Abstract
The first tracking of the dynamics of a natural invasion by a transposable element (TE) provides unprecedented details on the establishment of host defense mechanisms against TEs. We captured a D. simulans population at an early stage of a P-element invasion and studied the spread of the TE in replicated experimentally evolving populations kept under hot and cold conditions. We analyzed the factors controlling the invasion by NGS, RNA-FISH, and gonadal dysgenesis assays. Under hot conditions, the P-element spread rapidly for 20 generations, but no further spread was noted later on. This plateauing of the invasion was mediated by the rapid emergence of P-element-specific piRNAs. Under cold conditions, we observed a lower expression of the P-element and a slower emergence of the piRNA defense, resulting in a three times slower invasion that continued beyond 40 generations. We conclude that the environment is a major factor determining the evolution of TEs in their host.
Transposable elements (TEs) are DNA sequences that selfishly multiply within genomes. TEs also frequently invade naïve species via horizontal transfer (Loreto et al. 2008; El Baidouri et al. 2014; Peccoud et al. 2017). Hosts have evolved a variety of defense mechanisms such as zinc-finger proteins, small RNA-based silencing strategies, DNA methylation, and chromatin modifications (Brennecke et al. 2007; Gunawardane et al. 2007; Slotkin and Martienssen 2007; Jacobs et al. 2014). Despite these elaborate defense mechanisms, TEs comprise a large portion of eukaryotic genomes, ranging from 3% in yeast to 85% in maize (Biémont and Vieira 2006; Schnable et al. 2009). It remains unclear how TEs have been able to attain such high proportions of genomes (Kazazian 2011). The issue is best addressed by quantifying the spread of TEs within a genome during an invasion before effective defense mechanisms are established.
Earlier studies introduced TEs into naïve genomes and monitored the dynamics of the ensuing TE invasion (Kidwell et al. 1988; Good et al. 1989; Montchamp-Moreau 1990). This work provided important insights into the biology of mobile DNA but suffers from certain limitations. First, the experiments were performed before the discovery of the piRNA host defense mechanism, such that this crucial component of the invasion dynamics was not monitored. Second, it is unclear whether artificially introduced TEs recapitulate natural invasions: Horizontal transfer of a TE may not only introduce naked TE DNA, but also other factors influencing TE dynamics (e.g., piRNAs, nonautonomous elements). Third, artificial introduction of TEs usually requires helper factors, such as transformation markers or sources of transposase (e.g., piggyBac) (Rozhkov et al. 2013) that are not transmitted during natural horizontal transfer but may influence the resulting TE invasion. The aberrant courtship behavior in Drosophila induced by the transformation marker mini-white is a classic example (Zhang and Odenwald 1995). Finally, many earlier studies relied on fairly coarse methods for monitoring TEs, so individual TE variants could not be distinguished. This is particularly important as nonautonomous TEs, such as internally deleted copies, could have profound effects on the invasion (Black et al. 1987; Robillard et al. 2016). Even recent work suffers to varying degrees from the same limitations: No natural TE invasion was investigated and the abundance of different TE variants was not considered (Khurana et al. 2011; Rozhkov et al. 2013; Robillard et al. 2016). One study did not investigate the role of small RNAs (Robillard et al. 2016), whereas another did not follow the invasion over multiple generations (Khurana et al. 2011).
Despite advances in the understanding of piRNA biology and the ability to characterize TEs by Next-Generation Sequencing (NGS), the dynamics of natural TE invasions and the accompanying response of the host defense system remain unknown. Natural TE invasions are extremely difficult to observe because successful horizontal transfer is probably very rare (Loreto et al. 2008), and the time from the horizontal transfer to the silencing of a TE is very short. Against these odds, in November 2010 we sampled a D. simulans population in Florida at the early stages of a P-element invasion, with 25%–44% of the flies having at least one P-element insertion (Kofler et al. 2015a; Hill et al. 2016).
The P-element is one of the best studied eukaryotic TEs (Kidwell et al. 1977; Engels 1983; Kelleher 2016). In the 1970s, a novel genetic syndrome known as “hybrid dysgenesis” (HD) was discovered (Kidwell et al. 1977). Crosses of males from wild-caught populations of D. melanogaster with females of laboratory strains led to offspring with spontaneous male recombination, mutations, and high rates of gonadal sterility, whereas reciprocal crosses produced offspring that were genetically identical but fertile (Kidwell et al. 1977; Bingham et al. 1982; Rubin et al. 1982). The non-reciprocality of HD led to the suggestion that it was caused by both chromosomal and cytoplasmic components (Kidwell and Kidwell 1975). Although the cytoplasmic component remained elusive, the chromosomal component was soon identified as a TE: the P-element, a 2907-bp DNA transposon encoding a single protein, the transposase (Bingham et al. 1982; O'Hare and Rubin 1983). The P-element invaded wild-caught populations but was absent in old laboratory strains (Kidwell 1983). Critically, although the P-element is transcribed in all tissues, it is only functional in the germline, where functional transposase is generated by splicing of the third intron (Engels 1983; Laski et al. 1986). Retention of this intron in somatic tissues results in a nonfunctional transposase that may even suppress P-element activity (Laski et al. 1986). This germline-restricted transposition minimizes damage to the host and maximizes the invasive properties of the P-element (Burt and Trivers 2008). The P-element transposes by a cut-and-paste mechanism (Engels et al. 1990; Kaufman and Rio 1992) that does not inherently increase copy numbers. A copy number increase is accomplished by sister chromatid–mediated gap repair resulting from the excision of a P-element (Engels et al. 1990) combined with preferential insertion into unreplicated DNA (Spradling et al. 2011). The gap repair of excised P-elements is occasionally interrupted, resulting in internally deleted P-elements (Engels et al. 1990). Some internally deleted P-elements, such as the KP-element, suppress P-element activity (similarly to unspliced P-element transcripts) (Black et al. 1987; Rio 1990). This raises the possibility that a P-element invasion may be stopped by accumulating internally deleted P-elements. The P-element is highly invasive, resulting in the invasion of D. melanogaster and D. simulans within the past 100 years (Daniels et al. 1990; Kofler et al. 2015a; Hill et al. 2016). After a horizontal transfer from D. melanogaster, it spread in D. simulans populations in the last 15 years (Kofler et al. 2015a; Hill et al. 2016). The sequence of the P-element in D. melanogaster and D. simulans is very similar, differing only by a single nucleotide replacement at position 2040 (Kofler et al. 2015a). The high rate of horizontal transfer may ensure the long-term survival of the P-element in the face of accumulating mutations, such as internally deleted P-elements (Schaack et al. 2010).
The cytoplasmic component of HD was discovered only recently: the piRNA-based host defense against TEs (Brennecke et al. 2007; Gunawardane et al. 2007). PIWI clade proteins are preferentially expressed in gonads and interact with 23- to 29-nt small RNAs (called piRNAs). In Drosophila, RNA-induced silencing complexes (RISC) of the Piwi family repress TE activity by two different mechanisms. In the primary pathway, piRNAs bound to Piwi guide the complex to nascent TE transcripts where the piRNA/Piwi complex transcriptionally silences TE insertions by locally modifying chromatin marks (transcriptional gene silencing [TGS]) (Sienski et al. 2012; Le Thomas et al. 2013). In the secondary pathway, piRNAs bound to the cytoplasmic proteins Aubergine (Aub) and Argonaute3 (Ago3) guide the complexes to cytoplasmic TE transcripts and direct their cleavage. The reciprocal and continuous cleavage cycle of sense and antisense TE transcripts emerging from piRNA clusters leads to amplification of piRNAs and post-transcriptional gene silencing (PTGS) (Brennecke et al. 2007; Gunawardane et al. 2007; Czech and Hannon 2016). Secondary piRNAs produced by this ping-pong cycle have a 10-nt overlap between piRNAs bound to Aub and Ago3. In addition to cleaving TE transcripts, the secondary pathway has two other main functions. First, during oogenesis, piRNA/Aub and piRNA/Piwi complexes are deposited in the egg by the mother, providing the offspring with an inherited defense against TEs (Brennecke et al. 2008). Maternally deposited piRNAs complementary to the P-element offer an explanation for the maternally transmitted cytoplasmic component of HD. Females with P-elements provide their offspring with piRNAs, which suppress P-element activity, and offspring of females lacking the P-element suffer from a high P-element mobility, leading to hybrid dysgenesis. Next, secondary piRNAs direct the biogenesis of primary piRNAs in the germline (piRNAs loaded into Piwi) (Han et al. 2015; Mohn et al. 2015; Senti et al. 2015). Consequently, the primary piRNA pathway in the germline is controlled by the secondary pathway. Because the secondary pathway already requires some mature piRNAs as input (presumably the maternally transmitted piRNAs) (Le Thomas et al. 2014), it remains unclear how piRNAs against a novel TE are actually established.
Results
Natural P-element invasion in D. simulans
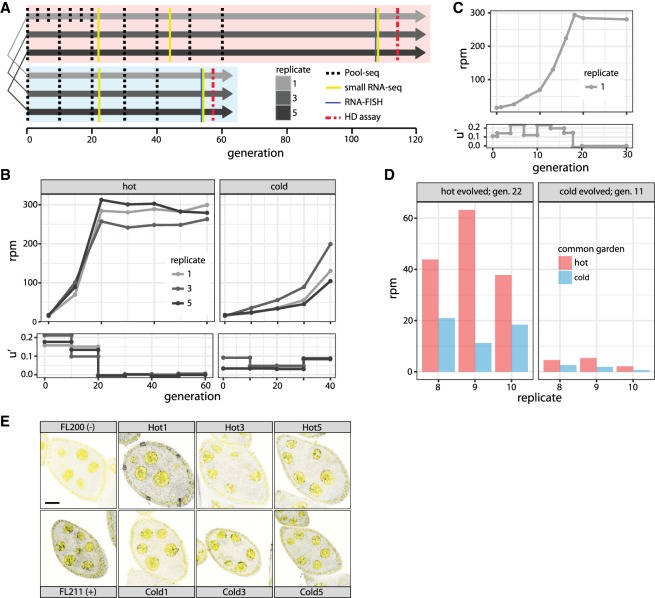
We collected 202 isofemale lines from a natural D. simulans population that was at the beginning of a P-element invasion (Kofler et al. 2015a). Using five females from each isofemale line, we established three replicate populations for each hot (cycling between 18°C and 28°C) and cold (cycling between 10°C and 20°C) conditions. Hot and cold replicates were paired, so that replicates with the same ID had identical parents (Fig. 1A). All populations were maintained at a population size of 1000 with nonoverlapping generations. We characterized the invasion by monitoring multiple molecular and phenotypic traits throughout the experiment (Fig. 1A).
Figure 1.
Overview of a natural P-element invasion in experimentally evolving D. simulans populations. (A) Experimental design. Three replicates were kept at hot and cold conditions using nonoverlapping generations. Replicates with the same index in hot and cold conditions are derived from the same parents. Note that flies in cold conditions develop slower. (B) P-element abundance in rpm (reads mapping to the P-element out of a million mapped reads; upper panel) and effective transposition rate (u′; lower panel) during the hot and cold invasion. (C) High-resolution view of the hot invasion in replicate 1. The invasion plateaus at generation 18. (D) RNA-seq shows differences in P-element expression between hot and cold conditions. We transferred flies from the hot invasion at generation 22 and from the cold invasion at generation 11 to both a hot and a cold common garden. P-element expression is consistently lower under cold conditions. (E) Sense transcripts of the P-element are expressed in D. simulans ovaries. We dissected flies from the hot invasion (Hot; index indicates replicate) at generation 108 and from the cold invasion (Cold) at generation 54 and performed single RNA-FISH using a set of Stellaris oligo probes to detect sense transcripts of the P-element (black dots). DAPI is shown in yellow. As a positive (+) and a negative control (−), we included ovaries from flies with and without the P-element, respectively (Hill et al. 2016). Scale bar, 20 µm.
We estimated P-element abundance by sequencing genomic DNA in intervals of no more than 10 generations for pools of individuals (Pool-seq) (Fig. 1A; Schlötterer et al. 2014). The P-element rapidly spread in all three replicates under hot conditions (ANOVA, effect of generations, P = 8.6 × 10−6) (Fig. 1B; Supplemental Tables S1, S2), increasing within 20 generations from an average of 16.1 rpm (reads per million, i.e., reads mapping to the P-element out of a million mapped reads; ≈1.79 insertions per diploid genome) to 280.6 rpm (≈31.7 insertions per diploid genome). The invasion plateaued in all three replicates around generation 20, and P-element abundance remained stable for the next 40 generations. Correspondingly, the effective transposition rate (u′ = transpositions minus losses, e.g., due to excisions or negative selection) was high over the first 20 generations (u′ = 0.1552; average over all three replicates) before it effectively dropped to zero (u′ = −0.0003) (Fig. 1B).
Because we inferred P-element abundance from the number of short reads mapping to the P-element, the increase in P-element abundance (Fig. 1B) may be due to either a few P-element insertions increasing in population frequency or to many novel insertions by transposition. To distinguish between the possibilities, we estimated the position and population frequency of TE insertions based on the fraction of paired-end reads supporting the presence/absence of a TE insertion (Kofler et al. 2016). We used a physical coverage of 15 (corresponding to sampling 15 haploid genomes from the total population) in all data sets to ensure an unbiased comparison of TE abundance among samples. Consistent with a high transposon activity, the number of low-frequency insertions rapidly increased during the early phase of the invasion under hot conditions (henceforth, “hot invasion”) (Supplemental Figs. S1–S3). The average population frequency of TE insertions also increased during the hot invasion (ANOVA, effect of generations P = 2.3 × 10−10) (Supplemental Fig. S8), possibly due to a loss of low-frequency insertions by genetic drift. To pinpoint when the hot invasion plateaued, we sampled more time points for hot replicate 1. The invasion rate did not slow down gradually but progressed at a constant high rate (u′ between 0.109 and 0.23) until generation 18, after which no further increase in copy number was noticed (Fig. 1C; Supplemental Table S3).
The P-element invasion under cold conditions (henceforth “cold invasion”) was much slower (Fig. 1B; Supplemental Table S1). P-element abundance increased from an average of 16.1 rpm (≈1.8 insertions per diploid genome) to an average of 144.7 rpm (≈16.1 insertions per diploid genome; ANOVA, effect of generations P = 2.9 × 10−5) by generation 40, but no plateau was observed. The effective transposition rate was much lower under cold conditions (u′ = 0.056), although it increased over the last 10 generations (u′ = 0.0865) compared to the first 30 generations (u′ = 0.0457). As with the hot invasion, the P-element increase under cold conditions reflects a proliferation of low-frequency insertions (Supplemental Figs. S4–S6). In contrast to the situation under hot conditions, the average population frequency of P-elements remained stable during the cold invasion (ANOVA, effect of generations P = 0.9) (Supplemental Fig. S9). This could be explained by an ongoing transposition at cold conditions, which generated novel low-frequency insertions, compensating for the loss of low-frequency insertions by drift. For this analysis, we solely considered P-element insertions on major chromosome arms (X, 2L, 2R, 3L, 3R, 4). Only very few P-element insertions were found in unmapped contigs (hot: 33 of 1696; cold: three of 204; population frequency <0.498).
Because all base substitutions within the P-element were at low frequencies (Supplemental Results S1), the P-element did not evolve during the invasion. The P-element had a similar insertion bias in all replicates (Supplemental Results S2).
The difference in the invasion dynamics under hot and cold conditions (Fig. 1B) is striking. As hybrid dysgenesis is only observed at temperatures above 24°C (Kidwell and Novy 1979), we hypothesized that an increased expression of the P-element under hot conditions may drive the faster invasion. To test this notion, we kept flies that had evolved at hot and cold conditions for 22 and 11 generations, respectively, under two different regimes (constant 23°C and constant 15°C) for two generations, sequenced RNA from whole bodies of virgin females [poly(A) selected] and measured the expression of the P-element. Expression of the P-element is significantly lower under cold than under hot conditions (ANOVA, effect of common garden P = 0.0075) (Fig. 1D). This may account for the slower invasion under cold conditions, although we cannot rule other factors, such as temperature-dependent efficiency of the transposase. Furthermore, it is not clear to what extent P-element expression in whole bodies and ovaries is correlated.
To test whether the P-element is transcribed in the ovarian germline, we performed single-molecule RNA-FISH, using ovaries dissected from flies of the hot invasion at generation 108 and from the cold invasion at generation 54. The presence of P-element transcripts (sense) in nurse cells (Fig. 1E) suggests that the P-element is expressed in the germline of the experimental populations.
Production of a functional transposase requires splicing of the third intron, which is restricted to the germline. In the RNA-seq data, we only found evidence for excision of the third intron in cold-evolved populations (generation 11) raised at constant 23°C (Supplemental Table S4). Despite many more reads mapping to the P-element, we found no evidence for splicing of the third intron in hot-evolved populations at generation 22 (Supplemental Table S4). This suggests that a functional P-element transposase is generated in cold-evolved populations at generation 11 but not in hot-evolved populations at generation 22, which is consistent with plateauing of the hot invasion by generation 20 (Fig. 1B). P-element expression at generation 22 (or later) at hot conditions may therefore be mostly somatic, or splicing of the third intron may be suppressed by the piRNA pathway (Teixeira et al. 2017).
The influence of TE activity on resident TE families has been studied in dysgenic crosses. In some studies, TE activity reactivated dormant families (Petrov et al. 1995; Khurana et al. 2011), but others could not confirm this result (Woodruff et al. 1987; Eggleston et al. 1988). Dysgenic crosses may not reflect the dynamics during a TE invasion, because they do not yield fertile offspring and novel TE insertions are not passed to the next generation. Our study provides insights on the effect of TE activity on resident TE families during a natural TE invasion. Only the fraction of reads mapping to the P-element increased during the hot as well as the cold invasion, while the other TE families remained constant (Supplemental Fig. S7). We conclude that other TE families are not influenced by the dynamics of the P-element.
Dynamics of internally deleted P-elements
P-elements with internal deletions are common in natural populations and in laboratory strains (O'Hare and Rubin 1983; Kofler et al. 2015a). Some of the internally deleted elements, such as the KP-element, repress P-element activity (Black et al. 1987; Rasmusson et al. 1993). P-elements with internal deletions express nonfunctional transposases that retain DNA-binding capacity and prevent functional transposases from accessing the transposase-binding sites and thus from mobilizing the P-element (Lee et al. 1998). Because of the tight coupling between P-element activity and the generation of internally deleted copies (Engels et al. 1990), it seems feasible that internally deleted P-elements emerge rapidly during an invasion and are responsible for the plateauing.
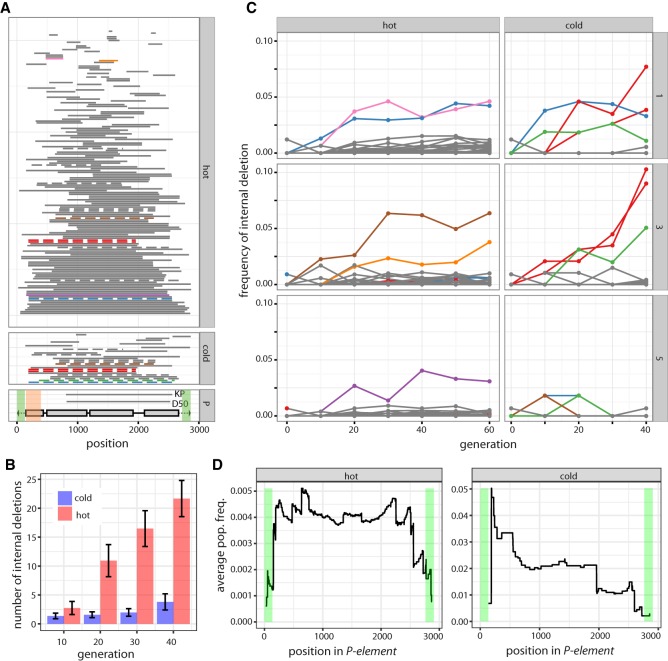
To address this idea, we searched for internally deleted P-elements in the experimentally evolving populations and realigned all reads mapping to the P-element allowing for large gaps. We identified 155 different internal deletions in hot populations (summed over all generations and replicates) and 27 in cold populations (Fig. 2A). Fifteen of the internal deletions were found in more than one of the six experimental populations. The most parsimonious explanation is that these internal deletions were present in the base population. Hence, about 140 internal deletions emerged during the hot invasion and 12 in the cold invasion. Consistent with deletions arising from interruption of sister chromatid–mediated gap repair (Engels et al. 1990), sites in the center of the P-element are most frequently deleted (Supplemental Fig. S13): As gap repair proceeds from both ends of a double-stranded break, interruptions will mostly happen before central parts of the P-element can be replicated.
Figure 2.
Dynamics of internally deleted P-elements during the invasion. (A) All internal deletions identified under hot and cold conditions are shown (sum over all replicates and generations). Horizontal bars represent the deleted sequence. Internal deletions that were probably present in the base population are shown in dashed lines, and internal deletions that increase in frequency (more than 3%) are indicated in color. The position of the two red internal deletions differs by a single nucleotide, so they likely refer to the same internal deletion. The lower panel shows the structure of the P-element, including the four ORFs (gray boxes) and the TIRs (black triangle). The position of two internal deletions with documented P-element repression (KP and D50) are indicated (Black et al. 1987; Ramussson et al. 1993). Regions required for mobilization of the P-element are shaded in green, and regions required for repressing P-element activity are shaded in orange (Majumdar and Rio 2015). (B) An unbiased comparison of the abundance of internally deleted P-elements between hot and cold conditions. The physical coverage of the P-element has been 100 times randomly sampled to 52. Internal deletions are consistently more frequent in hot-evolved populations. (C) Frequency of internally deleted P-elements during the hot and cold invasion for all three replicates (right panel). The color of the trajectories is as in panel A, allowing identification of the position (within the P-element) of internal deletions. (D) Fitness landscape of internally deleted P-elements: average population frequency of internal deletions covering a given site. A high average frequency indicates that deletion of a site is advantageous and leads, on average, to a frequency increase of an internally deleted P-element. Note that few internal deletions were observed under cold conditions.
The higher number of internally deleted P-elements under hot conditions could be an artifact of (1) more advanced generations sequenced, (2) the higher abundance of P-elements, and (3) differences in coverage between samples. To ensure an unbiased comparison, we analyzed the abundance of internally deleted P-elements at the same generations in the two environments and subsampled the coverage of the P-element to 52 (corresponding to randomly sampling 52 P-elements from every sample). There were consistently more internally deleted P-elements under hot than under cold conditions (Fig. 2B), probably due to the higher transposition rate (Fig. 1B), which results in more internally deleted copies generated from interrupted sister chromatid–mediated gap repair (Engels et al. 1990). We found that microhomology extends 2 nt upstream of and downstream from deletion breakpoints, which is consistent with nonhomologous end joining repairing double-stranded breaks resulting from interrupted gap repair (Supplemental Fig. S11; Engels et al. 1990; Chang et al. 2017).
P-elements with internal deletions rapidly emerged in our experimental populations. We estimated the frequency of internally deleted P-elements using short read data. By considering all reads mapping to the P-element (without inference of the genomic position), the abundance of a given internal deletion relative to the total population of P-elements can be assessed. We found that several internal deletions dramatically increased in frequency during both hot and cold invasions, with some reaching frequencies as high as 6.3% under hot conditions and 10.3% at cold conditions (Fig. 2C). The frequency increase of internally deleted copies could be explained by (1) genetic drift, (2) hitchhiking with positively selected variants, (3) positive selection because internally deleted copies repress P-element activity, and/or (4) preferential mobilization of internally deleted copies over full-length elements (Itoh et al. 2007). We used a “fitness landscape,” in which the frequency of an internal deletion reflects its relative fitness, to distinguish between the explanations. For every position in the P-element, we averaged the frequency of all internal deletions covering the site. Fitness landscapes were computed across all replicates using the frequency at generation 60 and 40 for hot- and cold-evolved populations, respectively. The four explanations of the increased frequency of internally deleted P-elements predict distinct fitness landscapes (Supplemental Fig. S12). Neutral deletions or deletions linked to positively selected variants will result in a similar average frequency across the entire P-element (Supplemental Fig. S12A). In the case of positive selection for internally deleted copies suppressing P-element activity, the DNA-binding domain of the P-element transposase (required for repression) (Fig. 2A; Lee et al. 1998) must be present (low frequency of deletions), and the transposase activity must be interrupted by deletions as seen for the KP-element (high frequency of deletions) (Fig. 2A; Supplemental Fig. S12B). If internally deleted P-elements are more readily mobilized than full-length elements (Itoh et al. 2007), we expect a low average frequency in regions necessary for mobilization of the P-element (Fig. 2A; Supplemental Fig. S12C; Majumdar and Rio 2015). Our analysis revealed a low average frequency at sites necessary for mobilization (Fig. 2D; Supplemental Fig. S13), implying that the internally deleted P-elements increase in frequency because they are more readily mobilized. We note, however, that the average frequency estimates at the ends of the P-element are based on few internal deletions and may thus be less reliable than in central regions.
The preferential mobilization of internally deleted elements may stop the invasion as a “side-effect” if they are sufficiently abundant. Lee et al. (1998) suggested that KP-like repressors need to be 30 times as abundant as full-length insertions for complete suppression of the P-element. Under the conservative assumption that a single P-element has no more than one internal deletion, we estimated at least 87% full-length P-elements in the hot populations at generation 20 (H1 87.4%; H3 90.2%; H5 94.1%) (Supplemental Fig. S10). With a frequency of 7.2%, 8.9%, and 2.6% for replicates 1, 3, and 5, respectively, KP-like repressors (first breakpoint of an internal deletion occurring at position >416 of the P-element) are less abundant than full-length copies (ratio of full-length to KP-like repressors of H1 12:1, H3 10:1, H5 36:1), making it very unlikely that internally deleted copies are responsible for halting the hot invasion.
Rapid emergence of a piRNA-based defense system
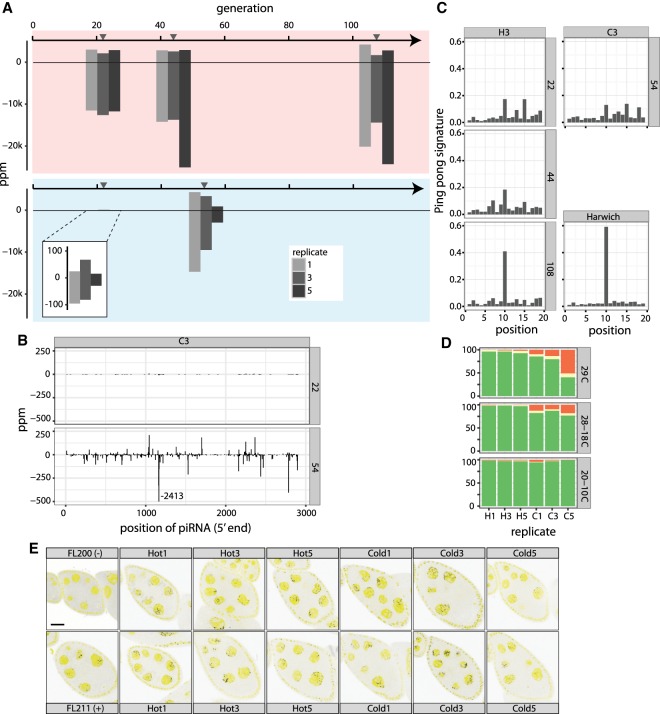
Because piRNAs are powerful regulators of TE activity, the piRNA pathway may be responsible for the plateauing of the hot invasion. To test this idea, we measured the abundance of piRNAs complementary to the P-element in our experimental populations by sequencing small RNAs at multiple time points (Fig. 1A). The majority of small RNAs are miRNAs and TE-derived piRNAs, with smaller fractions derived from tRNAs, rRNAs, and mRNAs (Supplemental Table S5). The vast majority of small RNAs complementary to the P-element had the expected length (23 and 29 nt) of piRNAs (Supplemental Figs. S14, S15). In generation 22 of the hot invasion, there was a substantial number of piRNAs complementary to the P-element (14,612 piRNAs per million miRNAs [ppm]; average over all replicates) (Fig. 3A). This level is within the range of piRNA abundance of other TE families in D. simulans (quantiles: 25% = 2774 ppm; 50% = 12,550 ppm; 75% = 26,642 ppm). The abundance of P-element piRNAs increased only slightly but not significantly different from generation 22 in the hot invasion (generations [g] g22 = 14,612 ppm; g44 = 20,440 ppm; g108 = 22,516 ppm; ANOVA, effect of generations, P = 0.0597). Only a few P-element piRNAs were detected at generation 22 in the cold invasion (104 ppm), with much larger amounts being present at generation 54 (11,888 ppm; ANOVA, effect of generations P = 0.013) (Fig. 3A). In both cases, the majority of the piRNAs are derived from the antisense strand (hot invasion: g22 = 81.2%, g44 = 86.1%, g108 = 86.6%; cold invasion: g22 = 67.0%, g54 = 75.4%). The dramatic increase in piRNA levels during the cold invasion is not an artifact of normalization because the piRNA abundance of all other TE families is stable during the hot and the cold invasion (Supplemental Fig. S20). In both temperature regimes, piRNAs emerged from the entire P-element (Fig. 3B; Supplemental Figs. S16, S17), with the most pronounced peaks of piRNA abundance at positions 1162 and 1164 (Fig. 3B; Supplemental Figs. S16, S17). It is unlikely that these two peaks are sequencing or mapping artifacts, as neither was found in the cold invasion at generation 22 (Fig. 3B; Supplemental Fig. S17).
Figure 3.
Dynamics of piRNAs during the P-element invasion. (A) Abundance of piRNAs complementary to the P-element during the hot and cold invasion in ppm (piRNAs per million miRNAs). Sense piRNA are on the positive y-axis, and antisense piRNAs on the negative y-axis. Gray triangles indicate the time of sequencing of small RNA libraries. (B) Abundance of piRNAs along the P-element during the cold invasion (replicate 3). Only the 5′ position of a piRNA is considered. A large peak at position 1164 was truncated. (C) Ping-pong signature of P-element piRNAs during the invasion (replicate 3). The ping-pong signature of D. melanogaster strain Harwich is shown for comparison (data from Brennecke et al. 2008). (D) Abundance of dysgenic ovaries at different temperature regimes for hot (H1, H3, H5) and cold (C1, C3, C5) evolved populations. A constant temperature of 29°C is generally used to test for dysgenic ovaries. Temperatures cycling between 28°C and 18°C and between 20°C and 10°C reflect conditions of our experimentally evolving populations: (green) normal ovaries; (red) dysgenic ovaries; (yellow) intermediate. (E) Antisense transcripts of the P-element are expressed in D. simulans ovaries. We dissected flies from the hot invasion (Hot) at generation 108 and from the cold invasion (Cold) at generation 54 and used single RNA-FISH to detect antisense transcripts of the P-element (black dots). DAPI is shown in yellow. As positive (+) and negative controls (−), we included ovaries from flies with and without the P-element (Hill et al. 2016). Scale bar, 20 µm.
Consistent with observed plateauing of the hot invasion between generation 18 and 20, a functional piRNA-based defense system was established by generation 22 (Fig. 1B,C). As the P-element was not entirely silenced by generation 40 in the cold invasion (Figs. 1B, 3D), the rate of establishment of a piRNA-based defense system depends on temperature. The temperature effect may be mediated by different numbers of insertions in the hot and cold invasion resulting in a more rapid silencing of abundant TEs.
The piRNA pathway–mediated silencing of TEs is more effective when a few initial piRNAs complementary to the TE are amplified by the ping-pong cycle (secondary piRNA pathway), which depends on both sense and antisense transcripts. Using single molecule RNA-FISH, we detected both sense and antisense transcripts in the ovaries of females from the hot and the cold invasion (Figs. 1E, 3E). Further evidence for the buildup of an active secondary piRNA pathway comes from the observation that the weakest signal of antisense transcripts was found under cold conditions in replicate 5, which has the fewest piRNAs (Fig. 3A,E). This suggests that piRNA production requires a sufficient supply of antisense transcripts.
During the ping-pong cycle, RNA cleavage products of Aub are loaded onto Ago3 and vice versa, where the Aub cleavage site is shifted by 10 bp from the Ago3 cleavage site. When plotting the average distance between sense and antisense piRNAs (using the 5′ ends of piRNA), a characteristic peak at position 10 can be found, i.e., the ping-pong signature (Brennecke et al. 2008). The size of the peak indicates the contribution of the secondary piRNA pathway to the total piRNA population. We found a peak at position 10 for the hot invasion at generations 22, 44, and 108, and for the cold invasion at generation 54 (Fig. 3C; Supplemental Fig. S19; excluding the antisense peaks at positions 1162 and 1164; Supplemental Fig. S18). The small number of piRNAs at generation 22 of the cold invasion did not result in a reliable ping-pong signature. Even after 54 generations, the ping-pong signature was still weak, suggesting that most piRNAs in the cold invasion did not derive from the secondary piRNA pathway (Fig. 3C; Supplemental Fig. S19). In the hot invasion, the ping-pong signature was weak at the start (generation 22) but became more pronounced at later generations (generations 44 and 108) (Fig. 3D; Supplemental Fig. S19), where it resembled the ping-pong signature of the P-element in the D. melanogaster strain Harwich (Fig. 3C). This suggests that the defense system is optimized by an increasing fraction of secondary piRNAs even after the hot invasion plateaued.
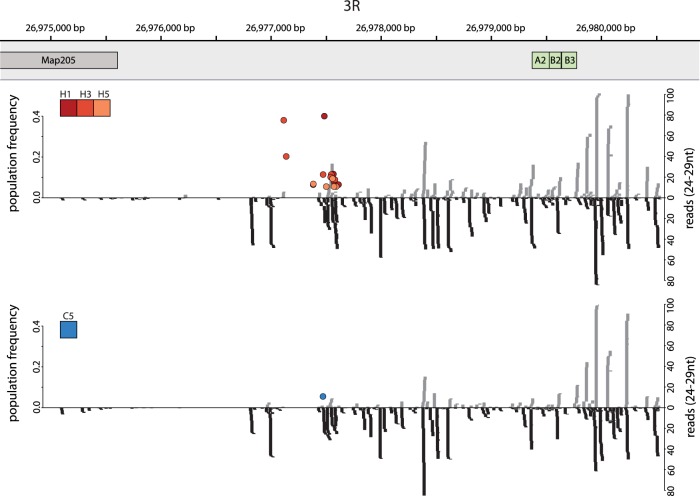
The presence of piRNAs complimentary to the P-element suggests that some P-elements transposed into piRNA producing loci, i.e., piRNA cluster. To find these insertions, we first identified the positions of piRNA clusters in D. simulans. We mapped small RNA reads of 24–29 bp to the reference genome, filtered ambiguously mapped reads, and determined piRNA clusters as regions with a high read density using a local score approach (Fariello et al. 2017). Combining all time points and replicates, we found 20 (out of 1698) P-element insertions in piRNA clusters for the hot invasion and one (out of 204) for the cold invasion (Supplemental Figs. S1–S6). The majority mapped to Chromosome 3R (16 hot, 1 cold) (Fig. 4), and only two were located on an unmapped contig (hot: two insertions on U). All of the the piRNA-cluster insertions were segregating (f < 0.41), with the vast majority segregating at low frequency (80% with f < 0.12). One piRNA-cluster insertion in the cold invasion (Fig. 4, blue dot) was already present in the base population, raising the possibility that this insertion has been positively selected in some replicates. The high abundance of P-element insertions at the telomeric end of Chromosome 3R may reflect the strong insertion bias of the P-element into a subset of telomere associated regions (TAS) as described by Karpen and Spradling (1992). To test whether these P-element insertions are near TAS regions, we aligned three TAS specific repeats (TLL) sequences of D. simulans (Asif-Laidin et al. 2017) to the genome. These TAS repeats are very close to the P-element insertions (Fig. 4), suggesting that multiple independent P-element insertions could have occurred in this region. Furthermore, with up to 496 bp between P-elements (Fig. 4), the insertion spacing exceeds the imprecision of the method for TE identification, which supports multiple independent P-element insertions in the piRNA cluster on 3R. Every individual carried, on average, 0.16 P-element insertions in piRNA clusters after the hot invasion plateaued. Assuming that a single piRNA-cluster insertion is sufficient to suppress the P-element, ∼84% of piRNA producing insertions have not been identified. It is likely that our approach does not identify all piRNA-cluster insertions, because only a small fraction of the low-frequency insertions were sampled. Additionally, insertions in other TEs were also missed. Using paired-end reads spanning P-element insertions, we estimate that ∼5% of the P-element insertions (4.9% at cold and 4.98% at hot conditions) are within another TE (mostly 1360, hobo and Juan) and may thus have been missed by our approach. Another explanation for the missing fraction of cluster insertions are euchromatic insertions that have been converted into piRNA producing loci by paramutations (de Vanssay et al. 2012; Mohn et al. 2014).
Figure 4.
P-element insertions in a piRNA cluster at the distal end of Chromosome 3R. The positions of P-elements (colored dots, with color indicating the replicate) are shown for hot (top panel; replicates H1, H3 and H5) and cold conditions (bottom panel; replicate C5). The positions of the telomere proximal gene Map205 and of TAS specific repeats (A2, B2, B3) (Asif-Laidin et al. 2017) are indicated. The abundance of piRNAs is shown on the right y-axis, and the population frequency of P-element insertions is shown on the left y-axis. Sense piRNAs are shown in gray and antisense piRNAs in black.
Given the lower piRNA abundance, the P-element could still be active in the cold invasion. High activity of the P-element results in dysgenic ovaries, whereas low activity results in mostly normal ovaries (Kidwell et al. 1977; Montchamp-Moreau 1990; Rio 1990; Kelleher 2016). In the hot invasion, we detected only few dysgenic ovaries at generation 114 (2.1%–4.6%) (Fig. 3D; Supplemental Table S6), whereas cold-evolved populations were highly dysgenic at generation 57 (11.9%–55.1%) (Fig. 3D; Supplemental Table S6). Cold replicate 5, with the lowest piRNA abundance (984 ppm), had the most dysgenic ovaries (55.1%). Our results suggest that the P-element is still active in cold populations at generation 57, while it is silenced in hot populations at generation 114. The abundance of piRNAs correlates with the severity of gonadal dysgenesis: Most dysgenic ovaries are found when piRNA abundance is low. The low frequency of dysgenic ovaries at cold conditions can be explained by a low P-element activity at cold conditions, which is also supported by the lower P-element expression at cold than at hot conditions (Fig. 1D).
Discussion
For the first time, we document the dynamics of a TE invasion after a naturally occurring horizontal transfer. Using experimental evolution, we show that the invasion dynamics are strongly affected by the environment and distinguish between stochastic and deterministic processes.
Origin of de novo piRNAs
The plateauing of P-element abundance around generation 20 in the hot invasion suggests that the activity of P-elements is suppressed around this time point. Because too few P-elements have internal deletions to silence the P-element and piRNAs complementary to the P-element emerge rapidly, we conclude that the establishment of a piRNA-based defense system is responsible for the plateauing of the hot invasion. piRNAs are derived from discrete genomic loci, i.e., piRNA clusters (Brennecke et al. 2007; Malone et al. 2009; Czech and Hannon 2016). Mature piRNAs suppress TE activity either through the primary pathway (Piwi-bound piRNAs; TGS), or the secondary pathway (Aub- and Ago3-bound piRNAs; PTGS) (Czech and Hannon 2016). In the germline, the primary piRNA pathway operates downstream from the secondary pathway, and piRNAs produced by the secondary pathway define the targets of the primary pathway (Han et al. 2015; Mohn et al. 2015; Senti et al. 2015). The secondary piRNA pathway thus requires mature piRNAs as input. Maternally deposited piRNAs induce the secondary pathway in flies with established TEs (Senti et al. 2015). In the case of an invasion of a novel TE family, the secondary piRNA pathway needs to be initiated by de novo piRNAs. Four sources of de novo piRNAs against the P-element are conceivable: (1) piRNAs may be derived from a different TE with partial sequence similarity to the P-element; (2) P-element transcripts may be endolytically cleaved to give fragments with a 5′-phosphate that are loaded onto Piwi and Aub (Garneau et al. 2007); (3) siRNAs (or their cleavage products) from the cleavage of double-stranded P-element transcripts may be loaded onto Piwi and Aub (siRNAs may appear before piRNAs during a TE invasion) (Rozhkov et al. 2013) and there is a link between piRNA and siRNA abundance (Erwin et al. 2015); and (4) a P-element inserted into a germline piRNA cluster may be the origin of phased piRNAs, trailing ping-pong amplification of neighboring TE insertions (Han et al. 2015; Mohn et al. 2015). The last possibility is consistent with the observation that a single P-element insertion into subtelomeric regions (likely a piRNA cluster) is sufficient to repress P-element activity (Ronsseray et al. 1991; Josse et al. 2007). Our observation of P-element insertions in piRNA clusters is consistent with this view. Nevertheless, more work is needed to identify the source of de novo piRNAs during a TE invasion.
The influence of temperature on the dynamics of the P-element invasion
Temperature has a pronounced effect on the dynamics of a P-element invasion. P-element expression is positively correlated with temperature, resulting in more insertions at higher temperatures. Despite a probable occurrence of P-elements at different positions in each replicate (the invasion occurred independently in each replicate), P-element expression was consistently lower under cold conditions. This suggests that the expression differences are not caused by the position of P-element insertion, but rather result from trans-acting factors. Many TEs contain transcription factor binding sites (TFBS) (Feschotte 2008), and the expression of transcription factors varies with temperature (Chen et al. 2015a). We thus propose that the P-element has internal TFBS (Supplemental Table S7) that bind temperature-dependent transcription factors (Jakšić et al. 2017). Alternatively, stress associated with hot conditions may activate the P-element. Nevertheless, as cold temperatures are also stressful (Petavy et al. 2001; Chen et al. 2015b), but do not induce hybrid dysgenesis, which is caused by a high P-element activity (Kidwell and Novy 1979), we argue that temperature-dependent P-element regulation is a more likely explanation for the higher P-element expression at hot conditions. Although the P-element defense system is more rapidly established under hot conditions, it is not clear whether the difference is directly linked to temperature or reflects the higher activity of the P-element. The analysis of replicate populations evolved for more generations under cold conditions will be required to shed light on the temperature dependence of the piRNA-based defense system.
Shotgun silencing model
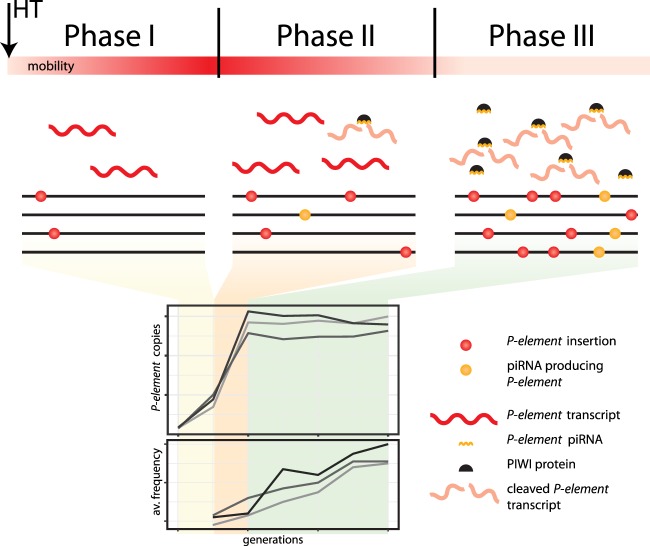
We distinguish three different phases of a P-element invasion. Initially, the piRNA pathway is not active and the P-element multiplies rapidly within populations, possibly at an increasing rate (Fig. 5). In the second phase, the first P-elements insert into piRNA clusters, resulting in piRNAs complementary to the P-element (Fig. 5; Saint-Leandre et al. 2017). piRNAs are not yet sufficiently abundant to repress P-element activity fully, as seen in the cold invasion at generation 54. The third phase sees an increasing number of piRNAs and complete silencing of the P-element (Fig. 5), mediated by increasing numbers of P-element insertions within piRNA clusters; an increasing efficiency of the secondary piRNA pathway, possibly due to larger amounts of maternally inherited P-element piRNAs (Josse et al. 2007); and increasing conversion of euchromatic P-element insertions into piRNA-generating loci (Mohn et al. 2014; Shpiz et al. 2014). The plateau of the hot invasion, at 31.7 P-element copies per diploid genome, may be interpreted as the average number of insertions when every fly has at least one P-element insertion in a piRNA cluster. We propose that the highly reproducible invasion dynamics arises from the availability of many piRNA clusters. High P-element activity results in many independent de novo insertions into piRNA clusters. This view is supported by our finding that most piRNA-cluster insertions segregate at a low frequency. Consequently, most flies in a population obtain a functional defense system in a very narrow time interval. We call this novel, rapid adaptation to invading TEs “Shotgun Silencing” to signify (1) a rapid silencing response, with most individuals acquiring a piRNA-cluster insertion within a short period of time; and (2) the insertions are distributed over many piRNA clusters, with most individuals having different insertion sites. This model does not require negative selection against TEs because we assume that piRNAs stop the invasion. It is, however, an important open question to which extent negative selection against TEs (e.g., insertions causing hybrid dysgenesis) (Kelleher 2016) influences the dynamics of TE invasions.
Figure 5.
Shotgun silencing model of the P-element invasion. The heatmap indicates mobility of the P-element. (av. frequency) average population frequency of the P-element insertions.
The lag time between invasion and silencing of a TE is an important determinant of genome size
Our model of the P-element invasion dynamics suggests that a large number of TEs are inserted into the genome before the piRNA defense system silences TE activity. We propose that the lag time between invasion and silencing is an important determinant of genome size. Because the P-element is silenced at 31.7 copies per diploid genome, the haploid genome size of D. simulans is increased by about 46 kb (2907 × 31.7/2) or 0.02% (based on a genome of 162 Mb) (Bosco et al. 2007). D. simulans has about 120 TE families (Kofler et al. 2015b), so TE insertions accumulating during the lag time could account for ∼3.4% (120 × 0.02) of the genome, assuming a similar lag time for all TE families. D. simulans has a repeat content of between 2.73% and 12.5% (Drosophila 12 Genomes Consortium 2007; Kofler et al. 2015b), so 27%–100% of the TE content of D. simulans can be accounted for by TE proliferation in the time before silencing. The time between initial TE transmission and its silencing, may thus account for a substantial fraction of the TE content of each species, explaining how TEs have reached such high proportions in genomes (Kazazian 2011). However, it is clear that other factors also influence genome size such as genetic drift, chromosomal duplications, deletion bias, microsatellites, and physiological constraints (Petrov 2001). We expect that future research will shed light on the factors that influence this lag time, thereby revealing some of the major mechanisms governing TE content and genome evolution across species.
Methods
Experimental populations
We established 202 isofemale lines from a D. simulans population collected in November 2010. After six to seven generations in the laboratory, replicate populations were established. Three replicates each were exposed to hot (cycling between 18°C and 28°C) and cold (cycling between 10°C and 20°C) conditions using a census size of about 1000 and nonoverlapping generations.
Estimating P-element abundance
The evolved populations were sequenced each tenth generation as pools (Pool-seq) (Schlötterer et al. 2014) using Illumina paired-end technology. Reads were aligned with bwa aln (Li and Durbin 2010) to the D. simulans reference genome (Palmieri et al. 2015), and P-element abundance was estimated with PoPoolationTE2 (Kofler et al. 2016).
Estimating the abundance of P-elements with internal deletions
Reads mapping to the P-element were realigned with GSNAP, a tool allowing large indels (Wu and Nacu 2010), and the frequency of internally deleted P-elements was estimated as the ratio between reads supporting an internal deletion and the coverage of the P-element.
RNA-seq
We estimated the expression of the P-element in replicates 8, 9, and 10 of hot-evolved (generation 22) and cold-evolved (generation 11) populations. We sampled 50 virgin females from populations that were kept in a hot (constant 23°C) and a cold common garden (constant 15°C) for two generations and sequenced poly(A) RNA libraries with the Illumina paired-end technology.
Small RNA-seq
Total RNA was extracted from virgin females reared in under hot (constant 23°C) conditions for two generations. The small RNA libraries were sequenced by Fasteris (https://www.fasteris.com/dna/).
Single molecule RNA-FISH
CAL Fluor Red 590-labeled Stellaris oligo probes were used to detect P-element transcripts in D. simulans ovaries. One probe set was designed for sense mRNA, and two probe sets were designed for antisense mRNA. Confocal sections of egg chambers were acquired with a Zeiss LSM780 microscope.
Gonadal dysgenesis assays
Fly eggs were kept until eclosion under three temperature regimes (29°C constant; hot fluctuating 18°C–28°C; cold fluctuating 10°C–20°C). Eclosed flies were kept for 2 d, at constant 23°C on apple juice agar with live yeast, before dissection in PBS. The presence of dysgenic ovaries was scored.
Data access
The data from this study have been submitted to the European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena) under accession numbers PRJEB20533 and PRJEB20780. The scripts used for data analysis, the positions of piRNA clusters, and TE insertions are available from Sourceforge (https://sourceforge.net/projects/te-tools/) and as Supplemental Data S1.zip.
Supplementary Material
Acknowledgments
We thank all members of the Institute of Population Genetics for feedback and support and Justin Blumenstiel for helpful discussions. The work was supported by the European Research Council Grant “ArchAdapt.”
Author contributions: R.K., K.-A.S., and C.S. conceived the study. R.K. analyzed the data and performed gonadal dysgenesis assays. K.-A.S. performed single molecule RNA-FISH and RNA extractions. R.T. provided fly strains, set up the experimental cages, and contributed to writing. V.N. prepared Pool-seq and RNA-seq libraries. R.K., K.-A.S., and C.S. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.228627.117.
References
- Asif-Laidin A, Delmarre V, Laurentie J, Miller WJ, Ronsseray S, Teysset L. 2017. Short and long-term evolutionary dynamics of subtelomeric piRNA clusters in Drosophila. DNA Res 24: 459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biémont C, Vieira C. 2006. Junk DNA as an evolutionary force. Nature 443: 521–524. [DOI] [PubMed] [Google Scholar]
- Bingham PM, Carolina N, Kiclwell G, Rubin GM. 1982. The molecular basis of P-M hybrid dysgenesis: the role of the P element, a P-strain-specific transposon family. Cell 29: 995–1004. [DOI] [PubMed] [Google Scholar]
- Black DM, Jackson MS, Kidwell MG, Dover GA. 1987. KP elements repress P-induced hybrid dysgenesis in Drosophila melanogaster. EMBO J 6: 4125–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco G, Campbell P, Leiva-Neto JT, Markow TA. 2007. Analysis of Drosophila species genome size and satellite DNA content reveals significant differences among strains as well as between species. Genetics 177: 1277–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. 2008. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322: 1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A, Trivers R. 2008. Genes in conflict: the biology of selfish genetic elements. Belknap Press, Cambridge, MA. [Google Scholar]
- Chang HH, Pannunzio NR, Adachi N, Lieber MR. 2017. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol 18: 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Nolte V, Schlötterer C. 2015a. Temperature related reaction norms of gene expression: regulatory architecture and functional implications. Mol Biol Evol 32: 2393–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Nolte V, Schlötterer C. 2015b. Temperature stress mediates decanalization and dominance of gene expression in Drosophila melanogaster. PLoS Genet 11: e1004883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. 2016. One loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem Sci 41: 324–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels SB, Peterson KR, Strausbaugh LD, Kidwell MG, Chovnick A. 1990. Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics 124: 339–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vanssay A, Bougé AL, Boivin A, Hermant C, Teysset L, Delmarre V, Antoniewski C, Ronsseray S. 2012. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature 490: 112–115. [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218. [DOI] [PubMed] [Google Scholar]
- Eggleston W, Schlitz D, Engels W. 1988. P–M hybrid dysgenesis does not mobilize other transposable element families in D. melanogaster. Nature 331: 368–370. [DOI] [PubMed] [Google Scholar]
- El Baidouri M, Carpentier MC, Cooke R, Gao D, Lasserre E, Llauro C, Mirouze M, Picault N, Jackson SA, Panaud O. 2014. Widespread and frequent horizontal transfers of transposable elements in plants. Genome Res 24: 831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels WR. 1983. The P family of transposable elements in Drosophila. Annu Rev Genet 17: 315–344. [DOI] [PubMed] [Google Scholar]
- Engels WR, Johnson-Schlitz DM, Eggleston WB, Sved J. 1990. High-frequency P element loss in Drosophila is homolog dependent. Cell 62: 515–525. [DOI] [PubMed] [Google Scholar]
- Erwin AA, Galdos MA, Wickersheim ML, Harrison CC, Marr KD, Colicchio JM, Blumenstiel JP. 2015. piRNAs are associated with diverse transgenerational effects on gene and transposon expression in a hybrid dysgenic syndrome of D. virilis. PLoS Genet 11: e1005332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fariello MI, Boitard S, Mercier S, Robelin D, Faraut T, Arnould C, Recoquillay J, Bouchez O, Salin G, Dehais P, et al. 2017. Accounting for linkage disequilibrium in genome scans for selection without individual genotypes: the local score approach. Mol Ecol 26: 3700–3714. [DOI] [PubMed] [Google Scholar]
- Feschotte C. 2008. Transposable elements and the evolution of regulatory networks. Nat Rev Genet 9: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. 2007. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8: 113–126. [DOI] [PubMed] [Google Scholar]
- Good AG, Meister GA, Brock HW, Grigliatti T, Hickey DA. 1989. Rapid spread of transposable P elements in experimental populations of Drosophila melanogaster. Genetics 122: 387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590. [DOI] [PubMed] [Google Scholar]
- Han BW, Wang W, Li C, Weng Z, Zamore PD. 2015. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348: 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T, Schlötterer C, Betancourt AJ. 2016. Hybrid dysgenesis in Drosophila simulans associated with a rapid invasion of the P-element. PLoS Genet 12: e1005920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Takeuchi N, Yamaguchi M, Yamamoto MT, Boussy IA. 2007. Prevalence of full-size P and KP elements in North American populations of Drosophila melanogaster. Genetica 131: 21–28. [DOI] [PubMed] [Google Scholar]
- Jacobs FM, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. 2014. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516: 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakšić AM, Kofler R, Schlötterer C. 2017. Regulation of transposable elements: interplay between TE-encoded regulatory sequences and host-specific trans-acting factors in Drosophila melanogaster. Mol Ecol 26: 5149–5159. [DOI] [PubMed] [Google Scholar]
- Josse T, Teysset L, Todeschini AL, Sidor CM, Anxolabéhère D, Ronsseray S. 2007. Telomeric trans-silencing: an epigenetic repression combining RNA silencing and heterochromatin formation. PLoS Genet 3: 1633–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Spradling AC. 1992. Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single P element insertional mutagenesis. Genetics 132: 737–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman PD, Rio DC. 1992. P element transposition in vitro proceeds by a cut-and-paste mechanism and uses GTP as a cofactor. Cell 69: 27–39. [DOI] [PubMed] [Google Scholar]
- Kazazian HH. 2011. Mobile DNA: finding treasure in junk. FT Press, Upper Saddle River, NJ. [Google Scholar]
- Kelleher ES. 2016. Reexamining the P-element invasion of Drosophila melanogaster through the lens of piRNA silencing. Genetics 203: 1513–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana JS, Wang J, Xu J, Koppetsch BS, Thomson TC, Nowosielska A, Li C, Zamore PD, Weng Z, Theurkauf WE. 2011. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell 147: 1551–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG. 1983. Evolution of hybrid dysgenesis determinants in Drosophila melanogaster. Proc Natl Acad Sci 80: 1655–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, Kidwell JF. 1975. Cytoplasm-chromosome interactions in Drosophila melanogaster. Nature 253: 755–756. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Novy JB. 1979. Hybrid dysgenesis in Drosophila melanogaster: sterility resulting from gonadal dysgenesis in the P-M system. Genetics 92: 1127–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, Kidwell JF, Sved JA. 1977. Hybrid dysgenesis in Drosophila melanogaster: a syndrome of aberrant traits including mutations, sterility and male recombination. Genetics 86: 813–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell M, Kimura K, Black D. 1988. Evolution of hybrid dysgenesis potential following P element contamination in Drosophila melanogaster. Genetics 119: 815–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Hill T, Nolte V, Betancourt A, Schlötterer C. 2015a. The recent invasion of natural Drosophila simulans populations by the P-element. Proc Natl Acad Sci 112: 6659–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Nolte V, Schlötterer C. 2015b. Tempo and mode of transposable element activity in Drosophila. PLoS Genet 11: e1005406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Gómez-Sánchez D, Schlötterer C. 2016. PoPoolationTE2: comparative population genomics of transposable elements using pool-seq. Mol Biol Evol 33: 2759–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laski FA, Rio DC, Rubin GM. 1986. Tissue specificity of Drosophila P element transposition is regulated at the level of mRNA splicing. Cell 44: 7–19. [DOI] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Tóth KF. 2013. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev 27: 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Stuwe E, Li S, Du J, Marinov G, Rozhkov N, Chen YC, Luo Y, Sachidanandam R, Toth KF, et al. 2014. Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev 28: 1667–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC, Beall EL, Rio DC. 1998. DNA binding by the KP repressor protein inhibits P-element transposase activity in vitro. EMBO J 17: 4166–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreto EL, Carareto CM, Capy P. 2008. Revisiting horizontal transfer of transposable elements in Drosophila. Heredity 100: 545–554. [DOI] [PubMed] [Google Scholar]
- Majumdar S, Rio DC. 2015. P transposable elements in Drosophila melanogaster. Microbiol Spectr 3: 484–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Sienski G, Handler D, Brennecke J. 2014. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 157: 1364–1379. [DOI] [PubMed] [Google Scholar]
- Mohn F, Handler D, Brennecke J. 2015. piRNA-guided slicing specifies transcripts for Zucchini-dependent phased piRNA biogenesis. Science 348: 812–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montchamp-Moreau C. 1990. Dynamics of P-M hybrid dysgenesis in P-transformed lines of Drosophila simulans. Evolution 44: 194–203. [DOI] [PubMed] [Google Scholar]
- O'Hare K, Rubin GM. 1983. Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell 34: 25–35. [DOI] [PubMed] [Google Scholar]
- Palmieri N, Nolte V, Chen J, Schlötterer C. 2015. Genome assembly and annotation of a Drosophila simulans strain from Madagascar. Mol Ecol Resour 15: 372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccoud J, Loiseau V, Cordaux R, Gilbert C. 2017. Massive horizontal transfer of transposable elements in insects. Proc Natl Acad Sci 114: 4721–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petavy G, David JR, Gibert P, Moreteau B. 2001. Viability and rate of development at different temperatures in Drosophila: a comparison of constant and alternating thermal regimes. J Therm Biol 26: 29–39. [DOI] [PubMed] [Google Scholar]
- Petrov D. 2001. Evolution of genome size: new approaches to an old problem. Trends Genet 17: 23–28. [DOI] [PubMed] [Google Scholar]
- Petrov DA, Schutzman JL, Hartl DL, Lozovskaya ER. 1995. Diverse transposable elements are mobilized in hybrid dysgenesis in Drosophila virilis. Proc Natl Acad Sci 92: 8050–8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmusson KE, Raymond JD, Simmons MJ. 1993. Repression of hybrid dysgenesis in Drosophila melanogaster by individual naturally occurring P elements. Genetics 133: 605–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio DC. 1990. Molecular mechanisms regulating Drosophila P element transposition. Annu Rev Genet 24: 543–578. [DOI] [PubMed] [Google Scholar]
- Robillard E, Le Rouzic A, Zhang Z, Capy P, Hua-Van A. 2016. Experimental evolution reveals hyperparasitic interactions among transposable elements. Proc Natl Acad Sci 113: 14763–14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronsseray S, Lehman M, Anxolabéhère D. 1991. The maternally inherited regulation of P elements in Drosophila melanogaster can be elicited by two P copies at cytological site 1A on the X chromosome. Genetics 129: 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhkov NV, Schostak NC, Zelentsova ES, Yushenova IA, Zatsepina OG, Evgen'ev MB. 2013. Evolution and dynamics of small RNA response to a retroelement invasion in Drosophila. Mol Biol Evol 30: 397–408. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Kidwell MG, Bingham PM. 1982. The molecular basis of P-M hybrid dysgenesis: the nature of induced mutations. Cell 29: 987–994. [DOI] [PubMed] [Google Scholar]
- Saint-Leandre B, Claverau I, Hua-Van A, Capy P. 2017. Transcriptional polymorphism of piRNA regulatory genes underlies the mariner activity in Drosophila simulans testes. Mol Ecol 38: 42–49. [DOI] [PubMed] [Google Scholar]
- Schaack S, Gilbert C, Feschotte C. 2010. Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol 25: 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlötterer C, Tobler R, Kofler R, Nolte V. 2014. Sequencing pools of individuals—mining genome-wide polymorphism data without big funding. Nat Rev Genet 15: 749–763. [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, Minx P, et al. 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Senti KA, Jurczak D, Sachidanandam R, Brennecke J. 2015. piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes Dev 29: 1747–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpiz S, Ryazansky S, Olovnikov I, Abramov Y, Kalmykova A. 2014. Euchromatic transposon insertions trigger production of novel pi- and endo-siRNAs at the target sites in the Drosophila germline. PLoS Genet 10: e1004138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Dönertas D, Brennecke J. 2012. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151: 964–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. 2007. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Bellen HJ, Hoskins RA. 2011. Drosophila P elements preferentially transpose to replication origins. Proc Natl Acad Sci 108: 15948–15953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira FK, Okuniewska M, Malone CD, Coux RX, Rio DC, Lehmann R. 2017. piRNA-mediated regulation of transposon alternative splicing in the soma and germ line. Nature 552: 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff R, Blount J, Thompson J Jr. 1987. Hybrid dysgenesis in D. melanogaster is not a general release mechanism for DNA transpositions. Science 237: 1206–1209. [DOI] [PubMed] [Google Scholar]
- Wu TD, Nacu S. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26: 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SD, Odenwald WF. 1995. Misexpression of the white (w) gene triggers male–male courtship in Drosophila. Proc Natl Acad Sci 92: 5525–5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.