Abstract
Radiation biodosimetry based on transcriptomic analysis of peripheral blood is a valuable tool to detect radiation exposure after a radiological/nuclear event and obtain useful biological information that could predict tissue and organismal injury. However, confounding factors, including chronic inflammation or immune suppression, can potentially obscure the predictive power of the method. Members of the p38 mitogen-activated protein kinase (MAPK) family respond to pro-inflammatory signals and environmental stresses, whereas genetic ablation of the p38 signaling pathway in mice leads to reduced susceptibility to collagen-induced arthritis and experimental autoimmune encephalomyelitis that model human rheumatoid arthritis and multiple sclerosis, respectively. p38 is normally regulated by the MAP3K-MAP2K pathway in mammalian cells. However, in T cells there is an alternative pathway for p38 activation that plays an important role in antigen-receptor-activated T cells and participates in immune and inflammatory responses. To examine the role of p38 in response to radiation, we used two mouse models expressing either a p38α dominant negative (DN) mutation that globally suppresses p38 signaling or a p38αβ double-knock-in (DKI) mutant, which inhibits specifically T-cell receptor activation. We exposed p38 wild-type (p38WT) and mutant male mice to 7 Gy X rays and 24 h later whole blood was isolated subjected to whole-genome microarray and gene ontology analysis. Irradiation of p38WT mice led to a significant overrepresentation of pathways associated with morbidity and mortality, as well as organismal cell death. In contrast, these pathways were significantly underrepresented in p38DN and p38DKI mutant mice, suggesting that p38 attenuation may protect blood cells from the deleterious effects of radiation. Furthermore, radiation exposure in p38 mutant mice resulted in an enrichment of phagocytosis-related pathways, suggesting a role for p38 signaling in restricting phagocytosis of apoptotic cells after irradiation. Finally, despite the significant changes in gene expression, it was still feasible to identify a panel of genes that could accurately distinguish between irradiated and control mice, irrespective of p38 status.
INTRODUCTION
Radiation exposure due to the detonation of an improvised nuclear device or nuclear accident constitutes a major threat to public health. In such a case, large numbers of people will be exposed to different doses of radiation that will need a rapid and reliable assessment of radiation exposure. The purpose of radiation biodosimetry is to accurately predict radiation dose, as a surrogate of radiological injury. Large-scale transcriptomic analysis using whole blood has been used to select genes that may detect radiation exposure. Furthermore, gene ontology analysis of the differentially expressed genes has helped to identify critical biological functions that could be used to rationally design new mitigators against the deleterious effects of radiation (1,2). However, gene expression is affected by a number of pre-existing conditions, including inflammation, which has the potential to obscure detection of radiation exposure. The p38 pathway plays a central role in the cellular response to stress and contributes to inflammatory responses (3).
The p38 mitogen-activated protein kinase (MAPK) family comprises four members (α, β, γ and δ), which share a 12-amino acid activation loop containing a Thr-Gly-Tyr motif located at amino acid position 180 and 182. The p38α and p38β isoforms are ubiquitously expressed, whereas p38γ is largely restricted to skeletal muscle and p38δ is mainly found in the testes, pancreas, small intestine (4), and detected in CD4+ T cells, macrophages, monocytes, and neutrophils (5). In mammalian cells, p38 kinases are activated by environmental stresses, including osmotic shock, ultraviolet and ionizing radiation exposure, and pro-inflammatory cytokines, such as TNFα and IL-1β, and regulates a multitude of substrates, including other protein kinases, transcriptional factors, transcriptional regulators, cell cycle proteins, and cell death regulators (3). In response to stress, p38 participate in a cascade that involves the initiating MAPK kinase kinases MEKK4, TGF-β activated kinase 1 (TAK1), and apoptosis signal-regulating kinase 1 (ASK1), which activate the MAPK kinases MKK3, MKK4, and MKK6 that in turn phosphorylate p38 at the conserved Thr180-X-Tyr182 motif (6). Although p38 activation has generally pro-survival functions, stress-induced p38 activation can sometimes induce cell death, which tends to correlate with higher/sustained levels of pathway activity (3). In the immune system, the p38 signaling cascade has been implicated in the regulation of both innate immunity and adaptive immunity by controlling T-cell activation and differentiation (7), and regulates the production of pro-inflammatory cytokines, such as IL-1β, IFNγ and TNFα, in different immune cell types However, p38 can be activated downstream of several pro-inflammatory cytokines, such as IL-1β and TNFα, as well (8–13).
Besides the “canonical” MKK-dependent phosphorylation cascade, p38 can be activated by an alternative pathway in T lymphocytes (4). Stimulation of the T-cell receptor results in the phosphorylation of p38 at Tyr323 by the non-receptor protein tyrosine kinase ZAP-70 [zeta-chain (TCR) associated protein kinase-70], which leads to autophosphorylation of both p38α and p38β, preferentially at Thr180 (14). Although monophosphorylated p38 shares most substrates with dual-phospho p38 (e.g., STAT4, MK2, and MEF2A), it does not phosphorylate them as efficiently as the dual-phospho form (15). However, ZAP-70 is an exception, being preferentially phosphorylated on Thr293 by monophosphorylated p38 (16).
TCR signaling in the presence of CD28-mediated costimulation can activate p38 in nonstress conditions (17) through induction of the alternative p38 activation pathway (18) and it is required for pro-inflammatory cytokine production (18–21). Genetic ablation of the p38 signaling pathway by constructing a p38α and p38β Y323F double knock-in (p38DKI) transgenic mouse (22, 23) leads to reduction in T-cell proliferation, diminished pro-inflammatory cytokine production, and reduced susceptibility to collagen-induced arthritis and experimental autoimmune encephalomyelitis (24), two widely used murine models for human rheumatoid arthritis and multiple sclerosis, respectively. However, it is believed that p38DKI mice have an intact stress-induced p38 activity (23).
In this study we have investigated the impact of p38 signaling on gene expression-based radiation biodosimetry of mouse peripheral blood exposed to radiation and identified canonical pathways and biological functions differentially affected, using two p38 transgenic mouse models. In the first model, the alternative p38 signaling pathway has been blocked using the p38DKI transgene, and, thus, T-cell receptor signaling is impaired (22, 23). In the second model, both the alternative and the canonical p38 signaling pathways are suppressed by generating a dominant negative p38α construct in which Thr180 and Tyr182 have been mutated to alanine and phenylalanine, respectively, resulting in the global attenuation of immune cell response to pro-inflammatory cytokines and cellular stress (24). Our results show that both pathways have a substantial impact on radiation-induced gene expression compared with p38 wild-type mice.
METHODS
Animals and Irradiation
Wild-type C57BL/6 male mice were obtained from Charles River Laboratories (Frederick, MD) and p38DKI (p38αβY323F) mice were kindly provided from the Laboratory of Immune Cell Biology, National Cancer Institute (P.I. Jonathan D. Ashwell, M.D.) (22, 23).
To generate the p38DN mutation, a mouse p38 (p38α or Mapk14) DNA sequence was modified via site-directed mutagenesis to harbor two amino acid substitutions at activating phosphorylation sites, threonine 180 to alanine (T180A) and tyrosine 182 to phenylalanine (Y182F) (24). Mice heterozygous for the p38DN allele were bred together or to wild-type siblings or to C57BL/6J inbred mice for many generations prior to sending to The Jackson Laboratory Repository. Upon arrival to Georgetown University, mice were bred to C57BL/6J inbred mice (Stock No. 000664) for at least one generation to establish the colony.
Animals were bred at Georgetown University, supplied with water and food ad libitum (12 h light-dark cycle conditions) according to Georgetown University Institutional Animal Care and Use Committee (GUACUC) protocols (2016–1152). Male mice that were 8–10 weeks old were exposed to a total-body X-ray dose (~1.67 Gy/min; X-Rad 320, Precision X-Ray Inc., Branford, CT; filter, 0.75 mm tin/0.25 mm copper/1.5 mm aluminum, 320 kV, 12.5 mA) of 0 or 7 Gy. Five (5) mice were used per experimental group.
Blood Collection and RNA Isolation
Blood was collected via cheek bleed from the submandibular vein at the time of euthanasia at 24 h postirradiation. Each sample (~0.2–0.3 ml blood) was added to a 15 ml centrifuge tube that contained 1.6 ml of PAXgene Blood RNA stabilization and lysis solution (PreAnalytix GmBH, catalog no. 762165), mixed thoroughly, incubated at room temperature for 2–3 h, and stored at −80°C. Blood samples were allowed to reach room temperature for 2 h before proceeding to RNA isolation. RNA was purified following the PAXgene RNA kit recommendations with on-column DNase I treatment. Globin RNA was reduced using the Ambion GLOBINclear-mouse/rat kit (Thermofisher). RNA yields were quantified using the NanoDrop ND1000 spectrophotometer (Thermofisher) and RNA quality was checked by the 2100 Bioanalyzer (Agilent). High-quality RNA with an RNA integrity number of at least 7.0 was used for microarray hybridization.
Microarray Hybridization and Data Analysis
Cyanine-3 labeled cRNA was prepared using the One-Color low-input quick amp labeling kit (Agilent). Dye incorporation and cRNA yield were measured with a NanoDrop ND1000 spectrophotometer (Thermofisher). Labeled cRNA was fragmented and hybridized to Agilent Mouse Gene Expression 4×44K v2 Microarray Kit (G4846A). Slides were scanned with the Agilent DNA microarray scanner (G2505B) and the images were analyzed with Feature Extraction software (Agilent) using default parameters for background correction and flagging non-uniform features.
Background-corrected hybridization intensities were imported into BRB-ArrayTools, version 4.5.0, log2-transformed and median normalized. Non-uniform outliers or features not significantly above background intensity in 25% or more of the hybridizations were filtered out. An additional filter requiring a minimum 1.5-fold change in at least 20% of the hybridizations was also applied. Furthermore, probes were averaged to one probe per gene and duplicate features were reduced by selecting the one with maximum signal intensity. The microarray data is available through the Gene Expression Omnibus with accession number GSE184361.
Class comparison was conducted in BRB-ArrayTools to identify genes differentially expressed (P < 0.001) between irradiated samples and time-matched unirradiated controls using a random variance t-test. Genes with P values < 0.001 were considered statistically significant. The false discovery rate (FDR) was estimated for each gene using the method of Benjamini and Hochberg (25), to control for false positives. All genes used in this analysis had an FDR of less than 0.05.
Class Prediction
The class prediction tool in BRB-ArrayTools was used to select genes and build predictors of radiation exposure status. The greedy pairs method was used to select the 12 pairs of genes that best discriminated between control and irradiated samples in each training set. After feature selection, seven classification methods (compound covariate predictor, linear discriminant analysis, 1- and 3-nearest neighbors, nearest centroid, support vector machines, and Bayesian compound covariate predictor) were used with the selected feature sets to predict the irradiation status of the remaining samples. The percentage correct classification was calculated for each approach.
Gene Ontology Analysis
Lists of genes that were either significantly overexpressed or underexpressed compared with controls were analyzed using the Ingenuity Pathway Analysis core pathway (Qiagen Ingenuity Systems) to identify canonical pathways and diseases and functions. Benjamini-Hochberg adjusted P values of < 0.05 were considered significant. Furthermore, a pathway with z-score equal or higher than 2 or equal or lower than −2 was considered activated or inactivated, respectively.
Quantitative RT-PCR
cDNA was prepared from total globin-cleared RNA using the High-Capacity cDNA Reverse Transcription Kit with RNase Inhibitor (Thermofisher). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed for four genes (Axl, Gas6, Marco and Cd93) using Taqman assays (Thermofisher). Axl (Mm00437221_m1), Gas6 (Mm00490378_m1), Marco (Mm00440265_m1), and Cd93 (Mm00440239_g1) were pre-designed validated assays. A β-actin (Actb) assay (Mm00607939_s1) was also performed alongside for normalization. The gene expression validation experiments were conducted with 20 ng cDNA using TaqMan™ Universal PCR Master Mix, no AmpErase™ UNG (Thermofisher) in a QuantStudio 7 Flex Real Time PCR system (Thermofisher). Relative fold-induction was calculated by the 2−ΔΔCT method (26) using QuantStudio™ Design and Analysis Software (Thermofisher). Data were normalized to β-actin gene expression levels.
RESULTS
Microarray Analysis
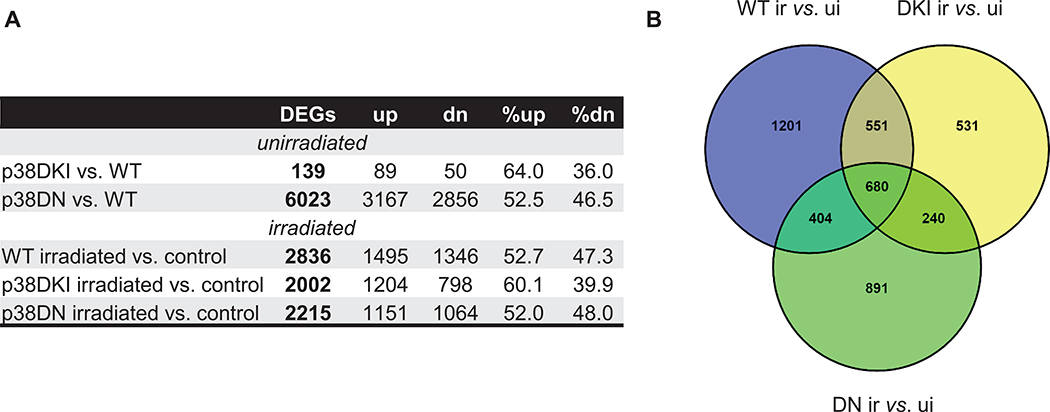
Mice were exposed to 7 Gy of X-ray radiation and analyzed at 24 h postirradiation. A radiation dose of 7 Gy in C57BL/6 mice, which have a LD50/30 ~8 Gy and, thus, almost all mice survive without treatment, was selected to represent a murine radiation model for a priority group for triage in humans after a nuclear emergency (27). We focused on transcriptomic changes at 24 h postirradiation as representing the earliest practical time for application of large-scale biodosimetry (1). The responses at this time may also indicate genes and/or pathways that can be exploited to develop future countermeasures. Global gene expression was measured in the blood of p38 p38WT and mutant (p38DKI and p38DN) C57BL/6J male mice using five mice per group and Agilent’s whole mouse genome microarrays. Class comparison using BRB-ArrayTools (28) identified a total of 139 and 6,023 differentially expressed genes (DEGs) [P < 0.001, FDR < 5%] in p38DKI vs. p38WT (control) mice and p38DN vs. p38WT mice, respectively, under basal conditions (Fig. 1A; fold changes, along with P values and FDR are listed for all DEGs from the microarray analysis in Supplementary File 1; https://doi.org/10.1667/RADE-21-00240.1.S1). Among the DEGs in p38DKI mice under basal conditions, 64% were upregulated and 36% were downregulated, whereas 52.5% of DEGs were upregulated in p38DN with the rest downregulated. These data suggest that whereas p38DKI has limited effect on gene expression under basal conditions, p38DN has a profound impact on immune cell physiology. Exposure to radiation resulted in 2,836, 2,002, and 2,215 DEGs in p38WT, p38DKI, and p38DN, respectively. A total of 4,498 genes were responsive to ionizing radiation in at least one genotype. Of those genes, 680 responded to irradiation in mice of all three genotypes and the majority of the genes also displayed similar fold-changes, whereas 1,201 genes (42%) in the p38WT cohort, 531 genes (26.5%) in the p38DKI cohort, and 891 genes (40%) in the p38DN cohort were unique to the respective p38 genotype (Fig. 1B).
FIG. 1.
Differentially expressed genes (DEGs) between mutant (p38DKI, p38DN) vs. p38WT and between irradiated vs. unirradiated samples. Panel A: Significantly differentially expressed genes (DEGs) in mouse blood or after 7 Gy X-ray irradiation relative to unirradiated controls (P < 0.001, FDR < 0.05). Absolute numbers and percentages of upregulated and downregulated genes are shown. Panel B: Venn diagram showing overlap patterns of genes that are differentially expressed in the control and irradiated p38 mutant mice. Abbreviations, WT: p38 wild-type; DKI: p38 double knock-in mutant; DN: p38 dominant negative; up: upregulated; dn: downregulated; ui: unirradiated; ir: irradiated.
Impact of p38 on Prediction of Radiation Exposure
We used the class prediction tool in BRB-ArrayTools (28) to develop a gene signature that would correctly detect radiation exposure. The greedy pairs method (29) was used to identify the 12 top-performing pairs of genes for discrimination between irradiated and control samples (Supplementary File 2; https://doi.org/10.1667/RADE-21-00240.1.S2). Each p38 genotype was used in turn as the training set for the gene selection process with the remaining samples serving as a test set. Although only two genes, Chst3 (carbohydrate sulfotransferase 3) and Dapl1 (death-associated protein-like 1), were common among the three p38 genotype gene sets, each training set was capable of distinguishing between irradiated and unirradiated samples with 100% accuracy (10 of 10 mice) in all three p38 genotypes, using seven independent classifier algorithms [Supplementary Table S1 (https://doi.org/10.1667/RADE-21-00240.1.S3) and Supplementary File 2 (https://doi.org/10.1667/RADE-21-00240.1.S2)]. In a previous report (30), we constructed a gene expression signature that combined samples from wild-type and two DNA repair mutant (Atm−/− and Prkdcscid) mouse models that was able to detect exposure to a LD50/30 radiation dose with a 100% accuracy. We tested the ability of that signature to detect radiation exposure in p38WT, p38DKI, and p38DN mice. Although there was no gene overlap between the earlier signature and any of the p38 signatures reported here, the so-called “mixed” (i.e., wild-type plus DNA repair deficient mutants) gene set was also able to detect irradiated mice in this study with 100% efficiency (P < 0.001), irrespective of p38 status (Supplementary Table S2; https://doi.org/10.1667/RADE-21-00240.1.S4).
Gene Ontology Analysis
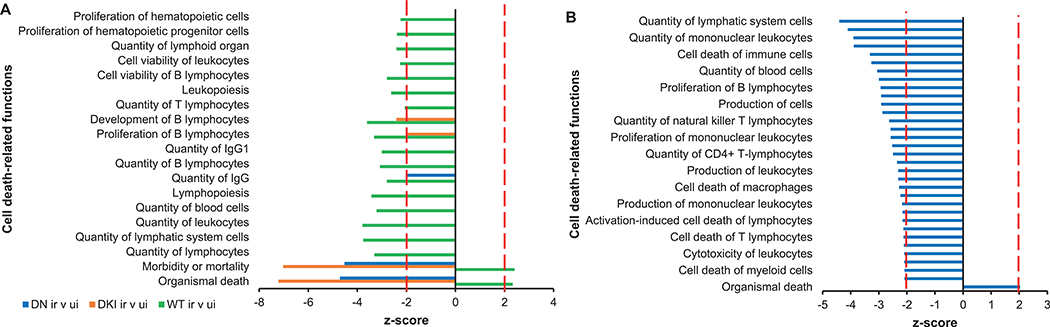
We used Ingenuity Pathway Analysis (IPA) to identify the most significantly enriched canonical pathways and diseases or functions among DEGs, derived from irradiated and unirradiated mice and then compared gene expression in p38DKI, and p38DN mice with p38WT mice. We also compared differential gene expression between unirradiated p38DN and p38WT mice. The complete lists of canonical pathways and diseases or functions are reported in Supplementary File 3 (https://doi.org/10.1667/RADE-21-00240.1.S5). DEGs in irradiated vs. unirradiated p38WT mice were enriched in functions such as organismal death (z = 2.335) and morbidity or mortality (z = 2.408) (Fig. 2A). Concordantly, quantities of various cell types, including lymphocytes, were underrepresented, whereas apoptosis-related pathways were over-represented. In contrast to the irradiated p38WT mice, DEGs from irradiated p38DKI or p38DN transgenic mice were negatively associated with functions such as organismal cell death (z = −7.217 and −4.709 for p38DKI and p38DN, respectively) and morbidity and mortality (z = −7.023 and −4.524 for p38DKI and p38DN, respectively), whereas functions related to quantities of various immune cells were not significantly different from unirradiated mice [Fig. 2A and Supplementary Table S3 (https://doi.org/10.1667/RADE-21-00240.1.S6) for complete list of cell death-related functions and z-scores]. p38p38The impact of p38DKI under basal conditions was minimal with only 139 DEGs compared with p38WT mice, whereas p38DN had a large number of DEGs compared with p38WT. However, unlike irradiated p38DN mice that showed 94.5% of the canonical pathways to be overrepresented, most significant canonical pathways in p38DN under basal levels were underrepresented compared with p38WT mice (Supplementary File 3; https://doi.org/10.1667/RADE-21-00240.1.S5). Interestingly, downregulation of p38 resulted in cell death-related functions with a negative z-score [Fig. 2B and Supplementary Table S4 (https://doi.org/10.1667/RADE-21-00240.1.S7) for complete list of cell-death related functions and z-scores], suggesting that p38 activity is required to preserve survival of immune cells under basal conditions.
FIG. 2.
Cell death-related functions. Panel A: Significant cell death-related diseases and functions among genes differentially expressed in irradiated WT, DKI, and DN mice as identified by Ingenuity Pathway Analysis (IPA). Panel B: Significant cell-death functions among genes differentially expressed in p38DN vs. p38WT mice under basal conditions as identified by IPA. Biological functions displaying a z-score ≥ 2 or ≤ −2 and showing Benjamini-Hochberg adjusted P value < 0.05 were considered significant. Abbreviations, WT: p38 wild-type; DKI: p38 double knock-in mutant; DN: p38 dominant negative.
Further analysis revealed that phagocytosis-related functions, defined by terms “phagocytosis”, “engulfment” and “endocytosis”, were over-represented in irradiated mice [Fig. 3 and Supplementary Table S5 (https://doi.org/10.1667/RADE-21-00240.1.S8) for complete list of phagocytosis-related functions and z-scores]. However, the number of phagocytosis-related functions was significantly greater in irradiated p38 transgenic mice, compared with irradiated p38WT mice, suggesting that p38 exerts an inhibitory effect on phagocytosis in response to stress. Genes with known roles in phagocytosis, including Axl and its ligand Gas6 (growth arrest specific 6) (31), the apoptotic recognition receptor on phagocytes, Marco (Macrophage receptor with collagenous structure) (32) and Cd93, a transmembrane receptor that mediates enhancement of phagocytosis in monocytes (33), were markedly upregulated in irradiated p38 mutant mice. In contrast, none of these genes were differentially expressed under basal conditions and neither phagocytosis-associated functions were over-represented in p38DKI or p38DN compared with p38WT mice. To validate the microarray findings, we analyzed expression of Axl, Gas6, Marco, and Cd93 genes by quantitative-PCR and normalized their expression relative to β-actin. We determined that all these genes were upregulated in irradiated p38WT mice, but they were expressed at considerably higher levels in the p38 mutant samples (Fig. 4). The fold-changes of these genes in response to X rays were similar to the fold-changes from the microarray experiment, although some values in the p38WT samples did not reach statistical significance by microarray measurement (Supplementary Table S6; https://doi.org/10.1667/RADE-21-00240.1.S9).
FIG. 3.
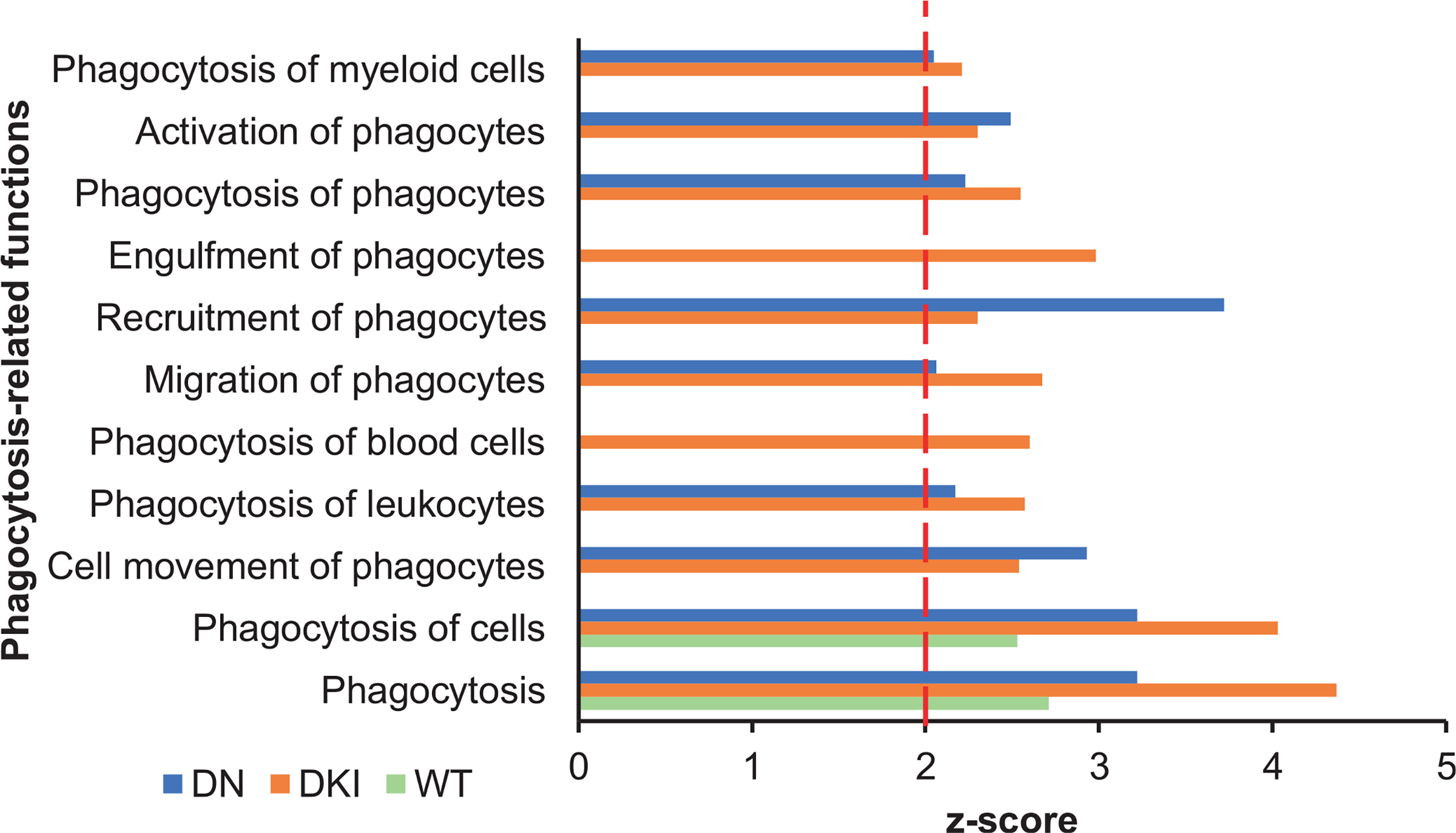
Phagocytosis-related biological functions are overrepresented in irradiated p38 transgenic mice. Significant phagocytosis-related diseases or functions among genes differentially expressed in irradiated WT, DKI, and DN mice as identified by Ingenuity Pathway Analysis. Biological functions displaying a z-score ≥ 2 and showing Benjamini-Hochberg adjusted P value < 0.05 were considered significant. Abbreviations, WT: p38 wild-type; DKI: p38 double knock-in mutant; DN: p38 dominant negative.
FIG. 4.
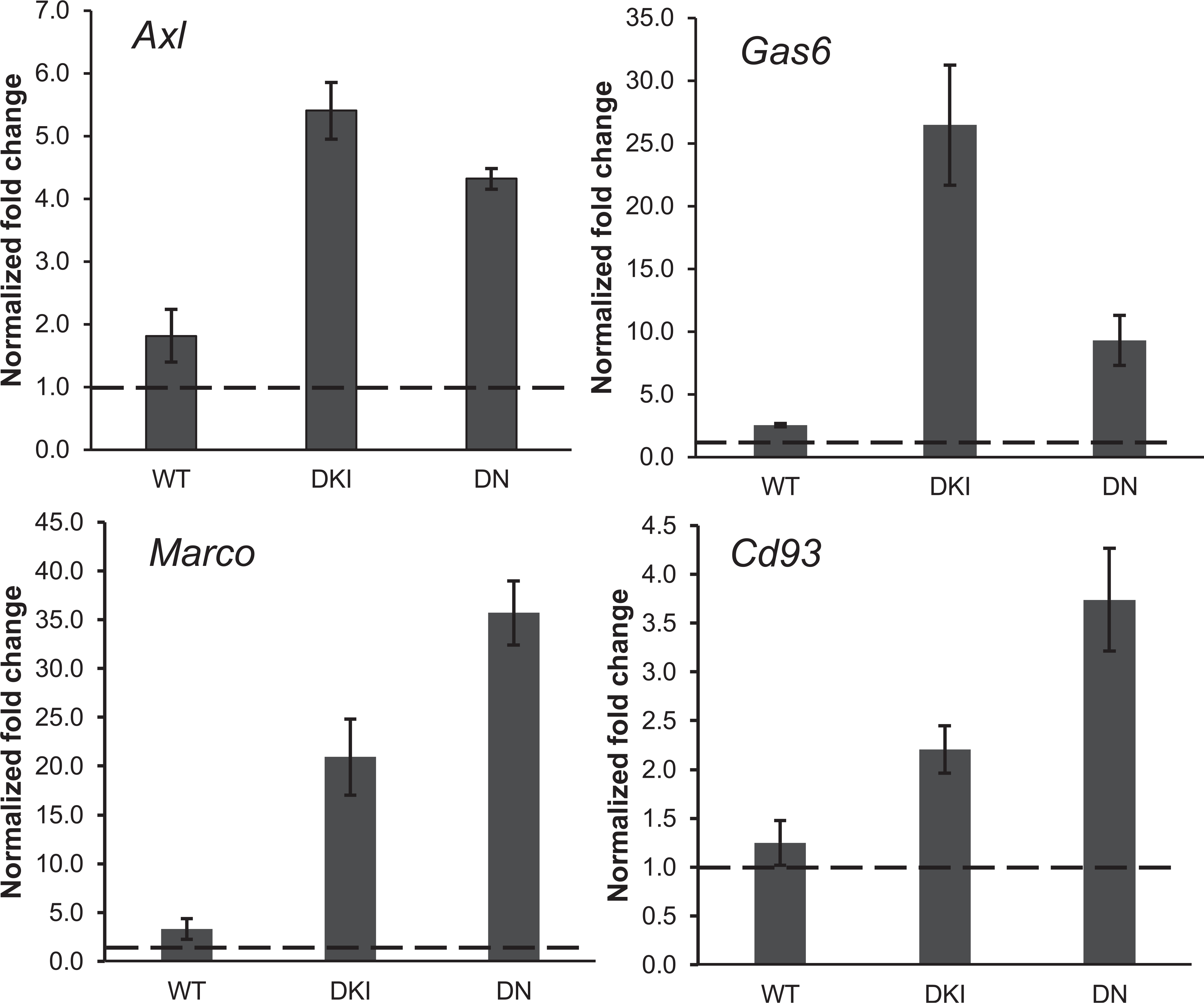
Phagocytosis-related gene expression measured by RT-qPCR. Gene expression levels of 4 phagocytosis-related genes (Axl, Gas6, Marco and Cd93) analyzed by qRT-PCR and normalized to Actb expression. Data represent the mean ± S.E.M. (n = 3). The dashed line represents the level in unirradiated controls. Abbreviations, WT: p38 wild-type; DKI: p38 double knock-in mutant; DN: p38 dominant negative.
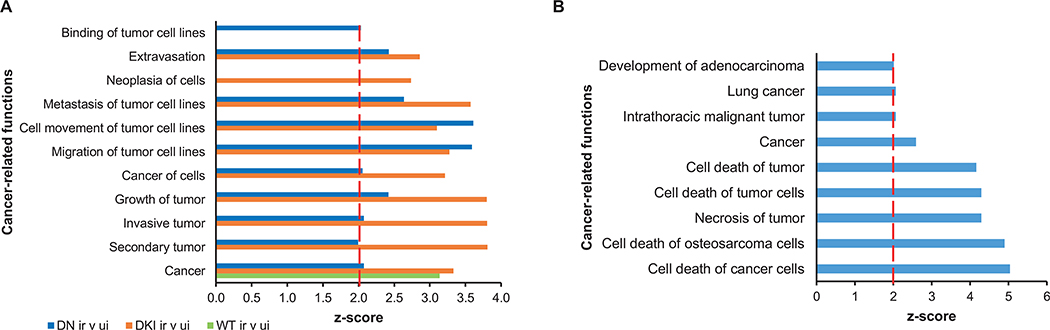
Dysregulation of the p38 signaling pathway has been linked to several diseases, including cancer (34). Exposure to ionizing radiation did not enrich for cancer-related functions in p38WT mice. In contrast, several cancer-related functions were significantly over-represented among irradiated p38DKI and p38DN mice (Fig. 5A and Supplementary Table S7; https://doi.org/10.1667/RADE-21-00240.1.S10). Importantly, irradiation of animals with mutant p38 resulted in an overrepresentation of tumor metastasis, migration, invasion, and extravasation functions (Fig. 5A), typically associated with advanced stage tumors. In contrast, DEGs in p38DN transgenic mice under basal (unirradiated) conditions were associated with an enrichment of functions related to cancer cell death (Fig. 5B and Supplementary Table S8; https://doi.org/10.1667/RADE-21-00240.1.S11), suggesting that p38 signaling under non-stressed conditions is required for the survival of cancer cells.
FIG. 5.
Cancer-related functions. Panel A: Enriched cancer-related functions in irradiated p38WT, p38DKI, and p38DN mouse blood compared with control animals. Panel B: Enriched cancer-related functions in p38DN versus p38WT under basal conditions. A P value < 0.05 and a z-score ≥ 2 (marked by the red line) were considered significant. Abbreviations, DKI: p38 double knock-in mutant; DN: p38 double dominant; WT: p38 wild-type.
DISCUSSION
The long-term goal of our studies is to develop gene expression signatures robust enough to detect radiation exposure, irrespective of confounding factors, such as radiation quality, age, and inflammatory status Recently, we demonstrated that age could have a profound impact on gene expression and diminish the predictive ability of a previously created radiation biodosimetry gene list (35, 36). Another significant factor that can have an impact on radiation biodosimetry is inflammation. Ionizing radiation is known to induce inflammation (37) and gene ontology and pathway analyses indicate that many pro-inflammatory processes are upregulated upon irradiation (38). Furthermore, radiation exposure triggers a long-lasting inflammatory response that leads to immune dysfunction (39). Therefore, a pre-existing chronic inflammatory condition, such as an autoimmune disease, or anti-inflammatory medication could affect gene expression-based biodosimetry. Recently, we investigated the impact of inflammatory signaling on radiation biodosimetry using the IL-10−/− transgenic mouse model. IL-10 is a well-known anti-inflammatory cytokine that plays a major role in various aspects of immune system regulation (40) and genetic ablation of IL-10 in mice results in an exaggerated inflammatory response to enteric pathogens that re-capitulate many symptoms of the human inflammatory bowel disease (IBD) (40). It was shown that generation of an inflammatory condition modified the expression of many radiation-responsive genes. Nevertheless, a panel of genes was identified that could predict radiation exposure irrespective of inflammation condition (41).
In the current study, we used the p38DKI and p38DN transgenic mice as models for immunosuppression and examined the impact of p38 on radiation biodosimetry and the identification of pathways over- or under-represented in response to high-dose ionizing radiation. Our results showed that it is possible to select panels of genes that were able to discriminate with 100% accuracy between irradiated samples from unirradiated-control samples, regardless of p38 status. In a previous study, we used male wild-type C57BL/6 mice and genetically engineered mouse models deficient in pathways known to affect radiation survival by impairing double-strand break repair by homologous recombination (Atm−/−) or non-homologous end joining (Prkdc−/−, SCID) and explored the potential effect of DNA repair deficiency on the gene expression response to radiation and implication for radiation biodosimetry. The inclusion of these DNA repair mutants resulted in an improved detection of radiation exposure (30). We tested the ability of a 24-gene classifier derived from the “mixed” gene expression data to detect radiation exposure in p38WT, DKI and DN mice and we found that it was still possible to discriminate irradiated mice from unirradiated mice, irrespective of p38 status. Future studies will examine the performance of these gene signatures at detecting a range of radiation doses, including low-dose, exposures.
The p38 signaling pathway plays a major role in the regulation of cell survival in response to stress. However, higher and/or sustained p38 activation may lead to cell death (3). Our results imply that radiation induces organismal and cell death with a concomitant decrease in the quantities of multiple immune cell types. In contrast, blood cells from p38DKI and p38DN mice may be protected against the adverse effects of radiation, since we find that organismal and cell death-related functions are underrepresented in the mutant p38 mice compared to p38WT animals. Mechanistically, these data support the idea that radiation induces immune cell death through the activation of the p38 pathway and inhibition of this pathway confers protection to immune cells exposed to radiation. Importantly, p38 seems to be required for basal survival of immune cells, since downregulation of p38 signaling in p38DN mice results in the enrichment of immune cell death functions. We postulate that under non-challenged conditions, p38 acts as a pro-survival kinase to sustain viability and numbers of various immune cell types. However, exposure to ionizing radiation leads to a high and/or sustained increase of p38 signaling that leads to increased cell death (42, 43). A novel finding of this study is that not only the canonical, but also the alternative p38 signaling pathway shows an altered pattern of gene expression that suggests a contribution to increased cell death in response to radiation.
Previously, we demonstrated that phagocytosis-related functions were significantly overrepresented among differentially expressed genes in young mice in response to 4 Gy X-ray irradiation (35). Here, we discovered that suppression of p38 signaling pathways by either p38DKI or p38DN enriches the DEG profile in phagocytosis-related functions to a greater extent than p38WT. Individual phagocytosis genes, such as Axl, Ga6, Marco and Cd93 (31–33, 44, 45), were upregulated to a much greater degree in irradiated p38 mutant mice than irradiated p38WT mice. In contrast, under basal conditions, neither p38DKI nor p38DN mice displayed significant differences in phagocytosis compared with p38WT mice.
Phagocytosis is crucial in maintaining tissue homeostasis and innate immune balance and it is responsible for the clearance of apoptotic cells, a process called efferocytosis, as well as invading pathogens such as bacteria and microbes (46). Although many cell types are endowed with phagocytotic activity and can clear apoptotic cell debris, phagocytosis is predominantly carried out by the so-called professional phagocytes, including macrophages, dendritic cells, and neutrophils. Dysfunctional phagocytosis leads to accumulation of unphagocytosed debris with subsequent accumulation of secondary necrotic debris that promotes chronic inflammation and autoimmune diseases (47) and exacerbates tissue damage. Therefore, phagocytosis has been considered a key requirement for inflammatory resolution and for the preservation of immune tolerance (48). Phagocytosis has also been associated with inflammaging. Specifically, it has been suggested that inflammaging is largely due to the imbalance between the production and disposal of cellular debris and misfolded and/or misplaced self-molecules, which develop with age (49). In support of this hypothesis, we have demonstrated that phagocytosis-related functions are enriched only in young mice exposed to radiation, but not old irradiated mice (35). Moreover, it has been shown that blocking elevated p38 restores efferocytosis and inflammatory resolution in the elderly (50). In that report, it was shown that p38α can control the resolution of inflammation by impairing the engulfment of apoptotic bodies (efferocytosis) in macrophages (50).
Intriguingly, our gene ontology and pathway analyses demonstrate that phagocytosis-related pathways are associated not only with p38DN that suppresses both the canonical and the alternative p38 pathways, but also with p38DKI, which abrogates the alternative pathway in T cells. Since the alternative pathway has definitely been demonstrated in T cells, a possible explanation for the observed overrepresentation of phagocytosis functions is the existence of crosstalk between T cells and phagocytes, presumably through the production of lymphokines. Specifically, we postulate that abrogation of the alternative pathway leads to increased survival of T lymphocytes in response to radiation, which, in turn, increases the activity of phagocytes that are induced to clear apoptotic cells from the circulation and peripheral tissues and promote inflammation resolution. This line of argument is supported by recent findings that show that CD4+ regulatory T (Treg) cells promote efferocytosis during inflammation resolution, whereas depletion of Treg cells disrupts apoptotic cell internalization in phagocytes and impair inflammation resolution (51). Furthermore, it has been shown that CD4+ T helper (Th1) cells produce lymphokines that promote the microbicidal activities of infected phagocytes (52).
Radiotherapy is an important modality in the management of various tumors (53). Moreover, many p38 small molecule inhibitors have been tested for their efficacy to control tumor growth (3). Here, we demonstrated that irradiated p38WT animals were not associated with an enrichment of cancer-related functions. In contrast, both irradiated p38DN and p38DKI mice displayed a significant enrichment of cancer-associated functions, including functions associated with tumor invasion and metastasis, implying that p38 may have tumor and/or metastasis suppressive properties in response to radiation. An implication of this observation would be that combination of radiotherapy with p38 inhibitors may be counter-indicated.
Notwithstanding the extensive similarities between p38DKI and p38DN in the response to radiation, p38DN is a key player in homeostasis and the classical pathway is required. Comparison of the gene expression profiles of p38WT mice with those of unirradiated p38DN and p38DKI reveals that p38DN has a profound effect on the transcriptome of mice with over 6,000 genes being differentially expressed between p38WT and p38DN mice. In contrast the number of DEGs between p38WT and p38DKI is very small (approximately 140 genes). Similarly, a low number of DEGs was also found in the IL-10 knockout mice (41). These data underscore the significance of the canonical p38 signaling pathway under unirradiated (control) conditions.
In conclusion, we show that radiation-induced immune cell death may, at least partially, be driven by p38 activation. In addition, p38 pathway inhibition leads to an enrichment of various phagocytosis functions in response to radiation. Thus, we propose a novel pathway by which radiation may promote inflammation that is dependent on p38 activation, whereas inhibition of p38 leads not only to diminished immune cell apoptosis but also to increased phagocytosis, thus diminishing inflammation and alleviating tissue injury. A small molecule p38 inhibitor could potentially be administered after irradiation to mitigate the detrimental effects of radiation on immune cells. However, careful design of administering a p38 inhibitor will be required due to the pleotropic functions of p38, as they control many processes in a cell type and context-specific manner and their pro- or anti-apoptotic effect is dependent on type, strength, or duration of stimulus. Our study suggests that p38DKI may increase immune cell viability, but has no impact under basal conditions, thus offering an opportunity to target the alternative p38 pathway instead of the canonical p38 pathway to mitigate the detrimental effects of radiation exposure on the hematopoietic system. However, further studies, including functional cell death and phagocytosis assays, will be required to substantiate the effects suggested by the microarray data. Finally, despite the extensive impact on gene expression, it was still possible to derive a panel of genes that could correctly distinguish between irradiated and control mice, irrespective of p38 status.
Supplementary Material
Supplementary File 1: Excel file with 5 tabs containing a summary of differentially expressed genes by p38 genotype.
Supplementary File 2: Composition of the classifiers trained using data from irradiated vs. unirradiated WT (tab1), p38DKI (tab2), and p38DN (tab3).
Supplementary Table S1: Impact of p38 pathways on prediction of radiation exposure. Performance of seven classifier algorithms using greedy pairs gene selection with WT, DKI, or DN used as training sets.
Supplementary Table S2: Performance of seven classifier algorithms using greedy pairs gene selection with the “mixed” training set.
Supplementary File 3: Excel file containing the complete output of Ingenuity Pathway Analysis canonical pathways (tab1) and Diseases and functions (tab2).
Supplementary Table S3: Cell death-related diseases and functions among genes differentially expressed in irradiated WT, DKI and DN mice.
Supplementary Table S4: Diseases and functions in p38DN vs. p38WT mice under unirradiated conditions.
Supplementary Table S5: Phagocytosis-related functions among genes differentially expressed in irradiated WT, DKI and DN mice as identified by IPA.
Supplementary Table S6: Comparison of relative fold-changes in the expression of phagocytosis genes in DNA microarray and qPCR.
Supplementary Table S7: Over-representation of cancer-related functions in irradiated p38 wild-type or transgenic mice compared with unirradiated-control animals.
Supplementary Table S8: Over-representation of cancer-related functions in p38DN transgenic mice compared with p38WT control animals, under basal conditions.
ACKNOWLEDGMENTS
The authors thank Ms. Pelagie Ake, Ms. Linze Song, and Mr. Jerry Angdisen for their help in animal breeding and sample collection. We are grateful to Dr. Jonathan Ashwell for kindly providing the p38 double knock-in transgenic mouse and for his critical comments on the manuscript. Analyses were performed using BRB-ArrayTools developed by Dr. Richard Simon and the BRB-ArrayTools Development Team at the National Cancer Institute. This work was supported by the Center for High-Throughput Minimally-Invasive Radiation Biodosimetry, National Institute of Allergy and Infectious Diseases grant number U19AI067773. The funding body did not play any role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Footnotes
Editor’s note. The online version of this article (DOI: https://doi.org/10.1667/RADE-21-00240.1) contains supplementary information that is available to all authorized users.
REFERENCES
- 1.Sullivan JM, Prasanna PG, Grace MB, Wathen LK, Wallace RL, Koerner JF, et al. Assessment of biodosimetry methods for a mass-casualty radiological incident: medical response and management considerations. Health Phys. 2013; 105:540–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amundson SA. Transcriptomics for radiation biodosimetry: progress and challenges. Int J Radiat Biol. 2021. May 21:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canovas B, Nebreda AR. Diversity and versatility of p38 kinase signalling in health and disease. Nat Rev Mol Cell Biol. 2021; 22:346–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashwell JD. The many paths to p38 mitogen-activated protein kinase activation in the immune system. Nat Rev Immunol. 2006; 6:532–40. [DOI] [PubMed] [Google Scholar]
- 5.Hale KK, Trollinger D, Rihanek M, Manthey CL. Differential expression and activation of p38 mitogen-activated protein kinase alpha, beta, gamma, and delta in inflammatory cell lineages. J Immunol. 1999; 162:4246–52. [PubMed] [Google Scholar]
- 6.Brancho D, Tanaka N, Jaeschke A, Ventura JJ, Kelkar N, Tanaka Y, et al. Mechanism of p38 MAP kinase activation in vivo. Genes Dev. 2003; 17:1969–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rincón M, Flavell RA, Davis RA. The JNK and P38 MAP kinase signaling pathways in T cell-mediated immune responses. Free Radic Biol Med. 2000; 28:1328–37. [DOI] [PubMed] [Google Scholar]
- 8.Lee JC, Laydon JT, McDonnell PC, Gallagher TF, Kumar S, Green D, et al. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature. 1994; 372:739–46. [DOI] [PubMed] [Google Scholar]
- 9.Winzen R, Kracht M, Ritter B, Wilhelm A, Chen CY, Shyu AB, et al. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 1999; 18:4969–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rousseau S, Morrice N, Peggie M, Campbell DG, Gaestel M, Cohen P. Inhibition of SAPK2a/p38 prevents hnRNP A0 phosphorylation by MAPKAP-K2 and its interaction with cytokine mRNAs. EMBO J. 2002; 21:6505–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoecklin G, Stoeckle P, Lu M, Muehlemann O, Moroni C. Cellular mutants define a common mRNA degradation pathway targeting cytokine AU-rich elements. RNA. 2001; 7:1578–88. [PMC free article] [PubMed] [Google Scholar]
- 12.Westra J, Doornbos-van der Meer B, de Boer P, van Leeuwen MA, van Rijswijk MH, Limburg PC. Strong inhibition of TNF-alpha production and inhibition of IL-8 and COX-2 mRNA expression in monocyte-derived macrophages by RWJ 67657, a p38 mitogen-activated protein kinase (MAPK) inhibitor. Arthritis Res Ther. 2004; 6:R384–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waetzig GH, Seegert D, Rosenstiel P, Nikolaus S, Schreiber S. p38 mitogen-activated protein kinase is activated and linked to TNF-alpha signaling in inflammatory bowel disease. J Immunol. 2002; 168:5342–51. [DOI] [PubMed] [Google Scholar]
- 14.Salvador JM, Hollander MC, Nguyen AT, Kopp JB, Barisoni L, Moore JK, et al. Mice lacking the p53-effector gene Gadd45a develop a lupus-like syndrome. Immunity. 2002; 16:499–508. [DOI] [PubMed] [Google Scholar]
- 15.Mittelstadt PR, Yamaguchi H, Appella E, Ashwell JD. T cell receptor-mediated activation of p38{alpha} by mono-phosphorylation of the activation loop results in altered substrate specificity. J Biol Chem. 2009; 284:15469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giardino Torchia ML, Dutta D, Mittelstadt PR, Guha J, Gaida MM, et al. Intensity and duration of TCR signaling is limited by p38 phosphorylation of ZAP-70T293 and destabilization of the signalosome. Proc Natl Acad Sci U S A. 2018; 115:2174–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudd CE. MAPK p38: alternative and nonstressful in T cells. Nat Immunol. 2005; 6:368–70. [DOI] [PubMed] [Google Scholar]
- 18.Salvador JM, Mittelstadt PR, Guszczynski T, Copeland TD, Yamaguchi H, Appella E, et al. Alternative p38 activation pathway mediated by T cell receptor-proximal tyrosine kinases. Nat Immunol. 2005; 6:390–5. [DOI] [PubMed] [Google Scholar]
- 19.Salvador JM, Mittelstadt PR, Belova GI, Fornace AJ Jr, Ashwell JD. The autoimmune suppressor Gadd45alpha inhibits the T cell alternative p38 activation pathway. Nat Immunol. 2005; 6:396–402. [DOI] [PubMed] [Google Scholar]
- 20.Alam MS, Gaida MM, Ogawa Y, Kolios AG, Lasitschka F, Ashwell JD. Counter-regulation of T cell effector function by differentially activated p38. J Exp Med. 2014; 211:1257–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alam MS, Gaida MM, Bergmann F, Lasitschka F, Giese T, Giese NA, et al. Selective inhibition of the p38 alternative activation pathway in infiltrating T cells inhibits pancreatic cancer progression. Nat Med. 2015; 21:1337–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shreeram S, Hee WK, Demidov ON, Kek C, Yamaguchi H, Fornace AJ Jr, et al. Regulation of ATM/p53-dependent suppression of myc-induced lymphomas by Wip1 phosphatase. J Exp Med. 2006; 203:2793–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jirmanova L, Giardino Torchia ML, Sarma ND, Mittelstadt PR, Ashwell JD. Lack of the T cell-specific alternative p38 activation pathway reduces autoimmunity and inflammation. Blood. 2011; 118:3280–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong ES, Le Guezennec X, Demidov ON, Marshall NT, Wang ST, Krishnamurthy J, et al. p38MAPK controls expression of multiple cell cycle inhibitors and islet proliferation with advancing age. Dev Cell. 2009; 17:142–9. [DOI] [PubMed] [Google Scholar]
- 25.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990; 9:811–8. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001; 25:402–8. [DOI] [PubMed] [Google Scholar]
- 27.Ossetrova NI, Ney PH, Condliffe DP, Krasnopolsky K, Hieber KP. Acute radiation syndrome severity score system in mouse total-body irradiation model. Health Phys. 2016; 111:134–44. [DOI] [PubMed] [Google Scholar]
- 28.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003; 19:2448–55. [DOI] [PubMed] [Google Scholar]
- 29.Bø T, Jonassen I. New feature subset selection procedures for classification of expression profiles. Genome Biol. 2002; 3, RESEARCH0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rudqvist N, Laiakis EC, Ghandhi SA, Kumar S, Knotts JD, Chowdhury M, et al. Global Gene Expression Response in Mouse Models of DNA Repair Deficiency after Gamma Irradiation. Radiat Res. 2018; 189:337–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothlin CV, Carrera-Silva EA, Bosurgi L, Ghosh S. TAM receptor signaling in immune homeostasis. Annu Rev Immunol. 2015; 33:355–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gordon S. Phagocytosis: An immunobiologic process. Immunity. 2016; 44:463–75. [DOI] [PubMed] [Google Scholar]
- 33.Steinberger P, Szekeres A, Wille S, Stöckl J, Selenko N, Prager E, et al. Identification of human CD93 as the phagocytic C1q receptor (C1qRp) by expression cloning. J Leukoc Biol. 2002; 71:133–40. [PubMed] [Google Scholar]
- 34.Gupta J, Nebreda AR. Roles of p38a mitogen-activated protein kinase in mouse models of inflammatory diseases and cancer. FEBS J. 2015; 282:1841–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broustas CG, Duval AJ, Amundson SA. Impact of aging on gene expression response to x-ray irradiation using mouse blood. Sci Rep. 2021; 11:10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broustas CG, Xu Y, Harken AD, Garty G, Amundson SA. Comparison of gene expression response to neutron and x-ray irradiation using mouse blood. BMC Genomics. 2017; 18:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukherjee D, Coates PJ, Lorimore SA, Wright EG. Responses to ionizing radiation mediated by inflammatory mechanisms. J Pathol. 2014; 232:289–99. [DOI] [PubMed] [Google Scholar]
- 38.Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001; 19:683–765. [DOI] [PubMed] [Google Scholar]
- 39.Neriishi K, Nakashima E, Delongchamp RR. Persistent subclinical inflammation among A-bomb survivors. Int J Radiat Biol. 2001; 77:475–82. [DOI] [PubMed] [Google Scholar]
- 40.Li MC, He SH. IL-10 and its related cytokines for treatment of inflammatory bowel disease. World J Gastroenterol. 2004; 10:620–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mukherjee S, Laiakis EC, Fornace AJ Jr, Amundson SA. Impact of inflammatory signaling on radiation biodosimetry: mouse model of inflammatory bowel disease. BMC Genomics. 2019; 20:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tobiume K, Matsuzawa A, Takahashi T, Nishitoh H, Morita K, Takeda K, et al. ASK1 is required for sustained activations of JNK/p38 MAP kinases and apoptosis. EMBO Rep. 2001; 2:222–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yue J, López JM. Understanding MAPK signaling pathways in apoptosis. Int J Mol Sci. 2020; 21:2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lemke G, Burstyn-Cohen T. TAM receptors and the clearance of apoptotic cells. Ann N Y Acad Sci. 2010; 1209:23–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lemke G. How macrophages deal with death. Nat Rev Immunol. 2019;19(9):539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li W. Phagocyte dysfunction, tissue aging and degeneration. Ageing Res Rev. 2013; 12:1005–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.A-Gonzalez N, Bensinger SJ, Hong C, Beceiro S, Bradley MN, Zelcer N, et al. Apoptotic cells promote their own clearance and immune tolerance through activation of the nuclear receptor LXR. Immunity. 2009; 31:245–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Savill J Apoptosis in resolution of inflammation. J Leukoc Biol. 1997; 61:375–80. [DOI] [PubMed] [Google Scholar]
- 49.Franceschi C, Garagnani P, Vitale G, Capri M, Salvioli S. Inflammaging and ‘Garb-aging’. Trends Endocrinol Metab. 2017; 28:199–212. [DOI] [PubMed] [Google Scholar]
- 50.De Maeyer RPH, van de Merwe RC, Louie R, Bracken OV, Devine OP, Goldstein DR, et al. Blocking elevated p38 MAPK restores efferocytosis and inflammatory resolution in the elderly. Nat Immunol. 2020; 21:615–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Proto JD, Doran AC, Gusarova G, Yurdagul A Jr, Sozen E, Subramanian M, et al. Regulatory T Cells Promote Macrophage Efferocytosis during Inflammation Resolution. Immunity. 2018; 49:666–77.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tubo NJ, Jenkins MK. CD4+ T Cells: guardians of the phagosome. Clin Microbiol Rev. 2014; 27:200–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Howlader NNA, Krapcho M, Miller D, Bishop K, Kosary CL, Yu M, et al. (eds). SEER Cancer Statistics Review, 1975–2014. National Cancer Institute. November 2016. ed: Bethesda, MD; 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1: Excel file with 5 tabs containing a summary of differentially expressed genes by p38 genotype.
Supplementary File 2: Composition of the classifiers trained using data from irradiated vs. unirradiated WT (tab1), p38DKI (tab2), and p38DN (tab3).
Supplementary Table S1: Impact of p38 pathways on prediction of radiation exposure. Performance of seven classifier algorithms using greedy pairs gene selection with WT, DKI, or DN used as training sets.
Supplementary Table S2: Performance of seven classifier algorithms using greedy pairs gene selection with the “mixed” training set.
Supplementary File 3: Excel file containing the complete output of Ingenuity Pathway Analysis canonical pathways (tab1) and Diseases and functions (tab2).
Supplementary Table S3: Cell death-related diseases and functions among genes differentially expressed in irradiated WT, DKI and DN mice.
Supplementary Table S4: Diseases and functions in p38DN vs. p38WT mice under unirradiated conditions.
Supplementary Table S5: Phagocytosis-related functions among genes differentially expressed in irradiated WT, DKI and DN mice as identified by IPA.
Supplementary Table S6: Comparison of relative fold-changes in the expression of phagocytosis genes in DNA microarray and qPCR.
Supplementary Table S7: Over-representation of cancer-related functions in irradiated p38 wild-type or transgenic mice compared with unirradiated-control animals.
Supplementary Table S8: Over-representation of cancer-related functions in p38DN transgenic mice compared with p38WT control animals, under basal conditions.