Abstract
Background
LncRNA FGF14-AS2 is a critical suppressor in breast cancer (BCa) metastasis. However, whether FGF14-AS2 plays a role in the bone metastasis of BCa remains unknown.
Methods
TRAP assay and intratibial injection were carried out to evaluate the role of FGF14-AS2 in BCa bone metastasis in vitro and in vivo. Polyribosome profiling was done to examine the translation level. RNA pulldown combined with LC/MS was performed to identify the lncRNA-binding partner, RIP, dual-luciferase assay, and Co-IP assays as well to testify these physical interactions. The prognostic value of FGF14-AS2 expression level in BCa patients was analysed using Kaplan–Meier Plotter.
Results
We found that FGF14-AS2 suppresses osteoclast differentiation and osteolytic metastasis of BCa. Mechanistically, FGF14-AS2 suppresses the translation of RUNX2 by inhibiting the assembly of eIF4E/eIF4G complex and the phosphorylation of eIF4E, thereby reducing the transcription of RANKL, an essential regulator of osteoclast differentiation. Moreover, FGF14-AS2 is downregulated by YTHDF2-mediated RNA degradation in an m6A-dependent manner. Clinically, patients with high YTHDF2 and low FGF14-AS2 expression levels showed worse distant metastasis-free survival (DMFS).
Conclusions
FGF14-AS2 plays a crucial role in osteolytic metastasis, and may serve as a promising prognostic biomarker and therapeutic target for BCa bone metastasis.
Subject terms: Bone metastases, Breast cancer, Gene silencing, Cancer epigenetics, Diagnostic markers
Background
Breast cancer (BCa) is one of the most common cancers and is the leading cause of cancer-related deaths among women worldwide [1]. Although great advances have been made in the diagnosis and treatment, the cure for BCa awaits better understanding of the pathophysiology of metastasis [2]. Metastasis accounts for more than 90% of the death, among which bone is the most common site of BCa metastasis [3, 4]. The symptoms of bone metastasis include severe bone pain, pathological fractures, hypercalcaemia, which reduce the life quality and the survival time of the patients. Therefore, a more comprehensive understanding of the mechanisms of bone metastasis is of great significance for improving the prognosis of BCa.
Bone metastasis is a complex process in which tumour cells detach from the primary site, transport in the blood, and reside in the bone microenvironment. Once cancer cells spread to the bone, the dynamic balance between osteoblasts and osteoclasts is broken, thus interrupting the normal bone remodelling process and causing bone destruction [5]. Bone metastases are usually divided into three types: osteolytic, osteoblastic and mixed [6]. Osteolytic metastasis accounts for the majority of BCa metastasis, which is quite different from the osteoblastic bone metastasis of prostate cancer [7]. In the process of osteolytic metastasis, the activation of osteoclasts plays a decisive role. The differentiation and maturation of osteoclasts are induced by several key cytokines secreted by BCa cells, such as receptor activator for nuclear factor-κB ligand (RANKL), macrophage colony-stimulating factor (M-CSF), and parathyroid hormone-related protein (PTHrP) [8]. Among these cytokines, the binding of RANKL to receptor activator of nuclear factor-κB (RANK) on the surface of preosteoclasts is essential for osteoclastogenesis and subsequent bone resorption [9]. The interaction of RANKL and RANK is proposed as a novel and effective therapeutic target in bone metastasis [10, 11]. Moreover, RUNX2, a transcription factor essential for osteoblast differentiation, can transcriptionally up-regulate RANKL expression [12] and plays an important role in osteolytic metastasis of BCa [13, 14].
Long noncoding RNAs (lncRNAs) are a type of noncoding RNA (ncRNA) longer than 200 nucleotides. Only a few of lncRNAs is capable of encoding micropeptides, peptides, or proteins [15]. A growing body of evidence increasingly validates that lncRNAs play critical roles in human tumorigenesis and progression by serving as tumour oncogenes or suppressors. For example, the lncRNA Uc003xsl.1 promotes triple-negative breast cancer (TNBC) tumorigenesis by binding with nuclear transcriptional factor NFκB-repressing factor (NKRF) to activate the NFκB/IL8 axis [16]. The lncRNA LINC00926 suppresses breast tumour growth and metastasis through inhibition of the PGK1-mediated Warburg effect [17]. LncRNAs have become an important class of molecules in the diagnosis, treatment, and prognosis of various types of cancers; however, our understanding of the mechanism of lncRNAs remains limited [18, 19]. FGF14-AS2 is an antisense lncRNA transcribed from the opposite strand of the FGF14 (fibroblast growth factor 14, FGF14) gene. Recently, we revealed that FGF14-AS2, which was predominantly distributed in the cytoplasm, inhibits BCa metastasis by acting as a competing endogenous RNA (ceRNA) for miR-370-3p [20]. However, the role of FGF14-AS2 in BCa bone metastasis remains unknown.
N6-methyladenosine (m6A) is the most prevalent modification of eukaryotic mRNA and is generally mediated by “writers” and “erasers”. The writer complex, comprising methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14) and Wilms tumour 1-associating protein (WTAP), catalyses the m6A methylation of mRNA [21, 22]. Eraser enzymes, such as AlkB homologue H5 (ALKBH5) and fat mass and obesity-associated protein (FTO), mediate the demethylation of mRNA [23, 24]. In addition, there are “readers” that recognise and bind to the m6A motif of the transcript, and regulate mRNA stability, splicing, export, localisation, or translation. The most common readers include YTH N6-methyladenosine RNA-binding protein (YTHDF) 1/2/3, eukaryotic translation initiation factor 3 (eIF3), insulin-like growth factor 2 mRNA binding protein (IGF2BP) 1/2/3 [25–27]. Notably, abundant noncoding RNAs, including microRNAs, lncRNAs, circular RNAs, small nuclear RNAs, and ribosomal RNAs, are also highly modified with m6A [28]. Aberrant m6A modification has been reported to be involved in the progression of various cancers [29]. For example, elevated METTL3 expression stimulates the m6A modification of heparin-binding growth factor (HDGF) mRNA and enhances its stability, thereby promoting angiogenesis and glycolysis in gastric cancer [30]. YTHDF1 augments the translation of eIF3C in an m6A-dependent manner and concomitantly promotes the overall translational output, thereby facilitating tumorigenesis and metastasis in ovarian cancer [31].
In this study, we revealed that FGF14-AS2 suppresses osteoclast differentiation in vitro and osteolytic bone metastasis of BCa in vivo. Mechanistically, FGF14-AS2 blocked RUNX2 mRNA translation by interfering with the formation of eIF4E/eIF4G complex and inhibiting the phosphorylation of eIF4E in BCa cells, and consequently reduced the transcription of RANKL, a key regulator of osteoclast differentiation. Moreover, YTHDF2 promoted FGF14-AS2 degradation in an m6A-dependent manner in BCa cells. High levels of YTHDF2 and low levels of FGF14-AS2 were significantly correlated with decreased DMFS in patients with BCa. Collectively, our data suggested that FGF14-AS2 might serve as a promising prognostic biomarker and therapeutic target for BCa bone metastasis.
Methods
Cell lines and cell culture
The human breast cancer cell line MDA-MB-231, the human embryonic kidney cell line HEK 293T, and the mouse macrophage cell line RAW264.7 were obtained from the American Type Culture Collection (Manassas, VA, USA). The human breast cancer cell line MDA-MB-468 was obtained from the Type Culture Collection, Chinese Academy of Sciences (Shanghai, China). Cells were maintained in Dulbecco’s modified Eagle’s medium (Gibco, Carlsbad, CA, USA) supplemented with 10% foetal bovine serum (Wisent, Nanjing, China) in an incubator at 37 °C with 5% CO2. In this study, the absence of mycoplasma or bacterial contamination of the cells was detected.
Plasmids, chemicals and transfection
The primers used for plasmid construction are shown in Supplementary Table S1. Additional information about the plasmid construction and the source of other plasmids and chemicals is presented in Supplementary methods.
Quantitative real-time reverse transcription PCR (qRT-PCR)
Total RNA from tissue samples or cancer cells was isolated using TRIzol (Takara, Dalian, China), and complementary DNA was synthesised from 1 μg total RNA using the PrimeScript RT reagent (Vazyme, Nanjing, China) following the manufacturer’s instructions. The qRT-PCR was performed using Hieff® qPCR SYBR Green Master Mix (Yeasen) in a Roche LightCycler 96 Real-Time PCR System. The RNA expression values were normalised to that of the β-actin gene and calculated using the 2-ΔΔCt method. The primers used in qRT-PCR are listed in Supplementary Table S2.
Western blotting
Western blotting was performed according to a previously reported protocol [32]. The following primary antibodies were used: anti-p-p38 (AF4001, Affinity, Cincinnati, OH, USA), anti-p38 (AF6456, Affinity), anti-p-Mnk1 (AF4488, Affinity), anti-p-eIF4E (AF3110, Affinity), and anti-RANKL (DF7006, Affinity), anti-EIF2AK2 (ab32052, Abcam, Shanghai, China), anti-eIF4E (ab33766, Abcam), anti-RUNX2 (ab236639, Abcam), anti-Mnk1 (10136-1-AP, Protech, Wuhan, China), anti-eIF4G (15704-1-AP, Protech), anti-β-actin (20536-1-AP, Protech).
Conditioned medium (CM) preparation
When the confluence reached 60–70%, the MDA-MB-231 and MDA-MB-468 cells were deprived of growth factors by overnight incubation in the presence of 0.1% serum before treatment with 10 ng/mL TGF-β1 for 20 h (Peprotech, Suzhou, China). The conditioned medium (CM) was prepared by filtering the supernatant medium through 0.22-µm of the filter.
TRAP staining for osteoclast differentiation
RAW264.7 cells were plated in 12-well plates (104 cells/well) and treated with RANKL (Peprotech, Suzhou, China; 50 ng/mL) for 3 days. Then, the cells were cultured in CM. Two days later, the RAW264.7 cells were fixed and stained for osteoclast-derived TRAP (tartrate-resistant acid phosphatase) (Wako, Osaka, Japan). Multinucleated cells that contained at least three nuclei were considered as TRAP-positive cells.
RNA pulldown and liquid chromatograph/mass spectrometer (LC/MS) analysis
FGF14-AS2 was transcribed in vitro using a MEGAscript™ T7 Transcription Kit (Thermo Fisher Scientific, Waltham, MA, USA) and then biotinylated using a Pierce RNA 3′ End Desthiobiotinylation Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. RNA pull-down assays were performed using a Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific). Briefly, biotinylated FGF14-AS2 was captured with streptavidin magnetic beads and incubated with cell lysates at 4 °C for 6 h. The mixture was washed, eluted and then separated using an SDS-polyacrylamide gel. Silver staining was performed using a Rapid Silver Staining Kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions. The lanes were extracted by cutting and then subjected to liquid chromatograph/mass spectrometer (LC/MS) analysis.
RNA immunoprecipitation (RIP) assay
RIP assay was performed according to a previously reported protocol [32]. The following primary antibodies were used: anti-N6-methyladenosine antibody (9010006, EpiGentek, Farmingdale, NY, USA), anti-EIF2AK2 antibody (ab32052, Abcam), anti-eIF4E antibody (ab33766, Abcam), or anti-YTHDF2 antibody (ab220163, Abcam).
Dual-luciferase reporter assay
The YTHDF2 or YTHDF2-5A-mut expression plasmids, together with psiCHECK2-FGF14-AS2 plasmids, were co-transfected into MDA-MB-231 cells using Lipofectamine 2000 Reagents (Invitrogen). At 48 h after transfection, the cells were lysed, and luciferase activities were measured using a Dual-Luciferase Reporter Assay Kit (Promega, Beijing, China).
Polysome profiling
Polysome profiling assay was performed according to a previously reported protocol [33].
Animal model construction
All animal procedures were approved by the Committee on the Ethics of Animal Experiments of Nanjing Medical University, Nanjing, China (Approval No: IACUC-1909010). Four-week-old female nude (SPF grade BALB/c) mice were obtained from Shanghai SLAC Laboratory Animal Co., Ltd. All mice were in good health status. Before the experiments, the mice were acclimatised to the new environment for one week. Mice were randomised into two groups (10 total, 5 in each group) before tumour cell inoculation (no repeat operation). The investigator was blinded to the group allocation of the animals during the experiment. Briefly, cells (5 × 105) in PBS (100 μl) were injected into the tibias of mice. Metastasis development was monitored by micro-CT. Five weeks later, the mice were sacrificed, and tibia tissues were used for subsequent qRT-PCR, western blotting, HE and immunohistochemical analysis.
Bone histological analysis
The tibias were fixed with 10% neutral formalin for 24 h, decalcified with 10% EDTA for 4 weeks, dehydrated by a graded alcohol series, and embedded in paraffin. Tibia tissue sections (5 μm) were stained with haematoxylin and eosin using standard protocols. For immunohistochemical studies, the sections were incubated with primary antibodies at 4 °C overnight and then incubated with secondary antibodies at 37 °C for 1 h. Signals were visualised using the chromogen 3′-diaminobenzidine (DAB), and then the sections were counterstained with haematoxylin. For TRAP staining, osteoclasts were detected using a TRAP staining kit (Wako) according to the manufacturer’s protocol.
Tissue samples
Thirty-nine pairs of BCa and adjacent normal tissues were collected from the First Affiliated Hospital of Nanjing Medical University. None of the patients had undergone treatment prior to surgery. The clinicopathological characteristics for all of the patients are shown in Supplementary Table S3. The study was approved by the Human Research Ethics Committee of Nanjing Medical University, and written consent was obtained from each patient enrolled in the study.
Statistical analysis
All data are expressed as the mean ± standard deviation. Comparisons between two groups were analysed using Student’s t test (two-tailed) or the Wilcoxon signed-rank test. The relationship between YTHDF2 and FGF14-AS2 expression levels was assessed using Spearman correlation analysis. Kaplan–Meier analysis with log-rank test was applied for survival analysis using the online bioinformatics tool (http://kmplot.com/analysis/). The FPKM-normalised data (level-3) of YTHDF2 in breast cancer were downloaded from The Cancer Genome Atlas (TCGA) database and the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds/, GSE109169). Differential expression analysis on BCa osteolytic metastasis genes was performed using “limma” package with a filter as P < 0.05 and |log2 FC | > 1 (GSE137842, using R version 3.6.0). GraphPad Prism v6.01 (GraphPad Inc., La Jolla, CA, USA) was used for the statistical analysis and P value less than 0.05 was considered statistically significant. All statistical tests are justified as appropriate, and the data meet the assumptions of the tests. The variance is similar between the groups that are being statistically compared (Student’s t test).
Results
FGF14-AS2 suppresses osteolytic metastasis
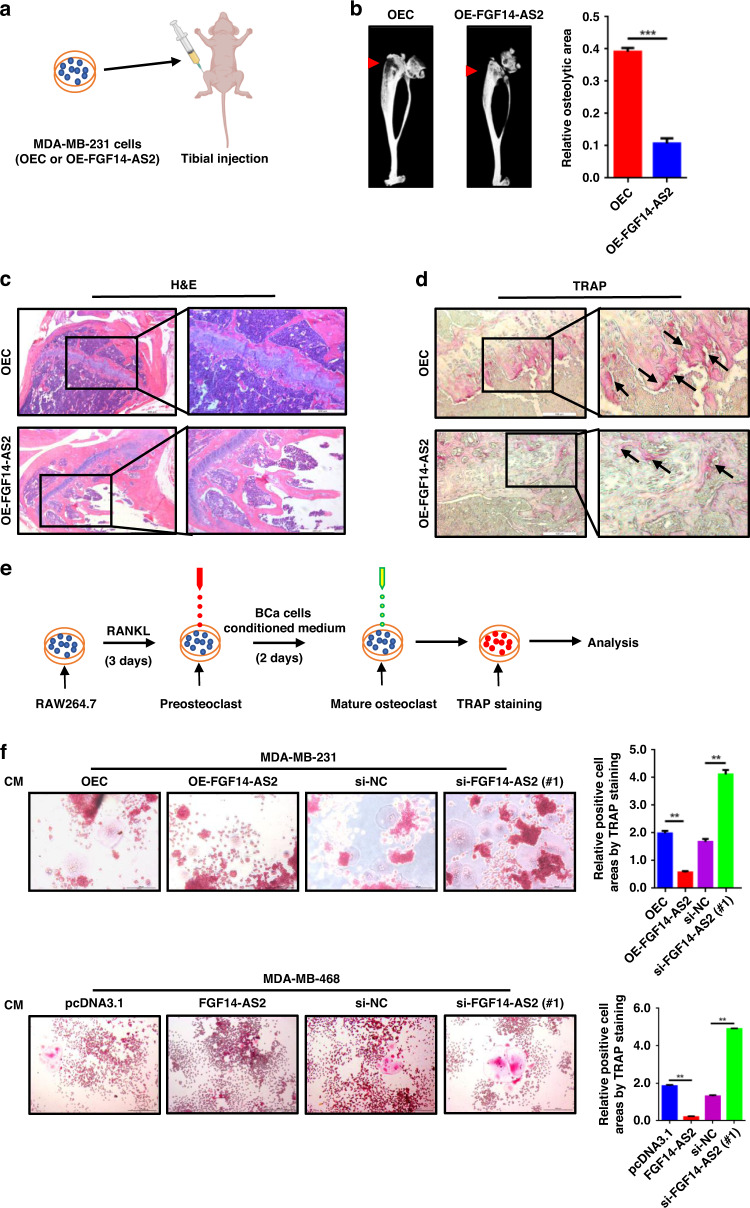
To explore the role of FGF14-AS2 in bone metastasis of BCa, we firstly established stable cell lines with FGF14-AS2 overexpression (FGF14-AS2 OE) and the vector control in MDA-MB-231 cells (Supplementary Fig. 1a), and then injected the cells into the tibiae of nude mice to mimic the later stage of BCa bone metastasis (Fig. 1a). Osteolysis was markedly weakened in the FGF14-AS2 OE mice as shown by micro-CT examination and H&E staining of tibial sections after five weeks since transplantation (Fig. 1b, c). Moreover, osteoclast activity was reduced in the tibiae of FGF14-AS2 OE mice compared with the control group, as shown by TRAP staining (arrows) (Fig. 1d). These results revealed that elevated expression of FGF14-AS2 in cancer cells markedly suppressed BCa osteolytic metastasis.
Fig. 1. FGF14-AS2 suppresses breast cancer osteolytic metastasis.
a FGF14-AS2-overexpressing MDA-MB-231 cells (OE-FGF14-AS2) and control cells (OEC) were injected into the tibiae of female nude mice. n = 5 for each group. b Representative micro-CT images of bone lesions in mice from each group (left panel) and quantitative analyses (right panel). c Representative images of H&E staining of the tibia sections. d Representative images of TRAP staining of the tibiae. e Schematic diagram of osteoclast differentiation of induced RAW264.7 cells. f RAW264.7 cells were stimulated with the indicated conditioned medium (CM) in the presence of RANKL (50 ng/mL). Cells were fixed and stained for TRAP activity (left panel), and the areas of TRAP-positive cells (multinucleated cells) were analysed (right panel). Scale bar = 200 µm. The data are shown as the mean ± s.d. of at least three independent experiments. **P < 0.005, ***P < 0.001.
To evaluate the function of FGF14-AS2 in osteoclast differentiation, we performed TRAP assays in RAW264.7 cells treated with CM from MDA-MB-231 and MDA-MB-468 cells with FGF14-AS2 overexpression (OE) or knockdown (KD) (Fig. 1e). Consistent with the strong inhibitory effect exhibited in vivo, CM from FGF14-AS2 OE cells dramatically inhibited the osteoclast differentiation of RAW264.7 cells. On the contrary, CM from FGF14-AS2 KD cells significantly enhanced the osteoclast differentiation (Fig. 1f). The overexpression and knockdown of FGF14-AS2 in MDA-MB-231 and MDA-MB-468 cells were confirmed by qRT-PCR (Supplementary Fig. 1a). These data suggest that FGF14-AS2 functions as a suppressor in osteoclast differentiation.
We also validated the specificity of the two siRNAs against FGF14-AS2 used in the experiments aforementioned to exclude the possible off-target effects. We constructed two plasmids containing FGF14-AS2 gene with point mutation within the respective regions targeted by the two siRNAs (Supplementary Fig. 1b). As shown in Supplementary Fig. 1c, FGF14-AS2 siRNAs efficiently reduced the expression levels of endogenous and exogenous FGF14-AS2, whereas, the two mutations were sufficient to fully abolish any siRNA knockdown of the expression level of the FGF14-AS2 (Supplementary Fig. 1d), indicating that the siRNAs were highly selective for endogenous FGF14-AS2.
FGF14-AS2 downregulates RUNX2/RANKL axis to suppress osteolytic metastasis
To identify the target gene of FGF14-AS2 in BCa osteolytic metastasis, we analysed the differentially expressed genes between primary tumour tissues (n = 3) and bone metastatic tissues of BCa patients (n = 3) from the GEO dataset (GSE137842). In total, 6097 differentially expressed mRNAs (P < 0.05 and |log2-fold change (FC) | > 1) were obtained (Supplementary Fig. 2a). RANKL/RANK signalling pathway plays an essential role in osteoclastogenesis, which is indispensable for osteolytic metastasis [10, 11]. Notably, these differentially expressed genes including RANKL were preferentially enriched in the signalling pathway involved in osteoclast differentiation (Supplementary Fig. 2b). Moreover, a high level of RANKL expression was significantly associated with worse DMFS (distant metastasis-free survival, DMFS) (Supplementary Fig. 2c).
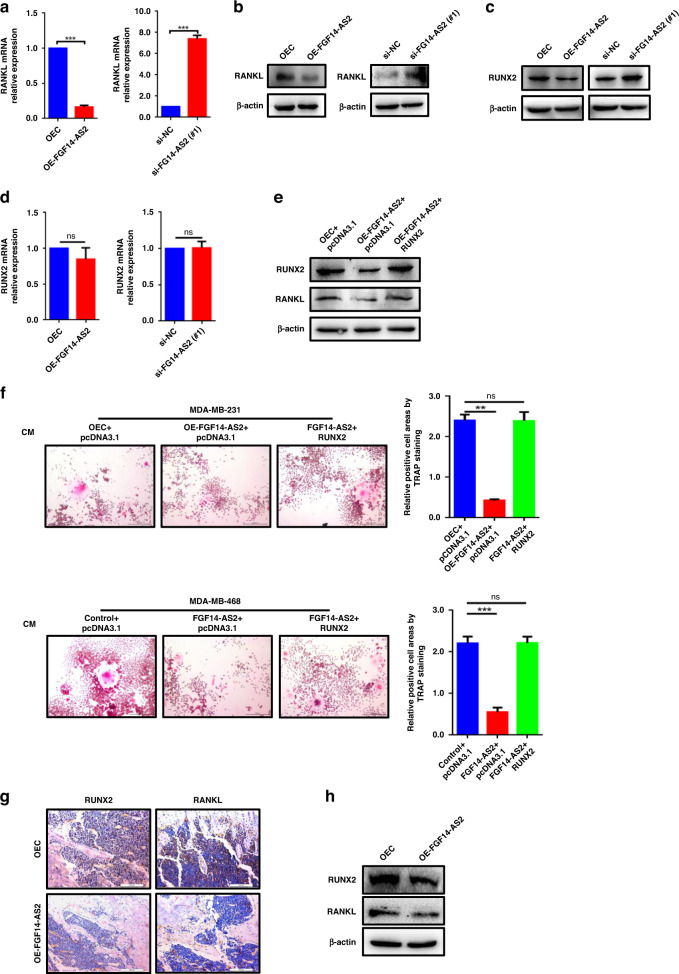
We then determined whether the expression of RANKL was regulated by FGF14-AS2 in breast cancer cells. As shown in Fig. 2a and b, the mRNA and protein levels of RANKL were markedly reduced upon FGF14-AS2 overexpression, and increased upon FGF14-AS2 knockdown. RUNX2, a key transcription factor in osteoblast differentiation, is an important contributor to BCa bone metastasis [34, 35]. Galea et al. reported that RUNX2 transcriptionally regulates RANKL expression by directly binding to the promoter of RANKL gene [12]. Intriguingly, we found that RUNX2 was downregulated in protein levels upon FGF14-AS2 overexpression, and upregulated upon FGF14-AS2 knockdown (Fig. 2c). However, FGF14-AS2 did not affect the mRNA levels of RUNX2 in MDA-MB-231 cells (Fig. 2d). These data suggest that FGF14-AS2 may regulate RUNX2 expression at the translational or posttranslational level. Furthermore, we demonstrated that overexpression of RUNX2 rescued the decrease of RANKL expression induced by FGF14-AS2 overexpression in MDA-MB-231 cells (Fig. 2e), suggesting that FGF14-AS2 downregulated RANKL expression through inhibiting RUNX2-mediated transcription in BCa cells.
Fig. 2. FGF14-AS2 suppresses osteolytic metastasis via the RUNX2/RANKL axis.
a, b RANKL levels in FGF14-AS2-overexpressing and FGF14-AS2-knockdown cells were detected using qRT‐PCR (a) and western blotting (b). c, d RUNX2 levels in FGF14-AS2-overexpressing and FGF14-AS2-knockdown cells were detected using western blotting (c) and qRT‐PCR (d). e RANKL levels in FGF14-AS2-overexpressing cells transfected with or without RUNX2 expression plasmids were detected using western blotting. f FGF14-AS2 expression plasmid was transiently transfected into MDA-MB-231 or MDA-MB-468 cells together with RUNX2 expression plasmid or pcDNA3.1 plasmid. RAW264.7 cells were stimulated with the indicated CM in the presence of RANKL (50 ng/mL). Cells were fixed and stained for TRAP activity (left), and the areas of TRAP-positive cells were analysed (right). Scale bar = 200 µm. g, h RUNX2 and RANKL levels in the tibiae of mice injected with FGF14-AS2 OE and control cells were detected by immunohistochemistry analysis (g) and western blotting (h). The data are shown as the mean ± s.d. of at least three independent experiments. **P < 0.005, ***P < 0.001 and ns, no significance.
To further determine the role of FGF14-AS2/RUNX2/RANKL axis in the inhibition of osteoclast differentiation, we performed TRAP assays in RAW264.7 cells using CM collected from FGF14-AS2 OE cells with or without RUNX2 overexpression. As shown in Fig. 2f, overexpression of RUNX2 successfully rescued the decrease of osteoclast differentiation induced by FGF14-AS2 overexpression in MDA-MB-231 and MDA-MB-468 cells. Moreover, the downregulation of RUNX2 and RANKL was also observed in the tibiae of FGF14-AS2 OE mice by using immunohistochemistry (IHC) and western blot assays (Fig. 2g, h). Collectively, these data suggest that FGF14-AS2 suppresses osteolytic metastasis via the RUNX2/RANKL axis.
FGF14-AS2 downregulates RUNX2 expression through inhibiting its translation
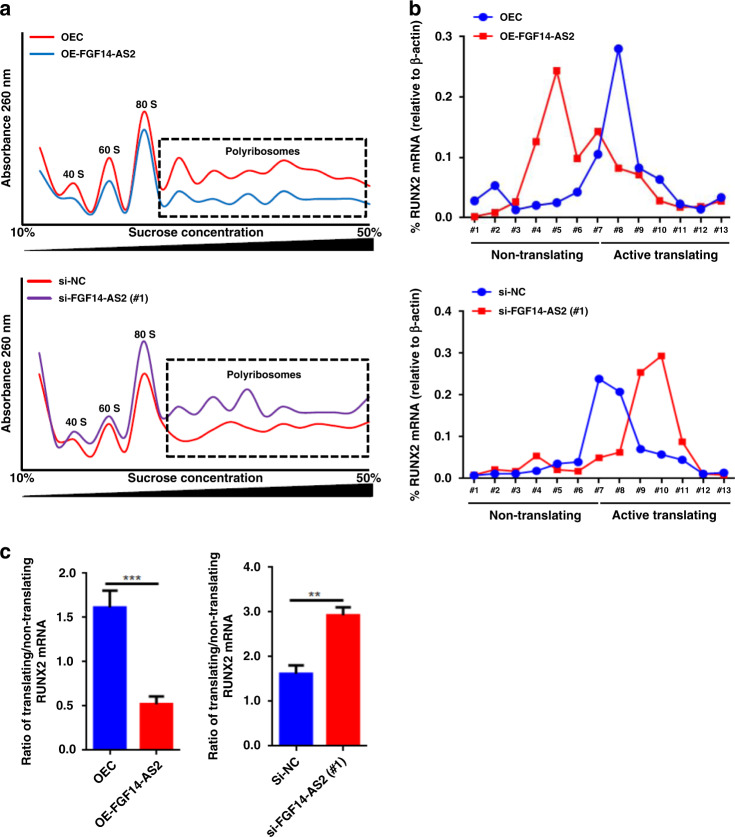
Next, we wonder whether FGF14-AS2 regulates the expression of RUNX2 at the translational level. To test this, polyribosome profiling followed by qRT-PCR was carried out using FGF14-AS2 OE and FGF14-AS2 KD cells. As shown in Fig. 3a, global cellular translation was slightly inhibited in FGF14-AS2 OE cells (A reduction of polyribosomes peak shown in the box) and stimulated in FGF14-AS2 KD cells (A rise of polyribosomes peak shown in the box), compared to the controls. However, the active translation of RUNX2 mRNA bound by heavy polyribosomes was significantly decreased in FGF14-AS2 OE cells, and increased in FGF14-AS2 KD cells (Fig. 3b). Meanwhile, qRT-PCR assays showed that the active translation of RUNX2 mRNA was decreased in FGF14-AS2 OE cells, and increased in FGF14-AS2 KD cells (Fig. 3c). Taken together, these data revealed that FGF14-AS2 decreased RUNX2 expression by inhibiting its mRNA translation.
Fig. 3. FGF14-AS2 inhibits RUNX2 translation.
a General polysome profiles in FGF14-AS2-overexpressing and FGF14-AS2-knockdown MDA-MB-231 cells. Polyribosomes within the box indicated by the dotted line represent active translation. b Percentage of RUNX2 mRNA in each fraction in FGF14-AS2-overexpressing and FGF14-AS2-knockdown MDA-MB-231 cells. Fractions #1–7 represent non-translated mRNA, whereas fractions #8–13 represent actively translated mRNA. c Ratios of actively translated RUNX2 mRNA to non-translated RUNX2 mRNA in FGF14-AS2-overexpressing and FGF14-AS2-knockdown MDA-MB-231 cells. Fractions #1–7 (representing non-translated RUNX2 mRNA group) were pooled, as were fractions #8–13 (representing translated RUNX2 mRNA group). The data are shown as the mean ± s.d. of at least three independent experiments. **P < 0.005, ***P < 0.001.
FGF14-AS2 interferes with the assembly of the eIF4G/eIF4E complex
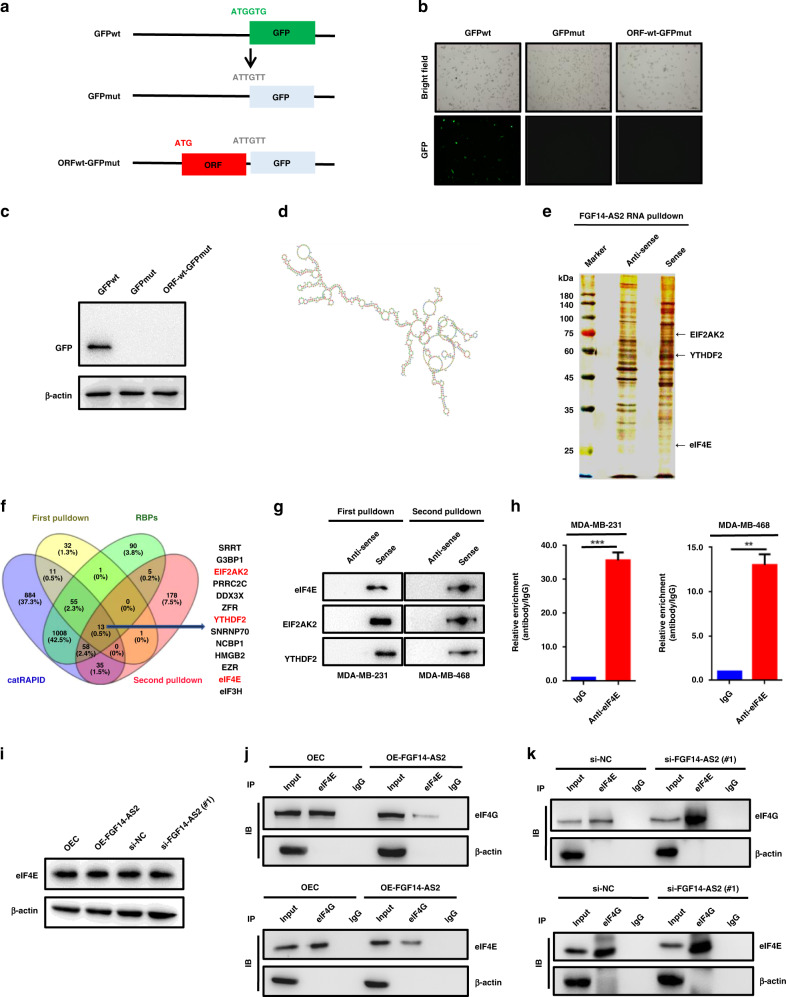
To reveal how FGF14-AS2 inhibits RUNX2 translation, we first determined whether FGF14-AS2 encodes a peptide. Potential ribosome occupancy of FGF14-AS2 was found using the Cistrome Data Browser database, although an ORF (342 nts) was predicted with no ribosome binding site using RegRNA 2.0 online software (Supplementary Fig. 3a, b). We then constructed a GFP fusion plasmid by inserting the potential ORF (339 bp, with stop codon deletion) of FGF14-AS2 into the upstream of GFP coding sequence with start codon mutated (Fig. 4a). We observed the expression of GFP in GFPwt-transfected MDA-MB-231 cells by fluorescence microscopy and western blot assays. However, no GFP was observed when the start codon of GFP was mutated. Notably, when the ORF of FGF14-AS2 was fused with GFPmut, no GFP expression was observed (Fig. 4b, c). Taken together, these data demonstrated that FGF14-AS2 is a noncoding RNA (ncRNA) without coding capacity. The functions of lncRNAs are usually determined by their structures [36]. Using omicX (a web server for RNA secondary structure prediction) analysis, we found that FGF14-AS2 has a complex secondary structure, implying its important role in regulating gene expression (Fig. 4d).
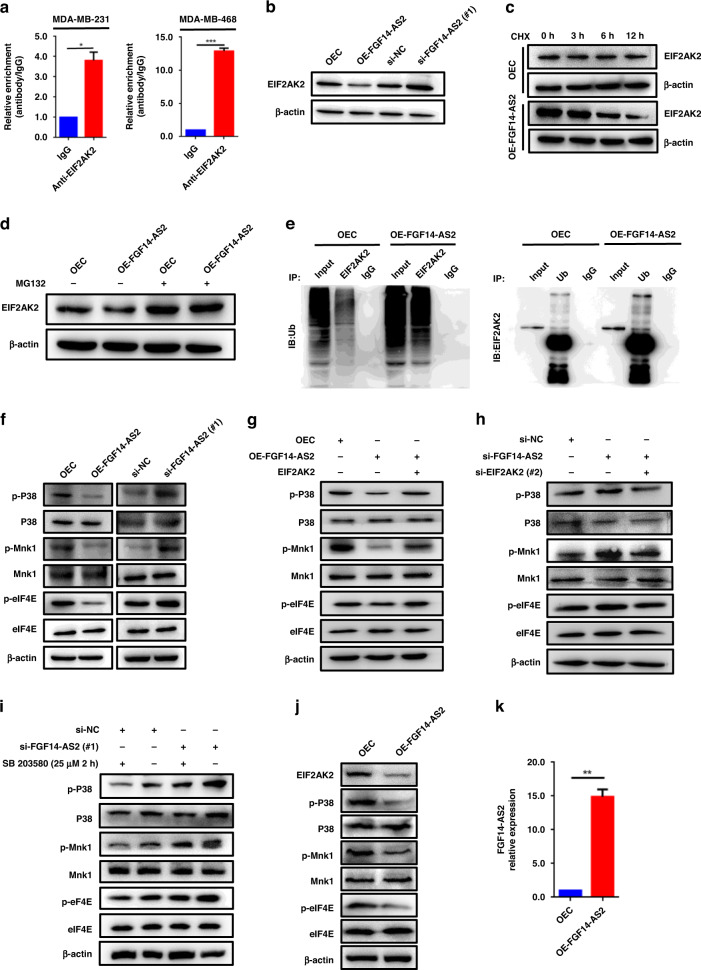
Fig. 4. FGF14-AS2 interferes with eIF4E/eIF4G complex formation.
a Schematic diagram of GFP constructs (GFPwt, GFPmut and ORFwt-GFPmut). b, c Expression of GFP was detected using fluorescence microscopy (b) and western blotting (c). d Secondary structure of FGF14-AS2 predicted by omicX (https://omictools.com/rnastructure-tool). e MDA-MB-231 cell lysates were incubated with biotin-labelled oligonucleotides, and the pull-down proteins were subjected to SDS-PAGE followed by silver staining. The lanes were extracted by cutting and then subjected to LC-MS analysis. f Venn diagram shows the 13 proteins potentially binding to FGF14-AS2 based on LC-MS data (two independent RNA pull-down assays), website predictions and classic RNA-binding protein dataset. g RNA pulldown followed by western blotting was performed with the indicated cell lysates and anti-eIF4E, anti-EIF2AK2 or anti-YTHDF2 antibodies. h RIP assay was performed using the indicated cell lysates and anti-eIF4E antibodies. The coprecipitated RNAs were subjected to qRT-PCR to detect FGF14-AS2. The fold enrichment of FGF14-AS2 in the eIF4E pellet is shown relative to its matching IgG control. i eIF4E levels in FGF14-AS2-overexpressing or FGF14-AS2-knockdown cells were detected using western blotting. j, k Co-IP assays were performed to detect the interaction between eIF4E and eIF4G in FGF14-AS2-overexpressing (j) and FGF14-AS2 knockdown cells (k). The data are shown as the mean ± s.d. of at least three independent experiments. ***P < 0.001.
Cytoplasmic lncRNAs usually exert their biological functions by binding to RNA-binding proteins (RBPs) to regulate mRNA stability, translation and protein stability, or acting as a miRNA sponge. Next, we sought to identify the proteins that interact with FGF14-AS2 using RNA pulldown combined with LC-MS/MS assays (Fig. 4e). A total of 13 candidate proteins were identified through the integration of our two independent RNA pull-down datasets, website predictions (catRAPID, http://service.tartaglialab.com/page/catrapid_omics2_group) and classic RNA-binding protein dataset (Fig. 4f and Supplementary Tables S4–7). eIF4E (eukaryotic translation initiation factor 4E) attracted our attention because it is a critical component of the translation initiation complex eIF4F. Besides, eIF4E acts as an important modulator of tumour growth and metastasis by mediating a rate-limiting process that drives the selective translation of many oncogenic proteins [37]. Then, the binding of eIF4E with FGF14-AS2 was validated by RNA pulldown followed by western blot assays with an anti-eIF4E antibody. As shown in Fig. 4g, labelled FGF14-AS2 sense RNA, but not antisense FGF14-AS2, specifically retrieved eIF4E from MDA-MB-231 and MDA-MB-468 cell extracts. Subsequent RIP assays further confirmed the physical interaction between eIF4E and FGF14-AS2 (Fig. 4h). However, neither FGF14-AS2 overexpression nor FGF14-AS2 knockdown had an obvious impact on the protein level of eIF4E (Fig. 4i). The binding of eIF4E to eIF4G is essential for the assembly of the initiation complex eIF4F. Interestingly, we found that the binding of eIF4E to eIF4G was reduced upon FGF14-AS2 overexpression (Fig. 4j), and enhanced upon FGF14-AS2 knockdown (Fig. 4k). Collectively, these data indicate that FGF14-AS2 interacts with eIF4E to abrogate the formation of the eIF4E/eIF4G complex, thereby inhibits RUNX2 translation.
FGF14-AS2 inhibits eIF4E phosphorylation via the EIF2AK2/p38 signalling pathway
Phosphorylated eIF4E has a higher binding affinity for the cap and contributes to form a more stable eIF4F complex. Another candidate, EIF2AK2, attracted our attention as well in the RNA pulldown followed LC-MS assays aforementioned (Fig. 4e, f), because (1) EIF2AK2 is an activator of the p38/Mnk1 signalling pathway; (2) eIF4E is phosphorylated by Mnk1 at Ser209. Thus, we hypothesised that FGF14-AS2 may interact with EIF2AK2 to regulate the p38/Mnk1 signalling pathway and subsequent phosphorylation of eIF4E. Firstly, we found that EIF2AK2 was presented in FGF14-AS2 sense RNA probe pull-down samples by western blotting (Fig. 4g). Moreover, RIP assays further confirmed the physical interaction between EIF2AK2 and FGF14-AS2 (Fig. 5a). Next, we examined the effect of FGF14-AS2 on the protein levels of EIF2AK2. As shown in Fig. 5b, EIF2AK2 protein levels were decreased upon FGF14-AS2 overexpression and increased upon FGF14-AS2 knockdown. Furthermore, we found that the degradation of EIF2AK2 protein was accelerated when FGF14-AS2 was overexpressed (Fig. 5c). Meanwhile, the proteasome inhibitor MG132 suppressed FGF14-AS2 overexpression-induced EIF2AK2 degradation (Fig. 5d). In addition, we found that overexpression of FGF14-AS2 significantly increased the ubiquitination level of EIF2AK2 (Fig. 5e). Taken together, these data revealed that the interaction of FGF14-AS2 with EIF2AK2 promoted the proteasome-dependent degradation of EIF2AK2.
Fig. 5. FGF14-AS2 inhibits eIF4E phosphorylation via the EIF2AK2/p38 signalling pathway.
a RIP assay was performed using indicated cell lysates and anti-EIF2AK2 antibodies, and the coprecipitated RNAs were subjected to qRT-PCR to detect FGF14-AS2. The fold enrichment of FGF14-AS2 in the EIF2AK2 pellet is shown relative to its matching IgG control. b EIF2AK2 levels in FGF14-AS2-overexpressing and FGF14-AS2-knockdown MDA-MB-231 cells were detected using western blotting. c, d EIF2AK2 levels in FGF14-AS2-overexpressing and control cells treated with Cycloheximide (CHX) (50 μg/ml) (c) or MG132 (25 µM, 4 h) (d) were detected using western blotting. e Co-IP assays were performed with either anti-EIF2AK2 or anti-ubiquitin (Ub) antibodies in FGF14-AS2-overexpressing and control cells. f p-p38, p38, p-Mnk1, Mnk1, p-eIF4E and eIF4E levels were examined using western blotting in MDA-MB-231 cells with FGF14-AS2 overexpression or knockdown. g, h p-p38, p38, p-Mnk1, Mnk1, p-eIF4E and eIF4E levels were examined using western blotting in MDA-MB-231 cells with both FGF14-AS2 and EIF2AK2 overexpression (g) and with both FGF14-AS2 and EIF2AK2 knockdown (h). i p-p38, p38, p-Mnk1, Mnk1, p-eIF4E and eIF4E levels were examined using western blotting in MDA-MB-231 cells with FGF14-AS2 knockdown and SB 203580 treatment (25 µM) for 2 h. j EIF2AK2, p-p38, p38, p-Mnk1, Mnk1, p-eIF4E and eIF4E AS2 levels in the tibiae of mice injected with FGF14-AS2 OE and control cells were detected by western blotting. k FGF14-AS2 levels in the tibiae of mice injected with FGF14-AS2 OE and control cells were detected by qRT-PCR. The data are shown as the mean ± s.d. of at least three independent experiments. *P < 0.05, **P < 0.005, ***P < 0.001.
Then, we examined the effect of FGF14-AS2 on the p38/Mnk1/eIF4E signalling pathway. As shown in Fig. 5f and Supplementary Fig. 4a, overexpression of FGF14-AS2 inhibited the phosphorylation of p38, Mnk1 and eIF4E in MDA-MB-231 and MDA-MB-468 cells, while FGF14-AS2 knockdown enhanced the phosphorylation of these proteins. Moreover, we found that overexpression of EIF2AK2 successfully rescued the decrease of the phosphorylation of p38, Mnk1 and eIF4E due to FGF14-AS2 overexpression (Fig. 5g and Supplementary Figs. 4b and 5a, b), while EIF2AK2 knockdown attenuated the enhanced phosphorylation of these proteins due to FGF14-AS2 knockdown (Fig. 5h and Supplementary Figs. 4c and 5a, b). Furthermore, we verified whether FGF14-AS2 inhibits the phosphorylation of eIF4E via the p38/Mnk1 signalling pathway. As shown in Fig. 5i and Supplementary Fig. 4d, the enhanced phosphorylation of p38, Mnk1 and eIF4E by FGF14-AS2 knockdown was weakened by treatment with the p38 MAPK inhibitor SB 203580. Finally, the decrease of EIF2AK2, p-P38, p-Mnk1 and p-eIF4E were observed in the tibiae of FGF14-AS2 OE mice by western blotting (Fig. 5j). qRT-PCR showed the overexpression of FGF14-AS2 in the tibiae of mice compared to the control group (Fig. 5k). Collectively, these data suggest that the interaction of FGF14-AS2 with EIF2AK2 promotes EIF2AK2 degradation, and consequently decreases eIF4E phosphorylation via the p38/Mnk1 signalling pathway. Therefore, in addition to interfering with eIF4E/eIF4G complex formation, FGF14-AS2 inhibits RUNX2 translation by influencing p38/Mnk1-mediated eIF4E phosphorylation.
m6A reader YTHDF2 mediates the degradation of FGF14-AS2
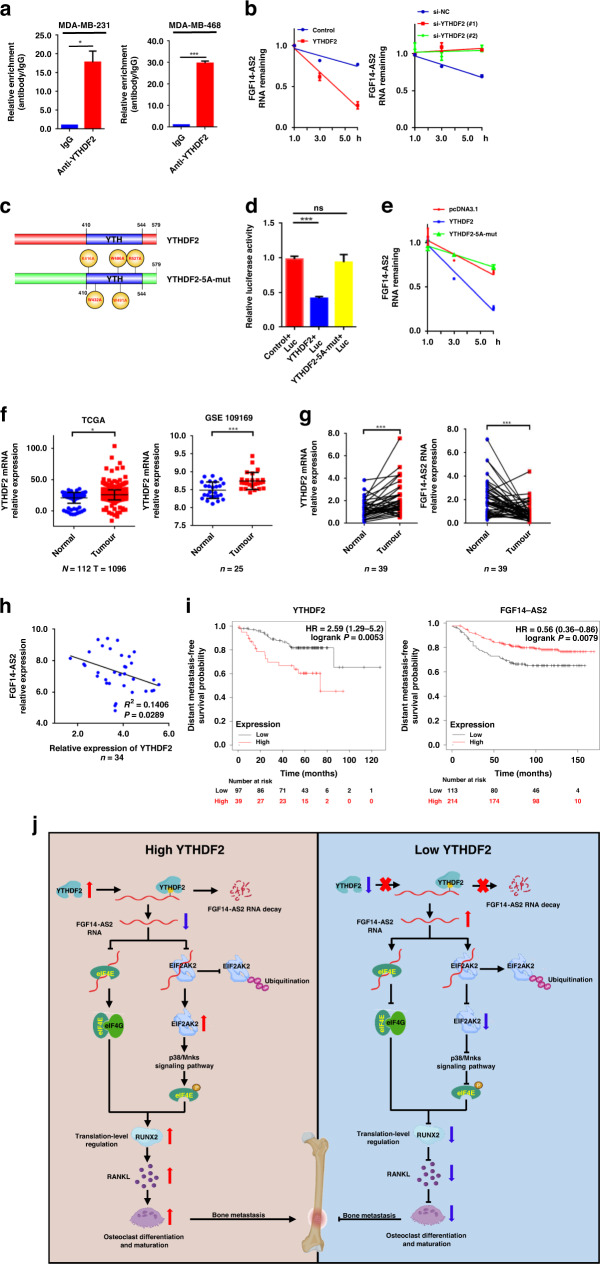
It has been reported that FGF14-AS2 is downregulated in BCa tissues [20, 38], albeit the mechanism is unclear. In the RNA pulldown followed LC-MS assays of FGF14-AS2 aforementioned, YTHDF2 was identified to interact with FGF14-AS2 (Fig. 4e, f). YTHDF2 is an m6A reader, which promotes the degradation of m6A-modified RNA. Therefore, we postulated that YTHDF2 might promote the degradation of m6A-modified FGF14-AS2 in BCa cells. To test this hypothesis, we firstly demonstrated the binding of FGF14-AS2 to YTHDF2. We found that labelled FGF14-AS2 sense RNA, but not antisense FGF14-AS2, specifically retrieved YTHDF2 from MDA-MB-231 and MDA-MB-468 extracts (Fig. 4g). In addition, RIP assays further confirmed the physical interaction between YTHDF2 and FGF14-AS2 (Fig. 6a). More importantly, we observed that overexpression of YTHDF2 promoted the rapid degradation of FGF14-AS2, on the contrary, YTHDF2 knockdown delayed the degradation of FGF14-AS2 (Fig. 6b).
Fig. 6. YTHDF2 mediates the downregulation of FGF14-AS2.
a RIP assay was performed using indicated cell lysates and anti-YTHDF2 antibodies, and the coprecipitated RNAs were subjected to qRT-PCR to detect FGF14-AS2. The fold enrichment of FGF14-AS2 in the YTHDF2 pellet is shown relative to its matching IgG control. b FGF14-AS2 levels were detected by qRT-PCR in MDA-MB-231 cells transfected with YTHDF2 expression plasmids, YTHDF2 siRNA and their respective controls in the presence of actinomycin D (2 μg/mL). c Schematic description of the YTHDF2 domains and mutant construction. d MDA-MB-231 cells were transiently transfected with luciferase reporter constructs containing m6A modification sites of FGF14-AS2 together with wild-type or mutated YTHDF2 expression plasmids (YTHDF2-5A-mut), and Firefly and Renilla luciferase activities were evaluated using the Dual-Luciferase Reporter Assay system (Promega). Normalised data were calculated as the quotient of Renilla/Firefly luciferase activities. e FGF14-AS2 levels were detected by qRT-PCR in MDA-MB-231 cells transfected with YTHDF2 wild-type, YTHDF2-5A-mut expression plasmids, or pcDNA3.1 plasmids (control) in the presence of actinomycin D (2 μg/mL). f Expression levels of YTHDF2 in breast cancer tissues and normal tissues were analysed using TCGA and GSE109169 datasets. g YTHDF2 and FGF14-AS2 levels in 39 pairs of breast cancer tissues and corresponding normal tissues were examined using qRT-PCR, respectively. h Spearman correlation analysis of the association between YTHDF2 and FGF14-AS2 expression in 39 breast cancer tissues. i Kaplan–Meier analysis of the correlation of the expression levels of YTHDF2 and FGF14-AS2 with distant metastasis-free survival (DMFS) based on TCGA data. The data are shown as the mean ± s.d. of at least three independent experiments. *P < 0.05, **P < 0.005, ***P < 0.001 and ns, no significance. j Diagram of the proposed mechanism showing the role of FGF14-AS2 as a suppressor in BCa osteolytic metastasis. The upregulated YTHDF2 promotes the degradation of m6A-modified FGF14-AS2. The decrease of FGF14-AS2 promotes eIF4E/eIF4G complex formation and inhibits the ubiquitination-mediated degradation of EIF2AK2, which leads to the increase of eIF4E phosphorylation via the p38/Mnk1 pathway. The enhanced eIF4E/eIF4G complex formation and eIF4E phosphorylation results in an increase in RUNX2 mRNA translation, thereby increasing RANKL transcription and eventually promoting osteoclast differentiation and bone metastasis of BCa.
Next, we proceeded to determine whether FGF14-AS2 is m6A-modified. The presence of m6A modification in FGF14-AS2 was predicted using m6A2Target (Supplementary Fig. 6a). meRIP-qPCR assay further confirmed that FGF14-AS2 was m6A modified (Supplementary Fig. 6b). To identify the m6A modification sites in the FGF14-AS2 sequence, we performed dual-luciferase reporter assays. SRAMP prediction server analysis revealed that FGF14-AS2 possessed m6A consensus sequences (GGACU, GGACA and AGACU) (Supplementary Fig. 6c). As shown in Supplementary Fig. 6d, e, two of the four sites (red) with high confidence in prediction were identified as m6A methylation sites. Additionally, the m6A-modified RNA levels were significantly higher in BCa tissues and cells than in normal breast tissues and cells by dot blot assays (Supplementary Fig. 6f, g).
Then we sought to determine whether the YTHDF2-induced downregulation of FGF14-AS2 depends on the m6A-modification of FGF14-AS2. It has been shown that K416 and R527 in the YTH domain of YTHDF2 are essential for binding of the RNA backbone and that W432, W486 and W491 are responsible for the recognition of m6A modification sites [39, 40]. Therefore, we constructed YTHDF2 expression plasmid containing K416A/R527A/W432A/W486A/W491A 5 A mutations (Fig. 6c). As shown in Fig. 6d, wild-type YTHDF2, but not mutated YTHDF2, markedly decreased the luciferase activity of FGF14-AS2 constructs. Simultaneously, we found that mutated YTHDF2 had no obvious effect on the degradation of FGF14-AS2 (Fig. 6e). Together, these data suggest that FGF14-AS2 expression is downregulated in BCa through YTHDF2-mediated degradation of m6A-modified FGF14-AS2.
Finally, we determined the clinical value of YTHDF2 and FGF14-AS2 dysregulation in human BCa tissues. As shown in Fig. 6f, YTHDF2 mRNA levels were significantly upregulated in BCa patients according to The Cancer Genome Atlas (TCGA) and GEO dataset analysis. Moreover, we detected the expression of FGF14-AS2 and YTHDF2 in our own cohort of 39 BCa tissues and paired adjacent normal tissues using qRT-PCR. We found that YTHDF2 was significantly upregulated in BCa tissues, while FGF14-AS2 was downregulated (Fig. 6g). Furthermore, we found that the expression level of FGF14-AS2 was negatively correlated with that of YTHDF2 in BCa tissues (R2 = 0.1406, P = 0.0289) (Fig. 6h). Notably, patients with high YTHDF2 and low FGF14-AS2 levels had worse DMFS according to the Kaplan–Meier plot (Fig. 6i). Taken together, these data revealed that FGF14-AS2 was negatively regulated by YTHDF2 and that the dysregulation of FGF14-AS2 and YTHDF2 was correlated with the prognosis of patients with BCa.
Discussion
Osteolytic metastasis represents a frequent and severe complication of advanced BCa and is associated with pathological fractures, pain, hypercalcaemia, spinal cord compression, and decreased mobility [41]. The mechanism of BCa osteolytic metastasis remains poorly understood. In this study, we found that (1) FGF14-AS2 functions as a suppressor in BCa osteolytic metastasis; (2) FGF14-AS2 suppresses the translation of RUNX2 by inhibiting the assembly of eIF4E/eIF4G complex and the phosphorylation of eIF4E, thereby reducing the transcription of RANKL, the crucial regulator of osteoclast differentiation; (3) YTHDF2 mediates the degradation of m6A-modified FGF14-AS2 in BCa cells; (4) low levels of FGF14-AS2 was correlated with worse DMFS in BCa patients.
Despite an increasing number of lncRNAs were reported to play important roles in cancer initiation and progression [42], our understanding of lncRNAs in tumour bone metastasis is still very limited. Lang et al. reported that lncRNA PCAT7 was involved in the SMAD3/SP1 complex-mediated constitutive active loop and promoted prostate bone metastasis [43]. Lnc34a promoted the bone metastasis of hepatocellular carcinoma by epigenetically suppressing miR-34a expression [44]. LncRNA MAYA promoted BCa bone metastasis by mediating the crosstalk between ROR1-HER3 and the Hippo-YAP pathway [45]. In this study, we revealed that lncRNA FGF14-AS2 suppresses BCa osteolytic metastasis via the RUNX2/RANKL axis, indicating that FGF14-AS2 functions as a tumour suppressor in osteolytic metastasis.
eIF4E is an important component of eIF4F (which also consists of a scaffold protein eIF4G and an RNA helicase, eIF4A) [46]. The binding of eIF4E to eIF4G and the phosphorylation of eIF4E are crucial steps in translation initiation [47]. Notably, eIF4E mediates a rate-limiting process that drives selective translation of many oncogenic proteins such as cyclin D1, survivin and VEGF, thereby contributing to tumour growth, metastasis and therapy resistance [37]. RUNX2 is a critical transcription factor for the upregulation of RANKL expression [12]. In this study, we found that FGF12-AS2 suppressed RUNX2 translation, and consequently inhibited RANKL expression in BCa cells. Furthermore, we uncovered that eIF4E and EIF2AK2 are FGF14-AS2 binding proteins using RNA pulldown combined with LC-MS/MS assays. FGF14-AS2 impeded eIF4G/eIF4E complex formation by interacting with eIF4E. EIF2AK2 is an important activator of the p38/MAPK signalling pathway, and excessive activation of p38 phosphorylates eIF4E via Mnk1 [48]. In addition, we found that the interaction of FGF14-AS2 with EIF2AK2 promoted the proteasome-dependent degradation of EIF2AK2, thereby inhibiting the p38 signalling pathway and eIF4E phosphorylation. Therefore, our data suggest that FGF14-AS2 suppressed the translation of RUNX2 by reducing eIF4E/eIF4G complex formation and eIF4E phosphorylation, thereby inhibiting RANKL transcription.
In recent years, m6A modification has attracted much attention because of its pivotal role in the regulation of various stages of the RNA life cycle, such as RNA processing, nuclear export, and translation. In addition, it has been reported that some lncRNAs are m6A modified. For instance, Wu et al. demonstrated that m6A modification increased the accumulation of RP11 in the nucleus [49]. Zuo et al. reported that m6A modification led to an increase in the stability of LINC00958 RNA [50]. Here, we demonstrated that FGF14-AS2 was m6A modified using meRIP-qPCR assay. Moreover, YTHDF2, which is an FGF14-AS2 binding protein identified by RNA pull-down and RIP assays, significantly accelerated the degradation of FGF14-AS2. Therefore, YTHDF2-mediated degradation of FGF14-AS2 is at least partially responsible for the downregulation of FGF14-AS2 in BCa.
In summary, our results revealed that FGF14-AS2 suppresses RUNX2 translation by reducing eIF4E/eIF4G complex formation and eIF4E phosphorylation, thereby inhibits RANKL transcription and osteolytic metastasis of BCa. Decreased FGF14-AS2 caused by YTHDF2-mediated degradation is significantly correlated with poor prognosis in BCa patients (Fig. 6j). Our findings suggested that FGF14-AS2 might serve as a promising prognostic biomarker and therapeutic target for BCa bone metastasis.
Supplementary information
Author contributions
MZ, MX and YT performed the experiments. WJ provided the clinical samples. MZ performed the statistical analysis. YM and YJ performed the bioinformatic analyses. HW and QZ reviewed the data. LL, BY, YJ and CM designed the research and drafted the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81872389, 81570804 and 82072484); the Priority Academic Program Development of Jiangsu Higher Education Institutions, and the Key Project of Science; the Major Projects of Science and Technology Development Fund of Nanjing Medical University (No. NMUD2019004).
Data availability
All the data that support the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Ethics Committee of Nanjing Medical University (Approval number: 2018-487) and with the Declaration of Helsinki and its later amendments or comparable ethical standards, and all patients provided written informed consent.
Consent to publish
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-022-02006-y.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Solanki M, Visscher D. Pathology of breast cancer in the last half century. Hum Pathol. 2020;95:137–48. doi: 10.1016/j.humpath.2019.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Coleman RE, Croucher PI, Padhani AR, Clézardin P, Chow E, Fallon M, et al. Bone metastases. Nat Rev Dis Primers. 2020; 10.1038/s41572-020-00216-3. [DOI] [PubMed]
- 4.Liang Y, Zhang H, Song X, Yang Q. Metastatic heterogeneity of breast cancer: molecular mechanism and potential therapeutic targets. Semin Cancer Biol. 2020;60:14–27. doi: 10.1016/j.semcancer.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Chen YC, Sosnoski DM, Mastro AM. Breast cancer metastasis to the bone: mechanisms of bone loss. Breast Cancer. Res. 2010; 10.1186/bcr2781. [DOI] [PMC free article] [PubMed]
- 6.Macedo F, Ladeira K, Pinho F, Saraiva N, Bonito N, Pinto L, et al. Bone metastases: an overview. Oncol Rev. 2017; 10.4081/oncol.2017.321. [DOI] [PMC free article] [PubMed]
- 7.Pathi SP, Lin DD, Dorvee JR, Estroff LA, Fischbach C. Hydroxyapatite nanoparticle-containing scaffolds for the study of breast cancer bone metastasis. Biomaterials. 2011;32:5112–22. doi: 10.1016/j.biomaterials.2011.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Futakuchi M, Fukamachi K, Suzui M. Heterogeneity of tumor cells in the bone microenvironment: mechanisms and therapeutic targets for bone metastasis of prostate or breast cancer. Adv Drug Deliv Rev. 2016;99:206–11. doi: 10.1016/j.addr.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 9.Schramek D, Sigl V, Penninger JM. RANKL and RANK in sex hormone-induced breast cancer and breast cancer metastasis. Trends Endocrinol Metab. 2011;22:188–94. doi: 10.1016/j.tem.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 10.Azim H, Azim HA., Jr Targeting RANKL in breast cancer: bone metastasis and beyond. Expert Rev Anticancer Ther. 2013;13:195–201. doi: 10.1586/era.12.177. [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Li F, Dang L, Liang C, Lu A, Zhang G. RANKL/RANK system-based mechanism for breast cancer bone metastasis and related therapeutic strategies. Front Cell Dev Biol. 2020; 10.3389/fcell.2020.00076. [DOI] [PMC free article] [PubMed]
- 12.Galea GL, Paradise CR, Meakin LB, Camilleri ET, Taipaleenmaki H, Stein GS, et al. Mechanical strain-mediated reduction in RANKL expression is associated with RUNX2 and BRD2. Gene. 2020; 10.1016/j.gene.2020.100027. [DOI] [PubMed]
- 13.Barnes GL, Hebert KE, Kamal M, Javed A, Einhorn TA, Lian JB, et al. Fidelity of Runx2 activity in breast cancer cells is required for the generation of metastases-associated osteolytic disease. Cancer Res. 2004;64:4506–13. doi: 10.1158/0008-5472.CAN-03-3851. [DOI] [PubMed] [Google Scholar]
- 14.Javed A, Barnes GL, Pratap J, Antkowiak T, Gerstenfeld LC, van Wijnen AJ, et al. Impaired intranuclear trafficking of Runx2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo. Proc Natl Acad Sci USA. 2005;102:1454–9. doi: 10.1073/pnas.0409121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi SW, Kim HW, Nam JW. The small peptide world in long noncoding RNAs. Brief Bioinform. 2019;20:1853–64. doi: 10.1093/bib/bby055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y, Ren W, Li Q, Duan C, Lin X, Bi Z, et al. LncRNA Uc003xsl.1-mediated activation of the NFκB/IL8 axis promotes progression of triple-negative breast cancer. Cancer Res. 2022;82:556–70. doi: 10.1158/0008-5472.CAN-21-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu Z, Huo N, Zhu X, Liu H, Cong R, Ma L, et al. FOXO3A-induced LINC00926 suppresses breast tumor growth and metastasis through inhibition of PGK1-mediated Warburg effect. Mol Ther. 2021;29:2737–53. doi: 10.1016/j.ymthe.2021.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajagopal T, Talluri S, Akshaya RL, Dunna NR. HOTAIR LncRNA: a novel oncogenic propellant in human cancer. Clin Chim Acta. 2020;503:1–18. doi: 10.1016/j.cca.2019.12.028. [DOI] [PubMed] [Google Scholar]
- 19.Ghafouri-Fard S, Dashti S, Taheri M. PCAT1: an oncogenic lncRNA in diverse cancers and a putative therapeutic target. Exp Mol Pathol. 2020; 10.1016/j.yexmp.2020.104429. [DOI] [PubMed]
- 20.Jin Y, Zhang M, Duan R, Yang J, Yang Y, Wang J, et al. Long noncoding RNA FGF14-AS2 inhibits breast cancer metastasis by regulating the miR-370-3p/FGF14 axis. Cell Death Discov. 2020; 10.1038/s41420-020-00334-7. [DOI] [PMC free article] [PubMed]
- 21.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–5. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Chem Biol. 2014;16:191–8. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Wu K, Quan W, Yu L, Chen S, Cheng C, et al. The dynamics of FTO binding and demethylation from the m(6)A motifs. Rna Biol. 2019;16:1179–89. doi: 10.1080/15476286.2019.1621120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315–28. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng J, Zhang J, Ye Y, Liu K, Zeng L, Huang J, et al. N(6) -methyladenosine-mediated upregulation of WTAPP1 promotes WTAP translation and Wnt signaling to facilitate pancreatic cancer progression. Cancer Res. 2021;81:5268–83. doi: 10.1158/0008-5472.CAN-21-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B, Zhu L, Lu C, Wang C, Wang H, Jin H, et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat Commun. 2021; 10.1038/s41467-020-20527-z. [DOI] [PMC free article] [PubMed]
- 28.Chen Y, Lin Y, Shu Y, He J, Gao W. Interaction between N(6)-methyladenosine (m(6)A) modification and noncoding RNAs in cancer. Mol Cancer. 2020; 10.1186/s12943-020-01207-4. [DOI] [PMC free article] [PubMed]
- 29.Lan Q, Liu PY, Haase J, Bell JL, Hüttelmaier S, Liu T. The critical role of RNA m(6)A methylation in cancer. Cancer Res. 2019;79:1285–92. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69:1193–205. doi: 10.1136/gutjnl-2019-319639. [DOI] [PubMed] [Google Scholar]
- 31.Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020;48:3816–31. doi: 10.1093/nar/gkaa048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Zhang M, Yang D, Ma Y, Tang Y, Xing M, et al. m(6)A-mediated upregulation of AC008 promotes osteoarthritis progression through the miR-328-3p‒AQP1/ANKH axis. Exp Mol Med. 2021;53:1723–34. doi: 10.1038/s12276-021-00696-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lou WP, Baser A, Klußmann S, Martin-Villalba A. In vivo interrogation of central nervous system translatome by polyribosome fractionation. J Vis Exp. 2014; 10.3791/51255. [DOI] [PMC free article] [PubMed]
- 34.Shore P. A role for Runx2 in normal mammary gland and breast cancer bone metastasis. J Cell Biochem. 2005;96:484–9. doi: 10.1002/jcb.20557. [DOI] [PubMed] [Google Scholar]
- 35.Li XQ, Lu JT, Tan CC, Wang QS, Feng YM. RUNX2 promotes breast cancer bone metastasis by increasing integrin α5-mediated colonization. Cancer Lett. 2016;380:78–86. doi: 10.1016/j.canlet.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Novikova IV, Hennelly SP, Sanbonmatsu KY. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 2012;40:5034–51. doi: 10.1093/nar/gks071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu C, Makala L, Wu D, Cai Y. Targeting translation: eIF4E as an emerging anticancer drug target. Expert Rev Mol Med. 2016; 10.1017/erm.2015.20. [DOI] [PubMed]
- 38.Yang Y, Xun N, Wu JG. Long non-coding RNA FGF14-AS2 represses proliferation, migration, invasion, and induces apoptosis in breast cancer by sponging miR-205-5p. Eur Rev Med Pharmacol Sci. 2019;23:6971–82. doi: 10.26355/eurrev_201908_18737. [DOI] [PubMed] [Google Scholar]
- 39.Li F, Zhao D, Wu J, Shi Y. Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res. 2014;24:1490–2. doi: 10.1038/cr.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu T, Roundtree IA, Wang P, Wang X, Wang L, Sun C, et al. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 2014;24:1493–6. doi: 10.1038/cr.2014.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tiedemann K, Hussein O, Komarova SV. Role of altered metabolic microenvironment in osteolytic metastasis. Front Cell Dev Biol. 2020; 10.3389/fcell.2020.00435. [DOI] [PMC free article] [PubMed]
- 42.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–81. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lang C, Dai Y, Wu Z, Yang Q, He S, Zhang X, et al. SMAD3/SP1 complex-mediated constitutive active loop between lncRNA PCAT7 and TGF-β signaling promotes prostate cancer bone metastasis. Mol Oncol. 2020;14:808–28. doi: 10.1002/1878-0261.12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L, Niu H, Ma J, Yuan BY, Chen YH, Zhuang Y, et al. The molecular mechanism of LncRNA34a-mediated regulation of bone metastasis in hepatocellular carcinoma. Mol Cancer. 2019; 10.1186/s12943-019-1044-9. [DOI] [PMC free article] [PubMed]
- 45.Li C, Wang S, Xing Z, Lin A, Liang K, Song J, et al. A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat Cell Biol. 2017;19:106–19. doi: 10.1038/ncb3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grüner S, Peter D, Weber R, Wohlbold L, Chung MY, Weichenrieder O, et al. The structures of eIF4E-eIF4G complexes reveal an extended interface to regulate translation initiation. Mol Cell. 2016;64:467–79. doi: 10.1016/j.molcel.2016.09.020. [DOI] [PubMed] [Google Scholar]
- 47.Bramham CR, Jensen KB, Proud CG. Tuning specific translation in cancer metastasis and synaptic memory: control at the MNK-eIF4E axis. Trends Biochem Sci. 2016;41:847–58. doi: 10.1016/j.tibs.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 48.Silva AM, Whitmore M, Xu Z, Jiang Z, Li X, Williams BR. Protein kinase R (PKR) interacts with and activates mitogen-activated protein kinase kinase 6 (MKK6) in response to double-stranded RNA stimulation. J Biol Chem. 2004;279:37670–6. doi: 10.1074/jbc.M406554200. [DOI] [PubMed] [Google Scholar]
- 49.Wu Y, Yang X, Chen Z, Tian L, Jiang G, Chen F, et al. m(6)A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer. 2019; 10.1186/s12943-019-1014-2. [DOI] [PMC free article] [PubMed]
- 50.Zuo X, Chen Z, Gao W, Zhang Y, Wang J, Wang J, et al. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol. 2020; 10.1186/s13045-019-0839-x. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data that support the findings of this study are available from the corresponding author upon reasonable request.