Abstract
Eukaryotes often form symbioses with microorganisms. Among these, associations between plants and nitrogen-fixing bacteria are responsible for the nitrogen input into various ecological niches. Plants of many different families have evolved the capacity to develop root or stem nodules with diverse genera of soil bacteria. Of these, symbioses between legumes and rhizobia (Azorhizobium, Bradyrhizobium, Mesorhizobium, and Rhizobium) are the most important from an agricultural perspective. Nitrogen-fixing nodules arise when symbiotic rhizobia penetrate their hosts in a strictly controlled and coordinated manner. Molecular codes are exchanged between the symbionts in the rhizosphere to select compatible rhizobia from pathogens. Entry into the plant is restricted to bacteria that have the “keys” to a succession of legume “doors”. Some symbionts intimately associate with many different partners (and are thus promiscuous), while others are more selective and have a narrow host range. For historical reasons, narrow host range has been more intensively investigated than promiscuity. In our view, this has given a false impression of specificity in legume-Rhizobium associations. Rather, we suggest that restricted host ranges are limited to specific niches and represent specialization of widespread and more ancestral promiscuous symbioses. Here we analyze the molecular mechanisms governing symbiotic promiscuity in rhizobia and show that it is controlled by a number of molecular keys.
Studies of nitrogen-fixing symbioses began in Europe, largely on Northern hemisphere plants. Thus, in 1542, the German physician and botanist Leonhard Fuchsius (88) published drawings of nodulated legumes. During the 17th century, Malpighi (167), who worked mostly in Bologna, Italy, observed nodules on the roots of beans (Phaseolus vulgaris and Vicia faba, members of the Leguminosae). Almost 200 years later, the Russian botanist Woronin (280) noted that the nodules of Alnus glutinosa (Betulaceae) and Lupinus mutabilis (Leguminosae) were filled with minute bodies resembling bacteria. Although the observations that both legumes and nonlegumes possess nodules were historically important, the origin of nodules was controversial (86). Frank (85) found nodules on the roots of all healthy legumes in a study in Germany and demonstrated that incinerating soil prevented the nodulation of Pisum sativum. Hellriegel (120), and Hellriegel and Wilfarth (121) showed, in a study in Germany, that nodule formation results from external infection of Lupinus spp., P. vulgaris, P. sativum, Ornithopus sativa, Trifolium spp., and Vicia sativa. However, it was Beyerinck (13–17), working in The Netherlands, who furnished the first proof that bacteria provoke nodules; he did this by preparing pure cultures of nodule organisms from V. faba and using them to infect Faba beans growing in sterile soil (18). Prazmowski (204, 205), working in Poland, inoculated P. sativum with pure cultures and showed that the bacteria penetrate legumes via infection threads in root hairs. In fact, it is now clear that many diverse soil bacteria harbor symbiotic loci (28), including the genera Azorhizobium, Bradyrhizobium, Mesorhizobium, and Rhizobium. Collectively, they are called rhizobia.
Root hairs, which develop near the root apex, are unicellular extensions of root epidermal cells. Most root hairs emerge at the apical end of particular epidermal cells and elongate by polar growth of the tip (43). The tips of root hairs have high exocytotic and cell wall assembly activities (221). In the soil, they are extensively colonized by soil-borne microorganisms, including nitrogen-fixing bacteria. In the rhizosphere, young, growing root hairs play an important role in symbiotic recognition. Rhizobia and the Nod factors they secrete (for reviews, see references 42, 63, 115, 173, 240, and 256) stimulate the reorientation of root hair cell wall growth (47, 139), resulting in curled root hairs. Within these curled root hairs, Nod factors promote the formation of infection threads, and it is through these tubular structures that the bacteria enter most (but not all) plants (Fig. 1 and 2).
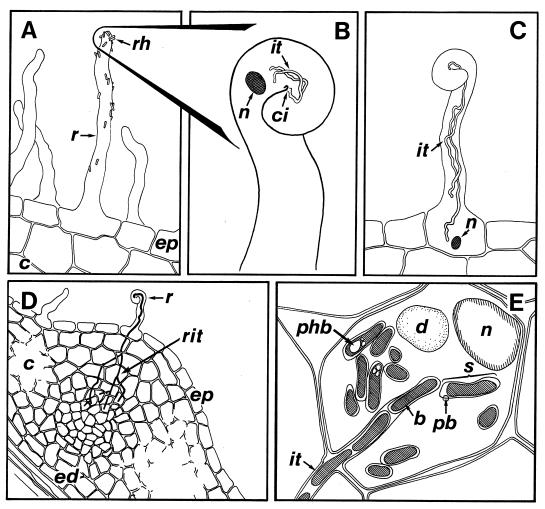
FIG. 1.
Invasion of legume root hairs by Rhizobium. (A) Rhizobia (rh) colonize the rhizosphere and attach to the root hairs (r). (B) Opening the “outer door.” Nod factors induce root hair curling and permit bacterial penetration at the center of infection (ci). The plant nucleus (n) precedes the growing infection thread(s) (it). (C) Crossing the inner doors. Still accompanied by the nucleus (n) an elongating infection thread (it) reaches the base of the root hair cell. (D) A developing infection thread ramifies (rit) near the nodule primordia formed by dividing cortical cells. (E) Bacteroids (b) are released from the infection thread (it) and form symbiosomes (s) in nodule cells. Granules of poly-β-hydroxybutarate (phb) accumulate in bacteroids surrounded by the peribacteroid membrane (pb). Other abbreviations: c, cortex; d, digestive vacuole; ep, epidermis; ed, endodermis.
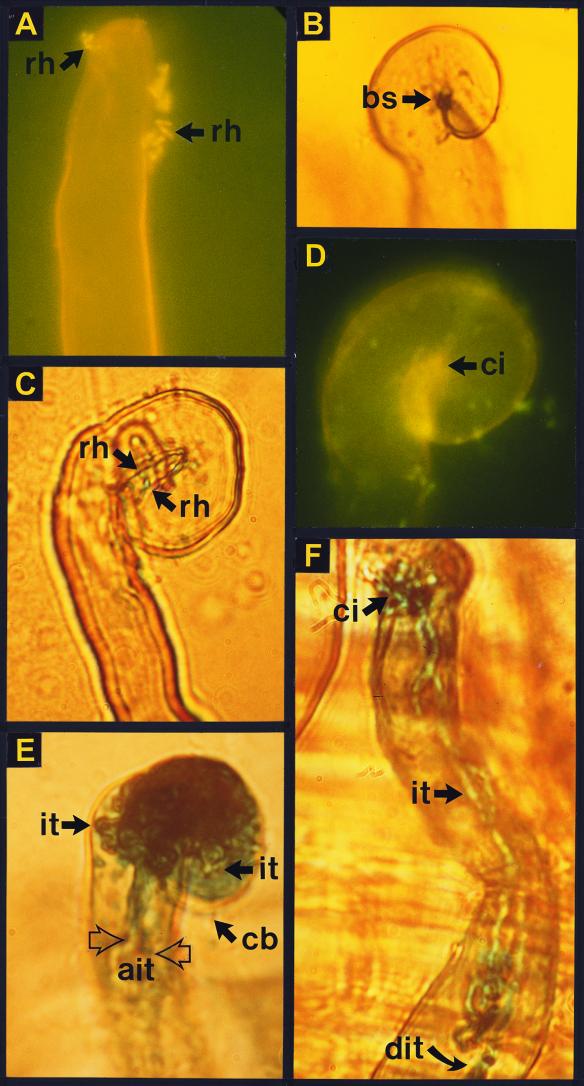
FIG. 2.
Early steps in nodulation of legumes showing that continued development of infection threads is under the control of the nodD1 gene in Rhizobium sp. NGR234. (A) Fluorescent image of a Vigna unguiculata root hair inoculated with a mutant incapable of producing NodNGR factors (NGRΔnodABC) and concomitantly treated with 10−7 M NodNGR(S) factors. Arrows point to rhizobia (rh). (B) Commencement of curling of a root hair stained with methylene blue. Extreme curling leads to the formation of a bright spot (bs), where rhizobia are often entrapped (same treatment as in panel A). (C) A bright-field image of a root hair inoculated with NGRΔnodABC::GUS3, treated with 10−7 M NodNGR(S) factors, and stained for β-glucuronidase activity. Arrows point to the entrapped rhizobia within the curl. (D) Fluorescent image of a root hair curled in the shape of a shepherd's crook, showing the center of infection (ci). (E) Experiment in which the nodABC mutant was replaced with NGRΔnodD1::GUS3, but incubated with 10−7 M NodNGR(S) factors, and stained for β-glucuronidase activity. Apparently, the nodD1 mutant lacks a factor(s) that is necessary for the continued development of infection threads (it). In its absence, infection threads abort (ait), forming a structure that resembles a cerebellum (cb). (F) Photomicrograph of root hairs inoculated with wild-type NGR234 marked with GUS3. Infection threads develop along the length of the root hair (dit). B. Relić and W. J. Broughton (unpublished results; see reference 215 for further details).
Work on the molecular basis of host specificity also began at the end of the last century. In a study in Germany, Hiltner (123) prepared aqueous, bacterium-free filtrates from mature P. sativum nodules and demonstrated that they contain a substance that induces root hair formation (Hai) and deformation of the root hairs (Had) in this plant. Many attempts to define the structure of these deformation factors followed (213), but these investigations yielded fruit only in 1990, when Lerouge et al. (154), working in France, showed that the substances responsible are N-acylated oligomers of N-acetyl-d-glucosamine. Since then, the Nod factor structures (the products of nod genes) of a number of rhizobia have been elucidated (see “Baroque decorations to the Nod factor core” below).
In other words, research on symbiotic nitrogen fixation was concentrated on European legumes, and although this has become less so with time, the focus on genera such as Medicago, Pisum, Trifolium, and Vicia has prevailed. Of course, model plants have advantages, but it is our contention that this bias toward European plants for historic reasons may have given a false impression of symbiotic specificity. What follows is an analysis of the molecular mechanisms governing broad host range (i.e., promiscuity) in rhizobia that may be regarded as a paradigm for other microbial symbioses. Readers are referred to the reviews by Irving et al. (128) and Downie and Walker (64) for more information on the role of the plant in nodulation.
BACTERIAL PROMISCUITY
Doubts that bacteria were faithful to the legume from which they were isolated also surfaced early. In experiments that are probably unrepeatable (Table 1), Bréal (25) induced nodule formation on Lupinus, Medicago sativa, and P. sativum by bacteria isolated from nodules of alfalfa (M. sativa). In similar, questionable experiments, Laurent (149) suggested that nodules could be produced on P. sativum by bacteria isolated from 36 species of plants. Perhaps the first reproducible data of this kind was obtained by Nobbe et al. (183, 184), who showed in experiments performed in Germany at the end of the 19th century that bacteria from P. sativum nodules were unable to nodulate plants of the tribes Genisteae and Hedysareae. Indeed, rhizobia which nodulate Lupinus, Medicago, Pisum, Vicia, etc., are now known to have restricted host ranges (Table 1).
TABLE 1.
Nodulation capacities of symbiotic members of the bacterial family Rhizobiaceaea
| Species | Typical host(s) | Host range | Examples of nonhosts | Reference(s) |
|---|---|---|---|---|
| Azorhizobium caulinodans | Robinieae (P), Sesbania spp. | Compatible only with Sesbania punctata and S. rostrata | MIMOSOIDEAE, PAPILIONOIDEAE (except Sesbania) | M. Holsters, personal communication |
| Bradyrhizobium spp. | Genisteae (P), Lupinus spp. | Acacieae (M), Mimoseae (M), Desmodieae (P), Loteae (P), Psoraleeae (P), Phaseoleae (P), Aeschynomeneae (P) | Vicieae (P), Cicereae (P), Trifolieae (P) | C. E. Pankhurst and C. W. Ronson, personal communication; H. Meyer z. A. and W. J. Broughton, unpublished data |
|
Lupinus isolates Vigna isolates |
Phaseoleae (P), Macroptilium, Vigna spp. | |||
| Bradyrhizobium elkanii | Phaseoleae (P), Glycine spp. | Phaseoleae (P), Macroptilium, Vigna spp. | Vicieae (P), Cicereae (P), Trifolieae (P) | G. Stacey, personal communication |
| Bradyrhizobium japonicum | Phaseoleae (P), Glycine spp. | Aeschynomeneae (P), Arachis spp., Phaseoleae (P), Macroptilium, Vigna spp. | Vicieae (P), Cicereae (P), Trifolieae (P) | G. Stacey, personal communication |
| Mesorhizobium huakuii | Astragalus sinicus | Galegeae (P), Astragalus spp. | MIMOSOIDEAE, PAPILIONOIDEAE | 282 |
| Mesorhizobium loti | Loteae (P), Lotus spp., Genisteae (P), Lupinus spp. | Mimoseae (M), Mimosa pudica, Leuceana leucocephala; Phaseoleae (P), Macroptilium atropurpureum | Vicieae (P), Cicereae (P), Trifolieae (P) | G. Stacey, personal communication |
| Rhizobium sp. strain NGR234 | Phaseoleae (P), Desmodieae (P) | Mimoseae (M), Acacieae (M), Ingeae (M), Sophoreae (P), Dalbergieae (P), Amorpheae (P), Millettieae (P), Robinieae (P), Indigofereae (P), Loteae (P), Galegeae (P), Bossiaeae (P), Mirbelieae (P), Podalyrieae (P), Crotalarieae (P), Thermopsideae (P), Genisteae (P), Psoraleeae (P) | Vicieae (P), Cicereae (P), Trifolieae (P) | 207 |
| Rhizobium etli | Phaseoleae (P), Phaseolus spp. | Ingeae (M), Crotalarieae (P), Galegeae (P), Mimoseae (M), Desmodieae (P), Robinieae (P) | Robinieae (P) | 122; E. Martínez-Romero, personal communication |
| Rhizobium fredii | Phaseoleae (P), Desmodieae (P) | Mimoseae (M), Ingeae (M), Sophoreae (P), Amorpheae (P), Millettieae (P), Robinieae (P), Indigofereae (P), Loteae (P) | Vicieae (P), Cicereae (P), Trifolieae (P) | 207 |
| Rhizobium galegae | Galegeae (P), Galega spp. | Compatible only with Galega spp. | MIMOSOIDEAE, PAPILIONOIDEAE (except Galega) | K. Lindström, personal communication |
| Rhizobium leguminosarum bv. phaseoli | Phaseoleae (P), Phaseolus spp. | Phaseoleae (P) | MIMOSOIDEAE, Trifolieae (P) | E. Martínez-Romero, personal communication |
| Rhizobium leguminosarum bv. trifolii | Trifolieae (P), Trifolium spp. | Phaseoleae (P), Desmodieae (P), Trifolieae (P), Medicago spp. | MIMOSOIDEAE, Phaseoleae (P) | H. P. Spaink, personal communication |
| Rhizobium leguminosarum bv. viciae | Vicieae (P), Pisum sativum, Vicia spp. | Lathyrus spp. (e.g., L. sativa) | MIMOSOIDEAE, Phaseoleae (P), Desmodieae (P) | J. A. Downie, personal communication |
| Rhizobium meliloti | Trifolieae (P), Medicago spp., Melilotus spp., Trigonella spp. | Acacieae (M), Desmodieae (P), Mimoseae (M), Phaseoleae (P), Vicieae (P) | J. Dénarié, personal communication | |
| Rhizobium saheli | Robinieae (P), Sesbania spp. | Acacieae (M), Mimoseae (M) | 165; C. Boivin, personal communication | |
| Rhizobium teranga bv. acaciae | Acacieae (M), Acacia spp. | Mimoseae (M), Leucaena leucocephala, Prosopis juliflora | Robinieae (P), Sesbania spp. | 165; C. Boivin, personal communication |
| Rhizobium teranga bv. sesbaniae | Robinieae (P), Sesbania spp. | Acacieae (M), Mimoseae (M) | 165; C. Boivin, personal communication | |
| Rhizobium tropici | Phaseoleae (P), Phaseolus spp. | Galegeae (P), Crotalarieae (P), Desmodieae (P), Mimoseae (M), Robinieae (P), Loteae (P) | Vicieae (P), Cicereae (P), Trifolieae (P) | E. Martinez-Romero, personal communication |
Plants are listed by their subfamilies, tribes, genera, and species. Associations are listed only where fully effective nodulation (i.e., Fix+) has been reported. Subfamily names are listed in capital letters (Caesalpinioideae [C], Mimosoideae [M], and Papilionoideae [P]) (201).
The U.S. researcher J. K. Wilson was one of the first non-Europeans to publish extensive information on different symbiotic associations (279). He tested the host range properties of rhizobia isolated from 31 genera of legumes on 160 different legume species. All isolates, including those from Vicia spp., nodulated legumes of different tribes. The average number of plant species nodulated by a particular strain was 33%, but it ranged from 6% (for an isolate from Spartium scoparium) to 66% (for a slow-growing, and therefore probably Bradyrhizobium, strain isolated from Mucuna nodules). Indeed, four of Wilson's isolates nodulated more than half the legumes tested. These data are included in Table 2 to show the patterns of nodulation provoked by broad-host-range rhizobia. Both Bradyrhizobium and Rhizobium species can be promiscuous, as shown in Table 2, and broad-host-range rhizobia were even isolated from Albizia julibrissin (Mimosoideae). Isolates from P. vulgaris, which include R. etli and R. tropici, were shown to nodulate 17 and 18 different legume genera, respectively (122). It thus seems as if rhizobia associated with most tribes of the Leguminosae have various degrees of promiscuity. The principal exceptions to this rule are the more specialized rhizobia associated with the tribes Cicereae, Trifolieae, and Viceae, which have restricted host ranges (28).
TABLE 2.
Nodulation capacities of some broad-host-range rhizobia isolated from various legumes
| Plant nodulateda | Nodulationab by rhizobia isolated from:
|
|||||
|---|---|---|---|---|---|---|
| Mucuna (P10)c | Albizia (M5) | Desmodium (P11) | Glycine (P10) | Lablab (P10) NGR234 | Sesbania (P8) | |
| Amorpheae (P6) | ||||||
| Amorpha spp. | + | + | + | + | + | + |
| Mirbelieae (P24) | ||||||
| Chorizema spp. | + | + | + | − | + | + |
| Phaseoleae (P10) | ||||||
| Centrosema spp. | + | + | + | − | + | + |
| Phaseolus spp. | + | + | + | + | + | + |
| Robinieae (P8) | ||||||
| Robinia spp. | + | + | + | + | + | + |
| Sesbania spp. | + | + | + | + | + | + |
Letters in brackets represent the subfamily to which the legume belongs (M, Mimosoideae; P, Papilionoideae), and the numbers represent the tribe (201).
+, effective (Fix+) nodulation; −, failure to nodulate (Nod−). Data for Mucuna, Albizia, Desmodium, and Sesbania rhizobia from reference 279; data for Glycine (rhizobial strain USDA257) and Lablab (rhizobial strain NGR234) rhizobia from reference 207.
We assigned this rhizobial isolate to Bradyrhizobium because of its slow growth.
LEGUME FIDELITY
As expected, hosts also vary greatly in their ability to enter into symbiosis with different rhizobia. In Wilson's study (279), six plants formed nodules with at least 90% of the isolates (Table 2). At the other extreme, Hippocrepis sp. and Ornithopus sativa formed nodules with only one other isolate. In further studies of this kind, numerous legumes have been inoculated with large collections of rhizobia and the capacity of the legumes to nodulate have been evaluated. Again, promiscuity exists among diverse legumes (Table 3). Mimosoid legumes not only harbor broad-host-range rhizobia but also are capable of interacting with diverse bacteria; Leucaena leucocephala, for example, is nodulated by a moderately broad spectrum of symbionts. The fact that Azorhizobium caulinodans, a microsymbiont of Sesbania rostrata, has a very restricted host range (Table 1) gives the impression that Sesbania spp. have very defined rhizobial requirements. However, Wilson (279) showed that S. drummondii, along with another tree (Robinia pseudoacacia), had the broadest capacity to nodulate among the legumes he tested.
TABLE 3.
Responses of known promiscuous legumes to diverse rhizobia
| Legumea | Geographical originb | % Nod+ | Reference |
|---|---|---|---|
| Mimosoideae (M3) | I | ||
| Leucaena leucocephala | Tropical America | 44 | 157 |
| Amorpheae (P6) | I | ||
| Amorpha fruticosa | North America | 91 | 279 |
| Mirbelieae (P24) | I | ||
| Chorizema ilicifolium | Southeast Australia | 91 | 279 |
| Phaseoleae (P10) | D | ||
| Centrosema virgininianum | Warm America | 91 | 279 |
| Lablab purpureus | Tropical Africa | 50 | 157 |
| Macroptilium atropurpureum | Tropical America | 41 | H. Meyer z. A. and W. J. Broughton, unpublished |
| Phaseolus coccineus | Tropical and warm America | 91 | 279 |
| Vigna unguiculata | Old World Tropical | 56 | 157 |
| Robinieae (P8) | I | ||
| Robinia pseudoacacia | Tropical and warm America | 94 | 279 |
| Sesbania drummondii | Old World Tropical | 94 | 279 |
Letters in parentheses represent the subfamily to which the legume belongs (M, Mimosoideae; P, Papilionoideae), and the numbers represent the tribe (201). D = determinate nodules; I, indeterminate nodules.
Data from reference 166. The data were taken from the reports of Wilson (279), who tested the nodulation capacity of 32 rhizobial isolates (from 31 legume genera) on the nodulation capacities of 160 species (78 genera), and Meyer z. A. and Broughton (see reference 157), who assayed the ability of 50 rhizobial isolates to nodulate 16 species (13 genera) of legumes.
However, most broad-spectrum hosts known are annual and climbing legumes of the tribe Phaseoleae. These include Macroptilium atropurpureum, which has been widely used as a test plant for nodulation by uncharacterized rhizobia (27, 248); Lablab purpureus, the legume from which Rhizobium sp. strain NGR234 was isolated (269); and P. vulgaris (175). Equally, the other plants listed in Table 3 are considered to have broad host ranges (185). The capacity of Vigna unguiculata to nodulate with many different rhizobia has been exploited in the search for nod genes (156, 157). Taken together, these data suggest that symbiotic promiscuity is widely dispersed in nature. It is not associated with a particular bacterial or plant taxonomic group and is not correlated with the growth habit of the legume. Broad-host-range legumes are also almost equally distributed between those that form indeterminate as opposed to determinate nodules (Table 3). Perhaps the only underlying similarity is that promiscuous bacteria and plants are found mostly in warm or tropical parts of the world.
MOLECULAR BASES
Nodulation leads to the colonization of plant cells by invading bacteria. Although many host plants and effective rhizobia have the ability to enter into symbiosis with more than one partner, only certain combinations of symbionts result in the formation of nitrogen-fixing nodules. Ineffective associations lead to empty or nonfixing bacteroid-containing nodules. Specificity among compatible partners minimizes the chances of infection by pathogens and the formation of ineffective associations that are detrimental to both symbionts. Experimental evidence suggests that the progression of invasive rhizobia towards nodule primordia is challenged at various “doors” (Fig. 1). Codes contained in molecular signals open these checkpoints. During the initial phases of nodulation (root hair curling and bacterial entry), these codes are given by flavonoids and Nod factors. In both cases, NodD proteins are the chief interlocutors of molecular traffic in the rhizosphere.
Flavonoids and Regulation of Nodulation Genes
Plants jettison surprisingly large amounts of organic matter into the soil, most of which supports the growth of rhizospheric microorganisms. By labelling whole plants with 14CO2, Helal and Sauerbeck (119) showed that 19% of the photosynthate was released as organic material into the rhizosphere. These compounds include carbohydrates, organic acids, vitamins, amino acids, and phenolic derivatives. Of these, flavonoids (2-phenyl-1,4-benzopyrone derivatives) are the most important from the symbiotic perspective. Although found throughout the plant kingdom, flavonoids specifically trigger the expression of the rhizobial genes required for nodulation (nod, nol, and noe). The inducing capacity varies with flavonoids and rhizobia; and in some cases flavonoids may inhibit induction (61, 79, 196, 197, 199). Other molecules, such as the betaines and erythronic and tetronic acids, may also act as inducers (90, 198).
Regulation of nod gene expression in rhizobia varies from strain to strain but is almost always mediated by NodD (for a review, see reference 237). NodD proteins belong to a family of LysR-like transcriptional regulators that bind to highly conserved 47-bp DNA motifs (nod boxes) found in the promoter regions of many nodulation loci (81, 227). Although NodD proteins bind to nod boxes even in the absence of an inducer, flavonoids are generally required for the expression of nod genes (81, 82, 101, 110). Thus, NodD acts both as a sensor of the plant signal and as an activator of transcription of nod loci (236). In R. leguminosarum bv. viciae, NodD proteins are localized in the cytoplasmic membrane (234), where the inducing flavonoid, naringenin, also accumulates (211). Direct binding of inducers to NodD has not been demonstrated, but point mutations in nodD affect the recognition of inducing molecules and cause an extension of the host range (30, 174). Furthermore, comparison of the NodD structure with various nuclear receptors has shown that the two types of proteins share conserved ligand-interacting domains located at the C-terminal end of their respective DNA-binding motifs (111).
Although nodD genes are ubiquitous in rhizobia, their symbiotic characteristics vary from one species to another. Some strains, such as R. leguminosarum bv. trifolii, have only one nodD gene, and in these cases, its mutation renders the strain incapable of nodulation (Nod−). In contrast, B. japonicum, Rhizobium sp. strain NGR234, R. meliloti, and R. tropici possess two to five copies of nodD (71, 105, 194, 269c). In R. meliloti, mutation of all three copies of nodD is required to abolish nodulation (124), whereas inactivation of nodD1 is sufficient to render strain NGR234 Nod− (213). nodD products of various Rhizobium species vary in that they respond to different sets of flavonoids (72, 249). Moreover, NodD homologues from the same strain may have different flavonoid preferences (112, 117, 198). In R. meliloti, NodD1 is activated when cells are supplied with a complex plant seed extract or the flavonoid luteolin; NodD2 is activated only with the complex extract, not with purified luteolin; and NodD3 apparently modulates the expression of nod genes even in the absence of any plant factor (180). Together with syrM, nodD3 of R. meliloti constitutes a self-amplifying positive regulatory circuit that is involved in the regulation of nod genes within the developing nodule (266). In contrast, NodD1 of strain NGR234 responds to a wide range of inducing molecules that include flavonoids known to be inhibitors in other rhizobia (e.g., vanillin and isovanillin) (4, 72, 155) and several estrogenic compounds (112). Transfer of the nodD1 gene of strain NGR234 to restricted-host-range rhizobia extends the nodulation capacity of the recipients to new hosts, including the nonlegume Parasponia andersonii (4, 11, 126). Although a number of correlations exist between the spectrum of flavonoids able to interact with NodD proteins and the breadth of the host range (11, 54, 126, 249, 250, 263, 269d), variations in the ability of these proteins to differentially sense inducing molecules (and regulate the expression of nod genes) are insufficient to explain the phenomenon of host specificity.
Nonetheless, NodD genes represent a molecular interface between the bacterium and the plant. However, other species-specific sensor-activator systems also contribute to the control of bacterial host range. For instance, nodV and nodW of B. japonicum are essential for the nodulation of M. atropurpureum, Vigna radiata, and V. unguiculata but contribute only marginally to the symbiosis with G. max (104). Modulation of two-component regulatory systems such as nodV and nodW is mediated by a series of phosphorylation steps. NodV is thought to be a membrane-bound protein that senses signals from the plant and transduces the signal to NodW, which in turn activates the expression of nod genes (230). Both in vitro and in vivo studies have confirmed that phosphorylation of NodW is induced by genistein and depends on both acetyl phosphate and its cognate kinase, NodV (159). Also, comparison of the biological activities of the wild-type and mutant proteins indicates that phosphorylation of NodW is essential for nod gene activation (159). A search for genes whose transcription is dependent on NodW led to the identification of two suppressor genes, nwsA and nwsB. Together, these genes also form a dual-component regulatory system (109). When overexpressed, nwsB can complement a nodW mutant as well as a nodD1 nodD2 double mutant, suggesting that in B. japonicum at least, alternatives to NodD- and flavonoid-sensing regulatory systems coexist (105).
nod gene expression is also subject to negative control. After initial induction by flavonoids, repression of several nodulation genes is required for optimal nodulation of M. sativa by R. meliloti (142) and of P. sativum by R. leguminosarum bv. viciae strain Tom (141). In R. meliloti, repression of several nod genes is controlled by NolR, a 13-kDa product that contains a helix-turn-helix (HTH) motif which is homologous to other regulators of the LysR family such as NodD and SyrM (143). In the dimeric form, NolR binds to conserved (A/T)TTAGN9A(T/A) target sequences found in the promoter regions it controls (46) and thus represses the expression of the nodD genes as well as those necessary for the synthesis of the core Nod factor structures (nodABC). However, it does not affect host-specific nod loci such as nodH. Interestingly, the absence of NolR repressor activity in R. meliloti stain 1021 is due to a single insertional mutation in the C-terminal coding sequence, which abolishes the DNA binding ability of the protein without affecting its HTH motif (45).
With a few exceptions, transcription of most inducible nod loci is repressed before rhizobia differentiate into functional bacteroids (235, 244), although transcription of nodD and nodE of R. leguminosarum bv. viciae in P. sativum nodules and nodA of A. caulinodans in stem nodules of S. rostrata still occurs (56, 235). R. meliloti is no exception, although NolR does not play a direct role in the down regulation of flavonoid-inducible nodulation genes in bacteroids (46). In B. japonicum, repression of nod gene expression by NolA is probably an indirect effect, mediated perhaps by nodD2 (91). B. japonicum nolA and R. meliloti nolR mutants retain the ability to nodulate their respective hosts, albeit at lower efficiency, suggesting that fine-tuning of nod gene expression is required for optimal nodulation. Similarly, a nodD2 mutant of strain NGR234 fails to repress the expression of the nodABCIJnolOnoeI operon after initial flavonoid induction and, in contrast to the wild-type strain, is unable to form nitrogen-fixing nodules on V. unguiculata and Cajanus cajan (73).
nod Boxes: Variations on a Theme
Sequence analysis showed that pNGR234a carries 19 sequences homologous to conserved NodD-dependent promoter elements (nod boxes), 5 of which control the expression of known nod operons (87, 195). No significant difference was found between the consensus sequence derived for nod boxes of pNGR234a and those published for other rhizobia (269d, 278). Interestingly, analysis of the promoter regions of the nodABCIJnolOnoeI, nodSU, nodZ, noeE, and nolL loci showed that nod boxes with up to 11 base mismatches (compared to the consensus sequence) are still functional and regulate gene expression in a NodD1-dependent manner (195). It is possible, however, that some of these nod boxes have higher affinities for certain NodD1-inducer complexes than do others.
Thus, a microsymbiont such as strain NGR234 has many possibilities for fine-tuning nod gene expression. First, NodD1 may interact with a broad spectrum of inducing molecules to produce complexes that may preferentially trigger transcription from certain nod boxes over others. Second, supplementary activators of transcription (e.g., SyrM1 [114] and y4xI [274]) could, together with NodD1, form a regulatory network which exerts interlaced control over dispersed nodulation loci. Third, repressors such as NodD2 and NolR (73) could further modulate nod gene expression. Together, these systems control the expression of NGR234 nodulation genes in a host-specific way, possibly resulting in the secretion of different sets of molecular signals in response to the macrosymbiont. Nod factors are obviously the most important of these signals, and their synthesis depends upon the timely and coordinated expression of many nod genes.
nod Enzymes and Nod Factor Synthesis
Nod factor core.
In response to the release by host plants of appropriate inducers, rhizobia synthesize and secrete a family of lipochitooligosaccharides (LCOs) called Nod factors. The first step in Nod factor assembly is performed by an N-acetylglucosaminyltransferase encoded by nodC (95). Chain elongation by NodC takes place at the nonreducing terminus (134, 135, 170). Then the deacetylase NodB removes the N-acetyl moiety from the nonreducing terminus of the N-acetylglucosamine oligosaccharides (132, 253). Finally, an acyltransferase encoded by nodA links the acyl chain to the acetyl-free C-2 carbon of the nonreducing end of the oligosaccharide (51, 226). Although not essential, NodI and NodJ seem to be involved in the export of Nod factors (34, 74, 254).
However, recent work suggests that nodA and nodC are also components of host-specific nodulation (hsn) (225). NodA varies in its specificity for different fatty acid substrates, thus contributing to the host range. For instance, replacement of R. meliloti nodA by R. tropici nodA leads to the production of Nod factors acylated with vaccenic acid instead of C16:2 (51), whereas Bradyrhizobium sp. strain ANU289 NodA is unable to direct the transfer of the R. leguminosarum bv. viciae nodFE-dependent multiunsaturated fatty acids to the chitin oligosaccharide acceptor (222). NodC is also a determinant of the length of the Nod factor backbone and thus of host specificity (133, 135).
Baroque decorations to the Nod factor core.
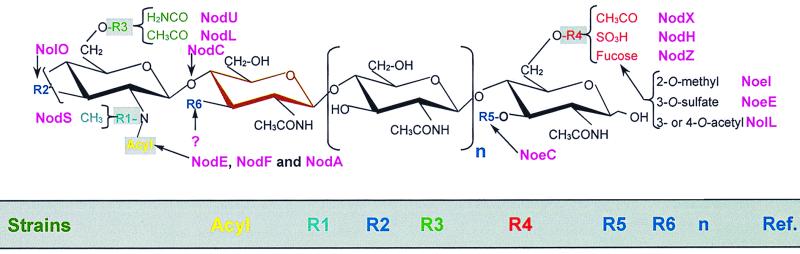
Properly expressed nodABC genes are all that is needed for the synthesis of acylated dimers to pentamers of N-acetyl-d-glucosamine that possess symbiotic activity on certain plants (3, 200, 252). Thus all other Nod factor substituents must play more subtle roles in nodulation, perhaps by permitting interaction with certain plants or by protecting Nod factors from degradation. In this sense, these substituents can be likened to baroque decorations—they enhance rather than support the basic structure (Table 4). By definition, the loci that control these additions are unique to one or a few rhizobia and are thus hsn genes.
TABLE 4.
Nod factors and their baroque decorationsa
| A. caulinodans ORS571 | C18:1, C16:0 | Me | OH | Cb, H | Fuc, H | Ara, H | OH | 1, 2 | 172 |
| B. elkanii USDA61 | C18:1 | Me, H | Cb, H | Ac, H | MeFuc | H | OH | 1, 2 | 35 |
| B. japonicum USDA110 | C16:0, C16:1, C18:1 | H | OH | H | MeFuc | H | OH | 2 | 229 |
| Rhizobium sp. strain GRH2 | C16:0, C18:0, C18:1, C20:1 | Me, H | OH | H | S, H | H | OH | 1, 2, 3 | 160 |
| Rhizobium sp. strain ORS1645 | C18:0, C18:1 | Me | Cbc | Cbc | S, H | H | OH | 2 | 164 |
| Rhizobium sp. strain NGR234 | C16:1, C18:0, C18:1 | Me | Cb, OH | Cb, H | MeFuc, AcMeFuc, SMeFuc | H | OH | 2 | 206 |
| R. etli CFN42 | C18:1 | Me | Cb, OH | H | AcFuc | H | OH | 2 | 33 |
| R. etli CE3 | C18:0, C18:1 | Me | Cb, OH | H | AcFuc | H | OH | 2 | 203 |
| R. fredii USDA191b | C18:1, C18:0, C16:1 | H | H | H | Fuc, MeFuc | H | H | 0, 1, 2 | 7 |
| R. fredii USDA257 | C18:1 | H | OH | H | Fuc, MeFuc | H | OH | 0, 1, 2 | 6 |
| R. galegae | C18:1, C18:2, C18:3, C20:2, C20:3 | H | OH | Cb | H | H | Ac | 1 | 282 |
| R. huakuiid | C18:4 | H | OH | H | S | H | OH | 2 | 282 |
| R. leguminosarum | |||||||||
| bv. trifolii ANU843 | C16:0, C16:1, C18:0, C18:1, C18:2, C20:3 | H | OH | H, Ac | H | H | OH | 0, 1, 2 | 188 |
| bv. trifolii LPR5045 | C18:0, C18:1, C20:3, C20:4 | H | OH | Ac | H | H | OH | 2 | 269b |
| bv. viciae RBL5560 | C18:1, C18:4 | H | OH | Ac | H | H | OH | 1, 2 | 252 |
| bv. viciae TOM | C18:1, C18:4 | H | OH | Ac | Ac | H | OH | 1, 2 | 80 |
| R. loti NZP2213 | C16:0, C16:1, C18:0, C18:1, C20:0, C20:1, C22:1 | H | Cb | H | Fuc, AcFuc | H | Fuc | −1, 0, 1, 2 | 187 |
| R. loti NZP2037 | C18:0, C18:1 | Me | Cb | Cb | AcFuc | H | OH | 2 | 161 |
| R. meliloti RCR2011 | C16:1, C16:2, C16:3 | H | OH | H, Ac | S | H | OH | 1, 2 | 2, 154 |
| R. tropici CFN299 | C18:1 | Me | OH | H | S, H | H | OH | 2 | 202 |
| S. saheli ORS611 | C16:0, C18:1 | Me | Cbc | Cbc | Fuc, H | Ara, H | OH | 2 | 163 |
| S. teranga bv. acaciae ORS1602 | C16:0, C18:0, C18:1 | Me | Cbc | Cbc | S, H | H | OH | 2 | 164 |
Abbreviations: Ac, acetyl; Ara, arabinosyl; Cb, carbamoyl; Fuc, fucosyl; H, hydrogen; Me, methyl; S, sulfate; MeFuc, methylfucose; AcMeFuc, acetylated methylfucose; SMeFuc, sulfated methylfucose.
In USDA191, a minor fraction of the N-acetylglucosamine marked in brown is replaced by a glucose.
Carbamoyl group is either on R2 or R3.
R. huakuii Nod factors are partially glycolylated at the C-2 position of the reducing terminus.
(i) Fatty acids (nodEF).
Two basic types of fatty acids are N-linked to the terminal nonreducing sugar of the chitomeric core by NodA (Table 4): (i) the relatively common saturated or monounsaturated fatty acids (e.g., stearic and vaccenic acids) and (ii) the rarer, highly unsaturated compounds containing two to four double bonds as found in R. leguminosarum and R. meliloti strains containing nodEF (54, 252). NodF has homology to acyl carrier proteins, while NodE is a β-acetoacetylsynthase (21, 22, 49, 245). Transfer of R. meliloti nodEFGHPQ into R. leguminosarum bv. trifolii or bv. viciae confers on these strains the ability to nodulate M. sativa but prevents nodulation of the normal hosts, Trifolium repens and V. sativa, respectively (50, 70). Inactivation of R. leguminosarum bv. viciae nodE leads to the synthesis of vaccenic acid rather than the polyunsaturated fatty acids (C18:4) produced by the parent strain (252). Insertional inactivation of R. leguminosarum bv. trifolii nodE severely inhibits the nodulation of several Trifolium species and simultaneously enhances the nodulation of P. sativum and V. sativa (59, 251). Mutation of R. leguminosarum bv. viciae nodE renders the strain Nod− on V. sativa (32). Replacement of R. meliloti nodEF with R. leguminosarum bv. viciae nodEF leads to the synthesis of Nod factors containing a polyunsaturated C18 fatty acid side chain which resembles those produced by R. leguminosarum bv. viciae (52). Thus, nodEF and the unsaturated Nod factors they produce appear to be specializations necessary for nodulation of the legume tribes Trifolieae and Vicieae.
(ii) 6-O glycosylation.
An additional sugar, which can be either arabinose or fucose, represents another type of decoration.
(a) Arabinosylation (noeC).
noeC and/or downstream genes are essential for arabinosylation of A. caulinodans Nod factors (171). In these Nod factors, the reducing terminus can be fucosylated, arabinosylated, or both (172). Perhaps this suggests that both arabinosylated and fucosylated Nod factors are necessary for nodule formation on the stems of Sesbania rostrata (163, 164). d-Arabinosylated Nod factors result in larger numbers of nodules on the roots of S. rostrata, whereas the presence or absence of l-fucose has no effect. In contrast, other hosts of A. caulinodans prefer fucosylated Nod factors (75).
(b) Fucosylation (nodZ, nolK).
Fucose is a frequent substitution, and its addition to Nod factors is encoded by nodZ in many rhizobia (162, 171, 208, 210, 261). Since all the usual Glycine max symbionts (B. elkanii, B. japonicum, R. fredii, and strain NGR234) secrete fucosylated Nod factors, the expectation that mutation of nodZ would prevent the nodulation of soybeans was strong. In fact, B. japonicum nodZ is required for nodulation of M. atropurpureum but not of G. max (261). Similarly, nodZ mutants of strain NGR234 produce nonfucosylated Nod factors and are unable to nodulate Pachyrhizus tuberosus but retain the capacity to nodulate soybeans (208). Since transconjugants of R. leguminosarum containing B. japonicum nodZ acquire the capacity to nodulate Macroptilium (but not to fix nitrogen) (162), it seems as if fucosylated Nod factors play a role in nodulation of only a few legumes. In A. caulinodans, NolK is involved in the synthesis of GDP-fucose, which is used by NodZ as the fucosyl donor in fucosylation of A. caulonidans Nod factors (171).
(iii) Sulfation (nodH, noeE).
NodH and NoeE, the only two characterized sulfotransferases, are specific to the reducing terminus of Nod factors (68, 154, 209, 224, 239). Both enzymes use 3′-phosphoadenosine-5′-phosphosulfate as the sulfate donor. 3′-Phosphoadenosine-5′-phosphosulfate is synthesized by the enzymes encoded by nodPQ (242, 243). NodH-dependent sulfation of the reducing terminus at C-6 of R. meliloti Nod factors is a major determinant of host range in R. meliloti (154, 224). nodH mutants which fail to produce sulfated Nod factors were thought to completely lose the capacity to nodulate M. sativa but gain the ability to nodulate the nonhost V. sativa (49, 224). Although other data indicate that nodulation of alfalfa by some nodH mutants is strongly impaired rather than abolished (125, 186), these results confirm that optimal nodulation of M. sativa requires sulfated Nod factors whereas V. sativa does not tolerate them (70).
In contrast, NoeE is a fucose-specific sulfotransferase (113, 209). Mutation of strain NGR234 noeE blocks the nodulation of P. tuberosus, while its introduction into the closely related strain USDA257 extends the host range of R. fredii to include Calopogonium caeruleum (113). Surprisingly, these transconjugants do not acquire the ability to nodulate P. tuberosus (M. Hanin, unpublished results).
(iv) Acetylation (nodL, nodX, nolL).
Unlike sulfate groups, acetyl residues may be found at both extremities of Nod factors, i.e., on C-6 of the reducing or nonreducing termini, as well as on the fucose (Table 4). Furthermore, acetate groups on the fucose can move from one α-hydroxyl group to another (12). Mutation of nodX provokes a drastic reduction in Nod factor production by R. leguminosarum strain TOM and the ability to nodulate primitive cultivars of P. sativum (48). In R. leguminosarum bv. viciae however, a fucosyl group added by NodZ can functionally replace the missing acetyl group in a nodX mutant (189). Although NodL is responsible for acetylation of C-6 of the nonreducing terminus in R. meliloti Nod factors (2), R. meliloti nodL mutants are impaired in their ability to elicit infection thread formation on M. sativa but form nodules with only a moderate delay (96). A similar phenotype was observed when the corresponding mutant of R. leguminosarum bv. viciae was used to inoculate V. hirsuta, while nodL nodF double mutants of R. meliloti are unable to penetrate their hosts (2). In strain NGR234, disruption of the flavonoid-inducible nolL gene results in the synthesis of NodNGR factors that lack the 3-O- or 4-O-acetate group (12). Interestingly, the nodulation capacity of the mutant NGRΩnolL is not impaired whereas transconjugants of USDA257 containing nolL of NGR234 produce acetylated Nod factors and nodulate the nonhosts C. caerulum, L. leucocephala, and Lotus halophilus (12). Acetylation of the fucose on Nod factors of R. etli also conditions efficient nodulation of some P. vulgaris cultivars and of the alternate host Vigna umbellata (44).
(v) N methylation and carbamoylation (nodS, nodU, nolO).
NodS is an N-methyltransferase (93, 94, 129), while NodU and NolO control carbamoylation at C-6 and C-3 (or C-4), respectively, on the nonreducing N-acetyl-d-glucosamine (57, 129, 131). Inactivation of nodS in A. caulinodans, strain NGR234, and R. tropici abolishes the nodulation of L. leucocephala and P. vulgaris (158, 276). Introduction of either nodS or nodU into R. fredii extends its host range to include Leucaena spp. (129, 144).
(vi) 2-O methylation (noeI).
The fucose group of Nod factors of B. japonicum USDA110, NGR234, and USDA257 is mostly 2-O methylated (Table 4). In strains NGR234, NoeI controls this function, and mutation of noeI leads to the production of fucosylated Nod factors that are not 2-O methylated (131). Introduction of noeI into R. loti allows the production of 2-O-methylated NodNGR factors. However, nodulation tests using L. leucocephala, Lotus japonicus, M. atropurpureum, and V. unguiculata and the NGRΔnoeI mutant have failed to identify a host which requires 2-O-methylated NodNGR factors (S. Jabbouri and M. Hanin, unpublished results).
Nod enzymes, which are responsible for the elaboration of Nod factors, are among the principal determinants of host specificity. Indeed, a number of them were called hsn determinants before the unified nod gene nomenclature was established. Most of them catalyze the adjunction of baroque decorations to the core Nod factor molecule (which is synthesized by NodC, NodB, and NodA); the effect of these different families of molecules on legumes is discussed below.
Nod Factors and Host Specificity
Although much information on the influence of Nod factor substituents on host range exists, no strict correlation can be drawn between the types of LCOs produced by rhizobia and the plants they nodulate. For example, although the Nod factors produced by R. etli and R. loti are identical, the two species have distinct host ranges (Phaseolus spp. and Lotus spp., respectively) (33). Also, the major Nod factors secreted by R. leguminosarum bv. trifolii are the same as two of the major LCOs produced by R. leguminosarum bv. viciae (188, 255), yet the two biovars nodulate distinct plants. Although the inability of NodD proteins to recognize the host flavonoids (and thus to activate nod gene expression) might be responsible for some of these phenotypes, these data emphasize the point that Nod factor structures alone cannot be used to predict host range. Furthermore, two rhizobia that nodulate the same plant may secrete different Nod factors. R. tropici and R. etli produce sulfated and acetylfucosylated Nod factors, respectively (Table 4), but both effectively nodulate P. vulgaris (202, 203). B. elkanii, B. japonicum, strain NGR234, and strain USDA257 have a number of common hosts (207), but their Nod factors vary considerably (Table 4).
The amounts of Nod factors released by rhizobia are also important in determining the host range. For instance, introduction of strain NGR234 nodD1 into R. meliloti increases Nod factor production by about twofold and permits the nodulation of V. unguiculata, a nonhost (214). Similarly, conjugation of the nodSU genes of strain NGR234 into R. fredii USDA257 not only extends the host range of the transconjugant to include L. leucocephala (144) but also vastly increases Nod factor production (Table 5). In contrast, the NGRΔnodSU mutant, which secretes only 1/10 the amount of Nod- factor produced by the wild type, is incapable of nodulating L. leucocephala (Table 5). One interpretation of these data is that higher levels of Nod factors (produced when a functional NodS or NodU is present) (129), not the presence of N-methylated or 6-O-carbamoylated LCOs, is necessary for nodulation of Leucaena.
TABLE 5.
Effects of apigenin, nodD1, and nodSU on Nod factor production and nodulation of L. leucocephala by strains NGR234 and USDA257
| Straina | Nod factor productionb
|
Nodulation of L. leucocephala | |
|---|---|---|---|
| − Apigenin | + Apigenin | ||
| NGR234 | 0.4 ± 0.2 | 61.8 ± 9.7 | Nod+ Fix+ |
| NGRΔnodD1 | 0.4 ± 0.3 | 0.3 ± 0.2 | Nod− |
| NGRΔnodSU | 0.3 ± 0.3 | 5.7 ± 2.5 | Nod− |
| USDA257 | 0.2 ± 0.2 | 1.5 ± 1.1 | Nod− |
| USDA257(nodSU) | 0.1 ± 0.1 | 38.3 ± 9.8 | Nod+ Fix+ |
NGRΔnodD1, nodD1 mutant of Rhizobium sp. strain NGR234; NGRΔnodSU, nodSU mutant of strain NGR234; USDA257 (nodSU), R. fredii USDA257 transconjugant containing the functional nodSU genes of NGR234.
Although the supporting evidence is lacking, it also seems likely that in addition to variations in the levels of Nod factors, different members and various proportions of the respective Nod factor families are secreted into the rhizospheres of specific legumes. There, only certain combinations of Nod factors, present at optimal levels, are capable of inducing deformation (Had) and curling (Hac) in homologous legumes (118, 148, 154, 169, 206, 213, 238, 252, 269a).
Cooperation among Nod factors and related signal molecules has also been demonstrated. Minami et al. (177, 178) examined the effects of LCOs on changes in the expression of early nodulin genes in G. soja roots and found that any combination of one active Nod factor with a nonspecific LCO was sufficient to induce mRNA expression of early nodulin genes such as Enod2. In contrast, both synthetic Nod factors and the chitin pentamer PACT, which is unable to provoke Had on G. soja, induced the transient accumulation of Enod40 mRNA. It would thus seem that not only are absolute levels of Nod-factors important but also the composition and relative proportions of the mixtures excreted by rhizobia are necessary for the induction of different components of the nodulation pathway (190).
Nod factor binding proteins.
The extent to which specific Nod factors bind to their respective plant receptors is still unknown. In Medicago, two putative Nod factor binding sites (NFSB) have been characterized (23, 181). Some properties of NFSB1 of M. truncatula suggest that it may also be involved in processes other than nodulation. First, its affinity and specificity for LCOs are low. Second, a similar site exists in particulate fractions of tomato roots (23). In contrast, NFSB2, which was isolated from the microsomal fraction of Medicago varia cell suspension cultures, is sensitive to proteases and shows high affinity for Nod factors (181). Covalent linkage of both the lipid and the chitooligosaccharide moieties, as well as O acetylation of the nonreducing sugars of Nod factors, is required for high-affinity binding to NFSB2 (108). In competition experiments in which the ligand specificity of NFSB2 was assayed by titrating 35S-labelled R. meliloti Nod factors against synthetic LCOs, those with short (C8) acyl chains were poor competitors compared to those with C16 and C18 fatty acid side chains (108). However, NFSB2 does not select for the presence of sulfate on the reducing sugar or for the number and/or positions of double bonds in the acyl chain. Since sulfated R. meliloti Nod factors are required for optimal nodulation of Medicago (see above) and as synthetic LCOs containing at least two double bonds are more active biologically than are monounsaturated compounds (53), it seems doubtful that these putative receptors are host-specific determinants of the Medicago-R. meliloti interaction.
Similarly, a root surface lectin of Dolichos biflorus has apyrase (Ca2+-dependent ATP diphosphatase) activity and binds Nod factors from both heterologous and homologous rhizobia (69). Reverse PCR-based techniques using mRNAs of L. japonicus and M. sativa root confirmed the presence of orthologues in these two legumes. Similar experiments were also performed with D. biflorus and Arabidopsis, leading to a second lectin in the former and a nonlegume lectin in the latter (223). Since much work still has to be done on these Nod factors binding lectin/nucleotide phosphohydrolases (LNPs), it is too early to pronounce on their exact symbiotic role. It is intriguing, however, that D. biflorus has two LNPs. Close orthologues of LNP1 are found only in legumes, while D. biflorus LNP2 is more closely related to apyrase sequences of nonleguminous plants (223).
Although Nod factors are bacterial products, they share many characteristics with plant hormones (213). One of these is that perturbations in the auxin-cytokinin balance mimic Nod factors in inducing meristematic activity of nodule meristems (128). Another is that the range of concentrations over which both hormones and Nod factors are active can vary by a millionfold. This suggests a desensitisation or an adaptation by the plant to increasing hormone concentrations (137). In turn, this implies that covalent and reversible modification to the “receptors” occurs; these suggestions are supported by the fact that the ethylene receptor is homologous to two-component chemoreceptors of bacteria (38). Perhaps this explains why the search for Nod factor receptors in direct Nod factor binding studies has been only moderately successful.
Hydrolysis of Nod factors.
In principle, the chemical stability of Nod factors should influence the biological activity of the LCO mixtures secreted by rhizobia. In fact, chitinases and other enzymes rapidly cleave purified Nod factors in the host rhizosphere. The degradation products formed are only weakly active on their respective hosts (118, 258). Plant chitinases have been studied extensively in the context of plant-pathogen interactions. Among these are isoforms that are induced in symbiotic and/or pathogenic interactions (103, 192, 257, 273, 281). Distinct isoforms of chitinases and related hydrolases cleave specific β-1,4-linkages in the carbohydrate moiety of Nod factors (Tables 6 and 7), resulting in the formation of acylated degradation products that are either di-, tri- or tetrameric. In general, leguminous and nonleguminous chitinases that belong to the same chitinase class have similar substrate specificities. Interestingly, the baroque decorations on certain Nod factors have a strong influence on the stability of these molecules (241, 258, 259) and modify the cleavage specificity of a given chitinase. For example, pentameric LCOs are hydrolyzed in two positions by an M. sativa class I chitinase, but only one of these positions is susceptible in sulfated Nod factors (Table 7). Furthermore, the sulfate group at the reducing end and the O-acetyl group at the nonreducing terminus of R. meliloti Nod factors dramatically increase the stability of these LCOs, rendering tetrameric molecules resistant to degradation by a large number of different chitinases (Table 6). Novel enzymes have been identified in Medicago, and their cleavage preferences are different from those of known chitinases (Tables 6 and 7). One of them is an extracellular lipodisaccharide-releasing Nod factor hydrolase that cleaves all tetra- and pentameric R. meliloti Nod factors. Preincubation with R. meliloti Nod factors stimulated the activity of this enzyme in Medicago roots, suggesting that Nod factor inactivation is an early feedback response by the host plant.
TABLE 6.
Hydrolysis of Nod factors by chitinases and related enzymesa
| Enzyme | Plant isozymesb | Nod factors used as substratesbc | Acylated products in hydrolysatebd | Reference(s) |
|---|---|---|---|---|
| Chitinase class I | Ms, Pv, Vs, Nt | NodRm-V(C16:2, S) | III | 241, 258 |
| Ms, Vs | NodRm-V(C16:2) | IV, III | 258 | |
| Ms, Pv, Vs, Nt | NodRm-IV(C16:2, S) | None | 241, 258 | |
| Ms, Pv, Vs, Nt | NodRm-IV(C16:2) | III | 241, 258 | |
| Vu | NodNGR-V(AcMeFuc) | Unknown structures | 259 | |
| Vu | NodNGR-V(SMeFuc) | Unknown structures | 259 | |
| Ps, Pv, Vu | NodNGR-V(MeFuc) | Unknown structures | 259 | |
| Nt | NodRm-IV(Ac, C16:2) | None | 241 | |
| Nt | NodRlv-V(Ac, C18:4) | AcIV, AcIII | Ovtsyna et al., submitted | |
| Nt | NodRlv-V(Ac, C18:4, Fuc) | AcIII | Ovtsyna et al., submitted | |
| Nt | NodRlv-IV(Ac, C18:4) | None | Ovtsyna et al., submitted | |
| Nt | NodRlv-IV(Ac, C18:4, Fuc) | None | Ovtsyna et al., submitted | |
| Chitinase class II | Nt | NodRm-V(C16:2, S) | III | 241 |
| Nt | NodRm-IV(C16:2, S) | None | 241 | |
| Nt | NodRm-IV(C16:2) | III | 241 | |
| Chitinase class III | Ca, Bv, Nt | NodRm-V(C16:2, S) | III, II | 241 |
| Ca, Bv, Nt | NodRm-IV(C16:2, S) | II | 241 | |
| Ca, Bv, Nt | NodRm-IV(C16:2) | III, II | 241 | |
| Sr | NodAc factors | Unknown structures | 103 | |
| Nt | NodRm-IV(Ac, C16:2, S) | None | 241 | |
| Nt | NodRm-V(Ac, C16:2, S) | AcIII | 241 | |
| Nt | NodRlv-V(Ac, C18:4) | AcIV, AcIII | Ovtsyna et al., submitted | |
| Nt | NodRlv-V(Ac, C18:4, Fuc) | AcIII | Ovtsyna et al., submitted | |
| Nt | NodRlv-IV(Ac, C18:4) | AcIII | Ovtsyna et al., submitted | |
| Nt | NodRlv-IV(Ac, C18:4, Fuc) | None | Ovtsyna et al., submitted | |
| Chitinase class IV | Bv, Dc | NodRm-V(C16:2, S) | III | 241 |
| Bv, Dc | NodRm-IV(C16:2, S) | None | 241 | |
| Bv, Dc | NodRm-IV(C16:2) | III | 241 | |
| Chitinase class V | Nt | NodRm-V(C16:2, S) | III | 241 |
| Nt | NodRm-IV(C16:2, S) | None | 241 | |
| Nt | NodRm-IV(C16:2) | III | 241 | |
| Chitinase class VI | Nt | NodRm-V(C16:2, S) | III | 241 |
| Nt | NodRm-IV(C16:2, S) | None | 241 | |
| Nt | NodRm-IV(C16:2) | III | 241 | |
| Novel chitinase/lysozyme | Ms | NodRm-V(C16:2, S) | III | 179 |
| Ms | NodRm-IV(C16:2, S) | III | 179 | |
| Ms | NodRm-IV(C16:2) | III | 179 | |
| Ms | NodRm-IV(Ac, C16:2, S) | AcIII | 179 | |
| Nod factor hydrolase | Ms | NodRm-V(C16:2, S) | II | 258, 260 |
| Ms | NodRm-V(C16:2) | II | 258 | |
| Ms | NodRm-IV(C16:2, S) | II | 258, 260 | |
| Ms | NodRm-IV(C16:2) | II | 258, 260 | |
| Ms | NodRm-IV(Ac, C16:2, S) | AcII | 260 | |
| Ms | NodRm-V(Ac, C16:2, S) | AcII | 260 | |
| Roots | Ms, Vs | NodRm-V(C16:2, S) | III, II | 258, 260 |
| Ms, Vs | NodRm-V(C16:2) | IV, III, II | 258 | |
| Ms, Vs | NodRm-IV(C16:2, S) | II | 258, 260 | |
| Ms, Vs | NodRm-IV(C16:2) | III, II | 258 | |
| Ms | NodRm-IV(C16:0, S) | II | 258 | |
| Ms, Ps, Vs | NodRlv-V(Ac, C18:4) | AcIV, AcIII, AcII | 97, 118 | |
| Vs | NodRlv-IV(Ac, C18:4) | AcIII, AcII | 118 | |
| Vs | NodRlv-V(Ac, C18:0) | AcIV, AcIII, AcII | 118 | |
| Sr | NodAc factors | Unknown structures | 103 |
Purified enzymes (as well as whole roots) of both legumes (bold) and nonlegumes were incubated with Nod factors, and the reaction products were examined for the presence of acylated chitin fragments.
Abbreviations are as follows. Hydrolases were isolated from the following legumes: Cicer arietinum (Ca), Medicago sativa (Ms), Phaseolus vulgaris (Pv), Pisum sativum (Ps), Sesbania rostrata (Sr), Vicia sativa (Vs), and Vigna unguiculata (Vu) and the nonlegumes Beta vulgaris (Bv), Daucus carota (Dc), and Nicotiana tabacum (Nt). Chemical groups are abbreviated as follows: acetyl (Ac), fucosyl (Fuc), methyl (Me), sulfate (S), methylfucose (MeFuc), acetylated methylfucose (AcMeFuc), sulfated methylfucose (SMeFuc).
Nod factors were purified from A. caulinodans (NodAc), R. leguminosarum bv. viciae (NodRlv), Rhizobium sp. strain NGR234 (NodNGR), and R. meliloti (NodRm). Details of their structure, such as the presence of certain baroque decorations (for abbreviations, see footnote b), the length of the acyl chain, and the number of double bonds, are shown in brackets. Tetramers and pentamers of N-acetylglucosamine are marked as IV and V, respectively. The nonsulfated NodRm factors and fucosylated NodRlv factors are derivatives of the wild type obtained by desulfation and fucosylation with NodZ, respectively. NodRm-IV(C16:0, S) was prepared from NodRm-IV(C16:2, S) by catalytic reduction.
Acylated products were separated from the substrate by reverse-phase high-pressure liquid chromatography (179, 241, 258, 260; A. O. Ovtsyna, M. Schultze, I. A. Tikhonovich, H. P. Spaink, É. Kondorosi, Á. Kondorosi, and C. Staehelin, submitted for publication) and thin-layer chromatography (97, 103, 118, 258). Abbreviations: lipotetrasaccharide (IV); lipotrisaccharide (III); lipodisaccharide (II); O-acetylated lipotetrasaccharide (AcIV); O-acetylated lipotrisaccharide (AcIII); O-acetylated lipodisaccharide (AcII); Nod factors resistant to hydrolysis by the given enzyme (None).
TABLE 7.
Sites of hydrolytic cleavage of Nod factors of Rhizobium meliloti by enzymes isolated from the roots of Medicago sativaa
| Hydrolase (reference) | No. of N-acetylglucosamine residues | R1 | R2 | Cleavage site(s) |
|---|---|---|---|---|
| Intact roots (258, 260) | IV | Sulfatyl | H | a |
| IV | H | H | a, b | |
| IV | Sulfatyl | O-Acetyl | a | |
| V | Sulfatyl | H | c, d | |
| V | H | H | c, d, e | |
| Chitinase class I (241, 258) | IV | Sulfatyl | H | None |
| IV | H | H | b | |
| IV | Sulfatyl | O-Acetyl | None | |
| V | Sulfatyl | H | d | |
| V | H | H | d, e | |
| Novel chitinase/lysozyme (179) | IV | Sulfatyl | H | b |
| IV | H | H | b | |
| IV | Sulfatyl | O-Acetyl | b | |
| V | Sulfatyl | H | d | |
| Nod factor hydrolase (258, 260) | IV | Sulfatyl | H | a |
| IV | H | H | a | |
| IV | Sulfatyl | O-Acetyl | a | |
| V | Sulfatyl | H | c | |
| V | H | H | c | |
| V | Sulfatyl | O-Acetyl | c |
Distinct cleavage site preferences exist toward tetrameric (IV) and pentameric (V) Nod factors of R. meliloti as well as their desulfated and deacetylated derivatives, respectively.
However, two different types of observations suggest that chitinases may not play major roles in the determination of host specificity. First, the class I, II, III, V, and VI chitinases isolated from Nicotiana tabacum, as well as the class III and IV chitinases of Beta vulgaris and Daucus carota (all nonlegumes), degrade Nod factors in similar ways to that used by legume chitinases (Table 6). One interpretation of these data is that chitinases form part of a generalized plant response system rather than tailoring Nod factor degradation to specific legume-Rhizobium associations. A second important consideration is that root hydrolases take hours to degrade Nod factors (260), while the first responses of root hairs to inoculation by rhizobia (or to the presence of purified Nod factors) can be measured in seconds (64). Nevertheless, expression of a chitinase from Serratia marcescens by R. fredii USDA191 transconjugants resulted in delayed nodulation and a marked decrease in total nodule number on soybean cultivar McCall in comparison with the wild-type strain (146). What is undisputed, however, is that chitinases modulate the levels of Nod factors in the rhizosphere and, in so doing, help suppress defense responses stimulated by high concentrations of LCOs (231, 260). Since defense reactions also occur in aborted infection threads (273), it is possible that plant hydrolases help regulate Nod factor levels even after the bacterium has entered the plant. In other words, chitinases and related hydrolases cleave and so inactivate Nod factors. Local induction or down-regulation of these enzymes in leguminous roots would thus modulate local LCO concentrations. Since Nod factor levels seem to be host-range determinants, plant hydrolases might help tailor host specificity.
NOD FACTORS OPEN THE LEGUME OUTER DOOR
Although the mechanism of root hair deformation is poorly understood, it probably resembles that of root hair development (118). The tips of Nod factor-treated V. sativa root hairs swell before polar growth is induced. Induction of tip growth is preceded by local hydrolysis of the cell wall and followed by polar growth (118). Schiefelbein and Somerville (233) showed that the phenotype of root hair-specific mutants of Arabidopsis thaliana resembles root hair deformations caused by rhizobia. Thus, initiation and deformation of root hairs appear to be ontogenically related.
Nod factors are the signals required for entry of rhizobia into some legumes. Concomitant treatment of G. max and V. unguiculata roots with NodNGR factors and nodABC mutants of strain NGR234 or B. japonicum USDA110 permits these strains to nodulate and fix nitrogen on their respective hosts (215). NodNGR factors also allow the entry of R. fredii USDA257 into the roots of the nonhost Calopogonium caeruleum (215) and of the nodABC mutant of NGR234 into Macroptilium atropurpureum (213). Similarly, coapplication of purified A. caulinodans Nod factors and a nodA mutant of the same strain restores the formation of outer cortical infection pockets in Sesbania rostrata (56). These data leave little doubt that at least in some symbioses, Nod factors act like keys to legume doors. Interestingly, a recent report suggests that one of the “latches” that is opened by a Nod factor “key” has been found. A transposon-tagged mutant of Lotus japonicus, which is arrested at the stage of bacterial recognition, was identified (232). The mutation, which has been named nin (for nodule inception), is required for the formation of infection threads and is a transcription factor.
Once Nod factors have opened the root hair door to the invading rhizobia, additional bacterial signals appear necessary for continued development of the infection threads. This is clearly seen in variations of the Nod factor complementation experiments described above. These experiments work well if the strain deficient in Nod factor production is mutated in genes essential for Nod factor synthesis (nodABC) but not if the strain is mutated in nodD1. In this latter case, rhizobia bunch up in the curled part of the root hair and the infection thread fails to develop toward the root cortex, resulting in contortions that resemble a cerebellum (Fig. 2E). It thus seems probable that products of genes other than those needed for the synthesis of Nod factors, but also under the control of NodD1, are required for the continued development of infection threads.
DO INFECTION THREADS HAVE AN INNER DOOR?
Other Host Range Keys
NodD proteins, with their ability to recognize different inducers, the complex mixtures of Nod factors as well as the various levels at which they are maintained, are not the only determinants of host specificity. Although Nod factors permit rhizobia to enter the outer door of the legume host and may play a role during nodule development (56, 267), additional “keys” are necessary for the formation of symbiotically proficient nodules. In fact, later steps of the infection process such as infection thread formation and propagation, as well as bacterial release into the cytoplasm of infected cells, require constituents of the cell wall and in some cases secretion of specific proteins (for a review, see reference 275).
Polysaccharides and surface components.
Symbiotically relevant components of the rhizobial cell wall include extracellular polysaccharides (also known as exopolysaccharides) (EPS), lipopolysaccharides (LPS), capsular polysaccharides (CPS and KPS), and cyclic β-glucans (Fig. 3). Surface polysaccharides (SPS) form a complex macromolecular structure at the bacterium-plant interface. They accumulate on the surface of the prokaryote as a capsular layer but are also released into the extracellular space as bacterial slime.
FIG. 3.
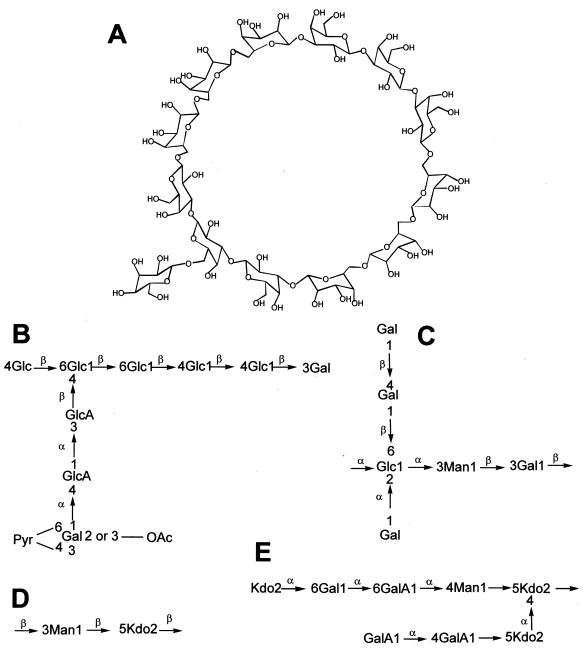
Examples of the five classes of rhizobial polysaccharides. (A) Structure of cyclic β-(1,6)-β-(1,3)-glucans common to B. japonicum. Redrawn from reference 26. (B) Acidic EPS of Rhizobium sp. strain NGR234 (60). EPS I (succinoglycan) of R. meliloti resembles the EPS of strain NGR234. (C) KPS of R. leguminosarum bv. trifolii (98). (D) The somatic K antigen of R. fredii USDA257 (83). (E) Core structure of the LPS of R. etli (84). Abbreviations: Gal, galactose; GalA, galacturonic acid; Glc, glucose; GlcA, glucoronic acid; Man, mannose; Kdo, 3-deoxy-d-manno-2-octulosonic acid; OAc, acetate group; Pyr, pyridine.
EPS I and EPS II represent the two major classes of EPS synthesized by R. meliloti. EPS I members are polymers of an octasaccharide repeating unit of succinoglycan ranging in size from 106 to 107 Da for the high-molecular-weight (HMW) fraction (152). In contrast, the low-molecular-weight (LMW) fraction is formed of monomers, dimers, and trimers (277). Although mutants of R. meliloti Rm1021 incapable of synthesizing succinoglycan (EPS I) produce normal root hair curling on M. sativa, the nodules are devoid of bacteria and bacteroids and thus are ineffective (76, 151). Addition of the LMW fraction, specifically trimers of succinoglycan, to roots of alfalfa restored the nodule invasion capability of the exo mutant (5, 277). Interestingly, EPS I mutant strains of R. meliloti derepressed for the synthesis of another class of EPS are capable of forming normal nodules on alfalfa (99). EPS II members are polymers of modified glucose-(β-1,3)-galactose (99), and only small amounts (as little as 7 pmol per plant) of the purified LMW fraction are sufficient to restore nodule invasion by noninfective strains (102). Thus, although EPS II members can replace succinoglycan in nodule invasion, they are not required when wild-type EPS I is produced.
Fluorescence microscopy analyses show that nodule invasion is aborted at various stages in different exo mutants (40). Cells that lack ExoR, a negative regulator of exo gene expression (212), vastly overproduce succinoglycan and are unable to colonize curled root hairs and form infection threads. In contrast, a mutant with a mutation in exoY that is incapable of synthesizing succinoglycan (since it lacks the first enzyme in the biosynthetic pathway) (217) colonizes curled root hairs but forms few, very short infection threads. Although the exoH mutant that produces symbiotically dysfunctional succinoglycan forms infection threads longer than those produced by the exoY mutant, they never extend as far as the base of the root hair cell. Thus, in R. meliloti, succinoglycan could be regarded as a symbiotic signal (or key) required for opening the inner root hair door(s). Mutants that fail to produce the symbiotically active forms of these EPSs in adequate quantities are incapable of penetrating the adjacent plant cell and thus remain blocked or trapped in the infected root hair (40, 153). Also, since nodules induced by EPS I mutants of R. meliloti show pronounced symptoms of plant defense, it is possible that a molecule derived from the EPS I biosynthetic pathway functions as a suppressor of plant defense reactions (182). It has been postulated that in other plants, EPS of R. leguminosarum bv. viciae accelerates root hair curling and infection in such a way that invasion by rhizobia precedes the plant defense response (272). Similarly, it is thought that the wild-type cyclic β-glucans of B. japonicum (Fig. 3) may function as suppressors of host defense responses (20).
Formation of symbiosomes (which involves the release of rhizobia from infection threads into the cytoplasm of infected nodule cells) in plants other than M. sativa requires different components including EPS, LPS, CPS, KPS, and cyclic β-glucans (Fig. 3, Table 8). Although EPS− mutants of M. loti are fully effective on Lotus pedunculatus, they provoke small, ineffective, tumor-like growths on L. leucocephala (127). Similarly, Exo− mutants of R. leguminosarum bv. trifolii (271), R. leguminosarum bv. viciae (24), and strain NGR234 (39) are ineffective on their respective hosts, T. repens, P. sativum, and L. leucocephala. In contrast, Exo− mutants of R. leguminosarum bv. phaseoli (58) and R. fredii (140), are fully effective on the determinate legumes P. vulgaris and G. max. One possible explanation of these data is that EPS is required for the formation of fully effective symbioses on plants that produce indeterminate nodules but not on legumes which form determinate nodules. However, Parniske et al. (192) showed that an EPS-defective mutant of B. japonicum which forms nitrogen-fixing nodules on G. max was ineffective on G. soja. Furthermore, a mutant of Rhizobium strain TAL1145 that is deficient in EPS synthesis still nodulates various hosts, independently of their nodule type (193).
TABLE 8.
Effects of mutations in nonnodulation genes on Fix phenotypes in various legumes
| Rhizobium strain | Locus or gene(s) | Producta | Regulator | Phenotypea | Plant | Reference(s) |
|---|---|---|---|---|---|---|
| A. caulinodans ORS571 | dTDP-d-gluc. synth. | Deoxy sugars | Fix− | Sesbania rostrata | 56 | |
| B. japonicum USDA110 | exoP | EPS export | Fix− | Glycine max | 10 | |
| ndvC | Cyclic β-glucans | Fix− | Glycine max | 19 | ||
| ndvB | Cyclic β-glucans | Fix− | Glycine max | 65 | ||
| M. loti NZP2037 | EPS | Fix− | Leucaena leucocephala | 127 | ||
| Fix+ | Lotus pedunculatus | |||||
| NGR234 | exoY | EPS | Fix− | Leucaena leucocephala | 106 | |
| fixF | Rhamnose-rich SPS | nodD1 | Fix− | Vigna unguiculata | 130 | |
| R. etli CFN42 | lpsβ | LPS | Fix− | Phaseolus vulgaris | 92 | |
| R. leguminosarum strain 300 | lps | LPS | Fix− | Pisum sativum | 136 | |
| bv. trifolii AR5 | LPS | Fix− | Trifolium pratense | 100 | ||
| bv. trifolii 24.1 | EPS | Fix− | Trifolium pratense | 247 | ||
| bv. trifolii LPR5 | pssD | EPS | Fix− | Trifolium repens | 270, 271 | |
| bv. trifolii TA1 | pssD | EPS | Inf+ Fix− | Trifolium pratense | 147 | |
| R. meliloti 2011 | exoAMONP | EPS 1 | Fix− | Medicago sativa | 8 | |
| exoUVWTI | EPS 1 | Fix− | Medicago sativa | 9 | ||
| R. meliloti 1021 | exoY | SG | exoS, chv1 | Inf+ Fix− | Medicago sativa | 40, 41 |
| ndvA | β-(1→2)Glucan | Fix− | Medicago sativa | 262 | ||
| R. meliloti AK631 | rkpK | KPS | Fix+ | Medicago sativa | 138 | |
| rkpK1 | KPS | exoB (galE) | Fix− | Medicago sativa | 31 | |
| R. tropici CIAT899::Tn5 | EPS | Exo− Fix+ | Macroptilium atropurpureum | 176 |
Abbreviations: SPS, surface polysaccharide; SG, succinoglycans; Inf, formation of infection threads; Fix, nitrogen fixation; Exo, EPS synthesis.
A number of mutations that encode similar phenotypes but that occur in a variety of genes have been found (Table 8). For example, where the production of cyclic-β-(1→2)glucans, rhamnose-rich SPS, LPS, SG, β-(1→2)glucans, and KPS is impaired, Nod+ Fix− phenotypes are also observed. Different regulators appear to control the expression of these diverse genes, and the resultant phenotype depends on the plant. Quite often an Inf+ Fix− phenotype is observed, suggesting that these genes play similar roles. One possibility is that such surface components are necessary for or form part of the developing infection thread. Hence, a plant that could not itself supply the missing carbohydrate would give a Fix− phenotype while the rhizobial mutant in a plant that normally synthesizes the compound would have no effect.
Secreted proteins.
Several strains of Rhizobium secrete symbiotically active proteins. Among these, the nodO product is required for nodulation of V. hirsuta by mutants of R. leguminosarum bv. viciae with nodFELMNT deleted (62), as well as for extension of the host range of an R. trifolii nodE mutant to include V. sativa (67). nodO encodes a Ca2+ binding protein that is thought to form cation-specific channels in membranes of leguminous plants (66, 264). Although NodO and NodS have distinct biochemical functions, a nodO homologue of Rhizobium sp. strain BR816 was shown to complement a nodS mutant of NGR234 for nodulation of L. leucocephala (269e), suggesting that secreted proteins may supplement some Nod factor deficiencies. Secretion of NodO is dependent on a C-terminal signal of about 24 residues (265) and on the prsDE genes, which encode two type I-like inner membrane proteins (77). In addition to NodO, at least three other proteins are secreted via this system, two of which (PlyA and PlyB) are glycanases involved in the processing of bacterial EPSs (78).
Sequencing the symbiotic plasmid of NGR234 revealed flavonoid-inducible genes encoding components of a type III secretion system (TTSS) (87). In a number of bacterial pathogens, TTSSs are induced upon contact with host cells (of plants or animals) and deliver virulence proteins directly into the eukaryotic cytosol (150). Strain NGR234, as well as several strains of R. fredii, was shown to excrete at least three to five proteins in a NodD1- and TTSS-dependent manner (145, 274). In USDA257, the nolXWBTUV cluster (corresponding to the nolX, rhcC1, nolB, rhcJ, nolU and nolV genes of NGR234) (274) regulates the nodulation of G. max in a cultivar-specific manner (168), whereas the TTSS of NGR234 profoundly affects the nodulation of various legumes such as P. tuberosus and Tephrosia vogelii (274). The absence of conserved nod box regulatory elements in the promoter regions of the nol and rhc operons indicates that NodD1-dependent transcriptional regulation of the TTSS genes is mediated by another factor. y4xI, an HrpG homologue which is under the control of a functional nod box, is thought to be the key intermediary in the regulatory cascade between flavonoids and activation of the TTSS machinery in NGR234 (274). The delayed induction of rhc genes in comparison with loci involved in the elaboration of the Nod factors suggests that the TTSS machinery is assembled after Nod factors have been elaborated and that protein export begins when intimate contact between bacteria and root hairs has been established. Thus, the most important role of TTSS-secreted proteins such as NolX and y4xL of NGR234 would occur after the bacteria have entered the plant, possibly during the development of infection threads.
It has been suggested that bacterial invasion of plant cells triggers nonspecific defense reactions and that successful pathogens overcome these defenses (89). Similarly, invading symbionts have probably evolved different strategies to lower host plant defenses. TTSS proteins and polysaccharides may contribute to this phenomenon. Some plants would perceive these compounds as part of the infection pathway and react to their presence by increased nodulation, as occurs in alfalfa inoculated with Exo+ R. meliloti and in T. vogelii, which favors strains of NGR234 with a functional TTSS. An alternate explanation is that these secreted substances function as suppressers of plant defense reactions, as proposed for various polysaccharides (20, 182). In contrast, P. tuberosus which is poorly nodulated by wild-type NGR234, produces many effective nodules when inoculated with TTSS mutants, suggesting that secreted proteins have detrimental effects (elicitors of defense reactions?) on certain hosts (274). Other rhizobia may have different keys to the inner door. Since apparently functional TTSSs are absent from R. leguminosarum and R. meliloti, other factors probably play similar roles. Some of these are undoubtedly polysaccharides, but it seems likely that a variety of keys exist. When they are used sequentially and in the correct combinations, infection thread development continues beyond the epidermal cells and a home for the invading rhizobia is constructed in the cortex of the root. Improper use of the keys results in abortion of the infection threads, as shown in Fig. 2E.
Clearly, various components of nodulating rhizobia, which include the NodD proteins, the spectrum of inducers, the palette of Nod factor substituents, levels of Nod factors, polysaccharides, and secreted proteins, contribute to the control of host specificity. Although some of these may act synergistically (e.g., complementation of nodS mutants by NodO), most are part of a developmental process that leads to nodule invasion. Host plants are thus capable of controlling the successive steps of the rhizobial infection through checkpoints demanding specific keys. The absence of a suitable key does not necessarily result in a Nod− or Fix− phenotype, however. Most determinants of symbiotic specificity optimize the chances of forming effective nodules. Consecutive host specificity barriers are thus like hurdles of different heights that allow the host to select among many strains for the one best suited to its requirements.
EVOLUTION OF HOST SPECIFICITY
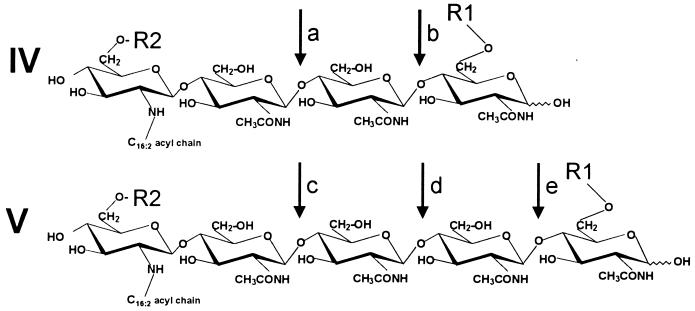
Broad-host-range rhizobia most probably have keys which fit many legume doors. Experiments with two different but closely related bacteria, Rhizobium sp. strain NGR234 and R. fredii USDA257, and 452 different species of legumes showed that NGR234 and USDA257 form fully functional nodules on 136 and 66 species of legumes, respectively (207). Exactly which symbiotic determinants are shared by these two rhizobia is not known, yet both produce similar Nod factors. Those of USDA257 lack the baroque decorations encoded by nodS, nodU, nolO, nolL, noeE, and noeI of NGR234 (Table 4). Similarly, nodZ mutants of NGR234 still nodulate most plants tested, although the Nod factors lack all substituents carried on the fucose group (Table 4). In other words, it seems as if these additional Nod factor substituents extend an already broad host range but that they are not necessary for basic promiscuity.
If this logic reflects evolution, then ancestral Nod factors were unmodified N-acylated oligomers of N-acetyl-d-glucosamine, possessing simple, unsaturated fatty acid side chains. It should be remembered that polymers of N-acetyl-d-glucosamine, i.e., chitin, are abundant in nature where they form an important part of the cell wall of many fungi, including mycorrhizae. Vesicular arbuscular mycorrhizae establish symbiotic associations with a broad range of vascular plants (116, 246). The existence of mycorrhizal-like structures in fossils strongly suggests that fungal-plant associations developed before rhizobial symbioses (216). Since highly sensitive recognition systems for chitin oligomers are found in diverse plants, it is not unreasonable to suggest that evolution of rhizobial perception in legumes resulted in a perception apparatus that is specific to N-acylated chitin oligomers (i.e., Nod factors) (259).
Presumably, a bacterium producing ancestral Nod factors had the ability to nodulate many plants. Later, modifications followed which, in the case of NGR234, allowed it to nodulate even more plants. Acquisition of these broad-host-range genes was probably the result of lateral transfer. Concomitantly, Nod factor-cleaving chitinases coevolved in the legume host, thus directing selective pressure toward more stable Nod factors. In NGR234, this was provided through the acquisition of protective baroque decorations.
Morphological and molecular data suggest that both NGR234 and USDA257 have acquired the ability to nodulate three distinct subfamilies of plants, i.e., the Caesalpiniodieae, Mimosoideae, and Papilionoideae (207). In the Papilionoideae, this ability arose early and was maintained through to the Amorpheae, Robinieae, Indigofereae, Phaseoleae, and Desmodieae. Once acquired, the capacity to form symbioses with promiscuous rhizobia was only apparently lost in five tribes including the Cicereae, Vicieae, and Trifolieae. Since the microsymbionts of these plants generally secrete fewer, less highly modified Nod factors (albeit possessing polyunsaturated fatty acids), this implies that narrow host range is a specialization which developed for certain plants in restricted niches.
In other words, symbiotic promiscuity is probably ancestral to restricted host range. Support for this hypothesis comes from the observation that NGR234 and USDA257 also nodulate Parasponia andersonii (Ulmaceae) (207, 269). Parasponia spp. are small trees (up to 15 m high) which grow as pioneer plants in mountainous areas of Indonesia, Malaysia, and Papua New Guinea (1). Since Trinick isolated NGR234 in Papua New Guinea (268), this means that Parasponia-Rhizobium symbioses evolved in the same habitat. Most probably, the direct correlation that exists between solar energy input and species diversity (228) is driving evolution in tropical regions. Thus, ancestors of bacteria such as NGR234 could have been the first to intimately associate with legumes.
ACKNOWLEDGMENTS
We thank C. Boivin, J. Dénarié, W. D'Haeze, J. A. Downie, M. Holsters, S. Jabbouri, T. Laeremans, K. Lindström, E. Martínez-Romero, G. Stacey, and J. Vanderleyden for helpful comments during the preparation of the manuscript. We are grateful to B. Relić for providing photographs of infected root hairs. Dora Gerber is thanked for her unfailing support of our work.
Financial assistance was provided by the Fonds National de la Recherche Scientifique and the Université de Genève.
REFERENCES
- 1.Akkermans A D L, van Dijk C. Non-leguminous root-nodule symbiosis with actinomycetes and Rhizobium. In: Broughton W J, editor. Nitrogen fixation. 1. Ecology. Oxford, United Kingdom: Oxford University Press; 1981. pp. 57–103. [Google Scholar]
- 2.Ardourel M, Demont N, Debellé F, Maillet F, DeBilly F, Promé J-C, Dénarié J, Truchet G. Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell. 1994;6:1357–1374. doi: 10.1105/tpc.6.10.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banfalvi Z, Kondorosi Á. Production of root hair deformation factors by Rhizobium meliloti nodulation genes in Escherichia coli: HsnD (NodH) is involved in the plant-specific modification of the NodABC factor. Plant Mol Biol. 1989;13:1–12. doi: 10.1007/BF00027330. [DOI] [PubMed] [Google Scholar]
- 4.Bassam B J, Djordjevic M A, Redmond J W, Batley M, Rolfe B G. Identification of a nodD-dependent locus in the Rhizobium strain NGR234 activated by phenolic factors secreted by soybeans and other legumes. Mol Plant-Microbe Interact. 1988;1:161–168. doi: 10.1094/mpmi-1-161. [DOI] [PubMed] [Google Scholar]
- 5.Battisti L, Lara J C, Leigh J A. Specific oligosaccharide form of the Rhizobium meliloti exopolysaccharide promotes nodule invasion in alfalfa. Proc Natl Acad Sci USA. 1992;89:5625–5629. doi: 10.1073/pnas.89.12.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bec-Ferté M P, Krishnan H B, Promé D, Savagnac A, Pueppke S G, Promé J-C. Structures of nodulation factors from the nitrogen-fixing soybean symbiont Rhizobium fredii USDA257. Biochemistry. 1994;33:11782–11788. doi: 10.1021/bi00205a014. [DOI] [PubMed] [Google Scholar]
- 7.Bec-Ferté M P, Krishnan H B, Savagnac A, Pueppke S G, Promé J-C. Rhizobium fredii synthesizes an array of lipooligosaccharides, including a novel compound with glucose inserted into the backbone of the molecule. FEBS Lett. 1996;393:273–279. doi: 10.1016/0014-5793(96)00903-9. [DOI] [PubMed] [Google Scholar]
- 8.Becker A, Kleickmann A, Keller M, Arnold W, Pühler A. Identification and analysis of the Rhizobium meliloti exoAMONP genes involved in exopolysaccharide biosynthesis and mapping of promoters located on the exoHKLAMONP fragment. Mol Gen Genet. 1993;241:367–379. doi: 10.1007/BF00284690. [DOI] [PubMed] [Google Scholar]
- 9.Becker A, Kleickmann A, Küster H, Keller M, Arnold W, Pühler A. Analysis of the Rhizobium meliloti genes exoU, exoV, exoW, exoT, and exoI involved in exopolysaccharide biosynthesis and nodule invasion: exoU and exoW probably encode glucosyltransferases. Mol Plant-Microbe Interact. 1993;6:735–744. doi: 10.1094/mpmi-6-735. [DOI] [PubMed] [Google Scholar]
- 10.Becker B U, Kosch K, Parniske M, Müller P. Exopolysaccharide (EPS) synthesis in Bradyrhizobium japonicum: sequence, operon structure and mutational analysis of an exo gene cluster. Mol Gen Genet. 1998;259:161–171. doi: 10.1007/s004380050801. [DOI] [PubMed] [Google Scholar]
- 11.Bender G L, Nayudu M, Strange K K L, Rolfe B G. The nodD1 gene from Rhizobium strain NGR234 is a key determinant in the extension of host range to the nonlegume Parasponia. Mol Plant-Microbe Interact. 1988;1:259–266. [Google Scholar]
- 12.Berck S, Perret X, Quesada-Vincens D, Promé J-C, Broughton W J, Jabbouri S. nolL of Rhizobium sp. strain NGR234 is required for O-acetyltransferase activity. J Bacteriol. 1999;181:957–964. doi: 10.1128/jb.181.3.957-964.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beyerinck M W. Die Bacterien der Papilionaceen-Knöllchen. Bot Zeitung. 1888;46:725–740. [Google Scholar]
- 14.Beyerinck M W. Die Bacterien der Papilionaceen-Knöllchen. Bot Zeitung. 1888;46:741–750. [Google Scholar]
- 15.Beyerinck M W. Die Bacterien der Papilionaceen-Knöllchen. Bot Zeitung. 1888;46:757–771. [Google Scholar]
- 16.Beyerinck M W. Die Bacterien der Papilionaceen-Knöllchen. Bot Zeitung. 1888;46:781–790. [Google Scholar]
- 17.Beyerinck M W. Die Bacterien der Papilionaceen-Knöllchen. Bot Zeitung. 1888;46:797–804. [Google Scholar]
- 18.Beyerinck M W. Künstliche Infection von Vicia Faba mit Bacillus radicicola. Ernährungsbedingungen dieser Bacterie. Bot Zeitung. 1890;52:837–843. [Google Scholar]
- 19.Bhagwat A A, Gross K C, Tully R E, Keister D. β-Glucan synthesis in Bradyrhizobium japonicum: characterization of a new locus (ndvC) influencing β-(1→6) linkages. J Bacteriol. 1996;178:4635–4642. doi: 10.1128/jb.178.15.4635-4642.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhagwat A A, Mithofer A, Pfeffer P E, Kraus C, Spickers N, Hotchkiss A, Ebel J, Keister D L. Further studies of the role of cyclic β-glucans in symbiosis. An ndvC mutant of Bradyrhizobium japonicum synthesizes cyclodecakis-(1→3)-β-glucosyl. Plant Physiol. 1999;119:1057–1064. doi: 10.1104/pp.119.3.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bibb M J, Biro S, Motamedi H, Collins J F, Hutchinson C R. Analysis of the nucleotide sequence of the Streptomyces glaucescens tcmI genes provides key information about the enzymology of polypeptide antibiotic biosynthesis. EMBO J. 1989;8:2727–2736. doi: 10.1002/j.1460-2075.1989.tb08414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bloemberg G V, Kamst E, Harteveld M, van Der Drift K M, Haverkamp J, Thomas-Oates J E, Lugtenberg B J J, Spaink H P. A central domain of Rhizobium NodE protein mediates host specificity by determining the hydrophobicity of fatty acyl moieties of nodulation factors. Mol Microbiol. 1995;16:1123–1136. doi: 10.1111/j.1365-2958.1995.tb02337.x. [DOI] [PubMed] [Google Scholar]
- 23.Bono J J, Riond J, Nicolaou K C, Bockovich N J, Estevez V A, Cullimore J V, Ranjeva R. Characterization of a binding site for chemically synthesized lipo-oligosaccharidic NodRm factors in particulate fractions prepared from roots. Plant J. 1995;7:253–260. doi: 10.1046/j.1365-313x.1995.7020253.x. [DOI] [PubMed] [Google Scholar]
- 24.Borthakur D, Barbur C E, Lamb J W, Daniels M J, Downie J A, Johnston A W B. A mutation that blocks exopolysaccharide synthesis prevents nodulation of peas by Rhizobium leguminosarum but not of beans by R. phaseoli and is corrected by cloned DNA from Rhizobium or the phytopathogen Xanthomonas. Mol Gen Genet. 1986;203:320–323. [Google Scholar]
- 25.Bréal E. Observations sur les tubercules à bactéries qui se développent sur les racines des légumineuses. Ann Agron. 1888;14:481–495. [Google Scholar]
- 26.Breedveld M W, Miller K J. Cyclic β-glucans of members of the family Rhizobiaceae. Microbiol Rev. 1994;58:145–161. doi: 10.1128/mr.58.2.145-161.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Broughton W J, John C K. Rhizobia in tropical legumes. III. Experimentation and supply in Malaysia 1927–1976. In: Broughton W J, John C K, Rajaro J C, Lim B, editors. Soil microbiology and plant nutrition. Kuala Lumpur: University of Malaya Press; 1979. pp. 113–136. [Google Scholar]
- 28.Broughton W J, Perret X. Genealogy of Legume-Rhizobium symbioses. Curr Opinion in Plant Biol. 1999;2:305–311. doi: 10.1016/S1369-5266(99)80054-5. [DOI] [PubMed] [Google Scholar]
- 29.Reference deleted.
- 30.Burn J, Rossen L, Johnston A W B. Four classes of mutations in the nodD gene of Rhizobium leguminosarum biovar. viciae that affect its ability to autoregulate and/or activate other nod genes in the presence of flavonoid inducers. Genes Dev. 1987;1:456–464. [Google Scholar]
- 31.Campbell G R O, Reuhs B L, Walker G C. Different phenotypic classes of Sinorhizobium meliloti mutants defective in synthesis of K. antigen. J Bacteriol. 1998;180:5432–5436. doi: 10.1128/jb.180.20.5432-5436.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Canter-Cremers H C J, Spaink H P, Wijfjes A H M, Pees E, Wijffelman C A, Okker R J H, Lugtenberg B J J. Additional nodulation genes on the Sym plasmid of Rhizobium leguminosarum biovar viciae. Plant Mol Biol. 1989;13:163–174. doi: 10.1007/BF00016135. [DOI] [PubMed] [Google Scholar]
- 33.Cardenas L, Dominguez J, Quinto C, Lopez-Lara I M, Lugtenberg B J J, Spaink H P, Rademaker G J, Haverkamp J, Thomas-Oates J E. Isolation, chemical structure and biological activity of the lipo-chitin oligosaccharide nodulation signals from Rhizobium etli. Plant Mol Biol. 1995;29:453–464. doi: 10.1007/BF00020977. [DOI] [PubMed] [Google Scholar]
- 34.Cardenas L, Dominguez J, Santana O, Quinto C. The role of nodI and nodJ genes in the transport of Nod metabolites in Rhizobium etli. Gene. 1996;173:183–187. doi: 10.1016/0378-1119(96)00166-7. [DOI] [PubMed] [Google Scholar]
- 35.Carlson R W, Sanjuan J, Bhat U R, Glushka J, Spaink H P, Wijfjes A H, van Brussel A A N, Stokkermans T J, Peters N K, Stacey G. The structures and biological activities of the lipo-oligosaccharide nodulation signals produced by type I and II strains of Bradyrhizobium japonicum. J Biol Chem. 1993;268:18372–18381. [PubMed] [Google Scholar]
- 36.Cava J R, Elias P M, Turowski D A, Noel K D. Rhizobium leguminosarum CFN42 genetic regions encoding lipopolysaccharide structures essential for complete nodule development on bean plants. J Bacteriol. 1989;171:8–15. doi: 10.1128/jb.171.1.8-15.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chakravorty A K, Zurowski W, Shine J, Rolfe B G. Symbiotic nitrogen fixation: molecular cloning of Rhizobium genes involved in exopolysaccharide synthesis and effective nodulation. J Mol Appl Genet. 1982;1:585–596. [PubMed] [Google Scholar]
- 38.Chang C, Kwok S F, Bleecker A B, Meyerowitz E M. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- 39.Chen H, Batley M, Redmond J, Rolfe B G. Alteration of the effective nodulation properties of a fast-growing broad host range Rhizobium due to changes in exopolysaccharide synthesis. J Plant Physiol. 1985;120:331–349. [Google Scholar]
- 40.Cheng H-P, Walker G C. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J Bacteriol. 1998;180:5183–5191. doi: 10.1128/jb.180.19.5183-5191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng H-P, Walker G C. Succinoglycan production by Rhizobium meliloti is regulated through the ExoS-Chv1 two-component regulatory system. J Bacteriol. 1998;180:20–26. doi: 10.1128/jb.180.1.20-26.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohn J, Day R B, Stacey G. Legume nodule organogenesis. Trends Plant Sci. 1998;3:105–110. [Google Scholar]
- 43.Cormack R G H. Development of root hairs in angiosperms. Bot Rev. 1962;28:446–464. [Google Scholar]
- 44.Corvera A, Promé D, Promé J-C, Martínez-Romero E, Romero D. The nolL gene from Rhizobium etli determines nodulation efficiency by mediating the acetylation of the fucosyl residue in the nodulation factor. Mol Plant-Microbe Interact. 1999;12:236–246. doi: 10.1094/MPMI.1999.12.3.236. [DOI] [PubMed] [Google Scholar]
- 45.Cren M, Kondorosi Á, Kondorosi É. An insertional point mutation inactivates NolR repressor in Rhizobium meliloti 1021. J Bacteriol. 1994;176:518–519. doi: 10.1128/jb.176.2.518-519.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cren M, Kondorosi Á, Kondorosi É. NolR controls expression of the Rhizobium meliloti nodulation genes involved in the core Nod factor synthesis. Mol Microbiol. 1995;15:733–747. doi: 10.1111/j.1365-2958.1995.tb02381.x. [DOI] [PubMed] [Google Scholar]
- 47.Dart P. Infection and development of leguminous nodules. In: Hardy R W F, Silver W S, editors. A treatise on dinitrogen fixation, sect. 1977. pp. 367–342. III. Cambridge University Press, London, United Kingdom. [Google Scholar]
- 48.Davis E O, Evans I J, Johnston A W B. Identification of nodX, a gene that allows Rhizobium leguminosarum biovar viciae strain TOM to nodulate Afghanistan peas. Mol Gen Genet. 1988;212:531–535. doi: 10.1007/BF00330860. [DOI] [PubMed] [Google Scholar]
- 49.Debellé F, Sharma S B. Nucleotide sequence of Rhizobium meliloti RC2011 genes involved in host specificity of nodulation. Nucleic Acids Res. 1986;14:7453–7472. doi: 10.1093/nar/14.18.7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Debellé F, Maillet F, Vasse J, Rosenberg C, De Billy F, Truchet G, Dénarié J, Ausubel F M. Interference between Rhizobium meliloti and Rhizobium trifolii nodulation genes: genetic basis of R. meliloti dominance. J Bacteriol. 1988;170:5718–5727. doi: 10.1128/jb.170.12.5718-5727.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Debellé F, Plazanet C, Roche P, Pujol C, Savagnac A, Rosenberg C, Promé J-C, Dénarié J. The NodA proteins of Rhizobium meliloti and Rhizobium tropici specify the N-acylation of Nod factors by different fatty acids. Mol Microbiol. 1996;22:303–314. doi: 10.1046/j.1365-2958.1996.00069.x. [DOI] [PubMed] [Google Scholar]
- 52.Demont N, Debellé F, Aurelle H, Dénarié J, Promé J-C. Role of the Rhizobium meliloti nodF and nodE genes in the biosynthesis of lipooligosaccharidic nodulation factors. J Biol Chem. 1993;268:20134–20142. [PubMed] [Google Scholar]
- 53.Demont-Caulet N, Maillet F, Tailler D, Jacquinet J-C, Promé J-C, Nicolaou K C, Truchet G, Beau J-M, Dénarié J. Nodule-inducing activity of synthetic Sinorhizobium meliloti nodulation factors and related lipo-chitooligosaccharides on alfalfa. Importance of the acyl chain structure. Plant Physiol. 1999;120:83–92. doi: 10.1104/pp.120.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dénarié J, Debellé F, Rosenberg C. Signaling and host range variation in nodulation. Annu Rev Microbiol. 1992;46:497–531. doi: 10.1146/annurev.mi.46.100192.002433. [DOI] [PubMed] [Google Scholar]
- 55.Reference deleted.
- 56.D'Haeze W, Gao M, De Rycke R, van Montagu M, Engler G, Holsters M. Roles for azorhizobial Nod factors and surface polysaccharides in intercellular invasion and nodule penetration, respectively. Mol Plant-Microbe Interact. 1998;11:999–1008. [Google Scholar]
- 57.D'Haeze W, van Montagu M, Promé J-C, Holsters M. Carbamoylation of azorhizobial Nod factors is mediated by NodU. Mol Plant-Microbe Interact. 1999;12:68–73. doi: 10.1094/MPMI.1999.12.1.68. [DOI] [PubMed] [Google Scholar]
- 58.Diebold R, Noel K D. Rhizobium leguminosarum exopolysaccharide mutants: biochemical and genetic analyses and symbiotic behavior on three hosts. J Bacteriol. 1989;171:4821–4830. doi: 10.1128/jb.171.9.4821-4830.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Djordjevic M A, Schofield P R, Rolfe B G. Tn-5 mutagenesis of Rhizobium trifolii host specific nodulation genes results in mutants with altered host range ability. Mol Gen Genet. 1985;200:463–471. [Google Scholar]
- 60.Djordjevic S P, Rolfe B G. The structure of the exopolysaccharide from Rhizobium sp. strain ANU280 (NGR234) Carbohydr Res. 1986;148:87–99. [Google Scholar]
- 61.Djordjevic M A, Redmond J W, Batley M, Rolfe B G. Clovers secrete specific phenolic compounds which either stimulate or repress nod gene expression in Rhizobium trifolii. EMBO J. 1987;6:1173–1178. doi: 10.1002/j.1460-2075.1987.tb02351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Downie J A, Surin B. Either of two nod gene loci can complement the nodulation defect of a nod deletion mutant of Rhizobium leguminosarum bv. viciae. Mol Gen Genet. 1990;222:81–86. doi: 10.1007/BF00283027. [DOI] [PubMed] [Google Scholar]
- 63.Downie J A. Functions of rhizobial nodulation genes. In: Spaink H P, Kondorosi Á, Hooykaas P J J, editors. The Rhizobiaceae. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 387–402. [Google Scholar]
- 64.Downie J A, Walker S W. Plant responses to nodulation factors. Curr Opin Plant Biol. 1999;2:483–489. doi: 10.1016/s1369-5266(99)00018-7. [DOI] [PubMed] [Google Scholar]
- 65.Dunlap J, Minami E, Bhagwat A A, Keister D L, Stacey G. Nodule development induced by mutants of Bradyrhizobium japonicum defective in cyclic β-glucan synthesis. Mol Plant-Microbe Interact. 1996;9:546–555. doi: 10.1094/mpmi-9-0546. [DOI] [PubMed] [Google Scholar]
- 66.Economou A, Hamilton W D O, Johnston A W B, Downie J A. The Rhizobium nodulation gene nodO encodes a Ca2+-binding protein that is exported without N-terminal cleavage and is homologous to haemolysin and related proteins. EMBO J. 1990;9:349–354. doi: 10.1002/j.1460-2075.1990.tb08117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Economou A, Davies A E, Johnston A W B, Downie J A. The Rhizobium leguminosarum biovar viciae nodO gene can enable a nodE mutant of Rhizobium leguminosarum biovar trifolii to nodulate vetch. Microbiology. 1994;140:2341–2347. [Google Scholar]
- 68.Ehrhardt D W, Atkinson E M, Faull K F, Freedberg D I, Sutherlin D P, Armstrong R, Long S R. In vitro sulfotransferase activity of NodH, a nodulation protein of Rhizobium meliloti required for host-specific nodulation. J Bacteriol. 1995;177:6237–6245. doi: 10.1128/jb.177.21.6237-6245.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Etzler M E, Kalsi G, Ewing N N, Roberts N J, Day R B, Murphy J B. A nod factor binding lectin with apyrase activity from legume roots. Proc Natl Acad Sci USA. 1999;96:5856–5861. doi: 10.1073/pnas.96.10.5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Faucher C, Camut S, Dénarié J, Truchet G. The nodH and nodQ host range genes of Rhizobium meliloti behave as avirulence genes in R. leguminosarum bv. viciae and determine changes in the production of plant-specific extracellular signals. Mol Plant-Microbe Interact. 1989;2:291–300. [Google Scholar]
- 71.Fellay R, Perret X, Viprey V, Broughton W J, Brenner S. Organisation of host-inducible transcripts on the symbiotic plasmid of Rhizobium sp. NGR234. Mol Microbiol. 1995;16:657–667. doi: 10.1111/j.1365-2958.1995.tb02428.x. [DOI] [PubMed] [Google Scholar]
- 72.Fellay R, Rochepeau P, Relić B, Broughton W J. Signals to and emanating from Rhizobium largely control symbiotic specificity. In: Singh U S, Singh R P, Kohmoto K, editors. Pathogenesis and host specificity in plant diseases. Histopathological, biochemical, genetic and molecular bases. Oxford, United Kingdom: Pergamon Elsevier Science Ltd.; 1995. pp. 199–220. [Google Scholar]
- 73.Fellay R, Hanin M, Montorzi G, Frey J, Freiberg C, Golinowski W, Staehelin C, Broughton W J, Jabbouri S. nodD2 of Rhizobium sp. NGR234 is involved in repression of the nodABC operon. Mol Microbiol. 1998;27:1039–1050. doi: 10.1046/j.1365-2958.1998.00761.x. [DOI] [PubMed] [Google Scholar]
- 74.Fernandez-Lopez M, D'Haeze W, Mergaert P, Verplancke C, Promé J-C, van Montagu M, Holsters M. Role of nodI and nodJ in lipochitooligosaccharide secretion in Azorhizobium caulinodans and Escherichia coli. Mol Microbiol. 1996;20:993–1000. doi: 10.1111/j.1365-2958.1996.tb02540.x. [DOI] [PubMed] [Google Scholar]
- 75.Fernandez-Lopez M, D'Haeze W, van Montagu M, Holsters M. Changes in the glycosylation pattern at the reducing end of azorhizobial Nod factors affect nodulation efficiency. FEMS Microbiol Lett. 1998;158:237–242. [Google Scholar]
- 76.Finan T M, Hirsch A M, Leigh J A, Johansen E, Kuldau G A, Deegan S, Walker G C, Signer E R. Symbiotic mutants of Rhizobium meliloti that uncouple plant from bacterial differentiation. Cell. 1985;40:869–877. doi: 10.1016/0092-8674(85)90346-0. [DOI] [PubMed] [Google Scholar]
- 77.Finnie C, Hartley N M, Findlay K C, Downie J A. The Rhizobium leguminosarum prsDE genes are required for secretion of several proteins, some of which influence nodulation, symbiotic nitrogen fixation and exopolysaccharide modification. Mol Microbiol. 1997;25:135–146. doi: 10.1046/j.1365-2958.1997.4471803.x. [DOI] [PubMed] [Google Scholar]
- 78.Finnie C, Zorreguieta A, Hartley N M, Downie J A. Characterization of Rhizobium leguminosarum exopolysaccharide glycanases that are secreted via a type I exporter and have a novel heptapeptide repeat motif. J Bacteriol. 1998;180:1691–1699. doi: 10.1128/jb.180.7.1691-1699.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Firmin J L, Wilson K E, Rossen L, Johnston A W B. Flavonoid activation of nodulation genes in Rhizobium reversed by other compounds present in plants. Nature. 1986;324:90–92. [Google Scholar]
- 80.Firmin J L, Wilson K E, Carlson R W, Davies A E, Downie J A. Resistance to nodulation of cv. Afghanistan peas is overcome by nodX, which mediates an O-acetylation of the Rhizobium leguminosarum lipooligosaccharide nodulation factor. Mol Microbiol. 1993;10:351–360. doi: 10.1111/j.1365-2958.1993.tb01961.x. [DOI] [PubMed] [Google Scholar]
- 81.Fisher R F, Egelhoff T T, Mulligan J T, Long S R. Specific binding of proteins from Rhizobium meliloti cell-free extracts containing NodD to DNA sequences upstream of inducible nodulation genes. Genes Dev. 1988;2:282–293. doi: 10.1101/gad.2.3.282. [DOI] [PubMed] [Google Scholar]
- 82.Fisher R F, Long S R. Interactions of NodD at the nod box: NodD binds to two distinct sites on the same face of the helix and induces a bend in the DNA. J Mol Biol. 1993;233:336–348. doi: 10.1006/jmbi.1993.1515. [DOI] [PubMed] [Google Scholar]
- 83.Forsberg L S, Reuhs B L. Structural characterization of the K antigens from Rhizobium fredii USDA257: evidence for a common structural motif, with strain-specific variation, in the capsular polysaccharides of Rhizobium spp. J Bacteriol. 1997;179:5366–5371. doi: 10.1128/jb.179.17.5366-5371.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Forsberg L S, Carlson R W. The structures of the lipopolysaccharides from Rhizobium etli strains CE358 and CE359. The complete structure of the core region of R. etli lipopolysaccharides. J Biol Chem. 1998;273:2747–2757. doi: 10.1074/jbc.273.5.2747. [DOI] [PubMed] [Google Scholar]
- 85.Frank B. Über die Parasiten in den Wurzelanschwellungen der Papilionaceeen. Bot Zeitung. 1879;24:377–388. [Google Scholar]
- 86.Fred E B, Baldwin I L, McCoy E. Root nodule bacteria and leguminous plants. University of Wisconsin. Studies in Science. Vol. 5. University of Wisconsin, Madison; 1932. [Google Scholar]
- 87.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 88.Fuchsius L. De historia stirpium commentarii isignes. 1542. Michael Isingrin, Basel, Switzerland. [Google Scholar]
- 89.Gabriel D W, Rolfe B G. Working models of specific recognition in plant-microbe interactions. Annu Rev Phytopathol. 1990;28:365–391. [Google Scholar]
- 90.Gagnon H, Ibrahim R K. Aldonic acids: a novel family of nod gene inducers of Mesorhizobium loti, Rhizobium lupini, and Sinorhizobium meliloti. Mol Plant-Microbe Interact. 1998;11:988–998. [Google Scholar]
- 91.Garcia M, Dunlap J, Loh J, Stacey G. Phenotypic characterization and regulation of the nolA gene of Bradyrhizobium japonicum. Mol Plant-Microbe Interact. 1996;9:625–635. doi: 10.1094/mpmi-9-0625. [DOI] [PubMed] [Google Scholar]
- 92.Garcia-de los Santos A, Brom S. Characterization of two plasmid-borne lpsβ loci of Rhizobium etli required for lipopolysaccharide synthesis and for optimal interaction with plants. Mol Plant-Microbe Interact. 1997;7:891–902. doi: 10.1094/MPMI.1997.10.7.891. [DOI] [PubMed] [Google Scholar]
- 93.Geelen D, Mergaert P, Geremia R A, Goormachtig S, van Montagu M, Holsters M. Identification of nodSUIJ genes on Nod locus 1 of Azorhizobium caulinodans: evidence that nodS encodes a methyltransferase involved in Nod factor modification. Mol Microbiol. 1993;9:145–154. doi: 10.1111/j.1365-2958.1993.tb01676.x. [DOI] [PubMed] [Google Scholar]
- 94.Geelen D, Leyman B, Mergaert P, Klarskov K, van Montagu M, Geremia R A, Holsters M. NodS is an S-adenosyl-l-methionine-dependent methyltransferase that methylates chitooligosaccharides deacetylated at the non-reducing end. Mol Microbiol. 1995;17:387–397. doi: 10.1111/j.1365-2958.1995.mmi_17020387.x. [DOI] [PubMed] [Google Scholar]
- 95.Geremia R A, Mergaert P, Geelen D, van Montagu M, Holsters M. The NodC protein of Azorhizobium caulinodans is an N-acetylglucosaminyltransferase. Proc Natl Acad Sci USA. 1994;91:2669–2673. doi: 10.1073/pnas.91.7.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gerhold D, Stacey G, Kondorosi Á. Use of promoter-specific probe to identify two loci from the Rhizobium meliloti nodulation regulon. Plant Mol Biol. 1989;12:181–188. doi: 10.1007/BF00020503. [DOI] [PubMed] [Google Scholar]
- 97.Geurts R, Heidstra R, Hadri A E, Downie J A, Franssen H, van Kammen A, Bisseling T. Sym2 of pea is involved in a nodulation factor-perception mechanism that controls the infection process in the epidermis. Plant Physiol. 1997;115:351–359. doi: 10.1104/pp.115.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gidley M J, Dea I C M, Eggleston G, Morris E R. Structure and gelation of Rhizobium capsular polysaccharide. Carbohydr Res. 1987;160:381–396. [Google Scholar]
- 99.Glazebrook J, Walker G C. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell. 1989;56:661–672. doi: 10.1016/0092-8674(89)90588-6. [DOI] [PubMed] [Google Scholar]
- 100.Glowacka M, Urbanik T, Derylo M. Rhizobium trifolii mutant defective in the structure of lipopolysaccharide. Arch Microbiol. 1989;152:539–541. [Google Scholar]
- 101.Goethals K, van Montagu M, Holsters M. Conserved motifs in a divergent nod box of Azorhizobium caulinodans ORS571 reveal a common structure in promoters regulated by LysR-type proteins. Proc Natl Acad Sci USA. 1992;89:1646–1650. doi: 10.1073/pnas.89.5.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gonzalez J E, Reuhs B L, Walker G C. Low molecular weight EPS II of Rhizobium meliloti allows nodule invasion in Medicago sativa. Proc Natl Acad Sci USA. 1996;93:8636–8641. doi: 10.1073/pnas.93.16.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Goormachtig S, Lievens S, van de Velde W, van Montagu M, Holsters M. Srchi13, a novel early nodulin from Sesbania rostrata, is related to acidic class III chitinases. Plant Cell. 1998;10:905–915. doi: 10.1105/tpc.10.6.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Göttfert M, Grob P, Hennecke H. Proposed regulatory pathway encoded by the nodV and nodW genes, determinants of host specificity in Bradyrhizobium japonicum. Proc Natl Acad Sci USA. 1990;87:2680–2684. doi: 10.1073/pnas.87.7.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Göttfert M, Holzhauser D, Bani D, Hennecke H. Structural and functional analysis of two different nodD genes in Bradyrhizobium japonicum USDA110. Mol Plant-Microbe Interact. 1992;5:257–265. doi: 10.1094/mpmi-5-257. [DOI] [PubMed] [Google Scholar]
- 106.Gray J X, Zhan H, Levery S B, Battisti L, Rolfe B G, Leigh J A. Heterologous exopolysaccharide production in Rhizobium sp. strain NGR234 and consequences for nodule development. J Bacteriol. 1991;173:3066–3077. doi: 10.1128/jb.173.10.3066-3077.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gressent F, Bono J J, Niebel A, Canut H, Cullimore J V, Ranjeva R. Characterization of a high affinity binding site for NodRm factors in Medicago varia cell culture extracts. In: Elmerich C, Kondorosi Á, Newton W E, editors. Biological nitrogen fixation for the 21st century. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 213–214. [Google Scholar]
- 108.Gressent F, Drouillard S, Mantegazza N, Samain E, Geremia R A, Canut H, Niebel A, Driguez H, Ranjeva R, Cullimore J, Bono J J. Ligand specificity of a high-affinity binding site for lipo-chitooligosaccharidic nod factors in Medicago cell suspension cultures. Proc Natl Acad Sci USA. 1999;96:4704–4709. doi: 10.1073/pnas.96.8.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Grob P, Michel P, Hennecke H, Göttfert M. A novel response-regulator is able to suppress the nodulation defect of a Bradyrhizobium japonicum nodW mutant. Mol Gen Genet. 1993;241:531–541. doi: 10.1007/BF00279895. [DOI] [PubMed] [Google Scholar]
- 110.Györgypal Z, Kiss G B, Kondorosi Á. Transduction of plant signal molecules by the Rhizobium NodD proteins. Bioessays. 1991;13:575–581. doi: 10.1002/bies.950131106. [DOI] [PubMed] [Google Scholar]
- 111.Györgypal Z, Kondorosi Á. Homology of the ligand-binding regions of Rhizobium symbiotic regulatory protein NodD and vertebrate nuclear receptors. Mol Gen Genet. 1991;226:337–340. doi: 10.1007/BF00273624. [DOI] [PubMed] [Google Scholar]
- 112.Györgypal Z, Kondorosi É, Kondorosi Á. Diverse signal sensitivity of NodD protein homologs from narrow and broad host range rhizobia. Mol Plant-Microbe Interact. 1991;4:356–364. [Google Scholar]
- 113.Hanin M, Jabbouri S, Quesada-Vincens D, Freiberg C, Perret X, Promé J-C, Broughton W J, Fellay R. Sulphation of Rhizobium sp. NGR234 Nod factors is dependent on noeE, a new host-specificity gene. Mol Microbiol. 1997;24:1119–1129. doi: 10.1046/j.1365-2958.1997.3981777.x. [DOI] [PubMed] [Google Scholar]
- 114.Hanin M, Jabbouri S, Broughton W J, Fellay R. SyrM1 of Rhizobium sp. NGR234 activates transcription of symbiotic loci and controls the level of sulfated Nod factors. Mol Plant-Microbe Interact. 1998;11:343–350. [Google Scholar]
- 115.Hanin M, Jabbouri S, Broughton W J, Fellay R, Quesada-Vincens D. Molecular aspects of host-specific nodulation. In: Stacey G, Keen N T, editors. Plant-microbe interactions. Vol. 4. St. Paul, Minn: American Phytopathological Society; 1999. pp. 1–37. [Google Scholar]
- 116.Harrison M J. Development of the arbuscular mycorrhizal symbiosis. Curr Opin Plant Biol. 1998;1:360–365. doi: 10.1016/1369-5266(88)80060-8. [DOI] [PubMed] [Google Scholar]
- 117.Hartwig U A, Maxwell C A, Joseph C M, Phillips D A. Effects of alfalfa nod gene-inducing flavonoids on nodABC transcription in Rhizobium meliloti strains containing different nodD genes. J Bacteriol. 1990;172:2769–2773. doi: 10.1128/jb.172.5.2769-2773.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Heidstra R, Geurts R, Franssen H, Spaink H P, van Kammen A, Bisseling T. Root hair deformation activity of nodulation factors and their fate in Vicia sativa. Plant Physiol. 1994;105:787–797. doi: 10.1104/pp.105.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Helal H M, Sauerbeck D. Carbon turnover in the rhizosphere. Z Pflanzenernaehr Bodenkd. 1989;152:211–216. [Google Scholar]
- 120.Hellriegel H. Welche Stickstoffquellen stehen der Pflanze zu Gebote? Landwirtsch Versuchstat Dresden. 1886;33:464–465. [Google Scholar]
- 121.Hellriegel H, Wilfarth H. Untersuchungen über die Stickstoff-nahrung der Gramineen und Leguminosen. Beilageheft zu der Zeitschrift des Vereins für die Rübenzucker-Industrie des Deutschen Reiches, Buchdruckerei der “Post.”. 1888. Kayssler & Co., Berlin, Germany. [Google Scholar]
- 122.Hernandez-Lucas I, Segovia L, Martínez-Romero E, Pueppke S G. Phylogenetic relationships and host range of Rhizobium spp. that nodulate Phaseolus vulgaris. Appl Environ Microbiol. 1995;61:2775–2779. doi: 10.1128/aem.61.7.2775-2779.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hiltner, L. 1900. Über die Ursachen, welche die Grösse, Zahl, Stellung und Wirkung der Wurzelknöllchen der Leguminosen bedingen. Arbeiten aus der Biologischen Abteilung für Land- und Forstwirthschaft am Kaiserlichen Gesundheitsamte, Berlin 1:177–222.
- 124.Honma M A, Asomaning M, Ausubel F M. Rhizobium meliloti nodD genes mediate host-specific activation of nodABC. J Bacteriol. 1990;172:901–911. doi: 10.1128/jb.172.2.901-911.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Horvath B, Kondorosi É, John M, Schmidt J, Török I, Györgypal Z, Barabas I, Wieneke U, Schell J, Kondorosi Á. Organization, structural and symbiotic function of Rhizobium meliloti nodulation genes determining host specificity for alfalfa. Cell. 1986;46:335–343. doi: 10.1016/0092-8674(86)90654-9. [DOI] [PubMed] [Google Scholar]
- 126.Horvath B, Bachem C W, Schell J, Kondorosi Á. Host-specific regulation of nodulation genes in Rhizobium is mediated by a plant signal interacting with the nodD gene product. EMBO J. 1987;6:841–848. doi: 10.1002/j.1460-2075.1987.tb04829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hotter G S, Scott D B. Exopolysaccharide mutants of Rhizobium loti are fully effective on a determinate nodulating host but are ineffective on an indeterminate nodulating host. J Bacteriol. 1991;173:851–859. doi: 10.1128/jb.173.2.851-859.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Irving, H. I., N. M. Boukli, M. N. Kelly, and W. J. Broughton. Nod-factors in the symbiotic development of root hairs. In R. W. Ridge and A. M. Emons (ed.), Cell and molecular biology of plant root hairs, in press. Springer-Verlag KG, Berlin, Germany.
- 129.Jabbouri S, Fellay R, Talmont F, Kamalaprija P, Burger U, Relić B, Promé J-C, Broughton W J. Involvement of nodS in N-methylation and nodU in 6-O-carbamoylation of Rhizobium sp. NGR234 Nod factors. J Biol Chem. 1995;270:22968–22973. doi: 10.1074/jbc.270.39.22968. [DOI] [PubMed] [Google Scholar]
- 130.Jabbouri S, Hanin M, Fellay R, Quesada-Vincens D, Reuhs B, Carlson R W, Perret X, Freiberg C, Rosenthal A, Leclerc D, Broughton W J, Relić B. Rhizobium species NGR234 host-specificity of nodulation locus III contains nod- and fix-genes. In: Stacey G, Mullen B, Gresshoff P, editors. Biology of plant-microbe interactions. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 319–324. [Google Scholar]
- 131.Jabbouri S, Relić B, Hanin M, Burger U, Promé J-C, Broughton W J. nolO and noeI (HsnIII) of Rhizobium sp. NGR234 are involved in 3-O-carbamoylation and 2-O-methylation of Nod-factors. J Biol Chem. 1998;273:12047–12055. doi: 10.1074/jbc.273.20.12047. [DOI] [PubMed] [Google Scholar]
- 132.John M, Röhrig H, Schmidt J, Wieneke U, Schell J. Rhizobium NodB protein involved in nodulation signal synthesis is a chitooligosaccharide deacetylase. Proc Natl Acad Sci USA. 1993;90:625–629. doi: 10.1073/pnas.90.2.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kamst E, van der Drift K M, Thomas-Oates J E, Lugtenberg B J J, Spaink H P. Mass spectrometric analysis of chitin oligosaccharides produced by Rhizobium NodC protein in Escherichia coli. J Bacteriol. 1995;21:6282–6285. doi: 10.1128/jb.177.21.6282-6285.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kamst E, Pilling J, Raamsdonk L M, Lugtenberg B J J, Spaink H P. Rhizobium nodulation protein NodC is an important determinant of chitin oligosaccharide chain length in Nod factor biosynthesis. J Bacteriol. 1997;179:2103–2108. doi: 10.1128/jb.179.7.2103-2108.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kamst E, Bakkers J, Quaedvlieg N E, Pilling J, Kijne J W, Lugtenberg B J J, Spaink H P. Chitin oligosaccharide synthesis by rhizobia and zebrafish embryos starts by glycosyl transfer to O4 of the reducing-terminal residue. Biochemistry. 1999;38:4045–4052. doi: 10.1021/bi982531u. [DOI] [PubMed] [Google Scholar]
- 136.Kannenberg E L, Rathburn E A, Brewin N J. Molecular dissection of structure and function in the lipopolysaccharide of Rhizobium leguminosarum strain 3841 using monoclonal antibodies and genetic analysis. Mol Microbiol. 1992;6:2477–2487. doi: 10.1111/j.1365-2958.1992.tb01424.x. [DOI] [PubMed] [Google Scholar]
- 137.Kende H, Gardner G. Hormone binding in plants. Annu Rev Plant Physiol. 1976;27:267–290. [Google Scholar]
- 138.Kereszt A, Kiss E, Reuhs B L, Carlson R W, Kondorosi Á, Putnoky P. Novel rkp gene clusters of Sinorhizobium meliloti involved in capsular polysaccharide production and invasion of the symbiotic nodule: the rkpK gene encodes a UDP-glucose dehydrogenase. J Bacteriol. 1998;180:5426–5431. doi: 10.1128/jb.180.20.5426-5431.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kijne J W. The Rhizobium infection process. In: Stacey G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman & Hall; 1992. pp. 349–398. [Google Scholar]
- 140.Kim C H, Tully R E, Keister D L. Exopolysaccharide-deficient mutants of Rhizobium fredii HH303 which are symbiotically effective. Appl Environ Microbiol. 1989;55:1852–1854. doi: 10.1128/aem.55.7.1852-1854.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kiss E, Mergaert P, Olàh B, Kereszt A, Staehelin C, Davies A E, Downie J A, Kondorosi Á, Kondorosi É. Conservation of nolR in the Sinorhizobium and Rhizobium genera of the Rhizobiaceae family. Mol Plant-Microbe Interact. 1998;11:1186–1195. [Google Scholar]
- 142.Kondorosi É, Gyuris J, Schmidt J, John M, Duda E, Hoffman B, Schell J, Kondorosi Á. Positive and negative control of nod gene expression in Rhizobium meliloti is required for optimal nodulation. EMBO J. 1989;8:1331–1340. doi: 10.1002/j.1460-2075.1989.tb03513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kondorosi É, Pierre M, Cren M, Haumann U, Buire M, Hoffmann B, Schell J, Kondorosi Á. Identification of NolR, a negative transacting factor controlling the nod regulon in Rhizobium meliloti. J Mol Biol. 1991;222:885–896. doi: 10.1016/0022-2836(91)90583-r. [DOI] [PubMed] [Google Scholar]
- 144.Krishnan H B, Lewin A, Fellay R, Broughton W J, Pueppke S G. Differential expression of nodS accounts for the varied abilities of Rhizobium fredii USDA257 and Rhizobium sp. strain NGR234 to nodulate Leucaena spp. Mol Microbiol. 1992;6:3321–3330. doi: 10.1111/j.1365-2958.1992.tb02200.x. [DOI] [PubMed] [Google Scholar]
- 145.Krishnan H B, Kuo C-I, Pueppke S G. Elaboration of flavonoid-induced proteins by the nitrogen-fixing soybean symbiont Rhizobium fredii is regulated by both nodD1 and nodD2, and is dependent on the cultivar-specificity locus, nolXWBTUV. Microbiology. 1995;141:2245–2251. [Google Scholar]
- 146.Krishnan H B, Kim K Y, Krishnan A H. Expression of a Serratia marcescens chitinase gene in Sinorhizobium fredii USDA191 and Sinorhizobium meliloti RCR2011 impedes soybean and alfalfa nodulation. Mol Plant-Microbe Interact. 1999;12:748–751. doi: 10.1094/MPMI.1999.12.8.748. [DOI] [PubMed] [Google Scholar]
- 147.Król J, Wielbo J, Mazur A, Kopciñska J, Lotocka B, Golinowski W, Skorupska A. Molecular characterisation of pssCDE genes of Rhizobium leguminosarum bv. trifolii strain TA1: pssD mutant is affected in exopolysaccharide synthesis and endocytosis of bacteria. Mol Plant-Microbe Interact. 1998;11:1142–1148. doi: 10.1094/MPMI.1998.11.11.1142. [DOI] [PubMed] [Google Scholar]
- 148.Kurkdjian A C. Role of differentiation of root epidermal cells in Nod factor (from Rhizobium meliloti)-induced root-hair depolarization of Medicago sativa. Plant Physiol. 1995;107:783–790. doi: 10.1104/pp.107.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Laurent E. Recherches sur les nodosités des légumineuses. Inst Pasteur Ann. 1891;5:105–139. [Google Scholar]
- 150.Lee C A. Type III secretion systems: machines to deliver bacterial proteins into eukaryotic cells? Trends Microbiol. 1997;5:149–156. doi: 10.1016/S0966-842X(97)01029-9. [DOI] [PubMed] [Google Scholar]
- 151.Leigh J A, Signer E R, Walker G C. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc Natl Acad Sci USA. 1985;82:6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Leigh J A, Lee C C. Characterization of polysaccharides of Rhizobium meliloti exo mutants that form ineffective nodules. J Bacteriol. 1988;170:3327–3332. doi: 10.1128/jb.170.8.3327-3332.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Leigh J A, Walker G C. Exopolysaccharides of Rhizobium: synthesis, regulation and symbiotic function. Trends Genet. 1994;10:63–67. doi: 10.1016/0168-9525(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 154.Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé J-C, Dénarié J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990;344:781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- 155.Le Strange K K, Bender G L, Djordjevic M A, Rolfe B G, Redmond J W. The Rhizobium strain NGR234 nodD1 gene product responds to activation by simple phenolic compounds vanillin and isovanillin present in wheat seedling extracts. Mol Plant-Microbe Interact. 1990;3:214–220. [Google Scholar]
- 156.Lewin A, Rosenberg C, Meyer z. A. H, Wong C-H, Nelson L, Manen J-F, Stanley J, Dowling D N, Dénarié J, Broughton W J. Multiple host-specificity loci of the broad host range Rhizobium sp. NGR234 selected using the widely compatible legume Vigna unguiculata. Plant Mol Biol. 1987;8:447–459. doi: 10.1007/BF00017990. [DOI] [PubMed] [Google Scholar]
- 157.Lewin A, Rosenberg C, Stanley J, Dowling D N, Manen J-F, Debellé F, Broughton W J. Multiple host-specificity loci in the broad host-range Rhizobium NGR234. In: Verma D P S, Brisson N, editors. Molecular genetics of plant-microbe interactions. Dordrecht, The Netherlands: Martinus Nijhoff; 1987. pp. 232–237. [Google Scholar]
- 158.Lewin A, Cervantes E, Wong C-H, Broughton W J. nodSU, two new nod-genes of the broad host range Rhizobium strain NGR234, encode host-specific nodulation of the tropical tree leucaena leucocephala. Mol Plant-Microbe Interact. 1990;3:317–326. doi: 10.1094/mpmi-3-317. [DOI] [PubMed] [Google Scholar]
- 159.Loh J, Garcia M, Stacey G. NodV and NodW, a second flavonoid recognition system regulating nod gene expression in Bradyrhizobium japonicum. J Bacteriol. 1997;179:3013–3020. doi: 10.1128/jb.179.9.3013-3020.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.López-Lara I M, van der Drift K M, van Brussel A A, Haverkamp J, Lugtenberg B J J, Thomas-Oates J E, Spaink H P. Induction of nodule primordia on Phaseolus and Acacia by lipo-chitin oligosaccharide nodulation signals from broad-host-range Rhizobium strain GRH2. Plant Mol Biol. 1995;29:465–477. doi: 10.1007/BF00020978. [DOI] [PubMed] [Google Scholar]
- 161.López-Lara I M, van der Berg J D J, Thomas-Oates J E, Glushka J, Lugtenberg B J J, Spaink H P. Structural identification of the lipo-chitin oligosaccharide nodulation signals of Rhizobium loti. Mol Microbiol. 1995;15:627–638. doi: 10.1111/j.1365-2958.1995.tb02372.x. [DOI] [PubMed] [Google Scholar]
- 162.López-Lara I M, Blok-Tip L, Quinto C, Garcia M L, Bloemberg G V, Lamers G E M, Kafetzopoulos D, Stacey G, Lugtenberg B J J, Thomas-Oates J E, Spaink H P. NodZ of Bradyrhizobium extends the nodulation host range of Rhizobium by adding a fucosyl residue to nodulation factors. Mol Microbiol. 1996;21:397–408. doi: 10.1046/j.1365-2958.1996.00644.x. [DOI] [PubMed] [Google Scholar]
- 163.Lorquin J, Lortet G, Ferro M, Méar N, Dreyfus B, Promé J-C, Boivin C. Nod factors from Sinorhizobium saheli and S. teranaga bv. sesbania are both arabinosylated and fucosylated, a structural feature specific to Sesbania rostrata. Mol Plant-Microbe Interact. 1997;10:879–890. [Google Scholar]
- 164.Lorquin J, Lortet G, Ferro M, Méar N, Promé J-C, Boivin C. Sinorhizobium teranga bv. acaciae ORS1073 and Rhizobium sp. strain ORS1001, two distantly related Acacia-nodulating strains, produce similar Nod factors that are O carbamoylated, N methylated, and mainly sulfated. J Bacteriol. 1997;179:3079–3083. doi: 10.1128/jb.179.9.3079-3083.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lortet G, Méar N, Lorquin J, Dreyfus B, de Lajudie P, Rosenberg C, Boivin C. Nod factor thin-layer chromatography profiling as a tool to characterize symbiotic specificity of rhizobial strains: applications to Sinorhizobium saheli, S. teranga, and Rhizobium sp. strains isolated from Acacia and Sesbania. Mol Plant-Microbe Interact. 1996;9:736–747. [Google Scholar]
- 166.Mabberley D J. The plant book. Cambridge, United Kingdom: Cambridge University Press; 1997. [Google Scholar]
- 167.Malpighi M. Anatome plantarum. London, United Kingdom: Regiae Societati (J. Martyn); 1675. [Google Scholar]
- 168.Meinhardt L W, Krishnan H B, Balatti P A, Pueppke S G. Molecular cloning and characterization of a sym plasmid locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol Microbiol. 1993;9:17–29. doi: 10.1111/j.1365-2958.1993.tb01665.x. [DOI] [PubMed] [Google Scholar]
- 169.Mergaert P, van Montagu M, Promé J-C, Holsters M. Three unusual modifications, a d-arabinosyl, an N-methyl, and a carbamoyl group, are present on the Nod factors of Azorhizobium caulinodans strain ORS71. Proc Natl Acad Sci USA. 1993;90:1551–1555. doi: 10.1073/pnas.90.4.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mergaert P, D'Haeze W, Geelen D, Promé D, van Montagu M, Geremia R, Promé J-C, Holsters M. Biosynthesis of Azorhizobium caulinodans Nod factor: study of the activity of the NodABC proteins by expression of the genes in Escherichia coli. J Biol Chem. 1995;270:29217–29223. doi: 10.1074/jbc.270.49.29217. [DOI] [PubMed] [Google Scholar]
- 171.Mergaert P, D'Haeze W, Fernandez-Lopez M, Geelen D, Goethals K, Promé J-C, van Montagu M, Holsters M. Fucosylation and arabinosylation of Nod factors in Azorhizobium caulinodans: involvement of nolK, nodZ as well as noeC and/or downstream genes. Mol Microbiol. 1996;21:409–419. doi: 10.1046/j.1365-2958.1996.6451366.x. [DOI] [PubMed] [Google Scholar]
- 172.Mergaert P, Ferro M, D'Haeze W, van Montagu M, Holsters M, Promé J-C. Nod factors of Azorhizobium caulinodans strain ORS571 can be glycosylated with an arabinosyl group, a fucosyl group or both. Mol Plant-Microbe Interact. 1997;10:683–687. doi: 10.1094/MPMI.1997.10.5.683. [DOI] [PubMed] [Google Scholar]
- 173.Mergaert P, van Montagu M, Holsters M. Molecular mechanisms of Nod factor diversity. Mol Microbiol. 1997;25:811–817. doi: 10.1111/j.1365-2958.1997.mmi526.x. [DOI] [PubMed] [Google Scholar]
- 174.McIver J, Djordjevic M A, Weinman J J, Bender G L, Rolfe B G. Extension of host range of Rhizobium leguminosarum bv. trifolii caused by point mutations in nodD that result in alterations in regulatory function and recognition of inducer molecules. Mol Plant-Microbe Interact. 1989;2:97–106. doi: 10.1094/mpmi-2-097. [DOI] [PubMed] [Google Scholar]
- 175.Michiels J, Dombrecht B, Vermeiren N, Xi C-W, Luyten E, Vanderleyden J. Phaseolus vulgaris is a non-selective host for nodulation. FEMS Microbiol Ecol. 1998;26:193–205. [Google Scholar]
- 176.Milner J L, Araujo R S, Handelsman J. Molecular and symbiotic characterization of exopolysaccharide-deficient mutants of Rhizobium tropici strain CIAT899. Mol Microbiol. 1992;6:3137–3147. doi: 10.1111/j.1365-2958.1992.tb01770.x. [DOI] [PubMed] [Google Scholar]
- 177.Minami E, Kouchi H, Carlson R W, Cohn J R, Kolli V K, Day R B, Ogawa T, Stacey G. Cooperative action of lipo-chitin nodulation signals on the induction of the early nodulin, ENOD2 in soybean roots. Mol Plant-Microbe Interact. 1996;9:574–583. doi: 10.1094/mpmi-9-0574. [DOI] [PubMed] [Google Scholar]
- 178.Minami E, Kouchi H, Cohn J R, Ogawa T, Stacey G. Expression of the early nodulin, ENOD40, in soybean roots in response to various lipo-chitin signal molecules. Plant J. 1996;10:23–32. doi: 10.1046/j.1365-313x.1996.10010023.x. [DOI] [PubMed] [Google Scholar]
- 179.Minic Z, Brown S, de Kouchkovsky Y, Schultze M, Staehelin C. Purification and characterisation of a novel chitinase-lysozyme, of another chitinase, both hydrolysing Rhizobium meliloti Nod factors, and of a pathogenesis-related protein from Medicago sativa roots. Biochem J. 1998;332:329–335. doi: 10.1042/bj3320329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mulligan J T, Long S R. A family of activator genes regulates expression of Rhizobium meliloti nodulation genes. Genetics. 1989;122:7–18. doi: 10.1093/genetics/122.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Niebel A, Bono J J, Ranjeva R, Cullimore J V. Identification of a high affinity binding site for lipo-oligosaccharidic NodRm factors in the microsomal fraction of Medicago cell suspension cultures. Mol Plant-Microbe Interact. 1997;10:132–134. [Google Scholar]
- 182.Niehaus K, Kapp D, Pühler A. Plant defence and delayed infection of alfalfa pseudonodules induced by an exopolysaccharide (EPS I)-deficient Rhizobium meliloti mutant. Planta. 1993;190:415–425. [Google Scholar]
- 183.Nobbe F, Schmid E, Hiltner L, Hotter E. Versuche über die Stickstoff-Assimilation der Leguminosen. Landwirtsch Versuchstat Dresden. 1891;39:327–359. [Google Scholar]
- 184.Nobbe F, Hiltner L, Schmid E. Versuche über die Biologie der Knöllchenbakterien der Leguminosen, insbesondere über die Frage der Arteinheit derselben. Landwirtsch Versuchstat Dresden. 1895;45:1–27. [Google Scholar]
- 185.Norris D O. Legumes and the Rhizobium symbiosis. Empire J Exp Agric. 1956;24:247–270. [Google Scholar]
- 186.Ogawa J, Brierley H L, Long S R. Analysis of Rhizobium meliloti nodulation mutant WL131: novel insertion sequence ISRm3 in nodG and altered nodH protein product. J Bacteriol. 1991;173:3060–3065. doi: 10.1128/jb.173.10.3060-3065.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Olsthoorn M M A, Lopez-Lara I M, Petersen B O, Bock K, Haverkamp J, Spaink H P, Thomas-Oates J E. Novel branched Nod factor structure results from alpha-(1→3) fucosyl transferase activity: the major lipo-chitin oligosaccharides from Mesorhizobium loti strain NZP2213 bear an alpha-(1→3) fucosyl substituent on a nonterminal backbone residue. Biochemistry. 1998;37:9024–9032. doi: 10.1021/bi972937r. [DOI] [PubMed] [Google Scholar]
- 188.Orgambide G G, Lee J I, Hollingsworth R I, Dazzo F B. Structurally diverse chitolipooligosaccharide Nod factors accumulate primarily in membranes of wild-type Rhizobium leguminosarum biovar trifolii. Biochemistry. 1995;34:3832–3840. doi: 10.1021/bi00011a041. [DOI] [PubMed] [Google Scholar]
- 189.Ovtsyna A O, Geurts R, Bisseling T, Lugtenberg B J J, Tikhonovich A I, Spaink H P. Restriction of host range by the sym2 allele of afghan pea is nonspecific for the type of modification at the reducing terminus of nodulation signals. Mol Plant-Microbe Interact. 1998;11:418–422. [Google Scholar]
- 190.Ovtsyna A O, Rademaker G J, Esser E, Weinman J, Rolfe B G, Tikhonovich I A, Lugtenberg B J J, Thomas-Oates J E, Spaink H P. Comparison of characteristics of the nodX genes from various Rhizobium leguminosarum strains. Mol Plant-Microbe Interact. 1999;12:252–258. doi: 10.1094/MPMI.1999.12.3.252. [DOI] [PubMed] [Google Scholar]
- 191.Reference deleted.
- 192.Parniske M, Schmidt P E, Kosch K, Müller P. Plant defense responses of host plants with determinate nodules induced by EPS-defective exoB mutants of Bradyrhizobium japonicum. Mol Plant-Microbe Interact. 1994;7:631–638. [Google Scholar]
- 193.Parveen N, Webb D T, Borthakur D. The symbiotic phenotypes of exopolysaccharide-defective mutants of Rhizobium sp. strain TAL1145 do not differ on determinate- and indeterminate-nodulating tree legumes. Microbiology. 1997;143:1959–1967. doi: 10.1099/00221287-143-6-1959. [DOI] [PubMed] [Google Scholar]
- 194.Perret X, Broughton W J, Brenner S. Canonical ordered cosmid library of the symbiotic plasmid of Rhizobium species NGR234. Proc Natl Acad Sci USA. 1991;88:1923–1927. doi: 10.1073/pnas.88.5.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Perret X, Freiberg C, Rosenthal A, Broughton W J, Fellay R. High resolution transcriptional analysis of the symbiotic replicon of Rhizobium sp. NGR234. Mol Microbiol. 1999;32:415–425. doi: 10.1046/j.1365-2958.1999.01361.x. [DOI] [PubMed] [Google Scholar]
- 196.Peters N K, Frost J W, Long S R. A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science. 1986;233:977–980. doi: 10.1126/science.3738520. [DOI] [PubMed] [Google Scholar]
- 197.Peters N K, Long S R. Alfalfa root exudates and compounds which promote or inhibit induction of Rhizobium meliloti nodulation genes. Plant Physiol. 1988;88:396–402. doi: 10.1104/pp.88.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Phillips D A, Joseph C M, Maxwell C A. Trigonelline and stachydrine released from alfalfa seeds activate NodD2 protein in Rhizobium meliloti. Plant Physiol. 1992;99:1526–1531. doi: 10.1104/pp.99.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Phillips D A, Kapulnik Y. Plant isoflavonoids, pathogens and symbionts. Trends Microbiol. 1995;3:58–64. doi: 10.1016/s0966-842x(00)88876-9. [DOI] [PubMed] [Google Scholar]
- 200.Plazinski J, Ridge R W, McKay I A, Djordjevic M A. The nodDABC genes of Rhizobium leguminosarum biovar trifolii confer root-hair curling ability to a diverse range of soil bacteria and the ability to induce novel swellings on beans. Aust J Plant Physiol. 1994;21:311–325. [Google Scholar]
- 201.Polhill R M. Classification of the Leguminosae, p. xvi–xxxvii. In: Bisby F A, Buckingham J, Harborne J B, editors. Phytochemical dictionary of the Leguminosae. 1. Plants and their constituents. London, United Kingdom: Chapman & Hall; 1994. [Google Scholar]
- 202.Poupot R, Martínez-Romero E, Promé J-C. Nodulation factors from Rhizobium tropici are sulfated or non-sulfated chitopentasaccharides containing an N-methyl-N-acylglucosamine terminus. Biochemistry. 1993;32:10430–10435. doi: 10.1021/bi00090a019. [DOI] [PubMed] [Google Scholar]
- 203.Poupot R, Martínez-Romero E, Gautier N, Promé J-C. Wild-type Rhizobium etli, a bean symbiont, produces acetyl-fucosylated, N-methylated, and carbamoylated nodulation factors. J Biol Chem. 1995;270:6050–6055. doi: 10.1074/jbc.270.11.6050. [DOI] [PubMed] [Google Scholar]
- 204.Prazmowski A. Das Wesen und die biologische Bedeutung der Wurzelknöllchen der Erbse. Bot Centbl. 1889;39:356–362. [Google Scholar]
- 205.Prazmowski A. Die Wurzelknöllchen der Erbse. Landwirtsch Versuchsstat Berlin. 1890;37:161–238. [Google Scholar]
- 206.Price N P J, Relić B, Talmont F, Lewin A, Promé D, Pueppke S G, Maillet F, Dénarié J, Promé J-C, Broughton W J. Broad-host-range Rhizobium species strain NGR234 secretes a family of carbamoylated and fucosylated, nodulation signals that are O-acetylated or sulphated. Mol Microbiol. 1992;6:3575–3584. doi: 10.1111/j.1365-2958.1992.tb01793.x. [DOI] [PubMed] [Google Scholar]
- 207.Pueppke S G, Broughton W J. Rhizobium sp. NGR234 and R. fredii USDA257 share exceptionally broad, nested host-ranges. Mol Plant-Microbe Interact. 1999;12:293–318. doi: 10.1094/MPMI.1999.12.4.293. [DOI] [PubMed] [Google Scholar]
- 208.Quesada-Vincens D, Fellay R, Nassim T, Viprey V, Burger U, Promé J-C, Broughton W J, Jabbouri S. Rhizobium sp. NGR234 NodZ protein is a fucosyltransferase. J Bacteriol. 1997;179:5087–5093. doi: 10.1128/jb.179.16.5087-5093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Quesada-Vincens D, Hanin M, Fellay R, Broughton W J, Jabbouri S. In vitro sulfotransferase activity of NoeE, a nodulation protein of Rhizobium sp. NGR234. Mol Plant-Microbe Interact. 1998;11:592–600. doi: 10.1094/MPMI.1998.11.7.592. [DOI] [PubMed] [Google Scholar]
- 210.Quinto C, Wijfjes A H M, Bloemberg G V, Blok-Tip L, Lopez-Lara I M, Lugtenberg B J J, Thomas-Oates J E, Spaink H P. Bacterial nodulation protein NodZ is a chitin oligosaccharide fucosyltransferase which can also recognize related substrates of animal origin. Proc Natl Acad Sci USA. 1997;94:4336–4341. doi: 10.1073/pnas.94.9.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Recourt K, van Brussel A A N, Driessen A J, Lugtenberg B J J. Accumulation of a nod gene inducer, the flavonoid naringenin, in the cytoplasmic membrane of Rhizobium leguminosarum biovar viciae is caused by the pH-dependent hydrophobicity of naringenin. J Bacteriol. 1989;171:4370–4377. doi: 10.1128/jb.171.8.4370-4377.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Reed J W, Glazebrook J, Walker G C. The exoR gene of Rhizobium meliloti affects RNA levels of other exo genes but lacks homology to known transcriptional regulators. J Bacteriol. 1991;173:3789–3794. doi: 10.1128/jb.173.12.3789-3794.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Relić B, Talmont F, Kopcinska J, Golinowsky W, Promé J-C, Broughton W J. Biological activity of Rhizobium sp. NGR234 Nod-factors on Macroptilium atropurpureum. Mol Plant-Microbe Interact. 1993;6:764–774. doi: 10.1094/mpmi-6-764. [DOI] [PubMed] [Google Scholar]
- 214.Relić B, Staehelin C, Fellay R, Jabbouri S, Boller T, Broughton W J. Do Nod-factor levels play a role in host-specificity? In: Kiss G B, Endre G, editors. Proceedings of the 1st European Nitrogen Fixation Conference. Szeged, Hungary: Officina Press; 1994. pp. 69–75. [Google Scholar]
- 215.Relić B, Perret X, Estrada-Garcia M T, Kopcinska J, Golinowski W, Krishnan H B, Pueppke S G, Broughton W J. Nod factors of Rhizobium are a key to the legume door. Mol Microbiol. 1994;13:171–178. doi: 10.1111/j.1365-2958.1994.tb00412.x. [DOI] [PubMed] [Google Scholar]
- 216.Remy W, Taylor T N, Hass H, Kerp H. Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci USA. 1994;91:11841–11843. doi: 10.1073/pnas.91.25.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Reuber T L, Walker G C. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell. 1993;74:269–280. doi: 10.1016/0092-8674(93)90418-p. [DOI] [PubMed] [Google Scholar]
- 218.Reference deleted.
- 219.Reference deleted.
- 220.Reference deleted.
- 221.Ridge R W. Freeze-substitution improves the ultrastructural preservation of legume root hairs. Bot Mag. 1988;101:427–441. [Google Scholar]
- 222.Ritsema T, Wijfjes A H M, Lugtenberg B J J, Spaink H P. Rhizobium nodulation protein NodA is a host-specific determinant of the transfer of fatty acids in Nod factor biosynthesis. Mol Gen Genet. 1996;251:44–51. doi: 10.1007/BF02174343. [DOI] [PubMed] [Google Scholar]
- 223.Roberts N J, Brigham J, Wu B, Murphy J B, Volpin H, Phillips D A, Etzler M E. A Nod factor-binding lectin is a member of a distinct class of apyrases that may be unique to the legumes. Mol Gen Genet. 1999;262:261–267. doi: 10.1007/s004380051082. [DOI] [PubMed] [Google Scholar]
- 224.Roche P, Debellé F, Maillet F, Lerouge P, Faucher C, Truchet G, Dénarié J, Promé J-C. Molecular basis of symbiotic host specificity in Rhizobium meliloti: nodH and nodPQ genes encode the sulfation of lipochitooligosaccharide signals. Cell. 1991;67:1131–1143. doi: 10.1016/0092-8674(91)90290-f. [DOI] [PubMed] [Google Scholar]
- 225.Roche P, Maillet F, Plazanet C, Debellé F, Ferro M, Truchet G, Promé J-C, Dénarié J. The common nodABC genes of Rhizobium meliloti are host-range determinants. Proc Natl Acad Sci USA. 1996;93:15305–15310. doi: 10.1073/pnas.93.26.15305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Röhrig H, Schmidt J, Wieneke U, Kondorosi É, Barlier I, Schell J, John M. Biosynthesis of lipooligosaccharide nodulation factors-Rhizobium NodA protein is involved in N-acylation of the chitooligosaccharide backbone. Proc Natl Acad Sci USA. 1994;91:3122–3126. doi: 10.1073/pnas.91.8.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Rostas K, Kondorosi É, Horvath B, Simoncsits A, Kondorosi Á. Conservation of extended promoter regions of nodulation genes in Rhizobium. Proc Natl Acad Sci USA. 1986;83:1757–1761. doi: 10.1073/pnas.83.6.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Roy K, Jablonski D, Valentine J W, Rosenberg G. Marine latitudinal diversity gradients: tests of causal hypotheses. Proc Natl Acad Sci USA. 1998;95:3699–3702. doi: 10.1073/pnas.95.7.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Sanjuan J, Carlson R W, Spaink H P, Bhat U R, Barbour W M, Glushka J, Stacey G. A 2-O-methylfucose moiety is present in the lipo-oligosaccharide nodulation signal of Bradyrhizobium japonicum. Proc Natl Acad Sci USA. 1992;89:8789–8793. doi: 10.1073/pnas.89.18.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sanjuan J, Grob P, Göttfert M, Hennecke H, Stacey G. NodW is essential for full expression of the common nodulation genes in Bradyrhizobium japonicum. Mol Plant-Microbe Interact. 1994;7:364–369. [Google Scholar]
- 231.Savouré A, Sallaud C, El-Turk J, Zuanazzi J, Ratet P, Schultze M, Kondorosi Á, Esnault R, Kondorosi É. Distinct response of Medicago suspension cultures and roots to Nod factors and chitin oligomers in the elicitation of defense-related responses. Plant J. 1997;11:277–287. [Google Scholar]
- 232.Schauser L, Roussis A, Stiller J, Stougaard J. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402:191–195. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- 233.Schiefelbein J W, Somerville C. Genetic control of root-hair development in Arabidopsis thaliana. Plant Cell. 1990;2:235–243. doi: 10.1105/tpc.2.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Schlaman H R, Spaink H P, Okker R J, Lugtenberg B J J. Subcellular localization of the nodD gene product in Rhizobium leguminosarum. J Bacteriol. 1989;171:4686–4693. doi: 10.1128/jb.171.9.4686-4693.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Schlaman H R, Horvath B, Vijgenboom E, Okker R J, Lugtenberg B J J. Suppression of nodulation gene expression in bacteroids of Rhizobium leguminosarum biovar viciae. J Bacteriol. 1991;173:4277–4287. doi: 10.1128/jb.173.14.4277-4287.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Schlaman H R, Okker R J, Lugtenberg B J J. Regulation of nodulation gene expression by NodD in rhizobia. J Bacteriol. 1992;174:5177–5182. doi: 10.1128/jb.174.16.5177-5182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Schlaman H R, Phillips D A, Kondorosi É. Genetic organization and transcriptional regulation of rhizobial nodulation genes. In: Spaink H P, Kondorosi Á, Hooykaas P J J, editors. The Rhizobiaceae. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 361–386. [Google Scholar]
- 238.Schultze M, Quiclet-Sire B, Kondorosi É, Virelizier H, Glushka J N, Endre G, Géro S D, Kondorosi Á. Rhizobium meliloti produces a family of sulfated lipooligosaccharides exhibiting different degrees of plant host specificity. Proc Natl Acad Sci USA. 1992;89:192–196. doi: 10.1073/pnas.89.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Schultze M, Staehelin C, Röhrig H, John M, Schmidt J, Kondorosi É, Schell J, Kondorosi Á. In vitro sulfotransferase activity of Rhizobium meliloti NodH protein: lipochitooligosaccharide nodulation signals are sulfated after synthesis of the core structure. Proc Natl Acad Sci USA. 1995;92:2706–2709. doi: 10.1073/pnas.92.7.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Schultze M, Kondorosi Á. Regulation of symbiotic root nodule development. Annu Rev Genet. 1998;32:33–57. doi: 10.1146/annurev.genet.32.1.33. [DOI] [PubMed] [Google Scholar]
- 241.Schultze M, Staehelin C, Brunner F, Genetet I, Legrand M, Fritig B, Kondorosi É, Kondorosi Á. Plant chitinase/lysozyme isoforms show distinct substrate specificity and cleavage site preference towards lipochitooligosaccharide Nod signals. Plant J. 1998;16:571–580. [Google Scholar]
- 242.Schwedock J S, Long S R. ATP sulphurylase activity of the nodP and nodQ gene products of Rhizobium meliloti. Nature. 1990;348:644–647. doi: 10.1038/348644a0. [DOI] [PubMed] [Google Scholar]
- 243.Schwedock J S, Liu C, Leyh T S, Long S R. Rhizobium meliloti NodP and NodQ form a multifunctional sulfate-activating complex requiring GTP for activity. J Bacteriol. 1994;176:7055–7064. doi: 10.1128/jb.176.22.7055-7064.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Sharma S B, Signer E R. Temporal and spatial regulation of the symbiotic genes of Rhizobium meliloti in planta revealed by transposon Tn5-gusA. Genes Dev. 1990;4:344–356. doi: 10.1101/gad.4.3.344. [DOI] [PubMed] [Google Scholar]
- 245.Shearman C A, Rossen L, Johnston A W B, Downie J A. The Rhizobium leguminosarum nodulation gene nodF encodes a polypeptide similar to acyl-carrier protein and is regulated by nodD plus a factor in pea root exudate. EMBO J. 1986;5:647–652. doi: 10.1002/j.1460-2075.1986.tb04262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Simon L, Bousquet J, Lévesque R C, Lalonde M. Origin and diversification of endomycorrhizal fungi and coincidence with vascular land plants. Nature. 1993;363:67–69. [Google Scholar]
- 247.Skorupska A, Bialek U, Urbanik-Sypniewska T, van Lammeren A. Two types of nodules induced on Trifolium pratense by mutants of Rhizobium leguminosarum bv. trifolii deficient in exopolysaccharide production. J Plant Physiol. 1995;147:93–100. [Google Scholar]
- 248.Somasegaran P, Hoben H J. Methods in legume-Rhizobium technology. Maui: NifTAL, MIRCEN, University of Hawaii Press; 1985. [Google Scholar]
- 249.Spaink H P, Wijffelman C A, Pees E, Okker R J H, Lugtenberg B J J. Rhizobium nodulation gene nodD as a determinant of host specificity. Nature. 1987;328:337–340. [Google Scholar]
- 250.Spaink H P, Okker R J H, Wijffelman C A, Tak T, Goosen-de-Roo L, Pees E, van Brussel A A N, Lugtenberg B J J. Symbiotic properties of rhizobia containing a flavonoid-independent hybrid nodD product. J Bacteriol. 1989;171:4045–4053. doi: 10.1128/jb.171.7.4045-4053.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Spaink H P, Weinman J, Djorjevic M A, Wijffelman C A, Okker R J H, Lugtenberg B J J. Genetic analysis and cellular localisation of the Rhizobium host specificity determining NodE protein. EMBO J. 1989;8:2811–2818. doi: 10.1002/j.1460-2075.1989.tb08427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Spaink H P, Sheeley D M, van Brussel A A N, Glushka J, York W S, Tak T, Geiger O, Kennedy E P, Reinhold V N, Lugtenberg B J J. A novel highly unsaturated fatty acid moiety of lipo-oligosaccharide signals determines host-specificity of Rhizobium. Nature. 1991;354:125–130. doi: 10.1038/354125a0. [DOI] [PubMed] [Google Scholar]
- 253.Spaink H P, Wijfjes A H M, van der Drift K M G M, Haverkamp J, Thomas-Oates J E, Lugtenberg B J J. Structural identification of metabolites produced by the NodB and NodC proteins of Rhizobium leguminosarum. Mol Microbiol. 1994;13:821–831. doi: 10.1111/j.1365-2958.1994.tb00474.x. [DOI] [PubMed] [Google Scholar]
- 254.Spaink H P, Wijfjes A H M, Lugtenberg B J J. Rhizobium NodI and NodJ proteins play a role in the efficiency of secretion of lipochitin oligosaccharides. J Bacteriol. 1995;177:6276–6281. doi: 10.1128/jb.177.21.6276-6281.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Spaink H P, Bloemberg G V, van Brussel A A N, Lugtenberg B J J, van der Drift K M G M, Haverkamp J, Thomas-Oates J E. Host specificity of Rhizobium leguminosarum is determined by the hydrophobicity of highly unsaturated fatty acyl moieties of the nodulation factors. Mol Plant-Microbe Interact. 1995;8:155–164. [Google Scholar]
- 256.Spaink H P. Regulation of plant morphogenesis by lipo-chitin oligosaccharides. Crit Rev Plant Sci. 1996;15:559–582. [Google Scholar]
- 257.Staehelin C, Müller J, Mellor R B, Wiemken A, Boller T. Chitinase and peroxidase in effective (Fix+) and ineffective (Fix−) soybean nodules. Planta. 1992;187:295–300. doi: 10.1007/BF00195651. [DOI] [PubMed] [Google Scholar]
- 258.Staehelin C, Schultze M, Kondorosi É, Mellor R B, Boller T, Kondorosi Á. Structural modifications in Rhizobium meliloti Nod factors influence their stability against hydrolysis by root chitinases. Plant J. 1994;5:319–330. [Google Scholar]
- 259.Staehelin C, Granado J, Müller J, Wiemken A, Mellor R B, Felix G, Regenass M, Broughton W J, Boller T. Perception of Rhizobium nodulation factors by tomato cells and inactivation by root chitinases. Proc Natl Acad Sci USA. 1994;91:2196–2200. doi: 10.1073/pnas.91.6.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Staehelin C, Schultze M, Kondorosi É, Kondorosi Á. Lipochitooligosaccharide nodulation signals from Rhizobium meliloti induce their rapid degradation by the host plant alfalfa. Plant Physiol. 1995;108:1607–1614. doi: 10.1104/pp.108.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Stacey G, Luka S, Sanjuan J, Banfalvi Z, Nieuwkoop A J, Chun J Y, Forsberg L S, Carlson R W. nodZ, a unique host-specific nodulation gene, is involved in the fucosylation of the lipopolysaccharide nodulation signal of Bradyrhizobium japonicum. J Bacteriol. 1994;176:620–633. doi: 10.1128/jb.176.3.620-633.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Stanfield S W, Ielpi L, O'Brochta D, Helinski D, Ditta G S. The ndvA gene product of Rhizobium meliloti is required for β-(1→2)glucan production and has homology to the ATP-binding export protein HlyB. J Bacteriol. 1988;170:3523–3530. doi: 10.1128/jb.170.8.3523-3530.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Surin B P, Downie J A. Rhizobium leguminosarum genes required for expression and transfer of host specific nodulation. Plant Mol Biol. 1989;12:19–29. doi: 10.1007/BF00017444. [DOI] [PubMed] [Google Scholar]
- 264.Sutton J M, Lea E J, Downie J A. The nodulation-signaling protein NodO from Rhizobium leguminosarum biovar viciae forms ion channels in membranes. Proc Natl Acad Sci USA. 1994;91:9990–9994. doi: 10.1073/pnas.91.21.9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Sutton J M, Peart J, Dean G, Downie J A. Analysis of the C-terminal secretion signal of the Rhizobium leguminosarum nodulation protein NodO; a potential system for the secretion of heterologous proteins during nodule invasion. Mol Plant-Microbe Interact. 1996;9:671–680. doi: 10.1094/mpmi-9-0671. [DOI] [PubMed] [Google Scholar]
- 266.Swanson J A, Mulligan J T, Long S R. Regulation of syrM and nodD3 in Rhizobium meliloti. Genetics. 1993;134:435–444. doi: 10.1093/genetics/134.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Timmers A C, Auriac M C, de Billy F, Truchet G. Nod factor internalization and microtubular cytoskeleton changes occur concomitantly during nodule differentiation in alfalfa. Development. 1998;125:339–349. doi: 10.1242/dev.125.3.339. [DOI] [PubMed] [Google Scholar]
- 268.Trinick M J. Symbiosis between Rhizobium and the non-legume, Trema aspera. Nature. 1973;244:459–460. [Google Scholar]
- 269.Trinick M J. Relationships amongst the fast-growing rhizobia of Lablab purpureus, Leucaena leucocephala, Mimosa spp., Acacia farnesiana and Sesbania grandiflora and their affinities with other rhizobial groups. J Appl Bact. 1980;49:39–53. [Google Scholar]
- 269a.van Brussel A A N, Bakhuizen R, van Spronsen P C, Spaink H P, Tak T, Lugtenberg B J J, Kijne J W. Induction of preinfection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharides of Rhizobium. Science. 1992;257:70–72. doi: 10.1126/science.257.5066.70. [DOI] [PubMed] [Google Scholar]
- 269b.van der Drift K M, Spaink H P, Bloemberg G V, van Brussel A A N, Lugtenberg B J J, Haverkamp J, Thomas-Oates J E. Rhizobium leguminosarum bv. trifolii produces lipo-chitin oligosaccharides with nodE-dependent highly unsaturated fatty acyl moieties. An electrospray ionization and collision-induced dissociation tandem mass spectrometric study. J Biol Chem. 1996;271:22563–22569. doi: 10.1074/jbc.271.37.22563. [DOI] [PubMed] [Google Scholar]
- 269c.van Rhijn P J, Feys B, Verreth C, Vanderleyden J. Multiple copies of nodD in Rhizobium tropici CIAT899 and BR816. J Bacteriol. 1993;175:438–447. doi: 10.1128/jb.175.2.438-447.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269d.van Rhijn P, Vanderleyden J. The Rhizobium-plant symbiosis. Microbiol Rev. 1995;59:124–142. doi: 10.1128/mr.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269e.van Rhijn P, Luyten E, Vlassak K, Vanderleyden J. Isolation and characterization of a pSym locus of Rhizobium sp. BR816 that extends nodulation ability of narrow host range Phaseolus vulgaris symbionts to Leucaena leucocephala. Mol Plant-Microbe Interact. 1996;9:74–77. doi: 10.1094/mpmi-9-0074. [DOI] [PubMed] [Google Scholar]
- 270.van Workum W A T, van Brussel A A N, Tak T, Wijffelman C A, Kijne J W. Ethylene prevents nodulation of Vicia sativa ssp. nigra by exopolysaccharide-deficient mutants of Rhizobium leguminosarum bv. viciae. Mol Plant-Microbe Interact. 1995;8:278–285. [Google Scholar]
- 271.van Workum W A T, Canter-Cremers H C J, Wijfjes A H M, van der Kolk C, Wijffelman C A, Kijne J W. Cloning and characterization of four genes of Rhizobium leguminosarum bv. trifolii involved in exopolysaccharide production and nodulation. Mol Plant-Microbe Interact. 1997;10:290–301. doi: 10.1094/MPMI.1997.10.2.290. [DOI] [PubMed] [Google Scholar]
- 272.van Workum W A T, van Slageren S, van Brussel A A N, Kijne J W. Role of exopolysaccharides of Rhizobium leguminosarum bv. viciae as host plant-specific molecules required for infection thread formation during nodulation of Vicia sativa. Mol Plant-Microbe Interact. 1998;11:1233–1241. [Google Scholar]
- 273.Vasse J, de Billy F, Truchet G. Abortion of infection during the Rhizobium meliloti-alfalfa symbiotic interaction is accompanied by a hypersensitive reaction. Plant J. 1993;4:555–566. [Google Scholar]
- 274.Viprey V, Del Greco A, Golinowski W, Broughton W J, Perret X. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol Microbiol. 1998;28:1381–1389. doi: 10.1046/j.1365-2958.1998.00920.x. [DOI] [PubMed] [Google Scholar]
- 275.Viprey V, Perret X, Broughton W J. Host-plant invasion by rhizobia. Subcell Biochem. 1999;33:437–456. doi: 10.1007/978-1-4757-4580-1_17. [DOI] [PubMed] [Google Scholar]
- 276.Waelkens F, Voets T, Vlassak K, Vanderleyden J, van Rhijn P. The nodS gene of Rhizobium tropici CIAT899 is necessary for nodulation of Phaseolus vulgaris and Leucaena leucocephala. Mol Plant-Microbe Interact. 1995;8:147–154. doi: 10.1094/mpmi-8-0147. [DOI] [PubMed] [Google Scholar]
- 277.Wang L X, Wang Y, Pellock B, Walker G C. Structural characterization of the symbiotically important low-molecular-weight succinoglycan of Sinorhizobium meliloti. J Bacteriol. 1999;181:6788–6796. doi: 10.1128/jb.181.21.6788-6796.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Wang S-P, Stacey G. Studies of the Bradyrhizobium japonicum nodD1 promoter: a repeated structure for the nod box. J Bacteriol. 1991;173:3356–3365. doi: 10.1128/jb.173.11.3356-3365.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Wilson J K. Leguminous plants and their associated organisms. Cornell University Agricultural Experiment Station memoir 221. Ithaca, N.Y: Cornell University Press; 1939. [Google Scholar]
- 280.Woronin M S. Über die bei der Schwarzerle (Alnus glutinosa) und bei der gewöhnlichen Gartenlupine (Lupinus mutabilis) auftretenden Wurzelanschwellungen. 1866. Mémoires de l'Academie Impériale des Sciences de St. Pétersbourg, VII Series, vol. X. [Google Scholar]
- 281.Xie Z-P, Staehelin C, Wiemken A, Broughton W J, Müller J, Boller T. Symbiosis-stimulated chitinase isoenzymes of soybean (Glycine max (L.) Merr.) J. Exp Bot. 1999;50:327–333. [Google Scholar]
- 282.Yang G P, Debellé F, Savagnac A, Ferro M, Schiltz O, Maillet F, Promé D, Treilhou M, Vialas C, Lindstrom K, Dénarié J, Promé J-C. Structure of the Mesorhizobium huakuii and Rhizobium galegae Nod factors: a cluster of phylogenetically related legumes are nodulated by rhizobia producing Nod factors with α-,β-unsaturated N-acyl substitutions. Mol Microbiol. 1999;34:227–237. doi: 10.1046/j.1365-2958.1999.01582.x. [DOI] [PubMed] [Google Scholar]