Abstract
Background
Dynamic chromatin remodeling is associated with changes in the epigenetic pattern of histone acetylations and methylations required for processes based on dynamic chromatin remodeling and implicated in different nuclear functions. These histone epigenetic modifications need to be coordinated, a role that may be mediated by chromatin kinases such as VRK1, which phosphorylates histones H3 and H2A.
Methods
The effect of VRK1 depletion and VRK1 inhibitor, VRK-IN-1, on the acetylation and methylation of histone H3 in K4, K9 and K27 was determined under different conditions, arrested or proliferating cells, in A549 lung adenocarcinoma and U2OS osteosarcoma cells.
Results
Chromatin organization is determined by the phosphorylation pattern of histones mediated by different types of enzymes. We have studied how the VRK1 chromatin kinase can alter the epigenetic posttranslational modifications of histones by using siRNA, a specific inhibitor of this kinase (VRK-IN-1), and of histone acetyl and methyl transferases, as well as histone deacetylase and demethylase. Loss of VRK1 implicated a switch in the state of H3K9 posttranslational modifications. VRK1 depletion/inhibition causes a loss of H3K9 acetylation and facilitates its methylation. This effect is similar to that of the KAT inhibitor C646, and to KDM inhibitors as iadademstat (ORY-1001) or JMJD2 inhibitor. Alternatively, HDAC inhibitors (selisistat, panobinostat, vorinostat) and KMT inhibitors (tazemetostat, chaetocin) have the opposite effect of VRK1 depletion or inhibition, and cause increase of H3K9ac and a decrease of H3K9me3. VRK1 stably interacts with members of these four enzyme families. However, VRK1 can only play a role on these epigenetic modifications by indirect mechanisms in which these epigenetic enzymes are likely targets to be regulated and coordinated by VRK1.
Conclusions
The chromatin kinase VRK1 regulates the epigenetic patterns of histone H3 acetylation and methylation in lysines 4, 9 and 27. VRK1 is a master regulator of chromatin organization associated with its specific functions, such as transcription or DNA repair.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13072-023-00494-7.
Keywords: Histone H3, Acetylation, Methylation, Kinase, Chromatin, VRK-IN-1
Introduction
Chromatin remodeling is a basic process required for the adaptation of specific genomic regions to different biological processes, such as gene transcription, gene silencing, replication, chromosome condensation in mitosis, recombination, and DNA damage responses [1]. All these processes require a dynamic adaptation of chromatin to different functional requirements, as these processes progress, and may also differ among different chromosomal regions such as euchromatin or heterochromatin. The dynamic reorganization of chromatin is associated with epigenetic modification of DNA and histones, gene expression or silencing. Furthermore, chromatin remodeling also underlies cell fate and identity [2]
Some epigenetic modifications of histones are associated with the regulation of genes and its methylation with transcriptional silencing [3, 4]. The H3K9ac and H3K27ac marks are mainly surrounding promoters and enhancers [5, 6], occur in gene transcription, and their methylation is also associated with gene silencing [6], and affects cell identity [7]. The two alternative epigenetic modifications in these two H3 lysine residues require the coordination of the different epigenetic enzymes implicated that add, or remove, acetyl and methyl groups. Furthermore, the methylation of H3 in K4 is associated with gene transcription [6, 8].
The different and alternative epigenetic modifications of histones, such as acetylation or methylation in the same lysine residue, requires writers and erasers that belong to different enzyme families, which need to be coordinated based on their particular functional role. These epigenetic modifications control the degree of chromatin compaction or relaxation as well as the recruitment of proteins associated with specific functions [9, 10]. The coordination of the epigenetic enzymes implicated is likely to be mediated by kinases, which indirectly control the pattern of these histone modifications. All of these epigenetic enzymes are coordinated or regulated by phosphorylation in different specific enzymes, which may differ depending on their function.
Among the candidates to play a regulatory role of epigenetic modifications is VRK1, a chromatin kinase that appeared late in evolution in pluricellular eukaryotes. It is not present in yeasts, and in Drosophila melanogaster this gene is unique and known as NHK-1 (nucleosomal kinase-1) [11]. The VRK1 kinase domain is distantly related to casein kinases and insensitive to inhibitors targeting different kinase families [12–14]. The VRK1 protein has characteristics that make it distinct and candidate for development of highly specific inhibitors [13], one of which has been recently developed [15, 16]. VRK1 has been associated with the control of cell proliferation [17] and mitosis [18, 19], and is expressed at high levels in proliferating and tumor cells [17, 20], which is an indicator of a poorer prognosis in many tumor types [21–25]. VRK1 is also necessary to maintain genomic stability and its loss facilitates DNA damage by a defective DNA damage response [20, 26].
Among VRK1 direct nucleosomal phosphorylation targets are histone H3 in Thr3 [19, 27] and Ser10 [28], and H2A in Thr120 [11]. H4K16 acetylation controls chromatin organization and protein interactions [29]. The phosphorylation of histone H3 in Thr3 is required for phosphorylating and recruiting KAT5/Tip60 to chromatin [30, 31]. VRK1 phosphorylates and activates Tip60/KAT5 and regulates the acetylation of H4K16 [30, 31]. In DNA damage responses (DDR), VRK1 also phosphorylates H2AX in Ser139 (γH2AX) [32]. Furthermore, VRK1 also directly phosphorylates several transcription factors such as p53 [33, 34], c-Jun [35], ATF2 [36] and CREB [37], and all of phosphorylation are lost by VRK1 depletion. Furthermore, VRK1 inhibition also impairs H3Thr3 phosphorylation in a dose-dependent manner [38]. These phosphorylations, mediated by VRK1, are functionally associated with gene transcription, cell cycle progression, mitosis, and DNA damage responses (DDR) [26], and it is of note that all these processes require a dynamic and sequential remodeling of chromatin [20, 39]. VRK1 impairs G0 exit, because is required for CCND1 gene (cyclin D1) expression [17], and G2/M in cell cycle progression [19].
Because of the roles that VRK1 has on the regulation of several chromatin-associated proteins, we have studied whether VRK1 is indeed affecting the epigenetic pattern of histone posttranslational modifications (PTMs), mainly acetylation and methylation, in specific histone lysine residues. For this aim we used depletion of VRK1 with siRNA, or its inhibition with VRK-IN-1, to determine how histone H3 epigenetic PTM patterns are modified. Modifications of the histone epigenetic patterns can have pathological and therapeutic implications because of their role in DDR and other functions requiring a dynamic chromatin remodeling in tumor biology.
Results
VRK1 depletion alters the epigenetic pattern of histone H3K9 acetylation and methylation
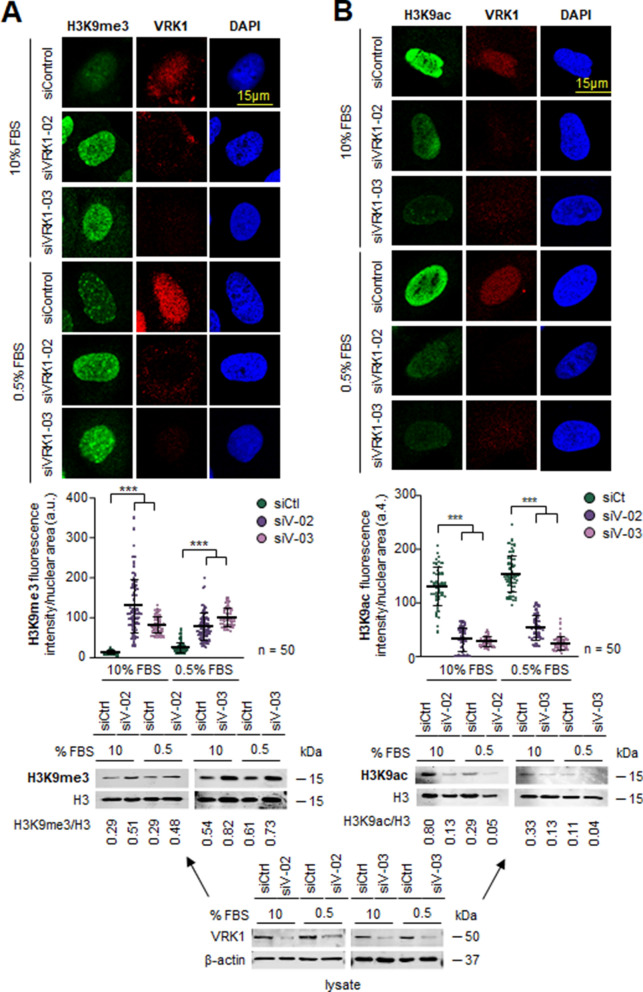
The acetylation and methylation of H3K9 is associated with gene transcription and repression, and they are mainly located in the gene body. The effect of VRK1 depletion, using two different siRNA on epigenetic modifications of histone H3K9 was determined in two different cell lines, A549 lung adenocarcinoma and U2OS osteosarcoma, under two conditions, in the presence and absence of serum. VRK1 depletion caused an increase in H3K9 methylation (Fig. 1a; Additional file 1: Fig. S1A) and a decrease in H3K9 acetylation (Fig. 1b; Additional file 1: Fig. S1B) in both A549 (Fig. 1) and U20S (Additional file 2: Fig. S2) cells cell lines independently of serum, which is consistent with a reduction in transcription. H3K9 methylation is associated with repressive chromatin [40].
Fig. 1.
Effect of VRK1 depletion on epigenetic modifications of H3K9 in the presence or absence of serum in A549 cells. The field figure is shown in Additional file 1: Fig. S1. A Effect of VRK1 depletion on histone H3K9 methylation in A549 cells. B Effect of VRK1 depletion on histone H3K9 acetylation in A549 cells. The quantification of H3K9 methylation (left) and acetylation (right) from 50 cells are shown below. The immunoblots of the cell extracts from the experiment are shown at the bottom. The lysate was divided in two for loading in gels to determine each of the H3K9 modifications. ***p < 0.001. siCt, siControl; siV-02, siVRK1-02; siV-03, siVRK1-03. The same lysate was used for both determinations
VRK1 depletion alters the epigenetic pattern of histone H3K27 acetylation and methylation
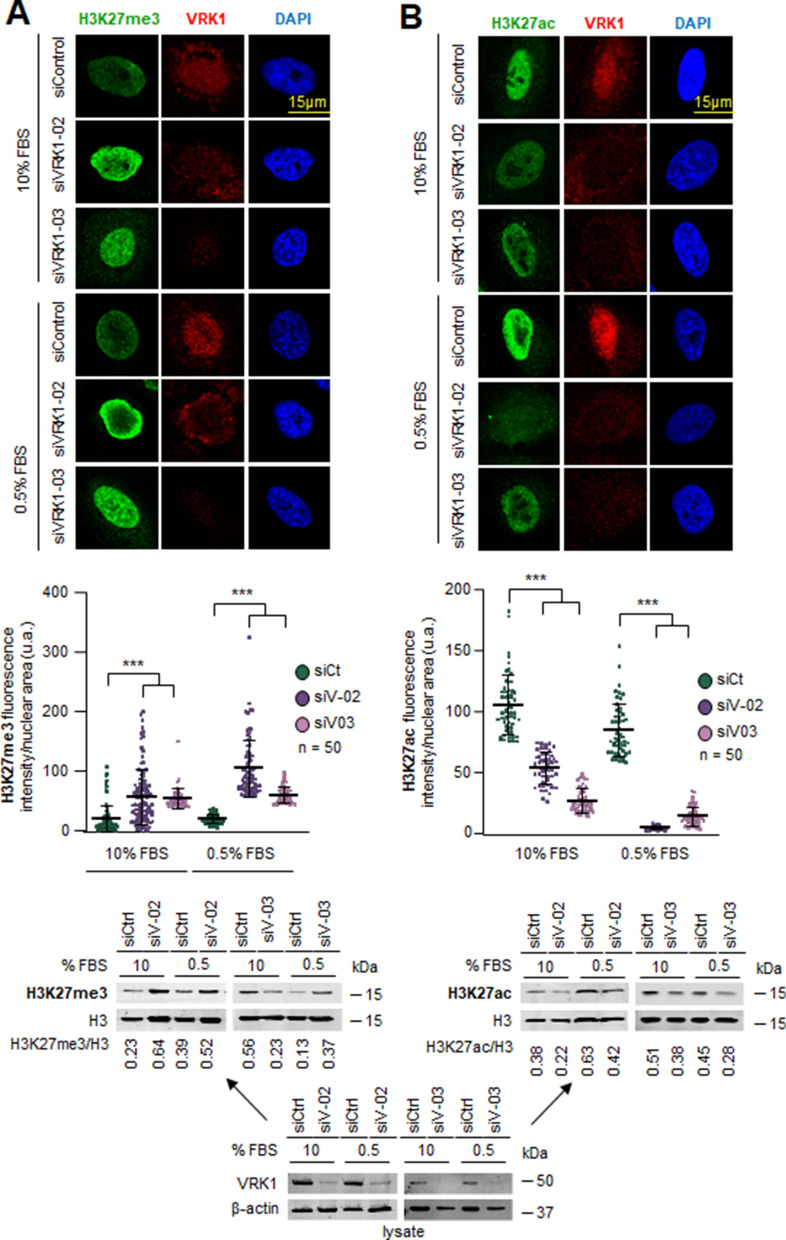
H3K27 acetylation is associated with active enhancers [5, 41, 42] and its methylation with repression of transcription [42–44]. The effect of VRK1 depletion, using two different siRNA on epigenetic modifications of histone H3K27 was determined in two different cell lines, A549 and U2OS. VRK1 depletion caused an increase in H3K27 methylation (Fig. 2a; Additional file 3: Fig. S3A) and a decrease in H3K27 acetylation (Fig. 2b; Additional file 3: Fig. S3B) in both A549 (Fig. 2) and U2OS cells (Additional file 4: Fig. S4) independently of the presence or absence of serum.
Fig. 2.
Effect of VRK1 depletion of epigenetic modifications of H3K27 in the presence or absence of serum in A549 lung adenocarcinoma cells. The field figure is shown in Additional file 3: Fig. S3. A Effect of VRK1 depletion on histone H3K27 methylation in A549 cells. B Effect of VRK1 depletion on histone H3K9 acetylation in A549 cells. The quantification of H3K27 methylation (left) and acetylation (right) from 50 cells is shown below. The immunoblots of the cell extracts from the experiment are shown at the bottom. The lysate was divided in two for loading in gels to determine each of the H3K27 modifications. ***p < 0.001. siCt, siControl; siV-02, siVRK1-02; siV-03, siVRK1-03
The VRK-IN-1 inhibitor alters the epigenetic pattern of histone H3K4, H3K9 and H3K27 modifications
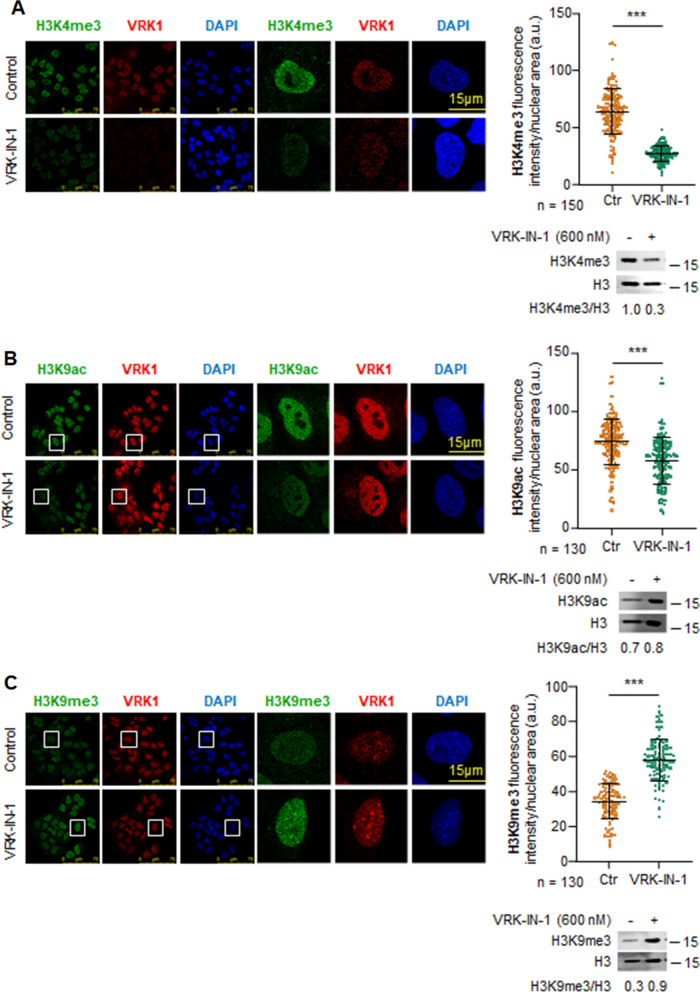
A novel and specific inhibitor of VRK1 (VRK-IN-1), based on a pyridine structure, has been recently developed [15, 16]. Therefore, we tested the effect of VRK-IN-1 inhibitor in A549 and U2OS cells either in the presence and absence of serum to determine whether it can mimic the effect of VRK1 depletion on H3 posttranslational epigenetic modifications. Under both conditions, VRK1 inhibition in A549 cells resulted in a loss of H3K4me3 (Fig. 3a), a mark associated with promoters and gene transcription [45, 46]. Inhibition of VRK1 resulted in a loss of H3K9 acetylation (Fig. 3b) and a gain of H3K9me3 (Fig. 3c). Regarding H3K27, a similar but less pronounced result was obtained by inhibition with VRK-IN-1, a loss of H3K27 acetylation (Additional file 5: Fig. S5) and a gain of H3K27me3 (Additional file 5: Fig. S5). Similar results were obtained in U2OS cells treated with VRK-IN-1 for H4K16ac (Additional file 6: Fig. S6A), H3K4me3 (Additional file 6: Fig. S6B), H3K9ac (Additional file 7: Fig. S7A), H3K9me3 (Additional file 7: Fig. S7B), H3K27ac (Additional file 8: Fig. S8A) and H3K27me3 (Additional file 8: Fig. S8B). The effect of VRK1 inhibition or depletion on histone posttranslational modifications (PTMs) has to be indirect, and mediated by the enzymes performing these specific modifications.
Fig. 3.
Effect of VRK1 depletion or inhibition with VRK1-IN-1 on epigenetic modifications of H3K4 and H3K9 in A549 cells the presence or absence of serum. A Effect of VRK-IN-1 on histone H3K4m3 levels. B Effect of VRK-IN-1 on histone H3K9ac levels. C Effect of VRK-IN-1 on histone H3K9m3 levels. Ctr: control. ***p < 0.001
Inhibitors of epigenetic enzymes mimic the effects of either VRK1 inhibition or depletion on H3K9 acetylation and methylation
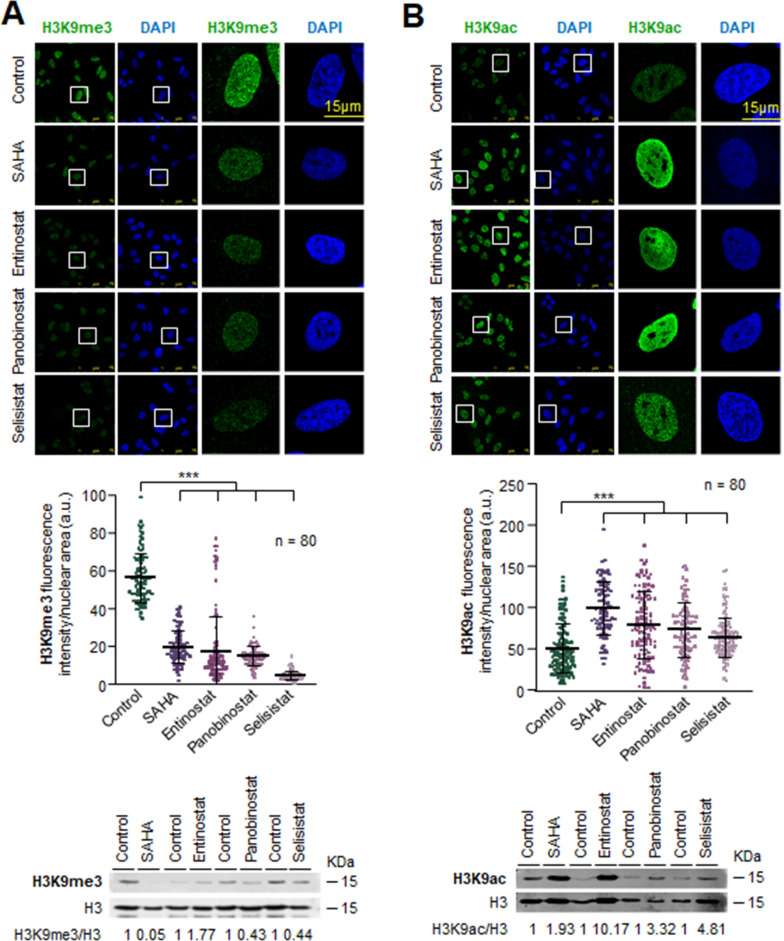
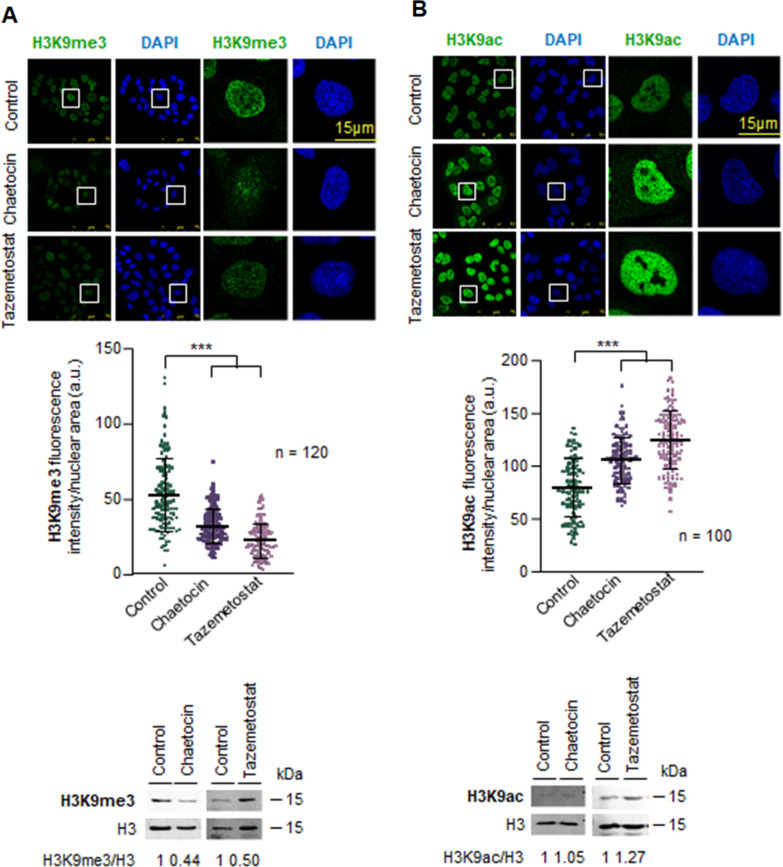
The effect of VRK1 depletion, or theVRK-IN-1 inhibitor, on histones PTMs is indirect and has to me mediated by the epigenetic enzymes that add or remove acetyl or methyl marks. Therefore, we determined whether the effects on H3K9 epigenetic posttranslational modifications by the use of inhibitors targeting the four types of enzymes directly involved in these histone PTMs and that might mimic the effect of VRK1 loss. Inhibition of HDAC1 with vorinostat, entinostat, panobinostat or selisistat resulted in an accumulation of acetylation and a consequent reduction of methylation in A549 cells (Fig. 4) and U2OS cells (Additional file 9: Fig. S9). KMT1 inhibition with either chaetocin or tazemetostat facilitated H3K9 acetylation and inhibited H3K9 methylation in both A549 (Fig. 5) and U2OS cell lines (Additional file 10: Fig. S10).
Fig. 4.
Effect of HDAC inhibitors on histone H3K9 epigenetic modifications in serum-deprived A549 cells. A Effect of HDAC inhibitors on H3K9m3 levels. B Effect of HDAC inhibitors on H3K9ac levels. The middle panel shows the quantification of the fluorescence. The immunoblots of the two modifications are shown at the bottom. ***p < 0.001
Fig. 5.
Effect of KMT inhibitors, tazemetostat and chaetocin, on histone H3K9 epigenetic modifications in serum-deprived A549 cells. A Effect of KMT inhibitors on H3K9m3 levels. B Effect of KMT inhibitors on H3K9ac levels. The middle panel shows the quantification of the fluorescence. The immunoblots of the two modifications are shown at the bottom. ***p < 0.001
Alternatively, the inhibition of KAT with the C646 inhibitor of p300/CBP acetyl transferase caused a reduction of H3K9ac and an increase in H3K9me3 levels in both cell lines [47] (Additional file 11: Fig. S11). Alternatively, inhibition of KDM1A with ORY-1001 (iadademstat) [48], or JMJD2i caused an accumulation of H3K9 methylation and a loss of H3K9 acetylation (Additional file 12: Fig. S12) in both A549 and U2OS cell lines.
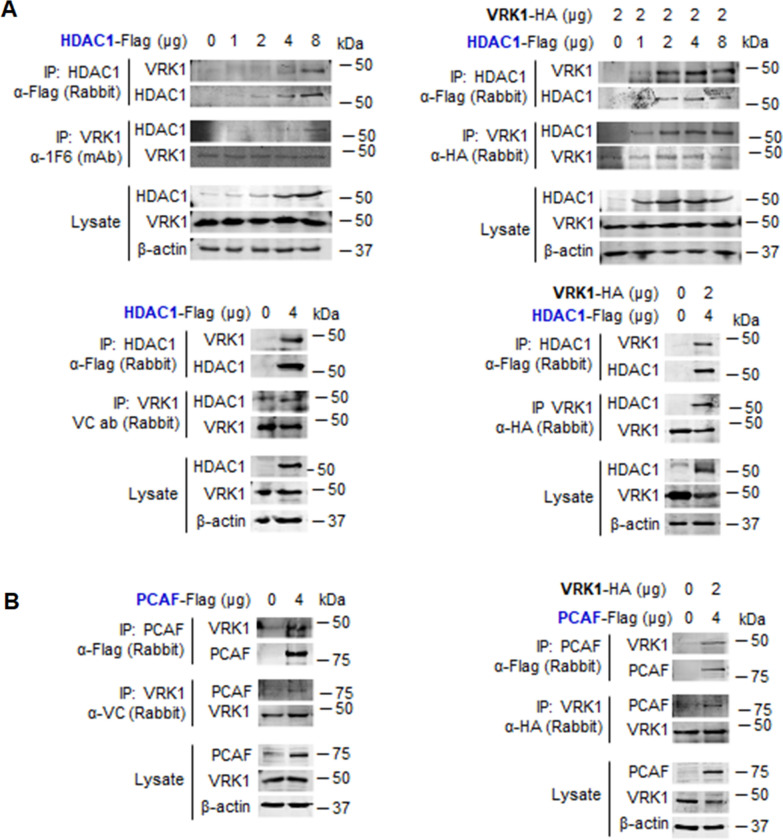
VRK1 stably interacts with lysine acetyl transferases (KAT) and histone deacetylases (HDAC)
The role of VRK1 in the modulation of epigenetic histone modifications is likely to be due to its interaction with members of these protein families. It is known that VRK1 interacts with the Tip60/KAT5 and regulates its translocation from nucleoplasm to chromatin and its activity [30, 31]. Therefore, it is likely that VRK1 might also form complexes with other members of these epigenetic enzyme families. To test this possibility, we studied the interaction of endogenous VRK1 with tagged HDAC1 transfected in HEK293T cells. The VRK1–HDAC1 interaction was detected in reciprocal immunoprecipitations (Fig. 6a, left), but in the case of immunoprecipitation with an antibody targeting the endogenous VRK1 protein, the detection is weaker because of the interference between its binding to either the antibody or the HDAC1 protein. Therefore, the direct interaction was confirmed by using both tagged proteins (Fig. 6a, right).
Fig. 6.
Interaction of HDAC and KAT with VRK1. A Left: reciprocal interaction of endogenous VRK1 with increasing amount of transfected HDAC-Flag. Right: dose-dependent interaction between VRK1-HA and increasing amounts of HDAC1-Flag. B Left: interaction of endogenous VRK1 with transfected PCAF-Flag. Right: reciprocal interaction of transfected VRK1-HA and PCAF-Flag. Transfections were performed in HEK293T cells
Next, we performed a similar experiment to detect the interaction between endogenous VRK1 and the PCAF (KAT2B) acetyl transferase. PCAF interacted with tagged and endogenous VRK1 (Fig. 6b, left). When both PCAF and VRK1 were tagged, both proteins were detected in reciprocal immunoprecipitations (Fig. 6b, right). The direct PCAF–VRK1 interaction was further confirmed in a dose-dependent pull-down in vitro experiments using GST-PCAF and His-VRK1 that were bacterially expressed and purified proteins (Additional file 13: Fig. S13A). This direct interaction was confirmed by transfecting tagged PCAF-flag, which immunoprecipitated the endogenous VRK1 protein in a dose-dependent manner (Additional file 13: Fig. S13B).
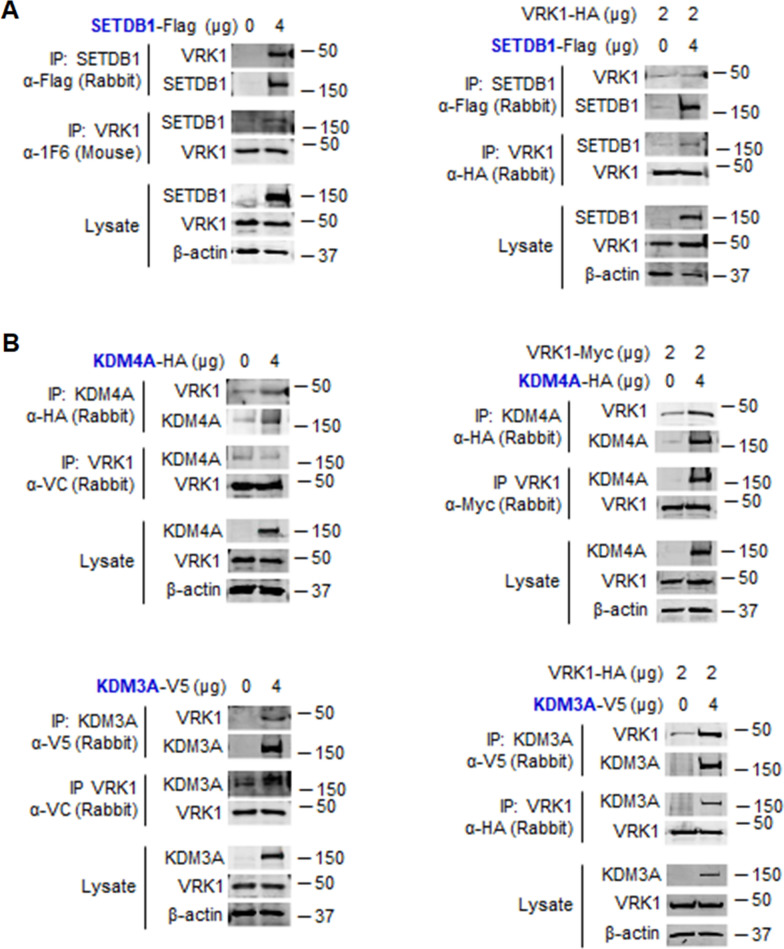
VRK1 stably interacts with histone methylases (KMT) and histone demethylases (KDM)
Next, we performed similar experiments to detect direct interaction of VRK1 with KMT and KDM proteins. First the interaction with the SETDB1 (KMT1E) enzyme was determined. SETDB1 interacted with endogenous VRK1 (Fig. 7a, left) or transfected VRK1-HA (Fig. 7b, right) in reciprocal immunoprecipitations.
Fig. 7.
Interaction of SETDB1 (KMT1E) and KDM with VRK1. A Left: reciprocal interaction of SETDB1 with endogenous VRK1. Right: reciprocal interaction of SETDB1-Flag (KMT1E) with transfected VRK1-HA. B Left: reciprocal interaction of KDM4A (JMJD2A)-HA (top) or KDM3A (JMJD1A)-V5 (bottom) with endogenous VRK1. Right: reciprocal interaction of JMJD2A-HA (top) or KDM3A-V5 (bottom) with transfected VRK1-HA. Transfections were performed in HEK293T cells
Also, similar experiments were performed with two different KDMs, JMJD2A (KDM4A) and KDM3A. These two KDMs interacted with endogenous (Fig. 7b, left) or transfected VRK1-HA (Fig. 7b, right) in reciprocal immunoprecipitations. VRK1 forms stable protein complexes with SETDB1 (KMT1E), JMJD2A (KDM4A) and JMJD1A (KDM3A).
Discussion
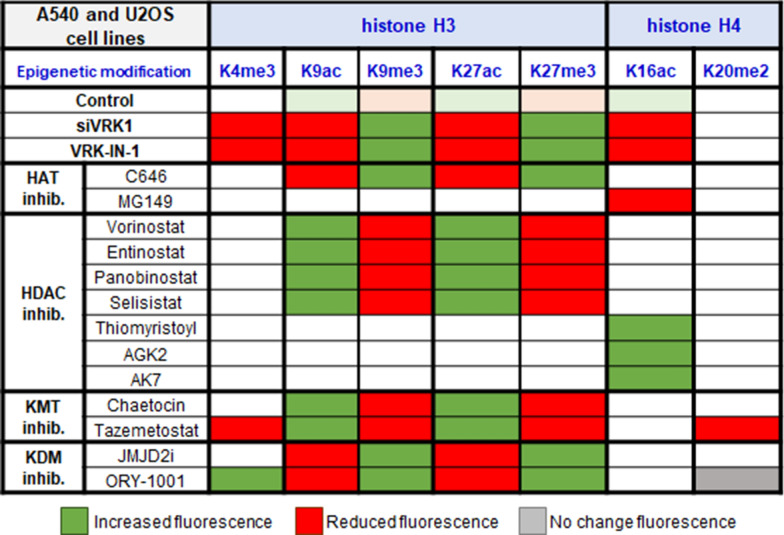
The different functional states of chromatin organization are associated with different combinations of epigenetic posttranslational modifications of histones, which are mediated by enzymes belonging to different protein families, each with multiple members. Among them are kinases, KATs, HDACs, KMTs and KDMs. Furthermore, as biological processes such as transcription, replication, recombination or DDR progression, there is a dynamic change in the local pattern of PTMs, which can vary depending on the stage of the biological process, and can determine its functional role. This requires the coordination of different enzymes among which are KATs, HDACs, KMTs, and KDMs, which introduce different PTMs that are involved in these dynamic processes, in which kinases can play a relevant coordinating role. In this context, the switch in histone epigenetic PTMs caused by VRK1 depletion, or the specific inhibition of its kinase activity (Fig. 8), indicates that there is a significant alteration in chromatin organization, which can be exploited for therapeutic applications. This blockade in the dynamic regulation of chromatin mediated, or coordinated, by VRK1 impairs processes associated with preventing activation of transcription factors such as c-Jun [35], CREB [37], ATF2 [36], p53 [33, 49], or proteins implicated in DDR such as H2AX [32], NBS1 [50], or 53BP1 in non-homologous-end joining (NHEJ) [51], which is recruited H4K20me2 [52, 53], a specific histone modifications that is also involved in nucleotide excision repair (NER) [54].
Fig. 8.
Alteration of the balance and pattern of histone epigenetic posttranslational modifications caused by drugs targeting different enzymes in A549 and U2OS cells
VRK1 depletion or inhibition caused a reduction of gene expression in different cell lines, which is consistent with the modification in the PTMs patterns of H3K9 and H3K27 associated with these treatments. The effect of VRK1 depletion on H3K9 has also been detected in human glioblastoma cell lines [55]. Cells blocked in a histone H3K9 and H3K27 methylated state are unlikely to be able to permit a correct DNA damage response, and thus sensitize cells to treatments with these drugs. This configuration will also impair gene transcription, which is an effect of VRK1 depletion or inhibition independent of cell type [21, 24, 35, 36, 56]. Furthermore, the transcriptional H3K4me3 epigenetic mark [57] is also impaired by either VRK1 inhibition or depletion. These data are consistent with the known role of VRK1 in the regulation of cell proliferation [21, 58, 59]. Inhibition of KDM impairs the acetylation of methylated lysine residues. In pancreatic cancer cells the expression of PD-L1 is very high [60], and in these tumors the loss of H3K4me3 reduces the expression of PD1-L1 [60]. In this context, targeting VRK1 might be a potentially useful synthetic lethality strategy in combination with immunotherapy approaches targeting the PD1/PD1-L1 axis, which need to be explored. Alteration in epigenetic patterns have also been associated with allergic diseases [61], in which the role of VRK1 has not been studied.
Different drugs targeting enzymes that introduce histone PTMs such as KATs, HDACs, KMTs or KDMs are currently in use as therapeutic drugs. Among them is iadademstat (ORY-1001) targeting KDM1A/LSD1 and used in AML [48, 62], Ewing sarcoma [63], and triple negative breast cancer [64], but their effectiveness can be improved if combinatorial strategies with other drugs are identified. Specific pharmacological targeting of VRK1 can mimic the combined effects of drugs targeting specific epigenetic enzymes. Therefore, the potential use of a kinase inhibitor combined with specific inhibitors of histone epigenetic enzymes can permit a reduction in their doses.
A potential synthetic lethality strategy can be based on the simultaneous targeting of KMT inhibition with tazemetostat that impairs H4K20me2 required for the recruitment of 53BP1 in response to DNA damage in sarcomas [65]. Its combination with a VRK1 inhibitor targeting different pathways, such as proliferation signals, as well as DNA responses such as those mediated by p53, or those associated with different and early steps in DDR such as the formation of γH2AX foci in the response to doxorubicin or ionizing radiation [51, 66] regulated by VRK1 or olaparib, a PARP inhibitor [67]. Furthermore, inhibition of histone H4 acetylation impairs DDR, which by itself may not be enough [68], but can cooperate with VRK1 inhibition in the elimination of tumor cells [30, 31].
The consequence of the drug combination can have two main effects, a reduction of their individual dose and thus of toxic side effects, as well as a reduction of the selective pressure to develop resistance to a specific drug. Drug combinations also permit the targeting of difference pathways. In the case of the VRK-IN-1 inhibitor, it not only targets VRK1, but also the cytosolic and membrane-bound VRK2 [16], which can impair mitogenic signaling [69, 70], and thus can simultaneously block two different tumorigenic pathways. Targeting VRK1 has been shown to be a suitable target in glioblastomas in which the VRK2 gene is silenced [71, 72]. A strategy that is likely to be applicable to other tumors, and even improved if combined with other drugs targeting other epigenetic enzymes from different families.
An example of the relation between VRK1 and epigenetic enzymes is represented by its interaction with Tip60 and HDAC1, two enzymes that regulate the acetylation of histone H4.
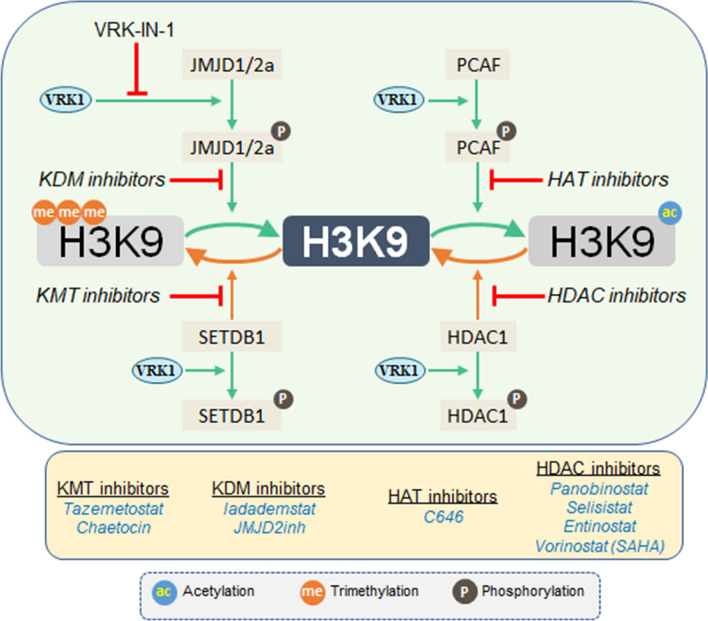
Previously, it has been shown that VRK1 specifically phosphorylates Tip60/KAT5 facilitating its translocation to chromatin and activating the acetylation of H4K16 in the response to DNA damage [30, 31]. The reversal of this acetylation requires histone deacetylases (HDACs). In this context, it has been shown that the deacetylase SIRT2, by interacting with VRK1, inhibits its kinase activity [73], and facilitates the de-acetylation mediated by SIRT2, and thus preventing the re-acetylation of the target protein. HDAC inhibition also facilitates the impairment of DDR by preventing the dynamic remodeling of chromatin associated with the progression of DNA damage responses (DDR) [74]. In this context, the crosstalk in the regulation of H4K16 acetylation by different enzymes supports its potential for pharmacological intervention. The modification in the pattern of H3 and H4 histone acetylation, resulting from by the loss of VRK1, is similar to that caused by KAT inhibitors. Both VRK1 or KAT inhibition result in a defective DDR [31, 75, 76]. Alternatively, VRK1 depletion facilitates histone methylation, a similar effect to that of KDM inhibitors. The maintenance of a histone methylated state also impairs progression of DDR [77, 78]. Thus, the patterns of different combinations of histone epigenetic acetylations and methylations determine the functional situation at specific chromatin locations on genes and chromosomes, which need to be identified in order to associate specific PTM patterns and functions. The use of inhibitors targeting these epigenetics marks blocks chromatin in a specific configuration that is unable to remodel in order to perform specific functions. The complex regulation of the coordination of the epigenetic PTMs on an individual histone is exemplified for H3K9 (Fig. 9). There are four different enzyme activities requiring their coordination, among which VRK1 is a candidate, but other kinases can also be implicated, but need to be identified. Therefore, the formation of stable complexes between VRK1 and several different epigenetic types of enzymes suggests that it is likely that they participate in the regulation of different PTMs, which need to be individually characterized in a manner similar to that of the VRK1-KAT5/Tip60 and HDAC1 [30, 31, 38], in order to determine their role and therapeutic potential.
Fig. 9.
Diagram illustrating the complex regulation of the epigenetic modifications of histone H3K9, and its manipulation by the use of different types of inhibitors
We concluded that the pattern of epigenetic histone posttranslational modifications is regulated by the chromatin kinase VRK1. Loss of VRK1 activity by depletion or pharmacological inhibition causes a change in the pattern of histone PTMs associated with chromatin remodeling. A similar effect can be achieved by the inhibition of KAT and KDM inhibitors. Therefore, the combination of drugs targeting proteins belonging to different types of epigenetic enzymes can be useful for development of novel therapeutic approaches based on synthetic lethality strategies for cancer treatments.
Methods
Reagents and inhibitors
The VRK1 inhibitor (VRK-IN-1) [16] was obtained from MedChemExpress (Monmouth Junction, NJ, USA; Cat. No. HY-126542) and used as recommended by the manufacturer.
The inhibitors used in this work are listed in Table 1.
Table 1.
Inhibitors of histone PTMs
| Inhibitor | Target | Concentration/time (24 h) | Supplier | Catalog reference |
|---|---|---|---|---|
| VRK-IN-1 | VRK1 | 600 nM | MedChem | HY-126542 |
| C646 | P300/CBP/PCAF | 5 μM | Selleck | S7152 |
| Vorinostat (SAHA) | HDAC1 | 5 μM | Axon | 1785 |
| Entinostat | HDAC1/3 | 5 μM | Selleck | S1053 |
| Panobinostat | HDAC1 | 50 nM | Selleck | S1030 |
| Selisistat | HDAC1 | 50 nM | Selleck | S1541 |
| Chaetocin | KMT1E (SETDB1) | 100 nM | Sigma | C9492 |
| Tazemetostat | KMT6A (EZH2) | 50 nM | Selleck | S7128 |
| Iadademstat (ORY1001) | KDM1A (LSD1) | 50 nM | Selleck | S7795 |
| JMJD2i (Jumonji) | KDM4A (JMJD2A/2E) | 100 µM | Millipore | 420,201 |
Plasmids
The plasmids detailed in Table 2 were used for mammalian cell transfection to express human proteins, or expression in E. coli for protein purification. GeneJET Plasmid Maxiprep kit (Thermo Fisher Scientific; Waltham, MA, USA) was used for plasmid purification.
Table 2.
Plasmids
| Protein or tag | Plasmid | Tag | Expression | Supplier, reference |
|---|---|---|---|---|
| EZH2 | pCMV6 | Myc-DDK | Mammalian | Origene, #RC202054 |
| Flag-Ø | pCEFL | Flag | Mammalian | Gift from JS Gutkind (NIH) |
| GST-Ø | pGEX-2TK | GST | Bacterial | GE Healthcare GE28-9546-53 |
| HA-Ø | pCEFL | HA | Mammalian | Gift from JS Gutkind (NIH) |
| HDAC1 | pcDNA3.1+ | Flag | Mammalian | Addgene, #13820 |
| KDM1A (LSD1) | pCMV6 | Myc-DDK | Mammalian | Origene, #RC208480 |
| KDM3A (JMJD1A) | pLenti | V5-Myc | Mammalian | Addgene, #82331 |
| KDM4A (JMJD2A) | pCMV | HA | Mammalian | Addgene, #24180 |
| PCAF | pCl | Flag | Mammalian | Nakatani [81] |
| SETDB1 | pIRES2-EGFP | Flag | Mammalian | Rodriguez Paredes [82] |
| VRK1 | pCEFL | HA | Mammalian | Valbuena et al. [83] |
| VRK1 | pcDNA3.1 | Myc | Mammalian | Valbuena et al. [83] |
Antibodies
The antibodies used in this work and their applications and conditions of primary antibodies are listed in Table 3. Secondary antibodies are in Table 4. Antibodies were diluted in TBS-T buffer (25 mM Tris HCl pH 8.0, 50 mM NaCl and 2.5 mM KCl, 0.1% Tween-20) or PBS-1% BSA for immunoblots or immunofluorescence assays, respectively.
Table 3.
Primary antibodies
| Antibody | Type | Dilution (WB/IF) | Clon, reference | Supplier |
|---|---|---|---|---|
| H3K4me3 | Rabbit polyclonal | 1:800 | 9727 | Cell signaling |
| 1:1000 | ||||
| H3K9ac | Rabbit polyclonal | 1:2000 | 07-352 | Millipore |
| 1:1000 | ||||
| H3K9me3 | Rabbit polyclonal | 1:800 | 07-442 | Millipore |
| 1:800 | ||||
| H3K27ac | Rabbit polyclonal | 1:1000 | 07-360 | Millipore |
| 1:1000 | ||||
| H3K27me3 | Rabbit polyclonal | 1:800 | 07-449 | Millipore |
| 1:800 | ||||
| Histone H3 | Rabbit polyclonal | 1:1000 | 9715 | Cell signaling |
| – | ||||
| VRK1 | Mouse monoclonal | 1:1000 | 1F6 | [83] |
| – | ||||
| VRK1 | Rabbit polyclonal | 1:1000 | VC | [83] |
| – | ||||
| HA.11 tag | Mouse monoclonal | 1:1000 | 901514, 16B12 | BioLegend |
| – | ||||
| HA tag | Rabbit polyclonal | 1:1000 | H6908 | Sigma-Aldrich |
| – | ||||
| Myc tag | Mouse monoclonal | 1:1000 | 4A | Millipore |
| – | 05-724 | |||
| Myc tag | Rabbit polyclonal | 1:1000 | 06-549 | Millipore |
| – | ||||
| V5 tag | Mouse monoclonal | 1:1000 | V5-10 | Sigma-Aldrich |
| – | V8012 | |||
| V5 tag | Rabbit polyclonal | 1:1000 | V8137 | Sigma-Aldrich |
| – | ||||
| β-actin | Mouse monoclonal | 1:2000 | AC15 | Sigma-Aldrich |
| – | A5441 | |||
| Flag tag | Mouse monoclonal | 1:1000 | M5 | Sigma-Aldrich |
| – | F4042 | |||
| Flag tag | Rabbit polyclonal | 1:1000 | F7425/ab1162 | Sigma-Aldrich/Abcam |
| – |
Table 4.
Secondary antibodies
| Antibody | Fluorophore | Use dilution | Reference | Supplier |
|---|---|---|---|---|
| CyTM2-goat anti-rabbit | Cy2 (green) | IF | 111-225-144 | Jackson Immunoresearch |
| 1:1000 | ||||
| CyTM3-goat anti-mouse | Cy3 (red) | IF | 15-165-146 | Jackson Immunoresearch |
| 1:1000 | ||||
| CyTM5-goat anti-mouse | Cy5 (far red) | IF | 115-175-146 | Jackson Immunoresearch |
| 1:1000 | ||||
| Goat anti-mouse IgG | DyLight 680 (red) | WB 1:10,000 | 35,518 | Thermo Fisher Scientific |
| Goat anti-rabbit IgG | DyLight 800 (green) | WB 1:10,000 | 35,571 | Thermo Fisher Scientific |
Cell lines, culture and transfections
The following cell lines were used and purchased from the American Type Culture Collection (ATCC): A549 (CCL-185), U2OS (HTB-96) and HEK 293T (CRL-3216). Cells were grown with DMEM (Gibco-Life Technologies-Invitrogen; Waltham, MA, USA) containing 10% fetal bovine serum (FBS), 2 mM glutamine (l-glutamine) and 1% penicillin–streptomycin (Pen/Strep), all of them from Gibco-Life Technologies (Waltham, MA, USA). The cell lines were placed in a humidified atmosphere containing 5% CO2 at 37 °C. Twenty-four hours later, cells were transfected with the corresponding expression plasmid. Plasmid transfections were performed mixing 4–8 µg DNA with two volumes of polyethylenimine (PEI; Polysciences; Warrington; PA, USA) reagent, incubated for 30 min and added to the cells, which were assayed 48 h after transfection. Serum starvation (DMEM supplemented with 0.5% FBS, 2 mM L-glutamine, 1% Pen/Strep) was performed for 48 h when indicated.
VRK1 depletion by siRNA
Two specific siRNAs were used for human VRK1 depletion: siVRK1-02 (5′-CAAGGAACCTGGTGTTGAA-3′) and siVRK1-3 (5′-GGAAUGGAAAGUAGGAUUA-3′). ON-TARGET plus siControl non-targeting siRNA (siControl) was used as a negative control. VRK1 depletion was performed as previously reported [50, 79]. All RNAi were from GE-Healthcare-Dharmacon. Both lipotransfectin (Solmeglas, Madrid, Spain) and 200 nM siRNA was diluted in Opti-MEM (GIBCO-Life Technologies) according to manufacturer guidelines. After 30 min of incubation, the mix was added to the cells. Cells were maintained with antibiotic-free medium.
Cell lysates and histone extraction
Protein extraction was carried out at 4 °C. Cells were resuspended in lysis buffer (50 mM Tris HCl pH 8.0, 150 mM NaCl, 1% triton X-100 and 1 mM EDTA) supplemented by phosphatases inhibitors (1 mM sodium fluoride and 1 mM sodium orthovanadate) and proteases inhibitors (1 mM PMSF, 10 mg/mL aprotinin, and 10 mg/mL leupeptin). The cell lysate suspension was incubated for 15 min and then centrifuged for 15 min (16,000 × g). Histones were isolated by acidic extraction, as previously described [80]. BCA assay (Thermo Fisher Scientific; Waltham, MA, USA) was used to determine the total protein concentration. 40 µg of protein and 5–10 µg of acidic extracts of histones were used for immunoblots.
Pull-down and immunoprecipitation assays
Pull-down assay was carried out with purified VRK1-His and GST-PCAF (aa 352–832) in the amounts indicated in the experiment. For this aim, purified proteins were incubated in pull-down buffer (20 mM Tris–HCl pH 7.5, 5 mM MgCl2, 0.5 mM DTT and 150 mM KCl) at 37 °C and gentle agitation for 45 min. After that, 40 µL of Glutathione Sepharose 4B beads (GE Healthcare; Chicago, IL, USA) was added, and the mix was incubated overnight at 4 °C in rotation. The pull-down was performed by centrifugation (500 × g, 2 min, 4 °C) and the beads were washed in the same buffer for three times. Then, the protein complexes were resuspended in the sample loading buffer, loaded in polyacrylamide gel to separate proteins via SDS-PAGE and visualized by Coomassie Blue staining (3 g/L Coomassie Brilliant Blue R250, 45% methanol, and 10% glacial acetic acid).
The co-immunoprecipitations were performed using 0.5–1 mg of the whole-cells extracts. The protocol consists in incubating the proteins with the specific antibody for 6–8 h at 4 and then adding 40 µL of Protein G–Agarose Resin 4 Rapid Run (4RRPG, Agarose Bead Technologies; Madrid, Spain) overnight at 4 °C in rotation. Finally, the immunoprecipitate was collected by centrifugation (500 × g, 2 min, 4 °C) [30, 31, 38].
Western blot analysis
Western blot technique was performed according to standardized protocols. Briefly, protein samples were boiled and loaded into a polyacrylamide gel to separate proteins via SDS-PAGE. Then, proteins were transferred to PVDF Immobilon-FL (0.22 or 0.45 µm pore size; Millipore; Burlington, MA, USA) membranes and blocked for 1 h at room temperature with 5% nonfat milk or 5% of BSA in TBS-T buffer. Membranes were incubated with the primary antibody overnight at 4 °C (Table 3). The following day, they were incubated in darkness with their corresponding secondary antibodies (Table 4) diluted 1:10,000 in TBS-T for 1 h. Lastly, fluorescence signals were detected using a LI-COR Odyssey Infrared Imaging System (LI-COR Biosciences; Lincoln, NE, USA). To analyze differences in proteins levels, densitometric analysis was done using ImageJ 1.52p software (https://imagej.nih.gov). All western blots were performed in triplicate and correspond to the accompanying immunofluorescence image.
Immunofluorescence and confocal microscopy
In this study, cells were cultured with glass coverslips (Thermo Fisher Scientific; Waltham, MA, USA). Cells were fixed with 3% paraformaldehyde (PFA) in PBS for 15 min, and treated with 200 mM glycine solution to remove the PFA. Subsequently, cells were incubated with 0.2% triton X-100 permeabilization for 15 min and then blocked with PBS–1% BSA with 0.1% sodium azide for 1 h at room temperature, or overnight at 4 °C, to block unspecific binding of the antibodies. Cells were consecutively incubated with two primary antibodies for concurrently protein detection. The first primary antibody was incubated between 5 and 6 h at room temperature and next the second primary antibody overnight at 4 °C. Cells were washed in PBS three times for 5 min. Next, cells were incubated with the secondary antibodies at 1:1000 dilution for 1 h at room temperature in the dark. Cells were washed in PBS three times for 5 min in the dark. Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole) at 1:1000 dilution for 5 min. Finally, cells were washed three times for 5 min in PBS and slides were mounted with Mowiol. Fluorescent images were acquired with a LEICA SP5 DMI-6000B confocal microscope (Leica; Wetzlar, Germany), using the following lasers: Argon (488 nm), DPSS (561 nm) and UV Diode (405 nm). Images were analyzed using the ImageJ software.
Statistical analysis
GraphPad Prism 8.0 was used to analyze and computed graphs. Experiments were set up with 3 replicates per experiment. Data are presented as dot plots with the median, first and third quartiles and whiskers. Statistical analysis was performed by Mann–Whitney U-test test for two-group comparisons after confirming samples did not adjust to a normal distribution (non-parametric distributions) according to two-tailed Kolmogorov test. In all cases, the level of significance was 0.05 (*, p < 0.05; **, p < 0.01; and ***, p < 0.001).
Supplementary Information
Additional file 1. Fig. S1: Effect of VRK1 depletion on the epigenetic modifications of H3K9 in the presence (top) or absence (bottom) of serum in A549 lung adenocarcinoma cells.
Additional file 2. Fig. S2: Effect of VRK1 depletion on epigenetic modifications of H3K9 in the presence or absence of serum in U2OS osteosarcoma cells.
Additional file 3. Fig. S3: Effect of VRK1 depletion on the epigenetic modifications of H3K27 in the presence or absence of serum in A549 lung adenocarcinoma cells.
Additional file 4. Fig. S4: Effect of VRK1 depletion of epigenetic modifications of H3K27 in the presence or absence of serum in U2OS osteosarcoma cells.
Additional file 5. Fig. S5: Effect of the VRK-IN-1 inhibitor on the levels of H3K27 acetylation and methylation in A549 cells.
Additional file 6. Fig. S6: Effect of the VRK1 depletion and the VRK-IN-1 inhibitor on the levels of H4K16 acetylation and H3K4me3 associated with transcription in U2OS cells.
Additional file 7. Fig. S7: Effect of the VRK-IN-1 inhibitor on the levels of H3K9 acetylation and methylation in U2OS cells.
Additional file 8. Fig. S8: Effect of the VRK-IN-1 inhibitor on the levels of H3K27 acetylation and methylation in U2OS cells.
Additional file 9. Fig. S9: Effect of HDAC inhibitors on H3K9 acetylation and methylation in U2OS cells.
Additional file 10. Fig. S10: Effect of KMT inhibitors on H3K9 acetylation and methylation in U2OS cells.
Additional file 11. Fig. S11: Effect of the KAT inhibitor C646 on H3K9 acetylation and methylation in A549 and U2OS cells.
Additional file 12. Fig. S12: Effect of KDM inhibitors on H3K9 acetylation and methylation in A549 and U2OS cells.
Additional file 13. Fig. S13: Direct interaction between VRK1 and the PCAF acetyl transferase.
Author contributions
Conceptualization: PAL, EM-S, IC-M; formal analysis and funding acquisition: PAL; supervision: PAL; investigation: EM-S, PM-C, IC-M, AC-D, EN-C; writing: PAL. All authors read and approved the final manuscript.
Funding
This work was supported by grants from Agencia Estatal de Investigación (10.13039/501100011033) AEI/Ministerio de Ciencia e Innovación-“FEDER Una manera de hacer Europa” (PID2019-105610RB-I00, RED2018-102801-T), and Consejería de Educación de la Junta de Castilla y León-FEDER (CSI264P20, CLC-2017-01) to P. A. L. E. M.-S was funded by predoctoral contract (CSI004-18) from Consejería de Educación-Junta de Castilla y León-Fondo Social Europeo (FSE). Ministerio de Educación-FSE predoctoral FPU contracts funded P. M.-G. (FPU16/01883) and A. C.-D. (FPU20/02978). I.C.-M. was supported by FPI-MINECO-FSE contract (BES-2014-067721). E. N-C. was funded by Agencia Estatal de Investigación-MINECO predoctoral contract (BES-2017-080543).
Availability of data and materials
All data and reagents are available upon request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Woodcock CL, Ghosh RP. Chromatin higher-order structure and dynamics. Cold Spring Harb Perspect Biol. 2010;2(5):a000596. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yadav T, Quivy JP, Almouzni G. Chromatin plasticity: a versatile landscape that underlies cell fate and identity. Science. 2018;361(6409):1332–1336. doi: 10.1126/science.aat8950. [DOI] [PubMed] [Google Scholar]
- 3.Nicolas E, Roumillac C, Trouche D. Balance between acetylation and methylation of histone H3 lysine 9 on the E2F-responsive dihydrofolate reductase promoter. Mol Cell Biol. 2003;23(5):1614–1622. doi: 10.1128/mcb.23.5.1614-1622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16(8):919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raisner R, Kharbanda S, Jin L, Jeng E, Chan E, Merchant M, Haverty PM, Bainer R, Cheung T, Arnott D, et al. Enhancer activity requires CBP/P300 bromodomain-dependent histone H3K27 acetylation. Cell Rep. 2018;24(7):1722–1729. doi: 10.1016/j.celrep.2018.07.041. [DOI] [PubMed] [Google Scholar]
- 6.Audia JE, Campbell RM. Histone modifications and cancer. Cold Spring Harb Perspect Biol. 2016;8(4):a019521. doi: 10.1101/cshperspect.a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lavarone E, Barbieri CM, Pasini D. Dissecting the role of H3K27 acetylation and methylation in PRC2 mediated control of cellular identity. Nat Commun. 2019;10(1):1679. doi: 10.1038/s41467-019-09624-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol. 2010;339(2):240–249. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker PB, Workman JL. Nucleosome remodeling and epigenetics. Cold Spring Harb Perspect Biol. 2013;5(9):a017905. doi: 10.1101/cshperspect.a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehrenhofer-Murray AE. Chromatin dynamics at DNA replication, transcription and repair. Eur J Biochem. 2004;271(12):2335–2349. doi: 10.1111/j.1432-1033.2004.04162.x. [DOI] [PubMed] [Google Scholar]
- 11.Aihara H, Nakagawa T, Mizusaki H, Yoneda M, Kato M, Doiguchi M, Imamura Y, Higashi M, Ikura T, Hayashi T, et al. Histone H2A T120 phosphorylation promotes oncogenic transformation via upregulation of cyclin D1. Mol Cell. 2016;64(1):176–188. doi: 10.1016/j.molcel.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 12.Eswaran J, Patnaik D, Filippakopoulos P, Wang F, Stein RL, Murray JW, Higgins JM, Knapp S. Structure and functional characterization of the atypical human kinase haspin. Proc Natl Acad Sci USA. 2009;106(48):20198–20203. doi: 10.1073/pnas.0901989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fedorov O, Marsden B, Pogacic V, Rellos P, Muller S, Bullock AN, Schwaller J, Sundstrom M, Knapp S. A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc Natl Acad Sci USA. 2007;104(51):20523–20528. doi: 10.1073/pnas.0708800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vazquez-Cedeira M, Barcia-Sanjurjo I, Sanz-Garcia M, Barcia R, Lazo PA. Differential inhibitor sensitivity between human kinases VRK1 and VRK2. PLoS ONE. 2011;6(8):e23235. doi: 10.1371/journal.pone.0023235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Counago RM, Allerston CK, Savitsky P, Azevedo H, Godoi PH, Wells CI, Mascarello A, de Souza Gama FH, Massirer KB, Zuercher WJ, et al. Structural characterization of human vaccinia-related kinases (VRK) bound to small-molecule inhibitors identifies different P-loop conformations. Sci Rep. 2017;7(1):7501. doi: 10.1038/s41598-017-07755-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serafim RAM, de Souza Gama FH, Dutra LA, Dos Reis CV, Vasconcelos SNS, da Silva SA, Takarada JE, Di Pillo F, Azevedo H, Mascarello A, et al. Development of pyridine-based inhibitors for the human vaccinia-related kinases 1 and 2. ACS Med Chem Lett. 2019;10(9):1266–1271. doi: 10.1021/acsmedchemlett.9b00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valbuena A, López-Sánchez I, Lazo PA. Human VRK1 is an early response gene and its loss causes a block in cell cycle progression. PLoS ONE. 2008;3(2):e1642. doi: 10.1371/journal.pone.0001642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valbuena A, Sanz-Garcia M, Lopez-Sanchez I, Vega FM, Lazo PA. Roles of VRK1 as a new player in the control of biological processes required for cell division. Cell Signal. 2011;23(8):1267–1272. doi: 10.1016/j.cellsig.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Moura DS, Campillo-Marcos I, Vazquez-Cedeira M, Lazo PA. VRK1 and AURKB form a complex that cross inhibit their kinase activity and the phosphorylation of histone H3 in the progression of mitosis. Cell Mol Life Sci. 2018;76:2591–2611. doi: 10.1007/s00018-018-2746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campillo-Marcos I, García-González R, Navarro-Carrasco E, Lazo PA. The human VRK1 chromatin kinase in cancer biology. Cancer Lett. 2021;503:117–128. doi: 10.1016/j.canlet.2020.12.032. [DOI] [PubMed] [Google Scholar]
- 21.Santos CR, Rodriguez-Pinilla M, Vega FM, Rodriguez-Peralto JL, Blanco S, Sevilla A, Valbuena A, Hernandez T, van Wijnen AJ, Li F, et al. VRK1 signaling pathway in the context of the proliferation phenotype in head and neck squamous cell carcinoma. Mol Cancer Res. 2006;4(3):177–185. doi: 10.1158/1541-7786.MCR-05-0212. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Zhai R, Shen H, Song G, Wan F, Li Q. VRK1 promotes proliferation, migration, and invasion of gastric carcinoma cells by activating β-catenin. Neoplasma. 2021 doi: 10.4149/neo_2021_210304N278. [DOI] [PubMed] [Google Scholar]
- 23.Ryu HG, Jung Y, Lee N, Seo JY, Kim SW, Lee KH, Kim DY, Kim KT. HNRNP A1 promotes lung cancer cell proliferation by modulating VRK1 translation. Int J Mol Sci. 2021. 10.3390/ijms22115506. [DOI] [PMC free article] [PubMed]
- 24.Colmenero-Repiso A, Gómez-Muñoz MA, Rodríguez-Prieto I, Amador-Álvarez A, Henrich KO, Pascual-Vaca D, Okonechnikov K, Rivas E, Westermann F, Pardal R, et al. Identification of VRK1 as a new neuroblastoma tumor progression marker regulating cell proliferation. Cancers (Basel). 2020. 10.3390/cancers12113465. [DOI] [PMC free article] [PubMed]
- 25.Liu J, Wang Y, He S, Xu X, Huang Y, Tang J, Wu Y, Miao X, He Y, Wang Q, et al. Expression of vaccinia-related kinase 1 (VRK1) accelerates cell proliferation but overcomes cell adhesion mediated drug resistance (CAM-DR) in multiple myeloma. Hematology. 2016;21(10):603–612. doi: 10.1080/10245332.2016.1147678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campillo-Marcos I, Lazo PA. Implication of the VRK1 chromatin kinase in the signaling responses to DNA damage: a therapeutic target? Cell Mol Life Sci. 2018;75(13):2375–2388. doi: 10.1007/s00018-018-2811-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Budziszewski GR, Zhao Y, Spangler CJ, Kedziora KM, Williams MR, Azzam DN, Skrajna A, Koyama Y, Cesmat AP, Simmons HC, et al. Multivalent DNA and nucleosome acidic patch interactions specify VRK1 mitotic localization and activity. Nucl Acids Res. 2022;50(8):4355–4371. doi: 10.1093/nar/gkac198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang TH, Park DY, Choi YH, Kim KJ, Yoon HS, Kim KT. Mitotic histone H3 phosphorylation by vaccinia-related kinase 1 in mammalian cells. Mol Cell Biol. 2007;27(24):8533–8546. doi: 10.1128/MCB.00018-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311(5762):844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Gonzalez R, Morejon-Garcia P, Campillo-Marcos I, Salzano M, Lazo PA. VRK1 phosphorylates Tip60/KAT5 and is required for H4K16 acetylation in response to DNA damage. Cancers (Basel) 2020;12(10):2986. doi: 10.3390/cancers12102986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.García-González R, Monte-Serrano E, Morejón-García P, Navarro-Carrasco E, Lazo PA. The VRK1 chromatin kinase regulates the acetyltransferase activity of Tip60/KAT5 by sequential phosphorylations in response to DNA damage. Biochim Biophys Acta Gene Regul Mech. 2022;1865(8):194887. doi: 10.1016/j.bbagrm.2022.194887. [DOI] [PubMed] [Google Scholar]
- 32.Salzano M, Sanz-Garcia M, Monsalve DM, Moura DS, Lazo PA. VRK1 chromatin kinase phosphorylates H2AX and is required for foci formation induced by DNA damage. Epigenetics. 2015;10(5):373–383. doi: 10.1080/15592294.2015.1028708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez-Borges S, Lazo PA. The human vaccinia-related kinase 1 (VRK1) phosphorylates threonine-18 within the mdm-2 binding site of the p53 tumour suppressor protein. Oncogene. 2000;19(32):3656–3664. doi: 10.1038/sj.onc.1203709. [DOI] [PubMed] [Google Scholar]
- 34.Vega FM, Sevilla A, Lazo PA. p53 Stabilization and accumulation induced by human vaccinia-related kinase 1. Mol Cell Biol. 2004;24(23):10366–10380. doi: 10.1128/MCB.24.23.10366-10380.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sevilla A, Santos CR, Barcia R, Vega FM, Lazo PA. c-Jun phosphorylation by the human vaccinia-related kinase 1 (VRK1) and its cooperation with the N-terminal kinase of c-Jun (JNK) Oncogene. 2004;23(55):8950–8958. doi: 10.1038/sj.onc.1208015. [DOI] [PubMed] [Google Scholar]
- 36.Sevilla A, Santos CR, Vega FM, Lazo PA. Human vaccinia-related kinase 1 (VRK1) activates the ATF2 transcriptional activity by novel phosphorylation on Thr-73 and Ser-62 and cooperates with JNK. J Biol Chem. 2004;279(26):27458–27465. doi: 10.1074/jbc.M401009200. [DOI] [PubMed] [Google Scholar]
- 37.Kang TH, Park DY, Kim W, Kim KT. VRK1 phosphorylates CREB and mediates CCND1 expression. J Cell Sci. 2008;121(Pt 18):3035–3041. doi: 10.1242/jcs.026757. [DOI] [PubMed] [Google Scholar]
- 38.Monte-Serrano E, Lazo PA. VRK1 kinase activity modulating histone H4K16 acetylation inhibited by SIRT2 and VRK-IN-1. Int J Mol Sci. 2023 doi: 10.3390/ijms24054912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huertas D, Sendra R, Munoz P. Chromatin dynamics coupled to DNA repair. Epigenetics. 2009;4(1):31–42. doi: 10.4161/epi.4.1.7733. [DOI] [PubMed] [Google Scholar]
- 40.Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, Price BD. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci USA. 2014;111(25):9169–9174. doi: 10.1073/pnas.1403565111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pelham-Webb B, Polyzos A, Wojenski L, Kloetgen A, Li J, Di Giammartino DC, Sakellaropoulos T, Tsirigos A, Core L, Apostolou E. H3K27ac bookmarking promotes rapid post-mitotic activation of the pluripotent stem cell program without impacting 3D chromatin reorganization. Mol Cell. 2021;81(8):1732–1748.e1738. doi: 10.1016/j.molcel.2021.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ott CJ, Federation AJ, Schwartz LS, Kasar S, Klitgaard JL, Lenci R, Li Q, Lawlor M, Fernandes SM, Souza A, et al. Enhancer architecture and essential core regulatory circuitry of chronic lymphocytic leukemia. Cancer Cell. 2018;34(6):982–995.e987. doi: 10.1016/j.ccell.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conway E, Healy E, Bracken AP. PRC2 mediated H3K27 methylations in cellular identity and cancer. Curr Opin Cell Biol. 2015;37:42–48. doi: 10.1016/j.ceb.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Al-Ghabkari A, Narendran A. Targeting EZH2-mediated methylation of histone 3 inhibits proliferation of pediatric acute monocytic leukemia cells in vitro. Cancer Biol Ther. 2021;22(4):333–344. doi: 10.1080/15384047.2021.1902913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kusch T, Mei A, Nguyen C. Histone H3 lysine 4 trimethylation regulates cotranscriptional H2A variant exchange by Tip60 complexes to maximize gene expression. Proc Natl Acad Sci U S A. 2014;111(13):4850–4855. doi: 10.1073/pnas.1320337111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130(1):77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santer FR, Hoschele PP, Oh SJ, Erb HH, Bouchal J, Cavarretta IT, Parson W, Meyers DJ, Cole PA, Culig Z. Inhibition of the acetyltransferases p300 and CBP reveals a targetable function for p300 in the survival and invasion pathways of prostate cancer cell lines. Mol Cancer Ther. 2011;10(9):1644–1655. doi: 10.1158/1535-7163.MCT-11-0182. [DOI] [PubMed] [Google Scholar]
- 48.Maes T, Mascaró C, Tirapu I, Estiarte A, Ciceri F, Lunardi S, Guibourt N, Perdones A, Lufino MMP, Somervaille TCP, et al. ORY-1001, a potent and selective covalent KDM1A inhibitor, for the treatment of acute leukemia. Cancer Cell. 2018;33(3):495–511.e412. doi: 10.1016/j.ccell.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 49.Valbuena A, Suarez-Gauthier A, Lopez-Rios F, Lopez-Encuentra A, Blanco S, Fernandez PL, Sanchez-Cespedes M, Lazo PA. Alteration of the VRK1-p53 autoregulatory loop in human lung carcinomas. Lung Cancer. 2007;58(3):303–309. doi: 10.1016/j.lungcan.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 50.Monsalve DM, Campillo-Marcos I, Salzano M, Sanz-Garcia M, Cantarero L, Lazo PA. VRK1 phosphorylates and protects NBS1 from ubiquitination and proteasomal degradation in response to DNA damage. Biochim Biophys Acta Molecular Cell Res. 2016;1863(4):760–769. doi: 10.1016/j.bbamcr.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 51.Sanz-Garcia M, Monsalve DM, Sevilla A, Lazo PA. Vaccinia-related Kinase 1 (VRK1) is an upstream nucleosomal kinase required for the assembly of 53BP1 foci in response to ionizing radiation-induced DNA damage. J Biol Chem. 2012;287(28):23757–23768. doi: 10.1074/jbc.M112.353102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jorgensen S, Schotta G, Sorensen CS. Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucl Acids Res. 2013;41(5):2797–2806. doi: 10.1093/nar/gkt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lou J, Priest DG, Solano A, Kerjouan A, Hinde E. Spatiotemporal dynamics of 53BP1 dimer recruitment to a DNA double strand break. Nat Commun. 2020;11(1):5776. doi: 10.1038/s41467-020-19504-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chitale S, Richly H. H4K20me2: orchestrating the recruitment of DNA repair factors in nucleotide excision repair. Nucleus. 2018;9(1):212–215. doi: 10.1080/19491034.2018.1444327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Navarro-Carrasco E, Lazo PA. VRK1 depletion facilitates the synthetic lethality of temozolomide and olaparib in glioblastoma cells. Front Cell Dev Biol. 2021;9:683038. doi: 10.3389/fcell.2021.683038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu ZC, Cao K, Xiao ZH, Qiao L, Wang XQ, Shang B, Jia Y, Wang Z. VRK1 promotes cisplatin resistance by up-regulating c-MYC via c-Jun activation and serves as a therapeutic target in esophageal squamous cell carcinoma. Oncotarget. 2017;8(39):65642–65658. doi: 10.18632/oncotarget.20020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen J, Horton J, Sagum C, Zhou J, Cheng X, Bedford MT. Histone H3 N-terminal mimicry drives a novel network of methyl-effector interactions. Biochem J. 2021;478(10):1943–1958. doi: 10.1042/bcj20210203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mon AM, MacKinnon AC, Jr, Traktman P. Overexpression of the VRK1 kinase, which is associated with breast cancer, induces a mesenchymal to epithelial transition in mammary epithelial cells. PLoS ONE. 2018;13(9):e0203397. doi: 10.1371/journal.pone.0203397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu J, Li T, Ji H, Chen Z, Zhai B. VRK1 predicts poor prognosis and promotes bladder cancer growth and metastasis in vitro and in vivo. Front Pharmacol. 2022;13:874235. doi: 10.3389/fphar.2022.874235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu C, Paschall AV, Shi H, Savage N, Waller JL, Sabbatini ME, Oberlies NH, Pearce C, Liu K. The MLL1-H3K4me3 axis-mediated PD-L1 expression and pancreatic cancer immune evasion. J Natl Cancer Inst. 2017 doi: 10.1093/jnci/djw283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alaskhar Alhamwe B, Khalaila R, Wolf J, von Bülow V, Harb H, Alhamdan F, Hii CS, Prescott SL, Ferrante A, Renz H, et al. Histone modifications and their role in epigenetics of atopy and allergic diseases. Allergy Asthma Clin Immunol. 2018;14:39. doi: 10.1186/s13223-018-0259-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bose P, Konopleva MY. ORY-1001: overcoming the differentiation block in AML. Cancer Cell. 2018;33(3):342–343. doi: 10.1016/j.ccell.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 63.Romo-Morales A, Aladowicz E, Blagg J, Gatz SA, Shipley JM. Catalytic inhibition of KDM1A in Ewing sarcoma is insufficient as a therapeutic strategy. Pediatr Blood Cancer. 2019;66(9):e27888. doi: 10.1002/pbc.27888. [DOI] [PubMed] [Google Scholar]
- 64.Wang T, Zhang F, Sun F. ORY-1001, a KDM1A inhibitor, inhibits proliferation, and promotes apoptosis of triple negative breast cancer cells by inactivating androgen receptor. Drug Dev Res. 2022;83(1):208–216. doi: 10.1002/ddr.21860. [DOI] [PubMed] [Google Scholar]
- 65.Campillo-Marcos I, Monte-Serrano E, Navarro-Carrasco E, García-González R, Lazo PA. Lysine methyltransferase inhibitors impair H4K20me2 and 53BP1 foci in response to DNA damage in sarcomas, a synthetic lethality strategy. Front Cell Dev Biol. 2021;9:715126. doi: 10.3389/fcell.2021.715126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salzano M, Vazquez-Cedeira M, Sanz-Garcia M, Valbuena A, Blanco S, Fernandez IF, Lazo PA. Vaccinia-related kinase 1 (VRK1) confers resistance to DNA-damaging agents in human breast cancer by affecting DNA damage response. Oncotarget. 2014;5(N7):1770–1778. doi: 10.18632/oncotarget.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Campillo-Marcos I, Lazo PA. Olaparib and ionizing radiation trigger a cooperative DNA-damage repair response that is impaired by depletion of the VRK1 chromatin kinase. J Exp Clin Cancer Res. 2019;38(1):203. doi: 10.1186/s13046-019-1204-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.López-Bañuelos L, Vega L. Inhibition of acetylation, is it enough to fight cancer? Crit Rev Oncol Hematol. 2022;176:103752. doi: 10.1016/j.critrevonc.2022.103752. [DOI] [PubMed] [Google Scholar]
- 69.Fernandez IF, Blanco S, Lozano J, Lazo PA. VRK2 inhibits mitogen-activated protein kinase signaling and inversely correlates with ErbB2 in human breast cancer. Mol Cell Biol. 2010;30(19):4687–4697. doi: 10.1128/MCB.01581-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fernandez IF, Perez-Rivas LG, Blanco S, Castillo-Dominguez AA, Lozano J, Lazo PA. VRK2 anchors KSR1-MEK1 to endoplasmic reticulum forming a macromolecular complex that compartmentalizes MAPK signaling. Cell Mol Life Sci. 2012;69(22):3881–3893. doi: 10.1007/s00018-012-1056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shields JA, Meier SR, Bandi M, Mulkearns-Hubert EE, Hajdari N, Dam Ferdinez M, Engel JL, Silver DJ, Shen B, Zhang W, et al. VRK1 is a synthetic lethal target in VRK2-deficient Glioblastoma. Cancer Res. 2022 doi: 10.1158/0008-5472.Can-21-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.So J, Mabe NW, Englinger B, Chow KH, Moyer SM, Yerrum S, Trissal MC, Marques JG, Kwon JJ, Shim B, et al. VRK1 as a synthetic lethal target in VRK2 promoter-methylated cancers of the nervous system. JCI Insight. 2022 doi: 10.1172/jci.insight.158755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Monte-Serrano E, Lazo PA. VRK1 kinase activity modulating histone H4K16 acetylation inhibited by SIRT2 and VRK-IN-1. Int J Mol Sci. 2023;24(5):4912. doi: 10.3390/ijms24054912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robert C, Nagaria PK, Pawar N, Adewuyi A, Gojo I, Meyers DJ, Cole PA, Rassool FV. Histone deacetylase inhibitors decrease NHEJ both by acetylation of repair factors and trapping of PARP1 at DNA double-strand breaks in chromatin. Leuk Res. 2016;45:14–23. doi: 10.1016/j.leukres.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oike T, Komachi M, Ogiwara H, Amornwichet N, Saitoh Y, Torikai K, Kubo N, Nakano T, Kohno T. C646, a selective small molecule inhibitor of histone acetyltransferase p300, radiosensitizes lung cancer cells by enhancing mitotic catastrophe. Radiother Oncol. 2014;111(2):222–227. doi: 10.1016/j.radonc.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 76.Yan G, Eller MS, Elm C, Larocca CA, Ryu B, Panova IP, Dancy BM, Bowers EM, Meyers D, Lareau L, et al. Selective inhibition of p300 HAT blocks cell cycle progression, induces cellular senescence, and inhibits the DNA damage response in melanoma cells. J Invest Dermatol. 2013;133(10):2444–2452. doi: 10.1038/jid.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ekstrom TL, Pathoulas NM, Huehls AM, Kanakkanthara A, Karnitz LM. VLX600 disrupts homologous recombination and synergizes with PARP inhibitors and cisplatin by inhibiting histone lysine demethylases. Mol Cancer Ther. 2021;20(9):1561–1571. doi: 10.1158/1535-7163.Mct-20-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bayo J, Tran TA, Wang L, Peña-Llopis S, Das AK, Martinez ED. Jumonji inhibitors overcome radioresistance in cancer through changes in H3K4 methylation at double-strand breaks. Cell Rep. 2018;25(4):1040–1050.e1045. doi: 10.1016/j.celrep.2018.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cantarero L, Sanz-Garcia M, Vinograd-Byk H, Renbaum P, Levy-Lahad E, Lazo PA. VRK1 regulates Cajal body dynamics and protects coilin from proteasomal degradation in cell cycle. Sci Rep. 2015;5:10543. doi: 10.1038/srep10543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2(6):1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 81.Masumi A, Wang IM, Lefebvre B, Yang XJ, Nakatani Y, Ozato K. The histone acetylase PCAF is a phorbol-ester-inducible coactivator of the IRF family that confers enhanced interferon responsiveness. Mol Cell Biol. 1999;19(3):1810–1820. doi: 10.1128/MCB.19.3.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rodriguez-Paredes M, Martinez de Paz A, Simo-Riudalbas L, Sayols S, Moutinho C, Moran S, Villanueva A, Vazquez-Cedeira M, Lazo PA, Carneiro F, et al. Gene amplification of the histone methyltransferase SETDB1 contributes to human lung tumorigenesis. Oncogene. 2014;33(21):2807–2813. doi: 10.1038/onc.2013.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Valbuena A, Lopez-Sanchez I, Vega FM, Sevilla A, Sanz-Garcia M, Blanco S, Lazo PA. Identification of a dominant epitope in human vaccinia-related kinase 1 (VRK1) and detection of different intracellular subpopulations. Arch Biochem Biophys. 2007;465(1):219–226. doi: 10.1016/j.abb.2007.06.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Fig. S1: Effect of VRK1 depletion on the epigenetic modifications of H3K9 in the presence (top) or absence (bottom) of serum in A549 lung adenocarcinoma cells.
Additional file 2. Fig. S2: Effect of VRK1 depletion on epigenetic modifications of H3K9 in the presence or absence of serum in U2OS osteosarcoma cells.
Additional file 3. Fig. S3: Effect of VRK1 depletion on the epigenetic modifications of H3K27 in the presence or absence of serum in A549 lung adenocarcinoma cells.
Additional file 4. Fig. S4: Effect of VRK1 depletion of epigenetic modifications of H3K27 in the presence or absence of serum in U2OS osteosarcoma cells.
Additional file 5. Fig. S5: Effect of the VRK-IN-1 inhibitor on the levels of H3K27 acetylation and methylation in A549 cells.
Additional file 6. Fig. S6: Effect of the VRK1 depletion and the VRK-IN-1 inhibitor on the levels of H4K16 acetylation and H3K4me3 associated with transcription in U2OS cells.
Additional file 7. Fig. S7: Effect of the VRK-IN-1 inhibitor on the levels of H3K9 acetylation and methylation in U2OS cells.
Additional file 8. Fig. S8: Effect of the VRK-IN-1 inhibitor on the levels of H3K27 acetylation and methylation in U2OS cells.
Additional file 9. Fig. S9: Effect of HDAC inhibitors on H3K9 acetylation and methylation in U2OS cells.
Additional file 10. Fig. S10: Effect of KMT inhibitors on H3K9 acetylation and methylation in U2OS cells.
Additional file 11. Fig. S11: Effect of the KAT inhibitor C646 on H3K9 acetylation and methylation in A549 and U2OS cells.
Additional file 12. Fig. S12: Effect of KDM inhibitors on H3K9 acetylation and methylation in A549 and U2OS cells.
Additional file 13. Fig. S13: Direct interaction between VRK1 and the PCAF acetyl transferase.
Data Availability Statement
All data and reagents are available upon request.