Abstract
The model bryophyte Physcomitrella patens is unique among plants in supporting the generation of mutant alleles by facile homologous recombination-mediated gene targeting (GT). Reasoning that targeted transgene integration occurs through the capture of transforming DNA by the homology-dependent pathway for DNA double-strand break (DNA-DSB) repair, we analysed the genome-wide transcriptomic response to bleomycin-induced DNA damage and generated mutants in candidate DNA repair genes. Massively parallel (Illumina) cDNA sequencing identified potential participants in gene targeting. Transcripts encoding DNA repair proteins active in multiple repair pathways were significantly up-regulated. These included Rad51, CtIP, DNA ligase 1, Replication protein A and ATR in homology-dependent repair, Xrcc4, DNA ligase 4, Ku70 and Ku80 in non-homologous end-joining and Rad1, Tebichi/polymerase theta, PARP in microhomology-mediated end-joining. Differentially regulated cell-cycle components included up-regulated Rad9 and Hus1 DNA-damage-related checkpoint proteins and down-regulated D-type cyclins and B-type CDKs, commensurate with the imposition of a checkpoint at G2 of the cell cycle characteristic of homology-dependent DNA-DSB repair. Candidate genes, including ATP-dependent chromatin remodelling helicases associated with repair and recombination, were knocked out and analysed for growth defects, hypersensitivity to DNA damage and reduced GT efficiency. Targeted knockout of PpCtIP, a cell-cycle activated mediator of homology-dependent DSB resection, resulted in bleomycin-hypersensitivity and greatly reduced GT efficiency.
Introduction
The moss Physcomitrella patens is the pre-eminent experimental model for comparative analysis of the evolution of gene function in plants. As a bryophyte, P. patens occupies a basal position in the land plant phylogeny. The bryophytes diverged from the land plant lineage approximately 450–500 million years ago and were the first group of plants to colonise terrestrial habitats [1, 2]. Many of the features present in extant bryophytes represent ancient adaptations necessary for the conquest of dry land, including properties of resilience to a wide range of abiotic stresses. Experimentally, P. patens is highly amenable to genetic analysis and manipulation. The determination of the complete genome sequence of the moss and the development of a well-marked sequence-anchored linkage map provide the opportunity for the forward-genetic identification of genes responsible for key developmental transitions and responses to environmental and hormonal cues [3, 4]. Most significantly, P. patens has emerged as an excellent model for the reverse-genetic analysis of gene function due to its remarkable ability to integrate transgenes at predefined loci through homologous recombination-mediated “gene targeting” (GT) [5, 6, 7].
Gene targeting in P. patens enables precise allele replacement at high frequency. Only relatively short (500-1000bp) lengths of homology are required for efficient GT, so that a range of gene modifications are possible [6]. These include gene disruption and deletion (gene knockout), precise insertion of reporter genes or epitope and affinity tags to native loci (gene knock-in), and sequence alteration by as little as a single base (directed point mutation). Such efficient GT is not possible in other model plant species. Alternative approaches such as stringent counter-selection to recover low frequency targeting events [8] or the deployment of complex protein engineering procedures to design site specific endonucleases capable of introducing DNA breaks at selected sites for transgene insertion have been described [9], but currently remain of limited use for plant genetic manipulation. Targeted mutagenesis through inaccurate repair of CRISPR/Cas9-induced DNA breaks can be used to generate mutant alleles [10], but its potential to enable high-frequency precision gene editing is uncertain and likely to be limited by the low frequency with which homology-dependent repair occurs in angiosperms.
The insertion of transgenes in the genomes of eukaryotic organisms is believed to occur through the capture of transforming DNA by the endogenous mechanisms of DNA double-strand break (DNA-DSB) repair [11]. DNA-DSBs occur with high frequency as a result of exposure to environmental insults such as ionising radiation or chemical mutagens, and (most commonly) through the frequent collapse of replication forks during DNA synthesis, when the replication machinery encounters a single-strand break. It is therefore essential that organisms deploy a range of efficient procedures to repair DNA-DSBs if they are not to suffer catastrophic consequences of genetic loss. There are three principal paths by which DNA-DSBs are repaired. These are the non-homologous end-joining (NHEJ) pathway, the microhomology-mediated end-joining pathway (MMEJ) and the homology-dependent pathway (homologous recombination: HR). NHEJ is typically activated during the G1 phase of the cell-cycle, when the ATM protein kinase initiates a phosphorylation-based signalling cascade culminating in a cell-cycle checkpoint [12]. The broken ends are successively bound by the proteins Ku70/Ku80, and religated through the action of DNA ligase 4. This mechanism appears to be highly conserved throughout the Eukaryota. HR is activated during S and G2, and involves resection of one strand of the broken DNA to leave a long 3’-single-stranded overhang. The ATR protein kinase induces a G2-specific cell-cycle checkpoint [12], and the single-strand end is successively modified by protein interactions, finally becoming coated with the Rad51 recombinase–the eukaryotic homologue of the E. coli RecA protein–to form an invasive nucleoprotein strand that can invade a complementary sequence (usually the adjacent, undamaged, replicated strand) that acts as a template for the accurate resynthesis of the damaged DNA [13]. MMEJ also occurs principally during S-phase. This is a rapid but highly inaccurate mechanism, the broken ends being processed by only a short resection, and the end-joining characterised by insertions or deletions [14].
When a transgene is delivered to a cell, multiple copies of exogenous DNA are taken up, and free ends of these molecules are most likely recognised by endogenous DNA repair enzymes that incorporate the transgenic sequence into the genome. The fate of the transforming DNA will depend on the dominant DNA repair pathway operating in the target cell. Thus in yeasts (S. cerevisiae and S. pombe) the HR-DNA repair pathways are highly active, and transgenes will be integrated at targeted sites, if they contain short lengths of homologous sequence [15]. By contrast, transgene integration into the genomes of flowering plants occurs essentially at random, even when transgenes contain long (several kilobases) of genome homology. This most likely reflects a dominant NHEJ pathway capturing the incoming DNA and inserting it adventitiously at available DSBs [11].
Understanding how HR-mediated transgene insertion occurs in plant cells would be of value in enabling the development of knowledge-based strategies to increase the frequency of GT for genetic manipulation of flowering plants–particularly in the development of precision-engineered crop species. To this end, it is important to study the process of HR-mediated DNA repair and transgene capture in a model plant species that is highly competent to undertake this process. P. patens is such a model [7, 16]. In order to identify potential components of the plant HR-mediated DNA repair/GT pathway, we have undertaken a genome-wide analysis of the transcriptional response to the induction of DNA-DSBs in P. patens. This identifies a number of candidate genes whose contribution to efficient GT can now be tested.
Materials and Methods
Plant material
Physcomitrella patens ssp. patens (accession Villersexel K3) [4,17] was vegetatively propagated as protonemal homogenates on cellophane-overlaid BCD agar medium supplemented with ammonium tartrate (BCDAT) or as individual plants regenerated from protonemal tissue explants (“spot inocula”) on BCDAT-agar as described previously[18].
Responses to DNA damage were measured following either chronic or acute treatment with Bleomycin (Bleocin inj., Euro Nippon Kayaku GmbH, Germany). Chronic treatment was by growth of explants on BCDAT-agar containing bleomycin at concentrations indicated in the text. For acute treatment, protonemal tissue (7d following subculture) was harvested from cellophane overlays, briefly blotted with sterile filter paper to remove residual water, then dispersed in 5ml liquid BCDAT medium containing bleomycin as indicated. Recovery from acute treatment was measured by inoculating explants on drug-free BCDAT agar.
Sensitivity to bleomycin treatment was assessed by measuring the rate of plant growth as previously described [19]. Digital photographs of plants were taken at intervals following inoculation, and the area of each plant determined as the number of pixels using the ImageJ suite of image analysis programs [20]. Variation between photographs was minimised by normalising the area occupied by each plant with relative to the area of the culture dish in each image to generate a “growth index”.
Gene targeting
For gene targeted knockouts of helicases, sequences comprising approx. 1kb from each of the 5’- and 3’- sequences of target genes were amplified by PCR and cloned into EcoRV and Ecl136II sites, located on either side of an NPTII selection cassette by blunt-end ligation in a pMBL5-derived [6] plasmid vector. For the PpCtIP gene, a 3.9 kb fragment comprising the entire gene was amplified and the selection cassette was cloned between MunI and ClaI sites, thereby replacing 1018bp of the 1335bp polypeptide coding sequence. Linear targeting fragments were amplified by PCR for delivery to protoplasts. Primers used are listed in S1 Table. DNA delivery, regeneration of protoplasts, selection of transformants on medium containing G418 and analysis of targeted transformants by PCR and Southern blot analysis were all carried out as previously described [6]. The nature and verification of mutant genotypes is provided in S1–S7 Figs. The ability of wild-type and mutant strains to undertake targeted transgene integration was determined by targeted knockout of the PpAPT locus (Pp3c8_16590V3.1), encoding adenosine phosphoribosyl transferase. Targeted replacement of this gene confers resistance to the nucleotide analogue 2-fluoroadenine (2-FA). Protoplasts were transformed with a PpAPT targeting construct comprising 1419bp (5’) and 1001bp (3’) genomic sequences flanking a 35S-driven hygromycin resistance cassette, cloned between two XbaI sites located in intron 4 and exon 5 of the PpAPT gene, respectively. The proportion of hygromycin-resistant transformants exhibiting 2-FA resistance represented the gene targeting frequency [21].
RNA extraction and analysis
Total RNA was extracted from protonemal tissue propagated for 7d following subculture using aqueous phenol extraction and selective precipitation with 2.5M NaCl as previously described [18].
For the isolation of RNA from a polyribosome-enriched fraction, protonemal tissue was harvested and residual liquid was squeezed from the tissue by pressing between two layers of Whatman 3MM chromatography paper. Squeeze-dried tissue was frozen in liquid nitrogen and homogenized. Three replicates of approx. 0.4g squeeze-dried protonemata, each corresponding to four 9cm Petri dish cultures were ground to a powder in liquid N2 in a mortar and pestle and homogenised in 30 ml 200mM sucrose– 200mM Tris-Cl, pH8.5 – 60mM KCl– 30mM MgCl2−1% (v/v) Triton X-100 – 5mM β-mercaptoethanol. The homogenates were centrifuged (20 min x 30,000 x g: Sorvall RC5B SS-34 rotor) and the supernatants aspirated and layered over 5ml sucrose cushions (1M sucrose– 40mM Tris-Cl, pH8.5 – 20mM KCl– 10mM MgCl2) and centrifuged at 150,000 x g (3h, Beckman Sw28 rotor). Following centrifugation, the pellets were each resuspended in 0.5 ml RNA extraction buffer for isolation of RNA by phenol-chloroform extraction.
All RNA samples were dissolved in sterile water and digested with 1 unit RQ Dnase 1 (Promega) (10’ at room temperature) in a total volume of 60 microlitres before addition of Na2EDTA to 5mM and a further phenol-chloroform extraction. The final pellets were dissolved in sterile water for quantification using a Nanodrop spectrophotometer, and the integrity of the RNA monitored by electrophoretic analysis.
For gene expression profiling by quantitative real-time PCR, RNA (1μg) was reverse-transcribed using a Promega reverse transcription system. The 20μl reaction mixture was diluted 25-fold and 2.5 μl aliquots were used for real-time PCR amplifications (10μl) used the KAPA Biosystems SybrFast reaction kit in a BioRad CFX96 Real Time Detection System. Three biological replicate samples were analysed in duplicate. Transcript abundance was estimated by reference to both internal and external reference sequences. As an external reference, Physcomitrella RNA samples were “spiked” with tenfold serial dilutions (10−1–10−4) of an in vitro transcript from a full-length wheat “Em” cDNA [22, 23] prior to reverse transcription. These were used to test a number of candidate internal reference sequences, corresponding to Physcomitrella gene models Pp3c10_13820V3.1 (SAND family endocytosis protein), Pp3c27_2250V3.1 (Clathrin adapter complex subunit: “CAP-50”)), Pp3c4_32050V3.1 (Acyltransferase) and Pp3c13_7750V3.1 (Ribosomal protein S4) for stability of expression in response to bleomycin treatment. CAP-50 was found to have the most stable expression levels under all conditions tested and was subsequently used as the internal reference standard throughout. Primers used for PCR amplification are detailed in S2 Table.
Illumina mRNA-sequencing was used for digital expression analysis of the response to bleomycin treatment. Polyribosome-enriched RNA derived from three replicate control and bleomycin-treated tissue samples was used. For each treatment, equal quantities of the three replicate samples were mixed and submitted for Illumina short-read (36 base) sequence analysis by GATC Biotech AG, Konstanz, Germany. Aliquots (1μg) were used for cDNA synthesis using the Clontech SMART cDNA protocol (Clontech Protocol No. PT3041-1; Cat No. 634902). Libraries were prepared according to the Illumina protocol accompanying the DNA Sample Kit (Cat No. 0801–0303). Briefly, DNA was end-repaired using a combination of T4 DNA polymerase, E. coli DNA Pol I large fragment (Klenow polymerase) and T4 polynucleotide kinase. The blunt, phosphorylated ends were treated with Klenow polymerase and dATP to yield a protruding 3’- 'A' base for ligation of the Illumina adapters which have a single 'T' base overhang at the 3’ end. After adapter ligation, DNA was PCR-amplified with Illumina primers for 12 cycles and library fragments of ~375 bp (insert plus adaptor and PCR primer sequences) were band-isolated from an agarose gel. The purified DNA was captured on an Illumina flow cell for cluster generation. Libraries were sequenced on the Genome Analyzer following the manufacturer's protocols.
Transcriptomic analysis
The transcriptome analysis was performed using Version 1.2.1 of the P. patens genome and filtered cleaned gene models, which were obtained from http://www.cosmoss.org/. The Illumina reads were mapped to the P. patens genome using the program TopHat [24, 25]. When mapping the reads to the P. patens genome, TopHat was run using default settings and a list of existing potential exon junctions was provided. Reads that mapped to unique positions during the first stage of the TopHat mapping process were used to identify stretches of continuous read coverage. This resulted in 309,118 blocks of reads. Next adjacent blocks were then merged if they were within 500bps of each other. After the initial merging any remaining blocks that contained only 1 read were deleted. This resulted in 30,976 blocks of reads. When sequence blocks were joined using TopHat putative splice junctions, a total of 75,461 possible splice junctions were predicted. Only junctions separated by less than 10kbps were used, a cut-off resulting in 99.94% of the junctions being used and only 45 junctions were ignored. This resulted in 29,303 blocks of reads. Next the available gene models were used to further refine the blocks in two ways: (i) if two blocks were within the same gene model then the blocks were joined; (ii) if a block crossed two of the current gene models then it was split into two parts at the midpoint between the two gene models. This resulted in 28,802 sequence blocks, of which 20,347 overlapped with the existing gene models. For manual curation of the alignment, uniquely mapping reads were converted into binary format using SAMtools [26] and then visualised using the Integrative Genomic Viewer (http://www.broadinstitute.org/igv).
Differential expression values for sequence blocks were calculated by counting the number of reads that were present within each block from each sample. These were then divided by the number of uniquely mapping reads (divided by 1 million) present in the sample. This gave an expression for every block from each sample as the number of reads per million reads in the sample. The log-fold change was then calculated and the statistical significance of this change was assessed using a Bayesian method developed for digital gene expression profiling and the assignment of P-values [27], which were corrected for multiple testing [28]. Manual curation and annotation of gene models used BLASTP similarity and conserved domain searches using NCBI conserved domain and SMART (http://smart.embl.de/). Putative signal and transit peptides were identified using TargetP (www.cbs.dtu.dk/services/TargetP).
Results
Bleomycin sensitivity
In order to determine a level of DNA damage that would result in the most effective transcriptional induction of DNA-DSB repair components, we first established the sensitivity and responses of Physcomitrella protonemal tissue to treatment with the radiomimetic agent, bleomycin. Bleomycin induces DNA-DSBs at high frequency, and Physcomitrella is highly sensitive to this drug [29]. We first determined the sensitivity of protonemata to chronic exposure to bleomycin by transferring protonemal explants as a series of “spot inocula” to medium supplemented with a range of bleomycin concentrations.
Bleomycin treatment caused impaired growth in a concentration-dependent manner, lethality occurring at bleomycin concentrations of 200ng.ml-1 or greater (Fig 1A and 1B). Since we sought to establish the parameters for the induction of the DNA damage-response leading to recovery, we then assessed the effects of acute treatment with the drug. Protonemal tissue was incubated in liquid medium containing a range of bleomycin concentrations for 24 hours before explants were subcultured on drug-free medium in order to determine the ability of tissue to recover. Colony growth was monitored at intervals over a 3-week period. Whilst the initial treatment was not lethal, even with relatively high concentrations of bleomycin (40μg.ml-1), the recovery of protonemal growth showed a clear concentration-dependence, with the first sign of impaired growth becoming apparent at a concentration of 200ng.ml-1 (Fig 1C).
Fig 1. Growth responses of P. patens to treatment with bleomycin.
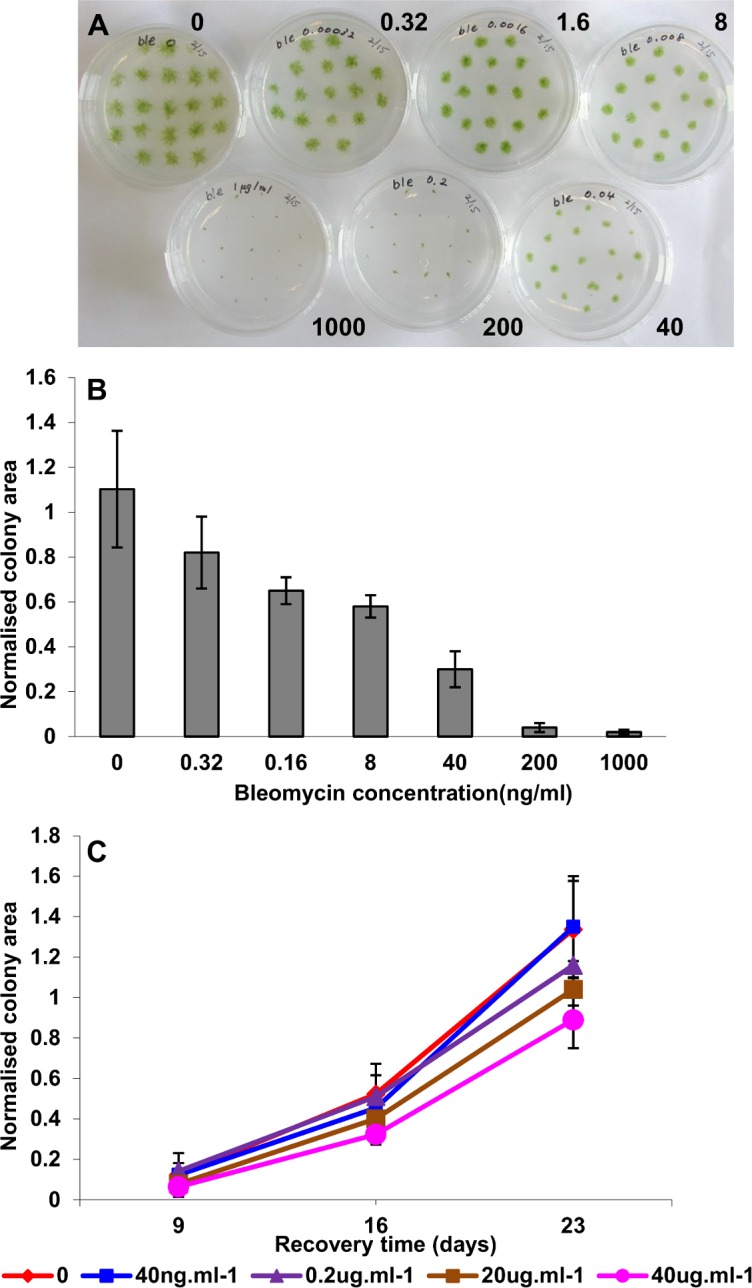
(A) Chronic exposure. Tissue explants were inoculated on BCDAT medium supplemented with bleomycin at 0, 0.32, 1.6, 8, 40, 200 and 1000 ng.ml-1 bleomycin as indicated, and incubated for 10 days under standard growth conditions. (B) Chronic exposure. Colony areas (mean ± SD) for plants exposed to bleomycin. Colony areas in the plates illustrated in Fig 1a were determined by image analysis of digital photographs of the individual plates, and are presented as the mean colony areas normalized to the area of the culture dish for each treatment. (C) Acute treatment. Tissue was incubated for 24h in BCDAT liquid medium supplemented with bleomycin at the concentrations indicated. Explants were then inoculated on drug-free medium and incubated under standard growth conditions for 23d and photographed at 9, 16 and 23d for image analysis. Mean colony areas (± SD, n = 12) were normalized to the area of the culture dish.
Transcriptional induction of HR-mediated repair genes
We next monitored HR-mediated DNA repair gene expression following induction of DNA-DSBs. Protonemal tissue was incubated in liquid medium containing a sublethal concentration of bleomycin for 24 hours, after which accumulation of the two Physcomitrella Rad51 transcripts was measured by quantitative real-time PCR. Since the Rad51 proteins are key components of the HR-mediated DNA-DSB repair process, and absolutely required for gene targeting in Physcomitrella [21], induction of their transcripts is indicative of the transcriptional induction of this pathway, which would additionally be expected to encompass other components required for HR-mediated DNA repair and transgene integration. Both Rad51 transcripts were induced by bleomycin treatment, with expression of the RAD51-1 gene being significantly more strongly induced than that of the RAD51-2 gene, whose transcript level remained relatively low (Fig 2). Accumulation of the Rad51-1 transcript showed a clear correlation between the level of accumulation and the concentration of bleomycin (and by extension, the extent of DNA-DSB induction) peaking at the previously determined sublethal acute doses of 200ng.ml-1 and 1μg.ml-1.
Fig 2. Transcriptional responses of PpRad51 genes to bleomycin treatment.
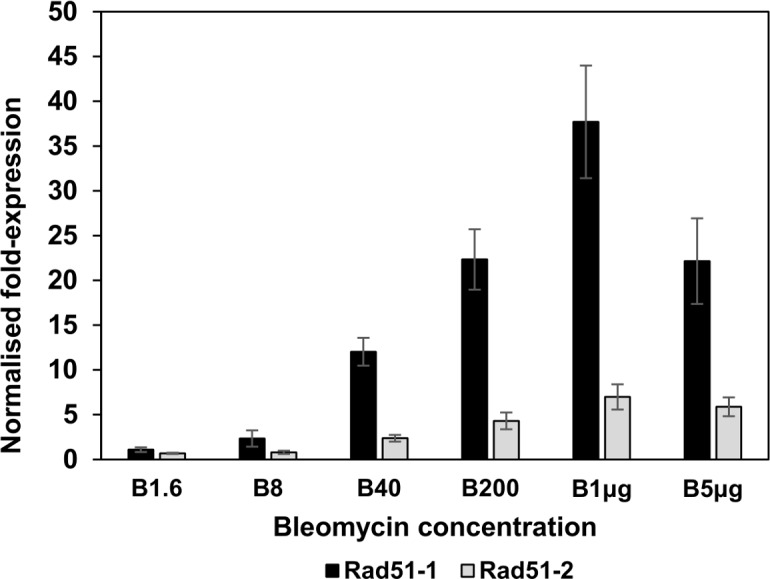
Tissue was incubated for 24 hours in BCDAT liquid medium supplemented with bleomycin at concentrations of 1.6, 8, 40 and 200ng.ml-1 and 1 and 5 μg.ml-1 respectively, as indicated. RNA was extracted for determination of transcript levels by real-time qPCR. The fold changes in transcript abundance relative to control (drug-free) treatments are shown (means ± SEM). Black bars: Rad51-1 mRNA; Grey bars: Rad51-2 mRNA.
Transcriptomic analysis
We incubated protonemal tissue in the presence or absence of 200ng.ml-1 bleomycin for 24 hours, before isolating a polyribosome-enriched fraction from triplicate samples for the extraction of RNA actively engaged in translation. Each sample was analysed by real-time PCR for the PpRAD51-1 transcript, which was found to be reproducibly enhanced approx. 30-fold (30.4 ± 3.8) in each bleomycin-treated sample. For transcriptomic analysis, equal quantities of the three control (untreated) RNA samples were pooled, as were the three bleomycin-treated RNA samples, in order to further minimise variation between replicates before being submitted to cDNA synthesis and Illumina short-read sequence analysis.
A total of 30,487,204 Illumina short reads (40 bases) were obtained, corresponding to a total of 1,219,488,160 bases. Of these, 15,942,360 reads were obtained from the control treatment and 14,544,845 from the bleomycin treatment. In total, 21,399,712 of the reads (70%) were mapped to the Physcomitrella genome assembly, Version 1.2.1 obtained from http://www.cosmoss.org, with 20,931,284 (69%) of them mapping to unique positions (11,183,882 from the control treatment and 9,747,402 from the bleomycin treatment). Assembly of reads into contiguous blocks, aligned with the genome assembly generated 28,802 sequence blocks, of which 20,347 overlapped with the Version 1.2.1 filtered gene models. From this, we estimate that approximately two-thirds of P. patens genes are expressed in chloronemal filaments. These alignments have been integrated into the Cosmoss genome browser, where they have been used in the generation of the Version 1.6 and 3.3 gene models, and are available for further structural gene annotation.
To identify genes up-regulated in response to the induction of DNA-DSBs, we used an automated procedure for digital gene expression analysis. The entire dataset, filtered by Log fold-change and P-value, has been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo) as Accession GSE25237. We undertook a detailed manual curation of those genes identified as significantly up- and down-regulated in response to bleomycin. In order to make such a task manageable, candidates were limited (i) to those genes exhibiting a change in expression level of 3-fold or greater and (ii) applying a cutoff based on selecting only those genes represented by at least 50 sequence traces in one or the other treatment, to exclude stochastic variation. This latter criterion excludes a significant number of genes expressed at a low level (approx. 40% of all genes identified in the experiment), but still provided a list of 500 up-regulated and 380 down-regulated sequences. These genes are listed in the Supporting Information files (S1 Dataset: up-regulated genes and S2 Dataset: down-regulated genes). Because the Physcomitrella genome has been undergoing continual refinement and reassembly, the list is annotated with a range of accession numbers (Locus identifiers, protein IDs) used in the successive assemblies.
Verification of transcriptomic analysis
We used real-time PCR to verify the results of the global transcriptome analysis, by monitoring the time-course of accumulation of selected transcripts in response to 200ng.ml-1 bleomycin. Target genes were selected from among those identified as significantly up-regulated in the transcriptome-wide analysis. As exemplars of the HR-mediated DNA-DSB repair pathway, we analysed the Rad51-1 and Rad51-2 transcripts (Fig 3A), for the NHEJ pathway, we selected the Ku70 and Ku80 transcripts (Fig 3B). We also selected two genes encoding polyADP ribose polymerase (PARP) (Fig 3C) and two genes encoding DNA helicases (Fig 3D). In each case, a general trend is clear. First, gene induction is rapid: significant accumulation of each transcript relative to that in the control treatment is detectable after 1 hour of bleomycin treatment, with a peak of expression seen at the 4-hour time-point. Second, although the levels of gene induction relative to the control treatments at the 24-hour time-point do not exactly replicate those determined in the transcriptome analysis for each gene, the trends are generally similar. Third, for each category of gene analysed by real-time PCR, the levels of induction of the selected gene pairs relative to each other is similar to that identified in the transcriptomic analysis. We are thus confident that the genome-wide assay of gene induction provides a true reflection of the protonemal response to bleomycin-induced DNA damage.
Fig 3. Time-course of the transcriptional response of selected DNA repair genes.
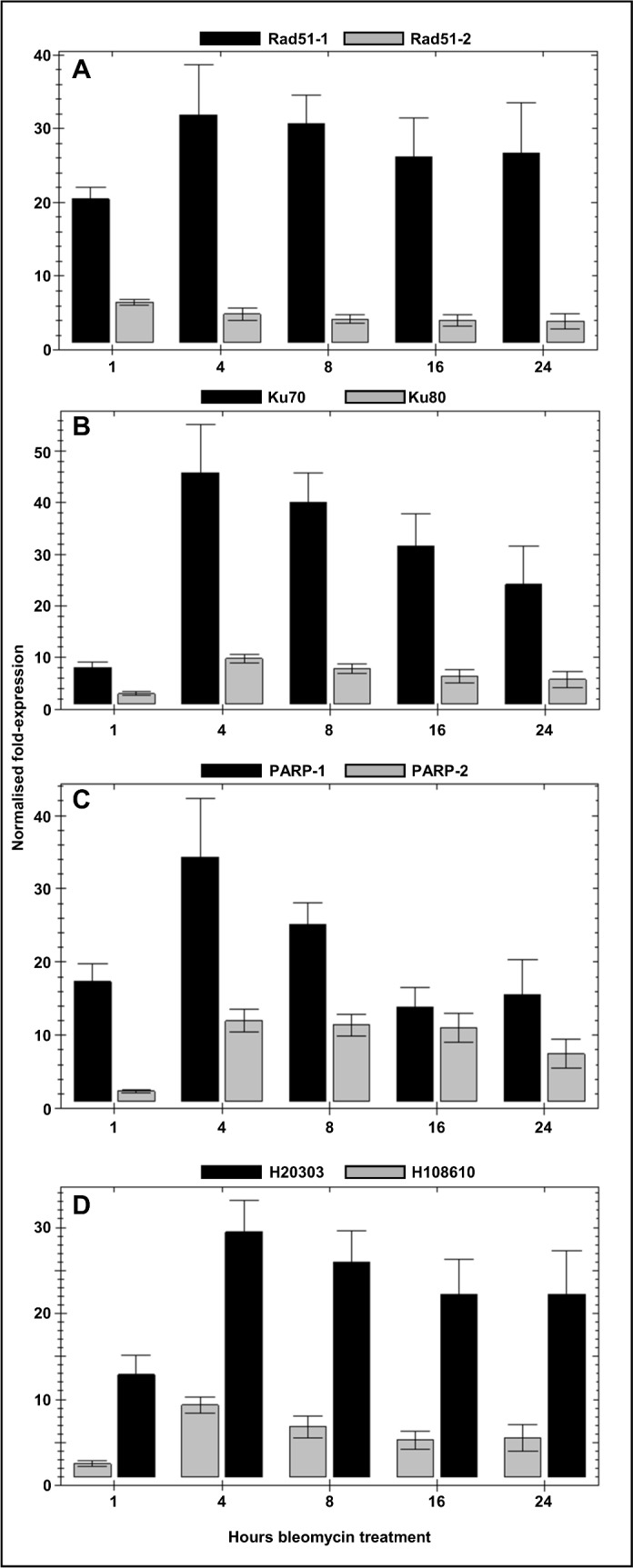
Tissue was incubated in BCDAT medium supplemented with 200ng.ml-1 for 1, 4, 8, 16 and 24 hours prior to harvest. RNA was extracted for determination of transcript levels by real-time qPCR. The fold changes in transcript abundance relative to control (drug-free) treatments are shown (means ± SEM). (A) Rad51-1 mRNA (black bars); Rad51-2 mRNA (grey bars). (B) Ku70 (black bars); Ku80 (grey bars). (C) PARP-1 (Pp3c22_13240V3.1) (black bars); PARP-2 (Pp3c8_17220V3.1) (grey bars); (D) SRS2-like helicase (Pp3c1_29170V3.1) (black bars); Alc1-like helicase (Pp3c10_6710V3.1) (grey bars).
Functional analysis of selected genes
The catalogue of genes significantly induced in response to DNA damage includes those implicated in all DNA repair pathways, not only the HR-mediated repair pathway that is responsible for mediating high-efficiency gene targeting in P. patens. Table 1 lists the up-regulated genes that function in DNA-DSB repair by the NHEJ, and MMEJ pathways, as well as components associated with nucleotide-excision repair and base-excision repair.
Table 1. Up-regulated DNA repair and replication genes.
| Fold change | V3.3 ID | Phypa1_1:ID and annotation |
|---|---|---|
| 149x | Pp3c1_38430V3.1 | 86560: NHEJ DNA-repair protein XRCC4 |
| 52x | Pp3c22_13240V3.1 | 150949 NAD+ ADP-ribosyltransferase PARP-1 |
| 27x | Pp3c16_1250V3.1 | 190334: HhH-GPD base-excision repair family protein |
| 21x | Pp3c8_19560V3.1 | 166702: Chromosome segregation ATPase |
| 20x | Pp3c24_12060V3.1 | 20647: minichromosome maintenance family protein 8 |
| 19x | Pp3c14_5920V3.1 | 228728: NHEJ-associated DNA ligase 4 |
| 15x | Pp3c13_17960V3.1 | 86100: Fanconi anemia, complementation group I gene involved in interstrand crosslink repair. |
| 14x | Pp3c2_30860V3.1 | 200726: UV DNA repair protein: Flap endonuclease 1a |
| 14x | Pp3c18_7140V3.1 | 60909: PpKu70 NHEJ DNA repair protein |
| 13x | Pp3c12_3560V3.1 | 173451: Similar to telomere maintenance component 1 |
| 11x | Pp3c6_22830V3.1 | 82469: SMC class chromosome segregation protein |
| 9x | Pp3c6_17700V3.1 | 203240: HhH-GPD base excision DNA repair family protein |
| 8x | Pp3c13_20209V3.1 | 40610: DNA ligase 1 |
| 8x | Pp3c11_22000V3.1 | 206066 HR DNA repair PpRad51-1 |
| 8x | Pp3c4_21450V3.1 | 171541: DNA polymerase delta, subunit 4 |
| 8x | Pp3c16_2140V3.1 | 235804: PpCtIP (yeast Sae2) HR End resection nuclease |
| 7x | Pp3c6_3460V3.1 | 129584: PpATR HR checkpoint activation kinase |
| 7x | Pp3c5_18670V3.1 | 226781: Metallo-beta-lactamase (β-CASP) interstrand DNA cross-link pair protein-related |
| 7x | Pp3c22_7360V3.1 | 115005: Rad4-like nucleotide excision repair protein |
| 7x | Pp3c18_11670V3.1 | 201653: ScRAD1 DNA repair endonuclease (hsERCC4) |
| 6x | Pp3c8_21590V3.1 | 231531: Topoisomerase-interacting protein |
| 6x | Pp3c10_1920V3.1 | 93627: Excinucleases ABC, C subunit protein |
| 6x | Pp3c8_17250V3.1 | 134789: UMUC-like DNA-repair protein: Rad30 |
| 6x | Pp3c11_21910V3.1 | 161308: Sister chromatid cohesion protein (cohesin) |
| 6x | Pp3c17_7580V3.1 | 95275: HMG1/2 protein (high mobility group b2) |
| 5x | Pp3c17_18800V3.1 | 164103: DNA polymerase B: delta subunit |
| 5x | Pp3c4_1630V3.1 | 167899: C-terminal BRCT domain–containing protein |
| 5x | Pp3c8_17220V3.1 | 188096: poly(ADP-ribose) polymerase (PARP-2) |
| 5x | Pp3c27_130V3.1 | 148594: Nse4: Smc5/6 DNA repair complex member |
| 5x | Pp3c8_16660V3.1 | 188047: "damaged DNA -binding protein 2" (DDB2) |
| 5x | Pp3c22_11100V3.1 | 188592: PpKu80 NHEJ DNA repair protein. |
| 4x | Pp3c2_33440V3.1 | 147728: DNA polymerase B zeta subunit |
| 4x | Pp3c20_10450V3.1 | 11886: Replication protein A subunit 1: PpRPA-1a |
| 4x | Pp3c23_16880V3.1 | 144964: Replication Protein A subunit 1: PpRPA-1b |
| 3x | Pp3c22_21030V3.1 | 145270: DNA replication factor C complex subunit 1 |
| 3x | Pp3c14_15380V3.1 | 107931: Uracil DNA glycosidase |
We tested the contributions made by some of these genes to the DNA-damage response by constructing targeted knockout mutants. Genes were selected either because their homologues had been implicated in DNA-DSB repair in other organisms, or because they appeared functionally interesting. As a gene known to be important for homology-dependent repair, encoding the initiator of end-resection that characterises this pathway, we generated a targeted deletion of PpCtIP (Pp3c16_2140V3.1) (S1 Fig), encoding the orthologue of the human and yeast genes, CtIP and SAE2, respectively [30–32]. Additionally, we noted that a small group of chromatin-remodelling DNA helicases were represented among the strongly damage-induced genes (Table 2). These included PpTEBICHI, (Pp3c5_12930V3.1) (S2 Fig)–a helicase with a DNA polymerase theta domain implicated in replication-related MMEJ [33, 34], a helicase/nuclease PpZRL (for “ZRANB3-Like”–a member of the SMARCA L1 Snf2 helicase subfamily, Pp3c1_13308V3.1) (S3 Fig), PpALC1 (Pp3c10_6710V3.1) (S4 Fig) a homologue of human “Amplified in Liver Cancer 1” [35], a chromodomain-helicase gene PpCHD5 (Pp3c20_11500V3.1) (S5 Fig) similar to the Arabidopsis helicase AtCHR5 and the developmental regulator, PICKLE [36, 37], the Rad3-like gene PpRTEL1 “Regulator of telomere elongation”: Pp3c18_12550V3.1) (S6 Fig), PpSRS2 (Pp3c1_29170V3.1) and pERCC6 (Pp3c15_23590V3.1) (S7 Fig). Significantly, among the Arabidopsis orthologues of these genes, the ERCC6 (AT2G18760) gene was the only one to be significantly up-regulated in response to DNA damage [38], suggesting that the differential inducibility of these genes by DNA damage might reflect the relative competences to undertake targeted transgene integration between moss and angiosperms.
Table 2. Up-regulated genes annotated as DNA helicases.
| Fold change | V3.3 ID | Arabidopsisorthologue | Fold change (Arabidopsis) | Functional annotation |
|---|---|---|---|---|
| 25x | Pp3c1_29170V3.1 | AT4G25120 | 1.4x down | ATP-dependent DNA helicase similar to yeast SRS2 PpSRS2 |
| 16x | Pp3c10_6710V3.1 | AT2G44980 | 1.1x down | Alc1-like SNF2 family DNA- dependent ATPase/helicase |
| 13x | Pp3c8_17910V3.1 | none | - | SNF2 family DNA-dependent ATPase/helicase Rad54B |
| 11x | Pp3c15_23590V3.1 | AT2G18760 | 6.7x up | ERCC6-like SNF2 helicase PpERCC6 |
| 11x | Pp3c5_12930V3.1 | At4G32700 | 1.1x down | Tebichi ortholog: polymerase with DEAD/DEAH box and polymerase theta helicase domains PpTEB |
| 11x | Pp3c1_13308V3.1 | AT5G07810 | 1.4x down | SNF2-related DNA Helicase/endonuclease PpZRL |
| 8x | Pp3c20_11500V3.1 | AT2G13370 | 1.2x down | SNF2 family chromodomain-helicase PpCHD5 |
| 6x | Pp3c18_12550V3.1 | AT1G79950 | No change (1x) | RTEL1-like helicase PpRTEL |
| 5x | Pp3c10_16570V3.1 | AT1G03190 | 1.2x down | ERCC2-like helicase PpERCC2 |
Up-regulated P. patens helicases compared with the DNA-damage (100Gy γ-irradiation) induced changes in expression of their orthologues in wild-type A. thaliana, as reported by Culligan et al. [38].
All mutants subsequently used for phenotypic analysis were verified as being correctly targeted gene replacements, in which the native gene was replaced by a single copy of the gene disruption cassette (S1–S7 Figs). The mutant lines were analyzed in two ways. First, by determining whether they displayed hypersensitivity to DNA damage caused by the genotoxin bleomycin, and secondly by analyzing the efficiency with which targeted transgene integration occurred following protoplast transfection.
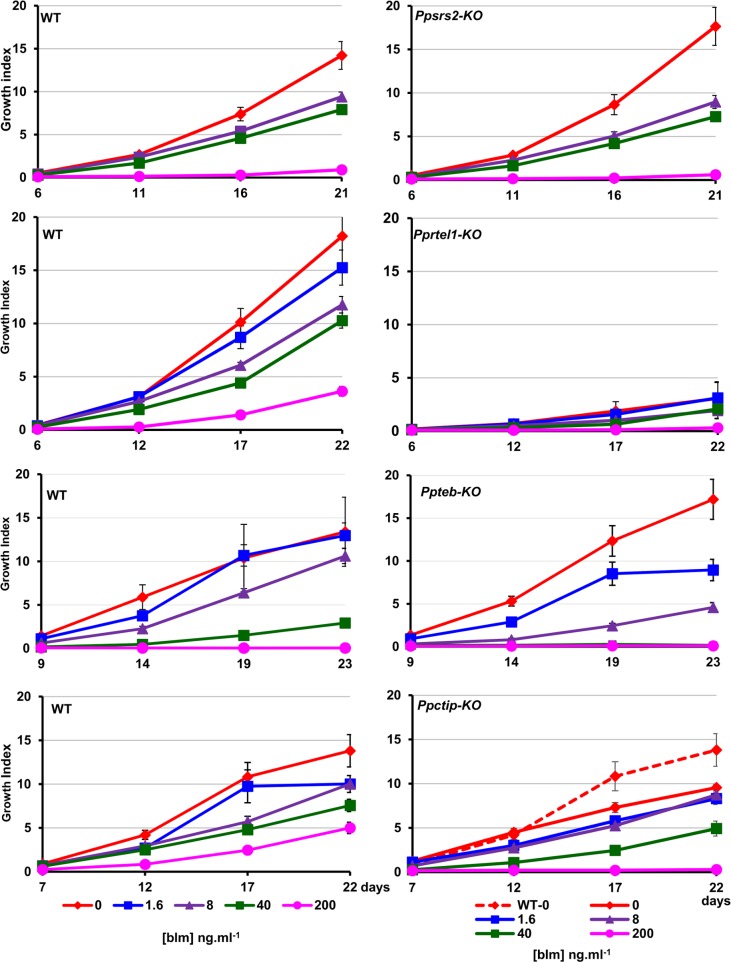
In growth tests, the Ppercc6, Ppzrl, Ppchd5, and Ppalc1 mutant lines displayed no discernible growth defects or hypersensitivity to DNA damage (S8 Fig). Neither did the Ppsrs2 mutant, although the Pprtel1 mutant exhibited a severe growth defect, plants achieving only one-sixth of the size of the corresponding wild-type. However, this mutant showed no discernible hypersensitivity to genotoxin (Fig 4; S9 Fig). By contrast, the Ppctip mutants grew slightly less rapidly under normal conditions, and were sensitised to DNA damage, being susceptible to 200ng.ml-1 bleomycin, whereas the wild-type plants survived this dose. The Ppteb mutant plants also exhibited a similar hypersensitivity to bleomycin, but showed no growth defects on unsupplemented growth medium (Fig 4; S9 Fig).
Fig 4. Bleomycin sensitivity of P. patens mutants.
Tissue explants of wild-type and mutant protonemal tissue were inoculated on BCDAT agar medium, supplemented with a 5-fold dilution series of bleomycin (1.6ng.ml-1 – 200ng.ml-1). Plants were grown for a period of approximately 3 weeks, and plant growth was determined by image analysis of digital photographs taken at intervals to generate the Growth Index. WT and mutant plants (8–12 explants) were inoculated on the same plates for direct comparison. Because individual experiments took place at intervals over a period of 2 years, direct comparisons cannot be made between each pair of panels, as different batches of bleomycin with varying potency were used during this time. Mutants analysed (from top) were Ppsrs2-KO; Pprtel1-KO (note the 5-fold difference in the amplitudes of the y-axes); Ppteb-KO and Pctip-KO. Representative images of plant growth are shown in S9 Fig.
These mutants were also tested for their ability to undertake targeted transgene integration (with the exception of the Pprtel mutant line, which grew too slowly for the isolation of protoplasts suitable for transformation). Only the Ppctip mutant exhibited a clear severe gene targeting deficiency (Table 3), a finding consistent with the conserved role of the CtIP/Sae function in initiating resection of double-strand breaks during homology-dependent repair. The Ppchd5, Ppzrl and Ppercc6 all undertook targeted integration at a frequency some 5–10% below that of the wild-type, but this is not thought to represent a significant difference between the lines. The Ppalc1 and Ppteb mutants showed no defects in transformation efficiency or transgene integration.
Table 3. Gene targeting efficiency in mutant lines.
| Strain | RTF (%) | HygR | 2-FAR | %GT |
|---|---|---|---|---|
| WT | 1.14 | 1183 | 843 | 73.3% (72.1 ± 5.2: n = 3) |
| Ppalc1-KO | n.d. | 506 | 397 | 78.4% (79.3 ± 5.6: n = 3) |
| Ppchd5-KO | 1.77 | 1600 | 1081 | 67.6% (67.6 ± 7.9: n = 3) |
| Ppzrl-KO | 0.68 | 345 | 220 | 63.7% (65 ± 11.2: n = 3) |
| Ppteb-KO | 1.65 | 1149 | 910 | 79.2% (79.3 ± 3.9: n = 3) |
| Ppctip-KO | n.d. | 285 | 64 | 22.5% (22.2 ± 4.7: n = 6) |
| Ppercc6-KO | 2.87 | 2680 | 1739 | 64.9% (61.7 ± 12.8: n = 3) |
| Ppsrs2-KO | 1.33 | 1266 | 923 | 72.9% (72 ± 3: n = 3) |
RTF (Relative transformation frequency) is the proportion of viable protoplasts that regenerate into hygromycin-resistant plants following transformation. HygR (hygromycin resistant) is the number of stably transformed plants containing the APT targeting construct. 2-FAR (2-fluoroadenine resistant) is the number of plants resistant to 2-fluoroadenine. %GT (% gene targeting) is the proportion of stable transformants that are resistant to 2-fluoroadenine: (2-FAR/HygR)x100.
n.d.: Not determined.
PpSRS2 is a homologue of the yeast SRS2 helicase, often termed an “anti-recombinase”, which interacts with and is activated by Rad51 in HR-mediated DNA repair. It is posited to be important in preventing unwanted gene conversion events in homology-dependent repair of DSBs through disassembling Rad51-ssDNA complexes [39, 40]. We have previously demonstrated that targeted knockout of PpSRS2 has no phenotypic effects in Physcomitrella, either for DNA damage-sensitivity or gene targeting efficiency, and neither did its deletion relieve a GT deficiency resulting from the knockout of the RAD51 paralogue RAD51B [41]. We therefore analyzed whether deletion of this gene would render P. patens susceptible to targeted integration of mismatched transgenes, by analyzing the integration of a hygromycin resistance marker into the PpAPT locus by targeting sequences containing a 1%, 2% or 3% complement of mismatches [42]. Unlike the msh2 mutation, deficient in mismatch repair [42], the srs2-KO mutant did not enhance the targeted integration of mismatched sequences (Table 4). The frequencies of targeted transgene integration obtained with these constructs were essentially identical to those previously observed in wild-type P. patens [42], and the relative transformation frequency obtained with these constructs showed a substantial decrease, proportional to the extent of mismatch, demonstrating the very strong preference for the integration of transforming DNA at homologous sites over illegitimate recombination [5, 16, 21].
Table 4. Gene targeting by Ppapt-KO mismatch vectors in Ppsrs2-KO.
| Strain | Mismatch | HygR | 2-FAR | %GT | RTF(%) |
|---|---|---|---|---|---|
| Ppsrs2-KO | 0% mismatch | 248 | 52 | 41.9* | 1.12 |
| Ppsrs2-KO | 1% mismatch | 48 | 13 | 16.7 | 0.10 |
| Ppsrs2-KO | 2% mismatch | 17 | 5 | 5.9 | 0.10 |
| Ppsrs2-KO | 3% mismatch | 26 | 8 | 0 | 0.006 |
*GT efficiencies vary, depending on the nature of the targeting construct (Kamisugi et al., 2005). Thus the GT frequency obtained with this vector cannot be compared with that obtained with the different PpAPT targeting vector used for the experiments reported in Table 3.
Discussion
Bleomycin induces changes in gene expression of not only DNA-repair components but a range of other functions reflecting a general stress-response (S1 Dataset and S2 Dataset).
DNA repair components
Several up-regulated DNA-repair genes (Table 1) have a highly conserved role in HR-mediated repair [43]. These include the PpRAD51-1 recombinase, the ATR kinase that executes a cell-cycle checkpoint at G2/M to facilitate HR-mediated DNA-DSB repair [12] and that co-activates the up-regulated end-resection protein CtIP (Sae2 in yeast) [30–32, 44, 45] and both genes encoding the largest subunit of the single-strand DNA-binding Replication Protein A complex. Unsurprisingly, targeted knockout of PpCtIP resulted in enhanced sensitivity to genotoxic stress and a significant reduction in GT efficiency–although GT was not abolished as occurs following deletion of RAD51 paralogues [21, 41].
Other DNA repair functions are also represented. The RAD1 exonuclease (with RAD10 and the MSH2/3 mismatch repair system) trims non-homologous sequences from the ends of transforming DNA prior to insertion by HR in yeast [46], and is implicated in plant somatic recombination [47]. Two polyADP-ribose polymerase (PARP) genes are strongly induced. PARPs participate in a range of DNA repair activities, including MMEJ and base-excision repair, and transcriptional activation of PARP genes by genotoxic stress is characteristic of mammalian cells [48]. In Physcomitrella, two PARP genes (PARP-1: Pp3c22_13240V3.1 and PARP-2: Pp3c8_17220V3.1) are strongly up-regulated. The third (PARP-3: Pp3c1_22640V3.1) is not significantly expressed, being represented by only a single sequence read. PpTEBICHI encodes a helicase-DNA polymerase theta, an enzyme that in metazoans processes MMEJ ends [49]. In P. patens, MMEJ has been implicated in transgene concatenation prior to “one-ended” targeted insertion into the genome [50], a process analogous to the targeted integration of multiple tandem transgenes in yeast [51]. The Ppteb-KO mutant showed enhanced bleomycin sensitivity, but no recombination defect or serious developmental phenotype. By contrast, the corresponding teb mutant in Arabidopsis displays constitutively activated DNA-damage responses, an inability of meristematic cells to progress through the G2/M transition, and a deficiency in intrachromosomal recombination [33, 34]. This contrast likely reflects the greater prominence of the HR repair pathway in P. patens, compared with A. thaliana.
Up-regulated NHEJ-related genes include Ku70, Ku80, XRCC4 and DNA ligase 4. Interestingly several of these are up-regulated to a greater extent than are HR-related genes. This may reflect a preference for the HR pathway in normal growth and development of P. patens, such that NHEJ-associated gene products are generally maintained at a relatively low constitutive cellular concentration, and are only required to be produced in response to the induction of extensive DNA damage.
Cell-cycle regulation
Cell-cycle checkpoint imposition is a critical part of the DNA damage response. Differentially regulated genes associated with cell-cycle checkpoint control include PpATR and homologues of the S. pombe RAD9 and HUS1 genes whose products combine (with that of SpRad1, in the “9-1-1” complex) to execute a G2/M checkpoint [52]. The down-regulated gene set includes both D-type cyclin and B-type cyclin-dependent kinase genes required for cell-cycle progression (Table 5).
Table 5. Differentially regulated genes annotated as cell-cycle associatedFold change.
| V3.3 ID | Phypa1_1:ID and annotation | |
|---|---|---|
| Up-regulated | ||
| 50x | Pp3c7_570V3.1 | 234322: Checkpoint 9-1-1 complex, RAD9 component |
| 24x | Pp3c21_200V3.1 | 197198: AAA-type ATPase (cell-division related) |
| 11x | Pp3c1_35610V3.1 | 160969: AAA-ATPase |
| 8x | Pp3c10_18160V3.1 | 219794: cdc20 anaphase promoting complex protein |
| 4x | Pp3c1_24880V3.1 | 106047: Cell cycle checkpoint protein Hus1 |
| 3x | Pp3c9_2440V3.1 | 10529: Retinoblastoma-related protein |
| Down-regulated | ||
| 5x | Pp3c15_17470V3.1 | 226408: Cyclin D |
| 5x | Pp3c16_3910V3.1 | 107101: B-type cyclin-dependent kinase |
| 6x | Pp3c15_21520V3.1 | 196659: Cyclin B |
| 44x | Pp3c27_6070V3.1 | 60060: B-type cyclin-dependent kinase cdc2 |
Housekeeping genes
Many housekeeping functions are downregulated (S2 Dataset), in particular protein synthesis, photosynthesis and cell growth: 186 of the downregulated genes encode ribosomal proteins, probably reflecting a checkpoint-mediated reduction in mitotic activity with which the synthesis and assembly of ribosomes are closely correlated. Down-regulation of photosynthetic functions is likely a general abiotic stress response, since loss of photosynthetic function is one of the most striking features of salt- and drought-stress in Physcomitrella [53]. However, the responses to bleomycin-induced DNA damage are distinct from those associated with drought stress, in that a number of characteristically drought-responsive genes [53, 54] are down-regulated following bleomycin treatment (S3 Table).
Reduction of mitotic activity and cell elongation, would explain the reduced growth rate seen in response to bleomycin treatment, and a number of genes with growth-regulating cell wall-modifying properties are also represented in both the up- and down-regulated gene sets. These include expansins [55], xyloglucan endotransglycosidases [56], pectin modifying enzymes, arabinogalactan proteins and germin-like proteins.
Transcription factors
Bleomycin treatment induces significant changes in the expression profiles of transcription factors that implement essential cellular damage control measures. Thirty-six transcription factor genes appear in the up-regulated set and fourteen in the down-regulated set (S4 Table and S5 Table).
Members of the AP2/EREBP-domain family are most prominent (16 up-regulated and 4 down-regulated). A highly expanded plant transcription factor family, this comprises nearly 150 members in Arabidopsis [57] and a similar number (approx. 135) in Physcomitrella, so these might be expected to comprise a significant fraction of the differentially regulated transcription factor subset. Additionally, many AP2/EREBP transcription factors regulate responses to both biotic and abiotic stresses that involve the intracellular generation of active oxygen species (AOS) (pathogen attack, drought, wounding), and their induction in response to bleomycin may reflect secondary effects of AOS generated by the drug’s mode of action. Notably, genes whose products ameliorate the cytotoxic consequences of AOS are also up-regulated, including lipases (14 up-regulated) and three tetratricopeptide-repeat containing thioredoxins, as well as genes known to be responsive to AOS in pathogen responses, such as the 8 chalcone synthase genes and 3 phenylalanine ammonia lyase genes identified in this study. These enzymes are components of the phenylpropanoid biosynthetic pathway, regulated by R2R3 myb-family transcription factors [58–60] and we note that two such transcription factor genes are highly up-regulated.
Candidate modulators of gene targeting
A rationale for this study was to determine whether genes up-regulated in response to DNA-DSB induction were associated with the high frequency of HR-mediated GT. We therefore compared the DNA-damage transcriptomes of GT-competent Physcomitrella and GT-incompetent Arabidopsis to identify potential candidates. Several genome-wide studies of the Arabidopsis DNA-DSB response have been carried out, utilising macro- and microarray platforms to identify genes responsive to genotoxic chemicals (bleomycin, mitomycin C and methane methylsulphonate: [61, 62] and to ionising radiation (X- and γ-irradiation, [38, 63].
We identified a number of up-regulated moss-specific genes encoding chromatin remodeling DNA helicase functions (Table 2) and selected these for further analysis since DNA helicases of the Swi2/Snf2 family are implicated in intrachromosomal recombination between inverted repeats in Arabidopsis [37], and different helicases are associated with the two different mechanisms of HR-mediated DSB repair: the single-strand annealing mechanism and the synthesis-dependent strand annealing pathway [64].
The Alc1 helicase relocates within mammalian nuclei to the site of DNA-DSBs within seconds of their induction [35] in a process involving activation of its ATPase activity by PARylation by nucleosome-bound PARP to mediate DNA repair [65]. Since two PARP genes were also upregulated by bleomycin, this suggested a potential role in DNA-DSB repair and gene targeting. In mammalian cells, both overexpression and RNA-mediated knock-down of ALC1 result in sensitivity to genotoxins. By contrast, knockout of ALC1 appeared neither to compromise growth of P. patens nor affect its ability to undertake targeted transgene integration.
Shaked et al. [37] reported that Arabidopsis mutants of the PpERCC6 orthologue, At2g18760, exhibited sensitivity to UV-C but not to γ-irradiation: a response consistent with its predicted role in nucleotide excision repair. We found the Ppercc6-KO mutant to show no growth deficiency or bleomycin sensitivity, and although GT efficiency in the mutant was lower than that in the wild-type, it is doubtful that this is a significant difference. Alone among the genes we tested, PpERCC6 is represented by additional paralogues in the P. patens genome (there are 3 other closely related family members, none of which were up-regulated following bleomycin treatment), so genetic redundancy likely accounts for the lack of a clearly observable deleterious phenotype.
By contrast, knockout of the PpRTEL1 orthologue generated a clear, general growth deficiency. However this was unrelated to the presence or absence of bleomycin, and most likely represents a deficiency in telomere maintenance: mutations of this gene in different organisms consistently result in severe phenotypic consequences, such as the human degenerative condition Dyskeratosis congenita. In Arabidopsis, mutation of AtRTEL1 has severe pleiotropic effects that include growth retardation, suppressed homologous recombination and defects in both intra- and interstrand crosslink repair [66] and cell-cycle arrest [67]. Due to the difficulty of generating viable protoplasts from this mutant, we were unable to determine its impact on gene targeting.
Physcomitrella patens is extraordinarily robust in its ability to withstand genotoxic stress. This may be a necessary characteristic for an organism that spends most of its life cycle in the haploid phase. Although it has been postulated that the dominant form of DNA-DSB repair is the homology-dependent pathway, analysis of the DNA-damage transcriptome reveals the rapid induction of genes encoding components active in most forms of DNA repair process, including nucleotide excision repair, base excision repair, transcription coupled repair, inter- and intra-strand crosslink repair, NHEJ- and HR-mediated DNA-DSB repair and in DNA-damage associated checkpoint signalling. Targeted knockout of a number of individual genes identified in this analysis had surprisingly little adverse effect on either growth, sensitivity to genotoxin or to targeted transgene integration, unlike the consequences of their mutation in other organisms, suggesting a high level of built-in redundancy in genome maintenance.
Supporting Information
(XLSX)
(XLSX)
A: Schematic of gene structure and knockout construct. B: Identification of targeted loci by PCR amplification with cassette-specific “outward” and gene-specific “inward” primers C: Identification of single-copy targeted transformants with external gene-specific primers D: Southern blot (HindIII digest) to identify transformants containing only a single, targeted selection cassette. In S1–S7 Figs, Red boxes = protein-coding sequence, Blue boxes = 5’- and 3’-UTRs, Green boxes = NPTII selection cassette, Red triangles = LoxP sites. In Southern blots, the NPTII sequence was used as a probe.
(PDF)
A: Schematic of gene structure and knockout construct. B: Identification of targeted loci by PCR amplification with cassette-specific “outward” and gene-specific “inward” primers. C: Identification of single-copy targeted transformants with external gene-specific primers (track “P” = plasmid control). D: Southern blot (EcoRI digest) to identify transformants containing only a single, targeted selection cassette.
(PDF)
A: Schematic of gene structure and knockout construct. B: Identification of targeted loci by PCR amplification with cassette-specific “outward” and gene-specific “inward” primers. C: Identification of single-copy targeted transformants with external gene-specific primers (Track “P” = plasmid control). D: Southern blot (HindIII digest) to identify transformants containing only a single, targeted selection cassette. (Analysed on same gel as PptebKO: see S2 Fig for WT control).
(PPTX)
A: Schematic of gene structure and knockout construct. B: Identification of targeted loci by PCR amplification with cassette-specific “outward” and gene-specific “inward” primers C: Identification of single-copy targeted transformants with external gene-specific primers (Tracks “P” and “wt” = plasmid and wild-type genomic DNA controls) D: Southern blot (HindIII digest) to identify transformants containing only a single, targeted selection cassette. Analysed on same gel as Ppchd5-KO: see S5 Fig for wild-type control).
(PPTX)
A: Schematic of gene structure and knockout construct. B: Identification of targeted loci by PCR amplification with cassette-specific “outward” and gene-specific “inward” primers C: Identification of single-copy targeted Transformants with external gene-specific primers (Tracks “P” and “wt” = plasmid and wild-type genomic DNA controls). D: Southern blot (BglII digest) to identify transformants containing only a single, targeted selection cassette.
(PPTX)
A: Schematic of gene structure and knockout construct. B: Identification of targeted loci by PCR amplification with cassette-specific “outward” and gene-specific “inward” primers. C: Identification of single-copy targeted transformants with external gene-specific primers (Track “P” = plasmid control). D: Southern blot (EcoRV digest) to identify transformants containing only a single, targeted selection cassette.
(PPTX)
A: Schematic of gene structure and knockout construct. B: Identification of targeted loci by PCR amplification with cassette-specific “outward” and gene-specific “inward” primers C: Identification of single-copy targeted transformants with external gene-specific Primers primers (Tracks “P” and “wt” = plasmid and wild-type genomic DNA controls). D: Southern blot (BglII digest) to identify transformants containing only a single, targeted selection cassette
(PPTX)
(PPTX)
Each photograph represents the final time-point presented in the growth test analysis in Fig 4. For Ppteb-KO the growth on 40ng.ml-1 bleomycin is shown. For all other mutants, growth on 200ng.ml-1 bleomycin is shown.
(PPTX)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
The authors are grateful to the UK Biotechnology and Biological Sciences Research Council for its support through BBSRC Research Grant BB/I006710/1. We are grateful to Dr. Fabien Nogué (INRA, Versailles) and Dr. Didier Schaefer (Univ. Neuchâtel) for providing us with the mismatched PpAPT targeting vectors. We also thank Stefan Rensing, Daniel Lang and Andreas Zimmer (Freiburg University) for integrating the sequence reads into the Cosmoss Physcomitrella genome browser.
Data Availability
The complete transcriptomic dataset has been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo) as Accession GSE25237, and in the NCBI SRA as accessions SRX031155 and SRX031156. Alignment of all sequence reads with the Physcomitrella genome can be viewed on the Cosmoss genome browser (http://www.cosmoss.org).
Funding Statement
The authors are grateful to the UK Biotechnology and Biological Sciences Research Council (http://www.bbsrc.ac.uk) for its support through BBSRC Research Grant BB/I006710/1. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kenrick P. Crane PR. The origin and early evolution of plants on land. Nature 1997;389: 33–39. [Google Scholar]
- 2.Zimmer A, Lang D, Richardt S, Frank W, Reski R, Rensing SA. Dating the early evolution of plants: detection and molecular clock: analyses of orthologs. Mol. Genet. Genomics. 2007;278: 393–402. [DOI] [PubMed] [Google Scholar]
- 3.Rensing SA, Lang D, Zimmer A, Terry A, Salamov A, Shapiro H, et al. The Physcomitrella genome reveals insights into the conquest of land by plants. Science. 2008;319: 64–69. [DOI] [PubMed] [Google Scholar]
- 4.Kamisugi Y, von Stackelberg M, Lang D, Care M, Reski R, Rensing SA, et al. A sequence-anchored genetic linkage map for the moss Physcomitrella patens. Plant J. 2008;56: 855–866. 10.1111/j.1365-313X.2008.03637.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schaefer DG, Zryd JP. Efficient gene targeting in the moss Physcomitrella patens. Plant J. 1997;11: 1195–1206. [DOI] [PubMed] [Google Scholar]
- 6.Kamisugi Y, Cuming AC, Cove DJ. Parameters determining the efficiency of homologous recombination mediated gene targeting in the moss Physcomitrella patens. Nucleic Acids Res. 2005;33: e173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamisugi Y, Cuming AC. Gene targeting In. Perroud P-F, Cove DJ, Knight CD, editors. The moss Physcomitrella. Annual Plant Reviews Vol. 36 Blackwell Publishing Ltd; 2009;pp. 76–112. [Google Scholar]
- 8.Terada R, Johzuka-Hisatomi Y, Saito M, Asao H, Iida S. Gene targeting by homologous recombination as a biotechnological tool for rice functional genomics. Plant Physiol. 2007;144: 846–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, et al. High frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459: 442–445. 10.1038/nature07845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiml S, Puchta H. Revolutionizing plant biology: multiple ways of genome engineering by CRISPR/Cas. Plant Methods 2016;12: 8 10.1186/s13007-016-0103-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Britt AM, May GD. Re-engineering plant gene targeting. Trends Plant Sci. 2003;8: 90–95. [DOI] [PubMed] [Google Scholar]
- 12.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2006;8: 37–45. [DOI] [PubMed] [Google Scholar]
- 13.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77: 229–257. 10.1146/annurev.biochem.77.061306.125255 [DOI] [PubMed] [Google Scholar]
- 14.Sfeir A, Symington LS. Microhomology-Mediated End Joining: A back-up survival mechanism or dedicated pathway? Trends Biochem. Sci. 2015; 40, 701–714. 10.1016/j.tibs.2015.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orr-Weaver TL, Szostak JW, Rothstein RJ. Yeast transformation: a model system for the study of recombination. Proc. Natl. Acad. Sci. USA. 1981;78: 6354–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaefer DG. A new moss genetics: targeting mutagenesis in Physcomitrella patens. Annu. Rev. Plant Biol. 2002;53: 477–501. [DOI] [PubMed] [Google Scholar]
- 17.von Stackelberg M., Rensing S.A. and Reski R. Identification of genic moss SSR markers and a comparative search in twenty-four algal and plant gene indices reveal species-specific rather than group-specific characteristics of microsatellites. BMC Plant Biology. 2006;6: 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knight CD, Cove DJ, Cuming AC, Quatrano RS. Moss gene technology In: Gilmartin PM, Bowler C, editors. Molecular Plant Biology Vol. 2: Oxford University Press; 2002;pp. 285–299. [Google Scholar]
- 19.Kamisugi Y, Schaefer DG, Kozak J, Charlot F, Vrielynk N, Hola M, et al. MRE11 and RAD50, but not NBS1, are essential for DNA repair and gene targeting in the moss Physcomitrella patens. Nucleic Acids Res. 2012; 40: 3496–3510. 10.1093/nar/gkr1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11: 36–42. [Google Scholar]
- 21.Schaefer DG, Delacote F, Charlot F, Vrielynck N, Guyon-Debast A, Le Guin S. RAD51 loss of function abolishes gene targeting and de-represses illegitimate integration in the moss Physcomitrella patens. DNA Repair 2010;9: 526–533. 10.1016/j.dnarep.2010.02.001 [DOI] [PubMed] [Google Scholar]
- 22.Futers TS, Onde S, Turet M, Cuming AC. Sequence analysis of 2 tandemly linked Em genes from wheat. Plant Mol. Biol. 1993;23: 1067–1072. [DOI] [PubMed] [Google Scholar]
- 23.Taylor RM, Cuming AC. Selective proteolysis of the wheat Em polypeptide - identification of an endopeptidase activity in germinating wheat embryos. FEBS Lett. 1993;331: 71–75. [DOI] [PubMed] [Google Scholar]
- 24.Trapnell C, Patcher L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25: 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7: 986–995. [DOI] [PubMed] [Google Scholar]
- 28.Benjamini Y, and Hochberg Y. Controlling the False Discovery Rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B, 1995;57: 289–300. [Google Scholar]
- 29.Markmann-Mulisch U, Wendeler E, Schween G, Steinbiss H-H, Reiss B. Differential requirements for RAD51 in Physcomitrella patens and Arabidopsis thaliana development and DNA damage repair. Plant Cell. 2007;19: 3080–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clerici M, Mantiero D, Lucchini G, Longhese MP. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J. Biol. Chem. 2005;280: 38631–38638. [DOI] [PubMed] [Google Scholar]
- 31.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, et al. Human CtIP promotes DNA end resection. Nature 2007;450: 509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yun MH, Hiom K. 2009. CtIP-BRCA1 modulates the choice of DNA double-strand break repair pathway throughout the cell cycle. Nature 459;460–463. 10.1038/nature07955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inagaki S, Suzuki T, Ohto M, Urawa H, Horiuchi T, Nakamura K, et al. Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell. 2006;18: 879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inagaki S, Nakamura K, Morikami A. A link among DNA replication, recombination and gene expression revealed by genetic and genomic analysis of TEBICHI gene of Arabidopsis thaliana. PLoS Genetics 2009;5: e1000613 10.1371/journal.pgen.1000613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahel D, Horejsi Z, Wiechens N, Polo S, Garcia-Wilson E, Ahel I, et al. Poly(ADP-ribose)–dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325: 1240–1243. 10.1126/science.1177321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogas J, Kaufmann S, Henderson J, Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA. 1999;96: 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaked H, Avivi-Ragolsky N, Levy AA. Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics. 2006;173: 985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AM. ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 2006;48: 947–961. [DOI] [PubMed] [Google Scholar]
- 39.Antony E, Tomko EJ, Xiao Q, Krecji L, Lohman TM, Ellenberger T. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Molec. Cell. 2009;35: 105–115. 10.1016/j.molcel.2009.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colavito S, Macris-Kiss M, Seong C, Gleeson O, Greene EC, Klein HL, et al. Functional significance of the Rad51-Srs2 complex in Rad51 presynaptic filament disruption. Nucleic Acids Res. 2009;37: 6754–6764. 10.1093/nar/gkp748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charlot F, Chelysheva L, Kamisugi Y, Vrielynk N, Guyon A, Epert A, et al. RAD51B plays an essential role during somatic and meiotic recombination in Physcomitrella. Nucleic Acids Res. 2014;42: 11965–11978. 10.1093/nar/gku890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trouiller B, Schaefer DG, Charlot F, Nogué F. MSH2 is essential for the preservation of genome integrity and prevents homeologous recombination in the moss Physcomitrella patens. Nucleic Acids Res. 2006;34, 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krogh BO, Symington LS. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38: 233–271. [DOI] [PubMed] [Google Scholar]
- 44.Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 2009;284: 9558–9565. 10.1074/jbc.M808906200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peterson SE, Li Y, Wu-Baer F, Chait BT, Baer R, Yan H, et al. Activation of DSB processing requires phosphorylation of CtIP by ATR. Molec. Cell. 2013;49: 657–667. 10.1016/j.molcel.2012.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colaiacovo MP, Paques F, Haber JE. Removal of one nonhomologous DNA end during gene conversion by a RAD1- and MSH2-independent pathway. Genetics. 1999;151: 1409–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dubest S, Gallego ME, White CI. Role of the AtRad1p endonuclease in homologous recombination in plants. EMBO Rep. 2002;3: 1049–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woodhouse BC, Dianov GL. PolyADP-ribose polymerase-1: an international molecule of mystery. DNA Repair. 2008;7: 1077–1086. 10.1016/j.dnarep.2008.03.009 [DOI] [PubMed] [Google Scholar]
- 49.Roerink SF, van Schendel R, Tijsterman M. Polymerase theta-mediated end joining of replication-associated DNA breaks in C. elegans. Genome Res. 2014;24: 954–962. 10.1101/gr.170431.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamisugi Y, Schlink K, Rensing SA, Schween G, von Stackelberg M, Cuming AC, et al. The mechanism of gene targeting in Physcomitrella patens: homologous recombination, concatenation and multiple integration. Nucleic Acids Res. 2006;34: 6205–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Plessis A, Dujon B. 1993. Multiple tandem integrations of transforming DNA sequences in yeast chromosomes suggest a mechanism for integrative transformation by homologous recombination. Gene. 134;41–50. [DOI] [PubMed] [Google Scholar]
- 52.Kai M. Role of the Checkpoint Clamp in DNA Damage Response. Biomolecules 2013;3: 75–84. 10.3390/biom3010075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cuming AC, Cho SH, Kamisugi Y, Graham H, Quatrano RS. Microarray analysis of transcriptional responses to Abscisic acid and osmotic, salt, and drought stress in the moss, Physcomitrella patens. New Phytol. 2007;176: 275–287. [DOI] [PubMed] [Google Scholar]
- 54.Frank W, Ratnadewi D, Reski R. Physcomitrella patens is highly tolerant against drought, salt and osmotic stress. Planta 2005;220: 384–394. [DOI] [PubMed] [Google Scholar]
- 55.Cosgrove DJ. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005;6: 850–861 [DOI] [PubMed] [Google Scholar]
- 56.Thompson JE, Fry SC. Restructuring of wall-bound xyloglucan by transglycosylation in living plant cells. Plant Journal. 2001;26: 23–34. [DOI] [PubMed] [Google Scholar]
- 57.Feng JX, Liu D, Pan Y, Gong W, Ma LG, Luo JC, et al. An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Molec. Biol. 2005;59: 853–868. [DOI] [PubMed] [Google Scholar]
- 58.Dixon RA, Paiva NL. Stress-induced phenylpropanoid metabolism. Plant Cell 1995;7: 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jin H, Martin CR. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 1999;41: 577–585. [DOI] [PubMed] [Google Scholar]
- 60.Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes Plant Mol. Biol 2005;57: 155–171. [DOI] [PubMed] [Google Scholar]
- 61.Chen IP, Haehnel U, Altschmied L, Schubert I, Puchta H. The transcriptional response of Arabidopsis to genotoxic stress–a high-density colony array study (HDCA). Plant J. 2003;35: 771–786. [DOI] [PubMed] [Google Scholar]
- 62.Kim YS. Analysis of gene expression upon DNA damage in Arabidopsis Plant Biology 2005;49: 298–302. [Google Scholar]
- 63.West CE, Waterworth WM, Bray CM. Arabidopsis DNA double-strand break repair pathways. Biochem. Soc. Trans. 2004;32: 964–966. [DOI] [PubMed] [Google Scholar]
- 64.Manuss A, Dukowicz-Schulze S, Hartung F, Pacher M, Puchta H. RAD5A, RECQ4A, and MUS81 have specific functions in homologous recombination and define different pathways of DNA repair in Arabidopsis thaliana. Plant Cell. 2010;22: 3318–3330. 10.1105/tpc.110.078568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gottschalk AJ, Trivedi RD, Conaway JW, Conaway RC. Activation of the SNF2 family ATPase ALC1 by Poly(ADP-ribose) in a stable nucleosome-PARP1-ALC1 intermediate. J. Biol. Chem. 2012;287: 43527–43532. 10.1074/jbc.M112.401141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Recker J, Knoll A, Puchta H. 2014. The Arabidopsis thaliana homolog of the helicase RTEL1 plays multiple roles in preserving genome stability. Plant Cell. 26: 4889–4902.67. 10.1105/tpc.114.132472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu Z, Cools T, Kalhorzadeh P, Heyman J, De Veylder L. Deficiency of the Arabidopsis helicase RTEL1 triggers a SOG1-dependent replication checkpoint in response to DNA cross-Links. Plant Cell 2015;27: 149–161. 10.1105/tpc.114.134312 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
A: Schematic of gene structure and knockout construct. B: Identification of targeted loci by PCR amplification with cassette-specific “outward” and gene-specific “inward” primers C: Identification of single-copy targeted transformants with external gene-specific primers D: Southern blot (HindIII digest) to identify transformants containing only a single, targeted selection cassette. In S1–S7 Figs, Red boxes = protein-coding sequence, Blue boxes = 5’- and 3’-UTRs, Green boxes = NPTII selection cassette, Red triangles = LoxP sites. In Southern blots, the NPTII sequence was used as a probe.
(PDF)
A: Schematic of gene structure and knockout construct. B: Identification of targeted loci by PCR amplification with cassette-specific “outward” and gene-specific “inward” primers. C: Identification of single-copy targeted transformants with external gene-specific primers (track “P” = plasmid control). D: Southern blot (EcoRI digest) to identify transformants containing only a single, targeted selection cassette.
(PDF)
A: Schematic of gene structure and knockout construct. B: Identification of targeted loci by PCR amplification with cassette-specific “outward” and gene-specific “inward” primers. C: Identification of single-copy targeted transformants with external gene-specific primers (Track “P” = plasmid control). D: Southern blot (HindIII digest) to identify transformants containing only a single, targeted selection cassette. (Analysed on same gel as PptebKO: see S2 Fig for WT control).
(PPTX)
A: Schematic of gene structure and knockout construct. B: Identification of targeted loci by PCR amplification with cassette-specific “outward” and gene-specific “inward” primers C: Identification of single-copy targeted transformants with external gene-specific primers (Tracks “P” and “wt” = plasmid and wild-type genomic DNA controls) D: Southern blot (HindIII digest) to identify transformants containing only a single, targeted selection cassette. Analysed on same gel as Ppchd5-KO: see S5 Fig for wild-type control).
(PPTX)
A: Schematic of gene structure and knockout construct. B: Identification of targeted loci by PCR amplification with cassette-specific “outward” and gene-specific “inward” primers C: Identification of single-copy targeted Transformants with external gene-specific primers (Tracks “P” and “wt” = plasmid and wild-type genomic DNA controls). D: Southern blot (BglII digest) to identify transformants containing only a single, targeted selection cassette.
(PPTX)
A: Schematic of gene structure and knockout construct. B: Identification of targeted loci by PCR amplification with cassette-specific “outward” and gene-specific “inward” primers. C: Identification of single-copy targeted transformants with external gene-specific primers (Track “P” = plasmid control). D: Southern blot (EcoRV digest) to identify transformants containing only a single, targeted selection cassette.
(PPTX)
A: Schematic of gene structure and knockout construct. B: Identification of targeted loci by PCR amplification with cassette-specific “outward” and gene-specific “inward” primers C: Identification of single-copy targeted transformants with external gene-specific Primers primers (Tracks “P” and “wt” = plasmid and wild-type genomic DNA controls). D: Southern blot (BglII digest) to identify transformants containing only a single, targeted selection cassette
(PPTX)
(PPTX)
Each photograph represents the final time-point presented in the growth test analysis in Fig 4. For Ppteb-KO the growth on 40ng.ml-1 bleomycin is shown. For all other mutants, growth on 200ng.ml-1 bleomycin is shown.
(PPTX)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
The complete transcriptomic dataset has been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo) as Accession GSE25237, and in the NCBI SRA as accessions SRX031155 and SRX031156. Alignment of all sequence reads with the Physcomitrella genome can be viewed on the Cosmoss genome browser (http://www.cosmoss.org).