Abstract
Cryptophytes are an ecologically important group of largely photosynthetic unicellular eukaryotes. This lineage is of great interest to evolutionary biologists because their plastids are of red algal secondary endosymbiotic origin and the host cell retains four different genomes (host nuclear, mitochondrial, plastid, and red algal nucleomorph). Here, we report a comparative analysis of plastid genomes from six representative cryptophyte genera. Four newly sequenced cryptophyte plastid genomes of Chroomonas mesostigmatica, Ch. placoidea, Cryptomonas curvata, and Storeatula sp. CCMP1868 share a number of features including synteny and gene content with the previously sequenced genomes of Cryptomonas paramecium, Rhodomonas salina, Teleaulax amphioxeia, and Guillardia theta. Our analysis of these plastid genomes reveals examples of gene loss and intron insertion. In particular, the chlB/chlL/chlN genes, which encode light-independent (dark active) protochlorophyllide oxidoreductase (LIPOR) proteins have undergone recent gene loss and pseudogenization in cryptophytes. Comparison of phylogenetic trees based on plastid and nuclear genome data sets show the introduction, via secondary endosymbiosis, of a red algal derived plastid in a lineage of chlorophyll-c containing algae. This event was followed by additional rounds of eukaryotic endosymbioses that spread the red lineage plastid to diverse groups such as haptophytes and stramenopiles.
Keywords: plastid genome, cryptophyte, horizontal gene transfer
Introduction
The cryptophyte algae (= cryptomonads) are an evolutionarily distinct and ecologically important unicellular eukaryotic lineage inhabiting marine, brackish water, and freshwater environments (Graham and Wilcox 2000; Shalchian-Tabrizi et al. 2008). Cryptophytes are mostly photosynthetic with plastids that contain chlorophyll-a and -c, as well as phycobilins as accessary pigments. They are comprised of brown-, red-, or blue-green-colored photosynthetic groups (Hill and Rowan 1989; Deane et al. 2002; Hoef-Emden 2008), colorless nonphotosynthetic groups including Cryptomonas paramecium with a secondarily reduced plastid genome (Donaher et al. 2009), and heterotrophic Goniomonas species that lack plastids (McFadden et al. 1994; Hoef-Emden et al. 2002; Hoef-Emden and Melkonian 2003; von der Heyden et al. 2004; Hoef-Emden 2008).
Cryptophyte plastids are bounded by two inner and two outer envelope membranes. The outermost membrane is continuous with the endoplasmic reticulum (i.e., chloroplast ER; CER), which is connected to the outer membrane of the nuclear envelope. The nucleomorph, the remnant nucleus derived from the red algal progenitor of the cryptophyte plastid, is located between the two pairs of inner and outer plastid membranes (Gilson et al. 1997; Archibald 2007; Curtis et al. 2012). Cryptophyte cells contain four genomes: host-derived nuclear and mitochondrial genomes, and plastid and nucleomorph genomes of red algal endosymbiotic origin. Given this unusual feature, cryptophytes provide direct evidence of secondary endosymbiotic events occurring between phagotrophic and photoautotrophic eukaryotes (Douglas et al. 1991; McFadden 1993), a process that presumably occurred in several other protist lineages (e.g., euglenoids, chlorarachniophytes; Bhattacharya and Medlin 1995; Delwiche and Palmer 1996).
To date, the plastid genomes of three photosynthetic cryptophytes (Douglas and Penny 1999; Khan et al. 2007; Kim et al. 2015), and one colorless cryptophyte (Donaher et al. 2009) have been reported. The overall organization of these genomes is conserved and comprises a large single copy region (LSC), a small single copy region (SSC), and two inverted repeats (IR) with ribosomal RNA operons. The plastid genomes of cryptophytes range in size from ∼77 kilobase pairs (Kbp) in the colorless, nonphotosynthetic cryptophyte Cryptomonas paramecium to ∼135 Kbp in the phototrophs, and have a rich gene content (177–180 genes). These numbers are comparable to the plastid genomes of the chorophyll-c containing haptophytes (144 genes) and stramenopiles (137–197 genes) but less than the gene-rich red algae (232–251 genes) (Lee et al. 2016). Introns are rare in cryptophyte plastid genomes, with reports of an intron in the psbN and groEL genes in the genus Rhodomonas (Maier et al. 1995; Khan et al. 2007).
Here we present complete plastid genome sequences from the blue-green colored cryptophytes Chroomonas placoidea and Ch. mesostigmatica, the brown colored Cyptomonas curvata, and the red colored Storeatula sp. CCMP 1868. We carried out a detailed analysis of their genome structures and coding capacities relative to four published cryptophyte plastid genome sequences (Cryptomonas paramecium, Guillardia theta, Rhodomonas salina, and Teleaulax amphioxeia). Furthermore, to better understand the phylogenetic relationships and evolutionary history of algae with red alga-derived plastids, we reconstructed a phylogenetic tree using 88 protein coding genes from the currently available plastid genome data from a total of 56 species including 8 cryptophytes, 5 haptophytes, 20 stramenopiles, 2 alveolates, and 14 red algae. We also investigated the extent to which the phylogeny of plastid genes is congruent with those previously inferred from nuclear genes. Our results highlight both conserved and variable features of plastid genomes amongst the cryptophyte algae. Whereas genome architecture and gene composition are generally conserved, several examples of gene loss and intron gain were identified. Our results provide important general insights into the evolutionary history of organelle genomes and a more fine-scale understanding of cryptophyte evolution.
Materials and Methods
DNA Isolation and Sequencing
Cultures of Chroomonas placoidea CCAP 978/8, Ch. mesostigmatica CCMP 1168 and Storeatula sp. CCMP 1868 were obtained from the Culture Collection of Algae and Protozoa (CCAP) and the National Center for Marine Algae and Microbiota (NCMA), respectively. Cryptomonas curvata was collected from Cheongyang Pond, Cheongyang, Korea (36° 30′ N, 126° 47′ E), established as a clonal culture and available strain FBCC300012D (= strain CNUKR) from the Freshwater Bioresources Culture Collection at the Nakdong-Gang National Institute of Biological Resources, Korea. All cultures were grown in AF-6 medium (Watanabe and Hiroki 1997) with distilled water for the freshwater strain (Cryptomonas curvata) or distilled seawater for marine strains, and were maintained at 20 °C under conditions of a 14:10 light:dark cycle with 30 µmol photons·m−2 s−1 from cool white fluorescent tubes. All cultures were derived from a single-cell isolate for unialgal cultivation and genomic DNA extraction and sequencing. For Ch. placoidea and Storeatula sp. CCMP 1868, DNA was extracted using the QIAGEN DNEasy Blood Mini Kit (QIAGEN, Valencia, CA) following the manufacturer’s instructions, and next-generation sequencing (NGS) was carried out using the Ion Torrent PGM platform (ThermoFisher Scientific, San Francisco, CA) in the lab of Sungkyunkwan University. Sequencing libraries were prepared using the Ion Xpress Plus gDNA Fragment Library Preparation Kit for 200- or 400-bp-sized sequencing library preparation and the Ion OneTouch 200 or 400 Template Kit (ThermoFisher Scientific, San Francisco, CA) according to the manufacturer’s protocol. The genomes were sequenced on an Ion Torrent Personal Genome Machine (PGM) using the Ion PGM sequencing 200 or 400 Kit (ThermoFisher Scientific, San Francisco, CA). For Ch. mesostigmatica, DNA extraction was carried out as described in Moore et al. (2012); the plastid genome was sequenced using a combination of 454 pyrosequencing (GS FLX titanium reagents) and Illumina (GAIIx) technologies.
Genome Assembly and Annotation
Plastid genome assemblies and annotations followed procedures used by Song et al. (2016). The data were trimmed (i.e., base = 80 bp, error threshold = 0.05, n ambiguities = 2) prior to de novo assembly with the default option (automatic bubble size, minimum contig length =1,000 bp). The raw reads were assembled using the MIRA4 (http://mira-assembler.sourceforge.net/docs/DefinitiveGuideToMIRA.html) and SPAdes 3.7 (http://bioinf.spbau.ru/spades) assembler. Raw reads were then mapped to the assembly contigs (similarity = 95%, length fraction = 75%), and regions with no evidence of short-read data were removed (up to 1,000 bp). The assembled contigs were determined to correspond to the plastid genome according to several criteria: 1) BLAST searches of commonly known plastid genes against the entire assembly resulted in hits to these contigs and 2) a genome size consistent with other photosynthetic cryptophyte plastid genomes, which range from 121 Kbp (Guillardia theta NC000926) to 136 Kbp (Rhodomonas salina NC009573). Plastid genome-derived contigs were then manually aligned in the Genetic Data Environment (MacGDE2.5) program (Smith et al. 1994) to produce a consensus sequence.
A database of protein coding genes, rRNA, and tRNA genes was created using all previously sequenced cryptophyte plastid genomes. Preliminary annotation of protein coding genes was performed using GeneMarkS (http://opal.biology.gatech.edu/genemarks.cgi). The final annotation file was checked in Geneious Pro 9.1.3 (http://www.geneious.com/) using the ORF Finder with genetic code 11 (Bacterial, Archaeal and Plant Plastid Code). The predicted ORFs were checked manually and the corresponding ORFs (and predicted functional domains) in the genome sequence were annotated.
To identify tRNA sequences, the plastid genome was submitted to the tRNAscan-SE version 1.21 server (http://lowelab.ucsc.edu/tRNAscan-SE/). The genome was searched with the default settings using the “Mito/Chloroplast” model. To identify rRNA sequences, a set of known plastid rRNA sequences was extracted from the published plastid genomes of cryptophytes and used as a query sequence to search in the new genome data using BLASTn. We used RNAweasel (http://megasun.bch.umontreal.ca/cgi-bin/RNAweasel/RNAweaselInterface.pl) to determine the types of introns that were present. Physical maps were designed with the OrganellarGenomeDRAW program (http://ogdraw.mpimp-golm.mpg.de/).
Gene Arrangement Comparisons
Four published cryptophyte plastid genome sequences (Cryptomonas paramecium CCAP 977/2a, Donaher et al. 2009; Guilardia theta CCMP 2712, Douglas and Penny 1999; Rhodomonas salina CCMP 1319, Khan et al. 2007; Teleaulax amphioxeia HACCP CR01, Kim et al. 2015) were downloaded from GenBank. An additional plastid genome is available from GenBank under the name of Guillardia theta CCMP 2712 (KT428890, Tang and Bi 2016), however, the gene sequences are very different from those of the previously reported G. theta CCMP 2712 (Douglas and Penny 1999; Curtis et al. 2012). Therefore, we did not include this genome in our study. For structural and synteny comparisons, the genomes were aligned using Mauve Genome Alignment version 2.2.0 (Darling et al. 2004) with default settings. To aid in visualization, we arbitrarily designated the beginning of the trnY gene marker to rpl19 direction as position 1 in each genome.
Phylogenetic Analysis
Phylogenetic analyses were carried out on data sets created by combining 88 proteins encoded by 56 plastid genomes, including those of 8 cryptophytes, 5 haptophytes, 20 stramenopiles, 2 alveolates, and 14 red algae (supplementary table S1, Supplementary Material online). The sequences of six Viridiplantae and one glaucophyte species were used as outgroup taxa to root the tree. The data were concatenated (16,878 amino acid sequences) and manually aligned using MacGDE2.5 (Smith et al. 1994). For the RNA operon (16S-trnI-trnA-23S rDNA) phylogeny, the data were concatenated into 4,046 nucleotides from plastid genome sequences in 38 taxa including 8 cryptophytes, 5 haptophytes, 1 rappemonad, 13 stramenopiles, 12 rhodophytes, and 16 outgroup taxa including 2 glaucophytes, 9 chlorophytes, and 5 cyanobacteria.
Maximum likelihood (ML) phylogenetic analyses were conducted using RAxML version 8.0.0 (Stamatakis 2014) with the Le and Gascuel gamma (LG + GAMMA) model (Le and Gascuel 2008) for amino acid data chosen by ProtTest 3 (Darriba et al. 2011) and the general time-reversible plus gamma (GTR + GAMMA) model for nucleotide data. We used 1,000 independent tree inferences using the -# option to identify the best tree. The model parameters with gamma correction values and the proportion of invariable sites in the combined data set were obtained automatically by the program. ML bootstrap support values (MLB) were calculated using 1,000 replicates with the same substitution model. To reduce calculation time, ML phylogenetic trees for lineage-specific genes were inferred using IQ-TREE Ver. 1.5.2 (Nguyen et al. 2015) with 1,000 bootstrap replications (e.g., supplementary figs. S2–S12, Supplementary Material online). Evolutionary models for each tree were automatically selected by the –m LG + I+G option incorporated in IQ-TREE.
Bayesian analyses were run using MrBayes 3.2.6 (Ronquist et al. 2012) with a random starting tree, two simultaneous runs (nruns = 2) and four Metropolis-coupled Markov chain Monte Carlo (MC3) algorithms for 2×106 generations, with one tree retained every 1,000 generations (e.g., supplementary fig. S13, Supplementary Material online). The burn-in point was identified graphically by tracking the likelihoods (Tracer v.1.6; http://tree.bio.ed.ac.uk/software/tracer/). The first 500 trees were discarded, and the remaining 1,501 trees were used to calculate the posterior probabilities (PP) for each clade. Additionally, the “sump” command in MrBayes was used to confirm convergence. This analysis was repeated twice independently; identical topologies were obtained. Trees were visualized using FigTree v.1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Results and Discussion
General Features of Cryptophyte Plastid Genomes
Four new plastid genomes (ptDNA) were sequenced from representatives of three groups of differentially colored cryptophyte algae: red Storeatula, blue-green Chroomonas, and brown-colored Cryptomonas species (table 1). These ptDNAs were then compared with previously reported data from three red-colored cryptophytes, Guillardia, Teleaulax, Rhodomonas, and the colorless Cryptomonas paramecium (which has a secondarily reduced plastid genome). The plastid genome sizes of the cryptophyte algae ranged from ∼77 Kbp (Cr. paramecium) to ∼141 Kbp (Storeatula sp. CCMP 1868). The overall GC content ranged from 32% to 38.1%, similar to those of other chromists and red algae (Kowallik et al. 1995; Douglas and Penny 1999; Ohta et al. 2003; Sánchez Puerta et al. 2005). Photosynthetic cryptophyte plastid genomes shared a core set of 143 protein-coding genes, 3 rRNAs, and 30 tRNAs. All cryptophyte plastid genomes encode inverted repeat (IR) regions with 2 rRNA operons and 44 ribosomal genes, except for the loss of 1 rRNA operon and the rps6 and rpl32 genes in the colorless Cr. paramecium (Donaher et al. 2009). Percentages of intergenic sequences in the ptDNAs ranged from 20.3% (Storeatula sp. CCMP 1868) to 12.2% (G. theta). Minimal variation was found in the tRNA gene content. However, trnL(CAA) appears to be absent in Storeatula sp., whereas Ch. placoidea and Ch. mesostigmatica have a unique isotype trnV(GAC) (see table 2).
Table 1.
Characteristics of Cryptophyte Plastid Genomes Analyzed in This Study
| General Characteristics | Guilardia theta CCMP 2712 | Teleaulax amphioxeia HACCP CR01 | Rhodomonas salina CCMP1319 | Storeatula species CCMP1868 | Chroomonas placoidea CCAP978/8 | Chroomonas mesostigmatica CCMP 1168 | Cryptomonas curvata FBCC300012D | Cryptomonas paramecium CCAP977/2a |
|---|---|---|---|---|---|---|---|---|
| Plastid color | Red | Red | Red | Red | Green | Green | Brown | Colorless |
| Size (bp) | 121,524 | 129,774 | 135,854 | 140,953 | 139,432 | 139,403 | 128,285 | 77,717 |
| G+C (%) | 33.0% | 34.2% | 34.8% | 32.0% | 35.8% | 35.8% | 35.3% | 38.1% |
| Intergenic space (%) | 14,792 (12.2%) | 20,108 (15.5%) | 23,767 (17.5%) | 28,638 (20.3%) | 27,796 (19.9%) | 25289 (18.1%) | 20,442 (15.9%) | 9,917 (12.8%) |
| Total gene (include RNAs) | 183 | 179 | 183 | 187 | 186 | 189 | 184 | 114 |
| No. of protein-coding genes | 147 | 143 | 146 | 149 | 149 | 150 | 147 | 82 |
| Unknown ORFs | 1 (orf91, orf555, orf147, orf164/orf335 | 2 (orf77=orf282, ycf20) | 4 | 5 | 1 | |||
| Ribosomal proteins | 44 | 44 | 44 | 44 | 44 | 44 | 44 | 42 |
| LIPOR gene | – | – | chlB a, chlL a, chlN a | chlB, chlL, chlN, | chlB a, chlL a, chlN a, | chlB a, chlL a, chlN a, | chlB, chlN, chlL | – |
| tRNAs | 30 | 30 | 31 | 30 | 32 | 32 | 31 | 29 |
| rRNA operons | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 |
| Introns | – | – | 1 | 4 | 4 | 4 | 1 | – |
| Type | – | – | Group II | Group II | Group II | Group II | Group II | |
| Gene with intron | psbN | groEL/ftrB/orf27/ psbN | groEL/minD/ petG/psbN | groEL/ minD/petG/psbN | psbN | |||
| Pseudogene | – | – | – | – | – | – | – | atpF |
| Missing | – | – | – | – | dnaB, dnaX, ycf26 | dnaB, dnaX, ycf26 | dnaB, dnaX, ycf26 | pet, psa, psb |
| GenBank accession | NC000926 | NC027589 | NC009573 | KY856940 | KY856941 | KY860574 | KY856939 | NC013703 |
Note.—(a) present as pseudogene. LIPOR; the light-independent protochlorophyllide oxidoreductases gene in plastid genome.
Table 2.
tRNAs Present in Cryptophyte Plastid Genomes
| trnA (TGC) | trnC (GCA) | trnD (GTC) | trnE (TTC) | trnF (GAA) | trnG (GCC) | trcG (TCC) | trnH (GTG) | trnI (GAT) | trnK (TTT) | trnL (CAA) | trnL (TAA) | trnL (TAG) | trnfM (CAT) | trnM (CAT) | trnN (GTT) | trnP (TGG) | trnQ (TTG) | trnR (ACG) | trnR (CCG) | trnR (TCT) | trnS (GCT) | trnS (GGA) | trnS (TGA) | trnT (TGT) | trnV (GAC) | trnV (TAC) | trnW (CCA) | trnY (GTA) | Total | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rhodomonas salina | CCMP1319 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 31 | ||||
| Guillardia theta | CCMP2712 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 30 | |||
| Teleaulax amphioxeia | HACCP-CR01 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 30 | ||||
| Storeatula sp. | CCMP1868 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 30 | ||||
| Chroomonas mesostigmatica | CCMP1168 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 32 | ||
| Chroomonas placoidea | CCAP978/8 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 32 | ||
| Cryptomonas curvata | FBCC300012D | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 31 | |||
| Cryptomonas paramecium | CCAP977/2a | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 29 |
Highly Conserved Cryptophyte Plastid Genome Structure
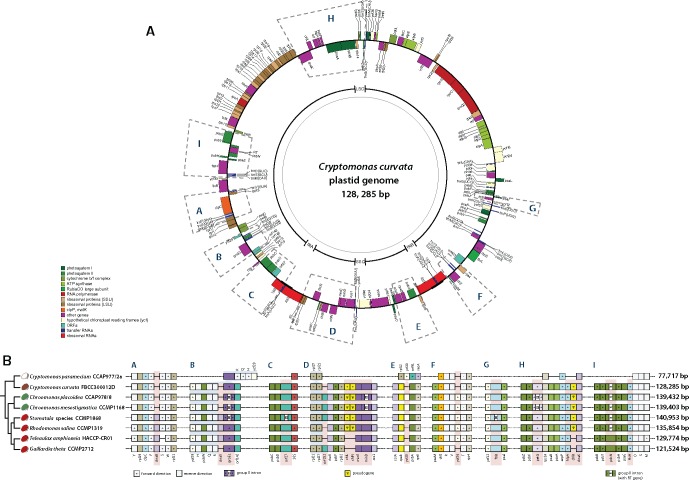
The four newly determined plastid genomes showed a high degree of structural conservation when compared with the representative species, Cryptomonas curvata FBCC300012D (fig. 1). Gene order among the photosynthetic cryptophyte ptDNAs was constrained. The most obvious feature in this regard was the regions of ribosomal protein genes and the rpo and atp gene clusters that were identical among all compared cryptophyte genomes. Based on the Mauve pairwise genome alignment analysis, additional regions of synteny were detected; gene order was found to be essentially identical amongst all cryptophytes, including the genome of the colorless Cr. paramecium, which has undergone numerous gene losses (fig. 2 and supplementary fig. S1, Supplementary Material online).
Fig. 1.
—Circular map of the plastid genome of Cryptomonas curvata FBC300012D showing nine syntenic regions. (A) The gene content and arrangement of the photosynthetic cryptophyte genomes are identical with the exception of the nine syntenic regions shown as (A–I). The protein coding genes, rRNA and tRNA genes are labeled inside or outside of the circle. The genes are color-coded according to the functional categories in the index. (B) Linear form of the nine syntenic regions indicates the lineage-specific gene losses and intron insertions among cryptophytes.
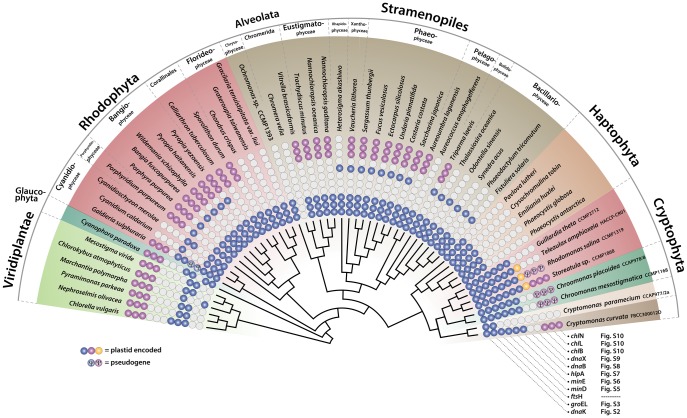
Fig. 2.
—Distribution of 11 patchily distributed genes in the major photosynthetic eukaryotic groups. These genes show outright gene losses, pseudogenes, and intron insertions. The cladogram was built using a concatenated data set of 88 plastid genes (fig. 3 and supplementary table S1, Supplementary Material online). The 11 target genes are color-coded: purple = three LIPOR genes, yellow = dnaX gene, and dark blue = dnaK, groEL, ftsH, minD/E, hlpA, and dnaB genes. The open white circles indicate gene absence in plastid genomes.
Distribution of Gene Losses, Pseudogenes, and Intron Insertions in Photosynthetic Eukaryotes
Although the plastid genomes of cryptophytes are highly conserved in structure and content, nine variable syntenic regions were identified (fig. 1). Among these regions, gene losses, pseudogenes, and intron insertions were found, particularly in 11 genes (dnaB, dnaX, dnaK, groEL, hlpA, minD, minE, and ftsH) within the cryptophyte species (fig. 2). To extend the distribution of these genes, we surveyed the plastid genomes of major photosynthetic eukaryotic groups including primary (rhodophytes, glaucophytes, and Viridiplantae) and red algal derived secondary plastids (stramenopiles, alveolates, haptophytes, and cryptophytes) from GenBank. Presence/absence of these genes varies widely, suggesting that deletions involving these 11 genes occurred not only in cryptophytes, but also in other major photosynthetic eukaryotic groups. For instance, several pseudogenes within minD/E and chlB/L/N suggest an ongoing process of plastid gene loss.
dnaK and groEL
The chaperone protein coding genes dnaK (a member of the hsp70 family; Wang and Liu 1991) and groEL (the chaperonin family; Ellis and van der Vies 1991) were found in the plastid genomes of cryptophytes, haptophytes, stramenopiles, rhodophytes, and glaucophytes (fig. 2 and supplementary figs. S2 and S3, Supplementary Material online). In contrast, these two genes are absent from the chloroplast genome of green algae and land plants and have been transferred to the nucleus via endosymbiotic gene transfer (EGT), with the evolution of one additional homolog (cpn60) (fig. 2 and supplementary fig. S3, Supplementary Material online). EGT or outright gene loss from the plastid genome is common among algae and plants. The nucleomorph genome of cryptophytes encodes a cpn60 homolog (Douglas et al. 2001; Tanifuji et al. 2011; Moore et al. 2012), a feature that is shared with the green algal derived nucleomorph genome of the chlorarachniophyte Bigelowiella natans (Gilson et al. 2006).
The groEL gene in cryptophytes contains group II introns in three strains of the genus Rhodomonas (Maier et al. 1995; Khan and Archibald 2008). These introns are inserted in three different locations in Rhodomonas sp. CCMP1178, R. salina Maier strain, and cryptophyte species CCMP2045, but three other Rhodomonas species, R. salina CCMP1319, R. baltica RCC350, and Rhodomonas sp. CCMP1170, lack the intron (Khan and Archibald 2008). We found similar results with different intron locations in other genera (supplementary fig. S3C, Supplementary Material online). The groEL gene of the two blue-green-colored cryptophytes Ch. placoidea and Ch. mesostigmatica has a group II intron (with a reverse transcriptase gene) in the same position (after amino acid 41 of the groEL gene) as that of Rhodomonas sp. CCMP1178, whereas Storeatula sp. CCMP 1868 has a group II intron (apparently ORF-free) in the same position (again, after amino acid 41 of groEL). The ORFs of group II introns in groEL showed sequence similarity with the N-terminal domain of putative reverse transcriptases (supplementary fig. S3C, Supplementary Material online, RVT_N, pfam13655) and shared similarity with an intron-encoded protein (IEP) in mat1a of the red alga Bangiopsis subsimplex (supplementary fig. S4, Supplementary Material online). With respect to the evolution of cryptophyte introns, it is noteworthy that the groEL introns are distinct from one another, with or without ORFs and in different locations in various organisms including stramenopiles, rhodophytes, Viridiplantae, euglenophytes, cyanobacteria, and bacteria (Khan and Archibald 2008; this study), suggesting multiple independent origins.
ftsH
The ftsH gene encodes a AAA metalloprotease that degrades membrane-bound proteins (Chiba et al. 2000; Lindahl et al. 2000). With the exception of glaucophytes, ftsH is present in most plastid genomes; i.e., rhodophytes, stramenopiles, haptophytes, cryptophytes (except the colorless Cryptomonas paramecium, figs. 1 and 2), and most green plant lineages (Prasinophyceae, Ulvophyceae, Trebouxiophyceae, Chlorophyceae, and Charophyceae). It is, however, absent in the plastid genomes of land plants (Embryophyta) (Martin et al. 1998), where ftsH is encoded in the nuclear genome. This gene was likely transferred to the host nucleus from the plastid genome after the Charophyta–Embryophyta split (de Vries et al. 2013).
minD and minE
The min genes are required to prevent formation of DNA-less “minicells” during division. The minD and minE genes are present in several algal plastid genomes (Turmel et al. 1999; Lemieux et al. 2000), but absent from most red algae, with the exception of Galdieria sulphuraria where it is a plastid-encoded pseudogene (fig. 2; Jain et al. 2014). Interestingly, both protein-coding genes are located in the plastid genome of all photosynthetic cryptophytes (fig. 1D), suggesting that plastid minD and minE were present in the red-algal derived endosymbiont. In cryptophytes, haptophytes and Viridiplantae, minD is encoded in the plastid genome, and minE is only found in cryptophytes and Chlorella vulgaris (sole case in green algae), as a remnant of the ancestral minCDE operon (fig. 2 and supplementary figs. S5 and S6, Supplementary Material online). Both minD and E genes are missing in Cryptomonas paramecium (Donaher et al. 2009). In other algal groups, with the exception of cryptophytes and haptophytes, the distribution pattern of minD suggests this gene was transferred to the nuclear genome on independent occasions in the ancestors of algae with red algal-derived plastids, similar to the situation in green algae and land plants (Miyagishima et al. 2012). However, minE has thus far not been found in plant or most algal nuclear genomes, implying that its function has been replaced or lost. The plastid-encoded minD gene with an intron is present in two blue-green-colored cryptophytes Chroomonas placoidea and Ch. mesostigmatica (fig. 1D). These Chroomonas minD intron sequences show 72% nucleotide similarity (e value = 2e-47) to the groEL intron of Rhodomonas sp. CCMP1178 (GenBank EU305621), suggesting a common origin.
hlpA
The hlpA gene (encoding a chromatin associated architectural protein) behaves as a functional homolog of E. coli DNA-binding (Grasser et al. 1997) and DNA-packaging proteins (histone-protein DNA binding and -bending HU- and HMG1-like proteins). Among algae containing red-algal derived plastids, the gene is uniquely found in the plastid genomes of all photosynthetic cryptophytes (figs. 1H and 2 and supplementary fig. S7, Supplementary Material online). This gene is also found in Cyanidioschyzon merolae and Galdieria sulphuraria as a hupA gene (Hu homologous), but not in Cyanidium caldarium. The apicomplexan hlpA gene is present in the nucleus (Hall et al. 2002; Nierman 2005; Pain 2005). Given this distribution pattern, we postulate that the hlpA gene was most likely located in the plastid genome of the red algal progenitor.
dnaB
The dnaB (a DNA helicase) gene is involved in organelle division (Douglas and Penny 1999; Ohta et al. 2003) and found in the plastid genome of cryptophytes, stramenopiles, and nonflorideophycean red algae (supplementary fig. S8, Supplementary Material online). In cryptophytes, dnaB was restricted to the red-colored G. theta, T. amphioxeia, R. salina, and Storeatula sp. CCMP 1868 (figs. 1A and 2 and supplementary fig. S8, Supplementary Material online). In phylogenetic analyses, the red alga Galdieria sulphuraria (Cyanidiophyceae) diverged at the base of the algal dnaB gene tree. The cryptophyte clade formed a sister group relationship with the red algal clade (i.e., Bangiophyceae and Porphyridiophyceae). In stramenopiles, dnaB is present only in Bacillariophyceae (except Synedra acus), Phaeophyceae, Raphidophyceae, and Xanthophyceae, but absent in Pelagophyceae, Eustigmatophyceae, and Chrysophyceae (fig. 2). The dnaB gene is located in the plastid genome of Cyanidium caldarium and Cyanidioschyzon merolae, but the sequence similarity is very low, suggesting nonorthologous replacement of the gene (data not shown). Taken together, the dnaB gene appears to have been present in the red algal common ancestor, but lost independently in many red-algal derived plastid genomes.
dnaX
The dnaX gene encodes the tau/gamma components of bacterial DNA polymerase III (Blinkova et al. 1993; Dallmann and McHenry 1995). Considering all sequenced cryptophyte plastid genomes, we found that the dnaX gene is restricted to the three red-colored cryptophytes, T. amphioxeia, R. salina, and Storeatula sp. CCMP 1868 (fig. 2). A recent phylogenetic analysis (Kim et al. 2015) reported that the dnaX gene was directly acquired from a bacterial lineage through horizontal gene transfer (HGT) (i.e., the donor is related to a termite symbiont, Endomicrobium proavitum, WP_052570901) in the ancestor of the red-colored Storeatula/Rhodomonas/Teleaulax lineage (supplementary fig. S9, Supplementary Material online).
LIPOR
The light-independent (or “dark active”) protochlorophyllide oxidoreductase (LIPOR) genes that are involved in the light-independent synthesis of chlorophyll (Shi and Shi 2006) are present in some cryptophyte plastid genomes. LIPOR arose in anoxygenic photosynthetic bacteria, likely evolving from a nitrogenase (Fujita and Bauer 2003; Muraki et al. 2010). In extant cyanobacteria, both POR (light-dependent protochlorophyllide oxidoreductase) and LIPOR genes are present. In eukaryotic algae, the gene encoding POR appears to have been transferred to the host nucleus, whereas LIPOR genes remain in the plastid (Hunsperger et al. 2015). However, the three LIPOR genes (chlB, chlL, and chlN) are not universally distributed in plastids and have been independently lost in many cases (fig. 2 and supplementary fig. S8, Supplementary Material online). In cryptophytes, these genes occur as pseudogenes (ΨchlB, ΨchlL, and ΨchlN) in R. salina, Ch. placoidea, and Ch. mesostigmatica (Khan et al. 2007; Fong and Archibald 2008; this study). However, we found putatively functional chlB, chlL, and chlN in Cr. curvata, and Storeatula sp. CCMP 1868 (figs. 1D and H and 2). The discovery of LIPOR subunit genes in cryptophyte plastid genomes is an example of gene deletion in action: some cryptophyte species retain a full gene set (e.g., Cr. curvata and Storeatula sp. CCMP 1868), some species are in the process of losing these genes (e.g., R. salina, Ch. placoidea, and Ch. mesostigmatica), whereas others have completely lost them (e.g., G. theta, T. amphioxeia, and Cr. paramecium). Nuclear genome data could provide evidence of EGT and help explain cases of plastid gene loss.
Intron Insertion
Ferredoxin thioreductase (ftrB), a photosynthetic regulator and electron transfer protein, is present in all photosynthetic cryptophyte plastid genomes (fig. 1G). The ftrB gene of Storeatula sp. CCMP 1868 was unique in containing an intron (672 nt) (fig. 1G). This intron nucleotide sequence showed no significant similarity to genes of other organisms, but showed 71.5% similarity (e value = 2.6e-13) to 189 nt of intron sequence in the groEL gene of Rhodomonas sp. CCMP1178 (GenBank EU305621).
The petG gene encodes cytochrome b6/f complex subunit V that mediates electron transfer between photosystem II and photosystem I; It is present in most algal plastid genomes; i.e., red-algal derived plastids (including photosynthetic cryptophytes), glaucophytes and green algae. Interestingly, Chroomonas placoidea and Ch. mesostigmatica (but not other cryptophytes) have a group II intron in their petG gene (fig. 1H) that shares high nucleotide similarity (e value = 7e-63) to maturase/reverse-transcriptase domains (cd01651, pfam01348). Furthermore, the IEP product shares sequence similarity with reverse transcriptases encoded in the genomes of firmicute bacteria and genes of fungi, rhodophytes, stramenopiles, and Viridiplantae as intergenic ORFs in mitochondrial genomes (supplementary fig. S11, Supplementary Material online). Interestingly, the IEPs in the Ch. placoidea and Ch. mesostigmatica petG genes also appear closely related to ORFs in the plastid genes of green algae including Pyramimonas parkeae (atpB), Stichococcus bacillaris (psbB), Caulerpa filiformis, Gloeotilopsis sarcinoidea and Tydemania expeditionis (psaC) and Netrium digitus (psbE) (supplementary fig. S11, Supplementary Material online). Therefore, this “patchy” distribution of the petG group II intron is suggestive of several independent HGT events in different genic regions from diverse organisms.
In most cryptophyte species, with the exception of G. theta and T. amphioxeia, a reverse transcriptase coding region was found in an intronic region in the psbN gene that encodes one of the smaller subunits of photosystem II (fig. 1I; Khan et al. 2007; this study). Apart from cryptophytes, an intron-containing psbN gene has only been reported in the rhodophyte Porphyridium purpureum (Tajima et al. 2014). This intron is a remnant (or “pseudo”) ORF structure that has lost its IEP via sequence degeneration or excision (Perrineau et al. 2015). The psbN group II intron of cryptophytes is a unique feature among the red-algal derived plastids and shows strong similarity to the IEP-containing intron (mat1e in psbN) of the red alga Bangiopsis subsimplex (Lee et al. 2016, NC_031173). In our phylogenies, the reverse transcriptase gene within psbN grouped together with the groEL intron of cryptophyte species CCMP2045 and appears to have been acquired via HGT in a common ancestor of the Rhodomonas/Storeatula/Chroomonas/Cryptomonas lineages (supplementary fig. S12, Supplementary Material online). Based on the predicted relationships between these organisms (see below), this would imply one or more secondary losses of the intron in the psbN genes of Guillardia and Teleaulax.
Conserved Plastid ORFs
We found four conserved plastid ORFs in cryptophytes which showed lineage specific distributions. Of these, orf27 is located between the psbD and 16S rRNA genes of all cryptophytes (albeit, absent in the colorless Cr. paramecium), but only the Storeatula sp. CCMP1868 homolog contained a group II intron (fig. 1C). Using BLASTn, this intron had 68% similarity (e value = 2e-15) over 347 nt (from nucleotide 567 to 221 in reverse) to intron sequences in the groEL gene of Rhodomonas sp. CCMP1178 (GenBank EU305621). In the case of orf252, which is located between the rpl27 and rbcR genes in four species and is 252 amino acids long in G. theta, it is only 77 amino acids in Storeatula sp. CCMP1868, and 290 amino acids in Ch. placoidea and Ch. mesostigmatica (fig. 1D). These ORFs were located at the same position (fig. 1D) but with different amino acid compositions. The ycf20 gene is located between cpeB and psbA in Ch. placoidea, Ch. mesostigmatica, Storeatula sp. CCMP1868 and G. theta, but in Cr. paramecium the gene is between rpl12 and secA (fig. 1E). The ycf26 gene (uncharacterized sensor-like histidine kinase) is located between the chlI-trnR-trnV and trnT-rps4 gene clusters in Ch. placoidea, Ch. mesostigmatica, Storeatula sp. CCMP1868, R. salina and T. amphioxeia (fig. 1F). Taken together, these gene loses and intron insertions suggest independent evolutionary events in each algal plastid genome.
Phylogenetic Relationships among Major Red-Algal-Derived Plastids
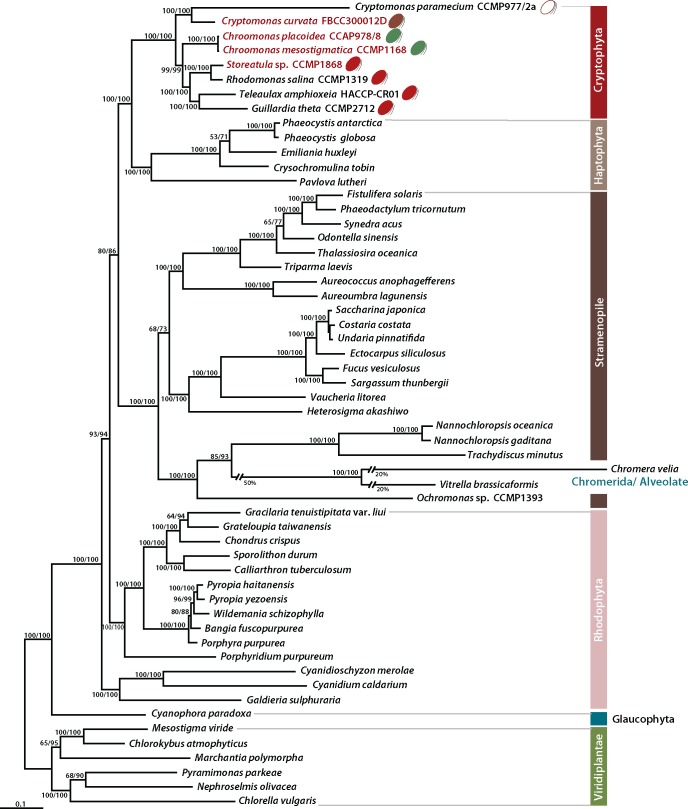
Phylogenomic analyses were done using a concatenated data set of 88 proteins encoded on 56 complete plastid genomes from alveolates, cryptophytes, haptophytes, and stramenopiles with 7 outgroup species (i.e., 6 Viridiplantae and 1 glaucophyte). The sequences of dinoflagellates were not included in this analysis due to the limited number of plastid-encoded genes on the distinctive mini-circular chromosomes in these species (Zhang et al. 1999; Howe et al. 2008). An ML tree was reconstructed using 16,878 amino acids (fig. 3) and a Bayesian tree was inferred from nucleotide sequences using the RNA operon (16S-trnA-trnI-23S, supplementary fig. S13, Supplementary Material online). A monophyletic cryptophyte clade was found to be strongly supported (MLB = 100%), and internal relationships among the species were also well resolved. The ML phylogeny using plastid genome data clearly shows that the brown-colored Cryptomonas curvata and the colorless Cryptomonas parmecium form a monophyletic group that is sister to the remaining taxa, whereas the two blue-green Chroomonas species were separated from four red-colored taxa (i.e., Storeatula, Rhodomonas, Teleaulax, and Guillardia) (fig. 3). Although our taxon sampling is limited, the phylogenomic analysis is consistent with published taxon-rich, single gene analyses of nuclear SSU rDNA, that recover three groups of phycoerythrin bearing red-colored cryptophyte species (I; Teleaulax, Geminigera, Plagioselmis, II; Hanusia, Guillardia, and III; Rhodomonas, Storeatula, Proteomonas) (Deane et al. 2002; Hoef-Emden 2008). Based on this grouping, group I (Teleaulax) and II (Guillardia) are separated from group III (Rhodomonas and Storeatula) in our tree with strong bootstrap support values. Because single gene phylogenies typically show low support (ca. 60% bootstrap) for the deepest branches (e.g., Deane et al. 2002; Hoef-Emden 2008), plastid genome data from a broader sampling of taxa will be needed to better resolve internal cryptophyte relationships.
Fig. 3.
—Phylogenetic tree of cryptophyte plastids and those of other photosynthetic eukaryotes. This tree was constructed using a data set of 88 concatenated proteins (16,878 amino acids). The numbers on each node represent RAxML bootstrap values (left) and ultrafast bootstrap approximation (UFBoot) using IQ-Tree (right). The scale bar indicates the number of substitutions/site.
The plastid genome phylogeny shows a strong monophyletic relationship of cryptophytes and haptophytes (MLB = 100/100%), which together, form a sister group relationship with the stramenopiles and alveolates (MLB = 80/86%). Two alveolate species (Chromera vella and Vitrella brassicaformis) are positioned inside of the stramenopiles that form a clade with two eustigmatophycean taxa (Nannochloropsis and Trachydiscus) and one chrysophycean species (Ochromonas sp.), consistent with previous results (Ševčíková et al. 2015). When we performed phylogenetic analysis of plastid rRNA sequence data, the cryptophytes grouped together with the haptophyte and rappemonad lineages (pp = 0.99, MLB = 66%, supplementary fig. S13, Supplementary Material online and Kim et al. 2011). As seen in earlier multigene analyses (e.g., Yoon et al. 2002a; 2002b), the monophyletic clade of chlorophyll-c containing lineages (cryptophytes/haptophytes/alveolates/stramenopiles) clustered together with one red algal subphylum, Rhodophytina (MLB = 93%), that is evolutionarily distinct from the early diverging Cyanidiophytina (Yoon et al. 2006). This result suggests that the red algal secondary endosymbiosis occurred after the divergence of the cyanidiophycean lineage (Khan et al. 2007; Donaher et al. 2009; Janouškovec et al. 2010; Kim et al. 2015; Ševčíková et al. 2015; Muñoz-Gómez et al. 2017).
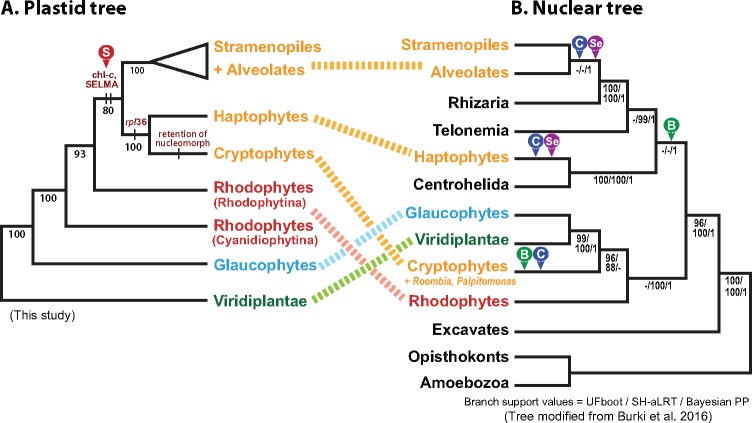
Based on the monophyletic relationship of chlorophyll-c containing groups and red algal plastids, the hypothesis of a single red alga-derived secondary endosymbiosis has been widely adopted (Stoebe and Kowallik 1999; Yoon 2002a; 2002b; Bhattacharya et al. 2004, 2007; Keeling 2004; Archibald and Keeling 2005; Archibald 2009; Kim et al. 2015). However, recent studies using nuclear genome data suggest the existence of multiple secondary and serial endosymbioses (Baurain et al. 2010; Burki et al. 2012, 2016; Stiller et al. 2014). Our plastid genome-based phylogeny was compared with a previously published phylogeny using 250 nuclear genes (fig. 4; Burki et al. 2016). In our plastid genome tree, the monophyly of chlorophyll-c groups and Rhodophytina (Yoon et al. 2006) supports the single secondary endosymbiosis scenario, whereby an ancestor of chlorophyll-c containing groups engulfed a red alga (event marked with “S” in the red arrow, fig. 4A). Within these lineages, the cryptophytes and haptophytes consistently cluster together as close relatives, which is a relationship supported by the presence of a unique bacterium-derived rpl36 gene in their plastid genomes (Rice and Palmer 2006). However, in recently published nuclear genome-based multi-gene phylogenies (which are far more controversial due to the poor resolution of protist relationships; e.g., Burki et al. 2016), cryptophytes and haptophytes appear to be distantly related to each other. The former is apparently associated with Archaeplastida, whereas the latter is sister to the SAR lineage (stramenopiles, alveolates, and Rhizaria) and their nonphotosynthetic relatives (fig. 4B, Baurain et al. 2010; Burki et al. 2012, 2016). Although the clade of haptophytes + SAR + other relatives is supported only by Bayesian analysis (see Burki et al. 2016), if true, the independent origin of the cryptophyte plastid from other chlorophyll-c containing groups is a possible conclusion. Because the nonphotosynthetic relatives (i.e., Centrohelida, Rhizaria, and Telonemia) are intermingled with the haptophytes and stramenopiles/alveolates, two independent origins of plastids in the ancestors of cryptophytes and haptophytes + stramenopiles + relatives (events marked with “B” in the green arrows, fig. 4B), or three independent origins only for the plastid-containing groups of cryptophytes, haptophytes, and stramenopiles (events marked with “C” in the blue arrows) are theoretically possible. These multiple independent secondary endosymbioses, however, are difficult to explain because all chlorophyll-c containing lineages share a unique protein import machinery referred to as SELMA [symbiont-specific ERAD (endoplasmic reticulum associated degradation)-like machinery] (see Zimorski et al. 2014 and references therein). SELMA is a multi-protein system that is integrated in the second outermost plastid membrane (i.e., inner face of the host ER membrane). Because SELMA genes are encoded in the nucleomorph (the former red algal nucleus) of cryptophytes and are homologous to genes in other chlorophyll-c containing lineages (Zimorski et al. 2014), this result provides strong evidence in support of a single red algal secondary endosymbiosis as depicted in figure 4A. If two or three independent secondary endosymbioses from closely related red algal species are posited, then there must have been independent origins of the SELMA system, as shown in the nuclear gene tree (cases B and C, fig. 4B), which seems highly unlikely.
Fig. 4.
—Secondary endosymbiotic origin of red alga-derived plastids and the nuclear multigene tree of eukaryotes. (A) The plastid-genome based tree of algae with the red arrow (marked with “S”) showing the putative single origin of the red alga-derived plastid in chlorophyll-c containing algae. (B) The nuclear multigene tree of life showing the putative relationships of chlorophyll-c containing algae in the context of nonphotosynthetic lineages. A possible explanation for the conflict in topology between the plastid and nuclear gene trees is that there were multiple independent red algal secondary endosymbioses in the photosynthetic lineages (either two events [marked with “B” and green arrows] or three events [marked with “C” and blue arrows]). A more likely scenario is that there was a single red algal secondary endosymbiosis (perhaps in the cryptophyte ancestor), followed by serial eukaryotic endosymbioses involving chlorophyll-c lineages that spread the plastid to other groups (marked with “Se” and magenta arrows).
The discrepancy between the plastid (fig. 4A) and nuclear tree (fig. 4B) may therefore be explained by serial endosymbioses. For example, the red algal endosymbiont could have become a secondary plastid with the SELMA machinery in an ancestor of chlorophyll-c containing algae. This could have been followed by tertiary (e.g., involving engulfment of the initial red algal plastid host) and/or quaternary endosymbioses in different host(s) (events marked with “Se” in the magenta arrows in figure 4B, see also Gould et al. 2015). Apart from the well-supported cryptophyte–haptophyte monophyly in plastid phylogenies, Stiller et al. (2014) suggested a model of serial plastid endosymbioses based on linear regression analysis of genome data from chlorophyll-c containing groups; i.e., that the ancestor of modern-day cryptophytes engulfed a red alga, whose plastid was subsequently transferred to the ancestor of photosynthetic stramenopiles by tertiary endosymbiosis, and perhaps finally to haptophytes by quaternary endosymbiosis (fig. 4B). Taken together, our results suggest that secondary endosymbiosis occurred a single time, involving a Rhodophytina donor, and then this plastid and its associated machinery were spread to different lineages through additional rounds of eukaryotic endosymbiosis. This hypothesis is of course tentative and awaits a better resolved host tree of eukaryotes. This could be precipitated by greater taxon sampling and a better understanding of how EGT, HGT, and other forces that bias gene-based trees may have (or not) impacted the topologies produced thus far.
Conclusions
We have sequenced four cryptophyte plastid genomes with a wide range of plastid pigmentation: the red-colored Storeatula sp. CCMP1868, the blue-green Chroomonas placoidea and Ch. mesostigmatica, and the brown Cryptomonas curvata. These newly sequenced genomes increase the breadth of data available from algae and will aid in the identification of general trends in organellar genome evolution, particularly in organisms with red-algal derived plastids. Within cryptophytes, most of the genomes are highly conserved with respect to genome structure and coding capacity, however, lineage specific gene content (e.g., dnaX, chlB/L/N) was identified. In addition, examples of lineage-specific gene losses and intron insertions were found for 18 genes (dnaB, dnaK, dnaX, groEL, ftsH, hlpA, minD, minE, chlB/L/N, ftrB, petG, psbN, orf27, orf252, ycf20, and ycf26). The distribution patterns of these genes suggest independent HGT events and/or intra-genomic transfers during the evolutionary history of these ecologically important algae.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This research was supported by the National Research Foundation (NRF) of Korea funded by the Ministry of Science, ICT & Future Planning, Basic Science Research Program (MSIP; NRF-2013R1A1A3012539) and the Ministry of Education (2015R1D1A1A01057899) to J.I.K.; NRF (2016R1D1A1A09919318) to G.Y.; NRF (2017R1A2B3001923), Korean Rural Development Administration Next-Generation BioGreen21 program (PJ011121), and the Collaborative Genome Program (20140428) funded by the Ministry of Oceans and Fisheries, Korea to H.S.Y.; NRF (MSIP; 2015R1A2A2A01003192 and 2015M1A5A1041808) and the 2014 CNU research fund of Chungnam National University to W.S.; and an operating grant from the Canadian Institutes of Health Research—Nova Scotia Regional Partnership Program (ROP85016) to J.M.A. C.E.M. held a Doctoral Student Award from the Natural Sciences and Engineering Research Council of Canada. J.M.A. acknowledges support from the Canadian Institute for Advanced Research, Program in Integrated Microbial Biodiversity. We thank B. Curtis for bioinformatic assistance.
Literature Cited
- Archibald JM. 2007. Nucleomorph genomes: structure, function, origin and evolution. BioEssays 29:392–402. [DOI] [PubMed] [Google Scholar]
- Archibald JM. 2009. The puzzle of plastid evolution. Curr Biol. 19:R81–R88. [DOI] [PubMed] [Google Scholar]
- Archibald JM, Keeling PJ.. 2005. On the origin and evolution of plastids In: Saap J, editor. Microbial phylogeny and evolution. New York: Oxford University Press; p. 238–260. [Google Scholar]
- Baurain D, et al. 2010. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol Biol Evol. 27:1698–1709. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Medlin L.. 1995. The phylogeny of plastids: a review based on comparisons of small-subunit ribosomal RNA coding regions. J Phycol. 31:489–498. [Google Scholar]
- Bhattacharya D, Archibald JM, Weber APM, Reyes-Prieto A.. 2007. How do endosymbionts become organelles? Understanding early events in plastid evolution. Bioessays 29:1239–1246. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Yoon HS, Hackett JD.. 2004. Photosynthetic eukaryotes unite: endosymbiosis connects the dots. Bioessays 26:50–60. [DOI] [PubMed] [Google Scholar]
- Blinkova A, et al. 1993. The Escherichia coli DNA polymerase III holoenzyme contains both products of the dnaX gene, tau and gamma, but only tau is essential. J Bacteriol. 175:6018–6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, et al. 2016. Untangling the earch diversification of eukaryotes: a phylogeneomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista. Proc R Soc B. 283:20152802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Okamoto N, Pombert J-F, Keeling PJ.. 2012. The evolutionary history of haptophytes and cryptophytes: phylogenomic evidence for separate origins. Proc Biol Sci. 279:2246–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S, Akiyama Y, Mori H, Matsuo E, Ito K.. 2000. Length recognition at the N-terminal tail for the initiation of FtsH-mediated proteolysis. EMBO Rep. 1:47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis BA, et al. 2012. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 492:59–65. [DOI] [PubMed] [Google Scholar]
- Dallmann HG, McHenry CS.. 1995. DnaX complex of Escherichia coli DNA polymerase III holoenzyme. Physical characterization of the DnaX subunits and complexes. J Biol Chem. 270:29563–29569. [PubMed] [Google Scholar]
- Darling AC, Mau B, Blattner FR, Perna NT.. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. BioInformatics 27:1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, et al. 2013. Is ftsH the key to plastid longevity in sacoglossan slugs?. Genome Biol Evol. 5:2540–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane JA, Hill DRA, Brett SJ, McFadden GI.. 2002. Cryptomonad evolution: nuclear 18S rDNA phylogeny versus cell morphology and pigmentation. J Phycol. 38:1236–1244. [Google Scholar]
- Delwiche CF, Palmer JD.. 1996. Rampant horizontal transfer and duplication of rubisco genes in eubacteria and plastids. Mol Biol Evol. 13:873–882. [DOI] [PubMed] [Google Scholar]
- Donaher N, et al. 2009. The complete plastid genome sequence of the secondarily nonphotosynthetic alga Cryptomonas paramecium: reduction, compaction, and accelerated evolutionary rate. Genome Biol Evol. 1:439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas SE, Penny SL.. 1999. The plastid genome of the cryptophyte alga, Guillardia theta: complete sequence and conserved synteny groups confirm its common ancestry with red algae. J Mol Evol. 48:236–244. [DOI] [PubMed] [Google Scholar]
- Douglas SE, Murphy CA, Spencer DF, Gray MW.. 1991. Cryptomonad algae are evolutionary chimaeras of two phylogenetically distinct unicellular eukaryotes. Nature 350:148–151. [DOI] [PubMed] [Google Scholar]
- Douglas S, et al. 2001. The highly reduced genome of an enslaved algal nucleus. Nature 410:1091–1096. [DOI] [PubMed] [Google Scholar]
- Ellis RJ, van der Vies SM.. 1991. Molecular chaperones. Annu Rev Biochem. 60:321–347. [DOI] [PubMed] [Google Scholar]
- Fong A, Archibald JM.. 2008. Evolutionary dynamics of light-independent protochlorophyllide oxidoreductase (LIPOR) genes in the secondary plastids of cryptophyte algae. Eukaryot Cell 7:550–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Bauer CE.. 2003. The light-dependent protochlorophyllide reductase: a nitrogenase-like enzyme catalyzing a key reaction for greening in the dark In: Kadish K, Smith K, Guilard R, editors. The porphyrin handbook. Vol. 13. San Diego: Elsevier Science; p. 109–156. [Google Scholar]
- Gilson PR, Maier UG, McFadden GI.. 1997. Size isn’t everything: lessons in genetic miniaturisation from nucleomorphs. Curr Opin Genet Dev. 7:800–806. [DOI] [PubMed] [Google Scholar]
- Gilson PR, et al. 2006. Complete nucleotide sequence of the chlorarachniophyte nucleomorph: nature’s smallest nucleus. Proc Natl Acad Sci U S A. 103:9566–9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SB, Maier U-G, Martin WF.. 2015. Protein import and the origin of red complex plastids. Curr Biol. 25:R515–R521. [DOI] [PubMed] [Google Scholar]
- Graham LK, Wilcox LW.. 2000. The origin of alteration of generations in land plants: a focus on matrotrophy and hexose transport. Philos Trans R Soc Lond B Biol Sci. 255:757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasser KD, et al. 1997. The recombinant product of the Cryptomonas plastid gene hlpA is an architectural HU-like protein that promotes the assembly of complex nucleoprotein structures. Eur J Biochem. 249:70–76. [DOI] [PubMed] [Google Scholar]
- Hall N, et al. 2002. Sequence of Plasmodium falciparum chromosomes 1, 3-9 and 13. Nature 419:527–531. [DOI] [PubMed] [Google Scholar]
- Hill DRA, Rowan KS.. 1989. The biliproteins of the Cryptophyceae. Phycologia 28:455–463. [Google Scholar]
- Hoef-Emden K, Marin B, Melkonian M.. 2002. Nuclear and nucleomorph SSU rDNA phylogeny in the Cryptophyta and the evolution of cryptophyte diversity. J Mol Evol. 55:161–179. [DOI] [PubMed] [Google Scholar]
- Hoef-Emden K, Melkonian M.. 2003. Revision of the genus Cryptomonas (Cryptophyceae): a combination of molecular phylogeny and morphology provides insights into a long-hidden dimorphism. Protist 154:371–409. [DOI] [PubMed] [Google Scholar]
- Hoef-Emden K. 2008. Molecular phylogeny of phycocyanin-containing cryptophytes: evolution of biliproteins and geographical distribution. J Phycol. 44:985–993. [DOI] [PubMed] [Google Scholar]
- Howe CJ, Nisbet ER, Barbrook AC.. 2008. The remarkable chloroplast genome of dinoflagellates. J Exp Bot. 59:1035–1045. [DOI] [PubMed] [Google Scholar]
- Hunsperger HM, Randhawa T, Cattolico RA.. 2015. Extensive horizontal gene transfer, duplication, and loss of chlorophyll synthesis genes in the algae. BMC Evol Biol. 15:16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain K, et al. 2014. Extreme features of the Galdieria sulphuraria organellar genomes: a consequence of polyextremophily? Genome Biol Evol. 7:367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouškovec J, Horák A, Oborník M, Lukeš J, Keeling PJ.. 2010. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci U S A. 107:10949–10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ. 2004. Diversity and evolutionary history of plastids and their hosts. Am J Bot. 91:1481–1493. [DOI] [PubMed] [Google Scholar]
- Khan H, Archibald JM.. 2008. Lateral transfer of introns in the cryptophyte plastid genome. Nucleic Acids Res. 36:3043–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H, et al. 2007. Plastid genome sequence of the cryptophyte alga Rhodomonas salina CCMP1319: lateral transfer of putative DNA replication machinery and a test of chromist plastid phylogeny. Mol Biol Evol. 24:1832–1842. [DOI] [PubMed] [Google Scholar]
- Kim E, et al. 2011. Newly identified and diverse plastid-bearing branch on the eukaryotic tree of life. Proc Natl Acad Sci U S A. 108:1496–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, et al. 2015. The plastid genome of the cryptomonad Teleaulax amphioxeia. PLoS One 10:e0129284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowallik KV, Stoebe B, Schaffran I, Kroth-Pancic P, Freier U.. 1995. The chloroplast genome of chlorophyll a+c-containing alga, Odontella sinensis. Plant Mol Biol Rep. 13:336–342. [Google Scholar]
- Le SQ, Gascuel O.. 2008. An improved general amino acid replacement matrix. Mol Biol Evol. 25:1307–1320. [DOI] [PubMed] [Google Scholar]
- Lee JM, et al. 2016. Parallel evolution of highly conserved plastid genome architecture in red seaweeds and seed plants. BMC Biol. 14:75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux C, Otis C, Turmel M.. 2000. Ancestral chloroplast genome in Mesostigma viride reveals an early branch of green plant evolution. Nature 403:649–652. [DOI] [PubMed] [Google Scholar]
- Lindahl M, et al. 2000. The thylakoid FtsH protease plays a role in the lightinduced turnover of the photosystem II D1 protein. Plant Cell 12:419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier UG, Rensing SA, Igloi GL, Maerz M.. 1995. Twintrons are not unique to the Euglena chloroplast genome: structure and evolution of a plastome cpn60 gene from a cryptomonad. Mol Gen Genet. 246:128–131. [DOI] [PubMed] [Google Scholar]
- Martin W, et al. 1998. Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393:162–165. [DOI] [PubMed] [Google Scholar]
- McFadden GI. 1993. Second-hand chloroplasts: evolution of cryptomonad algae In: Callow JA, editor. Advances in botanical research. London: Academic Press Limited; p. 189–230. [Google Scholar]
- McFadden GI, Gilson PR, Hill DRA.. 1994. Goniomonas: rRNA sequences indicate that that this phagotrophic flagellate is a close relative to the host component of cryptomonads. Eur J Phycol. 29:29–32. [Google Scholar]
- Miyagishima SY, Suzuki K, Okazaki K, Kabeya Y.. 2012. Expression of the nucleus-encoded chloroplast division genes and proteins regulated by the algal cell cycle. Mol Biol Evol. 29:2957–2970. [DOI] [PubMed] [Google Scholar]
- Moore CE, Curtis B, Mills T, Tanifuji G, Archibald JM.. 2012. Nucleomorph genome sequence of the cryptophyte alga Chroomonas mesostigmatica CCMP1168 reveals lineage-specific gene loss and genome complexity. Genome Biol Evol. 4:1162–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Gómez SA, et al. 2017. The new red algal subphylum Proteorhodophytina comprises the largest and most divergent plastid genomes known. Curr Biol. 27:1677–1684. [DOI] [PubMed] [Google Scholar]
- Muraki N, et al. 2010. X-ray crystal structure of the light-independent protochlorophyllide reductase. Nature 465:110–114. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman WC, et al. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156. [DOI] [PubMed] [Google Scholar]
- Ohta N, et al. 2003. Complete sequence and analysis of the plastid genome of the unicellular red alga Cyanidioschyzon merolae. DNA Res. 10:67–77. [DOI] [PubMed] [Google Scholar]
- Pain A, et al. 2005. Genome of the host-cell transforming parasite Theileria annulata compared with T. parva. Science 309:131–133. [DOI] [PubMed] [Google Scholar]
- Perrineau M, Price DC, Mohr G, Bhattacharya D.. 2015. Recent mobility of plastid encoded group II introns and twintrons in five strains of the unicellular red alga Porphyridium. PeerJ PrePrints 2:e729v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice DW, Palmer JD.. 2006. An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are siters. BMC Biol. 4:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Puerta MV, Bachvaroff TR, Delwiche CF.. 2005. The complete plastid genome sequence of the haptophyte Emiliania huxleyi: a comparison to other plastid genomes. DNA Res. 12:151–156. [DOI] [PubMed] [Google Scholar]
- Ševčíková T, et al. 2015. Updating algal evolutionary relationships through plastid genome sequencing: did alveolate plastids emerge through endosymbiosis of an ochrophyte? Sci Rep. 5:10134.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, et al. 2008. Multigene phylogeny of choanozoa and the origin of animals. PLoS One 3:e2098.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, Shi X.. 2006. Characterization of three genes encoding the subunits of light-independent protochlorophyllide reductase in Chlorella protothecoides CS-41. Biotechnol Prog. 22:1050–1055. [DOI] [PubMed] [Google Scholar]
- Smith SW, Overbeek R, Woese CR, Gilbert W, Gillevet PM.. 1994. The genetic data environment: an expandable GUI for multiple sequence analysis. Comput Appl Biosci. 10:671–675. [DOI] [PubMed] [Google Scholar]
- Song HJ, et al. 2016. A novice’s guide to analyzing NGS-derived organelle and metagenome data. Algae 31:137–154. [Google Scholar]
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller JW, et al. 2014. The evolution of photosynthesis in chromist algae through serial endosymbioses. Nat Commun. 5:5764.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoebe B, Kowallik KV.. 1999. Gene-cluster analysis in chloroplast genomics. Trends Genet. 15:344–347. [DOI] [PubMed] [Google Scholar]
- Tajima N, et al. 2014. Analysis of the complete plastid genome of the unicellular red alga Porphyridium purpureum. J Plant Res. 127:389–397. [DOI] [PubMed] [Google Scholar]
- Tang X, Bi G.. 2016. The complete chloroplast genome of Gullardia theta strain CCMP2712. Mitochondrial DNA 27:4423–4424. [DOI] [PubMed] [Google Scholar]
- Tanifuji G, et al. 2011. Complete nucleomorph genome sequence of the nonphotosynthetic alga Cryptomonas paramecium reveals a core nucleomorph gene set. Genome Biol Evol. 3:44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C.. 1999. The complete chloroplast DNA sequence of the green alga Nephroselmis olivacea: insights into the architecture of ancestral chloroplast genomes. Proc Natl Acad Sci U S A. 96:10248–10253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Heyden S, Chao E, Cavalier-Smith T.. 2004. Genetic diversity of goniomonads: an ancient divergence between marine and freshwater species. Eur J Phycol. 39:343–350. [Google Scholar]
- Wang SL, Liu X-Q.. 1991. The plastid genome of Cryptomonas phi encodes an hsp70-like protein, a histone-like protein, and an acyl carrier protein. Proc Natl Acad Sci U S A. 88:10783–10787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe MM, Hiroki M.. 1997. NIES-collection list of strains. 5th ed.Tsukuba: National Institute for Environmental Studies; 127pp. [Google Scholar]
- Yoon HS, Hackett JD, Bhattacharya D.. 2002a. A single origin of the peridinin—and fucoxanthin-containing plastids in dinoflagellates through tertiary endosymbiosis. Proc Natl Acad Sci U S A. 99:11724–11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Pinto G, Bhattacharya D.. 2002b. The single, ancient origin of chromist plastids. Proc Natl Acad Sci U S A. 99:15507–15512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Müller KM, Sheath RG, Ott FD, Bhattacharya D.. 2006. Defining the major lineages of red algae (Rhodophyta). J Phycol. 42:482–492. [Google Scholar]
- Zhang Z, Green BR, Cavalier-Smith T.. 1999. Single gene circles in dinoflagellate chloroplast genomes. Nature 400:155–159. [DOI] [PubMed] [Google Scholar]
- Zimorski V, Ku C, Martin WF, Gould SB.. 2014. Endosymbiotic theory for organelle origins. Curr Opin Microbiol. 22:38–48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.