Abstract
Background
Development of phylogenetic methods that do not rely on fossils for the study of evolutionary processes through time have revolutionized the field of evolutionary biology and resulted in an unprecedented expansion of our knowledge about the tree of life. These methods have helped to shed light on the macroevolution of many taxonomic groups such as the placentals (Mammalia). However, despite the increase of studies addressing the diversification patterns of organisms, no synthesis has addressed the case of the most diversified mammalian clade: the Rodentia.
Results
Here we present a rodent maximum likelihood phylogeny inferred from a molecular supermatrix. It is based on 11 mitochondrial and nuclear genes that covers 1,265 species, i.e., respectively 56% and 81% of the known specific and generic rodent diversity. The inferred topology recovered all Rodentia clades proposed by recent molecular works. A relaxed molecular clock dating approach provided a time framework for speciation events. We found that the Myomorpha clade shows a greater degree of variation in diversification rates than Sciuroidea, Caviomorpha, Castorimorpha and Anomaluromorpha. We identified a number of shifts in diversification rates within the major clades: two in Castorimorpha, three in Ctenohystrica, 6 within the squirrel-related clade and 24 in the Myomorpha clade. The majority of these shifts occurred within the most recent familial rodent radiations: the Cricetidae and Muridae clades. Using the topological imbalances and the time line we discuss the potential role of different diversification factors that might have shaped the rodents radiation.
Conclusions
The present glimpse on the diversification pattern of rodents can be used for further comparative meta-analyses. Muroid lineages have a greater degree of variation in their diversification rates than any other rodent group. Different topological signatures suggest distinct diversification processes among rodent lineages. In particular, Muroidea and Sciuroidea display widespread distribution and have undergone evolutionary and adaptive radiation on most of the continents. Our results show that rodents experienced shifts in diversification rate regularly through the Tertiary, but at different periods for each clade. A comparison between the rodent fossil record and our results suggest that extinction led to the loss of diversification signal for most of the Paleogene nodes.
Background
A fundamental question in evolutionary biology is understanding why some clades are highly diverse. Species diversity is the result of the balance between speciation and extinction whereas morphological disparity is primarily a consequence of adaptation [1-3]. For a long time, only the study of the fossil record provided a direct view of the patterns of diversification revealing major speciation and extinction trends through time [4]. Development of methods that do not rely on fossils for the study of the evolutionary processes through time [5] have dramatically changed the way we study diversification patterns. The use of dated phylogenetic trees in combination with knowledge on species assemblies allows us to (1) estimate speciation and extinction rates [6], (2) detect shifts in diversification rates [7,8], (3) test diversification constancy through time [9,10] and (4) study the link between biological traits and diversification within clades [11,12].
Throughout the Cenozoic, rodents underwent an extraordinary adaptive radiation. As a result rodents represent nearly half of the current mammalian diversity with more than 2,261 species organized into 474 genera [13]. These small to medium-sized placentals have spread over all continents (except Antarctica) and most islands, where they occupy virtually all terrestrial ecosystems from tropical rainforests and deserts to the arctic tundra. New species and genera are being described each year, such as Laonastes aenigmamus[14], the sole extant representative of a morphologically and phylogenetically distinct family, the Diatomyidae [15,16]. Rodents also display a wide range of life histories and ecomorphological adaptations including fossorial, arboreal, subaquatic, jumping and gliding capacities. Their outstanding diversity among mammals, combined with the richness of their fossil record, makes rodents a suitable model to study the factors that promote morphological diversity and trigger evolutionary radiations.
Repetitive bursts of speciation and a high level of homoplasy in morphological characters [17-19] have hindered delimitation of inter- and intra-familial relationships within rodents. However, a resurgence of interest in rodent phylogeny using molecular markers, most notably mitochondrial markers [20-23], nuclear genes [16,24-30] and retroposed elements [31], has provided a new insight of familial relationships and has challenged traditional classifications based on myological (e.g.[32], [33] and [18]) and cranio-dental characters [34]. Simultaneously, many molecular studies have addressed phylogenetic relationships at lower taxonomic levels, releasing a large number of sequences from a variety of loci [21,35-54]. These recent developments make now possible the construction of a large phylogeny based on DNA data for this mammalian order. Such a phylogenetic framework is the basis for macroevolutionary and comparative meta-analyses that aim to address questions about rodent evolutionary history.
Two approaches have been proposed to reconstruct large evolutionary trees from partially overlapping character and taxon datasets: the supertree, and the supermatrix. In the supertree approach, independent data sets are analysed separately to yield source topologies which are subsequently combined to produce a larger phylogenetic tree [55,56]. In contrast, supermatrix analyses use characters gathered from the widest possible range of taxa in a single analysis to provide a “large tree”. Gatesy et al.[57,58] compared the two approaches and brought attention to the methodological constraints of the supertree approach e.g. (i) source data which contain non-cladistic characters such as taxonomy lists, (ii) duplication of homologous characters or (iii) robustness values of tree nodes that are difficult to interpret. Gatesy et al.[58] supported the use of a supermatrix as the combination of independent features could reveal hidden relationships [59]. To date, the only large-scale comprehensive phylogeny available for rodents is part of the family-level supertree of Beck et al.[60], and the species-level supertree published by Bininda-Emonds et al.[61] which included nearly all extant families and species of mammals. Furthermore due to lack of phylogenetic data for many of the rodent groups at that time, their final topologies contain a large amount of polytomies (less than < 40% of the branches are fully resolved at the genus level) and do not reflect our current knowledge of rodent systematics. We therefore expect that a more robust framework for rodent molecular phylogeny may benefit from a gene concatenation approach as illustrated by the family-level supermatrix tree of Meredith et al.[62]. Here, we present the first large-scale phylogenetic analysis which includes the most representative molecular markers for rodents. The inferred topology is subsequently used to provide divergence date estimates with a relaxed molecular clock. Our species level rodent phylogeny allows us to address specifically the following questions: (1) Is the rate of diversification constant over all lineages ? (2) Within which lineages, if any, do shifts in diversification rate occur ? (3) When did major rodent diversification events occur during the Tertiary ? (4) Can we connect potential shifts in diversification rate to macroevolutionary events ?
Results and Discussion
Phylogenetic results and systematics
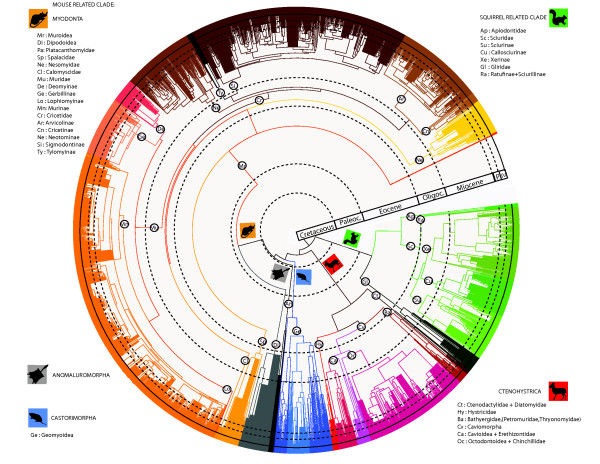
Since the first mammal supertree [63], there has been no integrated, molecular-based synthesis of rodent systematics. However, several extensive mitochondrial molecular studies of other mammalian orders have been performed for Primates [64-66], Carnivora [67], Cetartiodactyla [68,69] and Chiroptera [70]. The molecular supermatrix presented here is the first attempt to include all Rodentia taxa for which mitochondrial and nuclear DNA sequences are available in public databases within a common phylogenetic framework. The supermatrix concatenates 11 genes, and contains 1,265 taxon sequences aligned for 15,535 sites, with 75% of missing character states. Maximum likelihood analysis yields a phylogenetic hypothesis for Rodentia, with bootstrap values (BP) greater than 70% for 64% of the nodes summarized in Figures 1 and 2 (see also Additional file 1: Figure S1, Additional file 2: Figure S2, Additional file 3: Figure S3, Additional file 4: Figure S4, Additional file 5: Figure S5, Additional file 6: Figure S6, Additional file 7: Figure S7, Additional file 8: Figure S8, Additional file 9: Figure S9, Additional file 10: Figure S10, Additional file 11: Figure S11, Additional file 12: Figure S12, Additional file 13: Figure S13 for details about the species-level topology). Figure 1, Figure 2 and Additional file 1: Figure S1 represents a topological summary of all other topological figures.
Figure 1.
Rodent species-level evolutionary dated tree. The species-level chronogram is based on the ML topologies from 8 supermatrix trees. Stratigraphic scale : P : Paleocene, E : Eocene, O : Oligocene, M : Miocene, P : Pliocene, Pl : Pleistocene. Bootstrap and divergence time estimates for all nodes are detailed in Supplementary Data.
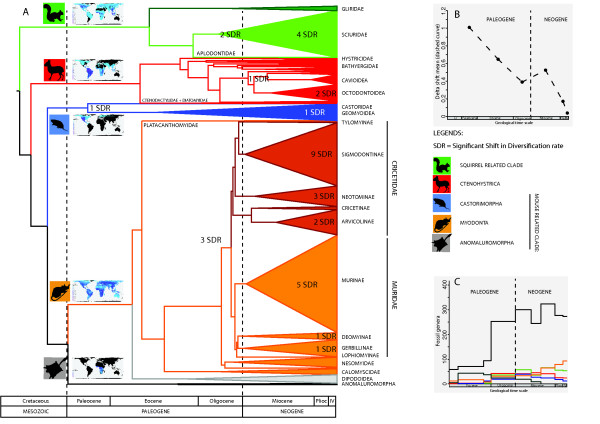
Figure 2.
Diversification of rodents through time. Left part (A): Simplified family-level phylogenetic dated tree of rodents. Stratigraphic scale is in the lower part. Significant shifts in diversification rate (SDR) are indicated (see also Table 1 and Additional file 2: Figure S2, Additional file 3: Figure S3, Additional file 4: Figure S4, Additional file 5: Figure S5, Additional file 6: Figure S6, Additional file 7: Figure S7, Additional file 8: Figure S8, Additional file 9: Figure S9, Additional file 10: Figure S10, Additional file 11: Figure S11, Additional file 12: Figure S12 to Additional file 13: Figure S13). Upper right part (B): Variation through each epoch bin of the mean of absolute nodal Δshift statistics values calculated from the overall 2,263-taxon topology. Lower right part (C): Histogram of the number of rodents genera through Tertiary (McKenna and Bell, 1997). This illustrates the evolution of genus diversity for all rodents (black), extinct stem rodents (darkgreen), Muroidea+Anomaluroidea (orange), Castorimorpha (blue), Sciuroidea (green) and Ctenohystrica (red).
The gene supermatrix supports rodent monophyly (BP = 100%). Four main clades (Figure 1 and Additional file 1: Figure S1) are recovered : (1) the Ctenohystrica (BP = 100% , Additional file 2: Figure S2 and Additional file 3: Figure S3), (2) a squirrel-related clade (BP = 100%, Additional file 4: Figure S4 and Additional file 5: Figure S5), (3) the Castorimorpha (BP = 100%, Additional file 6: Figure S6) and (4) the Myodonta + Anomaluroidea (BP = 66%, Additional file 7: Figure S7, Additional file 8: Figure S8, Additional file 9: Figure S9, Additional file 10: Figure S10, Additional file 11: Figure S11, Additional file 12: Figure S12 and Additional file 13: Figure S13). The Myodonta + Anomaluroidea and the Castorimorpha clade are grouped in the mouse-related clade (BP =57%).
The Guinea-Pig related clade
Ctenohystrica (Additional file 2: Figure S2 and Additional file 3: Figure S3) is composed by the well-supported Ctenodactylidae + Diatomyidae (BP = 99%) and Hystricognathi sensu stricto(BP = 81%)[16]. Phiomorpha (Old World Hystricognathi) is paraphyletic (BP = 74%) with the Bathyergidae clade (mole rats, BP = 98%) being more closely related to Caviomorpha (South American Hystricognathi, BP = 100%) than to Hystricidae (Old World Porcupines, BP = 100%) [30]. Within the Caviomorpha, we recovered the dichotomy between Cavioidea + Erethizontoidea (BP = 84%) and Octodontoidea + Chinchilloidea (BP = 97%) [26]. The monophyly of these 4 superfamilies is also supported (BP > 95%). The intergeneric relationships within caviomorphs are in agreement with recent molecular phylogenies [35,39,44,54,71].
The squirrel-related clade
Within the squirrel-related lineage (Additional file 4: Figure S4 and Additional file 5: Figure S5), we recovered the reciprocal monophyly of Sciuridae (BP = 99%) and Gliridae (BP=100%). In agreement with the most recent phylogenetic analyses of rodents [28-30,35], our results corroborate the Aplodontidae + Sciuridae clade (BP = 100%). Within Gliridae, we found the same clades as those inferred by Montgelard et al.[37] and Nunome et al.[72]. Among Sciuridae, relationships are also well-resolved and the following lineages are recognized: the south-east Asian Callosciurinae (BP = 100%), the Xerinae (BP = 99%), and the Sciurinae (BP= 99%), that includes the Sciurini (BP = 100%) and Pteromyini (BP = 99%) tribes. These results are in agreement with previous hypotheses of Sciuridae composition and relationships [36,42].
The mouse-related clade
The monophyly of the mouse-related clade (Additional file 6: Figure S6 to Additional file 7: Figure S7, Additional file 8: Figure S8, Additional file 9: Figure S9, Additional file 10: Figure S10, Additional file 11: Figure S11, Additional file 12: Figure S12, Additional file 13: Figure S13) is poorly supported (BP = 57%). It contains three major clades: Anomaluroidea (BP = 100%), Castorimorpha (Castoridae + Geomyoidea ; BP = 100%) and Myodonta (BP = 100%) [21,27,29,30]. Myodonta is divided into Muroidea (BP = 100%) and Dipodidae (BP = 100%). Platacanthomyidae is the sister-group to all other Muroidea [49]. Our phylogeny corroborates the monophyly of Spalacidae (BP = 95%), Nesomyidae (BP = 100%), Cricetidae (BP = 99%) and Muridae (BP = 95%). Cricetidae subfamilies are also recovered as monophyletic in our analysis: Sigmodontinae (BP = 99%), Cricetinae (BP = 100%), Arvicolinae (BP = 99%), Tylomyinae (BP = 98%) and Neotominae (BP = 98%). Muridae as defined by recent analyses [40,41,74] is recovered monophyletic and includes the Acomyinae, Gerbillinae, Lophiomyinae and Murinae subfamilies. The monophyly of the Murinae (BP = 93%), Gerbillinae (BP = 100%) and Deomyinae (BP = 96%) subfamilies is also recovered [41,74]. Relationships and support within the muroids agree with those identified in the previous molecular phylogenies of [41,43], [46] and [47]. Our results suggested a sister clade relationship of Myodonta + Anomaluroidea with Castorimorpha. Castorimorpha (BP = 100%) is divided into the Castoridae (BP = 100%) and the Geomyoidea (BP = 99%). Our results also support the paraphyly of Heteromyidae with respect to the Geomyidae [28,30].
Impact of missing data
Molecular marker coverage is uneven among different taxa and between genomes. For example, sequencing effort for the Muridae has been very significant due to medical importance and genomic interests of model species (cf.Mus musculusRattus norvegicus). Furthermore, at the species level the mitochondrial genome has been better studied than the nuclear genome. Thus, mitochondrial genes have been sequenced for most of the available species within our dataset, and mitochondrial markers like CYTB (with 1152 sequenced taxa; Table 2) constitute the backbone of our phylogenetic inference. The single gene analysis of the CYTB provides a relatively similar topology at lower taxonomic (species level) but leads to either unresolved or conflicting results at higher taxonomic level (suborder, family, genus) compared to multigene topologies including nuclear genes, as attested by significant approximately unbiased (AU) [75] and Shimodaira and Hasegawa (SH) tests [76] (P < 0.05). By contrast, there have been relatively few nuclear gene studies addressing the phylogeny of lower level rodent relationships, except for some subfamilies, tribes and genera (e.g. Neotominae, Cricetinae, Oryzomyini, MicrotusMusApodemusRattus, and Phyllotis). At the species and subspecies level, Murinae is undersampled and only the higher-level taxonomic diversity (i.e. genus and family level) is represented by both nuclear and mitochondrial markers [41,43,46,47]. Capromyidae, Dipodidae, Gerbillinae, and African and Indonesian murines are understudied and not included in the present study (Table 3).
Table 2.
Mitochondrial (mtDNA) and nuclear (nucDNA) loci used in this study
| Gene | Model | N taxa | N sites |
|---|---|---|---|
|
12S rRNA [mtDNA] |
GTR+I+Γ |
391 |
724 |
| Breast and ovarian cancer susceptibility protein exon 11 (BRCA1) [nucDNA] |
TVM+I+Γ |
99 |
2977 |
| Control region (DLOOP) [mtDNA] |
HKY+I+Γ |
45 |
996 |
| Cytochrome oxydase 3 (COX3) [mtDNA] |
GTR+I+Γ |
105 |
784 |
| Cytochrome b (CYTB) [mtDNA] |
GTR+I+Γ |
1152 |
1140 |
| Interphotoreceptor retinoid-binding protein exon 1 (RBP3) [nucDNA] |
GTR+I+Γ |
536 |
1302 |
| Growth hormone receptor (GHR) exon 10 [nucDNA] |
HKY+I+Γ |
282 |
974 |
|
NADH dehydrogenase 4 (NADH4) [mtDNA] |
GTR+I+Γ |
99 |
1389 |
|
NADH dehydrogenase 1 (NADH1) [mtDNA] |
TVM+I+Γ |
45 |
961 |
| Recombination activating protein 1 exon 1 (RAG1) [nucDNA] |
GTR+I+Γ |
238 |
3044 |
| von Willebrand gene (vWF) exon 28 [nucDNA] | TrN+I+Γ | 110 | 1272 |
The abbreviated models are the following: HKY: Hasegawa, Kishino, Yano [79]; GTR: General Time Reversible [80,81]; TrN: Tamura-Nei [82]; TVM: Transversion Model; + Γ: variation in rates among sites modeled using a gamma distribution [83]; +I; a proportion of sites modeled as invariant [79]. N taxa = number of available taxa on public databases. N sites = Number of aligned nucleotides.
Table 3.
Summary statistics for gene sequences available for rodent genera and species
| CLADE | Ngenera | gGENBANK | Percentage | Nspecies | spGENBANK | Percentage |
|---|---|---|---|---|---|---|
| Rodentia |
474 |
387 |
81 |
2261 |
1265 |
56 |
| SCIUROIDEA |
61 |
58 |
95,1 |
307 |
200 |
65 |
| Aplodontidae |
1 |
1 |
100 |
1 |
1 |
100 |
| Gliridae |
9 |
7 |
78 |
28 |
15 |
54 |
| Sciuridae |
51 |
50 |
98 |
278 |
184 |
66 |
| CASTORIMORPHA |
14 |
14 |
100 |
102 |
90 |
88 |
| Castoridae |
2 |
2 |
100 |
2 |
2 |
100 |
| Geomyidae |
6 |
6 |
100 |
40 |
33 |
83 |
| Heteromyidae |
6 |
6 |
100 |
60 |
55 |
91 |
| CTENOHYSTRICA |
72 |
59 |
82 |
275 |
158 |
57 |
| Abrocomidae |
2 |
1 |
50 |
10 |
2 |
20 |
| Bathyergidae |
5 |
5 |
100 |
15 |
14 |
77 |
| Capromyidae |
6 |
1 |
17 |
8 |
1 |
7 |
| Caviidae |
6 |
6 |
100 |
18 |
13 |
89 |
| Chinchillidae |
3 |
3 |
100 |
7 |
6 |
86 |
| Ctenodactylidae |
4 |
2 |
50 |
5 |
3 |
60 |
| Ctenomyidae |
1 |
1 |
100 |
60 |
38 |
65 |
| Cuniculidae |
1 |
1 |
100 |
2 |
2 |
100 |
| Dasyproctidae |
2 |
2 |
100 |
13 |
5 |
38 |
| Diatomyidae |
1 |
1 |
100 |
1 |
1 |
100 |
| Dinomyidae |
1 |
1 |
100 |
1 |
1 |
100 |
| Echimyidae |
21 |
17 |
81 |
86 |
44 |
54 |
| Erethizontidae |
5 |
4 |
80 |
17 |
7 |
44 |
| Hystricidae |
3 |
3 |
100 |
11 |
7 |
64 |
| Myocastoridae |
1 |
1 |
100 |
1 |
1 |
100 |
| Octodontidae |
8 |
8 |
100 |
13 |
11 |
85 |
| Petromuridae |
1 |
1 |
100 |
1 |
1 |
100 |
| Thryonomyidae |
1 |
1 |
100 |
2 |
1 |
50 |
| ANOMALUROMORPHA |
4 |
3 |
75 |
9 |
4 |
44 |
| Pedetidae |
1 |
1 |
100 |
2 |
1 |
50 |
| Anomaluridae |
3 |
2 |
67 |
7 |
2 |
29 |
| MYOMORPHA |
324 |
253 |
78 |
1568 |
813 |
52 |
| Dipodidae |
16 |
7 |
44 |
51 |
10 |
20 |
| MUROIDEA |
308 |
246 |
79 |
1516 |
803 |
53 |
| Platacanthomyidae |
2 |
1 |
50 |
2 |
1 |
50 |
| Arvicolinae |
28 |
25 |
89 |
151 |
111 |
74 |
| Calomyscidae |
1 |
1 |
100 |
8 |
2 |
25 |
| Cricetinae |
7 |
6 |
86 |
18 |
14 |
78 |
| Deomyinae |
4 |
4 |
100 |
42 |
33 |
48 |
| Gerbillinae |
16 |
14 |
88 |
103 |
40 |
39 |
| Leimacomyinae |
1 |
0 |
0 |
1 |
0 |
0 |
| Lophiomyinae |
1 |
1 |
100 |
1 |
1 |
100 |
| Murinae |
124 |
88 |
70 |
560 |
231 |
41 |
| Neotominae |
16 |
16 |
100 |
124 |
103 |
83 |
| Nesomyidae |
21 |
17 |
81 |
61 |
28 |
43 |
| Otomyinae |
3 |
3 |
100 |
23 |
16 |
52 |
| Sigmodontinae |
74 |
61 |
82 |
377 |
231 |
61 |
| Spalacidae |
6 |
6 |
100 |
36 |
9 |
25 |
| Tylomyinae | 4 | 3 | 75 | 10 | 3 | 30 |
Ngenera and Nspecies represent the number of genera and species described in Wilson and Reeder (2005); gGENBANK and spGENBANK represent the number of genera and species available in GENBANK; Percentage represents the proportion of genera and species included in our study.
The present phylogeny is the most comprehensive hypothesis for rodent species and generic relationships up to date and provide substantial improvement in comparison with previous studies (Bininda-Emonds et al[61]). Despite the 75% of missing data, the ML trees (summarized in Additional file 1: Figure S1 and Additional file 2: Figure S2, Additional file 3: Figure S3, Additional file 4: Figure S4, Additional file 5: Figure S5, Additional file 6: Figure S6, Additional file 7: Figure S7, Additional file 8: Figure S8, Additional file 9: Figure S9, Additional file 10: Figure S10, Additional file 11: Figure S11, Additional file 12: Figure S12, and Additional file 13: Figure S13) corroborate recent findings [16,29,30,35,36,38,40-43,47,77,78] with bootstrap values (BP) > 70% for 64% of the nodes. This suggests that despite a large proportion of missing data the present molecular character sample provides information about rodent evolutionary affinities. Simulations and large scale analyses have shown that missing data may not lead to inaccuracies in phylogeny reconstruction. As an example, Wiens [84] concluded that “the reduced accuracy associated with including incomplete taxa is caused by these taxa bearing too few complete characters rather than too many missing data cells”. Philippe et al[85] came to the same conclusion using a eukaryote protein supermatrix and computational simulations, and remarked that as much as 75% of the data could be missing without significantly decreasing the reliability of the phylogeny produced. AU [75] and SH tests [76] were used to compare our best topology with trees inferred from two reduced datasets containing 56% (i.e., 1254 taxa and 4130 sites) and 39% (i.e., 371 taxa and 4130 sites) of missing data respectively. Topological tests did not find significant difference (P > 0.05) between the best tree and the topological hypothesis obtained from both reduced datasets. Our findings corroborate results of [84] and [85] as we recovered most relationships inferred in previous works at lower taxonomic levels, an indication that enough informative characters were present to mitigate the effect of missing data. Of course, we acknowledge that the rodent phylogeny here presented has to be ameliorated because of the suboptimal gene and taxon coverage, but we really think it is a reasonable approximation of the rodent phylogeny which accuracy is sufficient to allow for diversification analyses.
Imbalance and shifts in diversification rate within rodents
Whole-tree tests conducted on the complete species sampling indicate significant variation in diversification rates among rodent lineages (Table 4, Figure 3). Except for Castorimorpha and Anomaluromorpha, all P-values of the 4 topology-based indices of whole-tree symmetry (IC, MΠ*, Mσ*, B1) within rodent subclades (Myomorpha, Sciuroidea, Ctenohystrica) ranged from significant (P <0.05) to highly significant (P <0.001). Myomorpha is the most imbalanced clade followed by Sciuroidea, Ctenohystrica, Castorimorpha and Anomaluromorpha (Table 4). The significant shifts in diversification rate (at P < 0.05 level) within rodent taxa under the delta shift statistics (Δ1) are reported in the Table 1, Figure 2 and Additional file 1: Figure S1, Additional file 2: Figure S2, Additional file 3: Figure S3, Additional file 4: Figure S4, Additional file 5: Figure S5, Additional file 6: Figure S6, Additional file 7: Figure S7, Additional file 8: Figure S8, Additional file 9: Figure S9, Additional file 10: Figure S10, Additional file 11: Figure S11, Additional file 12: Figure S12, and Additional file 13: Figure S13. The (Δ1) statistics suggests that unequivocal shifts in diversification rate (SDR) occurred within the 4 major rodent clades, with two significant SDR (0.01 < P < 0.05) in Castorimorpha, 3 significant SDR in Ctenohystrica, 6 significant SDR within the squirrel-related clade, and 24 significant SDR in Myomorpha. We also detected one, 4, 3, and 9 marginally significant SDR (0.05 < P < 0.10) in Castorimorpha, Ctenohystrica, the squirrel-related clade and in Myomorpha respectively.
Table 4.
Tests of among-clade diversification rate using 4 topology-based indices of whole-tree symmetry in Rodentia
|
Clade |
IC |
MΠ |
Mσ |
B1 |
|---|---|---|---|---|
| |
Min (0.025) |
Min (0.025) |
Min (0.025) |
Min (0.025) |
| Max (0.975) | Max (0.975) | Max (0.975) | Max (0.975) | |
| Rodentia |
35928−35412 |
(−)0.912(−)0.905 |
0.574−0.575 |
1087.040−1084.840 |
| |
0.001−0.001 |
0.001−0.001 |
0.001−0.001 |
0.001−0.001 |
| Myomorpha |
23741−23739 |
(−)0.911(−)0.904 |
0.578−0.571 |
761.502−760.302 |
| |
0.001−0.001 |
0.001−0.001 |
0.001−0.001 |
0.001−0.001 |
| Sciuridae |
2778−2655 |
(−)0.813(−)0.754 |
0.607−0.628 |
150.188−156.420 |
| |
0.001−0.001 |
0.001−0.001 |
0.001−0.001 |
0.001−0.006 |
| Ctenohystrica |
2233−2093 |
(−)0.707−(−)0.630 |
0.628−0.660 |
136.493−140.953 |
| |
0.001−0.001 |
0.001−0.001 |
0.001−0.002 |
0.001−0.010 |
| Castorimorpha |
549−507 |
(−)0.705−(−)0.618 |
0.620−0.669 |
49.591−52.810 |
| |
0.020−0.040 |
0.004−0.030 |
0.002−0.060 |
0.004−0.170 |
| Anomaluromopha |
5−0 |
(−)0.080−(−)0.802 |
0.901−1.100 |
3.302−4.000 |
| 0.800−1.000 | 0.800−0.001 | 0.600−0.800 | 0.620−1.000 |
Values represent the test statistics with the P-value on the second line for each clade. The range of values represents the upper and lower bounds generated when the analyses were repeated with 1,000,000 random resolutions of polytomies with different degrees of symmetry. Indices are Colless Index (IC), Shao and Sokal Index (B1) and the M statistics MΠand Mσ.
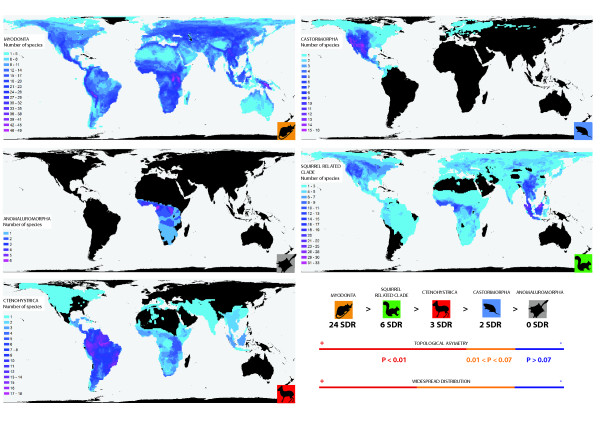
Figure 3.
Current distribution maps of the major rodent clades. Color gradient represents species richness - a warmer color indicates higher richness. Black corresponds to areas where the group is not present. The maximum number of species in a cell (warmest color) for each clade is 49 (Myodonta), 16 (Castorimorpha), 6 (Anomaluromorpha), 33 (squirrel-related clade), and 18 (Ctenohystrica) respectively. Distribution width and topological asymmetry are indicated on the lower right part of the figure, together with the number of significant SDR (shifts in diversification rate).
Table 1.
Rodent sister-clades with significant (P < 0.05) and marginal (0.05 < P < 0.10) shifts in diversification rate using Δ1shift statistics
| Clade | Δ1 | P-value |
|---|---|---|
| CTENOHYSTRICA |
|
|
| (1) Bathyergidae: Heliophobius / Bathyergus,CryptomysClade |
1.99 |
0.09 |
| (2) Caviomorpha: Dasyproctidae / Caviidae |
2.04 |
0.06 |
| (3) Octodontoidea: Base of Octodontidae+Ctenomyidae / Echimyidae |
2.40 |
0.05 |
| (4) Echimyidae : Base Proechimys |
2.75 |
0.05 |
| (5) Ctenomyidae: Ctenomys leucodon/ Sister clade |
3.46 |
0.01 |
| (6) Ctenomyidae: Ctenomys maulinusclade / Sister clade |
2.23 |
0.06 |
| (7) Ctenomyidae: Ctenomys mendocinus / Sister clade |
2.54 |
0.06 |
| SCIUROIDEA |
|
|
| (8) Sciuridae: Aplodontidae / Sciuridae |
2.25 |
0.05 |
| (9) Sciuridae: Sciurillus / other Sciuridae |
5.36 |
0.01 |
| (10) Sciuridae: Flying squirrel clade (Pteromyini tribe) |
2.82 |
0.01 |
| (11)Sciuridae: Nannosciurus / Sister clade |
3.28 |
0.02 |
| (12) Sciuridae: Dremomys clade / Callosciurus clade |
3.28 |
0.03 |
| (13) Sciuridae:Paraxerus clade / (Tamias, Spermophilus) clade |
1.73 |
0.08 |
| (14) Sciuridae: Protoxerus clade / Funisciurusclade |
1.96 |
0.07 |
| (15) Sciuridae: Tamias sibiricus / Sister clade |
1.54 |
0.10 |
| (16) Sciuridae: Spermophilus francklini / Sister clade |
2.17 |
0.05 |
| CASTORIMORPHA |
|
|
| (17) Geomyidae: Castoridae / Geomyioidea clade |
2.17 |
0.05 |
| (18) Geomyidae: Zyzogeomys / Orthogeomys clade |
3.04 |
0.01 |
| (19) Geomyidae: Chaetodipius fallax / Chaetodipius penicillatusclade |
2.27 |
0.07 |
| MYOMORPHA |
|
|
| (20) Anomaluromorpha / Myomorpha |
1.81 |
0.07 |
| (21) Typhlomys / other Muroidea |
3.18 |
0.02 |
| (22) Spalacidae / Eumuroidea |
3.18 |
0.02 |
| (23) Calomyscus / other Muroidea |
1.97 |
0.06 |
| (24) Nesomyidae / other Muroidea |
1.97 |
0.06 |
| (25) Cricetidae / Muridae |
2.18 |
0.05 |
| (26) Murinae: Batomys division / other Murinae |
3.42 |
0.05 |
| (27) Murinae : Micromys / rest ofRattusclade |
4.08 |
0.01 |
| (28) Murinae : Chiropodomys / Sahul + Philippine murine sister-clade |
2.27 |
0.07 |
| (29) Murinae :Vandeularia / Sister clade |
4.03 |
0.01 |
| (30) Murinae : Golunda / Sister clade |
3.03 |
0.02 |
| (31) Deomyinae / Gerbillinae |
4.69 |
0.01 |
| (32) Gerbillinae: Taterillus clade / Gerbillus,Meriones clade |
1.66 |
0.09 |
| (33) Gerbillinae: Gerbillusclade |
3.38 |
0.02 |
| (34) Deomyinae : Deomys / Acomys,Lophuromys clade |
3.65 |
0.01 |
| (35) Arvicolinae : Arvicolinae without Prometheomys |
4.29 |
0.01 |
| (36) Arvicolinae : Dicrostonyx clade / Microtusclade |
2.07 |
0.05 |
| (37) Arvicolinae : Microtus xanthognathus / Sister clade |
2.23 |
0.06 |
| (38) Neotominae : Ochrotomys / Sister clade |
2.26 |
0.05 |
| (39) Neotominae :Baiomys/Scotinomys clade / Sister clade |
2.26 |
0.05 |
| (40) Neotominae : Peromyscus crinitusclade / Sister clade |
2.17 |
0.05 |
| (41) Sigmodontinae : Sigmodontini tribe / Oryzomyala |
1.55 |
0.10 |
| (42) Sigmodontinae : Oxymycterus clade / Akodonclade |
1.98 |
0.08 |
| (43) Sigmodontinae : Akodon cursor clade / Akodon lutescensclade |
2.61 |
0.03 |
| (44) Sigmodontinae : Aepeomys / Sister clade |
2.41 |
0.04 |
| (45) Sigmodontinae : Thomasomysclade |
3.05 |
0.03 |
| (46) Sigmodontinae : Eremoryzomys polius / Sister clade |
2.29 |
0.05 |
| (47) Sigmodontinae : Cerradomysclade / Sister clade |
2.29 |
0.05 |
| (48) Sigmodontinae : Sooretamys angouya / Sister clade |
2.56 |
0.05 |
| (49) Sigmodontinae : Phyllotini and some Akodontini taxa / Oryzomyini tribe sensu lato |
1.93 |
0.06 |
| (50) Sigmodontinae Wiedomys / Sister clade |
2.64 |
0.04 |
| (51) Sigmodontinae : Andinomys / Sister clade |
3.44 |
0.02 |
| (52) Sigmodontinae : Calomys clade / Phyllotis clade | 3.45 | 0.02 |
Clades can be found in the Additional file 2: Figure S2, Additional file 3: Figure S3, Additional file 4: Figure S4, Additional file 5: Figure S5, Additional file 6: Figure S6, Additional file 7: Figure S7, Additional file 8: Figure S8, Additional file 9: Figure S9, Additional file 10: Figure S10, Additional file 11: Figure S11, Additional file 12: Figure S12, and Additional file 13: Figure S13. Δ1represents the delta shift-statistics of [7] and [73].
Relationships significant at P = 0.05 are shown in bold. Clades in bold are the most diversified ones.
Diversification rate varies significantly in rodents, a pattern also observed in bats and lagomorphs ([86], Figure 3 and Table 4) and other vertebrate clades [87,88]. Within Rodentia, Myomorpha displayed higher imbalance in comparison to Sciuroidea, Caviomorpha, Castorimorpha and Anomaluromorpha. Our results suggest that rodents underwent a number of significant SDR, especially within the Cricetidae, Muridae, Sciuridae and Octodontoidae. These different imbalance signatures suggest distinct diversification processes among rodent lineages. Many hypotheses have been proposed to explain these evolutionary radiations. The most common explanations are key innovations (e.g. hypsodonty or teeth patterns like the murine or cricetine dental plans, [40]), events related to biogeographical history (e.g. colonization of south America by Sigmodontinae; [41,89]), extinction of competitors (e.g. multituberculate and plesiadapid extinction through the Paleogene; [90]), a predator absence (e.g. insular New Guinea / Papua murine diversification), and / or environnemental changes (e.g. opening of habitats). All these factors could have played a role during the rodent radiations.
The most imbalanced clade is the Myodonta (Muroidea + Dipodidae). Most shifts in diversification rates (Table 1, Figure 2) are located within the two most speciose muroid families: the Cricetidae (681 species) and the Muridae (728 species). Of the 24 significant SDR, 7 and 14 shifts are located within Cricetidae and Muridae respectively. For Cricetidae, accelerations of the diversification rates were found for 3 clades of Neotominae (North-American Cricetidae), 10 for Sigmodontinae (South-American Cricetidae) and two within Arvicolinae (voles). Within the Muridae, accelerations in the diversification rates are found for one clade of Deomyinae, two of Gerbillinae and 4 within Murinae. The outstanding muroid diversity in both tropical and boreal habitats is peculiar within the evolutionary history of the placentals (Figure 3). Muroid rodents comprise 28% of mammal species and this superfamily [13] is larger than any other non-rodent orders. Our analyses agree with three conclusions of Steppan et al.[41], who delineated 4 bursts of speciation within their Muroidea timetree: (i) the initial radiation of the Eumuroidea (SDR 22), (ii) the radiation among cricetid families (SDR 25), (iii) the initial radiation among Oryzomyala sigmodontines (SDR 41) [89,91], and (iv) the initial radiation among the Murinae (at the exception of Batomys division (SDR 26)). To explain these speciation bursts, they referred to an increase in speciation rate due to evolutionary and biogeographic events. In fact, the major centers of the muroid diversification overlapped most continents in both hemispheres: America and Palearctic for Cricetidae, and Old World and Sahul for Muridae. Key opportunities, such as colonization of new areas are well-known for driving speciation and have contributed significantly to the diversification of organisms [92,93]. These dispersal events led some organisms to exceptional evolutionary [94-97] or adaptive radiations [2,98-100]. The importance of colonization is considered essential for the radiations and acceleration of the speciation rates within Muroidea. Some works on murinae have recently confirmed the role of biogeographic events in driving shifts in diversification, for example the colonization of the Sahul by Murinae [47] (SDR 28), the radiation of Rattus in South East Asia and Sahul [51] (SDR 27) and the colonization of Africa by the PraomysMus and Arvicanthis lineages (SDR 29) [46,101]. The radiation of Sigmodontinae have also been related to the colonization of the South American continent [91]. From the beginning of the Miocene (24.7 +/- 1.1 Mya cf.[41]), the radiation of Muroidea have caused a major turnover in the composition of rodent lineages as suggested by the fossil record [102,103] and by our topological-shape results. Our study (Figure 3), as well as recent phylogenetic works on Eumuroidea clades strongly support the role of colonization processes in SDR. In addition, lineages that originated during these radiations exhibited a broad array of both ecological generalist and specialists within different colonized areas. Compared to the other rodent clades, Muroidea includes smaller sized and less “specialized” taxa [104]. The high diversity pattern in such small size taxa has been linked to the shortest generation time among terrestrial mammals [105] and to a better partition of ecological niches [106]. Evidence so far is consistent with these hypotheses, for instance previous works on primates and carnivores have found marginally significant association between diversification and body mass [11,12]. Muroids displays the highest molar diversity among Rodentia associated to a wide number of dental vicariants due to convergent evolution [40,107-109]. Their small size, their teeth diversity and their “generalist” morphology could be linked to their recent success. They have succeeded to colonize new areas and to diversify in more habitats than their more specialized sister clades (e.g., arboreal squirrels, porcupines, mole rats, and ancient American endemics like Caviomorpha and Castorimorpha).
Within Sciuridae, two significant SDR (P < 0.05) occurred at the origin of the family, and along the branch leading to the Sciurillini tribe and the rest of the Sciuridae, two within the Callosciurinae subfamily, one within the Pteromyini tribe, and then one significant SDR within the Xerinae subfamily. The Sciuridae is characterized by a wide geographic distribution (Figure 3) and a high specific diversity (278 species) associated to many adaptive trends (terrestrial, arboreal and gliding). Mercer and Roth [36] showed that Cenozoic global changes mediated their diversification history. After the Eocene, the colonization of major land masses by the Sciuridae have led to their diversification within forest or open habitats. The squirrel-related clade is widespread like Myomorpha but compared to Muroidea its members display more constrained adaptations and morphologies. These differences could explain why they have higher imbalance index (Table 4, Figure 3) than Ctenohystrica and Castorimorpha and less than Myomorpha. Most of the Ctenohystrica radiation is represented by Caviomorpha which have undergone endemic evolution in South America. Caviomorpha have colonized South America from Africa [35,110,111]. We did not detect a significant shift at the root of Caviomorpha in our analysis despite their high diversity. Such a result could be a consequence of the extinction of taxa of the earliest Caviomorpha radiations [112-115]. Octodontoidea are the most speciose suborder within the Caviomorpha comprising the Echimyidae (South American spiny rats), the Ctenomyidae (tuco-tucos), the Abrocomidae and the Octodontidae. They underwent an adaptive radiation in South America during the Miocene with scansorial (Capromys), fossorial (Ctenomyidae), terrestrial (Trinomys), semi aquatic (Myocastor) and arboreal (Echimys) representatives. Concerning Echimyidae, Galewski et al.[44] did not resolve the origin of this clade with one nuclear gene, a pattern possibly associated with rapid diversification events. They invoked the role of paleoclimatic variation as a driving force through their radiation in the Miocene. Our results converge on the same conclusion with two shifts occurring at (1) the split between Caviidae vs. Dasyproctidae (SDR 2) and (2) the divergence between Echimyidae and (Ctenomyidae+Octodontidae) (SDR 3). These clades display adaptations to open habitats (Caviidae and (Ctenomyidae + Octodontidae)) or forest habitats (Dasyproctidae and Echimyidae) where they subsequently diversified. Miocene climatic changes in South America may have played a major role in the diversification of Caviomorpha as suggested by the fossil record [116,117], molecular dating results (herein and also [26,44,54,111]) and our SymmeTREE results. Castorimorpha and Anomaluromorpha clades display high morphological and ecological constraints with fossorial (Geomyioidea), gliding (Anomaluridae) or jumping (Pedetidae and Heteromyidae) adaptations. Moreover they display high endemism like Caviomorpha (Figure 3). Anomaluromorpha are only found in Africa and Castorimorpha are mainly distributed in North America (except Castor fiber) (Figure 3). Their geographical distribution and their specialized morphology could explained the difference in the imbalance analyses and the low number of inferred SDR in comparison to other rodent clades.
Investigating correlates of diversification shifts for the Rodentia remains a challenge, and a variation in a single trait is unlikely to explain all shifts detected. In this framework, methods incorporating paleoclimatic and biogeographic information would be informative. Such an approach could be useful for clades such as the Cricetidae or Muridae where numerous shifts in diversification were recorded.
The Paleogene / Neogene contrast of the rodent timetree
Calibrating phylogenetic trees is a difficult problem for data with a patchy taxonomic sampling and markers with heterogeneous patterns of molecular evolution. Likelihood ratio tests [118] rejected the molecular clock for the 11 genes. This result is not surprising as rates variations have been evidenced for rodent mitochondrial and nuclear genes. To get maximum dating signal, genes were analyzed in combination to infer divergence times. Calibration of our ML trees using the partitioned Bayesian relaxed clock model of [119,120] provides an estimate of the rodent timetree (Figure 1). All analyses with different MCMC sampling converged to the same divergence time estimates.
Our supermatrix-based molecular clock approach simultaneously calibrated by multiple fossil constraints provides an alternative to previous dated supertrees [61] because we use the concatenated information of independent molecular markers rather than averaging over independent source analyses.
Molecular dating here suggests that many extant families originated during the Paleogene. The divergence dates of rodent families indicate that they were all established before the end of Oligocene (Median family age= 31 Mya). The majority of radiations leading to extant rodent diversity seems to have occurred during the Neogene (Median age of generic radiation = 22 Mya) with some exceptions such as the older diversification of the Sciuroidea or the Phiomorpha families. Analysis of diversification rates shows that statistically significant (P < 0.05) and substantial diversification shifts (0.05 < P < 0.1) were concentrated in the Neogene, and that the majority of SDR occurred around 10 Mya during the middle Miocene. Means of the absolute value of the delta shift statistic for nodes of the rodent clades in each geological epoch are presented in the Figure 2 (Upper Part: B). We obtained the largest values from Paleocene intervals (65.5-55.8 Mya) (Figure 2 B). Mean values in the SDR are significantly different among time intervals (one-way ANOVA, F 5,1259 = 13.42, P < 0.01). The mean value for the Paleocene (65.5-55.8 Mya) is significantly larger than in the Pliocene and Quaternary (Tukey test P < 0.01 and P < 0.01), and is not significantly different from the Eocene, Oligocene and Miocene time intervals (Tukey test P < 0.40, P < 0.07, P < 0.09). We examined the distribution of species in each clades that were present since 65.5 Mya to identify which lineages were responsible for the large SDR within different geological periods. The lineages leading to Myodonta, Sciuroidea+Gliridae, Castorimorpha and Ctenohystrica were present before 65.5 Mya and displays most of the extant diversity of rodents. During the 60-40 Mya period, the first rodent families emerged in the fossil record and explosive radiations took place [121]. Because there is no significant difference in SDR from the Paleocene to the Miocene, it seems that rodent clades have diversified at a fairly constant rate during these epochs.
Rodents have undergone regular Shifts in Diversification Rate (SDR) through the Cenozoic (Figure 2). Among the 35 significant SDR (see previous section), only six took place during the Paleogene. However, the fossil record has revealed that the Paleogene was a period of intensive rodent diversification with the appearance of 9 new families (i.e. Cylindrodontidae, Eutypomyidae, Sciuravidae, Gliridae, Zegdoumyidae, Chapattimyidae, Cocomyidae, Ivanantomyidae, and Yuomyidea) [122]. According to their period of diversification (i.e. Paleogene or Neogene), two groups emerged from the timeline analysis: the first included the sciurid-related clade and the Castorimorpha, whereas the second included Myodonta, Anomaluromorpha and Ctenohystrica. The first group is characterized by older generic divergences and a higher density SDR through the Paleogene and this is also attested by the richness and occurence of the fossil record of Gliroidea [123-125], Aplodontoidea [126,127] and Castorimorpha [102]. In the second group, the mouse-related clade and Ctenohystrica have the majority of generic divergences and SDR through the Neogene. Within Muroidea, even if stem Cricetidae occurred in the Eocene and Oligocene fossil records [128], it is now clear that the extant subfamilies diversified during the Neogene. Numerous cladogenesis events are identified during the Neogene within the Muroidea, especially in the Cricetidae and Muridae (Figure 1 and 2) that represent the most important and recent evolutionary radiations. This result is congruent with the richness of their fossil record during the Neogene (Figure 2 - [103,128-130]).
Comparisons between results from our diversification analyses and the available fossil record point to a late Paleogene or Neogene radiation of extant rodent lineages. The extinction of stem lineages could also explain the low number of speciation events detected in most stem branches. These results corroborate the macroevolutionary study of Bininda-Emonds et al [61] who observed a delay between the KT boundary and the Neogene regarding the diversification of placentals (see also [62,131]). The long branches leading to Geomyoidea or extant Ctenodactyloidea (Ctenodactylidae + Diatomyidae) could be explained by the extinction of stem Castorimorpha and Ctenodactyloidea. The diversification of crown rodents from the late Eocene onwards coincides with the extinction or decline of the major Paleogene fossil groups (Figure 2C - [103]). Several extinct groups, without extant relatives (e.g. Theridomorpha, Ischyromyoidea, Ctenodactyloidea, and Sciuravida), disappeared or declined in the Oligocene and the Neogene (Figure 2C). Simultaneously, most of the relatives of extant species played a major role in rodent communities during that period, in particular the Muridae and Cricetidae (Figure 2 and 3). Because extinction processes may have biased the interpretation of SDR, future studies should incorporate fossil data in supermatrix/supertree inferences.
Conclusions and Perpectives
The present study is a first attempt to provide a phylogenetic synthesis to be used for comparative meta-analyses of rodent evolution (topology are available in the Additional file 14). We demonstrated that the diversification rates of rodent taxa were not constant through time and some clades have experienced significant shifts in diversification rates. Our results show that most widespread and diversified clades (Myodonta and the squirrel-related clade) display a higher degree of topological asymmetry and more SDR. Recent opportunities to colonize new geographical areas must have driven speciation and contributed significantly to the diversification of both groups. Numerous SDR are evidenced through the Tertiary, but at different periods for each clade. The majority of these shifts occurred for the most recent familial rodent radiations: the Cricetidae and Muridae clades. Comparison between the rodent fossil record and our results suggest that extinctions led to the loss of diversification signal for the Paleogene nodes. The main perspective of this study is to provide a framework for comparative studies of rodents and an update of large scale phylogenies of this order. The ML trees (summarized in Additional file 1: Figure S1 and Additional file 2: Figure S2, Additional file 3: Figure S3, Additional file 4: Figure S4, Additional file 5: Figure S5, Additional file 6: Figure S6, Additional file 7: Figure S7, Additional file 8: Figure S8, Additional file 9: Figure S9, Additional file 10: Figure S10, Additional file 11: Figure S11, Additional file 12: Figure S12, and Additional file 13: Figure S13) corroborate recent multigene analysis with bootstrap values (BP) > 70% for 64% of the nodes. The occurence of taxa not studied in a phylogenetic framework and lack of DNA data for many of the genetic markers, however, constitute the main challenge for the further clarification of rodent evolution.
One avenue for further research is to explore the morphological / biogeographical drivers of diversification. The use of ancestral character reconstruction methods will be required to test if there are correlations between phenotypic innovations or biogeographic events and diversification in rodents. The exploration of macroevolutionary patterns and their link with morphological innovations, biogeography or climatic events is a key for a better understanding of the mammalian Cenozoic radiations.
Methods
Taxonomy
All species names followed the rodents classification of Carleton and Musser [132]. We chose this classification – recognizing about 2,261 rodent species – because it is the most recent update, and it is widely used and cited in the mammalian biology literature. We added the newly discovered genus Laonastes [14,16] which had not been described in reference [132]. The Carleton and Musser [132] taxonomy provides the most recent and accepted species list for Rodentia that also includes species synonyms. Tracing synonyms is essential for establishing congruence among different gene datasets that have used different names for the same taxa. Synonyms that coud not be traced in public databases for available molecular markers were excluded from subsequent analyses.
Sequence data
In order to collect suitable candidate genes for the supermatrix assembly, DNA sequences of rodents were downloaded from EMBL / GenBank / DDBJ databases. Keyword frequency searches were performed to collect genes that were sequenced over a large taxonomic range using rodent species and genus names [132]. For these searches we focused on genes that have been previously used to infer rodent phylogenies. Refined searches were then performed using the rodent section of the NCBI taxonomic browser and BLAST [133] searches on euarchontan assembled genomes (mouse, rat, rabbit, human and rhesus macaque). This cross-search allowed for the retrieval of an extensive dataset of all rodents DNA sequence data available in public repositories. If multiple DNA sequences were available for the same taxon we checked its monophyly by using literrature and keep the most complete of the fragments prior to subsequent analyses. During the course of our study some additional sequences become available (e.g. [53,134]) but were not included in the analyses.
In this study we focused on the 11 nuclear and mitochondrial markers which allow us to maximise rodent species sampling (Table 2 and Table 3). Following this procedure we harvested 1,265 DNA sequences. The resulting dataset represents 100% of the families (33 families), 81% of the genera (387 of 474 genera), and 56% of the species (1,265 of 2,261 species) of rodents currently recognized in Wilson and Reeder [132] and recent phylogenetic works were also taken into account (eg. [50,89,135-141]). The rodent taxonomy adopted for the present study followed references [25,27,132] and is provided as Additional file 15. Due to the size of this dataset many taxa suffer from large amount of missing data, but all share at least one mitochondrial or nuclear gene, thus avoiding the problem of non overlapping sequences [142].
The rodent outgroups were chosen among the Euarchontoglires [143] for which genomes were available (OryctolagusMacacaHomo). If available, one Scandentia (Tupaia), one Dermoptera (Cynocephalus) and two additional Lagomorpha (OchotonaLepus) outgroups were added to each gene. DNA sequences were aligned with MUSCLE [144] and subsequently checked by eye with SEAVIEW [145]. For the 12S rRNA and 16S rRNA alignments, ambiguous positions were eliminated using the Gblocks program (version 0.91b, [146]) with the following options: a minimum of half the number of sequences for a conserved position and for a flank position, a maximum of 8 contiguous non-conserved positions, a minimum of 2 sites for the block length after gap cleaning, and all gap positions can be selected. The supermatrix concatenate contains 1265 rodent taxon sequences aligned for 15,535 sites, with 75% of missing character states. If necessary, non overlapping sequences (e.g. sequences available for different species of the same genus) were eliminated from the matrix. All genes are described in Table 2 and all datasets are available online (also see additional file 16 and Additional file 17 for accession numbers).
Phylogenetic analyses
The general time reversible (GTR) model plus invariable sites and Gamma (Γ) distribution [81] was selected as the best fit under the AIC criterion using Modeltest 3.04 [147]. The dataset was partitioned by codon positions for exons. Maximum likelihood (ML) analyses were run with RaXML version 7.0.4 [148]. For the dataset partitioned only by gene and codons, we applied to each partition the GTRGAMMA (GTR+Γ) + Invariant site option. For the gene-codon-partition dataset, we used the GTRMIX option of RAxML. The GTRMIX option assumes the faster GTRCAT model for the topological search, but then uses the GTRGAMMA model when computing the likelihood value of the topology. Each RAxML run comprised 100 tree search replicates (with the default parameters).
Node support for codon/gene-partioned datasets was estimated by the means of non-parametric bootstrap resampling [149]. Bootstrap proportions (BP) were calculated with the following procedure: 100 pseudoreplicates for the supermatrix and 1000 pseudoreplicates for each single-gene matrix. Pseudoreplicate trees were inferred using the ML method in RAxML under a GTRMIX model.
In order to evaluate the impact of missing data on our inference we built two additional matrices: (1) a supermatrix containing the 4 genes with best taxonomic coverage (cf. 12S rRNA + CYTB + RBP3 + GHR ; 56% of missing character states) and (2) a supermatrix containing the same 4 genes (39% of missing data) but maximizing the taxon sampling at the genus level. These supermatrices were subsequently analysed with RaxML following the same procedure as described for the 11-gene supermatrix. The two inferred topologies were compared to the 11-gene topology after restriction to the subset of shared taxa and using the approximately unbiased (AU) [75] test as implemented in CONSEL [150]. PAUP* version 4.0b10 [151] was used to calculate the site likelihoods for each of the test topologies with the GTR + I + Γ model as specified using the output from Modeltest 2.2. The CONSEL analyses employed 10 batches of 106 bootstrap replicates.
Diversification rate analysis
To estimate diversification rates we used the phylogeny with the complete taxon sampling according to the classification of reference [132]. Species for which no DNA data were available were grafted to the most recent common ancestor of the closest relative taxon available within our molecular framework, i.e., a species of the same genus, or tribe, or family. In this way, one composite topology was generated from the supermatrix analyses and the taxonomic list.
To study species diversification patterns, 4 topology-based indices of whole-tree symmetry were employed [7,152,153]. All 4 methods (IC, MΠ*, Mσ*, B1) use an equal rates Markov (ERM) model of clade growth [154] to test how well a tree fits to the equal-rates null hypothesis. A taxonomic imbalance in extant lineages is found if nonrandom diversification has taken place. Each topological-based statistic was calculated using a Monte Carlo simulation of its null distribution using 1,000,000 tree topologies of the same size as our rodent phylogeny, but generated under an ERM model. We used this approach on the complete topology. Analyses of tree symmetry and identification of diversifying clades were performed with SymmeTREE version 1.1 [7,73]. Because polytomies in the tree may bias SymmeTREE analysis [7], they were treated as soft.
To identify the nodes of the tree that show significant imbalance, the delta-shift method (Δ1) was used [7]. This likelihood topological-based method searches for significant shifts in diversification rates (SDR), and incorporates information on the distribution of taxonomic diversity over the entire tree. The delta shift-statistics determines the diversification rate shift probability along the internal branch of a local triplet tree that includes the two basal-most ingroup clades and a local outgroup. The three-taxon computations are replicated over all internal branches to check for diversification rate shifts within the whole tree [7]. The Δ1distribution was obtained by means of Monte Carlo simulation of its null distribution, using 1,000,000 topologies of the same size as the final tree, but generated under an ERM model.
Estimating divergence times within rodents
Ideally, all the 1,265 species would have been analysed simultaneously within a single molecular dating analysis. However probabilistic search algorithms become prohibitively slow for a large number of taxa and are less likely to identify an optimal dated topology. In an attempt to approach this problem and to reduce computational time, a compartimentalization approach [155] was used. The global chronogram was constructed after analysis of hierarchically nested supermatrices. Ultimately, 8 supermatrices (Sciuroidea; Ctenohystrica; Castorimorpha; Anomaluromorpha + Dipodoidea + Platacanthomyidae + Spalacidae; Sigmodontinae + Tylomyinae; Neotominae; Arvicolinae + Cricetidae; Murinae; Gerbillinae + Acomyinae + Lophiomyinae) were built with subsamples of genes as indicated in Additional file 18. We used BEAST v1.6 [119,120] to estimate the divergence dates within our 8 supermatrices, by applying the best fitting model, as estimated by MODELTEST 2.0 to each of the partitions. We assumed prior Yule speciation process and an uncorrelated lognormal distribution for the molecular clock model [156]. Default prior distributions were used for all other parameters, and two independant MCMC chains were ran for 200 million generations. The program Tracer [157] was used to assess convergence diagnostics, and showed that each run reached similar date estimates for all nodes.
Three calibration constraints based on paleontological estimates and previously used for rodent molecular dating studies [16,41,71] were incorporated: the Caviomorpha radiation (28.5 to 37 Mya ; [158,159]), the Aplodontidae / Sciuridae divergence (37 to 50 ; Myr, [103]), and the Glis - Dryomys split (28.5 to 50 Mya ; [122]). Based on the 61.5 to 100.5 Mya estimate for the divergence between Lagomorpha and Rodentia [160], the a priori expected root height was set to 100 Mya with a standard deviation of 50 Myr. Finally, the overall dated tree was reconstructed by combining results from the hierarchically nested supermatrices.
The resulting chronogram has been used to study the occurrences of significant SDR throughout the Tertiary. To do so we followed the methodology of Jones et al. (2005), using speciation date estimates to calculate the mean of the absolute value of the delta shift statistic (Δ1) in each geological epoch (1 : Paleocene (65.5-55.8 Mya), 2 : Eocene (55.8-33.9 Mya), 3 : Oligocene (33.9-23.03 Mya), 4 : Miocene (23.0-5.3 Mya), 5 : Pliocene (5.3-1.8 Mya), 6 : Quaternary (1.8-0 Mya)).
Clades distributions and richness
Clades distributions / species richness maps were created using gridded species distribution data from Fritz and collaborators [161,162]. Grid cells with equal surface of 9309.6 square kilometers were used. Species presence/absence was recorded for each species and each cell for all the species in every major lineage. Species richness was then calculated as the total number of species co-occurring in every cell. The overlap of the species distributions is used to represent the distribution of the higher level taxon to which they belong and the color gradient within its range represents species richness. Areas where the lineage is not present are left black. Resulting maps were drawn using the Behrmann projection and manipulated in ArcGIS 9.3 computer program (ESRI Inc.).
Authors contributions
P-HF designed the project, planned and conducted analysis and wrote the manuscript. All authors analysed data and contributed to the writing and editing of the manuscript. All authors approved the final manuscript.
Supplementary Material
Figure S1. Rodent species level evolutionary tree. Species-level phylogenetic topology based on the highest-likelihood tree inferred from the 11-gene supermatrix, and combined with the taxonomic information of Wilson and Reeder (2005).
Figure S2. Cladogram depicting the highest-likelihood topology for the Ctenohystrica. Circles at nodes represent bootstrap support (circles: black 100-95%, white 95-70%, gray 70-50%). Maximum likekihood tree (lnL=-714749.2). Displayed clade are highlighted using the simplified full ML topology on the left side of the figure. Molecular marker sampling is depicted for each taxa to the right of the tree. Names of the genes are given. Genes sampled for our dataset (Table 2) are marked in black square while missing genes are symbolized by white squares. Triangles indicate significant (P < 0.05) and marginal (0.05 < P < 0.1) shifts in diversification rate (SDR) as inferred by Δ1. LA = Diatomyidae, CT = Ctenodactylidae, TH = Thryonomyidae, PE = Petromuridae, CU = Cuniculidae, DASYPROC = Dasyproctidae, DI = Dinomyidae, CHINCHILLID = Chinchillidae. Bootstrap for all nodes and topology can be found in Additional file 14.
Figure S3. Cladogram depicting the highest-likelihood topology for the Octodontoidea. See Additional file 2: Figure S2 for details of the legend. AB = Abrocomidae, CA = Capromyidae.
Figure S4. Cladogram depicting the highest-likelihood topology for the Sciuroidea. See Additional file 2: Figure S2 for details of the legend. AP = Aplodontidae, RA = Ratufinae, SI = Sciurillinae.
Figure S5. Cladogram depicting the highest-likelihood topology for the Xerinae. See Additional file 2: Figure S2 for details of the legend.
Figure S6. Cladogram depicting the highest-likelihood topology for the Castorimorpha. See Additional file 2: Figure S2 for details of the legend. CAS = Castoridae.
Figure S7. Cladogram depicting the highest-likelihood topology for the mouse-related clade. See Additional file 2: Figure S2 for details of the legend. PED = Pedetidae, ANO = Anomaluroidea, PLA = Platacanthomyidae, Rhy = Rhyzomyidae, Spalac = Spalacidae, CAL = Calomyscidae, LOP = Lophiomyinae.
Figure S8. Cladogram depicting the highest-likelihood topology for Sigmodontinae [part 1] + Tylomyinae. See Additional file 2: Figure S2 for details of the legend. Tyl = Tylomyinae, Ich = Ichthyomyini, Rei = Reithrodontini.
Figure S9. Cladogram depicting the highest-likelihood topology for Sigmodontinae [part 2]. See Additional file 2: Figure S2 for details of the legend. Tyl = Tylomyinae, Ich = Ichthyomyini, Rei = Reithrodontini, Phy = Phyllotini.
Figure S10. Cladogram depicting the highest-likelihood topology for Neotominae. See Additional file 2: Figure S2 for details of the legend. Och = Ochrotomyini, Baiom = Baiomyini.
Figure S11. Cladogram depicting the highest-likelihood topology for Arvicolinae. See Additional file 2: Figure S2 for details of the legend. Pro = Prometheomyini, Dic = Dicrostonychini, Ond = Ondatrini, Plio = Pliomyini, Arv = Arvicolini, Ell = Ellobiusini, Lag = Lagurini.
Figure S12. Cladogram depicting the highest-likelihood topology for Murinae [part 1]. See Additional file 2: Figure S2 for details of the legend. Mic = Micromys division, Cru = Crunomys division, Max = Maxomys division, Mel = Melasmothrix division, Hydrom = Hydromyines division, Con = Conilurines division, Urom = Uromyines division.
Figure S13. Cladogram depicting the highest-likelihood topology for Murinae [part 2]. See Additional file 2: Figure S2 for details of the legend. Mic = Micromys division, Mil = Millardia division, Col = Colomys division, Cre = Cremnomys division, Gol = Golunda, Oen = Oenomys division, Hyb = Hybomys division, Mi = Micaelamys division, Das = Dasymys division, Mal = Malacomys division, Stenocep = Stenocephalemys division.
ML RaxML topology.
Rodentia species list names.
Rodentia accession numbers by gene.
Rodentia accession numbers by taxonomic group (Sheet 1: MYODONTA + ANOMALUROMORPHA; Sheet 2: SCIUROIDEA; Sheet 3: CTENOHYSTRICA; Sheet 4: CASTORIMORPHA).
Loci used in each molecular dating analysis (see Material and Methods).
Contributor Information
Pierre-Henri Fabre, Email: phfmourade@gmail.com.
Lionel Hautier, Email: ljh75@cam.ac.uk.
Dimitar Dimitrov, Email: dimitard.gwu@gmail.com.
Emmanuel J P Douzery, Email: Emmanuel.Douzery@univ-montp2.fr.
Acknowledgements
We thank Laurent Marivaux, Frédéric Delsuc, Kenneth Puliafico, Andrew Rodrigues, Thomas Galewski, two anonymous reviewers and Hervé Philippe for their discussion and constructive comments on this paper. We also thanks Carsten Rahbek and Michael Krabbe Borregaard for providing us with the rodent distribution datasets. This work has been supported by the Agence Nationale de la Recherche “Domaines Emergents” (ANR-08-EMER-011 “PhylAriane”) and “Bio-Informatique” (ANR-10-BINF-01 “Ancestrome”). This publication is the contribution No2012-064 of the Institut des Sciences de l’Évolution de Montpellier (UMR 5554 - CNRS). Lionel Hautier acknowledges the Leverhulme trust and Sidney Sussex College for financial support. P-HF and DD were supported by a grant from the Danish National Research Foundation.
References
- Reeve H, Sherman P. Adaptation and the goals of evolutionary research. Q Rev Biol. 1993;68:1–32. doi: 10.1111/j.1469-185X.1993.tb00731.x. [DOI] [Google Scholar]
- Grant P, Grant B. How and why species multiply: The radiation of Darwin’s finches. Princeton: Princeton University Press; 2008. [Google Scholar]
- Butlin R, Bridle J, D S. Speciation and Patterns of Diversity. Cambridge: Cambridge University Press; 2009. [Google Scholar]
- Alroy J, Aberhan M, Bottjer DJ, Foote M, Fürsich FT, Harries PJ, Hendy AJW, Holland SM, Ivany LC, Kiessling W, Kosnik MA, Marshall CR, McGowan AJ, Miller AI, Olszewski TD, Patzkowsky ME, Peters SE, Villier L, Wagner PJ, Bonuso N, Borkow PS, Brenneis B, Clapham ME, Fall LM, Ferguson CA, Hanson VL, Krug AZ, Layou KM, Leckey EH, Nürnberg S, Powers CM, Sessa JA, Simpson C, Tomasovych A, Visaggi CC. Phanerozoic trends in the global diversity of marine invertebrates. Science. 2008;321:97–100. doi: 10.1126/science.1156963. [DOI] [PubMed] [Google Scholar]
- Barraclough TG, Nee S. Phylogenetics and speciation. Trends Ecol Evol. 2001;16:391–399. doi: 10.1016/S0169-5347(01)02161-9. [DOI] [PubMed] [Google Scholar]
- Nee S, May R, Harvey P. The reconstructed evolutionnary process. Philos Trans R Soc Lond B Biol Sci. 1994;344:305–311. doi: 10.1098/rstb.1994.0068. [DOI] [PubMed] [Google Scholar]
- Moore B, Chan K, Donoghue M. In: Phylogenetic supertrees: combining information to reveal the, Tree of Life. Bininda-Emonds O, editor. Dordrecht, the Netherlands: Kluwer Academic; 2004. Detecting diversification rate variation in supertrees; pp. 487–533. [Google Scholar]
- Rabosky DL. Likelihood methods for detecting temporal shifts in diversification rates. Evolution. 2006;60:1152–1164. [PubMed] [Google Scholar]
- Paradis E. Assessing temporal variations in diversification rates from phylogenies: estimation and hypothesis testing. Proc R Soc Lond B. 1997;264:1141–1147. doi: 10.1098/rspb.1997.0158. [DOI] [Google Scholar]
- Pybus OG, Harvey PH. Testing macro-evolutionary models using incomplete molecular phylogenies. Proc R Soc Lond B. 2000;267:2267–2272. doi: 10.1098/rspb.2000.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittleman J, Purvis A. Body size and species-richness in carnivores and primates. Proc R Soc Lond B. 1998;265:113–119. doi: 10.1098/rspb.1998.0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E. Statistical analysis of diversification with species traits. Evolution. 2005;59:1–12. [PubMed] [Google Scholar]
- Wilson D, Reeder D (Eds) Mammal species of the world. Baltimore: Johns Hopkins University Press; 2005. [Google Scholar]
- Jenkins P, Kilpatrick C, Robinson M, Timmins R. Morphological and molecular investigations of a new family, genus and species of rodent (M, ammalia: Rodentia: Hystricognatha) from Laos. Syst Biodiv. 2005;2:419–454. doi: 10.1017/S1477200004001549. [DOI] [Google Scholar]
- Dawson M, Marivaux L, Li CK, Beard K, Métais G. Laonastes and the “Lazarus effect” in recent mammals. Science. 2006;311:1456–1458. doi: 10.1126/science.1124187. [DOI] [PubMed] [Google Scholar]
- Huchon D, Chevret P, Jordan U, Kilpatrick C. Multiple molecular evidences for a living mammalian fossil. Proc Natl Acad Sci USA. 2007;104:7495–7499. doi: 10.1073/pnas.0701289104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenberger JL. In: Evolutionary relationships among rodents: a multidisciplinary analysis. Luckett W, Hartenberger JL, editor. New York and London: Plenum Press; 1985. The order Rodentia: Major questions on their evolutionary origin, relationships and suprafamilial systematics; pp. 1–33. [Google Scholar]
- Wood A. Grades and clades among rodents. Evolution. 1965;19:115–130. doi: 10.2307/2406300. [DOI] [Google Scholar]
- Hautier L, Michaux J, Marivaux L, Vianey-Liaud M. The evolution of the zygomasseteric construction in Rodentia, as revealed by a geometric morphometric analysis of the mandible of Graphiurus (Rodentia, Gliridae) Zool J Linn Soc. 2008;154:801–821. [Google Scholar]
- Catzeflis F, Hänni C, Sourrouille P, Douzery E. Molecular systematics of hystricognath rodents: the contribution of sciurognath mitochondrial 12S rRNA sequences. Mol Phylogenet Evol. 1995;4:357–360. doi: 10.1006/mpev.1995.1032. [DOI] [PubMed] [Google Scholar]
- Horner DS, Lefkimmiatis K, Reyes A, Gissi C, Saccone C, Pesole G. Phylogenetic analyses of complete mitochondrial genome sequences suggest a basal divergence of the enigmatic rodent Anomalurus. BMC Evol Biol. 2007;7:16. doi: 10.1186/1471-2148-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgelard C, Bentz S, Tirard C, Verneau O, Catzeflis FM. Molecular systematics of Sciurognathi (Rodentia): the mitochondrial cytochrome b and 12S rRNA genes support the Anomaluroidea (Pedetidae and Anomaluridae) Mol Phylogenet Evol. 2002;22:220–233. doi: 10.1006/mpev.2001.1056. [DOI] [PubMed] [Google Scholar]
- Nedbal M, Allard M, Honeycutt R. Molecular systematics of hystricognath rodents: evidence from the mitochondrial 12S rRNA gene. Mol Phylogenet Evol. 1994;3:206–220. doi: 10.1006/mpev.1994.1023. [DOI] [PubMed] [Google Scholar]
- Huchon D, Catzeflis FM, Douzery EJP. Molecular evolution of the nuclear von Willebrand Factor gene in mammals and the phylogeny of rodents. Mol Biol Evol. 1999;16:577–589. doi: 10.1093/oxfordjournals.molbev.a026140. [DOI] [PubMed] [Google Scholar]
- Adkins R, Gelke E, Rowe D, Honeycutt R. Molecular phylogeny and divergence time estimates for major rodent groups: evidence from multiple genes. Mol Biol Evol. 2001;18:777–791. doi: 10.1093/oxfordjournals.molbev.a003860. [DOI] [PubMed] [Google Scholar]
- Huchon D, Madsen O, Sibbald M, Ament K, Stanhope M, Catzeflis F, de Jong W, Douzery E. Rodent phylogeny and a timescale for the evolution of Glires: evidence from an extensive taxon sampling using three nuclear genes. Mol Biol Evol. 2002;19:1053–1065. doi: 10.1093/oxfordjournals.molbev.a004164. [DOI] [PubMed] [Google Scholar]
- Adkins R, Walton A, Honeycutt R. Higher-level systematics of rodents and divergence time estimates based on two congruent nuclear genes. Mol Phylogenet Evol. 2003;26:409–420. doi: 10.1016/S1055-7903(02)00304-4. [DOI] [PubMed] [Google Scholar]
- DeBry R. Identifying conflicting signal in a multigene analysis reveals a highly resolved tree: the phylogeny of Rodentia (Mammalia) Syst Biol. 2003;52:604–617. doi: 10.1080/10635150390235403. [DOI] [PubMed] [Google Scholar]
- Montgelard C, Forty E, Arnal V, Matthee CA. Suprafamilial relationships among Rodentia and the phylogenetic effect of removing fast-evolving nucleotides in mitochondrial, exon and intron fragments. BMC Evol Biol. 2008;8:321. doi: 10.1186/1471-2148-8-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanga-Kanfi S, Miranda H, Penn O, Pupko T, DeBry R, Huchon D. Rodent phylogeny revised: analysis of six nuclear genes from all major rodent clades. BMC Evol Biol. 2009;9:71. doi: 10.1186/1471-2148-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churakov G, Sadasivuni M, Rosenbloom K, Huchon D, Brosius J, Schmitz J. Rodent evolution: back to the root. Mol Biol Evol. 2010;27:1315–1326. doi: 10.1093/molbev/msq019. [DOI] [PubMed] [Google Scholar]
- Brandt J. Beiträge zur nähern Kenntniss der Säugethiere Russlands. Mem Acad Imp St Petersbourg Ser. 1855;69:1–375. [Google Scholar]
- Tullberg T. Über das System der Nagetiere: eine phylogenetische Studie. Nova Acta Reg Soc Sci Upsala (Ser. 3) 1899;18:1–514. [Google Scholar]
- Luckett W, Hartenberger JL. Monophyly or polyphyly of the order Rodentia: possible conflict between morphological and molecular interpretations. J Mammal Evol. 1993;1:127–147. doi: 10.1007/BF01041591. [DOI] [Google Scholar]
- Huchon D, Douzery E. From the Old World to the New World: a molecular chronicle of the phylogeny and biogeography of hystricognath rodents. Mol Phylogenet Evol. 2001;20:239–251. doi: 10.1006/mpev.2001.0961. [DOI] [PubMed] [Google Scholar]
- Mercer J, Roth V. The effects of Cenozoic global change on squirrel phylogeny. Science. 2003;299:1568–1572. doi: 10.1126/science.1079705. [DOI] [PubMed] [Google Scholar]
- Montgelard C, Matthee CA, Robinson TJ. Molecular systematics of dormice (Rodentia: Gliridae) and the radiation of Graphiurus in Africa. Proc R Soc Lond B. 2003;270:1947–55. doi: 10.1098/rspb.2003.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weksler M. Phylogeny of Neotropical oryzomyine rodents (Muridae: Sigmodontinae) based on the nuclear IRBP exon. Mol Phylogenet Evol. 2003;29:331–349. doi: 10.1016/S1055-7903(03)00132-5. [DOI] [PubMed] [Google Scholar]
- Honeycutt R, Rowe D, Gallardo M. Molecular systematics of the South American caviomorph rodents: Relationships among species and genera in the family Octodontidae. Mol Phylogenet Evol. 2003;26:476–489. doi: 10.1016/S1055-7903(02)00368-8. [DOI] [PubMed] [Google Scholar]
- Jansa S, Weksler M. Phylogeny of muroid rodents: relationships within and among major lineages as determined by IRBP gene sequences. Mol Phylogenet Evol. 2004;31:256–276. doi: 10.1016/j.ympev.2003.07.002. [DOI] [PubMed] [Google Scholar]
- Steppan S, Adkins R, Anderson J. Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst Biol. 2004;53:533–553. doi: 10.1080/10635150490468701. [DOI] [PubMed] [Google Scholar]
- Steppan S, Storz B, Hoffmann R. Nuclear DNA phylogeny of the squirrels (Mammalia: Rodentia) and the evolution of arboreality from c-myc and RAG1. Mol Phylogenet Evol. 2004;30:703–719. doi: 10.1016/S1055-7903(03)00204-5. [DOI] [PubMed] [Google Scholar]
- Steppan SJ, Adkins RM, Spinks PQ, Hale C. Multigene phylogeny of the Old World mice, Murinae, reveals distinct geographic lineages and the declining utility of mitochondrial genes compared to nuclear genes. Mol Phylogenet Evol. 2005;37:370–388. doi: 10.1016/j.ympev.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Galewski T, Mauffrey JF, Leite YL, Patton JL, Douzery EJP. Ecomorphological diversification among South American spiny rats (Rodentia; Echimyidae): a phylogenetic and chronological approach. Mol Phylogenet Evol. 2005;34:601–615. doi: 10.1016/j.ympev.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Galewski T, Tilak MK, Sanchez S, Chevret P, Paradis E, Douzery EJ. The evolutionary radiation of Arvicolinae rodents (voles and lemmings): relative contribution of nuclear and mitochondrial DNA phylogenies. BMC Evol Biol. 2006;6:80. doi: 10.1186/1471-2148-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecompte E, Aplin K, Denys C, Catzeflis F, Chades M, Chevret P. Phylogeny and biogeography of African Murinae based on mitochondrial and nuclear gene sequences, with a new tribal classification of the subfamily. BMC Evol Biol. 2008;8:199. doi: 10.1186/1471-2148-8-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe K, Reno M, Richmond D, Adkins R, Steppan S. Pliocene colonization and adaptive radiations in Australia and New Guinea (Sahul): Multilocus systematics of the old endemic rodents (Muroidea : Murinae) Mol Phylogenet Evol. 2008;47:84–101. doi: 10.1016/j.ympev.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Patterson BD, Velazco PM. Phylogeny of the rodent genus Isothrix (Hystricognathi, Echimyidae) and its diversification in Amazonia and the eastern Andes. J Mammal Evol. 2008;15:181–201. doi: 10.1007/s10914-007-9070-6. [DOI] [Google Scholar]
- Jansa S, Giarla T, Lim B. The phylogenetic position of the rodent genus Typhlomys and the geographic origin of Muroidea. J Mammal. 2009;90:1083–1094. doi: 10.1644/08-MAMM-A-318.1. [DOI] [Google Scholar]
- Pages M, Chaval Y, Herbreteau V, Waengsothorn S, Cosson JF, Hugot JP, Morand S, Michaux J. Revisiting the taxonomy of the Rattini tribe: a phylogeny-based delimitation of species boundaries. BMC Evol Biol. 2010;10:184. doi: 10.1186/1471-2148-10-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins JH, McLenachan PA, Phillips MJ, McComish BJ, Matisoo-Smith E, Ross HA. Evolutionary relationships and divergence times among the native rats of Australia. BMC Evol Biol. 2010;10:375. doi: 10.1186/1471-2148-10-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S, Durka W, Wolf R, Ermala A, Stubbe A, Stubbe M, Hofreiter M. Mitochondrial genomes reveal slow rates of molecular evolution and the timing of speciation in beavers (Castor), one of the largest rodent species. PLoS One. 2011;6:e14622. doi: 10.1371/journal.pone.0014622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe KC, Aplin KP, Baverstock PR, Moritz C. Recent and rapid speciation with limited morphological disparity in the genus Rattus. Syst Biol. 2011;60:188–203. doi: 10.1093/sysbio/syq092. [DOI] [PubMed] [Google Scholar]
- Upham N, Patterson B. Diversification and biogeography of the Neotropical caviomorph lineage Octodontoidea (Rodentia: Hystricognathi) Mol Phylogenet Evol. 2012;63:417–429. doi: 10.1016/j.ympev.2012.01.020. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ, Purvis A, Henze C. Phylogenetic supertrees: assembling the trees of life. Trends Ecol Evol. 1998;13:105–109. doi: 10.1016/S0169-5347(97)01242-1. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds O. The evolution of supertrees. Trends Ecol Evol. 2004;19:315–322. doi: 10.1016/j.tree.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Gatesy J, Matthee C, DeSalle R, Hayashi C. Resolution of a supertree/supermatrix paradox. Syst Biol. 2002;51:652–664. doi: 10.1080/10635150290102311. [DOI] [PubMed] [Google Scholar]
- Gatesy J, Baker RH, Hayashi C. Inconsistencies in arguments for the supertree approach: supermatrices versus supertrees of Crocodylia. Syst Biol. 2004;53:342–355. doi: 10.1080/10635150490423971. [DOI] [PubMed] [Google Scholar]
- Bapteste E, Brinkmann H, Lee JA, Moore DV, Sensen CW, Gordon P, Durufle L, Gaasterland T, Lopez P, Muller M, Philippe H. The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba. Proc Natl Acad Sci USA. 2002;99:1414–1419. doi: 10.1073/pnas.032662799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck R, Bininda-Emonds O, Cardillo M, Liu F, Purivs A. A higher-level MRP supertree of placental mammals. BMC Evol Biol. 2006;6:93. doi: 10.1186/1471-2148-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bininda-Emonds ORP, Cardillo M, Jones KE, MacPhee RDE, Beck RMD, Grenyer R, Price SA, Vos RA, Gittleman JL, Purvis A. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- Meredith RW, Janecka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simao TLL, Stadler T, Rabosky DL, Honeycutt RL, Flynn JJ, Ingram CM, Steiner C, Williams TL, Robinson TJ, Burk-Herrick A, Westerman M, Ayoub NA, Springer MS, Murphy WJ. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science. 2011;334(6055):521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- Liu FG, Miyamoto MM, Freire NP, Ong PQ, Tennant MR, Young TS, Gugel KF. Molecular and morphological supertrees for eutherian (placental) mammals. Science. 2001;291:1786–9. doi: 10.1126/science.1056346. [DOI] [PubMed] [Google Scholar]
- Fabre PH, Rodrigues A, Douzery EJP. Patterns of macroevolution among Primates inferred from a supermatrix of mitochondrial and nuclear DNA. Mol Phylogenet Evol. 2009;53:808–25. doi: 10.1016/j.ympev.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Chatterjee HJ, Ho SYW, Barnes I, Groves C. Estimating the phylogeny and divergence times of primates using a supermatrix approach. BMC Evol Biol. 2009;9:259. doi: 10.1186/1471-2148-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelman P, Johnson WE, Roos C, Seuánez HN, Horvath JE, Moreira MAM, Kessing B, Pontius J, Roelke M, Rumpler Y, Schneider MPC, Silva A, O’Brien SJ, Pecon-Slattery J. A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnarsson I, Kuntner M, May-Collado LJ. Dogs, cats, and kin: a molecular species-level phylogeny of Carnivora. Mol Phylogenet Evol. 2010;54:726–45. doi: 10.1016/j.ympev.2009.10.033. [DOI] [PubMed] [Google Scholar]
- Agnarsson I, May-Collado LJ. The phylogeny of Cetartiodactyla: the importance of dense taxon sampling, missing data, and the remarkable promise of cytochrome b to provide reliable species-level phylogenies. Mol Phylogenet Evol. 2008;48:964–85. doi: 10.1016/j.ympev.2008.05.046. [DOI] [PubMed] [Google Scholar]
- Steeman ME, Hebsgaard MB, Fordyce RE, Ho SYW, Rabosky DL, Nielsen R, Rahbek C, Glenner H, Sørensen MV, Willerslev E. Radiation of extant cetaceans driven by restructuring of the oceans. Syst Biol. 2009;58:573–85. doi: 10.1093/sysbio/syp060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnarsson I, Zambrana-Torrelio CM, Flores-Saldana NP, May-Collado LJ. A time-calibrated species-level phylogeny of bats (Chiroptera, Mammalia) PLoS Curr. 2011;3:RRN1212. doi: 10.1371/currents.RRN1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC. A molecular timescale for caviomorph rodents (Mammalia, Hystricognathi) Mol Phylogenet Evol. 2005;37:932–7. doi: 10.1016/j.ympev.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Nunome M, Yasuda SP, Sato JJ, Vogel P, Suzuki H. Phylogenetic relationships and divergence times among dormice (Rodentia, Gliridae) based on three nuclear genes. Zool Scripta. 2007;36:537–546. doi: 10.1111/j.1463-6409.2007.00296.x. [DOI] [Google Scholar]
- Chan K, Moore B. SymmeTREE: Whole-tree analysis of differential diversification rates. Bioinformatics. 2005;21:1709–1710. doi: 10.1093/bioinformatics/bti175. [DOI] [PubMed] [Google Scholar]
- Michaux J, Reyes A, Catzeflis F. Evolutionary history of the most speciose mammals: molecular phylogeny of muroid rodents. Mol Biol Evol. 2001;18:2017–31. doi: 10.1093/oxfordjournals.molbev.a003743. [DOI] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihood with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. doi: 10.1093/oxfordjournals.molbev.a026201. [DOI] [Google Scholar]
- Douzery EJP, Delsuc F, Stanhope MJ, Huchon D. Local molecular clocks in three nuclear genes: divergence times for rodents and other mammals and incompatibility among fossil calibrations. J Mol Evol. 2003;57(Suppl 1):S201–13. doi: 10.1007/s00239-003-0028-x. [DOI] [PubMed] [Google Scholar]
- Jansa SA, Barker FK, Heaney LR. The pattern and timing of diversification of Philippine endemic rodents: evidence from mitochondrial and nuclear gene sequences. Syst Biol. 2006;55:73–88. doi: 10.1080/10635150500431254. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–74. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Rodríguez F, Oliver JL, Marín A, Medina JR. The general stochastic model of nucleotide substitution. J Theor Biol. 1990;142:485–501. doi: 10.1016/S0022-5193(05)80104-3. [DOI] [PubMed] [Google Scholar]
- Yang Z. Estimating the pattern of nucleotide substitution. J Mol Evol. 1994;39:105–11. doi: 10.1007/BF00178256. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Yang Z. Among-site rate variation and its impact on phylogenetic analyses. Trends Ecol Evol. 1996;11:367–372. doi: 10.1016/0169-5347(96)10041-0. [DOI] [PubMed] [Google Scholar]
- Wiens J. Missing data, incomplete taxa, and phylogenetic accuracy. Syst Biol. 2003;52:528–538. doi: 10.1080/10635150390218330. [DOI] [PubMed] [Google Scholar]
- Philippe H, Snell E, Bapteste E, Lopez P, Holland P, Casane D. Phylogenomics of eukaryotes: impact of missing data on large alignments. Mol Biol Evol. 2004;9:1740–1752. doi: 10.1093/molbev/msh182. [DOI] [PubMed] [Google Scholar]
- Jones KE, Bininda-Emonds ORP, Gittleman JL. Bats, clocks, and rocks: diversification patterns in Chiroptera. Evolution. 2005;59:2243–55. [PubMed] [Google Scholar]
- Guyer O, Slowinski J. Comparison of observed phylogenetic topologies with null expectations among three monophyletic lineages. Evolution. 1991;45:340–350. doi: 10.2307/2409668. [DOI] [PubMed] [Google Scholar]
- Purvis A, Agapow PM. Phylogeny imbalance: taxonomic level matters. Syst Biol. 2002;51:844–54. doi: 10.1080/10635150290102546. [DOI] [PubMed] [Google Scholar]
- Engel SR, Hogan KM, Taylor JF, Davis SK. Molecular systematics and paleobiogeography of the South American sigmodontine rodents. Mol Biol Evol. 1998;15:35–49. doi: 10.1093/oxfordjournals.molbev.a025845. [DOI] [PubMed] [Google Scholar]
- van Valen L, Sloan L. The extinction of the multituberculates. Syst Zool. 1966;15:261–278. doi: 10.2307/2411985. [DOI] [Google Scholar]
- Smith M, Patton J. Phylogenetic relationships and the radiation of sigmodontine rodents in South America: evidence from cytochrome b. J Mammal Evol. 1999;6:89–128. doi: 10.1023/A:1020668004578. [DOI] [Google Scholar]
- Simpson G. The Major Features of Evolution. New York: Columbia University Press; 1953. [Google Scholar]
- Mayr E. Animal species and their evolution. Harvard Univ: Mus Comp Zool; 1963. [Google Scholar]
- Rosenzweig M. Species diversity in space and time. Cambridge: Cambridge University Press; 1995. [Google Scholar]
- Moore BR, Donoghue MJ. Correlates of diversification in the plant clade Dipsacales: geographic movement and evolutionary innovations. Am Nat. 2007;170(Suppl 2):S28–55. doi: 10.1086/519460. [DOI] [PubMed] [Google Scholar]
- Moore BR, Donoghue MJ. A Bayesian approach for evaluating the impact of historical events on rates of diversification. Proc Natl Acad Sci USA. 2009;106:4307–12. doi: 10.1073/pnas.0807230106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jønsson KA, Fabre PH, Ricklefs RE, Fjeldså J. Major global radiation of corvoid birds originated in the proto-Papuan archipelago. Proc Natl Acad Sci USA. 2011;108:2328–33. doi: 10.1073/pnas.1018956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth J. Molecular phylogeny of the cardueline finches and Hawaiian honeycreepers. Ostrich. 1998;69:401. [Google Scholar]
- Schluter D. The ecology of adaptive radiation. Oxford, UK: Oxford University Press; 2000. [Google Scholar]
- Losos J. Lizards in an evolutionary tree: ecology and adaptive radiation of anoles. Berkeley: Univ Calif Press; 2010. [Google Scholar]
- Lecompte E, Denys C, Granjon L. Confrontation of morphological and molecular data: the Praomys group (Rodentia, Murinae) as a case of adaptive convergences and morphological stasis. Mol Phylogenet Evol. 2005;37:899–919. doi: 10.1016/j.ympev.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Korth W. The tertiary record of rodents in North America, Volume 12 of Topics in paleobiology. New York: Plenum Publishing Corporation; 1994. [Google Scholar]
- McKenna M, S B. Classification of mammals above the species level. New York: Columbia University Press; 1997. [Google Scholar]
- Eisenberg J. The mammalian radiations: analysis of trends in evolution, adaptation, and behaviour. Chicago: The University of Chicago Press; 1981. [Google Scholar]
- Marzluff J, Dial K. Life history correlates of taxonomic diversity. Ecology. 1991;72:428–439. doi: 10.2307/2937185. [DOI] [Google Scholar]
- Hutchinson G, MacArthur R. A theoritical ecological model of size distributions among species of animals. Am Nat. 1959;93:117–125. doi: 10.1086/282063. [DOI] [Google Scholar]
- Jernvall J. Mammalian molar cusp patterns: Developmental mechanisms of diversity. Acta Zool Fenn. 1995;198:1–61. [Google Scholar]
- Hunter JP, Jernvall J. The hypocone as a key innovation in mammalian evolution. Proc Natl Acad Sci USA. 1995;92:10718–22. doi: 10.1073/pnas.92.23.10718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price S, Hopkins S, Smith K, Roth V. Tempo of trophic evolution and its impact on mammalian diversification. Proc Natl Acad Sci USA. 2012;109:7008–7012. doi: 10.1073/pnas.1117133109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poux C, Chevret P, Huchon D, de Jong WW, Douzery EJP. Arrival and diversification of caviomorph rodents and platyrrhine primates in South America. Syst Biol. 2006;55:228–44. doi: 10.1080/10635150500481390. [DOI] [PubMed] [Google Scholar]
- Rowe D, Dunn K, Adkins R, Honeycutt R. Molecular clocks keep dispersal hypotheses afloat: evidence for trans-Atlantic rafting by rodents. J Biogeography. 2010;37:305–324. doi: 10.1111/j.1365-2699.2009.02190.x. [DOI] [Google Scholar]
- Candela A. A new porcupine (Rodentia, Hystricognathi, Erethizontidae) from the Early and Middle Miocene of Patagonia. Ameghiniana. 2003;40:483–494. [Google Scholar]
- Carvalho G, Salles L. Relationships among extant and fossil echimyids (Rodentia: Hystricognathi) Zool J Linn Soc. 2004;142:445–477. doi: 10.1111/j.1096-3642.2004.00150.x. [DOI] [Google Scholar]
- Hoffstetter R. Relationships, origins, and history of the ceboid monkeys and caviomorph rodents: a modern reinterpretation. Evol Biol. 1972;6:322–347. [Google Scholar]
- Vucetich M, Verzi D, Hartenberger JL. Review and analysis of the radiation of the South American Hystricognathi (Mammalia, Rodentia) C R Acad Sci Paris, Earth and Planetary Sciences. 1999;329:763–769. [Google Scholar]
- Janis C. Tertiary mammal evolution in the context of changing climates, vegetation, and tectonic events. Ann Rev Ecol Syst. 1993;24:467–500. doi: 10.1146/annurev.es.24.110193.002343. [DOI] [Google Scholar]
- MacFadden B. Extinct mammalian biodiversity of the ancient New World tropics. Trends Ecol Evol. 2006;21:157–165. doi: 10.1016/j.tree.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161:1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alroy J. New methods for quantifying macroevolutionary patterns and processes. Paleobiology. 2000;26:707–733. doi: 10.1666/0094-8373(2000)026<0707:NMFQMP>2.0.CO;2. [DOI] [Google Scholar]
- Hartenberger JL. Description de la radiation des Rodentia (Mammalia) du Paléocène supérieur au Miocène ; incidences phylogénétiques. C. R. Acad. Sci. Paris, Sciences de la terre et des planètes. 1998;326:439–444. [Google Scholar]
- Hartenberger JL. Contribution à l’étude des genres Gliravus and Microparamys (Rodentia) de l’Eocène d’Europe. Palaeovertebrata. 1971;4:97–135. [Google Scholar]
- Hartenberger JL. In: Rodent and lagomorph families of Asian origins and diversification, Volume 8 of Natl. Science Mus. Monogr., national science museum monographs edition. Tomida Y, Li CK, Setoguchi T, editor. Tokyo: National Science Museum Monographs; 1994. The evolution of the Gliroidea; pp. 19–33. [Google Scholar]
- Vianey-Liaud M. La radiation des Gliridae (Rodentia) à l’Eocene superieur en Europe Occidentale, et sa descendance Oligocene. Munch Geowissen Abh. 1994;26:117–160. [Google Scholar]
- Hopkins SSB. Causes of lineage decline in the Aplodontidae: testing for the influence of physical and biological change. Palaeogeogr Palaeocl Palaeoecol. 2006;246:331–353. [Google Scholar]
- Hopkins SSB. Phylogeny and evolutionary history of the Aplodontoidea (Mammalia : Rodentia) Zool J Linn Soc. 2008;153:769–838. doi: 10.1111/j.1096-3642.2008.00399.x. [DOI] [Google Scholar]
- Lindsay E. The fossil record of Asian Cricetidae with emphasis on Siwalik cricetids. Kyoto: National Science Museum, No. 131-147 in Monographs; 1994. [Google Scholar]
- Vianey-Liaud M. Contribution l’étude des Cricetidae Oligocenes d’Europe occidentale. Palaeovertebrata. 1972;5:1–44. [Google Scholar]
- Martin L. The early evolution of the Cricetidae in North America. Univ Kansas Paleont Contrib. 1980;102:1–42. [Google Scholar]
- Stadler Ta. Inferring speciation and extinction processes from extant species data. Proc Natl Acad Sci USA. 2011;108:16145–16146. doi: 10.1073/pnas.1113242108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton M, Musser G. Order Rodentia, Volume 2 of Mammal species of the world: A taxonomic and geographic reference. Baltimore: Johns Hopkins University Press; 2005. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Parada A, D’Elia G, Bidau CJ, Lessa EP. Species groups and the evolutionary diversification of tuco-tucos, genus Ctenomys (Rodentia: Ctenomyidae) J Mammal. 2011;92:671–682. doi: 10.1644/10-MAMM-A-121.1. [DOI] [Google Scholar]
- Steppan SJ, Akhverdyan MR, Lyapunova EA, Fraser DG, Vorontsov NN, Hoffmann RS, Braun MJ. Molecular phylogeny of the marmots (Rodentia: Sciuridae): tests of evolutionary and biogeographic hypotheses. Syst Biol. 1999;48:715–34. doi: 10.1080/106351599259988. [DOI] [PubMed] [Google Scholar]
- Herron M, Castoe T, Parkinson C. Sciurid phylogeny and the paraphyly of Holarctic ground squirrels (Spermophilus) Mol Phylogenet Evol. 2004;31:1015–1030. doi: 10.1016/j.ympev.2003.09.015. [DOI] [PubMed] [Google Scholar]
- D’Elía G, Luna L, González EM, Patterson BD. On the Sigmodontinae radiation (Rodentia, Cricetidae): an appraisal of the phylogenetic position of Rhagomys. Mol Phylogenet Evol. 2006;38:558–64. doi: 10.1016/j.ympev.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Reeder SA, Carroll DS, Edwards CW, Kilpatrick CW, Bradley RD. Neotomine-peromyscine rodent systematics based on combined analyses of nuclear and mitochondrial DNA sequences. Mol Phylogenet Evol. 2006;40:251–258. doi: 10.1016/j.ympev.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Miller JR, Engstrom MD. The relationships of major lineages within peromyscine rodents: a molecular phylogenetic hypothesis and systematic reappraisal. J Mammal. 2008;89:1279–1295. doi: 10.1644/07-MAMM-A-195.1. [DOI] [Google Scholar]
- Chambers RR, Sudman PD, Bradley RD. A phylogenetic assessment of pocket gophers (Geomys): evidence from nuclear and mitochondrial genes. J Mammal. 2009;90:537–547. doi: 10.1644/08-MAMM-A-180R1.1. [DOI] [Google Scholar]
- Belfiore NM, Liu L, Moritz C. Multilocus phylogenetics of a rapid radiation in the genus thomomys (Rodentia : Geomyidae) Syst Biol. 2008;57:294–310. doi: 10.1080/10635150802044011. [DOI] [PubMed] [Google Scholar]
- Springer M, Stanhope M, Madsen O, de Jong W. Molecules consolidate the placental mammal tree. Trends Ecol Evol. 2004;19:430–438. doi: 10.1016/j.tree.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Janecka JE, Miller W, Pringle TH, Wiens F, Zitzmann A, Helgen KM, Springer MS, Murphy WJ. Molecular and genomic data identify the closest living relative of primates. Science. 2007;318:792–794. doi: 10.1126/science.1147555. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall K. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony(and Other Methods), 4. Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]
- Shao KT, Sokal RR. Tree balance. Syst Zool. 1990;39:266–276. doi: 10.2307/2992186. [DOI] [Google Scholar]
- Heard S. Patterns in tree balance among cladistic, phenetic, and randomly generated phylogenetic trees. Evolution. 1992;46:1818–1826. doi: 10.2307/2410033. [DOI] [PubMed] [Google Scholar]
- Yule G. A mathematical theory of evolution, based on the conclusions of Dr. J. C. Willis. Trans R Soc Lond B Biol Sci. 1924;213:21–87. [Google Scholar]
- de Queiroz A, Gatesy J. The supermatrix approach to systematics. Trends Ecol Evol. 2007;22:34–41. doi: 10.1016/j.tree.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Ho SYW, Kolokotronis SO, Allaby RG. Elevated substitution rates estimated from ancient DNA sequences. Biol Lett. 2007;3:702–705. doi: 10.1098/rsbl.2007.0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer v1.4, Available from http://beast. bio.ed.ac.uk/Tracer 2007.
- Wyss AR, Flynn JJ, Norell MA, Swisher IIICC, Charrier R, Novacek MJ, McKenna MC. South America’s earliest rodent and recognition of a new interval of mammalian evolution. Nature. 1993;365:434–437. doi: 10.1038/365434a0. [DOI] [Google Scholar]
- Walton A. In: Vertebrate paleontology in the neotropics. The Miocene fauna of La Venta, Columbia. Kay R, Madden R, Ciffelli R, Flynn J, editor. Washington and London: Smithsonian Institution Press; 1997. Rodents; pp. 392–409. [Google Scholar]
- Benton M, Donoghue P. Paleontological evidence to date the Tree of Life. Mol Biol Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- Davies TJ, Fritz SA, Grenyer R, Orme CDL, Bielby J, Bininda-Emonds ORP, Cardillo M, Jones KE, Gittleman JL, Mace GM, Purvis A. Colloquium paper: phylogenetic trees and the future of mammalian biodiversity. Proc Natl Acad Sci USA. 2008;105(Suppl 1):11556–63. doi: 10.1073/pnas.0801917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis A, Fritz SA, Rodriguez J, Harvey PH, Grenyer R. The shape of mammalian phylogeny: patterns, processes and scales. Philos Trans R Soc Lond B Biol Sci. 2011;366:2462–2477. doi: 10.1098/rstb.2011.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Rodent species level evolutionary tree. Species-level phylogenetic topology based on the highest-likelihood tree inferred from the 11-gene supermatrix, and combined with the taxonomic information of Wilson and Reeder (2005).
Figure S2. Cladogram depicting the highest-likelihood topology for the Ctenohystrica. Circles at nodes represent bootstrap support (circles: black 100-95%, white 95-70%, gray 70-50%). Maximum likekihood tree (lnL=-714749.2). Displayed clade are highlighted using the simplified full ML topology on the left side of the figure. Molecular marker sampling is depicted for each taxa to the right of the tree. Names of the genes are given. Genes sampled for our dataset (Table 2) are marked in black square while missing genes are symbolized by white squares. Triangles indicate significant (P < 0.05) and marginal (0.05 < P < 0.1) shifts in diversification rate (SDR) as inferred by Δ1. LA = Diatomyidae, CT = Ctenodactylidae, TH = Thryonomyidae, PE = Petromuridae, CU = Cuniculidae, DASYPROC = Dasyproctidae, DI = Dinomyidae, CHINCHILLID = Chinchillidae. Bootstrap for all nodes and topology can be found in Additional file 14.
Figure S3. Cladogram depicting the highest-likelihood topology for the Octodontoidea. See Additional file 2: Figure S2 for details of the legend. AB = Abrocomidae, CA = Capromyidae.
Figure S4. Cladogram depicting the highest-likelihood topology for the Sciuroidea. See Additional file 2: Figure S2 for details of the legend. AP = Aplodontidae, RA = Ratufinae, SI = Sciurillinae.
Figure S5. Cladogram depicting the highest-likelihood topology for the Xerinae. See Additional file 2: Figure S2 for details of the legend.
Figure S6. Cladogram depicting the highest-likelihood topology for the Castorimorpha. See Additional file 2: Figure S2 for details of the legend. CAS = Castoridae.
Figure S7. Cladogram depicting the highest-likelihood topology for the mouse-related clade. See Additional file 2: Figure S2 for details of the legend. PED = Pedetidae, ANO = Anomaluroidea, PLA = Platacanthomyidae, Rhy = Rhyzomyidae, Spalac = Spalacidae, CAL = Calomyscidae, LOP = Lophiomyinae.
Figure S8. Cladogram depicting the highest-likelihood topology for Sigmodontinae [part 1] + Tylomyinae. See Additional file 2: Figure S2 for details of the legend. Tyl = Tylomyinae, Ich = Ichthyomyini, Rei = Reithrodontini.
Figure S9. Cladogram depicting the highest-likelihood topology for Sigmodontinae [part 2]. See Additional file 2: Figure S2 for details of the legend. Tyl = Tylomyinae, Ich = Ichthyomyini, Rei = Reithrodontini, Phy = Phyllotini.
Figure S10. Cladogram depicting the highest-likelihood topology for Neotominae. See Additional file 2: Figure S2 for details of the legend. Och = Ochrotomyini, Baiom = Baiomyini.
Figure S11. Cladogram depicting the highest-likelihood topology for Arvicolinae. See Additional file 2: Figure S2 for details of the legend. Pro = Prometheomyini, Dic = Dicrostonychini, Ond = Ondatrini, Plio = Pliomyini, Arv = Arvicolini, Ell = Ellobiusini, Lag = Lagurini.
Figure S12. Cladogram depicting the highest-likelihood topology for Murinae [part 1]. See Additional file 2: Figure S2 for details of the legend. Mic = Micromys division, Cru = Crunomys division, Max = Maxomys division, Mel = Melasmothrix division, Hydrom = Hydromyines division, Con = Conilurines division, Urom = Uromyines division.
Figure S13. Cladogram depicting the highest-likelihood topology for Murinae [part 2]. See Additional file 2: Figure S2 for details of the legend. Mic = Micromys division, Mil = Millardia division, Col = Colomys division, Cre = Cremnomys division, Gol = Golunda, Oen = Oenomys division, Hyb = Hybomys division, Mi = Micaelamys division, Das = Dasymys division, Mal = Malacomys division, Stenocep = Stenocephalemys division.
ML RaxML topology.
Rodentia species list names.
Rodentia accession numbers by gene.
Rodentia accession numbers by taxonomic group (Sheet 1: MYODONTA + ANOMALUROMORPHA; Sheet 2: SCIUROIDEA; Sheet 3: CTENOHYSTRICA; Sheet 4: CASTORIMORPHA).
Loci used in each molecular dating analysis (see Material and Methods).