Abstract
The tumor suppressor p16INK4A induces cell cycle arrest and senescence in response to oncogenic transformation and is therefore frequently lost in cancer. p16INK4A is also known to accumulate under conditions of oxidative stress. Thus, we hypothesized it could potentially be regulated by reversible oxidation of cysteines (redox signaling). Here we report that oxidation of the single cysteine in p16INK4A in human cells occurs under relatively mild oxidizing conditions and leads to disulfide-dependent dimerization. p16INK4A is an all α-helical protein, but we find that upon cysteine-dependent dimerization, p16INK4A undergoes a dramatic structural rearrangement and forms aggregates that have the typical features of amyloid fibrils, including binding of diagnostic dyes, presence of cross-β sheet structure, and typical dimensions found in electron microscopy. p16INK4A amyloid formation abolishes its function as a Cyclin Dependent Kinase 4/6 inhibitor. Collectively, these observations mechanistically link the cellular redox state to the inactivation of p16INK4A through the formation of amyloid fibrils.
Keywords: Amyloids, Protein aggregation, Redox signaling, Cysteine oxidation, Structural biology
Graphical abstract
Highlights
-
•
The only cysteine in human p16INK4A is prone to oxidation in vitro and in vivo.
-
•
p16INK4A forms disulfide-dependent homodimers upon oxidation.
-
•
The all α-helical p16INK4A protein aggregates as β-amyloids upon oxidation.
-
•
Oxidation and subsequent β-amyloid formation impair p16INK4A function.
1. Introduction
The CDKN2A gene-product p16INK4A is an important cell-cycle regulator and acts as a tumor suppressor. It inhibits the D-type cyclin-dependent kinases CDK4 and CDK6 and hence prevents the downstream phosphorylation of the retinoblastoma (Rb) pocket protein [1]. This prevents release of E2 promoter binding factor 1 (E2F1), which is otherwise required for the transcriptional regulation of proteins that control controlentry into S-phase of the cell cycle [2]. Accumulation of p16INK4A is observed upon exposure of cells to several stressors such as oxidative stress and is one of the earliest markers of oncogenic transformation [3]. The loss of p16INK4A function, or loss of Rb downstream of CDK4/6, are some of the most frequently observed mutations in tumors [4]. Additionally, p16INK4A plays an important role in aging, as clearance of p16INK4A-expressing senescent cells has been shown to prolong lifespan in mice [5,6].
The molecular basis of p16INK4A-mediated CDK4/6 inhibition is well established. p16INK4A is a small, globular all-α-helical protein, that tightly binds into one side of the catalytic cleft of the CDK4/6 kinases. It efficiently distorts the cyclin D binding site, thereby blocking formation of the active CDK4/6-cyclin D complex, and preventing Rb phosphorylation [7,8]. Of particular relevance for this study, the single cysteine residue (C72) present in p16INK4A is located on its surface and points away from the CDK4/6 kinase in the bound state; the residue is fully solvent accessible.
Reversible cysteine oxidation is the lynchpin in redox signaling, a form of signal transduction that is regulated by the cellular redox state. A more oxidizing cellular redox state, either due to elevated reactive oxygen species or a lack of reducing power, leads to oxidative modification of specific cysteine-thiols to form reversible disulfide (S–S) bridges. These oxidative modifications can lead to structural rearrangements and can both negatively and positively regulate protein function (for a review see Ref. [9]).
A number of observations spurred us to hypothesize that oxidation of p16INK4A C72 could play a role in the regulation of p16INK4A activity at the molecular level. Firstly, several studies have implicated a role for increased ROS in the oncogene-induced accumulation of p16INK4A [10], but cysteine oxidation as the underlying mechanism has thus far not been considered nor excluded. Secondly, we identified p16INK4A as prone to cysteine oxidation in a large mass-spectrometry based screen for redox sensitive proteins [11]. Here, we provide evidence that p16INK4A itself is indeed sensitive to cysteine oxidation. We find that p16INK4A is readily oxidized both in vitro and in cultured human cells to form a disulfide-dependent homodimer, and the oxidizing conditions required are well within the physiological range. Surprisingly, disulfide-dependent dimerization of p16INK4A subsequently leads to the rapid formation of β-sheet amyloid fibril structures, a state that has not been previously described for p16INK4A. This transition subsequently leads to loss of CDK4/6 inhibitory capacity. Redox signaling-induced reversible disulfides have not previously been shown to induce β-sheet amyloid fibrils in other proteins and hence adds to the repertoire of redox dependent protein regulation.
2. Materials & methods
2.1. Cell lines, plasmids, antibodies and compounds
Cells (HEK293T, COLO829) were cultured in DMEM low glucose supplemented with FCS (10%), l-glutamine (2 mM) and Penicillin-Streptomycin (100 Units-0.1 mg per ml medium) (all from Lonza). Cells were transfected using PEI (Sigma Aldrich) or X-tremeGENE 9 (Roche) according to manufacturer's instruction.
The vector containing the truncated form (Δ1–8) of wild type p16INK4A was a gift from R. Medema [12]. The cysteine mutant of p16INK4A was created by site-directed pcr mutagenesis. mCherry- (backbone pLV-CMV-bc) and FLAG-His-tagged (backbone pCDNA3) and doxycycline-inducible (backbone pINDUCER20, Addgene #44012) p16INK4A constructs were created using Gateway technology (Life Technologies). COLO-829 melanoma cells stably expressing doxycyline-inducible constructs were transduced with lentivirus containing pINDUCER20-p16INK4A or -p16INK4AC72A. Cells were selected for stable transduction using geneticin and p16INK4A expression was confirmed by qPCR and western blotting. Subsequently, cells were transduced with lentivirus containing the FUCCI cell cycle probes mAG-hGeminin(1/110)/pCSII-EF (#RDB15268) and Fucci-G1 phase probe mKO2-hCdt1(30/120)/pCSII-EF (#RDB15267) (RIKEN BioResource Center), which were a kind gift from A. Miyawaki [13]. Cells were selected for stable transduction using zeocin and made monoclonal by limiting dilution.
The following antibodies were used in this study: FLAG (M2 F3165 Sigma Aldrich), p16INK4A (10883 Proteintech), p16INK4A (ab16123 Abcam), CDK4 (C-22 Santa Cruz), CDK6 (C-21 Santa Cruz), tubulin (CP06 Merck Millipore), Phospho-Rb Ser807/811 (9308 Cell Signaling).
Solutions/compounds not mentioned elsewhere: 35% hydrogen peroxide (Merck).
2.2. Co-immunoprecipitation, SDS-PAGE and western blotting
Immuno-precipitation of endogenous p16 was done with rabbit anti-p16INK4A (10883 Proteintech) using three 10 cm dishes of HEK293T cells per sample. Transfection was performed two days prior to sample preparation. Cells were scraped in 100 mM N-ethyl-maleimide in PBS (37 °C) to prevent post-lysis oxidation and collected by centrifugation at 1200 rpm for 3 min. Cells were lysed in a standard lysis buffer containing 50 mM Tris pH 7.5, 1% Triton X-100, 1.5 mM MgCl2, 5 mM EDTA, 100 mM NaCl supplemented with Aprotinin, Leupeptin and NaF. 100 mM iodoacetamide was added to prevent post-lysis oxidation of cysteines. The cell lysates were subsequently centrifuged at 14000 rpm for 10 min. 50 μl of the supernatant was kept as input. For immunoprecipitation of endogenous p16INK4A the pellet was resuspended in lysis buffer, sonicated and supplemented with 125 U of benzonase and incubated for 1 h at 37°. The remaining supernatant (and where indicated also the resolubilized pellet) was used for immunoprecipitation with the indicated antibody coupled to protein A-Agarose (Roche) or in case of transfected FLAG-tagged constructs with anti-FLAG M2 Affinity gel (Sigma Aldrich). After 2 h of incubation, beads were washed 3 times with standard lysis buffer supplemented with extra NaCl (1 M final concentration). After washing, samples were split into two parts, and sample buffer with or without a reducing agent (β-mercaptoethanol) was added. Samples were boiled for 5 min and separated on a 15% polyacrylamide gel. For Western Blotting, proteins were transferred to immobilon-FL membranes. For diagonal electrophoresis, non-reducing samples were separated on a 15% polyacrylamide gel, after which the entire protein containing lane was excised, reduced, and separated on a new 15% polyacrylamide gel. For visualization of diagonal electrophoresis proteins were stained by SimplyBlue.
2.3. Filter trap assay
HEK293T cells were transfected with FLAG-p16INK4A, FLAG-p16INK4AC72A or GFP as a control. After two days, cells were scraped in 100 mM N-ethyl-maleimide in PBS (37 °C) to prevent post-lysis oxidation and collected by centrifugation at 1200 rpm for 3 min. Lysis buffer (50 mM Tris pH 7.5, 1% v/v Triton X-100, 1.5 mM MgCl2, 5 mM EDTA, 100 mM NaCl, Protease inhibitors (Aprotinin + Leupeptin), NaF, 100 mM iodoacetamide) added for 10 min followed by centrifugation for 15 min at 14000 rpm in an Eppendorf microfuge. The pellet was resuspended in benzonase buffer (50 mM Tris-HCL pH 8.0, 1 mM MgCl2) and 125 U of benzonase was added followed by incubation for 1 h at 37°. The reaction was stopped by adding 2x termination buffer (40 mM EDTA, 0.2% SDS, with or without 10 mM DTT). Protein content was measured using the BCA kit and 50 μg of protein per sample were loaded on a Bio Dot apparatus (Biorad) equipped with a 0.2 μm pore size nitrocellulose membrane soaked in buffer B (10 mM Tris-HCL pH 8.0, 150 mM NaCl, 2% SDS) on top of two Whatmann paper filters soaked in Buffer A (10 mM Tris-HCL pH 8.0, 150 mM NaCl, 1% SDS), after pre-washing the membrane with 100 μl of buffer A. Samples were pulled through the membrane using vacuum, followed by washing thrice with 100 μl of buffer A. The nitrocellulose membrane was further processed as for Western Blotting to detect trapped p16INK4A.
2.4. Kinase assay
The kinase assays in this study were performed with immunoprecipitated FLAG-p16INK4A or FLAG-p16INK4AC72A and the attached binding-partners. Beads were washed three times with standard lysis buffer followed by two washes with kinase buffer containing 25 mM Tris pH 7.5 and 10 mM MgCl2. The beads were subsequently incubated for 30 min at 30 °C in 20 μl kinase buffer supplemented with 1 mM DTT, 1 μg human GST-Rb (48–378 Sigma Aldrich) as substrate and 0.2 mM ATP. Directly after the assay beads were boiled for 5 min in reducing sample buffer and loaded on a 15% polyacrylamide gel followed by Western Blotting.
2.5. RNA extraction, cDNA synthesis and qPCR
Doxycylcine-inducible p16INK4A expressing Colo-829 cells were plated, and 24 h before harvesting doxycline was added to induce expression of p16INK4A or p16INK4AC72A. Cells were treated as indicated. Cells were washed once with PBS and collected in RLT buffer. RNA was extracted using the RNeasy kit (Qiagen) according to the manufacturer's instructions. 1 μg RNA was used as input for cDNA synthesis using the iScript cDNA synthesis kit (Biorad). qPCR was performed with FastStart SYBR Green Master mix (Roche) using the CFX96 Touch Real-Time PCR Detection System. Relative gene expression was calculated using the ΔΔCt method [14] by normalization to HNRNPA1. Statistics were done with Graphpad Prism 8. Primer sequences: p16INK4A forward (5′-CAACGCACCGAATAGTTACG-3′), p16INK4A reverse (5′-ACCAGCGTGTCCAGGAAG-3′), CCNE2 forward (5′-GGGGGATCAGTCCTTGCATT-3′), CCNE2 reverse (5′-TCCCCAGCTTAAATCAGGCA-3′), HNRNPA1 forward (5′-GGAAGCTACAGGTTACAACA-3′), HNRNPA1 reverse (5′-AGTCACAAATACAGTCCTCG-3′).
2.6. Protein expression and purification
An Escherichia coli codon-optimized gene of Homo sapiens p16INK4A (UniProt ID: P42771) was cloned into a modified version of pETM-11 that includes a 6xHis, a protein A tag and a tobacco etch virus (TEV) protease cleavage site, leaving an additional glycine residue at the N-terminal cleavage site. For a glycerol stock solution, competent E. coli BL21 (DE3) cells were transformed with the p16INK4A harboring gene and inoculated in minimal medium in the presence of kanamycin (50 μg/l). After overnight incubation at 37 °C the cell suspension was mixed with glycerol to 50% (v/v) glycerol and stored at −80 °C. Uniformly 15N- and/or 13C-labeled protein samples were prepared from this stock by growing the cells in minimal medium containing 15NH4Cl and/or 13C-glucose as the only nitrogen or carbon source. The protein synthesis was induced by addition of 0.5 mM isopropyl-1-thio-d-galactopyranoside (IPTG) at an OD600 of 0.8 and cells were harvested after 14 h of induction at 19 °C. Cells were resuspended in purification buffer (110 mM potassium acetate, 20 mM HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid], pH 8.0, 2 mM MgCl2, 2 mM β-mercaptoethanol (BME), 5% (v/v) glycerol and 20 mM imidazole) and ultrasonicated for lysis. The solution was applied to a gravity Ni-NTA agarose (QIAGEN) column following the manufacturer's instructions. After washing with purification buffer including 20 mM imidazole, the protein was eluted by using purification buffer including 200 mM imidazole and further purified by gel chromatography on an ÄKTA pure system equipped with a HiLoad 16/600 Superdex 75 pg column (GE Healthcare) running with purification buffer. The fractions containing the protein were pooled, incubated with 0.2 mg TEV protease overnight at 4 °C. On the following day, the protein solution was applied to the Ni-NTA agarose column again to remove the cleaved tag, remaining undigested protein and the His6-tagged TEV protease. The eluate was then buffer exchanged into purification buffer where BME was substituted with 1 mM DTT using dialysis. The protein solution was aliquoted, frozen in liquid nitrogen and stored at −80 °C. Solution NMR spectroscopy showed that no differences were observed after the freezing and thawing of the samples.
2.7. Solution NMR spectroscopy
The protein stock was freshly buffer exchanged into NMR buffer (4 mM HEPES, 5 mM EDTA, pH 7.5) before measurement. Samples for protein characterization contained 1 mM DTT (dithiothreitol) whereas samples for redox-experiments did not. The protein concentration was 0.15 mM and contained 7% (v/v) 2H2O for the lock signal. Solution 1H15N HSQC NMR spectra were measured on an Avance III 600 Bruker NMR spectrometer equipped with a cryogenic triple resonance probe with gradients in the z-direction. The recycle delay of the redox experiments performed was set to 1 s with a spectral window of 11/34 ppm in 1H/15N dimensions, 2048 points in the direct 1H dimension, 128 complex data points in the indirect 15N dimension, and with 8 or 16 scans per increment. Spectra were processed with the NMRPipe package [15] and analyzed by CcpNmr Analysis [16].
For determination of the S-glutathionylation redox potential, the concentration of GSSG in the sample buffer was gradually increased (0.2; 1.0; 2.3; 3.3; 3.9; 5.9; 7.8; 8.8; 9.8; 11.7 mM) by titrating from a 65 mM pH corrected stock solution while the concentration of the reduced form of GSH was kept constant at 4 mM. At each concentration point, a 1H15N HSQC NMR spectrum was recorded immediately after addition of the oxidation agent. For assignment of the S-GSHylated protein, HNCA and CBCACONH spectra were acquired. Determination of the redox potential was performed following the protocol of Piotukh et al. [17]. In summary, peak intensities of well isolated peaks (N71, C72, D74, T79) of the oxidized form were normalized to a non-affected asparagine side chain resonance and plotted against the redox potential of the buffer. This potential was determined from the standard half-cell potential of the glutathione redox couple (−240 mV at pH 7.0, 25 °C) after pH corrections [18]. The data were fitted to the sigmoidal decay function (equation (1)):
| (1) |
where A1 and A2 are the initial and final plateaus of the function, dx is the slope and x0 refers to the desired parameter of the redox potential.
For oxidized samples of p16INK4, a final concentration of 50 mM hydrogen peroxide (H2O2, from a 30% solution diluted with NMR buffer) or 100 μM diamide (from a freshly prepared 10 mM stock solution in H2O) was added to the 15N-labeled protein solution. 1H15N HSQC NMR spectra were acquired after the addition of the oxidizing agent in subsequent manner. After 10 h, no spectral changes were observed for WT protein anymore and the sample was applied to a size exclusion column (HiLoad 10/300 Superdex 75 pg column, GE Healthcare) and the aggregated fraction was pooled. The protein was applied to SDS gel electrophoresis in the presence and absence of BME.
2.8. CD spectroscopy
Far-UV CD experiments were recorded on a J-715 spectropolarimeter (Jasco, Tokyo, Japan). Spectra were acquired at room temperature with a response time of 1 s and a step resolution of 0.3 nm. Five scans were averaged and the signal was background corrected by subtraction of the buffer signal. The CD spectra were obtained from a 1 μM p16INK4A sample before and after oxidation with 200 μM hydrogen peroxide for 8 h at room temperature and data were converted to molar ellipticity. Samples were freshly prepared and transferred into a 10 mM sodium phosphate buffer at pH 7.4 in the absence of reducing agents.
2.9. Small-angle X-ray scattering measurements
SAXS data was recorded on an in-house SAXS instrument (SAXSess mc2, Anton Paar, Graz, Austria) equipped with a Kratky camera, a sealed X-ray tube and a two-dimensional Princeton Instruments PI•SCX:4300 (Roper Scientific) CCD camera. Measurements were performed with 90 min exposure time (540 frames of 10 s each) of three concentrations of 2.5 mg/ml, 1.25 mg/ml and 0.625 mg/ml in NMR buffer. The sample of reduced p16INK4A was measured immediately after purification and buffer exchange. The oxidized sample of p16INK4A was treated overnight with 50 m M H2O2 at room temperature. Next day, the sample was applied to size exclusion chromatography and the peak from 9 to 10 ml elution volume was pooled (see Figs. S6 and B) and concentrated. Individual frames of the 90 min exposure were compared and no radiation damage was observed. A range of momentum transfer of 0.012 < s < 0.63 Å−1 was covered (s = 4π sin(θ)/λ, where 2θ is the scattering angle and λ = 1.5 Å is the X-ray wavelength). SAXS data were analyzed with the package ATSAS version 2.8.1 [19]. Desmearing of the oxidized sample could not be performed because the scattering curve showed strong aggregation.
2.10. Thioflavin-T fluorescence assay
Samples of p16INK4A (20 μM) were prepared in buffer (4 mM HEPES, pH 7.5) with 20 μM ThT. Three replicates were measured for each sample. Samples were subjected to 1 mm orbital shaking for 2 min of every 5 min, in a 96-well, half-area, fluorescence plate (Corning) and measurements were taken every 5 min. Samples were excited at 440 nm and fluorescence emission was measured at 480 nm. Measurements were made using a Tecan GENios microplate reader.
2.11. Negative-stain transmission electron microscopy
Protein samples were prepared by incubation of p16INK4A samples with or without oxidizing agents for 6–24 h at RT. Copper grids with 300 meshes coated with formvar/carbon film (Electron Microscopy Sciences, Hatfield, USA) were glow-discharged in argon atmosphere for 30 s at 3 mA. Grids were floated on a 5 μl drop of a 50 μM protein sample and incubated for 60 s. Grids were washed once with water, and then floated on 5 μl of uranyl acetate solution (2% w/v) for 30 s. Micrographs were taken on either a JEOL JEM 100CX transmission electron microscope or a JEOL JEM 1400 Plus transmission electron microscope (JEOL, Tokyo, Japan).
2.12. Prediction of aggregation-prone regions
Four different aggregation prediction programs, TANGO [20], PASTA [21], Zyggregator [6] and Aggrescan [22] were used to identify putative regions responsible for the beta-aggregation of p16INK4A using the default settings.
2.13. Congo-red absorbance
Absorbance spectra were measured on samples of p16INK4A (20 μM) with Congo red (20 μM) in buffer (4 mM HEPES, pH 7.5). Spectra were recorded from 400 to 650 nm, with a resolution of 2 nm, on a Tecan Infinite 200Pro microplate reader in a 96-well plate (Corning).
2.14. Fourier-transform infrared spectroscopy
p16INK4A (80 μM) samples were treated with 50 mM H2O2 overnight in buffer (4 mM HEPES pH 7.5), then dialysed against water overnight. Spectra were recorded on a JASCO FT/IR-4100 FT-IR spectrometer with attenuated total reflectance (ATR) attachment. The samples were measured with 128 scans at a resolution of 2 cm−1 at room temperature.
2.15. Solid-state NMR spectroscopy
10 mg (150 μM) of 13C–15N labeled p16INK4A fibrils were prepared in buffer (4 mM HEPES pH 7.5) by addition of 50 m M H2O2 and incubation at 37 °C overnight. The sample was packed into a 3.2 mm MAS rotor by ultracentrifugation. Spectra were recorded on a Bruker Avance III 750 MHz spectrometer (Bruker BioSpin) equipped with a 3.2 mm triple-resonance MAS probe. A proton-driven spin diffusion (PDSD) spectrum [23] was recorded at 16.5 kHz MAS, with 50 ms mixing time, at a set temperature of 273 K, with 352 scans and 22 ms and 8 ms acquisition time in the direct and indirect dimensions, respectively. Experiments were acquired using Topspin 3.2 (Bruker Biospin) and analyzed using CCPN Analysis 2 [16] The secondary chemical shift was calculated as [δ(Cαobserved) – δ(Cαrandom coil)] – [δ(Cβobserved) – δ (Cβrandom coil)] with random coil chemical shifts taken from Ref. [24].
2.16. Sample preparation for mass spectrometry
HEK293T cells were transfected with FLAG-p16INK4A or FLAG-p16INK4A C72A. After two days, half of the dishes (3 full 15 cm dishes per sample) were treated for 10 min with diamide (250 μM) (Sigma Aldrich) and subsequently harvested for immunoprecipitation with FLAG beads as described before. After washing 2x with 1 M salt buffer and 3x with PBS to remove all soap, proteins were eluted from the beads by two times 5 min incubation with 75 μl 0.1 M glycine pH 2. The protein containing supernatant was transferred to a new tube and incubated for 20 min with 10 mM DTT and 2 M Urea (dissolved in 100 mM Tris pH 7.5), followed by 10 min incubation with iodoacetamide (50 mM). To digest the proteins, 0.25 μg trypsin (Promega) was added per sample and samples were incubated ON at 25 °C. The next day C18‐stagetips were used for filtering and loading of the protein digest.
2.17. Mass spectrometry and data analysis
The mass spectrometry was performed as previously described [11].
For the analysis Maxquant software version 1.5.1.0 [25] was used. During the analysis, oxidation of methionine, alkylation of cysteines with iodoacetamide were set as variable modification. Proteins were identified by the IPI human V3.68 database and the relative amounts of protein in the separate experiments, the Intensity Based Absolute Quantification (IBAQ) [26], as well as the label free quantification [27] were calculated.
Further analysis was done using R version 3.4.0. The used code and description is currently being uploaded to GitHub. Proteins identified with two or more unique peptides were filtered for reverse hits, decoy hits and standard contaminants and samples that show aberrant clustering or low protein count were excluded. Subsequently the IBAQ data was Log2 transformed and normalized for IP efficiency. For p-value calculations, left-censored missing data was first imputed using a stochastic minimal value approach. Imputation was performed by random draws from a Gaussian distribution centered in the 10−4 quantile of the known data with a standard deviation that is same as the observed values [28] A standard t-test was performed on the imputed data and p-values were adjusted for multiple testing using the Benjamini-Hochberg correction.
2.18. Mass spectrometry of GS-p16INK4A
The intact mass of p16INK4A before and after oxidation by GSSG was obtained from LC-MS analysis, that was performed on recombinantly expressed protein (see protein expression and purification section). The sample was desalted using 10 kDa Amicon Ultra-0.5 centrifugal filter devices prior to measurement. 150 pmol of protein were injected into an Agilent 1200 system (Vienna, Austria) coupled to an LTQ-FT mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). Separation was carried out on a monolithic PepSwift® column (500 μm × 5 cm Monolithic PS – DVB) at a flow rate of 20 μl/min using the following gradient, where solvent A is 0.05% trifluoroacetic acid in water and solvent B is 0.05% trifluoroacetic acid in acetonitrile: 0–5 min: 10% B; 5–55 min: 10–100% B; 55–65 min: 100% B, 65–80 min: 10% B. The sample was ionized in the electrospray source and analyzed using FT-MS operated in positive ion mode applying full scan MS (m/z 300 to 2000) with a resolution of 400,000. Acquired data were processed with the software Protein Deconvolution 2.0 SP2 (Thermo Fisher Scientific) using a S/N threshold of 5 and a relative abundance threshold of 20%.
2.19. Flow cytometry
Cell cycle analysis was performed using a combination of the FUCCI [13] system and DNA content staining. COLO-829 cells expressing the FUCCI constructs as well as pINDUCER20-p16INK4A or p16INK4A-C72A were seeded and treated with doxycycline and LEE-011 for the indicated times and samples. Nocodazole was added 16 h before harvest to capture cycling cells in mitosis to distinguish transient G1 from G1/G0 arrested cells. Cell culture medium was collected and cells were washed in PBS before trypsinization. Cells were fixed for 10 min at room temperature with 2% formalin in PBS. After 2 washes with PBS, cells were permeabilized with 70% ethanol for 30 min. Cells were resuspended in PBS with 0.1% BSA with DAPI and RNAse to stain the DNA. mKO2-hCtd1(30/120), mAG-hGeminin(1/110) and DAPI were measured using a BD FACSCelesta Flow Cytometer (BD Bioscience).
2.20. Evolutionary analysis
Amino acid sequences of vertebrate orthologues of human p16INK4A and p15INK4B were downloaded from the ENSEMBL database [29]. Alignment and visualization of conservation was performed using JalView 2.10.1 software [30], with the Clustal plugin with default settings.
3. Results
3.1. p16INK4A is oxidized to form disulfide dependent homodimers in human cells
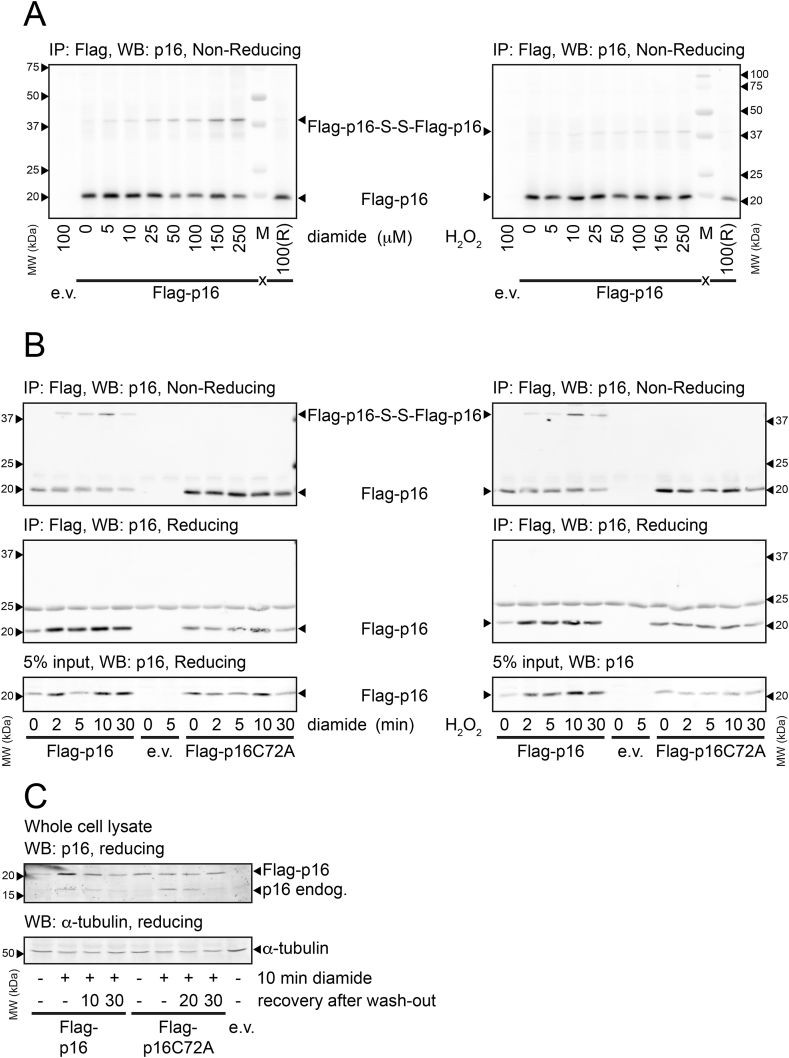
To test whether p16INK4A forms disulfide-dependent complexes under oxidizing conditions, immunoprecipitation followed by non-reducing SDS-PAGE was performed. Oxidation was induced using a concentration series of either hydrogen peroxide or the thiol-specific oxidant diamide (tetramethylazodicarboxamide) that ranged from subtoxic (5 μM) to mildly toxic (250 μM) in HEK293T cells. FLAG-p16INK4A readily formed intermolecular disulfide-dependent complexes (Fig. 1A), as judged by the large mobility shift under non-reducing conditions, that was abolished when the samples were reduced prior to SDS-PAGE (last lanes). Note that only the FLAG-p16INK4A disulfide-dependent dimer band is observed and that its intensity increases upon exposure to increasing oxidant concentration. We did not observe a “smear”, which would occur if p16INK4A were to undergo random, nonspecific crosslinking to proteins. We then created p16INK4A mutants in which the only cysteine was replaced by alanine (C72A) or serine (C72S). We opted to use p16INK4A-C72A for our further experiments in human cells in order to circumvent the potential effects of the introduction of a novel phosphorylation site in the C72S mutant. The C72A mutant is still functional as a cell-cycle inhibitor and hence the mutation does not grossly affect protein function per se (Fig. S1). To explore whether the observed intermolecular disulfide-dependent complexes are FLAG-p16INK4A homodimers, Co-IP experiments were performed using combinations of wild-type (WT) p16INK4A and C72A with short (FLAG) and long (mCherry) N-terminal tags. Under non-reducing conditions, bands could be observed corresponding to disulfide-dependent dimers of FLAG-p16INK4A-S-S-FLAG-p16INK4A and of mCherry-p16INK4A-S-S-FLAG-p16INK4A (Fig. S2A) after FLAG pull down. Furthermore, diagonal electrophoresis showed that after reduction (2nd dimension) FLAG-p16INK4A drops out of the diagonal as a single dot, whereas a heterodimer would have revealed a second Simply Blue stained protein with similar intensity dropping out of the diagonal (Fig. S2B). To test the dynamics of the intermolecular disulfide-dependent dimerization of p16INK4A, a time course of oxidant treatment was performed, using p16INK4A-C72A as a control. The oxidation of p16INK4A was rapid, peaked after 10 min, was fully dependent on the only cysteine and was resolved upon boiling in reducing sample buffer (Fig. 1B).
Fig. 1.
p16INK4Aforms intermolecular disulfides upon exposure to oxidants.
(A) Analysis of Immuno-precipitated FLAG-p16INK4A by non-reducing SDS-PAGE and Western Blot shows that part of the FLAG-p16INK4A migrates at about double the molecular weight upon 5 min treatment with low amounts of the thiol-specific oxidant diamide (left panel) or H2O2 (right panel). Reduction (R) prior to SDS-PAGE abolishes the shift in molecular weight, indicating that it is indeed due to an intermolecular disulfide (see also Figs. S2A and S2B for confirmation that the high molecular weight form of p16INK4A is an S–S-dependent homodimer).
(B) S–S-dependent p16INK4A homodimerization upon 200 μM diamide (left) or 200 μM H2O2 (right) occurs rapidly, coincides with accumulation of p16INK4A protein levels and oxidation as well as accumulation are fully dependent on C72.
(C) Endogenous p16INK4A and over-expressed FLAG-p16INK4A accumulate in response to 200 μM diamide with similar kinetics. Note that p16INK4AC72A does not accumulate whereas endogenous p16INK4A does, suggesting that endogenous p16INK4A levels are also regulated by Cys-oxidation. (IP: immunoprecipitation, WB: Western Blot). All Western blots shown in Fig. 1 are typical results of several repeats (N ≥ 3 for all experiments).
Concomitantly with its oxidation, p16INK4A protein abundance increased, and this was again dependent on its single cysteine residue. Protein levels already increased after a few minutes of diamide treatment, excluding a role for gene transcription but suggestive of a role for decreased protein breakdown. The accumulation is best observed using reducing SDS-PAGE because all p16INK4A then migrates in a single band (Fig. 1B and C). The increased p16INK4A protein levels rapidly returned to basal upon diamide wash-out with fresh media (Fig. 1C). Endogenous p16INK4A levels follow the same trend as FLAG-p16INK4A indicating that endogenous p16INK4A is also regulated through cysteine oxidation. Taken together, both the increase in p16INK4A levels as well as the oxidation are dependent on C72.
3.2. C72 of p16INK4A is readily oxidized in vitro
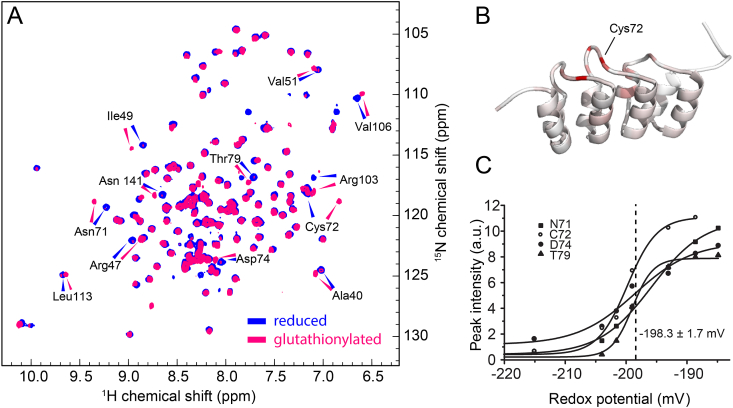
To study the reactivity of the p16INK4A cysteine residue directly, we expressed and purified the isotope-labeled recombinant protein and confirmed its correct folding by solution nuclear magnetic resonance (NMR) spectroscopy. Upon treatment of the purified protein with oxidized glutathione (GSSG), we observed chemical shift changes that occurred most strongly in proximity to the cysteine residue of p16INK4A (Fig. 2A and B, Fig. S3). These changes were reversed by addition of the reducing agent DTT, confirming the involvement of reversible cysteine oxidation. Mass spectrometric analysis of the intact protein showed that p16INK4A was S-glutathionylated (S-GSHylated) after treatment with GSSG (Fig. S4). S-GSHylation of the protein does not change its monomeric state or overall fold, as indicated by identical peak line shapes and unchanged elution times in size exclusion chromatography. By titrating the reduced p16INK4A sample with increasing GSSG in a GSH/GSSG redox buffer we determined the redox potential of C72 using the Nernst equation [17] to be −198.3±1.7 mV (Fig. 2C).
Fig. 2.
In vitrooxidation of p16INK4A
(A)1H15N HSQC solution NMR spectrum of recombinant p16INK4A in the reduced state (blue) and after S-glutathionylation (magenta). Amino acids with large chemical shift changes are labeled.
(B) Cartoon representation of the p16INK4A structure. A color gradient from white (unaffected) to red (strongly affected) shows the influence of S-glutathionylation on the chemical shift.
(C) The redox potential of C72 is 198.3 ± 1.7 mV, as determined from intensity changes of four well-separated amino acids by titration of the reduced protein with oxidized glutathione. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To study the structural role of the cysteine residue, we used the p16INK4A-C72S mutant, which is the most structurally conservative mutation, differing only by replacement of the sulfhydryl group with a hydroxyl group. This mutant showed monomeric behavior during purification and largely identical chemical shifts as compared to the wild-type protein (Fig. S5), indicating that the mutation does not affect p16INK4A protein structure per se. Addition of GSSG to this mutant did not show any indications of interaction or covalent modification. Collectively, these experiments indicate that the p16INK4A C72 can be directly oxidized in vitro at physiologically occurring redox states.
3.3. Disulfide-dependent homo-dimerization of p16INK4A triggers aggregation
The use of GSSG as an oxidant resulted in p16INK4A S-GSHylation in vitro, whereas experiments in human cells showed S–S-linked homodimerization (Fig. 1) upon treatment with diamide or H2O2, even though both these agents lead to increased GSSG in cells. We therefore assessed whether H2O2 and diamide can directly oxidize p16INK4A in vitro. SDS-PAGE analysis revealed that disulfide-linked protein homodimers are also formed in vitro, similar to the pattern observed in the experiments in human cells (Fig. S6A). Comparison of size exclusion chromatography traces of reduced and oxidized samples show that oxidation of p16INK4A causes a shift to high molecular mass, with the oxidized sample eluting in the void volume of the column (Fig. S6B). In contrast to oxidation by GSSG, the addition of either H2O2 (Fig. 3A) or diamide (Figs. S6C and S6D) led to strong NMR chemical shift changes that indicated protein unfolding and aggregation. This indicates the formation of a multimeric species and the absence of monomeric p16INK4A. Characterization of reduced and oxidized WT p16INK4A by Circular Dichroism (CD) spectroscopy revealed a transition to a less structured species, with some alpha-helical characteristics remaining (Fig. S7A). Small-angle X-ray scattering (SAXS) on the oxidized samples indicates that the aggregates are larger than the detection limit of about 400 nm diameter (Fig. S7B). S-Glutathionylation of p16INK4A C72 would abolish its reactivity towards diamide or H2O2. To exclude that the observed aggregation of p16INK4A stems from modifications other than C72, we also performed solution NMR spectroscopy on the S-glutathionylated p16INK4A (GS-p16INK4A), as well as on the p16INK4A-C72A mutant, treated with diamide or H2O2. No chemical shift changes were observed and the monomeric, S-glutathionylated protein sample as well as the p16INK4A-C72A mutant were stable in presence of oxidizing agents (Fig. S8). Together, these experiments indicate the formation of an aggregated p16INK4A species upon intermolecular disulfide bond formation in vitro.
Fig. 3.
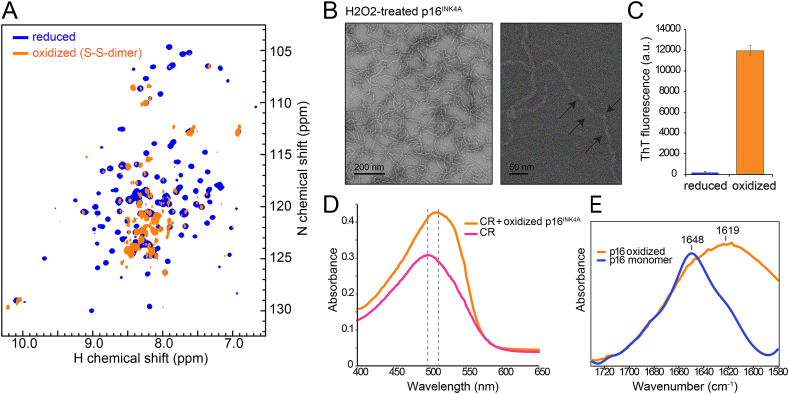
p16INK4Aforms amyloid fibrils under oxidizing conditions
(A)1H,15N HSQC solution NMR spectra of recombinant p16INK4A before (blue) and after (orange) oxidation with 50 mM H2O2 for 10 h at room temperature (orange). The collapse of the peaks to the center of the spectrum suggests formation of unstructured or aggregated protein.
(B) Negative-stained transmission electron micrographs of diamide-treated p16INK4A showing the presence of amyloid fibrils. Right panel is at higher magnification, arrows highlight the twisted morphology which is typical of amyloid fibrils.
(C) Fibril formation of p16INK4A monitored by thioflavin-T fluorescence measurement. Error bars represent standard deviation from four measurements.
(D) Fibril formation of p16INK4A monitored by Congo red absorption. The absorption maximum shifts when bound to oxidized p16INK4A, indicating the presence of amyloid fibrils.
(E) Amide I region of the attenuated-total-reflectance Fourier-transform infrared spectra of p16INK4A monomer and fibrils. Peak maxima indicate that the structure contains primarily α-helix for monomers and cross-β sheet for fibrils. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Disulfide-linked homodimers of p16INK4A form β-amyloid fibrils
Based on the indications of aggregation, transmission electron microscopy (EM) was employed to study the nature of the p16INK4A aggregates that formed upon oxidation. Interestingly, we found that the oxidized p16INK4A samples produced fibrillar structures (Fig. 3B). One prominent group of fibrillar proteins are amyloid fibrils, which are characterized by a cross-β sheet core structure, meaning that β-strands run perpendicular to the fibril long axis [31,32]. To determine if p16INK4A fibrils are amyloid, we used the amyloid-specific dyes Thioflavin T (ThT) and Congo Red [[33], [34], [35]]. ThT assays showed an increase in fluorescence, which is characteristic for amyloids, when bound to oxidized p16INK4A (Fig. 3C), while Congo Red absorbance measurements showed the red shift and increased absorbance characteristic of amyloid binding (Fig. 3D). Fourier-transform infrared spectroscopy can distinguish cross β-sheets found in amyloid fibrils from β-sheets found in globular monomeric proteins [36,37]; in line with the dye-binding data, p16INK4A fibrils had a maximum at 1620 cm−1, which falls within the typical amyloid β-sheet region, confirming the amyloid structure of the fibrils (Fig. 3E). We performed a computational analysis using four fibril prediction algorithms [[20], [21], [22],38] and all suggested a high propensity for β-amyloid formation in the region from residue no. 91–99 (Fig. S9A).
Additionally, we recorded solid state NMR spectra on a uniformly 13C-labeled p16INK4A sample to confirm the secondary structure of the aggregated state. Samples were prepared using the same conditions that were used for other experiments including electron microscopy, to ensure that results are comparable between experiments. In 13C–13C correlation spectra, only amino acid residues within a structured core are visible (Fig. S9). The spectrum shows relatively few peaks compared to the 156 residues of full-length p16INK4A, suggesting that only specific parts of the sequence form the structural core of the amyloid fibrils. This is also indicated by the absence of certain residue types from the spectrum, for example isoleucine (Fig. S9B). Many of the peaks could be assigned to their amino-acid type due to their distinctive Cα and Cβ peak positions, which allowed for determination of the secondary structure. Nearly all assigned peaks have peak positions that are characteristic for β-sheet or random coil (Fig. S9C), confirming that p16INK4A aggregates have a β-sheet core and are therefore amyloid. These findings emphasize the dramatic structural rearrangement from the monomeric eight-α-helical bundle that is devoid of β-sheets.
3.5. Intermolecular p16INK4A disulfide bond formation is essential for amyloid fibril assembly
We further investigated the role of oxidation of the cysteine residue in fibril formation by comparing the p16INK4A WT protein to p16INK4A-C72S. Electron microscopy revealed that upon treatment with as little as 200 μM H2O2 or diamide, WT samples formed large fibrillar structures (Fig. 4A, top panels). There was no discernible difference between the morphology of fibrils formed with diamide or H2O2 treatment, or when higher concentrations of oxidizing agent were used. p16INK4A-C72S samples displayed a very different morphology, instead forming unordered, amorphous aggregates, as well as a small amount of short fibrillar species (Fig. 4A, middle panels).
Fig. 4.
C72 is important for fibrillar morphology and ThT binding.
(A) Negative-stain transmission electron micrographs of p16INK4A treated with 200 μM hydrogen peroxide (left) and 200 μM diamide (right). Top: WT p16INK4A. Middle: p16INK4A-C72S. Bottom: GS-p16INKA.
(B) Thioflavin-T fluorescence kinetics assay of p16INK4A samples (WT, C72S and GS) with 200 μM diamide addition (top) 200 μM H2O2 addition (middle) and without oxidizing agents (bottom). Three replicates were measured per sample and each are separately plotted.
Glutathionylated-p16INK4A (GS-p16INK4A) samples were also analyzed by electron microscopy, and were found to form large, granular aggregates without any fibrils upon treatment with either H2O2 or diamide (Fig. 4A, bottom panels). Although aggregates are seen by electron microscopy, the solution NMR spectra of GS-p16INK4A indicate the sample is largely soluble and monomeric (Fig S8A, S8B), suggesting that the aggregates seen by electron microscopy constitute only a small proportion of the total GS-p16INK4A protein.
We next investigated the influence of intermolecular disulfide bond formation on the kinetics of fibril formation, by measuring the time- and oxidation-dependence of p16INK4A WT, p16INK4A C72S and GS-p16INK4A variants using the ThT assay (Fig. 4B). Upon treatment of WT p16INK4A with oxidizing agents, the sample fluorescence rapidly increased as ThT-positive fibrils were formed, with maximum signal being achieved within 2 h upon 200 μM diamide treatment (Fig. 4B, top) and more than 16 h for 200 μM H2O2 treatment (Fig. 4B, middle), while no fibrils were formed in the absence of oxidizing agents (Fig. 4B, bottom). The C72S mutant displayed only a very slight increase in fluorescence over time, and this was not affected by the presence or absence of oxidizing agents. GS-p16INK4A did not form ThT-positive aggregates under any condition. The amyloids formed in vitro were reversible: boiling in non-reducing sample buffer for 5 min prior to SDS-PAGE resulted mainly in detection of the S–S-dependent dimers (Fig. S10). The EM and ThT-kinetics data emphasize that the amorphous aggregates observed by EM for the p16INK4A-C72S and GS-p16INK4A are not amyloid, both by morphology and by dye-binding properties. Therefore, we conclude that the intermolecular disulfide bond formation of p16INK4A through C72 is critical for the formation of p16INKA amyloid fibrils.
3.6. p16INK4A cysteine oxidation triggers its aggregation in cultured cells
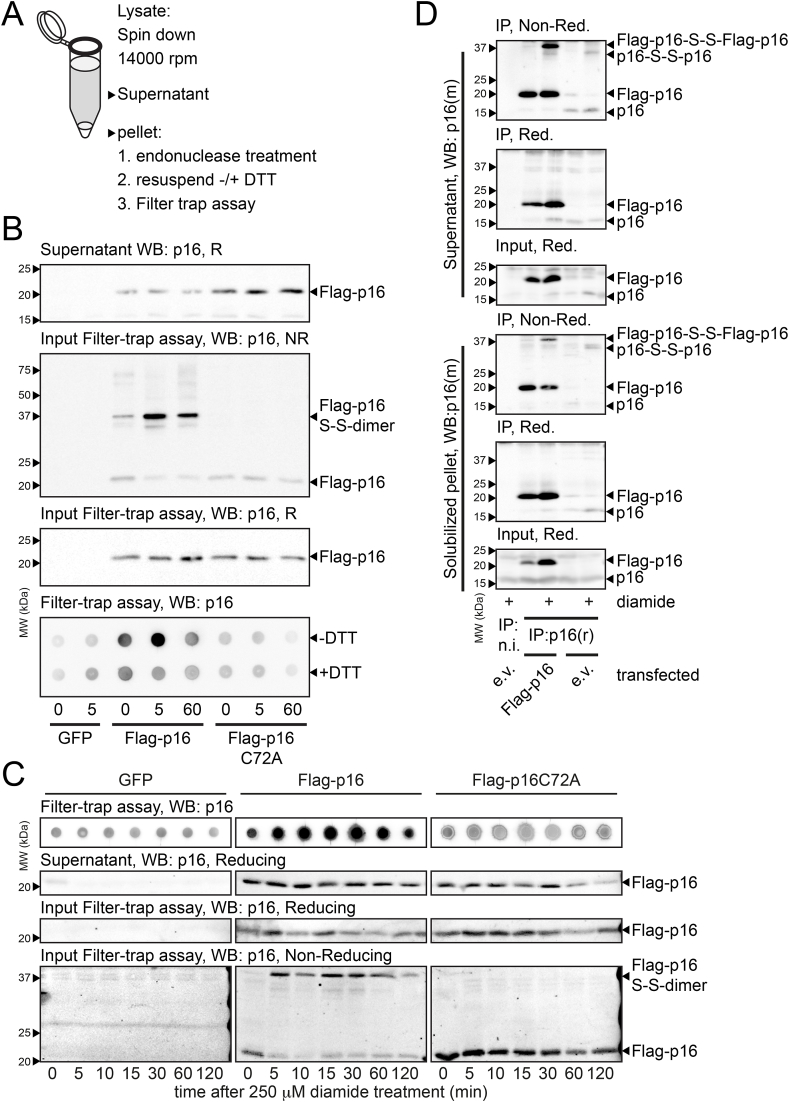
Having shown that p16INK4A forms amyloid fibrils upon oxidation in vitro, we investigated to what extent this behavior was conserved in cultured human cells. HEK293T cells expressing FLAG-tagged p16INK4A WT and p16INK4A C72A mutant were exposed to diamide and processed for a filter trap assay: an assay that is commonly used to detect amyloids in cell extracts (Scheme top of Fig. 5A). Exposure to diamide prior to lysis rapidly led to accumulation of the p16INK4A homodimer in the solubilized pellet fraction and subsequent trapping of p16INK4A on the filter membrane, and both were dependent on C72. Reduction of the protein lysates with DTT prior to washing abolished trapping, suggesting that the aggregation was reversible (Fig. 5B). Clearance of p16INK4A aggregates was observed over time after diamide treatment. After 120 min the aggregates were largely resolved (Fig. 5C), reminiscent of turnover of the disulfide-linked homodimer in Fig. 1C.
Fig. 5.
p16INK4Aaggregates in live cells in response to oxidation.
(A) Set up for the Filter Trap assay for the detection of aggregates in cell lysates.
(B) Results for the Filter Trap assay. Note that the majority of p16INK4A in the pellet is in the form of S-S-linked homodimers after lysis. p16INK4A, but not p16INK4AC72A, was trapped on the filter membrane upon treatment with 200 μM diamide and trapping was prevented by pretreatment with DTT. Equal amounts of p16INK4A and p16INK4AC72A were used as input for the filter trap assay. Note that the input for the Filter Trap assay runs mainly as a S–S-dependent dimer after boiling in non-reducing SDS-PAGE sample buffer. R: reducing, NR: non-reducing.
(C) Aggregates of p16INK4A peak around 30 min and are then cleared over time.
(D) Endogenous p16INK4A also forms S–S-linked dimers that (partially) form aggregates in the insoluble pellet fraction. Note that also mixed S–S-linked dimers of endogenous and overexpressed p16INK4A can be observed in lanes where FLAG-p16INK4A was transfected. n.i.: IP with non-immune serum as a control. p16(r): rabbit anti-p16INK4A, p16(m): mouse anti-p16INK4A, e.v.: empty vector, Red: reducing, Non-Red: non-reducing. All experiments were performed at least 3 times. Representative experiments are shown.
We also studied whether oxidation induced the aggregation of endogenous p16INK4A, for which the expression levels are generally lower than those of transfected FLAG-p16INK4A. We found that endogenous p16INK4A also forms disulfide-linked dimers upon oxidation and that part of these accumulate in the insoluble pellet fraction (Fig. 5D). Similar amounts of endogenous p16INK4A are found in the solubilized pellet fraction in the presence and absence of transfected FLAG-p16INK4A, suggesting that endogenous p16INK4A levels are sufficient to form homodimers and aggregates (see Reducing IP panels). The phase-transition from monomeric to the amyloid form of p16INK4A would likely change its biochemical properties and protein-protein interactions. To test this, the interactomes of immunoprecipitated FLAG-p16INK4A and FLAG-p16INK4AC72A were investigated and compared by label-free quantitative mass spectrometry. Under basal conditions the interactomes were largely identical, but this changed dramatically upon diamide treatment (Fig. S11). Diamide-treated WT p16INK4A indeed showed a large number of binding partners not identified in diamide-treated p16INK4A-C72A pull-downs. The interaction with CDK4 and CDK6 was not affected by diamide treatment or the C72A mutation. Proteins that form amyloids are known to bind to chaperones and indeed several Heat Shock Proteins were found to interact with WT p16INK4A upon diamide treatment, again indicative of aggregate formation.
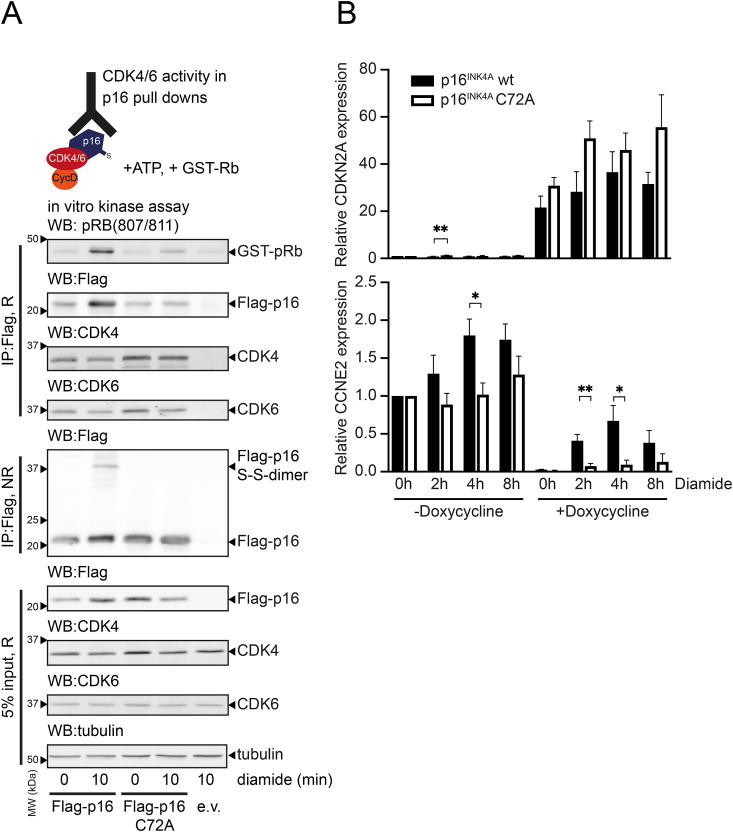
3.7. p16INK4A cysteine oxidation restores CDK4/6 kinase activity
We then investigated how the p16INK4A function as a CDK4/6 inhibitor would be affected by amyloid formation. CDK4/6 was co-immunoprecipitated with FLAG-tagged p16INK4A, and in vitro kinase assays were performed using a GST-tagged Rb fragment as a substrate (Scheme Fig. 6A). These experiments revealed that oxidation of WT p16INK4A or p16INK4A-C72S did not greatly affect the binding to CDK4/6 (Fig. 6A), which agrees with our observations in the MS experiments (Fig. S11). The in vitro kinase assay showed that p16INK4A-associated CDK4/6 was, as expected, inactive under basal conditions. CDK4/6 activity towards the GST-Rb fragment was regained when cells were exposed to diamide prior to lysis, despite the increased amount of p16INK4A that was pulled down. The reactivation was strictly dependent on p16INK4A C72. Taken together, these observations suggest that the amyloid form of p16INK4A still interacts with CDK4/6 but in a manner that does not lead to kinase inhibition.
Fig. 6.
Oxidation of p16INK4A impairs its inhibitory function towards CDK4/6.
(A) In vitro CDK4/6 kinase assay on WT p16INK4A pulldowns shows that the oxidation of p16INK4A-C72 impairs its inhibitory function. Note that oxidation or mutation of Cys72 does not affect the amount of CDK4/6 that is co-immunoprecipitated. (R: Reducing, NR: Non-reducing). All Western blots shown in Fig. 6 are typical results of several repeats (N ≥ 3 for all experiments).
(B) qPCR analysis showing the expression of WT p16INK4AWT or p16INK4A-C72A (top) in COLO-829 cells upon induction of expression from the respective pINDUCER20-p16INK4A construct. CCNE2 expression (bottom) is strongly repressed both by WT p16INK4A and p16INK4A-C72A. Diamide treatment relieves this repression only in the WT p16INK4 expressing cell line. Note that the inducible system is somewhat leaky, which could explain induction of CCNE2 in the p16INK4AWT cells by diamide in the absence of doxycycline. The mean plus SEM is displayed from n = 5 biological replicates. *p < 0.05, **p < 0.01, pairwise comparison using a two-sided t-test comparing cells carrying the inducible WT p16INK4 or p16INK4A-C72A construct in each condition.
To test how p16INK4A oxidation and aggregation impacts on the total CDK4/6 activity we performed qPCR analysis on the expression of Cyclin E2 (CCNE2), which is a transcriptional target of E2F1 and hence should be upregulated by active CDK4/6. CCNE2 (Fig. 6B, lower panel) was strongly repressed upon the doxycycline-induced p16INK4A or p16INK4A-C72A expression (Fig. 6B, top panel) in otherwise p16INK4A deficient COLO829 melanoma cells. CCNE2 transcription was partially restored upon diamide treatment, but not in the cells expressing the p16INK4A-C72A redox-insensitive variant. The induction of CCNE2 by diamide in the absence of doxycycline in the p16INK4A WT cell line is likely due to the fact that the inducible system is somewhat leaky. Although we did find evidence that CDK4/6 is indeed reactivated upon oxidation of p16INK4A, and that this partially relieves E2F1 repression, we did not find evidence of perturbation of the S-phase checkpoint in cell-cycle analysis experiments using flow cytometry and video time-lapse microscopy (data not shown). This might be explained by the notion that substantial amounts of monomeric p16INK4A persist under conditions where we find aggregated p16INK4A in the insoluble pellet fraction of our lysates. The observed reactivation of CDK4/6 might therefore not be strong enough for full re-entry of the cell cycle under the tested conditions, but we cannot exclude that this might occur in other situations. Furthermore, treatment with oxidizing agents also affects the cell cycle through several other pathways, possibly obscuring p16INK4A oxidation dependent effects.
3.8. Evolutionary analysis of the redox sensitive cysteine in p16INK4A
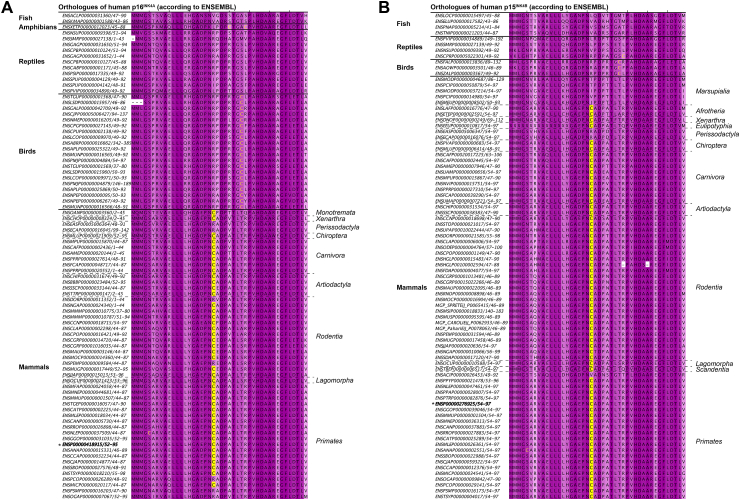
Aggregation and inactivation of a tumor suppressor protein may seem a maladaptive response to oxidizing conditions, although we cannot exclude that it may support another currently unknown biological function. Conservation of evolutionarily-acquired cysteines at the surface of proteins can be indicative for a role in redox signaling, whereas the loss of acquired cysteines may indicate the cysteine hampers protein function [11,39]. We therefore investigated to what extent p16INK4A Cys72 is conserved throughout evolution. The genes encoding p16INK4A (CDKN2A) and p15INK4B (CDKN2B) are the product of a gene duplication that occurred somewhere in evolution around the time that the mammalian branch of the evolutionary tree was formed [40] (see Fig. S12A for reference). Strikingly, Cys72 in the human protein is not conserved in any of the protein sequences of the CDKN2 orthologues in fish, birds, reptiles and amphibians. Yet, this cysteine is present in both p16INK4A and p15INK4B in almost all mammalian species for which sequence information is available in the ENSEMBL database [29], suggesting that acquisition of this cysteine occurred just before or coincided with the gene duplication of the ancestral CDKN2 gene (Fig. 7A and B). Further investigation of the protein sequences of the p16INK4A and p15INK4B orthologues revealed that the cysteine homologous to the human p16INK4A-Cys72 was substituted later in evolution in several branches of mammalian species including some primates, rodentia and perissodactyla (see Fig. 7A and B and Fig. S12B). The latter could indicate that redox regulation of the acquired cysteine homologous to human p16INK4ACys72 is not absolutely required for a p16INK4A function and that Cys72 might be slightly deleterious and displays a weak purifying selection.
Fig. 7.
Evolutionary analysis of p16INK4Aand its paralogue p15INK4B.
(A) Conservation of Cys72 in human p16INK4A and (B) Cys74 in its paralogue p15INK4B throughout vertebrate evolution. Note that ENSEMBL [29] assigns some non-mammalian orthologues to either p16INK4A or p15INK4B whereas gene duplication to yield these paralogues did not take place until mammalian species had branched off [40]. Sequences are named by ENSEMBL ID [29]. The human orthologues are indicated with * and in bold. Cysteines conserved at the position homologous to human p16INK4ACys72 are indicated on yellow background, other cysteines are in yellow text. The darker the purple shading the more conserved the amino acids. Note that several distant branches of mammals have lost the cysteine homologous to human p16INK4ACys72. See Fig. S12 for a schematic tree of vertebrate and mammalian evolution for reference. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Amyloid fibrils are structures that can be formed by a wide variety of protein sequences [41]. These fibrils share common features, including an unbranched fibrillar morphology and a cross-β sheet core structure. Amyloids were previously thought to be largely disease-related, especially involved in neurodegenerative diseases, but there are increasing numbers of amyloid fibrils discovered that have a physiological function. Several mammalian examples have been reported in recent years, including the melanosome protein pMeL, amyloids involved in hormone storage, and the RIPK proteins, whose fibril formation appears to trigger necroptotic signaling pathways [[42], [43], [44], [45]]. In this work we show that p16INK4A can form fibrils under physiological conditions and in cell-based models, and that the formation is triggered by oxidation of the single cysteine residue and homodimerization. We presented evidence that p16INK4A can form aggregates that have the typical features of amyloid fibrils, including binding of diagnostic dyes, presence of cross-β sheet structure, and typical dimensions found in EM. The critical dependence of a disulfide cross-linked dimer as a subunit has not been observed so far and highlights the role of the cellular redox state as an important regulator of fibril formation.
The role of cysteine chemistry in amyloid formation has been discussed in detail previously [46]. Most commonly, disulfide bonds are found to stabilize a soluble, usually monomeric, form of the protein, preventing its un- or misfolding and aggregation. In many cases, blocking of disulfide bond formation by mutagenesis or chemistry generates proteins that are more prone to amyloid formation, such as has been demonstrated for lysozyme [47], SOD1 [48,49], insulin [50] and prion protein [51,52]. In contrast, oxidation of the single cysteine of p16INK4A is important for the transition of the monomeric protein to form amyloid fibrils, and modification of the cysteine by mutagenesis or S-glutathionylation prevents its amyloid formation. Proteins in which disulfide bonds have been postulated to be involved in stabilization of amyloid fibrils include the prion protein and RIPK fibrils, however disulfide bond formation itself is not required for conversion to amyloid in either of these cases [[51], [52], [53]].
Several properties of p16INK4A fibrils fit with those expected for functional amyloids, including their rapid formation (instead, disease-related amyloids can take days to form fibrils) [54,55], and their lack of clear polymorphism (no evidence of peak doubling in the solid-state NMR spectrum) [56,57]. We have shown that p16INK4A fibrils can be formed in vitro under relatively mildly oxidizing conditions at physiological pH. In contrast to many other amyloid-forming proteins, p16INK4A does not require harsh treatments, such as extreme pH, high temperatures, high pressure or organic solvents, that are frequently used to destabilize folded proteins in order to convert them to amyloid fibrils in a practical time scale. One striking difference in the properties of p16INK4A amyloid fibrils, compared to classical disease-linked amyloids like amyloid-β and α-synuclein, is the apparent reversibility of the fibrillar state. Typical amyloids are highly stable and are not disaggregated by SDS buffers and do not run into SDS-PAGE gels [41,58,59]. Our results show that SDS is able to disaggregate p16INK4A fibrils into dimers, and that reducing-agent treatment further returns p16INK4A to a monomeric state. This was shown for both recombinantly-produced p16INK4A amyloid in vitro, as well as p16INK4A amyloid formed in a cellular model. Furthermore, we find that p16INK4A amyloids are transiently formed and do not persist in the cellular environment as judged by the filter trap assay. Clearance of the amyloids in cells could in principle proceed through degradation or disaggregation into monomers. There does not seem to be a clear increase of the monomeric form upon disappearance of the aggregates, which could suggest that the aggregates are being degraded rather than disaggregated. A recent study showed that p16INK4A also forms aggregates upon inhibition of autophagy [60], which is in line with this notion. The observation that p16INK4A amyloids are reversible and can be cleared from cells potentially adds to an emerging pattern of lability of amyloid fibrils that have functional roles in the cell. For example, RIPK1/3, and the phase-separating proteins FUS and hnRNPA2 are all reported to form functional amyloid fibrils, and are apparently SDS soluble [50,53]. This may be a hallmark for functional fibrils, which must be regulated and removed when the function is no longer needed. In contrast, highly stable, persistent aggregates may have toxic properties, such as in the case of Tau or TDP-43 aggregates, which are pathogenically related to various neurodegenerative diseases [61,62].
It is now generally accepted that the cellular redox environment contributes to various signaling pathways and that H2O2 can act as a secondary messenger molecule [9]. Cysteine residues are frequently found to be modified in large-scale analysis of the cysteinome and thousands of proteins are affected by redox regulation [63,64]. Redox potentials vary widely between cell types and specific compartments, ranging from −374 mV for the strongly reducing NADPH/NADP+ redox couple in the liver cytosol to −180 mV from the GSH/GSSG couple in the oxidizing endoplasmic reticulum of B-cells [18]. In addition, the average redox potential varies greatly between cell cycle stages [65]. The redox potential of −198 mV that we found for the oxidation of C72 of p16INK4A is well within the physiological range. One difference between in vitro and cell culture oxidation of p16INK4A is that in vitro p16INK4A could be stably GSH-ylated whereas in cell cultures we only observed the disulfide-bonded homodimer. This might seem surprising, because there is ample GSH available in the cell and addition of micromolar ranges of diamide will likely react to form GSSG. Therefore, it may be unlikely that in cells diamide and H2O2 oxidize p16INK4A directly, but rather that its oxidation is the result of a redox relay or disulfide exchange mechanism, as has been proposed for other redox-sensitive proteins [66]. However, we cannot exclude that transient GSH-ylation of p16INK4A occurs in cells. Of note, tumor cells have been frequently reported to exhibit a more oxidizing milieu [67]. This could mean that p16INK4A would be more often in the dimerized and aggregated state, which could contribute to reactivation of CDK4/6 in tumors that express p16INK4A. One might argue that p16INK4A inactivation by oxidizing conditions would be an inadequate response, that would be lost through evolutionary pressure. Evolutionary analysis indeed shows that the cysteine homologous to human p16INK4A-Cys72 was acquired recently: near or coinciding with the appearance of mammals and was then substituted again multiple times in several branches of mammalian evolution. However, this analysis would have to be further extended to be able to reject a potential beneficial biological role for p16INK4A amyloid formation, for instance to temporarily reactivate CDK4/6 in cells with high p16INK4A levels. Whether and how this indeed could also contribute to tumor progression remains to be established. Our pull-down experiments indicate that oxidation of p16INK4A leads to loss of its inhibitory activity towards CDK4/6 but that oxidized p16INK4A and CDK4/6 still interact. We speculate that the CDK4/6 interaction that we observe is of a different nature, which could underlie loss of inhibition of CDK4/6 upon oxidation of p16INK4A. Amyloid formation-dependent inactivation of tumor suppressor proteins has been described before for p53 and also in this case amyloid formation was reversible [68].
Taken together, the presented work shows that redox signaling can trigger the formation of amyloid fibrils from an otherwise alpha-helical protein. Conversion of a protein from a soluble, monomeric to an insoluble, β-amyloid fibrillar phase dependent on the cellular redox state has not been reported before. This could pose a more general mechanism in amyloid formation, as was recently shown for redox-dependent liquid-liquid phase-separation [69]. It also adds to the repertoire of structural alterations known to occur in response to redox signaling.
Funding
This work was supported by the President’s International Fellowship Initiative of CAS (No. 2015VBB045, to TM), the National Natural Science Foundation of China (No. 31450110423, to TM, KLI425 and KLI645), the Austrian Science Fund (FWF: P28854, I3792, DK-MCD W1226 to TM), the Austrian Research Promotion Agency (FFG: 864690, 870454), the Integrative Metabolism Research Center Graz, the Austrian infrastructure program 2016/2017, BioTechMed-Graz, the Styrian government (Zukunftsfonds) and the OMICS center Graz. CG was supported by a PFP fellowship of the Helmholtz Zentrum München. VM was supported by a CJ Martin Early Career Fellowship from the National Health and Medical Research Council of Australia. TD and MV were supported by a grant from the Dutch Cancer Society (KWF Kankerbestrijding). HV was supported by the ProteinsAtWork research programme.
Author contributions
C.G., V.M., L.D., T.M. and T.D. planned the study, performed experiments and wrote the manuscript with input from the other authors. P.P., L.D., M.V., and H.R. performed immuno-precipitation and Western blot experiments in cultured cells. M.V. and P.P. did the in vitro kinase assays. The filter trap assays were done by P.P. and L.D. Mass spectrometry sample preparation, measurement and analysis for the experiments using cultured cells was done by L.D., M.V. and H.V. C.G. produced recombinant protein samples, C.G., C.H. and T.M. performed solution NMR experiments, V.M. and M.H. performed electron microscopic analysis, V.M and B.R. measured solid-state NMR experiments, and V.M. performed all other amyloid characterization experiments. L.L. and R.B.G. performed intact mass spectrometry measurements. T.D. performed the evolutionary analysis. R.B.G and B.R. edited the manuscript. L.D. sketched the graphical abstract.
Declaration of interests
The authors declare no competing interests.
Data availability
The datasets generated and/or analyzed in the current study are available from the corresponding authors upon reasonable request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [70] partner repository with the dataset identifier PXD012353. reviewer89417@ebi.ac.uk.
Acknowledgements
We would like to thank Marc Pagès-Gallego, Vladimir Hristov, Dr. Holger Rehmann, Dr. Marten Hornsveld, Dr. Richard Loeser and dr. John Collins, Zoë Alexiou and Kim van Dorenmalen for assistance with preparation and validation of DNA constructs and cell lines, additional experiments, analysis or discussion. We thank Dr. Mark Hipp and Dr. Willianne Vonk for discussions on aggregation and help with the Filter trap assay protocol. Berend Snel and Bas Dutilh helped with the evolutionary analysis. We are grateful for input of the UMCU Molecular Cancer Research lab members on this project. We would like to gratefully acknowledge the assistance of Dr. Carsten Peters and Prof. Sevil Weinkauf with electron microscopy measurements, and the assistance of the groups of Prof. Johannes Buchner and Prof. Stephan Sieber with the use of the fluorescence plate readers. We would also like to thank Dr. Riddhiman Sarkar with assistance with solid-state NMR measurements, and Dr. Ralf Stehle for assistance with FTIR measurements.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101316.
Contributor Information
Tobias Madl, Email: tobias.madl@medunigraz.at.
Tobias B. Dansen, Email: T.B.Dansen@umcutrecht.nl.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Sherr C.J. Cancer cell cycles. Science (New York, N.Y.) 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 2.Johnson D.G., Schwarz J.K., Cress W.D., Nevins J.R. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 3.Burd C.E. Monitoring tumorigenesis and senescence in vivo with a p16(INK4a)-luciferase model. Cell. 2013;152:340–351. doi: 10.1016/j.cell.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forbes S.A. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker D.J. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker D.J. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530:184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russo A.A., Tong L., Lee J.O., Jeffrey P.D., Pavletich N.P. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature. 1998;395:237–243. doi: 10.1038/26155. [DOI] [PubMed] [Google Scholar]
- 8.Stepanova L., Leng X., Parker S.B., Harper J.W. Mammalian p50Cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes Cdk4. Genes Dev. 1996;10:1491–1502. doi: 10.1101/gad.10.12.1491. [DOI] [PubMed] [Google Scholar]
- 9.Holmstrom K.M., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014;15:411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 10.Vurusaner B., Poli G., Basaga H. Tumor suppressor genes and ROS: complex networks of interactions. Free Radic. Biol. Med. 2012;52:7–18. doi: 10.1016/j.freeradbiomed.2011.09.035. [DOI] [PubMed] [Google Scholar]
- 11.Putker M. Redox-dependent control of FOXO/DAF-16 by transportin-1. Mol. Cell. 2013;49:730–742. doi: 10.1016/j.molcel.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 12.Medema R.H., Herrera R.E., Lam F., Weinberg R.A. Growth suppression by p16ink4 requires functional retinoblastoma protein. Proc. Natl. Acad. Sci. U.S.A. 1995;92:6289–6293. doi: 10.1073/pnas.92.14.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakaue-Sawano A. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 14.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Delaglio F. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 16.Vranken W.F. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 17.Piotukh K., Kosslick D., Zimmermann J., Krause E., Freund C. Reversible disulfide bond formation of intracellular proteins probed by NMR spectroscopy. Free Radic. Biol. Med. 2007;43:1263–1270. doi: 10.1016/j.freeradbiomed.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Schafer F.Q., Buettner G.R. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 19.Franke D. ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 2017;50:1212–1225. doi: 10.1107/S1600576717007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez-Escamilla A.-M., Rousseau F., Schymkowitz J., Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 21.Walsh I., Seno F., Tosatto S.C.E., Trovato A. PASTA 2.0: an improved server for protein aggregation prediction. Nucleic Acids Res. 2014;42:W301–W307. doi: 10.1093/nar/gku399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conchillo-Solé O. AGGRESCAN: a server for the prediction and evaluation of "hot spots" of aggregation in polypeptides. BMC Bioinf. 2007;8 doi: 10.1186/1471-2105-8-65. 65–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szeverenyi N.M., Sullivan M.J., Maciel G.E. Observation of spin exchange by two-dimensional fourier transform 13C cross polarization-magic-angle spinning. J. Magn. Reson. 1969;47:462–475. 1982. [Google Scholar]
- 24.Wang Y., Jardetzky O. Probability-based protein secondary structure identification using combined NMR chemical-shift data. Protein Sci. 2002;11:852–861. doi: 10.1110/ps.3180102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 26.Schwanhäusser B. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 27.Luber C.A. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity. 2010;32:279–289. doi: 10.1016/j.immuni.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 28.Chich J.F. Statistics for proteomics: experimental design and 2-DE differential analysis. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;849:261–272. doi: 10.1016/j.jchromb.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 29.Zerbino D.R. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sunde M. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 32.Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 33.LeVine H. Quantification of beta-sheet amyloid fibril structures with thioflavin T. Methods Enzymol. 1999;309:274–284. doi: 10.1016/s0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]
- 34.Klunk W.E., Pettegrew J.W., Abraham D.J. Quantitative evaluation of Congo red binding to amyloid-like proteins with a beta-pleated sheet conformation. J. Histochem. Cytochem. : official journal of the Histochemistry Society. 1989;37:1273–1281. doi: 10.1177/37.8.2666510. [DOI] [PubMed] [Google Scholar]
- 35.Nilsson M.R. Techniques to study amyloid fibril formation in vitro. Methods. 2004;34:151–160. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 36.Zandomeneghi G., Krebs M.R.H., McCammon M.G., Fändrich M. FTIR reveals structural differences between native beta-sheet proteins and amyloid fibrils. Protein Sci. : a publication of the Protein Society. 2004;13:3314–3321. doi: 10.1110/ps.041024904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarroukh R., Goormaghtigh E., Ruysschaert J.-M., Raussens V. ATR-FTIR: A "rejuvenated" tool to investigate amyloid proteins. Biochim. Biophys. Acta. 2013;1828:2328–2338. doi: 10.1016/j.bbamem.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 38.Tartaglia G.G. Prediction of aggregation-prone regions in structured proteins. J. Mol. Biol. 2008;380:425–436. doi: 10.1016/j.jmb.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 39.Marino S.M., Gladyshev V.N. Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J. Mol. Biol. 2010;404:902–916. doi: 10.1016/j.jmb.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Regneri J. Analysis of the putative tumor suppressor gene cdkn2ab in pigment cells and melanoma of Xiphophorus and medaka. Pigment Cell Melanoma Res. 2019;32:248–258. doi: 10.1111/pcmr.12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiti F., Dobson C.M. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu. Rev. Biochem. 2017;86:27–68. doi: 10.1146/annurev-biochem-061516-045115. [DOI] [PubMed] [Google Scholar]
- 42.Fowler D.M. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040006. e6-e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maji S.K. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science (New York, N.Y.) 2009;325:328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang S., Tang M.-B., Luo H.-Y., Shi C.-H., Xu Y.-M. Necroptosis in neurodegenerative diseases: a potential therapeutic target. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.286. e2905-e2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y., Yan J., Zhang X., Huang K. Disulfide bonds in amyloidogenesis diseases related proteins. Proteins. 2013;81:1862–1873. doi: 10.1002/prot.24338. [DOI] [PubMed] [Google Scholar]
- 47.Mossuto M.F. Disulfide bonds reduce the toxicity of the amyloid fibrils formed by an extracellular protein. Angew. Chem. 2011;50:7048–7051. doi: 10.1002/anie.201100986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chattopadhyay M. Initiation and elongation in fibrillation of ALS-linked superoxide dismutase. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18663–18668. doi: 10.1073/pnas.0807058105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Furukawa Y., Kaneko K., Yamanaka K., O'Halloran T.V., Nukina N. Complete loss of post-translational modifications triggers fibrillar aggregation of SOD1 in the familial form of amyotrophic lateral sclerosis. J. Biol. Chem. 2008;283:24167–24176. doi: 10.1074/jbc.M802083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kato M. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maiti N.R., Surewicz W.K. The role of disulfide bridge in the folding and stability of the recombinant human prion protein. J. Biol. Chem. 2001;276:2427–2431. doi: 10.1074/jbc.M007862200. [DOI] [PubMed] [Google Scholar]
- 52.Honda R. Role of the disulfide bond in prion protein amyloid formation: a thermodynamic and kinetic analysis. Biophys. J. 2018;114:885–892. doi: 10.1016/j.bpj.2017.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mompeán M. The structure of the necrosome RIPK1-RIPK3 core, a human hetero-amyloid signaling complex. Cell. 2018;173:1244–1253. doi: 10.1016/j.cell.2018.03.032. e1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ono K., Takahashi R., Ikeda T., Yamada M. Cross-seeding effects of amyloid β-protein and α-synuclein. J. Neurochem. 2012;122:883–890. doi: 10.1111/j.1471-4159.2012.07847.x. [DOI] [PubMed] [Google Scholar]
- 55.Ehrnhoefer D.E. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat. Struct. Mol. Biol. 2008;15:558–566. doi: 10.1038/nsmb.1437. [DOI] [PubMed] [Google Scholar]
- 56.Deshmukh M., Evans M.L., Chapman M.R. Amyloid by design: intrinsic regulation of microbial amyloid assembly. J. Mol. Biol. 2018;430:3631–3641. doi: 10.1016/j.jmb.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murray D.T. Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell. 2017;171:615–627. doi: 10.1016/j.cell.2017.08.048. e616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Makin O.S., Atkins E., Sikorski P., Johansson J., Serpell L.C. Molecular basis for amyloid fibril formation and stability. Proc. Natl. Acad. Sci. U.S.A. 2005;102:315–320. doi: 10.1073/pnas.0406847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baldwin A.J. Metastability of native proteins and the phenomenon of amyloid formation. J. Am. Chem. Soc. 2011;133:14160–14163. doi: 10.1021/ja2017703. [DOI] [PubMed] [Google Scholar]
- 60.Coryell P.R. 2019. Autophagy Regulates the Localization and Degradation of p16INK4a. bioRxiv; p. 521682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanger D.P., Anderton B.H., Noble W. Tau phosphorylation: the therapeutic challenge for neurodegenerative disease. Trends Mol. Med. 2009;15:112–119. doi: 10.1016/j.molmed.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 62.Capitini C. TDP-43 inclusion bodies formed in bacteria are structurally amorphous, non-amyloid and inherently toxic to neuroblastoma cells. PLoS One. 2014;9 doi: 10.1371/journal.pone.0086720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van der Reest J., Lilla S., Zheng L., Zanivan S., Gottlieb E. Proteome-wide analysis of cysteine oxidation reveals metabolic sensitivity to redox stress. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-04003-3. 1581-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang J., Carroll K.S., Liebler D.C. The expanding landscape of the thiol redox proteome. Mol. Cell. Proteom. 2016;15:1–11. doi: 10.1074/mcp.O115.056051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Menon S.G., Goswami P.C. A redox cycle within the cell cycle: ring in the old with the new. Oncogene. 2007;26:1101–1109. doi: 10.1038/sj.onc.1209895. [DOI] [PubMed] [Google Scholar]
- 66.Stöcker S., Maurer M., Ruppert T., Dick T.P. A role for 2-Cys peroxiredoxins in facilitating cytosolic protein thiol oxidation. Nat. Chem. Biol. 2018;14:148–155. doi: 10.1038/nchembio.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gorrini C., Harris I.S., Mak T.W. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 2013;12:931. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 68.Lee A.S. Reversible amyloid formation by the p53 tetramerization domain and a cancer-associated mutant. J. Mol. Biol. 2003;327:699–709. doi: 10.1016/s0022-2836(03)00175-x. [DOI] [PubMed] [Google Scholar]
- 69.Kato M. Redox state controls phase separation of the yeast ataxin-2 protein via reversible oxidation of its methionine-rich low-complexity domain. Cell. 2019;177:711–721. doi: 10.1016/j.cell.2019.02.044. e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vizcaino J.A. Update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed in the current study are available from the corresponding authors upon reasonable request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [70] partner repository with the dataset identifier PXD012353. reviewer89417@ebi.ac.uk.