Posttranscriptional regulation shifts the metabolic flux of flavonoids toward coumarin and general phenylpropanoids.
Abstract
The flavonoids are phenylpropanoid-derived metabolites that are ubiquitous in plants, playing many roles in growth and development. Recently, we observed that fruit rinds of yellow casaba muskmelons (Cucumis melo ‘Inodorous Group’) accumulate naringenin chalcone, a yellow flavonoid pigment. With RNA-sequencing analysis of bulked segregants representing the tails of a population segregating for naringenin chalcone accumulation followed by fine mapping and genetic transformation, we identified a Kelch domain-containing F-box protein coding (CmKFB) gene that, when expressed, negatively regulates naringenin chalcone accumulation. Additional metabolite analysis indicated that downstream flavonoids are accumulated together with naringenin chalcone, whereas CmKFB expression diverts the biochemical flux toward coumarins and general phenylpropanoids. These results show that CmKFB functions as a posttranscriptional regulator that diverts flavonoid metabolic flux.
Muskmelons (Cucumis melo; Cucurbitaceae) exhibit extreme diversity in fruit traits, varying in size, shape, external color, aroma, sugar content, acidity, pigmentation, and climacteric or nonclimacteric fruit ripening (Burger et al., 2010; Cohen et al., 2014). Various classifications of muskmelons have been proposed over the years. The most comprehensive yet easily understood classification distinguishes two subspecies: Cucumis agrestis and C. melo, with 5 and 11 groups, respectively (Pitrat et al., 2000).
The phenylpropanoid pathway gives rise to a large range of phenolic compounds, including flavonoids, lignins, lignans, coumarins, stilbenes, and sinapate esters (Veitch and Grayer, 2008; Petersen et al., 2010), which are involved in many aspects of plant physiology. Phenylpropanoids have roles related to photoprotection (Kim et al., 2008), cross talk with hormones (Besseau et al., 2007; Yuan et al., 2013), Rhizobium spp. symbiosis (Wasson et al., 2006), pollination (Mo et al., 1992), structural components (Pizzi and Cameron, 1986), and plant-animal interactions (Haribal and Renwick, 1996). Recently, flavonoids have gained increasing interest as health benefit agents in the human diet (Butelli et al., 2008; Romano et al., 2013).
Because of the wide interest in flavonoids, considerable efforts have focused on understanding the regulatory mechanisms of the different branches of their synthesis pathway. Our general understanding is that flavonoid accumulation is largely controlled at the transcriptional level by transcription factors regulating the expression of the pathway structural genes (Hartmann et al., 2005; Koes et al., 2005). However, pathway regulation downstream to the transcriptional level is less understood (Tanaka and Uritani, 1977; Pairoba and Walbot, 2003; Deguchi et al., 2013). Lately, Kelch domain-containing F-box (KFB) proteins were shown to be involved in regulation of the phenylpropanoid pathway (Shao et al., 2012; Zhang et al., 2013, 2015). F-box proteins serve as substrate recruitment domains in the S-phase kinase-associated protein1, Cullin, F-box (SCF) E3 ubiquitin ligase complexes, targeting protein substrates to 26S proteasome-dependent degradation, in which the F box serves as the target recognition factor, mediating the specificity of the complex (Jonkers and Rep, 2009). In plants, the F-box superfamily seems to have gone through vast duplication and specialization. Although the human and Drosophila spp. genomes contain only 68 and 33 F-box genes, respectively (Ou et al., 2003; Jin et al., 2004), the Arabidopsis (Arabidopsis thaliana) genome contains approximately 700 members of this superfamily (Gagne et al., 2002). One subgroup of this family, the KFB, found as a single ortholog in the human genome is highly represented in the Arabidopsis genome, comprising at least 103 KFB sequences, most of them still uncharacterized (Schumann et al., 2011). A rice (Oryza sativa) KFB homolog was found to negatively regulate flavonoid accumulation (Shao et al., 2012). Four members of the KFB subfamily in Arabidopsis were shown to physically interact with different Phe ammomia-lyases (PAL) isozymes, mediating their degradation, subsequently decreasing accumulation of an array of various phenylpropanoids, indicating these KFBs serve as general repressors of the phenylpropanoid pathway (Zhang et al., 2013, 2015).
Little is known about the regulation of the flavonoid pathway in the Cucurbitaceae. Flavone derivatives are accumulated in leaves of cucumber (Cucumis sativus) and muskmelon (Krauze-Baranowska and Cisowski, 2001). Kaempferol and quercetin glycosides (flavonol derivatives) accumulate in the reproductive organs of some cucurbits (Imperato, 1980). Recently, we showed that naringenin chalcone (NarCh) is accumulated as the main flavonoid in the fruit rind of some muskmelon varieties, including the yellow casaba muskmelon ‘Noy Amid’ (NA; Tadmor et al., 2010).
In this study, we used a combination of RNA sequencing (RNA-Seq) of pools of F3 segregants together with fine mapping to identify CmKFB, a protein coding gene that negatively regulates flavonoid accumulation in muskmelon. The repression of the flavonoid pathway results in biochemical flux alternation toward coumarin and other general phenylpropanoids derivatives. Functional verification of CmKFB was performed by its transient expression in muskmelon leaves and stable transformation of tomato (Solanum lycopersicum), resulting in altered phenylpropanoid patterns. Our results provide experimental evidence of regulation that alters the flow between the phenylpropanoid pathway branches.
RESULTS
Characterization of Biochemical Differences between Two Muskmelon Genotypes Differentiating in NarCh Accumulation
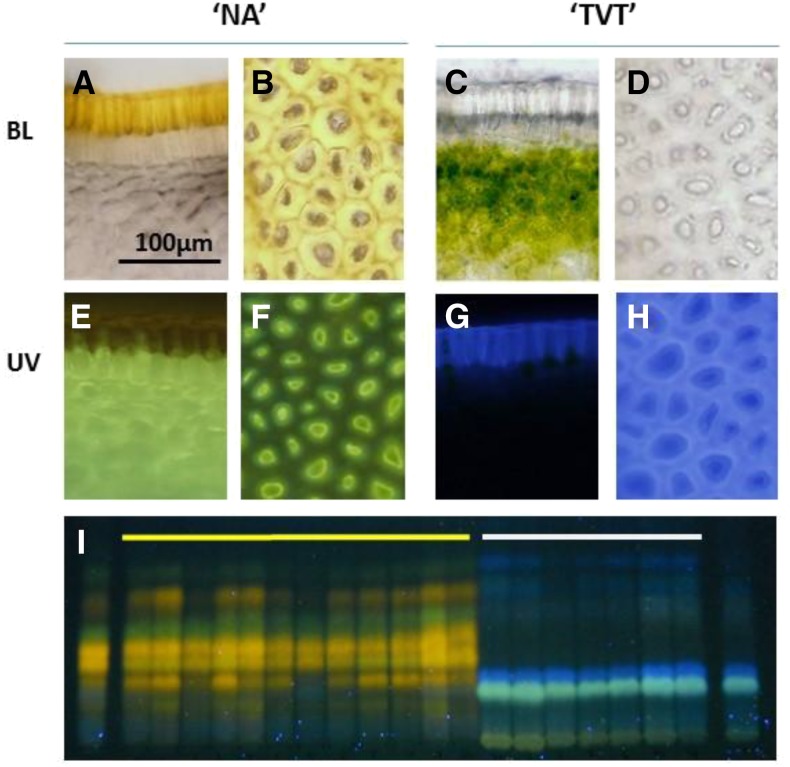
To obtain a general view of the biochemical differences between the NarCh-accumulating (NA) and NarCh-nonaccumulating (‘Tendral Verde Tardio’ [TVT]) muskmelons, microscopic examinations were performed. Slices of muskmelon rind were stained with diphenyl boric acid aminoethyl ester (DPBA), which fluorescently dyes flavonoid glycosides (Stracke et al., 2007). Under visible light, the upper cuticle layer of NA was yellow, probably because of the accumulation of NarCh, whereas in TVT, this layer seems transparent. After DPBA staining and under UV illumination, NA samples exhibited a greenish fluorescence in the tissue that is located at approximately 2 mm under the upper cuticle layer, whereas the cuticle of TVT exhibited blue fluorescence and lacked the greenish fluorescence under the cuticle (Fig. 1, A–H). This differential fluorescence display suggests that both NA and TVT accumulate different flavonoid compounds in addition to NarCh. The cuticle of NA exhibited no fluorescence after DPBA staining, suggesting the NarCh is accumulated in the cuticle as aglycone and not as a glycoside that would be fluorescent under UV light. To further investigate whether the additional fluorescent compounds accumulated in different tissues in NA and TVT are associated with NarCh accumulation, methanol extracts of mature fruit rinds of 19 F3 families were fractionated by thin-layer chromatography (TLC), stained with DPBA, and visualized under UV light. The 19 families were generated from a cross between NA and TVT (12 yellow and 7 white F3 families in “Materials and Methods”; Supplemental Fig. S1). Under these conditions, the fluorescence pattern of the yellow F3 families (NarCh accumulating) was identical with that of the NA parent, whereas the white F3 families exhibited the same fluorescence pattern as TVT, the non-NarCh-accumulating parent, showing association of these fluorescent compounds with NarCh accumulation after two generations (Fig. 1I).
Figure 1.
DPBA staining of muskmelon fruit rinds. Microscopic view of fruit rinds of the parental lines NA (A, B, E, and F) and TVT (C, D, G, and H). A, C, E, and G are fruit rind cross sections, and B, D, F, and H are upper views of fruit rind. BL, Bright visible light. I, TLC of methanol extract stained with DPBA of NA (left lane) and TVT (right lane) marked by yellow and white lines are the 12 F3 yellow families and 7 F3 white families, respectively.
DPBA-mediated fluorescence of additional tissues of the two muskmelon types indicated that leaves and flowers, regardless the presence/absence of NarCh in their fruits, exhibited similar fluorescence patterns (Supplemental Fig. S2, A and B). The differences in fruit rind fluorescence between the two muskmelon types were evident from 10 d after anthesis (DAA). During fruit development, the fluorescence intensity of yellow/green compounds decreased in white and increased in yellow F3s, whereas blue fluorescent compounds gradually increased during white fruit development and were absent in yellow fruit (Supplemental Fig. S2, C–E).
Similar staining experiments performed in Arabidopsis identified yellow and green DPBA-mediated fluorescent compounds as quercetin and kaempferol (flavonols) and blue fluorescence-emitting compounds as sinapate glycoside derivatives (Stracke et al., 2007, 2009). Application of additional staining, including Shinoda’s test and Zn-HCl (Pew’s test) on yellow and white F3 methanol extracts (Supplemental Fig. S3), showed that color accumulation in yellow was correlated with the accumulation of flavonols and dihydroflavonols (Pew, 1948), respectively, in contrast to white, which was negative to this staining, suggesting that additional flavonoids were accumulated in addition to NarCh.
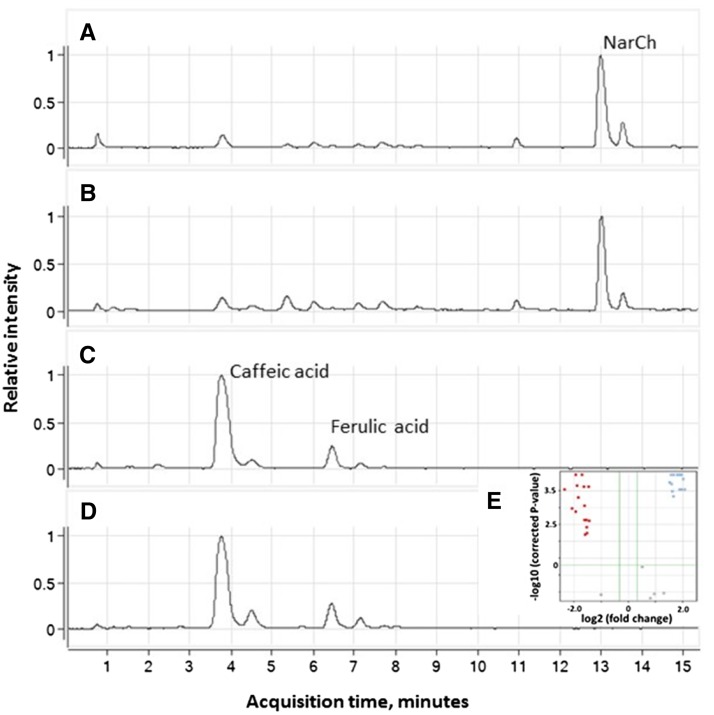
To further identify these fluorescent compounds, methanolic extracts of both parents (NA and TVT) and bulks of F3 families (white and yellow) were analyzed with liquid chromatography (LC)-mass spectrometry (MS; Supplemental Fig. S4, A–D). A similar pattern to the previously presented fluorescence on TLC was observed: the NarCh-accumulating parent and the yellow F3 bulk possessed similar characteristics, which are distinctive from the non-NarCh-accumulating TVT and the white F3 groups. An untargeted metabolic analysis was conducted to identify the main compounds that were differently accumulated in these two populations using Agilent’s Mass Profiler Professional Metabolic Tools. The means of the total ion counts of the different compounds found in yellow and white groups (each group is comprised of a parental line and a bulk of F3 families) were compared by a volcano plot using a threshold of at least 10-fold difference between the means and a P value of 0.05. In total, 99 masses that significantly differentiated between these two groups were identified (Supplemental Fig. S4E). The software could predict molecular formulas for 64 compounds of these 99 masses, many of them matching different flavonoid glycosides (Supplemental Table S1). To further identify the aglycone backbones that underlined these differences, enzymatic hydrolysis was performed on the samples, after which aglycones were isolated and analyzed with LC-MS. A corresponding similar pattern of statistically significant differential accumulation of compounds between the two groups was observed (Fig. 2; Supplemental Table S2). Identification of the aglycones was performed by comparison with authentic standards. Eight compounds were identified, among which caffeic acid, ferulic acid, and aesculetin showed significantly higher accumulation in the non-NarCh-accumulating fruit rinds (TVT and white F3 families), whereas naringenin, luteolin, kaempferol, quercetin, and traces of dihydrokaempferol were found to specifically accumulate in the NarCh-accumulating tissues (NA and yellow F3 families; Supplemental Fig. S5; Supplemental Table S3). Thus, a clear change of the pathway flux could be noted, which was evidenced by the metabolic patterns observed between the two groups (Fig. 3).
Figure 2.
LC-MS analysis of fruit rind methanol extracts after enzymatic hydrolysis. A to D, Base peak chromatograms of mass range mass-to-charge ratio 100 to 1,000 in ESI negative mode. A, NA parent. B, Bulk of yellow F3. C, TVT parent. D, Bulk of white F3. E, A volcano plot indicating specific compounds that differentially accumulated (P < 0.05; minimum 10-fold difference) in the NA and yellow F3 group (red) or the TVT and white F3 group (blue).
Figure 3.
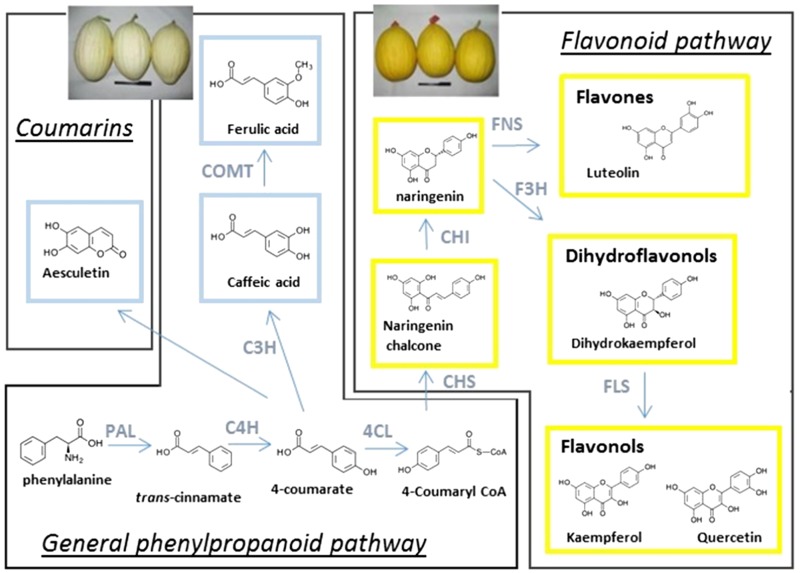
Phenylpropanoid compounds accumulated in yellow and white muskmelon fruit rinds and their corresponding biosynthesis pathway. Six compounds were specifically accumulated in the NarCh-accumulating lines (framed in yellow): NarCh, naringenin, luteolin, dihydrokaempferol, kaempferol, and quercetin. Three compounds were found to be specifically accumulated in the non-NarCh-accumulating lines (framed in blue): caffeic acid, ferulic acid, and aesculetin. C3H, 4-Coumarate 3-hydroxylase; C4H, cinnamate 4-hydroxylase; CHI, chalcone isomerase; COMT, caffeic acid 3-O-methyltransferase; F3H, naringenin 3-hydroxylase; FNS, flavone synthase; FLS, flavonol synthase.
NarCh-accumulating lines accumulate additional downstream flavonoids, including luteolin (a flavone), dihydrokaempferol (a dihydroflavonol), quercetin (a flavonol), and kaempferol (a flavonol). The non-NarCh-accumulating lines overaccumulate mainly caffeic and ferulic acids in addition to aesculetin (a coumarin), which are branched before the flavonoid biosynthetic pathway. Although clear differences in accumulation of caffeic and ferulic acids were observed between the NarCh-accumulating types, smaller levels of caffeic and ferulic acids were detectable in the NarCh-accumulating lines (retention times, 3.7 and 6.3 min in Fig. 2, A and B; Supplemental Table S3), whereas flavonoids were not detected in the non-NarCh-accumulating types.
Genetic Mapping of the Locus Governing Flavonoid Accumulation
Previous studies on fruit rind NarCh accumulation in an F2 population derived from the cross NA × TVT suggested that this trait is governed by a single gene (Tadmor et al., 2010). To better identify the genetic factor regulating flavonoid accumulation in muskmelon fruit rind, we comparatively analyzed the transcriptome of the yellow and white bulks of F3 families, a methodology that is based on the classical bulk segregant analysis (BSA) approach (Michelmore et al., 1991; Supplemental Fig. S1; “Materials and Methods”). RNA extracted from developing fruits of yellow and white F3 families was bulked and sequenced (Supplemental Table S4). Because at the time that we carried out this portion of the research, the muskmelon genome sequence was not yet available, a de novo-assembled transcriptome was developed that included 87,336 unigenes. All RNA-Seq reads of the two bulks at three developmental stages were aligned to the unigenes for the identification of single-nucleotide polymorphisms (SNPs) that existed between the yellow and white bulks. Upon the availability of the muskmelon genome (Garcia-Mas et al., 2012), SNPs were mapped to the muskmelon genome; 179 SNPs were found to differentiate between the two bulks (Supplemental Table S5), 164 of them are located on scaffold 16 (linkage group 10) of the muskmelon genome, and the majority of them is localized in a region of approximately 0.5 Mb (Fig. 4A), suggesting that the gene controlling NarCh accumulation in muskmelon fruit rind is located on scaffold 16 of chromosome 10. The remaining 15 SNPs were scattered randomly throughout the genome, suggesting that they are not specifically related to flavonoid accumulation.
Figure 4.
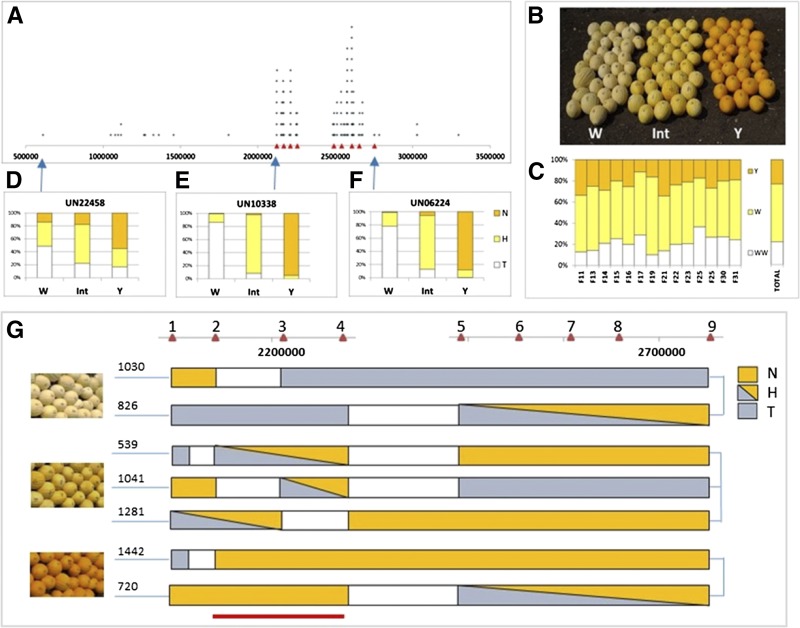
Genetic mapping of the locus governing flavonoid accumulation. A, Physical positions of the 164 of 179 SNPs that significantly differentiated between the bulks on scaffold 16 of the muskmelon genome. Red triangles represent positions of nine selected HRM markers. B, Phenotypes of individual F3 fruits included in the recombinants analysis: white (W), pale yellow (Int), and yellow (Y) fruit. C, Segregation (percentage) of Y, Int, and W phenotypes within each of the 14 F3 families and among all analyzed individual F3 plants (total). D to F, Distribution of TVT (T), heterozygote (H), and NA (N) alleles of the three fluorescent SNP probes (UN22458, UN10338, and UN06224) within W, Int, and Y phenotypes of individual F3 plants; the physical position of each probe is indicated by a blue arrow. G, Fine mapping with the nine HRM markers (red triangles 1–9). Below are individual plants segregating to these HRM markers and exhibiting defined phenotypes (white, pale yellow, and yellow). The bottom red line indicates the 100-kb genomic region that shows 100% cosegregation of genotype and phenotype.
For a more detailed characterization of the genetic locus that governs NarCh accumulation, analysis was performed on an additional 784 plants derived from 14 F3 families segregating for fruit rind NarCh accumulation. Mature fruit of this additional plant material exhibited three phenotypes in the field: yellow rind, pale yellow rind, and white rind (Fig. 4B). Phenotypic distribution of these F3 individuals supported a 1:2:1 ratio (yellow:pale yellow:white), expected for a single incomplete dominant gene (χ2 = 2.593, P = 0.27; Fig. 4C). DNA extracted from these F3 individuals was used to map the locus governing NarCh accumulation by the analysis of recombinants.
At first, three SNPs, UN22458, 06224, and 10338, were selected for genotyping of the segregating population (SNP locations are marked with arrows in Fig. 4A; Supplemental Tables S5 and S6) to screen the segregating population. UN10338 showed the highest genotype to phenotype association (Fig. 4E). Fine mapping using recombinants, which were identified by these three markers, was carried out using nine selected SNPs dispersed in the genomic region restricted by UN10338 and UN06224 (Supplemental Fig. S6; Supplemental Table S6). A number of informative recombinants were identified, including two plants with white fruit (nos. 1,030 and 826), three pale yellow fruit plants (nos. 539, 1,041, and 1,281), and two plants with yellow fruits (nos. 1,442 and 720; Fig. 4G). Combined phenotypic and genotypic data of the plants indicated that the genetic factor regulating flavonoid accumulation in the muskmelon segregating population is located between high-resolution melt (HRM) markers numbers 2 and 4 (Fig. 4G). All of the recombinant F3 plants that were genotyped showed a complete cosegregation between their genotype at this chromosomal region and their fruit color phenotype. Seventeen protein coding genes, from MELO3C01178 to MELO3C01194 (melon gene names come from the melon genome project at https://melonomics.net/genome/; Supplemental Table S7), are located within this approximately 100-kb chromosomal region from position 2,156,094 to position 2,255,982 on scaffold 16 of chromosome 10.
Characterization of a Candidate Gene Regulating Flavonoid Accumulation
MELO3C011980 is 1 of the 17 genes that are located in the region restricted by HRM markers numbers 2 and 4. This gene is similar to F-box/Kelch repeat protein coding gene At1g23390 (Arabidopsis), which has not been functionally characterized (Supplemental Fig. S7). MELO3C011980 was the most significantly differentially expressed gene between the two bulks; whereas thousands of raw reads were generated in white bulks during all three analyzed developmental stages, only a few reads were detected in yellow bulks (Supplemental Fig. S8A). We named MELO3C011980 as CmKFB. The RNA-Seq data could not detect any sequence polymorphism between CmKFB yellow and white alleles because of the extremely low number of reads derived from the yellow allele. To identify the DNA sequence polymorphism that might affect CmKFB differential expression, sequencing of the upstream portion of the gene was performed. A deletion of 12 bp in the 5′ untranslated region (UTR) of CmKFB was found in TVT parental lines (Supplemental Fig. S8B). To indicate prevalence of this deletion, we screened a range of muskmelon accessions differing in peel color (Supplemental Table S8). This deletion was present in eight analyzed muskmelon accessions that do not accumulate NarCh and is absent in all analyzed muskmelon accessions that accumulate NarCh (Supplemental Fig. S8C; Supplemental Table S8).
As indicated earlier, DPBA staining of leaves exhibited similar yellow UV fluorescence patterns in both white and yellow F3 plants, suggesting that both types accumulate flavonoids in their leaves (Supplemental Fig. S2A). To further associate the role of CmKFB with flavonoid accumulation, we conducted quantitative reverse transcription (qRT)-PCR analysis of CmKFB in leaves. As expected, a very low expression of CmKFB was found in leaves irrelevant to fruit rind NarCh accumulation (Supplemental Fig. S8D).
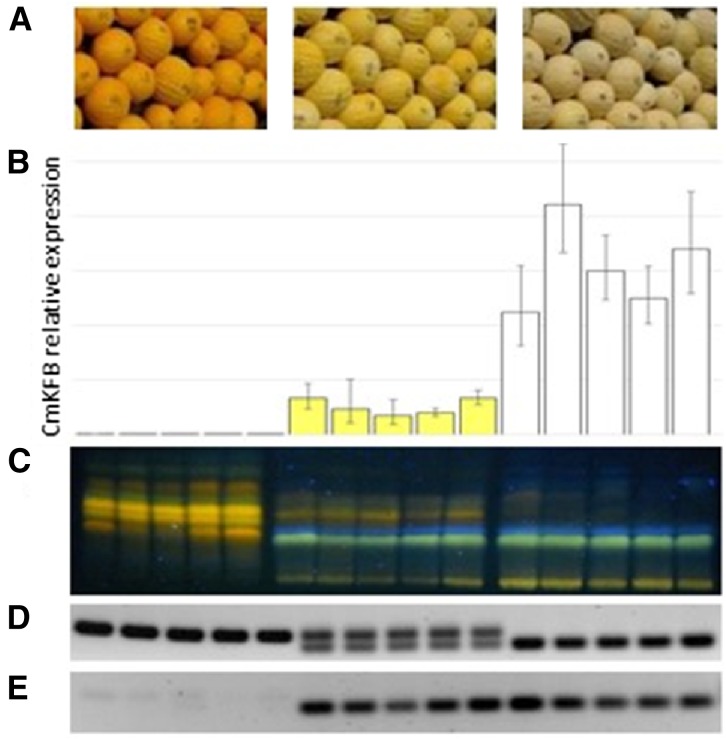
Further analysis of the effect of CmKFB expression on the flavonoid pathway was performed on individual F3 plants derived from the self-pollination of CmKFB heterozygous F2 plants. The fruit rinds of five F3 individuals from each of the visual phenotypic groups, white, pale yellow, and yellow, were analyzed for CmKFB expression by qRT-PCR. All white fruits showed strong expression of CmKFB, and all yellow fruits showed very weak expression, similar to its expression in the F3 bulks derived from the RNA-Seq analysis (Supplemental Fig. S8A). The pale yellow fruits showed intermediate expression of CmKFB gene (Fig. 5B), which is correlated with the intermediate color phenotype and intermediate fluorescence pattern of DPBA-stained fruit rind methanol extracts fractionated on TLC and visualized under UV light (Fig. 5C).
Figure 5.
Analysis of CmKFB gene and biochemical differences in representative segregating F3 plants. A, Three visual phenotype groups: yellow, light yellow, and white fruit. B, qRT-PCR of CmKFB performed on five individual F3 plants within each phenotypic group. C, Methanol extraction of each individual fractionated by TLC and visualized under UV light after DPBA staining. D, Agarose gel of PCR products of genomic DNA fragments containing the 5′ UTR deletion. E, Similar PCR analysis performed on cDNA.
PCR analysis of the CmKFB 5′ UTR deletion area was performed on genomic DNA of the analyzed F3 plants, and it verified that white plants are homozygotes to the TVT allele, that yellow plants are homozygotes to the NA allele, and that the pale yellow plants are heterozygotes (Fig. 5D). Interestingly, similar PCR analysis with complementary DNA (cDNA) suggests heterozygotes to accumulate only the TVT nonflavonoid-accumulating parent allele (Fig. 5E).
We used the RNA-Seq data to analyze the expression level (reads per kilobase of exon model per million mapped reads) of all gene family members of the genes coding for major enzymes of the flavonoid pathway, including PAL, cinnamate 4-hydroxylase, and chalcone synthase (CHS). Only one major ortholog in each family was found to be significantly up-regulated during development (MELO3C025786, MELO3C019585, and MELO3C014767, respectively; Supplemental Fig. S9) in correlation with NarCh accumulation pattern in NA (Supplemental Fig. S1) and phenylpropanoid accumulation in TVT as observed by the increased blue fluorescence (Supplemental Fig. S2), suggesting that only a single ortholog of these gene families participated in the fruit rind pathway in both muskmelon types. However, 4-Coumarate:CoA ligase (4CL) gene family members exhibited various expression patterns during fruit development, one of which matched the expression pattern of the previous three genes (MELO3C024724; Supplemental Fig. S9). No significant differences in transcript abundance were found between white and yellow that could explain the biochemical differences between these two bulks.
The AtKFBs homologs (KFB01, KFB20, KFB39, and KFB50), which were reported to regulate the phenylpropanoid pathway, have been shown to be involved in PAL degradation, resulting in down-regulation of the phenylpropanoid pathway (Zhang et al., 2013, 2015). The biochemical evidence presented in this study suggests CmKFB to regulate a downstream point to these Arabidopsis KFBs. To access this point, muskmelon KFB homologs (Supplemental Table S9) and the Arabidopsis KFBs (Zhang et al., 2015) were phylogenetically analyzed (Supplemental Fig. S10). This analysis shows the four AtKFBs to cluster with the muskmelon MELO3C014678 and MELO3C009596 homologs, whereas CmKFB clusters with the Arabidopsis At1g23390 in a subtree that is not tightly linked to the previous one.
Functional Analysis of CmKFB
To functionally validate the ability of CmKFB to divert the flavonoid pathway biochemical flux, two different transformation systems were applied: a virus-induced gene expression (VIGE) of CmKFB in muskmelon leaves and a stable transformation of tomato ‘MP-1’ plants with CmKFB under the constitutive 35S promoter.
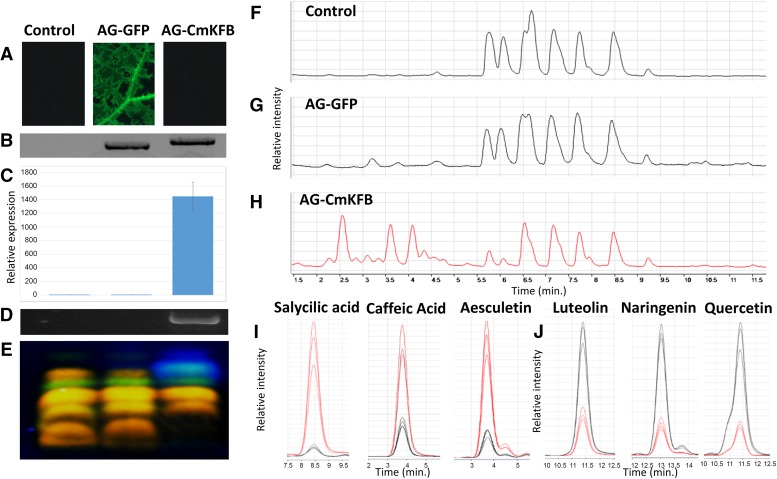
VIGE is an established experimental procedure to analyze the effect of gene overexpression in cucurbits (Harpaz-Saad et al., 2007). Our chemical analysis of muskmelon leaves suggested that flavonoid accumulation in leaves is irrelevant to flavonoid accumulation in muskmelon fruit rind (Supplemental Fig. S2A), which is associated with very low expression of CmKFB (Supplemental Fig. S8D); thus, overexpression of CmKFB in leaves seemed like a suitable experimental system to validate CmKFB function. Fourteen days after VIGE infection, CmKFB expression was significantly increased as indicated by qRT-PCR analysis (Fig. 6C). Leaf methanol extracts of VIGE CmKFB and the two controls (virus only and VIGE GFP) were fractionated with TLC, stained with DPBA, and observed under UV light. CmKFB overexpression exhibited enhanced blue fluorescence and reduction in yellow-green fluorescence accumulation compared with the controls (Fig. 6E). Further LC-MS analysis of leaf methanol extracts showed that VIGE CmKFB accumulated higher amounts of compounds (retention time, 2.2–4.5 min) but lower amounts of other compounds eluting from the column during the time range of 5.5 to 9.5 min compared with the two controls (Fig. 6, F–H). To identify the main aglycones differentiating between the controls and VIGE CmKFB, enzymatic hydrolysis was performed, and the hydrolyzed samples were analyzed by LC-MS and compared with authentic standards. Three aglycones were identified to be up-regulated by VIGE CmKFB: caffeic acid, salicylic acid, and aesculetin, and three were identified to be down-regulated: luteolin, naringenin, and quercetin (Fig. 6, I and J). These differences are less significant than those observed in fruits, most probably because of smaller difference in CmKFB expression and incomplete infection of the virus, which was observed in the GFP control (Fig. 6A). VIGE CmKFB down-regulated compounds were flavonoids, whereas the up-regulated compounds belong to side chains of the phenylpropanoid and coumarin pathways upstream of the flavonoid pathway, indicating that CmKFB expression in muskmelon leaves mediated a metabolic flux alternation as was observed in muskmelon fruits.
Figure 6.
Virus-induced CmKFB expression in muskmelon leaves. A, Control (uninfected and VIGE GFP) and VIGE CmKFB leaves observed through a GFP filter under UV light. B, PCR amplification of the inserted cDNA products. C, CmKFB expression analyzed by qRT-PCR. D, PCR amplification of CmKFB CDS cDNA. E, TLC fractionation of leaf methanol extracts observed under UV after DPBA staining. F to H, Base peak chromatograms of mass range mass-to-charge ratio 100 to 1,000 measured in LC-MS. F, Uninfected control leaf. G, VIGE GFP leaf. H, VIGE CmKF leaf. I and J, Quantitative comparison of differentiating compounds in leaf methanol extract after enzymatic hydrolysis. Compounds were identified by comparisons with authentic standards. I, Up-regulated in VIGE CmKFB (red) compared with controls (black). J, Down-regulated in VIGE CmKFB (red) compared with controls (black).
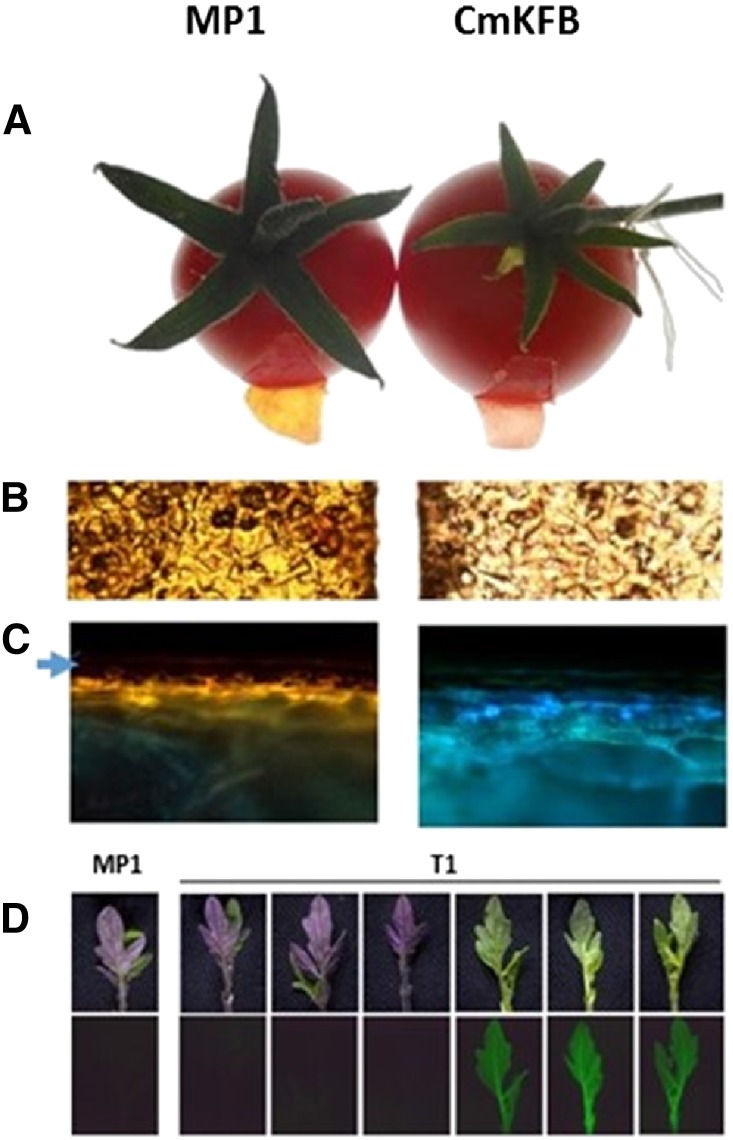
To functionally analyze CmKFB in a heterologous system, we used a stable transformation of tomato ‘MP-1’. Tomato plants accumulate anthocyanins in vegetative tissues in response to stress and accumulate NarCh, kaempferol, and quercetin in their fruit peels (Mintz-Oron et al., 2008). A binary vector harboring CmKFB and a GFP reporter gene was used for stable transformation. Two transgenic plants were isolated. These plants did not accumulate visible amounts of anthocyanins in any observed tissues: leaves, stems, or flowers (Supplemental Fig. S11). At the ripe fruit stage, fruit peels of both transformed T0 plants accumulated significantly reduced NarCh in the cuticle compared with the cv MP-1 control (Fig. 7, A and B). In addition, fruit sections under UV light after DPBA staining exhibited a shift from yellow to blue fluorescence in the CmKFB transgenic fruit (Fig. 7C), similar to VIGE CmKFB leaves (Fig. 6E) and muskmelon fruit rinds (Fig. 1I). T1 seeds from both transgenic plants were germinated and grown under cold stress conditions. After 15 d, a clear difference could be observed between anthocyanin-accumulating seedlings and seedlings that did not accumulate anthocyanins. The T1 seedlings with anthocyanins did not exhibit GFP fluorescence, whereas those that did not accumulate anthocyanins exhibited GFP fluorescence (Fig. 7D), indicating overexpression of CmKFB in tomato to repress flavonoid accumulation in a similar manner as observed in muskmelon.
Figure 7.
Overexpression of CmKFB under the control of the Cauliflower mosaic virus 35S promoter in stable transformed tomato plants. A, Visual comparison of control and T0 fruit peels. B, Microscopic view of control and T0 cuticles. C, Microscopic view of control and T0 fruit cut under UV light after DPBA staining (arrow marks the cuticle upper layer that did not emit any fluorescence). D, Leaves of T1 segregants of the first transgenic plant and ‘MP-1’ control grown under cold stress conditions: upper indicates visible light, and lower indicates GFP fluorescence.
DISCUSSION
BSA of RNA-Seq and Genetic Mapping
BSA is a method developed to identify genetic loci that are linked to a specific trait (Michelmore et al., 1991). BSA has been widely used ever since, uncovering genetic markers and genes in a wide variety of plants. The rapid development of new sequencing technologies and computation systems allows a very efficient use of BSA while using next generation sequencing technologies to discover polymorphism that discriminate between the bulks (Liu et al., 2012; Trick et al., 2012). Combining BSA with RNA-Seq and detailed metabolite analyses enabled a relatively rapid and accurate isolation and characterization of CmKFB, the gene of interest in this work.
A high level of recombination occurred in the 2-Mb region in the F3 segregating population (Fig. 4, d–f), Such a recombination rate in this genomic region was a significant factor enabling high-resolution mapping, which resulted in less than 200 SNPs that differentiate between the bulks, most of them localized in a 0.5-Mb genomic region in scaffold 16 of chromosome 10 (Fig. 4A), while using a relatively small number of participants in each bulk (12 F3 families in the yellow bulk and only 7 in the white). Recombinant analysis of the segregating population enabled further fine mapping of the genetic loci associated with NarCh accumulation to a genomic region of 100 kb, which consisted of 17 protein coding genes. CmKFB was 1 of these 17 coding genes and showed the most significant difference in expression between the yellow and the white F3 bulks, suggesting CmKFB as a primary candidate gene.
RNA-Seq analysis of yellow (accumulating NarCh) and white (do not accumulate NarCh) developing fruit of F3 segregant pools indicated that CmKFB is expressed at a very low level in fruits that accumulate NarCh, whereas it is highly expressed in NarCh-nonaccumulating fruits. Because of the low abundance of CmKFB RNA-Seq reads obtained in the yellow bulks, we could not define sequence variation between the yellow and white alleles of this gene. DNA sequence analysis identified a 12-bp deletion in the 5′ UTR of the white bulk and TVT. The 5′ UTR 12-bp deletion was consistently associated with flavonoid accumulation in our segregating population (Fig. 5D) and different muskmelon accessions (Supplemental Fig. S8C). Interestingly, monitoring allelic expression on cDNA (Fig. 5E) showed heterozygous F3 to accumulate only the TVT nonflavonoid-accumulating parent allele, suggesting CmKBF allelic expression to be affected by allelic variation. Motif analysis using JASPAR (Mathelier et al., 2014) suggested this deletion to disrupt a putative binding site of High Mobility Group box1 protein, which is a chromatin remodeling factor (Grosschedl et al., 1994; Webster et al., 1997; Ikeda et al., 2011), suggesting differential epigenetic allelic states to result in differential allelic CmKFB expression. Nevertheless, the association of the 12-bp deletion with CmKFB expression, the tissue specificity of this expression, and the codominant nature of CmKFB expression need further studies.
Metabolites Analysis and CmKFB Function
There is growing interest in manipulating the metabolic flux through different phenylpropanoid pathway branches for a number of reasons (for example, their association with plant tolerance to different biotic and abiotic stresses, commercial and consumer health impact of increased bioactive compounds in agricultural products, and alteration of the lignin biosynthesis pathway; Liu et al., 2014). An example of such metabolic flux alternation is the silencing of Arabidopsis hydroxycinnamoyl CoA:shikimate hydroxycinnamoyl transferase that led to repression of lignin biosynthesis and redirection of the metabolic flux toward the flavonoid pathway (Besseau et al., 2007).
Various transcription factors were shown to divert the phenylpropanoid metabolic flux. Zea mays R2R3-MYB transcription factor (ZmMYB31), a direct repressor of caffeic acid 3-O-methyltransferase and Arabidopsis ferulate-5-hydroxylase genes, overexpression in Arabidopsis results in reduction in sinaopylmalate and lignin accumulation while enhancing accumulation of ferulic acid, coumaric acid, and anthocyanins (Fornalé et al., 2010). In addition, the colorless fruit peel y mutant in tomato was found to be a mutation causing down-regulation of SlMYB12, a transcription factor that acts as a positive regulator of an array of phenylpropanoid structural genes. Interestingly, this mutation exhibits elevated accumulation of caffeic and ferullic compounds accompanied by reduction in flavonoid content (Adato et al., 2009), similar to the flux alteration observed in this work.
Biochemical analysis of muskmelon fruit rinds showed that NarCh accumulation is accompanied by the accumulation of downstream flavonoids as observed by DPBA staining (Fig. 1), Pew’s and Shinoda’s tests (Supplemental Fig. S3), and LC-MS analysis (Fig. 2). This accumulation is associated with significant decline in CmKFB transcription in a dosage-dependent manner (Fig. 5) and reduction in caffeic acid, ferulic acid, and aesculetin accumulation, which are branched from the phenylpropanoids, prior to flavonoid pathway (Fig. 3). No significant differences in the expression pattern of structural genes of the flavonoid pathway were found based on our RNA-Seq data analysis (Supplemental Fig. S9), which could explain the biochemical differences between yellow and white. The flux alternation showed here can be explained by a negative posttranslational regulation of CmKFB at the entry point of the flavonoid pathway. This biochemical flux alternation was observed both when CmKFB was transiently expressed in muskmelon leaves, resulting in elevation of salicylic acid, caffeic acid, and aesculetin accumulation accompanied by reduction in naringenin, luteolin, and quercetin accumulation (Fig. 6), and by stable expression in tomato, resulting in elevated blue fluorescence accompanied by reduction in NarCh accumulation in fruits and anthocyanins in leaves and flowers (Fig. 7; Supplemental Fig. S11). The functionality of CmKFB in the tomato system indicates the possible general role of this gene in plants. CmKFB is most probably involved in a proteasome-dependent protein degradation mechanism mediated by the SCF E3 ubiquitin ligase complex, which has been previously reported in Arabidopsis (Zhang et al., 2013, 2015). In these studies, four closely related KFB orthologs (AtKFB01, AtKFB20, AtKFB39, and AtKFB50) were found to target different PAL isozymes, which serve as the entry point to the phenylpropanoid pathway, to proteasome-dependent degradation, resulting in reduced accumulation of a wide array of compounds: flavonols, anthocyanins, condensed tannis, sinapic acid esters, and lignin. In this study, PAL does not seem to be a suitable candidate as a target of CmKFB. The expression pattern of the single PAL ortholog (MELO3C025786; Supplemental Fig. S9), which matches both the flavonoid and phenylpropanoid accumulation patterns during development (Supplemental Figs. S1 and S2), suggests that this gene is functionally active in both muskmelon types. Phylogenetic analysis of muskmelon and Arabidopsis KFB proteins shows the four characterized AtKFBs and CmKFB to cluster in two distinct phylogenetic clades (Supplemental Fig. S10), suggesting the presence of two distinctive functional groups, which are conserved between the Brassicales spp. and the Cucurbitales spp. orders, further suggesting different substrate specificity. Observing the biochemical branch point between the two muskmelon types, a natural candidate as a target of CmKFB is 4CL. This enzyme is encoded in all plants by multiple genes, which differ in substrate specificity of the paralogs, and it is considered one of the main regulatory points in directing the phenylpropanoid metabolic pathway (Ehlting et al., 1999; Hamberger and Hahlbrock, 2004). This option is further supported by significant expression of multiple paralogs of 4CL in muskmelon fruit (Supplemental Fig. S9). However, the degradation of MELO3C014767, which is the only significant ortholog of CHS expressed in muskmelon fruit rind, might also result in the biochemical differences that were observed. This work suggests that CmKFB regulates either 4CL or CHS enzymatic function by a protein degradation mechanism either directly by targeting one of these enzymes to proteolytic degradation or by modulating an additional factor, which regulates one of these enzymes function.
In summary, we have identified CmKFB, which negatively regulates flavonoid accumulation in muskmelon fruit by shifting the metabolic flux toward general phenylpropanoid products, including caffeic and ferulic acids and coumarin (aesculetin). Characterization of this natural variation in muskmelon might prove useful for plant breeders in tailoring new cultivars.
MATERIALS AND METHODS
Plant Material
A set of 18 muskmelon (Cucumis melo) cultivars was selected to represent rind color variation in muskmelon. They included the parental lines of the analyzed segregating population and representatives of five major taxonomic groups of muskmelon, which accumulate or lack NarCh, with both climacteric and nonclimacteric modes of fruit ripening (Supplemental Table S8). Plants were grown under standard conditions in the field and greenhouse at the Newe Ya‘ar Research Center.
F3 Population Establishment
A population segregating for fruit NarCh accumulation was established by crossing two casaba muskmelons, NA (an NarCh-accumulating muskmelon inbred line with a canary yellow fruit rind) and TVT (an NarCh-lacking inbred line with a green fruit rind). Two hundred thirteen F2 plants derived from this cross visually segregated to four fruit rind color phenotypes (green-yellow, green, yellow, and white), fitting the expected 9:3:3:1 (green-yellow:green:yellow:white) Mendelian ratio for two independent genes, green and yellow. All F2 fruit rinds were analyzed by HPLC for NarCh content (Tadmor et al., 2010). To define F3 families that are homozygous to either NarCh fruit accumulation or nonaccumulation, 50 F3 families were selected: 30 with yellow fruit rind F2 progenitors and 20 with white fruit rind F2 progenitors. Thirty plants from each family were grown in the winter of 2010 at Ein Tamar in the northern Arava Valley in Israel. Of these 50 families, 12 accumulating NarCh were selected phenotypically, the fruits of all plants in each family having yellow rinds. In addition, seven F3 families nonaccumulating NarCh were selected phenotypically for the fruits of all plants in each family having white rinds. Thirty plants of each of these 19 families were grown in an open field of Newe Ya‘ar Research Center in the spring of 2010. Female flowers were tagged on the day of anthesis, and three biological replicates of fruit rind from each F3 family were sampled at three fruit developmental stages: 10 DAA, 20 DAA, and ripe.
RNA-Seq, Data Processing, and De Novo Assembly
Fruit rind RNA extraction for RNA-Seq analysis was performed according to the work by Portnoy et al. (2011). Twelve samples (two replicates of three developmental stages: 10 DAA, 20 DAA, and mature fruit rind of yellow and white F3 families) were used to construct RNA-Seq libraries following the standard Illumina protocol. The libraries were sequenced on the Illumina HiSeq2000 System with the paired-end mode. The sequencing was performed at the Biotechnology Center at the University of Illinois at Urbana-Champaign. The raw Illumina RNA-Seq reads were first processed to remove low-quality regions and adaptor sequences using an in-house perl script. The RNA-Seq reads were further aligned to the ribosomal RNA database (Quast et al., 2013) using Bowtie (Langmead et al., 2009) and allowing two mismatches, and the mappable reads were discarded. The resulting high-quality cleaned reads were assembled de novo into contigs using Trinity with min_kmer_cov set to two (Grabherr et al., 2011). To remove the redundancy of Trinity-generated contigs, they were further assembled de novo using iAssembler with minimum percentage identify (−p) set to 99 (Zheng et al., 2011).
Gene Expression Quantification and Differential Expression Analysis
The high-quality cleaned RNA-Seq reads were aligned to the assembled muskmelon transcripts with the Bowtie program (Langmead et al., 2009) allowing one mismatch. After alignments, raw counts for each muskmelon transcript and each sample were derived and normalized to reads per kilobase of exon model per million mapped reads. Differentially expressed genes between white and yellow fruits at the same stages were identified with the DESeq Package (Anders and Huber, 2010). Raw P values of multiple tests were corrected using false discovery rate (Benjamini and Hochberg, 1995).
SNP and Small-Indel Identification from the RNA-Seq Data
To identify SNPs and small indels between yellow and white genotypes, identical RNA-Seq reads from each library were first collapsed into a single sequence. The resulting unique reads were then aligned to the assembled muskmelon transcripts using BWA (Li and Durbin, 2009) and allowing one mismatch, one gap opening, and one gap extension. After mapping, SNPs and small indels were identified based on the mpileup files generated by SAMtools (Li et al., 2009). The identified SNPs and small indels were supported by at least five independent reads and had an allele frequency of at least 0.8.
Phylogenetic Analysis
Muskmelon KFB protein sequences were retrieved through BLAST searches on the muskmelon genome Web site (https://melonomics.net/), and conserved domains were verified against the IterPro Protein Database. Arabidopsis (Arabidopsis thaliana) KBF homologs were adopted from Zhang et al., 2015. Protein sequences were aligned by the ClustalW Program. Phylogenetic analysis was performed using the MEGA Package, version 6 (Tamura et al., 2013) using the neighbor-joining method, with the pairwise deletion option.
Fluorescent Probes
UN22458 SNP (SNP no. 86; Supplemental Table S5) genotyping was performed in an ABI PRISM 7000 Sequence Detection System (Applied Biosystems). Each reaction contained 0.25 µL of assay mix (SNP Genotyping Mix; Biosearch Technologies), 5 µL of reaction mix (Genotyping ToughMix, ROX; Quanta Biosysyems), 1 µL of DNA, and 3.75 µL of water. UN06224 and UN10338 SNPs (SNPs nos. 39 and 54; Supplemental Table S5) genotyping was performed in a StepOnePlus Real-Time PCR System (Applied Biosystems), and each reaction contained 4 µL of mix (TaqMan Genotyping Master Mix; Applied Biosystems), 0.4 µL of assay mix (TaqMan SNP Genotyping Assay; Applied Biosystems), 0.8 µL of DNA, and 2.8 µL of water. Primers are listed in Supplemental Table S6.
HRM Analysis
DNA samples were diluted into equal concentrations of 100 ng µL−1 using NanoDrop. Samples were amplified on an Eco Real-Time PCR System (Illumina). Each sample contained 1 µL of DNA, 0.2 µL of 10 mm each primer, 5 µL of reaction mix (FastSYBR Green Master Mix; Applied Biosystems), and 3.6 µL of distilled, deionized water programmed as specified by the enzyme manufacturer and analyzed with the Eco, version 4 software. Primers for each HRM SNP marker are specified in Supplemental Table S6.
VIGE
Zucchini Yellow Mosaic Virus-AGII is a potyvirus-based vector system used to transiently overexpress genes in muskmelon (Arazi et al., 2001). To overexpress CmKFB, a GFP AGII vector was digested with PstI and SalI enzymes to remove the GFP fragment. CmKFB coding sequence (CDS) was amplified by PCR from TVT muskmelon cDNA with primers containing linkers to the corresponding PstI and SalI restriction sites (Supplemental Table S6) and ligated to the digested vector (T4 DNA Ligase; Thermo Scientific). Particle bombardment inoculation of AG-GFP and AG-CmKFB cDNA was performed using a handgun device (Gal-On et al., 1997) on cotyledons of 14-d-old NA muskmelon seedlings. After inoculation, seedlings were placed in a growth chamber with a 16-h photoperiod at 25°C for 14 d.
Solanum lycopersicum Stable Transformation
CmKFB was amplified from TVT muskmelon cDNA in two PCR steps, isolating the CDS with additional 39-bp linkers on each side of the gene and generating truncated AttL sites for direct recombination to pK7WG2D Gateway Binary Destination Vector (Invitrogen) by LR clonase (Fu et al., 2008; Ischebeck et al., 2011; Supplemental Table S6). Cotyledons of the ‘MP-1’ tomato (Solanum lycopersicum) were transformed with Agrobacterium tumefaciens GV3101 that harbored the vector. After 48 h of cocultivation in the dark (3% [w/v] Glc, 0.1 mg L−1 indole-3-acetic acid, 1 mg L−1 zeatin, and 100 µm acetosyringone), explants were transferred to a shoot-induction medium (3% [w/v] Glc, 0.1 mg L−1 indole-3-acetic acid, 1 mg L−1 zeatin, 100 mg L−1 kanamycin, and 500 mg L−1 claforan) under light. Regenerated shoots were moved to a shoot-elongation medium (3% [w/v] Glc, 0.1 mg L−1 zeatin, 100 mg L−1 kanamycin, and 500 mg L−1 claforan). Elongated shoots were transferred to a rooting medium (one-half-strength Murashige and Skoog medium, 2% [w/v] Suc, and 2 mg L−1 indole-butyric acid).
qRT-PCR
Reactions were performed on an Eco RT-PCR System (Illumina). Each sample contained 1 µL of cDNA, 0.2 µL of each primer (10 mm), 5 µL of reaction mix (FastSYBR Green Master Mix; Applied Biosystems), and 3.6 µL of distilled, deionized water programmed as specified by manufacturer and analyzed with Eco, version 4 software.
Flavonoid Extraction and Staining Methods
Fresh fruit rind or leaf samples were collected, immediately frozen in liquid nitrogen, and stored at −80°C. Before extraction, samples were ground to a fine powder with a mortar and pestle in the presence of liquid nitrogen. Two grams of ground tissue was extracted in 5 mL of methanol (MeOH), vortexed for 10 s, incubated for 10 min at 60°C, and vortexed again. Samples were then centrifuged at 10,000g for 5 min, and the upper methanol phase was collected. For DPBA staining, 1 µL of methanolic extract was applied on an HPTLC (Silica Gel 60; Merck) and developed with ethyl acetate:acetic acid:water at a ratio of 67:17.1:17.1 (v/v). After separation, the plate was dried with a hairdryer, sprayed with DPBA (Sigma) dissolved in MeOH (1% [w/v]), dried again, and visualized under a UV light (320 nm).
Additional flavonoid staining protocols performed on the methanol extracts included the zinc-HCl test (Pew’s test), for which zinc powder and a drop of 5 n HCl were added to the plant flavonoid extract, and the magnesium-HCl test (Shinoda’s test), which was applied in the same way as the zinc-HCl test, except that magnesium wire was used instead of zinc powder.
LC-MS
The method used was modified from De Vos et al., 2007. Methanol extraction was carried out as described above. Enzymatic hydrolysis was conducted using 1 mL of evaporated methanol extracts. Samples were speed vacuum evaporated, supplemented with an enzymatic solution containing 1 mg mL−1 cellulase, 1 mg mL−1 pectinase, and 1 mg mL−1 β-glucosidase (Sigma) dissolved in 0.1 m NaAc (pH 5.3) solution, and incubated at 37°C overnight. Aglycones were extracted with 500 mL of ethyl acetate. The upper ethyl acetate phase was removed and evaporated, and the pellet was redissolved in 300 µL of MeOH. Both samples, before and after hydrolysis, were supplemented with distilled, deionized water and formic acid (final concentrations of 25% and 0.1% [v/v], respectively) and filtered through a PFE 0.2-µm Filter (Pall Life Sciences).
Mass spectral analysis was conducted using an LC-MS-time-of-flight (TOF; LC, 1290 Infinity System; Agilent Technologies; MS-TOF, 6224-TOF-LC-MS; Agilent Technologies) in electrospray ionization negative mode. Separation of compounds was performed using Zorbax Extended C18 RRHT 2.1 × 50 mm and 1.8-µm column. The mobile phase consisted of 0.1% (v/v) formic acid in water (phase A) and 0.1% (v/v) formic acid in acetonitrile (phase B). A gradient program was used as follows: 95% to 65% (v/v) A over 15 min, 65% to 25% (v/v) A over 40 s, 25% A held over 2 min, 25% to 95% (v/v) A over 1.5 min, and 95% (v/v) A held over 2 min at constant flow rate of 0.19 mL min−1. Analysis of the masses was performed with the MassHunter Qualitative Analysis software (version B.05.00; Agilent Technologies). Determination of differential mass expression between samples was performed using the Mass Profiler Professional software (Agilent Technologies).
Microscopy
DPBA-stained tissue of handmade slices was observed with an Olympus BX61 Microscope coupled with a U-HGLGPS Illumination System and Olympus 20×/0.50 Objective using a U-MNBV2 Filter (excitation = 420–440 nm; emission > 475 nm). Images were collected with a digital camera (DP73; Olympus) and processed with cellSens Dimension software (Olympus).
Tomato seedling fluorescence was observed with a Leica M205FA Stereomicroscope and a Leica 10472649 Planapo 0.63× Objective under a GFP3 Filter. Images were collected with a digital camera (DFC495; Leica) and processed with the Leica Application Suit V3.8 software.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number SRP060002.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. F3 population establishment.
Supplemental Figure S2. Visualization of DPBA stained TLC under UV light of methanol extracts.
Supplemental Figure S3. Additional staining methods.
Supplemental Figure S4. LC-MS analysis of fruit rind methanol extracts.
Supplemental Figure S5. Fruit rind compound identification.
Supplemental Figure S6. The melting curves of HRM markers.
Supplemental Figure S7. CmKFB and At1g23390 protein sequence alignment.
Supplemental Figure S8. Molecular analysis of the candidate gene CmKFB.
Supplemental Figure S9. Digital expression pattern of the gene families leading to the flavonoid pathway.
Supplemental Figure S10. Phylogenetic analysis of KFB proteins in muskmelon and Arabidopsis.
Supplemental Figure S11. Overexpression of CmKFB in stable transgenic T0 tomato plants.
Supplemental Table S1. List of differentiating compounds between the two muskmelon groups.
Supplemental Table S2. List of differentiating compounds after hydrolysis between the two muskmelon groups.
Supplemental Table S3. List of fruit rind compounds identified by comparison with authentic standards.
Supplemental Table S4. RNA-Seq statistics.
Supplemental Table S5. List of 179 SNPs differentiating between yellow and white bulks.
Supplemental Table S6. List of primers and probes.
Supplemental Table S7. List of genes expressed in the physical region governing flavonoid accumulation.
Supplemental Table S8. List of muskmelon varieties included in this study.
Supplemental Table S9. List of F-box/Kelch repeat protein coding genes in muskmelon.
Supplementary Material
Acknowledgments
We thank Uzi Sa‘ar and Fabian Baumkoler for expert maintenance of the plants in the field and the greenhouse, Dalia Wolf for preparation of the transgenic tomato plants, and Harry Paris for editing the article. Publication no. 105/2015 of the Agricultural Research Organization, Bet Dagan, Israel.
Glossary
- BSA
bulk segregant analysis
- cDNA
complementary DNA
- CDS
CmKFB coding sequence
- DAA
days after anthesis
- DPBA
diphenyl boric acid aminoethyl ester
- HRM
high-resolution melt
- LC
liquid chromatography
- MS
mass spectrometry
- NA
Noy Amid
- NarCh
naringenin chalcone
- qRT
quantitative reverse transcription
- RNA-Seq
RNA sequencing
- SNP
single-nucleotide polymorphism
- TLC
thin-layer chromatography
- TVT
Tendral Verde Tardio
- UTR
untranslated region
- VIGE
virus-induced gene expression
Footnotes
This work was supported by the Center for the Improvement of Cucurbit Fruit Quality, Agricultural Research Organization (ARO).
Articles can be viewed without a subscription.
References
- Adato A, Mandel T, Mintz-Oron S, Venger I, Levy D, Yativ M, Domínguez E, Wang Z, De Vos RCH, Jetter R, et al. (2009) Fruit-surface flavonoid accumulation in tomato is controlled by a SlMYB12-regulated transcriptional network. PLoS Genet 5: e1000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arazi T, Shiboleth YM, Gal-On A (2001) A nonviral peptide can replace the entire N terminus of zucchini yellow mosaic potyvirus coat protein and permits viral systemic infection. J Virol 75: 6329–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Met 57: 289–300 [Google Scholar]
- Besseau S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M (2007) Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19: 148–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger Y, Paris HS, Cohen R, Katzir N, Tadmor Y, Lewinsohn E, Schaffer AA (2010) Genetic diversity of Cucumis melo. Hortic Rev 36: 165–198 [Google Scholar]
- Butelli E, Titta L, Giorgio M, Mock HP, Matros A, Peterek S, Schijlen EGWM, Hall RD, Bovy AG, Luo J, et al. (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26: 1301–1308 [DOI] [PubMed] [Google Scholar]
- Cohen S, Itkin M, Yeselson Y, Tzuri G, Portnoy V, Harel-Baja R, Lev S, Sa’ar U, Davidovitz-Rikanati R, Baranes N, et al. (2014) The PH gene determines fruit acidity and contributes to the evolution of sweet melons. Nat Commun 5: 4026. [DOI] [PubMed] [Google Scholar]
- Deguchi A, Ohno S, Hosokawa M, Tatsuzawa F, Doi M (2013) Endogenous post-transcriptional gene silencing of flavone synthase resulting in high accumulation of anthocyanins in black dahlia cultivars. Planta 237: 1325–1335 [DOI] [PubMed] [Google Scholar]
- De Vos RCH, Moco S, Lommen A, Keurentjes JJB, Bino RJ, Hall RD (2007) Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat Protoc 2: 778–791 [DOI] [PubMed] [Google Scholar]
- Ehlting J, Büttner D, Wang Q, Douglas CJ, Somssich IE, Kombrink E (1999) Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J 19: 9–20 [DOI] [PubMed] [Google Scholar]
- Fornalé S, Shi X, Chai C, Encina A, Irar S, Capellades M, Fuguet E, Torres JL, Rovira P, Puigdomènech P, et al. (2010) ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J 64: 633–644 [DOI] [PubMed] [Google Scholar]
- Fu C, Wehr DR, Edwards J, Hauge B (2008) Rapid one-step recombinational cloning. Nucleic Acids Res 36: e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99: 11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-On A, Meiri E, Elman C, Gray DJ, Gaba V (1997) Simple hand-held devices for the efficient infection of plants with viral-encoding constructs by particle bombardment. J Virol Methods 64: 103–110 [DOI] [PubMed] [Google Scholar]
- Garcia-Mas J, Benjak A, Sanseverino W, Bourgeois M, Mir G, González VM, Hénaff E, Câmara F, Cozzuto L, Lowy E, et al. (2012) The genome of melon (Cucumis melo L.). Proc Natl Acad Sci USA 109: 11872–11877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R, Giese K, Pagel J (1994) HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet 10: 94–100 [DOI] [PubMed] [Google Scholar]
- Hamberger B, Hahlbrock K (2004) The 4-coumarate:CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc Natl Acad Sci USA 101: 2209–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haribal M, Renwick JAA (1996) Oviposition stimulants for the monarch butterfly: flavonol glycosides from Asclepias curassavica. Phytochemistry 41: 139–144 [DOI] [PubMed] [Google Scholar]
- Harpaz-Saad S, Azoulay T, Arazi T, Ben-Yaakov E, Mett A, Shiboleth YM, Hörtensteiner S, Gidoni D, Gal-On A, Goldschmidt EE, et al. (2007) Chlorophyllase is a rate-limiting enzyme in chlorophyll catabolism and is posttranslationally regulated. Plant Cell 19: 1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B (2005) Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol Biol 57: 155–171 [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Kinoshita Y, Susaki D, Ikeda Y, Iwano M, Takayama S, Higashiyama T, Kakutani T, Kinoshita T (2011) HMG domain containing SSRP1 is required for DNA demethylation and genomic imprinting in Arabidopsis. Dev Cell 21: 589–596 [DOI] [PubMed] [Google Scholar]
- Imperato F. (1980) Five plants of the family Cucurbitaeeae with flavonoid patterns of pollens different from those of corresponding stigmas. Experientia 36: 1136–1137 [Google Scholar]
- Ischebeck T, Stenzel I, Hempel F, Jin X, Mosblech A, Heilmann I (2011) Phosphatidylinositol-4,5-bisphosphate influences Nt-Rac5-mediated cell expansion in pollen tubes of Nicotiana tabacum. Plant J 65: 453–468 [DOI] [PubMed] [Google Scholar]
- Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW (2004) Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev 18: 2573–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers W, Rep M (2009) Lessons from fungal F-box proteins. Eukaryot Cell 8: 677–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kang KA, Zhang R, Piao MJ, Ko DO, Wang ZH, Chae SW, Kang SS, Lee KH, Kang HK, et al. (2008) Protective effect of esculetin against oxidative stress-induced cell damage via scavenging reactive oxygen species. Acta Pharmacol Sin 29: 1319–1326 [DOI] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10: 236–242 [DOI] [PubMed] [Google Scholar]
- Krauze-Baranowska M, Cisowski W (2001) Flavonoids from some species of the genus Cucumis. Biochem Syst Ecol 29: 321–324 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CJ, Cai Y, Zhang X, Gou M, Yang H (2014) Tailoring lignin biosynthesis for efficient and sustainable biofuel production. Plant Biotechnol J 12: 1154–1162 [DOI] [PubMed] [Google Scholar]
- Liu S, Yeh CT, Tang HM, Nettleton D, Schnable PS (2012) Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLoS One 7: e36406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, Buchman S, Chen CY, Chou A, Ienasescu H, et al. (2014) JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res 42: D142–D147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88: 9828–9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz-Oron S, Mandel T, Rogachev I, Feldberg L, Lotan O, Yativ M, Wang Z, Jetter R, Venger I, Adato A, et al. (2008) Gene expression and metabolism in tomato fruit surface tissues. Plant Physiol 147: 823–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Y, Nagel C, Taylor LP (1992) Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc Natl Acad Sci USA 89: 7213–7217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou CY, Pi H, Chien CT (2003) Control of protein degradation by E3 ubiquitin ligases in Drosophila eye development. Trends Genet 19: 382–389 [DOI] [PubMed] [Google Scholar]
- Pairoba CF, Walbot V (2003) Post-transcriptional regulation of expression of the Bronze2 gene of Zea mays L. Plant Mol Biol 53: 75–86 [DOI] [PubMed] [Google Scholar]
- Petersen M, Hans J, Matern U (2010) Biosynthesis of phenylpropanoids and related compounds. Annu Plant Rev 40: 182–257 [Google Scholar]
- Pew JC. (1948) A flavonone from Douglas-fir heartwood. J Am Chem Soc 70: 3031–3034 [DOI] [PubMed] [Google Scholar]
- Pitrat M, Hanelt P, Hammer K (2000) Some comments on infraspecific classification of cultivars of melon. Acta Hortic 510: 29–36 [Google Scholar]
- Pizzi A, Cameron FA (1986) Flavonoid tannins - structural wood components for drought-resistance mechanisms of plants. Wood Sci Technol 20: 119–124 [Google Scholar]
- Portnoy V, Diber A, Pollock S, Karchi H, Lev S, Tzuri G, Harel-Beja R, Forer R, Portnoy VH, Lewinsohn E, et al. (2011) Use of non-normalized, non-amplified cDNA for 454-based RNA-seq of fleshy melon fruit. Plant Genome 4: 36–46 [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano B, Pagano E, Montanaro V, Fortunato AL, Milic N, Borrelli F (2013) Novel insights into the pharmacology of flavonoids. Phytother Res 27: 1588–1596 [DOI] [PubMed] [Google Scholar]
- Schumann N, Navarro-Quezada A, Ullrich K, Kuhl C, Quint M (2011) Molecular evolution and selection patterns of plant F-box proteins with C-terminal kelch repeats. Plant Physiol 155: 835–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao T, Qian Q, Tang D, Chen J, Li M, Cheng Z, Luo Q (2012) A novel gene IBF1 is required for the inhibition of brown pigment deposition in rice hull furrows. Theor Appl Genet 125: 381–390 [DOI] [PubMed] [Google Scholar]
- Stracke R, De Vos RCH, Bartelniewoehner L, Ishihara H, Sagasser M, Martens S, Weisshaar B (2009) Metabolomic and genetic analyses of flavonol synthesis in Arabidopsis thaliana support the in vivo involvement of leucoanthocyanidin dioxygenase. Planta 229: 427–445 [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50: 660–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadmor Y, Burger J, Yaakov I, Feder A, Libhaber SE, Portnoy V, Meir A, Tzuri G, Sa’ar U, Rogachev I, et al. (2010) Genetics of flavonoid, carotenoid, and chlorophyll pigments in melon fruit rinds. J Agric Food Chem 58: 10722–10728 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Uritani I (1977) Synthesis and turnover of phenylalanine ammonia-lyase in root tissue of sweet potatoe injured by cutting. Eur J Biochem 73: 255–260 [DOI] [PubMed] [Google Scholar]
- Trick M, Adamski NM, Mugford SG, Jiang CC, Febrer M, Uauy C (2012) Combining SNP discovery from next-generation sequencing data with bulked segregant analysis (BSA) to fine-map genes in polyploid wheat. BMC Plant Biol 12: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitch NC, Grayer RJ (2008) Flavonoids and their glycosides, including anthocyanins. Nat Prod Rep 25: 555–611 [DOI] [PubMed] [Google Scholar]
- Wasson AP, Pellerone FI, Mathesius U (2006) Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell 18: 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster CI, Packman LC, Pwee KH, Gray JC (1997) High mobility group proteins HMG-1 and HMG-I/Y bind to a positive regulatory region of the pea plastocyanin gene promoter. Plant J 11: 703–715 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Wu C, Liu Y, Yang J, Huang L (2013) The Scutellaria baicalensis R2R3-MYB transcription factors modulates flavonoid biosynthesis by regulating GA metabolism in transgenic tobacco plants. PLoS One 8: e77275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gou M, Guo C, Yang H, Liu CJ (2015) Down-regulation of Kelch domain-containing F-box protein in Arabidopsis enhances the production of (poly)phenols and tolerance to ultraviolet radiation. Plant Physiol 167: 337–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gou M, Liu CJ (2013) Arabidopsis Kelch repeat F-box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell 25: 4994–5010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Zhao L, Gao J, Fei Z (2011) iAssembler: a package for de novo assembly of Roche-454/Sanger transcriptome sequences. BMC Bioinformatics 12: 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.