Abstract
Mango (Mangifera indica L.) is an important commercial fruit that shows a noticeable loss of firmness during ripening. Polygalacturonase (PG, E.C. 3.2.1.15) is a crucial enzyme for cell wall loosening during fruit ripening since it solubilizes pectin and its activity correlates with fruit softening. Mango PGs were mapped to a genome draft using seventeen PGs found in mango transcriptomes and 48 bonafide PGs were identified. The phylogenetic analysis suggests that they are related to Citrus sinensis, which may indicate a recent evolutive divergence and related functions with orthologs in the tree. Gene expression analysis for nine PGs showed differential expression for them during post-harvest fruit ripening, MiPG21-1, MiPG14, MiPG69-1, MiPG17, MiPG49, MiPG23-3, MiPG22-7, and MiPG16 were highly up-regulated. PG enzymatic activity also increased during maturation and these results correlate with the loss of firmness observed in mango during post-harvest ripening, between the ethylene production burst and the climacteric peak. The analysis of PGs promoter regions identified regulatory sequences associated to ripening such as MADS-box, ethylene regulation like ethylene insensitive 3 (EIN3) factors, APETALA2-like and ethylene response element factors. During mango fruit ripening the action of at least these nine PGs contribute to softening, and their expression is regulated at the transcriptional level. The prediction of the tridimensional structure of some PGs showed a conserved parallel beta-helical fold related to polysaccharide hydrolysis and a modular architecture, where exons correspond to structural elements. Further biotechnological approaches could target specific softening-related PGs to extend mango post-harvest shelf life.
Keywords: polygalacturonase, Mangifera indica L., gene expression, enzymatic activity, firmness, ripening
Introduction
Fruit ripening is a physiological, biochemical, and genetically programmed process. This process is characterized by rheological and texture changes as the cell wall is disassembled by the action of hydrolytic enzymes (Li et al., 2010). Ripening leads to desirable sensorial characteristics of the fruit besides softening, such as aroma and color development (White, 2002). Accelerated mesocarp softening that characterizes mango fruit ripening occurs at the climacteric peak (Litz, 2009). The cell wall is a scaffold made of complex polysaccharides (pectins, cellulose, hemicelluloses, among others) that during ripening are hydrolyzed by enzymes such as polygalacturonases (PGs), pectate lyases, β-galactosidases, xylanases, glucosidases, among others (Brummell and Harpster, 2001; Brummell, 2006). Genetic modifications like suppression or over-expression of genes that encode for these hydrolytic enzymes have provided information about their function as well as the redundancy in the function of several isoforms that participate during fruit ripening (Goulao and Oliveira, 2008). Recently, mango mesocarp transcriptomes have been obtained (Dautt-Castro et al., 2015, 2018); thus family members encoding these hydrolases can be identified and further gene expression studies performed to understand more about their roles in the quick softening of mango fruit.
Polygalacturonases are cell wall disassembling enzymes with profound influence in fleshy fruit softening during ripening. Their specific function is pectin degradation, which is a structural polysaccharide of the primary cell wall and middle lamella, composed mainly of α-1, 4 D-galacturonic acid sugars (Lang and Dörnenburg, 2000). Several PG genes have been identified in fruits like banana (Musa accuminata) (Asif and Nath, 2005), fleshy fruit of oil palm (Roongsattham et al., 2012), papaya (Carica papaya) (Fabi et al., 2014), cucumber (Cucumis sativus) (Yu et al., 2014), among others. Also, transcriptomic analysis of nectarine (Prunus persica L.), orange (Citrus sinensis) and melon (Cucumis melo L.), among others, have revealed PG genes associated to fruit ripening (Ziliotto et al., 2008; Corbacho et al., 2013; Yu et al., 2014). Recently, with the sequencing of genomes from different plants, it has been possible to uncover whole families of PGs. Ke et al. (2018), identified and characterized 54 PGs in tomato, which were classified into seven clades. These clades have been related to specific functions and tissues in plants, for example, members from clades A and B have a role in fruit and abscission zone development, while clades C, D, and F members are involved in flowering development (Liang et al., 2015).
Efforts have been made to understand more about mango softening and few reports have focused on PG activity under different ripening stages or treatments. For example, PG isoforms have been identified in mango “Alphonso” (Prasanna et al., 2006), “Dasheari” (Singh and Dwivedi, 2008) and “Nam Dok Mai” (Suntornwat et al., 2000). In mango “Kent” and “Ataulfo” enzymatic activity of PG has been reported as well as the effect of the ethylene-antagonist 1-methyl cyclopropene (1-MCP) (Muy Rangel et al., 2009; Islas-Osuna et al., 2010). Only few gene expression studies were addressed using “next generation sequencing (NGS)” (Dautt-Castro et al., 2015, 2018) plus access to a mango genome draft from Tommy Atkins cultivar (Kuhn, personal communication). Therefore, the present study aimed to identify PG family members through transcriptomes and within the mango genome, to uncover their gene structure and phylogenetic relationship as well as to evaluate the expression of some PGs in mango mesocarp at different ripening stages, and PG enzymatic activity.
Materials and Methods
Plant Material
Mango (Mangifera indica L.) fruit cultivar “Kent” were hand-harvested in a commercial orchard located in Navojoa, Sonora, México (27°03′49.33″ N and 109°30′11.42″ W). Fruits were selected at physiological maturity stage according to fruit shape, peel color, size and at 125 days after anthesis. Mangos were transported to the laboratory where they were disinfected with chlorinated water and stored at 20°C with 60–65% humidity during 16 days.
CO2, Ethylene Measurement
CO2 and ethylene production were measured by gas chromatography (Varian Star 3400, Varian United States) equipped with thermal conductivity (TCD) and flame ionization detectors (FID) and a 2 m × 1/82″ metal column filled with Hayesep N 800/100. These measurements were done after 24 h of harvesting mango. The fruit was placed in sealed plastic containers (2 L) for 2 h at 20°C, then CO2 and ethylene head-space concentration were analyzed by withdrawing 1 mL sample from the container and injecting them into the gas chromatograph (Muy Rangel et al., 2009). A standard of 5% of CO2 and 0.1% for ethylene was used. Gas concentrations were estimated using the following equations:
Fruit Firmness
Firmness was measured using a digital texturometer (Chatillon Model TCM200). Mango pulp firmness was measured in two sites of the fruit, and the average was reported (Cárdenas-Coronel et al., 2012). The loss of firmness was reported as newtons (N).
Identification of Mango PG Family Genes in Transcriptome and Mapping to the Mango Genome
Two mango mesocarp transcriptomes (GenBank accessions PRJNA258477 and PRJNA286253) were used to identify candidate PG transcripts, and they were mapped into the mango cv. Tommy Atkins genome (DK, personal communication) to obtain information like the number of exons, and chromosomal localization of each gene. The deduced amino acid sequences were analyzed using the BLAST algorithm against the GenBank database. Also, other bioinformatics tools like gene ontology (GO) to know the biological process, molecular function and cellular component of the PG genes, as well as the clusters of orthologous groups (COG) to identify Endo and Exo PGs were used. To predict the theoretical molecular weight and the isoelectric point of the deduced PGs proteins, the compute pl/Mw tool of ExPASy1 was used. Multiple sequence alignments of the PGs sequences were done with CLUSTAL W, and the figures were made using BoxShade software2, to show the sequence similarity of enzymes within this family. The prediction of signal peptides in PG sequences was carried out using SignalP 5.03.
Phylogenetic, Gene Structure of PG Genes and Cis-Regulatory Elements Analysis
All 48 PG protein sequences from the mango genome were aligned along 69 PG from Arabidopsis thaliana, using the Neighbor-Joining method, with a previous alignment using the algorithm MUSCLE. The bootstrap consensus tree inferred from 2000 replicates is taken to represent the evolutionary relationship of the taxa analyzed. Based on the phylogenetic tree, mango PG sequences were named according to most nearby A. thaliana sequences. All phylogenetic inferences were conducted in MEGA (Kumar et al., 2016).
The full-length amino acid sequences of the PGs encoded in the mango genome were compared among them using multiple sequence alignments with MUSCLE and using the default settings (Edgar, 2004). The phylogenetic tree was constructed using the neighbor-joining method (Saitou and Nei, 1987) with a bootstrap value of 2000 replicates in MEGA (Kumar et al., 2016). To assign clades, a neighbor-joining tree was obtained comparing Solanum lycopersicum PG amino acid sequences with mango (M. indica L.) PGs. Trees were drawn using iTOL server (Letunic and Bork, 2016). The structure of the introns and exons of the MiPG genes were obtained using GSDS 2.0 (Hu et al., 2015)4.
A total of 1.5 Kb of genomic DNA sequences upstream of the initiation codon (ATG) of each MiPG gene was obtained from the mango genome database. These promoter sequences were used to investigate the regulatory elements in the MatInspector program, using general core promoter elements and plants matrix groups, with 0.90 of core and matrix similarity. Also, analysis of overrepresented transcription factor binding sites was carried out using genomic and promoter background of A. thaliana, TAIR 10 (Cartharius et al., 2005).
RNA Isolation, cDNA Synthesis and Relative Expression of PGs by qRT-PCR
Total RNA was isolated from the mango mesocarp tissue according to López-Gómez and Gómez-Lim (1992), at days 1, 4, 7, 10, and 16 of post-harvest storage. The RNA quantity was estimated at 260 nm using a Nano-Drop ND-1000 UV-Vis spectrophotometer (Nano Drop Technologies Inc., Wilmington, DE, United States). RNA integrity was assessed using agarose gel electrophoresis under denaturing conditions. RNA was treated with RNA-free DNase I (Roche, CA, United States) to eliminate the genomic DNA. Then, the cDNA synthesis was performed by reverse transcription from 2.5 μg of total RNA, using the SuperScript II kit (Invitrogen, CA, United States) according to manufacturer conditions.
Quantitative PCR was carried out using iQTM SYBR® Green Supermix (Bio-Rad, CA, United States). All samples were PCR-amplified by triplicates in reactions which included 100 ng of cDNA template, 12.5 μL of SYBR® Green qRT-PCR Master Mix, 1 μL of 5 μM sense primer, 1 μL of 5 μM antisense primer and water to 25 μL of final volume. Specific primers to amplify the nine PG genes and the reference gene GAPDH (glyceraldehyde 3-phosphate dehydrogenase) are shown in Supplementary Table S1. The PCR products were amplified in a Step-OneTM Real-time PCR System (Applied Biosystems). Amplification conditions were one cycle of 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 1 min. PCR product specificity was confirmed by constructing a melt curve after amplification raising the temperature from 95°C for 15 s, 60°C for 1 min and 95°C for 15 s. Non-template controls were included during each gene amplification. The method 2–ΔΔCT was used to evaluate changes in the relative mRNA amount of target genes (Livak and Schmittgen, 2001). The results are expressed as relative mRNA steady-state levels of the target gene and normalized to the GAPDH (Dautt-Castro et al., 2015) and ACT 7 (Tafolla-Arellano et al., 2017) expression levels. The differential expression obtained by RNA-Seq was also used to compare the expression patterns of PG genes and qPCR was used to validate those.
PG Enzymatic Activity
PG enzymatic activity was determined according to Gross (1982). Mesocarp tissue (25 g) was homogenized with 75 ml of 1% sodium bisulfite pH 6. The homogenized was filtered, and the residue was suspended in 75 ml of 1% sodium bisulfite (pH 6), filtered again and suspended in 37.5 ml of sodium chloride 1 M. The pH was adjusted at 6 with NaOH 1 N, and the solution was stirred for 3 h at 4°C. After agitation, the solution was filtrated and centrifuged at 9000 × g for 15 min. Aliquots of 2.5 ml of supernatant were desalted in a Sephadex G25 column equilibrated with sodium acetate 50 mM (pH 4). The enzymatic extract (50 μl) was incubated for 2 h at 30°C in a 0.2 ml solution containing sodium acetate 37.5 mM (pH 4.4) and 0.2% polygalacturonic acid previously washed with 80% ethanol. The reaction was stopped by addition of 1 ml of cold borate buffer 100 mM (pH 9). Then, 0.2 ml of 1% 2-cianoacetamide was added, and the samples were placed in boiling water by 10 min. Finally, the samples were cooled at room temperature and the absorbance was read at 276 nm. Protein was quantitated by the method of Bradford (1976). The results were expressed by nmol of reducing groups produced for mg of protein for an hour.
Molecular Modeling of PGs
The amino acid sequences of MiPG14, MiPG21-1, MiPG23-3, MiPG49, MiPG46-3, and MiPG69-1 were modeled by using Phyre2 server (Kelley et al., 2015). The crystallographic model used as a template was the rhamnogalacturonase A from Aspergillus aculeatus (PDB 1RMG). Figures were build using PyMol (DeLano, 2002; Schrödinger, 2019) the structures were colored according to the color code used for exons in the sequence alignments.
Statistical Analysis
Statistical analysis was performed using one-way ANOVA, with a 0.05 significance level (P < 0.05). Fisher’s test was used to detect statistical differences between means, using the XLSTAT software.
Results and Discussion
CO2 and Ethylene Levels Are Characteristic of a Climateric Fruit
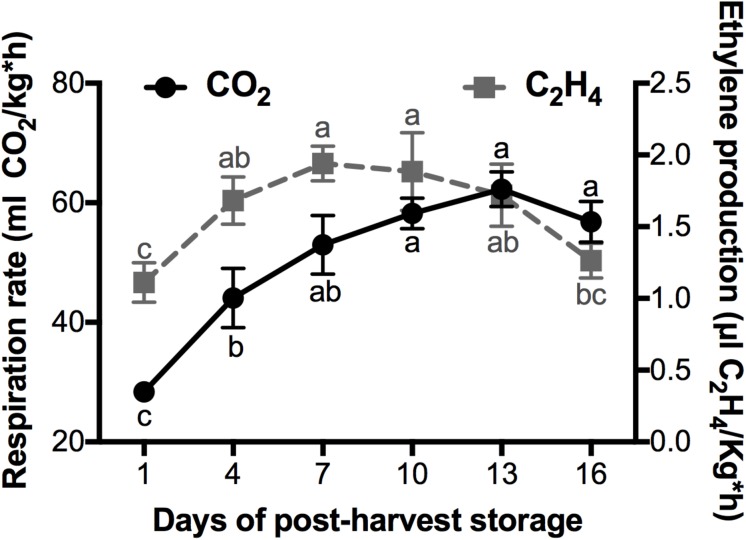
Mango belongs to the climacteric fruits, in which the CO2 and ethylene present a peak of production during post-harvest (Bouzayen et al., 2010). The respiration rate and the ethylene production observed in mango cv. Kent fruits during their ripening at a storage temperature of 20°C is shown in Figure 1. The maximum ethylene production occurred at day 7 of post-harvest storage, with a value of 1.9 uL C2H4/Kg*h. This ethylene accumulation preceded the climacteric peak that occurred at day 13 with a CO2 production of 62.29 mL CO2/kg*h. It is well documented that ethylene production during climacteric ripening triggers signal transduction for the activation of several transcription factors that in turn activate the expression of genes that encode enzymes that catalyze ripening changes such as color, flavor, texture, aroma, among others (Grierson, 2013). In agreement with this, most of the results that we discuss later are related to the increase of ethylene production.
FIGURE 1.
Evaluation of CO2 and Ethylene production (black circles and gray squares, respectively) of mango fruits cultivar “Kent” harvested at the stage of physiological maturity and stored at 20°C for 16 days at an RH of 60–65%. Data shown represent the average of five biological replicates (n = 5). Error bar indicates SE of the means and different letters indicate significant differences. Statistical significance between the control and treatment groups was determined by one-way ANOVA with the Tukey-Kramer test (P < 0.05).
Mango Suffered a Drastic Loss of Firmness by Day 10 of Post-harvest at 20°C
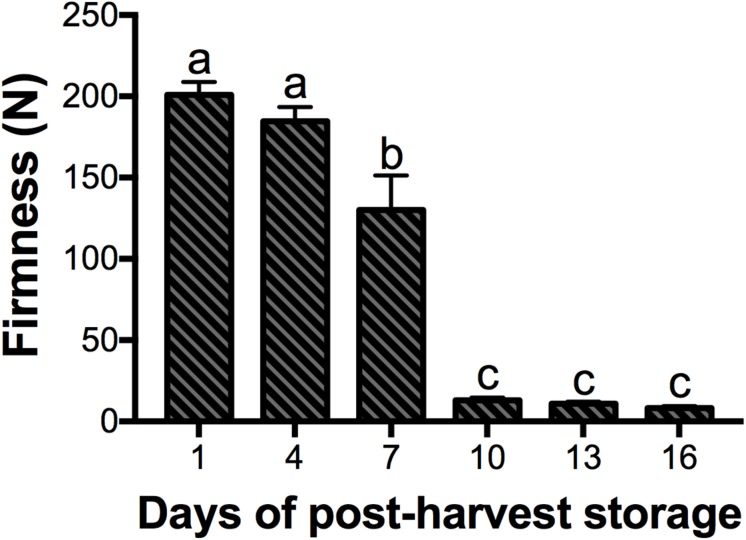
Among other changes that occur during ripening of mango fruit, the texture is essential in terms of its relationship with consumer preference, storage, transportation, shelf life and resistance to pathogens (Li et al., 2010). The firmness of the mango cv. Kent fruit is shown in Figure 2. Fruits from days 1 and 4 of post-harvest storage were similar in their firmness. However, at day 7, the firmness of the fruits decreased significantly by 30%, and at day 10, the firmness of fruits was drastically reduced by 90%, remaining constant until the end of ripening (P < 0.05). Mango cv. Kent is highly appreciated in the market; however, it has a very fast softening and a short shelf life (Jiménez et al., 2004). Similar results to ours have been reported in other mango varieties such as Harumanis, Kensington Pride and Ataulfo, where reductions of 50, 80, and 75% in mesocarp firmness were observed between 4.5, 6, and 6 days of post-harvest storage, respectively (Ali et al., 2004; Muy Rangel et al., 2009; Razzaq et al., 2016). These textural changes are the result of alterations in the structure and composition of the cell wall, mediated by enzymes related to softening (Brummell, 2006). From the plant cell wall components, the pectins are the most structurally complex and they are essential for cell-to-cell adhesion (Wang et al., 2018). One of the most important enzymes that hydrolyze pectin is PG, which has been extensively studied in a large number of fruits, particularly in tomato (Giovannoni et al., 1989). That is the importance of identifying these enzymes at the molecular level, and in the future, the use of this information might allow quality improvements.
FIGURE 2.
Mango fruit firmness. Data shown represent the average of five biological replicates (n = 5). Error bar indicates SE of the means and different letters indicate significant differences. Statistical significance between the control and treatment groups was determined by one-way ANOVA with the Tukey-Kramer test (P < 0.05).
Transcriptome-Wide and Genome-Wide Identification of PG Genes of Mango Fruit
A total of seventeen PG cDNAs were found in the mango mesocarp cv. Kent transcriptome (Dautt-Castro et al., 2015) and they were mapped to the mango cv. Tommy Atkins genome (Dr. DK, personal communication). A total of 48 encoded PGs were found in the mango genome and named after their Arabidopsis counterparts (González-Carranza et al., 2007). Those PGs are presented in Table 1, where some characteristics are shown for each identified PG like name, genome identification, GenBank accession number, exon number, protein length and theoretical molecular weight. As summarized in Table 1, the lengths of the mango PGs ranged from 83 aa (MiPG66) to 1916 aa (MiPG69-2) with molecular weights of 8.77 kDa to 210.08 kDa, respectively. PG isoelectric points ranged from 4.77 (MiPG6-1) to 9.27 (MiPG59). Fourty three PGs presented a Cellular Component of “cell wall” and “extracellular region”; meanwhile, five of them were “integral to the membrane.” About the Biological Process, 45 PGs presented “carbohydrate metabolic process,” five also showed “fruit ripening,” one PG showed cell wall organization and one was annotated as “Microsporogenesis/pollen exine formation.” For molecular function, 47 PG genes were annotated with “PG activity” (see Supplementary Table S2A). Also, the COG annotation showed that 41 PGs are endo-PGs (COG5434) and seven are exo-PGs (Supplementary Table S2). Interestingly, endo-PGs, whose activity is to endo-degrade homogalacturonans, the primary type of pectins, have been more associated to fruit ripening and softening compared to exo-PGs (Wakasa et al., 2006; Gu et al., 2016; Qian et al., 2016; Wang et al., 2018). On the other hand, 32 of 48 MiPGs (66%) were predicted to have a signal peptide with lengths ranging from 17 to 39 residues (Supplementary Table S2B). Moreover, those MiPGs with a signal peptide were located in the secretory pathway. In tomato, 38 of 54 PGs presented a signal peptide with lengths between 17 to 31 residues, similar to our results (Ke et al., 2018). These signal peptides present in PG amino acid sequences support the fact that they are guided to the cell wall to carry out their hydrolytic function.
TABLE 1.
Family of polygalacturonases in mango (Mangifera indica L.).
| MiPG | Genome ID | GenBank | Exons | Localization Chr | aa | kDa |
| 40-1 | Manin00g002060.1 | MK936539 | 16 | Chr 0:2,034,254 – 2,069,122 | 1420 | 153.28 |
| 40-2 | Manin00g002070.1 | MK936540 | 3 | Chr 0:2,075,240 – 2,076,583 | 388 | 41.9 |
| 22-2 | Manin00g014940.1 | MK936541 | 5 | Chr 0:40,127,372 – 40,129,482 | 361 | 38.77 |
| 22-3 | Manin00g014950.1 | MK936542 | 9 | Chr 0:40,149,927 – 40,158,394 | 697 | 74 |
| 22-7 | Manin02g003450.1 | MK936543 | 4 | Chr 2:8,085,713 – 8,087,455 | 388 | 41.01 |
| 22-8 | Manin02g003460.1 | MK936544 | 8 | Chr 2:8,121,984 – 8,135,247 | 717 | 75.52 |
| 22-6 | Manin02g003470.1 | MK936545 | 4 | Chr 2:8,164,464 – 8,165,561 | 222 | 23.35 |
| 22-5 | Manin02g006220.1 | MK936546 | 4 | Chr 2:12,050,377 – 12,052,051 | 395 | 41.51 |
| 44-2 | Manin02g010300.1 | MK936547 | 6 | Chr 2:16,207,991 – 16,209,888 | 430 | 46.24 |
| 46-1 | Manin02g010380.1 | MK936548 | 5 | Chr 2:16,272,348 – 16,276,078 | 455 | 49.49 |
| 6-1 | Manin02g011060.1 | MK936549 | 6 | Chr 2:16,947,539 – 16,949,777 | 474 | 51.36 |
| 71-1 | Manin03g008820.1 | MK936550 | 3 | Chr 3:14,644,395 – 14,646,459 | 506 | 55.11 |
| 49 | Manin04g001200.1 | MK936551 | 5 | Chr 4:840,755 – 845,219 | 444 | 48.29 |
| 56-2 | Manin04g001870.1 | MK936552 | 6 | Chr 4:1,283,689 – 1,286,480 | 467 | 51.42 |
| 69-1 | Manin04g008520.1 | MK936553 | 9 | Chr 4:6,449,559 – 6,452,110 | 401 | 43.57 |
| 69-2 | Manin04g008530.1 | MK936554 | 51 | Chr 4:6,453,359 – 6,506,648 | 1916 | 210.08 |
| 6-2 | Manin04g015430.1 | MK936555 | 6 | Chr 4:15,164,191 – 15,166,702 | 481 | 52.04 |
| 46-4 | Manin04g016040.2 | MK936556 | 2 | Chr 4:17,641,143 – 17,641,804 | 126 | 13.92 |
| 46-2 | Manin04g016050.1 | MK936557 | 5 | Chr 4:17,661,281 – 17,664,562 | 455 | 49.58 |
| 44-3 | Manin04g016150.1 | MK936558 | 10 | Chr 4:17,785,523 – 17,798,068 | 812 | 87.45 |
| 58 | Manin05g004560.1 | MK936559 | 5 | Chr 5:9,153,656 – 9,159,588 | 495 | 55.28 |
| 66 | Manin05g011290.1 | MK936560 | 3 | Chr 5:14,468,486 – 14,468,882 | 83 | 8.77 |
| 31 | Manin05g011300.1 | MK936561 | 3 | Chr 5:14,469,030 – 14,469,864 | 146 | 15.58 |
| 52 | Manin06g006140.1 | MK936562 | 5 | Chr 6:8,842,198 – 8,853,462 | 490 | 53.59 |
| 59 | Manin09g006660.1 | MK936563 | 6 | Chr 9:12,050,364 – 12,052,270 | 265 | 30.13 |
| 11-1 | Manin09g009540.1 | MK936564 | 6 | Chr 9:14,987,782 – 14,989,946 | 421 | 45.97 |
| 16 | Manin09g015600.1 | MK936565 | 9 | Chr 9:20,563,521 – 20,567,215 | 439 | 47.67 |
| 51-1 | Manin11g003610.1 | MK936566 | 7 | Chr 11:2,576,944 – 2,581,060 | 449 | 50.07 |
| 46-3 | Manin11g004580.1 | MK936567 | 8 | Chr 11:3,180,214 – 3,183,340 | 508 | 55.04 |
| 42 | Manin11g009310.1 | MK936568 | 12 | Chr 11:6,507,441 – 6,517,554 | 1195 | 126.7 |
| 10 | Manin11g013560.1 | MK936569 | 14 | Chr 11:9,949,786 – 9,957,811 | 630 | 68.86 |
| 22-4 | Manin12g011650.1 | MK936570 | 4 | Chr 12:14,098,917 – 14,100320 | 369 | 39.51 |
| 23-2 | Manin12g011660.1 | MK936571 | 4 | Chr 12:14,119,764 – 14,121,825 | 381 | 40.32 |
| 23-3 | Manin12g011670.1 | MK936572 | 4 | Chr 12:14,130,968 – 14,132,912 | 396 | 42.04 |
| 23-1 | Manin12g011680.1 | MK936573 | 2 | Chr 12:14,164,835 – 14,165,661 | 211 | 21.8 |
| 22-1 | Manin12g011690.1 | MK936574 | 13 | Chr 12:14,198,558 – 14,220,187 | 983 | 106.04 |
| 21-1 | Manin12g011700.1 | MK936575 | 3 | Chr 12:14,234,467 – 4,236,214 | 320 | 34.69 |
| 21-2 | Manin12g011710.1 | MK936576 | 4 | Chr 12:14,253,498 – 14,255,464 | 361 | 38.43 |
| 11-2 | Manin16g003570.1 | MK936577 | 7 | Chr 16:9,024,035 – 9,026,214 | 448 | 49.13 |
| 71-3 | Manin16g003910.1 | MK936578 | 2 | Chr 16:9,392,985 – 9,395,675 | 628 | 69.05 |
| 51-2 | Manin16g013070.1 | MK936579 | 6 | Chr 16:18,234,739 – 18,238,211 | 466 | 51.3 |
| 56-1 | Manin16g014550.1 | MK936580 | 6 | Chr 16:19,497,634-19,499,847 | 481 | 52.66 |
| 56-3 | Manin17g000810.1 | MK936581 | 6 | Chr 17:687,147 – 690,549 | 488 | 53.87 |
| 17 | Manin18g005790.1 | MK936582 | 9 | Chr 18:4,566,836 – 4,570,385 | 463 | 50.55 |
| 71-2 | Manin18g012040.1 | MK936583 | 3 | Chr 18:15,695,841 – 15,704,321 | 470 | 51.18 |
| 44-1 | Manin19g015630.1 | MK936584 | 7 | Chr 19:21,223,404 – 21,227,980 | 733 | 79.91 |
| 14 | Manin20g006090.1 | MK936585 | 9 | Chr 20:11,233,232 – 11,235,842 | 457 | 49.8 |
| 53 | Manin20g008040.1 | MK936586 | 9 | Chr 20:2,034,254 – 2,069,122 | 751 | 82.41 |
The ORFs from the 48 mango PG amino acid sequence were aligned to characterize their primary structure. A total of four conserved domains (motifs I, II, III, and IV) that are proposed as essential for PG hydrolysis activity were reported in PG amino acid sequences for several plants (Torki et al., 2000). These different characteristic motifs were found in 29 M. indica PGs in their primary structure, within the deduced amino acid sequences (Supplementary Figures S1B,C). Meanwhile, the motif III was absent in the primary structure of 16 PGs (Supplementary Figure S1A), and three sequences showed no domains. Most of the reported PGs for several species do have those four domains conserved. Nonetheless, one or more domains have been lost in some of these proteins, having the third domain turned into the most scarcely conserved (Chen et al., 2016). Aspartic acids in NTD and DD regions are contained in domains I and II, respectively; their carboxylate groups may represent a component of the catalytic site, in any case. The domain III (CGPGHG) seems to participate in the reaction by a histidine as the catalytic residue, whereas the domain IV, formed by residues positively charged, act in ionic interactions with carboxylate groups present in the substrate (Torki et al., 2000; Chen et al., 2016; Ke et al., 2018).
Phylogenetic, Gene Structure, and Regulatory Regions Analysis of Mango Polygalacturonases
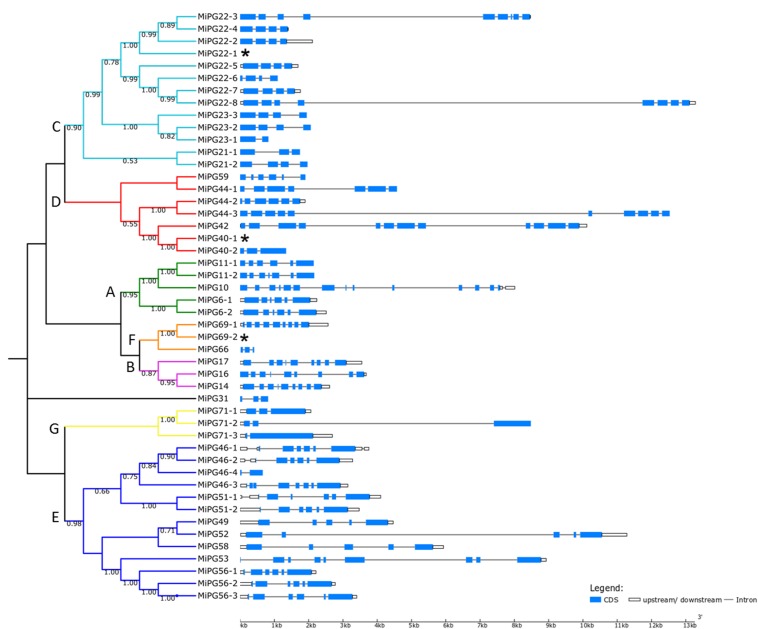
A rooted phylogenetic tree composed of 48 mango PG amino acid sequences, previously named according to A. thaliana orthologs, was obtained with the Neighbor-Joining tree method and MUSCLE as alignment algorithm (Figure 3 and Supplementary Figure S2). In agreement with Ke et al. (2018), as well as other authors (Yu et al., 2014; Liang et al., 2015), the tree was divided in seven main clades (Clade A to G), except for MiPG31 sequence that appear as an external branch. Interestingly, PG sequences that only presented three functional domains were correctly grouped between clades E, F, and G. It is worth noting that previous studies have shown that clade F members may be associated with flower and probably fruit development (Park et al., 2010; Liang et al., 2015). On the other hand, PG sequences containing all four domains were distributed relatively in the rest of the clades. Remarkably, all PGs classified as exo-PGs (MiPG40-1, MiPG40-2, MiPG44-1, MiPG44-2, MiPG44-3, MiPG42, and MiPG59) were grouped into clade D (see Figure 3), probably because different types of PG have evolved at different times in the history of plants, being the group of exo-PGs one of the most recent and only present in angiosperms (Park et al., 2010).
FIGURE 3.
Phylogenetic analysis of PG proteins sequences found in M. indica L. Tommy Atkins genome and exon/intron organization of mango PG genes. Left part shows the phylogenetic tree of 48 PG amino acid sequences in M. indica L. constructed using MEGA 7 by the NJ method. The number of branches indicates the bootstrap percentage based on 2000 replicates. Values under 50 are not shown in the tree. At the right part, gene structure including exon/intron configuration of each PG is shown and tagged with its respective name. Gene structures for MiPG22-1, MiPG40-1, and MiPG69-2 (∗) can be seen at Supplementary Figure S3.
The gene structures shown in Figure 3 and Supplementary Figure S3 correspond to their full genomic sequences, which show variability in exon/intron numbers within similar or different clades. According to the literature, the number of exons/introns is consistent within the different PGs groups in the tree, resulting in greater exons average number for clades A, B, and F (7.8, 9, and 21, respectively), as it was described before for other species such as S. lycopersicum, Brassica rapa and C. sativus (Yu et al., 2014; Liang et al., 2015; Ke et al., 2018). These results may suggest that across plant evolution PGs sequences have differentiated in each clade, resulting in common and specialized gene structures and biological functions in plants.
Detailed analysis of PG genes in mango cv. Tommy Atkins genome alongside the phylogenetic analysis revealed that all 48 PG genes are present in different chromosomal localizations (Table 1), distributed in all seven clades. Similar to tomato PGs (Ke et al., 2018) all mango PG genes are distributed in most of the chromosomes (Chr 0, 2, 4, 5, 6, 9, 10, 11, 12, 16 y 17). When we compared these findings with the phylogenetic tree (Figure 3), it was found that most of the PG genes that are located in chromosomes 2 and 0 were found in clades C and D. Interestingly, most of the PGs located in chromosome 12 were grouped under clade C, and the rest of the PG chromosomal localizations were equitably distributed around all clades. Some PG genes were found to be localized side to side, for example, MiPG22-2 and MiPG22-3 or MiPG22-4, MiPG23-2, MiPG23-3, MiPG23-1, MiPG22-1, MiPG21-1, and MiPG21-2 set, all of those are found under the same clade C that contains all four specific domains. On the other hand, PGs located in other clades, like MiPG71-1, MiPG72-2, and MiPG72-3, which only contain three of the domains (I, II, and IV) are located in clade G. These results are consistent, for the most part, with gene structure, protein structure, biological function and grouping within the clades.
To further analyze the mango PG sequences, we selected 1.5 Kb upstream of each PG gene and analyzed these regulatory regions. According to MatInspector results, the 48 PGs have in common 20 regulatory regions. Within these, the Arabidopsis homeobox protein (P$AHBP), high mobility group factor (P$HMGF), DNA binding with one finger (P$DOFF), CCAAT binding factors (P$CAAT) and soybean embryo factor 4 (P$SEF4) were the most abundant with more than 800 matches (Supplementary Table S3A). Also, the ARID/BRIGHT DNA-binding domain-containing transcription factors (P$ARID), Plant TATA binding protein factor and Yeast TATA binding protein factor (O$YTBP) were the most overrepresented TF in the 48 PGs, compared to Arabidopsis genome (Supplementary Table S3B). It is known that some TF are directly related to fruit ripening. In this sense, some MADS-box, like LeMADS-RIN in tomato are necessary for fruit ripening (Vrebalov et al., 2002). A total of 39 intergenic sequences of mango PGs showed hit with MADS box proteins. Moreover, MADS-box also is implicated in ethylene regulation, which in turn activates several ripening-related genes (Vrebalov et al., 2002). A total of four ethylene insensitive3-like factors (P$EINL), four APETALA2-like transcription factors (P$AP2L) and one ethylene response element factors (P$EREF) were also found within the 48 PGs. Other important TF associated with plant hormone signaling like auxin response element (P$AREF) and auxin response factor 3 (P$ARF3) were also found in 38 and 10 PGs sequences, respectively.
Additionally, the 9 PGs which relative expression was evaluated shared 15 cis-regulatory elements in the 1500 pb promoter region analyzed (Figure 4 and Supplementary Table S3C). Among these, P$DOFF family was one of the most abundant with 285 matches. Interestingly, in banana fruit (Musa acuminata) it has been shown that four Dof transcription factors (MaDof10, 23, 24, and 25) that preferentially bind to the core sequence 5′-(T/A)AAAG-3′ belonging to P$DOFF elements, are ethylene-inducible, and their transcripts are accumulated during ripening. Also, MaDof23 interact with MaERF9 (regulator of fruit ripening), acting as a transcriptional repressor, whereas MaERF9 is a transcriptional activator. This antagonistic relationship, lead to the regulation of ripening-related genes, including MaPG1 (Yanagisawa, 2002; Feng et al., 2016). On the other hand, the presence of NAC cis-regulatory sites may play a main role in PGs expression. In the 9 PGs, several cis elements with NAC domain were found (i.e., P$NTMF, P$CNAC, P$NACD, P$NACF, and P$SWNS). In this sense, NAC gene family has been recognized as important transcription factors involved in ripening and softening in fruits like citrus, banana, tomato and peach (de Oliveira et al., 2011; Shan et al., 2012; Zhu et al., 2014; Zhou et al., 2015). Also, in Fragaria chiloensis FcNAC1 interact with DNA sequences containing NAC binding elements in the promoter of a pectate lyase FcPL, activating its transcription (Carrasco-Orellana et al., 2018). Otherwise, each of 9 mango PGs showed at least 4 elements of P$GARP family in their promoter regions. Evidence point out that a transcription factor of the GARP family acts together with bZIP transcription factor PERIANTHIA to activate the transcription of AGAMOUS, an important member of fruit ripening regulation (Maier et al., 2009). Together, these results suggest that PGs whose gene expression was evaluated, may be regulated by members of these transcription factors that bind to the predicted cis elements; moreover, their expression occurs during the advance of the ripening state. However, further studies must be performed in order to prove these asseverations.
FIGURE 4.
Identification of cis-regulatory elements in the 1500 pb promoter region analyzed for the 9 PGs which relative expression was evaluated. The MatInspector program was used, core promoter elements and plant matrix groups, with 0.90 of core and matrix similarity.
Nine PGs Were Differentially Expressed During Post-harvest Ripening and That Correlated With PG Enzymatic Activity
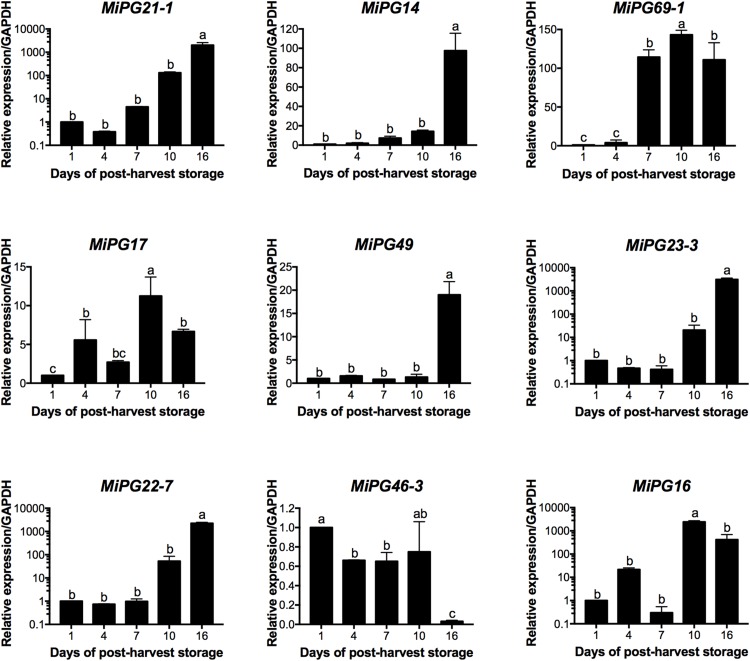
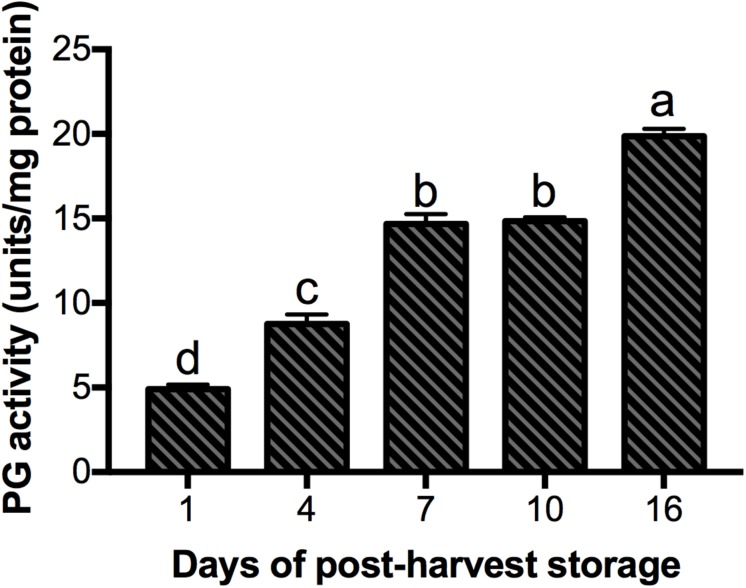
According to the transcriptome data (Dautt-Castro et al., 2015), nine PG genes were differentially expressed during fruit ripening and selected to validate their expression by qRT-PCR (Figure 5). For this validation, we used two reference genes (GAPDH and ACT 7) in order to normalize the data. In both analyses the expression patters for eight PGs evaluated followed the same trends with similar expression levels, which support our results (Figure 5 and Supplementary Figure S4). In this regard, five PG genes (MiPG21-1, MiPG14, MiPG49, MiPG23-3, MiPG22-7) presented their maximum level of expression at 16 days of post-harvest storage; meanwhile for MiPG69-1, MiPG17, and MiPG16 maximum levels were at day 10. MiPG46-3 was down-regulated in mango from day 4, 7, and 16; however, in fruit from day 1 and 10 it remained at similar levels. The transcript with highest relative expression was MiPG23-3 with 3131-fold at day 16 followed by MiPG16 with 2426-fold at day 10. These results were very similar to the expression ratio found in transcriptome by RNA-seq (Supplementary Figure S5). PGs from clade C (MiPG21-1, MiPG22-7, and MiPG23-3) are among those that presented the highest relative expression levels in mango from day 16 of post-harvest (2,000-fold). MiPG16 (clade B) also presented very high relative expression levels (3,000-fold). Meanwhile, MiPG69-1 (clade F) presented relative expression levels of about 200-fold in mango from day 7 of post-harvest when firmness was reduced by 30%. Also, MiPG14 (clade B) that is closely related to MiPG69-1 (see Figure 3) was expressed at 100-fold but in fruit from day 16 after post-harvest. MiPG49 (also from clade B) was 20-fold in mango from day 16, and MiPG17 was 12-fold in fruit from day 10 after post-harvest. These results suggest that these PGs could be related to the loss of firmness observed in the mango fruit. The up-regulation of these PGs during the advance of mango ripening correlates with the activity of PG (Figure 6), where the activity increased with the advance of ripening. In mango from day 1 and 4 of post-harvest, PG activity was 5 and 9 units/mg protein; while for mango of days 7 and 10 it was 15 units/mg protein and in mango from day 16 of post-harvest the maximum PG activity was reached (19.8 units/mg protein).
FIGURE 5.
Relative expression of MiPG21-1, MiPG14, MiPG69-1, MiPG17, MiPG49, MiPG23-3, MiPG22-7, MiPG46-3, and MiPG16 in mango mesocarp. Data of the nine genes represent the average of three biological replicates (n = 3) with three experimental replicates each. Error bars indicate SE of the means and different letters indicate significant differences. Statistical significance of differences between days of post-harvest storage was determined by one-way ANOVA with the Fisher test (P < 0.05).
FIGURE 6.
Polygalacturonase (PG) activity in “Kent” mango fruit during storage at 20°C. Error bars indicate SD of the means (n = 3). Different letters indicate statistically significant differences between days of post-harvest storage using one-way ANOVA and Tuckey-Kramer test (P < 0.05).
The expression of PG genes has been extensively studied in other fruits. Furthermore, it has been observed that PG genes related to fruit ripening, often are induced by the endogenous production of ethylene and the application of exogenous ethylene. As found in the expression of almost all evaluated mango PGs (except for MiPG46-3), their expressions levels increased with the advance of ripening. The differential expression of PG cDNAs has also been observed in fruits such as bananas, where four PGs associated with maturation (MaPG1, MaPG2, MaPG3, and MaPG4) were evaluated in the fruit peel. Maximum expression levels were observed at days 4, 2, 7, and 7 of post-harvest, respectively; featuring the highest levels of gene expression in MaPG4 (values of about 2300-fold, 2–ΔΔCt) and the lowest in MaPG3 (values of approximately 1.3-fold, 2–ΔΔCt) (Mbéguié-A-Mbéguié et al., 2009). The expression of the apricot ortholog gene of peach PRF5 named PaPG was evaluated in three different varieties (Goldrich, Currot, and Canino) and its expression correlated with fruit softening and ethylene release; moreover, PaPG responded to exogenous ethylene (Leida et al., 2011). The results obtained for mango PGs suggest that these enzymes could be ethylene-dependent, which correlates with the ethylene-response elements found in many PG promoter regions (Supplementary Table S3) and suggest that them could be associated with ripening and responsible in part for the softening on mango fruits.
As described above, the maximum loss of firmness of mango fruit was registered at day 10 of post-harvest storage. Different studies have shown that the silencing of independent cell wall-associated genes including PGs (Smith et al., 1988), pectin methylesterases (Phan et al., 2007), pectate lyases (Uluisik et al., 2016) and expansins (Brummell et al., 1999) have resulted in a range of effects of softening, and in some cases with zones still rich in de-esterified pectins (Yang et al., 2017). This evidence suggests that the loss of firmness is dependent on the action of different families of cell wall-degrading enzymes that act in distinct ripening stages. In this sense, we could infer that the evaluated mango PGs are related to loss of firmness in the fruit. Therefore, the relationship of the gene expression data found and analyzed in this study as well as the physiological parameters of mango cv. “Kent,” during their maturation process, show evidence of the role that PG genes associated with this process, specifically in fruit softening. Thereby also demonstrating the importance of these genes in the ripening process of fruits.
PG Structural Models
Mango PGs are structurally conserved (Figure 7) and have a single-stranded right-handed beta helix structure, also known as a pectin lyase-like CATH superfamily5 (Dawson et al., 2017; Lewis et al., 2018). This superfamily is mostly found in bacteria, plant and fungus, and scarcely on invertebrates and environmental samples. According to biochemical functions, this fold encompasses 63% of pectin esterases (E.C. 3.1.1.11), followed by pectate lyases (16%, E.C. 4.2.2.2). All enzymes having the parallel beta-helical fold can recognize and hydrolyze large polysaccharides (Fujimoto, 2013).
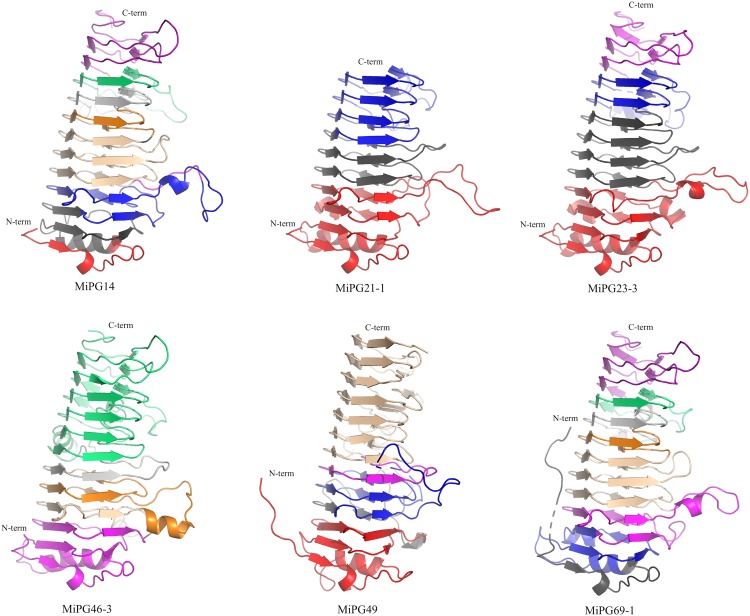
FIGURE 7.
Structural models for selected mango PGs. The structures are colored according to the exon color used in Supplementary Figure S6. MiPG14 (clade B), MiPG69-1 (clade F), MiPG21-1 and MiPG23-3 (clade C), MiPG46-3, and MiPG49 (clade E) were modeled using rhamnogalacturonase A from Aspergillus aculeatus (PDB 1RMG) as a template. Figures were built using PyMol.
The structures for the six PGs are very similar; for example, MiPG14 (clade B) and MiPG69-1 (clade F) have 9 exons in their gene structures, and they share a similar exon distribution and structural arrangement (Supplementary Figure S6). Interestingly, this result supports the hypothesis that exons code for functional structural units in proteins (Traut, 1988), and structural models may help to understand the evolutionary history of PGs in mango. MiPG21-1 and MiPG23-3 are from clade C, with three and four exons, respectively; their structural models are similar; although, MiPG23-3 is 76 residues longer and the fourth exon leads to a larger structure. MiPG46-3 and MiPG49 pertain to clade E, and have 8 and 5 exons, respectively; their models present differences in exon distribution, although overall the structural models are very similar. Until now, there is no crystallographic structure for a plant PG; all solved structures are mostly from fungi and one from bacteria. One important aspect to further explore the structural study of these proteins is the accommodation of the polysaccharide into each active site. Another area of interest would be the biochemical characterization of the PGs in terms of their kinetic properties.
Our results add knowledge about this multigenic family of enzymes involved in structural changes in the cell wall and the concomitant softening of the fruit. Beyond physiological aspects, molecular information about gene expression and evolutive relationships of mango PGs, this work add some perspectives about the study of PG in climacteric fruits, that could be used to improve also shelf-life of other plants species.
Data Availability
The datasets generated for this study can be accessed from GenBank MK936539-MK936586. The datasets analyzed for this study can be found deposited in the NCBI (Accesion PRJNA258477) (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA258477).
Author Contributions
MI-O and MD-C conceived and designed the experiments. MD-C performed the experiments. MD-C, AL-V, AO-L, MI-O, CC-V, DK, AS-S, and AS-B analyzed the data. MI-O, AO-L, GG-A, JC-F, MM-T, and AS-B contributed to the reagents, materials, and analysis tools. MD-C, MI-O, CC-V, AL-V, AS-S, and DK wrote the manuscript. All authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Rosalba Contreras-Martínez, Rosabel Velez de la Rocha, Célida I. Martínez-Rodríguez, Mónica A. Villegas-Ochoa, Emmanuel Hernández-Aispuro, and Francisco Soto for their technical support. We also thank Dr. Rogerio R. Sotelo-Mundo for critical reading of the manuscript and preparation of PG structural models.
Funding. MI-O thanks CONACyT for grant CB2012-01-178296. MD-C, AL-V, and AS-S thank CONACyT for the scholarship obtained for their Graduate Studies. AO-L thanks for an academic exchange CIAD-UNAM 2019 grant. DK thanks the BARD grant from USDA for publication fees.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00969/full#supplementary-material
Multiple sequence alignment analysis of the polygalacturonases (PGs) from mango (Mangifera indica L.). Partial sequences where the characteristic domains are located are shown, (1A) Shows PGs that contain motifs I, II and IV; (1B) and (1C) Show PGs that have motifs I, II, II and IV. Red boxes show the four functional motifs. Identical amino acids are shown in black, and gray shading represents conserved amino acid residues.
Phylogenetic analysis of Mangifera indica L. and Arabidopsis thaliana polygalacturonases. The evolutionary history was inferred using the Neighbor-Joining method, while distances were computed using the Poisson correction method. The analysis involved 117 amino acid sequences, of which 48 sequences belong to mango (MiPGs). Taxa clustered together in the bootstrap test (2000 replicates). Evolutionary analyses were conducted in MEGA X.
Gene structure of the longest mango PG genes including exon/intron configuration. MiPG40-1 contains 16 exons, MiPG69-2 contains 51 exons and MiPG22-1 has 13 exons. MiPG40-1 encodes an enzyme of 1420 aa, MiPG69-2 and enzyme of 1916 aa and MiPG22-1 an enzyme of 983 aa long. The structure of the introns and exons of the MiPG genes were obtained using GSDS 2.0 (http://gsds.cbi.pku.edu.cn/).
Relative expression of MiPG21-1, MiPG14, MiPG69-1, MiPG17, MiPG49, MiPG23-3, MiPG22-7, MiPG46-3, and MiPG16 in mango mesocarp, using ACT 7 as reference gene. Data of the nine genes represent the average of three biological replicates (n = 3) with three experimental replicates each. Error bars indicate SE of the means and different letters indicate significant differences. Statistical significance of differences between days of postharvest storage was determined by one-way ANOVA with the Fisher test (P < 0.05).
Expression ratio (Log2) for the 9 polygalacturonases was obtained by RNA-seq and qPCR. GAPDH was the constitutive gene used for normalization of qPCR data. Bars represent the standard error (n = 3).
Sequence alignment of polygalacturonases that were modeled to obtain theoretical models, the exons were colored with different colors. MiPG21-1 and MiPG23-3 are from clade C, MiPG14 from clade B, MiPG69-1 from clade F; while MiPG46-3 and MiPG49 are from clade E.
Primer sequences used for qRT-PCR.
PG superfamilies identified in mango polygalacturonases. (A) Genome ID, sequence name, biological process, cellular component, molecular function, classification, enzyme kind (endo/exo), isoelectric point and GenBank accession. (B) SignalP analysis of polygalacturonases from mango fruit.
Analysis of regulatory regions in 5′UTR polygalacturonases genes. (A) Common regulatory regions identified in 48 PGs. (B) Overrepresented TF families. (C) Common regulatory regions identified in 9 PGs which expression was evaluated.
References
- Ali Z. M., Chin L. H., Lazan H. (2004). A comparative study on wall degrading enzymes, pectin modifications and softening during ripening of selected tropical fruits. Plant Sci. 167 317–327. 10.1016/J.PLANTSCI.2004.03.030 [DOI] [Google Scholar]
- Asif M. H., Nath P. (2005). Expression of multiple forms of polygalacturonase gene during ripening in banana fruit. Plant Physiol. Biochem. 43 177–184. 10.1016/j.plaphy.2005.01.011 [DOI] [PubMed] [Google Scholar]
- Bouzayen M., Latché A., Nath P., Pech J. C. (2010). “Mechanism of fruit ripening,” in Plant Developmental Biology-Biotechnological Perspectives, eds Pua E. C., Davey M. R. (Berlin: Springer; ), 319–339. [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. 10.1006/abio.1976.9999 [DOI] [PubMed] [Google Scholar]
- Brummell D. A. (2006). Cell wall disassembly in ripening fruit. Funct. Plant Biol. 33 103–119. [DOI] [PubMed] [Google Scholar]
- Brummell D. A., Harpster M. H. (2001). Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plants. Plant Mol. Biol. 47 311–339. [PubMed] [Google Scholar]
- Brummell D. A., Harpster M. H., Civello P. M., Palys J. M., Bennett A. B., Dunsmuir P., et al. (1999). Modification of expansin protein abundance in tomato fruit alters softening and cell wall polymer metabolism during ripening. Plant Cell 11 2203–2216. 10.1105/TPC.11.11.2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas-Coronel W. G., Velez de la Rocha R., Siller-Cepeda J. H., Osuna-Enciso T., Muy-Rangel M. D., Sañudo-Barajas J. A., et al. (2012). Changes in the composition of starch, pectins and hemicelluloses during the ripening stage of mango (Mangifera indica cv. KENT). Rev. Chapingo Ser. Hortic. 18 5–19. 10.5154/R.RCHSH.18.001 [DOI] [Google Scholar]
- Carrasco-Orellana C., Stappung Y., Mendez-Yañez A., Allan A. C., Espley R. V., Plunkett B. J., et al. (2018). Characterization of a ripening-related transcription factor FcNAC1 from Fragaria chiloensis fruit. Sci. Rep. 8:10524. 10.1038/s41598-018-28226-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., et al. (2005). MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21 2933–2942. 10.1093/bioinformatics/bti473 [DOI] [PubMed] [Google Scholar]
- Chen H., Shao H., Fan S., Ma J., Zhang D., Han M., et al. (2016). Identification and phylogenetic analysis of the polygalacturonase gene family in apple. Hortic. Plant J. 2 241–252. 10.1016/J.HPJ.2017.01.004 [DOI] [Google Scholar]
- Corbacho J., Romojaro F., Pech J.-C., Latché A., Gomez-Jimenez M. C. (2013). Transcriptomic events involved in melon mature-fruit abscission comprise the sequential induction of cell-wall degrading genes coupled to a stimulation of endo and exocytosis. PLoS One 8:e58363. 10.1371/journal.pone.0058363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dautt-Castro M., Ochoa-Leyva A., Contreras-Vergara C. A., Muhlia-Almazán A., Rivera-Domínguez M., Casas-Flores S., et al. (2018). Mesocarp RNA-Seq analysis of mango (Mangifera indica L.) identify quarantine postharvest treatment effects on gene expression. Sci. Hortic. 227 146–153. 10.1016/j.scienta.2017.09.031 [DOI] [Google Scholar]
- Dautt-Castro M., Ochoa-Leyva A., Contreras-Vergara C. A., Pacheco-Sanchez M. A., Casas-Flores S., Sanchez-Flores A., et al. (2015). Mango (Mangifera indica L.) cv. kent fruit mesocarp de novo transcriptome assembly identifies gene families important for ripening. Front. Plant Sci. 6:62. 10.3389/fpls.2015.00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson N. L., Lewis T. E., Das S., Lees J. G., Lee D., Ashford P., et al. (2017). CATH: an expanded resource to predict protein function through structure and sequence. Nucleic Acids Res. 45 D289–D295. 10.1093/nar/gkw1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira T. M., Cidade L. C., Gesteira A. S., Coelho Filho M. A., Soares Filho W. S., Costa M. G. C., et al. (2011). Analysis of the NAC transcription factor gene family in citrus reveals a novel member involved in multiple abiotic stress responses. Tree Genet. Genomes 7 1123–1134. 10.1007/s11295-011-0400-8 [DOI] [Google Scholar]
- DeLano W. L. (2002). The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Sci. [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabi J. P., Broetto S. G., Silva S. L. G. L., da Zhong S., Lajolo F. M., do Nascimento J. R. O., et al. (2014). Analysis of papaya cell wall-related genes during fruit ripening indicates a central role of polygalacturonases during pulp softening. PLoS One 9:e105685. 10.1371/journal.pone.0105685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B., Han Y., Xiao Y., Kuang J., Fan Z., Chen J., et al. (2016). The banana fruit Dof transcription factor MaDof23 acts as a repressor and interacts with MaERF9 in regulating ripening-related genes. J. Exp. Bot. 67 2263–2275. 10.1093/jxb/erw032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto Z. (2013). Structure and function of carbohydrate-binding module families 13 and 42 of glycoside hydrolases, comprising a β-trefoil fold. Biosci. Biotechnol. Biochem. 77 1363–1371. 10.1271/bbb.130183 [DOI] [PubMed] [Google Scholar]
- Giovannoni J. J., DellaPenna D., Bennett A. B., Fischer R. L. (1989). Expression of a chimeric polygalacturonase gene in transgenic rin (ripening inhibitor) tomato fruit results in polyuronide degradation but not fruit softening. Plant Cell 1, 53–63. 10.1105/tpc.1.1.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Carranza Z. H., Elliott K. A., Roberts J. A. (2007). Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thaliana. J. Exp. Bot. 58 3719–3730. 10.1093/jxb/erm222 [DOI] [PubMed] [Google Scholar]
- Goulao L. F., Oliveira C. M. (2008). Cell wall modifications during fruit ripening: when a fruit is not the fruit. Trends Food Sci. Technol. 19 4–25. 10.1016/j.tifs.2007.07.002 [DOI] [Google Scholar]
- Grierson D. (2013). “Ethylene and the control of fruit ripening,” in The Molecular Biology and Biochemistry of Fruit Ripening, eds Seymour G., Tucker G. A., Poole M., Giovannoni J. (Hoboken, NJ: Wiley-Blackwell; ), 43–68. [Google Scholar]
- Gross K. C. (1982). A rapid and sensitive spectrophotometric method for assaying polygalacturonase using 2-cyanoacetamide [Tomato, fruit softening]. HortScience 17 933–934. [Google Scholar]
- Gu C., Wang L., Wang W., Zhou H., Ma B., Zheng H., et al. (2016). Copy number variation of a gene cluster encoding endopolygalacturonase mediates flesh texture and stone adhesion in peach. J. Exp. Bot. 67 1993–2005. 10.1093/jxb/erw021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Jin J., Guo A.-Y., Zhang H., Luo J., Gao G., et al. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31 1296–1297. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islas-Osuna M. A., Stephens-Camacho N. A., Contreras-Vergara C. A., Rivera-Dominguez M., Sanchez-Sanchez E., Villegas-Ochoa M. A., et al. (2010). Novel postharvest treatment reduces ascorbic acid losses in mango (Mangifera indica L.) var. kent. Am. J. Agric. Biol. Sci. 5 342–349. 10.3844/ajabssp.2010.342.349 [DOI] [Google Scholar]
- Jiménez D. P., Taddei E. B., Ruiz J. M., Robles J. G., Aguilar G. G., Rojas R. T., et al. (2004). Efecto del calcio y cera comestible en la calidad de mangos”~ kent”TM durante el almacenamiento. Rev. la Fac. Agron 21, 351–358. [Google Scholar]
- Ke X., Wang H., Li Y., Zhu B., Zang Y., He Y., et al. (2018). Genome-wide identification and analysis of polygalacturonase genes in Solanum lycopersicum. Int. J. Mol. Sci. 19:2290. 10.3390/ijms19082290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10 845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang C., Dörnenburg H. (2000). Perspectives in the biological function and the technological application of polygalacturonases. Appl. Microbiol. Biotechnol. 53, 366–375. 10.1007/s002530051628 [DOI] [PubMed] [Google Scholar]
- Leida C., Ríos G., Soriano J. M., Pérez B., Llácer G., Crisosto C. H., et al. (2011). Identification and genetic characterization of an ethylene-dependent polygalacturonase from apricot fruit. Postharvest Biol. Technol. 62 26–34. 10.1016/J.POSTHARVBIO.2011.04.003 [DOI] [Google Scholar]
- Letunic I., Bork P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44 W242–W245. 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis T. E., Sillitoe I., Dawson N., Lam S. D., Clarke T., Lee D., et al. (2018). Gene3D: extensive prediction of globular domains in proteins. Nucleic Acids Res. 46 D435–D439. 10.1093/nar/gkx1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Xu C., Korban S. S., Chen K. (2010). Regulatory mechanisms of textural changes in ripening fruits. CRC Crit. Rev. Plant Sci. 29 222–243. 10.1080/07352689.2010.487776 [DOI] [Google Scholar]
- Liang Y., Yu Y., Shen X., Dong H., Lyu M., Xu L., et al. (2015). Dissecting the complex molecular evolution and expression of polygalacturonase gene family in Brassica rapa ssp. chinensis. Plant Mol. Biol. 89 629–646. 10.1007/s11103-015-0390-2 [DOI] [PubMed] [Google Scholar]
- Litz R. E. (2009). Mango. Compend. Transgenic Crop Plants. Hoboken NJ: Wiley. [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 402–408. 10.1006/METH.2001.1262 [DOI] [PubMed] [Google Scholar]
- López-Gómez R., Gómez-Lim M. A. (1992). A method for extracting intact RNA from fruits rich in polysaccharides using ripe mango mesocarp. HortScience 27, 440–442. 10.21273/HORTSCI.27.5.440 [DOI] [Google Scholar]
- Maier A. T., Stehling-Sun S., Wollmann H., Demar M., Hong R. L., Haubeiß S., et al. (2009). Dual roles of the bZIP transcription factor PERIANTHIA in the control of floral architecture and homeotic gene expression. Development 136 1613–1620. 10.1242/dev.033647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbéguié-A-Mbéguié D., Hubert O., Baurens F. C., Matsumoto T., Chillet M., Fils-Lycaon B., et al. (2009). Expression patterns of cell wall-modifying genes from banana during fruit ripening and in relationship with finger drop. J. Exp. Bot. 60 2021–2034. 10.1093/jxb/erp079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muy Rangel D., Espinoza Valenzuela B., Siller Cepeda J., Sañudo Barajas J. A., Valdez Torres B., Osuna Enciso T., et al. (2009). Efecto del 1-metilciclopropeno (1-MCP) y de una película comestible sobre la actividad enzimática y calidad poscosecha del mango’ataulfo’. Rev. Fitotec. Mex. 32 53–60. [Google Scholar]
- Park K. C., Kwon S. J., Kim N. S. (2010). Intron loss mediated structural dynamics and functional differentiation of the polygalacturonase gene family in land plants. Genes Genomics 32 570–577. 10.1007/s13258-010-0076-78 [DOI] [Google Scholar]
- Phan T. D., Bo W., West G., Lycett G. W., Tucker G. A. (2007). Silencing of the major salt-dependent isoform of pectinesterase in tomato alters fruit softening. Plant Physiol. 144 1960–1967. 10.1104/pp.107.096347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanna V., Prabha T. N., Tharanathan R. N. (2006). Multiple forms of polygalacturonase from mango (Mangifera indica L. cv Alphonso) fruit. Food Chem. 95 30–36. 10.1016/J.FOODCHEM.2004.12.014 26047433 [DOI] [Google Scholar]
- Qian M., Zhang Y., Yan X., Han M., Li J., Li F., et al. (2016). Identification and expression analysis of polygalacturonase family members during peach fruit softening. Int. J. Mol. Sci. 17:1933. 10.3390/ijms17111933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzaq K., Singh Z., Khan A. S., Khan S. A. K. U., Ullah S. (2016). Role of 1-MCP in regulating ‘Kensington Pride’ mango fruit softening and ripening. Plant Growth Regul. 78 401–411. 10.1007/s10725-015-0101-107 [DOI] [Google Scholar]
- Roongsattham P., Morcillo F., Jantasuriyarat C., Pizot M., Moussu S., Jayaweera D., et al. (2012). Temporal and spatial expression of polygalacturonase gene family members reveals divergent regulation during fleshy fruit ripening and abscission in the monocot species oil palm. BMC Plant Biol. 12:150. 10.1186/1471-2229-12-150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Schrödinger L. (2019). The {PyMOL} Molecular Graphics System, Version 2.3. Available at: https://pymol.org/2/ (accessed March 22, 2019). [Google Scholar]
- Shan W., Kuang J., Chen L., Xie H., Peng H., Xiao Y., et al. (2012). Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. J. Exp. Bot. 63 5171–5187. 10.1093/jxb/ers178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Dwivedi U. N. (2008). Effect of Ethrel and 1-methylcyclopropene (1-MCP) on antioxidants in mango (Mangifera indica var. Dashehari) during fruit ripening. Food Chem. 111 951–956. 10.1016/J.FOODCHEM.2008.05.011 [DOI] [PubMed] [Google Scholar]
- Smith C. J. S., Watson C. F., Ray J., Bird C. R., Morris P. C., Schuch W., et al. (1988). Antisense RNA inhibition of polygalacturonase gene expression in transgenic tomatoes. Nature 334 724–726. 10.1038/334724a0 [DOI] [Google Scholar]
- Suntornwat O., Lertwikoon N., Bungaruang L., Chaimanee P., Speirs J. (2000). Cloning and characterization of a putative endopolygalacturonase cDNA from ripening mango (Mangifera indica Linn CV. Nam Dok Mai). Acta Hortic. 509 153–158. 10.17660/ActaHortic.2000.509.14 [DOI] [Google Scholar]
- Tafolla-Arellano J. C., Zheng Y., Sun H., Jiao C., Ruiz-May E., Hernández-Oñate M. A., et al. (2017). Transcriptome analysis of mango (Mangifera indica L.) fruit epidermal peel to identify putative cuticle-associated genes. Sci. Rep. 7:46163. 10.1038/srep46163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torki M., Mandaron P., Mache R., Falconet D. (2000). Characterization of a ubiquitous expressed gene family encoding polygalacturonase in Arabidopsis thaliana. Gene 242 427–436. 10.1016/S0378-1119(99)00497-497 [DOI] [PubMed] [Google Scholar]
- Traut T. W. (1988). Do exons code for structural or functional units in proteins? Proc. Natl. Acad. Sci. U.S.A. 85 2944–2948. 10.1073/pnas.85.9.2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uluisik S., Chapman N. H., Smith R., Poole M., Adams G., Gillis R. B., et al. (2016). Genetic improvement of tomato by targeted control of fruit softening. Nat. Biotechnol. 34 950–952. 10.1038/nbt.3602 [DOI] [PubMed] [Google Scholar]
- Vrebalov J., Ruezinsky D., Padmanabhan V., White R., Medrano D., Drake R., et al. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296 343–346. 10.1126/science.1068181 [DOI] [PubMed] [Google Scholar]
- Wakasa Y., Kudo H., Ishikawa R., Akada S., Senda M., Niizeki M., et al. (2006). Low expression of an endopolygalacturonase gene in apple fruit with long-term storage potential. Postharvest Biol. Technol. 39 193–198. 10.1016/j.postharvbio.2005.10.005 [DOI] [Google Scholar]
- Wang D., Yeats T. H., Uluisik S., Rose J. K. C., Seymour G. B. (2018). Fruit softening: revisiting the role of pectin. Trends Plant Sci. 23 302–310. 10.1016/j.tplants.2018.01.006 [DOI] [PubMed] [Google Scholar]
- White P. J. (2002). Recent advances in fruit development and ripening: an overview. J. Exp. Bot. 53 1995–2000. 10.1093/jxb/erf105 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. (2002). The dof family of plant transcription factors. Trends Plant Sci. 7 555–560. 10.1016/s1360-1385(02)02362-2 [DOI] [PubMed] [Google Scholar]
- Yang L., Huang W., Xiong F., Xian Z., Su D., Ren M., et al. (2017). Silencing of SlPL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould. Plant Biotechnol. J. 15 1544–1555. 10.1111/pbi.12737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Liang Y., Lv M., Wu J., Lu G., Cao J. (2014). Genome-wide identification and characterization of polygalacturonase genes in Cucumis sativus and Citrullus lanatus. Plant Physiol. Biochem. 74 263–275. 10.1016/J.PLAPHY.2013.11.022 [DOI] [PubMed] [Google Scholar]
- Zhou H., Lin-Wang K., Wang H., Gu C., Dare A. P., Espley R. V., et al. (2015). Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 82 105–121. 10.1111/tpj.12792 [DOI] [PubMed] [Google Scholar]
- Zhu M., Chen G., Zhou S., Tu Y., Wang Y., Dong T., et al. (2014). A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant Cell Physiol. 55, 119–135. 10.1093/pcp/pct162 [DOI] [PubMed] [Google Scholar]
- Ziliotto F., Begheldo M., Rasori A., Bonghi C., Tonutti P. (2008). Transcriptome profiling of ripening nectarine (Prunus persica L. Batsch) fruit treated with 1-MCP. J. Exp. Bot. 59 2781–2791. 10.1093/jxb/ern136 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple sequence alignment analysis of the polygalacturonases (PGs) from mango (Mangifera indica L.). Partial sequences where the characteristic domains are located are shown, (1A) Shows PGs that contain motifs I, II and IV; (1B) and (1C) Show PGs that have motifs I, II, II and IV. Red boxes show the four functional motifs. Identical amino acids are shown in black, and gray shading represents conserved amino acid residues.
Phylogenetic analysis of Mangifera indica L. and Arabidopsis thaliana polygalacturonases. The evolutionary history was inferred using the Neighbor-Joining method, while distances were computed using the Poisson correction method. The analysis involved 117 amino acid sequences, of which 48 sequences belong to mango (MiPGs). Taxa clustered together in the bootstrap test (2000 replicates). Evolutionary analyses were conducted in MEGA X.
Gene structure of the longest mango PG genes including exon/intron configuration. MiPG40-1 contains 16 exons, MiPG69-2 contains 51 exons and MiPG22-1 has 13 exons. MiPG40-1 encodes an enzyme of 1420 aa, MiPG69-2 and enzyme of 1916 aa and MiPG22-1 an enzyme of 983 aa long. The structure of the introns and exons of the MiPG genes were obtained using GSDS 2.0 (http://gsds.cbi.pku.edu.cn/).
Relative expression of MiPG21-1, MiPG14, MiPG69-1, MiPG17, MiPG49, MiPG23-3, MiPG22-7, MiPG46-3, and MiPG16 in mango mesocarp, using ACT 7 as reference gene. Data of the nine genes represent the average of three biological replicates (n = 3) with three experimental replicates each. Error bars indicate SE of the means and different letters indicate significant differences. Statistical significance of differences between days of postharvest storage was determined by one-way ANOVA with the Fisher test (P < 0.05).
Expression ratio (Log2) for the 9 polygalacturonases was obtained by RNA-seq and qPCR. GAPDH was the constitutive gene used for normalization of qPCR data. Bars represent the standard error (n = 3).
Sequence alignment of polygalacturonases that were modeled to obtain theoretical models, the exons were colored with different colors. MiPG21-1 and MiPG23-3 are from clade C, MiPG14 from clade B, MiPG69-1 from clade F; while MiPG46-3 and MiPG49 are from clade E.
Primer sequences used for qRT-PCR.
PG superfamilies identified in mango polygalacturonases. (A) Genome ID, sequence name, biological process, cellular component, molecular function, classification, enzyme kind (endo/exo), isoelectric point and GenBank accession. (B) SignalP analysis of polygalacturonases from mango fruit.
Analysis of regulatory regions in 5′UTR polygalacturonases genes. (A) Common regulatory regions identified in 48 PGs. (B) Overrepresented TF families. (C) Common regulatory regions identified in 9 PGs which expression was evaluated.
Data Availability Statement
The datasets generated for this study can be accessed from GenBank MK936539-MK936586. The datasets analyzed for this study can be found deposited in the NCBI (Accesion PRJNA258477) (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA258477).