Abstract
Genome-wide association studies have implicated thousands of noncoding variants across common human phenotypes. However, they cannot directly inform the cellular context in which disease-associated variants act. Here, we use open chromatin profiles from discrete mouse cell populations to address this challenge. We applied stratified linkage disequilibrium score regression and evaluated heritability enrichment in 64 genome-wide association studies, emphasizing schizophrenia. We provide evidence that mouse-derived human open chromatin profiles can serve as powerful proxies for difficult to obtain human cell populations, facilitating the illumination of common disease heritability enrichment across an array of human phenotypes. We demonstrate that signatures from discrete subpopulations of cortical excitatory and inhibitory neurons are significantly enriched for schizophrenia heritability with maximal enrichment in cortical layer V excitatory neurons. We also show that differences between schizophrenia and bipolar disorder are concentrated in excitatory neurons in cortical layers II-III, IV, and V, as well as the dentate gyrus. Finally, we leverage these data to fine-map variants in 177 schizophrenia loci nominating variants in 104/177. We integrate these data with transcription factor binding site, chromatin interaction, and validated enhancer data, placing variants in the cellular context where they may modulate risk.
Although genome-wide association studies (GWAS) have implicated thousands of variants in an array of human phenotypes, the variation underlying these signals and cellular contexts in which variants act have remained largely unclear (Visscher et al. 2017). Discernment of disease-relevant variants and cell populations is essential for comprehensive functional investigation of the mechanisms of disease.
Schizophrenia (SCZ) has been robustly investigated through GWAS, with the number of associated loci increasing from 12 to 179 independent associations in the last decade (O'Donovan et al. 2008; Pardiñas et al. 2018). However, this has not been accompanied by the elucidation of disease mechanisms or an increase in the identification of causal variants. To date, support for mechanisms and/or causal variants has been established for only two loci (Sekar et al. 2016; Song et al. 2018). Testing potential mechanisms underlying SCZ has been impeded by the challenge of distinguishing risk variants from those in linkage disequilibrium (LD) and from the lack of knowledge about the cells in which variants may act.
Recent studies have begun to identify cell populations for SCZ by leveraging GWAS summary statistics and stratified linkage disequilibrium score regression (S-LDSC) (Finucane et al. 2018; Skene et al. 2018). These studies have focused on human and rodent transcriptional data, with the finest resolution of cell populations provided by mouse single-cell RNA-seq data (scRNA-seq). The results from these studies have supported a role for cortical excitatory and inhibitory neurons in SCZ risk (Finucane et al. 2018; Skene et al. 2018). However, these studies only capture signals driven by variants residing in selected windows, excluding much of the regulatory landscape. As most variants identified through GWAS occur in noncoding DNA (Maurano et al. 2012), these studies systematically overlook the capacity to use these biological signatures to construct hypotheses indicting putative, cis-regulatory elements.
Ideally, human chromatin data with the same cell population resolution as transcriptome data would be used to provide a regulatory context for variants. However, human chromatin data analyzed with S-LDSC have been limited to easy-to-access cell populations (Ulirsch et al. 2019) or heterogeneous adult tissues, broad cell types, and in vitro cell lines (The ENCODE Project Consortium 2012; Finucane et al. 2018; Fullard et al. 2018; Tansey and Hill 2018). Mouse data have the potential to overcome these barriers by providing chromatin data for the same populations as scRNA-seq. Recently, mouse single-cell ATAC-seq was used to annotate variants and explore the heritability of a variety of traits, including SCZ (Cusanovich et al. 2018). This study implicated many of the same populations in SCZ as previous studies that leveraged expression data. However, which variants are relevant to disease and in which cells those variants may act was not explored.
We have successfully used mouse chromatin data to prioritize common human variants for pigmentation and Parkinson's disease (Praetorius et al. 2013; McClymont et al. 2018). Here, we set out to address whether mouse-derived human open chromatin profiles could be used to prioritize cell populations and variants important to SCZ. In this way, data from narrowly defined cell populations that are inaccessible in humans could be used to provide context for variants. We evaluate a limited number of strategies for converting mouse open chromatin peaks to human peaks and use heritability enrichment analyses to prioritize 27 (25 mouse and two human) cell populations across 64 GWAS with an emphasis on schizophrenia. Ultimately, we combine statistical fine-mapping of variants with mouse-derived human open chromatin data, transcription factor binding site data, chromatin interaction data, and validated enhancer libraries in order to prioritize variants in SCZ loci and predict a cellular context in which those variants may act.
Results
A uniform pipeline for processing of mouse ATAC-seq data
We obtained publicly available ATAC-seq data derived from cell types sorted ex vivo and brain single-nuclei analyses from mice (Supplemental Table S1; Mo et al. 2015; Matcovitch-Natan et al. 2016; Gray et al. 2017; Hughes et al. 2017; Hosoya et al. 2018; McClymont et al. 2018; Preissl et al. 2018). In total, we obtained 25 mouse ATAC-seq open chromatin region (OCR) data sets encompassing subclasses of six broader cell types (dopaminergic neurons, excitatory neurons, glia, inhibitory neurons, retina cells, and T cells) (Supplemental Table S1).
All ATAC-seq data were processed in a uniform manner. Sequencing for each cell population was aligned to the mouse genome (mm10), replicates were combined, and peak summits were called. This resulted in 165,143 summits called per sample (range: 54,880–353,125; median: 130,464), with profiles derived from the single-nuclei data having fewer summits in general (Supplemental Table S2).
To mitigate the potential for biases resulting from variable sequencing depths, we employed a filtering method used by The Cancer Genome Atlas (Corces et al. 2018; see Methods). We added 250 base pairs (bp) to either side of each summit, and the uniform peaks were merged within each population, yielding 78,115 filtered summits per cell population (range: 38,685–119,870) and an average of 62,309 peaks (range: 30,791–99,119 peaks) (Supplemental Table S2).
To ensure that the OCR profiles reflected expected cell population identities, read counts for each cell population for the union set of peaks (433,555 peaks) were compared using principal component analysis (PCA) and hierarchical clustering. PCA revealed that the majority of variation (70.29%) in the data could be explained by whether the ATAC-seq data were single-nuclei or bulk, not the experiment or cell population (Supplemental Fig. S1A–C). Stepwise quantile normalization and batch correction abolished the variation caused by this technical effect (Supplemental Fig. S1A).
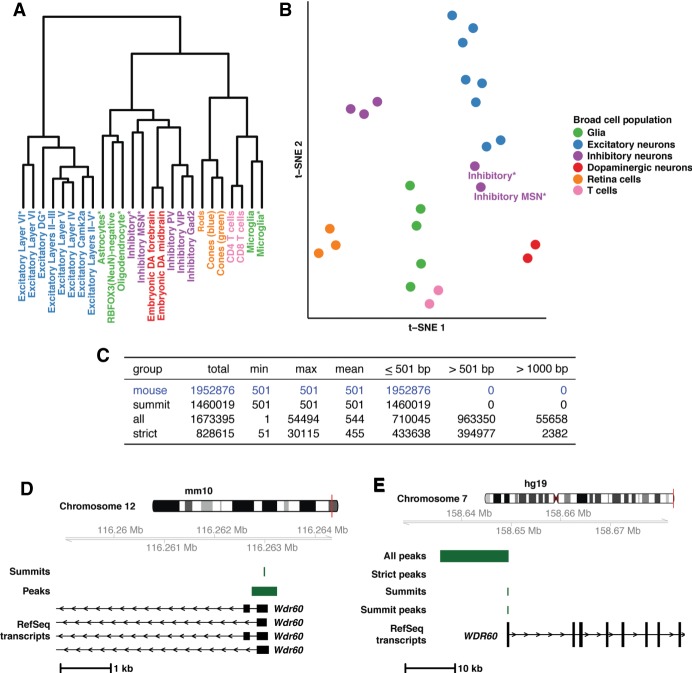
In general, broad cell types clustered together within hierarchical clustering of correlation (Fig. 1A) and when PCA results were projected into two-dimensional, t-distributed Stochastic Neighbor Embedding (t-SNE) space (Fig. 1B). Only single-nuclei data from inhibitory medium spiny neurons (Inhibitory MSN*) and broad inhibitory neurons (Inhibitory*) were separated from the bulk inhibitory neurons in both hierarchical clustering of correlation and t-SNE space (Fig. 1A,B), consistent with prior analysis (Preissl et al. 2018). This separation could be due to the different tissues and methods used to isolate these cells (cortex vs. whole forebrain or sorting vs. single-nuclei) (Mo et al. 2015; Gray et al. 2017; Preissl et al. 2018). Overall, these results establish that the uniformly processed OCR profiles appropriately reflect cell-dependent biology.
Figure 1.
Mouse open chromatin profiles show expected relationships (A,B) and liftOver of mouse peaks to human is best done with summits (C–E). (A) Dendrogram displaying results of hierarchical clustering of the peak count correlations of public, mouse ATAC-seq data. Asterisks in the cell population name indicate single-nuclei data sets. (B) t-SNE plot displaying relationships between the peak counts of mouse cell populations. (C) Table containing the summary of three liftOver strategies applied to public mouse ATAC-seq data. (D) Mouse ATAC-seq data at the Wdr60 promoter region in the mouse genome (mm10; Chr 12: 116,258,915–116,264,450). As an example of the data at this locus, summits and peaks from Excitatory Layers II-III are displayed along with RefSeq transcripts. (E) Mouse-derived human open chromatin data at the WDR60 promoter region in the human genome (hg19; Chr 7: 158,626,991–158,682,822). As an example of data at this locus, data from Excitatory Layers II-III are displayed along with human RefSeq transcripts. Data include results from all three liftOver strategies employed (“All peaks”, “Strict peaks”, and “Summits”) along with the peak created after summit liftOver.
Converting OCR summits from mouse to human provides the most accurate open chromatin profile
We lifted over all mouse OCR profiles to syntenic sequences in the human genome (hg19). We compared three methods, seeking to retain the maximum number of peaks while ensuring human profiles resembled mouse profiles. First, peaks were lifted over “as is” including 250-bp extensions with default parameters (“all”). Second, peaks were lifted over “as is” with much stricter parameters, limiting gap sizes to 20 bp to match previous studies (“strict”) (Vierstra et al. 2014). Finally, we converted the single-bp summits with default parameters and added 250 bp on each side after conversion (“summit”).
The first method (“all”) resulted in retention of the most peaks from mouse to human (∼86%); however, the resulting peak size range (1–54,494 bp) was vastly different from the uniform input size of 501 bp (Fig. 1C). Further, ∼58% of lifted over peaks were >501 bp, with 55,658 peaks doubling in size (>1000 bp) (Fig. 1C). Our second strategy (“strict”) led to the conversion of ∼42% of peaks, with 2382 peaks doubling in size (Fig. 1C). Finally, the third strategy (“summits”) led to ∼75% of peaks being converted while controlling for size (Fig. 1C), resulting in mouse-derived human peaks that better represent mouse peaks. This is exemplified at the WDR60 promoter (Fig. 1D,E). In mouse, an open chromatin summit in Excitatory Layers II-III neurons is identified, creating a peak directly over the Wdr60 promoter (Fig. 1D). When lifted over as a peak, it expands from 501 bp to ∼13 kb (“All peaks”, Fig. 1E), and when controlling for gaps, the peak fails to lift over (“Strict peaks”, Fig. 1E). Neither result is representative of the regulatory landscape in mice. The single-bp summit lifts over and produces a 501-bp peak that encompasses the WDR60 promoter, accurately representing the mouse data (“Summits” and “Summit peaks”, Fig. 1E). Ultimately, converting summits proved the most robust method.
Mouse-derived human peaks serve as robust proxies for cognate human tissues
Next, we sought to compare our mouse-derived human OCR data to existing human open chromatin data. However, since most cell populations included in our study do not have orthologous human data by design, we compared profiles to imputed, tissue-level open chromatin data from the Roadmap Epigenomics Project (Ernst and Kellis 2015; Roadmap Epigenomics Consortium et al. 2015). We also included human T cell ATAC-seq profiles (Corces et al. 2016) for direct comparison to our mouse T cell profiles (CD4 and CD8).
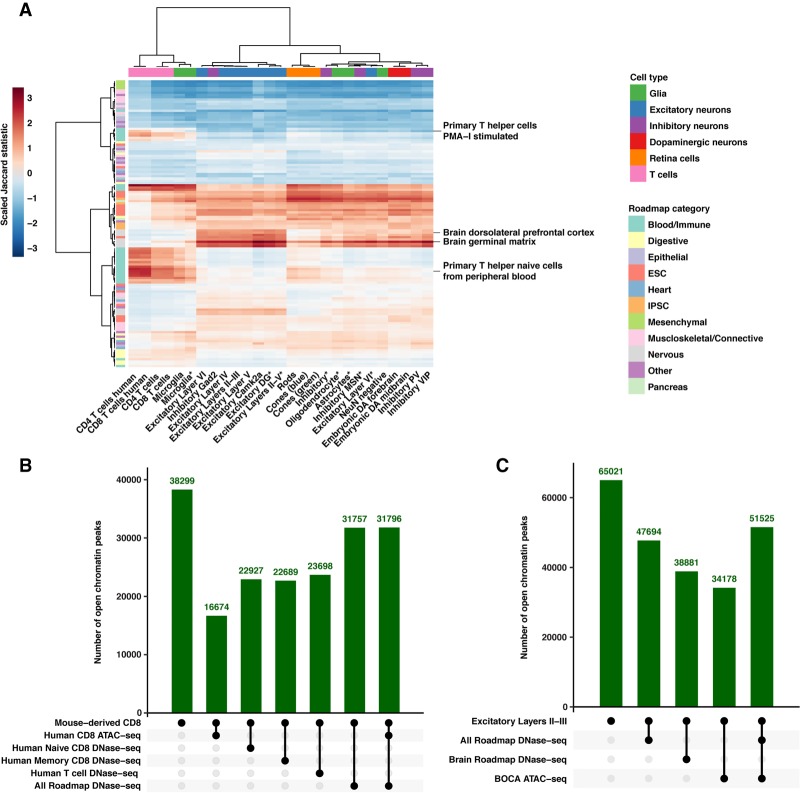
Through the use of pairwise Jaccard statistics scaled by ATAC-seq sample and hierarchical clustering, we observe immune ATAC-seq samples (T cells, microglia) cluster together and are most closely related to blood and immune tissues in Roadmap, especially “Primary T-cell” tissues (Fig. 2A). We also observe that all other ATAC-seq samples are most closely related to Roadmap brain-derived tissues, including “Brain Dorsolateral Prefrontal Cortex” and “Brain Germinal Matrix” (Fig. 2A). This general pattern holds when the comparisons are limited to Roadmap OCRs categorized as enhancers, promoters, or dyadic sequences (Supplemental Fig. S2A–C). Although this analysis is informative, the subtypes of cells (excitatory, inhibitory, etc.) are not perfectly grouped, potentially due to the use of tissue-level data or the use of imputed data. Adding to this, we compare mouse-derived human profiles to each other and observe that cell populations tend to be most related to other samples in their same category (Supplemental Fig. S3A–C). However, differences between orthologous T cell populations indicate that, although mouse-derived human OCR data can serve as good proxies for human data at the base pair level, they are imperfect (Supplemental Results).
Figure 2.
Mouse-derived human peaks serve as robust proxies for cognate human tissues. (A) A heatmap displaying the pairwise relationships between 27 mouse-derived ATAC-seq samples and imputed Roadmap Epigenomics Project open chromatin data from 127 tissues. Data displayed are the pairwise Jaccard statistic scaled by column (ATAC-seq sample). Data are hierarchical clustered by ATAC-sample and Roadmap tissue. Representative Roadmap tissues illustrating groups of immune and brain-derived tissues that are highlighted in the text are displayed. (IPSC) Induced pluripotent stem cells, (ESC) embryonic stem cell. (B) Plot displaying the intersection of mouse-derived CD8 T cell open chromatin peaks with publicly available human data sets. All numbers displayed are the number of mouse-derived peaks that meet the intersecting criteria below the plot. (C) Plot displaying the intersection of mouse-derived Excitatory Layers II-III open chromatin peaks with publicly available human data sets. All numbers displayed are the number of mouse-derived peaks that meet the intersecting criteria below the plot.
We further explored whether mouse-derived human peaks have regulatory potential in humans. We find 43.5% (16,674/38,299) (Fig. 2B) of mouse-derived CD8 ATAC-seq peaks overlap with human CD8 ATAC-seq peaks (40,916 peaks), slightly higher than previous studies (Vierstra et al. 2014). Using T cell Roadmap data, 60% (22,927/38,299) and 59% (22,689/38,299) overlap with naive (77,770 peaks) and memory CD8 T cell peaks (80,049 peaks) with a slight improvement (61%; 23,698/38,299) when combined (Fig. 2B) (90,267 peaks). Further, ∼83% overlap (31,757/38,299) with peaks found in any Roadmap tissue (493,894 total peaks) or the combination of Roadmap and ATAC-seq data (31,796/38,299) (Fig. 2B) (624,749 peaks). We observe similar results for mouse-derived CD4 T cells (Supplemental Table S3). We also evaluated how OCRs from other samples overlapped with human sequences, including brain-related Roadmap samples only (208,021 peaks) and ATAC-seq data from neurons from the Brain Open Chromatin Atlas (BOCA) (255,977 peaks) (Fullard et al. 2018). As with T cell data, all evaluated profiles including Excitatory Layers II-III (Fig. 2C) exhibit the highest overlap with combined data (Supplemental Table S4).
Overall, this data shows that mouse-derived human open chromatin profiles are most similar to tissues for which they would serve as proxies and that the vast majority of mouse-derived human peaks (average: 81%, range: 72%–92%) (Supplemental Table S4) show regulatory potential in at least one human tissue.
Mouse-derived human profiles recapitulate cell population disease enrichments and reveal new biology
We sought to determine whether mouse-derived OCR data could be used to inform cell-dependent heritability enrichment for common phenotypes. We employed S-LDSC using open chromatin data from 27 cell populations across 64 GWAS. We included open chromatin data from human T cells to allow for direct comparison to mouse-derived data. Traits studied included a selection of common neuropsychiatric, neurological, immunological, and behavioral traits, as well as traits from GWAS performed on UK Biobank data (Supplemental Table S5; The Brainstorm Consortium 2018; Bycroft et al. 2018).
To explore these enrichment patterns, we used hierarchical clustering of LDSC regression coefficient Z-scores. Z-scores greater than zero indicate that a cell population has increased heritability for a trait when accounting for the S-LDSC baseline model. This comparison revealed three clusters of samples and four clusters of phenotypes (Fig. 3A; Supplemental Table S6). Overall, S-LDSC results for all 64 GWAS established that mouse-derived human ATAC-seq profiles displayed increased heritability enrichment in cell populations consistent with the known biology and reveal new biological insights (see Supplemental Results).
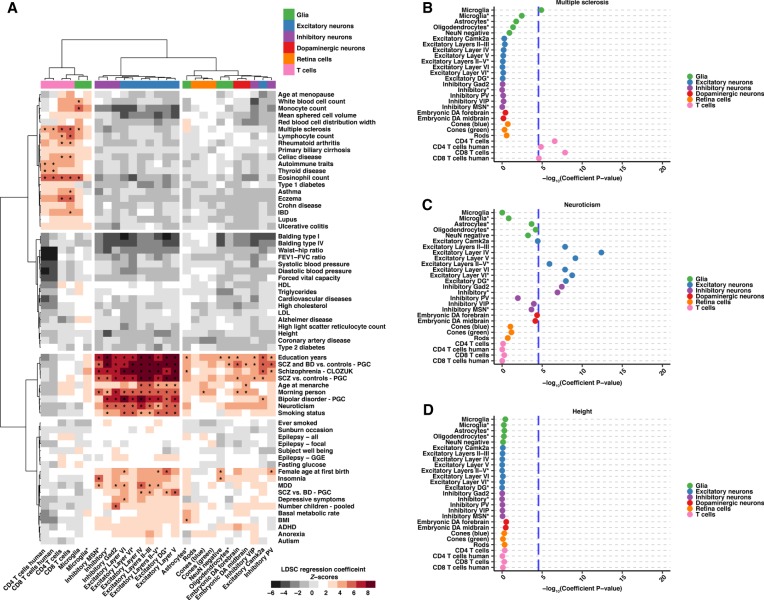
Figure 3.
S-LDSC results from 64 GWAS show heritability enrichment in expected cell populations and reveal further insight into disease. (A) A heatmap displaying regression coefficient Z-scores for 27 cell populations across 64 GWAS analyzed. Data are hierarchical clustered by GWAS and cell population. Cell populations that met the across trait significance level (Z-score = 4.02133, which is equivalent to a −log10[coefficient P-value] = 4.53857) are indicated with an asterisk. (B–D) Example dotplots displaying −log10(heritability coefficient P-values) S-LDSC results for GWAS indicative of the observed clustering groups: (B) multiple sclerosis, (C) neuroticism, and (D) height. Across trait significance levels are shown (−log10[coefficient P-value] = 4.53857; blue dashed line). Populations are colored and ordered by broader cell-type category. Asterisks in the cell population name indicate single-nuclei ATAC-seq data. All results can be found in Supplemental Table S6.
First, we observe that ATAC-seq immune cell samples (T cells and microglia) cluster together and demonstrate heritability enrichment for immune-related traits, including lupus, eczema, Crohn disease, and general autoimmune traits from the UK Biobank (Fig. 3A). We detect significant enrichment for many immune traits, including multiple sclerosis (Fig. 3A,B; Supplemental Fig. S4A). Many of these enrichments are consistent with prior analyses using human tissue (Finucane et al. 2018).
Next, we observe a collection of broadly defined inhibitory neurons, inhibitory MSNs, excitatory neurons in all cortical layers, and excitatory dentate gyrus (DG) neurons (Fig. 3A). This group shows consistently higher heritability enrichment for neuropsychiatric, neurological, and behavioral phenotypes with many showing significant heritability enrichment including neuroticism (Fig. 3A,C; Supplemental Fig. S4B).
Lastly, grouped together are a broadly defined collection of retinal (rods, cones) and nervous system populations (excitatory neurons, glial cells, inhibitory PV and VIP neurons, embryonic dopaminergic neurons) (Fig. 3A). Although not clustering with the second group of cells, the central nervous system-derived cell populations in this group show enrichment in neurological phenotypes (education years, bipolar disorder [BD], and SCZ) that also show enrichment in the second group of cells (Fig. 3A).
In contrast to the traits mentioned above, we observe no enrichment in a set of traits including blood pressure measurements, balding, and anatomical measurements (Fig. 3A). This is exemplified by height in which most cell populations show a negative Z-score (Supplemental Fig. S4C), and no populations reach significance for enrichment (Fig. 3D). It may be expected that traits like height, fasting glucose, and balding type I would not reveal significant enrichments in the cell populations we evaluate.
In order to explore how mouse-derived human data recapitulate observations in orthologous human data, we compared the Z-scores between human and mouse T cells. We observed that in both CD4 T cells (Spearman's rho = 0.6144) and CD8 T cells (Spearman's rho = 0.6832), the human and mouse-derived data show strong correlation (Supplemental Fig. S5A,B). These observations between T cells can be extended to the correlation between S-LDSC results for all populations, where we observe that samples from the same categories are highly correlated (Supplemental Fig. S5C). We also observe that S-LDSC results for related traits tend to be highly correlated, similar to what is seen in previous analyses (Supplemental Fig. S6; Watanabe et al. 2019).
Collectively, these results highlight that mouse-derived human profiles broadly recapitulate known biology across a wealth of human phenotypes and thus can serve as suitable proxies for orthologous human cell populations in this context.
Schizophrenia heritability is most enriched in cortical layer excitatory neurons
Having established the power of mouse-derived human profiles for studying common traits, we restricted our focus to schizophrenia. To facilitate comparison with transcription-based analyses (Skene et al. 2018), we used the recent CLOZUK SCZ GWAS (Pardiñas et al. 2018). Of 27 chromatin profiles, 13 achieved significance when corrected for all traits tested (Fig. 4A; Supplemental Table S6). Our analyses largely indict cortical neurons, with open chromatin profiles from both excitatory and inhibitory populations displaying significant enrichment (Fig. 4A; Supplemental Table S6).
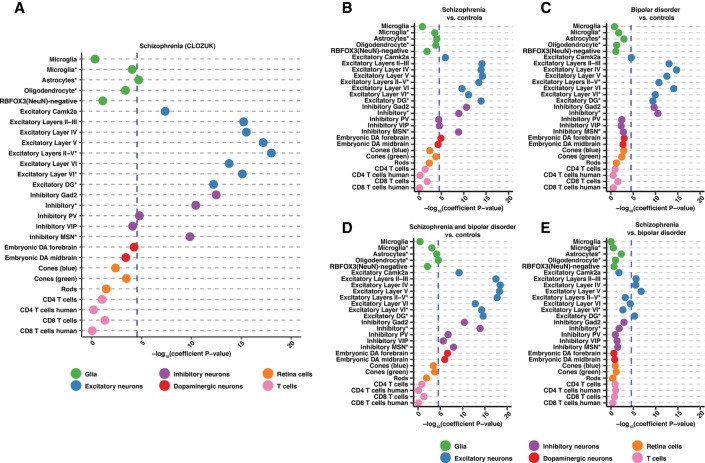
Figure 4.
S-LDSC results for CLOZUK and PGC schizophrenia studies as well as bipolar disorder GWAS reveal excitatory cortical neuron enrichment. (A–E) Dotplots displaying the −log10(heritability coefficient P-values) S-LDSC results for: (A) CLOZUK schizophrenia GWAS, (B) PGC schizophrenia GWAS, (C) PGC bipolar disorder GWAS, (D) schizophrenia and bipolar disorder GWAS, and (E) PGC schizophrenia versus bipolar disorder GWAS. Across trait significance levels are shown (−log10[coefficient P-values] = 4.53857; blue dashed line). Populations are colored and ordered by broader cell-type category. Asterisks in the cell population name indicate single-nuclei ATAC-seq data. All results can be found in Supplemental Table S6.
Within subsets of cortical excitatory neurons, we detect a progressive increase in enrichment when moving from layers II-III to layer V, reaching an apex with layer V OCRs, and then diminishing slightly in layer VI (Supplemental Fig. S7A; Supplemental Table S6). This pattern is mirrored in single-nuclei data wherein enrichment in layer III/IV/V cortical excitatory neurons (Excitatory Layers II-V*) exceeds that for layer VI cortical excitatory neurons (Excitatory Layer VI*) (Fig. 4A; Supplemental Fig. S7A; Supplemental Table S6). Significant enrichment for profiles derived from excitatory neurons of the dentate gyrus (Excitatory DG*) provides evidence of additional contribution arising from hippocampal excitatory neurons. The highest levels of enrichment in inhibitory neurons are seen in the broadly defined Gad2 GABAergic population with parvalbumin-positive neurons (Inhibitory PV) also reaching significance (Fig. 4A; Supplemental Table S6). We also detect significant enrichment in Drd1-positive medium spiny neurons (Inhibitory MSN*) and astrocytes (Fig. 4A; Supplemental Table S6).
In order to determine the extent to which the differences in heritability between correlated cell populations are a consequence of noise, we tested for SCZ enrichment in subsets of peaks from each annotation. We observe that distal elements in each annotation have a higher enrichment for SCZ heritability than peaks falling in promoter regions (Supplemental Fig. S8A,B; Supplemental Table S7). We also observe that annotations significantly enriched for SCZ (Fig. 4A) continue to show enrichment when peaks that overlap with peaks in the most enriched annotation are removed and when unique peaks in each annotation are tested (Supplemental Fig. S8A,B; Supplemental Table S7). Thus, although it is difficult to exclude the potential impact of noise, our analyses provide evidence that differences in heritability enrichment may reflect true independent signals.
Excitatory neurons in the cortex and hippocampus are enriched for differences between schizophrenia and bipolar disorder
Leveraging our success in analyzing SCZ, we set out to determine which cell populations may differentiate SCZ and BD. Although BD is related to SCZ and their genetics are highly correlated, they are unique disorders (The Brainstorm Consortium 2018). We took advantage of a recent study that not only performed traditional GWAS for SCZ and BD (affected vs. controls) but also performed GWAS for SCZ and BD compared to controls and SCZ compared to BD (Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium 2018). These comparisons allowed us to use S-LDSC to pinpoint what cell populations may be modulating disease differences.
The analysis of “SCZ versus controls” from this study showed similar results to the CLOZUK GWAS (Fig. 4B; Supplemental Fig. S7A,B; Supplemental Table S6; Supplemental Results), and the analysis of BD revealed enrichment in all excitatory neuron populations and broadly defined inhibitory neurons. The highest enrichment for BD was seen in individual excitatory layers (Fig. 4C; Supplemental Fig. S7C; Supplemental Table S6). In contrast to SCZ, subsets of cortical inhibitory neurons and inhibitory MSNs were not enriched (Fig. 4C; Supplemental Fig. S7C; Supplemental Table S6). Furthermore, the combined SCZ and BD cohort displayed significant enrichment in the same excitatory and inhibitory neurons as well as embryonic DA populations and oligodendrocytes (Fig. 4D; Supplemental Fig. S7D; Supplemental Table S6). Finally, we analyzed the SCZ versus BD cohort. Only four excitatory neuronal populations reach significance: cortical layers II-III, IV, V, and the dentate gyrus (Fig. 4E; Supplemental Fig. S7E; Supplemental Table S6). Overall, through the wealth of GWAS data available, we are able to begin to tease apart the complex relationship between SCZ and BD.
Statistical fine-mapping of 177 schizophrenia loci highlights novel complex biological hypotheses
Ultimately, our goal was to prioritize variants in SCZ GWAS loci. We incorporated significantly enriched open chromatin annotations into statistical fine-mapping of 177 independent schizophrenia loci using the fine-mapping program, PAINTOR (Kichaev et al. 2014, 2017; Kichaev and Pasaniuc 2015). SCZ loci were fine-mapped both with and without annotation and without specifying the number of causal SNPs in each locus. In total, 62,994 unique SNPs were fine-mapped with an average of 370 SNPs per locus (Supplemental Tables S8, S9). By using independent signals, not amalgamated loci, 2317 SNPs were fine-mapped in more than one locus (Supplemental Results; Supplemental Table S10; Supplemental Fig. S9).
When combining the results, 1512 SNPs in 166 loci reach a posterior inclusion probability (PIP) of ≥ 0.1, 82 SNPs in 56 loci reach a PIP ≥ 0.5, and 30 SNPs in 23 loci reach a PIP ≥ 0.9 (Supplemental Tables S8, S9). Although adding annotation to fine-mapping did not reduce the number of SNPs in 95% credible sets (Supplemental Fig. S10A), it did alter the PIP of many SNPs (Supplemental Fig. S10B), with 411 SNPs only reaching a PIP ≥ 0.1 when annotation is incorporated (Supplemental Fig. S10C). We then explored how all variants with a PIP ≥ 0.1 impact OCRs in SCZ-enriched cell populations; this cutoff was previously reported to provide a high benefit-to-cost ratio for follow-up experiments (Kichaev et al. 2014). Across 104 loci, 281 unique SNPs achieve a PIP ≥ 0.1 and overlap with an OCR present in at least one SCZ-enriched cell population (Supplemental Table S11). These SNPs are prime candidates for functional SNPs within these loci.
Whereas a myriad of hypotheses can be generated from an overlap of SNPs with OCRs, we sought to use additional functional data to annotate SNPs. These data included transcription factor (TF) binding motifs (Kulakovskiy et al. 2018), promoter capture Hi-C (PCHi-C) interactions (Song et al. 2019), and validated enhancers from the VISTA enhancer database (Visel et al. 2007). HOCOMOCO TF motif data predicts 163 SNPs across 88 loci disrupt transcription factor binding sites (Supplemental Table S12), most commonly impacting ARID3A, FOXJ3, and MAZ motifs (Supplemental Table S13). No TF motifs were significantly overrepresented for being disrupted by SNPs (exact binomial test) (Supplemental Table S13; Supplemental Methods). Across 43 loci, 113 SNPs fall into significant promoter interactions in neural, induced pluripotent stem cell (IPSC)-derived cell populations (Supplemental Tables S8, S14), with 59 SNPs also disrupting a TF binding motif (Supplemental Table S8). Two SNPs in two loci fell in a validated regulatory element from the VISTA enhancer database (Supplemental Table S15), with one of those SNPs disrupting a TF binding motif. The totality of these data allowed us to construct hypotheses for many SNPs and loci. We describe an example below.
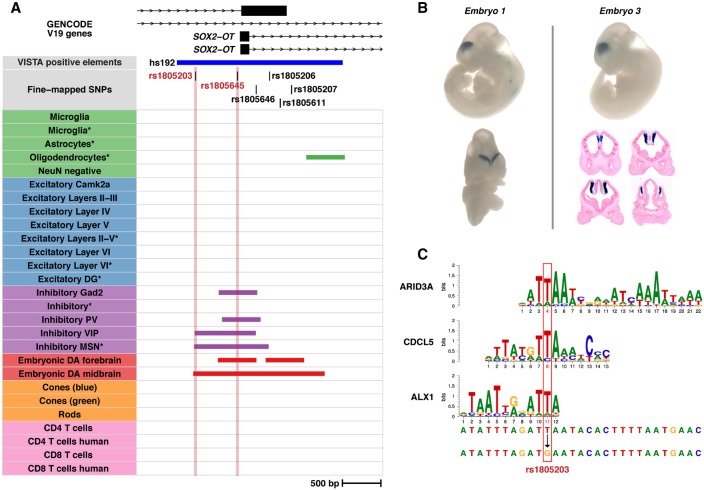
Two SNPs (rs1805203 and rs1805645) fell within the VISTA positive element, hs192, which encompasses the promoter region of the SOX2-OT gene transcript designated as SOX2DOT (Fig. 5A; Supplemental Table S15; Amaral et al. 2009; Shahryari et al. 2015). Hs192 drives strong LacZ expression in the forebrain of embryonic mice, specifically in the ventricular zone of medial pallium (Fig. 5B; Visel et al. 2013). rs1805203 falls in the locus tagged by rs55672338 which contains 210 fine-mapped SNPs, of which eight achieve a PIP ≥ 0.1 (Supplemental Table S8). Of those eight SNPs, rs1804203 is the only SNP that overlaps an OCR in a SCZ-enriched population and disrupts a TF binding motif. rs1805203 resides in open chromatin found in inhibitory VIP neurons, inhibitory MSNs, and embryonic DA neurons (Fig. 5A; Supplemental Table S11) and strongly disrupts ARID3A, CDCL5, and ALX1 binding sites (Fig. 5C; Supplemental Table S12). rs1805645 falls in the locus tagged by rs34796896 which contains 365 fine-mapped SNPs, wherein rs1805645 is the only SNP that lies within an OCR for a SCZ-enriched population (Supplemental Table S8). rs1805645 intersects inhibitory Gad2, inhibitory PV, inhibitory VIP, as well as embryonic DA neurons in the forebrain and midbrain (Fig. 5A; Supplemental Table S11). These two SNPs are in low LD (r2 ∼ 0.0127 in the 1000 Genomes European population). SOX2-OT has been shown to repress the expression of SOX2, which plays roles in development, in maintaining pluripotent stem cell populations, and in neural differentiation in the cortex especially impacting GABAergic neurons (Ferri et al. 2004; Cavallaro et al. 2008; Zhang and Cui 2014; Shahryari et al. 2015; Knauss et al. 2018; Messemaker et al. 2018; Mercurio et al. 2019). This leads us to construct a hypothesis where both SNPs (rs1805203 and rs1805645) impact regulatory DNA sequences in inhibitory GABAergic neurons leading to aberrant SOX2-OT expression and eventually to perturbation of SOX2 expression. We expand upon our observations at two additional loci, containing the CHRNA2 and NGEF genes, in the Supplemental Results (Supplemental Figs. S11–S13).
Figure 5.
Fine-mapping prioritizes SNPs in the schizophrenia-associated locus surrounding the SOX2-OT gene. (A) A visualization of two prioritized SNPs (rs1805203 and rs1805645) in the promoter region of a SOX2-OT transcript promoter. The plot displays transcripts, the prioritized SNPs, a VISTA positive element (hs192), and the mouse-derived human peaks from all ATAC-seq samples. The prioritized SNPs are highlighted. (B) Representative VISTA mouse embryo LacZ staining data for the element hs192. Downloaded from VISTA database (https://enhancer.lbl.gov/). (C) Transcription factor binding motifs derived from HOCOMOCO v10 data that are disrupted by rs1805203. The nucleotides impacted are highlighted in red.
Discussion
Despite the capacity of GWAS to inform genetic architecture, connecting variants to disease mechanisms remains a challenge. This challenge is stark in SCZ, where 179 independent loci implicate thousands of noncoding variants, without providing an informed strategy to construct hypotheses. We demonstrate the power of obtaining cellular surrogates from mice which may not be readily obtained from humans, highlighting their application to inform human trait heritability and functional dissection. Our results reinforce mice as a lens through which to study the genetics underlying common human phenotypes (Cusanovich et al. 2018; Hook et al. 2018; McClymont et al. 2018).
Our study confirms the contribution of cortical and interneuron populations in neuropsychiatric disorders, establishing that OCR signatures from cortical excitatory and inhibitory populations are most enriched for SCZ heritability. We demonstrate that enrichment of heritability for SCZ in excitatory neurons is not uniform and is maximal in discrete layer V excitatory neurons and single-nuclei populations containing layer V neurons. This is consistent with prior data implicating medium spiny neurons, all layers of cortical excitatory neurons, cortical inhibitory neurons, as well as hippocampal CA1 excitatory neurons (Skene et al. 2018). We find enrichment in all but hippocampal CA1 excitatory neurons, for which we did not have data. However, we observe enrichment in excitatory neurons derived from the dentate gyrus, which mirror significant SCZ enrichment seen in mouse dentate granule cells (Skene et al. 2018).
Our data also illuminate cell-dependent differences and similarities between SCZ and BD. We find that cortical excitatory neurons and excitatory DG neurons harbor heritability enrichment for the difference between SCZ and BD. This provides support for work that has shown layer-specific neuronal differences between SCZ and BD (Benes et al. 2001; Rajkowska et al. 2001; Chana et al. 2003) as well as differences in DG neuronal maturation in these diseases (Yu et al. 2014). We also observe that inhibitory MSNs consistently show enrichment in SCZ but not BD, pointing toward potentially important biological differences. However, in analyzing the “SCZ versus BD” association data, enrichment for MSN fails to reach significance (Fig. 4). This observation perhaps highlights that the estimation of heritability enrichment is inherently dependent upon annotation size and trait heritability (Hormozdiari et al. 2018), both of which vary across our study.
Although powerful, detecting enrichment remains dependent on the availability of relevant data sets including cell types, developmental stages, or physiological states. We recognize that conclusions cannot be drawn about the relevance of any cell type that was not tested here. Additionally, open chromatin profiles lack the biological interpretation provided by histone marks when trying to identify functional regulatory DNA as enhancers, promoters, or insulators (Nord and West 2019). We anticipate the increasing resolution, quality, and completeness of histone data over time in both human and mouse will allow for further functional delineation of OCRs.
We also prioritize SCZ variants through the fine-mapping of variants using SCZ-enriched open chromatin profiles. We identify SNPs in 104/177 tested loci (∼59%) that may now be considered prime candidates. By using functional data to annotate these candidates, we are able to construct straightforward (SOX2-OT locus) and complex (CHRNA2 and NGEF loci) hypotheses. We note that both our S-LDSC and fine-mapping analyses focus on common SNPs assayed in GWAS. We, therefore, cannot make any conclusions about SNPs not assayed in GWAS or about the contribution of rare variation to SCZ. Further, more complex variation has been shown to be important in SCZ loci (Sekar et al. 2016; Song et al. 2018) and is not assayed here.
Further, our results rely on converting mouse data to the human sequence. Although we optimize identification of orthologous human sequences and demonstrate that the vast majority of mouse peaks have a human syntenic ortholog, mouse-derived OCR data incompletely represent the human OCR spectra they aim to inform. This is highlighted by the strong but imperfect correlation of S-LDSC results between orthologous T cell open chromatin data. The observed differences may reflect the different locations from which T cells were collected (mouse T cells, thymus; human T cells, bone marrow/peripheral blood). They may also reflect the power resulting from more homogeneous and less challenged immune cell populations that may be obtained from laboratory mice or truly different regulatory profiles in the different organisms. Although any limitation this presents is difficult to quantify in cells without orthologous data, an average of 81% of mouse-derived human peaks are OCRs in human tissue, and they recapitulate heritability enrichment results for a variety of phenotypes. This gives us confidence in our approach; however, efforts to obtain single-nuclei data from human brain samples may provide clarity by making future human data sets a possibility (Lake et al. 2018).
Overall, our data define immediately testable hypotheses implicating specific variants as potentially modulating the activity of cis-regulatory elements in discrete cellular contexts in SCZ. The capacity to move directly from GWAS to the design of functional tests by using mouse-derived data represents a significant step forward in the dissection of common human phenotypes.
Methods
Obtaining ATAC-seq data
Raw ATAC-seq sequencing data were obtained from the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) except for single-nuclei ATAC-seq data (Preissl et al. 2018), which was obtained from the author's website. All details regarding downloaded sequencing data can be found in Supplemental Table S1. Additional steps were needed in order to aggregate ATAC-seq reads from individual nuclei into clusters using BBMap (https://sourceforge.net/projects/bbmap/) (see Supplemental Methods).
Alignment and peak calling
Paired-end reads were aligned to the mouse genome (mm10/GRCm38) using Bowtie 2 (version 2.2.5; http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) (Langmead and Salzberg 2012) with the following parameters: “-p 15 ‐‐local -X2000”. Paired-end reads aligning to the mitochondrial genome as well as random and unknown chromosomes were removed. SAMtools (Li et al. 2009) was used to remove duplicate reads (v0.1.9), improperly paired reads (v1.3.1), and reads with a mapping quality score of ≤30 (v1.3.1).
Replicates for each cell population were merged into a single BAM file, and peak summits were called for each mouse cell population (25 in total) using the MACS2 (v. 2.1.1.20160309) (Zhang et al. 2008) “macs2 callpeak” function with the following parameters: “‐‐seed 24 ‐‐nomodel ‐‐nolambda ‐‐call-summits ‐‐shift -100 ‐‐extsize 200 ‐‐keep-dup all ‐‐gsize mm”. Peaks overlapping with blacklisted regions in the mm10 genome (Amemiya et al. 2019) were removed prior to further analysis (Supplemental Methods). ATAC-seq data from CD4 and CD8 T cells were also obtained (Corces et al. 2016), aligned to hg19, and processed as above (Supplemental Table S1).
Since we were comparing ATAC-seq data sets with vastly different sequencing depths and numbers of called summits, we applied a recently introduced filtering strategy for ATAC-seq peaks (Corces et al. 2018). For each data set, we summed the MACS2 peak scores and divided that number by one million (total score per million). We then divided each individual peak score by the total score per million for that data set to produce a “score per million” (Corces et al. 2018). Ultimately we chose a “score per million” cutoff of two as that would equate to a P-value per million of 0.01.
Relationship between public mouse sets
Summits called in each population were made into uniform 501-bp peaks by adding 250 bp to each side of the summit. Peaks were then merged into a union set of peaks using BEDTools “merge” with default parameters. This final set of filtered and merged peaks contained a total of 433,555 peaks.
In order to obtain a count matrix for cell population comparison, featureCounts (v1.6.1) was used (Liao et al. 2014). The command “featureCounts” was used with the “-T 10 -F SAF” parameters in order to obtain a count matrix. BEDTools “nuc” was used with a FASTA file of combined mm10 chromosome sequences obtained from the UCSC Genome Browser (http://hgdownload.cse.ucsc.edu/goldenPath/mm10/chromosomes/) in order to calculate GC content for each peak.
The count matrix, the count matrix summary file, and the peak GC content file were read into the R statistical environment (R Core Team 2019). Data were transformed into log2(count + 1) counts, and the CQN R package (Hansen et al. 2012) and ComBat from the SVA R package (Leek et al. 2012) were used to quantile normalize counts and correct CQN normalized counts for the type of experiment (single-nuclei or bulk). Principal component analysis was performed using all peak counts with the R function “prcomp()” with default settings and “scale. = TRUE” setting. t-SNE was performed with the first six principal components from PCA using the “tsne” package (https://github.com/jdonaldson/rtsne) with the “tsne()” function with the following parameters: “perplexity = 5 max_iter = 10000 whiten = T”. Additionally, the Pearson's correlation between corrected peak counts was used to cluster the data. Correlations were converted to distances by subtracting the absolute value of the correlations from 1. Clustering was performed using the R function “hclust” with “method = ‘ward.D2’” and figures were produced with custom R scripts.
liftOver
All strategies used the liftOver script “bnMapper.py” from the bx-python software package (Denas et al. 2015) (https://github.com/bxlab/bx-python) along with the “reciprocal best” mm10 to hg19 chain file (mm10.hg19.rbest.chain.gz) from the UCSC Genome Browser (https://hgdownload-test.gi.ucsc.edu/goldenPath/hg19/vsMm10/reciprocalBest/). Three liftOver strategies were compared: one using the called summits and two using the uniform, unmerged 501-bp peaks. The first strategy lifted over the single-bp summit sets with the settings: “-f BED12” and added 250 bp to each side. The second strategy lifted over the 501-bp peak sets again with the settings: “-f BED12”. The third strategy again used the 501-bp peaks with the settings: “-f BED12 -g 20 -t 0.1”. This third strategy has been employed previously (Vierstra et al. 2014). Note that, for this comparison, peaks were not merged and no regions were removed. Ultimately, the liftOver of the peak summits was used for all subsequent analyses. After liftOver of the peak summits to hg19, 250 bp were added on to both sides of each summit to create peaks. For this final set, overlapping peaks for each annotation were merged using BEDTools “merge” with default parameters. Peaks overlapping with regions that are blacklisted in the hg19 genome were removed (Supplemental Methods; Amemiya et al. 2019).
Comparisons to publicly available human open chromatin data
Human open chromatin profiles derived from mouse data were compared to imputed Roadmap Epigenetic Project DNase I hypersensitivity data from 127 human tissues and cell populations (Ernst and Kellis 2015; Roadmap Epigenomics Consortium et al. 2015) and ATAC-seq data from neurons isolated from 14 human brain regions (Fullard et al. 2018) (https://bendlj01.u.hpc.mssm.edu/multireg/resources/boca_peaks.zip).
All imputed Roadmap DNase peaks as well as imputed Roadmap peaks designated as enhancers, promoters, or dyadic sequences were downloaded (Supplemental Methods). Custom scripts adapted from Aaron Quinlan (http://quinlanlab.org/tutorials/bedtools/bedtools.html#a-jaccard-statistic-for-all-400-pairwise-comparisons) using GNU parallel (https://zenodo.org/record/1146014) and BEDTools (v2.27.0 “jaccard”) (Quinlan and Hall 2010) were used to calculate pairwise comparisons between samples. Custom R scripts were used to produce heatmaps with the R package “pheatmap” (https://CRAN.R-project.org/package=pheatmap).
In order to explore how many lifted over peaks have been shown to have regulatory potential in human tissues, comparisons between ATAC-seq samples, Roadmap open chromatin, and BOCA open chromatin were made using the BEDTools “jaccard” command with default parameters in order to calculate overlaps (Supplemental Table S4).
Partitioning heritability with linkage disequilibrium score regression
Summary statistics for 64 GWAS were obtained from a variety of sources in either “raw” or preprocessed forms. “Raw” GWAS summary statistics were downloaded and processed using the “munge_sumstats.py” script (LDSC v1.0.0). Data sources and specific command parameters used to process the data are listed in Supplemental Table S5. Note, processed summary statistics from the CLOZUK SCZ GWAS (Pardiñas et al. 2018) needed minor modifications after processing, and the GWAS summary statistics for Alzheimer disease (Marioni et al. 2018) have been modified since analysis (see Marioni et al. 2018 and Supplemental Table S5). Annotation files and LD score files needed for analysis were created using the “make_annot.py” and “ldsc.py” scripts included in the LDSC software using standard parameters. The source for software and data downloaded to run S-LDSC can be found in Supplemental Table S16. Each ATAC-seq sample was added onto the baseline model and heritability enrichment was calculated individually (also referred to as S-LDSC). The analysis was performed with the “ldsc.py” script using standard parameters with the “‐‐h2” flag.
Results for each phenotype were aggregated, and the P-values for each annotation were calculated in R. The P-values for regression Z-scores are based on a one-sided test for the regression coefficient being greater than 0, so the P-values for each annotation were calculated using the regression coefficient Z-scores and the “pnorm” function with the following parameters: “lower.tail = FALSE”. For more information, see LDSC publications (Finucane et al. 2015, 2018) and the LDSC website (https://github.com/bulik/ldsc). Partitioned heritability calculations for all traits were combined and analyzed in R. Plots were created using custom R scripts. The level of significance was set for LDSC results as the Bonferroni corrected P-value when taking into account all summary statistics and cell populations tested (0.05/[27*64] = 0.00002894; −log10[P] = 4.53857). For more details and a description of peak subset analyses, see Supplemental Methods.
Fine-mapping SNPs in schizophrenia loci
Finding proxy SNPs
A total of 179 genome-wide significant, independent index SNPs were extracted from the CLOZUK SCZ GWAS (Pardiñas et al. 2018). The function “get_proxies()” from the R package “proxysnps” (https://github.com/slowkow/proxysnps) was used with the following parameters: “window_size = 2e6 pop = “EUR””. This was used with the 1000 Genomes reference VCF processed by BEAGLE (Browning et al. 2018) (http://bochet.gcc.biostat.washington.edu/beagle/1000_Genomes_phase3_v5a/). Only SNPs with an r2 ≥ 0.1 with an index SNP and a minor allele frequency (MAF) ≥ 1% were retained for fine-mapping. This method obtained 71,344 unique SNPs with reference SNP (RS) numbers across 177 loci (see Supplemental Methods; Supplemental Table S17).
File setup for fine-mapping
We fine-mapped all 177 SCZ loci using PAINTOR (v3.1) (Kichaev et al. 2014, 2017; Kichaev and Pasaniuc 2015) (https://github.com/gkichaev/PAINTOR_V3.0). PAINTOR was chosen for its ability to use summary statistics, run simulations on multiple loci at once, and incorporate chromatin annotation data. Proxy SNPs were merged with summary statistics from the CLOZUK SCZ GWAS, leaving 62,994 unique SNPs across 177 loci. The number of SNPs in each locus ranged from seven to 1919 (Supplemental Table S18). These loci were used to create both the LD and annotation files needed to run PAINTOR using the “CalcLD_1KG_VCF.py” and “AnnotateLocus.py” scripts (see Supplemental Methods). As suggested by the PAINTOR authors, the correlations between annotations found to be significant in LDSC were calculated using custom R scripts. All significant annotations had a Pearson's correlation ≥ 0.2 (the cutoff suggested by the authors), so all annotations were merged before running “AnnotateLocus.py”.
Running PAINTOR fine-mapping
Estimated enrichments for the baseline model and the annotation model (see Supplemental Methods) were used as input to PAINTOR Monte Carlo Markov Chain (MCMC) simulations with the following key parameters: “-mcmc -burn_in 100000 -max_ samples 1000000 -num_chains 5 -set_seed 3 -MI 1”. Fine-mapping was run both with and without merged annotations with the parameter for supplying enrichment estimates set at “-gamma_intial 3.79521” for the no annotation simulation and “-gamma_initial 3.79521, -0.939523” set for the simulation including annotation. The number of samples used for “-burn_in” and “-max_samples” parameters were chosen based on parameters set for MCMC fine-mapping with other methods (Banerjee et al. 2018). Visualizations of fine-mapping results and loci were created with custom R scripts and the R package “gviz” (v1.28.3) (Hahne and Ivanek 2016).
Functional annotation of fine-mapped SNPs
In order to explore the functional impact of fine-mapped SNPs on TF motifs, the R package motifbreakR was used (Coetzee et al. 2015) along with TF motifs as defined by the HOCOMOCO v10 database (Kulakovskiy et al. 2018). The overrepresentation of TF motifs disrupted by SNPs was tested in R. Results can be found in Supplemental Table S13. SNPs were additionally annotated using data from the VISTA enhancer browser (Visel et al. 2007) and significant PCHI-C interactions in excitatory neurons, motor neurons, and hippocampal dentate gyrus-like neurons derived from IPSCs as well as primary astrocytes (Song et al. 2019). Details for these analyses can be found in the Supplemental Methods.
Genome assembly versions
Mouse data were aligned to the most recent major mouse genome assembly version (mm10/GRCm38). Mouse data were lifted over to the human hg19 (GRCh37) assembly mainly due to the use of that assembly in the publicly available GWAS summary statistics analyzed. Additionally, the tools used in the paper (primarily S-LDSC) are configured for use with hg19, the publicly available open chromatin peaks (Roadmap, BOCA) were published in hg19, and the published annotations used (PCHI-C and VISTA enhancers) are in hg19. We do not anticipate that the use of GRCh38 (hg38) would significantly impact our results due to our removal of regions flagged in genomic blacklists. These blacklists have been shown to remove regions in hg19 that have assembly issues, including gaps and regions that were fixed in more recent assemblies (Amemiya et al. 2019).
Data access
The sources for publicly available ATAC-seq data can be found in Supplemental Table S1 and are described in the Methods. Documentation of code is available on GitHub (https://github.com/pwh124/open_chromatin) and in Supplemental Code 1. Access to data including peaks and all files for heritability enrichment analyses and fine-mapping is available via Zenodo (https://doi.org/10.5281/zenodo.3253180).
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank Sebastian Preissl, David U. Gorkin, and Rongxin Fang for providing the information necessary to process single-nuclei ATAC-seq data. This research, undertaken at Johns Hopkins University School of Medicine, was supported in part by awards from the National Institutes of Health (NS62972 and MH106522) to A.S.M.
Author contributions: P.W.H. and A.S.M. designed the study and wrote the manuscript. P.W.H. implemented the computational algorithms to process the raw data and conduct analyses. P.W.H. and A.S.M. analyzed and interpreted the resulting data. P.W.H. contributed novel computational pipeline development.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.256578.119.
References
- Amaral PP, Neyt C, Wilkins SJ, Askarian-Amiri ME, Sunkin SM, Perkins AC, Mattick JS. 2009. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA 15: 2013–2027. 10.1261/rna.1705309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya HM, Kundaje A, Boyle AP. 2019. The ENCODE blacklist: identification of problematic regions of the genome. Sci Rep 9: 9354 10.1038/s41598-019-45839-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Zeng L, Schunkert H, Söding J. 2018. Bayesian multiple logistic regression for case-control GWAS. PLoS Genet 14: e1007856 10.1371/journal.pgen.1007856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes FM, Vincent SL, Todtenkopf M. 2001. The density of pyramidal and nonpyramidal neurons in anterior cingulate cortex of schizophrenic and bipolar subjects. Biol Psychiatry 50: 395–406. 10.1016/S0006-3223(01)01084-8 [DOI] [PubMed] [Google Scholar]
- Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium. 2018. Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell 173: 1705–1715.e16. 10.1016/j.cell.2018.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Brainstorm Consortium. 2018. Analysis of shared heritability in common disorders of the brain. Science 360: eaap8757 10.1126/science.aap8757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Zhou Y, Browning SR. 2018. A one-penny imputed genome from next-generation reference panels. Am J Hum Genet 103: 338–348. 10.1016/j.ajhg.2018.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O'Connell J, et al. 2018. The UK Biobank resource with deep phenotyping and genomic data. Nature 562: 203–209. 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallaro M, Mariani J, Lancini C, Latorre E, Caccia R, Gullo F, Valotta M, DeBiasi S, Spinardi L, Ronchi A, et al. 2008. Impaired generation of mature neurons by neural stem cells from hypomorphic Sox2 mutants. Development 135: 541–557. 10.1242/dev.010801 [DOI] [PubMed] [Google Scholar]
- Chana G, Landau S, Beasley C, Everall IP, Cotter D. 2003. Two-dimensional assessment of cytoarchitecture in the anterior cingulate cortex in major depressive disorder, bipolar disorder, and schizophrenia: evidence for decreased neuronal somal size and increased neuronal density. Biol Psychiatry 53: 1086–1098. 10.1016/S0006-3223(03)00114-8 [DOI] [PubMed] [Google Scholar]
- Coetzee SG, Coetzee GA, Hazelett DJ. 2015. motifbreakR: an R/Bioconductor package for predicting variant effects at transcription factor binding sites. Bioinformatics 31: 3847–3849. 10.1093/bioinformatics/btv470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Greenleaf WJ, et al. 2016. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 48: 1193–1203. 10.1038/ng.3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Granja JM, Shams S, Louie BH, Seoane JA, Zhou W, Silva TC, Groeneveld C, Wong CK, Cho SW, et al. 2018. The chromatin accessibility landscape of primary human cancers. Science 362: eaav1898 10.1126/science.aav1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, Filippova GN, Huang X, Christiansen L, DeWitt WS, et al. 2018. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174: 1309–1324. e18. 10.1016/j.cell.2018.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denas O, Sandstrom R, Cheng Y, Beal K, Herrero J, Hardison RC, Taylor J. 2015. Genome-wide comparative analysis reveals human-mouse regulatory landscape and evolution. BMC Genomics 16: 87 10.1186/s12864-015-1245-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2015. Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat Biotechnol 33: 364–376. 10.1038/nbt.3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri ALM, Cavallaro M, Braida D, Di Cristofano A, Canta A, Vezzani A, Ottolenghi S, Pandolfi PP, Sala M, DeBiasi S, et al. 2004. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development 131: 3805–3819. 10.1242/dev.01204 [DOI] [PubMed] [Google Scholar]
- Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh P-R, Anttila V, Xu H, Zang C, Farh K, et al. 2015. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 47: 1228–1235. 10.1038/ng.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane HK, Reshef YA, Anttila V, Slowikowski K, Gusev A, Byrnes A, Gazal S, Loh P-R, Lareau C, Shoresh N, et al. 2018. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet 50: 621–629. 10.1038/s41588-018-0081-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullard JF, Hauberg ME, Bendl J, Egervari G, Cirnaru M-D, Reach SM, Motl J, Ehrlich ME, Hurd YL, Roussos P. 2018. An atlas of chromatin accessibility in the adult human brain. Genome Res 28: 1243–1252. 10.1101/gr.232488.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray LT, Yao Z, Nguyen TN, Kim TK, Zeng H, Tasic B. 2017. Layer-specific chromatin accessibility landscapes reveal regulatory networks in adult mouse visual cortex. eLife 6: e21883 10.7554/eLife.21883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne F, Ivanek R. 2016. Visualizing genomic data using Gviz and Bioconductor. Methods Mol Biol 1418: 335–351. 10.1007/978-1-4939-3578-9_16 [DOI] [PubMed] [Google Scholar]
- Hansen KD, Irizarry RA, Wu Z. 2012. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics 13: 204–216. 10.1093/biostatistics/kxr054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook PW, McClymont SA, Cannon GH, Law WD, Morton AJ, Goff LA, McCallion AS. 2018. Single-cell RNA-Seq of mouse dopaminergic neurons informs candidate gene selection for sporadic Parkinson disease. Am J Hum Genet 102: 427–446. 10.1016/j.ajhg.2018.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F, Gazal S, van de Geijn B, Finucane HK, Ju CJ-T, Loh P-R, Schoech A, Reshef Y, Liu X, O'Connor L, et al. 2018. Leveraging molecular quantitative trait loci to understand the genetic architecture of diseases and complex traits. Nat Genet 50: 1041–1047. 10.1038/s41588-018-0148-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya T, D'Oliveira Albanus R, Hensley J, Myers G, Kyono Y, Kitzman J, Parker SCJ, Engel JD. 2018. Global dynamics of stage-specific transcription factor binding during thymocyte development. Sci Rep 8: 5605 10.1038/s41598-018-23774-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AEO, Enright JM, Myers CA, Shen SQ, Corbo JC. 2017. Cell type-specific epigenomic analysis reveals a uniquely closed chromatin architecture in mouse rod photoreceptors. Sci Rep 7: 43184 10.1038/srep43184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichaev G, Pasaniuc B. 2015. Leveraging functional-annotation data in trans-ethnic fine-mapping studies. Am J Hum Genet 97: 260–271. 10.1016/j.ajhg.2015.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichaev G, Yang W-Y, Lindstrom S, Hormozdiari F, Eskin E, Price AL, Kraft P, Pasaniuc B. 2014. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet 10: e1004722 10.1371/journal.pgen.1004722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichaev G, Roytman M, Johnson R, Eskin E, Lindström S, Kraft P, Pasaniuc B. 2017. Improved methods for multi-trait fine mapping of pleiotropic risk loci. Bioinformatics 33: 248–255. 10.1093/bioinformatics/btw615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauss JL, Miao N, Kim S-N, Nie Y, Shi Y, Wu T, Pinto HB, Donohoe ME, Sun T. 2018. Long noncoding RNA Sox2ot and transcription factor YY1 co-regulate the differentiation of cortical neural progenitors by repressing Sox2. Cell Death Dis 9: 799 10.1038/s41419-018-0840-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakovskiy IV, Vorontsov IE, Yevshin IS, Sharipov RN, Fedorova AD, Rumynskiy EI, Medvedeva YA, Magana-Mora A, Bajic VB, Papatsenko DA, et al. 2018. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res 46: D252–D259. 10.1093/nar/gkx1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake BB, Chen S, Sos BC, Fan J, Kaeser GE, Yung YC, Duong TE, Gao D, Chun J, Kharchenko PV, et al. 2018. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat Biotechnol 36: 70–80. 10.1038/nbt.4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. 2012. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28: 882–883. 10.1093/bioinformatics/bts034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Marioni RE, Harris SE, Zhang Q, McRae AF, Hagenaars SP, Hill WD, Davies G, Ritchie CW, Gale CR, Starr JM, et al. 2018. GWAS on family history of Alzheimer's disease. Transl Psychiatry 8: 99 10.1038/s41398-018-0150-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, Ben-Yehuda H, David E, Zelada González F, Perrin P, et al. 2016. Microglia development follows a stepwise program to regulate brain homeostasis. Science 353: aad8670 10.1126/science.aad8670 [DOI] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337: 1190–1195. 10.1126/science.1222794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClymont SA, Hook PW, Soto AI, Reed X, Law WD, Kerans SJ, Waite EL, Briceno NJ, Thole JF, Heckman MG, et al. 2018. Parkinson-associated SNCA enhancer variants revealed by open chromatin in mouse dopamine neurons. Am J Hum Genet 103: 874–892. 10.1016/j.ajhg.2018.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercurio S, Serra L, Nicolis SK. 2019. More than just stem cells: functional roles of the transcription factor Sox2 in differentiated glia and neurons. Int J Mol Sci 20: 4540 10.3390/ijms20184540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messemaker TC, van Leeuwen SM, van den Berg PR, ‘t Jong AEJ, Palstra R-J, Hoeben RC, Semrau S, Mikkers HMM. 2018. Allele-specific repression of Sox2 through the long non-coding RNA Sox2ot. Sci Rep 8: 386 10.1038/s41598-017-18649-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, et al. 2015. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86: 1369–1384. 10.1016/j.neuron.2015.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord AS, West AE. 2019. Neurobiological functions of transcriptional enhancers. Nat Neurosci 23: 5–14. 10.1038/s41593-019-0538-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, et al. 2008. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet 40: 1053–1055. 10.1038/ng.201 [DOI] [PubMed] [Google Scholar]
- Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, Legge SE, Bishop S, Cameron D, Hamshere ML, et al. 2018. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet 50: 381–389. 10.1038/s41588-018-0059-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praetorius C, Grill C, Stacey SN, Metcalf AM, Gorkin DU, Robinson KC, Van Otterloo E, Kim RSQ, Bergsteinsdottir K, Ogmundsdottir MH, et al. 2013. A polymorphism in IRF4 affects human pigmentation through a tyrosinase-dependent MITF/TFAP2A pathway. Cell 155: 1022–1033. 10.1016/j.cell.2013.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissl S, Fang R, Huang H, Zhao Y, Raviram R, Gorkin DU, Zhang Y, Sos BC, Afzal V, Dickel DE, et al. 2018. Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat Neurosci 21: 432–439. 10.1038/s41593-018-0079-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowska G, Halaris A, Selemon LD. 2001. Reductions in neuronal and glial density characterize the dorsolateral prefrontal cortex in bipolar disorder. Biol Psychiatry 49: 741–752. 10.1016/S0006-3223(01)01080-0 [DOI] [PubMed] [Google Scholar]
- R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van Doren V, et al. 2016. Schizophrenia risk from complex variation of complement component 4. Nature 530: 177–183. 10.1038/nature16549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahryari A, Jazi MS, Samaei NM, Mowla SJ. 2015. Long non-coding RNA SOX2OT: expression signature, splicing patterns, and emerging roles in pluripotency and tumorigenesis. Front Genet 6: 196 10.3389/fgene.2015.00196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene NG, Bryois J, Bakken TE, Breen G, Crowley JJ, Gaspar HA, Giusti-Rodriguez P, Hodge RD, Miller JA, Muñoz-Manchado AB, et al. 2018. Genetic identification of brain cell types underlying schizophrenia. Nat Genet 50: 825–833. 10.1038/s41588-018-0129-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JHT, Lowe CB, Kingsley DM. 2018. Characterization of a human-specific tandem repeat associated with bipolar disorder and schizophrenia. Am J Hum Genet 103: 421–430. 10.1016/j.ajhg.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Yang X, Ren X, Maliskova L, Li B, Jones IR, Wang C, Jacob F, Wu K, Traglia M, et al. 2019. Mapping cis-regulatory chromatin contacts in neural cells links neuropsychiatric disorder risk variants to target genes. Nat Genet 51: 1252–1262. 10.1038/s41588-019-0472-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansey KE, Hill MJ. 2018. Enrichment of schizophrenia heritability in both neuronal and glia cell regulatory elements. Transl Psychiatry 8: 7 10.1038/s41398-017-0053-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulirsch JC, Lareau CA, Bao EL, Ludwig LS, Guo MH, Benner C, Satpathy AT, Kartha VK, Salem RM, Hirschhorn JN, et al. 2019. Interrogation of human hematopoiesis at single-cell and single-variant resolution. Nat Genet 51: 683–693. 10.1038/s41588-019-0362-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J, Rynes E, Sandstrom R, Zhang M, Canfield T, Hansen RS, Stehling-Sun S, Sabo PJ, Byron R, Humbert R, et al. 2014. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science 346: 1007–1012. 10.1126/science.1246426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. 2007. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res 35: D88–D92. 10.1093/nar/gkl822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, et al. 2013. A high-resolution enhancer atlas of the developing telencephalon. Cell 152: 895–908. 10.1016/j.cell.2012.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, Yang J. 2017. 10 years of GWAS discovery: biology, function, and translation. Am J Hum Genet 101: 5–22. 10.1016/j.ajhg.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Umićević Mirkov M, de Leeuw CA, van den Heuvel MP, Posthuma D. 2019. Genetic mapping of cell type specificity for complex traits. Nat Commun 10: 3222 10.1038/s41467-019-11181-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DX, Marchetto MC, Gage FH. 2014. How to make a hippocampal dentate gyrus granule neuron. Development 141: 2366–2375. 10.1242/dev.096776 [DOI] [PubMed] [Google Scholar]
- Zhang S, Cui W. 2014. Sox2, a key factor in the regulation of pluripotency and neural differentiation. World J Stem Cells 6: 305–311. 10.4252/wjsc.v6.i3.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.