Abstract
Fifty-three strains belonging to the pathogenic species Leptospira interrogans and Leptospira kirschneri were analyzed by multilocus sequence analysis. The species formed two distinct branches. In the L. interrogans branch, the phylogenetic tree clustered the strains into three subgroups. Genogroups and serogroups were superimposed but not strictly.
Leptospira spp. belong to the bacterial phylum “Spirochaetes.” Members of the genus Leptospira are generally divided into a pathogenic species, Leptospira interrogans sensu lato, and a nonpathogenic species, Leptospira biflexa sensu lato (3, 12). Pathogenic members are the causal agents of leptospirosis, a widespread zoonosis that is a major public health dilemma. In the natural reservoirs of the bacteria, such as rodents, infection produces chronic and persistent asymptomatic shedding in the renal tubules, and bacteria are then excreted in urine.
A wide range of molecular methods to type leptospiral isolates, including PCR-restriction endonuclease analysis (4, 19), pulsed-field gel electrophoresis (7), and random amplification (6, 17), have been applied with more or less success. The genomic DNA-DNA hybridization method has been widely used for the determination of phylogenetic relationships between closely related strains (3, 5, 14). Most procedures are time-consuming, and depending on the method, various limitations that include low degrees of reproducibility and high background levels have been pointed out.
The availability of an increasing number of sequenced genomes has favored the application of sequence-based approaches that can yield much deeper information than previous methods about relationships between strains (13). Nucleotide sequence-based methods are more suitable than conventional procedures, as they facilitate direct, unambiguous comparison between isolates typed in different locations (15).
Multilocus sequence typing (MLST) and multilocus sequence analysis (MLSA) have been recently proposed as alternative ways for defining species or recognizing distinct strains of named species (8, 20). These techniques require identification of loci that evolve more rapidly than rRNA genes and analyses of multiple genes to provide a buffer against the distorting effects of recombination at a single locus. The diversity and relationship of different isolates across related taxa are then assessed by using an appropriate phylogenetic or cladistic approach. This strategy has been used recently to obtain new perspectives on Listeria monocytogenes evolution and to characterize the genomic diversity of Enterobacter cloacae (16), Streptococcus agalactiae (10), and Campylobacter jejuni (11). In this study, we have applied an MLSA typing schema to pathogenic isolates of Leptospira.
A total of 51 strains, including 11 reference strains, from different sources and geographical regions were analyzed in this study (Table 1). Species identification was performed by rRNA gene sequencing using LeptoA (5′-GGCGGCGCGTCTTAAACATG-3′) and LeptoB (5′-TTCCCCCCATTGAGCAAGATT-3′) nucleotide primers. In addition, all strains were checked for the presence of the hap1 fragment (262 bp) by amplification of pathogenic Leptospira DNA with the specific primers described previously (2).
TABLE 1.
Characteristics of Leptospira strains
| No. | Species | Serogroup | Serovar | Origin of isolate | Geographical area | Source |
|---|---|---|---|---|---|---|
| 1 | L. interrogans | Australis | Bratislava | Animal (Erinaceus sp.) | Czechoslovakia | ATCC (ATCC 23578) |
| 2 | L. interrogans | Australis | Bratislava | Unknown | United States | Queensland Laboratory |
| 3 | L. interrogans | Australis | ND | Animal | France | National Veterinary School of Nantes (France) |
| 4 | L. interrogans | Australis | Australis | Environment | Malaysia | ATCC (ATCC 23605) |
| 5 | L. interrogans | Australis | Muenchen | Animal | France | National Veterinary School of Nantes (France) |
| 6 | L. interrogans | Australis | Muenchen | Animal (dog) | France | National Veterinary School of Nantes (France) |
| 7 | L. interrogans | Australis | ND | Human | France | National Veterinary School of Nantes (France) |
| 8 | L. interrogans | Autumnalis | Autumnalis | Animal | France | Adiagène (France) |
| 9 | L. interrogans | Autumnalis | Autumnalis | Human | Japan | ATCC (ATCC 23476) |
| 10 | L. interrogans | Autumnalis | Autumnalis | Human | France | Pasteur Institute (France) |
| 11 | L. interrogans | Autumnalis | Autumnalis | Animal | France | National Veterinary School of Nantes (France) |
| 12 | L. interrogans | Bataviae | Bataviae | Environment | Malaysia | ATCC (ATCC 23602) |
| 13 | L. interrogans | Canicola | Canicola | Animal (dog) | ND | ATCC (ATCC 23606) |
| 14 | L. interrogans | Canicola | Canicola | Animal | France | Adiagène (France) |
| 15 | L. interrogans | Canicola | Canicola | Unknown | France | Virbac (France) |
| 16 | L. interrogans | Canicola | ND | Animal (dog) | France | National Veterinary School of Nantes (France) |
| 17 | L. interrogans | Icterohaemorrhagiae | Copenhageni | Animal | France | Adiagène (France) |
| 18 | L. interrogans | Icterohaemorrhagiae | Icterohaemorrhagiae | Animal | France | Adiagène (France) |
| 19 | L. interrogans | Icterohaemorrhagiae | Icterohaemorrhagiae | Human | France | Pasteur Institute (France) |
| 20 | L. interrogans | Icterohaemorrhagiae | Icterohaemorrhagiae | Human | France | Pasteur Institute (France) |
| 21 | L. interrogans | Icterohaemorrhagiae | IH CF1 | Unknown | France | Merieux (France) |
| 22 | L. interrogans | Icterohaemorrhagiae | ND | Environment | France | Mérial (France) |
| 23 | L. interrogans | Icterohaemorrhagiae | Icterohaemorrhagiae | Human | France | ATCC (ATCC 23581) |
| 24 | L. interrogans | Icterohaemorrhagiae | Lai | Animal | France | National Veterinary School of Nantes (France) |
| 25 | L. interrogans | Icterohaemorrhagiae | ND | Animal (Rattus norvegicus) | France | National Veterinary School of Nantes (France) |
| 26 | L. interrogans | Icterohaemorrhagiae | ND | Animal (Ondatra zibethicus) | France | National Veterinary School of Nantes (France) |
| 27 | L. interrogans | Icterohaemorrhagiae | ND | Animal (Myocastor coypus) | France | National Veterinary School of Nantes (France) |
| 28 | L. interrogans | Pomona | Pomona | Human | Australia | ATCC (ATCC 23478) |
| 29 | L. interrogans | Pomona | Pomona | Animal | France | Adiagène (France) |
| 30 | L. interrogans | Pyrogenes | Pyrogenes | Human | Indonesia | ATCC (ATCC 23480) |
| 31 | L. interrogans | Pyrogenes | Pyrogenes | Animal | France | National Veterinary School of Nantes (France) |
| 32 | L. interrogans | Sejroe | Wolfii | Human | Indonesia | ATCC (ATCC 23482) |
| 33 | L. interrogans | Unknown | Unknown | Animal (Myocastor coypus) | France | Frank Duncombe Laboratory (France) |
| 34 | L. interrogans | Unknown | Unknown | Human | France | National Veterinary School of Nantes (France) |
| 35 | L. interrogans | Unknown | Unknown | Animal (Rattus norvegicus) | France | National Veterinary School of Nantes (France) |
| 36 | L. interrogans | Unknown | Unknown | Animal (Rattus norvegicus) | France | National Veterinary School of Nantes (France) |
| 37 | L. interrogans | Unknown | Unknown | Animal (Mus musculus) | France (Guadeloupe) | National Veterinary School of Nantes (France) |
| 38 | L. interrogans | Unknown | Unknown | Animal (Rattus norvegicus) | France | National Veterinary School of Nantes (France) |
| 39 | L. interrogans | Unknown | Unknown | Animal (bovine) | France | National Veterinary School of Nantes (France) |
| 40 | L. kirschneri | Cynopteri | ND | Animal | France | National Veterinary School of Nantes (France) |
| 41 | L. kirschneri | Grippotyphosa | Grippotyphosa | Environment | Malaysia | ATCC (ATCC 23604) |
| 42 | L. kirschneri | Grippotyphosa | Grippotyphosa | Animal | France | Adiagène (France) |
| 43 | L. kirschneri | Grippotyphosa | Grippotyphosa | Animal | France | Adiagène (France) |
| 44 | L. kirschneri | Grippotyphosa | Grippotyphosa | Human | France | Pasteur Institute (France) |
| 45 | L. kirschneri | Grippotyphosa | Grippotyphosa | Human | France | Pasteur Institute (France) |
| 46 | L. kirschneri | Grippotyphosa | Grippotyphosa | Human | Russia | ATCC (ATCC 23469) |
| 47 | NDa | Pomona | ND | Animal (pig) | France | New Caledonia Laboratory |
| 48 | ND | Pomona | ND | Animal (pig) | France | New Caledonia Laboratory |
| 49 | ND | Pomona | ND | Animal (pig) | France | New Caledonia Laboratory |
| 50 | ND | Unknown | Unknown | Animal | France | National Veterinary School of Nantes (France) |
| 51 | ND | Unknown | Unknown | Animal (Rattus norvegicus) | France | National Veterinary School of Nantes (France) |
ND, not determined.
For MLSA, 14 housekeeping genes were selected in a preliminary study by comparison between two Leptospira genomic sequences available in GenBank (http://www.ncbi.nlm.nih.gov): those of L. interrogans serovar Copenhageni strain Fiocruz L1-130 chromosome I (accession number AE016823.1) and L. interrogans serovar Lai strain 56601 chromosome I (accession number AE010300.1). Since studies by Haake et al. revealed that outer membrane protein sequences have mosaic compositions consistent with horizontal transfer of DNA between related bacterial species (9), genes encoding surface proteins were excluded from the initial choice of gene targets. Primers were evaluated against the collection of reference strains. Based on the performance of primers and the number of alleles at a given locus, seven pairs of primers were selected. The nucleotide primers and the sizes of amplified DNA fragments from internal gene regions are shown in Table 2. PCRs were carried out using the following conditions: initial denaturation at 94°C for 5 min, followed by 45 cycles of 94°C for 30 s, 46°C (for recF) or 50°C (for accA2, ccmF, czcA, gcvP, groEL, and polA) for 45 s, and 72°C for 60 s. The samples were maintained at 72°C for another 10 min. Both strands of each fragment were then sequenced.
TABLE 2.
Details of gene loci and corresponding primer sequences used for MLSA
| Gene | Product | Locusa | Gene size (bp) | Coordinates | Size of analyzed polymorphic sequence (bp) | G+C content (%) | No. of alleles | Primer sequence (5′-3′)b |
|---|---|---|---|---|---|---|---|---|
| accA2 | Acetyl coenzyme A | LIC11517 | 1,667 | 1874272-1875939 | 452 | 41.59 | 23 | + TTGATGCTTATGTTTGGGTTCAAT |
| carboxylase α subunit | − AGAATCGAATAAATCTAGTTCCT | |||||||
| ccmF | Cytochrome c biogenesis | LIC10813 | 2,189 | 983052-985241 | 295 | 40.31 | 17 | + GATATTCCTGTACAATTCCGG |
| factor | − GGCAGTTTTCTTTTTAATAAC | |||||||
| czcA | Heavy-metal efflux pump | LIC11937 | 3,320 | 2340958-2344278 | 448 | 41.75 | 15 | + TCTTTTGGGAGAAATGATCGG |
| − TAAGTTTGAAAGACTACAACG | ||||||||
| gcvP | Glycine cleavage system P | LIC10309 | 2,894 | 350674-353568 | 275 | 41.28 | 14 | + AAGAGCTAAAAGATCTGCAGC |
| protein | − CTGGCCTTGAAATTTCGAACG | |||||||
| groEL | 60-kDa chaperonin (Hsp60) | LIC11335 | 1,640 | 1646985-1648625 | 553 | 42.2 | 26 | + TCGCCTCGTCTCCATCCA |
| − TGACGCAGAATCCATGGTAG | ||||||||
| polA | DNA polymerase I | LIC10586 | 2,753 | 711562-714315 | 432 | 38.11 | 12 | + TCATAGTCGTTTGAATCTACGGTAAAA |
| − ATTAAAGATTCCGGTGTAGAACCAA | ||||||||
| recF | DNA replication and repair | LIC10003 | 1,097 | 5062-6159 | 400 | 31.94 | 22 | + CGGCGGAAAGAAGAAAGTT |
| protein RecF | − CTCGACTTTTCCTTGTCGAA |
L. interrogans serovar Copenhageni strain Fiocruz L1-130 has been used as a reference.
+, foward primer; −, reverse primer.
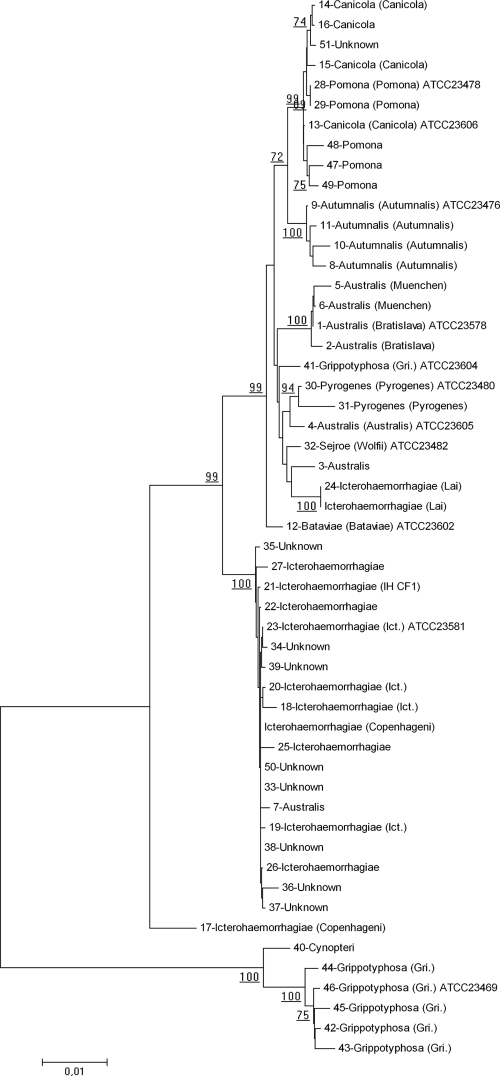
Sequence data for the 51 study strains, L. interrogans serovar Copenhageni strain Fiocruz L1-130, and L. interrogans serovar Lai strain 56601 were used to construct a phylogenetic tree (Fig. 1). Sequenced DNA fragments were concatenated in the following order to generate a single 2,855-bp sequence: accA2, ccmF, czcA, gcvP, groEL, polA, and recF. Phylogenetic analysis was conducted with MEGA4 (21). The neighbor-joining method (18) was used for sequence comparison. The evolutionary distances were computed using the maximum composite likelihood method (22).
FIG. 1.
Phylogenetic tree constructed from concatenated sequences (2,855 bp) of seven housekeeping genes of Leptospira strains. The identification numbers of the strains, the serogroups, and the serovars (in parentheses) are indicated. Gri., Grippotyphosa; Ict., Icterohaemorrhagiae. The neighbor-joining method (18) was used for sequence comparison. Phylogenetic analysis was conducted with MEGA4 (21). The evolutionary distances were computed using the maximum composite likelihood method (22). Only bootstrap values greater than 70 are shown.
16S RNA sequencing identified 39 strains of various serogroups (Icterohaemorrhagiae, Australis, Autumnalis, Canicola, Pomona, Pyrogenes, and Bataviae) as L. interrogans genomospecies and 7 strains (serogroups Grippotyphosa and Cynopteri) as L. kirschneri genomospecies. For the five remaining strains, the genomospecies could not be determined because of insufficient quantity of genomic material (Table 1).
The seven loci selected were suitable for MLSA, as they could be amplified from all isolates and sequenced, irrespective of the serogroup or species. Depending on the gene considered, 12 to 26 alleles could be observed, with the groEL gene being the most discriminant.
Phylogenetic analysis clustered the sequences into two main clades. The L. interrogans strains grouped into one clade, and the L. kirschneri strains grouped into the other clade, except for one strain, L. kirschneri serogroup Grippotyphosa ATCC 23604, which was classified in the L. interrogans clade, in contrast to the other reference strain, L. kirschneri serogroup Grippotyphosa ATCC 23469.
The major L. interrogans branch was further divided into several subgroups. A major subgroup comprised most Icterohaemorrhagiae serogroup strains (n = 10), although it also included one Australis serogroup strain. Eight strains of unknown serogroups belonged to this subgroup. The three remaining Icterohaemorrhagiae serogroup strains were placed into distant branches. Autumnalis (n = 4), Canicola (n = 4), and Pomona (n = 5) serogroup strains were clustered into one group. The other subgroups were more heterogeneous, comprising Australis, Pyrogenes, and Bataviae serogroup strains (n = 9). Individual gene analysis (see the figures in the supplemental material) showed that, in general, strains which clustered together in the multilocus analysis shared the same allele for each individual gene.
Noticeably, strain 17 (L. interrogans serogroup Icterohaemorrhagiae serovar Copenhageni) was placed alone on a distinct branch. Individual gene analysis placed this strain in the Icterohaemorrhagiae clade, except for the recF allele, which was closely related to that of the Grippotyphosa strains (see the figures in the supplemental material). This finding may be explained by horizontal gene transfer events that are observed in Leptospira and lead to mosaic compositions of sequences (9, 25). The positioning of L. kirschneri serogroup Grippotyphosa ATCC 23604 in the L. interrogans clade was unexplained. In this case, clustering using individual gene analysis was similar to that using MLSA. To exclude the possibility of contamination, we analyzed another sample of the isolate provided by the ATCC and found identical sequences. Possibly this strain was misidentified.
An MLSA technique for Leptospira species was developed previously by Ahmed et al. (1). Genotyping was based on DNA sequencing of six genes and allowed differentiation of the major species of Leptospira, L. interrogans, L. kirschneri, L. noguchii, L. santarosai, and L. borgpetersenii. The set of primers used permitted analysis of the genetic relationships among species and the respective evolution of the species. However, the clustering was limited to the species level. In contrast, the primers that we have used distinguished clusters at the serogroup level for the pathogenic species L. interrogans and L. kirschneri.
Recently, an MLST schema based on the sequences of seven loci was used for typing of Leptospira isolates from humans and rodents in Thailand (23). Twelve sequence types (ST) were identified among the isolates. ST34 was predominant in human and rodent populations, representing 71 and 88% of isolates, respectively. In contrast, strains from a reference collection had various STs. This study showed that ST34 may have a greater propensity than the other clones to cause human diseases in Thailand. Analysis of our sequences by the MLST method (15) showed a great diversity of STs and no clustering (data not shown). This result was not surprising, since our collection was composed of epidemiologically unrelated isolates.
In another study, Victoria et al. (24) showed that the S10-spc-α locus, which encodes ribosomal proteins and is highly conserved in L. interrogans, is a useful tool to elucidate evolutionary patterns. This analysis provided phylogenetic insights into the genus Leptospira at the species level but only limited information on subspecies discrimination.
Development of various schemas for molecular classification of Leptospira will probably allow better characterization of subtypes of Leptospira with particular epidemicity or propensity to cause diseases in humans or animals. More strains in various ecological niches need to be tested to determine which loci among those proposed in this study and those by Ahmed et al. (1) and Thaipadungpanit et al. (23) are suitable for a definitive typing schema.
Nucleotide sequence accession numbers.
Nucleotide sequences determined in this study have been deposited in the GenBank data library under accession numbers GU113162 to GU113532.
Supplementary Material
Acknowledgments
This work received financial support from the Conseil Général du Calvados, the Conseil Régional de Basse-Normandie, and the Association pour le Développement de la Recherche Equine en France (ADREF).
We thank the team of Mathieu Picardeau for identification of strains by rrs gene sequencing, and Béatrice Blanchard (Adiagène) and Denise Dessouter (New Caledonia Laboratory) for generous provision of Leptospira strains. This work was performed within the Competitiveness Cluster of the Horse Industry of Basse-Normandie.
Footnotes
Published ahead of print on 2 December 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Ahmed, N., S. M. Devi, M. L. Valverde, P. Vijayachari, R. S. Machang'u, W. A. Ellis, and R. A. Hartskeerl. 2006. Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann. Clin. Microbiol. Antimicrob. 5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Branger, C., B. Blanchard, C. Fillonneau, I. Suard, F. Aviat, B. Chevallier, and G. Andre-Fontaine. 2005. Polymerase chain reaction assay specific for pathogenic Leptospira based on the gene hap1 encoding the hemolysis-associated protein-1. FEMS Microbiol. Lett. 243:437-445. [DOI] [PubMed] [Google Scholar]
- 3.Brenner, D. J., A. F. Kaufmann, K. R. Sulzer, A. G. Steigerwalt, F. C. Rogers, and R. S. Weyant. 1999. Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int. J. Syst. Bacteriol. 49:839-858. [DOI] [PubMed] [Google Scholar]
- 4.Brown, P. D., and P. N. Levett. 1997. Differentiation of Leptospira species and serovars by PCR-restriction endonuclease analysis, arbitrarily primed PCR and low-stringency PCR. J. Med. Microbiol. 46:173-181. [DOI] [PubMed] [Google Scholar]
- 5.Chang, H. W., Y. D. Nam, M. Y. Jung, K. H. Kim, S. W. Roh, M. S. Kim, C. O. Jeon, J. H. Yoon, and J. W. Bae. 2008. Statistical superiority of genome-probing microarrays as genomic DNA-DNA hybridization in revealing the bacterial phylogenetic relationship compared to conventional methods. J. Microbiol. Methods 75:523-530. [DOI] [PubMed] [Google Scholar]
- 6.Corney, B. G., J. Colley, and G. C. Graham. 1997. Simplified analysis of pathogenic leptospiral serovars by random amplified polymorphic DNA fingerprinting. J. Med. Microbiol. 46:927-932. [DOI] [PubMed] [Google Scholar]
- 7.Galloway, R. L., and P. N. Levett. 2008. Evaluation of a modified pulsed-field gel electrophoresis approach for the identification of Leptospira serovars. Am. J. Trop. Med. Hyg. 78:628-632. [PubMed] [Google Scholar]
- 8.Gevers, D., F. M. Cohan, J. G. Lawrence, B. G. Spratt, T. Coenye, E. J. Feil, E. Stackebrandt, Y. Van de Peer, P. Vandamme, F. L. Thompson, and J. Swings. 2005. Opinion: re-evaluating prokaryotic species. Nat. Rev. Microbiol. 3:733-739. [DOI] [PubMed] [Google Scholar]
- 9.Haake, D. A., M. A. Suchard, M. M. Kelley, M. Dundoo, D. P. Alt, and R. L. Zuerner. 2004. Molecular evolution and mosaicism of leptospiral outer membrane proteins involves horizontal DNA transfer. J. Bacteriol. 186:2818-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honsa, E., T. Fricke, A. J. Stephens, D. Ko, F. Kong, G. L. Gilbert, F. Huygens, and P. M. Giffard. 2008. Assignment of Streptococcus agalactiae isolates to clonal complexes using a small set of single nucleotide polymorphisms. BMC Microbiol. 8:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levesque, S., E. Frost, R. D. Arbeit, and S. Michaud. 2008. Multilocus sequence typing of Campylobacter jejuni isolates from humans, chickens, raw milk and environmental water in Quebec. J. Clin. Microbiol. 46:3404-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levett, P. N. 2001. Leptospirosis. Clin. Microbiol. Rev. 14:296-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levett, P. N. 2007. Sequence-based typing of Leptospira: epidemiology in the genomic era. PLoS Negl. Trop. Dis. 1:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levett, P. N., R. E. Morey, R. L. Galloway, and A. G. Steigerwalt. 2006. Leptospira broomii sp. nov., isolated from humans with leptospirosis. Int. J. Syst. Evol. Microbiol. 56:671-673. [DOI] [PubMed] [Google Scholar]
- 15.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paauw, A., M. P. Caspers, F. H. Schuren, M. A. Leverstein-van Hall, A. Deletoile, R. C. Montijn, J. Verhoef, and A. C. Fluit. 2008. Genomic diversity within the Enterobacter cloacae complex. PLoS ONE 3:e3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramadass, P., S. Meerarani, M. D. Venkatesha, A. Senthilkumar, and K. Nachimuthu. 1997. Characterization of leptospiral serovars by randomly amplified polymorphic DNA fingerprinting. Int. J. Syst. Bacteriol. 47:575-576. [DOI] [PubMed] [Google Scholar]
- 18.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 19.Savio, M. L., C. Rossi, P. Fusi, S. Tagliabue, and M. L. Pacciarini. 1994. Detection and identification of Leptospira interrogans serovars by PCR coupled with restriction endonuclease analysis of amplified DNA. J. Clin. Microbiol. 32:935-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. Grimont, P. Kampfer, M. C. Maiden, X. Nesme, R. Rossello-Mora, J. Swings, H. G. Truper, L. Vauterin, A. C. Ward, and W. B. Whitman. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 52:1043-1047. [DOI] [PubMed] [Google Scholar]
- 21.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 22.Tamura, K., M. Nei, and S. Kumar. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101:11030-11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thaipadungpanit, J., V. Wuthiekanun, W. Chierakul, L. D. Smythe, W. Petkanchanapong, R. Limpaiboon, A. Apiwatanaporn, A. T. Slack, Y. Suputtamongkol, N. J. White, E. J. Feil, N. P. Day, and S. J. Peacock. 2007. A dominant clone of Leptospira interrogans associated with an outbreak of human leptospirosis in Thailand. PLoS. Negl. Trop. Dis. 1:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Victoria, B., A. Ahmed, R. L. Zuerner, N. Ahmed, D. M. Bulach, J. Quinteiro, and R. A. Hartskeerl. 2008. Conservation of the S10-spc-alpha locus within otherwise highly plastic genomes provides phylogenetic insight into the genus Leptospira. PloS ONE 3:e2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue, F., J. Yan, and M. Picardeau. 2009. Evolution and pathogenesis of Leptospira spp.: lessons learned from the genomes. Microbes Infect. 11:328-333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.