Abstract
High-throughput sequencing is a revolutionary technology for the analysis of metagenomic samples. However, querying large volumes of reads against comprehensive DNA/RNA databases in a sensitive manner can be compute-intensive. Here, we present taxMaps, a highly efficient, sensitive, and fully scalable taxonomic classification tool. Using a combination of simulated and real metagenomics data sets, we demonstrate that taxMaps is more sensitive and more precise than widely used taxonomic classifiers and is capable of delivering classification accuracy comparable to that of BLASTN, but at up to three orders of magnitude less computational cost.
Microbial communities of unknown composition can be collected from a wide array of locations. The examination of these microbial communities, known as metagenomics, has become increasingly prominent, with many recent studies focusing on the communities of the human body (The Human Microbiome Project Consortium 2012a,b; Zhang et al. 2015) or from our environment—for example, hospitals (Smith et al. 2013), subway stations (Afshinnekoo et al. 2015), or even ATM keypads (Bik et al. 2016). High-throughput sequencing enables the unbiased profiling of these communities as well as the ability to investigate clinical samples containing pathogens that are unable to be cultured using traditional laboratory techniques. Although the emergence of these technologies has also resulted in more comprehensive databases, querying them in a sensitive manner has become computationally more expensive.
Whether the goal is to estimate the relative abundance or to merely confirm the presence of particular organisms in a given sample, taxonomic classification of each sequence is an essential first step in many metagenomics experiments. Older strategies, based on machine-learning techniques such as the Naïve Bayes Classifier (NBC) (Rosen et al. 2008) and PhymmBL (Brady and Salzberg 2009) or on alignment tools such as BLAST (Altschul et al. 1990), like MEGAN (Huson et al. 2007), are slow and do not scale well to the size of today's experiments. More recently, a new class of faster taxonomic classifiers has emerged. Programs like Kraken (Wood and Salzberg 2014) and CLARK (Ounit et al. 2015) are based on alignment-free strategies in which k-mers extracted from the read data are compared to a set of preclassified k-mers in the database. Although these programs can classify millions of reads in just a few minutes, their memory requirements are usually high. By the use of a FM-index (Ferragina and Manzini 2000), which allows for efficient storage and querying of the database, Kaiju (Menzel et al. 2016), a protein homology-based classifier, and Centrifuge (Kim et al. 2016) have addressed the issue of memory consumption.
Estimation of the relative abundance of taxa in a sample is most often accomplished by comparing the number of classifications to expected relative values, based on the composition of the database. Programs such as MetaPhlAn (Segata et al. 2012) and mOTU (Sunagawa et al. 2013) rely on BLAST for taxonomic classification of raw or assembled reads to clade-specific marker gene databases, in which coverage of the marker gene is directly related to abundance. Centrifuge and Bracken (Lu et al. 2017), a program that uses Kraken taxonomic assignments, rely on statistical models to estimate abundance based on the number of reads classified as a given taxon and information about the genomes present in the database. Accurate abundance estimations therefore rely on first performing correct taxonomic classifications. Although more recent programs allow for taxonomic classification at an unprecedented speed, no significant improvements in classification accuracy have been reported over MegaBLAST (Zhang et al. 2000), the least sensitive BLAST program.
Here, we describe taxMaps, an ultra-sensitive, customizable, and fully scalable taxonomic mapping tool for short-read data designed to deal with large DNA/RNA metagenomics data. taxMaps is designed to facilitate the taxonomic classification operation, featuring thorough preprocessing, the ability to prioritize mapping to multiple indexes, detailed mapping reports, and interactive results visualization. Most importantly, by using a novel database compression algorithm that eliminates database redundancy, which improves querying performance and reduces the number of post-querying computations, and an optimal nonexact match mapping strategy using the state-of-the-art mapper GEM (Marco-Sola et al. 2012), taxMaps delivers classification accuracy that approximates that of BLASTN but in orders of magnitude less time.
Results
Database compression
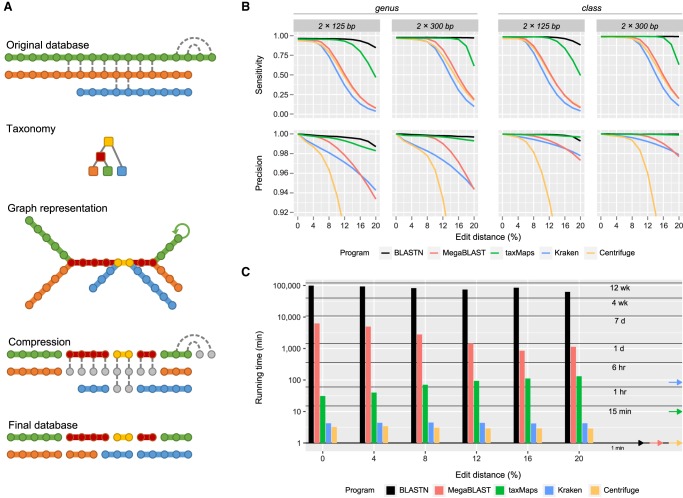
To taxonomically classify short-read data in a comprehensive manner, millions of reads must be compared against DNA/RNA databases, which contain sequences from thousands to millions of organisms. This operation is compute-intensive, because querying performance is highly dependent on database size and redundancy. This is particularly true when all best hits are to be exhaustively retrieved—something required to ensure maximum classification accuracy. With that in mind, we developed a compression algorithm that eliminates database redundancy by performing a Lowest Common Ancestor (LCA) preassignment and collapse for k-mers of length greater than a specified read length (Fig. 1A). This allows GEM mapper to conduct nonexact searches in the same manner as it would against the original database, resulting in compression that, for the purpose of taxonomic classification, is lossless. The use of compressed databases in taxMaps results in more stable GEM mapper runtimes for reads that align to highly redundant sequences in the original database (Supplemental Fig. S1). Moreover, the fact that BLASTN runtimes are also improved when using databases generated from FASTA files that have been compressed using the same algorithm, suggests that this approach could be applied to other alignment methods with the purpose of improving performance and scalability. Making use of that algorithm, we built several databases, some including millions of sequences from more than a million different taxonomic entities (Supplemental Table S1). Compression ratios varied from 1.08 to 4.67, with higher values obtained when using shorter k-mers, and usually for databases containing many bacterial genomes, due to the presence of multiple highly homologous strains. Databases compressed at shorter k-mers, despite better compression ratios, require more RAM to be loaded. This may be due to a higher probability of homology between short k-mers, compared to longer ones, which leads to more pronounced sequence fragmentation.
Figure 1.
Database compression and classification accuracy and performance on simulated metagenomics paired-end data sets. (A) Visual representation of the taxMaps database compression algorithm. Each sequence is represented as an array of k-mers (circles), colored according to their taxon (colored squares). Identical k-mers are linked by a dashed line. During compression, the first instance of every k-mer is reclassified to the Lowest Common Ancestor of all instances of that k-mer in the database, while the remaining (gray circles) are disregarded. New sequences, composed of k-mers that share the same taxonomic classification, are assembled on the fly as the algorithm traverses the database. A graph representation of the database is also shown. (B) Classification sensitivity and precision as a function of average sequence divergence and read length at the genus and class ranks. For visualization purposes, Centrifuge's precision series have been truncated. For complete results, see Supplemental Figure S3. (C) Wall clock time required for the classification of six different data sets, each consisting of 10 million read pairs of 125 bp of length, depending on average sequence divergence. The arrows on the right indicate the database loading time for each program.
Classification accuracy and performance on simulated metagenomes
We compared taxMaps to BLAST (in its two variants: MegaBLAST and the more sensitive BLASTN), Kraken, and Centrifuge. In this benchmarking exercise, we used NCBI's nucleotide database (NCBI Resource Coordinators 2016) as reference for all four methods to ensure that differences in classification accuracy and speed can only be attributed to algorithmic differences between classifiers and not to reference database differences. Given that classification accuracy strongly depends on factors such as sequence quality, distance to the closest available sequences in the database, and read length, we have generated 55 simulated paired-end read sets of increasing length (from 75 to 300 bp) and divergence (from 0% to 20%) from the reference sequences of more than 4000 different taxonomic units (Supplemental Fig. S2).
Classification accuracy results at the genus and class ranks for paired-end reads of length 125 and 300 bp are shown in Figure 1B. It is possible to observe that, although incapable of matching BLASTN accuracy for the most divergent read sets, taxMaps clearly outperforms MegaBLAST, Kraken, and Centrifuge in both sensitivity and precision. This is particularly striking when sequence divergence is >8%. For instance, on a highly divergent 300-bp paired-end data set (average edit distance: 16%), taxMaps sensitivity and precision at the genus level are 0.951 and 0.995, respectively. On the same data set, MegaBLAST, Kraken, and Centrifuge are incapable of classifying more than half of the reads, with sensitivity values of 0.470, 0.303, and 0.414, at a precision of 0.971, 0.961, and 0.817, respectively. Although Kraken and MegaBLAST are still capable of high precision on divergent data sets, we observe a significant drop in Centrifuge starting at 8% edit distance. These results are particularly relevant when choosing the right classifier for metagenomics samples containing organisms that are likely not represented in any database or in situations in which the error-rate is high. This trend was observed for all tested read lengths, at virtually all taxonomic ranks for both paired-end (Supplemental Fig. S3) and single-end classification (Supplemental Fig. S4). Regarding computational performance, Centrifuge and Kraken were the fastest methods, being capable of classifying 10 million 125-bp read pairs in <5 min, followed by taxMaps that, depending on the average sequence divergence, takes between 31 and 131 min to execute the same task (Fig. 1C). Nevertheless, this range is comparable to other NGS pipelines (e.g., mapping and variant calling)—and one to two orders of magnitude faster than MegaBLAST and up to three orders faster than BLASTN on data sets of low sequence divergence to the closest match in the database (Supplemental Fig. S5). For taxMaps, the computational cost is positively associated with the average sequence divergence to the reference database, whereas the inverse is true for MegaBLAST, probably because for extreme edit distance values, fewer reads have a seed hit in the database and therefore, no extension operation is performed.
By making use of simulated reads, we have also evaluated the strain-level classification accuracy of taxMaps, given as rank-level sensitivity, precision, and F-score (Supplemental Fig. S6). More prominently than at other ranks, strain-level accuracy when profiling metagenomics communities is expected to be highly dependent on the proportion of strain-specific sequence in the database given that, for the usually large amount of conserved regions between strains, it is only possible to classify reads at higher ranks, such as species or genus. To test this, we have selected 500 bacterial strain genomes, uniformly distributed along the spectrum of percentage of strain-specific sequence on the refseq_complete_genomes database (see Supplemental Table S1) and simulated paired-end reads that were then classified using taxMaps. Our results show that sensitivity is directly correlated with the amount of strain-specific sequence and that, for bacterial strains with at least 20% of strain-specific sequence, taxMaps’ precision is on average >90%, meaning that those assignments can be trusted.
Mock communities
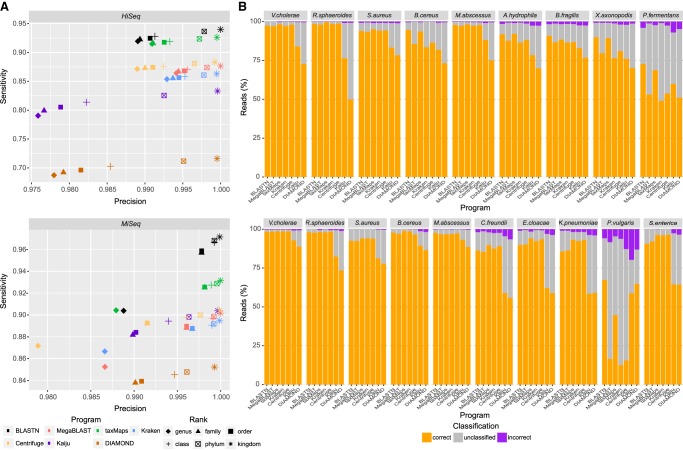
To test whether results observed on simulated data would hold when classifying real sequencing data, all five classifiers were tested on two data sets, HiSeq and MiSeq (Wood and Salzberg 2014), containing reads from 9 and 10 different bacterial species, respectively. All methods relied on the same database, refseq_complete_ genomes, that consists of complete bacterial, archaea, and viral genomes. In this exercise, we also included Kaiju and DIAMOND (Buchfink et al. 2015) followed by LCA—two strategies relying on protein homology for taxonomic classification. For those, the reference database consisted of all protein sequences annotated on the same set of genomes. For the BLAST methods in this analysis, database search hits were filtered using similar criteria to those used in Huson et al. (2007). This reduced the number of false-positive classifications originating from small partial alignments, at the cost of some sensitivity (Supplemental Fig. S7). As in the results observed for simulated data, BLASTN is the most sensitive method at all the taxonomic ranks considered, with the exception of genus-level classification in the MiSeq data set, for which taxMaps is marginally more sensitive (Fig. 2A). However, it is the second least precise, after Centrifuge, of all five nucleotide homology-based methods on the HiSeq data set. This discrepancy can be explained by the fact that, on simulated data sets, all reads originate from sequences that are already present in the database, therefore reducing the probability of incorrect classification, whereas for real sequencing data, that is not necessarily the case. Apart from the potential lack of complete genomes in the database, there may be other sequencing artifacts that were not captured in our simulation. After BLASTN, taxMaps is the second most sensitive method at all taxonomic ranks. In fact, for both the HiSeq and MiSeq data sets, taxMaps correctly assigns at least as many reads to the right genus (sensitivity of 0.914 and 0.904, respectively) as any of the remaining programs (MegaBLAST, Kraken, Centrifuge, Kaiju, and DIAMOND) assign to the right kingdom.
Figure 2.
Taxonomic classification accuracy on two mock metagenomics communities. (A) Classification sensitivity and precision at six major taxonomic ranks for two real data sets. For visualization purposes, genus-level accuracy values for Kaiju and DIAMOND on the MiSeq data set (sensitivity: 0.7414 and 0.7177; precision: 0.9526 and 0.9530, respectively) have been omitted. (B) The corresponding breakdown per species of the percentage of correct, incorrect, and unclassified reads at the genus level.
For the HiSeq data set, with the default parameter maximum edit distance, e = 0.2, taxMaps was slightly less precise at the genus level (0.991) than Kraken (0.993) and MegaBLAST (0.994) with default parameters. It is, however, possible to find values of e (e ≤ 0.12), where taxMaps is simultaneously more precise and more sensitive than these two methods. On the MiSeq data set, running taxMaps with e = 0.12 drops the genus-level sensitivity to that of Kraken (default k = 31), but with ∼60% fewer incorrect classifications. A similar tradeoff between sensitivity and precision is also observed for Kraken and Centrifuge when using different values of k and min-hitlen, respectively (Supplemental Fig. S7). Regarding the two protein homology-based classifiers, they were the least sensitive and least precise on both data sets at virtually all ranks considered. This result is rather surprising given that protein homology is usually higher than nucleotide homology.
Although in aggregate, BLASTN was the most sensitive method, when further breaking down the results by species (Fig. 2B), taxMaps has the highest number of correctly classified reads at the genus level in five of nine species in the HiSeq data set. For the remaining four species, BLASTN obtained the most correct classifications. In the MiSeq data set, taxMaps obtained the highest number of correct classifications in 7 of 10 species, three of which tie with either Kraken, Centrifuge, or both. A few species (Xanthomonas axonopodis, Pelosinus fermentans, and Proteus vulgaris), which are divergent from the species in the database, explain most of the differences in overall sensitivity between methods. In those cases, classification performance of taxMaps and BLASTN was significantly higher than that of Kraken, MegaBLAST, and Centrifuge, being comparable or superior to the protein homology-based methods Kaiju and DIAMOND, traditionally expected to perform well in that situation.
We also decided to explore, for these two data sets, the taxMaps feature that allows the use and prioritization of multiple databases/indexes. For that, we used the refseq_complete_genomes database with a strict value for maximum edit distance (e = 0.1) followed by either the blast_nt, refseq_microbial, or combined_ncbi databases, with e = 0.2 (Supplemental Fig. S8). Although the combination including the blast_nt database led to accuracy values similar to those of refseq_complete_genomes with e = 0.2, the use of refseq_microbial and combined_ncbi raised the genus-level sensitivity to values over 0.975 in the HiSeq and 0.92 in the MiSeq data sets, at precision values above 0.991 and 0.989, respectively. The increase in classification sensitivity when using more comprehensive databases also led to more accurate genus abundance estimation on these samples and on the recently published mock community data sets HC/LC and ZymoBIOMICS (Supplemental Fig. S9; McIntyre et al. 2017).
Human microbiome and environmental samples
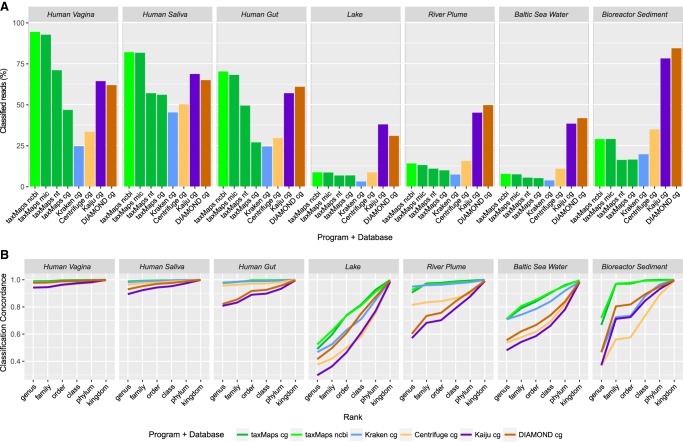
Although the two mock communities allow for comparisons of classifier accuracy based on real data, they represent a relatively simple classification task, given that most species are well represented in the database used. To assess classifier behavior in a more realistic scenario, we considered three human microbiome and four environmental metagenomics samples (Supplemental Table S2) as input for taxMaps, Kraken, Centrifuge, Kaiju, and DIAMOND. In this case, due to the large number of reads per sample, we did not consider the slower BLAST methods, because they would not represent a practical classification solution. When using the refseq_complete_genomes database, DIAMOND classified the largest number of reads on four samples and Kaiju on three. These two methods were followed either by Centrifuge (five samples) or by taxMaps (two samples). With the sole exception of the Bioreactor Sediment sample, Kraken classified the least number of reads on all samples (Fig. 3A). Although this suggests that DIAMOND, Kaiju, and even Centrifuge, may be more sensitive than taxMaps and Kraken on these data sets, the ground truth for these samples is unknown, and therefore it is impossible to assess the classification accuracy of each method. To address this problem, we developed a novel rank-level metric called classification concordance that, for a given taxonomic rank, can be defined as the percentage of read pairs for which the independent classification of both mates is concordant at that particular rank (for details, see Methods). On the simulated data set described previously, this metric shows a high correlation with classification precision at all ranks individually and in aggregate (ρ = 0.994) (Supplemental Fig. S10). Therefore, it has the potential to be used as proxy for classification accuracy. In Figure 3B it is possible to observe that both taxMaps and Kraken show significantly higher classification concordance than Centrifuge, Kaiju, and DIAMOND on most data sets with the exception of the Lake and Bioreactor Sediment samples, where Kraken classification concordance is comparable to that of DIAMOND and Kaiju. On this last sample, against the general trend, Kraken classified more reads than taxMaps. These results suggest that although DIAMOND, Kaiju, and Centrifuge up to some extent, may classify more reads, they likely do so with much lower precision than taxMaps and Kraken. This is particularly striking on the human microbiome samples and the River Plume sample where, for instance, classification concordance at the phylum level for Kaiju is lower than that of taxMaps and Kraken at the genus level.
Figure 3.
Percentage of classified reads and classification concordance for seven real metagenomics data sets. (A) Percentage of classified read pairs for three human microbiome and four environmental samples. (B) Classification concordance between paired mates, as proxy of precision, for six major taxonomic ranks.
Finally, we wanted to investigate how the use of more comprehensive databases in taxMaps would affect the percentage of classified reads and whether there would be a negative effect in classification concordance. We ran taxMaps using blast_nt, refseq_microbial, and combined_ncbi databases (Supplemental Table S1), and for all samples, the use of these more comprehensive databases resulted in a higher percentage of classified reads. This was particularly clear when using refseq_microbial and combined_ncbi. The use of this last database, comprising 374 Gb of sequence, did not have a negative effect on classification concordance compared to refseq_complete_genomes, suggesting that taxMaps precision was not affected by the significant increase in the number of sequences in the database. By using very large databases, taxMaps can classify more human microbiome reads than DIAMOND and Kaiju and, taking classification concordance as proxy, potentially at much higher precision. As such, taxMaps is particularly appropriate for microbiome studies where maximum classification accuracy at lower taxonomic ranks is desired.
Discussion
As genomic databases become more comprehensive, so grows the challenge of how to efficiently utilize such resources to accurately classify the large number of reads generated by high-throughput sequencing technologies. Although other recently published methods rely on alignment-free strategies to improve the computational performance of this task, taxMaps’ approach can be considered as an intermediate between that and the more sensitive alignments of BLASTN. By relying on a novel database compression algorithm, taxMaps can utilize the GEM mapper to conduct very sensitive searches on very large databases while maintaining good performance. Our results using simulated data sets show that the sensitivity and precision of taxMaps approximate that of BLASTN and are superior to those of Kraken, MegaBLAST, and Centrifuge, especially as read sequences diverge from the corresponding database reference. These results were further confirmed on the two mock community data sets, for which taxMaps delivered the highest number of correct classifications for the majority of the species included. Regarding real metagenomics samples (human microbiome and environmental), when using the same database, both taxMaps and Kraken classified fewer reads than Centrifuge and Kaiju. Although in a previous benchmark (Menzel et al. 2016), the number of classified reads has been interpreted as proxy for sensitivity, the ground truth for those data sets is unknown. Based on a novel rank-level metric called classification concordance, our results suggest that both Kraken and taxMaps are significantly more precise than Centrifuge, Kaiju, and DIAMOND. Moreover, we show that taxMaps classification concordance is not affected when using more comprehensive databases that, in the case of the human microbiome samples, led to a significant increase in the number of classified reads.
In summary, our results show that taxMaps offers class-leading accuracy and comprehensiveness while balancing performance, making it uniquely suitable for unbiased contamination detection in large-scale sequencing operations, microbiome studies comprising a large number of samples, and applications for which the analysis turnaround time is a critical factor, such as pathogen identification from clinical or environmental samples.
Methods
Database creation
Data from the RefSeq Genomes and BLAST nt databases were retrieved through the NCBI FTP server and organized in various databases (see Supplemental Table S1). For each database, duplicate sequence entries were removed, and all ambiguous nucleotides converted to N characters. Then, for every distinct k-mer, we computed the LCA between all taxonomic IDs of the sequences containing it, derived from the NCBI Taxonomy database (NCBI Resource Coordinators 2016). K-mers were assembled, through extension, into sequences that share the same LCA. This procedure is done on the fly as the algorithm traverses the database and k-mers are read and classified. For every sequence record in the database, compression is initialized by the creation of a sequence, corresponding to the first k-mer of the record and classified as its LCA. Then, for every k-mer, if the LCA classification matches that of the sequence, the sequence is extended with the last base of the k-mer. Otherwise, a new sequence consisting of that k-mer and corresponding classification is initiated. The newly assembled sequences are then indexed (FM-index) using GEM (Marco-Sola et al. 2012). This reassembly process and use of the FM-index result in a reduction of the memory footprint, allowing for very large databases to be merged and simultaneously queried. Although the overall strategy is similar, in essence, to the one used in Kraken, the fact that the operation is performed on k-mers of length equal or greater than a target read length allows for nonexact searches to be conducted in the same manner as they would against the original database, meaning that for every alignment in the raw database, there will be at least one k-mer in the compressed database, where the same alignment is possible, thus rendering this compression lossless for the purpose of taxonomic classification. This not only eliminates most of the database sequence redundancy, consequently improving mapping performance stability, but it also significantly reduces the number of post-mapping computations to be performed. This is particularly true for samples containing DNA or RNA from organisms that are highly represented in the databases (e.g., E. coli) or for which the repeat content is particularly high. Compressed indexes can be downloaded from ftp://ftp.nygenome.org/taxmaps.
Classification algorithm
Reads are mapped in single-end mode to an indexed database (k ≥ read length) using GEM mapper, which guarantees that all optimal alignments are retrieved, up to the user-defined maximum edit distance (-e, default = 0.2) parameter. Each read is then taxonomically classified as the LCA of all database sequences returned. For paired-end classification, reads are classified independently. If the classification of the two ends is discordant, meaning that they are different and the root-to-leaf (RTL) path of one end is not fully included in the RTL path of the other end, the pair is classified as the LCA of both single-end classifications. If the RTL path of one end is contained in the RTL path of the other end, the pair is then classified as the lower taxon of the longest RTL path. In situations in which no database match is found for one of the two reads, the pair is classified solely on one read. taxMaps also has a stricter paired-end classification scheme, for which both ends are required to have database hits. In that scheme, the pair is always classified as the LCA of both single-end classifications, even when one RTL path is contained in the other, ensuring maximum precision at the expense of a higher rank classification.
Implementation
taxMaps is fully implemented in Python and works as a transparent pipeline-generating script upon user-defined parameters. It reads data in FASTQ format but can also extract unmapped reads from BAM files through SAMtools (Li et al. 2009). Processing steps such as adapter removal, low-quality end trimming, and low complexity filtering are carried out by Cutadapt (Martin 2011) and PRINSEQ lite (Schmieder and Edwards 2011) as part of the taxMaps pipeline, upon user-specified options. Users can also specify multiple indexes to be queried and define, on an index-specific basis, the maximum edit distance and number of threads used by GEM (Marco-Sola et al. 2012). Apart from that, taxMaps offers one single-end and two paired-end classification modes (described above). Mapping and classification results are given as tab-delimited files, including full mapping information for each read in GEM format along with the corresponding taxonomic classification. Finally, results for all represented taxa are summarized in a table and an interactive report is generated using Krona (Ondov et al. 2011).
Simulated metagenomics data sets
To build the simulated data sets, we first selected taxa for which the RTL path included all the major taxonomic ranks and had at least one contiguous sequence longer than 100 kb in NCBI's nucleotide database (NCBI Resource Coordinators 2016) and then, for each of the 4089 selected taxa (Supplemental Fig. S2), we randomly extracted a single 100-kb sequence chunk. From these sequences, 55 simulated data sets, each consisting of 10 million read pairs, were generated using a version of wgsim forked from SAMtools (Li et al. 2009) (https://github.com/lh3/wgsim), by combining five different read lengths (75, 125, 150, 250, and 300 bp) with 11 edit distances (0.0, 0.02, 0.04, 0.06, 0.08, 0.10, 0.12, 0.14, 0.16, 0.18, and 0.20) and the following additional parameters: fragment length of 550 bp, indel fraction of 0.15 and a maximum fraction of ambiguous bases allowed of 0.003. Interleaved FASTQ files were converted to FASTA files for BLASTN and MegaBLAST, since these programs were not designed to handle the FASTQ format. Each read ID contains the taxonomic identifier of the sequence from which it was simulated as well as the read length and edit distance of the data set. All data sets are available at ftp://ftp.nygenome.org/taxmaps/Benchmark/Datasets. We additionally selected 1 million read pairs (2 × 125 bp, edit distance = 0.08) that were clustered into bins of increasing multiplicity (number of alignment best hits) on the uncompressed NCBI's nucleotide database. These were used to compare GEM mapper and BLASTN performance on compressed and uncompressed databases.
We ran taxMaps, Kraken, Centrifuge, BLASTN, and MegaBLAST on each of the 55 simulated data sets using default parameters and the NCBI's nucleotide database as reference for all methods. For taxMaps databases, the choice of k-mer depended on the read length (k = read length). For BLASTN, the number of read pairs analyzed was reduced to 100,000 by random sampling due to time constraints. Given that BLASTN and MegaBLAST are not taxonomic classifiers per se, the LCA of all best hits for each read was determined. For paired-end classification, the criteria used in taxMaps was applied. To estimate sensitivity and precision, classifications were split into four distinct categories: (1) correct, if the correct taxon is included in the RTL path of the assigned taxon; (2) concordant, if the assigned taxon is different from the correct taxon, but it is included in the RTL path of the correct taxon; (3) incorrect, if the assigned taxon is not included in the RTL path of the correct taxon, nor is the correct taxon included in the RTL path of the assigned taxon; and (4) unclassified, if no taxon was assigned. Rank-level sensitivity is then given by the number of correct classifications at a particular rank over the total number of possible classifications, and rank-level precision corresponds to the number of correct classifications at a particular rank over the number of correct and incorrect classifications at that same rank. Paired-end rank-level sensitivity and precision of each program was calculated at eight major taxonomic ranks (species, genus, family, order, class, phylum, kingdom, and root), for every edit distance and read length combination (Supplemental Fig. S3). Similarly, single-end rank-level sensitivity and precision data were also collected for each program's output in single-end mode (Supplemental Fig. S4). All corresponding F-scores can be found in Supplemental Table S3.
In addition to the sensitivity and precision metrics, wall clock time data was collected for each program on all paired-end data sets (Supplemental Fig. S5). taxMaps, MegaBLAST, Centrifuge, and BLASTN were run on a computer cluster running CentOS 7.1 on either Intel Xeon E5-2697 2.60 GHz CPUs or Intel Xeon CPU E5-2680 2.80 GHz CPUs. Due to the high memory requirements, Kraken was run on a large-memory shared host running CentOS 6.5 on Intel Xeon CPU E7-8830 2.13 GHz CPUs. All programs were run using 16 CPUs per job, except for BLASTN, which was run on eight CPUs given the long-term commitment required of these resources. The wall clock time reported for BLASTN was then extrapolated to match the number of reads classified and numbers of CPUs used by the other programs.
Strain-level accuracy assessment
For all of the bacterial strain genomes available in the refseq_complete_genomes database, we have determined the number of k-mers (k = 125) that are unique to each strain. This allowed us to select 500 bacterial strains that are evenly distributed along the spectrum of percentage of strain-specific sequence and, for each, simulate 1 million 125-bp read pairs with an average divergence of 4% (ftp://ftp.nygenome.org/taxmaps/Benchmark/Datasets). Reads were then classified using taxMaps with default parameters on both paired-end and single-end modes against the original database. For each strain, strain-level sensitivity, precision, and corresponding F-score were computed.
Mock community data sets
To assess the classification accuracy on real data, we used two mock community single-end data sets, HiSeq and MiSeq, from a previously published benchmark (Wood and Salzberg 2014). Each data set was originally composed of 10,000 single-end reads from 10 different bacterial species. After adapter clipping using Cutadapt (Martin 2011), removal of sequences shorter than 31 bp and the complete removal of Streptococcus pneumoniae from the HiSeq data set due to the presence of chimeric reads that were likely artifacts, there were 8850 and 9953 reads left on the HiSeq and MiSeq data sets, respectively. For each data set, apart from running taxMaps (k = 125 and k = 300 for HiSeq and MiSeq, respectively), Kraken, Centrifuge, MegaBLAST, and BLASTN, we additionally ran the protein homology-based classifiers Kaiju and DIAMOND. DIAMOND classification followed the same criteria as BLASTN and MegaBLAST. Moreover, a filtering strategy was implemented for both BLAST programs, using the criteria (minimum bit score, win-score, and top-percent) described by the authors of MEGAN (Huson et al. 2007). We selected a win-score of 100 and minimum bit score cutoffs of 60 and two values, 5% and 10%, were explored for the top-percent cutoff (Supplemental Fig. S7). All methods used the refseq_complete_genomes database, with the exception of Kaiju and DIAMOND that used the correspondent set of annotated proteins (available at ftp://ftp.nygenome.org/taxmaps/Benchmark/Refseq_complete_genomes_DB). For each tool, rank-level sensitivity and precision were computed. Corresponding F-scores are given in Supplemental Table S4. We have also estimated genus abundance based on read classification for the HiSeq, MiSeq, and the two recently published data sets HC/LC and ZymoBIOMICS (McIntyre et al. 2017). For every genus, abundance estimates Aobs from each tool were then compared to the truth set value Aexp and the relative difference between the two Drel was calculated as
Real metagenomics samples
We downloaded seven Illumina data sets of real metagenomics samples from the Sequence Read Archive (SRA) (Leinonen et al. 2011). Their description and corresponding accession numbers can be found in Supplemental Table S2. On all data sets, adapter sequences were clipped, and low-quality end bases trimmed (Q < 20). Reads were classified with paired-end and single-end modes using taxMaps (k = 300), Kraken, Centrifuge, Kaiju, and DIAMOND. For each data set, apart from determining the number of classified reads by each method, we computed a novel rank-level metric called classification concordance. This metric is defined as the percentage of read pairs for which the independent classification of both ends is either the same or concordant at that particular rank, as long as one of the ends has been classified at that rank or below. For instance, if one end is classified as Escherichia coli and the other as Enterobacteriaceae, the classification for that read pair is considered to be concordant at the species level and at all ranks above. If the second end had been classified as Proteus vulgaris instead, the classification would be concordant at the family level and at all ranks above. To assess whether classification concordance could be used as a proxy for precision, we calculated the Spearman's rank correlation ρ between the two metrics on the simulated data sets, for all methods and at all ranks with the exception of “root.”
Software availability
taxMaps is freely available for academic and noncommercial research purposes from https://github.com/nygenome/taxmaps and it is provided, along with the scripts to compute the classification accuracy metrics described in the Methods (sensitivity, precision, F-score, and classification concordance), in Supplemental Data S1.
Supplementary Material
Acknowledgments
This work was partially supported by the Alfred P. Sloan Foundation.
Author contributions: A.C. developed the software. W.E.C. and A.C. performed the experiments and analysis. A.C., W.E.C., N.R., and M.C.Z. designed the experiment and wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.225276.117.
References
- Afshinnekoo E, Meydan C, Chowdhury S, Jaroudi D, Boyer C, Bernstein N, Maritz JM, Reeves D, Gandara J, Chhangawala S, et al. 2015. Geospatial resolution of human and bacterial diversity with city-scale metagenomics. Cell Syst 1: 72–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Bik HM, Maritz JM, Luong A, Shin H, Dominguez-Bello MG, Carlton JM. 2016. Microbial community patterns associated with automated teller machine keypads in New York City. mSphere 1: e00226-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady A, Salzberg SL. 2009. Phymm and PhymmBL: metagenomic phylogenetic classification with interpolated Markov models. Nat Methods 6: 673–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12: 59–60. [DOI] [PubMed] [Google Scholar]
- Ferragina P, Manzini G. 2000. Opportunistic data structures with applications. In Proceedings of the 41st Annual Symposium on Foundations of Computer Science, p. 390 IEEE Computer Society, Washington, DC. [Google Scholar]
- The Human Microbiome Project Consortium. 2012a. A framework for human microbiome research. Nature 486: 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Human Microbiome Project Consortium. 2012b. Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. 2007. MEGAN analysis of metagenomic data. Genome Res 17: 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Song L, Breitwieser FP, Salzberg SL. 2016. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res 26: 1721–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R, Sugawara H, Shumway M, International Nucleotide Sequence Database Collaboration. 2011. The sequence read archive. Nucleic Acids Res 39: D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Breitwieser FP, Thielen P, Salzberg SL. 2017. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci 3: e104. [Google Scholar]
- Marco-Sola S, Sammeth M, Guigo R, Ribeca P. 2012. The GEM mapper: fast, accurate and versatile alignment by filtration. Nat Methods 9: 1185–1188. [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17: 10–12. [Google Scholar]
- McIntyre AB, Ounit R, Afshinnekoo E, Prill RJ, Henaff E, Alexander N, Minot SS, Danko D, Foox J, Ahsanuddin S, et al. 2017. Comprehensive benchmarking and ensemble approaches for metagenomic classifiers. Genome Biol 18: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel P, Ng KL, Krogh A. 2016. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun 7: 11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Resource Coordinators. 2016. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 44: D7–D19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondov BD, Bergman NH, Phillippy AM. 2011. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics 12: 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ounit R, Wanamaker S, Close TJ, Lonardi S. 2015. CLARK: fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers. BMC Genomics 16: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen G, Garbarine E, Caseiro D, Polikar R, Sokhansanj B. 2008. Metagenome fragment classification using N-mer frequency profiles. Adv Bioinformatics 2008: 205969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27: 863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. 2012. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 9: 811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D, Alverdy J, An G, Coleman M, Garcia-Houchins S, Green J, Keegan K, Kelley ST, Kirkup BC, Kociolek L, et al. 2013. The Hospital Microbiome Project: Meeting Report for the 1st Hospital Microbiome Project Workshop on sampling design and building science measurements, Chicago, USA, June 7th–8th 2012. Stand Genomic Sci 8: 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S, Mende DR, Zeller G, Izquierdo-Carrasco F, Berger SA, Kultima JR, Coelho LP, Arumugam M, Tap J, Nielsen HB, et al. 2013. Metagenomic species profiling using universal phylogenetic marker genes. Nat Methods 10: 1196–1199. [DOI] [PubMed] [Google Scholar]
- Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15: R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. 2000. A greedy algorithm for aligning DNA sequences. J Comput Biol 7: 203–214. [DOI] [PubMed] [Google Scholar]
- Zhang C, Cleveland K, Schnoll-Sussman F, McClure B, Bigg M, Thakkar P, Schultz N, Shah MA, Betel D. 2015. Identification of low abundance microbiome in clinical samples using whole genome sequencing. Genome Biol 16: 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.