Abstract
Background
Lipid transporters play an essential role in lipid delivery and distribution, but their influence on seed oil production in oilseed crops is not well studied.
Results
Here, we examined the effect of two lipid transporters, FAX1 (fatty acid export1) and ABCA9 (ATP-binding cassette transporter subfamily A9) on oil production and lipid metabolism in the oilseed plant Camelina sativa. Overexpression (OE) of FAX1 and ABCA9 increased seed weight and size, with FAX1-OEs and ABCA9-OEs increasing seed length and width, respectively, whereas FAX1/ABCA9-OEs increasing both. FAX1-OE and ABCA9-OE displayed additive effects on seed oil content and seed yield. Also, OE of FAX1 and ABCA9 affected membrane lipid composition in developing pods, especially on phosphatidylcholine, phosphatidylethanolamine, and phosphatidylglycerol. The expression of some genes involved in seed oil synthesis, such as DGAT2, PDAT1, and LEC1, was increased in developing seeds of FAX1- and/or ABCA9-OEs.
Conclusion
These results indicate that increased expression of FAX1 and ABCA9 can potentially be applied to improving camelina oil production.
Keywords: Camelina, Lipid metabolism, Oil production, Seed weight, Transporters
Background
Fatty acids (FAs) are the major and essential component of membrane lipids and important energy stores for metabolism and cellular energy homeostasis [1, 2]. In addition, FAs participate in many regulatory processes in organismal growth, development, and stress responses. In plants, FAs are synthesized in plastids and exported out of plastids and to endoplasmic reticulum (ER) for elongation, desaturation, and other embellishments [3, 4]. One transporter, FAX1 (fatty acid export 1), a membrane protein in chloroplast inner envelopes, was identified to transport FAs out of chloroplasts in Arabidopsis thaliana [5]. Overexpression (OE) of FAX1 led to an increase in ER-derived lipids and a decrease in several plastid-produced lipids in flowers and leaves [5]. FAX1 overexpression also increased the biomass production and seed oil content in Arabidopsis [5, 6]. After acyl-CoA transported to cytosol, it bound to acyl-CoA-binding proteins (ACBPs), which might contribute to drive plastid FA-export [7].
FA associated with ER is involved in the biosynthesis of triacylglycerol (TAG). An ER-localized ATP-binding cassette (ABC) transporter subfamily A, ABCA9, was described to transport FA/acyl-CoA to the ER [8]. Developing seeds of ABCA9-knockout mutant (abca9) incorporated less 14C-oleoyl-CoA into TAG compared with WT seeds. OE of ABCA9 enhanced TAG deposition by up to 40%, with enlarged seeds, larger embryo, more densely packed with oil bodies [8]. When ABCA9 was overexpressed, the seed size and seed oil content were increased, and abca9 had opposite effects [8]. However, the function of FAX1 and ABCA9 and influence on seed oil accumulation in oilseed crops remain to be tested.
Camelina sativa, an archaic oilseed crop had cultivation more than 3000 years [9, 10]. Camelina was an important oilseed crop in Europe and Asia for centuries [11, 12], but its cultivation declined in the past century and was replaced by higher-yielding crops, such as rapeseed (Brassica napus) [9, 13]. Camelina has several advantages as an oilseed crop. Camelina seed oil contains a high level of unsaturated FAs (> 90%), with the level of polyunsaturated α-linolenic acid being 30–40% of the total oil [14–16]. In addition, camelina seed meals contain a low level of glucosinolates, toxic for feed use [9, 16]. Furthermore, camelina is a low-input crop with a low requirement for water and nutrients, and it is resistant to common Brassicaceae pests and pathogens and adaptable to hostile environmental conditions [11, 17, 18]. Moreover, camelina has a short life cycle with 85–100 days from seeds to seeds, and it is easy to transform genetically [19, 20]. In recent years, biotechnological manipulations of one or multiple genes have been employed to modify camelina seed oil compositions. Those include the production of fish oil-like levels (> 12%) of polyunsaturated fatty acids to achieve a high ω3/ω6 ratio [21–25], oleyl oleate wax esters [26], acetyl glyceride oils [27], poly-3-hydroxybutyrate [28], ω-7 monounsaturated fatty acids [29], cyclopropane fatty acids [30], and oleic acid levels from 16% to over 50% [31], as well as decreased levels of α-linolenic acid levels [32] and of C20–C24 very long-chain fatty acids to less than 2% of total fatty acids [33]. However, low seed and oil yield relative to other oil crop, such as canola, are the major concerns for camelina production. In this study, we overexpressed the two Arabidopsis transporters, FAX1 and ABCA9, in camelina to test their functions on oil accumulation, yield and other agronomic traits, and lipid metabolism in developing pods.
Results
Overexpression of FAX1 and ABCA9 in camelina
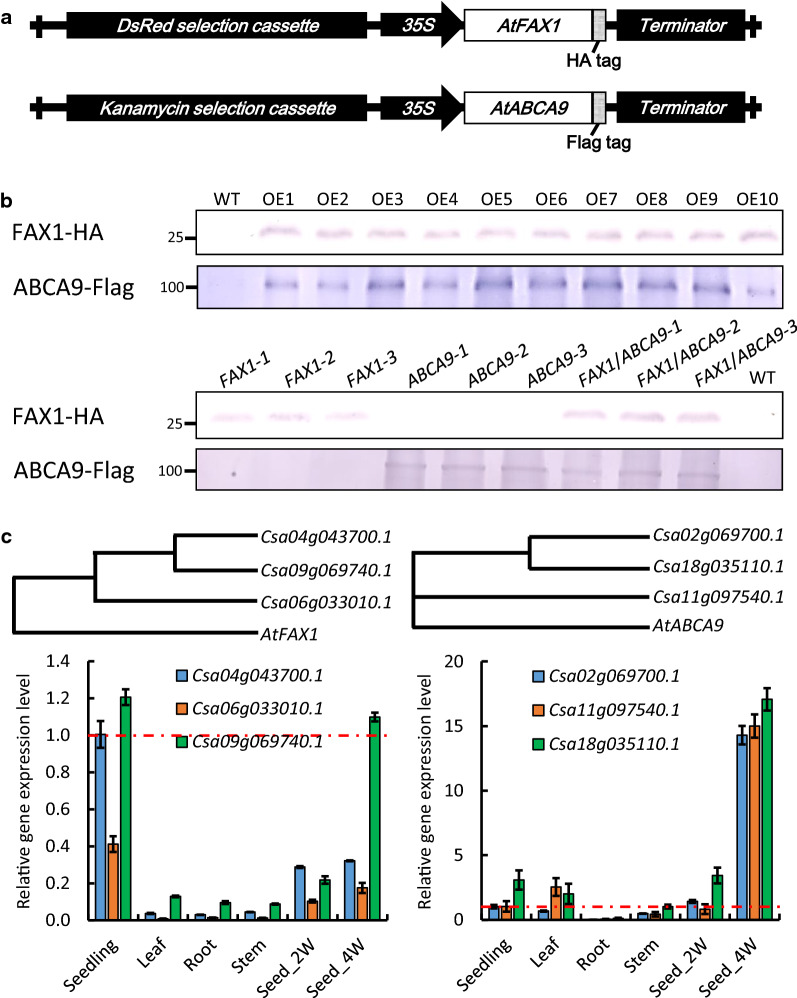
To investigate the function of FAX1 and ABCA9 in camelina, we overexpressed Arabidopsis FAX1 and ABCA9 genomic DNAs under the control of the cauliflower mosaic virus (CaMV)-35S promoter (Fig. 1a). The constitutive promoter was used because constitutive expression of these genes were reported to promote plant growth in Arabidopsis in addition to lipid production [5, 8]. FAX1 and ABCA9 were fused with a HA-tag and a Flag-tag at the C-terminus, respectively. The production of FAX1-HA and ABCA9-Flag in camelina was confirmed by immunoblotting using anti-HA and anti-Flag antibodies, respectively (Fig. 1b, upper panel). The homozygous OE plants producing FAX1-HA and ABCA9-Flag were identified. In addition, we generated camelina lines overexpressing both FAX1 and ABCA9 by transforming the FAX1-HA construct into homozygous ABCA9-OE plants. The production of FAX1-HA and ABCA9-Flag in FAX1/ABCA9 single and double OEs was confirmed by immunoblotting (Fig. 1b, lower panel). Camelina has 3 homeologs each of FAX1 and ABCA9 (Fig. 1c), and in WT camelina the transcript levels of the three FAX1s were relatively high in seedlings and developing seeds, but low in leaves, roots, and stems (Fig. 1c, left panel). The transcript levels of the three ABCA9 were relatively high in 4-week-old developing seeds (Fig. 1c, right panel).
Fig. 1.
Overexpression of AtFAX1 and AtABCA9 in camelina. a The overexpression (OE) constructs of AtFAX1 and AtABCA9. b Immunoblotting of HA-tagged FAX1 and Flag-tagged ABCA9 in AtFAX1-OE, AtABCA9-OE, and AtFAX1/ABCA9-OE camelina leaves. Total proteins (10 µg/lane) were extracted from leaves of 3-week-old plants, separated by 10% SDS-PAGE, and transferred to a polyvinylidene difluoride membrane. The membrane was blotted with anti-HA or anti-Flag antibody conjugated with alkaline phosphatase. Lanes OE1 through OE10 represent different transgenic lines harboring the FAX1-HA or ABCA9-Flag OE construct. Numbers on the left of each panel mark protein molecular mass standards in kilodaltons. c The phylogenetic tree of FAX1 (left) and ABCA9 (right) in camelina, and their expression pattern in seedlings, leaves, roots, stems, and 2- and 4-week (W) old developing seeds. ACT2 (actin2) was used as the internal standard for cDNA input adjustment. The expression level is relative to the value of Csa04g043700.1 for FAX1 and Csa02g069700.1 for ABCA9 in seedlings (red dashed line). Values are means ± SE (n = 3)
FAX1 and ABCA9 increase seed and oil yield
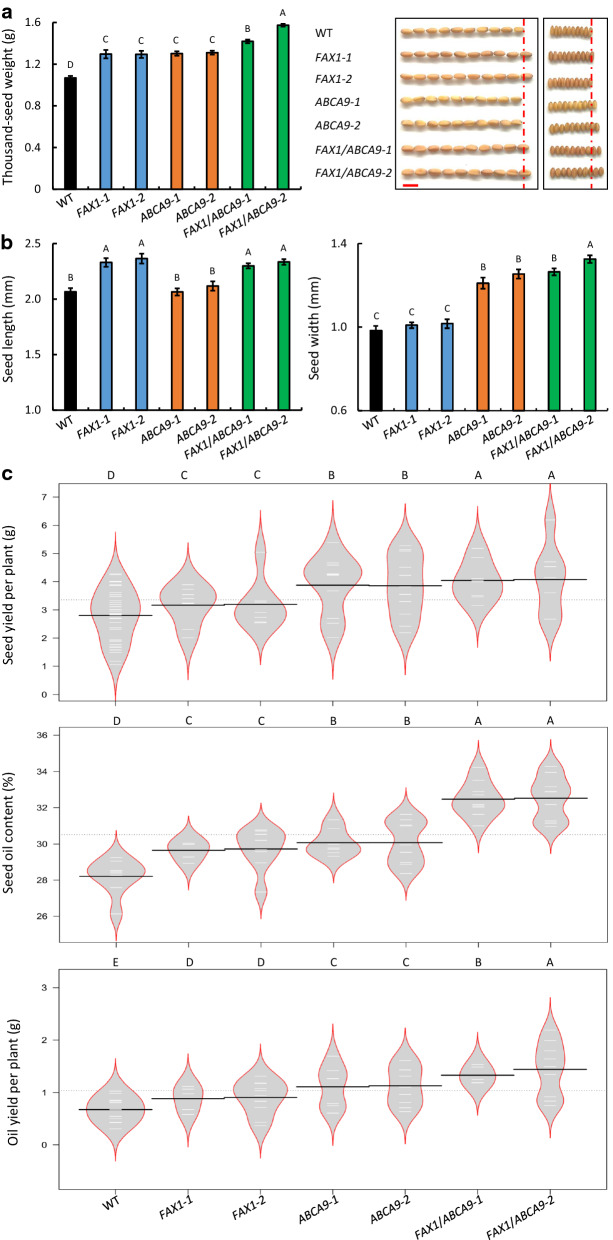
FAX1-OEs, ABCA9-OEs, FAX1/ABCA9-OEs, and WT plants were grown side by side to determine the effect of FAX1 and ABCA9 on camelina growth and production. The OE lines had similar flowering time, plant height, and branch number as WT (Additional file 1: Fig. S1). However, the thousand-seed weights (TSWs) of FAX1-OEs and ABCA9-OEs were 21% and 22% higher on average than that of WT, respectively, and FAX1/ABCA9-OEs were 40% higher on average than that of WT (Fig. 2a). Interestingly, FAX1-OEs and ABCA9-OEs had distinctive effect on seed length and seed width. FAX1-OEs increased seed length by 14%, whereas ABCA9-OEs increased seed width by 25%, while the seed length and width of FAX1/ABCA9-OEs were increase by 13% and 32%, respectively (Fig. 2a, b). The seed width of FAX1-OEs and seed length of ABCA9-OEs were similar with those of WT (Fig. 2b). The above results indicate that FAX1 and ABCA9 increase seed length and width, respectively, to increase seed weight.
Fig. 2.
Effect of AtFAX1- and AtABCA9-OEs on seed weight, seed size, seed oil content, and oil yield. a Thousand-seed weight and seed morphology of camelina dry seeds. Values are means ± SE (n = 12). Bar = 3 mm. b Seed length and width of camelina dry seeds. Values are means ± SE (n = 30). c Seed yield per plant (g), Seed oil content (w/w), and oil yield per plant (g) of different camelina lines. AtFAX1- and AtABCA9-single and double OEs were grown side by side together with WT. Seeds from whole plant were harvested and dried at room temperature for at least one month before measurement. Seed oil content was measured by gas chromatography, and calculated based on the internal standard (C17:0) peak area. Seeds from each plant were measured with 3 technical replicates, and the mean values are represented here as the seed oil content of each plant. Black lines show the averages, white lines represent individual data points (n = 18), and polygons represent the estimated density of the data. Homozygous, T3-generation OE lines were used for this and subsequent characterization. Capital letters indicate a significant difference (P < 0.01) based on Duncan-test
To determine the effect of FAX1 and ABCA9 on oil production in camelina, we grew the single and double OE plants together with WT side by side with multiple replicates (n = 18). Compared to WT, the seed yield per plant on average was 14% higher for FAX1-OE, 38% higher for ABCA9-OE, and 45% higher for FAX1/ABCA9-OEs (Fig. 2c). Moreover, the average oil content of WT seeds was 28%, whereas that FAX1-OE and ABCA9-OE seeds were 29% and 30%, respectively, and that of FAX1/ABCA9-OE seed was 32% (Fig. 2c). The average increase of seed oil content over WT was 4% for FAX1-OE, 6% for ABCA9-OE, and 13% for FAX1/ABCA9-OE seeds. The results indicate that FAX1 and ABCA9 increase seed oil content and seed yield simultaneously and these two genes had an additive effect on seed oil content and seed yield, leading a substantial improvement on overall oil yield. Combining the increases in seed oil content and seed yield, the oil production per plant of FAX1-OEs and ABCA9-OEs was 22% and 55% higher than that of WT, respectively, whereas that of FAX1/ABCA9-OEs was 75% higher than that of WT (Fig. 2c).
The FAX1-OE and ABCA9-OE seeds displayed altered FA composition from WT seeds. Compared with that of WT seeds, the level of C16:0 of FAX1-OEs and ABCA9-OEs was decreased by 6% and 4%, respectively, whereas that of FAX1/ABCA9-OEs was decreased by 14% (Additional file 2: Fig. S2). Conversely, the level of C18:0 was increased by 5% for FAX1-OEs and ABCA9-OEs, and 12% for FAX1/ABCA9-OEs. The level of C18:1 was increased by 13% and 16% for FAX1-OEs and ABCA9-OEs, respectively, and 29% for FAX1/ABCA9-OEs. The level of C18:2 was increased by 6% and 7% for FAX1-OEs and ABCA9-OEs, respectively, and 10% for FAX1/ABCA9-OEs whereas that of C18:3 was decreased by 4% for FAX1-OEs and ABCA9-OEs and 10% for FAX1/ABCA9-OEs comparing to that of WT. In addition, the level of C20:1 was decreased by 4% and 11% in FAX1-OE and ABCA9-OE seeds, and 10% in FAX1/ABCA9-OE seeds (Additional file 2: Fig. S2). The above results indicate that increased FAX1 and ABCA9 expressions positively affect the level of C18:0, C18:1 and C18:2, but negatively affect the level of C16:0, C18:3 and C20:1.
To test the seed performance, we monitored seed germination rates of the above lines. The seed germination rate at 12 h after imbibition of FAX1 and ABCA9 single and double OEs was slightly faster than that of WT while the final seed germination rate at 60 h after imbibition of all the lines tested was about 99% (Additional file 3: Fig. S3). In addition, we examined the vegetative growth of the above lines by growing the FAX1-OEs, ABCA9-OEs, and FAX1/ABCA9-OEs with WT side by side. The plant size appeared to be bigger in the single and double OE plants compared with WT (Additional file 4: Fig. S4a). The overground fresh weight of 3-week-old FAX1-OE and ABCA9-OE plants was 24% and 27% higher on average than that of WT, respectively, whereas that of FAX1/ABCA9-OE was 57% higher on average than WT (Additional file 4: Fig. S4b). Also, FAX1-OEs had one more, ABCA9-OEs had two more, FAX1/ABCA9-OEs had 2–3 more leaves of 3-week-old plants compared with WT (Additional file 4: Fig. S4b).
FAX1 and ABCA9 affect membrane glycerolipid composition
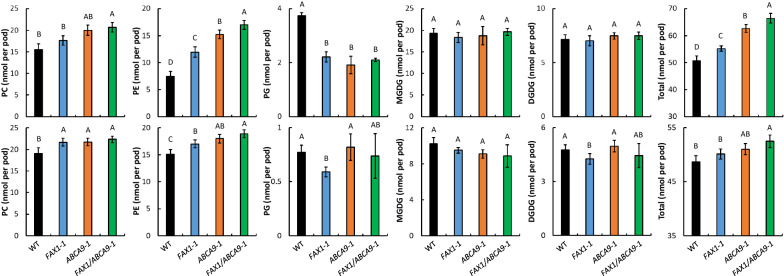
We further examined the effect of FAX1-OE and ABCA9-OE on membrane glycerolipid composition in seed pods containing developing seeds. Whole pods, instead of dissected seeds, were used because seed dissections would lead to wounding and activation of lipolytic activities. Total lipids were extracted from developing pods of 2 and 4 weeks after flowering from WT, and FAX1-1, ABCA9-1, and FAX1/ABCA9-1 OE lines, and analyzed using electrospray ionization tandem mass spectrometry (ESI–MS/MS). In 2-week-old pods (WOPs), which were considered as the outburst stage of lipid synthesis characterized by rapid lipid synthesis and oil production [34], compared to WT, the amount of phosphatidylcholine (PC) was increased 13%, 28%, and 33% in FAX1-1, ABCA9-1, and FAX1/ABCA9-1, respectively (Fig. 3). The amount of phosphatidylethanolamine (PE) was increased 60%, 104%, and 127% in FAX1-1, ABCA9-1, and FAX1/ABCA9-1. The total membrane glycerolipid level was 9%, 24%, and 31% higher in FAX1-1, ABCA9-1, and FAX1/ABCA9-1 than WT at the early stage of developing pods. However, the amount of phosphatidylglycerol (PG) was decreased approximately 45% in all three OE lines, and the level of monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG) had no obvious changes (Fig. 3). When the lipid data were calculated as mol% of total lipids analyzed, PE in FAX1-1, ABCA9-1, and FAX1/ABCA9-1 was 6, 12, and 13 mol% higher, respectively, than that of WT. However, the major plastidic lipids, phosphatidylglycerol (PG), MGDG, and DGDG were all lower in three OE lines than WT (Additional file 5: Fig. S5). Phosphatidic acid (PA) constituted less than 0.2 mol% in the developing pod of all lines tested and the PA mol% in FAX1-OE, ABCA9-OE, and FAX1/ABCA9-OE lines was all lower than that of WT (Additional file 5: Fig. S5).
Fig. 3.
Alterations of membrane glycerolipid levels (nmol per pod) in developing pods of AtFAX1- and AtABCA9-OEs. 2-week (upper panels) and 4-week (lower panels) old developing pods were sampled for lipid profiling by ESI–MS/MS. The total lipid levels referred to the total amount of major phospholipids (PC, PE, and PG) and galactolipids (MGDG and DGDG) measured. Values are means ± SE with 5 biological replicates. Capital letters on the top of each panel indicate a significant difference (P < 0.01) based on Duncan-test. PC: phosphatidylcholine; PE: phosphatidylethanolamine; PG: phosphatidylglycerol; MGDG: monogalactosyldiacylglycerol; and DGDG: digalactosyldiacylglycerol
In 4-WOPs, which was considered as the plateau stage of lipid synthesis [34], the level of PC in FAX1-1, ABCA9-1, and FAX1/ABCA9-1 was 14%, 14%, and 17% higher than that of WT (Fig. 3). The level of PE in FAX1-1, ABCA9-1, and FAX1/ABCA9-1 was 12%, 19%, and 25% higher than that of WT. The total membrane glycerolipid content in FAX1-1, ABCA9-1, and FAX1/ABCA9-1 was 3%, 5%, and 8% higher than that of WT (Fig. 3). However, the level of major plastidic lipids, PG and DGDG was decreased in FAX1-1 whereas was comparable among WT, ABCA9-1 and FAX1/ABCA9-1 at this stage (Fig. 3). PC in FAX1-1, ABCA9-1, and FAX1/ABCA9-1 was 2, 3, and 4 mol% higher than WT whereas mol% of MGDG and DGDG in the three OE lines was slightly lower than that of WT. The mol% of PE and PG was comparable between WT and OE lines at this stage (Additional file 5: Fig. S5). Also, the PA mol% was lower all OE lines than WT (Additional file 5: Fig. S5).
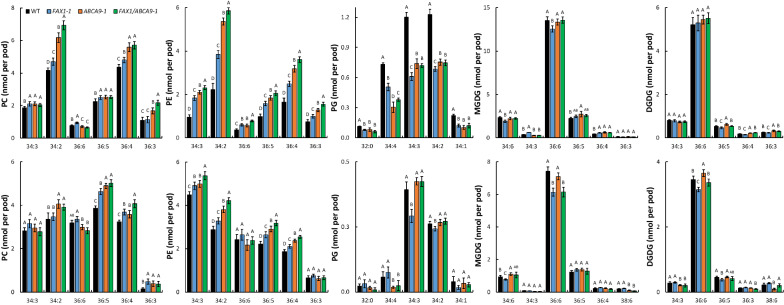
The major PC species (34:3, 34:2, 36:5, and 36:4 PC) in FAX1-1, ABCA9-1, and FAX1/ABCA9-1 were increased in 2-WOPs, and 36:5, 36:4, and 36:3 PC were increased in 4-WOPs compared with those of WT (Fig. 4). Similarly, the major PE species (34:3, 34:2, 36:6, 36:5, 36:4, and 36:3 PE) of FAX1-1, ABCA9-1, and FAX1/ABCA9-1 were all increased in 2-WOPs, and only 34:3, 34:2, 36:5, and 36:4 PE were increased in 4-WOPs compared to those of WT (Fig. 4). Also, the effect of FAX1 and ABCA9 exhibited additive effect, especially on 34:2 and 36:4 PC, and 34:3, 34:2, 36:5, and 36:4 PE (Fig. 4). In contrast, the major PG species (32:0, 34:4, 34:3, 34:2, and 34:1 PG) of FAX1-1, ABCA9-1, and FAX1/ABCA9-1 were all decreased compared to those of WT in 2-WOPs, while only 34:3 PG was decreased in FAX1-1, and 34:4 PG decreased in ABCA9-1, and FAX1/ABCA9-1 in 4-WOPs (Fig. 4). Although the total amount of MGDG was similar between WT and OE lines (Fig. 3), the level of some MGDG species, especially in FAX1-1, was different from that of WT. For instance, two major MGDG spices, 34:6 and 36:6 MGDG, in FAX1-1 was decreased both in 2- and 4-WOPs compared with WT (Fig. 4). For DGDG, only 36:5 DGDG in 2-WOPs, and 36:6 and 36:5 DGDG in 4-WOPs of FAX1-1 were lower than WT (Fig. 4). Similarly, the mol% of 34:2 PC of ABCA9-1 and FAX1/ABCA9-1 in 2-WOPs, and 36:5 PC of ABCA9-1 and FAX1/ABCA9-1 in 4-WOPs were higher than that of WT (Additional file 6: Fig. S6); the mol% of 34:2 PE of ABCA9-1 and FAX1/ABCA9-1 was higher than WT both in 2- and 4-WOPs; whereas the mol% of 34:2 PG of FAX1-1 was lower than WT; the major MGDG species 36:6 was lower in FAX1-1 and FAX1/ABCA9-1; and the major DGDG species 36:6 was lower in FAX1-1 in 4-WOPs (Additional file 6: Fig. S6). The above lipid data indicate that overexpression of FAX1 and ABCA9 also affect membrane lipid composition in developing pods, and the effect differs at different developmental stage.
Fig. 4.
Alterations of PC, PE, PG, MGDG, and DGDG species (nmol per pod) in developing pods of AtFAX1- and AtABCA9-OEs. 2-week (upper panels) and 4-week (lower panels) old developing pods were sampled for lipid profiling by ESI–MS/MS. The species levels were measured by internal standards. Values are means ± SE with 5 biological replicates. Capital letters on the top of each panel indicate a significant difference (P < 0.01) based on Duncan-test
FAX1 and ABCA9 alter the expression of genes in oil production in seeds
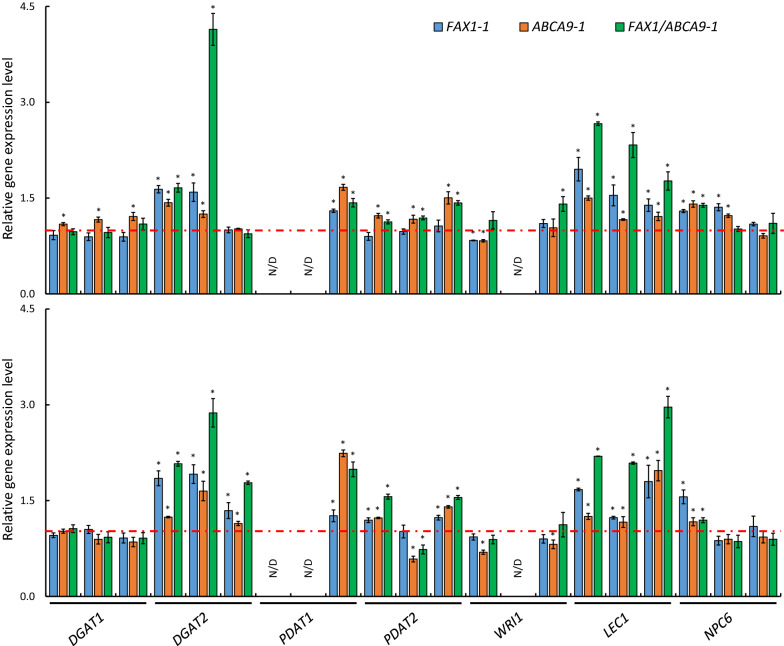
To gain insights into the enhancing oil production, we compared the transcript level of genes that are involved in oil accumulation in developing seeds of the FAX1- and ABCA9-OE lines with WT. The transcript level of AtFAX1 in 2- and 4-week-old developing seeds (WODS) was high in FAX1-OE and FAX1/ABCA9-OE lines, but was not detected in ABCA9-OE lines, as expected. Similarly, the level of AtABCA9 was high in ABCA9-OE and FAX1/ABCA9-OE lines, but was not detected in FAX1-OE lines (Additional file 7: Fig. S7). The transcript level of three DGAT1s (Acyl-CoA: diacylglycerol acyltransferase 1) was higher in ABCA9-1, and slightly lower in FAX1-1 in 2-WODS whereas that of two DGAT2s was higher in 2-WODS, and all three DGAT2 were higher in 4-WODS in all OE lines than WT (Fig. 5). The transcript of one PDAT1 (phospholipid: diacylglycerol acyltransferase 1) was detected, and its level was higher than that of WT in all OE lines at both stages. The level of three PDAT2s transcripts was higher in ABCA9-1 and FAX1/ABCA9-1 in 2-WODS, whereas that of two PDAT2s transcripts was higher in three OE lines but the other PDAT2 was lower in ABCA9-1 and FAX1/ABCA9-1 in 4-WODS. The transcript level of one WRI1 was lower in FAX1-1 and ABCA9-1 whereas another WRI1 was higher in FAX1/ABCA9-1 in 2-WODS (Fig. 5). At 4-WODS, the level of two WRI1s was lower in ABCA9-1 than WT. The transcript levels of LEC1 (leaf cotyledon 1) homeologs were higher in 2- and 4-WODS in all OE lines than WT. One NPC6 (nonspecific phospholipase C 6) displayed a higher level in FAX1- and ABCA9-OEs in 2- and 4-WODS, and another had a higher level in FAX1-1 and ABCA9-1 in 2-WODS (Fig. 5).
Fig. 5.
Effect of AtFAX1- and AtABCA9-OEs on the expression of genes involved in oil accumulation. Samples were collected with 2-week (upper panel) and 4-week (lower panel) old developing seeds for RNA extraction and real-time PCR. ACT2 (actin2) was used as the internal standard for cDNA input adjustment. The three bars of each genes indicated the three copies of each gene in camelina. Values are the relative expression level compared with WT (red dashed line) and are means ± SE (n = 3). *Significant difference (P < 0.05) based on Student’s t test compared with WT. N/D: not detected. DGAT: Acyl-CoA: diacylglycerol acyltransferase; PDAT: phospholipid: diacylglycerol acyltransferase; WRI1: wrinkled 1; LEC1: leaf cotyledon 1; and NPC6: nonspecific phospholipase C 6
Discussion
In this study, we show that OE of two Arabidopsis transporters, FAX1 and ABCA9, in camelina improves seed and oil production in the emerging oil crop. While the positive effect of FAX1-OE and ABCA9-OE in camelina is generally consistent with prior reports on the two genes being overexpressed individually in Arabidopsis [5, 6], the effect of simultaneously overexpressing the two transporters was previously unknown. A key new finding of the present study is that co-OE of FAX1 and ABCA9 has an additive effect on enhancing seed and oil production. In addition, our comparison of the two gene effects side by side in camelina led to other new findings. One is that FAX1 and ABCA9 exhibited different effects on seed size; whereas FAX1-OE increased seed length, ABCA9 -OE increased seed width. Several genes that affect differently only seed length or width were reported in rice [35–42], but those genes, such as those encoding a heterotrimeric G protein subunit and an E3 ubiquitin ligase, share no apparent common function with the two transporters. It would be of great interests in future studies to probe how the two transporter genes have such distinctive effects in camelina seed shape.
Another finding from the comparative analysis is that OE of the two transporters has different effects on lipid composition. FAX1-OE was lower than WT and ABCA9-OE in both the amount (nmol per pod) and mol% of plastidic lipids, such as 34:3- and 34:2-PG, 34:6- and 36:6-MGDG in 2- and 4-WOPs. In comparison, ABCA9-OE was higher than WT and FAX1-OE in the amount and mol% of extra-plastidic lipids, including 34:2-PC, and 34:2-PE in 2-WOPs, and 34:2- and 36:5-PC, and 34:2-, 36:5-, and 36:4-PE in 4-WOPs. FAX1 was localized at the inner envelope of the chloroplasts transporting FAs out of chloroplasts [5], whereas ABCA9 was associated with ER and proposed to transport FAs to ER [8].The decrease of plastidic lipids in FAX1-OE may result from an enhanced transport of FAs from plastids to cytosol/ER by FAX1, whereas the increase of extra-plastidic lipids in ABCA9-OE may result from enhanced FA transport to ER by ABCA9 for glycerolipid synthesis. The distinctive effects of FAX1 and ABCA9 on FA transport and lipid metabolism may help to explain the additive enhancements of FAX1 and ABCA9 on seed and oil production.
Furthermore, FAX1- and ABCA9-OE developing camelina seeds also displayed increased level of expression of specific genes related to TAG production, such as DGAT2, PDAT1, and LEC1. Those increases may result from an increase in supply of fatty acids and metabolic demand as the overall activity for lipid production and/or more substrates are available for those enzymes. ABCA9-OE and FAX1-OE exhibited different impacts on the expression of these genes. The expression level of DGAT1 and PDAT2 in 2-WODS was increased in ABCA9-1 and/or FAX1/ABCA9-1 but not in FAX1-1 compared to that in WT, which may explain the reason of a higher seed oil content in ABCA9-OEs than that in FAX1-OEs. It should be noted that all the observations of the present study are based on laboratory conditions and application of the promising results awaits further testing in field conditions, including the response of the transporter-altered lines under field growth environments.
Conclusion
Here, we show that OE of FAX1 and ABCA9 increased seed weight, size, and oil production in camelina and co-OE of the two genes has an additive effect on the enhancement. FAX1- and ABCA9-OEs had different effects on seed length and seed width. FAX1-OEs and ABCA9-OEs increase seed length and width, respectively, whereas co-OE of FAX1 and ABCA9 increases both seed length and width. The results indicate that simultaneous OE of FAX1 and ABCA9 may potentially be applied to improving camelina oil production.
Methods
Plant materials and growth conditions
To overexpress Arabidopsis ABCA9 transporter in camelina, the genomic sequence of ABCA9 (AT5G61730) was amplified by PCR using Col-0 Arabidopsis genomic DNA as a template and by forward primer with KpnI site and reverse primer with PacI site. The C-terminal Flag tag was fused upstream of the terminator manually by adding its coding sequence to the reverse primer. Transgenic T1 plants were selected on a medium containing 50 mg/L kanamycin. The putative transgenic seedlings were transferred to soil and leaves were collected for PCR confirmation of the presence of AtABCA9. The PCR reaction condition was pre-incubated at 94 °C for 4 min, 35 cycles of 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 1 min, and final extension at 72 °C for 10 min.
To overexpress Arabidopsis FAX1 transporter in camelina, the genomic sequence of FAX1 (AT3G57280) was amplified by PCR using Col-0 Arabidopsis genomic DNA as a template and by forward primer with EcoRI site and reverse primer with SmaI site. The C-terminal HA tag was fused upstream of the terminator manually by adding its coding sequence to the reverse primer. Putative FAX1-OE seeds were first identified by selecting red seeds under green flashlight with a red-light filter. Plants derived from putative transformed seeds were further verified by PCR, using the cloning primers and PCR condition as described above.
The details for gene cloning, plant transformation, putative transgenic plants identification were performed as described previously [43, 44]. Camelina plants were grown in greenhouse at 21 °C with approximately 16 h light (566 μmol/m2/s).
Homozygous lines of T3 OE lines were used to compare plant growth and yield traits among these OE lines and WT. We used 2.5-gallon pots with BM7-35% soil (Berger) and each pot had 4 plants with one as WT as control in greenhouse. There were 18 biological replicates with a completely randomized block design for each transgenic line. The greenhouse condition and plants managements were as described previously [43, 44]. Measurements of flowering time, plant height, branch number, thousand-seed weight, and plant yield per plant were performed as described [45]. All mother plants of the investigated lines were grown in the same condition and harvested at the same time, and seeds were stored under the same environment.
Phylogenetic analysis
The coding sequences of AtFAX1 and AtABCA9 were used as queries to search for homologous genes in the camelina reference genome [12] using BLASTn program with an E-value of 1E-50 and an identity of 50% set as thresholds. The phylogenetic tree was drawn by Phylodendron (http://iubio.bio.indiana.edu/treeapp/treeprint-sample1.html).
Immunoblotting and transcript analysis
Total protein extraction and immunoblotting were performed as described previously [44]. Protein concentrations were measured using the Bradford assay (Bio-Rad, 500–0205). RNA extraction, real-time PCR analysis, and semi-quantitative RT (reverse transcription)-PCR of transcript levels were performed as described previously [46, 47]. Total RNA was extracted from 2- and 4-week old developing seeds. Camelina ACT2 (Csa19g026200.1) were used as internal standard and for cDNA input adjustment. All primers used in RT-PCR and real-time PCR are listed in Additional file 8: Table S1.
Seed oil content and fatty acid composition analyses
Seed oil content and FA composition were determined as described previously [43, 44]. Fatty acid methyl esters (FAMEs) from TAG were identified by comparing their retention times with known standards. The FA composition was calculated as mol %.
Lipid extraction and profiling
Polar lipids were extracted and analyzed by ESI–MS/MS based on a method described previously [44, 48]. The mass spectrometry data for lipids were processed using the software Analyst 1.5.1.
Accession numbers
Sequence data from this article can be found in the following database under the accession numbers: Arabidopsis Genome Initiative database: FAX1, AT3G57280; and ABCA9, AT5G61730. Camelina sativa Genome Resources (http://www.camelinadb.ca/): CsACT2, Csa19g026200.1; CsFAX1: Csa09g069740.1, Csa06g033010.1, Csa04g043700.1; CsABCA9: Csa02g069700.1, Csa18g035110.1, Csa11g097540.1; CsDGAT1: Csa01g042590.1, Csa19g056370.1, Csa15g084220.1; CsDGAT2: Csa04g037310.1, Csa09g058550.1, Csa06g025650.1; CsPDAT1: Csa13g016300.1, Csa08g005560.1, Csa20g019000.1; CsPDAT2: Csa04g024660.1, Csa06g018480.1, Csa09g035780.1; CsWRI1: Csa06g028810.1, Csa09g064030.1, Csa04g040400.1; CsLEC1: Csa17g028800.1, Csa03g025850.1, Csa14g027200.1; and CsNPC6: Csa09g050690.1, Csa06g022410.1, Csa04g033750.1.
Supplementary Information
Additional file 1: Figure S1. Effect of AtFAX1- and AtABCA9-OEs on other major agronomic traits.
Additional file 2: Figure S2. Effect of AtFAX1- and AtABCA9-OEs on seed fatty acid composition.
Additional file 3: Figure S3. Seed germination rate of AtFAX1- and AtABCA9-OEs.
Additional file 4: Figure S4. Effect of AtFAX1- and AtABCA9-OEs on vegetative tissues.
Additional file 5: Figure S5. Alterations of membrane glycerolipid levels (mol%) in developing pods of AtFAX1- and AtABCA9-OEs.
Additional file 6: Figure S6. Alterations of PC, PE, PG, MGDG, and DGDG species (mol%) in developing pods of AtFAX1- and AtABCA9-OEs.
Additional file 7: Figure S7. Gene expression level of AtFAX1 and AtABCA9 in developing seeds of camelina OE lines.
Additional file 8: Table S1. Primers (5′ to 3′) used for cloning and RT-PCR.
Acknowledgements
Not applicable.
Abbreviations
- FA
Fatty acid
- ABC
ATP-binding cassette
- FAX
Fatty acid export
- TAG
Triacylglycerol
- PC
Phosphatidylcholine
- PE
Phosphatidylethanolamine
- PG
Phosphatidylglycerol
- PA
Phosphatidic acid
- MGDG
Monogalactosyldiacylglycerol
- DGDG
Digalactosyldiacylglycerol
Authors’ contributions
GC designed and performed most of the experiments, and wrote and revised the manuscript. GW performed ABCA9 cloning and transformation. SK did ABCA9 protein analysis and edited the manuscript. JL helped grow plants and edit manuscript. YZ co-supervised GC and revised the manuscript. XW proposed, designed, and supervised the study and revised the manuscript. All authors read and approved the final manuscript.
Funding
The work is supported by the National Natural Science Foundation of China (31801029), National Key Research and Development Program of China (2016YFD0100506, 2017YFE0104800), the US Department of Energy (DE-AR0000202), and the International Postdoctoral Exchange Fellowship Program 2016 by the Office of China Postdoctoral Council (20160034).
Availability of data and materials
All data generated or analyzed during this study are included in the article and its additional files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Guangqin Cai, Email: cgq6688@163.com.
Geliang Wang, Email: geliangwang@gmail.com.
Sang-Chul Kim, Email: skim@danforthcenter.org.
Jianwu Li, Email: lijw555@henau.edu.cn.
Yongming Zhou, Email: ymzhou@mail.hzau.edu.cn.
Xuemin Wang, Email: swang@danforthcenter.org.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13068-021-01899-w.
References
- 1.Patterson E, Wall R, Fitzgerald G, Ross R, Stanton C. Health implications of high dietary omega-6 polyunsaturated fatty acids. J Nutrit Metabol. 2012;2012:539426. doi: 10.1155/2012/539426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, DeBono A, Durrett TP. Acyl-lipid metabolism. Arabidopsis Book Am Soc Plant Biol. 2013;11:e0161. doi: 10.1199/tab.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchanan BB, Gruissem W, Jones RL. Biochemistry and molecular biology of plants. Hoboken: John; 2015. [Google Scholar]
- 4.Thelen JJ, Ohlrogge JB. Metabolic engineering of fatty acid biosynthesis in plants. Metab Eng. 2002;4:12–21. doi: 10.1006/mben.2001.0204. [DOI] [PubMed] [Google Scholar]
- 5.Li N, Gügel IL, Giavalisco P, Zeisler V, Schreiber L, Soll J, Philippar K. FAX1, a novel membrane protein mediating plastid fatty acid export. PLoS Biol. 2015;13:e1002053. doi: 10.1371/journal.pbio.1002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian Y, Lv X, Xie G, Zhang J, Xu Y, Chen F. Seed-specific overexpression of AtFAX1 increases seed oil content in Arabidopsis. Biochem Biophys Res Commun. 2018;500:370–375. doi: 10.1016/j.bbrc.2018.04.081. [DOI] [PubMed] [Google Scholar]
- 7.Du Z-Y, Arias T, Meng W, Chye M-L. Plant acyl-CoA-binding proteins: an emerging family involved in plant development and stress responses. Prog Lipid Res. 2016;63:165–181. doi: 10.1016/j.plipres.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Kim S, Yamaoka Y, Ono H, Kim H, Shim D, Maeshima M, Martinoia E, Cahoon EB, Nishida I, Lee Y. AtABCA9 transporter supplies fatty acids for lipid synthesis to the endoplasmic reticulum. Proc Natl Acad Sci. 2013;110:773–778. doi: 10.1073/pnas.1214159110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zubr J. Oil-seed crop: Camelina sativa. Ind Crops Prod. 1997;6:113–119. doi: 10.1016/S0926-6690(96)00203-8. [DOI] [Google Scholar]
- 10.Berti M, Gesch R, Eynck C, Anderson J, Cermak S. Camelina uses, genetics, genomics, production, and management. Ind Crops Prod. 2016;94:690–710. doi: 10.1016/j.indcrop.2016.09.034. [DOI] [Google Scholar]
- 11.Putnam D, Budin J, Field L, Breene W. Camelina: a promising low-input oilseed. New Crops. 1993;314:322. [Google Scholar]
- 12.Kagale S, Koh C, Nixon J, Bollina V, Clarke WE, Tuteja R, Spillane C, Robinson SJ, Links MG, Clarke C. The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nature Commun. 2014;5:3706. doi: 10.1038/ncomms4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knorzer K. Evolution and spreading of gold of pleasure (Camelina-sativa S1) Berichte der Deutschen Botanischen Gesellschaft. 1978;91:187–195. [Google Scholar]
- 14.Budin JT, Breene WM, Putnam DH. Some compositional properties of camelina (Camelina sativa L. Crantz) seeds and oils. J Am Oil Chem Soc. 1995;72:309–315. doi: 10.1007/BF02541088. [DOI] [Google Scholar]
- 15.Moser BR. Biodiesel from alternative oilseed feedstocks: camelina and field pennycress. Biofuels. 2012;3:193–209. doi: 10.4155/bfs.12.6. [DOI] [Google Scholar]
- 16.Pilgeram AL. Camelina sativa, a Montana omega-3 and fuel crop. 2007.
- 17.Séguin-Swartz G, Eynck C, Gugel R, Strelkov S, Olivier C, Li J, Klein-Gebbinck H, Borhan H, Caldwell C, Falk K. Diseases of Camelina sativa (false flax) Can J Plant Path. 2009;31:375–386. doi: 10.1080/07060660909507612. [DOI] [Google Scholar]
- 18.Bramm A, Dambroth M, Schulte-Körne S. Analysis of yield components of linseed, false flax and poppy. Landbauforschung Voelkenrode. 1990;40:107–114. [Google Scholar]
- 19.Lu C, Kang J. Generation of transgenic plants of a potential oilseed crop Camelina sativa by Agrobacterium-mediated transformation. Plant Cell Rep. 2008;27:273–278. doi: 10.1007/s00299-007-0454-0. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen HT, Silva JE, Podicheti R, Macrander J, Yang W, Nazarenus TJ, Nam JW, Jaworski JG, Lu C, Scheffler BE. Camelina seed transcriptome: a tool for meal and oil improvement and translational research. Plant Biotechnol J. 2013;11:759–769. doi: 10.1111/pbi.12068. [DOI] [PubMed] [Google Scholar]
- 21.Petrie JR, Shrestha P, Belide S, Kennedy Y, Lester G, Liu Q, Divi UK, Mulder RJ, Mansour MP, Nichols PD. Metabolic engineering Camelina sativa with fish oil-like levels of DHA. PLoS ONE. 2014;9:e85061. doi: 10.1371/journal.pone.0085061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruiz-Lopez N, Haslam RP, Napier JA, Sayanova O. Successful high-level accumulation of fish oil omega-3 long-chain polyunsaturated fatty acids in a transgenic oilseed crop. Plant J. 2014;77:198–208. doi: 10.1111/tpj.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Usher S, Haslam RP, Ruiz-Lopez N, Sayanova O, Napier JA. Field trial evaluation of the accumulation of omega-3 long chain polyunsaturated fatty acids in transgenic Camelina sativa: making fish oil substitutes in plants. Metabolic Eng Commun. 2015;2:93–98. doi: 10.1016/j.meteno.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han L, Usher S, Sandgrind S, Hassall K, Sayanova O, Michaelson LV, Haslam RP, Napier JA. High level accumulation of EPA and DHA in field-grown transgenic Camelina–a multi-territory evaluation of TAG accumulation and heterogeneity. Plant Biotechnol J. 2020;18:2280–2291. doi: 10.1111/pbi.13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruiz-Lopez N, Haslam RP, Usher S, Napier JA, Sayanova O. An alternative pathway for the effective production of the omega-3 long-chain polyunsaturates EPA and ETA in transgenic oilseeds. Plant Biotechnol J. 2015;13:1264–1275. doi: 10.1111/pbi.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iven T, Hornung E, Heilmann M, Feussner I. Synthesis of oleyl oleate wax esters in Arabidopsis thaliana and Camelina sativa seed oil. Plant Biotechnol J. 2016;14:252–259. doi: 10.1111/pbi.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Rice A, McGlew K, Shaw V, Park H, Clemente T, Pollard M, Ohlrogge J, Durrett TP. Metabolic engineering of oilseed crops to produce high levels of novel acetyl glyceride oils with reduced viscosity, freezing point and calorific value. Plant Biotechnol J. 2015;13:858–865. doi: 10.1111/pbi.12325. [DOI] [PubMed] [Google Scholar]
- 28.Malik MR, Yang W, Patterson N, Tang J, Wellinghoff RL, Preuss ML, Burkitt C, Sharma N, Ji Y, Jez JM. Production of high levels of poly-3-hydroxybutyrate in plastids of C amelina sativa seeds. Plant Biotechnol J. 2015;13:675–688. doi: 10.1111/pbi.12290. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen HT, Park H, Koster KL, Cahoon RE, Nguyen HT, Shanklin J, Clemente TE, Cahoon EB. Redirection of metabolic flux for high levels of omega-7 monounsaturated fatty acid accumulation in camelina seeds. Plant Biotechnol J. 2015;13:38–50. doi: 10.1111/pbi.12233. [DOI] [PubMed] [Google Scholar]
- 30.Yu XH, Cahoon RE, Horn PJ, Shi H, Prakash RR, Cai Y, Hearney M, Chapman KD, Cahoon EB, Schwender J. Identification of bottlenecks in the accumulation of cyclic fatty acids in camelina seed oil. Plant Biotechnol J. 2018;16:926–938. doi: 10.1111/pbi.12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang WZ, Henry IM, Lynagh PG, Comai L, Cahoon EB, Weeks DP. Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol J. 2017;15:648–657. doi: 10.1111/pbi.12663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Na G, Mu X, Grabowski P, Schmutz J, Lu C. Enhancing micro RNA 167A expression in seed decreases the α-linolenic acid content and increases seed size in Camelina sativa. Plant J. 2019;98:346–358. doi: 10.1111/tpj.14223. [DOI] [PubMed] [Google Scholar]
- 33.Ozseyhan ME, Kang J, Mu X, Lu C. Mutagenesis of the FAE1 genes significantly changes fatty acid composition in seeds of Camelina sativa. Plant Physiol Biochem. 2018;123:1–7. doi: 10.1016/j.plaphy.2017.11.021. [DOI] [PubMed] [Google Scholar]
- 34.Abdullah HM, Akbari P, Paulose B, Schnell D, Qi W, Park Y, Pareek A, Dhankher OP. Transcriptome profiling of Camelina sativa to identify genes involved in triacylglycerol biosynthesis and accumulation in the developing seeds. Biotechnol Biofuels. 2016;9:136. doi: 10.1186/s13068-016-0555-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theoret Appl Gen. 2006;112:1164–1171. doi: 10.1007/s00122-006-0218-1. [DOI] [PubMed] [Google Scholar]
- 36.Qi P, Lin Y-S, Song X-J, Shen J-B, Huang W, Shan J-X, Zhu M-Z, Jiang L, Gao J-P, Lin H-X. The novel quantitative trait locus GL3. 1 controls rice grain size and yield by regulating Cyclin-T1; 3. Cell Res. 2012;22:1666–1680. doi: 10.1038/cr.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song X-J, Huang W, Shi M, Zhu M-Z, Lin H-X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet. 2007;39:623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- 38.Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M. Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet. 2008;40:1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- 39.Weng J, Gu S, Wan X, Gao H, Guo T, Su N, Lei C, Zhang X, Cheng Z, Guo X. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008;18:1199–1209. doi: 10.1038/cr.2008.307. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Fan C, Xing Y, Jiang Y, Luo L, Sun L, Shao D, Xu C, Li X, Xiao J. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet. 2011;43:1266–1269. doi: 10.1038/ng.977. [DOI] [PubMed] [Google Scholar]
- 41.Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q. Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet. 2012;44:950–954. doi: 10.1038/ng.2327. [DOI] [PubMed] [Google Scholar]
- 42.Xing Y, Zhang Q. Genetic and molecular bases of rice yield. Annu Rev Plant Biol. 2010;61:421–442. doi: 10.1146/annurev-arplant-042809-112209. [DOI] [PubMed] [Google Scholar]
- 43.Li M, Wei F, Tawfall A, Tang M, Saettele A, Wang X. Overexpression of patatin-related phospholipase AIII δ altered plant growth and increased seed oil content in camelina. Plant Biotechnol J. 2015;13:766–778. doi: 10.1111/pbi.12304. [DOI] [PubMed] [Google Scholar]
- 44.Cai G, Fan C, Liu S, Yang Q, Liu D, Wu J, Li J, Zhou Y, Guo L, Wang X. Nonspecific phospholipase C6 increases seed oil production in oilseed Brassicaceae plants. New Phytol. 2020;226:1055–1073. doi: 10.1111/nph.16473. [DOI] [PubMed] [Google Scholar]
- 45.Cai G, Yang Q, Chen H, Yang Q, Zhang C, Fan C, Zhou Y. Genetic dissection of plant architecture and yield-related traits in Brassica napus. Sci Rep. 2016;6:21625. doi: 10.1038/srep21625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li M, Bahn SC, Guo L, Musgrave W, Berg H, Welti R, Wang X. Patatin-related phospholipase pPLAIIIβ-induced changes in lipid metabolism alter cellulose content and cell elongation in Arabidopsis. Plant Cell. 2011;23:1107–1123. doi: 10.1105/tpc.110.081240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai G, Kim S-C, Li J, Zhou Y, Wang X. Transcriptional Regulation of Lipid Catabolism during Seedling Establishment. Molecular Plant. 2020;13:984–1000. doi: 10.1016/j.molp.2020.04.007. [DOI] [PubMed] [Google Scholar]
- 48.Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou H-E, Rajashekar C, Williams TD, Wang X. Profiling membrane lipids in plant stress responses role of phospholipase Dα in freezing-induced lipid changes in Arabidopsis. J Biol Chem. 2002;277:31994–32002. doi: 10.1074/jbc.M205375200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Effect of AtFAX1- and AtABCA9-OEs on other major agronomic traits.
Additional file 2: Figure S2. Effect of AtFAX1- and AtABCA9-OEs on seed fatty acid composition.
Additional file 3: Figure S3. Seed germination rate of AtFAX1- and AtABCA9-OEs.
Additional file 4: Figure S4. Effect of AtFAX1- and AtABCA9-OEs on vegetative tissues.
Additional file 5: Figure S5. Alterations of membrane glycerolipid levels (mol%) in developing pods of AtFAX1- and AtABCA9-OEs.
Additional file 6: Figure S6. Alterations of PC, PE, PG, MGDG, and DGDG species (mol%) in developing pods of AtFAX1- and AtABCA9-OEs.
Additional file 7: Figure S7. Gene expression level of AtFAX1 and AtABCA9 in developing seeds of camelina OE lines.
Additional file 8: Table S1. Primers (5′ to 3′) used for cloning and RT-PCR.
Data Availability Statement
All data generated or analyzed during this study are included in the article and its additional files.