Abstract
Purpose:
Interactions with tumor-associated microglia and macrophages (TAM) are critical for glioblastoma progression. Polysialic acid (polySia) is a tumor-associated glycan, but its frequency of occurrence and its prognostic value in glioblastoma are disputed. Through interactions with the opposing immune receptors Siglec-11 and Siglec-16, polySia is implicated in the regulation of microglia and macrophage activity. However, due to a nonfunctional SIGLEC16P allele, SIGLEC16 penetrance is less than 40%. Here, we explored possible consequences of SIGLEC16 status and tumor cell-associated polySia on glioblastoma outcome.
Experimental Design:
FFPE specimens of two independent cohorts with 70 and 100 newly diagnosed glioblastoma patients were retrospectively analyzed for SIGLEC16 and polySia status in relation to overall survival. Inflammatory TAM activation was assessed in tumors, in heterotypic tumor spheroids consisting of polySia-positive glioblastoma cells and Siglec-16-positive or –negative macrophages, and by exposing Siglec-16-positive or –negative macrophages to glioblastoma cell-derived membrane fractions.
Results:
Overall survival of SIGLEC16 carriers with polySia-positive tumors was increased. Consistent with proinflammatory Siglec-16 signaling, levels of TAM positive for the M2 marker CD163 were reduced, whereas the M1 marker CD74 and TNF expression were increased, and CD8+ T cells enhanced in SIGLEC16/polySia double-positive tumors. Correspondingly, TNF production was elevated in heterotypic spheroid cultures with Siglec-16-expressing macrophages. Furthermore, a higher, mainly M1-like cytokine release and activating immune signaling was observed in SIGLEC16-positive as compared to SIGLEC16-negative macrophages confronted with glioblastoma cell-derived membranes.
Conclusions:
Collectively, these results strongly suggest that proinflammatory TAM activation causes the better outcome in glioblastoma patients with a functional polySia-Siglec-16 axis.
Keywords: Tumor-associated microglia and macrophages (TAM), glioblastoma survival, immune balance, tumor microenvironment, sialic acid-binding immunoglobulin-like lectins (Siglecs)
Introduction
Glioblastoma (GB) is one of the most aggressive primary brain tumor entities that still lacks curative therapy. With standard therapeutic intervention, GB patients have a median overall survival (OS) of less than 15 months (1–3). The immune response towards malignancies like GB is a key predictor of clinical outcome and concordantly, tumor progression is linked to the ability of malignant cells to suppress immune responses in the tumor microenvironment, including the activation of inhibitory immune checkpoints (4). Hence, the instructive interactions between tumor cells and tumor-associated microglia and macrophages (TAM) are increasingly recognized as a crucial factor in GB pathogenesis (5). Overall, an anti-inflammatory TAM phenotype seems positively correlated with malignancy (6), whereas a proinflammatory immune milieu is associated with increased patient OS (7). Accordingly, the TAM fraction with an anti-inflammatory (M2-like) signature is reduced in GB of long-term survivors (8).
A characteristic post-translational modification found in a number of tumor entities, including GB, is the glycan polysialic acid (polySia) (9–12). In neuroendocrine lung tumors, neuroblastoma, or medulloblastoma, the presence of polySia has been linked to elevated aggressiveness and poor prognosis (13–15), but the two reports on incidence and prognostic value in GB are contradictory (10,11). PolySia is synthesized in the Golgi compartment on a limited number of protein carriers, most prominently on the neural cell adhesion molecule (NCAM), and presented on the cell surface to regulate cellular interactions (12,16). In addition to cell surface presentation of polySia-NCAM by other cells, polySia attached to the proteins neuropilin-2 and E-selectin-ligand-1 can be released by activated microglia and macrophages themselves, and, independent of its protein carrier, polySia has the potential to modulate the activation state of microglia and macrophages by interacting with immune receptors of the sialic acid-binding immunoglobulin-like lectin (Siglec) family (17–21).
Siglec receptors contribute to immune balance by either inhibitory or activating signaling (22). In murine microglia, polySia interacts with the inhibitory immune receptor Siglec-E to dampen inflammatory activation (21) but in humans, microglia and macrophages can sense polySia by the paired immune receptors Siglec-11 and Siglec-16, which have almost identical extracellular receptor domains, but differ in their intracellular signaling (17,23–26). Like murine Siglec-E and most of the other Siglecs, Siglec-11 contributes to the inhibition of immune cells through an immunoreceptor tyrosine-based inhibitory motif (23). In contrast, Siglec-16-mediated signaling is initiated by the adaptor protein DAP12, containing an immunoreceptor tyrosine-based activation motif (25). By their opposing signal transduction, the paired receptors may balance responses to polySia-presenting pathogens (26). However, only a part of the human population has a SIGLEC16 allele coding for functional protein expression, whereas the majority carries an inactive pseudogene, SIGLEC16P, characterized by a four-nucleotide deletion disrupting the open-reading frame (24,25). Based on database information, the estimated SIGLEC16 allele frequency worldwide is 0.22 and, because homozygous SIGLEC16 carriers (SIGLEC16+/+) are rare (4.4%), 38.7% of the overall population are capable of expressing functional Siglec-16 (27). Importantly, the SIGLEC16 to SIGLEC16P gene conversion is uniquely human and Siglec-16 has no counterpart in rodents (25,28). This largely impedes studies on the role of Siglec-16 in animal models and particularly its functional assessment in GB.
To the best of our knowledge, the occurrence and cellular distribution of Siglec-16 in GB, as well as its potential influence on tumor progression have not yet been investigated. The current study explores the hypothesis that Siglec-16 and its ligand polySia jointly influence TAM activation and thereby GB outcome.
Materials and methods
Tumor specimens and inclusion criteria
Formalin-fixed paraffin embedded (FFPE) GB specimens were obtained from intracranial primary tumors resected from treatment-naïve patients between 2002 and 2011 (MHH cohort) or between 1997 and 2011 (UMG cohort) (29). For qPCR analysis, additional 16 primary FFPE GB specimens from treatment-naïve patients were available that were archived between 2017 and 2018 at MHH. The study was conducted in accordance with the Declaration of Helsinki and approved by the institutional review boards (ethic committees) of MHH (vote number: 1707-2013) and Göttingen University (vote number: 20/12/19). All patients gave written informed consent according to local, national, and international guidelines. Tumor samples were microscopically re-evaluated by two independent certified neuropathologists (MHH cohort CH and RB or CH and FF, respectively, UMG cohort CS and CH) and classified as glioblastomas WHO grade IV (GB) according to the WHO classification of brain tumors, 4th edition (MHH cohort) or revised 4th edition (UMG cohort) (30,31). Based on the operative reports, the extent of resection was categorized into gross total resection, subtotal resection and stereotactic biopsy. For the UMG cohort, MGMT promoter methylation and mutations of IDH1 and IDH2 were analyzed as described (29).
Inclusion criteria were valid results for SIGLEC16 genotyping, polySia immunofluorescence, CD68 and CD163 immunohistochemistry (IHC), and availability of clinical records on OS (time between primary resection and endpoint by death), age at tumor resection, sex, extent of primary resection and therapeutic intervention, leaving n=70 (MHH cohort) and n=100 (UMG cohort) GB patient samples for analysis.
Cells and spheroid cultures
Culturing primary GB cells and peripheral blood mononuclear cells (PBMCs) was approved by the ethic committee of MHH (vote numbers 1707-2013, 388-2008, and 9783-2021).
A polySia-positive GB cell line was established from a WHO grade IV glioblastoma without IDH mutation (IDH1/IDH2 wildtype), and lacking MGMT promoter methylation. Initially, cells were cultivated in DMEM (BioWest, Nuaillé, France, #L0103) with 10% FBS (Merck, #S0615) and cryopreserved. After reconstitution in the same medium, cells were seeded at 1x103 cells/cm2 in defined serum-free DMEM/F12 (PAN-Biotech, Aidenbach, Germany, #P04-41154) with N2 supplement (ThermoFisher, #17502048), to eliminate FBS-derived exogenous mitogens. To obtain clonal spheroids, GB cells were grown for at least eight weeks.
PBMCs were isolated as described (32), except that harvested PBMCs were washed with RPMI 1640 (PAN-Biotech, #1P04-18525) containing 2% FBS (Merck, #F9665) and centrifuged at 250xg. Subsequent purification by negative selection of monocytic cells was performed by the Pan Monocyte Isolation Kit (Miltenyi Biotec, #130-096-537) following the manufacturer’s instructions with an additional washing step with mono-PBS (5% FBS #F9665, 1.9 mM EDTA in PBS) after the magnetic labeling of the cells. Collected cells in the flow-through were pelleted, resuspended in ice-cold mono-PBS, and counted. From two independent purifications of n=7 and n=6 different donors, purities of 96±1.4% and 96±1.6% (SD) were determined by a Sysmex XN-9100 Hematology Analyzer (XN-10 module) using the body fluid mode.
PM-derived (PMd-) macrophages were generated from purified monocytes dissolved in N2-supplemented DMEM/F12. 120 μl with 9x105 monocytes were added to 600 μl GB cell-conditioned medium (GB spheroid culture supernatants filtered through 0.22 μm syringe filters) in 24-well plates. After ten days, 180 μl N2-supplemented DMEM/F12 with or without crude membrane fractions (33) obtained from 9x105 GB cells per well were added for 6h, before cells and supernatants were collected. Supernatants were cleared at 2,000xg and stored at −20°C. Cells were washed in pre-warmed PBS (37°C), pelleted at 450xg, snap-frozen in liquid nitrogen, and stored at −80°C.
For generation of heterotypic spheroid cultures, GB spheroids were disintegrated in Accumax® (Pan-Biotech, #P10-21250) for 15min at room temperature (RT) and centrifuged (400xg, 1min). Cells were resuspended in fresh N2-supplemented DMEM/F12 mixed 1:1 with GB cell-conditioned medium. Purified monocytes were dissolved in the same medium, before GB cells and monocytes were mixed 1:1, seeded as hanging drop cultures with 1x105 cells in 25 μl GB cell-conditioned medium, and cultivated for 10 days. Resulting spheroids were processed for RNA extraction and qPCR (see below), or fixed overnight with 4% paraformaldehyde/PBS at 4°C and embedded in paraffin using standard procedures. Per spheroid culture, 10 μl of medium were collected, pooled according to the individual PMBC donors, cleared at 2,000xg, and stored at −20°C.
Immunostaining
Seven or 3 μm thick paraffin sections were used for immunostaining of GB specimens or spheroids, respectively. For immunofluorescence, sections were processed as described (21), except for 1h antigen retrieval and 30min initial permeabilization. For Siglec-11 or Siglec-16 immunostaining biotin-tyramide signal amplification was applied. CD68/CD163, IBA1/CD74, and CD3/CD8 colorimetric double-stainings were performed on a Ventana Benchmark XT autostainer (Ventana Medical Systems, Tuscon, AZ, USA). For antibodies (with vendor, catalog number, and RRID) and details on staining protocols, see Supplementary Methods.
Genomic PCR and quantitative real-time RT-PCR (qPCR)
DNA and RNA from deparaffinized and rehydrated FFPE tumor samples was extracted as described (34), except for xylene incubation at 30°C, 4h proteinase K digestion, and inactivation for 15min at 85°C. Genotyping of SIGLEC16 and SIGLEC16P was performed by PCR with published primers (25) on template DNA from FFPE samples (see above and Supplementary Methods), or from PBMCs and THP-1 macrophages, obtained as outlined in (19). RNA was isolated from FFPE extracts by centrifugation at 11,000xg and purification with the NucleoSpin kit (Macherey-Nagel, Düren, Germany, #740955.50, see Supplementary Methods), or from spheroids by trizol extraction (19). qPCR was conducted as described (21), with PCR conditions and primers detailed in Supplementary Methods. Primary qPCR data were processed with the relative quantification tool of the ThermoFisher Connect Platform (ThermoFisher Cloud, v1.5.2, RRID:SCR_023441).
TNF ELISA, cytokine and phospho-kinase analysis
TNF was determined in 100 μl cell culture supernatants using a human TNFalpha ELISA kit (Invitrogen, #88-7346-88) according to the manufacturer’s instructions. Cytokines and related factors in supernatants, as well as phosphorylated kinases and related signaling molecules in cell pellets from PMd-macrophage cultures were determined with multiplex antibody arrays (human cytokine and phospho-kinase array kits, R&D systems, ARY005B and ARY003C) according to the manufacturer’s instructions. Chemiluminescence detection and densitometric quantification was performed with an Amersham imager 680 (GE Healthcare).
Immunoprecipitation and immunoblotting
Rehydrated FFPE material was dried at RT and incubated in 5 μl buffer (400 mM Tris-HCl pH 9.0 with 8 mM EDTA) per mg dry weight for 1h at 90°C. Thereafter, the buffer was diluted to 20μl of 150 mM Tris-HCl pH 7.5, 2 mM EDTA, 10 mM 2-mercaptoethanol, 1% NP-40, and 1 mM phenylmethylsulfonyl fluoride per mg of initial dry weight. 1200 μl of this lysate was processed for immunoprecipitation (IP) and Western blot detection of polySia and NCAM as described (21).
Microscopic image acquisition and evaluation
Microscopy was performed with Axio Observer.Z1 equipped with a motorized stage, ApoTome module for structured illumination, AxioCam MRm and MRc digital cameras, and Zen 2012 (blue edition) software (Carl Zeiss Microscopy, RRID:SCR_021725), or, for evaluation of entire sections, with Olympus BX46 equipped withXC50 digital camera and cellSens Entry software version 1.6 (Olympus, RRID:SCR_014551). Microscope settings and image acquisition are detailed in Supplementary Methods. Classifications were performed by two observers blinded to the clinical data and to the outcome of SIGLEC16 genotyping. A GB section was classified “polySia-negative” when polySia cell surface signals were absent from at least 10 randomly selected observation frames (20x magnification) and “positive” when strong polySia cell surface signals were observed in at least 50% of the frames. To assess CD163 or CD74 immunostaining of CD68- or IBA1-positive TAM, complete GB tissue sections were evaluated. A specimen was classified as CD163low, when at least 10% of the CD68-positive cells displayed no discernible CD163 signals, or CD74high, when at least 5% of the IBA1-positive cells displayed discernible CD74 signals. All of the investigated GB specimens could be unequivocally assigned to one of the categories defined above. CD8 staining of CD3-positive T cells was analyzed on 7 frames (40x magnification) per section on sections of six tumor specimens for each of the four combinations of polySia and SIGLEC16 status. Inclusion criteria were gross total resection and a cutting area of at least 2 cm2. Images of IHC-stained sections were acquired by an experimenter blind to the polySia/SIGLEC16 status and, because CD3-positive cells were scarce but enriched around blood vessels, frames with discernible vasculature were selected for evaluation. The overall 168 frames were randomized and coded before CD3-positive and CD8/CD3 double-positive cells were counted by visual inspection assisted by the event counting tool of the ZEN software. Eight frames with less than six CD3-positive cells were omitted from further analysis.
Statistics
Statistical analysis was performed using GraphPad Prism 9.3.0 (GraphPad, San Diego, CA, USA, RRID:SCR_002798). Applied tests are specified in Supplementary Methods and indicated with the respective results.
Data availability
The data generated in this study are available upon request from the corresponding author.
Results
Patient cohort characteristics
As summarized in Supplementary Table S1, the median age at primary resection was 60 (range 32-85) and 65 years (range 25-80), and the median OS (defined as the time in months between primary resection and endpoint by death) was 10 (range 0-76) and 13 months (range 1-55) for the MHH and UMG cohort, respectively. All patients of the UMG cohort, but only 45 patients (64%) of the MHH cohort received adjuvant chemotherapy, significantly improving survival (median OS 4 months without and 12 months with chemotherapy, P<0.0001, log-rank test). The female to male ratios were 1:1.26 and 1:1.33 for the MHH and UMG cohort, respectively, which is not significantly different from, e.g., the ratio of 1:1.35 reported for 54,980 GB cases diagnosed in the US between 2009 and 2013 (35) (P=0.82 for MHH and P>0.99 for UMG, Fisher’s exact test), but in contrast to the slightly increased OS of female patients detected by analyses of large cohorts (n>5,000) (36,37), the patients’ sex was not significantly associated with OS (median OS of female compared to male patients was 8 versus 11, and 12 versus 13 months for the MHH and UMG cohort, respectively; P=0.19 and P=0.76, log-rank tests). With a cutoff at 69 years of age, determined by CART analysis, the younger patient groups in both cohorts survived significantly longer (median OS was 11 versus 7, and 14 versus 9 months for the MHH and UMG cohort, respectively; P=0.033 and P=0.029, log-rank tests). For the extent of surgical resection, another established prognostic factor (37), categorization according to the operative reports (see Supplementary Table S1) revealed increased survival of patients with gross total resection, which however missed the level of significance in the log-rank test (median OS was 13 versus 10 months for the two cohorts combined, P=0.086). For the UMG cohort, MGMT promotor methylation and IDH1/2 status were available. The group of patients with methylated MGMT-promoters but no IDH1/2 mutations (IDH-wildtype) showed higher OS compared to IDH–wildtype patients without a methylated MGMT-promoter (median OS was 13.5 and 12 months, n=32 and n=65, respectively), but in contrast to a previous report focusing on other molecular markers in a different subset of the same patient collective (29), the difference was not significant (P=0.299, log-rank test).
SIGLEC16 genotyping and detection of Siglec-11 and Siglec-16 on TAM
For all GB specimens of the two cohorts, the SIGLEC16 genotype was determined by PCR on tumor-derived DNA (Fig. 1A). Ninety-six (56.5%) out of 170 GB were homozygous for SIGLEC16P (SIGLEC16P/P), all of the other 74 cases were heterozygous (SIGLEC16+/P, 43.5%). The resulting allele frequency and penetrance of SIGLEC16 were 0.218 and 0.436, which is not significantly different from values calculated for the overall population from n=993 human samples in the HapMap database (27) (0.220 and 0.387, respectively; P>0.999 and P=0.235, Fisher’s exact test). However, no subjects homozygous for functional SIGLEC16 (SIGLEC16+/+) were detected in any of the GB cases. Thus, compared to the 4.4% of SIGLEC16+/+ individuals in the overall population, this genotype was significantly underrepresented in the two GB cohorts (P=0.002, Fisher’s exact test). Immunodetection with Siglec-11- or Siglec-16-specific antibodies was performed by co-staining with IBA1 (gene name AIF1) to identify macrophages and microglia, and revealed the presence of both Siglecs on TAM in 3 out of 3 investigated SIGLEC16+/P GB (Fig. 1B). As expected, no Siglec-16 was detected in 3 out of 3 SIGLEC16P/P cases, while Siglec-11 was still localized to the surface of IBA1-positive TAM (Fig. 1C).
Figure 1. SIGLEC16 genotyping and immunodetection of Siglec-16 protein and polySia in GB.
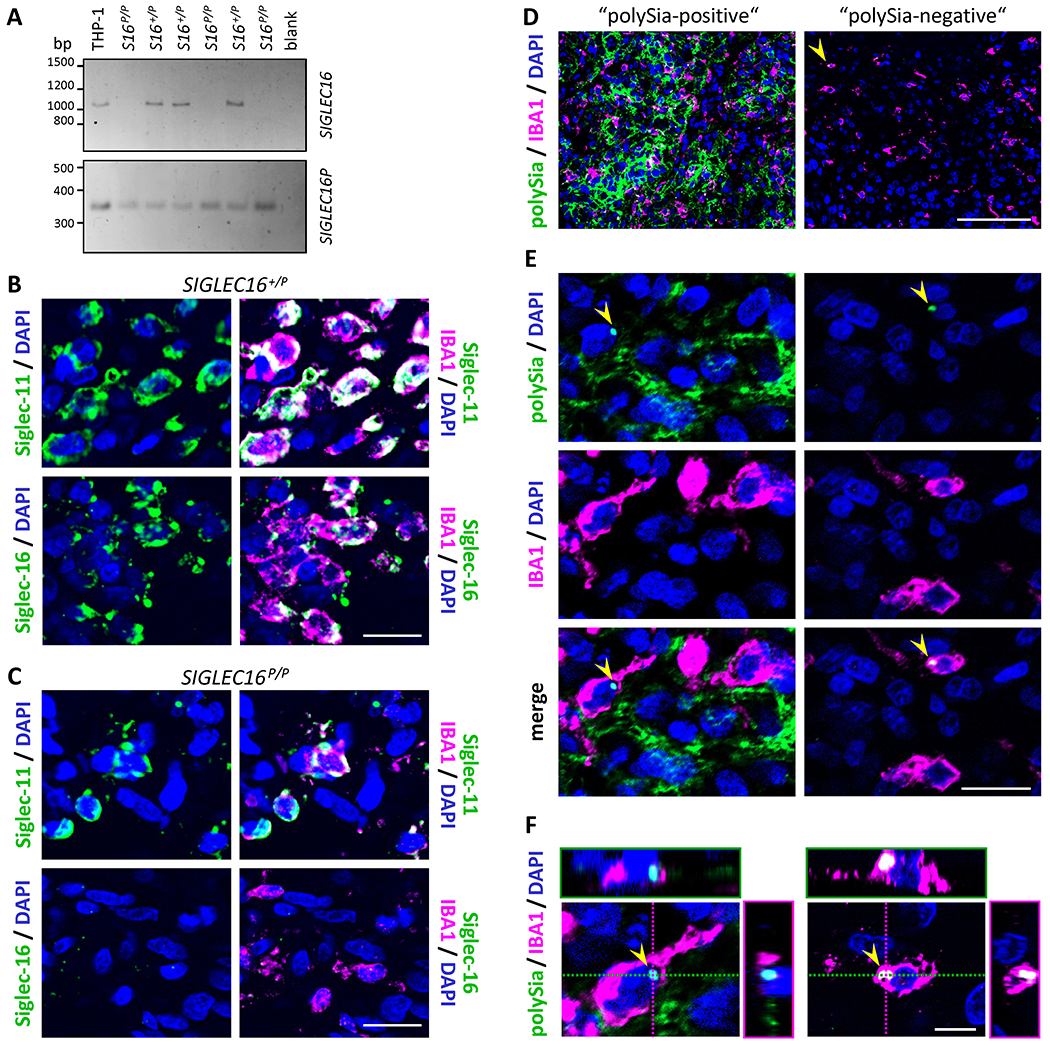
A, Genomic PCR with primers specific for SIGLEC16 (upper panel) or SIGLEC16P (lower panel). Representative results from six GB specimens, three heterozygous (S16 +/P) and three homozygous for SIGLEC16P (S16 P/P). PCR without DNA (blank) or with DNA from THP-1 macrophages with the genotype SIGLEC16 +/P were used as negative and positive controls, respectively. B and C, Immunofluorescence staining of Siglec-11 or Siglec-16 (green) together with IBA1 (magenta), as indicated. Representative examples of a SIGLEC16-positive (SIGLEC16 +/P, B) and a SIGLEC16-negative tumor (SIGLEC16 P/P, C) are shown. D-F, Representative examples of immunofluorescence staining for polySia (green) together with IBA1 (magenta). Specimens with or without strong polySia signals (“polySia-positive”, left; “polySia-negative”, right). Overviews (D), higher magnification views (E) and 3D reconstructions of IBA1-positive cells with perinuclear polySia signals (arrowheads, F). In B-F, nuclei were counterstained with DAPI (blue). Scale bars, 100 μm (D), 20 μm (B,C,E), and 5 μm (F).
Heterogeneous polySia patterns in GB
Occurrence and cellular distribution of polySia as a potential ligand of the immune receptors Siglec-11 and Siglec-16 were evaluated by double immunofluorescence staining with IBA1. Strong polySia-signals with heterogeneous distributions within the tumors were detected in 142 (83.5%) of the 170 GB investigated (Fig. 1D). As for other polySia-positive cancers and tumor cell lines (12), the abundance of the strong polySia signals in dense tumor areas suggested an association with tumor cells. This was supported by polySia staining of cells positive for the astrocytic glioma marker GFAP, but lacking morphologies of GB-associated reactive astrocytes (38, Supplementary Fig. S1A). In GB with established R132H mutation in the isocitrate dehydrogenase 1 gene (IDH1; 39), strong polySia signals were detected on mIDH1R132H-positive tumor cells (Supplementary Fig. S1B).
PolySia was never detected at the surface of IBA1-positive cells (Fig. 1E), but in GB with or without polySia on other cells, a small population of less than 0.1% of the IBA1-positive cells showed intracellular polySia signals (Fig. 1E and F). In rare cases, polySia was also detected on the surface of CD11c-positive or on IBA1-negative, CD45-positive cells in the vicinity of small intratumoral hemorrhages (Supplementary Fig. S2). These cells may represent infiltrating dendritic cells and blood monocytes prior to their differentiation into TAM, as polySia has been detected on the surface of mature dendritic cells as well as on freshly isolated blood monocytes and monocytic THP-1 cells in vitro (19,40). Co-staining for CD3 and CD34 demonstrated the absence of polySia from infiltrating T cells and endothelial cells (Supplementary Fig. S3).
Together, these results indicate that in GB with strong polySia staining the vast majority of polySia is presented at the surface of tumor cells. Furthermore, the presence of polySia-NCAM in GB, as suggested by previous studies (10,11), was corroborated by co-staining with NCAM-specific antibodies and by Western blot analysis of protein obtained by immunoprecipitation with polySia-specific antibody from a polySia-positive FFPE GB specimen (Supplementary Fig. S4).
SIGLEC16 and polySia correlate with overall survival
Both cohorts were stratified for the presence of a SIGLEC16 allele coding for functional protein expression, or for the presence of polySia on tumor cells. In the following, for the sake of better legibility, patients with or without an allele coding for functional Siglec-16 will be referred to as “SIGLEC16-positive” (SIGLEC16+/P) and “SIGLEC16-negative” (SIGLEC16P/P) and GB with polySia present or absent on tumor cells as “polySia-positive” (polySia+) or “polySia-negative” (polySia−), respectively. The two parameters were independent from each other as well as from age at tumor resection, sex, extent of resection, adjuvant chemotherapy, and, with data available for the UMG cohort only, MGMT-promotor methylation (Supplementary Table S2). Retrospective analyses of the two cohorts, individually or combined, showed significantly increased OS of SIGLEC16-positive (SIGLEC16+/P) cases (Fig. 2A–C). OS of patients with polySia-positive tumor cells was also higher and analyses by the log-rank test indicated significance for the MHH cohort and for the combined evaluation (Fig. 2D–F).
Figure 2. OS analysis of GB cases stratified for SIGLEC16 genotype and polySia on tumor cells.
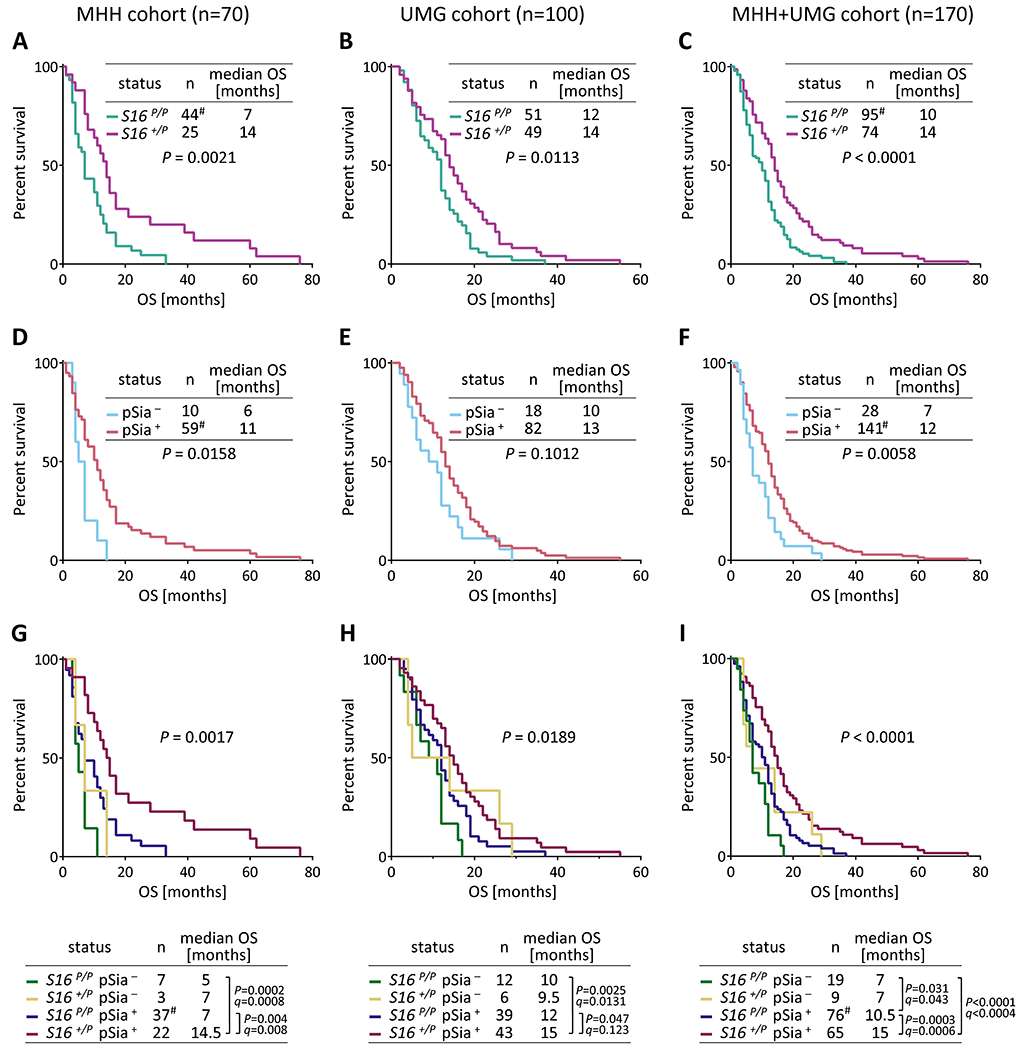
Kaplan-Meier survival plots with log-rank test results and median OS of the two GB cohorts, individually or combined, stratified for SIGLEC16 (S16, A-C), polySia on tumor cells (pSia, D-F), or both parameters (G-I), as indicated. The three cases with identified IDH1/2 mutations in the UMG cohort were SIGLEC16P/P/polySia−, SIGLEC16P/P/polySia+, and SIGLEC16+/P/polySia+ with OS = 3, 5, and 7 months, respectively. For comparisons of subgroups in G-I, P values < 0.05 and respective FDR adjusted q values are reported. # denotes that one SIGLEC16 P/P, polySia + case in the MHH cohort was censored because OS was zero.
Combined stratification for SIGLEC16 and polySia disclosed that the group of double-positive cases (SIGLEC16+/P, polySia+) had a significantly better clinical outcome than the groups of SIGLEC16- and polySia-negative or, except for the individual evaluation of the UMG cohort, SIGLEC16-negative, polySia-positive cases (SIGLEC16P/P, polySia− or SIGLEC16P/P, polySia+; Fig. 2G–I). OS of the SIGLEC16+/P, polySia+ patients was also significantly higher, when compared to all SIGLEC16- and/or polySia-negative cases together (log-rank tests with P=0.0008, P=0.019, and P<0.0001 for MHH, UMG, and both cohorts combined). Furthermore, for both cohorts combined, univariate and multivariate Cox regression analysis, adjusted for cohort, age at tumor resection, sex, extent of resection, and chemotherapy, revealed that the SIGLEC16 status alone or in combination with polySia was significantly associated with increased OS (Table 1 and Supplementary Table S3). Further analyzing a possible impact of age, extent of resection, or chemotherapy on SIGLEC16- and polySia-dependent differences, imbalanced age distributions could be excluded (Supplementary Table S4). OS was separately analyzed for patients receiving or not receiving gross total resection or chemotherapy. The stratifications for treatment indicated a loss of SIGLEC16-dependent group differences for patients receiving either no gross total resection or no chemotherapy, while the beneficial effect of SIGLEC16 alone or in combination with polySia was still pronounced in patients receiving any of the two or both treatments (Supplementary Tables S5–S7).
Table 1.
Cox uni- and multivariate analysis of SIGLEC16 and polySia status in relation to OS (MHH and UMG cohorts combined)
| Univariate |
Multivariate a |
|||
|---|---|---|---|---|
| Variable | HR b (95% CI) | P | HR (95% CI) | P |
| SIGLEC16 | 0.51 (0.31 - 0.70) | P < 0.0001 c | 0.53 (0.38 - 0.74) | P = 0.0002 |
|
| ||||
| polySia | 0.56 (0.38 - 0.87) | P = 0.0064 | 0.69 (0.46 - 1.07) | P = 0.086 |
|
| ||||
| SIGLEC16 / polySia double-positive | 0.50 (0.36 - 0.69) | P < 0.0001 | 0.55 (0.39 - 0.77) | P = 0.0007 |
Multivariate analysis adjusted for cohort, age at tumor resection, sex, gross total resection, and chemotherapy. For covariates, see Supplementary Table S3
Abbreviations: HR, hazard ratio; CI, confidence interval
P-values < 0.05 are shown in bold
It is worth noting that nine long-term survivors, defined by an OS of at least 36 months (8), were among the n=170 GB patients of both cohorts. Eight of them were SIGLEC16 and polySia double-positive, indicating a disproportionately high occurrence of long-term survivors in this group (P=0.002, Fisher’s exact test).
Due to the low numbers of the SIGLEC16-positive, polySia-negative cases (SIGLEC16+/P, polySia−) the outcome of this group could not be assessed adequately. However, even the joint evaluation of both cohorts indicated no differences between the SIGLEC16-negative, polySia-negative and corresponding SIGLEC16-positive cases (Fig. 2I, P=0.17, q=0.16), indicating that the ability to express functional Siglec-16 affects OS only in the presence of polySia on tumor cells. By contrast, the presence or absence of polySia on tumor cells affected OS in the group of SIGLEC16P/P patients (Fig. 2I), pointing towards some effect of polySia independent from functional Siglec-16.
For the UMG cohort, comparisons of OS data stratified for SIGLEC16 and/or polySia status were also performed after omission of the three cases with IDH1/2 mutations, yielding essentially the same results, i.e. significantly increased survival of the SIGLEC16-positive as well as the SIGLEC16 and polySia double-positive cases (Supplementary Table S8). Moreover, stratification for SIGLEC16 or polySia in combination with MGMT-promoter methylation status indicated significantly improved OS exclusively in the group of SIGLEC16-positive (SIGLEC16+/P) patients with a methylated MGMT-promoter (Supplementary Table S9).
Low ratios of CD163- and high ratios of CD74-positive TAM are associated with increased OS and overrepresented in SIGLEC16 and polySia double-positive GB
Ratios of CD163- and CD74-positive TAM, indicating M2- and M1-like polarization, respectively (6,7), were determined for both cohorts. Overall, 66% of GB were classified as CD163high, 69% as CD74low, and each of the two parameters was independent from age at tumor resection, sex, extent of resection, adjuvant chemotherapy, and, with data available for the UMG cohort only, MGMT-promotor methylation (Table 2 for CD163; Supplementary Table S10 for CD74; Fig. 3A, B). Consistent with previous reports (6,7), CD163low and CD74high were associated with significantly increased OS (Fig. 3C, D; Supplementary Fig. S5A–D). Combined evaluation of CD163 or CD74 with the SIGLEC16 status revealed that the SIGLEC16-positive patients with CD163low or CD74high tumors had the best clinical outcome (Fig. 3E, F; Supplementary Fig. S5E–H), and SIGLEC16-positive cases were significantly overrepresented in the CD163low and CD74high groups, i.e. in GB with low levels of pro-tumorigenic M2 and high levels of anti-tumoral M1 polarized TAM (Table 2; Supplementary Table S10). These unequal distributions and the survival benefit were maintained or even more pronounced, when the SIGLEC16 and polySia double-positive cases were considered (Table 2; Supplementary Table S10; Supplementary Fig. S5I–L). Again, imbalanced age distributions could be excluded (Supplementary Table S11) and stratifications for treatment indicated a loss of the CD163- or CD74-dependent group differences for patients receiving either no gross total resection or no chemotherapy, while CD163low and CD74high alone or combined with SIGLEC16 were still associated with improved survival in patients receiving any of the two or both treatments (Supplementary Tables S12–S14).
Table 2.
Characteristics of the two cohorts of n = 70 (MHH) and n = 100 (UMG) GB patients stratified for CD163high and CD163low
| CD163high | CD163low | Statistics | |||
|---|---|---|---|---|---|
| Distribution n (%) | MHH | 42 (60%) | 28 (40%) | P = 0.192 (MHH versus UMG) | Fisher’s exact test |
| UMG | 70 (70%) | 30 (30%) | |||
| MHH+UMG | 112 (66%) | 58 (34%) | |||
|
| |||||
| Age [years] a median ± SD (range) | MHH | 59 ± 13 (32-85) | 60 ± 12 (35-76) | P = 0.718 | Mann-Whitney test |
| UMG | 65 ± 10 (35-79) | 66.5 ± 14 (25-80) | P = 0.506 | ||
|
| |||||
| Sex female /male (ratio) | MHH | 16 / 26 (1 : 1.63) | 15 / 13 (1: 0.87) | P = 0.228 | Fisher’s exact test |
| UMG | 30 / 40 (1 : 1.33) | 13 / 17 (1 : 1.31) | P > 0.999 | ||
|
| |||||
| Gross total resection n (%) c | MHH | 23 (55%) | 14 (50%) | P = 0.801 | Fisher’s exact test |
| UMG | 51 (73%) | 24 (80%) | P = 0.615 | ||
|
| |||||
| Chemotherapy b n (%) c | MHH | 24 (57%) | 21 (75%) | P = 0.203 | Fisher’s exact test |
| UMG | 70 (100%) | 30 (100%) | P > 0.999 | ||
|
| |||||
| MGMT promotor methylated d n (%) c | UMG | 22 (31%) | 13 (43%) | P = 0.263 | Fisher’s exact test |
|
| |||||
| SIGLEC16 +/P cases n (%) c | MHH | 10 (24%) | 15 (54%) | P = 0.021 e | Fisher’s exact test |
| UMG | 29 (41%) | 20 (67%) | P = 0.029 | ||
| MHH+UMG | 39 (35%) | 35 (60%) | P = 0.002 | ||
|
| |||||
| SIGLEC16 +/P with polySia-positive tumor cells n (%) c | MHH | 7 (17%) | 15 (54%) | P = 0.002 | Fisher’s exact test |
| UMG | 25 (36%) | 18 (60%) | P = 0.029 | ||
| MHH+UMG | 32 (29%) | 33 (57%) | P = 0.0004 | ||
Age at primary resection
MHH: temozolomide, UMG: temozolomide, nimustine, or carmustine
% of all CD74low or CD74high, respectively
For MHH cohort not determined
P-values < 0.05 are shown in bold
Figure 3. OS analysis of GB cases stratified for ratios of CD163- or CD74-positive TAM and CD8-positive T cells related to SIGLEC16 and polySia status.
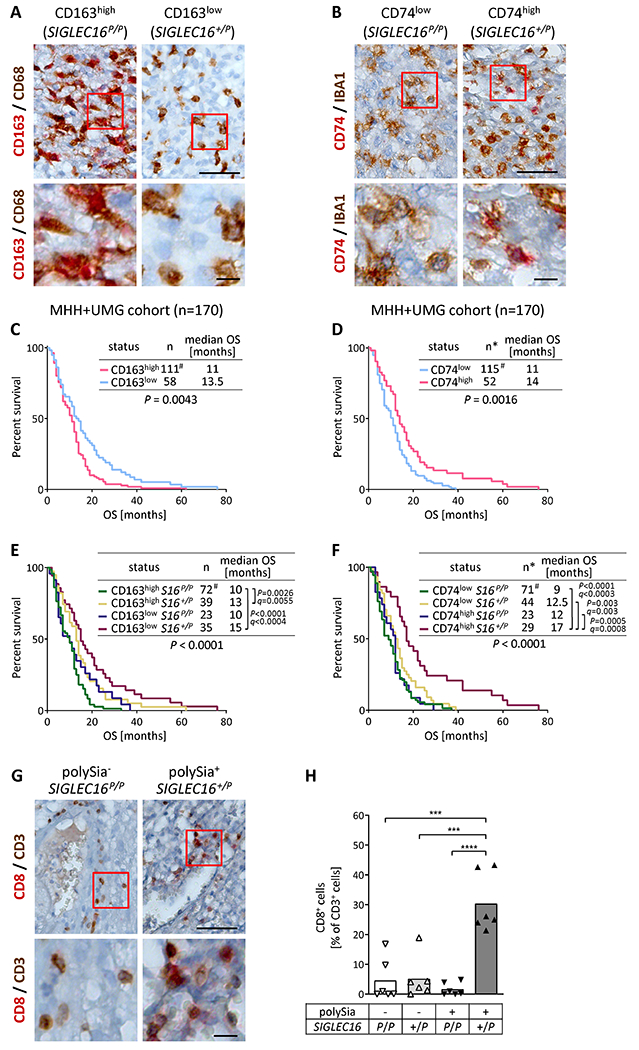
A and B, Detection of CD163 or CD74 (red) and CD68 or IBA1 (brown) on GB sections, as indicated. Representative examples of SIGLEC16P/P and SIGLEC16+/P GB specimens classified as CD163high or CD163low (A), and CD74low or CD74high (B), respectively. Scale bars, 50 μm. C-F, Kaplan-Meier survival plots with log-rank test results and median OS of the two GB cohorts combined, stratified for CD163high and CD163low (C), for CD74low and CD74high (D), or for different combinations of SIGLEC16 status with CD163 (E), or CD74 (F), as indicated. For the comparisons of subgroups in E and F, P values < 0.05 and respective FDR adjusted q values are reported. # denotes that one case was censored because OS was zero. G, Detection of CD8 (red) and CD3 (brown) on GB sections, as indicated. Representative examples of SIGLEC16P/P/polySia− and SIGLEC16+/P/polySia+ GB specimens. H, Ratios of CD8- relative to CD3-positive cells in GB with known polySia and SIGLEC16 status. Square root transformation was performed to meet the assumption of equal variances. Two-way ANOVA revealed significant differences (interaction: F(1,20)=14.42, P=0.001; polySia: F(1,20)=8.65, P=0.008; SIGLEC16 genotype: F(1,20)=21.33, p=0.0002) and Tukey’s post hoc tests were applied. Significant group differences are indicated (***, P<0.001; ****, P<0.0001). In A, B, and G, boxed areas in upper panels are shown at higher magnification (lower panels). Scale bars, 50 μm (upper panels), 10 μm (lower panels).
CD3-positive T cells were scarce but enriched around blood vessels. Analysis of CD3/CD8 double staining (Fig. 3G, H) indicated increased ratios of CD8-positive T cells in the SIGLEC16+/P/polySia+ group, meeting the expectation, because lower levels of M2 and higher levels of M1-like TAM polarization are linked to recruitment of CD8-positive T cells (41). Average numbers of CD3-positive cells per frame and GB specimen were not affected (means ±SD were 32.8±19.9, 26.2±17.5, 29.0±10.7, and 21.9±7.0 for the n=6 evaluated SIGLEC16P/P/polySia−, SIGLEC16P/P/polySia+, SIGLEC16+/P/polySia−, and SIGLEC16+/P/polySia+ specimens, respectively; P=0.70, Kruskal Wallis test).
TNF increase in SIGLEC16 and polySia double-positive GB and corresponding heterotypic spheroid cultures
We next sought to address whether the better outcome of SIGLEC16-positive GB patients with polySia-positive tumor cells is linked to proinflammatory activation. To explore the interaction of polySia-positive GB cells with Siglec-16-positive or -negative macrophages under controlled in vitro conditions, we established a serum-free hanging drop co-culture system, in which GB cells assemble into spheroids that incorporate monocytes from either SIGLEC16P/P or SIGLEC16+/P donors. GB cells for these heterotypic spheroid cultures were obtained by dissociation of spheroids grown from cells newly derived from a resected polySia-positive tumor). Co-culture of monocytes with the polySia- and GFAP-positive GB cells induced their differentiation into IBA1-positive PMd-TAM (Fig. 4A–C), which were Siglec-11-positive and, according to their genotype, positive or negative for Siglec-16 (stained against the monocyte lineage marker CD11b in Fig. 4D). Hence, concerning the interactions between polySia on tumor cells and PMd-TAM with or without Siglec-16, the heterotypic spheroid cultures were comparable to the parental tumor (Fig. 4B). Heterotypic tumor spheroids were obtained with PM from SIGLEC16P/P and SIGLEC16+/P donors and analyzed for expression and release of the key proinflammatory cytokine TNF (42,43). Relative TNF mRNA levels and release of spheroids with SIGLEC16+/P PMd-TAM were almost fivefold of those with SIGLEC16P/P PMd-TAM (Fig. 4E, F). For a respective qPCR analysis of GB samples, cDNA of sufficient quality could be obtained from nine GB specimens of the two cohorts, which all were archived before 2012, and from 16 FFPE GB specimens, archived between 2017 and 2018. Despite a considerable variance, the relative TNF mRNA levels were significantly increased only in the group of SIGLEC16-positive GB (SIGLEC16+/P) with tumor cell-localized polySia (Fig. 4G). Although not significant, a slight increase was also noted for the group of SIGLEC16+/P GB patients without tumor cell-localized polySia.
Figure 4. TNF production in polySia-positive GB and heterotypic GB spheroids as well as cytokine release and immune signaling in PMd-macrophages exposed to GB membranes depends on the SIGLEC16 status.
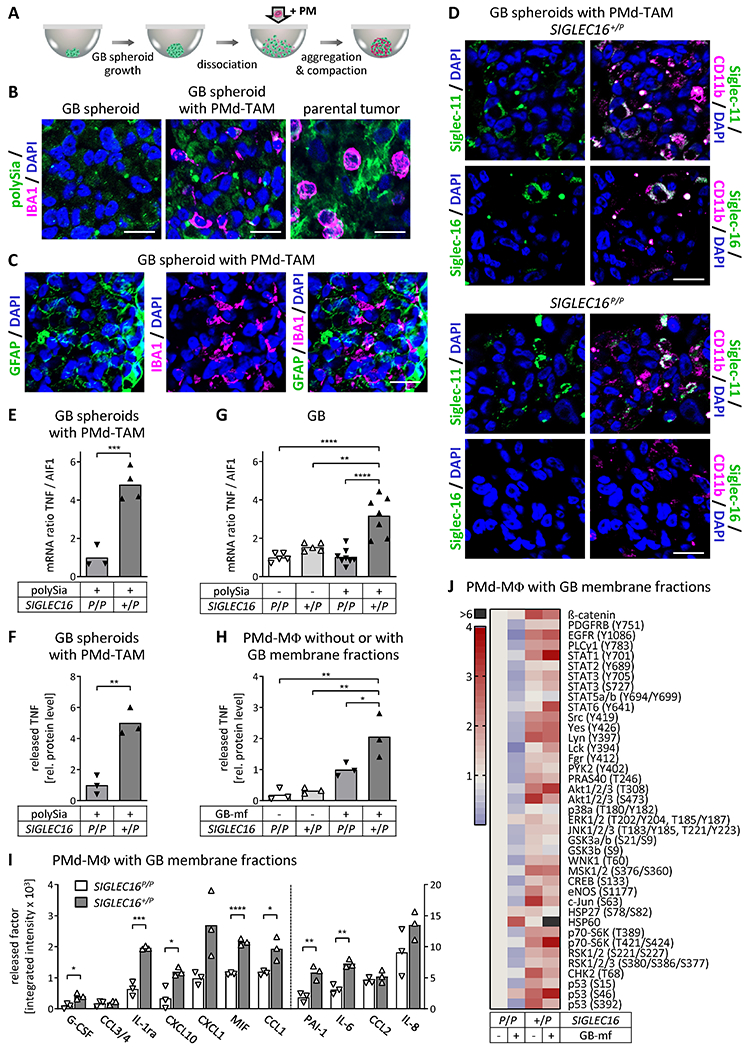
A, Schematic of heterotypic GB spheroid generation. PM, PBMC-derived monocytes. B and C, Immunofluorescence staining for polySia or GFAP (green) and IBA1 (magenta) on FFPE sections of GB cell spheroids, of heterotypic spheroids consisting of GB cells and PMd-TAM, and of the parental tumor from which the GB cell line was established, as indicated. D, FFPE sections of heterotypic GB spheroids with SIGLEC16 +/P or SIGLEC16 P/P PMd-TAM stained for Siglec-11 or Siglec-16 (green) together with CD11b (magenta), as indicated. In B-D, nuclei were counterstained with DAPI (blue). Scale bars, 20 μm. E-H, Relative TNF mRNA (E, G) and released protein levels (F, H) in heterotypic spheroid cultures with SIGLEC16 P/P or SIGLEC16 +/P PMd-TAM (E, F), in GB with known polySia and SIGLEC16 status (G), and in SIGLEC16 P/P or SIGLEC16 +/P PMd-macrophages not exposed or exposed to GB cell-derived membranes (GB-mf) obtained from GB spheroids (H), as indicated. Ratios of TNF relative to AIF1 mRNA levels were normalized to the mean value of the SIGLEC16 P/P/polySia+ (E) or SIGLEC16 P/P/polySia− group (G). Levels of released TNF were determined by ELISA and normalized to the mean value of the SIGLEC16 P/P/polySia+ (F) or SIGLEC16 P/P/GB-mf+ group (H). No TNF mRNA was detected in GB spheroids without PMd-TAM and background values of the TNF ELISA obtained with GB membrane fractions in GB cell-conditioned medium (<10% of the reference group) were subtracted to obtain values in H. I and J, Antibody array detection of the indicated cytokines, chemokines and related factors in supernatants (I) or phosphorylated kinases and related proteins in homogenates (J) of SIGLEC16P/P and SIGLEC16+/P PMd-macrophages exposed to GB cell-derived membrane fractions. Densitometric evaluation by integrated intensities of immune signals obtained in duplicate per array and sample. CXCL1, PAI1, IL-6, CCL2, IL-8 were detected in GB cell-conditioned medium with GB membrane fractions and the respective densitometric values were subtracted (I). Dashed line in I separates left and right y-axes. In E-I, means and individual values are plotted. Heatmap in J represents data from one experiment. Data in E, F, H and I were obtained from n=3 or n=4 heterotypic spheroid cultures consisting of polySia-positive GB cells with PMd-TAM (E, F), from n=3 PMd-macrophage cultures (H, I) derived from PBMCs of three or four different SIGLEC16-negative or –positive donors, respectively (SIGLEC16 P/P or SIGLEC16 +/P). Data in G were obtained from n=5 polySia- and SIGLEC16-negative (polySia - / SIGLEC16 P/P), n=5 polySia-negative, SIGLEC16-positive (polySia - / SIGLEC16 +/P), n=8 polySia-positive, SIGLEC16-negative (polySia + / SIGLEC16 P/P), and n=7 polySia-positive, SIGLEC16-positive GB specimen (polySia + / SIGLEC16 +/P). For statistical assessment of data in E and G, square root transformation was performed to meet the assumption of equal variances. Two-tailed t tests were applied to compare the two groups in E and F (t(5) = 7.02, P=0.0009, t(4) = 6.50, P=0.003) and for the group comparisons in I. For data in G and H, two-way ANOVA revealed significant differences (interaction: F(1,21)=9.65, P=0.005; polySia: F(1,21)=9.96, P=0.005; SIGLEC16 genotype: F(1,21)=36.82, p<0.0001 for G; interaction: F(1,8)=4.42, P=0.07; GB-mf treatment: F(1,8)=33.04, P=0.0004; SIGLEC16 genotype: F(1,8)=7.31, P=0.027 for H) and Tukey’s post hoc tests were applied. Significant group differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001).
M1-like profile of cytokine release and activating immune signaling in SIGLEC16-positive PMd-macrophages confronted with GB cell membranes
To corroborate higher TNF production and a shift towards an M1-like polarization of SIGLEC16-positive TAM, PMd-macrophages, differentiated by incubation with GB cell-conditioned media were exposed to crude GB cell-derived membrane fractions. Compared to PMd-macrophages alone, only the SIGLEC16-positive cells showed a significant increase of TNF release, when confronted with GB cell membranes (Fig. 4H). Membrane-exposed SIGLEC16-positive PMd-macrophages also displayed an enhanced release of various cytokines, chemokines, and related factors, such as IL-6, G-CSF, PAI-1, CCL1, CXCL10, implicated in CD8+ T cell recruitment (41), the CD74 ligand MIF (7), and the interleukin-1 receptor antagonist IL-1ra, a potent inhibitor of tumor growth (44) (Fig. 4I; Supplementary Fig. S6). Furthermore, several phosphorylated kinases and related signaling molecules were elevated in this group (Fig. 4J). This includes AKT phosphorylation at tyrosine 308, a target of phosphoinositide 3-kinase signaling downstream of DAP12, activated by, e.g., Siglec-16 (25,43), and tyrosine 701-phosphorylated STAT1, a major mediator of interferon-γ-induced signaling and transcriptional regulation of M1-like polarization (42). The most prominent upregulation, however, was found for HSP60, known to activate NF-κB signaling and the NLRP3 inflammasome, and to enhance the generation of reactive oxygen species in microglia (45). Of note, many of the signaling factors were elevated in Siglec-16 positive PMd-macrophages prior to membrane exposure. This might be caused by the presence of polySia or other, yet unknown Siglec-16 ligands in the differentiation medium. For example, polysialylated proteins could be released through ectodomain shedding by the differentiating monocytes or by the polySia-positive GB spheroids used to obtain the conditioned differentiation medium (16,19).
Most of the observed cytokine changes, such as the increased release of TNF and IL-6 are compatible with the secretion profile of M1-like polarized macrophages in vitro (42,43). However, all of these pleiotropic cytokines exert multiple, often contradictory context-specific functions. In GB, for example, IL-6 has been linked to pro- and anti-tumorigenic mechanisms, and, together with PAI-1, to the promotion of M2-like TAM polarization (46,47). Therefore, the Siglec-16-dependent changes in PMd-macrophages are not entirely compatible with the assumed role of proinflammatory TAM in SIGLEC16 and polySia double-positive GB, but it has to be considered that the responses after acute stimulation in vitro may not reflect altered TAM profiles in a tissue environment, precluding predictions on the situation in vivo. Despite these limitations, the in vitro experiments demonstrate a clearly different reactivity of SIGLEC16-positive and –negative PMd-macrophages towards polySia-positive GB cell membranes.
Overall, the presented data indicate that proinflammatory TAM activation by the polySia-Siglec-16 axis is linked to the better prognosis of GB patients with a functional SIGLEC16-gene in combination with polySia-positive tumor cells.
Discussion
Reciprocal interactions of tumor cells with the tumor microenvironment are important determinants of survival in GB. The current study demonstrates for the first time the presence of Siglec-11 and Siglec-16 on TAM and, based on retrospective analysis of two independent patient cohorts, provides strong evidence that the presence of polySia on tumor cells is a favorable prognostic marker particularly for those GB patients that are capable of expressing functional Siglec-16. Tumors with low CD163- and high CD74-positive TAM ratios were overrepresented in the group of patients with SIGLEC16 and polySia double-positive GB, indicating reduced pro-tumorigenic M2-like and enhanced M1-like polarization. Consistently, increased expression of TNF, a key cytokine of proinflammatory macrophage polarization (42,43), was detected in these tumors as well as in tumor spheroids consisting of polySia-positive GB cells and Siglec-16-positive PMd-TAM. Furthermore, the profile of cytokine and chemokine release as well as the enhanced activating immune signaling of SIGLEC16-positive as compared to SIGLEC16-negative PMd-macrophages confronted with GB cell membranes corroborates a function of Siglec-16 in driving polarization of TAM. Together, the data imply that interactions of polySia on tumor cells with Siglec-16 on TAM lead to prolonged survival of GB patients by promoting a more proinflammatory immune milieu. Siglec-16 therefore may represent a costimulatory immune checkpoint receptor on TAM with polySia as an endogenous ligand that antagonizes anti-inflammatory signaling.
Evaluation of GB tissue samples by immunofluorescence and immunopreciptation indicated the presence of polySia-NCAM on the surface of the tumor cells. This is consistent with the detection of polySia-NCAM by sandwich ELISA and the histochemical co-localization of polySia and NCAM in GB as shown in two previous studies (10,11). With the immunofluorescence staining method used in the current study, however, we detected tumor cell-associated polySia in 83.5% of the GB patients, which is somewhat above the 70% of polySia-positive cases identified by ELISA (10) and considerably higher than the 19% of polySia-positive GB discovered by histochemical staining with a fluorescently labelled, enzymatically inactive endosialidase variant (11). Apparently, the immunohistochemical method used for the detection of polySia in the current study has a higher sensitivity than detection by ELISA or by inactive endosialidase. The latter may be due to different polySia binding properties of the inactive endosialidase (11) as compared to mAb 735 (current study), or caused by differences in the staining procedure. Nevertheless, despite the conflicting results on the incidence of GB with polySia-positive tumor cells, the two studies consistently established an association of polySia-positive GB with increased survival and contrast with the study of Amoureux et al. (10) describing polySia-NCAM as an unfavorable marker. The outcome also differs from the majority of studies on a number of other tumor entities, characterizing polySia as an adverse prognostic factor by promoting migration, invasiveness and metastatic growth of the tumor cells (12–15,33). In advanced stage neuroblastoma however, the absence of polySia-NCAM on tumor cells has been linked to unfavorable prognosis and decreased OS (48).
Independent of polySia status, a positive association with survival was found for the SIGLEC16-positive cases (SIGLEC16+/P), but the additional stratification for the presence of tumor cell-associated polySia revealed that the better outcome is restricted to the SIGLEC16 and polySia double-positive cases. Furthermore, in SIGLEC16 and polySia double-positive GB specimens as well as in heterotypic spheroids consisting of polySia-positive tumor cells and SIGLEC16-positive macrophages, the expression of the proinflammatory cytokine TNF was increased. This is consistent with the assumed function of Siglec-16 as an activating immune receptor for polySia, leading to enhanced proinflammatory cytokine expression. This mechanism may have evolved to counteract the engagement of the inhibitory receptor Siglec-11 by bacterial pathogens that use their polySia capsules as a molecular mimic to escape immune defense (25,26).
In contrast to the 4.4% of SIGLEC16+/+ individuals in the overall population (27), none of the 170 GB cases studied was homozygous for SIGLEC16. In keeping with the model proposed above, this observation could be explained by a lower risk of SIGLEC16+/+ individuals for developing high grade glioma due to a more efficient immune surveillance (5). Along the same lines, a larger proportion of inflammatory TAM has been detected in GB patients with an OS of at least 36 months (8) and eight out of the nine long-term survivors in the current study were heterozygous for SIGLEC16 (SIGLEC16+/P) with polySia-positive tumors. Despite substantial progress in the identification of prognostic factors (3), molecular genetic determinants of the small group of long-term survivors among GB patients remain enigmatic. Considering that well over 80% of GB were polySia-positive, the increased OS of heterozygous SIGLEC16+/P patients together with the complete absence of SIGLEC16+/+ cases in the investigated patient cohorts suggests that homozygosity for SIGLEC16 may be a determinant of GB long-term survival. This possibility should be investigated in the near future.
Even in the absence of functional SIGLEC16, the presentation of polySia on GB tumor cells was associated with extended survival, although the effect was significantly weaker than in the SIGLEC16 and polySia double-positive cases. However, the TNF expression levels in GB with this constellation (SIGLEC16P/P, polySia+), were not elevated and not different from the SIGLEC16-negative cases with polySia-negative tumor cells (SIGLEC16P/P, polySia−). This implies that potential interactions of polySia with the inhibitory Siglec-11 as the remaining polySia receptor on TAM in SIGLEC16-negative GB have no impact on TNF expression and are not involved in the mechanisms that lead to improved survival in the group of SIGLEC16-negative patients with polySia-positive tumor cells. In the SIGLEC16-negative, polySia-positive cases therefore, polySia seems to have an additional effect independent of inflammatory regulation by interactions with Siglec-16 on TAM. Such a Siglec-independent role of polySia in GB could involve any of the other extensively studied mechanisms of polySia (for review, see 16). These include the well described modulation of cell adhesion, as well as interactions of polySia with growth factors or, as shown more recently, with chemokines (49), which may differ between GB and those tumors for which polySia has been firmly established as an adverse factor. Clearly, this needs to be explored in future studies.
Another interesting finding of the current study was the detection of TAM with intracellular, perinuclear polySia immunoreactivity. These cells occurred independently of the presence of polySia on tumor cells but were extremely rare. Their polySia pattern, however, is reminiscent of the pool of polysialylated proteins accumulating in the Golgi compartment of cultured microglia and macrophages (19), and to the perinuclear polySia staining observed in a transient activation state of microglia surrounding the lesion site in a mouse model of traumatic brain injury (21). Based on the observation that inflammatory activation of cultured microglia leads to a rapid depletion of polySia signals detectable by immunostaining and to a long-lasting shedding of polysialylated proteins by immuno-negative cells, it has been inferred that the amount of polySia released by injury-induced microglia is much higher than anticipated based on immunohistochemistry (21). Correspondingly, the scarce population of polySia-positive TAM detected by immunohistochemistry on GB sections may be indicative for the release of significant amounts of polySia-bearing proteins by inflammatory activated TAM that are immuno-negative for polySia. In this regard, it is noteworthy that the group of SIGLEC16-positve cases without polySia presentation on tumor cells (SIGLEC16+/P, polySia−) showed slightly increased TNF expression when compared to the corresponding SIGLEC16-negative cases (SIGLEC16P/P, polySia−). Hence, there is a possibility that not only tumor cell-associated polySia-NCAM but also polysialylated proteins released by TAM interact with Siglec-16 and thereby affect the inflammatory state of the tumor microenvironment. Alternatively, other yet unknown ligands for Siglec-16 might be presented in GB. To resolve these issues, prospective clinical studies with larger groups of GB patients as well as further investigations in cellular models are needed.
Other limitations of the current retrospective study are that IDH1/2 mutations and MGMT promoter methylation were only determined for patients of the UMG cohort and that only this cohort was treated homogeneously. On the other hand, despite these differences, the combination of functional SIGLEC16 and polySia was associated with increased survival in both cohorts, indicating a robust effect. Together with the observed synergistic effect of functional SIGLEC16 and MGMT promoter methylation in GB patients that had received chemotherapy with alkylating agents, the data raise hope that activation of Siglec-16 by pharmaceutical intervention could lead to improvement in outcomes for patients with a functional SIGLEC16 allele. Possibly, this strategy could be combined with established immune checkpoint inhibition to overcome the disappointing results of current immunotherapy in GB (3,50).
So far, our knowledge on the mechanisms of Siglec-16 activation by polySia and its impact on microglia and macrophage polarization is limited, mainly because this receptor has no comparable counterpart in rodents. To the best of our knowledge, the current study provides the first in vivo evidence for a polySia-Siglec-16 immune checkpoint axis in human malignancies and offers a mechanistic explanation for the association of functional SIGLEC16 with increased survival of GB patients. Together, our findings indicate prognostic value of the SIGLEC16 gene status combined with polySia expression in GB and possibly other cancers, and may enable new approaches in GB immune checkpoint therapy.
Supplementary Material
Translational relevance.
It is a remarkable feature of the microglia- and macrophage-specific activating immune receptor Siglec-16 that not all humans are equipped with a gene coding for functional Siglec-16, because approximately 60% of the population are homozygous for the non-functional pseudogene SIGLEC16P. Here we demonstrate that the presence of a functional SIGLEC16 allele is linked to increased survival of glioblastoma patients and a shift towards a proinflammatory marker profile if the tumor is positive for the Siglec-16 ligand polysialic acid, proposing prognostic relevance of the polysialic acid-Siglec-16 axis. Enhanced expression and release of TNF and other inflammatory cytokines, as well as increased activating immune signaling by Siglec-16-positive macrophages exposed to polysialic acid-positive glioblastoma cells or membranes provide a mechanistic explanation for the association with better survival, implicating Siglec-16 as a costimulatory immune checkpoint receptor on tumor-associated macrophages. Therefore, targeting the polySia-Siglec-16 axis may enable new approaches in immune checkpoint therapy of glioblastoma.
Acknowledgements
This work was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), project numbers 324633948, 43223629, 409784463 to H. Hildebrandt, and 435430032 to C. Stadelmann (DFG grants Hi 678/9-1; Hi 678/10-1 and Hi 678/10-2, FOR2953; Sta 1389/5-1), Deutsche Krebshilfe, project number 70112188 (to C. Hartmann) and NIH grant R01GM032373 (to A. Varki). L. Hoffmeister was supported by the Hannover Biomedical Research School (HBRS) and the MD/PhD program Molecular Medicine at MHH. We thank Nissi Varki (University of California, San Diego) for sharing her Siglec-16 staining protocol, Rita Gerardy-Schahn (MHH) for NCAM-specific antibody 123C3, the company BioLegend for donating Siglec-11-specific antibody, Patricia Zarnovican, Wiebke Schulze, Monika van Iterson, Vanessa Wittek and Henri Wedekind for technical support, and Martin Stangel and Dirk Hoffmann for helpful comments.
Footnotes
Conflict of interest
The authors declare no potential conflicts of interest.
References
- 1.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 2009;10:459–66. [DOI] [PubMed] [Google Scholar]
- 2.Molinaro AM, Hervey-Jumper S, Morshed RA, Young J, Han SJ, Chunduru P, et al. Association of maximal extent of resection of contrast-enhanced and non-contrast-enhanced tumor with survival within molecular subgroups of patients with newly diagnosed glioblastoma. JAMA Oncol 2020;6:495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weller M, van den Bent M, Preusser M, Le Rhun E, Tonn JC, Minniti G, et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat Rev Clin Oncol 2021;18:170–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marin-Acevedo JA, Dholaria B, Soyano AE, Knutson KL, Chumsri S, Lou Y. Next generation of immune checkpoint therapy in cancer: new developments and challenges. J Hematol Oncol 2018;11:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci 2016;19:20–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu S, Zhang C, Maimela NR, Yang L, Zhang Z, Ping Y, et al. Molecular and clinical characterization of CD163 expression via large-scale analysis in glioma. Oncoimmunology 2019;8:1601478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeiner PS, Preusse C, Blank AE, Zachskorn C, Baumgarten P, Caspary L, et al. MIF receptor CD74 is restricted to microglia/macrophages, associated with a M1-polarized immune milieu and prolonged patient survival in gliomas. Brain Pathol 2015;25:491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geisenberger C, Mock A, Warta R, Rapp C, Schwager C, Korshunov A, et al. Molecular profiling of long-term survivors identifies a subgroup of glioblastoma characterized by chromosome 19/20 co-gain. Acta Neuropathol 2015;130:419–34. [DOI] [PubMed] [Google Scholar]
- 9.Petridis AK, Wedderkopp H, Hugo HH, Mehdorn MH. Polysialic acid overexpression in malignant astrocytomas. Acta Neurochir (Wien) 2009;151:601–4. [DOI] [PubMed] [Google Scholar]
- 10.Amoureux MC, Coulibaly B, Chinot O, Loundou A, Metellus P, Rougon G, et al. Polysialic Acid Neural Cell Adhesion Molecule (PSA-NCAM) is an adverse prognosis factor in glioblastoma, and regulates olig2 expression in glioma cell lines. BMC Cancer 2010;10:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mäkelä K, Nordfors K, Finne J, Jokilammi A, Paavonen T, Haapasalo H, et al. Polysialic acid is associated with better prognosis and IDH1-mutation in diffusely infiltrating astrocytomas. BMC Cancer 2014;14:623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hildebrandt H, Mühlenhoff M, Gerardy-Schahn R. Polysialylation of NCAM. Adv Exp Med Biol 2010;663:95–109. [DOI] [PubMed] [Google Scholar]
- 13.Figarella-Branger D, Dubois C, Chauvin P, Devictor B, Gentet JC, Rougon G. Correlation between polysialic-neural cell adhesion molecule levels in CSF and medulloblastoma outcomes. J Clin Oncol 1996;14:2066–72. [DOI] [PubMed] [Google Scholar]
- 14.Glüer S, Schelp C, Madry N, Von Schweinitz D, Eckhardt M, Gerardy-Schahn R. Serum polysialylated neural cell adhesion molecule in childhood neuroblastoma. Br J Cancer 1998;78:106–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka F, Otake Y, Nakagawa T, Kawano Y, Miyahara R, Li M, et al. Prognostic significance of polysialic acid expression in resected non-small cell lung cancer. Cancer Res 2001;61:1666–70. [PubMed] [Google Scholar]
- 16.Schnaar RL, Gerardy-Schahn R, Hildebrandt H. Sialic acids in the brain: gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol Rev 2014;94:461–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shahraz A, Kopatz J, Mathy R, Kappler J, Winter D, Kapoor S, et al. Anti-inflammatory activity of low molecular weight polysialic acid on human macrophages. Sci Rep 2015;5:16800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Werneburg S, Mühlenhoff M, Stangel M, Hildebrandt H. Polysialic acid on SynCAM 1 in NG2 cells and on neuropilin-2 in microglia is confined to intracellular pools that are rapidly depleted upon stimulation. Glia 2015;63:1240–55. [DOI] [PubMed] [Google Scholar]
- 19.Werneburg S, Buettner FF, Erben L, Mathews M, Neumann H, Mühlenhoff M, et al. Polysialylation and lipopolysaccharide-induced shedding of E-selectin ligand-1 and neuropilin-2 by microglia and THP-1 macrophages. Glia 2016;64:1314–30. [DOI] [PubMed] [Google Scholar]
- 20.Karlstetter M, Kopatz J, Aslanidis A, Shahraz A, Caramoy A, Linnartz-Gerlach B, et al. Polysialic acid blocks mononuclear phagocyte reactivity, inhibits complement activation, and protects from vascular damage in the retina. EMBO Mol Med 2017;9:154–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thiesler H, Beimdiek J, Hildebrandt H. Polysialic acid and Siglec-E orchestrate negative feedback regulation of microglia activation. Cell Mol Life Sci 2021;78:1637–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol 2007;7:255–66. [DOI] [PubMed] [Google Scholar]
- 23.Angata T, Kerr SC, Greaves DR, Varki NM, Crocker PR, Varki A. Cloning and characterization of human Siglec-11 - A recently evolved signaling molecule that can interact with SHP-1 and SHP-2 and is expressed by tissue macrophages, including brain microglia. J Biol Chem 2002;277:24466–74. [DOI] [PubMed] [Google Scholar]
- 24.Hayakawa T, Angata T, Lewis AL, Mikkelsen TS, Varki NM, Varki A. A human-specific gene in microglia. Science 2005;309:1693. [DOI] [PubMed] [Google Scholar]
- 25.Cao H, Lakner U, de Bono B, Traherne JA, Trowsdale J, Barrow AD. SIGLEC16 encodes a DAP12-associated receptor expressed in macrophages that evolved from its inhibitory counterpart SIGLEC11 and has functional and non-functional alleles in humans. Eur J Immunol 2008;38:2303–15. [DOI] [PubMed] [Google Scholar]
- 26.Schwarz F, Landig CS, Siddiqui S, Secundino I, Olson J, Varki N, et al. Paired Siglec receptors generate opposite inflammatory responses to a human-specific pathogen. EMBO J 2017;36:751–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Mitra N, Cruz P, Deng L, Program NCS, Varki N, et al. Evolution of Siglec-11 and Siglec-16 genes in hominins. Mol Biol Evol 2012;29:2073–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayakawa T, Khedri Z, Schwarz F, Landig C, Liang SY, Yu H, et al. Coevolution of Siglec-11 and Siglec-16 via gene conversion in primates. BMC Evol Biol 2017;17:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Behling F, Barrantes-Freer A, Behnes CL, Stockhammer F, Rohde V, Adel-Horowski A, et al. Expression of Olig2, Nestin, NogoA and AQP4 have no impact on overall survival in IDH-wildtype glioblastoma. PLoS One 2020;15:e0229274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, editors. WHO classification of tumours of the central nervous system. 4th ed. Volume 1. Lyon, France: IARC Press; 2007.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, editors. WHO classification of tumours of the central nervous system. revised 4th ed. Volume 1. Lyon, France: IARC Press; 2016.. [Google Scholar]
- 32.Alberts A, Klingberg A, Hoffmeister L, Wessig AK, Brand K, Pich A, et al. Binding of macrophage receptor MARCO, LDL, and LDLR to disease-associated crystalline structures. Front Immunol 2020;11:596103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seidenfaden R, Krauter A, Schertzinger F, Gerardy-Schahn R, Hildebrandt H. Polysialic acid directs tumor cell growth by controlling heterophilic neural cell adhesion molecule interactions. Mol Cell Biol 2003;23:5908–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Körbler T, Grsković M, Dominis M, Antica M. A simple method for RNA isolation from formalin-fixed and paraffin-embedded lymphatic tissues. Exp Mol Pathol 2003;74:336–40. [DOI] [PubMed] [Google Scholar]
- 35.Ostrom QT, Gittleman H, Xu J, Kromer C, Wolinsky Y, Kruchko C, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2009-2013. Neuro Oncol 2016;18:v1–v75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ostrom QT, Rubin JB, Lathia JD, Berens ME, Barnholtz-Sloan JS. Females have the survival advantage in glioblastoma. Neuro Oncol 2018;20:576–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trifiletti DM, Alonso C, Grover S, Fadul CE, Sheehan JP, Showalter TN. Prognostic Implications of Extent of Resection in Glioblastoma: Analysis from a Large Database. World Neurosurg 2017;103:330–40. [DOI] [PubMed] [Google Scholar]
- 38.Heiland HD, Ravi VM, Behringer SP, Frenking JH, Wurm J, Joseph K, et al. Tumor-associated reactive astrocytes aid the evolution of immunosuppressive environment in glioblastoma. Nat Commun 2019;10:2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Capper D, Zentgraf H, Balss J, Hartmann C, von Deimling A. Monoclonal antibody specific for IDH1 R132H mutation. Acta Neuropathol 2009;118:599–601. [DOI] [PubMed] [Google Scholar]
- 40.Curreli S, Arany Z, Gerardy-Schahn R, Mann D, Stamatos NM. Polysialylated neuropilin-2 is expressed on the surface of human dendritic cells and modulates dendritic cell-T lymphocyte interactions. J Biol Chem 2007;282:30346–56. [DOI] [PubMed] [Google Scholar]
- 41.Petty AJ, Li A, Wang X, Dai R, Heyman B, Hsu D, et al. Hedgehog signaling promotes tumor-associated macrophage polarization to suppress intratumoral CD8+ T cell recruitment. J Clin Invest 2019;129:5151–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000prime reports 2014;6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kerneur C, Cano CE, Olive D. Major pathways involved in macrophage polarization in cancer. Front Immunol 2022;13:1026954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krelin Y, Voronov E, Dotan S, Elkabets M, Reich E, Fogel M, et al. Interleukin-1beta-driven inflammation promotes the development and invasiveness of chemical carcinogen-induced tumors. Cancer Res 2007;67:1062–71. [DOI] [PubMed] [Google Scholar]
- 45.Swaroop S, Mahadevan A, Shankar SK, Adlakha YK, Basu A. HSP60 critically regulates endogenous IL-1β production in activated microglia by stimulating NLRP3 inflammasome pathway. J Neuroinflammation 2018;15:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kubala MH, Punj V, Placencio-Hickok VR, Fang H, Fernandez GE, Sposto R, et al. Plasminogen activator inhibitor-1 promotes the recruitment and polarization of macrophages in cancer. Cell Rep 2018;25:2177–91.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang F, He Z, Duan H, Zhang D, Li J, Yang H, et al. Synergistic immunotherapy of glioblastoma by dual targeting of IL-6 and CD40. Nat Commun 2021;12:3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korja M, Jokilammi A, Salmi TT, Kalimo H, Pelliniemi TT, Isola J, et al. Absence of polysialylated NCAM is an unfavorable prognostic phenotype for advanced stage neuroblastoma. BMC Cancer 2009;9:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiermaier E, Moussion C, Veldkamp CT, Gerardy-Schahn R, de VI, Williams LG, et al. Polysialylation controls dendritic cell trafficking by regulating chemokine recognition. Science 2016;351:186–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brahm CG, van Linde ME, Enting RH, Schuur M, Otten RHJ, Heymans MW, et al. The current status of immune checkpoint inhibitors in neuro-oncology: a systematic review. Cancers (Basel) 2020;12:586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available upon request from the corresponding author.


