Abstract
Despite advances in DNA methylome analyses of cells and tissues, current techniques for genome-scale profiling of DNA methylation in circulating cell-free DNA (ccfDNA) remain limited. Here we describe a methylated CpG tandems amplification and sequencing (MCTA-Seq) method that can detect thousands of hypermethylated CpG islands simultaneously in ccfDNA. This highly sensitive technique can work with genomic DNA as little as 7.5 pg, which is equivalent to 2.5 copies of the haploid genome. We have analyzed a cohort of tissue and plasma samples (n = 151) of hepatocellular carcinoma (HCC) patients and control subjects, identifying dozens of high-performance markers in blood for detecting small HCC (≤ 3 cm). Among these markers, 4 (RGS10, ST8SIA6, RUNX2 and VIM) are mostly specific for cancer detection, while the other 15, classified as a novel set, are already hypermethylated in the normal liver tissues. Two corresponding classifiers have been established, combination of which achieves a sensitivity of 94% with a specificity of 89% for the plasma samples from HCC patients (n = 36) and control subjects including cirrhosis patients (n = 17) and normal individuals (n = 38). Notably, all 15 alpha-fetoprotein-negative HCC patients were successfully identified. Comparison between matched plasma and tissue samples indicates that both the cancer and noncancerous tissues contribute to elevation of the methylation markers in plasma. MCTA-Seq will facilitate the development of ccfDNA methylation biomarkers and contribute to the improvement of cancer detection in a clinical setting.
Keywords: circulating cell-free DNA, DNA methylation, next-generation sequencing, hepatocellular carcinoma
Introduction
Aberrant DNA methylation changes, including the hypermethylation of CpG islands (CGIs) concomitant with global hypomethylation, are hallmarks of nearly all human cancer types including hepatocellular carcinoma (HCC)1,2,3. Detecting hypermethylated CGIs of circulating cell-free DNA (ccfDNA) has emerged as a promising non-invasive approach for the diagnosis, prognosis and monitoring of cancers4,5. However, this is technically challenging because ccfDNA is highly fragmented and the cancer-associated ccfDNA makes up only a minority of the total ccfDNA4. Remarkably, despite rapid advances in next-generation sequencing (NGS) technologies for DNA methylome analysis6, no genome-scale method for detecting ccfDNA hypermethylation has (to our knowledge) been reported. Many genome-scale methods, such as the Infinium methylation array7 and reduced representation bisulfite sequencing (RRBS)8, have been successfully used for cell and tissue samples. Their applications to ccfDNA, however, are hampered by the fact that they require a relatively large amount of DNA or include a size selection step that is not suitable for severely fragmented ccfDNA9.
HCC is one of the most common and lethal malignant tumors worldwide10. Although early HCC responds to curative therapy, current tumor markers of HCC such as the serum alpha-fetoprotein (AFP) lack sufficient sensitivity and specificity for cancer detection11,12. Here we report a novel DNA methylation analysis technique named MCTA-Seq (methylated CpG tandems amplification and sequencing) for genome-wide detection of hypermethylated CGIs in ccfDNA. We applied this approach to a total of 151 clinical samples including 57 tissue samples and 94 plasma samples from HCC patients and control subjects, obtaining a first comprehensive view of the ccfDNA methylation pattern of HCC patients and identifying dozens of high-performance HCC-detecting markers.
Results
Design and validation of the MCTA-Seq technique
MCTA-Seq is based on the fact that there are a large number of CpG tandems that are highly enriched in the CGIs of human genomes; out of all 26 889 CGCGCGG sequences used in this study, 20 525 (76.3%) are located within the 9 373 (34.2% of all 27 435) CGIs in the human genome (Supplementary information, Figure S1). The key procedure of this technique is a single-tube three-step amplification of very short DNA fragments adjacent to the methylated CGCGCGG sequences from bisulfite-treated DNA (Figure 1). In the first step, a pool of CG-containing semi-random primers connected to a unique molecular identifier (UMI) sequence is used for linear amplification of CG-enriched genomic regions13. Then, a primer starting with CGCGCGG at the 3′-end is added to the reaction for amplification of the CpG tandem sites. Last, PCR amplification is performed using indexed primers against the anchor sequences. All three steps are integrated into a single-tube reaction and the amplified product is gel purified as the final library for high-throughput sequencing using a next-generation sequencer. We applied MCTA-Seq to genomic DNA samples with or without sonication (at an average size of 250 bp) and obtained similar amounts of PCR products, indicating that this technique is highly efficient for amplification of heavily fragmented DNA samples (Supplementary information, Figure S2).
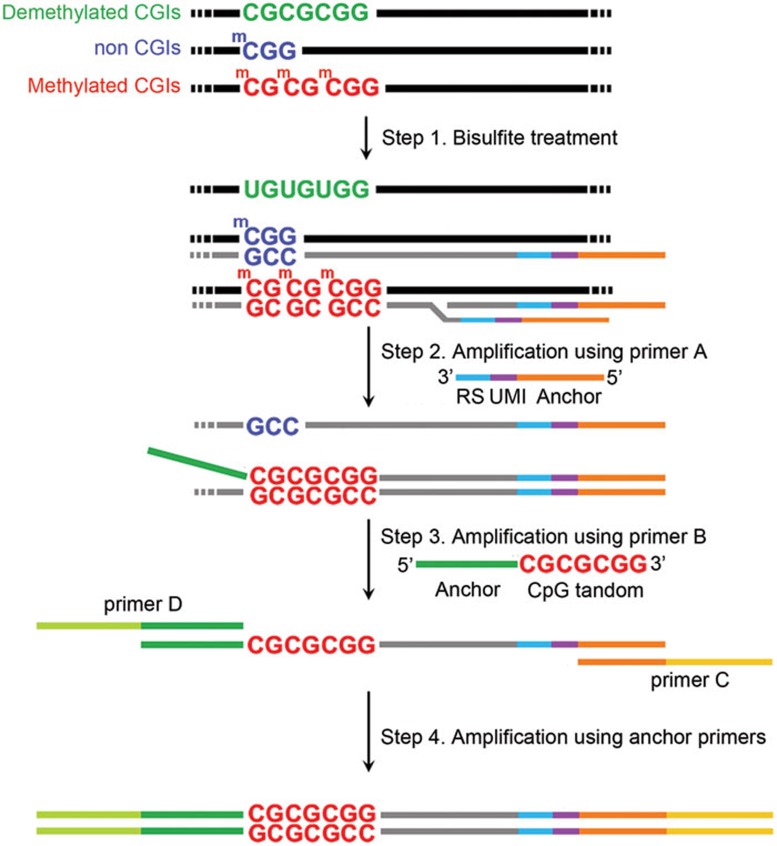
Figure 1.
Schematic of MCTA-Seq. First, the CpG tandems in the unmethylated CGIs are converted to UpG tandems via bisulfite treatment, whereas those in the methylated CGIs are unaffected. Second, the MCTA-Seq primer A binds semi-randomly to the converted DNA at the CpG site and is extended by a polymerase with displacement activity. The MCTA-Seq primer A consists of the following three parts: a semi-random sequence (RS) containing one CpG site (see Materials and Methods) is at the 3′-end, a unique molecular identifier (UMI) sequence is in the middle, and an anchor sequence is at the 5′-end. The methylated CGIs are expected to be amplified to a higher degree since they have high density of methylated CpG sites. Third, the MCTA-Seq primer B, which contains the CpG tandem sequence “CGCGCGG” at the 3′-end followed by a 4-bp “DDDD” sequence (D represents A or T or G), selectively amplifies the methylated CpG tandem sites. Last, exponential PCR amplification is performed using primer C and primer D, which is indexed, against the anchor sequences.
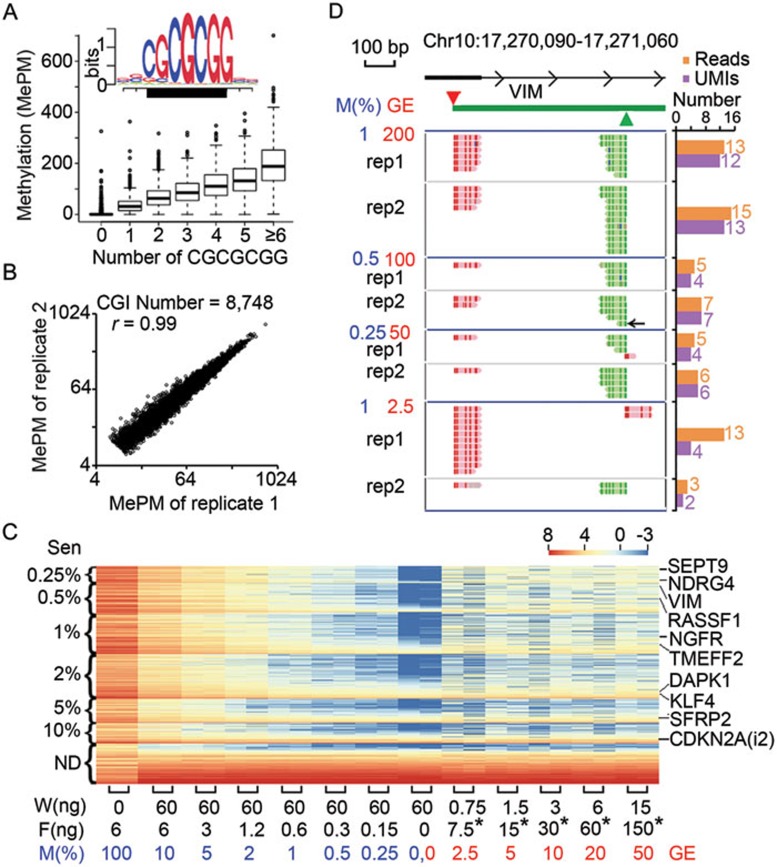
We validated the MCTA-Seq method by applying it to the fully methylated human genomic DNA (FMG), genomic DNA extracted from human white blood cells (WBCs) and two cancer cell lines (HepG2 and HeLa cells). For these and subsequent samples, the MCTA-Seq libraries were sequenced using the Illumina HiSeq2000/2500 system, obtaining on average of 8 million pair-end raw reads per library (Supplementary information, Table S1). The results demonstrate that the aligned reads predominantly started from genomic CGCGCGG sequences (Figure 2A) and our analysis suggest that MCTA-Seq gives > 50-fold enrichment on the fully methylated CGCGCGG sites over the partially methylated ones (Supplementary information, Figure S3). The data of FMG show that, out of all 9 373 CGIs containing one or more CGCGCGG sequences, 8 748 (93.3%) were efficiently detected (average methylated alleles per million mapped reads (MePM) > 8, Figure 2B); we focused on these in the subsequent analysis. The detection efficiency of a CGI is positively correlated with the number of CGCGCGG sequences within the CGI (Figure 2A). MCTA-Seq detected on average 2 849, 3 726 and 3 773 methylated CGIs in WBCs, HepG2 and HeLa cells, respectively. The methylation profiles were highly reproducible between technical replicates (Pearson's r: 0.99, 0.996, 0.96 and 0.96 for FMG, WBCs, HePG2 and Hela, respectively, Figure 2B and Supplementary information, Figure S4). In addition, the differentially methylated CGIs were clearly revealed when comparing the results of the cancer cell lines with those of WBCs, as exemplified by the CDKN2A locus (Supplementary information, Figure S4).
Figure 2.
Validation of the MCTA-Seq technique. (A) The detection values are shown for CGIs with different number of CGCGCGG sequence. The sequence logos of the targeted genomic sites are shown (black box indicates the CGCGCGG sequence). (B) The throughput and reproducibility of MCTA-Seq. A total of 8 748 CGIs were detected in FMGs (average MePM > 8). The Pearson correlation coefficient (r) is shown. (C) Heat map showing the sensitivity of MCTA-Seq. The 8 748 CGIs are divided into seven groups ranked by their analytic sensitivities. For each group (two technical replicates), the heat map is rank-ordered by the average methylation values of the CGIs in WBCs from the lowest to the highest. The amounts of FMG and WBC of each dilution experiment are shown along with the percentage [M(%)] or the haploid genomic equivalent (GE) of FMG. Representative CGIs that are frequently hypermethylated in human cancers are shown. In the heat map, blue color indicates low, white and yellow intermediate and red high DNA methylation values [Log2(MePM)]. Asterisks indicate that the mass unit is picogram (pg). (D) Genomic view of the promoter CGI of the VIM gene. All aligned reads in technical replicates of four dilution experiments are shown by the Integrative Genomics Viewer in the bisulfite mode. The green box indicates the CGI. The red and green triangles indicate two CGCGCGG sequences positioned at the forward and the reverse strand, respectively. Comparison between the UMI counts and the read counts is shown. The arrow indicates the shortest amplicon (30 bp).
Analytical sensitivity of the MCTA-Seq technique
We next analyzed the sensitivity of MCTA-Seq, which is crucial in the specific detection for cancer-associated ccfDNA from patient plasma or other bodily fluids, where the cancer-associated ccfDNA is present in extremely low amounts and constitutes a minor proportion of the total ccfDNA.
We first spiked FMG into WBC gDNA in serial ratios of 1:10, 1:20, 1:50, 1:100, 1:200 and 1:400 (Figure 2C). The data show that MCTA-Seq is highly sensitive in detecting methylated CGI alleles with frequencies as low as 0.25%. A total of 668 CGI loci showed significantly higher methylation in the 1:400 (0.25% methylation frequency) dilution groups comparing with the WBCs (one-tailed t-test, false discovery rate (FDR) < 0.05, Figure 2C). The cumulative number of detectable CGIs was 1 183 at 0.5%, 3 530 at 1%, 5 312 at 2%, 6 289 at 5% and 7 101 at 10%. The sensitivity of the remaining 1 647 CGIs was not determined because they were already highly methylated in WBCs. It should be noted that the sensitivities of many loci were underestimated due to their background methylation in WBCs. In addition, the sensitivity should depend on the low detection limit (LoD) of the method, and could be further increased as a function of sequencing depth.
To determine the LoD of MCTA-Seq, we performed another dilution experiment in which the absolute amounts of FMG were serially decreased to 150, 60, 30, 15 and 7.5 pg (Figure 2C). The results demonstrate that MCTA-Seq can detecte as little as 7.5 pg (equivalent of ∼2.5 haploid genome) of methylated DNA. Eighty-four percent (3 005 of 3 579) of the CGIs that were not detected in WBCs were detectable in at least one 7.5 pg replicate; 47% (1 687 of 3 579) of these were detectable in both 7.5 pg replicates (Supplementary information, Figure S5). The detection ratios substantially increased when the amount of methylated DNA rose to 15 pg (94% (3 367 of 3 579) in at least one replicate and 71% (2 582 of 3 579) in both), which is consistent with high sampling stochasticity in the 7.5 pg groups.
Importantly, a large number of markers known to be frequently hypermethylated in human cancers, including VIM14, SEPT915,16, NDRG217 and RASSF118, were detected with high sensitivity (Figure 2C). An example is the promoter CGI of VIM, which has been well studied for diagnosis of the colorectal cancer14. The amplicons predominantly initiated from two CGCGCGG sequences located within the CGI; the shortest fragment size was only 30 bp (Figure 2D). The technical duplicates were highly reproducible, and the methylation value was linearly quantitative over the lowest dilution range (r = 0.99, Supplementary information, Figure S6).
We also tested the use of UMIs to reduce the PCR amplification bias13. A 5-bp “HHHHH” sequence (H represents A or T or C) was tagged to each DNA molecule at the first amplification step and served as the UMI. The UMI could be used to distinguish up to 243 different molecules, which is sufficient to quantify the ccfDNA present at a low-copy number, particularly at the early cancer stage. The increase in reproducibility was evident in the 7.5 pg experiments, which were PCR over-amplified to the most extent, as exemplified at the VIM locus (Figure 2D and Supplementary information, Figure S7).
MCTA-Seq of HCC tissue samples
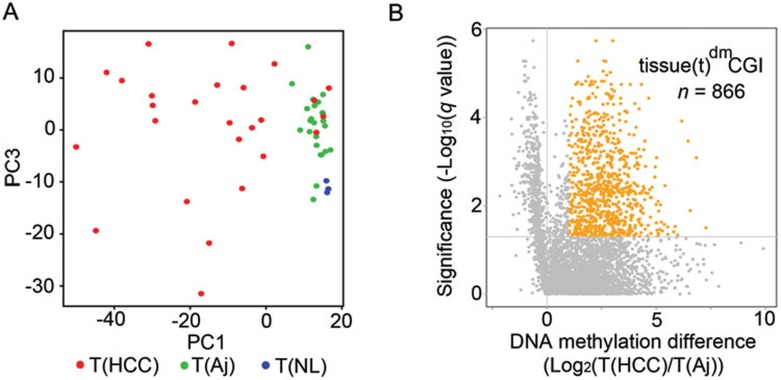
To assess the ability of the MCTA-Seq technique to distinguish between cancer and noncancerous tissues and to detect cancer-specific hypermethylated CGIs, we performed MCTA-Seq in 27 pairs of HCCs and matched adjacent noncancerous liver samples and three normal liver samples (Supplementary information, Table S2). The principal component analysis (PCA) demonstrate that MCTA-Seq can successfully distinguish most cancerous tissues (23 of 27) from noncancerous tissues (Figure 3A). Although tumor-adjacent cirrhosis tissues appear to differ from those of normal livers, the major hypermethylation changes clearly occurred during HCC formation. A total of 866 hypermethylated CGIs in HCC tissues were identified (referred to as tissue dmCGIs or tdmCGIs, two-tailed Mann-Whitney-Wilcoxon (MWW) test, FDR < 0.05, average methylation fold changes > 2, Figure 3B). We also found that nearly all (799 out of 866, 92%) tdmCGIs were pre-marked by H3K27me3 in the normal liver, which is in agreement with the hypothesis that cancer-associated CGI hypermethylation is primarily targeted to Polycomb-repressed genes19. Taken together, these data demonstrate that the MCTA-Seq technique can be used to profile aberrant CGI hypermethylation in HCC tissues.
Figure 3.
Analysis of HCC and noncancerous liver tissues. (A) Principal component analysis of 27 pairs of HCCs (T(HCC), red) and adjacent tissues (T(Aj), green) and 3 normal liver tissues (T(NL), blue) distinguishes most (23 of 27) HCCs from the noncancerous tissues. PC, principal component. (B) Volcano plot for differentially methylated CGIs between the HCCs and the adjacent noncancerous liver tissues. The x axis shows the fold changes of the average methylation value between the HCCs and the adjacent tissues, and the y axis shows the q-value as the FDR analogue of the P value (−Log10(q-value)) for a two-tailed MWW test of differences between two groups. The dashed line indicates statistical significance (FDR < 0.05). A total of 866 CGIs (tdmCGIs) were differentially hypermethylated in HCC tissues.
MCTA-Seq identifies novel markers for detecting HCC in blood
The tissue study gave us confidence in detecting tumor-specific CGI methylation by the MCTA-seq method. To investigate whether MCTA-Seq can be successfully applied to plasma samples to distinguish between HCC patients and cancer-free individuals, we applied MCTA-Seq to plasma ccfDNA obtained from HCC patients (n = 27) and 45 cancer-free individuals including cirrhosis patients (n = 17) and normal individuals (n = 28) (Supplementary information, Tables S2 and S3. Three groups of subjects were not statistically different in age (P > 0.05, two-tailed t-test) and gender (P > 0.1, χ2 test). The concentration of plasma ccfDNA varied among the HCC patients and was unrelated to the tumor size (Spearman's rho = −0.02, Supplementary information, Figure S8).
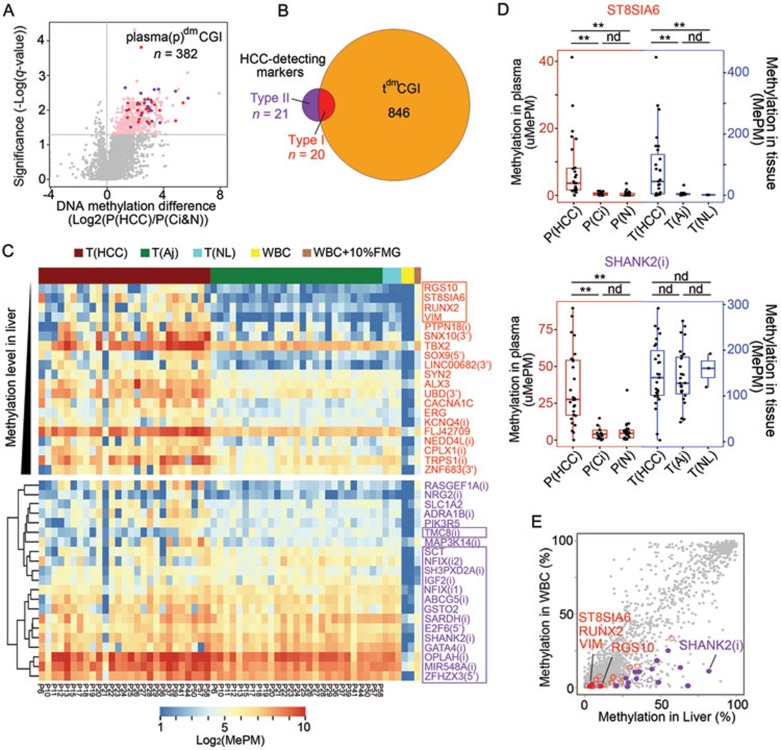
The overall number of methylated CGIs in HCC patients was significantly higher than that in cancer-free individuals (UMI-adjusted MePM (uMePM) > 1, HCC vs cirrhosis patients: median 3 381 vs 2 686, P < 0.01; HCC patients vs normal individuals: median 3 382 vs 2 862, P < 0.01, two-tailed MWW test, Supplementary information, Figure S9), whereas no differences were found between the cirrhosis patients and the normal controls (P = 0.1). Comparison between all HCC patients and cancer-free individuals identified 2 166 differentially hypermethylated CGIs in plasma of HCC patients (FDR < 0.05, average methylation fold changes > 1, two-tailed MWW test). To identify markers for detecting early stage HCC, we focused on plasma samples of nine patients with small HCC tumors (≤ 3 cm) and this resulted in 382 differentially methylated loci (referred to as plasma dmCGIs or pdmCGIs, FDR < 0.05, average methylation fold changes > 1, two-tailed MWW test, Figure 4A). Then, we applied the receiver operating characteristic (ROC) curve analysis to find markers with best diagnostic performance. A total of 41 top HCC-detecting markers were identified using an area under the curve (AUC) cutoff as ≥ 0.9 for tumor > 5 and 3–5 cm, and ≥ 0.80 for tumor ≤ 3 cm. Detailed information and data of these markers are provided in Supplementary information, Data S1.
Figure 4.
Identification of novel HCC-detecting markers in blood. (A) Volcano plot showing CGIs in plasma that are differentially methylated between the HCC patients with small tumors (n = 9, tumor size ≤ 3 cm) and cancer-free individuals (n = 45). The x axis shows the fold changes of the average methylation between the HCC patients and the cancer-free individuals, and the y axis shows the q-value as the FDR analogue of the P value (−Log10(q-value)) for a two-tailed MWW test of differences between two groups. The dashed line indicates statistical significance (FDR < 0.05). A total of 382 differentially methylated plasma CGIs (pdmCGIs) are identified (pink). (B) Venn plot view of the HCC-detecting markers (n = 41) and tdmCGIs (n = 866). (A, B) The type I and type II high-performance HCC-detecting markers are indicated in red and purple, respectively. (C) A heat map showing methylation of the HCC-detecting markers in the tissue samples. Each column represents a tissue sample of HCCs (T(HCC), red), adjacent livers (T(Aj), green), normal livers (T(NL), blue), WBCs (two biological replicates, yellow) or 10% FMG diluted in WBCs (brown), and each row represents a marker. The type I markers (n = 20) are ranked by their methylation levels in the liver calculated by their MePM values in the normal liver relative to the 10% FMG diluted in WBCs from the lowest to the highest. The type II markers (n = 21) are clustered using the hierarchical clustering. In the heat map, blue color indicates low, white and yellow intermediate and red high DNA methylation values, shown by log2(MePM). The markers for the classifiers are boxed. (D) Boxplots of the representative markers of the type I (ST8SIA6) and the type II [SHANK2(i)] HCC-detecting markers showing their methylation in plasma (red) or tissues (blue) samples. **P < 0.01; ND, no statistical difference. Two-tailed MWW test. (E) Scatter plots of the methylation levels of WBCs vs the normal liver. The RRBS results of 3 886 CGI loci are shown. Red circles indicate the type I markers (n = 19); purple circles indicate the type II markers (n = 20), and solid circles indicate the markers for the classifiers.
Comparison between the pdmCGIs and the tdmCGIs revealed that only 20 (2.3% of 866 loci) tdmCGIs emerged as the high-performance HCC-detecting markers (referred to as type I markers, Figure 4B). Unexpectedly, we found that half of high-performance pdmCGIs markers (21, referred to as type II markers) were not included in the list of tdmCGIs (Figure 4B).
To obtain insight into the nature of the type II markers, we examined their methylation status in tissue. The results demonstrate that most of the type II loci (14 of 21 loci) were highly methylated not only in HCC but also in the adjacent noncancerous and normal liver tissues, explaining why they were not identified as tdmCGIs (Figure 4C). SHANK2 (i), IGF2 (i) and ZFHX3 (5′) (where (i) indicates an intragenic CGI and (5′) indicates an intergenic CGI upstream of the gene) were representative of these markers (Figure 4D and Supplementary information, Figure S10). While they were significantly more methylated in the plasma of the HCC patients than the cancer-free individuals, they were similarly methylated in HCC, tumor-adjacent tissues and normal livers. One locus, the TMC8 (i), showed higher methylation levels in the cancer-adjacent tissues comparing with both the cancer and the normal liver tissues, suggesting that its methylation levels elevated specially during the cirrhosis stage (Supplementary information, Figure S10). All 15 markers exhibited low methylation levels in the WBCs, displaying a tissue-specific methylation pattern. We performed RRBS which confirmed that they were significantly more methylated in the normal liver tissues than the WBCs (median 40.1% vs 8.3%, P = 1E–9, two-tailed MWW test, Figure 4E and Supplementary information, Figure S11). Comparison between MCTA-Seq and RRBS revealed a concordance between two methods (Supplementary information, Figure S12). We found that the type II markers are likely to be intragenic CGIs (73%, 11 of 15 markers), which is in agreement with previous reports that tissue-specifically methylated CGIs are often intragenic20. Together, our data demonstrate that the type II markers are a set of CGI loci that are tissue-specifically hypermethylated in the liver, while hypomethylated in WBCs. They appear to be contained in liver cells under normal circumstances but are released into the blood when malignance occurs.
Of the type I markers, four CGIs (RGS10, ST8SIA6, VIM and RUNX2) showed the lowest methylation levels in the normal and cirrhosis liver tissues, but strong hypermethylation in HCC tissues; thus, they are the most tumor-specific markers (Figure 4C and 4D, Supplementary information, Figure S10). The RRBS confirmed that they had the lowest methylation levels in the liver among the type I markers (Figure 4E, on average 10.8%, 4.1%, 4.8%, and 3.1% for RGS10, ST8SIA6, RUNX2 and VIM, respectively). These four CGIs also had the extremely low methylation levels in WBC (0.6%, 0.8%, 2.3% and 0.6% for RGS10, ST8SIA6, RUNX2 and VIM, respectively, Figure 4E), and gave excellent quantitative performance (Supplementary information, Figure S6), thus providing bases for sensitive detection. Their methylation levels were positively correlated with the tumor size, indicating that they were derived from the tumor tissues (Spearman's rho = 0.69, Supplementary information, Figure S13). The other 16 type I markers showed relatively higher methylation levels in the normal liver; thus, they are less tumor-specific.
We also compared our list with the data of Infinium HumanMethylation450 BeadChip from a recent HCC tissue study21 and the TCGA (The Cancer Genome Atlas) database22, which confirmed the tissue results of MCTA-Seq (Supplementary information, Figure S14). Together, our data demonstrate that the MCTA-Seq method can efficiently identify a small number of well-performing ccfDNA methylation markers from hundreds of candidates that show differential hypermethylation in cancer tissues. Furthermore, novel types of markers can be uncovered without preconception.
ccfDNA released from the non-cancerous liver tissues of HCC patients
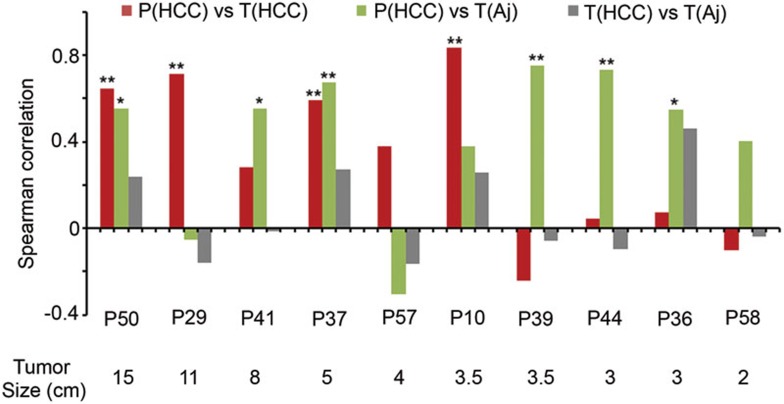
Previous investigations of cancer-associated ccfDNA generally focused on the tumor origin, but a complete picture of the genesis of cancer-associated ccfDNA is lacking23. For 10 HCC patients (P10, P29, P36, P37, P39, P41, P44, P50, P57 and P58), we have analyzed the matched HCC, the adjacent noncancerous liver tissues and plasma samples. We next used these data and asked whether the elevated ccfDNA markers came directly from the HCC tissues in these cases. We calculated the correlation of methylation levels of the HCC-detecting markers between the matched plasma and HCC, the plasma and the adjacent liver, as well as the HCC and the adjacent liver tissues. High correlations between the plasma and the HCC were found in 5 of 10 cases, with 4 showing statistical significance (Spearman's rho with P value: 0.65 (3.8E-3), 0.71 (8.9E-4), 0.59 (9.4E-3), 0.83 (1.8E-5) for P50, P29, P37 and P10, respectively, Figure 5), indicating that the HCC tissues were indeed the main source of the cancer-associated ccfDNA in these cases. However, low correlations were found in 4 cases (rho with P value: −0.24 (0.33), 0.04 (0.87), 0.07 (0.77) and −0.1 (0.68) for P39, P44, P36 and P58, respectively). Surprisingly, we found high correlations between the plasma and the adjacent noncancerous liver tissues in these cases, with three showing statistical significance (rho with P value: 0.75 (3.0E-4), 0.73 (5.1E-4) and 0.54 (0.018) for P39, P44 and P36, respectively, Figure 5).
Figure 5.
Comparison between matched tissue and plasma samples. The correlations (Spearman's rho) between the matched plasma and HCC tissue (red), the plasma and the adjacent liver tissue (green), as well as the HCC and the adjacent liver tissues (blue) from nine HCC patients as ordered by the tumor size. **P < 0.01; *P < 0.05.
To confirm these results, we examined RGS10, ST8SIA6, RUNX2 and VIM as the tumor-specific markers and IGF2(i) as a liver-general marker. Taking the case of P39 as an example, all four tumor-specific markers showed hypermethylation in the HCC tissue, with the methylation values being higher than IGF2(i) (MePM 46.2, 280.3, 136.2 and 130.6 for RGS10, ST8SIA6, RUNX2 and VIM, respectively, vs 45.8 for IGF2(i)), whereas their methylation values were significantly lower than IGF2(i) in the adjacent noncancerous tissues (MePM 3.7, 6.1, 7.7 and 2.9 for RGS10, ST8SIA6, RUNX2 and VIM, respectively, vs 96.0 for IGF2(i)). In plasma from this patient, all four markers were at the very low levels, while IGF2(i) was detected at a high level (uMePM 2.9, 2.6, 3.5 and 1.0 for RGS10, ST8SIA6, RUNX2 and VIM, respectively, vs 28.3 for IGF2(i)). Thus, the methylation pattern of plasma from this patient indeed resembled that of the adjacent noncancerous liver tissues, but not the cancer. The other three cases (P44, P36 and P58) displayed similar results. Therefore, the data suggest that the noncancerous liver tissues, instead of the cancer tissues, can serve as the main source of the elevated methylation markers in plasma. It is noteworthy that all four cases were small HCCs, which suggest that, at early stage of HCC, release of ccfDNA from the noncancerous liver cells exceeds that from the cancer cells.
HCC classifier development
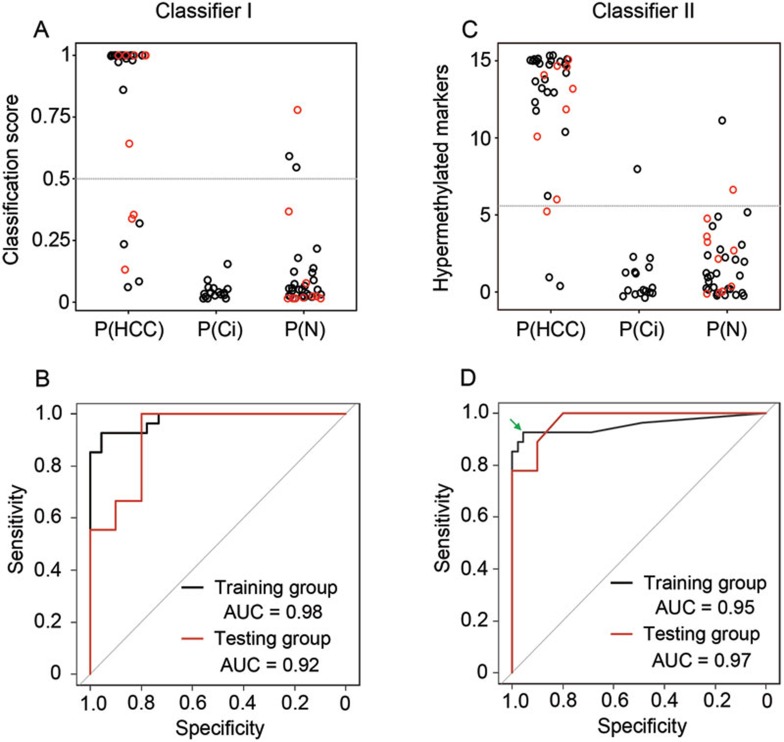
Next, we aimed to establish diagnostic classifiers to distinguish HCC patients from cancer-free individuals. Given that the type I and type II markers provided distinct information for detecting HCC, we developed two corresponding classifiers. For the classifier I, we focused on four type I markers (RGS10, ST8SIA6, RUNX2 and VIM), elevation of which gave the most specific indication for the presence of a cancer. Hypermethylation of four markers in the HCC tissues was weakly correlated, suggesting that a combination will recover more cancer cases than an individual marker (Supplementary information, Figure S15). A classifier was established by fitting to a logistic regression model to generate a score ranging from 0 to 1 with the discriminating value at 0.5. This classifier successfully identified 23 out of 27 (i.e., the sensitivity of 85%) HCC patients at a specificity of 96% (Figure 6A). The AUC of the ROC curve was 0.98 (95% confidence interval (CI): 0.95-1, Figure 6B), which was higher than any individual marker (AUC (95% CI): 0.91 (0.83–0.99), 0.95 (0.89–1), 0.95 (0.90–1) and 0.91 (0.83–0.98) for RGS10, ST8SIA6, RUNX2 and VIM, respectively).
Figure 6.
Development of HCC classifiers. (A, B) For classifier I, the classifier scores of HCC patients, cirrhosis patients and normal individuals (A) and AUC curves (B) are shown. The dash line in A indicates the cutoff at 0.5. (C, D) For classifier II, the number of hypermethylated markers (C) and AUC curve (B) are shown. The dash line in C indicates the cutoff at 5.5. The green arrow in D indicates the point nearest the upper left corner of the ROC curve that gives the cutoff. Black: the training group; red: the testing group.
The classifier II was built on 15 type II markers, elevation of which should indicate excessive death of liver cells. Since these markers were highly associated with each other, we established the classifier based on the number of hypermethylated markers. Cross-validation analysis demonstrated that this model performed better than the logistic regression model (Supplementary information, Figure S16). A marker was defined as hypermethylated if its methylation level was above the 90th percentile of 28 healthy subjects. We set the classifier cutoff to be 5.5 hypermethylated markers since this criterion corresponded to the upper left corner of the ROC curve. This classifier allowed us to identify 25 out of 27 (i.e., the sensitivity of 93%) HCC patients at a specificity of 96%, with the AUC of the ROC curve being 0.95 (95% CI: 0.89-1) (Figure 6C and 6D).
To test the performance of two classifiers, we then analyzed a new set of 19 plasma samples obtained from 9 HCC patients and 10 normal individuals, with the specimen being blinded to the experimental operator and the data analyzer. The classifier I and II identified 6 and 8 out of 9 HCC patients for a sensitivity of 67% and 89%, respectively. For either classifier, 1 out of 10 control subjects was positively detected, giving a false-positive rate of 10%. The AUCs of the ROC curve were 0.92 and 0.97 for classifier I and II, respectively (Figure 6).
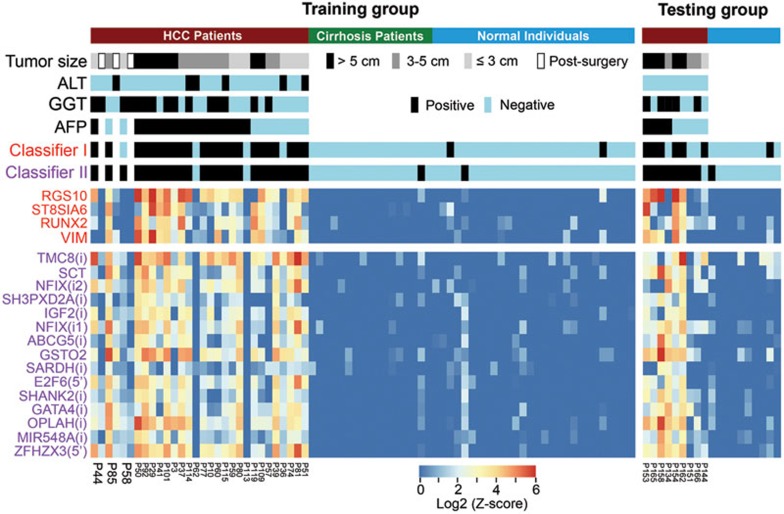
Figure 7 and Supplementary information, Table S3 and Figure S17 summarize the performance of two classifiers for all the training and testing cases, and it appears that the classifier II is more sensitive than the classifier I for detecting small HCCs (≤ 3 and 3–5 cm, Supplementary information, Figure S17). Two classifiers can be combined to further improve the sensitivity using an “OR” operation: the plasma is defined as positive if either classifier I or II is positive. This operation gives a sensitivity of 93% at a specificity of 91% in the training group and a sensitivity of 100% at a specificity of 80% in the testing group, with a sensitivity of 94% at a specificity of 89% as the combined scores. Alternatively, they can be combined to strengthen the specificity using an “AND” operation: the plasma is defined as positive if both classifier I and II are positive. This operation gives a sensitivity of 85% at a specificity of 100% in the training group, and a sensitivity of 56% at a specificity of 100% in the testing group, with a sensitivity of 78% at a specificity of 100% for all samples.
Figure 7.
Performance comparison of ccfDNA methylation classifiers, AFP and ALT. For plasma samples of the HCC patients, cirrhosis patients and normal individuals in the training group (n = 72) and the testing group (n = 19) and post-surgery HCC patients (n = 3), detection results of two ccfDNA methylation classifiers, AFP, gamma-glutamyl transferase (GGT) and ALT are shown as positive (black bars, AFP: > 20 ng/ml, GGT: > 60 U/l, ALT: > 50 U/l) versus negative (blue bars). The MCTA-Seq data of the type I and type II markers in the classifiers are shown in a heat map using a Z-score approach (see Materials and Methods). The tumor sizes are also indicated.
Comparison with serum HCC biomarkers
We compared the ccfDNA methylation markers with AFP, which is the most commonly used blood-based biomarker for HCC. This marker, however, exhibits false-negative results in about 40% of HCC patients12. Forty-two percentage (15 cases) of 36 HCC patients in our study exhibited false-negative results in the AFP assay (< 20 ng/ml, Figure 7). Notably, all of these AFP-negative patients were identified using the “OR” operation. We calculated the total Z-score (the sums of the Z-scores of the classifier I and II markers) to evaluate the overall level of the methylation markers. The results showed that the total Z-scores of the AFP-negative HCC patients were not significantly different from those of the AFP-positive patients (AFP-positive vs AFP-negative: median 164 vs 129, P = 0.36, two-tailed MWW tests, Supplementary information, Figure S18). Therefore, our data demonstrate that the ccfDNA methylation markers are largely independent of AFP for detecting HCC. It should be noted that the AFP levels were elevated in two patients (P62 and P113) who were negative for the methylation markers. Together, these results suggest that a combination of AFP and the methylation markers can significantly improve HCC diagnosis. Of note, we also examined another HCC biomarker, the gamma-glutamyl transferase12, which detected only 50% (18 cases) of 36 HCC patients.
Since elevation of the type II markers should indicate excessive death of liver cells, we examined the levels of serum alanine aminotransferase (ALT), a commonly used marker for liver cell injury, for a comparison. The ALT levels were elevated (> 50 U/l) in 14% (5 cases) of 36 HCC patients (Figure 7). This comparison suggests that the type II markers are more sensitive than ALT for detecting the HCC-associated liver cell death.
Comparison between the pre-surgery and post-surgery samples
In three cases (P44, P58 and P85), we analyzed both the pre-surgery and post-surgery plasma samples to explore the dynamics of the ccfDNA markers relating to the hepatectomy (Figure 7). All post-surgery samples were obtained three days after the hepatectomy. We have shown earlier that elevated ccfDNA in P44 originated from the noncancerous liver tissue. The levels of the ccfDNA markers dropped sharply after the surgery with the total Z-score of the type II markers showed a fivefold reduction from 165 to 30. Other two cases showed similar results. These data suggest that the abnormally increased ccfDNA methylation markers, though seem not directly derive from the HCC tissue, were tightly associated with the tumor.
Discussion
The current genome-wide DNA methylation technologies enable us to investigate the entire methylome using cell or tissue samples, but examination of the methylation markers in peripheral blood samples is low throughput and biased6. Here we describe a novel DNA methylation method, MCTA-Seq, and show this high-throughput platform can be used to investigate thousands of CGIs in one blood-based experiment.
The detection sensitivity is the most important benchmark for a ccfDNA methylation technique that will be applied in the early detection of cancer. MCTA-Seq has a low detection limit of 2.5 haploid genome equivalents, which is near the limit of a PCR-based assay. At a relatively low sequencing depth, MCTA-Seq can detect a methylated allele at a frequency as low as 0.25%, which is higher than any existing genome-wide DNA methylation analysis method6. It is possible to further increase the analytical sensitivity and throughput of the assay by simultaneously targeting several types of CpG tandems. Given that MCTA-Seq is highly efficient for amplification of short DNA fragments and can target multiple sites even in a single CGI locus, its detection sensitivity has the potential to compete with the most sensitive locus-specific methylation assays such as the digital Methyl-BEAMing14. Comparing with RRBS and Infinium methylation array, it appears that MCTA-Seq reduces the methylation background because it only detects fully methylated CpG tandem sequences, but not partially methylated sequences and individual methylated CpGs. The MCTA-Seq assay is also simple and cost-effective as the single-tube three-step library construction takes only a few hours and a relatively low sequencing depth is required.
Using the MCTA-Seq technique, we acquired comprehensive views of hypermethylated CGIs in ccfDNA of HCC patients, cirrhosis patients and normal individuals and identified dozens of high-performance HCC-detecting markers. We show that only a very small portion (2.3%, 20 of 866 loci) of CGIs hypermethylated in the HCC tissues are good markers for detecting HCC in blood (type I markers), and only 4 CGIs (RGS10, ST8SIA6, RUNX2 and VIM) further fit the tumor-specific criteria. These findings demonstrate the high efficiency of MCTA-Seq. In addition, identification of the type II markers reveals the advantage of the MCTA-Seq screening in uncovering novel types of markers without preconception. These markers have been overlooked in previous studies comparing HCC and noncancerous tissues because they are already tissue-specifically hypermethylated in the normal liver.
We established two classifiers for HCC detection corresponding to the type I and type II markers, of which the type II marker-based classifier seems to offer higher sensitivity. This can be explained by our finding that the noncancerous liver cells contribute to a greater proportion of elevated ccfDNA than the cancer cells in many cases of small HCCs (e.g., P39, P44, P36 and P58). In such cases, the type II markers in the blood in fact mainly come from the noncancerous liver cells. It is noteworthy that the elevated type II markers drop promptly after tumor resection as shown in three cases (P44, P58 and P85), suggesting that they are caused by the cancer. Further investigation is needed to elucidate the mechanisms that lead to the HCC-induced release of ccfDNA from the noncancerous liver cells.
Our data show that the type II markers are elevated in most HCC cases while the ALT levels are normal; thus, ALT is actually not a HCC marker. The higher sensitivity of the methylation markers for detecting HCC may be due to its much lower background than ALT. It is also possible that the ccfDNA is released in a manner different from ALT. Apoptosis has been thought as a major cause of ccfDNA4, but it has been shown to contribute very little to the release of ALT24. Thus, the type II markers may be sensitive indicators for excessive apoptosis of liver cells.
In contrast to the classifier II, a positive score of the classifier I indicates the presence of tumor DNA in blood more specially. Our data have shown that a combination of two classifiers using an “OR” operation further improves the sensitivity, which can be used for surveillance of a population with high risk for HCC such as cirrhosis patients for whom sensitive detection is crucial. A combination using an “AND” operation exhibits high specificity, which may be preferable for screening healthy populations for whom specificity is more important to reduce unnecessary clinical examinations and psychological stress.
Regarding that DNA methylation has both tissue- and tumor-specific patterns, the rich information provided by MCTA-Seq may make it possible to determine the tissue origin of a cancer in blood25. For example, elevation of the type II markers may indicate that a cancer is located in or originates from the liver. Further comparative studies between HCC and other cancer types are required to test this hypothesis.
Taken together, the MCTA-Seq technique described here represents valuable progress in the field of ccfDNA and offers great promise for clinical research and medical diagnostics using information on DNA hypermethylation.
Materials and Methods
Tissue and plasma sample collection
Twenty-seven paired HCC tumor and adjacent liver tissue samples and a total of 39 peripheral blood samples (36 pre-surgery samples including 27 for the training group and 9 for the testing group and 3 post-surgery samples) were obtained from HCC patients who underwent surgical resection and were pathologically diagnosed as HCC at the Department of Surgery, Beijing Shijitan Hospital, Capital Medical University and the Department of Hepatobiliary Surgery, Peking University People's Hospital, China. Three normal liver tissue samples adjacent to hemangioma were also obtained from the liver hemangioma patients who underwent surgical resection in these two hospitals. Seventeen peripheral blood samples were obtained from cirrhosis patients treated in above two hospitals and in the Center of Therapeutic Research for Liver Cancer, Beijing 302 Hospital, and the Department of Hepatobiliary Surgery, Beijing Ditan Hospital, Capital Medical University, China. Thirty-eight peripheral blood samples (28 samples for the training group and 10 for the testing group) were obtained from individuals who had no signs of cancer, hepatitis infection or cirrhosis. The present study was approved by the Ethics Committee of Beijing Shijitan Hospital, Capital Medical University. Written informed consent for the collection of samples and subsequent analysis was obtained from all subjects before inclusion in the study.
Genomic DNA isolation
Genomic DNA of tissue samples, WBCs or two cancer cell lines (the human liver HCC cell line (HePG2) and the human cervical cancer cell line (HeLa), which were obtained from China Infrastructure of Cell Line Resources) were extracted using the DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer's protocol. WBC genomic DNA of three normal individuals including two females and one male were mixed. The fully methylated human genomic DNA is purchased (Chemicon/Millipore, CpGenome Universal Methylated DNA, S7821).
Blood sample processing and cell-free DNA isolation
To prepare plasma, 5 ml peripheral blood was collected using EDTA anticoagulant tubes and the plasma samples were prepared within 6 h by centrifuging the blood tube at 1350× g for 12 min at room temperature, and transferring the plasma to a 15-ml tube, and recentrifuging at 1350× g for 12 min, and transferring to 1.5- or 2-ml tubes, and recentrifuging at 13500× g for 5 min and transferring to a new tube. The prepared plasma samples (about 2 ml) were then stored at −80 °C immediately. The plasma cell-free DNAs were extracted using the QIAamp DNA Blood Midi Kit (Qiagen) according to the manufacturer's protocol. Concentration of ccfDNA was quantified using the Qubit HsDNA Kits (invitrogen).
Bisulfite conversion and DNA quantification
Fully methylated Lambda DNA was added in a ratio of 0.5% as a spike-in DNA control to calculate the non-conversion rates of unmodified cytosine. Bisulfite conversion was performed by using the MethyCode bisulfite conversion kit (Invitrogen) according to the manufacturer's protocol. Briefly, the reaction containing the genomic or cell-free DNAs and the conversion reagent were incubated at 98 °C for 10 min and then at 65 °C for 2.5 h. After this, the converted DNAs along with 100 ng carrier tRNA (Roche) were purified using the Zymo-Spin columns (Zymo) with a step of on-column desulfonation, and were eluted in 10 μl elution buffer. For the dilution experiments, concentration of the converted FMG and WBC gDNAs was quantified using the Qubit ssDNA Kits (Invitrogen) before starting the experiments and the average values of duplicates were used.
MCTA-Seq library preparation
The MCTA-Seq library was prepared in a single-tube three-step reaction. In the first step, the bisulfite-converted DNA was linearly amplified in a 15-μl reaction containing 1× NEBuffer 2 (New England Biolabs, NEB), 250 μM each dNTP, 0.33 μM MCTA-Seq primer A and 2.5 units Klenow fragment with no 3′ to 5′ exonuclease activity (NEB) to obtain the semi-amplicon. The MCTA-Seq primer A is a mixture of four primers: (i) 5′-TTTCCCTACACGACGCTCTTCCGATCTHHHHHHHHCGCH-3′, (ii) 5′-TTTCCCTACACGACGCTCTTCCGATCTHHHHHHHCGHCH-3′, (iii) 5′-TTTCCCTACACGACGCTCTTCCGATCTHHHHHHCGHHCH-3′, and (iv) 5′-TTTCCCTACACGACGCTCTTCCGATCTHHHHHCGHHHCH-3′ the underlined portions correspond to the UMI sequences (H = A/T/C). The primers were designed to maximize their binding efficiency to a CGI and minimize the dimer formation. The reaction was assembled except the klenow fragment and incubated at 95 °C for 2 min before hold at 4 °C. The klenow fragment was then added. Then, the reaction was subjected to the following conditions: 4 °C for 50 s, 10 °C for 1 min, 20 °C for 4 min, 30 °C for 4 min, 37 °C for 4 min and 75 °C for 20 min (to inactivate the Klenow fragment). In the second step, the CpG tandem regions were selectively amplified in a 20-μl reaction by adding a 5-μl solution containing 1× Ex Taq Buffer, 1.5 units Hot Start Ex Taq (Takara) and 1 μM MCTA-Seq primer B(5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTDDDDCGCGCGG-3′, D = A/T/G) to obtain the full-amplicon. The reaction was subjected to the following conditions: 95 °C for 3 min (to activate the hot-start polymerase), followed by 50 °C for 2 min and 72 °C for 1 min. In the third step, the full-amplicon was exponentially amplified in a 50-μl reaction by adding a 30-μl solution containing 1× Ex Taq Buffer, 250 μM each dNTP, 1.5 units Hot Start Ex Taq (Takara), 2 μM primer C(5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′) and 2 μM primer D(5′-CAAGCAGAAGACGGCATACGAGATCTGATCGTGACTGGAGTTCAGACGTGTGCT-3′); the underlined portion in primer D corresponds to the Illumina index sequences (the sequence corresponding to index 9 is shown here). The reaction was subjected to the following cycling conditions: 95 °C for 3 min, 17 cycles of 95 °C for 30 s, 65 °C for 30 s, 72 °C for 1 min and a final cycle of 72 °C for 5 min. The resulting amplified product was resolved on a 4% agarose gel and the 180-a-250 bp fraction was excised and then purified to serve as the library for sequencing on an Illumina HiSeq2000 or HiSeq2500 sequencers as paired-end 100-bp reads.
Data processing and calculation of MePM and uMePM
FASTQ format R2 reads generated by the Illumina HiSeq2000 or HiSeq2500 platform were processed and filtered as follows: (i) whole or any subsets of adaptor sequences were trimmed at 3′-end of the reads; (ii) the MCTA-Seq primer sequences including 6 bp at the 5′-end and 12 bp at the 3′-end of reads were trimmed; (iii) low-quality reads, as > 10% bases being N, or Phred quality of > 50% bases being < 5 or having > 3 unmethylated CHs, were discarded; (iv) cytosines in a read were computationally replaced with thymines and then mapped to computationally converted hg19 references using the Bismark and Bowtie programs; the lambda genome was also included in the reference sequence as an extra chromosome for assessing the conversion rate, and (v) the reads were discarded if there was < 2 genomic CpG sites ± 3 bp surrounding the position corresponding to the 5′-end of the aligned reads.
The methylation value of a CGI is calculated as the number of reads mapped to the CGI normalized by the total number of reads uniquely mapped to the whole human genome and is expressed as methylated alleles per million mapped reads (MePM). For the UMI-adjusted counting, the UMIs were extracted from the R1 reads and a read was removed if the Phred score of the first 5 bp at 5′-end of the read was < 20. Then, a duplication index for the average number of reads per UMI is calculated using a set of 3 024 CGIs with low detection efficiency (to avoid saturation of the UMIs). The UMI-adjusted methylation value of a CGI is calculated as the number of UMIs mapped the CGI plus the duplication index then normalized by the total number of reads uniquely mapped to the whole human genome (after the R1 reads quality control) and is expressed as uMePM. We only calculated the uMePM value for the plasma samples; for the tissue samples, we calculated the MePM value to avoid saturation of the UMIs.
CpG island annotation
The CGIs were retrieved from the cpgIslandExt table in UCSC database. The promoter CGIs were defined as overlapping with the region 1-kb upstream and 300-bp downstream of a RefGene transcription start site (TSS). The intragenic CGIs were defined as starting after 300-bp downstream of a TSS and end before 300-bp downstream of a RefGene transcription end site. The other CGIs were defined as the intergenic CGIs.
HCC classifier development
For the establishment of classifier I, four tumor-specific markers (RGS10, ST8SIA6, RUNX2 and VIM) were trained to a logistic regression model using the R package “glm”. For development of classifier II, since methylation of the type II markers was highly correlated, we counted the number of the hypermethylated markers as the expressed results. The performance of this model was compared with the logistic regression model by cross-validation. We randomly split the plasma samples of both 27 HCC cases and 45 cancer-free cases into two sets: two-third as the training set and one-third as the test set. Two kinds of models were built on the training set and then applied to predict the testing set, and the Brier Scores were calculated. The procedure was repeated 50 times to generate 50 Brier Scores. The result was shown in Supplementary information, Figure S16.
Bioinformatics and statistical analysis
Custom R scripts were used to perform PCA, hierarchical clustering and ROC curve analysis and to calculate the AUC values, as well as to construct box plots, volcano plots, correlation plots and histograms. The R statistical package was used to perform the statistical analyses.
To determine the analytical sensitivity for each of 8 748 CGIs, one-tailed t-test was used to identify the CGI that was differentially methylated between two technical replicates of each dilution group (0.25%, 0.5%, 1%, 2%, 5% and 10% methylation) and two technical replicates of WBCs. P value thresholds were selected such that the number of false positives was < 5% using the Benjamini-Hochberg method.
The PCA analysis of tissue samples was performed using 5 014 CGIs that exhibited a minimum of 1 MePM in at least one tissue sample and a maximum of 100 MePM in at least one sample.
The two-tailed MWW test was used to identify the differentially methylated CGIs between HCC and adjacent noncancerous liver tissues. To avoid the influence of gender, 231 (2.6% of 8 748) CGIs that are located at the sex chromosomes were omitted for analysis of clinical samples. A total of 4 260 autosomal CGIs that exhibited a minimum of 16 MePM in at least one tissue sample were selected for analysis. The P values were adjusted to the FDR analogue q-values using the Benjamini-Hochberg methods. The same statistical approach and the same set of 4 260 CGIs were used to determine the differentially methylated CGIs between plasma obtained from HCC patients and cancer-free individuals.
The Spearman's rank correlation coefficient (rho) was used to examine the correlation between matched tissue and plasma samples. To distinguish the cancer-associated hypermethylation from the background methylation in plasma, we selected 18 out of 41 HCC-detecting markers that have lowest methylation values in plasma of normal individuals (90th percentile < 4 uMePM), and the cancer-associated hypermethylation was defined as the uMePM values above the 90th percentiles.
A Z-score approach was used to normalize the plasma MCTA-Seq data. The Z-score of each HCC-detecting marker was defined as the number of SDs above the mean, where the SD and the mean were calculated according to the plasma samples from normal individuals.
All MCTA-Seq and RRBS data have been deposited to the NCBI under accession number GSE63775.
Acknowledgments
We thank Yun Zhang, Jing Sun, Yang Xu and Yin Jiang at the Peking University High-throughput Sequencing Center for excellent NGS supports. This work was supported by the National Natural Science Foundation of China (81472857 to LW, 81372604 to JRP and 21327808 to YYH).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Information of different CpG tandems.
Gel results of MCTA-Seq.
Selective amplification of the CGCGCGG sequence.
MCTA-Seq reproducibility and differential methylation.
Low detection limits of MCTA-Seq.
Quantitative performance of MCTA-Seq.
Comparison of MePM and uMePM.
Plasma ccfDNA amount.
The number of CGI loci detected in plasma cfDNA.
Type I and type II HCC-detecting markers.
Tissue-specific methylation of the type II markers.
Comparison of MCTA-Seq and RRBS.
Tumor size and the methylation marker comparison.
Comparison with the methylation array data.
Type I markers correlation.
Comparison of different models for classifier II.
Sensitivity of MCTA-Seq for detecting HCC.
Methylation marker and AFP Comparison.
Sequencing information.
Clinical characteristic of HCC patients and control subjects.
Clinical, histological data and MCTA-Seq analysis of HCC patients.
References
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 2011; 11:726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer 2004; 4:143–153. [DOI] [PubMed] [Google Scholar]
- Pogribny IP, Rusyn I. Role of epigenetic aberrations in the development and progression of human hepatocellular carcinoma. Cancer Lett 2014; 342:223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011; 11:426–437. [DOI] [PubMed] [Google Scholar]
- Wong IH, Lo YM, Zhang J, et al. Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Res 1999; 59:71–73. [PubMed] [Google Scholar]
- Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet 2010; 11:191–203. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Lin Z, Zhou L, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res 2006; 16:383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008; 454:766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc 2011; 6:468–481. [DOI] [PubMed] [Google Scholar]
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65:87–108. [DOI] [PubMed] [Google Scholar]
- Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology 2011; 53:1020–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefaniuk P, Cianciara J, Wiercinska-Drapalo A. Present and future possibilities for early diagnosis of hepatocellular carcinoma. World J Gastroenterol 2010; 16:418–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivioja T, Vaharautio A, Karlsson K, et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods 2012; 9:72–74. [DOI] [PubMed] [Google Scholar]
- Li M, Chen WD, Papadopoulos N, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat Biotechnol 2009; 27:858–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofton-Day C, Model F, Devos T, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem 2008; 54:414–423. [DOI] [PubMed] [Google Scholar]
- Warren JD, Xiong W, Bunker AM, et al. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med 2011; 9:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusis EA, Watson MA, Chicoine MR, et al. Integrative genomic analysis identifies NDRG2 as a candidate tumor suppressor gene frequently inactivated in clinically aggressive meningioma. Cancer Res 2005; 65:7121–7126. [DOI] [PubMed] [Google Scholar]
- Patra SK, Szyf M. DNA methylation-mediated nucleosome dynamics and oncogenic Ras signaling: insights from FAS, FAS ligand and RASSF1A. FEBS J 2008; 275:5217–5235. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer 2009; 9:773–784. [DOI] [PubMed] [Google Scholar]
- Illingworth R, Kerr A, Desousa D, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol 2008; 6:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MA, Tiirikainen M, Kwee S, Okimoto G, Yu H, Wong LL. Elucidating the landscape of aberrant DNA methylation in hepatocellular carcinoma. PLoS One 2013; 8:e55761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA Data Portal Overview. Available at https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014; 6:224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedde T, Kaplowitz N, Schwabe RF. Cell death and cell death responses in liver disease: mechanisms and clinical relevance. Gastroenterology 2014; 147:765–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproul D, Kitchen RR, Nestor CE, et al. Tissue of origin determines cancer-associated CpG island promoter hypermethylation patterns. Genome Biol 2012; 13:R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information of different CpG tandems.
Gel results of MCTA-Seq.
Selective amplification of the CGCGCGG sequence.
MCTA-Seq reproducibility and differential methylation.
Low detection limits of MCTA-Seq.
Quantitative performance of MCTA-Seq.
Comparison of MePM and uMePM.
Plasma ccfDNA amount.
The number of CGI loci detected in plasma cfDNA.
Type I and type II HCC-detecting markers.
Tissue-specific methylation of the type II markers.
Comparison of MCTA-Seq and RRBS.
Tumor size and the methylation marker comparison.
Comparison with the methylation array data.
Type I markers correlation.
Comparison of different models for classifier II.
Sensitivity of MCTA-Seq for detecting HCC.
Methylation marker and AFP Comparison.
Sequencing information.
Clinical characteristic of HCC patients and control subjects.
Clinical, histological data and MCTA-Seq analysis of HCC patients.