Abstract
The advent of high-throughput RNA sequencing (RNA-seq) has led to the discovery of unprecedentedly immense transcriptomes encoded by eukaryotic genomes. However, the transcriptome maps are still incomplete partly because they were mostly reconstructed based on RNA-seq reads that lack their orientations (known as unstranded reads) and certain boundary information. Methods to expand the usability of unstranded RNA-seq data by predetermining the orientation of the reads and precisely determining the boundaries of assembled transcripts could significantly benefit the quality of the resulting transcriptome maps. Here, we present a high-performing transcriptome assembly pipeline, called CAFE, that significantly improves the original assemblies, respectively assembled with stranded and/or unstranded RNA-seq data, by orienting unstranded reads using the maximum likelihood estimation and by integrating information about transcription start sites and cleavage and polyadenylation sites. Applying large-scale transcriptomic data comprising 230 billion RNA-seq reads from the ENCODE, Human BodyMap 2.0, The Cancer Genome Atlas, and GTEx projects, CAFE enabled us to predict the directions of about 220 billion unstranded reads, which led to the construction of more accurate transcriptome maps, comparable to the manually curated map, and a comprehensive lncRNA catalog that includes thousands of novel lncRNAs. Our pipeline should not only help to build comprehensive, precise transcriptome maps from complex genomes but also to expand the universe of noncoding genomes.
Comprehensive transcriptome maps enhance understanding of gene expression regulation in both coding and noncoding genomic regions (Wang et al. 2009; Martin and Wang 2011). Recently, large-scale high-throughput RNA sequencing (RNA-seq) data from the ENCODE Project were used to characterize highly complex, overlapping transcription units on both strands, revealing that more than 60% of the human genome is reproducibly transcribed in at least two different cell types (Djebali et al. 2012; Harrow et al. 2012). Intriguingly, a significant portion of these extensive transcription signals, mostly from intergenic regions, turned out to be unannotated. To identify the unannotated transcriptome, gene annotation projects, such as GENCODE (Harrow et al. 2012), Human BodyMap 2.0 (Cabili et al. 2011), and MiTranscriptome (Iyer et al. 2015), have massively reconstructed whole transcriptomes by assembling large-scale RNA-seq data and have characterized transcriptome-wide noncoding RNAs (ncRNAs). The majority of RNAs in the noncoding transcriptome were long ncRNAs (lncRNAs), such as repeat-associated ncRNAs, enhancer-associated ncRNAs, long intervening ncRNAs (lincRNAs), antisense ncRNAs, and so on, (Ulitsky et al. 2011; Derrien et al. 2012; Nam and Bartel 2012; Pauli et al. 2012; Hangauer et al. 2013; Luo et al. 2013; Brown et al. 2014; Iyer et al. 2015).
Unknown transcripts can be identified via assembly of RNA-seq data by two approaches: the genome-guided approach (known as reference-based assembly) (Yassour et al. 2009; Guttman et al. 2010; Trapnell et al. 2010; Boley et al. 2014; Mangul et al. 2014; Maretty et al. 2014); and the de novo approach (Martin et al. 2010; Grabherr et al. 2011; Schulz et al. 2012; Safikhani et al. 2013; Xie et al. 2014; Chang et al. 2015; Tjaden 2015). Because the de novo approach assembles RNA-seq reads without a guide genome, it generally requires RNA-seq data with strand information (called stranded RNA-seq data). However, for the reference-based approach, the stranded RNA-seq data had been regarded as dispensable because the sense-orientation of some reads spanning exon junctions could be predicted based on the splicing signal. For that reason, the ENCODE Project (The ENCODE Project Consortium 2012), the modENCODE Project (Gerstein et al. 2010; The modENCODE Project Consortium et al. 2010), the Human BodyMap 2.0 Project (Cabili et al. 2011), the Genotype-Tissue Expression (GTEx) Project (http://www.gtexportal.org) (The GTEx Consortium 2013), the Human Protein Atlas (Uhlen et al. 2010, 2015), and The Cancer Genome Atlas (TCGA) (Ciriello et al. 2013; Kandoth et al. 2013) Consortium produced large-scale unstranded RNA-seq data that lack strand information; genome-wide gene annotation projects have proceeded using these data. For instance, the MiTranscriptome reused 6810 publicly available unstranded RNA-seq data from ENCODE, TCGA, and other studies to reconstruct a comprehensive map of the noncoding transcriptome (Iyer et al. 2015). Despite such applications, transcriptome assembly using unstranded RNA-seq data often results in erroneous transcript models, including chimeras, particularly when there are convergent, divergent, or antisense overlaps between two genes (Garber et al. 2011; Martin and Wang 2011; Nam and Bartel 2012). Stranded RNA-seq data can benefit reference-based assembly in those genomic regions (Martin and Wang 2011). Nevertheless, the reuse of the large amount of publicly available unstranded data with the stranded data could not only enhance detection of new transcripts but also reduce the generation of erroneous transcript models.
RNA-seq-based transcriptome assembly is also challenged by the imprecise ends of assembled transcripts (Steijger et al. 2013). Early methods roughly defined the transcription structures with the support of histone modification signals, such as H3K4me3 for active promoters and H3K36me3 for active gene bodies (Guttman et al. 2010; Ulitsky et al. 2011). Later, specialized RNA sequencing techniques, such as cap analysis gene expression by sequencing (CAGE-seq) (Yamashita et al. 2011; Brown et al. 2014; Kawaji et al. 2014) and poly(A) position profiling followed by sequencing (3P-seq) (Ulitsky et al. 2011; Nam and Bartel 2012), have been successfully applied to define the ends of transcripts at single-base resolution. Integrative analysis of the specialized RNA-seq data including CAGE-seq and poly(A)-seq enabled the identification of more complete gene structures (Boley et al. 2014), valuable information for functional studies of the genes. However, despite the precise boundary information, the data have been generated from a limited number of cell types. Recently, a computational method, GETUTR, that estimates 3′ UTR ends from general RNA-seq data, was introduced (Kim et al. 2015), and it is now possible to predict the 3′ end of transcripts more accurately in any cell type using RNA-seq data.
Results
Unstranded RNA-seq causes error-prone assembly
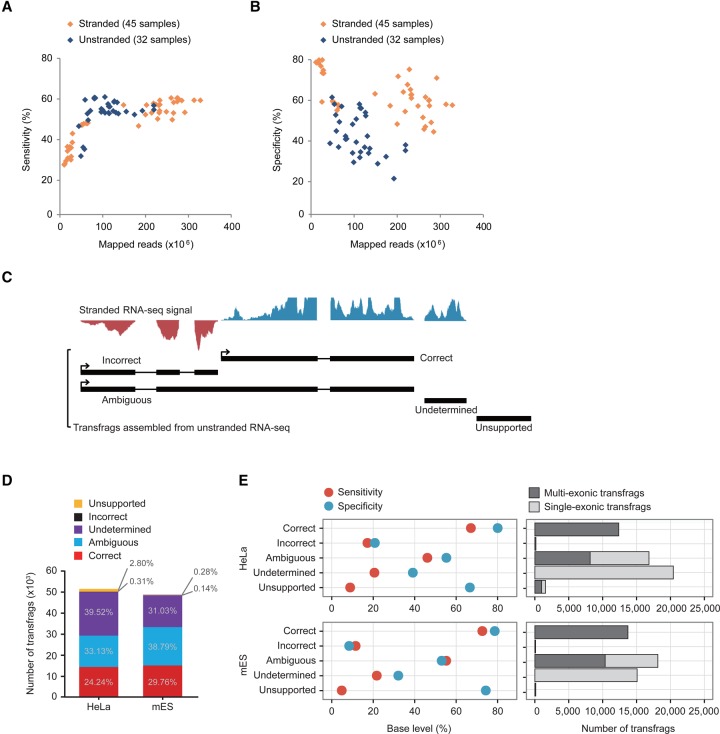
To investigate the inaccuracy of transcriptome assemblies reconstructed from unstranded RNA-seq data (unstranded assemblies) relative to that of assemblies from stranded RNA-seq data (stranded assemblies), we reconstructed 45 stranded and 32 unstranded assemblies from publicly available RNA-seq data from the ENCODE Project using Cufflinks (Trapnell et al. 2010). The resulting assemblies were evaluated based on the protein-coding genes of the GENCODE V19 annotations at the base level using Cuffcompare (Trapnell et al. 2010). The evaluation was done by counting false negative (FN), false positive (FP), and true positive (TP) bases upon agreement between the reference and the resulting assembly at the base level (Supplemental Fig. S1A; see the section “Evaluation of transcriptome assembly” in Supplemental Methods for more details). The sensitivity (TP/[TP + FN]) of the resulting assemblies appeared to be correlated with the size of mapped reads up to about 100 million mapped reads, but converged beyond that size (Fig. 1A), which suggests that many samples from the ENCODE Project still need more data to reach their maximum sensitivity. On the other hand, the specificity (TP/[TP + FP]) of the unstranded assemblies was much less than that of the stranded assemblies when the resulting assemblies were evaluated with their directionality considered, regardless of the size of the mapped reads (Fig. 1B). This result indicates that stranded reads provide more accurate information for transcriptome assembly. Previous studies failed to recognize the low accuracy of unstranded assemblies because they used a default option of Cuffcompare that ignores the directionality of the resulting assemblies during evaluation. In fact, the overall specificity, when the directionality of the resulting assemblies is not considered, neither correlates with the size of mapped reads nor differs between stranded and unstranded assemblies (Supplemental Fig. S1B,C).
Figure 1.
Error-prone unstranded transcriptome assembly. (A,B) Sensitivities (A) and specificities (B) of stranded (orange diamond) and unstranded (navy diamond) assemblies constructed from ENCODE RNA-seq data are shown over the number of mapped reads. (C) Classification of transfrags assembled from unstranded RNA-seq data. Graphs on the top are signals from stranded RNA-seq data (blue is the signal in the forward direction, and red is the signal in the reverse direction). (D) Shown are the percentages of transfrags belonging to the five groups—correct (red), ambiguous (blue), undetermined (purple), incorrect (black), and unsupported (yellow)—in HeLa and mES cells. (E) The specificity (light blue) and sensitivity (red) of the five groups compared to the reference protein-coding genes in HeLa (left, top) and mES cells (left, bottom). The number of multiexonic (dark gray) and single-exonic (gray) transfrags are indicated in each group (right).
To examine the nature and cause of the errors in unstranded assembly, we next sequenced both stranded and unstranded RNA-seq libraries that were simultaneously prepared in mouse embryonic stem (mES) cells. We also obtained a pair of publicly available stranded and unstranded RNA-seq data sets of human HeLa cells from the NCBI Gene Expression Omnibus (GEO). These reads were mapped to reference genomes (hg19 for human and mm9 for mouse) using TopHat (Supplemental Table S1; Trapnell et al. 2009), and unstranded reads (∼68 million mapped reads for mES cells and ∼40 million mapped reads for HeLa cells) were assembled using Cufflinks (Supplemental Table S1). In total, 51,045 and 48,509 transcript fragments (transfrags) whose full lengths were not examined were assembled from HeLa and mES cells, respectively (Supplemental Table S2). The resulting transfrags were divided into five groups based on their directions validated by stranded RNA-seq signals: correct, incorrect (those with an RNA-seq signal on the opposite strand), ambiguous (those with RNA-seq signals on both strands), undetermined (those with no direction), and unsupported (those with no stranded RNA-seq signals in either direction) (Fig. 1C). The criteria for the presence of antisense RNA-seq signals were determined by systematic analyses (Supplemental Fig. S2; see “Antisense RNA-seq signals” in Supplemental Methods for more details). All transfrags in the correct group (24.24% for HeLa cells and 29.76% for mES cells) were multiexonic (Fig. 1D,E); this high accuracy was the result of exon-junction reads that define the direction of the resulting intron with the splice-signal ‘GU-AG’ at the ends of the intron (Fig. 1E). The remainder were regarded as problematic transfrags (75.76% for HeLa cells and 70.24% for mES cells). They displayed low accuracies and were placed in the incorrect (0.31% and 0.14%), ambiguous (33.13% and 38.79%), undetermined (39.52% and 31.03%), and unsupported (2.8% and 0.28%) groups (Fig. 1D,E). They appeared to be severely defective in their structures and/or directions (Supplemental Fig. S3), and the majority in the undetermined group were single-exonic transfrags (Fig. 1E). However, except for those in the unsupported group (Fig. 1C), the defective transfrags (72.96% for HeLa cells and 69.96% for mES cells) could be corrected using the guide of the matched, stranded RNA-seq data.
Probabilistic estimation of the directions of unstranded RNA-seq reads
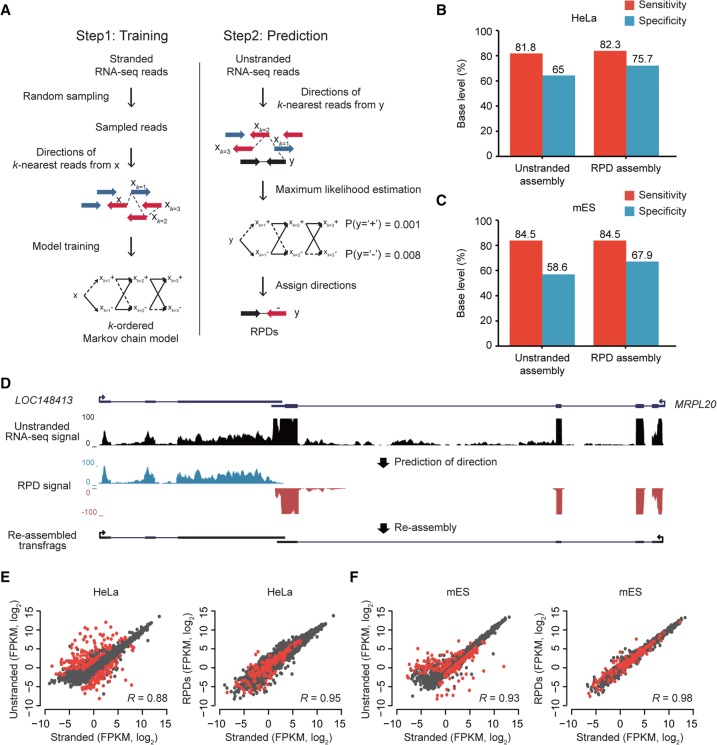
To facilitate stranded assemblies with additional stranded reads, we sought to predict the directions of unstranded RNA-seq reads using k-order Markov chain models (kMC) whose transition probabilities were estimated with the directions of a current read x and its k-nearest stranded reads, xk. In the prediction step, the direction of a read with an unknown direction, y, was determined using maximum likelihood estimation (MLE) (Fig. 2A). A read with a predicted direction (RPD) was treated as a stranded read and was used in the downstream assembly. For unstranded paired-end reads, the direction of a fragment was independently predicted. If the predicted direction of a fragment was not consistent with that of another fragment, the direction with a greater probability was chosen. Merely <1% (0.28% for HeLa and 0.14% for mES) were paired-end reads with discordantly predicted strands (Supplemental Fig. S4). Performing systematic analyses while increasing k, we found the optimum to be k = 3, a value at which the accuracy is maximized and the computational cost is minimized (Supplemental Fig. S5A,B). Compared to a simple majority voting method with k-nearest stranded reads, kMC performed better as k increased (Supplemental Fig. S5C,D). Thus, we predicted the directions of all unstranded RNA-seq data using the Markov chain model with the optimum k-order and assembled all stranded read-like RPDs. Compared to the original assembly (unstranded assembly), those that were reassembled from RPDs (RPD assembly) were significantly improved by 9.3%–10.7% in their specificity without compromising their sensitivity (Fig. 2B for HeLa cells and Fig. 2C for mES cells). For instance, unstranded reads from a genomic locus where LOC148413 and MRPL20 are convergently transcribed were assembled into an erroneous annotation, but their RPDs led to correction of the erroneous gene structure (Fig. 2D). The prediction of strand information also significantly improved the specificity of the annotations of antisense-overlapped loci without compromising the sensitivity at the base level (Supplemental Fig. S6A,B).
Figure 2.
Prediction of read directions using MLE. (A) Overview of kMC training and MLE of read direction. (Left) S base reads randomly sampled from stranded RNA-seq reads and their matched step-wise k-nearest reads (xk=1, xk = 2, xk=3,…) were used for training kMC. Blue arrows are reads in the forward (+) direction, and red arrows are reads in the reverse (−) direction. (Right) Prediction of read direction using MLE. Step-wise k-nearest stranded reads (xk=1, xk = 2, xk=3,…) from a query unstranded read (black arrow) were extracted and used to calculate two likelihoods at (+) and (−). A direction with the maximum likelihood is finally assigned to the query read. (B,C) Accuracies of transcriptomes assembled with RPDs (k = 3) and unstranded reads in HeLa (B) and mES cells (C). (D) An example of resulting transfrags reassembled with RPDs. LOC148413 and MRPL20 are convergently overlapped at a locus where unstranded RNA-seq signals (black) are not separated, but blue and red RPD signals are clearly separated in the forward and reverse directions, respectively. (E,F). Comparisons of gene expression values (FPKM, log2) estimated by stranded (x-axis) and unstranded reads (y-axis, left) or RPDs (y-axis, right) in HeLa (E) and mES cells (F). The correlation coefficients were calculated with Pearson's correlation between the x- and y-axis values. The red dots indicate genes with antisense-overlapped genes.
The use of stranded RNA-seq data leads not only to better transcriptome assembly but also, in principle, to better gene expression quantification. To test whether the expression quantification benefits from the prediction of strand information, the gene expression values were calculated with unstranded and corresponding RPDs and then were compared to those calculated with stranded reads (Fig. 2E,F; Supplemental Fig. S6C). Overall, the unstranded reads overestimated the expression level of genes in the loci with antisense-overlapping transcripts, but RPDs corrected the overestimation, leading to better correlation with those of stranded reads.
To test the general usage of the kMC model, we predicted the directions of unstranded reads from HeLa cells using the kMC model trained in mES cells, and vice versa. The species-mismatched models were comparable to the species-matched models (Supplemental Fig. S7), suggesting that the kMC model can be generalized to other cell types and species.
Refining boundaries and finding new exon junctions between transfrags
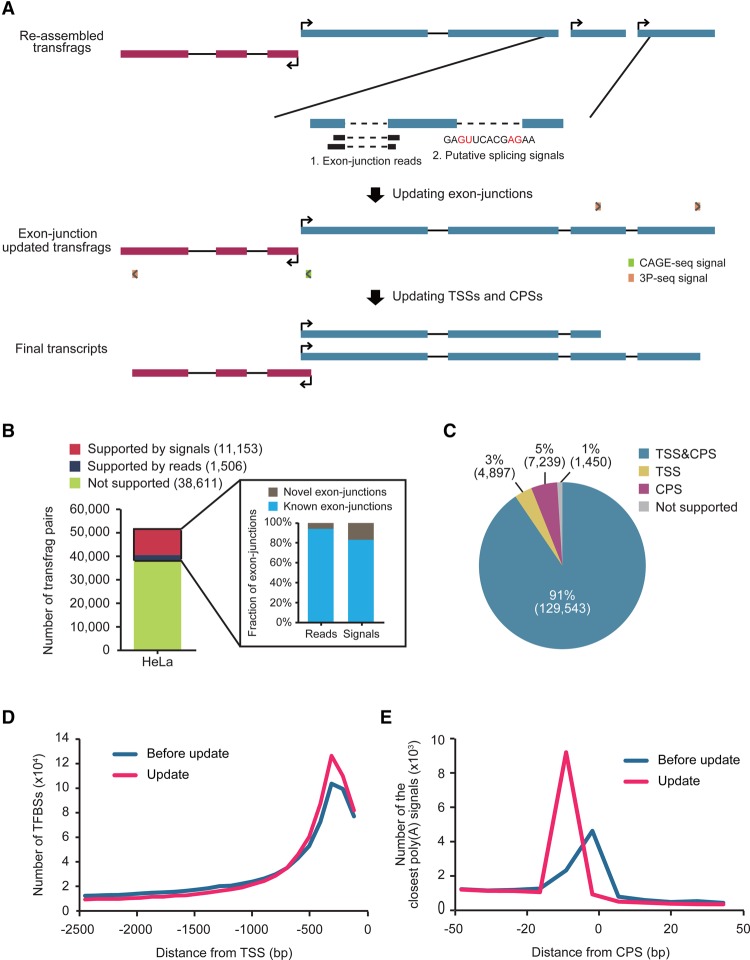
Shallow sequencing depth and short read length often cause transcript fragmentation in transcriptome assembly, mainly due to missing exon-junction reads and discontinuity of read overlaps. To improve the integrity of the assembled transcriptome, the missed exon junctions were examined by either experimental (Clark et al. 2015) or computational approaches (Fig. 3A; see Methods for more details; Shapiro and Senapathy 1987; Reese et al. 1997; Pertea et al. 2001; Yeo and Burge 2004; Desmet et al. 2009). Of 51,270 potential exon junctions, 1506 (3%) were additionally supported by the method in HeLa cells (Fig. 3B) and a similar fraction of potential junctions were supported in mES cells (Supplemental Fig. S8A). Of the newly connected exon junctions, 91.0%–94.4% were present in GENCODE annotations (V19) and the remainder were novel (Fig. 3B; Supplemental Fig. S8A). The unconnected potential exon junctions were examined further with the program MaxEntScan (Yeo and Burge 2004) to determine whether the most likely putative splicing signal, ‘GU-AG,’ existed in the region between two neighboring transfrags (Fig. 3A). Using that approach, 11,153 potential junctions for HeLa cells and 7634 for mES cells were newly connected (Fig. 3B; Supplemental Fig. S8A); 84.7%–85.2% were present in GENCODE gene annotations and the remainder were novel (Fig. 3B; Supplemental Fig. S8A).
Figure 3.
Updating exon junctions, TSSs, and CPSs in transfrag models. (A) Shown is a workflow for updating transfrag models, which comprises two steps: (1) updating exon junctions, and (2) updating TSSs and CPSs. (B) The number of neighboring transfrag pairs supported by putative splicing signals (red), by exon-junction reads (navy), and by neither (olive) in HeLa cells. The numbers in parentheses in the key indicate the number of pairs in each group. Among exon junctions supported by either exon-junction reads or putative splicing signals, the fractions of known (cyan) and novel (gray) exon junctions in GENCODE annotations are shown in the inset. (C) The fraction of transfrags updated with both TSS and CPS (blue), with only TSS (yellow), with only CPS (magenta), and with neither TSS or CPS (gray) in HeLa cells. (D) The number of TFBSs upstream of the original 5′ end (blue) and of the 5′ end updated with a TSS (pink) in HeLa cells. (E) The number of transfrags with a close poly(A) signal, AAUAAA, over the relative distances from the original 3′ end (blue) and the 3′ end updated with a CPS (pink) of transfrags in HeLa cells.
RNA-seq-based transcriptome assembly often results in imprecise transcript boundaries (Supplemental Fig. S9). To improve transfrag boundary annotation, transcription start sites (TSSs), determined from publicly available CAGE-seq (The FANTOM Consortium and the RIKEN PMI and CLST [DGT] 2014), and cleavage and polyadenylation sites (CPSs), determined from poly(A) position profiling by sequencing (3P-seq) (Nam et al. 2014), were incorporated into relevant transfrags. For TSSs and CPSs, respectively, 93%–94% and 96%–98% of transfrags were either confirmed or revised (Fig. 3C; Supplemental Fig. S8B). Transfrags updated for both TSSs and CPSs (91%–92%) were regarded as full-length transcripts (Fig. 3C; Supplemental Fig. S8B). Updating TSSs improved the definition of the upstream promoter regions in which transcription factor binding sites (TFBSs) are significantly enriched (Fig. 3D). Similarly, transfrags with CPSs displayed an enriched poly(A) signal, AAUAAA within 15–30 nt upstream of the cleavage site, compared to those without CPS updates (Fig. 3E).
CAFE improves transcriptome annotations
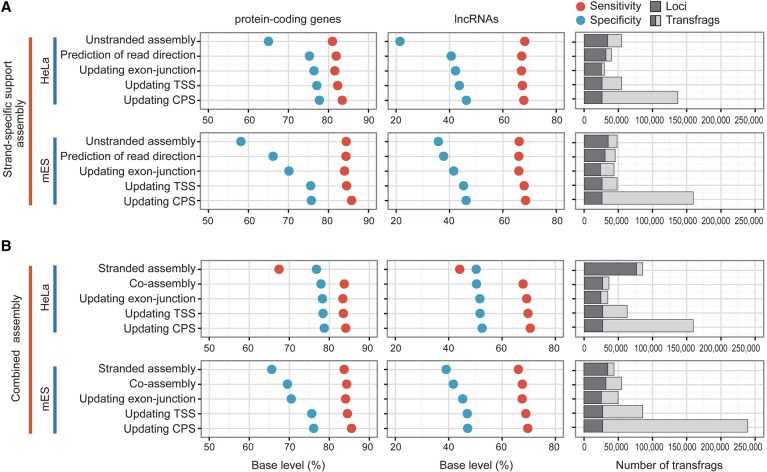
We developed a pipeline, CAFE, which utilizes both stranded and unstranded RNA-seq data to reconstruct full-length transcripts effectively (Supplemental Fig. S10). To evaluate the pipeline, we first sought to reassemble only RPDs (named strand-specific support assembly) from HeLa and mES cells and measured the accuracy of intermediate assembly at each step by comparing our results to GENCODE protein-coding genes in the base level (Fig. 4A). After updating TSSs and CPSs, the evaluation proceeded with only transfrags with a major TSS and CPS, while the count of transfrags took account of all isoforms. In total, 143,129 transfrags from 25,118 loci were assembled from HeLa cells; the quality of the resulting assembly for protein-coding genes was improved by ∼14% for specificity and ∼1.6% for sensitivity, compared to the original unstranded assembly (Fig. 4A). Similarly, CAFE assembled 164,423 transfrags from 24,605 loci in mES cells and improved the quality of protein-coding gene assembly by 18.4% for specificity and 1.3% for sensitivity (Fig. 4A). Although the resulting transfrags that overlapped with GENCODE lncRNAs were relatively less accurate than those of protein-coding genes, partly because of their low and condition-specific expression patterns, CAFE also improved the quality of such transfrags by 22.1% and 8.3% for specificity in HeLa and mES cells, respectively, without compromising sensitivity. A major factor behind the increased specificity for both protein-coding and lncRNA genes was the prediction of read direction and reassembly (Fig. 4A).
Figure 4.
Step-wise evaluation of transcriptomes reassembled by CAFE. (A) Shown are the accuracies and sizes of strand-specific support transcriptomes (RPD assembly) at each step of CAFE in HeLa (top) and mES cells (bottom). The sensitivity (red solid circle) and specificity (blue) of the assemblies are measured by comparing to GENCODE protein-coding genes (left panel) and lncRNAs (middle panel). The number of assembled transfrags and their loci are indicated at each step (right panel). (B) Shown are the accuracies and sizes of combined transcriptome assemblies of both stranded reads and RPDs. The low sensitivity of the stranded assembly from HeLa cells is presumably because the stranded reads are of the single-end type and are 36 or 72 nt long. Otherwise, as in A.
We next performed combined assembly (coassembly) of both stranded reads and RPDs using CAFE. The resulting assemblies included 166,227 transfrags from 25,591 loci in HeLa cells, of which 93.26% had their own TSSs and 94.62% had their own CPSs, and 244,085 transfrags from 26,332 loci in mES cells, of which 94.83% had their own TSSs and 98.08% had their own CPSs (Fig. 4B; Supplemental Fig. S11). Both the sensitivity and specificity of the final resulting transcriptome that overlapped with the GENCODE genes were greatly improved at the base level, compared to both of the original assemblies, and were slightly improved compared to the strand-specific support assembly (Fig. 4; Supplemental Fig. S12).
Benchmarking other transcriptome assemblers
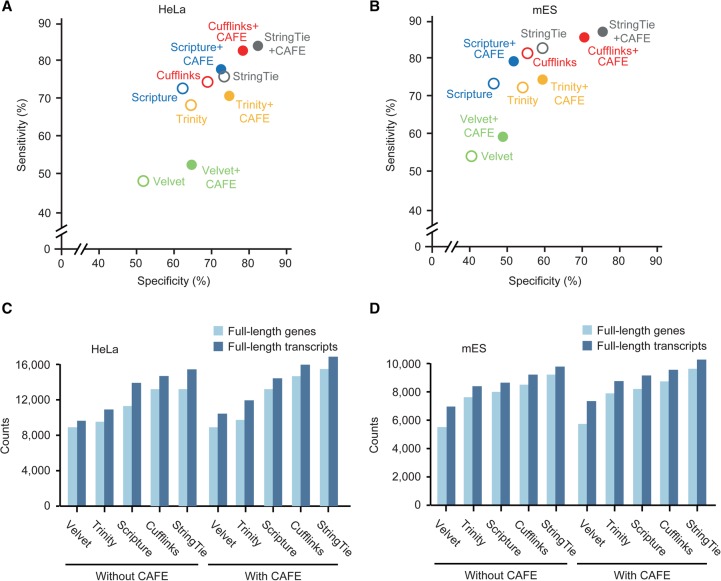
To check whether the improvement in transcriptome assembly depends on a specific base assembler (originally, Cufflinks+CAFE), other reference-based assemblers, Scripture (Guttman et al. 2010) and StringTie (Pertea et al. 2015), were benchmarked using the same data set (Scripture+CAFE and StringTie+CAFE). The resulting assemblies were more accurate for both HeLa (8.6%∼9.9% greater sensitivity and 11.4%∼12.9% greater specificity) (Fig. 5A) and mES cells (3.2%∼4.9% greater sensitivity and 10.2%∼10.6% greater specificity) (Fig. 5B) than the original assemblies in the base level. Because the recently published reference-based assembly pipeline GRITS utilizes only strand-specific paired-end reads, we excluded it from the benchmarking. Additionally, two available de novo assemblers, Trinity and Velvet (Zerbino and Birney 2008), were also benchmarked by predicting the strand information of unstranded reads using CAFE, and the resulting de novo assemblies of RPDs and stranded reads were more accurate than the original de novo assemblies (Fig. 5A,B). Taken together, CAFE was able to improve initial assemblies robustly regardless of the base assembler used.
Figure 5.
Benchmarking other base assemblers. (A,B) The accuracies of combined transcriptome assemblies (solid circles) reconstructed by CAFE with base assemblers and of the original transcriptome assemblies (open circles) reconstructed by respective base assemblers, such as Cufflinks (red), Scripture (blue), StringTie (gray), Velvet (green), and Trinity (yellow), in HeLa (A) and mES cells (B). The accuracies of the original assemblies were calculated by averaging the accuracies of stranded and unstranded assemblies reconstructed by each base assembler. Velvet and Trinity were used as de novo assemblers, and Scripture, StringTie, and Cufflinks were used as reference-based assemblers. (C,D) The numbers of full-length genes (light blue) and transcripts (blue) in the coassemblies were compared to those in the original assemblies from HeLa (C) and mES cells (D). For the original assemblies, the higher number of full-length genes in the stranded and unstranded original assemblies was chosen.
The number of full-length transcripts is another important aspect in the quality of transcriptome assembly. We thus compared the number of full-length transcripts assembled by CAFE to the number in the original and de novo assemblies. For these comparisons, transcripts that simultaneously included a TSS in the first exon and a CPS in the last exon were considered to be full-length transcripts. Trinity+CAFE and Velvet+CAFE assembled 8.8%∼10.4% more full-length transcripts than in the original de novo assemblies (Fig. 5C). Cufflinks+CAFE, StringTie+CAFE, and Scripture+CAFE assembled 14.6%, 10.1%, and 13.9% more full-length transcripts than in the original assembly, respectively (Fig. 5C). Similarly, CAFE constructed more full-length transcripts than in the original and de novo assemblies from mES cells (Fig. 5D). All source codes and a detailed manual for using CAFE can be found in Supplemental Materials File 1 and on our website (http://big.hanyang.ac.kr/CAFE).
High-confidence human transcriptome map
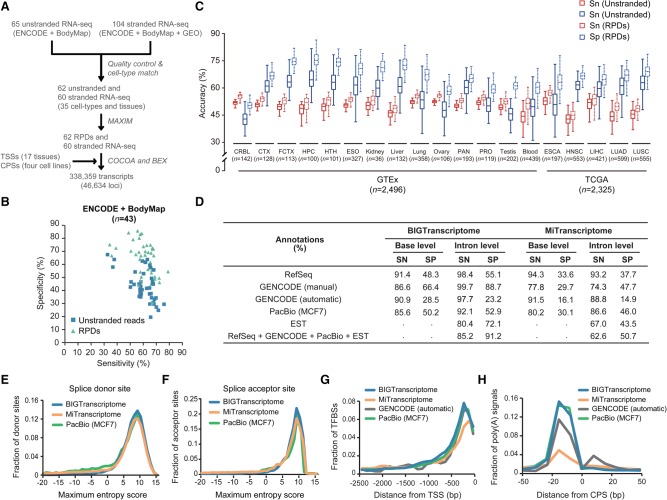
To construct a comprehensive human transcriptome map, large-scale transcriptome data were collected from the ENCODE Project, the Human BodyMap 2.0 Project, and NCBI GEO human cell lines; these data included 65 unstranded and 104 stranded RNA-seq data, TSS profiles across 17 human tissues, and CPS profiles from four human cell lines (Nam et al. 2014). We first predicted the directions of approximately six billion reads from 62 unstranded RNA-seq data sets using 60 cell-type–matched stranded RNA-seq data sets from 35 different cell types (Fig. 6A; Supplemental Table S3). The transcriptome assembly of the RPDs was more accurate than the unstranded transcriptome assembly at the base level (Fig. 6B), suggesting that the prediction of read directions significantly reduced erroneous transfrag assemblies. The coassembly of RPDs and stranded reads with TSS and CPS profiles (Fig. 6A) reconstructed 338,359 transcripts from 46,634 loci, referred to here as BIGTranscriptome. To expand our transcriptome map, we additionally predicted the strand information of >4800 individual unstranded RNA-seq data from 19 different tissues and tumors from the GTEx (The GTEx Consortium 2013) and TCGA Projects (Ciriello et al. 2013; Kandoth et al. 2013) and reconstructed more accurate transcriptome maps using the RPDs rather than using the unstranded reads at the base level (Fig. 6C). The coassembly of RPDs and stranded reads with TSS and CPS profiles using the same pipeline shown in Figure 6A reconstructed tissue-specific transcriptome maps, referred to here as BIGTranscriptome-TS (Supplemental Table S4). To examine their quality, all annotations were compared to those of RefSeq, GENCODE (manual), GENCODE (automatic), Pacific Biosciences (PacBio) long read assembly, and MiTranscriptome in terms of the number of full-length independent transcripts. Although BIGTranscriptome reconstructed fewer transcripts than did MiTranscriptome (Supplemental Table S4), it contained more (16,376, 35.11%) independent genes that had at least one transcript with boundaries defined by TSSs and CPSs than did MiTranscriptome (5741, 6.30%) and GENCODE (manual: 6522, 13.59%; automatic: 1301, 7.34%) (Supplemental Table S5). Moreover, BIGTranscriptome included 6000 full-length independent single-exonic transcripts with a direction (∼32.24% of single-exonic transcripts), whereas other annotations included tens of thousands of single-exonic transcripts, only 1.04%∼20.91% of which were full-length independent single-exonic transcripts (Supplemental Table S6A). Thousands of those that remained appeared to be partial fragments that were included in BIGTranscriptome annotations (Supplemental Table S6B).
Figure 6.
Comprehensive human transcriptome map. (A) A schematic flow for the reconstruction of the BIGTranscriptome map using large-scale RNA-seq samples from human cell lines, ENCODE, and Human BodyMap 2.0 Projects. (B) Accuracies of unstranded (blue) and RPD assemblies (mint) from the ENCODE and Human BodyMap 2.0 projects. (C) Sensitivities (red) and specificities (blue) of unstranded assemblies (solid line box) and RPD assemblies (dotted line box) are shown in box plots. The unstranded RNA-seq data are from GTEx (14 tissues) and TCGA Project (five tumor types). The numbers (n) indicate the sample numbers in each group. (CRBL) Brain cerebellum, (CTX) brain cortex, (FCTX) brain frontal cortex, (HPC) brain hippocampus, (HTH) brain hypothalamus, (ESO) esophagus-mucosa, (PAN) pancreas, (PRO) prostate, (ESCA) esophageal carcinoma, (HNSC) head and neck squamous cell carcinoma, (LIHC) liver hepatocellular carcinoma, (LUAD) lung adenocarcinoma, and (LUSC) lung squamous cell carcinoma. (D) Shown are the accuracies of BIGTranscriptome and MiTranscriptome at the base and intron levels based on four different sets of annotations (RefSeq, manual and automatic GENCODE, PacBio, and EST), and a combined set of annotations. (SN) Sensitivity, (SP) specificity. (E,F) Maximum entropy scores of the putative splice donor sites (E) and of putative splice acceptor sites (F). Blue lines are from BIGTranscriptome, green lines are from PacBio assembly, and orange lines are from MiTranscriptome. (G) The fraction of TFBSs upstream of the 5′ end of BIGTranscriptome transcripts (blue) was compared to those of MiTranscriptome (orange), GENCODE (automatic) (black), and PacBio assembly (green). (H) The fraction of the closest poly(A) signals, AAUAAA, in the region just upstream of the 3′ end of BIGTranscriptome annotations (blue) compared to those of MiTranscriptome (orange), GENCODE (automatic) (black), and PacBio assembly (green).
The accuracy of BIGTranscriptome annotations was also evaluated at the base level in terms of sensitivity and specificity based on RefSeq, GENCODE (manual), GENCODE (automatic), or PacBio (MCF7) annotations. BIGTranscriptome annotations were found to be 14.7%∼36.7% more specific for the RefSeq and GENCODE (manual) transcripts than were MiTranscriptome annotations, without compromising sensitivity (Fig. 6D). We also checked if the intron structures of BIGTranscriptome agreed with those of the RefSeq, GENCODE, expression sequence tags (ESTs), PacBio, and combined annotations (RefSeq + GENCODE + EST + PacBio) and compared the results to those of MiTranscriptome. Overall, our BIGTranscriptome annotations were superior to those of MiTranscriptome for both sensitivity (22.6% greater) and specificity (40.5% greater) in the combined annotations (Fig. 6D), indicating that BIGTranscriptome transcripts are less likely to be fragmented. Eighty-seven percent of the 29,274 putative BIGTranscriptome introns, not detected in the combined annotations, included a canonical splicing signal, ‘GU’-‘AG’, two nucleotides away from both ends; the remainder lacked the canonical splice signal (Supplemental Table S7). Although the putative introns of MiTranscriptome also included the canonical splice signals at a similar level as BIGTranscriptome (Supplemental Table S7), the putative splice sites of MiTranscriptome showed significantly lower maximum entropy scores than those of BIGTranscriptome at both splice donor and acceptor sites (Fig. 6E,F). We also evaluated tissue-specific BIGTranscriptome-TS annotations at the base and intron levels in terms of sensitivity and specificity based on RefSeq, GENCODE, EST, or PacBio annotations and found similar levels of accuracy in the transcriptome maps (Supplemental Fig. S13A).
To evaluate the accuracy of BIGTranscriptome transcript boundaries, we counted TFBSs in the regions upstream of the TSSs and canonical poly(A) signals in the regions around the CPSs. A higher fraction of TFBSs within 500 nt upstream of a TSS (Fig. 6G) and poly(A) signals within 15–30 nt upstream of a CPS (Fig. 6H) were observed for BIGTranscriptome transcripts than for MiTranscriptome and GENCODE (automatic), indicating that BIGTranscriptome includes transcripts with more precise ends. In addition, BIGTranscriptome agreed with PacBio and GENCODE about the 5′ and 3′ end positions of assembled transcripts better than did MiTranscriptome (Supplemental Fig. S13B–E). However, because the CPS information was profiled from only four human cell types, we additionally updated the cell-type–specific 3′ ends of transcripts using GETUTR, which predicts the 3′ end of a transcript from RNA-seq data (Kim et al. 2015).
Based on BIGTranscriptome and BIGTranscriptome-TS annotations, all ENCODE, Human BodyMap, TCGA, and GTEx RPDs were used to quantify the gene expression values (read counts) with featureCounts version 1.5.1 (Liao et al. 2014) (see “Expression profiling using RPD BAM files” in Supplemental Methods for more details). All BIGTranscriptome and BIGTranscriptome-TS annotations, as well as the expression data, can be downloaded from the NCBI GEO by following the pointers summarized in Supplemental Table S9.
A confident catalog of human lncRNAs
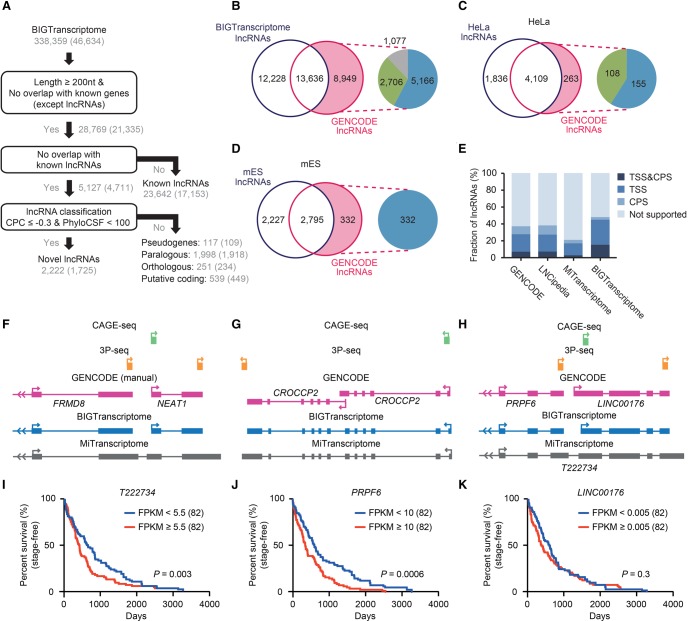
With our BIGTranscriptome map, we next sought to identify novel and known lncRNAs using the following lncRNA annotation pipeline, slightly modified from a previous method (Nam and Bartel 2012). Of 338,359 transcripts from 46,634 loci, 28,769 (8.5%) were longer than 200 nt in length and did not overlap with exons of known protein-coding genes or ncRNA genes, excepting lncRNAs. These transcripts were separated into known and putative lncRNAs. Transcripts totaling 23,642 from 17,153 genomic loci were previously annotated lncRNAs (Fig. 7A). Putative lncRNAs were subsequently subjected to coding-potential classifiers: (1) coding potential calculator (CPC) (Supplemental Fig. S14; Kong et al. 2007); and (2) PhyloCSF (Lin et al. 2011) (see the “Classification of lncRNAs” section in Supplemental Methods for more details). Novel lncRNAs totaling 2222 (from 1725 loci) (Supplemental Table S8) were identified as our human lncRNA catalog (Fig. 7A). Although the novel lncRNAs predominantly consisted of two exons, similar to known lncRNAs, their median length (890 nt) was longer than that (722 nt) of known lncRNAs (Supplemental Fig. S15), suggesting that our lncRNA catalog included more intact gene models. Transcripts totaling 2905 (from 2735 loci) that do not meet the lncRNA criteria were classified into pseudogenes (117 transcripts), paralogous transcripts (1998), orthologous transcripts (251), and putative coding transcripts (539) (see “Classification of unknown transcripts” in Supplemental Methods for more details).
Figure 7.
BIGTranscriptome includes known and novel noncoding genes. (A) A schematic flow for annotating novel and known noncoding genes in BIGTranscriptome. (B) The Venn diagrams display the fraction of BIGTranscriptome lncRNAs that are published GENCODE lncRNAs. The inset indicates that GENCODE lncRNAs (8949) not detected in BIGTranscriptome were classified as overlapping with known genes (blue), overlapping with falsely fused genes (green), or truly missed in our catalog (gray). (C,D) Transcriptomes of HeLa (C) and mES cells (D) were compared to GENCODE lncRNAs, expressed over 1 FPKM in the matched cell types. The insets indicate that HeLa- and mES-expressed lncRNAs not detected in our lncRNA set were filtered by either overlap with known genes (blue) or misannotation (green). (E) The fractions of the indicated lncRNA sets with both TSS and CPS, either site, or neither site are shown in bar graphs. (F–H) Examples of misannotated gene models in public databases (MiTranscriptome and GENCODE). (F) The gene for a well-studied lncRNA, NEAT1, has been combined with a protein-coding gene, FRMD8, leading to misannotation as a protein-coding gene. (G) CROCCP2 is annotated in GENCODE (automatic) as having two independent isoforms whereas it is annotated as a single transcript in BIGTranscriptome and MiTranscriptome. (H) Gene models of BIGTranscriptome and MiTranscriptome, and CAGE-seq and 3P-seq data, at a locus. A fused single form, T222734, was annotated in MiTranscriptome whereas two independent genes, PRPF6 and LINC00176, were annotated in BIGTranscriptome. (I–K) Survival analyses for TCGA liver cancer samples based on the resulting gene models. One hundred sixty-four patient samples including termination events were divided into two groups, the top 50% (red) and bottom 50% (blue), by the median FPKM values of T222834 (I), PRPF6 (J), and LINC00176 (K).
To evaluate our human lncRNA catalogs, genomic loci encoding GENCODE lncRNAs were compared to those of our catalogs. A majority (60.37% for GENCODE) were detected in our human lncRNA catalogs (Fig. 7B). Of 8949 GENCODE lncRNAs that were not in our catalogs, 7872 (87.96%) were transcripts that overlapped with annotated genes in RefSeq, GENCODE, Ensembl, or MiTranscriptome (Fig. 7B). We determined that 5166 (65.63%) of 7872 undetected GENCODE lncRNAs had been filtered out because they overlapped with known genes, and the remainder, 2706, overlapped with a falsely fused transcript of a protein-coding gene and an lncRNA, mostly originating from MiTranscriptome (Fig. 7B). For example, NEAT1, a well-studied lncRNA, is annotated as a protein-coding gene in MiTranscriptome because it was fused with FRMD8, an upstream protein-coding gene (Fig. 7F). Only 4.76% (1077/22,585) of GENCODE lncRNAs were truly missed in our catalog. To verify this notion, the genomic loci encoding lncRNAs annotated from HeLa (Fig. 7A) and mES cells (Supplemental Fig. S16) were respectively compared to those of GENCODE lncRNAs expressed at greater than one fragment per kilobase of exons per million mapped fragments (FPKM) in corresponding cells. We found that our HeLa and mES lncRNA sets included a majority of GENCODE lncRNAs (93.98% for HeLa and 89.38% for mES cells) (Fig. 7C,D). All of the lncRNAs not included in our sets were transcripts that overlapped with known genes or that were misannotated in the public databases (Fig. 7C,D). Similarly, the cell-type–specific lncRNA sets included more transcripts (∼67% for HeLa and ∼76% for mES cells) with fully or partially evident ends than did the non-cell-type–specific lncRNA catalog (∼48%) (Supplemental Fig. S17). Nevertheless, our lncRNA catalog (13.45%) included many more intact gene models with fully evident ends than those of GENCODE, LNCipedia, and MiTranscriptome (6.61%, 6.71%, and 2.22%, respectively) (Fig. 7E). For example, CROCCP2 is shown to exist in two independent isoforms in GENCODE; however, it actually exists in a single form, as shown in both BIGTranscriptome and MiTranscriptome (Fig. 7G).
We next sought to examine whether our BIGTranscriptome annotations could benefit the expression profiling of genes and their downstream analysis. T222734 was annotated as a single form in MiTranscriptome, but this sequence turned out to be an independent protein-coding gene, PRPF6, and an lncRNA, LINC00176, evident with CAGE-seq and 3P-seq, in BIGTranscriptome (Fig. 7H). Using the single and the two independent forms of the genes, we performed survival analyses for 164 liver cancer samples from TCGA (see the “Survival analysis” section in Supplemental Methods for more details). We found that the PRPF6 gene is a more significant marker (log rank test, P = 0.0006) (Fig. 7J) for the prognosis of the liver cancer patients than T222734 (log rank test, P = 0.003) (Fig. 7I), whereas LINC00176 is expressed at a low level and is not a significant marker (log rank test, P = 0.3) (Fig. 7K). Similarly, AC15645 (lncRNA) and MLXIP (protein-coding gene) were annotated in BIGTranscriptome, but they were annotated as a single form (T087998) in MiTranscriptome (Supplemental Fig. S18A). The MLXIP annotated in our BIGTranscriptome appeared to be a more significant prognosis marker (Supplemental Fig. S18E) than T087998 and T088004 annotated in MiTranscriptome (Supplemental Fig. S18B,C), but the lncRNA, AC15645, turned out to be expressed at a low level (Supplemental Fig. S18D).
Discussion
Our new transcriptome assembly pipeline, CAFE, enabled us to significantly improve the quality of the resulting assemblies by resurrecting large-scale unstranded RNA-seq data, which was formerly used for less informative or less specific transcriptome assembly. The reuse of the large-scale unstranded RNA-seq data could be valuable in three ways. For example, other public transcriptome databases, such as the TCGA Consortium (Ciriello et al. 2013; Kandoth et al. 2013), the GTEx Project (The GTEx Consortium 2013), the Human Protein Atlas (Uhlen et al. 2010, 2015), and NCBI GEO, include large-scale unstranded RNA-seq data. Hence, determining the direction of unstranded sequence reads enables the construction of highly accurate transcriptome maps, which is necessary for highly qualitative downstream analyses. Although determining the direction of unstranded reads requires stranded data in the corresponding cell type or tissue, the use of pooled stranded data can still be of benefit to the prediction of transcript direction and the following assembly. In fact, the RPDs of unstranded Human BodyMap 2.0, GTEx, and TCGA data were predicted using pooled stranded RNA-seq data and showed better performance for specificity (Fig. 6B,C). Secondly, in the case of genes with low expression such as those encoding lncRNAs, additional RPDs benefit transcriptome assembly by increasing the read-depth of those genes. Although the targeted capture of low-abundance transcripts like lncRNAs using antisense oligonucleotides enabled an increase in the copy number of the target transcripts (Clark et al. 2015), this approach is only applicable to known transcripts. Thirdly, additional RPDs could increase the detection of missed exon junctions, resulting in the connection of fragmented transfrags.
In this study, we utilized CAGE-seq and 3P-seq data to profile transcript TSSs and CPSs, which detect unambiguous ends at single-base resolution as well as transcript alternative forms. However, the assignment of multiple TSSs and CPSs raises a question: Which pairs of ends, in all possible combinations, are relevant? Moreover, if a gene has alternative splicing isoforms, the number of possible isoforms is exponentially increased by multiple TSSs and CPSs. CAFE now generates all possible but unique isoforms, some of which would not actually exist in cells. Therefore, a precise way to determine a TSS-CPS pair simultaneously would provide biologically relevant isoforms directly. One approach is to integrate paired-end ditag (PET) data that contain both 5′ and 3′ end sequence tags of transcripts (Djebali et al. 2012), and an alternative is to sequence full-length RNAs using third-generation sequencing methods such as Iso-seq (Sharon et al. 2013).
Methods
mES cell culture
mES cells were cultured in regular media containing 15% FBS, 1% penicillin-streptomycin, 1% glutamine, 1% NEAA, and leukemia inhibitory factor (LIF). For mES cell maintenance, dishes were coated with 0.2% gelatin, and irradiated CF1 mouse embryonic fibroblasts were plated as a confluent layer of feeder cells. mES cells were seeded at a density of 50,000 cells/6-well plate and were split every 2–3 days.
K-ordered Markov chain models for read direction
To predict the direction of unstranded reads mapped to the genome, kMC models were trained with the directions of the k-nearest stranded reads relative to a target read and with the direction of the target read. We built a training data set including S base reads randomly selected from stranded reads mapped to genomes and their matched k-nearest reads. To acquire the k-nearest reads, we used a step-wise k-nearest method, in which the read xk=1 nearest to a query read xk=0 was first selected, then the read xk=2 nearest to the current read xk=1 was selected, then the read xk=3 nearest to the current read xk=2 was selected, and so on. To train unbiased models, we used 10 million as S, a large enough sampling number that is proportional to the k (also proportional to the number of states and edges to train). Practically, 2 × K matrix M+ or M− for each emission value (+ and −) were constructed from the training data and each cell m+i,j or m−i,j in the matrix indicates the fraction of + or − direction of the jth-nearest read xk=j when the emission value (direction) of the previous state is i.
Maximum likelihood estimation of read direction
The direction of an unstranded read, r, was inferred from the trained kMC models given step-wise k-nearest stranded reads of a query unstranded read mapped to a genome locus using MLE as in the following equation:
where i is the direction of the kth-nearest read, L is a set of possible directions that are {+,−}, and is the maximum likelihood direction of the unstranded read r. Using the MLE, all maximum likelihood directions were predicted for all unstranded reads. If an unstranded read was paired-end, then its direction was determined differently, as follows. If a fragment of a paired-end read spanned an exon junction, the direction of the read was directly determined by the splice signal without MLE. If the directions of two fragments of a paired-read were inconsistent, the direction with greater likelihood was chosen for the read.
Updating exon junctions
To update exon-junction signals missed in the original assembly, all pairs of neighboring transfrags on the same strand within a distance ranging from 50 bp to the 99th percentile of the lengths of all known introns (50–265,006 bp for human and 50–240,764 bp for mouse) were re-examined. The neighboring transfrags within a distance of 50 bp were combined using a default Cufflinks parameter, ‘--overlap-radius = 50’. If more than two exon-junction reads in at least two samples were detected, the neighbored transfrags were connected by the junction. Otherwise, the gaps between two neighboring transfrags were further scrutinized to detect cis-splicing signals. The gaps including splice donor ‘GU’ and acceptor ‘AG’ signals, but not TSSs or CPSs, between two neighboring transfrags were scanned by MaxEntScan (version 20040420), which calculates entropy scores for splice donor and acceptor sites. If the maximum entropy scores of both the splice donor and acceptor sites were above 0.217, a cutoff used in previous studies (Jian et al. 2014), then the interspace between the ‘GU’ and ‘AG’ was regarded as an intron and the two transfrags were connected by the intron.
Updating TSSs and CPSs
The method for TSS identification from CAGE-seq tags was modified from the method for CPS identification from 3P-seq tags (Nam et al. 2014). Of the identified sites, those located in either the first exon or in the 3-kb upstream region of a gene, without overlapping the upstream gene, were regarded as TSSs of the gene. Similarly, of the CPSs identified from 3P-seqs, those assigned to either the 3′ UTR or the 5-kb downstream region of a gene, without overlapping the downstream gene, were regarded as CPSs of the gene. After updating TSSs and CPSs, we removed all redundant transcripts or inclusive transfrags.
Evaluation of transcriptome assembly
To evaluate the quality of transcriptome assembly, we compared the resulting assembly with the reference gene annotations (protein-coding genes and lncRNAs, respectively) using Cuffcompare (version 2.1.1). The sensitivity and specificity were estimated at the base and intron levels of the assembled transfrags (Supplemental Fig. S1; see the “Evaluation of transcriptome assembly” section in Supplemental Methods for more details).
Evaluation of full-length genes and isoforms
To evaluate how many full-length genes and isoforms were assembled, we collected the transcripts that simultaneously included a TSS in the first exon and a CPS in the last exon of the resulting transfrags. In addition, the transcripts aligned to the reference transcripts with at least a 95% match were regarded as full-length transcripts. At the gene level, gene models that unified all isoform exons were compared.
Data access
Raw RNA-seq data from mES cells from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/; GSE84946) under SuperSeries accession number GSE97212. All RPDs (ENCODE and Human BodyMap), gene annotations, lncRNA catalogs, and expression tables from this study have been also submitted to the NCBI GEO (GSE97211) under the same SuperSeries accession number GSE97212. The TCGA and GTEx RPD BAM files cannot be redistributed due to the Data User Certification Agreements. However, the source code and a detailed manual for reproducing the RPD BAM files are provided in Supplemental Materials File 1, and the simplified RPD files including read ID, strand information, and mapping position have been submitted to the NCBI GEO (GSE97211). All data and files submitted to the NCBI GEO can also be downloaded from our website (http://big.hanyang.ac.kr/downloads/datasets/).
Supplementary Material
Acknowledgments
We thank Drs. Youngsoo Song (Hanyang University) and Jiwon Shim (Hanyang University) for helpful comments. We also thank all members of the BIG lab for helpful comments and discussions, and Daehyun Baek and Jinman Park for providing resources. The results shown here are in part based upon data generated by TCGA, GTEx, and FANTOM consortia, and ENCODE and Human BodyMap 2.0 Projects. This work was supported by the Basic Science Research Program through NRF, funded by the Ministry of Science, ICT & Future Planning (NRF-2014M3C9A3063541 and NRF-2012M3A9D1054516), supported by the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), which was funded by the Ministry of Health and Welfare (grant number: HI15C3224 and HI15C1578), and supported by the Program for Agriculture Science & Technology Development (Project No. PJ01045303) of the Rural Development Administration.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.214288.116.
Freely available online through the Genome Research Open Access option.
References
- Boley N, Stoiber MH, Booth BW, Wan KH, Hoskins RA, Bickel PJ, Celniker SE, Brown JB. 2014. Genome-guided transcript assembly by integrative analysis of RNA sequence data. Nat Biotechnol 32: 341–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JB, Boley N, Eisman R, May GE, Stoiber MH, Duff MO, Booth BW, Wen J, Park S, Suzuki AM, et al. 2014. Diversity and dynamics of the Drosophila transcriptome. Nature 512: 393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z, Li G, Liu J, Zhang Y, Ashby C, Liu D, Cramer CL, Huang X. 2015. Bridger: a new framework for de novo transcriptome assembly using RNA-seq data. Genome Biol 16: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. 2013. Emerging landscape of oncogenic signatures across human cancers. Nat Genet 45: 1127–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Mercer TR, Bussotti G, Leonardi T, Haynes KR, Crawford J, Brunck ME, Cao KA, Thomas GP, Chen WY, et al. 2015. Quantitative gene profiling of long noncoding RNAs with targeted RNA sequencing. Nat Methods 12: 339–342. [DOI] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. 2012. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. 2009. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37: e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature 489: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The FANTOM Consortium and the RIKEN PMI and CLST (DGT). 2014. A promoter-level mammalian expression atlas. Nature 507: 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M, Grabherr MG, Guttman M, Trapnell C. 2011. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods 8: 469–477. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium. 2013. The Genotype-Tissue Expression (GTEx) project. Nat Genet 45: 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. 2010. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28: 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangauer MJ, Vaughn IW, McManus MT. 2013. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet 9: e1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. 2012. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22: 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. 2015. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47: 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian X, Boerwinkle E, Liu X. 2014. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res 42: 13534–13544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. 2013. Mutational landscape and significance across 12 major cancer types. Nature 502: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H, Lizio M, Itoh M, Kanamori-Katayama M, Kaiho A, Nishiyori-Sueki H, Shin JW, Kojima-Ishiyama M, Kawano M, Murata M, et al. 2014. Comparison of CAGE and RNA-seq transcriptome profiling using clonally amplified and single-molecule next-generation sequencing. Genome Res 24: 708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, You BH, Nam JW. 2015. Global estimation of the 3′ untranslated region landscape using RNA sequencing. Methods 83: 111–117. [DOI] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. 2007. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35: W345–W349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. [DOI] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M. 2011. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27: i275–i282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Sun S, Li P, Bu D, Cao H, Zhao Y. 2013. Comprehensive characterization of 10,571 mouse large intergenic noncoding RNAs from whole transcriptome sequencing. PLoS One 8: e70835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangul S, Caciula A, Al Seesi S, Brinza D, Mndoiu I, Zelikovsky A. 2014. Transcriptome assembly and quantification from Ion Torrent RNA-Seq data. BMC Genomics 15: S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretty L, Sibbesen JA, Krogh A. 2014. Bayesian transcriptome assembly. Genome Biol 15: 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JA, Wang Z. 2011. Next-generation transcriptome assembly. Nat Rev Genet 12: 671–682. [DOI] [PubMed] [Google Scholar]
- Martin J, Bruno VM, Fang Z, Meng X, Blow M, Zhang T, Sherlock G, Snyder M, Wang Z. 2010. Rnnotator: an automated de novo transcriptome assembly pipeline from stranded RNA-Seq reads. BMC Genomics 11: 663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The modENCODE Project Consortium, Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, et al. 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam JW, Bartel DP. 2012. Long noncoding RNAs in C. elegans . Genome Res 22: 2529–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam JW, Rissland OS, Koppstein D, Abreu-Goodger C, Jan CH, Agarwal V, Yildirim MA, Rodriguez A, Bartel DP. 2014. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol Cell 53: 1031–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, et al. 2012. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res 22: 577–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Lin X, Salzberg SL. 2001. GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res 29: 1185–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese MG, Eeckman FH, Kulp D, Haussler D. 1997. Improved splice site detection in Genie. J Comput Biol 4: 311–323. [DOI] [PubMed] [Google Scholar]
- Safikhani Z, Sadeghi M, Pezeshk H, Eslahchi C. 2013. SSP: an interval integer linear programming for de novo transcriptome assembly and isoform discovery of RNA-seq reads. Genomics 102: 507–514. [DOI] [PubMed] [Google Scholar]
- Schulz MH, Zerbino DR, Vingron M, Birney E. 2012. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28: 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MB, Senapathy P. 1987. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 15: 7155–7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon D, Tilgner H, Grubert F, Snyder M. 2013. A single-molecule long-read survey of the human transcriptome. Nat Biotechnol 31: 1009–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steijger T, Abril JF, Engström PG, Kokocinski F, Hubbard TJ, Guigó R, Harrow J, Bertone P; RGASP Consortium. 2013. Assessment of transcript reconstruction methods for RNA-seq. Nat Methods 10: 1177–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden B. 2015. De novo assembly of bacterial transcriptomes from RNA-seq data. Genome Biol 16: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. 2010. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 28: 1248–1250. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. 2015. Proteomics. Tissue-based map of the human proteome. Science 347: 1260419. [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. 2011. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147: 1537–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Wu G, Tang J, Luo R, Patterson J, Liu S, Huang W, He G, Gu S, Li S, et al. 2014. SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 30: 1660–1666. [DOI] [PubMed] [Google Scholar]
- Yamashita R, Sathira NP, Kanai A, Tanimoto K, Arauchi T, Tanaka Y, Hashimoto S, Sugano S, Nakai K, Suzuki Y. 2011. Genome-wide characterization of transcriptional start sites in humans by integrative transcriptome analysis. Genome Res 21: 775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassour M, Kaplan T, Fraser HB, Levin JZ, Pfiffner J, Adiconis X, Schroth G, Luo S, Khrebtukova I, Gnirke A, et al. 2009. Ab initio construction of a eukaryotic transcriptome by massively parallel mRNA sequencing. Proc Natl Acad Sci 106: 3264–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB. 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11: 377–394. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.