Abstract
Abnormal biology of α-synuclein (α-Syn) is directly implicated in the pathogenesis of Parkinson's disease and other α-synucleinopathies. Herein, we demonstrate that C-terminally truncated α-Syn (α-SynΔC), enriched in the pathological α-Syn aggregates, is normally generated from full-length α-Syn independent of α-Syn aggregation in brains and in cultured cells. The accumulation of α-SynΔC is enhanced in neuronal cells as compared with nonneuronal cells. Significantly, the expression of familial Parkinson's disease-linked mutant α-Syn is associated with the enhanced cellular accumulation of α-SynΔC. Moreover, substoichiometric amounts of α-SynΔC enhance the in vitro aggregation of the more abundant full-length α-Syn. Finally, cases of α-synucleinopathy exhibit increases in the total soluble α-Syn and a higher proportion of soluble α-SynΔC, a condition favoring the aggregation of α-Syn. Collectively, our results indicate that the biology behind the generation and accumulation of α-SynΔC is likely to have relevance for the initiation and the progression of α-Syn aggregation in vivo.
Keywords: α-synucleinopathy, mass spectrometry, proteolysis, Lewy body
Parkinson's disease (PD) is a common progressive neurodegenerative disease characterized by the loss of dopaminergic neurons of substantia nigra and the presence of the fibrillar cytoplasmic aggregates of α-synuclein (α-Syn) in multiple brain regions (1, 2). Mutations in the α-Syn gene (3-7) and the abnormal aggregation of α-Syn are implicated in the pathogenesis of PD, and other related diseases are classified as α-synucleinopathies (1, 8-10). α-Syn is a highly conserved protein of 140 amino acids that is predominantly expressed in neurons, particularly in presynaptic terminals (11), and may have a role in synaptic plasticity and modulation of dopaminergic neurotransmission (11).
Although the bulk of previous studies focused on the aggregation and the biology of the full-length α-Syn (α-SynFL) (12, 13), the conspicuous presence of lower molecular mass α-Syn species in α-Syn aggregates (14, 15), and the enhanced in vitro fibril assembly of recombinant C-terminally truncated α-Syn (16, 17) suggests that the low-molecular mass α-Syn species may be of pathogenic significance. However, because postpathogenic and/or postmortem processes could potentially generate a variety of α-Syn species, the significance of low-molecular mass α-Syn species to the development of α-synucleinopathy is uncertain.
Herein, we demonstrate that C-terminally truncated low-molecular mass α-Syn species (α-SynΔC) with aggregation-promoting properties are normally generated in vivo. The expression of familial PD (FPD)-linked mutant human (Hu) α-Syn is associated with the higher cellular accumulation of α-SynΔC. Moreover, human cases with α-Syn lesions show preferential accumulation of α-SynΔC in aggregates and higher relative levels of soluble α-SynΔC. Our findings show that α-SynΔCs are not an artifact of postpathologic processes and are likely to participate in the disease-linked aggregation of α-Syn.
Materials and Methods
Additional details are provided in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Subjects. MoPrP-Huα-Syn transgenic (Tg) mice, as described in ref. 14, were used. The experimental protocols involving mice were in strict adherence to the National Institutes of Health Animal Care Guidelines and were approved by the Animal Care and Use Committee at Johns Hopkins University School of Medicine. Human brain tissues were obtained from the Brain Resource Center (Department of Pathology, Johns Hopkins University School of Medicine).
Antibodies. The α-Syn variants were characterized by using antibodies with the following defined epitopes: NAC2 (18), Syn-1 (BD Biosciences, San Diego), N1/2 (18), Pan-Syn (Abcam, Cambridge, U.K.), and HuSyn1 (14), and NACP-C recognizing the last nine amino acids of Huα-Syn (19, 20) (Fig. 1A). Locations of epitopes for all antibodies, except Syn-1, were determined from the sequence of the peptide antigens. The approximate location of epitope for Syn-1 was determined from the immunoblot analysis showing that Syn-1 reacts with α-Syn encoding amino acids 1-100, but not 1-90.
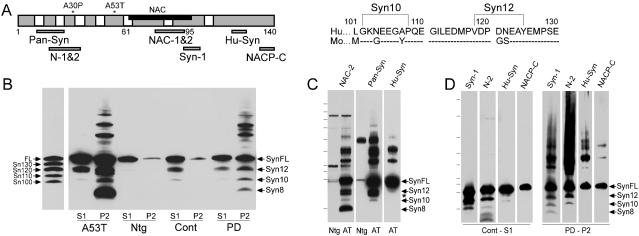
Fig. 1.
Epitope mapping of Huα-Syn variants in brain. (A) The locations of the epitopes for the anti-α-Syn antibodies used, FPD mutations (*), and the sequence for amino acids 101-140 of human and mouse α-Syn. Brackets show the potential sites of truncations for α-Syn12 and α-Syn10. (B) Syn-1 immunoblot analysis of nonionic detergent-soluble (S1) and -insoluble (P2) α-Syn species in brains of A53T Huα-Syn Tg mouse (A53T), non-Tg (Ntg) littermate, normal human (Cont), and a PD case (PD). The apparent molecular masses of the α-Syn species (SynFL, Syn12, Syn10, and Syn8) were compared with various α-SynΔCs (Sn130, Sn120, Sn110, and Sn100) expressed in transfected COS-1 cells. Because equal amounts of proteins were loaded, the analysis overrepresents the total amount of α-Syn in the P2 fraction (14). (C) Immunoblot analysis of P2 fractions from A53T Tg and non-Tg (Ntg) mice. (D) Immunoblot analysis of S1 fraction from normal human cingulate cortex (Cont-S1) and P2 fraction from cingulate cortex of a PD case (PD-P2). Syn12 and Syn10 do not react with antibodies to C-terminal epitopes (HuSyn, NACP-C). * Indicates an ≈12-kDa α-Syn variant that is only seen in P2 fractions from Tg mice. The N2-reactive bands with molecular mass below Syn8 also react to Pan-Syn Ab (data not shown) and may represent degradation intermediates containing the N-terminal portion of α-Syn. Molecular mass markers are 6.7, 16.1, 20.5, 27.5, 38.6, 50.1, 65.8, 83.8, 120, and 179.3 kDa (C) and 10, 15, 20, 25, 30, 40, 50, 80, and 120 kDa (D).
Expression Constructs and Cell Culture. cDNAs encoding Huα-Syn (WT, A30P, and A53T), Moα-Syn, and various truncated Huα-Syn were cloned into the pcDNA3.1+ (pCD)or pIND expression vectors (Invitrogen). For transient transfections, N2a, Ltk-, or COS-1 cells were transfected with the pCD constructs by using Lipofectamine 2000 (Invitrogen), and the cells were analyzed 2 days after transfection. Polyclonal SH-SY5Y cell lines and inducible 293T cell lines expressing Huα-Syn (21) also were used.
Protein Extraction and Immunoblot Analysis of α-Syn. Protein extraction, fractionation, and quantitative immunoblot analyses were performed as described in refs. 14 and 21. The soluble oligomers and insoluble aggregates in transfected cells were examined by the differential extraction of cells as described in ref. 22. In all cases, the buffers used contained 5 mM EDTA and the complete protease inhibitor mixture (Roche, Indianapolis). For the immunoblot analysis, carbonate transfer buffer (pH 10) (23) was used to significantly improve the efficiency of protein transfer onto nitrocellulose membranes.
Ciphergen Surface-Enhanced Laser Desorption and Ionization (SELDI)-MS Analysis of α-Syn Variants. α-Syn variants were immunoprecipitated by using established protocols (21, 24) with the Seize-X Protein G Immunoprecipitation Kit (Pierce), applied to Ciphergen H-50 (reverse-phase surface) ProteinChip (Ciphergen, Fremont, CA), and analyzed by using the Ciphergen SELDI-TOF MS (25). For the analysis of the acidic C-terminal fragments, tryptic digests of α-SynFL and α-Syn12, purified from human brains, were analyzed by using the Ciphergen strong anion exchange chip (SAX2). Trypsin-digested α-Syn also was analyzed by MALDI-TOF MS A detailed protocol is available in Supporting Materials and Methods.
Cell-Free in Vitro Assembly of α-Syn. Self-assembly of α-Syn was examined by using cell-free in vitro aggregation assay (26) by using Huα-Syn variants expressed in COS1 cells. The modifications to the assay are described in Supporting Materials and Methods.
Results
C-Terminally Truncated α-Syn Are Normally Present in Brain. To biochemically define the nature of the major low-molecular mass α-Syn species, a variety of anti-α-Syn antibodies with defined epitopes (Fig. 1 A) were used to map α-Syn species in our Huα-Syn Tg mice and in human postmortem material. The immunoblot analyses show that the antibodies to the nonamyloid component (NAC) domain of α-Syn recognizes the full ranges of high-molecular mass α-Syn, α-SynFL, and low-molecular mass α-Syn (α-Syn12, α-Syn10, and α-Syn8) (Fig. 1B). Anti-α-Syn antibodies with N- or C-terminal epitopes reacts to distinct subsets of α-Syn species (Fig. 1 C and D). The results show that both α-Syn12 and α-Syn10 contain the N-terminal and the central NAC region but lack the C-terminal region (Fig. 1 C and D). Analysis by using the NACP-C antibody (Ab), specific to both the α-SynFL and the alternatively spliced α-Syn112 (20) shows that α-Syn112 does not contribute to α-Syn variants in human brain (Fig. 1D). In contrast to α-Syn12 and α-Syn10, α-Syn8 lacks both N- and C-terminal regions (Fig. 1 C and D).
Semiquantitative analysis of the α-Syn species showed that the levels of α-Syn12 and α-Syn10 are ≈10-25% and ≈1-5%, respectively, of α-SynFL (see also Fig. 5). Moreover, α-SynFL, α-Syn12, and α-Syn10 are present in both detergent-soluble and -insoluble fractions, whereas α-Syn8 is exclusively associated with the insoluble α-Syn aggregates in mouse and human brains (Fig. 1B). Thus, the accumulation of α-Syn8ΔNC appears to require antecedent aggregations of α-Syn, whereas α-Syn12 and α-Syn10 accumulate independent of α-Syn aggregation.
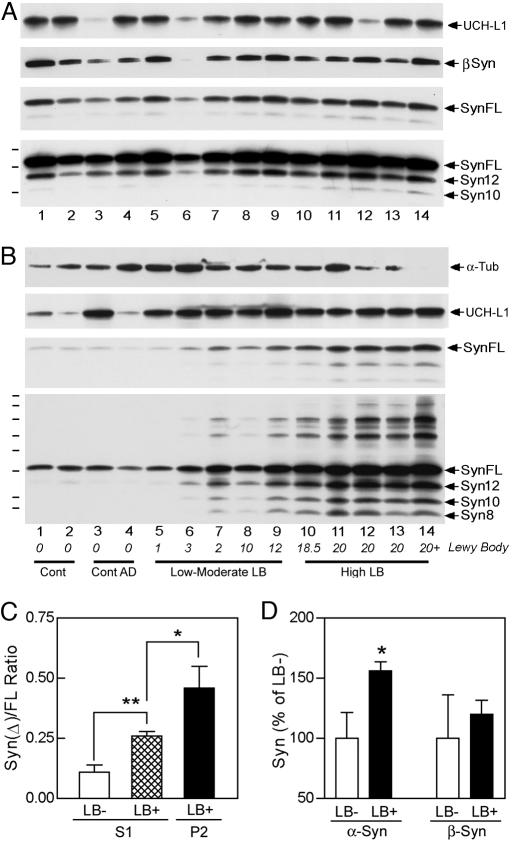
Fig. 5.
Human α-synucleinopathy is associated with enrichment of α-SynΔC in aggregates and higher soluble α-SynΔC levels. (A and B) Nonionic detergent-soluble (A) and -insoluble (B) brain fractions from human brain regions without α-synucleinopathy (lanes 1-4) with increasing Lewy body (LB) density (LB and LNs/3.14 mm2) (lanes 5-14) were analyzed for α-Syn, β-Syn, tubulin, and UCH-L1. The short and long exposures for α-Syn show the quantitative and the qualitative differences. The samples are from cingulate cortex (lanes 1-4, 8-12, and 14), midfrontal gyrus (lanes 5-7), and frontal cortex (lane 13). The final diagnosis, brain regions, LB density, age, and postmortem delay for the sample are listed in Table 2. (C) Quantitative analysis of α-Syn species in the cingulate cortices of PD cases (LB+) and controls (LB-). The levels of α-SynΔC are normalized to α-SynFL (ratio of α-SynΔC/α-SynFL). In addition to enrichment of α-SynΔC with the detergent-insoluble aggregates (P2) of LB+ cases, the relative level of α-SynΔC higher in the detergent-soluble (S1) fractions from the cingulate cortices with LBs (LB+, 0.26 ± 0.05, n = 6) than the controls (LB-, 0.11 ± 0.05, n = 4) (*, P < 0.05; **, P < 0.01, t test). (D) Quantitative analysis of α-SynFL and β-Syn levels in the S1 fractions. Presence of LBs (LB+) is associated with the significantly higher levels of soluble α-SynFL (*, P < 0.05, t test) but not β-Syn. The levels of α-Syn species do not correlate with the postmortem delay, sex, or age of the subjects. Also note that whereas the soluble UCH-L1 levels are comparable between LB- and LB+ samples, the LB+ samples are associated with higher levels of insoluble UCH-L1.
α-SynΔCs Are Generated from α-SynFL, and Accumulation of α-SynΔC Is Enhanced by Expression of FPD-Linked Mutant Huα-Syn. To establish that α-SynΔCs are derived from α-SynFL, a variety of cell lines were transfected with the pCD-α-SynFL, and the appearance of α-Syn species was examined (Fig. 2A). Analyses of the transfected cells showed the presence of both α-SynFL and the C-terminally truncated α-Syn12 (Fig. 2A and Fig. 6, which is published as supporting information on the PNAS web site). Because of the lower abundance of α-Syn10, documentation of α-Syn10 was variable. Although the expression of Huα-SynFL resulted in the accumulation of α-SynΔC in all cell lines (Fig. 2A), the levels of α-SynΔC, normalized to α-SynFL, were significantly higher (≈3- to 5-fold) in the neuronal cell lines (mouse N2a and human SH-SY5Y) as compared with the nonneuronal cell lines (mouse Ltk- and human HEK293) (Fig. 2A).
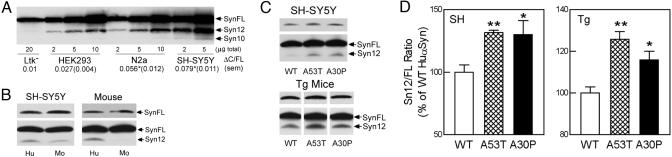
Fig. 2.
α-SynΔC accumulates in α-SynFL-transfected cells, and the accumulation shows cell type and α-Syn sequence preferences. (A) Steady-state levels of Huα-Syn variants in neural (SH-SY5Y, N2a) and nonneural (HEK293, Ltk-) cells transfected with Huα-SynFL cDNA were examined by Syn-1. The relative abundance of truncated α-Syn (ΔC/FL ratio) for the cell lines are shown (n = 3, P < 0.05 vs. HEK293 cells, t test). (B) Expression of Huα-SynFL leads to higher levels of α-SynΔC than with Moα-SynFL. Immunoblot analyses of α-Syn species in the SH-SY5Y cell lines and the brains (Mouse) of non-Tg (Ntg; Mo) and WT Huα-Syn+/Moα-Syn-null Tg mice (Hu) are shown. The protein loadings were normalized for the levels of α-SynFL (Upper). (C) SDS-soluble extracts from SH-SY5Y cell lines and brainstem from Huα-Syn Tg mice (12 months old) expressing WT, A53T, and A30P Huα-Syn were subjected to semiquantitative immunoblot analysis. Shorter exposure (SH-SY5Y, Syn-1 mAb) and HuSyn polyclonal Ab (Mouse) were used to determine the levels of α-SynFL. (D) The relative levels of Syn12 (Sn12/FL ratio) are plotted as the percent of WT Huα-Syn samples. To minimize skewing of results from the contribution of Moα-Syn (by underestimating the relative levels of α-SynΔCin the Tg mice with lower levels of Huα-Syn expression), the levels of Huα-SynFL in mice were determined by using the HuSyn polyclonal Ab (Tg mice) (mean and SEM from four to six independent samples; *, P < 0.05; **, P < 0.01, t test).
The cell type differences in the levels of α-SynΔC are reminiscent of our previous study showing the differential metabolism of α-Syn as functions of cell type and α-Syn sequence (21). Thus, we examined whether the primary sequence differences in α-SynFL also influence the levels of α-SynΔC in cells and brain. First, we compared the accumulation of α-SynΔC as a function of expressing either Huα-SynFL or mouse (Mo) α-SynFL (Fig. 2B), which differ at the C-terminal regions (Fig. 1A). The results showed that the expression of Huα-Syn is associated with higher relative levels of α-SynΔC than with Moα-Syn (Fig. 2B). We also examined whether the FPD-linked α-Syn mutations influence the accumulation of α-SynΔC (Fig. 2 C and D). Quantitative analyses of α-Syn species in SH-SY5Y cell lines and Tg mice expressing either WT or FPD-linked mutant Huα-SynFL showed that the expression of the FPD-linked mutant α-SynFL leads to higher relative levels of α-SynΔC (Fig. 2 C and D).
Although differential aggregation/oligomerization of α-Syn may be responsible for the differential accumulations of α-SynΔC, our analyses of the SH-SY5Y cell lines for soluble oligomers and the aggregates of α-Syn (22) showed very little soluble oligomers and insoluble aggregates in our cell lines (Fig. 7, which is published as supporting information on the PNAS web site), and the Tg mice used in our studies did not develop α-Syn aggregation at the ages used. Moreover, the size-exclusion chromatography of the soluble brain proteins showed that the majority of α-SynΔC resolved with the α-SynFL monomers (Fig. 7).
MS Characterization of α-Syn12 Truncations. To better define the sites of α-Syn truncations, two major α-Syn variants (α-SynFL and α-Syn12) were examined by using the Ciphergen SELDI-TOF MS (25) (Fig. 3A and Table 1, which is published as supporting information on the PNAS web site). Significantly, the apparent average masses of α-SynFL (≈14,540 Da) from cells and brains were significantly higher than the mass (14,460 Da) predicted from the amino acid sequence and that observed with the bacterial recombinant α-Syn (Fig. 3A). Thus, mammalian cell-derived Huα-Syn contains posttranslational modifications. Despite the posttrans-lational modifications, the apparent masses of α-Syn species were very consistent regardless of the source (Fig. 3A). The mass differences between the α-Syn12 and the α-SynFL (≈1,990 Da) suggested that α-Syn12 is missing the last 17 C-terminal amino acids (Fig. 3A).
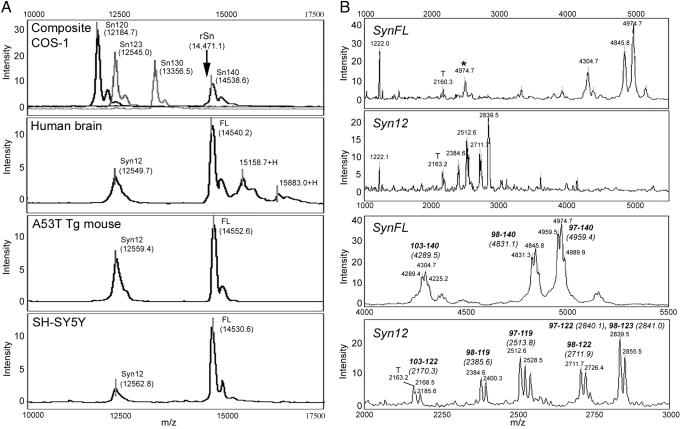
Fig. 3.
Ciphergen SELDI-MS analysis of Huα-Syn species. (A) Representative mass traces from the Ciphergen SELDI-TOF MS analysis of Huα-Syn from COS-1 cells, neurologically normal human brain, brain from A53T Huα-Syn Tg mice, and SH-SY5Y cells show the peaks corresponding to α-SynFL and α-Syn12. Analysis of artificially truncated Huα-Syn species expressed in COS-1 cells also are shown as follows: COS-1 composite, Sn120 (amino acids 1-120), Sn123 (amino acids 1-123), Sn130 (amino acids 1-130), and Sn FL (amino acids 1-140). The arrow indicate the apparent mass of the recombinant α-SynFL obtained with SELDI-TOF-MS (rSn, 14,471.1 Da). (B) Representative mass traces and the average masses of the α-Syn tryptic peptides analyzed by the SAX2 chip. Human brain-derived α-SynFL and α-Syn12 show distinct patterns of the acidic tryptic peptides. The peptides (and predicted average mass) corresponding to the peaks are shown. The secondary peaks result from the oxidation of methionines (+16 Da/MSO). MS peaks corresponding to trypsin-only control (T) and the double-charged ions (*) also are indicated.
Because the posttranslational modification(s) introduces uncertainties regarding the truncation site(s), human brain-derived α-Syn12 and α-SynFL was individually digested with trypsin, and the masses of the acidic C-terminal peptides captured on the SAX2 chips were determined (Fig. 3B). The analyses of α-SynFL-derived peptides showed the presence of unmodified C-terminal fragments with zero (103-140), one (98-140), and two (97-140) missed cleavages (Fig. 3B). The analyses of α-Syn12 revealed a series of peptides between 2,000 and 3,000 Da. Because these were not observed with the α-SynFL, the peptides are specifically associated with α-Syn12. The masses of these peptides correspond to the C-terminal peptides ending at Asp-119, Asn-122, and possibly Glu-123 (Fig. 3B). The MALDI-TOF-MS analysis of trypsin-digested α-Syn also identified similarly truncated C-terminal peptides (Table 2, which is published as supporting information on the PNAS web site). The low abundance of α-Syn10 precluded accurate mass analysis of the peptide.
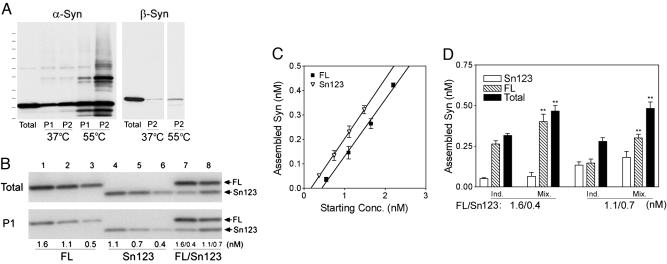
α-SynΔC Enhances the Aggregation of α-SynFL. Because abnormal aggregation and oligomerization of α-Syn are implicated in the pathogenesis of α-synucleinopathies (27), cell-free in vitro self-assembly assay (26) was used to determine whether α-SynΔC influences α-Syn aggregation. To model the assembly properties of α-SynΔC, we used α-Syn encoding amino acids 1-123 (α-Syn123) (Fig. 4) and 1-120 (α-Syn120) (Fig. 8, which is published as supporting information on the PNAS web site). Consistent with the previous studies using bacterially derived recombinant α-Syn (16, 17), α-SynΔC aggregated more readily than α-SynFL (Fig. 4 B and C).
Fig. 4.
α-SynΔC enhances the aggregation of α-SynFL. (A) Immunoblot analysis of pellet fractions (aggregates) taken at 1 day (P1) or 2 days (P2) of assembly (at either 37°C or 55°C) shows the time- and temperature-dependent self-assembly of Huα-Syn but not Huβ-Syn. (B) Known concentrations of α-SynFL and α-Syn12(Sn123) were assembled (37°C, 24 h) independently (lanes 1-6) or in a mixture containing both variants (FL/Sn123) (lanes 7 and 8). The amount of assembled α-Syn was determined by quantitative immunoblot analysis of the pellet fractions. (C) The amount of α-SynFL or α-Syn12 (Sn123) assembled, as determined from B, was plotted as the function of the initial α-Syn concentrations (mean and SEM, three independent reactions). Linear regression analysis shows that more α-Syn12 is assembled than α-SynFL [y = 0.221x - 0.092 (r2 = 0.997) for α-SynFL and y = 0.245x - 0.042 (r2 = 0.999) for α-Syn123] at 24 h. The amount of assembly is comparable between α-SynFL and α-Syn123 with longer aggregation times (>48 h). (D) The extents of assembled α-Syn species in the coassembly mixture (Mix.; see lanes 7-8 in B). Also shown is the assembly predicted by the simple addition of the two individual (Ind.) components. The amount of total (FL+Sn123) and α-SynFL in the pellet fraction from the coassembly mixtures are significantly higher than amounts predicted from the simple addition of each individual components (**, P < 0.01, t tests).
Significantly, when the assembly mixture contained both α-SynFL and substoichiometric amounts of α-Syn123, analogous to the relative abundance of α-Syn species in vivo, the total amounts of aggregated α-Syn were significantly higher than those predicted by the simple addition of the two components (Fig. 4 B and D). This increase in the aggregation is largely due to the enhanced assembly of α-SynFL, suggesting that the assembly of α-SynFL is enhanced by the presence of α-SynΔC (Fig. 4 B and D). Our results are different from those described in a previous report where the bacteria-derived recombinant α-Syn120 did not facilitate the assembly of α-SynFL (17). The discrepancy is not due to the inherent differences between α-Syn123 and α-Syn120 because the aggregation properties of α-Syn120 are very similar to α-Syn123 in our assay (Fig. 8). Possibly, the cell-free aggregation assay, involving mammalian cell-derived α-Syn and other cellular components, may reveal properties of α-Syn variants that are not apparent with the in vitro assays using bacterially expressed α-Syn.
Human α-Synucleinopathy Is Associated with the Preferential Aggregation of α-SynΔC and the Increased Levels of Soluble α-SynΔC. The enhancement of α-SynFL aggregation by α-SynΔC (Fig. 4) suggests that α-SynΔC may promote α-Syn aggregation in vivo. If α-SynΔC is involved in the initiation and/or progression of α-Syn aggregation in humans, we should observe alterations in the levels of α-SynΔC and the selective aggregation of α-SynΔC in the human α-synucleinopathy cases.
Analysis of human brain extracts from the cases with and without α-Syn lesions and demonstrated that all human brains contain significant levels of α-Syn variants in the detergent-soluble (S1) fractions (Fig. 5A) (Table 3, which is published as supporting information on the PNAS web site). The cases with α-Syn lesions also accumulated α-Syn variants in the detergent insoluble (P2) fractions (Fig. 5B). The severity of α-Syn histopathology (28) correlated with the levels of detergent-insoluble α-Syn (Fig. 5B). Consistent with the enhanced aggregation properties of α-SynΔC in vitro, the detergent-insoluble α-Syn aggregates consisted of higher proportion of α-SynΔC(≈100% increase) than the soluble fraction (Fig. 5C).
Because the potential proteolysis of α-Syn in aggregates could lead to the increased levels of the insoluble α-SynΔC, we also examined whether the α-synucleinopathy is associated with the increased levels of soluble α-SynΔC, a condition predicted to facilitate the aggregation of α-Syn. Semiquantitative analyses of the S1 fractions from the cingulate cortices showed that higher relative levels of α-SynΔC (Fig. 5C) and α-SynFL (Fig. 5D) were found in human cases with α-Syn pathology. Thus, human α-synucleinopathies are associated with increase in the steady-state levels of soluble α-Syn with a higher fraction consisting of α-SynΔC, a situation that would predispose neurons to developing α-Syn aggregates. Higher α-Syn levels found in human cases with the α-Syn pathology are consistent with the increased transcriptional affects of PD-associated α-Syn promoter polymorphism (29) and the potential role of protein stabilization in PD (21).
Discussion
The present report demonstrates that α-SynΔC, which promotes aggregation of α-Syn, is generated by the normal cellular processing of α-SynFL in cells and brain. The pathological significance α-SynΔC is supported by the enhanced accumulation of α-SynΔC with FPD-linked mutants, the enrichment of α-SynΔC in α-Syn aggregates, and the increased levels of soluble α-SynΔC in human brains with α-Syn pathology. Our studies show that α-SynΔC species are not mere postmortem artifacts or result from postpatho-logic events. Further, given that α-SynΔC is largely present as soluble monomers, α-SynΔC could, regardless of the origin, alter the biology of α-Syn monomers in neurons.
Although more detailed analyses of the posttranslational modifications and the truncations are required, current partial characterization of the C-terminal peptides allows us to identify Asp-119-Pro-120, Asn-122-Glu-123, and possibly Glu-123-Ala-124 as the sites of cleavages for α-Syn12. The sequence differences between Huα-Syn and Moα-Syn at this region and the putative sites for α-Syn10 (Fig. 1A), and the results that Moα-Syn expression leads to lower levels of α-SynΔC, suggest that the truncations exhibit amino acid sequence preferences. Notably, in vitro studies have shown endoproteolytic cleavage of α-Syn by proteasome (30) at Asp-119-Pro-120 (Phil Thomas, University of Texas Southwestern Medical Center, personal communication) and cleavage of α-Syn by Calpain I at Asn-122-Glu-123 (31). Thus, α-Syn truncation may be mediated by more than one proteolytic system. The differences between the in vitro and in vivo sites may arise from the interaction of α-Syn with other cellular components (e.g., membranes). Our preliminary studies indicate that the inhibition of caspases, proteasomes, and lysosomal proteases do not significantly affect the relative accumulation of α-SynΔC in cells (Fig. 6C). However, the conclusions from these inhibitor studies are premature because of the potential complicating effects of these and other inhibitors on the overall α-Syn levels and the stability of α-SynΔC.
In other neurodegenerative diseases such as Alzheimer's (32) and Huntington's (33) diseases, proteolytically cleaved proteins are implicated in the pathogenesis. In particular, α-Syn species exhibit an interesting parallel to the Aβ species in Alzheimer's disease (32). Although Aβ40 is usually more abundant than Aβ42/43 (≈10:1 ratio), the presence of certain familial Alzheimer's disease mutations can influence this ratio, and the Aβ aggregation is initiated and driven by the less abundant Aβ42/43. Similarly, our results are consistent with the hypothesis that the less abundant α-SynΔC influences the threshold for α-Syn aggregation. However, the basis for the enhanced accumulation of α-SynΔC with FPD-linked mutants must be different from the enhanced generation of Aβ species from familial Alzheimer's disease-linked mutant amyloid precursor protein (APP) (32). Although the APP mutations are located near the sites of Aβ-cleavage (32), the mutations in Huα-Syn are much further N-terminal to the truncation sites. Thus, the FPD-linked mutations may influence the levels of α-SynΔC by altering the cellular localization of α-Syn (34, 35) and/or differential interaction with other cellular components.
Although the current study only examined the overt aggregation properties of α-SynΔC, peptides with the propensity to aggregate also are prone to form potentially toxic nonfibrillar oligomers (36-38). Thus, we believe it is likely that α-SynΔC also facilitates the formation of the potentially pathogenic nonfibrillar oligomers of α-Syn (36, 37). In addition to influencing α-Syn aggregation/oligomerization, the C-terminal region of α-Syn appears to be important for the chaperone activity of α-Syn (39-41) and the stability of α-Syn (42). Further, the expression of α-Syn120 has been shown to increase cellular vulnerability to oxidative stress (43). Thus, normal presence of α-SynΔCs in neurons may have other significant cellular effects.
There are clearly multiple factors, such as oxidative modifications of α-Syn (44) and/or phosphorylation of α-Syn (45), that may be important in the pathogenesis of α-synucleinopathies. We propose that α-SynΔC is one of the factors that contributes to promote the pathologic transformation of α-Syn in brain. Overall, the level of experimental support for the involvement of α-SynΔCin α-synucleinopathy is similar to the other potential contributors to α-synucleinopathy (36, 37, 44, 45). Thus, our study provides a strong rationale for more detailed examinations regarding the pathogenic nature of α-SynΔC. Moreover, studies about α-Syn-dependent neurodegeneration will have to consider the effects of α-SynΔC. Finally, if the cell biological basis for α-Syn truncation can be identified, α-Syn truncation could be a potential target for development of novel therapeutic compounds for α-synucleinopathies.
Supplementary Material
Acknowledgments
We thank Ms. Yanqun Xu for technical assistance and Ms. Rebecca Lurz for editorial assistance. We also thank Dr. Robert Katzman (University of California at San Diego, La Jolla) for generously providing us with the anti-NACP-C Ab and Dr. Philip Wong for helpful comments on the manuscript. This research was supported by National Institutes of Health Grants NS38065 (to M.K.L.) and NS38377 (to J.C.T., M.K.L., and T.M.D.), grants from Dystonia Medical Research Foundation (to M.K.L.), and a National Institutes of Health postdoctoral training grant (to N.W.).
Author contributions: W.L., N.W., and M.K.L. designed research; W.L., N.W., E.C., O.P., and M.K.L. performed research; W.L., N.W., and M.K.L. analyzed data; and W.L., T.M.D., D.L.P., and M.K.L. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PD, Parkinson's disease; FPD, familial PD; α-Syn, α-Synuclein; α-SynFL, full-length α-Syn; Hu, human; Mo, mouse; Tg, transgenic; LB, Lewy body; NAC, nonamyloid component; SELDI, surface-enhanced laser desorption and ionization.
References
- 1.Dickson, D. W. (2001) Curr. Opin. Neurol. 14, 423-432. [DOI] [PubMed] [Google Scholar]
- 2.Fahn, S. & Przedborski, S. (2000) in Merritt's Neurology, ed. Rowland, L. P. (Lippincott, New York), pp. 679-695.
- 3.Polymeropoulos, M. H., Lavedan, C., Leroy, E., Ide, S. E., Dehejia, A., Dutra, A., Pike, B., Root, H., Rubenstein, J., Boyer, R., et al. (1997) Science 276, 2045-2047. [DOI] [PubMed] [Google Scholar]
- 4.Kruger, R., Kuhn, W., Muller, T., Woitalla, D., Graeber, M., Kosel, S., Przuntek, H., Epplen, J. T., Schols, L. & Riess, O. (1998) Nat. Genet. 18, 106-108. [DOI] [PubMed] [Google Scholar]
- 5.Kitada, T., Asakawa, S., Hattori, N., Matsumine, H., Yamamura, Y., Minoshima, S., Yokochi, M., Mizuno, Y. & Shimizu, N. (1998) Nature 392, 605-608. [DOI] [PubMed] [Google Scholar]
- 6.Bonifati, V., Rizzu, P., van Baren, M. J., Schaap, O., Breedveld, G. J., Krieger, E., Dekker, M. C., Squitieri, F., Ibanez, P., Joosse, M., et al. (2003) Science 299, 256-259. [DOI] [PubMed] [Google Scholar]
- 7.Singleton, A. B., Farrer, M., Johnson, J., Singleton, A., Hague, S., Kachergus, J., Hulihan, M., Peuralinna, T., Dutra, A., Nussbaum, R., et al. (2003) Science 302, 841. [DOI] [PubMed] [Google Scholar]
- 8.Galvin, J. E., Uryu, K., Lee, V. M. & Trojanowski, J. Q. (1999) Proc. Natl. Acad. Sci. USA 96, 13450-13455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goedert, M. & Spillantini, M. G. (1998) Mol. Psychiatry 3, 462-465. [DOI] [PubMed] [Google Scholar]
- 10.Spillantini, M. G., Schmidt, M. L., Lee, V. M., Trojanowski, J. Q., Jakes, R. & Goedert, M. (1997) Nature 388, 839-840. [DOI] [PubMed] [Google Scholar]
- 11.George, J. M. (2002) Genome Biol. 3, reviews3002.1-3002.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Souza, J. M., Giasson, B. I., Chen, Q., Lee, V. M. & Ischiropoulos, H. (2000) J. Biol. Chem. 275, 18344-18349. [DOI] [PubMed] [Google Scholar]
- 13.Paxinou, E., Chen, Q., Weisse, M., Giasson, B. I., Norris, E. H., Rueter, S. M., Trojanowski, J. Q., Lee, V. M. & Ischiropoulos, H. (2001) J. Neurosci. 21, 8053-8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, M. K., Stirling, W., Xu, Y., Xu, X., Qui, D., Mandir, A. S., Dawson, T. M., Copeland, N. G., Jenkins, N. A. & Price, D. L. (2002) Proc. Natl. Acad. Sci. USA 99, 8968-8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giasson, B. I., Duda, J. E., Quinn, S. M., Zhang, B., Trojanowski, J. Q. & Lee, V. M. (2002) Neuron 34, 521-533. [DOI] [PubMed] [Google Scholar]
- 16.Serpell, L. C., Berriman, J., Jakes, R., Goedert, M. & Crowther, R. A. (2000) Proc. Natl. Acad. Sci. USA 97, 4897-4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray, I. V., Giasson, B. I., Quinn, S. M., Koppaka, V., Axelsen, P. H., Ischiropoulos, H., Trojanowski, J. Q. & Lee, V. M. (2003) Biochemistry 42, 8530-8540. [DOI] [PubMed] [Google Scholar]
- 18.Bayer, T. A., Jakala, P., Hartmann, T., Havas, L., McLean, C., Culvenor, J. G., Li, Q. X., Masters, C. L., Falkai, P. & Beyreuther, K. T. (1999) Neurosci. Lett. 266, 213-216. [DOI] [PubMed] [Google Scholar]
- 19.Iwai, A., Masliah, E., Yoshimoto, M., Ge, N., Flanagan, L., de Silva, H. A. R., Kittel, A. & Saitoh, T. (1995) Neuron 14, 467-475. [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto, M., Yoshimoto, M., Sisk, A., Hsu, L. J., Sundsmo, M., Kittel, A., Saitoh, T., Miller, A. & Masliah, E. (1997) Biochem. Biophys. Res. Commun. 237, 611-616. [DOI] [PubMed] [Google Scholar]
- 21.Li, W., Lesuisse, C., Xu, Y., Troncoso, J. C., Price, D. L. & Lee, M. K. (2004) J. Neurosci. 24, 9400-9409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, H. J., Khoshaghideh, F., Patel, S. & Lee, S. J. (2004) J. Neurosci. 24, 1888-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunn, S. D. (1986) Anal. Biochem. 157, 144-153. [DOI] [PubMed] [Google Scholar]
- 24.Li, W., Hoffman, P. N., Stirling, W., Price, D. L. & Lee, M. K. (2004) J. Neurochem. 88, 401-410. [DOI] [PubMed] [Google Scholar]
- 25.Cai, H., Wang, Y., McCarthy, D., Wen, H., Borchelt, D. R., Price, D. L. & Wong, P. C. (2001) Nat. Neurosci. 4, 233-234. [DOI] [PubMed] [Google Scholar]
- 26.Uversky, V. N., Lee, H. J., Li, J., Fink, A. L. & Lee, S. J. (2001) J. Biol. Chem. 276, 43495-43498. [DOI] [PubMed] [Google Scholar]
- 27.Trojanowski, J. Q. & Lee, V. M. (2003) Ann. NY Acad. Sci. 991, 107-110. [DOI] [PubMed] [Google Scholar]
- 28.Pletnikova, O., West, N., Lee, M. K., Rudow, G., Dawson, T. M., Marsh, L. & Troncoso, J. C. (December 28, 2004) Neurobiol. Aging, 10.1016/j.neurobiolaging.2004.10.006. [DOI] [PubMed]
- 29.Chiba-Falek, O. & Nussbaum, R. L. (2001) Hum. Mol. Genet. 10, 3101-3109. [DOI] [PubMed] [Google Scholar]
- 30.Liu, C. W., Corboy, M. J., DeMartino, G. N. & Thomas, P. J. (2003) Science 299, 408-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishizen-Eberz, A. J., Guttmann, R. P., Giasson, B. I., Day, G. A., III, Hodara, R., Ischiropoulos, H., Lee, V. M., Trojanowski, J. Q. & Lynch, D. R. (2003) J. Neurochem. 86, 836-847. [DOI] [PubMed] [Google Scholar]
- 32.Sisodia, S. S. & St George-Hyslop, P. H. (2002) Nat. Rev. Neurosci. 3, 281-290. [DOI] [PubMed] [Google Scholar]
- 33.Ross, C. A. (2002) Neuron 35, 819-822. [DOI] [PubMed] [Google Scholar]
- 34.McLean, P. J., Kawamata, H., Ribich, S. & Hyman, B. T. (2000) J. Biol. Chem. 275, 8812-8816. [DOI] [PubMed] [Google Scholar]
- 35.Outeiro, T. F. & Lindquist, S. (2003) Science 302, 1772-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caughey, B. & Lansbury, P. T. (2003) Annu. Rev. Neurosci. 26, 267-298. [DOI] [PubMed] [Google Scholar]
- 37.Volles, M. J. & Lansbury, P. T., Jr. (2003) Biochemistry 42, 7871-7878. [DOI] [PubMed] [Google Scholar]
- 38.Kayed, R., Head, E., Thompson, J. L., McIntire, T. M., Milton, S. C., Cotman, C. W. & Glabe, C. G. (2003) Science 300, 486-489. [DOI] [PubMed] [Google Scholar]
- 39.Park, S. M., Jung, H. Y., Kim, T. D., Park, J. H., Yang, C. H. & Kim, J. (2002) J. Biol. Chem. 277, 28512-28520. [DOI] [PubMed] [Google Scholar]
- 40.Uversky, V. N. & Fink, A. L. (2002) FEBS Lett. 522, 9-13. [DOI] [PubMed] [Google Scholar]
- 41.Kim, T. D., Paik, S. R. & Yang, C. H. (2002) Biochemistry 41, 13782-13790. [DOI] [PubMed] [Google Scholar]
- 42.Choi, J. Y., Sung, Y. M., Park, H. J., Hur, E. H., Lee, S. J., Hahn, C., Min, B. R., Kim, I. K., Kang, S. & Rhim, H. (2002) Biotechnol. Appl. Biochem. 36, 33-40. [DOI] [PubMed] [Google Scholar]
- 43.Kanda, S., Bishop, J. F., Eglitis, M. A., Yang, Y. & Mouradian, M. M. (2000) Neuroscience 97, 279-284. [DOI] [PubMed] [Google Scholar]
- 44.Giasson, B. I., Duda, J. E., Murray, I. V., Chen, Q., Souza, J. M., Hurtig, H. I., Ischiropoulos, H., Trojanowski, J. Q. & Lee, V. M. (2000) Science 290, 985-989. [DOI] [PubMed] [Google Scholar]
- 45.Fujiwara, H., Hasegawa, M., Dohmae, N., Kawashima, A., Masliah, E., Goldberg, M. S., Shen, J., Takio, K. & Iwatsubo, T. (2002) Nat. Cell Biol. 4, 160-164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.