Abstract
Objectives:
Synovium is acutely affected following joint trauma and contributes to post-traumatic osteoarthritis (PTOA) progression. Little is known about discrete cell types and molecular mechanisms in PTOA synovium. We aimed to describe synovial cell populations and their dynamics in PTOA, with a focus on fibroblasts. We also sought to define mechanisms of synovial Wnt/β-catenin signaling, given its emerging importance in arthritis.
Methods:
We subjected mice to non-invasive anterior cruciate ligament rupture as a model of human joint injury. We performed single-cell RNA-sequencing to assess synovial cell populations, subjected Wnt-GFP reporter mice to joint injury to study Wnt-active cells, and performed intra-articular injections of the Wnt agonist R-spondin 2 (Rspo2) to assess whether gain-of-function induced pathologies characteristic of PTOA. Lastly, we used cultured fibroblasts, macrophages, and chondrocytes to study how Rspo2 orchestrates crosstalk between joint cell types.
Results:
We uncovered seven distinct functional subsets of synovial fibroblasts in healthy and injured synovium, and defined their temporal dynamics in early and established PTOA. Wnt/β-catenin signaling was overactive in PTOA synovium, and Rspo2 was strongly induced after injury and secreted exclusively by Prg4hi lining fibroblasts. Trajectory analyses predicted that Prg4hi lining fibroblasts arise from a pool of Dpp4+ mesenchymal progenitors in synovium, with SOX5 identified as a potential regulator of this emergence. We also showed that Rspo2 orchestrated pathological crosstalk between synovial fibroblasts, macrophages, and chondrocytes.
Conclusions:
Synovial fibroblasts assume distinct functional identities during PTOA in mice, and Prg4hi lining fibroblasts secrete Rspo2 that may drive pathological joint crosstalk after injury.
Keywords: Fibroblasts, Osteoarthritis, Synovitis
INTRODUCTION
Inflammation, fibrosis, and mineralization of synovium are key pathological features of post-traumatic osteoarthritis (PTOA)(1–3). Despite major recent advances in our understanding of synovial biology in inflammatory conditions such as rheumatoid arthritis (RA)(4–7), little is known about mechanisms of PTOA pathogenesis.
Synovium is a heterogeneous connective tissue that exhibits intrinsic pathologies during PTOA and promotes disease throughout the joint(8). Synovial fibroblasts (SFs) are a mixed population of stromal cells that have been classified into pathological subsets in RA, based on their proliferative capacity, invasiveness, and localization, along with their ability to orchestrate inflammation or bone and cartilage damage(6, 7). However, in PTOA, functional SF subsets have not been defined beyond an anatomical delineation between Thy1+ sublining and Prg4hi lining SFs(9). Given the strong focus on cartilage in osteoarthritis (OA), single-cell level analyses have shed significant light on chondrocyte subsets(10–12), but little attention has been given to disease-associated synovial cell populations.
Targeting overactive canonical Wnt/β-catenin signaling has emerged as a strong candidate for treating OA(13–17). However, the mediators and cell types responsible for overactive Wnt signaling in OA are not well described. R-spondin 2 (Rspo2) is a secreted, matricellular agonist of canonical Wnt signaling(18, 19), and recent reports have implicated its role in OA progression(20, 21).
Herein, to study synovial dynamics during PTOA, we employed a non-invasive anterior cruciate ligament rupture-based model of joint injury (ACLR) which recapitulates traumatic joint overloading and instability that occur in human joint injury without confounding effects from surgery(22–25). Transcriptomic profiling by single-cell RNA sequencing (scRNA-seq) uncovered distinct functional subsets of SFs in healthy and injured tissue. Further interrogation revealed overactive Wnt/β-catenin signaling throughout the SF niche in synovium, and pathological secretion of Rspo2 exclusively by Prg4hi lining SFs. These insights offer new avenues for specific therapeutic targeting in PTOA.
RESULTS
Single-cell transcriptomic profiling of synovium reveals cellular heterogeneity in PTOA
The cellular composition of synovium has been characterized in inflammatory models of arthritis(6, 26), but little is known about synovial cell dynamics in PTOA. We used flow cytometry to demonstrate that synovial tissue digestion yielded viable (>90%) cells representing major resident subpopulations – synovial fibroblasts (SFs; CD31− CD45−), endothelial cells (CD31+ CD45−), and hematopoietic cells (CD31− CD45+) (Figure S1A). To study PTOA synovium, we employed a non-invasive, joint-overloading ACLR model, which induces key clinical features of PTOA including cartilage degradation, synovial inflammation, fibrosis, and osteophyte formation(23–25). We performed scRNA-seq on synovium from mice 7d ACLR, 28d ACLR, or Sham controls (Figure S1B). Given inherent sex differences reported in pre-clinical PTOA models including ACLR(27, 28) we combined male and female synovium for each sample. After initial quality control, 20,422 cells were integrated across conditions by canonical correlation analysis(29) (Figure S1C–D), then unsupervised dimensionality reduction and clustering were performed (Figure 1A–B and Figure S2A–B). SFs (sublining and lining), myeloid, and endothelial cells predominated numerically in healthy and PTOA synovium, with SFs and myeloid cells undergoing the most drastic increase in abundance at 7d ACLR (Figure 1C–D). We also detected rarer cell populations including pericytes, T cells, and Schwann cells, which are not well characterized in synovium (Figure S2C). Profound injury-induced changes in synovial cellular composition were underpinned by changes in global transcriptomic profile among Sham, 7d, and 28d ACLR (Figure 1E). To examine intercellular communication between synovial cells, we used CellChat, which infers cell-cell signaling pathways based on curated interaction networks between ligands, receptors, agonists, and inhibitors(30). Outgoing and incoming communication patterns for each major synovial cell type were calculated based on relative contributions of ligand-receptor interactions, displayed as river plots (Figure 1F and Figure S3). These data demonstrate distinct, cell- and direction-specific communication signatures that orchestrate cell-cell signaling in synovium, allowing for subsequent analysis of networks of interest. Together these results highlight the plasticity of synovium following joint injury and shed light on the diverse cell types that populate PTOA synovium.
Figure 1. Single-cell transcriptomic profiling of synovium reveals cellular heterogeneity in PTOA.
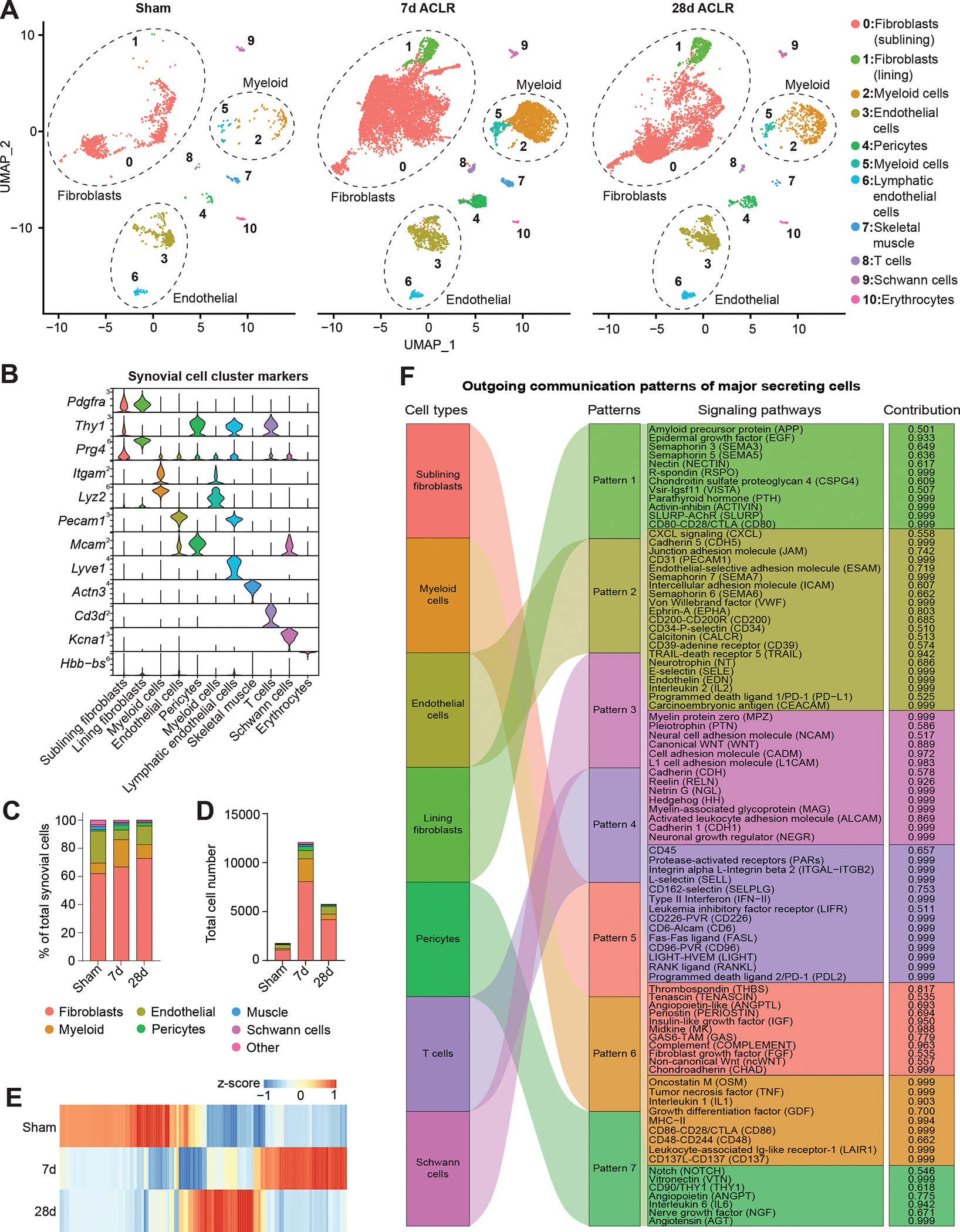
(A) UMAP plots showing reduced dimensionality projections of all synovial cells from Sham, 7d ACLR or 28d ACLR mice (20,422 cells, n=2 biological replicates per condition, each composed of one male and one female). Cell types are grouped by color and designated numbers, with annotations on the right. Major cell groups have dashed outlines. (B) Violin plots of synovial cell type marker genes. (C) Proportional breakdown of major synovial cell types in each condition. (D) Total abundance of major synovial cell types in each condition. (E) Global gene expression across conditions from pseudo-bulk analysis of scRNA-seq data of all synovial cells. (F) River plot from CellChat analysis showing outgoing signaling patterns from major synovial cell types and the pathways comprising each pattern (contribution score for each pathway is shown on the right). ACLR: anterior cruciate ligament rupture (injury model).
Identification of functionally distinct synovial fibroblast subsets
For more granular interrogation of SF subpopulations, we computationally subset SFs and related mesenchymal cells, excluding pericytes. Re-clustering uncovered seven distinct subsets present in healthy synovium (Angptl7+, IL-6+, Dpp4+, Prg4hi) or induced after injury (αSMA+, Sox9+, Mki67+), each designated by highly confined expression of marker genes (Figure 2A–C and Figure S4). Established SF markers, such as Vim, Pdgfra, and Pdpn were broadly present across subsets, with a distinction between Thy1+ SFs (expressed in Angptl7+, αSMA+/Acta2+, IL-6+, Dpp4+, and Mki67+ clusters) and Prg4hi Thy1- lining SFs (Figure 2D). Profound injury-induced changes in both proportion and abundance of subsets occurred by 7d ACLR; all clusters expanded numerically post-injury but the most robust increases were evident in subsets absent from healthy synovium – the αSMA+ and Sox9+ clusters (Figure 2E–G). Notably, the abundance of all subsets diminished by 28d ACLR, apart from Prg4hi lining SFs, consistent with persistent lining hyperplasia observed in human PTOA(31, 32). Having defined distinct SF clusters and their temporal abundance, we assessed whether their unique transcriptomic profiles underpinned distinct functional roles. All clusters, except for Mki67+ cells, exhibited hallmark features of SFs, with roles in extracellular matrix and collagen organization (Figure S5A). Gene Ontology and Reactome analyses uncovered unique functions of each subset (Figure 2H and Figure S5B). Fibroblast subsets were also subjected to CellChat analysis to characterize broad signaling patterns (Figure 2I and Figure S5C–I). Angptl7+ cells, present in healthy and injured synovium, were enriched for pathways relating to neurogenesis and were important secretors of ligands for glial-derived neurotrophic factor signaling. Injury-induced αSMA+ cells were enriched for pathways including myofibril assembly, smooth muscle contraction, and response to mechanical stimulus – reminiscent of myofibroblasts. IL-6+ cells, present in Sham and expanded by injury, were enriched for pro-inflammatory and chemotactic signaling, suggesting a role in orchestrating immune-SF crosstalk. Dpp4+ cells, which expressed previously reported mesenchymal progenitor markers Dpp4, Wnt2, Anxa3, Cd34 and Pi16(33, 34), were present both in Sham and ACLR. Prg4hi SFs, which constitute the thin lining layer in healthy synovium that exhibits dramatic hyperplasia in OA, were enriched for cell junction regulation, R-spondin and syndecan signaling, and were overrepresented for incoming signals from other SF subsets. Sox9+ cells, which also express Col2a1, Osx/Sp7, and Acan, were enriched for chondrogenic and osteogenic pathways, along with glycolytic metabolism, consistent with an osteochondral progenitor (OCP) phenotype. These cells may give rise to osteophytes, bony growths that constitute an important clinical feature of PTOA(35). And lastly, Mki67+ cells exhibited a distinct proliferating phenotype, expressing known markers such as Birc5 and Top2a. We then compared these fibroblast subsets to published scRNA-seq datasets from human OA and RA(4, 5). As expected, Prg4 was most highly expressed in lining SFs across disease states (Figure S6). Strikingly, Dpp4+ and IL-6+ subpopulations of SFs were observed across datasets and clearly segregated, suggesting a degree of conserved function in both OA and RA synovium. Together these data reveal the diversity of stromal subpopulations in healthy and PTOA synovium and point towards their distinct functions and signaling patterns.
Figure 2. Identification of functionally distinct synovial fibroblast subsets.
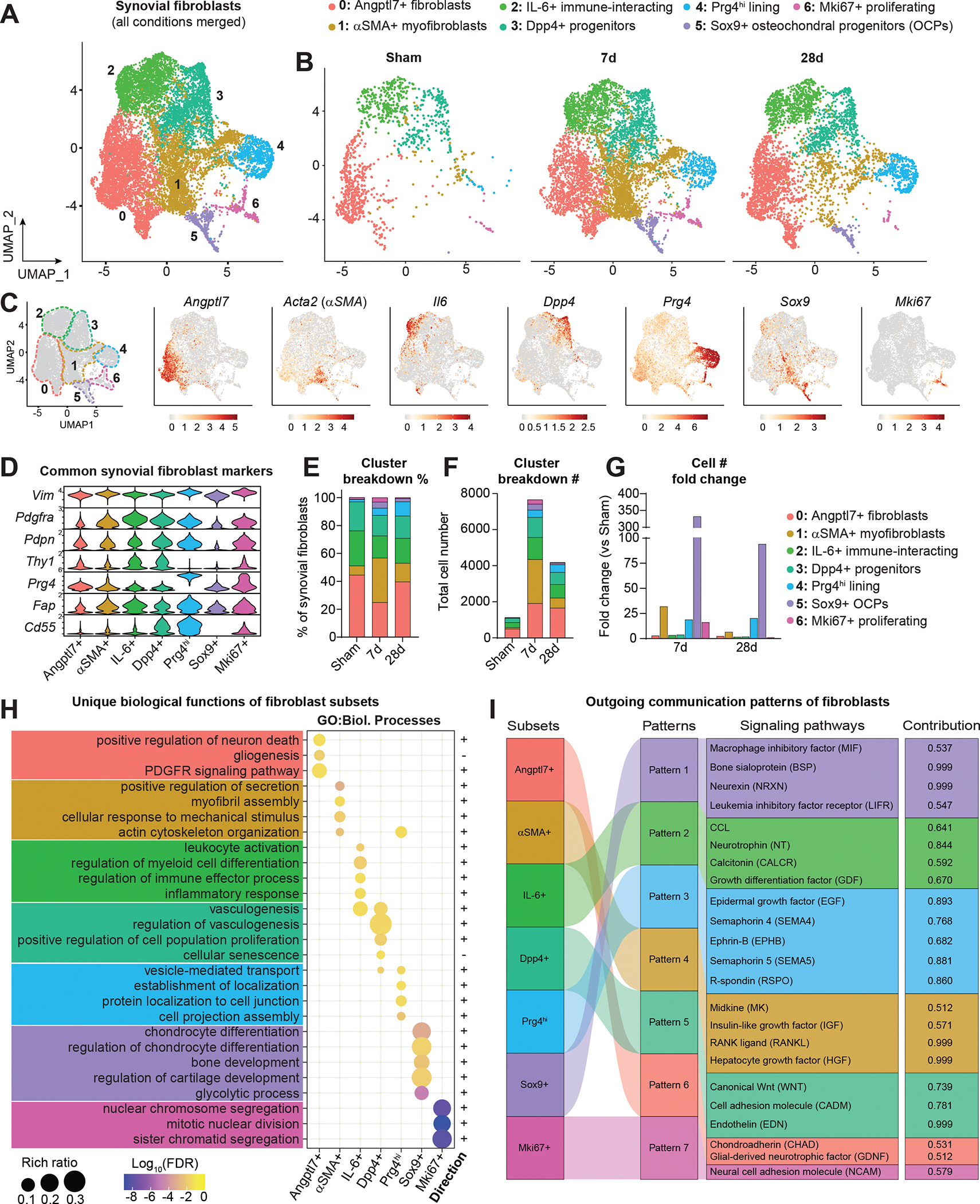
(A) UMAP plot showing SFs and related cell types integrated across conditions using canonical correlation analysis (n=2 biological replicates per condition, as in Figure 1). Clusters have designated numbers and colors corresponding to their annotation (top). (B) SF UMAP plots split by condition. (C) UMAP plot of SFs showing cluster borders with colored, dashed outlines (left), and feature plots of marker genes used to designate the naming of each cluster. Color scales represent relative expression for each gene individually. (D) Violin plots showing expression levels of common SF marker genes across clusters. (E) Proportional breakdown (%) of each cluster across condition. (F) Total abundance (#) of cells in each cluster across conditions. (G) Fold change (relative to Sham) of cell number (#) in each cluster at 7d and 28d ACLR. (H) Pathway analysis using Gene Ontology (GO) Biological Processes showing unique functional terms for each cluster and the direction of each term. (I) River plot showing CellChat analysis of outgoing signaling patterns from SF subsets and the pathways comprising each pattern, and the contribution score of each pathway on the right. FDR: false discovery rate.
Canonical Wnt/β-catenin signaling is activated in synovium during PTOA
Targeting canonical Wnt/β-catenin signaling has therapeutic promise in OA given its reported overactivation in joint disease(14, 36, 37), and we found that the Wnt pathway was an important element of broad synovial signaling patterns in our study (Figure 1F and 2I). We showed Schwann cells to be important secretors of Wnt ligands in synovium, especially Wnt6 and Wnt9a, while Dpp4+ mesenchymal progenitors were important secretors within the SF niche (Figure 3A–B and Figure S7A–B). Beyond just outgoing Wnt signals, we showed that each of the SF subsets (except for Mki67+ proliferating SFs) were enriched for canonical Wnt signaling terms compared to non-SF cell types in synovium (Figure 3C). Inspection of individual Wnt ligand expression patterns and regulation after injury showed that Wnt5a, Wnt5b, Wnt6, Wnt9a and Wnt11 were the most highly expressed (Figure 3D–E). While some Wnt ligands were down-regulated, most were induced after joint injury, and this observation is consistent more broadly with up-regulation of Wnt pathway genes seen in whole synovium and in only SFs, at 7d and 28d ACLR (Figure S7C–D). To assess synovial cells actively undergoing canonical Wnt/β-catenin signaling, we used Wnt-GFP reporter mice which express TCF/LEF response element-driven EGFP(38). The abundance, but not total proportion, of Wnt-GFP+ cells increased in 7d ACLR compared to contralateral synovium, and we observed increased GFP intensity – a proxy for increased Wnt/β-catenin signaling activity (Figure 3F–G and Figure S8A–B). Wnt-GFP+ cells were predominantly CD31− CD45−, indicative of SFs (Figure 3H), and total number and GFP intensity increased within this population in ACLR versus contralateral synovium (Figure 3I–J and Figure S8C). This corroborates our finding from scRNA-seq pathway analysis that SFs are the predominant Wnt-active cells in synovium (Figure 3C). These results demonstrate the importance of canonical Wnt/β-catenin signaling in synovial crosstalk and its overactivation in PTOA, with SFs being key cells undertaking Wnt signaling.
Figure 3. Canonical Wnt/β-catenin signaling is activated in synovium during PTOA.
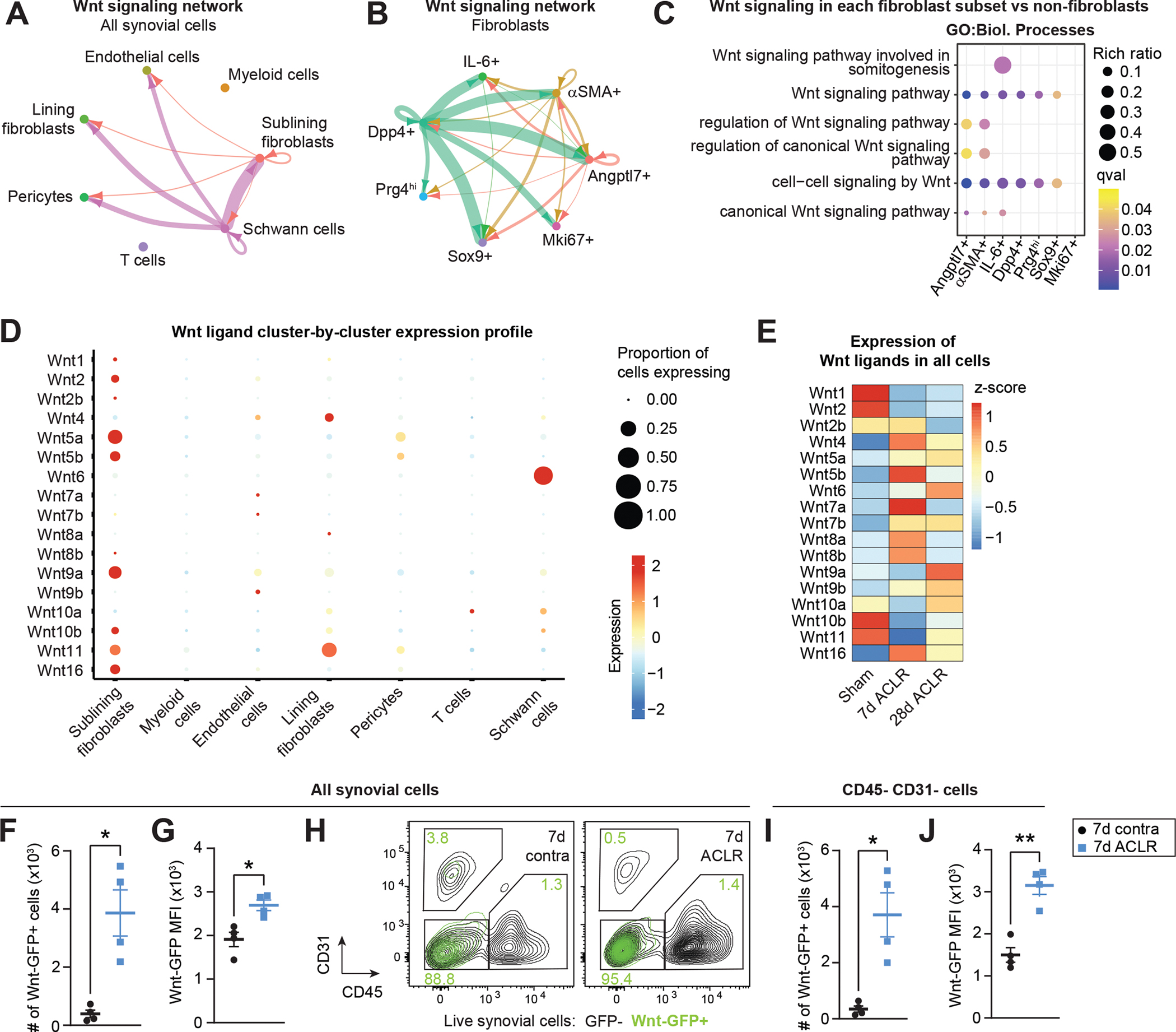
(A-B) Circle plots showing canonical Wnt signaling communication between all synovial cell types (A) or SFs only (B), with directionality indicated by arrowheads. Lines are color-coded by source cell type and line width is proportional to interaction strength. (C) Pathway analysis using GO Biological Processes showing enriched canonical Wnt signaling terms in each SF cluster compared to all non-SFs. (D) Bubble plot showing expression levels of each Wnt ligand gene across major synovial cell types, and the proportion of cells in each cluster expressing the gene. Some Wnt ligand genes were not detected. (E) Heatmap showing Wnt ligand gene expression (z-score) in all synovial cells from Sham, 7d ACLR or 28d ACLR conditions. (F-J) Flow cytometry assessment of injured or contralateral (contra) synovium from Wnt-GFP reporter mice 7d ACLR (n=4 mice). (F) Total number of Wnt-GFP+ synovial cells. (G) Median fluorescence intensity (MFI) of GFP for all synovial cells. (H) Overlay of Wnt-GFP+ cells (green) from contra and 7d ACLR synovium showing their expression of CD31 and CD45. CD31− CD45− cells represent predominantly SFs in synovium. Green values represent the percentage of Wnt-GFP+ cells in each gate. (I) Total number of CD31− CD45− synovial cells that were Wnt-GFP+. (J) MFI of GFP for CD31− CD45− synovial cells. Paired two-tailed student’s t-tests were used for F, G, I, and J, where *P<0.05, **P<0.01. Error bars are mean ± SEM. qval: P value adjusted for false discovery rate.
The canonical Wnt agonist R-spondin 2 is strongly and selectively induced during PTOA
To understand potential Wnt modulators activated during PTOA, we focused on R-spondin 2 (Rspo2), a secreted, matricellular agonist of canonical Wnt signaling that has been implicated in PTOA(20, 21). Immunostaining of whole joint sections from Sham and ACLR limbs revealed widespread up-regulation of Rspo2 in synovium and meniscus after injury, with strong expression evident in the osteophyte (Figure 4A–B). We detected elevated Rspo2 in synovial fluid, up-regulation of Rspo2 transcript by qPCR, and increased abundance of RSPO2+ cells by flow cytometry, in synovium of injured mice (Figure 4C–D and Figure S9A). Despite a previous report that synovial M1 macrophages express Rspo2 in OA(21), we did not detect any CD45+ hematopoietic cells expressing Rspo2 by flow cytometry, in healthy or injured synovium, nor did scRNA-seq indicate Rspo2 expression in myeloid cells (Figure 4E–F). Rspo2 transcript and protein were only detected in SFs, where it was significantly more highly expressed than other R-spondin family members and strongly induced in PTOA (Figure 4E–F and Figure S9B–D). This observation was consistent in primary cultured cells, where Rspo2 was highly expressed in knee-derived SFs compared to bone marrow-derived macrophages (BMDM) (Figure S9E–F). Primary cultured SFs are a well-established in vitro model(39), and we confirmed that our cells expressed classical SF markers (Podoplanin, THY1/CD90.2, CD55) and had <1% contamination from CD11b+ synovial macrophages (Figure S9G–J). Within SFs, Rspo2 was confined to the Prg4hi lining subset, not Thy1+ sublining SFs (Figure 4G), consistent with Rspo2 immunostaining localization to the synovial lining (Figure 4A). Cell-cell signaling analysis revealed that outgoing R-spondin signaling was dominated by interactions between lining-derived Rspo2 and its receptors Lgr4–6 on other SF subsets (Figure 4H–I). These results demonstrate that Rspo2 is induced in PTOA and, within synovium, is exclusively secreted by lining SFs.
Figure 4. The canonical Wnt agonist R-spondin 2 is strongly and selectively induced during PTOA.
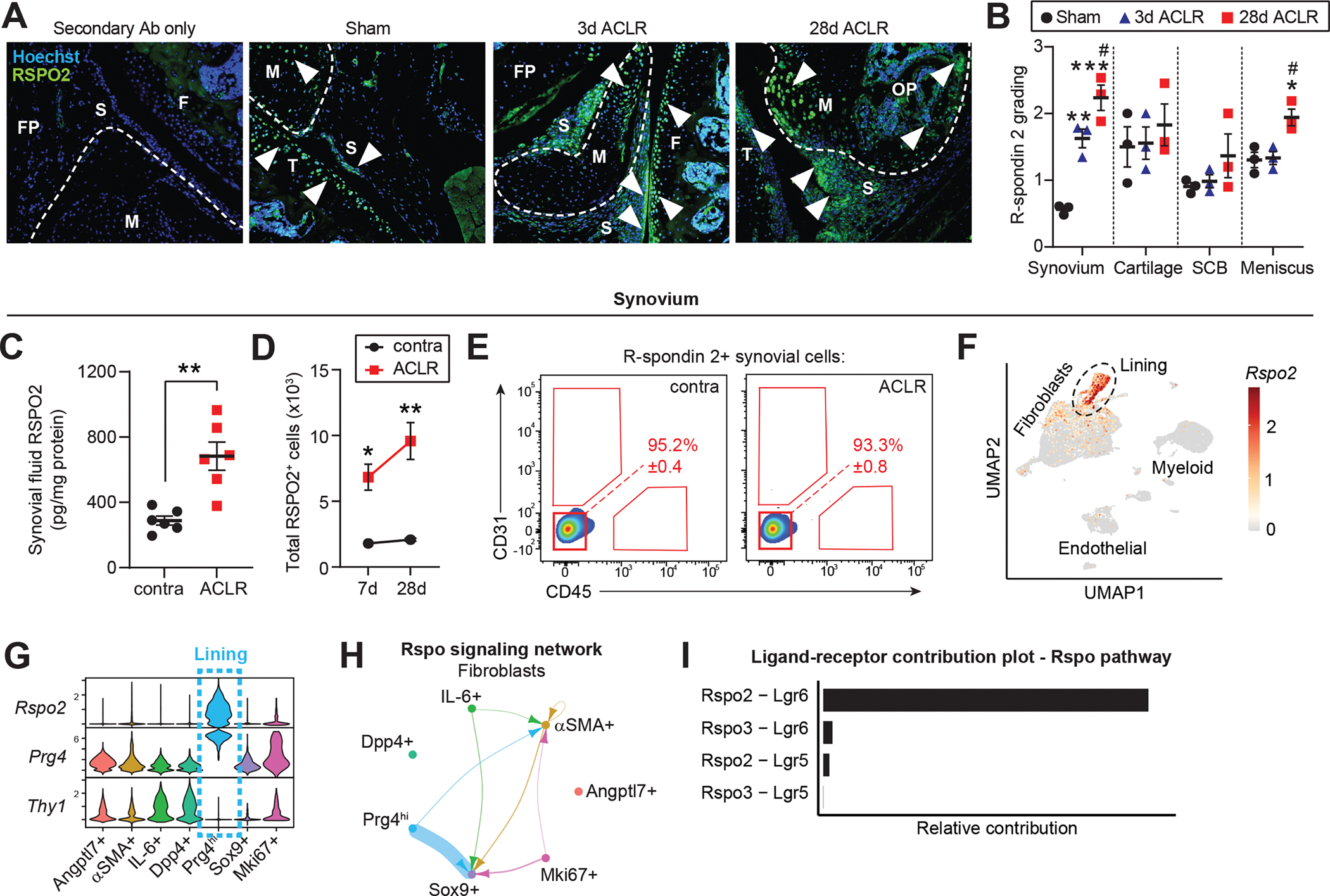
(A) Immunofluorescent staining of R-spondin 2 (RSPO2) in joint sections from Sham, 3d or 28d ACLR. A negative control with only secondary antibody (Ab) staining is included (left). Nuclei were counterstained with Hoechst 33342. White arrowheads indicate areas of RSPO2 expression. Representative images are shown from n=3 mice per condition. FP: fat pad; S: synovium; F: femur; T: tibia; M: meniscus; OP: osteophyte. (B) Qualitative grading of RSPO2 staining in (A) for synovium, cartilage, subchondral bone (SCB), and meniscus. (C) RSPO2 protein levels in synovial fluid from contralateral (contra) and 28d ACLR (n=6 mice), measured by ELISA. Amount of RSPO2 was normalized to total protein in each synovial fluid sample. (D-E) Assessment of RSPO2+ cells from contra, 7d ACLR or 28d ACLR synovium by flow cytometry (n=3 mice per condition). (D) Total number of RSPO2+ cells in synovium. (E) Expression of CD31 and CD45 in RSPO2+ synovial cells. Mean percentage of RSPO2+ cells in the CD31− CD45− gate is shown in red ± SEM. (F) Feature plot showing expression of Rspo2 in all synovial cells by scRNA-seq. Lining SFs are outlined. (G) Violin plots of Rspo2, the sublining SF marker Thy1, and the lining marker Prg4 across SF subsets. (H) Circle plot showing Rspo signaling communication between SF subsets, with directionality indicated by arrowheads. Lines are color-coded by source cell type and line width is proportional to interaction strength. (I) Relative contribution of Rspo pathway ligand-receptor pairs for the circle plot of SFs in (H). For (B), one-way ANOVA with multiple comparisons and Tukey’s post-hoc correction was used to assess significance in each tissue compartment (*P<0.05, **P<0.01, ***P<0.001 compared to Sham; #P<0.05 compared to 3d ACLR). For (C), a paired two-tailed student’s t-test was used with **P<0.01. For (D), a two-way ANOVA with multiple comparisons and Tukey’s post-hoc correction was used, where *P<0.05, **P<0.01 compared to the corresponding contralateral. For B-D, error bars are mean ± SEM.
Synovial lining fibroblasts arise from Dpp4+ mesenchymal progenitors
To assess the injury-associated emergence of Prg4hi Rspo2-expressing lining SFs, we performed trajectory analysis within the SF niche using Monocle3. This analysis identified Dpp4+ progenitors as precursors for Prg4hi lining SFs across pseudotime, via an αSMA+ intermediate akin to that seen in other musculoskeletal injury models such as fracture healing(40, 41) (Figure 5A–B). This differentiation was supported by pseudotime regression analysis of genes unique to the Dpp4+ root (Dpp4, Pi16, Wnt2) or the Prg4hi terminus (Prg4, Col22a1, Rspo2) of the proposed trajectory (Figure 5C–D). Transcription factor motif analysis of genes derived from co-regulation network analysis along the trajectory revealed SOX5 as a putative regulator of Rspo2 in lining SFs (Figure 5E–F and Figure S10A–C). siRNA-mediated knockdown of SOX5 in hindpaw-derived SFs reduced Sox5 transcript by ~50%, and we observed a concomitant ~65% to 80% reduction in Rspo2 transcript, pointing towards a direct role for SOX5 in regulation of Rspo2 levels in SFs (Figure 5G and Figure S10D). In summary, we propose that Prg4hi lining SFs that secrete Rspo2 arise from a Dpp4+ progenitor pool within synovium, and this differentiation trajectory, regulated in part by SOX5, may provide insights into mechanisms of chronic lining hyperplasia in PTOA.
Figure 5. Synovial lining fibroblasts arise from Dpp4+ mesenchymal progenitors.
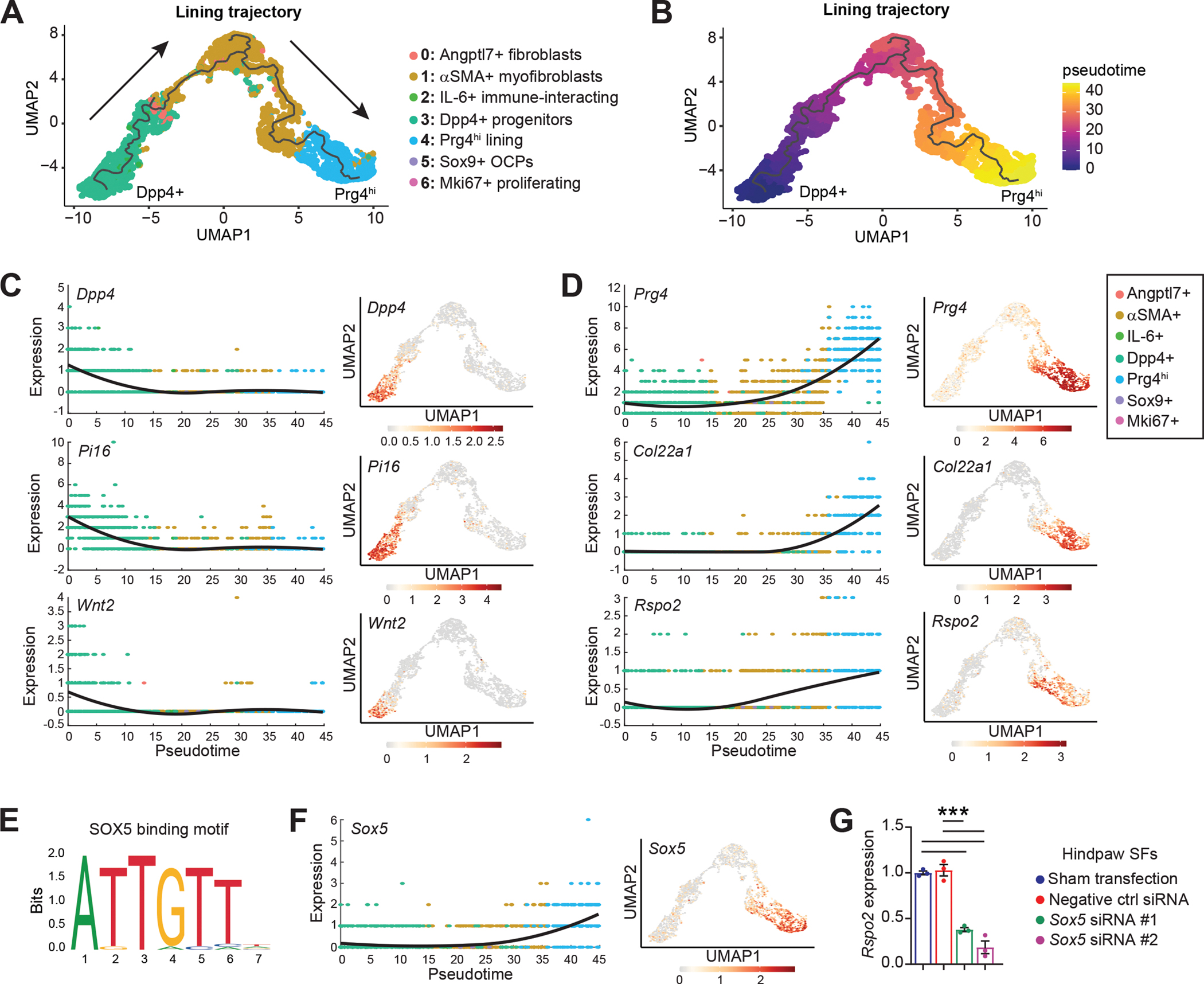
(A) Trajectory analysis of Prg4hi lining SFs. The black line represents the trajectory calculated by Monocle3 and cells are color-coded according to their cluster. Arrows indicate progression of pseudotime derived from (B). (B) Pseudotime plot for the Dpp4+ to Prg4hi lining SF trajectory in (A), calculated using Monocle3 (C-D) Pseudotime regression plots (left) and the corresponding feature plots mapped onto the lining trajectory UMAP (right), for genes associated with the (C) Dpp4+ root (Dpp4, Pi16, Wnt2) or (D) Prg4hi terminus (Prg4, Col22a1, Rspo2) of the trajectory. (E-F) Transcription factor motif analysis in Module 2 (from Figure S10A) of the Prg4hi lining trajectory, using RcisTarget. (E) Binding motif for SOX5 (JASPAR_MA0087.1). (F) Pseudotime regression plot (left) and feature plot (right) for Sox5 in the Dpp4+ to Prg4hi trajectory.(G) Hindpaw SFs were transfected with a negative control siRNA, siRNA for Sox5 (#1 and #2), or untransfected (Sham), then Rspo2 expression assessed after 48 h (n=3 biological replicates). Atp5b was used as the housekeeper gene, and the Sham transfection condition was set to 1. For G, one-way ANOVA with multiple comparisons and Tukey’s post hoc testing was performed where ***P<0.001. Error bars are mean ± SEM.
R-spondin 2 is sufficient to induce some pathological features in healthy joints of mice
To study gain-of-function effects in vivo, we performed intra-articular injections of RSPO2 (right hindlimbs) or PBS (left hindlimbs) into mice and assessed knee hyperalgesia, matrix metalloproteinase (MMP) activity, and histological features of OA and synovitis (Figure 6A). Limbs injected with RSPO2 had a lower pain threshold, indicative of RSPO2-induced hyperalgesia, and higher MMP activity compared to PBS-injected limbs (Figure 6B–C). RSPO2-injected joints had elevated PTOA and synovitis severity scores, driven by chondrocyte hypertrophy and structural damage in articular cartilage, synovial fibrosis, and inflammatory cell infiltration (Figure 6D–G, Figure S11 and Figure S12). These data clearly demonstrate that in healthy mouse joints, exogenous delivery of RSPO2 is sufficient to induce some of the pathological features observed in various joint conditions, including inflammatory arthritis and PTOA.
Figure 6. R-spondin 2 is sufficient to induce some pathological features in healthy joints of mice.
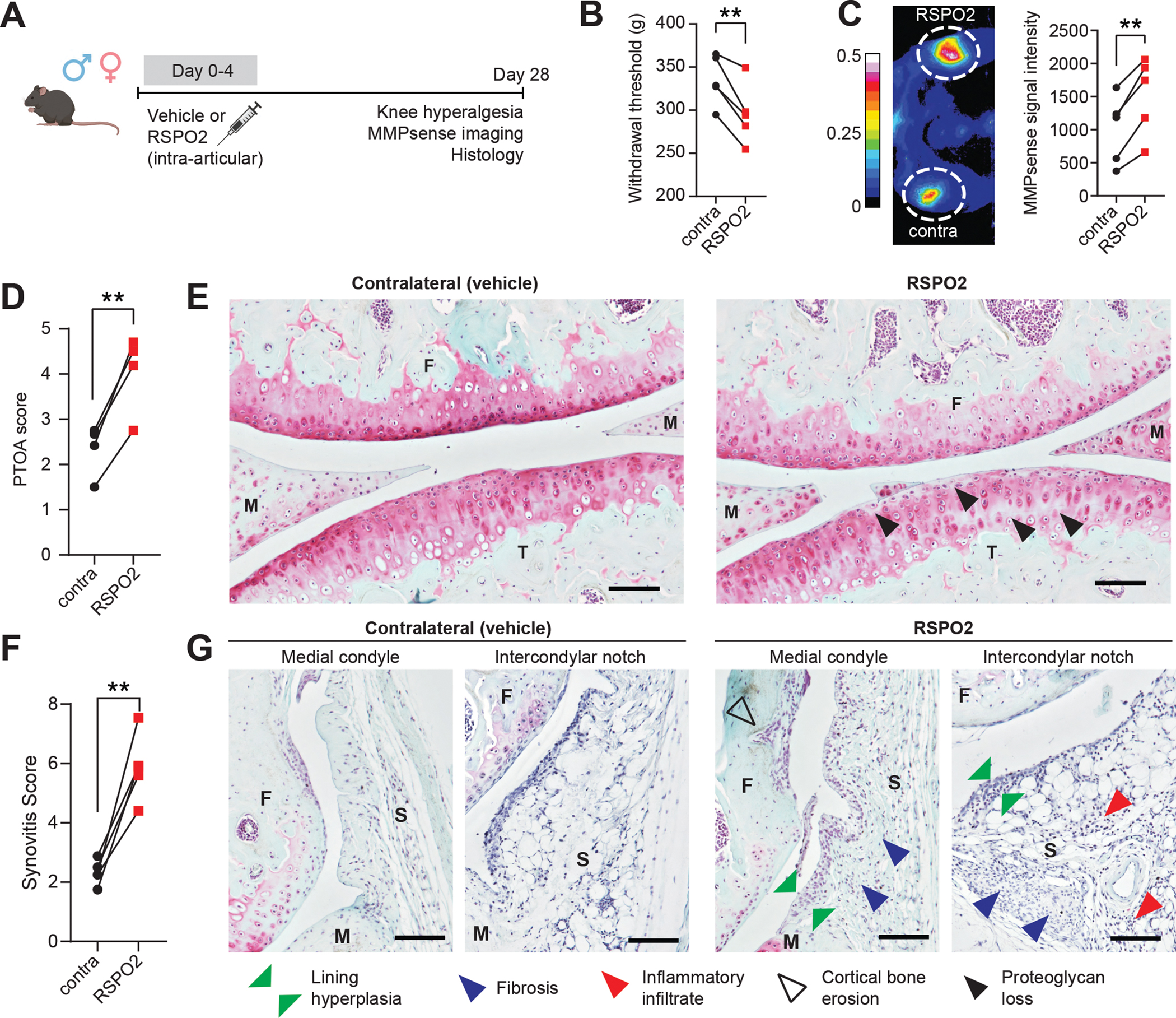
(A) Experimental design. Male and female C57Bl6/J mice (n=5) were given intra-articular injections with R-spondin 2 (RSPO2, 200 ng/mL, right limb) or PBS (left limb) for five consecutive days. 28 days later, knee hyperalgesia and matrix metalloproteinase (MMP) activity were assessed, then whole joint histological sections were graded for OA severity and synovitis. (B) Knee hyperalgesia testing of contralateral (contra, PBS-treated) or RSPO2-treated limbs 28d after first injection. Paired limbs are connected by lines. (C) MMP activity as measured by near-infrared imaging of MMPSense680 probe. Representative intensity image (left) and quantitation of signal (right). Paired limbs are connected by lines. (D-G) PTOA (D) and synovitis (F) severity scoring(63, 64) for contralateral versus RSPO2-injected limbs, with paired limbs connected by lines. Safranin O/Fast Green stained sections imaged at 10x magnification that are representative of (E) PTOA score or (G) synovitis score. Highlighted features are proteoglycan and matrix loss (black arrows), synovial lining hyperplasia (green arrows), synovial fibrosis (blue arrows), sub-synovial inflammatory infiltrate (red arrows), and cortical bone erosion (black outline arrows). Scale bar: 100 μm. For B, C, D and F, paired two-tailed student’s t-tests were used, where **P<0.01. F: femur; T: tibia; M: meniscus; S: synovium.
R-spondin 2 orchestrates pathological crosstalk between joint-resident cell types
The joint is a complex organ with diverse cell and tissue types capable of signaling to one another via synovial fluid. Having shown that SFs produce Rspo2 and that it is enriched in synovial fluid of injured joints, we sought to assess how SF-derived Rspo2 interacts with other joint-resident cell types. Treatment of knee-derived SFs with inflammatory cytokines TNFα or IL-1β, known to be activated in OA(42, 43), did not increase secretion of Rspo2 into conditioned media (Figure 7A). In vivo, SFs expressed Lgr4, Lgr5 and Lgr6, the receptors for Rspo2; Lgr4 was broadly expressed by SFs, whereas Lgr5 and Lgr6 were confined to the αSMA+ and IL-6+, and the Sox9+ clusters, respectively, and were induced after injury (Figure 7B and Figure S13A). SFs derived from murine hindpaws and from knees expressed high levels of Lgr4 and Lgr5, thus serving as valid models for studying Rspo2 signaling in SFs (Figure S13B). Treatment of Wnt-GFP reporter-derived hindpaw SFs with RSPO2 increased GFP intensity (a proxy for Wnt activity) and the percentage of Wnt-GFP+ cells (Figure 7C). Hindpaw SFs cultured on soft substrate (to reduce basal Wnt activity) were treated with RSPO2 and showed higher nuclear localization of active β-catenin compared to vehicle (Figure 7D and Figure S13C). Reduced translocation was seen after treatment with the Wnt inhibitor C59, whereas the Wnt activator CHIR99021 induced high levels of β-catenin in the nucleus. We observed induction of Wnt-responsive genes Axin2 and Lef1, and of Rspo2 itself, after treating hindpaw and knee-derived SFs with RSPO2 (Figure 7E and Figure S13D). This induction was mitigated by treatment with the Lgr inhibitor Mianserin(20). These data demonstrate that SFs secrete and respond to RSPO2, at least in part via Lgr signaling. Macrophages, on the other hand, did not express Lgr receptors (Figure S13E). We saw no induction of Wnt genes (Axin2, Lef1) or macrophage functional genes (Il1b, Tnf, Mrc1/Cd206, Il10) upon treatment of BMDM with RSPO2 (Figure 7F). Therefore, we treated BMDM with conditioned media from vehicle- or RSPO2-treated hindpaw SFs, to ask whether SFs secrete secondary mediators that affect macrophage polarization. SF conditioned media was given to BMDM polarized towards M1 (IL-1β), M2 (IL-4) or M0 (PBS) (Figure 7G). In each condition, media from RSPO2-treated SFs skewed macrophage polarization towards an inflammatory, M1-like phenotype, based on composite gene expression of M1/M2 markers (Figure 7H and Figure S13F). This was mitigated when SFs were co-treated with Mianserin, reinforcing that RSPO2-induced activation of SF-derived mediators that activate macrophages is Lgr-dependent. We then employed this conditioned media treatment protocol to chondrogenic ATDC5 cells, which express Lgr receptors (Figure S14A). Treatment of differentiated ATDC5 cells with media from RSPO2-treated hindpaw SFs increased their expression of osteogenic genes Bsp and Ocn (Figure S14B). Direct treatment of ATDC5 cells with RSPO2 during and after chondrogenic differentiation suppressed chondrogenic genes, but elevated hypertrophic, osteogenic, and Wnt genes (Figure S14C–D), consistent with previous reports of a pro-osteogenic and anti-chondrogenic role for Rspo2(20, 44). Together, these results suggest that Rspo2 may orchestrate pathological crosstalk in the joint via pro-inflammatory signaling between SFs and macrophages, and pro-hypertrophic signaling between SFs and chondrocytes.
Figure 7. R-spondin 2 orchestrates pathological crosstalk between joint-resident cell types.
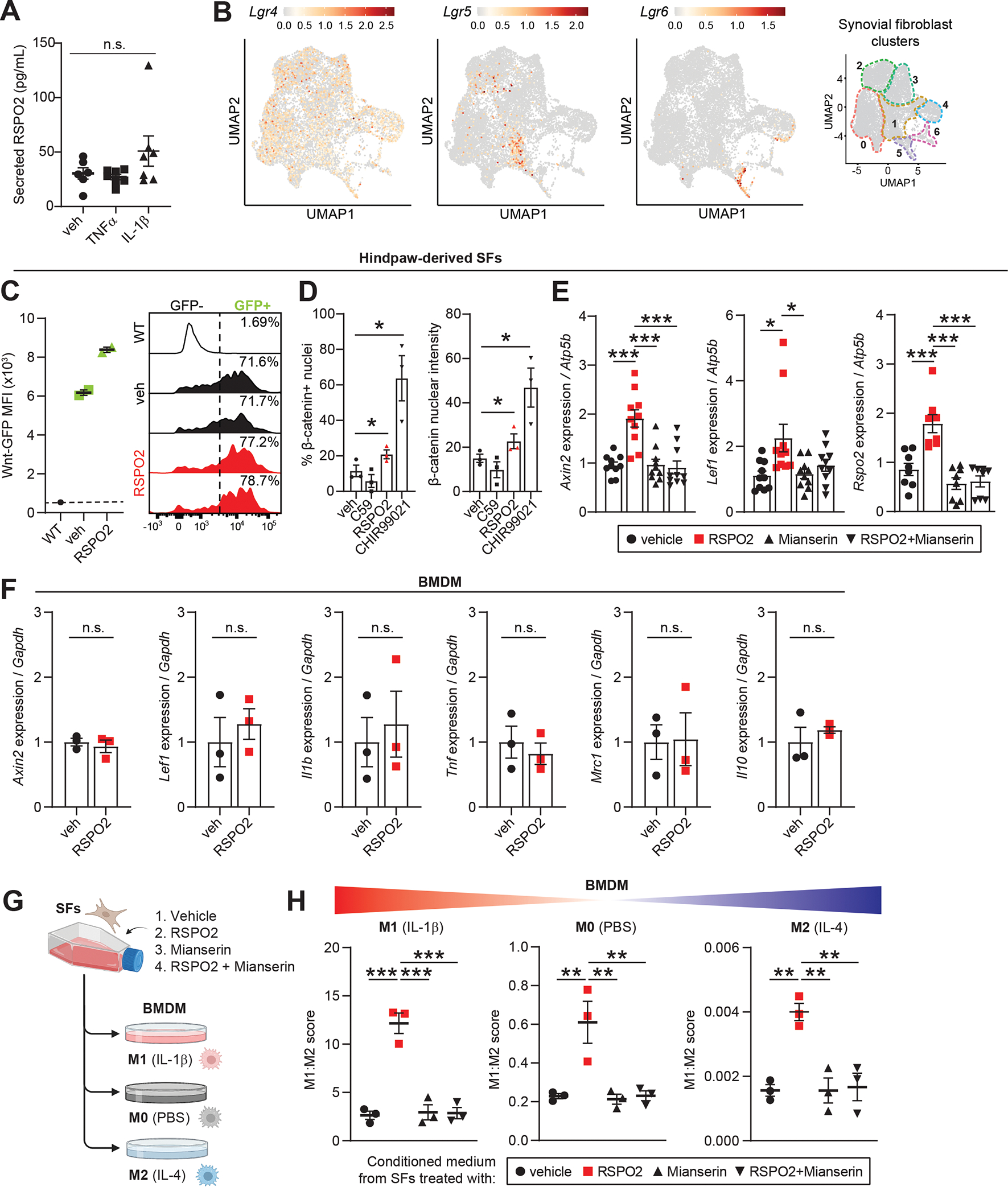
(A) Secreted RSPO2 protein levels in conditioned medium from cultured knee-derived SFs treated for 48 h with vehicle (veh), TNFα (10 ng/mL) or IL-1β (10 ng/mL). (B) Feature plots of Lgr receptors (Lgr4–6) in SFs. Color scales are not equivalent between plots. Cluster borders are shown on the right. (C) Hindpaw-derived SFs from Wnt-GFP reporter mice were treated with veh or RSPO2 (200 ng/mL) for 24 h. SFs were analyzed by flow cytometry to assess Wnt signaling activity (Wnt-GFP MFI, left) and the total percentage of Wnt-GFP+ cells (right). WT SFs (no reporter) were used as a negative control. Two biological replicates were performed. (D) Hindpaw-derived SFs were treated for 4 h with veh, C59 (0.25 μM), RSPO2 (200 ng/mL), or CHIR99021 (10 μM) on 1.5 kPa Ibidi soft substrate dishes (n=3 biological replicates). Immunocytochemical staining of non-phosphorylated/active β-catenin was performed to assess nuclear localization. Left: percentage of cells with nuclei positively stained for β-catenin. Right: Nuclear β-catenin staining intensity. (E) Hindpaw-derived SFs were treated for 24 h with veh, RSPO2 (200 ng/mL), Mianserin (20 μM), or RSPO2+Mianserin (n=8–10 biological replicates). Expression of Axin2, Lef1 and Rspo2 was normalized to Atp5b levels, and veh samples were set to 1. (F) Bone marrow-derived macrophages (BMDM) were treated for 8 h with veh or RSPO2 (200 ng/mL) (n=3 biological replicates). Expression of Axin2, Lef1, Il1b, Tnf, Mrc1 and Il10 were normalized to Gapdh levels, and veh samples were set to 1. (G) Schematic showing experimental design of SF-BMDM conditioned media treatment in (H). (H) Hindpaw-derived SFs were treated with vehicle, RPSO2 (200 ng/mL), Mianserin (20 μM), or RSPO2+Mianserin. After 6 h, treatment media was removed, cells were washed and replenished with normal SF media for another 18 h (24 h total). SF conditioned media was harvested and directly treated to BMDM for 8 h in tandem with M0 polarization (PBS, middle), M1 polarization (10 ng/mL IL-1β, left), or M2 polarization (10 ng/mL IL-4, right) (n=3 biological replicates per condition). A composite M1:M2 polarization score was calculated based on the expression of M1 (Il1b, Il6, Nos2) and M2 (Mrc1, Il10, Arg1) genes by qPCR. Gapdh was used as the housekeeper gene. For A, E and H, one-way ANOVA with multiple comparisons and Tukey’s post hoc testing was performed where *P<0.05, **P<0.01, ***P<0.001. For D and E, paired two-tailed student’s t-tests were performed where *P<0.05. Error bars are mean ± SEM. n.s.: not significant.
DISCUSSION
Synovium undergoes dramatic changes in composition and function after joint injury; however, underlying molecular mechanisms are not well understood. Here, we report a hierarchy of functionally distinct SF subsets in healthy and injured joints, and demonstrate that lining SFs secrete the Wnt/β-catenin agonist R-spondin 2, which promotes PTOA.
Recent studies utilizing ‘omics approaches have shed light on SF and macrophage subpopulations in RA synovium, in mice and humans(5–7, 26, 45). Single-cell studies of OA, in addition to their paucity, have largely focused on cartilage(9–12, 46), given its historical and prevailing position as the focal point of OA research. Reports are now emerging focused on the role of synovium specifically in OA, including recent evidence from Nanus et al showing a SF gene signature of inflammation and neurotrophic factors linked to early OA pain(47). Studies in human(9) and mouse(10) defined the broad distinction between sublining and lining SFs in OA. Here, we have extended these findings to propose distinct functional subsets within the SF niche, including Dpp4+ mesenchymal progenitors and IL-6+ immune-interacting SFs, along with αSMA+ myofibroblast-like cells and osteochondral progenitors that arise after injury. These functional subsets are worthy of future in vitro and in vivo characterization.
Human GWAS findings first linked canonical Wnt/β-catenin signaling to OA(48–50). Lorecivivint (SM04690), a small molecule Wnt inhibitor, has shown therapeutic promise in pre-clinical studies(51, 52) and Phase I/II clinical trials(15–17, 53, 54), with ongoing Phase III trials aiming to harness broad Wnt inhibition for treatment of moderate to severe OA patients. An important consideration when modulating Wnt activity is the deleterious effects of both overactive and absent Wnt signaling in chondrocytes(20, 55–59); finetuning Wnt activity is thus a warranted avenue of inquiry. R-spondins are potent Wnt agonists that facilitate Frizzled/LRP receptor stability to permit Wnt activation(19). Okura et al showed that Rspo2 is elevated in synovial fluid of OA patients(20), and identified the antidepressant Mianserin as a putative Rspo/Lgr inhibitor that mitigated anti-chondrogenic actions of Rspo2. Another study showed that intra-articular injection of RSPO2 exacerbated collagenase-induced OA in mice, consistent with our results in uninjured mice, and demonstrated efficacy of anti-Rspo2 treatment in suppressing β-catenin overactivation(21). The authors, however, proposed that Rspo2 was secreted by synovial macrophages, for which we found no evidence in healthy or injured synovium, nor in vitro. Instead, we used multiple approaches to demonstrate that Prg4hi lining SFs express and secrete Rspo2. Differentiation trajectory analysis pointed towards a Dpp4+ mesenchymal progenitor pool giving rise to Rspo2-expressing lining SFs, via an αSMA+ intermediate. Advances in multi-omic single-nucleus sequencing, combining gene expression and chromatin accessibility for refined trajectory inference(60), will facilitate further interrogation of lining SF differentiation and provide insights into pathological lining hyperplasia seen in PTOA. SOX5 has been shown to activate MMP9 and promote an invasive phenotype in RA SFs(61), reminiscent of the hyperplastic lining phenomenon seen in our model, which we propose is regulated in part by SOX5. Additionally, SFs were shown to up-regulate SOX5 in response to pro-inflammatory cytokine treatment(62), which may represent one mechanism by which SOX5-driven lining hyperplasia is initiated.
Given our observation of Rspo2 expression in multiple joint tissues, further work examining how Rspo2 and its cellular sources drive PTOA-relevant pathologies such as fibrosis, osteophytes, and inflammation is warranted. Rspo2 is a soluble matricellular protein that we and others(20) have shown to be detectable in healthy synovial fluid and elevated in OA. Our intra-articular injections delivered Rspo2 directly into the synovial fluid, and this resulted in features characteristic of arthritis pathology throughout the joint. These findings, in conjunction with our mechanistic in vitro results showing Rspo2-mediated interactions between joint-resident cell types, suggest that Rspo2 may indeed play a role in orchestrating pathological joint crosstalk after injury. This justifies further investigation of in vivo mechanisms to better understand how targeting Rspo2 signaling may be harnessed as a potential therapeutic approach. Building on promising findings of Rspo/Lgr inhibition by Mianserin(20), the development of specific and sustained-release OA therapies that target overactive Wnt and Rspo signaling should be prioritized.
The lack of longitudinal samples and bias towards end-stage donors are inherent drawbacks of human OA tissue samples, complicating studies focused on early disease etiology, especially post-traumatic. Therefore, the clinically-relevant ACLR model of PTOA in mice provides a useful means of studying pathophysiological events that precede established disease. Future investigations must discern the immediate molecular events after joint trauma from the chronic downstream sequelae that arise after initial injury via joint destabilization and inflammation. Further, it will be critical to assess the extent to which our findings, especially distinct SF subsets, are conserved in human PTOA. Changes in gene expression and preferential cell type depletion are shortcomings of common tissue dissociation methods, as used here. We observed very low numbers of contaminating skeletal muscle cells in our scRNA-seq dataset (<1%) and an absence of capsular cells, confirming the precision and reproducibility of our knee synovial dissection. However, a limitation of this procedure is that the posterior synovium and posterior fat pad are not captured. The rapid advancement of spatial transcriptomics may remedy these concerns and allow scientists to capture both the phenotypic and anatomical dimension, which is essential for an organ as architecturally complex as the joint. Meta-analysis of both mouse and human single-cell studies from OA synovium and cartilage will further help to bridge gaps in knowledge, harmonizing existing data for better utility and interpretation.
In summary, we defined distinct functional subsets of SFs in PTOA, showed that lining SFs secrete Rspo2, and that Rspo2 can promote pathological crosstalk within the joint. We also identified a Dpp4+ progenitor pool that gives rise to Prg4hi Rspo2-expressing lining SFs, in part via SOX5. These findings highlight the importance of SF speciation and canonical Wnt/β-catenin signaling in PTOA, which together serve as promising therapeutic targets in pursuit of disease-modifying OA drugs.
Supplementary Material
Key messages.
What is already known about this subject?
Wnt/β-catenin signaling is overactive in mouse and human OA joints, but the mechanism of action and identity of agonists are currently unknown. Lining and sublining synovial fibroblasts are known but specific functional subsets of these cells are undescribed in OA.
What does this study add?
We uncovered distinct functional subsets of synovial fibroblasts in OA, and showed that Prg4hi lining fibroblasts secrete the Wnt/β-catenin signaling agonist R-spondin 2 which orchestrates joint pathology.
How might this impact on clinical practice or future developments?
While Wnt signaling is recognized to be important in OA pathogenesis, very little is known about Wnt signaling in synovium nor the key modulators of Wnt signaling following joint injury. These findings may lead to more specific targeting of Wnt signaling via Rspo2 inhibition.
Acknowledgements
We would like to acknowledge the assistance of the University of Michigan Advanced Genomics Core, Flow Cytometry Core, Orthopaedic Research Laboratories Histology Core, and Unit for Laboratory Animal Medicine. We appreciate the intellectual input and critical feedback from Drs. Ormond MacDougald and Jan Stegemann. BioRender.com was used to generate cartoons and schematics. Data in this manuscript were presented at the Orthopaedic Research Society’s Annual Meetings in 2021 and 2022, and the Gordon Musculoskeletal Biology and Bioengineering conference in 2022.
Funding
This work was supported by funding from the National Institutes of Health (R21AR076487 to TM, R01DE030716 and R01AR066028 to KDH, R00AR075899 to CLW), a Catalyst Award from the Dr. Ralph and Marian Falk Medical Research Trust to TM, and from the Orthopaedic Research and Education Foundation (OREF) to CLW. Histology, imaging work, and pilot funding to TM was supported by the Michigan Integrative Musculoskeletal Health Core Center (P30AR069620, National Institutes of Health). Single-cell experiments utilized resources that are funded by the University of Michigan Comprehensive Cancer Center (P30CA046592, National Institutes of Health). AJK was supported by a Michigan Pioneer Postdoctoral Fellowship from the University of Michigan. PMR was supported by a T32 training grant (T32AR007080–40, National Institutes of Health). LL was supported by a Graduate Research Fellowship Program award from the National Science Foundation. RFB was supported by the J. Griswold and Margery H. Ruth Alpha Omega Alpha Research Fellowship and a research fellowship by the Office of Health Equity & Inclusion. DF was supported by the Community College Summer Fellowship Program from the University of Michigan.
Footnotes
Competing interests
None to declare.
Ethics approval
All animal experiments were performed in accordance with approved IACUC protocols.
Data availability statement
All scRNA-seq data are publicly available via the NCBI Gene Expression Omnibus (GEO) using the accession number GSE211584. Other data are available upon reasonable request.
References
- 1.Lieberthal J, Sambamurthy N, Scanzello CR. Inflammation in joint injury and post-traumatic osteoarthritis. Osteoarthr. Cartil. 2015;23(11):1825–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L, Xing R, Huang Z, Ding L, Zhang L, Li M, et al. Synovial Fibrosis Involvement in Osteoarthritis. Front. Med. 2021;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blom AB, van Lent PL, Holthuysen AE, van der Kraan PM, Roth J, van Rooijen N, et al. Synovial lining macrophages mediate osteophyte formation during experimental osteoarthritis. Osteoarthr. Cartil. 2004;12(8):627–35. [DOI] [PubMed] [Google Scholar]
- 4.Wei K, Korsunsky I, Marshall JL, Gao A, Watts GFM, Major T, et al. Notch signalling drives synovial fibroblast identity and arthritis pathology. Nature. 2020;582(7811):259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang F, Wei K, Slowikowski K, Fonseka CY, Rao DA, Kelly S, et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 2019;20(7):928–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Croft AP, Campos J, Jansen K, Turner JD, Marshall J, Attar M, et al. Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature. 2019;570(7760):246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizoguchi F, Slowikowski K, Wei K, Marshall JL, Rao DA, Chang SK, et al. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat. Commun. 2018;9(1):789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathiessen A, Conaghan PG. Synovitis in osteoarthritis: current understanding with therapeutic implications. Arthritis Res. Ther. 2017;19(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou C-H, Jain V, Gibson J, Attarian DE, Haraden CA, Yohn CB, et al. Synovial cell cross-talk with cartilage plays a major role in the pathogenesis of osteoarthritis. Sci. Rep. 2020;10(1):10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sebastian A, McCool JL, Hum NR, Murugesh DK, Wilson SP, Christiansen BA, et al. Single-Cell RNA-Seq Reveals Transcriptomic Heterogeneity and Post-Traumatic Osteoarthritis-Associated Early Molecular Changes in Mouse Articular Chondrocytes. Cells. 2021;10(6):1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Ning Y, Zhang P, Poulet B, Huang R, Gong Y, et al. Comparison of the major cell populations among osteoarthritis, Kashin–Beck disease and healthy chondrocytes by single-cell RNA-seq analysis. Cell Death Dis. 2021;12(6):551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji Q, Zheng Y, Zhang G, Hu Y, Fan X, Hou Y, et al. Single-cell RNA-seq analysis reveals the progression of human osteoarthritis. Ann. Rheum. Dis. 2019;78(1):100–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Fan X, Xing L, Tian F. Wnt signaling: a promising target for osteoarthritis therapy. Cell Commun. Signal. 2019;17(1):97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lane NE, Corr M, Baer N, Yazici Y. Wnt Signaling in Osteoarthritis: a 2017 Update. Curr. Treat. Options Rheumatol 2017;3(2):101–11. [Google Scholar]
- 15.Yazici Y, McAlindon TE, Gibofsky A, Lane NE, Lattermann C, Skrepnik N, et al. A Phase 2b randomized trial of lorecivivint, a novel intra-articular CLK2/DYRK1A inhibitor and Wnt pathway modulator for knee osteoarthritis. Osteoarthr. Cartil. 2021;29(5):654–66. [DOI] [PubMed] [Google Scholar]
- 16.Yazici Y, McAlindon TE, Gibofsky A, Lane NE, Clauw DJ, Jones MH, et al. Results from a 52-week randomized, double-blind, placebo-controlled, phase 2 study of a novel, intra-articular wnt pathway inhibitor (SM04690) for the treatment of knee osteoarthritis. Osteoarthr. Cartil. 2018;26:S293–S4. [Google Scholar]
- 17.Yazici Y, McAlindon TE, Fleischmann R, Gibofsky A, Lane NE, Kivitz AJ, et al. A novel Wnt pathway inhibitor, SM04690, for the treatment of moderate to severe osteoarthritis of the knee: results of a 24-week, randomized, controlled, phase 1 study. Osteoarthr. Cartil. 2017;25(10):1598–606. [DOI] [PubMed] [Google Scholar]
- 18.Knight MN, Karuppaiah K, Lowe M, Mohanty S, Zondervan RL, Bell S, et al. R-spondin-2 is a Wnt agonist that regulates osteoblast activity and bone mass. Bone Res. 2018;6(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Lau WB, Snel B, Clevers HC. The R-spondin protein family. Genome Biol. 2012;13(3):242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okura T, Ohkawara B, Takegami Y, Ito M, Masuda A, Seki T, et al. Mianserin suppresses R-spondin 2-induced activation of Wnt/β-catenin signaling in chondrocytes and prevents cartilage degradation in a rat model of osteoarthritis. Sci. Rep. 2019;9(1):2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H, Lin C, Zeng C, Wang Z, Wang H, Lu J, et al. Synovial macrophage M1 polarisation exacerbates experimental osteoarthritis partially through R-spondin-2. Ann. Rheum. Dis. 2018;77(10):1524–34. [DOI] [PubMed] [Google Scholar]
- 22.Rzeczycki P, Rasner C, Lammlin L, Junginger L, Goldman S, Bergman R, et al. Cannabinoid receptor type 2 is upregulated in synovium following joint injury and mediates anti-inflammatory effects in synovial fibroblasts and macrophages. Osteoarthr. Cartil. 2021;29(12):1720–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christiansen BA, Anderson MJ, Lee CA, Williams JC, Yik JHN, Haudenschild DR. Musculoskeletal changes following non-invasive knee injury using a novel mouse model of post-traumatic osteoarthritis. Osteoarthr. Cartil. 2012;20(7):773–82. [DOI] [PubMed] [Google Scholar]
- 24.Maerz T, Newton MD, Kurdziel MD, Altman P, Anderson K, Matthew HWT, et al. Articular cartilage degeneration following anterior cruciate ligament injury: a comparison of surgical transection and noninvasive rupture as preclinical models of post-traumatic osteoarthritis. Osteoarthr. Cartil. 2016;24(11):1918–27. [DOI] [PubMed] [Google Scholar]
- 25.Maerz T, Kurdziel M, Newton MD, Altman P, Anderson K, Matthew HWT, et al. Subchondral and epiphyseal bone remodeling following surgical transection and noninvasive rupture of the anterior cruciate ligament as models of post-traumatic osteoarthritis. Osteoarthr. Cartil. 2016;24(4):698–708. [DOI] [PubMed] [Google Scholar]
- 26.Culemann S, Grüneboom A, Nicolás-Ávila JÁ, Weidner D, Lämmle KF, Rothe T, et al. Locally renewing resident synovial macrophages provide a protective barrier for the joint. Nature. 2019;572(7771):670–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blaker CL, Ashton DM, Doran N, Little CB, Clarke EC. Sex- and injury-based differences in knee biomechanics in mouse models of post-traumatic osteoarthritis. J Biomech. 2021;114:110152. [DOI] [PubMed] [Google Scholar]
- 28.Bergman R, Rzeczycki P, Junginger L, Lammlin L, Goldman S, Rasner C, et al. Pain-, tissue degeneration-, and synovial transcriptome-based sex differences in murine post-traumatic osteoarthritis. Osteoarthr. Cartil. 2021;29:S111–S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotech. 2018;36(5):411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan C-H, et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12(1):1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanchez-Lopez E, Coras R, Torres A, Lane NE, Guma M. Synovial inflammation in osteoarthritis progression. Nat. Rev. Rheumatol. 2022;18(5):258–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krenn V, Morawietz L, Burmester GR, Kinne RW, Mueller-Ladner U, Muller B, et al. Synovitis score: discrimination between chronic low-grade and high-grade synovitis. Histopathology. 2006;49(4):358–64. [DOI] [PubMed] [Google Scholar]
- 33.Merrick D, Sakers A, Irgebay Z, Okada C, Calvert C, Morley MP, et al. Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science. 2019;364(6438):eaav2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buechler MB, Pradhan RN, Krishnamurty AT, Cox C, Calviello AK, Wang AW, et al. Cross-tissue organization of the fibroblast lineage. Nature. 2021;593(7860):575–9. [DOI] [PubMed] [Google Scholar]
- 35.Kortekaas MC, Kwok WY, Reijnierse M, Huizinga TW, Kloppenburg M. Osteophytes and joint space narrowing are independently associated with pain in finger joints in hand osteoarthritis. Ann. Rheum. Dis. 2011;70(10):1835–7. [DOI] [PubMed] [Google Scholar]
- 36.Zhou Y, Wang T, Hamilton JL, Chen D. Wnt/β-catenin Signaling in Osteoarthritis and in Other Forms of Arthritis. Curr. Rheumatol. Rep. 2017;19(9):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stampella A, Monteagudo S, Lories R. Wnt signaling as target for the treatment of osteoarthritis. Best Pract. Res. Clin. Rheumatol. 2017;31(5):721–9. [DOI] [PubMed] [Google Scholar]
- 38.Ferrer-Vaquer A, Piliszek A, Tian G, Aho RJ, Dufort D, Hadjantonakis AK. A sensitive and bright single-cell resolution live imaging reporter of Wnt/-catenin signaling in the mouse. BMC Dev. Biol. 2010;10(1):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Armaka M GV, Kontoyiannis D, Kollias G. A standardized protocol for the isolation and culture of normal and arthritogenic murine synovial fibroblasts. Protoc. Exch. 2009. [Google Scholar]
- 40.Grcevic D, Pejda S, Matthews BG, Repic D, Wang L, Li H, et al. In vivo fate mapping identifies mesenchymal progenitor cells. Stem Cells. 2012;30(2):187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matthews BG, Novak S, Sbrana FV, Funnell JL, Cao Y, Buckels EJ, et al. Heterogeneity of murine periosteum progenitors involved in fracture healing. eLife. 2021;10:e58534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Calich AL, Domiciano DS, Fuller R. Osteoarthritis: can anti-cytokine therapy play a role in treatment? Clin. Rheumatol. 2010;29(5):451–5. [DOI] [PubMed] [Google Scholar]
- 43.McNulty AL, Rothfusz NE, Leddy HA, Guilak F. Synovial fluid concentrations and relative potency of interleukin-1 alpha and beta in cartilage and meniscus degradation. J. Orthop. Res. 2013;31(7):1039–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakajima M, Kou I, Ohashi H, Ikegawa S. Identification and Functional Characterization of RSPO2 as a Susceptibility Gene for Ossification of the Posterior Longitudinal Ligament of the Spine. Am. J. Hum. Genet. 2016;99(1):202–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alivernini S, MacDonald L, Elmesmari A, Finlay S, Tolusso B, Gigante MR, et al. Distinct synovial tissue macrophage subsets regulate inflammation and remission in rheumatoid arthritis. Nat. Med. 2020;26(8):1295–1306. [DOI] [PubMed] [Google Scholar]
- 46.Rios-Arce ND, Murugesh DK, Hum NR, Sebastian A, Jbeily EH, Christiansen BA, et al. Preexisting Type 1 Diabetes Mellitus Blunts the Development of Posttraumatic Osteoarthritis. JBMR Plus. 2022;6(5):e10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nanus DE, Badoume A, Wijesinghe SN, Halsey AM, Hurley P, Ahmed Z, et al. Synovial tissue from sites of joint pain in knee osteoarthritis patients exhibits a differential phenotype with distinct fibroblast subsets. EBioMedicine. 2021;72:103618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loughlin J, Dowling B, Chapman K, Marcelline L, Mustafa Z, Southam L, et al. Functional variants within the secreted frizzled-related protein 3 gene are associated with hip osteoarthritis in females. Proc. Natl. Acad. Sci. U.S.A.. 2004;101(26):9757–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Min JL, Meulenbelt I, Riyazi N, Kloppenburg M, Houwing-Duistermaat JJ, Seymour AB, et al. Association of the Frizzled-related protein gene with symptomatic osteoarthritis at multiple sites. Arthritis Rheum. 2005;52(4):1077–80. [DOI] [PubMed] [Google Scholar]
- 50.Nakamura Y, Nawata M, Wakitani S. Expression profiles and functional analyses of Wnt-related genes in human joint disorders. Am. J. Pathol. 2005;167(1):97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deshmukh V, Hu H, Barroga C, Bossard C, Kc S, Dellamary L, et al. A small-molecule inhibitor of the Wnt pathway (SM04690) as a potential disease modifying agent for the treatment of osteoarthritis of the knee. Osteoarthr. Cartil. 2018;26(1):18–27. [DOI] [PubMed] [Google Scholar]
- 52.Deshmukh V, O’Green AL, Bossard C, Seo T, Lamangan L, Ibanez M, et al. Modulation of the Wnt pathway through inhibition of CLK2 and DYRK1A by lorecivivint as a novel, potentially disease-modifying approach for knee osteoarthritis treatment. Osteoarthr. Cartil. 2019;27(9):1347–60. [DOI] [PubMed] [Google Scholar]
- 53.Yazici Y, McAlindon TE, Gibofsky A, Lane NE, Clauw D, Jones M, et al. Lorecivivint, a Novel Intraarticular CDC-like Kinase 2 and Dual-Specificity Tyrosine Phosphorylation-Regulated Kinase 1A Inhibitor and Wnt Pathway Modulator for the Treatment of Knee Osteoarthritis: A Phase II Randomized Trial. Arthritis Rheumatol. 2020;72(10):1694–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tambiah JRS, Kennedy S, Swearingen CJ, Simsek I, Yazici Y, Farr J, et al. Individual Participant Symptom Responses to Intra-Articular Lorecivivint in Knee Osteoarthritis: Post Hoc Analysis of a Phase 2B Trial. Rheumatol Ther. 2021;8(2):973–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao X, Wang X, Zhang W, Xia G, Zhang L, Wen Z, et al. WNT10A induces apoptosis of senescent synovial resident stem cells through Wnt/calcium pathway-mediated HDAC5 phosphorylation in OA joints. Bone. 2021;150:116006. [DOI] [PubMed] [Google Scholar]
- 56.Lietman C, Wu B, Lechner S, Shinar A, Sehgal M, Rossomacha E, et al. Inhibition of Wnt/β-catenin signaling ameliorates osteoarthritis in a murine model of experimental osteoarthritis. JCI insight. 2018;3(3):e96308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Den Bosch MH, Blom AB, Sloetjes AW, Koenders MI, Van De Loo FA, Van Den Berg WB, et al. Induction of Canonical Wnt Signaling by Synovial Overexpression of Selected Wnts Leads to Protease Activity and Early Osteoarthritis-Like Cartilage Damage. Am J Pathol. 2015;185(7):1970–80. [DOI] [PubMed] [Google Scholar]
- 58.Zhu M, Chen M, Zuscik M, Wu Q, Wang YJ, Rosier RN, et al. Inhibition of beta-catenin signaling in articular chondrocytes results in articular cartilage destruction. Arthritis Rheum. 2008;58(7):2053–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu M, Tang D, Wu Q, Hao S, Chen M, Xie C, et al. Activation of β-Catenin Signaling in Articular Chondrocytes Leads to Osteoarthritis-Like Phenotype in Adult β-Catenin Conditional Activation Mice. J Bone Min. Res. 2009;24(1):12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li C, Virgilio M, Collins KL, Welch JD. Single-cell multi-omic velocity infers dynamic and decoupled gene regulation. bioRxiv. 2021:2021.12.13.472472. [Google Scholar]
- 61.Shi Y, Wu Q, Xuan W, Feng X, Wang F, Tsao BP, et al. Transcription Factor SOX5 Promotes the Migration and Invasion of Fibroblast-Like Synoviocytes in Part by Regulating MMP-9 Expression in Collagen-Induced Arthritis. Front Immunol. 2018;9:749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feng X, Shi Y, Xu L, Peng Q, Wang F, Wang X, et al. Modulation of IL-6 induced RANKL expression in arthritic synovium by a transcription factor SOX5. Sci Rep. 2016;6(1):32001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jackson MT, Moradi B, Zaki S, Smith MM, McCracken S, Smith SM, et al. Depletion of protease-activated receptor 2 but not protease-activated receptor 1 may confer protection against osteoarthritis in mice through extracartilaginous mechanisms. Arthritis Rheumatol. 2014;66(12):3337–48. [DOI] [PubMed] [Google Scholar]
- 64.Little CB, Barai A, Burkhardt D, Smith SM, Fosang AJ, Werb Z, et al. Matrix metalloproteinase 13-deficient mice are resistant to osteoarthritic cartilage erosion but not chondrocyte hypertrophy or osteophyte development. Arthritis Rheum. 2009;60(12):3723–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All scRNA-seq data are publicly available via the NCBI Gene Expression Omnibus (GEO) using the accession number GSE211584. Other data are available upon reasonable request.


