Abstract
A large listeriosis outbreak occurred in Austria, Germany and the Czech Republic in 2009 and 2010. The outbreak was traced back to a traditional Austrian curd cheese called “Quargel” which was contaminated with two distinct serovar 1/2a Listeria monocytogenes strains (QOC1 and QOC2). In this study we sequenced and analysed the genomes of both outbreak strains in order to investigate the extent of genetic diversity between the two strains belonging to MLST sequence types 398 (QOC2) and 403 (QOC1). Both genomes are highly similar, but also display distinct properties: The QOC1 genome is approximately 74 kbp larger than the QOC2 genome. In addition, the strains harbour 93 (QOC1) and 45 (QOC2) genes encoding strain-specific proteins. A 21 kbp region showing highest similarity to plasmid pLMIV encoding three putative internalins is integrated in the QOC1 genome. In contrast to QOC1, strain QOC2 harbours a vip homologue, which encodes a LPXTG surface protein involved in cell invasion. In accordance, in vitro virulence assays revealed distinct differences in invasion efficiency and intracellular proliferation within different cell types. The higher virulence potential of QOC1 in non-phagocytic cells may be explained by the presence of additional internalins in the pLMIV-like region, whereas the higher invasion capability of QOC2 into phagocytic cells may be due to the presence of a vip homologue. In addition, both strains show differences in stress-related gene content. Strain QOC1 encodes a so-called stress survival islet 1, whereas strain QOC2 harbours a homologue of the uncharacterized LMOf2365_0481 gene. Consistently, QOC1 shows higher resistance to acidic, alkaline and gastric stress. In conclusion, our results show that strain QOC1 and QOC2 are distinct and did not recently evolve from a common ancestor.
Introduction
Listeria (L.) monocytogenes is a Gram-positive facultative intracellular food-borne pathogen, which can survive in multiple habitats like soil, vegetation, food processing plants, food, domestic and wild animals as well as humans [1]. L. monocytogenes has a remarkable ability to resist environmental stresses such as heavy metal ions, high salt concentration, low pH-values, low temperature, as well as low water activity [2]–[4].
In humans, mammals and birds L. monocytogenes can cause listeriosis, a rare but severe disease. The vast majority of listeriosis cases and outbreaks have been associated with the consumption of contaminated food, mainly dairy products, ready-to-eat deli meats and produce [5]. In recent years, a number of listeriosis outbreaks have been linked to contaminated cheese, including those made from pasteurized milk: e.g. hard cheese in Belgium 2011 (12 cases, 4 deaths) [6], Ricotta salata cheese in the USA in 2012 (22 cases, 4 deaths) [7], and Les Frères Cheese also in the USA in 2013 (6 cases, 1 death) [8]. Although L. monocytogenes is classified into 13 serotypes, the majority of sporadic cases and listeriosis outbreaks were caused by strains of 4b, 1/2a and 1/2b [9].
In healthy individuals listeriosis is usually restricted to a self-limiting febrile gastroenteritis, whereas in immunocompromised individuals an invasive and systemic infection can occur leading to meningitis, encephalitis and septicaemia with a high mortality rate of 25–30% [10], [11]. In addition, infection during pregnancy can lead to abortion, still-birth or septicaemia of the neonate [12].
A large listeriosis outbreak occurred in Austria, Germany and the Czech Republic in 2009/2010 due to consumption of a traditional Austrian cheese called “Quargel” [13]. Quargel is an acid curd cheese with a red smear made from skimmed pasteurized milk. Some recalled Quargel lots were highly contaminated with up to 106–108 colony forming units (CFU) of L. monocytogenes per gram of cheese [14], [15]. Molecular typing, including pulsed-field gel electrophoresis, revealed that two different L. monocytogenes strains, both serotype 1/2a, have been involved in this outbreak [16]. From June 2009 to January 2010 Quargel outbreak clone 1 (hereafter: QOC1) was the cause of 14 cases, including 5 with a fatal outcome, while between December 2009 and February 2010, clone 2 (hereafter: QOC2) accounted for 20 cases, which resulted in 3 fatalities. No maternal or neonatal case had been reported. The median age of the cases was 72 years (range 57 to 89) and 76% of the patients were male. Of the 34 patients, 25 were Austrian, 8 were German and one was from the Czech Republic. The underlying diseases did not differ from those generally described [17].
In recent years, genome sequencing of several L. monocytogenes strains have elucidated serotype- and strain-specific features of L. monocytogenes and have given new insights into the genetic determinants underlying virulence, pathogenicity and survival in food and food processing environments [18], [19].
Genome sequencing is a feasible tool for retrospective epidemiological analyses. Recently, Gilmour and co-authors showed that two closely related L. monocytogenes strains were responsible for a large food-borne listeriosis outbreak in Canada in 2008 using high-throughput genome sequencing [20]. Differences were almost exclusively due to the presence or absence of a prophage. In addition, Orsi et al. revealed that the human listeriosis outbreak in 2000 in the USA was caused by a L. monocytogenes strain that persisted in a food processing plant for over 12 years, and which has also been responsible for a sporadic case in 1988. Both strains had highly similar genomic backbone sequences with very few single nucleotide polymorphisms (SNPs); the main differences were found in the comK prophage region [21].
In this study we sequenced and analysed the genomes of the two Quargel outbreak strains QOC1 and QOC2, responsible for the multinational listeriosis outbreak in 2009/2010. Sequence comparison of the genomes enabled us to show that the two strains are related but distinct and did not evolve recently from a common ancestor. In addition, we determined the stress response of both outbreak strains and analyzed the in vitro virulence of strains QOC1 and QOC2 using human intestinal, hepatocytic and macrophage-like cell lines and primary mouse bone marrow-derived macrophages to investigate the correlation between genetic determinants, stress response and virulence.
Materials and Methods
Bacterial Strains
The two L. monocytogenes Quargel outbreak strains QOC1 and QOC2, both serotype 1/2a, were isolated from blood cultures of two male patients, who were hospitalized for 15 and 35 days, respectively. Both strains were isolated in Austria in January 2010 and were received from the Austrian Agency for Health and Food Safety (AGES). AGES IDs for QOC1∶930009/10, QOC2∶930014/10. The L. monocytogenes type strain EGDe (ATCC BAA-679) was used for comparison in cell culture virulence assays.
DNA Isolation and Genome Sequencing
Both L. monocytogenes strains were cultivated under aerobic conditions at 37°C in brain heart infusion broth (BHI, Merck; with 125 rpm shaking) and harvested by centrifugation. The resulting pellet was used for DNA isolation using the QIAGEN genomic-tip columns and buffers (QIAGEN), in accordance with the recommendations of the manufacturer. Genome sequencing was performed using an Illumina GAII genome analyzer at the University of Veterinary Medicine Vienna, Austria. Sequencing was performed using paired-end sequencing technology and 101 bp read-length using Illumina standard protocols. Ten million reads were used for a de novo assembly using SeqManNGen (DNASTAR). For strain QOC1, the assembly of sequenced data resulted in generation of 17 contigs (>500 bp) for which there was an average coverage of 310×, while for strain QOC2, assembly of the reads resulted in 13 contigs (>500 bp), for which the average coverage was 330×. For ordering, the contigs were aligned to the L. monocytogenes EGDe genome using the “move contigs” option in MAUVE [22]. Some of the remaining gaps could be closed by PCR and Sanger sequencing - resulting in seven and four contigs in the final assembly for strain QOC1 and QOC2, respectively.
Sequence Analyses
The genomes were analyzed and automatically annotated using the RAST server (http://rast.nmpdr.org ) [23] and the Microbial Genome Analysis and Annotation Platform MaGe/MicroScope (https://www.genoscope.cns.fr/agc/microscope/home) [24]. Genome comparisons and determination of homologous proteins were done using BlastP, BlastN, and tBlastN [25]. Similar to a previous study [26] we used a similarity cut-off of 60% amino acid identity and 80% coverage for identification of homologous proteins. Phylogenetic analyses of MLST genes of 13 L. monocytogenes strains of different serotypes were carried out using full-length MLST genes retrieved from GenBank. For each strain, the MLST genes were concatenated and aligned using Muscle implemented in MEGA5 [27]. Maximum likelihood phylogenetic trees using the Tamura-Nei model were calculated in MEGA5 with 500×resampling. All positions containing gaps and missing data were eliminated.
Survival and Growth Experiments under Stress Conditions
Overnight cultures of QOC1 and QOC2 grown in BHI were centrifuged and adjusted to an optical density of 600 (OD600) of 0.1 in defined minimal medium (MM) consisting of RPMI-1640, 1% L-glutamine (both PAA, Austria) and 0.08 g/l ferric citrate (Merck) adjusted to pH 2 and pH 3 with 3% HCl and to pH 11 and pH 12 with 1 M NaOH, and synthetic gastric fluid according Cotter et al. [28]. CFU were determined by serial plating on tryptic soya agar (TSA) plates in triplicate after 2 h of incubation at 37°C. Survival (%) was calculated as CFU after 2 h of incubation under stress conditions divided by inoculated CFU multiplied by 100.
Growth of QOC1 and QOC2 under mild stress conditions (in MM adjusted to pH 5 and MM supplemented with 7.5% NaCl) was determined by measuring the OD600 at different time points. Each experiment was performed at least three times.
Cell Lines
We used four different human cell lines: intestinal epithelial Caco2 (ATCC® HTB-37™), hepatocytic HepG2 (ATCC® 77400™), macrophage-like U937 (ATCC® CRL-1593.2™) and THP1 (ATCC® TIB-202™) cells. Cells were cultivated either in Eagle’s minimum essential medium (MEM, for Caco2 and HepG2) or in RPMI-1640 (for U937 and THP1) containing 2 mM L-glutamine, 10% fetal bovine serum (FBS), 100 units/ml penicillin, 100 µg/ml streptomycin sulphate and 0.25 µg/ml amphotericin B (all PAA).
In addition, primary mouse bone marrow-derived macrophages (mBMDM) were obtained by culturing mouse bone marrow in DMEM high glucose supplemented with 10% FBS (both Gibco, Invitrogen) and L cell-derived colony stimulating factor (CSF)-1 as described by Baccarini et al. [29].
Virulence Assays
Cells were seeded in a 24-well plate at a mean cell density of 105 cells per well for Caco2 and HepG2 and 106 cells per well for U937, THP1 and mBMDM. U937 and THP1 were incubated for 48 h with 100 ng/ml phorbol 12-myristate 13-acetate prior the virulence assay (PMA, Sigma-Aldrich).
The in vitro virulence assay was performed as recently described by Pricope et al. [30]. Briefly, bacteria were grown to the mid-logarithmic growth phase at 37°C. Cells were infected for 1 h at 37°C at a multiplicity of infection of 25. Bacterial numbers were confirmed by plating serial dilutions on TSA agar plates. The infected cells were washed with Dulbecco’s Phosphate Buffered Saline (PBS, PAA), and incubated in MEM or RPMI containing 100 µg/ml gentamicin (PAA) in order to kill extracellular bacteria. After 45 min (invasion efficiency) or 4 h (intracellular growth) the cells were lysed with 0.1% TritonX-100 (Merck). For mBMDM we used DMEM-high glucose supplemented with 3% FBS, L cell-derived CSF-1 and 50 µg/ml gentamycin. After 45 min mBMDM were either lysed to determine the invasion efficiency or media was changed to media containing 10 µg/ml gentamycin to determine the intracellular proliferation rate. Intracellular bacteria were determined by serial plating on TSA plates. CFUs were counted after incubation at 37°C for 24 h. Each experiment was done in duplicate and repeated on four separate independent occasions.
Invasion efficiency was calculated as the number of intracellular bacteria divided by the number of inoculated bacteria, multiplied by 100. The intracellular growth coefficient (IGC) was calculated as follows: IGC = (IBt = 4 h−IBt = 0 h)/IBt = 0 h (IB = intracellular bacteria).
Ethics Statement
For obtaining mBMDM, C57BL/6N (WT, wild-type) mice were purchased from Charles River Laboratories. All animal experiments were discussed and approved by the Ethics and Animal Welfare Committee of the University of Veterinary Medicine Vienna, conform to the National Authority (Austrian Federal Ministry for Science and Research; Tierversuchsgesetz – TVG ref BMWF-68.205/0243-II/3b/2011), and to the guidelines of FELASA, which match those of ARRIVE.
Statistical Analysis
Microsoft Excel® 2007 was used for statistical analysis. Mean value and standard deviation of invasion efficiency, IGC and survival were calculated from four biological replicates, performed in duplicate for invasion efficiency and IGC, and in triplicate for survival. Values were compared statistically using two-tailed t-tests (independent variables). P-values <0.05 were considered to be significant.
Accession Numbers
The genome sequences have been deposited in the EMBL European nucleotide archive under accession numbers CBVZ010000001 to CBVZ010000007 for L. monocytogenes QOC1 contigs and CBVW010000001 to CBVW010000004 for QOC2 contigs, respectively.
Results and Discussion
Genome sequencing, assembly and subsequent gap closing resulted in a total of 7 contigs for strain QOC1, and 4 contigs for strain QOC2, respectively. With the exception of contig 7 from QOC1 all contigs were aligned successfully to the L. monocytogenes EGDe reference genome. Contig 7 (9498 bp) has an average coverage of 480× and is predicted to encode 14 genes; however, only five of the genes show significant matches in GenBank, where three of them have highest similarity (38 to 52% amino acid identity) to phage genes (locus_tags: LMQOC1_70001–70014, Table S1). Thus we cannot deduce a putative function or genomic localization for contig 7 within the QOC1 genome.
The main general features of both genomes are shown in Table 1. GC content (38.0%) and genome size are in the range found typical for L. monocytogenes genomes. The genome of strain QOC1 is approx. 74 kbp larger than the QOC2 genome.
Table 1. General features of the L. monocytogenes QOC1 and QOC2 genomes.
| QOC1 | QOC2 | |
| Genome size (bp)* | 2,931,460 | 2,857,445 |
| No of contigs | 7 | 4 |
| G+C content (%) | 38.0 | 38.0 |
| No. of predicted codingsequences (CDS) | 2890 | 2841 |
| Average length of CDS | 906 | 902 |
| Coding density (%) | 88.5 | 89.0 |
| No. of rRNA operons | 6 | 6 |
| No. of tRNA genes | 67 | 67 |
| No. of phages# | – | – |
| Sequence type | 403 | 398 |
*genomes are not closed.
excluding the monocin regions and contig 7 of strain QOC1.
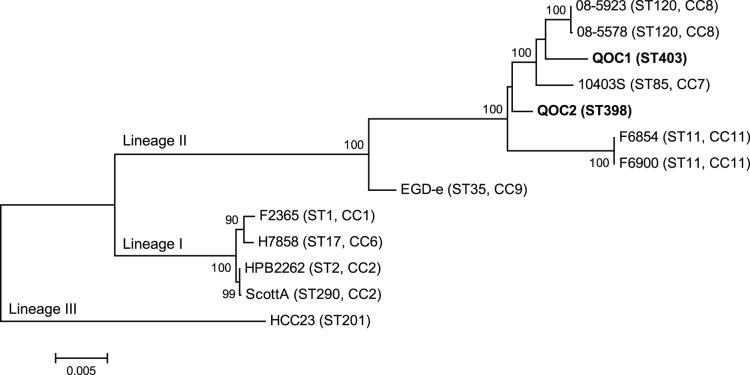
We extracted the MLST determinants from the genome sequences according to Ragon et al. [31] and performed in silico MLST. Strain QOC1 belongs to sequence type (ST) 403 (encoding the following alleles: abcZ 7, bglA 7, cat 10, dapE 4, dat 5, ldh 24, lhkA 1); whereas QOC2 is a ST398 strain (abcZ 7, bglA 13, cat 19, dapE 6, dat 1, ldh 7, lhkA 1). Both strains share only two alleles. Phylogenetic analyses based on MLST genes revealed distinct grouping of the two Quargel outbreak strains within evolutionary lineage II (Figure 1). Only two other ST403 strains (both human isolates, Germany) and eight ST398 strains (from food production, environmental, and animal samples, Europe) are currently found in the L. monocytogenes MLST database matching with QOC1 and QOC2.
Figure 1. Maximum likelihood phylogenetic tree of Listeria monocytogenes strains based on MLST loci.
L. monocytogenes sequence types are indicated („ST“); „CC“ denotes clonal complexes. The tree is based on concatenated full-length MLST gene sequences and was calculated with MEGA 5 [27] using the Tamura-Nei model. Bootstrap values (500× resampling) are indicated at the respective nodes.
We could not find evidence for plasmids and prophages in either of the genomes, except for the monocin region (the lma operon) - representing a cryptic prophage – which is present in both strains and possibly also contig 7 of strain QOC1 (see above).
In L. monocytogenes different restriction modification (RM) systems have been shown to be involved in phage resistance [32]–[34]. Both genomes harbour a putative type II Sau3AI-like RM system, consisting of a Sau3AI-like restriction enzyme and a DNA methylase, showing 32% and 50% amino acid sequence identity to the recently described Sau3AI-like RM system of L. monocytogenes F2365 (serovar 4b) [32] (Figure S1). Interestingly, no homologues of the DNA binding protein lmof2365_0326 or the recombinase lmof2365_0328 are present in the Quargel outbreak strains (verified by blastN and tblastN searches). The Sau3AI-like RM systems, while found in the same relative genomic region (hypervariable hotspot 4), differ in their location. In QOC1 and QOC2, these systems are located between the homologues of lmo0305 and lmo0314, while in F2365 and FSL N3–165, they are located between the homologues of lmo0301 and lmo0305. The Sau3AI-like RM systems in QOC1 and QOC2 thus most likely represent novel Sau3AI-like RM system variants. Furthermore, the QOC2 variant harbours an additional putative helicase and nucleotidase upstream of the restriction enzyme (Figure S1). Highly similar proteins to the QOC2 RM system (99% amino acid identity) are only found in one additional L. monocytogenes genome: SLCC7179 (serovar 3a, ST91) isolated 1986 in Austria from cheese potentially involved in a listeriosis outbreak [35], [36]. All other homologues were from other Firmicutes (amino acid identity up to 63%). The GC content of the RM system genes was noticeably lower (29 to 33%) than the average L. monocytogenes GC content (38%), suggesting a relatively recent evolutionary horizontal gene transfer event. The presence of novel Sau3AI-like RM system variants in this particular chromosomal region emphasizes the important role of hypervariable hotspot 4 in the dissemination of RM systems among L. monocytogenes. No homologues of other described L. monocytogenes RM systems were found in QOC1 and QOC2.
Another mechanism for phage resistance are CRISPR (clustered regularly interspaced short palindromic repeats) systems [26]. Both genomes harbour one CRISPR system at locus 2 (inserted between lmo0517 and lmo0518 homologues), differing in their spacer content. Additionally, the CRISPR system in strain QOC2 harbours a cas4 gene copy which is absent in QOC1.
The presence of RM and CRISPR systems might therefore be one reason for the absence of phages in the Quargel outbreak strains.
Genes Related to Stress Resistance
L. monocytogenes encounters various stress conditions during its extracellular lifestyle within food, the food environment, or human hosts, as well as during intracellular replication inside eukaryotic cells. L. monocytogenes QOC1 encodes a homologue of the so-called “stress survival islet 1” (SSI-1), whereas strain QOC2 harbours a homologue of the (uncharacterized) L. monocytogenes F2365 LMOf2365_0481 gene. SSI-1 is a 8.7 kbp region inserted between lmo0443 and lmo0449, consisting of five genes (lmo0444, lmo0445, pva, gadD1 and gadT1), which have been linked to tolerance towards acidic, salt, bile and gastric stress [37]–[39]. Therefore, strains harbouring SSI-1 might not only better survive in food or food production environments, but also the passage through the stomach and gut which could lead to a higher infection rate. Strains devoid of SSI-1 either encode a LMOf2365_0481 homologue, or of the (uncharacterized) L. innocua genes lin0464 and lin0465 [40].
We therefore analyzed the stress tolerance of QOC1 and QOC2 by incubating both strains for 2 h in acid (pH 2, pH 3), alkaline (pH 11, pH 12) and in gastric fluid conditions. The survival rate of QOC1 was significantly higher under all tested conditions compared to QOC2 (Figure 2). Our results are in accordance with previous studies which demonstrate a role for SSI-1 in stress response [39]. In addition, growth of QOC1 was significantly increased under mild acidic stress (pH 5), whereas no difference was observed under salt stress (7.5% NaCl, Figure S2).
Figure 2. Survival of L. monocytogenes QOC1 and QOC2 in minimal media adjusted to pH 2, pH 3, pH 11 and pH 12; and in gastric fluid.
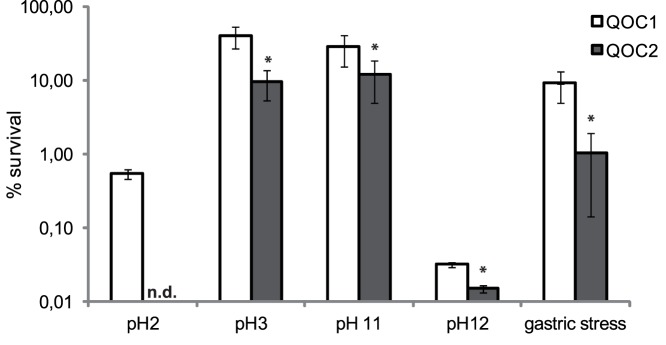
Values, given as percentage of survival, represent mean values ± SD of four biological replicates performed in triplicate. *indicates statistical significant differences (P<0.05) between QOC1 and QOC2. n.d.: not detectable.
Genome Comparison with Respect to Virulence
In general, both genomes are highly similar with respect to virulence genes, and have typical features of virulent L. monocytogenes genomes, such as a functional virulence gene cluster (Listeria pathogenicity island 1, LIPI-1), or full-length inlAB genes. We searched the genomes of the two Quargel outbreak strains for homologues of 81 virulence genes of L. monocytogenes EGDe [41]. With the exception of lmo1099 and lmo1102– which are absent from both Quargel strains – and lmo0320 (vip, which is missing in QOC1), homologues of all other virulence proteins were present (Table S2). Vip is an LPXTG surface protein which has been shown to be essential for the entry into host cells by recruiting Gp96 as a receptor [42], [43]. In addition, the QOC2 autolysin amidase homologue (ami, lmo2558) has a C-terminal truncation, and is thus a putative pseudogene. Ami contributes in adhesion to several eukaryotic cells and promotes colonization of hepatocytes [44], [45]. Interestingly, the QOC2 Listeria nuclear targeted protein A (LntA) shows only 89% amino acid and 95% nucleic acid identity to LntA from EGDe, whereas QOC1 LntA shares 99.5% amino acid identity with EGDe LntA (Table S2). LntA is secreted into the host cell nucleus and targets the chromatin repressor BAHD1 which is involved in immune response to L. monocytogenes infection [46]. The QOC2 LntA variant is currently found in three other sequenced L. monocytogenes genomes (strains: C1–387, Finland 1998, & J2–031).
Internalins and internalin-like proteins - whether secreted or located in the membrane - are essential to the virulence of L. monocytogenes, mainly for host cell interaction [47], [48]. Both strains encode the same set of 11 internalins (inlA, inlB, inlC, inlC2, inlD, inlE, inlF, inlG, inlI, inlJ, inlK). In addition, the organization of inlAB and inlGC2DE regions is identical in both strains (Figure S3, S4). Overall, strain QOC1 encodes 30 internalins or internalin-like proteins, while QOC2 encodes 26 such proteins.
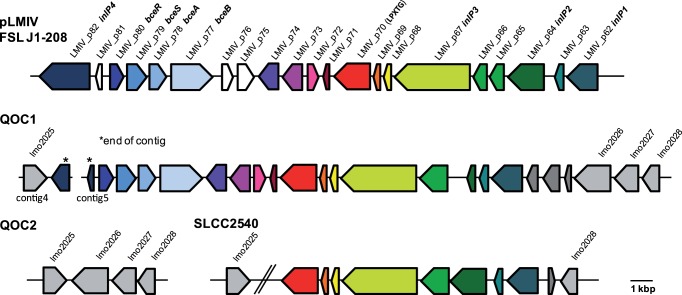
Four putative internalins and one internalin-like protein, present only in QOC1, show highest similarity to proteins of the internalin family found on plasmid pLMIV, carried by L. monocytogenes strain FSL J1–208. Strain FSL J1–208, belonging to serotype 4a, was isolated from a listeriosis outbreak in goats [49]. After a more detailed analysis we identified an approx. 21 kbp region in the QOC1 genome with a GC content of 33.5% showing highest similarity to a region of pLMIV (locus_tags: LMIV_p062 to LMIV_p082; Table 2, Figure S5). This region is integrated between the L. monocytogenes EGDe homologues lmo2025 (nadA, quinolinate synthetase) and lmo2026 (internalin-like protein) (Figure 3), and has previously been assigned as hypervariable hotspot 9 [26]. Thus, our results strongly suggest that a part of plasmid pLMIV is integrated into the QOC1 genome, due to the significantly lower GC content of this region (33.5%) compared to the average genomic GC content (38%) and due to the high similarity to pLMIV homologues. This is most likely a relatively recent evolutionary gene transfer event.
Table 2. Predicted genes in the hypervariable hotspot 9 region of the L. monocytogenes QOC1 genome.
| LMQOC1 locus_tag | Length (bp) | Description | Best Blast hit (GenBank accession no., amino acid identity) |
| LMQOC1_40229# | 588 | Internalin P4# | LMIV_p082 L. monocytogenes FSL J1–208 (EHY61417, 96%) |
| LMQOC1_50001# | 150 | Internalin P4# | LMIV_p082 L. monocytogenes FSL J1–208 (EHY61417, 90%) |
| LMQOC1_50002 | 669 | Two-component response regulator bceR | LMIV_p080 L. monocytogenes FSL J1–208 (EHY61415, 99%) |
| LMQOC1_50003 | 333 | Protein of unknown function | No hit |
| LMQOC1_50004 | 1014 | Sensor histidine kinase bceS | LMIV_p079 L. monocytogenes pLMIV FSL J1–208 (EHY61414, 98%) |
| LMQOC1_50005 | 768 | ABC transporter, ATP-binding protein bceA | LMIV_p078 L. monocytogenes pLMIV FSL J1–208 (EHY61413, 99%) |
| LMQOC1_50006 | 2031 | ABC transporter, permease protein bceB | LMIV_p077 L. monocytogenes pLMIV FSL J1–208 (EHY61412, 100%) |
| LMQOC1_50007 | 876 | Peptidase family M23 protein | LMIV_p074 L. monocytogenes pLMIV FSL J1–208 (EHY61409, 99%) |
| LMQOC1_50008 | 954 | Alpha/beta hydrolase fold protein | LMIV_p073 L. monocytogenes pLMIV FSL J1–208 (EHY61408, 98%) |
| LMQOC1_50009 | 528 | AcrR family transcriptional regulator | LMIV_p072 L. monocytogenes pLMIV FSL J1–208 (EHY61407, 98%) |
| LMQOC1_50010 | 153 | Conserved protein of unknown function | LMIV_p071 L. monocytogenes pLMIV FSL J1–208 (EHY61406, 97%) |
| LMQOC1_50011 | 1746 | Leucine-rich repeat domain protein(LPXTG motif) | LMOSLCC2540_2112 L. monocytogenes SLCC2540 (YP_006679655, 96%) |
| LMQOC1_50012 | 366 | Transposase | LMIV_p069 L. monocytogenes pLMIV FSL J1–208 (EHY61404, 95%) |
| LMQOC1_50013 | 267 | Transposase | LMIV_p068 L. monocytogenes pLMIV FSL J1–208 (EHY61403, 95%) |
| LMQOC1_50014 | 3612 | Internalin P3 | LMIV_p067 L. monocytogenes pLMIV FSL J1–208 (EHY61402, 94%) |
| LMQOC1_50015 | 1368 | Conserved protein of unknown function | LMOSLCC2540_2116 L. monocytogenes SLCC2540 (YP_006679659, 96%) |
| LMQOC1_50016* | 327 | Internalin P2 | LMIV_p064 L. monocytogenes pLMIV FSL J1–208 (EHY61399, 83%) |
| LMQOC1_50017 | 267 | Transposase | LMIV_p063 L. monocytogenes pLMIV FSL J1–208 (EHY61398, 97%) |
| LMQOC1_50018 | 1521 | Internalin P1 | LMIV_p063 L. monocytogenes pLMIV FSL J1–208 (EHY61398, 82%) |
on two contigs.
*putative pseudogene.
Figure 3. Genomic organization of the hypervariable hotspot 9 region in L. monocytogenes genomes harbouring homologues to proteins from plasmid pLMIV of L. monocytogenes FSL J1–208.
Homologous proteins are shown as the same color. pLMIV and L. monocytogenes EGDe locus_tags are indicated.
The pLMIV-like region in strain QOC1 is part of two contigs in the assembly. However, the fact that the largest fragment of this region is part of contig 5 and that one part of an inlP4 homologue (LMQOC1_40229) is located on contig 4, strongly suggests that this region is integrated in the QOC1 genome. We performed PCR for gap closing and obtained PCR products in the expected size. In addition, sequencing of the PCR products showed >99.7% similarity to the flanking regions of contig 4 and contig 5. However, due to the high number of identical repeat units within the inlP4-like gene (LMQOC1_40229, LMQOC1_50001), we were unable to unambiguously close this gap (Figure S6). The pLMIV-like region in QOC1 encodes four putative internalins (inlP1 (LMQOC1_50018), inlP3 (LMQOC1_50014), inlP4 (LMQOC1_40229, LMQOC1_50001, whereas inlP2 is putative pseudogene), one internalin-like protein (LMQOC1_50011) and a small cluster encoding a putative two-component regulatory system, the ABC transporter and a few other uncharacterized proteins (Table 2). Sequence analysis revealed the highest similarity among functionally characterized proteins of the two-component regulatory system and the ABC transporter subunits (21 to 47% amino acid sequence identity) to the bceRSAB system of Bacillus subtilis, which is responsible for bacitracin resistance [50]. Bacitracin is a non-ribosomally synthesized peptide antibiotic produced by various bacteria. Thus, the presence of an additional putative bacitracin resistance locus in strain QOC1 may be advantageous in coping with bacterial competitors in complex microbial communities. Whether or not the presence of bceRSAB homologues in the FSL J1–208 and QOC1 genomes results in an increased bacitracin tolerance has not yet been tested. In general, most L. monocytogenes strains, among them QOC1 and QOC2, harbour various additional genes responsible for bacitracin resistance e.g. telA, lisK and the ABC transporter anrB [51], [52].
The function of the four putative internalins and the additional internalin-like protein in the pLMIV-like region is still unknown. In vitro virulence assays using human epithelial Caco-2 cells revealed that the invasion efficiency of the plasmid-cured L. monocytogenes FSL J1–208 strain was not significantly different from the parental strain, suggesting that in this experimental setting genes encoded on pLMIV are not essential for virulence [49]. However, in vivo virulence, the invasion efficiency and intracellular proliferation of the plasmid-cured strain FSL J1–208 have not been tested in other cell types like hepatocytes and macrophages.
More detailed sequence analysis revealed that homologues of one part of this region - including inlP1, inlP2, inlP3 and an internalin-like protein (LMIV_p070 homologue) – are also present in the hypervariable hotspot 9 region of the L. monocytogenes strain SLCC2540 (CLIP74906) genome, which is a serotype 3b human isolate from 1956 in the USA (Figure 3).
An additional difference between the two Quargel outbreak strains is, that only QOC2 harbours a Vip (lmo0320) homologue (Table S2), a surface protein covalently attached to the bacterial cell wall. Vip, which binds to the eukaryotic Gp96 surface protein, is PrfA-dependent and is essential for the entry into certain mammalian cells e.g. fibroblasts and epithelial cells. In addition, infection studies suggest a role for Vip in Listeria virulence, mainly in the later stages of infection [42], [43].
We also identified differences in the monocin locus, a cryptic prophage region, found in most L. monocytogenes genomes. In the genome of the type strain EGDe, this locus consists of the genes lmo0115 to lmo0129 including the lma operon (lmaDCBA) [53]. Interestingly, strain QOC2 lacks lmaBA homologues as well as homologues of the genes lmo0119–0128 (Figure S7). Recently, it has been described that the genes of the lma operon and the surrounding prophage genes of the monocin locus are highly expressed during intracellular growth in macrophages [54]. The secreted protein LmaA provokes a delayed-type hypersensitivity response in Listeria immune mice, suggesting that LmaA is an immunologically relevant antigen in Listeria infection [55]. In addition, chromosomal deletion of lmaB resulted in attenuated growth of L. monocytogenes in the spleen and liver in a murine infection model [54]. These data suggest a role for the lma operon in the virulence of L. monocytogenes strains that harbour it. However, the presence of a truncated lma operon in QOC2, a feature also found in other L. monocytogenes strains [54], suggests that the lma operon is, at least in strain QOC2, not essential for virulence.
Another region with distinct differences between the two strains is the region between the lmo0061 and lmo0075 homologues, termed hypervariable hotspot 1 according to Kuenne et al. [26]. Interestingly, this region harbours the putative Listeria WSS (type VII/WXG100) secretion system. This secretion system has been shown to be important for virulence in Staphylococcus (S.) aureus, Bacillus (B.) anthracis and Mycobacterium tuberculosis [56]–[59], and homologues of this system have also previously been found in the L. monocytogenes EGDe genome [60], [61]. However, experimental evidence for the functionality as a secretion system is still missing for L. monocytogenes [62]. Experimental data showed that chromosomal deletion of lmo0056 (esxA), a putative WXG100 protein, did not result in attenuated virulence potential in L. monocytogenes 10403S [63]. However, strain 10403S does not encode homologues of the whole S. aureus WSS region, only esxA to essC homologues are present. The putative WSS secretion system of L. monocytogenes 10403S might therefore be non-functional or may display different functionality compared to those putative Listeria WSS secretion systems harbouring esxA to esaD homologues.
The genome of strain QOC1 encodes homologues of the complete WSS region (esxA to esaD) of S. aureus and B. cereus [59] (Figure S8), whereas the genome of strain QOC2 (and those of many other Listeria genomes) encodes only a subset of the S. aureus WSS secretion system: the esxA to essC homologues. Particularly the regions downstream of essC (lmo0061 homologues) and esaD (lmo0066 homologues) show a high level of diversity and encode mostly uncharacterized proteins, although some proteins show weak similarities to predicted polymorphic toxin systems [64].
In vitro Virulence
The two Quargel outbreak strains are highly pathogenic. Both were responsible for causing a number of reported listeriosis cases, some of which included fatal outcomes. However, based on genome sequence analyses, pronounced strain-specific differences with respect to virulence genes between QOC1 and QOC2 are evident. We therefore performed virulence assays in order to determine similarities and differences in the in vitro virulence potential of the two strains.
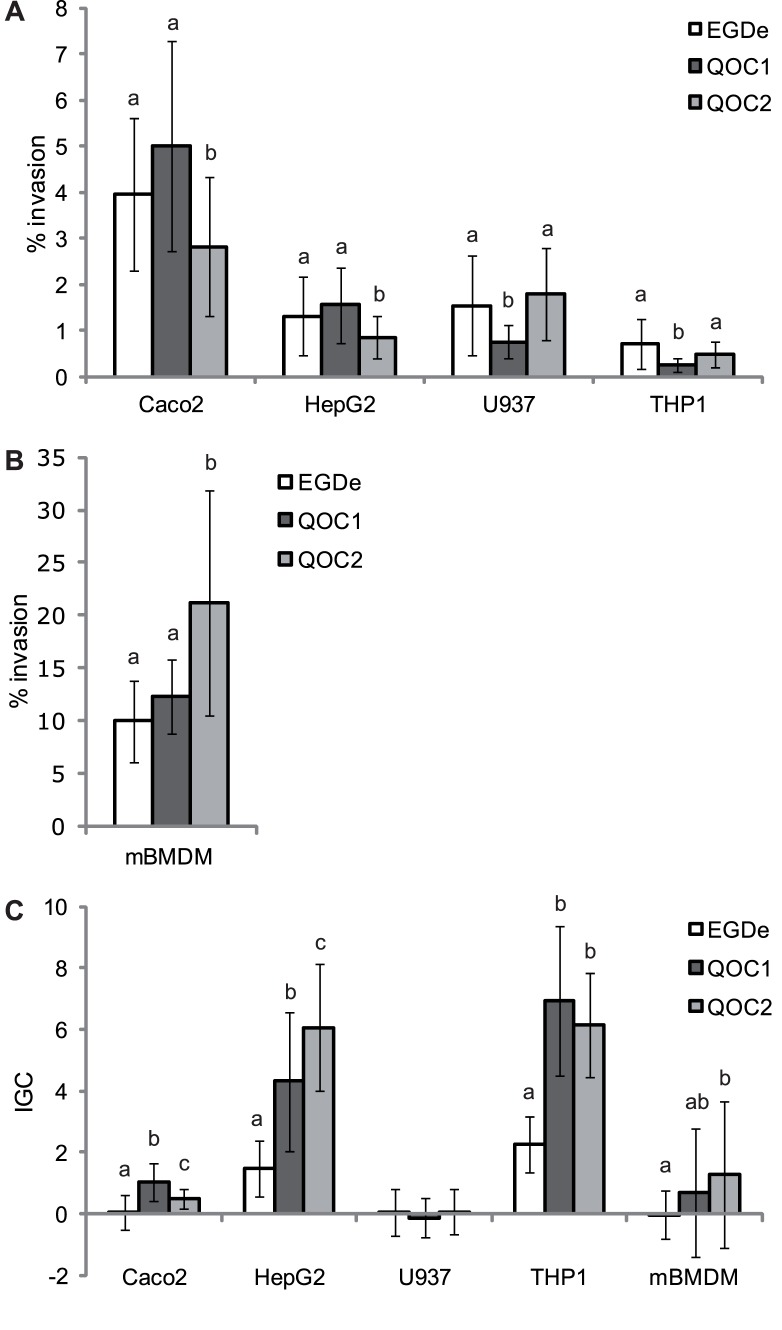
We evaluated invasion efficiency and intracellular growth of both Quargel outbreak strains and strain EGDe using different cell types involved in the infection pathway of L. monocytogenes. Invasion efficiencies of QOC1 and EGDe were significantly higher using human intestinal epithelial Caco2 and hepatocytic HepG2 cells compared to QOC2. In contrast, QOC2 showed higher invasion efficiency than QOC1 in macrophage-like cells (Figure 4). In addition, L. monocytogenes EGDe showed higher invasion efficiencies compared to QOC1 in U937 and THP1 cell lines. Considerable differences in intracellular growth between both Quargel outbreak strains could only be observed in non-phagocytic cells. QOC1 showed higher intracellular proliferation in Caco2, but reduced proliferation in HepG2 compared to QOC2. Interestingly, intracellular growth of EGDe in all tested host cells was low.
Figure 4. Invasion efficiency and intracellular proliferation of L. monocytogenes strains.
Invasion efficiency (panel A and B) and intracellular growth coefficient (IGC, panel C) of type strain EGDe, QOC1 and QOC2 using four different human cell lines (intestinal epithelial Caco2, hepatocytic HepG2 and macrophage-like U937 and THP1 cells) and primary mouse bone-marrow derived macrophages (mBMDM). Values represent mean values ± SD of four biological replicates performed in duplicate. Different letters indicate statistically significant differences (P<0.05).
Our data show that both Quargel outbreak strains are virulent, however with host cell type dependent differences. The higher invasion efficiency of QOC1 observed in non- phagocytic cells could be due to the presence of the four putative additional pLMIV-like internalins, a full-length ami gene, the complete lma operon, or the complete putative Listeria WSS secretion system - all of which are missing or truncated in QOC2. In contrast, both QOC2 and EGDe, which show a higher number of intracellular bacteria in phagocytic cells, harbour the surface protein Vip. There is some evidence that Vip could play a role in host cell interaction between L. monocytogenes and macrophages due to presence of the Gp96 receptor on macrophages [65]. However, experimental data are missing. Whether or not the presence of Vip in QOC2 is responsible for increased intracellular proliferation observed in hepatocytes compared to QOC1 is unclear. A vip deletion mutant strain showed reduced numbers of bacteria in the liver, spleen, intestine and brain of mice [42]. The autolysin amidase Ami has been shown to promote adhesion and internalization into hepatocytic cells [44]. In line with this, QOC2, which harbors a truncated ami gene, shows reduced invasion into HepG2 cells compared to EGDe and QOC1 (Figure 4). However, the intracellular proliferation of QOC2 in HepG2 cells is higher compared to EGDe and QOC1, suggesting that Ami is not required for intracellular proliferation in HepG2 cells.
One explanation for the higher level of intracellular growth of QOC1 in intestinal epithelial Caco2 cells might come down to the presence of the monocin locus. Genes of the monocin locus, including the lma operon, which are completely present in the QOC1genome, have been described to be highly expressed during intracellular growth in macrophages [54]. Unfortunately, no data on the expression level of this locus during intracellular proliferation in other host cells like Caco2 cells are currently available.
Comparison between Outbreak Strains
Currently, only a few genome sequences of L. monocytogenes strains from outbreaks with more than one strain involved are available. Orsi and coworkers analysed the genomes of four L. monocytogenes strains (two food and two human isolates - all serovar 1/2a) involved in a sporadic listeriosis case in 1988 (due to contaminated turkey frankfurters) and an outbreak in 2000 caused by deli turkey meat. The same food processing plant was responsible for cases in 1988 and 2000 [21]. The genomic backbone of all strains was almost identical (1 to 8 SNPs) when comparing the isolates from the same year as well as the isolates from 1988 and 2000. The only major rearrangements and variations were found in the prophage inserted into the comK gene. A large listeriosis outbreak occurred in Canada in 2008 due to the consumption of contaminated ready-to-eat meat. The genomes of two strains involved in this outbreak were subsequently sequenced and analysed [20]. Comparison of the two sequenced strains revealed that the genomic backbones were almost identical (28 SNPs), however, similar to the 1988–2000 listeriosis outbreak strains, the strains also differed in their prophage content: strain 08–5578 harboured a phage (φLMC1) which is absent in strain 08–5923. In addition, strain 08–5578 contained a plasmid (pLM5578), which is missing in 08–5923. Also, a comparison of four different L. monocytogenes strains involved in the recent cantaloupe listeriosis outbreak in the US [66], [67] using microarrays revealed strain-specific differences in phage-related genes [67]. These studies show the importance of bacteriophages in the evolution of closely related outbreak-associated L. monocytogenes strains. Similarly, Verghese et al. showed that phages inserted into the comK gene might provide fitness advantages in food production environments [68]. Unlike to the aforementioned outbreaks, no phages are found in QOC1 and QOC2. In addition, each of the Quargel strain harbours a considerable number of strain-specific genes, most of them with unknown function: L. monocytogenes QOC1 encodes 93 strain-specific genes, whereas strain QOC2 harbours 45 strain-specific genes (Table S1 and S3).
Conclusion
In conclusion, our results show that strains QOC1 and QOC2 are distinct and did not recently evolve from a common ancestor. Most likely, strain QOC1 was replaced by strain QOC2, probably by changing the commercial ripening culture used for smearing [14], [15], [17]. Recent studies analysing the L. monocytogenes strains isolated from the Quargel lots from the Austrian producer during the last period of the outbreak (December 2009 to January 2010) revealed that all strains isolated belong to either of the two PFGE types and showed the same PFGE patterns as the human outbreak isolates. PFGE typing revealed that PFGE type 2 (the PFGE type of QOC2) was highly dominant among the food isolates from this period [69]. Similarly, QOC2 (PFGE type2) was also predominant among the human isolates in this time period [17]. Two different types of Quargel were produced in this particular plant: either red-smear-ripened, or mold-ripened. Interestingly, only the red-smear Quargel lots were L. monocytogenes positive [14], [69]. Furthermore, the production lines for both Quargel types were identical except for the smearing process, which was performed in different machines in physically separated areas of the production plant [14]. This strongly suggests smearing as the main contamination source with L. monocytogenes. Based on the available data and the pronounced genomic differences between the two L. monocytogenes strains involved in this outbreak, the most likely explanation is that the 2009/2010 multinational Quargel listeriosis outbreak might actually represent two overlapping outbreaks caused by separate contamination events within the same food processing plant.
Supporting Information
Genomic organization of the Sau3AI restriction system locus in L. monocytogenes strains of different serovar.
(PDF)
Growth under stress conditions.
(PDF)
Genomic organization of the inlAB locus in L monocytogenes serovar 1/2a outbreak strains.
(PDF)
Genomic organization of the inlGHE/C2DE locus in L. monocytogenes serovar 1/2a outbreak strains.
(PDF)
DNA-based alignment (dotplot) of pLMIV from L. monocytogenes FSL J1–208 with the L. monocytogenes QOC1 genome.
(PDF)
Genomic region surrounding the gap between contig 4 and contig 5 in the L. monocytogenes QOC1 genome.
(PDF)
Organization of the monocin region/ lmaDCBA locus in L. monocytogenes serovar 1/2a outbreak strains.
(PDF)
Genomic organization of the putative WSS/type VII secretion system in L. monocytogenes serovar 1/2a outbreak strains.
(PDF)
Strain-specific proteins of L. monocytogenes QOC1 compared with L. monocytogenes QOC2.
(PDF)
Presence of 81 virulence-associated genes in L. monocytogenes Quargel outbreak strains.
(PDF)
Strain-specific proteins of L. monocytogenes QOC2 compared with L. monocytogenes QOC1.
(PDF)
Acknowledgments
We thank Dr. Birgit Strobl for providing the mBMDM. We thank the team of curators of the Institut Pasteur MLST system (Paris, France) for importing novel alleles, profiles and/or isolates at http:/www.pasteur.fr/mlst.
Funding Statement
This work was supported by the Christian-Doppler Society (https://www.cdg.ac.at/). Moreover, the work was partly supported by a grant from the Austrian Science Fund (FWF, http://www.fwf.ac.at/) to SSE (grant no. P22703-B17) and by a start-up project from the University of Veterinary Medicine Vienna to KR. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ivanek R, Grohn YT, Wiedmann M (2006) Listeria monocytogenes in multiple habitats and host populations: review of available data for mathematical modeling. Foodborne Pathog Dis 3: 319–336. [DOI] [PubMed] [Google Scholar]
- 2. Milillo SR, Friedly EC, Saldivar JC, Muthaiyan A, O’Bryan C, et al. (2012) A review of the ecology, genomics, and stress response of Listeria innocua and Listeria monocytogenes . Crit Rev Food Sci Nutr 52: 712–725. [DOI] [PubMed] [Google Scholar]
- 3. Ryan S, Hill C, Gahan CG (2008) Acid stress responses in Listeria monocytogenes . Adv Appl Microbiol 65: 67–91. [DOI] [PubMed] [Google Scholar]
- 4. Tasara T, Stephan R (2006) Cold stress tolerance of Listeria monocytogenes: A review of molecular adaptive mechanisms and food safety implications. J Food Prot 69: 1473–1484. [DOI] [PubMed] [Google Scholar]
- 5. Anonymous (2012) The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2010. Euro Surveill 17: 2597. [PubMed] [Google Scholar]
- 6.Yde M, Naranjo M, Mattheus W, Stragier P, Pochet B, et al.. (2012) Usefulness of the European epidemic intelligence information system in the management of an outbreak of listeriosis, Belgium, 2011. Euro Surveill 17. [PubMed]
- 7.CDC CfDCaP (2012) Centers for Disease Control and Prevention website. Multistate outbreak of listeriosis linked to imported Frescolina Marte brand Ricotta salata cheese. Available: http://www.cdc.gov/listeria/outbreaks/cheese-09-12/advice-consumers.html. Accessed 2014 Feb.
- 8.CDC CfDCaP (2013) Centers for Disease Control and Prevention website. Multistate outbreak of listeriosis linked to Crave Brothers Farmstead cheeses (final update). Available: http://www.cdc.gov/listeria/outbreaks/cheese-07-13/index.html. Accessed 2014 Feb.
- 9. Orsi RH, den Bakker HC, Wiedmann M (2011) Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol 301: 79–96. [DOI] [PubMed] [Google Scholar]
- 10. Ooi ST, Lorber B (2005) Gastroenteritis due to Listeria monocytogenes . Clin Infect Dis 40: 1327–1332. [DOI] [PubMed] [Google Scholar]
- 11. Allerberger F, Wagner M (2010) Listeriosis: a resurgent foodborne infection. Clin Microbiol Infect 16: 16–23. [DOI] [PubMed] [Google Scholar]
- 12. Lamont RF, Sobel J, Mazaki-Tovi S, Kusanovic JP, Vaisbuch E, et al. (2011) Listeriosis in human pregnancy: a systematic review. J Perinat Med 39: 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fretz R, Sagel U, Ruppitsch W, Pietzka A, Stoger A, et al.. (2010) Listeriosis outbreak caused by acid curd cheese Quargel, Austria and Germany 2009. Euro Surveill 15. [PubMed]
- 14. Schoder D, Rossmanith P, Glaser K, Wagner M (2012) Fluctuation in contamination dynamics of L. monocytogenes in quargel (acid curd cheese) lots recalled during the multinational listeriosis outbreak 2009/2010. Int J Food Microbiol 157: 326–331. [DOI] [PubMed] [Google Scholar]
- 15. Schoder D, Skandamis P, Wagner M (2013) Assessing in-house monitoring efficiency by tracing contamination rates in cheese lots recalled during an outbreak of listeriosis in Austria. Int J Food Microbiol 167: 353–358. [DOI] [PubMed] [Google Scholar]
- 16. Pichler J, Appl G, Pietzka A, Allerberger F (2011) Lessons to be learned from an outbreak of foodborne Listeriosis, Austria 2009–2010. Food Protection Trends 31: 268–273. [Google Scholar]
- 17.Fretz R, Pichler J, Sagel U, Much P, Ruppitsch W, et al. (2010) Update: Multinational listeriosis outbreak due to ‘Quargel’, a sour milk curd cheese, caused by two different L. monocytogenes serotype 1/2a strains, 2009–2010. Euro Surveill 15. [PubMed]
- 18. Buchrieser C (2007) Biodiversity of the species Listeria monocytogenes and the genus Listeria . Microbes Infect 9: 1147–1155. [DOI] [PubMed] [Google Scholar]
- 19.Buchrieser C, Glaser P (2011) Genomics of Listeria monocytogenes and other members of the genus Listeria. Genomes of foodborne and waterborne pathogens. ASM Press, Washington, DC. 125–145.
- 20. Gilmour MW, Graham M, Van Domselaar G, Tyler S, Kent H, et al. (2010) High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics 11: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Orsi RH, Borowsky ML, Lauer P, Young SK, Nusbaum C, et al. (2008) Short-term genome evolution of Listeria monocytogenes in a non-controlled environment. BMC Genomics 9: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Darling AE, Mau B, Perna NT (2010) progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5: e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, et al. (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vallenet D, Engelen S, Mornico D, Cruveiller S, Fleury L, et al. (2009) MicroScope: a platform for microbial genome annotation and comparative genomics. Database (Oxford) 2009: bap021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuenne C, Billion A, Mraheil MA, Strittmatter A, Daniel R, et al. (2013) Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics 14: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cotter PD, Gahan CG, Hill C (2001) A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol Microbiol 40: 465–475. [DOI] [PubMed] [Google Scholar]
- 29. Baccarini M, Bistoni F, Lohmann-Matthes ML (1985) In vitro natural cell-mediated cytotoxicity against Candida albicans: macrophage precursors as effector cells. J Immunol 134: 2658–2665. [PubMed] [Google Scholar]
- 30. Pricope L, Nicolau A, Wagner M, Rychli K (2013) The effect of sublethal concentrations of benzalkonium chloride on invasiveness and intracellular proliferation of Listeria monocytogenes . Food Control 31: 230–235. [Google Scholar]
- 31. Ragon M, Wirth T, Hollandt F, Lavenir R, Lecuit M, et al. (2008) A new perspective on Listeria monocytogenes evolution. PLoS Pathog 4: e1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yildirim S, Elhanafi D, Lin W, Hitchins AD, Siletzky RM, et al. (2010) Conservation of genomic localization and sequence content of Sau3AI-like restriction-modification gene cassettes among Listeria monocytogenes epidemic clone I and selected strains of serotype 1/2a. Appl Environ Microbiol 76: 5577–5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim JW, Dutta V, Elhanafi D, Lee S, Osborne JA, et al. (2012) A novel restriction-modification system is responsible for temperature-dependent phage resistance in Listeria monocytogenes ECII. Appl Environ Microbiol 78: 1995–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee S, Ward TJ, Siletzky RM, Kathariou S (2012) Two novel type II restriction-modification systems occupying genomically equivalent locations on the chromosomes of Listeria monocytogenes strains. Appl Environ Microbiol 78: 2623–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haase JK, Murphy RA, Choudhury KR, Achtman M (2011) Revival of Seeliger’s historical ‘Special Listeria Culture Collection’. Environ Microbiol 13: 3163–3171. [DOI] [PubMed] [Google Scholar]
- 36. Allerberger F, Guggenbuchler JP (1986) Listeriosis in Austria–report of an outbreak in 1986. Acta Microbiol Hung 36: 149–152. [PubMed] [Google Scholar]
- 37. Begley M, Cotter PD, Hill C, Ross RP (2010) Glutamate decarboxylase-mediated nisin resistance in Listeria monocytogenes . Appl Environ Microbiol 76: 6541–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Begley M, Sleator RD, Gahan CG, Hill C (2005) Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes . Infect Immun 73: 894–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ryan S, Begley M, Hill C, Gahan CG (2010) A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J Appl Microbiol 109: 984–995. [DOI] [PubMed] [Google Scholar]
- 40. Hein I, Klinger S, Dooms M, Flekna G, Stessl B, et al. (2011) Stress survival islet 1 (SSI-1) survey in Listeria monocytogenes reveals an insert common to Listeria innocua in sequence type 121 L. monocytogenes strains. Appl Environ Microbiol 77: 2169–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. den Bakker HC, Cummings CA, Ferreira V, Vatta P, Orsi RH, et al. (2010) Comparative genomics of the bacterial genus Listeria: Genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics 11: 688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cabanes D, Sousa S, Cebria A, Lecuit M, Garcia-del Portillo F, et al. (2005) Gp96 is a receptor for a novel Listeria monocytogenes virulence factor, Vip, a surface protein. EMBO J 24: 2827–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martins M, Custodio R, Camejo A, Almeida MT, Cabanes D, et al. (2012) Listeria monocytogenes triggers the cell surface expression of Gp96 protein and interacts with its N terminus to support cellular infection. J Biol Chem 287: 43083–43093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Asano K, Sashinami H, Osanai A, Asano Y, Nakane A (2011) Autolysin amidase of Listeria monocytogenes promotes efficient colonization of mouse hepatocytes and enhances host immune response. Int J Med Microbiol 301: 480–487. [DOI] [PubMed] [Google Scholar]
- 45. Milohanic E, Jonquieres R, Cossart P, Berche P, Gaillard JL (2001) The autolysin Ami contributes to the adhesion of Listeria monocytogenes to eukaryotic cells via its cell wall anchor. Mol Microbiol 39: 1212–1224. [DOI] [PubMed] [Google Scholar]
- 46. Lebreton A, Lakisic G, Job V, Fritsch L, Tham TN, et al. (2011) A bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response. Science 331: 1319–1321. [DOI] [PubMed] [Google Scholar]
- 47. Bierne H, Sabet C, Personnic N, Cossart P (2007) Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes . Microbes Infect 9: 1156–1166. [DOI] [PubMed] [Google Scholar]
- 48.Pizarro-Cerda J, Kuhbacher A, Cossart P (2012) Entry of Listeria monocytogenes in mammalian epithelial cells: an updated view. Cold Spring Harb Perspect Med 2. [DOI] [PMC free article] [PubMed]
- 49. den Bakker HC, Bowen BM, Rodriguez-Rivera LD, Wiedmann M (2012) FSL J1–208, a virulent uncommon phylogenetic lineage IV Listeria monocytogenes strain with a small chromosome size and a putative virulence plasmid carrying internalin-like genes. Appl Environ Microbiol 78: 1876–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ohki R, Giyanto, Tateno K, Masuyama W, Moriya S, et al. (2003) The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis . Mol Microbiol 49: 1135–1144. [DOI] [PubMed] [Google Scholar]
- 51. Collins B, Curtis N, Cotter PD, Hill C, Ross RP (2010) The ABC transporter AnrAB contributes to the innate resistance of Listeria monocytogenes to nisin, bacitracin, and various beta-lactam antibiotics. Antimicrob Agents Chemother 54: 4416–4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Collins B, Joyce S, Hill C, Cotter PD, Ross RP (2010) TelA contributes to the innate resistance of Listeria monocytogenes to nisin and other cell wall-acting antibiotics. Antimicrob Agents Chemother 54: 4658–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schaferkordt S, Chakraborty T (1997) Identification, cloning, and characterization of the Ima operon, whose gene products are unique to Listeria monocytogenes . J Bacteriol 179: 2707–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hain T, Ghai R, Billion A, Kuenne CT, Steinweg C, et al. (2012) Comparative genomics and transcriptomics of lineages I, II, and III strains of Listeria monocytogenes . BMC Genomics 13: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gohmann S, Leimeister-Wachter M, Schiltz E, Goebel W, Chakraborty T (1990) Characterization of a Listeria monocytogenes-specific protein capable of inducing delayed hypersensitivity in Listeria-immune mice. Mol Microbiol 4: 1091–1099. [DOI] [PubMed] [Google Scholar]
- 56. Burts ML, DeDent AC, Missiakas DM (2008) EsaC substrate for the ESAT-6 secretion pathway and its role in persistent infections of Staphylococcus aureus . Mol Microbiol 69: 736–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen YH, Anderson M, Hendrickx AP, Missiakas D (2012) Characterization of EssB, a protein required for secretion of ESAT-6 like proteins in Staphylococcus aureus . BMC Microbiol 12: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Garufi G, Butler E, Missiakas D (2008) ESAT-6-like protein secretion in Bacillus anthracis . J Bacteriol 190: 7004–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schneewind O, Missiakas DM (2012) Protein secretion and surface display in Gram-positive bacteria. Philos Trans R Soc Lond B Biol Sci 367: 1123–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Anderson M, Chen YH, Butler EK, Missiakas DM (2011) EsaD, a secretion factor for the Ess pathway in Staphylococcus aureus . J Bacteriol 193: 1583–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pallen MJ (2002) The ESAT-6/WXG100 superfamily – and a new Gram-positive secretion system? Trends Microbiol 10: 209–212. [DOI] [PubMed] [Google Scholar]
- 62. Renier S, Micheau P, Talon R, Hebraud M, Desvaux M (2012) Subcellular localization of extracytoplasmic proteins in monoderm bacteria: rational secretomics-based strategy for genomic and proteomic analyses. PLoS One 7: e42982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Way SS, Wilson CB (2005) The Mycobacterium tuberculosis ESAT-6 homologue in Listeria monocytogenes is dispensable for growth in vitro and in vivo. Infect Immun 73: 6151–6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang DP, de Souza RF, Anantharaman V, Iyer LM, Aravind L (2012) Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biology Direct 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yang Y, Liu B, Dai J, Srivastava PK, Zammit DJ, et al. (2007) Heat shock protein gp96 is a master chaperone for toll-like receptors and is important in the innate function of macrophages. Immunity 26: 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. McCollum JT, Cronquist AB, Silk BJ, Jackson KA, O’Connor KA, et al. (2013) Multistate outbreak of listeriosis associated with cantaloupe. N Engl J Med 369: 944–953. [DOI] [PubMed] [Google Scholar]
- 67. Laksanalamai P, Joseph LA, Silk BJ, Burall LS, C LT, et al. (2012) Genomic characterization of Listeria monocytogenes strains involved in a multistate listeriosis outbreak associated with cantaloupe in US. PLoS One 7: e42448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Verghese B, Lok M, Wen J, Alessandria V, Chen Y, et al. (2011) comK prophage junction fragments as markers for Listeria monocytogenes genotypes unique to individual meat and poultry processing plants and a model for rapid niche-specific adaptation, biofilm formation, and persistence. Appl Environ Microbiol 77: 3279–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schoder D, Stessl B, Szakmary-Brandle K, Rossmanith P, Wagner M (2014) Population diversity of Listeria monocytogenes in quargel (acid curd cheese) lots recalled during the multinational listeriosis outbreak 2009/2010. Food Microbiol 39: 68–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomic organization of the Sau3AI restriction system locus in L. monocytogenes strains of different serovar.
(PDF)
Growth under stress conditions.
(PDF)
Genomic organization of the inlAB locus in L monocytogenes serovar 1/2a outbreak strains.
(PDF)
Genomic organization of the inlGHE/C2DE locus in L. monocytogenes serovar 1/2a outbreak strains.
(PDF)
DNA-based alignment (dotplot) of pLMIV from L. monocytogenes FSL J1–208 with the L. monocytogenes QOC1 genome.
(PDF)
Genomic region surrounding the gap between contig 4 and contig 5 in the L. monocytogenes QOC1 genome.
(PDF)
Organization of the monocin region/ lmaDCBA locus in L. monocytogenes serovar 1/2a outbreak strains.
(PDF)
Genomic organization of the putative WSS/type VII secretion system in L. monocytogenes serovar 1/2a outbreak strains.
(PDF)
Strain-specific proteins of L. monocytogenes QOC1 compared with L. monocytogenes QOC2.
(PDF)
Presence of 81 virulence-associated genes in L. monocytogenes Quargel outbreak strains.
(PDF)
Strain-specific proteins of L. monocytogenes QOC2 compared with L. monocytogenes QOC1.
(PDF)