Abstract
Interferon induced transmembrane protein 1 (IFITM1) belongs to a family of interferon stimulated genes (ISGs) that is associated with tumor progression and DNA damage resistance, however, its role in endocrine resistance is not known. Here, we correlate IFITM1 expression with clinical stage and poor response to endocrine therapy in a tissue microarray consisting of 94 estrogen receptor (ER)-positive breast tumors. IFITM1 overexpression is confirmed in the AI-resistant MCF-7:5C cell line and not found in AI-sensitive MCF-7 cells. In this study, the orthotopic (mammary fat pad) and mouse mammary intraductal (MIND) models of breast cancer are used to assess tumor growth and invasion in vivo. Lentivirus-mediated shRNA knockdown of IFITM1 in AI-resistant MCF-7:5C cells diminished tumor growth and invasion and induced cell death, whereas, overexpression of IFITM1 in wild-type MCF-7 cells promoted estrogen-independent growth and enhanced their aggressive phenotype. Mechanistic studies indicated that loss of IFITM1 in MCF-7:5C cells markedly increased p21 transcription, expression and nuclear localization which was mediated by JAK/STAT activation. These findings suggest IFITM1 overexpression contributes to breast cancer progression and that targeting IFITM1 may be therapeutically beneficial to patients with endocrine-resistant disease.
Keywords: Aromatase inhibitor-resistance, interferon stimulated genes, mouse mammary intraductal model, p21, JAK/STAT signaling
Graphical abstract

1 INTRODUCTION
Roughly 75% of breast cancers are estrogen receptor positive (ER+) and rely on estrogen signaling to stimulate their growth and survival [46]. As such, many therapies for ER+ breast cancer target the estrogen receptor, estrogen production or estrogen signaling (i.e. endocrine therapy). The introduction of endocrine therapy has resulted in significant reductions in ER+ breast cancer-related mortality. One class of drugs, the aromatase inhibitors (AIs), prevents estrogen production from precursor hormones by targeting the aromatase enzyme [46, 47]. The AIs used in the clinic include non-steroidal anastrozole and letrozole as well as steroidal exemestane [47]. All three compounds deplete circulating estrogen, effectively starving the breast cancer cells and stimulating cell cycle arrest and cell death, which results in tumor shrinkage and prevention of metastasis [6, 15]. Unfortunately approximately 30% of women receiving AI treatment eventually develop resistance and the tumors continue to grow and metastasize despite the absence of estrogen [6, 15, 48, 67]. These AI-resistant tumors are typically more aggressive and are less responsive to other types of endocrine therapy, leaving radiation and chemotherapy as the main treatments available to these patients [67]. The mechanism by which AI resistance develops in breast cancer and the molecular factors driving its aggressive phenotype are still not completely known.
We recently published that type 1 interferon (IFN) signaling is hyperactivated in AI-resistant breast cancer cells and that the resistant cells and AI-resistant tumors constitutively overexpress several IFNα-stimulated genes (ISGs) which promote an aggressive phenotype in these cells [14]. Notably, we have identified interferon induced transmembrane protein 1 (IFITM1) as a critical downstream target of the IFN signaling pathway and have found that it is overexpressed in AI-resistant breast cancer cells and AI-resistant patient tumors [14, 53]. IFITM1 is a 17-kDa transmembrane protein coded on the short arm of chromosome 11 [66]. It belongs to a family of proteins known as interferon stimulated genes (ISGs) that are known to aid in resisting viral infection and to mediate cell survival and cell death decisions [63, 66, 68, 70]. ISGs are induced by type 1 IFN signaling through the type 1 IFN(α/β) receptor (IFNAR), which stimulates JAK/STAT activation and transcription [5, 23, 57]. We have shown that IFITM1-expressing breast cancer cells produce type 1 IFNs and stimulate IFITM1 production in an autocrine manner, but outside of DNA damage resistance, little is understood about the mechanism by which IFITM1 influences the breast cancer cell phenotype [14, 53].
Traditionally, the orthotopic model is used to evaluate tumor growth. The breast cancer cells are injected directly into the mammary fat pad and allowed to grow over time, allowing for assessment of proliferation, cell death, and/or metastasis. Breast cancer, however, originates inside the mammary duct where the cells must invade out of the duct prior to establishing a solid tumor. The mouse mammary intraductal (MIND) model has been developed to study breast cancer cell invasion in vivo [4, 69]. In the MIND model, breast cancer cells are injected into the mammary duct through the nipple, where they populate the duct and can invade into the surrounding mammary gland. This model provides a tumor microenvironment that permits the in vivo study of previously difficult to grow ER+ breast cancer cell lines and faithfully mirrors the behavior of primary breast cancer cells in patients with regard to aggression and response to therapy [65].
In this study, we reveal that high IFITM1 expression correlates with higher clinical stage and rate of recurrence for 94 ER+ breast cancer patients. In vivo studies using the orthotopic and MIND models of breast cancer reveal that IFITM1 overexpression enhances tumor progression and invasion. Gain and loss of function studies demonstrate that IFITM1 contributes directly to cell survival, proliferation and invasion. We also report that loss of IFITM1 markedly increases p21 expression and nuclear localization which promotes cell death in AI-resistant cells. Our preclinical data suggests that targeting IFITM1 in AI-resistant breast cancer may have therapeutic benefit in the clinic.
2 MATERIALS AND METHODS
2.1 Cell lines and culture conditions
The MCF-7 cell line [32, 58] was obtained from Dr. V. Craig Jordan (University of Texas MD Anderson Cancer Center, Houston) and maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum, 2 mM glutamine, Antibiotic/Antimitotic mix, MEM Non-Essential Amino Acids (Invitrogen, Waltham, MA), and bovine insulin at 6 ng/mL (Sigma Aldrich, St. Louis, MO). The long-term estrogen deprived human breast cancer cell lines; MCF-7:5C and MCF-7:2A [41, 58] were cloned from parental MCF-7 cells following long term (> 12 months) culture in estrogen-free medium composed of phenol red-free RPMI-1640, 10% fetal bovine serum treated three times with dextran-coated charcoal (SFS), 2 mM glutamine, bovine insulin at 6 ng/mL, Antibiotic/Antimitotic mix, and MEM Non-Essential Amino Acids (Invitrogen). The MCF10A cell line was purchased from the American Type Tissue Culture Collection. They are maintained in Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F12) in a 1:1 mixture and supplemented with 5% horse serum, Antibiotic/Antimitotic mix (100 IU/mL penicillin, 100 μg/mL streptomycin, 25 μg/mL of Fungizone® from Invitrogen, Grand Island, NY), 20ng/ml EGF (Millipore), 0.5mg/ml hydrocortisone, 100ng/ml cholera toxin (Sigma Aldrich). All cell lines were cultured at 37°C under 5% CO2.
2.2 Western blotting
Cells were seeded in 6-well plates, collected using a cell scraper and suspended in RIPA buffer (Thermo Scientific, Pittsburgh, PA) supplemented with protease inhibitor cocktail and phosphatase inhibitor (Sigma Aldrich). Cells were homogenized over ice by sonication. After purification of the sample by centrifugation, protein concentration was determined by protein assay (Bio-Rad, Hercules, CA). The proteins were separated by 4–12% SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and electrically transferred to a polyvinylidene difluoride membrane (Santa Cruz Biotechnology). After blocking the membrane using 5% non-fat milk, target proteins were detected using anti-IFITM1, anti-PARP, anti-ERα, anti-phospho-STAT1 (ser701), anti-STAT1, anti-p21, anti-p53 or anti-laminin B (Santa Cruz Biotechnology) antibodies. Membranes were stripped and re-probed for β-actin (Cell Signaling). The appropriate horseradish peroxidase (HRP)-conjugated secondary antibody was applied and the positive bands were detected using Amersham ECL Plus Western blotting detection reagents (GE Health care, Piscataway, NJ) and exposed to autoradiography film (Midwest Scientific).
2.3 RNA Isolation and Real Time PCR
Cells were harvested by cell scraping in RLT lysis buffer and total RNA was isolated using the Qiagen RNeasy kit (Venlo, Limburg). First strand cDNA synthesis was performed from 3 μg total RNA using MulV Reverse Transcriptase (Applied Biosystems, Carlsbad, CA) on a Bio Rad MyCycler™. RT-PCR was conducted using the ViiA™ 7 Real-Time PCR system (Applied Biosystems) and SYBR Green Reagent (Life Technologies, Carlsbad, CA) with 25 pmol primers specific for human PLSCR1 (sense: 5′-CATTCACCGGGCTCTCTAC-3′; antisense: 5′-GGCAGCTGGGCA ATCTTGCA-3′), IFITM1 (sense: 5′-GGATTTCGGCTTGTCCCGAG-3′; antisense: 5′-CCATGTGGAAGGGAGGGCTC-3′). Relative mRNA expression level was determined as the ratio of the signal intensity to that of PUM1 using the formula: 2−ΔCT. When cells were treated, fold change in ERα expression was normalized to PUM1 and then compared to the untreated value for that cell line using the formula: 2−ΔΔCT.
2.4 Survival analysis
Survival data were obtained from the 2014 version of the Kaplan-Meier Plotter breast cancer survival database (http://kmplot.com/analysis/index.php?p=service&cancer=breast).[26] The 131 patients with grade 1 ER+ disease as determined by gene expression data were included in the analysis. Patients were stratified by the average of IFITM1 expression (probe sets 214022_s_at and 201601_x_at) relative to median. The P-value was calculated using a log-rank test.
2.5 Human breast cancer specimens and IHC data analysis
This retrospective study was approved by the Institutional Review Committee at the University of Kansas Medical Center (KUMC). A total of 94 primary ER+ invasive ductal carcinomas diagnosed and removed surgically between 2001 and 2010, and for whom follow-up information was available, were examined. Samples were taken from 6 normal breast tissue samples from routine reduction mammoplasties were also examined. These tissues were formalin-fixed and paraffin-embedded (FFPE). Clinicopathological data including age, race, clinical stage and Her2 staining are shown in Table 1. At diagnosis, tissue blocks containing the most representative and well-preserved tumor areas were selected for IHC. IHC analysis was performed on tissue fixed with 10% neutral buffered formalin. IHC analyses for ER was performed at the time of diagnosis on all specimens in the Department of Pathology and Laboratory Medicine at KUMC. Samples with greater than 1% of the tumors staining for ER were considered positive and qualified the sample to be included in the tissue microarray. After review of the hematoxylin and eosin slides and marking of tumor areas, 2-mm tissue cores of representative tumor areas were extracted and inserted in recipient blocks. IHC analysis for IFITM1 (Santa Cruz) was performed on tissue microarrays obtained from the same samples (see IHC Staining protocol below). Two cores from each tumor were analyzed in an attempt to account for the impact of tumor heterogeneity on IFITM1 expression. IFITM1 staining intensity was quantified manually on a scale of 0–3 where 0 means no staining, 1+ is faint staining, 2+ is moderate staining and 3+ is strong staining. Cores were scored by three independent individuals prior to accessing patient medical records. Final distribution of IFITM1 expression is shown in Table 2. Histopathologic parameters were extracted from patient pathology records and clinical parameters were obtained from electronic medical records.
Table 1.
| Frequency | Percent | |
|---|---|---|
| Age | ||
|
| ||
| 30–39 | 10 | 11% |
| 40–49 | 19 | 20% |
| 50–59 | 23 | 24% |
| 60–69 | 19 | 20% |
| 70–79 | 13 | 14% |
| 80+ | 10 | 11% |
|
| ||
| Total | 94 | |
|
| ||
| Race | ||
|
| ||
| European American | 86 | 91% |
| African American | 5 | 5% |
| Asian/Pacific Islander | 3 | 3% |
|
| ||
| Total | 94 | |
|
| ||
| Clinical Stage | ||
|
| ||
| 1 | 20 | 21% |
| 2 | 22 | 23% |
| 3 | 10 | 11% |
| 4 | 5 | 5% |
|
| ||
| Total (available) | 57 | |
|
| ||
| Her2/Neu | ||
|
| ||
| 0+ | 38 | 40.43% |
| 1+ | 15 | 15.96% |
| 2+ | 4 | 4.26% |
Table 2.
IFITM1 staining intensity in normal and cancerous breast tissues
| Tissue Type | Intensity | Frequency | Percent |
|---|---|---|---|
| Normal Breast | |||
| 0 | 6 | 100% | |
| 1+ | 0 | 0% | |
| 2+ | 0 | 0% | |
| 3+ | 0 | 0% | |
|
| |||
| Total | 6 | ||
|
| |||
| Breast Tumors | |||
| 0 | 7 | 7% | |
| 1+ | 24 | 26% | |
| 2+ | 48 | 51% | |
| 3+ | 22 | 23% | |
|
| |||
| Total | 94 | ||
2.6 Immunohistochemical (IHC) Staining Protocol
IHC staining was performed after tissue deparaffinization by clearance in xylene and hydration through graded ethanol series. Antigen retrieval was conducted at 99°C in Dako Target retrieval solution (S1700) for 20 min per manufacturer’s instructions (Agilent Technologies, Copenhagen, Denmark). For human samples, blocking was performed using 5% normal horse serum and antibody dilution was performed in 0.01% Triton-X. For mouse xenografts, blocking and antibody dilution were performed using the Mouse on Mouse (MOM™) Kit following manufacturer’s instructions (Vector Labs, Burlingame, CA). Sections were stained using primary human antibodies targeted against IFITM1, ERα, phospo-STAT1 (ser701), CD31, MMP1 (Santa Cruz) and Ki67 (Dako) and HRP-conjugated biotinylated secondary antibodies (Vector Labs). Immunoperoxidase signal was produced using 3,3′-Diaminobenzidine (DAB) and amplified using the Vectastain® Elite ABC Kit (Vector Laboratories). Tissue sections were counter stained using hematoxylin and mounted in xylene. Slides were imaged on a Nikon Eclipse 80i Upright Microscope in the Imaging Core of KUMC.
2.7 Small interfering RNA (siRNA) transfections
Cells were transiently transfected with siRNA for IFITM1 (cat# sc-44549A/B/C), p21, MUC1 or a scrambled negative control (cat# sc-37007) (Santa Cruz Biotechnology, Santa Cruz, CA). All of the siRNAs were pools of three target specific 20 to 25 nt siRNAs. Cells were seeded the night before transfection and allowed to reach 60% confluence by the time of transfection. Twenty nmol of each siRNA was introduced using Lipofectamine 2000 in OptiMEM Reduced-Serum Medium (Invitrogen, San Diego, CA, USA) according to the manufacturer’s instructions. After overnight incubation, the transfection mixture was replaced with normal culture medium containing 1 μg/mL doxycycline, only where indicated.
2.8 TUNEL Staining
TUNEL staining was conducted using the Invitrogen Click-iT™ Plus TUNEL assay kit on either methanol fixed cell lines grown on chamber slides or deparaffinized tissue (See below) following manufacturer’s instructions (Thermo Fisher Scientific). The average TUNEL intensity was quantified using the red color channel on Image J software from a minimum of three images.
2.9 Annexin V Apoptosis Assay
The annexin V–FITC-labeled Apoptosis Detection Kit I (BD Pharmingen, San Diego, CA) was used to detect and quantify apoptotic cells by flow cytometry according to the manufacturer’s instructions. In brief, cells were seeded in 6-well plates and cultured for up to 118 hours in their regular media or with 1 μg/mL doxycycline, where indicated. Media was changed at 72 hours for longer experiments. Cells were then harvested by trypsinization and resuspended in 1 × binding buffer (HEPES buffer, 10 mM, pH 7.4, 150 mM NaCl, 5 mM KCl, 1 mM MgCl2, and 1.8 mM CaCl2). Samples were stained simultaneously with FITC-labeled annexin V (25 ng/mL) and propidium iodide (PI) (50 ng/mL). Cells were analyzed using the BD FACSAria™ II Flow Cytometer (BD, Franklin Lakes, NJ) in the Flow Cytometry Core Facility at the University of Kansas Medical Center, and the data was analyzed with FlowJo software (Ashland, OR).
2.10 Plasmid DNA construction
shRNA cloning was performed utilizing Gateway® Technology (Invitrogen Life Technologies). Double stranded IFITM1 shRNA was generated as previously described.[7] The vectors used for cloning were a kind gift from Eric Campeau. Briefly, double stranded IFITM1 shRNA (5′-GATCGCTGTGACAGTCTACCATATTTCAAGAGAATATGGTAGACTGTCACAG-3′) and scrambled shRNA sequences (5′-CATCGCCTAAGGTTAAGTCGCCCTCGCTCG AGCGAGGGCGACTTAACCTTAGG-3′,) were inserted into the pENTR/pTER+ vector (430–1) (Addgene #17453), placing their expression under the control of the inducible H1/TO promoter which contains a Tet-operator (TetO). The pENTR/pTER+ shRNA constructs were then incubated with LR clonase II enzyme mix (Invitrogen) and pLentiX2-Hygro-DEST (w17-1) (Addgene #17295) vector to generate pLentiX2 DEST/shIFITM1 and pLentiX2 DEST/shCon constructs.
2.11 Virus preparation and transduction
Lentivirus preparation was performed as previously described.[7] Briefly, pLentiX2 DEST constructs were co-transfected with pVSVG packaging vectors into HEK293T cells using Lipofectamine 2000 (Invitrogen). Medium was changed after 24 hours and viral supernatant was harvested 48 hours later. Virus-containing medium was then filtered using a 0.45μM syringe filter (Millipore). MCF-7:5C cells were then seeded in 6-well plates and allowed to reach 70% confluence overnight. 400 μL of viral supernatant of the filtered supernatant and 1mL
Lentivirus expressing pLenti CMV rtTA3 BLAST (TetR) vector (w756-1) was another kind gift from Eric Campeau purchased from Addgene.[7] First, transduction of MCF-7:5C cells with the construct was accomplished using 100 μL virus-containing medium in 2 mL normal medium and 3 μg/mL polybrene which generated MCF-7:5C/TetR cells which express the TetR gene under a CMV promoter. TetR expression occupies TetO sites, keeping target-genes silenced until tetracycline or doxycycline exposure. MCF-7:5C/TetR cells were then transduced with the pLentiX2 DEST constructs which produced the MCF-7:5C/TetR/shCon (MCF-7:5C/shCon) and MCF-7:5C/TetR/shIFITM1 (MCF-7:5C/shIF) cell lines.
2.12 Animals
Recipients were 8- to 10-week-old virgin female NOD-SCID IL2Rgammanull (NSG) mice which were purchased from Jackson Laboratories or bred (a gift from Thomas Yankee, PhD at The University of Kansas Medical Center). Animal experiments were conducted following protocols approved by the University of Kansas School of Medicine Animal Care and Use.
2.13 Orthotopic Cell Line Transplantation
Cells were suspended in 50:50 PBS/Matrigel (Corning) were bilaterally into 4th mammary fat pads of NSG mice, as described previously. [42] 3 x 106 cells were delivered per injection in a volume of 100μL. The length (L) and width (W) of tumors was measured weekly with digital calipers, and the tumor volume was calculated by the formula L2/(2W). When tumors reached a mean volume of 0.20 cm3, groups of 5–15 mice were randomly assigned to treatment groups. For doxycycline treatment, mice were provided with 50 μg/mL doxycycline in dark water bottles to induce IFITM1 shRNA expression. Water was replaced weekly to maintain efficacy. Where indicated, mice were given 50 μg/kg body weight of Ruxolitinib suspended in methylcellulose by oral gavage every other day. When using MCF-7 cells, capsules made of medical grade silastic tubing (Dow-Corning) containing 1:4 estradiol/cholesterol (Sigma Aldrich) were implanted subcutaneously in the mice where indicated. These capsules produce a mean serum estradiol level of ~80 pg/mL, which is similar to postmenopausal serum levels of estradiol. For the control group, the mice did not receive any treatment.
2.14 Mammary intraductal transplantation (MIND) method
MIND injections were conducted as previously described using 8–10 weeks old ovariectomized NSG mice. [4, 37] Briefly, cells were resuspended as single cells in PBS and counted. A 30-gauge Hamilton syringe, 50-μl capacity, with a blunt-ended 1/2-inch needle was used to deliver the cells. The mice were anesthetized by ketamine/xylene injection, and a Y-incision was made on the abdomen, allowing exposure of the inguinal mammary fat pads. The nipple of the inguinal gland is snipped so that the needle can be directly inserted through the nipple. Two microliters of cell-culture medium (with 0.1% trypan blue) containing cells at a concentration of 2,500 to 5,000 cells/μl were injected. Successfully injection was confirmed by visual detection of trypan blue in the ductal tree branches. The skin flaps were then repositioned normally and held together with wound clips. When required, mice were provided with 50 μg/mL doxycycline in dark water bottles to induce IFITM1 shRNA expression. Water was replaced weekly to maintain efficacy.
2.15 Tissue Harvesting and Preparation
Mammary glands and solid tumors were removed and fixed in 4% paraformaldehyde for 72 hours and then 70% ethanol. Fixed glands were then processed, embedded and sectioned by the Biospecimen Repository at the University of Kansas Medical Center.
2.16 Immunofluorescent (IF) staining of tissue
IF was performed as previously described [69] after tissue deparaffinization by clearance in xylene and hydration through graded ethanol series. Antigen retrieval was conducted at 99°C in Dako Target retrieval solution (S1700) for 20 min per manufacturer’s instructions (Agilent Technologies, Copenhagen, Denmark). Washes were performed in IF buffer (130 mM NaCl, 7 mM Na2HPO4, 3.5 mM NaH2PO4, 7.7 mM NaN3, 0.1% bovine albumin, 0.2% Triton X-100, 0.5% Tween-20). Due to use of mouse antibodies on mouse tissue, blocking and antibody dilution were performed using the Mouse on Mouse (MOM™) Kit following manufacturer’s instructions (Vector Labs, Burlingame, CA). Sections were stained using human antibodies targeted against Keratin 19 (Neomarkers #MS-198-P1), anti- actin smooth muscle (Spring Biosciences #E2464), anti-ki67 (Dako #M7240), or anti-IFITM1 (Santa Cruz Technology). Secondary antibodies were FITC or Texas Red conjugated (Santa Cruz). Sections were mounted using ProLong® Gold Antifade Reagent with DAPI (Cell Signaling) and visualized on a Leica TCS SPE confocal microscope in the Confocal Imaging Core at The University of Kansas Medical Center. Images were collected and analyzed using the Leica LAS AF Lite software (Leica Biosystems, Nussloch, Germany). For quantification, mean fluorescent intensity was determined using Image J software on single color channel images.
2.17 Cell Viability
Cells were assayed for viability in 24-well plates in triplicate in either estrogen or estrogen-free medium (see Cell Culture Conditions). At each time point, a measure of the viable cells was taken using the Cell-Titer Blue Assay Kit (Promega, Madison, WI) per the manufacturer’s instructions. Assay plates were kept at 37°C in 5% CO2 for 3 hours and read at 560–590 nM on a BioTek Synergy 4 microplate reader using the Gen 5 data analysis software (BioTek Instruments, Winooski, VT).
2.18 Soft Agar Anchorage-independent Growth Assay
6-well plates were coated with 1 mL of 0.8% agarose in the appropriate culture media. Cells were then suspended in 0.48% agarose and immediately overlaid on the pre-coated plates. Once the agarose was solid, 1mL of culture medium was added to the wells and changed every 4 days for 20 days (see Cell Culture Conditions). Cultures were then stained with 50 μl of MTT solution (5 mg/mL) was added to each well at a final concentration of 500 μg/mL, and the plates were further incubated for 2 h at 37°C. Plates were imaged at 1X in a Bio-Rad ChemiDoc™ XRS+ System with Image Lab™ Software (Bio-Rad). Colonies were counted and measured using Image J software (NIH).
2.19 Wound healing assay
Cell lines were seeded in 6-well culture plates so that they would reach 70% confluence overnight. The next day, a single wound was made on the plates for each cell line by scratching the attached cells using a 200 μl sterile pipette tip. The plates were washed with complete medium to remove cellular debris from the scraped surface. The images of the cells were taken at 10X immediately, and after 24 and 48 hours using phase-contrast microscope. The area of the wound was quantified using the MRI Wound Healing Tool in Image J Software (NIH).
2.20 Cell Cycle Analysis
Cells were incubated in the appropriate cell culture media with and without doxycycline treatment. Cells were harvested at the indicated time points by trypsinization and then fixed with 0.9% NaCl and ice cold ethanol. Once all samples were collected, DNA was stained with 50 μg/mL Propidium Iodide and 100 μg/mL RNase A in PBS (Invitrogen). Samples were analyzed using a BD FACSAria™ II Flow Cytometer in the flow cytometry core at The University of Kansas Medical Center (BD, Franklin Lakes, NJ). The data were analyzed with FlowJo software (Ashland, OR).
2.21 CRISPR /Cas9 Lentiviral Activation of IFITM1
IFITM1 Lentiviral Activation Particles (Santa Cruz sc-416878-LAC) is a synergistic activation mediator (SAM) transcription activation system designed to specifically and efficiently upregulate gene expression via lentiviral transduction of cells. IFITM1 Lentiviral Activation Particles (h) contain the following SAM Activation elements: a deactivated Cas9 (dCas9) nuclease (D10A and N863A) fused to the transactivation domain VP64, an MS2-p65-HSF1 fusion protein and a target-specific 20 nt. guide RNA. Upon transduction, the SAM complex binds to a site-specific region approximately 200–250 nt. upstream of the transcriptional start site and provides robust recruitment of transcription factors for highly efficient gene activation. Viral transduction and cloning was conducted per manufacturer’s protocol.
2.22 Luciferase Assays
For IFITM1 and p21 promoter assays, 0.8 μg of plasmid DNA and the pRL CMV Renilla vector were used as previously described.[53] For analysis of IFITM1 promoter activity, the pGL3 plasmid with the first 750 nucleotides of the IFITM1 promoter inserted (pGL3-IFITM1 [−750/−1]), was used.[53] The pGL3-Basic-IRES was a kind gift from Joshua Mendell (Addgene #64784) [8]. For analysis of p21 promoter activity, the pGL2-p21 promoter-Luc plasmid was used, which as a kind gift from Martin Walsh (Addgene #33021).[51] After 24 hours, transfection reagent was replaced with normal cell culture media containing ruxolitinib where indicated. Luciferase and Renilla activities were measured 24 h later using the Dual-Luciferase® reporter assay kit (Promega) according to the manufacturer’s instructions on a BioTek Synergy 4 microplate reader using the Gen 5 data analysis software (BioTek Instruments).
For Gamma Associated Sequence (GAS/STAT1) assays, the Cignal™ Reporter Assay (#CCS-009L) for analysis of STAT1 signaling was used according to manufacturer’s instructions. Briefly, the reverse transcription protocol was used where cells were suspended in Opti-MEM and seeded in 96-well plates on top of already aliquoted transfection cocktail. This cocktail contained Lipofectamine 2000™ and the GAS Reporter, which is a mixture of an inducible reporter plasmid containing tandem GAS-responsive elements upstream of a firefly luciferase construct and a constitutively expressing Renilla luciferase construct. After overnight incubation, the mixture was replaced with normal media containing Ruxolitinib where indicated. Luciferase and Renilla activities were measured 24 h later using the Dual-Luciferase® reporter assay kit (Promega) according to the manufacturer’s instructions on a BioTek Synergy 4 microplate reader using the Gen 5 data analysis software (BioTek Instruments).
2.23 Chromatin Immunoprecipitation (ChIP) Assay
ChIP was performed using the ChIP-IT Express Kit (Active Motif, Carlsbad, CA) according to the manufacturer’s instructions using sonication as the method for chromatin shearing. MCF-7:5C/shIF cells were treated with 1μg/mL doxycycline for 24 hours and harvested according to manufacturer’s instructions. Lysates were immunoprecipitated (IP) overnight (18 h) with 3μg of the following antibodies STAT1, STAT2, STAT3 (Santa Cruz) or an equal amount of rabbit IgG. Resulting DNA was analyzed using qPCR as described previously [27], and data are represented as a percentage of input DNA. Primer set used for the −640 STAT motif was Forward: 3′-TTTTGGTAGTCTCTCCAATTCC-5′ and Reverse: 3′-CCGTTTATTTCACAGATGAGG -5′.
3 RESULTS
3.1 Clinical significance of IFITM1 overexpression in ER-positive breast cancer
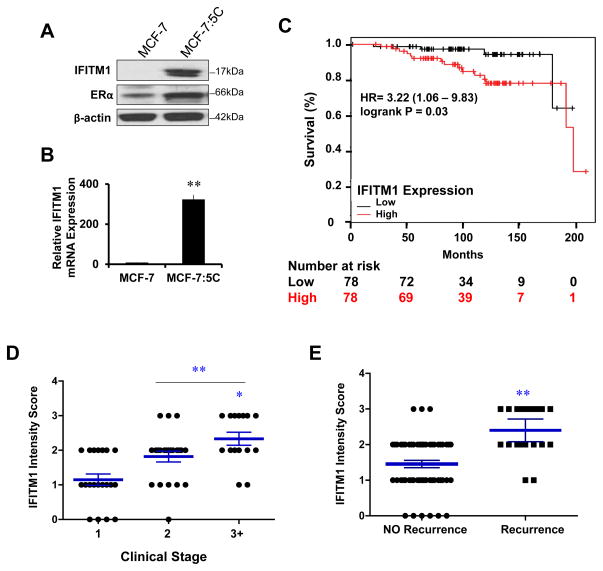
We have previously shown that AI-resistant MCF-7:5C cells overexpress a panel of ISGs as compared to the AI-sensitive parental cell line MCF-7 [14]. We confirmed IFITM1 overexpression in MCF-7:5C cells at the protein (Figure 1A) and mRNA level (Figure 1B). The Kaplan-Meier Plotter breast cancer survival database revealed that high IFITM1 expression correlated with poor overall survival for patients with grade 1 ER+ disease (Figure 1C) [26]. Next, we conducted in-house analysis of IFITM1 expression in 94 ER+ breast cancer patient samples using immunohistochemistry (Table 1). Clinical stage and Her2/Neu expression information was available for only 57 of the 94 patients. The 94 samples represented the full range of IFITM1 expression (0–3) (Supplementary Figure S1) and all of the normal breast tissue samples were negative for IFITM1 expression (Table 2). High IFITM1 intensity score correlated with increased clinical stage (Figure 1D) and increased risk of recurrence during endocrine therapy (Figure 1E). These two separate analyses suggest that high IFITM1 expression in ER+ breast cancer is significantly associated with poor clinical outcome and poor response to endocrine therapy.
Figure 1. IFITM1 overexpression is associated with aromatase inhibitor-resistant breast cancer cells and correlates with poor clinical outcome.
(a) Whole cell lysates from 80% confluent MCF-7 and MCF-7:5C cells were immunoblotted for IFITM1 and ERα protein expression. (b) IFITM1 and ERα mRNA expression was determined by RT-PCR and is displayed relative to PUM1. Data represents three independent experiments run in triplicate. (c) The hazard ratio (HR) and 95 % confidence interval for high IFITM1 expression in ER+ Luminal A breast cancer patients was analyzed using The Kaplan-Meier Plotter database. Patients were stratified by average IFITM1 expression (probe sets 214022_s_at and 201601_x_at) relative to median. The P-value was calculated using a log-rank test. (d) Immunohistochemical staining for IFITM1 expression was conducted in 94 ER+ breast cancer patient samples and 6 normal breast tissue samples. Staining intensity was graded on a scale of 0–3. IFITM1 staining intensity was correlated with clinical stage (d) and recurrence (e). *p < 0.05, ** p< 0.01
3.2 Loss of IFITM1 inhibits AI-resistant breast tumor growth and promotes cell death in vivo
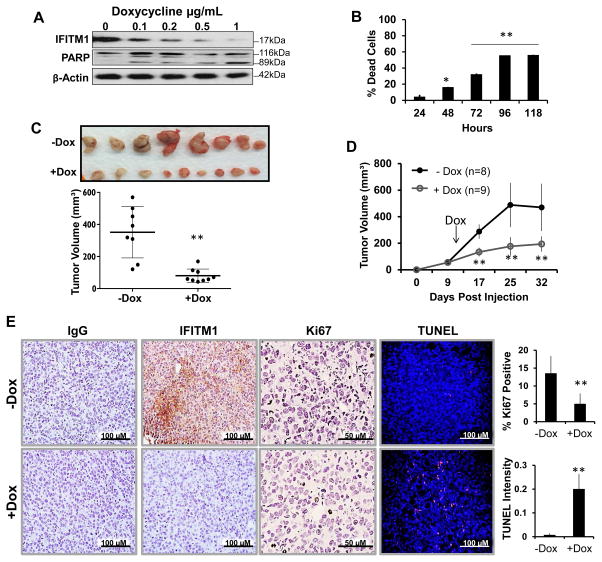
To investigate the effects of IFITM1 loss on proliferation and survival in AI-resistant cells, we developed the MCF-7:5C/shIF cell line which expresses IFITM1 shRNA under the control of a tetracycline/doxycycline-inducible promoter. Our lentivirus-inducible system showed that upon doxycycline (Dox) exposure, IFITM1 expression was reduced in MCF-7:5C/shIF cells in a concentration-dependent manner, with maximum reduction at 1 μg/mL Dox (Figure 2A). Loss of IFITM1 using Dox-inducible shRNA initiated cell death in a concentration-dependent and time-dependent manner which were demonstrated by PARP cleavage (Figure 2A) and annexin-PI staining (Figure 2B). These findings were consistent with our siRNA in vitro data (Supplementary Figure S2). Notably, Dox treatment had no effect on the survival of un-transduced MCF-7:5C cells or MCF-7:5C cells expressing a scrambled control vector (MCF-7:5C/shCon) (Supplementary Figure S3).
Figure 2. Loss of IFITM1 inhibits MCF-7:5C tumor growth and promotes cell death in vivo.
(a) MCF-7:5C/shIF cells were treated with the indicated concentrations of doxycycline to induce IFITM1 shRNA expression. IFITM1 and PARP protein expression levels were determined by immunoblotting. (b) MCF-7:5C/shIF cells were treated with 10μg/mL doxycycline over a 118 hour period. Cells were harvested on the last day and Annexin V/ Propidium Iodide staining quantified the percent of dead cells over the accompanying untreated (control) sample. Data represents two independent experiments conducted in duplicate. (c) 3 million MCF-7:5C/shIF cells were injected into the 4th mammary fat pad of ovariectomized female NSG mice. 9 days after orthotopic injection, mice were randomized to treatment groups and half were provided doxycycline (Dox)-treated drinking water. Tumors were measured by digital calipers and tumor volume (mm3) displayed over time. (d) At the end of the experiment tumors were excised and are shown. Tumor volumes were determined by digital caliper measurement. (e) IFITM1 and Ki67 expression was determined by immunohistochemical staining. The percent of Ki67 positive cells was determined by counting four separate 40X fields. Cell death was analyzed by TUNEL staining and quantified by Image J software. ** p< 0.01
Generation of the MCF-7:5C/shCon and MCF-7:5C/shIF cells allowed us to assess the effect of IFITM1 loss on the growth of MCF-7:5C cells in the orthotopic model of breast cancer. We found that Dox exposure (+Dox) reduced IFITM1 expression which significantly inhibited MCF-7:5C/shIF tumor growth compared to the control group (−Dox) (Figure 2C and 2D). Reduction in tumor growth correlated with a decrease in proliferation (Ki67 positivity) and an increase in cell death (TUNEL staining) (Figure 2E). Notably, Dox exposure had no effect on the growth of MCF-7:5C/shCon cell tumors (Supplementary Figure S2C) or on the expression of IFITM1, Ki67 or TUNEL (Supplementary Figure S4). This data confirm that IFITM1 overexpression is important for the growth and survival of AI-resistant MCF-7:5C tumors in vivo.
3.3 Loss of IFITM1 blocks aromatase inhibitor-resistant tumor cell invasion out of the mammary duct
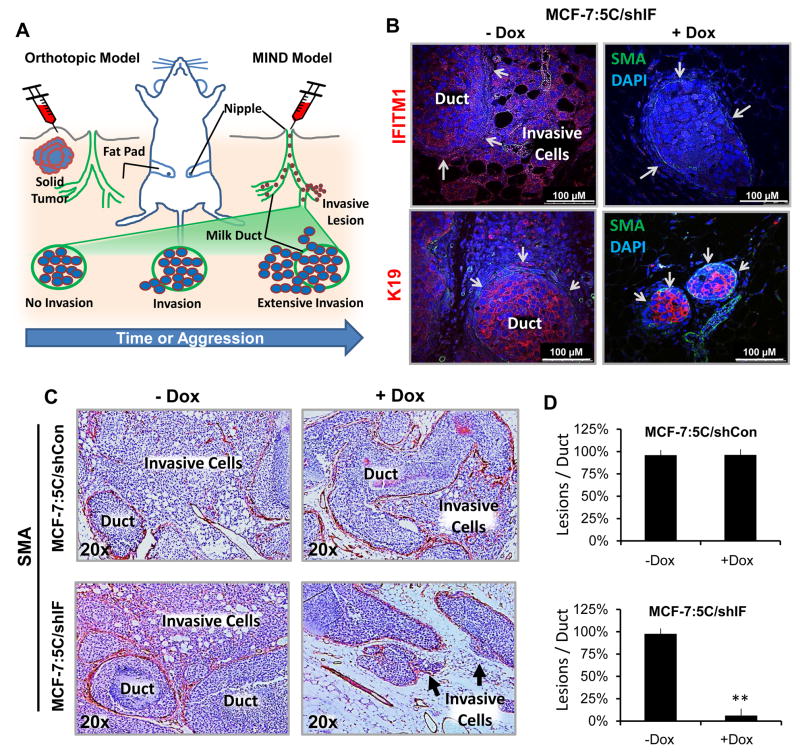
We used the mammary intraductal (MIND) model of breast cancer to investigate the effect of reduced IFITM1 expression on AI-resistant tumor cell invasion in vivo (Figure 3A) [14, 53]. After three weeks of Dox exposure, we confirmed that the MIND model was able to accurately assess tumor cell invasion out of the mammary duct using immunofluorescent staining (Figure 3B). Immunofluorescent staining verified that Dox exposure resulted in IFITM1 loss in the MIND model (Figure 3B). Smooth muscle actin (SMA) was used to stain the myoepithelial layer of the mammary duct and human keratin 19 (K19) was used to label the breast cancer cells (Figure 3B). Hematoxylin and eosin (H&E) staining further revealed the architecture of the mammary glands, demonstrating that invasive IFITM1-expressing cells breach the mammary duct and invade into the mammary gland stroma (Figure 3C and Supplementary Figure S5A). Immunohistochemical staining for SMA was used to visualize the extent of tumor burden in the mammary gland parenchyma (Figure 3C) and to quantify the extent of invasion (Lesions/Duct) (Figure 3D). The MIND model revealed that MCF-7:5C/shIF cells were highly invasive, however, loss of IFITM1 (+Dox) completely blocked the ability of these cells to invade out of the mammary duct as demonstrated by the lack of K19-stained cells outside of the duct (Figure 3B and 3D). Using Ki67 and TUNEL staining, we found that loss of IFITM1 expression also reduced cell proliferation and increased cell death in the MIND model of breast cancer (Supplementary Figure S5B). Dox exposure had no effect on the invasion, proliferation or survival of MCF-7:5C/shCon cells in either in vivo model (Figure 3C and Supplementary Figure S6). This experiment demonstrates that the MIND model can be used to assess the impact of individual gene expression on ERα+ tumor cell invasion in vivo and that it faithfully replicates effects on cell survival and proliferation seen in vitro or the orthotopic model of breast cancer.
Figure 3. Loss of IFITM1 reduces aromatase inhibitor-resistant tumor cell invasion in the MIND model.
5,000 MCF-7:5C/shIF cells were injected into the duct of the 4th mammary gland of female ovariectomized NSG mice use the MIND injection protocol. Four glands were also injected with PBS (sham). After 10 days, mice were randomized to treatment groups and half were provided doxycycline (Dox)-treated drinking water. After 3 weeks, mammary glands were removed. (a) A diagram of the two breast cancer models where cells are injected into the mammary fat pad (Orthotopic Model) or through the nipple (MIND Model). (b) Mammary glands were fixed and processed onto glass slides. Immunofluorescent staining of the breast cancer cells for IFITM1, human keratin 19 (HuK19-red) and the mammary duct for smooth muscle actin (SMA-green) was conducted. Arrows indicate the mammary duct wall. (c) The milk duct was identified by immunohistochemistry. SMA was used to delineate invasive areas in MCF-7:5C/shCon and MCF-7:5C/shIF cells. (d) Number of invasive lesions were quantified per duct in a 10X field. Numbers represent averages of 5 fields in 3 different mammary glands for each treatment group. ** p< 0.01
3.4 Overexpression of IFITM1 promotes breast cancer cell aggression in vitro and in vivo
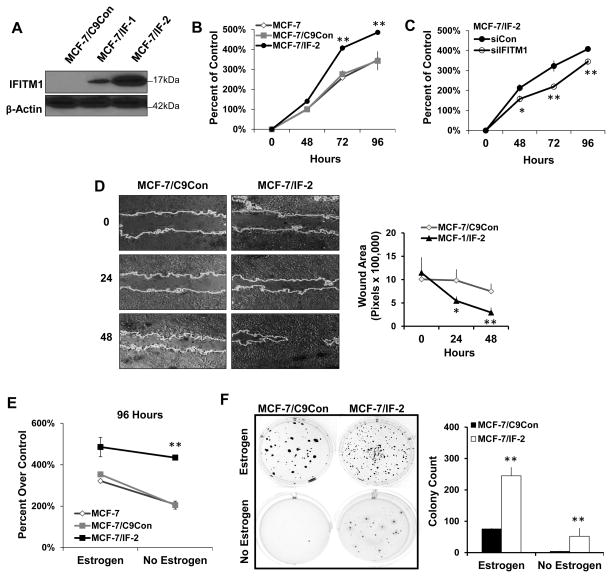
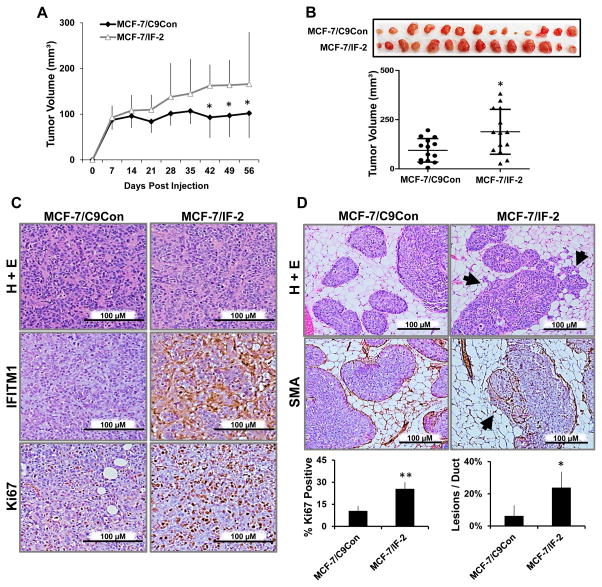
To validate the importance of IFITM1 in the ER+ breast cancer cell phenotype, we overexpressed IFITM1 in parental MCF-7 cells using CRISPR/Cas9 guided transcription. Two clones, MCF-7/IF-1 and MCF-7/IF-2 were generated which expressed moderate and high levels of IFITM1 respectively (Figure 4A), as compared to control scrambled guiding-RNA expressing MCF-7/C9Con cells which lack IFITM1. IFITM1 overexpression significantly enhanced MCF-7 cell proliferation in a 96 hour cell viability assay (Figure 4B); however, knockdown of IFITM1 reduced the proliferation of MCF-7/IF-2 cells to that of wild type MCF-7 cells (Figure 4C). Additionally, IFITM1 overexpression enhanced the aggressive phenotype and tumorigenic potential of MCF-7 cells as demonstrated by wound healing cell migration assays (Figure 4D) and soft agar anchorage-independent growth (Figure 4F). To determine whether IFITM1 overexpression contributes directly to AI-resistance, we cultured the MCF-7/IF clones (MCF-7/IF1 and MCF-7/IF2) in estrogen-deprived conditions to mimic AI treatment, and found that overexpression of IFITM1 in MCF-7 cells increased their proliferation, tumorigenicity, and estrogen-independent growth (Figure 4E and 4F) compared to MCF-7 and MCF-7/C9Con cells (Supplementary Figure S7).
Figure 4. Overexpression of IFITM1 enhances the aggressive phenotype of MCF-7 breast cancer cells in vitro.
(a) IFITM1 expression in MCF-7 cells expressing scrambled control (MCF-7/C9Con) or an IFITM1 guided Cas9 overexpression vector (MCF-7/IF-1 and MCF-7/IF-2) was determined by immunoblotting and compared to expression in MCF-7:5C cells. (b) The proliferation of parental MCF-7, MCF-7/C9Con and MCF-7/IF-2 cells in estrogen containing media was measured over 96 hours by Cell Viability assay. (c) siRNA knockdown of IFITM1 in MCF-7/IF-2 cells over 96 hours demonstrated the specificity of IFITM1 overexpression. (d) Scratch assay was conducted on 70 % confluent plates of MCF-7/C9Con and MCF-7/IF-2 cells. Plates were imaged at 0, 24 and 48 hours and the size of the wound was quantified by image J (right panel). Values represent two independent experiments conducted in triplicate. (e) A direct comparison of proliferation in estrogen and estrogen free media is shown at 96 hours. (f) MCF-7/C9Con and MCF-7/IF-2 were subjected to soft agar anchorage independent growth assay. Plates were imaged after 21 days and the number of colonies was quantified using Image J (lower panel). Values represent two independent experiments conducted in duplicate. *p < 0.05, ** p< 0.01
To assess the role of IFITM1 overexpression in vivo, we transplanted the MCF-7/C9Con, MCF-7/IF-1 and MCF-7/IF-2 cells into intact (not ovariectomized) female NSG mice using both the orthotopic and MIND models of breast cancer. Overexpression of IFITM1 resulted in the enhanced growth of MCF-7/IF-2 solid tumors (Figure 5A and B). Immunohistochemistry revealed that the MCF-7/IF-2 tumors continued to express IFITM1 in vivo and had increased ki67 positivity (Figure 5C). Additionally, MCF-7/IF-2 cells were significantly more invasive than MCF-7/C9Con cells in the MIND model (Figure 5D). Similar results were seen in moderately IFITM1-overexpressing MCF-7/IF-1 cells (Supplementary Figure S7 and S8), further demonstrating that IFITM1 expression alone can promote the proliferation, migration and invasion of MCF-7 cells in vitro and in vivo.
Figure 5. Overexpression of IFITM1 promoted breast cancer cell proliferation and invasion in vivo.
(a) Intact female NSG mice were orthotopically injected with MCF-7/C9Con and MCF-7/IF-2 cells and tumors measured by digital calipers weekly. Tumor volume (mm3) was calculated and charted over time. (b) The tumors were excised and imaged at day 56 and final tumor volume (mm3) determined. (c) IFITM1 and Ki67 expression was determined by immunohistochemical staining. The percent of Ki67 positive cells was determined by counting four separate 40X fields (lower panel). (d) Female NSG mice were injected with MCF-7/C9Con and MCF-7/IF-2 cells using the MIND model. Smooth muscle actin (SMA) staining by immunohistochemistry elucidated the milk duct. Arrow heads indicate invasive lesions which were quantified per duct in a 10X field. Numbers represent averages of 5 fields in 3 different mammary glands for each group (lower panel). *p < 0.05, ** p< 0.01
3.5 High IFIM1 expression is associated with increased blood vessel density and MMP1 expression
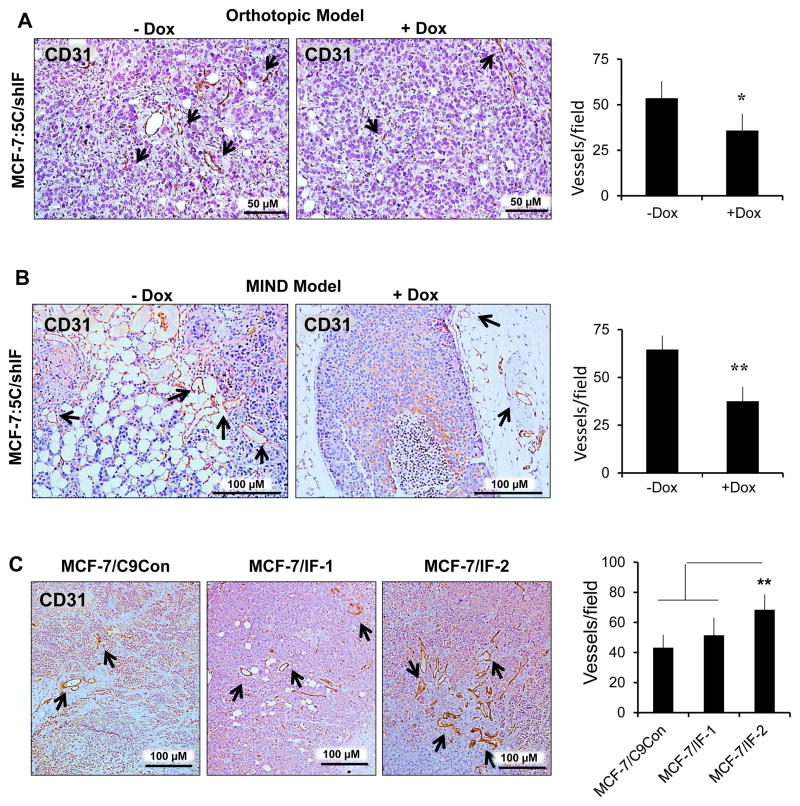
Notably, we found that loss of IFITM1 significantly reduced angiogenesis in both the MIND and orthotopic models of breast cancer, as demonstrated by CD31 staining (Figure 6A and 6B). This was seen in the doxycycline-treated MCF-7:5C/shIF groups in both the orthotopic and MIND models of breast cancer. Immunohistochemistry for CD31 expression demonstrated a significant decrease in blood vessel density in both models. Increased blood vessel density was also seen in MCF-7 tumors overexpressing IFITM1 as compared to control (Figure 6C).
Figure 6. IFITM1 overexpression enhances blood vessel density.
MCF-7:5C/shIF xenografts from the orthotopic (a) and MIND model experiments were stained for CD31 expression using immunohistochemistry. Blood vessel density was determined by counting the number of CD31 positive blood vessels in at least 4 random 10X fields in each group from three separate samples. (c) Similar analysis was conducted on MCF-7/C9Con, MCF-7/IF-1 and MCF-7/IF-2 MIND xenografts. Graphs represent the average number of vessels per field. Arrows indicate CD31+ blood vessels.
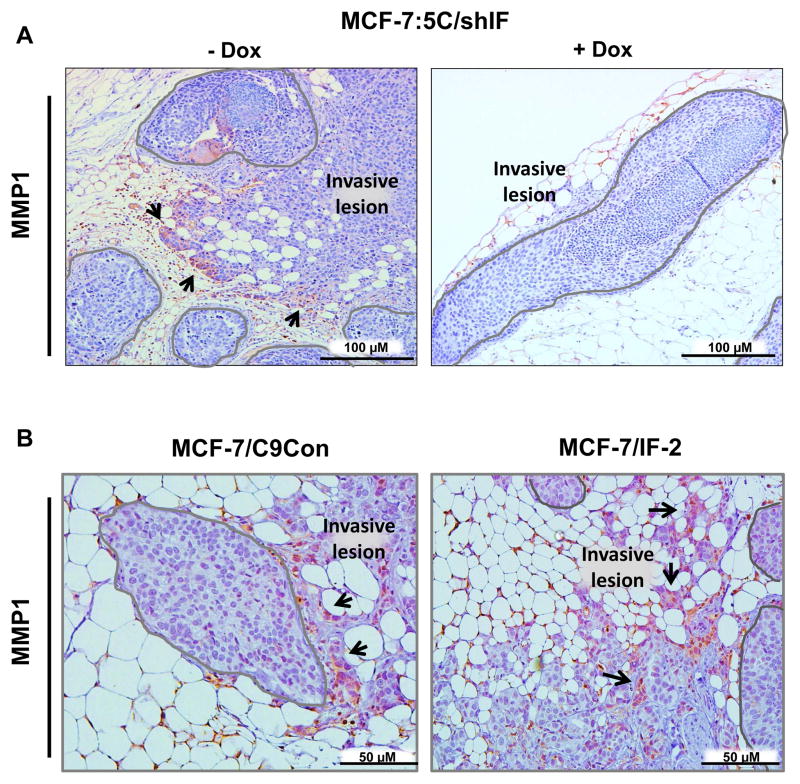
Since IFITM1 expression positively correlated with invasion we also measured matrix metalloproteinase 1 (MMP1) expression in our MIND xenografts. We found that MMP1 was upregulated in AI-resistant MCF-7:5C/shIF cells at the leading edge of invasion, while the non-invasive cells inside of the duct remain MMP1 negative (Figure 7A, left panel). Notably, loss of IFITM1 (+Dox) reduced the number and extent of invasive lesions and was associated with lower MMP1 expression (Figure 7A, right panel). Similarly, overexpression of IFITM1 in the MCF-7 cells enhanced tumor invasion and MMP1 expression in invasive lesions (Figure 7B). The upregulation of MMP1 likely facilitates colonization of the mammary gland parenchyma.
Figure 7. Invasive lesions express MMP1 in the MIND model of breast cancer.
MIND xenografts from the MCF-7:5C/shIF (a), MCF-7/C9Con and MCF-7/IF-2 (b) cell lines were stained for matrix metalloproteinase 1 (MMP1) expression by immunohistochemistry and representative images are shown. Mammary ducts are outlined in gray and MMP1 positive invasive cells indicated with arrows. ** p< 0.01
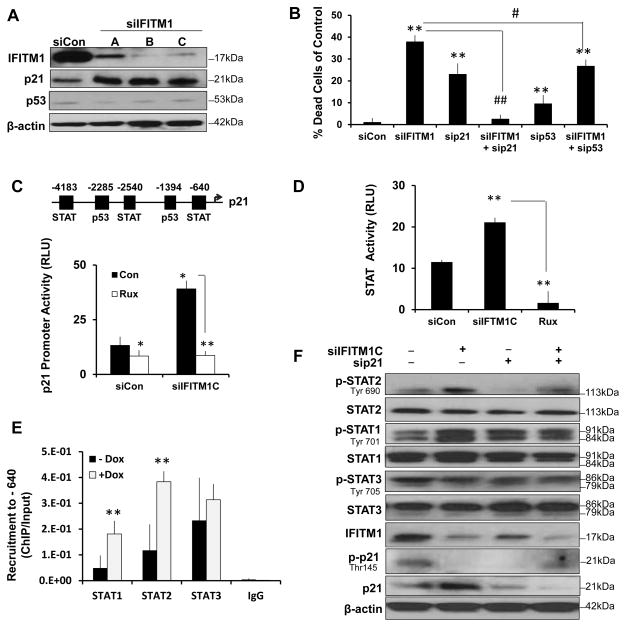
3.6 Loss of IFITM1 induces p21 expression in AI-resistant cells through enhanced STAT1 activity
IFN signaling and ISGs modulate cell cycle progression and cell survival, hence, we assessed the mechanism by which loss of IFITM1 induces cell death in AI-resistant MCF-7:5C cells. We found that siRNA/shRNA knockdown of IFITM1 in the MCF-7:5C cells increased p21 protein expression with no effect on p53 expression (Figure 8A). Additionally, blocking p21, but not p53, prevented the apoptotic effect of IFITM1 knockdown (Figure 8B). Using a p21 promoter luciferase reporter, we found that loss of IFITM1 increased p21 transcription (Figure 8C). The p21 promoter is known to have binding motifs for a variety of transcription factors including p53, SP1, AP2 and STAT proteins [1, 24]. To demonstrate that the increase in p21 transcription was mediated by STAT activation, we knocked down IFITM1 in the presence and absence of the JAK1/2 inhibitor ruxolitinib/Jakafi™ (Rux). Notably, inhibition of STAT phosphorylation with Rux prevented the increase in p21 expression and transcription (Figure 8C and Supplementary Figure S9D) and protected the cells from death (Supplementary Figure S9C and S9E). We confirmed that IFITM1 knockdown enhanced JAK/STAT signaling using a STAT luciferase reporter and that Rux blocks STAT signaling (Figure 8D). Chromatin immunoprecipitation (ChIP) showed that in MCF-7:5C cells STAT1, STAT2 and STAT3 bind to the STAT motif in the p21 promoter at the −640 position. Significantly, IFITM1 knockdown enhanced the recruitment of STAT1 and STAT2 to the p21 promoter (Figure 8E) and it increased STAT1 (Tyr 701) and STAT2 (Tyr 690) phosphorylation (Figure 8F); however, there was no effect on STAT3 recruitment or phosphorylation (Tyr 705) (Figure 8E and 8F). Interestingly, IFITM1 knockdown also inhibited the phosphorylation of p21 (Thr145), which suggests a loss of cytoplasmic p21 (Figure 8F).
Figure 8. IFITM1 knockdown increases p21 expression through enhanced STAT1 activity, which mediates cell death.
(a) Cell lysates from MCF-7:5C cells transfected with scrambles control siRNA (siCon) or three different IFITM1 siRNA sequences (siIFITM1 A, B, C) were immunoblotted for IFITM1, p53 and p21 expression. (b) Dual Annexin V/Propidium Iodide staining was used to quantify cell death in each transfection group as compared to siCon. Data represent means ± SD from two experiments conducted in duplicate. (c) Activity at the p21 promoter was determined by luciferase assay. The motifs of interest are indicated in the promoter map (upper panel). MCF-7:5C cells were transfected with p21 luciferase reporter and renilla reporter constructs with siCon or siIFITM1. Cells were treated with 10μM Rux or vehicle (Con) and assayed after 24 hours of Rux treatment. Values represent means ± SD of two independent experiments done in triplicate. (d) MCF-7:5C cells were transfected with STAT/GAS luciferase cocktail and with siCon or siIFITM1 or treated with 10μM ruxolitinib (Rux). (e) Binding of STAT proteins to p21 promoter. Fixed cell lysates from MCF-7:5C/shIF cells treated with vehicle (−Dox) or 1μg/mL doxycycline (+Dox) for 24 hours and subjected to chromatin immunoprecipitation (ChIP) with antibodies against STAT1, STAT2, STAT3 or rabbit IgG control. qPCR was performed on the isolated DNA using primers designed to amplify the −640 STAT regulatory region in the p21 promoter. Recruitment was compared to input DNA and displayed as mean ± SD of technical triplicates. (f) MCF-7:5C cells were transfected with siCon, siIFITM1C, p21 siRNA (siP21) or siIFITM1-C with sip21 and lysates were immunoblotted for phospho-STAT2 (p-STAT2), STAT2, phospho-STAT1 (p-STAT1), STAT1, phospho-STAT3 (p-STAT3), STAT3 IFITM1, phopsoh-p21 (p-p21) and p21. * p<0.05, ** p<0.01
3.7 Loss of IFITM1 is associated with nuclear translocation of p21
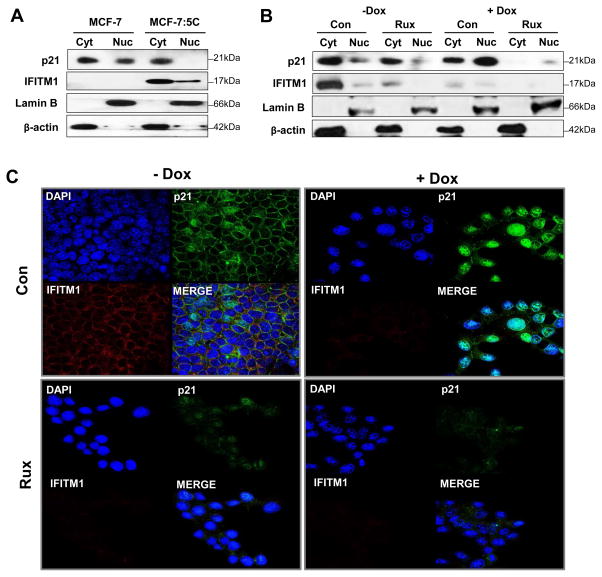
The cellular localization of p21 also plays a role in cell survival. We fractionated the cytoplasmic and nuclear fractions of parental MCF-7 cells and AI-resistant MCF-7:5C cells and found significantly diminished p21 expression in the nuclear fraction of MCF-7:5C cells (Figure 9A). We used the Dox-inducible MCF-7:5C/shIF cells to determine the effect of IFITM1 loss on p21 localization. Both cell lysate fractionation and immunofluorescent staining revealed that loss of IFITM1 (+Dox) was associated with increased nuclear translocation of p21 (Figure 9B and 8C). Interestingly, treatment with Rux also prevented induction of p21 translocation to the nucleus (Figure 9B and 8C). Our data suggest that IFITM1 expression is inversely related to p21 nuclear localization and that p21 nuclear translocation may be a key step in IFITM1 knockdown-induced cell death.
Figure 9. Loss of IFITM1 induces p21 nuclear translocation.
(a) Fresh whole cell lysates from 80% confluent MCF-7, and MCF-7:5C were separated into nuclear (laminin B) and cytoplasmic (β-actin) fractions. Protein expression of p21 and IFITM1 was determined by immunoblotting. (b) MCF-7:5C/shIF cells with and without doxycycline (± Dox) exposure were also treated with 10μM ruxolitinib (Rux). After 48 hours Rux treatment, cells were harvested and fresh whole cell lysates were fractionated and immunoblotted for p21 and IFITM1 protein expression. B-actin and Lamin B expression indicate the purity of the cytoplasmic and nuclear fractions respectively. (c) MCF-7:5C/shIF cells with and without doxycycline (± Dox) exposure were also treated with 10μM ruxolitinib (Rux). After 48 hours Rux treatment, p21 and IFITM1expression and localization as determined by immunofluorescent staining.
4 DISCUSSION
IFITM1 expression has been linked to the progression of several aggressive cancer types, including cancers of the cervix, esophagus, colon, ovary and brain [9, 25, 39, 52, 55, 64, 77]. The mechanism by which IFITM1 promotes tumor progression is not understood, however, IFITM1 is known to play a role in DNA damage resistance which contributes to treatment failure in cancer [12, 72, 78]. Recently, our laboratory has found an association between IFITM1 overexpression and aromatase inhibitor (AI)-resistance in breast cancer [14]. The present study demonstrated the importance of IFITM1 expression in predicting the survival and response to endocrine therapy for breast cancer patients with ER+ disease. We utilized the IFITM1-overexpressing AI-resistant MCF-7:5C and IFITM1-null AI-sensitive MCF-7 cell lines to investigate the potential mechanisms by which IFITM1 drives tumor aggression and demonstrate the possible benefit of targeting IFITM1 in the clinic.
IFITM1 is a transmembrane interferon stimulated gene (ISG) that is normally induced in response to IFN exposure to aid in resistance to viral infections and to modulate the decision between cell survival and death [11, 12, 63, 72]. Estrogen deprivation is known to promote inflammatory gene signatures in the breast tumor microenvironment and expression of several cytokine and chemokine receptors, including IFNAR which drives IFITM1 expression, are known to be associated with resistance to endocrine therapy [22, 62]. Notably, one study has found that the expression of ISGs increases in mouse models of endocrine resistance [19]. While the exact structure and functional domains of IFITM1 remain unknown, recently identified binding partners include caveolin-1 (CAV-1) and CD81/TAPA-1. CAV-1 is a cell surface protein that is necessary to form invaginations in the cell membrane which impacts intracellular vesicle transport, cell cycle control and apoptosis [44, 56, 77]. CD81 is a member of the tetraspan family and is known to control proliferation, adhesion, and migration through association with many cell signaling molecules and transmembrane receptors [31, 49, 74]. Both CAV-1 and CD81 have been linked to cancer progression and metastasis [30, 31, 52]. Investigation into the interaction of IFITM1 with CAV-1 and CD81 in breast cancer cells may reveal novel roles for these proteins in driving AI-resistance.
Previous work in our lab demonstrated that AI-resistant cells and tumors express IFITM1 at a high level due to enhanced JAK/STAT signaling [14]. It is not known what drives AI-resistance, however, alterations in ER, growth factor, PI3K/Akt/mTOR, apoptosis and autophagy signaling have been identified as potential mechanisms in both preclinical and clinical studies [2, 17, 48, 54]. Notably, we found that overexpression of IFITM1 in MCF-7 cells (MCF-7/IF-1 and MCF-7/IF-2) significantly enhanced the proliferation and tumorigenicity of these cells both in vitro and in vivo. IFITM1 overexpression also improved breast cancer cell survival and aggression under estrogen-deprived conditions. IFITM1 expression is driven by type 1 IFN mediated JAK/STAT signaling [11, 57, 63]. Notably, constitutively enhanced STAT expression and activation have been linked to breast cancer development, progression and therapeutic resistance [12, 34, 35, 60]. Hence, IFITM1 overexpression may be one of the mechanisms by which activated JAK/STAT contributes to breast cancer development, aggression and therapeutic resistance.
Using the orthotopic model we found that overexpressing IFITM1 increased tumor proliferation in the mammary fat pad and that loss of IFITM1 in AI-resistant cells reduced cell proliferation and induced death in vivo. The effect of IFITM1 expression on tumor proliferation and survival was likely due in part to the effect that IFITM1 had an effect on CD31+ blood vessel density in vivo. IFITM1 in endothelial cells is known to promote angiogenesis but the mechanism by which IFITM1 expression in cancer cells affects the tumor microenvironment and drives angiogenesis is not known and warrants further study in our system [61]. The MIND model was developed to study the progression of ductal carcinoma in situ (DCIS) to invasive breast cancer [4, 69]. While the orthotopic model is the most common method of studying breast cancer in animals we used and validated the use of the MIND model as in vivo invasion assay. We discovered that IFITM1 expression can increase MCF-7 cell invasion while loss of IFITM1 can inhibit invasion out of the mammary duct. Recently, it has been shown that the MIND model can be used to study ER+ breast cancer invasion and metastasis and that the model maintains the phenotype of ER+ breast cancer cells more faithfully than the orthotopic model [65]. We found that results from the MIND model also reflected the differences in MCF-7 and MCF-7:5C cells aggression. AI-resistant MCF-7:5C MIND xenografts were significantly more invasive than AI-sensitive MCF-7 xenografts. Notably, invasive lesions showed an induction in matrix metalloproteinase 1 (MMP1) at the leading edge of invasion. IFITM1 is thought to promote invasion and inflammation through control of the expression of several matrix metalloproteinases (MMPs) [21, 28, 36]. The mechanism of IFITM1 control over MMP expression and activity remains unknown and warrants further study in our IFITM1-overexpression breast cancer cell lines. It should be noted that transplantation of human breast cancer cell lines requires immunocompromised mice, which does not allow for analysis of the interaction between IFITM1-overexpressing tumor cells and the immune system and is a limitation of our current animal model.
Mechanistic studies revealed that loss of IFITM1 markedly induced p21 expression which was dependent on STAT1 and STAT2 activation but not p53. While p53 is a well-known transcriptional regulator of p21, several other transcription factors, including the STAT proteins, also activate p21 expression in response to stress signals [33]. Here, we propose that MCF-7:5C cells are dependent on IFITM1 overexpression and that an attempt to compensate for IFITM1 loss by enhancing stress-induced JAK/STAT signaling drives p21 expression and cell death. In the present study, increased p21 expression was associated with cell cycle arrest and cell death (Supplementary Figure S9B), which is consistent with similar studies in other cancer models [33]. High expression of cytoplasmic p21 is known to promote cell survival and cell cycling while higher nuclear p21 stimulates cell death [1, 13, 18]. p21 is phosphorylated by Akt1 and murine leukemia virus (Pim-1) at threonoine-145 in order to keep it in the cytoplasm [1, 71] and cancers with high p21 cytoplasmic/nuclear ratio are thought to be more aggressive. Therefore, our finding that AI-resistant MCF-7:5C cells have very high expression of IFITM1 but very low nuclear p21 expression is consistent with their highly aggressive phenotype both in vitro and in vivo [20, 40, 75].
Notably, IFITM1 knockdown facilitated p21 nuclear translocation which correlated with loss of p21 phosphorylation (Thr 145). High Pim-1 expression and p21 phosphorylation are associated with poor prognosis in various cancers [29, 73, 76]. Specifically Pim-1 is thought to promote inflammation induced tumorigenesis, and promote cancer cell survival of chemotherapy and radiation treatment [43, 50, 59, 80]. In breast cancer, high Pim-1 expression is associated with PI3K/Akt mediated drug resistance [38]. The PI3K/Akt pathway has long been associated with endocrine resistance in ER+ breast cancer [3, 10, 16, 45, 79]. Significantly, in this study, overexpression of IFITM1 promoted MCF-7 cell survival in estrogen-deprived conditions. Whether IFITM1 modulates Akt1 and Pim-1 activity requires further study.
In conclusion, this study demonstrated that IFITM1 overexpression promotes AI-resistant breast cancer cell aggression and survival. The MIND model served as a tool for assessing invasion in vivo in addition to reflecting the effect of IFITM1 on proliferation and survival seen in the orthotopic model. Loss of IFITM1 caused cell death through an induction of p21 transcription and nuclear translocation. Future studies are needed to determine the exact role IFITM1 plays in p21 cellular localization. Our data suggests that IFITM1 may be a targetable marker of aggressive breast cancer and that development of an IFITM1 specific inhibitor may hold promise for treatment refractory breast cancer.
Supplementary Material
HIGHLIGHTS.
Interferon induced transmembrane protein 1 (IFITM1) overexpression is correlated with poor clinical outcome in breast cancer, including clinical stage, lack of response to endocrine therapy, and poor overall survival.
FITM1 overexpression is confirmed in the aromatase–inhibitor (AI) resistant breast cancer cell line, MCF-7:5C, however, it is not expressed in AI-sensitive MCF-7 cells which makes these models ideal for investigating the mechanisms by which IFITM1 drives AI-resistance.
IFITM1 knockdown diminishes AI-resistant tumor growth and invasion in vivo which is associated with changes in CD31+ blood vessel density and matrix metalloproteinase 1 (MMP1) expression.
IFITM1 overexpression in parental MCF-7 cells promotes aggression and estrogen-independent proliferation in vitro and tumor growth and invasion in vivo.
Loss of IFITM1 induces cell death in AI-resistant MCF-7:5C cells due to JAK/STAT mediated increases in p21 transcription, expression, and nuclear localization.
Acknowledgments
We would like to thank the Flow Cytometry Core at KUMC which is sponsored, in part, by the NIH/NIGMS COBRE grant P30 GM103326. We also wish to acknowledge the KUMC Confocal Imaging Facility and Imaging Core. The confocal core is supported, in part, by NIH/NIGMS COBRE grant P20GM104936. We acknowledge support from the University of Kansas (KU) Cancer Center’s Biospecimen Repository Core Facility staff for helping obtain human specimens and performing histological work. The authors also acknowledge support from the KU Cancer Center’s Cancer Center Support Grant (P30 CA168524)
7 FUNDING SOURCES
This study was supported, in part, by grants from the Department of Defense (W81XWH-12-1-0139 to JLW), the National Cancer Institute (K01CA120051 to JLW), start-up funds from the University of Kansas Medical Center (KUMC) (to JLW), the KUMC Biomedical Research Training Program (BRTP to AL), the National Cancer Institute (1F30CA203160-01 to AL), the American Medical Association (AMA) Foundation (to AL) and the University of Kansas Cancer Center (CCSG grants P30 CA168524-0 to JLW).
ABBREVIATIONS
- AI
Aromatase inhibitor
- CAV-1
Calveolin-1
- ChIP
Chromatin Immunoprecipitation
- Dox
doxycycline
- ERα
Estrogen receptor alpha
- ER+
estrogen receptor-positive
- IFITM1
interferon induced transmembrane protein1
- IFNα/β
interferon alpha and beta
- IFNAR
type 1 interferon receptor
- ISGs
interferon stimulated genes
- ISRE
interferon stimulated response element
- JAK1/2
Janus kinase 1 and 2
- MIND
mouse mammary intraductal model
- mTOR
Mammalian target of rapamycin
- p21
Cyclin dependent kinas inhibitor 1
- PARP
poly ADP ribose polymerase
- PI
propidium iodide
- PI3K
Phosphoinositide 3-kinase
- Pim-1
murine leukemia virus
- Rux
ruxolitinib (Jakafi™)
- STAT1/2
signal transducer and activator of transcription 1 and 2
Footnotes
The authors declare that they have no conflicts of interest to disclose.
8 AUTHOR CONTRIBUTIONS
AL, FB and JLW designed the research. AL performed the in vitro experiments. AL, JO, EG, and KV performed and analyzed the in vivo experiments. AL, JM, WJ and OT collected, stained, scored and analyzed the human tissue microarray. AL wrote the manuscript. AL, EG and JLW edited the manuscript. All authors have read, revised and approved the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nature reviews Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nature reviews Cancer. 2002;2:101–112. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- 3.Barone I, Cui Y, Herynk MH, Corona-Rodriguez A, Giordano C, Selever J, Beyer A, Ando S, Fuqua SA. Expression of the K303R estrogen receptor-alpha breast cancer mutation induces resistance to an aromatase inhibitor via addiction to the PI3K/Akt kinase pathway. Cancer research. 2009;69:4724–4732. doi: 10.1158/0008-5472.CAN-08-4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behbod F, Kittrell FS, LaMarca H, Edwards D, Kerbawy S, Heestand JC, Young E, Mukhopadhyay P, Yeh HW, Allred DC, Hu M, Polyak K, Rosen JM, Medina D. An intraductal human-in-mouse transplantation model mimics the subtypes of ductal carcinoma in situ. Breast cancer research: BCR. 2009;11:R66. doi: 10.1186/bcr2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bekisz J, Schmeisser H, Hernandez J, Goldman ND, Zoon KC. Human interferons alpha, beta and omega. Growth factors (Chur, Switzerland) 2004;22:243–251. doi: 10.1080/08977190400000833. [DOI] [PubMed] [Google Scholar]
- 6.Brueggemeier RW. Aromatase, aromatase inhibitors, and breast cancer. American journal of therapeutics. 2001;8:333–344. doi: 10.1097/00045391-200109000-00007. [DOI] [PubMed] [Google Scholar]
- 7.Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, Campisi J, Yaswen P, Cooper PK, Kaufman PD. A versatile viral system for expression and depletion of proteins in mammalian cells. PloS one. 2009;4:e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Molecular cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chattopadhyay I, Phukan R, Singh A, Vasudevan M, Purkayastha J, Hewitt S, Kataki A, Mahanta J, Kapur S, Saxena S. Molecular profiling to identify molecular mechanism in esophageal cancer with familial clustering. Oncology reports. 2009;21:1135–1146. doi: 10.3892/or_00000333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X, Zhao M, Hao M, Sun X, Wang J, Mao Y, Zu L, Liu J, Shen Y, Wang J, Shen K. Dual Inhibition of PI3K and mTOR Mitigates Compensatory AKT Activation and Improves Tamoxifen Response in Breast Cancer. Molecular cancer research: MCR. 2013;11:1269–1278. doi: 10.1158/1541-7786.MCR-13-0212. [DOI] [PubMed] [Google Scholar]
- 11.Cheon H, Borden EC, Stark GR. Interferons and their stimulated genes in the tumor microenvironment. Seminars in oncology. 2014;41:156–173. doi: 10.1053/j.seminoncol.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheon H, Holvey-Bates EG, Schoggins JW, Forster S, Hertzog P, Imanaka N, Rice CM, Jackson MW, Junk DJ, Stark GR. IFNbeta-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. The EMBO journal. 2013;32:2751–2763. doi: 10.1038/emboj.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Child ES, Mann DJ. The intricacies of p21 phosphorylation: protein/protein interactions, subcellular localization and stability. Cell cycle (Georgetown, Tex) 2006;5:1313–1319. doi: 10.4161/cc.5.12.2863. [DOI] [PubMed] [Google Scholar]
- 14.Choi HJ, Lui A, Ogony J, Jan R, Sims PJ, Lewis-Wambi J. Targeting interferon response genes sensitizes aromatase inhibitor resistant breast cancer cells to estrogen-induced cell death. Breast cancer research: BCR. 2015;17:6. doi: 10.1186/s13058-014-0506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chumsri S, Howes T, Bao T, Sabnis G, Brodie A. Aromatase, aromatase inhibitors, and breast cancer. The Journal of steroid biochemistry and molecular biology. 2011;125:13–22. doi: 10.1016/j.jsbmb.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cidado J, Park BH. Targeting the PI3K/Akt/mTOR pathway for breast cancer therapy. Journal of mammary gland biology and neoplasia. 2012;17:205–216. doi: 10.1007/s10911-012-9264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke R, Liu MC, Bouker KB, Gu Z, Lee RY, Zhu Y, Skaar TC, Gomez B, O’Brien K, Wang Y, Hilakivi-Clarke LA. Antiestrogen resistance in breast cancer and the role of estrogen receptor signaling. Oncogene. 2003;22:7316–7339. doi: 10.1038/sj.onc.1206937. [DOI] [PubMed] [Google Scholar]
- 18.Cmielova J, Rezacova M. p21Cip1/Waf1 protein and its function based on a subcellular localization [corrected] Journal of cellular biochemistry. 2011;112:3502–3506. doi: 10.1002/jcb.23296. [DOI] [PubMed] [Google Scholar]
- 19.Dabydeen SA, Kang K, Diaz-Cruz ES, Alamri A, Axelrod ML, Bouker KB, Al-Kharboosh R, Clarke R, Hennighausen L, Furth PA. Comparison of tamoxifen and letrozole response in mammary preneoplasia of ER and aromatase overexpressing mice defines an immune-associated gene signature linked to tamoxifen resistance. Carcinogenesis. 2015;36:122–132. doi: 10.1093/carcin/bgu237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Renty C, DePamphilis ML, Ullah Z. Cytoplasmic localization of p21 protects trophoblast giant cells from DNA damage induced apoptosis. PloS one. 2014;9:e97434. doi: 10.1371/journal.pone.0097434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deraz EM, Kudo Y, Yoshida M, Obayashi M, Tsunematsu T, Tani H, Siriwardena SB, Keikhaee MR, Qi G, Iizuka S, Ogawa I, Campisi G, Lo Muzio L, Abiko Y, Kikuchi A, Takata T. MMP-10/stromelysin-2 promotes invasion of head and neck cancer. PloS one. 2011;6:e25438. doi: 10.1371/journal.pone.0025438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunbier AK, Ghazoui Z, Anderson H, Salter J, Nerurkar A, Osin P, A’Hern R, Miller WR, Smith IE, Dowsett M. Molecular profiling of aromatase inhibitor-treated postmenopausal breast tumors identifies immune-related correlates of resistance. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:2775–2786. doi: 10.1158/1078-0432.CCR-12-1000. [DOI] [PubMed] [Google Scholar]
- 23.Ferrantini M, Capone I, Belardelli F. Interferon-alpha and cancer: mechanisms of action and new perspectives of clinical use. Biochimie. 2007;89:884–893. doi: 10.1016/j.biochi.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Gartel AL, Tyner AL. Transcriptional regulation of the p21((WAF1/CIP1)) gene. Experimental cell research. 1999;246:280–289. doi: 10.1006/excr.1998.4319. [DOI] [PubMed] [Google Scholar]
- 25.Gyorffy B, Dietel M, Fekete T, Lage H. A snapshot of microarray-generated gene expression signatures associated with ovarian carcinoma. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society. 2008;18:1215–1233. doi: 10.1111/j.1525-1438.2007.01169.x. [DOI] [PubMed] [Google Scholar]
- 26.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast cancer research and treatment. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 27.Hagan CR, Regan TM, Dressing GE, Lange CA. ck2-dependent phosphorylation of progesterone receptors (PR) on Ser81 regulates PR-B isoform-specific target gene expression in breast cancer cells. Molecular and cellular biology. 2011;31:2439–2452. doi: 10.1128/MCB.01246-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He JD, Luo HL, Li J, Feng WT, Chen LB. Influences of the interferon induced transmembrane protein 1 on the proliferation, invasion, and metastasis of the colorectal cancer SW480 cell lines. Chinese medical journal. 2012;125:517–522. [PubMed] [Google Scholar]
- 29.Herzog S, Fink MA, Weitmann K, Friedel C, Hadlich S, Langner S, Kindermann K, Holm T, Bohm A, Eskilsson E, Miletic H, Hildner M, Fritsch M, Vogelgesang S, Havemann C, Ritter CA, Meyer zu Schwabedissen HE, Rauch B, Hoffmann W, Kroemer HK, Schroeder H, Bien-Moller S. Pim1 kinase is upregulated in glioblastoma multiforme and mediates tumor cell survival. Neuro-oncology. 2015;17:223–242. doi: 10.1093/neuonc/nou216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho CC, Huang PH, Huang HY, Chen YH, Yang PC, Hsu SM. Up-regulated caveolin-1 accentuates the metastasis capability of lung adenocarcinoma by inducing filopodia formation. Am J Pathol. 2002;161:1647–1656. doi: 10.1016/S0002-9440(10)64442-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong IK, Byun HJ, Lee J, Jin YJ, Wang SJ, Jeoung DI, Kim YM, Lee H. The tetraspanin CD81 protein increases melanoma cell motility by up-regulating metalloproteinase MT1-MMP expression through the pro-oncogenic Akt-dependent Sp1 activation signaling pathways. The Journal of biological chemistry. 2014;289:15691–15704. doi: 10.1074/jbc.M113.534206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang SY, Wolf DM, Yingling JM, Chang C, Jordan VC. An estrogen receptor positive MCF-7 clone that is resistant to antiestrogens and estradiol. Molecular and cellular endocrinology. 1992;90:77–86. doi: 10.1016/0303-7207(92)90104-e. [DOI] [PubMed] [Google Scholar]
- 33.Jung YS, Qian Y, Chen X. Examination of the expanding pathways for the regulation of p21 expression and activity. Cellular signalling. 2010;22:1003–1012. doi: 10.1016/j.cellsig.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khodarev N, Ahmad R, Rajabi H, Pitroda S, Kufe T, McClary C, Joshi MD, MacDermed D, Weichselbaum R, Kufe D. Cooperativity of the MUC1 oncoprotein and STAT1 pathway in poor prognosis human breast cancer. Oncogene. 2010;29:920–929. doi: 10.1038/onc.2009.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khodarev NN, Beckett M, Labay E, Darga T, Roizman B, Weichselbaum RR. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1714–1719. doi: 10.1073/pnas.0308102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JY, Kim H, Suk K, Lee WH. Activation of CD147 with cyclophilin a induces the expression of IFITM1 through ERK and PI3K in THP-1 cells. Mediators of inflammation. 2010;2010:821940. doi: 10.1155/2010/821940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kittrell F, Valdez K, Elsarraj H, Hong Y, Medina D, Behbod F. Mouse Mammary Intraductal (MIND) Method for Transplantation of Patient Derived Primary DCIS Cells and Cell Lines. Bio-protocol. 2016;6 doi: 10.21769/bioprotoc.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le X, Antony R, Razavi P, Treacy DJ, Luo F, Ghandi M, Castel P, Scaltriti M, Baselga J, Garraway LA. Systematic Functional Characterization of Resistance to PI3K Inhibition in Breast Cancer. Cancer discovery. 2016;6:1134–1147. doi: 10.1158/2159-8290.CD-16-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J, Goh SH, Song N, Hwang JA, Nam S, Choi IJ, Shin A, Kim IH, Ju MH, Jeong JS, Lee YS. Overexpression of IFITM1 Has Clinicopathologic Effects on Gastric Cancer and Is Regulated by an Epigenetic Mechanism. The American Journal of Pathology. 2012;181:43–52. doi: 10.1016/j.ajpath.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 40.Leisibach P, Schneiter D, Soltermann A, Yamada Y, Weder W, Jungraithmayr W. Prognostic value of immunohistochemical markers in malignant thymic epithelial tumors. Journal of thoracic disease. 2016;8:2580–2591. doi: 10.21037/jtd.2016.08.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis-Wambi JS, Kim HR, Wambi C, Patel R, Pyle JR, Klein-Szanto AJ, Jordan VC. Buthionine sulfoximine sensitizes antihormone-resistant human breast cancer cells to estrogen-induced apoptosis. Breast cancer research: BCR. 2008;10:R104. doi: 10.1186/bcr2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis JS, Meeke K, Osipo C, Ross EA, Kidawi N, Li T, Bell E, Chandel NS, Jordan VC. Intrinsic mechanism of estradiol-induced apoptosis in breast cancer cells resistant to estrogen deprivation. Journal of the National Cancer Institute. 2005;97:1746–1759. doi: 10.1093/jnci/dji400. [DOI] [PubMed] [Google Scholar]
- 43.Lilly M, Sandholm J, Cooper JJ, Koskinen PJ, Kraft A. The PIM-1 serine kinase prolongs survival and inhibits apoptosis-related mitochondrial dysfunction in part through a bcl-2-dependent pathway. Oncogene. 1999;18:4022–4031. doi: 10.1038/sj.onc.1202741. [DOI] [PubMed] [Google Scholar]
- 44.Liu P, Rudick M, Anderson RG. Multiple functions of caveolin-1. The Journal of biological chemistry. 2002;277:41295–41298. doi: 10.1074/jbc.R200020200. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, Zhang X, Liu J, Hou G, Zhang S, Zhang J. Everolimus in combination with letrozole inhibit human breast cancer MCF-7/Aro stem cells via PI3K/mTOR pathway: an experimental study. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:1275–1286. doi: 10.1007/s13277-013-1170-8. [DOI] [PubMed] [Google Scholar]
- 46.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA: a cancer journal for clinicians. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 47.Miller WR. Aromatase inhibitors: mechanism of action and role in the treatment of breast cancer. Seminars in oncology. 2003;30:3–11. doi: 10.1016/s0093-7754(03)00302-6. [DOI] [PubMed] [Google Scholar]
- 48.Miller WR, Larionov AA. Understanding the mechanisms of aromatase inhibitor resistance. Breast cancer research: BCR. 2012;14:201. doi: 10.1186/bcr2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narayana SK, Helbig KJ, McCartney EM, Eyre NS, Bull RA, Eltahla A, Lloyd AR, Beard MR. The Interferon-induced Transmembrane Proteins, IFITM1, IFITM2, and IFITM3 Inhibit Hepatitis C Virus Entry. The Journal of biological chemistry. 2015;290:25946–25959. doi: 10.1074/jbc.M115.657346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Narlik-Grassow M, Blanco-Aparicio C, Cecilia Y, Perez M, Munoz-Galvan S, Canamero M, Renner O, Carnero A. Conditional transgenic expression of PIM1 kinase in prostate induces inflammation-dependent neoplasia. PloS one. 2013;8:e60277. doi: 10.1371/journal.pone.0060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishio H, Walsh MJ. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11257–11262. doi: 10.1073/pnas.0401343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Novita Sari I, Yang YG, Hanh Phi LT, Kim H, Jun Baek M, Jeong D, Young Kwon H. Interferon-induced transmembrane protein 1 (IFITM1) is required for the progression of colorectal cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogony J, Choi HJ, Lui A, Cristofanilli M, Lewis-Wambi J. Interferon-induced transmembrane protein 1 (IFITM1) overexpression enhances the aggressive phenotype of SUM149 inflammatory breast cancer cells in a signal transducer and activator of transcription 2 (STAT2)-dependent manner. Breast cancer research: BCR. 2016;18:25. doi: 10.1186/s13058-016-0683-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Annual review of medicine. 2011;62:233–247. doi: 10.1146/annurev-med-070909-182917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pan Z, Chen S, Pan X, Wang Z, Han H, Zheng W, Wang X, Li F, Qu S, Shao R. Differential gene expression identified in Uigur women cervical squamous cell carcinoma by suppression subtractive hybridization. Neoplasma. 2010;57:123–128. doi: 10.4149/neo_2010_02_123. [DOI] [PubMed] [Google Scholar]
- 56.Parton RG, Simons K. The multiple faces of caveolae. Nature reviews Molecular cell biology. 2007;8:185–194. doi: 10.1038/nrm2122. [DOI] [PubMed] [Google Scholar]
- 57.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunological reviews. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 58.Pink JJ, Jiang SY, Fritsch M, Jordan VC. An estrogen-independent MCF-7 breast cancer cell line which contains a novel 80-kilodalton estrogen receptor-related protein. Cancer research. 1995;55:2583–2590. [PubMed] [Google Scholar]
- 59.Pircher TJ, Zhao S, Geiger JN, Joneja B, Wojchowski DM. Pim-1 kinase protects hematopoietic FDC cells from genotoxin-induced death. Oncogene. 2000;19:3684–3692. doi: 10.1038/sj.onc.1203684. [DOI] [PubMed] [Google Scholar]
- 60.Pitroda SP, Wakim BT, Sood RF, Beveridge MG, Beckett MA, MacDermed DM, Weichselbaum RR, Khodarev NN. STAT1-dependent expression of energy metabolic pathways links tumour growth and radioresistance to the Warburg effect. BMC medicine. 2009;7:68. doi: 10.1186/1741-7015-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Popson SA, Ziegler ME, Chen X, Holderfield MT, Shaaban CI, Fong AH, Welch-Reardon KM, Papkoff J, Hughes CC. Interferon-induced transmembrane protein 1 regulates endothelial lumen formation during angiogenesis. Arteriosclerosis, thrombosis, and vascular biology. 2014;34:1011–1019. doi: 10.1161/ATVBAHA.114.303352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ribas R, Ghazoui Z, Gao Q, Pancholi S, Rani A, Dunbier A, Dowsett M, Martin LA. Identification of chemokine receptors as potential modulators of endocrine resistance in oestrogen receptor-positive breast cancers. Breast cancer research: BCR. 2014;16:447. doi: 10.1186/s13058-014-0447-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Current opinion in virology. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seyfried NT, Huysentruyt LC, Atwood JA, 3rd, Xia Q, Seyfried TN, Orlando R. Up-regulation of NG2 proteoglycan and interferon-induced transmembrane proteins 1 and 3 in mouse astrocytoma: a membrane proteomics approach. Cancer letters. 2008;263:243–252. doi: 10.1016/j.canlet.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sflomos G, Dormoy V, Metsalu T, Jeitziner R, Battista L, Scabia V, Raffoul W, Delaloye JF, Treboux A, Fiche M, Vilo J, Ayyanan A, Brisken C. A Preclinical Model for ERalpha-Positive Breast Cancer Points to the Epithelial Microenvironment as Determinant of Luminal Phenotype and Hormone Response. Cancer cell. 2016;29:407–422. doi: 10.1016/j.ccell.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 66.Smith S, Weston S, Kellam P, Marsh M. IFITM proteins-cellular inhibitors of viral entry. Current opinion in virology. 2014;4:71–77. doi: 10.1016/j.coviro.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suba Z. The pitfall of the transient, inconsistent anticancer capacity of antiestrogens and the mechanism of apparent antiestrogen resistance. Drug design, development and therapy. 2015;9:4341–4353. doi: 10.2147/DDDT.S89536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tymoszuk P, Charoentong P, Hackl H, Spilka R, Muller-Holzner E, Trajanoski Z, Obrist P, Revillion F, Peyrat JP, Fiegl H, Doppler W. High STAT1 mRNA levels but not its tyrosine phosphorylation are associated with macrophage infiltration and bad prognosis in breast cancer. BMC cancer. 2014;14:257. doi: 10.1186/1471-2407-14-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Valdez KE, Fan F, Smith W, Allred DC, Medina D, Behbod F. Human primary ductal carcinoma in situ (DCIS) subtype-specific pathology is preserved in a mouse intraductal (MIND) xenograft model. The Journal of pathology. 2011;225:565–573. doi: 10.1002/path.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vladimer GI, Gorna MW, Superti-Furga G. IFITs: Emerging Roles as Key Anti-Viral Proteins. Frontiers in immunology. 2014;5:94. doi: 10.3389/fimmu.2014.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Z, Bhattacharya N, Mixter PF, Wei W, Sedivy J, Magnuson NS. Phosphorylation of the cell cycle inhibitor p21Cip1/WAF1 by Pim-1 kinase. Biochimica et biophysica acta. 2002;1593:45–55. doi: 10.1016/s0167-4889(02)00347-6. [DOI] [PubMed] [Google Scholar]
- 72.Weichselbaum RR, Ishwaran H, Yoon T, Nuyten DS, Baker SW, Khodarev N, Su AW, Shaikh AY, Roach P, Kreike B, Roizman B, Bergh J, Pawitan Y, van de Vijver MJ, Minn AJ. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weirauch U, Beckmann N, Thomas M, Grunweller A, Huber K, Bracher F, Hartmann RK, Aigner A. Functional role and therapeutic potential of the pim-1 kinase in colon carcinoma. Neoplasia (New York, NY) 2013;15:783–794. doi: 10.1593/neo.13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilkins C, Woodward J, Lau DT, Barnes A, Joyce M, McFarlane N, McKeating JA, Tyrrell DL, Gale M., Jr IFITM1 is a tight junction protein that inhibits hepatitis C virus entry. Hepatology (Baltimore, Md) 2013;57:461–469. doi: 10.1002/hep.26066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xia X, Ji T, Liu R, Weng Y, Fang Y, Wang Z, Xu H. Cytoplasmic p21 is responsible for paclitaxel resistance in ovarian cancer A2780 cells. European journal of gynaecological oncology. 2015;36:662–666. [PubMed] [Google Scholar]
- 76.Xu J, Xiong G, Cao Z, Huang H, Wang T, You L, Zhou L, Zheng L, Hu Y, Zhang T, Zhao Y. PIM-1 contributes to the malignancy of pancreatic cancer and displays diagnostic and prognostic value. Journal of experimental & clinical cancer research: CR. 2016;35:133. doi: 10.1186/s13046-016-0406-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu F, Xie D, Ng SS, Lum CT, Cai MY, Cheung WK, Kung HF, Lin G, Wang X, Lin MC. IFITM1 promotes the metastasis of human colorectal cancer via CAV-1. Cancer letters. 2015;368:135–143. doi: 10.1016/j.canlet.2015.07.034. [DOI] [PubMed] [Google Scholar]
- 78.Yu Q, Katlinskaya YV, Carbone CJ, Zhao B, Katlinski KV, Zheng H, Guha M, Li N, Chen Q, Yang T, Lengner CJ, Greenberg RA, Johnson FB, Fuchs SY. DNA-damage-induced type I interferon promotes senescence and inhibits stem cell function. Cell reports. 2015;11:785–797. doi: 10.1016/j.celrep.2015.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zardavas D, Fumagalli D, Loi S. Phosphatidylinositol 3-kinase/AKT/mammalian target of rapamycin pathway inhibition: a breakthrough in the management of luminal (ER+/HER2−) breast cancers? Current opinion in oncology. 2012;24:623–634. doi: 10.1097/CCO.0b013e328358a2b5. [DOI] [PubMed] [Google Scholar]
- 80.Zemskova M, Sahakian E, Bashkirova S, Lilly M. The PIM1 kinase is a critical component of a survival pathway activated by docetaxel and promotes survival of docetaxel-treated prostate cancer cells. The Journal of biological chemistry. 2008;283:20635–20644. doi: 10.1074/jbc.M709479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.