Abstract
Background:
The use of biomarkers that allow early therapeutic intervention or intensive follow-up is expected to be powerful means to reduce breast cancer mortality. MicroRNAs (miRNAs) are known to play major roles in cancer biology including metastasis. The aim of this study is to develop novel miRNA signature score to predict patient survival and metastasis in breast cancer.
Method:
An integrated unbiased approach was applied to derive a composite risk score for prognosis based on miRNA expression in primary breast tumors in 1,051 breast cancer patients from The Cancer Genome Atlas (TCGA). Further analysis of the risk score with metastasis/recurrence was conducted in the TCGA dataset and validated in a separate patient population using small RNA-sequencing.
Results:
The three-miRNA signature score (miR-19a, miR-93, and miR-106a) was developed using the TCGA cohort that predicted poor prognosis (p=0.0005), independent of known clinical risk factors. The prognostic value was validated in another three independent cohorts (GSE19536, p=0.0009; GSE22220, p=0.0003; and METABRIC, p=0.0023, respectively). The three-miRNA signature score predicted bone recurrence in TCGA (p=0.0052), and the findings were validated in another independent patient population of those who developed bone recurrence and age/stage-matched patients without any recurrence. The three-miRNA signature score enriched multiple metastasis-related gene sets such as angiogenesis and epithelial mesenchymal transition in Gene Set Enrichment Analysis.
Conclusions:
We developed novel miRNA-based risk score which is a promising biomarker to predict worse survival and bone recurrence potential in breast cancer.
Keywords: breast cancer, microRNA, risk score, biomarker, prognosis, bone metastasis, small RNA-Seq
Introduction
Intrinsic subtype classification utilizing gene expression microarray has revolutionized the way we understand breast cancer1. Differences across breast cancer subtypes in response to different treatments and prognosis are now well established1–3. Targeted therapy based on each patient’s unique cancer biology, in conjunction with more rational use of cytotoxic systemic therapy, has contributed to the decreasing mortality rate of breast cancer over recent decades in the U.S. However, over 40,000 women still die of breast cancer every year in the US alone4,5, highlighting the need of a deeper understanding of breast cancer biology to further improve treatment.
Breast cancer intrinsic subtypes were first discovered based primarily on the expression profile of coding genes, which predated our current appreciation of the versatile roles played by non-coding RNAs, such as long non-coding RNA and microRNA (miRNA), which impacts almost every aspect of cancer, from etiology to progression and response to treatment. MiRNAs, a class of small non-coding RNA, constitute an important epigenetic mechanism fine tuning the transcription and translation of protein-coding genes. Since its discovery6,7, dysregulated miRNA expression has been identified in various cancers including breast cancer8–13.
The primary aims of the current study were to evaluate the prognostic value of miRNA expression profiles of primary breast cancer and to develop a miRNA-based risk score for patient prognosis. Three public available datasets were used: The Cancer Genome Atlas (TCGA), the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC), and the Gene Expression Omnibus (GEO). Our integrated and unbiased approach identified a composite score based on three miRNAs from primary breast cancer that was significantly associated with poor prognosis. We also found that this score was associated with enrichment of metastasis related gene sets and it can be used to predict bone recurrence in breast cancer patients.
Materials and Methods
Patient populations
MiRNAs and RNA expression data with linked clinical data were available on a total of 2,580 breast cancer cases from TCGA (n=1,051)14, METABRIC (n=1,223)15,16 and two GEO datasets (GEO22220 n=210; GEO19536 n=96)17,18. TCGA was used as the discovery cohort, and the other three as validation cohorts. Patient demographics of those cohorts are summarized in Supplemental Tables S1 and S2. In all datasets, breast cancer subtypes were classified according to immunohistochemistry markers19,20. For TCGA data, because PAM50 subtype classification was not publically available from all breast tumors from XENA, the “genefu” package21 was applied to classify all 1051 samples in the cohort, which uses the same algorithm as TCGA.
Development of a miRNA-based risk score
Among the 1051 breast cancer patients who have microRNA expression data and required clinical information registered in TCGA dataset, two subsets of patients were defined based on overall survival (OS) status: “long survival” (those survived greater than 5 years after diagnosis, n=240) and “short survival” (those deceased within 3 years of diagnosis, n=65). In order to emphasize the biological feature that could impact patient survival, we utilized these two “long survival” and “short survival” subsets for initial development of the miRNA-based risk score.
We identified 1881 miRNAs annotated in TCGA dataset, and after excluding those with low miRNA counts, 1,549 miRNAs were analyzed for the developmental settings. Differentially expressed miRNAs between the two groups were identified using “DEseq2” based on the negative binomial distribution22. Of the top 19 miRNAs (miR-103a-1, miR-103a-2, miR-93, miR-92a-1, miR-92a-2, miR-1307, miR-17, miR-196b, miR-20a, miR-500a, miR-128–2, miR-19b-1, miR-19b-2, miR-20b, miR-106a, miR-19a, miR-660, miR-184, and miR-187) identified after adjusting for multiple comparing (adjusted p-value <0.1), one miRNA (mir-103a-2, mir-92a-2, mir-19b-1, miR-19b-2, and miR-17) was randomly dropped from highly correlated pairs (r2 >0.85) in order to reduce multicollinearity and improve stability for further model selection. Stepwise selection was then used to select 3 miRNAs (miR-19a, miR-93, and miR-106a) in a final multivariable model based on Akaike information criterion (AIC). Details were presented in Supplementary File S1. A composite risk scores based on the three miRNAs was derived as a weighted linear combination of their expression levels.
Patients were subsequently dichotomized into low-risk group and high-risk group based on the risk score. To determine an optimal cutoff point, a series of candidate points were evaluated in Cox proportional hazard models23, and the final cutoff was chosen based on model significance24–27. The same classification method based the miRNA risk score was applied to the three independent validation datasets from METABRIC15,16 and GEO17,18. To test whether the prognostic value of the risk score was independent from tumor histopathological characteristics, tumor stage (according to the TNM classification of the 7th Edition AJCC)20, ER, PR, and HER2 status were adjusted in a multivariable model.
Identification of subtypes utilizing Prosigna Breast Cancer Prognostic Gene Signature Assay (PAM50) gene signature in TCGA cohort
To classify all the TCGA patients into subtypes defined by PAM50 gene signature, we established a novel computational algorithm. The PAM50 subtypes were called using the Bioconductor genefu package and using RNA-Seq expression data of the TCGA breast cancer Primary Solid Tumor samples retrieved from Broad Institute Firehose (https://gdac.broadinstitute.org/). The subsequent classification was mostly consistent with the PAM50 calls made by the TCGA analysis Working Group (AWG), retrieved from the XENA browser (https://xenabrowser.net/).
Validation study of the miRNA-based risk score with bone metastasis using small RNA-sequencing
To independently validate the prognostic value of the miRNA-based risk score for bone metastasis, fresh frozen primary tumors were obtained from age- and stage-matched breast cancer patients who eventually developed bone recurrence as an initial metastatic site during follow-up (bone recurrence group, n=10) and who did not have any tumor recurrence at least for 5 years after primary tumor resection (control breast cancer group, n=10) from the Roswell Park Cancer Institute Pathology Network Shared Resource (patients’ details are not shown). Two pathologists independently evaluated all samples and confirmed that they included more than 80% neoplastic cell component. Total and small RNAs were isolated using the miRNeasy mini kit (Qiagen) as per manufacturer recommendations. The small RNA sequencing libraries were prepared with the TruSeq Small RNA kit (Illumina Inc) from 1ug total RNA. Validated libraries were pooled with equal molar in final concentration and sequenced using the Illumina HiSeq 2500 using 50 cycle single read sequencing (Illumina, Inc.). The miRNA expression level was normalized and log2 transformed using DEseq2 package. Ridge regression was used to derive a score for these samples based on the expression levels of the same three miRNAs and the predicting performance for bone metastatic status was evaluated using the area under curve (AUC) analysis28. The study was approved by the Institutional Review Board of Roswell Park Cancer Institute for human subject protection.
Statistical analysis
All statistical analyses were performed using R software and Bioconductor. In the TCGA and GEO cohorts, OS was defined as the time from date of diagnosis to the date of death by any cause. In the METABRIC cohort, disease specific survival (DSS) was defined as the time from date of diagnosis to the date of death by a cancer-specific cause. Disease free survival (DFS) was defined as the time from date of diagnosis to the date of relapse. To compare the survival curves between subgroups groups, the Kaplan-Meier method with log-rank test was used. As we previously reported, Cox proportional hazard models were used for multivariable analysis to derive hazard ratios (HR) and 95% confidence intervals (CI), and the proportional hazard assumption was tested using Schoenfeld residuals25–27,29–31. Cumulative Incidence Functions (CIFs) were estimated to assess the probability of metastatic to different sites, and tested for statistically significant in TCGA cohort. Gene Set Enrichment Analysis (GSEA) was performed using software provided by the Broad Institute (http://software.broadinstitute.org/gsea/index.jsp)32. In all analysis, a two-sided p<0.05 was considered statistically significant, unless otherwise specified. This “prognostic marker” study is conducted according to the REMARK guidelines33.
Results
Development of a miRNA-based risk score for breast cancer prognosis in TCGA
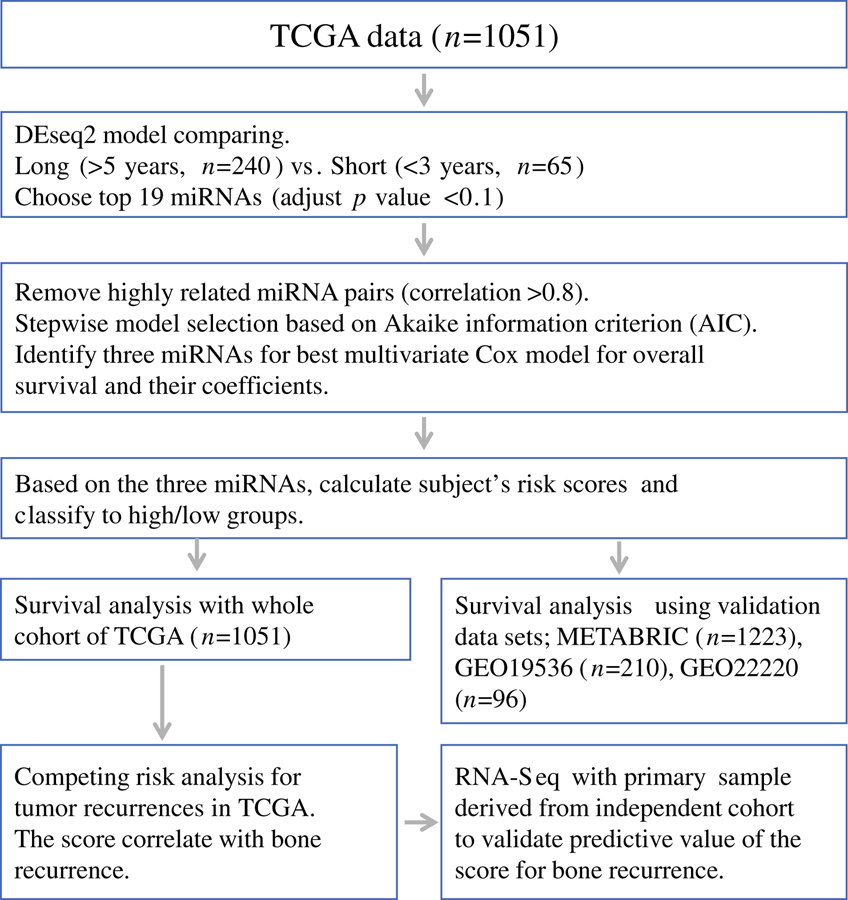
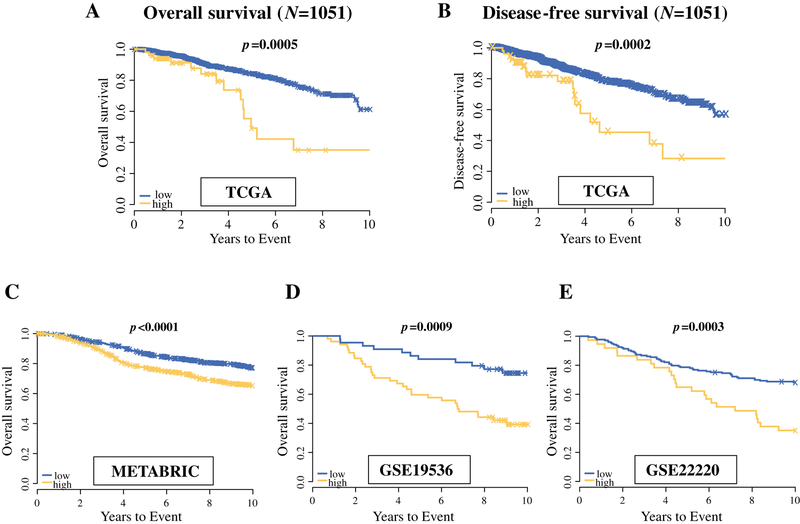
The overall study design is shown in Figure 1. We initially identified 19 miRNAs that were most differentially expressed between the “long survival” and “short survival” groups (BH fdr <0.1 after adjusting for multiple comparison) (Supplemental Table S3 and S4). After removing 5 miRNAs from clusters where miRNAs were highly correlated, stepwise selection retained three miRNAs in the final multivariable model, which included miR-19a, miR-93, and miR-106a. More details can be seen in Supplementary file. High miRNA-based risk score using the expression of these three miRNAs associated with poor survival of breast cancer patients with minimum p-value among the two distinct groups, and when miRNA-based score were calculated on the all TCGA patients (n=1051), the group with high miRNA-based score had significantly shorter OS compared to those with the low score (HR: 2.62, 95% CI: 1.53 – 4.49, p=0.0005) and DFS (HR: 2.51, 95% CI: 1.54 – 4.10, p=0.0002) (Figure 2A and 2B).
Figure 1.
Study strategy for selecting miRNA signature and generating risk scores to predict poor prognosis. Patients from two representative overall survival groups; “Long survival” (those survived greater than 5 years after diagnosis, n=240) vs. “Short survival” (deceased within 3 years of diagnosis, n=65); were used to identify the top miRNAs with differential expression using a model implemented in DEseq2 package based on the negative binomial distribution. First, we identified the top 19 miRNAs as our candidates, which showed most different expression levels in the two groups (adjust p-value <0.1). Next, highly related miRNA pairs (correlation>0.85) were excluded in order to reduce the multicollinearity and improve stability for further model selection. Finally, using stepwise model selection based on Akaike information criterion (AIC), we identified three miRNAs signature for best multivariate Cox proportional hazard model for overall survival and their coefficients (miR-19a, miR-93, and miR-106a). Calculated subject’s risk scores using three miRNAs signature and classify all patients of TCGA breast cancer into high score/low risk score groups. The same classification was made in the three independent validation datasets from METABRIC and GEO using these miRNAs.
Figure 2.
Overall and disease-free survival analyses with multivariate Cox model and Kaplan Meier curve for the three-miRNA signature in TCGA dataset. A, the patients with high score of the three miRNAs signature (n=52) have significantly poor prognosis than the patients with low score (n=999) for overall survival (p=0.0004), as well as for disease-free survival (p<0.0001). Validation analyses for survival using three independent data sets; METABRIC cohort (C), GSE19536 (D); GSE22220 (E) showed that the three miRNAs signature have a significant impact on patient survival in the three independent cohorts derived from cBioPortal and GEO dataset.
Validation of miRNA-based risk score in METABRIC and GEO datasets
Consistently across all the three validation cohorts, patients with the high score had a significantly poorer overall survival compared to those with the low score in METABRIC (p=0.0023), GSE19536 (p=0.0009), and GSE22220 (p=0.0003) (Figure 2C, 2D, 2E).
Independence of the miRNA-based risk score from known breast cancer prognostic markers
The miRNA-based risk score was not associated with any known breast cancer prognostic markers in TCGA, including PAM50 subtype (Supplemental Table S5). In METABRIC, patients with the high-risk score tended to have tumors of advanced tumor stage, ER/PR negativity, HER2 positivity, and TNBC subtype (p<0.01) (Supplemental Table S5). Regardless, adjusting for those prognostic markers in multivariable models did not substantially change the associations of OS with the miRNA-based risk score in either TCGA cohort or the METABRIC cohort (Table 1). In further subgroup analyses stratified by ER, PR, or HER2, no apparent difference in the associations of OS with the risk score was observed in TCGA (Supplemental Figure S2).
Table 1.
Univariate and multivariate Cox regression analyses with or without each clinical factor in TCGA and METABRIC dataset
| TCGA | METABRIC | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate model | Multivariate mode | Univariate model | Multivariate model | |||||||||
| Variables | HR | 95% CI of HR |
p-value | HR | 95% CI of HR |
p-value | HR | 95% CI of HR |
p-value | HR | 95% CI of HR |
p-value |
| miRNA risk score | 2.45 | 1.56–3.83 | 9.36E-05 | 2.56 | 1.62–4.02 | 4.96E-05 | 2.29 | 1.31–4.01 | 0.0036 | 2.17 | 1.25–3.77 | 0.0060 |
| Stage | 2.21 | 1.76–2.77 | 5.71E-12 | 2.26 | 1.80–2.84 | 2.27E-12 | 2.02 | 1.73–2.37 | <0.0001 | 1.99 | 1.70–2.32 | <0.0001 |
| miRNA risk score | 2.43 | 1.53–3.88 | 0.00019 | 2.42 | 1.51–3.88 | 0.00023 | 2.14 | 1.24–3.70 | 0.0065 | 2.1 | 1.21–3.63 | 0.0082 |
| ER | 0.64 | 0.44–0.94 | 0.02088 | 0.62 | 0.43–0.91 | 0.01331 | 0.65 | 0.52–0.81 | 0.00012 | 0.73 | 0.57–0.92 | 0.0091 |
| miRNA risk score | 2.42 | 1.53–3.83 | 0.00016 | 2.35 | 1.48–3.73 | 0.00031 | 2.29 | 1.35–3.88 | 0.0021 | 2.16 | 1.27–3.65 | 0.0042 |
| PR | 0.68 | 0.48–0.97 | 0.03312 | 0.68 | 0.48–0.97 | 0.03109 | 0.67 | 0.55–0.81 | <0.0001 | 0.72 | 0.59–0.88 | 0.0013 |
| miRNA risk score | 2.17 | 1.26–3.75 | 0.00535 | 2.07 | 1.20–3.56 | 0.00902 | 2.43 | 1.44–4.10 | 0.00089 | 2.38 | 1.41–4.02 | 0.0011 |
| HER2 | 1.03 | 0.67–1.56 | 0.90571 | 1.01 | 0.67–1.55 | 0.94718 | 1.86 | 1.44–2.41 | <0.0001 | 1.74 | 1.34–2.27 | <0.0001 |
| miRNA risk score | 2.13 | 1.22–3.71 | 0.00751 | 2.17 | 1.24–3.77 | 0.00626 | 2.45 | 1.43–4.19 | 0.0011 | 2.47 | 1.44–4.23 | 0.0010 |
| TNBC | 1.68 | 0.99–2.84 | 0.05592 | 1.78 | 1.05–3.03 | 0.03335 | 1.35 | 1.04–1.74 | 0.023 | 1.17 | 0.89–1.53 | 0.2590 |
1 HR, hazard ratio; 2 CI, Confidence interval
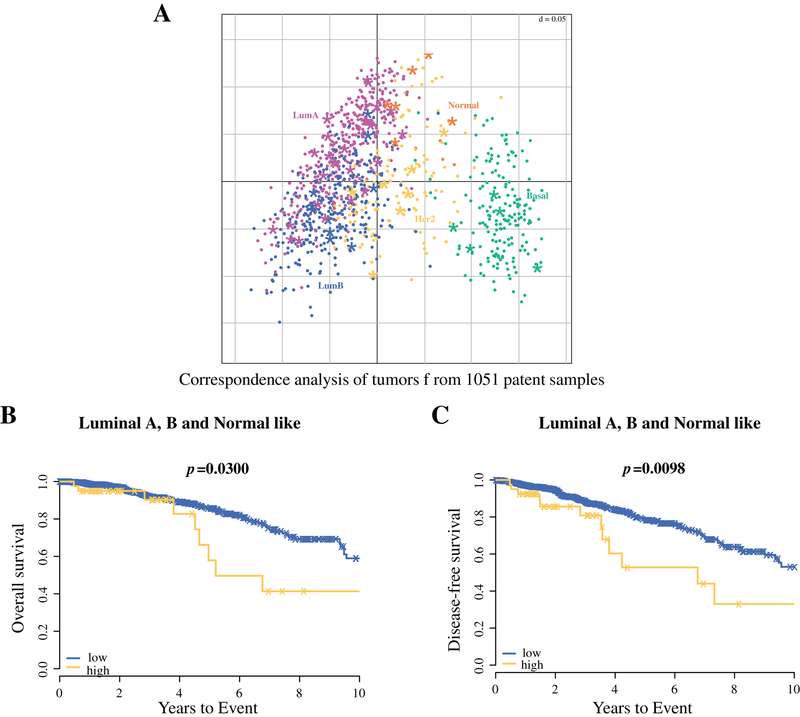
We also conducted stratified prognostic analysis for the miRNAs score using PAM50, which classified TCGA samples into the major breast cancer subtypes. All breast cancers in TCGA were divided into three representative populations using the PAM50 analysis; Luminal A or B or Normal-like; HER2-enriched; and Basal-like34. Although there was no significant separation of distribution by subtype based upon three miRNA score with corresponding analysis of the tumors from 1051 patient samples in TCGA (Figure 3A, Supplementary Table S6), patients with Luminal A, B and Normal like subtypes with a high miRNAs score had significantly worse OS (p=0.0300) and DFS (p=0.0098) in population, in contrast to the Basal-like and HER2-enriched population (Figures 3B and 3C, Supplemental Figure S2). Together, even when stratified by tumor stage or PAM50 subtype, the prognostic value of the risk score appeared to be limited to patients with stage ≥ II or those with luminal A, luminal B and normal-like subtype.
Figure 3.
(A) Application of PAM50 classification in TCGA cohort utilizing RNAseq data. Each dot represents an individual tumor, colored based on PAM50 profiling. Each asterisk represents patient with high risk score of the three miRNAs signature. Patients with high three-miRNA signature score show significantly worse overall survival (B) and disease-free survival (C) in Luminal A, B and Normal like subtypes, but neither Basal-like nor HER2-enriched subtypes.
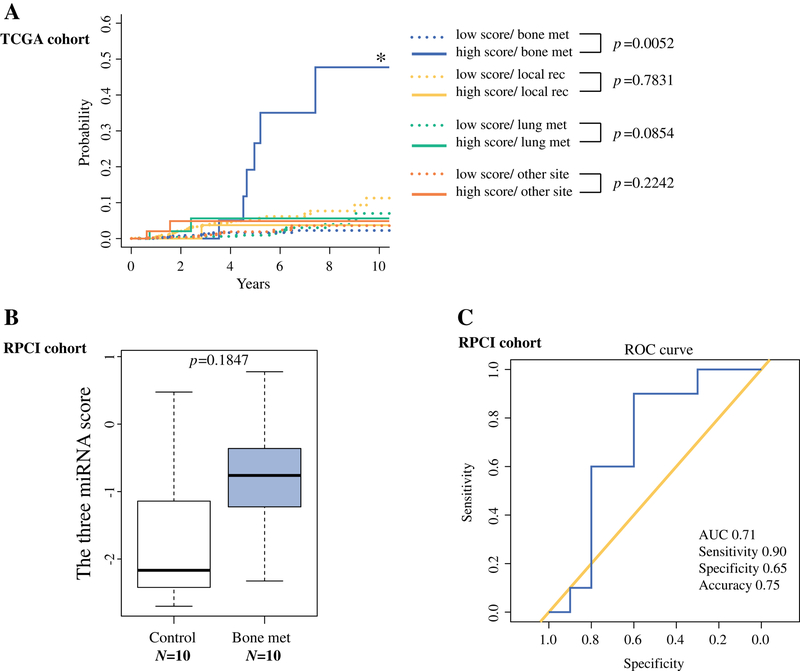
High miRNA-based risk score of primary breast tumors significantly associated with bone metastasis/recurrence
In analysis of local recurrence and distant recurrence at bone, lung and other sites in TCGA, patients with a high score were significantly more likely to develop bone metastasis (p=0.0052) (Figure 4A). To confirm the predictive potential for bone recurrence, we compared the risk score based on the expression of the three miRNAs in primary breast tumor tissues from patients who developed bone recurrence in the course of the follow-up (bone recurrence group, n=10) and from patients who never developed any recurrence for more than 5 years after diagnosis (control breast cancer group, n=10). The bone recurrence group showed higher miRNA-based risk score than the control group, although the statistical test was not significant (p=0.18), possibly due to small sample size (Figure 4B). Receiver operating characteristic (ROC) curve showed that the risk score could distinguish breast cancer patients who later had bone recurrence with relatively high diagnostic power (AUC, 0.71; Sensitivity, 0.90; specificity, 0.65; accuracy 0.75) (Figure 4C).
Figure 4.
Cumulative incidence rate analysis using the three-miRNA signature score for each metastatic site (bone, n=41; local recurrence, n=15; lung, n=12, other, n=15). (A) Bone met associated significantly with high three-miRNA signature score. (B) Patients that developed recurrent bone metastasis showed higher levels of the three-miRNA signature score compared from matched control patients that did not (control breast cancer group, n=10; bone recurrence group, n=10; p=0.1847). (C) ROC curve using the three-miRNA signature score derived from miRNA-Seq data of the 20 primary tissue samples showed AUC of 0.71; Sensitivity, 0.90; specificity, 0.65; accuracy 0.75.
Identification of gene sets enriched with high miRNA-based risk score
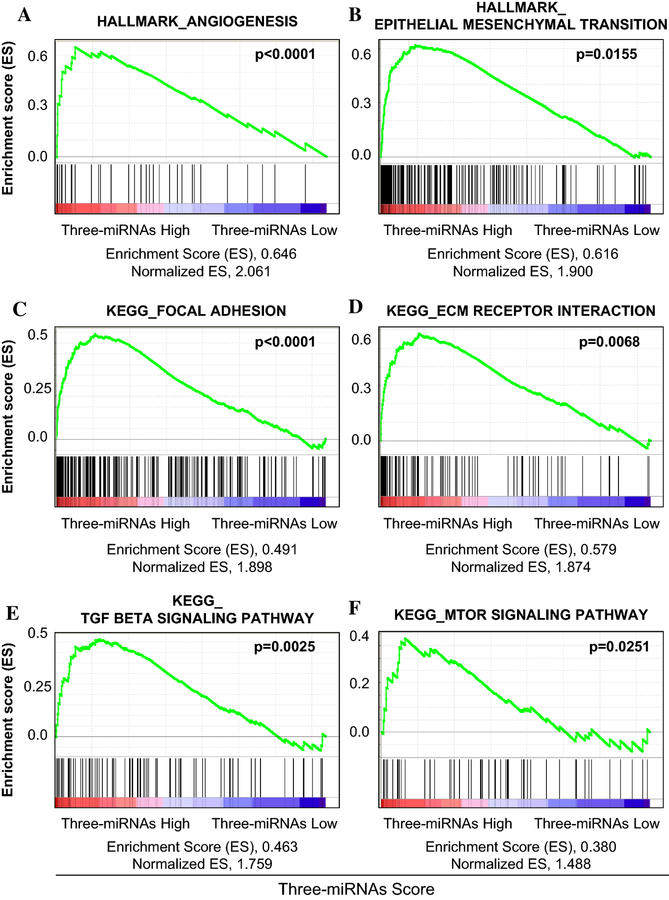
To identify pathways and gene sets enriched with the miRNA-based risk score, GSEA was conducted using RNA expression data from TCGA. GSEA with hallmark gene sets (the most essential data set35) revealed that several pathways and gene sets critical for cancer tumorigenesis and progression were associated with the risk score. Of these, angiogenesis (p<0.0001) and epithelial mesenchymal transition (EMT) (p=0.0155) gene sets were most significant (Figure 5A and 5B, Supplemental Table S7–S9). In addition, GSEA with C2 curated gene sets (including KEGG and GO pathway gene sets) highlighted a number of pathways, including focal adhesion (p<0.0001), TGF-beta signaling pathway (p=0.0025), ECM receptor interaction (p=0.0068), and mTOR pathway (p=0.0251), which are important for tumor progression, tumor invasion, or metastatic formation (Figure 5C–F, Supplemental Table S10–S12).
Figure 5.
Identification of gene sets enriched with high three-miRNA signature score using Gene Set Enrichment Analysis (GSEA) with TCGA data set. (A) Angiogenesis and (B) epithelial mesenchymal transition (EMT) among the Hallmark gene sets were significantly associated with high three miRNAs signature score. Focal adhesion; TGF-beta signaling pathway; ECM receptor interaction; and mTOR, which are important for cancer progression were found to significantly associate with high three-miRNA signature score in GSEA of Curated gene sets (C2, including KEGG and GO pathway gene sets).
Discussion
In the present study, we developed and subsequently validated a composite risk score based on expression of three miRNAs with prognostic values for breast cancer independent from conventional clinical predictors. Further analyses demonstrated that the score was associated specifically with bone metastasis.
In the initial report of TCGA14, very few data were presented regarding miRNA expression profiles in breast cancer tissues. Using clustering analysis of miRNA expression, there was little correlation between miRNA subtype and mutation status, with the exception of two of the seven miRNA subsets overlapping with basal-like subtype and showing a strong positive correlation with TP53 mutation and negative correlation with PIK3CA and GATA3 mutations14. Several later studies explored miRNA expression patterns with breast cancer prognosis using publically available TCGA dataset36–39. Volinia et al identified a prognostic microRNA/mRNA signature using TCGA dataset and a validation cohort36. However, they utilized only 247 miRNAs in 466 breast cancer samples and the prognostic signature included 10 mRNAs and 2 miRNAs. It was thus unclear whether the prognostic effects of the miRNAs were independent from mRNAs36. In another report, Zhou et al based on 915 patients from TCGA showed that a signature consisted of 14 miRNAs was prognostic in patients with ER positive cancers, while the significance of the signature in other subtypes was unclear39. The most important limitations in these previous TCGA-based studies were that the clinical data were obtained before 2015 when follow-up data were only integrated in TCGA datasets and that these studies relied solely on TCGA cohort without independent validation cohorts. In order to overcome these limitations, we utilized TCGA dataset with the latest clinical information as the discovery cohort, followed by validation using independent cohorts from the METABRIC and GEO datasets which had sufficient numbers of case.
Each of the three miRNAs selected in our risk score, miR-19a, miR-93, and miR-106a, is located in a miRNA cluster region, including miR17–92 cluster in 13q31, miR-106b-25 cluster in 7q22, and miR-106a-363 cluster in Xq26, respectively. In previous reports, these clusters have been demonstrated to play critical roles in various cancers8,40–42. MiRNA clusters are where miRNAs are frequently transcribed together as polycistronic primary transcripts that are processed into multiple individual mature miRNAs43,44. The genomic organization of miRNA clusters is often highly conserved, suggesting that it may play biologically important roles for coordinated regulations and functions. For instance, the miR-17–92 cluster encodes six miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b, and miR-92), which are tightly located within an 800 bp region of human chromosome 13. Ancient gene duplications have given rise to two miR-17–92 cluster paralogs, the miR-106b-25 cluster in chromosome 7 and the miR-106a-363 cluster in X chromosome, both of which contain homologous miRNAs to a subset of miR-17–92 components44,45. In previous reports, miR-17–92 cluster regulates multiple cellular processes and functions as a strong “oncogene” favoring malignant transformation, promoting cell survival, cell proliferation, and increased angiogenesis through critical molecules or pathway such as c-MYC or TGFβ signaling40,41,46,47. In this regard, our result from GSEA analysis showing that a high-risk score was in a significant association with angiogenesis or EMT is compatible with the reports described above.
Bone is the most frequent metastatic site in breast cancer, which remains an incurable disease. Therefore, novel prognostic biomarkers specific for bone recurrence of breast cancer, including the miRNA-based risk score we developed in this study, may improve risk assessment to guide adjuvant therapy, allowing patients with a high-risk of bone recurrence to receive more aggressive follow-up and/or more effective adjuvant therapy. The biomarkers may also identify early bone metastases.
Some studies have reported therapeutic potentials of the miRNAs we have used for the scoring. For instance, miR-106a can negatively regulate ZBTB4 expression, and overexpression or restoration of ZBTB4 by antagomir of miR-106a inhibits growth and invasion of breast cancer48. Another report demonstrated that suppressed miR-19a expression upregulate Fra-1 expression and induces M2 macrophage polarization, and miR-19a inhibits breast cancer progression and metastasis49. Together, these miRNAs can be potential therapeutic targets in addition to be diagnostic markers in breast cancer patients. Further investigation is warranted.
There are two major limitations in the present study. 1) First, the follow-up data in TCGA seems to be insufficient. We believe that our methodology in the development of the miRNA-based risk score, utilizing two distinct survivor groups; long- and short-term survivor, could emphasize the biological feature that could impact patient survival, and overcome this limitation for the “short follow-up” issue. Another limitation of our study is that the three miRNAs score was derived from women who received standard-of-care based on intrinsic subtype and stage. This study cannot discern if it is a general prognostic marker or also a predictive marker for response to treatment. Although we should recognize the limitation listed above, we believe the current study is in line with the important concept of “Building Bridges between Basic and Clinical Genomic Research” in translational research, which has been recently stressed in some commentaries50–52. It is a good example of utilizing bioinformatics approach on established large cohorts to clarify the clinical relevance of genomic and/or epigenomic biomarkers. As it has been emphasized by many, the utilization of Big Data is expected to become a common modality of analyses in very near future.
In conclusions, on the basis of unbiased integrated analysis utilizing large publically available cohorts, we identified a promising risk score based on three miRNAs in primary breast cancer for prognosis of survival outcomes and bone metastasis. Utilizing this score may allow for more aggressive intervention or intensive follow-up tailored to patients at high risk following initial breast cancer diagnosis.
Supplementary Material
Supplemental Table S8 HALLMARK_ANGIOGENE
Supplemental Table S3 Top 19 miRNAs selected based on BH fdr < 0.1 from DE results
Supplemental Table S4 Identification of miRNAs signature associated with prognosis using stepwise model selection based on Akaike Information Criterion on a multivariate Cox proportional hazard model with the rest of 14 miRNAs candidates
Suppletmental Table S5 Association between the three miRNA score (high and low) and patient characteristics in TCGA
Supplemental Table S6
Supplemental Table S7
Supplemental Table S9 HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION
Supplemental Table S10
Supplemental Table S11
Supplemental Table S12
Supplemental Figure S1 Hierarchical clustering of 19 miRNAs as our candidates, which showed most different expression levels in the two groups (“long” or “short” survival group) using TCGA.
Supplemental Figure S2 Overall survival curve using multivariate Cox model with the three-miRNA signature indicated that the its score showed prognostic impact on different stages and subtypes.
Supplemental Table S1 Summary of patient demographics and clinic characteristics.
Supplemental Table S2 Summary of patient demographics and clinical characteristics in the validation cohorts.
Synopsis.
Utilizing integrated analyses with multiple large cohorts, novel miRNA-based risk score was developed to predict bone recurrence potential and worse survival in breast cancer.
Acknowledgements
The authors acknowledge Bioinformatics and Biostatistics Shared Resources supported by National Cancer Institute (NCI) grant P30CA016056, as well as Pathology Resource Network and Clinical Data Delivery and Honest Broker services provided by the Clinical Data Network, which is funded by the National Cancer Institute and Roswell Park Cancer Institute Cancer Center Support Grant shared resource.
Funding
Kazuaki Takabe is supported by NIH/NCI grant R01CA160688 and Susan G. Komen Investigator Initiated Research Grant IIR12222224, as well as Institutional Grant 71-4085-01.
Footnotes
Ethics approval and consent to participate
The study was approved by the Institutional Review Board of Roswell Park Cancer Institute for human subject protection.
Consent for publication
Not applicable
Availability of data and materials
The datasets generated during the current study are available from the corresponding author on reasonable request
Competing Interests section
The authors declare that they have no competing interests.
References
- 1.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. August 17 2000;406(6797):747–752. [DOI] [PubMed] [Google Scholar]
- 2.Sparano JA, Gray RJ, Makower DF, et al. Prospective Validation of a 21-Gene Expression Assay in Breast Cancer. The New England journal of medicine. November 19 2015;373(21):2005–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America. September 11 2001;98(19):10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.SEER Stat Fact Sheets: Female Breast Cancer. 2016; http://seer.cancer.gov/statfacts/html/breast.html, 2016.
- 5.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA: a cancer journal for clinicians. Jan-Feb 2014;64(1):52–62. [DOI] [PubMed] [Google Scholar]
- 6.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. December 3 1993;75(5):843–854. [DOI] [PubMed] [Google Scholar]
- 7.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science (New York, N.Y.). October 26 2001;294(5543):862–864. [DOI] [PubMed] [Google Scholar]
- 8.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. June 9 2005;435(7043):828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. June 9 2005;435(7043):834–838. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews. Cancer. November 2006;6(11):857–866. [DOI] [PubMed] [Google Scholar]
- 11.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. June 28 2007;447(7148):1130–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer research. August 15 2005;65(16):7065–7070. [DOI] [PubMed] [Google Scholar]
- 13.Corcoran C, Friel AM, Duffy MJ, Crown J, O’Driscoll L. Intracellular and extracellular microRNAs in breast cancer. Clinical chemistry. January 2011;57(1):18–32. [DOI] [PubMed] [Google Scholar]
- 14.Comprehensive molecular portraits of human breast tumours. Nature. October 4 2012;490(7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pereira B, Chin SF, Rueda OM, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nature communications. May 10 2016;7:11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. April 18 2012;486(7403):346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enerly E, Steinfeld I, Kleivi K, et al. miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PloS one. February 22 2011;6(2):e16915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buffa FM, Camps C, Winchester L, et al. microRN A - a ssociated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer research. September 01 2011;71(17):5635–5645. [DOI] [PubMed] [Google Scholar]
- 19.Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn HJ. Strategies for subtypes--dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Annals of oncology ·: official, journal of the European Society for Medical Oncology / ESMO. August 2011;22(8):1736–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sobin LH GM, Wittekind C. TNM Classification of Malignant Tumours seventh edition. In: Cancer IUA, ed. New York: Wiley-Blackwell; 2009. [Google Scholar]
- 21.Gendoo DM, Ratanasirigulchai N, Schroder MS, et al. Genefu: an R/Bioconductor package for computation of gene expression-based signatures in breast cancer. Bioinformatics (Oxford, England). April 01 2016;32(7):1097–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crowley JLM, Jacobson J, Salmon S. Proceedings of the First Seattle Symposium in Biostatistics Survival Analysis. Vol 123 New York: Springer; 1997. [Google Scholar]
- 24.Kim SY, Kawaguchi T, Yan L, Young J, Qi Q, Takabe K. Clinical Relevance of microRNA Expressions in Breast Cancer Validated Using the Cancer Genome Atlas (TCGA). Ann Surg Oncol. August 01 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramanathan R, Olex AL, Dozmorov M, Bear HD, Fernandez LJ, Takabe K. Angiopoietin pathway gene expression associated with poor breast cancer survival. Breast cancer research and treatment. February 2017;162(1):191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young J, Kawaguchi T, Yan L, Qi Q, Liu S, Takabe K. Tamoxifen sensitivity-related microRNA-342 is a useful biomarker for breast cancer survival. Oncotarget. November 21 2017;8(59):99978–99989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawaguchi T, Yan L, Qi Q, et al. Overexpression of suppressive microRNAs, miR-30a and miR-200c are associated with improved survival of breast cancer patients. Scientific reports. November 21 2017;7(1):15945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoerl AK R Ridge Regression: Biased Estimation for Nonorthogonal Problems. TECHNOMETRICS. 1970;12(1):55–67. [Google Scholar]
- 29.Kim SY, Kawaguchi T, Yan L, Young J, Qi Q, Takabe K. Clinical Relevance of microRNA Expressions in Breast Cancer Validated Using the Cancer Genome Atlas (TCGA). Ann Surg Oncol. October 2017;24(10):2943–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narayanan S, Kawaguchi T, Yan L, Peng X, Qi Q, Takabe K. Cytolytic Activity Score to Assess Anticancer Immunity in Colorectal Cancer. Ann Surg Oncol. May 16 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terakawa T, Katsuta E, Yan L, et al. High expression of SLCO2B1 is associated with prostate cancer recurrence after radical prostatectomy. Oncotarget. March 6 2018;9(18):14207–14218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. October 25 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK). Journal of the National Cancer Institute. August 17 2005;97(16):1180–1184. [DOI] [PubMed] [Google Scholar]
- 34.McBryan J, Fagan A, McCartan D, et al. Transcriptomic Profiling of Sequential Tumors from Breast Cancer Patients Provides a Global View of Metastatic Expression Changes Following Endocrine Therapy. Clinical, cancer research : an official journal of the American Association for Cancer Research. December 1 2015;21(23):5371–5379. [DOI] [PubMed] [Google Scholar]
- 35.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell systems. December 23 2015;1(6):417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Volinia S, Croce CM. Prognostic microRNA/mRNA signature from the integrated analysis of patients with invasive breast cancer. Proceedings of the National Academy of Sciences of the United States of America. April 30 2013;110(18):7413–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng F, Zhang Y, Wang R, et al. Identification of differentially expressed miRNAs in individual breast cancer patient and application in personalized medicine. Oncogenesis. 2016;5:e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu X, Zeng R, Wu S, Zhong J, Yang L, Xu J. Comprehensive expression analysis of miRNA in breast cancer at the miRNA and isomiR levels. Gene. February 25 2015;557(2):195–200. [DOI] [PubMed] [Google Scholar]
- 39.Zhou X, Wang X, Huang Z, Xu L, Zhu W, Liu P. An ER-associated miRNA signature predicts prognosis in ER-positive breast cancer. Journal of experimental & clinical cancer research : CR. 2014;33:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dews M, Homayouni A, Yu D, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nature genetics. September 2006;38(9):1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dews M, Fox JL, Hultine S, et al. The myc-miR-17~92 axis blunts TGF{beta} signaling and production of multiple TGF{beta}-dependent antiangiogenic factors. Cancer research. October 15 2010;70(20):8233–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z, Yang CS, Nakashima K, Rana TM. Small RNA-mediated regulation of iPS cell generation. The EMBO journal. March 2 2011;30(5):823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nature reviews. Molecular cell biology. March 2008;9(3):219–230. [DOI] [PubMed] [Google Scholar]
- 44.Mendell JT. miRiad roles for the miR-17–92 cluster in development and disease. Cell. April 18 2008;133(2):217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17–92 clusters in the control of transforming growth factor beta signaling. Cancer research. October 15 2008;68(20):8191–8194. [DOI] [PubMed] [Google Scholar]
- 46.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. June 9 2005;435(7043):839–843. [DOI] [PubMed] [Google Scholar]
- 47.Dal Bo M, Bomben R, Hernandez L, Gattei V. The MYC/miR-17–92 axis in lymphoproliferative disorders: A common pathway with therapeutic potential. Oncotarget. August 14 2015;6(23):19381–19392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim K, Chadalapaka G, Lee SO, et al. Identification of oncogenic microRNA-17–92/ZBTB4/specificity protein axis in breast cancer. Oncogene. February 23 2012;31(8):1034–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang J, Zhang Z, Chen C, et al. MicroRNA-19a-3p inhibits breast cancer progression and metastasis by inducing macrophage polarization through downregulated expression of Fra-1 proto-oncogene. Oncogene. June 5 2014;33(23):3014–3023. [DOI] [PubMed] [Google Scholar]
- 50.Conley RB, Dickson D, Zenklusen JC, et al. Core Clinical Data Elements for Cancer Genomic Repositories: A Multi-stakeholder Consensus. Cell. November 16 2017;171(5):982–986. [DOI] [PubMed] [Google Scholar]
- 51.Manolio TA, Fowler DM, Starita LM, et al. Bedside Back to Bench’ Building Bridges between Basic and Clinical Genomic Research. Cell. March 23 2017;169(1):6–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodriguez H, Pennington SR. Revolutionizing Precision Oncology through Collaborative Proteogenomics and Data Sharing. Cell. April 19 2018;173(3):535–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S8 HALLMARK_ANGIOGENE
Supplemental Table S3 Top 19 miRNAs selected based on BH fdr < 0.1 from DE results
Supplemental Table S4 Identification of miRNAs signature associated with prognosis using stepwise model selection based on Akaike Information Criterion on a multivariate Cox proportional hazard model with the rest of 14 miRNAs candidates
Suppletmental Table S5 Association between the three miRNA score (high and low) and patient characteristics in TCGA
Supplemental Table S6
Supplemental Table S7
Supplemental Table S9 HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION
Supplemental Table S10
Supplemental Table S11
Supplemental Table S12
Supplemental Figure S1 Hierarchical clustering of 19 miRNAs as our candidates, which showed most different expression levels in the two groups (“long” or “short” survival group) using TCGA.
Supplemental Figure S2 Overall survival curve using multivariate Cox model with the three-miRNA signature indicated that the its score showed prognostic impact on different stages and subtypes.
Supplemental Table S1 Summary of patient demographics and clinic characteristics.
Supplemental Table S2 Summary of patient demographics and clinical characteristics in the validation cohorts.