF positive cells are observed: 8 cells are VRI positive and one cell displays low intensity of VRI immunostaining. Flies were collected and dissected at ZT15. Arrows indicate the position of each PDF positive cell. c. Number of PDF-positive cell bodies in ClkWT and ClkΔban brains. Data is taken from the ClkΔban (3-7) and ClkWT (1-1) fly strains. d. Comparison of left and right hemispheres in ClkWT and ClkΔban flies.
Discussion
Here we found that the circadian master regulator Clk is under very strong post-transcriptional control. One consequence of this tight control is to provide a threshold-like regulation that inhibits ectopic expression of Clk and CLK-transcriptional targets. Indeed, flies devoided of this regulation have more circadian cells in the brain and aberrant and variable circadian rhythms. Interestingly, in these flies, the levels of CLK on a per cell basis are not altered compared to flies with wild-type CLK, suggesting that the main role of Clk post-transcriptional regulation is to limit the variability of CLK activity rather than CLK protein levels. Moreover, we observed an increase in the number of pdf-expressing cells in ClkSV40 flies and pupae. To understand these results we formulated a mathematical model of the circadian CLK-VRI feedback loop. Our model predicts that Clk post-transcriptional control diminishes the noise in this transcriptional feedback loop. Indeed, we found that in ClkSV40 flies there was not only more pdf-expressing cells, but also that their numbers were variable among individuals or between the two lobes of the same brain, demonstrating that in absence of Clk post-transcriptional control LNvs development and maintenance is stochastic. Last, flies carrying Clk transgenes with mutation in the putative binding sites for the miRNA bantam also displayed stochastic number of pdf-expressing cells between and within individuals suggesting that the phenotypes observed in ClkSV40 flies are mediated through this miRNA.
Clk is strongly regulated at the post-transcriptional level. Indicative of regulation by miRNAs, Clk mRNA is strongly bound to AGO1 and does not bind efficiently to oligo-dT beads. In addition, we observed significant differences between the CLK protein to Clk mRNA ratios in ClkWT and ClkSV40 flies (compare Figure 3h to Figure 3e), which suggests that an important part of this regulation is at the translational level. Interestingly, mRNA turnover accounts at least partially for the post-translational control, as Clk mRNA is short lived when compared with other circadian mRNAs like cry (Figure 1f). Therefore, Clk post-transcriptional regulation appears to be complex and redundant. Our results suggest that the Clk 3’ UTR is responsible for the decrease in CLK levels in most (if not all) cell types, likely by miRNAs. Bantam seems to have a predominant role but it might not be unique. The presence of this type of regulation is not surprising given the large amount of reports demonstrating the importance of RNA metabolism for circadian timekeeping28,41,56–60.
The general assumption in the field is that Clk transcriptional oscillations are not essential for behavioral rhythms. This is based on the fact that the circadian behavior of ClkAR mutants can be rescued using the cry-GAL4 driver and a UAS-Clk transgene29 and that there are negligible effects on expression of CLK under the per or tim promoters (ARK flies21). However, the same UAS-Clk transgene expressed under control of other gal4 drivers like tim-gal4 leads to developmental lethality29 and the behavioral deficits of ClkAR mutants are not rescued by Clk ARK transgenes (our own unpublished observation). Moreover, addition of extra Clk gene copies leads to a copy number dependent shortening of the period28, demonstrating that CLK levels cannot be regulated solely at the post-translational level. Interestingly, the ARK transgenes carry a per 3’ UTR, which is strongly regulated post-transcriptionally (our unpublished data). This could explain why flies carrying this transgene are rhythmic in a wild-type background.
Our S2 cell and fly brain experiments demonstrate a key role of Clk 3’ UTR in avoiding ectopic CLK-driven transcriptional activity. Moreover, these results suggest that the Clk 3’ UTR is necessary for establishment of a threshold for meaningful Clk mRNA expression. This type of threshold might be important not only in circadian cell determination but also to prevent stochastic or random pulses of CLK-driven activity that could alter or shift the circadian clock. miRNA-mediated thresholding of master developmental regulators has been previously demonstrated for the miR-9a/senseless 61 and for the miR-263a/hid pairs62. We believe that our failure to detect signal from the Clk 3’ UTR reporters is not related to a sensitivity issue. This is based on our observation of the abnormally high number of circadian cells in the brains of ClkSV40 flies, which demonstrates that the threshold has a biological meaning.
Our work demonstrates a central role for Clk in the development of the pdf-expressing LNvs cells, as three of the fly lines with ClkSV40 insertions as well as ClkΔban flies display larger number of LNvs than observed in wild-type brains. Although this work constitutes the first report of flies with more than 16 LNvs per brain, a previous report suggests that Clk affects the development of these cells: Clk and cyc mutants have reduced numbers of LNvs, and Clk can activate (albeit weakly) a pdf-luciferase reporter31. From our results it is not clear whether these cells are part of the neuronal circadian network or whether their development and maintenance is independent. Indeed, the transgenic line with the largest number of extra LNvs (SV40 2-8) was fairly rhythmic at the behavioral level, suggesting that these extra cells do not dramatically influence the timekeeping process, at least early in constant darkness. The abnormal behavior cannot be explained neither by the presence of additional VRI-expressing cells, as we see no correlation between the number of them and any of the behavioral anomalies (Supplementary Figure 3b). We favor the possibility that the behavioral defects are due to stochastic changes in CLK-driven transcription. Stochastic variation in CLK-driven transcription could also explain the brain-to-brain variation in VRI levels in ClkSV40 flies and the observed variations in VRI levels between pdf-expressing cells within the same brain in the ClkSV40 pupae and ClkΔban fly brains (i.e. see Figure 5e and Figure 6b bottom respectively). Moreover, we observed that ClkSV40 pupae have generally larger numbers of extra LNvs, suggesting that many of these cells might lose pdf expression during adulthood (as we did not observed high levels of lethality in ClkSV40 flies). We believe that in the future and given the advances in CRISPR technology, it would be interesting to repeat some of the experiments in flies in which the deletion the 3’-UTR is in the endogenous Clk locus.
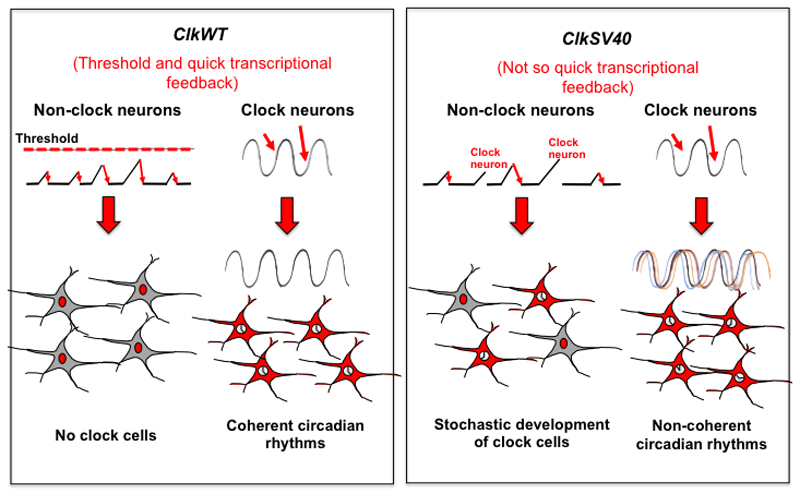
Our results suggest that the main role of Clk post-transcriptional control is to limit the variability in CLK activity rather than to control the total levels of CLK. This is based on the fact that the levels of CLK per cell were not altered in ClkSV40 flies despite the large increase in the total number of CLK-positive cells. We argue that the normal CLK levels in individual cells are due to an increase in VRI levels that reduce Clk transcription and hence total CLK levels (see model in Figure 7). Indeed, heads of ClkSV40 flies have lower levels of ClkV5 mRNA than do ClkWT flies (Figure 3h) despite much higher protein levels (Figure 3e). Therefore, our data suggests that the main difference between ClkWT and ClkSV40 flies is that the latter produce CLK by a low transcription/low degradation rather than the high transcription/high degradation profile observed in wild-type flies (Figure 7). We speculate that the increase in VRI levels keeps the overall levels of CLK per cell constant. On the other hand, the low transcription/low turnover Clk mRNA profiles observed in ClkSV40 flies lead to more random, less fixed levels of Clk mRNA and protein (Figure 7). We can then imagine a model in which under wild-type conditions (either no Clk transgene or one copy of the ClkWT transgene) stochastic pulses of Clk transcription in non-circadian neurons are filtered by the transcriptional threshold and do not lead to the development of circadian cells (Figure 7). This is essential during development as well as in adult animals, as ectopic clocks in the fly brain are dependent on continuous expression of Clk63. In theory, Clk post-transcriptional control could help stabilize CLK-driven transcription in the circadian cells both by thresholding and by diminishing cell-to cell variability in CLK-driven oscillations (Figure 7).
Figure 7. Role of Clk post-transcriptional regulation in clock and non-clock neurons.
Small red arrows represent random or environmentally driven fluctuations in Clk gene expression.
In sum, here we found that post-transcriptional control of a master transcriptional regulator provides an efficient mechanism that assures canalized development and that stabilizes a transcriptional network during development and in adult animals. The role of post-transcriptional control in stabilizing the activities and levels of key activators of systems based on feedback loops have been previously proposed46,47,51, but to the best of our knowledge, our work constitutes the first theoretical and experimental example of this control principle.
Methods
Fly strains
The CantonS (CS) strain used as wild type is CSIso3H (Bloomington Stock Center, Indiana). ClkAR mutants were described29. ClkSV40 flies were constructed by injecting ClkSV40 transgene into yw embryos (service provided by Best Gene, Inc.). ClkΔban flies were described 32.
Analysis of gene expression by real-time PCR
Total RNA was prepared from adult fly heads using TRI Reagent (SIGMA) according to the manufacturer’s protocol. RNA was DNase treated (DNaseI, NEB), and cDNA derived from this RNA (using iScript, Bio-Rad) was utilized as a template for quantitative real-time PCR performed with the C1000 Thermal Cycler Bio-Rad. The utilized primers are summarized in Supplementary Methods. mRNA values from heads were normalized to levels of mRNAs encoding ribosomal proteins rp49 and rpS18.
Assessment of post-transcriptional regulation
We utilized a mixture of RNA extracted at six different time points from fly heads. The three biological replicas were generated by three independent RNA extractions from each of the six time points. After extraction, the RNA was DNase treated, ethanol precipitated, and retrotranscribed using iScript (Bio-Rad) with random primers. We performed real-time PCR using the diluted cDNA template and an equimolar mixture of the DNA products. This DNA mixture was utilized in experiments to determine the absolute levels of the pre-mRNA and mature mRNA. We generated this mixture by pooling equimolar concentrations of the purified PCR products. We designed two sets of primers for each gene, one intronic and one exonic. For intronic PCRs, real-time PCR was performed using the cDNA as a template, and amounts were plotted on a standard curve constructed using different concentrations of the genomic DNA as templates to normalize the different efficiencies of the primers. For each gene a pre-mRNA/mRNA ratio was calculated. The primer sequences are detailed in Supplementary Information. Three independent biological replicates were performed.
Isolation of total, oligo-dT bound, and unbound RNAs
Oligo-dT bound fractions were isolated from total RNA using Invitrogen (dT)25 Dynabeads following the manufacturer’s protocol. The RNA not bound was precipitated with ethanol and dissolved in RNase-free water. For quantification, the unbound fraction was normalized to histone H1 and the bound fraction to rp49. Data was obtained from a pooled mixture of six time points, and three biological replicates were performed.
RNA measurements from cultured fly wings
Thirty adult male Canton-S fly wings were dissected and cultured as described 64 under 12 h light/12 h dark (LD) conditions for three days. In mRNA half-life measurements actinomycin D was added in ZT3 (10 µg/ml), wings were transferred into TRI Reagent (SIGMA) at relevant time points and mRNA was extracted. For each time point, the experiments were performed in triplicate.
Plasmids
To generate the tim-YFP reporter, we amplified by PCR the YFP coding sequence and the tim promoter region (760 bp from the timwt760-luciferase construct65). The fragments were ligated sequentially into pBluescript. pAc-Cherry and pAc-CFP were constructed by amplifying the Cherry or CFP coding sequences by PCR and ligating these products into pAcB V5/His6 (Invitrogen). The ClkSV40 plasmid was generated through steps similar to those used to construct the ClkWT (described in 28) with the following modification: a fragment containing the SV40 3’ UTR sequence was amplified by PCR from pAc V5/His6 (Invitrogen) and cloned by overlap PCR, exactly downstream the stop codon, into a plasmid containing a Sph-NotI fragment derived from a plasmid containing the last portion of the Clk transgene (from position 7741779 to 7736982, pdClk4). The Sph-NotI fragment containing the SV40 3’ UTR was re-cloned into the pdClk4, which was then used to generate the whole ClkV5 transgene as described previously28. The resulting plasmid was fully sequenced.
S2 cells maintenance and transfection
S2 cells were maintained in 10% fetal bovine serum (Invitrogen) insect tissue culture medium (HyClone). Cells were seeded in a six-well plate. Transfection was performed using 6 µl of MIRUS transfection reagent TransIT 2020 and 2 µg of total DNA as per the manufacturer’s instruction. For the 3’ reporter experiments, 250 ng each of pMT-Cherry and pMT-YFP reporters (one carrying the SV40 3’ UTR and one carrying the Clk 3’ UTR) were co-transfected into Drosophila S2 cells along a pAc-CFP transcriptional control. In order to avoid bias due to the different fluorophores, we performed the analysis comparing the levels of the same fluorescent protein under the different 3’ UTRs (SV40 vs. Clk 3’ UTR). Shown are data obtained with the Cherry reporter, but we obtained similar results when YFP reporters were compared. For the experiments to follow CLK-driven activity, we utilized 150 ng of pAcCherry, 150 ng of the tim-YFP reporter, and 500 ng of the ClkWT or ClkSV40 plasmids. pBS-KS (Stratagene) was used to bring the total amount of DNA to 2 µg.
High-throughput reporter expression analysis in S2 cells
One day after transfection, cells were transferred to optic-suitable 96-well plates and visualized over 24 hours using Scan^R high-throughput fluorescent microscope (Olympus). The intensity from the transfection control reporter (CFP for 3’ UTR and Cherry for the CLK-activity experiments), cell size, and roundness were evaluated. Cherry or YFP levels were assessed only for cell populations that met intensity and cell morphology criteria.
AGO-1 immunoprecipitation
AGO-1 immunoprecipitations were performed as previously described32. The transcripts were quantified by RT-PCR using primers that exclusively identify the endogenous (endo) or the V5-tagged Clk mRNA (V5). Data is shown as ratio of immunoprecipitate to input.
Immunofluorescence
Male flies were entrained for at least 3 days in LD and then transferred to DD for the indicated time. 3rd stage Larvae and last stage pupae were collected after 3 days entrainment in LD. Whole flies/larvae/pupae were collected into fixative solution, PBS 4% paraformaldehyde 0.1% Triton X-100, and incubated 30 minutes at 4 °C followed by 2 hours rotation at room temperature. After fixation, flies/larvae/pupae were transfer to PBS, and the brains were dissected. The brains were washed three times (PBS 0.1% Triton X-100) and transferred to blocking solution (PBS, 0.1% Triton X-100, 2% horse serum). After three washes, primary antibody solution was added and samples were incubated at 4 °C overnight. The primary antibody solution contained PBS, 0.1% Triton X-100, 2% horse serum, and 1:3000 guinea pig anti-VRI (gift from Paul Hardin), 1:1000 mouse anti-PDF (gift from the Justin Blau lab), 1:1000 mouse anti-V5 (Sigma), 1:1000 rat anti-TIM (a gift from Michael Rosbash), or 1:1000 guinea pig anti-CLK (a gift from Paul Hardin). Brains were washed three times and then exposed to secondary antibody solution: PBS, 0.1% Triton X-100, 2% horse serum and 1:500 Alexa Fluor 488 goat anti-guinea pig (Invitrogen), 1:500 Dylight 550 donkey anti-mouse (Abcam), or 1:500 Alexa Fluor donkey anti-rat (Jackson) for 1 hour in room temperature. Brains were washed three times and mounted in VECTASHIELD mounting medium (VECTOR) on microscope slides. Images were captured using a Nikon A1R confocal microscope and analyzed with NIS-Elements (Nikon). Quantification of brain cell expression of V5 or VRI proteins was performed as follows: Stained brains were placed between spacers to avoid pressure on the tissue. Confocal images were taken every 3 µm (on the Z axis) over the entire brain depth.
Western blotting
Fly heads (20 heads per sample) were collected on dry ice. Heads were homogenized in RIPA lysis buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, and 1 mM DTT, supplemented by protease inhibitor cocktail and phosphatase inhibitors) using a motorized pestle. Head lysates were then centrifuged at top speed for 10 minutes. The supernatant was boiled with protein sample buffer (Bio-Rad). Samples were resolved on Criterion XT Bis-Tris gels (Bio-Rad). Antibodies used for western blotting were those described above and anti-tubulin (DM1A, SIGMA).The complete blots of Figure 3e are provided as Supplementary Figure 9.
Locomotor activity
Male flies were monitored using Trikinetics Drosophila Activity Monitors. Analyses were performed with a signal processing toolbox44. Flies were considered rhythmic if the rhythm index was greater than 0.15 for the first 5 days in DD. Rhythmic flies after the 5 day period were followed for 5 more days and, if alive after this period, were classified as normal or atypical rhythmic by visual inspection after examination of the behavioral pattern and the MESA analysis.
Supplementary Material
Acknowledgements
We thank Paul Hardin and Justin Blau for reagents and Justin Blau, Rachel Green, Eran Hornstein, Laura Lande-Diner, and Daniel Kaganovich for reading the manuscript. SK is funded by the European Research Council Starting Grant (ERC #260911). OB is funded by ICRF. NF is funded by the European Research Council (ERC Advanced Investigator Award).
Footnotes
Author contributions: IL, OB, VW and UW performed the experiments. SA analyzed the single cell microscopy data. CG generated the ClkSV40 flies and performed the initial characterization. DH and NF formulated the mathematical model; MR supported JM, discussed the experiments and help writing the manuscript. SK designed the experiments and wrote the manuscript.
Competing Financial Interest Statement: The authors declare that there is no conflict of interest.
References
- 1.Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol. 2010;72:605–24. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall JC. Genetics and molecular biology of rhythms in Drosophila and other insects. Adv Genet. 2003;48:1–280. doi: 10.1016/s0065-2660(03)48000-0. [DOI] [PubMed] [Google Scholar]
- 3.Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 4.Rutila JE, et al. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–14. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 5.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–40. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 6.Myers MP, Wager-Smith K, Wesley CS, Young MW, Sehgal A. Positional cloning and sequence analysis of the Drosophila clock gene, timeless. Science. 1995;270:805–8. doi: 10.1126/science.270.5237.805. [DOI] [PubMed] [Google Scholar]
- 7.Kadener S, Stoleru D, McDonald M, Nawathean P, Rosbash M. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev. 2007;21:1675–86. doi: 10.1101/gad.1552607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim C, et al. Clockwork orange encodes a transcriptional repressor important for circadian-clock amplitude in Drosophila. Curr Biol. 2007;17:1082–9. doi: 10.1016/j.cub.2007.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsumoto A, et al. A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev. 2007;21:1687–700. doi: 10.1101/gad.1552207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bae K, Lee C, Hardin PE, Edery I. dCLOCK is present in limiting amounts and likely mediates daily interactions between the dCLOCK-CYC transcription factor and the PER-TIM complex. J Neurosci. 2000;20:1746–53. doi: 10.1523/JNEUROSCI.20-05-01746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cyran SA, et al. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–41. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- 12.Glossop NR, et al. VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron. 2003;37:249–61. doi: 10.1016/s0896-6273(03)00002-3. [DOI] [PubMed] [Google Scholar]
- 13.Benito J, et al. The circadian output gene takeout is regulated by Pdp1epsilon. Proc Natl Acad Sci U S A. 2010;107:2544–9. doi: 10.1073/pnas.0906422107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reischl S, Kramer A. Kinases and phosphatases in the mammalian circadian clock. FEBS Lett. 2011;585:1393–9. doi: 10.1016/j.febslet.2011.02.038. [DOI] [PubMed] [Google Scholar]
- 15.Lim C, Allada R. Emerging roles for post-transcriptional regulation in circadian clocks. Nat Neurosci. 2013;16:1544–50. doi: 10.1038/nn.3543. [DOI] [PubMed] [Google Scholar]
- 16.Bae K, Edery I. Regulating a circadian clock’s period, phase and amplitude by phosphorylation: insights from Drosophila. J Biochem. 2006;140:609–17. doi: 10.1093/jb/mvj198. [DOI] [PubMed] [Google Scholar]
- 17.Plautz JD, Kaneko M, Hall JC, Kay SA. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278:1632–5. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- 18.Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- 19.Sheeba V. The Drosophila melanogaster circadian pacemaker circuit. J Genet. 2008;87:485–93. doi: 10.1007/s12041-008-0071-x. [DOI] [PubMed] [Google Scholar]
- 20.Shafer OT, Helfrich-Forster C, Renn SC, Taghert PH. Reevaluation of Drosophila melanogaster’s neuronal circadian pacemakers reveals new neuronal classes. J Comp Neurol. 2006;498:180–93. doi: 10.1002/cne.21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim EY, et al. Drosophila CLOCK protein is under posttranscriptional control and influences light-induced activity. Neuron. 2002;34:69–81. doi: 10.1016/s0896-6273(02)00639-6. [DOI] [PubMed] [Google Scholar]
- 22.Luo W, et al. CLOCK deubiquitylation by USP8 inhibits CLK/CYC transcription in Drosophila. Genes Dev. 2012;26:2536–49. doi: 10.1101/gad.200584.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 2006;20:723–33. doi: 10.1101/gad.1404406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szabo A, et al. The CK2 kinase stabilizes CLOCK and represses its activity in the Drosophila circadian oscillator. PLoS Biol. 2013;11:e1001645. doi: 10.1371/journal.pbio.1001645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirayama J, et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–90. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 26.Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M. Dynamic PER repression mechanisms in the Drosophila circadian clock: from on-DNA to off-DNA. Genes Dev. 2010;24:358–67. doi: 10.1101/gad.1883910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fathallah-Shaykh HM, Bona JL, Kadener S. Mathematical model of the Drosophila circadian clock: loop regulation and transcriptional integration. Biophys J. 2009;97:2399–408. doi: 10.1016/j.bpj.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kadener S, Menet JS, Schoer R, Rosbash M. Circadian transcription contributes to core period determination in Drosophila. PLoS Biol. 2008;6:e119. doi: 10.1371/journal.pbio.0060119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allada R, Kadener S, Nandakumar N, Rosbash M. A recessive mutant of Drosophila Clock reveals a role in circadian rhythm amplitude. EMBO J. 2003;22:3367–75. doi: 10.1093/emboj/cdg318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J, et al. Drosophila clock can generate ectopic circadian clocks. Cell. 2003;113:755–66. doi: 10.1016/s0092-8674(03)00400-8. [DOI] [PubMed] [Google Scholar]
- 31.Park JH, et al. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci U S A. 2000;97:3608–13. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kadener S, et al. A role for microRNAs in the Drosophila circadian clock. Genes Dev. 2009;23:2179–91. doi: 10.1101/gad.1819509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Relogio A, et al. Tuning the mammalian circadian clock: robust synergy of two loops. PLoS Comput Biol. 2011;7:e1002309. doi: 10.1371/journal.pcbi.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erzberger A, Hampp G, Granada AE, Albrecht U, Herzel H. Genetic redundancy strengthens the circadian clock leading to a narrow entrainment range. J R Soc Interface. 2013;10:20130221. doi: 10.1098/rsif.2013.0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ukai H, Ueda HR. Systems biology of mammalian circadian clocks. Annu Rev Physiol. 2010;72:579–603. doi: 10.1146/annurev-physiol-073109-130051. [DOI] [PubMed] [Google Scholar]
- 36.Hogenesch JB, Ueda HR. Understanding systems-level properties: timely stories from the study of clocks. Nat Rev Genet. 2011;12:407–16. doi: 10.1038/nrg2972. [DOI] [PubMed] [Google Scholar]
- 37.Peng Y, Stoleru D, Levine JD, Hall JC, Rosbash M. Drosophila free-running rhythms require intercellular communication. PLoS Biol. 2003;1:E13. doi: 10.1371/journal.pbio.0000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hogenesch JB, Herzog ED. Intracellular and intercellular processes determine robustness of the circadian clock. FEBS Lett. 2011;585:1427–34. doi: 10.1016/j.febslet.2011.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiss R, Bartok O, Mezan S, Malka Y, Kadener S. Synergistic interactions between the molecular and neuronal circadian networks drive robust behavioral circadian rhythms in Drosophila melanogaster. PLoS Genet. 2014;10:e1004252. doi: 10.1371/journal.pgen.1004252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–26. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez J, et al. Nascent-Seq analysis of Drosophila cycling gene expression. Proc Natl Acad Sci U S A. 2013;110:E275–84. doi: 10.1073/pnas.1219969110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hughes ME, Grant GR, Paquin C, Qian J, Nitabach MN. Deep sequencing the circadian and diurnal transcriptome of Drosophila brain. Genome Res. 2012;22:1266–81. doi: 10.1101/gr.128876.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fabian MR, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:868–80. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levine JD, Funes P, Dowse HB, Hall JC. Resetting the circadian clock by social experience in Drosophila melanogaster. Science. 2002;298:2010–2. doi: 10.1126/science.1076008. [DOI] [PubMed] [Google Scholar]
- 45.Bae K, Lee C, Sidote D, Chuang KY, Edery I. Circadian regulation of a Drosophila homolog of the mammalian Clock gene: PER and TIM function as positive regulators. Mol Cell Biol. 1998;18:6142–51. doi: 10.1128/mcb.18.10.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hornstein E, Shomron N. Canalization of development by microRNAs. Nat Genet. 2006;38(Suppl):S20–4. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- 47.Siciliano V, et al. MiRNAs confer phenotypic robustness to gene networks by suppressing biological noise. Nat Commun. 2013;4:2364. doi: 10.1038/ncomms3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen SM, Brennecke J, Stark A. Denoising feedback loops by thresholding--a new role for microRNAs. Genes Dev. 2006;20:2769–72. doi: 10.1101/gad.1484606. [DOI] [PubMed] [Google Scholar]
- 49.Balazsi G, van Oudenaarden A, Collins JJ. Cellular decision making and biological noise: from microbes to mammals. Cell. 2011;144:910–25. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mukherji S, et al. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011;43:854–9. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fritsche-Guenther R, et al. Strong negative feedback from Erk to Raf confers robustness to MAPK signalling. Mol Syst Biol. 2011;7:489. doi: 10.1038/msb.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hansen AS, O’Shea EK. Promoter decoding of transcription factor dynamics involves a trade-off between noise and control of gene expression. Mol Syst Biol. 2013;9:704. doi: 10.1038/msb.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Munsky B, Neuert G, van Oudenaarden A. Using gene expression noise to understand gene regulation. Science. 2012;336:183–7. doi: 10.1126/science.1216379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suter DM, et al. Mammalian genes are transcribed with widely different bursting kinetics. Science. 2011;332:472–4. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- 55.Hornung G, et al. Noise-mean relationship in mutated promoters. Genome Res. 2012;22:2409–17. doi: 10.1101/gr.139378.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morf J, et al. Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science. 2012;338:379–83. doi: 10.1126/science.1217726. [DOI] [PubMed] [Google Scholar]
- 57.Gatfield D, et al. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009;23:1313–26. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lim C, Allada R. ATAXIN-2 activates PERIOD translation to sustain circadian rhythms in Drosophila. Science. 2013;340:875–9. doi: 10.1126/science.1234785. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, Ling J, Yuan C, Dubruille R, Emery P. A role for Drosophila ATX2 in activation of PER translation and circadian behavior. Science. 2013;340:879–82. doi: 10.1126/science.1234746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo W, Sehgal A. Regulation of circadian behavioral output via a MicroRNA-JAK/STAT circuit. Cell. 2012;148:765–79. doi: 10.1016/j.cell.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hilgers V, Bushati N, Cohen SM. Drosophila microRNAs 263a/b confer robustness during development by protecting nascent sense organs from apoptosis. PLoS Biol. 2010;8:e1000396. doi: 10.1371/journal.pbio.1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kilman VL, Allada R. Genetic analysis of ectopic circadian clock induction in Drosophila. J Biol Rhythms. 2009;24:368–78. doi: 10.1177/0748730409343761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krishnan B, et al. A new role for cryptochrome in a Drosophila circadian oscillator. Nature. 2001;411:313–7. doi: 10.1038/35077094. [DOI] [PubMed] [Google Scholar]
- 65.McDonald MJ, Rosbash M, Emery P. Wild-type circadian rhythmicity is dependent on closely spaced E boxes in the Drosophila timeless promoter. Mol Cell Biol. 2001;21:1207–17. doi: 10.1128/MCB.21.4.1207-1217.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.