Abstract
Evidence suggests that increased level/aggregation of beta-amyloid (Aβ) peptides initiate neurodegeneration and subsequent development of Alzheimer's disease (AD). At present, there is no effective treatment for AD. In this study, we reported the effects of gold nanoparticles surface-functionalized with a plant-based amino acid mimosine (Mimo-AuNPs), which is found to cross the blood-brain barrier, on the Aβ fibrillization process and toxicity. Thioflavin T kinetic assays, fluorescence imaging and electron microscopy data showed that Mimo-AuNPs were able to suppress the spontaneous and seed-induced Aβ1-42 aggregation. Spectroscopic studies, molecular docking and biochemical analyses further revealed that Mimo-AuNPs stabilize Aβ1-42 to remain in its monomeric state by interacting with the hydrophobic domain of Aβ1-42 (i.e., Lys16 to Ala21) there by preventing a conformational shift towards the β-sheet structure. Additionally, Mimo-AuNPs were found to trigger the disassembly of matured Aβ1-42 fibers and increased neuronal viability by reducing phosphorylation of tau protein and the production of oxyradicals. Collectively, these results reveal that the surface-functionalization of gold nanoparticles with mimosine can attenuate Aβ fibrillization and neuronal toxicity. Thus, we propose Mimo-AuNPs may be used as a potential treatment strategy towards AD-related pathologies.
Keywords: Alzheimer's disease, β-amyloid aggregation, Gold nanoparticles, Mimosine, Neuroprotection
Graphical abstract
Highlights
-
•
Mimosine functionalized with gold nanoparticles (Mimo-AuNPs) can cross blood-brain barrier.
-
•
Mimo-AuNPs inhibit aggregation of Aβ peptides by interacting with its hydrophobic domain.
-
•
Mimo-AuNPs can trigger disassembly of pre-aggregated Aβ fibers.
-
•
Mimo-AuNPs can protect neurons against Aβ toxicity by attenuating intracellular signaling.
Abbreviations
- Aβ
amyloid β
- AD
Alzheimer's disease
- ANS
8-Anilion-1-Naphthalene Sulfonate
- AuNPs
gold nanoparticles
- BBB
blood-brain barrier
- BCA
bicinchoninic acid
- CD
circular dichroism
- DLS
dynamic light scattering
- DMEM
Dulbecco's modified Eagle's medium
- DMSO
dimethyl sulfoxide
- ECL
enhanced chemiluminescence
- ERK1/2
extracellular signal-regulated kinases
- FBS
fetal bovine serum
- FCC
Face Centered Crystalline
- FTIR
Fourier transform infrared spectroscopy
- GSK-3β
glycogen synthase kinase-3β
- HBSS
Hanks' balanced salt solution
- HFIP
hexafluoro-2-propanol
- HRP
horseradish peroxidase
- LDH
lactate dehydrogenase
- MAP
mitogen-activated protein
- MTT
3-(4,5-dimethylthiazolyl)-2,5-diphenyltetrazolium bromide
- PAGE
polyacrylamide gel electrophoresis
- Papp
apparent permeability co-efficient
- PBS
phosphate-buffered saline
- PFA
paraformaldehyde
- ROS
reactive oxygen species
- SPR
surface plasma resonance
- TEER
trans endothelial electrical resistance
- TEM
transmission electron microscopy
- ThT
Thioflavin-T.
1. Introduction
Alzheimer's disease (AD), the most common form of late-life dementia, is a progressive neurodegenerative disorder characterized by a gradual loss of memory followed by deterioration of higher cognitive functions. Most AD cases are sporadic, whereas only a minority (<10%) of familial cases segregate with mutations in three known genes; amyloid precursor protein (APP), presenilin 1 (PSEN1) and PSEN2 [1,2]. At present, there is no effective treatment to either delay or arrest the progression of AD. The major pathological features associated with the disease include the presence of tau-positive intracellular neurofibrillary tangles, extracellular neuritic plaques and the loss of neurons in selected regions of the brain. Structurally, neuritic plaques comprise a compact deposit of β-amyloid (Aβ) peptides derived from the precursor APP by sequential processing mediated via β-secretase and the tetrameric γ-secretase complex [1,3]. Although various Aβ fragments containing 39–43 amino acids are evident physiologically, the two most prevalent isoforms found in the brain are Aβ1-40 and Aβ1-42 [1,4]. Of the two isoforms, Aβ1-42 constitutes ~10% of the total Aβ peptides but, due to its amphiphilic nature and high hydrophobicity, it tends to aggregate faster and is more toxic to neurons than the Aβ1-40 [5,6]. It is generally believed that conversion of Aβ from physiological monomeric forms into a variety of structures ranging from oligomers to amyloid fibrils may underlie the cause of neuronal dysfunction/death leading to the development of AD pathology [7,8]. While the C-terminal domain of Aβ determines the rate of fibril formation, the N-terminus promotes Aβ-Aβ interaction for polymerization, leading to a random coil or α-helix to β-sheet transition. The extended β-sheet formation promotes homophilic interactions creating Aβ oligomers that serve as seeds/nuclei for accelerated fibril growth [9,10]. Many studies are being pursued currently to develop small organic molecules, peptides and nanoparticles functionalized with drugs/phytochemicals/metal chelators to interfere with the Aβ aggregation process as a treatment strategy for AD [9,[11], [12], [13]].
Mimosine [β-[N-(3-hydroxy-4-oxypyridyl)]-α-aminopropionic acid], a non-protein amino acid found in a variety of tropical and subtropical plants, has been shown to display a wide range of biological properties significant to medicinal fields such as anti-cancer, anti-viral, anti-inflammatory and anti-fibrosis [[14], [15], [16]]. It has also been reported to suppress cell death in a variety of experimental paradigms partly by regulating the production of mitochondria-mediated reactive oxygen species (ROS) [17,18]. Structurally, like other phytochemicals such as capsaicin [19] and piperine [20], mimosine comprises an alanine side chain bound to the nitrogen atom of a hydroxypyridone ring with a resemblance to dihydroxyphenylalanine (DOPA) (Fig. 1a) [14]. Since hydrophobicity plays a critical role in Aβ aggregation [[21], [22], [23], [24]] and nanoparticles coated with hydrophobic molecules are capable of inhibiting the aggregation process [19,25], we wanted to determine if mimosine in a surface-functionalized form will be effective in inhibiting aggregation of Aβ peptide and protect mouse cultured neurons against Aβ-mediated toxicity.
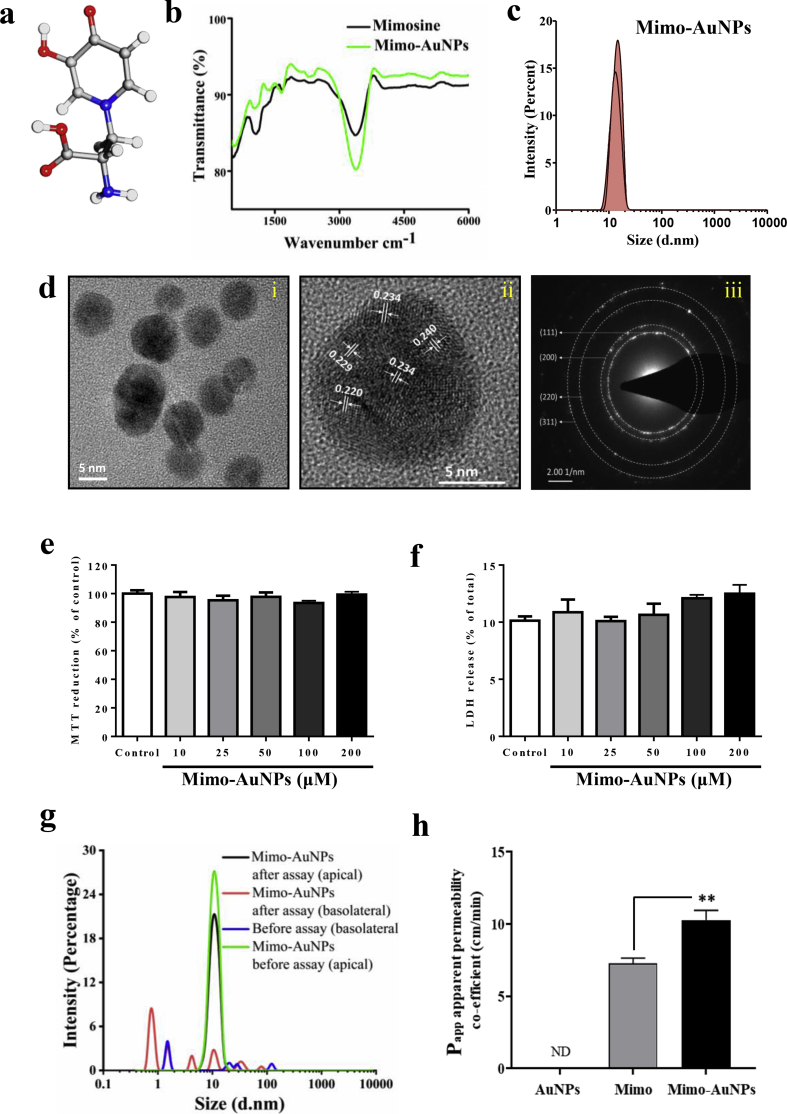
Fig. 1.
Characterization of Mimo-AuNPs. (a) Graphical image showing the chemical structure of mimosine obtained from Pubchem (ID: 440473). (b) FTIR spectra of Mimo (black) and Mimo-AuNPs (olive) displaying their respective vibrations before and after functionalization. (c) DLS peak of Mimo-AuNPs depicting an average diameter of ~8–12 nm. (d) TEM images revealing the evenly sized (~6 nm) spherical Mimo-AuNPs (i) and the high-resolution data show a fringe spacing of 2.2, 2.3 and 2.4 Å confirming its polycrystalline nature (ii) and selected area electron diffraction pattern (SAED) for Mimo-AuNPs showing the Scherrer ring patterns indicating the (111), (200) (220) and (311) nanocrystalline nature of FCC gold (iii). (e & f) Histograms depicting that 10–200 μM Mimo-AuNPs following 24h exposure did not alter the viability of cortical cultured neurons as evaluated using MTT (e) and LDH (f) assays. (g & h) DLS analysis showing Mimo-AuNPs in the apical side (before assay) and basolateral side (after assay) (g) and a histogram showing the apparent permeability coefficient (Papp) values after incubation of AuNPs, mimosine and Mimo-AuNPs for 30min in the activated BBB kit (h). All data expressed as mean ± SEM were obtained from three to five separate experiments, each performed in triplicate. ND, not detected. **p < 0.01.
Among various engineered nanoparticles, gold nanoparticles (AuNPs) are one of the most extensively studied nanoparticles due to their excellent biocompatibility, optical properties, easy fabrication and surface functionalization. Some surface-coated AuNPs depending on their intrinsic physicochemical properties (i.e., size, shape, surface modification and charge) have been shown to inhibit Aβ fibrillization and/or cell toxicity under in vitro paradigms [[26], [27], [28], [29]]. In the present study, we have successfully functionalized mimosine onto AuNPs (i.e., Mimo-AuNPs) and demonstrated their ability to cross the blood-brain barrier (BBB) and inhibit spontaneous as well as seed-induced aggregation of both normal and familial mutant human Aβ peptides using a Thioflavin-T (ThT) assay. The nature of the interaction between Aβ and Mimo-AuNPs is further characterized using a variety of structural and biophysical methods including Transmission Electron Microscopy (TEM), Dynamic light scattering (DLS), Circular dichroism (CD) as well as Fourier transform Infrared (FTIR) spectroscopy. Additionally, we revealed that Mimo-AuNPs can trigger the disassembly of preformed mature Aβ fibers and can protect mouse cortical cultured neurons against Aβ-induced toxicity by attenuating cell death mechanisms.
2. Experimental
2.1. Materials
Dulbecco's modified Eagle's medium (DMEM), neurobasal medium, Hanks' balanced salt solution (HBSS), fetal bovine serum (FBS), B27 and N2 supplement were purchased from Life Technology (NY, USA). The bicinchoninic acid (BCA) protein assay kit, enhanced chemiluminescence (ECL) kit were from Thermo Fisher Scientific Inc. (ON, Canada), whereas the RBT-24 BBB kit was from PharmaCo-Cell (Nagasaki, Japan). Isoforms of Aβ1-42 and Aβ1-40, Tau 04NR and the reverse sequence of Aβ1-42 (i.e. Aβ42-1) were purchased from R Peptide (CA, USA), whereas D23 N Iowa mutant, as well as E22Q Dutch mutant Aβ1-42, were procured from Ana Spec (CA, USA). ThT, Hexafluoro-2-Propanol (HFIP), Nile red, 8-Anilion-1-Naphthalene Sulfonate (ANS), mimosine, auric chloride (HAuCl3), trisodium citrate and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), potassium hydroxide (KOH) were obtained from Sigma-Aldrich (MO, USA) and the lactate dehydrogenase (LDH)-based cytotoxicity assay kit was purchased from Promega (OH, USA). Electron microscopy grids (Formvar/Carbon 200 mesh copper grids) and phosphotungstic acid stains were purchased from TedPella (CA, USA). Sources of primary antibodies used in the study are listed in Table 1. All horseradish peroxidase-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology (CA, USA). All other chemicals were obtained from either Sigma-Aldrich or Thermo Fisher Scientific.
Table 1.
Details of the primary antibodies used in this study.
| Antibody Type | Type | WB/FT dilution | Source |
|---|---|---|---|
| A11 oligomer specific | Polyclonal | 1:5000 | Invitrogen |
| Amyloid Fibrils OC | Polyclonal | 1:1000 | Sigma-Aldrich |
| Phospho-Thr202/Tyr204 ERK | Polyclonal | 1:1000 | Cell Signaling |
| Total-ERK | Monoclonal | 1:1000 | Cell Signaling |
| Phospho-Tyr216 GSK | Polyclonal | 1:1000 | Abcam |
| Total-GSK | Monoclonal | 1:1000 | Abcam |
| Tau (AT270) | Monoclonal | 1:1000 | Thermo Fisher Scientific |
| β-actin | Monoclonal | 1:5000 | Sigma-Aldrich |
| β-amyloid, 1–15 (3A1) | Monoclonal | 1:1000 | BioLegend |
| β-amyloid, 1–16 (6E10) | Monoclonal | 1:1000 | BioLegend |
| β-amyloid, 17–24 (4G8) | Monoclonal | 1:1000 | BioLegend |
| β-amyloid, 23-29 | Polyclonal | 1:1000 | Anaspec |
WB: western blotting; FT: Filter-trap.
2.2. Preparation of mimosine functionalized nanoparticles
The mimosine molecule was functionalized onto AuNPs utilizing the KOH reduction method using an established protocol [20,30]. As for control, we used citrate capped AuNPs which were synthesized by boiling 0.2 mM of HAuCl3 under the reflux condition followed by the addition of trisodium citrate as described earlier [20]. Finally, these nanoparticles were characterized by measuring their absorption spectra using a nanodrop spectrophotometer.
2.3. BBB permeability assay
The ability of the Mimo-AuNPs to cross the BBB was measured by determining the BBB permeability co-efficient using the established RBT-24 kit according to manufacturer's instruction. In brief, rat brain astrocytes, pericytes and vascular endothelial cells were co-cultured for 4 days (see Fig. S1c) and then the integrity of the formed endothelial barrier and tight junctions was evaluated by measuring the transendothelial resistance (TEER) of the cells using EVOM2-Epithelial Voltohmmeter. The TEER value was calculated in the following manner as reported earlier [31].
TEER (Ω x cm2) = [(relative resistance of experimental wells - relative resistance of blank wells) x (0.33) Membrane Surface Area (cm2)]
The wells with TEER value > 150 Ω x cm2, were subjected to 900 μl of phosphate-buffered saline (PBS)-based assay buffer on the brain side and 200 μl of the drugs (i.e, mimosine, AuNPs and Mimo-AuNPs) on the blood side of the trans-wells. The plate was incubated for 30min with mild agitation and the samples from both sides were collected to detect the concentrations of mimosine, AuNPs and Mimo-AuNPs using UV visible spectra and DLS. The Papp value was calculated according to the kit's protocol.
2.4. Preparation of Aβ peptides
All lyophilized Aβ1-42, Aβ1-40, Aβ42-1, D23 N Iowa mutant Aβ1-42 and E22Q Dutch mutant Aβ1-42 stored at −80 °C were first equilibrated to room temperature for 30min before dissolving in HFIP to obtain a 1 mM solution. Once dissolved, peptide aliquots were dried to remove HFIP and then restored at −80 °C for subsequent use as described earlier [32]. For the preparation of Aβ fibrils, diluted peptides were incubated at 37 °C overnight in PBS (pH 7.4).
2.5. Aβ aggregation kinetics
The aggregation kinetics of Aβ1-42, Aβ1-40, D23 N mutant Aβ1-42 and E22Q mutant Aβ1-42 (10 μM) were carried out in the presence or absence of different concentrations of AuNPs, mimosine and Mimo-AuNPs using the ThT assay as described earlier [33]. In the case of seed-induced aggregation studies, preformed Aβ1-42 fibers (~15% w/v) were used as a primary seed. For Aβ1-42 disassembly experiments, the ThT signal was monitored for the mature fibers in the presence of Mimo-AuNPs at regular time intervals for 120hrs. The fluorescence signal for spontaneous Aβ aggregation was measured every 15min for 24h with excitation at 440 nm and emission at 480 nm using a Flurostar omega BMG Labtech (Aylesbury, UK) or a Spectra max M5 (Molecular Devices, USA). All kinetic experiments were repeated nine times with three technical replicates and the graphs were plotted using ORIGIN 2020.
2.6. Fluorescence microscopy
After incubation of Aβ samples with or without mimosine, AuNPs and Mimo-AuNPs for different periods (0–24h), 100 μl of the sample was transferred to glass slides, stained with ThT as described earlier [23,34]. The samples were then examined and images were captured using a Nikon 90i fluorescence microscope.
2.7. Transmission electron microscopy (TEM)
The Aβ samples with or without Mimo-AuNPs (10 μL) were loaded onto freshly glow-discharged 200 mesh carbon-coated copper grids and adsorbed onto grid surface for ~1min. Grids were then washed with 50 μL 0.1 M and 0.01 M ammonium acetate and negatively stained with 2% phosphotungstic acid solution. After drying, the stained samples were analyzed with a FEI Tecnai G20 electron microscope (FEI Company, The Netherlands). An acceleration voltage of 200 kV was used to record micrographs on an Eagle 4k x 4k CCD camera.
2.8. Circular dichroism (CD) spectroscopy
The changes in the secondary structure of Aβ1-42 in the presence and absence of Mimo-AuNPs were determined using a Chirascan CD spectrophotometer as described earlier [23,24]. After incubating 10 μM Aβ1-42 in the presence or absence of 200 μM Mimo-AuNPs, each reaction mixture is subjected to spectroscopic studies and the CD spectra were recorded at room temperature in a CD cuvette of 2 mm path length. The reported spectra were the average of nine different acquisitions between 200-260 nm. The alteration in the secondary structure of Aβ1-42 was determined by analyzing the ellipticity values of the samples taken from the aggregation reactions.
2.9. Dynamic light scattering (DLS)
The hydrodynamic radius of various AuNPs with or without Aβ1-42 was measured using a Malvern Zetasizer-Nano ZS (Malvern Instruments, USA) equipped with a back-scattering detector (173°). The samples (AuNPs and Mimo-AuNPs) were prepared by filtering it through a pre-rinsed 0.2 μm filter and all readings were recorded after equilibrating the samples for 5min at 25 °C. Particle size was calculated by the manufacturer's software through the Stokes-Einstein equation assuming spherical shapes of the particles [35].
2.10. Zeta potential measurement
The surface zeta potential measurement was carried out using a Nano ZS (Malvern) equipped with MPT-2 titrator. The Mimo-AuNPs and AuNPs samples were resuspended in deionized water and the Zeta potentials were calculated from the electrophoretic mobility using a Smoluchowski relationship as described earlier [35].
2.11. Native polyacrylamide gel electrophoresis (PAGE) analysis
We used Native PAGE to determine the formation of higher-ordered aggregates of Aβ with or without Mimo-AuNPs [23,24]. The Native PAGE was performed using a 12% polyacrylamide gel in an Invitrogen novex Mini cell system. The gels were silver stained and visualized by a FluorChem E system (CA, USA). The images taken from native gels were subsequently processed by using Image J software.
2.12. Fourier transform Infrared (FTIR) spectroscopy
The FTIR spectroscopy study was performed to identify the presence of functional groups in the mimosine that characteristically bind AuNPs. The FTIR spectra for mimosine and Mimo-AuNPs were obtained by using the KBr pellet method, whereas the ATR mode was used for obtaining the secondary derivative (1700 cm−1 and 1600 cm−1) for mature Aβ1-42 fibers with or without Mimo-AuNPs. All spectra were collected with a Bruker Vertor 70 spectrometer equipped with a silicon carbide source and an MCT (Mercury-Cadmium-Telluride) detector. The spectra were processed using the OPUS 6.5 software as described previously [36].
2.13. 8-Anilion-1-Naphthalene sulfonate (ANS) assay
ANS is a specific assay to detect Aβ aggregation [37]. The reaction samples containing Aβ1-42 aggregates in the presence or absence of Mimo-AuNPs were mixed with ANS, incubated for 30min at 37 °C in the dark and then recorded at 380 nm using a spectramax M5 multi-plate reader. The experiments were repeated three to four times and the average values were plotted using ORIGIN 2020.
2.14. Nile red assay
Nile red, a hydrophobic dye, was used to detect the presence of amyloid fibers [37]. After the Aβ1-42 aggregation reaction, 150 μl of samples with or without Mimo-AuNPs were mixed with 5 μM Nile red and incubated for 60min at room temperature. The samples were then excited at 530 nm and the emission spectra were recorded from 540 to 700 nm using a spectramax M5 multi-plate reader. The experiments were repeated three times and the average values were plotted using ORIGIN 2020.
2.15. Filter-trap assay for epitope mapping
The site of interaction between Aβ1-42 monomers and Mimo-AuNPs was detected by a filter-trap assay using various site-specific Aβ antibodies (see Table 1) [38]. The Aβ samples after aggregation in the presence or absence of Mimo-AuNPs were spotted on a nitrocellulose membrane (0.02 μm), subjected to vacuum filtration through a 96-well Bio-Dot Microfiltration apparatus, washed with Tris-buffer with 0.1% Tween 20 and then incubated at 4 °C for 12h with Aβ antibodies. The membranes were then washed, incubated with appropriate secondary antibodies (1:5000) and developed with an ECL kit. All blots were examined using a Fluor Chem E system and the images were processed by using Image J software.
2.16. Molecular docking
The molecular interaction between Aβ and mimosine ligand was studied by using AutoDock Vina wizard of PyRx(v0.8) [39]. The structure of Aβ1-42 monomers (PDB ID 1IYT, 6SZF and 1ZOQ), Aβ1-42 fibers (PDB ID 5OQV), Aβ1-40 monomer (PDB ID 1BA4) and Tau monomer fragment (PDB ID 2MZ7) were obtained from RSCB [40], whereas the three-dimensional structure for mimosine CID 3862 was obtained from PubChem [23,41]. All protein molecules were pre-processed and protonated before conducting a docking experiment using an AutoDock Vina platform in which each protein molecule was docked with the mimosine in a manually defined grid box. After docking, results were screened based on energy values and the selected pose cluster was analyzed using Discovery studio visualizer 2019 and Chimera 14.1 [36].
2.17. Mouse cortical neuronal cultures
Timed pregnant BALB/c mice purchased from Charles River (St. Constant, Canada) were maintained according to Institutional guidelines. Primary cortical cultures were prepared from 18-day-old embryos of pregnant mice as described previously [42]. At first, the frontal cortex from pup brains was dissected in HBSS supplemented with 15 mM HEPES (hydroxyethyl piperazineethanesulfonic acid), 10units/mL penicillin and 10 μg/ml streptomycin and then digested with 0.25% trypsin-EDTA. The cell suspension was filtered through a cell strainer and plated on either 96-well plates (for survival/death assay) or 6-well plates (for biochemical analysis). The cultures were grown at 37 °C in a 5%CO2 atmosphere in Neurobasal medium supplemented with B27/N2, 50 μM l-glutamine, 15 mM HEPES, 10units/ml penicillin, 10 mg/ml streptomycin and 1% FBS. The medium was replaced 1 day later without FBS, and all experiments were performed on day 6 after plating. In brief, cultured neurons were treated with different concentrations of Mimo-AuNPs (10–200 μM) or Aβ1-42 (1–10 μM) for 24h. In parallel, cultured neurons were exposed to Aβ samples treated with or without 200 μM Mimo-AuNPs for 24h. The control and treated cultures were processed for cell viability/toxicity as well as western blotting as described earlier [42].
2.18. Neuronal viability and toxicity assays
Neuronal viability following various experimental paradigms was assessed using MTT and LDH-based cytotoxicity assays as described earlier [32,43]. For the MTT assay, control and Aβ-treated culture plates were replaced with new media containing 0.25% MTT and then incubated for 4h at 37 °C with 5% CO2/95% air. The formazan was dissolved in DMSO and absorbance was measured at 570 nm with a microplate reader. To validate the MTT data, control and Aβ-treated cultured neurons were assayed for cytotoxicity based on the activity of LDH that is released into conditioned medium from the cytosol of damaged cells. The absorbance was measured at 490 nm with a Spectramax M5 spectrophotometer. Both assays were repeated three to five times with three technical replicates per sample.
2.19. Western blotting
Western blotting of cultured cells from various experimental paradigms was performed using different primary antibodies as described earlier [44]. In brief, control and treated mouse cortical neuronal cultures were homogenized in Radio-immunoprecipitation assay (RIPA) lysis buffer containing protease inhibitor cocktail and proteins were quantified using BCA kit. Denatured samples were resolved on 10% or 12% gradient sodium dodecyl sulfate polyacrylamide gels, transferred to polyvinylidene difluoride membranes, blocked with 5% milk and incubated overnight at 4 °C with various primary antibodies at dilutions listed in Table 1. The membranes were washed and incubated with horseradish peroxidase-conjugated secondary antibodies (1:5000) and immunoreactive proteins were detected with ECL kit. All blots were re-probed with β-actin antibody and quantified using ImageJ.
2.20. Statistical analysis
All data were expressed as mean ± SEM. Kinetics of peptide aggregation as well as cell viability data from cultured neurons were analyzed by one-way ANOVA followed by Bonferroni's post-hoc analysis for multiple comparisons with a significance threshold set at p < 0.05. All statistical analysis was performed using GraphPad Prism (GraphPad Software, Inc., USA).
3. Results
3.1. Synthesis and characterization of Mimo-AuNPs
Using an established protocol [20], we have successfully synthesized Mimo-AuNPs displaying a surface plasma resonance (SPR) peak around ~525 nm. These nanoparticles are found to be quite stable without any notable aggregation or self-assembly over a one-year period (Fig. S1a). To characterize the surface functionalization of Mimo-AuNPs we used FTIR which revealed the distinctive vibration spectra at 1163 and 3355 cm−1 for the C-N aliphatic amines and OH stretching vibration, due to the phenolic hydroxyl groups, over the benzene ring [45]. Additionally, the vibration between 1300-1400 cm−1 is possibly due to C-N stretching of mimosine [46]. From these spectra, it is evident that the functionalization of the nanoparticles was mediated by the amine group leaving the aromatic phenyl group exposed for interactions (Fig. 1a and b). DLS data for the Mimo-AuNPs showed a homogeneous population with an average hydrodynamic radius of ~10 nm (Fig. 1c) and zeta potential of about -6mV (Fig. S1b). TEM images confirmed that these nanoparticles are homogeneous with spheroidal morphology of ~6 nm radius (Fig. 1d). The difference in nanoparticle size as measured by TEM and DLS was because DLS, unlike TEM, includes the hydration layer that covers the surface of the nanoparticles [47]. To further understand the atomistic arrangements, we used high-resolution TEM which revealed the polycrystalline nature of Mimo-AuNPs with fringe patterns of 2.2 Å, 2.3 Å and 2.4 Å respectively, suggesting the presence of (111) and (200) index in Face Centered Crystalline (FCC) lattice (Fig. 1d) [20,28]. The cytotoxicity profile showed that, Mimo-AuNPs did not affect the viability of primary cortical neurons even at 200 μM over 24h exposure, suggesting that these nanoparticles are non-toxic to the neurons (Fig. 1e and f). Additionally, assessment of BBB permeability using a well-established in vitro kit depicted that Mimo-AuNPs exhibit a higher permeability with an apparent permeability coefficient (Papp) value of 10x10−6 cm/s compared to the mimosine alone with a Papp value of 7.3x10−6 cm/s. AuNPs, on the other hand, were not detected over the 30min incubation period (Fig. 1g and h; Fig. S1c).
For control experiments, we synthesized AuNPs in the absence of mimosine using trisodium citrate. These AuNPs also showed an SPR peak at ~530 nm (Fig. S2a). The TEM data revealed that these nanoparticles exhibited a polycrystalline nature of ~7–10 nm in radius (Fig. S2b) with a d-spacing of 2.1 Å, 2.3 Å, 2.4 Å, and 2.5 Å respectively, indicating the presence of (111), (220) and (400) index in FCC lattice (Fig. S2c). The hydrodynamic radius of the AuNPs analyzed by DLS was found to be ~30 nm (Fig. S2d) with a total surface charge of -1mV (Fig. S2e).
3.2. Mimo-AuNPs and spontaneous Aβ aggregation
ThT, an amyloid-specific dye, is usually used to detect the molecular conversion of Aβ peptide from monomeric to fibrillar state (Fig. 2) [48]. It is well known that Aβ1-42 has a high tendency to form aggregates and it starts forming aggregates as soon as it gets into an aqueous media [49]. The ThT aggregation kinetic assay revealed that 10 μM Aβ1-42 displayed a 3h lag phase, 10h log phase followed by an 11h stationary phase (Fig. 2a). The propensity of Aβ aggregation was validated by fluorescence imaging which showed the presence of Aβ fibrillar entities following ThT labeling (Fig. 2g). Incubation of 10 μM Aβ1-42 with 10–300 μM Mimo-AuNPs dose-dependently attenuated fibril formation over 24h likely due to alteration kinetics and its conformation state (Fig. 2a, Figs. S3a–c). An initial delay in the lag time was achieved at 30 μM Mimo-AuNPs, which increases gradually with increasing concentrations of Mimo-AuNPs reaching the highest lag time of ~10h at 300 μM Mimo-AuNPs. The aggregation kinetic was attenuated to 50% at 30 μM Mimo-AuNPs (1:3) and reached more than 90% in the presence of 300 μM Mimo-AuNPs (1:30) at 24h relative to that of pure Aβ peptide (Fig. S3a). In contrast to Mimo-AuNPs, the aggregation kinetic of Aβ1-42 was not altered by either 200 μM AuNPs or mimosine alone, indicating that surface functionalization of AuNPs is critical to interfere with Aβ fibril formation (Fig. 2a).
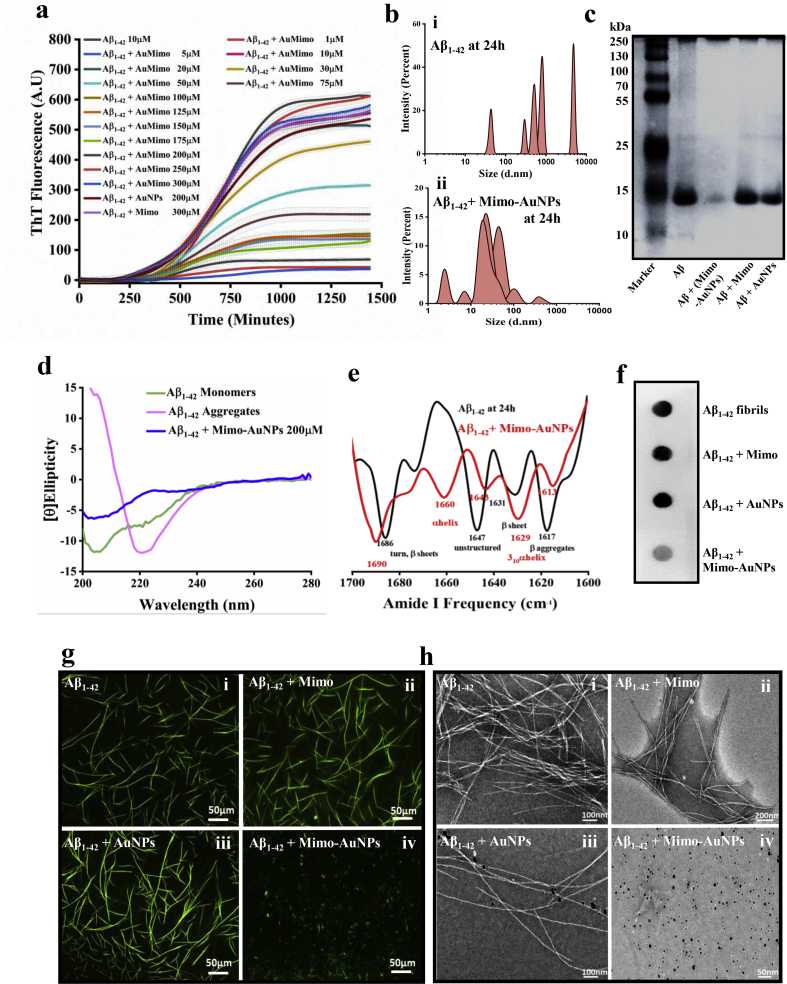
Fig. 2.
Suppression of Aβ1-42aggregation in presence of Mimo-AuNPs. (a) Thioflavin T assay showing the aggregation curves of 10 μM Aβ1-42 in the absence or presence of 200 μM Mimo, 200 μM AuNPs and different doses (1–300 μM) of Mimo-AuNPs over 24h incubation. Note that Mimo-AuNPs, but not Mimo or AuNPs, can attenuate the spontaneous aggregation Aβ1-42. (b) DLS analysis revealing the hydrodynamic radius of ~10 μM Aβ1-42 in the absence (i) and presence of Mimo-AuNPs (ii). Note the decrease in the hydrodynamic radius of Aβ1-42 in the presence of Mimo-AuNPs. (c) Native-PAGE analysis of Aβ1-42 samples after 24h reaction in the absence or presence of Mimo, AuNPs and Mimo-AuNPs. Note the decreased level of higher-ordered Aβ entities in the presence of Mimo-AuNPs but not Mimo or AuNPs. (d) CD spectra for Aβ1-42 monomers (green) undergoing aggregation in the presence (blue) or absence (magenta) of 200 μM Mimo-AuNPs. Note the decreased beta-sheet formation following incubation with Mimo-AuNPs. (e) FTIR secondary derivative spectra of the aggregating Aβ1-42 samples in the presence (red) and absence (black) of Mimo-AuNPs. Note the dominance of beta rich signals after aggregation of Aβ1-42 and the occurrence of 310 α-helix in presence of Mimo-AuNPs. (f) Filter-trap assay of Aβ1-42 samples after 24h reaction in the absence or presence of Mimo, AuNPs and Mimo-AuNPs revealing the decreased formation of Aβ1-42 fibers in presence of Mimo-AuNPs as detected by a fibril specific OC antibody. (g) Thioflavin T-stained fluorescence images of Aβ1-42 samples after 24h reaction in the absence (i) or presence of 200 μM Mimo (ii), 200 μM AuNPs (iii) and 200 μM Mimo-AuNPs (iv). Note the attenuation of Aβ1-42 aggregation in presence of Mimo-AuNPs. (h) TEM images of Aβ1-42 samples after 24h reaction in the absence (i) or presence of 200 μM Mimo (ii), 200 μM AuNPs (iii) and 200 μM Mimo-AuNPs (iv). Note the lack of Aβ fibrils in presence of Mimo-AuNPs at both magnification scales (fluorescence and TEM). All ThT kinetic data expressed as mean ± SEM were obtained from three to nine separate experiments each performed in triplicate.
To evaluate the size distribution of Aβ1-42 after 24h incubation with or without Mimo-AuNPs, we performed DLS analysis. While Aβ1-42 displayed loss of monomeric state and formation of higher-order aggregates, the presence of Mimo-AuNPs rendered Aβ peptide to retain its monomeric structures possibly by stabilizing the native conformers or hindering the on-pathway aggregation cycle of Aβ peptide (Fig. 2b). This was substantiated by native-PAGE data which showed the lack of higher-ordered aggregates in Aβ samples treated with 200 μM Mimo-AuNPs compared to samples treated with either mimosine or AuNPs (Fig. 2c). The CD spectra of soluble Aβ revealed the presence of an unstructured random coil with a negative peak around 195 nm, whereas the spectra of aggregated samples displayed a positive peak and negative valley around 195 and 215 nm, respectively suggesting a conformational change towards beta structure (Fig. 2d). These results were consistent with previous studies demonstrating the transition from monomeric to the aggregated state of Aβ1-42 [8,32,50]. Interestingly, in presence of Mimo-AuNPs, the transition of Aβ1-42 was evident but much less than that of the aggregated sample (Fig. 2d), suggesting that Mimo-AuNPs influence secondary structural changes during Aβ1-42 assembly. Further analysis revealed the presence of 39% alpha-helical, 15% anti-parallel and 46% unstructured entities in Aβ monomers but after aggregation, 58% anti-parallel and 43% unstructured conformers were evident in Aβ samples. Conversely, aggregating Aβ samples with Mimo-AuNPs showed the presence of 15% alpha-helical, 12% anti-parallel and 65% unstructured conformers. These data suggest that Mimo-AuNPs may have an inherent tendency to stabilize the native structure of Aβ monomers by preventing its aggregation pathway. To confirm this effect, we used FTIR which showed that secondary derivate spectra generated through ATR mode depicted the characteristic vibrations at 1616, 1635, 1646, 1667 and 1684 cm−1 indicating the presence of β-sheet enriched structures in Aβ1-42 aggregates, whereas in presence of Mimo-AuNPs we observed the vibration signals for the 310α helix – suggesting retention of monomeric state (Fig. 2e). Additionally, filter-trap analysis using the Aβ1-42 fiber specific OC antibody depicted a significant decrease in the fibrillar form of Aβ1-42 in presence of Mimo-AuNPs compared to samples treated with mimosine or AuNPs alone (Fig. 2f). In order to identify the aggregation state of Aβ1-42 fibrils, 10 μM Aβ1-42 after incubation for 24h with or without 200 μM mimosine, AuNPs or Mimo-AuNPs were analyzed using fluorescence microscopy as well as TEM. The fluorescence imaging data revealed the presence of bundles of mature fibrils in all Aβ preparations except the sample treated with Mimo-AuNPs, which is associated mostly with the amorphous unstructured aggregates (Fig. 2g). This is validated by TEM showing the absence of mature fibrillar entities in Aβ1-42 sample treated with Mimo-AuNPs but not with mimosine or AuNPs (Fig. 2h). It is noteworthy to mention that the TEM images (Fig. 2h) do not represent the large Aβ fibril bundles seen in the fluorescent images (Fig. 2g) due to their size. However, the presence of individual Aβ fibrils, or small bundles thereof, in the electron micrographs reinforces the observation that Mimo-AuNPs disassembles both individual Aβ fibrils and the bundles they form.
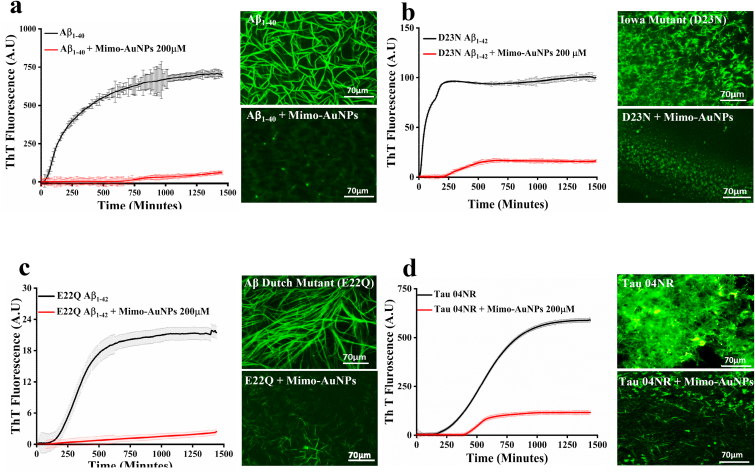
To determine if Mimo-AuNPs can influence the spontaneous aggregation of other isoforms of Aβ peptide, we performed ThT kinetic assays using 10 μM Aβ1-40 in the presence or absence of 200 μM Mimo-AuNPs at 37 °C over 24h period (Fig. 3a). In parallel, we also measured the effects of 200 μM Mimo-AuNPs on 10 μM D23 N Iowa and 10 μM E22Q Dutch mutant Aβ1-42 (Fig. 3b and c), which are known to aggregate faster and more toxic to neurons than normal Aβ1-42 [51]. Our data revealed that Mimo-AuNPs, as observed with Aβ1-42, can time-dependently decrease spontaneous aggregation of Aβ1-40 as well as the two mutant Aβ1-42 peptides (Fig. 3a–c). This is validated by our fluorescence microscopy results (Fig. 3a–c), highlighting the significance of Mimo-AuNPs in attenuating the aggregation of both normal and mutant Aβ peptides. Accompanying Aβ, our ThT kinetic and fluorescence microscopy data showed that Mimo-AuNPs can suppress spontaneous aggregation of 4-repeat tau protein over a 24h period (Fig. 3d). Molecular docking results also revealed a strong interaction between mimosine and tau protein (PDB ID: 2MZ7) via hydrogen and hydrophobic bonds with a binding energy of −4.0 kcal mol−1 (Fig. S4).
Fig. 3.
Mimo-AuNPs attenuates aggregation kinetics of different Aβ isoforms. (a–d) Thioflavin T aggregation kinetics and respective fluorescence images showing the effects of 200 μM Mimo-AuNPs on the aggregation of 100 μM Aβ1-40 (a), 10 μM mutant D23 N Aβ1-42 (b), 10 μM mutant E22Q Aβ1-42 (c) and 10 μM tau (d). Note that Mimo-AuNPs, as observed for Aβ1-42, can attenuate the spontaneous aggregation of various other Aβ fragments. All ThT kinetic data expressed as mean ± SEM were obtained from three to nine separate experiments each performed in triplicate.
3.3. Molecular interaction of Mimo-AuNPs with Aβ1-42
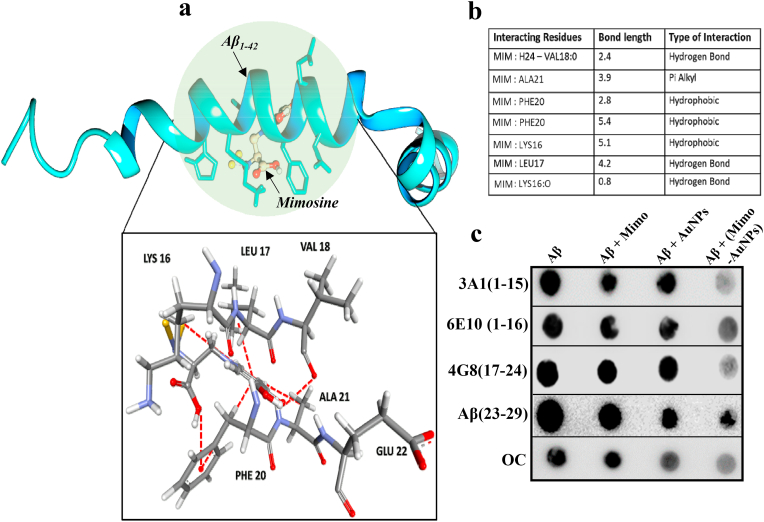
To understand the molecular basis of interaction between Mimo-AuNPs and Aβ1-42, we conducted molecular docking studies using Auto dock Vina software as described earlier [52]. Our data indicated a strong interaction between Aβ1-42 and Mimo-AuNPs with the formation of 7 contacts mediated via 3 hydrogens (i.e., Leu16, Lys17, Val18), 3 hydrophobic bonds with two amino acids (i.e., Lys16, Phe20) and 1 pi-alkyl with Ala21, mostly facilitated through the aromatic groups present in the mimosine (Fig. 4a and b). This interaction is highly specific as the Mimo-AuNPs bind the aggregation-prone region (i.e., Lys16 to Glu22) in the Aβ domain (1–42 PDB ID:1IYT) with a binding energy value of −4.2 kcal mol−1 (Fig. 4b). We have also performed molecular docking studies with the recently reported structure of Aβ1-42 (PDB ID: 6SZF and 1Z0Q), which are known as the solution and aqueous solution structure of Alzheimer's Disease Aβ1-42. Further analysis revealed that the Mimo-AuNPs display a strong affinity to the amino acids spanning from Glu11 to Phe19 of Aβ1-42 with a binding energy of 3.6 kcal mol−1 for 6SZF (6 viable interactions including 4 hydrogen and 2 hydrophobic interactions) and 3.3 kcal mol−1 for 1ZOQ (3 viable interactions with 3 hydrogen bonds) (see Figs. S5 and S6). These data also reveal that Mimo-AuNPS have a strong affinity to the aggregation prone structure of Aβ monomer. In addition to Aβ1-42, Mimo-AuNPs were able to interact with the aggregation-prone domain (i.e., 15–25 amino acids) of Aβ1-40 (PDB ID: 1BA4) with a binding energy of −4.8 kcal mol−1 (Fig. S7). To verify further the site of interaction, we carried out filter-trap-based epitope mapping analysis of Aβ1-42 samples treated with or without mimosine, AuNPs and Mimo-AuNPs using various Aβ antibodies. Our data revealed that Mimo-AuNPs, but not mimosine or AuNPs, markedly decreased the interaction of Aβ1-42 with various antibodies including fiber-specific OC antibody. Intriguingly, the interaction of multiple Aβ antibodies recognizing Gln15 and Gly29 region of the peptide was decreased in Aβ samples treated with Mimo-AuNPs indicating that the site of potential interaction may reside in the aggregation prone hydrophobic domain (i.e., Lys16 to Ala21) of Aβ peptide (Fig. 4c). The hydrophobic interaction between Mimo-AuNPs and Aβ1-42 is further substantiated by ANS and Nile red assays which showed fluorescence intensity of Aβ1-42 aggregates decreased markedly in the presence Mimo-AuNPs (Figs. S8a and b).
Fig. 4.
Molecular interactions of Mimo-AuNPs with Aβ1-42. (a) Molecular docking data using Auto dock Vina revealing the interaction of Mimo-AuNPs with the monomeric Aβ1-42 representing 7 viable interactions including 3 hydrogen, 3 hydrophobic and 1 pi-alkyl interaction with amino acid residues such as Valine, Alanine, Phenylalanine, Lysine, and Leucine, respectively with an interaction energy of −4.2 kcal mol−1. (b) Table listing all the viable interactions between Mimo-AuNPs and Aβ1-42 PDB ID:1IYT showing its respective bond length in (Å). (c) Epitope mapping of Aβ1-42 conformers using filter-trap assay revealing that Mimo-AuNPs interact with the hydrophobic domain of Aβ1-42.
3.4. Mimo-AuNPs and seed-induced Aβ1-42 aggregation
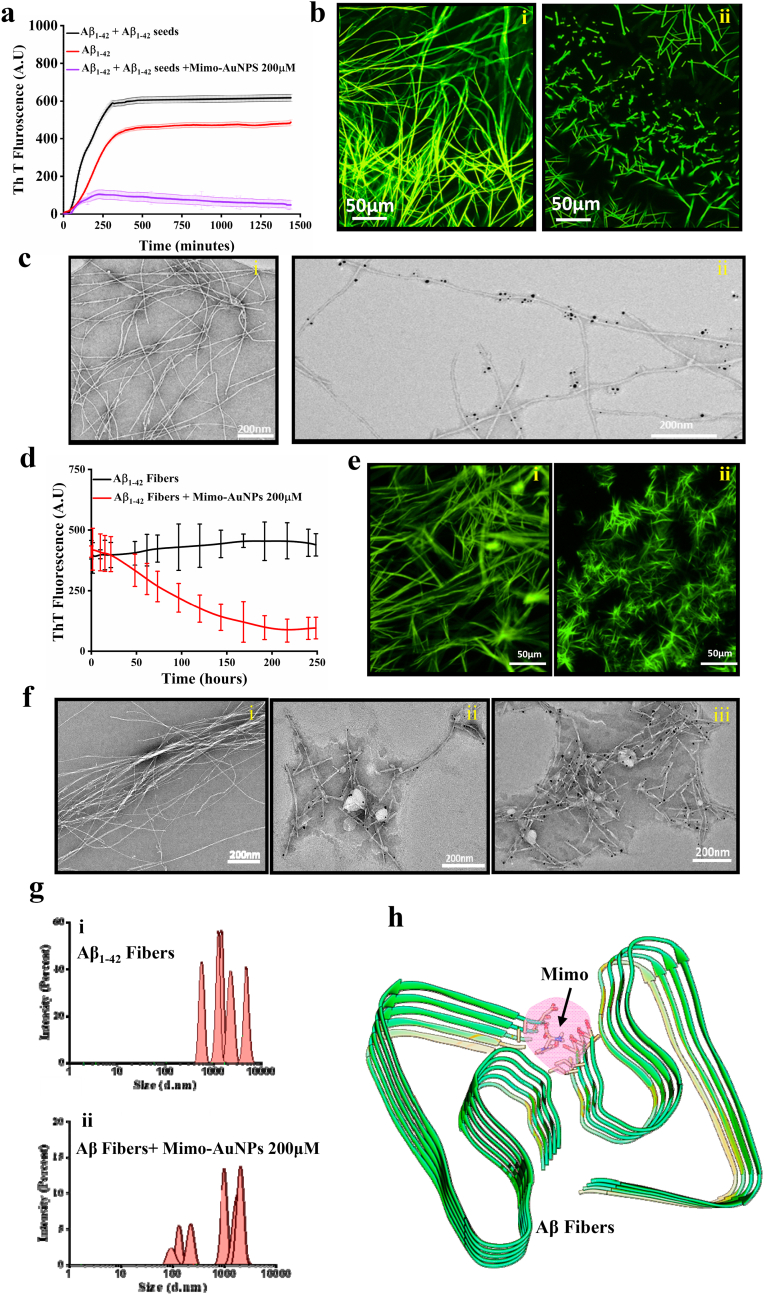
Since seed-induced aggregation plays a major role in the secondary nucleation process [41,53,54], we evaluated the effect of Mimo-AuNPs on seed-induced fibril assembly of Aβ1-42. As expected, we observed faster aggregation kinetics of Aβ1-42 in the presence of preformed Aβ seeds prepared by incubating Aβ1-42 (~15%w/v) for 24h. The presence of Mimo-AuNPs, as evident by ThT fluorescence assay, markedly attenuated seed-induced fibril formation (Fig. 5a), which is confirmed by analysis of Aβ fibrils using fluorescence microscopy (Fig. 5b) as well as TEM (Fig. 5c). It is possible that Mimo-AuNPs interfere with the aggregation process by suppressing monomer recruitment as well as interaction with the hydrophobic surfaces on Aβ fibrils to prevent the further seeding effect. As before, the differences in magnification between the fluorescent images and the electron micrographs reinforce the efficacy by which Mimo-AuNPs interfere with the Aβ fibrillization process.
Fig. 5.
Mimo-AuNPs on seed-induced Aβ1-42aggregation and disassembly of Aβ1-42fibers. (a) Thioflavin T kinetic data showing aggregation curves of Aβ1-42 (red), 15% (w/v) seed-induced reaction of Aβ1-42 (black) and seed-induced Aβ1-42 reaction in presence of 200 μM Mimo-AuNPs. (blue) (b) Fluorescence images of Thioflavin T staining showing seed-induced Aβ1-42 aggregation in the absence (i) and presence (ii) of Mimo-AuNPs. Note the suppressing effects of Mimo-AuNPs on the seed-induced aggregation of Aβ1-42. (c) TEM images of matured Aβ fibers from uninhibited and inhibited reactions depicting the interaction of Mimo-AuNPs with the seeded fibers. (d & e) Thioflavin T kinetic data (d) and corresponding fluorescence images (e) showing disassembly of matured Aβ1-42 fibers in the absence (i) or presence (ii) of 200 μM Mimo-AuNPs. Note the suppressing effects of Mimo-AuNPs on preformed Aβ aggregates. (f) TEM images of matured Aβ1-42 fibers (i) and disassembled fiber conjugated with Mimo-AuNPs (ii and iii). (g) DLS histograms revealing hydrodynamic radius of aggregated Aβ1-42 in the absence (i) and presence of 200 μM Mimo-AuNPs (ii) indicating the presence of disassembled fragments. (h) Molecular docking data using Auto dock Vina revealing the interaction of Mimo-AuNPs with the steric zipper domain present in Aβ1-42 (PDB ID:5OQV) fibers. All ThT kinetic data expressed as mean ± SEM were obtained from three to nine separate experiments each performed in triplicate.
3.5. Mimo-AuNPs and disassembly of Aβ1-42 fibrils
To establish if Mimo-AuNPs can promote disassembly of mature Aβ fibers, preformed Aβ1-42 fibers were incubated with 200 μM Mimo-AuNPs over 120h at 37 °C. The ThT assay revealed that Mimo-AuNPs can time-dependently disassemble the mature Aβ1-42 fibers (Fig. 5d). This is supported by fluorescence imaging (Fig. 5e) as well as TEM (Fig. 5f) data showing the presence of smaller fragments of Aβ fibrils following Mimo-AuNPs treatment. Analysis of Mimo-AuNPs-treated Aβ1-42 fibers by DLS also revealed a shift towards the lower ordered entities with a hydrodynamic radius between 0.1-5 μM compared with untreated (>1–10 μM) Aβ1-42 fibers (Fig. 5g). It is likely that Mimo-AuNPs trigger disassembly by disrupting the β-sheet structure or by interaction with the steric zippers present in Aβ1-42 fibers as well as salt bridges between Aβ1-42 fibers as suggested by the molecular docking studies (Fig. 5h; Fig. S9).
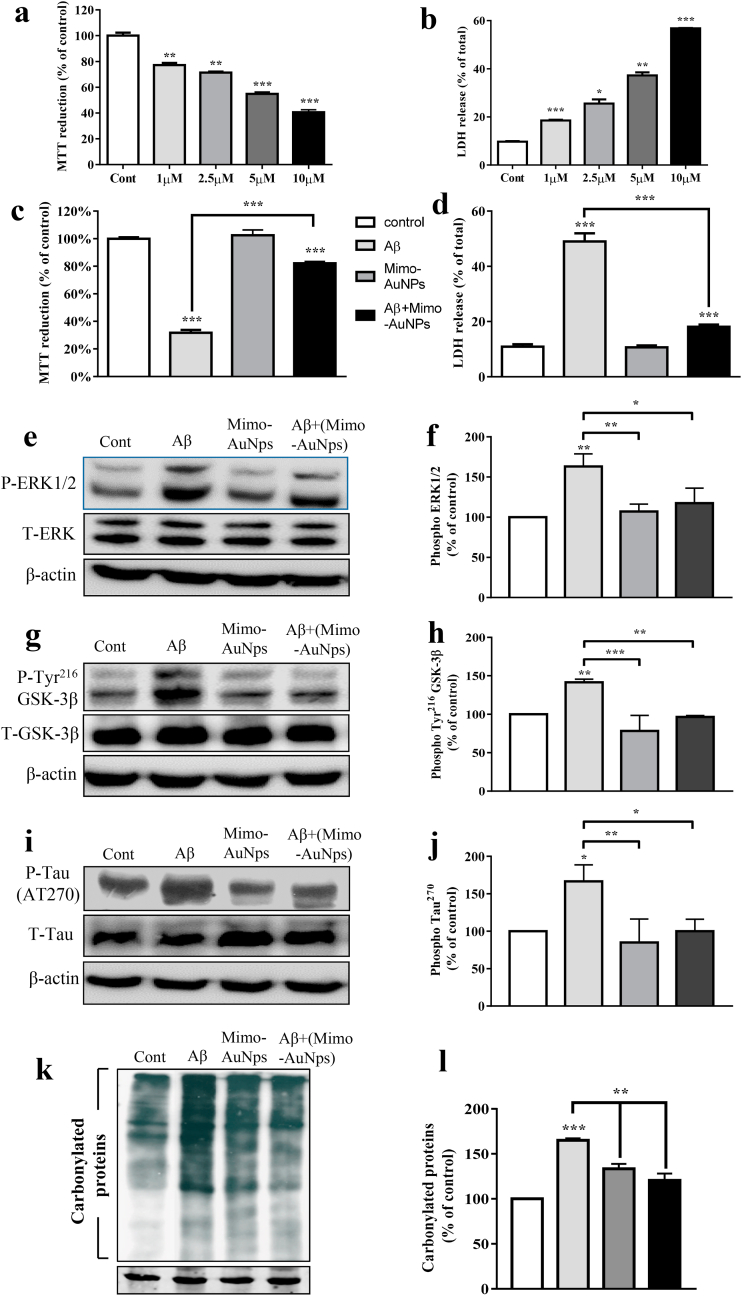
3.6. Mimo-AuNPs and Aβ-mediated toxicity
To determine if suppression of Aβ1-42 aggregation can influence cell viability, primary mouse cortical neurons were exposed to Aβ samples treated with or without 200 μM Mimo-AuNPs and their viability was assessed using MTT and LDH assays. Indeed, suppression of Aβ1-42 aggregation by Mimo-AuNPs increased the viability of cultured neurons (Fig. 6a–d). The toxicity induced by Aβ peptide, as expected [8,55], is associated with enhanced levels of phospho-Thr202/Tyr204 extracellular-signal related kinase 1⁄2 (ERK1/2), phospho-Tyr216 glycogen synthase kinase (GSK-3β), phospho-tau (Fig. 6e–j) and oxidative stress marker carbonylated proteins (Fig. 6k,l). Interestingly, the protective effect of Mimo-AuNPs is partly mediated by attenuating activation of ERK1⁄2 and GSK-3β as well as reducing phosphorylation of tau protein (Fig. 6e–j). Additionally, Mimo-AuNPs reduced the levels of protein carbonyl groups in Aβ-treated neurons suggesting an attenuation of oxidative stress (Fig. 6k,l).
Fig. 6.
Disaggregation of Aβ1-42 by Mimo-AuNPs protects cultured neurons. (a & b) Histogram depicting the dose-dependent decrease in the viability of mouse cortical cultured neurons following 24h treatment with oligomeric human Aβ1-42 compared to neurons treated with 10 μM Aβ42-1 (Cont) as revealed by MTT and LDH assays. (c & d) Histograms showing protection of cortical neurons following disassembly of preformed Aβ1-42 aggregates by Mimo-AuNPs as detected with MTT (c) and LDH (d) assays. (e–l) Immunoblots and corresponding histograms showing that the protective effect of Mimo-AuNPs is mediated by decreasing the levels of Phospho-ERK1/2 (e, f), Phospho-Tyr216 GSK-3β (g, h), Phospho-tau (i, j) and carbonylated protein (k, l) levels induced by preformed aggregated Aβ1-42. All data expressed as mean ± SEM were obtained from three to five separate experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
4. Discussion
The present study, using a variety of experimental approaches, revealed that AuNPs surface-functionalized with mimosine can suppress not only spontaneous and seed-induced Aβ aggregation but also can trigger the disassembly of matured Aβ fibers. Additionally, surface functionalization of AuNPs with mimosine enables them to cross an in vitro model of the BBB efficiently and provide protection to cultured neurons against Aβ-induced toxicity, suggesting their therapeutic potential in the treatment of AD pathology. Since neither mimosine nor AuNPs alone was able to attenuate Aβ fibril formation, it appears that surface functionalization was vital for mimosine to interact with the aggregation prone domain to suppress Aβ fibril assembly and its toxic potency. This is supported by the recent studies involving amino acids, natural products and certain peptides which exhibit enhanced inhibitory efficacy on Aβ fibrillation due to structural limitations following surface functionalization [[56], [57], [58], [59]].
Previous studies have shown that, mimosine can have protective effects by chelating metals such as iron as well as attenuating oxidative stress, which plays a critical role in a variety of neurodegenerative disorders including AD [[14], [15], [16]]. Nevertheless, it remains unknown if mimosine can affect Aβ fibril formation or toxicity. Recently, mimosine has been shown to interact with bovine serum albumin and disrupt its conformation by reducing the α-helix component and promoting an unfolding of the protein. Additionally, it can bind DNA by electrostatic attractions via phosphate groups and grooves [60]. The binding affinity of a molecule for the native or aggregate-prone intermediate structures of peptides plays a crucial role in inhibiting protein aggregation [18,28,46]. Mimo-AuNPs appears to be effective in targeting the amyloidogenic protein because of its aromatic moiety which may have a role in initiating interactions with the hydrophobic domain of Aβ1-42. The mode of inhibition was achieved by altering the aggregation kinetics notably by delaying the lag time as a function of Mimo-AuNPs concentrations. This could be due to the interaction of Mimo-AuNPs with Aβ monomers precluding monomer-monomer hydrogen bonding and noncovalent interactions leading to a delay in aggregation kinetics. This may lead to an induction of an energy barrier by the nanoparticles to the aggregating Aβ peptides preventing them to form fibrillar nuclei or spheroidal oligomers [58,61]. Our ThT kinetic assay showed that Mimo-AuNPs, but not mimosine or AuNPs alone, can inhibit fibrillization of both normal and mutant human Aβ peptides. This is confirmed by fluorescence imaging and TEM showing the generation of shorter and fragmented fibrils as well as DLS analyses in which large fibrils dominating the scattered lights are reduced to smaller species with a hydrodynamic radius of ~2–10 nm. It is well known that the conformational changes that occur during Aβ aggregation from a random coil to the beta-sheeted structure are one of the most important phenomena during fibril formation. Interestingly, the CD and FTIR data suggest that Mimo-AuNPs allow Aβ peptides to remain in the monomeric state as demonstrated by the respective signals for α-helical structure. It is possible that the chirality of Mimo-AuNPs can trigger binding with the Aβ peptide resulting in the formation of a stable α-helical structure through π-π and hydrophobic interactions. This interaction will consequently prevent Aβ to adopt a well-organized cross beta structure by inducing viable contacts between the residues that would form parallel β-sheets in Aβ and the respective side chains of the mimosine.
It is well known that amino acid residues Leu17 to Ala21 in Aβ1-42 are essential for the β-sheet formation and stabilization [22,61]. For the molecular docking studies, we selected the solution structure of the Alzheimer's disease Aβ1-42 PDB ID:1IYT because this structure exhibits the helical conformation known to promote membrane rupture/cell death and thus we believed this could be one of the best model systems for screening the molecular interaction between Mimo-AuNPs and cytotoxic Aβ1-42. Since mimosine is hydrophobic, it is found to interact directly with the Lys16 to Ala21 region of the Aβ1-42 and prevent it to aggregate further. Reports also suggest that the His14, Gln15, Phe20, Ala21, Ala30, Ile31, Met35 and Val36 are the unique residues in Aβ1-42 that are responsible for oligomerization and fibril formation. It is therefore possible that interaction with these residual domains may prevent the Aβ fibril formation by stabilizing its native structure. Supporting the notion, the docking data showed a strong interaction of Mimo-AuNPs with Lys16, Leu17, Val18, Phe20, and Ala21 of Aβ1-42 which enables it to stabilize the N-terminal region of the peptide. This may allow Mimo-AuNPs to induce structural changes in Aβ oligomers resulting in the inhibition of Aβ-Aβ interactions leading to the formation of irregular aggregates. It has also been shown that monomeric units in Aβ1-42 fibrils have intermolecular side-chain contacts between Phe19 and Gly38 as well as between Met35 and Ala42 which may have a role in stabilizing the structural conformer of the peptide [62]. Our Molecular interaction snapshot (Fig. 4) revealed that Mimo-AuNPs can interact with Aβ monomers which may prevent the Aβ-Aβ monomer interactions. This effect can interfere with the nucleation phase of the Aβ aggregation kinetics as apparent by decreased levels of ThT fluorescence. Interestingly Mimo-AuNPs were able to interact with the key amino acid residues present in the fibrillar structure of the Aβ1-42 structure and were able to induce disassembly. It is well known that the C terminal domain involves in the formation of the protofilament structures due to its high affinity to the solvents [63,64]. Such interface may lead to the formation of a hydrophobic core between the residues such as Ile41 and Val39 with the adjacent fibril. The N terminal domain of a fibril close to the C terminal domain of another fibril may induce the formation of salt bridge by intermolecular interaction between Asp1 and Lys28. This complimentary interaction stabilizes the interface between the two protofilaments. A vital information gained from the molecular docking analysis revealed that Mimo-AuNPs were able to interfere with the salt bridges and with the residues present in the steric zipper domain. This viable interaction may in turn promote disassembly of the preformed fibrillar structure. It is likely that Mimo-AuNPs once interact with Asp1 (as shown in Fig. S9) can hinder the preformed salt bridge between two fibrillar units leading to the formation of disrupted fibrils. Similar phenomenon has been reported using a variety of phytochemicals, peptides and polymers including curcumin, clioquinol, epigallocatechin gallate, polyproline and myricetin [[65], [66], [67], [68], [69], [70]]. Intriguingly, Mimo-AuNPs are also found to suppress aggregation of tau protein via interactions with hydrogen and hydrophobic bonds indicating their potential to influence tau pathology in AD.
Evidence suggests that the hydrophobicity of the Aβ peptide increases once it undergoes aggregation forming oligomers and fibrils [8,71]. A decreased fluorescence signal at ~625 nm in the Nile red assay for Aβ samples in the presence of Mimo-AuNPs indicates its potential interaction with the hydrophobic domain of Aβ1-42 to inhibit ongoing fibril formation. Additionally, the ANS fluorescence intensity decreased with Mimo-AuNPs suggesting the presence of less solvent-exposed hydrophobic groups. These results also indicate that Mimo-AuNPs possibly bind to critical hydrophobic residues, thus stabilizing the native state of Aβ1-42. This is supported by the filter-trap assay, which showed that presence of Mimo-AuNPs decreased the interaction of Aβ1-42 with its antibodies recognizing Gln15 and Gly29 region of the peptide. Apart from suppressing Aβ aggregation, Mimo-AuNPs were found to protect cultured neurons against Aβ-induced toxicity. This is likely due to the retention of Aβ monomers or sequestration of Aβ peptides into amorphous aggregates, which are less toxic than Aβ oligomers [55,62,[71], [72], [73]]. The protective response is mediated by decreased phosphorylation of the tau protein and its associated signaling pathway. Although the significance of reduced ROS production in Mimo-AuNPs treated cultured neurons remains unclear, it may have a role in the restitution of lysosomal integrity which is known to be severely compromised following Aβ treatment due to enhanced intra-lysosomal ROS production [[74], [75], [76]]. Since neurons can be protected against Aβ toxicity by inhibiting the formation of soluble oligomer and/or higher-order aggregates, many studies have pursued small organic molecules, peptides and functionalized nanoparticles as a treatment strategy for AD to prevent Aβ aggregation/toxicity [12,13]. One of the limiting factors, however, is the BBB preventing the entry of most drugs/agents into the brain [72,77]. Over the last decade, various surface-functionalized AuNPs of 10–50 nm size have been used for the targeted delivery of drugs/molecules that can interfere with Aβ aggregation/toxicity because of their ability i) to conjugate readily with a wide spectrum of biomolecules including amino acids, proteins/enzymes and drugs/molecules via thiol, amines or disulfide functional groups without altering their biological activity and ii) to overcome the protective BBB following systemic administration [47]. Earlier studies suggest that the ability of the functionalized AuNPs to penetrate into cells depends partly on the size, surface charge and surface chemical composition. While permeability across the BBB may correlate inversely with the size, negatively charged AuNPs are known to exhibit better penetrability than positively charged nanoparticles. The cellular uptake and the BBB permeability of functionalized AuNPs, depending on their characteristic features, are known to be mediated via carrier-mediated transport, receptor-mediated transport or absorptive-mediated transport mechanisms [78,79]. Consistent with this notion, our in vitro BBB assay show that functionalization of mimosine onto AuNPs, in contrast to mimosine alone, enhances their permeability across BBB, thus enabling them to overcome one of the major impediments associated with most small molecule drug candidates. At present, the precise mechanism by which Mimo-AuNPs are taken up by cells or cross the BBB remains unclear. Nevertheless, the ability of Mimo-AuNPs to cross BBB and inhibit/destabilize Aβ as well as tau fibril formation and to protect neurons against Aβ-induced toxicity, bolster their potential as a promising therapeutic strategy for the treatment of AD. Additionally, mimosine due to its anti-inflammatory, metal chelating and anti-oxidative properties may have added advantages over other molecules to influence AD pathology.
5. Conclusion
In the current study we successfully functionalized plant-based amino acid mimosine onto AuNPs (i.e., Mimo-AuNPs) and revealed their ability to cross the BBB under in vitro paradigm. These Mimo-AuNPs can suppress not only spontaneous and seed-induced Aβ aggregation but also can induce the disassembly of pre-aggregated Aβ fibers. It appears that Mimo-AuNPs by interacting with the hydrophobic domain of Aβ1-42 (i.e., Lys16 to Ala21) can enable the peptide to remain in its monomeric state thereby preventing a conformational shift towards the β-sheet structure. Additionally, Mimo-AuNPs can able to protect mouse cortical neurons against Aβ-induced toxicity by attenuating intracellular signaling mechanism. These results, in keeping with the established beneficial effects of mimosine in various medicinal fields, reveal a unique therapeutic potential of Mimo-AuNPs in the treatment of AD pathology.
Data availability
The data in this work are available in the manuscript or Supplementary Information, or available from the corresponding author upon reasonable request.
CRediT authorship contribution statement
Bibin G. Anand: did the experiments and analyzed the data included in the manuscript. Qi Wu: did the experiments and analyzed the data included in the manuscript. Govindarajan Karthivashan: did the experiments and analyzed the data included in the manuscript. Kiran P. Shejale: did the experiments and analyzed the data included in the manuscript. Sara Amidian: did the experiments and analyzed the data included in the manuscript. Satyabrata Kar: helped in analyzing the data and wrote the manuscript with help/input from Bibin G. Anand, Qi Wu, Govindarajan Karthivashan, Sara Amidian and Holger Wille.
Declaration of competing interest
The authors declare no conflict of interest or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by grants from APRI-ASANT and CIHR (MOP-84480) to SK and from APRI (APRI 201600028) to HW. SynAD, University of Alberta provided a part of the postdoctoral fellowships for BGA, QW and KG.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.04.029.
Contributor Information
Bibin G. Anand, Email: banand@ualberta.ca.
Qi Wu, Email: qw6@ualberta.ca.
Govindarajan Karthivashan, Email: karthiva@ualberta.ca.
Kiran P. Shejale, Email: shejalekp@gmail.com.
Sara Amidian, Email: samidian@ualberta.ca.
Holger Wille, Email: wille@ualberta.ca.
Satyabrata Kar, Email: skar@ualberta.ca, skar@ualberta.ca.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Selkoe D.J., Hardy J. The amyloid hypothesis of Alzheimer's disease at 25 years. EMBO Mol. Med. 2016;8(6):595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen X.Q., Mobley W.C. Alzheimer disease pathogenesis: insights from molecular and cellular biology studies of oligomeric abeta and tau species. Front. Neurosci. 2019;13:659. doi: 10.3389/fnins.2019.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perl D.P. Neuropathology of Alzheimer's disease. Mt. Sinai J. Med. 2010;77(1):32–42. doi: 10.1002/msj.20157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrew R.J., Kellett K.A., Thinakaran G., Hooper N.M. A Greek tragedy: the growing complexity of alzheimer amyloid precursor protein proteolysis. J. Biol. Chem. 2016;291(37):19235–19244. doi: 10.1074/jbc.R116.746032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Straub J.E., Thirumalai D. Toward a molecular theory of early and late events in monomer to amyloid fibril formation. Annu. Rev. Phys. Chem. 2011;62:437–463. doi: 10.1146/annurev-physchem-032210-103526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bisceglia F., Natalello A., Serafini M.M., Colombo R., Verga L., Lanni C., De Lorenzi E. An integrated strategy to correlate aggregation state, structure and toxicity of Ass 1-42 oligomers. Talanta. 2018;188:17–26. doi: 10.1016/j.talanta.2018.05.062. [DOI] [PubMed] [Google Scholar]
- 7.Chen G.F., Xu T.H., Yan Y., Zhou Y.R., Jiang Y., Melcher K., Xu H.E. Amyloid beta: structure, biology and structure-based therapeutic development. Acta Pharmacol. Sin. 2017;38(9):1205–1235. doi: 10.1038/aps.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S.J., Nam E., Lee H.J., Savelieff M.G., Lim M.H. Towards an understanding of amyloid-beta oligomers: characterization, toxicity mechanisms, and inhibitors. Chem. Soc. Rev. 2017;46(2):310–323. doi: 10.1039/c6cs00731g. [DOI] [PubMed] [Google Scholar]
- 9.Kumar S., Walter J. Phosphorylation of amyloid beta (Abeta) peptides - a trigger for formation of toxic aggregates in Alzheimer's disease. Aging (Albany NY) 2011;3(8):803–812. doi: 10.18632/aging.100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarrett J.T., Lansbury P.T., Jr. Seeding "one-dimensional crystallization" of amyloid: a pathogenic mechanism in Alzheimer's disease and scrapie? Cell. 1993;73(6):1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 11.Hajipour M.J., Santoso M.R., Rezaee F., Aghaverdi H., Mahmoudi M., Perry G. Advances in Alzheimer's diagnosis and therapy: the implications of nanotechnology. Trends Biotechnol. 2017;35(10):937–953. doi: 10.1016/j.tibtech.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Cao J., Hou J., Ping J., Cai D. Advances in developing novel therapeutic strategies for Alzheimer's disease. Mol. Neurodegener. 2018;13(1):64. doi: 10.1186/s13024-018-0299-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tosi G., Pederzoli F., Belletti D., Vandelli M.A., Forni F., Duskey J.T., Ruozi B. Nanomedicine in Alzheimer's disease: amyloid beta targeting strategy. Prog. Brain Res. 2019;245:57–88. doi: 10.1016/bs.pbr.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen B.C., Tawata S. The chemistry and biological activities of mimosine: a review. Phytother Res. 2016;30(8):1230–1242. doi: 10.1002/ptr.5636. [DOI] [PubMed] [Google Scholar]
- 15.Singh B., Bhat T.K., Singh B. Potential therapeutic applications of some antinutritional plant secondary metabolites. J. Agric. Food Chem. 2003;51(19):5579–5597. doi: 10.1021/jf021150r. [DOI] [PubMed] [Google Scholar]
- 16.Hallak M., Vazana L., Shpilberg O., Levy I., Mazar J., Nathan I. A molecular mechanism for mimosine-induced apoptosis involving oxidative stress and mitochondrial activation. Apoptosis. 2008;13(1):147–155. doi: 10.1007/s10495-007-0156-7. [DOI] [PubMed] [Google Scholar]
- 17.Park D.S., Morris E.J., Greene L.A., Geller H.M. G1/S cell cycle blockers and inhibitors of cyclin-dependent kinases suppress camptothecin-induced neuronal apoptosis. J. Neurosci. 1997;17(4):1256–1270. doi: 10.1523/JNEUROSCI.17-04-01256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaman K., Ryu H., Hall D., O'Donovan K., Lin K.I., Miller M.P., Marquis J.C., Baraban J.M., Semenza G.L., Ratan R.R. Protection from oxidative stress-induced apoptosis in cortical neuronal cultures by iron chelators is associated with enhanced DNA binding of hypoxia-inducible factor-1 and ATF-1/CREB and increased expression of glycolytic enzymes, p21(waf1/cip1), and erythropoietin. J. Neurosci. 1999;19(22):9821–9830. doi: 10.1523/JNEUROSCI.19-22-09821.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anand B.G., Dubey K., Shekhawat D.S., Kar K. Capsaicin-coated silver nanoparticles inhibit amyloid fibril formation of serum albumin. Biochemistry. 2016;55(24):3345–3348. doi: 10.1021/acs.biochem.6b00418. [DOI] [PubMed] [Google Scholar]
- 20.Anand B.G., Shekhawat D.S., Dubey K., Kar K. Uniform, polycrystalline, and thermostable piperine-coated gold nanoparticles to target insulin fibril assembly. ACS Biomater. Sci. Eng. 2017;3(6):1136–1145. doi: 10.1021/acsbiomaterials.7b00030. [DOI] [PubMed] [Google Scholar]
- 21.Fandrich M., Fletcher M.A., Dobson C.M. Amyloid fibrils from muscle myoglobin. Nature. 2001;410(6825):165–166. doi: 10.1038/35065514. [DOI] [PubMed] [Google Scholar]
- 22.Chiti F., Dobson C.M. Amyloid formation by globular proteins under native conditions. Nat. Chem. Biol. 2009;5(1):15–22. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- 23.Anand B.G., Prajapati K.P., Dubey K., Ahamad N., Shekhawat D.S., Rath P.C., Joseph G.K., Kar K. Self-assembly of artificial sweetener aspartame yields amyloid-like cytotoxic nanostructures. ACS Nano. 2019;13(5):6033–6049. doi: 10.1021/acsnano.9b02284. [DOI] [PubMed] [Google Scholar]
- 24.Anand B.G., Prajapati K.P., Shekhawat D.S., Kar K. Tyrosine-generated nanostructures initiate amyloid cross-seeding in proteins leading to a lethal aggregation trap. Biochemistry. 2018;57(35):5202–5209. doi: 10.1021/acs.biochem.8b00472. [DOI] [PubMed] [Google Scholar]
- 25.Skaat H., Chen R., Grinberg I., Margel S. Engineered polymer nanoparticles containing hydrophobic dipeptide for inhibition of amyloid-beta fibrillation. Biomacromolecules. 2012;13(9):2662–2670. doi: 10.1021/bm3011177. [DOI] [PubMed] [Google Scholar]
- 26.Gao G., Zhang M., Gong D., Chen R., Hu X., Sun T. The size-effect of gold nanoparticles and nanoclusters in the inhibition of amyloid-beta fibrillation. Nanoscale. 2017;9(12):4107–4113. doi: 10.1039/c7nr00699c. [DOI] [PubMed] [Google Scholar]
- 27.Gao H. Progress and perspectives on targeting nanoparticles for brain drug delivery. Acta Pharm. Sin. B. 2016;6(4):268–286. doi: 10.1016/j.apsb.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao N., Sun H., Dong K., Ren J., Qu X. Gold-nanoparticle-based multifunctional amyloid-beta inhibitor against Alzheimer's disease. Chemistry. 2015;21(2):829–835. doi: 10.1002/chem.201404562. [DOI] [PubMed] [Google Scholar]
- 29.Xiong N., Zhao Y., Dong X., Zheng J., Sun Y. Design of a molecular hybrid of dual peptide inhibitors coupled on AuNPs for enhanced inhibition of amyloid beta-protein aggregation and cytotoxicity. Small. 2017;13(13) doi: 10.1002/smll.201601666. [DOI] [PubMed] [Google Scholar]
- 30.Anand B.G., Dubey K., Shekhawat D.S., Prajapati K.P., Kar K. Strategically designed antifibrotic gold nanoparticles to prevent collagen fibril formation. Langmuir. 2017;33(46):13252–13261. doi: 10.1021/acs.langmuir.7b01504. [DOI] [PubMed] [Google Scholar]
- 31.Wnuk A., Rzemieniec J., Staron J., Litwa E., Lason W., Bojarski A., Kajta M. Prenatal exposure to benzophenone-3 impairs autophagy, disrupts RXRs/PPARgamma signaling, and alters epigenetic and post-translational statuses in brain neurons. Mol. Neurobiol. 2019;56(7):4820–4837. doi: 10.1007/s12035-018-1401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foroutanpay B.V., Kumar J., Kang S.G., Danaei N., Westaway D., Sim V.L., Kar S. The effects of N-terminal mutations on beta-amyloid peptide aggregation and toxicity. Neuroscience. 2018;379:177–188. doi: 10.1016/j.neuroscience.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 33.Kodali R., Williams A.D., Chemuru S., Wetzel R. Abeta(1-40) forms five distinct amyloid structures whose beta-sheet contents and fibril stabilities are correlated. J. Mol. Biol. 2010;401(3):503–517. doi: 10.1016/j.jmb.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prajapati K.P., Singh A.P., Dubey K., Ansari M., Temgire M., Anand B.G., Kar K. Myricetin inhibits amyloid fibril formation of globular proteins by stabilizing the native structures. Colloids Surf. B Biointerfaces. 2019;186:110640. doi: 10.1016/j.colsurfb.2019.110640. [DOI] [PubMed] [Google Scholar]
- 35.Sze A., Erickson D., Ren L., Li D. Zeta-potential measurement using the Smoluchowski equation and the slope of the current-time relationship in electroosmotic flow. J. Colloid Interface Sci. 2003;261(2):402–410. doi: 10.1016/S0021-9797(03)00142-5. [DOI] [PubMed] [Google Scholar]
- 36.Anand B.G., Dubey K., Shekhawat D.S., Kar K. Intrinsic property of phenylalanine to trigger protein aggregation and hemolysis has a direct relevance to phenylketonuria. Sci. Rep. 2017;7(1):11146. doi: 10.1038/s41598-017-10911-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hawe A., Sutter M., Jiskoot W. Extrinsic fluorescent dyes as tools for protein characterization. Pharm Res. 2008;25(7):1487–1499. doi: 10.1007/s11095-007-9516-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X., Zhang X., Ladiwala A.R., Du D., Yadav J.K., Tessier P.M., Wright P.E., Kelly J.W., Buxbaum J.N. Mechanisms of transthyretin inhibition of beta-amyloid aggregation in vitro. J. Neurosci. 2013;33(50):19423–19433. doi: 10.1523/JNEUROSCI.2561-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dallakyan S., Olson A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- 40.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dubey K., Anand B.G., Temgire M.K., Kar K. Evidence of rapid coaggregation of globular proteins during amyloid formation. Biochemistry. 2014;53(51):8001–8004. doi: 10.1021/bi501333q. [DOI] [PubMed] [Google Scholar]
- 42.Amritraj A., Wang Y., Revett T.J., Vergote D., Westaway D., Kar S. Role of cathepsin D in U18666A-induced neuronal cell death: potential implication in Niemann-Pick type C disease pathogenesis. J. Biol. Chem. 2013;288(5):3136–3152. doi: 10.1074/jbc.M112.412460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu Q., Qiao L., Yang J., Zhou Y., Liu Q. Stronger activation of SREBP-1a by nucleus-localized HBx. Biochem. Biophys. Res. Commun. 2015;460(3):561–565. doi: 10.1016/j.bbrc.2015.03.069. [DOI] [PubMed] [Google Scholar]
- 44.Wu Q., Li Z., Mellor P., Zhou Y., Anderson D.H., Liu Q. The role of PTEN - HCV core interaction in hepatitis C virus replication. Sci. Rep. 2017;7(1):3695. doi: 10.1038/s41598-017-03052-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu T., Liu Y., Ye Y., Li J., Yang F., Zhao H., Wang L. Corrosion protective properties of epoxy coating containing tetraaniline modified nano-α-Fe2O3. Prog. Org. Coating. 2019;132:455–467. doi: 10.1016/j.porgcoat.2019.04.010. [DOI] [Google Scholar]
- 46.Singh V., Snigdha K., Singh C., Sinha N., Thakur A.K. Understanding the self-assembly of Fmoc-phenylalanine to hydrogel formation. Soft Matter. 2015;11(26):5353–5364. doi: 10.1039/c5sm00843c. [DOI] [PubMed] [Google Scholar]
- 47.Xiong N., Dong X.Y., Zheng J., Liu F.F., Sun Y. Design of LVFFARK and LVFFARK-functionalized nanoparticles for inhibiting amyloid beta-protein fibrillation and cytotoxicity. ACS Appl. Mater. Interfaces. 2015;7(10):5650–5662. doi: 10.1021/acsami.5b00915. [DOI] [PubMed] [Google Scholar]
- 48.Biancalana M., Koide S. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim. Biophys Acta 1804. 2010;(7):1405–1412. doi: 10.1016/j.bbapap.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haass C., Selkoe D.J. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat. Rev. Mol. Cell Biol. 2007;8(2):101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 50.Battisti A., Piccionello A.P., Sgarbossa A., Vilasi S., Ricci C., Ghetti F., Spinozzi F., Gammazza A.M., Giacalone V., Martorana A., Lauria A., Ferrero C., Bulone D., Mangione M.R., Biagio P.L.S., Ortore M.G. Curcumin-like compounds designed to modify amyloid beta peptide aggregation patterns. RSC Adv. 2017;7(50):31714–31724. doi: 10.1039/c7ra05300b. [DOI] [Google Scholar]
- 51.Van Nostrand W.E., Melchor J.P., Cho H.S., Greenberg S.M., Rebeck G.W. Pathogenic effects of D23N Iowa mutant amyloid beta -protein. J. Biol. Chem. 2001;276(35):32860–32866. doi: 10.1074/jbc.M104135200. [DOI] [PubMed] [Google Scholar]
- 52.Trott O., Olson A.J., Vina AutoDock. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sowade R.F., Jahn T.R. Seed-induced acceleration of amyloid-beta mediated neurotoxicity in vivo. Nat. Commun. 2017;8(1):512. doi: 10.1038/s41467-017-00579-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Linse S. Monomer-dependent secondary nucleation in amyloid formation. Biophys. Rev. 2017;9(4):329–338. doi: 10.1007/s12551-017-0289-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reiss A.B., Arain H.A., Stecker M.M., Siegart N.M., Kasselman L.J. Amyloid toxicity in Alzheimer's disease. Rev. Neurosci. 2018;29(6):613–627. doi: 10.1515/revneuro-2017-0063. [DOI] [PubMed] [Google Scholar]
- 56.Shiraki K., Kudou M., Fujiwara S., Imanaka T., Takagi M. Biophysical effect of amino acids on the prevention of protein aggregation. J. Biochem. 2002;132(4):591–595. doi: 10.1093/oxfordjournals.jbchem.a003261. [DOI] [PubMed] [Google Scholar]
- 57.Alvarez Y.D., Fauerbach J.A., Pellegrotti J.V., Jovin T.M., Jares-Erijman E.A., Stefani F.D. Influence of gold nanoparticles on the kinetics of alpha-synuclein aggregation. Nano Lett. 2013;13(12):6156–6163. doi: 10.1021/nl403490e. [DOI] [PubMed] [Google Scholar]
- 58.Rajasekhar K., Suresh S.N., Manjithaya R., Govindaraju T. Rationally designed peptidomimetic modulators of abeta toxicity in Alzheimer's disease. Sci. Rep. 2015;5:8139. doi: 10.1038/srep08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stefani M., Rigacci S. Protein folding and aggregation into amyloid: the interference by natural phenolic compounds. Int. J. Mol. Sci. 2013;14(6):12411–12457. doi: 10.3390/ijms140612411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baltazar C.J., Mun R., Tajmir-Riahi H.A., Bariyanga J. Spectroscopic studies on the interaction of mimosine with BSA and DNA. J. Mol. Struct. 2018;1161:273–278. doi: 10.1016/j.molstruc.2018.01.039. [DOI] [Google Scholar]
- 61.Fezoui Y., Teplow D.B. Kinetic studies of amyloid beta-protein fibril assembly. Differential effects of alpha-helix stabilization. J. Biol. Chem. 2002;277(40):36948–36954. doi: 10.1074/jbc.M204168200. [DOI] [PubMed] [Google Scholar]
- 62.Ahmed M., Davis J., Aucoin D., Sato T., Ahuja S., Aimoto S., Elliott J.I., Van Nostrand W.E., Smith S.O. Structural conversion of neurotoxic amyloid-beta(1-42) oligomers to fibrils. Nat. Struct. Mol. Biol. 2010;17(5):561–567. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hayden E.Y., Hoi K.K., Lopez J., Inayathullah M., Condron M.M., Teplow D.B. Identification of key regions and residues controlling Abeta folding and assembly. Sci. Rep. 2017;7(1):12434. doi: 10.1038/s41598-017-10845-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gremer L., Scholzel D., Schenk C., Reinartz E., Labahn J., Ravelli R.B.G., Tusche M., Lopez-Iglesias C., Hoyer W., Heise H., Willbold D., Schroder G.F. Fibril structure of amyloid-beta(1-42) by cryo-electron microscopy. Science. 2017;358(6359):116–119. doi: 10.1126/science.aao2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kanchi P.K., Dasmahapatra A.K. Polyproline chains destabilize the Alzheimer's amyloid-beta protofibrils: a molecular dynamics simulation study. J. Mol. Graph. Model. 2019;93:107456. doi: 10.1016/j.jmgm.2019.107456. [DOI] [PubMed] [Google Scholar]
- 66.Zhang L., Yagnik G., Peng Y., Wang J., Xu H.H., Hao Y., Liu Y.N., Zhou F. Kinetic studies of inhibition of the amyloid beta (1-42) aggregation using a ferrocene-tagged beta-sheet breaker peptide. Anal. Biochem. 2013;434(2):292–299. doi: 10.1016/j.ab.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Andarzi Gargari S., Barzegar A., Tarinejad A. The role of phenolic OH groups of flavonoid compounds with H-bond formation ability to suppress amyloid mature fibrils by destabilizing beta-sheet conformation of monomeric Abeta17-42. PloS One. 2018;13(6) doi: 10.1371/journal.pone.0199541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andrade S., Ramalho M.J., Loureiro J.A., Pereira M.D.C. Natural compounds for Alzheimer's disease therapy: a systematic review of preclinical and clinical studies. Int. J. Mol. Sci. 2019;20(9) doi: 10.3390/ijms20092313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stefanescu R., Stanciu G.D., Luca A., Paduraru L., Tamba B.I. Secondary metabolites from plants possessing inhibitory properties against beta-amyloid aggregation as revealed by thioflavin-T assay and correlations with investigations on transgenic mouse models of Alzheimer's disease. Biomolecules. 2020;10(6) doi: 10.3390/biom10060870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thapa A., Jett S.D., Chi E.Y. Curcumin attenuates amyloid-beta aggregate toxicity and modulates amyloid-beta aggregation pathway. ACS Chem. Neurosci. 2016;7(1):56–68. doi: 10.1021/acschemneuro.5b00214. [DOI] [PubMed] [Google Scholar]
- 71.Lee J., Culyba E.K., Powers E.T., Kelly J.W. Amyloid-beta forms fibrils by nucleated conformational conversion of oligomers. Nat. Chem. Biol. 2011;7(9):602–609. doi: 10.1038/nchembio.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fazil M., Shadab, Baboota S., Sahni J.K., Ali J. Nanotherapeutics for Alzheimer's disease (AD): past, present and future. J. Drug Target. 2012;20(2):97–113. doi: 10.3109/1061186X.2011.607499. [DOI] [PubMed] [Google Scholar]
- 73.Sonzini S., Stanyon H.F., Scherman O.A. Decreasing amyloid toxicity through an increased rate of aggregation. Phys. Chem. Chem. Phys. 2017;19(2):1458–1465. doi: 10.1039/c6cp06765d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Amritraj A., Peake K., Kodam A., Salio C., Merighi A., Vance J.E., Kar S. Increased activity and altered subcellular distribution of lysosomal enzymes determine neuronal vulnerability in Niemann-Pick type C1-deficient mice. Am. J. Pathol. 2009;175(6):2540–2556. doi: 10.2353/ajpath.2009.081096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Umeda T., Tomiyama T., Sakama N., Tanaka S., Lambert M.P., Klein W.L., Mori H. Intraneuronal amyloid beta oligomers cause cell death via endoplasmic reticulum stress, endosomal/lysosomal leakage, and mitochondrial dysfunction in vivo. J. Neurosci. Res. 2011;89(7):1031–1042. doi: 10.1002/jnr.22640. [DOI] [PubMed] [Google Scholar]
- 76.Ditaranto K., Tekirian T.L., Yang A.J. Lysosomal membrane damage in soluble Abeta-mediated cell death in Alzheimer's disease. Neurobiol. Dis. 2001;8(1):19–31. doi: 10.1006/nbdi.2000.0364. [DOI] [PubMed] [Google Scholar]
- 77.Karthivashan G., Ganesan P., Park S.Y., Lee H.W., Choi D.K. Lipid-based nanodelivery approaches for dopamine-replacement therapies in Parkinson's disease: from preclinical to translational studies. Biomaterials. 2020;232 doi: 10.1016/j.biomaterials.2019.119704. 119704. [DOI] [PubMed] [Google Scholar]
- 78.Lombardo S.M., Schneider M., Tureli A.E., Gunday Tureli N. Key for crossing the BBB with nanoparticles: the rational design. Beilstein J. Nanotechnol. 2020;11:866–883. doi: 10.3762/bjnano.11.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thangudu S., Cheng F.Y., Su C.H. Advancements in the blood-brain barrier penetrating nanoplatforms for brain related disease diagnostics and therapeutic applications. Polymers. 2020;12(12) doi: 10.3390/polym12123055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data in this work are available in the manuscript or Supplementary Information, or available from the corresponding author upon reasonable request.