Abstract
Drosophila females are larger than males. In this article, we describe how X-chromosome dosage drives sexual dimorphism of body size through two means: first, through unbalanced expression of a key X-linked growth-regulating gene, and second, through female-specific activation of the sex-determination pathway. X-chromosome dosage determines phenotypic sex by regulating the genes of the sex-determining pathway. In the presence of two sets of X-chromosome signal elements (XSEs), Sex-lethal (Sxl) is activated in female (XX) but not male (XY) animals. Sxl activates transformer (tra), a gene that encodes a splicing factor essential for female-specific development. It has previously been shown that null mutations in the tra gene result in only a partial reduction of body size of XX animals, which shows that other factors must contribute to size determination. We tested whether X dosage directly affects animal size by analyzing males with duplications of X-chromosomal segments. Upon tiling across the X chromosome, we found four duplications that increase male size by >9%. Within these, we identified several genes that promote growth as a result of duplication. Only one of these, Myc, was found not to be dosage compensated. Together, our results indicate that both Myc dosage and tra expression play crucial roles in determining sex-specific size in Drosophila larvae and adult tissue. Since Myc also acts as an XSE that contributes to tra activation in early development, a double dose of Myc in females serves at least twice in development to promote sexual size dimorphism.
Keywords: Drosophila melanogaster, Myc, tra, dosage compensation, sexual dimorphism
MOST animals display striking sexual dimorphism that evolved for success in sexual reproduction (Hedrick and Temeles 1989; Cline and Meyer 1996; Fairbairn 1997). This involves not only the development of sex-specific structures like genitalia, but also distinct behaviors, pigmentation, and body sizes. In the fruit fly Drosophila melanogaster, as in most species of insects, females attain larger body sizes than males (Darwin 1871; Shine 1990; Stillwell and Davidowitz 2010). Although this aspect of sexual dimorphism in Drosophila has long been appreciated, how the sex-specific difference in body size is genetically controlled is still unknown. In principle, there are two possibilities: genes of the sex-determining pathway might control the sex-specific difference in body size, or the different chromosomal constitution of males (XY) and females (XX) could initiate a process that leads to sex-specific size irrespective of the genes of the sex-determining pathway.
In Drosophila, Sex-lethal (Sxl) is the master switch of the sex-determination pathway, a cascade of sex-determining genes that includes transformer (tra) and doublesex (dsx) (McKeown et al. 1988; Nagoshi et al. 1988; Burtis and Baker 1989; Cline and Meyer 1996). Although Sxl is transcribed in both sexes, only XX animals produce a protein product with defined roles (Bell et al. 1988; Cline 1988; Duffy and Gergen 1991; Torres and Sanchez 1991; Kappes et al. 2011). Sxl activates tra, which ensures that dsx is expressed in the female mode. XX flies lacking tra expression are physically and behaviorally transformed into males, with the exception of their body size. They are larger than their XY brothers (Sturtevant 1945), having adult sex-specific traits that are intermediate in size between male and female traits (Brown and King 1961). Recently it has been reported that XX pupae that are homozygous mutants for tra are 10% larger than XY pupae (Rideout et al. 2015). Control XX females, however, are 30% larger than males. This indicates that tra contributes to male-female differences in body size, but also that tra is not the sole regulator of this trait.
In agreement with the observation that XX animals lacking tra are intermediate in size, XX animals carrying hypomorphic alleles of Sxl are phenotypically male and are smaller than wild-type females (Cline 1984; Evans and Cline 2013). In addition to activating tra, Sxl also controls dosage compensation; a process by which X-chromosomal transcription is equalized in animals where females have two X chromosomes and males have only one (Lucchesi and Kuroda 2015). Due to this essential function, females with homozygous null mutations in Sxl are lethal, which explains the use of viable hypomorphic Sxl alleles that do not completely remove the gene function. Due to this difficulty, it has not yet been possible to distinguish between two alternatives: Sxl might be the sole regulator of sex-specific size acting through multiple genes, one of which is tra. Alternatively, factors that are not controlled by Sxl might contribute to regulation of sex-specific size in concert with Sxl acting through tra.
We wondered whether genes outside the sex-determining pathway participate in regulating sex-specific body size. In particular, we were curious whether X-chromosome gene dosage plays a direct role in determining body size. This idea was based on the fact that, while penetrant, the process of dosage compensation in Drosophila is not absolute, with a substantial fraction of X-linked genes being under- or overexpressed in males compared to females (Smith and Lucchesi 1969; Roberts and Evans-Roberts 1979; Legube et al. 2006; Chang et al. 2011; Lott et al. 2011; Sun et al. 2013). Our rationale was that XX animals might be larger than XY animals because they express two copies of a growth gene that is not, or is only partly, dosage compensated. If such a gene exists, it should fulfill four criteria: (1) A duplication of this gene should lead to an increase in body size in males, (2) the increase in size seen with a duplication should be eliminated in a male carrying a mutation of the gene, (3) females with only one functional copy of the gene should be smaller than control females with two functional copies, and (4) females should express the gene at higher levels than males during the critical phase of female-specific growth.
Through a combination of genetic and molecular analyses, we identified several X-linked genes that lead to an increase in male size upon duplication. However, only one of our candidates, the Myc gene, fulfilled all our criteria. Mutations in Myc restored males with a Myc-containing duplication to normal male size, and heterozygous females carrying one mutation of Myc were smaller than control females. Furthermore, Myc displays a sex-specific difference in expression in the third larval instar stage. Our experiments identify Myc as a potent regulator of sex-specific body size in Drosophila, both in larvae and in adult tissue.
Materials and Methods
Drosophila stocks
Oregon R (OR), white1118 (w1118), and white1 (w1) lines were used as controls. Dp(1;Y) and Dp(1;3) duplication lines, including Df(1)Exel6233 (stock 7707), Cg-Gal4 (stock 7011) and UAS-tra (stock 4590), were obtained from the Bloomington Stock Center; CanAdKO (stock 109-862) and SLIRP1GS1189 (stock 200-175) lines from the Drosophila Genetic Resource Center (Kyoto, Japan). MycP0 (Johnston et al. 1999), Mnt1 (Loo et al. 2005), cycD1 (Emmerich et al. 2004), CanA-14FKO (Nakai et al. 2011), shi4C (Kasprowicz et al. 2014), tra1 (Sturtevant 1945),UAS-Desat1 (Kohler et al. 2009), da-Gal4 (Wodarz et al. 1995), and genomicSmc3-ha (Heidmann et al. 2004) were as described. UAS-lacZ86 and UAS-lacZ75c flies were a gift from Johannes Bischof. Flies were raised at 25 ± 1° and 60–70% humidity with a 12:12 light:dark photoperiod on standard medium (100 g/l yeast, 75 g/l dextrose, 55 g/l cornmeal, 10 g/l flour, 8 g/l agar, 7.5 ml/l nipagin).
Experimental design
All crosses were performed at 25° in bottles containing 45 ml standard medium. In the first round of screening, Dp(1;Y) males were backcrossed to OR females twice and F2-sibling wandering larvae were weighed individually. For fine-scale mapping using Dp(1;3) duplication lines, Dp(1;3) males were crossed with w1118 females. Heterozygous Dp(1;3) males were then crossed to w1118 females and wandering-male sibling larvae were weighed individually.
Larval measurements
Wandering third instar larvae were rinsed with water, blotted dry, and weighed on a Mettler Toledo MX5 Microbalance. Next, for the Dp(1;3) screen, individual larvae were placed into apple juice-/agar-containing wells of 64-well nucleon dishes, sealed with punctured Parafilm M, and maintained until adulthood at which point genotype was assessed by eye color. At least 20 wandering larvae of each genotype from each cross were analyzed in this manner, except for larvae with Dp(1;3)DC271 for which only 10 male larvae were tested due to weak viability. For tra experiments, tra1 females were first mated with Df(3L)st-E52 males for 48 hr and then cocultured with OR females, which serve as internal controls. Offspring third instar wandering larvae were weighed and maintained as described above. Gonads from adult tra1/Df(3L)st-E52 flies were dissected to determine genotype. Testes develop fully in XY;tra1/Df(3L)st-E52 flies, whereas only rudimentary testes form in XX;tra1/Df(3L)st-E52 flies.
Adult trait measurements
Between 10 and 32 flies were measured for each genotype, dependent upon availability due to genotype fitness. A minimum of two data sets was collected for each genotype. For interocular distance (IOD) measurements, flies were positioned on apple agar plates and photographed using a Leica MZ FLIII Stereomicroscope in combination with a Moticam 2300 camera and Motic Images Plus software. ImageJ 2.0.0 provided by Fiji was used for manual measurement of IODs, defined by the distance from eye edge to eye edge across the anterior orbital bristles.
Quantitative PCR analysis
We assessed RNA levels in feeding larvae in their third instar stage using reverse transcriptase-quantitative PCR (RT-qPCR). Total RNA was isolated from OR larvae dissected 68–72 hr after egg laying using TRI Reagent (Sigma Chemical, St. Louis, MO). Dnase I digestion (Ambion) was carried out twice for 30 min at 37°. Reverse transcription was performed using a Transcriptor High Fidelity Complementary DNA (cDNA) Synthesis Kit on 150 ng total RNA using oligo(dT)16, as described by the manufacturer (Roche). For each sex, at least 10 larvae were dissected. A minimum of three biological replicates was used for each experimental point. cDNA generated from 150-ng RNA was used as the template for amplification, using validated primers in MESA Green qPCR Mastermix for SYBR Assay (Eurogentec). Gene expression was normalized to the expression of two reference genes: tubulin binding protein and α-tubulin. Primers used for qPCR analysis were as follows: Myc, 5′-GAC GGA TAC GGA AAC TAT GTT-3′/5′-GTA AAG GGC CAT TGC GAT TA-3′; Mnt, 5′-CCA CAA TGG AAC CAC AAT TC-3′/5′-CGG TTG CTC CTC CAT TAG TT-3′; Max, 5′-CAA GGA GAG CTT CAC CAA CC-3′/5′-TCT GTA TGC ATT CGG TGG TC-3′; rpl1, 5′-CAA CAA CCG ACA GGC CTA C-3′/5′-GAA TAC GGG CCA CAG CAC-3′; rpl135, 5′-AGA CCA TTC CAG TGC TGA CC-3′/5′-CTC GTC GAA GGA ATC CAC AT-3′; cycD, 5′-CAC GGA CAA CAG CAT CTA CAA-3′/5′-GGA AGT CCA AGG GAG TCA CA-3′; CanA/Pp2B, 5′-CGA CTT CCT GCA GAA CAA CA-3′/5′-GTC TGG CTT TTT GCG GTA CAT-3′; and Desat1, 5′-ACG TAA CCT GGC TGG TGA AC-3′/5′-TAT GCC ATC CCT CTC CAA AG-3′.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. All supporting data and reagents used in this work will be made available upon request.
Results
At the third instar larval stage, females consistently weigh 30% more than males
We started with the hypothesis that sex-specific differences in body size might be controlled, not only by genes of the sex-determining pathway, but also by other genes whose dosage could regulate growth. We sought to identify factors that contribute to the growth difference between males and females. To assess sex-specific size, we chose to measure body size of larvae in their wandering stage, because weight differences between the sexes have reached a maximum in this phase lasting several hours, with females weighing 30% more than males (Bakker 1959; Okamoto et al. 2009). By comparison, adult traits are more complex and female traits are, on average, a mere 10.5% larger than those of adult males, making smaller changes in adult size between sexes more laborious to detect. Further complicating adult measurements, the degree of sexual-size dimorphism is not uniform across traits, reflecting the autonomous and specialized nature of growth regulation in imaginal disks (Cowley and Atchley 1988; David et al. 2003).
Growth in Drosophila is extremely sensitive to environmental conditions including nutrition, temperature, oxygen levels, and humidity (Nijhout et al. 2014). We raised all flies in a climate-controlled room on normal food to reduce environmental effects. Furthermore, we considered only weight ratios of flies cocultured within the same vial for our screen. Whereas a weight ratio of 1.0 would be expected if females and males were of equal size, in Drosophila larvae the weight ratio of wild-type females to males is 1.3. To examine possible crowding effects on sexual size dimorphism, we examined a range of population densities for control w1118 and wild-type OR fly strains used in our screens. We found that the female:male weight ratio was consistently within a range of 1.26–1.32 and 1.29–1.32 for w1118 and wild-type OR flies, respectively, across a wide range of densities (Supplemental Material, Table S1).
X-chromosome screen for dose-dependent modifiers of body size
We were interested in determining what, other than the sex-determining gene tra, could account for size differences between sexes in Drosophila. To test whether X dosage plays a role, we analyzed the effect of duplicating X-chromosome segments on male size. We chose to analyze males because they express neither Sxl nor tra, and would therefore allow us to identify growth effects regulated by genes that do not act through the sex-determining pathway.
For our initial assessment, we selected and analyzed 22 isogenic stocks with Y-linked X duplications that together cover >91% of the X chromosome (Cook et al. 2010). Due to severely reduced viability and fertility caused by two duplications, we were unable to generate sufficient offspring to assess the effects of these duplications on growth, reducing our coverage of the X chromosome to 81%. The remaining 19% of the X chromosome not covered in this initial screen was distributed across five gaps, which we will discuss later.
Of the 20 duplications we examined, 4 had a positive effect (Table 1 and Table S2). A striking increase in growth was observed, as these four duplication lines yielded XX/XY, DpX weight ratios of 1.20 or less. In each of these cases, males carrying a duplication experienced a growth increase of at least 8% compared to control males. We chose to focus on these for further study.
Table 1. Dp(1;Y) duplications spanning the X chromosome used in the first round of screening.
| Duplication | First replicate | Second replicate | Third replicate | XX/XY,DpX |
|---|---|---|---|---|
| BSC296 | ♀ 1.93 ± 0.02 | 1.33 | ||
| ♂ 1.45 ± 0.02 | ||||
| BSC74 | ♀ 1.96 ± 0.02 | ♀ 2.02 ± 0.02 | ♀ 2.13 ± 0.02 | 1.19 |
| ♂ 1.66 ± 0.02 | ♂ 1.70 ± 0.01 | ♂ 1.78 ± 0.01 | ||
| BSC158 | ♀ 1.95 ± 0.02 | 1.31 | ||
| ♂ 1.49 ± 0.02 | ||||
| BSC91 | ♀ 2.08 ± 0.02 | ♀ 2.07 ± 0.02 | 1.26 | |
| ♂ 1.69 ± 0.02 | ♂ 1.61 ± 0.01 | |||
| BSC277 | ♀ 2.01 ± 0.02 | 1.27 | ||
| ♂ 1.58 ± 0.02 | ||||
| BSC178 | ♀ 2.18 ± 0.04 | 1.29 | ||
| ♂ 1.69 ± 0.02 | ||||
| BSC33 | ♀ 1.90 ± 0.02 | ♀ 1.98 ± 0.02 | 1.30 | |
| ♂ 1.47 ± 0.01 | ♂ 1.51 ± 0.01 | |||
| BSC170 | ♀ 1.99 ± 0.02 | 1.34 | ||
| ♂ 1.49 ± 0.01 | ||||
| BSC58 | ♀ 2.03 ± 0.01 | ♀ 2.04 ± 0.02 | 1.33 | |
| ♂ 1.51 ± 0.03 | ♂ 1.54 ± 0.02 | |||
| BSC47 | ♀ 2.07 ± 0.02 | ♀ 2.06 ± 0.02 | ♀ 2.16 ± 0.03 | 1.18 |
| ♂ 1.75 ± 0.02 | ♂ 1.72 ± 0.03 | ♂ 1.81 ± 0.03 | ||
| BSC185 | ♀ 2.06 ± 0.01 | 1.33 | ||
| ♂ 1.55 ± 0.02 | ||||
| BSC231 | ♀1.97 ± 0.02 | 1.29 | ||
| ♂ 1.53 ± 0.01 | ||||
| BSC223 | ♀ 2.05 ± 0.02 | ♀ 1.98 ± 0.02 | ♀ 2.12 ± 0.02 | 1.20 |
| ♂ 1.73 ± 0.02 | ♂ 1.66 ± 0.02 | ♂ 1.71 ± 0.02 | ||
| BSC240 | ♀ 2.00 ± 0.02 | ♀ 1.94 ± 0.02 | ♀ 2.04 ± 0.02 | 1.19 |
| ♂ 1.71 ± 0.01 | ♂ 1.56 ± 0.03 | ♂ 1.78 ± 0.02 | ||
| BSC200 | ♀ 2.04 ± 0.02 | ♀ 2.05 ± 0.04 | 1.29 | |
| ♂ 1.58 ± 0.01 | ♂ 1.60 ± 0.03 | |||
| BSC67 | ♀ 2.09 ± 0.02 | 1.25 | ||
| ♂ 1.67 ± 0.02 | ||||
| BSC14 | ♀ 2.05 ± 0.02 | 1.30 | ||
| ♂ 1.58 ± 0.01 | ||||
| BSC129 | ♀ 1.90 ± 0.01 | 1.33 | ||
| ♂ 1.42 ± 0.03 | ||||
| BSC329 | ♀ 2.12 ± 0.03 | 1.37 | ||
| ♂ 1.55 ± 0.02 | ||||
| BSC276 | ♀ 2.12 ± 0.02 | ♀ 2.06 ± 0.02 | 1.27 | |
| ♂ 1.63 ± 0.02 | ♂ 1.66 ± 0.02 |
Mean weights ± SE of Dp(1;Y) male larvae (XY,DpX) and sibling females (XX). Duplications yielding XX/XY, DpX ratios of 1.20 or less are in boldface font. ♀, female; ♂, male.
Narrowing down the four X-chromosomal regions that increase male size in a dose-dependent manner
Our regions of interest defined by duplications Dp(1;Y)BSC74, Dp(1;Y)BSC47, Dp(1;Y)BSC223, and Dp(1;Y)BSC240 (hereafter referred to as region A, B, C, and D, respectively) range between 681 kbp and 1.7 Mbp in size. To narrow our regions of interest and confirm our results, we analyzed a second set of smaller Y-linked X-chromosomal duplications specific to regions A–D in a similar manner as described above (Table 2 and Table S3).
Table 2. Dp(1;Y) duplications within regions of interest identified in the primary screen.
| Duplication | Region | First replicate | Second replicate | XX/XY,DpX |
|---|---|---|---|---|
| BSC76 | A | ♀ 2.03 ± 0.03 | 1.19 | |
| ♂ 1.70 ± 0.03 | ||||
| BSC82 | A | ♀ 1.98 ± 0.03 | 1.15 | |
| ♂ 1.72 ± 0.03 | ||||
| BSC49 | B | ♀ 2.13 ± 0.02 | ♀ 1.99 ± 0.02 | 1.23 |
| ♂ 1.73 ± 0.02 | ♂ 1.63 ± 0.02 | |||
| BSC52 | B | ♀ 2.01 ± 0.03 | ♀ 1.97 ± 0.03 | 1.22 |
| ♂ 1.63 ± 0.04 | ♂ 1.63 ± 0.02 | |||
| BSC55 | B | ♀ 2.05 ± 0.02 | 1.33 | |
| ♂ 1.54 ± 0.02 | ||||
| BSC105 | B | ♀ 2.07 ± 0.02 | 1.37 | |
| ♂ 1.51 ± 0.02 | ||||
| BSC225 | C | — | — | |
| BSC226 | C/D | ♀ 2.09 ± 0.03 | 1.23 | |
| ♂ 1.70 ± 0.02 | ||||
| BSC232 | D | ♀ 2.07 ± 0.03 | 1.22 | |
| ♂ 1.70 ± 0.02 | ||||
| BSC244 | D | ♀ 2.20 ± 0.03 | 1.24 | |
| ♂ 1.77 ± 0.02 |
Mean weights ± SE of Dp(1;Y) male larvae (XY,DpX) and sibling females (XX). ♀, female; ♂, male.
Region A, defined by Dp(1;Y)BSC74, was analyzed further using two small duplications, Dp(1;Y)BSC76 and Dp(1;Y)BSC82, which map within the region (Figure S1A). We found that Dp(1;Y)BSC76 reproduced results found, with the original duplication giving rise to a ratio of XX/XY,DpX of 1.19, which corresponds to an 8% increase in growth of test males compared to wild-type males. Dp(1;Y)BSC82, a smaller duplication that corresponds to the proximal end of Dp(1;Y)BSC74 and Dp(1;Y)BSC76, had a far stronger impact. The weight ratio of wild-type females to males with Dp(1;Y)BSC82 was 1.15, indicating a 13% weight increase in males compared to wild type (Figure S1A and Table 2). Our attempt to narrow down our 1.7 Mbp region of interest [Dp(1;Y)BSC74] thus led us to identify a candidate within 1.0 Mbp defined by Dp(1;Y)BSC82.
Four additional duplications in region B, Dp(1;Y)BSC49, Dp(1;Y)BSC52, Dp(1;Y)BSC55, and Dp(1;Y)BSC105, were used to narrow our region of interest from 681 to 219 kbp. Region B was newly defined by the proximal ends of Dp(1;Y)BSC52, which replicates the male growth increase observed with Dp(1;Y)BSC47, and Dp(1;Y)BSC55, which does not (Figure S1B and Table 2).
Duplications that define regions C and D overlap one another, making it unclear whether a single growth-promoting sequence resides in their shared sequence or whether multiple sequences generate the observed effects. We employed four Dp(1;Y) lines whose X-chromosome duplications together tile across regions C and D. A large region within C was refractory to this method of analysis due to weak viability and sterility in males expressing Dp(1;Y)BSC225 (Figure S1C and Table 2). The complete region C duplication, Dp(1;Y)BSC223, must therefore contain one or more suppressors to the observed male sterility and fertility. Each of the duplications tested across region D produced larger males, but neither had as strong an effect on growth as Dp(1;Y)BSC240. These results suggest that multiple dosage-sensitive regions exist within region D (Figure S1D and Table 2). Thus, we next performed a finer-tuned analysis of our regions of interest using smaller duplications to individuate growth-promoting and possible growth-suppressing sequences.
Fine-scale mapping and gene identification
We selected 54 duplication lines that each have a small segment of the X chromosome translocated to the third chromosome and collectively cover the entirety of the narrowed regions A–D (Figure 1A and Table S4). An additional 50 duplication lines were selected to cover gaps 1–5, for which no large duplications were either available or amenable to screening (Figure 1B and Table S5). The selected duplication lines are part of an X-chromosome duplication kit prepared by insertion of BAC clones marked with the dominant mini-white eye marker into polytene band 65B2 on the third chromosome (Venken et al. 2006, 2010). Using these lines, we designed a genetic mating scheme to generate male larvae carrying duplications alongside sibling males that do not. Since all Dp(1;3) lines were prepared in a w1118 background, we used w1118 males as controls in this set of experiments.
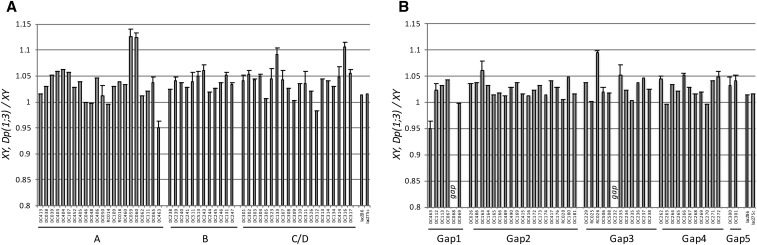
Figure 1.
Regions of interest (regions A–D) and gaps (gap 1–5) not covered in Dp(1;Y) screen were examined using smaller X duplications, Dp(1;3), translocated to the third chromosome. (A) Males with X duplications that span regions A–D were crossed with w females (allele w1118), sons with the duplication were again crossed with w females and F2 sibling males were analyzed. Bars represent weight ratios of third instar wandering w males with a duplication to w males with no duplication. Control lacZ transgene-expressing lines were examined in the same manner. (B) Males with X duplications spanning gaps 1–5 were examined as described for regions A–D above. The absence of bars for duplication lines Dp(1;3)DC068 and Dp(1;3)DC232 reflect persistent gaps in coverage, resulting from reduced viability and fertility in males with these duplications. Error bars indicate where experiments were repeated.
Of the duplications we analyzed, the vast majority (91%) provided male larvae a slight growth advantage over their w1118 siblings. We noticed that both male and female wandering larvae mutant for a w allele are strikingly smaller (8 and 11%, respectively) (Figure S2) than OR controls and asked if the mini-white marker in Dp(1;3) duplication lines contributed to larval size. Two transgenic lines carrying an insertion comprised of a UAS-lacZ transgene and the mini-white marker were a mere 1–2% larger than w1118 controls, leading us to conclude that the contribution of mini-white to size is modest at best.
In region A, 2 of the 22 duplications tested, Dp(1;3)DC059 and Dp(1;3)DC060, were exceptionally potent in promoting growth. Males harboring either of these were on average 12–14% larger than w1118 siblings lacking the duplication. A single gene locus, the Drosophila gene diminutive (Myc), is found in the large overlap of the duplications. We conclude that introduction of an extra copy of Myc is sufficient to substantially increase larval growth in males (Figure 1A, Figure 2A, and Table S4). Interestingly, the proximal-most duplication tested in region A repressed male growth, giving rise to males that were 5 ± 1% smaller than w1118 controls. We strongly suspected that expression of Mnt, a transcription factor known to antagonize Myc (Loo et al. 2005) which is also contained in this duplication, caused the observed reduced growth. Indeed, a null mutation in Mnt relieves the negative effect on growth imposed by the duplication Dp(1;3)DC463 (Figure 3A). Mnt and Myc dosage are thus both essential for regulation of growth in larvae.
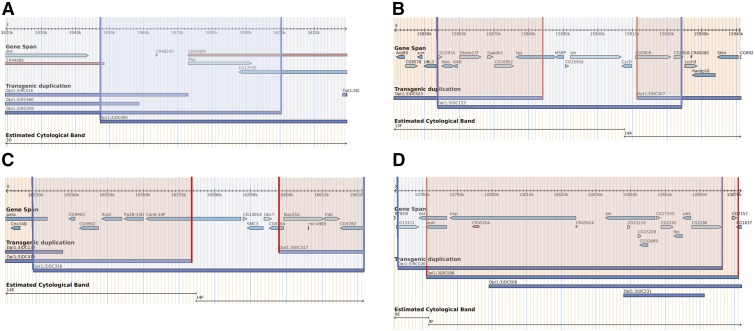
Figure 2.
Fine-scale mapping of duplication-sensitive regions using overlapping Dp(1;3) lines. Strong growth-promoting regions are highlighted in blue, regions with minor or no growth effects are shaded in pink. (A) Only the Myc locus is fully contained in growth-promoting duplications Dp(1;3)DC059 and Dp(1;3)DC060. (B) Candidates in region C narrowed to tay, MSBP, CG15916, shi, and cycD. (C) In region D, four candidates remain: CanA-14F, SMC3, Ubc7, and CG9784. (D) Three genes in Dp(1;3)RC026 are candidates for dose-sensitive growth promoters: CG15211, Ant2, and sesB. Maps were derived from GBrowse representations in FlyBase.
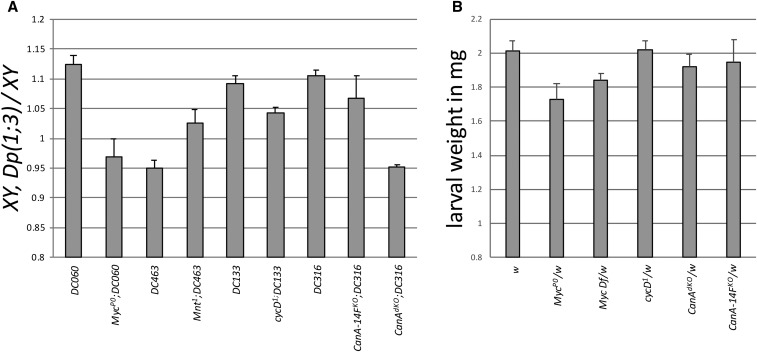
Figure 3.
(A) Effect of mutations in a candidate gene on male larvae with a duplication that induces or restricts growth. In each case, animals were heterozygous for the mutations described. The strong hypomorphic Myc mutation, MycP0, obliterates the growth promoting effects of Dp(1;3)DC060, indicating that duplication of Myc underlies the weight increase in Dp(1;3)DC060 males. The converse negative effect on growth imposed by Dp(1;3)DC463 is alleviated through a mutation of Mnt, implicating this gene as the growth suppressor. A partial suppression of the Dp(1;3)DC133’s growth promotion was observed by the null mutation cycD1. A second factor in Dp(1;3)DC133 likely contributes to the observed increase in male size. A partial suppression of the growth induced by Dp(1;3)DC316 was also found through elimination of CanA-14F (CanA-14FKO). Combined removal of CanA-14F and the neighboring gene calcineurin Pp2B-14D (CanAdKO) resulted in complete eradication of the duplication’s effects. (B) The effects of dosage of candidate genes were studied in females. Wandering female larvae heterozygous for mutations in candidate genes in a w background (allele w1118) were weighed. A significant weight reduction compared to w was only observed in females heterozygous for the hypomorphic Myc mutation, MycP0, or for a deficiency in Myc.
Of the 12 duplications tested in region B, several were able to elicit modest increases in male size, but none could replicate the 10 ± 1% weight increase resulting from duplication of the complete region B (Figure 1A). We conclude that the size increase observed in our initial screen by duplication of the entire region is a cumulative effect of multiple loci.
A total of 20 Dp(1;3) duplications were selected to tile across regions C and D. Of these, two duplications [Dp(1;3)DC133 and Dp(1;3)DC316] mimicked the increases in male size (9 ± 1% and 10 ± 1%, respectively) observed upon duplication of complete region C or D (Figure 1A and Table S4). We narrowed our regions of interest by analyzing neighboring duplications, which partially overlap Dp(1;3)DC133 in region C and Dp(1;3)DC316 in region D. Five candidate genes for region C were thus identified: tay bridge (tay), membrane steroid binding protein (MSBP), CG15916, shibire (shi), and cyclinD (cycD) (Figure 2B and Table 3). Likewise, using this approach, we identified five candidates within region D: Calcineurin A at 14F (CanA-14F), CG13014, structural maintenance of chromosome 3 (SMC3), ubiquitin conjugating enzyme 7, and CG9784 (Figure 2C and Table 3).
Table 3. Candidates identified in duplication screen.
| Candidates | Region |
|---|---|
| Myc | A |
| CG15211 | Gap 3 |
| Ant2 | Gap 3 |
| sesB | Gap 3 |
| Tay | C |
| MSBP | C |
| CG15916 | C |
| Shi | C |
| cycD | C |
| CanA-14F | D |
| CG13014 | D |
| SMC3 | D |
| Ubc7 | D |
| CG9784 | D |
Fourteen candidate genes were identified as a result of fine-scale mapping of regions of interest A–D and gap regions 1–5 using X-chromosome duplications translocated to the third chromosome. Myc, a known promoter of cell growth, was identified as the sole candidate in region A.
Of the duplications tested within the five gap regions, Dp(1;3)RC026 which covers part of gap 3, resulted in the most striking increase (9.5%) in male size compared to w1118 male siblings (Figure 1B and Table S5). A mere three candidate genes were identified upon subtracting away flanking regions without growth effects: CG15211, Ant2, and sesB (Figure 2D and Table 3). Finally, there remain two small regions, one in gap 1 and one in gap 3 that precluded analysis due to weak fertility and viability of duplications Dp(1;3)DC068 and Dp(1;3)DC232.
In conclusion, through a second round of screening, we succeeded in substantially trimming three (regions A, C, and D) of our four regions of interest (Figure 1A). More specifically, in region A we identify Myc and Mnt as dose-dependent modifiers of growth (Figure 1A and Figure 2A). We narrowed region C and region D so that five candidate genes remained in each (Figure 1A; Figure 2, B and C; and Table 3). Additionally, we detected a new region of interest in gap 3, one of the five gap regions not covered in the first round of screening (Figure 1B). In gap 3, we identify a short list of only three candidate genes (Figure 2D and Table 3).
Genetic analysis of candidate genes
We reasoned that if the additional copy of Myc provided by region A duplications [Dp(1;3)DC059 and Dp(1;3)DC060] was responsible for the observed growth increase in males, then reducing Myc dosage in flies with one of these duplications should return them to wild-type size. Indeed, reducing Myc levels through introduction of the hypomorphic MycP0 allele (Johnston et al. 1999) in males expunged the positive growth effects provided by Dp(1;3)DC060 (Figure 3A).
None of the small Dp(1;3) duplications tested across region B reproduced the growth increase observed in our original screen. Of note is that the region B-defining, Y-linked duplication chromosome harbors a segment of the distal X chromosome (19F4-h29) in addition to the one described in our fine-scale mapping. We had originally omitted the distal X sequence for further analysis because the entirety of this region was also represented in the terminal Y-linked duplication [Dp(1;Y)BSC276], a duplication that only modestly affected growth. We examined one candidate, SLIRP1, in this terminal region at 19F4 due to its described role in larval feeding behavior (Ryuda et al. 2011). Duplication of the SLIRP1 sequence did not increase growth in larvae (Figure S3A), leaving the question of what the growth promoter in region B is open.
Of the five candidates in region C (tay, MSBP, CG15916, shi, and cycD), we suspected that duplication of cycD was causing the observed larval-growth phenotype. We crossed Dp(1;3)DC133 flies into a cycD heterozygous null background and found a strong reduction (5 ± 1%) in growth (Figure 3A). The growth phenotype of the duplication is only partially suppressed, so it stands to reason that, while copy number of cycD can clearly play a role in male growth, a second factor expressed by the duplication contributes to the observed growth increase in region-C duplication males. Introduction of a genomic transgene expressing shi did not increase male weight (Figure S3B). Further analysis of candidates should uncover other growth promoter(s) in this region. Identification of these was beyond the scope of this article.
In region D, we were also left with five candidate genes: CanA-14F, CG13014, SMC3, Ubc7, and CG9784. We first assessed the impact of genomic SMC3 transgenes on size but found no significant weight changes (Figure S3C). Next, we made use of an available null mutation in CanA-14F and found that Dp(1;3)DC316 males lacking a copy of endogenous CanA-14F were ∼3% smaller than flies with the duplication alone (Figure 3A). Given that this duplication also contains Pp2B-14D, a second of the three calcineurin catalytic subunits found in Drosophila, we assessed the weights of larvae lacking a copy of each CanA-14F and Pp2B-14D genes (CanAdKO) in duplication males. Knocking down both aforementioned calcineurin catalytic subunits from Dp(1;3)DC316 males completely eliminated the duplication’s positive growth effect (Figure 3A). We deduce that duplication of CanA-14F and Pp2B-14D genes together are likely the root cause of growth increase in males with Dp(1;3)DC316.
Effects of Myc, cycD, and calcineurin subunit gene dosage on female size
We reasoned that if gene dosage plays a role in sexual dimorphism of body size, then reducing the dosage of our candidate genes in females should render them smaller than their wild-type counterparts. Indeed, we found that female larvae heterozygous for the strong Myc hypomorph MycP0 or the Myc deficiency Df(1)Exel6233 weighed ∼15% and 10% less than cocultured w1118 controls, respectively (Figure 3B). Reduction of cycD dosage by half, on the other hand, did not significantly affect female growth (Figure 3B). Finally, neither CanA-14FKO nor CanAdKO heterozygous female larvae were significantly smaller than w1118 controls (Figure 3B). These results suggest that, while Myc, cycD, and calcineurin expression levels have the potential to affect growth of Drosophila larvae, only Myc is likely to play a role in wild-type sexual dimorphism of body size.
Quantitative analysis of candidate gene expression
Our finding that Myc, cycD, and calcineurin dosage can affect larval growth made us curious if transcript levels differ between sexes of wild-type larvae. We performed qPCR analysis to determine transcript levels of our candidates in feeding third instar OR larvae. Synchronized male and female sibling larvae were collected between 68 and 72 hr after egg laying, a time period when females grow more rapidly than males (Okamoto et al. 2009). Given the central role attributed to the fat body in regulating organismal growth (Colombani et al. 2005; Delanoue et al. 2010; Parisi et al. 2013), we assessed transcript levels from isolated fat-body tissue in addition to determining whole larvae levels. We found Myc transcript levels to be 1.7 times higher in both female fat body and whole preparations as compared to sibling males (Figure 4A). Furthermore, introduction of a Myc duplication into males resulted in Myc transcript levels that were 1.6 times higher than wild-type males, while females heterozygous for a Myc deficiency accumulated less than half of what was detected in control females (Figure S4, A and B). On the other hand, no significant gender-specific differences in cycD or CanA-14F/Pp2B-14D transcript levels were detected in either whole or fat body tissues (Figure 4A). These results support our idea that Myc transcript levels play a key role in regulating sex-specific size differences in Drosophila larvae.
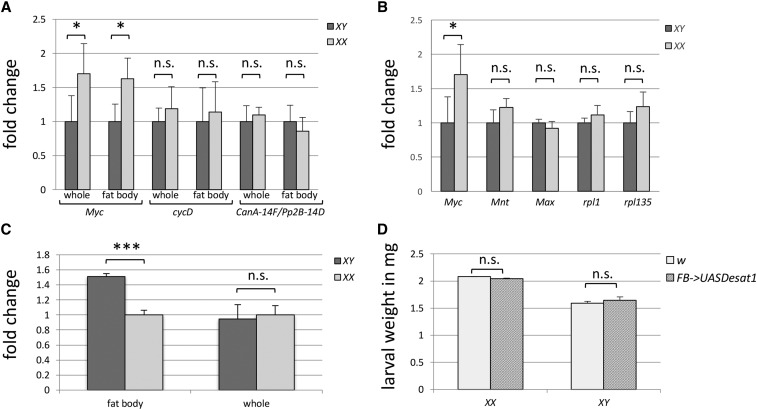
Figure 4.
Transcript levels of Myc and related genes were assessed by qRT-PCR in feeding third instar larvae. (A) Myc transcript levels in whole larvae, as well as in isolated fat body tissue, are 1.7 times higher in female than in sibling male larvae. No significant differences in cycD or CanA14-F/Pp2B-14D transcript levels were observed between the sexes. (B) Transcripts of neither Mnt nor Max, the two other components of the Myc-Mnt-Max interactome, amass in a sexually dimorphic manner. Transcripts for the ribosomal proteins Rpl1 and Rpl135, known targets of Myc, are expressed equally in males and females as well. (C) Overall levels of Desat1 RNA are the same in both sexes but transcript levels are 1.5 times higher in male fat-body tissue compared to that of sibling females. (D) Male and female larvae with Cg-Gal4-driven expression of Desat1 in fat-body tissue are not significantly different in size than cocultured w1118 control animals. * P < 0.02, *** P < 0.001; P-values were calculated using Student’s t-test. Error bars represent the standard deviation. n.s., not significant.
Examination of Myc cofactors and effectors
Myc is part of the conserved Myc-Max-Mnt network of transcription factors, which have been implicated in regulation of proliferation in vertebrates and Drosophila alike (Grandori et al. 2000; Bellosta and Gallant 2010). Max forms heterodimers with Myc, which together bind E-box sequences to activate transcription of nearby genes. Mnt, on the other hand, antagonizes Myc by heterodimerizing with Max and binding similar sequences and repressing transcription (Orian et al. 2003; Bellosta and Gallant 2010). We were curious whether Max or Mnt transcript levels were also sex-specifically regulated, but found no significant differences between female and male larvae (Figure 4B). Sex-specific Myc-Max-Mnt target gene expression therefore likely results from gender-specific imbalance in Myc transcript levels.
Ecdysone signaling in the larval fat body downregulates Myc expression as well as Myc target genes rpl1 or rpl135 (Grewal et al. 2005; Delanoue et al. 2010). Surprisingly, we found no upregulation of either rpl1 or rpl135 in whole female extracts, indicating that other Myc targets mediate body size dimorphism in Drosophila (Figure 4B).
The lipid desaturase Stearoyl-CoA desaturase (Desat1) is the Drosophila ortholog of vertebrate Stearoyl-CoA desaturase-1/SCD1 (Brock et al. 2007; Kohler et al. 2009). In mice, mutations in scd1 result in leaner and smaller animals (Ntambi et al. 2002). Similarly, reducing Desat1 in the Drosophila fat body results in smaller pupae and adults (Parisi et al. 2013). Furthermore, it was shown that Desat1 expression in the fat body is essential for fat body Myc-induced systemic growth. We asked whether Desat1 might be targeted by Myc to drive sex-specific differences in body size. Surprisingly, we found that Desat1 transcript levels were elevated in the male fat body compared to the female fat body, making it unlikely that Myc regulation of body size is mediated by Desat1 levels (Figure 4C). Finally, we overexpressed Desat1 in the fat body and found no significant difference in either female or male body size (Figure 4D). We conclude that Myc must regulate sex-specific growth via factors other than Desat1.
Myc and tra together promote larval and adult growth
It has been demonstrated that mutations in the tra gene hinder growth of XX animals, resulting in pupae and adults that are of an intermediate size between control female and male animals (Brown and King 1961; Rideout et al. 2015). We assessed the mass difference between male and female third instar larvae and tested the effect of tra mutations at this stage. In consonance with previous findings mentioned above, we found that XX larvae lacking tra were 12 ± 0.04% smaller than wild-type females, accounting for roughly half of the observed mass difference between control males and females. Lack of tra in XY larvae had no impact on size (Figure S5A).
Experiments presented up to this point describe two elements that ensure that female larvae weigh more than their male siblings. This sex-specific feature is regulated by the dosage of Myc and by female-specific expression of tra. We wondered whether these two elements together are sufficient to promote growth of males in such a way that they achieve the size of control females. Duplicating Myc in XY males by adding Dp(1:3)DC060 led to larvae that weighed 12% more than control larvae (Table 4). We expressed Tra ubiquitously in larvae using a da-Gal4 driver and obtained wandering XY larvae that attained ∼18% more mass than cocultured w1118 male controls. A second copy of Myc in addition to da-Gal4 and UAS-tra expression further increased the size of larvae, resulting in XY larvae that were 25% larger than control males, or in other words, not distinguishable from the size of control w1118 females having XX chromosomes (Table 4).
Table 4. Larval weight and IOD changes due to increase in Myc dosage and ectopic expression of Tra in males.
| Genotype | Larval weight | IOD |
|---|---|---|
| XX:XY | 1.28 ± 0.02 | 1.10 ± 0.02 |
| XY; Dp Myc:XY | 1.12 ± 0.01 | 1.06 ± 0.00 |
| XY; da-Gal4, UAS-tra:XY | 1.18 ± 0.01 | 1.06 ± 0.02 |
| XY; da-Gal4,UAS-tra; Dp Myc:XY | 1.25 ± 0.03 | 1.09 ± 0.01 |
Ratios ± SD of mean weights of wandering third instar larvae and of mean IODs of 1- to 5-D-old adults with respect to cocultured w1118 males.
In a complementary experiment, we measured the larval mass of XX animals that were heterozygous for the hypomorphic allele MycPO and homozygous for null mutations in tra. These animals were strikingly reduced in weight. Wild-type female larvae were 1.21 ± 0.02 larger than sibling MycP0/+; tra−/tra− larvae, nearing the 1.26 ± 0.01 weight ratio determined for wild-type females:males cultured in the same bottle. Put another way, MycP0/+; tra−/tra− larvae were 20% smaller than wild-type females, a significant reduction from the 12% difference measured between XX tra null larvae and wild-type females (Figure S5B).
We wondered whether the two genes, Myc and tra, also regulate sex-specific size of adult structures. We chose to measure the distance between eyes (IOD) because this trait has been demonstrated to robustly reflect adult body size (Vonesch et al. 2016). In males expressing an extra copy of Myc and da-driven tra, we found that IODs were increased to nearly the size of IODs of females. Individually, each element only led to a partial increase of the IOD (Table 4).
In conclusion, our results show that Myc dosage and tra are essential for the regulation of sexual size dimorphism in Drosophila larvae and adults.
Discussion
Sxl and tra expression in females contribute to size difference between sexes
Sex-specific differences in Drosophila body size result from an acceleration in female larval growth rate compared to that of males (Blanckenhorn 2007). The observation that hypomorphic mutations in Sxl hinder growth of XX animals led to the widely accepted model that, in addition to determining sex in dimorphic structures like the genitalia, expression of full-length Sxl protein is also responsible for the larger size of females (Cline 1984).
Results presented in this article concur with previous findings that tra is required in females to ensure that normal body size is attained (Brown and King 1961; Rideout et al. 2015). Since tra expression in the female is dependent on Sxl, these results support the model that Sxl plays a role in body size dimorphism in Drosophila. However, tra is only responsible for about half of the sex-specific size difference, begging the question of what else could contribute to regulating the differing sizes in males and females.
Several independent lines of study point to Sxl functions outside of the canonical sex-determination pathway (Horabin 2005; Penn and Schedl 2007; Chau et al. 2012; Evans and Cline 2013; Li et al. 2013). Given the abundance of putative Sxl binding sites in the genome, it is likely that still more targets of Sxl exist, with promoters of growth potentially among them (Kelley and Kuroda 1995; Robida et al. 2007).
There are, however, arguments against seeing Sxl as the only regulator of sexual dimorphism. The role of Sxl in regulation of tra expression in Drosophila is an evolutionarily recent adaptation (Bopp et al. 1996; Serna et al. 2004; Siera and Cline 2008). In most insects, sex is not determined by Sxl activity; rather it is directly regulated through sexually dimorphic expression of tra (Verhulst et al. 2010). And although it is well established that females are in general larger than males, to what extent tra regulates growth in other insects is not known. This leaves open the question of whether a Sxl-independent mechanism is responsible for the sexual dimorphism of body size.
Myc dosage contributes to sexual dimorphism of shape and size
In addition to regulating tra splicing, Sxl blocks splicing and represses translation of male specific lethal-2 (msl-2) messenger RNA in females (Bopp et al. 1991; Kelley et al. 1997; Gebauer et al. 1998). As an essential component of the dosage compensation complex, Msl-2 mediates the roughly twofold upregulation of gene expression from the single male X chromosome. This process relieves the inherent aneuploidy between sex chromosomes in Drosophila and thereby ensures viability of males. Interestingly, a number of genes including Lipid serum protein1-alpha, extradenticle, pvf1, l(1)G0193, and CG11750 escape dosage compensation (Roberts and Evans-Roberts 1979; Legube et al. 2006). Furthermore, whole transcriptome analysis has revealed sexually dimorphic expression of a large number of X-linked genes both upstream and downstream of tra (Chang et al. 2011).
These findings led us to investigate whether X-chromosome dosage contributes to body size differences between sexes. Indeed, we find that Myc appears to escape dosage compensation in males, putting them at a growth disadvantage compared to sibling females. Interestingly, Myc serves as one of several XSEs, albeit a weak one, whose early zygotic expression cues the establishment of sustained expression of full-length Sxl protein in females (Erickson and Quintero 2007; Kappes et al. 2011). Myc therefore helps to drive sexual dimorphism by (a) assisting in the activation of Sxl transcription in females, and (b) evading dosage compensation in males, a process activated in absence of the full-length Sxl protein.
Aneuploidy and position
The first screen for the effects of aneuploidy on Drosophila development was performed >40 years ago in a tremendous effort by Lindsley and Sandler et al. (Lindsley et al. 1972; Wolfner 2016). Upon generating translocations of second- and third-chromosome segments to the Y chromosome through X irradiation, the authors mapped breakpoints of the translocations and selected a collection of stocks with breakpoints spaced evenly across the autosomes. When two translocation stocks were crossed together, the progeny were either heterozygous for a deleted autosomal region or triploid for the same region. Of the 555 translocation crosses performed, 57 showed morphological phenotypes, only 3 of which showed a triploid phenotype. In our study of X duplications in males, we similarly find that the vast majority of genes are well tolerated when an additional copy is present. Only two regions of the X severely reduced male viability and fertility upon duplication.
Since relatively few regions showed a phenotype in Lindsley and Sandler et al.’s autosomal screen, it was concluded that additive effects of aneuploidy for a number of genes must underlie the observed disruptive phenotypes. In only one of the four growth-promoting regions we identified in our initial screen using 1;Y duplications, were we able to identify a single gene, Myc, whose duplication alone reproduced the phenotype of the 1;Y duplication. In two regions we were able to identify smaller duplications that partially phenocopied the growth effect of the large duplications. In the fourth region, we found no other duplications within that region that could reproduce the growth effects of the region-defining duplication. We, too, conclude that in most cases the growth phenotypes we observed are the composite effects of genes within a duplication. One caveat of our study is thus that individual positive or negative regulators of growth will have eluded our detection due to additive or synergistic effects of duplicating multiple genes at once. A second caveat to our screen is that gene expression from duplications used in this study might differ from expression of corresponding genes in their endogenous locations. Well-described position effects as well as regulation through feedback loops almost certainly impact effective expression levels of genes on the duplications used in this study (Birchler 2010).
Ploidy and growth
Accelerated tissue growth in females can be explained by increased rate of cell division, increased cell growth, or both. An often-used strategy in development in situations where growth is rapid or energy sources are limited is to increase cell volume rather than cell number (Kondorosi et al. 2000). Increasing DNA content is correlated with increased cell size and presents an effective means to promote growth (Maines et al. 2004). Drosophila larval tissues are highly polyploid, which strongly suggests that their growth results in large part from endoreplication. Myc has been shown to be required for endoreplication and cell growth, with ectopic expression leading to higher ploidy and larger cells (Pierce et al. 2004; Demontis and Perrimon 2009).
In this study, we find that Myc is more highly expressed in female than male larvae. It is tempting to speculate that sex-specific differences in Myc expression affect ploidy, possibly in a tissue-specific manner, and thereby contribute to sexual dimorphism of body size. Interestingly, it has been shown that Myc expression in the fat body alone results in similar increases in cell and larval size as found with ubiquitously expressed Myc (Johnston et al. 1999; Pierce et al. 2004). Increasing fat-body cell size through other means, however, such as local overexpression of CycD/Cdk4 or Rheb, does not impact larval size (Parisi et al. 2013). Therefore, Myc must act through other targets to promote organismal growth.
Parisi et al. (2013) reported further that expression of Myc in the fat body alone also drives growth of adult traits through increases in cell size and cell number, indicating that growth is stimulated not only in endoreplicative tissues but also in imaginal tissues of the larva. Our examination of IODs in adults shows that copy number of Myc likewise affects growth not only of larval tissue, but adult tissue as well.
The insulin signaling pathway: a common target for Myc and tra?
Myc plays an evolutionarily conserved role in promoting growth in Drosophila, as does insulin signaling (de la Cova and Johnston 2006). Partial loss-of-function mutations in components of the insulin pathway result in small but well-proportioned flies, while overexpression of insulin-like proteins lead to larger body sizes due to increased cell size and cell number (Brogiolo et al. 2001; Oldham et al. 2002; Rulifson et al. 2002). Similarly, hypomorphic mutations in Myc also yield small but normally proportioned flies (Johnston et al. 1999). Unlike insulin signaling, however, general overexpression of Myc accelerates growth of cell size but not cell division (Johnston et al. 1999; Grifoni and Bellosta 2015). Myc and the insulin signaling pathway therefore have distinct targets through which they regulate growth. In larval muscle tissue, both Myc and the insulin signaling pathway are required to autonomously increase cell size and to systemically promote organismal growth by affecting feeding behavior (Demontis and Perrimon 2009). Organismal growth resulting from Myc expression in the fat body, on the other hand, is mediated through humoral factors from the fat body that induce Drosophila insulin-like peptide 2 (Dilp2) release from the brain (Geminard et al. 2009; Parisi et al. 2013).
Rideout et al. (2015) presented evidence that tra expression in the female fat body is also instrumental to Dilp2 release from larval brain. Interestingly, two known targets of tra, dsx and fru, affect sexual dimorphism of body shape and behavior, respectively, but not size (Billeter et al. 2006; Camara et al. 2008). A novel branch of the sex-determination pathway, acting through an as yet unidentified target of tra, therefore contributes to dimorphism of body size in Drosophila. These results provide a clue as to how the sex-determination pathway and sex-specific growth regulation by Myc may be linked, possibly via the insulin signaling pathway.
Finally, in a recent study Hudry et al. (2016) reveal an unexpected role for tra in expression, rather than splicing, of genes involved in sexually dimorphic proliferation of adult intestinal stem cells. With this effect in mind, it seems likely that tra might regulate the expression of genes that drive sexual dimorphism during early development as well. The idea that Myc expression levels could be directly affected by tra opens the way to intriguing speculations that deserve further investigation.
Conclusion
In this study, we present two means by which sex-specific gene expression leads to sexual dimorphism of body size. As others have already shown, we find that body-size dimorphism depends in part on tra expression in the female (Brown and King 1961; Rideout et al. 2015). By screening the X chromosome for dose-dependent modifiers of growth, we identified a second factor that regulates growth differently in males and females. We find that a simple twofold difference in the copy number of Myc has a potent impact on growth. Furthermore, we find that we can increase male size further by expressing tra in addition to increasing Myc dosage. Females are thus larger than males not only because they express tra, but also because the growth-promoting gene Myc escapes dosage compensation. Since in early stages of embryogenesis Myc acts as an X-chromosome signal element that helps to activate Sxl and tra in females, we conclude that Myc acts twice in development to regulate dimorphism of body size. It acts first in the female embryo, where it contributes to the counting mechanism essential for activation of the sex-determining pathway. Second, the elevated expression of Myc in females directly promotes growth during larval stages of XX animals. Interestingly, the largest positive effect on growth is observed when Tra and elevated levels of Myc are present together in the larvae. Conversely, eliminating tra expression while simultaneously reducing Myc dosage in females results in XX animals of nearly XY animal size. It will be interesting to learn if tra and Myc act together through a single pathway or independently through distinct pathways to promote organismal growth.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.192260/-/DC1.
Acknowledgments
We thank Toshiro Aigaki, Konrad Basler, Johannes Bischof, Ernst Hafen, Christian Lehner, and Patrik Verstreken, as well as the Bloomington Drosophila Stock Center and the Drosophila Genetic Resource Center Kyoto for providing stocks. We are grateful to Martin Moser and Eliane Escher of the sequencing and robotics facility who executed the quantitative PCR analysis to determine transcript levels. Daniel Bopp kindly allowed us to use his microscope for adult fly measurements. We thank the Zurich fly community for numerous interactions and feedback in talks and presentations. We are especially grateful to Konrad Basler and Tripti Gupta for critically reading our manuscript. Our work was supported by the Swiss National Science Foundation grant number 3100-65333.01 to M.Z., a grant from the Swiss Bundesprogramm Chancengleichheit to M.Z., the Julius Klaus Foundation, and the University of Zürich.
Footnotes
Communicating editor: H. J. Bellen
Literature Cited
- Bakker K., 1959. Feeding period, growth, and pupation in larvae of Drosophila melanogaster. Entomol. Exp. Appl. 2: 171–186. [Google Scholar]
- Bell L. R., Maine E. M., Schedl P., Cline T. W., 1988. Sex-lethal, a Drosophila sex determination switch gene, exhibits sex-specific RNA splicing and sequence similarity to RNA binding proteins. Cell 55: 1037–1046. [DOI] [PubMed] [Google Scholar]
- Bellosta P., Gallant P., 2010. Myc function in Drosophila. Genes Cancer 1: 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billeter J. C., Rideout E. J., Dornan A. J., Goodwin S. F., 2006. Control of male sexual behavior in Drosophila by the sex determination pathway. Curr. Biol. 16: R766–R776. [DOI] [PubMed] [Google Scholar]
- Birchler J. A., 2010. Reflections on studies of gene expression in aneuploids. Biochem. J. 426: 119–123. [DOI] [PubMed] [Google Scholar]
- Blanckenhorn, W. U., A. F. Dixon, D. J. Fairbairn, M. W. Foellmer, P. Gilbert et al., 2007 Proximate causes of Rensch’s rule: does sexual size dimorphism in arthropods result from sex differences in development time? Am. Nat. 169: 245–257. [DOI] [PubMed] [Google Scholar]
- Bopp D., Bell L. R., Cline T. W., Schedl P., 1991. Developmental distribution of female-specific Sex-lethal proteins in Drosophila melanogaster. Genes Dev. 5: 403–415. [DOI] [PubMed] [Google Scholar]
- Bopp D., Calhoun G., Horabin J. I., Samuels M., Schedl P., 1996. Sex-specific control of Sex-lethal is a conserved mechanism for sex determination in the genus Drosophila. Development 122: 971–982. [DOI] [PubMed] [Google Scholar]
- Brock T. J., Browse J., Watts J. L., 2007. Fatty acid desaturation and the regulation of adiposity in Caenorhabditis elegans. Genetics 176: 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogiolo W., Stocker H., Ikeya T., Rintelen F., Fernandez R., et al. , 2001. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 11: 213–221. [DOI] [PubMed] [Google Scholar]
- Brown E. H., King R. C., 1961. Studies on the expression of the transformer gene of Drosophila melanogaster. Genetics 46: 143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtis K. C., Baker B. S., 1989. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 56: 997–1010. [DOI] [PubMed] [Google Scholar]
- Camara N., Whitworth C., Van Doren M., 2008. The creation of sexual dimorphism in the Drosophila soma. Curr. Top. Dev. Biol. 83: 65–107. [DOI] [PubMed] [Google Scholar]
- Chang P. L., Dunham J. P., Nuzhdin S. V., Arbeitman M. N., 2011. Somatic sex-specific transcriptome differences in Drosophila revealed by whole transcriptome sequencing. BMC Genomics 12: 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau J., Kulnane L. S., Salz H. K., 2012. Sex-lethal enables germline stem cell differentiation by down-regulating nanos protein levels during Drosophila oogenesis. Proc. Natl. Acad. Sci. USA 109: 9465–9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline T. W., 1984. Autoregulatory functioning of a Drosophila gene product that establish es and maintains the sexually determined state. Genetics 107: 231–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline T. W., 1988. Evidence that sisterless-a and sisterless-b are two of several discrete “numerator elements” of the X/A sex determination signal in Drosophila that switch Sxl between two alternative stable expression states. Genetics 119: 829–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline T. W., Meyer B. J., 1996. Vive la difference: males vs. females in flies vs. worms. Annu. Rev. Genet. 30: 637–702. [DOI] [PubMed] [Google Scholar]
- Colombani J., Bianchini L., Layalle S., Pondeville E., Dauphin-Villemant C., et al. , 2005. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science 310: 667–670. [DOI] [PubMed] [Google Scholar]
- Cook R. K., Deal M. E., Deal J. A., Garton R. D., Brown C. A., et al. , 2010. A new resource for characterizing X-linked genes in Drosophila melanogaster: systematic coverage and subdivision of the X chromosome with nested, Y-linked duplications. Genetics 186: 1095–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley D. E., Atchley W. R., 1988. Quantitative genetics of Drosophila melanogaster. II. Heritabilities and genetic correlations between sexes for head and thorax traits. Genetics 119: 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C., 1871. The Descent of Man and Selection in Relation to Sex. John Murray, London. [Google Scholar]
- David J. R., Gibert P., Mignon-Grasteau S., Legout H., Petavy G., et al. , 2003. Genetic variability of sexual size dimorphism in a natural population of Drosophila melanogaster: an isofemale-line approach. J. Genet. 82: 79–88. [DOI] [PubMed] [Google Scholar]
- de la Cova C., Johnston L. A., 2006. Myc in model organisms: a view from the flyroom. Semin. Cancer Biol. 16: 303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanoue R., Slaidina M., Leopold P., 2010. The steroid hormone ecdysone controls systemic growth by repressing dMyc function in Drosophila fat cells. Dev. Cell 18: 1012–1021. [DOI] [PubMed] [Google Scholar]
- Demontis F., Perrimon N., 2009. Integration of insulin receptor/foxo signaling and dMyc activity during muscle growth regulates body size in Drosophila. Development 136: 983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy J. B., Gergen J. P., 1991. The Drosophila segmentation gene runt acts as a position-specific numerator element necessary for the uniform expression of the sex-determining gene Sex-lethal. Genes Dev. 5: 2176–2187. [DOI] [PubMed] [Google Scholar]
- Emmerich J., Meyer C. A., de la Cruz A. F., Edgar B. A., Lehner C. F., 2004. Cyclin D does not provide essential Cdk4-independent functions in Drosophila. Genetics 168: 867–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson J. W., Quintero J. J., 2007. Indirect effects of ploidy suggest X chromosome dose, not the X:A ratio, signals sex in Drosophila. PLoS Biol. 5: e332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D. S., Cline T. W., 2013. Drosophila switch gene Sex-lethal can bypass its switch-gene target transformer to regulate aspects of female behavior. Proc. Natl. Acad. Sci. USA 110: E4474–E4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbairn D. J., 1997. Allometry for sexual dimorphism: pattern and process in the coevolution of body size in males and females. Annu. Rev. Ecol. Evol. Syst. 28: 659–687. [Google Scholar]
- Gebauer F., Merendino L., Hentze M. W., Valcarcel J., 1998. The Drosophila splicing regulator sex-lethal directly inhibits translation of male-specific-lethal 2 mRNA. RNA 4: 142–150. [PMC free article] [PubMed] [Google Scholar]
- Geminard C., Rulifson E. J., Leopold P., 2009. Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. 10: 199–207. [DOI] [PubMed] [Google Scholar]
- Grandori C., Cowley S. M., James L. P., Eisenman R. N., 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16: 653–699. [DOI] [PubMed] [Google Scholar]
- Grewal S. S., Li L., Orian A., Eisenman R. N., Edgar B. A., 2005. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat. Cell Biol. 7: 295–302. [DOI] [PubMed] [Google Scholar]
- Grifoni D., Bellosta P., 2015. Drosophila Myc: a master regulator of cellular performance. Biochim. Biophys. Acta 1849: 570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick A. V., Temeles E. J., 1989. The evolution of sexual dimorphism in animals: hypotheses and tests. Trends Ecol. Evol. 4: 136–138. [DOI] [PubMed] [Google Scholar]
- Heidmann D., Horn S., Heidmann S., Schleiffer A., Nasmyth K., et al. , 2004. The Drosophila meiotic kleisin C(2)M functions before the meiotic divisions. Chromosoma 113: 177–187. [DOI] [PubMed] [Google Scholar]
- Horabin J. I., 2005. Splitting the hedgehog signal: sex and patterning in Drosophila. Development 132: 4801–4810. [DOI] [PubMed] [Google Scholar]
- Hudry B., Khadayate S., Miguel-Aliaga I., 2016. The sexual identity of adult intestinal stem cells controls organ size and plasticity. Nature 530: 344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L. A., Prober D. A., Edgar B. A., Eisenman R. N., Gallant P., 1999. Drosophila myc regulates cellular growth during development. Cell 98: 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes G., Deshpande G., Mulvey B. B., Horabin J. I., Schedl P., 2011. The Drosophila Myc gene, diminutive, is a positive regulator of the Sex-lethal establishment promoter, Sxl-Pe. Proc. Natl. Acad. Sci. USA 108: 1543–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprowicz J., Kuenen S., Swerts J., Miskiewicz K., Verstreken P., 2014. Dynamin photoinactivation blocks Clathrin and alpha-adaptin recruitment and induces bulk membrane retrieval. J. Cell Biol. 204: 1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R. L., Kuroda M. I., 1995. Equality for X chromosomes. Science 270: 1607–1610. [DOI] [PubMed] [Google Scholar]
- Kelley R. L., Wang J., Bell L., Kuroda M. I., 1997. Sex lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature 387: 195–199. [DOI] [PubMed] [Google Scholar]
- Kohler K., Brunner E., Guan X. L., Boucke K., Greber U. F., et al. , 2009. A combined proteomic and genetic analysis identifies a role for the lipid desaturase Desat1 in starvation-induced autophagy in Drosophila. Autophagy 5: 980–990. [DOI] [PubMed] [Google Scholar]
- Kondorosi E., Roudier F., Gendreau E., 2000. Plant cell-size control: growing by ploidy? Curr. Opin. Plant Biol. 3: 488–492. [DOI] [PubMed] [Google Scholar]
- Legube G., McWeeney S. K., Lercher M. J., Akhtar A., 2006. X-chromosome-wide profiling of MSL-1 distribution and dosage compensation in Drosophila. Genes Dev. 20: 871–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang Q., Carreira-Rosario A., Maines J. Z., McKearin D. M., et al. , 2013. Mei-p26 cooperates with Bam, Bgcn and Sxl to promote early germline development in the Drosophila ovary. PLoS One 8: e58301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D. L., Sandler L., Baker B. S., Carpenter A. T., Denell R. E., et al. , 1972. Segmental aneuploidy and the genetic gross structure of the Drosophila genome. Genetics 71: 157–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo L. W., Secombe J., Little J. T., Carlos L. S., Yost C., et al. , 2005. The transcriptional repressor dMnt is a regulator of growth in Drosophila melanogaster. Mol. Cell. Biol. 25: 7078–7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott S. E., Villalta J. E., Schroth G. P., Luo S., Tonkin L. A., et al. , 2011. Noncanonical compensation of zygotic X transcription in early Drosophila melanogaster development revealed through single-embryo RNA-seq. PLoS Biol. 9: e1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi J. C., Kuroda M. I., 2015. Dosage compensation in Drosophila. Cold Spring Harb. Perspect. Biol. 7: a019398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines J. Z., Stevens L. M., Tong X., Stein D., 2004. Drosophila dMyc is required for ovary cell growth and endoreplication. Development 131: 775–786. [DOI] [PubMed] [Google Scholar]
- McKeown M., Belote J. M., Boggs R. T., 1988. Ectopic expression of the female transformer gene product leads to female differentiation of chromosomally male Drosophila. Cell 53: 887–895. [DOI] [PubMed] [Google Scholar]
- Nagoshi R. N., McKeown M., Burtis K. C., Belote J. M., Baker B. S., 1988. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell 53: 229–236. [DOI] [PubMed] [Google Scholar]
- Nakai Y., Horiuchi J., Tsuda M., Takeo S., Akahori S., et al. , 2011. Calcineurin and its regulator sra/DSCR1 are essential for sleep in Drosophila. J. Neurosci. 31: 12759–12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhout H. F., Riddiford L. M., Mirth C., Shingleton A. W., Suzuki Y., et al. , 2014. The developmental control of size in insects. Wiley Interdiscip. Rev. Dev. Biol. 3: 113–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntambi J. M., Miyazaki M., Stoehr J. P., Lan H., Kendziorski C. M., et al. , 2002. Loss of stearoyl-CoA desaturase-1 function protects mice against adiposity. Proc. Natl. Acad. Sci. USA 99: 11482–11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N., Yamanaka N., Yagi Y., Nishida Y., Kataoka H., et al. , 2009. A fat body-derived IGF-like peptide regulates postfeeding growth in Drosophila. Dev. Cell 17: 885–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S., Stocker H., Laffargue M., Wittwer F., Wymann M., et al. , 2002. The Drosophila insulin/IGF receptor controls growth and size by modulating PtdInsP(3) levels. Development 129: 4103–4109. [DOI] [PubMed] [Google Scholar]
- Orian A., van Steensel B., Delrow J., Bussemaker H. J., Li L., et al. , 2003. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 17: 1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi F., Riccardo S., Zola S., Lora C., Grifoni D., et al. , 2013. dMyc expression in the fat body affects DILP2 release and increases the expression of the fat desaturase Desat1 resulting in organismal growth. Dev. Biol. 379: 64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn J. K., Schedl P., 2007. The master switch gene sex-lethal promotes female development by negatively regulating the N-signaling pathway. Dev. Cell 12: 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce S. B., Yost C., Britton J. S., Loo L. W., Flynn E. M., et al. , 2004. dMyc is required for larval growth and endoreplication in Drosophila. Development 131: 2317–2327. [DOI] [PubMed] [Google Scholar]
- Rideout E. J., Narsaiya M. S., Grewal S. S., 2015. The sex determination gene transformer regulates male-female differences in drosophila body size. PLoS Genet. 11: e1005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D. B., Evans-Roberts S., 1979. The X-linked alpha-chain gene of Drosophila LSP-1 does not show dosage compensation. Nature 280: 691–692. [DOI] [PubMed] [Google Scholar]
- Robida M. D., Rahn A., Singh R., 2007. Genome-wide identification of alternatively spliced mRNA targets of specific RNA-binding proteins. PLoS One 2: e520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson E. J., Kim S. K., Nusse R., 2002. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science 296: 1118–1120. [DOI] [PubMed] [Google Scholar]
- Ryuda M., Tsuzuki S., Matsumoto H., Oda Y., Tanimura T., et al. , 2011. Identification of a novel gene, anorexia, regulating feeding activity via insulin signaling in Drosophila melanogaster. J. Biol. Chem. 286: 38417–38426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serna E., Gorab E., Ruiz M. F., Goday C., Eirin-Lopez J. M., et al. , 2004. The gene Sex-lethal of the Sciaridae family (order Diptera, suborder Nematocera) and its phylogeny in dipteran insects. Genetics 168: 907–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine R., 1990. Proximate determinants of sexual differences in adult body size. Am. Nat. 135: 278–283. [Google Scholar]
- Siera S. G., Cline T. W., 2008. Sexual back talk with evolutionary implications: stimulation of the Drosophila sex-determination gene sex-lethal by its target transformer. Genetics 180: 1963–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. D., Lucchesi J. C., 1969. The role of sexuality in dosage compensation in Drosophila. Genetics 61: 607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillwell R. C., Davidowitz G., 2010. Sex differences in phenotypic plasticity of a mechanism that controls body size: implications for sexual size dimorphism. Proc. Biol. Sci. 277: 3819–3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1945. A gene in Drosophila melanogaster that transforms females into males. Genetics 30: 297–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Johnson A. F., Donohue R. C., Li J., Cheng J., et al. , 2013. Dosage compensation and inverse effects in triple X metafemales of Drosophila. Proc. Natl. Acad. Sci. USA 110: 7383–7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M., Sanchez L., 1991. The sisterless-b function of the Drosophila gene scute is restricted to the stage when the X:A ratio determines the activity of Sex-lethal. Development 113: 715–722. [DOI] [PubMed] [Google Scholar]
- Venken K. J., He Y., Hoskins R. A., Bellen H. J., 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751. [DOI] [PubMed] [Google Scholar]
- Venken K. J., Popodi E., Holtzman S. L., Schulze K. L., Park S., et al. , 2010. A molecularly defined duplication set for the X chromosome of Drosophila melanogaster. Genetics 186: 1111–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst E. C., van de Zande L., Beukeboom L. W., 2010. Insect sex determination: it all evolves around transformer. Curr. Opin. Genet. Dev. 20: 376–383. [DOI] [PubMed] [Google Scholar]
- Vonesch S. C., Lamparter D., Mackay T. F., Bergmann S., Hafen E., 2016. Genome-wide analysis reveals novel regulators of growth in Drosophila melanogaster. PLoS Genet. 12: e1005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A., Hinz U., Engelbert M., Knust E., 1995. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell 82: 67–76. [DOI] [PubMed] [Google Scholar]
- Wolfner M. F., 2016. Lindsley and Sandler et al. on gene dosage and the Drosophila genome. Genetics 202: 1247–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. All supporting data and reagents used in this work will be made available upon request.