Abstract
Background
While our battle with the COVID-19 pandemic continues, a multitude of Omics data have been generated from patient samples in various studies. Translation of these data into clinical interventions against COVID-19 remains to be accomplished. Exploring host response to COVID-19 in the upper respiratory tract can unveil prognostic markers and therapeutic targets.
Methods
We conducted a meta-analysis of published transcriptome and proteome profiles of respiratory samples of COVID-19 patients to shortlist high confidence upregulated host factors. Subsequently, mRNA overexpression of selected genes was validated in nasal swabs from a cohort of COVID-19 positive/negative, symptomatic/asymptomatic individuals. Guided by this analysis, we sought to check for potential drug targets. An FDA-approved drug, Auranofin, was tested against SARS-CoV-2 replication in cell culture and Syrian hamster challenge model.
Findings
The meta-analysis and validation in the COVID-19 cohort revealed S100 family genes (S100A6, S100A8, S100A9, and S100P) as prognostic markers of severe COVID-19. Furthermore, Thioredoxin (TXN) was found to be consistently upregulated. Auranofin, which targets Thioredoxin reductase, was found to mitigate SARS-CoV-2 replication in vitro. Furthermore, oral administration of Auranofin in Syrian hamsters in therapeutic as well as prophylactic regimen reduced viral replication, IL-6 production, and inflammation in the lungs.
Interpretation
Elevated mRNA level of S100s in the nasal swabs indicate severe COVID-19 disease, and FDA-approved drug Auranofin mitigated SARS-CoV-2 replication in preclinical hamster model.
Funding
This study was supported by the DBT-IISc partnership program (DBT (IED/4/2020-MED/DBT)), the Infosys Young Investigator award (YI/2019/1106), DBT-BIRAC grant (BT/CS0007/CS/02/20) and the DBT-Wellcome Trust India Alliance Intermediate Fellowship (IA/I/18/1/503613) to ST lab.
KEYWORDS: COVID-19, Nasal swab/BALF, Transcriptome, Proteome, Meta-analysis, Prognostic marker, Auranofin
Graphical abstract
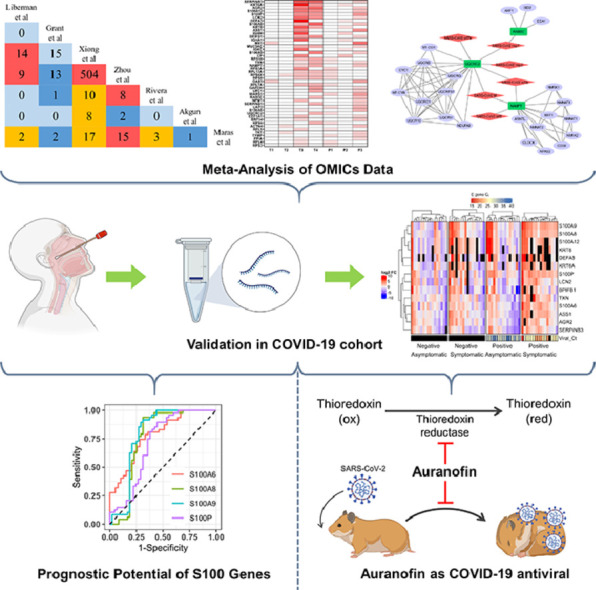
Research in context.
Evidence before this study
Some studies have demonstrated the prognostic value of host factors in COVID-19 patients, primarily in serum samples, however reliable biomarkers in nasal swab samples remain to be identified. Also, Auranofin has been described earlier to have anti-inflammatory and anti-infective properties, however, its antiviral effect against SARS-CoV-2 in preclinical animal models and potential mechanism of action was not described.
Added value of this study
The set of genes identified by meta-analysis and validation of Omics data in COVID-19 patient nasal swabs are all interferon regulated and may be involved in disease progression. In line with this hypothesis, we show that S100 family genes have significant sensitivity and specificity as COVID-19 prognostic markers in the nasal swabs. Furthermore, we also demonstrate TXN as a consistently upregulated host factor, which can be targeted by Auranofin to mitigate SARS-CoV-2 replication. The in vivo protective action of Auranofin was shown to involve reduction of IL-6 production and lung inflammation.
Implications of all the available evidence
The current most widely used diagnostic method for COVID-19 makes use of qRT-PCR to quantify viral RNA levels in nasal swabs. Our findings indicate that measuring S100 family genes in the same samples by qRT-PCR can inform the severity of the disease. Furthermore, we demonstrate antiviral efficacy of the FDA-approved drug Auranofin against SARS-CoV-2 in cell culture and preclinical hamster model.
Alt-text: Unlabelled box
1. Introduction
The COVID-19 pandemic has emerged as the biggest global public health crisis of this century. As of August 4, 2021, more than 200 million infections and 4.2 million deaths have been reported (https://www.worldometers.info/coronavirus/). The causative agent SARS-CoV-2 contains a single-stranded positive-sense RNA genome that encodes 29 proteins [1]. COVID-19 disease is quite heterogeneous, and its manifestation ranges from asymptomatic, mild, severe to lethal, depending on a variety of host, virus, and environmental factors [2]. Age, sex, ethnicity, and co-morbidities have all been implicated in determining disease outcomes [2, 3]. An effective and early interferon (IFN) response is critical in resolving viral infections [4], however, SARS-CoV-2 has multiple strategies to suppress host immune responses [5]. Disruption of immune homeostasis and induction of cytokine storm has been recognized as one of the underlying causes of severe COVID-19 [6], yet the molecular mechanisms underlying immune dysregulations are yet to be defined.
Several research groups have applied tour de force high throughput methodologies to profile the host responses upon viral infections [7], [8], [9], [10], [11], [12], [13], [14]. This has resulted in a wealth of virus-host interaction Big Data, which holds the key to novel therapeutic strategies and molecular markers of infection and disease progression. Examining host response at the primary site of infection in the upper respiratory tract is crucial to understanding viral pathogenesis. Various studies have utilized BALF and nasopharyngeal swabs to characterize the changes in transcripts and proteins during infection to understand COVID-19 pathogenesis [7], [8], [9], [10], [11], [12], [13], which have highlighted significantly upregulated genes and biological pathways altered during infection. While proinflammatory cytokines, chemokines, enzymes in neutrophil-mediated immunity, and several IFN stimulated genes (ISGs) have consistently shown up in their analysis, experimental validation and mechanistic studies are generally lacking [8], [9], [10], [11], [12], [13]. A detailed characterization of antiviral responses in the upper respiratory tract of patients, its variation with age and sex, and association with progression of disease severity remains to be accomplished.
The goal of our study was to identify genes that are consistently upregulated during SARS-CoV-2 infection in the upper respiratory tract of patients and understand their role in viral infection and disease progression. More specifically, we were interested in secreted signalling mediators which can serve as markers of disease progression or druggable proteins that can serve as therapeutic targets. For this, we surveyed the literature for Omics data from COVID-19 positive patient's nasal swab and BALF samples and selected 4 transcriptomic and 3 proteomic datasets. We performed a hypergeometric distribution-based overlap analysis followed by cumulative fold-change score-based prioritization to shortlist genes. This was followed by an examination of selected gene expression levels in nasal swab/ BALF samples from a cohort of COVID positive, negative, symptomatic, and asymptomatic individuals, ranging from 30-60 years in age and of mixed gender. Receiver operating characteristic (ROC) curve analysis of gene expression data in nasal swabs revealed S100 family genes (S100A6, S100A8, S100A9, and S100P) as high confidence markers of disease severity. Among other shortlisted genes, Thioredoxin (TXN) emerged as a significantly upregulated factor supported by multiple datasets. Thioredoxin is a proinflammatory protein that requires to be reduced by Thioredoxin reductase enzyme, which itself can be targeted by an FDA-approved gold drug Auranofin [15]. We tested the antiviral efficacy of Auranofin in cell culture and preclinical Syrian hamster challenge model and found that it can reduce SAR-CoV-2 replication over one order of magnitude at a well-tolerated non-toxic dosage. We also establish its mechanism of protection, which is through suppressing the expression of proinflammatory cytokine IL-6 expression. This drug is already in clinical use for inflammatory diseases and can have clinical implications in COVID-19 treatment based on our data.
Through collective global efforts, several COVID-19 vaccines have become available in an astonishingly short period, although new virus variants have emerged, some of which can escape vaccine-mediated immunity [16, 17]. Progress on the development of antivirals and disease prognostic markers has been lagging. Repurposing clinically approved drugs for use against SARS-CoV-2 has been an attractive option and has been explored by many research groups through different approaches [18]. Our study translates COVID-19 virus-host interaction and response Big Data into potential actionable clinical interventions, including the use of S100 genes as a prognostic marker in nasal swabs and repurposing the clinically approved drug, Auranofin for COVID-19 treatment.
2. Methods
2.1. Ethics statement
This study was conducted after approval from Institutional Human Ethics Committee (Approval Number: IHEC No. 13-11092020), Institutional Bio-Safety Committee (Approval Number: IBSC/IISc/ST/17/2020) and Institutional Animal Ethics Committee (Approval Number: IAEC/IISc/ST/784/2020), following the Indian Council of Medical Research and Department of Biotechnology recommendations. For use of human samples, informed consent was obtained from each participant, before the study. All experiments involving infectious SARS-CoV-2 were conducted in the Viral Biosafety level-3 facility at the Indian Institute of Science.
2.2. Cells and viruses
Authenticated (relevant documentation regarding authentication by suppliers is available in Supplemental Data) HEK 293T cells expressing human ACE2 (NR-52511, BEI Resources, NIAID, NIH, RRID: CVCL_A7UK) and VeroE6 cells (CRL-1586, ATCC, RRID:CVCL_0574) were cultured in complete media containing Dulbecco's modified Eagle medium (12100-038, Gibco) with 10% HI-FBS (16140-071, Gibco), 100 IU/ml Penicillin, 100 μg/ml Streptomycin and 0.25μg/ml Amphotericin-B (Penicillin-Streptomycin-Amphotericin B, ICN1674049, MP Biomedicals) supplemented with GlutaMAX™ (35050-061, Gibco). SARS-CoV2 (Isolate Hong Kong/VM20001061/2020, NR-52282, BEI Resources, NIAID, NIH) was propagated and titered by plaque assay in Vero E6 cells as described before [19].
2.3. Omics data collection and processing
Transcriptomics and protein abundance data from COVID-19 patient's naso- and oropharyngeal swabs, bronchoalveolar lavage fluid (BALF), and other respiratory specimens were chosen from PubMed, BioRxiv, and MedRxiv using different combinations of keywords like "COVID-19, SARS-CoV-2, Transcriptomics, Proteomics, BALF, swab". Studies dealing with gene expression profiles of SARS-CoV-2 infected non-human cell lines and tissues were not considered. The SARS-CoV-2 and COVID-19 collections in the EMBL-EBI PRIDE proteomics database [20] were retrieved and used without any modification. In the NCBI GEO database [21], the following combination of terms was used to collect relevant datasets: ((covid-19 OR SARS-COV-2) AND gse [entry type]) AND "Homo sapiens"[porgn: _txid9606]. The retrieved datasets were then filtered by their date of publication to collect the studies published between the 1st of January 2020 and the 15th of September 2020. The filtration of datasets was carried out using two parameters, fold-change, and its significance value. Genes and proteins with a fold-change value of ≥ 1.5 and q-value ≤ 0.05 were chosen for the overlap analysis. The raw p-value was used for filtering in cases where the adjusted p-value was not provided, albeit with a more stringent cut-off of ≤ 0.01. The UniProt IDs in filtered protein abundance datasets were converted to their corresponding primary Gene Symbols using UniProt [22].
2.4. Gene set overlap analysis
The GeneOverlap class of R package "GeneOverlap" [23] was used for testing whether two lists of genes are independent, which is represented as a contingency table, and then Fisher's exact test was used to find the statistical significance. Genes with less than 0.01 overlap p-value were selected for further analysis. The number of background genes for proteome-proteome pairwise study and the transcriptome-proteome pairwise study was 25,000, i.e., the number of protein-coding genes in Hg19. For the transcriptome-transcriptome overlap study, the number of background genes was taken to be the union of total expressed genes in both the datasets considered.
2.5. Gene ontology, Interferome, cellular and tissue localization analysis
Enriched GO terms were obtained by express analysis on Metascape [24] and plotted using ggplot2 [25]. The database Interferome v2.01 [26] was queried using gene symbols for identifying interferon-regulated genes (IRGs) in normal samples of the respiratory system from both in vitro and in vivo experiments in humans. For cellular localization, each gene was queried on UniProt annotation [27] and Human Protein Atlas ver20.0 [28, 29] and then manually annotated. The single-cell expression data of transcripts was also obtained from Human Protein Atlas ver20.0 (Available from http://www.proteinatlas.org/). They were further filtered to obtain cells that are associated with the immune system or respiratory tract.
2.6. Virus-Host protein-protein interaction network analysis
The interaction data for the selected 46 genes were retrieved from publicly available interaction datasets [14]. The retrieved information was then used to generate a network map. Cytoscape v3.8.0 [30] was used to construct the interaction network for virus-host protein-protein interaction (PPI). STRING database within the Cytoscape store was used to query the proteins to elucidate the interactions between the proteins significantly altered during SARS-CoV-2 infection. The resulting STRING interaction network (confidence ≥0.999 for all the proteins and confidence ≥0.90 for NAMPT; max number of interactors = 10) was merged with the virus-host PPI on Cytoscape.
2.7. qRT-PCR based measurement of cellular gene expression for patient samples
Nasopharyngeal swabs were collected from COVID-19 patients and healthy individuals for diagnostic purposes by hospitals from Bengaluru Urban city and brought to COVID-19 Diagnostic Facility at the Indian Institute of Science in viral transport media (VTM). RNA from patients was isolated using kits recommended and provided by the Indian Council of Medical Research. Samples were chosen to have an almost equal number of patients falling into categories of age, sex, COVID-19 status, and symptomatic status (Table 1). Demographic information was not used as an inclusion criterion. Although a priori sample size determination was not conducted, the number of samples were chosen based on technical constraints and previous publications [31, 32]. The de-identified patient data is available upon request (see Data Sharing statement).
Table 1.
Summary of individual and different categories in the COVID-19 cohort used for qRT-PCR based validation analysis. All samples were collected from Bangalore Urban area for diagnostic purposes.
| Patient Status | Number of patients | Average age | Number of males | Number of females | Number in the age group 30-40 | Number in the age group 41-50 | Number in the age group 51-60 |
|---|---|---|---|---|---|---|---|
| Negative Asymptomatic | 16 | 43.9 | 8 | 8 | 6 | 5 | 5 |
| Negative Symptomatic | 16 | 41.7 | 12 | 4 | 9 | 4 | 3 |
| Positive Asymptomatic | 15 | 44.3 | 7 | 8 | 6 | 5 | 4 |
| Positive Symptomatic | 16 | 45 | 8 | 8 | 5 | 5 | 6 |
Equal amounts of RNA were converted into cDNA using Prime Script™ RT Reagent Kit with gDNA Eraser (Perfect Real Time) (RR047A, Takara-Bio) and then diluted with 80μl nuclease-free water. The gene expression study was conducted using PowerUp™ SYBR™ Green Master Mix (A25778, Applied Biosystems™) with 18srRNA as the internal control and appropriate primers for the genes (Supplementary Table 3).
2.8. Cytotoxicity assay
HEK-ACE2 cells were seeded in a 96-well cell culture dish pre-coated with 0.1 mg/mL poly-L-lysine (P9155-5MG, Sigma-Aldrich) and 24 hr later, treated with 0, 1, 2, and 4 µM Auranofin (A6733, Sigma-Aldrich) in triplicates. Cells were incubated at 37°C, 5% CO2, and 48 hr later, cytotoxicity was measured using AlamarBlue™ Cell Viability Reagent (DAL 1025, Thermo Fisher) as per manufacturer's instructions.
2.9. Infection in HEK-ACE2 and VeroE6 cells
Cells were seeded in a 24-well cell culture dish (pre-coated with 0.1 mg/mL poly-L-lysine for HEK-ACE2) and 24 hr later, used for infection.
HEK-ACE2: Cells were first pretreated for 3 hr with 0, 0.125, 0.25, 0.5, and 1µM Auranofin in quadruplicates. Infection was done with 0.1 MOI SARS CoV-2 in 100 µl inoculum in DMEM supplemented with 10% FBS for 1hr at 37°C.
VeroE6: Cells were pretreated for 3 hr with 0 and 1 µM Auranofin in quadruplicates. Cells were infected with 0.001 MOI SARS CoV-2 in 100 µL inoculum in DMEM supplemented with 2% FBS for 1 hr at 37°C.
For both cell lines, complete medium (DMEM with 2% FBS for VeroE6) restoring the initial dose of the drug was added to the cells. After 48 hr, cells were processed separately for plaque assay, western blot analysis, and RNA extraction using TRIzol™ Reagent (15596018, Thermo Fisher).
2.10. Western blot
Cells were washed with 1X PBS (162528, MP Biomedicals) and lysed with 1X Laemmli buffer (1610747, BIO-RAD). Cell lysates were loaded and resolved using a 10% SDS-PAGE gel, and the separated proteins were transferred onto a PVDF membrane (IPVH00010, Immobilon-P; Merck). Blocking was performed using 5% Skimmed milk (70166, Sigma-Aldrich) in 1X PBS containing 0.05% Tween 20 (P1379, Sigma-Aldrich) (1X PBST) for 2 hr at room temperature with slow rocking. Primary antibody incubation was performed overnight (12 hr) at 4°C using SARS-CoV / SARS-CoV-2 (COVID-19) spike antibody (180 KDa) (GTX632604, GeneTex, RRID: AB_2864418 or NR-52947, BEI Resources, NIAID, NIH). Secondary antibody incubation was performed for 2 hr at room temperature with slow rocking using Goat Anti-Mouse IgG H&L (ab6789, Abcam, RRID: AB_955439) or Goat Anti-Rabbit IgG H&L (ab6721, Abcam, RRID:AB_955447). The blots were developed using Clarity Western ECL Substrate (1705061, BIO-RAD). Blots were probed for beta-actin (42 KDa) using mouse monoclonal antibody to beta Actin [AC-15] (HRP) (ab49900, Abcam, RRID: AB_867494). All antibodies were authenticated by the respective companies and relevant documentation is available in Supplemental Data.
2.11. Plaque assay
Plaque assay to measure infectious virus counts were performed as described before [19]. VeroE6 cells were seeded in 6-well cell culture dishes to reach complete confluency the next day. Cells were washed once with 2 mL warm PBS and incubated with dilutions of cell culture supernatants in 200 μL complete DMEM for 1 hr at 37°C. The virus inoculum was then removed, and cells were overlaid with DMEM containing 2% FBS and 0.8% agarose (MB002, Himedia). After 48 hr incubation, cells were fixed with 4% formalin, and plaques were visualized by crystal violet (C6158, Merck) staining.
2.12. Tissue-culture infectious dose 50 (TCID50)
HEK-ACE2 cells were seeded in a 96-well cell culture dish pre-coated with 0.1 mg/mL poly-L-lysine and 24 hr later, used for infection. Cells were first pretreated for 3 hr with 1 µM Auranofin and subsequently infected with two-fold serial dilutions of SARS-CoV-2 starting at 0.1 MOI. Each condition was performed in ten wells. Plates were incubated for 48 hr, and the presence or absence of cytopathic effects were recorded. TCID50 was estimated using methods described by Reed and Muench [33].
2.13. Cytopathic Effect (CPE) reduction
HEK-ACE2 cells were seeded in a 24-well cell culture dish pre-coated with 0.1 mg/mL poly-L-lysine and 24 hr later, used for infection. Cells were first pretreated for 3 hr with 1 µM Auranofin in triplicates and subsequently incubated with 0.1 MOI SARS CoV-2 in 100 µL inoculum for 1 hr at 37°C. Subsequently, 400 µL complete medium restoring the prior dose of the drug was added to the cells. After 48 hr, the percentage of viable cells was measured by Trypan blue (93595, Sigma-Aldrich) dye exclusion method.
2.14. Animal experiments
Animal Handling: All animal experiments were performed using 10 to 12-week-old male and female Syrian golden hamsters purchased from Biogen Laboratory Animal Facility (Karnataka, India). The animals were allowed to acclimatize for 3 days at the experimental location, and given access to pellet feed and water ad libitum. Males and females were housed separately and maintained on a 12-hr day/night light cycle at the Viral Biosafety level-3 facility at the Indian Institute of Science. Hamsters were euthanized by an overdose of Ketamine (Bharat Parenterals Limited) and Xylazine (21, Indian Immunologicals Ltd).
Toxicity and Infection assays: Toxicity of 1 and 5 mg/kg bodyweight Auranofin was tested on Syrian golden hamsters by once-daily oral administration of the drug in 200 µl PBS. This corresponds to a dosage of 1 mg/kg (Hamster) x 0.13 (conversion factor) = 0.13 mg/kg (Human equivalent dose) and 5 mg/kg (Hamster) x 0.13 (conversion factor) = 0.65 mg/kg (Human equivalent dose) Auranofin per day (conversions as desribed in https://www.fda.gov/media/72309/download). The total bodyweight of hamsters was monitored for up to 7 days (see Supplementary Fig 8). Infection experiments were performed by intranasal inoculation of animals with 105 PFU SARS-CoV-2 in 100 µL PBS. The animals were anesthetized using intraperitoneal injections of Ketamine (150 mg/kg) (Bharat Parenterals Limited) and Xylazine (10 mg/kg) (21, Indian Immunologicals Ltd) cocktail before infection. Prophylactic treatment involved oral administration of Auranofin (5 mg/kg/day) 3-, 2-, and 1-day before infection and followed by virus challenge at day 0. The therapeutic treatment regimen used oral administration of Auranofin (5 mg/kg/day) starting at 24-hr post-infection (hpi), followed by 2- and 3-days post-infection (dpi). Total body weight was recorded each day during the entire course of the experiment until the animals were sacrificed at 4 dpi. Viral RNA load in lung tissue specimens was detected by qRT-PCR. Sample size for hamster experiments were chosen based on previous studies [34, 35].
2.15. RT PCR for viral copy number calculation
For qRT-PCR, total RNA was isolated using TRIzol™ Reagent (15596018, Thermo Fisher) as per manufacturer's instructions and equal amounts of RNA was used to determine viral load using AgPath-ID™ One-Step RT-PCR kit (AM1005, Applied Biosystems) using primers and probes targeting the SARS CoV-2 N-1 gene (Forward primer: 5′GACCCCAAAATCAGCGAAAT3′ and Reverse primer: 5′ TCTGGTTACTGCCAGTTGAATCTG3′, Probe: (6-FAM / BHQ-1) ACCCCGCATTACGTTTGGTGGACC). Viral copy number was estimated by generating a standard curve using SARS-CoV-2 genomic RNA standard.
2.16. Histopathology of Lung tissue
Lung tissue specimens of hamsters were fixed in 4% paraformaldehydein PBS, and embedded in paraffin blocks. Tissue sections of 4-6 μm thickness were stained with Hematoxylin and Eosin (H&E) for examination by light microscopy as previously described [36].
2.17. Graphical representations and statistical analysis
Statistical analyses and overlaps were performed in the R statistical environment version 4.0.3 via RStudio version 1.3.1093. All statistical tests are two-sided unless specified otherwise. Plots were made using the ggplot2 package in R [25] and GraphPad Prism v8.0.2. In boxplots, the hinges of boxes represent the first and third quartiles. The whiskers of the boxplot extend to the value which is 1.5 times the distance between the first and third quartiles. Each data point in the boxplot represents one of the triplicates in qRT-PCR for a particular gene in a particular patient sample. Heatmaps were generated using the R package Complex Heatmap with Euclidean method used for clustering [37]. Receiver Operating Characteristic (ROC) curve analysis and Optimal cut-off determination were performed using the online tool easyROC (ver. 1.3.1) [38].
2.18. Role of funding source
Only financial support was provided by the funders. They have not participated in study design, data collection, data analyses, interpretation, or writing of the report.
3. Results
3.1. Compilation and overlap analysis of published transcriptomics and proteomics data from COVID-19 patient samples revealed 566 upregulated host factors
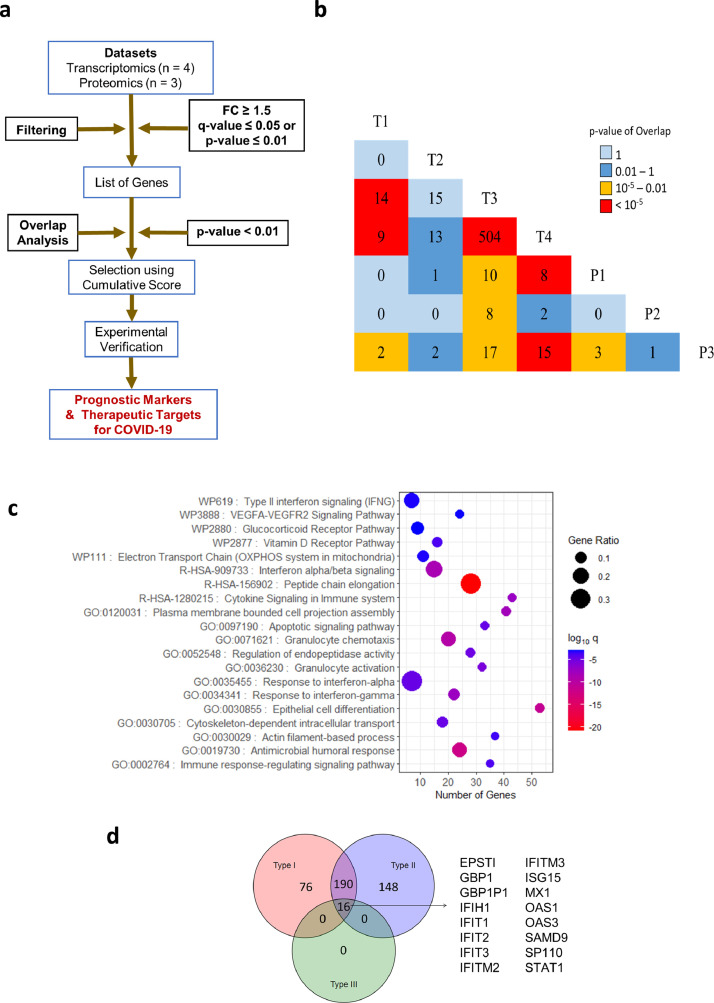
We started the study by compiling the host factors that are consistently and significantly upregulated in the upper respiratory tract of COVID-19 patients. For this 'top-down' approach to narrow down severity markers and drug targets from genome-wide data, we decided to use published transcriptomics and proteomics datasets derived from nasal swab or BALF samples of COVID-19 patients. We chose four transcriptomics (T), and three proteomics (P) datasets, and further analyses were performed according to a rationally designed workflow (Fig. 1a). All datasets included differentially expressed genes in infected patients with healthy individuals as control (see Supplementary Table 1). The selection criteria (described in materials and methods) included at least 1.5-fold gene upregulation at both mRNA and protein levels. The filtration of data was carried out to sort only significantly upregulated genes from all the datasets (see Supplementary Table 2). A pairwise overlap analysis was performed on the filtered genes/proteins from each study and significantly overlapping genes (p-value < 0.01 calculated using Fisher's exact test) between T1-T3 (14), T1-T4 (9), T1-P3 (2), T3-T4 (504), T3-P1 (10), T3-P2 (8), T3-P3 (17), T4-P1 (8). T4-P3 (15) and P1-P3 (3) were determined (Fig. 1b, Supplementary File 1). This method was adapted from similar overlap analysis conducted previously to compare multiple virus-host interaction datasets and to obtain the significance of intersections [39]. Union of significant intersections after the overlap analysis results in 566 genes (Fig. 1b). To reiterate the functional characteristics of the differentially expressed genes (DEGs), we examined the biological processes and signaling pathways they are involved in. Pathway enrichment of 566 genes from the union of all significant intersections from overlap analysis showed enrichment of biological processes like protein elongation, interferon (IFN) signaling, chemotaxis of granulocytes, and inflammatory pathways (Fig. 1c). The antiviral response to respiratory viral infections, including SARS-CoV-2, is driven by interferons (IFNs) [40]. Hence, we examined the shortlisted set of genes for their potential regulation by different categories of IFNs, using the Interferome tool [26]. We found that out of 566 genes, 76 were regulated by type I IFN, 148 genes by Type II IFN, 190 genes were regulated by both type I and type II IFN, while 16 genes were commonly regulated by all the three classes of IFNs (Fig. 1d). These 16 genes are well-characterized interferon-stimulated genes (ISGs), which include direct antiviral effector ISGs (IFITs, MX1, OAS3, and OAS1), as well as positive regulators (STAT1) of IFN response [41]. This indicated an active IFNmediated innate antiviral response in the upper respiratory tract cells during SARS-CoV-2 infection and highlighted potential antiviral factors.
Fig. 1.
Meta-analysis pipeline for gene prioritization and associated pathway analysis. a) Workflow used to obtain prognostic markers and therapeutic targets from proteomics and transcriptomics datasets. b) Triangular heatmap showing pairwise overlaps between transcriptomic and proteomic datasets. The number within each box denotes the number of genes that showed up between the corresponding intersections. The colour of a box denotes the significance of overlap determined by Fisher's exact test. c) Gene ontology of all genes (566) in the significant intersections obtained during the overlap analysis plotted with the number of genes in each term on the X-axis, proportion of genes enriched compared to the total number of genes in each term as the size of dots and the colour representing log10 p-adj value (q-value) of enrichment (calculated by a hypergeometric test with Benjamini-Hochberg correction). d) Venn diagram showing the number of genes that are induced by Type I, II, or III interferons. The analysis was performed on Interferome v2.01 using the union of significant intersections (566).
3.2. Rank ordering and shortlisting of upregulated host factors highlighted host factors regulating the antiviral and inflammatory immune response in COVID-19 patients
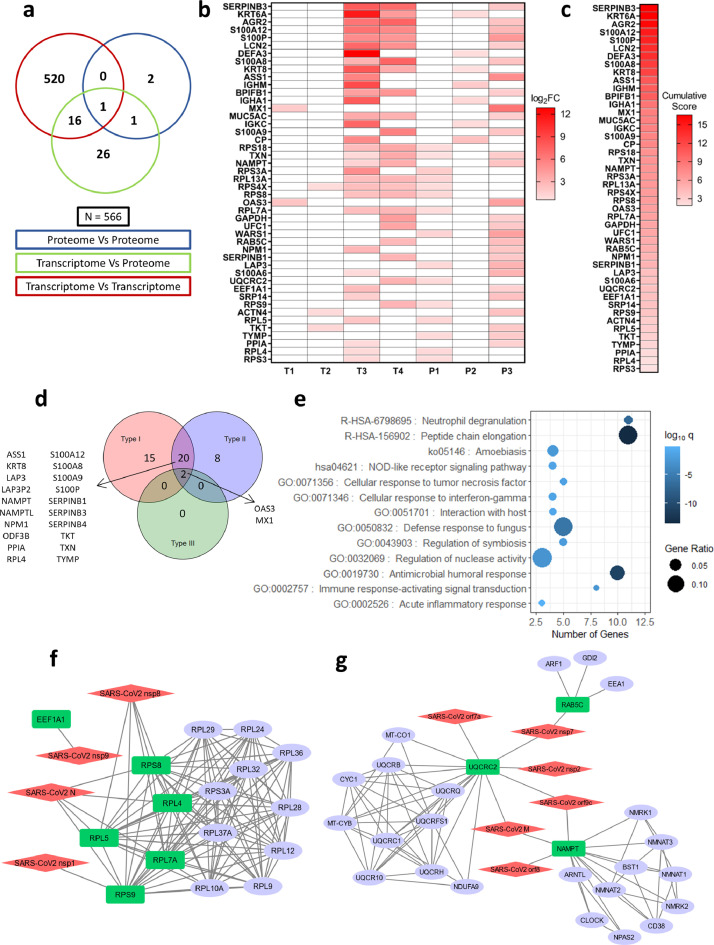
Since proteome dictates the outcome inside a cell, soluble factors are key in shaping the antiviral response. We focused on genes supported by orthogonal transcript (T) and protein (P) abundance data. For this, we chose genes from the union of intersections of T-T, T-P, and P-P overlaps, which was reported at least in one of the proteomics studies. This narrowed down the list to a total of 46 genes that were intersecting in T-P (26), P-P (2), TT-TP (16), TP-PP (1), and TT-TP-PP (1) overlaps (Fig. 2a and 2b, Supplementary File 1). A cumulative score for the 46 selected significantly upregulated genes was calculated using the sum of their log2 fold-change values in the parent datasets and ranked (Fig. 2c). The enrichment of these 46 genes in each of the datasets, where the expression is reported, is shown in Fig. 2b. Many of these genes are directly regulated by different classes of interferons. 15 genes are regulated by IFN-I, while 8 genes by IFN-II. 20 genes are regulated by both type-I and type-II IFNs, while only 2 genes by all the three types of IFNs (Fig. 2d). Most of the IFITs and other ISGs that were earlier determined in our analysis to be regulated by all the three types IFNs are no more in the list since those ISGs were only reported upregulated at transcriptome level (only in T-T overlap) and hence were lost when the genes were filtered for their upregulation at the protein level, leaving behind only MX1 and OAS3 (Fig. 1c and 2d). The biological functions of the selected 46 genes were also investigated to understand their roles in COVID-19 pathophysiology. The enriched pathways were mainly related to innate immune response and defense against microbes along with inflammatory and immune signaling, neutrophil degranulation, and cellular response to TNF and interferon-gamma (Fig. 2e).
Fig. 2.
Cumulative score ranking, pathway, and interactome analysis of selected host factors. a) Venn diagram of genes obtained from significant intersections among proteomic or transcriptomic datasets after pairwise overlap analysis. b) Genes in the Venn diagram that were found in at least one proteomic dataset with their log2FC values in the datasets where they are present. Boxes colored in white denote that the gene is not present in the filtered dataset. c) Genes arranged in descending order of cumulative scores obtained as a sum of log2FC values in the datasets where they are present. d) Venn diagram showing the number of interferon-induced genes performed using Interferome v2.01 for 46 selected genes. e) Gene ontology of 46 genes plotted with the number of genes in each term on the X-axis, the proportion of genes enriched compared to the total number of genes in each term as the size of dots and the color representing log10 p-adj value (q-value) of enrichment (calculated by a hypergeometric test with Benjamini-Hochberg correction). f, g) Virus-host protein-protein interactions among SARS-CoV2 proteins and significant genes in the overlap analysis that shows up in at least one proteomic dataset modeled using Cytoscape v3.8.0. Red: SARS-CoV-2 proteins, Green: Host proteins (primary interactor), blue: STRING interactors (other cellular proteins interacting with the primary interactors).
Further, to understand the potential role of shortlisted genes in COVID-19 pathophysiology, their interactions with SARS-CoV-2 proteins were inspected by analyzing the publicly available SARS-CoV-2 cellular interactome data [14]. For this, host protein-protein interactions were retrieved from the STRING database [42] and merged with the virus-host protein-protein interactions giving a discrete picture of how the viral proteins target various cellular processes during infection. Other than NAMPT, UQCRC2, and RAB5C, it was mainly proteins associated with ribosomes that were primary interactors to the SARS-CoV-2 proteins (Fig. 2f and 2g). We also examined the intracellular, cellular, tissue, and organ-specific expression for shortlisted genes using publicly available data [28, 29]. Many upregulated proteins were predicted to localize in the intracellular organelles like endoplasmic reticulum, mitochondria, Golgi complex, and endosomes (see Supplementary Fig 1a), while 19 genes were predicted to be secretory. A thorough analysis of the list of 46 selected genes using Human Tissue Atlas revealed that they are expressed in the respiratory tract and in immune effector cells known to survey infection sites (see Supplementary Fig 1b). The relative expression levels show that genes associated with protein synthesis (ribosomal proteins and elongation factors) are highly expressed compared to any other genes and are enriched across all the tissues in the map (see Supplementary Fig 1b).
3.3. qRT-PCR based validation in a cohort of COVID-19 positive/negative, symptomatic/asymptomatic individuals reveals differential upregulation of selected genes in a disease-specific manner
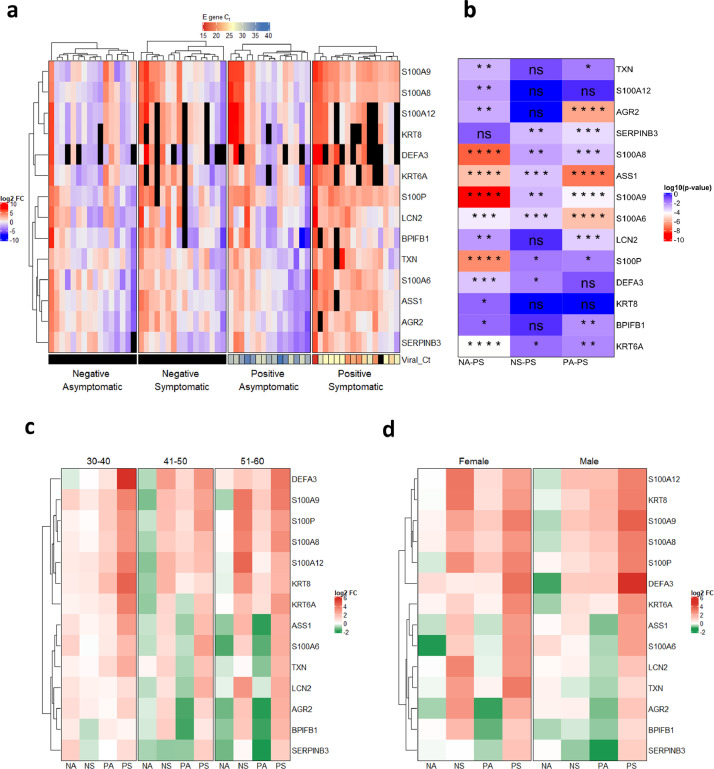
For validation using qRT-PCR and further analysis, we selected genes with a cumulative score greater than 10, except for IGHM due to the lack of compatible primers (Fig. 2c). Also, we considered genes belonging to the S100 family that came up within 46 shortlisted genes, since they are known regulators of inflammation [43, 44]. Furthermore, we also selected TXN since it was supported by multiple lines of evidence and appeared in the TT-TP-PP overlap in our study (Fig. 2a). The COVID-19 patient cohort used for qRT-PCR of genes included 63 individuals (both males and females, aged 30-60 years), out of which 16 each were COVID-19 positive symptomatic (PS), COVID-19 negative asymptomatic (NA), COVID-19 negative symptomatic (NS), and 15 were COVID-19 positive asymptomatic (PA) healthy category (Table 1). Total mRNA from the nasal swab was isolated and the upregulation of 14 selected genes was verified by qRT-PCR. The log2 fold-change expression with respect to the average of the negative asymptomatic group (Fig. 3a, see Supplementary Fig. 2) was calculated and plotted on a heatmap (Fig. 3a), which depicts the mRNA enrichment of the selected genes in different patient samples and categories. Next, we determined the correlation between the viral RNA load in COVID-19 patients (qRT-PCR of viral envelope (E) gene) and log2 fold-change of selected host genes in the patient samples. It was observed that the threshold cycle (Ct) value for the E gene was negatively correlated with log2 fold-change of genes showing that viral load and expression levels of the selected genes are positively correlated (see Supplementary Fig 3). Furthermore, the upregulation of selected host genes was more pronounced in positive symptomatic patients than positive asymptomatic individuals (Fig. 3a, see Supplementary Fig. 3). A comparative heatmap in Fig. 3b gives an insight into the genes that can be considered as COVID-19 disease and/or severity marker. While all the upregulated genes except SERPINB3 indicate infection (Fig. 3b; NA-PS), only a few genes showed significant upregulation in a COVID-19 specific manner (Fig. 3b; NS-PS).
Fig. 3.
qRT-PCR validated expression profile of selected genes in different categories of COVID-19 cohort. a) qRT-PCR was performed on RNA isolated from COVID-19 patients for 14 genes and average log2FC values (with respect to Negative Asymptomatic group) of PCR triplicates are shown in a heatmap. Each column represents a patient. The bottom annotation shows the Ct value for the viral gene encoding Envelope (E) protein with a corresponding legend on the top. Black boxes denote 'value unknown/undetermined'. b) Differences between groups for each gene were computed and the log10 (p-value) of comparisons is shown in the heatmap. The comparisons are Negative asymptomatic vs Positive symptomatic (NA-PS), Negative symptomatic vs Positive symptomatic (NS-PS), and Positive asymptomatic vs Positive symptomatic (PA-PS). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns – not significant (Kruskal-Wallis test followed by post hoc Dunn's test with Bonferroni corrections for multiple comparisons). c) log2FC values are grouped based on age groups 30-40, 41-50, and 51-60. Each row represents the average of log2FC values for patients falling into the particular age group and respective disease status. d) log2FC values are grouped according to sex. Each row represents the average of log2FC values for patients falling into the particular sex and respective disease status.
Multiple genes from the S100 family, including S100A8, S100A9, S100A6, and S100P, and few other genes such as ASS1 and SERPINB3 were significantly upregulated in positive symptomatic patients when compared to other three categories (NA, NS, PA), suggesting their potential diagnostic and prognostic value (Fig. 3b, NS-PS). Expression of neutrophil defensin alpha 3 (DEFA3) was upregulated in some of the positive symptomatic patients but remained undetermined in many cases. Furthermore, we examined the influence of age and sex on the upregulation of selected genes in patient samples by categorizing them based on age groups [30-40, 41-50 and 51-60] and gender (male and female) (Fig. 3c, Fig. 3d, see Supplementary Fig 4 and Supplementary Fig 5). The qRT-PCR data revealed that all the selected genes were induced in positive symptomatic patients, irrespective of age or gender. However, closer examination of the heatmap reveals S100 family genes (S100A8, S100A9, and S100P) being upregulated to a higher level in the 30–40-year age group and male individuals (Fig. 3c, 3d).
3.4. ROC analysis of mRNA expression of shortlisted significant genes in the COVID-19 cohort unveils the prognostic potential of the S100 family of genes
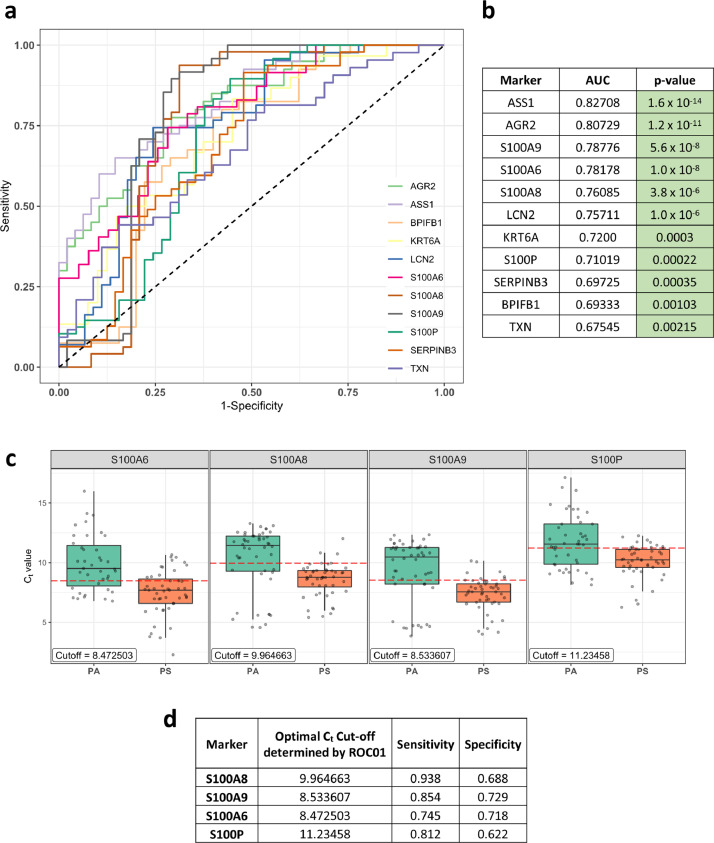
The COVID-19 symptomatic group of patients included individuals with breathing difficulty, fever, hospitalization, and SARI (severe acute respiratory infections), whereas asymptomatic patients had none of these features (Table 1). To evaluate the prognostic value of selected genes in differentiating asymptomatic vs symptomatic COVID-19 cases, we conducted a non-parametric ROC curve analysis [38] for the 11 genes that were significant after comparison between positive symptomatic and asymptomatic groups (Fig. 3b, PA-PS). For this, we used their Ct values for COVID-19 positive cases to plot the curve and the area under the curve (AUC) was computed (Fig. 4a). All genes were found to significantly differ (AUC > 0.5) from the line where True positive rate = False positive rate, indicating their potential to differentiate between asymptomatic and symptomatic individuals (Fig. 4b). The optimal Ct value cut-off was determined for significant genes using the ROC01 method which finds the point in the ROC curve closest to (0,1) corresponding to 100% specificity and sensitivity. Since the prognostic marker should correctly identify symptomatic patients from asymptomatic ones, we looked at the genes with maximum sensitivity while not compromising on specificity at the optimal cut-off. S100A8 (Cut-off = 9.964663, Sensitivity = 0.938, Specificity = 0.688) had the highest sensitivity at the optimal cut-off. Other S100 family members like S100A9 (Cut-off = 8.533607, Sensitivity = 0.854, Specificity = 0.729), S100A6 (Cut-off =8.472503, Sensitivity = 0.745, Specificity = 0.718) and S100P (Cut-off = 11.23458, Sensitivity = 0.812, Specificity = 0.622) also showed good prognostic potential (Fig. 4c and 4d). Genes like LCN2 (Cut-off = 11.23362, Sensitivity = 0.744, Specificity = 0.756), AGR2 (Cut-off = 11.19266, Sensitivity = 0.775, Specificity = 0.708) and ASS1 (Cut-off = 12.70913, Sensitivity = 0.7, Specificity =0.771) were also found to have desired sensitivity and specificity values (see Supplementary Fig 6).
Fig. 4.
ROC analysis of genes in COVID-19 positive patients to identify prognostic markers. a) ROC curve for Ct value of genes in COVID-19 positive patients. The black dashed line corresponds to no prognostic potential where True positive rate (Sensitivity) and False positive rate (1-Specificity) are equal. b) The AUC value for each ROC curve along with the p-value (calculated non-parametrically (DeLong's estimate) using the Wald test statistic). c) Boxplot of Ct values (technical replicates) for significant S100 family of genes in Positive asymptomatic (PA) and Positive symptomatic (PS) patients. The red dashed line shows the optimal Ct cut-off determined by the ROC01 method (also shown in the label in each graph). d) Optimal Ct cut-off, sensitivity, and specificity values for significant S100 family of genes.
3.5. Thioredoxin reductase inhibitor drug Auranofin significantly mitigates SARS-CoV-2 replication in vitro, and in vivo in the hamster challenge model
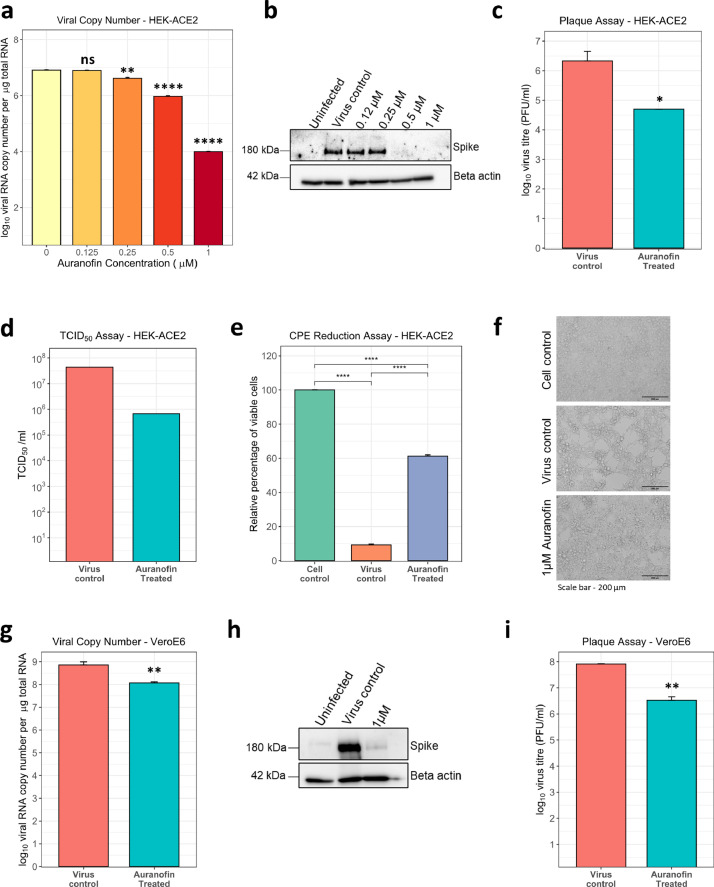
Thioredoxin (TXN) was a single hit that appeared in the TT-TP-PP overlap in our study and remained in the shortlisted gene set at the end of the meta-analysis. Although its expression upregulation or the prognostic value was not the highest, it is part of a druggable pathway. Thioredoxin is known to promote inflammatory cytokine induction, apoptosis, and regulate redox status, for which it switches between oxidized and reduced forms through the action of thioredoxin reductase, which can be inhibited by an FDA approved orphan drug Auranofin (2,3,4,6-tetra-o-acetyl-L-thio-β-D-glycopyranp-sato-S-(triethyl-phosphine)-gold) [15, 45, 46]. We sought to check the effect of Auranofin on SARS-CoV-2 infection and replication in cell culture and animal models. To begin, cell viability assay performed in HEK-ACE2 and VeroE6 cells using increasing doses of Auranofin showed minimal cytotoxicity at the lowest concentration (1 µM) and had predicted CC50 of 9.659µM (see Supplementary Fig. 7a, 7b). The effects of increasing doses of Auranofin, up to 1 µM (∼0.67 mg/L of media), was then tested on SARS-CoV-2 replication in vitro. For this, HEK ACE2 cells were pretreated with the drug, which remained present during the entire course of infection. Analysis of viral RNA 48 hr post-infection showed a reduction of more than one order of magnitude, starting at treatment with 0.25 µM Auranofin (Fig. 5a). With a calculated EC50 = 0.29 µM, the selectivity index (CC50/EC50) of Auranofin was determined to be 33.3. The potent antiviral effect of Auranofin was confirmed by western blot for the full-length viral spike protein (Fig. 5b). Treatment with 1 µM Auranofin showed a significant reduction of infectious virus titer in cell culture supernatants at 48 hr post-infection (Fig. 5c) and this was supported by a ∼2-log reduction by TCID50 assay (Fig. 5d). Virus-induced cytopathic effects (CPE) was also mitigated significantly and cell viability increased by ∼6 fold in the presence of Auranofin (Fig. 5e and 5f), as observed by microscopy and measured by trypan-blue exclusion assay. Furthermore, the anti-viral effect of Auranofin was also demostrated in VeroE6 cells, wherein, analysis of viral RNA 48 hr post-infection revealed ~1-log reduction in drug-treated cells (Fig. 5g). This was reflected in western blot analysis of infected cells, where we observed almost complete inhibition of viral spike protein expression (Fig. 5h). Plaque assay quantification of infectious virus particles from cell culture supernatants revealed >1 log reduction in the presence of Auranofin (Fig. 5i).
Fig. 5.
Auranofin inhibits SARS-CoV-2 replication in cell culture. a-c) HEK-ACE2 cells were pre-treated with the indicated amount of drug for 3 hr, infected with 0.1 MOI SARS-CoV-2, and incubated for 48 hr. a) Viral RNA copy number in cells was determined by qRT-PCR. b) Cell lysates were analyzed by western blot, probed for spike (180 kDa) and beta-actin (42 kDa). c) Cell culture supernatant was collected from virus control and 1µM drug-treated, and infectivity titers were measured using plaque assay. d). HEK-ACE2 cells were infected with serial dilutions of SARS-CoV-2 in the presence or absence of 1 µM Auranofin and TCID50 was estimated 48 hr post-infection. e-f) HEK-ACE2 cells were pre-treated with 1µM drug for 3 hr and then infected with SARS-CoV-2 at 0.1 MOI for 48 hr. e) Fold change of viable cells was measured by trypan blue dye exclusion and data was analyzed by normalizing the values to uninfected cell control. f) Brightfield images of representative images (scale bar - 200 µm) g-i) VeroE6 cells were pre-treated with 1µM of Auranofin for 3 hr and then infected with SARS-CoV-2 at 0.001 MOI for 48 hr. g) Viral RNA copy number was measured by qRT-PCR. h) Cell lysates were analyzed by western blot and probed for spike (180 kDa) and beta-actin (42 kDa). i) Infectivity titers were measured by plaque assay with cell culture supernatant. For all comparisons, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns – not significant (using t-test with Bonferroni corrections for multiple comparisons wherever necessary). Error bars represent mean + standard error.
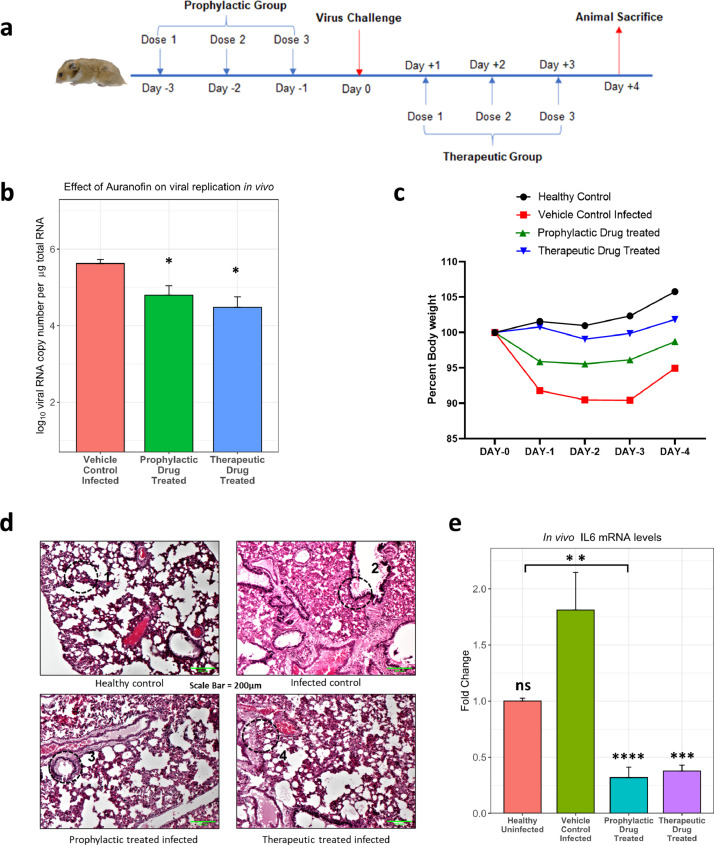
Next, we proceeded to confirm the antiviral activity of Auranofin in Syrian golden hamsters, which are currently considered as the animal model of choice to evaluate vaccines and antivirals [36]. Auranofin (PubChem CID 6333901) toxicity and bioavailability in rodents have been described before [47], based on which we first tested its oral toxicity in hamsters at 1mg/kg and 5mg/kg body weight, which showed the drug was well tolerated at the tested doses (see Supplementary Fig 8). For infection studies, the drug was orally administered in prophylactic and therapeutic formats; before and after infection, respectively (Fig. 6a). The viral titers in lungs of animals at Day 4 revealed that both prophylactic and therapeutic administration of Auranofin with a non-toxic concentration of 5mg/kg body weight was more effective at mitigating virus replication in lung tissue, compared to the vehicle control group (Fig. 6b). Bodyweight loss results were also indicative of the same when compared to the virus challenge group (Fig. 6c). Also, we found that the TXN gene was upregulated in cell culture as well as in the lungs of infected animals compared to the mock group, which correlates to our findings from patient sample gene expression data (Fig. S9). Examination of H&E stained histological sections of the lung showed evident damage of alveolar epithelial lining and cellular infiltration in infected animal lungs. The lung damage and inflammation were clearly reduced in the case of both therapeutic as well as prophylactically treated animals (Fig. 6d). Furthermore, TXN has been reported to increase proinflammatory cytokine induction [48], especially IL-6, which is a well-established mediator of COVID-19 severity [49]. We tested the effect of Auranofin on IL-6 production in infected hamster lungs and found it to be significantly diminished (Fig. 6e). This is likely to be a mechanism of action of Auranofin against SARS-CoV-2 infection and disease.
Fig. 6.
Auranofin inhibits SARS-CoV-2 replication in the preclinical hamster challenge model. a) Scheme for animal experiments involving 10–12-week-old hamsters (n=4). b) Total RNA was isolated from the lung tissue of infected animals and viral RNA copy number was measured by qRT-PCR. c) Body weight of hamsters was measured from D0 to D4, considering weight on D0 as 100% (n=4). Differences between test groups and control groups were computed using the t-test with Bonferroni corrections for multiple comparisons. d) H&E-stained section of lung tissue from indicated group of animals. The areas marked in the circle show following (1) Normal healthy alveolar lining and morphology (2) Alveolar damage, cellular infiltration, inflammation (3,4) Protected alveolar morphology and reduced infiltration, inflammation. (scale bar - 200µm). e) IL-6 mRNA levels were determined by qRT-PCR from total RNA isolated from the lungs of hamsters. Comparisons against the "vehicle control infected" infected groups are shown above bar plots. The bracket within the figure shows other significant comparisons. Differences were computed using the Kruskal-Wallis test followed by a post hoc Dunn's test with Bonferroni corrections for multiple comparisons. n=4 for hamster experiments except in healthy uninfected group where n=2. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns – not significant. Error bars represent mean + standard error.
4. DISCUSSION
Several studies have analysed changes in global transcriptome and proteome in COVID-19 patient samples of various kinds [7], [8], [9], [10], [11], [12], [13]. These studies have given an overview of the biological processes that are modulated during SARS-CoV-2 infection; however, translation of this knowledge into antiviral interventions requires validation and mechanistic studies. Meta-analysis of virus-host interaction Big Data is a useful approach to narrow down key host factors and processes involved in viral replication and pathogenesis [39, 50]. In our study, we focussed on transcriptomics and proteomics data from COVID-19 positive nasal swab and BALF samples and performed an integrative analysis to identify host factors involved in SARS-CoV-2 infection and disease progression. We reasoned that changes at mRNA levels must also be manifested at the protein level to bring out phenotypic differences in the infected individuals. Hence, we designed our meta-analysis pipeline to shortlist genes that were represented in orthogonal transcriptomics as well as proteomics datasets. Expression of the genes selected through meta-analysis was examined in nasal swab/BALF samples collected for COVID-19 diagnosis from a cohort of individuals that were COVID-19 negative or positive and within those two categories either symptomatic or asymptomatic. The cohort design was to ensure the identification of genes that are overexpressed in a COVID-19 specific manner and those which indicate disease severity. The initial compilation of upregulated factors had 566 genes, of which 46 genes passed through the selection pipeline (Fig. 2b). Most of these genes turned out to be IFN regulated and among them, the major category was ribosomal proteins (RPs), including RSP3A, RPL4, RPL5, RPL18, RPL13A, RPS4X, RPL7A, RPS9, and RPS3 (Fig. 2b). RPs have been reported to be hijacked by different viruses, including SARS-CoV-2, during infection to shut off host translation and facilitate IRES-mediated translation of viral proteins [51], [52], [53]. Inspection for reported interactions between shortlisted RPs with the SARS-CoV-2 proteins revealed that nsp1, nsp8, nsp9, and nucleocapsid (N) proteins of SARS-CoV-2 are potential interactors (Fig. 2f). This suggests extensive targeting of host translational machinery by multiple SARS-CoV-2 proteins in the upper respiratory tract cells. Other shortlisted cellular proteins with reported interactions with viral proteins were NAMPT, UQCRC2, and RAB5C (Fig. 2g). These are involved in cellular processes like ATP production, NAD synthesis, and vesicular fusion respectively, all of which have been reported to be influenced during viral infections [54], [55], [56], [57], [58].
Subsequent ranking of genes based on cumulative upregulation score across different datasets, with dual support from transcriptomic and proteomic evidences, shortlisted 14 high confidence upregulated genes (Fig 2b). To confirm their upregulation during SARS-CoV-2 infection and the effect of patient age, sex, disease severity on the same, their expression was measured in a cohort of patients described earlier (Table 1). The data revealed that 11 genes were upregulated significantly in the PS category when compared to PA and hence had prognostic value. Whereas, 8 genes were upregulated when compared to the NS category, hence had diagnostic value (Fig. 3b). The data indicated higher levels of selected gene expression in younger male patients, which is consistent with previous reports of age and sex-dependent differences in COVID-19 induced gene expression and disease severity [7, 59]. Among host factors that appeared at the end of meta-analysis and validation in the COVID-19 cohort, the S100 family of genes (S100A6, S100A8, S100A9, S100A12, and S100P) emerged as a major group. An upregulation of S100 proteins is reported previously as an indication of viral or bacterial infections [43]. The extracellularly secreted S100 proteins include S100A12, S100A8, and S100A9 (see Supplementary Fig 1a), all of which have been shown to serve as a danger signal and in regulating immune response [44]. They activate NF-kB signalling through RAGE and TLR4 pathways stimulating the cells to produce proinflammatory cytokines at the site of infection [44]. Several studies have explored serum diagnostic and prognostic markers by evaluating transcriptomic and proteomic changes in mild, severe, and fatal cases of COVID-19 [60, 61]. An increase in S100A8/A9 (calprotectin) levels in serum have been correlated with severe forms of the disease [62]. Transcriptomic studies on lung tissue of fatal COVID-19 cases have also reported an upregulation in S100A12, S100A8, S100A9, and S100P in patients [63]. In our study, the ROC curve analysis of the PA and PS group qRT-PCR data showed that all shortlisted S100s (except S100A12) had significant sensitivity as a prognostic marker of symptomatic COVID-19 (Fig. 4c, d). Overall, taking our data and published information together, the S100 family of genes can be considered as reliable prognostic markers of COVID-19 infection and disease progression. Another host factor LCN2, which came up in our study was previously shown to be an important biomarker for viral infection [64], and was also reported to be upregulated in transcriptomic and proteomic studies in COVID-19 patients [65, 66]. Furthermore, Serine protease inhibitor (SERPIN) family genes SERPINB3 and SERPINB1 were present among the initially selected 46 upregulated genes. SERPINB3 was at the top of cumulative upregulation ranking (Fig. 2c) and in the COVID-19 cohort, it was significantly upregulated in the PS category. It is an inhibitor of papain-like cysteine proteases such as cathepsin [67], which is required for Spike cleavage during SARS CoV-2 entry [68]. Interestingly SERPINA1 deficiencies or mutations in populations were found to be associated with severe forms of COVID-19 [69]. Taken together, this indicates a potential antiviral role for SERPINs against SARS-CoV-2, which needs further exploration.
Finally, one gene of interest which passed the rigor of meta-analysis was TXN. Although its cumulative upregulation or prognostic values were not very high, we explored its potential as a therapeutic target. Thioredoxin is a small redox protein that plays an active role in keeping the intracellular compartment in a reduced state, which is important to prevent protein aggregation [70]. The thioredoxin system consists of three components, namely thioredoxin, thioredoxin reductase, and the reducing agent nicotinamide adenine dinucleotide phosphate (NADPH). Thioredoxin reductase is a homeostatic redox enzyme that can be inhibited by FDA-approved, gold-containing triethyl phosphine drug Auranofin [15]. This drug has been shown to have inhibitory activity against rheumatoid arthritis, cancer, HIV/AIDS, parasitic, and bacterial infections [71], albeit with side effects like diarrhea (45-50%), rashes (24%), abdominal cramping (14%), stomatitis (13%) and nausea (10%) [47]. The drug is sold under the brand name RIDAURA® in the USA and Goldar in India, where it cost INR 112 (∼1.5 USD) for 10 tablets of 3 mg, making it an economically viable option. Auranofin is a metallodrug, which may have implications in its mechanism of action. Another metallodrug, ranitidine bismuth citrate, was found to exert SARS-CoV2 antiviral activity by sequestering Zinc ions, necessary for helicase function [72]. A recent study by Rothan et al. showed Auranofin to inhibit SARS-CoV-2 in Huh-7 cells at an EC50 of 1.4µM [73]. In comparison, our data in HEK-ACE2 cells showed improved antiviral activity at much lower concentrations of the drug (EC50 of 0.29 µM or 197 µg/L of the medium; selectivity index - 33.3, versus 4.07), as evidenced by decrease in viral infectious counts, viral RNA, protein, and cytopathic effects (Fig. 5a-f). This effect was also confirmed in VeroE6 cells. In terms of gold concentrations, the EC50 value would be 13.3 µg/L which is dominated by steady-state serum gold concentrations of 300 µg/L after oral administration of 3 mg dose of auranofin and 500-700 µg/L after oral administration of 6 mg in humans [47]. Since the EC50 value can be easily achieved in humans, it hints at the translational potential of this study. Furthermore, we went on to validate the antiviral activity of Auranofin for the first time in the preclinical hamster challenge model. Results showed a significant reduction in the lung viral load and rescue of animal body weight, when the drug was orally administered, which may be attributed to the anti-inflammatory activity of the compound [74]. Notably, Auranofin has been shown to decrease proinflammatory cytokines IL-6, IL1β, and TNFα mRNA levels during SARS-CoV-2 infection in vitro, which are known mediators of disease severity [73]. In Auranofin-treated animals, lung tissue damage, cellular infiltration, and inflammation, as well as IL-6 expression, was significantly reduced compared to control infected animals. This substantiates our proposed mechanism of action of Auranofin against SARS-CoV-2. Similar immunosuppressive medications like dexamethasone and IL-6 receptor inhibitor tocilizumab are being used for COVID-19, which could aid in reducing the inflammation that leads to poor prognosis in severe COVID-19 cases [75, 76]. Furthermore, TXN mRNA levels were upregulated in cell culture, hamsters as well as nasal swabs of COVID-19 patients, which confirms it as a reliable phenotype of infection and target for therapy. Auranofin also has inhibitory effects on the PI3K/AKT/mTOR pathway [77], which is required for SARS-CoV-2 viral protein translation [78, 79]. This may also contribute to its mechanism of action, however, that needs to be further investigated.
There are a few limitations to our study, which can be alleviated with follow-up experiments. This includes a small sample size of human nasal swabs samples and limited experimentation in the hamster model. With a greater number of human samples in a larger cohort, a detailed categorization of patients (such as hospitalized vs. non-hospitalized, ICU vs. non-ICU, survived vs. deceased) can confirm the utility of S100 markers in predicting the diseases severity with high confidence. Also, with detailed experimentation in hamsters, especially with changing the Auranofin dosage and treatment intervals, a more effective regimen can be identified. Furthermore, a detailed characterization of the mode of action of Auranofin against SARS-CoV-2 needs to be conducted. Nonetheless, this study highlights the value of comprehensive analyses of Omics datasets to gain insight into infection biology and identify avenues for potential therapeutic targeting. The selected gene expression data obtained with the COVID-19 cohort reaffirmed the heterogeneity of individual immune response, the role of age, sex, and the effect of viral load, all of which are in coherence with observations made by other research groups. We especially uncover the prognostic value of S100 family genes in nasal swabs, many of which are soluble secretory factors. They can be easily tested by RT-PCR or ELISA-based methods in nasal swabs that are routinely collected for diagnostic purposes. Finally, the identification of Auranofin (already in clinical use for other medical conditions) as a drug that can be further explored as a potential COVID-19 treatment option culminates the importance of our study and meta-analysis approach in translating virus-host interaction Big Data into clinical interventions.
CONTRIBUTORS
ST conceived the study. AB, OK, RN, RSR performed the experiments. ST, AB, OK, RN, SS, RS, DS, DG analysed the data. SM, HB, MJ, DKS, AS provided patient samples. SS, AB, ST, OK, RN wrote the manuscript. AB, OK and ST have verified the underlying data. All authors read and approved the final version of the manuscript.
Declaration of Competing Interest
Dr. Tripathi and Ms. Oyahida have a patent, Indian Patent 'A gene expression signature in nasopharyngeal swab samples for highly sensitive and specific COVID-19 prognosis' pending, and a patent, Indian Patent 'Use of Auranofin and its combination with other antiviral agents for COVID-19 treatment' pending. Mr. Biji has a patent, Indian Patent 'A gene expression signature in nasopharyngeal swab samples for highly sensitive and specific COVID-19 prognosis' pending. Dr. Narayan has a patent, Indian Patent 'Use of Auranofin and its combination with other antiviral agents for COVID-19 treatment' pending. The other authors have nothing to disclose.
Acknowledgments
Acknowledgments
We thank the funding from the DBT-IISc partnership program (DBT (IED/4/2020-MED/DBT)), the Infosys Young Investigator award (YI/2019/1106), DBT-BIRAC grant (BT/CS0007/CS/02/20) and the DBT-Wellcome Trust India Alliance Intermediate Fellowship (IA/I/18/1/503613) to ST lab. AB is supported by KVPY (Kishore Vaigyanik Protsahan Yojana) fellowship from DST, India. SS is supported by PMRF (Prime Minister's Research Fellowship) from the Ministry of Education, India. We thank Prof. Umesh Varshney and Prof. K.N.Balaji for their administrative guidance.
Data sharing statement
Data that support the findings of this study (deidentified participant data, study protocol, raw data etc.) will be available from the corresponding author, Dr Shashank Tripathi (shashankt@iisc.ac.in) upon request following publication of this study.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103525.
Appendix. Supplementary materials
Reference
- 1.Alanagreh L., Alzoughool F., Atoum M. The human coronavirus disease COVID-19: its origin, characteristics, and insights into potential drugs and its mechanisms. Pathogens. 2020;9(5) doi: 10.3390/pathogens9050331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paces J., Strizova Z., Smrz D., Cerny J. COVID-19 and the immune system. Physiol Res. 2020;69(3):379–388. doi: 10.33549/physiolres.934492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vepa A., Bae J.P., Ahmed F., Pareek M., Khunti K. COVID-19 and ethnicity: a novel pathophysiological role for inflammation. Diabetes Metab Syndr. 2020;14(5):1043–1051. doi: 10.1016/j.dsx.2020.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samuel C.E. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14(4):778–809. doi: 10.1128/CMR.14.4.778-809.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z. Activation and evasion of type i interferon responses by SARS-CoV-2. Nat Commun. 2020;11(1):3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mangalmurti N., Hunter C.A. Cytokine storms: understanding COVID-19. Immunity. 2020;53(1):19–25. doi: 10.1016/j.immuni.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lieberman N.A.P., Peddu V., Xie H., Shrestha L., Huang M.L., Mears M.C. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLoS Biol. 2020;18(9) doi: 10.1371/journal.pbio.3000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grant R.A., Morales-Nebreda L., Markov N.S., Swaminathan S., Querrey M., Guzman E.R. Circuits between infected macrophages and t cells in SARS-CoV-2 pneumonia. Nature. 2021 doi: 10.1038/s41586-020-03148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27(6):883–90e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivera B., Leyva A., Portela M.M., Moratorio G., Moreno P., Duran R. Quantitative proteomic dataset from oro- and naso-pharyngeal swabs used for COVID-19 diagnosis: detection of viral proteins and host's biological processes altered by the infection. Data Brief. 2020;32 doi: 10.1016/j.dib.2020.106121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akgun E., Tuzuner M.B., Sahin B., Kilercik M., Kulah C., Cakiroglu H.N. Proteins associated with neutrophil degranulation are upregulated in nasopharyngeal swabs from SARS-CoV-2 patients. PLoS One. 2020;15(10) doi: 10.1371/journal.pone.0240012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maras JS, Sharma S, Bhat A, Rooge S, Aggrawal R, Gupta E. Multi-omics analysis of respiratory specimen characterizes baseline molecular determinants associated with SARS-CoV-2 outcome. iScience. 2021;24(8):102823. doi: 10.1016/j.isci.2021.102823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gromer S., Arscott L.D., Williams C.H., Jr., Schirmer R.H., Becker K. Human placenta thioredoxin reductase. isolation of the selenoenzyme, steady state kinetics, and inhibition by therapeutic gold compounds. J Biol Chem. 1998;273(32):20096–20101. doi: 10.1074/jbc.273.32.20096. [DOI] [PubMed] [Google Scholar]
- 16.Izda V., Jeffries M.A., Sawalha A.H. COVID-19: a review of therapeutic strategies and vaccine candidates. Clin Immunol. 2021;222 doi: 10.1016/j.clim.2020.108634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021 doi: 10.1016/j.cell.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh T.U., Parida S., Lingaraju M.C., Kesavan M., Kumar D., Singh R.K. Drug repurposing approach to fight COVID-19. Pharmacol Rep. 2020;72(6):1479–1508. doi: 10.1007/s43440-020-00155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Case J.B., Bailey A.L., Kim A.S., Chen R.E., Diamond M.S. Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology. 2020;548:39–48. doi: 10.1016/j.virol.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J. The pride database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–DD50. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H., McGarvey P.B., Suzek B.E., Mazumder R., Zhang J., Chen Y. A comprehensive protein-centric id mapping service for molecular data integration. Bioinformatics. 2011;27(8):1190–1191. doi: 10.1093/bioinformatics/btr101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.L. S, ISoMaM. S. GeneOverlap: test and visualize gene overlaps. R package version 1.26.0 ed2020.
- 24.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wickham H. Elegant graphics for data analysis. Springer-Verlag; New York: 2016. ggplot2. [Google Scholar]
- 26.Rusinova I., Forster S., Yu S., Kannan A., Masse M., Cumming H. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41(Database issue):D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.UniProt C. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–DD15. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thul P.J., Akesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H. A subcellular map of the human proteome. Science. 2017;356(6340) doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 29.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A. Proteomics. tissue-based map of the human proteome. Science. 2015;347(6220) doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 30.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N. Impaired type i interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369(6504):718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., Hou G., Zhou H., Wang Y., Tun H.M., Zhu A. Multi-platform omics analysis reveals molecular signature for COVID-19 pathogenesis, prognosis and drug target discovery. Signal Transduct Target Ther. 2021;6(1):155. doi: 10.1038/s41392-021-00508-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reed L.J., Muench H. A simple method of estimating fifty-percent endpoints. Am J Epidemiol. 1938;27(3):493–497. [Google Scholar]
- 34.Liesenborghs L., Spriet I., Jochmans D., Belmans A., Gyselinck I., Teuwen L.A. Itraconazole for COVID-19: preclinical studies and a proof-of-concept randomized clinical trial. EBioMedicine. 2021;66 doi: 10.1016/j.ebiom.2021.103288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du S., Cao Y., Zhu Q., Yu P., Qi F., Wang G. Structurally resolved SARS-CoV-2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy. Cell. 2020;183(4):1013–23e13. doi: 10.1016/j.cell.2020.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan J.F., Zhang A.J., Yuan S., Poon V.K., Chan C.C., Lee A.C. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020;71(9):2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 38.Goksuluk. D., Korkmaz. S., Zararsiz. G., Karaagaoglu A.E. easyROC: an interactive web-tool for roc curve analysis using r language environment. The R Journal. 2016;8:213–230. [Google Scholar]
- 39.Bushman F.D., Malani N., Fernandes J., D'Orso I., Cagney G., Diamond T.L. Host cell factors in hiv replication: meta-analysis of genome-wide studies. PLoS Pathog. 2009;5(5) doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim Biophys Acta Mol Basis Dis. 2020;1866(10) doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–DD13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donato R., Cannon B.R., Sorci G., Riuzzi F., Hsu K., Weber D.J. Functions of S100 proteins. Curr Mol Med. 2013;13(1):24–57. [PMC free article] [PubMed] [Google Scholar]
- 44.Xia C., Braunstein Z., Toomey A.C., Zhong J., Rao X. S100 proteins as an important regulator of macrophage inflammation. Front Immunol. 2017;8:1908. doi: 10.3389/fimmu.2017.01908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holmgren A., Bjornstedt M. Thioredoxin and thioredoxin reductase. Methods Enzymol. 1995;252:199–208. doi: 10.1016/0076-6879(95)52023-6. [DOI] [PubMed] [Google Scholar]
- 46.Sido B., Giese T., Autschbach F., Lasitschka F., Braunstein J., Meuer S.C. Potential role of thioredoxin in immune responses in intestinal lamina propria t lymphocytes. Eur J Immunol. 2005;35(2):408–417. doi: 10.1002/eji.200424500. [DOI] [PubMed] [Google Scholar]
- 47.McEvoy GKASoH-SP. American Society of Health-System Pharmacists; 2008. AHFS drug information 2008. [Google Scholar]
- 48.Schenk H., Vogt M., Droge W., Schulze-Osthoff K. Thioredoxin as a potent costimulus of cytokine expression. J Immunol. 1996;156(2):765–771. [PubMed] [Google Scholar]
- 49.Tufan A., Avanoglu Guler A., Matucci-Cerinic M. COVID-19, immune system response, hyperinflammation and repurposing antirheumatic drugs. Turk J Med Sci. 2020;50(SI-1):620–632. doi: 10.3906/sag-2004-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tripathi S., Pohl M.O., Zhou Y., Rodriguez-Frandsen A., Wang G., Stein D.A. Meta- and Orthogonal integration of influenza "OMICs" data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015;18(6):723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dong H.J., Zhang R., Kuang Y., Wang X.J. Selective regulation in ribosome biogenesis and protein production for efficient viral translation. Arch Microbiol. 2020 doi: 10.1007/s00203-020-02094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fukushi S., Okada M., Stahl J., Kageyama T., Hoshino F.B., Katayama K. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis c virus. J Biol Chem. 2001;276(24):20824–20826. doi: 10.1074/jbc.C100206200. [DOI] [PubMed] [Google Scholar]
- 53.Schubert K., Karousis E.D., Jomaa A., Scaiola A., Echeverria B., Gurzeler L.A. SARS-CoV-2 nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol. 2020;27(10):959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- 54.Fu X., Jiang X., Chen X., Zhu L., Zhang G. The differential expression of mitochondrial function-associated proteins and antioxidant enzymes during bovine herpesvirus 1 infection: a potential mechanism for virus infection-induced oxidative mitochondrial dysfunction. Mediators Inflamm. 2019;2019 doi: 10.1155/2019/7072917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Derakhshan M., Willcocks M.M., Salako M.A., Kass G.E.N., Carter M.J. Human herpesvirus 1 protein US3 induces an inhibition of mitochondrial electron transport. J Gen Virol. 2006;87(Pt 8):2155–2159. doi: 10.1099/vir.0.81949-0. [DOI] [PubMed] [Google Scholar]
- 56.Garten A., Schuster S., Penke M., Gorski T., de Giorgis T., Kiess W. Physiological and pathophysiological roles of nampt and nad metabolism. Nat Rev Endocrinol. 2015;11(9):535–546. doi: 10.1038/nrendo.2015.117. [DOI] [PubMed] [Google Scholar]
- 57.Ziegler C.M., Bruce E.A., Kelly J.A., King B.R., Botten J.W. The use of novel epitope-tagged arenaviruses reveals that rab5c-positive endosomal membranes are targeted by the lcmv matrix protein. J Gen Virol. 2018;99(2):187–193. doi: 10.1099/jgv.0.001004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heer C.D., Sanderson D.J., Voth L.S., Alhammad Y.M.O., Schmidt M.S., Trammell S.A.J. Coronavirus infection and parp expression dysregulate the nad metabolome: an actionable component of innate immunity. J Biol Chem. 2020;295(52):17986–17996. doi: 10.1074/jbc.RA120.015138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O'Driscoll M., Ribeiro Dos Santos G., Wang L., Cummings D.A.T., Azman A.S., Paireau J. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature. 2021;590(7844):140–145. doi: 10.1038/s41586-020-2918-0. [DOI] [PubMed] [Google Scholar]
- 60.Velavan T.P., Meyer C.G. Mild versus severe COVID-19: laboratory markers. Int J Infect Dis. 2020;95:304–307. doi: 10.1016/j.ijid.2020.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pan F., Yang L., Li Y., Liang B., Li L., Ye T. Factors associated with death outcome in patients with severe coronavirus disease-19 (COVID-19): a case-control study. Int J Med Sci. 2020;17(9):1281–1292. doi: 10.7150/ijms.46614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen L., Long X., Xu Q., Tan J., Wang G., Cao Y. Elevated serum levels of S100A8/A9 and HMGB1 at hospital admission are correlated with inferior clinical outcomes in COVID-19 patients. Cell Mol Immunol. 2020;17(9):992–994. doi: 10.1038/s41423-020-0492-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu M., Chen Y., Xia H., Wang C., Tan C.Y., Cai X. Transcriptional and proteomic insights into the host response in fatal COVID-19 cases. Proc Natl Acad Sci U S A. 2020;117(45):28336–28343. doi: 10.1073/pnas.2018030117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bogorodskaya M., Fitch K.V., Burdo T.H., Maehler P., Easly R.M., Murray G.R. Serum lipocalin 2 (Neutrophil gelatinase-associated lipocalin) in relation to biomarkers of inflammation and cardiac stretch during activation of the renin-angiotensin-aldosterone system in human immunodeficiency virus. J Infect Dis. 2019;220(9):1420–1424. doi: 10.1093/infdis/jiz346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li G., Wang J., He X., Zhang L., Ran Q., Xiong A. An integrative analysis identifying transcriptional features and key genes involved in COVID-19. Epigenomics. 2020;12(22):1969–1981. doi: 10.2217/epi-2020-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeng H.L., Chen D., Yan J., Yang Q., Han Q.Q., Li S.S. Proteomic characteristics of bronchoalveolar lavage fluid in critical COVID-19 patients. Febs J. 2020 doi: 10.1111/febs.15609. [DOI] [PubMed] [Google Scholar]
- 67.Schick C., Pemberton P.A., Shi G.P., Kamachi Y., Cataltepe S., Bartuski A.J. Cross-class inhibition of the cysteine proteinases cathepsins K, L, and s by the serpin squamous cell carcinoma antigen 1: a kinetic analysis. Biochemistry. 1998;37(15):5258–5266. doi: 10.1021/bi972521d. [DOI] [PubMed] [Google Scholar]
- 68.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with sars-cov. Nat Commun. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang C., Chapman K.R., Wong A., Liu M. alpha1-Antitrypsin deficiency and the risk of COVID-19: an urgent call to action. Lancet Respir Med. 2021 doi: 10.1016/S2213-2600(21)00018-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.May H.C., Yu J.J., Guentzel M.N., Chambers J.P., Cap A.P., Arulanandam BP. Repurposing auranofin, ebselen, and PX-12 as antimicrobial agents targeting the thioredoxin system. Front Microbiol. 2018;9:336. doi: 10.3389/fmicb.2018.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roder C., Thomson MJ. Auranofin: repurposing an old drug for a golden new age. Drugs R D. 2015;15(1):13–20. doi: 10.1007/s40268-015-0083-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuan S., Wang R., Chan J.F., Zhang A.J., Cheng T., Chik K.K. Metallodrug ranitidine bismuth citrate suppresses SARS-CoV-2 replication and relieves virus-associated pneumonia in syrian hamsters. Nat Microbiol. 2020;5(11):1439–1448. doi: 10.1038/s41564-020-00802-x. [DOI] [PubMed] [Google Scholar]
- 73.Rothan H.A., Stone S., Natekar J., Kumari P., Arora K., Kumar M. The FDA-approved gold drug auranofin inhibits novel coronavirus (SARS-COV-2) replication and attenuates inflammation in human cells. Virology. 2020;547:7–11. doi: 10.1016/j.virol.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim N.H., Lee M.Y., Park S.J., Choi J.S., Oh M.K., Kim I.S. Auranofin blocks interleukin-6 signalling by inhibiting phosphorylation of JAK1 and STAT3. Immunology. 2007;122(4):607–614. doi: 10.1111/j.1365-2567.2007.02679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Group R.C. Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet. 2021;397(10285):1637–1645. doi: 10.1016/S0140-6736(21)00676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Group R.C., Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L. Dexamethasone in hospitalized patients with covid-19. N Engl J Med. 2021;384(8):693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li H., Hu J., Wu S., Wang L., Cao X., Zhang X. Auranofin-mediated inhibition of PI3K/AKT/mTOR axis and anticancer activity in non-small cell lung cancer cells. Oncotarget. 2016;7(3):3548–3558. doi: 10.18632/oncotarget.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Karam B.S., Morris R.S., Bramante C.T., Puskarich M., Zolfaghari E.J., Lotfi-Emran S. mTOR inhibition in COVID-19: a commentary and review of efficacy in rna viruses. J Med Virol. 2021;93(4):1843–1846. doi: 10.1002/jmv.26728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Terrazzano G., Rubino V., Palatucci A.T., Giovazzino A., Carriero F., Ruggiero G. An open question: is it rational to inhibit the mTor-Dependent pathway as COVID-19 therapy? Front Pharmacol. 2020;11:856. doi: 10.3389/fphar.2020.00856. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.