Abstract
Phosphatidylinositol-5-phosphate (PtdIns5P) 4-kinase beta (PIP4K2B) directly regulates the levels of two important phosphoinositide (PI) second messengers PtdIns5P and phosphatidylinositol-(4,5)-bisphosphate (PtdIns(4,5)P2). PIP4K2B has been linked to the regulation of gene transcription, TP53 and AKT activation and to the regulation of cellular reactive oxygen accumulation. However its role in human tumour development and on patient survival is not known. Here we have interrogated the expression of PIP4K2B in a cohort (489) of breast tumour patients using IHC staining and by a meta-analysis of gene expression profiles from 2999 breast tumours, both with associated clinical outcome data. Low PIP4K2B expression was associated with increased tumour size, high Nottingham histological grade (NHG), Ki67 expression and distant metastasis, while high PIP4K2B expression strongly associated with ERBB2 expression. Kaplan Meier curves showed that both high and low PIP4K2B expression correlated with poorer patient survival compared to intermediate expression. In normal (MCF10A) and tumour (MCF7) breast epithelial cell lines, mimicking low PIP4K2B expression, using sh-RNAi mediated knockdown, led to a decrease in the transcription and expression of the tumour suppressor protein E-cadherin (CDH1). In MCF10A cells knockdown of PIP4K2B also enhanced TGFβ induced epithelial to mesenchymal transition (EMT), a process required during the development of metastasis. Analysis of gene expression datasets confirmed the association between low PIP4K2B and low CDH1expression. Decreased CDH1 expression and enhancement of TGFβ-induced EMT by reduced PIP4K2B expression might in part explain the association between low PIP4K2B expression and poor patient survival.
Keywords: Phosphoinositides, E-cadherin, Breast Cancer, Phosphatidylinositol-5-phosphate, Phosphatidylinositol-5-phosphate-4-kinase, PIP4K2B
Introduction
Alterations in components of various signalling pathways can lead to the generation of neoplasms derived from breast epithelium which through the acquisition of further somatic mutations and epigenetic changes develop into malignant tumours which can metastasise and invade and colonise new tissues eventually leading to patient demise. The acquisition of invasive characteristics is often associated with a decrease in the level of the tumour suppressor CDH1 and a subsequent decrease in cell-cell adhesion and increase in cell motility and degradation of the stromal matrix. CDH1 is an epithelial Ca+-dependent adhesion molecule which facilitates cell-cell contact through homophilic interactions with adjacent CDH1 molecules expressed on neighbouring cells (1;2). Loss of CDH1 contributes to invasiveness and the development of metastasis (3) and can occur through mutation, gene deletion, silencing of its promoter and by proteolytic degradation. CDH1 loss facilitates and is also a hall mark of EMT (4). EMT frequently occurs during normal development and is coordinated by transcriptional regulators such as the Snail and Twist family, Goosecoid and members of the ZFH family (zinc-finger homeodomain, ZEB1 and ZEB2) (5). Characteristics of EMT are normally associated with metastatic cells (6;7). Extracellular factors such as TGFβ, which suppresses early tumour development, can also induce EMT at later stages to enhance tumour metastasis.
Phosphoinositides are signalling lipids whose levels are regulated by an array of kinases and phosphatases (8). PtdIns(4,5)P2 is a substrate for both receptor stimulated phospholipase C (PLC) and PtdIns-3-kinase family members and also acts as a signalling moiety itself by recruiting and regulating the activity of specific interacting proteins (8). In breast tumours PtdIns(3,4,5)P3 signalling is enhanced by gain of function mutations in PIK3CA (30%) and loss of function or deletion negative regulators such as PTEN and INPP4A (9;10). Up-regulation of the PI-3-kinase pathway is associated with resistance to therapeutic ERBB2 inhibitors (11) and PI-3-kinase inhibitors sensitise triple negative breast tumour cells to PARP inhibition (12). Increased PI-3-kinase signalling also decreases CDH1 mRNA and protein levels (13). PtdIns(4,5)P2 signalling can also impact on breast cancer. Two families of lipid kinases generate PtdIns(4,5)P2. Three isoforms of PIP5K, 1A, 1B and 1C regulate the bulk synthesis of PtdIns(4,5)P2 and modulate membrane trafficking, cytoskeletal dynamics and focal adhesion turnover (14). The PIP5K1C splice variant, PIP5K1γi2, interacts with talin, localises to focal adhesions and regulates integrin activation (15;16). PIP5Kγi2 also regulates trafficking and expression of CDH1 (17) and high expression in breast tumours is associated with poor clinical prognosis (18). Also loss of Breast cancer metastasis suppressor 1 (BRMS1) is associated with reduced survival of subsets of breast tumour patients (19), increased metastasis (20) and interestingly increases PIP5K expression and PtdIns(4,5)P2 synthesis (21). PIP4Ks phosphorylate PtdIns5P on the 4-position to generate PtdIns(4,5)P2 and in vivo they regulate cellular pools of both PtdIns5P (22;23) and PtdIns(4,5)P2. The three isoforms of PIP4K, 2A (24;25), 2B (26), and 2C have distinct subcellular localisations; PIP4K2A is predominantly cytosolic (27); PIP4K2B localises in the plasma membrane, the cytoplasm and in nuclear speckles (27) and PIP4K2C localises to endo-membranes (28).
PIP4K2B regulates AKT activation (29;30), reactive oxygen accumulation (31) and in the nucleus controls PtdIns5P levels (22), which can regulate gene transcription (32;33). PIP4K2B is also required for vitamin-D3 induced CDH1 transcription (34). PIP4K2B is highly expressed in several breast cancer cell-lines such as UACC-812, BT474 and T47D, and the gene encoding PIP4K2B, which is located at 17q21.2 can be amplified as part of the ERBB2 amplicon (35).
Here using two independent methodologies we show that both high and low expression compared to intermediate PIP4K2B expression levels are associated with poor patient survival. High PIP4K2B expression was associated with high ERBB2 expression, while low PIP4K2B expression correlated with increased tumour size, proliferation and distant metastasis.
Suppression of PIP4K2B in MCF7 and in MCF10A led to decreased transcription and expression of CDH1 and increased TGFβ induced EMT. PIP4K2B signalling represents a novel pathway to regulate CDH1 expression and TGFβ induced EMT, which might in part underlie the poorer survival observed in patients with low PIP4K2B expression.
Materials and Methods
Cell lines and reagents
Cell lines (HEK293, MCF7 and MCF10A) were obtained from the American Type Culture Collection and cultured as recomended. TGFβ was from R&D systems. pGL3-E-cadherin-luciferase was a gift from J.A. Martignetti (Mount Sinai School of Medicine, New York, USA). The lentiviral vector pLKO.1 containing shRNA targeting sequences were used to knock down PIP4K2B in MCF7 and MCF10A cells. Viral particles were generated in HEK293FT cells using pLKO based vectors and plasmids encoding GAG-Pol and VSVG (4:2:1, respectively). Cells were transduced in the presence of polybrene (5μg/ml) and selected using puromycin (2μg/ml).
Patients and tumour samples
The TMA used included tumour cores from 489 consecutive breast cancer cases consisting mainly of ductal and lobular tumours diagnosed at the Department of Pathology, Malmö University Hospital, Sweden between 1988 and 1992 (36)
Immunocytochemistry
For immunocytochemistry HEK293 cells were harvested, washed in PBS and fixed for 4 hours in 4% paraformaldehyde. Cell pellets were dehydrated in a graded ethanol series and embedded in paraffin. The TMA slides were deparaffinised, rehydrated and microwave-treated in target retrieval solution citrate buffer (10mM, pH 6.0), Sections were incubated with the indicated antibodies and were visualised using DAB. The PIP4K2BP6 antibody was used at 1:500, while the anti-ERBB2 antibody was used according to manufactures instructions (Pathway CB-USA, 760-2694).
PIP4K2B staining intensities were subdivided into six categories (groups 0, 1, 2, 3, 4 and 5) and for further statistical analysis we combined groups 0 and 1, groups 2 and 3 and groups 4 and 5 to generate three intensity groupings. ERBB2 staining was scored semi-quantitatively by the intensity and percentage of staining as 0 and 1+ negative, 2+ equivocal and 3+ as positive. Evaluation was performed by two independent observers (one a pathologist), with the pathologist’s score superseding the other observer’s at consolidation. Conflicting observations were low (< 5%) for all evaluations made. All immunohistochemical evaluations were performed blind and randomised without prior knowledge of tumour characteristics.
Meta-analysis of gene expression data from primary breast tumours and breast cell lines
Raw .cel files from seventeen Affymetrix U133A/plus 2 primary breast tumour and three cell line gene expression datasets were downloaded from NCBI GEO (GSE12276, GSE21653, GSE3744, GSE5460, GSE2109, GSE1561, GSE17907, GSE2990, GSE7390, GSE11121, GSE16716, GSE2034, GSE1456, GSE6532, GSE3494, GSE10890, GSE12777), ArrayExpress (E-TABM-194) or caBIG (geral-00143) repositories, summarised with Ensembl alternative CDF (37), and normalised with RMA (38), before integrating using ComBat to remove dataset-specific bias as previously described (39). The intrinsic molecular subtypes were assigned for each dataset separately based upon the highest correlation to those defined in Sorlie et al. (40;41). Centred average linkage clustering of the integrated tumour datasets was performed using the Cluster and TreeView programs. Survival curves related to PIP4K2B expression were generated without prior knowledge of the data generated in the IHC study.
Cell viability/proliferation assay
MCF7 cells were plated (3000 cells/well) in 96-well plates and viability/proliferation was monitored 1, 3 and 5 days later using Alamar Blue reagent (Invitrogen)
Anchorage-dependent and -independent clonogenic growth assays
MCF7 cells were plated at low density (1000 cells/10cm diameter plate) grown for 10 days and colonies were stained with crystal violet (0.5% w/v in PBS-0.5% formaldehyde v/v) and quantified with Image J software. For colony density analysis, colonies were also stained with ethidium bromide (Sigma) and fluorescence and phase-contrast images were quantified with Image J software. Data were presented as number of colonies divided by colony area. Anchorage-independent growth was monitored in soft agar. Images were taken using an Axiocamera camera and colonies were quantified with Image J software.
Western immunoblot analysis
Protein expression was analysed by standard Western blotting procedures employing the following antibodies: PIP4K2BP6 (generated against the peptide CNLLSFPRFFGP), ERBB2 (Labvision Neomarkers), CDH1 (BD), Actin (Chemicon International), Fibronectin (BD), Vimentin (Novo Castro), B-catenin (Cell Signalling Technology) and Tubulin (Sigma).
Luciferase assay
Cells transfected with a vector encoding Renilla luciferase and pGL3-E-cadherin-luciferase were lysed and luciferase was measured using the Dual-GLO® Luciferase System (Promega).
RT-PCR analysis
RNA was isolated with an RNeasy® Plus Mini kit and QIAshredder spin columns (Qiagen). cDNA was generated using the high capacity reverse transcription kit (Applied Biosystems). qPCR assays were performed in MicroAmp® optical 384-well reaction plate (Applied Biosystems) and analysed using an Applied Biosystems 7900HT Sequence Detection System (SDS). Ribosomal protein L32 was used as a loading control.
Results
PIP4K2B antibody characterisation for IHC analysis
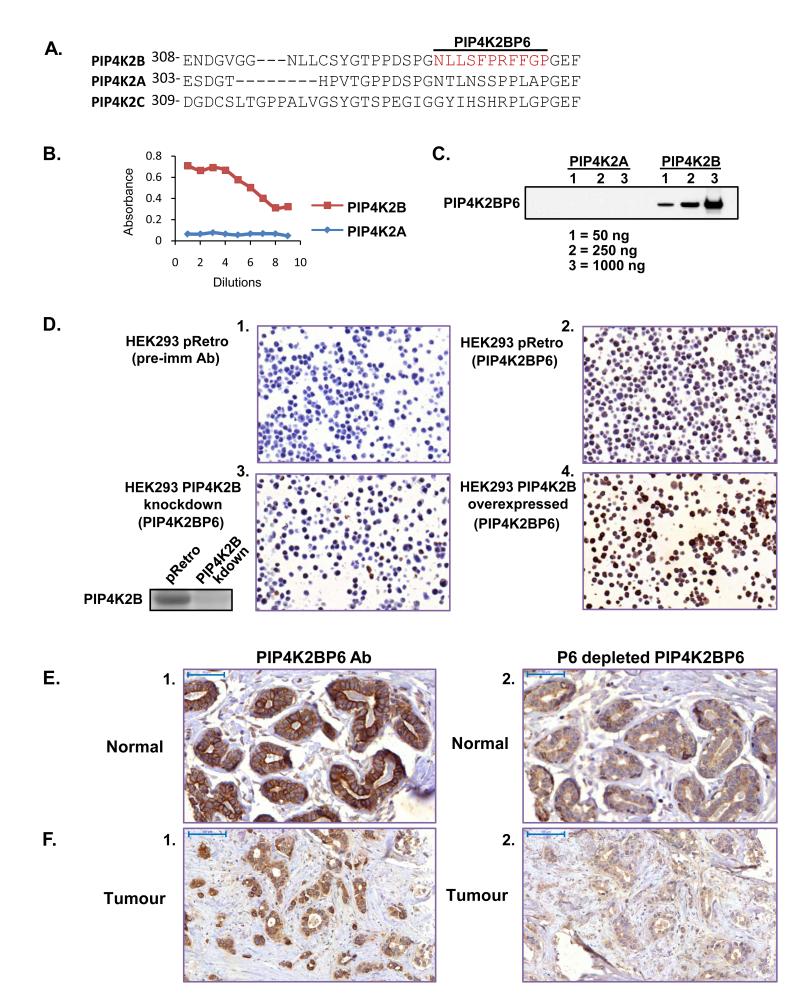
PIP4K2B expression was assessed using an specific antibody (PIP4K2BP6) (27) generated against the peptide sequence NLLSFPRFFGP which is absent in both the 2A and 2C isoforms (Figure 1A). PIP4K2BP6, recognised purified GST-PIP4K2B but not equivalent amounts of GST-PIP4K2A assessed by ELISA (Figure 1B) or by western blotting (Figure 1C) even though 1μg of purified PIP4K2A was loaded on the gels. PIP4K2BP6 recognised a 51KDa protein in total MCF10A lysates, which was absent after PIP4K2B knockdown (Supplementary Figure 1A). PIP4K2BP6 was then used to stain HEK293 cells fixed, embedded and sectioned in a similar manner to tissue sections. To generate control for the staining we suppressed or overexpressed PIP4K2B in cells and also used a pre-immune serum. PIP4K2BP6 strongly stained wild type HEK293 cells compared to the pre-immune serum (Figure 1D panels 1 and 2) and the staining was diminished upon knockdown of endogenous PIP4K2B (Figure 1D panel 3 and insert) and increased on overexpression of PIP4K2B (Figure 1D panel 4). We next stained sections of normal and tumour breast tissue. PIP4K2B was strongly expressed in the luminal epithelial cells of normal breast ducts and acini and was predominantly localised at the plasma membrane. Little staining was observed in the myoepithelial cell layer (Figure 1E panel 1). In tumours, heterogeneous staining was observed: strong plasma membrane staining was often lost and cytosolic and nuclear staining became more apparent (Figure 1F panel 1). Specific immuno-depletion of the antisera (Supplemental Figure 1B) decreased the staining of both normal and tumour tissue (Figure 1E and F panel 2). Heterogeneous cellular localisation of PIP4K2B observed in tumour tissue was also observed in several breast cancer cell-lines (Supplementary Figure 2). These data strongly support the use of the PIP4K2BP6 antibody to interrogate the expression of PIP4K2B in TMA.
Figure 1.
Characterisation of the PIP4K2BP6 antibody. A. Sequence alignment of the region surrounding the epitope (in red) for PIP4K2BP6 in PIP4K2A, PIP4K2B and PIP4K2C. B. ELISA of serial dilutions of PIP4K2BP6 against recombinant PIP4K2B and PIP4K2A protein. C. Western blot of recombinant PIP4K2B and PIP4K2A using PIP4K2BP6. D. HEK293 cells were transduced with a control vector (pRetro panels 1 and 2), or a shRNA construct targeting PIP4K2B (panel 3). The shRNA vector targeting PIP4K2B decreased PIP4K2B protein levels compared to control vector (inset next to panel 3). Cells were also transduced with a construct to overexpress PIP4K2B (panel 4). Cells were fixed and embedded in paraffin and stained with a control pre-immune antibody (panel 1) or the PIP4K2BP6 antibody (panels 2, 3 and 4). The images show that PIP4K2BP6 specifically recognises PIP4K2B. E, F. Normal breast tissue (E) or tumour tissue (F) was stained with PIP4K2BP6 before (panel 1) or after (panel 2) specific depletion of PIP4K2BP6 with the P6 peptide. Scale bars represent 50μm (E) or 100μm (F)
Low PIP4K2B expression correlates with adverse patient survival
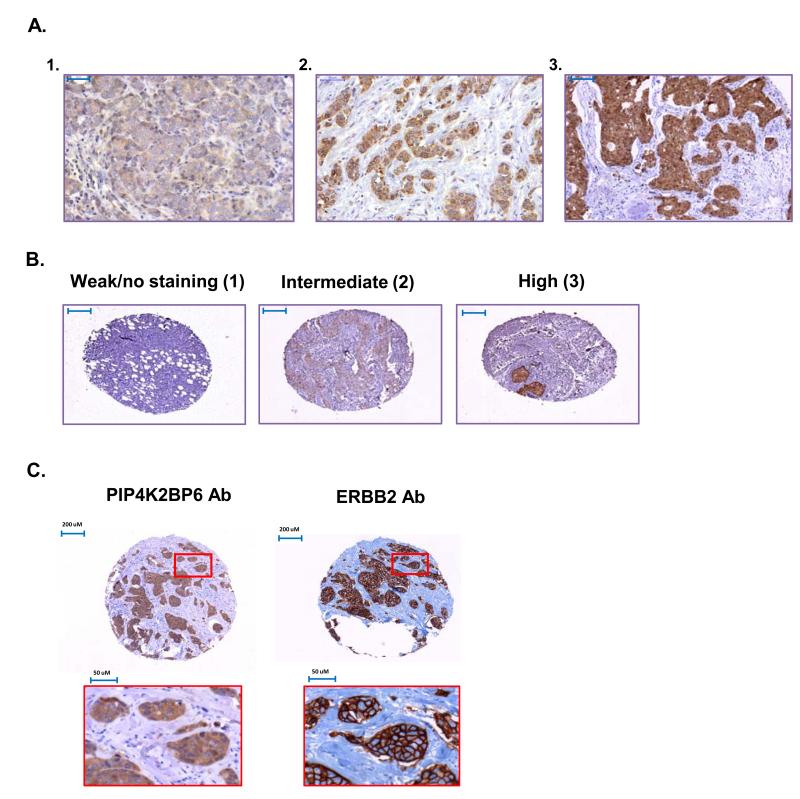
PIP4K2B expression varied dramatically across a TMA containing 489 advanced breast tumour samples (Figure 2A panel 1-3). We categorised PIP4K2B expression into three groups with 1 representing the lowest and 3 the highest intensity (Figure 2B). As the PIP4K2B gene can be co-amplified with the neighbouring ERBB2 gene (35), we also assessed the expression of ERBB2 protein. Figure 2C shows a tumour sample that expressed high levels of both PIP4K2B and ERBB2. ERBB2 staining was strong at the membrane while PIP4K2B appeared more cytosolic. Statistical analysis showed a strong correlation between high ERBB2 expression and high PIP4K2B expression (Table 1), which confirms and extends previous studies (35). We did not observe a significant correlation of high PIP4K2B or high ERBB2 with poor patient survival (Figure 3A).
Figure 2.
Expression of PIP4K2B and ERBB2 in breast tumour samples. A. Examples of typical Low (panel 1), intermediate (panel 2) and high intensity (panel 3) staining patterns observed with PIP4K2BP6. Scale bars represent 50μm (panel 1) 100μM (panels 2 and 3). B. Examples of the scoring for the three different categories of PIP4K2B staining intensities: group 1, weak or no staining; group 2, intermediate staining and group 3, high staining. C. Tumour core showing high expression of both PIP4K2B and ERBB2. The images in the red boxes show higher magnifications.
Table 1. High PIP4K2B expression correlates with ERBB2 expression and low PIP4K2B expression associates with worse clinical parameter.
Correlation between PIP4K2B staining and clinico-pathological and molecular parameters of advanced breast cancer. Significant P values are marked in bold.
| PIP4K2B | P-Chi-Sq | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Variable and categories | 1 | 2 | 3 | Total | P | |
|
| ||||||
| Her 2 | 0 | 188 | 151 | 58 | 397 | 0.000 |
| 2+ | 10 | 13 | 11 | 34 | ||
| 3+ | 15 | 18 | 16 | 49 | ||
| Median age (range) | 65(28-92) | 65(27-96) | 63(34-90) | |||
| 221 | 183 | 85 | 489 | 0.165 | ||
| Nhg | 1 | 43 | 57 | 22 | 122 | 0.007 |
| 2 | 91 | 68 | 44 | 203 | ||
| 3 | 87 | 57 | 19 | 163 | ||
| Size (median) | 19 | 15 | 15 | |||
| 221 | 183 | 85 | 489 | 0.021 | ||
| Lymph node status | 0 | 117 | 109 | 49 | 275 | 0.375 |
| 1 | 79 | 59 | 23 | 161 | ||
| ki67 | 0-10 | 65 | 76 | 34 | 175 | 0.011 |
| 11-25 | 67 | 60 | 28 | 155 | ||
| >25 | 74 | 38 | 16 | 128 | ||
| ER | 0 | 37 | 19 | 13 | 69 | 0.174 |
| 1 | 181 | 161 | 71 | 422 | ||
| PR | 0 | 53 | 30 | 17 | 102 | 0.332 |
| 1 | 162 | 146 | 67 | 375 | ||
| Tumour type | Ductal | 139 | 122 | 63 | 324 | |
| Lobular | 33 | 26 | 11 | 70 | ||
| Distant metastasis | no | 113 | 115 | 53 | 281 | 0.040 |
| during follow-up | yes | 52 | 23 | 13 | 88 | |
| unknown | 54 | 43 | 19 | 116 | ||
Figure 3.
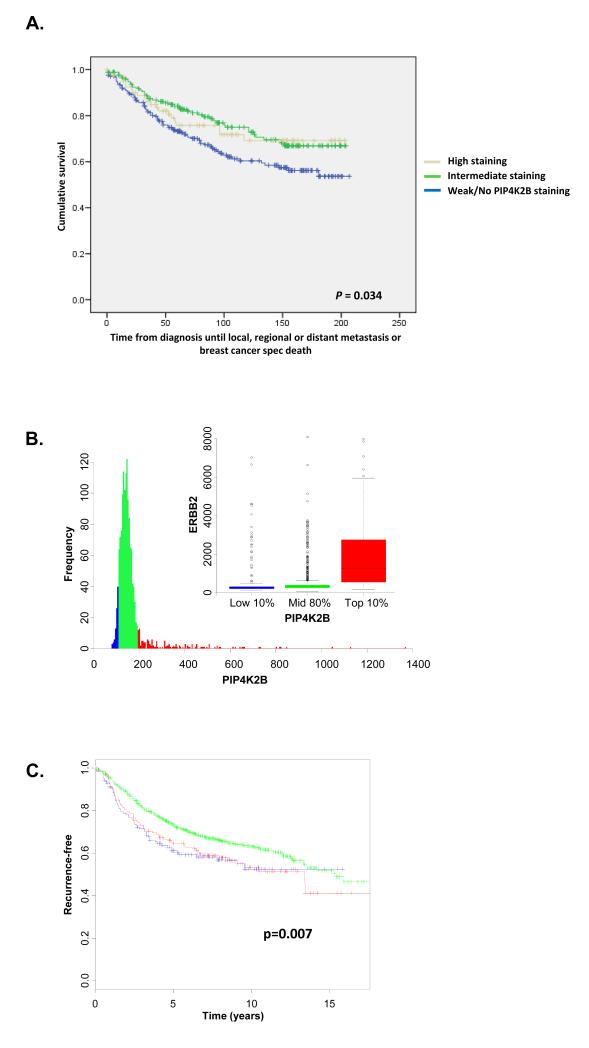
Low expression of PIP4K2B correlates with adverse parameters of tumour progression. A. Kaplan-Meier curve showing the effect of PIP4K2B protein expression levels on event-free survival as a function of time from diagnosis until local, regional or distant metastasis or breast cancer-specific death. Low PIP4K2B expression group 1 correlated with poor patient outcome in contrast to the intermediate and high expression of PIP4K2B (groups 2 and 3), (Log Rank chi-squared = 6.753, P = 0.034).
B. Distribution of PIP4K2B gene expression was assessed across 2999 primary breast cancer tumours integrated from 17 studies. The data are presented as a frequency distribution of PIP4K2B expression. The samples were amalgamated into three groups based on PIP4K2B expression for further analysis: Top 10% (red) middle 80% (green) and the lowest 10% (blue). High expression of PIP4K2B (Red) was clearly associated with high ERBB2 expression. C. Cumulative recurrence-free outcome was plotted for low, middle and high PIP4K2B expression as defined above.
Low PIP4K2B expression showed a significant correlation with increased NHG (P = 0.007, Pearson Chi Square test), tumour size (P = 0.021, Pearson Chi Square test) and expression of Ki67 (P = 0.011, Pearson Chi Square test) (Table 1). PIP4K2B expression did not correlate with ER or PR status, age or lymph node positivity or with histological sub type. Interestingly, low PIP4K2B expression correlated with increased distant metastasis during follow up (P = 0.040, Pearson Chi Square test). Together these data suggest that low PIP4K2B expression correlates with a more malignant grade of tumour and with worse clinico-pathological parameters.
In accordance a Kaplan Meier survival curve showed that low PIP4K2B expression correlated with decreased patient survival (Log Rank chi-squared = 6.753, P = 0.034) defined by time from diagnosis until local, regional or distant metastasis or breast cancer-specific death (Figure 3A).
Meta-analysis of gene expression profiles in breast tumours confirms an association between poor patient outcome and PIP4K2B expression
In order to independently verify the IHC results, we carried out a meta-analysis of gene expression profiles derived from 17 data sets representing 2999 breast tumour samples. Raw gene expression profiles were downloaded and normalised together as described (materials and methods). PIP4K2B expression was then divided into three groups (Figure 3B). As observed in the IHC analysis, high PIP4K2B expression strongly correlated with high ERBB2 expression (Figure 3B inset). The suggestion that they are co-amplified in the majority of tumours is clearly illustrated by plotting the expression of genes surrounding the ERBB2/PIP4K2B region of chromosome 17 and clustering the tumours (Supplementary figure 3A). PIP4K2B is most highly expressed when all the genes between its genomic position and the genomic position of ERBB2 are also overexpressed. Co-amplification of PIP4K2B and ERBB2 was confirmed by examining this region in a high resolution CGH dataset (42)(Supplementary figure 3B). Kaplan Meier survival curves showed that both high and low expression of PIP4K2B is associated with significantly worse prognosis than an intermediate level confirming and extending our IHC studies (Figure 3C). PIP4K2B expression is significantly lower in invasive ductal carcinoma (IDC) compared to normal breast tissue (Supplementary figure 4A) and the lowest PIP4K2B expression is associated with the more aggressive basal subtype compared to the other molecular subtypes (41) (Supplementary figure 4B). As expected, the average PIP4K2B expression was highest in the ERRB2 subtype. Finally in a panel of widely used breast tumour cells the average PIP4K2B expression was significantly lower in the more aggressive basal subtype cells, with the lowest expression being found in the highly aggressive “claudin low” subtype (Supplementary figure 5B). Western blotting confirmed that PIP4K2B expression was low in MCF7 tumour cells compared to the non-cancer cell breast cell line MCF10A and that PIP4K2B expression was highly up-regulated in the ERBB2 overexpressing cell line ZR-75 (Supplementary figure 5C). These data confirm and extend our IHC studies and show that both high and low PIP4K2B expression associate with decreased patient survival compared to intermediate expression.
PIP4K2B knock down in MCF7 cells affects colony formation in anchorage-dependent clonogenicity
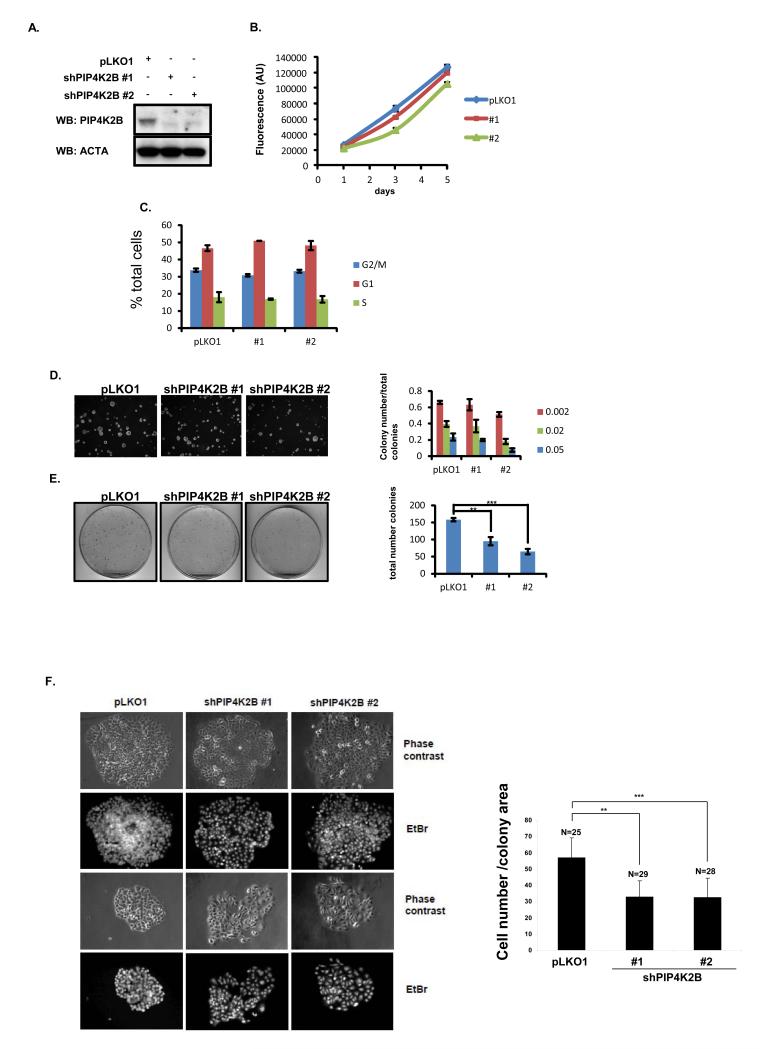
To understand if the association of low PIP4K2B expression with poor patient survival might be functional, using two separate lentiviral-driven shRNAs we knocked down PIP4K2B in MCF7 cells and examined their growth properties. Western blotting demonstrated that both shRNA constructs strongly reduced the level of PIP4K2B protein (Figure 4A). No significant differences between PIP4K2B knockdown cells and control cells were observed in growth rates (Figure 4B), cell cycle distribution (Figure 4C) or anchorage -independent clonogenic growth (Figure 4D). PIP4K2B knockdown, however, reduced the number of colonies observed in anchorage-dependent growth compared to control cells (Figure 4E). Microscopic analysis showed that PIP4K2B knock down colonies appeared less tightly packed than the control colonies. To quantify this, cell nuclei were stained with ethidium bromide and the cell number and the colony area was assessed using image J. The number of cells per colony area was reduced in PIP4K2B knock down cells compared to control cells (Figure 4F). This decrease might reflect changes in cell/cell contact-mediated growth inhibition, or surface area/cell number maintenance mechanisms, or may reflect an increase in cell migratory behaviour.
Figure 4.
PIP4K2B knock down in MCF7 does not affect normal cell growth, cell cycle distribution or growth in soft agar, but decreases anchorage-dependent clonogenic growth. MCF7 cells were transduced with a control lentiviral vector or two different shRNA vectors targeting PIP4K2B (#1 and #2) and then analysed as below. A. Cell lysates were subjected to western blotting to detect PIP4K2B and actin. B, MCF7 cells were plated and viability/proliferation was measured on days 1, 3 and 5 post plating using alamar blue. Data are representative of three experiments and represent the mean ± SD. C. Cells were fixed and labelled with propidium iodide and analysed by FACS. D. MCF7 cells were plated in soft agarose to assess anchorage-independent growth. The graph presents the numbers of three different sizes of colonies (Image J Arbitrary Units, from small 0.002 to big 0.05) divided by the total number of colonies. Data are representative of two experiments performed in triplicate and the data represent the mean ± SD. E. 1000 MCF7 cells were plated in a 10 cm plate and grown for 10 days. Typical images of the colonies are shown and quantification of colony number showed significant differences (** P <0.01, *** P <0.001 (student t-test)) between pLKO and PIP4K2B knockdown constructs (#1 and #2). F. Colony density from E was evaluated by staining cell nuclei with ethidium bromide (EtBr) to assess cell number per colony and phase contrast images were used to determine the colony area The data are presented as the number of cells per colony surface area (Image J, arbitrary units). ** P <0.01, *** P <0.001 (Students t-test).
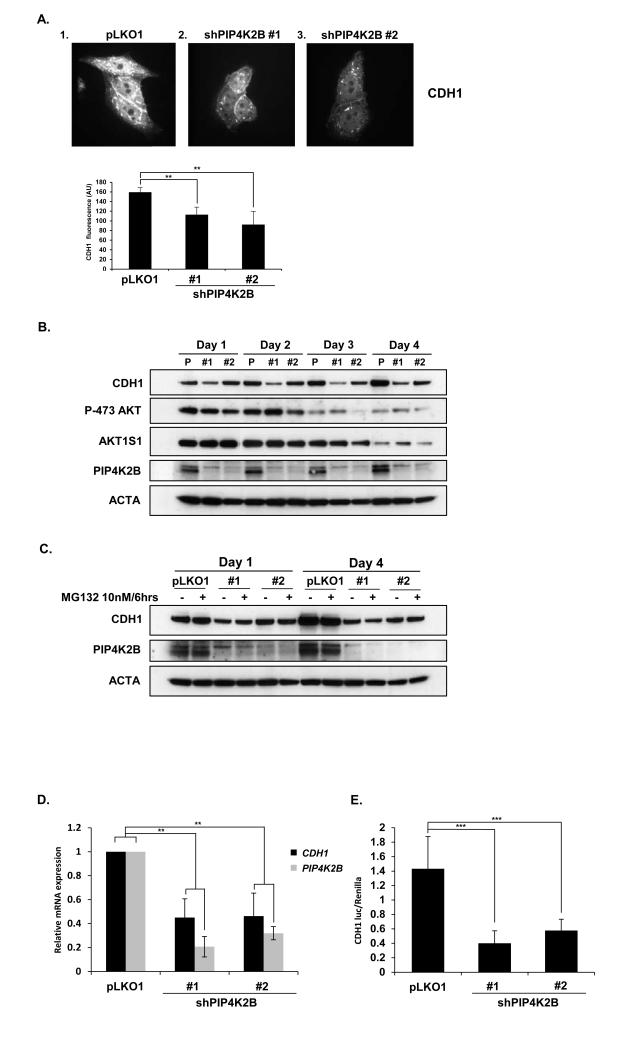
Lentiviral-mediated knock down of PIP4K2B leads to a decrease in CDH1 expression
PtdIns(4,5)P2 levels regulate trafficking of E-cadherin (17), and RNAi-mediated depletion of PIP4K2B decreases vitamin D3 induced expression of CDH1 (34). As the loss of cell-cell contacts and increased migration are often associated with loss of CDH1 (3, 9) and PIP4K2B knockdown cells appeared less tightly packed we investigated whether PIP4K2B knockdown changed the levels of CDH1. Knockdown of PIP4K2B in MCF7 cells reduced total CDH1 staining and staining at cell/cell junctions (Figure 5A). In the non-tumorigenic breast cell line MCF10A, PIP4K2B knock down also led to a reduction in CDH1 levels. shPIP4K2B #1 reduced PIP4K2B more efficiently than shPIP4K2B #2 correlating with the observed difference in inhibition of CDH1 expression (Figure 5B). PIP4K2B can regulate AKT activation which can impact CDH1 expression. However, AKT activation and the phosphorylation of its downstream target AKT1S1 (PRAS40) were not significantly different in PIP4K2B knockdown cells, although AKT activation decreased during the time course of the experiment (Figure 5B). Total AKT levels were not decreased during the four day period in control cells (Supplementary figure 6A). The decrease in CDH1 expression induced by PIP4K2B knockdown was not suppressed by pre-treatment with the proteasome inhibitor MG132 suggesting that ubiquitin mediated proteolytic cleavage of CDH1 is not increased by PIP4K2B knockdown (Figure 5C), although MG132 increased general polyubiquination (Supplementary figure 6B). QRT-PCR analysis demonstrated that PIP4K2B knock down decreased the levels of CDH1 mRNA (Figure 5D) and also decreased CDH1 promoter activity assessed using the CDH1 gene promoter coupled to the synthesis of luciferase (Figure 5E). These data show that PIP4K2B regulates the transcription of CDH1. We next assessed whether the knockdown of PIP4K2B impacted on the levels of cellular phosphoinositides. We observed a small but significant decrease in the levels of PtdIns(4,5)P2. No significant changes in the mass of PtdIns5P were observed (Supplementary figure 7) as measured using a specific mass assay(43;44).
Figure 5.
Lentiviral-mediated knock down of PIP4K2B leads to a decrease in CDH1 expression. A. MCF7 cells transduced with control (1) or with shRNA constructs targeting PIP4K2B (2 and 3) were fixed and immunostained with antibodies to CDH1 and analysed by fluorescence microscopy. Total immunofluorescence staining was determined and is graphically presented. Statistically, differences were determined using Students t-test between control and PIP4K2B knockdown cells (** P <0.01). B. MCF10A cells were transduced with control (pLKO1) or with lentiviral-mediated shRNAs against PIP4K2B (#1 and #2). Cell lysates were immunoprobed with the indicated antibodies. C. Control and PIP4K2B knockdown MCF10A cells were with MG132 as indicated. Cell lysates were western blotted with the indicated antibodies. D. Control and PIP4K2B knockdown MCF10A cells were assessed for the relative expression of PIP4K2B and CDH1 mRNA using qRT-PCR. The data are presented as fold change compared to control. E. MCF10A cells transduced with the indicated shRNAs were transfected with Renilla and E-cadherin-luc. A luciferase activity was measured and the data are presented after correction for the expression of the control Renilla luciferase.
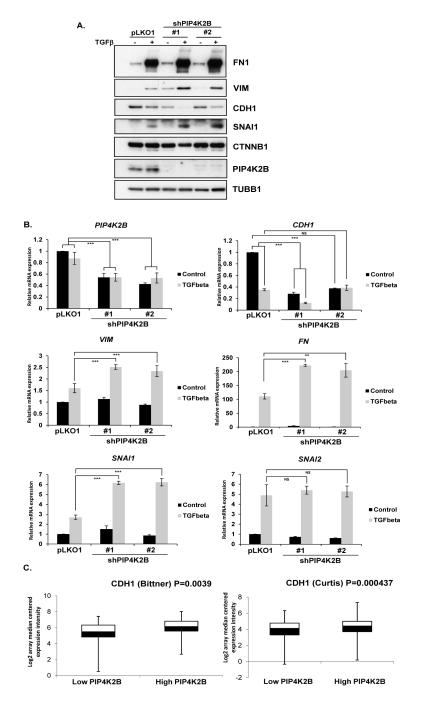
Lentiviral mediated knock down of PIP4K2B induces EMT characteristics in MCF10A cells
Deregulation and loss of CDH1 expression facilitates and is a hallmark of EMT. Therefore we assessed whether PIP4K2B knock down induced the EMT programme or enhanced TGFβ induced EMT. PIP4K2B was knocked down in MCF10A cells and the cells were maintained untreated or treated with TGFβ for 72 hours and cell lysates were analysed for markers of EMT (Figure 6A). Although PIP4K2B knock down decreased the basal level of CDH1 it did not induce the expression of the mesenchymal markers FN1 or VIM (Figure 6A), while TGFβ led to the expected decrease in CDH1 and increase in fibronectin (FN1) and vimentin (VIM).
Figure 6.
Knock down of PIP4K2B enhances EMT in response to TGFβ treatment. A. control (pLKO1) and PIP4K2B knockdown MCF10A cells were treated for 72 hours with TGFβ (10 ng/ml) as indicated . Cell Lysates were blotted with the indicated antibodies. B, RNA was extracted from a similar panel of cells described in A and the mRNA for epithelial (CDH1) or mesenchymal (VIM, FN1) markers or transcription factors that control EMT switching (SNAI1 and SNAI2) were measured using qRT-PCR. Relative mRNA expression was normalised against control cells. C. Expression data for PIP4K2B and CDH1 were downloaded from the Bittner et al array (336 samples) and the Curtis et al (2136 samples) breast cancer studies. The data were sorted for PIP4K2B expression and the CDH1 levels were determined in the bottom (Low) and top (High) 10% of PIP4K2B expressors. Statistical significance was determined using Students t-test and the probabilities are represented on the graphs as ** P <0.01, *** P <0.001.
Treatment of PIP4K2B knockdown cells with TGFβ further decreased CDH1 levels and augmented the increase in the mesenchymal markers FN1 and VIM compared to TGFβ treatment of control cells. SNAI1 is a master transcriptional regulator of the EMT programme and is induced by TGFβ treatment. PIP4K2B knockdown alone did not induce SNAI1 levels but augmented TGFβ-induced SNAI1 levels (Figure 6A). TGFβ induces EMT through regulating gene transcription programmes and therefore we assessed whether knockdown of PIP4K2B augmented TGFβ1 induced transcriptional regulation (Figure 6B). PIP4K2B knockdown decreased basal levels of CDH1 mRNA but did not increase VIM, FN1 and SNAI1 mRNA.
TGFβ1 treatment also reduced CDH1 mRNA and increased VIM, FN1 and SNAI1 expression. In accordance with the western blotting data, PIP4K2B knock down augmented increases in TGFβ-induced VIM, FN1 and SNAI1 mRNA. PIP4K2B knock down did not significantly change the expression of Slug (SNAI2) before or after treatment with TGFβ. These data suggest that although PIP4K2B regulates CDH1 expression, it does not induce the EMT programme. The data are consistent with a role for PIP4K2B in regulating the sensitivity of cells to TGFβ.
Reduced CDH1 expression is associated with more aggressive tumours and might in part explain poorer survival of patients with tumours that have low PIP4K2B expression. We therefore interrogated CDH1 and PIP4K2B expression levels in the unpublished Bittner et al breast tumour array and the published Curtis et al METABRIC study (45) using Oncomine. The data were sorted with respect to PIP4K2B expression and the CDH1 levels were determined in the top and bottom 10% of the PIP4K2B expressing population. We observed a significant decrease in the expression levels of CDH1 in the low PIP4K2B expressing tumours compared to the high PIP4K2B expressing populations in both data sets (P = 0.0039 in the Bittner array and P = 0.000437 in the Curtis array, Figure 6C). A similar trend was also observed in the tumour data sets used in this study. No significant changes were observed in SNAI1 or TWIST1, two upstream repressors of CDH1 expression.
Discussion
We describe the characterisation and use of a specific PIP4K2B antibody to interrogate its expression in a TMA of breast tumour samples, enabling the correlation between PIP4K2B expression and clinico-pathological parameters. The protein expression studies and their outcomes were supported by an independent meta-analysis of gene expression profiles from seventeen previously published studies representing 2999 breast tumours. We found that both high and low expression of PIP4K2B associates with poor survival compared to intermediate expression. High PIP4K2B levels strongly correlate with high ERBB2 expression which is likely to explain the associated poor patient survival. We discovered that low expression of PIP4K2B induced by shRNA in normal and breast tumour cell lines leads to a decrease in the expression of the tumour suppressor CDH1 and enhances TGFβ–induced EMT.
High PIP4K2B expression strongly correlated with high ERRB2 expression, and genomic analysis of a published breast cancer dataset (42) confirmed that PIP4K2B can be co-amplified with ERBB2 (35). The association between high PIP4K2B expression and decreased patient survival was only observed in the analysis of gene expression profiles and not in the TMA. It is unclear why the established association between expression of ERBB2 and poor patient survival was not evident in the TMA but is likely due to different patient cohorts. Whether high expression of PIP4K2B affects patient survival independently of its association with high ERBB2 expression remains to be determined. PIP4K2B can associate with members of the EGF receptor family (46), and therefore might play a role in regulating their activity during breast cancer development. Interestingly Luoh et al (35) observed one tumour with PIP4K2B but not ERRB2 amplification . Although we did not specifically study gene amplification, 58 tumours in the TMA showed strong PIP4K2B expression in the absence of ERRB2 expression.
IHC analysis and analysis of gene expression profiles both showed that low PIP4K2B expression correlated with increased malignancy and poorer patient outcome related to a greater incidence of metastasis. In accordance PIP4K2B levels were lower in tumours compared with normal breast tissue samples and the lowest levels of PIP4K2B were associated with more aggressive tumours and tumour-derived cell lines. PIP4K2B knockdown had little effect on the growth rate of MCF7 cells under normal conditions or in soft agar but decreased their growth in anchorage-dependent clonogenic assays contrary to what might be expected from the patient survival correlations. PIP4K2B regulates many pathways such as PKB signalling (29;47), reactive oxygen accumulation (31) and p53 activation and apoptosis (33) and the balance of these pathways is likely to underlie the survival of cells clonogenic growth assays. How this might relate to overall patient survival, which is more closely related to the onset of metastasis is unclear. However PIP4K2B knockdown decreased the expression of CDH1, a tumour suppressor that is a critical regulator of cell/cell adhesion and who’s loss can drive invasive carcinoma development in mice (3;4;48) and is associated with high grade tumours and poor prognosis (49;50).
How does PIP4K2B regulate CDH1 transcription? PIP4K2B could regulate the levels of both PtdIns(4,5)P2 and PtdIns5P in the nucleus or in the cytoplasm (27),which could impact directly or indirectly on CDH1 expression. In fact we observed a significant decrease in total PtdIns(4,5)P2 levels, without any significant changes in PtdIns5P. Interestingly decreased expression of PIP4K2B suppressed vitamin-D3 induced CDH1 transcription, which was suggested to occur through a decrease the synthesis of nuclear PtdIns(4,5)P2 (34). Whether the our observed changes in PtdIns(4,5)P2 occur in the nucleus, cytoplasm or both remains to be determined. As we did not observe an increase in PtdIns5P levels after knock down of PIP4K2B the decrease in PtdIns(4,5)P2 might be indirect. It should also be noted that PIP4K2B homo and heterodimerises with PIP4K2A and as PIP4K2B has approximately 1000 times less activity than PIP4K2A (27), it is difficult to predict exactly how changes in the expression of PIP4K2B might impact on subcellular pools of PtdIns5P/PtdIns(4,5)P2 . The recent identification of PHD fingers as receptors for nuclear phosphoinositides (32;33) and of nuclear proteins that interact with PtdIns(4,5)P2 (51) might provide plausible pathways for direct transcriptional regulation of CDH1 expression by nuclear phosphoinositides. PIP4K2B has also been implicated in EGF and TNF receptor function (26;52); (46), AKT activity (29), ubiquitination (53), and p38-regulated pathways (22), all of which can also impact CDH1 regulation. In addition it is possible that scaffolding functions of PIP4K2B unrelated to its PIP4K activity or non-coding regions within the mRNA encoding PIP4K2B (54) could be involved in regulating CDH1 expression.
CDH1 expression can be transcriptionally regulated by repressors such as ZEB1, ZEB2, SNAI1 and SNAI2 and TWIST family members and by epigenetic and chromatin remodelling factors such as EZH2(5), which also all induce EMT. Although knockdown of PIP4K2B alone reduced CDH1 it did not induce EMT, and is unlikely to regulate CDH1 expression through those transcriptional repressors. Understanding exactly how PIP4K2B regulates CDH1 expression remains a challenge. EMT is required for metastasis in vivo (55) and can be induced by a number of factors, including TGFβ1. TGFβ1 plays a complex role during tumour development, acting both as an early tumour suppressor and as a late oncogene. Although down regulation of PIP4K2B did not induce EMT, it did enhance TGFβ1-induced expression of mesenchymal markers suggesting a possible function for PIP4K2B in regulating TGFβ sensitivity.
Our data are consistent with an association between PIP4K2B expression and patient survival in breast cancer which might be related to PIP4K2B mediated-regulation of the expression of the tumour suppressor protein CDH1 and increased TGFβ induced EMT.
Supplementary Material
Acknowledgements
We would like to thank Elise Nilsson for excellent technical assistance and Prof.T. Howell for reading and editing the manuscript. We also acknowledge all members of the Inositide Laboratory and Leukaemia Biology for support and reagents and to funding from CRUK. AHS is very grateful for funding from the Breakthrough Breast Cancer.
Footnotes
There is no conflict of interests.
References
- 1.Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991;251:1451–5. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- 2.Baum B, Georgiou M. Dynamics of adherens junctions in epithelial establishment, maintenance, and remodeling. J Cell Biol. 2011;192:907–17. doi: 10.1083/jcb.201009141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perl AK, Wilgenbus P, Dahl U, Semb H, Christofori G. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 1998;392:190–3. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- 4.Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–54. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 5.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di PG, De CP. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–7. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 9.Gewinner C, Wang ZC, Richardson A, Teruya-Feldstein J, Etemadmoghadam D, Bowtell D, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. 2009;16:115–25. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–91. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 12.Ibrahim YH, Garcia-Garcia C, Serra V, He L, Torres-Lockhart K, Prat A, et al. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discov. 2012;2:1036–47. doi: 10.1158/2159-8290.CD-11-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larue L, Bellacosa A. Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene. 2005;24:7443–54. doi: 10.1038/sj.onc.1209091. [DOI] [PubMed] [Google Scholar]
- 14.van dB I, Divecha N. PIP5K-driven PtdIns(4,5)P2 synthesis: regulation and cellular functions. J Cell Sci. 2009;122:3837–50. doi: 10.1242/jcs.056127. [DOI] [PubMed] [Google Scholar]
- 15.Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA. Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature. 2002;420:89–93. doi: 10.1038/nature01082. [DOI] [PubMed] [Google Scholar]
- 16.Di Paolo G, Pellegrini L, Letinic K, Cestra G, Zoncu R, Voronov S, et al. Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the FERM domain of talin. Nature. 2002;420:85–9. doi: 10.1038/nature01147. [DOI] [PubMed] [Google Scholar]
- 17.Ling K, Bairstow SF, Carbonara C, Turbin DA, Huntsman DG, Anderson RA. Type Igamma phosphatidylinositol phosphate kinase modulates adherens junction and E-cadherin trafficking via a direct interaction with mu 1B adaptin. J Cell Biol. 2007;176:343–53. doi: 10.1083/jcb.200606023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y, Turbin DA, Ling K, Thapa N, Leung S, Huntsman DG, et al. Type I gamma phosphatidylinositol phosphate kinase modulates invasion and proliferation and its expression correlates with poor prognosis in breast cancer. Breast Cancer Res. 2010;12:R6. doi: 10.1186/bcr2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hicks DG, Yoder BJ, Short S, Tarr S, Prescott N, Crowe JP, et al. Loss of breast cancer metastasis suppressor 1 protein expression predicts reduced disease-free survival in subsets of breast cancer patients. Clin Cancer Res. 2006;12:6702–8. doi: 10.1158/1078-0432.CCR-06-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seraj MJ, Samant RS, Verderame MF, Welch DR. Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res. 2000;60:2764–9. [PubMed] [Google Scholar]
- 21.DeWald DB, Torabinejad J, Samant RS, Johnston D, Erin N, Shope JC, et al. Metastasis suppression by breast cancer metastasis suppressor 1 involves reduction of phosphoinositide signaling in MDA-MB-435 breast carcinoma cells. Cancer Res. 2005;65:713–7. [PubMed] [Google Scholar]
- 22.Jones DR, Bultsma Y, Keune WJ, Halstead JR, Elouarrat D, Mohammed S, et al. Nuclear PtdIns5P as a transducer of stress signaling: an in vivo role for PIP4Kbeta. Mol Cell. 2006;23:685–95. doi: 10.1016/j.molcel.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 23.Gupta A, Toscano S, Trivedi D, Jones DR, Mathre S, Clarke JH, et al. Phosphatidylinositol 5-phosphate 4-kinase (PIP4K) regulates TOR signaling and cell growth during Drosophila development. Proc Natl Acad Sci U S A. 2013;110:5963–8. doi: 10.1073/pnas.1219333110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boronenkov IV, Anderson RA. The sequence of phosphatidylinositol-4-phosphate 5-kinase defines a novel family of lipid kinases. J Biol Chem. 1995;270:2881–4. doi: 10.1074/jbc.270.7.2881. [DOI] [PubMed] [Google Scholar]
- 25.Divecha N, Truong O, Hsuan JJ, Hinchliffe KA, Irvine RF. The cloning and sequence of the C isoform of PtdIns4P 5-kinase. Biochem J. 1995;309:715–9. doi: 10.1042/bj3090715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castellino AM, Parker GJ, Boronenkov IV, Anderson RA, Chao MV. A novel interaction between the juxtamembrane region of the p55 tumor necrosis factor receptor and phosphatidylinositol-4- phosphate 5-kinase. J Biol Chem. 1997;272:5861–70. doi: 10.1074/jbc.272.9.5861. [DOI] [PubMed] [Google Scholar]
- 27.Bultsma Y, Keune WJ, Divecha N. PIP4Kbeta interacts with and modulates nuclear localization of the high-activity PtdIns5P-4-kinase isoform PIP4Kalpha. Biochem J. 2010;430:223–35. doi: 10.1042/BJ20100341. [DOI] [PubMed] [Google Scholar]
- 28.Clarke JH, Emson PC, Irvine RF. Localization of phosphatidylinositol phosphate kinase IIgamma in kidney to a membrane trafficking compartment within specialized cells of the nephron. Am J Physiol Renal Physiol. 2008;295:F1422–F1430. doi: 10.1152/ajprenal.90310.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carricaburu V, Lamia KA, Lo E, Favereaux L, Payrastre B, Cantley LC, et al. The phosphatidylinositol (PI)-5-phosphate 4-kinase type II enzyme controls insulin signaling by regulating PI-3,4,5-trisphosphate degradation. Proc Natl Acad Sci U S A. 2003;19(100):9867–72. doi: 10.1073/pnas.1734038100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamia KA, Peroni OD, Kim YB, Rameh LE, Kahn BB, Cantley LC. Increased insulin sensitivity and reduced adiposity in phosphatidylinositol 5-phosphate 4-kinase beta−/− mice. Mol Cell Biol. 2004;24:5080–7. doi: 10.1128/MCB.24.11.5080-5087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keune WJ, Jones DR, Bultsma Y, Sommer L, Zhou XZ, Lu KP, et al. Regulation of phosphatidylinositol-5-phosphate signaling by pin1 determines sensitivity to oxidative stress. Sci Signal. 2012;5:ra86. doi: 10.1126/scisignal.2003223. [DOI] [PubMed] [Google Scholar]
- 32.Ndamukong I, Jones DR, Lapko H, Divecha N, Avramova Z. Phosphatidylinositol 5-phosphate links dehydration stress to the activity of ARABIDOPSIS TRITHORAX-LIKE factor ATX1. PLoS One. 2010;5:e13396. doi: 10.1371/journal.pone.0013396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, et al. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- 34.Kouchi Z, Fujiwara Y, Yamaguchi H, Nakamura Y, Fukami K. Phosphatidylinositol 5-phosphate 4-kinase type II beta is required for vitamin D receptor-dependent E-cadherin expression in SW480 cells. Biochem Biophys Res Commun. 2011;408:523–9. doi: 10.1016/j.bbrc.2011.04.045. [DOI] [PubMed] [Google Scholar]
- 35.Luoh SW, Venkatesan N, Tripathi R. Overexpression of the amplified Pip4k2beta gene from 17q11-12 in breast cancer cells confers proliferation advantage. Oncogene. 2004;19(23):1354–63. doi: 10.1038/sj.onc.1207251. [DOI] [PubMed] [Google Scholar]
- 36.Svensson S, Jirstrom K, Ryden L, Roos G, Emdin S, Ostrowski MC, et al. ERK phosphorylation is linked to VEGFR2 expression and Ets-2 phosphorylation in breast cancer and is associated with tamoxifen treatment resistance and small tumours with good prognosis. Oncogene. 2005;24:4370–9. doi: 10.1038/sj.onc.1208626. [DOI] [PubMed] [Google Scholar]
- 37.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 39.Sims AH, Smethurst GJ, Hey Y, Okoniewski MJ, Pepper SD, Howell A, et al. The removal of multiplicative, systematic bias allows integration of breast cancer gene expression datasets - improving meta-analysis and prediction of prognosis. BMC Med Genomics. 2008;1:42. doi: 10.1186/1755-8794-1-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chin SF, Teschendorff AE, Marioni JC, Wang Y, Barbosa-Morais NL, Thorne NP, et al. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol. 2007;8:R215. doi: 10.1186/gb-2007-8-10-r215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones DR, Bultsma Y, Keune WJ, Divecha N. Methods for the determination of the mass of nuclear PtdIns4P, PtdIns5P, and PtdIns(4,5)P2. Methods Mol Biol. 2009;462:75–88. doi: 10.1007/978-1-60327-115-8_5. [DOI] [PubMed] [Google Scholar]
- 44.Clarke JH, Letcher AJ, D’Santos CS, Halstead JR, Irvine RF, Divecha N. Inositol lipids are regulated during cell cycle progression in the nuclei of murine erythroleukaemia cells. Biochem J. 2001;357:905–10. doi: 10.1042/0264-6021:3570905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castellino AM, Chao MV. Differential association of phosphatidylinositol-5-phosphate 4- kinase with the EGF/ErbB family of receptors. Cell Signal. 1999;11:171–7. doi: 10.1016/s0898-6568(98)00056-4. [DOI] [PubMed] [Google Scholar]
- 47.Jones DR, Foulger R, Keune WJ, Bultsma Y, Divecha N. PtdIns5P is an oxidative stress-induced second messenger that regulates PKB activation. FASEB J. 2012;27:1644–56. doi: 10.1096/fj.12-218842. [DOI] [PubMed] [Google Scholar]
- 48.Vleminckx K, Vakaet L, Jr., Mareel M, Fiers W, van RF. Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell. 1991;66:107–19. doi: 10.1016/0092-8674(91)90143-m. [DOI] [PubMed] [Google Scholar]
- 49.Gamallo C, Palacios J, Suarez A, Pizarro A, Navarro P, Quintanilla M, et al. Correlation of E-cadherin expression with differentiation grade and histological type in breast carcinoma. Am J Pathol. 1993;142:987–93. [PMC free article] [PubMed] [Google Scholar]
- 50.Siitonen SM, Kononen JT, Helin HJ, Rantala IS, Holli KA, Isola JJ. Reduced E-cadherin expression is associated with invasiveness and unfavorable prognosis in breast cancer. Am J Clin Pathol. 1996;105:394–402. doi: 10.1093/ajcp/105.4.394. [DOI] [PubMed] [Google Scholar]
- 51.Lewis AE, Sommer L, Arntzen MO, Strahm Y, Morrice NA, Divecha N, et al. Identification of nuclear phosphatidylinositol 4,5-bisphosphate-interacting proteins by neomycin extraction. Mol Cell Proteomics. 2011;10:M110. doi: 10.1074/mcp.M110.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Castellino AM, Chao MV. Differential association of phosphatidylinositol-5-phosphate 4-kinase with the EGF/ErbB family of receptors. Cell Signal. 1999;11:171–7. doi: 10.1016/s0898-6568(98)00056-4. [DOI] [PubMed] [Google Scholar]
- 53.Bunce MW, Boronenkov IV, Anderson RA. Coordinated activation of the nuclear ubiquitin ligase Cul3-SPOP by the generation of phosphatidylinositol 5-phosphate. J Biol Chem. 2008;283:8678–86. doi: 10.1074/jbc.M710222200. [DOI] [PubMed] [Google Scholar]
- 54.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–8. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–4. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.