Abstract
Purpose
Serine Peptidase Inhibitor, Kazal type-1 (SPINK1) overexpression defines the second most recurrent and aggressive prostate cancer (PCa) subtype. However, the underlying molecular mechanism and pathobiology of SPINK1 in PCa remains largely unknown.
Experimental Design
MicroRNA-prediction tools were employed to examine the SPINK1-3’UTR for miRNAs binding. Luciferase reporter assays were performed to confirm the SPINK1-3’UTR binding of shortlisted miR-338-5p/miR-421. Further, miR-338-5p/-421 overexpressing cancer cells (SPINK1-positive) were evaluated for oncogenic properties using cell-based functional assays and mice xenograft model. Global gene expression profiling was performed to unravel the biological pathways altered by miR-338-5p/-421. Immunohistochemistry and RNA in-situ hybridization was carried-out on PCa patients’ tissue microarray for SPINK1 and EZH2 expression respectively. Chromatin immunoprecipitation assay was performed to examine EZH2 occupancy on the miR-338-5p/-421 regulatory regions. Bisulfite sequencing and methylated DNA-immunoprecipitation was performed on PCa cell lines and patients’ specimens.
Results
We established a critical role of miRNA-338-5p/-421 in post-transcriptional regulation of SPINK1. Ectopic expression of miRNA-338-5p/-421 in SPINK1-positive cells abrogate oncogenic properties including cell-cycle progression, stemness and drug resistance, and show reduced tumor burden and distant metastases in mice model. Importantly, we show SPINK1-positive PCa patients exhibit increased EZH2 expression, suggesting its role in epigenetic silencing of miRNA-338-5p/-421. Furthermore, presence of CpG dinucleotide DNA methylation marks on the regulatory regions of miR-338-5p/-421 in SPINK1-positive PCa cells and patients’ specimens confirms epigenetic silencing.
Conclusion
Our findings revealed that miRNA-338-5p/-421 are epigenetically silenced in SPINK1-positive PCa, while restoring the expression of these miRNAs using epigenetic drugs or synthetic mimics could abrogate SPINK1-mediated oncogenesis.
Introduction
Prostate Cancer (PCa) is characterized by extensive molecular heterogeneity and varied clinical outcomes (1). Multiple molecular subtypes involving recurrent genetic rearrangements, DNA copy number alterations, and somatic mutations have been associated with this disease (1–3). Majority of these patients harbor gene rearrangements between members of the E26 transformation-specific (ETS) transcription factor family and the androgen-regulated transmembrane protease serine 2 (TMPRSS2), most recurrent (~50%) being TMPRSS2-ERG, involving the v-ets erythroblastosis virus E26-oncogene homolog (ERG) (3,4). The TMPRSS2-ERG encoded ERG transcription factor is known to drive cell invasion and metastases, DNA damage in-vitro and focal pre-cancerous prostatic intraepithelial neoplasia (PIN) lesions in transgenic mice (5).
While TMPRSS2-ERG fusion forms the most frequent molecular subtype, a significant subset of ETS-negative (–) PCa show overexpression of Serine Peptidase Inhibitor, Kazal type-1 (SPINK1) in ~10-15% of the total PCa patients, a distinct subtype defined by overall higher Gleason score, shorter progression-free survival and biochemical recurrence (6–8). SPINK1 promotes cell proliferation and invasion through autocrine/paracrine signaling and mediate its oncogenic effects in part through EGFR interaction by activating downstream signaling. Monoclonal EGFR antibody administered in SPINK1-positive xenografted mice showed only a marginal decrease in tumor burden, suggesting involvement of EGFR-independent oncogenic pathways (9).
Although, genomic events such as gene rearrangements and somatic mutations constitute most recurrent oncogenic aberrations, many could also be attributed to epigenetic alterations. Earlier studies have shown that aberrant expression of Enhancer of Zeste Homolog 2 (EZH2) owing to genomic loss of miRNA-101 (10) or hypermethylation of miR-26a (11) constitutes a common regulatory mechanism across several solid cancers. EZH2, being the key component of the Polycomb-Repressive Complex 2 (PRC2) mediates trimethylation on the histone 3 lysine 27 (H3K27me3), leading to gene silencing (12). However, phosphorylated form of EZH2 is known to switch its function from Polycomb repressor to transcriptional coactivator of androgen receptor in castration-resistant prostate cancers (CRPC) (13). Moreover, recent studies have shown PRC2 epigenetically suppresses the expression of several tumor suppressive miRNAs such as, miR-181a/b, miR-200b/c, and miR-203, while these miRNAs in turn directly target PRC1 members, namely BMI1 and RING2, thereby reinforcing the repressive molecular circuitry (14).
Although overexpression of SPINK1 forms the second most prevalent and aggressive subtype of PCa (4,6), the underlying mechanism involved in its upregulation is poorly understood and remains a matter of conjecture. Further, SPINK1 overexpression is not ascribed to chromosomal rearrangement, deletion, or amplification (6), and thus alludes to a possible transcriptional or post-transcriptional regulation. A recent study showed the transcriptional activation of SPINK1 along with gastrointestinal (GI) lineage signature genes in CRPC patients (15). Our study focuses on the post-transcriptional regulation of SPINK1 expression by miR-338-5p and miR-421, which are epigenetically silenced in SPINK1-positive prostate cancer. We also provide evidence that EZH2 acts as an epigenetic switch, thereby promoting transcriptional silencing of miR-338-5p/miR-421 by establishing H3K27me3 repressive marks, thus leading to SPINK1 overexpression. Collectively, our findings suggest potential benefits with epigenetic inhibitors or synthetic miR-338-5p/-421 mimics as adjuvant therapy for the treatment of aggressive SPINK1+ malignancies.
Materials and Methods
Human Prostate Cancer Specimens
Fresh-frozen PCa specimens used in this study were procured from King George's Medical University (KGMU), Lucknow, India. Clinical specimens were collected after obtaining written informed consent and Institutional Review Board approvals from KGMU and Indian Institute of Technology, Kanpur, India. A total of 20 PCa specimens were selected for this study based on SPINK1 and TMPRSS2-ERG status, confirmed by qPCR, immunohistochemistry and Fluorescent in situ hybridization (4). Tissue microarrays (TMA) comprising PCa specimens (n=238) were obtained from Dept. of Pathology, Henry Ford Health System, Detroit, Michigan, USA, after getting written informed consent and Institutional Review Board approval. All patients’ specimens used in this study were collected in accordance to the Declaration of Helsinki. TMAs were stained for SPINK1 and EZH2 by performing immunohistochemistry and RNA in situ hybridization (RNA-ISH) respectively.
Mice Xenograft Studies
For mice xenograft studies, five to six weeks old NOD.CB17-Prkdcscid/J (NOD/SCID) male mice (Jackson Laboratory) were randomized into three groups (n=8 for each experimental condition). All procedures involving mice were approved by the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA) and conform to all regulatory standards of the Institutional Animal Ethics Committee of the Indian Institute of Technology Kanpur. Detailed methodology of the xenograft studies is provided in the Supplementary Methods.
MicroRNA 3’UTR SPINK1 luciferase reporter assay
Full length SPINK1 3’ untranslated region (3’UTR) wild type, and mutant with altered residues in the binding sites of miR-338-5p and miR-421 was clone in Firefly/Renilla Dual-Luciferase reporter vector pEZX-MT01 (GeneCopoeia). Cells were seeded in a 24-well plate at 30-40% confluency, and co-transfected with 30pmol of miRNA mimics along with 25ng of pEZX-MT01 constructs using lipofectamine RNAiMax (Invitrogen). Luciferase assay was performed using Dual-Glo luciferase assay (Promega) 24 hours after the second transfection. Firefly luciferase activity was obtained by normalizing with Renilla for each sample.
Gene expression array analysis
Total RNA was isolated from stable 22RV1-miR-338-5p, 22RV1-miR-421 and 22RV1-CTL cells and subjected to Agilent Whole Human Genome Oligo Microarray profiling (dual color) using Agilent platform (8x60K format) according to the manufacturer’s protocol. Microarray hybridization was performed using three independent stable miRNA-overexpressing clones against control cells. Microarray data was normalized by following locally weighted linear regression (Lowess normalization). Further, details of differential expression analysis are provided in Supplementary Methods.
Statistical analysis
Statistical significance was determined by either two-tailed Student's t-test for independent samples or one-way Analysis of Variance (ANOVA), otherwise specified. The differences between the experimental groups were considered significant if the p-value was of less than 0.05. Error bars represent mean ± SEM. All experiments were repeated three times in triplicates.
Supplementary methods
Additional methodological details can be found in the Supplementary Methods.
Results
Identification of differentially expressed miRNAs in SPINK1+/ERG-fusion-negative prostate cancer
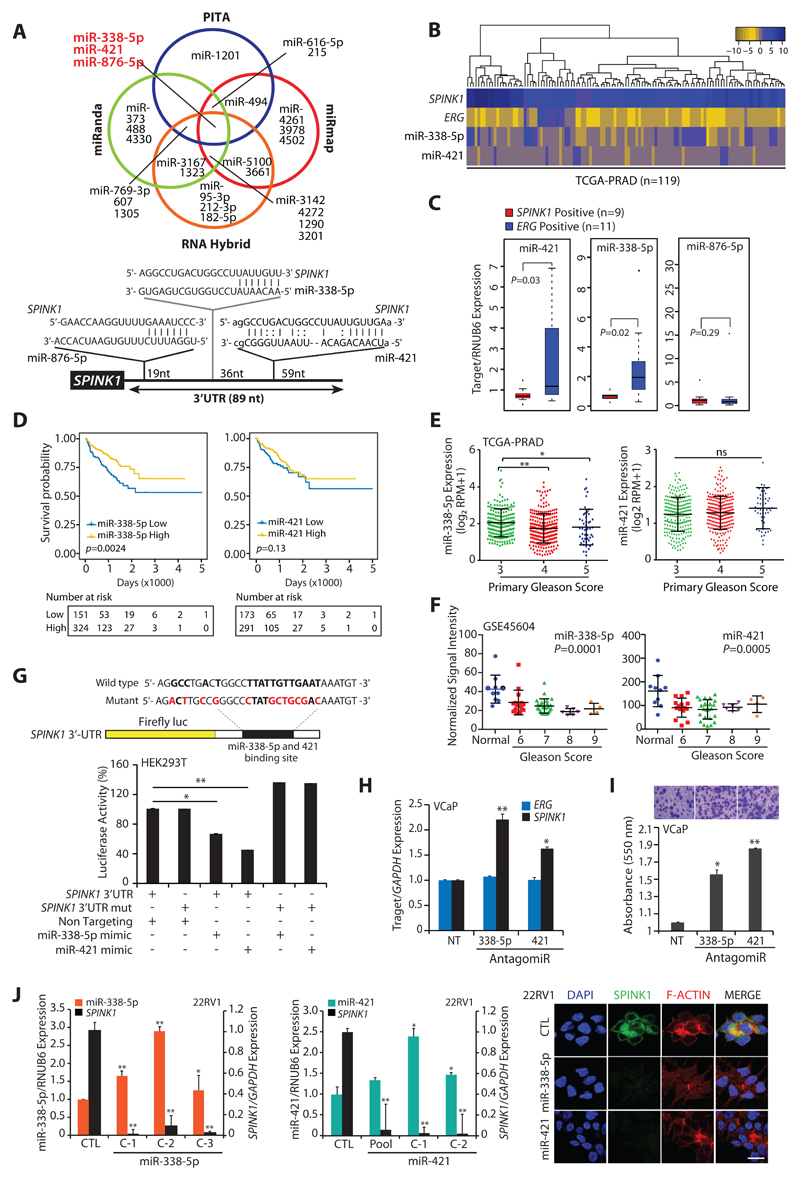
We employed four miRNA prediction algorithms, namely PITA (omictools.com), miRmap (mirmap.ezlab.org), miRanda (microrna.org) and RNAHybrid (BiBiserv2-RNAhybrid) to examine putative binding of miRNAs to the 3’UTR of SPINK1 transcript. Notably, three miR-338-5p, -421 and -876-5p were predicted as strong candidates by all four algorithms (Fig. 1A and Supplementary Table S1), and were taken forward for further investigation. To examine the differential expression of these miRNAs between SPINK1+ and ERG+ patients’ specimens, publicly available RNA-seq data, The Cancer Genome Atlas Prostate Adenocarcinoma (TCGA-PRAD) was analyzed. Interestingly, hierarchical clustering of TCGA-PRAD RNA-Seq data exhibit reduced expression of miR-338-5p and miR-421 (miR-338-5p/-421) in SPINK1+/ERG-negative patients (Fig. 1B). To validate further, we examined the expression of miR-338-5p/-421 and miR-876-5p in our PCa patients’ specimens. A significant lower expression of miR-338-5p and miR-421 was observed specifically in SPINK1+ as compared to ERG+ specimens, while no difference in miR-876-5p expression was noticed (Fig. 1C). To understand the clinical significance of miR-338-5p/-421, we stratified TCGA-PRAD data into high and low miRNAs expressing groups. Intriguingly, patients with low miR-338-5p expression showed significant (p=0.0024) association with decreased survival probability compared to high miRNAs group, while no such association was found in case of miR-421 (Fig. 1D). We speculate that this could be attributed due to narrow range of miRNA-421 expression in TCGA-PRAD cohort. Moreover, lower expression of miR-338-5p also associate with higher Gleason score (Fig. 1E). Similar association for both miRNAs was further confirmed in another independent cohort (GSE45604) (Fig. 1F). In summary, SPINK1+ subtype show lower expression of miR-338-5p/-421, which strongly associate with overall poor survival and aggressiveness of the disease.
Figure 1. MiR-338-5p and miR-421 are differentially expressed in SPINK1+/ERG-fusion-negative prostate cancer.
(A) Venn diagram displaying miRNAs computationally predicted to target SPINK1 by PITA, miRmap, miRanda and RNAHybrid (top panel). Schematic of predicted miR-338-5p, miR-421 and miR-876-5p binding sites on the 3′-UTR of SPINK1 (bottom panel).
(B) Heatmap depicting miR-338-5p and miR-421 expression in the SPINK1+/ERG-negative patients’ (n=119) in TCGA-PRAD dataset. Shades of blue and golden represents log2 (x+1), where x represents the gene expression value.
(C) Taqman assay showing relative expression for miR-338-5p, miR-421 and miR-876-5p in SPINK1+ and ERG+ PCa patients’ specimens (n=20). Data represents normalized expression values with respect to RNUB6 control. Error bars represent mean ± SEM. P-values were calculated using two-tailed unpaired Student's t test.
(D) Kaplan-Meier curve showing survival probability in TCGA-PRAD cohort stratified based on high versus low miR-338-5p and miR-421expression (n=475 and n=465 respectively).
(E) RNA-Seq data from TCGA-PRAD cohort showing expression of miR-338-5p (n=475) and miR-421 (n=465) in PCa patients categorized by varying primary Gleason Score.
(F) Expression of miR-338-5p and miR-421 in PCa patients (normal=10, PCa =50) categorized by Gleason grades (from GSE45604 dataset).
(G) Schematic of luciferase reporter construct with the wild-type or mutated (altered residues in red) SPINK1 3’ untranslated region (3’UTR) downstream of the Firefly luciferase reporter gene (top panel). Luciferase reporter activity in HEK293T cells co-transfected with wild-type or mutant 3′-UTR SPINK1 constructs with mimics for miR-338-5p or miR-421.
(H) QPCR data showing SPINK1 and ERG expression in VCaP cells transfected with antagomiRs as indicated (n=3 biologically independent samples; data represent mean ± SEM).
(I) Boyden chamber Matrigel invasion assay using same cells as in (E). Representative fields of the invaded cells are shown in the inset.
(J) QPCR analysis demonstrating SPINK1 and miRNAs expression in stable 22RV1-miR-338-5p (left panel) and 22RV1-miR-421 cells (middle panel) (n=3 biologically independent samples; data represent mean ± SEM). Immunostaining for SPINK1 (right panel). Scale bar represents 20µm.
Statistical significance was calculated by one-way ANOVA with Tukey’s post hoc test for multiple comparisons in the pane E and F. For all other panels *P ≤ 0.05 and **P ≤ 0.005 using two-tailed unpaired Student's t test.
MiR-338-5p and miR-421 directly target SPINK1 and modulate its expression
Having established an association between miR-338-5p/-421 and SPINK1 expression in PCa specimens, we next examined the ability of these miRNAs to bind to SPINK1-3'-UTR. The wild-type (3’-UTR-WT) and mutant (3’-UTR-mut) SPINK1-3'-UTR cloned in Firefly/Renilla dual-luciferase reporter vectors were co-transfected with synthetic mimics for miR-338-5p or miR-421 in HEK293T cells, a significant reduction in the luciferase activity was noted with 3’-UTR-WT, while no suppressive effect was observed in 3’-UTR-mut constructs (Fig. 1G). We next evaluated the expression of these miRNAs in various PCa cell lines including 22RV1 (SPINK1+), ETS-fusion positive VCaP (TMPRSS2-ERG+) and LNCaP (ETV1+) cells. Supporting our observation in clinical specimens, the cell line data also showed lower expression of miR-338-5p/-421 in 22RV1 cells relative to ETS-fusion-positive cell lines (Supplementary Fig. S1A). To further ascertain that miR-338-5p/-421 specifically regulates SPINK1, we used antagomiRs (anti-miRs) to abrogate miR-338-5p and miR-421 expression (anti-338-5p and anti-421, respectively) in VCaP cells (Supplementary Fig. S1B). As expected, anti-338-5p or anti-421 significantly induced SPINK1 expression with concomitant increase in cell invasion and migration (Fig. 1H, I and Supplementary Fig. S1C, D), while there was no change in the endogenous ERG expression (Fig. 1H and Supplementary Fig. S1C). We next established stable miR-338-5p or miR-421 overexpressing 22RV1 cells (22RV1-miR-338-5p and 22RV1-miR-421, respectively) and examined SPINK1 expression, a significant reduction in SPINK1 both at transcript (~80-90%) and protein (Fig. 1J) levels was observed. Since, SPINK1 overexpression has also been implicated in colorectal, lung, pancreatic, breast and ovarian cancers (16,17), we sought to examine if SPINK1 is regulated by a similar mechanism in cancers of different cellular/tissue origins. Thus, we determined the status of SPINK1 expression in multiple cancer cell lines (Supplementary Fig. S2A, B). Further, SPINK1+ cancer cell lines, namely, colorectal (WiDr), melanoma (SK-MEL-173), pancreatic (CAPAN-1) and prostate (22RV1) transfected with mimics for miR-338-5p or miR-421 showed a significant decrease in SPINK1 expression (Supplementary Fig. S2C, D). This provides irrevocable evidence that these two miRNAs modulate the expression of SPINK1 transcript irrespective of the tissue background. To ascertain whether decrease in oncogenic properties is indeed due to miR-338/-421 mediated reduction in SPINK1 expression, a rescue cell migration assay using human recombinant SPINK1 (rSPINK1) was performed. As expected, 22RV1-miR-338/-421 cells show decrease in migration, while adding rSPINK1 to these miRNAs-overexpressing cells rescued the invasive phenotype, indicating that miR-338/-421 mediated effects are indeed due to reduced SPINK1 expression (Supplementary Fig. S2E).
Ectopic expression of miR-338-5p and miR-421 attenuate SPINK1-mediated oncogenesis
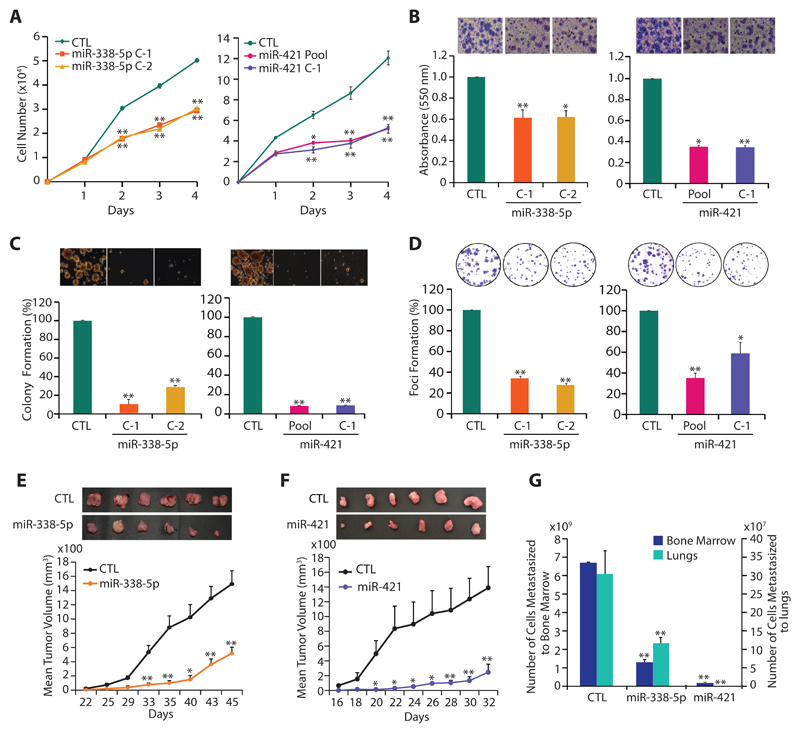
SPINK1 overexpression is known to contribute to cell proliferation, invasion, motility and distant metastases (4,6,18). Hence, to understand the functional relevance of miR-338-5p/-421, we examined 22RV1-miR-338-5p and 22RV1-miR-421 cells for any change in their oncogenic properties. Both 22RV1-miR-338-5p (C1 and C2) and 22RV1-miR-421 (pooled and C1) cells showed a significant decrease in cell proliferation compared to 22RV1-CTL cells (Fig. 2A). Similarly, a significant reduction in invasive properties of 22RV1-miR-338-5p and 22RV1-miR-421 cells were noted (~40% and 60% respectively) (Fig. 2B). While, only a modest decrease in cell proliferation and invasion was observed in pooled 22RV1-miR-338-5p cells (Supplementary Fig. S3A-C). To assess any change in neoplastic transformation, soft agar colony formation assay was performed, where both 22RV1-miR-338-5p and 22RV1-miR-421 cells exhibited marked reduction (~60% and ~80% respectively) in number and size of the colonies (Fig. 2C). Likewise, 22RV1-miR-338-5p and 22RV1-miR-421 cells demonstrate significantly lower numbers (~70% and ~60% respectively) of dense foci (Fig. 2D). While, overexpressing miR-338-5p/-421 in immortalized benign prostate epithelial RWPE-1 cells show no significant change in cell proliferation or migration with miR-338-5p mimics transfection, but a marginal decrease in proliferation and migration was noted with miR-421 (Supplementary Fig. S3D). Further, to demonstrate that miR-338-5p/-421 modulate SPINK1 expression and attenuate SPINK1-mediated oncogenicity irrespective of the tissue background, we performed functional assays using colorectal carcinoma WiDr cells (SPINK1+) stably overexpressing these miRNAs. As anticipated, a significant decrease in the oncogenic potential of the miR-338-5p/-421 overexpressing WiDr cells was observed (Supplementary Fig. S3E, F).
Figure 2. MiR-338-5p and miR-421 abrogates oncogenic properties of SPINK1-positive prostate cancer cells.
(A) Cell proliferation assay using 22RV1-miR-338-5p, 22RV1-miR-421 and 22RV1-CTL cells at the indicated time points.
(B) Boyden chamber Matrigel invasion assay using same cells as in (A). Representative fields with invaded cells are shown in the inset (n=3 biologically independent samples; data represent mean ± SEM).
(C) Soft agar assay for anchorage-independent growth using same cells as in (A). Representative soft agar colonies are shown in the inset (n=3 biologically independent samples; data represent mean ± SEM).
(D) Foci formation assay using same cells as in (A). Representative images depicting foci are shown in the inset (n=3 biologically independent samples; data represent mean ± SEM).
(E) Mean tumor growth in NOD/SCID mice (n=8) subcutaneously implanted with stable 22RV1-miR-338-5p and 22RV1-CTL cells.
(F) Same as (E), except stable 22RV1-miR-421 cells were implanted.
(G) Same as (E and F), except genomic DNA extracted from the lung and bone marrow of the xenografted mice.
Data represent mean ± SEM. *P≤ 0.05 and **P≤ 0.005 using two-tailed unpaired Student's t test.
To examine tumorigenic potential of 22RV1-miR-338-5p and 22RV1-miR-421 cells in-vivo, chick chorioallantoic membrane (CAM) assay was performed, and relative number of intravasated cancer cells was analyzed. Consistent with in-vitro results, 22RV1-miR-338-5p and 22RV1-miR-421 cells showed significant reduction in the number of intravasated cells and tumor weight, compared to control (Supplementary Fig. S4A-C). To evaluate distant metastases, lungs and liver excised from the chick-embryos were characterized for metastasized cancer cells. The groups implanted with miRNAs-overexpressing cells revealed ~80% reduction in cancer cell metastases to lungs (Supplementary Fig. S4D), while no sign of liver metastases was observed in either group. Further, tumor xenograft experiment was recapitulated in immunodeficient NOD/SCID mice (n=8 per group) by subcutaneously implanting 22RV1-miR-338-5p, 22RV1-miR-421 and 22RV1-CTL cells into flank region, and trend of tumor growth was recorded. A significant reduction in the tumor burden was observed in the mice bearing miR-338-5p and miR-421 overexpressing xenografts as compared to control (~70% and 85% reduction respectively) (Fig. 2E, F). To examine spontaneous metastases, lung, liver and bone marrow were excised from the xenografted mice, and genomic DNA was quantified for the presence of human specific Alu-sequences. A significant decrease (~85% for miR-338-5p and ~90% for miR-421) in cancer cell metastases was observed in the groups implanted with miRNA-338-5p/-421 overexpressing cells (Fig. 2G). Similar to CAM assay, cancer cells failed to metastasize to murine liver (data not shown). Furthermore, a significant drop (~50%) in Ki-67-positive cells in the miRNAs-overexpressing xenografts confirms that tumor regression was indeed due to decline in cell proliferation (Supplementary Fig. S4E). Taken together, our findings indicate that miR-338-5p/-421 downregulate the expression of SPINK1 and abrogate SPINK1-mediated oncogenic properties and tumorigenesis.
MiR-338-5p and miR-421 exhibit functional pleiotropy by regulating diverse biological processes
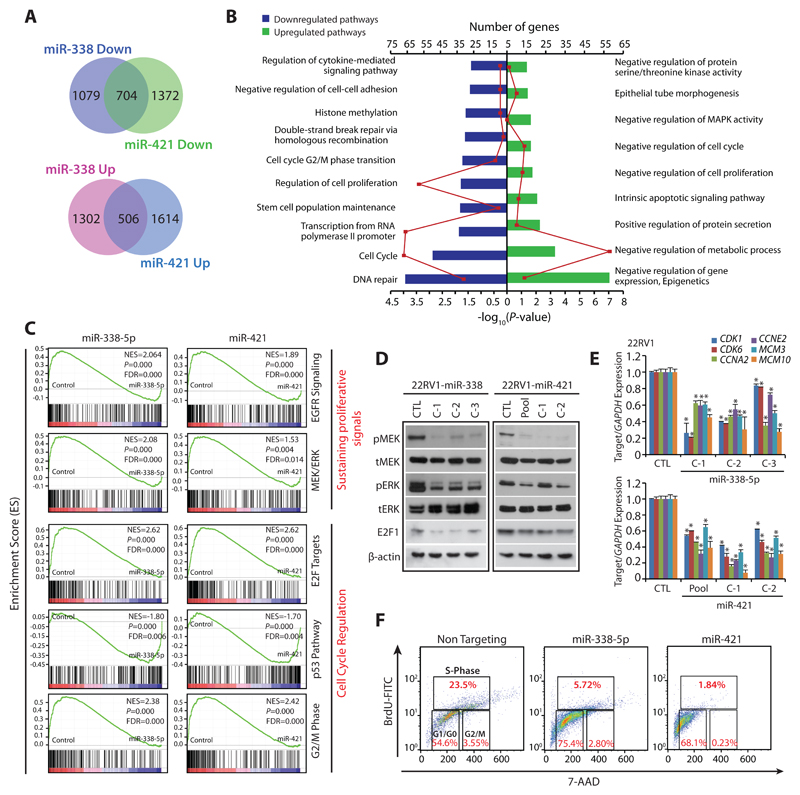
To explore critical biological pathways involved in the tumor-suppressive properties rendered by miR-338-5p/-421 in SPINK1+ cancers, we determined global gene expression profiles of miRNAs-overexpressing 22RV1 cells. Our analysis revealed 2,801 and 2,979 genes significantly dysregulated in 22RV1-miR-338-5p and 22RV1-miR-421 cells respectively relative to control (log2 fold change of 0.6, FDR<0.05 and p<0.05). Of these, ~22% (704 genes) of the downregulated and ~15% (506 genes) of the upregulated transcripts show an overlap in miRNAs-overexpressing cells (90% confidence interval) (Fig. 3A), indicating that these miRNAs regulate a significant number of common gene-sets and cellular processes. To examine biological processes commonly regulated by miR-338-5p/-421, we employed DAVID (Database for Annotation, Visualization and Integrated Discovery) and GSEA (Gene set enrichment analysis). Most of the downregulated genes were associated with DNA double-strand break repair by homologous recombination, cell-cycle regulation including G2/M-phase transition, stem-cell maintenance, histone methylation and negative regulation of cell-cell adhesion. Whereas, genes involved in negative regulation of gene expression or epigenetics, intrinsic apoptotic signaling pathways, negative regulation of metabolic process and cell-cycle were significantly upregulated (Fig. 3B, Supplementary Table S3). Moreover, GSEA also revealed a significant decrease in enrichment of genes associated with sustaining proliferative signaling (EGFR and MEK/ERK) and cell-cycle regulators (E2F targets and G2/M transition) in miRNA-overexpressing cells, while tumor suppressive p53 signaling was found to be positively enriched (Fig. 3C), indicating their role in reduced oncogenicity. Additionally, an overlapping network of pathways using Enrichment map revealed regulation of cell-cycle phase transition and DNA repair pathways (overlap coefficient=0.8, p<0.001, FDR=0.01), as one of the significantly enriched pathways for both miRNAs (Supplementary Fig. S5).
Figure 3. MiR-338-5p and miR-421 overexpression suppress oncogenic pathways and triggers G1/S arrest.
(A) Gene expression profiling data showing overlap of downregulated (upper panel) and upregulated genes (lower panel) in stable 22RV1-miR-338-5p and 22RV1-miR-421 cells relative to 22RV1-CTL cells (n=3 biologically independent samples).
(B) Same as in (A), except DAVID analysis showing various downregulated (left) and upregulated (right) pathways. Bars represent –log10 (P-values) and frequency polygon (line in red) represents the number of genes.
(C) Gene Set Enrichment Analysis (GSEA) plots showing various deregulated oncogenic gene signatures with the corresponding statistical metrics in the same cells as in (A).
(D) Western blot analysis for phosphor (p) and total (t) MEK1/2, ERK1/2 and cell-cycle regulator E2F1 levels. β-actin was used as a loading control.
(E) QPCR analysis showing expression of cell-cycle regulators for G1 and S phase as indicated. Expression level for each gene was normalized to GAPDH.
(F) BrdU/7-AAD cell-cycle analysis for S-phase arrest in 22RV1 cells transfected with miR-338-5p or miR-421 mimics relative to control cells.
In the panels (D), (E) and (F) biologically independent samples were used (n=3); data represents mean ± SEM *P≤ 0.05 and **P≤ 0.005 using two-tailed unpaired Student's t test.
Since MAPK signaling pathways involving a series of protein kinase cascades play a critical role in the regulation of cell proliferation, we examined the phosphorylation status of MEK (pMEK) and ERK (pERK), as a read-out of this pathway. In agreement with our in-silico analysis, a significant decrease in pMEK and pERK was observed in 22RV1-miR-338-5p and 22RV1-miR-421 cells (Fig. 3D). E2F transcription factors are known to interact with phosphorylated retinoblastoma, and positively regulate genes involved in S-phase entry and DNA synthesis (19), thus we next examined the E2F1 level in miRNAs-overexpressing cells, surprisingly a notable decrease in E2F1 was observed (Fig. 3D). Further, a significant decrease in the expression of genes involved in G1/S transition such as cyclin E2 (CCNE2), cyclin A2 (CCNA2) and cyclin-dependent kinase (CDK1 and CDK6), including mini-chromosome maintenance (MCM3 and MCM10), required for the initiation of eukaryotic replication machinery was recorded (Fig. 3E). Thus, these findings corroborate with previous literature that during DNA damage, CDKs being cell-cycle regulators crosstalk with the checkpoint activation network to temporarily halt the cell-cycle progression and promote DNA repair (20). Intriguingly, presence of putative miR-338-5p/miR-421 binding sites on the 3’UTRs of these cell-cycle regulators (Supplementary Table S4) further support that these targets could be directly controlled by these miRNAs. Next, to validate that miR-338-5p/-421 overexpression leads to cell-cycle arrest, cell-cycle analysis using miRNA mimics transfected 22RV1 cells, revealed a significant increase in cells arrested in S-phase (Supplementary Fig. S6A). To delineate that this increase in the S-phase cells is indeed due to cell-cycle arrest and not because of DNA replication, BrdU-7AAD-based cell-cycle analysis was performed, which revealed a significant decrease in the percentage of BrdU incorporated S-phase cells (Fig. 3F). Taken together, our findings strongly indicate that miR-338-5p/-421 overexpression led to S-phase arrest, thus elucidating the mechanism for reduced proliferation and dramatic regression in tumor growth.
Ectopic expression of miR-338-5p and miR-421 suppresses Epithelial-to-Mesenchymal Transition (EMT) and stemness
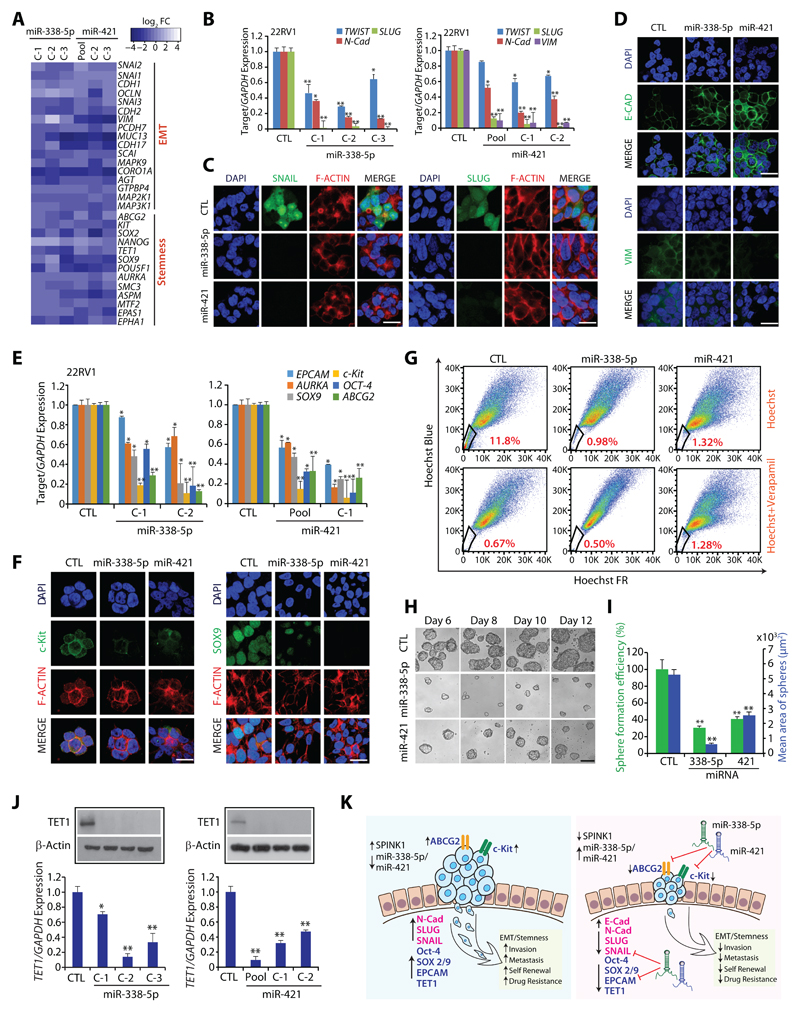
Association between EMT and cancer stem cells (CSCs) has been well-established, indicating that a subpopulation of neoplastic cells, which harbor self-renewal capacity and pluripotency, are associated with highly metastatic and drug-resistant cancers (21). To delineate the pathways involved in miR-338-5p/-421-mediated regression in tumor burden and metastases (Fig. 2E-G), we analyzed microarray data and noted a marked decrease in the expression of genes involved in EMT and stemness (Fig. 4A), including the EMT-inducing transcription factors namely, SNAI1 (SNAIL), SNAI2 (SLUG), and TWIST1 (Fig. 4B, C). Since, SNAIL and SLUG are known to negatively regulate CDH1 (E-Cadherin) (22), an epithelial marker involved in cell-cell adhesion, we next examined miR-338-5p/-421 overexpressing cells for any change in E-Cadherin expression, a prominent increase in the membrane localization of E-Cadherin, while a significant decrease in the expression of vimentin, a mesenchymal marker was observed (Fig. 4D).
Figure 4. MiR-338-5p and miR-421 overexpression attenuates EMT and Stemness.
(A) Heatmap depicting change in the expression of EMT and pluripotency markers in 22RV1-miR-338-5p and 22RV1-miR-421 cells. Shades of blue represents log2 fold-change in gene expression (n=3 biologically independent samples).
(B) QPCR analysis depicts expression of EMT markers in 22RV1-miR-338-5p, 22RV1-miR-421 and control cells. Expression for each gene was normalized to GAPDH.
(C) Immunostaining showing SLUG and SNAIL expression in the same cells as in (B).
(D) Same cells as in (B), except immunostained for E-cadherin and Vimentin.
(E) Same cells as in (B), except qPCR analysis for stem cell markers.
(F) Same cells as in (B), except immunostained for c-Kit and SOX-9.
(G) Hoechst 33342 staining for side population (SP) analysis using same cells as in (B). Percentages of SP were analyzed using the blue and far red filters, gated regions as indicated (red) in each panel.
(H) Phase contrast microscope images for the prostatospheres using same cells as in (B). Scale bar 100μm.
(I) Bar plot depicts percent sphere formation efficiency and mean area of the prostatosphere.
(J) Expression of TET1 by qPCR and Western blot using same cells as in (B).
(K) Schematic describing the role of miR-338-5p and miR-421 in regulating EMT, cancer stemness and drug resistance in SPINK1+ cancer.
For panels (C), (D) and (F), scale bar represents 20µm. In the panels (B), (E), (I) and (J) biologically independent samples were used (n=3); data represents mean ± SEM *P≤ 0.05 and **P≤ 0.005 using two-tailed unpaired Student's t test.
A sub-population (CD117+/ABCG2+) of 22RV1 cells, known as prostate carcinoma-initiating stem-like cells, has been shown to exhibit stemness and multi-drug resistance (23). Therefore, we examined the expression of genes associated with CSC-like properties in 22RV1-miR-338-5p/-421 cells. Strikingly, the expression of well-known pluripotency markers, such as AURKA, SOX9 and OCT-4, and stem-cell surface markers EPCAM, CD117 (c-Kit), and ABCG2, an ATP-binding cassette transporter, were markedly downregulated in miRNAs-overexpressing cells (Fig. 4E, F). Having confirmed that miR-338-5p/-421 downregulate expression of ABCG2 and c-Kit, we next evaluated the efflux of Hoechst dye via ABC transporters in the absence or presence of verapamil, a competitive inhibitor for ABC transporters (24). As expected, 22RV1-miR-338-5p/-421 cells show a significant reduction (~91% and 89% respectively) in the side population (SP) cells involved in Hoechst dye efflux (Fig. 4G). As expected, efflux assay performed in the presence of verapamil show substantial reduction in the SP cells due to inhibition of ABC transporters (Fig. 4G). Further, to confirm if miRNAs overexpression lead to decrease in CSC-like properties, prostatosphere assay, a surrogate model for testing enhanced stem cell-like properties was performed, a significant decrease in the size and prostatosphere formation efficiency was observed in 22RV1-miR-338-5p/-421 cells (Fig. 4H, I). Further, prostatospheres formed by miRNAs-overexpressing cells exhibit a significant reduction in the expression of genes implicated in self-renewal and stemness (Supplementary Fig. S6B). Intriguingly, miR-338-5p/-421 putative binding sites on the 3’UTR of EPCAM, c-Kit, SOX9, SOX2 and ABCG2 were also noticed (Supplementary Table S3), suggesting a possible mechanism involved in the downregulation of these genes.
Epigenetic regulators, such as ten-eleven-translocation (TET) family member, TET1, convert 5’-methylcytosine (5mC) to 5’-hydroxymethylcytosine (5hmC), are well-known to induce pluripotency and maintain self-renewal capacity (25). Thus, we analyzed the expression of TET family members, and a striking decrease in TET1 expression was observed in miRNAs-overexpressing cells (Fig. 4J). Since, major players involved in drug-resistance such as ABCG2 and c-Kit were downregulated in miRNAs-overexpressing cells, we examined the sensitivity of these cells to chemotherapeutic drugs. Interestingly, 22RV1-miR-338-5p/-421 cells show enhanced sensitivity to doxorubicin as compared to 22RV1-CTL (Supplementary Fig. S6C). Collectively, miR-338-5p/-421 downregulate the expression of genes implicated in multiple oncogenic pathways namely EMT, stemness and drug resistance, signifying that these two tumor suppressor miRNAs could represent alternative novel approaches for integrative cancer therapy (Fig. 4K).
Transcriptional repression of miR-338 and miR-421 by EZH2 drives SPINK1-positive prostate cancer
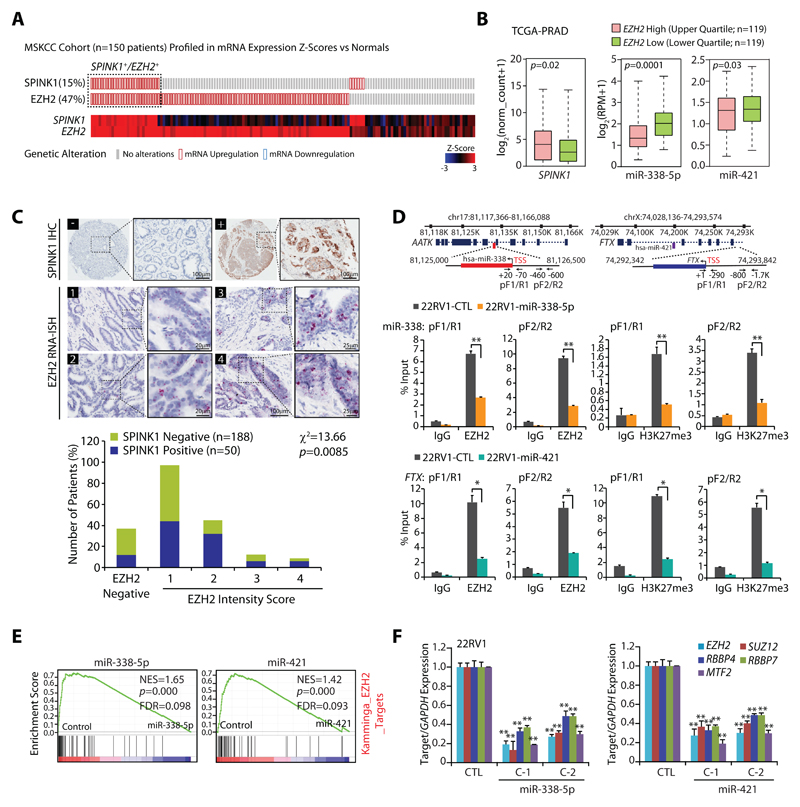
Aberrant transcriptional regulation, genomic loss or epigenetic silencing are well-known mechanisms involved in miRNAs deregulation (26,27). Since SPINK1+ patients exhibit reduced expression of miR-338-5p/-421, we sought to decipher the role of epigenetic silencing involved in reduced expression of these miRNAs. Moreover, one of the well-studied key players of epigenetic silencing is EZH2, implicated in many malignancies, thus, we interrogated for any plausible association between SPINK1 and EZH2 using Memorial Sloan Kettering Cancer Center (MSKCC) patients’ cohort on cBioPortal (http://cbioportal.org). Interestingly, most of the SPINK1+ specimens show concordance with EZH2 expression (Fig. 5A). Further, TCGA-PRAD patients harboring higher expression of EZH2 show increased levels of SPINK1 and decreased expression of miR-338-5p/-421 as compared to EZH2-low patients (Fig. 5B). To further confirm the association between these two oncogenes, we performed immunohistochemistry (IHC) and RNA in situ hybridization (RNA-ISH) for SPINK1 and EZH2 expression respectively using tissue microarrays (TMAs) comprising a total of 238 PCa specimens. In accordance with the previous reports (3,4), we found 21% PCa patients (50 out of 238 cases) were positive for SPINK1 expression. Interestingly, 88% (44 cases) of these SPINK1+ specimens show positive staining for EZH2, of which ~14% of the SPINK1+/EZH2+ patients fall into the high EZH2 range (score 3 and 4), ~36% in medium EZH2 (score 2) and ~50% into low EZH2 expression group (score 1) indicating a significant association between SPINK1 and EZH2 expression (Fig. 5C; χ2=13.66; p=0.008). Although, 75% (141 cases) of the SPINK1-negative patients were also found positive for EZH2, while majority of these cases (~71%) exhibit low EZH2 levels (score 1). Thus, in corroboration with the previous reports (1,11), our data indicate a more pronounced role of epigenetic alterations involved in ETS-fusion negative prostate cancer. Although, six SPINK1+ cases failed to show any expression of EZH2, pointing that an alternative mechanism may be involved in SPINK1 regulation or possibly miRNA-338/-421 genomic deletion could be a cause.
Figure 5. Epigenetic silencing of miR-338-5p and miR-421 via EZH2 in SPINK1 positive prostate cancer.
(A) OncoPrint depicting mRNA upregulation of EZH2 and SPINK1 in MSKCC cohort using cBioportal. In the lower panel shades of blue and red represents Z-score normalized expression for EZH2 and SPINK1.
(B) Box plot depicting SPINK1, miR-338-5p and miR-421 expression in EZH2 high (n=119) and EZH2 low (n=119) in PCa patients from TCGA-PRAD cohort
(C) Representative micrographs depicting PCa tissue microarray (TMA) cores (n=238) stained for SPINK1 by immunohistochemistry (IHC) and EZH2 by RNA in-situ hybridization (RNA-ISH). Top panel represents SPINK1 IHC in SPINK1 negative (–) and SPINK1 positive (+) patients. RNA-ISH intensity score for EZH2 expression was assigned on a scale of 0 to 4 according to visual criteria for the presence of transcript at 40X magnification. Bar plot show EZH2 expression in the SPINK1-negative and SPINK1+ patient specimens. P-value for Chi-square test is indicated.
(D) Genomic location for EZH2 binding sites on the miR-338 and FTX promoters and location of ChIP primers (top panel). ChIP-qPCR data showing EZH2 occupancy and H3K27me3 marks on the miR-338, FTX promoters, and MYT1 used as positive control in stable 22RV1-miR-338-5p, 22RV1-miR-421 and 22RV1-CTL cells.
(E) GSEA plots showing the enrichment of EZH2 interacting partners (Kamminga) in 22RV1-miR-338-5p and 22RV1-miR-421 cells.
(F) QPCR data showing expression of EZH2 and its interacting partners in the same cells as indicated.
Biologically independent samples (n=3) were used in panels (D) and (F); data represent mean ± SEM. *P≤ 0.05 and **P≤ 0.005 using two-tailed unpaired Student's t test.
To investigate whether epigenetic silencing of these miRNAs is mediated by EZH2, we screened the promoters of miR-338, miR-421 and FTX (miR-421 host gene) for putative transcription factor binding sites and identified MYC and MAX (Myc-Associated Factor X) elements within ~2 kb upstream of Transcription Start Site (TSS). MYC is known to form a repressive complex with EZH2 and HDACs, and downregulate multiple tumor suppressive miRNAs, which in turn target PRC2-interacting partners (28). Additionally, EZH2-silenced DU145 cells miRNA expression data (GSE26996) indicates an increase in the expression of numerous EZH2-regulated miRNAs including miR-338-5p and miR-421 (Supplementary Fig. S7A). We therefore examined the promoters of miR-338 and FTX for the recruitment of EZH2 in stable 22RV1-miR-338-5p, 22RV1-miR-421 and 22RV1-CTL cells. Interestingly, a significant enrichment of EZH2 over input was observed on the promoters of miR-338 and FTX in control cells, while substantial decrease in miR-338-5p/-421 overexpressing cells was noticed (Fig. 5D), suggesting the presence of negative feedback regulatory network between miR-338-5p/-421 and EZH2. Subsequently, we checked for EZH2 recruitment on miR-421 promoter and no enrichment was observed (Supplementary Fig. S7B), indicating that FTX promoter regulates the expression of this intronic miRNA. Further, to confirm EZH2-mediated methyltransferase activity, we sought to identify H3K27me3 marks on these promoters. A remarkable enrichment of H3K27me3 marks on the miR-338, miR-421 and FTX promoters were noted relative to IgG control (Fig. 5D and Supplementary Fig. S7B), confirming the role of EZH2-mediated epigenetic silencing of miRNA-338-5p/-421.
Comprehensive GSEA analysis revealed that miRNA-338/-421 overexpressing cells show an enrichment for EZH2 interacting partners, including PRC2 members (29) and EZH2 regulated genes (30,31) (Fig. 5E and Supplementary Fig. S7C), indicating that these two miRNAs in turn regulate EZH2 partners and their target genes. Thus, we next examined the putative binding of miRNA-338-5p/-421 on the 3’UTR of the PRC2 members, interestingly both miRNAs show negative mirSVR binding score indicating miRNAs binding strength (Supplementary Table S3). Moreover, a significant decrease in the transcript levels of EZH2, and its interacting partners SUZ12, RBBP4, RBBP7 and MTF2 were observed in miRNA-338-5p/-421 overexpressing cells (Supplementary Fig. S7D and Fig.5F). Collectively, our data indicates that overexpression of miR-338-5p/-421 downregulates EZH2 expression and its interacting members, leading to impaired histone methyltransferase activity of PRC2, thereby establishing a double-negative feedback loop.
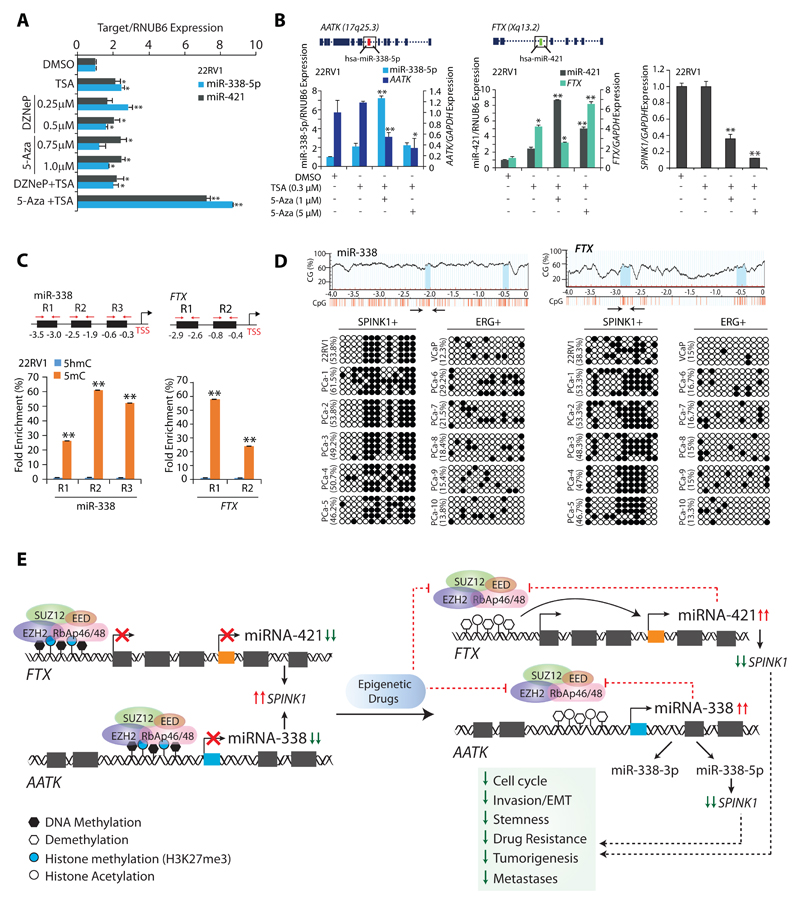
Since inhibitors for chromatin modifiers are known to erase epigenetic marks, we tested 3-Deazaneplanocin A (DZNep), a histone methyltransferase inhibitor; 2'-deoxy-5-azacytidine (5-Aza), a DNA methyltransferase (DNMT) inhibitor and Trichostatin A (TSA), a HDAC inhibitor in 22RV1 cells and examined miR-338-5p/-421 expression. Treatment with TSA, DZNep, 5-Aza alone or a combination of DZNep and TSA in 22RV1 cells showed a modest increase in miR-338-5p/-421 expression, while 5-Aza and TSA together resulted in ~9-fold increase (Fig. 6A). Intriguingly, 5-Aza and TSA in combination results in significant increase in miRNAs expression accompanied with a notable decrease (~60-80%) in SPINK1 levels (Fig. 6B). Since, 3’-arm of miR-338 (miR-338-3p) is known to negatively regulate Apoptosis Associated Tyrosine Kinase (AATK) expression (32), likewise a significant reduction in the AATK expression was noticed (Fig. 6B).Previously, a deletion construct of FTX showed decreased expression of miR-374/-421 cluster (33). Corroborating with this, we also observed a significant increase in the FTX and miR-421 expression upon 5-Aza and TSA combinatorial treatment, signifying the importance of FTX in the regulation of miR-421 (Fig. 6B).
Figure 6. Epigenetic drugs ablate EZH2-mediated silencing of the miR-338-5p and miR-421.
(A) TaqMan assay for miR-338-5p and miR-421 expression in 22RV1 cells treated with different combination of epigenetic drugs.
(B) QPCR showing relative expression of miR-338-5p, miR-421, AATK, FTX and SPINK1 in 22RV1 cells treated with 5-Aza or TSA as indicated.
(C) MeDIP-qPCR showing fold enrichment of 5-mC over 5-hmC in 22RV1 cells as indicated.
(D) Bisulfite–sequencing showing CpG methylation marks on the region upstream of miR-338-5p (left) and FTX (right) in 22RV1, VCaP cells and patients’ tumor specimens (PCa-1 to 5 are SPINK1 positive and PCa-6 to 10 are ERG fusion positive). PCR amplified regions are denoted by arrows. Data represents DNA sequence obtained from five independent clones. Hollow circles represent non-methylated CpG dinucleotides, whereas black solid circles show methylated-CpG sites.
(E) Illustration depicting the molecular mechanism involved in EZH2-mediated epigenetic silencing of miR-338-5p and miR-421 in SPINK1-positive prostate cancer.
In panels (A), (B) and (C) biologically independent samples (n=3) were used; data represent mean ± SEM. *P≤ 0.05 and **P≤ 0.005 using two-tailed unpaired Student's t test.
Additionally, EZH2 is also known to interact with DNMTs, thus enabling chromatin remodeling and DNA methylation (34). We next examined the presence of methylated CpG marks on miR-338-5p and FTX promoters. Interestingly, methylated DNA-immunoprecipitation (MeDIP) revealed locus-specific enrichment in 5mC levels over 5hmC on these regulatory regions (Fig. 6C). To ascertain the presence of DNA methylation marks, we performed bisulfite sequencing using PCa cell lines, a relative increase in the methylated CpG sites on miR-338 and FTX promoters was observed in 22RV1 (SPINK1-positive) as compared to VCaP (ERG-positive) cells (Fig. 6D). No significant difference in the methylated CpG sites on the AATK and miR-421 promoters was observed (Supplementary Fig. S7E, F). To recapitulate this finding in PCa patients, bisulfite sequencing was carried out on SPINK1-positive (n=5) and ERG fusion positive (n=5) patients’ specimens. Interestingly, all SPINK1-postive specimens exhibit increased methylation marks on the promoters of miR-338 and FTX as compared to ERG positive (Fig. 6D). Taken together, our results strongly indicate that epigenetic machinery comprising of EZH2 and its interacting partners play a critical role in the epigenetic silencing of miRNA-338-5p and miR-421 in SPINK1+ subtype, which in turn reaffirms its silencing by a positive feedback loop.
Discussion
In this study, we unraveled the underlying molecular mechanism involved in the overexpression of SPINK1 exclusively in ETS-fusion negative PCa. Our study provides a molecular basis for SPINK1 overexpression, brought about by epigenetic repression of the key post-transcriptional negative regulators of SPINK1 namely, miR-338-5p and miR-421. We demonstratemiR-338-5p/-421 exhibits functional anti-cancer pleiotropy in SPINK1+ subtype, by attenuating oncogenic properties, tumor growth and metastases in murine model. Conversely, miR-421 has also been reported to be a potential oncogenic miRNA in multiple cancers (35,36). However, in corroboration with our findings, a recent report suggested tumor suppressive role of miR-421 in prostate cancer (37). We also established that miR-338-5p/-421 overexpressing cells display perturbed cell-cycle machinery triggered by dysregulated cyclins and CDKs, subsequently leading to S-phase arrest. Recently miRNAs targeting multiple cyclins/CDKs are shown to be more effective than the FDA-approved CDK4/6 inhibitor in triple-negative breast cancer (38), thus supporting our findings that replenishing miRNA-338-5p/-421 may prove advantageous in SPINK1+ cancers.
Emerging evidences suggest a complex interaction between EMT and CSCs during cancer progression, and in developing resistance towards anti-cancer drugs. Previous studies have implicated role of several miRNAs, such as miR-200 family, and miR-34a (39,40) in regulating the expression of genes involved in metastases, stemness and drug-resistance. Furthermore, miR-338 exhibits tumor-suppressive role, and inhibits EMT by targeting ZEB2 (41) and PREX2a (42) in gastric cancer. Here, we identified miR-338-5p/-421 as critical regulators of EMT-inducing transcription factors and -associated markers, which in turn led to decreased stem-cells like features. Moreover, CSCs are known to express ABC transporters, which efflux the chemotherapeutic drugs during resistance (24). Remarkably, miR-338-5p/-421 overexpression shows decreased expression of ABCG2 and c-KIT, consequently a significant drop in the drug-resistant side population, indicating that miR-338-5p/-421 are highly effective in conferring drug-sensitivity and reducing the therapy-resistant CSCs. Collectively, our findings provide a solid foundation for qualifying these miRNAs as an adjuvant therapy for the SPINK1+ and other drug resistant malignancies.
Our findings were further strengthened by a TCGA study (1), wherein a subset of PCa patients’ harboring SPOP-mutation/CHD1-deletion exhibits elevated DNA methylation levels accompanied with frequent events of SPINK1 overexpression. Recently, a new subtype of ETS-fusion-negative tumors has been defined by frequent mutations in the epigenetic regulators and chromatin remodelers (43). Yet another study, using genome-wide methylated DNA-immunoprecipitation sequencing revealed higher number of methylation events in TMPRSS2-ERG fusion-negative as compared to normal and TMPRSS2–ERG fusion-positive PCa specimens (11). Collectively, these independent findings reaffirm the critical role of epigenetic pathways engaged in the pathogenesis of SPINK1+ subtype.
Interestingly, increased methylated regions in the ETS-fusion negative patients have been attributed to hypermethylation of miR-26a, a post-transcriptional regulator of EZH2 (11). Thus, given the central role played by EZH2 and the epigenetic mechanism involved in ETS-fusion negative cases, our findings rationalize the role of EZH2-mediated epigenetic regulation of miR-338-5p/-421 in SPINK1+/ETS-negative subtype. Hence, we propose a molecular model involving SPINK1, EZH2 and miR-338-5p/-421, wherein EZH2 acts as an epigenetic switch and by its histone methyltransferase activity establishes H3K27me3 repressive marks on the regulatory regions of miR-338 and FTX, a miR-421 host-gene (Fig. 6E).
In consonance with this, miR-338-5p/-421 overexpression also results in decreased Tet1 expression. Converging lines of evidences suggest dual-role of Tet1 in promoting transcription of pluripotency factors and recruitment of PRC2 on CpG rich promoters (44). Collectively, miR-338-5p/-421 mediated decrease in Tet1 expression might possibily contribute in reduced stemness and drug-resistance. We also conjecture that decrease in Tet1 expression may result in reduced PRC2 occupancy on the miRNA promoters, diminish epigenetic silencing marks, and consequently downregulate their targets including SPINK1.
Currently, there is no effective therapeutic intervention for SPINK1-positive malignancies including prostate, although use of monoclonal EGFR antibody has been suggested (9). Nevertheless, outcome of phase I/II clinical trials using cetuximab (45) and EGFR small molecules inhibitors was largely unsuccessful (46,47). For instance, in a phase Ib/IIa clinical trial using cetuximab and doxorubicin combination therapy, only a fraction of CRPC patients (~8%) showed >50% PSA decline (45), revealing its limited efficacy. Owing to the pleiotropic anti-cancer effects exhibited by miRNA-338-5p/-421, we propose miRNA-replacement therapy as one of the potential therapeutic approaches for SPINK1+ cancers; nonetheless in-vivo delivery methods and stability are some of the major challenges for successful translation into the clinic (48). While not restricted to miRNA-replacement therapy, the present study also suggests alternative therapeutic avenues for SPINK1+ malignancies, for instance adjuvant therapy using inhibitors against DNMTs, HDACs or EZH2, several of which are already in clinical trials (49). Conclusively, we moved the field forward by addressing an important question that how SPINK1 is aberrantly overexpressed in ETS-negative prostate cancer, and the stratification of patients based on SPINK1-positive and miRNA-338-5p/-421-low criteria could further improve therapeutic modalities and overall management strategies.
Supplementary Material
Translational Impact.
We establish a regulatory model involving the functional interplay between SPINK1, miRNA-338-5p/miRNA-421 and EZH2, thereby, revealing hitherto unknown mechanism of SPINK1 up-regulation in SPINK1-positive prostate cancer subtype. Our findings provide a strong rationale for the development of potential therapeutic strategies for SPINK1-positive malignancies. We demonstrate that restoring miRNA-338-5p/miRNA-421 expression using epigenetic drugs including DNMTs inhibitors in combination with HDACs or HKMTs inhibitors or miRNA synthetic mimics in SPINK1-positive prostate cancer abrogate SPINK1-mediated oncogenicity. The major findings of this study will not only advance the prostate cancer field, but will also be valuable for treatment and disease management of other SPINK1-positive malignancies.
Acknowledgements
B.A. is an Intermediate Fellow of the Wellcome Trust/DBT India Alliance. This work is supported by the Wellcome Trust/DBT India Alliance Fellowship [grant number: IA/I(S)/12/2/500635] awarded to B.A. We thank Yuping Zhang, Mahendra Palecha, Ayush Praveen for their technical support and Anjali Bajpai for critically reading the manuscript. We also thank Jonaki Sen for extending the use of fertilized eggs facility. The IIT Kanpur has filed a patent (IN 201611016564) on the therapeutic applicability of miR-338-5p and miR-421 described in this study in which B.A., V.B. and A.Y. are named as inventors.
Financial support: This work is supported by the Wellcome Trust/ DBT India Alliance grant (IA/I(S)/12/2/500635 to BA).
Footnotes
Data availability
The gene expression microarray data from this study has been submitted to the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE108558.
Conflict of interest: The authors declare no conflicts of interest or disclosures.
Authors’ contributions
V.B., A.Y., and B.A. designed and directed the experimental studies. V.B. and A.Y. performed in-vitro cell line-based studies. V.B., A.Y. and S.N. performed the bisulfite sequencing experiments and analysis. V.B., A.Y., R.T. and S.G. performed the gene expression studies, bioinformatics analysis and ChIP assays. A.Y. performed the immunofluorescence and RIP experiments. V.B., A.Y., and B.A. performed statistical analysis and interpreted the data. V.B., A.Y., and B.A. executed the in-vivo experiments. A.G. provided PCa patient specimens. S.C., N.G., and N.P. performed immunohistochemistry and RNA in situ staining on the PCa tissue microarrays. V.B., A.Y., and B.A. wrote the manuscript. B.A. directed the overall project.
References
- 1.Abeshouse A, Ahn J, Akbani R, Ally A, Amin S, Andry CD, et al. The molecular taxonomy of primary prostate cancer. Cell. 2015;163(4):1011–25. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palanisamy N, Ateeq B, Kalyana-Sundaram S, Pflueger D, Ramnarayanan K, Shankar S, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nature medicine. 2010;16(7):793. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun X-W, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. science. 2005;310(5748):644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 4.Ateeq B, Kunju LP, Carskadon SL, Pandey SK, Singh G, Pradeep I, et al. Molecular profiling of ETS and non-ETS aberrations in prostate cancer patients from northern India. The Prostate. 2015;75(10):1051–62. doi: 10.1002/pros.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, et al. Mechanistic rationale for inhibition of poly (ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer cell. 2011;19(5):664–78. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomlins SA, Rhodes DR, Yu J, Varambally S, Mehra R, Perner S, et al. The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer cell. 2008;13(6):519–28. doi: 10.1016/j.ccr.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leinonen KA, Tolonen TT, Bracken H, Stenman UH, Tammela TL, Saramaki OR, et al. Association of SPINK1 expression and TMPRSS2:ERG fusion with prognosis in endocrine-treated prostate cancer. Clin Cancer Res. 2010;16(10):2845–51. doi: 10.1158/1078-0432.CCR-09-2505. 1078-0432.CCR-09-2505 [pii] [DOI] [PubMed] [Google Scholar]
- 8.Ateeq B, Bhatia V, Goel S. Molecular discriminators of racial disparities in prostate cancer. Trends in cancer. 2016;2(3):116–20. doi: 10.1016/j.trecan.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ateeq B, Tomlins SA, Laxman B, Asangani IA, Cao Q, Cao X, et al. Therapeutic targeting of SPINK1-positive prostate cancer. Science translational medicine. 2011;3(72):72ra17–72ra17. doi: 10.1126/scitranslmed.3001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322(5908):1695–9. doi: 10.1126/science.1165395. 1165395 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borno ST, Fischer A, Kerick M, Falth M, Laible M, Brase JC, et al. Genome-wide DNA methylation events in TMPRSS2-ERG fusion-negative prostate cancers implicate an EZH2-dependent mechanism with miR-26a hypermethylation. Cancer Discov. 2012;2(11):1024–35. doi: 10.1158/2159-8290.CD-12-0041. [DOI] [PubMed] [Google Scholar]
- 12.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 13.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338(6113):1465–9. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao Q, Mani RS, Ateeq B, Dhanasekaran SM, Asangani IA, Prensner JR, et al. Coordinated regulation of polycomb group complexes through microRNAs in cancer. Cancer Cell. 2011;20(2):187–99. doi: 10.1016/j.ccr.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shukla S, Cyrta J, Murphy DA, Walczak EG, Ran L, Agrawal P, et al. Aberrant activation of a gastrointestinal transcriptional circuit in prostate cancer mediates castration resistance. Cancer cell. 2017;32(6):792–806. e7. doi: 10.1016/j.ccell.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasanen K, Itkonen O, Koistinen H, Stenman UH. Emerging Roles of SPINK1 in Cancer. Clinical chemistry. 2016;62(3):449–57. doi: 10.1373/clinchem.2015.241513. [DOI] [PubMed] [Google Scholar]
- 17.Soon WW, Miller LD, Black MA, Dalmasso C, Chan XB, Pang B, et al. Combined genomic and phenotype screening reveals secretory factor SPINK1 as an invasion and survival factor associated with patient prognosis in breast cancer. EMBO molecular medicine. 2011;3(8):451–64. doi: 10.1002/emmm.201100150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiwari R, Pandey SK, Goel S, Bhatia V, Shukla S, Jing X, et al. SPINK1 promotes colorectal cancer progression by downregulating Metallothioneins expression. Oncogenesis. 2015;4:e162. doi: 10.1038/oncsis.2015.23. oncsis201523 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin XQ, Livingston DM, Kaelin WG, Jr, Adams PD. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci U S A. 1994;91(23):10918–22. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nature reviews Molecular cell biology. 2008;9(4):297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 21.Mani SA, Guo W, Liao M-J, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno-Bueno G, Cubillo E, Sarrió D, Peinado H, Rodríguez-Pinilla SM, Villa S, et al. Genetic profiling of epithelial cells expressing E-cadherin repressors reveals a distinct role for Snail, Slug, and E47 factors in epithelial-mesenchymal transition. Cancer research. 2006;66(19):9543–56. doi: 10.1158/0008-5472.CAN-06-0479. [DOI] [PubMed] [Google Scholar]
- 23.Liu T, Xu F, Du X, Lai D, Liu T, Zhao Y, et al. Establishment and characterization of multi-drug resistant, prostate carcinoma-initiating stem-like cells from human prostate cancer cell lines 22RV1. Molecular and cellular biochemistry. 2010;340(1–2):265–73. doi: 10.1007/s11010-010-0426-5. [DOI] [PubMed] [Google Scholar]
- 24.Zhou S, Schuetz JD, Bunting KD, Colapietro A-M, Sampath J, Morris JJ, et al. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nature medicine. 2001;7(9):1028. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
- 25.Gao Y, Chen J, Li K, Wu T, Huang B, Liu W, et al. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell stem cell. 2013;12(4):453–69. doi: 10.1016/j.stem.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(24):15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bueno MJ, Perez de Castro I, Gomez de Cedron M, Santos J, Calin GA, Cigudosa JC, et al. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13(6):496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Zhao X, Fiskus W, Lin J, Lwin T, Rao R, et al. Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-Cell lymphomas. Cancer cell. 2012;22(4):506–23. doi: 10.1016/j.ccr.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Kamminga LM, Bystrykh LV, de Boer A, Houwer S, Douma J, Weersing E, et al. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood. 2006;107(5):2170–9. doi: 10.1182/blood-2005-09-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu C, Han HD, Mangala LS, Ali-Fehmi R, Newton CS, Ozbun L, et al. Regulation of tumor angiogenesis by EZH2. Cancer cell. 2010;18(2):185–97. doi: 10.1016/j.ccr.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nuytten M, Beke L, Van Eynde A, Ceulemans H, Beullens M, Van Hummelen P, et al. The transcriptional repressor NIPP1 is an essential player in EZH2-mediated gene silencing. Oncogene. 2008;27(10):1449. doi: 10.1038/sj.onc.1210774. [DOI] [PubMed] [Google Scholar]
- 32.Kos A, Olde Loohuis NF, Wieczorek ML, Glennon JC, Martens GJ, Kolk SM, et al. A potential regulatory role for intronic microRNA-338-3p for its host gene encoding apoptosis-associated tyrosine kinase. PLoS One. 2012;7(2):e31022. doi: 10.1371/journal.pone.0031022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chureau C, Chantalat S, Romito A, Galvani A, Duret L, Avner P, et al. Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region. Hum Mol Genet. 2011;20(4):705–18. doi: 10.1093/hmg/ddq516. [DOI] [PubMed] [Google Scholar]
- 34.Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439(7078):871–4. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 35.Hu H, Du L, Nagabayashi G, Seeger RC, Gatti RA. ATM is down-regulated by N-Myc–regulated microRNA-421. Proceedings of the National Academy of Sciences. 2010;107(4):1506–11. doi: 10.1073/pnas.0907763107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ge X, Liu X, Lin F, Li P, Liu K, Geng R, et al. MicroRNA-421 regulated by HIF-1α promotes metastasis, inhibits apoptosis, and induces cisplatin resistance by targeting E-cadherin and caspase-3 in gastric cancer. Oncotarget. 2016;7(17):24466. doi: 10.18632/oncotarget.8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meng D, Yang S, Wan X, Zhang Y, Huang W, Zhao P, et al. A transcriptional target of androgen receptor, miR-421 regulates proliferation and metabolism of prostate cancer cells. The international journal of biochemistry & cell biology. 2016;73:30–40. doi: 10.1016/j.biocel.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 38.Hydbring P, Wang Y, Fassl A, Li X, Matia V, Otto T, et al. Cell-Cycle-Targeting MicroRNAs as Therapeutic Tools against Refractory Cancers. Cancer cell. 2017;31(4):576–90 e8. doi: 10.1016/j.ccell.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nature medicine. 2011;17(2):211–5. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tellez CS, Juri DE, Do K, Bernauer AM, Thomas CL, Damiani LA, et al. EMT and stem cell-like properties associated with miR-205 and miR-200 epigenetic silencing are early manifestations during carcinogen-induced transformation of human lung epithelial cells. Cancer research. 2011 doi: 10.1158/0008-5472.CAN-10-3035. canres. 3035.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang N, Wu Z, Lin L, Zhou M, Wang L, Ma H, et al. MiR-338-3p inhibits epithelial-mesenchymal transition in gastric cancer cells by targeting ZEB2 and MACC1/Met/Akt signaling. Oncotarget. 2015;6(17):15222. doi: 10.18632/oncotarget.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo B, Liu L, Yao J, Ma R, Chang D, Li Z, et al. miR-338-3p suppresses gastric cancer progression through a PTEN-AKT axis by targeting P-REX2a. Molecular Cancer Research. 2014;12(3):313–21. doi: 10.1158/1541-7786.MCR-13-0507. [DOI] [PubMed] [Google Scholar]
- 43.Armenia J, Wankowicz SAM, Liu D, Gao J, Kundra R, Reznik E, et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet. 2018 doi: 10.1038/s41588-018-0078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473(7347):389–93. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slovin SF, Kelly WK, Wilton A, Kattan M, Myskowski P, Mendelsohn J, et al. Anti-epidermal growth factor receptor monoclonal antibody cetuximab plus Doxorubicin in the treatment of metastatic castration-resistant prostate cancer. Clinical genitourinary cancer. 2009;7(3):E77–82. doi: 10.3816/CGC.2009.n.028. [DOI] [PubMed] [Google Scholar]
- 46.Nabhan C, Lestingi TM, Galvez A, Tolzien K, Kelby SK, Tsarwhas D, et al. Erlotinib has moderate single-agent activity in chemotherapy-naive castration-resistant prostate cancer: final results of a phase II trial. Urology. 2009;74(3):665–71. doi: 10.1016/j.urology.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 47.Pezaro C, Rosenthal MA, Gurney H, Davis ID, Underhill C, Boyer MJ, et al. An open-label, single-arm phase two trial of gefitinib in patients with advanced or metastatic castration-resistant prostate cancer. American journal of clinical oncology. 2009;32(4):338–41. doi: 10.1097/COC.0b013e31818b946b. [DOI] [PubMed] [Google Scholar]
- 48.Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. The Journal of clinical investigation. 2017;127(3):761–71. doi: 10.1172/JCI84424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–12. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.