Abstract
Chondrocytes are responsible for maintaining the cartilage that helps joints bear load and move smoothly. These cells typically respond to physiological compression with pathways consistent with matrix synthesis, and chondrocyte mechanotransduction is essential for homeostasis. In osteoarthritis (OA), chondrocyte mechanotransduction appears to be dysregulated, yet the mechanisms remain poorly understood. The objective of this study is to document the phosphoproteomic responses of primary osteoarthritic chondrocytes to physiological sinusoidal compression. We show that OA chondrocytes respond to physiological compression by first activating proteins consistent with cytoskeletal remodeling and decreased transcription, and then later activating proteins for transcription. These results show that several microtubule-related proteins respond to compression. Our results demonstrate that compression is a relevant physiological stimulus for osteoarthritic chondrocytes. Future analyses may build on these results to find differences in compression-induced phosphoproteins between normal and OA cells that lead to druggable targets to restore homeostasis to diseased joints.
Introduction
Osteoarthritis (OA) is the most common joint disorder worldwide (Cicuttini et al., 2011; Cobb, 2011; Goekoop et al., 2011; Hayashi et al., 2011a; Woolf et al., 2012; Woolf and Pfleger, 2003; Zhang and Jordan, 2010), and is characterized by the breakdown of the articular cartilage covering the joint surfaces. Articular cartilage is composed of a dense extra cellular matrix (ECM), a less-dense pericellular matrix (PCM), and highly specialized cells, chondrocytes (Sophia Fox et al., 2009). At these joint surfaces (e.g. the hip and knee), the chondrocytes are subjected to repetitive mechanical loading, which can reach magnitudes as high as 10 times an individual’s body weight (Guilak, 2011). These loads alter the chondrocyte environment. In OA, the processes by which chondrocytes sense and respond to their mechanical environment, termed mechanotransduction (Vincent, 2013), are disrupted.
Chondrocytes (Haudenschild et al., 2008a; Kawakita et al., 2012; Neu et al., 2007; Vincent, 2013), and other mammalian cells (Fogh, 1986; Grygorczyk et al., 2013; Ko et al., 2013; Ward et al., 2013) can transduce mechanical inputs into biological signals, but the link between these two processes remains unclear (Jaalouk and Lammerding, 2009). Because most of the druggable genome resides in phosphoprotein-mediated interactions (Russ and Lampel, 2005), understanding how the chondrocyte phosphoproteome changes in response to compression is important for developing new treatments for joint disease. The objective of this study is to characterize changes in protein phosphorylation after dynamic compression of primary osteoarthritic chondrocytes.
Chondrocytes are the sole cell type in articular cartilage and play a critical role in maintaining cartilage homeostasis through anabolic and catabolic processes. Mechanical stimulation drives this delicate balance (Farnsworth et al., 2013; Ruiz-Romero et al., 2008). The role of healthy chondrocytes is primarily anabolic in nature. This anabolism includes protecting, maintaining, and repairing cartilage by synthesizing collagen (mostly type II), and proteoglycans (Melas et al., 2014) through the secretion of cytokines, growth factors, and protease inhibitors (Ruiz-Romero et al., 2008). However, in diseased cartilage (e.g. OA), catabolism dominates, and usually involves breakdown of ECM and PCM through secretion of proteases (e.g. matrix metalloproteinase (MMPs)). Dynamic loading, such as walking, promotes anabolic responses in diseased chondrocytes, whereas static loading inhibits matrix anabolism (e.g. by upregulation of catabolic enzymes such as MMP-13) (Bougault et al., 2012; Buschmann et al., 1995; Jones et al., 1982; Sah et al., 1989).
Previous studies of signaling in chondrocytes discovered pathways involving proliferation (Hayashi et al., 2011b), cell differentiation and dedifferentiation (Nishihara et al., 2003), matrix catabolism (via MMPs and ADAM/ADAM-TS gene expression) (Koshy et al., 2002), and programmed cell death (Notoya et al., 2000). These studies provide a detailed understanding of various processes for mechanically-induced signaling in chondrocytes. Studies of chondrocyte mechanotransduction show that chondrocytes respond to applied loading by remodeling their cytoskeleton (Campbell et al., 2007; Haudenschild et al., 2008a). Previous phosphoproteomic analysis of primary human chondrocytes provided insight into the pathophysiology of degradative diseases in cartilage (Melas et al., 2014). However, to our knowledge, no studies have used phosphoproteomics as a tool for elucidating signaling mechanisms of chondrocyte mechanotransduction.
In this paper, we move toward understanding the role of protein phosphorylation in chondrocyte mechanotransduction. To do this, we identify phosphorylated proteins in primary human OA chondrocytes that are dynamically compressed to physiological levels. The data show a phosphoproteomic signature consistent with mechanotransduction beginning at the cell membrane and proceeds through the cytoskeleton to the nucleus. This spatiotemporal sequence of pathways can inform studies aiming to harness chondrocyte mechanotransduction to repair and rebuild cartilage.
Materials and Methods
Chondrocyte Culture and Encapsulation
Primary human chondrocytes were harvested, isolated, and encapsulated using previously optimized methods (Jutila et al., 2014a; Jutila et al., 2014b; Zignego et al., 2013). Chondrocytes were harvested from the femoral heads of n=5 Grade IV OA patients undergoing total hip joint replacement surgery (mean age: 63 years (range: 54-80), and mean mass: 80.4 kg (range: 56.9-99 kg)). To harvest chondrocytes, cartilage shavings were digested in Type IV collagenase (2 mg/mL for 12-14 hrs. at 37°C), and cultured in DMEM with 10% fetal bovine serum and 1% antibiotics (10,000 I.U. /mL penicillin and 10000 μg/mL streptomycin) in 5% atmospheric CO2. After 1 passage, cells were encapsulated at a concentration of ~500,000 cells/gel, and equilibrated in tissue culture conditions for 72 hours.
Mechanical Stimulation
Mechanical stimulation was performed using established techniques (Jutila et al., 2014a; Zignego et al., 2015). For each donor (n=5, female), cell-seeded agarose gels were randomly assigned to one of three loading groups: unloaded controls (i.e. 0 minutes of loading), 15, or 30 minutes of dynamic, cyclic compression (n = 5 biological replicates for each loading group). Homogenous deformations (Zignego et al., 2013) were applied to cell-seeded gels using previously optimized methods (Jutila et al., 2014a; Zignego et al., 2015) with a custom-built bioreactor emulating physiological loading conditions (3.1-6.9% compressive strain, calculated from initial gel height and a frequency = 1.1 Hz (Umberger and Martin, 2007)).
Protein Preparation and Extraction
To extract proteins after compression, gels were flash frozen in liquid nitrogen, pulverized, and stored at −80°C prior to cell lysis (Jutila et al., 2014a; Zignego et al., 2015). Cells were lysed by sonication and vortexing in RIPA buffer (50mM Tris-HCL (pH 8.0), 50mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS (Ikeda et al., 2013)). Samples were then centrifuged at 21,000 x g at 4°C for 10 minutes. Proteins were precipitated from the supernatant using ice-cold acetone overnight at −20°C. Samples were centrifuged the next day at 21,000 x g at 4°C for 10 min, and the acetone supernatant was discarded. The purified protein pellet was then re-suspended in 0.5M triethylammonium bicarbonate (TEAB).
Proteomics
Proteins were digested and protein concentration quantified for each sample. Proteins were reduced before digestion with mass-spectrometry-grade trypsin. Samples were acidified and solvent was removed by vacuum centrifugation. Digested peptides were enrichched for phosphopeptides using graphite columns. After a second vacuum centrifugation, peptides were resuspended in 50 μL of mass spectrometry grade water, acetonitrile, and formic acid (98:2:0.1v/v/v). 5 μL of sample was analyzed via nano UPLC-MS (ultra high performance liquid chromatography mass spectrometry.) Complete proteomic methods are available in the supplement.
Data Processing
We converted the resulting data files with MSConvert (ProteoWizard (Kessner et al., 2008)) to 32-bit .mzML format. These files were then processed with a series of bioinformatics tools. OpenMS TOPPAS was used with published methods (Kenar et al., 2014) to create XTandem workflows to search two established databases (SwissProt and TrEMBL (UniProt, 2019)) to evaluate all possible matches . Both fixed (f) and variable (v) modifications were considered in the database search (carbamidomethyl modification of cysteine (f), oxidation of methionine (v), phosphorylation of serine (v), phosphorylation of threonine (v), and phosphorylation of tyrosine (v)), allowing for up to two missed cleavage sites. Precursor tolerance was 50 ppm and MS/MS fragmentation tolerance was 0.05 Da (Ruiz-Romero et al., 2008). Instrument type was set to ESI-quad-TOF, and peptide charges up to +3 were permitted. The search database included the reviewed, Homo sapiens database (Uniprot/Swissprot) modified to contain both targets and “reversed” decoys for FDR (false discovery rate) corrections for multiple comparisons (Käll et al., 2008; Moore et al., 2002). Protein-level quantification included all isoforms.
Processed data files were then exported into Excel (Microsoft, Redmond, WA) for further processing. Cluster analysis was performed in MATLAB (Natick, MA) using the clustergram function and complete linkage. Pathway analysis was performed IMPaLA (Kamburov et al., 2011) and domain analysis was performed using DAVID (Huang da et al., 2009a, b). The background protein set for searches was Uniprot Release 2019_02.
Statistical Analysis and Candidate Selection
To assess the effects of mechanical loading on chondrocyte protein phosphorylation, three randomly assigned groups of cell-seeded agarose hydrogels were established: unloaded control samples (UC), samples undergoing 15 minutes of dynamic compression (DL15), and samples undergoing 30 minutes of dynamic compression (DL30). Statistical analysis was performed using the combined samples for each loading group, from each donor (n=5 samples per group). We defined detected phosphoproteins as those present in the majority of samples (e.g. more than 60%). To determine loading induced differences between samples, each dataset was compared against the others to determine (1) unique phosphoproteins to each loading group and (2) overlapped or shared phosphoproteins between loading groups. Four separate comparisons were made: (1) UC vs. DL15, (2) UC vs. DL30, (3) DL15 vs. DL30, and (4) UC vs. DL15 vs. DL30. For phosphoproteins identified in all three groups (i.e. UC, DL15, and DL30), statistical comparisons were made using Wilcoxon signed rank tests and Kruskal-Wallis one-way analysis of variance.
Results
The objective of this study is to characterize the phosphoproteomic response of primary human OA chondrocytes stimulated by physiological dynamic compression. We analyze the untargeted phosphoproteomic profiles to minimize bias by the exclusion of important, but unexpected, data (i.e. if only a single signaling pathway were selected a priori). Primary chondrocytes from OA joint replacement tissues were harvested, encapsulated in physiologically stiff agarose, and subjected to physiological compression tests for 15 (DL15) or 30 (DL30) minutes prior to phosphoproteomic analysis (Figure 1). We also analyzed uncompressed control samples (UC). To our knowledge, the present study is the first application of phosphoproteomics to analyze mechanotransduction in primary OA chondrocytes. These data demonstrate that applied dynamic compression stimulates primary osteoarthritic chondrocytes to alter their phosphoproteomic profiles in a 3D culture system.
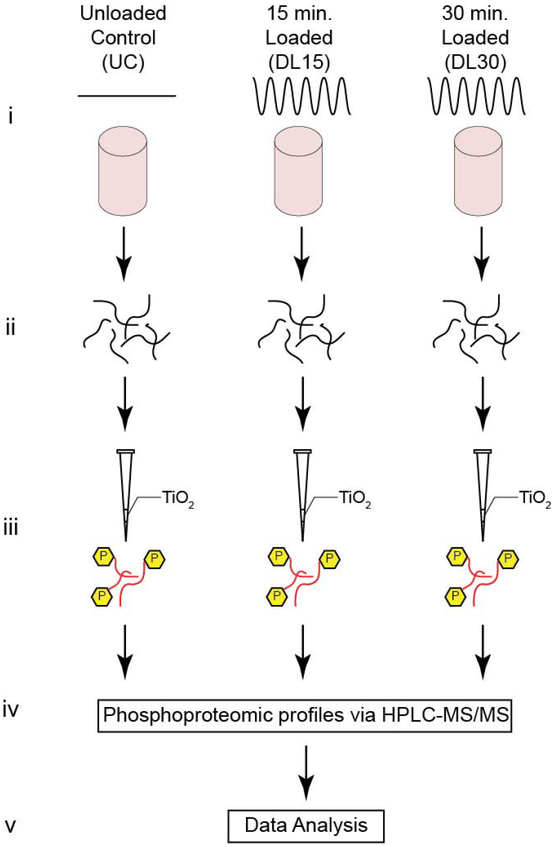
Figure 1. Experimental Design.
(A) Schematic for both untargeted experimental methods. (i) Primary human OA chondrocytes are encapsulated in physiologically stiff agarose (4.5% agarose, stiffness ~35 kPa), cultured for 72 hours, and then dynamically compressed in tissue culture for 0, 15, or 30 minutes (Control, DL15, or DL30) at 1.1 Hz. (ii) Proteins are extracted by flash freezing the samples, pulverizing, and lysing the cells followed by overnight enzymatic digestion. (iii) Samples are enriched for phosphopeptides using TiO2 enrichment, (iv) phosphoproteomic profiles identified via HPLC-MS/MS, and (v) the untargeted data analyzed.
Proteomic Analysis and Characterization
LCMS data for each of the individual samples (n=5 per group) from each experimental group (UC, DL15, and DL30) were processed individually through the TOPPAS pipeline (Junker et al., 2012). After false discovery rate (FDR) filtering with a 5% threshold and using the decoy protein hits and combining all replicates from each loading group, we identified 2858, 2246, and 2570 phosphoproteins including isomers for UC, DL15, and DL30, respectively (Supplementary Table 6). We further refined these data by focusing only on phosphoproteins detected in more than half of the samples for each loading group. The final dataset consisted of 767, 359, and 623 phosphoproteins detected for UC, DL15, and DL30, respectively (Figure 2). To analyze the effects of dynamic compression on chondrocyte metabolism, we made individual comparisons between unloaded control samples and samples loaded for either 15 or 30 minutes of dynamic compression, discussed below. We first report differences in phosphoprotein abundances before examining differences in pathways represented by various groups of proteins.
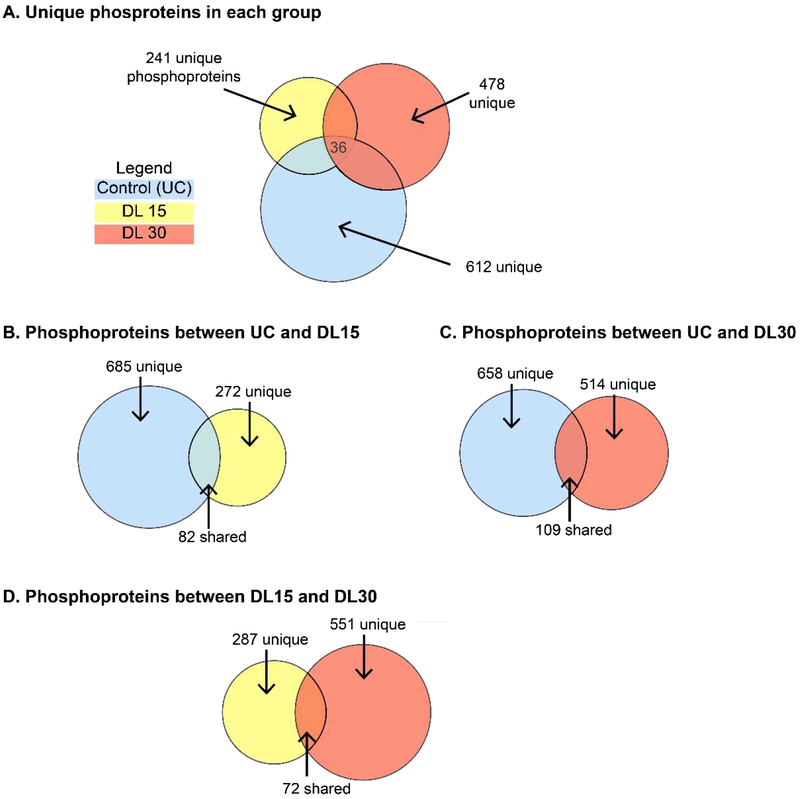
Figure 2. Dynamic compression alters phosphoprotein in 3D primary OA chondrocytes.
To determine signaling pathways activated by dynamic compression, 3D-encapsulated OA chondrocytes were encapsulated in agarose and subjected to 0 (control, UC), 15 minutes (DL15), or 30 minutes (DL30) of compression before analysis by LC-MS that examined phosphoprotein isomers based on detected peptides. Figure quantifies detected proteins using Venn Diagrams with circle areas proportional to the number of proteins. Unique refers to the number of proteins within the indicated area. (A) Unique patterns of phosphoproteins were observed in each group. (B) There were 277 unique phosphoproteins detected after 15 minutes of compression in comparison to unloaded controls. (C) There were 514 phosphoproteins detected after 30 minutes of compression in comparison to controls. (D) There were both unique and shared phosphoproteins detected between 15 and 30 minutes of compression. Proteins shown in File S5.
Differential phosphoprotein abundances after 0, 15, or 30 minutes of compression
By comparing samples stimulated by dynamic compression to the controls, we identified proteins phosphorylated in response to mechanotransduction. This group helped us to establish the breadth of compression as an environmental stimulus. Comparing all groups (e.g. UC, DL15, and DL30, Figure 2A), we find 612 phosphoproteins are unique to UC, 241 unique to DL15, 478 unique to DL30, and 36 common between all three of them. The first comparison between unloaded controls (UC) and DL15 samples reveals 685 phosphoproteins unique to UC, 277 unique to DL15, and 82 phosphoproteins common between the two samples (Figure 2B). Of the 82 shared phosphoproteins, 2 are significantly (p<0.05) up-regulated by mechanical loading (E3 ubiquitin-protein ligase UBR4 and Coiled-coil domain-containing protein 87) and 1 is downregulated (Protein PRR14L). Levels of the remaining 79 proteins are not significantly different. For the second comparison, we compare UC against DL30 samples (Figure 2C). 658 phosphoproteins are unique to UC, 514 unique to DL30, and 109 common between both samples. Of the 109, 4 are up-regulated and 2 down-regulated (all p<0.05). The remaining 103 that were shared, were not significantly different. For the third comparison (DL15 vs. DL30, Figure 2D), 287 phosphoproteins are unique to DL15, 551 unique to DL30, and were 72 common between them. Of the 72 common between DL15 and DL30, one phosphoprotein is down-regulated with increased loading in the DL30 group and the remaining 71 are not significantly different.
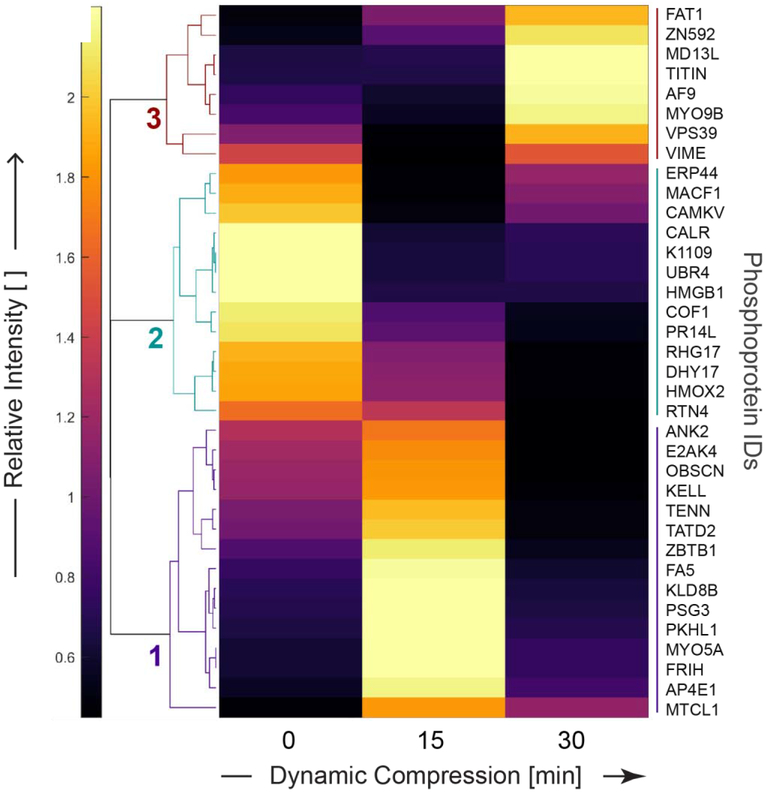
We identify three unique clusters of phosphoproteins that were co-regulated as a result of dynamic compression using unsupervised clustering (Figure 3, Table 1). These clusters describe the timecourse of protein phosphorylation stimulated by compressive loading. Cluster 1 includes phosphoproteins up-regulated with only 15 minutes of dynamic compression. Cluster 2 contains phosphoproteins down-regulated as a result of dynamic compression (either 15 or 30 minutes), and cluster 3 identified phosphoproteins up-regulated only after 30 minutes of dynamic compression. Phosphorylated proteins from cluster 1 include microtubule cross-linking factor 1 (MTCL1), unconventional myosin-Va (MYO5A), ankyrin-2 (ANK2), and obscurin (OBSCN). Cluster 2 phosphorylated proteins include cofilin-1 (COF1), microtubule-actin crosslinking-factor 1 (MACF1), E3 ubiquitin-protein ligase (UBR4), and calreticulin (CALR). Phosphorylated proteins from Cluster 3 include vimentin (VIME), unconventional myosin-IXb (MYO9B), Titin (TITIN), Protein AF-9 (AF9), mediator of RNA polymerase II transcription subunit 13 (MD13L), and Vam6/Vps39-like protein (VPS39). Taken together, these data show a broad response to compression. This includes cytoskeletal regulation, regulation of intracellular calcium, organelle trafficking, and transcription through RNA polymerase II.
Figure 3. Applied compression resulted in distinct untargeted phosphoproteomic profiles for primary OA chondrocytes.
Clustering of phosphoprotein intensities common to all groups (e.g. intersection from Fig. 2A) found three groups of mechanically regulated protein phosphorylation. Patterns of expression displayed by heatmap following unsupervised, hierarchical clustering on differentially phosphorylated proteins common to the dynamic compression (15 and 30 minutes) and unloaded control groups. Heatmap labels are protein identifiers, with complete names found in Supplementary Table 5.
Table 1. Key proteins common to all groups suggest mechanotransduction involves cytoskeletal, calcium regulation, organelle trafficking, and transcriptional elements.
Table summarizes key proteins detected in all groups from clusters in Figure 3.
| Component | Protein | Cluster (Fig. 3) |
|---|---|---|
| Cytoskeletal | ANK2 | 1 |
| COF1 | 2 | |
| DYH17 | 2 | |
| K1109 | 2 | |
| MACF1 | 2 | |
| MTCL1 | 1 | |
| MYO5A | 1 | |
| MYO9B | 1 | |
| OBSCN | 1 | |
| RHG17 | 2 | |
| TENN | 1 | |
| TITIN | 1 | |
| VIME | 1 | |
| Calcium Signaling | ||
| Inhibition | CALR | 2 |
| ERP44 | 2 | |
| Transcription | AF9 | 3 |
| MD13L | 3 | |
| ZBTB1 | 1 | |
| ZN592 | 3 | |
| VPS39 | 3 | |
| Organelle Trafficking | AP4E1 | 1 |
| VPS39 | 3 |
These data show that dynamic compression substantially alters the detected abundances of phosphorylated proteins, and that there are differences between cells stimulated by 15 and 30 minutes of compression. To further understand chondrocyte mechanotransduction, we analyzed pathways represented by these phosphoproteins.
Pathways stimulated by 0, 15, or 30 minutes of compression
To examine how compression regulated the chondrocyte phenotype, we use pathway enrichment analysis. The phosphoproteins identified after FDR-correction from each of the comparisons (UC vs. DL15, UC vs. DL30, and DL15 vs. DL30) identify key signaling pathways using enrichment analysis (Table 2, Supplemental Tables 2-4) (Kamburov et al., 2011). This analysis compares the relative enrichment of phosphoproteins between specific signaling pathways and the specific phosphoproteins identified in each group.
Table 2. Pathways affected by physiological compression of OA chondrocytes.
Over-representation analysis performed using IMPALA based on proteins detected in clusters in Figure 4. Pathways defined as significant when p-values were smaller than the FDR-corrected significance level of 0.05. PID = pathway interaction database. INOH = Integrating Network Objects with Hierarchies. Wiki = wikipathways. Reactome = reactome.
| Pathway proteins | ||||||
|---|---|---|---|---|---|---|
| Observation | Pathway (cluster from Fig 4) | Database | # Detected | Total | Proteins (Uniprot IDs) | P-value |
| Increased by 15 min compression | Regulation of CDC42 activity (p) Urokinase- | PID | 4 | 40 | GIT1; DOC10; BCAR3; DOCK9 | 7.48E-05 |
| type plasminogen activator (uPA) and uPAR-mediated signaling (o) | PID | 3 | 33 | DOCK1; FIBG; LRP1 | 1.67E-04 | |
| Increased by 30 min compression Inhibited by compression (higher in control) | Arginine Proline metabolism (h) | INOH | 3 | 54 | KCRS; ARGI1; AOC3 | 2.03E-04 |
| FTO Obesity Variant Mechanism (j) | Wiki. | 2 | 8 | PRD16; IRX3 | 3.01E-04 | |
| GP1b-IX-V activation signalling (a) | Reactome | 3 | 10 | VWF; GP1BA; FLNA | 6.31E-05 | |
| Peptide hormone biosynthesis (a) Hedgehog (c) | Reactome | 3 | 14 | INHBE; NEC1; COLI | 1.87E-04 | |
| INOH | 5 | 72 | PRS6B; GLI2; PRS4; PSMD2; KIF3A | 1.97E-04 | ||
| Senescence and Autophagy (s) | Wiki. | 6 | 105 | IBP5; CDN1B; LAMP1; SQSTM; CO1A1; IBP3 | 2.41E-04 | |
| FOXM1 transcription factor network (c) | PID | 4 | 42 | CCNA2; XRCC1; NFAC3; CENPF | 2.63E-04 | |
| IL-6 signaling (f) | INOH | 2 | 6 | IL6RB; JAK2 | 3.54E-04 | |
| Collagen chain trimerization (s) | Reactome | 4 | 44 | COIA1; CO1A1; CO1A2; COOA1 | 4.81E-04 | |
| RNA Polymerase III Transcription (d) | Reactome | 2 | 42 | TF3B; BDP1 | 9.65E-04 | |
| Intestinal lipid absorption (d) | Reactome | 1 | 1 | NPCL1 | 1.11E-03 | |
When comparing UC against DL15, we observe 201 signaling pathways (p<0.05) unique to UC samples. These pathways included collagen formation and biosynthesis (p<0.01). 92 significant pathways are enriched from phosphoproteins unique to DL15 samples, including Acetyl-CoA biosynthesis (p=0.0157) and Hedgehog signaling (p=0.0386). Phosphoproteins common to both UC and DL15 show 61 significant signaling pathways, including RhoA activity (p<0.01), and Rho GTPase signaling (p<0.01) that have previously been observed in chondrocyte mechanotransduction (Haudenschild et al., 2008b). These data indicate that 15 minutes of loading induces phosphoproteomic changes at the cell periphery.
For the UC vs. DL30 comparison, we identify 215 signaling pathways (p<0.05) unique to UC, 119 unique to DL30, and 71 shared by both UC and DL30. The pathways for UC include collagen formation and biosynthesis (p<0.01), and extracellular matrix organization (p=0.01). Pathways for DL30 samples included MAPK signaling (p<0.01), Rho GTPase signaling (p<0.01), hyaluronan biosynthesis (p=0.0252), and glucose-6-phosphate dehydrogenase deficiency (p=0.0473). Overlapping pathways between UC and DL30 included ECM proteoglycan synthesis (p=0.0374), and Erk2 activation (p=0.0489). These data indicate that 30 minutes of compression activate phosphoproteins representing a pattern of signals for matrix synthesis (e.g. hyaluronan) and nuclear signaling (e.g. MAPK).
To examine how compression changed over time, we compare DL15 to DL30. We find 80 signaling pathways unique to DL15, 100 unique to DL30, and 90 shared by both DL15 and DL30 (all p<0.05). Specific pathways to DL15 samples include Hedgehog (p<0.01), and calcium signaling (p=0.0453), consistent with prior studies (Erickson et al., 2001; Wu et al., 2001). Specific pathways to DL30 samples include MAPK signaling (p<0.01), fatty acid activation (p<0.01), and Rho GTPase signaling (p<0.01). Common pathways between both DL15 and DL30 samples included RhoA activity (p=0.0168) and nucleotide sugars metabolism (p=0.0342). These data emphasize the role that RhoA plays in responding to applied compression of chondrocytes. Taken together, these patterns of phosphoproteins support the concept of mechanotransduction beginning at the membrane (e.g. calcium signaling) and moving toward the nucleus (e.g. MAPK) with potential feedback for membrane remodeling by fatty acid activation through Acetyl-CoA metabolism.
Protein domains affected by compression
To assess sequence similarities between proteins affected by dynamic compression of chondrocytes, protein domain analysis was performed using DAVID (Huang da et al., 2009a, b) using FDR-correction. There were several structural domains common to the uncompressed controls (Table 3). These include several helicase domains indicating that dynamic compression deactivates helicases. Furthermore, domain analysis of proteins detected after either 15 or 30 minutes of compression indicates that Dynein, myosin, ATPase, and Calmodulin-binding domains are activated by compression.
Table 3. Protein domains absent after compression.
Proteins unique to uncompressed controls were analyzed for structural domains using DAVID. Helicase domains are enriched in these proteins, indicating that dynamic compression deactivates helicases, which are required for de novo transcription.
| Protein Domain | Accession # | Database | P- value |
|---|---|---|---|
| Armadillo-like fold. | IPR016024 | Interpro | 2.63E-02 |
| ATP-binding domain, helicase superfamily 1-2 | IPR014001 | Interpro | 6.77E-03 |
| C-terminal domain, helicase superfamily 1-2 | IPR001650 | Interpro | 5.44E-03 |
| DEAD-like helicases superfamily | SM00487 | SMART | 4.76E-02 |
| Helicase conserved C-terminal domain | PF00271 | Pfam | 2.74E-02 |
| Superfamilies 1 and 2 helicase ATP-binding type-1 domain profile | PS51192 | Prosite | 1.15E-02 |
| Superfamilies 1 and 2 helicase C-terminal domain profile | PS51194 | Prosite | 1.15E-02 |
Discussion
We used phosphoproteomics to identify pathways used by primary chondrocytes to respond to compression. During OA, cartilage deterioration proceeds through catabolic processes within the joint. Matrix degradation softens and weakens cartilage, and further joint loading through physiological processes (e.g. walking) promotes additional joint damage. Because chondrocytes and all cells respond to mechanical stimuli (Mofrad, 2010), these loads also provide an opportunity for therapeutic direction of mechanotransduction to restore cartilage and joint health. These data show that even chondrocytes from joints with advanced grade IV osteoarthritis can respond to applied compression with a broad phosphoproteomic response.
Here we found phosphorylation changes consistent with calcium signaling, cytoskeletal remodeling, and modulation of transcription and in response to applied compression of primary OA chondrocytes. We analyzed the short-term (<30 min) phosphoproteomic changes of these chondrocytes by comparing unloaded control samples (0 min. of loading), to samples subjected to either 15 (DL15) or 30 (DL30) minutes of dynamic compression.
In a prior metabolomic study, we found that this compression regime alters metabolites associated with lipid metabolism, synthesis, and transport, as well as the metabolism of glycine, serine, and threonine (Zignego et al., 2015). In the current study, we found that compression increased phosphorylation of proteins associated with arginine and proline metabolism and membrane receptor CDC42 activity regulation (Table 2). Both studies find changes associated with amino acid metabolism, but the specific differences likely result from the different methods of each study.
Changes in phosphoproteins representing the cytoskeleton mainly appeared after 15 minutes of loading whereas phosphoproteins representing transcriptional regulation followed after 30 minutes of loading. Importantly, these initial changes likely begin prior to the 15 minute timepoint observed in this study (Haudenschild et al., 2008b).
Calreticulin was de-phosphorylated in response to applied compression. Calcium signaling is a well-established mechanosensitive response (D'Andrea et al., 2000; O'Conor et al., 2014; Wann et al., 2012) and there are interactions between the actin cytoskeleton and calcium signaling (D'Andrea et al., 2000; Li et al., 2011; Wu et al., 2013).
The cytoskeleton is a dynamic structure that provides crucial physical linkages within the cytosol and across the plasma membrane. We observed phosphorylation of ankyrin-2 and actin after 15 minutes of dynamic compression. We further observed dephosphorylation of the E3 ubiquitin ligase UBR4 which promotes actin remodeling (Nakatani et al., 2005). After 30 minutes, we observed phosphorylation of vimentin, a perinuclear intermediate filament (Haudenschild et al., 2011). Finally, regulation of CDC42 activity was an enriched pathway after 15 minutes of compression. These changes in phosphoprotein abundances suggest “outside-in” mechanotransduction, wherein an externally applied load is first (e.g. after 15 minutes in this study) perceived at the plasma membrane and is then transduced through intermediate filaments which act as secondary transducers of mechanical loads.
Ankyrin not only affects chondrocyte cytoskeletal dynamics, but is important for maintaining cartilage homeostasis through CD44 (Knudson and Loeser, 2002). This provides a potential mechanism whereby CD44 binds extracelluar hyaluronan for ECM deformation to be transmitted intracellularly. The actin cytoskeleton is a key component of cell shape, which has been linked with matrix synthesis supporting the chondrocyte phenotype (Daniels and Solursh, 1991; Haudenschild et al., 2009), and prior studies found active remodeling of chondrocyte actin in response loading (Haudenschild et al., 2009; Haudenschild et al., 2008a; Knight et al., 2006). Disruption of vimentin networks in chondrocytes leads to decreased matrix synthesis and cell stiffness (Blain et al., 2006), and OA chondrocytes exhibit altered vimentin networks (Haudenschild et al., 2011).
The cytoskeleton contains microtubules. However, little is known about the role of microtubules in chondrocyte mechanotransduction. Here, we found that compression decreased phosphorylation of microtubule-actin crosslinking factor 1 (MACF1) and dynein heavy chain 17 (DYH17), while compression increased the abundance of microtubule crosslinking factor 1 (MTCL1.) DYH17 produces force at the minus end of microtubules and MACF1 crosslinks microtubules to actin (Ning et al., 2016). MTCL1 helps organize microtubules during polarization. Furthermore, several dynein heavy chain and ATPase domains were present in proteins detected after both 15 and 30 minutes of compression. Additionally, both adaptor protein complex 4ε1 (AP4E1) and VPS39 were increased after compression. These proteins are involved in organelle traffic and sorting, potentially indicating compression-dependence in these processes. Taken together, these results suggest that compression induces phosphoproteomic changes associated with microtubule reorganization. However, the effects of this potential reorganization (e.g. increased cellular stiffness, organelle trafficking, or other effects) remain unclear.
Cytoskeletal signaling regulates transcriptional activity (Alberts et al., 2002). Consistent with our interpretation that early (e.g. before 15 minutes) compression-induced cytoskeletal changes activate later transcriptional responses, after 30 minutes we observed phosphorylation of AF-9. AF-9 is a component of the RNA Pol II complex that accelerates elongation of nascent mRNA precursors (Lin et al., 2010). Also after 30 minutes of compression, Cam6/Cps39-like protein, a regulator of TGF-beta activity through activation of SMAD2-dependent transcription, was phosphorylated. Furthermore, we also observed compression-induced phosphorylation of zinc finger proteins (ZN592 and ZBTB1). ZBTB1 is a transcriptional repressor (Matic et al., 2010), which was elevated after 15, but not 30 minutes of compression. ZNF592 contains the KRAB (Kruppel-associated box) domain which also represses transcription (Peng et al., 2007; Urrutia, 2003). Taken together, these data suggest that compression induces phosphoproteomic changes that includes repression of target genes for ZN592 and ZBTB1, consistent with several prior studies (Bougault et al., 2012; Bougault et al., 2009).
To our knowledge, this is the first study to perform proteome-wide phosphorylation analysis following applied compression to chondrocytes encapsulated in a physiologically stiff 3D microenvironment (Jutila et al., 2014b; Zignego et al., 2013). There are important limitations to this study. First, this study focused on phosphoproteins, and therefore the expression of many unphosphorylated proteins is likely compression-dependent. This phosphoproteomic response is consistent with compression-dependent changes in transcription, and future transcriptional profiling may identify the specific genes induced by applied compression. Second, chondrocytes from only female donors were analyzed, and future studies should examine both male and female chondrocytes, as well as potential sex-dependent differences. Third, these methods required an in vitro monolayer passage to generate sufficient cell numbers, and extensive passaging in tissue culture can alter chondrocyte phenotype (Son and Levenston, 2017). Finally, compressive-sensitive phosphoproteomics may be affected by variables not considered in this study such as pericellular matrix, cell age, and other types of mechanical load such as shear.
In summary, this study demonstrates the power of using phosphoproteomics as a tool for probing the chondrocyte response to short-duration (<30 min), dynamic compression. By expanding upon prior metabolomic analysis of primary human OA chondrocytes in response to dynamic compression (Zignego et al., 2015), we identified 514 phosphoproteins unique to dynamically stimulated samples. To our knowledge, this was the first study to successfully identify phosphoproteomic profiles for OA human chondrocytes in response to mechanical loading. This work suggests the potential to use mechanical stimulation (i.e. short-duration, low-impact exercise) as a tool to promote cartilage repair in OA clinical populations.
Supplementary Material
Supplemental Table 1
Signaling pathways determined from pathway enrichment analysis comparing between unloaded controls (UC) and 15 minutes of dynamic compression (DL15).
Supplemental Table 2
Signaling pathways determined from pathway enrichment analysis comparing between unloaded controls (UC) and 30 minutes of dynamic compression (DL30).
Supplemental Table 3
Signaling pathways determined from pathway enrichment analysis comparing between 15 and 30 minutes of dynamic compression (DL15 and DL30, respectively).
Supplemental Table 4
Signaling pathways determined from pathway enrichment analysis comparing overlapping proteins between all conditions.
Supplemental Table 5
Complete proteomics dataset. Labels as follows: raw—detected proteins. refined—detected proteins after FDR filtering. majority—detected proteins from a majority of the samples in each group.
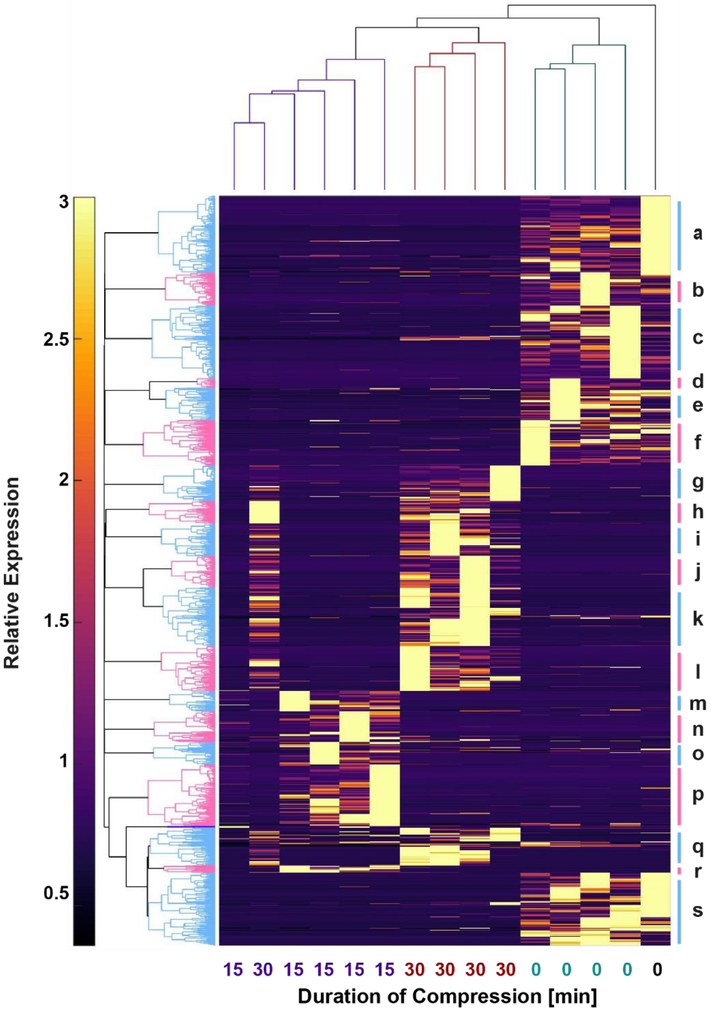
Figure 4. Clustering indicates strong effects of mechanical loading on global phosphoprotein expression.
Unsupervised clustering of all phosphoproteins detected in all samples conducted in MATLAB. Vertical dendrogram (top) indicates that the uncompressed controls (UC) were the most distinct samples, while there was similarity between the samples compressed 15 and 30 minutes. Individual clusters of phosphoproteins labeled alphabetically beginning on the top right. Pathways associated with these clusters shown in Supplementary File Pathway_Enrichment_Results.zip. Key cluster results shown in Table 3.
Table 4. Protein domains detected after compression.
Proteins present after either 15- or 30-minutes of compression were analyzed for structural domains using DAVID.
| Protein Domain | Accession # | Database | P- value |
|---|---|---|---|
| AAA - ATPases | SM00382 | SMART | 1.65E-03 |
| AAA domain (dynein-related subfamily) | PF07728 | PFAM | 2.46E-04 |
| AAA+ superfamily ATPase domain | IPR003593 | INTERPRO | 2.79E-04 |
| ATPase, dynein-related, AAA domain | IPR011704 | INTERPRO | 2.26E-04 |
| ATPase; molecular moto | SM00242 | SMART | 2.34E-02 |
| Calmodulin-binding motif. | SM00015 | SMART | 5.19E-03 |
| Dynein heavy chain | IPR026983 | INTERPRO | 2.76E-05 |
| Dynein heavy chain and region D6 of dynein motor | PF12777 | PFAM | 3.06E-05 |
| Dynein heavy chain region D6 P-loop domain | IPR004273 | INTERPRO | 2.76E-05 |
| Dynein heavy chain, coiled coil stalk | IPR024743 | INTERPRO | 2.76E-05 |
| Dynein heavy chain, domain-1 | IPR013594 | INTERPRO | 8.06E-03 |
| Dynein heavy chain, domain-2 | IPR013602 | INTERPRO | 2.76E-05 |
| Dynein heavy chain, N-terminal region 1 | PF08385 | PFAM | 8.39E-03 |
| Dynein heavy chain, N-terminal region 2 | PF08393 | PFAM | 3.06E-05 |
| Dynein heavy chain, P-loop containing D4 domain | IPR024317 | INTERPRO | 4.12E-04 |
| Dynein motor, microtubule-binding stalk | PF12777 | PFAM | 3.06E-05 |
| isoleucine-glutamine calmodulin-binding motif | PF00612 | PFAM | 1.26E-02 |
| isoleucine-glutamine motif | PS50096 | PROSITE | 1.46E-02 |
| isoleucine-glutamine motif, EF-hand binding site | IPR000048 | INTERPRO | 1.16E-02 |
| Myosin head (motor domain) | PF00063 | PFAM | 1.09E-02 |
| Myosin head, motor domain | IPR001609 | INTERPRO | 9.73E-03 |
| Myosin heavy chain signature | PR00193 | PRINTS | 7.91E-03 |
| Myosin motor domain | PS51456 | PROSITE | 1.25E-02 |
| P-loop containing dynein motor region D4 | PF12780 | PFAM | 4.47E-04 |
| P-loop containing nucleoside triphosphate hydrolases | SSF52540 | SUPFAM | 2.22E-06 |
| P-loop containing nucleotide triphosphate hydrolases | 3.40.50.300 | GENE3D | 9.23E-03 |
| P-loop containing nucleotide triphosphate hydrolases | IPR027417 | INTERPRO | 1.09E-07 |
Acknowledgements
Funding was provided by NIH R01AR073964, NIH P20GM10339405S1, NSF 1342420, NSF 1554708, Montana State University, and the Murdock Charitable Trust. The funding sources had no role in the design or execution of the study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interests
Dr. June owns stock in Beartooth Biotech, which was not involved in this study.
Supplemental Proteomics Methods
This file contains detailed methods for protein processing and proteomic analysis.
Pathway Enrichment Results
Zipped directory of all pathway results based on clusters in Figure 3.
References
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P, 2002. Molecular Biology of the Cell, Fourth Edition ed. Garland Science, New York, NY. [Google Scholar]
- Blain EJ, Gilbert SJ, Hayes AJ, Duance VC, 2006. Disassembly of the vimentin cytoskeleton disrupts articular cartilage chondrocyte homeostasis. Matrix Biol 25, 398–408. [DOI] [PubMed] [Google Scholar]
- Bougault C, Aubert-Foucher E, Paumier A, Perrier-Groult E, Huot L, Hot D, Duterque-Coquillaud M, Mallein-Gerin F, 2012. Dynamic compression of chondrocyte-agarose constructs reveals new candidate mechanosensitive genes. PloS one 7, e36964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougault C, Paumier A, Aubert-Foucher E, Mallein-Gerin F, 2009. Investigating conversion of mechanical force into biochemical signaling in three-dimensional chondrocyte cultures. Nat Protoc 4, 928–938. [DOI] [PubMed] [Google Scholar]
- Buschmann MD, Gluzband YA, Grodzinsky AJ, Hunziker EB, 1995. Mechanical compression modulates matrix biosynthesis in chondrocyte/agarose culture. J Cell Sci 108 ( Pt 4), 1497–1508. [DOI] [PubMed] [Google Scholar]
- Campbell JJ, Blain EJ, Chowdhury TT, Knight MM, 2007. Loading alters actin dynamics and up-regulates cofilin gene expression in chondrocytes. Biochem Biophys Res Commun 361, 329–334. [DOI] [PubMed] [Google Scholar]
- Cicuttini FM, Wluka AE, Urquhart D, Tanamas SK, Wang Y, 2011. Epidemiology should not be forgotten in osteoarthritis imaging Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society; 19, 1165–1166. [DOI] [PubMed] [Google Scholar]
- Cobb JP, 2011. Osteoarthritis excess mortality. Painful limping and early death. BMJ 342, d2137. [DOI] [PubMed] [Google Scholar]
- D'Andrea P, Calabrese A, Capozzi I, Grandolfo M, Tonon R, Vittur F, 2000. Intercellular Ca2+ waves in mechanically stimulated articular chondrocytes. Biorheology 37, 75–83. [PubMed] [Google Scholar]
- Daniels K, Solursh M, 1991. Modulation of chondrogenesis by the cytoskeleton and extracellular matrix. J Cell Sci 100 ( Pt 2), 249–254. [DOI] [PubMed] [Google Scholar]
- Erickson GR, Alexopoulos LG, Guilak F, 2001. Hyper-osmotic stress induces volume change and calcium transients in chondrocytes by transmembrane, phospholipid, and G-protein pathways. J Biomech 34, 1527–1535. [DOI] [PubMed] [Google Scholar]
- Farnsworth NL, Antunez LR, Bryant SJ, 2013. Dynamic compressive loading differentially regulates chondrocyte anabolic and catabolic activity with age. Biotechnol Bioeng. [DOI] [PubMed] [Google Scholar]
- Fogh J, 1986. Human tumor lines for cancer research. Cancer investigation 4, 157–184. [DOI] [PubMed] [Google Scholar]
- Goekoop RJ, Kloppenburg M, Kroon HM, Dirkse LEV, Huizinga TWJ, Westendorp RGJ, Gussekloo J, 2011. Determinants of absence of osteoarthritis in old age. Scandinavian Journal of Rheumatology 40. [DOI] [PubMed] [Google Scholar]
- Grygorczyk R, Furuya K, Sokabe M, 2013. Imaging and characterization of stretch-induced ATP release from alveolar A549 cells. J Physiol 591, 1195–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilak F, 2011. Biomechanical factors in osteoarthritis. Best Practice & Research in Clinical Rheumatology 25, 815–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudenschild DR, Chen J, Pang N, Steklov N, Grogan SP, Lotz MK, D'Lima DD, 2011. Vimentin contributes to changes in chondrocyte stiffness in osteoarthritis. J Orthop Res 29, 20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudenschild DR, Chen J, Steklov N, Lotz MK, D'Lima DD, 2009. Characterization of the chondrocyte actin cytoskeleton in living three-dimensional culture: response to anabolic and catabolic stimuli. Mol Cell Biomech 6, 135–144. [PMC free article] [PubMed] [Google Scholar]
- Haudenschild DR, D'Lima DD, Lotz MK, 2008a. Dynamic compression of chondrocytes induces a Rho kinase-dependent reorganization of the actin cytoskeleton. Biorheology 45, 219–228. [PubMed] [Google Scholar]
- Haudenschild DR, Nguyen B, Chen J, D'Lima DD, Lotz MK, 2008b. Rho kinase-dependent CCL20 induced by dynamic compression of human chondrocytes. Arthritis and rheumatism 58, 2735–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi D, Guermazi A, Hunter DJ, 2011a. Response to Letter to the Editor: 'Epidemiology should not be forgotten in osteoarthritis imaging' Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society; 19, 1167. [Google Scholar]
- Hayashi S, Nishiyama T, Miura Y, Fujishiro T, Kanzaki N, Hashimoto S, Matsumoto T, Kurosaka M, Kuroda R, 2011b. DcR3 induces cell proliferation through MAPK signaling in chondrocytes of osteoarthritis Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society; 19, 903–910. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA, 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA, 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Ikeda D, Ageta H, Tsuchida K, Yamada H, 2013. iTRAQ-based proteomics reveals novel biomarkers of osteoarthritis. Biomarkers 18, 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaalouk DE, Lammerding J, 2009. Mechanotransduction gone awry. Nat Rev Mol Cell Biol 10, 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones IL, Klämfeldt A, Sandström T, 1982. The effect of continuous mechanical pressure upon the turnover of articular cartilage proteoglycans in vitro. Clin Orthop Relat Res, 283–289. [PubMed] [Google Scholar]
- Junker J, Bielow C, Bertsch A, Sturm M, Reinert K, Kohlbacher O, 2012. TOPPAS: a graphical workflow editor for the analysis of high-throughput proteomics data. J Proteome Res 11, 3914–3920. [DOI] [PubMed] [Google Scholar]
- Jutila AA, Zignego DL, Hwang BK, Hilmer JK, Hamerly T, Minor CA, Walk ST, June RK, 2014a. Candidate mediators of chondrocyte mechanotransduction via targeted and untargeted metabolomic measurements. Arch Biochem Biophys 545, 116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutila AA, Zignego DL, Schell WJ, June RK, 2014b. Encapsulation of Chondrocytes in High-Stiffness Agarose Microenvironments for In Vitro Modeling of Osteoarthritis Mechanotransduction. Ann Biomed Eng. [DOI] [PubMed] [Google Scholar]
- Käll L, Storey JD, MacCoss MJ, Noble WS, 2008. Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J Proteome Res 7, 29–34. [DOI] [PubMed] [Google Scholar]
- Kamburov A, Cavill R, Ebbels TM, Herwig R, Keun HC, 2011. Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics 27, 2917–2918. [DOI] [PubMed] [Google Scholar]
- Kawakita K, Nishiyama T, Fujishiro T, Hayashi S, Kanzaki N, Hashimoto S, Takebe K, Iwasa K, Sakata S, Nishida K, Kuroda R, Kurosaka M, 2012. Akt phosphorylation in human chondrocytes is regulated by p53R2 in response to mechanical stress Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society; 20, 1603–1609. [DOI] [PubMed] [Google Scholar]
- Kenar E, Franken H, Forcisi S, Wörmann K, Häring HU, Lehmann R, Schmitt-Kopplin P, Zell A, Kohlbacher O, 2014. Automated label-free quantification of metabolites from liquid chromatography-mass spectrometry data. Mol Cell Proteomics 13, 348–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessner D, Chambers M, Burke R, Agus D, Mallick P, 2008. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight MM, Toyoda T, Lee DA, Bader DL, 2006. Mechanical compression and hydrostatic pressure induce reversible changes in actin cytoskeletal organisation in chondrocytes in agarose. J Biomech 39, 1547–1551. [DOI] [PubMed] [Google Scholar]
- Knudson W, Loeser RF, 2002. CD44 and integrin matrix receptors participate in cartilage homeostasis. Cell Mol Life Sci 59, 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko FC, Dragomir C, Plumb DA, Goldring SR, Wright TM, Goldring MB, van der Meulen MC, 2013. In vivo cyclic compression causes cartilage degeneration and subchondral bone changes in mouse tibiae. Arthritis and rheumatism 65, 1569–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshy PJ, Lundy CJ, Rowan AD, Porter S, Edwards DR, Hogan A, Clark IM, Cawston TE, 2002. The modulation of matrix metalloproteinase and ADAM gene expression in human chondrocytes by interleukin-1 and oncostatin M: a time-course study using real-time quantitative reverse transcription-polymerase chain reaction. Arthritis and rheumatism 46, 961–967. [DOI] [PubMed] [Google Scholar]
- Li F, Wang W, Gu M, Gyoneva S, Zhang J, Huang S, Traynelis SF, Cai H, Guggino SE, Zhang X, 2011. L-type calcium channel activity in osteoblast cells is regulated by the actin cytoskeleton independent of protein trafficking. J Bone Miner Metab 29, 515–525. [DOI] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A, 2010. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell 37, 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic I, Schimmel J, Hendriks IA, van Santen MA, van de Rijke F, van Dam H, Gnad F, Mann M, Vertegaal AC, 2010. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol Cell 39, 641–652. [DOI] [PubMed] [Google Scholar]
- Melas IN, Chairakaki AD, Chatzopoulou EI, Messinis DE, Katopodi T, Pliaka V, Samara S, Mitsos A, Dailiana Z, Kollia P, Alexopoulos LG, 2014. Modeling of signaling pathways in chondrocytes based on phosphoproteomic and cytokine release data Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society; 22, 509–518. [DOI] [PubMed] [Google Scholar]
- Mofrad MRKK, Roger D, 2010. Cellular Mechanotransduction: Diverse Perspectives from Molecules to Tissues. Cambridge University Press, New York, NY. [Google Scholar]
- Moore RE, Young MK, Lee TD, 2002. Qscore: an algorithm for evaluating SEQUEST database search results. J Am Soc Mass Spectrom 13, 378–386. [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Konishi H, Vassilev A, Kurooka H, Ishiguro K, Sawada J, Ikura T, Korsmeyer SJ, Qin J, Herlitz AM, 2005. p600, a unique protein required for membrane morphogenesis and cell survival. Proc Natl Acad Sci U S A 102, 15093–15098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu CP, Khalafi A, Komvopoulos K, Schmid TM, Reddi AH, 2007. Mechanotransduction of bovine articular cartilage superficial zone protein by transforming growth factor beta signaling. Arthritis and rheumatism 56, 3706–3714. [DOI] [PubMed] [Google Scholar]
- Ning W, Yu Y, Xu H, Liu X, Wang D, Wang J, Wang Y, Meng W, 2016. The CAMSAP3-ACF7 Complex Couples Noncentrosomal Microtubules with Actin Filaments to Coordinate Their Dynamics. Dev Cell 39, 61–74. [DOI] [PubMed] [Google Scholar]
- Nishihara A, Fujii M, Sampath TK, Miyazono K, Reddi AH, 2003. Bone morphogenetic protein signaling in articular chondrocyte differentiation. Biochem Biophys Res Commun 301, 617–622. [DOI] [PubMed] [Google Scholar]
- Notoya K, Jovanovic DV, Reboul P, Martel-Pelletier J, Mineau F, Pelletier JP, 2000. The induction of cell death in human osteoarthritis chondrocytes by nitric oxide is related to the production of prostaglandin E2 via the induction of cyclooxygenase-2. J Immunol 165, 3402–3410. [DOI] [PubMed] [Google Scholar]
- O'Conor CJ, Leddy HA, Benefield HC, Liedtke WB, Guilak F, 2014. TRPV4-mediated mechanotransduction regulates the metabolic response of chondrocytes to dynamic loading. Proceedings of the National Academy of Sciences of the United States of America 111, 1316–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Gibson LC, Capili AD, Borden KL, Osborne MJ, Harper SL, Speicher DW, Zhao K, Marmorstein R, Rock TA, Rauscher FJ, 2007. The structurally disordered KRAB repression domain is incorporated into a protease resistant core upon binding to KAP-1-RBCC domain. J Mol Biol 370, 269–289. [DOI] [PubMed] [Google Scholar]
- Ruiz-Romero C, Carreira V, Rego I, Remeseiro S, López-Armada MJ, Blanco FJ, 2008. Proteomic analysis of human osteoarthritic chondrocytes reveals protein changes in stress and glycolysis. Proteomics 8, 495–507. [DOI] [PubMed] [Google Scholar]
- Russ AP, Lampel S, 2005. The druggable genome: an update. Drug Discov Today 10, 1607–1610. [DOI] [PubMed] [Google Scholar]
- Sah RLY, Kim YJ, Doong JYH, Grodzinsky AJ, Plaas AHK, Sandy JD, 1989. BIOSYNTHETIC RESPONSE OF CARTILAGE EXPLANTS TO DYNAMIC COMPRESSION. J. Orthop. Res. 7, 619–636. [DOI] [PubMed] [Google Scholar]
- Son MS, Levenston ME, 2017. Quantitative tracking of passage and 3D culture effects on chondrocyte and fibrochondrocyte gene expression. Journal of tissue engineering and regenerative medicine 11, 1185–1194. [DOI] [PubMed] [Google Scholar]
- Sophia Fox AJ, Bedi A, Rodeo SA, 2009. The basic science of articular cartilage: structure, composition, and function. Sports Health 1, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umberger BR, Martin PE, 2007. Mechanical power and efficiency of level walking with different stride rates. The Journal of experimental biology 210, 3255–3265. [DOI] [PubMed] [Google Scholar]
- UniProt C, 2019. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47, D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia R, 2003. KRAB-containing zinc-finger repressor proteins. Genome Biol 4, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent TL, 2013. Targeting mechanotransduction pathways in osteoarthritis: a focus on the pericellular matrix. Curr Opin Pharmacol. [DOI] [PubMed] [Google Scholar]
- Wann AK, Zuo N, Haycraft CJ, Jensen CG, Poole CA, McGlashan SR, Knight MM, 2012. Primary cilia mediate mechanotransduction through control of ATP-induced Ca2+ signaling in compressed chondrocytes. FASEB J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CW, Prosser BL, Lederer WJ, 2013. Mechanical stretch induced activation of ROS/RNS signaling in striated muscle. Antioxid Redox Signal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf AD, Erwin J, March L, 2012. The need to address the burden of musculoskeletal conditions. Best Practice & Research in Clinical Rheumatology 26. [DOI] [PubMed] [Google Scholar]
- Woolf AD, Pfleger B, 2003. Burden of major musculoskeletal conditions. Bulletin of the World Health Organization 81, 646–656. [PMC free article] [PubMed] [Google Scholar]
- Wu M, Wu X, De Camilli P, 2013. Calcium oscillations-coupled conversion of actin travelling waves to standing oscillations. Proc Natl Acad Sci U S A 110, 1339–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Zhang Y, Chen Q, 2001. Indian hedgehog is an essential component of mechanotransduction complex to stimulate chondrocyte proliferation. J Biol Chem 276, 35290–35296. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Jordan JM, 2010. Epidemiology of osteoarthritis. Clinics in geriatric medicine 26, 355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zignego DL, Hilmer JK, Minor CA, Gelbke MK, June RK, 2015. Candidate Mediators of Osteoarthritic Human Chondrocyte Mechanotransduction via Targeted and Untargeted Metabolomic Measurements. Osteoarthritis and Cartilage. In review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zignego DL, Jutila AA, Gelbke MK, Gannon DM, June RK, 2013. The mechanical microenvironment of high concentration agarose for applying deformation to primary chondrocytes. J Biomech. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1
Signaling pathways determined from pathway enrichment analysis comparing between unloaded controls (UC) and 15 minutes of dynamic compression (DL15).
Supplemental Table 2
Signaling pathways determined from pathway enrichment analysis comparing between unloaded controls (UC) and 30 minutes of dynamic compression (DL30).
Supplemental Table 3
Signaling pathways determined from pathway enrichment analysis comparing between 15 and 30 minutes of dynamic compression (DL15 and DL30, respectively).
Supplemental Table 4
Signaling pathways determined from pathway enrichment analysis comparing overlapping proteins between all conditions.
Supplemental Table 5
Complete proteomics dataset. Labels as follows: raw—detected proteins. refined—detected proteins after FDR filtering. majority—detected proteins from a majority of the samples in each group.