Abstract
Glioblastoma (GBM) constitutes the most common and aggressive primary brain tumor. To better understand how GBM evolves we analyzed longitudinal genomic and transcriptomic data of 114 patients. The analysis reveals a highly branched evolutionary pattern in which 63% of patients experience expression-based subtype changes. The branching pattern together with estimates of evolutionary rates suggest that the relapse associated clone typically preexisted years before diagnosis. 15% of tumors present hypermutations at relapse in highly expressed genes with a clear mutational signature. We find that 11% of recurrent tumors harbor mutations in LTBP4, a protein binding to TGF-β. Silencing LTBP4 in GBM cells leads to TGF-β activity suppression and decreased proliferation. In IDH1-wild-type recurrent GBM, high LTBP4 expression is associated with worse prognosis, highlighting the TGF-β pathway as a potential therapeutic target in GBM.
INTRODUCTION
Glioblastoma (GBM) is the most common and most aggressive type of primary brain tumor in adults1. Therapeutic options are limited, consisting of surgery and treatment with radiotherapy plus an oral alkylating agent, temozolomide (TMZ). Despite TMZ’s benefits, the extension of patients’ survival averages ~2.5 months, and tumors invariably recur leading to fatal outcome2. Recent progress in large-scale sequencing techniques has revealed the genomic landscape of the untreated tumor3,4, yet very few studies have analyzed recurrent GBM, and patient cohorts are limited in size5–7.
The evolution of tumor cells under therapy can be viewed as a Darwinian process of clonal replacement8–10 in which treatment ablates vulnerable cells while positively selecting for resistant clones. Studies of spatially distinct tumor fragments indicate that treatment failure is frequently complicated by intratumor heterogeneity (ITH), a common phenomenon in low and high-grade glioma11–14. Mutations of TP53 gene were recently proposed to mark subclonal heterogeneity of GBM7, but a clear pattern of tumor evolution remains elusive. ITH and diversity in evolutionary trajectories preclude the identification of general evolutionary patterns in GBM, especially when only limited cohorts of patients are available.
To find genetic markers of progression and to elucidate the diverse evolutionary trajectories by which GBM can occur and recur, we performed whole-exome and transcriptome analysis of untreated and recurrent tumors from 114 GBM patients with corresponding matched normal tissue.
RESULTS
Longitudinal Mutational Landscape of GBM
To elucidate the mechanisms driving the evolution of high-grade glioma under therapy, we analyzed 293 whole-exomes and 141 transcriptomes from longitudinal tumor/matched normal samples in 114 GBM patients (Figure 1A). Recurrent GBM patients (89 diagnosed with primary GBM) were collected from Istituto Neurologico C. Besta (INCB, R001-R019), MD Anderson Cancer Center (R020-R029), The Cancer Genome Atlas (TCGA, R030-R042), University of California San Francisco (UCSF, R043-R052), Kyoto University (KU, R053-R055), and Samsung Medical Center (SMC, R056-R114). Whole-exome triplets of initial tumor sample, recurrent tumor sample, and normal genomic DNA were sequenced from 93 patients. Transcriptomes of initial and recurrent tumor were sequenced from 65 patients. All but 14 patients received standard treatment, including TMZ2. Greater than 200 fold mean target coverage was achieved in 84% of samples (246 out of 293). On average, 76% of coding bases within the exome were covered by at least 100 high-quality reads (Supplementary Table 1).
Figure 1. Mutation landscape of recurrent Glioblastoma.
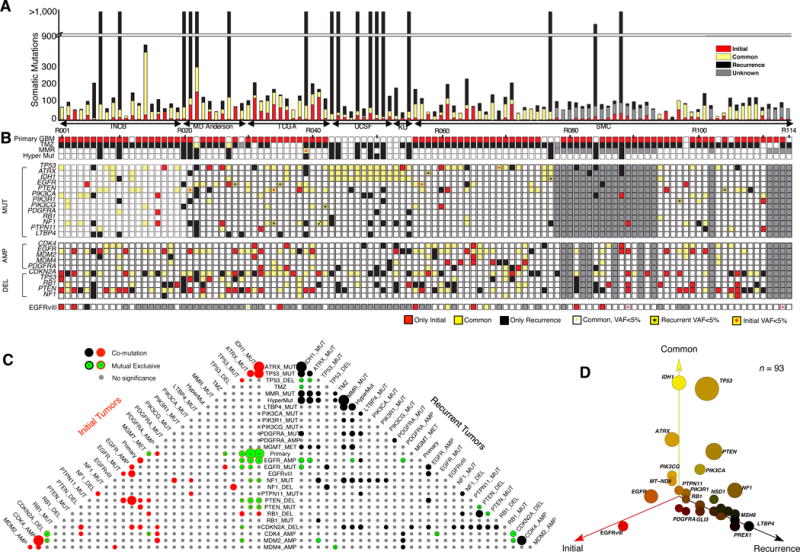
(A) Number of somatic mutations. 114 Patients from six sources (Instituto Neurologico C. Besta, MD Anderson Cancer Center, The Cancer Genome Atlas, University of California San Francisco, Kyoto University, and Samsung Medical Center). (B) Clinic and genetic profile of patients. TMZ indicates Temozolomide; MMR represents mismatch repair pathway (MSH6, MSH2, MSH4, MSH5, PMS1, PMS2, MLH1, MLH3 were considered). Hyper Mut represents hypermutation. MUT indicates somatic non-synonymous mutations with allele frequency >5% in at least one sample. AMP/DEL indicates copy number change with segmentation mean >0.5, computed either by SNP/CGH array data or by whole-exome sequencing data. TMZ represents Temozolomide. (C) Pyramids plot highlighting the correlation between different features. Hypergeometric test was performed for each pair of elements by considering Initial and recurrent tumors separately. The size of the circle indicates significance level of the correlation. Any associations with p-value < 0.1 were illustrated in this plot. (D) 3-D bubble plot illustrating the mutation frequency of somatic non-synonymous mutations in exclusively initial (red, left axis), exclusively recurrence (black, right axis), and in common (yellow, upper axis). 93 patients with exome-sequencing data in matched normal, initial tumor, and recurrent tumor were considered in this analysis.
To identify somatic single nucleotide variants (SNVs) as well as short insertions and deletions (INDELs), we utilized the variant-calling software SAVI215. We included as somatic variants only those with mutant allele frequency of 5% or more. From these variants we selected 40 mutations from the INCB cohort for validation. Sanger sequencing successfully validated 98% (39/40) of the mutational calls as well as changes in allele frequency between untreated and recurrent tumor (Supplementary Table 2 and Supplementary Data 1). Untreated tumor samples harbor an average of 60 somatic mutations. Recurrent tumor samples have 585 somatic mutations on average, but this figure is unrepresentative due to the presence of 17 patients (6 primary GBM and 11 secondary GBM) with hypermutated recurrent tumors (>500 mutated genes per tumor). The remaining non-hypermutated tumors have only 50 mutations on average. All hypermutated tumors originated within TMZ treated patients. 16 out of 17 hypermutated samples gained mutations in genes coding DNA mismatch repair proteins (MSH6, MSH2, MHS4, MSH5, PMS1, PMS2, MLH1, and MLH2) (Figure 1B).
We compared mutations found in the initial and recurrent samples for each of the 93 patients for whom whole-exome triplets were available. Appearance of the same mutation in both the initial and recurrent samples for a patient suggests that the mutation originated relatively early in that patient’s tumor development, while appearance only in one sample suggests that the mutation may have originated after the clonal lineages leading to the two samples diverged. We discovered that the mutations occurring in only one of a patient’s two GBM samples outnumber the common ones in more than half of all patients (57%, 53/93) (Figure 1A: single-sample mutations (red and black) versus common mutations (yellow)). We next assessed pairwise co-occurrence and mutual exclusivity of genomic/clinic features across all patients. In addition to previously reported association16–18 (Figure 1C), we observed a number of significant associations not previously reported for GBM that were exclusive to recurrence. These associations include co-occurrence of MGMT promoter methylation and hypermutation (p-value=4×10−3, Fisher’s exact test, only TMZ treated patients), co-deletion of RB1 and PTEN (p-value<10−4, Fisher’s exact test); and co-mutation of NF1 and TP53 (p-value=10−2, Fisher’s exact test).
Overall, our mutational analysis reveals both known and potentially novel driver gene mutations in GBM. We observed mutations in known drivers of GBM3 including TP53, PTEN, EGFR, PIK3CA, ATRX, IDH1, PIK3R1, and PDGFRA with similar frequency in both untreated and recurrent tumors (Figure 1D). We also identified hotspot mutations in unreported potential driver genes in GBM. In particular, we found seven patients with PTPN11 nonsynonymous mutations (SHP2 protein) in the first SH2 and PTP domains with a similar distribution to what has been found in juvenile myelomonocytic leukemia19. A few genes appear exclusively mutated and expressed in recurrent tumors (Figure 1D), including LTBP4 (10/93), DNA mismatch repair gene MutS Homolog 6, MSH6 (8/93), PRDM2 (10/93) and IGF1R (9/93) (for a complete list see Supplementary Table 2). Interestingly all eight cases with mutations in MSH6 occurred in hypermutated recurrences (p-value<10−4, Fisher’s exact test), and three of these cases include nonsense mutations in the gene, indicating that loss of function of MSH6 is related to genomic hypermutation in GBM. This finding is consistent with previous observations of the induction of a hypermutant genotype following treatment of glioma20–23. Recurrence-only mutated gene LTBP4 has been reported to be an activator of TGF-β signaling by promoting the assembly, secretion and targeting of sites where TGF-β1 is stored and/or activated24. Disruption of this gene causes abnormal lung development, cardiomyopathy, and colorectal cancer in mouse25.
To explore copy number variations (CNVs) of initial and recurrent GBM, recurrence-based analysis, GISTIC226 (Supplementary Figure 1 and Supplementary Table 3) and MutComFocal27 (Supplementary Figure 2 and Supplementary Table 4) were applied. We found copy number alterations in several well-known GBM drivers. EGFR amplification, which is frequently co-occurrent with EGFR SNVs and EGFRvIII, was observed in 42% of initial tumors (44/104) and 34% of recurrent tumors (35/102), whereas CDK4 amplification was detected in 19% of both initial and recurrent samples (20/104). Deletions in CDKN2A were the most frequent copy deletion in 47% (49/104) of initial samples and 52% of recurrent tumors (53/102). PTEN displayed a similar prevalence of loss in initial 37% (38/104) and recurrent 34% (35/102) samples.
We then defined a zygosity score (ZS) to identify regions of loss of heterozygosity (LOH) (Methods, Supplementary Figure 3, and Supplementary Table 5). The median ZS of a normal diploid chromosome is expected to be near to 0.25. To identify potential tumor suppressors associated to a two-hit mechanism, we analyzed genes with point mutations in regions with LOH in non-hypermutated recurrent tumors. This analysis recapitulated known tumor suppressors in GBM, including TP53 (14/78 samples), PTEN (9/78), and NF1 (3/78), and LOH encompassing inactivating mutations in other genes not previously reported in GBM including APC (R876*) (Supplementary Table 5).
Gene fusions detected from the RNA-seq analysis were summarized in Supplementary Table 6. We found gene fusions reported as recurrent alterations in GBM, such as FGFR3-TACC328 and EGFR fusions with multiple partners3. FGFR3-TACC3 fusions were highly expressed in both the untreated and matched recurrent tumors, thus confirming the clonal nature of these fusion events28,29. We also found rare fusions involving other Receptor Tyrosine Kinase (RTK)-coding genes such as PDGFRA, MET and ROS. Interestingly, two patients harbored in-frame gene fusions involving MGMT at relapse (see Supplementary Figure 4). Patient R114 harbors two highly expressed in-frame fusions NFYC-MGMT, BTRC-MGMT and patient R056 at recurrence presents the fusion SAR1A-MGMT. Of particular significance, these three fusion transcripts carry the same breakpoint in the MGMT gene, and the reconstructed open reading frame preserves the methyl-transferase and DNA binding domain (Supplementary Table 6). The fusion transcripts were further validated by RT-PCR (Methods and Supplementary Figure 5). MGMT is a gene that encodes for an O6-methylguanine–DNA methyltransferase and epigenetic silencing of this gene has been associated with longer overall survival in GBM patients under therapy30. Consistently, we observed that MGMT methylation at diagnosis predicts longer survival (p-value=0.018). We also observed a high correlation between MGMT methylation and expression both at initial and at relapse (p-value=6×10−3 in initial and 0.016 in relapse, Supplementary Figure 6). At recurrence, but not in the initial tumor, low expression of MGMT is significantly related to better prognosis (p-value=4×10−4).
Hypermutation Related to Temozolomide
As indicated in Figure 1A, 17% of TMZ treated GBM patients (17/100) relapsed with hypermutated tumors, yet there is no incidence of hypermutation in non-TMZ treated patients (0/14). The median survival of hypermutated IDH1-wild-type primary GBM patients is 24 months, a slight increase from other IDH1-wild-type primary GBM patients (18 months). The gain of mutations in the mismatch repair pathway as well as the accompanying hypermutations in glioma patients after treatment has been reported before20–23 but the pattern of the hypermutated genes and the mechanism causing the mutations remain unclear. To better explain patient mutational variation, we grouped all mutations into four types: those identified in recurrent samples without TMZ, those in untreated tumors, those in TMZ-treated but non-hypermutated cases, and mutations in TMZ-treated hypermutated cases. Hypermutated recurrent tumors are highly enriched with C>T (G>A) transitions (Figure 2A). To identify additional markers of hypermutation we extracted 10 bp of DNA sequence from the coding strand of hypermutated loci. The motif analysis31 shows that hypermutation occurs predominantly in the coding strand at the first cytosine of CpY elements (Figure 2B). By contrast, a pattern of mutations at CpR32 elements can be seen in all other tumor types tested. Although we found no significant association in ratios of silent/missense mutations in non-hypermutated tumors, recurrent hypermutated samples contained significantly greater numbers of silent mutations (Figure 2C). Moreover, hypermutation may be related to expression of genes involved in tumor recurrence: In hypermutated tumors, those genes containing hypermutated loci are more highly expressed than are mutated genes with no hypermutated loci and non-mutated genes (Figure 2D).
Figure 2. Temozolomide (TMZ) related Hypermutation (HM).
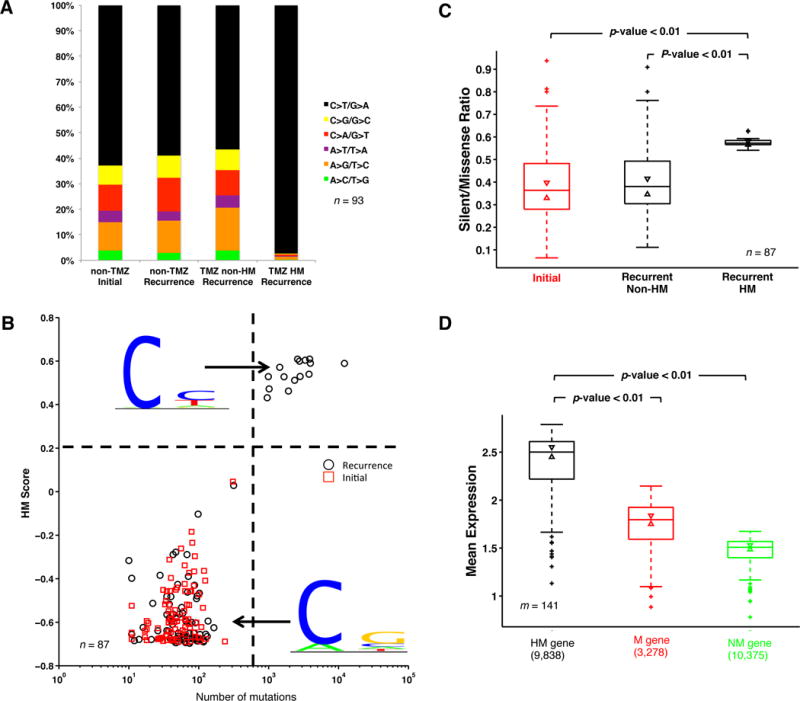
(A) Fraction of different types of nucleotide change. In this analysis, 93 patients with trios of normal, initial and recurrent DNA data were considered. (B) HM Score and mutation load. HM logo and non-HM logo were separately calculated based on all substitutions from HM and non-HM samples. Given this, HM score of each sample was defined based on its mutation pattern. If mutations in a sample follow the pattern of HM logo, the sample will have higher HM score. Patients with less than ten mutations in either initial or recurrent samples were not considered in the analysis of B and C. (C) Silent/missense ratio analysis. P-value was calculated by Ranksum test. (D) Expression comparison between three gene clusters: HM genes, mutated (M) genes, and non-mutated (NM) genes. Mean expression of three gene clusters in samples with expression data available (m=160) was calculated to generate the box plot. The bottom and top of the box indicate first and third quartiles, and the line inside is the median. Whiskers represent 1.5 IQR. P-values were calculated by Ranksum test.
Reconstruction of the main routes of GBM evolution
The number of mutations exclusive to untreated tumors, recurrent tumors, or those in common can be used to describe an evolutionary tree. We developed a method to perform statistics on the space of evolutionary trees33 by embedding each tree within a sphere (Figure 3A). Here, the upper corner represents the fraction of mutations that are common to both samples; the left corner represents the fraction exclusive to the untreated sample, and the right corner the fraction exclusive to recurrence. Unsupervised clustering of the different phylogenies identifies three clusters (Methods). The yellow group represents the limiting case where few mutations are lost from diagnosis, similar to the classical model of linear tumor evolution typified by previous treatment-naïve studies of colon cancer34. Treatment however can change linear patterns35. The abundance of points far from the right edge of the diagram suggests that in most patients, the dominant clones prior to treatment appear to be replaced by new clones that do not share many of the same mutations.
Figure 3. Mathematical model of tumor evolution.
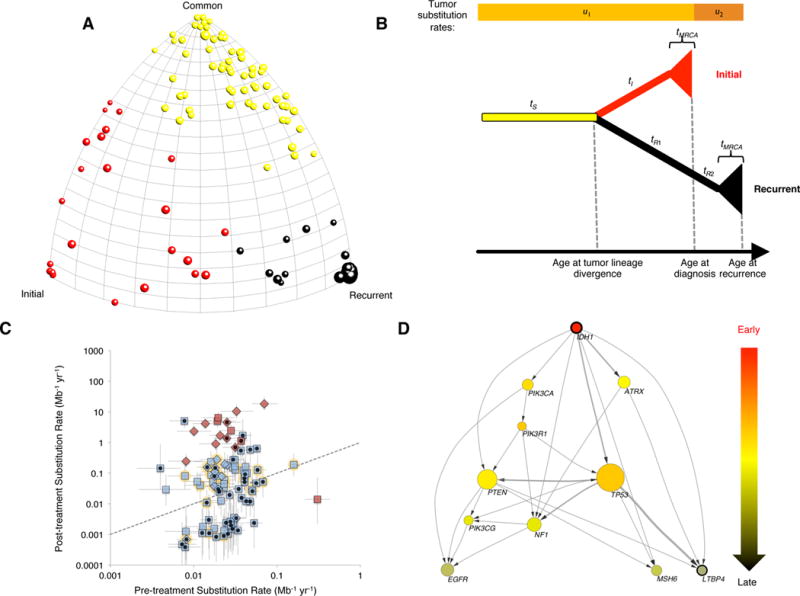
(A) Moduli space of GBM evolution trees. Each ball represents one patient, and different colors represent three clusters in moduli space. (B) Model of branching tumor evolution. This model assumes independent monophyletic origin for initial and recurrent tumors sharing an ancestral clonal lineage (duration tS), after which they branch off from one another (durations tI and tR1+tR2). After this clonal evolution, the lineage leading to each sample diversifies for a duration tMRCA, during which subclonal variants can accrue. Somatic variants accrue according to substitution rates u1 and u2 before and after treatment, respectively. (C) Relationship between estimated substitution rates before and after treatment, in substitutions per Mb-yr (median and interquartile range for each patient). Dashed line shows diagonal (pre- and post-substitution rates equal). Hypermutated tumors shown in red, non-hypermutated tumors in light blue. Primary GBM diagnoses shown as squares, secondary GBM diagnoses as diamonds. Black dot in center of symbol shows patients who fit the model well. Yellow halo shows patients with TP53 mutated in both the initial and recurrent samples. Patient R069 was not considered for evolutionary analysis as no valid mutations were detected in the initial sample. (D) Cross-sectional integration of longitudinal data by tumor evolutionary directed graph. Arrow represents time order of mutations. Wider arrows represent there are more independent patients containing the same order of mutation. The size of the node indicated the frequency of the mutations in our cohort.
If many mutations in the initial sample are lost at recurrence, this suggests that the clone dominant at recurrence originated (i.e., diverged from the clone dominant at diagnosis) relatively long before the initial sample was taken. Consistent with epidemiological observations and classical models of tumor evolution of Armitage-Doll10 and Nordling9, the number of mutations in the untreated tumor increases with the patient’s age at diagnosis (Supplementary Figure 7D, average of 0.6 protein changing mutations per year or 0.02 per Mb-year). Encouraged by this concordance, we developed a mathematical model of branching tumor evolution with independent monophyletic origin for diagnosis and relapse.
In our branching model (Figure 3B), if a mutation occurs along the lineage common to both the initial and recurrent samples, it will be clonal in both of these samples. If a mutation occurs along the lineage leading to the most recent common ancestor (MRCA) of a single sample, then it will be clonal in that sample and absent in the other. If a mutation occurs in a descendant of the MRCA of a sample, then it will be subclonal in that sample and absent in the other. Any other pattern — a mutation that appears subclonally in both samples, or one that appears clonally in one sample and subclonally in the other, would require either recurrent mutation or “back-mutation”. An alternate model is also possible, in which the recurrence stems from a lineage nested within the initial sample, perhaps selected by therapy. In this case, a single mutational event could produce a variant that is present subclonally at diagnosis and clonally at recurrence, but two events would be needed to explain loss of a clonal mutation (Supplementary Figure 7A versus 7B). We find that 59% of patients (54 / 92) have at least four clonal mutations at diagnosis that are lost in the recurrence, supporting the branching model as the typical scenario. The picture is nuanced, however, as 17 patients have at least four subclonal mutations at diagnosis that become clonal at recurrence, supporting the alternate model as a minority scenario (Supplementary Figure 7C).
By accounting for the likelihood of each mutational pattern within our branching model, we fit substitution rates before and after treatment, as well as the amount of time before diagnosis that the untreated and recurrence lineages diverged (see Methods). Using a collection of statistical criteria (see Methods), we found that 49% of patients analyzed (45/92) fit well to the model, without requiring an unrealistic frequency of recurrent mutation or “back-mutation”. The pre-treatment substitution rate was consistent among these 45 well-fitting patients (Figure 3C), having a median and interquartile range of 0.028 subs Mb-1 yr-1 and 0.018 – 0.041 subs Mb-1 yr-1. Statistics for all 92 patients were similar, with a median (IQR) of 0.024 (0.018–0.035) subs Mb-1 yr-1. No relationship was observed between the substitution rate and age of diagnosis (Supplementary Figure 8). Considerably more variation was observed in post-treatment substitution rates, with 15 of the 92 patients exhibiting significantly higher mutation rates after treatment (Supplementary Figure 9). All but one of these patients showed hypermutation, with over 500 variants found in the recurrent sample.
Estimates of divergence time suggest that the recurrent clone diverged from the untreated clone many years before disease was detected (Supplementary Figure 7E). The median among the 45 well-fitting patients had a divergence time of 12.6 years (range 2.3–50.5, IQR 7.2–22.6). Since the remaining 47 non-fitting patients may fail the model’s assumption that untreated and recurrent tumors be evolutionarily distinct (i.e., monophyletic), we caution that divergence time for these patients be interpreted only as a heuristic measure of genetic difference between the two tumor samples. In general, uncertainty in the divergence time was large, with the median well-fitting patient showing a 95% CI 24 years wide. Still, even the bottom of the 95% CI exceeded three years for a majority of patients.
To reveal the potential evolutionary trajectories of GBM under therapy a tumor evolutionary directed graph (TEDG)36 was constructed for the 93 triplet samples. As this analysis uses as input the fraction of cells harboring a particular mutation, we estimated the purity of the tumor using ABSOLUTE37 (Supplementary Table 7) and PyClone38 (Supplementary Table 8). The resulting TEDG indicates that mutations in IDH1, PIK3CA and ATRX are early events, mutations in TP53, NF1, and PTEN occur later, and mutations in MSH6 and LTBP4 are relapse-specific events (Figure 3D). A more complex set of possible evolutionary trajectories appears when copy number information is included in the analysis (Supplementary Figure 10).
Clonal Replacement Events are Frequent in GBM
To discover the pattern of alterations in recurrent GBM compared with untreated tumors, we performed in-depth investigations into any gains or losses of genetic alterations. The epidermal growth factor receptor (EGFR) gene is known to be frequently amplified, mutated, and rearranged in untreated gliomas39. To uncover the role of EGFR alterations in GBM evolution, we applied PRADA to detect EGFR structure variance from RNA sequencing data40. By calculating junction reads we have found at least one junction read of EGFRvIII in 18% (12/67) of initial tumors and 11% (8/76) of recurrent tumors (Supplementary Table 9). Interestingly, nine patients lost EGFRvIII and one patient gained EGFRvIII at relapse (transcribed allelic fractions>5%), indicating, first, that EGFRvIII is a late event originated after the clonal lineages leading to the two samples diverged and, second, that EGFRvIII is more common in initial tumors and lost during treatment (Figure 1D). An example is patient R005 whose untreated tumor harbors EGFR amplification and the S645C mutation. The EGFR S645C mutation was lost in the recurrent tumor and replaced by EGFRvIII (Supplementary Figure 11).
A switch between differently mutated versions of the same gene also occurs in platelet-derived growth factor receptor alpha polypeptide (PDGFRA), another RTK-coding gene frequently activated in GBM (Figure 4B, and Supplementary Figure 11). Mutation E229K, which is relatively common in cross-sectional mutation databases (e.g. TCGA41), appears to be a relatively late event, as it is exclusive to recurrence and replaces the initial mutation P443L (Figure 4B). Mutational replacement also occurs in the tumor suppressor TP53 (G105R to R337C in Patient R038, Figure 4C) in EGFR (A1201T to G598V in Patient R065, Figure 4D). In all, we found that 11% (10 out of 93) of recurrent GBM patients have clonal replacements within key drivers (Supplementary Figure 12). These clonal switching events within the same gene occur preferentially in genes known to play a role in GBM (Figure 4A, p-value<10−4). The strong association between switching alterations and key driver genes (EGFR, TP53, PDGFRA) suggests (1) some of these genes contribute to a late expansion both with treated and untreated tumors and (2) converging evolution is associated to these genes.
Figure 4. Clonal replacement in key driver genes.
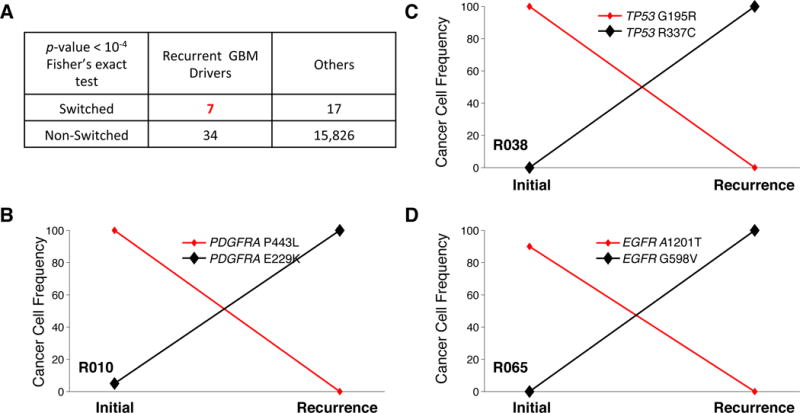
(A) Mutations of seven key GBM drivers (EGFR, TP53, PDGFRA, PTEN, ATRX, NF1, and RB1) were replaced by different mutations in the same genes. (B–C) Mutational replacement in three different patients. Cancer cell frequency was estimated by Pyclone.
Expression Analysis and Subtype Switching
Based on its pattern of gene expression, GBM is commonly divided into four subtypes, which display different responses to treatment4. To study evolution of gene expression in GBM, we followed the ssGSEA method4 to subtype each tumor sample (Figure 5A). As expected, we found that IDH1 mutated patients are mostly classified as proneural gliomas42; EGFR alterations are associated to classical subtype; and NF1 alterations to mesenchymal subtype (Figure 5B). We observed that all five hypermutated primary GBM cases switched their subtypes (two to mesenchymal, one to neural and two to proneural). Strikingly, we found that two-thirds of primary GBM cases (39/58) switch transcriptional subtype at relapse, while secondary GBM cases are more stable (2/7 switched) (Figure 5A). Interestingly, mesenchymal subtype is the most stable primary GBM, switching in 55% (12 of 22 primary GBM) of cases at recurrence; and mesenchymal subtype at recurrence is associated with worse overall survival (p-value=3×10−3, Supplementary Figure 13). As EGFRvIII is associated to the classical subtype (Figure 5B), loss of this alteration in the recurrent tumor is consistently associated to the transition from classical to other expression subtypes (Figure 5A and 5C, p-value=8×10−3, Fisher’s exact test).
Figure 5. Expression-based Subtyping of Recurrent GBM. (A) Expression based GBM subtyping.
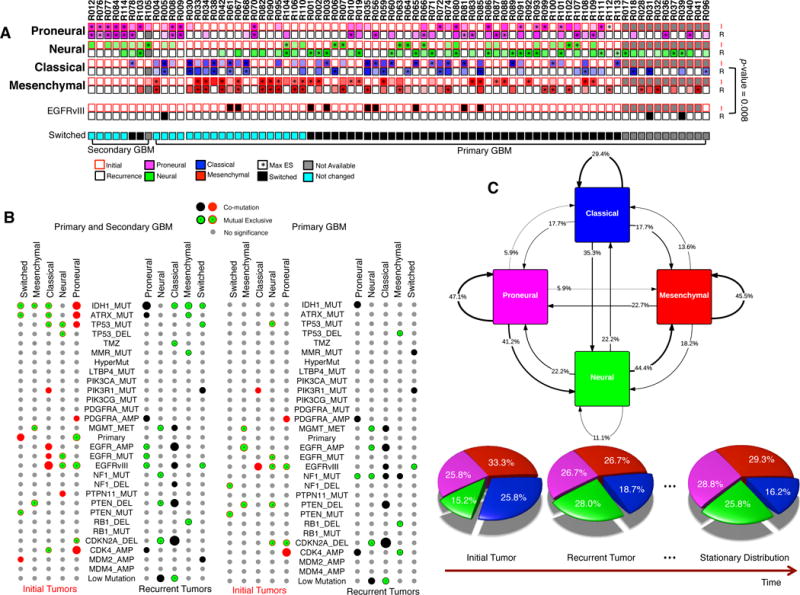
ssGSEA was performed to cluster each sample into four subtypes (proneural, neural, classical, and mesenchymal). “*” indicates subtypes with maximal enrichment score (ES). If the optimal subtype in initial and that in recurrent tumor is different, a patient was labeled as switched. P-value was calculated by Fisher’s exact test. (B) Association between expression-based subtype switching and genetic/clinic features. The same analysis as in Figure 1C had been performed. (C) The stochastic matrix of GBM subtypes. The large cohort of longitudinal GBM samples allows the construction of probability transition matrix between four subtypes. The arrows indicate the frequency of a patient to stay a subtype or to be switched from one subtype to another. A stationary distribution was calculated based on this stochastic matrix, indicating the proportion of these four subtypes after treatment.
LTBP4 promotes tumor growth and reduces survival
We found that the gene Latent transforming growth factor beta binding protein 4 (LTBP4) harbors significantly more mutations in recurrent than untreated GBM (Figure 1D, Supplementary Figure 14). The LTBP4 gene codes for a protein that belongs to the LTBP family, which is implicated in the regulation of the TGF-β pathway, typically acting as activator of TGF-β signaling24. Interestingly, activation of TGF-β is known to drive aggressiveness of malignant glioma43–46, and we found that high expression of LTBP4 in recurrent tumors is associated with worse prognosis in IDH1-wild-type primary GBM patients (p-value=7×10−3, Figure 6B). Furthermore, mutations of LTBP4 are correlated with higher expression of this gene (p-value<0.05, Figure 6A). Further strengthening the case that LTBP4 expression could drive tumor growth via TGF-β activation, elevated expression of LTBP4 in GBM is associated with elevated expression of genes implicated in the TGF-β pathway (Figure 6C, Gene Set Enrichment Analysis FDR<0.05).
Figure 6.
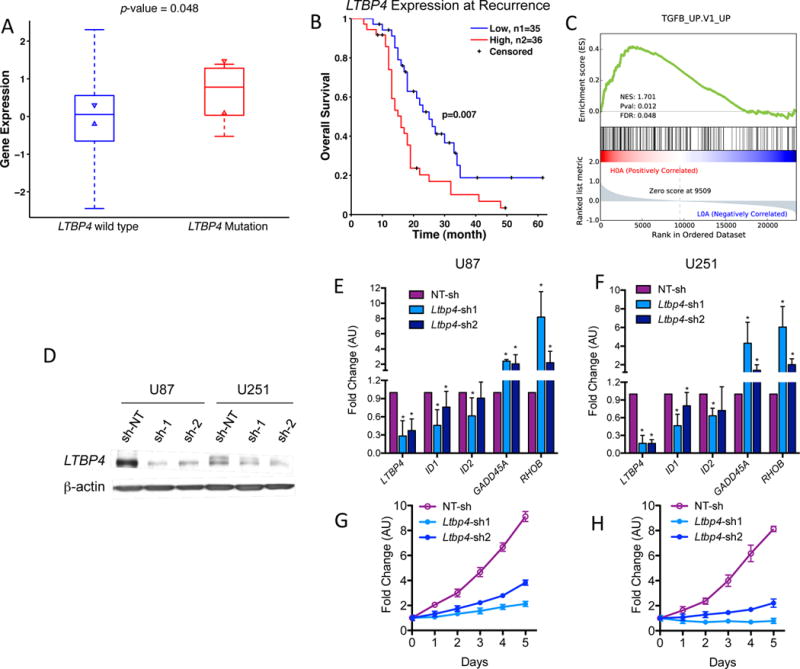
LTBP4 and TGF-β signaling pathway in recurrent GBM. (A) LTBP4 mutation was related to its high expression. P-value was calculated by Ranksum test. (B) Survival analysis of LTBP4 expression in IDH1-wild-type primary GBM patients. High indicates z-score of LTBP4>0, while low are LTBP4<0. P-value was calculated by log rank test. Only IDH1-wild-type primary GBM patients were considered in this analysis. (C) Gene set enrichment analysis. Recurrent tumor samples from IDH1-wild-type primary GBM were grouped according to LTBP4 expression. Samples with high LTBP4 expression (z score>0) were enriched with TGF-β activity. (D) Western blot of U87 and U251 glioma cells transduced with three independent sh-RNA, two against LTBP4 (sh1 and sh2), and one non-target-shRNA (sh-NT) as control. β-actin was used as loading control. (E–F) qRT-PCR of TGFβ target genes in U87 (E) and U251 (F); n = 9 (three biological replicates performed in triplicates) ± SD. Asterisk indicate statistical significance. (G–H) Growth curve of a representative experiment using U87 (G) and U251 (H) glioma cells treated as in D (means of six experimental replicates) ± SD.
To experimentally validate the functional link between LTBP4 and TGF-β, we used lentiviruses carrying two independent LTBP4 shRNA cassettes to silence the LTBP4 gene in the human glioma cell lines U87 and U251 (Figures 6D). LTBP4 silencing in both cell lines resulted in reduced expression of the ID genes ID1 and ID2, which are positively regulated by TGF-β in glioma. Conversely, LTBP4 silencing also led to the up-regulation of RhoB and GADD45a, two genes repressed by TGF-β in glioma (Figures 6E and 6F)43. Consistent with the pro-tumorigenic role of TGF-β in GBM, LTBP4 silencing markedly impaired proliferation of U87 and U251 glioma cells (Figures 6G and 6H).
DISCUSSION
Using longitudinal genomic and transcriptomic analysis of 114 GBM patients, we have detailed the major routes of GBM evolution under therapy. GBM evolution is highly branched, and specific alterations and evolutionary patterns are associated with treatment. Our first observation from this analysis is that, despite 45% of mutations (in non-hypermutated tumors) being shared between diagnosis and relapse samples, the dominant clone at diagnosis is generally not a lineal ancestor of the dominant clone at relapse. Instead, these two clones diverged from a common ancestor more than a decade before diagnosis in most patients (Supplementary Figure 7E).
Since 11% of patients (10/93) exhibit replacement of one mutated version of a gene (at diagnosis) with another, differently mutated version of the same gene (at relapse), it is conceivable that genes associated with undergoing clonal completion are late driver events. In fact, this mutational switching phenomenon is enriched ~200-fold in genes known to be implicated in GBM, including EGFR, TP53, and PDGFRA (Figure 4A, Supplementary Figure 13). This scenario of convergent evolution suggests that the common ancestor of diagnosis and relapse clones had fewer driver alterations and therefore a less aggressive phenotype. The accumulation of alterations in GBM cells therefore seems to occur over a decade(s)-long growing phase that leads to a highly diverse population, each clone experiencing a parallel series of expansions.
Related to mutational switching, we also find that two-thirds of primary GBM patients exhibit different transcriptional subtypes at diagnosis and relapse. Our observation of subtype switching, considered together with recent findings that different parts of the same tumor can exhibit different GBM subtypes12,47, also calls into question the significance of the expression-based classification as a prognostic marker prior to relapse.
Evolutionary dynamics generally appear similar before and after treatment: our mathematical model estimates typical substitution rates of ~0.03 substitutions per Mb per year during both periods, except in the 16% of cases that recur with hypermutated tumors. Hypermutated tumors, which are highly enriched for mutations at CpC dinucleotides21, harbor mutations in mismatch repair (MMR) genes, most commonly in MSH6, and can exhibit 100-fold higher substitution rates (~3 substitutions per Mb per year). We found that hypermutation preferentially targets highly expressed genes, suggesting that the mutagenic mechanisms related to TMZ treatment and subsequent MMR alteration act more efficiently in highly expressed regions of open chromatin.
Finally, and of particular relevance to discovery of novel GBM treatment, we uncovered unique alterations associated with relapsed GBM. In addition to previously reported mutations in MMR genes in 15% of patients (14/93), we found mutations in the LTBP4 gene in 11% of relapsed tumors (10/93). LTBP4 encodes a protein that binds to transforming growth factor beta (TGF-β). The TGF-β signaling pathway has been associated in a variety of biological contexts including proliferation, epithelial to mesenchymal transition21,48, and apoptosis. We have provided both clinical and in vitro evidence that LTBP4 activates this signaling pathway to drive tumor growth: Higher expression of LTBP4 in IDH1 wild-type primary GBM associated to poorer survival (Figure 6B, p-value=7×10−3), and silencing LTBP4 in two different cell lines decreases both proliferation and activity of TGF-β target genes. These results are consistent with recent animal studies showing that TGF-β inhibitors reduce viability and invasion of gliomas49 and advance the case for these molecules as potential anti-tumor therapeutics.
In conclusion, our study sketches the main routes of GBM evolution under therapy, identifying a highly branched process with specific alterations and evolutionary patterns associated to treated tumors.
METHODS
Patients and samples
Recurrent GBM patients were collected from Besta Brain Tumor Biobank (INCB, R001-R019), MD Anderson Cancer Center (MD Anderson, R020-R0297), The Cancer Genome Atlas (TCGA, R030-R0427), University of California San Francisco (UCSF, R043-R05213), Kyoto University (KU, R053-R05514), and Samsung Medical Center (SMC, R056-R0936, R094-R114).
The specimens in cohort INCB originate from the Besta Brain Tumor Biobank, which is partly funded by the Italian Minister of Health. All patients signed an informed consent for the use of their biological material for research purposes. One case (R012) from this cohort had a history of lower grade glioma prior to the first GBM. All patients were treated by standard Stupp treatment with surgery followed by radiotherapy plus concomitant and adjuvant TMZ2.
Samples from cohort MD Anderson were primary and recurrent paired tumor obtained from Henry Ford Hospital in accordance with institutional policies and all patients provided written consent, with approval from the Institutional Review Board (IRB protocol #402). Three cases had a history of lower grade astrocytoma prior to the first GBM (R022/R027/R029). All of the recurrent GBMs had been treated with radiochemotherapy plus TMZ. Cohort TCGA contains TCGA samples, following the publishing protocol of TCGA policies. All of the recurrent GBMs had been treated with chemotherapy or radiation. Six patients were not treated by TMZ (R031/R034-R038). Cohort MD Anderson and TCGA were initially published by Kim et al.7.
Cohort UCSF contains eight patients (R043-R050) collected from the Neurosurgery Tissue Bank at the University of California San Francisco (UCSF), approved by the Committee on Human Research at UCSF. Two patients (R051/R052) from this cohort were from University of Tokyo hospital and the study was approved by the Ethics Committee of the University of Tokyo. Initial tumors of all patients in this cohort were low-grade gliomas, and their recurrences were secondary GBM. This cohort was initially published by Johnson et al.13 Cohort KU makes use of data generated by Department of Pathology and Tumor Biology, Kyoto University. Initial tumors of patients from KU were low-grade gliomas, and their recurrences were secondary GBM. Those patents were initially published by Suzuki et al.14
Cohort SMC consists of GBM samples from Samsung Medical Center (SMC), Korea, following the prior publication (Kim et al, Cancer Cell 2015, R056-R0936) and additional unpublished samples (R094-R114). All samples from SMC had been collected with approval from the Institutional Review Board (IRB file #201004004 and #201310072). Initial tumors from R076-R078/R098/R105/R114 were secondary GBM, with history of low-grade gliomas. Patient R103 had cervical cancer three years prior to the first diagnosis of GBM.
Detailed clinical information of all cohorts was provided in Supplementary Table 10.
Sequencing and mapping
Genomic DNA from initial tumor/recurrent tumor/matched normal blood of patients R001-R016, and recurrent tumor of patients R017-R019 were extracted purified, quantitated, fragmented, quality controlled and used to create a library of genomic DNA fragments. gDNA fragmentation was performed using the Covaris S220 AFA instrument to reproducibly generate fragments of a precise length, while quality control of both gDNA samples and library fragments (at a later time) was performed using Agilent Bioanalyzer 2100 microfluidic device. Both untreated and treated tumor samples of R009, R011, and R014, plus recurrent samples of patients R017-R019 were sequenced by Agilent V3 50M kit, sequencing 90bp PE. Mapping files of untreated/normal samples of patients R017-R019 were obtained from TCGA through CG-hub. All other DNA samples from cohort INCB were sequenced by the protocol of Agilent SureSelect XT Human All Exon v4 Kit, PE, 80M reads, 150X on target coverage. High-quality reads of those samples were mapped by BWA50 to human genome assembly of hg1951 with default parameters. All mapped reads were then marked duplication by Picard to eliminate potential duplications. Total RNA of samples in cohort INCB was collected to investigate the transcriptional profiling by mRNASeq using Illumina technology. Upon quantification and quality controls, mRNAs were reverse transcribed to cDNA and a library of fragments was synthesized using Illumina TruSeq mRNA kits. Total RNA depleted of ribosomal RNA of patients R001-R005, R007-R008, R010 and R012 were sequenced by TrueSeq3 stranded prep (Illumina). RNA samples of R006, R009, R017-R019 were sequenced in BGI. All reads were mapped to human genome assembly of hg19 from UCSC genome browser51, using a fast splice junction mapper Tophat52.
Mapping files of TCGA samples but R039 were downloaded through CG-hub from TCGA. DNA mapping files of cohort UCSF, cohort MD Anderson, cohort KU, and R056-R093 from cohort SMC were all downloaded from European Genome-phenome Archive. Additional samples (R094-R114) from SMC followed the same sequencing protocols as the previous samples in the prior publication (Kim et al, Cancer Cell 2015, R056-R0936).
SAVI2 and driver gene selection
To identify somatic mutations from whole-exome sequencing data of triple samples (normal, initial tumor, and recurrent tumor) of GBM patients, we applied variance-calling software SAVI2 (statistical algorithm for variant frequency identification15) based on the empirical Bayesian method. Specifically, we first generated the candidate variant list by successively eliminating positions without variant reads, positions with low-depth, positions that were biased in one strand, and positions containing only low-quality reads. Then the number of high quality reads of forward ref alleles, reverse ref alleles, forward non-ref alleles, and reverse non-ref alleles were calculated in the remained candidate positions to build the prior and the posterior distribution of the mutation allele fraction. Finally somatic mutations were determined based on the posterior distribution of difference of the mutation allele fraction between normal and tumor samples15. SAVI2 was able to assess mutations by simultaneously considering multiple tumor samples, as well their corresponding RNA samples if available.
Common somatic mutations from Patients R078-R082 were unknown due to the lack of normal DNA. The initial and recurrence exclusive mutations were calculated based on the difference between initial and recurrent tumor DNA. Tumor DNA of Patient R083-R093, R102, R111-R114 were not complete. The somatic mutations of these patients were estimated based on RNA sequencing.
The known driver list (Supplementary Table 2) used in this manuscript was generated by combining GBM drivers from cancer gene census53 and our previous analysis of primary GBM2.
The Analysis of Loss of heterozygosity (LOH) and Copy Number Change
All common dbSNP variants of single samples were extracted to define Zygosity Score (ZS) as . The LOH rate of somatic mutations in a tumor sample was then defined by
where is zygosity score in tumor samples, while is that of the normal samples. If we thought the corresponding mutation is in a LOH region. Segmentation in Supplementary Figure 3 was performed based on CBS algorithm54.
The pipeline of EXCAVATOR55 was carried out to detect copy number alterations based on whole-exome sequencing data. EXCAVATOR considers mean number of reads per exon, and normalized the data by a three-step normalization procedure to eliminate the bias introduced by GC content, the genomic mappability and the exon size. Segmentation was then performed with a novel heterogeneous hidden Markov model algorithm, heterogeneous shifting level model (HSLM) algorithm, which considers the genomic distance between consecutive exons55. To confidently quantify variation arising in whole-exome sequencing (WES) data in each patient’s initial and recurrent sample compared to normal data we calibrated WES CNV calls to SNP array data in available samples (Supplementary Figure 15). In addition to WES we utilized segmentation data for TCGA samples from Broad Firehose platform and when available SNP6 data pre-processed with AROMA and normalized ArrayCGH obtained from Gene Expression Omnibus (GSE63035). To identify statistical significant regions, GISTIC26 was applied in initial and recurrent tumors respectively. GISTIC estimated the background rates for each amplification and deletion, and then summarize the input samples to score the significance of copy number altered regions. To integrate mutation and copy number data, MutComFocal27 was separately performed in initial and recurrent tumor. In the MutComFocal analysis, long proteins (with more than 3500 amino acids), not expressed genes (mutations were not expressed in any samples) and high-synonymous-rate genes (synonymous/non-synonymous>0.2) were not considered.
Gene Fusion Detection and Structure Rearrangement of EGFR
ChimeraScan56 was used to generate the starting set of gene fusion candidates. To reduce the false positive rate and nominate potential driving events, we applied the Pegasus annotation and prediction pipeline. We reconstructed the entire fusion sequence on the basis of the breakpoint coordinates and assigned a driver score to each candidate fusion via a machine learning model trained largely on GBM data57. All candidates reported in Supplementary Table 8 were selected according to four criteria: 1. Pegasus score was >0.5; 2. Either more than 400 span reads or at least two split reads supported fusion; 3. The two fusion partner was apart at least 50 kb.
To check the rearrangement of EGFR, we applied prada-guess-if from PRADA package. PRADA is a RNA sequencing analysis pipeline developed in MD Anderson40. Following the definition in Brennan et al. 2013, transcribed allelic fractions of EGFRvIII were defined as the fraction of junction reads between exon1 and exon8.
Gene expression analysis and expression-based subtyping analysis of GBM samples
Fragments Per Kilobase of exon model per Million mapped fragments (FPKMs) were calculated by Cufflinks58. To eliminate batch effect, we have normalized gene expression by calculating Z-score in each batch. The gene expression was assessed by their corresponding Z-scores. ssGSEA was applied to determine the subtype of GBM samples. For each sample, Z-score was used to rank all genes to generate the rnk.file as the input of GseaPreranked software. An enrichment score (ES) was generated for all four subtypes initially defined in Verhaak et al. 201042. The subtype with the maximal ES was selected as a representative subtype of each sample.
Moduli Space Analysis
Clustering analysis of the patient data was performed as follows. Each phylogenetic tree was represented as a point in the projective evolutionary moduli space, which in this case is a triple such that , by taking the raw mutation counts for the common, initial, and recurrent mutations and normalizing, setting . We discarded samples where any of the mutation counts were missing, leaving 93 points (out of 114 patients). The metric on the evolutionary moduli space was in this case simply the standard Euclidean metric. Note that for purposes of constructing this space, the “branch lengths” of each patient’s tree are simply mutation counts, in contrast to the evolutionary analysis described below, which estimates branch lengths in years.
We then applied three clustering algorithms to this metric space: -means clustering, spectral clustering, and density-based spatial clustering (DBSCAN). We used the code provided as part of the scikit Python package. For -means clustering and spectral clustering, we set the number of clusters at three; DBSCAN determines the number of clusters from the data, but we set the parameters to be and minimum cluster size =5. For spectral clustering, the affinity matrix was computed using the Gaussian kernel applied to the Euclidean distance.
In order to ensure stability of the results, we performed cross-validation using Monte Carlo simulations in which we sampled without replacement 95% of the data points and performed clustering.
Tumor Purity Estimation and Cellular Fraction
ABSOLUTE37 was used to infer tumor purities and ploidy for each WES sample by integrating mutational allele frequencies and copy number calls.
PyClone38 was run for each sample using default parameters. Briefly, we integrated both allele mutations, copy number calls and loh status for each sample as input to obtain cellular frequencies. Cellular frequencies were then rescaled by median adjustment and used as input for Tumor Evolutionary Directed Graph and Mathematical modeling of tumor evolution.
Evolutionary Model
We considered all 92 patients for whom mutations were sequenced in both the initial and recurrent tumor samples. To exclude false positives, only variants with an allele frequency of at least 5% were used. Variants occurring at a cellular fraction of at least 95% were classified as clonal in a sample, and others were considered subclonal. Check details of the model in Supplementary Note. In Supplementary Figure 16, we perform sensitivity analysis using alternate cutoffs for clonality. Related code is provided in Supplementary Code.
TEDG reconstruction
In order to reconstruct the order of events during tumor progression we followed the strategy in Wang et al. 201436. We selected genes that were recurrently mutated and expressed in our samples. In hypermutated cases, we only considered mutations of MSH6 and LTBP4. A mutation that was predicted to be clonal (cellular fraction>0.8) in both initial tumor and recurrent tumor was defined as an early event, while a mutation that was only present (variant allele fraction>5%) in one sample was defined as a late event. To represent the order of clonal mutations, for each sample, directed edges were added to connect early and late events. Then we combined all directed edges from different patients to show a global landscape of GBM evolution. A copy number alteration was defined as clonal if the absolute value of segmean was larger than one. A copy number alteration was defined as present at the threshold 0.5, and as absent at the threshold 0.1.
Hypermutation Score
Hypermutation (HM) score was defined as
where WM is the weight matrix of the DNA sequence logo of a given sample; WMH is the weight matrix of all mutations in hypermutated sampels; and WMN is the weight matrix of mutations from all non-hypermutation samples.
Validation of mutations
The genomic regions surrounding the predicted mutations were amplified using AccuPrime Taq DNA Polymerase High Fidelity (Invitrogen, USA). Primers were summarized in Supplementary Table 11.
The PCR products were purified with ExoSAP-IT (Affymetrix, USA) and subjected to Sanger Sequencing (Macrogen, USA). The amplicons containing the predicted genomic mutations were sequenced using BigDye Terminator Cycle Sequencing Kit v3.1 on the ABI Prism 3730×l DNA Analyzer (Applied Biosystems, USA). All Sanger validation figures are in Supplementary Data 1.
To assess the sensitivity of judging absence of a mutation in one phase that is present in the other phase, we studied 15 variants in the panel that are absent in one of the samples in WES, with median WES depth 117 [10-402]. Using CancerScan6, we found that no read reported the variant in the sample where it was deemed absent by WES (Supplementary Table 12), median CancerScan depth 563 [217-1377].
Cell culture, lentivirus production and cell growth analysis
U87 (ATCC HTB-14) cell line was acquired through American Type Culture Collection. U251 (Sigma, catalogue number 09063001) cell line was obtained through Sigma. Cell lines were cultured in DMEM supplemented with 10% fetal bovine serum (FBS, Sigma). Cells were routinely tested for mycoplasma contamination using Mycoplasma Plus PCR Primer Set (Agilent, Santa Clara, CA) and were found to be negative.
Lentivirus was generated by co-transfection of the lentiviral vectors with pCMV-ΔCMV-ors wpMD2.G plasmids into HEK293T cells as previously described (Niola et al. JCI 2013; Carro et al. Nature 2010). shRNA sequences for LTBP4 are in Supplementary Table 11.
After infection cells were selected with Puromycin (Sigma) at concentration of 2 mg/ml for 48 h. Cells were analysed by western blot, qRT-PCR and growth assay 3 days later.
Evaluation of cell growth was performed using the MTT assay. Cells were plated at density of 2.5×103 cells/well into 96 well plates in 6 replicates and allowed to adhere for 24 h. Viability was assessed daily by adding MTT ((3-[4,5-dimethylthiazol-2-yl]-2,5- diphenyltetrazolium, Sigma 5mg/ml in PBS). Following 4h incubation period, medium was removed and formazan crystal were solubilized with acidic isopropanol (0.1 N HCl in absolute isopropanol. The absorbance at 550 nm was measured with a plate reader.
RT–PCR
Total RNA was prepared with Trizol reagent (Invitrogen) and cDNA was synthesized using SuperScript II Reverse Transcriptase (Invitrogen) as described (Carro et al. Nature 2010; Zhao et al. Nature Cell Biol 2008). The quantitative RT–PCR was performed with 7500 Real-Time PCR system, using SYBR Green PCR Master Mix from Applied Biosystem. Primers used in qRT–PCR are summarized in Supplementary Figure 12.
Results are presented as the mean ± s.d. of three independent experiments each performed in triplicate (n=9). Statistical significance was determined by using unequal variance t-test (two-tailed).
Western Blot
Cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP40, 0.5% sodium dexoycholate, 0.1% sodium dodecyl sulphate, 1.5 mM Na3VO4, 50 mM sodium fluoride, 10 mM sodium pyrophosphate, 10 mM β-glycerolphosphate and EDTA-free protease inhibitor cocktail (Roche)). Lysates were cleared by centrifugation at 15,000 r.p.m. for 15 min at 4 °C. Protein samples were separated by SDS–PAGE and transferred to nitrocellulose membrane. Membranes were blocked in TBS with 5% non-fat milk and 0.1% Tween20, and probed with primary antibodies. Antibodies and working concentrations are: LTBP4 (1:200, sc-393666) obtained from Santa-Cruz Biotechnology; P-SMAD7 (1:1000, #3101) and SMAD7 (1:1000, #5339) obtained from Cell Signaling Technology; β-actin (1:2,000 dilution; A5441) obtained from Sigma.
Gene Fusion Validation
For validation of fusion transcripts and RT-PCR assays were performed. Total RNA was extracted from the tissues by AllPrep DNA/RNA Mini kit according to the manufacturer’s instructions (Qiagen). The total RNA (0.5 μg) was reverse transcribed to synthesize template cDNA by a random primer using the SuperScriptIII First-Strand System(Life Technologies), and 20 μl synthesized cDNA was diluted 10 times with DW. For RT-PCR, EzWay Taq PCR MasterMix (Komabiotech, KOREA) and 5 μl synthesized cDNA as template were used. Thermal cycling was carried out under the following conditions: 1 min at 95°C followed by 30 cycles of 30 sec at 95°C, 30 sec at 55°C, 30 sec at 72°C. The primer pairs used in this experiment were designed to make the amplification product including the breakpoints of the fusion genes. PCR products were analyzed by agarose gel electrophoresis. The primers were summarized in Supplementary Table 11.
Supplementary Material
Supplementary Data 1: Sanger validation of selected mutations.
Supplementary Note: All Supplementary Figures and Supplementary note of mathematical model for clonal evolution.
Supplementary Code: Commented Stan code for evolutionary model.
Supplementary Table 1: Coverage of whole-exome sequencing
Supplementary Table 10: Patient clinical data
Supplementary Table 11: Primer/oligo sequences for experimental validation
Supplementary Table 12: Validation of selected mutations by CancerSCAN. Variants deemed absent noted in red.
Supplementary Table 2: All somatic mutations of recurrent GBM patients
Supplementary Table 3: Significant regions and potential driver genes reported by GISTIC2
Supplementary Table 4: Significant driver genes predicted by MutComFocal
Supplementary Table 5: Analysis of Loss of Heterozygosity
Supplementary Table 6: Selected list of gene fusion candidates
Supplementary Table 7: Tumor purity estimation from ABSOLUTE
Supplementary Table 8: Cellular Frequency prediction from PyClone
Supplementary Table 9: EGFRvIII prediction from PRADA
Acknowledgments
R.R. acknowledges funding from the following sources: NIH (U54 CA193313, R01 CA185486, R01 CA179044). J.W. is supported by Precision Medicine Fellowship (UL1 TR000040). E.L. is supported by Cancer Biology Training Program (T32 CA09503). D.I.S.R. is supported by the U.S. National Library of Medicine (T15 LM007079). S.Z. is supported by a Personalized Medicine Fellowship (TL1 TR000082). This work is also supported by National Institute of Health grants to A. L. (R01CA101644 and R01CA131126), A.I. (R01CA178546 and R01NS061776 and a grant from The Chemotherapy Foundation). V.F. is supported by a fellowship from the American Brain Tumor Association (ABTA). G.F. is partly funded by the Italian Minister of Health. D.H.N. acknowledges funding from the Korea Healthcare Technology R&D project (HI14C3418) by Korea Health Industry Development Institute and Ministry for Health & Welfare Affairs.
Footnotes
The authors declare no competing financial interests.
URLs
Broad Institute Firehose platform for TCGA data, http://gdac.broadinstitute.org; AROMA for SNP6 data pre-processed, http://www.aroma-project.org; CG-hub for TCGA raw data, https://cghub.ucsc.edu.
Accession codes
Mapping files of TCGA samples but R039 were downloaded through CG-hub from TCGA. DNA mapping files of cohort UCSF, cohort MD Anderson, cohort KU, and R056-R093 from cohort SMC have been deposited in European Genome-phenome Archive (EGA) with accession number EGAS00001000579, EGAD00001001113, EGAD00001001213, and EGAD00001001424. Additional samples (R094-R114) from SMC have been deposited in EGAS00001001800. Additional samples from INCB have been deposited in Sequence Read Archive (SRP074425). ArrayCGH data for patients from MD Anderson have been deposited in Gene Expression Omnibus (GSE63035).
AUTHOR CONTRIBUTIONS
E.C., M.E. and G.F. started the collection of samples and prepared libraries for genomic and transcriptomic analysis. E.C., V.F., and A.L. performed LTBP4 experiments. V.F. performed Sanger validation. S.Z. and A.B. performed the statistical analysis of phylogenetic trees. D.N. provided additional data. F.A. and S.Z. ran Pegasus analysis. Z.L. ran ssGSEA analysis. D.R. constructed the mathematical model of tumor evolution before and after treatment. R.R., J.W., E.L. developed and carried out analysis of genomic data. A.L., R.R. and A.I conceived of the methodology. D.H.N, J.K.L., I.H.L., and W.Y.P provided clinical and genomic information for the SMC cohort. D.H.N. and Y.J.S. performed the validation of MGMT fusion. R.R., A.I and J.W. conceived project, discussed the results and implications and wrote the manuscript.
References
- 1.Ricard D, et al. Primary brain tumours in adults. Lancet. 2012;379:1984–1996. doi: 10.1016/S0140-6736(11)61346-9. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Frattini V, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45:1141–1149. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mazor T, et al. DNA Methylation and Somatic Mutations Converge on the Cell Cycle and Define Similar Evolutionary Histories in Brain Tumors. Cancer Cell. 2015;28:307–317. doi: 10.1016/j.ccell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J, et al. Spatiotemporal Evolution of the Primary Glioblastoma Genome. Cancer Cell. 2015;28:318–328. doi: 10.1016/j.ccell.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 7.Kim H, et al. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res. 2015;25:316–327. doi: 10.1101/gr.180612.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 9.Nordling CO. A New Theory on the Cancer-Inducing Mechanism. Br J Cancer. 1953;7:68–72. doi: 10.1038/bjc.1953.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Armitage P, Doll R. The Age Distribution of Cancer and a Multi-Stage Theory of Carcinogenesis. Br J Cancer. 1954;8:1–12. doi: 10.1038/bjc.1954.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sottoriva A, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gill BJ, et al. MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma. Proc Natl Acad Sci USA. 2014;111 doi: 10.1073/pnas.1405839111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson BE, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki H, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47:458–468. doi: 10.1038/ng.3273. [DOI] [PubMed] [Google Scholar]
- 15.Trifonov V, Pasqualucci L, Tiacci E, Falini B, Rabadan R. SAVI: a statistical algorithm for variant frequency identification. BMC Syst Biol. 2013;7(Suppl 2):S2. doi: 10.1186/1752-0509-7-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melamed RD, Wang JG, Iavarone A, Rabadan R. An information theoretic method to identify combinations of genomic alterations that promote glioblastoma. J Mol Cell Biol. 2015;7:203–213. doi: 10.1093/jmcb/mjv026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciriello G, Cerami E, Sander C, Schultz N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 2012;22:398–406. doi: 10.1101/gr.125567.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao JJ, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stieglitz E, et al. The genomic landscape of juvenile myelomonocytic leukemia. Nat Genet. 2015;47:1326–1333. doi: 10.1038/ng.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter C, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–3991. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yip S, et al. MSH6 Mutations Arise in Glioblastomas during Temozolomide Therapy and Mediate Temozolomide Resistance. Clin Cancer Res. 2009;15:4622–4629. doi: 10.1158/1078-0432.CCR-08-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cahill DP, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13:2038–2045. doi: 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chin L, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyazono K, O A, Colosetti P, Heldin CH. A role of the latent TGF-beta 1-binding protein in the assembly and secretion of TGF-beta 1. EMBO J. 1991;10:1091–1101. doi: 10.1002/j.1460-2075.1991.tb08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sterner-Kock A, et al. Disruption of the gene encoding the latent transforming growth factor-beta binding protein 4 (LTBP-4) causes abnormal lung development, cardiomyopathy, and colorectal cancer. Genes Dev. 2002;16:2264–2273. doi: 10.1101/gad.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trifonov V, Pasqualucci L, Dalla Favera R, Rabadan R. MutComFocal: an integrative approach to identifying recurrent and focal genomic alterations in tumor samples. BMC Syst Biol. 2013;7:25. doi: 10.1186/1752-0509-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh D, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–1235. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Stefano AL, et al. Detection, Characterization, and Inhibition of FGFR-TACC Fusions in IDH Wild-type Glioma. Clin Cancer Res. 2015;21:3307–3317. doi: 10.1158/1078-0432.CCR-14-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hegi ME, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 31.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakellarios Zairis HK, Blumberg Andrew, Rabadan Raul. Moduli Spaces of Phylogenetic Trees Describing Tumor Evolutionary Patterns. Lecture Notes in Computer Science. 2014;8609:528–839. [Google Scholar]
- 34.Vogelstein B, et al. Genetic Alterations during Colorectal-Tumor Development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 35.Yates LR, Campbell PJ. Evolution of the cancer genome. Nat Rev Genet. 2012;13:795–806. doi: 10.1038/nrg3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, et al. Tumor Evolutionary Directed Graphs and the History of Chronic Lymphocytic Leukemia. Elife. 2014;3:e02869. doi: 10.7554/eLife.02869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roth A, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods. 2014;11:396–398. doi: 10.1038/nmeth.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuan CT, Wikstrand CJ, Bigner DD. EGF mutant receptor vIII as a molecular target in cancer therapy. Endocr Relat Cancer. 2001;8:83–96. doi: 10.1677/erc.0.0080083. [DOI] [PubMed] [Google Scholar]
- 40.Torres-Garcia W, et al. PRADA: pipeline for RNA sequencing data analysis. Bioinformatics. 2014;30:2224–2226. doi: 10.1093/bioinformatics/btu169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerami E, et al. The Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2013;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anido J, et al. TGF-beta Receptor Inhibitors Target the CD44(high)/Id1(high) Glioma-Initiating Cell Population in Human Glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 44.Han J, Alvarez-Breckenridge CA, Wang QE, Yu J. TGF-beta signaling and its targeting for glioma treatment. Am J Cancer Res. 2015;5:945–955. [PMC free article] [PubMed] [Google Scholar]
- 45.Joseph JV, Balasubramaniyan V, Walenkamp A, Kruyt FA. TGF-beta as a therapeutic target in high grade gliomas - promises and challenges. Biochem Pharmacol. 2013;85:478–485. doi: 10.1016/j.bcp.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 46.Kaminska B, Kocyk M, Kijewska M. TGF beta signaling and its role in glioma pathogenesis. Adv Exp Med Biol. 2013;986:171–187. doi: 10.1007/978-94-007-4719-7_9. [DOI] [PubMed] [Google Scholar]
- 47.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fakhrai H, et al. Eradication of established intracranial rat gliomas by transforming growth factor beta antisense gene therapy. Proc Natl Acad Sci USA. 1996;93:2909–2914. doi: 10.1073/pnas.93.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khalili JS, Hanson RW, Szallasi Z. In silico prediction of tumor antigens derived from functional missense mutations of the cancer gene census. Oncoimmunology. 2012;1:1281–1289. doi: 10.4161/onci.21511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 55.Magi A, et al. EXCAVATOR: detecting copy number variants from whole-exome sequencing data. Genome Biol. 2013;14:R120. doi: 10.1186/gb-2013-14-10-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iyer MK, Chinnaiyan AM, Maher CA. ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics. 2011;27:2903–2904. doi: 10.1093/bioinformatics/btr467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abate F, et al. Pegasus: a comprehensive annotation and prediction tool for detection of driver gene fusions in cancer. BMC Syst Biol. 2014;8:97. doi: 10.1186/s12918-014-0097-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bedford T, Wapinski I, Hartl DL. Overdispersion of the molecular clock varies between yeast, Drosophila and mammals. Genetics. 2008;179:977–984. doi: 10.1534/genetics.108.089185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bedford T, Hartl DL. Overdispersion of the molecular clock: temporal variation of gene-specific substitution rates in Drosophila. Mol Biol Evol. 2008;25:1631–1638. doi: 10.1093/molbev/msn112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gelman A, Lee D, Guo JQ. Stan: A Probabilistic Programming Language for Bayesian Inference and Optimization. J Educ Behav Stat. 2015;40:530–543. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data 1: Sanger validation of selected mutations.
Supplementary Note: All Supplementary Figures and Supplementary note of mathematical model for clonal evolution.
Supplementary Code: Commented Stan code for evolutionary model.
Supplementary Table 1: Coverage of whole-exome sequencing
Supplementary Table 10: Patient clinical data
Supplementary Table 11: Primer/oligo sequences for experimental validation
Supplementary Table 12: Validation of selected mutations by CancerSCAN. Variants deemed absent noted in red.
Supplementary Table 2: All somatic mutations of recurrent GBM patients
Supplementary Table 3: Significant regions and potential driver genes reported by GISTIC2
Supplementary Table 4: Significant driver genes predicted by MutComFocal
Supplementary Table 5: Analysis of Loss of Heterozygosity
Supplementary Table 6: Selected list of gene fusion candidates
Supplementary Table 7: Tumor purity estimation from ABSOLUTE
Supplementary Table 8: Cellular Frequency prediction from PyClone
Supplementary Table 9: EGFRvIII prediction from PRADA


