Integration of epigenetic perturbations of cis-regulatory elements and their genetic profiling in matched relapsed patient cohort revealed the contribution of the noncoding genome to breast cancer evolution and dormancy escape upon estrogen deprivation.
Abstract
Only a handful of somatic alterations have been linked to endocrine therapy resistance in hormone-dependent breast cancer, potentially explaining ∼40% of relapses. If other mechanisms underlie the evolution of hormone-dependent breast cancer under adjuvant therapy is currently unknown. In this work, we employ functional genomics to dissect the contribution of cis-regulatory elements (CRE) to cancer evolution by focusing on 12 megabases of noncoding DNA, including clonal enhancers, gene promoters, and boundaries of topologically associating domains. Parallel epigenetic perturbation (CRISPRi) in vitro reveals context-dependent roles for many of these CREs, with a specific impact on dormancy entrance and endocrine therapy resistance. Profiling of CRE somatic alterations in a unique, longitudinal cohort of patients treated with endocrine therapies identifies a limited set of noncoding changes potentially involved in therapy resistance. Overall, our data uncover how endocrine therapies trigger the emergence of transient features which could ultimately be exploited to hinder the adaptive process.
Significance: This study shows that cells adapting to endocrine therapies undergo changes in the usage or regulatory regions. Dormant cells are less vulnerable to regulatory perturbation but gain transient dependencies which can be exploited to decrease the formation of dormant persisters.
Introduction
During development of multicellular organisms, cell fate is established through a series of heritable transcriptional changes (1, 2). These changes are orchestrated by the interaction of transcription factors (TF) with the regulatory portion of the noncoding genome [cis-regulatory elements (CRE); ref. 3]. CRE activity is largely tissue specific and contributes to many aspects of cancer etiology (4–6). A large fraction of cancer subtypes displays addiction to the activity of TFs. In line with this, active compounds against nuclear receptors, a targetable class of TFs, account for 16% of the total FDA-approved cancer drugs (7). Hormone-dependent breast cancer (HDBC) cells are strongly dependent on the activity of the nuclear receptor estrogen receptor (ERα), pioneer factors FOXA1 and PBX1, and the transcription factor YY1 (3, 8). These TFs collectively control many cancer hallmarks through their direct interaction with a subset of CREs, including distal enhancers and promoters (8–11). Continuous modulation of ERα activity after breast surgery (5 years of adjuvant endocrine therapy, ET) is one the most successful targeted strategies and it represents one of the first examples of precision medicine (12–14). Nevertheless, cancer returns in up to 50% of patients over the course of 20 years postsurgery, suggesting that residual tumor cells can undergo prolonged dormancy (Fig. 1A; refs. 15–17).
Figure 1.
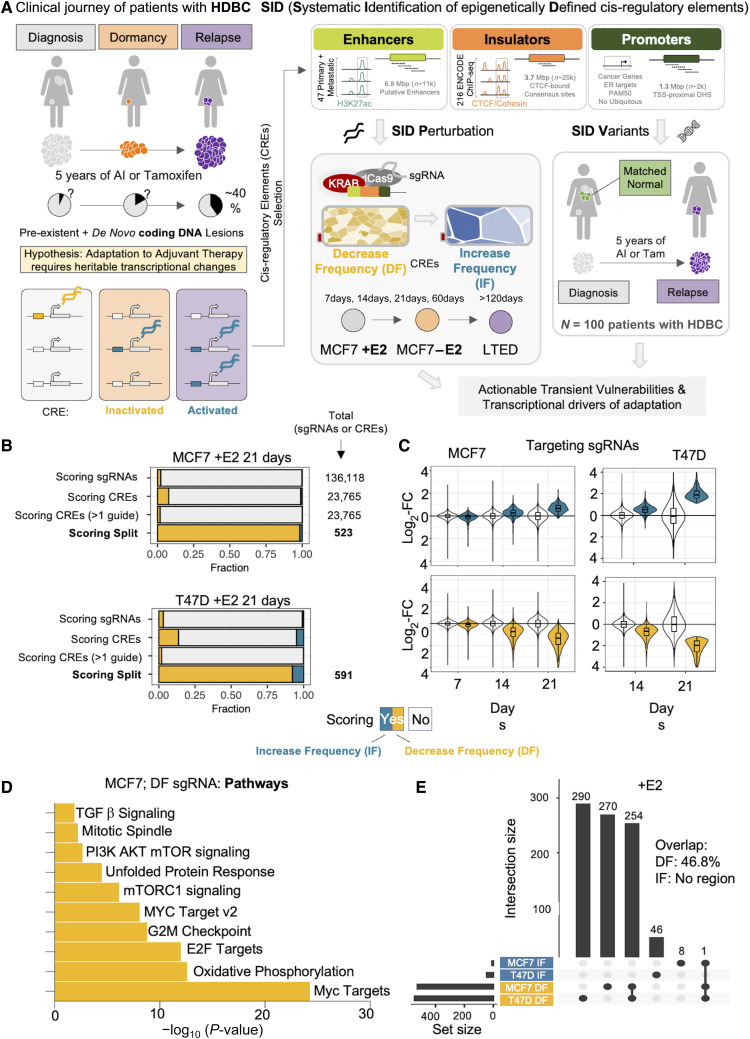
Defining a comprehensive strategy to functionally annotate the noncoding genome of HDBC. A, HDBC journey is characterized by distinct phases. Cells must adapt to different niches and treatments. Overcoming these stresses require profound, heritable transcriptional changes. Leveraging in vivo and in vitro data we develop SID, a strategy to prioritize HDBC-specific regulatory regions for functional (SID Perturbation) and genomic (SID Variants) annotation in cell line models and in patient samples. B, Bar plot showing the relative fraction of scoring sgRNAs and CREs bearing scoring sgRNAs, upon perturbation of noncoding genome of estrogen dependent MCF7 cells via SIDP. Scoring sgRNAs showing a significantly decreased frequency at 21 days postinfection are referred to as DF, whereas those with a significantly higher frequency as IF. C, Box plots showing the log2 fold change of both scoring (either blue or yellow) and nonscoring (white) sgRNAs at 21 days postinfection in estrogen-dependent MCF7 cells, at 7, 14, and 21 days, as compared with the initial library. D, Bar plot showing the top 10 hallmark gene sets enriched among the genes found in the proximity of the CREs with scoring sgRNAs showing a DF pattern at 21 days postinfection (P value estimated via hypergeometric test). E, UpSet plot showing the intersection between the SIDP loci showing two or more concordant significant sgRNAs after 21 days postinfection, in either MCF7 or T47D cells (+E2).
Despite HDBC cells being largely dependent on the activity of these TFs, previous perturbation screens focusing on ERα or FOXA1 bound CREs found that only a minority of binding sites seem to be essential for steady-state proliferation in vitro (18, 19). Yet, TF-centric perturbation likely missed CREs driven by additional TFs [i.e., YY1 and GATA3 (20–22)] and overlooked critical intermediate states in cancer evolution such as adaptive dormancy of persister cells (16, 17). To functionally explore the contribution of CREs to the evolution and adaptation of HDBC tumors exposed to ET, we developed a prioritized CREs panel [termed systematic identification of epigenetically defined loci (SID)] to investigate the role they play both in vitro and in vivo. The SID panel was built from a patient-derived epigenetic atlas (8) of putative enhancers with clonal or subclonal representation [i.e., clonal histone 3 lysine 27 acetylation (H3K27ac)] in primary and metastatic HDBC (see “Methods”). As disruption of chromatin topology can also contribute to disease evolution in both developmental and cancer models (23, 24), SID includes clusters of CTCF binding sites putatively controlling the integrity of topologically associating domain (TAD; Fig. 1A; “Methods”; refs. 25, 26).
Results
Perturbing SID Regions via CRISPRi
To increase the chances of perturbing entire CREs (promoters, enhancers, and TAD boundaries), which often extend over 1 to 2 kb and span several TF binding sites, we leveraged massively parallelized dCas9-KRAB repression [CRISPRi (27)]. We reasoned that KRAB-mediated repression predominantly mimics CRE loss of function potentially produced by somatic genetic alterations impinging on TF-binding affinity to these sites (28–30). We therefore designed 136,118 single guide RNAs (sgRNA) to interfere with the activity of 23,765 CREs in treatment-naïve MCF7 (HDBC cells grown with estrogen, +E2; Fig. 1A; Supplementary Tables S1 and S2; SID Perturbation or SIDP) SIDP covers more than 60% of the clonal enhancers active in MCF7 and almost every cluster of CTCF binding sites associated with TAD boundaries (Supplementary Fig. S1A). Nearly 100% of the sgRNAs were captured at high coverage (Supplementary Fig. S1B). These sgRNAs were then scored based on their relative change after 21 days postinfection considering both fold change and direction of the change in both replicates. This led to the identification of individual sgRNAs either increasing frequency (IF) across the population, corresponding to a potential fitness advantage after losing the activity of a CRE, decreasing frequency (DF), consistent with a fitness loss, or unchanged (neutral; Supplementary Table S3).
Both positive and nontargeting sgRNA controls showed highly concordant patterns after 21 days postinfection (Supplementary Fig. S1C and S1D) with 34% and 0.9% of positive controls and nontargeting sgRNAs significantly scored, respectively, demonstrating the robustness of the approach (FDR ≤ 0.05; fold change ≥ 1.5 or ≤ –1.5; Fig. 1B; Supplementary Table S3). Overall, 3,123 SID sgRNAs scored by day 21 (2.2%, Supplementary Table S3). Analysis of the temporal dynamics (7, 14, and 21 days) of the sgRNAs scoring at 21 days showed robust trends (Fig. 1C) with highly concordant replicates (Supplementary Fig. S1D). Interestingly, 98.4% of CREs showing multiple, reproducible scoring sgRNAs (including promoter, enhancers, and insulators) were associated with DF sgRNAs, indicating loss of fitness (Fig. 1B and C; Supplementary Fig. S1E). The regions scoring in our screen showed significant overlaps with observations from previous screens (Supplementary Table S3). Motif analysis on DF sgRNAs identified YY1 as the only enriched motif, in line with its critical role in shaping ERα transcriptional activity at clonal CRE in HDBC (Supplementary Fig. S1F; ref. 8). Scoring sgRNAs are also associated with many epigenetic features, including KDM5A binding (31, 32), promoter-specific H3K4me3, and enhancer-specific H3K4me1 (Supplementary Fig. S1G). DF sgRNAs were significantly associated with CREs near genes controlling metabolic processes (i.e., oxidative phosphorylation) and known MCF7 dependencies (MYC targets and PI3K and AKT signaling; Fig. 1D; Supplementary Table S3). Albeit many of these dependencies might be shared between models and patients, it is expected that a subset of these will be exclusive to MCF7 cells. To generalize our observations, we then applied SIP to a second independent cell line model (T47D, p53−/−), obtaining comparable, high-quality libraries (Supplementary Fig. S2A–S2C; Supplementary Table S4). With 92.2% of CREs showing multiple, reproducible scoring sgRNA promoting loss rather than gain of fitness, our results suggest that these cell lines have probably saturated their level of fitness to cell culture conditions (Fig. 1B). More importantly, SIDP exhibited significant overlap between the two ER+ cell lines, with ∼49% of robust DF sgRNAs (multiple hits within the same regions) from MCF7 being validated in T47D (Fig. 1E). Direct comparison of T47D and MCF7 libraries at 21 days highlighted only 26 of these regions as robustly and significantly different, with 23/26 showing lower frequencies in T47D. These genes tend to be related to RNA and protein metabolism. Collectively, these data establish SIDP as a powerful molecular tool for functional characterization of the noncoding genome and demonstrate that only a small fraction of CREs controls cellular proliferation in treatment-naïve HDBC cells.
SIDP Identifies De Novo Vulnerabilities in Cells Adapting to Treatment
Endocrine therapies target disseminated micrometastatic deposits by interfering with estrogen receptor activity, reducing the overall chance of relapse by half in patients followed over 20 years (13, 33). This effect is thought to be largely unpredictable at a single-patient level (17, 34) by virtue of ET ability to induce a transient dormant state in persister cells, a process mimicked in vitro by long-term estrogen deprivation (−E2; refs. 16, 17). Leveraging long-term linear tracing experiments, we have shown that that MCF7 and T47D evolve in a stochastic fashion, with each lineage randomly undergoing either cell death or cell state transition into a dormant state. More importantly, our data indicate that ET triggers these transitions via epigenetic changes that can be antagonized to hinder the formation of dormant cells. We then reasoned that the activity of specific CREs might contribute to the adaptive process occurring during the transition from growth to dormancy entrance. To test this, we run SIDP in MCF7 cells deprived of estrogen (−E2; Fig. 2; Supplementary Figs. S3–S9).
Figure 2.
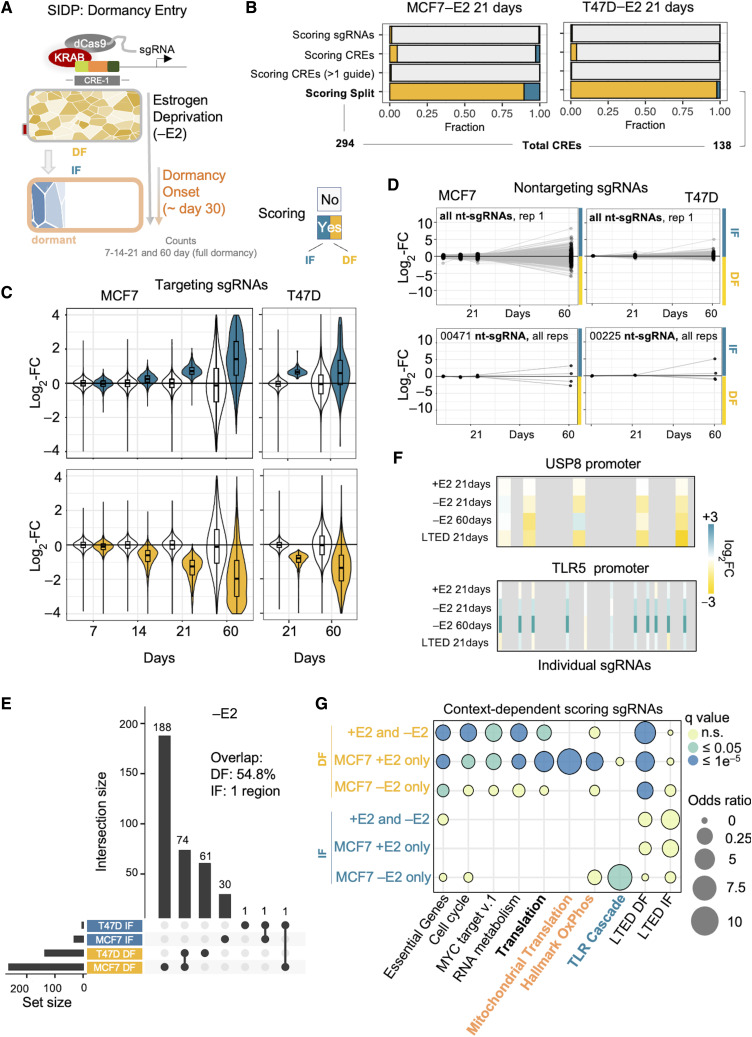
Adaptation to treatment exposes hidden roles for the noncoding genome. A, Experimental design. B, Bar plot showing the relative fraction of scoring sgRNAs and CREs bearing these sgRNAs, upon perturbation of the noncoding genome of estrogen deprived MCF7 cells via SIDP. Scoring sgRNAs showing a significantly decreased frequency at 21 days postinfection are referred to as DF, whereas those with a significantly higher frequency as IF. For the total numbers of sgRNAs and CREs, refer to Fig. 1B. C, Box plots showing the log2 fold change of both scoring (either blue or yellow) and nonscoring (white) sgRNAs at 21 days postinfection in estrogen-deprived MCF7 cells, at 7, 14, and 21 days, as compared with the initial library. D, Longitudinal tracking of individual non-targeting sgRNAs in four replicates during dormancy entrance (black dots highlight 7, 14, 21, and 60 days postinfection) support stochastic behavior of cells during dormancy entrance. E, UpSet plot showing the intersection between the SIDP loci showing two or more concordant significant sgRNAs after 21 days postinfection, in either MCF7 or T47D cells (−E2). F, Summary of the results for the sgRNAs targeting critical CREs of the USP8 and TLR5 genes. G, Bubble plot highlighting the enrichment of distinct biological functions, when considering sets of genes near CREs showing context-specific responses to perturbation.
To test if the stochastic process accompanying dormancy entrance and exit (17) also influences the readout of SIDP, we tracked individual nontargeting sgRNAs (n = 501; Supplementary Tables S5 and S6) for up to 60 days of hormone deprivation (full dormancy; Fig. 2A). Remarkably, 210/501 nontargeting sgRNAs (42%, compared with 0.9% in SIDP +E2) showed a nonneutral change in frequency at day 60. This behavior was completely unpredictable as shown by the evolution of individual nontargeting sgRNA in every replicate (two pools and two replicates; Fig. 2D) and by the overall divergent trajectories followed by the two replicates as highlighted by dimensionality reduction and correlograms (Supplementary Figs. S3A–S3D, S4A–S4D, and S5A–S5E). These data therefore confirm our lineage tracing results (17) and demonstrate that ET induces dormancy in a random subset of cells independently in each experiment, which makes the overall interpretation of the results at 60 days subject to extensive noise. Additionally, analysis on the long-term arm of the study (60 days) also identified stochastic awakenings and failed awakening (Supplementary Fig. S4D). This phenomenon progressively introduces stochastic deviations with time even in otherwise predictable perturbation (i.e., ESR1, and FOXA1; Supplementary Fig. S3E; ref.18). This again is expected to introduce noise in the system especially after day 30, when additional stochastic processes [failed awakening (17)] might inflate even further the noise created by dormancy entrance. These data indicate that investigating stochastic processes like dormancy process via classic CRISPR screens can be challenging, leading to a potential large number of false negative.
Nevertheless, our data uncovered a small but robust set of CREs (i.e., multiple scoring sgRNAs with consistent behavior across replicates; Fig. 2B–G) playing a role in the early phases of dormancy entrance. To identify those, we systematically compared +E2 and −E2 screens to identify regions showing context-specific behavior (Supplementary Figs. S6A–S6D and S7A–S7B; Supplementary Tables S7 and S8). During dormancy entrance, MCF7 seem to become independent of several metabolic dependencies, with CREs associated with genes involved in translation, mitochondrial function, and other metabolic processes switching from scoring to nonscoring (Fig. 2G; Supplementary Figs. S6A–S6D, S7A and S7B, e.g., MRPL58 and METTL17, Supplementary Fig. S8A and S8B). A significant proportion of these switches were recapitulated in the T47D model as well (Fig. 2E; Supplementary Figs. S5 and S6). Conversely, a small set of DF sgRNAs is specific to the −E2 condition, indicating de novo vulnerabilities emerging during hormone deprivation (−E2 >> +E2, e.g., USP8 and SYNV1; Fig. 2F; Supplementary Figs. S7 and S9A). Finally, the majority of sgRNAs expanding uniquely under therapy showed pronounced enrichment near genes from a single pathway, namely, the Toll-receptor activation of the NF-κB pathway (FDR = 0.0049; odds ratio = 13.3; Fig. 2F and G; Supplementary Figs. S7A, S7B, S9B and S9C; Supplementary Table S7). Perturbation of these CREs therefore seemed sufficient to influence the stochastic process controlling dormancy entrance.
Fully resistant clones emerge from a persister pool after extensive dormancy in both patients and HDBC cell lines models (17, 35, 36). Awakening clones exhibit extensive epigenetic reprogramming (35, 36), suggesting that the growth of therapy-resistant cells might be driven by a set of CREs distinct from that driving the proliferation of the primary tumor. To test this, we run SIDP in fully resistant long-term estrogen-deprived (LTED) cells (36, 37), which represent one fully awakened lineage that emerged from the matched parental MCF7 (Supplementary Fig. S10A–S10D; refs. 17, 36, 37). In line with the results of the screens in +E2 and −E2 MCF7, only a minority of CREs seem to control LTED fitness (Supplementary Fig. S10A and Supplementary Table S9). In stark contrast to proliferating MCF7, the DF subgroup does not dominate the scoring sgRNA landscape in LTED (55% vs. 98.4%, LTED vs. MCF7 +E2), suggesting that LTED have not yet fully adapted to cell culture conditions. Next, we examined if LTED inherited at least part of the CREs activity acquired during dormancy. Eighty percent of the dependencies acquired during dormancy seemed to be inherited in LTED (i.e., USP8, Fig. 2F; Supplementary Fig. S10D). Conversely, LTED fitness does not improve upon NF-κB suppression, suggesting that this signaling pathway plays a critical but transient role during dormancy entrance and exit (Fig. 2F; i.e., MYD88 and TLR5; Supplementary Fig. S10D). Overall, the application of SIDP showed that a relatively small subset of CREs can contribute to different phases of the adaptive process during breast cancer evolution in vitro.
Targeted CRE Perturbations Influences Adaptation to Treatment
SIDP demonstrated that cells entering dormancy generally decrease their dependencies (DF sgRNAs) on individual CRE activity (Figs. 2 vs. 1B; ref. 17) suggesting that adapting cells rely on a smaller regulatory network for their survival. These observations are consistent with our proteomic data which show that therapy induced dormancy involve a significant accumulation of heterochromatin (17). One notable exception was the USP8 locus, which seem to be either a de novo vulnerability in dormant clones or an essential gene for adaptation. The interpretation of IF sgRNAs is more complicated owing to the stochastic processes occurring during dormancy entrance (Fig. 2D; Supplementary Figs. S3–S5; ref. 17). We hypothesized that the frequency of these sgRNAs (i.e., TLR5 signaling) could have increased in the screen via three alternative scenarios: increased plasticity (a larger subset of lineages carrying the sgRNA become persister), early awakening and clonal expansion (17), or complete dormancy bypass (Supplementary Fig. S11A). To test these hypotheses, we developed assays to monitor the growth rates of edited cells (CRISPRi for IF TLR5, MYD88, UNC93B1, and DF USP8 vs. nontargeting sgRNA) by live imaging (Fig. 3A) under +E2 and −E2 conditions. To accommodate and quantify the underlying stochasticity of the process, all these experiments were run in 10 replicates in the absence of cell passaging (17). sgRNA-mediated recruitment of KRAB on promoter CREs efficiently led to downregulation of all targets (Supplementary Fig. S11B). Interestingly, the UNC93B1 locus was included in SIDP as a cluster of CTCFs and ChIP-seq profiling demonstrated that KRAB recruitment was sufficient to displace CTCF, leaving the possibility that the perturbation from the sgRNA either interfered with the 3D structure or with UNC93B1 expression or both (Supplementary Fig. S11C).
Figure 3.
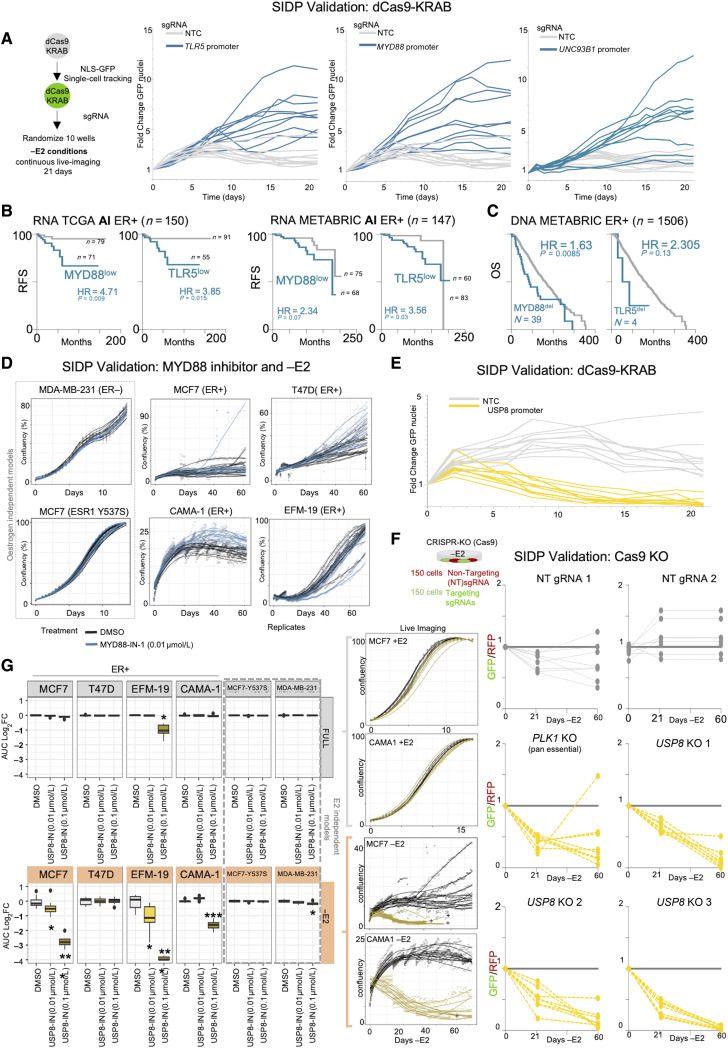
Targeted CRE perturbations facilitate or disturb the adaptive processes. A, Overview of the experimental design. A, Cell growth dynamics of MCF7 cells under estrogen deprivation (−E2) were monitored by tracking the total number of GFP-positive nuclei with continuous live imaging over the course of 21 days. Cells carrying sgRNA for MYD88, TLR5, and UNC93B1 have a significant higher chance of avoiding therapy induced dormancy B and C, Retrospective patient stratification based on RNA expression (B) or CNVs (C) for MYD88 and TLR5. Log-rank P values calculated with a Mantel–Cox test. D, Cell growth dynamics for a panel of estrogen dependent (MCF7, T47D, CAMA1, and EFM-19) and estrogen independent (MDA-MB231 and MCF7 Y537S) breast cancer cell lines under estrogen deprivation (−E2) were monitored with continuous live imaging over the course of 60 days in presence of a low dose of MYD88 inhibitor (MyD88-IN-1). Chemical MYD88 perturbation increased the number of dormant persister and in turn the chances of early awakening. The same concentration did not have any significant effect in +E2 condition. E, Same as A but targeting the USP8 gene promoter. Cell growth dynamics of MCF7 cells under estrogen deprivation (−E2) were monitored by tracking the total number of GFP-positive nuclei with continuous live imaging over the course of 21 days. Cells carrying sgRNA for USP8 have a lower chance of adapting to therapy. F, CRISPR-Cas9 knockout of USP8. FACS sorting was used to quantify green (USP8 sgRNAs carrying cells) and red (nontargeting sgRNAs). FACS analyses were carried out at three specific timepoints. G, Cell growth dynamics for a panel of estrogen dependent (MCF7, T47D, CAMA1, and EFM-19) and estrogen independent (MDA-MB231 and MCF7 Y537S) breast cancer cell lines under estrogen deprivation (−E2) were monitored with continuous live imaging over the course of 60 days in presence of low dose of USP8 inhibitor (DUB-IN-2). Area under the curve during the entire length of experiment was compared with the average of the controls to quantify the overall impact of USP8 inhibition. Chemical inhibition of USP8 significantly impact the survival of cells adapting to long term −E2 conditions. *, P < 0.01; **, P < 0.001; ***, P <10−5 (Mann–Whitney test).
We began by validating our live tracking analysis using sgRNAs targeting critical CREs for CCND1 in conjunction with a GFP-NLS tracker (Supplementary Fig. S12A–S12E). As expected, cells transfected with the targeting sgRNA (green) disappear more rapidly in +E2 conditions in competition assays (Supplementary Fig. S11D). Conversely, MYD88, TLR5, and USP8 targeting sgRNAs do not have any significant impact on the fitness of treatment naïve MCF7 (Supplementary Fig. S11D) in agreement with MCF7 and T47D +E2 SIDP. We next focused our attention on TLR5-mediated signaling in dormancy entrance (Fig. 3A). Competition experiment using sgRNA targeting CCND1 confirmed that our assay worked in –E2 conditions (Supplementary Fig. S12A). TLR5 and MYD88 suppressed cells exhibited altered pattern in dormancy entrance, with GFP-positive cells demonstrating clear fitness advantages in some replicates (Supplementary Fig. S12B–S12E). To gain a better understanding of the dynamics driving this process we switched to clonal populations (either edited with the target sgRNA or the nontargeting sgRNA). These experiments showed that cells with suppressed TLR5, MYD88, or UNC93B1 expression have increased fitness when exposed to the estrogen depleted conditions (Fig. 3A). Collectively, these live cell imaging experiments also confirmed the stochastic nature of the process and suggest that functional TLR signaling might be required for the formation of dormant persisters (Fig. 3A; Supplementary Fig. S12B–S12D). To explore the relevance of these observations in the clinical setting, we stratified independent retrospective cohorts containing only aromatase inhibitor (AI)-treated patients based on pretreatment expression of MYD88 and TLR5 expression. We found that patients with low MYD88 and TLR5 expression relapsed significantly earlier than those with high expression when treated with adjuvant endocrine therapy (AI; Fig. 3B). Of note, low expression of MYD88 and TLR5 was not significantly associated with shorter recurrence-free survival in untreated cohorts [Molecular Taxonomy of Breast Cancer International Consortium (METABRIC), MYD88 HR = 1.56, P = 0.29; TLR5 HR = 1.42, P = 0.35, log-rank, Mantel–Cox test] or ER negative cohorts (The Cancer Genome Atlas (TCGA): MYD88 HR = 0.74, P = 0.47; TLR5 HR = 2.22, P = 0.03, log-rank, Mantel–Cox test). On the other hand, TRAF6 was also associated with earlier relapse in AI-treated cohorts (Supplementary Fig. S9D). One caveat of this analysis is that bulk RNA sequencing profiles are derived from heterogeneous tissues. We therefore tested if MYD88 and TLR5 expression are driven by different levels of immune infiltration, because immune cells are known to express high levels of these transcriptions. Using de-convolved bulk RNA sequencing from TCGA we show however that MYD88 and UNC93B1 levels do not track immune infiltration (as opposed to CD19 and CD69, two markers associated with immune cells, Supplementary Fig. S12F). Interestingly, UNC93B1 is more strongly associated with epithelial cells, suggesting that the prognostic signal might be compatible with a cell intrinsic mechanism originating from ER+ breast cancer cells. This conclusion was supported by H3K27ac epigenetic profiling of primary and metastatic ER+ patient samples (8), which shows that MYD88, TRAF6, UNC93B1, and TLR5 promoters are active in most patients (Supplementary Fig. S13). Additionally, TLR5 expression is most abundant in breast cancer (TCGA dataset) and ER+ cells from normal breast (Supplementary Fig. S14A–S14B). Although MYD88 and TLR5 gene CNAs are rare, patients characterized by heterozygous deletion also show shorter responses to endocrine treatment (Fig. 3C). To further support the role of TLR-MYD88 signaling, we leverage chemical probes which interfere with TLR–MYD88 complex formation [MyD88-IN-1(38)]. This inhibitor has no significant impact on cell proliferation in treatment naïve conditions in six independent breast cancer cell lines at concentrations below 100 nmol/L (Supplementary Fig. S15A and S15B). However, low dose of MyD88-IN-1 led to an increase formation of dormant persister or increase the chance of early awakening in a subset of replicates specifically in ER+ cells (Fig. 3D). Collectively, these data suggest that functional TLR signaling is important for therapy-induced dormancy.
Next, we became interested in the potential upstream drivers of TLR5/MYD88 in adapting ER+ cells. Cell-intrinsic activation of innate immune signaling is significantly associated with ER+ patients with residual disease after neoadjuvant therapy (39), suggesting a critical but unexpected association between innate immunity, dormancy, and persister cells. We find significant evidence that cell-intrinsic activation of this pathway is triggered during active dormancy and suppressed at final awakening in single lineages adapting to therapy (Supplementary Fig. S16A and S16B; ref. 17). In our system this signal can only be provided by other cancer cells, considering the absence of tumor microenvironment or immune system. Toll-like receptors (TLR) are essential components of the innate immune system that respond to endogenous molecules that are released during host tissue injury/death [damage-associated molecular patterns (DAMP); ref.40]. A recent report demonstrated that TLR5 can function as a receptor for HMGB1, a nuclear histone line protein with DAMP function (41–43). Absence of HMGB1 and HMGB2 is a critical feature of preadapted cells, a cell state which shares many features with therapy induced dormancy (16). We thus hypothesized a potential crosstalk between adapting cancer cells via HMGB1/2-TLR (Supplementary Fig. S17A). First, we looked for evidence of additional TLRs activity in ER+ cells in patients but extensive analysis of our epigenetic atlas shows that TLR5 promoter is the only clonal CRE specifically active in ER+ breast cancer (Supplementary Fig. S13; ref. 8). Meta-analysis of donor-derived single-cell datasets from normal breast cells show that TLR5 is expressed in ER+ glandular cells (Supplementary Fig. S14A), whereas patients with breast cancer display the highest TLR5 level among all cancers despite being generally resistant to immune infiltration (Supplementary Fig. S14B). Collectively these data suggest TLR5 might have a role in ER+ cancer cells. Next, we sought to understand the dynamic of HMGB1 loss in ER+ cells. Immunofluorescence analysis showed that HMGB1 loses nuclear localization in response to estrogen starvation (Supplementary Fig. S17B). HMGB1 is then released in the media in a population size–dependent manner (Supplementary Fig. S17C). Accumulation of HMGB1 begins around the time cells begin to either enter dormancy or become apoptotic (17). HMGB1 activity as a DAMP molecule is dependent on its redox status [fully oxidized = Off; disulfide = On (44)]. When we exposed adapting cells to increasing doses of both forms, only the disulfide HMGB1 led to increased formation of dormant persister in a dose-dependent manner (Supplementary Fig. S17D). Collectively, these data suggest that TLR5 activation via paracrine HMGB1 signaling contribute to therapy-induced dormancy.
Our screen showed that adapting cells lose most vulnerabilities while entering dormancy (Fig. 2G). Conversely, there was more limited evidence for dormancy-specific vulnerabilities (DF sgRNA in −E2 but not in +E2 conditions). Considering the experimental design, these hits should represent factors which are intrinsically important for cells to transition to the dormant cell state but not necessarily important for the maintenance of a dormant phenotype. The most significant SIDP region having multiple differentially scoring sgRNA was USP8 promoter (Fig. 2F; Supplementary Fig. S9A). Validation experiments confirmed that treatment-naïve MCF7 cells with heritably repressed USP8 transcription do not exhibit any decrease in fitness (Supplementary Fig. S11D). On the other hand, USP8 suppression significantly interferes with MCF7 adaptation to −E2 conditions leading to almost complete eradication (Fig. 3E; Supplementary Fig. S12C–S12E). Repeating the long-term competition experiment using a genetic CRISPR-Cas9 system to knock-out USP8 further confirms its vital role in MCF7 adaptation to endocrine therapies (Fig. 3F). To expand on these observations, we inhibited USP8 activity using a chemical probe (45) in a panel of ER+ and ER− cell lines. Low doses of USP8 inhibitor (10 and 100 nmol/L) did not affect the proliferation of treatment-naïve ER+ cells (Supplementary Fig. S15A). Conversely, 100 nmol/L completely blocked the formation of dormant persister in most MCF7 and EFM-1 replicates and severely impaired CAMA1 adaptation as well (Fig. 3G; Supplementary Fig. S15B). Of note, it neither affected persister formation in T47D, in agreement with T47D SID-P results, nor affected ER− cells (MDA-MB-231) and ER mutant MCF7 (Y537S). Finally, we stratified independent retrospective cohorts containing only AI-treated patients and found that tumors with low levels of USP8 mRNA pretreatment relapse significantly later (Supplementary Fig. S15C and S15D), in agreement with a potential need for USP8 during therapy-induced dormancy entrance or maintenance. Overall, SIDP data show that emergent but transient phenotypes can be exploited to disrupt or accelerate HDBC cells adaptation to treatment. In vitro, these transitions are not the results of Darwinian selection of preexistent epigenetic clones but are rather induced and become heritable through therapy-induced dormancy (8, 16, 17).
SID Variants Identifies Patterns of CRE Mutations in Longitudinal Cohorts
SIDP is designed to model CRE loss of function via heritable epigenetic repression of CRE activity [KRAB-mediated heterochromatin formation (46)]. Somatic genomic alterations can also strongly influence the activity of individual CREs as well as chromosomal architecture (23, 47). We reasoned that high-depth genomic sequencing of SID CREs in matched pretreatment and relapsed samples might shed some insight on the role of the noncoding genome during tumor evolution. For this purpose, we developed SID variants (SIDV, Fig. 4A; Supplementary Fig. S18A–S18F) and profiled 300 matched samples (normal, primary, and relapse biopsies). All patients received either adjuvant tamoxifen (a selective estrogen receptor modulator) or AI (Fig. 4A; Supplementary Table S10). The median age of diagnosis was 46 for tamoxifen and 58 for AI. Grade and Ki67 status of the primary lesions were similar between cohorts (Fig. 4B; Supplementary Fig. S18B, S18E, and S18F; Supplementary Table S10). For 58 patients we could also co-profile variants in protein-coding regions, which identified de novo drivers of treatment failure (by comparing primary vs. matched relapse) at frequencies comparable with previous studies [i.e., ESR1 mutations (48–50); Supplementary Fig. S19A–S19E; Supplementary Table S11]. Using a highly stringent computational pipeline (see “Methods” and Supplementary Fig. S18A), we identified a total of 3,369 single-nucleotide variants (SNV) and 2,311 INDELs across the cohort, with a median coverage of 117× (Supplementary Table S12). Relapsed samples covered a wide spectrum of anatomic sites and despite showing comparable purity with matched primaries (P value = 0.088, paired two-tailed t test), show significantly less genomic alterations (P value = 0.0007, paired two-tailed t test), potentially indicating decreased genetic intratumor heterogeneity due to the bottleneck induced by metastatic seeding (Supplementary Figs. S18 and S19). The mutational burden from SIDV regions is highly consistent with previous WGS (Supplementary Fig. S18D). Interestingly, the mutational burden is higher in tumors showing high Ki67 and lower in those positive for the progesterone receptor (Supplementary Fig. S18E and S18F). Therapy choice (AI vs. tamoxifen) did not seem to impact the number of SNVs at relapse (P value = 0.21; Mann–Whitney test; Supplementary Fig. S19D). We then extended and integrated several machine learning approaches to prioritize the identified SNVs and short INDELs based on their predicted effect on TF binding (51), chromatin state (52), accessibility (53), and splicing (54) using only models derived from relevant, HDBC-specific genome-wide measurements (Supplementary Fig. S18A and “Methods”). A model-specific P value for each prediction was derived either using permutation-based approaches or by generating a null distribution from the noncoding alterations across all cancer types available in COSMIC (see “Methods”; ref. 55).
Figure 4.
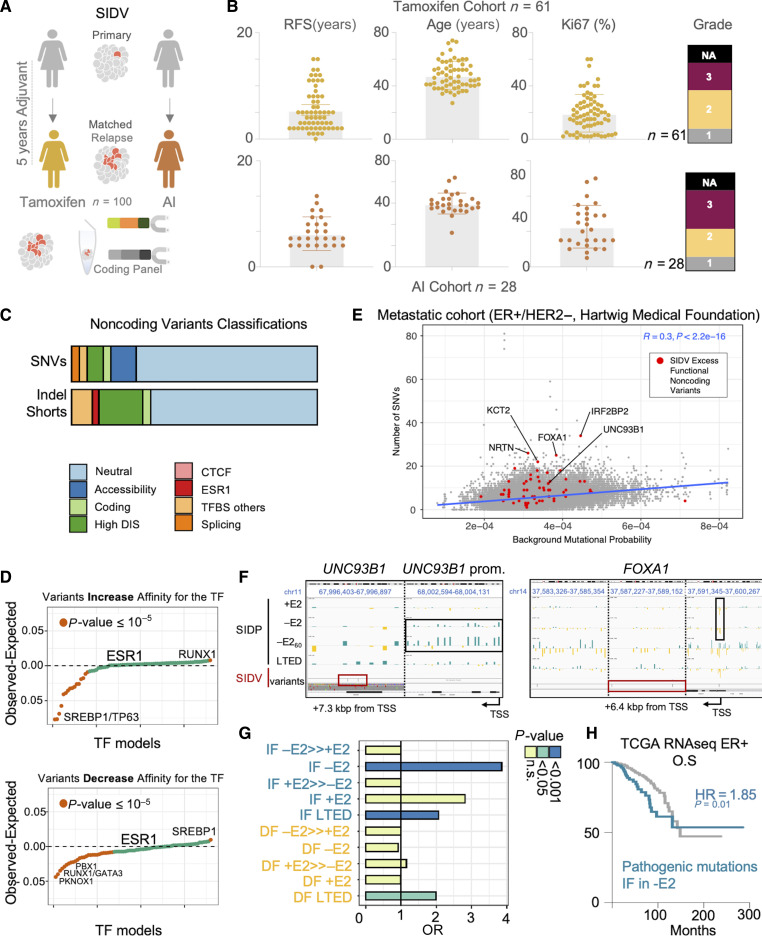
Noncoding variants contribute to heritable transcriptional changes during tumor progression. A, Schematic showing the rationale and implementation of SIDV. B, Overview of the clinical cohorts and the associated features. C, Pathogenic classification of noncoding variants identified by SIDV. D, Scatterplot summarizing the potential of the profiled SIDV variants to alter transcription factor binding. Each dot represents a TF. TFs are sorted based on their propensity to either increase (top) or decrease (bottom) the affinity to each TF. Values significantly larger than zero indicate a propensity to alter the binding that is higher than expected by chance. Those significantly smaller instead indicate a depletion of variants potentially altering the affinity for a given TF. P values estimated via χ2 test. E, Scatterplot showing the number of SNVs in the SID regions (each dot is a region) across 551 ER-positive, HER2-negative metastatic breast cancer samples, vs. the estimated background mutational rate. Regions showing an excess of functional variants are highlighted in red. The blue line represents a linear fit of the data. F, Integration of SIDV and SIDP identify critical regulators of HDBC biology. SIDP log2 fold changes (for the indicated samples, in black; blue fold changes indicate an increased frequency compared with the control library, yellow ones indicate a decrease; scale is [−3; +3]) and SIDV calls (in dark red) at the indicated loci are shown (IGV genome browser). Dark red and black boxes indicate regions with clusters of mutations or with multiple scoring sgRNAs, respectively. For both loci, different zoomed-in regions are shown, separate by vertical, black, dashed lines (precise coordinates of each region are indicated on top). G, Bar plot showing enrichment of SIDV-identified alterations at sets of regions showing condition-specific patterns upon perturbation (SIDP). P values estimated via χ2 Test. H, Kaplan–Meier plot showing that genes near CREs with an excess of SIDV mutations and overlapping IF sgRNAs upon estrogen deprivation (−E2) are associated with prognostic expression levels (HR = 1.85, P value = 0.01; log-rank Test).
We predict that ∼up to 30% of SIDV calls might have a functional impact on chromatin (Fig. 4C; Supplementary Table S13). The disease impact score [(DIS) as predicted by DeepSEA (56)] of called SIDV variants showed significantly higher values than noncoding variants across different cancer types in COSMIC (P value < 1e−16; KS test; Supplementary Fig. S19F). We also observe enrichment for SNVs with a negative impact on chromatin accessibility [as predicted by Sasquatch (53); Supplementary Fig. S19G]. Variants predicted to exert pathogenic impact on splicing seemed to be under negative selection (our set: 2.28% vs. expected: 4.71%, P value = 9.4e−15, χ2 test). We then focused on those alterations with predicted impact on HDBC-specific TF-binding [as predicted by deltaSVM (51); see Supplementary Table S14 for the complete information about the TFs considered]. Our data show that SNVs potentially altering the binding of several critical HDBC TFs are less frequent than expected (i.e., GATA3 and PBX1; Fig. 4D; Supplementary Table S15) with the notable exception of SNVs increasing the binding affinity of the HDBC cancer driver RUNX1 or decreasing SREBP1 binding. Interestingly, SNVs with predicted activity (increased or decreased) against ERα binding sites do not seem to be under any selective pressure, supporting the notion that most ESR1-bound CREs are not functionally significant (8, 9, 18). These data suggest that there is an overall negative selection on the binding sites of key TFs. However, when comparing the HDBC-specific alterations to those reported across different cancer types (COSMIC), a residual enrichment for functional alterations was spotted (Supplementary Fig. S19F and S19G).
Degeneration and redundancy in the genetic grammar governing cis-regulatory element activity have strongly limited our ability to spot recurrent noncoding mutations (57). Nevertheless, we hypothesized that by integrating the results from SIDV and SIDP we could gain more specific insights into the role of noncoding genetic alterations in HDBC (see Extended “Methods”). Using a lenient threshold (n > = 2; P value ≤ 0.05; binomial test), 63 SIDP CREs showed a significant excess of functional alterations (Supplementary Tables S16 and S17). These included one CRE falling in a cluster of CTCF binding sites within the UNC93B1 gene, which is part of the genes of the Toll receptor cascade in which downregulation leads to an advantage in −E2 (Fig. 4E). Interestingly, both UNC93B1-associated SNVs are predicted to alter splicing, whereas sgRNAs targeting this CRE or UNC93B1 promoter are significantly expanded in either −E2 or LTED screens (but not in +E2 conditions, Fig. 4F). Other regions showing both excesses of mutations and SIDP significant scores include CREs near FOXA1, a critical TF involved in many aspects of HDBC biology (Fig. 4E-F; ref. 9). Interestingly, integration with data from a large cohort of metastatic breast cancer samples (n = 551; ref. 58) confirmed an overall larger number of genetic alterations at the UNC93B1 and FOXA1 loci, than would be expected by chance (Fig. 4F; Supplementary Table S18). Intersection of the 63 loci mentioned above with SIDP results and previously identified noncoding putative driver loci highlighted once again FOXA1 (Supplementary Table S19). Furthermore, collapsing the predicted functional mutations at the level of pathways identified an interesting set of biological processes, suggesting that noncoding variants might contribute to promoting cancer evolution by suppressing differentiation and G1 arrest (Supplementary Table S16). Finally, we observed a significant overlap between SIDV mutations predicted as potentially pathogenic and SIDP but only when considering CREs bearing expanding sgRNAs under −E2 condition or in LTED cells, suggesting that mutations in these CREs have the potential of conferring a heritable fitness advantage to cells under treatment (Fig. 4G; Supplementary Table S16). Mutations found in these CREs tend to show a slight increase in cancer cell fraction (CCF) in matched metastatic deposits (P value = 0.08; paired Wilcoxon test). Low expression of genes associated with these CREs is associated with poorer prognosis in HDBC (Fig. 4H; HR = 1.85; P value = 0.01; log-rank test). This suggests that cells losing the expression of the target genes due to loss of function of the corresponding CREs might have increased fitness under the selective pressure imposed by endocrine therapies. In support of this, 4/6 of the SNVs in this set show a higher CCF in matched metastatic samples (P value = 0.03; χ2 test with Yates correction). Taken together, our results demonstrate that nongenetic and genetic mechanisms targeting CREs might significantly contribute to tumor evolution by modulating therapy-induced dormancy.
Discussion
The role of the noncoding genome in cancer has been under intense debate (30, 59, 60). In this work, we have (i) established a HDBC-specific cistrome (8); (ii) systematically perturbed it via targeted epigenetic repression, and (iii) profiled a large set of somatic alterations accumulated at these regions during tumor evolution. We ran three large-scale perturbation screens against the critical portion of the HDBC noncoding genome at an unprecedented depth and resolution. We also leveraged a unique patient cohort to profile noncoding genetic alterations longitudinally and at high coverage. Finally, we applied machine learning approaches to systematically dissect the functional consequences of these variants on regulatory potential. Systematic integration of results from these orthogonal experimental and computational strategies led to the conclusion that genetic variation at CREs do not display the strong signature associated with coding drivers and that noncoding variation, when taken in isolation, do not provide a strong fitness advantage to adapting cells. Conversely, our study highlights that nonmutational context-specific changes in the activity of a defined set of CREs might play a role during therapy-induced dormancy. Our results stand out considering the stochastic processes dominating dormancy entrance and exit (Fig. 2C; Supplementary Figs. S3–S5; ref. 17). For example, our SIDP screens strongly suggest that signaling converging on NF-κB activation plays a central role in acquiring long-term dormancy. This prediction is corroborated by our transcriptional tracking of single lineages, which shows NF-κB activity being induced in dormant cells but reversed in awakened lineages (Supplementary Fig. S16; ref. 17). We hypothesize that TLR signaling suppression increases the chance of escaping therapy induced dormancy. Of note, mutations on CREs associated with NF-κB regulation are surprisingly infrequent considering the potential benefit to cancer cells under AI pressure (Fig. 3B). This suggests that transcriptional switches are the preferred route to adaptation for HDBC cells, possibly because of their reversible nature. In agreement, we could not identify recurrent genetic mechanisms leading to awakening (17). Although profiling primary and secondary lesions as an evolutionary endpoint did not reveal many additional therapeutic entry points, transient dormancy might offer an attractive and unexplored stage with potentially actionable transient dependencies. As a proof of concept, we indeed show that targeting USP8 can actively eradicate HDBC once cells commit to dormancy. As such, we anticipate that our results will also have critical relevance for the design of future screens that will help expand our knowledge on the regulatory networks underlying therapy-induced dormancy, which we propose as the critical targetable bottleneck in the adaptive journey of breast cancer cells.
Methods
SID Panel Design
Previous epigenomic annotation of primary and metastatic luminal breast cancer tissues led to the identification of 326,729 putative enhancer regions (8). Most of these regions were private or poorly shared amongst individual tumors. However, an overall correlation between the activity of an enhancer in an individual tumor [low ranking index (RI)] and the pervasiveness of its activity across tumors (high sharing index, or SI) was observed. Thus, putative enhancer regions for the panel were biased for those showing a low RI. Starting from the ∼326 K regions mentioned above, we first excluded all the private enhancers (RI ≥ 80). 19,482 enhancers were retained and evaluated in terms of their delta of activity between primary and metastatic tumors. The average RI of each enhancer in the primary and metastatic cohorts was calculated (termed RI_Prim and RI_Met, respectively). These two numbers were then used to calculate a region-specific log2(RI_Met/RI_Prim). Putative enhancers showing either higher enrichment in the primary or metastatic samples were selected (regions with RI ≤ 50 in both primary and metastatic, and either in the top positive or negative log2(RI_Met/RI_Prim)). This resulted in 8.05 Mbps covering regions with higher RI in the metastatic samples and 3.7 Mbps showing higher RI in the primary samples. Finally, 2.5 Mbps was assigned to private enhancers being clonal in only one or two samples. As an internal control, 800 putative enhancer regions were randomly selected among those showing extremely low sharing (SI = 1) and ranking (RI = 100) index. To reduce the required coverage and to increase the enrichment for potentially functional regulatory regions, DNase-I accessible regions available in ENCODE (61) were then used to restrict the area of investigation to the subregions within the selected putative regulatory regions. These are more likely to represent clusters of TF-binding sites. To this aim, the regions resulting from the analysis described above were intersected with the DHS from HoneyBadger2 (https://personal.broadinstitute.org/meuleman/reg2map/), which effectively lowered the coverage to ∼9 Mbps. Based on an initial iteration of the capturing strategy, these 9 Mbps were further reduced to about 7, by excluding those regions with either a very low or an extremely high coverage (i.e., the bottom and top 1% in terms of normalized coverage, considering a previous iteration of the design that was applied to a small, pilot cohort). This resulted into a higher and more even coverage on the majority of the targeted elements Putative insulator regions were selected through a meta-analysis of previously published human ChIP-seq profiles, namely 161 for CTCF (in 89 cell lines or primary cells), 46 for subunits of cohesin (8 targeting SMC3 and 38 targeting RAD21, corresponding to multiple profiles across 5 and 11 cell lines or primary cells, respectively, for SMC3 and RAD21), and 8 for ZNF143 (in 4 cell lines or primary cells). ZNF143 has been shown to bind together with CTCF and cohesin and to be specifically enriched at domain boundaries (62). Briefly, to identify the strongest, most conserved insulator sites in the human genome, site-specific scoring and spatial clustering of CTCF, cohesin, and ZNF143 binding across different cell types were calculated and combined. First, consistently derived, enriched regions from ENCODE datasets (61) were downloaded from the UCSC genome browser on July 16, 2016. ChIP-seqs for the same protein in the same cell line (or primary cells) were considered as replicates. Narrow peaks from replicates were merged. The union of the peaks was then computed, and each peak was re-annotated to the sum of the corresponding −log10(P value) of the overlapping peaks across replicates. To compare the binding profiles across cell types, the obtained scores were converted to percentiles. Given a cell type, percentiles from overlapping CTCF, cohesin, and ZNF143 peaks were then summed, resulting in site-specific scores. Separately for each cell type, nearby CTCF-bound regions were then clustered together if found within 10 Kbp from each other. Given each cluster, site-specific scores for each constituent region were combined, first for each cell type, and eventually across all the cell types considered, obtaining an overall score for each cluster. For the final design, the clusters were sorted according to this score, and starting from the highest scoring cluster, the top clusters covering 3 Mbp of the genome were considered. This way, >95% of previously annotated TAD boundaries (63) were covered by one or more clusters (keeping in mind the resolution limit of the corresponding HiC datasets, namely 40 Kbp). Promoter regions were selected according to the following strategy. Genes that are either annotated as ERα targets [from the MSigDB Hallmark datasets (64)], found in the PAM50 signature (65) or being annotated as cancer genes [Network of Cancer Genes version 6.0 (66)] while showing an FPKM ≥ 50 (FPKM = Fragments Per Kilobase of Exons per Millions sequenced) in bulk RNA sequencing data from either LTED-, TamR-, or FulvR-resistant cell lines (36) were considered. From this initial list, genes annotated as housekeeping (67) were excluded. Promoter regions [(−750, +250) from annotated transcriptional start sites] were derived from the refGene table of the UCSC genome browser on December 13, 2018. Within these regions, only those DNA stretches overlapping DHS (as described above for the putative enhancer regions) were retained. Regions of low mappability along with those mapping to either chromosome Y or the mitochondrial chromosome, as well as those overlapping segmental duplications, were excluded from the design. Regions of unique mappability were defined according to the UCSC genome browser track k50.Unique.Mappability.bb in the Hoffman Mappability collection. After performing an initial, small set of captures, the overall design was further improved by excluding the top and bottom 1% regions. The top 1% regions were responsible for ∼21% of the signal, and the bottom 1% for just ∼0.03% of the signal. Omission of these regions resulted in a more uniform coverage.
SIDP Screens
Two oligo pools for the SIDP library (n = 67,839 and 69,569 oligos respectively, see design information below) were synthesized by Twist Bioscience. Each 60 bp ssDNA oligos contained a 20 bp sgRNA sequence flanked by these sequences 5′-gccatccagaagacttaccg-3′ and 5′-gtttccgtcttcacgactgc-3′ used for PCR amplification and BbsI restriction enzyme-mediated cloning. The oligo pools were cloned into a modified pLKO-TET-ON plasmid by the Golden Gate method and the resulting product was used to transform Endura electrocompetent cells (Lucigen) according to the manufacturer’s protocol. The transformation efficiency was ≈500 fold higher than the SIDP library size and complete and even oligos representation was confirmed by NGS. Large-scale preps of bacteria cultures containing the sgRNA plasmid library were harvested using the Genopure plasmid maxi kit (Roche). SIDP library was packaged in lentiviral particles by large scale co-transfection of HEK293T cells with CELLECTA ready-to-use packaging plasmid (Cellecta—cat.no CPCP-K2A) using TRANSIT-LT1 transfection reagent (Mirus Biologicals—cat. no. MIR 2300) according to manufacturer guidelines.
MCF7, LTED, and T47D cells were engineered to stably express dCas9-KRAB by lentiviral transduction and selected using 10 μg/mL blasticidin (Invitrogen) and initially maintained in EMEM (Amimed #1-31S01-I; for MCF7 and LTED) or RPMI (Amimed # 1-41F01-l), 10% FBS (Seradigm #1500-500, Lot:077B15), 2 mmol/L L-glutamine, 1 mmol/L sodium pyruvate, 10 mmol/L HEPES, and 1% P/S. Homogeneous dCas9-KRAB expression was confirmed by intracellular staining using Cas9 antibody (Cell Signaling Cat-14697) according to the manufacturer’s protocol.
MCF7-dCas9-KRAB, LTED-dCas9-KRAB, and T47D-dCas9-KRAB cells were then infected with SIDP lentiviral particles at low MOI (≈0.3) in two independent replicates. We transduced ≈1,000 cells per plasmid present in the library to guarantee a good representation of all sgRNAs in the population of cells under screening. The cells were selected using 2 μg/mL puromycin (Invitrogen) starting at 24 hours posttransduction and maintained in culture in CellStacks (Corning) in the described conditions and for the indicated time points. Cells were then harvested and gDNA isolated using the QIAamp DNA maxi kit (QIAGEN). Amplicons containing the sgRNA sequences were amplified using NEBNext High-Fidelity (NEB) and their representation was analyzed by next-generation sequencing (HiSeq2500, Illumina). During SIDP, for +E2 condition (full growth media +estrogen) cells were maintained in DMEM (Gibco #11885-084; for MCF7) or RPMI (Gibco #11875093) supplemented with 10% FBS (Seradigm #1500-500, Lot:077B15), 10 mmol/L HEPES, 1 mmol/L sodium pyruvate, and 1% P/S. For −E2 (estrogen-deprived media) cells were maintained in phenol-free DMEM (Gibco #11880-028; for MCF7 and LTED) or phenol-free RPMI (Gibco #11835030) supplemented with 10% FBS, charcoal-stripped, USDA-approved regions (Gibco #12676029), 2 mmol/L L-glutamine, 10 mmol/L HEPES, 1 mmol/L sodium pyruvate, and 1% P/S.
Flow Cytometry–Based Cell Competition Assays
MCF7-dcas9KRAB cells were infected with a modified pLKO-TET-ON lentiviral vector to deliver constitutively expressed sgRNAs in the target cells. Cells transduced with targeting sgRNAs (expressing mCherry) or nontargeting sgRNAs (expressing GFP) were mixed (ratio 2:1 mCherry: GFP) and maintained in culture as described above. At each time point, cells were harvested and analyzed by flow cytometry using CitoFLEX S (Beckman Coulter). We recorded at a minimum of 2,000 single cells for each condition, and the results were analyzed using FlowJo.
IncuCyte-Based Competition Assays
MCF7-dCas9-KRAB cells were engineered by lentiviral transduction containing a vector expressing NLS-eGFP (kindly provided by Dr. Chun Fui Lai, Imperial College London). Transduction efficiency was evaluated with EVOS XL Core Imaging System microscope (Thermo Fisher—AMEX100), and a population of bright GFP-positive cells was obtained by FACS. Sorting was performed by the Flow Cytometry facility at MRC London Institute of Medical Sciences. MCF7-NLS-eGFP-dCAS9KRAB cells were then transduced with lentiviral particles containing plasmids expressing individual sgRNAs and selected with puromycin (Sigma-Aldrich cat no. P8833). For each gene of interest, 150 eGFP-positive (targeting sgRNA) and 150 transparent (NTC-sgRNA) MCF7-dcas-9KRAB cells were seeded per well in a 96 wells ImageLock plate (Sartorius—cat no 4379) both in the presence and absence of estradiol [Complete medium with 10% FCS ± 17-ß estradiol 1 × 10–8 mol/L (Sigma-Aldrich cat no. E-060)] in parallel, for a total of 10 replicates per condition. The plate was routinely media changed and imaged daily with IncuCyte (IncuCyte ZOOM—Sartorius) using a Dual Color 10× 1.22 μm/pixel Nikon Air Objective (Sartorius cat no 4464). (Green filter: Ex 440/480 nm, Em 504/544 nm). The IncuCyte ZOOM Live-cell analysis system software was used to perform automated cell imaging over time and to calculate cell-by-cell segmentation employing a manually adjusted segmentation mask used to train the images taken at each time point. The total percentage of confluency and the total GFP-positive area percentage were automatically registered by the software and used to calculate the ratio between the two parameters normalized to day 0, to highlight an increase (>1: fitness) or a decrease (<1: vulnerability) in the trend of GFP-targeting representation over the non-targeting one. Numbers of green nuclei were also automatically counted by the software to obtain the GFP+ only cell count.
qPCR Analysis
RNA was extracted from dcas9-KRAB-MCF7 cells transduced with targeting and nontargeting sgRNA (Qiagen, cat no. 74016). RNA was retrotranscribed using iScript (Bio-Rad, cat no. 1708891). Quantitative PCR was performed with QuantStudio3 Real-Time PCR instrument (Applied Biosystems, cat.no A28567) using an SYBR-green PCR master mix reporter (Applied Biosystems, cat no. 4309155) and the following primers, designed around the promoter of the repressed genes. USP8 fwd: GGGTCTTGGGCCCTAGCA, rvrs: CAGAGCTTGTCTCCGGGGTA—MYD88 fwd: CTGCTCTCAACATGCGAGTG,rvs: CAGTTGCCGGATCTCCAAGT—TLR5 fwd: GCGCGAGTTGGACATAGACT, rvrs: GAGGTTTTCAGGAGCCCGAG).
Tissue Specimens
Longitudinal formalin-fixed paraffin-embedded (FFPE) HDBC samples were retrospectively collected from 100 patients. Samples from 61 patients were collected from Professor Giancarlo Pruneri at The European Institute for Oncology, Milan. Samples from 26 patients were collected from Professor Andrea Rocca at The Cancer Institute of Romagna, Meldola. The remaining 14 patient samples were collected from Professor Maria Vittoria Dieci at The Institute of Oncology Padova. We have obtained written informed consent from all patients. This study was conducted in accordance with recognized ethical guidelines (Declaration of Helsinki). Tissue collection was approved by each respective institutional review board. Germline DNA was extracted from normal lymph nodes (FFPE). The material was collected in the form of 10-µm slices. Detailed clinical notes were provided for each patient including age at diagnosis, tumor grade, percentage of ER-positive cells, percentage of PR-positive cells, percentage of Ki-67 high cells, percentage of HER2-positive cells, years until relapse, metastatic site, type of chemotherapy, and type of hormonal therapy. A full summary of the clinical data can be found in Supplementary Table S3.
Sample Preparation Workflow Extraction
DNA was extracted from 10-µm slices using the Qiagen GeneRead DNA FFPE extraction kit (Qiagen, Catalog no. 180134) which includes a Uracil N Glycosylase enzyme treatment to reduce FFPE artifacts. DNA quality and quantity were assessed using an Agilent Tapestation 2200 using the Genomic DNA screentape and reagents (Agilent, Catalog no. 5067-5365 and 5067-5366). Samples were sonicated custom number of cycles to achieve fragments of uniform length. Postsonication samples were quality controlled using the Tapestation 2200 instrument with a threshold set for samples to have at least 60% of fragments between 100 and 500 bp to proceed with processing. DNA underwent a second treatment with NEBNext FFPE DNA Repair Mix (NEB, Catalog no. M6630) to further reduce FFPE artifacts.
Library Preparation and Capture
DNA libraries were prepared from 30 ng to 1 μg of DNA using the NEBNext Ultra 2 DNA library kit for Illumina sequencing. Unique dual 8 bp indexes were used for each sample (a gift from Paolo Piazza of the Imperial British Research Council Genomics Facility). DNA libraries from 15 samples were pooled and captured with the SIDV capture probes produced by Twist Biosciences (ratio of 1.5 μg DNA libraries, 100 ng each, to 800 ng of capture probes). Noncaptured DNA was recovered using SPRI size selection beads to be used for a secondary capture. Postcapture amplification was performed using the KAPA HiFi Hot Start PCR ReadyMix Kit (KAPA Biosystems, Catalog no. KK2601). Postcapture amplified libraries were quality controlled and quantified using a Tapestation 2200 with the High Sensitivity reagents.
Sequencing
The initial 40 patients were sequenced on an Illumina HiSeq 4000 Instrument (Standard mode, 2 × 150 bp). After sequencing the initial 40 patients, sequencing was then performed by Novogene on an Illumina NovaSeq 6000 using paired-end 150bp reads. An average of 176 million reads per sample was achieved.
Raw Data Processing of the Captured DNA
First, paired-end reads from each sample were trimmed for adapter sequences and based on quality using Trim Galore (version 0.6.4; http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) in –paired mode. Alignment to the hg38 genome was then performed using bwa mem (version 0.7.15; https://arxiv.org/abs/1303.3997) using default parameters. The hg38 reference genome along with the corresponding annotation and known variant files mentioned in this and the following paragraphs were part of the Broad Institute Bundle, as per download from the Broad FTP on February 5th, 2018. Sambamba [version 0.7.1 (68)] was then used to convert the resulting SAM to a BAM file (using Sambamba view -S -h -F “not unmapped” -f bam). Sambamba sort and index were then used for sorting and indexing the resulting BAM file. The markdup function from Sambamba was used to mark potential PCR duplicates. Recalibration of base quality scores was performed using GATK4 [version 4.1.3.0 (69)]. The BaseRecalibrator function was run (providing dbSNP version 146 via the parameter –known-sites) followed by ApplyBQSR. The resulting BAM file with recalibrated scores was indexed using Sambamba. Final metrics for each sample were computed using the CollectHsMetrics function of the Picard tools (version 2.20.6; http://broadinstitute.github.io/picard/).
Mutational Calling Pipeline
To robustly identify SNVs and short INDELs, a pipeline deriving a consensus between three independent tools (Mutect2, Platypus, and Strelka) was deployed. Mutect2 [part of GATK4 version 4.1.3.0 (70)] was run individually on each primary and metastatic sample using the matched normal as reference. The -L option was used to specify the targeted regions. The file af-only-gnomad.hg38.vcf.gz acted as the source of germline variants with estimated allele frequency (as specified via the –germline-resource option). Parameters –af-of-alleles-not-in-resource 0.001, –disable-read-filter MateOnSameContigOrNoMappedMateReadFilter and –f1r2-tar-gz were also specified. The output from running the –f1r2-tar-gz option was then used to learn an orientation biased model (separately for each sample), leveraging the LearnReadOrientationModel function of GATK4. This allows estimating the substitution errors occurring because of damage induced by FFPE, by identifying residues showing a significant bias of substitutions on a single strand. The resulting model was then fed into the FilterMutectCalls function of GATK4 so that potentially affected residues can be flagged for subsequent filtering (see the bullet points later in this section for details about filtering).
Platypus [version 0.8.1.2 (71)] was run on each patient, jointly considering the normal as well the primary and metastatic profiles. The union of the variants called by Mutect2 separately on the primary and metastatic samples was used as prior (-source option). Option -minReads was set to 4.
Strelka [version 2.9.10 (72)] was run independently for each primary and metastatic sample using the matched normal as a reference, with default parameters. Although both Mutect2 and Platypus jointly identify SNVs and INDELs, Strelka relies on Manta [version 1.6.0 (73)] for the detection of INDELs. Manta was run first, and the resulting list of candidate INDELs was then provided to Strelka via the –indelCandidates option.
Considering the resulting lists of SNVs and INDELs, both common and tool-specific filters were applied to the lists generated by the different tools. General filters included:
-
•
A minimum depth of 20 reads was applied to both normal and tumor samples.
-
•
A minimum alternate allele coverage of two reads.
-
•
Exclusion of variant overlapping known SNPs (dbSNP version 146).
Tool-specific filters were set as follows:
-
•
Mutect2: after running FilterMutectCalls (GATK4) which also considered FFPE artifacts as estimated by the orientation bias model, only those variants marked as PASS were retained.
-
•
Platypus: all variants flagged by the tool were discarded, except those marked as PASS or including just one or more of the following flags: badReads, HapScore, alleleBias.
-
•
Strelka: only variants marked as PASS were kept for further analyses.
-
•
Of the resulting filtered variants, only those SNVs or short INDELs that were consistently identified by at least two out of three calling algorithms, very retained for further investigation.
Copy Number Calling Pipeline
CNVkit [version 0.9.7 (74)] was run in batch mode on the tumor bam files, using all normal bam files of each capturing-sequencing batch as input for the option -normal. SIDV3 intervals were specified under option—targets. The reference genome used for mutational calling was employed (Broad Bundle).
Purity and CCF estimation
To estimate the CCF of each SNV, only SNVs with an estimated copy number of two were considered. Separately for each sample, the SNVs fulfilling this criterion were hierarchically clustered based on their VAF (using Euclidean distance and complete linkage). The dendrogram was then cut at a fixed height of 0.15, and the cluster with the larger mean VAF was identified. This mean VAF was then used to estimate the purity of the sample: purity = VAFmean * 2. The CCF of each variant was then calculated starting from its VAF and the estimated purity for the sample, using the following formula: CCF = VAF * (2 * (1 − purity) + CNA_TOT * purity)/(CNA_MUT * purity) (75). Although CNA_TOT was known (2, see above), each variant was assumed to be heterozygous, with CNA_MUT set to be 1 (75).
Data Collection and Preprocessing to Train the DeltaSVM Models
A manually curated list of previously published, high-quality human ChIP-seq datasets from luminal breast cancer cell lines was compiled. Only those having a high-quality model (position weight matrix or PWM) describing their binding preferences were considered. The reason behind this choice is that knowing the binding preferences was a prerequisite to generate well-controlled negative sets for the deltaSVM models. Briefly, each PWM was used for genome-wide predictions of binding sites specific for each TF, to then derive a positive (predicted TF-binding site showing a ChIP-seq peak) and a negative (predicted TF-binding site, that could be in principle be contacted by the TF, but without a ChIP-seq peak) training set. This selection resulted in 72 ChIP-seq, corresponding to 43 transcription factors. Peaks in BED format were downloaded from the Gene Expression Omnibus (76). Regions in hg18 or hg19 coordinates were converted to hg38 using liftOver (77) and then filtered against the ENCODE blacklists (78) using BEDTools (79).
Predicting the Functional Effects of the Identified Variant
Available, precomputed genome-wide predictions were used to assess the impact of somatic variants on chromatin accessibility [Sasquatch (53)], mRNA splicing [Splicing Clinically Applicable Pathogenicity prediction or S-CAP (54)], and protein-coding sequence [Cancer Genome Interpreter or CGI (80)]. Available models based on deep learning [DeepSEA (56)] were used to compute the overall DIS of each variant. Support vector machines (SVM) were instead trained to predict the impact of somatic variants on the binding affinity of luminal breast cancer-relevant TFs. For each one of the different functional categories, the predictions were obtained as follows:
-
•
Chromatin Accessibility: The Sasquatch R package version 0.1 (https://github.com/Hughes-Genome-Group/sasquatch) was used to assess the impact of the identified somatic variants using the available model pre-trained with ENCODE_DUKE_MCF7_merged DNase-seq dataset. Briefly, hg38 coordinates were converted to hg19 using liftOver (77). Analysis of multiple reference-alternative alleles pairs was then performed using the RefVarBatch wrapper, using DNase as fragmentation type: (frag. type = “DNase”) and human as propensity source (pnorm.tag = “h_ery_1”). Empirical P values were estimated separately for observing a predicted increase or decrease in accessibility. A null distribution was derived from the COSMIC noncoding database (55), which contains millions of variants from different cancer types. Version 92 (08.2020) was downloaded as a flat file on October 12th, 2020. Sasquatch was run on the entire set of variants, but only those overlapping with the SIDV3 intervals were retained to compute the null.
-
•
mRNA splicing: Full S-CAP predictions (scap_COMBINED_v1.0.vcf) were downloaded from http://bejerano.stanford.edu/scap/ on August 27th, 2019. A custom Python script was prepared to annotate the somatic variants with these predictions.
-
•
Protein-coding sequence: The list of candidate somatic mutations was submitted to the CGI webserver on December 1, 2020 (https://www.cancergenomeinterpreter.org/). Also, in this case, hg38 coordinates were converted to hg19 using liftOver (77).
-
•
DIS: models from DeepSEA version 3 were used to estimate this. Hg38 coordinates were converted to hg19 using liftOver (77) and a corresponding null distribution leveraging COSMIC was computed as described above for chromatin accessibility.
-
•
TF-binding affinity: deltaSVM (51) was used to predict significant effects of a somatic variant in decreasing on increasing the affinity of the region for a given TF. First, for each considered PWM a genome-wide map of the high-affinity sites in the human genome (hg38) was predicted using FIMO((81)81). FIMO was run with the following parameters: –thresh 1e-4 –no-qvalue –max-stored-scores 10,000,000, separately for each motif. Regions of unique mappability (as defined according to the UCSC genome browser track k50.Unique.Mappability.bb in the hoffmanMappability collection) were defined using BEDTools (79), and only those were retained for the next steps. This information was coupled to the corresponding TF-ChIP-seq, to derive a positive (predicted TF-binding site showing a ChIP-seq peak) and a negative (predicted TF-binding site, that could be in principle be contacted by the TF, but without a ChIP-seq peak) training set. Each region in these two sets was defined as the 100 bps of genomic DNA centered on the predicted, high-affinity site. The actual training set used were randomly subsampled versions of these two sets (n = 10,000). Training of the SVM discriminating the positive from the negative examples was performed by running gkmsvm_kernel (with option -d set to 3) followed by gkmsvm_train. After that, gkmsvm_classify was used to generate a weighted list of all possible 10-mers, in which each 10-mer is assigned a SVM weight corresponding to its contribution to the prediction. With this list of weights, it was possible to predict (using the script deltasvm.pl) the impact of any sequence variant on the regulatory activity of a given region. One limitation of this approach when comparing models generated with very different data (like in this case for different TFs) is to define model-specific thresholds. To overcome this, the set of genomic regions under investigation was randomly mutagenized, resulting in a dataset in which every sequence was mutagenized at five residues (to all the three possible variants). The resulting values were used to compute model-specific null distributions that were used to estimate empirical P values for the predicted effects of the real set of mutations.
Variant Classification
A variant was classified as potentially pathogenic if meeting at least one of the following conditions:
-
•
Annotated as either Missense, Nonsense, or Frameshift by the CGI.
-
•
Showing an empirical P value equal or lower than 0.05 in terms of either DIS (DeepSEA) or predicted increase or decrease in chromatin accessibility (Sasquatch), or for the affinity of any of the 43 transcription factors considered in the deltaSVM models.
-
•
Showing any of the following S-CAP scores: (i) score ≥ 0.006 in case of mutations in the introns upstream of a 3′ SS or downstream of a 5′ SS; (ii) score ≥ 0.033 in case of a mutation in the 3′ AG (3′ SS core); (iii) score ≥ 0.009 in case of synonymous exonic mutation; (iv) score ≥ 0.034 for a mutation in the 5′ GT (5′ SS core); (v) score ≥ 0.005 in case of variants lying in the canonical U1 snRNA-binding site, excluding the 5′ SS core (5′ extended); (vi) score ≥ 0. 006.
Identification of Regions Showing an Excess of Regulatory Mutations in the Tumor Samples Cohort
Given a regulatory element targeted by the enrichment strategy, the probability of a given region to show an excess of mutations predicted as pathogenic was evaluated based on a binomial distribution. The expected probability P was estimated as the fraction of variants predicted as pathogenic in the entire datasets. The pbinom function from R was used to calculate the probability of seeing a better number of q pathogenic variants in the region, given the expected probability P and the total number of variants n identified in the region [pbinom (q, n, P, lower.tail = FALSE)].
Recurrence Analysis Using the HMF Metastatic, Breast Cancer Cohort
SNVs from 551 ER-positive, HER2-negative metastatic breast cancer samples from the Hartwig Medical Foundation (HMF; ref. 58) were used for the analysis. Each SID locus was enlarged by 1 kbp each side and then the mutational burden of each region was estimated as the total number of SNVs in the cohort overlapping the interval. To control for local differences in the propensity of each region to accumulate a different number of SNVs, the number of SNVs per base pair in a 1 Mbp window centered on the SID locus was used as proxy for the background (expected) mutational rate. After that, the SID regions were split into deciles based on their background mutational rate, and the number of SNVs at each locus was converted to a P value, as the fraction of loci in the decile showing an equal or higher number of SNVs. These P values were used as proxy for how good each region ranked in each respective decile, and then specifically to rank the 63 regions identified with an excess of alterations in SIDV that were predicted as functional.
Coding Variants Panel Design
To profile the coding genome in these patients, a refined panel of genes known as the Oncomine panel was utilized, specifically designed to cover key areas of mutation in luminal breast cancers (82). The panel targets 6,812 coding regions selected by compiling commonly mutated sites identified in up-to-date studies, sequencing both primary and metastatic luminal breast cancer tumors. The panel utilized data from an array of databases and studies including: TCGA database, the METABRIC database (83), Lefebvre and colleagues (84), the MSKCC IMPACTTM study (85), the AACR GENIE database (86), the COSMIC database, the Cancer Gene Census, and the Pharmacogenomics Knowledgebase (PharmKGB; ref. 87). In total, these datasets included 1,673 primary and 1,596 metastatic luminal breast cancer cases. Mutated genes identified in these datasets were compiled and refined using the following criteria. Sites that were mutated in at least 2% of primary or metastatic samples and CNVs with a frequency of more than 5% or with a fold change of more than 5% in either primary or metastatic tumors were compiled. All breast cancer genes reported in the Cancer Gene Census and all pharmacogenomic SNPs related to breast cancer in the PharmKGB database were compiled. Finally, some manual curation was included, adding in the CYP19A1 and SQLE amplification (88, 89). After refinement, the panel included 6,812 regions covering 134 genes, 27 CNV sites, 37 germline cancer genes, and 59 germline loci, with associations to pharmacogenomic interactions.
Sample Preparation and Sequencing
Secondary captures, on SIDV, captured DNA libraries, was carried out using the Oncomine panel. After hybridization of SIDV capture probes to complementary DNA and purification, noncaptured DNA was recovered and concentrated using SPRI size-selection beads. Quality control assessment using a Tapestation 2200 instrument was performed reporting that, in all cases, at least 50% recovery of initial DNA concentrations before the SIDV capture had been achieved. A custom set of capture probes for the Oncomine regions were produced by Twist Biosciences. Pools of DNA were captured using the Oncomine panel and quality controlled as previously described with the SIDV panel. Pools of 10 patients were sequenced at Novogene on an Illumina NovaSeq 6000 (150 bp paired-end), with 700 million reads per pool.
Computational Analysis of Coding Variants
Variant calling was initially performed for all 100 patients that were sequenced—matched normal, primary, and metastatic samples. Adapter trimming was performed using Trim Galore version 0.6.4 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Bwa-mem version 0.7.15 was used for alignment to the hg38 human genome reference. Sambamba (68) version 0.7.0 was used for conversion to binary, removal of PCR duplicates, sorting and indexing. Preprocessing before variant calling was performed using GATK (90), version 4.1.3.0: read groups were added using Picard version 2.20.6 (https://sourceforge.net/projects/picard/files/picard-tools/), base quality recalibration using GATK BaseRecalibrator and GATK ApplyBQSR. Mutect2 was used for somatic variant calling against the matched normal bam samples: using the germline resource from the GATK resource bundle af-only-gnomad.hg38.vcf.gz with option –af-of-alleles-not-in-resource set as 0.001 and with MateOnSameContigOrNoMappedMateReadFilter disabled. To flag possible FFPE artifacts, GATK LearnReadOrientationModel was run, using output during the filtering of variants with FilterMutectCalls. Only PASS mutations were further processed. Depth was checked at 500 mutated loci [variants with a FATHMM score ≥ 0.8 and a variant allele frequency (VAF) of at least 0.1 from the pool of de novo metastatic mutations] in all 100 patients—across normal, primary, and metastatic—using Samtools depth. This analysis revealed that in 42/100 patients, depth was lower than 10 in most of the loci, in at least one of the normal, primary, or metastatic bam files. As this low number of reads could affect variant detection generally or affect the identification of de novo metastatic variants (i.e., impossible to discern whether a mutation found in the metastatic sample was not present in the primary if the depth at that locus is low in the primary). As depth was sufficient across all variants in the other 58 patients, these were further processed. Variant annotation was performed using OpenCRAVAT, filtering for mutations only found in established breast cancer driver genes (91). To discover potential de novo driver variants of metastasis in these patients, we filtered for non-synonymous coding variants, with ≥0.1 VAF, private to metastasis or with an allele frequency at least five times higher than in the primary. ComplexHeatmap version 2.9.3 was used to generate an OncoPrint heatmap of these de novo, possibly pathogenic variants.
CRISPRi Screen: sgRNA Design
First, promoter-associated SIDV3 regions were excluded (a more tailored design of sgRNAs guided by available CAGE tags data in MCF7 was performed instead, see below for details). After enlarging each region to be at least 500 bps in size, the command-line version of the CRISPR-DO (92) tool [version 0.04 (93)] was then run separately for each one of the considered regions (with –spacer-len = 20), and the predicted sgRNAs stored. Only sgRNAs showing efficiency between 0.4 and 1.3, and specificity ≥ 80% were retained for further analyses. One G nucleotide was then added at both 5′ and 3′ of each sgRNA, and the resulting guides predicted to be digested by endonuclease BbsI were discarded. In silico digestion was performed using the digest package in R. After that, to obtain a more uniform distribution of sgRNAs, an iterative pruning procedure was applied until no two guides were found within 50 bps from each other. This resulted in 62.2% and 79.7% of the putative insulators and enhancers showing three or more sgRNAs targeting them, respectively. Only the sgRNAs targeting those regions were retained. hg19 coordinates for CAGE tags peaks from FANTOM5 (93) were downloaded from the consortium website (https://fantom.gsc.riken.jp/5/datafiles/latest/extra/CAGE_peaks/). Briefly, starting from hg19.cage_peak_phase1and2combined_tpm_ann.osc.txt.gz, only those expressed at least with a TPM ≥ 1 in unstimulated MCF7 were considered further. For each gene (after filtering for blacklisted regions in ENCODE and for promoters of antisense, noncoding RNAs) the dominant transcription start site (TSS; based on highest CAGE TPM) was identified. Only a single, dominant TSS for each expressed gene was retained. Of those, only those corresponding to promoters of genes with at least one overlapping putative insulator or enhancer in SIDV3 were considered for sgRNA design. Considering the directionality of transcription at each CAGE tags cluster, each region was standardized to [−100, +300] bps from the dominant position in the cluster. Design and filtering of the sgRNAs were then performed as described in the previous paragraph.
CRISPRi Screen: Data Analysis
Count data were normalized according to the weighted trimmed mean of the log expression ratios [trimmed mean of M values (TMM)] normalization (94), using the calcNormFactors function from edgeR (95). Initial principal component analysis and clustering analyses indicated high similarity between the 7 days samples and the initial library. For this reason, the replicated 7 -day samples were used as a reference to identify statistically significant changes in abundance of sgRNAs at later time points, using edgeR (95). Briefly, after estimating dispersion using the estimateDisp function, generalized linear models (GLM) were fit separately to each condition (full and estrogen-depleted medium), using the glmFit function. Coefficients were retrieved with glmLRT, and significant changes were retained as those showing a Benjamini–Hochberg corrected FDR ≤ 0.05 and a linear fold change of at least 1.5, in either direction. This procedure was applied to MCF7 and LTED samples, and also to T47D data with minor modifications, that is, a replicate of the initial library was used as baseline. The same computational strategy was applied to compare the sgRNAs counts in full (+E2) versus estrogen-depleted media (−E2), at any given time point, for both MCF7 and T47D.
Survival Analyses
Kaplan–Meier analysis was performed as described previously (96). Three main cohorts were considered for this manuscript. A meta-cohort including several Affymetrix profiled individual cohorts, which were reprocessed as a single cohort, the TCGA cohort and the METABRIC cohort (97). For the analysis, patients were dichotomized based on the median expression of MYD88, TLR5, or USP8 and a Cox regression analysis was run (where possible, using covariates). The Kaplan–Meier survival plot and hazard ratio with 95% confidence intervals and log-rank P value were calculated and plotted in R using Bioconductor packages.
Statistical Analyses and Plotting Using R
Unless indicated otherwise, all the described statistical analyses and preparation of plots were performed in the statistical computing environment R v4 (www.r-project.org).
Data Access
SIDP CRISPR screen results are accessible following this link: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE197504. SIDV sequencing data have been deposited at EGA: http://ega-archive.org/studies/EGAS00001006340.
Data Availability
The R, Python, and bash scripts to reproduce analyses and figures have been deposited in Zenodo: http://zenodo.org/record/8097853.
Supplementary Material
Supplementary Table S1: Regions defined by SID (Systematic Identification of epigenetically Defined loci). For each region (hg38 genomic coordinates including chromosome, starting, and ending positions), the table indicates whether the region was selected as a gene promoter, putative enhancer, or putative insulator. Whether the region is covered by designed oligo baits for SIDV profiling, and the number of sgRNAs targeting the region in SIDP, are also indicated.
Supplementary Table S2: sgRNAs sequences and metadata for the SIDP assays. S2.1: for each sgRNA targeting a region in the human genome, an identifier (using the corresponding hg38 genomic coordinates), the DNA sequence, the genomic coordinates (hg38) including the strand, along with efficiency and specificity scores as estimated by CRISPR-do, are provided. S2.2: for each positive control or non-targeting sgRNA, a custom identifier is shown along with the DNA sequence.
Supplementary Table S3: SIDP results in MCF7 grown in full (red; +E2) media. S3.1: results of the differential abundance analysis for the positive controls and the non-targeting sgRNAs (as indicated in the genome_partition field). For each sgRNA, an identifier, the pool, and the results from the edgeR analysis are shown. Average abundance of the sgRNA at day 7 and 21 post infection is indicated as logCPM (counts per million). The log2-fold changes (log2FC) between day 21 and 7, and between day 21 and the initial library, are indicated, along with the FDR (Benjamini-corrected p-value). Two further fields indicate whether the sgRNA was identified as showing a significant increase in frequency (IF; FDR <= 0.05 and linear fold-change >= 1.5) or decrease (DF; FDR <= 0.05 and linear fold-change <= -1.5). S3.2: like S3.1 but listing the results for the sgRNAs targeting the genomic regions of interest. Hg38 coordinates are also included in this case. S3.3: summary of the results at the level of each SID region. For each region, hg38 coordinates are listed, along with the symbol of the nearest gene, and the distance to its TSS in bp (positive or negative values indicate the region is either downstream or upstream the TSS, respectively). The table then indicates whether the region was selected as a gene promoter, putative enhancer, or putative insulator. The number of sgRNAs targeting the enlarged region (indicated coordinates +- 1 kbp), is followed by information on the overlapping sgRNAs that scored significantly, separately for DF and IF. In both cases, the total number of significant guides, the corresponding fraction, and the FDR and log2FC of the highest scoring sgRNA are reported. A column indicating significance of one or more sgRNAs is also provided. S3.4: enriched terms in the set of genes close to the regions showing scoring sgRNAs, separately for the DF and the IF sets. For each group, hallmark sets showing a p-value <= 0.05 are included in the table. Statistics of the hyper-geometric test are shown, along with the total number and identity of the overlapping genes. S3.5: overlap between the regions identified in our +E2 MCF7 SIDP assay and previously published screens in breast cancer cell lines (marcotte: Marcotte et al. 2012; fei: Fei at al. 2019; Korkmaz: Korkmaz et al. 2019; ggg: Rui Lopes et al. 2020).
Supplementary Table S4: SIDP results in T47D grown in full (red; +E2) media. S4.1-3: the tables follow the same structure of S3.1-3.
Supplementary Table S5: SIDP results in MCF7 grown in white media (-E2). S5.1-5: the tables follow the same structure of S3.1-5.
Supplementary Table S6: SIDP results in T47D grown in white media (-E2) media. S6.1-3: the tables follow the same structure of S3.1-3.
Supplementary Table S7: SIDP results (MCF7 & LTED) summary. S7.1: regions showing at least one overlapping sgRNA scoring in at least one of the different conditions assayed. For each region (hg38 genomic coordinates), the table indicates whether this was selected as a gene promoter, putative enhancer, or putative insulator. It also shows the symbol of the nearest gene, and the distance to its TSS in bp (positive or negative values indicate the region is either downstream or upstream the TSS, respectively). For each condition (MCF7 +E2, MCF7 -E2 or LTED) and direction of the change (DF vs IF), the table indicates whether the region overlaps one or more (columns labeled “single”) vs two or more (columns labeled “multiple”) sgRNAs. S7.2: summary of the overlaps between either scoring sgRNAs (“guides”), regions showing at least one scoring sgRNA (“regions_single”), or regions showing two or more consistently scoring sgRNAs (“regions_multiple”) between pairs of conditions (as indicated by columns assay_1 and assay_2). The nature of the change (either DF or IF), along with the total number of overlapping sgRNAs or regions, and the corresponding fraction, are also indicated. S7.3: results of gene set enrichment analysis using the indicated gene sets and the set of genes close to the regions showing scoring sgRNAs, according to the indicated pattern (SIDP_set). Statistics of the hyper-geometric test are shown, along with the total number of the overlapping genes (count), the observed and expected overlaps, and the odds ratio. S7.4: total number of regions showing multiple, consistent, scoring sgRNAs, and relative split-up based on the distance (bp) to the nearest TSS (0-0.5 kbp, 0.5-2.5 kbp, 2.5-5 kbp, 5-10 kbp, over 10 kbp), per condition (MCF7 +E2, MCF7 -E2 or LTED) and direction of the change (DF vs IF).
Supplementary Table S8: Summary of SIDP results in T47D vs MCF7. S8.1-2: the tables follow the same structure of S8.1-2. S8.3: the table has the same structure of S8.3. S8.4: summary of the edgeR comparison between T47D and MCF7, for each medium (+E2 or -E2) and direction of the change (DF or IF). Results are presented similarly to tables S3.3. S8.5: total number of significant guides, regions with at least of significant guide, or regions with multiple guides, per medium (+E2 or -E2) and direction of the change (DF or IF).
Supplementary Table S9: SIDP results in LTED. S9.1: results of the differential abundance analysis for the positive controls and the non-targeting sgRNAs. The structure of the table is like S3.1. S9.2: results for the sgRNAs targeting the genomic regions of interest. The structure of the table is like S3.2.
Supplementary Table S10: Metadata of the clinical cohort profiled by SIDV. S10.1: for each donor, from which genetic material from matched normal, primary, and metastatic samples were derived, the following information is provided: the identifier for the samples; the center where the samples were collected; the sequencing batch; the age of diagnosis; the clinical features of the primary tumors; the indication of the metastatic sites. Legend: ER = estrogen-receptor alpha; PR = progesterone receptor; pct = percentage; HR = hormone therapy. S10.2: for each triplet of matched normal, primary and metastasis derived material, and separately for each one of the 100 donors, sequencing statistics are provided. Sequencing depth, fraction of the reads mapping to oligo baits, mean coverage on baits and corresponding fold-enrichment, and on-target mean coverage, are shown. The percentages (pct) of targeted bases covered at least 10x, 30x, 50x or 100x are also indicates.
Supplementary Table S11: De novo coding variants. The table lists all the variants (SNVs and short INDELs) identified in the profiled sub-cohort of 58 SIDV metastatic samples. Chromosome and position on the chromosome (hg38 coordinates) are indicated for each variant, along with the reference, detected alternative allele and its frequency, the gene symbol and the ensemble transcripts, the type of alteration and the codon change, and the identifier of the sample in which the variant was identified.
Supplementary Table S12: Summary of SNVs and INDELs identified by SIDV. S12.1: full list of SNVs and INDELs. Chromosome and position on the chromosome (hg38 coordinates) are indicated for each variant, along with the reference and detected alternative allele. Also, the table indicates the donor, and whether the variant allele was directly detected in the primary (P_CALL) and/or the metastatic material (M_CALL). S12.2: tumor purity estimation for each sample and site (P = primary; M = metastasis) is listed, along with the size of the subset of SNVs used for the purity estimation analysis. S12.3: final annotation of the SNVs after sample-specific purity correction. For each SNV, genomic coordinates, reference and alternative alleles, donor identifier, and evidence (filtered read counts) supporting the different alleles in normal (N), primary (P) and metastatic (M) samples are provided. For both primary and metastatic samples, the variant allele frequency (VAF), along with the estimated purity for the sample, the estimated copy number alterations of the region bearing the variant (CNA) and the purity-corrected VAF, or cancer-cell fraction (CCF), are indicated. S12.4: regions showing an enrichment in either amplifications (amp) or deletions (del) across the metastatic samples as compared to the matched primary samples, are indicated.
Supplementary Table S13: Computational predictions of the functional impact of the SNVs and short INDELs identified through SIDV. S13.1: for each variant, the type (SNV or INDEL_short) and its hg38 coordinates are listed, along with the symbol of the nearest gene, and the distance to its TSS in bp (positive or negative values indicate the region is either downstream or upstream the TSS, respectively). Reference and alternative alleles are also provided, along with whether the variant is computationally predicted to alter the molecular function of the genomic element bearing it (indicated as different “pathogenic” classes; column mutation_class) or not (“benign”). The table is then indicating, for each one of the models considered, whether the variant is predicted to significantly affect the indicated molecular function. S13.2: extract of S12.1, for three regions of interest.
Supplementary Table S14: Datasets used for the training of the TF-specific deltaSVM models. For each TF, the corresponding gene symbol, along with information about the cells from which the ChIP-seq binding profile was obtained, the treatment the cells were exposed to (if any), and reference to the corresponding records on the Gene Expression Omnibus, are indicated. Information about the matched, high-quality position weight matrix (PWM) utilized as source of information to infer the binding affinities of each TF is also provided. For each PWM, an identifier is indicated, along with the corresponding reference database or publication (including Pubmed ID).
Supplementary Table S15: Results of the enrichment analyses looking for binding sites of specific TFs accumulating a greater or a smaller number of genetic variants than expected by change. For each TF and category (mutations significantly increasing or decreasing affinity) the observed and expected fraction of mutations overlapping the TF-bound sites are indicated, along with the difference between these two fractions, and the p-value of the corresponding χ2 test. Considering each TF and the mutations affecting the affinity to its target sites either positively or negatively (based on the p-value of the test) TFs could be either classified as showing significantly more or less mutations than expected, or not significant (ns).
Supplementary Table S16: Downstream analyses considering only the SIDV inferred genetic alterations with predicted impact on function. S16.1: results of the binomial enrichment test. SID regions overlapping at least 2 SNVs predicted as pathogenic are included. Along with genomic coordinates (hg38) the total number of SNVs as well as the number of predicted pathogenic SNVs overlapping the region are indicated. The p-value and the q-value (after Benjamini-Hochberg correction) of the binomial test are indicated, along with annotation to the closest gene. The 63 regions considered for further analyses are highlighted in red. S16.2: same as S13.1 but considering all the regions assigned to the genes annotated to the same ontological terms together. The number and identity of the genes contributing to the overlap are indicated, along with p-value of the binomial test, and the q-value (after Benjamini-Hochberg correction). Statistically significant terms (q-value <= 0.05) are highlighted in red. S16.3: results of the analyses testing for the enrichment of mutations (either SNVs, short INDELs, or both; mutation_type column) with computationally predicted pathogenic effects in the sets of regions also showing a certain behavior in SIDP (CRISPRi_hit_type column). Observed and expected overlap are indicated, along with the odds ratio and the p-value (Chi-squared test).
Supplementary Table S17: Downstream analyses considering only the SIDV inferred genetic alterations with predicted impact on function and stratifying them by cancer-cell fraction (CCF) increase and decrease in metastatic samples. S17.1: summary of the results of the statistical tests performed to identify differences in the predicted impact of mutations stratified by change in CCF in metastatic samples compared to matched primary. The fraction of variants predicted as pathogenic and either showing an increase or a decrease in CCF (+- 0.1) was compared to that of those showing no change. P-values for the indicated features are shown (Chi-squared test). S17.2: similarly, the distribution of the predicted molecular effects of variants in the three groups (increase, decrease or no change in CCF) were compared using the Kruskal-Wallis test. S17.3: like S13.3 but testing for the enrichment of mutations with both computationally predicted pathogenic effects and a certain CCF increase or decrease in metastatic samples, that also show a certain behavior in SIDP.
Supplementary Table S18: Estimation of excess of mutations in the hormone-receptor positive breast cancer cohorts from the Hartwig Medical Foundation (HMF). The table lists, for each one of the regions showing an excess of functional alterations in the SIDV cohort (highlighted in table S13.1) the number of mutations in the region in both the SIDV and HMF cohort, along with the estimated background probability of seeing a mutation for the regions (using the HMF cohort) and the corresponding, empirical p-value of seeing the observed number of mutations, given the background probability (see Methods).
Supplementary Table S19: Overlap between regions with excess of functional alterations. The regions considered in these overlaps are highlighted in table S13.1. S19.1: overlap with noncoding drivers identified by PCAWG (as listed in Supplementary Table 5 of Rheinbay et al. 2020). S19.2: overlap with regulatory mutations from breast cancer samples, as identified by HMF (as listed in Supplementary Table 5 of Dietlein et al. 2022). S19.3: overlap with noncoding mutational hotspots identified by HMF (as listed in Supplementary Table 21 of Dietlein et al. 2022). S19.4: overlap with genes associated with mutation events in non-coding regions from previous studies, as annotated by HMF (as listed in Supplementary Table 22 of Dietlein et al. 2022).
Supplementary Figure S1. SIDP identifies Cis Regulatory regions in MCF7 with high reproducibility.
Supplementary Figure 2. SIDP identifies Cis Regulatory regions in T47D with high reproducibility.
Supplementary Figure 3. Estrogen deprivation in MCF7 cells introduce stochastic selective pressure in long-term CRISPRi screens.
Supplementary Figure 4. Estrogen deprivation in MCF7 cells introduce stochastic selective pressure in long-term CRISPRi screens.
Supplementary Figure 5. Estrogen deprivation in T47D cells introduce stochastic selective pressure in long-term CRISPRi screens.
Supplementary Figure 6. SIDP results are consistent across ER+ BC cell lines.
Supplementary Figure 7. SIDP identifies regulatory regions with context-dependent activity in MCF7 cells.
Supplementary Figure 8. SIDP identifies regulatory regions with context-dependent activity in MCF7 cells.
Supplementary Figure 9. SIDP identifies regulatory regions with context-dependent activity in MCF7 cells.
Supplementary Figure 10. SIDP identifies regulatory regions with context-dependent activity in LTED cells.
Supplementary Figure 11. Validation of DF and IF SIDP hits.
Supplementary Figure 12. Validation of DF and IF SIDP hits.
Supplementary Figure 13. The promoters of genes belonging to the TLR5 cascade are active in ER+ breast cancer patients.
Supplementary Figure 14. TLR5 expression in ER+ cells from normal and cancer samples.
Supplementary Figure S15. Validation of SIDP via chemical perturbation.
Supplementary Figure 16. Genes involved in TLR signaling are upregulated in MCF7 during therapy induced dormancy.
Supplementary Figure 17. Activation of TLR5 via paracrine signaling.
Supplementary Figure 18. SIDV design and results.
Supplementary Figure 19. SIDV functional analyses.
Acknowledgments
All the authors acknowledge and thank all patients and their families for their support and for donating research samples. The authors gratefully acknowledge infrastructure support provided by Imperial Experimental Cancer Medicine Centre, Cancer Research UK Imperial Centre, National Institute for Health Research, Imperial Biomedical Research Centre, and Imperial College Healthcare NHS Trust Tissue Bank. We thank the NIBR CBT Genomics unit and Michelle Piquet and David Ruddy from NIBR ONC IT&T for sequencing support. This publication and the underlying research are partly facilitated by Hartwig Medical Foundation and the Centre for Personalized Cancer Treatment which have generated, analyzed, and made available data for this research. L. Magnani was supported by a CRUK fellowship (C46704/A23110). I. Barozzi was supported by CRUK funding (C46704/A23110), an Imperial College Research Fellowship, and the Medical University Vienna. Consent was collected at European Institute of Oncology, Milan; Istituto Oncologico Veneto; and Istituto Tumori della Romagna. Other investigators may have received samples from these same tissues. A special thanks to Xixuan Zhu and Rakshindh Sekhon for their help in the initial crunching of the data and Giacomo Corleone for help with the initial selection of the SID regions. The authors also thank F. Battiato, Z.I. Magnani, and A.F. Magnani for their continuous support.
Footnotes
Note Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
M.V. Dieci reports personal fees from Eli Lilly, Novartis, Pfizer, Roche, Gilead, Seagen, Daiichi Sankyo, AstraZeneca, MSD, and Exact Sciences outside the submitted work, as well as a patent for EP20382679.7 licensed to Università di Padova. G. Pruneri reports grants from Fondazione AIRC per la Ricerca sul Cancro ETS - Project ID: 26320 PI: G. Pruneri during the conduct of the study, as well as personal fees from Novartis, Illumina, and Eli Lilly and Company outside the submitted work. G.G. Galli reports being an employee and a shareholder of Novartis. No disclosures were reported by the other authors.
Authors’ Contributions
I. Barozzi: Conceptualization, resources, software, formal analysis, supervision, funding acquisition, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. N. Slaven: Software, formal analysis, investigation, visualization, methodology, writing–original draft. E. Canale: Investigation. R. Lopes: Investigation. I. Amorim Monteiro Barbosa: Validation. M. Bleu: Validation, investigation, visualization. D. Ivanoiu: Software, validation, investigation, methodology. C. Pacini: Software, formal analysis, investigation, methodology. E. Mensa: Validation, investigation, visualization. A. Chambers: Validation, investigation. S. Bravaccini: Resources, investigation. S. Ravaioli: Resources. B. Gyorffy: Resources, software, formal analysis. M.V. Dieci: Resources, software, formal analysis. G. Pruneri: Resources. G.G. Galli: Resources, investigation, visualization, methodology, writing–original draft. L. Magnani: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
Disclaimer
The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health.
References
- 1. Festuccia N, Gonzalez I, Owens N, Navarro P. Mitotic bookmarking in development and stem cells. Development 2017;144:3633–45. [DOI] [PubMed] [Google Scholar]
- 2. He P, Williams BA, Trout D, Marinov GK, Amrhein H, Berghella L, et al. The changing mouse embryo transcriptome at whole tissue and single-cell resolution. Nature 2020;583:760–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Magnani L, Eeckhoute J, Lupien M. Pioneer factors: directing transcriptional regulators within the chromatin environment. Trends Genet 2011;27:465–74. [DOI] [PubMed] [Google Scholar]
- 4. Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, et al. Cell-of-Origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 2018;173:291–304.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gaiti F, Chaligne R, Gu H, Brand RM, Kothen-Hill S, Schulman RC, et al. Epigenetic evolution and lineage histories of chronic lymphocytic leukaemia. Nature 2019;569:576–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Polak P, Karlić R, Koren A, Thurman R, Sandstrom R, Lawrence MS, et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature 2015;518:360–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG, et al. A comprehensive map of molecular drug targets. Nat Rev Drug Discov 2017;16:19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Patten DK, Corleone G, Győrffy B, Perone Y, Slaven N, Barozzi I, et al. Enhancer mapping uncovers phenotypic heterogeneity and evolution in patients with luminal breast cancer. Nat Med 2018;24:1469–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 2012;481:389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Magnani L, Ballantyne EB, Zhang X, Lupien M. PBX1 genomic pioneer function drives ERα signaling underlying progression in breast cancer. PLoS Genet 2011;7:e1002368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 2008;132:958–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Aromatase inhibitors versus tamoxifen in early breast cancer: patient-level meta-analysis of the randomised trials. Lancet 2015;386:1341–52. [DOI] [PubMed] [Google Scholar]
- 13. Early Breast Cancer Trialists’ Collaborative Group (EBCTCG); Davies C, Godwin J, Gray R, Clarke M, Cutter D, Darby S, et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet 2011;378:771–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beatson G. ON the treatment of inoperable cases of carcinoma of the mamma: suggestions for a new method of treatment, with illustrative CASES.1. Lancet 1896;148:104–7. [PMC free article] [PubMed] [Google Scholar]
- 15. Pan H, Gray R, Braybrooke J, Davies C, Taylor C, McGale P, et al. 20-Year risks of breast-cancer recurrence after stopping endocrine therapy at 5 years. N Engl J Med 2017;377:1836–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hong SP, Chan TE, Lombardo Y, Corleone G, Rotmensz N, Bravaccini S, et al. Single-cell transcriptomics reveals multi-step adaptations to endocrine therapy. Nat Commun 2019;10:3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rosano D, Sofyali E, Dhiman H, Ghirardi C, Ivanoiu D, Heide T, et al. Long-term multimodal recording reveals epigenetic adaptation routes in dormant breast cancer cells. Cancer Discov 2024;14:866–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lopes R, Sprouffske K, Sheng C, Uijttewaal ECH, Wesdorp AE, Dahinden J, et al. Systematic dissection of transcriptional regulatory networks by genome-scale and single-cell CRISPR screens. Sci Adv 2021;7:eabf5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fei T, Li W, Peng J, Xiao T, Chen C-H, Wu A, et al. Deciphering essential cistromes using genome-wide CRISPR screens. Proc Natl Acad Sci U S A 2019;116:25186–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Perone Y, Farrugia AJ, Rodríguez-Meira A, Győrffy B, Ion C, Uggetti A, et al. SREBP1 drives Keratin-80-dependent cytoskeletal changes and invasive behavior in endocrine-resistant ERα breast cancer. Nat Commun 2019;10:2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nagarajan S, Rao SV, Sutton J, Cheeseman D, Dunn S, Papachristou EK, et al. ARID1A influences HDAC1/BRD4 activity, intrinsic proliferative capacity and breast cancer treatment response. Nat Genet 2020;52:187–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu G, Chhangawala S, Cocco E, Razavi P, Cai Y, Otto JE, et al. ARID1A determines luminal identity and therapeutic response in estrogen-receptor-positive breast cancer. Nat Genet 2020;52:198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 2015;161:1012–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lambuta RA, Nanni L, Liu Y, Diaz-Miyar J, Iyer A, Tavernari D, et al. Whole-genome doubling drives oncogenic loss of chromatin segregation. Nature 2023;615:925–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nora EP, Goloborodko A, Valton A-L, Gibcus JH, Uebersohn A, Abdennur N, et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 2017;169:930–44.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, et al. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 2015;162:900–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013;154:442–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Katainen R, Dave K, Pitkänen E, Palin K, Kivioja T, Välimäki N, et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet 2015;47:818–21. [DOI] [PubMed] [Google Scholar]
- 29. Rheinbay E, Nielsen MM, Abascal F, Wala JA, Shapira O, Tiao G, et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature 2020;578:102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang X, Meyerson M. Illuminating the noncoding genome in cancer. Nat Cancer 2020;1:864–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hinohara K, Wu H-J, Vigneau S, McDonald TO, Igarashi KJ, Yamamoto KN, et al. KDM5 histone demethylase activity links cellular transcriptomic heterogeneity to therapeutic resistance. Cancer Cell 2018;34:939–53.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010;141:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pagani O, Regan MM, Walley BA, Fleming GF, Colleoni M, Láng I, et al. Adjuvant exemestane with ovarian suppression in premenopausal breast cancer. N Engl J Med 2014;371:107–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rueda OM, Sammut S-J, Seoane JA, Chin S-F, Caswell-Jin JL, Callari M, et al. Dynamics of breast-cancer relapse reveal late-recurring ER-positive genomic subgroups. Nature 2019;567:399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Magnani L, Stoeck A, Zhang X, Lánczky A, Mirabella AC, Wang T-L, et al. Genome-wide reprogramming of the chromatin landscape underlies endocrine therapy resistance in breast cancer. Proc Natl Acad Sci U S A 2013;110:E1490–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nguyen VTM, Barozzi I, Faronato M, Lombardo Y, Steel JH, Patel N, et al. Differential epigenetic reprogramming in response to specific endocrine therapies promotes cholesterol biosynthesis and cellular invasion. Nat Commun 2015;6:10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shaw LE, Sadler AJ, Pugazhendhi D, Darbre PD. Changes in oestrogen receptor-α and -β during progression to acquired resistance to tamoxifen and fulvestrant (Faslodex, ICI 182,780) in MCF7 human breast cancer cells. J Steroid Biochem Mol Biol 2006;99:19–32. [DOI] [PubMed] [Google Scholar]
- 38. Chen P, Zhou Y, Li X, Yang J, Zheng Z, Zou Y, et al. Design, synthesis, and bioevaluation of novel MyD88 inhibitor c17 against acute lung injury derived from the virtual screen. J Med Chem 2023;66:6938–58. [DOI] [PubMed] [Google Scholar]
- 39. Sammut S-J, Crispin-Ortuzar M, Chin S-F, Provenzano E, Bardwell HA, Ma W, et al. Multi-omic machine learning predictor of breast cancer therapy response. Nature 2022;601:623–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gong T, Liu L, Jiang W, Zhou R. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat Rev Immunol 2020;20:95–112. [DOI] [PubMed] [Google Scholar]
- 41. Das N, Dewan V, Grace PM, Gunn RJ, Tamura R, Tzarum N, et al. HMGB1 activates proinflammatory signaling via TLR5 leading to allodynia. Cell Rep 2016;17:1128–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yanai H, Ban T, Wang Z, Choi MK, Kawamura T, Negishi H, et al. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature 2009;462:99–103. [DOI] [PubMed] [Google Scholar]
- 43. Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 2002;418:191–5. [DOI] [PubMed] [Google Scholar]
- 44. Venereau E, Casalgrandi M, Schiraldi M, Antoine DJ, Cattaneo A, De Marchis F, et al. Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J Exp Med 2012;209:1519–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Colombo M, Vallese S, Peretto I, Jacq X, Rain J, Colland F, et al. Synthesis and biological evaluation of 9-oxo-9H-indeno[1,2-b]pyrazine-2,3-dicarbonitrile analogues as potential inhibitors of deubiquitinating enzymes. ChemMedChem 2010;5:552–8. [DOI] [PubMed] [Google Scholar]
- 46. Thakore PI, D’Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods 2015;12:1143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, et al. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 2014;346:1373–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Harrod A, Fulton J, Nguyen VTM, Periyasamy M, Ramos-Garcia L, Lai C-F, et al. Genomic modelling of the ESR1 Y537S mutation for evaluating function and new therapeutic approaches for metastatic breast cancer. Oncogene 2017;36:2286–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bertucci F, Ng CKY, Patsouris A, Droin N, Piscuoglio S, Carbuccia N, et al. Genomic characterization of metastatic breast cancers. Nature 2019;569:560–4. [DOI] [PubMed] [Google Scholar]
- 50. Angus L, Smid M, Wilting SM, van Riet J, Van Hoeck A, Nguyen L, et al. The genomic landscape of metastatic breast cancer highlights changes in mutation and signature frequencies. Nat Genet 2019;51:1450–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee D, Gorkin DU, Baker M, Strober BJ, Asoni AL, McCallion AS, et al. A method to predict the impact of regulatory variants from DNA sequence. Nat Genet 2015;47:955–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhou J, Theesfeld CL, Yao K, Chen KM, Wong AK, Troyanskaya OG. Deep learning sequence-based ab initio prediction of variant effects on expression and disease risk. Nat Genet 2018;50:1171–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schwessinger R, Suciu MC, McGowan SJ, Telenius J, Taylor S, Higgs DR, et al. Sasquatch: predicting the impact of regulatory SNPs on transcription factor binding from cell- and tissue-specific DNase footprints. Genome Res 2017;27:1730–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jagadeesh KA, Paggi JM, Ye JS, Stenson PD, Cooper DN, Bernstein JA, et al. S-CAP extends pathogenicity prediction to genetic variants that affect RNA splicing. Nat Genet 2019;51:755–63. [DOI] [PubMed] [Google Scholar]
- 55. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res 2018;47:D941–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhou J, Park CY, Theesfeld CL, Wong AK, Yuan Y, Scheckel C, et al. Whole-genome deep-learning analysis identifies contribution of noncoding mutations to autism risk. Nat Genet 2019;51:973–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Smith RP, Taher L, Patwardhan RP, Kim MJ, Inoue F, Shendure J, et al. Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nat Genet 2013;45:1021–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Priestley P, Baber J, Lolkema MP, Steeghs N, de Bruijn E, Shale C, et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature 2019;575:210–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cowper-Sal lari R, Zhang X, Wright JB, Bailey SD, Cole MD, Eeckhoute J, et al. Breast cancer risk–associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet 2012;44:1191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mazrooei P, Kron KJ, Zhu Y, Zhou S, Grillo G, Mehdi T, et al. Cistrome partitioning reveals convergence of somatic mutations and risk variants on master transcription regulators in primary prostate tumors. Cancer Cell 2019;36:674–89.e6. [DOI] [PubMed] [Google Scholar]
- 61. Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mourad R, Cuvier O. Computational identification of genomic features that influence 3D chromatin domain formation. PLoS Comput Biol 2016;12:e1004908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012;485:376–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009;27:1160–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Repana D, Nulsen J, Dressler L, Bortolomeazzi M, Venkata SK, Tourna A, et al. The Network of Cancer Genes (NCG): a comprehensive catalogue of known and candidate cancer genes from cancer sequencing screens. Genome Biol 2019;20:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lin Y, Ghazanfar S, Strbenac D, Wang A, Patrick E, Lin DM, et al. Evaluating stably expressed genes in single cells. GigaScience 2019;8:giz106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics 2015;31:2032–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. WGS500 Consortium; Rimmer A, Phan H, Mathieson I, Iqbal Z, Twigg SRF, Wilkie AOM, et al. Integrating mapping-assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet 2014;46:912–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kim S, Scheffler K, Halpern AL, Bekritsky MA, Noh E, Källberg M, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods 2018;15:591–4. [DOI] [PubMed] [Google Scholar]
- 73. Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 2016;32:1220–2. [DOI] [PubMed] [Google Scholar]
- 74. Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol 2016;12:e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jiang Y, Qiu Y, Minn AJ, Zhang NR. Assessing intratumor heterogeneity and tracking longitudinal and spatial clonal evolutionary history by next-generation sequencing. Proc Natl Acad Sci U S A 2016;113:E5528–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, et al. The UCSC genome browser database: update 2006. Nucleic Acids Res 2006;34:D590–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Amemiya HM, Kundaje A, Boyle AP. The ENCODE blacklist: identification of problematic regions of the genome. Sci Rep 2019;9:9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010;26:841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tamborero D, Rubio-Perez C, Deu-Pons J, Schroeder MP, Vivancos A, Rovira A, et al. Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med 2018;10:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics 2011;27:1017–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zoppoli G, Garuti A, Belfiore A, Bonizzi G, Ferrando L, Vingiani A, et al. Abstract PD8-04: Ultra-deep multigene profiling of matched primary and metastatic hormone receptor positive breast cancer patients relapsed after adjuvant endocrine treatment reveals novel aberrations in the estrogen receptor pathway. Cancer Res 2020;80:PD8-04. [Google Scholar]
- 83. Mukherjee A, Russell R, Chin S-F, Liu B, Rueda OM, Ali HR, et al. Associations between genomic stratification of breast cancer and centrally reviewed tumour pathology in the METABRIC cohort. NPJ Breast Cancer 2018;4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lefebvre C, Bachelot T, Filleron T, Pedrero M, Campone M, Soria J-C, et al. Mutational profile of metastatic breast cancers: a retrospective analysis. PLoS Med 2016;13:e1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. The AACR Project GENIE Consortium; The AACR Project GENIE Consortium; André F, Arnedos M, Baras AS, Baselga J, Bedard PL, Berger MFet al. AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov 2017;7:818–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 2012;92:414–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Magnani L, Frigè G, Gadaleta RM, Corleone G, Fabris S, Kempe H, et al. Acquired CYP19A1 amplification is an early specific mechanism of aromatase inhibitor resistance in ERα metastatic breast cancer. Nat Genet 2017;49:444–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Brown DN, Caffa I, Cirmena G, Piras D, Garuti A, Gallo M, et al. Squalene epoxidase is a bona fide oncogene by amplification with clinical relevance in breast cancer. Sci Rep 2016;6:19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Martínez-Jiménez F, Muiños F, Sentís I, Deu-Pons J, Reyes-Salazar I, Arnedo-Pac C, et al. A compendium of mutational cancer driver genes. Nat Rev Cancer 2020;20:555–72. [DOI] [PubMed] [Google Scholar]
- 92. Ma J, Köster J, Qin Q, Hu S, Li W, Chen C, et al. CRISPR-DO for genome-wide CRISPR design and optimization. Bioinformatics 2016;32:3336–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. FANTOM Consortium and the RIKEN PMI and CLST DGT; Forrest ARR, Kawaji H, Rehli M, Baillie JK, de Hoon MJL, Haberle V, et al. A promoter-level mammalian expression atlas. Nature 2014;507:462–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010;11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 2012;40:4288–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Győrffy B, Lánczky A, Szállási Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer 2012;19:197–208. [DOI] [PubMed] [Google Scholar]
- 97. Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1: Regions defined by SID (Systematic Identification of epigenetically Defined loci). For each region (hg38 genomic coordinates including chromosome, starting, and ending positions), the table indicates whether the region was selected as a gene promoter, putative enhancer, or putative insulator. Whether the region is covered by designed oligo baits for SIDV profiling, and the number of sgRNAs targeting the region in SIDP, are also indicated.
Supplementary Table S2: sgRNAs sequences and metadata for the SIDP assays. S2.1: for each sgRNA targeting a region in the human genome, an identifier (using the corresponding hg38 genomic coordinates), the DNA sequence, the genomic coordinates (hg38) including the strand, along with efficiency and specificity scores as estimated by CRISPR-do, are provided. S2.2: for each positive control or non-targeting sgRNA, a custom identifier is shown along with the DNA sequence.
Supplementary Table S3: SIDP results in MCF7 grown in full (red; +E2) media. S3.1: results of the differential abundance analysis for the positive controls and the non-targeting sgRNAs (as indicated in the genome_partition field). For each sgRNA, an identifier, the pool, and the results from the edgeR analysis are shown. Average abundance of the sgRNA at day 7 and 21 post infection is indicated as logCPM (counts per million). The log2-fold changes (log2FC) between day 21 and 7, and between day 21 and the initial library, are indicated, along with the FDR (Benjamini-corrected p-value). Two further fields indicate whether the sgRNA was identified as showing a significant increase in frequency (IF; FDR <= 0.05 and linear fold-change >= 1.5) or decrease (DF; FDR <= 0.05 and linear fold-change <= -1.5). S3.2: like S3.1 but listing the results for the sgRNAs targeting the genomic regions of interest. Hg38 coordinates are also included in this case. S3.3: summary of the results at the level of each SID region. For each region, hg38 coordinates are listed, along with the symbol of the nearest gene, and the distance to its TSS in bp (positive or negative values indicate the region is either downstream or upstream the TSS, respectively). The table then indicates whether the region was selected as a gene promoter, putative enhancer, or putative insulator. The number of sgRNAs targeting the enlarged region (indicated coordinates +- 1 kbp), is followed by information on the overlapping sgRNAs that scored significantly, separately for DF and IF. In both cases, the total number of significant guides, the corresponding fraction, and the FDR and log2FC of the highest scoring sgRNA are reported. A column indicating significance of one or more sgRNAs is also provided. S3.4: enriched terms in the set of genes close to the regions showing scoring sgRNAs, separately for the DF and the IF sets. For each group, hallmark sets showing a p-value <= 0.05 are included in the table. Statistics of the hyper-geometric test are shown, along with the total number and identity of the overlapping genes. S3.5: overlap between the regions identified in our +E2 MCF7 SIDP assay and previously published screens in breast cancer cell lines (marcotte: Marcotte et al. 2012; fei: Fei at al. 2019; Korkmaz: Korkmaz et al. 2019; ggg: Rui Lopes et al. 2020).
Supplementary Table S4: SIDP results in T47D grown in full (red; +E2) media. S4.1-3: the tables follow the same structure of S3.1-3.
Supplementary Table S5: SIDP results in MCF7 grown in white media (-E2). S5.1-5: the tables follow the same structure of S3.1-5.
Supplementary Table S6: SIDP results in T47D grown in white media (-E2) media. S6.1-3: the tables follow the same structure of S3.1-3.
Supplementary Table S7: SIDP results (MCF7 & LTED) summary. S7.1: regions showing at least one overlapping sgRNA scoring in at least one of the different conditions assayed. For each region (hg38 genomic coordinates), the table indicates whether this was selected as a gene promoter, putative enhancer, or putative insulator. It also shows the symbol of the nearest gene, and the distance to its TSS in bp (positive or negative values indicate the region is either downstream or upstream the TSS, respectively). For each condition (MCF7 +E2, MCF7 -E2 or LTED) and direction of the change (DF vs IF), the table indicates whether the region overlaps one or more (columns labeled “single”) vs two or more (columns labeled “multiple”) sgRNAs. S7.2: summary of the overlaps between either scoring sgRNAs (“guides”), regions showing at least one scoring sgRNA (“regions_single”), or regions showing two or more consistently scoring sgRNAs (“regions_multiple”) between pairs of conditions (as indicated by columns assay_1 and assay_2). The nature of the change (either DF or IF), along with the total number of overlapping sgRNAs or regions, and the corresponding fraction, are also indicated. S7.3: results of gene set enrichment analysis using the indicated gene sets and the set of genes close to the regions showing scoring sgRNAs, according to the indicated pattern (SIDP_set). Statistics of the hyper-geometric test are shown, along with the total number of the overlapping genes (count), the observed and expected overlaps, and the odds ratio. S7.4: total number of regions showing multiple, consistent, scoring sgRNAs, and relative split-up based on the distance (bp) to the nearest TSS (0-0.5 kbp, 0.5-2.5 kbp, 2.5-5 kbp, 5-10 kbp, over 10 kbp), per condition (MCF7 +E2, MCF7 -E2 or LTED) and direction of the change (DF vs IF).
Supplementary Table S8: Summary of SIDP results in T47D vs MCF7. S8.1-2: the tables follow the same structure of S8.1-2. S8.3: the table has the same structure of S8.3. S8.4: summary of the edgeR comparison between T47D and MCF7, for each medium (+E2 or -E2) and direction of the change (DF or IF). Results are presented similarly to tables S3.3. S8.5: total number of significant guides, regions with at least of significant guide, or regions with multiple guides, per medium (+E2 or -E2) and direction of the change (DF or IF).
Supplementary Table S9: SIDP results in LTED. S9.1: results of the differential abundance analysis for the positive controls and the non-targeting sgRNAs. The structure of the table is like S3.1. S9.2: results for the sgRNAs targeting the genomic regions of interest. The structure of the table is like S3.2.
Supplementary Table S10: Metadata of the clinical cohort profiled by SIDV. S10.1: for each donor, from which genetic material from matched normal, primary, and metastatic samples were derived, the following information is provided: the identifier for the samples; the center where the samples were collected; the sequencing batch; the age of diagnosis; the clinical features of the primary tumors; the indication of the metastatic sites. Legend: ER = estrogen-receptor alpha; PR = progesterone receptor; pct = percentage; HR = hormone therapy. S10.2: for each triplet of matched normal, primary and metastasis derived material, and separately for each one of the 100 donors, sequencing statistics are provided. Sequencing depth, fraction of the reads mapping to oligo baits, mean coverage on baits and corresponding fold-enrichment, and on-target mean coverage, are shown. The percentages (pct) of targeted bases covered at least 10x, 30x, 50x or 100x are also indicates.
Supplementary Table S11: De novo coding variants. The table lists all the variants (SNVs and short INDELs) identified in the profiled sub-cohort of 58 SIDV metastatic samples. Chromosome and position on the chromosome (hg38 coordinates) are indicated for each variant, along with the reference, detected alternative allele and its frequency, the gene symbol and the ensemble transcripts, the type of alteration and the codon change, and the identifier of the sample in which the variant was identified.
Supplementary Table S12: Summary of SNVs and INDELs identified by SIDV. S12.1: full list of SNVs and INDELs. Chromosome and position on the chromosome (hg38 coordinates) are indicated for each variant, along with the reference and detected alternative allele. Also, the table indicates the donor, and whether the variant allele was directly detected in the primary (P_CALL) and/or the metastatic material (M_CALL). S12.2: tumor purity estimation for each sample and site (P = primary; M = metastasis) is listed, along with the size of the subset of SNVs used for the purity estimation analysis. S12.3: final annotation of the SNVs after sample-specific purity correction. For each SNV, genomic coordinates, reference and alternative alleles, donor identifier, and evidence (filtered read counts) supporting the different alleles in normal (N), primary (P) and metastatic (M) samples are provided. For both primary and metastatic samples, the variant allele frequency (VAF), along with the estimated purity for the sample, the estimated copy number alterations of the region bearing the variant (CNA) and the purity-corrected VAF, or cancer-cell fraction (CCF), are indicated. S12.4: regions showing an enrichment in either amplifications (amp) or deletions (del) across the metastatic samples as compared to the matched primary samples, are indicated.
Supplementary Table S13: Computational predictions of the functional impact of the SNVs and short INDELs identified through SIDV. S13.1: for each variant, the type (SNV or INDEL_short) and its hg38 coordinates are listed, along with the symbol of the nearest gene, and the distance to its TSS in bp (positive or negative values indicate the region is either downstream or upstream the TSS, respectively). Reference and alternative alleles are also provided, along with whether the variant is computationally predicted to alter the molecular function of the genomic element bearing it (indicated as different “pathogenic” classes; column mutation_class) or not (“benign”). The table is then indicating, for each one of the models considered, whether the variant is predicted to significantly affect the indicated molecular function. S13.2: extract of S12.1, for three regions of interest.
Supplementary Table S14: Datasets used for the training of the TF-specific deltaSVM models. For each TF, the corresponding gene symbol, along with information about the cells from which the ChIP-seq binding profile was obtained, the treatment the cells were exposed to (if any), and reference to the corresponding records on the Gene Expression Omnibus, are indicated. Information about the matched, high-quality position weight matrix (PWM) utilized as source of information to infer the binding affinities of each TF is also provided. For each PWM, an identifier is indicated, along with the corresponding reference database or publication (including Pubmed ID).
Supplementary Table S15: Results of the enrichment analyses looking for binding sites of specific TFs accumulating a greater or a smaller number of genetic variants than expected by change. For each TF and category (mutations significantly increasing or decreasing affinity) the observed and expected fraction of mutations overlapping the TF-bound sites are indicated, along with the difference between these two fractions, and the p-value of the corresponding χ2 test. Considering each TF and the mutations affecting the affinity to its target sites either positively or negatively (based on the p-value of the test) TFs could be either classified as showing significantly more or less mutations than expected, or not significant (ns).
Supplementary Table S16: Downstream analyses considering only the SIDV inferred genetic alterations with predicted impact on function. S16.1: results of the binomial enrichment test. SID regions overlapping at least 2 SNVs predicted as pathogenic are included. Along with genomic coordinates (hg38) the total number of SNVs as well as the number of predicted pathogenic SNVs overlapping the region are indicated. The p-value and the q-value (after Benjamini-Hochberg correction) of the binomial test are indicated, along with annotation to the closest gene. The 63 regions considered for further analyses are highlighted in red. S16.2: same as S13.1 but considering all the regions assigned to the genes annotated to the same ontological terms together. The number and identity of the genes contributing to the overlap are indicated, along with p-value of the binomial test, and the q-value (after Benjamini-Hochberg correction). Statistically significant terms (q-value <= 0.05) are highlighted in red. S16.3: results of the analyses testing for the enrichment of mutations (either SNVs, short INDELs, or both; mutation_type column) with computationally predicted pathogenic effects in the sets of regions also showing a certain behavior in SIDP (CRISPRi_hit_type column). Observed and expected overlap are indicated, along with the odds ratio and the p-value (Chi-squared test).
Supplementary Table S17: Downstream analyses considering only the SIDV inferred genetic alterations with predicted impact on function and stratifying them by cancer-cell fraction (CCF) increase and decrease in metastatic samples. S17.1: summary of the results of the statistical tests performed to identify differences in the predicted impact of mutations stratified by change in CCF in metastatic samples compared to matched primary. The fraction of variants predicted as pathogenic and either showing an increase or a decrease in CCF (+- 0.1) was compared to that of those showing no change. P-values for the indicated features are shown (Chi-squared test). S17.2: similarly, the distribution of the predicted molecular effects of variants in the three groups (increase, decrease or no change in CCF) were compared using the Kruskal-Wallis test. S17.3: like S13.3 but testing for the enrichment of mutations with both computationally predicted pathogenic effects and a certain CCF increase or decrease in metastatic samples, that also show a certain behavior in SIDP.
Supplementary Table S18: Estimation of excess of mutations in the hormone-receptor positive breast cancer cohorts from the Hartwig Medical Foundation (HMF). The table lists, for each one of the regions showing an excess of functional alterations in the SIDV cohort (highlighted in table S13.1) the number of mutations in the region in both the SIDV and HMF cohort, along with the estimated background probability of seeing a mutation for the regions (using the HMF cohort) and the corresponding, empirical p-value of seeing the observed number of mutations, given the background probability (see Methods).
Supplementary Table S19: Overlap between regions with excess of functional alterations. The regions considered in these overlaps are highlighted in table S13.1. S19.1: overlap with noncoding drivers identified by PCAWG (as listed in Supplementary Table 5 of Rheinbay et al. 2020). S19.2: overlap with regulatory mutations from breast cancer samples, as identified by HMF (as listed in Supplementary Table 5 of Dietlein et al. 2022). S19.3: overlap with noncoding mutational hotspots identified by HMF (as listed in Supplementary Table 21 of Dietlein et al. 2022). S19.4: overlap with genes associated with mutation events in non-coding regions from previous studies, as annotated by HMF (as listed in Supplementary Table 22 of Dietlein et al. 2022).
Supplementary Figure S1. SIDP identifies Cis Regulatory regions in MCF7 with high reproducibility.
Supplementary Figure 2. SIDP identifies Cis Regulatory regions in T47D with high reproducibility.
Supplementary Figure 3. Estrogen deprivation in MCF7 cells introduce stochastic selective pressure in long-term CRISPRi screens.
Supplementary Figure 4. Estrogen deprivation in MCF7 cells introduce stochastic selective pressure in long-term CRISPRi screens.
Supplementary Figure 5. Estrogen deprivation in T47D cells introduce stochastic selective pressure in long-term CRISPRi screens.
Supplementary Figure 6. SIDP results are consistent across ER+ BC cell lines.
Supplementary Figure 7. SIDP identifies regulatory regions with context-dependent activity in MCF7 cells.
Supplementary Figure 8. SIDP identifies regulatory regions with context-dependent activity in MCF7 cells.
Supplementary Figure 9. SIDP identifies regulatory regions with context-dependent activity in MCF7 cells.
Supplementary Figure 10. SIDP identifies regulatory regions with context-dependent activity in LTED cells.
Supplementary Figure 11. Validation of DF and IF SIDP hits.
Supplementary Figure 12. Validation of DF and IF SIDP hits.
Supplementary Figure 13. The promoters of genes belonging to the TLR5 cascade are active in ER+ breast cancer patients.
Supplementary Figure 14. TLR5 expression in ER+ cells from normal and cancer samples.
Supplementary Figure S15. Validation of SIDP via chemical perturbation.
Supplementary Figure 16. Genes involved in TLR signaling are upregulated in MCF7 during therapy induced dormancy.
Supplementary Figure 17. Activation of TLR5 via paracrine signaling.
Supplementary Figure 18. SIDV design and results.
Supplementary Figure 19. SIDV functional analyses.
Data Availability Statement
The R, Python, and bash scripts to reproduce analyses and figures have been deposited in Zenodo: http://zenodo.org/record/8097853.