Abstract
This map is an update of the edition 9 map by Berlyn et al. (M. K. B. Berlyn, K. B. Low, and K. E. Rudd, p. 1715–1902, in F. C. Neidhardt et al., ed., Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2, 1996). It uses coordinates established by the completed sequence, expressed as 100 minutes for the entire circular map, and adds new genes discovered and established since 1996 and eliminates those shown to correspond to other known genes. The latter are included as synonyms. An alphabetical list of genes showing map location, synonyms, the protein or RNA product of the gene, phenotypes of mutants, and reference citations is provided. In addition to genes known to correspond to gene sequences, other genes, often older, that are described by phenotype and older mapping techniques and that have not been correlated with sequences are included.
Previously, Berlyn et al. (323) presented the traditional map, the EcoMap physical map, and a map by Singer and Low showing the distribution of the Gross-Singer transposon set around the chromosome. The map in this paper is a revision of that traditional map of Escherichia coli K-12, the linkage map of known genes and other functional sites (Fig. 1), and the physical map, EcoMap 10, of Kenneth Rudd is presented in the companion article (3763a).
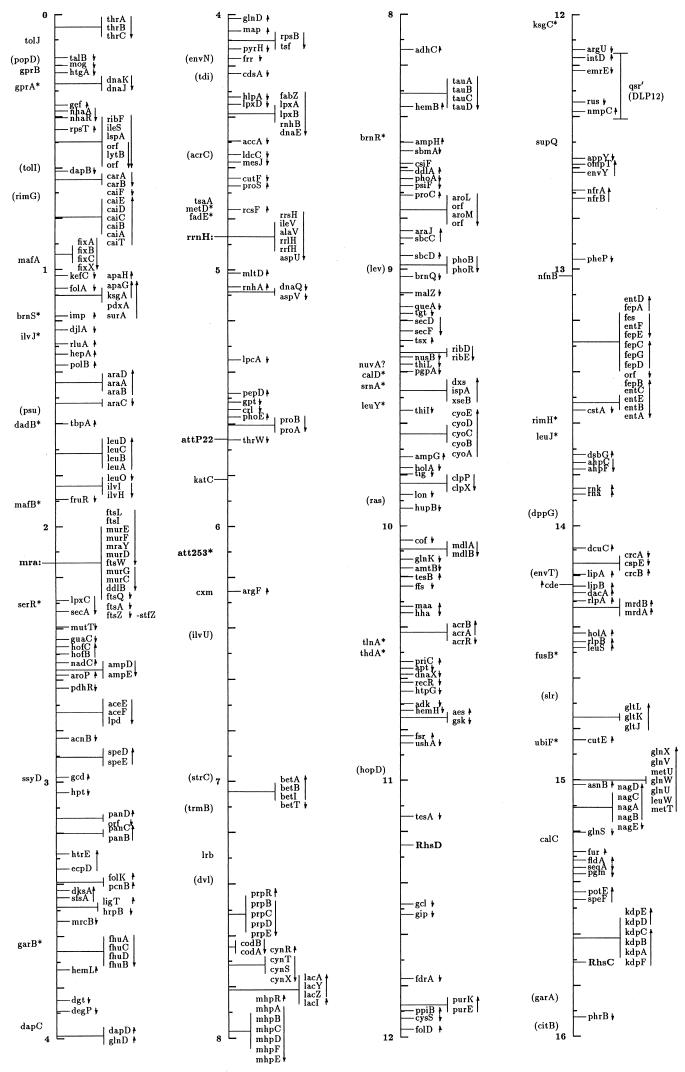
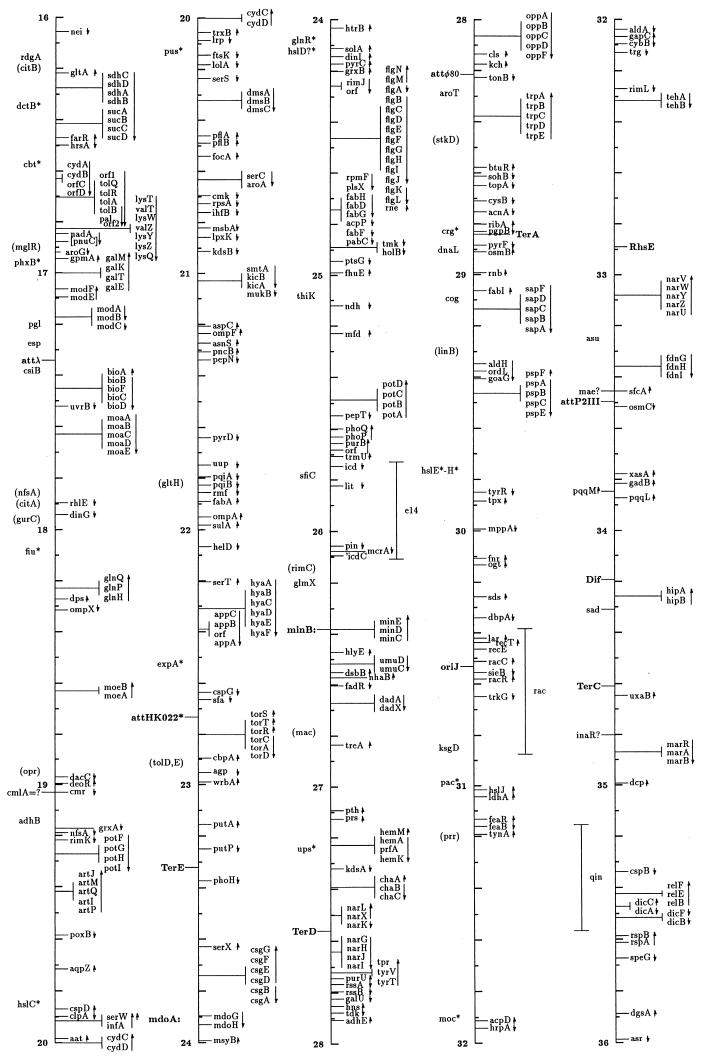
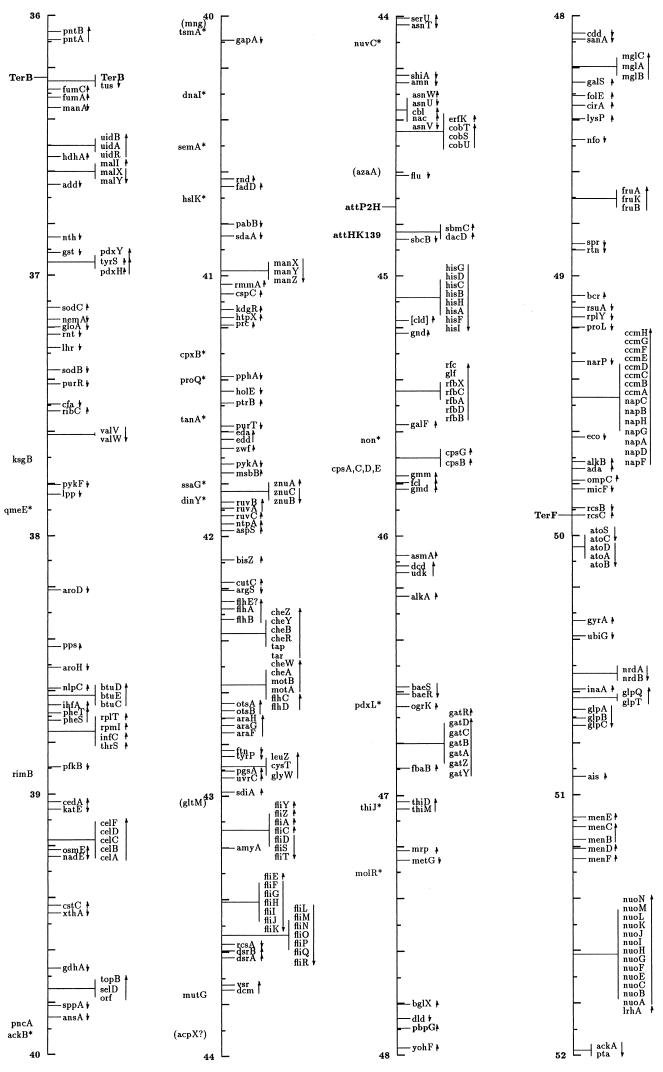
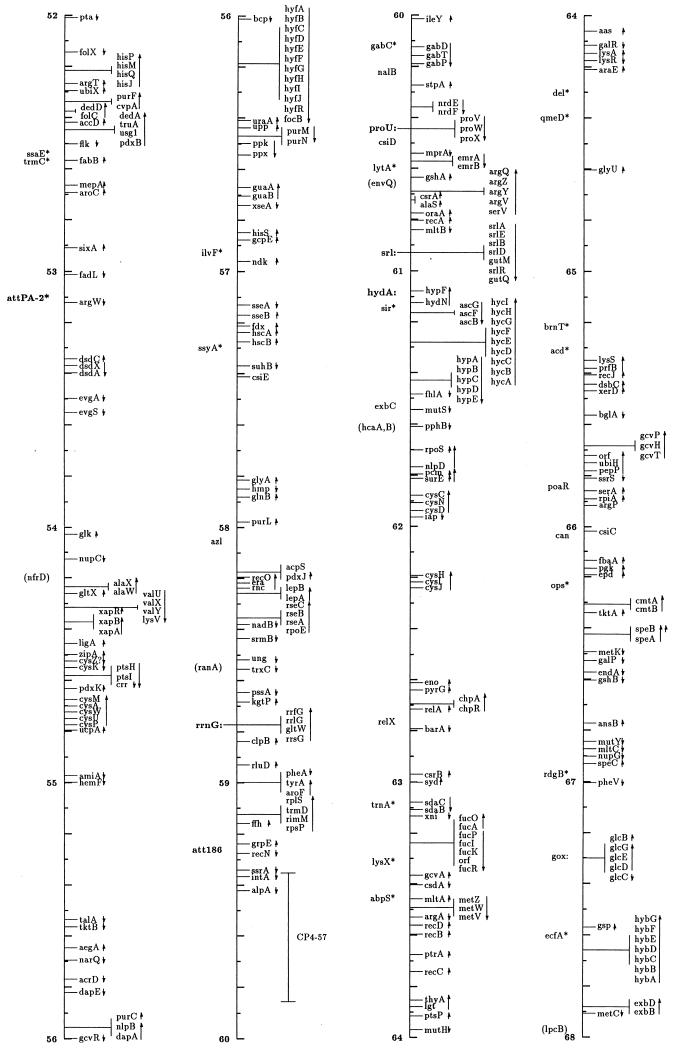
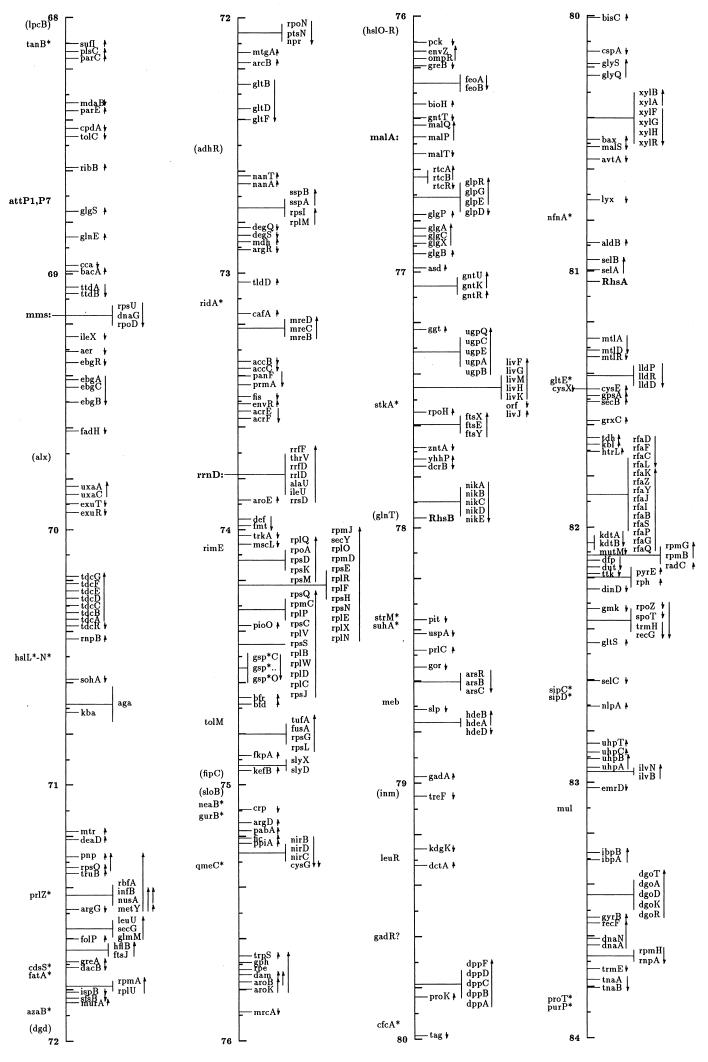
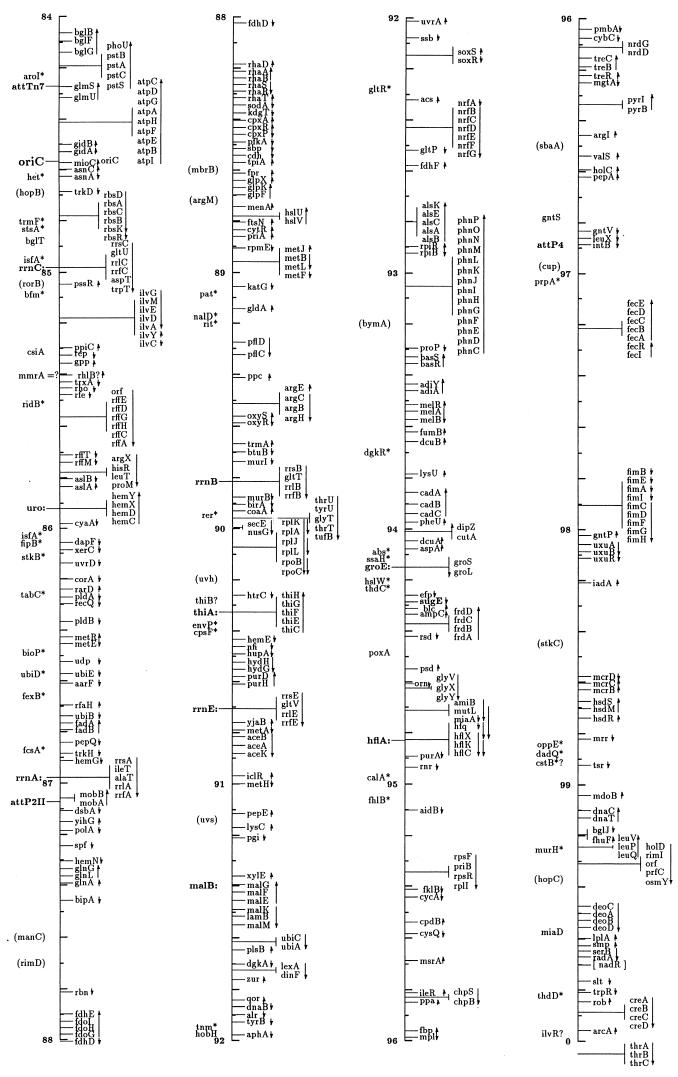
FIG. 1.
Linear drawing of circular linkage map of E. coli K-12. Symbols are defined in Table 1. Arrows show the direction of transcription. Where T-bars are used to display groups of genes, the length of the T shows the approximate length and position of the group in terms of the map coordinates, allowing visual ordering of closely packed groups.
The linkage map in this presentation includes genes located primarily by restriction, sequence, and cotransduction data reported in the literature and databases. It uses coordinates based on the complete sequence released by the Blattner laboratory. Obviously, the sequence is now the major resource for placing genes on the map. In some regions the placement represents a shift from the edition 9 map, which was based on coordinates of Rudd’s EcoMap 7 composite of sequenced genes and regions (27, 33, 395, 568, 569, 926, 3308, 3465, 4127, 4128), placed on the physical map of Escherichia coli (2291, 3763b) by restriction and sequence comparisons. Those map positions were based on the results in the literature and on EcoMap and GenBank database entries. EcoMap 10 coordinates are of course also based on the completed sequence, and cross-consulting this summary map and the EcoMap that follows should be straightforward.
The linkage map of Fig. 1 includes 2,220 genes and about 40 other chromosomal markers, such as phage attachment sites, defective-phage elements, replication origins and termini, and other features traditionally included on the published linkage map. It does not include open reading frames (ORFs) lacking evidence for expression, with unknown functions or putative functions inferred by sequence homologies only. A few exceptions occur for Salmonella genes where the inference is strong that they are also expressed in E. coli. The ORFs not included in this map can be found on EcoMap 10. The Fig. 1 map places the genes that can be found in sequence annotation and EcoMap 10 on the right side of the line. On the left side are genes not present on physical maps or the sequence, and in most cases these are not connected to a specific point on the axis to indicate that the localization is only approximate. As in previous editions of the E. coli linkage map (187, 188, 189, 190, 190a, 323, 4368, 4369, 4370, 4371), an asterisk indicates that the gene is not precisely located with respect to near neighbors and parentheses indicate that the location is even more uncertain and that the gene is located only within that general region. I have been very conservative about removing these from the map; even though the usefulness of some of these may be quite limited, there will probably be cases where the old, sometimes poorly characterized phenotype may be helpful in ascribing functions and phenotypic effects to ORFs. Also shown on the left side in boldface followed by colons are operon names that are distinct from any gene name within the operon and termination and attachment sites. The arrows indicate the direction of transcription and span genes included within a transcription unit.
Updates of map information are available in electronic form from several sites. These include the E. coli Genetic Stock Center’s (CGSC’s) World Wide Web server at URL http://cgsc.biology.yale.edu, which provides an interface for querying the database and retrieving formatted reports about genes, map regions, strains, and mutations, etc. (323a); the National Center for Biotechnology Information ftp site for EcoSeq and EcoMap, ncbi.nlm.nih.gov/repository/Eco/EcoMap7; the Colibri map at http://www.pasteur.fr/Bio/Colibri.html, the ECDC map at http://susi.bio.unigiessen.de/ecdc.html, the site for the sequencing project at the University of Wisconsin, http://www.genetics.wisc.edu, a gene-protein database, http://www.mbl.edu/html/ecoli.html, Genome Information Broker at http://mol.genes.nig.ac.jp/ecoli, and others. See also Rudd (3673). The references attempt to document map information, the basic definition of the gene’s function, and expression information and do not include information relating to detailed physical structure, active site in vitro mutagenesis, or enzyme mechanism. Earlier map papers contain additional references for some of the loci (188–190a, 323).
MAP UNITS
Since the 1976 recalibration of the linkage map in terms of minutes required for time of entry of markers in interrupted conjugation experiments, the standard representation of the map has used the basic units of minutes and a total length of 100 minutes (190a). This has been a convenient and accepted coordinate system for the map, and although the current map units are based on restriction and sequence data rather than time of entry, we retain the term minute for 1/100 of the length of the chromosome. Both the CGSC database and EcoMap use as “left endpoints” the counterclockwise boundary of the coding region, and genes in Fig. 1 are placed approximately at these coordinates, with the higher-resolution map of Rudd (3763a) providing more exact placement, showing nucleotide and minute coordinates for the physically mapped genes.
NOMENCLATURE
Gene Symbol Convention
The standard genetic nomenclature for E. coli is that of Demerec et al. (1016), as subsequently amended through use, and as described in Instructions to Authors for the Journal of Bacteriology (see also reference 3821). This map, like those preceding it, follows those nomenclatural conventions. Accordingly, we have adhered to a three-letter lowercase mnemonic symbol, with an uppercase letter added when there are two or more genes in that mnemonic category. If authors have added an uppercase letter for a gene in a single-instance category, we have used that published four-letter symbol. For attachment sites and noncoding features of the chromosome, etc., the same standard has not been used, and we have continued to use the variable-length symbols historically applied to these sites. We have continued the convention proposed for sites of termination of replication and repetitive sequences, by using italicized symbols with the first letter uppercase.
The Issue of Stability
Many names have been changed by investigators since the 1990 map was published. When those changes were part of a systematic revision of nomenclature (often aimed at clarifying usage and resolving conflicts) for a group of related genes and were in compliance with the current E. coli gene nomenclature system, or were changed for compelling mnemonic reasons or for resolution of redundancy or conflict, also in conformance with the standard system, we have adopted those changes. We have not adopted and we wish to discourage changes of valid preexisting names proposed by authors simply because they believe that theirs is a symbol signifying a more apt or accurate mnemonic. For example, a previously published name based on the pathway or phenotype is valid and should not, simply as a matter of course, be replaced by an alternate mnemonic based on the name of the enzyme that the gene codes for once that functional information has been determined. In general, the stability of a name has more value than improved nuances. In a few cases, we have been compelled to use a new name, despite the apparent validity of the original name, simply because the new name has been widely adopted in the literature. In a number of cases, a new gene has been assigned a symbol which has already been used or which is simultaneously proposed for another gene, with the two mnemonics having entirely different meanings. These names have had to be resolved, usually by changing the newer assignment. In a few cases, a uniquely named gene has been shown later to belong to a category for which a symbol already exists, and the latter symbol has been used instead of the earlier assignment. There is one case in this paper where use of a symbol already assigned to a different gene was strongly preferred by authors, and I was very reluctant to suggest a new symbol for the earlier, published gene name to the earlier authors, since that symbol has been used in a number of publications; for the interim I have broken convention to assign the newer genes temporary symbols with asterisks (gsp*), in order to show them on the map and in the hope of resolving that naming with the usual precedence custom in the near future. Some synonymy is unavoidable, since a gene under study may be named and described in print before its identity to a known gene is discovered. However, a common practice in the recent literature seems to allow publication of an author’s preliminary name for a gene even if its identity to a known gene has been discovered before publication, and that practice creates unnecessary synonymy. Alternate gene symbols are listed in Table 1, and Table 2 provides an alphabetized list of such symbols with cross-references to the symbols used in Table 1.
TABLE 1.
E. coli genes and replication- or phage-related sitesa
| Gene symbol | Map location (min) | Mnemonic for symbol | Synonyms and gene product—enzyme, RNA, or phenotype affected | CGSC no.d | Referencesc |
|---|---|---|---|---|---|
| aarF | 86.6 | Aminoglycoside acetyltransferase regulator | yigQR; regulator of 2′-N-acetyltransferase; involved in respiratory cofactor ubiquinone production | 53879 | 2696 |
| aas | 64.1 | Acyl-ACP synthase | 2-Acyl-glycerophosphoethanolamine acyltransferase; acyl-ACP synthetase; salvage pathway for reacylation; inner membrane; bifunctional for turnover/incorporation | 29780 | 1831, 1972 |
| aat | 20.0 | Amino acyl-tRNA-protein transferase | Aminoacyl-tRNA-protein-transferase (EC 2.3.2.6) | 1054 | 4045 |
| abpS | 63.5 | Arg binding protein | Low-affinity transport system for arginine and ornithine; periplasmic binding protein | 18562 | 664 |
| abs | 94.1 | Antibiotic sensitivity | Sensitivity and permeability to antibiotics and dyes | 18559 | 763 |
| accA | 4.5 | Acetyl-CoA carboxylase | Acetyl-CoA carboxylase α-carboxyltransferase subunit; (EC 6.4.1.2) | 29829 | 2536, 2537 |
| accB | 73.4 | Acetyl-CoA carboxylase | fabE; acetyl-CoA carboxylase, biotin carboxyl carrier protein (EC 6.4.1.2) | 796 | 2537, 2712, 3057, 4302, 4616 |
| accC | 73.4 | Acetyl-CoA carboxylase | fabG; acetyl-CoA carboxylase, biotin carboxylase (BC) subunit (EC 6.4.1.2) | 29834 | 2315, 2537, 3253 |
| accD | 52.4 | Acetyl-CoA carboxylase | dedB, usg; acetyl-CoA carboxylase β-carboxyltransferase subunit (EC 6.4.1.2) | 28570 | 2534, 2537, 3081 |
| acd | 65.1 | Acetaldehyde-CoA deHase | Acetaldehyde-CoA dehydrogenase (EC 1.2.1.10) | 1053 | 764 |
| aceA | 90.8 | Acetate | icl; isocitrate lyase (EC 4.1.3.1); acetate utilization | 1052 | 2744, 2811, 3674, 754, 841 |
| aceB | 90.8 | Acetate | mas; malate synthase A (EC 4.1.3.2) | 1051 | 2744, 591, 592, 840, 841 |
| aceE | 2.7 | Acetate | aceE1; pyruvate dehydrogenase (decarboxylase component) E1p; (EC 1.2.4.1); acetate requirement | 1050 | 1553, 1555, 1556, 1558, 2934, 4150, 4204, 655 |
| aceF | 2.7 | Acetate | aceE2; pyruvate dehydrogenase (dihydrolipoyltransacetylase component) E2p (EC 1.6.4.3, EC 2.3.1.12); acetate requirement | 26530 | 1554, 2934, 655 |
| aceK | 90.9 | Acetate | Isocitrate dehydrogenase kinase/phosphatase | 17770 | 1343, 1897, 2274, 2418, 2419, 753, 842, 754 |
| ackA | 52.0 | Acetate kinase | Acetate kinase (EC 2.7.2.1); mutants fluoroacetate resistant | 1048 | 1548, 2094, 2512, 2812, 530, 224 |
| ackB | 39.9 | Acetate kinase | Acetate kinase activity* (EC 2.7.2.1) | 1047 | 3379 |
| acnA | 28.8 | Aconitase | Aconitase A (EC 4.2.1.2) | 28218 | 3542, 3543 |
| acnB | 2.8 | Aconitase | Aconitase B (EC 4.2.1.2) | 36955 | 1538, 2141, 474 |
| acpD | 31.9 | Acyl carrier protein | ACP phosphodiesterase | 52896 | 1256, 394 |
| acpP | 24.8 | Acyl carrier protein | ACP (acyl carrier protein) | 31871 | 2183, 3621 |
| acpS | 58.2 | Acyl carrier protein | dpj; holo-ACP synthase (EC 2.7.8.7) | 32953 | 2397, 2400, 2401, 3471, 4340 |
| acpX | 43.9 | Acyl carrier protein | acpS; originally thought to be holo-ACP synthase; perhaps cryptic second gene or regulator | 1046 | 3471 |
| acrA | 10.4 | Acridine | Mb, lir, mbl, mtcA, sipB; AcrAB efflux system effects Mar multiple resistance | 1045 | 1291, 1726, 2684, 2685, 3282, 3563, 4723, 808 |
| acrB | 10.4 | Acridine | acrE; AcrAB system has major role in Mar multiple resistance to NAL, TET, AMP, etc.; beware renamings of acrE, acrB, and envC | 35806 | 1291, 2684, 2685, 3282, 4723 |
| acrC | 4.5 | Acridine | Sensitivity to acriflavine; transmembrane protein | 1044 | 3104 |
| acrD | 55.7 | Acridine | Sensitivity to acriflavine | 35697 | 3188 |
| acrE | 73.5 | Acridine | envC; anomolous cell division; chain formation; splits cross-wall to form new poles; see acrB | 813 | 2133, 2255, 2256, 2516, 3702 |
| acrF | 73.6 | Acridine | envD; encodes lipoprotein with signal peptide; osmotically remedial envelope defect | 33608 | 2255, 2256, 2516, 2684 |
| acrR | 10.5 | Acridine | Regulatory protein for acrA and acrB | 35809 | 2686 |
| acs | 92.3 | Acetyl-CoA synthetase | Acetyl CoA synthetase 2 (EC 6.2.1.1) | 34317 | 2367, 395 |
| ada | 49.7 | Adaptive (response) | O6-methylguanine-DNA methyltransferase, inducible; DNA repair against methylating and alkylating agents; transcription factor | 1043 | 1184, 2001, 2158, 2410, 2489, 2698, 2760, 2967, 3078, 3094, 3795, 3812, 3815, 3957, 3958, 3959, 4330, 4385 |
| add | 36.6 | Adenine deaminase | Adenosine deaminase (EC 3.5.4.4.); mutants affect growth on deoxyadenosine in purA, B mutants | 1042 | 2024, 765 |
| adhB | 19.1 | Alcohol dehydrogenase | Alcohol dehydrogenase | 36932 | E, 763a |
| adhC | 8.1 | Alcohol dehydrogenase | Alcohol-acetaldehyde dehydrogenase; adhC has also been used for adhE control region | 52901 | 2070, 394 |
| adhE | 27.9 | Alcohol/acetaldehyde dehydrogenase | ana; adhC; alcohol dehydrogenase, acetaldehyde dehydrogenase, CoA-linked (EC 1.1.1.1) allyl alcohol resistance; deactivase for PFL | 1041 | 129, 1457, 2203, 2495–2497, 2634, 710, 887, 1582 |
| adhR | 72.5 | Alcohol/acetaldehyde dehydrogenase | Regulatory gene for acd and adhE | 18556 | 723, 767 |
| adiA | 93.5 | Arginine decarboxylase, induced | adi; arginine decarboxylase, inducible by acid; homology with CadA, SpeC, SpeF | 34495 | 1665, 4022, 4023, 4024, 4223, 4224 |
| adiY | 93.4 | Arginine decarboxylase, induced | Sequence similarity with XylS/AraC family, including EnvY and AppY regulates adiA | 35597 | 4224 |
| adk | 10.7 | Adenylate kinase | dnaW, plsA; adenylate kinase (EC 2.7.4.3) pleiotropic effects on glycerol-3-phosphate acyltransferase activity | 1040 | 1176, 1725, 541 |
| aegA | 55.6 | Anaerobically expressed gene | air; induced by anaerobiosis, repressed by NO3; control mediated by Fnr, NarX, Q, L; nonessential in respiration, N source utilized | 50847 | 662 |
| aer | 69.3 | Aerotaxis | air; possibly flavoprotein, mediates positive aerotactic responses; signal transducer | 47273 | 347 |
| aes | 10.7 | Acetyl esterase | Esterase affecting maltose system expression | 53369 | 2114, 3393 |
| aga | 70.7 | Acetylgalactosamine | Cluster of putative N-acetylgalactosamine pathway genes, including the kba gene, and mannose permease homologs | 55305 | 3649 |
| agp | 23.0 | Acid glucose-1-phosphatase | Homology with appA; periplasmic | 31830 | 3521, 3524, 3525, 3526 |
| ahpC | 13.8 | Alkyl hydroperoxide | tpx; alkyl hydroperoxide reductase small subunit | 31190 | 4106, 4241, 667, 668 |
| ahpF | 13.8 | Alkyl hydroperoxide | Alkyl hydroperoxide reductase large subunit | 31194 | 4106, 4241, 668 |
| aidB | 95.1 | Alkylating agent induced | Adaptive response | 18553 | 2408, 2409, 4602, 4603 |
| ais | 50.9 | Aluminum inducible | An aluminum-inducible protein | 53490 | 1589 |
| alaS | 60.7 | Alanine | act, ala-act, lovB; alanyl-tRNA synthetase 1B (ligase) (EC 6.1.1.7) | 1039 | 2132, 2317, 3556, 3557, 3558, 458, 4590 |
| alaT | 87.0 | Alanine | talA; alanine tRNA 1B; rrnA operon | 1038 | 2313, 926 |
| alaU | 73.8 | Alanine | talD; alanine tRNA 1B; rrnD operon | 1037 | 2313 |
| alaV | 4.8 | Alanine | Alanine tRNA 1B; rrnH operon | 1036 | 1146 |
| alaW | 54.2 | Alanine | alaWα; alanine tRNA 2; tandemly duplicated; see alaX | 32851 | 539 |
| alaX | 54.2 | Alanine | alaWβ; alanine tRNA 2; tandemly duplicated alaW | 18547 | 2313, 539 |
| aldA | 32.0 | Aldehyde dehydrogenase | ald; aldehyde dehydrogenase, NAD linked | 17767 | 1749, 3571, 712 |
| aldB | 80.9 | Aldehyde dehydrogenase | Aldehyde dehydrogenase | 35668 | 4813, 4815 |
| aldH | 29.3 | Aldehyde dehydrogenase | Putative aldehyde dehydrogenase, by homology; transcribed in operon with goaG | 35320 | 1697, 2066 |
| alkA | 46.2 | Alkylation | aidA; 3-methyl-adenine DNA glycosylase II, inducible; repairs by single- and double-strand excision of 3-methyl adenine | 1035 | 1184, 3093, 3095, 365, 4385, 4602, 773 |
| alkB | 49.7 | Alkylation | aidD; DNA repair specific for alkylated DNA; mutants extremely sensitive to MMS | 18544 | 2157, 2158, 2316, 4602, 4689, 693 |
| alpA | 59.4 | Activation of Lon protease | Alp protease; cryptic prophage CP4-57 element; regulates intA (slp [suppressor of Lon]) | 33086 | 2241, 4455 |
| alr | 91.9 | Alanine racemase | Alanine racemase (EC 5.1.1.1) | 1034 | 2549, 395 |
| alsA | 92.8 | Allose | Allose transport ABC protein | 53362 | 2223 |
| alsB | 92.9 | Allose | Allose-binding protein | 53365 | 2223 |
| alsC | 92.8 | Allose | Allose transport, membrane component | 53357 | 2223 |
| alsE | 92.8 | Allose | Allulose-6-P 3-epimerase | 53353 | 2223 |
| alsK | 92.8 | Allose | Allose kinase | 53345 | 2223 |
| alx | 70.0 | Alkaline-induced expression | pH-regulated locus; induced in alkaline medium | 36513 | 356 |
| amiA | 55.0 | Amidase | N-Acetylmuramyl-l-alanine amidase activity | 18541 | 4425 |
| amiB | 94.7 | Amidase | Cell wall amidase (EC 3.5.1.28?); overexpression causes lysis, osmotic hypersensitivity, autolysis | 34432 | 4489, 4490 |
| amn | 44.3 | AMP nucleosidase | AMP nucleosidase (EC 3.2.2.4) | 17764 | 2503, 2504 |
| ampC | 94.3 | Ampicillin | ampA; β-lactamase; penicillin resistance; affects peptidoglycan synthesis; cell morphology | 1033 | 1116, 1117, 1167, 1540, 1541, 1713, 1993–1995, 3213, 796 |
| ampD | 2.6 | Ampicillin | ampC regulation | 30478 | 1794, 2561 |
| ampE | 2.6 | Ampicillin | Ampicillin resistance; membrane protein | 30481 | 1794, 2561 |
| ampG | 9.7 | Ampicillin | Ampicillin resistance; membrane protein | 31027 | 2560 |
| ampH | 8.5 | Ampicillin | Probable role in peptidoglycan, cell wall synthesis; cell morphology | 51873 | 1713 |
| amtB | 10.2 | Ammonia transport | Putative ammonia transporter | 47403 | 4557 |
| amyA | 43.2 | Amylase | α-amylase, cytoplasmic | 30745 | 3581, 3582 |
| ansA | 39.9 | Asparaginase | l-Asparaginase I; (EC 3.5.1.1) | 1030 | 1005, 4167 |
| ansB | 66.8 | Asparaginase | l-Asparaginase II; (EC 3.5.1.1) | 30045 | 427, 2008 |
| apaG | 1.1 | ad-P-ad | Expressed as part of complex ksgA operon | 30310 | 3683, 380 |
| apaH | 1.1 | ad-P-ad | Diadenosine tetraphosphatase; stress response; complex operon | 17761 | 1205, 2034, 2509, 2861, 3683, 380 |
| aphA | 92.0 | Acid phosphatase | Acid phosphatase/phosphotransferase, class B | 45401 | 4390 |
| appA | 22.4 | Acid (poly)phosphatase | Acid phosphatase, pH 2.5, exopolyphosphatase (EC 3.1.3.2, 3.6.1.11); agp homology; phytase P2 | 17758 | 1514, 432, 945, 946, 4446 |
| appB | 22.4 | Acid (poly)phosphatase | cyxB; cytochrome oxidase, putative additional one | 31813 | 161, 947 |
| appC | 22.4 | Acid (poly)phosphatase | cyxA; cytochrome oxidase, putative additional one | 31810 | 161, 947 |
| appY | 12.6 | Acid (poly)phosphatase | Regulates hya and appA operons; induced by PO4 starvation and stationary phase | 31146 | 160, 161, 162, 1786, 2195, 516 |
| apt | 10.6 | ad-P transferase | Adenine phosphoribosyltransferase (EC 2.4.2.7); adenine salvage, AMP from PRPP + Ad | 1029 | 1733, 1734, 577 |
| aqpZ | 19.7 | Aquaporin | Aquaporin Z, bacterial water channel | 40930 | 607, 608 |
| araA | 1.4 | Arabinose | l-Arabinose isomerase (EC 5.3.1.4) | 1028 | 2469 |
| araB | 1.5 | Arabinose | Ribulokinase (EC 2.7.1.16) | 1027 | 1102, 2169, 2469, 2470, 2970, 3247, 4110 |
| araC | 1.5 | Arabinose | Regulatory gene: activator and repressor | 1026 | 1102, 1158, 2169, 2300, 2470, 2971, 3247, 4018, 4110, 4237, 4642, 643, 651 |
| araD | 1.4 | Arabinose | l-Ribulosephosphate 4-epimerase (EC 5.1.3.4) | 1025 | 2469, 2949 |
| araE | 64.2 | Arabinose | Low-affinity l-arabinose transport; l-arabinose proton symport | 1024 | 2299, 2300, 2708, 2724, 4238 |
| araF | 42.7 | Arabinose | l-Arabinose-binding protein | 1023 | 2299, 2300, 2328, 3953, 3954, 758 |
| araG | 42.7 | Arabinose | High-affinity l-arabinose transport | 1022 | 2299, 2300, 2328, 3953 |
| araH | 42.7 | Arabinose | High-affinity l-arabinose transport, membrane protein | 18535 | 3953 |
| araJ | 8.8 | Arabinose | Function unknown, arabinose inducible, not affecting Ara transport or utilization | 29861 | 1716, 3636 |
| arcA | 100.0 | Aerobic pathways control | dye, fexA, msp, seg, sfrA; negative regulatory gene for aerobic path genes, anaerobic repression; activates cydAB | 831 | 1089, 1955, 333, 4069, 587, 588, 689 |
| arcB | 72.2 | Aerobic pathways control | Activates ArcA in response to anoxia; probable histidine kinase phosphorylating ArcA | 29063 | 1953, 1956, 1958, 3886, 689 |
| argA | 63.5 | Arginine | Arg1, Arg2, argB; N-acetylglutamate synthase (EC 2.3.1.1); growth on acetylornithine; see argE | 1021 | 1107, 3152, 3870, 528, 862 |
| argB | 89.5 | Arginine | Acetylglutamate kinase (EC 2.7.2.8); see argA | 1020 | 255, 3000, 3374, 862, 863 |
| argC | 89.5 | Arginine | Arg2, argH; N-acetyl-γ-glutamyl-phosphate reductase (EC 1.2.1.38) | 1019 | 255, 3000, 311, 3374, 3433, 862, 863 |
| argD | 75.2 | Arginine | Arg1, argG; acetylornithine aminotransferase (EC 2.6.1.11); see argF | 1018 | 2191, 315, 3676, 862 |
| argE | 89.5 | Arginine | Arg4, argA; acetylornithine deacetylase (EC 3.5.1.16); see argG | 1017 | 2187, 255, 3000, 311, 3433, 863 |
| argF | 6.2 | Arginine | Arg5, argD; ornithine transcarbamylase (EC 2.1.3.3); see duplicate locus, argI | 1016 | 1597, 1838, 2015, 2476, 2997, 2998, 3434, 4561, 4892, 863, 1717 |
| argG | 71.5 | Arginine | Arg6, argE; argininosuccinate synthetase (EC 6.3.4.5); see argD | 1015 | 4560, 1717 |
| argH | 89.5 | Arginine | Argininosuccinate lyase (EC 4.3.2.1); see argC | 1014 | 255, 3000, 863, 1717 |
| argI | 96.5 | Arginine | Ornithine transcarbamylase (EC 2.1.3.3); see argF | 1013 | 295, 3434, 3729 |
| argM | 88.7 | Arginine | Acetylornithine transaminase; cryptic gene; may be duplicate of argD | 18532 | 3676 |
| argP | 65.9 | Arginine | iciA (inhibitor of chromosome initiation); transport of arginine, ornithine, and lysine; canavanine sensitivity | 1012 | 1614, 1871, 1873, 1874, 2474, 2691, 3427, 3733, 665, 4405 |
| argQ | 60.7 | Arginine | Rarg; arginine tRNA2 tandem quadruple genes | 2313 | 35604 |
| argR | 72.9 | Arginine | xerA; repressor of Arg regulon; cer-mediated site-specific recombination | 1011 | 1114, 2187, 2481, 254, 2552, 2690, 315, 3399, 3427, 4225 |
| argS | 42.2 | Arginine | lov; arginyl-tRNA synthetase (EC 6.1.1.19); | 1010 | 1164, 1772, 3147, 4590 |
| argT | 52.3 | Arginine | Salmonella homolog codes for Lys-, Arg-, and Orn-binding proteins | 18529 | 3211 |
| argU | 12.1 | Arginine | dnaY, pin; arginine tRNA4 | 17755 | 1285, 1360, 1415, 2313, 2563, 3888, 4140, 505, 696 |
| argV | 60.7 | Arginine | argVα; arginine tRNA2 tandem quadruple genes | 2313 | 11650 |
| argW | 53.1 | Arginine | Arginine tRNA5 | 17752 | 2313 |
| argX | 85.8 | Arginine | Arginine tRNA3 | 17749 | 1832, 2313, 926 |
| argY | 60.7 | Arginine | argVβ; arginine tRNA2 tandem quadruple genes | 2313 | 35610 |
| argZ | 60.7 | Arginine | argVγ; arginine tRNA2 tandem quadruple genes | 2313 | 35607 |
| aroA | 20.7 | Aromatic | 3-Enolpyruvylshikimate-5-phosphate synthetase (EC 2.5.1.19) | 1008 | 1098, 1099 |
| aroB | 75.8 | Aromatic | 3-Dehydroquinate synthase (EC 4.6.1.3) | 1007 | 2937 |
| aroC | 52.7 | Aromatic | Chorismate synthase (EC 4.6.1.4) | 1006 | 683 |
| aroD | 38.2 | Aromatic | 3-Dehydroquinate dehydratase (EC 4.2.1.10) | 1005 | 2240, 433 |
| aroE | 73.9 | Aromatic | Dehydroshikimate reductase (EC 1.1.1.25) | 1004 | 116, 2865 |
| aroF | 59.0 | Aromatic | DAHP synthetase (tyrosine repressible) (EC 4.1.2.15) | 1003 | 1371, 1847, 4047 |
| aroG | 16.9 | Aromatic | DAHP synthetase (phenylalanine repressible) (EC 4.1.2.15); TyrR regulon | 1002 | 1189, 1704, 1903, 2819, 958, 959 |
| aroH | 38.5 | Aromatic | DAHP synthetase (tryptophan repressible) (EC 4.1.2.15) | 1001 | 1100, 1848, 3030, 433, 4968, 958 |
| aroI | 84.2 | Aromatic | Function unknown | 1000 | 1422 |
| aroK | 75.8 | Aromatic | Shikimate kinase I | 30224 | 1516, 2604, 2605, 4586, 4719 |
| aroL | 8.7 | Aromatic | Shikimate kinase II (EC 2.7.1.71) | 999 | 2436, 2936, 4719, 998, 999 |
| aroM | 8.8 | Aromatic | Function unknown; regulated by aroR | 18523 | 998, 999 |
| aroP | 2.6 | Aromatic | General aromatic amino acid transport; TyrR regulon | 998 | 1553, 1556, 1558, 1657, 2131, 2415, 2416, 3689, 529, 755, 756, 83, 1795 |
| aroT | 28.3 | Aromatic | trpR; aroR; indole acrylic acid resistant mutants; transport | 997 | 4410 |
| arsB | 78.6 | Arsenate resistance | arsF; resistance to arsenate, arsenite, and antimonite | 35996 | 1055, 4128, 600, 637 |
| arsC | 78.6 | Arsenate resistance | arsG; resistance to arsenate, arsenite, and antimonite | 35999 | 1055, 4128, 600, 637 |
| arsR | 78.6 | Arsenate resistance | arsE; resistance to arsenate, arsenite, and antimonite | 35993 | 1055, 4128, 4812, 600, 637 |
| artI | 19.4 | Arginine transport | Periplasmic binding protein of Arg transport system | 31674 | 4759, 4760 |
| artJ | 19.4 | Arginine transport | Periplasmic binding protein of Arg transport system | 31664 | 4759 |
| artM | 19.4 | Arginine transport | Arg periplasmic transport system; similarity to transmembrane proteins, BPC ATPases | 31667 | 4759, 4760 |
| artP | 19.4 | Arginine transport | Arg periplasmic transport system; similarity to transmembrane proteins, BPC ATPases | 31677 | 4759, 4760 |
| artQ | 19.4 | Arginine transport | Arg periplasmic transport system; similarity to transmembrane proteins, BPC ATPases | 31670 | 4759, 4760 |
| ascB | 61.2 | Arbutin, salicin, cellobiose | sac; cryptic; paralogous to cryptic bglFB; expressed when AscG is mutated | 33236 | 1606, 3365 |
| ascF | 61.2 | Arbutin, salicin, cellobiose | sac; cryptic; paralogous to cryptic bglFB; expressed when AscG is mutated | 33233 | 1606, 3365 |
| ascG | 61.1 | Arbutin, salicin, cellobiose | Repressor of cryptic asc operon; galR paralog | 33230 | 1606 |
| asd | 77.0 | Aspartate semialdehyde deHase | dap, hom; aspartate semialdehyde dehydrogenase (EC 1.2.1.11) | 996 | 1679, 1680, 3281, 3940 |
| aslA | 85.8 | Arylsulfatase-like | gppB; suppresses gpp mutants | 33957 | 3069 |
| aslB | 85.8 | Arylsulfatase-like | gppB; suppresses gpp mutants | 33960 | 3069 |
| asmA | 46.1 | Assembly suppressor mutant | Membrane protein; suppressor of ompF assembly mutants | 41068 | 1021; 2957, 4811 |
| asnA | 84.6 | Asparagine | Asparagine synthetase A (EC 6.3.1.1) | 995 | 1769, 1861, 2295, 3111, 4610, 4621, 556 |
| asnB | 15.0 | Asparagine | Asparagine synthetase B (EC 6.3.1.1) | 994 | 1861, 3454, 3952 |
| asnC | 84.6 | Asparagine | Regulatory gene for asnA, asnC, gidA | 18520 | 1033, 2294, 556, 729 |
| asnS | 21.3 | Asparagine | lcs, tss; asparaginyl-tRNA synthetase (EC 6.1.1.22) | 993 | 1917, 4433 |
| asnT | 44.0 | Asparagine | Asparagine tRNA | 992 | 1285, 2313, 3324 |
| asnU | 44.4 | Asparagine | Asparagine tRNA | 17746 | 2313 |
| asnV | 44.4 | Asparagine | Asparagine tRNA | 17743 | 2313 |
| asnW | 44.3 | Asparagine | Asparagine tRNA | 51238 | 2313 |
| aspA | 94.1 | Aspartate | Aspartate ammonia-lyase (aspartase) (EC 4.3.1.1) | 991 | 1557, 4327 |
| aspC | 21.2 | Aspartate | Aspartate aminotransferase (EC 2.6.1.1) | 990 | 1283, 2317, 2373, 2740 |
| aspS | 42.0 | Aspartate | tls; aspartyl-tRNA synthetase | 32508 | 1165, 1351, 4000 |
| aspT | 85.0 | Aspartate | tasC; aspartate tRNA1 triplicated gene | 989 | 2313, 2777, 4904, 926 |
| aspU | 4.8 | Aspartate | Aspartate tRNA1 triplicated gene, in rrnH operon | 988 | 1146, 2313, 2777 |
| aspV | 5.1 | Aspartate | Aspartate tRNA1 triplicated gene | 987 | 1806, 2313, 2777, 3324 |
| asr | 35.9 | Acid shock RNA | Acid shock RNA; expression controlled by phoBR | 53672 | 1986 |
| asue | 33.3 | Asparagine utilization | Utilizes asparagine as sole nitrogen source | 18517 | 723 |
| atoA | 50.1 | Acetoacetate | Acetate CoA-transferase (EC 2.8.3.-) | 986 | 2321 |
| atoB | 50.1 | Acetoacetate | Acetyl-CoA acetyltransferase (EC 2.3.1.9) | 985 | 2005 |
| atoC | 50.0 | Acetoacetate | Az; positive regulator in two-component system, with AtoS sensor kinase | 984 | 395a, 1949a, 3985a, 2005, 2006, 3382, 3817, 629 |
| atoD | 50.0 | Acetoacetate | Acetyl-CoA:acetoacetyl-CoA transferase β-subunit | 18514 | 2005 |
| atoS | 50.0 | Acetoacetate | AtoS sensor kinase, with AtoC response regulator in 2-component system | 37061 | 395a, 1949a, 3985a |
| atpA | 84.4 | ATP | papA, uncA; membrane-bound ATP synthase, F1 sector, α-subunit (EC 3.6.1.3) | 33 | 1390, 1570, 2275, 4187, 4635 |
| atpB | 84.5 | ATP | papD, uncB; membrane-bound ATP synthase, F0 sector, subunit a (EC 3.6.1.3) | 32 | 1570, 1830, 2993, 3180, 3860, 4609, 4635 |
| atpC | 84.3 | ATP | papG, uncC; membrane-bound ATP synthase, F1 sector, ɛ-subunit (EC 3.6.1.3) | 31 | 1570, 4635, 4809, 4810 |
| atpD | 84.4 | ATP | papB, uncD; membrane-bound ATP synthase, F1 sector, β-subunit (EC 3.6.1.3) | 30 | 1570, 3296, 4635 |
| atpE | 84.5 | ATP | papH, uncE; membrane-bound ATP synthase, F0 sector, subunit c; DCCD− (EC 3.6.1.3) | 29 | 1570, 1992, 2836, 2993, 3860, 3900, 4635 |
| atpF | 84.5 | ATP | papF, uncF; membrane-bound ATP synthase, F0 sector, subunit b (EC 3.6.1.3) | 28 | 1570, 2993, 4635 |
| atpG | 84.4 | ATP | papC, uncG; membrane-bound ATP synthase, F1 sector, γ-subunit (EC 3.6.1.3) | 27 | 1570, 2275, 2692, 3860, 4635 |
| atpH | 84.4 | ATP | papE, uncH; membrane-bound ATP synthase, F1 sector, δ-subunit (EC 3.6.1.3) | 26 | 1570, 2994, 3380, 4635 |
| atpI | 84.5 | ATP | uncI; membrane-bound ATP synthase subunit, F1-F0-type proton-ATPase (EC 3.6.1.34) | 18511 | 1830, 2040, 2156, 3180, 3491, 4609, 4635, 544, 3923 |
| att186 | 59.3 | Attachment | Integration site for phage HK186 | 972 | D |
| att253 | 6.1 | Attachment | Integration site for phage HK253 | 18508 | 3484 |
| attHK139 | 44.8 | Attachment | Attachment site for phage HK139 | 981 | 1035 |
| attHK022 | 22.7 | Attachment | atthtt; lambdoid prophage HK022 attachment site | 982 | 1034, 2301 |
| attλ | 17.3 | Attachment | att92, att434; lambda attachment site | 980 | 1470, 18, 1035, 1836, 2940 |
| attP1,P7 | 68.7 | Attachment | loxB; attachment site for phage P1 and P7 | 979 | 4205, 722 |
| attP22 | 5.6 | Attachment | ata; phage P22 attachment site, within thrW gene | 975 | 2562 |
| attP2H | 44.7 | Attachment | Integration site H for phage P2 | 978 | 241a, 4291, D |
| attP2II | 87.1 | Attachment | Integration site II for phage P2 | 977 | 241a, 37, D |
| attP2III | 83.5 | Attachment | Integration site III for phage P2 | 32179 | 241a |
| attP4 | 96.9 | Attachment | Integration site for phage P4 | 976 | 611, 3431 |
| attPA-2 | 53.1 | Attachment | Integration site for phage PA-2 | 974 | C |
| attφ80 | 28.2 | Attachment | Integration site for phage phi80 | 973 | 3794a |
| attTn7 | 84.3 | Attachment | Specific site for Tn7 insertion | 37363 | 1520, 2852 |
| avtA | 80.6 | Alanine-isoketovalerate transaminase | Alanine-α-ketoisovalerate transaminase, transaminase C | 971 | 1196, 2585, 4656, 4718 |
| azaA | 44.6 | Azaserine | Mutants azaserine resistant | 970 | 4744 |
| azaB | 71.9 | Azaserine | Mutants azaserine resistant | 969 | 4744 |
| azl | 58.1 | Azaleucine | Mutants azaleucine resistant; regulates ilv and leu | 967 | 3452 |
| bacA | 69.0 | Bacitracin resistance | A lipid kinase; may confer resistance by phosphorylation of undecaprenol | 29739 | 606 |
| baeR | 46.6 | Bacterial adaptive envZ regulator | Suppresses envZ and phoR/creC mutations | 30764 | 3083 |
| baeS | 46.6 | Bacterial adaptive envZ regulator | Suppresses envZ and phoR/creC mutations | 30761 | 3083 |
| barA | 62.8 | Bacterial adaptive response | Has sensory kinase and response regulator domains like OmpR and EnvZ | 33320 | 1924, 3084 |
| basR | 93.4 | Bacterial adaptive sensor | BasRS two-component regulatory system homologous with OmpR-EnvZ family | 28168 | 3083, 3716 |
| basS | 93.3 | Bacterial adaptive sensor | BasRS two-component regulatory system homologous with OmpR-EnvZ family | 28171 | 3083 |
| bax | 80.5 | Gene transcribed divergently from malS | 54736 | 3922 | |
| bcp | 56.0 | Bacterioferritin comigratory protein | Probable bacterioferritin | 33035 | 112 |
| bcr | 49.1 | Bicyclomycin resistance | bicA, bicR, sur, suxA; transmembrane; affects sulfathiazole-sulfonamide resistance | 32582 | 309 |
| betA | 7.0 | Betaine | Choline dehydrogenase | 17740 | 104, 2399, 4257, 470 |
| betB | 7.0 | Betaine | Betaine aldehyde dehydrogenase (EC 1.2.1.8) | 17737 | 104, 2399, 470 |
| betI | 7.1 | Betaine | Regulatory gene, perhaps repressor for choline regulation of bet genes | 30692 | 2399 |
| betT | 7.1 | Betaine | High-affinity choline transport | 18505 | 104, 2399 |
| bfd | 74.7 | Bfr regulating | Regulatory or redox component complexing with Bfr in iron storage and mobility [2Fe-2S] | 43017 | 1369 |
| bfm | 85.9 | BF23 multiplication | Controls phage BF23 multiplication | 966 | A, 4041a |
| bfr | 74.7 | Bacterioferritin | Bacterioferritin | 32528 | 111 |
| bglA | 65.6 | β-Glucoside | bglD; phospho-β-glucosidase A; growth on arbutin or salicin when activated | 965 | 2076, 3239 |
| bglB | 84.1 | β-Glucoside | blgA; phospho-β-glucosidase B; growth on arbutin or salicin when activated | 964 | 2718, 3528, 3658, 3924, 3925, 4611 |
| bglF | 84.1 | β-Glucoside | bglC, bglB; BglG kinase; transport | 18502 | 2718, 3924, 476, 704 |
| bglG | 84.1 | β-Glucoside | bglC, bglS; positive regulatory gene, RNA-binding protein; regulated by phosphorylation | 963 | 1818, 2718, 3658, 3659, 3924, 3825, 4077, 4506 |
| bglJ | 99.2 | β-Glucoside | Mutation bglJ4 activates silent bgl operon, allowing arbutin and salicin transport and utilization | 45483 | 1424, 4506 |
| bglT | 84.9 | β-Glucoside | bglE; regulatory gene for BglA | 961 | 3890 |
| bglX | 47.8 | β-Glucoside | Periplasmic β-glucosidase (EC 3.2.1.21) | 4858 | 4815,4 |
| bioA | 17.4 | Biotin | Diaminopelargonic acid synthetase | 959 | 18, 232, 2331, 235, 3312, 4246, 4319 |
| bioB | 17.4 | Biotin | Biotin synthetase; dethiobiotin to biotin pathway | 958 | 18, 232, 2331, 235, 3312, 4246, 4319 |
| bioC | 17.5 | Biotin | Blocked prior to pimeloyl CoA formation | 957 | 18, 2488, 4246, 4319 |
| bioD | 17.5 | Biotin | Dethiobiotin synthetase | 956 | 18, 2488, 4246, 4319, 4852 |
| bioF | 17.5 | Biotin | 7-Keto-8-aminopelargonic acid synthetase | 955 | 18, 2488, 4246, 4319 |
| bioH | 76.3 | Biotin | bioB; blocked prior to pimeloyl CoA formation | 954 | 232, 2488, 3303, 3940 |
| bioP | 86.6 | Biotin | bir, birB; biotin transport | 953 | 1298, 624 |
| bipA | 87.4 | BPI-induced protein | yihK; in EPEC strains, it mediates interactions with epithelial cells; tyrosine-phosphorylated GTPase | 37329 | 1206 |
| birA | 89.9 | Biotin retention | bioR, dhbB; biotin-[acetyl-CoA carboxylase] holoenzyme synthetase, and repressor | 952 | 1825, 232–234, 2743, 3337, 4817, 624 |
| bisC | 80.0 | Biotin sulfoxide | Biotin sulfoxide reductase, structural gene | 951 | 1003, 3430 |
| bisZ | 42.1 | Biotin sulfoxide | Responsible for background activity of biotin sulfoxide reductase in bisC mutants | 50496 | 1004 |
| blc | 94.3 | Bacterial lipocalin | Membrane protein, first prokaryotic lipocalin; cell division and growth, rpoS regulon; shares translation termination codons with sugE | 40887 | 361 |
| bolA | 9.8 | Bolus | Morphogene; overexpression produces osmotically stable spherical cells; FtsZ dependent | 31032 | 2413, 55–57 |
| brnQ | 9.0 | Branched chain | hrbA; mutants valine and o-methylthreonine resistant, glycylvaline sensitive; transport system I for Ile, Leu, and Val | 950 | 1542, 1543 |
| brnR | 8.5 | Branched chain | Mutants valine resistant, glycylvaline sensitive | 949 | 1542 |
| brnS | 1.2 | Branched chain | Mutants valine resistant, glycylvaline sensitive | 948 | 1542 |
| brnT | 65.2 | Branched chain | Low-affinity transport of Ile | 947 | 1876a |
| btuB | 89.7 | B12 uptake | bfe, cer; receptor for vitamin B12, E colicins, and phage BF23; also C1 phage absorption | 946 | 1545, 1546, 166, 167, 1705, 1850, 2082, 2430, 2546, 3000, 585 |
| btuC | 38.6 | B12 uptake | Vitamin B12 transport | 945 | 1029, 1031, 1315 |
| btuD | 38.6 | B12 uptake | B12 transport, membrane associated | 18499 | 1029, 1031, 1315 |
| btuE | 38.6 | B12 uptake | Not required for vitamin B12 transport, perhaps periplasmic protein | 18496 | 1029, 1315, 3679 |
| btuR | 28.6 | B12 uptake | Regulatory gene affecting btuB | 18493 | 2667 |
| bymA | 93.2 | Bypass maltose | Growth on maltose in MalT− cells | 944 | 1781 |
| cadA | 93.9 | Cadaverine | Lysine decarboxylase (EC 4.1.1.18) | 943 | 168, 169, 2886, 2887, 3144, 4022, 4325, 4680 |
| cadB | 93.9 | Cadaverine | Arginine/ornithine antiporter, probably | 34228 | 2886, 3144, 4680 |
| cadC | 93.9 | Cadaverine | Regulatory gene | 34231 | 2887, 3144, 4680 |
| cafA | 73.2 | Cytoplasmic axial filaments | Cell division and growth, overexpression forms minicells and chains with long axial structures | 31358 | 3278 |
| caiA | 0.8 | Carnitine inducible | Carnitine metabolism, oxidoreductase | 36825 | 1134, 550 |
| caiB | 0.8 | Carnitine inducible | Carnitine dehydratase | 36800 | 1135, 550 |
| caiC | 0.8 | Carnitine inducible | Crotonobetain/carnitine-CoA ligase | 36828 | 1134, 550 |
| caiD | 0.8 | Carnitine inducible | Putative enoyl hydratase/isomerase with carnitine racemase activity | 36831 | 1134, 550 |
| caiE | 0.8 | Carnitine inducible | Stimulates carnitine racemase activity of CaiD and CaiB activity | 36834 | 1134, 550 |
| caiF | 0.7 | Carnitine inducible | Regulatory gene; transcriptional activation of cai operon | 42995 | 1133, 550 |
| caiT | 0.9 | Carnitine inducible | Putative carnitine/betaine transport | 36822 | 1134, 550 |
| calA | 95.0 | Calcium | Calcium-proton antiport activity | 941 | 4417, 499 |
| calC | 15.2 | Calcium | Calcium transport; mutants defective in chemotaxis | 940 | 4417, 499 |
| calD | 9.4 | Calcium | Calcium transport; mutants defective in chemotaxis | 939 | 4417, 499 |
| can | 66.0 | Canavanine | Resistance to canavanine | 938 | A, 2691a |
| carA | 0.6 | Carbamoyl P | arg+ura, cap, pyrA; carbamoylphosphate synthase (glutamine-hydrolysing) light subunit (EC 6.3.5.5) | 936 | 1427, 3432, 465, 861 |
| carB | 0.7 | Carbamoyl P | arg+ura, cap, pyrA; carbamoylphosphate synthase (ammonia), heavy subunit (EC 6.3.4.16) | 935 | 1427, 3234, 861 |
| cbl | 44.4 | cysB-like | cys regulon member; perhaps an accessory regulatory circuit within the cys regulon | 50175 | 1964, 4550 |
| cbpA | 22.9 | Curved-DNA binding protein | Recognizes a curved DNA sequence; sequence similarity to DnaJ | 31822 | 4505 |
| cbt | 16.6 | Colicin B (and D) tolerance | Dicarboxylate binding protein production; ColB and ColD tolerance | 934 | 334 |
| cca | 69.0 | CCA tRNA terminus | tRNA nucleotidyl transferase | 933 | 880 |
| ccmA | 49.5 | Cytochrome c maturation | ABC transporter, ATPase subunit | 36574 | 1537, 4406, 4411, 739, 740 |
| ccmB | 49.5 | Cytochrome c maturation | ABC transporter, ATPase subunit | 36577 | 1537, 4406, 4411, 739, 740 |
| ccmC | 49.4 | Cytochrome c maturation | ABC transporter, heme binding | 36581 | 1537, 4406, 4411, 739, 740 |
| ccmD | 49.4 | Cytochrome c maturation | Cytochrome c related | 36584 | 1537, 4406, 739, 740 |
| ccmE | 49.4 | Cytochrome c maturation | Cytochrome c related | 36587 | 1537, 4406, 739, 740 |
| ccmF | 49.4 | Cytochrome c maturation | Required for synthesis of c-type cytochromes; similarity with NrfE | 36590 | 1536, 1537, 4406, 4411, 739, 740 |
| ccmG | 49.4 | Cytochrome c maturation | dsbE; thioredoxin homolog; thiol-disulfide interchange protein | 36594 | 1192, 1537, 4406, 4411, 739, 740 |
| ccmH | 49.3 | Cytochrome c maturation | Required for synthesis of c-type cytochromes; similarity with NrfF and NrfG | 36597 | 1536, 1537, 4406, 739, 740 |
| cdd | 48.1 | Deoxycytidine deaminase | Deoxycytidine deaminase (EC 3.5.4.5); mutants 5-fluorodeoxycytidine resistant | 932 | 1075, 2057, 2058, 2925, 3241, 429, 4325, 4851 |
| cde | 14.2 | Control of dam expression | Affects growth rate control of dam expression; near or within lipB | 37102 | 3616 |
| cdh | 88.5 | CDP diglyceride hydrolase | CDP-diglyceride hydrolase | 931 | 1710, 1884, 51, 559, 560 |
| cdsA | 4.2 | CDP diglyceride synthase | CDP-diglyceride synthase | 930 | 1356, 1886 |
| cdsS | 71.7 | CDP diglyceride synthase | Stability of CDP diglyceride synthase | 18490 | 1355 |
| cedA | 39.1 | Cell division | Modulates cell division, affects inhibition after overreplication of chromosome in dnaAcos mutants | 55374 | 2161 |
| celA | 39.2 | Cellobiose | chbB; member of cryptic cel operon | 34873 | 2205, 3366 |
| celB | 39.2 | Cellobiose | chbC; phosphotransferase system enzyme IIcel, PEP dependent; cryptic operon; cellobiose, arbutin, and salicin transport | 18487 | 2205, 2340, 2341, 3366, 3647 |
| celC | 39.2 | Cellobiose | chbA; phosphotransferase system enzyme IIIcel, PEP dependent; cryptic operon; cellobiose, arbutin, and salicin transport | 18484 | 2205, 2340, 2341, 3366, 3547 |
| celD | 39.2 | Cellobiose | chbR; Cel regulatory protein | 18481 | 2205, 2340, 2341, 3366 |
| celF | 39.1 | Cellobiose | chbF; phospho-β-glucosidase B; cryptic operon | 17734 | 2205, 2340, 2341, 3366 |
| cfa | 37.5 | Cyclopropane fatty acid | cdfA; cyclopropane fatty acid synthase | 10810 | 1527, 1528, 1529, 4649 |
| cfcA | 79.9 | Control frequency of cell division | Controls cell division frequency per round of DNA replication | 36615 | 3194 |
| chaA | 27.4 | Ca2+/H+ antiporter | Ca2+/H+ antiporter | 30293 | 1961, 3271, 3330 |
| chaB | 27.4 | Ca2+/H+ antiporter | Accessory and regulatory protein for chaA | 37193 | 4792a |
| chaC | 27.4 | Ca2+/H+ antiporter | Accessory and regulatory protein for chaA | 37196 | 4792a |
| cheA | 42.5 | Chemotaxis | Autophosphorylating histidine kinase of chemotactic response; clockwise and counterclockwise signals; Fla regulon | 928 | 2285, 2679, 3757, 3830, 4064, 4101, 4119, 4363, 4652, 84 |
| cheB | 42.4 | Chemotaxis | Protein methylesterase; flagellar regulon member | 927 | 2349, 3076, 4101, 4214, 468, 923 |
| cheR | 42.4 | Chemotaxis | cheX; protein methyltransferase (in chemotactic response); flagellar regulon | 926 | 2951, 3076, 4101, 468 |
| cheW | 42.5 | Chemotaxis | Signal transduction; couples CheA to chemoreceptor control by promoting CheW/CheA/Tsr; flagellar regulon | 925 | 3076, 3757, 4064, 4101 |
| cheY | 42.4 | Chemotaxis | Response regulator CheY for chemotactic signal transduction; flagellar regulon member | 924 | 2679, 2809, 3076, 3757, 3857, 4046, 4101, 468, 776 |
| cheZ | 42.3 | Chemotaxis | Chemotactic signal transduction; flagellar regulon member | 923 | 3076, 3856, 3857, 4101, 468 |
| chpA | 62.7 | Chromosomal homolog of pem | chpAK, mazF; ChpAB growth inhibitor, homology to R100 pemK gene, programmed cell death? toxic protein | 33287 | 2794, 35 |
| chpB | 95.8 | Chromosomal homolog of pem | chpBK, yjfB; ChpAB growth inhibitor, homology to pemK | 33290 | 2793, 2794 |
| chpR | 62.7 | Chromosomal homolog of pem | chpAI, mazE; homology to R100 pemI, which suppresses pemK; suppresses ChpA inhibition | 33283 | 2794, 35 |
| chpS | 95.8 | Chromosomal homolog of pem | chpBI, yjfA; homology to pemI; supresses ChpB | 33293 | 2793, 2794 |
| cirA | 48.3 | Colicin I resistance/receptor | feuA; colicin I receptor production | 916 | 1518, 2925, 428, 4783 |
| citA | 17.9 | Citrate | Cryptic gene for citrate transport system | 18469 | 1604 |
| citB | 16.0 | Citrate | Cryptic gene for citrate transport system | 18466 | 1604 |
| cld | 45.2 | Chain length determination | rol; regulator of lipopolysaccharide O-chain length; gene studied in Salmonella and non-K-12 strains | 56819 | 1061a, 1293a |
| clpA | 19.9 | Caseinolytic protease | Clp ATP-dependent protease, ATP-binding subunit | 31293 | 1472, 3972, 3973, 3990 |
| clpB | 58.8 | Caseinolytic protease | ClpB protease, ATP dependent (EC 1.17.4.–, 3.4.21.–) | 32875 | 1327, 2244, 3359, 3478, 4178, 4772 |
| clpP | 9.8 | Caseinolytic protease | F21.5, LopP; ClpP ATP-dependent protease proteolytic subunit | 31280 | 2344, 2822, 2824, 3973, 4765 |
| clpX | 9.8 | Caseinolytic protease | LopC; ClpX protease, which activates ClpP | 31287 | 1297, 1473, 2505, 4765, 4891 |
| cls | 28.1 | Cardiolipin synthase | nov; cardiolipin synthase; mutants dihydroxybutylphosphonate resistant; novobiocin sensitivity | 915 | 1690, 1765, 1766, 1876, 1960, 3193, 3267, 3600, 4462, 4463 |
| cmk | 20.7 | CMP kinase | mssA; multicopy suppressor; CMP kinase | 31736 | 1308, 4843 |
| cmlA | 19.0 | Chloramphenicol | Probably same as cmr; resistance or sensitivity to chloramphenicol, also tetracycline resistance | 914 | 187, 3637, 4043 |
| cmr | 19.0 | Chloramphenicol resistance | cmlA?, mdfA; transmembrane multidrug/chloramphenicol efflux transporter | 55066 | 1115, 3189, 3637 |
| cmtA | 66.3 | Cryptic mannitol | Similar to mannitol phosphotransferase enzymes | 33362 | 4163 |
| cmtB | 66.3 | Cryptic mannitol | tolM; protein-Nπ-phosphohistidine sugar P-transferase; enzyme III of PEP-PTS cryptic mannitol transport | 33365 | 4163 |
| coaA | 89.9 | CoA | panK, rts; pantothenate kinase | 17731 | 1259, 4134, 4135, 4538 |
| cobS | 44.5 | Cobalamin, coenzyme B12 | Partial cobalamin biosynthesis pathway present in E. coli | 40912 | 2439 |
| cobT | 44.4 | Cobalamin, coenzyme B12 | Partial cobalamin biosynthesis pathway present in E. coli | 40908 | 2439 |
| cobU | 44.5 | Cobalamin, coenzyme B12 | Partial cobalamin biosynthesis pathway present in E. coli | 40916 | 2439 |
| codA | 7.7 | Cytosine deaminase | Cytosine deaminase (EC 3.5.4.1) | 913 | 23, 4562, 927, 95 |
| codB | 7.6 | Cytosine deaminase | Cytosine transport | 912 | 23, 927, 95 |
| cof | 10.1 | Complementation of fur | Complements deletion mutant for growth on succinate | 53227 | 1640 |
| cog | 29.1 | Control of ompG | Probable repressor of ompG | 37297 | 2956 |
| corA | 86.2 | Cobalt resistance | Mg2+ transport system; mutants resistant to Co2+, Mn2+, and Ni2+, insensitive to Ca2+ | 911 | 2750, 3242, 3243, 3353, 4120, 4683, 58, 926 |
| CP4-57 | 59.4 | Cryptic prophage | Cryptic prophage; see intA and alpA | 33089 | 2241, 3653 |
| cpdA | 68.4 | Cyclic nucleotide P-diesterase | icc; affects cAMP requirement during growth on maltose; 3′,5′ cAMP phosphodiesterase | 37437 | 1907 |
| cpdB | 95.5 | Cyclic nucleotide P-diesterase | 2′,3′-Cyclic nucleotide 2′-phosphodiesterase (EC 3.1.4.16) | 909 | 1996, 2580, 259 |
| cpsA | 45.4 | Capsular polysaccharide synthesis | Colanic acid (CPS) biosynthesis | 18463 | 4460 |
| cpsB | 45.7 | Capsular polysaccharide synthesis | Colanic acid (CPS) biosynthesis; mannose 1-P guanyltransferase | 18460 | 4460 |
| cpsC | 45.8 | Capsular polysaccharide synthesis | Colanic acid (CPS) biosynthesis | 18457 | 4460 |
| cpsD | 45.8 | Capsular polysaccharide synthesis | Colanic acid (CPS) biosynthesis | 18454 | 4460 |
| cpsE | 45.8 | Capsular polysaccharide synthesis | Colanic acid (CPS) biosynthesis | 51241 | 4460 |
| cpsF | 90.2 | Capsular polysaccharide synthesis | Colanic acid (CPS) biosynthesis | 18448 | 4460 |
| cpsG | 45.7 | Capsular polysaccharide synthesis | Phosphomannomutase isozyme; colanic acid biosynthesis | 37429 | 1999 |
| cpxA | 88.4 | Conjugative plasmid expression | ecfB, eup, ssd; membrane sensor in two-component cpxAR signal transduction system; Kanr, phage Q resistant; l-serine growth | 908 | 1952, 2844, 2845, 3011, 3166, 3448, 3595, 3596, 4066–4068, 4408, 4684, 50, 52, 843 |
| cpxB | 41.3 | Conjugative plasmid expression | Phage Q resistance, membrane protein | 907 | 2844, 2845, 4066 |
| cpxP | 88.4 | Conjugative plasmid expression | Periplasmic protein, CpxA/R activated, induced in alkaline pH; suppresses toxic envelope protein effects | 54751 | 922 |
| cpxR | 88.4 | Conjugative plasmid expression | Regulator in two-component cpxAR | 34166 | 1065, 3596, 843 |
| crcA | 14.1 | Camphor resistance and chromosome condensation | High-copy crc-csp restores normal chromosome condensation in presence of camphor or mukB mutations | 41127 | 1837 |
| crcB | 14.2 | Camphor resistance and chromosome condensation | See crcA | 41131 | 1837 |
| creA | 99.9 | Catabolite regulation | Function unknown; transcribed with cre operon | 34809 | 81 |
| creB | 99.9 | Catabolite regulation | phoM-orf2; structurally homologous to creC (phoM) | 34803 | 81, 82 |
| creC | 99.9 | Catabolite regulation | phoM; sensor in Pho regulon | 395 | 2654, 2733, 2734, 4431, 4660, 4661, 4664, 4666, 4667, 81 |
| creD | 99.9 | Catabolite regulation | cet (colicin E2 tolerance), refII | 929 | 1090, 1284, 1462, 1568, 81 |
| crg | 28.8 | Cold resistant growth | Allows cold-resistant growth | 18445 | 2175 |
| crl | 5.6 | Curli | Regulatory protein for curli (cryptic csgA) | 30625 | 137, 1722, 3291, 3545 |
| crp | 75.1 | cAMP receptor protein | cap, csm; cAMP receptor protein | 906 | 1404, 147, 1559, 2233, 2365, 258, 2627, 32, 3610, 3279, 3559, 3617, 4108, 4394, 500, 8, 844, 924 |
| crr | 54.6 | Carrier? | gsr, iex, tgs; phosphocarrier protein for glucose of the PTS; IIIglc | 905 | 1023, 2326, 2859, 3150, 3369, 3370, 3793, 508, 539, 557, 562, 982, 983b |
| csdA | 63.4 | Cysteine, selenocysteine decomposition | Cysteine sulfinate desulfinase | 52073 | 2926 |
| csgA | 23.8 | Curlin ςS-dependent growth | Curlin, ςS (stationary phase) dependent, cryptic | 30620 | 137, 138, 1619, 2612, 3289–3291, 3725, 4584 |
| csgB | 23.8 | Curlin ςS-dependent growth | Curlin nucleator protein, homology with major curlin, CsgA | 36735 | 138, 2612, 3725 |
| csgD | 23.7 | Curlin ςS-dependent growth | csgD insertions eliminate ςS-dependent transcription from csgBA promoter | 50655 | 1619, 3725 |
| csgE | 23.7 | Curlin ςS-dependent growth | Possible secretion or assembly protein for bacterial fibers | 50658 | 1619, 3725 |
| csgF | 23.7 | Curlin ςS-dependent growth | Possible assembly or transport protein for curli | 50661 | 1619 |
| csgG | 23.7 | Curlin ςS-dependent growth | Possible assembly or transport protein for curli; novel lipoprotein | 50664 | 1619, 2612, 3725 |
| csiA | 85.3 | Carbon starvation induced | Stationary phase inducible protein | 36892 | 4690 |
| csiB | 17.4 | Carbon starvation induced | Stationary phase inducible protein | 36895 | 4690 |
| csiC | 66.0 | Carbon starvation induced | Stationary phase inducible protein | 36898 | 4690 |
| csiD | 60.5 | Carbon starvation induced | Stationary phase inducible protein | 36904 | 2775, 4690 |
| csiE | 57.4 | Carbon starvation induced | Stationary phase inducible protein | 36901 | 2774, 4690 |
| csiF | 8.6 | Carbon starvation induced | Stationary phase inducible protein | 36998 | 4690 |
| cspA | 80.1 | Cold shock protein | Cold shock protein CS7.4; similar to Y-box DNA binding proteins of eukaryotes; transcription factor | 29540 | 1199, 1200, 1443, 1452, 194, 2019, 2046, 2473, 478, 4841a, 4347 |
| cspB | 35.3 | Cold shock protein | Cold shock protein with similarity to CspA | 32231 | 1200, 2473 |
| cspC | 41.1 | Cold shock protein | msmB; multicopy suppresses mukB mutants | 35339 | 2043, 2473, 4842 |
| cspD | 19.9 | Cold shock protein | Similarity to CspA but not cold shock induced | 31688 | 2473, 4841 |
| cspE | 14.2 | Cold shock protein | msmC; with crcAB, high copy promotes or protects chromosome condensation | 31528 | 1837, 4841, 4842 |
| cspG | 12.6 | Cold shock protein | Cold-induced CspA/B analog | 53423 | 194, 3119 |
| csrA | 60.7 | Carbon storage regulator | zfiA, regulatory gene inhibiting glycogen biosynthesis; global regulatory protein | 34504 | 2586, 3061, 3723, 3724, 3787, 4854 |
| csrB | 62.9 | Carbon storage regulator | CsrA-binding RNA, antagonizing CsrA regulation | 2585a | |
| cstA | 13.5 | Carbon starvation | Starvation induced stress response protein | 31179 | 1523, 2798, 3937, 403 |
| cstCb | 39.4 | Carbon starvation | astC; starvation gene regulated by cAMP and RpoS, T; induced by ornithine; arginine succinyltransferase | 54626 | 1290a, 1290b, 1950a, 3921a |
| cup | 97.0 | Carbohydrate uptake | Mutants have defective carbohydrate uptake | 18442 | 2720 |
| cutA | 94.0 | Cu tolerance | cycY, cutA1; copper sensitivity; possible role in cytochrome c maturation; cytochrome c-like | 34216 | 1271, 1921, 875 |
| cutC | 42.2 | Cu tolerance | Copper sensitivity | 36974 | 1574 |
| cutE | 14.8 | Cu tolerance | lnt; copper sensitivity; apolipoprotein N-acetyltransferase | 31471 | 3710 |
| cutF | 4.6 | Cu tolerance | nlpE; copper sensitivity | 35748 | 1574, 4122 |
| cvpA | 52.3 | Colicin V production | dedE; member of purF operon; affects Col V production | 32727 | 1207 |
| cxm | 6.3 | Carbon-xylose metabolism | cxr; methyl glyoxal synthesis; d-xylose utilization | 903 | 8 |
| cyaA | 86.0 | Cyclase, adenylate | Adenylate cyclase (EC 4.6.1.1) | 902 | 1008, 219, 2233, 2275, 2321, 2365, 26, 29, 30, 3559, 3758, 3759, 3760, 4401, 500, 58, 924, 926 |
| cybB | 32.1 | Cytochrome b | Cytochrome b561 | 17728 | 3052, 3054, 3105 |
| cybC | 96.1 | Cytochrome b | Cytochrome b562 | 34583 | 4465 |
| cycA | 95.4 | Cycloserine | dagA; d-alanine, d-serine, glycine permease | 900 | 3685, 3686, 3939, 4671, 569 |
| cydA | 16.6 | Cytochrome d | Cytochrome d terminal oxidase, polypeptide subunit I | 10369 | 1060, 1447, 1491, 1492, 1493, 1494, 3039, 3611, 615, 847 |
| cydB | 16.6 | Cytochrome d | Cytochrome d terminal oxidase, polypeptide subunit II | 9469 | 1009, 1010, 1011, 1060, 1447, 1491, 1492, 1494, 1495, 3039, 3611, 4638, 615, 847 |
| cydC | 20.0 | Cytochrome d | mdrA, mdrH, surB; cytochrome d terminal oxidase, possibly heme d component | 17725 | 1009, 1405, 1447, 3763, 4054, 4055 |
| cydD | 20.0 | Cytochrome d | ATP-binding cassette membrane transporter; bd-type oxidase | 31720 | 1009, 1447, 262, 3479–3481 |
| cynR | 7.7 | Cyanase; cyanate metabolism | Transcriptional activator of cyn operon | 31255 | 102, 2403, 4288 |
| cynS | 7.7 | Cyanase; cyanate metabolism | Cyanate aminohydrolase (EC 3.5.5.3) | 15267 | 102, 1564, 1565, 2403, 4286, 4288, 4289 |
| cynT | 7.7 | Cyanase; cyanate metabolism | Carbonic anhydrase | 31258 | 102, 1564, 1566, 2333, 4286, 4287, 4289 |
| cynX | 7.7 | Cyanase; cyanate metabolism | Apparent hydrophobic protein, member of cyn operon | 31261 | 102, 1564 |
| cyoA | 9.7 | Cytochrome o oxidase | Cytochrome o oxidase subunit II; cytochrome bo3 ubiquinol oxidase subunit II | 18439 | 164, 165, 2687, 615 |
| cyoB | 9.7 | Cytochrome o oxidase | Cytochrome o oxidase subunit I | 30997 | 164, 3106, 615 |
| cyoC | 9.6 | Cytochrome o oxidase | Cytochrome o oxidase subunit III | 31005 | 164, 615, 720, 721 |
| cyoD | 9.6 | Cytochrome o oxidase | Cytochrome o oxidase subunit IV | 31008 | 164, 615, 720 |
| cyoE | 9.6 | Cytochrome o oxidase | Cytochrome o oxidase subunit, protoheme IX farnesyltransferase | 31014 | 164, 3799, 615, 720, 721 |
| cysA | 54.7 | Cysteine | Sulfate permease; chromate resistance | 898 | 3793, 4082, 4172, 47, 539 |
| cysB | 28.7 | Cysteine | Positive regulator for cys regulon | 897 | 1980, 3310, 4382 |
| cysC | 61.9 | Cysteine | Adenylylsulfate kinase (EC 2.7.1.25) | 896 | 2520, 3879, 4380 |
| cysD | 61.9 | Cysteine | Sulfate adenylyltransferase (EC 2.7.7.4) | 895 | 1862, 2742, 4380 |
| cysE | 81.5 | Cysteine | Serine acetyltransferase (EC 2.3.1.30) | 894 | 1021, 4381 |
| cysG | 75.3 | Cysteine | Uroporphyrinogen III methyltransferase; transcribed from nirB operon and cysG promoters | 893 | 1647, 1973, 2693, 2694, 3386, 3387, 4148, 4380, 792 |
| cysH | 62.2 | Cysteine | Adenylylsulfate reductase (EC 1.8.99.2) | 892 | 1862, 2345, 2346, 2522, 4380 |
| cysI | 62.2 | Cysteine | cysQ; sulfite reductase, α-subunit (EC 1.8.1.2) | 891 | 1175, 1862, 2522, 4380, 4795 |
| cysJ | 62.3 | Cysteine | cysP; sulfite reductase, β-subunit (EC 1.8.1.2) | 890 | 1175, 1862, 2522, 2637, 4380, 4795 |
| cysK | 54.5 | Cysteine | cysZ; cysteine synthase (EC 4.2.99.8); homodimer; selenate resistance, azaserine resistance | 889 | 1247, 3370, 3793, 4082, 438, 47, 4731, 508, 539 |
| cysM | 54.7 | Cysteine | o-Acetylserine sulfhydrolase B (EC 4.2.99.8) | 17722 | 3793, 4082, 47, 539 |
| cysN | 61.9 | Cysteine | ATP sulfurylase (ATP:sulfate adenylyltransferase) | 18436 | 2520 |
| cysP | 54.8 | Cysteine | Periplasmic sulfate binding protein; see cysJ | 27367 | 1828, 4081 |
| cysQ | 95.6 | Cysteine | amt, amtA; requirement for sulfite or cysteine during aerobic growth; see also cysI | 34409 | 1193, 1996, 1997, 3159 |
| cysS | 11.9 | Cysteine | Cysteinyl-tRNA synthetase (EC 6.1.1.16) | 888 | 176, 413, 1816 |
| cysT | 42.9 | Cysteine | Cysteine tRNA; see also cysU | 17719 | 1285, 2313 |
| cysU | 54.7 | Cysteine | Cysteine transport system; may also transport molybdate (see mod) | 37093 | 4081 |
| cysW | 54.7 | Cysteine | Membrane-bound sulfate transport protein; may also transport molybdate (see mod) | 27371 | 4081 |
| cysX | 81.5 | Cysteine | Reading frame in opposite orientation within cysE gene; polypeptide synthesized in maxicells | 55332 | 4381 |
| cysZ | 54.5 | Cysteine | putative; ORF upstream of cysK may be cysZ | 33018 | 593 |
| cytR | 88.8 | Cytosine resistant? | Regulatory gene for deo, udp, and cdd; mutants show improved growth on uridine | 887 | 226, 3048, 3617, 4128, 4529 |
| dacA | 14.3 | d-Alanine carboxypeptidase | pfv; d-alanine carboxypeptidase IA (EC 3.4.12.11); penicillin-binding protein; deletion suppresses ftsK mutant block | 886 | 2801, 2802, 3198, 3360, 4157, 4232, 520 |
| dacB | 71.7 | d-Alanine carboxypeptidase | d-Alanine carboxypeptidase IB; penicillin-binding protein (EC 3.4.12.11) | 885 | 2322, 2803, 3021, 3022, 4075, 4337 |
| dacC | 19.0 | d-Alanine carboxypeptidase | Penicillin-binding protein 6 (EC 3.4.17.8) | 34706 | 276, 3421, 520 |
| dacD | 44.8 | d-Alanine carboxypeptidase | phsE? Penicillin-binding protein 6b | 50486 | 223 |
| dadA | 26.7 | d-Amino acid dehydrogenase | dadR; d-amino acid dehydrogenase subunit | 884 | 2357, 2607, 2797, 4736, 4737, 4739, 4740 |
| dadB | 1.6 | d-Amino acid dehydrogenase | alnA; d-amino acid dehydrogenase subunit | 883 | 1298 |
| dadQ | 98.9 | d-Amino acid dehydrogenase | alnR; regulator of dad regulon | 882 | 1298 |
| dadX | 26.7 | d-Amino acid dehydrogenase | msuA? alanine racemase (EC 5.1.1.1) | 17716 | 1526, 2607, 2797, 4739 |
| dam | 75.7 | DNA adenine methylase | DNA adenine methylase | 881 | 145, 1609, 2851, 3411, 518 |
| dapA | 56.0 | Diaminopimelate (lysine path) | Dihydrodipicolinate synthase (EC 4.2.1.52) | 880 | 3667 |
| dapB | 0.6 | Diaminopimelate (lysine path) | Dihydrodipicolinate reductase (EC 1.3.1.26); see dapE and lspA | 879 | 2699, 464 |
| dapC | 3.9 | Diaminopimelate (lysine path) | Tetrahydropicolinate succinylase | 878 | 1359, 558 |
| dapD | 4.0 | Diaminopimelate (lysine path) | Succinyl-diaminopimelate aminotransferase | 877 | 1000, 1359, 296, 3665 |
| dapE | 55.8 | Diaminopimelate (lysine path) | dapB; N-succinyl-diaminopimelate deacylase | 876 | 3364, 3667 |
| dapF | 86.1 | Diaminopimelate (lysine path) | Diaminopimelate epimerase | 17713 | 3664, 3666, 926 |
| dbpA | 30.3 | DNA binding protein | Binds DNA, RNA, only hydrolyses ATP in presence of 23S rRNA | 32058 | 1337, 3177 |
| dcd | 46.1 | dCTP deaminase | paxA; dCTP deaminase (EC 3.5.4.13); mutants suppress lethal dut mutants | 875 | 1180, 3156, 4655 |
| dcm | 43.7 | DNA cytosine methylation | mec; DNA cytosine methylase; internal cytosine methylated | 874 | 1314, 1625, 337 |
| dcp | 35.0 | Dipeptidyl carboxypeptidase | Dipeptidyl carboxypeptidase II (EC 3.4.15.1) | 873 | 1027, 1723, 269 |
| dcrB | 77.8 | C resistance | Resistant to lytic phage C1; periplasmic protein perhaps anchored to inner membrane | 46606 | 2546 |
| dctA | 79.3 | Dicarboxylic acid transport | Uptake of C-4 dicarboxylic acids; 3-fluoromalate resistance, d-tartrate resistant | 872 | 206, 2181, 2602, 3897 |
| dctB | 16.4 | Dicarboxylic acid transport | Uptake of C-4 dicarboxylic acids; 3-fluoromalate resistance, d-tartrate resistant | 871 | 2181, 3897 |
| dcuA | 94.0 | Dicarboxylate uptake | genA; C4-dicarboxylate transporter, anaerobic | 34476 | 4084 |
| dcuB | 93.7 | Dicarboxylate uptake | genF; C4-dicarboxylate transporter, anaerobic | 34479 | 4084 |
| dcuC | 14.1 | Dicarboxylate uptake | C4-dicarboxylate carrier, anaerobic; W3110 has an IS5 insertion | 50968 | 4956 |
| ddlA | 8.6 | d-Alanine ligase | d-Alanine:d-alanine ligase, ADP-forming | 30966 | 3350, 4921 |
| ddlB | 2.2 | d-Alanine ligase | ddl; d-Alanine:d-alanine ligase | 870 | 1893, 2071, 2675, 2676, 3350, 3692 |
| deaD | 71.2 | Dead-box protein | csdA, mssB; gene dosage-dependent suppressor of rpsB(ts) mutations; putative RNA helicase | 33472 | 1200, 2045, 4437, 4843 |
| dedA | 52.4 | Downstream, expressed? | Temporary designation for genes in pdxB and folC operons; unknown function | 32742 | 3211 |
| dedD | 52.4 | Downstream, expressed? | Temporary designation for genes in pdxB and folC operons; unknown function | 32739 | 3211 |
| def | 74.0 | Deformylase | fms; peptide deformylase, N-formylmethionylaminoacyl-tRNA deformylase (EC 3.4.11.–, EC 3.5.1.27) | 33619 | 2830, 2869, 2871, 2872, 2873, 3597 |
| degP | 3.9 | Degradative protease | htrA; DegP periplasmic serine endoprotease (EC 3.4.99–), protease Do, required for high-temperature growth; ςE promoter | 30554 | 1767, 2570, 2571, 2572, 3974, 4092, 4251, 660 |
| degQ | 72.8 | Degradative protease | hhoA; periplasmic serineendoprotease | 36675 | 2296, 250, 4643 |
| degS | 72.8 | Degradative protease | htrH, hhoB; periplasmic serineendoprotease | 36678 | 250, 4643 |
| del | 64.3 | Deletion | Affects frequency of IS1-mediated deletions; 1,000-fold reduction in deletion frequency | 869 | 3164 |
| deoA | 99.5 | Deoxyribose | tpp-75; thymidine phosphorylase (EC 2.4.2.4) | 868 | 1255, 22, 4270, 4528, 4530, 4531, 4532 |
| deoB | 99.5 | Deoxyribose | drm, thyR, tlr; deoxyribouratase (EC 2.7.5.6), phosphopentomutase | 867 | 1255, 22, 2616, 3705, 4527, 4530, 4532 |
| deoC | 99.5 | Deoxyribose | dra, thyR, tlr; deoxyribose-phosphate aldolase (EC 4.1.2.4) | 866 | 1255, 2616, 3705, 4526, 4527, 4528, 4532, 54, 920 |
| deoD | 99.6 | Deoxyribose | pup; purine-nucleoside phosphorylase PNP (EC 2.4.2.1) | 865 | 1255, 22, 2282, 2422, 3705, 4528, 4532 |
| deoR | 19.0 | Deoxyribose | nucR, nupG, tse; regulatory gene for deo operon | 864 | 3017, 3048, 4533 |
| dfp | 82.1 | DNA synthesis flavoprotein | dnaS, dut; flavoprotein affecting DNA synthesis and pantothenate metabolism | 18430 | 4154, 4155 |
| dgd | 72.0 | d-Galactose dehydrogenase | d-Galactose dehydrogenase production | 863 | 4803 |
| dgkA | 91.7 | Diglyceride kinase | Diglyceride kinase | 862 | 2544, 2545 |
| dgkR | 93.7 | Diglyceride kinase | Regulatory | 861 | 3578 |
| dgoA | 83.4 | d-Galactonate | 2-Oxo-3-deoxygalactonate 6-phosphate aldolase (EC 4.1.2.21) | 36891 | 182 |
| dgoD | 83.4 | d-Galactonate | Galactonate dehydratase (EC 4.2.1.6) | 859 | 182 |
| dgoK | 83.5 | d-Galactonate | 2-Oxo-3-deoxygalactonate kinase (EC 2.7.1.58) | 858 | 182 |
| dgoR | 83.5 | d-Galactonate | Regulatory; growth on 2-keto-3-deoxygalactonate, dgoRc | 857 | 830 |
| dgoT | 83.4 | d-Galactonate | Galactonate transport | 856 | 182 |
| dgsA | 35.9 | d-Glucosamine | mlc (makes large colonies); affects function of phosphotransferase system enzyme IIA/IIB, anaerobic growth on glucosamine; binds NagC promoters; regulates manX | 855 | 1813, 3012, 3453, 3707 |
| dgt | 3.9 | dGTP triphosphohydrolase | optA; deoxyguanosine 5′-triphosphate triphosphohydrolase (EC 3.1.5.1) | 30546 | 261, 3572, 3573, 4806 |
| dicA | 35.5 | Division control | Regulatory for dicB | 18427 | 282, 283 |
| dicB | 35.5 | Division control | Control of cell division | 18424 | 282, 283, 622 |
| dicC | 35.5 | Division control | Regulatory for dicB | 18421 | 282, 283 |
| dicF | 35.5 | Division control | DicF antisense RNA; inhibits Qin | 32240 | 1208, 1209, 4389, 453 |
| dif | 34.2 | Deletion-induced filamentation | Recombination site in terminus, recA independent | 30208 | 1675, 2354, 2356, 376, 378, 835, 836, 4378 |
| dinB | 5.4 | Damage inducible | dinP; increased mutagenesis, apparently independent of umuDC; SOS related | 53389 | 2232, 526 |
| dinD | 82.2 | Damage inducible | orfY, pcsA, yicD; mutant cs phenotype filamentous with large nucleoid | 33582 | 2200, 2352, 2353, 2664, 3262, 4877 |
| dinF | 91.7 | Damage inducible | Induced by UV and mitomycin C; SOS, lexA regulon | 854 | 2200, 2348, 2928 |
| dinG | 17.9 | Damage inducible | LexA regulated (SOS) repair enzyme | 31247 | 2318, 2518, 2519 |
| dinI | 24.1 | Damage inducible | Multicopy suppresses phenotype of cold-sensitive dinD filamentous mutation | 53428 | 4877 |
| dinY | 41.9 | Damage inducible | Repair gene | 36880 | 3412 |
| dipZ | 94.0 | Disulfide isomerase | cycZ, dsbD, cutA2; may be involved in cytochrome maturation, see ccm genes; affects disulfide binding | 34213 | 1921, 268, 2910, 874, 875 |
| djlA | 1.2 | DnaJ-like | Proposed to dock and interact with variety of membrane proteins; mutants rapidly accumulate suppressors | 51192 | 2188, 769, 770 |
| dksA | 3.5 | dnaK suppressor | msmA; high copy suppresses muk and TS growth and filamentation of dnaK mutant | 30521 | 2122, 4842 |
| dld | 47.9 | d-Lactate dehydrogenase | ldh; d-lactate dehydrogenase (EC 1.1.1.28); vinylglycolate resistance, FAD enzyme | 852 | 2058, 3768, 4012, 626 |
| dmsA | 20.3 | DMSO reductase | DMSO reductase subunit A, anaerobic | 17710 | 353, 354, 3749 |
| dmsB | 20.3 | DMSO reductase | DMSO reductase subunit B; apparent Fe-S binding domain; anaerobic | 31724 | 354, 3749 |
| dmsC | 20.3 | DMSO reductase | DMSO reductase subunit C, membrane bound | 31727 | 354, 3749 |
| dnaA | 83.6 | DNA | DNA biosynthesis; initiation; binding protein | 851 | 1372, 1630, 1631, 1872, 1873, 1875, 2160, 2235, 2272, 2275, 2474, 2927, 2985, 2986, 3051, 3261, 3400, 3541, 3569, 3570, 3809, 3813, 4087, 4300, 4301, 4611, 471, 4910, 1790 |
| dnaB | 91.9 | DNA | groP, grpA, grpD; DNA biosynthesis; chain elongation | 850 | 1406, 2259, 2545, 3126, 3333, 3804, 3951, 4449, 561 |
| dnaC | 99.1 | DNA | dnaD; DNA biosynthesis; initiation and chain elongation | 849 | 2786, 3125, 3753 |
| dnaE | 4.4 | DNA | polC, sdgC (suppressor of dnaG mutation); DNA polymerase III, α-subunit | 373 | 1239, 1240, 2194, 296, 3889, 4016, 4424, 4708, 4761 |
| dnaG | 69.2 | DNA | dnaP, parB, sdgA; primase; primer synthesis for leading- and lagging-strand synthesis | 847 | 1531, 2671, 2672, 2673, 3055, 3113, 3125, 3216, 3753, 4105, 4376, 4449, 4578, 4766, 579 |
| dnaI | 40.3 | DNA | DNA biosynthesis | 846 | 334a |
| dnaJ | 0.3 | DNA | groP, grpC; chain elongation; stress-related DNA biosynthesis, responsive to heat shock; chaperone with DnaK | 845 | 1406, 2069, 231, 3256, 3257, 3258, 3333, 3804, 4252, 4293, 4830, 972 |
| dnaK | 0.3 | DNA | gro, groP, groPAB, groPC, groPF, grpC, grpF, seg; stress-related heat-shock DNA biosynthesis, ATP-regulated binding and release of polypeptide substrates; HSP-70-type molecular chaperone, with DnaJ | 844 | 1143, 1186, 1187, 1875, 2120, 227, 2837, 3035, 3158, 3257, 3332, 3807, 4252, 567, 3697 |
| dnaL | 28.9 | DNA | DNA biosynthesis | 843 | 3982 |
| dnaN | 83.6 | DNA | DNA biosynthesis; sliding clamp subunit, required for high processivity; DNA polymerase III β subunit | 842 | 132, 1630, 2194, 2235, 3261, 3626, 3627, 3809, 3810, 3813, 4910, 564 |
| dnaQ | 5.1 | DNA | mutD; DNA polymerase III ɛ-subunit; streptomycin, azaserine resistant; 3′ to 5′ proofreading, lexA regulon | 840 | 1047, 1113, 1239, 1706, 1804, 2194, 2730, 2783, 3209, 3568, 3903, 4052, 779, 855, 856, 4331 |
| dnaT | 99.1 | DNA | Primasomal protein i | 839 | 2786, 2787, 3125 |
| dnaX | 10.6 | DNA | mutH, dnaZ; subunit of DNA polymerase III holoenzyme; DNA elongation factor III; τ and γ subunits | 838 | 1264, 1265, 1266, 1669, 1845, 2194, 2228, 2229, 2283, 2421, 2471, 2472, 2511, 2731, 3043, 3753, 398, 399, 4485, 4486, 4885, 701, 912, 913 |
| dppA | 79.8 | Dipeptide permease | alu, tpp?; uptake of dipeptides | 35111 | 3294, 4, 4576 |
| dppB | 79.8 | Dipeptide permease | Uptake of dipeptides | 33771 | 4, 4118, 4576 |
| dppC | 79.8 | Dipeptide permease | Uptake of dipeptides | 33768 | 4, 4118, 4576 |
| dppD | 79.8 | Dipeptide permease | Uptake of dipeptides | 33765 | 4, 4118, 4576 |
| dppF | 79.7 | Dipeptide permease | Uptake of dipeptides | 33752 | 4 |
| dppG | 14.0 | Dipeptide permease | Uptake of dipeptides; dipeptide permease | 835 | 1453, 3384, 997 |
| dps | 18.3 | DNA-binding protein, stationary phase | pexB; stress response DNA-binding protein; starvation induced resistance to H2O2 | 31650 | 1226, 2620, 71 |
| dsbA | 87.1 | Disulfide bond | iarA, ppfA; disulfide oxidoreductase, periplasmic protein disulfide-isomerase; role in cytochrome c synthesis (EC 5.3.4.1) | 34063 | 1560, 2243, 229, 230, 2778, 287, 2910, 3549, 3550, 3823, 45, 4805, 4840, 67, 904 |
| dsbB | 26.6 | Disulfide bond | iarB; PDI or PDI-like protein; DTT-sensitive phenotype; periplasm/inner membrane | 31933 | 1560, 1989, 2243, 229, 288, 2960, 3823, 67 |
| dsbC | 65.4 | Disulfide bond | xprA; periplasmic disulfide oxidoreductase, protein disulfide isomerase | 33355 | 2639, 2641, 2962, 4019 |
| dsbG | 13.8 | Disulfide bond | Thiol-disulphide oxidase; multicopy resistance to DTT; mutants accumulate reduced proteins, corrected by DsbA/B overexpression | 53792 | 92 |
| dsdA | 53.4 | d-Serine deaminase | d-Serine deaminase | 834 | 2751, 2846, 2847, 3215, 3339, 436, 48, 642 |
| dsdC | 53.3 | d-Serine deaminase | LysR-type transcriptional regulator; previously cited as d-serine permease; dsdC/X order reversed in different sequence entries | 833 | 2847, 3215, 3339, 436, 48, 642 |
| dsdX | 53.4 | d-Serine deaminase | Homology with gluconate permease; d-serine tolerance; dsdC/A order reversed in different sequence entries | 35717 | 3215 |
| dsrA | 43.6 | Small RNA | Regulatory RNA; positive regulation of promoters sensitive to HNS negative regulation | 48173 | 2249, 4098 |
| dsrB | 43.6 | Small RNA | Regulatory RNA; regulated by DsrA and HNS, under control of RpoS | 48176 | 4099 |
| dut | 82.2 | dUTPase | dnaS, sof; deoxyuridinetriphosphatase (EC 3.6.1.23) | 832 | 2661, 2662, 4154, 4367 |
| dvl | 7.4 | Dye-visible light? | Sensitivity to SDS and toluidine blue plus light | 18418 | 4632 |
| dxs | 9.4 | Deoxy-xylulose-P synthase | DXP synthase; DXP is precursor to isoprenoids, thiamin, pyridoxol | 52930 | 2615, 4166 |
| e14 | 25.7 | Prophage element 14 | Defective prophage element; includes loci sfiC, lit, pin, mcrA | 18409 | 2128, 2715, 3447, 3448, 3604, 4544, 511, 512, 625 |
| ebgA | 69.4 | Evolved β-galactosidase | Cryptic β-galactoside utilization | 830 | 1607, 4234, 4235, 627 |
| ebgB | 69.5 | Evolved β-galactosidase | Cryptic β-galactoside utilization, possible paralog of lacY | 18415 | 1607, 4234 |
| ebgC | 69.5 | Evolved β-galactosidase | Phospho-β-d-galactosidase, β-subunit; cryptic gene | 18412 | 1607 |
| ebgR | 69.4 | Evolved β-galactosidase | Regulatory gene from ebg cryptic operon | 829 | 1607, 4234 |
| ecfA | 67.6 | Energy coupling factor | With metC mutation, ecf mutation abolishes coupling of energy with active transport | 828 | 4434 |
| eco | 49.6 | Ecotin | Ecotin, serine protease inhibitor | 32630 | 1173 |
| ecpD | 3.4 | E. coli papD homolog | Possible pilin chaperone | 30513 | 3593 |
| eda | 41.6 | Entner-Douderoff aldolase | hga, kdgA, kga; 2-keto-3-deoxygluconate 6-phosphate aldolase (EC 4.1.2.14); 2-keto-4-hydroxyglutarate aldolase | 826 | 1154, 1290 |
| edd | 41.6 | Entner-Douderoff dehydratase | Phosphogluconate dehydratase (EC 4.2.1.12); growth on gluconate | 825 | 1121, 1289, 1290, 4594 |
| efp | 94.3 | Elongation factor P | Elongation factors P and EF-P; prokaryotic | 34470 | 118, 119, 1357 |
| emrA | 60.6 | E-multidrug resistance | Multidrug resistance pump family | 33259 | 2619, 2621 |
| emrB | 60.6 | E-multidrug resistance | Hydrophobic, inner membrane-spanning domains; multidrug resistance pump family | 33262 | 2619, 2621 |
| emrD | 83.0 | E-multidrug resistance | Multidrug resistance pump family | 36938 | 3132 |
| emrE | 12.2 | E-multidrug resistance | envB, mvrC, mon, rodY; multidrug resistance pump family; cell shape; methylviologen sensitivity | 36935 | 2452, 4879, 4880, 300 |
| endA | 66.6 | Endonuclease | DNA-specific endonuclease I; extensive DNA breakdown | 824 | 2002, 4785 |
| eno | 62.6 | Enolase | Enolase (EC 4.2.1.11) | 823 | 158, 2257, 4709 |
| entA | 13.5 | Enterochelin | 2,3-Dihydro-2,3-dihydroxybenzoate dehydrogenase | 822 | 1262, 1777, 2394, 2395, 2581, 3090, 3091, 3424, 4180 |
| entB | 13.5 | Enterochelin | 2,3-Dihydro-2,3-dihydroxybenzoate synthetase | 821 | 1262, 2394, 2395, 3090, 3091, 3326, 5424 |
| entC | 13.5 | Enterochelin | Isochorismate synthetase | 820 | 1262, 2394, 2395, 2582, 3090, 3091, 3325, 3326, 3424, 4644 |
| entD | 13.1 | Enterochelin | Enterochelin synthetase, component D; facilitates secretion of enterobactin peptide | 819 | 1261, 133, 1534, 2394, 2395, 2400, 786, 787 |
| entE | 13.5 | Enterochelin | Enterochelin synthetase, component E | 818 | 1262, 2394, 2395, 3090, 3091, 3424 |
| entF | 13.2 | Enterochelin | Enterochelin synthetase, component F | 817 | 1262, 2394, 2395, 3415, 3770, 286 |
| envN | 4.2 | Envelope | Affects envelope; defects osmotically remedied | 811 | 1120 |
| envP | 90.4 | Envelope | Affects envelope; defects osmotically remedied | 810 | 1120 |
| envQ | 60.7 | Envelope | Affects envelope; defects osmotically remedied | 809 | 1120 |
| envR | 73.5 | Envelope | acrS; regulatory gene for envCD (acrEF) | 33605 | 2516, 2684 |
| envT | 14.2 | Envelope | Affects envelope; defects osmotically remedied | 808 | 1120 |
| envY | 12.6 | Envelope | Envelope protein involved with thermoregulation of porin | 18406 | 2666, 2668 |
| envZ | 76.1 | Envelope | ompB, perA, tpo; inner membrane osmosensor protein; regulates production of outer membrane proteins | 807 | 1373, 1374, 1610, 1611, 2665, 2978, 2979, 2983, 3349, 3769, 4647, 4662, 4681, 4807, 661, 816 |
| epd | 66.2 | Erythrose-4-P dehydrogenase | gapB; erythrose-4-P dehydrogenase | 32089 | 1071, 3980, 444, 4946, 61 |
| epp | E-pentapeptide | Minigene within 23S rRNA encoding functional pentapeptide; erythromycin resistance | 51135 | 4383 | |
| era | 58.2 | E. coli ras-like | sdgE; GTP-binding protein, essential gene | 29010 | 2498, 25, 4137, 708 |
| esp | 17.3 | Efficiency site for phage | Site for efficient packaging of phage T1 | 805 | 1085a |
| evgA | 53.5 | E. coli homolog of virulence gene | Multicopy on plasmid in envZ-deleted strain induces ompC expression; see evgS | 32763 | 4519 |
| evgS | 53.5 | E. coli homolog of virulence gene | With evgA, two-component regulatory system, environmentally responsive | 32766 | 4519 |
| exbB | 67.9 | Export | Uptake of enterochelin; resistance or sensitivity to colicins; similarity with TolQ | 804 | 1136, 1137, 1254, 1580, 2109, 2136, 2221, 24, 3554, 481, 482 |
| exbC | 61.5 | Export | Uptake of enterochelin; resistance or sensitivity to colicins | 803 | 1580, 3554 |
| exbD | 67.9 | Export | Uptake of enterochelin; resistance or sensitivity to colicins; similarity with TolR | 33384 | 1137, 1254, 1721, 2221, 24, 482 |
| expA | 22.2 | Export | Expression of a group of export proteins | 802 | 944 |
| exuR | 69.9 | Hexuronate | Negative regulatory gene for exu regulon (exu, uxu, uxa) | 801 | 1852, 1853, 2795, 3489, 3490, 3682, 3687 |
| exuT | 69.9 | Hexuronate | Transport of hexuronates | 800 | 1852, 2795, 2796, 3489, 3490, 3682, 382 |
| fabA | 21.9 | Fatty acid biosynthesis | β-hydroxydecanoylthioester dehydrase (EC 4.2.1.60) | 799 | 870, 872 |
| fabB | 52.6 | Fatty acid biosynthesis | fabC; β-ketoacyl-acyl carrier protein synthase I (EC 2.3.1.41) | 798 | 1380, 1381, 4056 |
| fabD | 24.8 | Fatty acid biosynthesis | Malonyl-CoA-acyl carrier protein transacylase (EC 2.3.1.39) | 797 | 1381, 2713, 4579 |
| fabF | 24.8 | Fatty acid biosynthesis | cvc, fabJ, vtr; β-ketoacyl-acyl carrier protein synthase II (EC 2.3.1.41) | 795 | 1380, 1381, 3621, 4057, 4512 |
| fabG | 24.8 | Fatty acid biosynthesis | 3-Ketoacyl-ACP reductase (EC 1.1.1.100); reuse of fabG synonym for accC | 31865 | 2315, 3253, 3621 |
| fabH | 24.7 | Fatty acid biosynthesis | ACP synthase III | 31860 | 3253, 3466, 4481 |
| fabI | 29.1 | Fatty acid biosynthesis | gts, envM, qmeA; enoyl-ACP reductase, NADH dependent (EC 1.3.1.9) | 812 | 1687, 2251, 321, 322, 4502, 4734, 4735 |
| fabZ | 4.4 | Fatty acid biosynthesis | sefA; 3R-hydroxymyristoyl acyl carrier protein dehydrase | 30597 | 2990, 3212 |
| fadA | 86.8 | Fatty acid degradation | Thiolase I (EC 2.3.1.16) | 794 | 1057, 3099, 4160, 4860, 4862, 762, 926 |
| fadB | 86.8 | Fatty acid degradation | oldB; 3-hydroxyacyl-CoA dehydrogenase: 3-hydroxyacyl-CoA epimerase; dodecenoyl-CoA-Δ-isomerase, enoyl-CoA hydratase; fatty acid oxidation complex, α-subunit (EC 1.1.1.35, 4.2.1.17, 5.1.2.3, and 5.3.3.8); four activities | 793 | 1057, 3099, 3098, 4160, 4860–4862, 926 |
| fadD | 40.7 | Fatty acid degradation | Acyl-CoA synthetase (EC 6.2.1.3) | 792 | 1336, 371 |
| fadE | 4.8 | Fatty acid degradation | Electron transport flavoprotein of β-oxidation | 791 | 762 |
| fadH | 69.6 | Fatty acid degradation | 2,4-Dienoyl CoA reductase | 54705 | 1686 |
| fadL | 53.0 | Fatty acid degradation | ttr; fatty acid transport protein, outer membrane | 790 | 3010, 3222, 370, 372, 3796 |
| fadR | 26.6 | Fatty acid degradation | dec, ole, thdB; negative regulatory gene for fad regulon, positive regulator of fabA; regulates aceBAK, glyoxylate shunt | 789 | 1056, 1058, 1204, 1729, 2744, 3, 4073, 4074, 4739, 762 |
| farR | 16.5 | Fatty acyl responsive | Fatty acyl responsive regulator | 36837 | 3566, 551 |
| fatA | 71.7 | Fatty acid | Utilization of trans unsaturated fatty acids | 18403 | 1030 |
| fbaA | 66.1 | Fructose bis-P aldolase | ald, fda; fructose-bisphosphate aldolase | 786 | 326, 3560, 4076, 62 |
| fbaB | 46.9 | Fructose bis-P aldolase | dhnA; fructose 1,6-bisphosphate aldolase | 54836 | 4404, 4076 |
| fbp | 96.0 | Fructose-bisphosphatase | fdp; fructose-bisphosphatase (EC 3.1.3.11) | 784 | 2275, 3961 |
| fcl | 45.8 | Fucose, FX-like | Colanic acid gene cluster, fucose synthetase | 55074 | 113 |
| fcsA | 86.9 | Filamentous, cold sensitive | Cold-sensitive cell division mutant | 787 | 2352 |
| fdhD | 88.0 | Formate dehydrogenase | Homologous to fdnC and -B of Salmonella spp. | 33972 | 2746, 3465, 3912, 4221 |
| fdhE | 87.9 | Formate dehydrogenase | Homologous to fdnC and -B of Salmonella spp. | 33978 | 2746, 3193, 3465, 3912, 4221 |
| fdhF | 92.6 | Formate dehydrogenase | chlF, fdh; formate dehydrogenase-N selenopolypeptide subunit molybdate dependence (EC 1.2.2.1), part of FHL complex | 18400 | 3388, 360, 3736, 4798, 4799, 4964, 4965, 697 |
| fdnG | 33.3 | Formate dehydrogenase-N | Formate dehydrogenase-N major subunit | 32160 | 2527, 312, 313, 314, 3575, 3678, 4221 |
| fdnH | 33.4 | Formate dehydrogenase-N | Formate dehydrogenase-N Fe-S subunit | 32164 | 2527, 312, 313, 314, 3575, 3678, 4221, 859 |
| fdnI | 33.4 | Formate dehydrogenase-N | Formate dehydrogenase-N cytochrome subunit | 32168 | 2527, 312, 313, 314, 3575, 3678, 4221 |
| fdoG | 88.0 | Formate dehydrogenase-O | Formate dehydrogenase-O subunit, major | 33985 | 2, 3465 |
| fdoH | 87.9 | Formate dehydrogenase-O | Formate dehydrogenase-O subunit, Fe-S | 33988 | 2, 3465 |
| fdoI | 87.9 | Formate dehydrogenase-O | Formate dehydrogenase-O subunit cytochrome b556 | 33991 | 2, 3465 |
| fdrA | 11.8 | ftsH dominant rescue | In multicopy, suppresses negative ftsH mutations | 40877 | 44 |
| fdx | 57.2 | Ferredoxin | Ferredoxin, cotranscribed with hscBA, presumably cold inducible | 32984 | 2484, 4320, 4321 |
| feaB | 31.2 | Phenylethylamine | padA; catabolism of phenylethylamine | 50925 | 1230, 1629 |
| feaR | 31.1 | Phenylethylamine | maoB; regulates expression of tynA and padA | 50928 | 1230, 1629 |
| fecA | 97.3 | Iron (citrate dependent) | Citrate-dependent iron transport, outer membrane receptor | 783 | 114, 1870, 3536, 4558, 4569, 4629, 4962 |
| fecB | 97.2 | Iron (citrate dependent) | Citrate-dependent iron transport, periplasmic protein | 782 | 1870, 3536, 4190, 4569, 4962 |
| fecC | 97.2 | Iron (citrate dependent) | Transport gene | 35620 | 4190, 4569, 4558 |
| fecD | 97.2 | Iron (citrate dependent) | Citrate-dependent iron transport, membrane bound | 18394 | 3536, 4190, 4569, 4558 |
| fecE | 97.2 | Iron (citrate dependent) | Transport gene | 35617 | 4190, 4569, 4558 |
| fecI | 97.3 | Iron (citrate dependent) | Transport gene mediating induction by iron | 35630 | 114, 4569, 4558 |
| fecR | 97.3 | Iron (citrate dependent) | Regulatory gene mediating induction by iron | 35627 | 4558, 4569 |
| feoA | 76.3 | Ferrous iron | Ferrous iron uptake system | 28964 | 1638, 2107 |
| feoB | 76.3 | Ferrous iron | Membrane protein of ferrous iron uptake system | 28967 | 1638, 2107 |
| fepA | 13.1 | Ferrienterobactin permease | cbr, cbt; outer membrane component of ferribactin transport system | 18388 | 1261, 1863, 2669, 3325, 3416, 3429, 3554, 4013, 4780, 786 |
| fepB | 13.4 | Ferrienterobactin permease | Periplasmic component of ferrienterobactin transport system | 18385 | 3325, 3428, 3429, 4013, 4202, 4780, 486, 501 |
| fepC | 13.3 | Ferrienterobactin permease | Cytoplasmic membrane component of ferrienterobactin transport | 4987 | 3325, 3326, 3428, 4013 |
| fepD | 13.4 | Ferrienterobactin permease | Ferrienterobactin permease, membrane-bound subunit | 18382 | 1863, 3325, 4013 |
| fepE | 13.3 | Ferrienterobactin permease | Ferric enterobactin uptake | 18379 | 3325, 4013 |
| fepG | 13.4 | Ferrienterobactin permease | Ferrienterobactin permease, membrane-bound subunit | 31162 | 717, 4013 |
| fes | 13.2 | Iron | Enterochelin esterase | 780 | 1261, 1777, 1863, 2394, 2395, 3415, 3416, 786 |
| fexB | 86.7 | F exclusion | Affects ArcA phenotype | 778 | 2499 |
| ffh | 59.2 | 54-kDa homolog (4.5S particle) | 4.5S-RNP protein; RNP has mammalian protein-targeting counterpart: SRP | 33006 | 2695, 325, 3407, 3419, 3488, 3520 |
| ffs | 10.2 | 4.5S | 4.5S rRNA; mammalian counterpart, SRP, includes 4.5S RNA; cotranslational integration of proteins into membrane | 18373 | 1064, 1834, 2695, 534 |
| fhlA | 61.5 | Formate hydrogen-lyase | Transcription factor, formate hydrogen lyase system activator, global regulator, hyc, hyp | 18370 | 1800, 1801, 1957, 3745, 3852, 3853, 3907, 3909 |
| fhlB | 95.1 | Formate hydrogen-lyase | Formate hydrogen-lyase; activated by fhlA | 36851 | 2821 |
| fhuA | 3.6 | Ferric hydroxamate uptake | T1, T5rec, tonA; OMP receptor for ferrichrome, colicin M, and phages T1, T5, and φ80; energy-coupled transport of Fe3+ via ferrichrome; mutants albomycin resistant | 777 | 1216, 2081, 2219, 2221, 2220, 2652, 2653, 457, 483, 485, 486, 849, 850 |
| fhuB | 3.7 | Ferric hydroxamate uptake | Hydroxamate-dependent iron uptake, cytoplasmic membrane component; mutants albomycin resistant | 776 | 1216, 2081, 2330, 3544, 486 |
| fhuC | 3.7 | Ferric hydroxamate uptake | Hydroxamate-dependent iron uptake, cytoplasmic membrane component; mutants albomycin resistant | 11187 | 1216, 486, 566, 848 |
| fhuD | 3.7 | Ferric hydroxamate uptake | Hydroxamate-dependent iron uptake, cytoplasmic membrane component; mutants albomycin resistant | 11184 | 1216, 3712, 486, 566, 848 |
| fhuE | 25.0 | Ferric hydroxamate uptake | Outer membrane receptor for ferric-rhodotorulic acid | 18367 | 1636, 3881, 3882 |
| fhuF | 99.2 | Ferric hydroxamate uptake | Ferric hydroxamate transport | 18364 | 1639, 3033 |
| fic | 75.2 | Filamentation-cAMP | Filamentation in presence of cyclic AMP, in mutants | 18361 | 2302, 4521 |
| fimA | 97.9 | Fimbriae | fimD, pilA; fimbrin type 1, major structural subunit; phase variation | 18358 | 1142, 1304, 1305, 1634, 2261, 2264, 5 |
| fimB | 97.8 | Fimbriae | pil; recombinase? regulatory gene for fimA | 18355 | 1072, 1142, 1347, 1348, 2262, 2264, 2838 |
| fimC | 97.9 | Fimbriae | pil, pilB; biosynthesis of fimbriae; periplasmic chaperone for type 1 fimbriae | 18352 | 2038, 2263, 2264 |
| fimD | 97.9 | Fimbriae | pil, pilC; export and assembly of type 1 fimbrial outer membrane protein | 18349 | 2264 |
| fimE | 97.9 | Fimbriae | pilH; recombinase? regulatory gene for expression of fimA | 18346 | 1072, 1142, 1347, 1348, 2262, 2838 |
| fimF | 98.0 | Fimbriae | pilD; fimbrin type 1 minor component; fimbrial morphology, assembly | 18343 | 2265, 3779 |
| fimG | 98.0 | Fimbriae | pilD; fimbrin type 1 minor component; pilus length; perhaps inhibits polymerization in pilus assembly | 18340 | 2265, 3779 |
| fimH | 98.0 | Fimbriae | pilE; membrane-specific adhesin (lectin); major fimbrial subunit; mediates mannose-binding | 18337 | 1634, 1655, 2265, 2343, 6 |
| fimI | 97.9 | Fimbriae | Fimbria related | 35633 | 2261 |
| fipB | 86.0 | F1 phage | Morphogenesis of phage F1 | 18334 | 2629 |
| fipC | 75.0 | F1 phage | Morphogenesis of phage F1 | 18331 | 2629 |
| fis | 73.5 | Factor for inversion stimulation | nbp; transcriptional activator for rRNA operons; bends DNA; interacts with RNAP; nucleoid-associated protein | 18328 | 1195, 1244, 1430, 1456, 1464, 1465, 1496, 170, 2280, 2779, 3403, 3742, 443, 4767, 4814, 4815, 4943 |
| fiu | 18.1 | Ferric iron uptake | Outer membrane protein, ferric iron uptake | 18325 | 1636 |
| fixA | 0.9 | Sequence similar to those of N-fixation genes | Related to carnitine metabolism; amino acid similarity to Rhizobium fix gene | 53111 | 1132, 550 |
| fixB | 0.9 | Sequence similar to those of N-fixation genes | Related to carnitine metabolism; amino acid similarity to Rhizobium fix gene | 53114 | 1132, 550 |
| fixC | 1.0 | Sequence similar to those of N-fixation genes | Related to carnitine metabolism; amino acid similarity to Rhizobium fix gene | 53117 | 1132, 550 |
| fixX | 1.0 | Sequence similar to those of N-fixation genes | Related to carnitine metabolism; amino acid similarity to Rhizobium fix gene | 53121 | 1132, 550 |
| flkB | 95.4 | FKBP-like | Periplasmic PPIase, of FK506-binding protein type (EC 5.2.1.8); contains pipecolic acid residue | 48137 | 3584 |
| fkpA | 74.9 | FK506-binding protein | Protein that binds immunosuppressive drug FK506, a peptidomacrolide, MIP-like | 36042 | 1809, 2964 |
| fldA | 15.3 | Flavodoxin | Flavodoxin | 31496 | 3306 |
| flgA | 24.3 | Flagella | flaU; flagellar synthesis; flagellar regulon member | 756 | 1892, 2305 |
| flgB | 24.4 | Flagella | flbA; flagellar synthesis; flagellar regulon member | 750 | 2309 |
| flgC | 24.4 | Flagella | flaW; flagellar synthesis; flagellar regulon member; basal body protein | 754 | 2309 |
| flgD | 24.4 | Flagella | flaV; flagellar synthesis; flagellar regulon member; basal body rod modification | 755 | 2309, 2307 |
| flgE | 24.4 | Flagella | flaK; flagellar hook subunit protein; flagellar regulon member | 766 | 2308, 2309, 2307 |
| flgF | 24.4 | Flagella | flaX; flagellar synthesis; flagellar regulon member; basal body rod protein | 753 | 2309 |
| flgG | 24.4 | Flagella | flaL; flagellar synthesis; flagellar regulon member; basal body rod protein | 765 | 2307, 2309 |
| flgH | 24.5 | Flagella | flaY; flagellar synthesis, basal body L-ring protein | 752 | 2039, 2309 |
| flgI | 24.5 | Flagella | flaM; flagellar regulon member; basal body P-ring protein | 764 | 2039, 2307, 2309 |
| flgJ | 24.5 | Flagella | flaZ; flagellar synthesis; flagellar regulon member | 751 | 2704 |
| flgK | 24.5 | Flagella | flaS; flagellar synthesis; flagellar regulon member; hook-associated protein I | 758 | 2307 |
| flgL | 24.6 | Flagella | flaT; flagellar synthesis; flagellar regulon member; hook-associated protein | 757 | 2307 |
| flgM | 24.3 | Flagella | anti-ς factor regulator of FlhD | 53436 | 532 |
| flgN | 24.3 | Flagella | FlgN flagellar synthesis protein | 53440 | 532 |
| flhA | 42.3 | Flagella | flaH; flagellar synthesis; flagellar regulon member | 768 | 2704 |
| flhB | 42.3 | Flagella | flaG; flagellar synthesis; flagellar regulon member | 769 | 2704, 86 |
| flhC | 42.6 | Flagella | flaI; flagellar synthesis; regulatory gene | 767 | 246, 2589, 2590, 40 |
| flhD | 42.6 | Flagella | flbB; regulatory gene, flagellum-specific ς factor, transcriptional activator of Fla class II operons | 749 | 246, 2589, 2590, 3547 |
| fliA | 43.1 | Flagella | flaD, rpoF; regulation of late gene expression; ς transcription factor for class 3a and 3b operons | 771 | 1925, 2083, 2305, 2369, 2591, 2623, 2704 |
| fliC | 43.1 | Flagella | flaF, hag; flagellin, structural gene; flagellar regulon member | 649 | 1623, 2382, 4316 |
| fliD | 43.2 | Flagella | flbC; hook-associated protein 2, axial family; flagellar regulon member | 748 | 2172 |
| fliE | 43.3 | Flagella | flaN; flagellar synthesis; basal body component | 763 | 2704, 3041 |
| fliF | 43.4 | Flagella | flaBI; flagellar basal body M-ring protein | 17707 | 1891, 2304, 2306, 247, 4065 |
| fliG | 43.4 | Flagella | flaBII; motor switching and energizing | 18322 | 1891, 2304, 2306, 247, 2601, 2785, 3720, 4065 |
| fliH | 43.4 | Flagella | flaBIII; flagellar biosynthesis | 18319 | 1891, 2304, 2306, 247, 4065 |
| fliI | 43.4 | Flagella | flaC; flagellar biosynthesis | 772 | 247 |
| fliJ | 43.5 | Flagella | flaO; flagellar biosynthesis | 762 | 247 |
| fliK | 43.5 | Flagella | flaE; hook filament junction; controls hook length | 770 | 2171, 2172, 247, 2704 |
| fliL | 43.5 | Flagella | flaAI; flagellar biosynthesis | 18316 | 2370, 247, 2739, 3580 |
| fliM | 43.5 | Flagella | cheC, flaA; flagellar synthesis, motor switching and energizing | 774 | 2370, 2785, 777 |
| fliN | 43.5 | Flagella | motD; flagellar switch protein | 18313 | 2739 |
| fliO | 43.5 | Flagella | flbD; flagellar synthesis; flagellar regulon member | 17510 | 2704, 2738, 2739 |
| fliP | 43.5 | Flagella | flaR; flagellar synthesis; flagellar regulon member | 759 | 2738, 2739 |
| fliQ | 43.6 | Flagella | flaQ; flagellar synthesis; flagellar regulon member | 760 | 2739 |
| fliR | 43.6 | Flagella | flaP; flagellar synthesis; flagellar regulon member | 761 | 2738, 2739 |
| fliS | 43.2 | Flagella | Flagellar synthesis; flagellar regulon member | 30736 | 2172 |
| fliT | 43.2 | Flagella | Flagellar synthesis; flagellar regulon member | 30739 | 2172 |
| fliY | 43.1 | Flagella | Not required for motility; may regulate FliA (ςF) | 49936 | 3080 |
| fliZ | 43.1 | Flagella | Not required for motility; may regulate FliA (ςF) | 49933 | 3080 |
| flk | 52.5 | Flagella | div; transcribed divergently from pdxB, promoters overlapping, Salmonella homolog functions in sensing flagellar assembly stage | 32751 | 2134, 3930 |
| flu | 44.6 | Flagella | Antigen 43, phase-variable bipartite outer membrane protein; affects surface properties, piliation, colonial morphology; unstable gene | 746 | 1712, 1712a, 2704 |
| fmt | 74.0 | fMet-tRNA formyltransferase | Methionyl-tRNA formyltransferase (EC 2.1.2.9) | 33616 | 1562, 1563, 2748, 2830, 2870, 3609 |
| fnr | 30.1 | Fumarate-nitrate-reductase | frdB, nirA, nirR, ossA; regulatory gene for nitrite and nitrate reductases, hydrogenase, fumarate reductase | 745 | 2878, 3611, 3863, 4008, 4009, 4010, 4153, 4216 |
| focA | 20.5 | Formate channel | Membrane protein | 31732 | 2088, 4294 |
| focB | 56.3 | Formate channel | Membrane protein, formate transporter of hyf operon | 55723 | 108a |
| folA | 1.1 | Folate | tmrA; trimethoprim resistance; dihydrofolate reductase (EC 1.5.1.3) | 744 | 106, 1962, 3437, 3728, 380, 4014, 4112, 4113, 4114 |
| folC | 52.4 | Folate | Dihydrofolate:folylpolyglutamate synthetase (EC 6.3.2.12) | 17704 | 1228, 3211, 409 |
| folD | 12.0 | Folate | ads; methenyltetrahydrofolate dehydrogenase/cyclohydrolase (EC 1.5.1.5, EC 3.5.4.9) | 31127 | 1052, 932 |
| folE | 48.3 | Folate | GTP cyclohydrolase I | 32718 | 2170 |
| folK | 3.4 | Folate | Dihydro-hydroxymethylpterin pyrophosphokinase | 29572 | 4341 |
| folP | 71.6 | Folate | Dihydropteroate synthase (EC 2.5.1.15) | 29566 | 4312, 910, 911 |
| folX | 52.1 | Folate | Dihydroneopterin triphosphate epimerase | 54773 | 1668 |
| fpr | 88.6 | Ferredoxin NADP+ reductase | mvrA; ferredoxin NADP+ reductase; anaerobic | 18127 | 3002, 344, 4474 |
| frdA | 94.4 | Fumarate reductase | Fumarate reductase flavoprotein subunit (EC 1.3.99.1) | 742 | 1116, 1540, 2041, 2614, 396, 793, 796, 797, 798, 801 |
| frdB | 94.4 | Fumarate reductase | Fumarate reductase iron-sulfur protein subunit (EC 1.3.99.1) | 741 | 1540, 2041, 2614, 663, 796, 797, 801 |
| frdC | 94.3 | Fumarate reductase | Fumarate reductase membrane anchor polypeptide (EC 1.3.99.1) | 740 | 1540, 2041, 2486, 663, 796 |
| frdD | 94.3 | Fumarate reductase | Fumarate reductase membrane anchor polypeptide (EC 1.3.99.1) | 739 | 1540, 2041, 2486, 663, 796 |
| frr | 4.2 | Factor for ribosome recycling | Ribosome recycling factor; essential gene; dissociates ribosomes from mRNA after termination of translation | 30590 | 1739, 1880, 1881, 1991, 4033 |
| fruA | 48.7 | Fructose | ptsF; fructosephosphotransferase enzyme II | 350 | 1396, 3538 |
| fruB | 48.7 | Fructose | fruF; fructosephosphotransferase enzyme III | 17515 | 1396, 3646 |
| fruK | 48.7 | Fructose | fpk, fruF; fructose-1-phosphate kinase (EC 2.7.1.3) | 743 | 1061, 1396, 2925, 428 |
| fruR | 1.9 | Fructose | fruC, shl, cra; regulatory gene for fru operon, other catabolite-regulated genes | 18304 | 1396, 1982, 2324, 3611, 3797, 397 |
| fsr | 10.8 | Fosmidomycin resistance | Amplification confers fosmidomycin resistance | 49279 | 1329 |
| ftn | 42.8 | Ferritin | gen-165, rsgA; ferritin | 32521 | 1846, 1970 |
| ftsA | 2.2 | Filamentation, temperature sensitive | divA; cell division, septation | 738 | 1024, 17, 2036, 2675, 2676, 2689, 279, 3691, 3693, 4274, 4589, 4882, 57, 621 |
| ftsE | 77.6 | Filamentation, temperature sensitive | Cell division; ATP-binding protein | 736 | 1429, 3821 |
| ftsI | 2.0 | Filamentation, temperature sensitive | pbpB, sep; peptidoglycan synthetase; penicillin-binding protein 3 | 423 | 1439, 1642, 2071, 279, 2889, 3107 |
| ftsJ | 71.7 | Filamentation, temperature sensitive | Cell division and growth; heat inducible | 33511 | 3250 |
| ftsK | 20.1 | Filamentation, temperature sensitive | Cell division and growth, septation; NA-binding membrane protein; homology with B. subtilis SpoIIIE protein; filamentous growth mutants, suppressed by dacA deletion; promoter region dinH | 40896 | 1046, 2519, 278, 4908 |
| ftsL | 2.0 | Filamentation, temperature sensitive | Cell division and growth; essential gene; cytoplasmic membrane protein | 30402 | 1588, 1929, 2071, 238 |
| ftsN | 88.8 | Filamentation, temperature sensitive | msgA; essential gene; cell division and growth; multicopy suppresses ftsA12 | 34153 | 16, 903 |
| ftsQ | 2.2 | Filamentation, temperature sensitive | Cell division and growth of wall at septum | 734 | 1024, 2071, 277, 279, 3691, 3692, 4240, 4882, 57 |
| ftsW | 2.1 | Filamentation, temperature sensitive | Cytoplasmic membrane required for PBP 2 expression; homology to rodA | 30409 | 1896, 2071, 2209, 2210 |
| ftsX | 77.6 | Filamentation, temperature sensitive | ftsS; cell division | 18298 | 1429 |
| ftsY | 77.6 | Filamentation, temperature sensitive | Cell division | 18295 | 1429, 2381, 2656, 3520 |
| ftsZ | 2.3 | Filamentation, temperature sensitive | sfiB, sulB; cell division and growth; initiation of septation; GTP binding protein | 143 | 1024, 1377, 17, 1978, 2036, 2071, 2674, 2677, 2689, 279, 339, 341, 3443, 3516, 3614, 3623, 4274, 4589, 4881, 4882, 57, 621, 901, 902 |
| fucA | 63.2 | Fucose | fucC; l-fuculose-1-phosphate aldolase | 17701 | 2650, 4089, 4955, 669, 713, 714 |
| fucI | 63.2 | Fucose | l-Fucose isomerase | 10878 | 2650, 4955, 713, 714 |
| fucK | 63.3 | Fucose | l-Fuculose kinase (EC 2.7.1.51) | 10881 | 2650, 4955, 713, 714 |
| fucO | 63.2 | Fucose | l-1,2-Propanediol oxidoreductase | 17698 | 2650, 4089, 4955, 713, 714 |
| fucP | 63.2 | Fucose | prd; fucose permease | 10875 | 2650, 4089, 4955, 669, 713, 714 |
| fucR | 63.3 | Fucose | Positive regulatory protein for fuc regulon | 10884 | 2650, 4090, 4955, 713, 714 |
| fumA | 36.3 | Fumarate | Fumarase A, aerobic | 18292 | 1555, 2932, 2933, 3357 |
| fumB | 93.6 | Fumarate | Fumarase B, anaerobic; regulatory gene? | 18289 | 1552, 1555, 292 |
| fumC | 36.3 | Fumarate | Fumarase C, aerobic; member of soxRS regulon | 15273 | 1552, 2568, 3357 |
| fur | 15.3 | Ferric uptake regulation | Ferric iron uptake, negative regulatory gene | 18286 | 1637, 1777, 198, 199, 3178, 3891, 978 |
| fusA | 74.8 | Fusidic acid | far; fusidic acid resistance; protein chain elongation factor G | 732 | 1815, 2383, 3498, 4929 |
| fusB | 14.5 | Fusidic acid | Fusidic acid resistance; pleiotropic effects on RNA synthesis and ribosomes, ribosomal protein S6 | 731 | 1923a, 4332a |
| gabC | 60.1 | γ-Aminobutyrate | Utilization of GABA as N source; regulatory for gab? | 730 | 1082, 2911, 2912 |
| gabD | 60.1 | γ-Aminobutyrate | Succinate-semialdehyde dehydrogenase; NADP dependent (EC 1.2.1.16) | 729 | 2911, 2912, 3179 |
| gabP | 60.2 | γ-Aminobutyrate | GABA permease, membrane protein | 728 | 2239, 2911, 2912 |
| gabT | 60.2 | γ-Aminobutyrate | Aminobutyrate aminotransferase (EC 2.6.1.19) | 727 | 249, 2911, 2912 |
| gadA | 79.0 | Glutamate deCase | gadS? glutamate decarboxylase (EC 4.1.1.15) | 32191 | 4111 |
| gadB | 33.8 | Glutamate deCase | Glutamate decarboxylase (EC 4.1.1.15) | 32194 | 4111 |
| gadR | 79.6 | Glutamate deCase | Regulatory gene for gat? | 726 | 2670, 2758 |
| galE | 17.0 | Galactose | UDP-galactose 4-epimerase; hexose-1-phosphate uridylyltransferase (EC 2.7.7.12) | 724 | 2456, 2485, 31, 4678, 4693, 581 |
| galF | 45.5 | Galactose | wcaN; not required for colanic acid synthesis; putative UTP-glucose-P-uridylyltransferase (galU) regulatory subunit | 53092 | 2771, 4209 |
| galK | 17.0 | Galactose | Galactose resistance; galactokinase (EC 2.7.1.6) | 723 | 225, 2456, 3792, 4285, 4693, 4876, 993, 994 |
| galM | 17.0 | Galactose | Aldose-1-epimerase (mutarotease) | 35171 | 456 |
| galP | 66.5 | Galactose | d-Galactose/H+ symporter | 722 | 4693 |
| galR | 64.1 | Galactose | Repressor of galETK operon; regulates low-affinity transport; Gal repressor | 721 | 1395, 2491, 2708, 3792, 4613, 4691, 4692, 4693, 4953, 584 |
| galS | 48.3 | Methyl-galactoside | mglD? utilization of methylgalactoside; negative regulation of high-affinity transport; Gal isorepressor; represses mgl | 499 | 1395, 3684, 3751, 4693 |
| galT | 17.0 | Galactose | galB; galactose-1-phosphate uridylyltransferase; (EC 2.7.7.10) | 720 | 2095, 2371, 2456, 2485, 2491, 4693, 839 |
| galU | 27.8 | Galactose | Glucose-1-P uridylyltransferase (UDP-glucose pyrophosphorylase) (EC 2.7.7.9), regulatory | 719 | 1168, 1299, 1547, 3989, 4678, 4704 |
| gapA | 40.1 | Glyceraldehyde phosphate dehydrogenase | gad; glyceraldehyde 3-P dehydrogenase A (EC 1.2.1.12); horizontal transfer? similar to eukaryotic GAPDH | 718 | 1071, 2438, 3149, 3980, 480 |
| gapC | 32.1 | Glyceraldehyde phosphate dehydrogenase | gad; glyceraldehyde 3-phosphate dehydrogenase C (EC 1.2.1.12) | 53105 | 1747 |
| garA | 15.8 | Glucarate | d-Glucarate utilization | 717 | 3690 |
| garB | 3.6 | Glucarate | d-Glucarate utilization | 716 | 3690 |
| gatA | 46.8 | Galactitol | Galactitol-specific enzyme IIA of phosphotransferase system (PTS) | 715 | 1015, 2058, 2565, 2925, 3202, 3644, 429 |
| gatB | 46.8 | Galactitol | Galactitol-specific enzyme IIB of PTS | 32559 | 3202 |
| gatC | 46.8 | Galactitol | Galactitol-specific enzyme IIC of PTS | 714 | 2058, 2565, 2925, 3202, 429 |
| gatD | 46.8 | Galactitol | Galactitol-1-phosphate dehydrogenase | 713 | 2058, 2565, 2925, 3202, 429 |
| gatR | 46.7 | Galactitol | Regulatory gene for gat | 32562 | 3202 |
| gatY | 46.8 | Galactitol | d-Tagatose-1,6-bisphosphate aldolase | 53142 | 3202 |
| gatZ | 46.8 | Galactitol | Function unknown | 53139 | 3202 |
| gcd | 3.0 | Glucose dehydrogenase | Glucose dehydrogenase (pyrroloquinoline-quinone) (EC 1.1.99.17); inner membrane | 30502 | 4825, 782 |
| gcl | 11.5 | Glyoxylate carboligase | Glyoxylate carboligase (EC 4.1.1.47) | 31107 | 677 |
| gcpE | 56.9 | Gene coding for protein E | Apparently essential gene | 32909 | 205 |
| gcvA | 63.4 | Glycine cleavage | trans-Acting protein, responsible for glycine-induced activation of gcv, perhaps repression in the presence of inosine | 28676 | 4756, 4757, 4758 |
| gcvH | 65.7 | Glycine cleavage | Glycine cleavage, carrier of aminomethyl group | 28664 | 4192, 4194 |
| gcvP | 65.6 | Glycine cleavage | Glycine dehydrogenase (decarboxylating) (EC 1.4.4.2) | 28661 | 3280 |
| gcvR | 56.0 | Glycine cleavage | Regulatory gene | 40204 | 1416, 1417 |
| gcvT | 65.7 | Glycine cleavage | Aminomethyl transferase, tetrahydrofolate dependent (EC 2.1.2.10) | 28667 | 3280, 4192, 4193 |
| gdhA | 39.7 | Glutamate dehydrogenase | Glutamate dehydrogenase | 712 | 1707, 265, 2857, 3844, 4539, 4540, 853, 968 |
| gef | 0.4 | Gene expression fatal | Small toxic membrane polypeptide; multimeric | 30252 | 2012, 3507, 3508, 3509, 3510 |
| ggt | 77.2 | γ-Glutamyltranspeptidase | γ-Glutamyltranspeptidase (EC 2.3.2.2) | 18280 | 1663, 4303, 4304, 4305, 4306 |
| gidA | 84.5 | Glucose-inhibited division | Glucose effects on cell division, perhaps replication | 18277 | 2275, 3245, 4635, 556 |
| gidB | 84.5 | Glucose-inhibited division | Glucose effects on cell division, perhaps replication | 18274 | 2275, 3180, 3245, 4635 |
| gip | 11.5 | Glyoxylate-induced protein | Induced by glyoxylate, downstream of gcl | 49283 | 677 |
| glcB | 67.2 | Glycolate | Malate synthase G (EC 4.1.3.2) | 711 | 3396 |
| glcC | 67.4 | Glycolate | Regulatory gene | 49985 | 3396 |
| glcD | 67.3 | Glycolate | Regulatory protein, transcriptional activator | 49982 | 3396 |
| glcE | 67.3 | Glycolate | glcF (frameshifted segment of glcE); glycolate oxidase subunit, FeS protein | 49979 | 3396 |
| glcG | 67.3 | Glycolate | Unknown function | 49974 | 3396 |
| gldA | 89.1 | Glycerol dehydrogenase | Glycerol dehydrogenase, NAD+ dependent | 34147 | 395, 4473 |
| glf | 45.4 | Galactofuranose | UDP-galactopyranose mutase (EC 5.4.99.9) | 54669 | 3137 |
| glgA | 76.8 | Glycogen | Glycogen synthase (EC 2.4.1.21) | 710 | 2084, 2364, 2432, 3281, 3721, 3722, 4854 |
| glgB | 76.9 | Glycogen | 1,4-α-Glucan branching enzyme (EC 2.4.1.18) | 709 | 196, 2432, 3721, 3722, 3820 |
| glgC | 76.9 | Glycogen | Glucose-1-phosphate adenylyltransferase (EC 2.7.7.27) | 708 | 195, 2432, 3721, 3722, 3281 |
| glgP | 76.8 | Glycogen | α-Glucan phosphorylase (EC 2.4.1.1) | 33653 | 4907, 742 |
| glgS | 68.8 | Glycogen | Glycogen synthesis protein, controlled by ςS (stationary phase) and cAMP | 29727 | 1719, 4854 |
| glgX | 76.9 | Glycogen | Glycogen debranching enzyme | 53576 | 4854 |
| glk | 54.0 | Glucokinase | Glucokinase; (EC 2.7.1.2) | 707 | 539, 890 |
| glmM | 71.6 | Glucosamine | mrsA; phosphoglucosamine mutase; essential gene; UDP-GlcNAc path, peptidoglycan, lipopolysaccharide synthesis; mRNA stability effects | 47201 | 2893, 4658 |
| glmS | 84.3 | Glucosamine | Glucosamine-6-phosphate synthase (EC 2.6.1.16) | 706 | 3464, 4611, 4635 |
| glmU | 84.3 | Glucosamine | Bifunctional glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-P uridyltransferase | 29106 | 2891, 2892 |
| glmX | 26.2 | Glucosamine | Affects suppression of glmS mutations by nagB | 36538 | 4601 |
| glnA | 87.4 | Glutamine | Glutamine synthetase (EC 6.3.1.2) | 705 | 1579, 192, 2705, 2706, 2710, 2849, 2952, 3072, 3336, 3645, 3695, 3750, 813, 852, 853 |
| glnB | 57.9 | Glutamine | PII regulatory protein for glutamine synthetase | 32933 | 1683, 2578, 4132, 4555, 555 |
| glnD | 4.0 | Glutamine | Uridylyltransferase | 704 | 1359, 4556 |
| glnE | 68.9 | Glutamine | GS adenylyl transferase (EC 2.7.7.42) | 33468 | 4555 |
| glnG | 87.3 | Glutamine | glnT, ntrC; nitrogen regulator I | 702 | 192, 2705, 2706, 2707, 2710, 2849, 2952, 3335, 3336 |
| glnH | 18.2 | Glutamine | Periplasmic glutamine-binding protein | 18271 | 3206, 3207 |
| glnK | 10.2 | Glutamine | Regulated through NRI/NRII two-component regulatory system | 38502 | 4554, 4557 |
| glnL | 87.4 | Glutamine | glnR, ntrB; bifunctional protein kinase/phosphatase nitrogen regulator II, NRII | 701 | 159, 192, 2706, 2707, 2710, 2848, 2849, 2952, 3207, 3336, 3645, 3695, 4510, 711 |
| glnP | 18.2 | Glutamine | l-Glutamate periplasmic binding protein; transport; methionine sulfoximine resistance | 700 | 2792, 3207, 4695, 4696 |
| glnQ | 18.2 | Glutamine | Glutamine high-affinity transport system | 18268 | 3207 |
| glnR | 24.1 | Glutamine | Glutamine transport | 699 | 724 |
| glnS | 15.2 | Glutamine | Glutaminyl-tRNA synthetase (EC 6.1.1.18) | 698 | 136, 3454, 3709, 4846, 1776 |
| glnT | 78.0 | Glutamine | Levels of glutamine tRNA1 and glutamine synthetase | 697 | 3001 |
| glnU | 15.0 | Glutamine | glnUα; suB, supB (ochre [UAA] suppression); glutamine tRNA1, tandem duplication | 696 | 1285, 1915, 2313, 3102, 3103, 3454 |
| glnV | 15.0 | Glutamine | glnVα; SuII, Su2, supE (amber suppression); glutamine tRNA2, tandem duplication (VX) | 695 | 1122a, 1285, 1915, 1916, 2313, 3102, 3103, 3454 |
| glnW | 15.0 | Glutamine | glnUβ; suB, supB (ochre [UAA] suppression); glutamine tRNA1, tandem duplication (UW) | 37440 | 1122a, 3001 |
| glnX | 15.0 | Glutamine | glnVβ; SuII, Su2, supE (amber suppression); glutamine tRNA2, tandem duplication (VX) | 37443 | 1122a, 1285, 1915, 1916, 2313, 3102, 3103, 3454 |
| gloA | 37.2 | Glyoxalase | Glyoxalase I | 39135 | 2703, 952 |
| glpA | 50.7 | Glycerol phosphate | Glycerol-3-phosphate dehydrogenase (anaerobic) large subunit (EC 1.1.99.5) | 694 | 1130, 2424, 3936, 799 |
| glpB | 50.7 | Glycerol phosphate | sn-Glycerol-3-phosphate dehydrogenase (anaerobic) subunit (EC 1.1.99.5); membrane anchor | 17692 | 1130, 4566, 799 |
| glpC | 50.7 | Glycerol phosphate | sn-Glycerol-3-phosphate dehydrogenase (anaerobic) small subunit (EC 1.1.99.5) | 17695 | 1130, 799 |
| glpD | 76.7 | Glycerol phosphate | glyD; glycerol-3-phosphate dehydrogenase (aerobic) (EC 1.1.99.5) | 693 | 171, 2491, 3940, 3942, 3947 |
| glpE | 76.7 | Glycerol phosphate | Gene of glp regulon | 18265 | 3942, 4926 |
| glpF | 88.7 | Glycerol phosphate | Glycerol facilitator | 692 | 4313 |
| glpG | 76.7 | Glycerol phosphate | Gene of glp regulon | 18262 | 3942, 4926 |
| glpK | 88.7 | Glycerol phosphate | Glycerol kinase (EC 2.7.1.30) | 691 | 324, 3413, 3414, 3798, 3940, 827 |
| glpQ | 50.6 | Glycerol phosphate | Glycerol-3-phosphate diesterase, periplasmic | 690 | 2424, 2426, 4428 |
| glpR | 76.7 | Glycerol phosphate | Repressor of glp operon | 688 | 3940, 3942, 3950, 4926, 743 |
| glpT | 50.6 | Glycerol phosphate | sn-Glycerol-3-phosphate permease | 689 | 1130, 1138, 1581, 1620, 2424, 2477, 2931, 334, 3936, 4571, 4753, 4827 |
| glpX | 88.6 | Glycerol phosphate | May be involved in glycerol metabolism but not required for growth on glycerol | 30884 | 4474 |
| gltA | 16.2 | Glutamate | glut, icdB; citrate synthase (EC 4.1.3.7) | 687 | 1549, 1708, 1859, 2396, 3151, 3356, 402, 4149, 4741, 4773 |
| gltB | 72.3 | Glutamate | aspB, ossB; glutamate synthase, large subunit (EC 1.4.1.13) | 686 | 1172, 1363, 2644, 3288, 3863, 657, 853 |
| gltD | 72.4 | Glutamate | aspB, ossB; glutamate synthase, small subunit | 17689 | 1363, 3288, 3863, 657 |
| gltE | 81.4 | Glutamate | Glutamyl-tRNA synthetase; possible regulatory subunit | 685 | 3062 |
| gltF | 72.4 | Glutamate | ossB; regulatory gene? | 18259 | 657 |
| gltH | 21.8 | Glutamate | Growth on glutamate | 684 | 2757, 2758, 3897 |
| gltJ | 14.8 | Glutamate | Membrane protein, transport gene | 49541 | 2659, 3308 |
| gltK | 14.8 | Glutamate | Membrane protein, transport gene | 49537 | 2659, 3308 |
| gltL | 14.7 | Glutamate | Membrane protein, transport gene | 49528 | 2659, 3308 |
| gltM | 43.0 | Glutamate | Affects glutamyl-tRNA synthetase level or activity? | 683 | 3062 |
| gltP | 92.5 | Glutamate | Proton-glutamate-aspartate transport protein | 34330 | 1001, 2660, 4423, 4639 |
| gltR | 92.3 | Glutamate | Growth on glutamate at 42°C | 682 | 2758 |
| gltS | 82.5 | Glutamate | Glutamate permease | 681 | 1001, 1002, 1078, 1178, 2098, 2655, 2757, 2758, 2950, 3897 |
| gltT | 89.8 | Glutamate | tgtB; glutamate tRNA2 | 680 | 2313, 3446, 3579, 522, 523 |
| gltU | 85.0 | Glutamate | tgtC; glutamate tRNA2 | 679 | 2313 |
| gltV | 90.7 | Glutamate | tgtE; glutamate tRNA2 | 678 | 2313 |
| gltW | 58.8 | Glutamate | Glutamate tRNA2 | 677 | 1146, 2313 |
| gltX | 54.3 | Glutamate | Glutamyl-tRNA synthetase (EC 6.1.1.17) | 676 | 2336, 3851, 498, 539 |
| glyA | 57.8 | Glycine | Serine hydroxymethyltransferase (EC 2.1.2.1) | 675 | 2633, 3444, 3445, 4191, 4522 |
| glyQ | 80.2 | Glycine | glySα; glycyl-tRNA synthetase, α-subunit (EC 6.1.1.14) | 33775 | 4127 |
| glyS | 80.2 | Glycine | act, gly, glySβ; glycyl-tRNA synthetase, β-subunit (EC 6.1.1.14) | 674 | 2196, 4127, 502 |
| glyT | 90.0 | Glycine | sumA; glycine tRNA2, UGA suppressionf | 673 | 1849, 2313, 2467, 3743, 90 |
| glyU | 64.6 | Glycine | suA36, sufD, sumA, sumB, supT; glycine tRNA1 | 672 | 1122a, 2313 |
| glyV | 94.6 | Glycine | ins, mutA; glycine tRNA3 (VXY) | 35636 | 1122a, 2921, 2313 |
| glyW | 42.9 | Glycine | ins, mutC; glycine tRNA3 | 670 | 2313, 2921, 4493, 4494 |
| glyX | 94.6 | Glycine | Glycine tRNA3 (VXY) | 35639 | 2313 |
| glyY | 94.6 | Glycine | Glycine tRNA3 (VXY) | 35642 | 2313 |
| gmd | 45.8 | GDP-d-mannose dehydratase | Fucose biosynthesis; GDP-d-mannose 4,6-dehydratase | 51902 | 4256 |
| gmk | 82.3 | GMP kinase | Guanylate kinase (EC 2.7.4.8) | 33871 | 1401 |
| gmm | 45.8 | GDP mannose mannosyl hydrolase | Guanosine diP mannose mannosyl hydrolase | 40934 | 1307 |
| gnd | 45.2 | Gluconate-P dehydrogenase | Gluconate-6-phosphate dehydrogenase (EC 1.1.1.43) | 669 | 3135, 3138, 3156, 4291, 4768 |
| gntK | 77.1 | Gluconate | Gluconokinase, thermoresistant | 35465 | 1969, 4436, 980 |
| gntP | 98.0 | Gluconate | Membrane protein, homologous to B. subtilis gluconate permease | 50203 | 2266 |
| gntR | 77.1 | Gluconate | Controls gntKU induction, also edd and eda; gluconate transport and P′n | 667 | 1194, 186, 1969, 3391, 3940, 4436, 4970 |
| gntS | 96.8 | Gluconate | Secondary gluconate transport system; putative regulator of GntV | 666 | 186, 1943, 569 |
| gntT | 76.4 | Gluconate | gntM, usgA; high-affinity gluconate transport | 668 | 1942, 3391, 3487, 4826, 981 |
| gntU | 77.0 | Gluconate | Low-affinity gluconate transport protein, membrane protein | 35461 | 1969, 4436, 4826, 980 |
| gntV | 96.8 | Gluconate | Glucokinase, thermosensitive | 17686 | 1942, 569 |
| goaG | 29.4 | GABA aminotransferase (EC 2.6.1.19) | 51935 | 2066 | |
| gor | 78.5 | GSH oxidoreductase | Glutathione oxidoreductase (EC 1.6.4.2) | 665 | 1150, 1513, 270, 964 |
| gph | 75.7 | Glycolate phosphatase | Phosphoglycolate phosphatase activity, in dam operon | 53568 | 2681 |
| gpmA | 16.9 | Glycerol P mutase | Phosphoglycerate mutase 1 | 35790 | 456, 958 |
| gpp | 85.4 | Guanosine pentaphosphatase | Guanosine pentaphosphatase activity; exopolyphosphatase | 664 | 289, 58, 926 |
| gprA | 0.3 | Growth of phage, replication | Replication of certain lambdoid phage | 15896 | 3240, 3829 |
| gprB | 0.2 | Growth of phage, replication | Replication of certain lambdoid phage | 15890 | 3240 |
| gpsA | 81.5 | Glycerol phosphate | sn-Glycerol-3-phosphate dehydrogenase [NAD(P)+] (EC 1.1.1.94) | 663 | 3700, 3701, 4032 |
| gpt | 5.5 | Guanine-xanthine phosphotransferase | glyD, gpp, gxu; guanine-xanthine phosphoribosyltransferase (EC 2.4.2.22) | 662 | 1469, 1722, 1979, 3221, 3531, 3662, 535 |
| greA | 71.7 | Growth restorer? | Transcription elongation factor | 33506 | 1166, 1835, 4141, 4142, 4195, 441, 442, 76 |
| greB | 76.2 | Growth restorer? | Transcription elongation factor | 35448 | 1835, 442, 76 |
| groE | 94.2 | Growth of phage | hdh, mop, tabB, groS; operon | 13587 | See groE, groL |
| groL | 94.2 | Growth of phage | groEL, mopB; hdh; tabB; chaperone for assembly of enzyme complexes; phage morphogenesis; large subunit | 492 | 1068, 1215, 1407, 1409, 2119, 2393, 2454, 3146, 4416, 573 |
| groS | 94.2 | Growth of phage | groES, mopA; hdh, tabB; small subunit of GroE chaperone | 493 | 1068, 1409, 2119, 2393, 2475, 3146, 4416 |
| grpE | 59.2 | Growth after phage induction | GrpE heat shock protein; mutant survives induction of prophage λ; stimulates DnaK ATPase; nucleotide exchange function | 660 | 2569, 3804, 4093, 4252, 4790, 4791 |
| grxA | 19.2 | Glutaredoxin | Glutaredoxin 1 | 17683 | 1796, 2337, 3771, 3778, 4350 |
| grxB | 24.2 | Glutaredoxin | Glutaredoxin 2 | 52078 | 154, 4595 |
| grxC | 81.6 | Glutaredoxin | Glutaredoxin 3 | 42921 | 154, 155 |
| gshA | 60.6 | GSH, glutathione | γ-Glutamyl-cysteine synthetase; (EC 6.3.2.2) | 659 | 1368, 1504, 1613, 2853, 3060, 4674 |
| gshB | 66.6 | GSH, glutathione | Glutathione synthetase (EC 6.3.2.3) | 33370 | 966 |
| gsk | 10.8 | Guanosine kinase | Guanosine kinase | 658 | 1653, 1824 |
| gsp | 67.6 | Glutathionylspermidine | Glutathionylspermidine synthetase/amidase; bifunctional protein | 36859 | 420 |
| gsp* | 74.6 | General secretory pathway operon | hopD, hopG; gsp previously used for glutathionylspermidine (see above), cryptic general secretory pathway; pul in Klebsiella spp. | 55310 | 1292 |
| gst | 36.9 | GSH S-transferase | Glutathione S-transferase | 37217 | 3192 |
| guaA | 56.7 | Guanine | GMP synthetase (EC 6.3.4.1) | 657 | 4414, 4503, 955, 4386 |
| guaB | 56.7 | Guanine | guaR; IMP dehydrogenase (EC 1.2.1.14) | 656 | 4399, 4400, 4413, 4503, 955, 4386 |
| guaC | 2.4 | Guanine | GMP reductase (EC 1.6.6.8) | 655 | 110, 2988, 3689 |
| gurB | 75.1 | Glucuronide | Utilization of glucuronides | 654 | 3219 |
| gurC | 18.0 | Glucuronide | Utilization of glucuronides | 653 | 3219 |
| gutM | 60.9 | Glucitol (sorbitol) | Part of srl operon | 33198 | 4824 |
| gutQ | 61.0 | Glucitol (sorbitol) | Part of srl operon; putative ATP-binding protein | 33206 | 4828, 757 |
| gyrA | 50.3 | Gyrase | hisW, nalA, parD, nfxA, norA; nalidixic acid resistance; cold shock regulon; DNA gyrase, subunit A | 651 | 1053, 1626, 1699, 1869, 2046, 2159, 2339, 2900, 2901, 3112, 4189, 4250, 4311, 4475, 4682, 4888, 4893 |
| gyrB | 83.5 | Gyrase | cou, acrB, himB, hisU, nalC, nalD, pcbA, parA; novobiocin, coumermycin resistance; DNA gyrase, subunit B | 650 | 10, 1053, 11, 1397, 2159, 2900, 2901, 2944, 3112, 3304, 4607, 4611, 4831, 9, 4438 |
| hcaA | 61.6 | HCA, hydrocinnamic acid | dig, phd; 3-phenylpropionate dioxygenase | 35007 | 570, 571 |
| hcaB | 61.7 | HCA, hydrocinnamic acid | dig, phd; 3-phenylpropionate-2′,3′-dihydrodiol deHase | 35010 | 570, 571 |
| hdeA | 78.8 | H-NS-determined expression | Periplasmic, unknown function, has ςS-dependent promoter | 33734 | 138, 4894 |
| hdeB | 78.8 | H-NS-determined expression | Periplasmic, unknown function, has ςS-dependent promoter | 33731 | 138, 4894 |
| hdeD | 78.8 | H-NS-determined expression | Periplasmic, unknown function, has ςS-dependent promoter | 33737 | 4894 |
| hdhA | 36.5 | Hydroxysteroid dehydrogenase | hsdH; 7-α-hydroxysteroid dehydrogenase (EC 1.1.1.159) | 32248 | 4897 |
| helD | 22.1 | Helicase | srjB; helicase IV | 31769 | 2642, 2883, 2884, 2885, 4774 |
| hemA | 27.2 | Hemin | Neomycin sensitivity; hemin biosynthesis; glutamyl tRNA dehydrogenase | 648 | 1087, 177, 1898, 2528, 4573, 709, 4575 |
| hemB | 8.4 | Hemin | ncf; 5-aminolevulinate dehydratase (EC 4.2.1.24) | 647 | 1111, 19, 2517, 2529, 2530, 2839, 2840, 3297 |
| hemC | 86.0 | Hemin | popE; porphobilinogen deaminase (EC 4.3.1.8); neomycin sensitivity | 646 | 1596, 2052, 2054, 2055, 2405, 2645, 2840, 3870, 3871, 4042, 4401, 60, 926 |
| hemD | 85.9 | Hemin | Uroporphyrinogen III cosynthase; neomycin sensitivity | 645 | 2052, 2053, 3870, 3871, 4042, 926 |
| hemE | 90.4 | Hemin | Uroporphyrinogen decarboxylase (EC 4.1.1.37) | 644 | 1912, 3196, 3874 |
| hemF | 55.0 | Hemin | popB, sec; coproporphyrinogen III oxidase (EC 1.3.3.3) | 643 | 2840, 3037, 4464 |
| hemG | 86.9 | Hemin | Protoporphyrinogen oxidase activity; neomycin sensitivity | 642 | 3197, 3872, 3873, 4853 |
| hemH | 10.7 | Hemin | popA, visA; ferrochelatase (EC 4.99.1.1) | 641 | 1323, 3100, 3101, 2974 |
| hemK | 27.3 | Hemin | Heme biosynthesis | 36992 | 3127 |
| hemL | 3.7 | Hemin | gsa, popC; glutamate-1-semialdehyde aminotransferase (EC 5.4.3.8) | 372 | 1519, 1899, 1900 |
| hemM | 27.2 | Hemin | lolB; glutamyl-tRNA dehydrogenase? novel outer membrane lipoprotein? | 31940 | 1898, 1981, 2815, 2816, 4573, 709 |
| hemX | 85.9 | Hemin | Uroporphyrinogen III methylase | 33965 | 3875, 926 |
| hemY | 85.9 | Hemin | Member of uro (hemC) operon | 33969 | 926 |
| hepA | 1.3 | Helicase-like protein | rapA; sequence similarity to helicases, downstream of polB, not under lexA control | 29491 | 1139, 2519, 4269a, 435 |
| het | 84.6 | Heterogeneous size | cop; possibly structural gene for DNA-binding protein; near ori | 640 | 3249, 4610, 4771 |
| hflA | 94.8 | High frequency lysogeny | Operon including hfq, hflX, -K, -C; part of complex mutL-hflC transcription unit | 34429 | 4487 |
| hflB | 71.6 | High-frequency lysogenization | ftsH, mrsC, tolZ; cell growth, septum formation, λ development, mRNA decay; essential inner membrane ATP-dependent protease, acting on SecY | 735 | 1486, 1727, 1728, 221, 3565, 41, 4435, 4657 |
| hflC | 94.9 | High-frequency lysogenization | HflA complex cleaves lambda cII; protease | 17520 | 220, 285 |
| hflK | 94.8 | High frequency lysogenization | HflA complex cleaves lambda cII; protease | 639 | 220, 285 |
| hflX | 94.8 | High frequency lysogenization | HflX GTPase, putative | 34423 | 220, 3203 |
| hfq | 94.8 | Host factor for Q β | HF-I, host factor for phage Qβ | 34450 | 2090, 2091, 4024, 4487–4490 |
| hha | 10.3 | High hemolysin activity | Histone-like; downregulates gene expression, stimulates transposition events | 31084 | 1438, 2929, 3182 |
| hipA | 34.2 | High persistence | Probable role in cell division | 18244 | 2882, 3028, 3029, 367, 368, 3902 |
| hipB | 34.3 | High persistence | Probable role in cell division | 32207 | 2882, 367, 368 |
| hisA | 45.1 | Histidine | N-(5′-phospho-l-ribosylformimino)-5-amino-1-(5′-phosphoribosyl)-4-imidazolecarboxamide isomerase (EC 5.3.1.16) | 636 | 1202, 65 |
| hisB | 45.1 | Histidine | Bifunctional enzyme imidazoleglycerolphosphate (IGP) dehydratase, histidinol phosphatase (EC 3.1.3.15, EC 4.2.1.19) | 635 | 1521, 730 |
| hisC | 45.1 | Histidine | Histidinol-phosphate aminotransferase (EC 2.6.1.9) | 634 | 1521, 1522 |
| hisD | 45.0 | Histidine | Histidinol dehydrogenase (EC 1.1.1.23) | 633 | 542, 65, 731 |
| hisF | 45.1 | Histidine | Cyclase component of IGP synthase complex | 631 | 1202, 1263, 1296, 1376, 1451, 2260, 65 |
| hisG | 45.0 | Histidine | ATP phosphoribosyltransferase (EC 2.4.2.17) | 630 | 1322, 3027, 4572, 542 |
| hisH | 45.1 | Histidine | Amidotransferase component of IGP synthase | 629 | 1263, 1296, 1376, 1451, 2260, 65 |
| hisI | 45.2 | Histidine | Formerly hisE and hisI; bifunctional enzyme PR-ATP pyrophosphatase PR-AMP cyclohydrolase (EC 3.5.4.19, EC 3.6.1.31) | 628 | 65, 731 |
| hisJ | 52.3 | Histidine | Histidine-binding protein of high-affinity histidine transport system | 627 | 126, 2512 |
| hisM | 52.2 | Histidine | Histidine transport membrane protein M | 25879 | 2335 |
| hisP | 52.2 | Histidine | Histidine permease | 626 | 126, 2335, 2512 |
| hisQ | 52.2 | Histidine | Histidine transport gene | 32721 | 126 |
| hisR | 85.8 | Histidine | hisT; histidine tRNA | 625 | 1832, 2313, 2535, 926 |
| hisS | 56.8 | Histidine | Histidyl-tRNA synthetase (EC 6.1.1.21) | 624 | 1140, 1141, 1301 |
| hlpA | 4.3 | Histone-like protein | ompH; firA, orf (skp), skp; histone-like protein HLP-I (cytoplasmic?); early confusion with adjacent lpxD gene | 18241 | 1, 1040, 1773, 1775, 1784, 2429, 2964, 4402, 4403, 706, 804, 1702 |
| hlyE | 26.5 | Hemolysin (latent) | Latent hemolysin expressed in presence of Actinobacillus glyX or fnr mutant | 53859 | 1498 |
| hmp | 57.9 | Hemoglobin-like | fsrB; hemoglobin-like flavoprotein | 32917 | 109, 2879, 2880, 2881, 3482, 3483, 4068 |
| hns | 27.8 | Histone-like protein, H-NS | bglY, cur, drc, drdX, drs, fimG, irk, msyA, osmZ, pilG, topX, virR; H-NS (H1a) DNA-binding protein, histone-like; diverse mutant phenotypes affecting transcription, transposition, inversion, cryptic-gene expression; involved in chromosome organization | 960 | 1041, 1195, 1458, 1750, 2160, 2385, 239, 2482, 2483, 2495, 2828, 327, 3476, 4022, 4146, 4173, 4504, 4507, 4821, 887, 2392, 3599 |
| hofB | 2.5 | Homologous to fim | hopB; homologous to PilB of Pseudomonas aeruginosa; function not established, insertion mutation gives no phenotype | 33679 | 4722 |
| hofC | 2.5 | Homologous to fim | hopC; homologous to PilC of P. aeruginosa; function not established, insertion mutation gives no phenotype | 33676 | 4722 |
| holA | 14.4 | Holoenzyme | DNA polymerase III, δ-subunit | 31431 | 1067, 2194, 3300, 645 |
| holB | 24.9 | Holoenzyme | DNA polymerase III, δ′-subunit | 31447 | 1067, 2194, 3300 |
| holC | 96.6 | Holoenzyme | DNA polymerase III, χ-subunit | 31456 | 2194, 3295, 4808, 644 |
| holD | 99.3 | Holoenzyme | DNA polymerase, ψ-subunit | 31460 | 2194, 3295, 4808, 646 |
| holE | 41.5 | Holoenzyme | DNA polymerase III, θ-subunit | 31439 | 2194, 2224, 4097, 4254, 647 |
| hopB | 84.7 | Host plasmid maintenance | hop; also used for hof loci (homologous to Pil genes in Pseudomonas); required for mini-F | 37343 | 3184 |
| hopC | 99.4 | Host plasmid maintenance | Required for maintenance of stable mini-F plasmid; see also hofC | 37346 | 3184 |
| hopD | 11 | Host plasmid maintenance | Required for maintenance of stable mini-F plasmid | 37349 | 3184 |
| hpt | 3.0 | HGPRT | Hypoxanthine-guanine phosphoribosyltransferase (EC 2.4.2.8) | 622 | 2024, 535 |
| hrpA | 31.9 | Helicase-related protein | RNA helicase-like; similarity to eukaryotic DEAH family | 36987 | 3005 |
| hrpB | 3.5 | Helicase-related protein | RNA helicase-like; similarity to eukaryotic DEAH family and araC | 36926 | 3005, 1331 |
| hrsA | 16.5 | Heat-responsive suppressor | Suppresses ompC mutation’s defective thermoresponse | 53146 | 4520 |
| hscA | 57.2 | Heat shock cognate | Stress response gene; Hsp70 family | 32977 | 2179, 2484, 3956, 4583 |
| hscB | 57.3 | Heat shock cognate | Stress response gene; heat shock regulon | 40196 | 2484, 4583 |
| hsdM | 98.7 | Host specificity | hs, hsm, hsp, rm; host DNA modification; DNA methylase M | 621 | 1014, 2608, 3800, 4909 |
| hsdR | 98.7 | Host specificity | hs, hsp, hsr, rm; host DNA restriction; endonuclease R | 620 | 1014, 2608, 3800, 4909 |
| hsdS | 98.7 | Host specificity | hss; specificity determinant for hsdM and hsdR | 619 | 1474, 2608, 3071, 3800, 4909 |
| hslC | 19.9 | Heat shock locus | Protein expressed as heat shock regulon member | 36762 | 749 |
| hslD | 24.1 | Heat shock locus | Protein expressed as heat shock regulon member | 36765 | 749 |
| hslE-H | 29.7 | Heat shock loci | Protein expressed as heat shock regulon member | 37190 | 749 |
| hslJ | 31.0 | Heat shock locus | Protein expressed as heat shock regulon member | 41193 | 323, 749 |
| hslK | 40.7 | Heat shock locus | Protein expressed as heat shock regulon member | 36945 | 749 |
| hslL-N | 70.5 | Heat shock loci | Protein expressed as heat shock regulon member | 37449 | 749 |
| hslO-R | 76.0 | Heat shock loci | Heat shock proteins; HslO is HspG21.0 | 36769 | 749 |
| hslU | 88.8 | Heat shock locus | htpI, clpY; heat shock protein D48.5, protease? required for peptide hydrolysis of HslVU; modulated by catabolite repression | 34157 | 1341, 2121, 2208, 2963, 3408, 3713, 4889, 749, 750 |
| hslV | 88.8 | Heat-shock locus | htpO; heat shock regulon | 34160 | 1341, 2121, 2208, 2963, 3713, 4889, 4890, 749, 750 |
| hslW | 94.2 | Heat shock locus | Heat-inducible; regulatory gene, near groE | 41190 | 749 |
| htgA | 0.2 | High-temperature growth | htpY; heat inducible | 30233 | 1987, 2961, 984 |
| htpG | 10.7 | Heat shock protein | Heat shock protein C62.5; chaperone | 17680 | 228 |
| htpX | 41.2 | Heat shock protein | Protein expressed as heat shock regulon member | 32320 | 2327 |
| htrB | 24.0 | High temperature (requirement) | Not under heat shock regulation; membrane protein affecting cell division, growth, and high-temperature survival | 31878 | 2137, 2140, 2141, 781 |
| htrC | 90.3 | High temperature (requirement) | Essential for growth at high temperature, under ς32 (heat shock) regulation | 34265 | 2141, 3589 |
| htrE | 3.3 | High temperature (requirement) | Sequence homology with pilin protein PapC | 30510 | 3593 |
| htrL | 81.7 | High temperature (requirement) | RfaH-regulated high-temperature gene | 54740 | 3591 |
| hupA | 90.5 | HU protein | HU-β, HU-2, histone-like protein | 34243 | 1977, 2125, 2126, 2292, 2391, 422 |
| hupB | 9.9 | HU protein | dpeA, hopD; HU-α, HU-1, histone-like protein | 18235 | 1463, 1977, 2124, 2127, 2292, 2391, 2749, 4239 |
| hyaA | 22.2 | Hydrogenase 1 | Hydrogenase 1 small subunit [NiFe] | 31781 | 2897, 2898 |
| hyaB | 22.3 | Hydrogenase 1 | Hydrogenase 1 large subunit [NiFe] | 31784 | 2897, 2898 |
| hyaC | 22.3 | Hydrogenase 1 | Possible membrane-spanning protein of hya operon | 31788 | 2897, 2898 |
| hyaD | 22.3 | Hydrogenase 1 | Processing of HyaA and HyaB | 31791 | 2897, 2898 |
| hyaE | 22.3 | Hydrogenase 1 | Processing of HyaA and HyaB | 31794 | 2897, 2898 |
| hyaF | 22.3 | Hydrogenase 1 | Nickel incorporation in hydrogenase 1 proteins | 31797 | 2897, 2898 |
| hybA | 67.7 | Hydrogenase 2 | hydL, hydC; hydrogenase 2 [Ni, Fe], small subunit, probably | 33407 | 2896 |
| hybB | 67.7 | Hydrogenase 2 | HYD2 cytochrome b type component, probably | 33414 | 2896 |
| hybC | 67.7 | Hydrogenase 2 | Hydrogenase 2 [Ni Fe] large subunit, probably | 33418 | 2896 |
| hybD | 67.7 | Hydrogenase 2 | Processing element for hydrogenase 2, probably | 33421 | 2896 |
| hybE | 67.6 | Hydrogenase 2 | Function undefined | 33424 | 2896 |
| hybF | 67.6 | Hydrogenase 2 | Regulatory gene | 33427 | 2896 |
| hybG | 67.6 | Hydrogenase 2 | May affect maturation of hydrogenase 2 large subunit | 33403 | 2896 |
| hycA | 61.4 | Hydrogenase 3 | Regulatory gene for hyc and hyp; counteracts activation by FhlA | 33143 | 1801, 2554, 3883, 412, 4294 |
| hycB | 61.4 | Hydrogenase 3 | Formate-hydrogenlyase system, formate regulon, small subunit of hydrogenase 3? | 33169 | 3883, 2554 |
| hycC | 61.3 | Hydrogenase 3 | Formate-hydrogenlyase system, formate regulon, small subunit of hydrogenase 3? | 33166 | 3883, 2554 |
| hycD | 61.3 | Hydrogenase 3 | Hydrogenase 3 subunit | 33162 | 3883, 2554 |
| hycE | 61.3 | Hydrogenase 3 | Hydrogenase 3 subunit, large, precursor | 33159 | 3746, 3883 |
| hycF | 61.3 | Hydrogenase 3 | Hydrogenase 3 subunit | 33154 | 3883 |
| hycG | 61.2 | Hydrogenase 3 | Hydrogenase 3 subunit | 33151 | 3883 |
| hycH | 61.2 | Hydrogenase 3 | Required for converting HycE precursor to Hyd-3 subunit | 33140 | 2678, 3883 |
| hycI | 61.2 | Hydrogenase 3 | Maturation protease for Ni-containing Hyd-3 | 50402 | 3747 |
| hydA | 61.1 | Hydrogenase | hydNF operon | 18232 | 2145, 2465, 2726, 3376, 3852, 4835 |
| hydG | 90.6 | Hydrogenase | Two-component regulation of Hyd-3 activity | 34252 | 3410, 4231 |
| hydH | 90.5 | Hydrogenase | Two-component regulation of Hyd-3 activity, sensor kinase | 34249 | 4231 |
| hydN | 61.1 | Hydrogenase | Iron-sulfur protein required for Hyd-3 activity | 33521 | 2726 |
| hyfA-J | 56.2 | Hydrogenase 4 | Part of 12-cistron operon encoding putative proton-translocating formate hydrogenlyase system | 55682 | 108a |
| hyfR | 56.3 | Hydrogenase 4 | Formate-sensing regulator for hyf operon | 55719 | 108a |
| hypA | 61.4 | Hydrogenase | Formate-hydrogenlyase system; guanine-nucleotide-binding protein, Ni donor for Hyd-3 large subunit | 33104 | 1801, 1976, 2678, 3745 |
| hypB | 61.4 | Hydrogenase | hydE; formate-hydrogenlyase system; formate regulon | 33110 | 1801, 1976, 2678, 2725, 3745 |
| hypC | 61.4 | Hydrogenase | Formate-hydrogenlyase system: formate regulon | 33113 | 1801, 1976, 2678, 3745 |
| hypD | 61.4 | Hydrogenase | hydF; formate-hydrogenlyase system: formate regulon | 33116 | 1801, 1976, 2678, 3745, 3855 |
| hypE | 61.5 | Hydrogenase | hydB; formate-hydrogenlyase system: formate regulon | 33119 | 1801, 1976, 2678, 3745, 3854 |
| hypF | 61.1 | Hydrogenase | Mutation results in loss of hydrogenase activity; affects maturation of all hydrogenases, formate regulon | 50120 | 2726 |
| iadA | 98.2 | Isoaspartyl dipeptidase | Isoaspartyl dipeptidase | 53593 | 1382 |
| iap | 62.0 | Isozymic alkaline P′ase | Aminopeptidase, presumably, that generates alkaline phosphatase isozyme | 616 | 1931, 3120 |
| ibpA | 83.3 | Inclusion body protein | hslT, htpN; chaperone, heat-inducible protein of HSP20 family | 33847 | 2427, 69, 750 |
| ibpB | 83.3 | Inclusion body protein | hslS, htpE; chaperone, heat-inducible protein of HSP20 family | 33843 | 2427, 69, 750 |
| icd | 25.7 | Isocitrate dehydrogenase | icdE; isocitrate dehydrogenase, NADP+-specific (EC 1.1.1.42) | 615 | 124, 1709, 1729, 1753, 625 |
| iclR | 91.0 | Isocitrate lyase | Isocitrate lyase (EC 4.1.3.1) | 614 | 1331, 2744, 3382, 4290, 840 |
| ihfA | 38.7 | Integration, host factor | himA, hid; host infection, mutant λ; site-specific recombination; sequence-specific DNA-binding transcriptional activator, IHF, α subunit | 638 | 1156, 1603, 2461, 2860, 2862, 2882, 2939, 2940, 2941, 2942, 2943, 2944, 3231, 4267, 685, 817 |
| ihfB | 20.8 | Integration, host factor | himD, hip; see ihfA, β subunit | 637 | 1156, 1258, 1603, 2216, 2461, 2941, 2943, 2944, 4687 |
| ileR | 95.8 | Isoleucine | avr, flrA; Ile repressor; regulation of thr and ilv operons; originally placed at 99.9 min; sequence puts it at 95.8 min | 15914 | 2028, 2029, 4702, 569 |
| ileS | 0.5 | Isoleucine | Isoleucyl-tRNA synthetase (EC 1.1.1.5) | 613 | 2104, 2945, 3640, 4317, 4422, 4820, 4830 |
| ileT | 87.0 | Isoleucine | Isoleucine tRNA1, triplicate (T, U, V) | 612 | 2313, 926 |
| ileU | 73.8 | Isoleucine | Isoleucine tRNA1, triplicate (T, U, V) | 611 | 2313 |
| ileV | 4.8 | Isoleucine | Isoleucine tRNA1, triplicate (T, U, V) | 610 | 1146, 2313 |
| ileX | 69.2 | Isoleucine | Isoleucine tRNA2 | 17677 | 3058 |
| ileY | 60.0 | Isoleucine | Isoleucine tRNA2 variant | 53508 | 4837 |
| ilvA | 85.2 | Isoleucine-valine (requirement) | ile; threonine deaminase (EC 4.2.1.16) | 609 | 1086, 1490, 1839, 2445, 3824, 4614, 58, 860, 926 |
| ilvB | 83.0 | Isoleucine-valine (requirement) | Acetolactate synthase I, valine sensitive (EC 4.1.3.18) | 608 | 1309, 1310, 1453, 1667, 2079, 3168, 3169, 3452, 3607, 4006, 4299, 4513, 4705, 681, 960 |
| ilvC | 85.3 | Isoleucine-valine (requirement) | ilvA; ketol-acid reductoisomerase (EC 1.1.1.86) | 607 | 257, 351, 4707, 58, 926 |
| ilvD | 85.2 | Isoleucine-valine (requirement) | Dihydroxyacid dehydrase (EC 4.2.1.9) | 606 | 1086, 1490, 1839, 2445, 317, 58, 860, 926 |
| ilvE | 85.1 | Isoleucine-valine (requirement) | ilvC, ilvJ; branched-chain amino acid aminotransferase (EC 2.6.1.42) | 605 | 1086, 149, 1490, 1839, 2374, 2441, 2445, 316, 4263, 4524, 4706, 613, 860, 926 |
| ilvF | 57.0 | Isoleucine-valine (requirement) | Production of valine-resistant acetolactate synthase activity | 604 | 3452, 960 |
| ilvG | 85.1 | Isoleucine-valine (requirement) | Acetolactate synthase II, valine insensitive (EC 4.1.3.18) | 603 | 1086, 1393, 1437, 1490, 1839, 2441, 2442, 2443, 2444, 2445, 2648, 3131, 316, 3348, 4262, 4263, 4524, 575, 58, 860, 926 |
| ilvH | 1.9 | Isoleucine-valine (requirement) | brnP; acetolactate synthase II, valine sensitive (EC 4.1.3.18) | 602 | 1437, 1453, 184, 1982, 3450, 3607, 4176, 4177, 4299, 4513, 960 |
| ilvI | 1.8 | Isoleucine-valine (requirement) | Acetolactate synthase II, valine sensitive (EC 4.1.3.18) | 601 | 1453, 1666, 184, 3450, 3607, 4176, 4177, 4299, 4513, 960 |
| ilvJ | 1.3 | Isoleucine-valine (requirement) | Acetolactate synthase IV, valine resistant (EC 4.1.3.18) | 600 | 1437, 1975, 3694, 960 |
| ilvM | 85.1 | Isoleucine-valine (requirement) | Acetohydroxy acid synthase II (EC 4.1.3.18); valine insensitive, small subunit | 1214 | 1839, 2445, 2628, 2648, 4706, 860, 926 |
| ilvN | 83.0 | Isoleucine-valine (requirement) | Acetohydroxy acid synthase I, small subunit (EC 4.1.3.18); valine sensitive small subunit; positive regulator for thr and ilv | 15441 | 1310, 4705, 681 |
| ilvR | 99.9 | Isoleucine-valine | 15917 | 2029 | |
| ilvU | 6.4 | Isoleucine-valine (requirement) | Regulation of ileS and modification of Ile tRNA2 and Val tRNA2 | 599 | 1214 |
| ilvY | 85.2 | Isoleucine-valine (requirement) | Positive regulator for ilvC | 598 | 351, 3824, 4707, 58, 926 |
| imp | 1.2 | Increased membrane permeability | ostA; permeability of outer membrane to large maltodextrins; antibiotic and detergent sensitivity, stress induced | 36754 | 3828 |
| inaA | 50.6 | Inducible by acid | Protein induced by acid, independent of SoxRS regulation | 32692 | 3737, 4727 |
| inaR | 34.8 | Inducible by acid | Regulates inaA (may be marA or soxZ?) | 32696 | 4727 |
| infA | 19.9 | Initiation factor | hypA1? protein chain initiation factor IF1 | 17674 | 3475, 3849, 885, 886, 899 |
| infB | 71.4 | Initiation factor | ssyG; protein chain initiation factor 2, IF2 | 597 | 1561, 1928, 2748, 3016, 3108, 3459, 3462, 4026, 4027, 4029 |
| infC | 38.8 | Initiation factor | fit?, srjA; protein chain initiation factor 3, IF3 | 596 | 1148, 1601, 2500, 2592, 2829, 3056, 3463, 3527, 3788, 4168, 4710, 4801, 514, 582, 583, 732, 942 |
| inm | 79.0 | Insensitive NG mutagenesis | Susceptibility to mutagenesis by nitrosoguanidine | 595 | 3767 |
| intA | 59.4 | Integrase | slpA; from defective prophage CP4-57 | 33082 | 2241, 4455 |
| intB | 96.9 | Integrase | Prophage P4 integrase, defective prophage derivative | 53585 | 394 |
| intD | 12.2 | Integrase | int (qsr′); integrase locus within defective prophage derivative qsr′ | 31133 | 3059 |
| isfA | 86.0 | Inhibits SOS function | Regulatory gene; SOS-related | 41044 | 263, 264 |
| ispA | 9.5 | Isoprenoid synthesis | Farnesyl diphosphate synthase (EC 2.5.1.1) | 30978 | 1328, 1330 |
| ispB | 71.8 | Isoprenoid synthesis | cel; octaprenyl diphosphate synthase | 35712 | 150, 3277 |
| katC | 6.0 | Catalase | IS1B and IS20B insertions sensitize to peroxide; deletion or Tn insertion increases resistance | 594 | 4605 |
| katE | 39.1 | Catalase | Catalase hydroperoxidase III | 593 | 2609, 2611, 3898, 3899, 4612 |
| katG | 89.1 | Catalase | Catalase hydrogen peroxidase I | 14983 | 2611, 3036, 3898, 4457, 4592, 4456 |
| kba | 71.8 | Ketose-bis-phosphate aldolase | Ketose-bis-phosphate aldolase, tagatose-bis-phosphate aldolase; part of aga cluster for K+ transport | 592 | 2490, 3649 |
| kbl | 81.7 | Ketobutyrate ligase | 2-Amino-3-ketobutyrate coenzyme A ligase (EC 2.3.1.29) | 18205 | 140, 3620 |
| kch | 28.2 | Potassium (K+) channel | Homology to potassium channel proteins | 35309 | 2026, 2935 |
| kdgK | 79.3 | Ketodeoxygluconate | Ketodeoxygluconokinase (EC 2.7.1.45) | 591 | 3515 |
| kdgR | 41.1 | Ketodeoxygluconate | Regulator of kdgK, kdgT, and eda | 590 | 2047, 3515 |
| kdgT | 88.4 | Ketodeoxygluconate | Ketodeoxygluconate transport system, structural gene | 589 | 2047, 2747, 3465, 3515 |
| kdpA | 15.7 | Potassium dependence | kac; K+-translocating ATPase Kdp subunit | 588 | 1225, 4266, 74 |
| kdpB | 15.6 | Potassium dependence | kac; high-affinity potassium transport | 587 | 4266, 74 |
| kdpC | 15.6 | Potassium dependence | kac; high-affinity potassium transport | 586 | 4266, 74 |
| kdpD | 15.5 | Potassium dependence | kac; sensor kinase for K+-kdp system | 585 | 153, 3117, 3118, 3472, 3555, 4266, 4597, 4633 |
| kdpE | 15.5 | Potassium dependence | Transcriptional effector of kdp operon | 31571 | 153, 3117, 3118, 3472, 4266, 4597, 4633 |
| kdpF | 15.7 | Potassium dependence | Inner membrane protein for K+ transport | 35751 | 1735, 74 |
| kdsA | 27.3 | KDO synthesis | 3-Deoxy-d-manno-octulosonic acid 8-P synthetase | 17671 | 4763, 4764 |
| kdsB | 20.9 | KDO synthesis | CMP-3-deoxy-d-manno-octulosonate cytidylyltransferase | 18202 | 1449, 1450, 3420 |
| kdtA | 82.0 | KDO transfer (to lipid A) | waaA; 3-deoxy-d-manno-octulosonate-lipid A transferase (EC 2.4.99.–) | 33819 | 293, 3921, 779, 780 |
| kdtB | 82.1 | KDO transfer (to lipid A) | CMP-deoxy-d-manno-octulosonate-lipid A transferase (EC 2.4.99.–) | 33900 | 3726, 4128 |
| kefB | 74.9 | K efflux | trkB; NEM-activatable K+/H+ antiporter | 83 | 1226, 209 |
| kefC | 1.0 | K efflux | trkC; NEM-activatable K+/H+ antiporter | 82 | 1226, 209, 2946, 3050 |
| kgtP | 58.7 | Ketoglutarate | witA; α-ketoglutarate permease | 32867 | 3977, 3975, 3976, 3978 |
| kicA | 21.0 | Kill cell | mukE; killing protein | 31751 | 1223, 4844, 4845 |
| kicB | 21.0 | Kill cell | mukF; Suppressor of killing protein | 31748 | 1223, 4845 |
| ksgA | 1.1 | Kasugamycin | S-Adenosylmethionine-6-N′,N′-adenosyl (rRNA) dimethyltransferase | 583 | 105, 106, 107, 1711, 380, 4542 |
| ksgB | 37.7 | Kasugamycin | High-level resistance to kasugamycin | 582 | 1042, 1286, 1529, 4144 |
| ksgC | 12.0 | Kasugamycin | Resistance to kasugamycin; affects ribosomal protein S2 | 581 | 4896 |
| ksgD | 30.9 | Kasugamycin | Resistance to kasugamycin | 580 | 1286 |
| lacA | 7.8 | Lactose | Thiogalactoside acetyltransferase; (EC 2.3.1.18) | 579 | 1696, 1805 |
| lacI | 7.9 | Lactose | Repressor protein of lac operon | 578 | 1203, 1461, 1811, 2202, 2417, 2492, 2501, 2625, 2626, 2767, 3042, 3217, 331, 335, 3798, 4121, 620 |
| lacY | 7.8 | Lactose | Galactoside permease (M protein) | 577 | 1127, 1685, 1805, 2033, 3042, 3293, 346, 404, 4121, 4647, 549 |
| lacZ | 7.8 | Lactose | β-d-Galactosidase (EC 3.2.1.23) | 576 | 1597, 1805, 2033, 2101, 2417, 2781, 4021, 4630 |
| lamB | 91.5 | Lambda | Phage lambda receptor protein; maltose high-affinity uptake system; in malB cluster | 575 | 1152, 1695, 1778, 1779, 1780, 3941, 4394, 488, 778, 814, 987 |
| lar | 30.4 | Ral inverse | rac locus; restriction alleviation | 37053 | 2238, 4439 |
| ldcC | 4.5 | Lysine decarboxylase | Lysine decarboxylase | 49257 | 2217, 4836, 4839 |
| ldhA | 31.0 | Lactate dehydrogenase | hslF, hslI, htpH; lactate dehydrogenase | 37446 | 2800, 749, 561a |
| lepA | 58.3 | Leader peptidase | GTP binding membrane protein, function unknown; lep | 18199 | 1039, 2754, 2755, 4062 |
| lepB | 58.3 | Leader peptidase | Signal peptidase I (for nonlipoproteins) | 573 | 2754, 4769, 906, 4062 |
| leuA | 1.8 | Leucine (biosynthesis) | α-Isopropylmalate synthase (EC 4.1.3.12) | 572 | 1399, 4711, 963, 245 |
| leuB | 1.7 | Leucine (biosynthesis) | β-Isopropylmalate dehydrogenase (EC 1.1.1.85) | 571 | 963 |
| leuC | 1.7 | Leucine (biosynthesis) | α-Isopropylmalate isomerase subunit | 570 | 963 |
| leuD | 1.7 | Leucine (biosynthesis) | α-Isopropylmalate isomerase subunit | 569 | 963 |
| leuJ | 13.7 | Leucine (biosynthesis) | flr; regulation of leu and ilv operons | 18196 | 2028 |
| leuO | 1.8 | Leucine (biosynthesis) | Affects expression of small regulatory Dsr-RNA, translational regulation of rpoS, relieves bgl silencing | 35918 | 2249, 4025, 4506 |
| leuP | 99.2 | Leucine | leuVβ tandemly triplicate leuVPQ, duplicate with leuT; leucine tRNA1 | 38293 | 2313 |
| leuQ | 99.2 | Leucine | leuVγ tandemly triplicate, and duplicate with leuT; leucine tRNA1 | 38296 | 2313 |
| leuR | 79.3 | Leucine | Regulates level of leucyl-tRNA synthetase | 567 | 4392 |
| leuS | 14.5 | Leucine | Leucyl-tRNA synthetase (EC 6.1.1.4) | 566 | 1658, 4157 |
| leuT | 85.8 | Leucine | Leucine tRNA1, duplicate with leuVPQ | 565 | 1832, 2313, 926 |
| leuU | 71.6 | Leucine | Leucine tRNA2 | 564 | 2313, 4631 |
| leuV | 99.2 | Leucine | leuVα leucine tRNA1, tandemly triplicate leuVPQ, duplicate with leuT | 563 | 1095, 1285, 2313 |
| leuW | 15.0 | Leucine | feeB; leucine tRNA3 | 562 | 2313, 3102, 3103, 3454, 460, 703 |
| leuX | 96.9 | Leucine | Su-6, supP; leucine tRNA5 (amber [UAG] suppressor) | 561 | 1122a, 135, 2313, 3210, 4407, 4409, 4899 |
| leuY | 9.5 | Leucine | Regulates level of leucyl-tRNA synthetase | 560 | 2420 |
| leuZ | 42.9 | Leucine | Leucine tRNA4 | 17668 | 2313 |
| lev | 9.0 | Levallorphan | Resistance to levallorphan | 559 | 916 |
| lexA | 91.7 | Lambda excision | exrA, recA, spr, umuA; global regulator for SOS regulon (represses ca. 20 genes) | 558 | 1654, 1755, 1802, 2575, 2576, 2765, 2928, 4017, 479, 496, 497 |
| lgt | 63.9 | (Pro)lipoprotein glyceryl transferase | umpA; phosphatidylglycerol:prolipoprotein diacylglycerol transferase | 33350 | 1352, 3561, 3562, 4743 |
| lhr | 37.2 | Long helicase related protein | Probable ATP-dependent helicase | 35720 | 3656 |
| ligA | 54.5 | Ligase | lig, dnaL, pdeC; DNA ligase | 557 | 1398, 1932, 1959 |
| ligT | 3.5 | Ligase tRNA | Name temporary, not published; 2′-5′ RNA ligase | 50132 | 134 |
| linB | 29.3 | Lincomycin | High-level lincomycin resistance | 556 | 120 |
| lipA | 14.2 | Lipoate | Lipoate biosynthesis | 31534 | 1672, 1673, 3632, 4563 |
| lipB | 14.2 | Lipoate | Lipoyl-protein ligase | 555 | 3014, 3632, 4157, 4563, 681 |
| lit | 25.8 | Late induced T4 | Locus within defective prophage e14; expression of T4 late genes | 554 | 2128, 2129, 829 |
| livF | 77.4 | Leucine, isoleucine, valine (transport) | Membrane protein | 33713 | 14 |
| livG | 77.4 | Leucine, isoleucine, valine (transport) | hrbB, hrbC, hrbD; high-affinity branched-chain amino acid transport system; membrane component | 553 | 14, 1657, 3141, 3323, 4850 |
| livH | 77.5 | Leucine, isoleucine, valine (transport) | hrbB, hrbC, hrbD; high-affinity branched-chain amino acid transport system; membrane component | 552 | 14, 3142, 3323, 4850, 98 |
| livJ | 77.5 | Leucine, isoleucine, valine (transport) | hrbB, hrbC, hrbD; binding protein, high-affinity branched-chain amino acid transport system | 551 | 14, 2406, 3317, 3323, 4418, 4850, 98 |
| livK | 77.5 | Leucine, isoleucine, valine (transport) | hrbB, hrbC, hrbD; leucine-specific periplasmic binding protein, high-affinity branched-chain amino acid transport | 550 | 14, 2406, 3141, 3142, 332, 3323, 4418, 4850, 98 |
| livM | 77.4 | Leucine, isoleucine, valine (transport) | High-affinity transport system | 18190 | 14, 3141, 3142 |
| lldD | 81.4 | l-Lactose dehydrogenase | lct, lctD; l-lactate dehydrogenase, FMN dependent (EC 1.1.1.27); Arc regulon | 574 | 1066, 1951, 2556, 4012 |
| lldP | 81.4 | l-Lactose dehydrogenase | lctP; l-lactate permease | 35161 | 1066, 1951, 2556, 4012, 4128 |
| lldR | 81.4 | l-Lactose dehydrogenase | lctR; regulatory gene | 35164 | 1066, 2556, 4012, 4128 |
| lolA | 20.2 | Localization OM lipoproteins | Periplasmic protein responsible for sorting and transporting lipoproteins to outer membrane | 53417 | 2815, 2816 |
| lon | 9.9 | Long form | capR, deg, dir, muc; DNA-binding, ATP-dependent protease LA; lon mutants form long cells | 547 | 1063, 1297, 1324, 1392, 20, 2768, 2769, 2823, 2992, 3929, 4924, 4925, 682, 733, 752, 4396 |
| lpcA | 5.4 | Lipopolysaccharide core | gmhA; phosphoheptose isomerase, T-phage resistance | 546 | 1698, 4562, 517 |
| lpcB | 68.0 | Lipopolysaccharide core | mrc, pon; T-phage resistance, novobiocin sensitivity | 545 | 1024,1 |
| lpd | 2.8 | Lipoamide dehydrogenase | dhl; lipoamide dehydrogenase (NADH) (EC 1.8.1.4) | 544 | 1553, 1556, 1558, 2415, 2416, 4199, 4150, 4203, 655, 866 |
| lplA | 99.6 | Lipoprotein ligase | slr; lipoate-protein ligase A; selenolipoate resistant | 33052 | 3013, 3014, 3633 |
| lpp | 37.8 | Lipoprotein | mlpA; murein lipoprotein structural gene | 543 | 1420, 1529, 1671, 3109, 3110, 4792, 4819, 487, 4971, 3748a |
| lpxA | 4.4 | Lipid A expression | UDP-N-acetylglucosamine acetyltransferase | 17665 | 1346, 4747, 804, 876, 877 |
| lpxB | 4.4 | Lipid A expression | pgsB; lipid A disaccharide synthase | 404 | 4424, 804, 876, 877 |
| lpxC | 2.3 | Lipid A expression | asmA; envA; essential gene; cell envelope and cell separation | 815 | 2071, 2273, 260, 2675, 2676, 2911, 3091, 3921, 4139, 4275, 4903 |
| lpxD | 4.3 | Lipid A expression | fir, firA, hlpA, omsA, skp, ssc; UDP-3-O-(R-3-hydroxymyristoyl)-glucosamine N-acyltransferase | 775 | 1, 1040, 1773, 1775, 2193, 3761, 3921, 804, 1702 |
| lpxK | 20.9 | Lipid A expression | Lipid A 4′ kinase | 51784 | 1375 |
| lrb | 7.3 | l-Ribose (utilization) | Affects NADPH-linked l-ribose reductase activity | 36791 | 4459 |
| lrhA | 51.8 | LysR homolog | genR; regulatory protein, similar to LysR family | 32685 | 421 |
| lrp | 20.1 | Leucine regulatory protein | mbf, ihb, livR, lrs, lss, lstR, oppI, rblA; high-affinity branched-chain amino acid transport system; global regulatory protein, Leu responsive; site-specific blocking of methylation | 549 | 100, 117, 1349, 2416, 2555, 2558, 3167, 3450, 3660, 4516, 4551, 473, 4748, 883, 1627 |
| lspA | 0.5 | Lipoprotein signal peptidase | dapB, ileS; prolipoprotein signal peptidase (EC 3.4.99.–) | 11326 | 1913, 2104, 2945, 3640, 4422, 466, 4820, 4829, 4830, 4905 |
| lysA | 64.1 | Lysine | Diaminopimelate decarboxylase (EC 4.1.1.20) | 540 | 4247, 4249, 716 |
| lysC | 91.2 | Lysine | apk; aspartokinase III (EC 1.1.1.3) | 539 | 467, 652, 653 |
| lysP | 48.4 | Lysine | cadR; lysine permease | 942 | 3143, 3485, 4198, 4325 |
| lysQ | 16.8 | Lysine | Lysine tRNA (multiple loci, see lysT) | 51037 | 2313 |
| lysR | 64.2 | Lysine | LysR, prototype of family of global transcriptional regulators | 18187 | 4248 |
| lysS | 65.3 | Lysine | asuD herC; lysyl tRNA synthetase, constitutive | 17662 | 1122a, 1151, 1351, 1383, 1771, 1946, 2173, 686, 768 |
| lysT | 16.8 | Lysine | Suβ, lysTα, supG, supL (ochre suppression); lysine tRNA (multiple loci, lysQTVWYZ) | 537 | 1122a, 1285, 2313, 3530, 4898 |
| lysU | 93.8 | Lysine | Lysyl tRNA synthetase, inducible | 16693 | 1394, 1946, 1948, 2508, 4541, 686, 268 |
| lysV | 54.3 | Lysine | supN; lysine tRNA (multiple loci, see lysT) | 132 | 1122a, 2313, 3238, 4509, 539 |
| lysW | 16.8 | Lysine | lysTβ; lysine tRNA (multiple loci, see lysT) | 28352 | 2313 |
| lysX | 63.3 | Lysine | Lysine excretion | 536 | 2007, 2008 |
| lysY | 16.8 | Lysine | lysTγ; lysine tRNA (multiple loci, see lysT) | 51031 | 2313 |
| lysZ | 16.8 | Lysine | lysine tRNA (multiple loci, see lysT) | 51034 | 2313 |
| lytA | 60.6 | Lytic | Tolerance to β-lactams; autolysis | 18184 | 1649, 1651, 2380, 4035 |
| lytB | 0.6 | Lytic | Penicillin tolerance and stringent response effects | 34696 | 1575, 1651, 2380, 3506, 466 |
| lyx | 80.7 | Lyxose | Novel pathway for utilization of lyxose via xylulose through rhamnose and pentose pathways; xylulose kinase | 36908 | 2437, 3843 |
| maa | 10.3 | Maltose acetylase | mac; maltose transacetylase | 36808 | 477 |
| mac | 26.5 | Macrolide | Macrolide resistance, erythromycin resistance; see also maa | 535 | 4143 |
| mae | 33.5 | Malic enzyme | May be sfcA; malic enzyme, NAD linked (EC 1.1.1.38) | 36710 | 2719 |
| mafA | 0.9 | Maintenance of F | Maintenance of F-like plasmids | 534 | 3249, 3313, 4625–4628 |
| mafB | 1.9 | Maintenance of F | Maintenance of F-like plasmids | 533 | 4625 |
| malA | 76.4 | Maltose | malQPT cluster | 23561 | 1781 |
| malB | 91.5 | Maltose | malG-M cluster | 18924 | |
| malE | 91.5 | Maltose | Maltose-binding protein, periplasmic; substrate recognition for transport and chemotaxis | 532 | 1103, 1224, 1293, 271, 272, 273, 274, 3265, 329, 330, 3644, 3668, 810 |
| malF | 91.4 | Maltose | Maltose transport complex, inner membrane-spanning subunit | 531 | 1128, 1320, 2850, 3798 |
| malG | 91.4 | Maltose | Maltose/maltodextrin transport complex, inner membrane-spanning subunit | 530 | 1224, 1295, 3798, 943 |
| malI | 36.6 | Maltose | Regulatory gene whose product has homology to repressors LacI, GalR, CytR | 18181 | 1129, 3642 |
| malK | 91.5 | Maltose | malB; maltose transport complex, ATP-binding subunit | 529 | 1224, 1431, 1701, 271, 272, 274, 3265, 3798, 4394, 814 |
| malM | 91.5 | Maltose | molA; periplasmic protein | 18178 | 1432 |
| malP | 76.5 | Maltose | blu; maltodextrin phosphorylase (EC 2.4.1.1) | 528 | 1778, 1779, 1781, 3340, 3588, 3706, 3940, 990 |
| malQ | 76.4 | Maltose | Amylomaltase (EC 2.4.1.25) | 527 | 1778, 1779, 1781, 3551, 3940 |
| malS | 80.5 | Maltose | α-Amylase, periplasmic, MalT dependent | 17659 | 1306, 3922 |
| malT | 76.5 | Maltose | Lambda sensitivity; positive regulator for mal regulon | 526 | 1224, 1778, 1779, 1780, 1781, 3587, 3588, 3669, 800, 928, 931, 988, 989, 990 |
| malX | 36.6 | Maltose | PTS enzyme II homolog; malI regulated | 32254 | 3643 |
| malY | 36.6 | Maltose | Affects induction of maltose system | 32257 | 3643, 4922 |
| malZ | 9.1 | Maltose | Maltodextrin glucosidase | 29868 | 2651, 3394, 4352 |
| manA | 36.4 | Mannose | pmi; mannosephosphate isomerase (EC 5.3.1.8) | 525 | 2024, 2491, 2932, 2933, 3207, 386 |
| manC | 87.6 | Mannose | mni; d-mannose isomerase | 524 | 4207 |
| manX | 41.0 | Mannose | gptB, mpt, ptsL, ptsM, ptsX; mannose phosphotransferase system, protein II-A (III) | 17656 | 1169, 1171, 3344, 4745 |
| manY | 41.0 | Mannose | pel, ptsM, ptsP | 18175 | 1169, 1171, 1844, 3344, 4745 |
| manZ | 41.0 | Mannose | gptB, mpt, ptsM, ptsX; mannosephosphotransferase enzyme IIB | 346 | 1844, 3344, 4745, 890 |
| map | 4.1 | Methionine aminopeptidase (EC 3.4.11.18) | 30569 | 294, 676 | |
| marA | 34.9 | Multiple antibiotic resistance | soxQ, cfxB? nfxC, norB? resistance to tetracycline, other antibiotics; transcription activator of multiple antibiotic resistance system | 18172 | 130, 1350, 1403, 1448, 1505, 1591, 1592, 1984, 2780, 3737, 4724, 790, 791 |
| marB | 34.9 | Multiple antibiotic resistance | Regulatory gene for mar | 32217 | 130, 1448, 3737, 790 |
| marR | 34.9 | Multiple antibiotic resistance | cfxB, soxQ; repressor of mar operon | 30935 | 130, 1448, 2780, 3737, 4271, 790 |
| mbrB | 88.6 | Mothball resistant | Resistance to camphor vapors; coupling of cell division and replication, growth rate and partitioning | 36929 | 4468, 4469 |
| mcrA | 26.1 | Methylcytosine restriction | rglA; within defective prophage e14; restriction of DNA at 5-methylcytosine residues | 10961 | 3602, 3603, 3604, 3618, 1759 |
| mcrB | 98.6 | Methylcytosine restriction | rglB; restriction of DNA at 5-methylcytosine residues | 4978 | 1050, 2350, 3602, 3603, 3604, 3739, 3740, 3741 |
| mcrC | 98.6 | Methylcytosine restriction | Modifies specificity of McrB restriction | 34613 | 1050, 2350, 3603, 3740 |
| mcrD | 98.6 | Methylcytosine restriction | Inhibits McrE restriction resistance to DMP840, adriamycin, etoposide; possibly modulates Topo IV activity | 34616 | 3603, 3740 |
| mdaB | 68.3 | Modulator of drug activity | 53544 | 688 | |
| mdh | 72.9 | MDH | Malate dehydrogenase (EC 1.1.1.37) | 523 | 1227, 2831, 4298, 4598 |
| mdoA | 23.9 | Membrane-derived oligosaccharide | modGH operon | 4970 | |
| mdoB | 99.0 | Membrane-derived oligosaccharide | Phosphoglycerol transferase I activity | 18169 | 1234, 1974, 569 |
| mdoG | 23.9 | Membrane-derived oligosaccharide | mdoA; periplasmic oligosaccharide synthesis | 31849 | 1235, 2387, 2388, 2636, 411 |
| mdoH | 23.9 | Membrane-derived oligosaccharide | mdoA; membrane glycosyltransferase | 31852 | 1235, 2387, 2388, 2636, 411 |
| meb | 78.7 | malE bypass | Suppressor of malE secB-defective transport of mal-binding protein | 28194 | 1293 |
| melA | 93.5 | Melibiose | mel-7; α-galactosidase (EC 3.2.1.22) | 522 | 1624, 2185, 2501, 2548, 4031, 4685 |
| melB | 93.6 | Melibiose | mel-4; thiomethylgalactoside permease II | 521 | 1624, 2185, 2501, 252, 4878 |
| melR | 93.5 | Melibiose | Regulatory | 18166 | 2501, 462, 4685, 659 |
| menA | 88.7 | Menaquinone (vitamin K2) | Dimethylmenaquinone formation in vitamin K2 biosynthesis | 520 | 3465, E |
| menB | 51.2 | Menaquinone | 1,4-Dihydroxy-2-naphthoate synthase | 519 | 1548, 1550, 3997, 4007, 4011 |
| menC | 51.2 | Menaquinone | o-Succinylbenzoate synthase II | 518 | 1548, 1550, 310, 4007, 4011 |
| menD | 51.2 | Menaquinone | o-Succinylbenzoate synthase I (EC 4.1.3.-) | 517 | 1548, 1550, 3338, 3486, 4007, 4011 |
| menE | 51.1 | Menaquinone | o-Succinylbenzoate-CoA synthase | 17653 | 2384, 3998, 4011 |
| menF | 51.2 | Menaquinone | Menaquinone pathway-specific isochorismate synthase | 35955 | 3040, 310, 936 |
| mepA | 52.7 | Murein peptidase | Murein dd-endopeptidase | 17650 | 1890, 2184 |
| mesJ | 4.6 | Cell cycle protein | 49262 | 3423a | |
| metA | 90.8 | Methionine | Homoserine transsuccinylase (EC 2.3.1.46) | 516 | 1094, 2919, 2920, 358, 35 |
| metB | 88.9 | Methionine | met1, met-1; cystathionine γ-synthase (EC 4.2.99.9) | 515 | 1093, 1508, 1509, 2242, 2347, 2547, 3802, 4917 |
| metC | 67.9 | Methionine | Cystathionine β-lyase; (EC 4.4.1.8) | 514 | 284, 2918 |
| metD | 4.8 | Methionine | Methionine sulfoximine sensitivity, d-methionine transport | 513 | 2078 |
| metE | 86.5 | Methionine | Tetrahydropteroyltriglutamate methyltransferase (EC 2.1.1.14) | 512 | 3284, 58, 66, 926 |
| metF | 89.0 | Methionine | 5,10-Methylenetetrahydrofolate reductase (EC 1.1.1.68) | 511 | 1509, 2242, 2347, 3802, 3803, 4454, 4917 |
| metG | 47.3 | Methionine | Methionyl-tRNA synthetase; ethionine effects | 510 | 125, 929, 930 |
| metH | 91.0 | Methionine | B12-dependent homocysteine-N5-methyltetrahydrofolate transmethylase | 509 | 218, 2756, 3283, 3284 |
| metJ | 88.9 | Methionine | Methionine sulfoximine plus methylmethionine sensitivity; repressor | 508 | 1509, 2078, 2242, 2336, 2347, 2547, 3802, 4109, 4917 |
| metK | 66.5 | Methionine | Methionine adenosyltransferase (EC 2.5.1.6); ethionine sensitivity | 507 | 1510, 1598, 2336, 2766, 3878, 472 |
| metL | 89.0 | Methionine | metM; aspartokinase II-homoserine dehydrogenase II | 506 | 1093, 1508, 1509, 2242, 2347, 3802, 3877, 4916, 4917, 789, 953 |
| metR | 86.4 | Methionine | Positive regulatory gene for metE and metH and autogenous regulation | 18163 | 2825, 4515, 4703, 601, 926, 4515 |
| metT | 15.0 | Methionine | metTα; duplicate gene; methionine tRNAm; see metU | 505 | 1285, 1915, 2313, 3102, 3103, 3454 |
| metU | 15.0 | Methionine | metTβ; duplicate gene; methionine tRNAm | 28349 | 2313 |
| metV | 63.5 | Methionine | metZβ; triplicate gene; initiator methionine tRNAf1 (metVWZ) | 35659 | 2198, 2199, 2313 |
| metW | 63.5 | Methionine | Triplicate gene; methionine tRNAf1 | 35656 | 2313, 2198 |
| metY | 71.5 | Methionine | Methionine tRNAf2 | 504 | 1487, 1927, 1928, 2197, 2313 |
| metZ | 63.5 | Methionine | metZα; triplicate gene; initiator methionine tRNAf1; metVWZ | 503 | 2198, 2199, 2313, 3085 |
| mfd | 25.2 | Mutation frequency decline | Transcription repair coupling factor | 35179 | 3964–3969 |
| mglA | 48.2 | Methyl-galactoside | mglP; PMG; methyl-galactoside transport and galactose taxis; cytoplasmic membrane protein | 502 | 1158, 1645, 1783, 2925, 3156, 3302, 3751, 3932, 428, 429, 758 |
| mglB | 48.2 | Methyl-galactoside | PMG; galactose-binding protein; receptor for galactose taxis | 501 | 1158, 1645, 1783, 2925, 3156, 3302, 3751, 3932, 3938, 3954, 758 |
| mglC | 48.2 | Methyl-galactoside | mglP; PMG; methyl-galactoside transport and galactose taxis | 500 | 1158, 1645, 1783, 2925, 3156, 3302, 3751, 758 |
| mglR | 16.9 | Methyl-galactoside | R-MG; regulatory gene | 498 | 1354 |
| mgsA | 22.1b | Methylglyoxal synthase | Methylglyoxal synthase (not shown on map) | 57077 | 4444a |
| mgtA | 96.2 | Magnesium transport | corB; cobalt resistance, magnesium transport | 497 | 3353, 569 |
| mhpA | 7.9 | m-Hydroxyphenylpropionic acid | 3-(3-Hydroxyphenyl)propionate 2-hydroxylase; utilizes MHP | 29024 | 1229, 570, 571 |
| mhpB | 8.0 | m-Hydroxyphenylpropionic acid | 3-(2,3-Dihydroxyphenyl)propionate dioxygenase; utilizes MHP | 29027 | 1229, 4147, 570, 571 |
| mhpC | 8.0 | m-Hydroxyphenylpropionic acid | Dihydroxyphenylpropionate-ring-fission-product hydrolase; utilizes MHP | 29030 | 1229, 570, 571 |
| mhpD | 8.0 | m-Hydroxyphenylpropionic acid | mhpS; 2-keto-4-pentenoate hydratase; utilizes MHP | 29033 | 1229, 570, 571 |
| mhpE | 8.0 | m-Hydroxyphenylpropionic acid | 4-Hydroxy-2-oxovalerate aldolase; utilizes MHP | 51197 | 1229 |
| mhpF | 8.0 | m-Hydroxyphenylpropionic acid | Acetaldehyde dehydrogenase, acylating; utilizes MHP | 53814 | 1228, 1229 |
| mhpR | 7.9 | m-Hydroxyphenylpropionic acid | Regulatory gene for MHP utilization | 29021 | 1228, 1229, 570, 571 |
| miaA | 94.8 | Me-isopentyl-adenine | trpX; 2-methylthio-N6-isopentyladenosine tRNA hypermodification | 18160 | 1345, 4487, 4489, 4490, 563, 605, 825, 826 |
| miaD | 99.6 | Me-isopentyl-adenine | Suppresses leaky miaA(Oc) mutation | 35254 | 826 |
| micF | 49.8 | mRNA interfering cRNA | stc; regulatory antisense RNA affecting ompF; member of soxRS regulon | 18157 | 2955, 2982, 3008, 3608, 3918, 4309, 4520, 93, 4335 |
| minB | 26.4 | Minicell | min operon; formation of minicells containing no DNA; positioning division septum | 495 | 3895, 954, 971a |
| minC | 26.4 | Minicell | Inhibition of FtsZ ring at division site | 31329 | 340, 3614, 970, 971, 971a |
| minD | 26.4 | Minicell | Affects cell division and growth; membrane ATPase that activates MinC | 31326 | 2386, 340, 3614, 948, 969, 970, 971a |
| minE | 26.4 | Minicell | Reverses inhibition by MinC of FtsZ ring | 31317 | 3614, 948, 970, 993, 971a |
| mioC | 84.6 | Minimal origin | Initiation of replication; transcription of 16-kDa protein proceeds through oriC | 18154 | 2603, 2606, 3245, 408 |
| mltA | 63.5 | Membrane-bound lytic transglycosylase | waaN; lipoprotein lytic transglycosylase; membrane-bound murein hydrolase, affecting sacculus maturation | 53524 | 1789, 2618, 4517 |
| mltB | 60.8 | Membrane-bound lytic transglycosylase | slt; murein hydrolase lipoprotein; Slt35, soluble lytic transglycosylase | 41040 | 1124, 1154, 1789, 1048a |
| mltC | 66.9 | Membrane-bound lytic transglycosylase | Peptidoglycan hydrolase activity, lytic transglycosylase family | 54700 | 1049 |
| mltD | 5.0 | Membrane-bound lytic transglycosylase | dniR (dissimilatory nitrite reductase); reduced amounts of hexaheme nitrite reductase; membrane transglycosylase | 1575 | 2089 |
| mmrA | 85.4 | Minimal medium recovery | Same as rhlB? RNA helicase motif | 18151 | 2099, 3993, 926, 4508 |
| mms | 69.2 | Macromolecular synthesis | Complex operon, macromolecular synthesis | 36863 | 3154 |
| mng | 40.0 | Manganese | Manganese resistance | 494 | 4063 |
| moaA | 17.6 | Molybdenate | bisA, chlA, narA; MPT synthesis; chlorate resistance protein A | 922 | 1003, 1245, 1434, 18, 187, 2030, 207, 2402, 3534, 3986, 4570, 72 |
| moaB | 17.6 | Molybdenate | MPT synthesis; chlorate resistance protein B | 31222 | 3986 |
| moaC | 17.6 | Molybdenate | MPT synthesis; chlorate resistance protein C | 31225 | 3986 |
| moaD | 17.6 | Molybdenate | chlM; MPT synthesis; chlorate resistance | 18475 | 2030, 3986, 4373 |
| moaE | 17.6 | Molybdenate | MPT synthesis; chlorate resistance | 31228 | 3986 |
| mobA | 87.1 | Molybdenum | chlB, narB; MPT guanine dinucleotide synthesis; chlorate resistance | 921 | 1003, 1245, 1434, 18, 187, 1922, 2402, 3342, 3343, 3465, 3534, 3749, 3986, 654 |
| mobB | 87.0 | Molybdenum | Molybdenum cofactor biosynthesis, putative nucleotide binding site | 43961 | 1922, 3343 |
| moc | 31.9 | Modification of CCA | Modification of CCA at 3′ end of tRNA | 36923 | 3237 |
| modA | 17.1 | Molybdenum | Molybdate uptake; chlorate resistance; periplasmic molybdate binding protein | 37372 | 2060, 2820, 2973, 4634 |
| modB | 17.1 | Molybdenum | chlJ; molybdate uptake; chlorate resistance; membrane-spanning ABC protein | 18478 | 2025, 2060, 2820, 2973, 3630, 3986, 4634 |
| modC | 17.2 | Molybdenum | chlD, narD; molybdate uptake; chlorate resistance | 920 | 1003, 1245, 1434, 18, 187, 2025, 2060, 2820, 2973, 3630, 3986, 4634 |
| modE | 17.1 | Molybdenum | chlD, modR, narD; molybdate uptake | 37366 | 1539, 2855, 4634 |
| modF | 17.1 | Molybdenum | chlD, narD, phrA; molybdate uptake | 37369 | 1539, 4634 |
| moeA | 18.6 | Molybdenum | bisB, chlE; MPT synthesis; chlorate resistance | 919 | 1003, 1245, 1434, 1664, 18, 187, 2030, 2402, 3208, 3378, 3534, 3986, 4219 |
| moeB | 18.6 | Molybdenum | chlN; MPT synthesis; chlorate resistance | 18472 | 2030, 3208, 3986, 4373 |
| mog | 0.2 | Molybdenum | bisD, chlG; unknown function; chlorate resistance | 917 | 1003, 1245, 1434, 18, 187, 2003, 2004, 2060, 3986, 4219 |
| molR | 47.3 | Molybdate | Unknown function, probably related to molybdate transport | 32567 | 2464, 3736 |
| motA | 42.6 | Motility | flaJ; flagellar-regulon member; flagellar rotation | 491 | 374, 375, 4064, 4101, 985 |
| motB | 42.5 | Motility | flaJ; flagellar-regulon member; flagellar rotation | 490 | 373, 4064, 4101, 4183 |
| mpl | 96.0 | meso-Diaminopimelate ligase | UDP-N-acetylmuramate:l-alanyl-γ-d-glutamyl-meso-diaminopimelate ligase; recycles cell wall peptidoglycan (EC 6.3.2.13) | 46610 | 2894 |
| mppA | 30.0 | Murein peptide permease | Murein tripeptide (l-Ala-γ-d-Glut-m-DAP) permease | 53908 | 3352 |
| mprA | 60.5 | Microcin peptide regulation | emrR; controls level of microcin synthesis; negative regulation of EmrAB | 33255 | 1006, 1007, 2622 |
| mraA | 2.0 | Murein cluster a | d-Alanine carboxypeptidase | 489 | |
| mraY | 2.1 | Murein cluster a | UDP-N-acetylmuramoyl-pentapeptide:undecaprenyl-PO4 phosphatase (EC 2.7.8.13) | 30466 | 1894, 1895 |
| mrcA | 75.9 | Murein cluster c | ponA; penicillin-binding protein 1A | 484 | 1930, 4337, 519 |
| mrcB | 3.6 | Murein cluster c | pbpF, ponB; peptidoglycan synthetase; penicillin-binding protein 1Bs | 483 | 2166, 3096, 3097, 3601, 3714, 4307, 4337, 4346, 4957, 519 |
| mrdA | 14.3 | Murein cluster d | pbpA; penicillin-binding protein PBP 2, mecillinam resistance | 18148 | 156, 157, 279, 4157, 4232, 4233, 4343, 4587, 545 |
| mrdB | 14.3 | Murein cluster d | rodA; affects cell shape; mecillinam sensitivity | 18145 | 156, 157, 279, 2818, 4232, 4233 |
| mreB | 73.2 | Murein cluster e; mecillinam resistance | envB, mon, rodY; mecillinam resistance; cell shape, affects division versus elongation | 31343 | 1062, 1968, 2624, 434, 4616, 4617, 4619, 4716 |
| mreC | 73.2 | Mecillinam resistance | Cell division and growth; mecillinam resistance; rod shape-determining protein | 31348 | 4617, 4618 |
| mreD | 73.2 | Mecillinam resistance | Mecillinam resistance; rod shape-determining protein | 31351 | 4617, 4618 |
| mrp | 47.2 | Methionine-related protein | Putative ATPase; in Salmonella, homolog is part of alternative pyrimidine pathway to Thi (apbC) | 32572 | 3409, 930 |
| mrr | 98.8 | Methyl-purine restriction | Restriction of methylated adenine | 18139 | 1700, 3603 |
| msbA | 20.8 | Multicopy suppressor of htrB | ABC transporter homology; biogenesis of outer membrane | 31743 | 2139, 3474 |
| msbB | 41.8 | Multicopy suppressor of htrB | Role in outer membrane structure or function | 32359 | 2138 |
| mscL | 74.1 | Mechanosensitive channel | Cytoplasmic membrane channel protein, opens large pore in response to mechanical stress | 35417 | 400, 401, 4267, 4268, 4269 |
| msrA | 95.7 | Methionine sulfoxide reductase | pmsR; methionine sulfoxide reductase | 34400 | 3018, 3585, 3586, 525 |
| msyB | 24.0 | Multicopy suppressor of secY | In multicopy restores growth and protein export functions of secY and secA mutants | 34718 | 4504 |
| mtgA | 72.1 | Monofunctional transglycolase | Monofunctional biosynthetic peptidoglycan transglycosylase | 54732 | 4159 |
| mtlA | 81.3 | Mannitol | mtlC (promoter/operator); mannitol-specific enzyme II of PTS | 481 | 2018, 2459, 2468, 965 |
| mtlD | 81.3 | Mannitol | Mannitol-1-phosphate dehydrogenase (EC 1.1.1.17) | 479 | 2018, 2347, 965 |
| mtlR | 81.3 | Mannitol | Mannitol repressor | 36774 | 1236 |
| mtr | 71.2 | Methyltryptophan | High-affinity Trp permease; TyR regulon | 478 | 1689, 1764, 1688, 3864 |
| mukB | 21.0 | Mukaku (anucleate) | Required for chromosome partitioning; DNA binding; kinesin-like motor protein? | 31516 | 1188, 1761, 1977, 3183, 1185, 3819, 4589 |
| mul | 83.1 | Mutability lambda | Mutability in UV-radiated lambda phage | 477 | 4620 |
| murA | 71.8 | Murein | mrbA, murZ; UDP-N-acetylglucosamine enoylpyruvyl transferase (EC 2.5.1.7); phosphomycin resistance | 33518 | 2773, 3548, 4571, 527 |
| murB | 89.9 | Murein | UDP-N-acetylglucosaminyl-3-enolpyruvate reductase (EC 1.1.1.158) | 34141 | 308, 3548, 395 |
| murC | 2.2 | Murein | l-Alanine adding enzyme | 476 | 1183, 1893, 2021, 2071, 2675, 2676 |
| murD | 2.1 | Murein | UDP-N-acetylmuramoyl-l-alanine:d-glutamate ligase (EC 6.3.2.9) | 30450 | 1894, 2889, 2890, 3533 |
| murE | 2.0 | Murein | meso-Diaminopimelate adding enzyme | 475 | 1183, 2071, 2889, 2923, 4349 |
| murF | 2.0 | Murein | mra; d-alanyl:d-alanine adding enzyme | 474 | 101, 1183, 2071, 2889, 3368 |
| murG | 2.1 | Murein | UDP-NAc-glucosamine: NAc-muramyl-(pentapeptide) pyrophosphoryl-undecaprenol NAc-glucosamine transferase | 473 | 1893, 2071, 2889, 2895, 3820 |
| murH | 99.3 | Murein | Terminal stage in peptidoglycan synthesis, incorporating disaccharide peptide units into wall | 18136 | 3204, 900 |
| murI | 89.7 | Murein | mbrC, dga, glr; glutamate racemase (EC 5.1.1.3); d-glutamate synthesis, essential for peptidoglycan | 29401 | 1076, 1077, 1078, 212, 2584, 4900, 523 |
| mutG | 43.7 | Mutator | Mutation causes high C-to-T mutation in second C of CCAGG; near but distinct from vsr; provisionally termed mutG | 28933 | 3766 |
| mutH | 64.0 | Mutator | mutR, prv; methyl-directed mismatch repair; see also dnaX | 471 | 1222, 1484, 1485, 1706, 4052, 4488, 856 |
| mutL | 94.7 | Mutator | Methyl-directed mismatch repair | 470 | 1088, 1222, 139, 1706, 4052, 4487, 4489, 4490, 856 |
| mutM | 82.1 | Mutator | fpg; repair; GC to TA; formamidopyrimidine-DNA glycosylase | 18133 | 1221, 1489, 1706, 2434, 3717, 4052, 4128, 416, 417, 568, 599, 805, 856 |
| mutS | 61.5 | Mutator | ant? fdv (formate dehydrogenase 2?); methyl-directed mismatch repair | 469 | 1222, 1706, 2587, 3615, 3853, 4052, 4488, 4802, 856 |
| mutT | 2.4 | Mutator | AT to GC transversions | 468 | 1288, 1706, 2729, 338, 39, 4052, 856 |
| mutY | 66.8 | Mutator | micA; GC to TA transversions; adenine glycosylase, G-A repair | 18130 | 1706, 2922, 3172, 3576, 4052, 856, 4476, 4477 |
| nac | 44.4 | Nitrogen assimilation control | Regulatory gene, binding ntrC | 50182 | 3073 |
| nadA | 16.8 | Nicotinamide adenine dinucleotide | Quinolinate synthetase, A protein | 467 | 1257, 2819, 4246 |
| nadB | 58.4 | Nicotinamide adenine dinucleotide | Quinolinate synthetase, B protein | 466 | 1257, 3015, 4379, 3963 |
| nadC | 2.5 | Nicotinamide adenine dinucleotide | nic; quinolinate phosphoribosyl transferase | 465 | 1553, 1556, 1558, 2415, 2416, 3689 |
| nadE | 39.2 | Nicotinamide adenine dinucleotide | efg, ntrL; NAD synthetase, ammonia dependent | 28576 | 4752, 70 |
| nagA | 15.1 | N-Acetylglucosamine | N-acetylglucosamine-6-phosphate deacetylase (EC 3.5.1.25) | 464 | 2491, 3402, 3454–3456, 4599, 4726 |
| nagB | 15.1 | N-Acetylglucosamine | glmD; glucosamine-6-phosphate deaminase (EC 5.3.1.10) | 463 | 3402, 3454, 3455, 3456, 3464, 3709, 4601, 4726 |
| nagC | 15.1 | N-Acetylglucosamine | nagR; transcriptional regulator of nag operon | 31479 | 3402, 3455, 3456, 3457, 4593, 3458 |
| nagD | 15.1 | N-Acetylglucosamine | Function unknown, expressed as part of nag operon | 36240 | 3455 |
| nagE | 15.2 | N-Acetylglucosamine | pstN; N-acetylglucosamine-specific enzyme II of phosphotransferase system | 462 | 2048, 2491, 3401, 3402, 3454, 3709, 4599, 4726, 4600 |
| nalB | 60.2 | Nalidixic acid | Sensitivity to nalidixic acid (NAL) | 460 | 1626 |
| nalD | 89.2 | Nalidixic acid | NAL sensitivity; NAL and glycerol penetration | 18124 | 1827 |
| nanA | 72.6 | N-Acetylneuraminate | N-Acetylneuraminate lyase (aldolase) (EC 4.1.3.3) | 17647 | 2782, 3270, 4585 |
| nanT | 72.6 | N-Acetylneuraminate | Sialic acid transport | 18121 | 2782, 4585 |
| napA | 49.5 | NitrAte reductase, periplasmic | Nitrate reductase homolog | 36550 | 1537, 739, 740 |
| napB | 49.5 | NitrAte reductase, periplasmic | Cytochrome c homolog | 36560 | 1537, 739, 740 |
| napC | 49.5 | NitrAte reductase, periplasmic | Cytochrome c homolog | 36566 | 1537, 739, 740 |
| napD | 49.6 | NitrAte reductase, periplasmic | Unknown function, nap operon | 36547 | 1537, 739, 740 |
| napF | 49.6 | NitrAte reductase, periplasmic | Ferredoxin homolog | 36544 | 1537, 739, 740 |
| napG | 49.5 | NitrAte reductase, periplasmic | Ferredoxin homolog | 36553 | 1537, 739, 740 |
| napH | 49.5 | Nitrate reductase, periplasmic | Ferredoxin homolog | 36556 | 1537, 739, 740 |
| narG | 27.6 | Nitrate reductase, nitrate regulation | chlC, narC; nitrate reductase α-subunit (EC 1.7.99.4) | 459 | 1118, 1595, 240, 2495, 2532, 2533, 2856, 3727, 391, 4125 |
| narH | 27.7 | Nitrate reductase, nitrate regulation | chlC; nitrate reductase β-subunit | 18118 | 1118, 1246, 1433, 240, 2495, 392, 4125 |
| narI | 27.7 | Nitrate reductase, nitrate regulation | chlI; cytochrome bNR, structural gene; γ subunit | 458 | 1595, 2495, 3305, 3727, 4125, 4126, 4219, 425, 735 |
| narJ | 27.7 | Nitrate reductase, nitrate regulation | Nitrate reductase δ-subunit; chaperone | 18115 | 1092, 2495, 2588, 3343, 392, 4125 |
| narK | 27.5 | Nitrate reductase, nitrate regulation | Nitrate/nitrite antiporter (probably) | 18112 | 1017, 2495, 391, 4220 |
| narL | 27.5 | Nitrate reductase, nitrate regulation | frdR, narR; regulatory protein | 18109 | 1122, 1569, 1954, 2096, 2097, 2495, 4008, 4215, 4220 |
| narP | 49.3 | Nitrate reductase, nitrate regulation | Regulatory protein | 32598 | 3575, 4217 |
| narQ | 55.7 | Nitrate reductase, nitrate regulation | Nitrate sensor-transmitter protein, anaerobic respiratory path; function redundant with narX | 36333 | 3574, 3575, 3935, 726–728 |
| narU | 33.2 | Nitrate reductase, nitrate regulation | Nitrate sensor-trasmitter protein, anaerobic respiratory path | 52770 | 423 |
| narV | 33.1 | Nitrate reductase, nitrate regulation | Cryptic nitrate reductase II, γ-subunit | 32136 | 391, 393 |
| narW | 33.1 | Nitrate reductase, nitrate regulation | Cryptic nitrate reductase II, δ-subunit | 32139 | 391, 393, 423 |
| narX | 27.5 | Nitrate reductase, nitrate regulation | frdR, narR; nitrate sensor-transmitter protein; functional redundance with narQ | 18106 | 2096, 2097, 2495, 4215, 4220, 4746, 728 |
| narY | 33.1 | Nitrate reductase, nitrate regulation | Cryptic NR II, β-subunit | 32142 | 391, 393, 423 |
| narZ | 33.1 | Nitrate reductase, nitrate regulation | Cryptic NR II, α-subunit | 18103 | 391, 393, 424 |
| ndh | 25.1 | NADH dehydrogenase | Respiratory NADH dehydrogenase | 457 | 1497, 2888, 4901, 615 |
| ndk | 57.0 | Nucleoside diphosphate kinase | Nucleoside diphosphate kinase (EC 2.7.4.6) | 32926 | 1615, 1616, 2649, 3985 |
| neaB | 75.0 | Neamine | Neamine sensitivity | 456 | 630 |
| nei | 16.0 | Endonuclease VIII | DNA glycosylase/apurinic lyase, deoxyinosine-specific; endonuclease VIII | 53314 | 1656, 2016, 2017, 2875, 3805 |
| nemA | 37.2 | N-Ethylmaleimide | N-Ethylmaleimide reductase | 52714 | 2968 |
| nfi | 90.5 | Endonuclease V | Endonuclease V, specific for single-stranded DNA or duplex DNA with damaged U | 45426 | 1018, 1571, 1572, 4874 |
| nfnA | 80.8 | Nitrofurantoin | Nitrofurantoin sensitivity | 18100 | 3876 |
| nfnB | 13.0 | Nitrofurantoin | nfsI, nfsB; resistance to nitrofurantoin; a nitroreductase | 18097 | 2832, 2917, 3876, 490, 4931, 4932 |
| nfo | 48.5 | Endonuclease IV | Endonuclease IV; member of soxRS regulon | 14161 | 889 |
| nfrA | 12.7 | N4 (phage) resistant | Outer membrane protein, putative structural receptor for N4 adsorption | 31154 | 2213, 2214 |
| nfrB | 12.7 | N4 (phage) resistant | Phage N4 susceptibility; membrane protein | 31157 | 2213, 2214 |
| nfrD | 54.2 | N4 (phage) resistant | Phage N4 susceptibility | 36113 | 2215 |
| nfsA | 21.9 | Nitrofurazone sensitivity | mdaA; nitrofuran reductase I activity B; overexpression results in resistance to cytotoxic drugs | 454 | 688, 2832, 490, 4930 |
| nhaA | 0.4 | Na+/H+ antiporter | ant; Na+/H+ antiporter; stress response to high salinity and pH | 15893 | 1081, 1410, 1440, 1909, 2142, 2143, 3272, 3329, 3330, 3583, 4326, 4393, 4577 |
| nhaB | 26.6 | Na+/H+ antiporter | Regulator of intracellular pH | 30269 | 2182, 3272, 3329, 3330, 3438, 3439, 4393, 4577 |
| nhaR | 0.4 | Na+/H+ antiporter | Positive regulator of Na+-dependent transcription of nhaA; DNA-binding; LysR family of regulatory proteins | 30281 | 3330, 3583, 641 |
| nikA | 77.8 | Nickel | hydC, hydD; affects formate hydrogen-lyase activity, hydrogenase, and hydrogenase-related fumarase | 18226 | 1340, 3139, 4230, 4796, 4797, 4800 |
| nikB | 77.9 | Nickel | hydC, hydD; activity as nikA | 33180 | 3139, 4796 |
| nikC | 77.9 | Nickel | hydC, hydD; activity as nikA | 33183 | 3139, 4796 |
| nikD | 77.9 | Nickel | hydC, hydD; activity as nikA | 33186 | 3139, 4796 |
| nikE | 77.9 | Nickel | hydD, lipP? formate hydrogen-lyase activity | 18223 | 3139, 4796, 4797 |
| nirB | 75.3 | Nitrite reductase | Nitrite reductase [NAD(P)H] subunit (EC 1.6.6.4) | 451 | 1647, 1973, 1998, 2693, 2694, 3386 |
| nirC | 75.3 | Nitrite reductase | Membrane protein affecting nitrite reductase [NAD(P)H] activity | 452 | 1647, 3386 |
| nirD | 75.3 | Nitrite reductase | Nitrite reductase [NAD(P)H] subunit (EC 1.6.6.4) | 35425 | 1647, 3386 |
| nlpA | 82.7 | New lipoprotein | Lipoprotein in outer membrane vesicles | 35704 | 2022, 3563, 4833, 4906 |
| nlpB | 56.0 | New lipoprotein | Nonessential lipoprotein in outer membrane vesicles | 33030 | 463 |
| nlpC | 38.6 | New lipoprotein | Lipoprotein | 35802 | 4792a |
| nlpD | 61.8 | New lipoprotein | May function in cell wall formation | 33225 | 1879, 2414 |
| nmpC | 12.4 | New membrane protein | phmA; locus of defective phage qsr′; porin, outer membrane, not expressed in K-12 due to IS5 insertion | 447 | 1751, 389, 390, 809 |
| non | 45.7 | Nonmucoid | Affects capsule formation | 446 | 3577 |
| npr | 72.1 | N-regulated protein | ptsO; NPr, N-regulated HPr-like protein | 43864 | 3517 |
| nrdA | 50.5 | Nucleotide reductase | dnaF; ribonucleoside diphosphate reductase subunit B1 (EC 1.17.4.1); class I enzyme, aerobic, physiologically active | 445 | 1326, 3190, 3451, 4085, 4278, 4279, 4280, 4495, 4496, 4827, 640, 1242, 1243, 1487, 1628 |
| nrdB | 50.6 | Nucleotide reductase | ftsB; ribonucleoside diphosphate reductase subunit B2 (EC 1.17.4.1); class I enzyme, aerobic | 444 | 1326, 2338, 3451, 4279, 4357, 4496, 4827, 640, 1242, 1243, 1487, 1628, 3822 |
| nrdD | 96.1 | Nucleotide reductase | Ribonucleotide reductase, class III, anaerobic | 34589 | 4284 |
| nrdE | 60.3 | Nucleotide reductase | Nonessential ribonucleoside-diP reductase 2, subunit α, class I, function unknown | 53512 | 2050, 2051 |
| nrdF | 60.3 | Nucleotide reductase | Nonessential ribonucleoside-diP reductase 2, subunit β, class I, function unknown | 53515 | 2050, 2051 |
| nrdG | 96.1 | Nucleotide reductase | NrdD activating enzyme, generating glycyl radical | 54766 | 4283 |
| nrfA | 92.4 | Nitrite reductase, formate-dependent | aidC; formate-dependent nitrite reduction; tetraheme cytochrome c552; aidC mutation complemented by nrfG | 34336 | 1868, 1921, 3334, 937, 11a, 4602 |
| nrfB | 92.4 | Nitrite reductase, formate-dependent | Formate-dependent nitrite reduction; pentaheme cytochrome c | 34364 | 1868, 938 |
| nrfC | 92.4 | Nitrite reductase, formate-dependent | Formate-dependent nitrite reduction; nonheme FE-S protein, probably transmembrane | 34352 | 1868, 938 |
| nrfD | 92.4 | Nitrite reductase, formate-dependent | Formate-dependent nitrite reduction; transmembrane protein similar to QOR | 34355 | 1868, 938 |
| nrfE | 92.5 | Nitrite reductase, formate-dependent | Formate-dependent nitrite reduction; membrane protein | 34358 | 1536, 1868, 938 |
| nrfF | 92.5 | Nitrite reductase, formate-dependent | Formate-dependent nitrite reduction; periplasmic protein; NrfA/B synthesis? | 34361 | 1536, 1868, 938 |
| nrfG | 92.5 | Nitrite reductase, formate-dependent | aidC; function unknown, required for Nrf pathway; aidC mutation complemented by nrfG | 34345 | 1536, 1868, 3334, 11a, 4602 |
| nth | 36.9 | Endonuclease III | DNA glycosylase/apyrimidinic (AP) lyase, specific for damaged pyrimidine sites, particularly Thy; endonuclease III | 13070 | 1656, 202, 2434, 3717, 3805, 4701, 888 |
| ntpA | 42.0 | Nucleoside triphosphatase | dATP-preferring nucleoside tri-P pyrophosphohydrolase | 46700 | 1950, 3254 |
| nuoA | 51.8 | NADH:ubiquinone oxidoreductase | NADH dehydrogenase I subunit (EC 1.6.99.–) | 32679 | 1197, 4694, 4918, 616 |
| nuoB | 51.8 | NADH:ubiquinone oxidoreductase | NADH dehydrogenase I subunit (EC 1.6.99.–) | 32676 | 1197, 4694, 4918, 616 |
| nuoC | 51.7 | NADH:ubiquinone oxidoreductase | NADH dehydrogenase I subunit (EC 1.6.99.–) | 32673 | 1197, 4694, 616 |
| nuoE | 51.7 | NADH:ubiquinone oxidoreductase | NADH dehydrogenase I subunit (EC 1.6.99.–) | 32667 | 4694, 616 |
| nuoF | 51.7 | NADH:ubiquinone oxidoreductase | NADH dehydrogenase I subunit (EC 1.6.99.–) | 32664 | 1197, 3546, 4694, 616 |
| nuoG | 51.6 | NADH:ubiquinone oxidoreductase | NADH dehydrogenase I subunit (EC 1.6.99.–) | 32661 | 1197, 3546, 4694, 616 |
| nuoH | 51.6 | NADH:ubiquinone oxidoreductase | NADH dehydrogenase I subunit (EC 1.6.99.–) | 32658 | 1197, 3546, 4694, 616 |
| nuoI | 51.6 | NADH:ubiquinone oxidoreductase | NADH dehydrogenase I subunit (EC 1.6.99.–) | 32655 | 1197, 3546, 4694, 616 |
| nuoJ | 51.6 | NADH:ubiquinone oxidoreductase | NADH dehydrogenase I subunit (EC 1.6.99.–) | 32652 | 3546, 4694, 616 |
| nuoK | 51.6 | NADH:ubiquinone oxidoreductase | NADH dehydrogenase I subunit (EC 1.6.99.–) | 32649 | 3546, 4694, 616 |
| nuoL | 51.5 | NADH:ubiquinone oxidoreductase | NADH dehydrogenase I subunit (EC 1.6.99.–) | 32646 | 3117, 3546, 616 |
| nuoM | 51.5 | NADH:ubiquinone oxidoreductase | NADH dehydrogenase I subunit (EC 1.6.99.–) | 32639 | 1197, 4694, 616 |
| nuoN | 51.5 | NADH:ubiquinone oxidoreductase | NADH dehydrogenase I subunit (EC 1.6.99.–) | 29354 | 1197, 4694, 616 |
| nupC | 54.1 | Nucleoside permease | cru; transport of nucleosides, except guanosine | 443 | 2303, 3046, 3047, 4944, 539, 586, 864 |
| nupG | 66.9 | Nucleoside permease | Transport of nucleosides | 442 | 3046, 3047, 3172, 4715 |
| nusA | 71.4 | N (λ protein) utilization substance | Survives lambda prophage induction; transcription termination/antitermination L factor | 441 | 1312, 1506, 1507, 1513, 1927, 1928, 2375, 2566, 3108, 3459, 3462, 4670, 4945, 939, 3804a |
| nusB | 9.4 | N (λ protein) utilization substance | groNB, ssaD, ssyB; survives prophage induction; transcription termination L factor | 440 | 1313, 1366, 1408, 1926, 3285, 3598, 4004, 4315, 4361, 4670, 851 |
| nusG | 90.0 | N (λ protein) utilization substance | Stabilizes λ-N-NusA-RNAP antitermination complex | 31308 | 2525, 2526, 2566, 4276, 576, 939 |
| nuvA | 9.4 | Near UV (sensitivity) | Uridine thiolation factor A activity | 439 | 1210, 2573, 3606, 609 |
| nuvC | 44.1 | Near UV (sensitivity) | Uridine thiolation factor C activity | 438 | 3781 |
| ogrK | 46.7 | P2 ogr gene | Positive regulator of P2 growth (insertion of P2 ogr gene) | 53483 | 4100 |
| ogt | 30.1 | O-methylguanine transferase | O6-methylguanine-DNA methyltransferase, constitutive | 31984 | 1990, 2698, 2761, 3006, 3503, 3504, 3505, 3629, 7 |
| ompA | 21.9 | Outer membrane protein | con, tolG, tut; outer membrane protein 3a (II*;G;d) structural gene | 437 | 1501, 1694, 267, 280, 3023, 3024, 3025, 493, 707, 795, 949, 986 |
| ompC | 49.8 | Outer membrane protein | meoA, par; outer membrane protein 1b (Ib, c) | 436 | 2954, 2980, 2981, 3880, 4520, 4574, 4811 |
| ompF | 21.2 | Outer membrane protein | cmlB, coa, cry, tolF; outer membrane protein 1a (Ia;b;F) | 435 | 1652, 1841, 1842, 1917, 1918, 258, 2740, 2954, 3075, 3077, 3225, 3550, 3637, 3738, 3880, 4375, 4433, 4492, 4574, 905 |
| ompG | 29.7 | Outer membrane protein | Novel porin; not shown in Fig. 1; see EcoMap10 (3763a) at 29.75 min | 57349 | 2987, 1194a |
| ompR | 76.2 | Outer membrane protein | cry, envZ, ompB; activator protein for osmoregulation of OmpC and OmpF | 434 | 1374, 1610, 1611, 2110, 1167, 2978, 2979, 2983, 3129, 3946, 4374, 4491, 4584, 4681, 4807, 816, 4038 |
| ompT | 12.6 | Outer membrane protein | Outer membrane protein 2b; protease VII; cleaves T7 RNA polymerases, Ada, SecY | 4984 | 1462, 1524, 1525, 1715, 2786, 3769, 4265 |
| ompX | 18.3 | Outer membrane protein | Outer membrane protein, with role in inducing RNAP-ςE production | 35913 | 2863 |
| oppA | 28.0 | Oligopeptide permease | Oligopeptide permease | 18094 | 108, 1782, 2494, 3351, 4188 |
| oppB | 28.0 | Oligopeptide permease | Oligopeptide transport | 18091 | 108, 1782, 2494 |
| oppC | 28.1 | Oligopeptide permease | Oligopeptide transport | 18088 | 108, 1782 |
| oppD | 28.1 | Oligopeptide permease | Oligopeptide transport | 18085 | 108, 1782 |
| oppE | 98.9 | Oligopeptide permease | Oligopeptide transport | 18082 | 108 |
| oppF | 28.1 | Oligopeptide permease | Oligopeptide transport; ATP hydrolysis | 35799 | 1344 |
| opr | 19.0 | rpo reversed | Rate of degradation of aberrant RNAP-subunit proteins | 18079 | 3983 |
| ops | 66.1 | Overproduction of polysaccharide | Level of exopolysaccharide production | 18076 | 4963 |
| oraA | 60.8 | orf-recA | Putative RecX regulatory protein | 33246 | 4914 |
| ordL | 29.4 | Oxidoreductase | Putative oxidoreductase | 51940 | 2066 |
| oriC | 84.6 | Origin of replication | poh; origin of DNA replication | 431 | 148, 1633, 1769, 2075, 2084, 2275, 253, 2635, 2711, 2805, 2905, 2985, 3273, 3275, 3276, 3981, 4131, 4255, 4322, 452, 4610, 4611, 4635, 556, 628 |
| oriJ | 30.5 | Origin of replication | Locus in defective prophage rac | 430 | 1037, 1038, 2086 |
| orn | 94.6 | Oligoribonuclease | 3′ to 5′ oligoribonuclease | 54868 | 4942 |
| osmB | 28.9 | Osmotically inducible | OsmB lipoprotein | 31965 | 1054, 2072, 2073 |
| osmC | 33.5 | Osmotically inducible | Nonessential gene | 32184 | 1460, 1584 |
| osmE | 39.2 | Osmotically inducible | anr; promoter overlaps nadE promoter; regulated by growth phase as well as osmotic pressure | 36628 | 1585, 828 |
| osmY | 99.3 | Osmotically inducible | csi-5; periplasmic, ςS dependent protein (stationary phase) | 34640 | 1054, 2412, 4883, 4884 |
| otsA | 42.6 | Osmoregulated trehalose synthesis | Trehalose phosphate synthase (EC 2.4.1.15) | 18073 | 1419, 2077 |
| otsB | 42.7 | Osmoregulated trehalose synthesis | Trehalose phosphate phosphatase (EC 3.1.3.12) | 18070 | 1419, 2077 |
| oxyR | 89.6 | Oxygen | mor, momR; bifunctional regulatory protein sensor for oxidative stress | 28841 | 1455, 1712, 2358, 2359, 4242, 4951, 4356 |
| oxyS | 89.6 | Oxygen | Regulatory RNA; activator for genes that detoxify oxidative damage; small RNA | 28844 | 77 |
| pabA | 75.2 | para-Aminobenzoate | Sulfonamide resistance; p-aminobenzoate biosynthesis | 429 | 2130, 4593, 535, 4450 |
| pabB | 40.8 | para-Aminobenzoate | Sulfonamide resistance; p-aminobenzoate biosynthesis | 428 | 1454, 3624, 4593, 535 |
| pabC | 24.8 | para-Aminobenzoate | Sulfonamide resistance; aminodeoxychorismate lyase | 31889 | 1499, 1500 |
| pac | 31.0 | Phenylacetate | Phenylacetate degradation | 18067 | 831 |
| pal | 16.8 | Peptidoglycan-associated lipoprotein | excC; essential lipoprotein associated with peptidoglycan | 35129 | 1270, 2448, 2450, 4581, 705 |
| panB | 3.2 | Pantothenate | Ketopantoate hydroxymethyltransferase (EC 4.1.2.12) | 427 | 2037, 2903, 871 |
| panC | 3.2 | Pantothenate | Pantothenate synthetase (EC 6.3.2.1) | 426 | 2903, 871 |
| panD | 3.2 | Pantothenate | Aspartate 1-decarboxylase (EC 4.1.1.11) | 425 | 2903, 871 |
| panF | 73.4 | Pantothenate | Pantothenate permease (symporter) | 10818 | 1971, 4537, 4565 |
| parC | 68.2 | Partition | Cell partitioning; topoisomerase IV subunit A | 33440 | 1699, 2163, 2164, 2165, 2211, 2360, 3397, 3398, 4923, 695, 4213 |
| parE | 68.4 | Partition | nfxD; topoisomerase IV subunit B | 33451 | 2163, 2165, 2211, 2511, 3397, 3398, 491, 4923 |
| pat | 89.1 | Putrescine aminotransferase | Putrescine aminotransferase | 18064 | 3984 |
| pbpG | 47.9 | Penicillin-binding protein | Penicillin-binding protein 7 (PBP7) | 36349 | 1714 |
| pck | 76.1 | PEP carboxykinase | Phosphoenolpyruvate carboxykinase (ATP); (EC 4.1.1.49) | 422 | 1444, 1445, 1446, 2864 |
| pcm | 61.8 | Protein carboxyl methyltransferase | l-Isoaspartyl protein carboxyl methyltransferase (EC 2.1.1.77); repair of isoaspartyl residues | 33221 | 1325, 2524, 4591 |
| pcnB | 3.4 | Plasmid copy number | Replication and cell division; poly(A) polymerase I; controls plasmid copy number | 13584 | 1358,4,2583, 2631, 2572, 2790, 2791, 3625, 631, 632, 1682 |
| pdhR | 2.6 | Pyruvate-dehydrogenase | Pyruvate-dehydrogenase repressor | 30488 | 1674, 3567, 655 |
| pdxA | 1.1 | Pyridoxine | Isoniazid resistance; pyridoxine biosynthesis | 420 | 1020, 105, 106 |
| pdxB | 52.5 | Pyridoxine | Isoniazid resistance; erythronate-4-phosphate dehydrogenase? pyridoxine biosynthesis | 419 | 1020, 143, 144, 3930 |
| pdxH | 37.0 | Pyridoxine | Isoniazid resistance; pyridoxine-phosphate oxidase | 417 | 1020, 1526, 1529, 2398, 4034 |
| pdxJ | 58.2 | Pyridoxine | Codon overlap with recO; complex operon | 416 | 2397, 4340 |
| pdxK | 54.6 | Pyridoxine | Vitamin B6 kinase | 51757 | 4866 |
| pdxL | 46.6 | Pyridoxine | Pyridoxine kinase | 50836 | 2977 |
| pdxY | 37 | Pyridoxine | Pyridoxal kinase | 53757 | 4867 |
| pepA | 96.6 | Peptidase | xerB, carP; amino-exopeptidase A | 30215 | 2842, 3732, 4226, 4227, 684 |
| pepD | 5.5 | Peptidase | Peptidase D, a dipeptidase | 415 | 1698, 1722, 2254, 2938, 3384, 724 |
| pepE | 91.1 | Peptidase | α-Aspartyl dipeptidase (EC 3.4.11.–) | 34261 | 822 |
| pepN | 21.3 | Peptidase | Aminopeptidase N | 414 | 1233, 1269, 213, 214, 215, 2431, 2833, 2834, 2835, 3384 |
| pepP | 65.8 | Peptidase | Proline aminopeptidase II | 28682 | 4897a, 3100 |
| pepQ | 86.8 | Peptidase | Proline dipeptidase (EC 3.4.13.9) | 34071 | 3098, 926 |
| pepT | 25.5 | Peptidase | Putative peptidase T | 31914 | 2617 |
| pfkA | 88.5 | Phosphofructokinase | 6-Phosphofructokinase I (EC 2.7.1.11) | 413 | 1710, 3706, 907 |
| pfkB | 38.9 | Phosphofructokinase | Suppresses pfkA mutations; phosphofructokinase, Pfk-2 | 412 | 817, 908, 909 |
| pflA | 20.5 | Pyruvate formate-lyase | act; pyruvate formate lyase I activase | 35839 | 1085, 3699 |
| pflB | 20.5 | Pyruvate formate-lyase | Pyruvate formate lyase I; induced anaerobically | 410 | 3389, 3699, 3884, 3885, 3886 |
| pflC | 89.3 | Pyruvate formate-lyase | Pyruvate formate lyase II activase | 35847 | 395, 396 |
| pflD | 89.3 | Pyruvate formate-lyase | Pyruvate formate lyase II | 35843 | 395, 396 |
| pfs | 3.8 | PNP similarity | 5′-Methylthioadenosine/S-adenosylhomocysteine nucleosidase | 54950 | 834 |
| pgi | 91.2 | Phosphoglucose isomerase | Glucosephosphate isomerase (EC 5.3.1.9) | 409 | 1311, 1319, 3706, 4678 |
| pgk | 66.2 | Phosphoglycerate kinase | Phosphoglycerate kinase (EC 2.7.2.3) | 408 | 3148 |
| pgl | 17.2 | Phosphogluconolactonase | blu; 6-phosphogluconolactonase (EC 3.1.1.31) | 407 | 3706 |
| pgm | 15.4 | Phophoglucomutase | blu; phosphoglucomutase (EC 5.4.4.2) | 406 | 2647, 3706 |
| pgpA | 9.5 | Phosphatidylglycerophosphate phosphatase | Phosphatidylglycerophosphate phosphatase, membrane bound, nonessential | 17644 | 1338, 1882, 1885 |
| pgpB | 28.8 | Phosphatidylglycerophosphate phosphatase | Phosphatidylglycerophosphate phosphatase, membrane bound, nonessential | 17641 | 1051, 1338, 1883, 1885 |
| pgsA | 42.9 | Phosphotidylglycerophosphate synthase | Phosphatidylglycerophosphate synthetase (EC 2.7.8.5) | 405 | 1459, 3266, 4493, 4518 |
| pheA | 59.0 | Phenylalanine | Chorismate mutase-P-prephenate dehydratase (EC 5.4.99.5, EC 4.2.1.51) FPA resistance | 403 | 1386, 1388, 1389, 1479, 1480, 1847 |
| pheP | 13.0 | Phenylalanine | Phenylalanine-specific permease | 402 | 3422, 4720 |
| pheS | 38.7 | Phenylalanine | phe-act; phenylalanyl-tRNA synthetase α-subunit (EC 6.1.1.20) | 400 | 1148, 1201, 1213, 2132, 2860, 2862, 3460, 3461, 3463, 4168, 4169, 4171, 4801 |
| pheT | 38.7 | Phenylalanine | Phenylalanyl-tRNA synthetase β-subunit (EC 6.1.1.20) | 399 | 1148, 1213, 2860, 2862, 3460, 3461, 3463, 4168, 4169, 4801 |
| pheU | 94.0 | Phenylalanine | pheR, pheW; phenylalanine tRNA | 18061 | 1345, 1387, 1388, 2313, 3938, 4755, 603, 3442 |
| pheV | 67.0 | Phenylalanine | Phenylalanine tRNA | 18058 | 2313, 4755, 602, 604, 3442 |
| phnC | 93.2 | Phosphonate | Phosphonate transporter subunit I (cryptic in K-12); member Pho regulon | 34553 | 2737, 2906, 2908, 4669, 694 |
| phnD | 93.1 | Phosphonate | psiD; phosphonate transporter subunit, periplasmic (cryptic in K-12) | 17638 | 2737, 2906, 2907, 2908, 2909, 4621, 4665, 4669, 694 |
| phnE | 93.1 | Phosphonate | Phosphonate transporter subunit, integral membrane component (cryptic in K-12) | 34550 | 2737, 2906, 2908, 4669, 694 |
| phnF | 93.1 | Phosphonate | Phosphonate utilization (cryptic in K-12); putative regulatory gene, member of pho regulon | 34547 | 2737, 2906, 2908, 4669, 694 |
| phnG | 93.1 | Phosphonate | Carbon-phosphorus lyase complex subunit (phosphonate utilization cryptic in K-12) | 34544 | 2737, 2906, 2908, 4669, 694 |
| phnH | 93.1 | Phosphonate | Carbon-phosphorus lyase complex subunit (phosphonate utilization cryptic in K-12) | 34541 | 2737, 2906, 2908, 4669, 694 |
| phnI | 93.1 | Phosphonate | Carbon-phosphorus lyase complex subunit (phosphonate utilization cryptic in K-12) | 34538 | 2737, 2906, 2908, 4669, 694 |
| phnJ | 93.0 | Phosphonate | Carbon-phosphorus lyase complex subunit (phosphonate utilization cryptic in K-12) | 34535 | 2737, 2906, 2908, 4669, 694 |
| phnK | 93.0 | Phosphonate | Carbon-phosphorus lyase complex subunit (phosphonate utilization cryptic in K-12) | 34532 | 2737, 2906, 2908, 4669, 694 |
| phnL | 93.0 | Phosphonate | Carbon-phosphorus lyase complex subunit (phosphonate utilization cryptic in K-12) | 34529 | 2737, 2906, 2908, 4669, 694 |
| phnM | 93.0 | Phosphonate | Carbon-phosphorus lyase complex subunit (phosphonate utilization cryptic in K-12) | 34526 | 2737, 2906, 2908, 4669, 694 |
| phnN | 93.0 | Phosphonate | Carbon-phosphorus lyase complex subunit (phosphonate utilization cryptic in K-12) | 34523 | 2737, 2906, 2908, 4669, 694 |
| phnO | 93.0 | Phosphonate | Probably regulatory for C-P lyase complex (phosphonate utilization cryptic in K-12) | 34520 | 2737, 2906, 2908, 4669, 694 |
| phnP | 92.9 | Phosphonate | Carbon-phosphorus lyase complex membrane-bound subunit (utilization cryptic in K-12) | 34514 | 2737, 2906, 2908, 4669, 694 |
| phoA | 8.6 | Phosphate | Alkaline phosphatase (EC 3.1.3.1) | 398 | 1303, 1597, 1740, 1919, 1920, 2105, 2218, 319, 3866, 3867, 4050, 415, 4443, 4667, 4668, 475, 4816, 674 |
| phoB | 9.0 | Phosphate | phoRc, phoT; positive response regulator for pho regulon, two-component system | 397 | 1597, 2230, 2732, 2734, 2735, 4040, 415, 4426, 4430, 4661, 4663, 4667, 4668 |
| phoE | 5.6 | Phosphate | ompE; outer membrane porin protein E | 396 | 1676, 1722, 3221, 3319, 4426, 4429, 4432, 4433, 4553 |
| phoH | 23.4 | Phosphate | psiH; member of pho regulon, P starvation induced | 31841 | 2231, 2320, 2909, 4661 |
| phoP | 25.6 | Phosphate | In Salmonella, a sensor in the two-component regulatory system, with phoQ | 31919 | 1530, 2146 |
| phoQ | 25.6 | Phosphate | In Salmonella two-component regulatory system with phoP | 31922 | 2146 |
| phoR | 9.0 | Phosphate | nmpB, phoR1, R1pho; positive and negative regulatory gene for pho regulon; sensor of two-component system for pho regulon | 394 | 1112, 2734, 2736, 4040, 4427, 4430, 4661, 4663, 4667, 4753 |
| phoU | 84.2 | Phosphate | phoT; P uptake, high-affinity P-specific transport system, regulatory gene | 18055 | 3121, 4196, 4295, 79, 80 |
| phrB | 15.9 | Photoreactivation | Deoxyribodipyrimidine photolyase (EC 4.1.99.3) | 391 | 1073, 1826, 1866, 2632, 2683, 3327, 3841, 3842 |
| phxB | 17.0 | Phage φX | Adsorption of φX154 | 389 | 3049 |
| pin | 26.1 | Prophage-derived inversion | Locus in defective prophage e14; calcium-binding protein required for initiation of replication | 18049 | 1159, 2168, 3447, 3448, 4544 |
| pioO | 74.4 | Protein, initiation? | pinO; calcium-binding protein, may have a role in initiation of replication | 53557 | 1586, 1587 |
| pit | 78.4 | Pi transport | Low-affinity Pi transport | 385 | 1149, 1150, 300, 4156 |
| pldA | 86.3 | Phospholipase, detergent resistant | Detergent-resistant phospholipase A activity | 384 | 1648, 1791, 1792, 2277, 4606, 513, 58, 926, 973, 974 |
| pldB | 86.4 | Phospholipase, detergent resistant | Lysophospholipase L2 | 5001 | 1792, 2277, 2278, 58, 926 |
| plsB | 91.6 | Phospholipid synthesis | Glycerolphosphate acyltransferase activity | 382 | 2544, 2545 |
| plsC | 68.1 | Phospholipid synthesis | 1-Acyl-sn-glycerol-3-phosphate acyltransferase (EC 2.3.1.51); affects partitioning | 33443 | 802, 803 |
| plsX | 24.7 | Phospholipid synthesis | Glycerol P auxotrophy in plsB background | 18046 | 2425 |
| pmbA | 96.0 | Peptide MccB17 | mcb, tldE; antibiotic peptide MccB17 | 34577 | 2619, 3061, 3703 |
| pncA | 39.9 | Pyridine nucleotide cycle | nam; nicotinamide deamidase (EC 3.5.1.19) | 381 | 1321, 2487, 3347 |
| pncB | 21.3 | Pyridine nucleotide cycle | Nicotinate phosphoribosyltransferase (EC 2.4.2.11) | 380 | 2487 |
| pnp | 71.3 | Polynucleotide phosphorylase | Polynucleotide phosphorylase (EC 2.7.7.8) | 379 | 1182, 1787, 2854, 290, 3492, 3493, 3494, 3638, 3639, 4333, 868, 869, 4940 |
| pntA | 36.1 | Pyridine nucleotide transhydrogenase | Pyridine nucleotide transhydrogenase α subunit (EC 1.6.1.1) | 18043 | 771, 772 |
| pntB | 36.1 | Pyridine nucleotide transhydrogenase | Pyridine nucleotide transhydrogenase β subunit (EC 1.6.1.1) | 18040 | 1436, 771, 772 |
| pnuC | 16.9 | Putative NMN uptake | Nicotinamide mononucleotide transporter, putative, by homology with Salmonella | 53409 | 1280, 4954 |
| poaR | 65.9 | Proline oxidase | Regulation of proline oxidase production | 377 | 819a |
| polA | 87.2 | Polymerase | resA; DNA polymerase I (EC 2.7.7.7) | 375 | 2068, 2189, 2190, 3072, 4869, 4941, 4966 |
| polB | 1.4 | Polymerase | dinA; DNA polymerase II (EC 2.7.7.7) | 374 | 1174, 1967, 2519, 3385, 4039, 426, 698, 699 |
| popD | 0.2 | Porphyrin | 5-Aminolevulinate dehydratase (EC 4.2.1.24) | 371 | 2839, 3518 |
| potA | 25.5 | Putrescine-ornithine transporter | ATP-binding membrane protein; putrescine/spermidine-ornithine transporter | 31899 | 1339, 2148, 2150, 2617 |
| potB | 25.5 | Putrescine-ornithine transporter | Membrane protein, channel-forming, for spermidine uptake | 31902 | 1339, 2150, 2617 |
| potC | 25.5 | Putrescine-ornithine transporter | Membrane protein, channel-forming, for spermidine uptake | 31905 | 1339, 2150, 2617 |
| potD | 25.5 | Putrescine-ornithine transporter | Spermidine-binding membrane protein | 31908 | 1339, 2150, 2617 |
| potE | 15.4 | Putrescine-ornithine transporter | Putrescine-lyase antiporter | 31552 | 2149, 2151, 2152 |
| potF | 19.2 | Putrescine-ornithine transporter | Apparent periplasmic putrescine-specific binding protein | 31694 | 3440 |
| potG | 19.3 | Putrescine-ornithine transporter | Apparent nucleotide-binding subunit of putrescine-ornithine transporter | 31697 | 3440 |
| potH | 19.3 | Putrescine-ornithine transporter | Transmembrane-spanning subunit | 31700 | 3440 |
| potI | 19.3 | Putrescine-ornithine transporter | Apparent transmembrane-spanning subunit | 31703 | 3440 |
| poxA | 94.5 | Pyruvate oxidase | Regulator of poxB | 370 | 679 |
| poxB | 19.6 | Pyruvate oxidase | Pyruvate oxidase (EC 1.2.2.2) | 369 | 1482, 1483, 678, 680, 681 |
| ppa | 95.8 | Pyrophosphatase | Inorganic pyrophosphatase (EC 3.6.1.1) | 34394 | 2389, 2390, 700 |
| ppc | 89.4 | PEP carboxylase | asp, glu; phosphoenolpyruvate carboxylase (EC 4.1.1.31) | 368 | 1332, 3000, 3786 |
| pphA | 41.4 | Phosphoprotein phosphatase | Phosphoprotein phosphatase involved in signalling protein misfolding; heat shock regulon | 54632 | 2966 |
| pphB | 61.5 | Phosphoprotein phosphatase | As above; heat shock regulon | 54636 | 2966 |
| ppiA | 75.2 | Peptidylprolyl isomerase | rot, a rotomase; peptidylprolyl-cis-trans-isomerase A | 31121 | 1670, 2252, 2579, 3214 |
| ppiB | 11.9 | Peptidylprolyl isomerase | A rotomase; peptidylprolyl-cis-trans-isomerase B | 31117 | 1670 |
| ppiC | 85.3 | Peptidylprolyl isomerase | parvA; peptidylprolyl-cis-trans isomerase C | 35829 | 3764 |
| ppk | 56.5 | Polyphosphate kinase | Polyphosphate (linear Pi linked by high-energy bonds) kinase | 32894 | 2323, 37, 873 |
| pps | 38.4 | PEP synthase | ppsA; phosphoenolpyruvate synthase | 367 | 3181 |
| ppx | 56.5 | Exopolyphosphatase | Exopolyphosphatase | 32899 | 2323, 38 |
| pqiA | 21.8 | Paraquat inducible | Induced by paraquat, regulated by SoxRS | 39521 | 2289, 2290 |
| pqiB | 21.8 | Paraquat inducible | Induced by paraquat, regulated by SoxRS | 39524 | 2289 |
| pqqL | 33.9 | Pyrroloquinoline quinone | Redox cofactor for pyrroloquinoline quinone synthesis (cryptic in K-12) | 35854 | 364, 4497 |
| pqqM | 33.9 | Pyrroloquinoline quinone | Pyrroloquinoline quinone synthesis (cryptic in K-12) | 35857 | 4497 |
| prc | 41.2 | PBP protease, C terminal | tsp; carboxy-terminal protease for penicillin-binding protein, PBP 3 | 32334 | 1643, 1644, 179, 2186, 3082, 3970, 4060, 4061 |
| prfA | 27.3 | Protein release factor | asuA?, sueB, uar, ups? peptide chain release factor 1 | 14922 | 1880, 2460, 2808, 3782, 3283, 4273, 650 |
| prfB | 65.4 | Protein release factor | supK; peptide chain release factor 2 | 17635 | 1880, 2174, 2178, 2460 |
| prfC | 99.3 | Protein release factor | tos; release factor 3 | 34633 | 1515, 1739, 2808, 2930, 4873 |
| priA | 88.9 | Primosome | srgA; primosome factor Y, also called protein n′ | 27611 | 2462, 3227, 3228, 3314, 3846, 59 |
| priB | 95.3 | Primosome | Primosomal protein n | 29153 | 4920, 68 |
| priC | 10.6 | Primosome | Primosomal protein n" | 29161 | 4920 |
| prlC | 78.5 | Protein localization | opdA in Salmonella; oligopeptidase A | 18031 | 1153, 4471, 4472, 820, 821 |
| prlZ | 71.4 | Protein localization | Suppresses export defects in signal sequence mutations | 36421 | 4688 |
| prmA | 73.4 | Posttranslation ribosomal protein modification | Methyltransferase for 50s subunit L11 | 366 | 4565 |
| prmB | 53.0 | Posttranslation ribosomal protein modification | Methylation of 50s subunit L3 | 365 | 815 |
| proA | 5.6 | Proline | pro1; γ-glutamyl phosphate reductase (EC 1.2.1.41) | 364 | 1026, 1214, 1597, 1676, 1677, 1768, 2721, 315, 3153, 4562, 1722 |
| proB | 5.6 | Proline | pro2; γ-glutamyl kinase (EC 2.7.2.11) | 363 | 1026, 1214, 1597, 1676, 1677, 1768, 2721, 315, 3153, 3612, 4562, 878 |
| proC | 8.7 | Proline | pro2, pro3; pyrroline-5-carboxylate reductase (EC 1.5.1.2) | 362 | 1025, 1597, 2839, 415, 999 |
| proK | 79.8 | Proline | proV; proline tRNA1 | 17632 | 1285, 2313, 2351, 4128 |
| proL | 49.2 | Proline | proW; proline tRNA2 | 17629 | 2313 |
| proM | 85.8 | Proline | proU; proline tRNA3 | 17626 | 2313, 926 |
| proP | 93.3 | Proline | Low-affinity transport; proline permease, minor | 361 | 1475, 1476, 2827, 2877, 2899, 3613, 4186, 4814, 884 |
| proQ | 41.4 | Proline | Sensitivity to toxic proline analogs and positive regulator of proline porter II | 36840 | 2947 |
| proS | 4.7 | Proline | drp; prolyl-tRNA synthetase (EC 1.1.1.15) | 360 | 2453, 414 |
| proT | 83.8 | Proline | Proline transport carrier protein, putative | 359 | 3020 |
| proU | 60.4 | Proline | osrA; operon proVWX for high-affinity transport for glycine; see also proM | 18025 | 1477, 1478, 1535, 2826, 3537, 4297, 951, 3599 |
| proV | 60.4 | Proline | proU; high-affinity transport for glycine; glycine betaine-binding protein; see also proK | 18022 | 1191, 1285, 1477, 1481, 1535, 2826, 4228, 4297, 951 |
| proW | 60.4 | Proline | proU; high-affinity transport for glycine, betaine, and proline; see also proL | 18019 | 1535, 1590, 4228 |
| proX | 60.5 | Proline | proU; high-affinity transport for glycine, betaine, and proline | 35653 | 1535, 4228 |
| prpA | 97.0 | Propionate | Growth on propionate | 358 | 2180, 4158 |
| prpB-E | 7.5 | Propionate | Propionate catabolism operon, Salmonella homology and some expression information | 55452 | 4389a |
| prpR | 7.5 | Propionate | Regulator, propionate catabolism operon, Salmonella homology | 55455 | 4389a |
| prr | 31.2 | Pyrroline | γ-Aminobutyraldehyde (pyrroline) dehydrogenase activity | 18016 | 3984 |
| prs | 27.1 | PRPP synthetase | dnaR; phosphoribosylpyrophosphate synthetase (EC 2.7.6.1) | 357 | 103, 1819, 1820, 1821, 1823, 3808, 3495 |
| psd | 94.6 | Phosphatidylserine decarboxylase | Phosphatidylserine decarboxylase | 356 | 2531, 4020 |
| psiF | 8.7 | P starvation induced | pho regulon member, requiring PhoRB system | 18013 | 2909, 4668, 674 |
| pspA | 29.4 | Phage shock protein | Negative regulatory gene for stress ς54 dependent phage-shock-protein operon | 32009 | 2250, 2253, 2276, 2987, 320, 4697, 4698, 506, 507 |
| pspB | 29.5 | Phage shock protein | Regulatory gene, with PspC activates expression of psp operon | 32012 | 2987, 4697, 4698, 506 |
| pspC | 29.5 | Phage shock protein | Positive regulatory gene, cooperatively with PspB | 32015 | 2987, 4697, 4698, 506 |
| pspE | 29.5 | Phage shock protein | Expressed in response to stress as part of psp operon, but also transcribed independently | 32018 | 4697, 506 |
| pspF | 29.4 | Phage shock protein | Transcriptional ς54-dependent activator of psp | 50349 | 1105, 2063, 2064, 2065, 2066 |
| pssA | 58.6 | Phosphatidylserine synthase | Phosphatidylserine synthase (EC 2.7.8.8) | 355 | 2806, 3269, 4020, 766, 995 |
| pssR | 85.0 | Phosphatidylserine synthase | Regulatory gene | 18010 | 4145, 832 |
| pstA | 84.2 | P-specific transport | R2pho, phoR2b, phoT; high-affinity P-specific transport | 18007 | 3734, 4196, 4296, 79, 80 |
| pstB | 84.2 | P-specific transport | phoT; high-affinity P-specific transport; cytoplasmic ATP-binding protein | 18004 | 2514, 3734, 4196, 4296, 672, 79, 80 |
| pstC | 84.2 | P-specific transport | phoW; high-affinity P-specific transport; cytoplasmic membrane component | 18001 | 3734, 4196, 4296, 79, 80 |
| pstS | 84.2 | P-specific transport | phoR2, nmpA, phoR2a, phoS, R2pho; high-affinity P-specific transport; periplasmic P binding | 17998 | 1112, 1963, 2513, 2515, 2714, 3004, 3734, 4152, 4196, 4295, 4296, 4611, 4667, 4753, 4818, 4967, 536, 79, 80, 857 |
| psu | 1.5 | Pleiotropic suppressor | Temporary designation for pleiotropic suppressor gene; oxolinic acid resistance | 37018 | 1160 |
| pta | 52.0 | Phosphotransacetylase | Phosphotransacetylase (EC 2.3.1.8) | 353 | 1548, 1573, 2094, 2512, 2813, 530 |
| pth | 27.1 | Peptidyl tRNA hydrolase | rap; peptidyl-tRNA hydrolase; required for phage λ growth | 352 | 1342, 1364, 1544, 1739, 1820, 3064, 976, 4269a |
| ptrA | 63.7 | Protease III | Protease III | 351 | 1106, 1107, 1253, 220, 718, 775 |
| ptrB | 41.5 | Protease III | Protease II | 32339 | 2112 |
| ptsG | 24.9 | Phosphotransferase system | CR, car, cat, gpt, umg; glucosephophotransferase enzyme II | 349 | 1170, 2233, 2925, 3765, 4095, 459, 461, 557, 890 |
| ptsH | 54.6 | Phosphotransferase system | Hpr, ctr, hpr; phosphohistidinoprotein-hexose phosphotransferase (EC 2.7.1.69) | 348 | 9836, 2031, 2679, 3369, 3793, 4391, 508, 509, 510, 982, 983 |
| ptsI | 54.6 | Phosphotransferase system | ctr; phosphotransferase system enzyme I | 347 | 3369, 983, 983a, 983b, 1163, 1168, 2491, 3793, 4571, 508, 509, 510, 85, 982 |
| ptsN | 72.1 | Phosphotransferase system | pts in the rpoN operon | 38380 | 3517 |
| ptsP | 63.9 | Phosphotransferase system | PEP-protein phosphotransferase (reuse of manY synonym) | 53527 | 1171, 3648 |
| purA | 94.9 | Purine | Adenylosuccinate synthetase (EC 6.3.4.4) | 345 | 1681, 2788, 304, 4770 |
| purB | 25.6 | Purine | Adenylosuccinate lyase (EC 4.3.2.2) | 344 | 12, 1503, 2788, 1668, 1682 |
| purC | 55.9 | Purine | adeg; phosphoribosylaminoimidazole-succinocarboxamide synthetase (EC 6.3.2.6) | 343 | 2788, 3364, 4415 |
| purD | 90.6 | Purine | adtha; phosphoribosylglycinamide synthetase (EC 6.3.4.13) | 342 | 1260, 2789, 28, 719 |
| purE | 11.9 | Purine | Pur2, ade3, adef; phosphoribosylaminoimidazole carboxylase, catalytic subunit (EC 4.1.1.21) | 341 | 1597, 2103, 4412 |
| purF | 52.3 | Purine | adeub, purC; amidophosphoribosyl transferase (EC 2.4.2.14) | 340 | 2727, 3211, 3719, 3826, 4117, 4483 |
| purH | 90.6 | Purine | Phosphoribosylaminoimidazolecarboxamide formyltransferase (EC 2.1.2.3) | 338 | 1260, 2789, 28, 3410 |
| purK | 11.9 | Purine | purE2; phosphoribosyl glucinamide formyltransferase | 17995 | 2103, 4412 |
| purL | 58.0 | Purine | purI; phosphoribosylformylglycinamide synthetase (EC 6.3.5.3) | 336 | 1817, 3827, 3901 |
| purM | 56.5 | Purine | purG; phosphoribosylaminoimidazole synthetase (EC 6.3.3.1) | 335 | 1817, 4115, 4116 |
| purN | 56.5 | Purine | adeC; 5′-phosphoribosylglycinamide transformylase 1; see purT | 17623 | 4116 |
| purP | 83.9 | Purine | High-affinity purine transport | 17992 | 578 |
| purR | 37.4 | Purine | Purine repressor | 17989 | 1681, 2222, 3718, 741 |
| purT | 41.6 | Purine | Glycinamide ribonucleotide transformylase 2, non-folate-requiring; see purN | 32348 | 2772, 3230 |
| purU | 27.7 | Purine | tgs; formyltetrahydrofolate hydrolase | 35231 | 3088, 3089, 444, 446, 447 |
| pus | 20.6 | Reverse of sup (amber) | Reverses accentuation effects of amber suppressor on relB mutations | 334 | 1044 |
| putA | 23.2 | Proline utilization | poaA; proline dehydrogenase (EC 1.5.99.8) | 333 | 1028, 2743, 2989, 3115, 3612, 4186, 4461, 4771, 4777 |
| putP | 23.2 | Proline utilization | Proline/Na+, Li+ symport protein | 332 | 2989, 3115, 3116, 3264, 3612, 4186, 4461, 4775, 4776, 4777, 4849 |
| pykA | 41.7 | Pyruvate kinase | Pyruvate kinase A (II); (EC 2.7.1.40) | 32363 | 1378, 3477, 4534 |
| pykF | 37.8 | Pyruvate kinase | Pyruvate kinase I (EC 2.7.1.40); fructose-stimulated | 17620 | 1378, 3255, 3477, 397 |
| pyrB | 96.3 | Pyrimidine | Aspartate transcarbamylase, catalytic subunit (EC 2.1.3.2) | 330 | 1797, 2275, 2510, 3140, 3383, 3715, 3729, 4499 |
| pyrC | 24.2 | Pyrimidine | Dihydro-orotase (EC 3.5.2.3) | 329 | 193, 2010, 4754 |
| pyrD | 21.6 | Pyrimidine | Dihydro-orotate oxidase (EC 1.3.3.1) | 328 | 2010, 2423 |
| pyrE | 82.2 | Pyrimidine | Orotate phosphoribosyltransferase (EC 2.4.2.10) | 327 | 2009, 3511–3514, 4367, 94 |
| pyrF | 28.9 | Pyrimidine | Orotidine-5′-phosphate decarboxylase (EC 4.1.1.23) | 326 | 1070, 2010, 407, 4498 |
| pyrG | 62.6 | Pyrimidine | CTP synthetase (EC 6.3.4.2) | 325 | 158, 3426, 4709 |
| pyrH | 4.1 | Pyrimidine | smbA; UMP kinase | 324 | 4104, 4843 |
| pyrI | 96.3 | Pyrimidine | Aspartate transcarbamylase, regulatory subunit (EC 2.1.3.2) | 323 | 1219, 1797, 2510, 3383, 3426, 3715, 3729, 4738 |
| qin | 35.1 | Q independent | kim; cryptic lambdoid phage | 17529 | 1177, 455 |
| qmeC | 75.3 | Q membrane | Glycine resistance; penicillin sensitivity; membrane defect | 321 | 4735 |
| qmeD | 64.4 | Q membrane | Glycine resistance; penicillin sensitivity; membrane defect | 320 | 4735 |
| qmeE | 37.9 | Q membrane | Glycine resistance; penicillin sensitivity; membrane defect | 319 | 4735 |
| qor | 91.8 | Quinone oxidoreductase | Quinone oxidoreductase, NADPH dependent | 36654 | 2549 |
| qsr′ | 12.2 | QSR replacement | DLP12; defective Q-independent lambdoid prophage, includes intD and nmpD | 19200 | 115, 1751, 2085, 3059, 390 |
| queA | 9.1 | Queuosine/queuine | S-Adenosylmethionine:tRNA ribosyltransferase- isomerase | 29889 | 2651, 3654, 4094 |
| rac | 30.4 | Recombination activation | Defective prophage Rac loci; see recE, oriJ, trkG, sieB, lar, racC, racR | 318 | 1036, 1037, 1181, 2086, 2087, 2643, 355, 4750, 4751, 503, 759a |
| racC | 30.5 | Recombination activation | Element of defective prophage Rac | 32076 | 748 |
| racR | 30.6 | Recombination activation | Element of defective prophage Rac; repressor | 37433 | 748, 759a |
| radA | 99.7 | Radiation | sms; sensitivity to gamma and UV radiation | 15905 | 1059, 3160, 4136 |
| radC | 82.1 | Radiation | Sensitivity to radiation | 13913 | 1220 |
| ranA | 58.3 | Affects RNA metabolism | 317 | 121 | |
| rarD | 86.2 | Recombination and repair | Chloramphenicol resistance | 34089 | 3259, 926 |
| ras | 9.9 | Radiation sensitive | Sensitive to X rays and UV | 316 | 4636, 4637 |
| rbfA | 71.4 | Ribosome binding factor | Overexpression suppresses cold-sensitive 16S rRNA; ribosome binding factor | 36919 | 2044, 590, 917 |
| rbn | 87.8 | RNase BN | o290; RNase BN, tRNA processing enzyme | 50482 | 3850, 618 |
| rbsA | 84.7 | Ribose | rbsP, rbsT; d-ribose high-affinity transport system (may have chemotaxis function) | 12082 | 1888, 242, 2630, 291 |
| rbsB | 84.8 | Ribose | prlB, rbsP; d-ribose periplasmic binding protein | 12092 | 1153, 1888, 2630 |
| rbsC | 84.8 | Ribose | rbsP, rbsT; d-ribose high-affinity transport system | 12089 | 1888, 2630, 291, 4915 |
| rbsD | 84.7 | Ribose | rbsP; d-ribose high-affinity transport system | 314 | 291 |
| rbsK | 84.8 | Ribose | Ribokinase (EC 2.7.1.15) | 315 | 1798, 1888, 2630, 4611 |
| rbsR | 84.8 | Ribose | Regulatory gene | 12086 | 2630 |
| rcsA | 43.6 | Regulation capsule synthesis | Positive regulatory gene for capsule (colanic acid) synthesis; two regulatory proteins from the same gene | 17980 | 1045, 1471, 4243, 4244, 4441 |
| rcsB | 49.9 | Regulation capsule synthesis | Positive regulatory gene for capsule (colanic acid) synthesis; when overexpressed, restores ftsZ84 growth on low-salt medium | 17977 | 1413, 1471, 2188, 4099, 4243–4245, 504 |
| rcsC | 49.9 | Regulation capsule synthesis | Negative regulatory gene for capsule (colanic acid) synthesis, controls sliminess; contains TerE; probable histidine kinase | 17974 | 1471, 2188, 3037, 4099, 4245, 504 |
| rcsF | 4.7 | Regulation capsule synthesis | Overexpression confers mucoid phenotype, increases capsule synthesis; restores colony formation of ftsZ84 mutants on low salt | 29845 | 1412 |
| rdgA | 16.1 | RecA-dependent growth | Dependence of growth upon recA gene product | 17971 | 1317 |
| rdgB | 67.0 | RecA-dependent growth | Dependence of growth and viability upon recA | 17968 | 784 |
| recA | 60.8 | Recombination | srf, lexB, umuB, zab; general recombination and DNA repair; pairing and strand exchange; role in cleavage of LexA repressor, SOS mutagenesis | 312 | 1104, 1123, 1239, 1654, 1803, 2177, 2574, 2575, 3833, 3991, 4395, 4604, 4749, 495, 497, 64, 671, 759 |
| recB | 63.6 | Recombination | rorA; recombination and repair; RecBCD enzyme (exonuclease V) subunit | 311 | 1107, 1180, 1249, 1743, 3870, 4365, 690, 759, 4785a, 406a |
| recC | 63.7 | Recombination | Recombination and repair; RecBCD enzyme (exonuclease V) subunit | 310 | 1107, 1252, 1262, 1743, 3870, 4365, 690, 759, 4785a |
| recD | 63.6 | Recombination | hopE; recombination and repair; RecBCD enzyme (exonuclease V) α-subunit | 4975 | 1248, 3187, 348, 4365, 759, 87 |
| recE | 30.5 | Recombination | Recombination and repair; in prophage rac locus; degrades one strand 5′–3′ in duplex DNA; exonuclease VIII | 309 | 1181, 1287, 2086, 2087, 362, 4751, 748, 759, 760 |
| recF | 83.6 | Recombination | uvrF; recombination and repair | 308 | 10, 11, 132, 1517, 152, 2709, 3626, 3627, 379, 3846, 3847, 3848, 4611, 671, 759 |
| recG | 82.4 | Recombination | spoV? branch migration of Holliday junctions, junction-specific DNA helicase (see ruvABC) | 307 | 1281, 2100, 2593, 2596, 2597, 2598, 2599, 3086, 4721, 59 |
| recJ | 65.4 | Recombination | Single-stranded DNA-specific exonuclease, 5′–3′ | 17965 | 2638, 2639, 2640, 2641, 2642, 4511, 759 |
| recN | 59.3 | Recombination | radB; lexA regulon; recombination and repair | 10872 | 2600, 2924, 3425, 3748, 3861, 3862 |
| recO | 58.2 | Recombination | Conjugational recombination and repair; DNA-binding protein; RecA-like strand assimilation | 17962 | 2297, 2657 |
| recQ | 86.3 | Recombination | Conjugational recombination and repair, presynaptic stage of recombination; lexA regulon; RecQ helicase | 17959 | 1923, 2883, 3123, 3124, 58, 926 |
| recR | 10.6 | Recombination | Recombination and DNA repair | 31049 | 2723 |
| recT | 30.4 | Recombination | Locus in defective prophage rac; activated by sbcA mutation; DNA-annealing protein | 32070 | 1612, 2298, 2378, 761 |
| relA | 62.7 | Relaxed | Required for ppGpp synthesis during stringent response to amino acid starvation; ATP:GTP 3′-pyrophosphotransferase (EC 2.7.6.5) | 306 | 1237, 1441, 1442, 2913, 2914, 2915, 3070, 3606, 3934, 4310, 4807a, 924 |
| relB | 35.4 | Relaxed | Stringent/relaxed response; regulation of RNA synthesis | 305 | 1042, 266 |
| relE | 35.4 | Relaxed | Function unknown | 17956 | 1411, 266 |
| relF | 35.4 | Relaxed | Function unknown; overproduction lethal | 17953 | 1411, 266 |
| relX | 62.8 | Relaxed | Control of ppGpp synthesis | 304 | 3346 |
| rep | 85.3 | Replicase | dasC, mbrA, mmrA; Rep helicase, a single-stranded DNA-dependent ATPase | 303 | 1086, 1428, 257, 2613, 342, 343, 4388, 58, 610, 926 |
| rer | 89.9 | Resistance to radiation | Resistance to UV and gamma radiation | 302 | 4179 |
| rfaB | 81.9 | Rough | waaB; UDP-galactose: (glucosyl)lipopolysaccharide-1,6-galactosyltransferase | 17617 | 3361, 3522, 3726, 3921, 4128, 867 |
| rfaC | 81.8 | Rough | waaC; LPS core biosynthesis; proximal hexose; UDB-galactose: (glucosyl)LPS-1,6-galactosyltransferase | 300 | 280, 3362, 3726, 3920, 3921, 4128, 702 |
| rfaD | 81.7 | Rough | htrM; heat-inducible, LPS; allows high-temperature growth; d-glycero-d-mannoheptose epimerase | 299 | 2269, 2728, 2964, 3392, 3590, 3726, 3920, 3921, 4128, 806, 807, 808 |
| rfaF | 81.8 | Rough | waaF; ADP-heptose;LPS heptosyltransferase 1 | 28434 | 2269, 3362, 3392, 3726, 3920, 3921, 4079, 4128, 702, 807 |
| rfaG | 82.0 | Rough | waaG; LPS core biosynthesis; glucosyltransferase I | 15583 | 172, 3361, 3362, 3522, 3726, 3920, 3921, 4128, 867 |
| rfaH | 86.7 | Rough | sfrB; regulates LPS core biosynthesis, transcriptional activator | 164 | 200, 201, 2959, 333, 3641, 3921, 867 |
| rfaI | 81.9 | Rough | UDP-d-galactose: (glucosyl)lipopolysaccharide-1,3-d-galactosyltransferase | 17614 | 3361, 3523, 3726, 3920, 3921, 4128, 867 |
| rfaJ | 81.9 | Rough | UDP-d-glucose: (galactosyl)lipopolysaccharide glucosyltransferase | 17611 | 3361, 3522, 3523, 3726, 3920, 3921, 4128, 867 |
| rfaK | 81.8 | Rough | waaK; not similar to Salmonella rfaK; adds terminal GlcNac side branch to the lipopolysaccharide core prior to attachment of the O antigen | 33791 | 1698a, 2269, 2271, 3726, 3920, 3921, 4128 |
| rfaL | 81.8 | Rough | LPS core biosynthesis; O-antigen ligase | 28438 | 2269, 2271, 3726, 3920, 3921, 4128, 1698a |
| rfaP | 82.0 | Rough | LPS core biosynthesis; phosphorylation of core | 298 | 280, 3361, 3362, 3522, 3726, 3920, 3921, 4128 |
| rfaQ | 82.0 | Rough | Heptose region of LPS core | 33801 | 2268, 2271, 3361, 3362, 3435, 3522, 3523, 3726, 3920, 3921, 4128, 779 |
| rfaS | 81.9 | Rough | LPS core, not affecting attachment of O antigen | 28949 | 172, 2268, 2270, 2271, 3523, 3726, 3920, 3921 |
| rfaY | 81.9 | Rough | LPS core biosynthesis | 33828 | 2269, 3920, 3921, 4128 |
| rfaZ | 81.8 | Rough | LPS core biosynthesis | 33824 | 2269, 2271, 3920, 3921 |
| rfbA | 45.4 | Rough | som; TDP-glucose pyrophosphorylase | 297 | 2267, 2728, 4208, 4875 |
| rfbB | 45.4 | Rough | som; TDP-glucose oxidoreductase-4,6 dehydratase | 296 | 2267, 2728, 4208, 4875 |
| rfbC | 45.4 | Rough | rfbD; dTDP-4-deoxyrhamnose-3,5-epimerase | 38129 | 4875 |
| rfbD | 45.5 | Rough | rfbC; TDP-rhamnose synthetase | 295 | 2267, 4208, 4875 |
| rfbX | 45.4 | Rough | Hydrophobic protein, O-antigen | 37353 | 2267, 4875 |
| rfc | 45.4 | Rough | O-antigen polymerase | 53474 | 2658, 4875 |
| rfe | 85.5 | Rough | UDP-GlcNAc:undecaprenylphosphate GlcNAc-1-P transferase; common and O-antigen synthesis, tunicamycin sensitivity | 294 | 2866, 2867, 2868, 3268, 926 |
| rffA | 85.6 | Rough | Lipid III biosynthesis in common antigen synthesis | 33935 | 241, 2867 |
| rffC | 85.6 | Rough | Synthesis of enterobacterial common antigen and chain elongation | 33938 | 241, 2867 |
| rffD | 85.5 | Rough | Synthesis of enterobacterial common antigen; UDP-ManNAcA dehydrogenase | 293 | 2866, 2867, 2868, 3268 |
| rffE | 85.5 | Rough | nfrC, wecB; synthesis of enterobacterial common antigen; UDP-GlcNAc-2-epimerase | 33925 | 2770, 2867, 2868, 926 |
| rffG | 85.6 | Rough | rffE? dehydratase activity | 40941 | 2770, 926 |
| rffH | 85.6 | Rough | Hypothetical protein in rffE operon | 52957 | 394 |
| rffM | 85.7 | Rough | UDP-ManNAcA transferase | 33948 | 2867, 2868 |
| rffT | 85.7 | Rough | Synthesis of enterobacterial common antigen; Fuc4NAc transferase | 33930 | 2867, 2868 |
| rhaA | 88.2 | Rhamnose | l-Rhamnose isomerase (EC 5.3.1.14) | 292 | 2999, 3465, 4421 |
| rhaB | 88.2 | Rhamnose | Rhamnulokinase (EC 2.7.1.5) | 291 | 3465, 4421 |
| rhaD | 88.2 | Rhamnose | Rhamnulosephosphate aldolase D (EC 4.1.2.19) | 289 | 3465, 4421 |
| rhaR | 88.3 | Rhamnose | rhaC; positive regulatory gene | 290 | 3465, 4421, 4580 |
| rhaS | 88.3 | Rhamnose | rhaC; positive regulatory gene | 17950 | 3465, 4421, 4580 |
| rhaT | 88.3 | Rhamnose | Rhamnose permease; l-rhamnose-H+ symporter, membrane protein | 34185 | 1362, 211, 4358, 4359, 4580 |
| rhlB | 85.4 | RNA helicase like | Same as mmrA? protein with RNA helicase-like motif | 36971 | 2099, 926 |
| rhlE | 17.9 | RNA helicase like | DEAD-box protein family; ATP-dependent RNA helicase-like protein | 33907 | 3260 |
| rho | 85.4 | Rho termination factor | nitA, nusD, psuA, rnsC, sun, tsu; transcription termination factor Rho | 288 | 1914, 2550, 257, 2717, 2807, 2969, 3436, 3663, 4229, 4240, 4388, 4524, 533, 58, 614, 926, 940 |
| RhsA | 81.0 | Recombination hotspot | Repetitive sequence responsible for duplications within chromosome | 17947 | 1232, 2557, 3790, 4596, 4950 |
| RhsB | 78.0 | Recombination hotspot | Repetitive sequence responsible for duplications within chromosome | 17944 | 2557, 3790, 4950 |
| RhsC | 15.7 | Recombination hotspot | Repetitive sequence responsible for duplications within chromosome | 17941 | 3790, 4950 |
| RhsD | 11.3 | Recombination hotspot | Repetitive sequence responsible for duplications within chromosome | 17938 | 3790, 3791, 4950 |
| RhsE | 32.9 | Recombination hotspot | Repetitive sequence responsible for duplications within chromosome | 33720 | 3791, 4950 |
| ribA | 28.8 | Riboflavin | Riboflavin biosynthesis; GTP-cyclohydrolase II (EC 3.5.4.20) | 287 | 216, 217, 2288, 3670, 4387 |
| ribB | 68.6 | Riboflavin | htrP; luxH-like; riboflavin biosynthesis; 3,4-dihydroxy-2-butanone 4-phosphate synthase | 286 | 216, 217, 3594, 3672, 4387 |
| ribC | 37.5 | Riboflavin | Riboflavin synthase α chain; see ribE | 11923 | 1110, 217, 4387 |
| ribD | 9.3 | Riboflavin | ribG; riboflavin biosynthese; a deaminase | 35685 | 3671, 4361 |
| ribE | 9.3 | Riboflavin | ribH; riboflavin synthase β chain; see ribC | 35688 | 4361 |
| ribF | 0.5 | Riboflavin | Flavokinase and FAD synthetase | 35833 | 185, E |
| ridA | 73.1 | Rifampin dependence | Rifampin resistance and dependence | 285 | 898 |
| ridB | 85.5 | Rifampin dependence | Transcription and translation; rifampin (rifamycin) sensitivity | 17935 | 895 |
| rimB | 38.9 | Ribosomal modification | 50S ribosomal subunit maturation | 284 | 546 |
| rimC | 26.2 | Ribosomal modification | 50S ribosomal subunit maturation | 283 | 546 |
| rimD | 87.7 | Ribosomal modification | 50S ribosomal subunit maturation | 282 | 546 |
| rimE | 74.1 | Ribosomal modification | Ribosomal protein modification | 281 | 2865 |
| rimF? | 0.8 | Ribosomal modification | res; ribosomal modification; may be same as rimG | 280 | 1379 |
| rimG | 0.7 | Ribosomal modification | ramB; modification of 30S ribosomal subunit protein S4; probably same as rimF (res) in E. coli B | 279 | 1379, 4961 |
| rimH | 13.6 | Ribosomal modification | stsB; ribosomal modification | 278 | |
| rimI | 99.3 | Ribosomal modification | Modification of 30S ribosomal subunit protein S18; acetylation of N-terminal alanine | 277 | 1939, 4895 |
| rimJ | 24.2 | Ribosomal modification | tcp; modification of 30S ribosomal subunit protein S5; acetylation of N-terminal alanine | 276 | 1988, 4728, 4729, 4895 |
| rimK | 19.2 | Ribosomal modification | nek; modification of 30S ribosomal subunit protein S6 by addition of glutamic acid residues | 31682 | 122, 1860, 2123 |
| rimL | 32.3 | Ribosomal modification | Modification of 50S ribosomal subunit protein L7/L12; acetylation of N-terminal serine | 275 | 1940, 4348 |
| rimM | 59.1 | Ribosomal modification | yfjA; 21-kDa protein essential for 16S RNA processing | 51908 | 589, 590 |
| rit | 89.2 | Ribosomal thermolability | Affects thermolability of 50S ribosomal subunit | 274 | 3298 |
| rlpA | 14.3 | Rare lipoprotein | Minor lipoprotein | 17932 | 4332 |
| rlpB | 14.5 | Rare lipoprotein | Minor lipoprotein | 17929 | 4332 |
| rluA | 1.3 | rRNA, large, uridine modification | Dual specificity pseudouridine synthase for 23S rRNA and tRNAphe | 36966 | 1578, 2106, 4787 |
| rluC | 24.66b | Pseudouridine synthase | Responsible for pseudouridine at three positions in 23S RNA (not on map, Fig. 1) | 57124 | 827a |
| rluD | 48.9 | Suppressor ftsH | sfhB; suppresses ftsH(ts) mutants; 23S rRNA pseudouridine synthase (pseudouridines at position 1911, 1915, and 1917) | 53497 | 3079, 689a |
| rmf | 21.9 | Ribosome modulation factor | Associated with the 100S dimers of 70S ribosomes observed in stationary phase cells | 31760 | 4622, 4624, 4832 |
| rna | 13.9 | RNase A | rnsA; cleaves phosphodiester bond between two nucleotides; RNase I | 273 | 2858 |
| rnb | 29.0 | RNase B | RNase II; mRNA degradation | 272 | 1069, 4958, 4959, 4960 |
| rnc | 58.2 | RNase C | RNase III; cleaves double-stranded RNA | 271 | 1910, 2753, 2810, 3134, 4679, 708, 4339 |
| rnd | 40.6 | RNase D | Rnase D; processes tRNA precursors | 270 | 1336, 4919, 4939, 4938 |
| rne | 24.6 | RNase E | ams; RNase E; enzyme complex for RNA processing, mRNA turnover and 5S RNA maturation | 269 | 183, 1840, 1983, 2663, 2702, 2843, 2876, 2958, 3031, 3299, 3622, 4354, 621, 648, 673, 691, 774, 833 |
| rnhA | 5.1 | RNase H | cer, dasF, herA, rnh, sdrA, sin; degrades RNA of DNA-RNA hybrids; replication; RNase HI; participates in DNA replication | 268 | 127, 1793, 1804, 2113, 2286, 2287, 2553, 2730, 3092, 3209, 3244, 4442, 633, 634, 636, 855 |
| rnhB | 4.4 | RNase H | Degrades RNA of DNA-RNA hybrids; RNase HII (EC 3.1.26.4) | 30604 | 1944 |
| rnk | 13.9 | Regulator nucleoside-diP kinase | Regulator of nucleoside diphosphate kinase; suppresses Pseudomonas algR2 | 36984 | 3911, 3985 |
| rnpA | 83.7 | RNase P | tRNA, 4.5S RNA-processing; RNase P, protein component | 267 | 1632, 2074, 2293, 3328 |
| rnpB | 70.4 | RNase P | RNase P, RNA component | 266 | 1064, 123, 2293, 2311, 2663, 3019, 3634, 3635, 3811 |
| rnr | 94.3 | RNase R | vacB; homology with Shigella virulence gene vacB; exoribonuclease R | 34417 | 569, 719a, 4417a |
| rnt | 37.2 | RNase T | Degrades tRNA; RNase T (EC 3.1.13–) | 32271 | 1843, 3331, 3656, 649 |
| rob | 99.8 | Right oriC binding | oriC-binding protein, binds to right border of oriC | 34661 | 131, 2093, 4086 |
| rorB | 85.1 | Roentgen resistance | Sensitivity to ionizing radiation, mitomycin C | 36611 | 991, 992 |
| rpe | 75.7 | Ribulose-5-phosphate epimerase | d-Ribulose-5-phosphate epimerase, in dam operon | 53572 | 2681 |
| rph | 82.2 | RNase PH | RNase PH | 33892 | 2009, 2011, 2192, 3309, 94 |
| rpiA | 65.9 | Ribose-P isomerase | Ribose phosphate isomerase (constitutive) (EC 5.3.1.6) | 264 | 1822 |
| rpiB | 92.9 | Ribose-P isomerase | Allose-6-P isomerase; ribose P isomerase? | 50228 | 2223, 4138 |
| rpiR | 92.9 | Ribose-P isomerase | alsR; repressor for als operon and rpiB | 50233 | 2223, 4138 |
| rplA | 90.0 | Ribosomal protein, large | 50S ribosomal subunit protein L1 | 263 | 1083, 1855, 2567, 2577, 2688, 3499, 3605, 537, 89 |
| rplB | 74.3 | Ribosomal protein, large | 50S ribosomal subunit protein L2 | 262 | 4969 |
| rplC | 74.4 | Ribosomal protein, large | 50S ribosomal subunit protein L3 | 261 | 4969 |
| rplD | 74.4 | Ribosomal protein, large | eryA; 50S ribosomal subunit protein L4; erythromycin sensitivity | 260 | 2538, 2559, 4293, 4762, 4927, 4969, 736 |
| rplE | 74.2 | Ribosomal protein, large | 50S ribosomal subunit protein L5 | 259 | 666 |
| rplF | 74.2 | Ribosomal protein, large | 50S ribosomal subunit protein L6; gentamicin sensitivity | 258 | 554, 666 |
| rplI | 95.4 | Ribosomal protein, large | 50S ribosomal subunit protein L9 | 257 | 15, 1938, 3928 |
| rplJ | 90.0 | Ribosomal protein, large | strA; streptomycin resistance; 50S ribosomal subunit protein L10 | 256 | 1083, 1238, 1788, 1855, 243, 244, 2567, 2577, 2688, 3165, 3499, 3605, 4210, 4377, 537, 783, 89 |
| rplK | 90.0 | Ribosomal protein, large | relC; 50S ribosomal subunit protein L11; kasugamycin sensitivity | 255 | 1083, 1855, 2567, 2577, 3065, 3165, 3499, 3605, 4210, 4377, 537, 89, 891, 918, 4398 |
| rplL | 90.1 | Ribosomal protein, large | 50S ribosomal subunit protein L7/L12 | 254 | 1083, 1788, 1855, 243, 244, 2567, 2577, 2688, 3165, 3605, 4210, 537, 89 |
| rplM | 72.8 | Ribosomal protein, large | 50S ribosomal subunit protein L13 | 253 | 1941, 894 |
| rplN | 74.3 | Ribosomal protein, large | 50S ribosomal subunit protein L14 | 252 | 666 |
| rplO | 74.2 | Ribosomal protein, large | 50S ribosomal subunit protein L15 | 251 | 1945, 666 |
| rplP | 74.3 | Ribosomal protein, large | 50S ribosomal subunit protein L16 | 250 | 4969 |
| rplQ | 74.1 | Ribosomal protein, large | 50S ribosomal subunit protein L17 | 249 | 275, 2865, 3496, 666 |
| rplR | 74.2 | Ribosomal protein, large | 50S ribosomal subunit protein L18 | 248 | 666 |
| rplS | 59.1 | Ribosomal protein, large | 50S ribosomal subunit protein L19 | 247 | 3407, 597 |
| rplT | 38.7 | Ribosomal protein, large | pdzA; 50S ribosomal subunit protein L20 | 17608 | 1213 |
| rplU | 71.8 | Ribosomal protein, large | 50S ribosomal subunit protein L21 | 246 | 2013, 2246 |
| rplV | 74.3 | Ribosomal protein, large | eryB; erythromycin sensitivity; 50S ribosomal subunit protein L22 | 245 | 4762, 4969, 736 |
| rplW | 74.3 | Ribosomal protein, large | 50S ribosomal subunit protein L23 | 244 | 4969 |
| rplX | 74.3 | Ribosomal protein, large | 50S ribosomal subunit protein L24 | 243 | 666, 896 |
| rplY | 49.2 | Ribosomal protein, large | 50S ribosomal subunit protein L25 | 242 | |
| rpmA | 71.8 | Ribosomal protein, large | 50S ribosomal subunit protein L27 | 241 | 2246 |
| rpmB | 82.1 | Ribosomal protein, large | 50S ribosomal subunit protein L28 | 240 | 1936, 2466, 2716, 4154 |
| rpmC | 74.3 | Ribosomal protein, large | 50S ribosomal subunit protein L29 | 239 | 4969 |
| rpmD | 74.2 | Ribosomal protein, large | 50S ribosomal subunit protein L30 | 238 | 666 |
| rpmE | 88.9 | Ribosomal protein, large | 50S ribosomal subunit protein L31 | 237 | 893 |
| rpmF | 24.7 | Ribosomal protein, large | 50S ribosomal subunit protein L32 | 17605 | 1988 |
| rpmG | 82.1 | Ribosomal protein, large | 50S ribosomal subunit protein L33 | 236 | 1936, 2466, 2716, 4154 |
| rpmH | 83.7 | Ribosomal protein, large | rimA, ssaF; 50S ribosomal subunit protein L34 | 235 | 1631, 3285, 4611, 546 |
| rpmI | 38.8 | Ribosomal protein, large | 50S ribosomal subunit protein A (L35) | 17602 | 4623 |
| rpmJ | 74.2 | Ribosomal protein, large | 50S ribosomal subunit protein X (L36) | 17599 | 4623, 666 |
| rpoA | 74.1 | RNA polymerase | phs, sez; phage P2 vir1 resistance; RNA polymerase, α-subunit (EC 2.7.7.6) | 234 | 1426, 2236, 275, 2865, 3318, 3496, 3497, 3754, 3755, 4292, 666 |
| rpoB | 90.1 | RNA polymerase | ftsR, groN, mbrD, nitB, rif, ron, stl, stv, tabD, sdgB; streptovar., rif., streptolyd. sensitivity; RNA polymerase, β-subunit (EC 2.7.7.6) | 233 | 174, 1013, 1083, 1108, 1190, 1406, 1418, 173, 1788, 1855, 2020, 2092, 2286, 243, 244, 2463, 2567, 2577, 2688, 3165, 3316, 3499, 3605, 3698, 3756, 3906, 4210, 4211, 4523, 4588, 4912, 537, 89 |
| rpoC | 90.2 | RNA polymerase | tabD; RNA polymerase, β-subunit (EC 2.7.7.6) | 232 | 1083, 1108, 1788, 2020, 2092, 243, 244, 2567, 2577, 2688, 3165, 3315, 3316, 3605, 4175, 4210, 537, 89 |
| rpoD | 69.2 | RNA polymerase | alt; most exponential phase transcription; RNA polymerase, ς-subunit, ς70 (EC 2.7.7.6) | 231 | 1367, 1531, 2230, 2542, 2671, 2673, 3113, 3114, 4053, 4070, 4376, 565, 579, 580 |
| rpoE | 58.4 | RNA polymerase | RNA polymerase ςE-subunit, high-temperature transcription (heat shock and oxidative stress) | 36181 | 1767, 3592 |
| rpoH | 77.5 | RNA polymerase | fam, hin, htpR; heat-shock transcription RNA polymerase ς32-subunit (EC 2.7.7.6) | 618 | 1532, 1533, 2120, 2407, 3145, 4418, 4484, 4913, 612 |
| rpoN | 72.0 | RNA polymerase | glnF, ntrA; RNA polymerase ς60-subunit, transcription of N-source-controlled genes | 17926 | 1770, 1864, 1904, 2904, 3517, 656, 657 |
| rpoS | 61.7 | RNA polymerase | abrD, dpeB, katF, nur, appR; ςS; RNA polymerase ς, stationary phase | 18208 | 1226, 1402, 1925, 2049, 2077, 2411, 2610, 2749, 3044, 3045, 3806, 3825, 3899, 4448, 4488, 4591, 4603, 4447, 3697, 2413a |
| rpoZ | 82.3 | RNA polymerase | spoS; RNA polymerase, ω subunit | 28445 | 1400, 2597 |
| rpsA | 20.7 | Ribosomal protein, small | ssyF; 30S ribosomal subunit protein S1 | 230 | 1099, 2247, 2248, 3390, 3926, 3927, 4027, 746 |
| rpsB | 4.1 | Ribosomal protein, small | 30S ribosomal subunit protein S2 | 229 | 2428, 296, 3133, 418, 88 |
| rpsC | 74.3 | Ribosomal protein, small | 30S ribosomal subunit protein S3 | 228 | 4969 |
| rpsD | 74.1 | Ribosomal protein, small | ramA, sud2; 30S ribosomal subunit protein S4 | 227 | 1662, 275, 3496, 3497, 666 |
| rpsE | 74.2 | Ribosomal protein, small | eps, spc, spcA; 30S ribosomal subunit protein S5 | 226 | 1662, 2975, 666, 956 |
| rpsF | 95.3 | Ribosomal protein, small | sdgH; 30S ribosomal subunit protein S6 | 225 | 1937, 3928 |
| rpsG | 74.8 | Ribosomal protein, small | K12; 30S ribosomal subunit protein S7 | 224 | 3498 |
| rpsH | 74.2 | Ribosomal protein, small | 30S ribosomal subunit protein S8 | 223 | 666, 4784 |
| rpsI | 72.8 | Ribosomal protein, small | 30S ribosomal subunit protein S9 | 17596 | 1941, 897 |
| rpsJ | 74.4 | Ribosomal protein, small | nusE; 30S ribosomal subunit protein S10 | 222 | 1012, 2559, 4927, 851 |
| rpsK | 74.1 | Ribosomal protein, small | 30S ribosomal subunit protein S11 | 221 | 275, 3496, 666 |
| rpsL | 74.8 | Ribosomal protein, small | asuB; strA; 30S ribosomal subunit protein S12 | 220 | 489, 3498 |
| rpsM | 74.1 | Ribosomal protein, small | 30S ribosomal subunit protein S13 | 219 | 1212, 275, 3496, 666 |
| rpsN | 74.2 | Ribosomal protein, small | 30S ribosomal subunit protein S14 | 218 | 666 |
| rpsO | 71.3 | Ribosomal protein, small | secC; 30S ribosomal subunit protein S15 | 217 | 1126, 1182, 1231, 2246, 2457, 3459, 3492, 3494, 3638, 3850, 4333, 4334 |
| rpsP | 59.1 | Ribosomal protein, small | 30S ribosomal subunit protein S16 | 216 | 1935, 3235, 3407, 599 |
| rpsQ | 74.3 | Ribosomal protein, small | neaA; 30S ribosomal subunit protein S17 | 215 | 4969, 630 |
| rpsR | 95.3 | Ribosomal protein, small | 30S ribosomal subunit protein S18 | 214 | 1938, 3928, 891 |
| rpsS | 74.3 | Ribosomal protein, small | 30S ribosomal subunit protein S19 | 213 | 1935, 4969 |
| rpsT | 0.4 | Ribosomal protein, small | sups20; 30S ribosomal subunit protein S20 | 212 | 2700, 2701, 4830 |
| rpsU | 69.2 | Ribosomal protein, small | 30S ribosomal subunit protein S21 | 211 | 2671, 2673, 3148, 579, 892 |
| rrfA | 87.0 | Ribosomal RNA, 5S | 5S rRNA of rrnA operon | 210 | 1146, 4044, 439 |
| rrfB | 89.9 | Ribosomal RNA, 5S | 5S rRNA of rrnB operon | 209 | 4044, 522, 523 |
| rrfC | 85.0 | Ribosomal RNA, 5S | 5S rRNA of rrnC operon | 208 | 4044, 4904 |
| rrfD | 73.7 | Ribosomal RNA, 5S | 5S rRNA of rrnD operon | 207 | 1096, 4044 |
| rrfE | 90.8 | Ribosomal RNA, 5S | 5S rRNA of rrnE operon | 206 | 1146, 2543, 4044, 439 |
| rrfF | 73.7 | Ribosomal RNA, 5S | rrfDβ, rrvD; 5S rRNA of rrnD operon | 33643 | 394 |
| rrfG | 58.7 | Ribosomal RNA, 5S | 5S rRNA of rrnG operon | 205 | 1146, 4044 |
| rrfH | 4.8 | Ribosomal RNA, 5S | 5S rRNA of rrnH operon | 204 | 1146 |
| rrlA | 87.0 | Ribosomal RNA, 23S | 23S rRNA of rrnA operon | 203 | 1146, 1502, 3236, 4383, 439, 915, 926 |
| rrlB | 89.8 | Ribosomal RNA, 23S | 23S rRNA of rrnB operon | 202 | 1502, 3063, 3236, 3785, 4383, 522, 523, 524, 915 |
| rrlC | 85.0 | Ribosomal RNA, 23S | 23S rRNA of rrnC operon | 201 | 1502, 3236, 4383, 915 |
| rrlD | 73.8 | Ribosomal RNA, 23S | 23S rRNA of rrnD operon | 200 | 1096, 1502, 3236, 4383, 915 |
| rrlE | 90.7 | Ribosomal RNA, 23S | 23S rRNA of rrnE operon | 199 | 1146, 1502, 2543, 3236, 4383, 439, 915 |
| rrlG | 58.7 | Ribosomal RNA, 23S | 23S rRNA of rrnG operon | 198 | 1146, 1502, 3236, 4383, 915 |
| rrlH | 4.8 | Ribosomal RNA, 23S | 23S rRNA of rrnH operon | 197 | 1146, 1502, 3236, 4383, 4384, 915 |
| rrmA | 41.1 | Ribosomal RNA modification | 23S rRNA m1G745 methyltransferase; mutant has slow growth rate, slow chain elongation rate, and viomycin resistance | 56816 | 1576 |
| rrnA | 96.9 | Ribosomal RNA (operon) | See Fig. 1 | 23593 | |
| rrnB | 89.8 | Ribosomal RNA (operon) | cqsE, rrnB1; see Fig. 1 | 195 | 1661, 248 |
| rrnC | 84.9 | Ribosomal RNA (operon) | cqsB; see Fig. 1 | 19503 | 2056 |
| rrnD | 73.7 | Ribosomal RNA (operon) | cqsD; see Fig. 1 | 193 | 1752 |
| rrnE | 90.7 | Ribosomal RNA (operon) | rrnD1; see Fig. 1 | 192 | 1752, 2543 |
| rrnG | 58.7 | Ribosomal RNA (operon) | See Fig. 1 | 10891 | 1661, 53 |
| rrnH | 4.8 | Ribosomal RNA (operon) | See Fig. 1 | 190 | |
| rrsA | 86.9 | Ribosomal RNA, 16S | 16S RNA of rrnA operon | 189 | 4458, 926 |
| rrsB | 89.8 | Ribosomal RNA, 16S | 16S RNA of rrnB operon | 188 | 3063, 318, 3341, 3535, 440, 4458, 522, 523, 879 |
| rrsC | 84.9 | Ribosomal RNA, 16S | 16S RNA of rrnC operon | 187 | 4458 |
| rrsD | 73.8 | Ribosomal RNA, 16S | 16S RNA of rrnD operon | 186 | 4458 |
| rrsE | 90.7 | Ribosomal RNA, 16S | 16S RNA of rrnE operon | 185 | 2543, 4458 |
| rrsG | 58.8 | Ribosomal RNA, 16S | 16S RNA of rrnG operon | 184 | 1146, 4015, 4458 |
| rrsH | 4.8 | Ribosomal RNA, 16S | 16S RNA of rrnH operon | 183 | 1146, 4058, 4458 |
| rsd | 94.4 | Regulation of ςD | Stationary phase protein, binds major ς-subunit | 54664 | 2023 |
| rseA | 58.3 | Regulation of ςE | mclA; membrane protein, negative regulator of ςE | 37241 | 2965, 977 |
| rseB | 58.3 | Regulation of ςE | Binds rseA, negative regulation of ςE | 37244 | 2965, 977 |
| rseC | 58.3 | Regulation of ςE | Deletion does not affect ςE activity | 37247 | 2965, 977 |
| rspA | 35.5 | Repressor of ςS production | Prevents homoserine lactone-induced synthesis of ςS, starvation induced | 36959 | 1856 |
| rspB | 35.5 | Repressor of ςS production | Function unknown, transcribed with rspA, starvation induced | 36962 | 1856 |
| rssA | 27.8 | Regulation of ςS | Two-component response regulator, affecting ςS-dependent proteins | 37468 | 1718 |
| rssB | 27.8 | Regulation of ςS | mviA, spreE, two-component response regulator, affecting ςS-dependent proteins | 37471 | 1718, 3034, 3532, 4952 |
| rsuA | 49.1 | rRNA, small pseudouridine | 16S RNA pseudouridine 516 synthase | 36034 | 4786 |
| rtcA | 76.6 | RNA terminal phosphate cyclase | Cyclase activity similar to human enzyme; RNA 3′-terminal phosphate cyclase | 56879 | 1399a |
| rtcB | 76.6 | RNA terminal phosphate cyclase | 56882 | 1399a | |
| rtcR | 76.6 | RNA terminal phosphate cyclase | 56885 | 1399a | |
| rtn | 48.9 | Resistance to N4 | Probable membrane protein; resistance to phages lambda and N4 | 54796 | 1605 |
| rus | 12.3 | ruv suppressor | Resolvase; resolves Holliday structures | 32471 | 2722, 2745, 4003 |
| ruvA | 41.9 | Repair UV? | lexA regulon; Holliday junction recognition | 17923 | 1281, 13, 163, 1966, 305, 306, 3373, 4049, 4478, 4479, 4712 |
| ruvB | 41.9 | Repair UV? | lexA regulon; branch migration of Holliday structures | 17920 | 1281, 13, 305, 306, 3373, 4712 |
| ruvC | 41.9 | Repair UV? | Not SOS regulated; resolves Holliday structures; RuvC endonuclease | 32474 | 1101, 151, 1965, 2593, 299, 307, 3999, 4001, 4002, 4328, 4364, 4712, 4713, 4714, 824 |
| sad | 34.3 | Succinate-semialdehyde deHase | Succinate-semialdehyde dehydrogenase, NAD dependent (EC 1.2.1.24) | 180 | 2759, 4088 |
| sanA | 48.1 | Sensitivity vancomycin | Amplification abolishes vancomycin-sensitive permeability defects of mutants | 50224 | 3673 |
| sapA | 29.2 | Sensitivity to antimicrobial peptides | Peptide transport system periplasmic protein | 51886 | 3371, 322a, 4792a |
| sapB | 29.2 | Sensitivity to antimicrobial peptides | Peptide transport system permease | 51883 | 322a, 4792a |
| sapC | 29.1 | Sensitivity to antimicrobial peptides | Peptide transport system permease | 51880 | I, H |
| sapD | 29.1 | Sensitivity to antimicrobial peptides | trkE; affects potassium transport; peptide transport system ATP-binding protein | 35326 | 1161, 3371, 3372, 3914 |
| sapF | 29.1 | Sensitivity to antimicrobial peptides | Peptide transport system ATP-binding protein | 35323 | 3372 |
| sbaA | 96.5 | Serine, branched-chain amino acids | Regulation of serine and branched-chain amino acid metabolism | 179 | 919 |
| sbcB | 44.9 | Suppression of recBC | xonA; suppresses recB recC mutations; exonuclease I | 178 | 3156, 3417, 3418, 3529, 3845, 4291, 4834 |
| sbcC | 8.9 | Suppression of recBC | Cosuppressor with sbcB of recB recC mutations | 17917 | 2451, 2594, 2595, 3128, 4834, 676, 823 |
| sbcD | 8.9 | Suppression of recBC | Cosuppressor with sbcB of recB recC mutations | 30972 | 1423, 2451, 823 |
| sbmA | 8.5 | Sensitivity B17 microcin | Methylmalonyl-CoA mutase (mcm) | 17914 | 2435 |
| sbmC | 44.8 | Sensitivity B17 microcin | gyrI; DNA gyrase inhibitor; in high copy protects cells from replication inhibitor MccB17; SOS induced; blocks MccB17 export | 50396 | 222, 3114a |
| sbp | 88.5 | Sulfate binding protein | Periplasmic sulfate-binding protein | 17911 | 1710 |
| sdaA | 40.8 | Serine deaminase | l-Serine deaminase | 32312 | 3987, 4258, 4260 |
| sdaB | 63.1 | Serine deaminase | l-Serine deaminase, l-SD2 | 33324 | 3988, 4259 |
| sdaC | 63.1 | Serine deaminase | dcrA; regulator of l-SD2; putative serine transporter | 33329 | 2546, 3988, 4259 |
| sdhA | 16.3 | Succinate dehydrogenase | Succinate dehydrogenase flavoprotein subunit (EC 1.3.99.1) | 17908 | 4741, 4773, 615, 866, 934, 3354 |
| sdhB | 16.3 | Succinate dehydrogenase | Succinate dehydrogenase iron-sulfur protein | 17905 | 4741, 4773, 615, 934, 3354 |
| sdhC | 16.3 | Succinate dehydrogenase | cybA; succinate dehydrogenase membrane anchor subunit, cytochrome b556 (EC 1.3.99.1) | 17902 | 3052, 3053, 615, 3354 |
| sdhD | 16.3 | Succinate dehydrogenase | Succinate dehydrogenase hydrophobic subunit (EC 1.3.99.1) | 17899 | 4741, 4773, 615, 3354 |
| sdiA | 43.0 | Suppress division inhibitors | Suppresses inhibitory effect of the MinC/MinD division inhibitor, positive regulator ftsQAZ | 30919 | 1361, 4659 |
| sds | 30.4 | Suppresses disulfide bond | Motility defects | 4792a | |
| secA | 2.3 | Secretory | azi, pea, prlD; translocation ATPase for protein export | 176 | 1079, 1241, 1279, 2329, 3286, 3287, 3358, 349, 3818, 3919, 598, 751, 921 |
| secB | 81.5 | Secretory | Protein export; chaperone SecB | 17896 | 1293, 2227, 2234, 2361, 2362, 2363, 2454, 260, 3519, 3919, 4675, 4676 |
| secD | 9.2 | Secretory | Membrane component of protein export complex | 17893 | 1366, 2329, 2651, 3469, 921 |
| secE | 90.0 | Secretory | prlG; inner membrane protein involved in protein secretion (with SecY) | 34108 | 1079, 1080, 1084, 1267, 181, 2035, 2814, 3067, 3068, 3200, 3468, 3470, 349, 350, 3553, 36, 3675, 3893, 3894, 395, 4182, 4264, 4362, 540, 921, 2329 |
| secF | 9.2 | Secretory | Membrane protein with protein secretion function | 29893 | 1198, 1365, 2329, 2651, 2817, 3467, 3469, 3794, 4264, 921 |
| secG | 71.6 | Secretory | prlH; p12 cytoplasmic membrane protein involved with protein export | 33484 | 1079, 1622, 3199, 3201, 450, 451, 4733 |
| secY | 74.2 | Secretory | prlA; multispanning membrane protein, translocator of proteins (with SecE) | 18037 | 1079, 1153, 1267, 1293, 180, 1945, 1947, 3218, 3292, 3307, 3470, 349, 350, 3553, 40, 4028, 4037, 4048, 42, 43, 46, 4868, 666, 921, 1949, 2329 |
| selA | 81.0 | Selenium | fdhA (formate dehydrogenase activity); with SelD, converts serine residue to selenocysteine on tRNA | 785 | 1276, 2480, 3887 |
| selB | 81.0 | Selenium | fdhA; novel elongation factor promoting selenocysteine incorporation (specific for selenocysteyl tRNA) | 17890 | 1274, 2480, 345, 3887 |
| selC | 82.6 | Selenium | fdhC; selenocysteyl tRNAUCA (converted from serine tRNA) | 17887 | 2313, 237, 2478, 2480, 3887 |
| selD | 39.8 | Selenium | Selenophosphate synthase; reduced selenium donor for selenocysteic tRNA and protein | 32292 | 1125, 1275, 2226, 2479, 3887, 4184 |
| semA | 40.5 | Sensitivity to microcin | Sensitivity to microcin E492 | 17884 | 3552 |
| seqA | 15.4 | Sequestration | hobH; DNA biosynthesis; negative modulator of replication initiation | 31506 | 208, 2646, 4096, 4608, 471 |
| serA | 65.9 | Serine | Phosphoglycerate dehydrogenase (EC 1.1.1.95) | 173 | 3239, 4420, 4444, 4949, 73 |
| serB | 99.6 | Serine | Phosphoserine phosphatase (EC 3.1.3.3) | 172 | 3705 |
| serC | 20.6 | Serine | pdxC, pdxF; phosphoserine aminotransferase (EC 2.6.1.52) | 171 | 1019, 1020, 1099, 1370, 3161, 4034a |
| serR | 2.3 | Serine | Regulates level of seryl-tRNA synthetase | 4392 | |
| serS | 20.2 | Serine | Serine hydroxamate resistance; seryl-tRNA synthetase (EC 6.1.1.11) | 169 | 1659, 4444 |
| serT | 22.2 | Serine | divE; serine tRNA1 | 168 | 2313, 34, 4345 |
| serU | 44.0 | Serine | su1, Su-1, ftsM, supD, supH (amber suppressor); serine tRNA2 | 167 | 2313, 2455, 3324, 4197, 4407, 1122a |
| serV | 60.7 | Serine | supD (ochre suppressor); serine tRNA3 | 166 | 2313, 1122a |
| serW | 19.9 | Serine | serWα; serWX duplicate genes; serine tRNA5 | 17881 | 2313, 3849, 1122a |
| serX | 23.6 | Serine | serWβ; serWX duplicate genes; serine tRNA5 | 17878 | 2313, 1122a |
| sfa | 22.7 | Suppressor of fabA | Suppresses fabA6 | 46624 | 3696 |
| sfcA | 33.5 | sbc fusion C-terminal | Probably maeA, malic enzyme, NAD linked (EC 1.1.1.38) | 32176 | 2719 |
| sfiC | 25.8 | Septum formation inhibition | Element of e14, inhibitor of cell division | 17875 | 1858, 1978, 2715, 933 |
| sfsA | 3.5 | Sugar fermentation stimulation | sfs1; regulatory for maltose metabolism; overexpression increases amylomaltase | 30524 | 2176 |
| sfsB | 71.8 | Sugar fermentation stimulation | nlp, sfs7; Ner-like regulatory protein | 35693 | 175, 2176, 744 |
| shiA | 44.1 | Shikimate | Shikimate and dehydroshikimate permease | 163 | 1185 |
| sipC | 82.6 | Suppressor of increased permeability | Mutations reverse the susceptibility to vancomycin and other hydrophobic antibiotics caused by TraT plasmid gene | 51306 | 3563 |
| sipD | 82.7 | Suppressor of increased permeability | Same phenotype as that produced by sipC | 36869 | 3563 |
| sir | 61.2 | SOS-independent repair | SOS-independent repair of mitomycin C-induced DNA damage | 36505 | 2366 |
| sixA | 52.9 | Phosphohistidine phosphatase affecting His-Asp phosphorelay of ArcB, etc. | 54917 | 3248 | |
| sloB | 75.0 | Slow growth | Tolerance for amidinopenicillin and nalidixic acid; slow growth rate | 162 | 4716 |
| slp | 78.7 | Stationary-phase lipoprotein | C starvation and stationary phase inducible; outer membrane lipoprotein | 33726 | 3971, 63 |
| slr | 14.7 | Selenolipoic acid resistant | Suppresses lipoate requirement of lipA; functions in lipoic acid synthesis | 36705 | 3633 |
| slt | 99.8 | Soluble lytic transglycosylase | Lytic transglycosylase, major autolysin | 34819 | 1155, 332 |
| slyD | 74.9 | Sensitivity to lysis | FK-506-BP-like lysis protein for φX174; metal ion-regulated peptidyl-prolyl cis/trans-isomerase, filamentation if overexpressed | 35441 | 1809, 1814, 3730, 3731, 4804 |
| slyX | 74.9 | Sensitivity to lysis | Required for φX174 lysis | 54726 | 3731 |
| smp | 99.6 | Serine B-contraposed membrane protein | Transcribed divergently from serB, overlapping promoter; membrane protein | 34649 | 3162, 3163 |
| smtA | 21.0 | SAM-dependent methyltransferase | Putative methyltransferase, SAM-dependent | 50317 | 4844 |
| sodA | 88.3 | SOD | Member of SoxRS regulon; superoxide dismutase, Mn | 17593 | 1217, 301, 3178, 3540, 3541, 4338, 4445, 638, 819, 4355 |
| sodB | 37.4 | SOD | Superoxide dismutase, Fe | 15256 | 1217, 1529, 2204, 301, 3155, 3178, 3904, 638, 639 |
| sodC | 37.1 | SOD | Superoxide dismutase, Cu, Zn | 50310 | 1908, 302, 303 |
| sohA | 70.6 | Suppressor of htr | prlF; suppressor of htrA; putative protease | 35411 | 204, 2212, 4123 |
| sohB | 28.6 | Suppressor of htr | Suppressor of hyr; homology with inner membrane protease IV, which digests cleaved signal peptides | 31951 | 203 |
| solA | 24.1 | Sarcosine oxidase-like | Homology with sarcosine oxidases, but stronger activity with methyltryptophan than with sarcosine | 50669 | 2332 |
| soxR | 92.2 | Superoxide | Regulatory protein of soxRS regulon; induces nine-protein sox regulon when superoxide levels increase | 27798 | 1384, 1748, 3223, 3224, 4480, 4793, 78 |
| soxS | 92.1 | Superoxide | Regulatory protein of soxRS regulon; induces nine-protein sox regulon when superoxide levels increase | 27801 | 1211, 1985, 2539, 3223, 3224, 4793, 4794, 78 |
| speA | 66.4 | Spermidine | Biosynthetic decarboxylase (EC 4.1.1.19) | 161 | 2996, 3877, 472 |
| speB | 66.4 | Spermidine | Agmatinase (EC 3.5.3.11) | 160 | 3877, 472, 4318 |
| speC | 66.9 | Spermidine | Ornithine decarboxylase (EC 4.1.1.17) | 159 | 3877, 472 |
| speD | 2.9 | Spermidine | S-Adenosylmethionine decarboxylase (EC 4.1.1.50) | 158 | 4323, 4324 |
| speE | 2.9 | Spermidine | Spermidine synthase | 17590 | 4323, 4324 |
| speF | 15.5 | Spermidine | Ornthine decarboxylase, inducible (EC 4.1.1.17) | 31559 | 2149, 2153 |
| speG | 35.7 | Spermidine | Spermidine acetyltransferase | 53446 | 1334 |
| spf | 87.2 | Spot 42 | Spot 42 RNA | 157 | 2067, 3473, 3661 |
| spoT | 82.3 | Spot (magic spot) | Guanosine 5′-diphosphate, 3′-diphosphate pyrophosphatase, ppGpp synthetase II activity | 156 | 2913, 3070, 3868, 3869, 4807a |
| sppA | 39.8 | Signal peptide peptidase | Protease IV; signal peptide peptidase | 13763 | 1877, 4308 |
| spr | 48.9 | Suppressor of prc | Suppresses thermosensitivity of prc mutants at low osmolality; in turn suppressed by multicopy expression of PBP 7 | 54675 | 1641 |
| srlA | 60.9 | Sorbitol | gutA, sbl; d-glucitol-specific enzyme II of phosphotransferase system | 155 | 3644, 437, 4749, 4822, 4823 |
| srlB | 60.9 | Sorbitol | gutB, sbl; d-glucitol (sorbitol)-specific enzyme III of the phosphotransferase system | 11886 | 4822, 4823 |
| srlD | 60.9 | Sorbitol | gutD, sbl; sorbitol-6-phosphate dehydrogenase (EC 1.1.1.140) | 153 | 4749, 4822, 4823 |
| srlE | 60.9 | Sorbitol | gutE; protein encoded by reading frame adjacent to srlA | 56893 | 3644a |
| slrR | 60.9 | Sorbitol | gutR; regulatory gene for srl | 152 | 4749 |
| srmB | 58.4 | Suppressor ribosomal mutant | rbaB, rhlA; ATP-dependent RNA helicase | 32859 | 3191 |
| srnA | 9.4 | Stable RNA | Degradation of stable RNA | 151 | 3263 |
| ssaE | 52.5 | Suppression secA | Suppresses secA mutations | 17869 | 3285 |
| ssaG | 41.8 | Suppression secA | Suppresses secA mutations | 17866 | 3285 |
| ssaH | 94.1 | Suppression secA | Suppresses secA mutations | 17863 | 3285 |
| ssb | 92.1 | Single-strand binding | exrB, lexC; single-strand DNA-binding protein; member of lex regulon | 150 | 1435, 2237, 2393, 2916, 3568, 3835, 3837, 3917 |
| sseA | 57.1 | Sensitivity to serine | Enhances serine sensitivity (inhibits homoserine deHase) on lactate; rhodanese-like protein | 33023 | 1617 |
| sseB | 57.2 | Sensitivity to serine | Enhances serine sensitivity (inhibits homoserine deHase) on lactate; weaker than sseA | 33026 | 1617 |
| sspA | 72.7 | Stringent starvation protein | pog; stress response protein | 17860 | 1333, 1335, 3979, 3985, 4742 |
| sspB | 72.7 | Stringent starvation protein | Stress response protein | 33594 | 4742 |
| ssrA | 59.3 | Small stable RNA | sipB; 10Sa RNA (nonribosomal); role in modulating DNA-binding proteins | 35783 | 103, 1218, 2310, 2312, 3074, 3251, 3252, 3652, 3653, 692 |
| ssrS | 65.8 | Small stable RNA | 6S RNA | 17857 | 1833, 2459 |
| ssyA | 57.1 | Suppression of secY | Suppressor of secY mutation | 17854 | 4026 |
| ssyD | 3.0 | Suppression of secY | Suppressor of secY mutation | 17848 | 4027 |
| stfZ | 2.3 | fts in reverse | Antisense RNA blocks ftsZ mRNA translation, inhibits cell division | 36124 | 1032 |
| stkA | 77.5 | Suppressor of transposase killing | Suppresses cell aberrations and death caused by Tn5 transposase overexpression | 36083 | 4699 |
| stkB | 86.1 | Suppressor of transposase killing | Suppresses cell aberrations and death caused by Tn5 transposase overexpression | 36093 | 4699 |
| stkC | 98.5 | Suppressor of transposase killing | Suppresses cell aberrations and death caused by Tn5 transposase overexpression | 36087 | 4699 |
| stkD | 28.6 | Suppressor of transposase killing | Suppresses cell aberrations and death caused by Tn5 transposase overexpression | 36090 | 4699 |
| stpA | 60.3 | Suppressor of td mutant | hnsB; hns-like protein, suppresses T4 td mutant | 34492 | 1300, 4024, 4133, 4933–4935 |
| strC | 7.0 | Streptomycin | strB; low-level streptomycin resistance; modifies ribosome structure | 149 | 3688 |
| strM | 78.3 | Streptomycin | asuF?; control of ribosomal ambiguity | 148 | 4272 |
| stsA | 84.7 | Altered RNase activity | 147 | 2493 | |
| sucA | 16.3 | Succinate | lys, met; α-ketoglutarate dehydrogenase (decarboxylase component) | 146 | 3355, 4149, 4741, 4773, 553, 934, 935 |
| sucB | 16.4 | Succinate | lys, met; dihydrolipoamide succinyltransferase component of α-ketoglutarate deHase (EC 2.3.1.61) | 145 | 4149, 4150, 4151, 553 |
| sucC | 16.4 | Succinate | Succinyl-CoA synthetase β-subunit | 17845 | 4150, 4151, 552, 553 |
| sucD | 16.5 | Succinate | Succinyl-CoA synthetase α-subunit | 17842 | 552, 553 |
| sufI | 68.1 | Suppressor of ftsI | sui; suppressor of ftsI | 33437 | 2164 |
| sugE | 94.3 | Suppressor of gro | Suppresses groL mutation and mimics effects of gro overexpression | 34460 | 1512 |
| suhA | 78.4 | Suppressor heat-shock proteins | Induction of heat shock genes | 17839 | 4419 |
| suhB | 57.4 | Suppressor heat-shock proteins | Inositol monophosphate (EC 3.1.3.25) | 32968 | 1910, 1911, 2804, 4870, 675 |
| sulA | 22.0 | Suppressor of lon | sfiA; inhibits cell division and ftsZ ring formation | 144 | 1391, 1857, 1978, 2027, 2984, 794 |
| sup | Suppressor | Various suppressor tRNAs, see Table 2 | |||
| supQ | 12.5 | Suppressor | Uncharacterized suppressor | 129 | 3780 |
| surA | 1.2 | Survival | Affects stationary-phase survival | 30326 | 2446, 2964, 3752, 4440 |
| surE | 61.8 | Survival | ygbC; required for stationary-phase survival | 33215 | 2523 |
| syd | 63.0 | secY-d1 suppression | Interacts with secY | 36980 | 4036 |
| tabC | 86.3 | T4 abortion | Mutants fail to support growth of T4 | 125 | 4329 |
| tag | 80.0 | 3-Methyl adenine glycosylase | 3-Methyladenine DNA glycosylase I, constitutive | 124 | 1184, 2144, 365, 3816, 3957, 4201, 773 |
| talA | 55.5 | Transaldolase | Transaldolase A | 54690 | 4164 |
| talB | 0.2 | Transaldolase | Transaldolase B | 36818 | 4165 |
| tanA | 41.6 | Trehalose, anaerobic | Anaerobic growth on trehalose | 37222 | 2799 |
| tanB | 68.1 | Trehalose, anaerobic | Anaerobic growth on trehalose | 37910 | 2799 |
| tap | 42.4 | Taxis protein | Flagellar regulon; chemotactic membrane receptor for aspartate; methyl-accepting protein IV; peptide receptor | 123 | 2342, 2951, 4101, 4102, 4651, 468 |
| tar | 42.4 | Taxis, aspartate and repellents | cheM; chemotactic signal transducer protein II, methyl accepting; aspartate chemoreceptor | 122 | 2206, 2342, 4101, 4102, 4650, 4651, 468 |
| tas | 64 | Tyrosine auxotrophy suppressor | Suppresses tyrosine requirement of tyrA14 O6 strain | 57111 | 4416a |
| tauA | 8.3 | Taurine | Uptake of taurine (probable S source) | 51775 | 4549, 4550 |
| tauB | 8.3 | Taurine | Taurine transport | 51772 | 4549, 4550 |
| tauC | 8.3 | Taurine | Taurine transport | 51769 | 4549, 4550 |
| tauD | 8.3 | Taurine | Taurine dioxygenase | 51766 | 1131, 4549, 4550 |
| tbpA | 1.6 | Thiamin binding protein | Periplasmic thiamin binding protein | 34691 | 1785 |
| tdcA | 70.4 | Threonine dehydratase, catabolic | Transcriptional activator of tdc operon | 29702 | 1353, 1466, 1600, 3944, 3945, 689 |
| tdcB | 70.3 | Threonine dehydratase, catabolic | tdc, mukA; threonine dehydratase (EC 4.2.1.16); LysR family of regulator proteins | 17587 | 1466, 1467, 1468, 3944, 3945, 950 |
| tdcC | 70.3 | Threonine dehydratase, catabolic | l-Threonine l-serine permease, membrane associated, anaerobically inducible | 29709 | 1466, 3246, 3944, 3945 |
| tdcD | 70.3 | Threonine dehydratase, catabolic | Propionate kinase, anaerobic | 54712 | 1736, 3944 |
| tdcE | 70.2 | Threonine dehydratase, catabolic | Pyruvate formate-lyase/ketobutyrate formate-lyase | 54717 | 1736, 3944 |
| tdcF | 70.2 | Threonine dehydratase, catabolic | Function unknown | 54720 | 1736 |
| tdcG | 70.2 | Threonine dehydratase, catabolic | Anaerobic pathway, l-serine deaminase, l-serine dehydratase | 54723 | 1736, 3944 |
| tdcR | 70.4 | Threonine dehydratase, catabolic | Positive regulatory protein for threonine dehydratase, TdcB | 29692 | 3943, 3944 |
| tdh | 81.7 | Threonine dehydrogenase | Threonine dehydrogenase | 17584 | 141, 3619, 3620, 715 |
| tdi | 4.3 | Transduction inhibition | Affects transduction, transformation, and rates of mutation | 121 | 4181 |
| tdk | 27.9 | Thymidine kinase | Deoxythymidine kinase | 120 | 1763, 1887, 369, 405 |
| tehA | 32.3 | Tellurite resistance | Multidrug resistance; K+-tellurite ethidium and proflavin transport; membrane protein | 32106 | 4372, 4500, 4501, 4645 |
| tehB | 32.3 | Tellurite resistance | Multidrug resistance; K+-tellurite ethidium and proflavin transport | 32109 | 4372, 4500, 4501 |
| TerA | 28.8 | Terminus | Terminus of DNA replication; replication fork inhibition | 17836 | 1294, 1744, 1754, 1756, 1757, 3395, 979 |
| TerB | 36.2 | Terminus | psrB; terminus of DNA replication; replication fork inhibition | 17833 | 1097, 1294, 1744, 1754, 1756, 1757, 281, 3395, 352, 979 |
| TerC | 34.6 | Terminus | psrA; terminus of DNA replication; replication fork inhibition | 17830 | 1294, 1744, 297, 298, 454 |
| TerD | 27.6 | Terminus | Terminus of DNA replication; replication fork inhibition | 17827 | 1294, 1744 |
| TerE | 23.3 | Terminus | Terminus of DNA replication; replication fork inhibition | 29805 | 1746 |
| TerF | 50.0 | Terminus | Terminus of DNA replication; replication fork inhibition | 29289 | 3992 |
| tesA | 11.2 | Thioesterase | apeA; acyl-CoA thioesterase I; also protease I | 31102 | 1878, 737, 738 |
| tesB | 10.2 | Thioesterase | Thioesterase II | 17581 | 3087, 3130 |
| tgt | 9.2 | tRNA-guanine transglycosylase | tRNA-guanine transglycosylase | 118 | 1358, 2651, 3205, 3654, 3655 |
| thdA | 10.5 | Thiophene degradation | Degradation of furans and thiophenes; may be tlnA? | 17824 | 3 |
| thdC | 94.2 | Thiophene degradation | Degradation of furans and thiophenes | 17821 | 3 |
| thdD | 99.8 | Thiophene degradation | Degradation of furans and thiophenes | 15908 | 3 |
| thiA | 90.3 | Thiamin cluster | Hydroxyethylthiazole synthesis, cluster of genes | 117 | 4564 |
| thiB | 90.3 | Thiamin | Unidentified, not THI-P synthase, thiamine pyrophosphate pyrophosphorylase activity | 116 | 4564 |
| thiC | 90.4 | Thiamin (and thiazole) | Hydroxymethylpyrimidine synthesis; thiamin pyridine moiety biosynthesis | 115 | 4564 |
| thiD | 47.2 | Thiamin (and thiazole) | Phosphomethylpyrimidine kinase | 114 | 1905 |
| thiE | 90.3 | Thiamin (and thiazole) | thiA; thiamin-thiazole moiety synthesis | 34301 | 4564 |
| thiF | 90.3 | Thiamin (and thiazole) | thiA; thiamin-thiazole moiety synthesis | 34298 | 4564 |
| thiG | 90.3 | Thiamin (and thiazole) | thiA; thiamin-thiazole moiety synthesis | 34295 | 4564 |
| thiH | 90.3 | Thiamin (and thiazole) | thiA; thiamin-thiazole moiety synthesis | 342952 | 4564 |
| thiI | 9.5 | Thiamin (and thiazole) | Thiamin biosynthesis; also 4-thiouridine generation in tRNAs | 56890 | 3032, 4681a |
| thiJ | 47.1 | Thiamin (and thiazole) | thiA, thiN; hydroxymethylpyrimidine kinase | 37230 | 2977, 3122 |
| thiK | 25.1 | Thiamin (and thiazole) | Thiamin kinase | 113 | 1906, 2425 |
| thiL | 9.4 | Thiamin (and thiazole) | thiJ; thiaminmonophosphate kinase | 112 | 1906, 4681a |
| thiM | 47.2 | Thiamin (and thiazole) | Hydroxyethylthiazole kinase (EC 2.7.1.50) | 34309 | 2976 |
| thrA | 0.0 | Threonine | thrD, Hs; aspartokinase I-homoserine dehydrogenase I (EC 1.1.1.3 and EC 2.7.2.4) | 111 | 1568, 2162, 2682, 4859, 845, 925 |
| thrB | 0.1 | Threonine | Homoserine kinase (EC 2.7.1.39) | 110 | 1568, 3801, 4948, 845, 846 |
| thrC | 0.1 | Threonine | Threonine synthase (EC 4.2.99.2) | 109 | 1568, 3375, 845, 846 |
| thrS | 38.8 | Threonine | Autogenously regulated threonyl-tRNA synthetase (EC 6.1.1.3) | 108 | 1318, 2743, 2829, 3136, 3333, 3461, 3463, 3527, 4168, 4170, 4710, 4801 |
| thrT | 90.0 | Threonine | Threonine tRNA3 | 107 | 1849, 2313, 2467, 3743, 90 |
| thrU | 89.9 | Threonine | Threonine tRNA4 | 106 | 1849, 2313, 2467, 3743, 90 |
| thrV | 73.7 | Threonine | Threonine tRNA1 in rrnD | 105 | 1096, 1146, 2313, 3814, 818 |
| thrW | 5.6 | Threonine | Threonine tRNA2 | 10363 | 2313, 818, 914 |
| thyA | 63.9 | Thymine | Aminopterin, trimethoprim resistance; thymidylate synthetase (EC 2.1.1.45) | 104 | 1107, 1730, 1742, 286, 328, 3762, 3870, 658 |
| tig | 9.8 | Trigger factor | Trigger factor; chaperone | 35681 | 1583, 1737, 1738, 2119, 4236, 4525, 619 |
| tktA | 66.3 | Transketolase | Transketolase (EC 2.2.1.1) | 103 | 2059, 4161, 4162, 4947 |
| tktB | 55.6 | Transketolase | Transketolase (EC 2.2.1.1) | 29992 | 1889, 4947 |
| tldD | 73.0 | Tolerance for letD | Tolerance for effects on DNA gyrase by sex factor F gene letD | 50304 | 3061 |
| tlnA | 10.5 | Thiolutin | tlnI; resistance or sensitivity to thiolutin; may be thdA? | 102 | 4083 |
| tmk | 24.9 | Thymidine kinase | Deoxythymidine kinase | 17578 | 357, 3657, 967, 405 |
| tnaA | 83.8 | Tryptophanase | ind, tnaR; tryptophanase (EC 4.1.99.1) | 101 | 2927, 3170, 3171, 4222, 4611, 4677, 4911, 568, 996 |
| tnaB | 83.8 | Tryptophanase | trpP; low-affinity Trp permease | 69 | 1119, 2102, 3865, 4218, 568, 996 |
| tnm | 92.0 | Tn migration | Transposition of Tn9 and other transposons, development of phage Mu | 100 | 1901, 1902, 4107, 4725 |
| tolA | 16.7 | Tolerance | cim, excC, lky, tol-2; tolerance to group A colicins, single-stranded filamentous DNA phage; required for OM integrity; membrane protein; bacteriocin tolerant | 99 | 1023, 1270, 1284, 2506, 2507, 3039, 4281, 4282, 4397, 4686, 482, 484, 635, 957 |
| tolB | 16.7 | Tolerance | lkyA (leakage of periplasmic proteins), tol-3; azaleucine resistant; tolerance to colicins E2, E, A, and K | 98 | 1270, 1284, 2447, 2449, 2506, 3039, 4281, 4282, 484, 635, 957, 99 |
| tolC | 68.5 | Tolerance | colE1-i, mtcB, refI, weeA, toc (topoisomerase compensation), mukA; specific tolerance to ColE1; affects chromosome segregation; OM porin | 97 | 1284, 1291, 1593, 1594, 1762, 3007, 3009, 3186, 3311, 4648, 484, 4863, 4864, 635, 957, 975 |
| tolD | 22.9 | Tolerance | Bacteriocin tolerant; tolerance to colicins E2, E3, and ampicillin | 96 | 1284, 484, 572, 957 |
| tolE | 22.9 | Tolerance | Bacteriocin tolerant; tolerance to colicins E2, E3, and ampicillin | 95 | 1284, 484, 635, 957 |
| tolI | 0.1 | Tolerance | Bacteriocin tolerant; sensitivity to colicins Ia and Ib | 94 | 484, 635, 957 |
| tolJ | 0.1 | Tolerance | Bacteriocin tolerant; sensitivity to L, A, S4, E, and K | 93 | 1284, 484, 635, 957 |
| tolM | 74.8 | Tolerance | cmt; high-level tolerance to colicin M | 92 | 1284, 1618, 1650, 2865, 3892, 484, 635, 957 |
| tolQ | 16.7 | Tolerance | fii, tolP? tolerance to group A colicins, single-stranded DNA filamentous phage; cell envelope integrity; inner membrane protein | 17815 | 2108, 3038, 3039, 4281, 4282, 4397, 4582, 4686, 481, 482 |
| tolR | 16.7 | Tolerance | Tolerance to group A colicins, single-stranded DNA filamentous phage; cell envelope integrity; inner membrane protein | 17812 | 1721, 2108, 3039, 4282, 4397, 4686, 482 |
| tonB | 28.2 | T-one | T1rec, exbA; sensitivity to T1, φ80, and colicins; uptake of chelated Fe and cyanocobalamin; energy transducer | 90 | 1254, 1470, 1488, 2080, 2135, 2136, 2221, 2502, 251, 3500, 3501, 3502, 4059, 4451, 4452, 482, 484 |
| topA | 28.6 | Topoisomerase | supX; topoisomerase I, omega protein I | 89 | 1091, 2494, 2680, 2762, 3301, 3310, 4206, 4466, 4467, 4482, 4654 |
| topB | 39.7 | Topoisomerase | mutR; topoisomerase III; mutR phenotype is decreased deletion formation between short repeats | 29988 | 1048, 3931, 4653, 4730, 4937 |
| torA | 22.8 | Trimethylamine oxide reductase | Molybdoprotein trimethylamine N-oxide reductase | 17575 | 2874, 3377 |
| torC | 22.8 | Trimethylamine oxide reductase | c-Type cytochrome | 29977 | 2874 |
| torD | 22.9 | Trimethylamine oxide reductase | Transcribed with tor; no similarity with other proteins in database | 29980 | 2874 |
| torR | 22.8 | Trimethylamine oxide reductase | Regulatory gene | 36408 | 4071, 4072 |
| torS | 22.7 | Trimethylamine oxide reductase | Sensor partner of TorRT, two-component system for trimethylamine N-oxide induction | 50902 | 2061 |
| torT | 22.8 | Trimethylamine oxide reductase | TorRT system | 50188 | 2062 |
| tpiA | 88.6 | Triose P isomerase | Triosephosphate isomerase | 88 | 1710, 3423 |
| tpr | 27.7 | tyrT region protamine | Protamine-like protein, apparently transcribed with tyrT | 87 | 75 |
| tpx | 29.9 | Thiol peroxidase | Thioredoxin-linked thiol peroxidase | 46706 | 2225, 668 |
| treA | 26.8 | Trehalose | tre; trehalase, periplasmic | 17572 | 2865, 3651, 430 |
| treB | 96.2 | Trehalose | IITre, translocation system, Tre-specific PTS enzyme II | 34601 | 4253, 431 |
| treC | 96.2 | Trehalose | Trehalose-6-phosphate hydrolase, osmoprotectant | 34597 | 3677, 4253, 431 |
| treF | 79.0 | Trehalose | Cytoplasmic trehalase | 35861 | 1807 |
| treR | 96.2 | Trehalose | Repressor | 51427 | 1807, 1808, 2258 |
| trg | 32.1 | Taxis to ribose and galactose | Methyl-accepting chemotaxis protein III, ribose acceptor, flagellar regulon | 85 | 1646, 1678, 2865, 2991, 362, 363, 419 |
| trkA | 74.0 | Transport K+ | Major constitutive K+ transport system; potassium transport inner membrane protein subunit | 84 | 1161, 1162, 1618, 2865, 3914, 448 |
| trkD | 84.7 | Transport K+ | kup; major constitutive K+ transport system | 81 | 1161, 3910, 449, 4611 |
| trkG | 30.6 | Transport K+ | Major constitutive K+ transport system; membrane protein, perhaps inactive during anaerobic growth | 29963 | 1074, 1162, 3915, 3916, 4453 |
| trkH | 86.9 | Transport K+ | Potassium transport membrane protein subunit; binding of TrkA to membrane; mutants require high K+ | 34066 | 1074, 1162, 3915, 448 |
| trmA | 89.7 | tRNA methyltransferase | rT; tRNA (uracil-5)-methyltransferase (EC 2.1.1.35) | 79 | 1577, 256, 2564, 3229, 3405 |
| trmB | 7.1 | tRNA methyltransferase | tRNA (guanine-7)-methyltransferase (EC 2.1.1.33) | 78 | 2763 |
| trmC | 52.6 | tRNA methyltransferase | 5-Methylaminoethyl-2-thiouridine in tRNA | 77 | 1599, 366 |
| trmD | 59.1 | tRNA methyltransferase | tRNA (guanine-7)-methyltransferase (EC 2.1.1.33) | 76 | 1599, 3407, 594–596 |
| trmE | 83.7 | tRNA methyltransferase | thdF (thiophene oxidation?); tRNA base modification, 5-methyl-aminoethyl, 2-thiouridine synthesis | 17806 | 1147, 49, 568 |
| trmF | 84.8 | tRNA methyltransferase | 5-Methyl aminoethyl-2-thiouridine biosynthesis | 17803 | 1147 |
| trmH | 82.4 | tRNA methyltransferase | spoU; tRNA (Gm18) 2′-O-methyltransferase | 33877 | 2319, 3406 |
| trmU | 25.7 | tRNA methyltransferase | asuE; antisuppressor; tRNA base-modifying enzyme; 2-thiouridine synthesis | 37613 | 1503, 3708, 4272 |
| trnA | 63.1 | tRNA | Level of several tRNAs | 75 | 724 |
| trpA | 28.3 | Tryptophan | Tryptophan synthase subunit A (EC 4.2.1.20) | 74 | 1157, 1470, 1551, 1760, 2819, 3173, 4314, 4788, 4789, 4871 |
| trpB | 28.4 | Tryptophan | Tryptophan synthase subunit B (EC 4.2.1.20) | 73 | 1157, 1470, 1760, 2819, 4871, 865 |
| trpC | 28.4 | Tryptophan | Bifunctional enzyme N-(5-phosphoribosyl)anthranilate isomerase, indole-3-glycerolphosphate synthetase (EC 4.1.1.48) | 72 | 1157, 1470, 1760, 1812, 2819, 4871, 747 |
| trpD | 28.4 | Tryptophan | Bifunctional enzyme anthranilate synthase component II, phosphoribosyl anthranilate transferase; (EC 4.1.3.27 and EC 2.4.2.18) | 71 | 1157, 1470, 1760, 1810, 1812, 2819, 3174, 4871 |
| trpE | 28.4 | Tryptophan | anth, tryD, tryp-4; anthranilate synthase component I (EC 4.1.3.27) | 70 | 1157, 1470, 1703, 1760, 2819, 3026, 3174, 3175, 3321, 4871 |
| trpR | 99.8 | Tryptophan | 5-Methyltryptophan resistance; regulator of trp operon and aroH; autogenously regulated tryptophan repressor protein | 68 | 1567, 1568, 210, 3705, 4078, 4872, 288 |
| trpS | 75.7 | Tryptophan | Tryptophanyl-tRNA synthetase (EC 6.1.1.2) | 67 | 1608, 1609, 410 |
| trpT | 85.0 | Tryptophan | su7 (UGA suppression), supU (UAG suppression), su8, supV (UAA suppression)f; tryptophan tRNA | 66 | 2313, 3324, 4904, 926, 1122a |
| truA | 52.4 | tRNA uridine (modification to pseudouridine | asuC, hisT, leuK; pseudouridine synthase, anticodon stem and loop specific | 623 | 142, 143, 144, 2784, 3211, 3226, 3363, 3955, 4523, 543 |
| truB | 71.3 | tRNA uridine (modification to pseudouridine) | Pseudouridine synthase, ψ55 specific | 35528 | 3226 |
| trxA | 85.4 | Thioredoxin | fipA, tsnC; thioredoxin | 65 | 1865, 2550, 2551, 2764, 2807, 3850, 3772, 3773, 3775, 3778, 3784, 4641, 58, 926 |
| trxB | 20.1 | Thioredoxin | Thioredoxin reductase | 17569 | 1009, 1613, 3774, 3776, 3777 |
| trxC | 58.6 | Thioredoxin | Thioredoxin 2 | 54694 | 2953 |
| tsaA | 4.8 | t6AtRNA-SAM-methyltransferase | N6-threonylcarbamoyladenosine(t6A) modification of tRNAThr, adenosine A37 to threonylated adenosine; tRNA(m6t6A37) methyltransferase | 53741 | 3564 |
| tsf | 4.1 | Ts elongation factor | EF-Ts, elongation factor for transcription, stable | 64 | 2428, 296, 418, 88 |
| tsmA | 40.1 | Thymine suppression modifier | Affects suppression efficiency for nonsense and frameshift mutations of Thy− strains | 36603 | 1731 |
| tsr | 98.9 | Taxis to serine and repellents | cheD; serine chemoreceptor; methyl accepting chemotaxis protein II, membrane receptor | 63 | 1693, 2206, 3367, 4650, 467, 469, 617, 84 |
| tsx | 9.3 | T-six | T6rec, nupA; T6, colicin K resistance; nucleoside channel | 62 | 1635, 2651, 492, 494 |
| ttdA | 69.1 | Tartrate dehydratase | l-Tartrate dehydratase subunit (EC 4.2.1.32) | 33459 | 3628 |
| ttdB | 69.1 | Tartrate dehydratase | l-Tartrate dehydratase subunit (EC 4.2.1.32) | 33462 | 3628 |
| ttk | 82.2 | Function unknown, TetR, AcrR family of regulators | 52724 | 1144, 568 | |
| tufA | 74.7 | Tu elongation factor | kirT, pulT; duplicate gene for EF-Tu subunit; elongation factor, unstable | 61 | 1561, 2042, 2334, 2379, 2433, 4342, 4543, 4548, 4732, 4887, 4928, 91 |
| tufB | 90.0 | Tu elongation factor | kirT, pulT; duplicate gene for EF-Tu subunit; elongation factor, unstable | 60 | 1561, 1849, 2334, 2433, 2467, 2567, 2972, 4336, 4342, 4377, 4543, 4546, 4548, 4732, 89, 90 |
| tus | 36.3 | Terminus utilization substance | tau; inhibits replication at Ter; Ter DNA-binding protein, blocking replication forks; DNA sequence-specific contrahelicase | 17800 | 1745, 1756, 1758, 2207, 2279, 2355, 2473, 3395, 3704, 941 |
| tynA | 31.2 | Tyramine | feaA, maoA; tyramine oxidase (EC 1.4.3.4) | 59 | 178, 3066, 4200, 4848 |
| tyrA | 59.0 | Tyrosine | Chorismate mutase T-prephenate dehydrogenase (EC 5.4.99.5 and EC 1.3.1.12) bifunctional; TyrR regulon | 58 | 1847, 2819 |
| tyrB | 91.9 | Tyrosine | Tyrosine aminotransferase (EC 2.6.1.5); TyrR regulon | 57 | 1283, 2372, 4856 |
| tyrP | 42.8 | Tyrosine | Tyrosine-specific transport system, TyrR regulon | 56 | 2154, 2155, 4720, 4781, 4782 |
| tyrR | 29.8 | Tyrosine | TyrR regulon repressor; regulates aroF, aroG, tyrA, and aromatic amino acid transport; autoregulation of tyrR not mediated by tyrosine | 55 | 2436, 2819, 3441, 4640, 4740, 4855, 4857, 531, 623, 734, 785, 837, 838, 881, 882 |
| tyrS | 36.9 | Tyrosine | Tyrosyl-tRNA synthetase (EC 6.1.1.1) | 54 | 1043, 236, 2398, 2938 |
| tyrT | 27.7 | Tyrosine | tyrTα, suIII, Su-3, Su-4, supC (ochre suppressorf), supF (amber suppressor), supO? tandemly duplicated tyrosine tRNA1 (tyrTV) | 53 | 2313, 3743, 3744, 445, 446, 447 |
| tyrU | 90.0 | Tyrosine | sup15B, supM (ochre suppressor), supZ (amber suppressorf); tyrosine tRNA2 | 52 | 1122a, 1849, 2313, 2467, 3743, 90 |
| tyrV | 27.7 | Tyrosine | tyrTβ; tandemly duplicated, tyrosine tRNA1 | 51 | 3743, 1122a |
| ubiA | 91.6 | Ubiquinone | 4-Hydroxybenzoate polyprenyltransferase | 50 | 2549, 2576, 3195, 725 |
| ubiB | 86.7 | Ubiquinone | fadI, fre, fsrC; 2-octaprenylphenol→2-octaprenyl-6-methoxyphenol; flavin reductase (EC 1.6.99.–)? | 49 | 1273, 1385, 4174, 926 |
| ubiC | 91.6 | Ubiquinone | Chorismate lyase | 48 | 2549, 3176, 4124 |
| ubiD | 86.7 | Ubiquinone | Reaction: 3-octaprenyl-4-hydroxybenzoate to 2-octaprenylphenol | 47 | 858 |
| ubiE | 86.7 | Ubiquinone | Reaction: 2-octaprenyl-6-methoxy-1,4-benzoquinone to 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinone | 46 | 4902 |
| ubiF | 14.8 | Ubiquinone | Reaction: 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinone to 2-octaprenyl-3-methyl-5-hydroxy-6-methoxy-1,4 benzoquinone | 45 | 811 |
| ubiG | 50.4 | Ubiquinone | Reaction: 1-octaprenyl-3-methyl-5-hydroxy-6-methoxy-1,4-benzoquinone to ubiquinone 8 | 44 | 1421, 1829, 3451 |
| ubiH | 65.8 | Ubiquinone | visB; reaction: 2-octaprenyl-6-methoxyphenol to 2-octaprenyl-6-methoxy-1,4-benzoquinone | 43 | 3100, 2974 |
| ubiX | 52.3 | Ubiquinone | dedF; sequence homologous to ubiX of S. typhimurium, which codes for 3-octaprenyl 4-hydroxybenzoate carboxy-lyase | 17797 | 3211 |
| ucpA | 54.8 | Upstream cysP | Temporary name; protein with homology to short-chain dehydrogenases/reductases | 51898 | 4080 |
| udk | 46.1 | Uridine kinase | Uridine/cytidine kinase (EC 2.7.1.48) | 42 | 2058, 3156, 3157 |
| udp | 86.5 | Uridine phosphorylase | Uridine phosphorylase (EC 2.4.2.3) | 41 | 1272, 3426, 3539, 58, 66, 926, 4646 |
| ugpA | 77.3 | Uptake glycerol phosphate | psiB, psiC; glycerol P transport system, integral membrane protein | 40 | 2147, 2931, 3320, 3539, 3946, 3948, 3949, 4261 |
| ugpB | 77.4 | Uptake glycerol phosphate | psiB, psiC; periplasmic binding protein of sn-glycerol-3-phosphate transport system | 39 | 2147, 2931, 3320, 3946, 3948, 3949, 4261 |
| ugpC | 77.3 | Uptake glycerol phosphate | sn-Glycerol-3-P transport system; ABC family permease | 17794 | 1701, 2147, 3320, 3948, 3949, 4261 |
| ugpE | 77.3 | Uptake glycerol phosphate | sn-Glycerol-3-P transport system; membrane protein | 17791 | 2147, 3320, 3948, 3949, 4261 |
| ugpQ | 77.3 | Uptake glycerol phosphate | Glycerophosphoryl diester phosphodiesterase (EC 3.1.4.-); cytosolic | 33697 | 548, 4261, 4428 |
| uhpA | 82.9 | Utilization hexose phosphate | Response regulator (2-component system) required for uhpT transcription | 15437 | 1316, 1933, 2079, 4006, 4717 |
| uhpB | 82.9 | Utilization hexose phosphate | Membrane protein controlling UhpA activity, sensor kinase | 15448 | 1316, 1933, 1934, 4717 |
| uhpC | 82.9 | Utilization hexose phosphate | Membrane protein controlling UhpA activity, in concert with UhpB | 15449 | 1316, 1933, 1934, 2079, 4006, 4717 |
| uhpT | 82.8 | Utilization hexose phosphate | Fosfomycin sensitivity; sugar P transport system; transport protein for hexose P’s | 37 | 1179, 1316, 1581, 2079, 2902, 2931, 4005, 4006, 4545, 4571, 4717 |
| uidA | 36.5 | HexUronIDes | gurA, gusA; β-d-glucuronidase (EC 3.2.1.31) | 36 | 2000, 386, 387, 3905 |
| uidB | 36.5 | HexUronIDes | gusB, uidP; glucuronide permease | 53467 | 33, 394 |
| uidR | 36.5 | HexUronIDes | gusR; regulatory gene | 35 | 384, 386, 387, 388 |
| umuC | 26.5 | UV mutator | uvm; UV induction of mutations, error-prone repair; forms complex with UmuD and UmuD′ | 34 | 1068, 1145, 1297, 2245, 2776, 3404, 4041, 4778, 4779, 2284 |
| umuD | 26.5 | UV mutator | uvm; inducible mutagenesis; error-prone repair; processed to UmuD′; UmuDC single-stranded binding protein with RecA-coated DNA; SOS | 17788 | 1068, 1145, 1297, 2245, 2281, 2776, 3404, 3960, 4041, 2284 |
| ung | 58.5 | Uracil nucleic acid glycosylase | Uracil-DNA-glycosylase | 25 | 4567 |
| upp | 56.4 | Uracil PRTase | uraP; uracil phosphoribosyltransferase (EC 2.4.2.9) | 24 | 1621, 1799, 97 |
| ups | 27.1 | Up-suppression? | Efficiency of nonsense suppressors; see prfA | 23 | 3782, 854 |
| uraA | 56.4 | Uracil | Uracil concentration dependence of pyr mutants; Ura ABC transporter | 33043 | 96 |
| uro | 85.9 | Uroporphyrinogen | hemC-Y operon | 37849 | 3871, 60 |
| usg1 | 52.5 | Upstream gene, temporary | Temporary name for gene in pdxB operon, unknown function | 3274,6 | 144 |
| ushA | 10.9 | UDP sugar hydrolase | UDP-glucose hydrolase (5′-nucleotidase) | 22 | 574, 854 |
| uspA | 78.4 | Universal stress protein | Global regulatory gene for stress response | 28127 | 1046, 1302, 3232, 3233 |
| uup | 21.7 | Precise excision of insertion elements | 17785 | 1799, 3631 | |
| uvh | 90.2 | UV hyperresistant to UVC | Resistance to UVC, peroxide and antibiotics; may affect SOS repair | 51073 | 21 |
| uvrA | 92.0 | Ultraviolet resistant | dar; lex regulon; repair of UV damage to DNA; excision nuclease subunit A | 21 | 1867, 2201, 3831, 3834, 3837, 3838, 3479 |
| uvrB | 17.5 | UV resistant | dar-1, dar-6; DNA repair; excision nuclease subunit B, ATPase I and helicase II | 20 | 1268, 128, 191, 3345, 1831, 3836, 3838, 3839, 4319, 4547 |
| uvrC | 42.9 | UV resistant | DNA repair; multicopy causes mucoidy; excision nuclease subunit C | 19 | 1277, 1278, 2995, 3220, 3831, 3832, 3838, 3840, 3994, 3995, 3996, 4493, 4559, 4886 |
| uvrD | 86.1 | UV resistant | dar-2, dda, mutU, pdeB, rad, recL, srjC, uvr502, uvrE; DNA-dependent ATPaseI-DNA helicase II; increased rates of spontaneous mutagenesis | 18 | 1109, 1250, 1251, 146, 1741, 2368, 2642, 2750, 2883, 2885, 3242, 3243, 4051, 4360, 4672, 4673, 4838, 4936, 521, 58, 926 |
| uvs | 91.1 | UV sensitivity | UV-sensitive mutants, locus linked to uvr | 51076 | 21 |
| uxaA | 69.8 | Utilization hexuronate galacturonate | Altronate hydrolase (EC 4.2.1.7) | 17 | 1851, 3489, 3682, 892 |
| uxaB | 34.6 | Utilization hexuronate galacturonate | Altronate oxidoreductase (EC 1.1.1.58); exu regulon | 16 | 1854, 2796, 381, 383, 394 |
| uxaC | 69.9 | Utilization hexuronate galacturonate | Uronate isomerase (EC 5.3.1.12) | 15 | 1851, 2795, 2796, 3489, 3682, 382, 892 |
| uxuA | 98.1 | Utilization hexuronide glucuronate | Mannonate hydrolase (EC 4.2.1.8) | 14 | 1305, 3681, 3687, 385 |
| uxuB | 98.1 | Utilization hexuronide glucuronate | Mannonate oxidoreductase (EC 1.1.1.57) | 13 | 1304, 3681, 3687, 385 |
| uxuR | 98.1 | Utilization hexuronide glucuronate | Regulatory gene for uxuBA operon | 12 | 3680, 3681, 3687 |
| valS | 96.5 | Valine | val-act; valyl-tRNA synthetase (EC 6.1.1.9) | 11 | 1660, 1691, 1692, 197, 4091 |
| valT | 16.8 | Valine | valTα; duplicate gene with triplicated valUXY; valine tRNA1 | 10 | 2313, 3324, 4898 |
| valU | 54.3 | Valine | valUα, valUγ; tandemly triplicate valUXY; valine tRNA1 | 17782 | 2313, 3238, 538, 539 |
| valV | 37.6 | Valine | Valine tRNA2B | 17563 | 2313 |
| valW | 37.6 | Valine | Valine tRNA2A | 17560 | 2313 |
| valX | 54.3 | Valine | valUγ; tandemly triplicate valUXY; valine tRNA1 | 28696 | 3238, 539 |
| valY | 54.3 | Valine | valUγ; tandemly triplicate valUXY; valine tRNA1 | 28699 | 3238, 539 |
| valZ | 16.8 | Valine | valTβ; valine tRNA1 | 51028 | 394 |
| vsr | 43.7 | Very short (patch) repair | Repairs mismatches; minimizes effects of C methylation | 28927 | 1282, 2540, 2541, 2697, 336, 4130, 1720 |
| wrbA | 23.0 | W(Trp) repressor binding | Affects association between Trp repressor and operators in stationary phase | 31836 | 4865 |
| xapA | 54.4 | Xanthosine P | pndA; xanthosine phosphorylase | 41105 | 539 |
| xapB | 54.3 | Xanthosine P | Xanthosine transport protein, NupG-like | 41101 | 3962 |
| xapR | 54.3 | Xanthosine P | pndR; regulatory gene for xapA | 8 | 2282, 3962, 539, 586 |
| xasA | 33.8 | Extreme acid survival | Glutamate-dependent enzyme, may function in protection against cytoplasmic acidification | 36518 | 1732 |
| xerC | 86.1 | cer-specific recombination | Recombinase, site specific | 30184 | 2841, 376, 377, 378, 812 |
| xerD | 41.7 | cer-specific recombination | xprB; recombinase, site specific | 30201 | 2639, 2641, 376, 377 |
| xni | 63.1 | Exonuclease nine | exo; exonuclease IX, a 3′–5′ exonuclease acting preferentially on single-stranded DNA, probable excision repair function | 56869 | 3983a |
| xseA | 56.7 | Exonuclease VII | Exonuclease VII, large subunit | 6 | 4535, 687 |
| xseB | 9.5 | Exonuclease VII | Exonuclease VII, small subunit | 17557 | 4535, 4536 |
| xthA | 39.4 | Exonuclease III | Exonuclease III | 7 | 3711, 3858, 4030 |
| xylA | 80.3 | Xylose | d-Xylose isomerase (EC 5.3.1.5) | 5 | 2377, 2437, 2741, 3735, 3896, 4128, 502 |
| xylB | 80.3 | Xylose | Xylulokinase (EC 2.7.1.17) | 4 | 2741, 3735, 4128, 502 |
| xylE | 91.4 | Xylose | Xylose-proton symport | 17776 | 1295, 2376, 4277, 961, 962 |
| xylF | 80.4 | Xylose | xylT; xylose-binding protein, transport system | 17773 | 2377, 3735, 4128, 4277, 961 |
| xylG | 80.4 | Xylose | Xylose transport gene | 33780 | 4128 |
| xylH | 80.4 | Xylose | Xylose permease protein | 33783 | 4128 |
| xylR | 80.5 | Xylose | Regulatory gene | 3 | 2741, 3735, 4128 |
| yhhP | 77.7 | Temporary name | sirA; provisional name for small protein required for cell growth | 55460 | 4847 |
| yihG | 87.1 | Temporary name | Poly(A) polymerase II; see also pcnB | 50490 | 631 |
| yjaB | 90.8 | Temporary name | f147; temporary names for protein transcribed divergently from a promoter that overlaps the metA promoter, divergent from metA | 51779 | 1414 |
| yohF | 47.9 | Temporary name | RpoS regulon, acid sensitive, induced by ALS inhibition, similarity to insect-type ADH | 55069 | 4552 |
| zipA | 54.5 | ftsZ-interacting protein | Essential gene, affects cell division and growth; septal ring structural protein | 51069 | 1602 |
| zntA | 77.7 | Zn translocation | Zn2+ translocating P-type ATPase; hypersensitive to Zn and Cd salts | 53884 | 3650 |
| znuA | 41.8 | Zn uptake | High-affinity ABC transport system for zinc | 52879 | 3381 |
| znuB | 41.8 | Zn uptake | High-affinity ABC transport system for zinc | 52882 | 3381 |
| znuC | 41.8 | Zn uptake | High-affinity ABC transport system for zinc | 52886 | 3381 |
| zur | 91.8 | Zn uptake regulation | Regulatory gene for high-affinity transport of zinc | 52889 | 3381 |
| zwf | 41.7 | Zwischenferment | Glucose 6-phosphate dehydrogenase (EC 1.1.1.49) | 2 | 1211, 3756a, 643a, 828a |
The following abbreviations are used in the table: ACP, acyl carrier protein; ad, adenine; PFL, pyruvate-formate lyase; PRPP, phosphoribosyl pyrophosphate; DAHP, 3-deoxy-d-arabinoheptulosonic acid 7-phosphate; BPC, binding protein carriers; FHL, formate hydrogen lyase; EPEC, enteropathogenic E. coli; ABC, ATP binding cassette, PEP, phosphoenolpyruvate; CMP, cytidine monophosphate; PTS, phosphotransferase system; cAMP, cyclic AMP; ORF, open reading frame; DMSO, dimethyl sulfoxide; PDI, protein disulfide isomerase; DTT, dithiothreitol; SDS, sodium dodecyl sulfate; CoA, coenzyme A; SRP, signal recognition particle; Me, methyl; OMP, outer membrane protein; RNAP, RNA polymerase; B. subtilis, Bacillus subtilis; GABA, γ-aminobutyric acid; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; FPA, fluorophenyl alanine; QOR, quinone oxidoreductase; PNP, purine nucleoside phosphorylase; GS, glutamine synthetase; GSH, glutathione; HCA, hydrocinnamic acid; IGP, imidazole glycerol phosphate; FMN, flavin mononucleotide; MPT, molybdopterin; QSR, phage QSR; LPS, lipopolysaccharide; FAD, flavin adenine dinucleotide; SAM, S-adenosylmethionine; SOD, superoxide dismutase; streptovar., streptovarycin; rif., rifampin (rifamycin); streptolyd., streptolydigin; NAL, nalidixic acid; TET, tetracycline; AMP, ampicillin.
Not shown on map since characterization was published very recently.
Letters used as references are identified as follows: A, Table 1 of reference 187; B, Table 1 of reference 189; C, Table 1 of reference 190; D, Table 1 of reference 190a; E, Table 1 of reference 323.
These are the unique identifiers of this site in the CGSC database. Although the database is easily queried by name or synonym, this unchanging identifier is useful in tracking gene name changes that may have resulted in homonyms among the alternative names or in following a complicated history of name changes.
Also used for “antisuppressor” phenotype for alleles at various known (or subsequently renamed) loci.
Only single-mutation suppression is indicated; double and triple mutations suppress other nonsense triplets. See reference 1122a.
TABLE 2.
Alternate gene symbols
| Synonyma | Gene symbol from Table 1 |
|---|---|
| abrD | rpoS |
| acpS | acpX |
| acrB | gyrB |
| acrE | acrB |
| acrS | envR |
| act | alaS, glyS, pflA |
| adec | purN |
| ade3 | purE |
| adef | purE |
| adeg | purC |
| adeub | purF |
| adhC | adhE |
| adi | adiA |
| ads | folD |
| adtha | purD |
| aeg-46.5 | ccm-nap operon |
| aidA | alkA |
| aidC | nrfA, nrfG |
| aidD | alkB |
| air | aer, aegA? |
| ala-act | alaS |
| alaWβ | alaX |
| alaWα | alaW |
| ald | fbaA, aldA |
| alnA | dadB |
| alnR | dadQ |
| alt | rpoD |
| alu | dpp |
| ampA | ampC |
| ams | rne |
| amt, amtA | cysQ |
| ana | adhE |
| anr | osmE |
| ant | nhaA, mutS |
| anth | trpE |
| apbC | mrp |
| apeA | tesA |
| apk | lysC |
| appR | rpoS |
| arg+ura | carA, carB |
| Arg1 | argA, argD |
| Arg2 | argA, argC |
| Arg4 | argE |
| Arg5 | argF |
| Arg6 | rargG |
| argA | argE |
| argB | argA |
| argD | argF |
| argE | argG |
| argG | argD |
| argH | argC |
| argVα | argV |
| argVβ | argY |
| argVγ | argZ |
| aroR | aroT |
| arsE | arsR |
| arsF | arsB |
| arsG | arsC |
| asmB | lpxC |
| asp | ppc |
| aspB | gltB, D |
| astC | cstC |
| asuA | prfA? |
| asuB | rpsL |
| asuC | truA |
| asuD | lysS |
| asuE | trmU |
| asuF | strM? |
| ata | attP22 |
| avr | ileR |
| Az | atoC |
| azi | secA |
| bfe | btuB |
| bglC | bglF, G |
| bglD | bglA |
| bglE | bglT |
| bglS | bglG |
| bglY | hns |
| bicA, bicR | bcr |
| bioB | bioH |
| bioR | birA |
| bir, birB | bioP |
| bisA | moaA |
| bisB | moeA |
| bisD | mog |
| blgA | bglB |
| blgB | bglF |
| blu | malP, pgl, pgm |
| brnP | ilvH |
| bypA1 | infA? |
| cadR | lysP |
| cap | crp, carA, carB |
| capR | lon |
| car | ptsG |
| carP | pepA |
| cat | ptsG |
| cbr | fepA |
| cbt | fepA |
| cdfA | cfa |
| cfxB | marA? R |
| cel | ispB |
| cer | btuB, rnhA |
| cet | creD |
| cfxB | marR, marA |
| chbA | celC |
| chbB | celA |
| chbF | celF |
| chbR | celD |
| cheC | fliM |
| cheD | tsr |
| cheM | tar |
| cheX | cheR |
| chl | moe |
| chlA | moaA |
| chlB | mobA |
| chlC | narG, H |
| chlD | modC, E, F |
| chlE | moeA |
| chlF | fdhF |
| chlG | mog |
| chlI | narI |
| chlJ | modB |
| chlM | moaD |
| chlN | moeB |
| chpAI | chpR |
| chpAK | chpA |
| chpBI | chpS |
| chpBK | chpB |
| cim | tolA |
| clpY | hslU |
| cmlA | cmr? |
| cmlB | ompF |
| cmt | tolM |
| coa | ompF |
| colE1-i | tolC |
| con | ompA |
| cop | het |
| corB | mgtA |
| cou | gyrB |
| cqsB | rrnC |
| cqsD | rrnD |
| cqsE | rrnB |
| CR | ptsG |
| cra | fruR |
| cru | nupC |
| cry | ompF, R |
| csdA | deaD |
| csi-5 | osmY |
| csm | crp |
| ctr | ptsH, I |
| cur | hns |
| cutA1 | cutA |
| cutA2 | dipZ |
| cvc | fabF |
| cxr | cxm |
| cybA | sdhC |
| cycY | cutA |
| cycZ | dipZ |
| cysZ | cysK |
| cyxA | appC |
| cyxB | appB |
| dadR | dadA |
| dagA | cycA |
| dap | asd |
| dar | uvrA |
| dar-1 | uvrB |
| dar-2 | uvrD |
| dar-6 | uvrB |
| dasC | rep |
| dasF | rnhA |
| dcrA | sdaC |
| dda | uvrD |
| ddl | ddlB |
| dec | fadR |
| dedB | accD |
| dedE | cvpA |
| dedF | ubiX |
| deg | lon |
| dga | murI |
| dhbB | birA |
| dhl | lpd |
| dhnA | fbaB |
| dig | hca |
| dinA | polB |
| dinH | ftsK regulatory region |
| dinP | dinB |
| dir | lon |
| div | flk |
| divA | ftsA |
| divE | serT |
| DLP12 | qsr′ |
| dnaD | dnaC |
| dnaF | nrdA |
| dnaL | ligA |
| dnaP | dnaG |
| dnaR | prs |
| dnaS | dfp, dut |
| dnaW | adk |
| dnaY | argU |
| dnaZ | dnaX |
| dniR | mltD |
| dpeA | hupB |
| dpeB | rpoS |
| dpj | acpS |
| dra | deoC |
| drc | hns |
| drdX | hns |
| drm | deoB |
| drp | proS |
| drs | hns |
| dsbD | dipZ |
| dsbE | ccmG |
| dut | dfp |
| dye | arcA |
| ecfB | cpxA |
| efg | nadE |
| emrR | mprA |
| envA | lpxC |
| envB | mreB |
| envC | acrE |
| envD | acrF |
| envM | fabI |
| envZ | ompR |
| eps | rpsE |
| eryA | rplD |
| eryB | rplV |
| eup | cpxA |
| exbA | tonB |
| excC | pal, tolA |
| exo | xni |
| exrA | lexA |
| exrB | ssb |
| fabC | fabB |
| fabE | accB |
| fabG | accC |
| fabJ | fabF |
| fadI | ubiB |
| fam | rpoH |
| far | fusA |
| fda | fbaA |
| fdhA | selA, B |
| fdhC | selC |
| fdp | fbp |
| fdv | mutS |
| feaA | tynA |
| feeB | leuW |
| feuA | cirA |
| fexA | arcA |
| fii | tolQ |
| fimD | fimA |
| fimG | hns |
| fipA | trxA |
| fir | lpxD |
| firA | hlpA, ileR, lpxD |
| fit | infC? |
| flaA | fliM |
| flaAI | fliL |
| flaBI | fliF |
| flaBII | fliG |
| flaBIII | fliH |
| flaC | fliI |
| flaD | fliA |
| flaE | fliK |
| flaF | fliC |
| flaG | flhB |
| flaH | flhA |
| flaI | flhC |
| flaJ | motA, B |
| flaK | flgE |
| flaL | flgG |
| flaM | flgI |
| flaN | fliE |
| flaO | fliJ |
| flaP | fliR |
| flaQ | fliQ |
| flaR | fliP |
| flaS | flgK |
| flaT | flgL |
| flaU | flgA |
| flaV | flgD |
| flaW | flgC |
| flaX | flgF |
| flaY | flgH |
| flaZ | flgJ |
| flbA | flgB |
| flbB | flhD |
| flbC | fliD |
| flbD | fliO |
| flr | leuJ |
| flrA | ileR |
| fms | def |
| fpg | mutM |
| fpk | fruK |
| frdB | fnr |
| frdR | narL, narX |
| fre | ubiB |
| fruC | fruR |
| fruF | fruB, fruK |
| fsrB | hmp |
| fsrC | ubiB |
| ftsB | nrdB |
| ftsH | hflB |
| ftsM | serU |
| ftsR | rpoB |
| ftsS | ftsX |
| fucC | fucA |
| gad | gapA, C |
| gadS | gadA? |
| galB | galT |
| gapB | epd |
| gen-165 | ftn |
| genA | dcuA |
| genF | dcuB |
| genR | lrhA |
| glcF | glcE |
| glmD | nagB |
| glnF | rpoN |
| glnR | glnL |
| glnT | glnG |
| glnUα, β | glnU, W |
| glnVα, β | glnV, X |
| glr | murI |
| glu | ppc |
| glut | gltA |
| gly | glyS |
| glyD | gpt, glpD |
| glySα, β | glyQ, S |
| gmhA | lpcA |
| gntM | gntT |
| gpp | gpt |
| gppB | aslA, B |
| gpt | ptsG |
| gptB | manX, manZ |
| groEL | groL |
| groES | groS |
| gro | dnaK |
| groN | rpoB |
| groNB | nusB |
| groP | dnaB, J, K |
| groPAB | dnaK |
| groPC | dnaK |
| groPF | dnaK |
| grpA | dnaB |
| grpC | dnaJ, K |
| grpD | dnaB |
| grpF | dnaK |
| gsa | hemL |
| gsr | crr |
| gts | fabI |
| guaR | guaB |
| gurA | uidA |
| gusA, B, R | uidA, B, R |
| gutA–E, R | srlA–E, R |
| gxu | gpt |
| gyrI | sbmC |
| hag | fliC |
| hdh | groE |
| herA | rnhA |
| herC | lysS |
| hga | eda |
| hhoA | degQ |
| hhoB | degS |
| hid | ihfA |
| himA | ihfA |
| himB | gyrB |
| himD | ihfB |
| hin | rpoH |
| hip | ihfB |
| hisE | hisI |
| hisT | hisR, truA |
| hisU | gyrB |
| hisW | gyrA |
| hnsB | stpA |
| hobH | seqA? |
| hom | asd |
| hopD | hupB, gsp* |
| hopE | recD |
| hopG | gsp* |
| Hpr, hpr | ptsH |
| hpr | ptsH |
| hrbA | brnQ |
| hrbB | livG, H, J, K |
| hrbC | livG, H, J, K |
| hrbD | livG, H, J, K |
| Hs | thrA |
| hs | hsdM, R |
| hsdH | hdhA |
| hslF | ldhA |
| hslI | ldhA |
| hslS | ibpB |
| hslT | ibpA |
| hsm | hsdM |
| hsp | hsdM, R |
| hsr | hsdR |
| hss | hsdS |
| htpE | ibpB |
| htpG21.0 | hslO-R |
| htpH | ldhA |
| htpI | hslU |
| htpN | ibpA |
| htpO | hslV |
| htpR | rpoH |
| htpY | htgA |
| htrA | degP |
| htrH | degS |
| htrM | rfaD |
| htrP | ribB |
| hyd | nik |
| hydB | hypE |
| hydC | hybA, nikA–D |
| hydD | nikA–E |
| hydF | hypD |
| hydL | hybA |
| iarA | dsbA |
| iarB | dsbB |
| icc | cpdA |
| icdB | gltA |
| icdE | icd |
| iciA | argP |
| icl | aceA |
| iex | crr |
| ihb | lrp |
| ile | ilvA |
| ileS | lspA |
| ind | tnaA |
| ins | glyV, W |
| int (qsr′) | intD |
| irk | hns |
| K12 | rpsG |
| kac | kdpA, B, C, D |
| katF | rpoS |
| kdgA | eda |
| kga | eda |
| kim | qin |
| kirT | tufA, B |
| kup | trkD |
| lcs | asnS |
| lct | lldD |
| lctD | lldD |
| lctP | lldP |
| lctR | lldR |
| ldh | dld |
| leuK | truA |
| leuVα, β, γ | leuV, P, Q |
| lexB | recA |
| lexC | ssb |
| lig | ligA |
| lipP | nike? |
| lir | acrA |
| livR | lrp |
| lky | tolA, B |
| lkyA | tolB |
| lnt | cutE |
| lolB | hemM |
| LopC | clpX |
| LopP | clpP |
| lov | argS |
| lovB | alaS |
| loxB | attP1,P7 |
| lrs | lrp |
| lss | lrp |
| lstR | lrp |
| luxH-like | ribB |
| lysTα, β, γ | lysT, W, Y |
| mac | maa |
| malB | malK |
| maoA | tynA |
| maoB | feaR |
| mas | aceB |
| mazE | chpR |
| mazF | chpA |
| Mb | acrA |
| mbf | lrp |
| mbl | acrA |
| mbrA | rep |
| mcb | pmbA |
| mclA | rseA |
| mdaA | nfsA |
| mdfA | cmr |
| mdrA | cydC |
| mdrH | cydC |
| mec | dcm |
| mel-4 | melB |
| mel-7 | melA |
| meoA | ompC |
| met-1 | metB |
| metM | metL |
| metTβ | metU |
| metZβ | metV |
| mglD | galS |
| mglP | mglA, C |
| mhpS | mhpD |
| micA | mutY |
| mlc | dgsA |
| mlpA | lpp |
| mmrA | rep, rhlB? |
| mni | manC |
| modR | modE |
| molA | malM |
| momR | oxyR |
| mon | mreB |
| mopA | groS |
| mopB | groL |
| mor | oxyR |
| motD | fliN |
| mpt | manX, Z |
| mra | murF |
| mrbA | murA |
| mrc | lpcB |
| mrsA | glmM |
| mrsC | hflB |
| msgA | ftsN |
| msmA | dksA |
| msmB | cspC |
| msmC | cspE |
| msp | arcA |
| mssA | cmk |
| mssB | deaD |
| msuA | dadX? |
| msyA | hns |
| mtcA | acrA |
| mtcB | tolC |
| muc | lon |
| mukA | tolC |
| mukE | kicA |
| mukF | kicB |
| murZ | murA |
| mutA | glyV |
| mutC | glyW |
| mutD | dnaQ |
| mutH | dnaX |
| mutR | topB |
| mutU | uvrD |
| mvrA | fpr |
| mvrC | emrE |
| nagR | nagC |
| nalA | gyrA |
| nalC | gyrB |
| nalD | gyrB |
| nam | pncA |
| narA | moaA |
| narB | mobA |
| narC | narG |
| narD | modC, E, F |
| narR | narL, X |
| nbp | fis |
| ncf | hemB |
| neaA | rpsQ |
| nek | rimK |
| nfrC | rffE |
| nfsB | nfnB |
| nfsI | nfnB |
| nfxA | gyrA |
| nfxC | marA |
| nfxD | parE |
| nic | nadB |
| nirA | fnr |
| nirR | fnr |
| nitA | rho |
| nitB | rpoB |
| nlp | sfsB |
| nlpE | cutF |
| nmpA | pst, pstS |
| nmpB | phoR |
| norA | gyrA |
| norB | marA? |
| nov | cls |
| ntr | nfnB |
| ntrA | rpoN |
| ntrB | glnL |
| ntrC | glnG |
| ntrL | nadE |
| nucR | deoR |
| nupA | tsx |
| nur | rpoS |
| nusD | rho |
| nusE | rpsJ |
| oldB | fadB |
| ole | fadR |
| ompB | envZ, ompR |
| ompE | phoE |
| ompH | hlpA |
| omsA | lpxD |
| opdA | prlC |
| oppI | lrp |
| optA | dgt |
| osmZ | hns |
| osrA | proU |
| ossA | fnr |
| ossB | gltB, D, F |
| ostA | imp |
| padA | feaB |
| panK | coaA |
| papA | atpA |
| papC | atpG |
| papD | atpB, D |
| papE | atpH |
| papF | atpF |
| papG | atpC |
| papH | atpE |
| par | ompC |
| parB | dnaG |
| parD | gyrA |
| parvA | ppiC |
| paxA | dcd |
| pbpA | mrdA |
| pbpB | ftsI |
| pbpF | mrcB |
| pcbA | gyrB |
| pcsA | dinD |
| pdeB | uvrD |
| pdeC | ligA |
| pdxC | serC |
| pdxF | serC |
| pdzA | rplT |
| pea | secA |
| pel | manY |
| perA | envZ |
| pexB | dps |
| pfv | dacA |
| pgsB | lpxB |
| phd | hcaA, B |
| phe-act | pheS |
| pheR | pheU |
| pheW | pheU |
| phmA | nmpC |
| phoM | creC |
| phoM-orf2 | creB |
| phoR1 | phoR |
| phoR2a | pstS |
| phoR2b | pstA |
| phoRc | phoB |
| phoS | pstS |
| phoT | phoB, U, pstA, B |
| phoW | pstC |
| phrA | modF |
| phs | rpoA |
| phsE? | dacD |
| pil | fim, fimB, C, D |
| pilA | fimA |
| pilB | fimC |
| pilC | fimD |
| pilD | fimF, G |
| pilE | fimH |
| pilG | hns |
| pilH | fimE |
| pin | argU |
| pinO | pioO |
| plsA | adk |
| PMG | mgl |
| pmi | manA |
| pmsR | msrA |
| pndR | xapR |
| poaA | putA |
| pog | sspA |
| poh | oriC |
| polC | dnaE |
| pon | lpcB |
| ponA | mrcA |
| ponB | mrcB |
| popA | hemH |
| popB | hemF |
| popC | hemL |
| popE | hemC |
| ppfA | dsbA |
| ppsA | pps |
| prd | fucP |
| prlA | secY |
| prlB | rbsB |
| prlD | secA |
| prlF | sohA |
| prlG | secE |
| prlH | secG |
| Pro2 | proC |
| pro1 | proA |
| pro2 | proB |
| pro3 | proC |
| prv | mutH |
| psiB | ugpA, B |
| psiC | ugpA, B |
| psiD | phn, phnD |
| psiH | phoH |
| psrA, B | TerC, B |
| psuA | rho |
| pts | man |
| ptsF | fruA |
| ptsL | manX |
| ptsM | manX, Y, Z |
| ptsN | nagE |
| ptsO | npr |
| ptsP | manY |
| ptsX | manX, Z |
| pulT | tufA, B |
| pup | deoD |
| Pur2 | purE |
| purC | purF |
| purE2 | purK |
| purG | purM |
| purI | purL |
| pyrA | carA, B |
| qmeA | fabI |
| R1pho | phoR |
| R2pho | pstA, S |
| rad | uvrD |
| radB | recN |
| ramA | rpsD |
| ramB | rimG |
| rap | pth |
| rapA | hepA |
| rbaB | srmB |
| rblA | lrp |
| rbsP | rbsA, B, C, D |
| rbsT | rbsA, C |
| recA | lexA |
| recL | uvrD |
| refI | tolC |
| refII | creD |
| relC | rplK |
| res | rimF |
| resA | polA |
| rfbC | rfbD |
| rfbD | rfbC |
| rffE | rffG? |
| rglA | mcrA |
| rglB | mcrB |
| rhaC | rhaR, S |
| rhlA | srmB |
| ribE | ribC |
| ribG | ribD |
| ribH | ribE |
| rif | rpoB |
| rimA | rpmH |
| rm | hsdM, R |
| rnh | rnhA |
| rnsA | rna |
| rnsC | rho |
| rodA | mrdB |
| rodY | mreB |
| ron | rpoB |
| rorA | recB |
| rot | ppiA |
| rpoF | fliA |
| rrfDβ | rrfF |
| rrnB1 | rrnB |
| rrnD1 | rrnE |
| rrvD | rrfF |
| rsgA | ftn |
| rT | trmA |
| rts | coaA |
| sac | ascB, F |
| sbl | srlA, B, D |
| sdgA | dnaG |
| sdgB | rpoB |
| sdgE | era |
| sdrA | rnhA |
| sec | hemF |
| secC | rpsO |
| sefA | fabZ |
| seg | arcA, dnaK |
| sep | ftsI |
| sez | rpoA |
| sfcA | mae? |
| sfhB | rluD |
| sfiA | sulA |
| sfiB | ftsZ |
| sfrA | arcA |
| sfrB | rfaH |
| sfs1 | sfsA |
| sfs7 | sfsB |
| shl | fruR |
| sin | rnhA |
| sipB | acrA, ssrA |
| sirA | yhhP |
| skp | hlpA, lpxD |
| slpA | intA |
| slr | lplA |
| slt | mltB |
| smbA | pyrH |
| sms | radA |
| sof | dut |
| som | rfbA, B |
| soxQ | marA, R |
| spcA | rpsE |
| spoS | rpoZ |
| spoU | trmH |
| spoV | recG? |
| spr | lexA |
| srf | recA |
| srgA | priA |
| srjA | infC |
| srjB | helD |
| srjC | uvrD |
| ssaD | nusB |
| ssaF | rpmH |
| ssc | lpxD |
| ssd | cpxA |
| ssyB | nusB |
| ssyF | rpsA |
| ssyG | infB |
| stc | micF |
| stl | rpoB |
| strA | rpsL |
| strB | strC |
| stsB | rimH |
| stv | rpoB |
| Su-1 | serU |
| Su-2 | glnV, X |
| Su-3 | tyrT |
| Su-4 | tyrT |
| Su-6 | leuX |
| Suβ | lysT |
| SuII | glnV, X |
| su1 | serU |
| suB | glnU |
| suIII | tyrT |
| su7 | trpT |
| su8 | trpT |
| suA36 | glyU |
| sud2 | rpsD |
| sueB | prfA |
| sufD | glyU |
| sui | sufI |
| sulB | ftsZ |
| sumA | glyT, U |
| sumB | glyU |
| sun | rho |
| supS20 | rpsT |
| sup15B | tyrU |
| supB | glnU, W |
| supC | tyrT |
| supD | serU, V |
| supE | glnV, X |
| supF | tyrT |
| supG | lysT |
| supH | serU |
| supK | prfB |
| supL | lysT |
| supM | tyrU |
| supN | lysV |
| supO | tyrT? |
| supP | leuX |
| supT | glyU |
| supU | trpT |
| supV | trpT |
| supX | topA |
| supZ | tyrU |
| sur | bcr |
| surB | cydC |
| suxA | bcr |
| T1 | fhuA |
| T1rec | tonB |
| T5rec | fhuA |
| T6rec | tsx |
| tabB | groE |
| tabD | rpoB, C |
| talA | alaT |
| talD | alaU |
| tasC | aspT |
| tau | tus |
| tcp | rimJ |
| tdc | tdcB |
| tgs | crr, purU |
| tgtB | gltT |
| tgtC | gltU |
| tgtE | gltV |
| thdB | fadR |
| thdF | trmE |
| thiA | thiE, F, G, H |
| thiJ | thiL |
| thiN | thiJ |
| thrD | thrA |
| thyR | deoB, C |
| tldE | pmbA |
| tlnI | tlnA |
| tlr | deoB, C |
| tls | aspS |
| tmrA | folA |
| tnaC, L | tna leader |
| tnaR | tnaA |
| toc | tolC |
| tol-2 | tolA |
| tol-3 | tolB |
| tolF | ompF |
| tolG | ompA |
| tolM | cmtB |
| tolP | tolQ? |
| tolZ | hflB |
| tonA | fhuA |
| topX | hns |
| tos | prfC |
| tpo | envZ |
| tpp | dppA? |
| tpp-75 | deoA |
| tpx | ahpC |
| tre | treA |
| trkB | kefB |
| trkC | kefC |
| trkE | sapD |
| trpP | tnaB |
| trpX | miaA |
| tryD | trpE |
| tryp-4 | trpE |
| tsnC | trxA |
| tsp | prc |
| tss | asnS |
| tsu | rho |
| ttr | fadL |
| tut | ompA |
| tyrTα, β | tyrT, V |
| uar | prfA |
| uidP | uidB |
| umg | ptsG |
| umpA | lgt |
| umuA | lexA |
| umuB | recA |
| uncA–I | atpA–I |
| ups? | prfA |
| ura | car |
| uraP | upp |
| usg | accD |
| usgA | gntT |
| uvm | umuC, D |
| uvr502 | uvrD |
| uvrE | uvrD |
| uvrF | recF |
| vacB | rnr |
| val-act | valS |
| valTα | valT |
| valTβ | valZ |
| valUα | valU |
| valUγ | valY |
| virR | hns |
| visA | hemH |
| visB | ubiH |
| vtr | fabF |
| waaA | kdtA |
| waaB | rfaB |
| waaC | rfaC |
| waaE | rfaE |
| waaF | rfaF |
| waaG | rfaG |
| waaI | rfaI |
| waaJ | rfaJ |
| waaK | rfaK |
| waaL | rfaL |
| waaM | htrB |
| waaN | mltA |
| waaP | rfaP |
| waaQ | rfaQ |
| waaS | rfaS |
| waaU | rfaK |
| waaY | rfaY |
| waaZ | rfaZ |
| wbbH | rfc |
| wcaN | galF |
| wec | rff |
| wecA | rfe |
| wecB | rffE |
| weeA | tolC |
| witA | kgtP |
| wzxB | rfbX |
| xerA | argR |
| xerB | pepA |
| xonA | sbcB |
| xprA | dsbC |
| xprB | xerD |
| xylT | xylF |
| zab | recA |
| zfiA | csrA |
Temporary names (y names, orfs) are not included.
There is a standing tradition of coordinating gene symbols between the CGSC and the Salmonella Genetic Stock Center to avoid the assignment of the same symbol to different genes in the two organisms and the assignment of different symbols to homologous genes, insofar as this coordination is feasible. We have not, however, changed names of E. coli genes in order to extend this tradition to other bacteria or other organisms. The desirability and feasibility of uniform nomenclature conventions for all bacteria or other microbial groupings are currently only topics of discussion and conjecture, and changes to enhance similarities in an ad hoc, piecemeal fashion seem counterproductive at the present time. Readers are reminded that symbol changes create discontinuity with previous literature concerning genes, with even more serious ramifications for allele designations, since unique allele numbers are assigned on the basis of the three letter mnemonic and changes in a symbol may necessitate renumbering of alleles as well.
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation grants BIR9315421 and BIR9010005.
I thank Stanley Letovsky and Peter Kalamarides for their work in implementation and system administration for the database, which allowed direct retrieval of all the various Table 1 data and the drawing of map segments directly from the CGSC database. Even so, extensive layout work was required for the final figure, and I am indebted to Elise Low for her skill and patience in doing that during all the revisions that went into this version of the map. A number of people have worked tirelessly to get this paper together. Special thanks again go to Elise Low for editing graphics files and for unending proofreading and cross-checks on the tables and references in addition to the layout work. Completion of this task owes much to her skill and her dedication to accuracy and timeliness and to Peter Kalamarides for writing and executing scripts that brought the tables together at every critical juncture. Special thanks also go to Linda Mattice and Narinder Whitehead for valuable help with proofreading and tracking down publications for both versions and for keeping the stock center on course during this work. I’m especially grateful to all of the above for their unstinting efforts at deadline-approaching time. I thank Graeme Berlyn for sharing and helping with use of his computer setup during the printing operation, and also James Bryan for table formatting help in the earlier edition. Brooks Low and Kenn Rudd coauthored the 1996 edition of the map and therefore were major contributors to this map as well; I thank them for that collaboration and their continued help and support. Kenn Rudd and I have attempted to keep this map and the physical map compatible in terms of names and annotations; Kenn has been crucial in keeping me updated with regard to his new information, displaying his characteristic dedication and generosity with regard to his sequence-to-gene detection and expertise. Special thanks this year go to Edward Adelberg for supplying the database with bibliographic updates based on his scanning and annotation of Medline references in the context of his own bibliographic database of current papers in E. coli genetics developed and kept up to date over the past 2 1/2 years. They have been enormously helpful in our attempts to keep the CGSC database current with respect to literature on E. coli genes. These maps continue the tradition of E. coli K-12 linkage map editions so capably constructed by Barbara J. Bachmann over the past 20 years and that of her predecessors for E. coli maps dating back to 1958. Our debt to these previous maps is obvious. The accuracy of the great majority of gene positions on the map is completely indebted to the American and Japanese sequencing projects, and for this particular version of the map, especially to GenBank access to the complete sequence submitted by Fred Blattner and his colleagues as part of the Genome Sequencing project in Wisconsin. And of course we are indebted to numerous scientists who have provided helpful map-related information in the form of discussion, corrections of database inaccuracies, preprints, and personal communications.
REFERENCES
- 1.Aasland R, Coleman J, Holck A L, Smith C L, Raetz C R H, Kleppe K. Identity of the 17-kilodalton protein, a DNA-binding protein from Escherichia coli, and the firA gene product. J Bacteriol. 1988;170:5916–5918. doi: 10.1128/jb.170.12.5916-5918.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abaibou H, Pommier J, Benoit S, Giordano G, Mandrand-Berthelot M-A. Expression and characterization of the Escherichia coli fdo locus and a possible role for aerobic formate dehydrogenase. J Bacteriol. 1995;177:7141–7149. doi: 10.1128/jb.177.24.7141-7149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdulrashid N, Clark D P. Isolation and genetic analysis of mutations allowing the degradation of furans and thiophenes by Escherichia coli. J Bacteriol. 1987;169:1267–1271. doi: 10.1128/jb.169.3.1267-1271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abouhamad W N, Manson M D, Gibson M M, Higgins C F. Peptide transport and chemotaxis in Escherichia coli and Salmonella typhimurium: characterization of the dipeptide permease (Dpp) and the dipeptide-binding protein. Mol Microbiol. 1991;5:1035–1047. doi: 10.1111/j.1365-2958.1991.tb01876.x. [DOI] [PubMed] [Google Scholar]
- 5.Abraham J M, Freitag C S, Clements J R, Eisenstein B I. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:5724–5727. doi: 10.1073/pnas.82.17.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abraham S N, Goguen J D, Sun D, Klemm P, Beachey E H. Identification of two ancillary subunits of Escherichia coli type 1 fimbriae by using antibodies against synthetic oligopeptides of fim gene products. J Bacteriol. 1987;169:5530–5535. doi: 10.1128/jb.169.12.5530-5536.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abril N, Hera C, Alejandre E, Rafferty J A, Margison G P, Pueyo C. Effect of ogt expression on mutation induction by methyl-, ethyl- and propylmethanesulphonate in Escherichia coli K12 strains. Mol Gen Genet. 1994;242:744–748. doi: 10.1007/BF00283431. [DOI] [PubMed] [Google Scholar]
- 8.Ackerman R S, Cozzarelli N R, Epstein W. Accumulation of toxic concentrations of methylglyoxal by wild-type Escherichia coli K-12. J Bacteriol. 1974;119:357–362. doi: 10.1128/jb.119.2.357-362.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adachi H, Ohta T, Matsuzawa H. A water-soluble form of penicillin-binding protein 2 of Escherichia coli constructed by site-directed mutagenesis. FEBS Lett. 1987;226:150–154. doi: 10.1016/0014-5793(87)80569-0. [DOI] [PubMed] [Google Scholar]
- 10.Adachi T, Mizuuchi M, Robinson A, Appella E, O’Dea M H, Gellert M, Mizuuchi K. DNA sequence of the E. coli gyrB gene: application of a new sequencing strategy. Nucleic Acids Res. 1987;15:771–784. doi: 10.1093/nar/15.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adachi T, Mizuuchi R, Menzel R, Gellert M. DNA sequence and transcription of the region upstream of the E. coli gyrB gene. Nucleic Acids Res. 1984;12:6389–6395. doi: 10.1093/nar/12.16.6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Adam E, Volkert M R, Blot M. Cytochrome c biogenesis is involved in the transposon Tn5-mediated bleomycin resistance and the associated fitness effect in Escherichia coli. Mol Microbiol. 1998;28:15–24. doi: 10.1046/j.1365-2958.1998.00755.x. [DOI] [PubMed] [Google Scholar]
- 12.Adams B S, Helling R B. Nalidixic acid-resistant auxotrophs of Escherichia coli. J Bacteriol. 1970;104:1027–1030. doi: 10.1128/jb.104.2.1027-1029.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams D E, West S C. Bypass of DNA heterologies during RuvAB-mediated three- and four-strand branch migration. J Mol Biol. 1996;263:582–596. doi: 10.1006/jmbi.1996.0600. [DOI] [PubMed] [Google Scholar]
- 14.Adams M D, Wagner L M, Graddis T J, Landick R, Antonucci T K, Gibson A L, Oxender D L. Nucleotide sequence and genetic characterization reveal six essential genes for the LIV-I and LS transport systems of Escherichia coli. J Biol Chem. 1990;265:11436–11443. [PubMed] [Google Scholar]
- 15.Adamski F M, Atkins J F, Gesteland R F. Ribosomal protein L9 interactions with 23 S rRNA: the use of a translational bypass assay to study the effect of amino acid substitutions. J Mol Biol. 1996;261:357–371. doi: 10.1006/jmbi.1996.0469. [DOI] [PubMed] [Google Scholar]
- 16.Addinall S G, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in Escherichia coli. Mol Microbiol. 1997;25:303–309. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- 17.Addinall S G, Lutkenhaus J. FtsA is localized to the septum in an FtsZ-dependent manner. J Bacteriol. 1996;178:7167–7172. doi: 10.1128/jb.178.24.7167-7172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adhya S, Cleary P P, Campbell A. A deletion analysis of prophage lambda and adjacent genetic regions. Proc Natl Acad Sci USA. 1968;61:956–962. doi: 10.1073/pnas.61.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adler H I, Mural R, Suttle B. Oxygen sensitivity of an Escherichia coli mutant. J Bacteriol. 1992;174:2072–2077. doi: 10.1128/jb.174.7.2072-2077.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adler H I, Fisher W D, Hardigree A A, Stapleton G E. Repair of radiation-induced damage to the cell division mechanism of Escherichia coli. J Bacteriol. 1966;91:737–742. doi: 10.1128/jb.91.2.737-742.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmad S I. A mutant of Escherichia coli hyper-resistant to a number of DNA damaging agents: location of the mutational site. J Photochem Photobiol B. 1996;36:47–53. doi: 10.1016/S1011-1344(96)07329-0. [DOI] [PubMed] [Google Scholar]
- 22.Ahmad S I, Pritchard R H. A map of four genes specifying enzymes involved in catabolism of nucleosides and deoxynucleosides in Escherichia coli. Mol Gen Genet. 1969;104:351–359. doi: 10.1007/BF00334234. [DOI] [PubMed] [Google Scholar]
- 23.Ahmad S I, Pritchard R H. Location of the gene specifying cytosine deaminase in Escherichia coli. Mol Gen Genet. 1972;118:323–325. doi: 10.1007/BF00333567. [DOI] [PubMed] [Google Scholar]
- 24.Ahmer B M, Thomas M G, Larsen R A, Postle K. Characterization of the exbBD operon of Escherichia coli and the role of ExbB and ExbD in TonB function and stability. J Bacteriol. 1995;177:4742–4747. doi: 10.1128/jb.177.16.4742-4747.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahnn J, March P E, Takiff H E, Inouye M. A GTP-binding protein of Escherichia coli has homology to yeast RAS proteins. Proc Natl Acad Sci USA. 1986;83:8849–8853. doi: 10.1073/pnas.83.23.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aiba H. Transcription of the Escherichia coli adenylate cyclase gene is negatively regulated by cAMP-cAMP receptor protein. J Biol Chem. 1985;260:3063–3070. [PubMed] [Google Scholar]
- 27.Aiba, H., T. Baba, K. Hayashi, T. Inada, K. Isono, T. Itoh, H. Kasai, K. Kashimoto, S. Kimura, M. Kitakawa, M. Kitagawa, K. Makino, T. Miki, K. Mizobuchi, H. Mori, T. Mori, K. Motomura, S. Nakade, Y. Nakamura, H. Nashimoto, Y. Nishio, T. Oshima, N. Saito, G. Sampei, Y. Seki, S. Sivasunddaram, H. Tagami, J. Takeda, K. Takemoto, Y. Takeuchi, C. Wada, Y. Yamamoto, and T. Horiuchi. 1996. GenBank submissions.
- 28.Aiba H, Mizobuchi K. Nucleotide sequence analysis of genes purH and purD involved in the de novo purine nucleotide biosynthesis of Escherichia coli. J Biol Chem. 1989;264:21239–21246. [PubMed] [Google Scholar]
- 29.Aiba H, Mori K, Tanaka M, Ooi T, Roy A M, Danchin A. The complete nucleotide sequence of the adenylate cyclase gene of Escherichia coli. Nucleic Acids Res. 1984;12:9427–9440. doi: 10.1093/nar/12.24.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aiba H, Kawamukai M, Ishihama A. Cloning and promoter analysis of the Escherichia coli adenylate cyclase gene. Nucleic Acids Res. 1983;11:3451–3465. doi: 10.1093/nar/11.11.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 32.Aiba H, Fujimoto S, Ozaki N. Molecular cloning and nucleotide sequencing of the gene for E. coli cAMP receptor protein. Nucleic Acids Res. 1982;10:1345–1361. doi: 10.1093/nar/10.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aiba H, Baba T, Fujita K, Hayashi K, Inada T, Isono K, Itoh T, Kasai H, Kashimoto K, Kimura S, Kitakawa M, Makino K, Miki T, Mizobuchi K, Mori H, Mori T, Motomura K, Nakade S, Nakamura Y, Nashimoto H, Nishio Y, Oshima T, Saito N, Sampei G, Seki Y, Sivasundaramm S, Tagami H, Takeda J, Takemoto K, Takeuchi Y, Wada C, Yamamoto Y, Horiuchi T. A 570-kb DNA sequence of the Escherichia coli K-12 genome corresponding to the 28.0–40.1 min region on the linkage map. DNA Res. 1996;3:363–377. doi: 10.1093/dnares/3.6.363. [DOI] [PubMed] [Google Scholar]
- 34.Aiso T, Ohki R. An rne-1 pnp-7 double mutation suppresses the temperature-sensitive defect of lacZ gene expression in a divE mutant. J Bacteriol. 1998;180:1389–1395. doi: 10.1128/jb.180.6.1389-1395.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal “addiction module” regulated by 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci USA. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akimaru J, Matsuyama S, Tokuda H, Mizushima S. Reconstitution of a protein translocation system containing purified SecY, SecE, and SecA from Escherichia coli. Proc Natl Acad Sci USA. 1991;88:6545–6549. doi: 10.1073/pnas.88.15.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akiyama M, Crooke E, Kornberg A. The polyphosphate kinase gene of Escherichia coli. Isolation and sequence of the ppk gene and membrane location of the protein. J Biol Chem. 1992;267:22556–22561. [PubMed] [Google Scholar]
- 38.Akiyama M, Crooke E, Kornberg A. An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J Biol Chem. 1993;268:633–639. [PubMed] [Google Scholar]
- 39.Akiyama M, Horiuchi T, Sekiguchi M. Molecular cloning and nucleotide sequence of the mutT mutator of Escherichia coli that causes A:T to C:G transversion. Mol Gen Genet. 1987;206:9–16. doi: 10.1007/BF00326530. [DOI] [PubMed] [Google Scholar]
- 40.Akiyama Y, Kihara A, Tokuda H, Ito K. FtsH (HflB) is an ATP-dependent protease selectively acting on SecY and some other membrane proteins. J Biol Chem. 1996;271:31196–31201. doi: 10.1074/jbc.271.49.31196. [DOI] [PubMed] [Google Scholar]
- 41.Akiyama Y, Kihara A, Ito K. Subunit a of proton ATPase F0 sector is a substrate of the FtsH protease in Escherichia coli. FEBS Lett. 1996;399:26–28. doi: 10.1016/s0014-5793(96)01283-5. [DOI] [PubMed] [Google Scholar]
- 42.Akiyama Y, Ito K. The secY membrane component of the bacterial protein export machinery: analysis by new electrophoretic methods for integral membrane proteins. EMBO J. 1985;4:3351–3356. doi: 10.1002/j.1460-2075.1985.tb04088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akiyama Y, Ito K. SecY protein, a membrane-embedded secretion factor of E. coli, is cleaved by the OmpT protease in vitro. Biochem Biophys Res Commun. 1990;167:711–715. doi: 10.1016/0006-291x(90)92083-c. [DOI] [PubMed] [Google Scholar]
- 44.Akiyama Y, Ito K. A new Escherichia coli gene, fdrA, identified by suppression analysis of dominant negative FtsH mutations. Mol Gen Genet. 1995;249:202–208. doi: 10.1007/BF00290367. [DOI] [PubMed] [Google Scholar]
- 45.Akiyama Y, Kamitani S, Kusukawa N, Ito K. In vitro catalysis of oxidative folding of disulfide-bonded proteins by the Escherichia coli dsbA (ppfA) gene product. J Biol Chem. 1992;267:22440–22445. [PubMed] [Google Scholar]
- 46.Akiyama Y, Inada T, Nakamura Y, Ito K. SecY, a multispanning integral membrane protein, contains a potential leader peptidase cleavage site. J Bacteriol. 1990;172:2888–2893. doi: 10.1128/jb.172.6.2888-2893.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akiyama Y, Shirai Y, Ito K. Involvement of FtsH in protein assembly into and through the membrane. II. Dominant mutations affecting FtsH functions. J Biol Chem. 1994;269:5225–5229. [PubMed] [Google Scholar]
- 48.Alaeddinoglu N G, Charles H P. Transfer of a gene for sucrose utilization into Escherichia coli K12, and consequent failure of expression of genes for d-serine utilization. J Gen Microbiol. 1979;110:47–59. doi: 10.1099/00221287-110-1-47. [DOI] [PubMed] [Google Scholar]
- 49.Alam K Y, Clark D P. Molecular cloning and sequence of the thdF gene, which is involved in thiophene and furan oxidation by Escherichia coli. J Bacteriol. 1991;173:6018–6024. doi: 10.1128/jb.173.19.6018-6024.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Albin R, Silverman P M. Identification of the Escherichia coli K-12 cpxA locus as a single gene: construction and analysis of biologically-active cpxA gene fusions. Mol Gen Genet. 1984;197:272–279. doi: 10.1007/BF00330973. [DOI] [PubMed] [Google Scholar]
- 51.Albin R, Silverman P M. Physical and genetic structure of the glpK-cpxA interval of the Escherichia coli K-12 chromosome. Mol Gen Genet. 1984;197:261–271. doi: 10.1007/BF00330972. [DOI] [PubMed] [Google Scholar]
- 52.Albin R, Weber R, Silverman P M. The Cpx proteins of Escherichia coli K-12. Immunologic detection of the chromosomal cpxA gene product. J Biol Chem. 1986;261:4698–4705. [PubMed] [Google Scholar]
- 53.Albrechtsen B, Ross B M, Squires C, Squires C L. Transcriptional termination sequence at the end of the Escherichia coli RNA G operon: complex terminators and antitermination. Nucleic Acids Res. 1991;19:1845–1852. doi: 10.1093/nar/19.8.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Albrechtsen H, Ahmad S I. Regulation of the synthesis of nucleoside catabolic enzymes in Escherichia coli: further analysis of a deoOc mutant strain. Mol Gen Genet. 1980;179:457–460. doi: 10.1007/BF00425477. [DOI] [PubMed] [Google Scholar]
- 55.Aldea M, Hernandez-Chico C, de la Campa A G, Kushner S R, Vicente M. Identification, cloning, and expression of bolA, an ftsZ-dependent morphogene of Escherichia coli. J Bacteriol. 1988;170:5169–5176. doi: 10.1128/jb.170.11.5169-5176.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aldea M, Garrido T, Hernandez-Chico C, Vicente M, Kushner S R. Induction of a growth-phase-dependent promoter triggers transcription of bolA, an Escherichia coli morphogene. EMBO J. 1989;8:3923–3931. doi: 10.1002/j.1460-2075.1989.tb08573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aldea M, Garrido T, Pla J, Vicente M. Division genes in Escherichia coli are expressed coordinately to cell septum requirements by gearbox promoters. EMBO J. 1990;9:3787–3794. doi: 10.1002/j.1460-2075.1990.tb07592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aldea M, Maples V F, Kushner S R. Generation of a detailed physical and genetic map of the ilv-metE-udp region of the Escherichia coli chromosome. J Mol Biol. 1988;200:427–438. doi: 10.1016/0022-2836(88)90533-5. [DOI] [PubMed] [Google Scholar]
- 59.Al-Deib A A, Mahdi A A, Lloyd R G. Modulation of recombination and DNA repair by the RecG and PriA helicases of Escherichia coli K-12. J Bacteriol. 1996;178:6782–6789. doi: 10.1128/jb.178.23.6782-6789.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alefounder P R, Abell C, Battersby A R. The sequence of hemC, hemD and two additional E. coli genes. Nucleic Acids Res. 1988;16:9871. doi: 10.1093/nar/16.20.9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alefounder P R, Perham R N. Identification, molecular cloning and sequence analysis of a gene cluster encoding the class II fructose 1,6-bisphosphate aldolase, 3-phosphoglycerate kinase and a putative second glyceraldehyde 3-phosphate dehydrogenase of Escherichia coli. Mol Microbiol. 1989;3:723–732. doi: 10.1111/j.1365-2958.1989.tb00221.x. [DOI] [PubMed] [Google Scholar]
- 62.Alefounder P R, Baldwin S A, Perham R N, Short N J. Cloning, sequence analysis and over-expression of the gene for class II fructose 1,6-bisphosphate aldolase of Escherichia coli. Biochem J. 1989;257:529–534. doi: 10.1042/bj2570529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alexander D M, St. John A C. Characterization of the carbon starvation-inducible and stationary phase-inducible gene slp encoding an outer membrane lipoprotein in Escherichia coli. Mol Microbiol. 1994;11:1059–1071. doi: 10.1111/j.1365-2958.1994.tb00383.x. [DOI] [PubMed] [Google Scholar]
- 64.Alexseyev A A, Bakhlanova I V, Zaitsev E N, Lanzov V A. Genetic characteristics of new recA mutants of Escherichia coli K-12. J Bacteriol. 1996;178:2018–2024. doi: 10.1128/jb.178.7.2018-2024.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alifano P, Carlomagno M S, Bruni C B. Location of the hisGDCBHAFI operon on the physical map of Escherichia coli. J Bacteriol. 1992;174:3830–3831. doi: 10.1128/jb.174.11.3830-3831.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alkhimova R A, Mironov A S, Sukhodolets V V. Regulation of the activity of the uridine phosphorylase of Escherichia coli K-12. I. Mapping of mutations for the structural gene and determination of the direction of transcription. Sov Genet (Engl Transl Genetika) 1981;17:1127–1133. [PubMed] [Google Scholar]
- 67.Alksne L E, Keeney D, Rasmussen B A. A mutation in either dsbA or dsbB, a gene encoding a component of a periplasmic disulfide bond-catalyzing system, is required for high-level expression of the Bacteroides fragilis metallo-β-lactamase, CcrA, in Escherichia coli. J Bacteriol. 1995;177:462–464. doi: 10.1128/jb.177.2.462-464.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Allen G C, Kornberg A. The priB gene encoding the primosomal replication n protein of Escherichia coli. J Biol Chem. 1991;266:11610–11613. [PubMed] [Google Scholar]
- 69.Allen S P, Polazzi J O, Gierse J K, Easton A M. Two novel heat shock genes encoding proteins produced in response to heterologous protein expression in Escherichia coli. J Bacteriol. 1992;174:6938–6947. doi: 10.1128/jb.174.21.6938-6947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Allibert P, Willison J C, Vignais P M. Complementation of nitrogen-regulatory (ntr-like) mutations in Rhodobacter capsulatus by an Escherichia coli gene: cloning and sequencing of the gene and characterization of the gene product. J Bacteriol. 1987;169:260–271. doi: 10.1128/jb.169.1.260-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Almiron M, Link A J, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 72.Alper M D, Ames B N. Positive selection of mutants with deletions of the gal-chl region of the Salmonella chromosome as a screening procedure for mutagens that cause deletions. J Bacteriol. 1975;121:259–266. doi: 10.1128/jb.121.1.259-266.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Al-Rabiee R, Zhang Y, Grant G A. The mechanism of velocity modulated allosteric regulation in d-3-phosphoglycerate dehydrogenase. Site-directed mutagenesis of effector binding site residues. J Biol Chem. 1996;271:23235–23238. doi: 10.1074/jbc.271.38.23235. [DOI] [PubMed] [Google Scholar]
- 74.Altendorf K, Siebers A, Epstein W. The KDP ATPase of Escherichia coli. Ann N Y Acad Sci. 1992;671:228–243. doi: 10.1111/j.1749-6632.1992.tb43799.x. [DOI] [PubMed] [Google Scholar]
- 75.Altman S, Model P, Dixon N E, Wosnick M A. An E. coli gene coding for a protamine-like protein. Cell. 1981;26:299–304. doi: 10.1016/0092-8674(81)90198-7. [DOI] [PubMed] [Google Scholar]
- 76.Altmann C R, Solow-Cordero D E, Chamberlin M J. RNA cleavage and chain elongation by Escherichia coli DNA-dependent RNA polymerase in a binary enzyme · RNA complex. Proc Natl Acad Sci USA. 1994;91:3784–3788. doi: 10.1073/pnas.91.9.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 78.Amabile-Cuevas C F, Demple B. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 1991;19:4479–4484. doi: 10.1093/nar/19.16.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Amemura M, Shinagawa H, Makino K, Otsuji N, Nakata A. Cloning of and complementation tests with alkaline phosphatase regulatory genes (phoS and phoT) of Escherichia coli. J Bacteriol. 1982;152:692–701. doi: 10.1128/jb.152.2.692-701.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Amemura M, Makino K, Shinagawa H, Kobayashi A, Nakata A. Nucleotide sequence of the genes involved in phosphate transport and regulation of the phosphate regulon in Escherichia coli. J Mol Biol. 1985;184:241–250. doi: 10.1016/0022-2836(85)90377-8. [DOI] [PubMed] [Google Scholar]
- 81.Amemura M, Makino K, Shinagawa H, Nakata A. Nucleotide sequence of the phoM region of Escherichia coli: four open reading frames may constitute an operon. J Bacteriol. 1986;168:294–302. doi: 10.1128/jb.168.1.294-302.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Amemura M, Makino K, Shinagawa H, Nakata A. Cross talk to the phosphate regulon of Escherichia coli by PhoM protein: PhoM is a histidine protein kinase and catalyzes phosphorylation of PhoB and PhoM-open reading frame 2. J Bacteriol. 1990;172:6300–6307. doi: 10.1128/jb.172.11.6300-6307.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ames G F-L. Uptake of amino acids by Salmonella typhimurium. Arch Biochem. 1964;104:1–18. doi: 10.1016/s0003-9861(64)80028-x. [DOI] [PubMed] [Google Scholar]
- 84.Ames P, Yu Y A, Parkinson J S. Methylation segments are not required for chemotactic signalling by cytoplasmic fragments of Tsr, the methyl-accepting serine chemoreceptor of Escherichia coli. Mol Microbiol. 1996;19:737–746. doi: 10.1046/j.1365-2958.1996.408930.x. [DOI] [PubMed] [Google Scholar]
- 85.Ammer J, Brennenstuhl M, Schindler P, Höltje J-V, Zähner H. Phosphorylation of streptozotocin during uptake via the phosphoenolpyruvate:sugar phosphotransferase system in Escherichia coli. Antimicrob Agents Chemother. 1979;16:801–807. doi: 10.1128/aac.16.6.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Amsler C D, Cho M, Matsumura P. Multiple factors underlying the maximum motility of Escherichia coli as cultures enter post-exponential growth. J Bacteriol. 1993;175:6238–6244. doi: 10.1128/jb.175.19.6238-6244.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Amundsen S K, Taylor A F, Chaudhury A M, Smith G R. recD: the gene for an essential third subunit of exonuclease V. Proc Natl Acad Sci USA. 1986;83:5558–5562. doi: 10.1073/pnas.83.15.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.An G, Bendiak D S, Mamelak L A, Friesen J D. Organization and nucleotide sequence of a new ribosomal operon in Escherichia coli containing the genes for ribosomal protein S2 and elongation factor Ts. Nucleic Acids Res. 1981;9:4163–4172. doi: 10.1093/nar/9.16.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.An G, Friesen J D. Characterization of promoter-cloning plasmids: analysis of operon structure in the rif region of Escherichia coli and isolation of an enhanced promoter mutant. J Bacteriol. 1980;144:904–916. doi: 10.1128/jb.144.3.904-916.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.An G, Friesen J D. The nucleotide sequence of tufB and four nearby tRNA structural genes for Escherichia coli. Gene. 1980;12:33–39. doi: 10.1016/0378-1119(80)90013-x. [DOI] [PubMed] [Google Scholar]
- 91.An G, Lee J S, Friesen J D. Evidence for an internal promoter preceding tufA in the str operon of Escherichia coli. J Bacteriol. 1982;149:548–553. doi: 10.1128/jb.149.2.548-553.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Andersen C L, Matthey-Dupraz A, Missiakas D, Raina S. A new Escherichia coli gene, dsbG, encodes a periplasmic protein involved in disulphide bond formation, required for recycling DsbA/DsbB and DsbC redox proteins. Mol Microbiol. 1997;26:121–132. doi: 10.1046/j.1365-2958.1997.5581925.x. [DOI] [PubMed] [Google Scholar]
- 93.Andersen J, Delihas N, Ikenaka K, Green P J, Pines O, Ilercil O, Inouye M. The isolation and characterization of RNA coded by the micF gene in Escherichia coli. Nucleic Acids Res. 1987;15:2089–2101. doi: 10.1093/nar/15.5.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Andersen J T, Poulsen P, Jensen K F. Attenuation in the rph-pyrE operon of Escherichia coli and processing of the dicistronic mRNA. Eur J Biochem. 1992;206:381–390. doi: 10.1111/j.1432-1033.1992.tb16938.x. [DOI] [PubMed] [Google Scholar]
- 95.Andersen L, Kilstrup M, Neuhard J. Pyrimidine, purine and nitrogen control of cytosine deaminase synthesis in Escherichia coli K12. Arch Microbiol. 1989;152:115–118. doi: 10.1007/BF00456087. [DOI] [PubMed] [Google Scholar]
- 96.Andersen P S, Frees D, Fast R, Mygind B. Uracil uptake in Escherichia coli K-12: isolation of uraA mutants and cloning of the gene. J Bacteriol. 1995;177:2008–2013. doi: 10.1128/jb.177.8.2008-2013.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Andersen P S, Smith J M, Mygind B. Characterization of the upp gene encoding uracil phosphoribosyltransferase of Escherichia coli K12. Eur J Biochem. 1992;204:51–56. doi: 10.1111/j.1432-1033.1992.tb16604.x. [DOI] [PubMed] [Google Scholar]
- 98.Anderson J J, Oxender D L. Escherichia coli transport mutants lacking binding protein and other components of the branched-chain amino acid transport system. J Bacteriol. 1977;130:384–392. doi: 10.1128/jb.130.1.384-392.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Anderson J J, Wilson J M, Oxender D L. Defective transport and other phenotypes of a periplastic “leaky” mutant of Escherichia coli K-12. J Bacteriol. 1979;140:351–358. doi: 10.1128/jb.140.2.351-358.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Anderson J J, Quay S C, Oxender D L. Mapping of two loci affecting the regulation of branched-chain amino acid transport in Escherichia coli K-12. J Bacteriol. 1976;126:80–90. doi: 10.1128/jb.126.1.80-90.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Anderson M S, Eveland S S, Onishi H R, Pompliano D L. Kinetic mechanism of the Escherichia coli UDPMurNAc-tripeptide d-alanyl-d-alanine-adding enzyme: use of a glutathione S-transferase fusion. Biochemistry. 1996;35:16264–16269. doi: 10.1021/bi961872+. [DOI] [PubMed] [Google Scholar]
- 102.Anderson P M, Sung Y-C, Fuchs J A. The cyanase operon and cyanate metabolism. FEMS Microbiol Rev. 1990;7:247–252. doi: 10.1111/j.1574-6968.1990.tb04920.x. [DOI] [PubMed] [Google Scholar]
- 103.Ando H, Kitabatake M, Inokuchi H. 10Sa RNA complements the temperature-sensitive phenotype caused by a mutation in the phosphoribosyl pyrophosphate synthetase (prs) gene in Escherichia coli. Genes Genet Syst. 1996;71:47–50. doi: 10.1266/ggs.71.47. [DOI] [PubMed] [Google Scholar]
- 104.Andresen P A, Kaasen I, Styrvold O B, Boulnois G, Strom A R. Molecular cloning, physical mapping and expression of the bet genes governing the osmoregulatory choline-glycine betaine pathway of Escherichia coli. J Gen Microbiol. 1988;134:1737–1746. doi: 10.1099/00221287-134-6-1737. [DOI] [PubMed] [Google Scholar]
- 105.Andresson O S, Davies J E. Genetic organization and restriction enzyme cleavage map of the ksgA-pdxA region of the Escherichia coli chromosome. Mol Gen Genet. 1980;179:211–216. doi: 10.1007/BF00268465. [DOI] [PubMed] [Google Scholar]
- 106.Andresson O S, Davies J E. Isolation and characterization of lambda transducing phages for the E. coli genes ksgA and pdxA. Mol Gen Genet. 1980;179:201–209. doi: 10.1007/BF00268464. [DOI] [PubMed] [Google Scholar]
- 107.Andresson O S, Davies J E. Some properties of the ribosomal RNA methyltransferase encoded by ksgA and the polarity of ksgA transcription. Mol Gen Genet. 1980;179:217–222. doi: 10.1007/BF00268466. [DOI] [PubMed] [Google Scholar]
- 108.Andrews J C, Short S A. Genetic analysis of Escherichia coli oligopeptide transport mutants. J Bacteriol. 1985;161:484–492. doi: 10.1128/jb.161.2.484-492.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108a.Andrews S C, Berks B C, McClay J, Ambler A, Quail M A, Golby P, Guest J R. A 12-cistron Escherichia coli operon (hyf) encoding a putative proton-translocating formate hydrogenylase system. Microbiology. 1997;143:3633–3647. doi: 10.1099/00221287-143-11-3633. [DOI] [PubMed] [Google Scholar]
- 109.Andrews S C, Shipley D, Keen J N, Findlay J B, Harrison P M, Guest J R. The haemoglobin-like protein (HMP) of Escherichia coli has ferrisiderophore reductase activity and its C-terminal domain shares homology with ferredoxin NADP+ reductases. FEBS Lett. 1992;302:247–252. doi: 10.1016/0014-5793(92)80452-m. [DOI] [PubMed] [Google Scholar]
- 110.Andrews S C, Guest J R. Nucleotide sequence of the gene encoding the GMP reductase of Escherichia coli K12. Biochem J. 1988;255:35–43. doi: 10.1042/bj2550035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Andrews S C, Harrison P M, Guest J R. Cloning, sequencing, and mapping of the bacterioferritin gene (bfr) of Escherichia coli K-12. J Bacteriol. 1989;171:3940–3947. doi: 10.1128/jb.171.7.3940-3947.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Andrews S C, Harrison P M, Guest J R. A molecular analysis of the 53.3 minute region of the Escherichia coli linkage map. J Gen Microbiol. 1991;137:361–367. doi: 10.1099/00221287-137-2-361. [DOI] [PubMed] [Google Scholar]
- 113.Andrianopoulos K, Wang L, Reeves P R. Identification of the fucose synthetase gene in the colanic acid gene cluster of Escherichia coli K-12. J Bacteriol. 1998;180:998–1001. doi: 10.1128/jb.180.4.998-1001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Angerer A, Enz S, Ochs M, Braun V. Transcriptional regulation of ferric citrate transport in Escherichia coli K-12. Fecl belongs to a new subfamily of sigma 70-type factors that respond to extracytoplasmic stimuli. Mol Microbiol. 1995;18:163–174. doi: 10.1111/j.1365-2958.1995.mmi_18010163.x. [DOI] [PubMed] [Google Scholar]
- 115.Anilionis A, Ostapchuk P, Riley M. Identification of a second cryptic lambdoid prophage locus in the E. coli K12 chromosome. Mol Gen Genet. 1980;180:479–481. doi: 10.1007/BF00425865. [DOI] [PubMed] [Google Scholar]
- 116.Anton I A, Coggins J R. Sequencing and over expression of the Escherichia coli aroE gene encoding shikimate dehydrogenase. Biochem J. 1988;249:319–326. doi: 10.1042/bj2490319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Antonucci T K, Wagner M, Oxender D L. Cloning, expression, and nucleotide sequence of livR, the repressor for high-affinity branched-chain amino acid transport in Escherichia coli. Proteins Struct Funct Genet. 1986;1:125–133. doi: 10.1002/prot.340010204. [DOI] [PubMed] [Google Scholar]
- 118.Aoki H, Dekany K, Adams S L, Ganoza M C. The gene encoding the elongation factor P protein is essential for viability and is required for protein synthesis. J Biol Chem. 1997;272:32254–32259. doi: 10.1074/jbc.272.51.32254. [DOI] [PubMed] [Google Scholar]
- 119.Aoki H, Adams S L, Chung D G, Yaguchi M, Chuang S E, Ganoza M C. Cloning, sequencing and overexpression of the gene for prokaryotic factor EF-P involved in peptide bond synthesis. Nucleic Acids Res. 1991;19:6215–6220. doi: 10.1093/nar/19.22.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Apirion D. Three genes that affect Escherichia coli ribosomes. J Mol Biol. 1967;30:255–275. [PubMed] [Google Scholar]
- 121.Apirion D, Watson N. Mapping and characterization of a mutation in Escherichia coli that reduces the level of ribonuclease III specific for double-stranded ribonucleic acid. J Bacteriol. 1975;124:317–324. doi: 10.1128/jb.124.1.317-324.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Apirion D, Schlessinger D. Coresistance to neomycin and kanamycin by mutations in an Escherichia coli locus that affects ribosomes. J Bacteriol. 1968;96:768–776. doi: 10.1128/jb.96.3.768-776.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Apirion D, Watson N. A second gene which affects the RNA processing enzyme ribonuclease P of Escherichia coli. FEBS Lett. 1980;110:161–163. doi: 10.1016/0014-5793(80)80062-7. [DOI] [PubMed] [Google Scholar]
- 124.Apostolakos D, Menter P A, Rampsch B J, Reeves H C, Birge E A. Genetic map position of the cistron coding for isocitrate dehydrogenase in Escherichia coli K-12. Curr Microbiol. 1982;7:45–47. [Google Scholar]
- 125.Archibold E R, Williams L S. Regulation of methionyl-transfer ribonucleic acid synthetase formation in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1973;114:1007–1013. doi: 10.1128/jb.114.3.1007-1013.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ardeshir F, Ames G F-L. Cloning of the histidine transport genes from Salmonella typhimurium and characterization of an analogous transport system in Escherichia coli. J Supramol Struct (Suppl) 1980;13:117–130. doi: 10.1002/jss.400130111. [DOI] [PubMed] [Google Scholar]
- 127.Arendes J, Carl P, Sugino A. A mutation in the rnh-locus of Escherichia coli affects the structural gene for RNase H. Examination of the mutant and wild type protein. J Biol Chem. 1982;257:4719–4722. [PubMed] [Google Scholar]
- 128.Arikan E, Kulkarni M S, Thomas D C, Sancar A. Sequences of the E. coli uvrB gene and protein. Nucleic Acids Res. 1986;14:2637–2650. doi: 10.1093/nar/14.6.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Aristarkhov A, Mikulskis A, Belasco J G, Lin E C C. Translation of the adhE transcript to produce ethanol dehydrogenase requires RNase III cleavage in Escherichia coli. J Bacteriol. 1996;178:4327–4332. doi: 10.1128/jb.178.14.4327-4332.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ariza R R, Cohen S P, Bachhawat N, Levy S B, Demple B. Repressor mutations in the marRAB operon that activate oxidative stress genes and multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1994;176:143–148. doi: 10.1128/jb.176.1.143-148.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ariza R R, Li Z-W, Ringstad N, Demple B. Activation of multiple antibiotic resistance and binding of stress-inducible promoters by Escherichia coli Rob protein. J Bacteriol. 1995;177:1655–1661. doi: 10.1128/jb.177.7.1655-1661.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Armengod M-E, Lambies E. Overlapping arrangement of the recF and dnaN operons of Escherichia coli: positive and negative control sequences. Gene. 1986;43:183–196. doi: 10.1016/0378-1119(86)90206-4. [DOI] [PubMed] [Google Scholar]
- 133.Armstrong S K, Pettis G S, Forrester L J, McIntosh M A. The Escherichia coli enterobactin biosynthesis gene, entD: nucleotide sequence and membrane localization of its protein product. Mol Microbiol. 1989;3:757–766. doi: 10.1111/j.1365-2958.1989.tb00224.x. [DOI] [PubMed] [Google Scholar]
- 134.Arn E A, Abelson J N. The 2′-5′ RNA ligase of Escherichia coli. Purification, cloning, and genomic disruption. J Biol Chem. 1996;271:31145–31153. doi: 10.1074/jbc.271.49.31145. [DOI] [PubMed] [Google Scholar]
- 135.Arnardóttir A, Thorbjarnardóttir S, Eggertsson G. Mapping of the supP (Su6+) amber suppressor gene in Escherichia coli. J Bacteriol. 1980;141:977–978. doi: 10.1128/jb.141.2.977-978.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Arnez J G, Steitz T A. Crystal structures of three misacylating mutants of Escherichia coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP. Biochemistry. 1996;35:14725–14733. doi: 10.1021/bi961532o. [DOI] [PubMed] [Google Scholar]
- 137.Arnqvist A, Olsen A, Pfeifer J, Russell D G, Normark S. The Crl protein activates cryptic genes for curli formation and fibronectin binding in Escherichia coli HB101. Mol Microbiol. 1992;6:2443–2452. doi: 10.1111/j.1365-2958.1992.tb01420.x. [DOI] [PubMed] [Google Scholar]
- 138.Arnqvist A, Olsen A, Normark S. Sigma S-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by sigma 70 in the absence of the nucleoid-associated protein H-NS. Mol Microbiol. 1994;13:1021–1032. doi: 10.1111/j.1365-2958.1994.tb00493.x. [DOI] [PubMed] [Google Scholar]
- 139.Aronshtam A, Marinus M G. Dominant negative mutator mutations in the mutL gene of Escherichia coli. Nucleic Acids Res. 1996;24:2498–2504. doi: 10.1093/nar/24.13.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Aronson B, Ravnikar P D, Somerville R L. Nucleotide sequence of the 2-amino-3-ketobutyrate coenzyme A ligase (kbl) gene of E. coli. Nucleic Acids Res. 1988;16:3586. doi: 10.1093/nar/16.8.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Aronson B D, Somerville R L, Epperly B R, Dekker E E. The primary structure of Escherichia colil-threonine dehydrogenase. J Biol Chem. 1989;264:5226–5232. [PubMed] [Google Scholar]
- 142.Arps P J, Marvel C C, Rubin B C, Tolan D A, Penhoet E E, Winkler M E. Structural features of the hisT operon of Escherichia coli K-12. Nucleic Acids Res. 1985;13:5297–5315. doi: 10.1093/nar/13.14.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Arps P J, Winkler M E. An unusual genetic link between vitamin B6 biosynthesis and tRNA pseudouridine modification in Escherichia coli K-12. J Bacteriol. 1987;169:1071–1079. doi: 10.1128/jb.169.3.1071-1079.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Arps P J, Winkler M E. Structural analysis of the Escherichia coli K-12 hisT operon by using a kanamycin resistance cassette. J Bacteriol. 1987;169:1061–1070. doi: 10.1128/jb.169.3.1061-1070.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Arraj J A, Marinus M G. Phenotypic reversal in dam mutants of Escherichia coli K-12 by a recombinant plasmid containing the dam+ gene. J Bacteriol. 1983;153:562–565. doi: 10.1128/jb.153.1.562-565.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Arthur H M, Eastlake P B. Transcriptional control of the uvrD gene of Escherichia coli. Gene. 1983;25:309–316. doi: 10.1016/0378-1119(83)90235-4. [DOI] [PubMed] [Google Scholar]
- 147.Artman M, Werthamer S. Use of streptomycin and cyclic adenosine 5′-monophosphate in the isolation of mutants deficient in CAP protein. J Bacteriol. 1974;120:542–544. doi: 10.1128/jb.120.1.542-544.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Asada K, Sugimoto K, Oko A, Takanami M, Hirota Y. Structure of replication origin of the Escherichia coli K-12 chromosome: the presence of spacer sequences in the ori region carrying information for autonomous replication. Nucleic Acids Res. 1982;10:3745–3754. doi: 10.1093/nar/10.12.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Asada K, Nakatani S, Takanami M. Cloning of the contiguous 165-kilobase-pair region around the terminus of Escherichia coli K-12 DNA replication. J Bacteriol. 1985;163:398–400. doi: 10.1128/jb.163.1.398-400.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Asai K, Fujisaki S, Nishimura Y, Nishino T, Okada K, Nakagawa T, Kawamukai M, Matsuda H. The identification of Escherichia coli ispB (cel) gene encoding the octaprenyl diphosphate synthase. Biochem Biophys Res Commun. 1994;202:340–345. doi: 10.1006/bbrc.1994.1933. [DOI] [PubMed] [Google Scholar]
- 151.Asai T, Sommer S, Bailone A, Kogoma T. Homologous recombination-dependent initiation of DNA replication from DNA damage-inducible origins in Escherichia coli. EMBO J. 1993;12:3287–3295. doi: 10.1002/j.1460-2075.1993.tb05998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Asai T, Kogoma T. The RecF pathway of homologous recombination can mediate the initiation of DNA damage-inducible replication of the Escherichia coli chromosome. J Bacteriol. 1994;176:7113–7114. doi: 10.1128/jb.176.22.7113-7114.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Asha H, Gowrishankar J. Regulation of kdp operon expression in Escherichia coli: evidence against turgor as signal for transcriptional control. J Bacteriol. 1993;175:4528–4537. doi: 10.1128/jb.175.14.4528-4537.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Åslund F, Zehnbauer B A, Miranda-Vizuete A, Pueyo C, Holmgren A. Two additional glutaredoxins exist in Escherichia coli: glutaredoxin 3 is a hydrogen donor for ribonucleotide reductase in a thioredoxin/glutaredoxin 1 double mutant. Proc Natl Acad Sci USA. 1994;91:9813–9817. doi: 10.1073/pnas.91.21.9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Åslund F, Nordstrand K, Berndt K D, Nikkola M, Bergman T, Ponstingl H, Jornvall H, Otting G, Holmgren A. Glutaredoxin-3 from Escherichia coli. Amino acid sequence, 1H and 15N NMR assignments, and structural analysis. J Biol Chem. 1996;271:6736–6746. doi: 10.1074/jbc.271.12.6736. [DOI] [PubMed] [Google Scholar]
- 156.Asoh S, Matsuzawa H, Ishino F, Strominger J L, Matsuhashi M, Ohta T. Nucleotide sequence of the pbpA gene and characteristics of the deduced amino acid sequence of penicillin-binding protein 2 of Escherichia coli K-12. Eur J Biochem. 1986;160:231–238. doi: 10.1111/j.1432-1033.1986.tb09961.x. [DOI] [PubMed] [Google Scholar]
- 157.Asoh S, Matsuzawa H, Matsuhashi M, Ohta T. Molecular cloning and characterization of the genes pbpA and rodA responsible for the rod shape of Escherichia coli K-12: analysis of gene expression with transposon Tn5 mutagenesis and protein synthesis directed by constructed plasmids. J Bacteriol. 1983;154:10–16. doi: 10.1128/jb.154.1.10-16.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Atherly A. Escherichia coli mutant containing a large deletion from relA to argA. J Bacteriol. 1979;138:530–534. doi: 10.1128/jb.138.2.530-534.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Atkinson M R, Ninfa A J. Mutational analysis of the bacterial signal-transducing protein kinase/phosphatase nitrogen regulator II (NRII or NtrB) J Bacteriol. 1993;175:7016–7023. doi: 10.1128/jb.175.21.7016-7023.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Atlung T, Nielsen A, Hansen F G. Isolation, characterization, and nucleotide sequence of appY, a regulatory gene for growth-phase-dependent gene expression in Escherichia coli. J Bacteriol. 1989;171:1683–1691. doi: 10.1128/jb.171.3.1683-1691.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Atlung T, Brondsted L. Role of the transcriptional activator AppY in regulation of the cyx appA operon of Escherichia coli by anaerobiosis, phosphate starvation, and growth phase. J Bacteriol. 1994;176:5414–5422. doi: 10.1128/jb.176.17.5414-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Atlung T, Sund S, Olesen K, Brøndsted L. The histone-like protein H-NS acts as a transcriptional repressor for expression of the anaerobic and growth phase activator AppY of Escherichia coli. J Bacteriol. 1996;178:3418–3425. doi: 10.1128/jb.178.12.3418-3425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Attfield P V, Benson F E, Lloyd R G. Analysis of the ruv locus of Escherichia coli K-12 and identification of the gene product. J Bacteriol. 1985;164:276–281. doi: 10.1128/jb.164.1.276-281.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Au D C-T, Gennis R B. Cloning of the cyo locus encoding the cytochrome o terminal oxidase complex of Escherichia coli. J Bacteriol. 1987;169:3237–3242. doi: 10.1128/jb.169.7.3237-3242.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Au D C-T, Lorence R M, Gennis R B. Isolation and characterization of an Escherichia coli mutant lacking the cytochrome o terminal oxidase. J Bacteriol. 1985;161:123–127. doi: 10.1128/jb.161.1.123-127.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Aufrere R, Tempete M, Bohin J P. Overlapping reading frames in Escherichia coli. The two promoters and the first 65 nucleotides of a gene related to the rrnB operon are localized in the coding sequence of btuB. C R Acad Sci Ser III. 1986;303:49–54. [PubMed] [Google Scholar]
- 167.Aufrere R, Tempete M, Bohin J P. Regulation of expression of the gene for vitamin B12 receptor cloned on a multicopy plasmid in Escherichia coli. Mol Gen Genet. 1986;205:358–365. doi: 10.1007/BF00430451. [DOI] [PubMed] [Google Scholar]
- 168.Auger E A, Bennett G N. Regulation of lysine decarboxylase activity in Escherichia coli. Arch Microbiol. 1989;151:466–468. doi: 10.1007/BF00416608. [DOI] [PubMed] [Google Scholar]
- 169.Auger E A, Redding K E, Plumb T, Childs L C, Meng S Y, Bennett G N. Construction of lac fusions to the inducible arginine and lysine decarboxylase genes of Escherichia coli K12. Mol Microbiol. 1989;3:609–620. doi: 10.1111/j.1365-2958.1989.tb00208.x. [DOI] [PubMed] [Google Scholar]
- 170.Augustin L B, Jacobson B A, Fuchs J A. Escherichia coli Fis and DnaA proteins bind specifically to the nrd promoter region and affect expression of an nrd-lac fusion. J Bacteriol. 1994;176:378–387. doi: 10.1128/jb.176.2.378-387.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Austin D, Larson T J. Nucleotide sequence of the glpD gene encoding aerobic sn-glycerol 3-phosphate dehydrogenase of Escherichia coli K-12. J Bacteriol. 1991;173:101–107. doi: 10.1128/jb.173.1.101-107.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Austin E A, Graves J F, Hite L A, Parker C T, Schnaitman C A. Genetic analysis of lipopolysaccharide core biosynthesis by Escherichia coli K-12: insertion mutagenesis of the rfa locus. J Bacteriol. 1990;172:5312–5325. doi: 10.1128/jb.172.9.5312-5325.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Austin S. Wild-type and mutant in vitro products of an operon for ribonucleic acid polymerase subunits. J Bacteriol. 1976;127:32–39. doi: 10.1128/jb.127.1.32-39.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Austin S, Scaife J G. A new method for selecting RNA polymerase mutants. J Mol Biol. 1970;49:263–267. doi: 10.1016/0022-2836(70)90394-3. [DOI] [PubMed] [Google Scholar]
- 175.Autexier C, DuBow M S. The Escherichia coli Mu/D108 phage ner homologue gene (nlp) is transcribed and evolutionarily conserved among the Enterobacteriaceae. Gene. 1992;114:13–18. doi: 10.1016/0378-1119(92)90701-p. [DOI] [PubMed] [Google Scholar]
- 176.Avalos J, Corrochano L M, Brenner S. Cysteinyl-tRNA synthetase is a direct descendant of the first aminoacyl-tRNA synthetase. FEBS Lett. 1991;286:176–180. doi: 10.1016/0014-5793(91)80968-9. [DOI] [PubMed] [Google Scholar]
- 177.Avissar Y J, Beale S I. Identification of the enzymatic basis for delta-aminolevulinic acid auxotrophy in the hemA mutant of Escherichia coli. J Bacteriol. 1989;171:2919–2924. doi: 10.1128/jb.171.6.2919-2924.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Azakami H, Yamashita M, Roh J H, Suzuki H, Kumagai H, Murooka Y. Nucleotide sequence of the gene for monamine oxidase (maoA) from Escherichia coli. J Ferment Bioeng. 1994;77:315–319. [Google Scholar]
- 179.Azizan A, Black P N. use of transposon TnphoA to identify genes for cell envelope proteins of Escherichia coli required for long-chain fatty acid transport: the periplasmic protein Tsp potentiates long-chain fatty acid transport. J Bacteriol. 1994;176:6653–6662. doi: 10.1128/jb.176.21.6653-6662.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Baba T, Jacq A, Brickman E, Beckwith J R, Taura T, Ueguchi C, Akiyama Y, Ito K. Characterization of cold-sensitive secY mutants of Escherichia coli. J Bacteriol. 1990;172:7005–7010. doi: 10.1128/jb.172.12.7005-7010.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Baba T, Taura T, Shimoike T, Akiyama Y, Yoshihisa T, Ito K. A cytoplasmic domain is important for the formation of a SecY-SecE translocator complex. Proc Natl Acad Sci USA. 1994;91:4539–4543. doi: 10.1073/pnas.91.10.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Babbitt P C, Mrachko G T, Hasson M S, Huisman G W, Kolter R, Ringe D, Petsko G A, Kenyon G L, Gerlt J A. A functionally diverse enzyme superfamily that abstracts the alpha protons of carboxylic acids. Science. 1995;267:1159–1161. doi: 10.1126/science.7855594. [DOI] [PubMed] [Google Scholar]
- 183.Babitzke P, Kushner S R. The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. Proc Natl Acad Sci USA. 1991;88:1–5. doi: 10.1073/pnas.88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Baccigalupi L, Marasco R, Ricca E, De Felice M, Sacco M. Control of ilvIH transcription during amino acid downshift in stringent and relaxed strains of Escherichia coli. FEMS Microbiol Lett. 1995;131:95–98. doi: 10.1111/j.1574-6968.1995.tb07760.x. [DOI] [PubMed] [Google Scholar]
- 185.Bacher A, Eberhardt S, Richter G. Biosynthesis of riboflavin. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 657–664. [Google Scholar]
- 186.Bachi B, Kornberg H L. Genes involved in the uptake and catabolism of gluconate by Escherichia coli. J Gen Microbiol. 1975;90:321–335. doi: 10.1099/00221287-90-2-321. [DOI] [PubMed] [Google Scholar]
- 187.Bachmann B J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983;47:180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bachmann B J. Linkage map of Escherichia coli K-12, edition 7. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 807–876. [Google Scholar]
- 189.Bachmann B J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990;54:130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bachmann B J, Low K B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980;44:1–56. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190a.Bachmann B J, Low K B, Taylor A L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976;40:116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Backendorf C, Spaink H, Barbiero A P, Van de Putte P. Structure of the uvrB gene of Escherichia coli. Homology with other DNA repair enzymes and characterization of the uvrB5 mutation. Nucleic Acids Res. 1986;14:2877–2890. doi: 10.1093/nar/14.7.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Backman K, Chen Y-M, Magasanik B. Physical and genetic characterization of the glnA-glnG region of the Escherichia coli chromosome. Proc Natl Acad Sci USA. 1981;78:3743–3747. doi: 10.1073/pnas.78.6.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Backstrom D, Sjoberg R-M, Lundberg L G. Nucleotide sequence of the structural gene for dihydroorotase of Escherichia coli K-12. Eur J Biochem. 1986;160:77–82. doi: 10.1111/j.1432-1033.1986.tb09942.x. [DOI] [PubMed] [Google Scholar]
- 194.Bae W, Jones P G, Inouye M. CspA, the major cold shock protein of Escherichia coli, negatively regulates its own gene expression. J Bacteriol. 1997;179:7081–7088. doi: 10.1128/jb.179.22.7081-7088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Baecker P A, Furlong C E, Preiss J. Biosynthesis of bacterial glycogen: primary structure of Escherichia coli ADP-glucose synthetase as deduced from the nucleotide sequence of the glgC gene. J Biol Chem. 1986;258:5084–5088. [PubMed] [Google Scholar]
- 196.Baecker P A, Greenberg J, Preiss J. Biosynthesis of bacterial glycogen: primary structure of Escherichia coli 1,4-a-d-glucan-6-a-d-(1,4-a-d-glucano)-transferase as deduced from the nucleotide sequence of the glgB gene. J Biol Chem. 1986;261:8738–8743. [PubMed] [Google Scholar]
- 197.Baer M F, Low K B, Soll D. Regulation of the biosynthesis of aminoacyl-transfer ribonucleic acid synthetases and of transfer-ribonucleic acid in Escherichia coli. V. Mutants with increased levels of valyl-transfer ribonucleic acid synthetase. J Bacteriol. 1979;139:165–175. doi: 10.1128/jb.139.1.165-175.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bagg A, Neilands J B. Mapping of a mutation affecting regulation of iron uptake systems in Escherichia coli K-12. J Bacteriol. 1985;161:450–453. doi: 10.1128/jb.161.1.450-453.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Bagg A, Neilands J B. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry. 1987;26:5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- 200.Bailey M J, Hughes C, Koronakis V. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol Microbiol. 1997;26:845–851. doi: 10.1046/j.1365-2958.1997.6432014.x. [DOI] [PubMed] [Google Scholar]
- 201.Bailey M J, Koronakis V, Schmoll T, Hughes C. Escherichia coli HlyT protein, a transcriptional activator of haemolysin synthesis and secretion, is encoded by the rfaH (sfrB) locus required for expression of sex factor and lipopolysaccharide genes. Mol Microbiol. 1992;6:1003–1012. doi: 10.1111/j.1365-2958.1992.tb02166.x. [DOI] [PubMed] [Google Scholar]
- 202.Bailly V, Verly W G. Escherichia coli endonuclease III is not an endonuclease but a beta-elimination catalyst. Biochem J. 1987;242:565–572. doi: 10.1042/bj2420565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Baird L, Lipinska B, Raina S, Georgopoulos C. Identification of the Escherichia coli sohB gene, a multicopy suppressor of the HtrA (DegP) null phenotype. J Bacteriol. 1991;173:5763–5770. doi: 10.1128/jb.173.18.5763-5770.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Baird L, Georgopoulos C. Identification, cloning, and characterization of the Escherichia coli sohA gene, a suppressor of the htrA (degP) null phenotype. J Bacteriol. 1990;172:1587–1594. doi: 10.1128/jb.172.3.1587-1594.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Baker J, Franklin D B, Parker J. Sequence and characterization of the gcpE gene of Escherichia coli. FEMS Microbiol Lett. 1992;73:175–180. doi: 10.1016/0378-1097(92)90604-m. [DOI] [PubMed] [Google Scholar]
- 206.Baker K E, Ditullio K P, Neuhard J, Kelln R A. Utilization of orotate as a pyrimidine source by Salmonella typhimurium and Escherichia coli requires the dicarboxylate transport protein encoded by dctA. J Bacteriol. 1996;178:7099–7105. doi: 10.1128/jb.178.24.7099-7105.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Baker K P, Boxer D H. Regulation of the chlA locus of Escherichia coli K12: involvement of molybdenum cofactor. Mol Microbiol. 1991;5:901–907. doi: 10.1111/j.1365-2958.1991.tb00764.x. [DOI] [PubMed] [Google Scholar]
- 208.Baker T A. Replication initiation. A new controller in Escherichia coli. Curr Biol. 1994;4:945–946. doi: 10.1016/s0960-9822(00)00214-1. [DOI] [PubMed] [Google Scholar]
- 209.Bakker E P, Booth I R, Dinnbier U, Epstein W, Gajewska A. Evidence for multiple K+ export systems in Escherichia coli. J Bacteriol. 1987;169:3743–3749. doi: 10.1128/jb.169.8.3743-3749.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Balbinder E, Callahan R, McCann P P, Cordero J C, Weber R, Smith A M, Angelosanto F. Regulatory mutants of the tryptophan operon of Salmonella typhimurium. Genetics. 1970;66:31–53. doi: 10.1093/genetics/66.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Baldoma L, Badia J, Sweet G, Aguilar J. Cloning, mapping and gene product identification of rhaT from Escherichia coli K12. FEMS Microbiol Lett. 1990;60:103–107. doi: 10.1016/0378-1097(90)90353-r. [DOI] [PubMed] [Google Scholar]
- 212.Baliko G, Venetianer P. An Escherichia coli gene in search of a function: phenotypic effects of the gene recently identified as murI. J Bacteriol. 1993;175:6571–6577. doi: 10.1128/jb.175.20.6571-6577.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Bally M, Foglino M, Bruschi M, Murgier M, Lazdunski A. Nucleotide sequence of the promoter and amino-terminal encoding region of the Escherichia coli pepN gene. Eur J Biochem. 1986;155:565–569. doi: 10.1111/j.1432-1033.1986.tb09525.x. [DOI] [PubMed] [Google Scholar]
- 214.Bally M, Murgier M, Lazdunski A. Cloning and orientation of the gene encoding aminopeptidase N in Escherichia coli. Mol Gen Genet. 1984;195:507–510. doi: 10.1007/BF00341454. [DOI] [PubMed] [Google Scholar]
- 215.Bally M, Murgier M, Tommassen J. Physical mapping of the gene for aminopeptidase N in Escherichia coli K12. Mol Gen Genet. 1984;193:190–191. doi: 10.1007/BF00327436. [DOI] [PubMed] [Google Scholar]
- 216.Bandrin S V, Rabinovich P M, Stepanov A I. Three linkage groups of genes involved in riboflavin biosynthesis in Escherichia coli. Genetika. 1983;19:1419–1425. . (In Russian.) [PubMed] [Google Scholar]
- 217.Bandrin S V, Rabinovich P M, Stepanov A I. Three linkage groups of genes for riboflavin biosynthesis. Sov Genet (Engl Transl Genetika) 1984;19:1103–1109. [PubMed] [Google Scholar]
- 218.Banerjee R V, Johnston N L, Sobeski J K, Datta P, Matthews R G. Cloning and sequence analysis of the Escherichia coli metH gene encoding cobalamin-dependent methionine synthase and isolation of a tryptic fragment containing the cobalamin-binding domain. J Biol Chem. 1989;264:13888–13895. [PubMed] [Google Scholar]
- 219.Bankaitis V A, Bassford P J. Regulation of adenylate cyclase synthesis in Escherichia coli: studies with cya-lac operon and protein fusion strains. J Bacteriol. 1982;151:1346–1357. doi: 10.1128/jb.151.3.1346-1357.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Banuett F, Herskowitz I. Identification of polypeptides encoded by an Escherichia coli locus (hflA) that governs the lysis-lysogeny decision of bacteriophage λ. J Bacteriol. 1987;169:4076–4085. doi: 10.1128/jb.169.9.4076-4085.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Banuett F, Hoyt M A, McFarlane L, Echols H, Herskowitz I. hflB, a new Escherichia coli locus regulating lysogeny and the level of bacteriophage lambda cII protein. J Mol Biol. 1986;187:213–224. doi: 10.1016/0022-2836(86)90229-9. [DOI] [PubMed] [Google Scholar]
- 222.Baquero M R, Bouzon M, Varea J, Moreno F. sbmC, a stationary-phase induced SOS Escherichia coli gene, whose product protects cells from the DNA replication inhibitor microcin B17. Mol Microbiol. 1995;18:301–311. doi: 10.1111/j.1365-2958.1995.mmi_18020301.x. [DOI] [PubMed] [Google Scholar]
- 223.Baquero M-R, Bouzon M, Quintela J C, Ayala J A, Moreno F. dacD, an Escherichia coli gene encoding a novel penicillin-binding protein (PBP6b) with dd-carboxypeptidase activity. J Bacteriol. 1996;178:7106–7111. doi: 10.1128/jb.178.24.7106-7111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Barak R, Abouhamad W N, Eisenbach M. Both acetate kinase and acetyl coenzyme A synthetase are involved in acetate-stimulated change in the direction of flagellar rotation in Escherichia coli. J Bacteriol. 1998;180:985–988. doi: 10.1128/jb.180.4.985-988.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Barak Z, Gilvarg C. Triornithine-resistant strains of Escherichia coli. Isolation, definition, and genetic studies. J Biol Chem. 1974;249:143–148. [PubMed] [Google Scholar]
- 226.Barbier C S, Short S A. Studies on deo operon regulation in Escherichia coli: cloning and expression of the cytR structural gene. Gene. 1985;36:37–44. doi: 10.1016/0378-1119(85)90067-8. [DOI] [PubMed] [Google Scholar]
- 227.Bardwell J C, Craig E A. Major heat shock gene of Drosophila and the Escherichia coli heat-inducible dnaK gene are homologous. Proc Natl Acad Sci USA. 1984;81:848–852. doi: 10.1073/pnas.81.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Bardwell J C, Craig E A. Eukaryotic Mr 83,000 heat shock protein has a homologue in Escherichia coli. Proc Natl Acad Sci USA. 1987;84:5177–5181. doi: 10.1073/pnas.84.15.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Bardwell J C, Lee J O, Jander G, Martin N, Belin D, Beckwith J R. A pathway for disulfide bond formation in vivo. Proc Natl Acad Sci USA. 1993;90:1038–1042. doi: 10.1073/pnas.90.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Bardwell J C, McGovern K, Beckwith J R. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 231.Bardwell J C, Tilly K, Craig E A, King J, Zylicz M, Georgopoulos C. The nucleotide sequence of the Escherichia coli K12 and dnaJ+ gene: a gene that encodes a heat shock protein. J Biol Chem. 1986;261:1782–1785. [PubMed] [Google Scholar]
- 232.Barker D F, Campbell A. Use of bio-lac fusion strains to study regulation of biotin biosynthesis in Escherichia coli. J Bacteriol. 1980;143:789–800. doi: 10.1128/jb.143.2.789-800.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Barker D F, Campbell A. Genetic and biochemical characterization of the birA gene and its product: evidence for a direct role of biotin holoenzyme synthetase in repression of the biotin in Escherichia coli. J Mol Biol. 1981;146:469–492. doi: 10.1016/0022-2836(81)90043-7. [DOI] [PubMed] [Google Scholar]
- 234.Barker D F, Campbell A. The birA gene of Escherichia coli encodes a biotin holoenzyme synthetase. J Mol Biol. 1981;146:451–467. doi: 10.1016/0022-2836(81)90042-5. [DOI] [PubMed] [Google Scholar]
- 235.Barker D F, Kuhn J, Campbell A. Sequence and properties of operator mutations in the bio operon of Escherichia coli. Gene. 1981;13:89–102. doi: 10.1016/0378-1119(81)90046-9. [DOI] [PubMed] [Google Scholar]
- 236.Barker D G, Bruton J, Winter G. The tyrosyl tRNA synthetase from Escherichia coli. Complete nucleotide sequence of the structural gene. FEBS Lett. 1982;150:419–423. doi: 10.1016/0014-5793(82)80781-3. [DOI] [PubMed] [Google Scholar]
- 237.Baron C, Heider J, Bock A. Mutagenesis of selC, the gene for the selenocysteine-inserting tRNA species in E. coli: effects on in vivo function. Nucleic Acids Res. 1990;19:6761–6766. doi: 10.1093/nar/18.23.6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Barondess J J, Carson M, Guzman L, Beckwith J R. Alkaline phosphatase fusions in the study of cell division genes. Res Microbiol. 1991;142:295–299. doi: 10.1016/0923-2508(91)90044-b. [DOI] [PubMed] [Google Scholar]
- 239.Barr G C, Bhriain N N, Dorman C J. Identification of two new genetically active regions associated with the osmZ locus of Escherichia coli: role in regulation of proU expression and mutagenic effect of cya, the structural gene for adenylate cyclase. J Bacteriol. 1992;174:998–1006. doi: 10.1128/jb.174.3.998-1006.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Barr G C, Palm-Nicholls S E. Cloning the chlC gene for nitrate reductase of Escherichia coli. FEMS Microbiol Lett. 1981;11:213–216. [Google Scholar]
- 241.Barr K, Rick P D. Physical map location of the rffC and rffA genes of Escherichia coli. J Bacteriol. 1993;175:5738–5739. doi: 10.1128/jb.175.17.5738-5739.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Barroga C F, Zhang H, Wajih N, Bouyer J H, Hermodson M A. The proteins encoded by the rbs operon of Escherichia coli: I. Overproduction, purification, characterization, and functional analysis of RbsA. Protein Sci. 1996;5:1093–1099. doi: 10.1002/pro.5560050611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Barry G, Squires C, Squires C L. Attenuation and processing of RNA from the rplJL-rpoBC transcription unit of Escherichia coli. Proc Natl Acad Sci USA. 1980;77:3331–3335. doi: 10.1073/pnas.77.6.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Barry G, Squires C L, Squires C. Control features within the rplJL-rpoBC transcription unit of Escherichia coli. Proc Natl Acad Sci USA. 1979;76:4922–4926. doi: 10.1073/pnas.76.10.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Bartkus J M, Tyler B, Calvo J M. Transcription attenuation-mediated control of leu operon expression: influence of the number of Leu control codons. J Bacteriol. 1991;173:1634–1641. doi: 10.1128/jb.173.5.1634-1641.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Bartlett D H, Frantz B B, Matsumura P. Flagellar transcriptional activators FlbB and FlaI: gene sequence and 5′ consensus sequences of operons under FlbB and FlaI control. J Bacteriol. 1988;170:1575–1581. doi: 10.1128/jb.170.4.1575-1581.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Bartlett D H, Matsumura P. Identification of Escherichia coli region III flagellar gene products and description of two new flagellar genes. J Bacteriol. 1984;160:577–585. doi: 10.1128/jb.160.2.577-585.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Bartlett M S, Gourse R L. Growth rate-dependent control of the rrnB P1 core promoter in Escherichia coli. J Bacteriol. 1994;176:5560–5564. doi: 10.1128/jb.176.17.5560-5564.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Bartsch K, von Johnn-Marteville A, Schulz A. Molecular analysis of two genes of the Escherichia coli gab cluster: nucleotide sequence of the glutamate:succinic semialdehyde transaminase gene (gabT) and characterization of the succinic semialdehyde dehydrogenase gene (gabD) J Bacteriol. 1990;172:7035–7042. doi: 10.1128/jb.172.12.7035-7042.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Bass S, Gu Q, Christen A. Multicopy suppressors of Prc mutant Escherichia coli include two HtrA (DegP) protease homologs (HhoAB), DksA, and a truncated RlpA. J Bacteriol. 1996;178:1154–1161. doi: 10.1128/jb.178.4.1154-1161.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Bassford P J, Jr, Bradbeer C, Kadner R J, Schnaitman C A. Transport of vitamin B12 in tonB mutants of Escherichia coli. J Bacteriol. 1976;128:242–247. doi: 10.1128/jb.128.1.242-247.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Bassilana M, Gwizdek C. In vivo membrane assembly of the E. coli polytopic protein, melibiose permease, occurs via a Sec-independent process which requires the protonmotive force. EMBO J. 1996;15:5202–5208. [PMC free article] [PubMed] [Google Scholar]
- 253.Bates D B, Asai T, Cao Y, Chambers M W, Cadwell G W, Boye E, Kogoma T. The DnaA box R4 in the minimal oriC is dispensable for initiation of Escherichia coli chromosome replication. Nucleic Acids Res. 1995;23:3119–3125. doi: 10.1093/nar/23.16.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Baumberg S. Acetylhistidine as substrate for acetylornithinase: a new system for the selection of arginine regulation mutants in Escherichia coli. Mol Gen Genet. 1970;106:162–173. doi: 10.1007/BF00323835. [DOI] [PubMed] [Google Scholar]
- 255.Baumberg S. Genetic control of arginine metabolism in prokaryotes. In: MacDonald K D, editor. Proceedings of the Second International Symposium on Genetics and Industrial Microorganisms. London, United Kingdom: Academic Press; 1976. pp. 369–389. [Google Scholar]
- 256.Baumstark B R, Spremulli L L, RajBhandary U L, Brown G M. Initiation of protein synthesis without formylation in a mutant of Escherichia coli that grows in the absence of tetrahydrofolate. J Bacteriol. 1977;129:457–471. doi: 10.1128/jb.129.1.457-471.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Baylor N W, Williams A L, Cofie N. Molecular characteristics of ilvC specialized transducing phages of Escherichia coli K-12. Mol Gen Genet. 1983;191:347–352. doi: 10.1007/BF00425744. [DOI] [PubMed] [Google Scholar]
- 258.Beacham I R, Kahana R, Levy L, Yagil E. Mutants of Escherichia coli K-12 “cryptic” or deficient in 5′-nucleotidase (uridine diphosphate-sugar hydrolase) and 3′-nucleotidase (cyclic phosphodiesterase) activity. J Bacteriol. 1973;116:957–964. doi: 10.1128/jb.116.2.957-964.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Beacham I R, Garrett S. Isolation of Escherichia coli mutants (cpdB) deficient in periplasmic 2′:3′-cyclic phosphodiesterase and genetic mapping of the cpdB locus. J Gen Microbiol. 1980;119:31–34. doi: 10.1099/00221287-119-1-31. [DOI] [PubMed] [Google Scholar]
- 260.Beall B, Lutkenhaus J. Sequence analysis, transcriptional organization, and insertional mutagenesis of the envA gene of Escherichia coli. J Bacteriol. 1987;169:5408–5415. doi: 10.1128/jb.169.12.5408-5415.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Beauchamp B B, Richardson C C. A unique deoxyguanosine triphosphatase is responsible for the OptA1 phenotype of Escherichia coli. Proc Natl Acad Sci USA. 1988;85:2563–2567. doi: 10.1073/pnas.85.8.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Bebbington K J, Williams H D. Investigation of the role of the cydD gene product in production of a functional cytochrome d oxidase in Escherichia coli. FEMS Microbiol Lett. 1993;112:19–24. doi: 10.1111/j.1574-6968.1993.tb06417.x. [DOI] [PubMed] [Google Scholar]
- 263.Bebenek A, Pietrzykowska I. A new mutation in Escherichia coli K12, isfA, which is responsible for inhibition of SOS functions. Mol Gen Genet. 1995;248:103–113. doi: 10.1007/BF02456619. [DOI] [PubMed] [Google Scholar]
- 264.Bebenek A, Pietrzykowska I. The isfA mutation inhibits mutator activity and processing of UmuD protein in Escherichia coli recA730 strains. Mol Gen Genet. 1996;250:674–680. doi: 10.1007/BF02172978. [DOI] [PubMed] [Google Scholar]
- 265.Becerril B, Valle F, Marino E, Riba L, Bolivar F. Repetitive extragenic palindromic (REP) sequences in the Escherichia coli gdhA gene. Gene. 1985;37:53–62. doi: 10.1016/0378-1119(85)90257-4. [DOI] [PubMed] [Google Scholar]
- 266.Bech F W, Jorgensen S T, Diderichsen B. Sequence of the relB transcription unit from Escherichia coli and identification of the relB gene. EMBO J. 1985;4:1059–1066. doi: 10.1002/j.1460-2075.1985.tb03739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Beck E, Bremer E. Nucleotide sequence of the gene ompA coding the outer membrane protein II* of Escherichia coli. Nucleic Acids Res. 1980;8:3011–3024. doi: 10.1093/nar/8.13.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Beck R, Crooke H, Jarsch M, Cole J A, Burtscher H. Mutation in dipZ leads to reduced production of active human placental alkaline phosphatase in Escherichia coli. FEMS Microbiol Lett. 1994;124:209–214. doi: 10.1016/0378-1097(94)90251-8. [DOI] [PubMed] [Google Scholar]
- 269.Becker S, Plapp R. Location of the dcp gene on the physical map of Escherichia coli. J Bacteriol. 1992;174:1698–1699. doi: 10.1128/jb.174.5.1698-1699.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Becker-Hapak M, Eisenstark A. Role of rpoS in the regulation of glutathione oxidoreductase (gor) in Escherichia coli. FEMS Microbiol Lett. 1995;134:39–44. doi: 10.1111/j.1574-6968.1995.tb07911.x. [DOI] [PubMed] [Google Scholar]
- 271.Bedouelle H. Mutations in the promoter regions of the malEFG and malK-lamB operons of Escherichia coli K12. J Mol Biol. 1983;170:861–882. doi: 10.1016/s0022-2836(83)80192-2. [DOI] [PubMed] [Google Scholar]
- 272.Bedouelle H, Hofnung M. A DNA sequence containing the control regions of the malEFG and malK-lamB operons in Escherichia coli K12. Mol Gen Genet. 1982;185:82–87. doi: 10.1007/BF00333794. [DOI] [PubMed] [Google Scholar]
- 273.Bedouelle H, Bassford P J, Fowler A, Zabin I, Beckwith J R, Hofnung M. Mutations which alter the function of the signal sequence of the maltose binding protein of Escherichia coli. Nature. 1980;285:78–81. doi: 10.1038/285078a0. [DOI] [PubMed] [Google Scholar]
- 274.Bedouelle H, Schmeissner U, Hofnung M, Rosenberg L. Promoters of the malEFG and malK-lamB operons in Escherichia coli K12. J Mol Biol. 1982;161:519–531. doi: 10.1016/0022-2836(82)90405-3. [DOI] [PubMed] [Google Scholar]
- 275.Bedwell D M, Davis G R, Gosink M, Post L, Nomura M, Kestler H, Zengel J M, Lindahl L. Nucleotide sequence of the alpha ribosomal protein operon of Escherichia coli. Nucleic Acids Res. 1985;13:3891–3903. doi: 10.1093/nar/13.11.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Begg K J, Takasuga A, Edwards D H, Dewar S J, Spratt B G, Adachi T, Ohta T, Matsuzawa H, Donachie W D. The balance between different peptidoglycan precursors determines whether Escherichia coli cells will elongate or divide. J Bacteriol. 1990;172:6697–6703. doi: 10.1128/jb.172.12.6697-6703.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Begg K J, Hatfull G F, Donachie W D. Identification of new genes in a cell envelope-cell division gene cluster of Escherichia coli: cell division gene ftsQ. J Bacteriol. 1980;144:435–437. doi: 10.1128/jb.144.1.435-437.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Begg K J, Dewar S J, Donachie W D. A new Escherichia coli cell division gene, ftsK. J Bacteriol. 1995;177:6211–6222. doi: 10.1128/jb.177.21.6211-6222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Begg K J, Donachie W D. Cell shape and division in Escherichia coli: experiments with shape and division mutants. J Bacteriol. 1985;163:615–622. doi: 10.1128/jb.163.2.615-622.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Beher M G, Schnaitman C A. Regulation of the OmpA outer membrane protein of Escherichia coli. J Bacteriol. 1981;147:972–985. doi: 10.1128/jb.147.3.972-985.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Bejar S, Bouché J P. Molecular cloning of the region of the terminus of Escherichia coli K-12 DNA replication. J Bacteriol. 1983;153:604–609. doi: 10.1128/jb.153.2.604-609.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Bejar S, Bouché J P. A new dispensable genetic locus of the terminus region involved in control of cell division in Escherichia coli. Mol Gen Genet. 1985;201:146–150. doi: 10.1007/BF00425651. [DOI] [PubMed] [Google Scholar]
- 283.Bejar S, Cam K, Bouché J P. Control of cell division in Escherichia coli. DNA sequence of dicA and of a second gene complementing mutation of dicA1, dicC. Nucleic Acids Res. 1986;14:6821–6833. doi: 10.1093/nar/14.17.6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Belfaiza J C, Parsot C, Martel A, Bouthier de la Tour C, Margarita D, Cohen G N, Saint-Girons I. Evolution in biosynthetic pathways: two enzymes catalyzing consecutive steps in methionine biosynthesis originate from a common ancestor and possess a similar regulatory region. Proc Natl Acad Sci USA. 1986;83:867–871. doi: 10.1073/pnas.83.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Belfort M, Wulff D. Genetic and biochemical investigation of the Escherichia coli mutant hfl-1 which is lysogenized at high frequency by bacteriophage lambda. J Bacteriol. 1973;115:299–306. doi: 10.1128/jb.115.1.299-306.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Belfort M, Maley G, Pederson-Lane J, Maley F. Primary structure of the Escherichia coli thyA gene and its thymidylate synthase product. Proc Natl Acad Sci USA. 1983;80:4914–4918. doi: 10.1073/pnas.80.16.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Belin P, Quemeneur E, Boquet P L. A pleiotropic acid phosphatase-deficient mutant of Escherichia coli shows premature termination in the dsbA gene. Use of dsbA::phoA fusions to localize a structurally important domain in DsbA. Mol Gen Genet. 1994;242:23–32. doi: 10.1007/BF00277344. [DOI] [PubMed] [Google Scholar]
- 288.Belin P, Boquet P L. A second gene involved in the formation of disulfide bonds in proteins localized in Escherichia coli periplasmic space. C R Acad Sci Ser III. 1993;316:469–473. . (In French.) [PubMed] [Google Scholar]
- 289.Belitskii B R, Kulakauskas S T, Sukhodolets V V, Shakulov R S. Precise mapping of the gap gene involved in guanosine tetraphosphate synthesis and deletion of the ilvC-gpp region of Escherichia coli chromosome. Sov Genet (Engl Transl Genetika) 1985;22:1539–1546. [PubMed] [Google Scholar]
- 290.Beljanski M, Pourgarel P. Showdomycine et biosynthese d’ARN non complementaires de ′ADN. Ann Inst Pasteur. 1970;118:253–276. [PubMed] [Google Scholar]
- 291.Bell A W, Buckel S D, Groarke J M, Hope J N, Kingsley D H, Hermodsen A. The nucleotide sequences of the rbsD, rbsA and rbsC genes of Escherichia coli K12. J Biol Chem. 1986;261:7652–7658. [PubMed] [Google Scholar]
- 292.Bell P J, Andrews S C, Sivak M N, Guest J R. Nucleotide sequence of the FNR-regulated fumarase gene (fumB) of Escherichia coli K-12. J Bacteriol. 1989;171:3494–3503. doi: 10.1128/jb.171.6.3494-3503.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Belunis C J, Clementz T, Carty S M, Raetz C R. Inhibition of lipopolysaccharide biosynthesis and cell growth following inactivation of the kdtA gene in Escherichia coli. J Biol Chem. 1995;270:27646–27652. doi: 10.1074/jbc.270.46.27646. [DOI] [PubMed] [Google Scholar]
- 294.Ben-Bassat A, Bauer K, Chang S-Y, Myambo K, Boosman A, Chang S. Processing of the initiation methionine from proteins: properties of the Escherichia coli methionine aminopeptidase and its gene structure. J Bacteriol. 1987;169:751–757. doi: 10.1128/jb.169.2.751-757.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Bencini D A, Houghton J E, Hoover T A, Folterman K F, Wild J R, O’Donovan G A. The DNA sequence of argI from Escherichia coli K12. Nucleic Acids Res. 1983;11:8509–8518. doi: 10.1093/nar/11.23.8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Bendiak D S, Friesen J D. Organization of genes in the four minute region of the Escherichia coli chromosome: evidence that rpsB and tsf are co-transcribed. Mol Gen Genet. 1981;181:356–362. doi: 10.1007/BF00425611. [DOI] [PubMed] [Google Scholar]
- 297.Ben-Neria T, Ron E Z. A DNA replication gene maps near terC in Escherichia coli K12. Mol Gen Genet. 1991;226:315–317. doi: 10.1007/BF00273619. [DOI] [PubMed] [Google Scholar]
- 298.Ben-Neria T, Ron E Z. A cluster of cell division genes maps to the terC region of the chromosome of Escherichia coli K-12. Mol Gen Genet. 1995;246:605–609. doi: 10.1007/BF00298967. [DOI] [PubMed] [Google Scholar]
- 299.Bennett R J, West S C. Resolution of Holliday junctions in genetic recombination: RuvC protein nicks DNA at the point of strand exchange. Proc Natl Acad Sci USA. 1996;93:12217–12222. doi: 10.1073/pnas.93.22.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Bennett R L, Malamy M. Arsenate resistant mutants of Escherichia coli and phosphate transport. Biochem Biophys Res Commun. 1970;40:469–503. doi: 10.1016/0006-291x(70)91036-3. [DOI] [PubMed] [Google Scholar]
- 301.Benov L, Fridovich I. A superoxide dismutase mimic protects sodA sodB Escherichia coli against aerobic heating and stationary-phase death. Arch Biochem Biophys. 1995;322:291–294. doi: 10.1006/abbi.1995.1465. [DOI] [PubMed] [Google Scholar]
- 302.Benov L, Fridovich I. Functional significance of the Cu,ZnSOD in Escherichia coli. Arch Biochem Biophys. 1996;327:249–253. doi: 10.1006/abbi.1996.0117. [DOI] [PubMed] [Google Scholar]
- 303.Benov L T, Beyer W F, Stevens R D, Fridovich I. Purification and characterization of the Cu,Zn SOD from Escherichia coli. Free Radic Biol Med. 1996;21:117–121. doi: 10.1016/0891-5849(95)02217-1. [DOI] [PubMed] [Google Scholar]
- 304.Benson C E, Love S H, Remy C N. Inhibition of de novo purine biosynthesis and interconversion by 6-methyl purine in Escherichia coli. J Bacteriol. 1970;101:872–880. doi: 10.1128/jb.101.3.872-880.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Benson F, Collier S, Lloyd R G. Evidence of abortive recombination in ruv mutants of Escherichia coli K12. Mol Gen Genet. 1991;225:266–272. doi: 10.1007/BF00269858. [DOI] [PubMed] [Google Scholar]
- 306.Benson F E, Illing G T, Sharples G J, Lloyd R G. Nucleotide sequencing of the ruv region of Escherichia coli K-12 reveals a LexA regulated operon encoding two genes. Nucleic Acids Res. 1988;16:1541–1549. doi: 10.1093/nar/16.4.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Benson F E, West S C. Substrate specificity of the Escherichia coli RuvC protein. Resolution of three- and four-stranded recombination intermediates. J Biol Chem. 1994;269:5195–5201. [PubMed] [Google Scholar]
- 308.Benson T, Marquardt J L, Marquardt A C, Etzkorn F A, Walsh C T. Overexpression, purification, and mechanistic study of UDP-N-acetylenolpyruvylglucosamine reductase. Biochemistry. 1993;32:2024–2030. doi: 10.1021/bi00059a019. [DOI] [PubMed] [Google Scholar]
- 309.Bentley J, Hyatt L S, Ainley K, Parish J H, Herbert R B, White G R. Cloning and sequence analysis of an Escherichia coli gene conferring bicyclomycin resistance. Gene. 1993;127:117–120. doi: 10.1016/0378-1119(93)90625-d. [DOI] [PubMed] [Google Scholar]
- 310.Bentley R, Meganathan R. Biosynthesis of the isoprenoid quinones ubiquinone and menaquinone. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 512–520. [Google Scholar]
- 311.Beny G, Boyen A, Charlier D, Lissens W, Feller A, Glansdorff N. Promoter mapping and selection of operator mutants by using insertion of bacteriophage Mu in the argECBH divergent operon of Escherichia coli K-12. J Bacteriol. 1982;151:62–67. doi: 10.1128/jb.151.1.62-67.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Berg B L, Baron C, Stewart V J. Nitrate-inducible formate dehydrogenase in Escherichia coli K-12. II. Evidence that a mRNA stem-loop structure is essential for decoding opal (UGA) as selenocysteine. J Biol Chem. 1991;266:22386–22391. [PubMed] [Google Scholar]
- 313.Berg B L, Li J, Heider J, Stewart V J. Nitrate-inducible formate dehydrogenase in Escherichia coli K-12. I. Nucleotide sequence of the fdnGHI operon and evidence that opal (UGA) encodes selenocysteine. J Biol Chem. 1991;266:22380–22385. [PubMed] [Google Scholar]
- 314.Berg B L, Stewart V J. Structural genes for nitrate-inducible formate dehydrogenase in Escherichia coli K-12. Genetics. 1990;125:691–702. doi: 10.1093/genetics/125.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Berg C M, Rossi J J. Proline excretion and indirect suppression in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1974;118:928–939. doi: 10.1128/jb.118.3.928-934.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Berg C M, Shaw K J, Berg D E. The ivlG gene is expressed in Escherichia coli K-12. Gene. 1980;12:165–170. doi: 10.1016/0378-1119(80)90028-1. [DOI] [PubMed] [Google Scholar]
- 317.Berg C M, Shaw K J, Vender J, Borucka-Mankiewicz M. Physiological characterization of polar Tn5-induced isoleucine-valine auxotrophs in Escherichia coli: evidence for an internal promoter in the ilvOGEDA operon. Genetics. 1979;93:309–319. [PMC free article] [PubMed] [Google Scholar]
- 318.Berg K L, Squires C L, Squires C. In vivo translation of a region within rrnB 16S rRNA gene of Escherichia coli. J Bacteriol. 1987;169:1691–1701. doi: 10.1128/jb.169.4.1691-1701.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Berg P E. Cloning and characterization of the Escherichia coli gene coding for alkaline phosphatase. J Bacteriol. 1981;146:660–667. doi: 10.1128/jb.146.2.660-667.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Bergler H, Abraham D, Aschauer H, Turnowsky F. Inhibition of lipid biosynthesis induces the expression of the pspA gene. Microbiology. 1994;140:1937–1944. doi: 10.1099/13500872-140-8-1937. [DOI] [PubMed] [Google Scholar]
- 321.Bergler H, Hogenauer G, Turnowsky F. Sequences of the envM gene and of two mutated alleles in Escherichia coli. J Gen Microbiol. 1992;138:2093–2100. doi: 10.1099/00221287-138-10-2093. [DOI] [PubMed] [Google Scholar]
- 322.Bergler H, Wallner P, Ebeling A, Leitinger B, Fuchsbichler S, Aschauer H, Kollenz G, Hogenauer G, Turnowsky F. Protein EnvM is the NADH-dependent enoyl-ACP reductase (FabI) of Escherichia coli. J Biol Chem. 1994;269:5493–5496. [PubMed] [Google Scholar]
- 322a.Bergler, H., et al. 1994. GenBank submission U08190.
- 323.Berlyn M K B, Low K B, Rudd K E. Linkage map of Escherichia coli K-12, edition 9. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1715–1902. [Google Scholar]
- 323a.Berlyn M K B. Accessing the E. coli Genetic Stock Center Database. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Rezerikoff W S, Riley M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2489–2495. [Google Scholar]
- 324.Berman-Kurtz M, Lin E C C, Richey D P. Promoter-like mutant with increased expression of the glycerol kinase operon of Escherichia coli. J Bacteriol. 1971;106:724–731. doi: 10.1128/jb.106.3.724-731.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Bernstein H D, Zopf D, Freymann D M, Walter P. Functional substitution of the signal recognition particle 54-kDa subunit by its Escherichia coli homolog. Proc Natl Acad Sci USA. 1993;90:5229–5233. doi: 10.1073/pnas.90.11.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Berry A, Marshall K E. Identification of zinc-binding ligands in the class II fructose-1,6-bisphosphate aldolase of Escherichia coli. FEBS Lett. 1993;318:11–16. doi: 10.1016/0014-5793(93)81317-s. [DOI] [PubMed] [Google Scholar]
- 327.Bertin P, Terao E, Lee E H, Lejeune P, Colson C, Danchin A, Collatz E. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J Bacteriol. 1994;176:5537–5540. doi: 10.1128/jb.176.17.5537-5540.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Bertino J B, Stacey K A. A suggested mechanism for the selective procedure for isolating thymine-requiring mutants of Escherichia coli. Biochem J. 1966;101:32–33. doi: 10.1042/bj1010032c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Betton J, Hofnung M. Folding of a mutant maltose-binding protein of Escherichia coli which forms inclusion bodies. J Biol Chem. 1996;271:8046–8052. doi: 10.1074/jbc.271.14.8046. [DOI] [PubMed] [Google Scholar]
- 330.Betton J M, Boscus D, Missiakas D, Raina S, Hofnung M. Probing the structural role of an alpha beta loop of maltose-binding protein by mutagenesis: heat-shock induction by loop variants of the maltose-binding protein that form periplasmic inclusion bodies. J Mol Biol. 1996;262:140–150. doi: 10.1006/jmbi.1996.0504. [DOI] [PubMed] [Google Scholar]
- 331.Betz J L. Cloning and characterization of several dominant-negative and tight-binding mutants of lac repressor. Gene. 1986;42:283–292. doi: 10.1016/0378-1119(86)90232-5. [DOI] [PubMed] [Google Scholar]
- 332.Betzner A S, Keck W. Molecular cloning, overexpression and mapping of the slt gene encoding the soluble lytic transglycosylase of Escherichia coli. Mol Gen Genet. 1989;219:489–491. doi: 10.1007/BF00259625. [DOI] [PubMed] [Google Scholar]
- 333.Beutin L, Manning P A, Achtman M, Willets N. sfrA and sfrB products of Escherichia coli K-12 are transcriptional control factors. J Bacteriol. 1981;145:840–844. doi: 10.1128/jb.145.2.840-844.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Bewick M A, Lo T C Y. Localization of the dicarboxylate binding protein in the cell envelope of Escherichia coli K12. Can J Biochem. 1980;58:885–897. doi: 10.1139/o80-123. [DOI] [PubMed] [Google Scholar]
- 334a.Beyersmann D, Messer W, Schlicht M. Mutants of Escherichia coli B/r defective in deoxyribonucleic acid initiation: dnaI, a new gene for replication. J Bacteriol. 1974;118:783–789. doi: 10.1128/jb.118.3.783-789.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Beyreuther K, Adler K, Fanning E, Murray C, Klemm A, Geisler N. Amino-acid sequence of lac repressor from Escherichia coli. Isolation, sequence analysis and sequence assembly of tryptic peptides and cyanogen-bromide fragments. Eur J Biochem. 1975;59:491–509. doi: 10.1111/j.1432-1033.1975.tb02477.x. [DOI] [PubMed] [Google Scholar]
- 336.Bhagwat A S, Sohail A, Lieb M. A new gene involved in mismatch correction in Escherichia coli. Gene. 1988;74:155–156. doi: 10.1016/0378-1119(88)90274-0. [DOI] [PubMed] [Google Scholar]
- 337.Bhagwat A S, Sohail A, Roberts R J. Cloning and characterization of the dcm locus of Escherichia coli K-12. J Bacteriol. 1986;166:751–755. doi: 10.1128/jb.166.3.751-755.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Bhatnagar S K, Bessman M J. Studies on the mutator gene, mutT, of Escherichia coli. Molecular cloning of the gene, purification of the gene product, and identification of a novel nucleoside triphosphatase. J Biol Chem. 1988;263:8953–8957. [PubMed] [Google Scholar]
- 339.Bi E, Lutkenhaus J. FtsZ regulates frequency of cell division in Escherichia coli. J Bacteriol. 1990;172:2765–2768. doi: 10.1128/jb.172.5.2765-2768.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Bi E, Lutkenhaus J. Interaction between the min locus and ftsZ. J Bacteriol. 1990;172:5610–5616. doi: 10.1128/jb.172.10.5610-5616.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Bi E, Lutkenhaus J. Isolation and characterization of ftsZ alleles that affect septal morphology. J Bacteriol. 1992;174:5414–5423. doi: 10.1128/jb.174.16.5414-5423.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Bialkowska-Hobrzanska H, Gilchrist C A, Denhardt D T. Escherichia coli rep gene: identification of the promoter and N terminus of the rep protein. J Bacteriol. 1985;164:1004–1010. doi: 10.1128/jb.164.3.1004-1010.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Bialkowska-Hobrzanska H, Denhardt D. The rep mutation. VII. Cloning and analysis of the functional rep gene of Escherichia coli K-12. Gene. 1984;28:93–102. doi: 10.1016/0378-1119(84)90091-x. [DOI] [PubMed] [Google Scholar]
- 344.Bianchi V, Reichard P, Eliasson R, Pontis E, Krook M, Jörnvall H, Haggård-Ljungquist E. Escherichia coli ferredoxin NADP+ reductase: activation of E. coli anaerobic ribonucleotide reduction, cloning of the gene (fpr), and overexpression of the protein. J Bacteriol. 1993;175:1590–1595. doi: 10.1128/jb.175.6.1590-1595.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Bianco P R, Kowalczykowski S C. The recombination hotspot Chi is recognized by the translocating RecBCD enzyme as the single strand of DNA containing the sequence 5′-GCTGGTGG-3′. Proc Natl Acad Sci USA. 1997;94:6706–6711. doi: 10.1073/pnas.94.13.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Bibi E, Kaback H R. Functional complementation of internal deletion mutants in the lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 1992;89:1524–1528. doi: 10.1073/pnas.89.5.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Bibikov S I, Biran R, Rudd K E, Parkinson J S. A signal transducer for aerotaxis in Escherichia coli. J Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Biek D P, Cohen S N. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J Bacteriol. 1986;167:594–603. doi: 10.1128/jb.167.2.594-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Bieker K L, Phillips G J, Silhavy T J. The sec and prl genes of Escherichia coli. J Bioenerg Biomembr. 1990;22:291–310. doi: 10.1007/BF00763169. [DOI] [PubMed] [Google Scholar]
- 350.Bieker K L, Silhavy T J. PrlA (SecY) and PrlG (SecE) interact directly and function sequentially during protein translocation in E. coli. Cell. 1990;61:833–842. doi: 10.1016/0092-8674(90)90193-i. [DOI] [PubMed] [Google Scholar]
- 351.Biel A J, Umbarger H E. Mutations in the ilvY gene of Escherichia coli K-12 that cause constitutive expression of ilvC. J Bacteriol. 1981;146:718–724. doi: 10.1128/jb.146.2.718-724.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Bierne H, Ehrlich S D, Michel B. Flanking sequences affect replication arrest at the Escherichia coli terminator TerB in vivo. J Bacteriol. 1994;176:4165–4167. doi: 10.1128/jb.176.13.4165-4167.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Bilous P T, Weiner J H. Molecular cloning and expression of the Escherichia coli dimethyl sulfoxide reductase operon. J Bacteriol. 1988;170:1511–1518. doi: 10.1128/jb.170.4.1511-1518.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Bilous P T, Cole S T, Anderson W F, Weiner J H. Nucleotide sequence of the dmsABC operon encoding the anaerobic dimethylsulphoxide reductase of Escherichia coli. Mol Microbiol. 1988;2:785–795. doi: 10.1111/j.1365-2958.1988.tb00090.x. [DOI] [PubMed] [Google Scholar]
- 355.Binding R, Romansky G, Bitner R M, Kuempel P. Isolation and properties of Tn10 insertions in the rac locus of Escherichia coli. Mol Gen Genet. 1981;183:333–340. doi: 10.1007/BF00270637. [DOI] [PubMed] [Google Scholar]
- 356.Bingham R J, Hall K S, Slonczewski J L. Alkaline induction of a novel gene locus, alx, in Escherichia coli. J Bacteriol. 1990;172:2184–2186. doi: 10.1128/jb.172.4.2184-2186.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Binkley J P, Kuempel P. Genetic mapping in Escherichia coli of tmk, the locus for dTMP kinase. J Bacteriol. 1986;168:1457–1458. doi: 10.1128/jb.168.3.1457-1458.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Biran D, Brot N, Weissbach H, Ron E Z. Heat shock-dependent transcriptional activation of the metA gene of Escherichia coli. J Bacteriol. 1995;177:1374–1379. doi: 10.1128/jb.177.5.1374-1379.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Biran D, Michaeli S, Segal G, Ron E Z. Location of the metA gene on the physical map of Escherichia coli. J Bacteriol. 1992;174:5753–5754. doi: 10.1128/jb.174.17.5753-5754.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Birkmann A, Zinoni F, Sawers G, Bock A. Factors affecting transcriptional regulation of the formate-hydrogen-lyase pathway of Escherichia coli. Arch Microbiol. 1987;148:44–51. doi: 10.1007/BF00429646. [DOI] [PubMed] [Google Scholar]
- 361.Bishop R E, Penfold S S, Frost L S, Holtje J V, Weiner J H. Stationary phase expression of a novel Escherichia coli outer membrane lipoprotein and its relationship with mammalian apolipoprotein D. Implications for the origin of lipocalins. J Biol Chem. 1995;270:23097–23103. doi: 10.1074/jbc.270.39.23097. [DOI] [PubMed] [Google Scholar]
- 362.Bitner R M, Kuempel P. P1 transduction map spanning the replication terminus of Escherichia coli K12. Mol Gen Genet. 1981;184:208–212. doi: 10.1007/BF00272906. [DOI] [PubMed] [Google Scholar]
- 363.Bitner R M, Kuempel P. P1 transduction mapping of the trg locus in rac+ and rac strains of Escherichia coli K-12. J Bacteriol. 1982;149:529–533. doi: 10.1128/jb.149.2.529-533.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Biville F, Turlin E, Gasser F. Mutants of Escherichia coli producing pyrroloquinoline quinone. J Gen Microbiol. 1991;137:1775–1782. doi: 10.1099/00221287-137-8-1775. [DOI] [PubMed] [Google Scholar]
- 365.Bjelland S, Seeberg E. Different efficiencies of the Tag and AlkA DNA glycosylases from Escherichia coli in the removal of 3-methyladenine from single-stranded DNA. FEBS Lett. 1996;397:127–129. doi: 10.1016/s0014-5793(96)01166-0. [DOI] [PubMed] [Google Scholar]
- 366.Bjork G R, Kjellin-Straby K. Escherichia coli mutants with defects in the biosynthesis of 5-methylamino-methyl-2-thiouridine or 1-methylguanosine in their tRNA. J Bacteriol. 1978;133:508–517. doi: 10.1128/jb.133.2.508-517.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Black D S, Kelly A J, Mardis M J, Moyed H S. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J Bacteriol. 1991;173:5732–5739. doi: 10.1128/jb.173.18.5732-5739.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Black D S, Irwin B, Moyed H S. Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J Bacteriol. 1994;176:4081–4091. doi: 10.1128/jb.176.13.4081-4091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Black M E, Hruby D. Nucleotide sequence of the Escherichia coli thymidine kinase gene provides evidence for conservation of functional domains and quaternary structure. Mol Microbiol. 1991;5:373–379. doi: 10.1111/j.1365-2958.1991.tb02119.x. [DOI] [PubMed] [Google Scholar]
- 370.Black P N. The fadL gene product of Escherichia coli is an outer membrane protein required for uptake of long-chain fatty acids and involved in sensitivity to bacteriophage T2. J Bacteriol. 1988;170:2850–2854. doi: 10.1128/jb.170.6.2850-2854.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Black P N, DiRusso C C, Metzger A K, Heimert T L. Cloning, sequencing, and expression of the fadD gene of Escherichia coli encoding acyl coenzyme A synthetase. J Biol Chem. 1992;267:25513–25520. [PubMed] [Google Scholar]
- 372.Black P N, Kianian S F, DiRusso C C, Nunn W D. Long-chain fatty acid transport in Escherichia coli. Cloning, mapping, and expression of the fadL gene. J Biol Chem. 1985;260:1780–1789. [PubMed] [Google Scholar]
- 373.Blair D F, Kim D Y, Berg H C. Mutant MotB proteins in Escherichia coli. J Bacteriol. 1991;173:4049–4055. doi: 10.1128/jb.173.13.4049-4055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Blair D F, Berg H C. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell. 1990;60:439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- 375.Blair D F, Berg H C. Mutations in the MotA protein of Escherichia coli reveal domains critical for proton conduction. J Mol Biol. 1991;221:1433–1442. doi: 10.1016/0022-2836(91)90943-z. [DOI] [PubMed] [Google Scholar]
- 376.Blakely G, Sherratt D. Determinants of selectivity in Xer site-specific recombination. Genes Dev. 1996;10:762–773. doi: 10.1101/gad.10.6.762. [DOI] [PubMed] [Google Scholar]
- 377.Blakely G, May G, McCulloch R, Arciszewska L K, Burke M E, Lovett S T, Sherratt D J. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell. 1993;75:351–361. doi: 10.1016/0092-8674(93)80076-q. [DOI] [PubMed] [Google Scholar]
- 378.Blakely G, Colloms S D, May G, Burke M E, Sherratt D J. Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol. 1991;3:789–798. [PubMed] [Google Scholar]
- 379.Blanar M A, Sandler S J, Armengod M-E, Ream L W, Clark A J. Molecular analysis of the recF gene of Escherichia coli. Proc Natl Acad Sci USA. 1984;81:4622–4626. doi: 10.1073/pnas.81.15.4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Blanchin-Roland S, Blanquet S, Schmitter J M, Fayet G. The gene for Escherichia coli diadenosine tetraphosphatase is located immediately clockwise to folA and forms an operon with ksgA. Mol Gen Genet. 1986;205:515–522. doi: 10.1007/BF00338091. [DOI] [PubMed] [Google Scholar]
- 381.Blanco C, Mata-Gilsinger M. A DNA sequence containing the control sites for the uxaB gene of Escherichia coli. J Gen Microbiol. 1986;132:697–705. doi: 10.1099/00221287-132-3-697. [DOI] [PubMed] [Google Scholar]
- 382.Blanco C, Mata-Gilsinger M. Identification of cyclic AMP-CRP binding sites in the intercistronic regulatory uxaCA-exuT region of Escherichia coli. FEMS Microbiol Lett. 1986;33:205–209. [Google Scholar]
- 383.Blanco C, Mata-Gilsinger M, Ritzenthaler P. Construction of hybrid plasmids containing the Escherichia coli uxaB gene: analysis of its regulation and direction of transcription. J Bacteriol. 1983;153:747–755. doi: 10.1128/jb.153.2.747-755.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Blanco C, Mata-Gilsinger M, Ritzenthaler P. The use of gene fusions to study the expression of uidR, a negative regulatory gene of Escherichia coli K-12. Gene. 1985;36:159–167. doi: 10.1016/0378-1119(85)90080-0. [DOI] [PubMed] [Google Scholar]
- 385.Blanco C, Ritzenthaler P, Kolb A. The regulatory region of the uxuAB operon in Escherichia coli K12. Mol Gen Genet. 1986;202:112–119. doi: 10.1007/BF00330526. [DOI] [PubMed] [Google Scholar]
- 386.Blanco C, Ritzenthaler P, Mata-Gilsinger M. Cloning and endonuclease restriction analysis of uidA and uidR genes in Escherichia coli K-12: determination of transcription direction for the uidA gene. J Bacteriol. 1982;149:587–594. doi: 10.1128/jb.149.2.587-594.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Blanco C, Ritzenthaler P, Mata-Gilsinger M. Nucleotide sequence of a regulatory region of the uidA gene in Escherichia coli K12. Mol Gen Genet. 1985;199:101–105. doi: 10.1007/BF00327517. [DOI] [PubMed] [Google Scholar]
- 388.Blanco C, Ritzenthaler P, Mata-Gilsinger M. Negative dominant mutations of the uidR gene in Escherichia coli: genetic proof for a cooperative regulation of uidA expression. Genetics. 1986;112:173–182. doi: 10.1093/genetics/112.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Blasband A J. Regulation in Escherichia coli of the porin protein gene encoded by lambdoid bacteriophages. J Bacteriol. 1987;169:2171–2176. doi: 10.1128/jb.169.5.2171-2176.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Blasband A J, Marcotte W R, Jr, Schnaitman C A. Structure of the lc and nmpC outer membrane porin protein genes of lambdoid bacteriophage. J Biol Chem. 1986;261:12723–12732. [PubMed] [Google Scholar]
- 391.Blasco F, Iobbi C. Nitrate reductases of Escherichia coli: sequence of the second nitrate reductase and comparison with that encoded by the narGHJI operon. Mol Gen Genet. 1990;222:104–111. doi: 10.1007/BF00283030. [DOI] [PubMed] [Google Scholar]
- 392.Blasco F, Iobbi C, Giordano G, Chippaux M, Bonnefoy V. Nitrate reductase of Escherichia coli: completion of the nucleotide sequence of the nar operon and reassessment of the role of the alpha and beta subunits in iron binding and electron transfer. Mol Gen Genet. 1989;218:249–256. doi: 10.1007/BF00331275. [DOI] [PubMed] [Google Scholar]
- 393.Blasco F, Pommier J, Augier V, Chippaux M, Giordano G. Involvement of the narJ or narW product in the formation of active nitrate reductase in Escherichia coli. Mol Microbiol. 1992;6:221–239. doi: 10.1111/j.1365-2958.1992.tb02003.x. [DOI] [PubMed] [Google Scholar]
- 394.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, McGregor J F, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 395.Blattner F R, Burland V D, Plunkett G, Sofia H J, Daniels D L. Analysis of the Escherichia coli genome. IV. DNA sequence of the region from 89.2 to 92.8 minutes. Nucleic Acids Res. 1993;21:5408–5417. doi: 10.1093/nar/21.23.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395a.Blattner, F. R., et al. GenBank submission U00096.
- 396.Blaut M, Whittaker K, Valdovinos A, Ackrell B A, Gunsalus R P, Cecchini G. Fumarate reductase mutants of Escherichia coli that lack covalently bound flavin. J Biol Chem. 1989;264:13599–13604. [PubMed] [Google Scholar]
- 397.Bledig S A, Ramseier T M, Saier M H., Jr FruR mediates catabolite activation of pyruvate kinase (pykF) gene expression in Escherichia coli. J Bacteriol. 1996;178:280–283. doi: 10.1128/jb.178.1.280-283.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Blinkowa A, Haldenwang W G, Ramsey J A, Henson J M, Mullin D A, Walker J R. Physiological properties of cold-sensitive suppressor mutations of a temperature-sensitive dnaZ mutant of Escherichia coli. J Bacteriol. 1983;153:66–75. doi: 10.1128/jb.153.1.66-75.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Blinkowa A L, Walker J R. Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III gamma subunit from within the tau subunit reading frame. Nucleic Acids Res. 1990;18:1725–1729. doi: 10.1093/nar/18.7.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Blount P, Sukharev S I, Schroeder M J, Nagle S K, Kung C. Single residue substitutions that change the gating properties of a mechanosensitive channel in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:11652–11657. doi: 10.1073/pnas.93.21.11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Blount P, Sukharev S I, Moe P C, Schroeder M J, Guy H R, Kung C. Membrane topology and multimeric structure of a mechanosensitive channel protein of Escherichia coli. EMBO J. 1996;15:4798–4805. [PMC free article] [PubMed] [Google Scholar]
- 402.Bloxham D P, Herbert C J, Giles I G, Ner S S. The use of bacteriophage M13 carrying defined fragments of the Escherichia coli gltA gene to determine the location and structure of the citrate synthase promoter region. Mol Gen Genet. 1983;191:499–506. doi: 10.1007/BF00425769. [DOI] [PubMed] [Google Scholar]
- 403.Blum P H, Jovanovich S B, McCann M P, Schultz J E, Lesley S A, Burgess R R, Matin A. Cloning and in vivo and in vitro regulation of cyclic AMP-dependent carbon starvation genes from Escherichia coli. J Bacteriol. 1990;172:3813–3820. doi: 10.1128/jb.172.7.3813-3820.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Blumenthal R M, Borst D W, Matthews R G. Experimental analysis of global gene regulation in Escherichia coli. Prog Nucleic Acid Res Mol Biol. 1996;55:1–86. doi: 10.1016/s0079-6603(08)60189-0. [DOI] [PubMed] [Google Scholar]
- 405.Bock A, Forchhammer K, Heider J, Baron C. Selenoprotein synthesis: an expansion of the genetic code. Trends Biochem Sci. 1991;16:463–467. doi: 10.1016/0968-0004(91)90180-4. [DOI] [PubMed] [Google Scholar]
- 406.Bockamp E O, Blasco R, Vinuela E. Escherichia coli thymidine kinase: nucleotide sequence and relationships to other thymidine kinases. Gene. 1991;101:9–14. doi: 10.1016/0378-1119(91)90218-z. [DOI] [PubMed] [Google Scholar]
- 406a.Boehmer P E, Emmerson P T. The RecB subunit of the Escherichia coli RecBCD enzyme couples ATP hydrolysis to DNA unwinding. J Biol Chem. 1992;267:4981–4987. [PubMed] [Google Scholar]
- 407.Boeke J, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 408.Bogan J A, Helmstetter C E. mioC transcription, initiation of replication, and the eclipse in Escherichia coli. J Bacteriol. 1996;178:3201–3206. doi: 10.1128/jb.178.11.3201-3206.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Bognar A L, Osborne C, Shane B. Primary structure of the Escherichia coli folC gene and its folypolyglutamate synthetase-dihydrofolate synthetase product and regulation of expression by an upstream gene. J Biol Chem. 1987;262:12337–12343. [PubMed] [Google Scholar]
- 410.Bogosian G, Haydock P V, Somerville R L. Indolmycin-mediated inhibition and stimulation of transcription at the trp promoter of Escherichia coli. J Bacteriol. 1983;153:1120–1123. doi: 10.1128/jb.153.2.1120-1123.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Bohin J P, Kennedy E P. Mapping of a locus (mdoA) that affects the biosynthesis of membrane-derived oligosaccharides in Escherichia coli. J Bacteriol. 1984;157:956–957. doi: 10.1128/jb.157.3.956-957.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Bohm R, Sauter M, Bock A. Nucleotide sequence and expression of an operon in Escherichia coli coding for formate hydrogenlyase components. Mol Microbiol. 1990;4:231–243. doi: 10.1111/j.1365-2958.1990.tb00590.x. [DOI] [PubMed] [Google Scholar]
- 413.Bohman K, Isaksson L A. Temperature-sensitive mutants in cysteinyl-tRNA ligase of Escherichia coli K-12. Mol Gen Genet. 1979;176:53–55. doi: 10.1007/BF00334295. [DOI] [PubMed] [Google Scholar]
- 414.Bohman K, Isaksson L A. A temperature-sensitive mutant in prolinyl-tRNA ligase of Escherichia coli K-12. Mol Gen Genet. 1980;177:603–605. doi: 10.1007/BF00272670. [DOI] [PubMed] [Google Scholar]
- 415.Boidol W, Simonis M, Topert M, Siewert G. Recombinant plasmids with genes for the biosynthesis of alkaline phosphatase of Escherichia coli. Mol Gen Genet. 1982;185:510–512. doi: 10.1007/BF00334150. [DOI] [PubMed] [Google Scholar]
- 416.Boiteux S, Huisman O. Isolation of a formamidopyrimidine-DNA glycosylase (fpg) mutant of Escherichia coli K12. Mol Gen Genet. 1989;215:300–305. doi: 10.1007/BF00339732. [DOI] [PubMed] [Google Scholar]
- 417.Boiteux S, O’Connor T R, Laval J. Formamidopyrimidine-DNA glycosylase (fpg) mutant of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 1987;6:3177–3183. doi: 10.1002/j.1460-2075.1987.tb02629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Bollen A, Lathe R, Herzog A, Denicourt D, Lecocq J-P, Desmarez L, Lavallé R. A conditionally lethal mutation of Escherichia coli affecting the gene coding for ribosomal protein S2 (rpsB) J Mol Biol. 1979;132:219–233. doi: 10.1016/0022-2836(79)90392-9. [DOI] [PubMed] [Google Scholar]
- 419.Bollinger J, Park C, Harayama S, Hazelbauer G L. Structure of the Trg protein: homologies with and differences from other sensory transducers of Escherichia coli. Proc Natl Acad Sci USA. 1984;81:3287–3291. doi: 10.1073/pnas.81.11.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Bollinger J M, Kwon D S, Huisman G W, Kolter R, Walsh C T. Glutathionylspermidine metabolism in Escherichia coli. Purification, cloning, overproduction and characterization of a bifunctional glutathionylspermidine synthetase/amidase. J Biol Chem. 1995;270:14031–14041. doi: 10.1074/jbc.270.23.14031. [DOI] [PubMed] [Google Scholar]
- 421.Bongaerts J, Zoske S, Weidner U, Unden G. Transcriptional regulation of the proton translocating NADH dehydrogenase genes (nuoA-N) of Escherichia coli by electron acceptors, electron donors and gene regulators. Mol Microbiol. 1995;16:521–534. doi: 10.1111/j.1365-2958.1995.tb02416.x. [DOI] [PubMed] [Google Scholar]
- 422.Bonnefoy E, Rouviere-Yaniv J. HU, the major histone-like protein of E. coli, modulates the binding of IHF to oriC. EMBO J. 1992;11:4489–4496. doi: 10.1002/j.1460-2075.1992.tb05550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Bonnefoy V, Ratouchniak J, Blasco F, Chippaux M. Organization of the nar genes at the chlZ locus. FEMS Microbiol Lett. 1997;147:147–149. doi: 10.1111/j.1574-6968.1997.tb10234.x. [DOI] [PubMed] [Google Scholar]
- 424.Bonnefoy V, Burini J-F, Giordano G, Pascal M C, Chippaux M. Presence in the ‘silent’ terminus region of the Escherichia coli K12 chromosome of cryptic gene(s) encoding a new nitrate reductase. Mol Microbiol. 1987;1:143–150. doi: 10.1111/j.1365-2958.1987.tb00506.x. [DOI] [PubMed] [Google Scholar]
- 425.Bonnefoy-Orth V, Lepelletier M, Pascal M C, Chippaux M. Nitrate reductase and cytochrome b (nitrate reductase) structural genes as parts of the nitrate reductase operon. Mol Gen Genet. 1981;181:535–540. doi: 10.1007/BF00428749. [DOI] [PubMed] [Google Scholar]
- 426.Bonner C A, Hays S, McEntee K, Goodman M F. DNA polymerase II is encoded by the DNA damage-inducible dinA gene of Escherichia coli. Proc Natl Acad Sci USA. 1990;87:7663–7667. doi: 10.1073/pnas.87.19.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Bonthron D T. l-Asparaginase II of Escherichia coli K-12: cloning, mapping and sequencing of the ansB gene. Gene. 1990;91:101–105. doi: 10.1016/0378-1119(90)90168-q. [DOI] [PubMed] [Google Scholar]
- 428.Boos W, Bantlow C, Benner D, Roller E. cir, a gene conferring resistance to colicin I, maps between mgl and fpk on the Escherichia coli chromosome. Mol Gen Genet. 1983;191:401–406. doi: 10.1007/BF00425754. [DOI] [PubMed] [Google Scholar]
- 429.Boos W, Steinacher I, Engelhardt-Altendorf D. Mapping of mglB, the structural gene of the galactose-binding protein of Escherichia coli. Mol Gen Genet. 1981;184:508–518. doi: 10.1007/BF00352531. [DOI] [PubMed] [Google Scholar]
- 430.Boos W, Ehmann U, Bremer E, Middendorf A, Postma P W. Trehalase of Escherichia coli. Mapping and cloning of its structural gene and identification of the enzyme as a periplasmic protein induced under high osmolarity growth conditions. J Biol Chem. 1987;262:13212–13218. [PubMed] [Google Scholar]
- 431.Boos W, Ehmann U, Forkl H, Klein W, Rimmele M, Postma P W. Trehalose transport and metabolism in Escherichia coli. J Bacteriol. 1990;172:3450–3461. doi: 10.1128/jb.172.6.3450-3461.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Boquet P L, Manoil C, Beckwith J. Use of TnphoA to detect genes for exported proteins in Escherichia coli: identification of the plasmid-encoded gene for a periplasmic acid phosphatase. J Bacteriol. 1987;169:1663–1669. doi: 10.1128/jb.169.4.1663-1669.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Borg-Olivier S A, Tarlinton D, Brown K D. Defective regulation of the phenylalanine biosynthetic operon in mutants of the phenylalanyl-tRNA synthetase operon. J Bacteriol. 1987;169:1949–1953. doi: 10.1128/jb.169.5.1949-1953.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci USA. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Bork P, Koonin E V. An expanding family of helicases within the ‘DEAD/H’ superfamily. Nucleic Acids Res. 1993;21:751–752. doi: 10.1093/nar/21.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Bornstein-Forst S M, McFall E, Palchaudhuri S. In vivo d-serine deaminase transcription start sites in wild-type Escherichia coli and in dsdA promoter mutants. J Bacteriol. 1987;169:1056–1060. doi: 10.1128/jb.169.3.1056-1060.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Boronat A, Jones-Mortimer M C, Kornberg H L. A specialized transducing phage, λpsrlA, for the sorbitol phosphotransferase of Escherichia coli K12. J Gen Microbiol. 1982;128:605–611. doi: 10.1099/00221287-128-3-605. [DOI] [PubMed] [Google Scholar]
- 438.Boronat A, Britton P, Jones-Mortimer M C, Kornberg H L, Lee L G, Murfitt D, Parr F. Location on the Escherichia coli genome of a gene specifying O-acetylserine (thiol)-lyase. J Gen Microbiol. 1984;130:673–685. doi: 10.1099/00221287-130-3-673. [DOI] [PubMed] [Google Scholar]
- 439.Boros I, Kiss A, Venetianer P. Physical map of the seven ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 1979;6:1817–1830. doi: 10.1093/nar/6.5.1817. . (Erratum, 6:2961, 1979.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Boros I, Csordas-Toth E, Kiss A, Kiss I, Torok I, Udvardy A, Udvardy K, Venetianer P. Identification of two new promoters probably involved in the transcription of a ribosomal RNA gene of Escherichia coli. Biochim Biophys Acta. 1983;739:173–180. doi: 10.1016/0167-4781(83)90027-1. [DOI] [PubMed] [Google Scholar]
- 441.Borukhov S, Polyakov A, Nikiforov V, Goldfarb A. GreA protein: a transcription elongation factor from Escherichia coli. Proc Natl Acad Sci USA. 1992;89:8899–8902. doi: 10.1073/pnas.89.19.8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- 443.Bosch L, Nilsson L, Vijgenboom E, Verbeek H. Fis-dependent trans-activation of tRNA and rRNA operons of Escherichia coli. Biochim Biophys Acta. 1990;1050:293–301. doi: 10.1016/0167-4781(90)90184-4. [DOI] [PubMed] [Google Scholar]
- 444.Boschi-Muller S, Azzi S, Pollastro D, Corbler C, Branlant G. Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and phosphorylating glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1997;272:15106–15112. doi: 10.1074/jbc.272.24.15106. [DOI] [PubMed] [Google Scholar]
- 445.Bosl M. Genetic map of the tyrT region of Escherichia coli from 27.1 to 27.7 minutes based exclusively on sequence data. J Bacteriol. 1993;175:7751–7753. doi: 10.1128/jb.175.23.7751-7753.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Bosl M, Kersten H. A novel RNA product of the tyrT operon of Escherichia coli. Nucleic Acids Res. 1991;19:5863–5870. doi: 10.1093/nar/19.21.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Bosl M, Kersten H. Organization and functions of genes in the upstream region of tyrT of Escherichia coli: phenotypes of mutants with partial deletion of a new gene (tgs) J Bacteriol. 1994;176:221–231. doi: 10.1128/jb.176.1.221-231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Bossemeyer D, Borchard A, Dosch D C, Helmer G L, Epstein W, Booth I R, Bakker E P. K+-transport protein TrkA of Escherichia coli is a peripheral membrane protein that requires other trk gene products for attachment to the cytoplasmic membrane. J Biol Chem. 1989;264:16403–16410. [PubMed] [Google Scholar]
- 449.Bossemeyer D, Schlosser A, Bakker E P. Specific cesium transport via the Escherichia coli Kup (TrkD) K+ uptake system. J Bacteriol. 1989;171:2219–2221. doi: 10.1128/jb.171.4.2219-2221.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Bost S, Belin D. A new genetic selection identifies essential residues in SecG, a component of the Escherichia coli protein export machinery. EMBO J. 1995;14:4412–4421. doi: 10.1002/j.1460-2075.1995.tb00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Bost S, Belin D. prl mutations in the Escherichia coli secG gene. J Biol Chem. 1997;272:4087–4093. doi: 10.1074/jbc.272.7.4087. [DOI] [PubMed] [Google Scholar]
- 452.Botello E, Jimenez-Sanchez A. A temperature upshift induces initiation of replication at oriC on the Escherichia coli chromosome. Mol Microbiol. 1997;26:133–144. doi: 10.1046/j.1365-2958.1997.5621924.x. [DOI] [PubMed] [Google Scholar]
- 453.Bouché F, Bouché J P. Genetic evidence that DicF, a second division inhibitor encoded by the Escherichia coli dicB operon, is probably RNA. Mol Microbiol. 1989;3:991–994. doi: 10.1111/j.1365-2958.1989.tb00249.x. [DOI] [PubMed] [Google Scholar]
- 454.Bouché J P. Physical map of a 470 × 103 base-pair region flanking the terminus of DNA replication in the Escherichia coli K12 genome. J Mol Biol. 1982;154:1–20. doi: 10.1016/0022-2836(82)90413-2. [DOI] [PubMed] [Google Scholar]
- 455.Bouché J P, Gelugne J P, Louarn J, Louarn J M, Kaiser C. Relationship between the physical and genetic maps of a 470 × 103 base-pair region around the terminus of Escherichia coli K12 DNA replication. J Mol Biol. 1982;154:21–23. doi: 10.1016/0022-2836(82)90414-4. [DOI] [PubMed] [Google Scholar]
- 456.Bouffard G, Rudd K E, Adhya S. Dependence of lactose metabolism upon mutarotase encoded in the gal operon in Escherichia coli. J Mol Biol. 1994;244:269–278. doi: 10.1006/jmbi.1994.1728. [DOI] [PubMed] [Google Scholar]
- 457.Boulanger P, le Maire M, Bonhivers M, Dubois S, Desmadril M, Letellier L. Purification and structural and functional characterization of FhuA, a transporter of the Escherichia coli outer membrane. Biochemistry. 1996;35:14216–14224. doi: 10.1021/bi9608673. [DOI] [PubMed] [Google Scholar]
- 458.Bouloc P, Jaffe A, D’Ari R. The Escherichia coli lov gene product connects peptidoglycan synthesis, ribosomes and growth rate. EMBO J. 1989;8:317–323. doi: 10.1002/j.1460-2075.1989.tb03379.x. . (Erratum, 8:1290.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Bouma C L, Meadow N D, Stover E W, Roseman S. II-BGlc, a glucose receptor of the bacterial phosphotransferase system: molecular cloning of ptsG and purification of the receptor from an overproducing strain of Escherichia coli. Proc Natl Acad Sci USA. 1987;84:930–934. doi: 10.1073/pnas.84.4.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Bouquin N, Chen M X, Kim S, Vannier F, Bernard S, Holland I B, Seror S J. Characterization of an Escherichia coli mutant, feeA, displaying resistance to the calmodulin inhibitor 48/80 and reduced expression of the rare tRNA3Leu. Mol Microbiol. 1996;20:853–865. doi: 10.1111/j.1365-2958.1996.tb02523.x. [DOI] [PubMed] [Google Scholar]
- 461.Bourd G, Erlagaeva R S, Bolshakova T N, Gershanovich V N. Glucose catabolite repression in Escherichia coli K12 mutants defective in methyl-a-d-glucose transport. J Biochem (Tokyo) 1975;53:419–427. doi: 10.1111/j.1432-1033.1975.tb04082.x. [DOI] [PubMed] [Google Scholar]
- 462.Bourgerie S J, Michan C M, Thomas M S, Busby S J, Hyde E I. DNA binding and DNA bending by the MelR transcription activator protein from Escherichia coli. Nucleic Acids Res. 1997;25:1685–1693. doi: 10.1093/nar/25.9.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Bouvier J, Pugsley A P, Stragier P. A gene for a new lipoprotein in the dapA-purC interval of the Escherichia coli chromosome. J Bacteriol. 1991;173:5523–5531. doi: 10.1128/jb.173.17.5523-5531.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Bouvier J, Richaud F, Richaud C, Patte J C, Stragier P. Nucleotide sequence and expression of the Escherichia coli dapB gene. J Biol Chem. 1984;259:14829–14834. [PubMed] [Google Scholar]
- 465.Bouvier J, Patte J C, Stragier P. Multiple regulatory signals in the control region of the Escherichia coli carAB operon. Proc Natl Acad Sci USA. 1984;81:4139–4143. doi: 10.1073/pnas.81.13.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Bouvier J, Stragier P. Nucleotide sequence of the lsp-dapB interval in Escherichia coli. Nucleic Acids Res. 1991;19:180. doi: 10.1093/nar/19.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Boy E, Borne F, Patte J C. Isolation and identification of mutants constitutive for aspartokinase III synthesis in Escherichia coli K-12. Biochimie. 1979;61:1151–1160. doi: 10.1016/s0300-9084(80)80228-8. [DOI] [PubMed] [Google Scholar]
- 468.Boyd A, Krikos A, Simon M. Sensory transducers of E. coli are encoded by homologous genes. Cell. 1981;26:333–343. doi: 10.1016/0092-8674(81)90202-6. [DOI] [PubMed] [Google Scholar]
- 469.Boyd A, Kendall K, Simon M I. Structure of the serine chemoreceptor in Escherichia coli. Nature. 1983;301:623–626. doi: 10.1038/301623a0. [DOI] [PubMed] [Google Scholar]
- 470.Boyd L A, Adam L, Pelcher L, McHughen A, Hirji R, Selvaraj G. Characterization of an Escherichia coli gene encoding betaine aldehyde dehydrogenase (BADH): structural similarity to mammalian ALDHs and a plant BADH. Gene. 1991;103:45–52. doi: 10.1016/0378-1119(91)90389-s. [DOI] [PubMed] [Google Scholar]
- 471.Boye E, Stokke T, Kleckner N, Skarstad K. Coordinating DNA replication initiation with cell growth: differential roles for DnaA and SeqA proteins. Proc Natl Acad Sci USA. 1996;93:12206–12211. doi: 10.1073/pnas.93.22.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Boyle S M, Markham G D, Hafner E W, Wright J M, Tabor H, Tabor C W. Expression of the cloned genes encoding the putrescine biosynthetic enzymes and methionine adenosyltransferase of Escherichia coli. Gene. 1984;30:129–136. doi: 10.1016/0378-1119(84)90113-6. [DOI] [PubMed] [Google Scholar]
- 473.Braaten B A, Platko J V, van der Woude M W, Simons B H, de Graaf F K, Calvo J M, Low D A. Leucine-responsive regulatory protein controls the expression of both the pap and fan pili operons in Escherichia coli. Proc Natl Acad Sci USA. 1992;89:4250–4254. doi: 10.1073/pnas.89.10.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Bradbury A J, Gruer M J, Rudd K E, Guest J R. The second aconitase (AcnB) of Escherichia coli. Microbiology. 1996;142:389–400. doi: 10.1099/13500872-142-2-389. [DOI] [PubMed] [Google Scholar]
- 475.Bradshaw R A, Cancedda F, Ericsson L H, Neumann P A, Piccoli S P, Schlesinger M J, Shriefer K, Walsh K A. Amino acid sequence of Escherichia coli alkaline phosphatase. Proc Natl Acad Sci USA. 1981;78:3473–3477. doi: 10.1073/pnas.78.6.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Bramley H F, Kornberg H L. Nucleotide sequence of bglC, the gene specifying enzyme IIbgl of the PEP:sugar phosphotransferase system in Escherichia coli K12 and overexpression of the gene product. J Gen Microbiol. 1987;133:563–573. doi: 10.1099/00221287-133-3-563. [DOI] [PubMed] [Google Scholar]
- 477.Brand B, Boos W. Maltose transacetylase of Escherichia coli. Mapping and cloning of its structural gene, mac, and characterization of the enzyme as a dimer of identical polypeptides with a molecular weight of 20,000. J Biol Chem. 1991;266:14113–14118. [PubMed] [Google Scholar]
- 478.Brandi A, Pietroni P, Gualerzi C O, Pon C L. Post-transcriptional regulation of CspA expression in Escherichia coli. Mol Microbiol. 1996;19:231–240. doi: 10.1046/j.1365-2958.1996.362897.x. [DOI] [PubMed] [Google Scholar]
- 479.Brandsma J A, Stoorvogel J, Van Sluis C A, Van de Putte P. Effect of lexA and ssb genes, present on a uvrA recombinant plasmid, on the UV survival of Escherichia coli K-12. Gene. 1982;18:77–85. doi: 10.1016/0378-1119(82)90058-0. [DOI] [PubMed] [Google Scholar]
- 480.Branlant G, Branlant C. Nucleotide sequence of the Escherichia coli gap gene. Different evolutionary behavior of the NAD+-binding domain and of the catalytic domain of d-glyceraldehyde-3-phosphate dehydrogenase. Eur J Biochem. 1985;150:61–66. doi: 10.1111/j.1432-1033.1985.tb08988.x. [DOI] [PubMed] [Google Scholar]
- 481.Braun V. The structurally related exbB and tolQ genes are interchangeable in conferring tonB-dependent colicin, bacteriophage, and albomycin sensitivity. J Bacteriol. 1989;171:6387–6390. doi: 10.1128/jb.171.11.6387-6390.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Braun V, Herrmann C. Evolutionary relationship of uptake systems for biopolymers in Escherichia coli: cross-complementation between the TonB-ExbB-ExbD and the TolA-TolQ-TolR proteins. Mol Microbiol. 1993;8:261–268. doi: 10.1111/j.1365-2958.1993.tb01570.x. [DOI] [PubMed] [Google Scholar]
- 483.Braun V, Killmann H, Benz R. Energy-coupled transport through the outer membrane of Escherichia coli: small deletions in the gating loop convert the FhuA transport protein into a diffusion channel. FEBS Lett. 1994;346:59–64. doi: 10.1016/0014-5793(94)00431-5. [DOI] [PubMed] [Google Scholar]
- 484.Braun V, Frenz J, Hantke K, Schaller H. Penetration of colicin M into cells of Escherichia coli. J Bacteriol. 1980;142:162–168. doi: 10.1128/jb.142.1.162-168.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Braun V, Hantke K, Stauder W. Identification of the sid outer membrane receptor protein in Salmonella typhimurium SL-1024. Mol Gen Genet. 1977;155:227–229. doi: 10.1007/BF00393164. [DOI] [PubMed] [Google Scholar]
- 486.Braun V, Gross R, Koster W, Zimmerman L. Plasmid and chromosomal mutants in the iron (III)-aerobactin transport system of Escherichia coli. Mol Gen Genet. 1983;192:131–139. doi: 10.1007/BF00327658. [DOI] [PubMed] [Google Scholar]
- 487.Braun V, Bosch V. Sequence of the murein-lipoprotein and the attachment site of the lipid. Eur J Biochem. 1972;28:51–69. doi: 10.1111/j.1432-1033.1972.tb01883.x. [DOI] [PubMed] [Google Scholar]
- 488.Braun-Breton C, Hofnung M. In vivo and in vitro functional alterations of the bacteriophage lambda receptor to lamB missense mutants of Escherichia coli K-12. J Bacteriol. 1981;148:845–852. doi: 10.1128/jb.148.3.845-852.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Breckenridge L, Gorini Genetic analysis of streptomycin resistance in Escherichia coli. Genetics. 1970;65:9–25. doi: 10.1093/genetics/65.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Breeze A S, Obaseiki-Ebor E E. Mutations in nitrofurantoin and nitrofurazone resistance in Escherichia coli K-12. J Gen Microbiol. 1983;129:99–103. doi: 10.1099/00221287-129-1-99. [DOI] [PubMed] [Google Scholar]
- 491.Breines D M, Ouabdesselam S, Ng E Y, Tankovic J, Shah S, Soussy C J, Hooper D C. Quinolone resistance locus nfxD of Escherichia coli is a mutant allele of the parE gene encoding a subunit of topoisomerase IV. Antimicrob Agents Chemother. 1997;41:175–179. doi: 10.1128/aac.41.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Bremer E, Middendorf A, Martinussen J, Valentin-Hansen P. Analysis of the tsx gene, which encodes a nucleoside-specific channel-forming protein (Tsx) in the outer membrane of Escherichia coli. Gene. 1990;96:59–65. doi: 10.1016/0378-1119(90)90341-n. [DOI] [PubMed] [Google Scholar]
- 493.Bremer E, Beck E, Hindennach I, Sonntag I, Henning U. Cloned structural gene (ompA) for an integral outer membrane protein of Escherichia coli K-12. Mol Gen Genet. 1980;179:13–20. doi: 10.1007/BF00268440. [DOI] [PubMed] [Google Scholar]
- 494.Bremer E, Gerlach P, Middendorf A. Double negative and positive control of tsx expression in Escherichia coli. J Bacteriol. 1988;170:108–116. doi: 10.1128/jb.170.1.108-116.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Brendel V, Brocchieri L, Sandler S J, Clark A J, Karlin S. Evolutionary comparisons of RecA-like proteins across all major kingdoms of living organisms. J Mol Evol. 1997;44:528–541. doi: 10.1007/pl00006177. [DOI] [PubMed] [Google Scholar]
- 496.Brent R, Ptashne M. The lexA gene product represses its own promoter. Proc Natl Acad Sci USA. 1980;77:1932–1936. doi: 10.1073/pnas.77.4.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Brent R, Ptashne M. Mechanism of action of the lexA gene product. Proc Natl Acad Sci USA. 1981;78:4204–4208. doi: 10.1073/pnas.78.7.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Breton R, Sanfacon H, Papayannopoulos I A, Biemann K, LaPointe J. Glutamyl-tRNA synthetase of Escherichia coli: isolation and primary structure of the gltX gene and homology with other aminoacyl-tRNA synthetases. J Biol Chem. 1986;261:10610–10617. [PubMed] [Google Scholar]
- 499.Brey R N, Rosen B P. Properties of Escherichia coli mutants altered in calcium/proton antiport activity. J Bacteriol. 1979;139:824–834. doi: 10.1128/jb.139.3.824-834.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Brickman E, Soll L, Beckwith J R. Genetic characterization of mutations which affect catabolite-sensitive operons in Escherichia coli, including deletions of the gene for adenyl cyclase. J Bacteriol. 1973;116:582–587. doi: 10.1128/jb.116.2.582-587.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Brickman T J, Ozenberger B A, McIntosh M A. Regulation of divergent transcription from the iron-responsive fepB-entC promoter-operator regions in Escherichia coli. J Mol Biol. 1990;212:669–682. doi: 10.1016/0022-2836(90)90229-F. [DOI] [PubMed] [Google Scholar]
- 502.Briggs K A, Lancashire W E, Hartley B S. Molecular cloning, DNA structure and expression of the Escherichia colid-xylose isomerase. EMBO J. 1984;3:611–616. doi: 10.1002/j.1460-2075.1984.tb01856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Brikun I, Suziedelis K, Berg D E. DNA sequence divergence among derivatives of Escherichia coli K-12 detected by arbitrary primer PCR (random amplified polymorphic DNA) fingerprinting. J Bacteriol. 1994;176:1673–1682. doi: 10.1128/jb.176.6.1673-1682.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Brill J A, Quinlan-Walshe C, Gottesman S. Fine-structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K-12. J Bacteriol. 1988;170:2599–2611. doi: 10.1128/jb.170.6.2599-2611.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Brinkmann U, Mattes R E, Buckel P. High-level expression of recombinant genes in Escherichia coli is dependent on the availability of the dnaY gene product. Gene. 1989;85:109–114. doi: 10.1016/0378-1119(89)90470-8. [DOI] [PubMed] [Google Scholar]
- 506.Brissette J L, Weiner L, Ripmaster T L, Model P. Characterization and sequence of the Escherichia coli stress-induced psp operon. J Mol Biol. 1991;220:35–48. doi: 10.1016/0022-2836(91)90379-k. [DOI] [PubMed] [Google Scholar]
- 507.Brissette J L, Russel M, Weiner L, Model P. Phage shock protein, a stress protein of Escherichia coli. Proc Natl Acad Sci USA. 1990;87:862–866. doi: 10.1073/pnas.87.3.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Britton P, Boronat A, Hartley D A, Jones-Mortimer M C, Kornberg H L, Parra F. Phosphotransferase-mediated regulation of carbohydrate utilization in Escherichia coli K12: location of the gsr (tgs) and iex (crr) genes by specialized transduction. J Gen Microbiol. 1983;129:349–356. doi: 10.1099/00221287-129-2-349. [DOI] [PubMed] [Google Scholar]
- 509.Britton P, Murfitt D, Parra F, Jones-Mortimer M C, Kornberg H L. Phosphotransferase-mediated regulation of carbohydrate utilization in Escherichia coli K12: identification of the products of genes on the specialized transducing phages lambda iex (crr) and lambda gsr (tgs) EMBO J. 1982;1:907–911. doi: 10.1002/j.1460-2075.1982.tb01270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Britton P, Lee L G, Murfitt D, Boronat A, Jones-Mortimer M C, Kornberg H L. Location and direction of transcription of the ptsH and ptsI genes on the Escherichia coli K12 genome. J Gen Microbiol. 1984;130:861–868. doi: 10.1099/00221287-130-4-861. [DOI] [PubMed] [Google Scholar]
- 511.Brody H, Greener A, Hill C W. Excision and reintegration of the Escherichia coli K-12 chromosomal element. J Bacteriol. 1985;161:1112–1117. doi: 10.1128/jb.161.3.1112-1117.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Brody H, Hill C W. Attachment site of the genetic element e14. J Bacteriol. 1988;170:2040–2044. doi: 10.1128/jb.170.5.2040-2044.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Brok R G P M, Brinkman E, van Boxtel R, Bekkers A C A P A, Verheij H M, Tommassen J. Molecular characterization of enterobacterial pldA genes encoding outer membrane phospholipase A. J Bacteriol. 1994;176:861–870. doi: 10.1128/jb.176.3.861-870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Brombach M, Pon C L. The unusual translational initiation codon AUU limits the expression of the infC (initiation factor IF3) gene of Escherichia coli. Mol Gen Genet. 1987;208:94–100. doi: 10.1007/BF00330428. [DOI] [PubMed] [Google Scholar]
- 515.Brondsted L, Atlung T. Anaerobic regulation of the hydrogenase 1 (hya) operon of Escherichia coli. J Bacteriol. 1994;176:5423–5428. doi: 10.1128/jb.176.17.5423-5428.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Brondsted L, Atlung T. Effect of growth conditions on expression of the acid phosphatase (cyx-appA) operon and the appY gene, which encodes a transcriptional activator of Escherichia coli. J Bacteriol. 1996;178:1556–1564. doi: 10.1128/jb.178.6.1556-1564.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Brooke J S, Valvano M A. Biosynthesis of inner core lipopolysaccharide in enteric bacteria: identification and characterization of a conserved phosphoheptose isomerase. J Biol Chem. 1996;271:3608–3614. doi: 10.1074/jbc.271.7.3608. [DOI] [PubMed] [Google Scholar]
- 518.Brooks J E, Blumenthal R M, Gingeras T R. The isolation and characterization of the Escherichia coli DNA adenine methylase (dam) gene. Nucleic Acids Res. 1983;11:837–851. doi: 10.1093/nar/11.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Broome-Smith J K, Edelman A, Yousif S, Spratt B G. The nucleotide sequences of the ponA and ponB genes encoding penicillin-binding proteins 1A and 1B of Escherichia coli K-12. Eur J Biochem. 1985;147:437–466. doi: 10.1111/j.1432-1033.1985.tb08768.x. [DOI] [PubMed] [Google Scholar]
- 520.Broome-Smith J K, Ioannidis I, Edelman A, Spratt B G. Nucleotide sequences of the penicillin-binding protein 5 and 6 genes of Escherichia coli. Nucleic Acids Res. 1988;16:1617. doi: 10.1093/nar/16.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Brosh R M, Matson S W. Mutations in motif II of Escherichia coli DNA helicase II render the enzyme nonfunctional in both mismatch repair and excision repair with differential effects on the unwinding reaction. J Bacteriol. 1995;177:5612–5621. doi: 10.1128/jb.177.19.5612-5621.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Brosius J, Ullrich A, Raker M A, Gray A, Dull T J, Gutell R R, Noller H F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981;6:112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- 523.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 524.Brosius J, Dull T J, Noller H F. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1980;77:201–204. doi: 10.1073/pnas.77.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Brot N, Rahman M A, Moskovitz J, Weissbach H. Escherichia coli peptide methionine sulfoxide reductase: cloning, high expression, and purification. Methods Enzymol. 1995;251:462–470. doi: 10.1016/0076-6879(95)51150-4. [DOI] [PubMed] [Google Scholar]
- 526.Brotcorne-Lannoye A, Maenhaut-Michel G. Role of RecA protein in untargeted UV mutagenesis of bacteriophage lambda: evidence for the requirement for the dinB gene. Proc Natl Acad Sci USA. 1986;83:3904–3908. doi: 10.1073/pnas.83.11.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Brown E D, Vivas E I, Walsh C T, Kolter R. MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J Bacteriol. 1995;177:4194–4197. doi: 10.1128/jb.177.14.4194-4197.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Brown K, Finch P W, Hickson I D, Emmerson P T. Complete nucleotide sequence of the Escherichia coli argA gene. Nucleic Acids Res. 1987;15:10586. doi: 10.1093/nar/15.24.10586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Brown K D. Formation of aromatic amino acid pools in Escherichia coli K-12. J Bacteriol. 1970;104:177–188. doi: 10.1128/jb.104.1.177-188.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Brown K D, Jones-Mortimer M C, Kornberg H L. The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J Gen Microbiol. 1977;102:327–336. doi: 10.1099/00221287-102-2-327. [DOI] [PubMed] [Google Scholar]
- 531.Brown K D, Somerville R L. Repression of aromatic amino acid biosynthesis in Escherichia coli K-12. J Bacteriol. 1971;108:386–399. doi: 10.1128/jb.108.1.386-399.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Brown K L, Hughes K T. The role of anti-sigma factors in gene regulation. Mol Microbiol. 1995;16:397–404. doi: 10.1111/j.1365-2958.1995.tb02405.x. [DOI] [PubMed] [Google Scholar]
- 533.Brown S, Albrechtsen B, Pedersen S, Klemm P. Localization and regulation of the structural gene for transcription-termination factor rho of Escherichia coli. J Mol Biol. 1982;162:283–298. doi: 10.1016/0022-2836(82)90527-7. [DOI] [PubMed] [Google Scholar]
- 534.Brown S, Fournier M J. The 4.5S RNA gene of Escherichia coli is essential for cell growth. J Mol Biol. 1984;178:533–550. doi: 10.1016/0022-2836(84)90237-7. [DOI] [PubMed] [Google Scholar]
- 535.Bruce I, Hardy J, Stacey K A. Potentiation by purines of the growth-inhibitory effects of sulphonamides on Escherichia coli K-12 and the location of the gene which mediates this effect. J Gen Microbiol. 1984;130:2489–2495. doi: 10.1099/00221287-130-10-2489. [DOI] [PubMed] [Google Scholar]
- 536.Brucker R, Levitz R, Yagil E, Friedberg I. Complementation tests between mutations in the phosphate-specific transport region of Escherichia coli. Curr Microbiol. 1984;10:303–308. [Google Scholar]
- 537.Bruckner R, Matzura H. In vivo synthesis of a polycistronic messenger RNA for the ribosomal proteins L11, L1, L10, and L7/12 in Escherichia coli. Mol Gen Genet. 1981;183:277–282. doi: 10.1007/BF00270629. [DOI] [PubMed] [Google Scholar]
- 538.Brun Y V, Lapointe J. Locations of genes in the 52-minute region on the physical map of Escherichia coli K-12. J Bacteriol. 1990;172:4746–4747. doi: 10.1128/jb.172.9.4746-4747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Brun Y V, Breton R, Lanouette P, LaPointe J. Precise mapping and comparison of two evolutionarily related regions of the Escherichia coli K-12 chromosome. Evolution of valU and lysT from an ancestral tRNA operon. J Mol Biol. 1990;214:825–843. doi: 10.1016/0022-2836(90)90339-N. [DOI] [PubMed] [Google Scholar]
- 540.Brundage L, Fimmel C J, Mizushima S, Wickner W. SecY, SecE and band 1 form the membrane-embedded domain of Escherichia coli preprotein translocase. J Biol Chem. 1992;267:4166–4170. [PubMed] [Google Scholar]
- 541.Brune M, Schumann R, Wittinghofer F. Cloning and sequencing of the adenylate kinase gene (adk) of Escherichia coli. Nucleic Acids Res. 1985;13:7139–7151. doi: 10.1093/nar/13.19.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Bruni C B, Musti A M, Frunzio R, Blasi F. Structural and physiological studies of the Escherichia coli histidine operon inserted into plasmid vectors. J Bacteriol. 1980;142:32–42. doi: 10.1128/jb.142.1.32-42.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Bruni C B, Colantuoni V, Sbordone L, Cortese R, Blasi F. Biochemical and regulatory properties of Escherichia coli K-12 hisT mutants. J Bacteriol. 1977;130:4–10. doi: 10.1128/jb.130.1.4-10.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Brusilow W S, Porter A C G, Simoni R D. Cloning and expression of uncI, the first gene of the unc operon of Escherichia coli. J Bacteriol. 1983;155:1265–1270. doi: 10.1128/jb.155.3.1265-1270.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Bryan S K, Moses R E. Map location of the pcbA mutation and physiology of the mutant. J Bacteriol. 1984;158:216–221. doi: 10.1128/jb.158.1.216-221.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Bryant R E, Sypherd P S. Genetic analysis of cold-sensitive ribosome maturation mutants of Escherichia coli. J Bacteriol. 1974;117:1082–1092. doi: 10.1128/jb.117.3.1082-1092.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Brzoska P, Rimmele M, Brzostek K, Boos W. The pho regulon-dependent Ugp uptake system for glycerol-3-phosphate in Escherichia coli is trans inhibited by Pi. J Bacteriol. 1994;176:15–20. doi: 10.1128/jb.176.1.15-20.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Brzoska P, Boos W. Characteristics of a ugp-encoded and phoB-dependent glycerophosphoryl diester phosphodiesterase which is physically dependent on the ugp transport system of Escherichia coli. J Bacteriol. 1988;170:4125–4135. doi: 10.1128/jb.170.9.4125-4135.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Buchel D E, Gronenborn B, Muller-Hill B. Sequence of the lactose permease gene. Nature. 1980;283:541–545. doi: 10.1038/283541a0. [DOI] [PubMed] [Google Scholar]
- 550.Buchet A, Eichler K, Mandrand-Berthelot M A. Regulation of the carnitine pathway in Escherichia coli: investigation of the cai-fix divergent promoter region. J Bacteriol. 1998;180:2599–2608. doi: 10.1128/jb.180.10.2599-2608.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Buck D, Guest J R. Overexpression and site-directed mutagenesis of the succinyl-CoA synthetase of Escherichia coli and nucleotide sequence of a gene (g30) that is adjacent to the suc operon. Biochem J. 1989;260:737–747. doi: 10.1042/bj2600737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Buck D, Spencer M E, Guest J R. Primary structure of the succinyl-CoA synthetase of Escherichia coli. Biochemistry. 1985;24:6245–6252. doi: 10.1021/bi00343a031. [DOI] [PubMed] [Google Scholar]
- 553.Buck D, Spencer M E, Guest J R. Cloning and expression of the succinyl-CoA synthetase genes of Escherichia coli K12. J Gen Microbiol. 1986;132:1753–1762. doi: 10.1099/00221287-132-6-1753. [DOI] [PubMed] [Google Scholar]
- 554.Buckel P, Buchberger A, Bock A, Wittmann H G. Alteration of ribosomal protein L6 in mutants of Escherichia coli resistant to gentamicin. Mol Gen Genet. 1977;158:47–54. doi: 10.1007/BF00455118. [DOI] [PubMed] [Google Scholar]
- 555.Bueno R, Pahel G, Magasanik B. Role of glnB and glnD gene products in regulation of the glnALG operon of Escherichia coli. J Bacteriol. 1985;164:816–822. doi: 10.1128/jb.164.2.816-822.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Buhk H-J, Messer W. The replication origin region of Escherichia coli: nucleotide sequence and functional units. Gene. 1983;24:265–279. doi: 10.1016/0378-1119(83)90087-2. [DOI] [PubMed] [Google Scholar]
- 557.Buhr A, Daniels G A, Erni B. The glucose transporter of Escherichia coli. Mutants with impaired translocation activity that retain phosphorylation activity. J Biol Chem. 1992;267:3847–3851. [PubMed] [Google Scholar]
- 558.Bukhari A I, Taylor A L. Genetic analysis of diaminopimelic acid- and lysine-requiring mutants of Escherichia coli. J Bacteriol. 1971;105:844–854. doi: 10.1128/jb.105.3.844-854.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.Bulawa C E, Raetz R H. Program and abstracts of the XIII International Congress of Microbiology. Washington, D.C: American Society for Microbiology; 1982. Function of CDP-diglyceride hydrolase in membranes of E. coli, abstr. P48:4; p. 131. [Google Scholar]
- 560.Bulawa C E, Raetz C R H. Isolation and characterization of Escherichia coli strains defective in CDP-diglyceride hydrolase. J Biol Chem. 1984;259:11257–11264. [PubMed] [Google Scholar]
- 561.Bull H J, Hayes S. The grpD55 locus of Escherichia coli appears to be an allele of dnaB. Mol Gen Genet. 1996;252:755–760. doi: 10.1007/BF02173984. [DOI] [PubMed] [Google Scholar]
- 562.Burd G I, Gabrielyan T R, Bol’shakova T N, Gershanovich V N. Study of linkage with pts genes of pleiotropic mutation affecting expression of catabolite-sensitive genes in Escherichia coli K-12. Sov Genet (Engl Transl Genetika) 1980;16:622–629. [PubMed] [Google Scholar]
- 563.Burdett V. tRNA modification activity is necessary for Tet(M)-mediated tetracycline resistance. J Bacteriol. 1993;175:7209–7215. doi: 10.1128/jb.175.22.7209-7215.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Burgers P M J, Kornberg A, Sakakibara Y. The dnaN gene codes for the β subunit of DNA polymerase III holoenzyme of Escherichia coli. Proc Natl Acad Sci USA. 1981;78:5391–5395. doi: 10.1073/pnas.78.9.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.Burgess R R, Gross C A, Walter W, Lowe P A. Altered chemical properties in three mutants of E. coli RNA polymerase sigma subunit. Mol Gen Genet. 1979;175:251–257. doi: 10.1007/BF00397224. [DOI] [PubMed] [Google Scholar]
- 566.Burkhardt R, Braun V. Nucleotide sequence of the fhuC and fhuD genes involved in iron (III) hydroxamate transport: domains in FhuC homologous to ATP-binding proteins. Mol Gen Genet. 1987;209:49–55. doi: 10.1007/BF00329835. [DOI] [PubMed] [Google Scholar]
- 567.Burkholder W F, Zhao X, Zhu X, Hendrickson W A, Gragerov A, Gottesman M E. Mutations in the C-terminal fragment of DnaK affecting peptide binding. Proc Natl Acad Sci USA. 1996;93:10632–10637. doi: 10.1073/pnas.93.20.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Burland V D, Plunkett G, Daniels D L, Blattner F R. DNA sequence and analysis of 136 kilobases of the Escherichia coli genome: organizational symmetry around the origin of replication. Genomics. 1993;16:551–561. doi: 10.1006/geno.1993.1230. [DOI] [PubMed] [Google Scholar]
- 569.Burland V D, Plunkett G, Sofia H J, Daniels D L, Blattner F R. Analysis of the Escherichia coli genome. VI. DNA sequence of the region from 92.8 through 100 minutes. Nucleic Acids Res. 1995;23:2105–2119. doi: 10.1093/nar/23.12.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Burlingame R P. The degradation of 3-phenylpropionic acid by Escherichia coli. Ph.D. thesis. Minneapolis-St. Paul: University of Minnesota; 1983. [Google Scholar]
- 571.Burlingame R P, Wyman L, Chapman P J. Isolation and characterization of Escherichia coli mutants defective for phenylpropionate degradation. J Bacteriol. 1986;168:55–64. doi: 10.1128/jb.168.1.55-64.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Burman L G, Nordstrom K. Colicin tolerance induced by ampicillin resistance in a strain of Escherichia coli K-12. J Bacteriol. 1971;106:1–13. doi: 10.1128/jb.106.1.1-13.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.Burnett B P, Horwich A L, Low K B. A carboxy-terminal deletion impairs the assembly of GroEL and confers a pleiotropic phenotype in Escherichia coli K-12. J Bacteriol. 1994;176:6980–6985. doi: 10.1128/jb.176.22.6980-6985.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574.Burns D M, Beacham I R. Nucleotide sequence and transcriptional analysis of the E. coli ushA gene, encoding periplasmic UDP-sugar hydrolase (5′-nucleotidase): regulation of the ushA gene, and the signal sequence of its encoded protein product. Nucleic Acids Res. 1986;14:4325–4342. doi: 10.1093/nar/14.10.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575.Burns D M, Burger M J, Beacham I R. Silent genes in bacteria: the previously designated ‘cryptic’ ilvHI locus of ‘Salmonella typhimurium LT2’ is active in natural isolates. FEMS Microbiol Lett. 1995;131:167–172. doi: 10.1111/j.1574-6968.1995.tb07772.x. [DOI] [PubMed] [Google Scholar]
- 576.Burova E, Hung S C, Sagitov V, Stitt B L, Gottesman M E. Escherichia coli NusG protein stimulates transcription elongation rates in vivo and in vitro. J Bacteriol. 1995;177:1388–1392. doi: 10.1128/jb.177.5.1388-1392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 577.Burton K. Transport of adenine, hypoxanthine and uracil into Escherichia coli. Biochem J. 1977;168:195–204. doi: 10.1042/bj1680195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578.Burton K. Transport of nucleic acid bases into Escherichia coli. J Gen Microbiol. 1983;129:3505–3513. doi: 10.1099/00221287-129-11-3505. [DOI] [PubMed] [Google Scholar]
- 579.Burton Z F, Gross C A, Watanabe K, Burgess R R. The operon that encodes the ς subunit of RNA polymerase also encodes ribosomal protein S21 and DNA primase in E. coli K12. Cell. 1983;32:335–349. doi: 10.1016/0092-8674(83)90453-1. [DOI] [PubMed] [Google Scholar]
- 580.Burton Z F, Burgess R R, Lin J, Moore D, Holder S, Gross C A. The nucleotide sequence of the cloned rpoD gene for the RNA polymerase sigma subunit from E. coli K12. Nucleic Acids Res. 1981;9:2889–2903. doi: 10.1093/nar/9.12.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 581.Busby S J, Aiba H, de Crombrugghe B. Mutations in the Escherichia coli operon that define two promoters and the binding site of the cyclic AMP receptor protein. J Mol Biol. 1982;154:211–227. doi: 10.1016/0022-2836(82)90061-4. [DOI] [PubMed] [Google Scholar]
- 582.Butler J S, Springer M, Dondon J, Graffe M, Grunberg-Manago M. Escherichia coli protein synthesis initiation factor IF3 controls its own gene expression at the translational level in vivo. J Mol Biol. 1986;192:767–780. doi: 10.1016/0022-2836(86)90027-6. [DOI] [PubMed] [Google Scholar]
- 583.Butler J S, Springer M, Grunberg-Manago M. AUU-to-AUG mutation in the initiator codon of the translation initiation factor IF3 abolishes translational autocontrol of its own gene (infC) in vivo. Proc Natl Acad Sci USA. 1987;84:4022–4025. doi: 10.1073/pnas.84.12.4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584.Buttin G. Mecanismes regulateurs dans la biosynthese des enzymes du metabolisme du galactose chez Escherichia coli K12. II. Le determinisme genetique de la regulation. J Mol Biol. 1963;7:183–205. doi: 10.1016/s0022-2836(63)80045-5. [DOI] [PubMed] [Google Scholar]
- 585.Buxton R S. Genetic analysis of Escherichia coli K-12 mutants resistant to bacteriophage BF23 and the E-group colicins. Mol Gen Genet. 1971;113:154–156. doi: 10.1007/BF00333188. [DOI] [PubMed] [Google Scholar]
- 586.Buxton R S, Hammer-Jespersen K, Valentin-Hansen P. A second purine nucleoside phosphorylase in Escherichia coli K12. I. Xanthosine phosphorylase regulatory mutants isolated as secondary-site revertants of a deoD mutant. Mol Gen Genet. 1980;179:331–340. doi: 10.1007/BF00425461. [DOI] [PubMed] [Google Scholar]
- 587.Buxton R S, Drury L S. Cloning and insertional inactivation of the dye (sfrA) gene, mutation of which affects sex factor F expression and dye sensitivity of Escherichia coli K-12. J Bacteriol. 1983;154:1309–1314. doi: 10.1128/jb.154.3.1309-1314.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 588.Buxton R S, Drury L S. Identification of the dye gene product, mutational loss of which alters envelope protein composition and also affects sex factor F expression in Escherichia coli. Mol Gen Genet. 1984;194:241–247. doi: 10.1007/BF00383523. [DOI] [PubMed] [Google Scholar]
- 589.Bylund G O, Persson B C, Lundberg L A, Wikstrom P M. A novel ribosome-associated protein is important for efficient translation in Escherichia coli. J Bacteriol. 1997;179:4567–4574. doi: 10.1128/jb.179.14.4567-4574.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 590.Bylund G O, Wipemo L C, Lundberg L A, Wikstrom P M. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli. J Bacteriol. 1998;180:73–82. doi: 10.1128/jb.180.1.73-82.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Bylund J E, Haines M A, Walsh K, Bouloc P, D’Ari R, Higgins M L. Buoyant density studies of several mecillinam-resistant and division mutants of Escherichia coli. J Bacteriol. 1991;173:5396–5402. doi: 10.1128/jb.173.17.5396-5402.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.Byrne C, Stokes H W, Ward K A. Nucleotide sequence of the aceB gene encoding malate synthase A in Escherichia coli. Nucleic Acids Res. 1989;16:9342. doi: 10.1093/nar/16.19.9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 593.Byrne C R, Monroe R S, Ward K A, Kredich N M. DNA sequences of the cysK regions of Salmonella typhimurium and Escherichia coli and linkage of the cysK regions to ptsH. J Bacteriol. 1988;170:3150–3157. doi: 10.1128/jb.170.7.3150-3157.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Bystrom A S, von Gabain A, Bjork G R. Differentially expressed trmD ribosomal protein operon of Escherichia coli is transcribed as a single polycistronic mRNA species. J Mol Biol. 1989;208:575–586. doi: 10.1016/0022-2836(89)90149-6. [DOI] [PubMed] [Google Scholar]
- 595.Bystrom A S, Bjork G R. Chromosomal location and cloning of the gene (trmD) responsible for the synthesis of tRNA (m1/G) methyltransferase in Escherichia coli K-12. Mol Gen Genet. 1982;188:440–446. doi: 10.1007/BF00330046. [DOI] [PubMed] [Google Scholar]
- 596.Bystrom A S, Bjork G R. The structural gene (trmD) for the tRNA(m1G)methyltransferase is part of a four polypeptide operon in Escherichia coli K-12. Mol Gen Genet. 1982;188:447–454. doi: 10.1007/BF00330047. [DOI] [PubMed] [Google Scholar]
- 597.Bystrom A S, Hjalmarsson K J, Wiksstrom P M, Bjork G R. The nucleotide sequence of an Escherichia coli operon containing genes for the tRNA(m1G)methyltransferase, the ribosomal proteins S16 and L19 and a 21-K polypeptide. EMBO J. 1983;2:899–905. doi: 10.1002/j.1460-2075.1983.tb01519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 598.Cabelli R J, Chen L, Tai P C, Oliver D B. SecA protein is required for secretory protein translocation into E. coli membrane vesicles. Cell. 1988;55:683–692. doi: 10.1016/0092-8674(88)90227-9. [DOI] [PubMed] [Google Scholar]
- 599.Cabrera M, Nghiem Y, Miller J H. mutM, a second mutator locus in Escherichia coli that generates G · C → T · A transversions. J Bacteriol. 1988;170:5405–5407. doi: 10.1128/jb.170.11.5405-5407.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 600.Cai J, DuBow M S. Expression of the Escherichia coli chromosomal ars operon. Can J Microbiol. 1996;42:662–671. doi: 10.1139/m96-091. [DOI] [PubMed] [Google Scholar]
- 601.Cai X Y, Maxon M E, Redfield B, Glass R, Brot N, Weissbach H. Methionine synthesis in Escherichia coli: effect of the MetR protein on metE and metH expression. Proc Natl Acad Sci USA. 1989;86:4407–4411. doi: 10.1073/pnas.86.12.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602.Caillet J. Genetic mapping of pheV, an Escherichia coli gene for tRNA(Phe) Mol Gen Genet. 1990;220:317–319. doi: 10.1007/BF00260501. [DOI] [PubMed] [Google Scholar]
- 603.Caillet J, Pages D. Precise physical mapping of the Escherichia coli pheU transcription unit. FEBS Lett. 1991;292:45–47. doi: 10.1016/0014-5793(91)80830-v. [DOI] [PubMed] [Google Scholar]
- 604.Caillet J, Plumbridge J A, Springer M. Evidence that pheV, a gene for tRNAPhe of E. coli, is transcribed from tandem promoters. Nucleic Acids Res. 1985;13:3699–3710. doi: 10.1093/nar/13.10.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 605.Caillet J, Droogmans L. Molecular cloning of the Escherichia coli miaA gene involved in the formation of the Δ2-isopentenyl adenosine in tRNA. J Bacteriol. 1988;170:4147–4152. doi: 10.1128/jb.170.9.4147-4152.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606.Cain B, Norton P J, Eubanks W, Nick H S, Allen C M. Amplification of the bacA gene confers bacitracin resistance to Escherichia coli. J Bacteriol. 1993;175:3784–3789. doi: 10.1128/jb.175.12.3784-3789.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607.Calamita G, Kempf B, Rudd K E, Bonhivers M, Kneip S, Bishai W R, Bremer E, Agre P. The aquaporin-Z water channel gene of Escherichia coli: structure, organization and phylogeny. Biol Cell. 1997;89:321–329. [PubMed] [Google Scholar]
- 608.Calamita G, Bishai W R, Preston G M, Guggino W B, Agre P. Molecular cloning and characterization of AqpZ, a water channel from Escherichia coli. J Biol Chem. 1995;270:29063–29066. doi: 10.1074/jbc.270.49.29063. [DOI] [PubMed] [Google Scholar]
- 609.Caldeira de Araujo A, Favre A. Near ultraviolet DNA damage induces the SOS responses in Escherichia coli. EMBO J. 1986;5:175–179. doi: 10.1002/j.1460-2075.1986.tb04193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610.Calendar R, Lindquist B, Sironi G, Clark A J. Characterization of REP− mutants and their interaction with P2 phage. Virology. 1970;40:72–83. doi: 10.1016/0042-6822(70)90380-6. [DOI] [PubMed] [Google Scholar]
- 611.Calendar R, Ljungquist E, Deho G, Usher D C, Goldstein R, Youderian P, Sironi G, Six E W. Lysogenization by satellite phage P4. Virology. 1981;113:20–38. doi: 10.1016/0042-6822(81)90133-1. [DOI] [PubMed] [Google Scholar]
- 612.Calendar R, Erickson J W, Halling C, Nolte A. Deletion and insertion mutations in the rpoH gene of Escherichia coli that produce functional ς32. J Bacteriol. 1988;170:3479–3484. doi: 10.1128/jb.170.8.3479-3484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 613.Calhoun D H, Wallen J W, Traub L, Gray J E, Kung H-F. Internal promoter in the ilvGEDA transcription unit of Escherichia coli K-12. J Bacteriol. 1985;161:128–132. doi: 10.1128/jb.161.1.128-132.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 614.Calhoun D H, Traub L, Wallen J W, Gray J E, Guterman S K. Location of the rho gene and characterization of λ ilv-gal derivatives of λ ilv-rho bacteriophage. Mol Gen Genet. 1984;193:205–209. doi: 10.1007/BF00330668. [DOI] [PubMed] [Google Scholar]
- 615.Calhoun M W, Newton G, Gennis R B. Physical map locations of genes encoding components of the aerobic respiratory chain of Escherichia coli. J Bacteriol. 1991;173:1569–1570. doi: 10.1128/jb.173.5.1569-1570.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 616.Calhoun M W, Gennis R B. Demonstration of separate genetic loci encoding distinct membrane-bound respiratory NADH dehydrogenases in Escherichia coli. J Bacteriol. 1993;175:3013–3019. doi: 10.1128/jb.175.10.3013-3019.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 617.Callahan A M, Frazier B L, Parkinson J S. Chemotaxis in Escherichia coli: construction and properties of λ tsr transducing phage. J Bacteriol. 1987;169:1246–1253. doi: 10.1128/jb.169.3.1246-1253.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 618.Callahan C, Deutscher M P. Identification and characterization of the Escherichia coli rbn gene encoding the tRNA processing enzyme RNase BN. J Bacteriol. 1996;178:7329–7332. doi: 10.1128/jb.178.24.7329-7332.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 619.Callebaut I, Mornon J P. Trigger factor, one of the Escherichia coli chaperone proteins, is an original member of the FKBP family. FEBS Lett. 1995;374:211–215. doi: 10.1016/0014-5793(95)01109-r. [DOI] [PubMed] [Google Scholar]
- 620.Calos M P, Miller J H. DNA sequence alteration resulting from a mutation impairing promoter function in the lac repressor gene. Mol Gen Genet. 1980;178:225–227. doi: 10.1007/BF00267233. [DOI] [PubMed] [Google Scholar]
- 621.Cam K, Rome G, Krisch H M, Bouché J P. RNase E processing of essential cell division genes mRNA in Escherichia coli. Nucleic Acids Res. 1996;24:3065–3070. doi: 10.1093/nar/24.15.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 622.Cam K, Bejar S, Gil D, Bouché J P. Identification and sequence of gene dicB: translation of the division inhibitor from an in-phase internal start. Nucleic Acids Res. 1988;16:6327–6338. doi: 10.1093/nar/16.14.6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623.Camakaris H, Pittard J. Autoregulation of the tyrR gene. J Bacteriol. 1982;150:70–75. doi: 10.1128/jb.150.1.70-75.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 624.Campbell A, Chang R, Barker D, Ketner G. Biotin regulatory (bir) mutations of Escherichia coli. J Bacteriol. 1980;142:1025–1028. doi: 10.1128/jb.142.3.1025-1028.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 625.Campbell A, Schneider S J, Song B. Lambdoid phages as elements of bacterial genomes. Genetica. 1992;86:259–267. doi: 10.1007/BF00133724. [DOI] [PubMed] [Google Scholar]
- 626.Campbell H D, Rogers B L, Young I G. Nucleotide sequence of the respiratory d-lactate dehydrogenase gene of Escherichia coli. Eur J Biochem. 1984;144:367–373. doi: 10.1111/j.1432-1033.1984.tb08473.x. [DOI] [PubMed] [Google Scholar]
- 627.Campbell J, Lengyel J, Langridge J. Evolution of a second gene for B-galactosidase in Escherichia coli. Proc Natl Acad Sci USA. 1973;70:1841–1845. doi: 10.1073/pnas.70.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 628.Campbell J L, Kleckner N. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell. 1990;62:967–979. doi: 10.1016/0092-8674(90)90271-f. [DOI] [PubMed] [Google Scholar]
- 629.Canellakis E S, Paterakis A A, Huang S C, Panagiotidis C A, Kyriakidis D A. Identification, cloning and nucleotide sequencing of the ornithine decarboxylase antizyme gene of Escherichia coli. Proc Natl Acad Sci USA. 1993;90:7129–7133. doi: 10.1073/pnas.90.15.7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 630.Cannon M, Cabezon T, Bollen A. Mapping of neamine resistance: identification of two genetic loci, neaA and neaB. Mol Gen Genet. 1974;130:321–326. doi: 10.1007/BF00333871. [DOI] [PubMed] [Google Scholar]
- 631.Cao G J, Pogliano J, Sarkar N. Identification of the coding region for a second poly(A) polymerase in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:11580–11585. doi: 10.1073/pnas.93.21.11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 632.Cao G J, Sarkar N. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc Natl Acad Sci USA. 1992;89:10380–10384. doi: 10.1073/pnas.89.21.10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 633.Cao Y, Rowland R R R, Kogoma T. DNA polymerase I and the bypassing of RecA dependence of constitutive stable DNA replication in Escherichia coli rnhA mutants. J Bacteriol. 1993;175:7247–7253. doi: 10.1128/jb.175.22.7247-7253.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 634.Cao Y, Kogoma T. Requirement for the polymerization and 5′→3′ exonuclease activities of DNA polymerase I in initiation of DNA replication at oriK sites in the absence of RecA in Escherichia coli rnhA mutants. J Bacteriol. 1993;175:7254–7259. doi: 10.1128/jb.175.22.7254-7259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 635.Cardelli J, Konisky J. Isolation and characterization of an Escherichia coli mutant tolerant to colicins Ia and Ib. J Bacteriol. 1974;119:379–385. doi: 10.1128/jb.119.2.379-385.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 636.Carl P. Isolation and mapping of a mutation in Escherichia coli with altered levels of ribonuclease H. J Bacteriol. 1980;144:28–35. doi: 10.1128/jb.144.1.28-35.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 637.Carlin A, Shi W, Dey S, Rosen B P. The ars operon of Escherichia coli confers arsenical and antimonial resistance. J Bacteriol. 1995;177:981–986. doi: 10.1128/jb.177.4.981-986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 638.Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 639.Carlioz A, Ludwig M L, Stallings W C, Fee J A, Steinman H M, Touati D. Iron superoxide dismutase: nucleotide sequence of the gene from Escherichia coli K12 and correlations with crystal structures. J Biol Chem. 1988;263:1555–1562. [PubMed] [Google Scholar]
- 640.Carlson J, Fuchs J A, Messing J. Primary structure of the Escherichia coli ribonucleoside diphosphate reductase operon. Proc Natl Acad Sci USA. 1984;81:4294–4297. doi: 10.1073/pnas.81.14.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 641.Carmel O, Dover N, Rahav-Manor O, Dibrov P, Kirsch D, Karpel R, Schuldiner S, Padan E. A single amino acid substitution (Glu134→Ala) in NhaR1 increases the inducibility by Na+ of the product of nhaA, a Na+/H+ antiporter gene in Escherichia coli. EMBO J. 1994;13:1981–1989. doi: 10.1002/j.1460-2075.1994.tb06467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 642.Carothers A M, McFall E, Palchaudhuri S. Physical mapping of a mutation in Escherichia coli with altered levels of ribonuclease. J Bacteriol. 1980;142:174–184. doi: 10.1128/jb.142.1.174-184.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 643.Carra J H, Schleif R F. Variation of half-site organization and DNA looping by AraC protein. EMBO J. 1993;12:35–44. doi: 10.1002/j.1460-2075.1993.tb05629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 643a.Carter A T, Pearson B M, Dickinson J R, Lancashire W E. Sequence of the Escherichia coli K-12 edd and eda genes of the Entner-Douderoff pathway. Gene. 1993;130:155–156. doi: 10.1016/0378-1119(93)90362-7. [DOI] [PubMed] [Google Scholar]
- 644.Carter J R, Franden M A, Lippincott J A, McHenry C S. Identification, molecular cloning and characterization of the gene encoding the chi subunit of DNA polymerase III holoenzyme of Escherichia coli. Mol Gen Genet. 1993;241:399–408. doi: 10.1007/BF00284693. [DOI] [PubMed] [Google Scholar]
- 645.Carter J R, Franden M A, Aebersold R, McHenry C S. Molecular cloning, sequencing, and overexpression of the structural gene encoding the δ subunit of Escherichia coli DNA polymerase III holoenzyme. J Bacteriol. 1992;174:7013–7025. doi: 10.1128/jb.174.21.7013-7025.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 646.Carter J R, Franden M A, Aebersold R, McHenry C S. Identification, isolation, and overexpression of the gene encoding the ψ subunit of DNA polymerase III holoenzyme. J Bacteriol. 1993;175:5604–5610. doi: 10.1128/jb.175.17.5604-5610.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 647.Carter J R, Franden M A, Aebersold R, Kim D R, McHenry C S. Isolation, sequencing and overexpression of the gene encoding the theta subunit of DNA polymerase III holoenzyme. Nucleic Acids Res. 1993;21:3281–3286. doi: 10.1093/nar/21.14.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 648.Casaregola S, Jacq A, Laoudj D, McGurk G, Margarson S, Tempete M, Norris V, Holland I B. Cloning and analysis of the entire Escherichia coli ams gene. ams is identical to hmp1 and encodes a 114 kDa protein that migrates as a 180 kDa protein. J Mol Biol. 1992;228:30–40. doi: 10.1016/0022-2836(92)90489-7. [DOI] [PubMed] [Google Scholar]
- 649.Case L M, Chen X N, Deutscher M P. Localization of the Escherichia coli rnt gene encoding RNase T by using a combination of physical and genetic mapping. J Bacteriol. 1989;171:5736–5737. doi: 10.1128/jb.171.10.5736-5737.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 650.Caskey C T, Forrester W C, Tate W, Ward C D. Cloning of the Escherichia coli release factor 2 gene. J Bacteriol. 1984;158:365–368. doi: 10.1128/jb.158.1.365-368.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 651.Cass L G, Horwitz A H, Miyada C G, Greenfield L, Wilcox G. The araC regulatory gene mRNA contains a leader sequence. Mol Gen Genet. 1980;180:219–226. doi: 10.1007/BF00267373. [DOI] [PubMed] [Google Scholar]
- 652.Cassan M, Parsot C, Cohen G N, Patte J C. Nucleotide sequence of lysC gene encoding the lysine-sensitive aspartokinase II of Escherichia coli. J Biol Chem. 1986;261:1052–1057. [PubMed] [Google Scholar]
- 653.Cassan M, Ronceray J, Patte J C. Nucleotide sequence of the promoter region of the E. coli lysC gene. Nucleic Acids Res. 1983;11:6157–6165. doi: 10.1093/nar/11.18.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 654.Casse F. Mapping of the gene chlB controlling membrane bound nitrate reductase and formic hydrogen-lyase activities in Escherichia coli K-12. Biochem Biophys Res Commun. 1970;39:429–436. doi: 10.1016/0006-291x(70)90596-6. [DOI] [PubMed] [Google Scholar]
- 655.Cassey B, Guest J R, Attwood M M. Environmental control of pyruvate dehydrogenase complex expression in Escherichia coli. FEMS Microbiol Lett. 1998;159:325–329. doi: 10.1111/j.1574-6968.1998.tb12878.x. [DOI] [PubMed] [Google Scholar]
- 656.Castano I, Bastarrachea F. glnF-lacZ fusions in Escherichia coli: studies on glnF expression and its chromosomal orientation. Mol Gen Genet. 1984;195:228–233. doi: 10.1007/BF00332751. [DOI] [PubMed] [Google Scholar]
- 657.Castano I, Bastarrachea F, Covarrubias A. gltBDF operon of Escherichia coli. J Bacteriol. 1988;170:821–827. doi: 10.1128/jb.170.2.821-827.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 658.Caster J H. Selection of thymine-requiring strains from Escherichia coli on solid medium. J Bacteriol. 1967;94:1804. doi: 10.1128/jb.94.5.1804-.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 659.Caswell R, Williams J, Lyddiatt A, Busby S. Overexpression, purification and characterization of the Escherichia coli MelR transcription activator protein. Biochem J. 1992;287:493–499. doi: 10.1042/bj2870493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 660.Cavard D, Lazdunski C, Howard S P. The acylated precursor form of the colicin A lysis protein is a natural substrate of the DegP protease. J Bacteriol. 1989;171:6316–6322. doi: 10.1128/jb.171.11.6316-6322.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 661.Cavard D, Pages J M, Lazdunski A. A protease as a possible sensor of environmental conditions in E. coli outer membrane. Mol Gen Genet. 1982;188:508–512. doi: 10.1007/BF00330057. [DOI] [PubMed] [Google Scholar]
- 662.Cavicchioli R, Kolesnikow T, Chiang R C, Gunsalus R P. Characterization of the aegA locus of Escherichia coli: control of gene expression in response to anaerobiosis and nitrate. J Bacteriol. 1996;178:6968–6974. doi: 10.1128/jb.178.23.6968-6974.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 663.Cecchini G, Sices H, Schröder I, Gunsalus R P. Aerobic inactivation of fumarate reductase from Escherichia coli by mutation of the [3Fe-4S]-quinone binding domain. J Bacteriol. 1995;177:4587–4592. doi: 10.1128/jb.177.16.4587-4592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 664.Celis R T F. Mapping of two loci affecting the synthesis and structure of a periplasmic protein involved in arginine and ornithine transport in Escherichia coli K-12. J Bacteriol. 1982;151:1314–1319. doi: 10.1128/jb.151.3.1314-1319.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 665.Celis, R. T. F. Submitted for publication.
- 666.Cerretti D P, Dean D, Davis G R, Bedwell D M, Nomura M. The spc ribosomal protein operon of Escherichia coli: sequence and cotranscription of the ribosomal protein genes and a protein export gene. Nucleic Acids Res. 1983;11:2599–2616. doi: 10.1093/nar/11.9.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 667.Cha M K, Kim H K, Kim I H. Thioredoxin-linked “thiol peroxidase” from periplasmic space of Escherichia coli. J Biol Chem. 1995;270:28635–28641. doi: 10.1074/jbc.270.48.28635. [DOI] [PubMed] [Google Scholar]
- 668.Cha M K, Kim H K, Kim I H. Mutation and mutagenesis of thiol peroxidase of Escherichia coli and a new type of thiol peroxidase family. J Bacteriol. 1996;178:5610–5614. doi: 10.1128/jb.178.19.5610-5614.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 669.Chakrabarti T, Chen Y-M, Lin E C C. Clustering of genes for l-fucose dissimilation by Escherichia coli. J Bacteriol. 1984;157:984–986. doi: 10.1128/jb.157.3.984-986.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 670.Chalker A F, Leach D R F, Lloyd R G. Escherichia coli sbcC mutants permit stable propagation of DNA replicons containing a long palindrome. Gene. 1988;71:201–205. doi: 10.1016/0378-1119(88)90092-3. [DOI] [PubMed] [Google Scholar]
- 671.Chan A, Nagel R. Involvement of recA and recF in the induced precise excision of Tn10 in Escherichia coli. Mutat Res. 1997;381:111–115. doi: 10.1016/s0027-5107(97)00157-7. [DOI] [PubMed] [Google Scholar]
- 672.Chan F Y, Torriani A. PstB protein of the phosphate-specific transport system of Escherichia coli is an ATPase. J Bacteriol. 1996;178:3974–3977. doi: 10.1128/jb.178.13.3974-3977.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 673.Chanda P K, Ono M, Kuwano M, Kung H-F. Cloning, sequence analysis, and expression of alteration of the mRNA stability gene (ams+) of Escherichia coli. J Bacteriol. 1985;161:446–449. doi: 10.1128/jb.161.1.446-449.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 674.Chang C N, Kuang W-J, Chen E Y. Nucleotide sequence of the alkaline phosphatase gene of Escherichia coli. Gene. 1986;44:121–125. doi: 10.1016/0378-1119(86)90050-8. [DOI] [PubMed] [Google Scholar]
- 675.Chang S F, Ng D, Baird L, Georgopoulos C. Analysis of an Escherichia coli dnaB temperature-sensitive insertion mutation and its cold-sensitive extragenic suppressor. J Biol Chem. 1991;266:3654–3660. [PubMed] [Google Scholar]
- 676.Chang S-Y, McGary E C, Chang S. Methionine aminopeptidase gene of Escherichia coli is essential for cell growth. J Bacteriol. 1989;171:4071–4072. doi: 10.1128/jb.171.7.4071-4072.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 677.Chang Y-Y, Wang A Y, Cronan J E., Jr Molecular cloning, DNA sequencing, and biochemical analyses of Escherichia coli glyoxylate carboligase. An enzyme of the acetohydroxy acid synthase-pyruvate oxidase family. J Biol Chem. 1993;268:3911–3919. [PubMed] [Google Scholar]
- 678.Chang Y-Y, Wang A Y, Cronan J E., Jr Expression of Escherichia coli pyruvate oxidase (PoxB) depends on the sigma factor encoded by the rpoS (katF) gene. Mol Microbiol. 1994;11:1019–1028. doi: 10.1111/j.1365-2958.1994.tb00380.x. [DOI] [PubMed] [Google Scholar]
- 679.Chang Y-Y, Cronan J E., Jr Mapping nonselectable genes of Escherichia coli by using transposon Tn10: location of a gene affecting pyruvate oxidase. J Bacteriol. 1982;151:1279–1289. doi: 10.1128/jb.151.3.1279-1289.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 680.Chang Y-Y, Cronan J E., Jr Genetic and biochemical analyses of Escherichia coli strains having a mutation in the structural gene (poxB) for pyruvate oxidase. J Bacteriol. 1983;154:756–762. doi: 10.1128/jb.154.2.756-762.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 681.Chang Y Y, Cronan J E, Li S J, Reed K, Vanden Boom T, Wang A Y. Locations of the lip, poxB, and ilvBN genes on the physical map of Escherichia coli. J Bacteriol. 1991;173:5258–5259. doi: 10.1128/jb.173.17.5258-5259.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 682.Charette M F, Henderson G W, Markovitz A. ATP hydrolysis-dependent protease activity of the lon (capR) protein of Escherichia coli K-12. Proc Natl Acad Sci USA. 1981;78:4728–4732. doi: 10.1073/pnas.78.8.4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 683.Charles I G, Lamb H K, Pickard D, Dougan G, Hawkins A R. Isolation, characterization and nucleotide sequences of the aroC genes encoding chorismate synthase from Salmonella typhi and Escherichia coli. J Gen Microbiol. 1990;136:353–358. doi: 10.1099/00221287-136-2-353. [DOI] [PubMed] [Google Scholar]
- 684.Charlier D, Hassanzadeh G, Kholti A, Gigot D, Pierard A, Glansdorff N. carP, involved in pyrimidine regulation of the Escherichia coli carbamoylphosphate synthetase operon, encodes a sequence-specific DNA-binding protein identical to XerB and PepA, also required for resolution of ColEI multimers. J Mol Biol. 1995;250:392–406. doi: 10.1006/jmbi.1995.0385. [DOI] [PubMed] [Google Scholar]
- 685.Charlier D, Roovers M, Gigot D, Huysveld N, Pierard A, Glansdorff N. Integration host factor (IHF) modulates the expression of the pyrimidine-specific promoter of the carAB operons of Escherichia coli K12 and Salmonella typhimurium LT2. Mol Gen Genet. 1993;237:273–286. doi: 10.1007/BF00282809. [DOI] [PubMed] [Google Scholar]
- 686.Charlier J, Sanchez R. Lysyl-tRNA synthetase from Escherichia coli K-12. Chromatographic heterogeneity and the lysU gene product. Biochem J. 1987;248:43–51. doi: 10.1042/bj2480043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 687.Chase J W, Rabin B A, Murphy J B, Stone K L, Williams K R. Escherichia coli exonuclease VII. Cloning and sequencing of the gene encoding the large subunit (xseA) J Biol Chem. 1986;261:14929–14935. [PubMed] [Google Scholar]
- 688.Chatterjee P K, Sternberg N L. A general genetic approach in Escherichia coli for determining the mechanisms of action of tumoricidal agents: application to DMP 840, a tumoricidal agent. Proc Natl Acad Sci USA. 1995;92:8950–8954. doi: 10.1073/pnas.92.19.8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 689.Chattopadhyay S, Wu Y, Datta P. Involvement of Fnr and ArcA in anaerobic expression of the tdc operon of Escherichia coli. J Bacteriol. 1997;179:4868–4873. doi: 10.1128/jb.179.15.4868-4873.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 689a.Chaudhuri, S. R., J. Conrad, B. G. Hall, and J. Ofengand. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for the normal growth of Escherichia coli. RNA, in press. [DOI] [PMC free article] [PubMed]
- 690.Chaudhury A M, Smith G R. Role of Escherichia coli RecBC enzyme in SOS induction. Mol Gen Genet. 1985;201:525–528. doi: 10.1007/BF00331350. [DOI] [PubMed] [Google Scholar]
- 691.Chauhan A K, Miczak A, Taraseviciene L, Apirion D. Sequencing and expression of the rne gene of Escherichia coli. Nucleic Acids Res. 1991;19:125–129. doi: 10.1093/nar/19.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 692.Chauhan A K, Apirion D. The gene for a small stable RNA (10Sa RNA) Mol Microbiol. 1989;3:1481–1485. doi: 10.1111/j.1365-2958.1989.tb00133.x. [DOI] [PubMed] [Google Scholar]
- 693.Chen B J, Carroll P, Samson L D. The Escherichia coli AlkB protein protects human cells against alkylation-induced toxicity. J Bacteriol. 1994;176:6255–6261. doi: 10.1128/jb.176.20.6255-6261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 694.Chen C M, Ye Q Z, Zhu Z M, Wanner B L, Walsh C T. Molecular biology of carbon-phosphorus bond cleavage. Cloning and sequencing of the phn (psiD) genes involved in alkylphosphonate uptake and C-P lyase activity in Escherichia coli B. J Biol Chem. 1990;265:4461–4471. [PubMed] [Google Scholar]
- 695.Chen C R, Malik M, Snyder M, Drlica K. DNA gyrase and topoisomerase IV on the bacterial chromosome: quinolone-induced DNA cleavage. J Mol Biol. 1996;258:627–637. doi: 10.1006/jmbi.1996.0274. [DOI] [PubMed] [Google Scholar]
- 696.Chen G F, Inouye M. Suppression of the negative effect of minor arginine codons on gene expression: preferential usage of minor codons within the first 25 codons of the Escherichia coli genes. Nucleic Acids Res. 1990;18:1465–1473. doi: 10.1093/nar/18.6.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 697.Chen G T, Axley M J, Hacia J, Inouye M. Overproduction of a selenocysteine-containing polypeptide in Escherichia coli: the fdhF gene product. Mol Microbiol. 1992;6:781–785. doi: 10.1111/j.1365-2958.1992.tb01528.x. [DOI] [PubMed] [Google Scholar]
- 698.Chen H, Bryan S K, Moses R E. Cloning the polB gene of Escherichia coli and identification of its product. J Biol Chem. 1989;264:20591–20595. [PubMed] [Google Scholar]
- 699.Chen H, Sun Y, Stark T F, Beattie W G, Moses R E. Nucleotide sequence and deletion analysis of the polB gene of Escherichia coli. DNA Cell Biol. 1990;9:631–635. doi: 10.1089/dna.1990.9.631. [DOI] [PubMed] [Google Scholar]
- 700.Chen J, Brevet A, Fromant M, Leveque F, Schmitter J M, Blanquet S, Plateau P. Pyrophosphatase is essential for growth of Escherichia coli. J Bacteriol. 1990;172:5686–5689. doi: 10.1128/jb.172.10.5686-5689.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 701.Chen K S, Saxena P, Walker J R. Expression of the Escherichia coli dnaX gene. J Bacteriol. 1993;175:6663–6670. doi: 10.1128/jb.175.20.6663-6670.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 702.Chen L S, Coleman W G. Cloning and characterization of the Escherichia coli K-12 rfa-2 (rfaC) gene, a gene required for lipopolysaccharide inner core synthesis. J Bacteriol. 1993;175:2534–2540. doi: 10.1128/jb.175.9.2534-2540.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 703.Chen M X, Bouquin N, Norris V, Casaregola S, Seror S J, Holland I B. A single base change in the acceptor stem of tRNA3Leu confers resistance upon Escherichia coli to the calmodulin inhibitor, 48/80. EMBO J. 1991;10:3113–3122. doi: 10.1002/j.1460-2075.1991.tb07865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 704.Chen Q, Arents J C, Bader R, Postma P W, Amster-Choder O. BglF, the sensor of the E. coli bgl system, uses the same site to phosphorylate both a sugar and a regulatory protein. EMBO J. 1997;16:4617–4627. doi: 10.1093/emboj/16.15.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 705.Chen R, Henning U. Nucleotide sequence of the gene for the peptidoglycan-associated lipoprotein of Escherichia coli K12. Eur J Biochem. 1987;163:73–77. doi: 10.1111/j.1432-1033.1987.tb10738.x. [DOI] [PubMed] [Google Scholar]
- 706.Chen R, Henning U. A periplasmic protein (Skp) of Escherichia coli selectively binds a class of outer membrane proteins. Mol Microbiol. 1996;19:1287–1294. doi: 10.1111/j.1365-2958.1996.tb02473.x. [DOI] [PubMed] [Google Scholar]
- 707.Chen R, Schmidmayr W, Kramer C, Chen-Schmeisser U, Henning U. Primary structure of major outer membrane protein II (OmpA protein) of Escherichia coli K-12. Proc Natl Acad Sci USA. 1980;77:4592–4596. doi: 10.1073/pnas.77.8.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 708.Chen S M, Takiff H E, Barber A M, Dubois G C, Bardwell J C, Court D L. Expression and characterization of RNase III and Era proteins. Products of the rnc operon of Escherichia coli. J Biol Chem. 1990;265:2888–2895. [PubMed] [Google Scholar]
- 709.Chen W, Russell C S, Murooka Y, Cosloy S D. 5-Aminolevulinic acid synthesis in Escherichia coli requires expression of hemA. J Bacteriol. 1994;176:2743–2746. doi: 10.1128/jb.176.9.2743-2746.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 710.Chen Y-M, Lin E C C. Regulation of the adhE gene, which encodes ethanol dehydrogenase in Escherichia coli. J Bacteriol. 1991;173:8009–8013. doi: 10.1128/jb.173.24.8009-8013.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 711.Chen Y-M, Backman K, Magasanik B. Characterization of a gene, glnL, the product of which is involved in the regulation of nitrogen utilization in Escherichia coli. J Bacteriol. 1982;150:214–220. doi: 10.1128/jb.150.1.214-220.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 712.Chen Y-M, Zhu Y, Lin E C C. NAD-linked aldehyde dehydrogenase for aerobic utilization of l-fucose and l-rhamnose by Escherichia coli. J Bacteriol. 1987;169:3289–3294. doi: 10.1128/jb.169.7.3289-3294.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 713.Chen Y-M, Zhu Y, Lin E C C. The organization of fuc regulon specifying l-fucose dissimilation in Escherichia coli K12 as determined by gene cloning. Mol Gen Genet. 1987;210:331–337. doi: 10.1007/BF00325702. [DOI] [PubMed] [Google Scholar]
- 714.Chen Y-M, Lu Z, Lin E C C. Constitutive activation of the fucAO operon and silencing of the divergently transcribed fucPIK operon by an IS5 element in Escherichia coli mutants selected for growth on l-1,2-propanediol. J Bacteriol. 1989;171:6097–6105. doi: 10.1128/jb.171.11.6097-6105.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 715.Chen Y W, Dekker E E, Somerville R L. Functional analysis of E. coli threonine dehydrogenase by means of mutant isolation and characterization. Biochim Biophys Acta. 1995;1253:208–214. doi: 10.1016/0167-4838(95)00162-2. [DOI] [PubMed] [Google Scholar]
- 716.Chenais J, Richaud C, Ronceray J, Cherest H, Surdin-Kerjan Y, Patte J C. Construction of hybrid plasmids containing the lysA gene of Escherichia coli: studies of expression in Escherichia coli and Saccharomyces cerevisiae. Mol Gen Genet. 1981;182:456–461. doi: 10.1007/BF00293935. [DOI] [PubMed] [Google Scholar]
- 717.Chenault S S, Earhart C F. Organization of genes encoding membrane proteins of the Escherichia coli ferrienterobactin permease. Mol Microbiol. 1991;5:1405–1413. doi: 10.1111/j.1365-2958.1991.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 718.Cheng Y S, Zipser D, Cheng C Y, Rosleth S J. Isolation and characterization of mutations in the structural gene for protease III (ptr) J Bacteriol. 1979;140:125–130. doi: 10.1128/jb.140.1.125-130.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 719.Cheng Y S, Shen Y, Rudolph J, Stern M, Stubbe J, Flannigan K A, Smith J M. Glycinamide ribonucleotide synthetase from Escherichia coli: cloning, overproduction, sequencing, isolation, and characterization. Biochemistry. 1990;29:218–227. doi: 10.1021/bi00453a030. . (Erratum, 29:5220.) [DOI] [PubMed] [Google Scholar]
- 719a.Cheng Z F, Zuo Y, Li Z, Rudd K E, Deutscher M P. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J Biol Chem. 1998;273:14077–14080. doi: 10.1074/jbc.273.23.14077. [DOI] [PubMed] [Google Scholar]
- 720.Chepuri V, Lemieux L, Hill J, Alben J O, Gennis R B. Recent studies of the cytochrome o terminal oxidase complex of Escherichia coli. Biochim Biophys Acta. 1990;1018:124–127. doi: 10.1016/0005-2728(90)90231-r. [DOI] [PubMed] [Google Scholar]
- 721.Chepuri V, Gennis R B. The use of gene fusions to determine the topology of all the subunits of the cytochrome o terminal oxidase complex of Escherichia coli. J Biol Chem. 1990;265:12978–12986. [PubMed] [Google Scholar]
- 722.Chesney R H, Adler E. Chromosomal location of attP7, the recA-independent integration site used in the suppression of Escherichia coli dnaA mutations. J Bacteriol. 1982;150:1400–1404. doi: 10.1128/jb.150.3.1400-1404.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 723.Chesney R H, Sollitti P, Vickery D R. Identification of a new locus in the Escherichia coli cotransduction gap that represents a new genetic component of the l-asparagine utilization system. J Gen Microbiol. 1985;131:2079–2085. doi: 10.1099/00221287-131-8-2079. [DOI] [PubMed] [Google Scholar]
- 724.Cheung A Y, Morgan S, Low K B, Soll D. Regulation of the biosynthesis of aminoacyl-transfer ribonucleic acid synthetases and of transfer ribonucleic acid in Escherichia coli. VI. Mutants with increased levels of glutaminyl-transfer ribonucleic acid synthetase and of glutamine transfer ribonucleic acid. J Bacteriol. 1979;139:176–184. doi: 10.1128/jb.139.1.176-184.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 725.Chi Y I, Yokota H, Kim S H. Apo structure of the ligand-binding domain of aspartate receptor from Escherichia coli and its comparison with ligand-bound or pseudoligand-bound structures. FEBS Lett. 1997;414:327–332. doi: 10.1016/s0014-5793(97)01027-2. [DOI] [PubMed] [Google Scholar]
- 726.Chiang R C, Cavicchioli R, Gunsalus R P. Identification and characterization of narQ, a second nitrate sensor for nitrate-dependent gene regulation in Escherichia coli. Mol Microbiol. 1992;6:1913–1923. doi: 10.1111/j.1365-2958.1992.tb01364.x. [DOI] [PubMed] [Google Scholar]
- 727.Chiang R C, Cavicchioli R, Gunsalus R P. Physical map location of the narQ gene of Escherichia coli. J Bacteriol. 1992;174:7882. doi: 10.1128/jb.174.23.7882.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 728.Chiang R C, Cavicchioli R, Gunsalus R P. ‘Locked-on’ and ‘locked-off’ signal transduction mutations in the periplasmic domain of the Escherichia coli NarQ and NarX sensors affect nitrate- and nitrite-dependent regulation by NarL and NarP. Mol Microbiol. 1997;24:1049–1060. doi: 10.1046/j.1365-2958.1997.4131779.x. [DOI] [PubMed] [Google Scholar]
- 729.Chiaramello A, Zyskind J W. Expression of Escherichia coli dnaA and mioC genes as a function of growth rate. J Bacteriol. 1989;171:4272–4280. doi: 10.1128/jb.171.8.4272-4280.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 730.Chiariotti L, Nappo A G, Carlomagno M S, Bruni C B. Gene structure in the histidine operon of Escherichia coli. Identification and nucleotide sequence of the hisB gene. Mol Gen Genet. 1986;202:42–47. doi: 10.1007/BF00330514. [DOI] [PubMed] [Google Scholar]
- 731.Chiariotti L, Alifano P, Carlomagno M S, Bruni C B. Nucleotide sequence of the Escherichia coli hisD gene and of the Escherichia coli and Salmonella typhimurium hisIE region. Mol Gen Genet. 1986;203:382–388. doi: 10.1007/BF00422061. [DOI] [PubMed] [Google Scholar]
- 732.Chiaruttini C, Milet M, Springer M. Translational coupling by modulation of feedback repression in the IF3 operon of Escherichia coli. Proc Natl Acad Sci USA. 1997;94:9208–9213. doi: 10.1073/pnas.94.17.9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 733.Chin D T, Goff S A, Webster T, Smith T, Goldberg A L. Sequence of the lon gene in Escherichia coli. A heat-shock gene which encodes the ATP-dependent protease La. J Biol Chem. 1988;263:11718–11728. [PubMed] [Google Scholar]
- 734.Chippaux M, Giudici D, Abou-Jaoude A, Pascal M C. A mutation leading to the total lack of nitrate reductase activity in Escherichia coli K-12. Mol Gen Genet. 1978;160:225–229. doi: 10.1007/BF00267485. [DOI] [PubMed] [Google Scholar]
- 735.Chippaux M, Bonnefoy-Orth V, Ratouchniak J, Pascal M C. Operon fusions in the nitrate reductase operon and study of the control gene nirR in Escherichia coli. Mol Gen Genet. 1981;182:477–479. doi: 10.1007/BF00293938. [DOI] [PubMed] [Google Scholar]
- 736.Chittum H S, Champney W S. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli. J Bacteriol. 1994;176:6192–6198. doi: 10.1128/jb.176.20.6192-6198.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 737.Cho H, Cronan J E., Jr Escherichia coli thioesterase I, molecular cloning and sequencing of the structural gene and identification as a periplasmic enzyme. J Biol Chem. 1993;268:9238–9245. [PubMed] [Google Scholar]
- 738.Cho H, Cronan J E., Jr “Protease I” of Escherichia coli functions as a thioesterase in vivo. J Bacteriol. 1994;176:1793–1795. doi: 10.1128/jb.176.6.1793-1795.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 739.Choe M, Reznikoff W S. Anaerobically expressed Escherichia coli genes identified by operon fusion techniques. J Bacteriol. 1991;173:6139–6146. doi: 10.1128/jb.173.19.6139-6146.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 740.Choe M, Reznikoff W S. Identification of the regulatory sequence of anaerobically expressed locus aeg-46.5. J Bacteriol. 1993;175:1165–1172. doi: 10.1128/jb.175.4.1165-1172.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 741.Choi K Y, Zalkin H. Role of the purine repressor hinge sequence in repressor function. J Bacteriol. 1994;176:1767–1772. doi: 10.1128/jb.176.6.1767-1772.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 742.Choi Y L, Kawamukai M, Utsumi R, Sakai H, Komano T. Molecular cloning and sequencing of the glycogen phosphorylase gene from Escherichia coli. FEBS Lett. 1989;243:193–198. doi: 10.1016/0014-5793(89)80128-0. [DOI] [PubMed] [Google Scholar]
- 743.Choi Y L, Kawase S, Nishida T, Sakai H, Komano T, Kawamukai M, Utsumi R, Kohara Y, Akiyama K. Nucleotide sequence of the glpR gene encoding the repressor for the glycerol-3-phosphate regulon of Escherichia coli K12. Nucleic Acids Res. 1988;16:7732. doi: 10.1093/nar/16.15.7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 744.Choi Y L, Nishida T, Kawamukai M, Utsumi R, Sakai H, Komano T. Cloning and sequencing of an Escherichia coli gene, nlp, highly homologous to the ner genes of bacteriophages Mu and D108. J Bacteriol. 1989;171:5222–5225. doi: 10.1128/jb.171.9.5222-5225.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 745.Chow J, Dennis P P. Coupling between mRNA synthesis and mRNA stability in Escherichia coli. Mol Microbiol. 1994;11:919–931. doi: 10.1111/j.1365-2958.1994.tb00371.x. [DOI] [PubMed] [Google Scholar]
- 746.Christiansen L, Pedersen S. Cloning, restriction endonuclease mapping and post-transcriptional regulation of rpsA, the structural gene for ribosomal protein S1. Mol Gen Genet. 1981;181:548–551. doi: 10.1007/BF00428751. [DOI] [PubMed] [Google Scholar]
- 747.Christie G E, Platt T. Gene structure in the tryptophan operon of Escherichia coli. Nucleotide sequence of trpC and the flanking intercistronic regions. J Mol Biol. 1980;142:519–530. doi: 10.1016/0022-2836(80)90261-2. [DOI] [PubMed] [Google Scholar]
- 748.Chu C C, Templin A, Clark A J. Suppression of a frameshift mutation in the recE gene of Escherichia coli K-12 occurs by gene fusion. J Bacteriol. 1989;171:2101–2109. doi: 10.1128/jb.171.4.2101-2109.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 749.Chuang S E, Blattner F R. Characterization of twenty-six new heat shock genes of Escherichia coli. J Bacteriol. 1993;175:5242–5252. doi: 10.1128/jb.175.16.5242-5252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 750.Chuang S E, Burland V D, Plunkett G, Daniels D L, Blattner F R. Sequence analysis of four new heat-shock genes constituting the hslTS/ibpAB and hslVU operons in Escherichia coli. Gene. 1993;134:1–6. doi: 10.1016/0378-1119(93)90167-2. [DOI] [PubMed] [Google Scholar]
- 751.Chun S-Y, Randall L L. In vivo studies of the role of SecA during protein export in Escherichia coli. J Bacteriol. 1994;176:4197–4203. doi: 10.1128/jb.176.14.4197-4203.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 752.Chung C H, Goldberg A L. The product of the lon (capR) gene in Escherichia coli is the ATP-dependent protease, protease La. Proc Natl Acad Sci USA. 1981;78:4931–4935. doi: 10.1073/pnas.78.8.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 753.Chung T, Klumpp D J, LaPorte D C. Glyoxylate bypass operon of Escherichia coli: cloning and determination of the functional map. J Bacteriol. 1988;170:386–392. doi: 10.1128/jb.170.1.386-392.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 754.Chung T, Resnik E, Stueland C S, LaPorte D C. Relative expression of the products of the glyoxylate bypass operon: contributions of transcription and translation. J Bacteriol. 1993;175:4572–4575. doi: 10.1128/jb.175.14.4572-4575.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 755.Chye M L, Pittard J. Transcription control of the aroP gene in Escherichia coli K-12: analysis of operator mutants. J Bacteriol. 1987;169:386–393. doi: 10.1128/jb.169.1.386-393.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 756.Chye M L, Guest J R, Pittard J. Cloning of the aroP gene and identification of its product in Escherichia coli K-12. J Bacteriol. 1986;167:749–753. doi: 10.1128/jb.167.2.749-753.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 757.Cieslewicz M J, Steenbergen S M, Vimr E. Cloning, sequencing, expression, and complementation analysis of the Escherichia coli K1 kps region 1 gene, kpsE, and identification of an upstream open reading frame encoding a protein with homology to GutQ. J Bacteriol. 1993;175:8018–8023. doi: 10.1128/jb.175.24.8018-8023.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 758.Clark A F, Hogg R W. High-affinity arabinose transport mutants of Escherichia coli: isolation and gene location. J Bacteriol. 1981;147:920–924. doi: 10.1128/jb.147.3.920-924.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 759.Clark A J. rec genes and homologous recombination proteins in Escherichia coli. Biochimie. 1991;73:523–532. doi: 10.1016/0300-9084(91)90124-j. [DOI] [PubMed] [Google Scholar]
- 759a.Clark, A. J. Personal communication.
- 760.Clark A J, Satin L H, Chu C C. Transcription of the Escherichia coli recE gene from a promoter in Tn5 and IS50. J Bacteriol. 1994;176:7024–7031. doi: 10.1128/jb.176.22.7024-7031.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 761.Clark A J, Sharma V, Brenowitz S, Chu C C, Sandler S, Satin L, Templin A, Berger I, Cohen A. Genetic and molecular analyses of the C-terminal region of the recE gene from the Rac prophage of Escherichia coli K-12 reveal the recT gene. J Bacteriol. 1993;175:7673–7682. doi: 10.1128/jb.175.23.7673-7682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 762.Clark D. Regulation of fatty acid degradation in Escherichia coli: analysis by operon fusion. J Bacteriol. 1981;148:521–526. doi: 10.1128/jb.148.2.521-526.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 763.Clark D P. Novel antibiotic hypersensitive mutants of Escherichia coli. Genetic mapping and chemical characterization. FEMS Microbiol Lett. 1984;21:189–195. [Google Scholar]
- 763a.Clark D P. The fermentation pathways of Escherichia coli. FEMS Microbiol Rev. 1989;63:223–224. doi: 10.1016/0168-6445(89)90033-8. [DOI] [PubMed] [Google Scholar]
- 764.Clark D P, Cronan J E., Jr Acetaldehyde coenzyme A dehydrogenase of Escherichia coli. J Bacteriol. 1980;144:179–184. doi: 10.1128/jb.144.1.179-184.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 765.Clark D P, Cronan J E., Jr Escherichia coli mutants with altered control of alcohol dehydrogenase and nitrate reductase. J Bacteriol. 1980;141:177–183. doi: 10.1128/jb.141.1.177-183.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 766.Clark D P, Beard J P. Altered phospholipid composition in mutants of Escherichia coli sensitive or resistant to organic solvents. J Gen Microbiol. 1979;113:267–274. doi: 10.1099/00221287-113-2-267. [DOI] [PubMed] [Google Scholar]
- 767.Clark D P, Rod M L. Regulatory mutations that allow the growth of Escherichia coli on butanol as carbon source. J Mol Evol. 1987;25:151–158. doi: 10.1007/BF02101757. [DOI] [PubMed] [Google Scholar]
- 768.Clark R L, Neidhardt F C. Roles of the two lysyl-tRNA synthetases of Escherichia coli: analysis of nucleotide sequences and mutant behavior. J Bacteriol. 1990;172:3237–3243. doi: 10.1128/jb.172.6.3237-3243.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 769.Clarke D J, Jacq A, Holland I B. A novel DnaJ-like protein in Escherichia coli inserts into the cytoplasmic membrane with a type III topology. Mol Microbiol. 1996;20:1273–1286. doi: 10.1111/j.1365-2958.1996.tb02646.x. [DOI] [PubMed] [Google Scholar]
- 770.Clarke D J, Holland I B, Jacq A. Point mutations in the transmembrane domain of DjlA, a membrane-linked DnaJ-like protein, abolish its function in promoting colanic acid production via the Rcs signal transduction pathway. Mol Microbiol. 1997;25:933–944. doi: 10.1111/j.1365-2958.1997.mmi528.x. [DOI] [PubMed] [Google Scholar]
- 771.Clarke D M, Bragg P D. Cloning and expression of the transhydrogenase gene of Escherichia coli. J Bacteriol. 1985;162:367–373. doi: 10.1128/jb.162.1.367-373.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 772.Clarke D M, Loo T W, Gillam S, Bragg P D. Nucleotide sequence of the pntA and pntB genes encoding the pyridine nucleotide transhydrogenase of Escherichia coli. Eur J Biochem. 1986;158:647–653. doi: 10.1111/j.1432-1033.1986.tb09802.x. [DOI] [PubMed] [Google Scholar]
- 773.Clarke N D, Kvaal M, Seeberg E. Cloning of Escherichia coli genes encoding 3-methyladenine DNA glycosylases I and II. Mol Gen Genet. 1984;197:368–372. doi: 10.1007/BF00329931. [DOI] [PubMed] [Google Scholar]
- 774.Claverie-Martin F, Diaz-Torres M R, Yancey S D, Kushner S R. Cloning of the altered mRNA stability (ams) gene of Escherichia coli K-12. J Bacteriol. 1989;171:5479–5486. doi: 10.1128/jb.171.10.5479-5486.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 775.Claverie-Martin F, Diaz-Torres M R, Kushner S R. Analysis of the regulatory region of the protease III (ptr) gene of Escherichia coli K-12. Gene. 1987;54:185–195. doi: 10.1016/0378-1119(87)90486-0. [DOI] [PubMed] [Google Scholar]
- 776.Clegg D O, Koshland D E. The role of a signaling protein in bacterial sensing: behavioral effects of increased gene expression. Proc Natl Acad Sci USA. 1984;81:5056–5060. doi: 10.1073/pnas.81.16.5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 777.Clegg D O, Koshland D E. Identification of a bacterial sensing protein and effects of its elevated expression. J Bacteriol. 1985;162:398–405. doi: 10.1128/jb.162.1.398-405.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 778.Clement J M, Hofnung M. Gene sequence of the LAM receptor, an outer membrane protein of E. coli K-12. Cell. 1981;27:507–514. doi: 10.1016/0092-8674(81)90392-5. [DOI] [PubMed] [Google Scholar]
- 779.Clementz T. The gene coding for 3-deoxy-manno-octulosonic acid transferase and the rfaQ gene are transcribed from divergently arranged promoters in Escherichia coli. J Bacteriol. 1992;174:7750–7756. doi: 10.1128/jb.174.23.7750-7756.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 780.Clementz T, Raetz C R H. A gene coding for 3-deoxy-d-manno-octulosonic-acid transferase in Escherichia coli. Identification, mapping, cloning, and sequencing. J Biol Chem. 1991;266:9687–9696. [PubMed] [Google Scholar]
- 781.Clementz T, Bednarski J J, Raetz C R. Function of the htrB high temperature requirement gene of Escherichia coli in the acylation of lipid A: HtrB catalyzed incorporation of laurate. J Biol Chem. 1996;271:12095–12102. doi: 10.1074/jbc.271.20.12095. [DOI] [PubMed] [Google Scholar]
- 782.Cleton-Jansen A M, Goosen N, Fayet O, Van de Putte P. Cloning, mapping, and sequencing of the gene encoding Escherichia coli quinoprotein glucose dehydrogenase. J Bacteriol. 1990;172:6308–6315. doi: 10.1128/jb.172.11.6308-6315.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 783.Climie S C, Friesen J D. Feedback regulation of the rplJL-rpoBC ribosomal protein operon of Escherichia coli requires a region of mRNA secondary structure. J Mol Biol. 1987;198:371–381. doi: 10.1016/0022-2836(87)90287-7. [DOI] [PubMed] [Google Scholar]
- 784.Clyman J, Cunningham R P. Escherichia coli K-12 mutants in which viability is dependent on recA function. J Bacteriol. 1987;169:4203–4210. doi: 10.1128/jb.169.9.4203-4210.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 785.Cobbett C S, Pittard J. Formation of a λ (Tn10) tyrR+ specialized transducing bacteriophage from Escherichia coli K-12. J Bacteriol. 1980;144:877–883. doi: 10.1128/jb.144.3.877-883.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 786.Coderre P E, Earhart C F. Characterization of a plasmid carrying the Escherichia coli K-12 entD, fepA, fes and entF genes. FEMS Microbiol Lett. 1984;25:111–116. [Google Scholar]
- 787.Coderre P E, Earhart C F. The entD gene of the Escherichia coli K12 enterobactin gene cluster. J Gen Microbiol. 1989;135:3043–3055. doi: 10.1099/00221287-135-11-3043. [DOI] [PubMed] [Google Scholar]
- 788.Cohen G, Jacob F. Sur la repression de la synthese des enzymes intervenant dans la formation du tryptophane chez Escherichia coli. C R Acad Sci Ser III. 1959;248:3490–3492. [PubMed] [Google Scholar]
- 789.Cohen G N, Patte J C, Truffa-Bachi P, Sawas C, Doudoroff M. Mecanismes de regulation des activities cellulaires chez les micro-organismes. Paris, France: Centre National de La Recherche Scientifique; 1965. Repression and end-product inhibition in a branched biosynthetic pathway; pp. 243–253. [Google Scholar]
- 790.Cohen S P, Hachler H, Levy S B. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993;175:1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 791.Cohen S P, McMurry L M, Levy S B. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J Bacteriol. 1988;170:5416–5422. doi: 10.1128/jb.170.12.5416-5422.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 792.Cole J A, Newman B M, White P. Biochemical and genetic characterization of nirB mutants of Escherichia coli K12 pleiotropically defective in nitrite and sulphite reduction. J Gen Microbiol. 1980;120:475–483. doi: 10.1099/00221287-120-2-475. [DOI] [PubMed] [Google Scholar]
- 793.Cole S T. Nucleotide sequence coding for the flavoprotein subunit of the fumarate reductase of Escherichia coli. Eur J Biochem. 1982;122:479–484. doi: 10.1111/j.1432-1033.1982.tb06462.x. [DOI] [PubMed] [Google Scholar]
- 794.Cole S T. Characterization of the promoter for the lexA regulated sulA gene of Escherichia coli. Mol Gen Genet. 1983;189:400–404. doi: 10.1007/BF00325901. [DOI] [PubMed] [Google Scholar]
- 795.Cole S T, Bremer E, Hindennach I, Henning U. Characterization of the promoters for the ompA gene which encodes a major outer membrane protein of Escherichia coli. Mol Gen Genet. 1982;188:472–479. doi: 10.1007/BF00330051. [DOI] [PubMed] [Google Scholar]
- 796.Cole S T, Guest J R. Production of a soluble form of fumarate reductase by multiple gene duplications in Escherichia coli K12. Eur J Biochem. 1979;102:65–71. doi: 10.1111/j.1432-1033.1979.tb06263.x. [DOI] [PubMed] [Google Scholar]
- 797.Cole S T, Guest J R. Amplification of fumarate reductase synthesis with frdA transducing phages and orientation of frdA gene expression. Mol Gen Genet. 1980;179:377–385. doi: 10.1007/BF00425468. [DOI] [PubMed] [Google Scholar]
- 798.Cole S T, Guest J R. Genetic and physical characterization of lambda transducing phages (lambda frdA) containing the fumarate reductase gene of Escherichia coli K-12. Mol Gen Genet. 1980;178:409–418. doi: 10.1007/BF00270492. [DOI] [PubMed] [Google Scholar]
- 799.Cole S T, Eiglmeier K, Ahmed S, Honore N, Elmes L, Anderson W F, Weiner J H. Nucleotide sequence and gene-polypeptide relationships of the glpABC operon encoding the anaerobic sn-glycerol-3-phosphate dehydrogenase of Escherichia coli K-12. J Bacteriol. 1988;170:2448–2456. doi: 10.1128/jb.170.6.2448-2456.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 800.Cole S T, Raibaud O. The nucleotide sequence of the malT gene encoding the positive regulator of the Escherichia coli maltose region. Gene. 1986;42:201–208. doi: 10.1016/0378-1119(86)90297-0. [DOI] [PubMed] [Google Scholar]
- 801.Cole S T, Grundstrom T, Jaurin B, Robinson J J, Weiner J H. Location and nucleotide sequence of frdB, the gene coding for the iron-sulphur protein subunit of the fumarate reductase of Escherichia coli. Eur J Biochem. 1982;126:211–216. doi: 10.1111/j.1432-1033.1982.tb06768.x. [DOI] [PubMed] [Google Scholar]
- 802.Coleman J. Characterization of Escherichia coli cells deficient in 1-acyl-sn-glycerol-3-phosphate acyltransferase activity. J Biol Chem. 1990;265:17215–17221. [PubMed] [Google Scholar]
- 803.Coleman J. Characterization of the Escherichia coli gene for 1-acyl-sn-glycerol-3-phosphate acyltransferase (plsC) Mol Gen Genet. 1992;232:295–303. doi: 10.1007/BF00280009. [DOI] [PubMed] [Google Scholar]
- 804.Coleman J, Raetz C R H. First committed step of lipid A biosynthesis in Escherichia coli: sequence of the lpxA gene. J Bacteriol. 1988;170:1268–1274. doi: 10.1128/jb.170.3.1268-1274.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 805.Coleman S H, Wild D G. Location of the fpg gene on the Escherichia coli chromosome. Nucleic Acids Res. 1991;19:3999. doi: 10.1093/nar/19.14.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 806.Coleman W G., Jr The rfaD gene codes for ADP-l-glycero-d-mannoheptose-6-epimerase. An enzyme required for lipopolysaccharide core biosynthesis. J Biol Chem. 1983;258:1985–1990. [PubMed] [Google Scholar]
- 807.Coleman W G, Jr, Deshpande K S. New cysE-pyrE-linked rfa mutation in Escherichia coli K-12 that results in a heptoseless lipopolysaccharide. J Bacteriol. 1985;161:1209–1214. doi: 10.1128/jb.161.3.1209-1214.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 808.Coleman W G, Jr, Leive L. Two mutations which affect the barrier function of the Escherichia coli K-12 outer membrane. J Bacteriol. 1979;139:899–910. doi: 10.1128/jb.139.3.899-910.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 809.Coll J L, Heyde M, Portalier R. Expression of the nmpC gene of Escherichia coli K-12 is modulated by external pH. Identification of cis-acting regulatory sequences involved in this regulation. Mol Microbiol. 1994;12:83–93. doi: 10.1111/j.1365-2958.1994.tb00997.x. [DOI] [PubMed] [Google Scholar]
- 810.Collier D N, Bassford P J. Mutations that improve export of maltose-binding protein in SecB− cells of Escherichia coli. J Bacteriol. 1989;171:4640–4647. doi: 10.1128/jb.171.9.4640-4647.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 811.Collis C M, Grigg G W. An Escherichia coli mutant resistant to phleomycin, bleomycin, and heat inactivation is defective in ubiquinone synthesis. J Bacteriol. 1989;171:4792–4798. doi: 10.1128/jb.171.9.4792-4798.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 812.Colloms S D, Sykora P, Szatmari G, Sherratt D J. Recombination at ColE2 cer requires the Escherichia coli xerC gene product, a member of the lambda integrase family of site-specific recombinases. J Bacteriol. 1990;172:6973–6980. doi: 10.1128/jb.172.12.6973-6980.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 813.Colombo G, Villafranca J J. Amino acid sequence of Escherichia coli glutamine synthetase deduced from the DNA nucleotide sequence. J Biol Chem. 1986;261:10587–10591. [PubMed] [Google Scholar]
- 814.Colonna B, Hofnung M. rho mutations restore lamB expression in E. coli K12 strains with an inactive malB region. Mol Gen Genet. 1981;184:479–483. doi: 10.1007/BF00352526. [DOI] [PubMed] [Google Scholar]
- 815.Colson C, Glover S W, Symonds N, Stacey K A. The location of the genes for host-controlled modification and restriction in Escherichia coli K-12. Genetics. 1965;52:1043–1050. doi: 10.1093/genetics/52.5.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 816.Comeau D E, Ikenaka K, Tsung K, Inoue M. Primary characterization of the protein products of the Escherichia coli ompB locus: structure and regulation of synthesis of the OmpR and EnvZ proteins. J Bacteriol. 1985;164:578–584. doi: 10.1128/jb.164.2.578-584.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 817.Comer M M. Gene organization around the phenylalanyl-transfer ribonucleic acid synthetase loci in Escherichia coli. J Bacteriol. 1981;146:269–274. doi: 10.1128/jb.146.1.269-274.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 818.Comer M M. Threonine tRNA’s and their genes in Escherichia coli. Mol Gen Genet. 1982;187:132–137. doi: 10.1007/BF00384396. [DOI] [PubMed] [Google Scholar]
- 819.Compan I, Touati D. Interaction of six global transcription regulators in expression of manganese superoxide dismutase in Escherichia coli K-12. J Bacteriol. 1993;175:1687–1696. doi: 10.1128/jb.175.6.1687-1696.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 819a.Condamine H. Sur la régulation de la production de proline chez E. coli K-12. Ann Inst Pasteur. 1971;120:126–143. [PubMed] [Google Scholar]
- 820.Conlin C A, Miller C G. Location of the prlC (opdA) gene on the physical map of Escherichia coli. J Bacteriol. 1993;175:5731–5732. doi: 10.1128/jb.175.17.5731-5732.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 821.Conlin C A, Trun N J, Silhavy T J, Miller C G. Escherichia coli prlC encodes an endopeptidase and is homologous to the Salmonella typhimurium opdA gene. J Bacteriol. 1992;174:5881–5887. doi: 10.1128/jb.174.18.5881-5887.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 822.Conlin C A, Knox T M, Miller C G. Cloning and physical map position of an alpha-aspartyl dipeptidase gene, pepE, from Escherichia coli. J Bacteriol. 1994;176:1552–1553. doi: 10.1128/jb.176.5.1552-1553.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 823.Connelly J C, Leach D R. The sbcC and sbcD genes of Escherichia coli encode a nuclease involved in palindrome inviability and genetic recombination. Genes Cells. 1996;1:285–291. doi: 10.1046/j.1365-2443.1996.23024.x. [DOI] [PubMed] [Google Scholar]
- 824.Connolly B, Parsons C A, Benson F E, Dunderdale H J, Sharples G J, Lloyd R G, West S C. Resolution of Holliday junctions in vitro requires the Escherichia coli ruvC gene product. Proc Natl Acad Sci USA. 1991;88:6063–6067. doi: 10.1073/pnas.88.14.6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 825.Connolly D M, Winkler M E. Genetic and physiological relationship among the miaA gene, 2-methylthio-N6-(Δ2-isopentenyl)-adenosine tRNA modification, and spontaneous mutagenesis in Escherichia coli K-12. J Bacteriol. 1989;171:3233–3246. doi: 10.1128/jb.171.6.3233-3246.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 826.Connolly D M, Winkler M E. Structure of Escherichia coli K-12 miaA and characterization of the mutator phenotype caused by miaA insertion mutations. J Bacteriol. 1991;173:1711–1721. doi: 10.1128/jb.173.5.1711-1721.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 827.Conrad C A, Stearns G W, Prater W E, Rheimer J A, Johnson J R. Characterization of a glpK transducing phage. Mol Gen Genet. 1984;193:376–378. doi: 10.1007/BF00330696. [DOI] [PubMed] [Google Scholar]
- 827a.Conrad J, Sun D, Englund N, Ofengand J. The rluC gene of Escherichia coli codes for a pseudouridine synthase which is solely responsible for synthesis of pseudouridine at positions 955, 2504 and 2580 in 23S ribosomal RNA. J Biol Chem. 1998;273:18562–18566. doi: 10.1074/jbc.273.29.18562. [DOI] [PubMed] [Google Scholar]
- 828.Conter A, Menchon C, Gutierrez C. Role of DNA supercoiling and rpoS sigma factor in the osmotic and growth phase-dependent induction of the gene osmE of Escherichia coli K12. J Mol Biol. 1997;273:75–83. doi: 10.1006/jmbi.1997.1308. [DOI] [PubMed] [Google Scholar]
- 828a.Conway T, Yi K C, Egan S E, Wolf R E, Jr, Rowley D L. Locations of the zwf, edd, and eda genes on an Escherichia coli physical map. J Bacteriol. 1991;173:5297–5298. doi: 10.1128/jb.173.17.5247-5248.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 829.Cooley W, Sirotkin K, Green R, Snyder L. A new gene of Escherichia coli K-12 whose product participates in T4 bacteriophage late gene expression: interaction of lit with the T4-induced polynucleotide 5′-kinase 3′-phosphatase. J Bacteriol. 1979;140:83–91. doi: 10.1128/jb.140.1.83-91.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 830.Cooper R. The utilization of d-galactonate and d-2-oxo-3-deoxygalactonate by Escherichia coli K-12. Arch Microbiol. 1978;118:199–206. doi: 10.1007/BF00415730. [DOI] [PubMed] [Google Scholar]
- 831.Cooper R A, Jones D C N, Parrott S. Isolation and mapping of Escherichia coli K12 mutants defective in phenylacetate degradation. J Gen Microbiol. 1985;131:2753–2757. doi: 10.1099/00221287-131-10-2753. [DOI] [PubMed] [Google Scholar]
- 832.Coppola G, Huang F, Riley J, Cox J L, Hantzopoulos P, Zhou L B, Calhoun D H. Sequence and transcriptional activity of the Escherichia coli K-12 chromosome region between rrnC and ilvGMEDA. Gene. 1991;97:21–27. doi: 10.1016/0378-1119(91)90005-v. [DOI] [PubMed] [Google Scholar]
- 833.Cormack R S, Genereaux J L, Mackie G A. RNase E activity is conferred by a single polypeptide: overexpression, purification, and properties of the ams/rne/hmp1 gene product. Proc Natl Acad Sci USA. 1993;90:9006–9010. doi: 10.1073/pnas.90.19.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 834.Cornell K, Riscoe M K. Cloning and expression of Escherichia coli 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase: identification of the pfs gene product. Biochim Biophys Acta. 1998;1396:8–14. doi: 10.1016/s0167-4781(97)00169-3. [DOI] [PubMed] [Google Scholar]
- 835.Cornet F, Mortier I, Patte J C, Louarn J M. Plasmid pSC101 harbors a recombination site, psi, which is able to resolve plasmid multimers and to substitute for the analogous chromosomal Escherichia coli site dif. J Bacteriol. 1994;176:3188–3195. doi: 10.1128/jb.176.11.3188-3195.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 836.Cornet F, Louarn J, Patte J, Louarn J M. Restriction of the activity of the recombination site dif to a small zone of the Escherichia coli chromosome. Genes Dev. 1996;10:1152–1161. doi: 10.1101/gad.10.9.1152. . (Comment.) [DOI] [PubMed] [Google Scholar]
- 837.Cornish E C, Davidson B E, Pittard J. Cloning and characterization of Escherichia coli K-12 regulator gene tyrR. J Bacteriol. 1982;152:1276–1279. doi: 10.1128/jb.152.3.1276-1279.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 838.Cornish E C, Argyropoulos V P, Pittard J, Davidson B E. Structure of the Escherichia coli K12 regulatory gene tyrR: nucleotide sequence and sites of initiation of transcription and translation. J Biol Chem. 1986;261:403–410. [PubMed] [Google Scholar]
- 839.Cornwell T L, Adhya S, Reznikoff W S, Frey P A. The nucleotide sequence of the galT gene of Escherichia coli. Nucleic Acids Res. 1987;15:8116. doi: 10.1093/nar/15.19.8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 840.Cortay J C, Negre D, Galinier A, Duclos B, Perriere G, Cozzone A J. Regulation of the acetate operon in Escherichia coli: purification and functional characterization of the IclR repressor. EMBO J. 1991;10:675–679. doi: 10.1002/j.1460-2075.1991.tb07996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 841.Cortay J C, Bleicher F, Duclos B, Cenatiempo Y, Gauthier C, Prato J L, Cozzone A J. Utilization of acetate in Escherichia coli: structural organization and differential expression of the ace operon. Biochimie. 1989;71:1043–1049. doi: 10.1016/0300-9084(89)90109-0. [DOI] [PubMed] [Google Scholar]
- 842.Cortay J-C, Bleicher F, Rieul C, Reeves P R, Cozzone A J, Klumpp D J. Nucleotide sequence and expression of the aceK gene coding for isocitrate dehydrogenase kinase/phosphatase in Escherichia coli. J Bacteriol. 1988;170:89–97. doi: 10.1128/jb.170.1.89-97.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 842a.Cosloy S D. d-Serine transport system in Escherichia coli K-12. J Bacteriol. 1973;114:679–684. doi: 10.1128/jb.114.2.679-684.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 843.Cosma C L, Danese P N, Carlson J H, Silhavy T J, Snyder W B. Mutational activation of the Cpx signal transduction pathway of Escherichia coli suppresses the toxicity conferred by certain envelope-associated stresses. Mol Microbiol. 1995;18:491–505. doi: 10.1111/j.1365-2958.1995.mmi_18030491.x. [DOI] [PubMed] [Google Scholar]
- 844.Cossart P, Gicquel-Sanzey B. Cloning and sequencing of the crp gene of Escherichia coli K12. Nucleic Acids Res. 1982;10:1363–1378. doi: 10.1093/nar/10.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 845.Cossart P, Katinka M, Yaniv M. Nucleotide sequence of the thrB gene of E. coli, and its two adjacent regions; the thrAB and thrBC junctions. Nucleic Acids Res. 1981;9:339–347. doi: 10.1093/nar/9.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 846.Cossart P, Katinka M, Yaniv M, Saint Girons I, Cohen G N. Construction and expression of a hybrid plasmid containing the Escherichia coli thrA and thrB genes. Mol Gen Genet. 1979;175:39–44. doi: 10.1007/BF00267853. [DOI] [PubMed] [Google Scholar]
- 847.Cotter P A, Melville S B, Albrecht J A, Gunsalus R P. Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: roles of Fnr and ArcA in repression and activation. Mol Microbiol. 1997;25:605–615. doi: 10.1046/j.1365-2958.1997.5031860.x. [DOI] [PubMed] [Google Scholar]
- 848.Coulton J W, Mason P, Allatt D D. fhuC and fhuD genes for iron (III)-ferrichrome transport into Escherichia coli K-12. J Bacteriol. 1987;169:3844–3849. doi: 10.1128/jb.169.8.3844-3849.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 849.Coulton J W, Mason P, Cameron D R, Carmel G, Jean R, Rode H N. Protein fusions of β-galactosidase to the ferrichrome-iron receptor of Escherichia coli K-12. J Bacteriol. 1986;165:181–192. doi: 10.1128/jb.165.1.181-192.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 850.Coulton J W, Mason P, DuBow M S. Molecular cloning of the ferrichrome-iron receptor of Escherichia coli K-12. J Bacteriol. 1983;156:1315–1321. doi: 10.1128/jb.156.3.1315-1321.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 851.Court D L, Patterson T A, Baker T, Costantino N, Mao X, Friedman D I. Structural and functional analyses of the transcription-translation proteins NusB and NusE. J Bacteriol. 1995;177:2589–2591. doi: 10.1128/jb.177.9.2589-2591.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 852.Covarrubias A, Bastarrachea F. Nucleotide sequence of the glnA control region of Escherichia coli. Mol Gen Genet. 1983;190:171–175. doi: 10.1007/BF00330342. [DOI] [PubMed] [Google Scholar]
- 853.Covarrubias A, Sanchez-Pescador R, Osorio A, Bolivar F, Bastarrachea F. ColE1 hybrid plasmids containing Escherichia coli genes involved in the biosynthesis of glutamate and glutamine. Plasmid. 1980;3:150–164. doi: 10.1016/0147-619x(80)90106-7. [DOI] [PubMed] [Google Scholar]
- 854.Cowman A, Beacham I R. Molecular cloning of the gene (ush) from Escherichia coli specifying periplasmic UDP-sugar hydrolase (5′-nucleotidase) Gene. 1980;12:281–286. doi: 10.1016/0378-1119(80)90111-0. [DOI] [PubMed] [Google Scholar]
- 855.Cox E C, Horner D L. DNA sequence and coding properties of mutD (dnaQ) a dominant Escherichia coli mutator gene. J Mol Biol. 1986;190:113–117. doi: 10.1016/0022-2836(86)90080-x. [DOI] [PubMed] [Google Scholar]
- 856.Cox E C, Gibson T. Selection for high mutation rates in chemostats. Genetics. 1974;77:169–184. doi: 10.1093/genetics/77.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 857.Cox G B, Rosenberg H, Downie J A, Silver S. Genetic analysis of mutants affected in the Pst inorganic phosphate transport system. J Bacteriol. 1981;148:1–9. doi: 10.1128/jb.148.1.1-9.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 858.Cox G B, Young I G, McCann L M, Gibson F. Biosynthesis of ubiquinone in Escherichia coli K-12; location of genes affecting the metabolism of 3-octaprenyl-4-hydroxybenzoic acid and 2-octaprenyl-phenol. J Bacteriol. 1969;99:450–458. doi: 10.1128/jb.99.2.450-458.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 859.Cox J C. Escherichia coli formate dehydrogenase mutants with altered selenopolymer profiles. Arch Microbiol. 1989;152:397–400. doi: 10.1007/BF00425180. [DOI] [PubMed] [Google Scholar]
- 860.Cox J L, Cox B J, Fidanza V, Calhoun D H. The complete nucleotide sequence of the ilvGMEDA cluster of Escherichia coli K-12. Gene. 1987;56:185–198. doi: 10.1016/0378-1119(87)90136-3. [DOI] [PubMed] [Google Scholar]
- 861.Crabeel M, Charlier D, Weyens G, Feller A, Pierard A, Glansdorff N. Use of gene cloning to determine polarity of an operon: genes carAB of Escherichia coli. J Bacteriol. 1980;143:921–925. doi: 10.1128/jb.143.2.921-925.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 862.Crabeel M, Charlier D, Cunin R, Bogen A, Glansdorff N, Pierard A. Accumulation of arginine precursors in Escherichia coli: effects on growth, enzyme repression, and application to the forward selection of arginine auxotrophs. J Bacteriol. 1975;123:898–904. doi: 10.1128/jb.123.3.898-904.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 863.Crabeel M, Charlier D, Cunin R, Glansdorff N. Cloning and endonuclease restriction analysis of argF and of the control region of the argECBH biopolar operon in Escherichia coli. Gene. 1979;5:207–231. doi: 10.1016/0378-1119(79)90079-9. [DOI] [PubMed] [Google Scholar]
- 864.Craig J E, Zhang Y, Gallagher M P. Cloning of the nupC gene of Escherichia coli encoding a nucleoside transport system, and identification of an adjacent insertion element, IS 186. Mol Microbiol. 1994;11:1159–1168. doi: 10.1111/j.1365-2958.1994.tb00392.x. [DOI] [PubMed] [Google Scholar]
- 865.Crawford I P, Nichols B, Yanofsky C. Nucleotide sequence of the trpB gene in Escherichia coli and Salmonella typhimurium. J Mol Biol. 1980;142:489–502. doi: 10.1016/0022-2836(80)90259-4. [DOI] [PubMed] [Google Scholar]
- 866.Creaghan I T, Guest J R. Suppression of the succinate requirement of lipoamide dehydrogenase mutants of Escherichia coli by mutations affecting succinate dehydrogenase activity. J Gen Microbiol. 1977;102:183–194. doi: 10.1099/00221287-102-1-183. [DOI] [PubMed] [Google Scholar]
- 867.Creeger E S, Schulte T, Rothfield L I. Regulation of membrane glycosyltransferases by the sfrB and rfaH genes of Escherichia coli and Salmonella typhimurium. J Biol Chem. 1984;259:3064–3069. [PubMed] [Google Scholar]
- 868.Crofton S, Dennis P P. Cloning and orientation of the gene encoding polynucleotide phosphorylase in Escherichia coli. J Bacteriol. 1983;156:479. doi: 10.1128/jb.154.1.58-64.1983. . (Author’s correction.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 869.Crofton S, Dennis P P. Cloning and orientation of the gene encoding polynucleotide phosphorylase in Escherichia coli. J Bacteriol. 1983;154:58–64. doi: 10.1128/jb.154.1.58-64.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 870.Cronan J E, Jr, Gelmann E P. An estimate of the minimum amount of unsaturated fatty acid required for growth of Escherichia coli. J Biol Chem. 1973;248:1188–1195. [PubMed] [Google Scholar]
- 871.Cronan J E, Jr, Littel K J, Jackowski S. Genetic and biochemical analyses of pantothenate biosynthesis in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1982;149:916–922. doi: 10.1128/jb.149.3.916-922.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 872.Cronan J E, Jr, Li W-B, Coleman R, Narasimhan M, de Mendoza D, Schwab J M. Derived amino acid sequence and identification of active site residues of Escherichia coli β-hydroxydecanoyl thioester dehydrase. J Biol Chem. 1988;263:4641–4646. [PubMed] [Google Scholar]
- 873.Crooke E, Akiyama M, Rao N N, Kornberg A. Genetically altered levels of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1994;269:6290–6295. [PubMed] [Google Scholar]
- 874.Crooke H, Cole J. The biogenesis of c-type cytochromes in Escherichia coli requires a membrane-bound protein, DipZ, with a protein disulphide isomerase-like domain. Mol Microbiol. 1995;15:1139–1150. doi: 10.1111/j.1365-2958.1995.tb02287.x. [DOI] [PubMed] [Google Scholar]
- 875.Crooke H, Cole J A. The biogenesis of c-type cytochromes in Escherichia coli requires an integral membrane protein with a protein disulphide isomerase-like domain. 1994. GenBank submission X77707. [DOI] [PubMed] [Google Scholar]
- 876.Crowell D N, Anderson M S, Raetz C R H. Molecular cloning of the genes for lipid A disaccharide synthase and UDP-N-acetylglucosamine acyltransferase in Escherichia coli. J Bacteriol. 1986;168:152–159. doi: 10.1128/jb.168.1.152-159.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 877.Crowell D N, Reznikoff W S, Raetz C R H. Nucleotide sequence of the Escherichia coli gene for lipid A disaccharide synthase. J Bacteriol. 1987;169:5727–5734. doi: 10.1128/jb.169.12.5727-5734.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 878.Csonka L N. Proline over-production results in enhanced osmotolerance in Salmonella typhimurium. Mol Gen Genet. 1981;182:82–86. doi: 10.1007/BF00422771. [DOI] [PubMed] [Google Scholar]
- 879.Csordas-Toth E, Boros I, Venetianer P. Structure of the promoter region for the rrnB gene in Escherichia coli. Nucleic Acids Res. 1979;7:2189–2197. doi: 10.1093/nar/7.8.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 880.Cudny H J, Lupski J R, Godson G N, Deutscher M P. Cloning, sequencing, and species relatedness of the Escherichia coli cca gene encoding the enzyme tRNA nucleotidyltransferase. J Biol Chem. 1986;261:6444–6449. [PubMed] [Google Scholar]
- 881.Cui J, Somerville R L. Physical map location and transcriptional orientation of the tyrR gene of Escherichia coli K-12. J Bacteriol. 1992;174:3832–3833. doi: 10.1128/jb.174.11.3832-3833.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 882.Cui J, Somerville R L. A mutational analysis of the structural basis for transcriptional activation and monomer-monomer interaction in the TyrR system of Escherichia coli. J Bacteriol. 1993;175:1777–1784. doi: 10.1128/jb.175.6.1777-1784.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 883.Cui Y, Wang Q, Stormo G D, Calvo J M. A consensus sequence for binding of Lrp to DNA. J Bacteriol. 1995;177:4872–4880. doi: 10.1128/jb.177.17.4872-4880.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 884.Culham D E, Lasby B, Marangoni A G, Milner J L, Steer B A, van Nues R W, Wood J M. Isolation and sequencing of Escherichia coli gene proP reveals unusual structural features of the osmoregulatory proline/betaine transporter, ProP. J Mol Biol. 1993;229:268–276. doi: 10.1006/jmbi.1993.1030. [DOI] [PubMed] [Google Scholar]
- 885.Cummings H S, Sands J F, Foreman P C, Fraser J, Hershey J W. Structure and expression of the infA operon encoding translational initiation factor IF1. Transcriptional control by growth rate. J Biol Chem. 1991;266:16491–16498. [PubMed] [Google Scholar]
- 886.Cummings H S, Hershey J W B. Translation initiation factor IF1 is essential for cell viability in Escherichia coli. J Bacteriol. 1994;176:198–205. doi: 10.1128/jb.176.1.198-205.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 887.Cunningham P R, Clark D P. The use of suicide substrates to select mutants of Escherichia coli lacking enzymes of alcohol fermentation. Mol Gen Genet. 1986;205:487–493. doi: 10.1007/BF00338087. [DOI] [PubMed] [Google Scholar]
- 888.Cunningham R P, Weiss B. Endonuclease III (nth) mutants of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:474–478. doi: 10.1073/pnas.82.2.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 889.Cunningham R P, Saporito S M, Spitzer S G, Weiss B. Endonuclease IV (nfo) mutant of Escherichia coli. J Bacteriol. 1986;168:1120–1127. doi: 10.1128/jb.168.3.1120-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 890.Curtis S J, Epstein W. Phosphorylation of d-glucose in Escherichia coli mutants defective in glucosephosphotransferase, mannosephosphotransferase, and glucokinase. J Bacteriol. 1975;122:1189–1199. doi: 10.1128/jb.122.3.1189-1199.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 891.Dabbs E R. Kasugamycin-dependent mutants of Escherichia coli. J Bacteriol. 1978;136:994–1001. doi: 10.1128/jb.136.3.994-1001.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 892.Dabbs E R. The gene for ribosomal protein S21, rpsU, maps close to dnaG at 66.5 min on the Escherichia coli chromosomal linkage map. J Bacteriol. 1980;144:603–607. doi: 10.1128/jb.144.2.603-607.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 893.Dabbs E R. The gene for ribosomal protein L31, rpmE, is located at 88.5 minutes on the Escherichia coli chromosomal linkage map. J Bacteriol. 1981;148:379–382. doi: 10.1128/jb.148.1.379-382.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 894.Dabbs E R. The gene for ribosomal protein L13, rplM, is located near argR, at about 70 minutes on the Escherichia coli chromosomal linkage map. J Bacteriol. 1982;149:779–782. doi: 10.1128/jb.149.2.779-782.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 895.Dabbs E R. Three additional loci of rifampicin dependence in Escherichia coli. Mol Gen Genet. 1982;187:519–522. doi: 10.1007/BF00332638. [DOI] [PubMed] [Google Scholar]
- 896.Dabbs E R. A spontaneous mutant of Escherichia coli with protein L24 lacking from the ribosome. Mol Gen Genet. 1983;187:453–458. doi: 10.1007/BF00332627. [DOI] [PubMed] [Google Scholar]
- 897.Dabbs E R. Escherichia coli kasugamycin dependence arising from mutation at the rpsI locus. J Bacteriol. 1983;153:709–715. doi: 10.1128/jb.153.2.709-715.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 898.Dabbs E R, Looman K. An antibiotic dependent conditional lethal mutant with a lesion affecting transcription and translation. Mol Gen Genet. 1981;184:224–229. doi: 10.1007/BF00272909. [DOI] [PubMed] [Google Scholar]
- 899.Dahlquist K, Puglisi J D. Investigating the structure and function of translation initiation factor 1 in Escherichia coli. Nucleic Acids Symp Ser. 1995;114:170–171. [PubMed] [Google Scholar]
- 900.Dai D, Ishiguro E E. murH, a new genetic locus in Escherichia coli involved in cell wall peptidoglycan biosynthesis. J Bacteriol. 1988;170:2197–2201. doi: 10.1128/jb.170.5.2197-2201.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 901.Dai K, Mukherjee A, Xu Y, Lutkenhaus J. Mutations in ftsZ that confer resistance to SulA affect the interaction of FtsZ with GTP. J Bacteriol. 1994;176:130–136. doi: 10.1128/jb.176.1.130-136.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 902.Dai K, Lutkenhaus J. ftsZ is an essential cell division gene in Escherichia coli. J Bacteriol. 1991;173:3500–3506. doi: 10.1128/jb.173.11.3500-3506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 903.Dai K, Xu Y, Lutkenhaus J. Cloning and characterization of ftsN, an essential cell division gene in Escherichia coli isolated as a multicopy suppressor of ftsA12(Ts) J Bacteriol. 1993;175:3790–3797. doi: 10.1128/jb.175.12.3790-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 904.Dailey F E, Berg H C. Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc Natl Acad Sci USA. 1993;90:1043–1047. doi: 10.1073/pnas.90.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 905.Dairi T, Inokuchi K, Mizuno T, Mizushima H. Positive control of transcription initiation in Escherichia coli. A base substitution at the Pribnow box renders ompF expression independent of a positive regulator. J Mol Biol. 1985;184:1–6. doi: 10.1016/0022-2836(85)90038-5. [DOI] [PubMed] [Google Scholar]
- 906.Dalbey R E. Leader peptidase. Mol Microbiol. 1991;5:2855–2860. doi: 10.1111/j.1365-2958.1991.tb01844.x. [DOI] [PubMed] [Google Scholar]
- 907.Daldal F. Molecular cloning of the gene for phosphofructokinase-2 of Escherichia coli and the nature of a mutation, pfkB1, causing a high level of the enzyme. J Mol Biol. 1983;168:285–305. doi: 10.1016/s0022-2836(83)80019-9. [DOI] [PubMed] [Google Scholar]
- 908.Daldal F. Nucleotide sequence of gene pfkB encoding the minor phosphofructokinase of Escherichia coli K-12. Gene. 1984;28:337–342. doi: 10.1016/0378-1119(84)90151-3. [DOI] [PubMed] [Google Scholar]
- 909.Daldal F, Fraenkel D. Tn10 insertions in the pfkB region of Escherichia coli. J Bacteriol. 1981;147:935–943. doi: 10.1128/jb.147.3.935-943.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 910.Dallas W S, Dev I K, Ray P H. The dihydropteroate synthase gene, folP, is near the leucine tRNA gene, leuU, on the Escherichia coli chromosome. J Bacteriol. 1993;175:7743–7744. doi: 10.1128/jb.175.23.7743-7744.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 911.Dallas W S, Gowen J E, Ray P H, Cox M J, Dev I K. Cloning, sequencing, and enhanced expression of the dihydropteroate synthase gene of Escherichia coli MC4100. J Bacteriol. 1992;174:5961–5970. doi: 10.1128/jb.174.18.5961-5970.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 912.Dallmann H G, McHenry C S. DnaX complex of Escherichia coli DNA polymerase III holoenzyme. Physical characterization of the DnaX subunits and complexes. J Biol Chem. 1995;270:29563–29569. [PubMed] [Google Scholar]
- 913.Dallmann H G, Thimmig R L, McHenry C S. DnaX complex of Escherichia coli DNA polymerase III holoenzyme. Central role of tau in initiation complex assembly and in determining the functional asymmetry of holoenzyme. J Biol Chem. 1995;270:29555–29562. [PubMed] [Google Scholar]
- 914.Dalrymple B, Mattick J S. Genes encoding threonine tRNA’s with the anticodon CGU from Escherichia coli and Pseudomonas aeruginosa. Biochem Int. 1986;13:547–553. [PubMed] [Google Scholar]
- 915.Dam M, Douthwaite S, Tenson T, Mankin A S. Mutations in domain II of 23 S rRNA facilitate translation of a 23 S rRNA-encoded pentapeptide conferring erythromycin resistance. J Mol Biol. 1996;259:1–6. doi: 10.1006/jmbi.1996.0296. [DOI] [PubMed] [Google Scholar]
- 916.Dame J B, Shapiro B M. Use of polymyxin G, levallorphan, and tetracaine to isolate novel mutants of Escherichia coli. J Bacteriol. 1976;127:961–972. doi: 10.1128/jb.127.2.961-972.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 917.Dammel C S, Noller H F. Suppression of a cold-sensitive mutation in 16S rRNA by overexpression of a novel ribosome-binding factor, RbfA. Genes Dev. 1995;9:626–637. doi: 10.1101/gad.9.5.626. [DOI] [PubMed] [Google Scholar]
- 918.Danchin A. A new technique for selection of sensitive and auxotrophic mutants of E. coli: isolation of a strain sensitive to an excess of one-carbon metabolites. Mol Gen Genet. 1977;150:293–299. doi: 10.1007/BF00268128. [DOI] [PubMed] [Google Scholar]
- 919.Danchin A, Dondon J. Serine sensitivity of Escherichia coli K12: partial characterization of a serine resistant mutant that is extremely sensitive to 2-ketobutyrate. Mol Gen Genet. 1980;178:155–164. doi: 10.1007/BF00267224. [DOI] [PubMed] [Google Scholar]
- 920.Dandanell G, Hammer K. Two operator sites separated by 599 base pairs are required for deoR repression of the deo operon of Escherichia coli. EMBO J. 1985;4:3333–3338. doi: 10.1002/j.1460-2075.1985.tb04085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 921.Danese P N, Murphy C K, Silhavy T J. Multicopy suppression of cold-sensitive sec mutations in Escherichia coli. J Bacteriol. 1995;177:4969–4973. doi: 10.1128/jb.177.17.4969-4973.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 922.Danese P N, Silhavy T J. CpxP, a stress-combative member of the Cpx regulon. J Bacteriol. 1998;180:831–839. doi: 10.1128/jb.180.4.831-839.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 923.Dang C V, Niwano M, Ryu J, Taylor B L. Inversion of aerotactic response in Escherichia coli deficient in cheB protein methylesterase. J Bacteriol. 1986;166:275–280. doi: 10.1128/jb.166.1.275-280.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 924.Daniel J, Danchin A. Involvement of cyclic AMP and its receptor protein in the sensitivity of Escherichia coli K12 toward serine. Mol Gen Genet. 1979;176:343–350. doi: 10.1007/BF00333096. [DOI] [PubMed] [Google Scholar]
- 925.Daniel J, Saint-Girons I. Attenuation in the threonine operon: effects of amino acids present in the presumed leader peptide in addition to threonine and isoleucine. Mol Gen Genet. 1982;188:225–227. doi: 10.1007/BF00332679. [DOI] [PubMed] [Google Scholar]
- 926.Daniels D L, Plunkett G, Burland V D, Blattner F R. Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science. 1992;257:771–778. doi: 10.1126/science.1379743. [DOI] [PubMed] [Google Scholar]
- 927.Danielsen S, Kilstrup M, Barilla K, Jochimsen B, Neuhard J. Characterization of the Escherichia coli codBA operon encoding cytosine permease and cytosine deaminase. Mol Microbiol. 1992;6:1335–1344. doi: 10.1111/j.1365-2958.1992.tb00854.x. [DOI] [PubMed] [Google Scholar]
- 928.Danot O, Vidal-Ingigliardi D, Raibaud O. Two amino acid residues from the DNA-binding domain of MalT play a crucial role in transcriptional activation. J Mol Biol. 1996;262:1–11. doi: 10.1006/jmbi.1996.0493. [DOI] [PubMed] [Google Scholar]
- 929.Dardel F, Fayat G, Blanquet S. Molecular cloning and primary structure of the Escherichia coli methionyl-tRNA synthetase gene. J Bacteriol. 1984;160:1115–1122. doi: 10.1128/jb.160.3.1115-1122.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 930.Dardel F, Panvert M, Blanquet S, Fayat G. Locations of the metG and mrp genes on the physical map of Escherichia coli. J Bacteriol. 1991;173:3273. doi: 10.1128/jb.173.11.3273.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 931.Dardonville B, Raibaud O. Characterization of malT mutants that constitutively activate the maltose regulon of Escherichia coli. J Bacteriol. 1990;172:1846–1852. doi: 10.1128/jb.172.4.1846-1852.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 932.D’Ari R, Rabinowitz J C. Purification, characterization, cloning, and amino acid sequence of the bifunctional enzyme 5,10-methylenetetrahydrofolate dehydrogenase/5,10-methenyltetrahydrofolate cyclohydrolase from Escherichia coli. J Biol Chem. 1991;266:23953–23958. [PubMed] [Google Scholar]
- 933.D’Ari R, Huisman O. Novel mechanism of cell division inhibition associated with the SOS response in Escherichia coli. J Bacteriol. 1983;156:243–250. doi: 10.1128/jb.156.1.243-250.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 934.Darlison M G, Guest J R. Nucleotide sequence encoding the iron-sulfur protein subunit of the succinate dehydrogenase of Escherichia coli. Biochem J. 1984;223:507–517. doi: 10.1042/bj2230507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 935.Darlison M G, Spencer M E, Guest J R. Nucleotide sequence of the sucA gene encoding the 2-oxoglutarate dehydrogenase of Escherichia coli K12. Eur J Biochem. 1984;141:351–359. doi: 10.1111/j.1432-1033.1984.tb08199.x. [DOI] [PubMed] [Google Scholar]
- 936.Daruwala R, Bhattacharyya D K, Kwon O, Meganathan R. Menaquinone (vitamin K2) biosynthesis: overexpression, purification, and characterization of a new isochorismate synthase from Escherichia coli. J Bacteriol. 1997;179:3133–3138. doi: 10.1128/jb.179.10.3133-3138.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 937.Darwin A, Hussain H, Griffiths L, Grove J, Sambongi Y, Busby S J, Cole J A. Regulation and sequence of the structural gene for cytochrome c552 from Escherichia coli: not a hexahaem but a 50 kDa tetrahaem nitrite reductase. Mol Microbiol. 1993;9:1255–1265. doi: 10.1111/j.1365-2958.1993.tb01255.x. [DOI] [PubMed] [Google Scholar]
- 938.Darwin A, Tormay P, Page L, Griffiths L, Cole J A. Identification of the formate dehydrogenases and genetic determinants of formate-dependent nitrite reduction by Escherichia coli K12. J Gen Microbiol. 1993;139:1829–1840. doi: 10.1099/00221287-139-8-1829. [DOI] [PubMed] [Google Scholar]
- 939.Das A. How the phage lambda N gene product suppresses transcription termination: communication of RNA polymerase with regulatory proteins mediated by signals in nascent RNA. J Bacteriol. 1992;174:6711–6716. doi: 10.1128/jb.174.21.6711-6716.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 940.Das A, Court D L, Adhya S. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc Natl Acad Sci USA. 1976;73:1959–1963. doi: 10.1073/pnas.73.6.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 941.Dasgupta S, Bernander R, Nordstrom K. In vivo effect of the tus mutation on cell division in an Escherichia coli strain where chromosome replication is under the control of plasmid R1. Res Microbiol. 1991;142:177–180. doi: 10.1016/0923-2508(91)90027-8. [DOI] [PubMed] [Google Scholar]
- 942.Dass S B, Jayaraman R. Intragenic suppression of the temperature sensitivity caused by a mutation in a gene controlling transcription (fit) in Escherichia coli. Mol Gen Genet. 1985;198:299–303. doi: 10.1007/BF00383010. [DOI] [PubMed] [Google Scholar]
- 943.Dassa E, Hofnung M. Sequence of gene malG in E. coli K12: homologies between integral membrane components from binding protein-dependent transport systems. EMBO J. 1985;4:2287–2293. doi: 10.1002/j.1460-2075.1985.tb03928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 944.Dassa E, Boquet P L. expA: a conditional mutation affecting the expression of a group of exported proteins in Escherichia coli K-12. Mol Gen Genet. 1981;181:192–200. doi: 10.1007/BF00268426. [DOI] [PubMed] [Google Scholar]
- 945.Dassa E, Boquet P L. Identification of the gene appA for the acid phosphatase (pH optimum 2.5) of Escherichia coli. Mol Gen Genet. 1985;200:68–73. doi: 10.1007/BF00383314. [DOI] [PubMed] [Google Scholar]
- 946.Dassa J, Marck C, Boquet P L. The complete nucleotide sequence of the Escherichia coli gene appA reveals significant homology between pH 2.5 acid phosphatase and glucose-1-phosphatase. J Bacteriol. 1990;172:5497–5500. doi: 10.1128/jb.172.9.5497-5500.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 947.Dassa J, Fsihi H, Marck C, Dion M, Kieffer-Bontemps M, Boquet P L. A new oxygen-regulated operon in Escherichia coli comprises the genes for a putative third cytochrome oxidase and for pH 2.5 acid phosphatase (appA) Mol Gen Genet. 1991;229:341–352. doi: 10.1007/BF00267454. [DOI] [PubMed] [Google Scholar]
- 948.Dassain M, Bouché J P. The min locus, which confers topological specificity to cell division, is not involved in its coupling with nucleoid separation. J Bacteriol. 1994;176:6143–6145. doi: 10.1128/jb.176.19.6143-6145.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 949.Datta D B, Arden B, Henning U. Major proteins of the Escherichia coli outer cell envelope membrane as bacteriophage receptors. J Bacteriol. 1977;131:821–829. doi: 10.1128/jb.131.3.821-829.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 950.Datta P, Gross T J, Omnass J R, Patil R V. Covalent structure of biodegradative threonine dehydratase of Escherichia coli: homology with other dehydratases. Proc Natl Acad Sci USA. 1987;84:393–397. doi: 10.1073/pnas.84.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 951.Dattananda C S, Gowrishankar J. Osmoregulation in Escherichia coli: complementation analysis and gene-protein relationships in the proU locus. J Bacteriol. 1989;171:1915–1922. doi: 10.1128/jb.171.4.1915-1922.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 952.Daub E, Kinach R, Miedema D, Barnard J F J, Clugston S L, Honek J F. Escherichia coli glyoxalase I. 1997. GenBank submission U57363. [DOI] [PubMed] [Google Scholar]
- 953.Dautry-Varsat A, Sibilli-Weil A L, Cohen G N. Subunit structure of the methionine-repressible aspartokinase II-homoserine dehydrogenase II from Escherichia coli K12. Eur J Biochem. 1977;76:1–6. doi: 10.1111/j.1432-1033.1977.tb11563.x. [DOI] [PubMed] [Google Scholar]
- 954.Davie E, Sydnor K, Rothfield L I. Genetic basis of minicell formation in Escherichia coli K-12. J Bacteriol. 1984;158:1202–1203. doi: 10.1128/jb.158.3.1202-1203.1984. . (Erratum, 160:831.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 955.Davies I J, Drabble W T. Stringent and growth-rate-dependent control of the gua operon of Escherichia coli K-12. Microbiology. 1996;142:2429–2437. doi: 10.1099/00221287-142-9-2429. [DOI] [PubMed] [Google Scholar]
- 956.Davies J E, Anderson P, Davis B D. Inhibition of protein synthesis by spectinomycin. Science. 1965;149:1096–1098. doi: 10.1126/science.149.3688.1096. [DOI] [PubMed] [Google Scholar]
- 957.Davies J K, Reeves P R. Genetics of resistance to colicins in Escherichia coli K-12: cross-resistance among colicins of group A. J Bacteriol. 1975;123:102–117. doi: 10.1128/jb.123.1.102-117.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 958.Davies W D, Davidson B E. The nucleotide sequence of aroG, the gene for 3-deoxy-d-arabinoheptulosonate-7-phosphate synthetase (phe) in Escherichia coli K-12. Nucleic Acids Res. 1982;10:4045–4048. doi: 10.1093/nar/10.13.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 959.Davies W D, Pittard J, Davidson B E. Cloning of aroG, the gene coding for phospho-2-keto-3-deoxy-heptonate aldolase (phe), in Escherichia coli K-12, and subcloning of the aroG promoter and operator in a promoter-detecting plasmid. Gene. 1985;33:323–331. doi: 10.1016/0378-1119(85)90240-9. [DOI] [PubMed] [Google Scholar]
- 960.Davis E J, Blatt J M, Henderson E K, Whittaker J J, Jackson J H. Valine-sensitive acetohydroxy acid synthetases in Escherichia coli K-12: unique regulation modulated by multiple genetic sites. Mol Gen Genet. 1977;156:239–249. doi: 10.1007/BF00267178. [DOI] [PubMed] [Google Scholar]
- 961.Davis E O, Jones-Mortimer M C, Henderson P J F. Location of a structural gene for xylose-H+ symport at 91 min. on the linkage map of Escherichia coli. J Biol Chem. 1984;259:1520–1525. [PubMed] [Google Scholar]
- 962.Davis E O, Henderson P J F. The cloning and DNA sequence of the gene xylE for xylose-proton symport in Escherichia coli K12. J Biol Chem. 1987;262:13928–13932. [PubMed] [Google Scholar]
- 963.Davis M G, Calvo J M. Isolation and characterization of λ pleu bacteriophages. J Bacteriol. 1977;129:1078–1090. doi: 10.1128/jb.129.2.1078-1090.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 964.Davis N K, Greer S, Jones-Mortimer M C, Perham R N. Isolation and mapping of glutathione reductase-negative mutants in Escherichia coli K-12. J Gen Microbiol. 1982;128:1631–1634. doi: 10.1099/00221287-128-7-1631. [DOI] [PubMed] [Google Scholar]
- 965.Davis T, Yamada M, Elgort M, Saier M H. Nucleotide sequence of the mannitol (mtl) operon in Escherichia coli. Mol Microbiol. 1988;2:405–412. doi: 10.1111/j.1365-2958.1988.tb00045.x. [DOI] [PubMed] [Google Scholar]
- 966.Daws T D, Lim C J, Fuchs J A. In vitro construction of gshB::kan in Escherichia coli and use of gshB::kan in mapping the gshB locus. J Bacteriol. 1989;171:5218–5221. doi: 10.1128/jb.171.9.5218-5221.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 967.Daws T D, Fuchs J A. Isolation and characterization of an Escherichia coli mutant deficient in dTMP kinase activity. J Bacteriol. 1984;157:440–444. doi: 10.1128/jb.157.2.440-444.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 968.De Biase D, Simmaco M, Sweeney G, Barra D, Bossa F, John R A. Isolation and cloning of the gene from Escherichia coli encoding glutamate decarboxylase. Biotechnol Appl Biochem. 1993;18:139–142. [PubMed] [Google Scholar]
- 969.de Boer P A, Crossley R E, Hand A R, Rothfield L I. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 1991;10:4371–4380. doi: 10.1002/j.1460-2075.1991.tb05015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 970.de Boer P A, Crossley R E, Rothfield L I. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 971.de Boer P A, Crossley R E, Rothfield L I. Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc Natl Acad Sci USA. 1990;87:1129–1133. doi: 10.1073/pnas.87.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 971a.de Boer P A, Crossley R E, Rothfield L I. Isolation and properties of minB, a complex genetic locus involved in correct placement of the division site in Escherichia coli. J Bacteriol. 1988;170:2106–2112. doi: 10.1128/jb.170.5.2106-2112.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 972.de Crouy-Chanel A, Kohiyama M, Richarme G. A novel function of Escherichia coli chaperone DnaJ. Protein-disulfide isomerase. J Biol Chem. 1995;270:22669–22672. doi: 10.1074/jbc.270.39.22669. [DOI] [PubMed] [Google Scholar]
- 973.de Geus P, Verheij H M, Riegman N H, Hoekstra W P M, de Haas G. The pro- and mature forms of the E. coli K-12 outer membrane phospholipase A are identical. EMBO J. 1984;3:1799–1802. doi: 10.1002/j.1460-2075.1984.tb02048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 974.de Geus P, van Die I, Bergmans H, Tommassen J, de Haas G. Molecular cloning of pldA, the structural gene for outer membrane phospholipase of E. coli K12. Mol Gen Genet. 1983;190:150–155. doi: 10.1007/BF00330338. [DOI] [PubMed] [Google Scholar]
- 975.de la Campa A G, Aldea M, Hernandez-Chico C, Tormo A, Vicente M. Segregation of elongation potential in Escherichia coli mediated by the wee genetic system. Curr Microbiol. 1988;17:315–319. [Google Scholar]
- 976.De La Vega F M, Galindo J M, Old I G, Guarneros G. Microbial genes homologous to the peptidyl-tRNA hydrolase-encoding gene of Escherichia coli. Gene. 1996;169:97–100. doi: 10.1016/0378-1119(95)00823-3. [DOI] [PubMed] [Google Scholar]
- 977.De Las Penas A, Connolly L, Gross C A. The ςE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of ςE. Mol Microbiol. 1997;24:373–385. doi: 10.1046/j.1365-2958.1997.3611718.x. [DOI] [PubMed] [Google Scholar]
- 978.De Lorenzo V, Herrero M, Giovannini F, Neilands J B. Fur (ferric uptake regulation) protein and CAP (catabolite-activator protein) modulate transcription of fur gene in Escherichia coli. Eur J Biochem. 1988;173:537–546. doi: 10.1111/j.1432-1033.1988.tb14032.x. [DOI] [PubMed] [Google Scholar]
- 979.de Massy B, Bejar S, Louarn J, Bouché J P. Inhibition of replication forks exiting the terminus region of the Escherichia coli chromosome occurs at two loci separated by 5 min. Proc Natl Acad Sci USA. 1987;84:1759–1763. doi: 10.1073/pnas.84.7.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 980.de Rekarte U D, Cortes M, Porco A, Nino G, Isturiz T. Mutations affecting gluconate catabolism in Escherichia coli. Genetic mapping of loci for the low affinity transport and the thermoresistant gluconokinase. J Basic Microbiol. 1994;34:363–370. doi: 10.1002/jobm.3620340602. [DOI] [PubMed] [Google Scholar]
- 981.De Rekarte U D, Isturiz T. A mutation affecting gluconate catabolism in Escherichia coli: the locus for the main high affinity transport. Acta Cient Venez. 1994;45:96–101. [PubMed] [Google Scholar]
- 982.De Reuse H, Danchin A. The ptsH, ptsI, and crr genes of the Escherichia coli phosphoenolpyruvate-dependent phosphotransferase system: a complex operon with several modes of transcription. J Bacteriol. 1988;170:3827–3837. doi: 10.1128/jb.170.9.3827-3837.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 983.De Reuse H, Kolb A, Danchin A. Positive regulation of the expression of the Escherichia coli pts operon. Identification of the regulatory regions. J Mol Biol. 1992;226:623–635. doi: 10.1016/0022-2836(92)90620-y. [DOI] [PubMed] [Google Scholar]
- 983a.De Reuse H, Roy A M, Danchin A. Analysis of the ptsH- ptsI- crr region in Escherichia coli K-12: nucleotide sequence of the ptsH gene. Gene. 1985;35:199–207. doi: 10.1016/0378-1119(85)90172-6. [DOI] [PubMed] [Google Scholar]
- 983b.De Reuse H, Hunter E, Danchin A. Analysis of the ptsH- ptsI- crr region in Escherichia coli K-12: evidence for the existence of a single transcriptional unit. Gene. 1984;32:31–40. doi: 10.1016/0378-1119(84)90029-5. [DOI] [PubMed] [Google Scholar]
- 984.Dean D O, James R. Identification of a gene, closely linked to dnaK, which is required for high-temperature growth of Escherichia coli. J Gen Microbiol. 1991;137:1271–1277. doi: 10.1099/00221287-137-6-1271. [DOI] [PubMed] [Google Scholar]
- 985.Dean G E, Macnab R M, Stader J, Matsumura P, Burks C. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J Bacteriol. 1984;159:991–999. doi: 10.1128/jb.159.3.991-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 986.Deana A, Ehrlich R, Reiss C. Synonymous codon selection controls in vivo turnover and amount of mRNA in Escherichia coli bla and ompA genes. J Bacteriol. 1996;178:2718–2720. doi: 10.1128/jb.178.9.2718-2720.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 987.Death A, Notley L, Ferenci T. Derepression of LamB protein facilitates outer membrane permeation of carbohydrates into Escherichia coli under conditions of nutrient stress. J Bacteriol. 1993;175:1475–1483. doi: 10.1128/jb.175.5.1475-1483.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 988.Debarbouille M, Schwartz M. The use of gene fusions to study the expression of malT, the positive regulator gene of the maltose regulon. J Mol Biol. 1979;132:521–534. doi: 10.1016/0022-2836(79)90273-0. [DOI] [PubMed] [Google Scholar]
- 989.Debarbouille M, Schwartz M. Mutants which make more malT product, the activator of the maltose regulon in Escherichia coli. Mol Gen Genet. 1980;178:589–595. doi: 10.1007/BF00337865. [DOI] [PubMed] [Google Scholar]
- 990.Debarbouille M, Cossart P, Raibaud O. A DNA sequence containing the control sites for gene malT and for the malPG operon. Mol Gen Genet. 1982;185:88–92. doi: 10.1007/BF00333795. [DOI] [PubMed] [Google Scholar]
- 991.Debenham P G, Webb M B T. The isolation and preliminary characterisation of a novel Escherichia coli mutant rorB with enhanced sensitivity to ionising radiation. Mol Gen Genet. 1988;215:161–164. doi: 10.1007/BF00331319. [DOI] [PubMed] [Google Scholar]
- 992.Debenham P G, Webb M B T, Law J. The cloning of the rorB gene of Escherichia coli. Mol Gen Genet. 1988;215:156–160. doi: 10.1007/BF00331318. [DOI] [PubMed] [Google Scholar]
- 993.Debouck, C. 1995. GenBank submission X02306.
- 994.Debouck C, Riccio A, Schumperli D, McKenney K, Jeffers J, Hughes C, Rosenberg M, Heusterspreute M, Davison J. Structure of the galactokinase gene of Escherichia coli, the last (?) gene of the gal operon. Nucleic Acids Res. 1985;13:1841–1853. doi: 10.1093/nar/13.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 995.DeChavigny A, Heacock P N, Dowhan W. Sequence and inactivation of the pss gene of Escherichia coli. Phosphatidylethanolamine may not be essential for cell viability. J Biol Chem. 1991;266:5323–5332. [PubMed] [Google Scholar]
- 996.Deeley M C, Yanofsky C. Nucleotide sequence of the structural gene for tryptophanase of Escherichia coli K-12. J Bacteriol. 1981;147:787–796. doi: 10.1128/jb.147.3.787-796.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 997.DeFelice M, Guardiola J, Lamberti A, Iaccarino M. Escherichia coli K-12 mutants altered in the transport systems for oligo- and dipeptides. J Bacteriol. 1973;116:751–756. doi: 10.1128/jb.116.2.751-756.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 998.DeFeyter R C, Davidson B E, Pittard J. Nucleotide sequence of the transcription unit containing the aroL and aroM genes from Escherichia coli K-12. J Bacteriol. 1986;165:233–239. doi: 10.1128/jb.165.1.233-239.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 999.DeFeyter R C, Pittard J. Genetic and molecular analysis of aroL, the gene for shikimate kinase II in Escherichia coli K-12. J Bacteriol. 1986;165:226–232. doi: 10.1128/jb.165.1.226-232.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000.Degryse E. Development of stable, genetically well-defined conditionally viable Escherichia coli strains. Mol Gen Genet. 1991;227:49–51. doi: 10.1007/BF00260705. [DOI] [PubMed] [Google Scholar]
- 1001.Deguchi Y, Yamato I, Anraku Y. Molecular cloning of gltS and gltP, which encode glutamate carriers of Escherichia coli. J Bacteriol. 1989;171:1314–1319. doi: 10.1128/jb.171.3.1314-1319.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1002.Deguchi Y, Yamato I, Anraku Y. Nucleotide sequence of gltS, the Na+ glutamate symport carrier gene of Escherichia coli B. J Biol Chem. 1990;265:21704–21708. [PubMed] [Google Scholar]
- 1003.del Campillo-Campbell A, Campbell A. Molydbenum cofactor requirement for biotin sulfoxide reduction in Escherichia coli. J Bacteriol. 1982;149:469–478. doi: 10.1128/jb.149.2.469-478.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1004.del Campillo-Campbell A, Campbell A. Alternative gene for biotin sulfoxide reduction in Escherichia coli K-12. J Mol Evol. 1996;42:85–90. doi: 10.1007/BF02198832. [DOI] [PubMed] [Google Scholar]
- 1005.Del Casale, Sollitti P, Chesney R H. Cytoplasmic l-asparaginase: isolation of a defective strain and mapping of ansA. J Bacteriol. 1983;154:513–515. doi: 10.1128/jb.154.1.513-515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1006.del Castillo I, Gonzalez-Pastor J E, San Millan J L, Moreno F. Nucleotide sequence of the Escherichia coli regulatory gene mprA and construction and characterization of mprA-deficient mutants. J Bacteriol. 1991;173:3924–3929. doi: 10.1128/jb.173.12.3924-3929.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1007.del Castillo I, Gomez J M, Moreno F. mprA, an Escherichia coli gene that reduces growth-phase-dependent synthesis of microcins B17 and C7 and blocks osmoinduction of proU when cloned on a high-copy-number plasmid. J Bacteriol. 1990;172:437–445. doi: 10.1128/jb.172.1.437-445.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1008.Delaney J M. A cya deletion mutant of Escherichia coli develops thermotolerance but does not exhibit a heat-shock response. Genet Res. 1990;55:1–6. doi: 10.1017/s001667230002512x. [DOI] [PubMed] [Google Scholar]
- 1009.Delaney J M, Georgopoulos C. Physical map locations of the trxB, htrD, cydC, and cydD genes of Escherichia coli. J Bacteriol. 1992;174:3824–3825. doi: 10.1128/jb.174.11.3824-3825.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1010.Delaney J M, Ang D, Georgopoulos C. Isolation and characterization of the Escherichia coli htrD gene, whose product is required for growth at high temperatures. J Bacteriol. 1992;174:1240–1247. doi: 10.1128/jb.174.4.1240-1247.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1011.Delaney J M, Wall D, Georgopoulos C. Molecular characterization of the Escherichia coli htrD gene: cloning, sequence, regulation, and involvement with cytochrome d oxidase. J Bacteriol. 1993;175:166–175. doi: 10.1128/jb.175.1.166-175.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1012.Delcuve G, Dennis P P. An amber mutation in a ribosomal protein gene: ineffective suppression stimulates operon-specific transcription. J Bacteriol. 1981;147:997–1001. doi: 10.1128/jb.147.3.997-1001.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1013.Delcuve G, Downing W L, Lewis H M, Dennis P P. Nucleotide sequence of the proximal portion of the RNA polymerase B subunit gene of Escherichia coli. Gene. 1980;11:367–373. doi: 10.1016/0378-1119(80)90076-1. [DOI] [PubMed] [Google Scholar]
- 1014.DelGiudice L. Method for isolating restriction- and modification-less mutants of Escherichia coli K-12. J Bacteriol. 1979;137:673–676. doi: 10.1128/jb.137.1.673-676.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1015.Delidakis C E, Jones-Mortimer M C, Kornberg H L. A mutant inducible for galactitol utilization in Escherichia coli K12. J Gen Microbiol. 1982;128:601–604. doi: 10.1099/00221287-128-3-601. [DOI] [PubMed] [Google Scholar]
- 1016.Demerec M, Adelberg E A, Clark A J, Hartman P E. A proposal for a uniform nomenclature in bacterial genetics. Genetics. 1966;54:61–76. doi: 10.1093/genetics/54.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1017.DeMoss J A, Hsu P-Y. NarK enhances nitrate uptake and nitrite excretion in Escherichia coli. J Bacteriol. 1991;173:3303–3310. doi: 10.1128/jb.173.11.3303-3310.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1018.Demple B, Linn S. On the recognition and cleavage mechanism of Escherichia coli endodeoxyribonuclease V, a possible DNA repair enzyme. J Biol Chem. 1982;257:2848–2855. [PubMed] [Google Scholar]
- 1019.Dempsey W B, Itoh H. Characterization of pyridoxine auxotrophs of Escherichia coli: serine and PdxF mutants. J Bacteriol. 1970;104:658–667. doi: 10.1128/jb.104.2.658-667.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1020.Dempsey W B, Arcement L J. Identification of the forms of vitamin B6 present in the culture media of “vitamin B6 control” mutants. J Bacteriol. 1971;107:580–582. doi: 10.1128/jb.107.2.580-582.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1021.Deng M, Misra R. Examination of AsmA and its effect on the assembly of Escherichia coli outer membrane proteins. Mol Microbiol. 1996;21:605–612. doi: 10.1111/j.1365-2958.1996.tb02568.x. [DOI] [PubMed] [Google Scholar]
- 1022.Denklau D, Bock A. l-Cysteine biosynthesis in Escherichia coli: nucleotide sequence and expression of the serine acetyltransferase (cysE) gene from the wild-type and a cysteine-excreting mutant. J Gen Microbiol. 1987;133:515–525. doi: 10.1099/00221287-133-3-515. [DOI] [PubMed] [Google Scholar]
- 1023.Derouiche, R., M. Gavioli, H. Benedetti, A. Prilipov, C. Lazdunski, and R. Lloubes. TolA central domain interacts with Escherichia coli porins. EMBO J. 15:6408–6415. [PMC free article] [PubMed]
- 1024.Descoteaux A, Drapeau G R. Regulation of cell division in Escherichia coli K-12: probable interactions among proteins FtsQ, FtsA, and FtsZ. J Bacteriol. 1987;169:1938–1942. doi: 10.1128/jb.169.5.1938-1942.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1025.Deutch A H, Smith C J, Rushlow K E, Kretschmer P J. Escherichia coli delta-1-pyrroline-5-carboxylate reductase: gene sequence, protein overproduction and purification. Nucleic Acids Res. 1982;10:7701–7714. doi: 10.1093/nar/10.23.7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1026.Deutch A H, Rushlow K E, Smith C J. Analysis of the Escherichia coli proBA locus by DNA and protein sequencing. Nucleic Acids Res. 1984;12:6337–6355. doi: 10.1093/nar/12.15.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1027.Deutch A H, Soffer R L. Escherichia coli mutants defective in dipeptidyl carboxypeptidase. Proc Natl Acad Sci USA. 1978;75:5998–6001. doi: 10.1073/pnas.75.12.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1028.Deutch C E. Oxidation of l-thiazolidine-4-carboxylate by l-proline dehydrogenase in Escherichia coli. J Gen Microbiol. 1992;138:1593. doi: 10.1099/00221287-138-8-1593. [DOI] [PubMed] [Google Scholar]
- 1029.DeVeaux L C, Clevenson D S, Bradbeer C, Kadner R J. Identification of the BtuCED polypeptides and evidence for their role in vitamin B12 transport in Escherichia coli. J Bacteriol. 1986;167:920–927. doi: 10.1128/jb.167.3.920-927.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1030.DeVeaux L C, Cronan J E, Jr, Smith T L. Genetic and biochemical characterization of a mutation (fatA) that allows trans unsaturated fatty acids to replace the essential cis unsaturated fatty acids of Escherichia coli. J Bacteriol. 1989;171:1562–1568. doi: 10.1128/jb.171.3.1562-1568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1031.DeVeaux L C, Kadner R J. Transport of vitamin B12 in Escherichia coli: cloning of the btuCD region. J Bacteriol. 1985;162:888–896. doi: 10.1128/jb.162.3.888-896.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1032.Dewar S J, Donachie W D. Antisense transcription of the ftsZ-ftsA gene junction inhibits cell division in Escherichia coli. J Bacteriol. 1993;175:7097–7101. doi: 10.1128/jb.175.21.7097-7101.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1033.deWind N, de Jong M, Meijer M, Stuitje A R. Site-directed mutagenesis of the Escherichia coli chromosome near oriC: identification and characterization of asnC, a regulatory element in E. coli asparagine metabolism. Nucleic Acids Res. 1985;13:8797–8811. doi: 10.1093/nar/13.24.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1034.Dhillon T S. Temperate coliphage HK022: virions, DNA, one-step growth, attachment site, and the prophage genetic map. J Gen Virol. 1981;55:487–492. doi: 10.1099/0022-1317-55-2-487. [DOI] [PubMed] [Google Scholar]
- 1035.Dhillon T S, Poon A P W, Hui Y W, Dhillon E K S. Lambdoid coliphage HK139 integrates between his and supD. J Virol. 1982;44:716–719. doi: 10.1128/jvi.44.2.716-719.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1036.Diaz R, Kaiser C. Rac−E. coli K12 strains carry a preferential attachment site for λrev. Mol Gen Genet. 1981;183:484–489. doi: 10.1007/BF00268769. [DOI] [PubMed] [Google Scholar]
- 1037.Diaz R, Barnsley P, Pritchard R H. Location and characterization of a new replication origin in the E. coli K12 chromosome. Mol Gen Genet. 1979;175:151–157. doi: 10.1007/BF00425531. [DOI] [PubMed] [Google Scholar]
- 1038.Diaz R, Pritchard R H. Cloning of replication origins from the E. coli K12 chromosome. Nature. 1978;275:561–564. doi: 10.1038/275561a0. [DOI] [PubMed] [Google Scholar]
- 1039.Dibb N J, Wolfe P B. lep operon proximal gene is not required for growth or secretion by Escherichia coli. J Bacteriol. 1986;166:83–87. doi: 10.1128/jb.166.1.83-87.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1040.Dicker I B, Seetharam S. Cloning and nucleotide sequence of the firA gene and the firA200(Ts) allele from Escherichia coli. J Bacteriol. 1991;173:334–344. doi: 10.1128/jb.173.1.334-344.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1041.Diderichsen B. cur-1, a mutation affecting the phenotype of sup+ strains of Escherichia coli. Mol Gen Genet. 1980;180:425–428. doi: 10.1007/BF00425858. [DOI] [PubMed] [Google Scholar]
- 1042.Diderichsen B. Improved mapping of ksgB and integration of transposons near rclB and terC in Escherichia coli. J Bacteriol. 1981;146:409–411. doi: 10.1128/jb.146.1.409-411.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1043.Diderichsen B, De Hauwer G. Improved mapping of the tyrS locus in Escherichia coli. Mol Gen Genet. 1980;178:647–650. doi: 10.1007/BF00337873. [DOI] [PubMed] [Google Scholar]
- 1044.Diderichsen B, Desmarez L. Variations in phenotype of relB mutants of Escherichia coli and the effect of pus and sup mutations. Mol Gen Genet. 1980;180:429–437. doi: 10.1007/BF00425859. [DOI] [PubMed] [Google Scholar]
- 1045.Dierksen K P, Trempy J E. Identification of a second RcsA protein, a positive regulator of colanic acid capsular polysaccharide genes, in Escherichia coli. J Bacteriol. 1996;178:5053–5056. doi: 10.1128/jb.178.16.5053-5056.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1046.Diez A A, Farewell A, Nannmark U, Nyström T. A mutation in the ftsK gene of Escherichia coli affects cell-cell separation, stationary-phase survival, stress adaptation, and expression of the gene encoding the stress protein UspA. J Bacteriol. 1997;179:5878–5883. doi: 10.1128/jb.179.18.5878-5883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1047.DiFrancesco R, Bhatnagar S K, Brown A, Bessman M J. The interaction of DNA polymerase III and the product of the Escherichia coli mutator gene, mutD. J Biol Chem. 1984;259:5567–5573. [PubMed] [Google Scholar]
- 1048.DiGate R J, Marians K J. Molecular cloning and DNA sequence analysis of Escherichia coli topB, the gene encoding topoisomerase III. J Biol Chem. 1989;264:17924–17930. [PubMed] [Google Scholar]
- 1048a.Dijkstra A J. Ph.D. thesis. Groningen, The Netherlands: University of Groningen; 1997. [Google Scholar]
- 1049.Dijkstra A J, Keck W. Identification of new members of the lytic transglycosylase family in Haemophilus influenzae and Escherichia coli. Microb Drug Resist. 1996;2:141–145. doi: 10.1089/mdr.1996.2.141. [DOI] [PubMed] [Google Scholar]
- 1050.Dila D, Sutherland E, Moran L, Slatko B, Raleigh E A. Genetic and sequence organization of the mcrBC locus of Escherichia coli K-12. J Bacteriol. 1990;172:4888–4900. doi: 10.1128/jb.172.9.4888-4900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1051.Dillon D A, Wu W I, Riedel B, Wissing J B, Dowhan W, Carman G M. The Escherichia coli pgpB gene encodes for a diacylglycerol pyrophosphate phosphatase activity. J Biol Chem. 1996;271:30548–30553. doi: 10.1074/jbc.271.48.30548. [DOI] [PubMed] [Google Scholar]
- 1052.Dimri G P, Ames G F-L, D’Ari L, Rabinowitz J C. Physical map location of the Escherichia coli gene encoding the bifunctional enzyme 5,10-methylene-tetrahydrofolate dehydrogenase/5,10-methenyl-tetrahydrofolate cyclohydrolase. J Bacteriol. 1991;173:5251. doi: 10.1128/jb.173.17.5251.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1053.DiNardo S, Voelkel K A, Sternglanz R, Reynolds A E, Wright A. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell. 1982;31:43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- 1054.Ding Q, Kusano S, Villarejo M, Ishihama A. Promoter selectivity control of Escherichia coli RNA polymerase by ionic strength: differential recognition of osmoregulated promoters by E sigma D and E sigma S holoenzymes. Mol Microbiol. 1995;16:649–656. doi: 10.1111/j.1365-2958.1995.tb02427.x. [DOI] [PubMed] [Google Scholar]
- 1055.Diorio C, Cai J, Marmor J, Shinder R, DuBow M S. An Escherichia coli chromosomal ars operon homolog is functional in arsenic detoxification and is conserved in gram-negative bacteria. J Bacteriol. 1995;177:2050–2056. doi: 10.1128/jb.177.8.2050-2056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1056.DiRusso C C. Nucleotide sequence of the fadR gene, a multifunctional regulator of fatty acid metabolism in Escherichia coli. Nucleic Acids Res. 1988;16:7995–8009. doi: 10.1093/nar/16.16.7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1057.DiRusso C C. Primary sequence of the Escherichia coli fadBA operon, encoding the fatty acid-oxidizing multienzyme complex, indicates a high degree of homology to eucaryotic enzymes. J Bacteriol. 1990;172:6459–6468. doi: 10.1128/jb.172.11.6459-6468.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1058.DiRusso C C, Nunn W D. Cloning and characterization of a gene fadR involved in regulation of fatty acid metabolism in Escherichia coli. J Bacteriol. 1985;161:583–588. doi: 10.1128/jb.161.2.583-588.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1059.Diver W P, Sargentini N J, Smith K C. A mutation (radA100) in Escherichia coli that selectively sensitizes cells grown in rich medium to X- or UV-radiation, or methyl methanesulphonate. Int J Radiat Biol. 1982;42:339–346. doi: 10.1080/09553008214551251. [DOI] [PubMed] [Google Scholar]
- 1060.D’Mello R, Hill S, Poole R K. The cytochrome bd quinol oxidase in Escherichia coli has an extremely high oxygen affinity and two oxygen-binding haems: implications for regulation of activity in vivo by oxygen inhibition. Microbiology. 1996;142:755–763. doi: 10.1099/00221287-142-4-755. [DOI] [PubMed] [Google Scholar]
- 1061.Dobrynina O Y, Bol’shakova T N, Gershanovich V N. The isolation and mapping of a mutation which impairs the function of the cytoplasm-specific component of the fructose transport system in Escherichia coli. Sov Genet (Engl Transl Genetika) 1986;21:981–986. [PubMed] [Google Scholar]
- 1061a.Dodgson C, Amor P, Whitfield C. Distribution of the rol gene encoding the regulator of lipopolysaccharide O-chain length in Escherichia coli and its influence on the expression of group I capsular K antigens. J Bacteriol. 1996;178:1895–1902. doi: 10.1128/jb.178.7.1895-1902.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1062.Doi M, Wachi M, Ishino F, Tomioka S, Ito M, Sakagami Y, Suzuki A, Matsuhashi M. Determinations of the DNA sequence of the mreB gene and of the gene products of the mre region that function in formation of the rod shape of Escherichia coli cells. J Bacteriol. 1988;170:4619–4624. doi: 10.1128/jb.170.10.4619-4624.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1063.Donch J, Greenberg J. Genetic analysis of lon mutants of strain K-12 of Escherichia coli. Mol Gen Genet. 1968;103:105–115. doi: 10.1007/BF00427138. [DOI] [PubMed] [Google Scholar]
- 1064.Dong H, Kirsebom L A, Nilsson L. Growth rate regulation of 4.5 S RNA and M1 RNA the catalytic subunit. J Mol Biol. 1996;261:303–308. doi: 10.1006/jmbi.1996.0461. [DOI] [PubMed] [Google Scholar]
- 1065.Dong J, Iuchi S, Kwan H S, Lu Z, Lin E C C. The deduced amino-acid sequence of the cloned cpxR gene suggests the protein is the cognate regulator for the membrane sensor, CpxA, in a two-component signal transduction system of Escherichia coli. Gene. 1993;136:227–230. doi: 10.1016/0378-1119(93)90469-j. [DOI] [PubMed] [Google Scholar]
- 1066.Dong J M, Taylor J S, Latour D J, Iuchi S, Lin E C C. Three overlapping lct genes involved in l-lactate utilization by Escherichia coli. J Bacteriol. 1993;175:6671–6678. doi: 10.1128/jb.175.20.6671-6678.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1067.Dong Z, Onrust R, Skangalis M, O’Donnell M E. DNA polymerase III accessory proteins. I. holA and holB encoding delta and delta′. J Biol Chem. 1993;268:11758–11765. [PubMed] [Google Scholar]
- 1068.Donnelly C E, Walker G C. Coexpression of UmuD′ with UmuC suppresses the UV mutagenesis deficiency of groE mutants. J Bacteriol. 1992;174:3133–3139. doi: 10.1128/jb.174.10.3133-3139.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1069.Donovan W P, Kushner S R. Amplification of ribonuclease II (rnb) activity in Escherichia coli K12. Nucleic Acids Res. 1983;11:265–275. doi: 10.1093/nar/11.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1070.Donovan W P, Kushner S R. Cloning and physical analysis of the pyrF gene (coding for orotidine-5′-phosphate decarboxylase) from Escherichia coli K-12. Gene. 1983;25:39–48. doi: 10.1016/0378-1119(83)90165-8. [DOI] [PubMed] [Google Scholar]
- 1071.Doolittle R F, Feng D F, Anderson K L, Alberro M R. A naturally occurring horizontal gene transfer from a eukaryote to a prokaryote. J Mol Evol. 1990;31:383–388. doi: 10.1007/BF02106053. [DOI] [PubMed] [Google Scholar]
- 1072.Dorman C J, Higgins C F. Fimbrial phase variation in Escherichia coli: dependence on integration host factor and homologies with other site-specific recombinases. J Bacteriol. 1987;169:3840–3843. doi: 10.1128/jb.169.8.3840-3843.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1073.Dorrell N, Ahmed A, Moss S H. Photoreactivation in a phrB mutant of Escherichia coli K-12: evidence for the role of a second protein in photorepair. Photochem Photobiol. 1993;58:831–835. doi: 10.1111/j.1751-1097.1993.tb04979.x. [DOI] [PubMed] [Google Scholar]
- 1074.Dosch D C, Helmer G L, Sutton S H, Salvacion F F, Epstein W. Genetic analysis of potassium transport loci in Escherichia coli: evidence for three constitutive systems mediating uptake of potassium. J Bacteriol. 1991;173:687–696. doi: 10.1128/jb.173.2.687-696.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1075.Doskocil J, Sorm F. The mode of action of 5-aza-2′-deoxycytidine in Escherichia coli. J Biochem (Tokyo) 1970;13:180–187. doi: 10.1111/j.1432-1033.1970.tb00916.x. [DOI] [PubMed] [Google Scholar]
- 1076.Doublet P, van Heijenoort J, Mengin-Lecreulx D. Identification of the Escherichia coli murI gene, which is required for the biosynthesis of d-glutamic acid, a specific component of bacterial peptidoglycan. J Bacteriol. 1992;174:5772–5779. doi: 10.1128/jb.174.18.5772-5779.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1077.Doublet P, van Heijenoort J, Bohin J P, Mengin-Lecreulx D. The murI gene of Escherichia coli is an essential gene that encodes a glutamate racemase activity. J Bacteriol. 1993;175:2970–2979. doi: 10.1128/jb.175.10.2970-2979.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1078.Dougherty T J, Thanassi J A, Pucci M J. The Escherichia coli mutant requiring d-glutamic acid is the result of mutations in two distinct genetic loci. J Bacteriol. 1993;175:111–116. doi: 10.1128/jb.175.1.111-116.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1079.Douville K, Price A, Eichler J, Economou A, Wickner W. SecYEG and SecA are the stoichiometric components of preprotein translocase. J Biol Chem. 1995;270:20106–20111. doi: 10.1074/jbc.270.34.20106. [DOI] [PubMed] [Google Scholar]
- 1080.Douville K, Leonard M, Brundage L, Nishiyama K, Tokuda H, Mizushima S, Wickner W. Band 1 subunit of Escherichia coli preprotein translocase and integral membrane export factor P12 are the same protein. J Biol Chem. 1994;269:18705–18707. [PubMed] [Google Scholar]
- 1081.Dover N, Higgins C F, Carmel O, Rimon A, Pinner E, Padan E. Na+-induced transcription of nhaA, which encodes an Na+/H+ antiporter in Escherichia coli, is positively regulated by nhaR and affected by hns. J Bacteriol. 1996;178:6508–6517. doi: 10.1128/jb.178.22.6508-6517.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1082.Dover S, Halpern Y S. Genetic analysis of the gamma-aminobutyrate utilization pathway in Escherichia coli K-12. J Bacteriol. 1974;117:494–501. doi: 10.1128/jb.117.2.494-501.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1083.Downing W L, Dennis P P. Transcription products from the rplKAJL-rpoBC gene cluster. J Mol Biol. 1987;194:609–620. doi: 10.1016/0022-2836(87)90238-5. [DOI] [PubMed] [Google Scholar]
- 1084.Downing W L, Sullivan S L, Gottesman M E, Dennis P P. Sequence and transcriptional pattern of the essential Escherichia coli secE-nusG operon. J Bacteriol. 1990;172:1621–1627. doi: 10.1128/jb.172.3.1621-1627.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1085.Drapal N, Sawers G. Promoter 7 of the Escherichia coli pfl operon is a major determinant in the anaerobic regulation of expression by ArcA. J Bacteriol. 1995;177:5338–5341. doi: 10.1128/jb.177.18.5338-5341.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1085a.Drexler H. Specialized transduction of the biotin region of Escherichia coli by phage T1. Mol Gen Genet. 1977;152:59–63. doi: 10.1007/BF00264940. [DOI] [PubMed] [Google Scholar]
- 1086.Driver R P, Lawther R P. Physical analysis of deletion mutations in the ilvGEDA operon of Escherichia coli K-12. J Bacteriol. 1985;162:598–606. doi: 10.1128/jb.162.2.598-606.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1087.Drolet M, Peloquin L, Echelard Y, Cousineau L, Sasarman A. Isolation and nucleotide sequence of the hemA gene of Escherichia coli K12. Mol Gen Genet. 1989;216:347–352. doi: 10.1007/BF00334375. [DOI] [PubMed] [Google Scholar]
- 1088.Drotschmann K, Aronshtam A, Fritz H J, Marinus M G. The Escherichia coli MutL protein stimulates binding of Vsr and MutS to heteroduplex DNA. Nucleic Acids Res. 1998;26:948–953. doi: 10.1093/nar/26.4.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1089.Drury L S, Buxton R S. DNA sequence analysis of the dye gene of Escherichia coli reveals amino acid homology between the Dye and OmpR proteins. J Biol Chem. 1985;260:4236–4242. [PubMed] [Google Scholar]
- 1090.Drury L S, Buxton R S. Identification and sequencing of the Escherichia coli cet gene which codes for an inner membrane protein, mutation of which causes tolerance of colicin E2. Mol Microbiol. 1988;2:109–119. doi: 10.1111/j.1365-2958.1988.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 1091.Dubnau E, Margolin P. Suppression of promoter mutations by the pleiotrophic supX mutations. Mol Gen Genet. 1972;117:91–112. doi: 10.1007/BF00267607. [DOI] [PubMed] [Google Scholar]
- 1092.Dubourdieu M, DeMoss J A. The narJ gene product is required for biogenesis of respiratory nitrate reductase in Escherichia coli. J Bacteriol. 1992;174:867–872. doi: 10.1128/jb.174.3.867-872.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1093.Duchange N, Zakin M M, Ferrara P, Saint-Girons I, Park I, Tran S V, Py M-C, Cohen G N. Structure of the metJBLF cluster in Escherichia coli K12. Sequence of the metB structural gene of the 5′- and 3′-flanking regions of the metBL operon. J Biol Chem. 1983;258:14868–14871. [PubMed] [Google Scholar]
- 1094.Duclos B, Cortay J C, Bleicher F, Ron E Z, Richaud C, Saint Girons I, Cozzone A J. Nucleotide sequence of the metA gene encoding homoserine trans-succinylase in Escherichia coli. Nucleic Acids Res. 1989;17:2856. doi: 10.1093/nar/17.7.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1095.Duester G, Campen R K, Holmes W M. Nucleotide sequence of an Escherichia coli tRNA (Leu 1) operon and identification of the transcription promoter signal. Nucleic Acids Res. 1981;9:2121–2139. doi: 10.1093/nar/9.9.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1096.Duester G, Holmes W M. The distal end of the ribosomal RNA operon rrnD of Escherichia coli contains a tRNA1thr gene, two 5s rRNA genes and a transcription terminator. Nucleic Acids Res. 1980;8:3793–3807. doi: 10.1093/nar/8.17.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1097.Duggan L J, Hill T M, Wu S, Garrison K, Zhang X, Gottlieb P A. Using modified nucleotides to map the DNA determinants of the Tus-TerB complex, the protein-DNA interaction associated with termination of replication in Escherichia coli. J Biol Chem. 1995;270:28049–28054. doi: 10.1074/jbc.270.47.28049. [DOI] [PubMed] [Google Scholar]
- 1098.Duncan K, Lewendon A, Coggins J R. The complete amino acid sequence of Escherichia coli 5-enolpyruvylshikimate 3-phosphate synthase. FEBS Lett. 1984;170:59–63. doi: 10.1016/0014-5793(84)80027-7. [DOI] [PubMed] [Google Scholar]
- 1099.Duncan K, Coggins J R. The serC-aroA operon of Escherichia coli. A mixed operon encoding enzymes from two different amino acid biosynthetic pathways. Biochem J. 1986;234:49–57. doi: 10.1042/bj2340049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1100.Duncan K, Chaudhuri S, Campbell M S, Coggins J R. The overexpression and complete amino acid sequence of Escherichia coli 3-dehydroquinase. Biochem J. 1986;238:475–483. doi: 10.1042/bj2380475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1101.Dunderdale H J, Sharples G J, Lloyd R G, West S C. Cloning, overexpression, purification and characterization of the Escherichia coli RuvC Holliday junction resolvase. J Biol Chem. 1994;269:5187–5194. [PubMed] [Google Scholar]
- 1102.Dunn T M, Schleif R. Deletion analysis of the Escherichia coli ara PC and PBAD promoters. J Mol Biol. 1984;180:201–204. doi: 10.1016/0022-2836(84)90437-6. [DOI] [PubMed] [Google Scholar]
- 1103.Duplay P, Bedouelle H, Fowler A, Zabin I, Saurin W, Hofnung M. Sequence of the malE gene and of its product, the maltose-binding protein of Escherichia coli K12. J Biol Chem. 1984;259:10606–10613. [PubMed] [Google Scholar]
- 1104.Dutreix M, Moreau P L, Bailone A, Galibert F, Battista J, Walker G C, Devoret R. New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J Bacteriol. 1989;171:2415–2423. doi: 10.1128/jb.171.5.2415-2423.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1105.Dworkin J, Jovanovic G, Model P. Role of upstream activation sequences and integration host factor in transcriptional activation by the constitutively active prokaryotic enhancer-binding protein PspF. J Mol Biol. 1997;273:377–388. doi: 10.1006/jmbi.1997.1317. [DOI] [PubMed] [Google Scholar]
- 1106.Dykstra C, Kushner S R. Physical characterization of the cloned protease III gene from Escherichia coli K-12. J Bacteriol. 1985;163:1055–1059. doi: 10.1128/jb.163.3.1055-1059.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1107.Dykstra C C, Prasher D, Kushner S R. Physical and biochemical analysis of the cloned recB and recC genes of Escherichia coli K-12. J Bacteriol. 1984;157:21–27. doi: 10.1128/jb.157.1.21-27.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1108.Dykxhoorn D M, St. Pierre R, Linn T. Synthesis of the beta and beta′ subunits of Escherichia coli RNA polymerase is autogenously regulated in vivo by both transcriptional and translational mechanisms. Mol Microbiol. 1996;19:483–493. doi: 10.1046/j.1365-2958.1996.384913.x. [DOI] [PubMed] [Google Scholar]
- 1109.Easton A M, Kushner S R. Transcription of the uvrD gene of Escherichia coli is controlled by the lexA repressor and by attenuation. Nucleic Acids Res. 1983;11:8625–8640. doi: 10.1093/nar/11.24.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1110.Eberhardt S, Richter G, Gimbel W, Werner T, Bacher A. Cloning, sequencing, mapping and hyperexpression of the ribC gene coding for riboflavin synthase of Escherichia coli. Eur J Biochem. 1996;242:712–719. doi: 10.1111/j.1432-1033.1996.0712r.x. [DOI] [PubMed] [Google Scholar]
- 1111.Echelard Y, Dymetryszyn J, Drolet M, Sasarman A. Nucleotide sequence of the hemB gene of Escherichia coli K12. Mol Gen Genet. 1988;214:503–508. doi: 10.1007/BF00330487. [DOI] [PubMed] [Google Scholar]
- 1112.Echols H, Garen A, Torriani A. Genetic control of repression of alkaline phosphatase in E. coli. J Mol Biol. 1961;3:425–438. doi: 10.1016/s0022-2836(61)80055-7. [DOI] [PubMed] [Google Scholar]
- 1113.Echols H, Lu C, Burgers P M J. Mutator strains of Escherichia coli, mutD and dnaQ, with defective exonucleolytic editing by DNA polymerase III holoenzyme. Proc Natl Acad Sci USA. 1983;80:2189–2192. doi: 10.1073/pnas.80.8.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1114.Eckhardt T. Isolation of plasmids carrying the arginine repressor gene argR of Escherichia coli K12. Mol Gen Genet. 1980;178:447–452. doi: 10.1007/BF00270498. [DOI] [PubMed] [Google Scholar]
- 1115.Edgar R, Bibi E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition J. Bacteriol. 1997;179:2274–2280. doi: 10.1128/jb.179.7.2274-2280.1997. . (Erratum, 179:5654.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1116.Edlund T, Normark S. Recombination between short DNA homologies causes tandem duplications. Nature. 1981;292:269–271. doi: 10.1038/292269a0. [DOI] [PubMed] [Google Scholar]
- 1117.Edlund T, Grundstrom T, Normark S. Isolation and characterization of DNA repetitions carrying the chromosomal β-lactamase gene of Escherichia coli K-12. Mol Gen Genet. 1979;173:115–125. doi: 10.1007/BF00330301. [DOI] [PubMed] [Google Scholar]
- 1118.Edwards E S, Rondeau S S, DeMoss J A. chlC (nar) operon of Escherichia coli includes structural genes for alpha and beta subunits of nitrate reductase. J Bacteriol. 1983;153:1513–1520. doi: 10.1128/jb.153.3.1513-1520.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1119.Edwards R M, Yudkin M. Location of the gene for the low-affinity tryptophan-specific permease of Escherichia coli. Biochem J. 1982;204:617–619. doi: 10.1042/bj2040617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1120.Egan A F, Russell R R B. Conditional mutations affecting the cell envelope of Escherichia coli K-12. Genet Res. 1973;21:139–152. doi: 10.1017/s001667230001332x. [DOI] [PubMed] [Google Scholar]
- 1121.Egan S E, Fliege R, Tong S, Shibata A, Wolf R E, Conway T. Molecular characterization of the Entner-Doudoroff pathway in Escherichia coli: sequence analysis and localization of promoters for the edd-eda operon. J Bacteriol. 1992;174:4638–4646. doi: 10.1128/jb.174.14.4638-4646.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1122.Egan S M, Stewart V. Mutational analysis of nitrate regulatory gene narL in Escherichia coli K-12. J Bacteriol. 1991;173:4424–4432. doi: 10.1128/jb.173.14.4424-4432.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1122a.Eggertsson G, Söll D. Transfer ribonucleic acid-mediated suppression of termination codons in Escherichia coli. Microbiol Rev. 1988;52:354–374. doi: 10.1128/mr.52.3.354-374.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1123.Eggleston A K, West S C. Exchanging partners: recombination in E. coli. Trends Genet. 1996;12:20–26. doi: 10.1016/0168-9525(96)81384-9. [DOI] [PubMed] [Google Scholar]
- 1124.Ehlert K, Holtje J V, Templin M F. Cloning and expression of a murein hydrolase lipoprotein from Escherichia coli. Mol Microbiol. 1995;16:761–768. doi: 10.1111/j.1365-2958.1995.tb02437.x. [DOI] [PubMed] [Google Scholar]
- 1125.Ehrenreich A, Forchhammer K, Tormay P, Veprek B, Bock A. Selenoprotein synthesis in E. coli. Purification and characterisation of the enzyme catalysing selenium activation. Eur J Biochem. 1992;206:767–773. doi: 10.1111/j.1432-1033.1992.tb16983.x. [DOI] [PubMed] [Google Scholar]
- 1126.Ehresmann C, Philippe C, Westhof E, Benard L, Portier C, Ehresmann B. A pseudoknot is required for efficient translational initiation and regulation of the Escherichia coli rpsO gene coding for ribosomal protein S15. Biochem Cell Biol. 1995;73:1131. doi: 10.1139/o95-122. [DOI] [PubMed] [Google Scholar]
- 1127.Ehring R, Beyreuther K, Wright J K, Overath P. In vitro and in vivo products of E. coli lactose permease gene are identical. Nature. 1980;283:537–540. doi: 10.1038/283537a0. [DOI] [PubMed] [Google Scholar]
- 1128.Ehrle R, Pick C, Ulrich R, Hofmann E, Ehrmann M. Characterization of transmembrane domains 6, 7, and 8 of MalF by mutational analysis. J Bacteriol. 1996;178:2255–2262. doi: 10.1128/jb.178.8.2255-2262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1129.Ehrmann M, Boos W. Identification of endogenous inducers of the mal regulon in Escherichia coli. J Bacteriol. 1987;169:3539–3545. doi: 10.1128/jb.169.8.3539-3545.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1130.Ehrmann M, Boos W, Ormseth E, Schweizer H P, Larson T J. Divergent transcription of the sn-glycerol-3-phosphate active transport (glpT) and anaerobic sn-glycerol-3-phosphate dehydrogenase (glpA glpC glpB) genes of Escherichia coli K-12. J Bacteriol. 1987;169:526–532. doi: 10.1128/jb.169.2.526-532.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1131.Eichhorn E, van der Ploeg J R, Kertesz M A, Leisinger T. Characterization of alpha-ketoglutarate-dependent taurine dioxygenase from Escherichia coli. J Biol Chem. 1997;272:23031–23036. doi: 10.1074/jbc.272.37.23031. [DOI] [PubMed] [Google Scholar]
- 1132.Eichler K, Buchet A, Bourgis F, Kleber H P, Mandrand-Berthelot M A. The fix Escherichia coli region contains four genes related to carnitine metabolism. J Basic Microbiol. 1995;35:217–227. doi: 10.1002/jobm.3620350404. [DOI] [PubMed] [Google Scholar]
- 1133.Eichler K, Buchet A, Lemke R, Kleber H P, Mandrand-Berthelot M A. Identification and characterization of the caiF gene encoding a potential transcriptional activator of carnitine metabolism in Escherichia coli. J Bacteriol. 1996;178:1248–1257. doi: 10.1128/jb.178.5.1248-1257.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1134.Eichler K, Bourgis F, Buchet A, Kleber H-P, Mandrand-Berthelot M A. Molecular characterization of the cai operon necessary for carnitine metabolism in Escherichia coli. Mol Microbiol. 1994;13:775–786. doi: 10.1111/j.1365-2958.1994.tb00470.x. [DOI] [PubMed] [Google Scholar]
- 1135.Eichler K, Schunck W-H, Kleber H-P, Mandrand-Berthelot M A. Cloning, nucleotide sequence, and expression of the Escherichia coli gene encoding carnitine dehydratase. J Bacteriol. 1994;176:2970–2975. doi: 10.1128/jb.176.10.2970-2975.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1136.Eick-Helmerich K, Hantke K, Braun V. Cloning and expression of the exbB gene of Escherichia coli K-12. Mol Gen Genet. 1987;206:246–251. doi: 10.1007/BF00333580. [DOI] [PubMed] [Google Scholar]
- 1137.Eick-Helmerich K, Braun V. Import of biopolymers into Escherichia coli: nucleotide sequences of the exbB and exbD genes are homologous to those of the tolQ and tolR genes, respectively. J Bacteriol. 1989;171:5117–5126. doi: 10.1128/jb.171.9.5117-5126.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1138.Eiglmeier K, Boos W, Cole S T. Nucleotide sequence and transcriptional startpoint of the glpT gene of E. coli: extensive sequence homology of the glycerol-3-phosphate transport protein with components of the hexose-6-phosphate transport system. Mol Microbiol. 1987;1:251–258. doi: 10.1111/j.1365-2958.1987.tb01931.x. [DOI] [PubMed] [Google Scholar]
- 1139.Eisen J A, Sweder K S, Hanawalt P C. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1140.Eisenbeis S, Parker J. Strains of Escherichia coli carrying the structural gene for histidyl-tRNA synthetase on a high copy-number plasmid. Mol Gen Genet. 1981;183:115–122. doi: 10.1007/BF00270148. [DOI] [PubMed] [Google Scholar]
- 1141.Eisenbeis S, Parker J. The nucleotide sequence of the promoter region of hisS, the structural gene for histidyl-tRNA synthetase. Gene. 1982;18:107–114. doi: 10.1016/0378-1119(82)90108-1. [DOI] [PubMed] [Google Scholar]
- 1142.Eisenstein B I, Sweet D S, Vaughn V, Friedman D I. Integration host factor is required for the DNA inversion that controls phase variation in Escherichia coli. Proc Natl Acad Sci USA. 1987;84:6506–6510. doi: 10.1073/pnas.84.18.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1143.el Yaagoubi A, Kohiyama M, Richarme G. Defect in export and synthesis of the periplasmic galactose receptor MglB in dnaK mutants of Escherichia coli, and decreased stability of the mglB mRNA. Microbiology. 1996;142:2595–2602. doi: 10.1099/00221287-142-9-2595. [DOI] [PubMed] [Google Scholar]
- 1144.el-Hajj H H, Wang L, Weiss B. Multiple mutant of Escherichia coli synthesizing virtually thymineless DNA during limited growth. J Bacteriol. 1992;174:4450–4456. doi: 10.1128/jb.174.13.4450-4456.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1145.Elledge S J, Walker G C. Proteins required for ultraviolet light and chemical mutagenesis. Identification of the products of the umuC locus of Escherichia coli. J Mol Biol. 1983;164:175–192. doi: 10.1016/0022-2836(83)90074-8. [DOI] [PubMed] [Google Scholar]
- 1146.Ellwood M, Nomura M. Chromosomal locations of the genes for rRNA in Escherichia coli K-12. J Bacteriol. 1982;149:458–468. doi: 10.1128/jb.149.2.458-468.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1147.Elseviers D, Petrullo L A, Gallagher P. Novel E. coli mutants deficient in biosynthesis of 5-methylaminomethyl-2-thiouridine. Nucleic Acids Res. 1984;12:3521–3533. doi: 10.1093/nar/12.8.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1148.Elseviers D, Gallagher P, Hoffman A, Weinberg B, Schwartz I. Molecular cloning and regulation of expression of the genes for initiation factor 3 and two aminoacyl-tRNA synthetases. J Bacteriol. 1982;152:357–362. doi: 10.1128/jb.152.1.357-362.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1149.Elvin C M, Hardy C M, Rosenberg H. Pi exchange mediated by the GlpT-dependent sn-glycerol-3-phosphate transport system in Escherichia coli. J Bacteriol. 1985;161:1054–1058. doi: 10.1128/jb.161.3.1054-1058.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1150.Elvin C M, Dixon N E, Rosenberg H. Molecular cloning of the phosphate (inorganic) transport (pit) gene of Escherichia coli K-12: identification of the pit+ gene product and physical mapping of the pit-gor region. Mol Gen Gen. 1986;204:477–484. doi: 10.1007/BF00331028. [DOI] [PubMed] [Google Scholar]
- 1151.Emmerich R V, Hirshfield I N. Mapping of the constitutive lysyl-tRNA synthetase gene of Escherichia coli K-12. J Bacteriol. 1987;169:5311–5313. doi: 10.1128/jb.169.11.5311-5313.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1152.Emr S D, Hedgepeth J, Clement J M, Silhavy T J, Hofnung M. Sequence analysis of mutations that prevent export of λ receptor, an Escherichia coli outer membrane protein. Nature. 1980;285:82–85. doi: 10.1038/285082a0. [DOI] [PubMed] [Google Scholar]
- 1153.Emr S D, Hanley-Way S, Silhavy T J. Suppressor mutations that restore export of a protein with a defective signal sequence. Cell. 1981;23:79–88. doi: 10.1016/0092-8674(81)90272-5. [DOI] [PubMed] [Google Scholar]
- 1154.Engel H, Smink A J, van Wijngaarden L, Keck W. Murein-metabolizing enzymes from Escherichia coli: existence of a second lytic transglycosylase. J Bacteriol. 1992;174:6394–6403. doi: 10.1128/jb.174.20.6394-6403.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1155.Engel H, Kazemier B, Keck W. Murein-metabolizing enzymes from Escherichia coli: sequence analysis and controlled overexpression of the slt gene, which encodes the soluble lytic transglycosylase. J Bacteriol. 1991;173:6773–6782. doi: 10.1128/jb.173.21.6773-6782.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1156.Engelhorn M, Boccard F, Murtin C, Prentki P, Geiselmann J. In vivo interaction of the Escherichia coli integration host factor with its specific binding sites. Nucleic Acids Res. 1995;23:2959–2965. [PubMed] [Google Scholar]
- 1157.Enger-Valk B, Heyneker H, Oosterbaan R A, Pouwels P H. Construction of new cloning vehicles with genes of the tryptophan operon of Escherichia coli as genetic markers. Gene. 1980;9:69–85. doi: 10.1016/0378-1119(80)90167-5. [DOI] [PubMed] [Google Scholar]
- 1158.Englesberg E, Irr J, Power J, Lee N. Positive control of enzyme synthesis by gene C in the l-arabinose system. J Bacteriol. 1965;90:946–957. doi: 10.1128/jb.90.4.946-957.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1159.Enomoto M, Oosawa K, Momota H. Mapping of the pin locus coding for a site-specific recombinase that causes flagellar-phase variation in Escherichia coli K-12. J Bacteriol. 1983;156:663–668. doi: 10.1128/jb.156.2.663-668.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1160.Ephrati-Elizur E. A mutation in a new gene of Escherichia coli, psu, requires secondary mutations for survival: psu mutants express a pleiotropic suppressor phenotype. J Bacteriol. 1993;175:207–213. doi: 10.1128/jb.175.1.207-213.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1161.Epstein W, Kim B S. Potassium transport loci in Escherichia coli K-12. J Bacteriol. 1971;108:639–644. doi: 10.1128/jb.108.2.639-644.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1162.Epstein W, Buurman E, McLaggan D, Naprstek J. Multiple mechanisms, roles and controls of K+ transport in Escherichia coli. Biochem Soc Trans. 1993;21:1006–1010. doi: 10.1042/bst0211006. [DOI] [PubMed] [Google Scholar]
- 1163.Epstein W, Jewett S, Fox C F. Isolation and mapping of phosphotransferase mutants in Escherichia coli. J Bacteriol. 1970;104:793–797. doi: 10.1128/jb.104.2.793-797.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1163a.Epstein, W., et al. 1996. GenBank submission X97282.
- 1164.Eriani G, Dirheimer G, Gangloff J. Isolation and characterization of the gene coding for Escherichia coli arginyl-tRNA synthetase. Nucleic Acids Res. 1989;17:5725–5736. doi: 10.1093/nar/17.14.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1165.Eriani G, Dirheimer G, Gangloff J. Aspartyl-tRNA synthetase from Escherichia coli: cloning and characterisation of the gene, homologies of its translated amino acid sequence with asparaginyl- and lysyl-tRNA synthetases. Nucleic Acids Res. 1990;18:7109–7118. doi: 10.1093/nar/18.23.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1166.Erie D A, Hajiseyedjavadi O, Young M C, von Hippel P H. Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science. 1993;262:867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- 1167.Eriksson-Grennberg K G, Boman H G, Jansson J A T, Thoren S. Resistance of Escherichia coli to penicillins. I. Genetic study of some ampicillin mutants. J Bacteriol. 1965;90:54–62. doi: 10.1128/jb.90.1.54-62.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1168.Eriksson-Grennberg K G, Nordstrom K, Englund P. Resistance of Escherichia coli to penicillins. IX. Genetics and physiology of class II ampicillin-resistant mutants that are galactose negative or sensitive to bacteriophage C21, or both. J Bacteriol. 1971;108:1210–1223. doi: 10.1128/jb.108.3.1210-1223.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1169.Erni B, Zanolari B. The mannose-permease of the bacterial phosphotransferase system. Gene cloning and purification of the enzyme IIMan/IIIMan complex of Escherichia coli. J Biol Chem. 1985;260:15495–15503. [PubMed] [Google Scholar]
- 1170.Erni B, Zanolari B. Glucose-permease of the bacterial phosphotransferase system. Gene cloning, overproduction and amino acid sequence of enzyme IIGlc. J Biol Chem. 1986;261:16398–16403. [PubMed] [Google Scholar]
- 1171.Erni B, Zanolari B, Kocher H P. The mannose permease of Escherichia coli consists of three different proteins: amino acid sequence and function in sugar transport, sugar phosphorylation, and penetration of phage λ DNA. J Biol Chem. 1987;262:5238–5247. [PubMed] [Google Scholar]
- 1172.Ernsting B R, Denninger J W, Blumenthal R M, Matthews R G. Regulation of the gltBDF operon of Escherichia coli: how is a leucine-insensitive operon regulated by the leucine-responsive regulatory protein? J Bacteriol. 1993;175:7160–7169. doi: 10.1128/jb.175.22.7160-7169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1173.Erpel T, Hwang P, Craik C S, Fletterick R J, McGrath M E. Physical map location of the new Escherichia coli gene eco, encoding the serine protease inhibitor ecotin. J Bacteriol. 1992;174:1704. doi: 10.1128/jb.174.5.1704.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1174.Escarceller M, Hicks J, Gudmundsson G, Trump G, Touati D, Lovett S T, Foster P L, McEntee K, Goodman M F. Involvement of Escherichia coli DNA polymerase II in response to oxidative damage and adaptive mutation. J Bacteriol. 1994;176:6221–6228. doi: 10.1128/jb.176.20.6221-6228.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1175.Eschenbrenner M, Coves J, Fontecave M. NADPH-sulfite reductase flavoprotein from Escherichia coli: contribution to the flavin content and subunit interaction. FEBS Lett. 1995;374:82–84. doi: 10.1016/0014-5793(95)01081-o. [DOI] [PubMed] [Google Scholar]
- 1176.Esmon B E, Kensil C R, Cheng C C, Glaser M. Genetic analysis of Escherichia coli mutants defective in adenylate kinase and sn-glycerol 3-phosphate acyltransferase. J Bacteriol. 1980;141:405–408. doi: 10.1128/jb.141.1.405-408.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1177.Espion D, Kaiser C, Dambly-Chaudiere C. A third defective lambdoid prophage of Escherichia coli K12 defined by the λ derivative. J Mol Biol. 1983;170:611–633. doi: 10.1016/s0022-2836(83)80124-7. [DOI] [PubMed] [Google Scholar]
- 1178.Essenberg R C. Use of homocysteic acid for selecting mutants at the gltS locus of Escherichia coli K12. J Gen Microbiol. 1984;130:1311–1314. doi: 10.1099/00221287-130-6-1311. [DOI] [PubMed] [Google Scholar]
- 1179.Essenberg R C, Kornberg H L. Location of the gene for hexose phosphate transport (uhp) on the chromosome of Escherichia coli. J Gen Microbiol. 1977;99:157–169. doi: 10.1099/00221287-99-1-157. [DOI] [PubMed] [Google Scholar]
- 1180.Estevenon A M, Kooistra J, Sicard N. An Escherichia coli strain deficient for both exonuclease V and deoxycytidine triphosphate deaminase shows enhanced sensitivity to ionizing radiation. Mol Gen Genet. 1995;246:514–518. doi: 10.1007/BF00290455. [DOI] [PubMed] [Google Scholar]
- 1181.Evans R, Seeley N R, Kuempel P. Loss of rac locus DNA in merozygotes of Escherichia coli K12. Mol Gen Genet. 1979;175:245–250. doi: 10.1007/BF00397223. [DOI] [PubMed] [Google Scholar]
- 1182.Evans S, Dennis P P. Promoter activity and transcript mapping in the regulatory region for genes encoding ribosomal protein S15 and polynucleotide phosphorylase of Escherichia coli. Gene. 1985;40:15–22. doi: 10.1016/0378-1119(85)90019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1183.Eveland S S, Pompliano D L, Anderson M S. Conditionally lethal Escherichia coli murein mutants contain point defects that map to regions conserved among murein and folyl poly-gamma-glutamate ligases: identification of a ligase superfamily. Biochemistry. 1997;36:6223–6229. doi: 10.1021/bi9701078. [DOI] [PubMed] [Google Scholar]
- 1184.Evensen G, Seeberg E. Adaptation to alkylation resistance involves the induction of a DNA glycosylase. Nature. 1982;296:773–775. doi: 10.1038/296773a0. [DOI] [PubMed] [Google Scholar]
- 1185.Ewart C D, Jude D A, Thain J L, Nichols W W. Frequency and mechanism of resistance to antibacterial action of ZM240401, (6S)-6-fluoro-shikimic acid. Antimicrob Agents Chemother. 1995;39:87–93. doi: 10.1128/aac.39.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1186.Ezaki B, Mori H, Ogura T, Hiraga S. Possible involvement of the ugpA gene product in the stable maintenance of mini-F plasmid in Escherichia coli. Mol Gen Genet. 1990;223:361–368. doi: 10.1007/BF00264441. [DOI] [PubMed] [Google Scholar]
- 1187.Ezaki B, Ogura T, Mori H, Niki H, Hiraga S. Involvement of DnaK protein in mini-F plasmid replication: temperature-sensitive seg mutations are located in the dnaK gene. Mol Gen Genet. 1989;218:183–189. doi: 10.1007/BF00331267. [DOI] [PubMed] [Google Scholar]
- 1188.Ezaki B, Ogura T, Niki H, Hiraga S. Partitioning of a mini-F plasmid into anucleate cells of the mukB null mutant. J Bacteriol. 1991;173:6643–6646. doi: 10.1128/jb.173.20.6643-6646.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1189.Ezekiel D H. False feedback inhibition of aromatic amino acid biosynthesis by B-2 thienyl-alanine. Biochim Biophys Acta. 1965;95:54–62. doi: 10.1016/0005-2787(65)90210-8. [DOI] [PubMed] [Google Scholar]
- 1190.Ezekiel D H, Hutchins J E. Mutations affecting RNA polymerase associated with rifampicin resistance in Escherichia coli. Nature. 1968;220:276–277. doi: 10.1038/220276a0. [DOI] [PubMed] [Google Scholar]
- 1191.Faatz E, Middendorf A, Bremer E. Cloned structural genes for the osmotically regulated binding-protein-dependent glycine betaine transport system (ProU) of Escherichia coli K-12. Mol Microbiol. 1988;2:265–279. doi: 10.1111/j.1365-2958.1988.tb00028.x. [DOI] [PubMed] [Google Scholar]
- 1192.Fabianek R A, Hennecke H, Thony-Meyer L. The active-site cysteines of the periplasmic thioredoxin-like protein CcmG of Escherichia coli are important but not essential for cytochrome c maturation in vivo. J Bacteriol. 1998;180:1947–1950. doi: 10.1128/jb.180.7.1947-1950.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1193.Fabiny J M, Jayakumar A, Chinault A C, Barnes E M., Jr Ammonium transport in Escherichia coli: localization and nucleotide sequence of the amtA gene. J Gen Microbiol. 1991;137:983–989. doi: 10.1099/00221287-137-4-983. [DOI] [PubMed] [Google Scholar]
- 1194.Faik P, Kornberg H L. Isolation and properties of E. coli mutants affected in gluconate uptake. FEBS Lett. 1973;32:260–263. doi: 10.1016/0014-5793(73)80847-6. [DOI] [PubMed] [Google Scholar]
- 1194a.Fajarto D A, Misra R. Escherichia coli porin (ompG) gene, complete cds. 1997. GenBank submission U49400. [Google Scholar]
- 1195.Falconi M, Brandi A, La Teana A, Gualerzi C O, Pon C L. Antagonistic involvement of FIS and H-NS proteins in the transcriptional control of hns expression. Mol Microbiol. 1996;19:965–975. doi: 10.1046/j.1365-2958.1996.436961.x. [DOI] [PubMed] [Google Scholar]
- 1196.Falkinham J O. Identification of a mutation affecting an alanine-α-ketoisovalerate transaminase activity in Escherichia coli K-12. Mol Gen Genet. 1979;176:147–149. doi: 10.1007/BF00334306. [DOI] [PubMed] [Google Scholar]
- 1197.Falk-Krzesinski H J, Wolfe A J. Genetic analysis of the nuo locus, which encodes the proton-translocating NADH dehydrogenase in Escherichia coli. J Bacteriol. 1998;180:1174–1184. doi: 10.1128/jb.180.5.1174-1184.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1198.Fandl J. Protein translocation in vitro: biochemical characterization of genetically defined translocation components. J Bioenerg Biomembr. 1990;22:369–387. doi: 10.1007/BF00763173. [DOI] [PubMed] [Google Scholar]
- 1199.Fang L, Jiang W, Bae W, Inouye M. Promoter-independent cold-shock induction of cspA and its derepression at 37 degrees C by mRNA stabilization. Mol Microbiol. 1997;23:355–364. doi: 10.1046/j.1365-2958.1997.2351592.x. [DOI] [PubMed] [Google Scholar]
- 1200.Fang L, Hou Y, Inouye M. Role of the cold-box region in the 5′ untranslated region of the cspA mRNA in its transient expression at low temperature in Escherichia coli. J Bacteriol. 1998;180:90–95. doi: 10.1128/jb.180.1.90-95.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1201.Fangman W L, Neidhardt F C. Demonstration of an altered aminoacyl ribonucleic acid synthetase in a mutant of Escherichia coli. J Biol Chem. 1964;239:1839–1843. [PubMed] [Google Scholar]
- 1202.Fani R, Lio P, Chiarelli I, Bazzicalupo M. The evolution of the histidine biosynthetic genes in prokaryotes: a common ancestor for the hisA and hisF genes. J Mol Evol. 1994;38:489–495. doi: 10.1007/BF00178849. [DOI] [PubMed] [Google Scholar]
- 1203.Farabaugh P J. Sequence of the lacI gene. Nature. 1978;274:765–769. doi: 10.1038/274765a0. [DOI] [PubMed] [Google Scholar]
- 1204.Farewell A, Diez A A, DiRusso C C, Nystrom T. Role of the Escherichia coli FadR regulator in stasis survival and growth phase-dependent expression of the uspA, fad, and fab genes. J Bacteriol. 1996;178:6443–6450. doi: 10.1128/jb.178.22.6443-6450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1205.Farr S B, Arnosti D N, Chamberlin M J, Ames B N. An apaH mutation causes AppppA to accumulate and affects motility and catabolite repression in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:5010–5014. doi: 10.1073/pnas.86.13.5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1206.Farris M, Grant A J, Richardson T B, O’Connor C D. BipA: a tyrosine-phosphorylated GTPase that mediates interactions between enteropathogenic Escherichia coli (EPEC) and epithelial cells. Mol Microbiol. 1998;28:265–279. doi: 10.1046/j.1365-2958.1998.00793.x. [DOI] [PubMed] [Google Scholar]
- 1207.Fath M J, Mahanty H K, Kolter R. Characterization of a purF operon mutation which affects colicin V production. J Bacteriol. 1989;171:3158–3161. doi: 10.1128/jb.171.6.3158-3161.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1208.Faubladier M, Bouché J P. Division inhibition gene dicF of Escherichia coli reveals a widespread group of prophage sequences in bacterial genomes. J Bacteriol. 1994;176:1150–1156. doi: 10.1128/jb.176.4.1150-1156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1209.Faubladier M, Cam K, Bouché J P. Escherichia coli cell division inhibitor DicF-RNA of the dicB operon. Evidence for its generation in vivo by transcription termination and by RNase III and RNase E-dependent processing. J Mol Biol. 1990;212:461–471. doi: 10.1016/0022-2836(90)90325-G. [DOI] [PubMed] [Google Scholar]
- 1210.Favre A, Chams V, Caldeira de Araujo A. Photosensitized UVA light induction of the SOS response of Escherichia coli. Biochimie. 1986;68:857–864. doi: 10.1016/s0300-9084(86)80101-8. [DOI] [PubMed] [Google Scholar]
- 1211.Fawcett W P, Wolf R E. Genetic definition of the Escherichia coli zwf “Soxbox,” the DNA binding site for SoxS-mediated induction of glucose 6-phosphate dehydrogenase in response to superoxide. J Bacteriol. 1995;177:1742–1750. doi: 10.1128/jb.177.7.1742-1750.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1212.Faxen M, Walles-Granberg A, Isaksson L A. Antisuppression by a mutation in rpsM(S13) giving a shortened ribosomal protein S13. Biochim Biophys Acta. 1994;1218:27–34. doi: 10.1016/0167-4781(94)90097-3. [DOI] [PubMed] [Google Scholar]
- 1213.Fayat G, Mayaux J-F, Sacerdot C, Fromant M, Springer M, Grunberg-Manago M, Blanquet S. Escherichia coli phenylalanyl-tRNA synthetase operon region: evidence for an attenuation mechanism. Identification of the gene for the ribosomal protein. J Mol Biol. 1983;171:239–261. doi: 10.1016/0022-2836(83)90092-x. [DOI] [PubMed] [Google Scholar]
- 1214.Fayerman J T, Vann M C, Williams L S, Umbarger H E. ilvU, a locus in Escherichia coli affecting the derepression of isoleucyl-tRNA synthetase in the RPC-5 chromatographic profiles of tRNAIle and tRNAVal. J Biol Chem. 1979;254:9429–9440. [PubMed] [Google Scholar]
- 1215.Fayet O, Ziegelhoffer T, Georgopoulos C. The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J Bacteriol. 1989;171:1379–1385. doi: 10.1128/jb.171.3.1379-1385.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1216.Fecker L, Braun V. Cloning and expression of the fhu genes involved in iron(III)-hydroxamate uptake by Escherichia coli. J Bacteriol. 1983;156:1301–1314. doi: 10.1128/jb.156.3.1301-1314.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1217.Fee J A. Regulation of sod genes in Escherichia coli: relevance to superoxide dismutase function. Mol Microbiol. 1991;5:2599–2610. doi: 10.1111/j.1365-2958.1991.tb01968.x. [DOI] [PubMed] [Google Scholar]
- 1218.Felden B, Himeno H, Muto A, Atkins J F, Gesteland R F. Structural organization of Escherichia coli tmRNA. Biochimie. 1996;78:979–983. doi: 10.1016/s0300-9084(97)86720-x. [DOI] [PubMed] [Google Scholar]
- 1219.Feller A, Pierard A, Glansdorff N, Charlier D, Crabeel M. Mutation of gene encoding regulatory polypeptide of aspartate carbamoyltransferase. Nature. 1981;292:370–373. doi: 10.1038/292370a0. [DOI] [PubMed] [Google Scholar]
- 1220.Felzenszwalb I, Sargentini N J, Smith K C. Characterization of a new radiation-sensitive mutant, Escherichia coli K-12 radC102. Radiat Res. 1984;97:615–625. [PubMed] [Google Scholar]
- 1221.Felzenszwalb I, Boiteux S, Laval J. Molecular cloning and DNA sequencing of the radC gene of Escherichia coli K-12. Mutat Res. 1992;273:263–269. doi: 10.1016/0921-8777(92)90088-k. [DOI] [PubMed] [Google Scholar]
- 1222.Feng G, Winkler M E. Single-step purifications of His6-MutH, His6-MutL and His6-MutS repair proteins of Escherichia coli K-12. BioTechniques. 1995;19:956–965. [PubMed] [Google Scholar]
- 1223.Feng J, Yamanaka K, Niki H, Ogura T, Hiraga S. New killing system controlled by two gene located immediately upstream of the mukB gene in Escherichia coli. Mol Gen Genet. 1994;243:136–147. doi: 10.1007/BF00280310. [DOI] [PubMed] [Google Scholar]
- 1224.Ferenci T. Methyl-α-maltoside and 5-thiomaltose: analogs transported by the Escherichia coli maltose transport system. J Bacteriol. 1980;144:7–11. doi: 10.1128/jb.144.1.7-11.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1225.Ferguson G P, Chacko A D, Lee C, Booth I R. The activity of the high-affinity K+ uptake system Kdp sensitizes cells of Escherichia coli to methylglyoxal J. Bacteriol. 1996;178:3957–3961. doi: 10.1128/jb.178.13.3957-3961.1996. . (Erratum, 179:568, 1997.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1226.Ferguson G P, Creighton R I, Nikolaev Y, Booth I R. Importance of RpoS and Dps in survival of exposure of both exponential- and stationary-phase Escherichia coli cells to the electrophile N-ethylmaleimide. J Bacteriol. 1998;180:1030–1036. doi: 10.1128/jb.180.5.1030-1036.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1227.Fernley R T, Lentz S R, Bradshaw R A. Malate dehydrogenase isolation from Escherichia coli and comparison with the eukaryotic mitochondrial and cytoplasmic forms. Biosci Rep. 1981;1:497–507. doi: 10.1007/BF01121583. [DOI] [PubMed] [Google Scholar]
- 1228.Ferone R, Singer S C, Hanlon M H, Roland S. Isolation and characterization of an E. coli mutant affected in dihydrofolate- and folylpolyglutamate-synthetase. In: Blair J, editor. Chemistry and biology of pteridines. New York, N.Y: W. de Gruyter; 1983. pp. 585–589. [Google Scholar]
- 1229.Ferrandez A, Garcia J L, Diaz E. Genetic characterization and expression in heterologous hosts of the 3-(3-hydroxyphenyl)propionate catabolic pathway of Escherichia coli K-12. J Bacteriol. 1997;179:2573–2581. doi: 10.1128/jb.179.8.2573-2581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1230.Ferrandez A, Prieto M A, Garcia J L, Diaz E. Molecular characterization of PadA, a phenylacetaldehyde dehydrogenase from Escherichia coli. FEBS Lett. 1997;406:23–27. doi: 10.1016/s0014-5793(97)00228-7. [DOI] [PubMed] [Google Scholar]
- 1231.Ferro-Novick S, Honma M, Beckwith J R. The product of gene secC is involved in the synthesis of exported proteins in E. coli. Cell. 1984;38:211–217. doi: 10.1016/0092-8674(84)90542-7. [DOI] [PubMed] [Google Scholar]
- 1232.Feulner G, Gray J A, Kirschman J A, Lehner A F, Sadosky A B, Vlazny D A, Zhang J, Zhao S, Hill C W. Structure of the rhsA locus from Escherichia coli K-12 and comparison of rhsA with other members of the rhs multigene family. J Bacteriol. 1990;172:446–456. doi: 10.1128/jb.172.1.446-456.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1233.Feutrier J, Lepelletier M, Pascal M C, Chippaux M. Tn10 insertions directed in the pyrD-serC region and improved mapping of pepN in Escherichia coli K12. Mol Gen Genet. 1982;185:518–519. doi: 10.1007/BF00334153. [DOI] [PubMed] [Google Scholar]
- 1234.Fiedler W, Rotering H. Characterization of an Escherichia coli mdoB mutant strain unable to transfer sn-1-phosphoglycerol to membrane-derived oligosaccharides. J Biol Chem. 1985;260:4799–4806. [PubMed] [Google Scholar]
- 1235.Fiedler W, Rotering H. Properties of Escherichia coli mutants lacking membrane-derived oligosaccharides. J Biol Chem. 1988;263:14684–14689. [PubMed] [Google Scholar]
- 1236.Figge R M, Ramseier T M, Saier M H. The mannitol repressor (MtlR) of Escherichia coli. J Bacteriol. 1994;176:840–847. doi: 10.1128/jb.176.3.840-847.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1237.Fiil N, Friesen D. Isolation of “relaxed” mutants of Escherichia coli. J Bacteriol. 1968;95:729–731. doi: 10.1128/jb.95.2.729-731.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1238.Fiil N, Friesen J D, Downing W L, Dennis P P. Post-transcriptional regulatory mutants in a ribosomal protein-RNA polymerase operon of E. coli. Cell. 1980;19:837–844. doi: 10.1016/0092-8674(80)90074-4. [DOI] [PubMed] [Google Scholar]
- 1239.Fijalkowska I J, Dunn R L, Schaaper R M. Genetic requirements and mutational specificity of the Escherichia coli SOS mutator activity. J Bacteriol. 1997;179:7435–7445. doi: 10.1128/jb.179.23.7435-7445.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1240.Fijalkowska I J, Schaaper R M. Effects of Escherichia coli dnaE antimutator alleles in a proofreading-deficient mutD5 strain. J Bacteriol. 1995;177:5979–5986. doi: 10.1128/jb.177.20.5979-5986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1241.Fikes J D, Bassford P J. Novel secA alleles improve export of maltose-binding protein synthesized with a defective signal peptide. J Bacteriol. 1989;171:402–409. doi: 10.1128/jb.171.1.402-409.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1242.Filpula D. Regulation of ribonucleoside diphosphate reductase synthesis in Escherichia coli: specific activity of the enzyme in relationship to perturbations of DNA replication. J Bacteriol. 1978;135:429–435. doi: 10.1128/jb.135.2.429-435.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1243.Filpula D, Fuchs J A. Regulation of ribonucleoside diphosphate reductase synthesis in Escherichia coli: increased enzyme synthesis as a result of inhibition of deoxyribonucleic acid synthesis. J Bacteriol. 1977;130:107–113. doi: 10.1128/jb.130.1.107-113.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1244.Filutowicz M, Ross W, Wild J R, Gourse R L. Involvement of Fis protein in replication of the Escherichia coli chromosome. J Bacteriol. 1992;174:398–407. doi: 10.1128/jb.174.2.398-407.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1245.Fimmel A L, Haddock B A. Use of chlC-lac fusions to determine regulation of gene chlC in Escherichia coli K-12. J Bacteriol. 1979;138:726–730. doi: 10.1128/jb.138.3.726-730.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1246.Fimmel A L, Haddock B A. Characterization of the Escherichia coli chlC regulatory region in a cloned chlC-lac gene fusion. FEMS Microbiol Lett. 1981;12:125–129. [Google Scholar]
- 1247.Fimmel A L, Loughlin R E. Isolation and characterization of cysK mutants of Escherichia coli. J Gen Microbiol. 1977;103:37–43. doi: 10.1099/00221287-103-1-37. [DOI] [PubMed] [Google Scholar]
- 1248.Finch P W, Storey A, Brown K, Hickson I D, Emmerson P T. Complete nucleotide sequence of recD, the structural gene for the alpha subunit of exonuclease V of Escherichia coli. Nucleic Acids Res. 1986;14:8583–8594. doi: 10.1093/nar/14.21.8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1249.Finch P W, Storey A, Chapman K E, Brown K, Hickson I D, Emmerson P T. Complete nucleotide sequence of the Escherichia coli recB gene. Nucleic Acids Res. 1986;14:8573–8582. doi: 10.1093/nar/14.21.8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1250.Finch P W, Emmerson P T. Nucleotide sequence of the regulatory region of the uvrD gene of Escherichia coli. Gene. 1983;25:317–323. doi: 10.1016/0378-1119(83)90236-6. [DOI] [PubMed] [Google Scholar]
- 1251.Finch P W, Emmerson P T. The nucleotide sequence of the uvrD gene of E. coli. Nucleic Acids Res. 1984;12:5789–5799. doi: 10.1093/nar/12.14.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1252.Finch P W, Wilson R E, Brown K, Hickson I D, Tomlinson A E, Emmerson P T. Complete nucleotide sequence of the Escherichia coli recC gene and of the thyA-recC intergenic region. Nucleic Acids Res. 1986;14:4437–4451. doi: 10.1093/nar/14.11.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1253.Finch P W, Wilson R E, Brown K, Hickson I D, Emmerson P T. Complete nucleotide sequence of Escherichia coli ptr gene encoding protease III. Nucleic Acids Res. 1986;14:7695–7703. doi: 10.1093/nar/14.19.7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1254.Fischer E, Gunter K, Braun V. Involvement of ExbB and TonB in transport across the outer membrane of Escherichia coli: phenotypic complementation of exb mutants by overexpressed tonB and physical stabilization of TonB by ExbB. J Bacteriol. 1989;171:5127–5134. doi: 10.1128/jb.171.9.5127-5134.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1255.Fischer M, Short S A. The cloning of the Escherichia coli K-12 deoxyribonucleoside operon. Gene. 1982;17:291–298. doi: 10.1016/0378-1119(82)90145-7. [DOI] [PubMed] [Google Scholar]
- 1256.Fischl A S, Kennedy E P. Isolation and properties of acyl carrier protein phosphodiesterase of Escherichia coli. J Bacteriol. 1990;172:5445–5449. doi: 10.1128/jb.172.9.5445-5449.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1257.Flachmann R, Kunz N, Seifert J, Gutlich M, Wientjes F J, Laufer A, Gassen H G. Molecular biology of pyridine nucleotide biosynthesis in Escherichia coli. Cloning and characterization of quinolinate synthesis genes nadA and nadB. Eur J Biochem. 1988;175:221–228. doi: 10.1111/j.1432-1033.1988.tb14187.x. [DOI] [PubMed] [Google Scholar]
- 1258.Flamm E L, Weisberg R. Primary structure of the hip gene of Escherichia coli and of its product, the β subunit of integration host factor. J Mol Biol. 1983;183:117–128. doi: 10.1016/0022-2836(85)90206-2. [DOI] [PubMed] [Google Scholar]
- 1259.Flamm J A, Friesen J D, Otsuka A J. The nucleotide sequence of the Escherichia coli rts gene. Gene. 1988;74:555–558. doi: 10.1016/0378-1119(88)90189-8. [DOI] [PubMed] [Google Scholar]
- 1260.Flannigan K A, Hennigan S H, Vogelbacker H H, Gots J S, Smith J M. Purine biosynthesis in Escherichia coli K12: structure and DNA sequence studies of the purHD locus. Mol Microbiol. 1990;4:381–392. doi: 10.1111/j.1365-2958.1990.tb00605.x. [DOI] [PubMed] [Google Scholar]
- 1261.Fleming T P, Nahlik M S, Neilands J B, McIntosh M A. Physical and genetic characterization of cloned enterobactin genomic sequences from Escherichia coli K-12. Gene. 1985;34:47–54. doi: 10.1016/0378-1119(85)90293-8. [DOI] [PubMed] [Google Scholar]
- 1262.Fleming T P, Nahlik M S, McIntosh M A. Regulation of enterobactin iron transport in Escherichia coli: characterization of ent::Mu d(Aprlac) operon fusions. J Bacteriol. 1983;156:1171–1177. doi: 10.1128/jb.156.3.1171-1177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1263.Flores A, Casadesus J. Suppression of the pleiotropic effects of HisH and HisF overproduction identifies four novel loci on the Salmonella typhimurium chromosome: osmH, sfiW, sfiX, and sfiY. J Bacteriol. 1995;177:4841–4850. doi: 10.1128/jb.177.17.4841-4850.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1264.Flower A M, McHenry C S. The adjacent dnaZ and dnaX genes of Escherichia coli are contained within one continuous open reading frame. Nucleic Acids Res. 1986;14:8091–8101. doi: 10.1093/nar/14.20.8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1265.Flower A M, McHenry C S. The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc Natl Acad Sci USA. 1990;87:3713–3717. doi: 10.1073/pnas.87.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1266.Flower A M, McHenry C S. Transcriptional organization of the Escherichia coli dnaX gene. J Mol Biol. 1991;220:649–658. doi: 10.1016/0022-2836(91)90107-h. [DOI] [PubMed] [Google Scholar]
- 1267.Flower A M, Doebele R C, Silhavy T J. PrlA and PrlG suppressors reduce the requirement for signal sequence recognition. J Bacteriol. 1994;176:5607–5614. doi: 10.1128/jb.176.18.5607-5614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1268.Fogliano M, Schendel P F. Evidence for the inducibility of the uvrB operon. Nature. 1981;289:196–198. doi: 10.1038/289196a0. [DOI] [PubMed] [Google Scholar]
- 1269.Foglino M, Gharbi S, Lazdunski A. Nucleotide sequence of the pepN encoding aminopeptidase N of Escherichia coli. Gene. 1986;49:303–309. doi: 10.1016/0378-1119(86)90366-5. [DOI] [PubMed] [Google Scholar]
- 1270.Fognini-Lefebvre N, Lazzarone J C, Portalier R. tolA, tolB and excC, three cistrons involved in the control of pleiotropic release of periplasmic proteins by Escherichia coli K12. Mol Gen Genet. 1987;209:391–395. doi: 10.1007/BF00329670. [DOI] [PubMed] [Google Scholar]
- 1271.Fong S T, Camakaris J, Lee B T. Molecular genetics of a chromosomal locus involved in copper tolerance in Escherichia coli K-12. Mol Microbiol. 1995;15:1127–1137. doi: 10.1111/j.1365-2958.1995.tb02286.x. [DOI] [PubMed] [Google Scholar]
- 1272.Fonstein M, Nikolskaya T, Zaporojets D, Nikolsky Y, Kulakauskas S T, Mironov A. Tn10-mediated inversions fuse uridine phosphorylase (udp) and rRNA genes of Escherichia coli. J Bacteriol. 1994;176:2265–2271. doi: 10.1128/jb.176.8.2265-2271.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1273.Fontecave M, Coves J, Pierre J L. Ferric reductases or flavin reductases? Biometals. 1994;7:3–8. doi: 10.1007/BF00205187. [DOI] [PubMed] [Google Scholar]
- 1274.Forchhammer K, Rucknagel K P, Bock A. Purification and biochemical characterization of SELB, a translation factor involved in selenoprotein synthesis. J Biol Chem. 1990;265:9346–9350. [PubMed] [Google Scholar]
- 1275.Forchhammer K, Leinfelder W, Bock A. Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature. 1989;342:453–456. doi: 10.1038/342453a0. [DOI] [PubMed] [Google Scholar]
- 1276.Forchhammer K, Leinfelder W, Boesmiller K, Veprek B, Bock A. Selenocysteine synthase from Escherichia coli. Nucleotide sequence of the gene (selA) and purification of the protein. J Biol Chem. 1991;266:6318–6323. [PubMed] [Google Scholar]
- 1277.Forster J W, Strike P. Organization and control of the Escherichia coli uvrC gene. Gene. 1985;35:71–82. doi: 10.1016/0378-1119(85)90159-3. [DOI] [PubMed] [Google Scholar]
- 1278.Forster J W, Strike P. Analysis of the regulatory elements of the Escherichia coli uvrC gene by construction of operon fusions. Mol Gen Genet. 1988;211:531–537. doi: 10.1007/BF00425712. [DOI] [PubMed] [Google Scholar]
- 1279.Fortin Y, Phoenix P, Drapeau G R. Mutations conferring resistance to azide in Escherichia coli occur primarily in the secA gene. J Bacteriol. 1990;172:6607–6610. doi: 10.1128/jb.172.11.6607-6610.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1280.Foster J W, Park Y K, Penfound T, Fenger T, Spector M P. Regulation of NAD metabolism in Salmonella typhimurium: molecular sequence analysis of the bifunctional nadR regulator and the nadA-pnuC operon. J Bacteriol. 1990;172:4187–4196. doi: 10.1128/jb.172.8.4187-4196.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1281.Foster P L, Trimarchi J M, Maurer R A. Two enzymes, both of which process recombination intermediates, have opposite effects on adaptive mutation in Escherichia coli. Genetics. 1996;142:25–37. doi: 10.1093/genetics/142.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1282.Foster P L, Rosche W A. Levels of the Vsr endonuclease do not regulate stationary-phase reversion of a Lac− frameshift allele in Escherichia coli. J Bacteriol. 1998;180:1944–1946. doi: 10.1128/jb.180.7.1944-1946.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1283.Fotheringham I G, Dacey S A, Taylor P P, Smith T J, Hunter M G, Finlay M E, Primrose S B, Parker D M, Edwards R M. The cloning and sequence analysis of the aspC and tyrB genes from Escherichia coli K-12. Biochem J. 1986;234:593–604. doi: 10.1042/bj2340593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1284.Foulds J, Barrett E L. Characterization of Escherichia coli mutants tolerant to bacteriocin JF246; two new classes of tolerant mutants. J Bacteriol. 1973;116:885–892. doi: 10.1128/jb.116.2.885-892.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1285.Fournier M J, Ozeki H. Structure and organization of the transfer ribonucleic acid genes of Escherichia coli K-12. Microbiol Rev. 1985;49:379–397. doi: 10.1128/mr.49.4.379-397.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1286.Fouts K E, Barbour S D. Transductional mapping of ksgB and a new Tn5-induced kasugamycin resistance gene, ksgD, in Escherichia coli K-12. J Bacteriol. 1981;145:914–919. doi: 10.1128/jb.145.2.914-919.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1287.Fouts K E, Wasie-Gilbert T, Willis D K, Clark A J, Barbour S D. Genetic analysis of transposon-induced mutations of the Rac prophage in Escherichia coli K-12 which affect expression and function of recE. J Bacteriol. 1983;156:718–726. doi: 10.1128/jb.156.2.718-726.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1288.Fowler R G, Erickson J A, Isbell R J. Activity of the Escherichia coli mutT mutator allele in an anaerobic environment. J Bacteriol. 1994;176:7727–7729. doi: 10.1128/jb.176.24.7727-7729.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1289.Fradkin J E, Fraenkel D. 2-Keto-3-deoxygluconate 6-phosphate aldolase mutants of Escherichia coli. J Bacteriol. 1971;108:1277–1283. doi: 10.1128/jb.108.3.1277-1283.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1290.Fraenkel D, Banerjee S. Deletion mapping of zwf, the gene for a constitutive enzyme, glucose 6-phosphate dehydrogenase, in Escherichia coli. Genetics. 1972;71:481–489. doi: 10.1093/genetics/71.4.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1290a.Fraley C D, Kim J H, McCann M P, Matin A. Cloning and characterization of catC, and Escherichia coli starvation gene regulated by cAMP, RpoS, and RpoN and induced by the presence of l-ornithine or N-alpha-acetyl-l-ornithine. 1997. GenBank submission U80416. [Google Scholar]
- 1290b.Fraley C, Kim J H, McCann P M, Matin A. The Escherichia coli starvation gene, cstC, is involved in amino acid catabolism. J Bacteriol. 1998;180:32–38. doi: 10.1128/jb.180.16.4287-4290.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1291.Fralick J A. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1292.Francetic O, Pugsley A P. The cryptic general secretory pathway (gsp) operon of Escherichia coli K-12 encodes functional proteins. J Bacteriol. 1996;178:3544–3549. doi: 10.1128/jb.178.12.3544-3549.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1293.Francetic O, Hanson M P, Kumamoto C A. prlA supression of defective export of maltose-binding protein in secB mutants of Escherichia coli. J Bacteriol. 1993;175:4036–4044. doi: 10.1128/jb.175.13.4036-4044.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1293a.Franco, A. V., D. Liu, and P. R. Reeves. 1998. The Wzz (Cld) protein in Escherichia coli: amino acid sequence variation determines O-antigen chain length specificity. 180:2670–2675. [DOI] [PMC free article] [PubMed]
- 1294.Francois V, Louarn J, Louarn J M. The terminus of the Escherichia coli chromosome is flanked by several polar replication pause sites. Mol Microbiol. 1989;3:995–1002. doi: 10.1111/j.1365-2958.1989.tb00250.x. [DOI] [PubMed] [Google Scholar]
- 1295.Francoz E, Dassa E. 3′ end of the malEFG operon in E. coli: localization of the transcription termination site. Nucleic Acids Res. 1988;16:4097–4109. doi: 10.1093/nar/16.9.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1296.Frandsen N, D’Ari R. Excess histidine enzymes cause AICAR-independent filamentation in Escherichia coli. Mol Gen Genet. 1993;240:348–354. doi: 10.1007/BF00280385. [DOI] [PubMed] [Google Scholar]
- 1297.Frank E G, Ennis D G, Gonzalez M, Levine A S, Woodgate R. Regulation of SOS mutagenesis by proteolysis. Proc Natl Acad Sci USA. 1996;93:10291–10296. doi: 10.1073/pnas.93.19.10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1298.Franklin F C H, Venables W A, Wijsman H J W. Genetic studies of d-alanine-dehydrogenase-less mutants of Escherichia coli K-12. Genet Res. 1981;38:197–208. doi: 10.1017/s0016672300020528. [DOI] [PubMed] [Google Scholar]
- 1299.Franklin N C. Mutation in galU gene of E. coli blocks phage P1 infection. Virology. 1969;38:189–191. doi: 10.1016/0042-6822(69)90144-5. [DOI] [PubMed] [Google Scholar]
- 1300.Free A, Dorman C J. The Escherichia coli stpA gene is transiently expressed during growth in rich medium and is induced in minimal medium and by stress conditions. J Bacteriol. 1997;179:909–918. doi: 10.1128/jb.179.3.909-918.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1301.Freedman R, Gibson B, Donovan D, Biemann K, Eisenbeis S, Parker J, Schimmel P. Primary structure of histidine-tRNA synthetase and characterization of hisS transcripts. J Biol Chem. 1985;260:10063–10068. [PubMed] [Google Scholar]
- 1302.Freestone P, Nystrom T, Trinei M, Norris V. The universal stress protein, UspA, of Escherichia coli is phosphorylated in response to stasis. J Mol Biol. 1997;274:318–324. doi: 10.1006/jmbi.1997.1397. [DOI] [PubMed] [Google Scholar]
- 1303.Freimuth P I, Taylor J W, Kaiser E T. Introduction of guest peptides into Escherichia coli alkaline phosphatase. Excision and purification of a dynorphin analogue from an active chimeric protein. J Biol Chem. 1990;265:896–901. [PubMed] [Google Scholar]
- 1304.Freitag C S, Eisenstein B I. Genetic mapping and transcriptional orientation of the fimD gene. J Bacteriol. 1983;156:1052–1058. doi: 10.1128/jb.156.3.1052-1058.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1305.Freitag C S, Abraham J M, Clements J R, Eisenstein B I. Genetic analysis of the phase variation control of expression of type 1 fimbriae in Escherichia coli. J Bacteriol. 1985;162:668–675. doi: 10.1128/jb.162.2.668-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1306.Freundlieb S, Boos W. Alpha-amylase of Escherichia coli, mapping and cloning of the structural gene, malS, and identification of its product as a periplasmic protein. J Biol Chem. 1986;261:2946–2953. [PubMed] [Google Scholar]
- 1307.Frick D N, Townsend B D, Bessman M J. A novel GDP-mannose mannosyl hydrolase shares homology with the MutT family of enzymes. J Biol Chem. 1995;270:24086–24091. doi: 10.1074/jbc.270.41.24086. [DOI] [PubMed] [Google Scholar]
- 1308.Fricke J, Neuhard J, Kelln R A, Pedersen S. The cmk gene encoding cytidine monophosphate kinase is located in the rpsA operon and is required for normal replication rate in Escherichia coli. J Bacteriol. 1995;177:517–523. doi: 10.1128/jb.177.3.517-523.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1309.Friden P, Donegan J, Mullen J, Tsui P, Freundlich M, Eoyang L, Weber R, Silverman P M. The ilvB locus of Escherichia coli K-12 is an operon encoding both subunits of acetohydroxy acid synthase I. Nucleic Acids Res. 1985;13:3979–3993. doi: 10.1093/nar/13.11.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1310.Friden P, Newman T, Freundlich M. Nucleotide sequence of the ilvB promoter-regulatory region: a biosynthetic operon controlled by attenuation and cyclic AMP. Proc Natl Acad Sci USA. 1982;79:6156–6160. doi: 10.1073/pnas.79.20.6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1311.Friedberg I. Localization of phosphoglucose isomerase in Escherichia coli and its relation to the induction of the hexose phosphate transport system. J Bacteriol. 1972;112:1201–1205. doi: 10.1128/jb.112.3.1201-1205.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1312.Friedman D I. A bacterial mutant affecting lambda development. Cold Spring Harbor Symp Quant Biol. 1971;1:733–738. [Google Scholar]
- 1313.Friedman D I, Baumann M, Baron L S. Cooperative effects of bacterial mutations affecting λ N gene expression. I. Isolation and characterization of a nusB mutant. Virology. 1976;73:119–127. doi: 10.1016/0042-6822(76)90066-0. [DOI] [PubMed] [Google Scholar]
- 1314.Friedman S. Bactericidal effect of 5-azacytidine on Escherichia coli carrying EcoRII restriction-modification enzymes. J Bacteriol. 1982;151:262–268. doi: 10.1128/jb.151.1.262-268.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1315.Friedrich M J, DeVeaux L C, Kadner R J. Nucleotide sequence of the btuCED genes involved in vitamin B12 transport in Escherichia coli and homology with components of periplasmic-binding-protein-dependent transport systems. J Bacteriol. 1986;167:928–934. doi: 10.1128/jb.167.3.928-934.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1316.Friedrich M J, Kadner R J. Nucleotide sequence of the uhp region of Escherichia coli. J Bacteriol. 1987;169:3556–3563. doi: 10.1128/jb.169.8.3556-3563.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1317.Froelich B, Epstein W. Escherichia coli mutants in which transcription is dependent on recA function. J Bacteriol. 1981;147:1117–1120. doi: 10.1128/jb.147.3.1117-1120.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1318.Frohler J, Rechenmacher A, Thomale J, Nass G, Bock A. Genetic analysis of mutations causing borrelidin resistance by overproduction of threonyl-transfer ribonucleic acid synthetase. J Bacteriol. 1980;143:1135–1141. doi: 10.1128/jb.143.3.1135-1141.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1319.Froman B E, Tait R C, Gottlieb L D. Isolation and characterization of the phosphoglucose isomerase gene from Escherichia coli. Mol Gen Genet. 1989;217:126–131. doi: 10.1007/BF00330951. [DOI] [PubMed] [Google Scholar]
- 1320.Froschauer S, Beckwith J R. The nucleotide sequence of the gene for malF protein, an inner membrane component of the maltose transport system of Escherichia coli. Repeated DNA sequences are found in the malE-malF intercistronic region. J Biol Chem. 1984;259:10896–10903. [PubMed] [Google Scholar]
- 1321.Frothingham R, Meeker O C W A, Talbot E A, George J W, Kreuzer K N. Identification, cloning, and expression of the Escherichia coli pyrazinamidase and nicotinamidase gene, pncA. Antimicrob Agents Chemother. 1996;40:1426–1431. doi: 10.1128/aac.40.6.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1322.Frunzio R, Bruni C B, Blasi F. In vivo and in vitro detection of the leader RNA of the histidine operon of Escherichia coli K-12. Proc Natl Acad Sci USA. 1981;78:2767–2771. doi: 10.1073/pnas.78.5.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1323.Frustaci J M, O’Brian M R. The Escherichia coli visA gene encodes ferrochelatase, the final enzyme of the heme biosynthetic pathway. J Bacteriol. 1993;175:2154–2156. doi: 10.1128/jb.175.7.2154-2156.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1324.Fu G K, Smith M J, Markovitz D M. Bacterial protease Lon is a site-specific DNA-binding protein. J Biol Chem. 1997;272:534–538. [PubMed] [Google Scholar]
- 1325.Fu J C, Ding L, Clarke S. Purification, gene cloning, and sequence analysis of an l-isoaspartyl protein carboxyl methyltransferase from Escherichia coli. J Biol Chem. 1991;266:14562–14572. [PubMed] [Google Scholar]
- 1326.Fuchs J A, Karlstrom H O. Mapping of nrdA and nrdB in Escherichia coli K-12. J Bacteriol. 1976;128:810–814. doi: 10.1128/jb.128.3.810-814.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1327.Fuge E K, Farr S B. AppppA-binding protein E89 is the Escherichia coli heat shock protein ClpB. J Bacteriol. 1993;175:2321–2326. doi: 10.1128/jb.175.8.2321-2326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1328.Fujisaki S, Hara H, Nishimura Y, Horiuchi K, Nishino T. Cloning and nucleotide sequence of the ispA gene responsible for farnesyl diphosphate synthase activity in Escherichia coli. J Biochem (Tokyo) 1990;108:995–1000. doi: 10.1093/oxfordjournals.jbchem.a123327. [DOI] [PubMed] [Google Scholar]
- 1329.Fujisaki S, Ohnuma S, Horiuchi T, Takahashi I, Tsukui S, Nishimura Y, Nishino T, Kitabatake M, Inokuchi H. Cloning of a gene from Escherichia coli that confers resistance to fosmidomycin as a consequence of amplification. Gene. 1996;175:83–87. doi: 10.1016/0378-1119(96)00128-x. [DOI] [PubMed] [Google Scholar]
- 1330.Fujisaki S, Nishino T, Katsuki H, Hara H, Nishimura Y, Hirota Y. Isolation and characterization of an Escherichia coli mutant having temperature-sensitive farnesyl diphosphate synthase. J Bacteriol. 1989;171:5654–5658. doi: 10.1128/jb.171.10.5654-5658.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1331.Fujita N, Mori H, Yura T, Ishihama A. Systematic sequencing of the Escherichia coli genome: analysis of the 2.4–4.1 min (110,917–193,643 bp) region. Nucleic Acids Res. 1994;22:1637–1639. doi: 10.1093/nar/22.9.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1332.Fujita N, Miwa T, Ishijima S, Izui K, Katsuki H. The primary structure of phosphoenolpyruvate carboxylase of Escherichia coli. Nucleotide sequence of the ppc gene and deduced amino acid sequence. J Biochem (Tokyo) 1984;95:909–916. doi: 10.1093/oxfordjournals.jbchem.a134718. [DOI] [PubMed] [Google Scholar]
- 1333.Fukuda R, Yano R, Fukui T, Hase T, Ishihama A, Matsubara H. Cloning of the Escherichia coli gene for the stringent starvation protein. Mol Gen Genet. 1985;201:151–157. doi: 10.1007/BF00425652. [DOI] [PubMed] [Google Scholar]
- 1334.Fukuchi J, Kashiwagi K, Yamagishi M, Ishihama A, Igarashi K. Decrease in cell viability due to the accumulation of spermidine in spermidine acetyltransferase-deficient mutant of Escherichia coli. J Biol Chem. 1995;270:18831–18835. doi: 10.1074/jbc.270.32.18831. [DOI] [PubMed] [Google Scholar]
- 1335.Fukuda R, Nishimura A, Serizawa H, Fukuda R. Genetic mapping of the Escherichia coli gene for the stringent starvation protein and its dispensability for normal cell growth. Mol Gen Genet. 1988;211:515–519. doi: 10.1007/BF00425709. [DOI] [PubMed] [Google Scholar]
- 1336.Fulda M, Heinz E, Wolter F P. The fadD gene of Escherichia coli K12 is located close to rnd at 39.6 min of the chromosomal map and is a new member of the AMP-binding protein family. Mol Gen Genet. 1994;242:241–249. doi: 10.1007/BF00280412. [DOI] [PubMed] [Google Scholar]
- 1337.Fuller-Pace F V, Nicol S M, Reid A D, Lane D P. DbpA: a DEAD box protein specifically activated by 23s rRNA. EMBO J. 1993;12:3619–3626. doi: 10.1002/j.1460-2075.1993.tb06035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1338.Funk C R, Zimniak L, Dowhan W. The pgpA and pgpB genes of Escherichia coli are not essential: evidence for a third phosphatidylglycerophosphate phosphatase. J Bacteriol. 1992;174:205–213. doi: 10.1128/jb.174.1.205-213.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1339.Furuchi T, Kashiwagi K, Kobayashi H, Igarashi K. Characteristics of the gene for a spermidine and putrescine transport system that maps at 15 min on the Escherichia coli chromosome. J Biol Chem. 1991;266:20928–20933. [PubMed] [Google Scholar]
- 1340.Furuya N, Komano T. Specific binding of the NikA protein to one arm of 17-base-pair inverted repeat sequences within the oriT region of plasmid R64. J Bacteriol. 1995;177:46–51. doi: 10.1128/jb.177.1.46-51.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1341.Gage D J, Neidhardt F C. Adaptation of Escherichia coli to the uncoupler of oxidative phosphorylation 2,4-dinitrophenol. J Bacteriol. 1993;175:7105–7108. doi: 10.1128/jb.175.21.7105-7108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1342.Galindo J M, Guarneros G, De La Vega F M. Open reading frames flanking the peptidyl-tRNA hydrolase-encoding gene of Escherichia coli. Gene. 1994;151:153–156. doi: 10.1016/0378-1119(94)90647-5. [DOI] [PubMed] [Google Scholar]
- 1343.Galinier A, Bleicher F, Negre D, Perriere G, Duclos B, Cozzone A J, Cortay J C. Primary structure of the intergenic region between aceK and iclR in the Escherichia coli chromosome. Gene. 1991;97:149–150. doi: 10.1016/0378-1119(91)90024-6. [DOI] [PubMed] [Google Scholar]
- 1344.Gallagher M P, Pearce S R, Higgins C F. Identification and localization of the membrane-associated, ATP-binding subunit of the oligopeptide permease of Salmonella typhimurium. Eur J Biochem. 1989;180:133–141. doi: 10.1111/j.1432-1033.1989.tb14623.x. [DOI] [PubMed] [Google Scholar]
- 1345.Gallagher P, Schwartz I, Elseviers D. Genetic mapping of pheU, an Escherichia coli gene for phenylalanine tRNA. J Bacteriol. 1984;158:762–763. doi: 10.1128/jb.158.2.762-763.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1346.Galloway A, Raetz C R H. A mutant of Escherichia coli defective in the first step of endotoxin biosynthesis. J Biol Chem. 1990;265:6394–6402. [PubMed] [Google Scholar]
- 1347.Gally D L, Leathart J, Blomfield I C. Interaction of FimB and FimE with the fim switch that controls the phase variation of type 1 fimbriae in Escherichia coli K-12. Mol Microbiol. 1996;21:725–738. doi: 10.1046/j.1365-2958.1996.311388.x. [DOI] [PubMed] [Google Scholar]
- 1348.Gally D L, Bogan J A, Eisenstein B I, Blomfield I C. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J Bacteriol. 1993;175:6186–6193. doi: 10.1128/jb.175.19.6186-6193.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1349.Gally D L, Rucker T J, Blomfield I C. The leucine-responsive regulatory protein binds to the fim switch to control phase variation of type 1 fimbrial expression in Escherichia coli K-12. J Bacteriol. 1994;176:5665–5672. doi: 10.1128/jb.176.18.5665-5672.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1350.Gambino L, Gracheck S J, Miller P F. Overexpression of the MarA positive regulator is sufficient to confer multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1993;175:2888–2894. doi: 10.1128/jb.175.10.2888-2894.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1351.Gampel A, Tzagoloff A. Homology of aspartyl- and lysyl-tRNA synthetases. Proc Natl Acad Sci USA. 1989;86:6023–6027. doi: 10.1073/pnas.86.16.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1352.Gan K, Sankaran K, Williams M G, Aldea M, Rudd K E, Kushner S R, Wu H C. The umpA gene of Escherichia coli encodes phosphatidylglycerol:prolipoprotein diacylglyceryl transferase (lgt) and regulates thymidylate synthase levels through translational coupling. J Bacteriol. 1995;177:1879–1882. doi: 10.1128/jb.177.7.1879-1882.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1353.Ganduri Y L, Sadda S R, Datta M W, Jambukeswaran R K, Datta P. TdcA, a transcriptional activator of the tdcABC operon of Escherichia coli, is a member of the LysR family of proteins. Mol Gen Genet. 1993;240:395–402. doi: 10.1007/BF00280391. [DOI] [PubMed] [Google Scholar]
- 1354.Ganesan A K, Rotman B. Transport systems for galactose and galactosides in Escherichia coli. I. Genetic determination and regulation of the methyl-galactoside permease. J Mol Biol. 1965;16:42–50. doi: 10.1016/s0022-2836(66)80261-9. [DOI] [PubMed] [Google Scholar]
- 1355.Ganong B R, Raetz C R H. pH-sensitive CDP-diglyceride synthetase mutants of Escherichia coli: phenotypic suppression by mutations at a second site. J Bacteriol. 1983;153:731–738. doi: 10.1128/jb.153.2.731-738.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1356.Ganong B R, Leonard J M, Raetz C R H. Phosphatidic acid accumulation in the membranes of Escherichia coli mutants defective in CDP-digylceride synthetase. J Biol Chem. 1980;255:1623–1629. [PubMed] [Google Scholar]
- 1357.Ganoza M C, Cunningham C, Green R M. A new factor from Escherichia coli affects translocation of mRNA. J Biol Chem. 1995;270:26377–26381. doi: 10.1074/jbc.270.44.26377. [DOI] [PubMed] [Google Scholar]
- 1358.Garcia G A, Koch K A, Chong S. tRNA-guanine transglycosylase from Escherichia. Overexpression, purification and quaternary structure. J Mol Biol. 1993;231:489–497. doi: 10.1006/jmbi.1993.1296. [DOI] [PubMed] [Google Scholar]
- 1359.Garcia G M, Federici M, Rhee S G, Berberich M A. Glutamine synthetase cascade: enrichment of uridylytransferase in Escherichia coli carrying hybrid ColE1 plasmids. Arch Biochem. 1980;203:181–189. doi: 10.1016/0003-9861(80)90167-8. [DOI] [PubMed] [Google Scholar]
- 1360.Garcia G M, Mar P K, Mullin D A, Walker J R, Prather N E. The E. coli dnaY gene encodes an arginine transfer RNA. Cell. 1986;45:453–459. doi: 10.1016/0092-8674(86)90331-4. [DOI] [PubMed] [Google Scholar]
- 1361.Garcia-Lara J, Shang L H, Rothfield L I. An extracellular factor regulates expression of sdiA, a transcriptional activator of cell division genes in Escherichia coli. J Bacteriol. 1996;178:2742–2748. doi: 10.1128/jb.178.10.2742-2748.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1362.Garcia-Martin C, Baldoma L, Badia J, Aguilar J. Nucleotide sequence of the rhaR-sodA interval specifying rhaT in Escherichia coli. J Gen Microbiol. 1992;138:1109–1116. doi: 10.1099/00221287-138-6-1109. [DOI] [PubMed] [Google Scholar]
- 1363.Garciarrubio A, Lozoya E, Covarrubias A, Bolivar F. Structural organization of the genes that encode two glutamate synthase subunits of Escherichia coli. Gene. 1983;26:165–170. doi: 10.1016/0378-1119(83)90186-5. [DOI] [PubMed] [Google Scholar]
- 1364.Garcia-Villegas M R, De La Vega F M, Galindo J M, Segura M, Buckingham R H, Guarneros G. Peptidyl-tRNA hydrolase is involved in lambda inhibition of host protein synthesis. EMBO J. 1991;10:3549–3555. doi: 10.1002/j.1460-2075.1991.tb04919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1365.Gardel C, Johnson K R, Jacq A, Beckwith J R. The secD locus of E. coli codes for two membrane proteins required for protein export. EMBO J. 1990;9:3209–3216. doi: 10.1002/j.1460-2075.1990.tb07519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1366.Gardel C, Benson S, Hunt J, Michaeli S, Beckwith J R. secD, a new gene involved in protein export in Escherichia coli. J Bacteriol. 1987;169:1286–1290. doi: 10.1128/jb.169.3.1286-1290.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1367.Gardella T, Moyle H, Susskind M M. A mutant Escherichia coli sigma 70 subunit of RNA polymerase with altered promoter specificity. J Mol Biol. 1989;206:579–590. doi: 10.1016/0022-2836(89)90567-6. [DOI] [PubMed] [Google Scholar]
- 1368.Gardner P R, Fridovich I. Effect of glutathione on aconitase in Escherichia coli. Arch Biochem. 1993;301:98–102. doi: 10.1006/abbi.1993.1120. [DOI] [PubMed] [Google Scholar]
- 1369.Garg R P, Vargo C J, Cui X, Kurtz D M. A [2Fe-2S] protein encoded by an open reading frame upstream of the Escherichia coli bacterioferritin gene. Biochemistry. 1996;35:6297–6301. doi: 10.1021/bi9600862. [DOI] [PubMed] [Google Scholar]
- 1370.Garnant M K, Stauffer G V. Construction and analysis of plasmids containing the Escherichia coli serB gene. Mol Gen Genet. 1984;193:72–75. doi: 10.1007/BF00327416. [DOI] [PubMed] [Google Scholar]
- 1371.Garner C C, Herrmann K M. Operator mutations of the Escherichia coli aroF gene. J Biol Chem. 1985;260:3820–3825. [PubMed] [Google Scholar]
- 1372.Garner J, Durrer P, Kitchen J, Brunner J, Crooke E. Membrane-mediated release of nucleotide from an initiator of chromosomal replication, Escherichia coli DnaA, occurs with insertion of a distinct region of the protein into the lipid bilayer. J Biol Chem. 1998;273:5167–5173. doi: 10.1074/jbc.273.9.5167. [DOI] [PubMed] [Google Scholar]
- 1373.Garrett S, Taylor R K, Silhavy T J. Isolation and characterization of chain-terminating nonsense mutations in a porin regulator gene, envZ. J Bacteriol. 1983;156:62–69. doi: 10.1128/jb.156.1.62-69.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1374.Garrett S, Taylor R K, Silhavy T J, Berman M L. Isolation and characterization of ΔompB strains of Escherichia coli by a general method based on gene fusions. J Bacteriol. 1985;162:840–844. doi: 10.1128/jb.162.2.840-844.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1375.Garrett T A, Kadrmas J L, Raetz C R H. Identification of the gene encoding the Escherichia coli lipid A 4′-kinase. Facile phosphorylation of endotoxin analogs with recombinant LpxK. J Biol Chem. 1997;272:21855–21864. doi: 10.1074/jbc.272.35.21855. [DOI] [PubMed] [Google Scholar]
- 1376.Garrick-Silversmith L, Hartman P E. Histidine-requiring mutants of Escherichia coli K12. Genetics. 1970;66:231–244. doi: 10.1093/genetics/66.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1377.Garrido T, Sanchez M, Palacios P, Aldea M, Vicente M. Transcription of ftsZ oscillates during the cell cycle of Escherichia coli. EMBO J. 1993;12:3957–3965. doi: 10.1002/j.1460-2075.1993.tb06073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1378.Garrido-Pertierra A, Cooper R A. Evidence for two distinct pyruvate kinase genes in Escherichia coli K-12. FEBS Lett. 1983;162:420–422. doi: 10.1016/0014-5793(83)80799-6. [DOI] [PubMed] [Google Scholar]
- 1379.Garvin R T, Gorini L. A new gene for ribosomal restriction in Escherichia coli. Mol Gen Genet. 1975;137:73–78. doi: 10.1007/BF00332540. [DOI] [PubMed] [Google Scholar]
- 1380.Garwin J L, Klages A L, Cronan J E., Jr β-Ketoacyl-acyl carrier protein synthase II of Escherichia coli. Evidence for function in the thermal regulation of fatty acid synthesis. J Biol Chem. 1980;255:3263–3265. [PubMed] [Google Scholar]
- 1381.Garwin J L, Klages A L, Cronan J E., Jr Structural, enzymatic, and genetic studies of β-ketoacylacyl carrier protein synthases I and II of Escherichia coli. J Biol Chem. 1980;255:11949–11956. [PubMed] [Google Scholar]
- 1382.Gary J D, Clarke S. Purification and characterization of an isoaspartyl dipeptidase from Escherichia coli. J Biol Chem. 1995;270:4076–4087. doi: 10.1074/jbc.270.8.4076. [DOI] [PubMed] [Google Scholar]
- 1383.Gatti D L, Tzagoloff A. Structure and evolution of a group of related amino acyl-tRNA synthetases. J Mol Biol. 1991;218:557–568. doi: 10.1016/0022-2836(91)90701-7. [DOI] [PubMed] [Google Scholar]
- 1384.Gaudu P, Weiss B. SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc Natl Acad Sci USA. 1996;93:10094–10098. doi: 10.1073/pnas.93.19.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1385.Gaudu P, Touati D, Niviere V, Fontecave M. The NAD(P)H:flavin oxidoreductase from Escherichia coli as a source of superoxide radicals. J Biol Chem. 1994;269:8182–8188. [PubMed] [Google Scholar]
- 1386.Gavini N, Davidson B E. pheAo mutants of Escherichia coli have a defective pheA attenuator. J Biol Chem. 1990;265:21532–21535. [PubMed] [Google Scholar]
- 1387.Gavini N, Davidson B E. The pheR gene of Escherichia coli encodes tRNA(Phe), not a repressor protein. J Biol Chem. 1990;265:21527–21531. [PubMed] [Google Scholar]
- 1388.Gavini N, Davidson B E. Regulation of pheA expression by the pheR product in Escherichia coli is mediated through attenuation of transcription. J Biol Chem. 1991;266:7750–7753. [PubMed] [Google Scholar]
- 1389.Gavini N, Pulakat L. Role of ribosome release in the basal level of expression of the Escherichia coli gene pheA. J Gen Microbiol. 1991;137:679–684. doi: 10.1099/00221287-137-3-679. [DOI] [PubMed] [Google Scholar]
- 1390.Gay N J, Walker J R. The atp operon: nucleotide sequence of the region encoding the α-subunit of Escherichia coli ATP-synthase. Nucleic Acids Res. 1981;9:2187–2194. doi: 10.1093/nar/9.9.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1391.Gayda R C, Yamamoto L T, Markovitz A. Second-site mutations in capR (lon) strains of Escherichia coli K-12 that prevent radiation sensitivity and allow bacteriophage lambda to lysogenize. J Bacteriol. 1976;127:1208–1216. doi: 10.1128/jb.127.3.1208-1216.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1392.Gayda R C, Stephens P E, Hewick R, Schoemaker J M, Dreyer W J, Markovitz A. Regulatory region of the heat shock-inducible capR (lon) gene: DNA and protein sequences. J Bacteriol. 1985;162:271–275. doi: 10.1128/jb.162.1.271-275.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1393.Gayda R C, Leathers T D, Noti J D, Smith F J, Subrahmanyam C S, Umbarger H E. Location of the multivalent control site for the ilvEDA operon of Escherichia coli. J Bacteriol. 1980;142:556–567. doi: 10.1128/jb.142.2.556-567.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1394.Gazeau M, Delort F, Dessen P, Blanquet S, Plateau P. Escherichia coli leucine-responsive regulatory protein (Lrp) controls lysyl-tRNA synthetase expression. FEBS Lett. 1992;300:254–258. doi: 10.1016/0014-5793(92)80857-d. [DOI] [PubMed] [Google Scholar]
- 1395.Geanacopoulos M, Adhya S. Functional characterization of roles of GalR and GalS as regulators of the gal regulon. J Bacteriol. 1997;179:228–234. doi: 10.1128/jb.179.1.228-234.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1396.Geerse R H, Ruig C R, Schuitema A R J, Postma P W. Relationship between pseudo-HPr and the PEP: fructose phosphotransferase system in Salmonella typhimurium and Escherichia coli. Mol Gen Genet. 1986;203:435–444. doi: 10.1007/BF00422068. [DOI] [PubMed] [Google Scholar]
- 1397.Gellert M, O’Dea M H, Itoh A, Tomizawa J-I. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci USA. 1976;73:4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1398.Gellert M, Bullock M L. DNA ligase mutant of Escherichia coli. Proc Natl Acad Sci USA. 1970;67:1580–1587. doi: 10.1073/pnas.67.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1399.Gemmill R M, Jones J W, Haughn G W, Calvo J M. Transcription initiation sites of the leucine operons of Salmonella typhimurium and Escherichia coli. J Mol Biol. 1983;170:39–59. doi: 10.1016/s0022-2836(83)80226-5. [DOI] [PubMed] [Google Scholar]
- 1399a.Genschik P, Billy E, Swianiewicz M, Filipowitz W. The human RNA 3′-terminal phosphate cyclase is a member of a new family of proteins conserved in Eucarya, Bacteria, and Archaea. EMBO J. 1997;16:2955–2967. doi: 10.1093/emboj/16.10.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1400.Gentry D R, Burgess R. rpoZ, encoding the omega subunit of Escherichia coli RNA polymerase, is in the same operon as spoT. J Bacteriol. 1989;171:1271–1277. doi: 10.1128/jb.171.3.1271-1277.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1401.Gentry D R, Bengra C, Ikehara K, Cashel M. Guanylate kinase of Escherichia coli K-12. J Biol Chem. 1993;268:14316–14321. [PubMed] [Google Scholar]
- 1402.Gentry D R, Hernandez V J, Nguyen L H, Jensen D B, Cashel M. Synthesis of the stationary-phase sigma factor ς s is positively regulated by ppGpp. J Bacteriol. 1993;175:7982–7989. doi: 10.1128/jb.175.24.7982-7989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1403.George A M, Levy S B. Gene in the major cotransduction gap of the Escherichia coli K-12 linkage map required for the expression of chromosomal resistance to tetracycline and other antibiotics. J Bacteriol. 1983;155:541–548. doi: 10.1128/jb.155.2.541-548.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1404.George S E, Melton T. Cloning and molecular characterization of csm mutations allowing expression of catabolite-repressible operons in the absence of exogenous cyclic AMP. J Bacteriol. 1986;166:533–540. doi: 10.1128/jb.166.2.533-540.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1405.Georgiou C D, Fang H, Gennis R B. Identification of the cydC locus required for expression of the functional form of the cytochrome d terminal oxidase complex in Escherichia coli. J Bacteriol. 1987;169:2107–2112. doi: 10.1128/jb.169.5.2107-2112.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1406.Georgopoulos C. Bacterial mutants in which the gene N function of bacteriophage lambda is blocked have an altered RNA polymerase. Proc Natl Acad Sci USA. 1971;68:2977–2981. doi: 10.1073/pnas.68.12.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1407.Georgopoulos C, Ang D. The Escherichia coli groE chaperonins. Semin Cell Biol. 1990;1:19–25. [PubMed] [Google Scholar]
- 1408.Georgopoulos C, Swindle J, Keppel F, Ballivet M, Bisig R, Eisen H A. Studies on the E. coli groNB (nusB) gene which affects bacteriophage λ N gene function. Mol Gen Genet. 1980;179:55–61. doi: 10.1007/BF00268446. [DOI] [PubMed] [Google Scholar]
- 1409.Georgopoulos C, Hendrix R W, Kaiser C. Role of the host cell in bacteriophage morphogenesis: effects of a bacterial mutation on T4 head assembly. Nat New Biol. 1972;239:38–41. doi: 10.1038/newbio239038a0. [DOI] [PubMed] [Google Scholar]
- 1410.Gerchman Y, Olami Y, Rimon A, Taglicht D, Schuldiner S, Padan E. Histidine-226 is part of the pH sensor of NhaA, a Na+/H+ antiporter in Escherichia coli. Proc Natl Acad Sci USA. 1993;90:1212–1216. doi: 10.1073/pnas.90.4.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1411.Gerdes K, Bech F W, Jorgensen S T, Lobner-Olesen A, Rasmussen P B, Atlung T, Boe L, Karlstrom H O, Molin S, von Meyenberg K. Mechanism of postreplicational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986;5:2023–2029. doi: 10.1002/j.1460-2075.1986.tb04459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1412.Gervais F G, Drapeau G R. Identification, cloning, and characterization of rcsF, a new regulator gene for exopolysaccharide synthesis that suppresses the division mutation ftsZ84 in Escherichia coli K-12. J Bacteriol. 1992;174:8016–8022. doi: 10.1128/jb.174.24.8016-8022.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1413.Gervais F G, Phoenix P, Drapeau G R. The rcsB gene, a positive regulator of colanic acid biosynthesis in Escherichia coli, is also an activator of ftsZ expression. J Bacteriol. 1992;174:3964–3971. doi: 10.1128/jb.174.12.3964-3971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1414.Gery S, Ron E Z. An Escherichia coli gene divergently transcribed from a promoter overlapping the metA promoter. FEMS Microbiol Lett. 1997;154:219–222. doi: 10.1111/j.1574-6968.1997.tb12647.x. [DOI] [PubMed] [Google Scholar]
- 1415.Ghisotti D, Zangrossi S, Sironi G. An Escherichia coli gene required for bacteriophage P2-λ interference. J Virol. 1983;48:616–626. doi: 10.1128/jvi.48.3.616-626.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1416.Ghrist A C, Stauffer G V. Characterization of the Escherichia coli gcvR gene encoding a negative regulator of gcv expression. J Bacteriol. 1995;177:4980–4984. doi: 10.1128/jb.177.17.4980-4984.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1417.Ghrist A C, Stauffer G V. Promoter characterization and constitutive expression of the Escherichia coli gcvR gene. J Bacteriol. 1998;180:1803–1807. doi: 10.1128/jb.180.7.1803-1807.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1418.Ghysen A, Pirionio M. Relationship between the N function of bacteriophage LAM and host RNA polymerase. J Mol Biol. 1972;65:259–272. doi: 10.1016/0022-2836(72)90281-1. [DOI] [PubMed] [Google Scholar]
- 1419.Giaever H M, Styrvold O B, Kassen I, Strom A R. Biochemical and genetic characterization of osmoregulatory trehalose synthesis in Escherichia coli. J Bacteriol. 1988;170:2841–2849. doi: 10.1128/jb.170.6.2841-2849.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1420.Giam C Z, Hayashi S, Wu H C. Characterization of a novel lipoprotein mutant in Escherichia coli. J Biol Chem. 1984;259:5601–5605. [PubMed] [Google Scholar]
- 1421.Gibert I, Llagostera M, Barbé J. Regulation of ubiG gene expression in Escherichia coli. J Bacteriol. 1988;170:1346–1349. doi: 10.1128/jb.170.3.1346-1349.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1422.Gibson F, Pittard J. Pathways of biosynthesis of aromatic amino acids and vitamins and their control in microorganisms. Bacteriol Rev. 1968;32:465–492. [PMC free article] [PubMed] [Google Scholar]
- 1423.Gibson F P, Leach D R F, Lloyd R G. Identification of sbcD mutations as cosuppressors of recBC that allow propagation of DNA palindromes in Escherichia coli K-12. J Bacteriol. 1992;174:1222–1228. doi: 10.1128/jb.174.4.1222-1228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1424.Giel M, Desnoyer M, Lopilato J. A mutation in a new gene bglJ, activates the bgl operon in Escherichia coli K-12. Genetics. 1996;143:627–635. doi: 10.1093/genetics/143.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1425.Gielow A, Diederich L, Messer W. Expression of the replication protein Arp of phasyl shows dual regulation by an antisense promoter. EMBO J. 1991;10:3061–3066. doi: 10.1002/j.1460-2075.1991.tb07857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1426.Giffard P M, Booth I R. The rpoA341 allele of Escherichia coli specifically impairs the transcription of a group of positively-regulated operons. Mol Gen Genet. 1988;214:148–152. doi: 10.1007/BF00340193. [DOI] [PubMed] [Google Scholar]
- 1427.Gigot D, Crabeel M, Feller A, Charlier D, Lissens W, Glansdorff N, Pierard A. Patterns of polarity in the Escherichia coli carAB gene cluster. J Bacteriol. 1980;143:914–920. doi: 10.1128/jb.143.2.914-920.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1428.Gilchrist C A, Denhardt D. Escherichia coli rep gene: sequence of the gene, the encoded helicase, and its homology with uvrD. Nucleic Acids Res. 1987;15:465–475. doi: 10.1093/nar/15.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1429.Gill D R, Hatfull G F, Salmond G P C. A new cell division operon in Escherichia coli. Mol Gen Genet. 1986;205:134–145. doi: 10.1007/BF02428043. [DOI] [PubMed] [Google Scholar]
- 1430.Gille H J, Egan J B, Roth A F, Messer W. The FIS protein binds and bends the origin of chromosomal DNA replication, oriC, of Escherichia coli. Nucleic Acids Res. 1991;19:4167–4172. doi: 10.1093/nar/19.15.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1431.Gilson E, Nikaido H, Hofnung M. Sequence of the malK gene in E. coli K12. Nucleic Acids Res. 1982;10:7449–7458. doi: 10.1093/nar/10.22.7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1432.Gilson E, Rousset J-P, Charbit A, Perrin D M, Hofnung M. malM, a new gene of the maltose regulon in Escherichia coli K12. I. malM is the last gene of the malK-lamB operon and encodes a periplasmic protein. J Mol Biol. 1986;191:303–311. doi: 10.1016/0022-2836(86)90127-0. [DOI] [PubMed] [Google Scholar]
- 1433.Giordani R, Buc J, Cornish-Bowden A, Cardenas M L. Kinetics of membrane-bound nitrate reductase A from Escherichia coli with analogues of physiological electron donors—different reaction sites for menadiol and duroquinol. Eur J Biochem. 1997;250:567–577. doi: 10.1111/j.1432-1033.1997.0567a.x. [DOI] [PubMed] [Google Scholar]
- 1434.Glaser J H, DeMoss J A. Comparison of nitrate reductase mutants of Escherichia coli selected by alternative procedures. Mol Gen Genet. 1972;116:1–10. doi: 10.1007/BF00334254. [DOI] [PubMed] [Google Scholar]
- 1435.Glassberg J, Meyer R R, Kornberg A. Mutant single-strand binding protein of Escherichia coli: genetic and physiological characterization. J Bacteriol. 1979;140:14–19. doi: 10.1128/jb.140.1.14-19.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1436.Glavas N A, Bragg P D. Evidence for the presence of two pyridine nucleotide-binding sites on the beta subunit of the Escherichia coli pyridine nucleotide transhydrogenase. Biochem Mol Biol Int. 1995;35:297–306. [PubMed] [Google Scholar]
- 1437.Glover S W. Valine-resistant mutants of Escherichia coli K-12. Genet Res. 1962;3:448–460. [Google Scholar]
- 1438.Godessart N, Munoa F J, Regue M, Juarez A. Chromosomal mutations that increase the production of a plasmid-encoded haemolysin in Escherichia coli. J Gen Microbiol. 1988;134:2779–2787. doi: 10.1099/00221287-134-10-2779. [DOI] [PubMed] [Google Scholar]
- 1439.Goffin C, Fraipont C, Ayala J, Terrak M, Nguyen-Distèche M, Ghuysen J M. The non-penicillin-binding module of the tripartite penicillin-binding protein 3 of Escherichia coli is required for folding and/or stability of the penicillin-binding module and the membrane-anchoring module confers cell septation activity on the folded structure. J Bacteriol. 1996;178:5402–5409. doi: 10.1128/jb.178.18.5402-5409.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1440.Goldberg E B, Arbel T, Chen J, Karpel R, Mackie G A, Schuldiner S. Characterization of a Na+/H+ antiporter gene of Escherichia coli. Proc Natl Acad Sci USA. 1987;84:2615–2619. doi: 10.1073/pnas.84.9.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1441.Goldemberg S H. A role for polyamines in the control of ppGpp levels in Escherichia coli. Cell Mol Biol (Noisy-Le-Grand) 1994;40:899–905. [PubMed] [Google Scholar]
- 1442.Goldemberg S H. In vitro studies on ppGpp synthetase I from polyamine-starved and unstarved Escherichia coli. Cell Mol Biol (Noisy-Le-Grand) 1996;42:719–727. [PubMed] [Google Scholar]
- 1443.Goldenberg D, Azar I, Oppenheim A B, Brandi A, Pon C L, Gualerzi C O. Role of Escherichia coli cspA promoter sequences and adaptation of translational apparatus in the cold shock response. Mol Gen Genet. 1997;256:282–290. doi: 10.1007/s004380050571. [DOI] [PubMed] [Google Scholar]
- 1444.Goldie H, Sanwal B D. Genetic and physiological characterization of Escherichia coli mutants deficient in phosphoenolpyruvate carboxykinase activity. J Bacteriol. 1980;141:1115–1121. doi: 10.1128/jb.141.3.1115-1121.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1445.Goldie H, Sanwal B D. Temperature-sensitive mutation affecting synthesis of phosphoenolpyruvate carboxykinase in Escherichia coli. J Bacteriol. 1981;148:720–723. doi: 10.1128/jb.148.2.720-723.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1446.Goldie H, Medina V. Physical and genetic analysis of the phosphoenolpyruvate carboxykinase (pckA) locus from Escherichia coli K12. Mol Gen Genet. 1990;220:191–196. doi: 10.1007/BF00260481. [DOI] [PubMed] [Google Scholar]
- 1447.Goldman B S, Gabbert K K, Kranz R G. The temperature-sensitive growth and survival phenotypes of Escherichia coli cydDC and cydAB strains are due to deficiencies in cytochrome bd and are corrected by exogenous catalase and reducing agents. J Bacteriol. 1996;178:6348–6351. doi: 10.1128/jb.178.21.6348-6351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1448.Goldman J D, White D G, Levy S B. Multiple antibiotic resistance (mar) locus protects Escherichia coli from rapid cell killing by fluoroquinolones. Antimicrob Agents Chemother. 1996;40:1266–1269. doi: 10.1128/aac.40.5.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1449.Goldman R C, Bolling T J, Kohlbrenner W E, Kim Y, Fox J L. Primary structure of CTP:CMP-3-deoxy-d-manno-octulosonate cytidylyl-transferase (CMP-KDO synthetase) from Escherichia coli. J Biol Chem. 1986;261:15831–15835. [PubMed] [Google Scholar]
- 1450.Goldman R C, Kohlbrenner W E. Molecular cloning of the structural gene coding for CTP:CMP-3-deoxy-d-manno-octulosonate cytidylytransferase from Escherichia coli K-12. J Bacteriol. 1985;163:256–261. doi: 10.1128/jb.163.1.256-261.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1451.Goldschmidt E P, Cater M S, Matney T S, Butler M A, Greene A. Genetic analysis of the histidine operon in Escherichia coli K12. Genetics. 1970;66:219–229. doi: 10.1093/genetics/66.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1452.Goldstein J, Pollitt N S, Inouye M. Major heat shock protein of Escherichia coli. Proc Natl Acad Sci USA. 1990;87:283–287. doi: 10.1073/pnas.87.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1453.Gollop N, Tavori H, Barak Z. Acetohydroxy acid synthase is a target for leucine-containing peptide toxicity in Escherichia coli. J Bacteriol. 1982;149:387–390. doi: 10.1128/jb.149.1.387-390.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1454.Goncharoff P, Nichols B. Nucleotide sequence of Escherichia coli pabB indicates a common evolutionary origin of p-aminobenzoate synthetase and anthranilate synthetase. J Bacteriol. 1984;159:57–62. doi: 10.1128/jb.159.1.57-62.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1455.Gonzalez-Flecha B, Demple B. Transcriptional regulation of the Escherichia coli oxyR gene as a function of cell growth. J Bacteriol. 1997;179:6181–6186. doi: 10.1128/jb.179.19.6181-6186.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1456.Gonzalez-Gil G, Bringmann P, Kahmann R. FIS is a regulator of metabolism in Escherichia coli. Mol Microbiol. 1996;22:21–29. doi: 10.1111/j.1365-2958.1996.tb02652.x. [DOI] [PubMed] [Google Scholar]
- 1457.Goodlove P E, Cunningham P R, Parker J, Clark D P. Cloning and sequence analysis of the fermentative alcohol-dehydrogenase-encoding gene of Escherichia coli. Gene. 1989;85:209–214. doi: 10.1016/0378-1119(89)90483-6. [DOI] [PubMed] [Google Scholar]
- 1458.Goosen N, van de Putte P. The regulation of transcription initiation by integration host factor. Mol Microbiol. 1995;16:1–7. doi: 10.1111/j.1365-2958.1995.tb02386.x. [DOI] [PubMed] [Google Scholar]
- 1459.Gopalakrishnan A S, Chen Y-C, Temkin M, Dowhan W. Structure and exression of the gene locus encoding the phosphatidylglycerophosphate synthase of Escherichia coli. J Biol Chem. 1986;261:1329–1338. [PubMed] [Google Scholar]
- 1460.Gordia S, Gutierrez C. Growth-phase-dependent expression of the osmotically inducible gene osmC of Escherichia coli K-12. Mol Microbiol. 1996;19:729–736. doi: 10.1046/j.1365-2958.1996.418945.x. [DOI] [PubMed] [Google Scholar]
- 1461.Gordon A J, Burns P A, Fix D F, Yatagai F, Allen F L, Horsfall M J, Halliday J A, Gray J, Bernelot-Moens C, Glickman B W. Missense mutation in the lacI gene of Escherichia coli. Inferences on the structure of the repressor protein. J Mol Biol. 1988;200:239–251. doi: 10.1016/0022-2836(88)90237-9. [DOI] [PubMed] [Google Scholar]
- 1462.Gordon A W, Gayda R C, Markovitz A. Sequence of the regulatory region of ompT, the gene specifying major outer membrane protein a (3b) of Escherichia coli K-12: implications for regulation and processing. Mol Gen Genet. 1984;193:414–421. doi: 10.1007/BF00382077. [DOI] [PubMed] [Google Scholar]
- 1463.Goshima N, Kohno K, Imamoto F, Kano Y. HU-1 mutants of Escherichia coli deficient in DNA binding. Gene. 1990;96:141–145. doi: 10.1016/0378-1119(90)90355-u. [DOI] [PubMed] [Google Scholar]
- 1464.Gosink K K, Gaal T, Bokal A J T, Gourse R L. A positive control mutant of the transcription activator protein FIS. J Bacteriol. 1996;178:5182–5187. doi: 10.1128/jb.178.17.5182-5187.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1465.Gosink K K, Ross W, Leirmo S, Osuna R, Finkel S, Johnson R C, Gourse R L. DNA binding and bending are necessary but not sufficient for Fis-dependent activation of rrnB P1. J Bacteriol. 1993;175:1580–1589. doi: 10.1128/jb.175.6.1580-1589.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1466.Goss T J, Schweizer H P, Datta P. Molecular characterization of the tdc operon of Escherichia coli K-12. J Bacteriol. 1988;170:5352–5359. doi: 10.1128/jb.170.11.5352-5359.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1467.Goss T J, Datta P. Escherichia coli K-12 mutation that inactivates biodegradative threonine dehydratase by transposon Tn5 insertion. J Bacteriol. 1984;158:826–831. doi: 10.1128/jb.158.3.826-831.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1468.Goss T J, Datta P. Molecular cloning and expression of the biodegradative threonine dehydratase gene (tdc) of Escherichia coli K12. Mol Gen Genet. 1985;201:308–314. doi: 10.1007/BF00425676. [DOI] [PubMed] [Google Scholar]
- 1469.Gots J S, Benson C E, Shumas S R. Genetic separation of hypoxanthine and guanine-xanthine phosphoribosyltransferase activities by deletion mutations in Salmonella typhimurium. J Bacteriol. 1972;112:910–916. doi: 10.1128/jb.112.2.910-916.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1470.Gottesman S, Beckwith J R. Directed transposition of the arabinose operon: a technique for the isolation of special transducing bacteriophages for any Escherichia coli gene. J Mol Biol. 1969;44:117–127. doi: 10.1016/0022-2836(69)90408-2. [DOI] [PubMed] [Google Scholar]
- 1471.Gottesman S, Trisler P, Torres-Cabassa A S. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. J Bacteriol. 1985;162:1111–1119. doi: 10.1128/jb.162.3.1111-1119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1472.Gottesman S, Clark W P, Maurizi M R. The ATP-dependent Clp-protease of Escherichia coli. Sequence of clpA and identification of a Clp-specific substrate. J Biol Chem. 1990;265:7886–7893. [PubMed] [Google Scholar]
- 1473.Gottesman S, Clark W P, de Crecy-Lagard V, Maurizi M R. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli. Sequence and in vivo activities. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- 1474.Gough J A, Murray N E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983;166:1–9. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- 1475.Gowrishankar J. Identification of osmoresponsive genes in Escherichia coli: evidence for participation of potassium and proline transport systems in osmoregulation. J Bacteriol. 1985;164:434–445. doi: 10.1128/jb.164.1.434-445.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1476.Gowrishankar J. proP-mediated proline transport also plays a role in Escherichia coli osmoregulation. J Bacteriol. 1986;166:331–333. doi: 10.1128/jb.166.1.331-333.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1477.Gowrishankar J. Nucleotide sequence of the osmoregulatory proU operon of Escherichia coli. J Bacteriol. 1989;171:1923–1931. doi: 10.1128/jb.171.4.1923-1931.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1478.Gowrishankar J, Manna D. How is osmotic regulation of transcription of the Escherichia coli proU operon achieved? A review and a model. Genetica. 1996;97:363–378. doi: 10.1007/BF00055322. [DOI] [PubMed] [Google Scholar]
- 1479.Gowrishankar J, Pittard J. Construction from Mu d1 (lac Apr) lysogens of lambda bacteriophage bearing promoter-lac fusions: isolation of λ ppheA-lac. J Bacteriol. 1982;150:1122–1129. doi: 10.1128/jb.150.3.1122-1129.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1480.Gowrishankar J, Pittard J. Regulation of phenylalanine biosynthesis in Escherichia coli K-12: control of transcription of the pheA operon. J Bacteriol. 1982;150:1130–1137. doi: 10.1128/jb.150.3.1130-1137.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1481.Gowrishankar J, Jayashree P, Rajkumari K. Molecular cloning of an osmoregulatory locus in Escherichia coli: increased proU gene dosage results in enhanced osmotolerance. J Bacteriol. 1986;168:1197–1204. doi: 10.1128/jb.168.3.1197-1204.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1482.Grabau C, Cronan J E., Jr Molecular cloning of the gene (poxB) encoding the pyruvate oxidase of Escherichia coli, a lipid-activated enzyme. J Bacteriol. 1984;160:1088–1092. doi: 10.1128/jb.160.3.1088-1092.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1483.Grabau C, Cronan J E., Jr Nucleotide sequence and deduced amino acid sequence of Escherichia coli pyruvate oxidase, a lipid-activated flavoprotein. Nucleic Acids Res. 1986;14:5449–5460. doi: 10.1093/nar/14.13.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1484.Graftstrom R H, Hoess R H. Cloning of mutH and identification of the gene product. Gene. 1983;22:245–253. doi: 10.1016/0378-1119(83)90109-9. [DOI] [PubMed] [Google Scholar]
- 1485.Graftstrom R H, Hoess R H. Nucleotide sequence of the Escherichia coli mutH gene. Nucleic Acids Res. 1987;15:3073–3083. doi: 10.1093/nar/15.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1486.Granger L L, O’Hara E B, Wang R F, Meffen F V, Armstrong K, Yancey S D, Babitzke P, Kushner S R. The Escherichia coli mrsC gene is required for cell growth and mRNA decay. J Bacteriol. 1998;180:1920–1928. doi: 10.1128/jb.180.7.1920-1928.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1487.Granston A E, Thompson D L, Friedman D I. Identification of a second promoter for the metY-nusA-infB operon of Escherichia coli. J Bacteriol. 1990;172:2336–2342. doi: 10.1128/jb.172.5.2336-2342.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1488.Gratia J. Studies on defective lysogeny due to chromosomal deletion in Escherichia coli. Biken J. 1966;9:77–87. [PubMed] [Google Scholar]
- 1489.Graves R J, Felzenszwalb I, Laval J, O’Connor T R. Excision of 5′-terminal deoxyribose phosphate from damaged DNA is catalyzed by the Fpg protein of Escherichia coli. J Biol Chem. 1992;267:14429–14435. [PubMed] [Google Scholar]
- 1490.Gray J E, Bennett D C, Umbarger H E, Calhoun D H. Physical and genetic localization of ilv regulatory sites in λ ilv bacteriophages. J Bacteriol. 1982;149:1071–1081. doi: 10.1128/jb.149.3.1071-1081.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1491.Green G N, Fang H, Lin R-J, Newton G, Mather M, Georgiou C D, Gennis R B. The nucleotide sequence of the cyd locus encoding the two subunits of the cytochrome d terminal oxidase complex of Escherichia coli. J Biol Chem. 1988;263:13138–13143. [PubMed] [Google Scholar]
- 1492.Green G N, Kranz J E, Gennis R B. Cloning the cyd locus coding for the cytochrome d complex of E. coli. Gene. 1984;32:99–106. doi: 10.1016/0378-1119(84)90037-4. [DOI] [PubMed] [Google Scholar]
- 1493.Green G N, Gennis R B. Isolation and characterization of an Escherichia coli mutant lacking cytochrome d terminal oxidase. J Bacteriol. 1983;154:1269–1275. doi: 10.1128/jb.154.3.1269-1275.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1494.Green G N, Kranz R G, Lorence R M, Gennis R B. Identification of subunit I as the cytochrome b558 component of the cytochrome d terminal oxidase complex of Escherichia coli. J Biol Chem. 1984;259:7994–7997. [PubMed] [Google Scholar]
- 1495.Green G N, Lorence R M, Gennis R B. Specific overproduction and purification of the cytochrome b558 component of the cytochrome d complex from Escherichia coli. Biochemistry. 1986;25:2309–2314. doi: 10.1021/bi00357a002. [DOI] [PubMed] [Google Scholar]
- 1496.Green J, Anjum M F, Guest J R. The ndh-binding protein (Nbp) regulates the ndh gene of Escherichia coli in response to growth phase and is identical to Fis. Mol Microbiol. 1996;20:1043–1055. doi: 10.1111/j.1365-2958.1996.tb02545.x. [DOI] [PubMed] [Google Scholar]
- 1497.Green J, Anjum M F, Guest J R. Regulation of the ndh gene of Escherichia coli by integration host factor and a novel regulator, Arr. Microbiology. 1997;143:2865–2875. doi: 10.1099/00221287-143-9-2865. [DOI] [PubMed] [Google Scholar]
- 1498.Green J, Baldwin M L. The molecular basis for the differential regulation of the hlyE-encoded haemolysin of Escherichia coli by FNR and HlyX lies in the improved activating region 1 contact of HlyX. Microbiology. 1997;143:3785–3793. doi: 10.1099/00221287-143-12-3785. [DOI] [PubMed] [Google Scholar]
- 1499.Green J M, Nichols B P. p-Aminobenzoate biosynthesis in Escherichia coli. Purification of aminodeoxychorismate lyase and cloning of pabC. J Biol Chem. 1991;266:12971–12975. [PubMed] [Google Scholar]
- 1500.Green J M, Merkel W K, Nichols B P. Characterization and sequence of Escherichia coli pabC, the gene encoding aminodeoxychorismate lyase, a pyridoxal phosphate-containing enzyme. J Bacteriol. 1992;174:5317–5323. doi: 10.1128/jb.174.16.5317-5323.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1501.Green P J, Inouye M. Roles of the 5′ leader region of the ompA mRNA. J Mol Biol. 1984;176:431–442. doi: 10.1016/0022-2836(84)90499-6. [DOI] [PubMed] [Google Scholar]
- 1502.Green R, Noller H F. In vitro complementation analysis localizes 23S rRNA posttranscriptional modifications that are required for Escherichia coli 50S ribosomal subunit assembly and function. RNA. 1996;2:1011–1021. [PMC free article] [PubMed] [Google Scholar]
- 1503.Green S M, Malik T, Giles I G, Drabble W T. The purB gene of Escherichia coli K-12 is located in an operon. Microbiology. 1996;142:3219–3230. doi: 10.1099/13500872-142-11-3219. [DOI] [PubMed] [Google Scholar]
- 1504.Greenberg J, Demple B. Glutathione in Escherichia coli is dispensable for resistance to H2O2 and gamma radiation. J Bacteriol. 1986;168:1026–1029. doi: 10.1128/jb.168.2.1026-1029.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1505.Greenberg J, Chou J H, Monach P A, Demple B. Activation of oxidative stress genes by mutations at the soxZ/cfxB/marA locus of Escherichia coli. J Bacteriol. 1991;173:4433–4439. doi: 10.1128/jb.173.14.4433-4439.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1506.Greenblatt J, Li J, Adhya S, Friedman D I, Baron L S, Redfield B, Kung H-F, Weissbach H. L factor that is required for β-galactosidase synthesis is the nusA gene product involved in transcription termination. Proc Natl Acad Sci USA. 1980;77:1991–1994. doi: 10.1073/pnas.77.4.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1507.Greenblatt J, McLimont M, Hanly S. Termination of transcription by nusA gene protein of Escherichia coli. Nature. 1981;292:215–220. doi: 10.1038/292215a0. [DOI] [PubMed] [Google Scholar]
- 1508.Greene R C, Smith A A. Insertion mutagenesis of the metJBLF gene cluster of Escherichia coli K-12: evidence for a metBL operon. J Bacteriol. 1984;159:767–769. doi: 10.1128/jb.159.2.767-769.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1509.Greene R C, Krueger J H, Johnson J R. Localization of the metJBLF gene cluster of Escherichia coli in λmet transducing phage. Mol Gen Genet. 1982;187:401–404. doi: 10.1007/BF00332618. [DOI] [PubMed] [Google Scholar]
- 1510.Greene R C, Hunter J V, Coch E H. Properties of metK mutants of Escherichia coli K-12. J Bacteriol. 1973;115:57–67. doi: 10.1128/jb.115.1.57-67.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1511.Greener A, Hill C W. Identification of a novel genetic element in Escherichia coli K-12. J Bacteriol. 1980;144:312–321. doi: 10.1128/jb.144.1.312-321.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1512.Greener T, Govezensky D, Zamir A. A novel multicopy suppressor of a groEL mutation includes two nested open reading frames transcribed from different promoters. EMBO J. 1993;12:889–896. doi: 10.1002/j.1460-2075.1993.tb05729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1513.Greer S, Perham R N. Glutathione reductase from Escherichia coli: cloning and sequence analysis of the gene and relationship to other flavoprotein disulfide oxidoreductases. Biochemistry. 1986;25:2736–2742. doi: 10.1021/bi00357a069. [DOI] [PubMed] [Google Scholar]
- 1514.Greiner R, Konietzny U, Jany K D. Purification and characterization of two phytases from Escherichia coli. Arch Biochem Biophys. 1993;303:107–113. doi: 10.1006/abbi.1993.1261. [DOI] [PubMed] [Google Scholar]
- 1515.Grentzmann G, Brechemier-Baey D, Heurgue V, Mora L, Buckingham R H. Localization and characterization of the gene encoding release factor RF3 in Escherichia coli. Proc Natl Acad Sci USA. 1994;91:5848–5852. doi: 10.1073/pnas.91.13.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1516.Griffin H G, Gasson M J. The gene (aroK) encoding shikimate kinase I from Escherichia coli. DNA Sequence. 1995;5:195–197. doi: 10.3109/10425179509029363. [DOI] [PubMed] [Google Scholar]
- 1517.Griffin T J, Kolodner R D. Purification and preliminary characterization of the Escherichia coli K-12 recF protein. J Bacteriol. 1990;172:6291–6299. doi: 10.1128/jb.172.11.6291-6299.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1518.Griggs D W, Tharp B B, Konisky J. Cloning and promoter identification of the iron-regulated cir gene of Escherichia coli. J Bacteriol. 1987;169:5343–5352. doi: 10.1128/jb.169.12.5343-5352.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1519.Grimm B, Bull A, Breu V. Structural genes of glutamate 1-semialdehyde aminotransferase for porphyrin synthesis in a cyanobacterium and Escherichia coli. Mol Gen Genet. 1991;225:1–10. doi: 10.1007/BF00282635. [DOI] [PubMed] [Google Scholar]
- 1520.Gringauz E, Orle K A, Waddell C S, Craig N L. Recognition of Escherichia coli attTn7 by transposon Tn7: lack of specific sequence requirements at the point of Tn7 insertion. J Bacteriol. 1988;170:2832–2840. doi: 10.1128/jb.170.6.2832-2840.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1521.Grisolia V, Riccio A, Bruni C B. Structure and function of the internal promoter (hisBp) of the Escherichia coli K-12 histidine operon. J Bacteriol. 1983;155:1288–1296. doi: 10.1128/jb.155.3.1288-1296.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1522.Grisolia V, Carlomagno M S, Nappo A G, Bruni C B. Cloning, structure and expression of the Escherichia coli K-12 hisC gene. J Bacteriol. 1985;164:1317–1323. doi: 10.1128/jb.164.3.1317-1323.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1523.Groat R G, Schultz J E, Zychlinsky E, Bockman A, Matin A. Starvation proteins in Escherichia coli: kinetics of synthesis and role in starvation survival [published erratum appears in J. Bacteriol. 1987, 169:3866] J Bacteriol. 1986;168:486–493. doi: 10.1128/jb.168.2.486-493.1986. . (Erratum, 169:3866, 1987.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1524.Grodberg J, Dunn J J. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J Bacteriol. 1988;170:1245–1253. doi: 10.1128/jb.170.3.1245-1253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1525.Grodberg J, Lundigan M D, Toledo D L, Mangel W F, Dunn J J. Complete nucleotide sequence and deduced amino acid sequence of the ompT gene of Escherichia coli K-12. Nucleic Acids Res. 1988;16:1209. doi: 10.1093/nar/16.3.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1526.Grogan D W. Temperature-sensitive murein synthesis in an Escherichia coli pdx mutant and the role of alanine racemase. Arch Microbiol. 1988;150:363–367. doi: 10.1007/BF00408308. [DOI] [PubMed] [Google Scholar]
- 1527.Grogan D W, Cronan J E., Jr Use of lambda plasmids for deletion mapping of non-selectable markers cloned in plasmids. Gene. 1983;22:75–83. doi: 10.1016/0378-1119(83)90066-5. [DOI] [PubMed] [Google Scholar]
- 1528.Grogan D W, Cronan J E., Jr Cloning and manipulation of the Escherichia coli cyclopropane fatty acid synthase gene: physiological aspects of enzyme overproduction. J Bacteriol. 1984;158:286–295. doi: 10.1128/jb.158.1.286-295.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1529.Grogan D W, Cronan J E., Jr Genetic characterization of Escherichia coli cyclopropane fatty acid (cfa) locus and neighboring loci. Mol Gen Genet. 1984;196:367–372. doi: 10.1007/BF00328074. [DOI] [PubMed] [Google Scholar]
- 1530.Groisman E A, Heffron F, Solomon F. Molecular genetic analysis of the Escherichia coli phoP locus. J Bacteriol. 1992;174:486–491. doi: 10.1128/jb.174.2.486-491.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1531.Gross C A, Blattner F R, Taylor W E, Lowe P A, Burgess R R. Isolation and characterization of transducing phage coding for a subunit of Escherichia coli RNA polymerase. Proc Natl Acad Sci USA. 1979;76:5789–5793. doi: 10.1073/pnas.76.11.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1532.Grossman A D, Erickson J, Gross C A. The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell. 1984;38:383–390. doi: 10.1016/0092-8674(84)90493-8. [DOI] [PubMed] [Google Scholar]
- 1533.Grossman A D, Zhou Y-N, Gross C, Heilig J, Christie G E, Calendar R. Mutations in the rpoH (htpR) gene of Escherichia coli K-12 phenotypically suppress a temperature-sensitive mutant defective in the ς70 subunit of RNA polymerase. J Bacteriol. 1985;161:939–943. doi: 10.1128/jb.161.3.939-943.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1534.Grossman T H, Tuckman M, Ellestad S, Osburne M S. Isolation and characterization of Bacillus subtilis genes involved in siderophore biosynthesis: relationship between B. subtilis sfp0 and Escherichia coli entD genes. J Bacteriol. 1993;175:6203–6211. doi: 10.1128/jb.175.19.6203-6211.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1535.Grothe S R, Krogsrud R L, McClellan D J, Milner J L, Wood J M. Proline transport and osmotic stress response in Escherichia coli K-12. J Bacteriol. 1986;166:253–259. doi: 10.1128/jb.166.1.253-259.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1536.Grove J, Busby S, Cole J. The role of the genes nrfEFG and ccmFH in cytochrome c biosynthesis in Escherichia coli. Mol Gen Genet. 1996;252:332–341. doi: 10.1007/BF02173779. [DOI] [PubMed] [Google Scholar]
- 1537.Grove J, Tanapongpipat S, Thomas G, Griffiths L, Crooke H, Cole J. Escherichia coli K-12 genes essential for the synthesis of c-type cytochromes and a third nitrate reductase located in the periplasm. Mol Microbiol. 1996;19:467–481. doi: 10.1046/j.1365-2958.1996.383914.x. [DOI] [PubMed] [Google Scholar]
- 1538.Gruer M J, Guest J R. Two genetically-distinct and differentially-regulated aconitases (AcnA and AcnB) in Escherichia coli. Microbiology. 1994;140:2531–2541. doi: 10.1099/00221287-140-10-2531. [DOI] [PubMed] [Google Scholar]
- 1539.Grunden A M, Ray R M, Rosentel J K, Healy F G, Shanmugam K T. Repression of the Escherichia coli modABCD (molybdate transport) operon by ModE. J Bacteriol. 1996;178:735–744. doi: 10.1128/jb.178.3.735-744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1540.Grundstrom T, Jaurin B. Overlap between ampC and frd operons on the Escherichia coli chromosome. Proc Natl Acad Sci USA. 1982;79:1111–1115. doi: 10.1073/pnas.79.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1541.Grundstrom T, Jaurin B, Edlund T, Normark S. Physical mapping and expression of hybrid plasmids carrying chromosomal β-lactamase genes of Escherichia coli K-12. J Bacteriol. 1980;143:1127–1134. doi: 10.1128/jb.143.3.1127-1134.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1542.Guardiola J, DeFelice M, Klopotowski T, Iaccarino M. Mutations affecting the different transport systems for isoleucine, leucine, and valine in Escherichia coli K-12. J Bacteriol. 1974;117:393–405. doi: 10.1128/jb.117.2.393-405.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1543.Guardiola J, Iaccarino M. Escherichia coli K-12 mutants altered in the transport of branched-chain amino acids. J Bacteriol. 1971;108:1034–1044. doi: 10.1128/jb.108.3.1034-1044.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1544.Guarneros G, Machado G, Guzman P, Garay E. Genetic and physical location of the Escherichia coli rap locus, which is essential for growth of bacteriophage lambda. J Bacteriol. 1987;169:5188–5192. doi: 10.1128/jb.169.11.5188-5192.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1545.Gudmundsdottir A, Bradbeer C, Kadner R J. Altered binding and transport of vitamin B12 resulting from insertion mutations in the Escherichia coli btuB gene. J Biol Chem. 1988;263:14224–14230. [PubMed] [Google Scholar]
- 1546.Gudmundsdottir A, Bell P E, Lundrigan M D, Bradbeer C, Kadner R J. Point mutations in a conserved region (TonB box) of Escherichia coli outer membrane protein BtuB affect vitamin B12 transport. J Bacteriol. 1989;171:6526–6533. doi: 10.1128/jb.171.12.6526-6533.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1547.Guest J R. Biochemical and genetic studies with nitrate reductase C-gene mutants of Escherichia coli. Mol Gen Genet. 1969;105:285–297. doi: 10.1007/BF00277583. [DOI] [PubMed] [Google Scholar]
- 1548.Guest J R. Anaerobic growth of Escherichia coli K12 with fumarate as terminal electron acceptor. Genetic studies with menaquinone and fluoracetate-resistant mutants. J Gen Microbiol. 1979;115:259–271. doi: 10.1099/00221287-115-2-259. [DOI] [PubMed] [Google Scholar]
- 1549.Guest J R. Hybrid plasmids containing the citrate synthase gene (gltA) of Escherichia coli K-12. J Gen Microbiol. 1981;124:17–23. doi: 10.1099/00221287-124-1-17. [DOI] [PubMed] [Google Scholar]
- 1550.Guest J R, Shaw D J. Molecular cloning of menaquinone biosynthetic genes of Escherichia coli K12. Mol Gen Genet. 1981;181:379–383. doi: 10.1007/BF00425615. [DOI] [PubMed] [Google Scholar]
- 1551.Guest J R, Drapeau G R, Carlton B C, Yanofsky C. The amino acid sequence of the A protein (alpha subunit) of the tryptophan synthetase of Escherichia coli. J Biol Chem. 1967;242:5442–5446. [PubMed] [Google Scholar]
- 1552.Guest J R, Miles J S, Roberts R E, Woods S A. The fumarase genes of Escherichia coli: location of the fumB gene and discovery of a new gene (fumC) J Gen Microbiol. 1985;131:2971–2984. doi: 10.1099/00221287-131-11-2971. [DOI] [PubMed] [Google Scholar]
- 1553.Guest J R, Stephens P E. Molecular cloning of the pyruvate dehydrogenase complex genes of Escherichia coli. J Gen Microbiol. 1980;121:277–292. doi: 10.1099/00221287-121-2-277. [DOI] [PubMed] [Google Scholar]
- 1554.Guest J R, Stephens P E, Darlison M G, Lewis H M. The pyruvate dehydrogenase complex of Escherichia coli K12. Nucleotide sequence encoding the dihydrolipoamide acetyltransferase component. Eur J Biochem. 1983;133:481–489. doi: 10.1111/j.1432-1033.1983.tb07490.x. [DOI] [PubMed] [Google Scholar]
- 1555.Guest J R, Roberts R E. Cloning, mapping, and expression of the fumarase gene of Escherichia coli K-12. J Bacteriol. 1983;153:588–596. doi: 10.1128/jb.153.2.588-596.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1556.Guest J R, Roberts R E, Stephens P E. Hybrid plasmids containing the pyruvate dehydrogenase complex genes and gene-DNA relationships in the 2 to 3 minute region of the Escherichia coli chromosome. J Gen Microbiol. 1983;129:671–680. doi: 10.1099/00221287-129-3-671. [DOI] [PubMed] [Google Scholar]
- 1557.Guest J R, Roberts R E, Wilde R J. Cloning of the aspartase gene (aspA) of Escherichia coli. J Gen Microbiol. 1984;130:1271–1278. doi: 10.1099/00221287-130-5-1271. [DOI] [PubMed] [Google Scholar]
- 1558.Guest J R, Cole S T, Jeyaseelan K. Organization and expression of the pyruvate dehydrogenase complex genes of Escherichia coli K12. J Gen Microbiol. 1981;127:65–79. doi: 10.1099/00221287-127-1-65. [DOI] [PubMed] [Google Scholar]
- 1559.Guidi-Rontani C, Danchin A, Ullmann A. Isolation and characterization of an Escherichia coli mutant affected in the regulation of adenylate cyclase. J Bacteriol. 1981;148:753–761. doi: 10.1128/jb.148.3.753-761.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1560.Guilhot C, Jander G, Martin N L, Beckwith J R. Evidence that the pathway of disulfide bond formation in Escherichia coli involves interactions between the cysteines of DsbB and DsbA. Proc Natl Acad Sci USA. 1995;92:9895–9899. doi: 10.1073/pnas.92.21.9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1561.Guillon J M, Heiss S, Soutourina J, Mechulam Y, Laalami S, Grunberg-Manago M, Blanquet S. Interplay of methionine tRNAs with translation elongation factor Tu and translation initiation factor 2 in Escherichia coli. J Biol Chem. 1996;271:22321–22325. doi: 10.1074/jbc.271.37.22321. [DOI] [PubMed] [Google Scholar]
- 1562.Guillon J-M, Mechulam Y, Schmitter J-M, Blanquet S, Fayat G. Disruption of the gene for Met-tRNAFMet formyltransferase severely impairs growth of Escherichia coli. J Bacteriol. 1992;174:4294–4301. doi: 10.1128/jb.174.13.4294-4301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1563.Guillon J-M, Mechulam Y, Blanquet S, Fayat G. Importance of formylability and anticodon stem sequence to give a tRNAMet an initiator identity in Escherichia coli. J Bacteriol. 1993;175:4507–4514. doi: 10.1128/jb.175.14.4507-4514.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1564.Guilloton M B, Lamblin A F, Kozliak E I, Gerami-Nejad M, Tu C, Silverman D, Anderson P M, Fuchs J A. A physiological role for cyanate-induced carbonic anhydrase in Escherichia coli. J Bacteriol. 1993;175:1443–1451. doi: 10.1128/jb.175.5.1443-1451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1565.Guilloton M B, Karst F. Isolation and characterization of mutants of Escherichia coli lacking inducible cyanase. J Gen Microbiol. 1987;133:645–653. doi: 10.1099/00221287-133-3-645. [DOI] [PubMed] [Google Scholar]
- 1566.Guilloton M B, Korte J J, Lamblin A F, Fuchs J A, Anderson P M. Carbonic anhydrase in Escherichia coli: a product of the cyn operon. J Biol Chem. 1993;267:3731–3734. [PubMed] [Google Scholar]
- 1567.Gunsalus R P, Yanofsky C. Nucleotide sequence and expression of Escherichia coli trpR, the structural gene for the trp aporepressor. Proc Natl Acad Sci USA. 1980;77:7117–7121. doi: 10.1073/pnas.77.12.7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1568.Gunsalus R P, Zurawski G, Yanofsky C. Structural and functional analysis of cloned deoxyribonucleic acid containing the trpR-thr region of the Escherichia coli chromosome. J Bacteriol. 1979;140:106–113. doi: 10.1128/jb.140.1.106-113.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1569.Gunsalus R P, Kalman L V, Stewart R R. Nucleotide sequence of the narL gene that is involved in global regulation of nitrate controlled respiratory genes of Escherichia coli. Nucleic Acids Res. 1989;17:1965–1975. doi: 10.1093/nar/17.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1570.Gunsalus R P, Brusilow W S, Simoni R D. Gene order and gene-polypeptide relationships of the proton-translocating ATPase operon (unc) of Escherichia coli. Proc Natl Acad Sci USA. 1982;79:320–324. doi: 10.1073/pnas.79.2.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1571.Guo G, Weiss B. Endonuclease V (nfi) mutant of Escherichia coli K-12. J Bacteriol. 1998;180:46–51. doi: 10.1128/jb.180.1.46-51.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1572.Guo G, Ding Y, Weiss B. nfi, the gene for endonuclease V in Escherichia coli K-12. J Bacteriol. 1997;179:310–316. doi: 10.1128/jb.179.2.310-316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1573.Gupta S, Clark D P. Escherichia coli derivatives lacking both alcohol dehydrogenase and phosphotransacetylase grow anaerobically by lactate fermentation. J Bacteriol. 1989;171:3650–3655. doi: 10.1128/jb.171.7.3650-3655.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1574.Gupta S D, Lee B T O, Camakaris J, Wu H C. Identification of cutC and cutF (nlpE) genes involved in copper tolerance in Escherichia coli. J Bacteriol. 1995;177:4207–4215. doi: 10.1128/jb.177.15.4207-4215.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1575.Gustafson C E, Kaul S, Ishiguro E E. Identification of the Escherichia coli lytB gene, which is involved in penicillin tolerance and control of the stringent response. J Bacteriol. 1993;175:1203–1205. doi: 10.1128/jb.175.4.1203-1205.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1576.Gustafsson C, Persson B C. Identification of the rrmA gene encoding the 23S rRNA m1G745 methyltransferase in Escherichia coli and characterization of an m1G745-deficient mutant. J Bacteriol. 1998;180:359–365. doi: 10.1128/jb.180.2.359-365.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1577.Gustafsson C, Lindstrom P H R, Hagervall T G, Esberg K B, Bjork G R. The trmA promoter has regulatory features and sequence elements in common with the rRNA P1 promoter family of Escherichia coli. J Bacteriol. 1991;173:1757–1764. doi: 10.1128/jb.173.5.1757-1764.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1578.Gustafsson C, Reid R, Greene P J, Santi D V. Identification of new RNA modifying enzymes by iterative genome search using known modifying enzymes as probes. Nucleic Acids Res. 1996;24:3756–3762. doi: 10.1093/nar/24.19.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1579.Guterman S K, Roberts G, Tyler B. Polarity in the glnA operon: suppression of the Reg− phenotype by rho mutations. J Bacteriol. 1982;150:1314–1321. doi: 10.1128/jb.150.3.1314-1321.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1580.Guterman S K, Dann L. Excretion of enterochelin by exbA and exbB mutants of Escherichia coli. J Bacteriol. 1973;114:1225–1230. doi: 10.1128/jb.114.3.1225-1230.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1581.Guth A, Engel R, Tropp B E. Uptake of glycerol 3-phosphate and some of its analogs by the hexose phosphate transport system of Escherichia coli. J Bacteriol. 1980;143:538–539. doi: 10.1128/jb.143.1.538-539.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1582.Gutheil W G, Holmquist B, Vallee B L. Purification, characterization and partial sequence of the glutathione-dependent formaldehyde dehydrogenase from Escherichia coli: a class III alcohol dehydrogenase. Biochemistry. 1992;31:475–481. doi: 10.1021/bi00117a025. [DOI] [PubMed] [Google Scholar]
- 1583.Guthrie B, Wickner W. Trigger factor depletion or overproduction causes defective cell division but does not block protein export. J Bacteriol. 1990;172:5555–5562. doi: 10.1128/jb.172.10.5555-5562.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1584.Gutierrez C, Devedjian J C. Osmotic induction of gene osmC expression in Escherichia coli K12. J Mol Biol. 1991;220:959–973. doi: 10.1016/0022-2836(91)90366-e. [DOI] [PubMed] [Google Scholar]
- 1585.Gutierrez C, Gordia S, Bonnassie S. Characterization of the osmotically inducible gene osmE of Escherichia coli K-12. Mol Microbiol. 1995;16:553–563. doi: 10.1111/j.1365-2958.1995.tb02418.x. [DOI] [PubMed] [Google Scholar]
- 1586.Guzman E C, Jimenez-Sanchez A. Location of pinO, a new gene located between tuf and rpsJ, on the physical map of Escherichia coli. J Bacteriol. 1991;173:7409. doi: 10.1128/jb.173.23.7409.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1587.Guzman E C, Pritchard R H, Jimenez-Sanchez A. A calcium-binding protein that may be required for the initiation of chromosome replication in Escherichia coli. Res Microbiol. 1991;142:137–140. doi: 10.1016/0923-2508(91)90021-2. [DOI] [PubMed] [Google Scholar]
- 1588.Guzman L, Barondess J J, Beckwith J R. FtsL, an essential cytoplasmic membrane protein involved in cell division in Escherichia coli. J Bacteriol. 1992;174:7716–7728. [PMC free article] [PubMed] [Google Scholar]
- 1589.Guzzo A, Macintyre G, Diorio C, Salmon K, DuBow M S. Identification, sequencing, and characterization of an aluminum-inducible Escherichia coli gene. 1995. GenBank submission X83874. [Google Scholar]
- 1590.Haardt M, Bremer E. Use of phoA and lacZ fusions to study the membrane topology of ProW, a component of the osmoregulated ProU transport system of Escherichia coli. J Bacteriol. 1996;178:5370–5381. doi: 10.1128/jb.178.18.5370-5381.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1591.Hachler H, Cohen S P, Levy S B. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1991;173:5532–5538. doi: 10.1128/jb.173.17.5532-5538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1592.Hachler H, Cohen S P, Levy S B. Untranslated sequence upstream of MarA in the multiple antibiotic resistance locus of Escherichia coli is related to the effector-binding domain of the XylS transcriptional activator. J Mol Evol. 1996;42:409–413. doi: 10.1007/BF02498634. [DOI] [PubMed] [Google Scholar]
- 1593.Hackett J, Reeves P R. Primary structure of the tolC gene that codes for an outer membrane protein of Escherichia coli K12. Nucleic Acids Res. 1983;11:6487–6495. doi: 10.1093/nar/11.18.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1594.Hackett J, Misra R, Reeves P R. The TolC protein of Escherichia coli K12 is synthesized in a precursor form. FEBS Lett. 1983;156:307–310. doi: 10.1016/0014-5793(83)80518-3. [DOI] [PubMed] [Google Scholar]
- 1595.Hackett N R, Bragg P D. Membrane cytochromes of Escherichia coli mutants. J Bacteriol. 1983;154:719–727. doi: 10.1128/jb.154.2.719-727.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1596.Hadener A, Alefounder P R, Hart G J, Abell C, Battersby A R. Investigation of putative active-site lysine residues in hydroxymethylbilane synthase. Biochem J. 1990;271:487–491. doi: 10.1042/bj2710487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1597.Hadley R G, Hu M, Timmons M, Yun K, Deonier R. A partial restriction map of the proA-purE region of the E. coli K-12 chromosome. Gene. 1983;22:281–287. doi: 10.1016/0378-1119(83)90113-0. [DOI] [PubMed] [Google Scholar]
- 1598.Hafner E W, Tabor C W, Tabor H. Isolation of a metK mutant with a temperature-sensitive S-adenosylmethionine synthetase. J Bacteriol. 1977;132:832–840. doi: 10.1128/jb.132.3.832-840.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1599.Hagervall T G, Bjork G R. Genetic mapping and cloning of the gene (trmC) responsible for the synthesis of tRNA (mnm5S2U) methyl transferase. Mol Gen Genet. 1984;196:201–207. doi: 10.1007/BF00328051. [DOI] [PubMed] [Google Scholar]
- 1600.Hagewood B T, Ganduri Y L, Datta P. Functional analysis of the tdcABC promoter of Escherichia coli: roles of TdcA and TdcR. J Bacteriol. 1994;176:6214–6220. doi: 10.1128/jb.176.20.6214-6220.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1601.Haggerty T J, Lovett S T. Suppression of recJ mutations of Escherichia coli by mutations in translation initiation factor IF3. J Bacteriol. 1993;175:6118–6125. doi: 10.1128/jb.175.19.6118-6125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1602.Hale C A, de Boer P A. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 1603.Hales L M, Gumport R I, Gardner J F. Examining the contribution of a dA+dT element to the conformation of Escherichia coli integration host factor-DNA complexes. Nucleic Acids Res. 1996;24:1780–1786. doi: 10.1093/nar/24.9.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1604.Hall B G. Chromosomal mutation for citrate utilization by Escherichia coli K-12. J Bacteriol. 1982;151:269–273. doi: 10.1128/jb.151.1.269-273.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1605.Hall B G. The rtn gene of Proteus vulgaris is actually from Escherichia coli. J Bacteriol. 1997;179:2433–2434. doi: 10.1128/jb.179.7.2433-2434.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1606.Hall B G, Xu L. Nucleotide sequence, function, activation, and evolution of the cryptic asc operon of Escherichia coli K12. Mol Biol Evol. 1992;9:688–706. doi: 10.1093/oxfordjournals.molbev.a040753. [DOI] [PubMed] [Google Scholar]
- 1607.Hall B G, Betts P W, Wootton J C. DNA sequence analysis of artificially evolved ebg enzyme and ebg repressor genes. Genetics. 1989;123:635–648. doi: 10.1093/genetics/123.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1608.Hall C V, van Cleemput M, Muench K H, Yanofsky C. The nucleotide sequence of the structural gene for Escherichia coli tryptophanyl-tRNA synthetase. J Biol Chem. 1982;257:6132–6136. [PubMed] [Google Scholar]
- 1609.Hall J, Yanofsky C. Cloning and characterization of the gene for Escherichia coli tryptophanyltransfer ribonucleic acid synthetase. J Bacteriol. 1981;148:941–949. doi: 10.1128/jb.148.3.941-949.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1610.Hall M N, Silhavy T J. Genetic analysis of the ompB locus in Escherichia coli K-12. J Mol Biol. 1981;151:1–15. doi: 10.1016/0022-2836(81)90218-7. [DOI] [PubMed] [Google Scholar]
- 1611.Hall M N, Silhavy T J. The ompB locus and the regulation of the major outer membrane porin proteins of Escherichia coli K12. J Mol Biol. 1981;146:23–43. doi: 10.1016/0022-2836(81)90364-8. [DOI] [PubMed] [Google Scholar]
- 1612.Hall S D, Kane M F, Kolodner R D. Identification and characterization of the Escherichia coli RecT protein, a protein encoded by the recE region that promotes renaturation of homologous single-stranded DNA. J Bacteriol. 1993;175:277–287. doi: 10.1128/jb.175.1.277-287.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1613.Haller B L, Fuchs J A. Mapping of trxB, a mutation responsible for reduced thioredoxin reductase activity. J Bacteriol. 1984;159:1060–1062. doi: 10.1128/jb.159.3.1060-1062.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1614.Halsall D M. Strains of E. coli with defective lysine transport systems. Biochem Genet. 1975;13:109–124. doi: 10.1007/BF00486010. [DOI] [PubMed] [Google Scholar]
- 1615.Hama H, Lerner C G, Inouye S, Inouye M. Location of the gene (ndk) for nucleoside diphosphate kinase on the physical map of the Escherichia coli chromosome. J Bacteriol. 1991;173:3276. doi: 10.1128/jb.173.11.3276.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1616.Hama H, Almaula N, Lerner C G, Inouye S, Inouye M. Nucleoside diphosphate kinase from Escherichia coli: its overproduction and sequence comparison with eukaryotic enzymes. Gene. 1991;105:31–36. doi: 10.1016/0378-1119(91)90510-i. [DOI] [PubMed] [Google Scholar]
- 1617.Hama H, Kayahara T, Ogawa W, Tsuda M, Tsuchiya T. Enhancement of serine-sensitivity by a gene encoding rhodanese-like protein in Escherichia coli. J Biochem (Tokyo) 1994;115:1135–1140. doi: 10.1093/oxfordjournals.jbchem.a124469. [DOI] [PubMed] [Google Scholar]
- 1618.Hamann A, Bossemeyer D, Bakker E P. Physical mapping of the K+ transport trkA gene of Escherichia coli and overproduction of the TrkA protein. J Bacteriol. 1987;169:3138–3145. doi: 10.1128/jb.169.7.3138-3145.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1619.Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 1620.Hammelburger J W, Orr G A. Interaction of sn-glycerol 3-phosphorothioate with Escherichia coli: effect on cell growth and metabolism. J Bacteriol. 1983;156:789–799. doi: 10.1128/jb.156.2.789-799.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1621.Hammer-Jespersen K, Munch-Petersen A. Mutants of Escherichia coli unable to metabolize cytidine: isolation and characterization. Mol Gen Genet. 1973;126:177–186. doi: 10.1007/BF00330992. [DOI] [PubMed] [Google Scholar]
- 1622.Hanada M, Nishiyama K, Tokuda H. SecG plays a critical role in protein translocation in the absence of the proton motive force as well as at low temperature. FEBS Lett. 1996;381:25–28. doi: 10.1016/0014-5793(96)00066-x. [DOI] [PubMed] [Google Scholar]
- 1623.Hanafusa T, Sakai A, Tominaga A, Enomoto M. Isolation and characterization of Escherichia coli hag operator mutants whose hag48 expression has become repressible by a Salmonella H1 repressor. Mol Gen Genet. 1989;216:44–50. doi: 10.1007/BF00332229. [DOI] [PubMed] [Google Scholar]
- 1624.Hanatani M, Yazyu H, Shiota-Niiya S, Moriya Y, Kanazawa H, Futai M, Tsuchiya T. Physical and genetic characterization of the melibiose operon and identification of the gene products in Escherichia coli. J Biol Chem. 1984;259:1807–1812. [PubMed] [Google Scholar]
- 1625.Hanck T, Gerwin N, Fritz H J. Nucleotide sequence of the dcm locus of Escherichia coli K12. Nucleic Acids Res. 1989;17:5844. doi: 10.1093/nar/17.14.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1626.Hane M W, Wood T H. Escherichia coli K-12 mutants resistant to nalidixic acid: genetic mapping and dominant studies. J Bacteriol. 1969;99:238–241. doi: 10.1128/jb.99.1.238-241.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1627.Haney S A, Platko J V, Oxender D L, Calvo J M. Lrp, a leucine-responsive protein, regulates branched-chain amino acid transport genes in Escherichia coli. J Bacteriol. 1992;174:108–115. doi: 10.1128/jb.174.1.108-115.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1628.Hanke P D, Fuchs J A. Regulation of ribonucleoside diphosphate reductase mRNA synthesis in Escherichia coli. J Bacteriol. 1983;154:1040–1045. doi: 10.1128/jb.154.3.1040-1045.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1629.Hanlon S P, Hill T K, Flavell M A, Stringfellow J M, Cooper R A. 2-Phenylethylamine catabolism by Escherichia coli K-12: gene organization and expression. Microbiology. 1997;143:513–518. doi: 10.1099/00221287-143-2-513. [DOI] [PubMed] [Google Scholar]
- 1630.Hansen E B, Hansen F G, von Meyenburg K. The nucleotide sequence of the dnaA gene and the first part of the dnaN gene of Escherichia coli K-12. Nucleic Acids Res. 1982;10:7373–7385. doi: 10.1093/nar/10.22.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1631.Hansen F G, Hansen E B, Atlung T. The nucleotide sequence of the dnaA gene promoter and of the adjacent rpmH gene, coding for the ribosomal protein L34, of Escherichia coli. EMBO J. 1982;1:1043–1048. doi: 10.1002/j.1460-2075.1982.tb01294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1632.Hansen F G, Hansen E B, Atlung T. Physical mapping and nucleotide sequence of the rnpA gene that encodes the protein component of ribonuclease P in Escherichia coli. Gene. 1985;38:85–93. doi: 10.1016/0378-1119(85)90206-9. [DOI] [PubMed] [Google Scholar]
- 1633.Hansen F G, Koefoid S, von Meyenburg K, Atlung T. Transcription and translation events in the oriC region of the E. coli chromosome. ICN-UCLA Symp Mol Cell Biol. 1981;22:37–55. [Google Scholar]
- 1634.Hanson M S. Identification and characterization of Escherichia coli type 1 pilus tip adhesion protein. Nature. 1988;332:265–268. doi: 10.1038/332265a0. [DOI] [PubMed] [Google Scholar]
- 1635.Hantke K. Phage T6-colicin K receptor and nucleoside transport in Escherichia coli. FEBS Lett. 1976;70:109–112. doi: 10.1016/0014-5793(76)80737-5. [DOI] [PubMed] [Google Scholar]
- 1636.Hantke K. Identification of an iron uptake system specific for coprogen and rhodotorulic acid in Escherichia coli K12. Mol Gen Genet. 1983;191:301–306. doi: 10.1007/BF00334830. [DOI] [PubMed] [Google Scholar]
- 1637.Hantke K. Cloning of the repressor protein gene of iron-regulated systems in Escherichia coli K12. Mol Gen Genet. 1984;197:337–341. doi: 10.1007/BF00330982. [DOI] [PubMed] [Google Scholar]
- 1638.Hantke K. Ferrous iron transport mutants in Escherichia coli K12. FEMS Microbiol Lett. 1987;44:53–57. [Google Scholar]
- 1639.Hantke K. Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K12: fur not only affects iron transport. Mol Gen Genet. 1987;210:135–139. doi: 10.1007/BF00337769. [DOI] [PubMed] [Google Scholar]
- 1640.Hantke, K., and S. I. Patzer. 1995. GenBank submission Z54355.
- 1641.Hara H, Abe N, Nakakouji M, Nishimura Y, Horiuchi K. Overproduction of penicillin-binding protein 7 suppresses thermosensitive growth defect at low osmolarity due to an spr mutation of Escherichia coli. Microb Drug Resist. 1996;2:63–72. doi: 10.1089/mdr.1996.2.63. [DOI] [PubMed] [Google Scholar]
- 1642.Hara H, Yasuda S, Horiuchi K, Park J T. A promoter for the first nine genes of the Escherichia coli mra cluster of cell division and cell envelope biosynthesis genes, including ftsI and ftsW. J Bacteriol. 1997;179:5802–5811. doi: 10.1128/jb.179.18.5802-5811.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1643.Hara H, Nishimura Y, Kato J, Suzuki H, Nagasawa H, Suzuki A, Hirota Y. Genetic analyses of processing involving C-terminal cleavage in penicillin-binding protein 3 of Escherichia coli. J Bacteriol. 1989;171:5882–5889. doi: 10.1128/jb.171.11.5882-5889.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1644.Hara H, Yamamoto Y, Higashitani Y, Suzuki H, Nishimura Y. Cloning, mapping, and characterization of the Escherichia coli prc gene, which is involved in C-terminal processing of penicillin-binding protein 3. J Bacteriol. 1991;173:4799–4813. doi: 10.1128/jb.173.15.4799-4813.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1645.Harayama S, Bollinger J, Iino T, Hazelbauer G L. Characterization of the mgl operon of Escherichia coli by transposon mutagenesis and molecular cloning. J Bacteriol. 1983;153:408–415. doi: 10.1128/jb.153.1.408-415.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1646.Harayama S, Engstrom S P, Wolf-Watz H, Iino T, Hazelbauer G L. Cloning of trg, a gene for a sensory transducer in Escherichia coli. J Bacteriol. 1982;152:372–383. doi: 10.1128/jb.152.1.372-383.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1647.Harborne N, Griffiths L, Busby S J, Cole J A. Transcriptional control, translation and function of the products of the five open reading frames of the Escherichia coli nir operon. Mol Microbiol. 1992;6:2805–2813. doi: 10.1111/j.1365-2958.1992.tb01460.x. [DOI] [PubMed] [Google Scholar]
- 1648.Hardaway K L, Buller C S. Effects of ethylenediaminetetraacetate on phospholipids and outer membrane function in Escherichia coli. J Bacteriol. 1979;137:62–68. doi: 10.1128/jb.137.1.62-68.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1649.Harkness R E, Ishiguro E E. Temperature-sensitive autolysis-defective mutants of Escherichia coli. J Bacteriol. 1983;155:15–21. doi: 10.1128/jb.155.1.15-21.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1650.Harkness R E, Braun V. In vitro peptidoglycan synthesis by envelopes from Escherichia coli tolM mutants is inhibited by colicin M. J Bacteriol. 1990;172:498–500. doi: 10.1128/jb.172.1.498-500.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1651.Harkness R E, Kusser W, Qi B J, Ishiguro E E. Genetic mapping of the lytA and lytB loci of Escherichia coli, which are involved in penicillin tolerance and control of the stringent response. Can J Microbiol. 1992;38:975–978. doi: 10.1139/m92-156. [DOI] [PubMed] [Google Scholar]
- 1652.Harlocker S L, Bergstrom L, Inouye M. Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J Biol Chem. 1995;270:26849–26856. doi: 10.1074/jbc.270.45.26849. [DOI] [PubMed] [Google Scholar]
- 1653.Harlow K W, Nygaard P, Hove-Jensen B. Cloning and characterization of the gsk gene encoding guanosine kinase of Escherichia coli. J Bacteriol. 1995;177:2236–2240. doi: 10.1128/jb.177.8.2236-2240.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1654.Harmon F G, Rehrauer W M, Kowalczykowski S C. Interaction of Escherichia coli RecA protein with LexA repressor. II. Inhibition of DNA strand exchange by the uncleavable LexA S119A repressor argues that recombination and SOS induction are competitive processes. J Biol Chem. 1996;271:23874–23883. [PubMed] [Google Scholar]
- 1655.Harris S L, Elliott D A, Blake M C, Must L M, Messenger M, Orndorff P E. Isolation and characterization of mutants with lesions affecting pellicle formation and erythrocyte agglutination by type 1 piliated Escherichia coli. J Bacteriol. 1990;172:6411–6418. doi: 10.1128/jb.172.11.6411-6418.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1656.Harrison L, Hatahet Z, Purmal A A, Wallace S S. Multiply damaged sites in DNA: interactions with Escherichia coli endonucleases III and VIII. Nucleic Acids Res. 1998;26:932–941. doi: 10.1093/nar/26.4.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1657.Harrison L I, Christensen H N, Handlogten M E, Oxender D L, Quay S C. Transport of l-4-azaleucine in Escherichia coli. J Bacteriol. 1975;122:957–965. doi: 10.1128/jb.122.3.957-965.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1658.Hartlein M, Madern D. Molecular cloning and nucleotide sequence of the gene for Escherichia coli leucyl-tRNA. Nucleic Acids Res. 1987;15:10199–10210. doi: 10.1093/nar/15.24.10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1659.Hartlein M, Madern D, Leberman R. Cloning and characterization of the gene for Escherichia coli seryl-tRNA synthetase. Nucleic Acids Res. 1987;15:1005–1017. doi: 10.1093/nar/15.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1660.Hartlein M, Frank R, Madern D. Nucleotide sequence of Escherichia coli valyl-tRNA synthetase gene valS. Nucleic Acids Res. 1987;15:9081–9082. doi: 10.1093/nar/15.21.9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1661.Harvey S, Hill C W, Squires C, Squires C L. Loss of the spacer loop sequence from the rrnB operon in the Escherichia coli K-12 subline that bears the relA1 mutation. J Bacteriol. 1988;170:1235–1238. doi: 10.1128/jb.170.3.1235-1238.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1662.Hasenbank R, Guthrie C, Stoffler G, Wittmann H G, Rosen L, Apirion D. Electrophoretic and immunological studies on ribosomal proteins of 100 Escherichia coli revertants from streptomycin dependence. Mol Gen Genet. 1973;127:1–18. doi: 10.1007/BF00267778. [DOI] [PubMed] [Google Scholar]
- 1663.Hashimoto W, Suzuki H, Nohara S, Tachi H, Yamamoto K, Kumagai H. Subunit association of gamma-glutamyltranspeptidase of Escherichia coli K-12. J Biochem (Tokyo) 1995;118:1216–1223. doi: 10.1093/oxfordjournals.jbchem.a125010. [DOI] [PubMed] [Google Scholar]
- 1664.Hasona A, Ray R M, Shanmugam K T. Physiological and genetic analyses leading to identification of a biochemical role for the moeA (molybdate metabolism) gene product in Escherichia coli. J Bacteriol. 1998;180:1466–1472. doi: 10.1128/jb.180.6.1466-1472.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1665.Hassani M, Pincus D H, Bennett G N, Hirshfield I N. Temperature-dependent induction of an acid-inducible stimulon of Escherichia coli in broth. Appl Environ Microbiol. 1992;58:2704–2707. doi: 10.1128/aem.58.8.2704-2707.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1666.Haughn G W, Squires C H, DeFelice M, Largo C T, Calvo J M. Unusual organization of the ilvIH promoter of Escherichia coli. J Bacteriol. 1985;163:186–198. doi: 10.1128/jb.163.1.186-198.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1667.Hauser C A, Hatfield G W. Nucleotide sequence of the ilvB multivalent attenuator region of Escherichia coli K-12. Nucleic Acids Res. 1983;11:127–139. doi: 10.1093/nar/11.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1668.Haussmann C, Rohdich F, Lottspeich F, Eberhardt S, Scheuring J, Mackamul S, Bacher A. Dihydroneopterin triphosphate epimerase of Escherichia coli: purification, genetic cloning, and expression. J Bacteriol. 1997;179:949–951. doi: 10.1128/jb.179.3.949-951.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1669.Hawker J R, McHenry C S. Monoclonal antibodies specific for the tau subunit of the DNA polymerase III holoenzyme of Escherichia coli. Use to demonstrate that tau is the product of the dnaZX gene and that both it and gamma, the dnaZ gene product, are integral. J Biol Chem. 1987;262:12722–12727. [PubMed] [Google Scholar]
- 1670.Hayano T, Takahashi N, Kato S, Maki N, Suzuki M. Two distinct forms of peptidylprolyl-cis-trans-isomerase are expressed separately in periplasmic and cytoplasmic compartments of Escherichia coli cells. Biochemistry. 1991;30:3041–3048. doi: 10.1021/bi00226a009. [DOI] [PubMed] [Google Scholar]
- 1671.Hayashi S, Chang S-Y, Chang S, Giam C Z, Wu H C. Modification and processing of internalized signal sequences of prolipoprotein in Escherichia coli and in Bacillus subtilis. J Biol Chem. 1985;260:5753–5759. [PubMed] [Google Scholar]
- 1672.Hayden M A, Huang I, Bussiere D E, Ashley G W. The biosynthesis of lipoic acid. Cloning of lip, a lipoate biosynthetic locus of Escherichia coli. J Biol Chem. 1992;267:9512–9515. [PubMed] [Google Scholar]
- 1673.Hayden M A, Huang I, Iliopoulos G, Orozco M, Ashley G W. Biosynthesis of lipoic acid: characterization of the lipoic acid auxotrophs Escherichia coli W1485-lip2 and JRG33-lip9. Biochemistry. 1993;32:3778–3782. doi: 10.1021/bi00065a033. [DOI] [PubMed] [Google Scholar]
- 1674.Haydon D J, Quail M A, Guest J R. A mutation causing constitutive synthesis of the pyruvate dehydrogenase complex in Escherichia coli is located within the pdhR gene. FEBS Lett. 1993;336:43–47. doi: 10.1016/0014-5793(93)81605-y. [DOI] [PubMed] [Google Scholar]
- 1675.Hayes F, Sherratt D J. Recombinase binding specificity at the chromosome dimer resolution site dif of Escherichia coli. J Mol Biol. 1997;266:525–537. doi: 10.1006/jmbi.1996.0828. [DOI] [PubMed] [Google Scholar]
- 1676.Hayzer D J. Sub-cloning of the wild-type proAB region of the Escherichia coli genome. J Gen Microbiol. 1983;129:3215–3225. doi: 10.1099/00221287-129-10-3215. [DOI] [PubMed] [Google Scholar]
- 1677.Hayzer D J, Leisinger T. The gene-enzyme relationships of proline biosynthesis in Escherichia coli. J Gen Microbiol. 1980;118:287–293. doi: 10.1099/00221287-118-2-287. [DOI] [PubMed] [Google Scholar]
- 1678.Hazelbauer G L, Engstrom S P, Harayama S. Methyl-accepting chemotaxis protein III and transducer gene trg. J Bacteriol. 1981;145:43–49. doi: 10.1128/jb.145.1.43-49.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1679.Haziza C, Cassan M, Patte J C. Identification of the promoter of the asd gene of Escherichia coli using in vitro fusion with the lac operon. Biochimie. 1982;64:227–230. doi: 10.1016/s0300-9084(82)80473-2. [DOI] [PubMed] [Google Scholar]
- 1680.Haziza C, Stragier P, Patte J C. Nucleotide sequence of the asd gene of Escherichia coli: absence of a typical attenuation signal. EMBO J. 1982;1:379–384. doi: 10.1002/j.1460-2075.1982.tb01178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1681.He B, Zalkin H. Regulation of Escherichia coli purA by purine repressor, one component of a dual control mechanism. J Bacteriol. 1994;176:1009–1013. doi: 10.1128/jb.176.4.1009-1013.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1682.He B, Smith J M, Zalkin H. Escherichia coli purB gene: cloning, nucleotide sequence, and regulation by purR. J Bacteriol. 1992;174:130–136. doi: 10.1128/jb.174.1.130-136.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1683.He B, Choi K Y, Zalkin H. Regulation of Escherichia coli glnB, prsA, and speA by the purine repressor. J Bacteriol. 1993;175:3598–3606. doi: 10.1128/jb.175.11.3598-3606.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1684.He L, Soderbom F, Wagner E G, Binnie U, Binns N, Masters M. PcnB is required for the rapid degradation of RNAI, the antisense RNA that controls the copy number of ColE1-related plasmids. Mol Microbiol. 1993;9:1131–1142. doi: 10.1111/j.1365-2958.1993.tb01243.x. [DOI] [PubMed] [Google Scholar]
- 1685.He M M, Sun J, Kaback H R. Cysteine-scanning mutagenesis of transmembrane domain XII and the flanking periplasmic loop in the lactose permease of Escherichia coli. Biochemistry. 1996;35:12909–12914. doi: 10.1021/bi960876b. [DOI] [PubMed] [Google Scholar]
- 1686.He X Y, Yang S Y, Schulz H. Cloning and expression of the fadH gene and characterization of the gene product 2,4-dienoyl coenzyme A reductase from Escherichia coli. Eur J Biochem. 1997;248:516–520. doi: 10.1111/j.1432-1033.1997.00516.x. [DOI] [PubMed] [Google Scholar]
- 1687.Heath R J, Rock C O. Enoyl-acyl carrier protein reductase (fabI) plays a determinant role in completing cycles of fatty acid elongation in Escherichia coli. J Biol Chem. 1995;270:26538–26542. doi: 10.1074/jbc.270.44.26538. [DOI] [PubMed] [Google Scholar]
- 1688.Heatwole V M, Somerville R L. Cloning, nucleotide sequence, and characterization of mtr, the structural gene for a tryptophan-specific permease of Escherichia coli K-12. J Bacteriol. 1991;173:108–115. doi: 10.1128/jb.173.1.108-115.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1689.Heatwole V M, Somerville R L. The tryptophan-specific permease gene, mtr, is differentially regulated by the tryptophan and tyrosine repressors in Escherichia coli K-12. J Bacteriol. 1991;173:3601–3604. doi: 10.1128/jb.173.11.3601-3604.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1690.Heber S, Tropp B E. Genetic regulation of cardiolipin synthase in Escherichia coli. Biochim Biophys Acta. 1991;1129:1–12. doi: 10.1016/0167-4781(91)90206-2. [DOI] [PubMed] [Google Scholar]
- 1691.Heck J D, Hatfield G W. Valyl-tRNA synthetase gene of Escherichia coli K-12. Molecular genetic characterization. J Biol Chem. 1988;263:857–867. [PubMed] [Google Scholar]
- 1692.Heck J D, Hatfield G W. Valyl-tRNA synthetase gene of Escherichia coli K12. Primary structure and homology within a family of aminoacyl-tRNA synthetases. J Biol Chem. 1988;263:868–877. [PubMed] [Google Scholar]
- 1693.Hedblom M L, Adler J. Genetic and biochemical properties of Escherichia coli mutants with defects in serine chemotaxis. J Bacteriol. 1980;144:1048–1060. doi: 10.1128/jb.144.3.1048-1060.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1694.Hedges R W, Shannon K P. Resistance to apramycin in Escherichia coli isolated from animals: detection of a novel aminoglycoside-modifying enzyme. J Gen Microbiol. 1984;130:473–482. doi: 10.1099/00221287-130-3-473. [DOI] [PubMed] [Google Scholar]
- 1695.Hedgpeth J, Clement J M, Marchal C, Perrin D M, Hofnung M. DNA sequence encoding the NH2-terminal peptide involved in transport of λ receptor, an Escherichia coli secretory protein. Proc Natl Acad Sci USA. 1980;77:2621–2625. doi: 10.1073/pnas.77.5.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1696.Hediger M A, Johnson D F, Nierlich D P, Zabin I. DNA sequence of the lactose operon: the lacA gene and the transcriptional termination region. Proc Natl Acad Sci USA. 1985;82:6414–6418. doi: 10.1073/pnas.82.19.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1697.Heim R, Strehler E E. Cloning an Escherichia coli gene encoding a protein remarkably similar to mammalian aldehyde dehydrogenases. Gene. 1991;99:15–23. doi: 10.1016/0378-1119(91)90028-a. [DOI] [PubMed] [Google Scholar]
- 1698.Heinrich B, Schroeder U, Frank R, Plapp R. Accurate mapping of the Escherichia coli pepD gene by sequence analysis of its 5′ flanking region. Mol Gen Genet. 1989;215:369–373. doi: 10.1007/BF00427031. [DOI] [PubMed] [Google Scholar]
- 1698a.Heinrichs D E, Monteiro M A, Perry M B, Whitfield C. The assembly system for the lipopolysaccharide R2 core-type of Escherichia coli is a hybrid of those found in Escherichia coli K-12 and Salmonella enterica. Structure and function of the WaaK and WaaL homologs. J Biol Chem. 1998;273:8849–8859. doi: 10.1074/jbc.273.15.8849. [DOI] [PubMed] [Google Scholar]
- 1699.Heisig P. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 1996;40:879–885. doi: 10.1128/aac.40.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1700.Heitman J, Model P. Site-specific methylases induce the SOS DNA repair response in Escherichia coli. J Bacteriol. 1987;169:3243–3250. doi: 10.1128/jb.169.7.3243-3250.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1701.Hekstra D, Tommassen J. Functional exchangeability of the ABC proteins of the periplasmic binding protein-dependent transport systems Ugp and Mal of Escherichia coli. J Bacteriol. 1993;175:6546–6552. doi: 10.1128/jb.175.20.6546-6552.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1702.Helander I M, Lindner B, Seydel U, Vaara M. Defective biosynthesis of the lipid A component of temperature-sensitive firA (omsA) mutant of Escherichia coli. Eur J Biochem. 1993;212:363–369. doi: 10.1111/j.1432-1033.1993.tb17670.x. [DOI] [PubMed] [Google Scholar]
- 1703.Held W A, Smith O H. Mechanism of 3-methyl-anthranilic acid derepression of the tryptophan operon in Escherichia coli. J Bacteriol. 1970;101:209–217. doi: 10.1128/jb.101.1.209-217.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1704.Held W A, Smith O H. Regulation of the Escherichia coli tryptophan operon by early reactions in the aromatic pathway. J Bacteriol. 1970;101:202–208. doi: 10.1128/jb.101.1.202-208.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1705.Heller K, Kadner R J. Nucleotide sequence of the gene for the vitamin B12 receptor protein in the outer membrane of Escherichia coli. J Bacteriol. 1985;161:904–908. doi: 10.1128/jb.161.3.904-908.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1706.Helling R B. Selection of a mutant of Escherichia coli which has high mutation rates. J Bacteriol. 1968;96:975–980. doi: 10.1128/jb.96.4.975-980.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1707.Helling R B. The glutamate dehydrogenase structural gene of Escherichia coli. Mol Gen Genet. 1990;223:508–512. doi: 10.1007/BF00264460. [DOI] [PubMed] [Google Scholar]
- 1708.Helling R B. icdB mutants of Escherichia coli. J Bacteriol. 1995;177:2592–2593. doi: 10.1128/jb.177.9.2592-2593.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1709.Helling R B, Kukora J. Nalidixic acid-resistant mutants of Escherichia coli deficient in isocitrate dehydrogenase. J Bacteriol. 1971;105:1224–1226. doi: 10.1128/jb.105.3.1224-1226.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1710.Hellinga H W, Evans P R. Nucleotide sequence and high-level expression of the major Escherichia coli phosphofructokinase. Eur J Biochem. 1985;149:363–373. doi: 10.1111/j.1432-1033.1985.tb08934.x. [DOI] [PubMed] [Google Scholar]
- 1711.Helser T L, Davies J E, Dahlberg J E. Mechanism of kasugamycin resistance in Escherichia coli. Nat New Biol. 1972;235:6–9. doi: 10.1038/newbio235006a0. [DOI] [PubMed] [Google Scholar]
- 1712.Henderson I R, Meehan M, Owen P. A novel regulatory mechanism for a novel phase-variable outer membrane protein of Escherichia coli. Adv Exp Med Biol. 1997;412:349–355. doi: 10.1007/978-1-4899-1828-4_56. [DOI] [PubMed] [Google Scholar]
- 1712a.Henderson I R, Meehan M, Owen P. Antigen 43, a phase-variable bipartite outer membrane protein, determines colony morphology and autoaggregation in Escherichia coli K-12. FEMS Microbiol Lett. 1997;149:115–120. doi: 10.1111/j.1574-6968.1997.tb10317.x. [DOI] [PubMed] [Google Scholar]
- 1713.Henderson T A, Young K D, Denome S A, Elf P K. AmpC and AmpH, proteins related to the class C β-lactamases, bind penicillin and contribute to the normal morphology of Escherichia coli. J Bacteriol. 1997;179:6112–6121. doi: 10.1128/jb.179.19.6112-6121.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1714.Henderson T A, Templin M F, Young K D. Identification and cloning of the gene encoding penicillin-binding protein 7 of Escherichia coli. J Bacteriol. 1995;177:2074–2079. doi: 10.1128/jb.177.8.2074-2079.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1715.Henderson T A, Dombrosky P M, Young K D. Artifactual processing of penicillin-binding proteins 7 and 1b by the OmpT protease of Escherichia coli. J Bacteriol. 1994;176:256–259. doi: 10.1128/jb.176.1.256-259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1716.Hendrickson W, Stoner C M, Schleif R. Characterization of the Escherichia coli araFGH and araJ promoters. J Mol Biol. 1990;215:497–510. doi: 10.1016/S0022-2836(05)80163-9. [DOI] [PubMed] [Google Scholar]
- 1717.Hendrickson W, Rudd K E. Physical map location of the argFGH operon of Escherichia coli. J Bacteriol. 1992;174:3836–3837. doi: 10.1128/jb.174.11.3836-3837.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1718.Hengge-Aronis R. Back to log phase: sigma S as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol Microbiol. 1996;21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- 1719.Hengge-Aronis R, Fischer D. Identification and molecular analysis of glgS, a novel growth-phase-regulated and rpoS-dependent gene involved in glycogen synthesis in Escherichia coli. Mol Microbiol. 1992;6:1877–1887. doi: 10.1111/j.1365-2958.1992.tb01360.x. [DOI] [PubMed] [Google Scholar]
- 1720.Hennecke F, Kolmar H, Brundl K, Fritz H J. The vsr gene product of E. coli K-12 is a strand- and sequence-specific DNA mismatch endonuclease. Nature. 1991;353:776–778. doi: 10.1038/353776a0. [DOI] [PubMed] [Google Scholar]
- 1721.Hennessey E S, Broome-Smith J K. Two related bacterial membrane proteins, ExbD and TolR, have opposite transmembrane charge dipolarity. Mol Microbiol. 1994;11:417. doi: 10.1111/j.1365-2958.1994.tb00321.x. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 1722.Henrich B, Plapp R. Locations of the genes from pepD through proA on the physical map of the Escherichia coli chromosome. J Bacteriol. 1991;173:7407–7408. doi: 10.1128/jb.173.23.7407-7408.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1723.Henrich B, Becker S, Schroeder U, Plapp R. dcp gene of Escherichia coli: cloning, sequencing, transcript mapping, and characterization of the gene product. J Bacteriol. 1993;175:7290–7300. doi: 10.1128/jb.175.22.7290-7300.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1724.Henrich B, Monnerjahn U, Plapp R. Peptidase D gene (pepD) of Escherichia coli K-12: nucleotide sequence, transcript mapping, and comparison with other peptidase genes. J Bacteriol. 1990;172:4641–4651. doi: 10.1128/jb.172.8.4641-4651.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1725.Henson J M, Blinkowa A, Walker J R. The Escherichia coli dnaW mutation is an allele of the adk gene. Mol Gen Genet. 1982;186:488–492. doi: 10.1007/BF00337953. [DOI] [PubMed] [Google Scholar]
- 1726.Henson J M, Walker J R. Genetic analysis of acrA and lir mutations of Escherichia coli. J Bacteriol. 1982;152:1301–1302. doi: 10.1128/jb.152.3.1301-1302.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1727.Herman C, Thevenet D, D’Ari R, Bouloc P. Degradation of sigma 32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc Natl Acad Sci USA. 1995;92:3516–3520. doi: 10.1073/pnas.92.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1728.Herman C, Ogura T, Tomoyasu T, Hiraga S, Akiyama Y, Ito K, Thomas R, D’Ari R, Bouloc P. Cell growth and lambda phage development controlled by the same essential Escherichia coli gene, ftsH/hflB. Proc Natl Acad Sci USA. 1993;90:10861–10865. doi: 10.1073/pnas.90.22.10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1729.Herold C, Birge E A. Location of icdA and fadR on the physical map of Escherichia coli. J Bacteriol. 1990;172:6618. doi: 10.1128/jb.172.12.6618.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1730.Herrington M B, Kohli A, Lapchak P H. Suppression by thymidine-requiring mutants of Escherichia coli K-12. J Bacteriol. 1984;157:126–129. doi: 10.1128/jb.157.1.126-129.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1731.Herrington M B, Basso J, Faraci M, Autexier C. Modification of the suppressor phenotype of thymine requiring strains of Escherichia coli. Genet Res. 1991;58:185–192. doi: 10.1017/s0016672300029931. [DOI] [PubMed] [Google Scholar]
- 1732.Hersh B M, Farooq F T, Barstad D N, Blankenhorn D L, Slonczewski J L. A glutamate-dependent acid resistance gene in Escherichia coli. J Bacteriol. 1996;178:3978–3981. doi: 10.1128/jb.178.13.3978-3981.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1733.Hershey H, Taylor M W. Nucleotide sequence and deduced amino acid sequence of Escherichia coli adenine phosphoribosyltransferase and comparison with other analogous enzymes. Gene. 1986;43:287–293. doi: 10.1016/0378-1119(86)90218-0. [DOI] [PubMed] [Google Scholar]
- 1734.Hershey J W B, Gutstein R, Taylor M W. Cloning and restriction map of the E. coli apt gene. Gene. 1982;19:89–92. doi: 10.1016/0378-1119(82)90192-5. [DOI] [PubMed] [Google Scholar]
- 1735.Hesse J E, Wieczorek L, Altendorf K, Reicin E, Dorus E, Epstein W. Sequence homology between two membrane transport ATPases, the Kdp-ATPase of Escherichia coli and the Ca2+-ATPase of sarcoplasmic reticulum. Proc Natl Acad Sci USA. 1984;81:4746–4750. doi: 10.1073/pnas.81.15.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1736.Hesslinger C, Fairhurst S, Sawers G. Novel keto acid formate-lyase and propionate kinase enzymes are components of an anaerobic pathway in Escherichia coli that degrades l-threonine to propionate. Mol Microbiol. 1998;27:477–492. doi: 10.1046/j.1365-2958.1998.00696.x. [DOI] [PubMed] [Google Scholar]
- 1737.Hesterkamp T, Bukau B. The Escherichia coli trigger factor. FEBS Lett. 1996;389:32–34. doi: 10.1016/0014-5793(96)00582-0. [DOI] [PubMed] [Google Scholar]
- 1738.Hesterkamp T, Hauser S, Lutcke H, Bukau B. Escherichia coli trigger factor is a prolyl isomerase that associates with nascent polypeptide chains. Proc Natl Acad Sci USA. 1996;93:4437–4441. doi: 10.1073/pnas.93.9.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1739.Heurgue-Hamard V, Karimi R, Mora L, MacDougall J, Leboeuf C, Grentzmann G, Ehrenberg M, Buckingham R H. Ribosome release factor RF4 and termination factor RF3 are involved in dissociation of peptidyl-tRNA from the ribosome. EMBO J. 1998;17:808–816. doi: 10.1093/emboj/17.3.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1740.Heyde M, Portalier R. New pleiotropic alkaline phosphatase-negative mutants of Escherichia coli K-12. J Bacteriol. 1982;151:529–533. doi: 10.1128/jb.151.2.529-533.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1741.Hickson I D, Arthur H M, Bramhill D, Emmerson P T. The E. coli uvrD gene product is DNA helicase II. Mol Gen Genet. 1983;190:265–270. doi: 10.1007/BF00330649. [DOI] [PubMed] [Google Scholar]
- 1742.Hickson I D, Atkinson K E, Emmerson P T. Molecular cloning and amplification of the gene for thymidylate synthetase. Gene. 1982;18:257–260. doi: 10.1016/0378-1119(82)90163-9. [DOI] [PubMed] [Google Scholar]
- 1743.Hickson I D, Emmerson P T. Identification of the Escherichia coli recB and recC gene products. Nature. 1981;294:578–580. doi: 10.1038/294578a0. [DOI] [PubMed] [Google Scholar]
- 1744.Hidaka M, Akiyama M, Horiuchi T. A consensus sequence of three DNA replication terminus sites on the E. coli chromosome is highly homologous to the terR sites of the R6K plasmid. Cell. 1988;55:467–475. doi: 10.1016/0092-8674(88)90033-5. [DOI] [PubMed] [Google Scholar]
- 1745.Hidaka M, Kobayashi T, Takenaka S, Takeya H, Horiuchi T. Purification of a DNA replication terminus (ter) site-binding protein in Escherichia coli and identification of the structural gene. J Biol Chem. 1989;264:21031–21037. [PubMed] [Google Scholar]
- 1746.Hidaka M, Kobayashi T, Horiuchi T. A newly identified DNA replication terminus site, TerE, on the Escherichia coli chromosome. J Bacteriol. 1991;173:391–393. doi: 10.1128/jb.173.1.391-393.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1747.Hidalgo E, Limon A, Aguilar J. A second Escherichia coli gene with similarity to gapA. Microbiologia. 1996;12:99–106. [PubMed] [Google Scholar]
- 1748.Hidalgo E, Demple B. Activation of SoxR-dependent transcription in vitro by noncatalytic or NifS-mediated assembly of [2Fe-2S] clusters into apo-SoxR. J Biol Chem. 1996;271:7269–7272. doi: 10.1074/jbc.271.13.7269. [DOI] [PubMed] [Google Scholar]
- 1749.Hidalgo E, Chen Y-M, Lin E C C, Aguilar J. Molecular cloning and DNA sequencing of the Escherichia coli K-12 ald gene encoding aldehyde dehydrogenase. J Bacteriol. 1991;173:6118–6123. doi: 10.1128/jb.173.19.6118-6123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1750.Higgins C F, Dorman C J, Stirling D A, Waddell L, Booth I R, May G, Bremer E. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988;52:569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- 1751.Highton P J, Chang Y, Marcotte W R, Jr, Schnaitman C A. Evidence that the outer membrane protein gene nmpC of Escherichia coli K-12 lies within the defective qsr′ prophage. J Bacteriol. 1985;162:256–262. doi: 10.1128/jb.162.1.256-262.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1752.Hill C W, Harnish B W. Transposition of a chromosomal segment bounded by redundant rRNA genes into other rRNA genes in Escherichia coli. J Bacteriol. 1982;149:449–457. doi: 10.1128/jb.149.2.449-457.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1753.Hill C W, Gray J A, Brody H. Use of the isocitrate dehydrogenase structural gene for attachment of e14 in Escherichia coli K-12. J Bacteriol. 1989;171:4083–4084. doi: 10.1128/jb.171.7.4083-4084.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1754.Hill T M, Pelletier A J, Tecklenburg M L, Kuempel P. Identification of the DNA sequence from the E. coli terminus region that halts replication forks. Cell. 1988;55:459–466. doi: 10.1016/0092-8674(88)90032-3. [DOI] [PubMed] [Google Scholar]
- 1755.Hill T M, Sharma B, Valjavec-Gratian M, Smith J. sfi-independent filamentation in Escherichia coli is lexA dependent and requires DNA damage for induction. J Bacteriol. 1997;179:1931–1939. doi: 10.1128/jb.179.6.1931-1939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1756.Hill T M, Kopp B J, Kuempel P. Termination of DNA replication in Escherichia coli requires a trans-acting factor. J Bacteriol. 1988;170:662–668. doi: 10.1128/jb.170.2.662-668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1757.Hill T M, Henson J M, Kuempel P. The terminus region of the Escherichia coli chromosome contains two separate loci that exhibit polar inhibition of replication. Proc Natl Acad Sci USA. 1987;84:1754–1758. doi: 10.1073/pnas.84.7.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1758.Hill T M, Tecklenburg M L, Pelletier A J, Kuempel P. tus, the trans-acting gene required for termination of DNA replication in Escherichia coli, encodes a DNA-binding protein. Proc Natl Acad Sci USA. 1989;86:1593–1597. doi: 10.1073/pnas.86.5.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1759.Hiom K, Sedgwick S G. Cloning and structural characterization of the mcrA locus of Escherichia coli. J Bacteriol. 1991;173:7368–7373. doi: 10.1128/jb.173.22.7368-7373.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1760.Hiraga S. Operator mutants of the tryptophan operon in Escherichia coli. J Mol Biol. 1969;39:159–179. doi: 10.1016/0022-2836(69)90340-4. [DOI] [PubMed] [Google Scholar]
- 1761.Hiraga S, Niki H, Imamura R, Ogura T, Yamanaka K, Feng J, Ezaki B, Jaffe A. Mutants defective in chromosome partitioning in E. coli. Res Microbiol. 1991;142:189–194. doi: 10.1016/0923-2508(91)90029-a. [DOI] [PubMed] [Google Scholar]
- 1762.Hiraga S, Niki H, Ogura T, Ichinose C, Mori H, Ezaki B, Jaffe A. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J Bacteriol. 1989;171:1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1763.Hiraga S, Igarashi K, Yura T. A deoxythymidine kinase-deficient mutant of Escherichia coli. I. Isolation and some properties. Biochim Biophys Acta. 1967;145:41–51. doi: 10.1016/0005-2787(67)90652-1. [DOI] [PubMed] [Google Scholar]
- 1764.Hiraga S, Ito K, Matsuyama S, Ozeki H, Yura T. 5-Methyltryptophan-resistant mutations linked with the arginine G marker in Escherichia coli. J Bacteriol. 1968;96:1880–1881. doi: 10.1128/jb.96.5.1880-1881.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1765.Hiraoka S, Matsuzaki H, Shibuya I. Active increase in cardiolipin synthesis in the stationary growth phase and its physiological significance in Escherichia coli. FEBS Lett. 1993;336:221–224. doi: 10.1016/0014-5793(93)80807-7. [DOI] [PubMed] [Google Scholar]
- 1766.Hiraoka S, Nukui K, Uetake N, Ohta A, Shibuya I. Amplification and substantial purification of cardiolipin synthase of Escherichia coli. J Biochem (Tokyo) 1991;110:443–449. doi: 10.1093/oxfordjournals.jbchem.a123600. [DOI] [PubMed] [Google Scholar]
- 1767.Hiratsu K, Amemura M, Nashimoto H, Shinagawa H, Makino K. The rpoE gene of Escherichia coli, which encodes ςE, is essential for bacterial growth at high temperature. J Bacteriol. 1995;177:2918–2922. doi: 10.1128/jb.177.10.2918-2922.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1768.Hirota S, Inuzuka M, Tomoeda M. Elective selection of proline-requiring mutants. J Bacteriol. 1966;91:2392. doi: 10.1128/jb.91.6.2392-.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1769.Hirota Y, Yasuda S, Yamada M, Nishimura A, Sugimoto K, Sugisaki H, Oka A, Takanami M. Structural and functional properties of the Escherichia coli origin of DNA replication. Cold Spring Harbor Symp Quant Biol. 1978;43:129–138. doi: 10.1101/sqb.1979.043.01.019. [DOI] [PubMed] [Google Scholar]
- 1770.Hirschman J, Wong P-K, Sei K, Keener J, Kustu S. Products of nitrogen regulatory genes ntrA and ntrC of enteric bacteria activate glnA transcription in vitro: evidence that the ntrA product is a sigma factor. Proc Natl Acad Sci USA. 1985;82:7525–7529. doi: 10.1073/pnas.82.22.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1771.Hirshfield I N, Zamecnik P C. Thiosine-resistant mutants of Escherichia coli K-12 with growth medium-dependent lysyl-tRNA synthetase activity. I. Isolation and physiological characterization. Biochim Biophys Acta. 1972;259:330–343. [PubMed] [Google Scholar]
- 1772.Hirshfield I N, Dedeken R, Horn P C, Hopwood D A, Maas W K. Studies on the mechanism of repression of arginine biosynthesis in Escherichia coli. III. Repression of enzymes of arginine biosynthesis in arginyl-tRNA synthetase mutants. J Mol Biol. 1968;35:83–93. doi: 10.1016/s0022-2836(68)80038-5. [DOI] [PubMed] [Google Scholar]
- 1773.Hirvas L, Coleman J, Koski P, Vaara M. Bacterial “histone-like protein I” (HLP-I) is an outer membrane constituent? FEBS Lett. 1990;262:123–126. doi: 10.1016/0014-5793(90)80169-j. [DOI] [PubMed] [Google Scholar]
- 1774.Hirvas L, Nurminen M, Helander I M, Vuorio R, Vaara M. The lipid A biosynthesis deficiency of the Escherichia coli antibiotic-supersensitive mutant LH530 is suppressed by a novel locus, ORF195. Microbiology. 1997;143:73–81. doi: 10.1099/00221287-143-1-73. [DOI] [PubMed] [Google Scholar]
- 1775.Hirvas L, Koski P, Vaara M. The ompH gene of Yersinia enterocolitica: cloning, sequencing, expression, and comparison with known enterobacterial ompH sequences. J Bacteriol. 1991;173:1223–1229. doi: 10.1128/jb.173.3.1223-1229.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1776.Hoben P, Royal N, Cheung A, Yamao F, Biemann K, Soll D. Escherichia coli glutaminyl-tRNA synthetase. II. Characterization of the glnS gene product. J Biol Chem. 1982;257:11644–11650. [PubMed] [Google Scholar]
- 1777.Hoerter J, Pierce A, Troupe C, Epperson J, Eisenstark A. Role of enterobactin and intracellular iron in cell lethality during near-UV irradiation in Escherichia coli. Photochem Photobiol. 1996;64:537–541. doi: 10.1111/j.1751-1097.1996.tb03102.x. [DOI] [PubMed] [Google Scholar]
- 1778.Hofnung M, Jezierska A, Braun-Breton C. lamB mutations in E. coli K 12: growth of lambda host range mutants and effect of nonsense suppressors. Mol Gen Genet. 1976;145:207–213. doi: 10.1007/BF00269595. [DOI] [PubMed] [Google Scholar]
- 1779.Hofnung M, Lepouce E, Braun-Breton C. General method for fine mapping of the Escherichia coli K-12 lamB gene: localization of missense mutations affecting bacteriophage lambda adsorption. J Bacteriol. 1981;148:853–860. doi: 10.1128/jb.148.3.853-860.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1780.Hofnung M, Schwartz M. Mutations allowing growth on maltose of Escherichia coli K12 strains with a deleted malT gene. Mol Gen Genet. 1971;112:117–132. doi: 10.1007/BF00267490. [DOI] [PubMed] [Google Scholar]
- 1781.Hofnung M, Schwartz M, Hatfield D. Complementation studies in the maltose-A region of the Escherichia coli K12 genetic map. J Mol Biol. 1971;61:681–694. doi: 10.1016/0022-2836(71)90072-6. [DOI] [PubMed] [Google Scholar]
- 1782.Hogarth B G, Higgins C F. Genetic organization of the oligopeptide permease (opp) locus of Salmonella typhimurium and Escherichia coli. J Bacteriol. 1983;153:1548–1551. doi: 10.1128/jb.153.3.1548-1551.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1783.Hogg R W, Voelker C, Von Carlowitz I. Nucleotide sequence and analysis of the mgl operon of Escherichia coli K12. Mol Gen Genet. 1991;229:453–459. doi: 10.1007/BF00267469. [DOI] [PubMed] [Google Scholar]
- 1784.Holck A L, Kleppe K. Cloning and sequencing of the gene for the DNA-binding 17K protein of Escherichia coli. Gene. 1988;67:117–124. doi: 10.1016/0378-1119(88)90014-5. [DOI] [PubMed] [Google Scholar]
- 1785.Hollenbach A D, Dickson K A, Washabaugh M W. E. coli periplasmic thiamin binding protein: cloning, overexpression, purification, and characterization. 1994. GenBank submission U09984. [DOI] [PubMed] [Google Scholar]
- 1786.Holmes Z, Henning U. Location of the ompT gene relative to that of appY on the physical map of the Escherichia coli chromosome. Mol Gen Genet. 1994;242:363–364. doi: 10.1007/BF00280427. [DOI] [PubMed] [Google Scholar]
- 1787.Holowachuk E W, Friesen J D. Isolation of a recombinant lambda phage carrying nusA and surrounding region of the Escherichia coli chromosome. Mol Gen Genet. 1982;187:248–253. doi: 10.1007/BF00331126. [DOI] [PubMed] [Google Scholar]
- 1788.Holowachuk E W, Friesen J D, Fiil N. Bacteriophage λ vehicle for the direct cloning of Escherichia coli promoter DNA sequences: feedback regulation of the rplJL-rpoBC operon. Proc Natl Acad Sci USA. 1980;77:2124–2128. doi: 10.1073/pnas.77.4.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1789.Holtje J V. From growth to autolysis: the murein hydrolases in Escherichia coli. Arch Microbiol. 1995;164:243–254. doi: 10.1007/BF02529958. [DOI] [PubMed] [Google Scholar]
- 1790.Holz A, Schaefer C, Gille H, Jueterbock W R, Messer W. Mutations in the DnaA binding sites of the replication origin of Escherichia coli. Mol Gen Genet. 1992;233:81–88. doi: 10.1007/BF00587564. [DOI] [PubMed] [Google Scholar]
- 1791.Homma H, Kobayashi T, Chiba N, Karasawa K, Mizushima H, Kudo I, Inoue K, Ikeda H, Sekiguchi M, Nojima S. The DNA sequence encoding pldA gene, the structural gene for detergent-resistant phospholipase A of E. coli. J Biochem (Tokyo) 1984;96:1655–1664. doi: 10.1093/oxfordjournals.jbchem.a134997. [DOI] [PubMed] [Google Scholar]
- 1792.Homma H, Kobayashi T, Ito Y, Kudo I, Inoue K, Ikeda H, Sekiguchi M, Nojima S. Identification and cloning of the gene coding for lysophospholipase L2 of E. coli K-12. J Biochem (Tokyo) 1983;94:2079–2081. doi: 10.1093/oxfordjournals.jbchem.a134566. [DOI] [PubMed] [Google Scholar]
- 1793.Hong X, Kogoma T. Absence of a direct role for RNase HI in initiation of DNA replication at the oriC site on the Escherichia coli chromosome. J Bacteriol. 1993;175:6731–6734. doi: 10.1128/jb.175.20.6731-6734.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1794.Honore N, Nicolas M, Cole S T. Regulation of enterobacterial cephalosporinase production—the role of a membrane-bound sensory transducer. Mol Microbiol. 1989;3:1121–1130. doi: 10.1111/j.1365-2958.1989.tb00262.x. [DOI] [PubMed] [Google Scholar]
- 1795.Honore N, Cole S T. Nucleotide sequence of the aroP gene encoding the general aromatic amino acid transport protein of Escherichia coli K-12: homology with yeast transport proteins. Nucleic Acids Res. 1990;18:653. doi: 10.1093/nar/18.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1796.Hoog J-O, von Bahr-Lindstrom H, Jornvall H, Holmgren A. Cloning and expression of the glutaredoxin (grx) gene of Escherichia coli. Gene. 1986;43:13–21. doi: 10.1016/0378-1119(86)90003-x. [DOI] [PubMed] [Google Scholar]
- 1797.Hoover T A, Roof W D, Folterman K F, O’Donovan G A, Bencini D A, Wild J R. Nucleotide sequence of the structural gene (pyrB) that encodes the catalytic polypeptide of aspartate transcarbamoylase of Escherichia coli. Proc Natl Acad Sci USA. 1983;80:2462–2466. doi: 10.1073/pnas.80.9.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1798.Hope J N, Bell A W, Hermodson M A, Groarke J M. Ribokinase from Escherichia coli K12. Nucleotide sequence and overexpression of the rbsK gene and purification of ribokinase. J Biol Chem. 1986;261:7663–7668. [PubMed] [Google Scholar]
- 1799.Hopkins J D, Clements M, Syvanen M. New class of mutations in Escherichia coli (uup) that affect precise excision of insertion elements and bacteriophage Mu growth. J Bacteriol. 1983;153:384–389. doi: 10.1128/jb.153.1.384-389.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1800.Hopper S, Bock A. Effector-mediated stimulation of ATPase activity by the ς54-dependent transcriptional activator FHLA from Escherichia coli. J Bacteriol. 1995;177:2798–2803. doi: 10.1128/jb.177.10.2798-2803.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1801.Hopper S, Babst M, Schlensog V, Fischer H M, Hennecke H, Bock A. Regulated expression in vitro of genes coding for formate hydrogenlyase components of Escherichia coli. J Biol Chem. 1994;269:19597–19604. [PubMed] [Google Scholar]
- 1802.Horii T, Ogawa T, Ogawa H. Nucleotide sequence of the lexA gene of E. coli. Cell. 1981;23:689–697. doi: 10.1016/0092-8674(81)90432-3. [DOI] [PubMed] [Google Scholar]
- 1803.Horii T, Ogawa T, Ogawa H. Organization of the recA gene of Escherichia coli. Proc Natl Acad Sci USA. 1981;77:313–317. doi: 10.1073/pnas.77.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1804.Horiuchi T, Maki H, Maruyana M, Sekiguchi M. Identification of the dnaQ gene product and location of the structural gene for RNase H of Escherichia coli by cloning of the genes. Proc Natl Acad Sci USA. 1981;78:3770–3774. doi: 10.1073/pnas.78.6.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1805.Horiuchi T, Horiuchi S, Novick A. The genetic basis of hypersynthesis of β-galactoside. Genetics. 1963;48:157–169. doi: 10.1093/genetics/48.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1806.Horiuchi T, Nagasawa T, Takano T, Sekiguchi M. A newly discovered tRNA1ASP gene (aspV) of Escherichia coli K12. Mol Gen Genet. 1987;206:356–357. doi: 10.1007/BF00333595. [DOI] [PubMed] [Google Scholar]
- 1807.Horlacher R, Uhland K, Klein W, Ehrmann M, Boos W. Characterization of a cytoplasmic trehalase of Escherichia coli. J Bacteriol. 1996;178:6250–6257. doi: 10.1128/jb.178.21.6250-6257.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1808.Horlacher R, Boos W. Characterization of TreR, the major regulator of the Escherichia coli trehalose system. J Biol Chem. 1997;272:13026–13032. doi: 10.1074/jbc.272.20.13026. [DOI] [PubMed] [Google Scholar]
- 1809.Horne S M, Young K D. Escherichia coli and other species of the Enterobacteriaceae encode a protein similar to the family of Mip-like FK506-binding proteins. Arch Microbiol. 1995;163:357–365. doi: 10.1007/BF00404209. [DOI] [PubMed] [Google Scholar]
- 1810.Horowitz H, Christie E, Platt T. Nucleotide sequence of the trpD gene, encoding anthranilate synthetase component II of Escherichia coli. J Mol Biol. 1982;156:245–256. doi: 10.1016/0022-2836(82)90326-6. [DOI] [PubMed] [Google Scholar]
- 1811.Horowitz H, Platt T. A termination site for Lac 1 transcription is between the CAP site and the lac promoter. J Biol Chem. 1982;257:11740–11746. [PubMed] [Google Scholar]
- 1812.Horowitz H, Platt T. Identification of trp-p2, an internal promoter in the tryptophan operon of Escherichia coli. J Mol Biol. 1982;156:257–267. doi: 10.1016/0022-2836(82)90327-8. [DOI] [PubMed] [Google Scholar]
- 1813.Hosono K, Kakuda H, Ichihara S. Decreasing accumulation of acetate in a rich medium by Escherichia coli on introduction of genes on a multicopy plasmid. Biosci Biotechnol Biochem. 1995;59:256–261. doi: 10.1271/bbb.59.256. [DOI] [PubMed] [Google Scholar]
- 1814.Hottenrott S, Schumann T, Pluckthun A, Fischer G, Rahfeld J U. The Escherichia coli SlyD is a metal ion-regulated peptidyl-prolyl cis/trans-isomerase. J Biol Chem. 1997;272:15697–15701. doi: 10.1074/jbc.272.25.15697. [DOI] [PubMed] [Google Scholar]
- 1815.Hou Y, Lin Y-P, Sharer J D, March P E. In vivo selection of conditional-lethal mutations in the gene encoding elongation factor G of Escherichia coli. J Bacteriol. 1994;176:123–129. doi: 10.1128/jb.176.1.123-129.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1816.Hou Y M, Shiba K, Mottes C, Schimmel P. Sequence determination and modeling of structural motifs for the smallest monomeric aminoacyl-tRNA synthetase. Proc Natl Acad Sci USA. 1991;88:976–980. doi: 10.1073/pnas.88.3.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1817.Houlberg U, Hove-Jensen B, Jochimsen B, Nygaard P. Identification of the enzymatic reactions encoded by the purG and purI genes of Escherichia coli. J Bacteriol. 1983;154:1485–1488. doi: 10.1128/jb.154.3.1485-1488.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1818.Houman F, Diaz-Torres M R, Wright A. Transcriptional antitermination in the bgl operon of E. coli is modulated by specific RNA binding protein. Cell. 1990;62:1153–1163. doi: 10.1016/0092-8674(90)90392-r. [DOI] [PubMed] [Google Scholar]
- 1819.Hove-Jensen B. Chromosomal location of the gene encoding phosphoribosylpyrophosphate synthetase in Escherichia coli. J Bacteriol. 1983;154:177–184. doi: 10.1128/jb.154.1.177-184.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1820.Hove-Jensen B. Cloning and characterization of the prs gene encoding phosphoribosylpyrophosphate synthetase of Escherichia coli. Mol Gen Genet. 1985;201:269–276. doi: 10.1007/BF00425670. [DOI] [PubMed] [Google Scholar]
- 1821.Hove-Jensen B, Harlow K W, King C J, Switzer R L. Phosphoribosylpyrphosphate synthetase of Escherichia coli. Properties of the purified enzyme and primary structure of the prs gene. J Biol Chem. 1986;261:6765–6771. [PubMed] [Google Scholar]
- 1822.Hove-Jensen B, Maigaard M. Escherichia coli rpiA gene encoding ribose phosphate isomerase A. J Bacteriol. 1993;175:5628–5635. doi: 10.1128/jb.175.17.5628-5635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1823.Hove-Jensen B, Nygaard P. Phosphoribosyl-pyrophosphate synthetase of Escherichia coli. Identification of a mutant enzyme. Eur J Biochem. 1982;126:327–332. doi: 10.1111/j.1432-1033.1982.tb06782.x. [DOI] [PubMed] [Google Scholar]
- 1824.Hove-Jensen B, Nygaard P. Role of guanosine kinase in the utilization of guanosine for nucleotide synthesis in Escherichia coli. J Gen Microbiol. 1989;135:1263–1273. doi: 10.1099/00221287-135-5-1263. [DOI] [PubMed] [Google Scholar]
- 1825.Howard P K, Shaw J, Otsuka A J. The nucleotide sequence of the birA gene encoding the biotin operon repressor and biotin holoenzyme synthetase functions of Escherichia coli. Gene. 1985;35:321–331. doi: 10.1016/0378-1119(85)90011-3. [DOI] [PubMed] [Google Scholar]
- 1826.Howard-Flanders P, Theriot L. A method for selecting radiation-sensitive mutants of Escherichia coli. Genetics. 1962;47:1219–1224. doi: 10.1093/genetics/47.9.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1827.Hrebenda J, Heleszko H, Brozostek K, Bielecki J. Mutation affecting resistance of Escherichia coli K12 to nalidixic acid. J Gen Microbiol. 1985;131:2285–2292. doi: 10.1099/00221287-131-9-2285. [DOI] [PubMed] [Google Scholar]
- 1828.Hryniewicz M, Sirko A E, Palucha A, Bock A, Hulanicka D. Sulfate and thiosulfate transport in Escherichia coli K-12: identification of a gene encoding a novel protein involved in thiosulfate binding. J Bacteriol. 1990;172:3358–3366. doi: 10.1128/jb.172.6.3358-3366.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1829.Hsu A Y, Poon W W, Shepherd J A, Myles D C, Clarke C F. Complementation of coq3 mutant yeast by mitochondrial targeting of the Escherichia coli UbiG polypeptide: evidence that UbiG catalyzes both O-methylation steps in ubiquinone biosynthesis. Biochemistry. 1996;35:9797–9806. doi: 10.1021/bi9602932. [DOI] [PubMed] [Google Scholar]
- 1830.Hsu D K, Brusilow W S. Effects of the uncI gene on expression of uncB, the gene coding for the a subunit of the F1F0 ATPase of Escherichia coli. FEBS Lett. 1995;371:127–131. doi: 10.1016/0014-5793(95)00867-9. [DOI] [PubMed] [Google Scholar]
- 1831.Hsu L, Jackowski S, Rock C O. Isolation and characterization of Escherichia coli K-12 mutants lacking both 2-acyl-glycerophosphoethanolamine acyltransferase and acyl-acyl carrier protein synthetase activity. J Biol Chem. 1991;266:13783–13788. [PubMed] [Google Scholar]
- 1832.Hsu L M, Klee H J, Zagorski J, Fournier M J. Structure of an Escherichia coli tRNA operon containing linked genes for arginine, histidine, leucine, and proline tRNAs. J Bacteriol. 1984;158:934–942. doi: 10.1128/jb.158.3.934-942.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1833.Hsu L M, Zagorski J, Wang J C, Fournier M J. Escherichia coli 6S RNA gene is part of a dual function transcription unit. J Bacteriol. 1985;161:1162–1170. doi: 10.1128/jb.161.3.1162-1170.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1834.Hsu L M, Zagorski J, Fournier M J. Cloning and sequence analysis of the Escherichia coli 4.5S RNA gene. J Mol Biol. 1984;178:509–531. doi: 10.1016/0022-2836(84)90236-5. [DOI] [PubMed] [Google Scholar]
- 1835.Hsu L M, Vo N V, Chamberlin M J. Escherichia coli transcript cleavage factors GreA and GreB stimulate promoter escape and gene expression in vivo and in vitro. Proc Natl Acad Sci USA. 1995;92:11588–11592. doi: 10.1073/pnas.92.25.11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1836.Hsu P L, Ross W, Landy A. The λ phage att site: functional limits and interaction with 1nt protein. Nature. 1980;285:85–91. doi: 10.1038/285085a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1837.Hu K H, Liu E, Dean K, Gringras M, DeGraff W, Trun N J. Overproduction of three genes leads to camphor resistance and chromosome condensation in Escherichia coli. Genetics. 1996;143:1521–1532. doi: 10.1093/genetics/143.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1838.Hu M, Deonier R. Mapping of Is1 elements flanking the argF gene region on the Escherichia coli K-12 chromosome. Mol Gen Genet. 1981;181:222–229. doi: 10.1007/BF00268430. [DOI] [PubMed] [Google Scholar]
- 1839.Huang F, Coppola G, Calhoun D H. Multiple transcripts encoded by the ilvGMEDA gene cluster of Escherichia coli K-12. J Bacteriol. 1992;174:4871–4877. doi: 10.1128/jb.174.15.4871-4877.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1840.Huang H, Liao J, Cohen S N. Poly(A)- and poly(U)-specific RNA 3′ tail shortening by E. coli ribonuclease E. Nature. 1998;391:99–102. doi: 10.1038/34219. [DOI] [PubMed] [Google Scholar]
- 1841.Huang K-J, Schieberl J L, Igo M M. A distant upstream site involved in the negative regulation of the Escherichia coli ompF gene. J Bacteriol. 1994;176:1309–1315. doi: 10.1128/jb.176.5.1309-1315.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1842.Huang K-J, Igo M M. Identification of the bases in the ompF regulatory region, which interact with the transcription factor OmpR. J Mol Biol. 1996;262:615–628. doi: 10.1006/jmbi.1996.0540. [DOI] [PubMed] [Google Scholar]
- 1843.Huang S, Deutscher M P. Sequence and transcriptional analysis of the Escherichia coli rnt gene encoding RNase T. J Biol Chem. 1992;267:25609–25613. [PubMed] [Google Scholar]
- 1844.Huber F, Erni B. Membrane topology of the mannose transporter of Escherichia coli K12. Eur J Biochem. 1996;239:810–817. doi: 10.1111/j.1432-1033.1996.0810u.x. [DOI] [PubMed] [Google Scholar]
- 1845.Hubscher U, Kornberg A. The delta subunit of Escherichia coli DNA polymerase III holoenzyme is the dnaX gene product. Proc Natl Acad Sci USA. 1979;76:6284–6288. doi: 10.1073/pnas.76.12.6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1846.Hudson A J, Andrews S C, Hawkins C, Williams J M, Izuhara M, Meldrum F C, Mann S, Harrison P M, Guest J R. Overproduction, purification, and characterization of the Escherichia coli ferritin. Eur J Biochem. 1993;218:985–995. doi: 10.1111/j.1432-1033.1993.tb18457.x. [DOI] [PubMed] [Google Scholar]
- 1847.Hudson G S, Davidson B E. Nucleotide sequence and transcription of the phenylalanine and tyrosine operons of Escherichia coli K-12. J Mol Biol. 1984;180:1023–1051. doi: 10.1016/0022-2836(84)90269-9. [DOI] [PubMed] [Google Scholar]
- 1848.Hudson G S, Rellos P, Davidson B E. Two promoters control the aroH gene of Escherichia coli. Gene. 1991;102:87–91. doi: 10.1016/0378-1119(91)90544-l. [DOI] [PubMed] [Google Scholar]
- 1849.Hudson L, Rossi J J, Landy A. Dual function transcripts specifying tRNA and mRNA. Nature. 1981;294:422–427. doi: 10.1038/294422a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1850.Hufton S E, Ward R J, Bunce N A C, Armstrong J T, Fletcher A J P, Glass R E. Structure-function analysis of the vitamin B12 receptor of Escherichia coli by means of informational suppression. Mol Microbiol. 1995;15:381–393. doi: 10.1111/j.1365-2958.1995.tb02251.x. [DOI] [PubMed] [Google Scholar]
- 1851.Hugouvieux-Cotte-Pattat N, Robert-Baudouy J M. Isolation of fusions between the lac genes and several genes of the exu regulon: analysis of their regulation, determination of the transcription direction of the uxaC-uxaA operon in Escherichia coli K-12. Mol Gen Genet. 1981;182:279–287. doi: 10.1007/BF00269671. [DOI] [PubMed] [Google Scholar]
- 1852.Hugouvieux-Cotte-Pattat N, Robert-Baudouy J M. Determination of the transcription direction of the exuT gene in Escherichia coli K-12; divergent transcription of the exuT-uxaCA operons. J Bacteriol. 1982;151:480–484. doi: 10.1128/jb.151.1.480-484.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1853.Hugouvieux-Cotte-Pattat N, Robert-Baudouy J M. Regulation and transcription of exuR, a self-regulated repressor in Escherichia coli K-12. J Mol Biol. 1982;156:221–228. doi: 10.1016/0022-2836(82)90468-5. [DOI] [PubMed] [Google Scholar]
- 1854.Hugouvieux-Cotte-Pattat N, Robert-Baudouy J M. Determination of the transcription direction of the uxaB gene, in Escherichia coli K12. Mol Gen Genet. 1983;189:334–336. doi: 10.1007/BF00337826. [DOI] [PubMed] [Google Scholar]
- 1855.Hui I, Maltman K, Little R, Hastrup S, Johnsen M, Fiil N, Dennis P P. Insertions of transposon Tn5 into ribosomal protein RNA polymerase operons. J Bacteriol. 1982;152:1022–1032. doi: 10.1128/jb.152.3.1022-1032.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1856.Huisman G W, Kolter R. Sensing starvation: a homoserine lactone-dependent signaling pathway in Escherichia coli. Science. 1994;265:537–539. doi: 10.1126/science.7545940. [DOI] [PubMed] [Google Scholar]
- 1857.Huisman O, D’Ari R. An inducible DNA replication-cell division coupling mechanism in E. coli. Nature. 1981;290:797–799. doi: 10.1038/290797a0. [DOI] [PubMed] [Google Scholar]
- 1858.Huisman O, D’Ari R, Gottesman S. Cell division control in Escherichia coli: specific induction of the SOS function SfiA protein is sufficient to block septation. Proc Natl Acad Sci USA. 1984;81:4490–4494. doi: 10.1073/pnas.81.14.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1859.Hull E P, Spencer M E, Wood D, Guest J R. Nucleotide sequence of the promoter region of the citrate synthase gene (gltA) of Escherichia coli. FEBS Lett. 1983;156:366–370. doi: 10.1016/0014-5793(83)80530-4. [DOI] [PubMed] [Google Scholar]
- 1860.Hull R, Klinger J D, Moody E E M. Isolation and characterization of mutants of Escherichia coli K12 resistant to the new aminoglycoside antibiotic, amikacin. J Gen Microbiol. 1976;94:389–394. doi: 10.1099/00221287-94-2-389. [DOI] [PubMed] [Google Scholar]
- 1861.Humbert R, Simoni R D. Genetic and biochemical studies demonstrating a second gene coding for asparagine synthetase in Escherichia coli. J Bacteriol. 1980;142:212–220. doi: 10.1128/jb.142.1.212-220.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1862.Hunt C L, Colless V, Smith M T, Molasky D O, Malo M S, Loughlin R E. Lambda transducing phage and clones carrying genes of the cysJIHDC gene cluster of Escherichia coli K-12. J Gen Microbiol. 1987;133:2707–2717. doi: 10.1099/00221287-133-10-2707. [DOI] [PubMed] [Google Scholar]
- 1863.Hunt M D, Pettis G S, McIntosh M A. Promoter and operator determinants for Fur-mediated iron regulation in the bidirectional fepA-fes control region of the Escherichia coli enterobactin gene system. J Bacteriol. 1994;176:3944–3955. doi: 10.1128/jb.176.13.3944-3955.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1864.Hunt T P, Magasanik B. Transcription of glnA by purified Escherichia coli components: core RNA polymerase and the products of glnF, glnG, and glnL. Proc Natl Acad Sci USA. 1985;82:8453–8457. doi: 10.1073/pnas.82.24.8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1865.Hupp T R, Kaguni J M. Suppression of the Escherichia coli dnaA46 mutation by a mutation in trxA, the gene for thioredoxin. Mol Gen Genet. 1988;213:471–478. doi: 10.1007/BF00339618. [DOI] [PubMed] [Google Scholar]
- 1866.Husain I, Sancar A. Photoreactivation in phr mutants of Escherichia coli K-12. J Bacteriol. 1987;169:2367–2372. doi: 10.1128/jb.169.6.2367-2372.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1867.Husain I, Van Houten B, Thomas D C, Sancar A. Sequences of Escherichia coli urvA gene and protein reveal two potential ATP binding sites. J Biol Chem. 1986;261:4895–4901. [PubMed] [Google Scholar]
- 1868.Hussain H, Grove J, Griffiths L, Busby S J, Cole J A. A seven-gene operon essential for formate-dependent nitrite reduction to ammonia by enteric bacteria. Mol Microbiol. 1994;12:153–163. doi: 10.1111/j.1365-2958.1994.tb01004.x. [DOI] [PubMed] [Google Scholar]
- 1869.Hussain K, Elliot E J, Salmond G P C. The parD− mutant of Escherichia coli also carries a gyrAam mutation. The complete sequence of gyrA. Mol Microbiol. 1987;1:259–273. doi: 10.1111/j.1365-2958.1987.tb01932.x. [DOI] [PubMed] [Google Scholar]
- 1870.Hussein S, Hantke K, Braun V. Citrate-dependent iron transport system in Escherichia coli. Eur J Biochem. 1981;117:431–437. doi: 10.1111/j.1432-1033.1981.tb06357.x. [DOI] [PubMed] [Google Scholar]
- 1871.Hwang D S, Kornberg A. A novel protein binds a key origin sequence to block replication of an E. coli minichromosome. Cell. 1990;63:325–331. doi: 10.1016/0092-8674(90)90165-b. [DOI] [PubMed] [Google Scholar]
- 1872.Hwang D S, Kornberg A. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J Biol Chem. 1992;267:23083–23086. [PubMed] [Google Scholar]
- 1873.Hwang D S, Kornberg A. Opposed actions of regulatory proteins, DnaA and IciA, in opening the replication origin of Escherichia coli. J Biol Chem. 1992;267:23087–23091. [PubMed] [Google Scholar]
- 1874.Hwang D S, Thony B, Kornberg A. IciA protein, a specific inhibitor of initiation of Escherichia coli chromosomal replication. J Biol Chem. 1992;267:2209–2213. [PubMed] [Google Scholar]
- 1875.Hwang D S, Crooke E, Kornberg A. Aggregated dnaA protein is dissociated and activated for DNA replication by phospholipase or dnaK protein. J Biol Chem. 1990;265:19244–19248. [PubMed] [Google Scholar]
- 1876.Hwang Y-W, Engel R, Troop B E. Correlation of 3,4-dihydroxybutyl 1-phosphonate resistance with a defect in cardiolipin synthesis in Escherichia coli. J Bacteriol. 1984;157:846–856. doi: 10.1128/jb.157.3.846-856.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1876a.Iaccarino M, Guardiola J, De Felice M. On the permeability of biological membranes. J Membr Sci. 1978;3:287–302. [Google Scholar]
- 1877.Ichihara S, Suzuki T, Mizushima H. Molecular cloning and sequencing of the sppA gene and characterization of the encoded protease IV, a signal peptide peptidase, of Escherichia coli. J Biol Chem. 1986;261:9405–9411. [PubMed] [Google Scholar]
- 1878.Ichihara S, Matsubara Y, Kato C, Akasaka K, Mizushima S. Molecular cloning, sequencing, and mapping of the gene encoding protease I and characterization of proteinase and proteinase-defective Escherichia coli mutants. J Bacteriol. 1993;175:1032–1037. doi: 10.1128/jb.175.4.1032-1037.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1879.Ichikawa J K, Li C, Fu J C, Clarke S. A gene at 59 minutes on the Escherichia coli chromosome encodes a lipoprotein with unusual amino acid repeat sequences. J Bacteriol. 1994;176:1630–1638. doi: 10.1128/jb.176.6.1630-1638.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1880.Ichikawa S, Kaji A. Molecular cloning and expression of ribosome releasing factor. J Biol Chem. 1989;264:20054–20059. [PubMed] [Google Scholar]
- 1881.Ichikawa S, Ryoji M, Siegfried Z, Kaji A. Localization of the ribosome-releasing factor gene in the Escherichia coli chromosome. J Bacteriol. 1989;171:3689–3695. doi: 10.1128/jb.171.7.3689-3695.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1882.Icho T. Membrane-bound phosphatases in Escherichia coli: sequence of the pgpA gene. J Bacteriol. 1988;170:5110–5116. doi: 10.1128/jb.170.11.5110-5116.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1883.Icho T. Membrane-bound phosphatases in Escherichia coli: sequence of the pgpB gene and dual subcellular localization of the pgpB product. J Bacteriol. 1988;170:5117–5124. doi: 10.1128/jb.170.11.5117-5124.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1884.Icho T, Bulawa C E, Raetz C R H. Molecular cloning and sequencing of the gene for CDP-diglyceride hydrolase of Escherichia coli. J Biol Chem. 1985;260:12092–12098. [PubMed] [Google Scholar]
- 1885.Icho T, Raetz C R H. Multiple genes for membrane-bound phosphatases in Escherichia coli and their action on phospholipid precursors. J Bacteriol. 1983;153:722–730. doi: 10.1128/jb.153.2.722-730.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1886.Icho T, Sparrow C P, Raetz C R H. Molecular cloning and sequencing of the gene for CDP-diglyceride synthetase of Escherichia coli. J Biol Chem. 1985;260:12078–12083. [PubMed] [Google Scholar]
- 1887.Igarishi K, Hiraga S, Yura T. A deoxythymidine kinase-deficient mutant of Escherichia coli. II. Mapping and transduction studies with phage phi80. Genetics. 1967;57:643–654. doi: 10.1093/genetics/57.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1888.Iida A, Harayama S, Iino T, Hazelbauer G L. Molecular cloning and characterization of genes required for ribose transport and utilization in Escherichia coli K-12. J Bacteriol. 1984;158:674–682. doi: 10.1128/jb.158.2.674-682.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1889.Iida A, Teshiba S, Mizobuchi K. Identification and characterization of the tktB gene encoding a second transketolase in Escherichia coli K-12. J Bacteriol. 1993;175:5375–5383. doi: 10.1128/jb.175.17.5375-5383.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1890.Iida K, Hirota Y, Schwarz U. Mutants of Escherichia coli defective in penicillin-insensitive murein dd-endopeptidase. Mol Gen Genet. 1983;189:215–221. doi: 10.1007/BF00337807. [DOI] [PubMed] [Google Scholar]
- 1891.Iino T. Genetics of structure and function of bacterial flagella. Annu Rev Microbiol. 1977;11:161–182. doi: 10.1146/annurev.ge.11.120177.001113. [DOI] [PubMed] [Google Scholar]
- 1892.Iino T, Komeda Y, Kutsukake K, Macnab R M, Matsumura P, Parkinson J S, Simon M I, Yamaguchi S. New unified nomenclature for the flagellar genes of Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1988;52:533–535. doi: 10.1128/mr.52.4.533-535.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1893.Ikeda H, Wachi M, Jung H K, Ishino F, Matsuhashi M. Nucleotide sequence involving murG and murC in the mra gene cluster region of Escherichia coli. Nucleic Acids Res. 1990;18:4014. doi: 10.1093/nar/18.13.4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1894.Ikeda M, Wachi M, Ishino F, Matsuhashi M. Nucleotide sequence involving murD and an open reading frame ORF-Y spacing murF and ftsW in Escherichia coli. Nucleic Acids Res. 1990;18:1058. doi: 10.1093/nar/18.4.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1895.Ikeda M, Wachi M, Jung H K, Ishino F, Matsuhashi M. The Escherichia coli mraY gene encoding UDP-N-acetylmuramoyl-pentapeptide:undecaprenyl-phosphate phospho-N-acetylmuramoyl-pentapeptide transferase. J Bacteriol. 1991;173:1021–1026. doi: 10.1128/jb.173.3.1021-1026.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1896.Ikeda M, Sato T, Wachi M, Jung H K, Ishino F, Kobayashi Y, Matsuhashi M. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J Bacteriol. 1989;171:6375–6378. doi: 10.1128/jb.171.11.6375-6378.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1897.Ikeda T, LaPorte D C. Isocitrate dehydrogenase kinase/phosphatase: aceK alleles that express kinase but not phosphatase activity. J Bacteriol. 1991;173:1801–1806. doi: 10.1128/jb.173.5.1801-1806.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1898.Ikemi M, Murakami K, Hashimoto M, Murooka Y. Cloning and characterization of genes involved in the biosynthesis of delta-aminolevulinic acid in Escherichia coli. Gene. 1992;121:127–132. doi: 10.1016/0378-1119(92)90170-t. [DOI] [PubMed] [Google Scholar]
- 1899.Ilag L L, Jahn D. Activity and spectroscopic properties of the Escherichia coli glutamate 1-semialdehyde aminotransferase and the putative active site mutant K265R. Biochemistry. 1992;31:7143–7151. doi: 10.1021/bi00146a016. [DOI] [PubMed] [Google Scholar]
- 1900.Ilag L L, Jahn D, Eggertsson G, Soll D. The Escherichia coli hemL gene encodes glutamate 1-semialdehyde aminotransferase. J Bacteriol. 1991;173:3408–3413. doi: 10.1128/jb.173.11.3408-3413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1901.Ilyina T S, Nechaeva E V, Romanova Y M, Smirnov Y V. Isolation and mapping of Escherichia coli K12 mutants defective in Tn9 transposition. Mol Gen Genet. 1981;181:384–389. doi: 10.1007/BF00425616. [DOI] [PubMed] [Google Scholar]
- 1902.Ilyina T S, Romanova Y M, Smirnov Y V. The effect of tnm mutations of Escherichia coli K12 on transposition of various movable genetic elements. Mol Gen Genet. 1981;183:376–379. doi: 10.1007/BF00270643. [DOI] [PubMed] [Google Scholar]
- 1903.Im S W K, Pittard J. Phenylalanine biosynthesis in Escherichia coli K-12: mutants derepressed for chorismate mutase P-prephenate dehydratase. J Bacteriol. 1971;106:784–790. doi: 10.1128/jb.106.3.784-790.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1904.Imaishi H, Gomada M, Inouye S, Nakazawa A. Physical map location of the rpoN gene of Escherichia coli. J Bacteriol. 1993;175:1550–1551. doi: 10.1128/jb.175.5.1550-1551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1905.Imamura N, Nakayama H. thiD locus of Escherichia coli. Experientia. 1981;37:1265–1266. doi: 10.1007/BF01948350. [DOI] [PubMed] [Google Scholar]
- 1906.Imamura N, Nakayama H. thiK and thiL loci of Escherichia coli. J Bacteriol. 1982;151:708–717. doi: 10.1128/jb.151.2.708-717.1982. . (Erratum, 152:1308, 1982.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1907.Imamura R, Yamanaka K, Ogura T, Hiraga S, Fujita N, Ishihama A, Niki H. Identification of the cpdA gene encoding cyclic 3′5′-adenosine monophosphate phosphodiesterase in Escherichia coli. J Biol Chem. 1996;271:25423–25429. doi: 10.1074/jbc.271.41.25423. [DOI] [PubMed] [Google Scholar]
- 1908.Imlay K R, Imlay J A. Cloning and analysis of sodC, encoding the copper-zinc superoxide dismutase of Escherichia coli. J Bacteriol. 1996;178:2564–2571. doi: 10.1128/jb.178.9.2564-2571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1909.Inaba K, Kuroda T, Shimamoto T, Kayahara T, Tsuda M, Tsuchiya T. Lithium toxicity and Na+(Li+)/H+ antiporter in Escherichia coli. Biol Pharm Bull. 1994;17:395–398. doi: 10.1248/bpb.17.395. [DOI] [PubMed] [Google Scholar]
- 1910.Inada T, Nakamura Y. Lethal double-stranded RNA processing activity of ribonuclease III in the absence of suhB protein of Escherichia coli. Biochimie. 1995;77:294–302. doi: 10.1016/0300-9084(96)88139-9. [DOI] [PubMed] [Google Scholar]
- 1911.Inada T, Nakamura Y. Autogenous control of the suhB gene expression of Escherichia coli. Biochimie. 1996;78:209–212. doi: 10.1016/0300-9084(96)89508-3. [DOI] [PubMed] [Google Scholar]
- 1912.Ineichen G, Biel A J. Location of the hemE gene on the physical map of Escherichia coli. J Bacteriol. 1993;175:7749–7750. doi: 10.1128/jb.175.23.7749-7750.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1913.Innis M A, Tokunaga M, Williams M E, Loranger J M, Chang S-Y, Chang S, Wu H C. Nucleotide sequence of the Escherichia coli prolipoprotein signal peptidase (lsp) gene. Proc Natl Acad Sci USA. 1984;81:3708–3712. doi: 10.1073/pnas.81.12.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1914.Inoko H, Shigesada K, Imai M. Isolation and characterization of conditional-lethal rho mutants of Escherichia coli. Proc Natl Acad Sci USA. 1977;74:1162–1166. doi: 10.1073/pnas.74.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1915.Inokuchi H, Yamao F, Sakano H, Ozeki H. Identification of transfer RNA suppressors in Escherichia coli. I. Amber suppressor su+2, and anticodon mutant of tRNA2Gln. J Mol Biol. 1979;132:649–662. doi: 10.1016/0022-2836(79)90380-2. [DOI] [PubMed] [Google Scholar]
- 1916.Inokuchi H, Kodaira M, Yamao F, Ozeki H. Identification of transfer RNA suppressors in Escherichia coli. II. Duplicate genes for tRNA2Gln. J Mol Biol. 1979;132:663–677. doi: 10.1016/0022-2836(79)90381-4. [DOI] [PubMed] [Google Scholar]
- 1917.Inokuchi K, Furukawa H, Nakamura H, Mizushima H. Characterization by deletion mutagenesis in vitro of the promoter region of ompF, a positively regulated gene of Escherichia coli. J Mol Biol. 1984;178:653–668. doi: 10.1016/0022-2836(84)90243-2. [DOI] [PubMed] [Google Scholar]
- 1918.Inokuchi K, Mutoh N, Matsuyama S, Mizushima H. Primary structure of the ompF gene that codes for a major outer membrane protein of Escherichia coli K-12. Nucleic Acids Res. 1982;10:6957–6968. doi: 10.1093/nar/10.21.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1919.Inouye M, Michaelis S, Wright A, Beckwith J R. Cloning and restriction mapping of the alkaline phosphatase structural gene (phoA) of Escherichia coli and generation of deletion mutants in vitro. J Bacteriol. 1981;146:668–675. doi: 10.1128/jb.146.2.668-675.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1920.Inouye M, Barnes W, Beckwith J R. Signal sequence of alkaline phosphatase of Escherichia coli. J Bacteriol. 1982;149:434–439. doi: 10.1128/jb.149.2.434-439.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1921.Iobbi-Nivol C, Crooke H, Griffiths L, Grove J, Hussain H, Pommier J, Mejean V, Cole J A. A reassessment of the range of c-type cytochromes synthesized by Escherichia coli K-12. FEMS Microbiol Lett. 1994;119:89–94. doi: 10.1111/j.1574-6968.1994.tb06872.x. [DOI] [PubMed] [Google Scholar]
- 1922.Iobbi-Nivol C, Palmer T, Whitty P W, McNairn E, Boxer D H. The mob locus of Escherichia coli K12 required for molybdenum cofactor biosynthesis is expressed at very low levels. Microbiology. 1995;141:1663–1671. doi: 10.1099/13500872-141-7-1663. [DOI] [PubMed] [Google Scholar]
- 1923.Irino N, Nakayama H, Nakayama K. The recQ gene of Escherichia coli K12: primary structure and evidence for SOS regulation. Mol Gen Genet. 1986;205:298–304. doi: 10.1007/BF00430442. [DOI] [PubMed] [Google Scholar]
- 1923a.Isaksson L A, Takata R. The temperature-sensitive mutant 72C. I. Pleiotropic growth behavior and changed response to some antibiotics and mutations. Mol Gen Genet. 1978;161:9–19. doi: 10.1007/BF00266609. [DOI] [PubMed] [Google Scholar]
- 1924.Ishige K, Nagasawa S, Tokishita S, Mizuno T. A novel device of bacterial signal transducers. EMBO J. 1994;13:5195–5202. doi: 10.1002/j.1460-2075.1994.tb06850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1925.Ishihama A. Protein-protein communication within the transcription apparatus. J Bacteriol. 1993;175:2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1926.Ishii S, Hatada E, Maekawa T, Imamoto F. Molecular cloning and nucleotide sequencing of the nusB gene of E. coli. Nucleic Acids Res. 1984;12:4987–4995. doi: 10.1093/nar/12.12.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1927.Ishii S, Kuroki K, Imamoto F. tRNAf2Met gene in the leader region of the nusA operon in Escherichia coli. Proc Natl Acad Sci USA. 1984;81:409–413. doi: 10.1073/pnas.81.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1928.Ishii S, Ihara M, Maekawa T, Nakamura H, Uchida H, Imamoto F. The nucleotide sequence of the cloned nusA gene and its flanking region of Escherichia coli. Nucleic Acids Res. 1984;12:3333–3342. doi: 10.1093/nar/12.7.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1929.Ishino F, Jung H K, Ikeda M, Doi M, Wachi M, Matsuhashi M. New mutations fts-36, lts-33, and ftsW clustered in the mra region of the Escherichia coli chromosome induce thermosensitive cell growth and division. J Bacteriol. 1989;171:5523–5530. doi: 10.1128/jb.171.10.5523-5530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1930.Ishino F, Mitsui K, Tamaki S, Matsuhashi M. Dual enzyme activities of cell wall peptidoglycan synthesis, peptidoglycan transglycosylase and penicillin transpeptidase, in purified preparations of Escherichia coli penicillin-binding protein 1A. Biochem Biophys Res Commun. 1980;97:287–293. doi: 10.1016/s0006-291x(80)80166-5. [DOI] [PubMed] [Google Scholar]
- 1931.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1932.Ishino Y, Shinagawa H, Makino K, Tsunasawa S, Sakiyama F, Nakata A. Nucleotide sequence of the lig gene and primary structure of DNA ligase of Escherichia coli. Mol Gen Genet. 1986;204:1–7. doi: 10.1007/BF00330179. [DOI] [PubMed] [Google Scholar]
- 1933.Island M D, Wei B Y, Kadner R J. Structure and function of the uhp genes for the sugar phosphate transport system in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1992;174:2754–2762. doi: 10.1128/jb.174.9.2754-2762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1934.Island M D, Kadner R J. Interplay between the membrane-associated UhpB and UhpC regulatory proteins. J Bacteriol. 1993;175:5028–5034. doi: 10.1128/jb.175.16.5028-5034.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1935.Isono K. Genes encoding ribosomal proteins S16 and L19 form a gene cluster at 56.4 min in Escherichia coli. Mol Gen Genet. 1978;165:265–268. doi: 10.1007/BF00332525. [DOI] [PubMed] [Google Scholar]
- 1936.Isono K, Schnier J, Kitakawa M. Genetic fine structure of the pyrE region containing the genes for ribosomal proteins L28 and L33 in Escherichia coli. Mol Gen Genet. 1980;179:311–317. doi: 10.1007/BF00425458. [DOI] [PubMed] [Google Scholar]
- 1937.Isono K, Kitakawa M. A new ribosomal protein locus in Escherichia coli: the gene for protein S6 maps at 97 min. Mol Gen Genet. 1977;153:115–120. doi: 10.1007/BF00264725. [DOI] [PubMed] [Google Scholar]
- 1938.Isono K, Kitakawa M. Cluster of ribosomal protein genes in Escherichia coli containing genes for proteins S6, S18, and L9. Proc Natl Acad Sci USA. 1978;75:6163–6167. doi: 10.1073/pnas.75.12.6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1939.Isono K, Isono S. Ribosomal protein modification in Escherichia coli. II. Studies of a mutant lacking the N-terminal acetylation of protein S18. Mol Gen Genet. 1980;177:645–651. doi: 10.1007/BF00272675. [DOI] [PubMed] [Google Scholar]
- 1940.Isono S, Isono K. Ribosomal protein modification in Escherichia coli. III. Studies of mutants lacking an acetylase activity specific for protein L12. Mol Gen Genet. 1981;183:473–477. doi: 10.1007/BF00268767. [DOI] [PubMed] [Google Scholar]
- 1941.Isono S, Thamm S, Kitagawa M, Isono K. Cloning and nucleotide sequencing of the genes for ribosomal proteins S9 (rpsI) and L13 (rplM) of Escherichia coli. Mol Gen Genet. 1985;198:279–282. doi: 10.1007/BF00383007. [DOI] [PubMed] [Google Scholar]
- 1942.Isturiz T, Palmero E, Vitelli-Flores J. Mutations affecting gluconate catabolism in Escherichia coli. Genetic mapping of the locus for the thermosensitive gluconokinase. J Gen Microbiol. 1986;132:3209–3219. doi: 10.1099/00221287-132-11-3209. [DOI] [PubMed] [Google Scholar]
- 1943.Isturiz T, Celaya J. The metabolism of gluconate in Escherichia coli. The subsidiary system and the nature of the gntS gene. J Basic Microbiol. 1997;37:105–114. doi: 10.1002/jobm.3620370205. [DOI] [PubMed] [Google Scholar]
- 1944.Itaya M. Isolation and characterization of a second RNase H (RNase HII) of Escherichia coli K-12 encoded by the rnhB gene. Proc Natl Acad Sci USA. 1990;87:8587–8591. doi: 10.1073/pnas.87.21.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1945.Ito K, Cerretti D P, Nashimoto H, Nomura M. Characterization of an amber mutation in the structural gene for ribosomal protein L15, which impairs the expression of the protein export gene, secY, in Escherichia coli. EMBO J. 1984;3:2319–2324. doi: 10.1002/j.1460-2075.1984.tb02133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1946.Ito K, Kawakami K, Nakamura Y. Multiple control of Escherichia coli lysyl-tRNA synthetase expression involves a transcriptional repressor and a translational enhancer element. Proc Natl Acad Sci USA. 1993;90:302–306. doi: 10.1073/pnas.90.1.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1947.Ito K, Wittekind M, Nomura M, Shiba K, Yura T, Miura A, Nashimoto H. A temperature-sensitive mutant of E. coli exhibiting slow processing of exported proteins. Cell. 1983;32:789–797. doi: 10.1016/0092-8674(83)90065-x. [DOI] [PubMed] [Google Scholar]
- 1948.Ito K, Oshima T, Mizuno T, Nakamura Y. Regulation of lysyl-tRNA synthetase expression by histone-like protein H-NS of Escherichia coli. J Bacteriol. 1994;176:7383–7386. doi: 10.1128/jb.176.23.7383-7386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1949.Ito K, Hirota Y, Akiyama Y. Temperature-sensitive sec mutants of Escherichia coli: inhibition of protein export at the permissive temperature. J Bacteriol. 1989;171:1742–1743. doi: 10.1128/jb.171.3.1742-1743.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1949a.Itoh, T., et al. GenBank submission D90851.
- 1950.Itoh T, Aiba H, Baba T, Fujita K, Hayashi K, et al. A 460-kb DNA sequence of the Escherichia coli K-12 genome corresponding to the 40.1-50.0 min region on the linkage map. DNA Res. 1996;3:379–392. doi: 10.1093/dnares/3.6.379. [DOI] [PubMed] [Google Scholar]
- 1950a.Itoh Y. Cloning and characterization of the aru genes encoding enzymes of the catabolic arginine succinyltransferase pathway in Pseudomonas aeruginosa. J Bacteriol. 1997;179:7280–7290. doi: 10.1128/jb.179.23.7280-7290.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1951.Iuchi S, Aristarkhov A, Dong J M, Taylor J S, Lin E C C. Effects of nitrate respiration on expression of the Arc-controlled operons encoding succinate dehydrogenase and flavin-linked l-lactate dehydrogenase. J Bacteriol. 1994;176:1695–1701. doi: 10.1128/jb.176.6.1695-1701.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1952.Iuchi S, Furlong D, Lin E C C. Differentiation of arcA, arcB, and cpxA mutant phenotypes of Escherichia coli by sex pilus formation and enzyme regulation. J Bacteriol. 1989;171:2889–2893. doi: 10.1128/jb.171.5.2889-2893.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1953.Iuchi S, Cameron D C, Lin E C C. A second global regulator gene (arcB) mediating repression of enzymes in aerobic pathways of Escherichia coli. J Bacteriol. 1989;171:868–873. doi: 10.1128/jb.171.2.868-873.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1954.Iuchi S, Lin E C C. The narL gene product activates the nitrate reductase operon and represses the fumarate reductase and trimethylamine N-oxide reductase operons in Escherichia coli. Proc Natl Acad Sci USA. 1987;84:3901–3905. doi: 10.1073/pnas.84.11.3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1955.Iuchi S, Lin E C C. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci USA. 1988;85:1888–1892. doi: 10.1073/pnas.85.6.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1956.Iuchi S, Lin E C C. Purification and phosphorylation of the Arc regulatory components of Escherichia coli. J Bacteriol. 1992;174:5617–5623. doi: 10.1128/jb.174.17.5617-5623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1957.Iuchi S, Lin E C C. Adaptation of Escherichia coli to redox environments by gene expression. Mol Microbiol. 1993;9:9–15. doi: 10.1111/j.1365-2958.1993.tb01664.x. [DOI] [PubMed] [Google Scholar]
- 1958.Iuchi S, Matsuda Z, Fujiwara T, Lin E C C. The arcB gene of Escherichia coli encodes a sensor-regulator protein for anaerobic repression of the arc modulon. Mol Microbiol. 1990;4:715–727. doi: 10.1111/j.1365-2958.1990.tb00642.x. [DOI] [PubMed] [Google Scholar]
- 1959.Ivanchenko M, van Holde K, Zlatanova J. Prokaryotic DNA ligases unwind superhelical DNA. Biochem Biophys Res Commun. 1996;226:498–505. doi: 10.1006/bbrc.1996.1384. [DOI] [PubMed] [Google Scholar]
- 1960.Ivanisevic R, Milic M, Ajdic D, Rakonjac J, Savic D J. Nucleotide sequence, mutational analysis, transcriptional start site, and product analysis of nov, the gene which affects Escherichia coli K-12 resistance to the gyrase inhibitor novobiocin. J Bacteriol. 1995;177:1766–1771. doi: 10.1128/jb.177.7.1766-1771.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1961.Ivey D M, Guffanti A A, Zemsky J, Pinner E, Karpel R, Padan E, Schuldiner S, Krulwich T A. Cloning and characterization of a putative Ca2+/H+ antiporter gene from Escherichia coli upon functional complementation of Na+/H+ antiporter-deficient strains by the overexpressed gene. J Biol Chem. 1993;268:11296–11303. [PubMed] [Google Scholar]
- 1962.Iwakura M, Shimura Y, Tsuda K. Cloning of dihydrofolate reductase gene of Escherichia coli K12. J Biochem (Tokyo) 1982;91:1205–1212. doi: 10.1093/oxfordjournals.jbchem.a133804. [DOI] [PubMed] [Google Scholar]
- 1963.Iwakura M, Shimura Y, Tsuda K. Isolation of DNA fragment containing phoS gene of Escherichia coli. J Biochem (Tokyo) 1982;92:615–622. doi: 10.1093/oxfordjournals.jbchem.a133972. [DOI] [PubMed] [Google Scholar]
- 1964.Iwanicka-Nowicka R, Hryniewicz M M. A new gene, cbl, encoding a member of the LysR family of transcriptional regulators, belongs to Escherichia coli cys regulon. Gene. 1995;166:11–17. doi: 10.1016/0378-1119(95)00606-8. [DOI] [PubMed] [Google Scholar]
- 1965.Iwasaki H, Takahagi M, Shiba T, Nakata A, Shinagawa H. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J. 1991;10:4381–4389. doi: 10.1002/j.1460-2075.1991.tb05016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1966.Iwasaki H, Shiba T, Nakata A, Shinagawa H. Involvement in DNA repair of the ruvA gene of Escherichia coli. Mol Gen Genet. 1989;219:328–331. doi: 10.1007/BF00261196. [DOI] [PubMed] [Google Scholar]
- 1967.Iwasaki H, Ishino Y, Toh H, Nakata A, Shinagawa H. Escherichia coli DNA polymerase II is homologous to alpha-like DNA polymerases. Mol Gen Genet. 1991;226:24–33. doi: 10.1007/BF00273583. [DOI] [PubMed] [Google Scholar]
- 1968.Iwaya M, Jones C W, Khorana J, Strominger J L. Mapping of the mecillinam-resistant, round morphological mutants of Escherichia coli. J Bacteriol. 1978;133:196–202. doi: 10.1128/jb.133.1.196-202.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1969.Izu H, Adachi O, Yamada M. Gene organization and transcriptional regulation of the gntRKU operon involved in gluconate uptake and catabolism of Escherichia coli. J Mol Biol. 1997;267:778–793. doi: 10.1006/jmbi.1996.0913. [DOI] [PubMed] [Google Scholar]
- 1970.Izuhara M, Takamune K, Takata R. Cloning and sequencing of an Escherichia coli K12 gene which encodes a polypeptide having similarity to the human ferritin H subunit. Mol Gen Genet. 1991;225:510–513. doi: 10.1007/BF00261694. [DOI] [PubMed] [Google Scholar]
- 1971.Jackowski S, Alix J H. Cloning, sequence, and expression of the pantothenate permease (panF) gene of Escherichia coli. J Bacteriol. 1990;172:3842–3848. doi: 10.1128/jb.172.7.3842-3848.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1972.Jackowski S, Jackson P D, Rock C O. Sequence and function of the aas gene in Escherichia coli. J Biol Chem. 1994;269:2921–2928. [PubMed] [Google Scholar]
- 1973.Jackson G S, Cornish-Bowden A, Cole J A. Prosthetic groups of the NADH-dependent nitrite reductase from Escherichia coli K12. Biochem J. 1981;193:861–867. doi: 10.1042/bj1930861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1974.Jackson G S, Bohin J P, Kennedy E P. Biosynthesis of membrane-derived oligosaccharides: characterization of mdoB mutants defective in phosphoglycerol transferase I activity. J Bacteriol. 1984;160:976–981. doi: 10.1128/jb.160.3.976-981.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1975.Jackson J H, Davis E J, Madu A C, Braxter S E. Three-factor reciprocal cross mapping of a gene that causes expression of feedback-resistant acetohydroxy acid synthase in Escherichia coli K-12. Mol Gen Genet. 1981;181:417–419. doi: 10.1007/BF00428729. [DOI] [PubMed] [Google Scholar]
- 1976.Jacobi A, Rossmann R, Bock A. The hyp operon gene products are required for the maturation of catalytically active hydrogenase isoenzymes in Escherichia coli. Arch Microbiol. 1992;158:444–451. doi: 10.1007/BF00276307. [DOI] [PubMed] [Google Scholar]
- 1977.Jaffé A, Vinella D, D’Ari R. The Escherichia coli histone-like protein HU affects DNA initiation, chromosome partitioning via MukB, and cell division via MinCDE. J Bacteriol. 1997;179:3494–3499. doi: 10.1128/jb.179.11.3494-3499.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1978.Jaffé A, D’Ari R, Norris V J. SOS-independent coupling between DNA replication and cell division in Escherichia coli. J Bacteriol. 1986;165:66–71. doi: 10.1128/jb.165.1.66-71.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1979.Jagadeeswaran P, Ashman C R, Roberts S, Langenberg J. Nucleotide sequence and analysis of deletion mutants of the Escherichia coli gpt gene in plasmids. Gene. 1984;31:309–313. doi: 10.1016/0378-1119(84)90228-2. [DOI] [PubMed] [Google Scholar]
- 1980.Jagura-Burdzy G, Hulanicka M D. Use of gene fusions to study expression of cysB, the regulatory gene of the cysteine regulon. J Bacteriol. 1981;147:744–751. doi: 10.1128/jb.147.3.744-751.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1981.Jahn D, Soll D. Two glutamyl-tRNA reductase activities in Escherichia coli. J Biol Chem. 1991;266:2542–2548. [PubMed] [Google Scholar]
- 1982.Jahreis K, Postma P W, Lengeler J W. Nucleotide sequence of the ilvH-fruR gene region of Escherichia coli K12 and Salmonella typhimurium LT2. Mol Gen Genet. 1991;226:332–336. doi: 10.1007/BF00273623. [DOI] [PubMed] [Google Scholar]
- 1983.Jain C, Belasco J G. Autoregulation of RNase E synthesis in Escherichia coli. Nucleic Acids Symp Ser. 1995;114:85–88. [PubMed] [Google Scholar]
- 1984.Jair K W, Martin R G, Rosner J L, Fujita N, Ishihama A, Wolf R E. Purification and regulatory properties of MarA protein, a transcriptional activator of Escherichia coli multiple antibiotic and superoxide resistance promoters. J Bacteriol. 1995;177:7100–7104. doi: 10.1128/jb.177.24.7100-7104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1985.Jair K W, Fawcett W P, Fujita N, Ishihama A, Ishihama A. Ambidextrous transcriptional activation by SoxS: requirement for the C-terminal domain of the RNA polymerase alpha subunit in a subset of Escherichia coli superoxide-inducible genes. Mol Microbiol. 1996;19:307–317. doi: 10.1046/j.1365-2958.1996.368893.x. [DOI] [PubMed] [Google Scholar]
- 1986.Jakubcionyte E, Apirion D, Normark S. Expression of the asr gene encoding acid shock RNA is controlled by the phoBR operon. 1993. GenBank submission L25410. [Google Scholar]
- 1987.James R, Dean D O, Debbage J. Five open reading frames upstream of the dnaK gene of E. coli. DNA Sequence. 1993;3:327–332. doi: 10.3109/10425179309020832. [DOI] [PubMed] [Google Scholar]
- 1988.Janda I, Kitagawa M, Isono K. Gene rpmF for ribosomal protein L32 and gene rimJ for a ribosomal protein acetylating enzyme are located near pyrC (23.4 min) in Escherichia coli. Mol Gen Genet. 1985;201:433–436. doi: 10.1007/BF00331335. [DOI] [PubMed] [Google Scholar]
- 1989.Jander G, Martin N L, Beckwith J R. Two cysteines in each periplasmic domain of the membrane protein DsbB are required for its function in protein disulfide bond formation. EMBO J. 1994;13:5121–5127. doi: 10.1002/j.1460-2075.1994.tb06841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1990.Janion C. Mechanisms of action of methyl methanesulfonate on Escherichia coli: mutagenesis, DNA damage and repair. Postepy Biochem. 1995;41:308–313. [PubMed] [Google Scholar]
- 1991.Janosi L, Shimizu I, Kaji A. Ribosome recycling factor (ribosome releasing factor) is essential for bacterial growth. Proc Natl Acad Sci USA. 1994;91:4249–4253. doi: 10.1073/pnas.91.10.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1992.Jans D A, Fimmel A L, Langman L, James L B, Downie J A, Senior A E, Ash G R, Gibson F, Cox G B. Mutations in the uncE gene affecting assembly of the c-subunit of the adenosine triphosphatase of Escherichia coli. Biochem J. 1983;211:717–726. doi: 10.1042/bj2110717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1993.Jaurin B, Grundstrom T. ampC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of β-lactamase of the penicillinase type. Proc Natl Acad Sci USA. 1981;78:4897–4901. doi: 10.1073/pnas.78.8.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1994.Jaurin B, Grundstrom T, Normark S. Sequence elements determining ampC promoter strength in E. coli. EMBO J. 1982;1:875–881. doi: 10.1002/j.1460-2075.1982.tb01263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1995.Jaurin B, Grundstrom T, Edlund T, Normark S. The E. coli β-lactamase attenuator mediates growth rate-dependent regulation. Nature. 1981;290:221–225. doi: 10.1038/290221a0. [DOI] [PubMed] [Google Scholar]
- 1996.Jayakumar A, Rudd K E, Fabiny J M, Barnes E M., Jr Localization of the Escherichia coli amtA gene to 95.8 minutes. J Bacteriol. 1991;173:1572–1573. doi: 10.1128/jb.173.5.1572-1573.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1997.Jayakumar A, Hwang S J, Fabiny J M, Chinault A C, Barnes E M., Jr Isolation of an ammonium or methylammonium ion transport mutant of Escherichia coli and complementation by the cloned gene. J Bacteriol. 1989;171:996–1001. doi: 10.1128/jb.171.2.996-1001.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1998.Jayaraman P S, Peakman T C, Busby S J, Quincy R V, Cole J A. Location and sequence of the promoter of the gene for the NADH-dependent nitrite reductase of Escherichia coli and its regulation by oxygen, the Fnr protein and nitrite. J Mol Biol. 1987;196:781–788. doi: 10.1016/0022-2836(87)90404-9. [DOI] [PubMed] [Google Scholar]
- 1999.Jayaratne P, Bronner D, MacLachlan P R, Dodgson C, Kido N, Whitfield C. Cloning and analysis of duplicated rfbM and rfbK genes involved in the formation of GDP-mannose in Escherichia coli 09:K30 and participation of rfb genes in the synthesis of the group I K30 capsular polysaccharide. J Bacteriol. 1994;176:3126–3139. doi: 10.1128/jb.176.11.3126-3139.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2000.Jefferson R A, Burgess S M, Hirsh D. β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci USA. 1986;83:8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2001.Jeggo P. Isolation and characterization of Escherichia coli K-12 mutants unable to induce the adaptive response to simple alkylating agents. J Bacteriol. 1979;139:783–791. doi: 10.1128/jb.139.3.783-791.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2002.Jekel M, Wackernagel W. Location of the endA gene coding for endonuclease I on the physical map of the Escherichia coli K-12 chromosome. J Bacteriol. 1994;176:1550–1551. doi: 10.1128/jb.176.5.1550-1551.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2003.Jenkins H E, Graham A, Haddock B A. Characterization of a chlG mutant of Escherichia coli K12. FEMS Microbiol Lett. 1979;6:169–173. [Google Scholar]
- 2004.Jenkins H E, Haddock B A. A specific method for the isolation of chlG mutants of Escherichia coli K12. FEMS Microbiol Lett. 1980;9:293–296. [Google Scholar]
- 2005.Jenkins L S, Nunn W D. Genetic and molecular characterization of the genes involved in short-chain fatty acid degradation in Escherichia coli: the ato system. J Bacteriol. 1987;169:42–52. doi: 10.1128/jb.169.1.42-52.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2006.Jenkins L S, Nunn W D. Regulation of the ato operon by the atoC gene in Escherichia coli. J Bacteriol. 1987;169:2096–2102. doi: 10.1128/jb.169.5.2096-2102.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2007.Jenkins S J, Sparkes C A, Jones-Mortimer M C. A gene involved in lysine excretion in Escherichia coli K-12. Heredity. 1974;32:409–412. doi: 10.1038/hdy.1974.51. [DOI] [PubMed] [Google Scholar]
- 2008.Jennings M P, Beacham I R. Analysis of the Escherichia coli gene encoding l-asparaginase II, ansB, and its regulation by cyclic AMP receptor and FNR proteins. J Bacteriol. 1990;172:1491–1498. doi: 10.1128/jb.172.3.1491-1498.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2009.Jensen K F. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J Bacteriol. 1993;175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2010.Jensen K F, Larsen J N, Schack L, Sivertsen A. Studies on the structure and expression of Escherichia coli pyrC, pyrD and pyrF using the cloned genes. Eur J Biochem. 1984;140:343–352. doi: 10.1111/j.1432-1033.1984.tb08107.x. [DOI] [PubMed] [Google Scholar]
- 2011.Jensen K F, Andersen J T, Poulsen P. Overexpression and rapid purification of the orfE/rph gene product, RNase PH of Escherichia coli. J Biol Chem. 1992;267:17147–17152. [PubMed] [Google Scholar]
- 2012.Jensen L B, Ramos J L, Kaneva Z, Molin S. A substrate-dependent biological containment system for Pseudomonas putida based on the Escherichia coli gef gene. Appl Environ Microbiol. 1993;59:3713–3717. doi: 10.1128/aem.59.11.3713-3717.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2013.Jeong J H, Kitakawa M, Isono S, Isono K. Cloning and nucleotide sequencing of the genes, rplU and rpmA, for ribosomal proteins L21 and L27 of Escherichia coli. DNA Sequence. 1993;4:59–67. doi: 10.3109/10425179309015624. [DOI] [PubMed] [Google Scholar]
- 2014.Jerlstrom P G, Bezjak D A, Jennings M P, Beacham I R. Structure and expression in Escherichia coli K-12 of the l-asparaginase I-encoding ansA gene and its flanking regions. Gene. 1989;78:37–46. doi: 10.1016/0378-1119(89)90312-0. [DOI] [PubMed] [Google Scholar]
- 2015.Jessop A P, Clugston C. Amplification of the ArgF region in strain HfrP4X of E. coli K-12. Mol Gen Genet. 1985;201:347–350. doi: 10.1007/BF00425683. [DOI] [PubMed] [Google Scholar]
- 2016.Jiang D, Hatahet Z, Blaisdell J O, Melamede R J, Wallace S S. Escherichia coli endonuclease VIII: cloning, sequencing, and overexpression of the nei structural gene and characterization of nei and nei nth mutants. J Bacteriol. 1997;179:3773–3782. doi: 10.1128/jb.179.11.3773-3782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2017.Jiang D, Hatahet Z, Melamede R J, Kow Y W, Wallace S S. Characterization of Escherichia coli endonuclease VIII. J Biol Chem. 1997;272:32230–32239. doi: 10.1074/jbc.272.51.32230. [DOI] [PubMed] [Google Scholar]
- 2018.Jiang W, Wu L F, Tomich J, Saier M H, Niehaus W G. Corrected sequence of the mannitol (mtl) operon in Escherichia coli. Mol Microbiol. 1990;4:2003–2006. doi: 10.1111/j.1365-2958.1990.tb02050.x. [DOI] [PubMed] [Google Scholar]
- 2019.Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 2020.Jin D J, Gross C A. Three rpoBC mutations that suppress the termination defects of rho mutants also affect the functions of nusA mutants. Mol Gen Genet. 1989;216:269–275. doi: 10.1007/BF00334365. [DOI] [PubMed] [Google Scholar]
- 2021.Jin H, Emanuele J J, Fairman R, Robertson J G, Hail M E, Ho H T, Falk P J, Villafranca J J. Structural studies of Escherichia coli UDP-N-acetylmuramate:l-alanine ligase. Biochemistry. 1996;35:1423–1431. doi: 10.1021/bi952334k. [DOI] [PubMed] [Google Scholar]
- 2022.Jin T, Rudd K E, Inouye M. The nlpA lipoprotein gene is located near the selC tRNA gene on the Escherichia coli chromosome. J Bacteriol. 1992;174:3822–3823. doi: 10.1128/jb.174.11.3822-3823.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2023.Jishage M, Ishihama A. A stationary phase protein in Escherichia coli with binding activity to the major sigma subunit of RNA polymerase. Proc Natl Acad Sci USA. 1998;95:4953–4958. doi: 10.1073/pnas.95.9.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2024.Jochimsen B, Nygaard P, Vestergaard T. Location on the chromosome of Escherichia coli of genes governing purine metabolism. Adenosone deaminase (add), guanosine kinase (gsk) and hypoxanthine phosphoribosyltransferase (hpt) Mol Gen Genet. 1975;143:85–91. doi: 10.1007/BF00269424. [DOI] [PubMed] [Google Scholar]
- 2025.Johann S, Hinton S M. Cloning and nucleotide sequence of the chlD locus. J Bacteriol. 1987;169:1911–1916. doi: 10.1128/jb.169.5.1911-1916.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2026.Johansson M, von Heijne G. Membrane topology of Kch, a putative K+ channel from Escherichia coli. J Biol Chem. 1996;271:25912–25915. doi: 10.1074/jbc.271.42.25912. [DOI] [PubMed] [Google Scholar]
- 2027.Johnson B F. Fine structure mapping and properties of mutations suppressing the lon mutation in Escherichia coli K-12 and B strains. Genet Res. 1977;30:273–286. doi: 10.1017/s0016672300017687. [DOI] [PubMed] [Google Scholar]
- 2028.Johnson D I, Somerville R L. Evidence that repression mechanisms can exert control over the thr, leu, and ilv operons of Escherichia coli K-12. J Bacteriol. 1983;155:49–55. doi: 10.1128/jb.155.1.49-55.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2029.Johnson D I, Somerville R L. New regulatory genes involved in the control of transcription initiation at the thr and ilv promoters in Escherichia coli K-12. Mol Gen Genet. 1984;195:70–76. doi: 10.1007/BF00332726. [DOI] [PubMed] [Google Scholar]
- 2030.Johnson M E, Rajagopalan K V. Involvement of chlA, E, M, and N loci in Escherichia coli molybdopterin biosynthesis. J Bacteriol. 1987;169:117–125. doi: 10.1128/jb.169.1.117-125.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2031.Johnson M S, Rowsell E H, Taylor B L. Investigation of transphosphorylation between chemotaxis proteins and the phosphoenolpyruvate:sugar phosphotransferase system. FEBS Lett. 1995;374:161–164. doi: 10.1016/0014-5793(95)01097-x. [DOI] [PubMed] [Google Scholar]
- 2032.Johnson R C, Ball C A, Pfeffer D, Simon M I. Isolation of the gene encoding the Hin recombinational enhancer binding protein. Proc Natl Acad Sci USA. 1988;85:3484–3488. doi: 10.1073/pnas.85.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2033.Johnston M A, Pivnick H. Use of autocytotoxic β-d-galactosides for selective growth of Salmonella typhimurium in the presence of coliforms. J Microbiol Serol. 1970;16:83–89. doi: 10.1139/m70-015. [DOI] [PubMed] [Google Scholar]
- 2034.Johnstone D B, Farr S B. AppppA binds to several proteins in Escherichia coli, including the heat shock and oxidative stress proteins DnaK, GroEL, E89, C45, and C40. EMBO J. 1991;10:3897–3904. doi: 10.1002/j.1460-2075.1991.tb04959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2035.Joly J C, Leonard M R, Wickner W. Subunit dynamics in Escherichia coli preprotein translocase. Proc Natl Acad Sci USA. 1994;91:4703–4707. doi: 10.1073/pnas.91.11.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2036.Jones C A, Holland I B. Inactivation of essential division genes ftsA, ftsZ, suppresses mutations at sfiB, a locus mediating division inhibition during the SOS response in E. coli. EMBO J. 1984;3:1181–1186. doi: 10.1002/j.1460-2075.1984.tb01948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2037.Jones C E, Brook J M, Buck D, Abell C, Smith A G. Cloning and sequencing of the Escherichia coli panB gene, which encodes ketopantoate hydroxymethyltransferase, and overexpression of the enzyme. J Bacteriol. 1993;175:2125–2130. doi: 10.1128/jb.175.7.2125-2130.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2038.Jones C H, Pinkner J S, Nicholes A V, Slonim L N, Abraham S N, Hultgren S J. FimC is a periplasmic PapD-like chaperone that directs assembly of type 1 pili in bacteria. Proc Natl Acad Sci USA. 1993;90:8397–8401. doi: 10.1073/pnas.90.18.8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2039.Jones C J, Homma M, Macnab R M. Identification of proteins of the outer (L and P) rings of the flagellar basal body of Escherichia coli. J Bacteriol. 1987;169:1489–1492. doi: 10.1128/jb.169.4.1489-1492.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2040.Jones H M, Brajkovich C M, Gunsalus R P. In vivo 5′ terminus and length of the mRNA for the proton-translocating ATPase (unc) operon of Escherichia coli. J Bacteriol. 1983;155:1279–1287. doi: 10.1128/jb.155.3.1279-1287.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2041.Jones H M, Gunsalus R P. Transcription of the Escherichia coli fumarate reductase genes (frdABCD) and their coordinate regulation by oxygen, nitrate, and fumarate. J Bacteriol. 1985;164:1100–1109. doi: 10.1128/jb.164.3.1100-1109.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2042.Jones M D, Petersen T E, Nielsen K M, Magnusson S, Sottrup-Jensen L, Gausing K, Clark B F. The complete amino-acid sequence of elongation factor Tu from Escherichia coli. Eur J Biochem. 1980;108:507–526. doi: 10.1111/j.1432-1033.1980.tb04748.x. [DOI] [PubMed] [Google Scholar]
- 2043.Jones P G, Inouye M. The cold-shock response—a hot topic. Mol Microbiol. 1994;11:811–818. doi: 10.1111/j.1365-2958.1994.tb00359.x. [DOI] [PubMed] [Google Scholar]
- 2044.Jones P G, Inouye M. RbfA, a 30S ribosomal binding factor, is a cold-shock protein whose absence triggers the cold-shock response. Mol Microbiol. 1996;21:1207–1218. doi: 10.1111/j.1365-2958.1996.tb02582.x. [DOI] [PubMed] [Google Scholar]
- 2045.Jones P G, Mitta M, Kim Y, Jiang W, Inouye M. Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:76–80. doi: 10.1073/pnas.93.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2046.Jones P G, Krah R, Tafuri S R, Wolffe A P. DNA gyrase, CS7.4, and the cold shock response in Escherichia coli. J Bacteriol. 1992;174:5798–5802. doi: 10.1128/jb.174.18.5798-5802.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2047.Jones-Mortimer M C, Kornberg H L. Genetical analysis of fructose utilization by Escherichia coli. Proc R Soc Lond Ser B. 1974;187:121–131. doi: 10.1098/rspb.1974.0066. [DOI] [PubMed] [Google Scholar]
- 2048.Jones-Mortimer M C, Kornberg H L. Amino-sugar transport systems of Escherichia coli K12. J Gen Microbiol. 1980;117:369–376. doi: 10.1099/00221287-117-2-369. [DOI] [PubMed] [Google Scholar]
- 2049.Joo D M, Nolte A, Calendar R, Zhou Y N, Jin D J. Multiple regions on the Escherichia coli heat shock transcription factor ς32 determine core RNA polymerase binding specificity. J Bacteriol. 1998;180:1095–1102. doi: 10.1128/jb.180.5.1095-1102.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2050.Jordan A, Pontis E, Atta M, Krook M, Gibert I, Barbé J, Reichard P. A second class I ribonucleotide reductase in Enterobacteriaceae: characterization of the Salmonella typhimurium enzyme. Proc Natl Acad Sci USA. 1994;91:12892–12896. doi: 10.1073/pnas.91.26.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2051.Jordan A, Aslund F, Pontis E, Reichard P, Holmgren A. Characterization of Escherichia coli NrdH.A glutaredoxin-like protein with a thioredoxin-like activity profile. J Biol Chem. 1997;272:18044–18050. doi: 10.1074/jbc.272.29.18044. [DOI] [PubMed] [Google Scholar]
- 2052.Jordan P M, Mgbeje B I A, Alwan A F, Thomas S D. Nucleotide sequence of hemD, the second gene in the hem operon of Escherichia coli K-12. Nucleic Acids Res. 1987;15:10583. doi: 10.1093/nar/15.24.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2053.Jordan P M, Mgbeje B I A, Thomas S D, Alwan A F. Nucleotide sequence for the hemD gene of Escherichia coli encoding uroporphyrinogen III synthase and initial evidence for a hem operon. Biochem J. 1988;249:613–616. doi: 10.1042/bj2490613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2054.Jordan P M, Woodcock S C. Mutagenesis of arginine residues in the catalytic cleft of Escherichia coli porphobilinogen deaminase that affects dipyrromethane cofactor assembly and tetrapyrrole chain initiation and elongation. Biochem J. 1991;280:445–449. doi: 10.1042/bj2800445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2055.Jordan P M, Thomas S D, Warren M J. Purification, crystallization and properties of porphobilinogen deaminase from a recombinant strain of Escherichia coli K12. Biochem J. 1988;254:427–435. doi: 10.1042/bj2540427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2056.Jorgensen P, Collins J, Fiil N, von Meyenbourg K. A ribosomal RNA gene, rrnC, of Escherichia coli, mapped by specialized transducing lambda dilv and lambda drbs phages. Mol Gen Genet. 1978;163:223–228. doi: 10.1007/BF00267413. [DOI] [PubMed] [Google Scholar]
- 2057.Josephsen J, Hammer-Jespersen K. Fusion of the lac genes to the promoter for the cytidine deaminase gene of Escherichia coli. Mol Gen Genet. 1981;182:154–158. doi: 10.1007/BF00422783. [DOI] [PubMed] [Google Scholar]
- 2058.Josephsen J, Hammer-Jespersen K, Hansen T D. Mapping of the gene for cytidine deaminase (cdd) in Escherichia coli K-12. J Bacteriol. 1983;154:72–75. doi: 10.1128/jb.154.1.72-75.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2059.Josephson B L, Fraenkel D G. Transketolase mutants of Escherichia coli. J Bacteriol. 1969;100:1289–1295. doi: 10.1128/jb.100.3.1289-1295.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2060.Joshi M S, Johnson J L, Rajagopalan K V. Molybdenum cofactor biosynthesis in Escherichia coli mod and mog mutants. J Bacteriol. 1996;178:4310–4312. doi: 10.1128/jb.178.14.4310-4312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2061.Jourlin C, Bengrine A, Chippaux M, Mejean V. An unorthodox sensor protein (TorS) mediates the induction of the tor structural genes in response to trimethylamine N-oxide in Escherichia coli. Mol Microbiol. 1996;20:1297–1306. doi: 10.1111/j.1365-2958.1996.tb02648.x. [DOI] [PubMed] [Google Scholar]
- 2062.Jourlin C, Simon G, Pommier J, Chippaux M, Méjean V. The periplasmic TorT protein is required for trimethylamine N-oxide reductase gene induction in Escherichia coli. J Bacteriol. 1996;178:1219–1223. doi: 10.1128/jb.178.4.1219-1223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2063.Jovanovic G, Dworkin J, Model P. Autogenous control of PspF, a constitutively active enhancer-binding protein of Escherichia coli. J Bacteriol. 1997;179:5232–5237. doi: 10.1128/jb.179.16.5232-5237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2064.Jovanovic G, Weiner L, Model P. Identification, nucleotide sequence, and characterization of PspF, the transcriptional activator of the Escherichia coli stress-induced psp operon. J Bacteriol. 1996;178:1936–1945. doi: 10.1128/jb.178.7.1936-1945.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2065.Jovanovic G, Model P. PspF and IHF bind co-operatively in the psp promoter-regulatory region of Escherichia coli. Mol Microbiol. 1997;25:473–481. doi: 10.1046/j.1365-2958.1997.4791844.x. [DOI] [PubMed] [Google Scholar]
- 2066.Jovanovic G, Model P. The RIB element in the goaG-pspF intergenic region of Escherichia coli. J Bacteriol. 1997;179:3095–3102. doi: 10.1128/jb.179.10.3095-3102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2067.Joyce C M, Grindley N. Identification of two genes immediately downstream from the polA gene of Escherichia coli. J Bacteriol. 1982;152:1211–1219. doi: 10.1128/jb.152.3.1211-1219.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2068.Joyce C M, Kelley W S, Grindley N. Nucleotide sequence of the Escherichia coli polA gene and primary structure of DNA polymerase I. J Biol Chem. 1982;257:1958–1964. [PubMed] [Google Scholar]
- 2069.Jubete Y, Maurizi M R, Gottesman S. Role of the heat shock protein DnaJ in the lon-dependent degradation of naturally unstable proteins. J Biol Chem. 1996;271:30798–30803. doi: 10.1074/jbc.271.48.30798. [DOI] [PubMed] [Google Scholar]
- 2070.Juhl M J, Clark D P. Thiophene-degrading Escherichia coli mutants possess sulfone oxidase activity and show altered resistance to sulfur-containing antibiotics. Appl Environ Microbiol. 1990;56:3179–3185. doi: 10.1128/aem.56.10.3179-3185.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2071.Jung H K, Ishino F, Matsuhashi M. Inhibition of growth of ftsQ, ftsA, and ftsZ mutant cells of Escherichia coli by amplification of a chromosomal region encompassing closely aligned cell division and cell growth genes. J Bacteriol. 1989;171:6379–6382. doi: 10.1128/jb.171.11.6379-6382.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2072.Jung J U, Gutierrez C, Martin F, Ardourel M, Villarejo M. Transcription of osmB, a gene encoding an Escherichia coli lipoprotein, is regulated by dual signals. Osmotic stress and stationary phase. J Biol Chem. 1990;265:10574–10581. [PubMed] [Google Scholar]
- 2073.Jung J U, Gutierrez C, Villarejo M. Sequence of an osmotically inducible lipoprotein gene. J Bacteriol. 1989;171:511–520. doi: 10.1128/jb.171.1.511-520.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2074.Jung Y H, Park K, Lee K H. Alteration of RNA I metabolism in a temperature-sensitive Escherichia coli rnpA mutant strain. Biochem Biophys Res Commun. 1992;186:1463–1470. doi: 10.1016/s0006-291x(05)81571-2. [DOI] [PubMed] [Google Scholar]
- 2075.Junker D E, Rokeach L A, Ganea D, Chiaramello A, Zyskind J W. Transcription termination within the Escherichia coli origin of DNA replication. Mol Gen Genet. 1986;203:101–109. doi: 10.1007/BF00330390. [DOI] [PubMed] [Google Scholar]
- 2076.Junne T, Schnetz K, Rak B. Location of the bglA gene on the physical map of Escherichia coli. J Bacteriol. 1990;172:6615–6616. doi: 10.1128/jb.172.12.6615-6616.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2077.Kaasen I, Falkenberg P, Styrvold O B, Strom A R. Molecular cloning and physical mapping of the otsBA genes, which encode the osmoregulatory trehalose pathway of Escherichia coli: evidence that transcription is activated by katF (AppR) J Bacteriol. 1992;174:889–898. doi: 10.1128/jb.174.3.889-898.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2078.Kadner R J. Transport and utilization of d-methionine and other methionine sources in Escherichia coli. J Bacteriol. 1977;129:207–216. doi: 10.1128/jb.129.1.207-216.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2079.Kadner R J, Shattuck-Eidens D M. Genetic control of the hexose phosphate transport system of Escherichia coli: mapping of deletion and insertion mutations in the uhp region. J Bacteriol. 1983;155:1052–1061. doi: 10.1128/jb.155.3.1052-1061.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2080.Kadner R J, Heller K. Mutual inhibition of cobalamin and siderophore uptake systems suggests their competition for TonB function. J Bacteriol. 1995;177:4829–4835. doi: 10.1128/jb.177.17.4829-4835.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2081.Kadner R J, Heller K, Coulton J W, Braun V. Genetic control of hydroxamate-mediated iron uptake in Escherichia coli. J Bacteriol. 1980;143:256–264. doi: 10.1128/jb.143.1.256-264.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2082.Kadner R J, Liggins G L. Transport of vitamin B12 in Escherichia coli: genetic studies. J Bacteriol. 1973;115:514–521. doi: 10.1128/jb.115.2.514-521.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2083.Kagawa H, Ono M, Enomoto M, Komeda Y. Bacteriophage chi sensitivity and motility of Escherichia coli and Salmonella typhimurium Fla− mutants possessing the hook structure. J Bacteriol. 1984;157:649–654. doi: 10.1128/jb.157.2.649-654.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2084.Kaidow A, Kataoka T, Wachi M, Takada A, Yamasaki M, Nagai K. The 55-kilodalton protein in an oriC complex fraction is glycogen synthase. J Bacteriol. 1992;174:5454–5456. doi: 10.1128/jb.174.16.5454-5456.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2085.Kaiser C. The origin of Q-independent derivatives of phage λ. Mol Gen Genet. 1980;179:547–554. doi: 10.1007/BF00271744. [DOI] [PubMed] [Google Scholar]
- 2086.Kaiser C, Murray N E. Physical characterization of the “Rac prophage” in E. coli K12. Mol Gen Genet. 1979;175:159–174. doi: 10.1007/BF00425532. [DOI] [PubMed] [Google Scholar]
- 2087.Kaiser C, Murray N E. On the nature of sbcA mutations in E. coli K12. Mol Gen Genet. 1980;179:555–563. doi: 10.1007/BF00271745. [DOI] [PubMed] [Google Scholar]
- 2088.Kaiser M, Sawers G. Overlapping promoters modulate Fnr- and ArcA-dependent anaerobic transcriptional activation of the focApfl operon in Escherichia coli. Microbiology. 1997;143:775–783. doi: 10.1099/00221287-143-3-775. [DOI] [PubMed] [Google Scholar]
- 2089.Kajie S, Ideta R, Yamato I, Anraku Y. Molecular cloning and DNA sequence of dniR, a gene affecting anaerobic expression of the Escherichia coli hexaheme nitrite reductase. FEMS Microbiol Lett. 1991;67:205–211. doi: 10.1016/0378-1097(91)90355-e. [DOI] [PubMed] [Google Scholar]
- 2090.Kajitani M, Ishihama A. Identification and sequence determination of the host factor gene for bacteriophage Q beta. Nucleic Acids Res. 1991;19:1063–1066. doi: 10.1093/nar/19.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2091.Kajitani M, Kato A, Wada A, Inokuchi Y, Ishihama A. Regulation of the Escherichia coli hfq gene encoding the host factor for phage Q beta. J Bacteriol. 1994;176:531–534. doi: 10.1128/jb.176.2.531-534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2092.Kajitani M, Fukuda R, Ishihama A. Autogenous and posttranslational regulation of Escherichia coli RNA polymerase synthesis in vitro. Mol Gen Genet. 1980;179:489–496. doi: 10.1007/BF00271738. [DOI] [PubMed] [Google Scholar]
- 2093.Kakeda M, Ueguchi C, Yamada H, Mizuno T. An Escherichia coli curved DNA-binding protein whose expression is affected by the stationary phase-specific sigma factor sigma S. Mol Gen Genet. 1995;248:629–634. doi: 10.1007/BF02423459. [DOI] [PubMed] [Google Scholar]
- 2094.Kakuda H, Hosono K, Shiroishi K, Ichihara S. Identification and characterization of the ackA (acetate kinase A)-pta (phosphotransacetylase) operon and complementation analysis of acetate utilization by an ackA-pta deletion mutant of Escherichia coli. J Biochem (Tokyo) 1994;116:916–922. doi: 10.1093/oxfordjournals.jbchem.a124616. [DOI] [PubMed] [Google Scholar]
- 2095.Kalckar H M, Kurahashi K, Jordan E. Hereditary defects in galactose metabolism in Escherichia coli mutants. I. Determination of enzyme activities. Proc Natl Acad Sci USA. 1959;45:1776–1786. doi: 10.1073/pnas.45.12.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2096.Kalman L V, Gunsalus R P. The frdR gene of Escherichia coli globally regulates several operons involved in anaerobic growth in response to nitrate. J Bacteriol. 1988;170:623–629. doi: 10.1128/jb.170.2.623-629.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2097.Kalman L V, Gunsalus R P. Identification of a second gene involved in global regulation of fumarate reductase and other nitrate-controlled genes for anaerobic respiration in Escherichia coli. J Bacteriol. 1989;171:3810–3816. doi: 10.1128/jb.171.7.3810-3816.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2098.Kalman M, Gentry D R, Cashel M. Characterization of the Escherichia coli K12 gltS glutamate permease gene. Mol Gen Genet. 1991;225:379–386. doi: 10.1007/BF00261677. [DOI] [PubMed] [Google Scholar]
- 2099.Kalman M, Murphy H, Cashel M. rhlB, a new Escherichia coli K-12 gene with an RNA helicase-like protein sequence motif, one of at least five such possible genes in a prokaryote. New Biol. 1991;3:886–895. [PubMed] [Google Scholar]
- 2100.Kalman M, Murphy H, Cashel M. The nucleotide sequence of recG, the distal spo operon gene in Escherichia coli K-12. Gene. 1992;110:95–99. doi: 10.1016/0378-1119(92)90449-y. [DOI] [PubMed] [Google Scholar]
- 2101.Kalnis A, Otto K, Ruther U, Muller-Hill B. Sequence of the lacZ gene of Escherichia coli. EMBO J. 1983;2:593–597. doi: 10.1002/j.1460-2075.1983.tb01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2102.Kamath A V, Gish K, Yanofsky C. A copy of insertion element IS5 is present within tnaB in the Kohara library of Escherichia coli W3110. J Bacteriol. 1994;176:1546–1547. doi: 10.1128/jb.176.5.1546-1547.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2103.Kamholz J, Keyhani J, Gots J S. Molecular cloning and characterization of the purE operon of Escherichia coli. Gene. 1986;44:55–62. doi: 10.1016/0378-1119(86)90042-9. [DOI] [PubMed] [Google Scholar]
- 2104.Kamio Y, Lin C-K, Regue M, Wu H C. Characterization of the ileS-lsp operon in Escherichia coli. Identification of an open reading frame upstream of the ileS gene and potential promoter(s) for the ileS-lsp operon. J Biol Chem. 1985;260:5616–5620. [PubMed] [Google Scholar]
- 2105.Kamitani S, Akiyama Y, Ito K. Identification and characterization of an Escherichia coli gene required for the formation of correctly folded alkaline phosphatase, a periplasmic enzyme. EMBO J. 1992;11:57–62. doi: 10.1002/j.1460-2075.1992.tb05027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2106.Kammen H O, Marvel C C, Hardy L, Penhoet E E. Purification, structure, and properties of Escherichia coli tRNA pseudouridine synthase I. J Biol Chem. 1988;263:2255–2263. [PubMed] [Google Scholar]
- 2107.Kammler M, Schon C, Hantke K. Characterization of the ferrous iron uptake system of Escherichia coli. J Bacteriol. 1993;175:6212–6219. doi: 10.1128/jb.175.19.6212-6219.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2108.Kampfenkel K, Braun V. Membrane topologies of the TolQ and TolR proteins of Escherichia coli: inactivation of TolQ by a missense mutation in the proposed first transmembrane segment. J Bacteriol. 1993;175:4485–4491. doi: 10.1128/jb.175.14.4485-4491.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2109.Kampfenkel K, Braun V. Topology of the ExbB protein in the cytoplasmic membrane of Escherichia coli. J Biol Chem. 1993;268:6050–6057. [PubMed] [Google Scholar]
- 2110.Kanamaru K, Mizuno T. Signal transduction and osmoregulation in Escherichia coli: a novel mutant of the positive regulator, OmpR, that functions in a phosphorylation-independent manner. J Biochem (Tokyo) 1992;111:425–430. doi: 10.1093/oxfordjournals.jbchem.a123773. [DOI] [PubMed] [Google Scholar]
- 2111.Kanatani A, Masuda T, Shimoda T, Misoka F, Lin X S, Yoshimoto T, Tsuru D. Protease II from Escherichia coli: sequencing and expression of the enzyme gene and characterization of the expressed enzyme. J Biochem (Tokyo) 1991;110:315–320. doi: 10.1093/oxfordjournals.jbchem.a123577. [DOI] [PubMed] [Google Scholar]
- 2112.Kanatani A, Yoshimoto T, Nagai H, Ito K, Tsuru D. Location of the protease II gene (ptrB) on the physical map of the Escherichia coli chromosome. J Bacteriol. 1992;174:7881. doi: 10.1128/jb.174.23.788.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2113.Kanaya S, Crouch R J. DNA sequence of the gene coding for Escherichia coli ribonuclease H. J Biol Chem. 1983;258:1276–1281. [PubMed] [Google Scholar]
- 2114.Kanaya S, Koyanagi T, Kanaya E. An esterase from Escherichia coli with a sequence similarity to hormone-sensitive lipase. Biochem J. 1998;332:75–80. doi: 10.1042/bj3320075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2115.Kanazawa H, Mabuchi K, Kayano T, Tamura F, Futai M. Nucleotide sequence of genes coding for dicyclohexylcarbodiimide-binding protein and the α subunit of proton-translocating ATPase of Escherichia coli. Biochem Biophys Res Commun. 1981;100:219–225. doi: 10.1016/s0006-291x(81)80085-x. [DOI] [PubMed] [Google Scholar]
- 2116.Kanazawa H, Mabuchi K, Kayano T, Noumi T, Sekiya T, Futai M. Nucleotide sequence of the genes for F0 components of the proton-translocating ATPase from Escherichia coli: prediction of the primary structure of F0 subunits. Biochem Biophys Res Commun. 1981;103:613–620. doi: 10.1016/0006-291x(81)90495-2. [DOI] [PubMed] [Google Scholar]
- 2117.Kanazawa H, Kayano T, Mabuchi K, Futai M. Nucleotide sequence of the genes coding alpha, beta and gamma subunits of the proton-translocating ATPase of Escherichia coli. Biochem Biophys Res Commun. 1981;103:604–612. doi: 10.1016/0006-291x(81)90494-0. [DOI] [PubMed] [Google Scholar]
- 2118.Kanazawa H, Kayano T, Kiyasu T, Futai M. Nucleotide sequence of the genes for beta and epsilon subunits of proton-translocating ATPase from Escherichia coli. Biochem Biophys Res Commun. 1982;105:1257–1264. doi: 10.1016/0006-291x(82)90922-6. [DOI] [PubMed] [Google Scholar]
- 2119.Kandror O, Sherman M, Rhode M, Goldberg A L. Trigger factor is involved in GroEL-dependent protein degradation in Escherichia coli and promotes binding of GroEL to unfolded proteins. EMBO J. 1995;14:6021–6027. doi: 10.1002/j.1460-2075.1995.tb00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2120.Kanemori M, Mori H, Yura T. Induction of heat shock proteins by abnormal proteins results from stabilization and not increased synthesis of ς32 in Escherichia coli. J Bacteriol. 1994;176:5648–5653. doi: 10.1128/jb.176.18.5648-5653.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2121.Kanemori M, Nishihara K, Yanagi H, Yura T. Synergistic roles of HslVU and other ATP-dependent proteases in controlling in vivo turnover of ς32 and abnormal proteins in Escherichia coli. J Bacteriol. 1997;179:7219–7225. doi: 10.1128/jb.179.23.7219-7225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2122.Kang P J, Craig E A. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J Bacteriol. 1990;172:2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2123.Kang W K, Icho T, Isono S, Kitakawa M, Isono K. Characterization of the gene rimK responsible for the addition of glutamic acid residues to the C-terminus of ribosomal protein S6 in Escherichia coli K12. Mol Gen Genet. 1989;217:281–288. doi: 10.1007/BF02464894. [DOI] [PubMed] [Google Scholar]
- 2124.Kano Y, Wada C, Nagase T, Imamoto F. Genetic characterization of the gene hupB encoding the HU-1 protein of Escherichia coli. Gene. 1986;45:37–44. doi: 10.1016/0378-1119(86)90129-0. [DOI] [PubMed] [Google Scholar]
- 2125.Kano Y, Osato K, Wada M, Imamoto F. Cloning and sequencing of the HU-2 gene of Escherichia coli. Mol Gen Genet. 1987;209:408–410. doi: 10.1007/BF00329674. [DOI] [PubMed] [Google Scholar]
- 2126.Kano Y, Wada M, Imamoto F. Genetic characterization of the gene hupA encoding the HU-2 protein of Escherichia coli. Gene. 1988;69:331–335. doi: 10.1016/0378-1119(88)90443-x. [DOI] [PubMed] [Google Scholar]
- 2127.Kano Y, Yoshino S, Wada C, Yokoyama K, Nobuhara M, Imamoto F. Molecular cloning and nucleotide sequence of the HU-1 gene of Escherichia coli. Mol Gen Genet. 1985;201:360–362. doi: 10.1007/BF00425687. [DOI] [PubMed] [Google Scholar]
- 2128.Kao C, Gumbs E, Snyder L. Cloning and characterization of the Escherichia coli lit gene, which blocks bacteriophage T4 late gene expression. J Bacteriol. 1987;169:1232–1238. doi: 10.1128/jb.169.3.1232-1238.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2129.Kao C, Snyder L. The lit gene product which blocks bacteriophage T4 late gene expression is a membrane protein encoded by a cryptic DNA element, e14. J Bacteriol. 1988;170:2056–2062. doi: 10.1128/jb.170.5.2056-2062.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2130.Kaplan J B, Nichols B. Nucleotide sequence of Escherichia coli pabA and its evolutionary relationship to trp(G)D. J Mol Biol. 1983;168:451–468. doi: 10.1016/s0022-2836(83)80295-2. [DOI] [PubMed] [Google Scholar]
- 2131.Kaplan L, Reilly H C, Stuck C C. Action of azaserine of Escherichia coli. J Bacteriol. 1959;78:511–519. doi: 10.1128/jb.78.4.511-519.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2132.Kaplan S, Anderson D. Selection of temperature-sensitive activating enzyme mutants in Escherichia coli. J Bacteriol. 1968;95:991–997. doi: 10.1128/jb.95.3.991-997.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2133.Karibian D, Michel P F, Starka J. The EnvC phenotype and its expression in various Escherichia coli K12 strains. FEMS Microbiol Lett. 1985;27:319–324. [Google Scholar]
- 2134.Karlinsey J E, Pease A J, Winkler M E, Bailey J L, Hughes K T. The flk gene of Salmonella typhimurium couples flagellar P- and L-ring assembly to flagellar morphogenesis. J Bacteriol. 1997;179:2389–2400. doi: 10.1128/jb.179.7.2389-2400.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2135.Karlsson M, Hannavy K, Higgins C F. A sequence-specific function for the N-terminal signal-like sequence of the TonB protein. Mol Microbiol. 1993;8:379–388. doi: 10.1111/j.1365-2958.1993.tb01581.x. [DOI] [PubMed] [Google Scholar]
- 2136.Karlsson M, Hannavy K, Higgins C F. ExbB acts as a chaperone-like protein to stabilize TonB in the cytoplasm. Mol Microbiol. 1993;8:389–396. doi: 10.1111/j.1365-2958.1993.tb01582.x. [DOI] [PubMed] [Google Scholar]
- 2137.Karow M, Georgopoulos C. Sequencing, mutational analysis, and transcriptional regulation of the Escherichia coli htrB gene. Mol Microbiol. 1991;5:2285–2292. doi: 10.1111/j.1365-2958.1991.tb02159.x. [DOI] [PubMed] [Google Scholar]
- 2138.Karow M, Georgopoulos C. Isolation and characterization of the Escherichia coli msbB gene, a multicopy suppressor of null mutations in the high-temperature requirement gene htrB. J Bacteriol. 1992;174:702–710. doi: 10.1128/jb.174.3.702-710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2139.Karow M, Georgopoulos C. The essential Escherichia coli msbA gene, a multicopy suppressor of null mutations in the htrB gene, is related to the universally conserved family of ATP-dependent translocators. Mol Microbiol. 1993;7:69–79. doi: 10.1111/j.1365-2958.1993.tb01098.x. [DOI] [PubMed] [Google Scholar]
- 2140.Karow M, Fayet O, Cegielska A, Ziegelhoffer T, Georgopoulos C. Isolation and characterization of the Escherichia coli htrB gene, whose product is essential for bacterial viability above 33°C in rich media. J Bacteriol. 1991;173:741–750. doi: 10.1128/jb.173.2.741-750.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2141.Karow M, Raina S, Georgopoulos C, Fayet O. Complex phenotypes of null mutations in the htr genes, whose products are essential for Escherichia coli growth at elevated temperatures. Res Microbiol. 1991;142:289–294. doi: 10.1016/0923-2508(91)90043-a. [DOI] [PubMed] [Google Scholar]
- 2142.Karpel R, Alon T, Glaser G, Schuldiner S, Padan E. Expression of a sodium proton antiporter (NhaA) in Escherichia coli is induced by Na+ and Li+ ions. J Biol Chem. 1991;266:21753–21759. [PubMed] [Google Scholar]
- 2143.Karpel R, Olami Y, Taglicht D, Schuldiner S, Padan E. Sequencing of the gene ant which affects the Na+/H+ antiporter activity in Escherichia coli. J Biol Chem. 1988;263:10408–10414. [PubMed] [Google Scholar]
- 2144.Karran P, Lindahl T, Ofsteng I, Evensen G, Seeberg E. Escherichia coli mutants deficient in 3-methyladenine-DNA glycosylase. J Mol Biol. 1980;140:101–127. doi: 10.1016/0022-2836(80)90358-7. [DOI] [PubMed] [Google Scholar]
- 2145.Karube I, Tomiyama M, Kikuchi Y. Molecular cloning and physical mapping of the hyd gene of Escherichia coli K-12. FEMS Microbiol Lett. 1984;25:165–168. [Google Scholar]
- 2146.Kasahara M, Nakata A, Shinagawa H. Molecular analysis of the Escherichia coli phoP-phoQ operon. J Bacteriol. 1992;174:492–498. doi: 10.1128/jb.174.2.492-498.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2147.Kasahara M, Makino K, Amemura M, Nakata A, Shinagawa H. Dual regulation of the ugp operon by phosphate and carbon starvation at two interspaced promoters. J Bacteriol. 1991;173:549–558. doi: 10.1128/jb.173.2.549-558.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2148.Kashiwagi K, Endo H, Kobayashi H, Takio K, Igarashi K. Spermidine-preferential uptake system in Escherichia coli. ATP hydrolysis by PotA protein and its association with membrane. J Biol Chem. 1995;270:25377–25382. doi: 10.1074/jbc.270.43.25377. [DOI] [PubMed] [Google Scholar]
- 2149.Kashiwagi K, Watanabe R, Igarashi K. Involvement of ribonuclease III in the enhancement of expression of the speF-potE operon encoding inducible ornithine decarboxylase and polyamine transport protein. Biochem Biophys Res Commun. 1994;200:591–597. doi: 10.1006/bbrc.1994.1489. [DOI] [PubMed] [Google Scholar]
- 2150.Kashiwagi K, Miyamoto S, Nukui E, Kobayashi H, Igarashi K. Functions of potA and potD proteins in spermidine-preferential uptake system in Escherichia coli. J Biol Chem. 1993;268:19358–19363. [PubMed] [Google Scholar]
- 2151.Kashiwagi K, Miyamoto S, Suzuki F, Kobayashi H, Igarashi K. Excretion of putrescine by the putrescine-ornithine antiporter encoded by the potE gene of Escherichia coli. Proc Natl Acad Sci USA. 1992;89:4529–4533. doi: 10.1073/pnas.89.10.4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2152.Kashiwagi K, Shibuya S, Tomitori H, Kuraishi A, Igarashi K. Excretion and uptake of putrescine by the PotE protein in Escherichia coli. J Biol Chem. 1997;272:6318–6323. doi: 10.1074/jbc.272.10.6318. [DOI] [PubMed] [Google Scholar]
- 2153.Kashiwagi K, Suzuki T, Suzuki F, Furuchi T, Kobayashi H, Igarashi K. Coexistence of the genes for putrescine transport protein and ornithine decarboxylase at 16 min on Escherichia coli chromosome. J Biol Chem. 1991;266:20922–20927. [PubMed] [Google Scholar]
- 2154.Kasian P A, Davidson B E, Pittard J. Molecular analysis of the promoter operator region of the Escherichia coli K-12 tyrP gene. J Bacteriol. 1986;167:556–561. doi: 10.1128/jb.167.2.556-561.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2155.Kasian P A, Pittard J. Construction of a tyrP-lac operon fusion strain and its use in the isolation and analysis of mutants derepressed for tyrP expression. J Bacteriol. 1984;160:175–183. doi: 10.1128/jb.160.1.175-183.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2156.Kasimoglu E, Park S J, Malek J, Tseng C P, Gunsalus R P. Transcriptional regulation of the proton-translocating ATPase (atpIBEFHAGDC) operon of Escherichia coli: control by cell growth rate. J Bacteriol. 1996;178:5563–5567. doi: 10.1128/jb.178.19.5563-5567.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2157.Kataoka H, Yamamoto H, Sekiguchi M. A new gene (alkB) of Escherichia coli that controls sensitivity to methyl methane sulfonate. J Bacteriol. 1983;153:1301–1307. doi: 10.1128/jb.153.3.1301-1307.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2158.Kataoka H, Sekiguchi M. Molecular cloning and characterization of the alkB gene of Escherichia coli. Mol Gen Genet. 1985;198:263–269. doi: 10.1007/BF00383004. [DOI] [PubMed] [Google Scholar]
- 2159.Kataoka K, Mizushima T, Ogata Y, Miki T, Sekimizu K. Heat shock-induced DNA relaxation in vitro by DNA gyrase of Escherichia coli in the presence of ATP. J Biol Chem. 1996;271:24806–24810. doi: 10.1074/jbc.271.40.24806. [DOI] [PubMed] [Google Scholar]
- 2160.Katayama T, Takata M, Sekimizu K. The nucleoid protein H-NS facilitates chromosome DNA replication in Escherichia coli dnaA mutants. J Bacteriol. 1996;178:5790–5792. doi: 10.1128/jb.178.19.5790-5792.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2161.Katayama T, Takata M, Sekimizu K. CedA is a novel Escherichia coli protein that activates the cell division inhibited by chromosomal DNA over-replication. Mol Microbiol. 1997;26:687–697. doi: 10.1046/j.1365-2958.1997.5941967.x. [DOI] [PubMed] [Google Scholar]
- 2162.Katinka M, Cossart P, Sibilli L, Saint-Girons I, Chalvignac M A, LeBras G, Cohen G N, Yaniv M. Nucleotide sequence of the thrA gene of Escherichia coli. Proc Natl Acad Sci USA. 1980;77:5730–5733. doi: 10.1073/pnas.77.10.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2163.Kato J, Suzuki H, Ikeda H. Purification and characterization of DNA topoisomerase IV in Escherichia coli. J Biol Chem. 1992;267:25676–25684. [PubMed] [Google Scholar]
- 2164.Kato J, Nishimura Y, Yamada M, Suzuki H, Hirota Y. Gene organization in the region containing a new gene involved in chromosome partition in Escherichia coli. J Bacteriol. 1988;170:3967–3977. doi: 10.1128/jb.170.9.3967-3977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2165.Kato J, Nishimura Y, Imamura R, Niki H, Hiraga S, Suzuki H. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 2166.Kato J-I, Hirota Y. Overlapping of the coding regions for alpha and gamma components of penicillin-binding protein 1b in Escherichia coli. Mol Gen Genet. 1984;196:449–457. doi: 10.1007/BF00436192. [DOI] [PubMed] [Google Scholar]
- 2167.Kato N, Tsuzuki M, Aiba H, Mizuno T. Gene activation by the Escherichia coli positive regulator OmpR: a mutational study of the DNA-binding domain of OmpR. Mol Gen Genet. 1995;248:399–406. doi: 10.1007/BF02191639. [DOI] [PubMed] [Google Scholar]
- 2168.Katsukake K, Nakao T, Iino T. A gene for DNA invertase and an invertible DNA in Escherichia coli K-12. Gene. 1985;34:343–350. doi: 10.1016/0378-1119(85)90143-x. [DOI] [PubMed] [Google Scholar]
- 2169.Katz L. Selection of araB and araC mutants of Escherichia coli B/r by resistance to ribitol. J Bacteriol. 1970;102:593–595. doi: 10.1128/jb.102.2.593-595.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2170.Katzenmeier G, Schmid C, Kellermann J, Lottspeich F, Bacher A. Biosynthesis of tetrahydrofolate. Sequence of GTP cyclohydrolase I from Escherichia coli. Biol Chem Hoppe-Seyler. 1991;372:991–997. doi: 10.1515/bchm3.1991.372.2.991. [DOI] [PubMed] [Google Scholar]
- 2171.Kawagishi I, Homma M, Williams A W, Macnab R M. Characterization of the flagellar hook length control protein FliK of Salmonella typhimurium and Escherichia coli. J Bacteriol. 1996;178:2954–2959. doi: 10.1128/jb.178.10.2954-2959.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2172.Kawagishi I, Muller V, Williams A W, Irikura V M, Macnab R M. Subdivision of flagellar region III of the Escherichia coli and Salmonella typhimurium chromosomes and identification of two additional flagellar genes. J Gen Microbiol. 1992;138:1051–1065. doi: 10.1099/00221287-138-6-1051. [DOI] [PubMed] [Google Scholar]
- 2173.Kawakami K, Naito S, Inoue N, Nakamura Y, Ikeda H, Uchida H. Isolation and characterization of herC, a mutation of Escherichia coli affecting maintenance of ColE1. Mol Gen Genet. 1989;219:333–340. doi: 10.1007/BF00259604. [DOI] [PubMed] [Google Scholar]
- 2174.Kawakami K, Jonsson Y H, Bjork G R, Ikeda H, Nakamura H. Chromosomal location and structure of the operon encoding peptide-chain-release factor 2 of Escherichia coli. Proc Natl Acad Sci USA. 1988;85:5620–5624. doi: 10.1073/pnas.85.15.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2175.Kawamoto S, Tokuyama S, Aoyama K, Yashima S, Eguchi Y. Genetic mapping of cold resistance gene of Escherichia coli. Agric Biol Chem. 1984;48:2067–2071. [Google Scholar]
- 2176.Kawamukai M, Utsumi R, Takeda K, Higashi A, Matsuda H, Choi Y L, Komano T. Nucleotide sequence and characterization of the sfs1 gene: sfs1 is involved in CRP*-dependent mal gene expression in Escherichia coli. J Bacteriol. 1991;173:2644–2648. doi: 10.1128/jb.173.8.2644-2648.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2177.Kawashima H, Horii T, Ogawa T, Ogawa H. Organization of the recA gene of Escherichia coli. Mol Gen Genet. 1984;193:288–292. doi: 10.1007/BF00330682. [DOI] [PubMed] [Google Scholar]
- 2178.Kawazu Y, Ito K, Matsumura K, Nakamura Y. Comparative characterization of release factor RF-3 genes of Escherichia coli, Salmonella typhimurium, and Dichelobacter nodosus. J Bacteriol. 1995;177:5547–5553. doi: 10.1128/jb.177.19.5547-5553.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2179.Kawula T H, Lelivelt M J. Mutations in a gene encoding a new Hsp70 suppress rapid DNA inversion and bgl activation, but not proU derepression, in hns-1 mutant Escherichia coli. J Bacteriol. 1994;176:610–619. doi: 10.1128/jb.176.3.610-619.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2180.Kay W W. Genetic control of the metabolism of propionate by Escherichia coli K12. Biochim Biophys Acta. 1972;264:508–521. doi: 10.1016/0304-4165(72)90014-1. [DOI] [PubMed] [Google Scholar]
- 2181.Kay W W, Kornberg H L. Genetic control of the uptake of C4-dicarboxylic acids by Escherichia coli. FEBS Lett. 1969;3:93–96. doi: 10.1016/0014-5793(69)80105-5. [DOI] [PubMed] [Google Scholar]
- 2182.Kayahara T, Thelen P, Ogawa W, Inaba K, Tsuda M, Goldberg E B, Tsuchiya T. Properties of recombinant cells capable of growing on serine without NhaB Na+/H+ antiporter in Escherichia coli. J Bacteriol. 1992;174:7482–7485. doi: 10.1128/jb.174.22.7482-7485.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2183.Keating D H, Carey M R, Cronan J E., Jr The unmodified (apo) form of Escherichia coli acyl carrier protein is a potent inhibitor of cell growth. J Biol Chem. 1995;270:22229–22235. doi: 10.1074/jbc.270.38.22229. [DOI] [PubMed] [Google Scholar]
- 2184.Keck W, van Leeuwen A M, Huber-Wunderlich M, Goodell E W. Cloning and characterization of mepA, the structural gene of the penicillin-insensitive murein endopeptidase from Escherichia coli. Mol Microbiol. 1990;4:209–219. doi: 10.1111/j.1365-2958.1990.tb00588.x. [DOI] [PubMed] [Google Scholar]
- 2185.Keen J, Williams J, Busby S J. Location of essential sequence elements at the Escherichia coli melAB promoter. Biochem J. 1996;318:443–449. doi: 10.1042/bj3180443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2186.Keiler K C, Silber K R, Downard K M, Papayannopoulos I A, Biemann K, Sauer R T. C-terminal specific protein degradation: activity and substrate specificity of the Tsp protease. Protein Sci. 1995;4:1507–1515. doi: 10.1002/pro.5560040808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2187.Kelker N E, Maas W K. Selection for genetically respressible (ArgR+) strains of Escherichia coli K12 from genetically derepressed (ArgR−) mutants using acetylnorvaline. Mol Gen Genet. 1974;131:131–136. doi: 10.1007/BF00272178. [DOI] [PubMed] [Google Scholar]
- 2188.Kelley W L, Georgopoulos C. Positive control of the two-component RcsC/B signal transduction network by DjIA: a member of the DnaJ family of molecular chaperones in Escherichia coli. Mol Microbiol. 1997;25:913–931. doi: 10.1111/j.1365-2958.1997.mmi527.x. [DOI] [PubMed] [Google Scholar]
- 2189.Kelley W S. Mapping of the polA locus of Escherichia coli K12: genetic fine structure of the cistron. Genetics. 1980;95:15–38. doi: 10.1093/genetics/95.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2190.Kelley W S, Whitfield H J., Jr Purification of an altered DNA polymerase from an E. coli strain with a pol mutation. Nature. 1971;230:33–36. doi: 10.1038/230033a0. [DOI] [PubMed] [Google Scholar]
- 2191.Kelln R A, O’Donovan G A. Isolation and partial characteristics of an argR mutant of Salmonella typhimurium. J Bacteriol. 1976;128:528–535. doi: 10.1128/jb.128.2.528-535.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2192.Kelly K O, Reuven N B, Li Z-W, Deutscher M P. RNase PH is essential for tRNA processing and viability in RNase-deficient Escherichia coli cells. J Biol Chem. 1992;267:16015–16018. [PubMed] [Google Scholar]
- 2193.Kelly T M, Stachula S A, Raetz C R H, Anderson M S. The firA gene of Escherichia coli encodes UDP-3-O-(R-3-hydroxymyristoyl)-glucosamine N-acyltransferase. The third step of endotoxin biosynthesis. J Biol Chem. 1993;268:19866–19874. [PubMed] [Google Scholar]
- 2194.Kelman Z, O’Donnell M A. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu Rev Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 2195.Kemp E H, Minton N P, Mann N H. Complete nucleotide sequence and deduced amino acid sequence of the M5 polypeptide gene of Escherichia coli. Nucleic Acids Res. 1987;15:3924. doi: 10.1093/nar/15.9.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2196.Keng T, Webster T A, Sauer R T, Schimmel P. Gene for Escherichia coli glycyl-tRNA synthetase has tandem subunit coding regions in the same reading frame. J Biol Chem. 1982;257:12503–12508. [PubMed] [Google Scholar]
- 2197.Kenri T, Imamoto F, Kano Y. Construction and characterization of an Escherichia coli mutant deficient in the metY gene encoding tRNAf2Met: either tRNAf1Met or tRNAf2Met is required for cell growth. Gene. 1992;114:109–114. doi: 10.1016/0378-1119(92)90715-2. [DOI] [PubMed] [Google Scholar]
- 2198.Kenri T, Imamoto F, Kano Y. Three tandemly repeated structural genes encoding tRNAf1Met in the metZ operon of Escherichia coli K-12. Gene. 1994;138:261–262. doi: 10.1016/0378-1119(94)90821-4. [DOI] [PubMed] [Google Scholar]
- 2199.Kenri T, Kohno K, Goshima N, Imamoto F, Kano Y. Construction and characterization of an Escherichia coli mutant with a deletion of the metZ gene encoding tRNAf1Met. Gene. 1991;103:31–36. doi: 10.1016/0378-1119(91)90387-q. [DOI] [PubMed] [Google Scholar]
- 2200.Kenyon C J, Walker G C. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci USA. 1980;77:2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2201.Kenyon C J, Walker G C. Expression of the E. coli uvrA gene is inducible. Nature. 1981;289:808–810. doi: 10.1038/289808a0. [DOI] [PubMed] [Google Scholar]
- 2202.Kessler D P, Rickenberg H V. A new method for the selection of mutants of Escherichia coli forming β-galactosidase constitutively. Biochim Biophys Acta. 1964;90:609–610. doi: 10.1016/0304-4165(64)90241-7. [DOI] [PubMed] [Google Scholar]
- 2203.Kessler D P, Leibrecht I, Knappe J. Pyruvate-formate-lyase-deactivase and acetyl-CoA reductase activities of Escherichia coli reside on a polymeric protein particle encoded by adhE. FEBS Lett. 1991;281:59–63. doi: 10.1016/0014-5793(91)80358-a. [DOI] [PubMed] [Google Scholar]
- 2204.Keyer K, Gort A S, Imlay J A. Superoxide and the production of oxidative DNA damage. J Bacteriol. 1995;177:6782–6790. doi: 10.1128/jb.177.23.6782-6790.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2205.Keyhani N O, Roseman S. Wild-type Escherichia coli grows on the chitin disaccharide, N,N′-diacetylchitobiose, by expressing the cel operon. Proc Natl Acad Sci USA. 1997;94:14367–14371. doi: 10.1073/pnas.94.26.14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2206.Khan S, Spudich J L, McCray J A, Trentham D R. Chemotactic signal integration in bacteria. Proc Natl Acad Sci USA. 1995;92:9757–9761. doi: 10.1073/pnas.92.21.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2207.Khatri G S, MacAllister T, Sista P R, Bastia D. The replication terminator protein of E. coli is a DNA sequence-specific contra-helicase. Cell. 1989;59:667–674. doi: 10.1016/0092-8674(89)90012-3. [DOI] [PubMed] [Google Scholar]
- 2208.Khattar M M. Overexpression of the hslVU operon suppresses SOS-mediated inhibition of cell division in Escherichia coli. FEBS Lett. 1997;414:402–404. doi: 10.1016/s0014-5793(97)01024-7. [DOI] [PubMed] [Google Scholar]
- 2209.Khattar M M, Begg K J, Donachie W D. Identification of FtsW and characterization of a new ftsW division mutant of Escherichia coli. J Bacteriol. 1994;176:7140–7147. doi: 10.1128/jb.176.23.7140-7147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2210.Khattar M M, Addinall S G, Stedul K H, Boyle D S, Lutkenhaus J, Donachie W D. Two polypeptide products of the Escherichia coli cell division gene ftsW and a possible role for FtsW in FtsZ function. J Bacteriol. 1997;179:784–793. doi: 10.1128/jb.179.3.784-793.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2211.Khodursky A B, Zechiedrich E L, Cozzarelli N R. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:11801–11805. doi: 10.1073/pnas.92.25.11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2212.Kiino D R, Phillips G J, Silhavy T J. Increased expression of the bifunctional protein PrlF suppresses overproduction lethality associated with exported β-galactosidase hybrid proteins in Escherichia coli. J Bacteriol. 1990;172:185–192. doi: 10.1128/jb.172.1.185-192.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2213.Kiino D R, Rothman-Denes L B. Genetic analysis of bacteriophage N4 adsorption. J Bacteriol. 1989;171:4595–4602. doi: 10.1128/jb.171.9.4595-4602.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2214.Kiino D R, Singer M S, Rothman-Denes L B. Two overlapping genes encoding membrane proteins required for bacteriophage N4 adsorption. J Bacteriol. 1993;175:7081–7085. doi: 10.1128/jb.175.21.7081-7085.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2215.Kiino D R, Licudine R, Wilt K, Yang D H, Rothman-Denes L B. A cytoplasmic protein, NfrC, is required for bacteriophage N4 adsorption. J Bacteriol. 1993;175:7074–7080. doi: 10.1128/jb.175.21.7074-7080.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2216.Kikuchi Y, Flamm E L, Weisberg R. An Escherichia coli mutant unable to support site-specific recombination of bacteriophage λ. J Mol Biol. 1985;183:129–140. doi: 10.1016/0022-2836(85)90207-4. [DOI] [PubMed] [Google Scholar]
- 2217.Kikuchi Y, Kojima H, Tanaka T, Takatsuka Y, Kamio Y. Characterization of a second lysine decarboxylase isolated from Escherichia coli. J Bacteriol. 1997;179:4486–4492. doi: 10.1128/jb.179.14.4486-4492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2218.Kikuchi Y, Yoda K, Yamasaki M, Tamura G. The nucleotide sequence of the promoter and the amino-terminal region of alkaline phosphatase structural gene (phoA) of Escherichia coli. Nucleic Acids Res. 1981;9:5671–5678. doi: 10.1093/nar/9.21.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2219.Killmann H, Benz R, Braun V. Conversion of the FhuA transport protein into a diffusion channel through the outer membrane of Escherichia coli. EMBO J. 1993;12:3007–3016. doi: 10.1002/j.1460-2075.1993.tb05969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2220.Killmann H, Braun V. An aspartate deletion mutation defines a binding site of the multifunctional FhuA outer membrane receptor of Escherichia coli K-12. J Bacteriol. 1992;174:3479–3486. doi: 10.1128/jb.174.11.3479-3486.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2221.Killmann H, Braun V. Energy-dependent receptor activities of Escherichia coli K-12: mutated TonB proteins alter FhuA receptor activities to phages T5, T1, phi80 and to colicin M. FEMS Microbiol Lett. 1994;119:71–76. doi: 10.1111/j.1574-6968.1994.tb06869.x. [DOI] [PubMed] [Google Scholar]
- 2222.Kilstrup M, Meng L M, Neuhard J, Nygaard P. Genetic evidence for a repressor of synthesis of cytosine deaminase and purine biosynthesis enzymes in Escherichia coli. J Bacteriol. 1989;171:2124–2127. doi: 10.1128/jb.171.4.2124-2127.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2223.Kim C, Song S, Park C. The d-allose operon of Escherichia coli K-12. J Bacteriol. 1997;179:7631–7637. doi: 10.1128/jb.179.24.7631-7637.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2224.Kim D R, McHenry C S. In vivo assembly of overproduced DNA polymerase III. Overproduction, purification, and characterization of the alpha, alpha-epsilon, and alpha-epsilon-theta subunits. J Biol Chem. 1996;271:20681–20689. doi: 10.1074/jbc.271.34.20681. [DOI] [PubMed] [Google Scholar]
- 2225.Kim H K, Kim S J, Lee J W, Lee J W, Cha M K, Kim I H. Identification of promoter in the 5′-flanking region of the E. coli thioredoxin-linked thiol peroxidase gene: evidence for the existence of oxygen-related transcriptional regulatory protein. Biochem Biophys Res Commun. 1996;221:641–646. doi: 10.1006/bbrc.1996.0649. [DOI] [PubMed] [Google Scholar]
- 2226.Kim I V, Veres Z, Stadtman T C. Escherichia coli mutant SELD enzymes. The cysteine 17 residue is essential for selenophosphate formation from ATP and selenide. J Biol Chem. 1992;267:19650–19654. [PubMed] [Google Scholar]
- 2227.Kim J, Lee Y, Kim C, Park C. Involvement of SecB, a chaperone, in the export of ribose-binding protein. J Bacteriol. 1992;174:5219–5227. doi: 10.1128/jb.174.16.5219-5227.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2228.Kim S, Dallmann H G, McHenry C S, Marians K J. tau couples the leading- and lagging-strand polymerases at the Escherichia coli DNA replication fork. J Biol Chem. 1996;271:21406–21412. doi: 10.1074/jbc.271.35.21406. [DOI] [PubMed] [Google Scholar]
- 2229.Kim S, Dallmann H G, McHenry C S, Marians K J. tau protects beta in the leading-strand polymerase complex at the replication fork. J Biol Chem. 1996;271:4315–4318. doi: 10.1074/jbc.271.8.4315. [DOI] [PubMed] [Google Scholar]
- 2230.Kim S K, Makino K, Amemura M, Nakata A, Shinagawa H. Mutational analysis of the role of the first helix of region 4.2 of the sigma 70 subunit of Escherichia coli RNA polymerase in transcriptional activation by activator protein PhoB. Mol Gen Genet. 1995;248:1–8. doi: 10.1007/BF02456607. [DOI] [PubMed] [Google Scholar]
- 2231.Kim S K, Makino K, Amemura M, Shinagawa H, Nakata A. Molecular analysis of the phoH gene, belonging to the phosphate regulon in Escherichia coli. J Bacteriol. 1993;175:1316–1324. doi: 10.1128/jb.175.5.1316-1324.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2232.Kim S R, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc Natl Acad Sci USA. 1997;94:13792–13797. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2233.Kimata K, Takahashi H, Inada T, Postma P, Aiba H. cAMP receptor protein-cAMP plays a crucial role in glucose-lactose diauxie by activating the major glucose transporter gene in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:12914–12919. doi: 10.1073/pnas.94.24.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2234.Kimsey H H, Dagarag M D, Kumamoto C A. Diverse effects of mutation on the activity of the Escherichia coli export chaperone SecB. J Biol Chem. 1995;270:22831–22835. doi: 10.1074/jbc.270.39.22831. [DOI] [PubMed] [Google Scholar]
- 2235.Kimura A, Miki T, Hiraga S, Nagata T, Yura T. Conditionally lethal amber mutations in the dnaA region of the Escherichia coli chromosome that affect chromosome replication. J Bacteriol. 1979;140:825–834. doi: 10.1128/jb.140.3.825-834.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2236.Kimura M, Ishihama A. Functional map of the alpha subunit of Escherichia coli RNA polymerase: amino acid substitution within the amino-terminal assembly domain. J Mol Biol. 1995;254:342–349. doi: 10.1006/jmbi.1995.0621. [DOI] [PubMed] [Google Scholar]
- 2237.Kinebuchi T, Shindo H, Nagai H, Shimamoto N, Shimizu M. Functional domains of Escherichia coli single-stranded DNA binding protein as assessed by analyses of the deletion mutants. Biochemistry. 1997;36:6732–6738. doi: 10.1021/bi961647s. [DOI] [PubMed] [Google Scholar]
- 2238.King G, Murray N E. Restriction alleviation and modification enhancement by the Rac prophage of Escherichia coli K-12. Mol Microbiol. 1995;16:769–777. doi: 10.1111/j.1365-2958.1995.tb02438.x. [DOI] [PubMed] [Google Scholar]
- 2239.King S C, Fleming S R, Brechtel C. Pyridine carboxylic acids as inhibitors and substrates of the Escherichia coli gab permease encoded by gabP. J Bacteriol. 1995;177:5381–5382. doi: 10.1128/jb.177.18.5381-5382.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2240.Kinghorn J R, Schweizer M, Giles N H, Kushner S R. The cloning and analysis of the aroD gene of E. coli K-12. Gene. 1981;14:73–80. doi: 10.1016/0378-1119(81)90149-9. [DOI] [PubMed] [Google Scholar]
- 2241.Kirby J E, Trempy J E, Gottesman S. Excision of a P4-like cryptic prophage leads to Alp protease expression in Escherichia coli. J Bacteriol. 1994;176:2068–2081. doi: 10.1128/jb.176.7.2068-2081.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2242.Kirby T W, Hindenach B R, Greene R C. Regulation of in vivo transcription of the Escherichia coli K-12 metJBLF gene cluster. J Bacteriol. 1986;165:671–677. doi: 10.1128/jb.165.3.671-677.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2243.Kishigami S, Kanaya E, Kikuchi M, Ito K. DsbA-DsbB interaction through their active site cysteines. Evidence from an odd cysteine mutant of DsbA. J Biol Chem. 1995;270:17072–17074. doi: 10.1074/jbc.270.29.17072. [DOI] [PubMed] [Google Scholar]
- 2244.Kitagawa M, Wada C, Yoshioka S, Yura T. Expression of ClpB, an analog of the ATP-dependent protease regulatory subunit in Escherichia coli, is controlled by a heat shock ς factor (ς32) J Bacteriol. 1991;173:4247–4253. doi: 10.1128/jb.173.14.4247-4253.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2245.Kitagawa Y, Akaboshi E, Shinagawa H, Horii T, Ogawa H, Kato T. Structural analysis of the umu operon required for inducible mutagenesis in Escherichia coli. Proc Natl Acad Sci USA. 1985;82:4336–4340. doi: 10.1073/pnas.82.13.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2246.Kitakawa M, Dabbs E R, Isono K. Genes coding for ribosomal proteins S15, L21, and L27 map near argG in Escherichia coli. J Bacteriol. 1979;138:832–838. doi: 10.1128/jb.138.3.832-838.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2247.Kitakawa M, Isono K. An amber mutation in the gene rpsA for ribosomal protein S1 in Escherichia coli. Mol Gen Genet. 1982;185:445–447. doi: 10.1007/BF00334137. [DOI] [PubMed] [Google Scholar]
- 2248.Kitakawa M, Blumenthal L, Isono K. Isolation and characterization of specialized transducing λ phages carrying ribosomal protein genes of Escherichia coli. Mol Gen Genet. 1980;180:343–349. doi: 10.1007/BF00425846. [DOI] [PubMed] [Google Scholar]
- 2249.Klauck E, Bohringer J, Hengge-Aronis R. The LysR-like regulator LeuO in Escherichia coli is involved in the translational regulation of rpoS by affecting the expression of the small regulatory DsrA-RNA. Mol Microbiol. 1997;25:559–569. doi: 10.1046/j.1365-2958.1997.4911852.x. [DOI] [PubMed] [Google Scholar]
- 2250.Kleerebezem M, Tommassen J. Expression of the pspA gene stimulates efficient protein export in Escherichia coli. Mol Microbiol. 1993;7:947–956. doi: 10.1111/j.1365-2958.1993.tb01186.x. [DOI] [PubMed] [Google Scholar]
- 2251.Kleerebezem M, Heutink M, de Cock H, Tommassen J. The qmeA (ts) mutation of Escherichia coli is localized in the fabI gene, which encodes enoyl-ACP reductase. Res Microbiol. 1996;147:609–613. doi: 10.1016/0923-2508(96)84016-2. [DOI] [PubMed] [Google Scholar]
- 2252.Kleerebezem M, Heutink M, Tommassen J. Characterization of an Escherichia coli rotA mutant, affected in periplasmic peptidyl-prolyl cis/trans isomerase. Mol Microbiol. 1995;18:313–320. doi: 10.1111/j.1365-2958.1995.mmi_18020313.x. [DOI] [PubMed] [Google Scholar]
- 2253.Kleerebezem M, Crielaard W, Tommassen J. Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the protonmotive force under stress conditions. EMBO J. 1996;15:162–171. [PMC free article] [PubMed] [Google Scholar]
- 2254.Klein G, Henrich B, Plapp R. Cloning and expression of the pepD gene of Escherichia coli. J Gen Microbiol. 1986;132:2337–2343. doi: 10.1099/00221287-132-8-2337. [DOI] [PubMed] [Google Scholar]
- 2255.Klein J R, Henrich B, Plapp R. Molecular analysis and nucleotide sequence of the envCD operon of Escherichia coli. Mol Gen Genet. 1991;230:230–240. doi: 10.1007/BF00290673. [DOI] [PubMed] [Google Scholar]
- 2256.Klein J R, Plapp R. Locations of the envCD genes on the physical map of the Escherichia coli chromosome. J Bacteriol. 1992;174:3828–3829. doi: 10.1128/jb.174.11.3828-3829.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2257.Klein M, Sprenger G A, Freudl R. Cloning, nucleotide sequence, and functional expression of the Escherichia coli enolase (eno) gene in a temperature-sensitive eno mutant strain. DNA Sequence. 1996;6:351–355. doi: 10.3109/10425179609047574. [DOI] [PubMed] [Google Scholar]
- 2258.Klein W, Horlacher R, Boos W. Molecular analysis of treB encoding the Escherichia coli enzyme II specific for trehalose. J Bacteriol. 1995;177:4043–4052. doi: 10.1128/jb.177.14.4043-4052.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2259.Kleinsteuber S, Quinones A. Expression of the dnaB gene of Escherichia coli is inducible by replication-blocking DNA damage in a recA-independent manner. Mol Gen Genet. 1995;248:695–702. doi: 10.1007/BF02191709. [DOI] [PubMed] [Google Scholar]
- 2260.Klem T J, Davisson V J. Imidazole glycerol phosphate synthase: the glutamine amidotransferase in histidine biosynthesis. Biochemistry. 1993;32:5177–5186. doi: 10.1021/bi00070a029. [DOI] [PubMed] [Google Scholar]
- 2261.Klemm P. The fimA gene encoding the type 1 fimbrial subunit of Escherichia coli: nucleotide sequence and primary structure of the protein. Eur J Biochem. 1984;143:395–399. doi: 10.1111/j.1432-1033.1984.tb08386.x. [DOI] [PubMed] [Google Scholar]
- 2262.Klemm P. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 1986;5:1389–1393. doi: 10.1002/j.1460-2075.1986.tb04372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2263.Klemm P. FimC, a chaperone-like periplasmic protein of Escherichia coli. Res Microbiol. 1992;143:831–838. doi: 10.1016/0923-2508(92)90070-5. [DOI] [PubMed] [Google Scholar]
- 2264.Klemm P, Jorgensen B J, van Die I, de Ree H, Bergmans H. The fim genes responsible for synthesis of type 1 fimbriae in Escherichia coli, cloning and genetic organization. Mol Gen Genet. 1985;199:410–414. doi: 10.1007/BF00330751. [DOI] [PubMed] [Google Scholar]
- 2265.Klemm P, Christiansen G. Three fim genes required for the regulation of length and mediation of adhesion of Escherichia coli type 1 fimbriae. Mol Gen Genet. 1987;208:439–445. doi: 10.1007/BF00328136. [DOI] [PubMed] [Google Scholar]
- 2266.Klemm P, Tong S, Nielsen H, Conway T. The gntP gene of Escherichia coli involved in gluconate uptake. J Bacteriol. 1996;178:61–67. doi: 10.1128/jb.178.1.61-67.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2267.Klena J D, Schnaitman C A. Function of the rfb gene cluster and rfe gene in the synthesis of O antigen by Shigella dysenteriae 1. Mol Microbiol. 1993;9:393–402. doi: 10.1111/j.1365-2958.1993.tb01700.x. [DOI] [PubMed] [Google Scholar]
- 2268.Klena J D, Schnaitman C A. Genes for TDP-rhamnose synthesis affect the pattern of lipopolysaccharide heterogeneity in Escherichia coli K-12. J Bacteriol. 1994;176:4003–4010. doi: 10.1128/jb.176.13.4003-4010.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2269.Klena J D, Pradel E, Schnaitman C A. Comparison of lipopolysaccharide biosynthesis genes rfaK, rfaL, rfaY, and rfaZ of Escherichia coli K-12 and Salmonella typhimurium. J Bacteriol. 1992;174:4746–4752. doi: 10.1128/jb.174.14.4746-4752.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2270.Klena J D, Pradel E, Schnaitman C A. The rfaS gene, which is involved in production of a rough form of lipopolysaccharide core in Escherichia coli K-12, is not present in the rfa cluster of Salmonella typhimurium LT2. J Bacteriol. 1993;175:1524–1527. doi: 10.1128/jb.175.5.1524-1527.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2271.Klena J D, Ashford R S, Schnaitman C A. Role of Escherichia coli K-12 rfa genes and the rfp gene of Shigella dysenteriae 1 in generation of lipopolysaccharide core heterogeneity and attachment of O antigen. J Bacteriol. 1992;174:7297–7307. doi: 10.1128/jb.174.22.7297-7307.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2272.Kline B C, Kogoma T, Tam J E, Shields M S. Requirement of the Escherichia coli danA gene product for plasmid F maintenance. J Bacteriol. 1986;168:440–443. doi: 10.1128/jb.168.1.440-443.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2273.Kloser A W, Laird M W, Misra R. asmB, a suppressor locus for assembly-defective OmpF mutants of Escherichia coli, is allelic to envA (lpxC) J Bacteriol. 1996;178:5138–5143. doi: 10.1128/jb.178.17.5138-5143.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2274.Klumpp D J, Plank D W, Bowdin L J, Stueland C S, Chung T, LaPorte D C. Nucleotide sequence of aceK, the gene encoding isocitrate dehydrogenase kinase/phosphatase. J Bacteriol. 1988;170:2763–2769. doi: 10.1128/jb.170.6.2763-2769.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2275.Knott V, Rees D J G, Cheng Z, Brownlee G G. Randomly picked cosmid clones overlap the pyrB and oriC gap in the physical map of the E. coli chromosome. Nucleic Acids Res. 1988;16:2601–2612. doi: 10.1093/nar/16.6.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2276.Kobayashi H, Yamamoto M, Aono R. Appearance of a stress-response protein, phage-shock protein A, in Escherichia coli exposed to hydrophobic organic solvents. Microbiology. 1998;144:353–359. doi: 10.1099/00221287-144-2-353. [DOI] [PubMed] [Google Scholar]
- 2277.Kobayashi T, Kudo I, Homma H, Karasawa K, Inoue K, Ikeda H, Nojima S. Gene organization of pldA and pldB, the structural genes for detergent-resistant phospholipase A and lysophospholipase L2 of Escherichia coli. J Biochem (Tokyo) 1985;98:1007–1016. doi: 10.1093/oxfordjournals.jbchem.a135346. [DOI] [PubMed] [Google Scholar]
- 2278.Kobayashi T, Kudo I, Karasawa K, Mizushima H, Inoue K, Nojima S. Nucleotide sequence of the pldB gene and characteristics of deduced amino acid sequence of lysophospholipase L2 in Escherichia coli. J Biochem (Tokyo) 1985;98:1017–1025. doi: 10.1093/oxfordjournals.jbchem.a135347. [DOI] [PubMed] [Google Scholar]
- 2279.Kobayashi T, Hidaka M, Horiuchi T. Evidence of a ter specific binding protein essential for the termination reaction of DNA replication in Escherichia coli. EMBO J. 1989;8:2435–2441. doi: 10.1002/j.1460-2075.1989.tb08374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2280.Koch J P, Vandekerckhove J, Kahmann R. Escherichia coli host factor for site-specific DNA inversion: cloning and characterization of the fis gene. Proc Natl Acad Sci USA. 1988;85:4237–4241. doi: 10.1073/pnas.85.12.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2281.Koch W H, Ennis D G, Levine A S, Woodgate R. Escherichia coli umuDC mutants: DNA sequence alterations and UmuD cleavage. Mol Gen Genet. 1992;233:443–448. doi: 10.1007/BF00265442. [DOI] [PubMed] [Google Scholar]
- 2282.Kocharyan S M, Melkumyan M A. Genetic study of Escherichia coli K-12 mutants defective for structural and regulatory genes of second purine nucleoside phosphorylase. Sov Genet (Engl Transl Genetika) 1986;22:963–972. [PubMed] [Google Scholar]
- 2283.Kodaira M, Biswas S B, Kornberg A. The dnaX gene encodes the DNA polymerase III holoenzyme T subunit, precursor of the y subunit, the dnaZ gene product. Mol Gen Genet. 1983;192:80–86. doi: 10.1007/BF00327650. [DOI] [PubMed] [Google Scholar]
- 2284.Koffel-Schwartz N, Coin F, Veaute X, Fuchs R P. Cellular strategies for accommodating replication-hindering adducts in DNA: control by the SOS response in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:7805–7810. doi: 10.1073/pnas.93.15.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2285.Kofoid E C, Parkinson J S. Tandem translation starts in the cheA locus of Escherichia coli. J Bacteriol. 1991;173:2116–2119. doi: 10.1128/jb.173.6.2116-2119.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2286.Kogoma T. Escherichia coli RNA polymerase mutants that enhance or diminish the SOS response constitutively expressed in the absence of RNase H1 activity. J Bacteriol. 1994;176:1521–1523. doi: 10.1128/jb.176.5.1521-1523.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2287.Kogoma T, Hong X, Cadwell G W, Barnard K G, Asai T. Requirement of homologous recombination functions for viability of the Escherichia coli cell that lacks RNase H1 and exonuclease V activities. Biochimie. 1993;75:89–99. doi: 10.1016/0300-9084(93)90029-r. [DOI] [PubMed] [Google Scholar]
- 2288.Koh Y S, Choih J, Lee J H, Roe J H. Regulation of the ribA gene encoding GTP cyclohydrolase II by the soxRS locus in Escherichia coli. Mol Gen Genet. 1996;251:591–598. doi: 10.1007/BF02173649. [DOI] [PubMed] [Google Scholar]
- 2289.Koh Y-S, Roe J-H. Isolation of a novel paraquat-inducible (pqi) gene regulated by the soxRS locus in Escherichia coli. J Bacteriol. 1995;177:2673–2678. doi: 10.1128/jb.177.10.2673-2678.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2290.Koh Y S, Roe J H. Dual regulation of the paraquat-inducible gene pqi-5 by SoxS and RpoS in Escherichia coli. Mol Microbiol. 1996;22:53–61. doi: 10.1111/j.1365-2958.1996.tb02655.x. [DOI] [PubMed] [Google Scholar]
- 2291.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 2292.Kohno K, Wada M, Kano Y, Imamoto F. Promoters and autogenous control of the Escherichia coli hupA and hupB genes. J Mol Biol. 1990;213:27–36. doi: 10.1016/S0022-2836(05)80119-6. [DOI] [PubMed] [Google Scholar]
- 2293.Kole R, Baer M F, Stark B C, Altman S. E. coli RNAase P has a required RNA component in vivo. Cell. 1980;9:881–887. doi: 10.1016/0092-8674(80)90079-3. [DOI] [PubMed] [Google Scholar]
- 2294.Kolling R, Gielow A, Seufert W, Kucherer C, Messer W. AsnC, a multifunctional regulator of genes located around the replication origin of Escherichia coli, oriC. Mol Gen Genet. 1988;212:99–104. doi: 10.1007/BF00322450. [DOI] [PubMed] [Google Scholar]
- 2295.Kolling R, Lother H. AsnC: an autogenously regulated activator of asparagine synthetase A transcription in Escherichia coli. J Bacteriol. 1985;164:310–315. doi: 10.1128/jb.164.1.310-315.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2296.Kolmar H, Waller P R, Sauer R T. The DegP and DegQ periplasmic endoproteases of Escherichia coli: specificity for cleavage sites and substrate conformation. J Bacteriol. 1996;178:5925–5929. doi: 10.1128/jb.178.20.5925-5929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2297.Kolodner R D, Fishel R A, Howard M. Genetic recombination of bacterial plasmid DNA: effect of recF pathway mutations on plasmid recombination in Escherichia coli. J Bacteriol. 1985;163:1060–1066. doi: 10.1128/jb.163.3.1060-1066.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2298.Kolodner R D, Hall S D, Luisi-DeLuca C. Homologous pairing proteins encoded by the Escherichia coli recE and recT genes. Mol Microbiol. 1994;11:23–30. doi: 10.1111/j.1365-2958.1994.tb00286.x. [DOI] [PubMed] [Google Scholar]
- 2299.Kolodrubetz D, Schleif R. l-Arabinose transport systems in Escherichia coli K-12. J Bacteriol. 1981;148:472–479. doi: 10.1128/jb.148.2.472-479.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2300.Kolodrubetz D, Schleif R. Regulation of the l-arabinose transport operons in Escherichia coli. J Mol Biol. 1981;151:215–227. doi: 10.1016/0022-2836(81)90512-x. [DOI] [PubMed] [Google Scholar]
- 2301.Kolot M, Yagil E. Position and direction of strand exchange in bacteriophage HK022 integration. Mol Gen Genet. 1994;245:623–627. doi: 10.1007/BF00282225. [DOI] [PubMed] [Google Scholar]
- 2302.Komano T, Utsumi R, Kawamukai M. Functional analysis of the fic gene involved in regulation of cell division. Res Microbiol. 1991;142:269–277. doi: 10.1016/0923-2508(91)90040-h. [DOI] [PubMed] [Google Scholar]
- 2303.Komatsu Y, Tanaka K. A showdomycin-resistant mutant of Escherichia coli K-12 with altered nucleoside transport character. Biochim Biophys Acta. 1972;288:390–403. doi: 10.1016/0005-2736(72)90260-x. [DOI] [PubMed] [Google Scholar]
- 2304.Komeda Y. Fusions of flagellar operons to lactose genes on a Mu lac bacteriophage. J Bacteriol. 1982;150:16–26. doi: 10.1128/jb.150.1.16-26.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2305.Komeda Y. Transcriptional control of flagellar genes in Escherichia coli K-12. J Bacteriol. 1986;168:1315–1318. doi: 10.1128/jb.168.3.1315-1318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2306.Komeda Y, Kutsukake K, Iino T. Definition of additional flagellar genes in Escherichia coli K-12. Genetics. 1980;94:277–290. doi: 10.1093/genetics/94.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2307.Komeda Y, Silverman M, Simon M. Genetic analysis of Escherichia coli K-12 region I flagellar mutants. J Bacteriol. 1977;131:801–808. doi: 10.1128/jb.131.3.801-808.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2308.Komeda Y, Silverman M, Simon M. Identification of the structural gene for the hook subunit protein of Escherichia coli flagella. J Bacteriol. 1978;133:364–371. doi: 10.1128/jb.133.1.364-371.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2309.Komeda Y, Silverman M, Matsumura P, Simon M. Genes for the hook-basal body proteins of the flagellar apparatus in Escherichia coli. J Bacteriol. 1978;134:655–667. doi: 10.1128/jb.134.2.655-667.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2310.Komine Y, Inokuchi H. Physical map locations of the genes that encode small stable RNAs in Escherichia coli. J Bacteriol. 1991;173:5252. doi: 10.1128/jb.173.17.5252.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2311.Komine Y, Inokuchi H. Precise mapping of the rnpB gene encoding the RNA component of RNase P in Escherichia coli K-12. J Bacteriol. 1991;173:1813–1816. doi: 10.1128/jb.173.5.1813-1816.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2312.Komine Y, Kitabatake M, Inokuchi H. 10Sa RNA is associated with 70S ribosome particles in Escherichia coli. J Biochem (Tokyo) 1996;119:463–467. doi: 10.1093/oxfordjournals.jbchem.a021264. [DOI] [PubMed] [Google Scholar]
- 2313.Komine Y, Adachi T, Inokuchi K, Ozeki H. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J Mol Biol. 1990;212:579–598. doi: 10.1016/0022-2836(90)90224-A. [DOI] [PubMed] [Google Scholar]
- 2314.Konan K V, Yanofsky C. Regulation of the Escherichia coli tna operon: nascent leader peptide control at the tnaC stop codon. J Bacteriol. 1997;179:1774–1779. doi: 10.1128/jb.179.5.1774-1779.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2315.Kondo H, Shiratsuchi K, Yoshimoto T, Masuda T, Kitazono A, Tsuru D, Anai M, Sekiguchi M, Tanabe T. Acetyl-CoA carboxylase from Escherichia coli: gene organization and nucleotide sequence of the biotin carboxylase subunit. Proc Natl Acad Sci USA. 1991;88:9730–9733. doi: 10.1073/pnas.88.21.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2316.Kondo H, Nakabeppu Y, Kataoka H, Kuhara S, Kawabata S, Sekiguchi M. Structure and expression of the alkB gene of Escherichia coli related to the repair of alkylated DNA. J Biol Chem. 1986;261:15772–15777. [PubMed] [Google Scholar]
- 2317.Kondo K, Wakabayashi S, Yagi T, Kagamiyama H. The complete amino acid sequence of aspartate aminotransferase from Escherichia coli: sequence comparison with pig isoenzymes. Biochem Biophys Res Commun. 1984;122:62–67. doi: 10.1016/0006-291x(84)90439-x. [DOI] [PubMed] [Google Scholar]
- 2318.Koonin E V. Escherichia coli dinG gene encodes a putative DNA helicase related to a group of eukaryotic helicases including Rad3 protein. Nucleic Acids Res. 1993;21:1497. doi: 10.1093/nar/21.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2319.Koonin E V, Rudd K E. SpoU protein of Escherichia coli belongs to a new family of putative rRNA methylases. Nucleic Acids Res. 1993;21:5519. doi: 10.1093/nar/21.23.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2320.Koonin E V, Rudd K E. Two domains of superfamily I helicases may exist as separate proteins. Protein Sci. 1996;5:178–180. doi: 10.1002/pro.5560050124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2321.Koop A H, Hartley M, Bourgeois S. Analysis of the cya locus of Escherichia coli. Gene. 1984;28:133–146. doi: 10.1016/0378-1119(84)90250-6. [DOI] [PubMed] [Google Scholar]
- 2322.Korat B, Mottl H, Keck W. Penicillin-binding protein 4 of Escherichia coli: molecular cloning of the dacB gene, controlled overexpression, and alterations in murein composition. Mol Microbiol. 1991;5:675–684. doi: 10.1111/j.1365-2958.1991.tb00739.x. [DOI] [PubMed] [Google Scholar]
- 2323.Kornberg A. Inorganic polyphosphate: toward making a forgotten polymer unforgettable. J Bacteriol. 1995;177:491–496. doi: 10.1128/jb.177.3.491-496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2324.Kornberg H L, Elvin C M. Location and function of fruC, a gene involved in the regulation of fructose utilization by Escherichia coli. J Gen Microbiol. 1987;133:341–346. doi: 10.1099/00221287-133-2-341. [DOI] [PubMed] [Google Scholar]
- 2325.Kornberg H L, Smith J. Genetic control of glucose uptake by Escherichia coli. FEBS Lett. 1972;20:270–272. doi: 10.1016/0014-5793(72)80084-x. [DOI] [PubMed] [Google Scholar]
- 2326.Kornberg H L, Watts P D. Roles of crr-gene products in regulating carbohydrate uptake by Escherichia coli. FEBS Lett. 1978;89:329–332. doi: 10.1016/0014-5793(78)80248-8. [DOI] [PubMed] [Google Scholar]
- 2327.Kornitzer D, Teff D, Altuvia S, Oppenheim A B. Isolation, characterization, and sequence of an Escherichia coli heat shock gene, htpX. J Bacteriol. 1991;173:2944–2953. doi: 10.1128/jb.173.9.2944-2953.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2328.Kosiba B E, Schleif R. Arabinose-inducible promoter from Escherichia coli. Its cloning from chromosomal DNA, identification as the araFG promoter and sequence. J Mol Biol. 1982;156:53–66. doi: 10.1016/0022-2836(82)90458-2. [DOI] [PubMed] [Google Scholar]
- 2329.Kosic N, Sugai M, Fan C K, Wu H C. Processing of lipid-modified prolipoprotein requires energy and sec gene products in vivo. J Bacteriol. 1993;175:6113–6117. doi: 10.1128/jb.175.19.6113-6117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2330.Koster W, Braun V. Iron hydroxamate transport of Escherichia coli: nucleotide sequence of the fhuB gene and identification of the protein. Mol Gen Genet. 1986;204:435–442. doi: 10.1007/BF00331021. [DOI] [PubMed] [Google Scholar]
- 2331.Kotval J, Campbell A, Konopa G, Szybalski W. Leftward transcription in the Escherichia coli bio operon does not require products of the rightward transcript. Gene. 1982;17:219–222. doi: 10.1016/0378-1119(82)90075-0. [DOI] [PubMed] [Google Scholar]
- 2332.Koyama Y, Ohmori H. Nucleotide sequence of the Escherichia coli solA gene encoding a sarcosine oxidase-like protein and characterization of its product. Gene. 1996;181:179–183. doi: 10.1016/s0378-1119(96)00500-8. [DOI] [PubMed] [Google Scholar]
- 2333.Kozliak E I, Guilloton M B, Gerami-Nejad M, Fuchs J A, Anderson P M. Expression of proteins encoded by the Escherichia coli cyn operon: carbon dioxide-enhanced degradation of carbonic anhydrase. J Bacteriol. 1994;176:5711–5717. doi: 10.1128/jb.176.18.5711-5717.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2334.Kraal B, Zeef L A, Mesters J R, Boon K, Vorstenbosch E L, Bosch L, Anborgh P H, Parmeggiani A, Hilgenfeld R. Antibiotic resistance mechanisms of mutant EF-Tu species in Escherichia coli. Biochem Cell Biol. 1995;73:1167–1177. doi: 10.1139/o95-126. [DOI] [PubMed] [Google Scholar]
- 2335.Kraft R, Leinwand A. Sequence of the complete P protein gene and part of the M protein gene from the histidine transport operon of Escherichia coli compared to that of Salmonella typhimurium. Nucleic Acids Res. 1987;15:8568. doi: 10.1093/nar/15.20.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2336.Kraus J, Soll D, Low K B. Glutamyl-gamma-methyl ester acts as a methionine analogue in Escherichia coli: analogue resistant mutants map at the metJ and metK loci. Genet Res. 1979;33:49–55. doi: 10.1017/s0016672300018152. [DOI] [PubMed] [Google Scholar]
- 2337.Kren B, Parsell D, Fuchs J A. Isolation and characterization of an Escherichia coli K-12 mutant deficient in glutaredoxin. J Bacteriol. 1988;170:308–315. doi: 10.1128/jb.170.1.308-315.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2338.Kren B, Fuchs J A. Characterization of the ftsB gene as an allele of the nrdB gene in Escherichia coli. J Bacteriol. 1987;169:14–18. doi: 10.1128/jb.169.1.14-18.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2339.Kreuzer K N, Cozzarelli N R. Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase subunit A: effects on deoxyribonucleic acid replication, transcription, and bacteriophage growth. J Bacteriol. 1979;140:424–435. doi: 10.1128/jb.140.2.424-435.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2340.Kricker M, Hall B G. Directed evolution of cellobiose utilization in Escherichia coli K12. Mol Biol Evol. 1984;1:171–182. doi: 10.1093/oxfordjournals.molbev.a040310. [DOI] [PubMed] [Google Scholar]
- 2341.Kricker M, Hall B G. Biochemical genetics of the cryptic gene system for cellobiose utilization in Escherichia coli K12. Genetics. 1987;115:419–429. doi: 10.1093/genetics/115.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2342.Krikos A, Mutoh N, Boyd A, Simon M I. Sensory transducers of E. coli are composed of discrete structural and functional domains. Cell. 1983;33:615–622. doi: 10.1016/0092-8674(83)90442-7. [DOI] [PubMed] [Google Scholar]
- 2343.Krogfelt K A, Bermans H, Klemm P. Direct evidence that the FimH protein is the mannose-specific adhesin of Escherichia coli type 1 fimbriae. Infect Immun. 1990;58:1995–1998. doi: 10.1128/iai.58.6.1995-1998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2344.Kroh H E, Simon L D. The ClpP component of Clp protease is the ς32-dependent heat shock protein F21.5. J Bacteriol. 1990;172:6026–6034. doi: 10.1128/jb.172.10.6026-6034.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2345.Krone F A, Westphal G, Meyer H E, Schwenn J D. PAPS-reductase of Escherichia coli. Correlating the N-terminal amino acid sequence with the DNA of gene cysH. FEBS Lett. 1990;260:6–9. doi: 10.1016/0014-5793(90)80052-k. [DOI] [PubMed] [Google Scholar]
- 2346.Krone F A, Westphal G, Schwenn J D. Characterisation of the gene cysH and of its product phospho-adenylylsulphate reductase from Escherichia coli. Mol Gen Genet. 1991;225:314–319. doi: 10.1007/BF00269864. [DOI] [PubMed] [Google Scholar]
- 2347.Krueger J H, Johnson J R, Greene R C, Dresser M. Structural studies of lambda transducing bacteriophage carrying bacterial deoxyribonucleic acid from the metBJLF region of the Escherichia coli chromosome. J Bacteriol. 1981;147:612–621. doi: 10.1128/jb.147.2.612-621.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2348.Krueger J H, Elledge S J, Walker G C. Isolation and characterization of Tn5 insertion mutations in the lexA gene of Escherichia coli. J Bacteriol. 1983;153:1368–1378. doi: 10.1128/jb.153.3.1368-1378.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2349.Krueger J K, Stock J, Schutt C E. Evidence that the methylesterase of bacterial chemotaxis may be a serine hydrolase. Biochim Biophys Acta. 1992;1119:322–326. doi: 10.1016/0167-4838(92)90220-8. [DOI] [PubMed] [Google Scholar]
- 2350.Kruger T, Grund C, Wild C, Noyer-Weidner M. Characterization of the mcrBC region of Escherichia coli K-12 wild-type and mutant strains. Gene. 1992;114:1–12. doi: 10.1016/0378-1119(92)90700-y. [DOI] [PubMed] [Google Scholar]
- 2351.Kuchino Y, Mori F, Nishimura S. Structure and transcription of the tRNA1Pro gene from Escherichia coli. Nucleic Acids Res. 1985;13:3213–3220. doi: 10.1093/nar/13.9.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2352.Kudo T, Nagai K, Tamura G. Characteristics of a cold-sensitive chromosome segregation mutant of Escherichia coli. Agric Biol Chem. 1977;41:89–95. [Google Scholar]
- 2353.Kudo T, Nagai K, Tamura G. Genetic analysis of a cold-sensitive chromosome segregation mutant of Escherichia coli. Agric Biol Chem. 1977;41:607–608. [Google Scholar]
- 2354.Kuempel P, Hogaard A, Nielsen M, Nagappan O, Tecklenburg M. Use of a transposon (Tndif) to obtain suppressing and nonsuppressing insertions of the dif resolvase site of Escherichia coli. Genes Dev. 1996;10:1162–1171. doi: 10.1101/gad.10.9.1162. . (Comment.) [DOI] [PubMed] [Google Scholar]
- 2355.Kuempel P, Pelletier A J, Hill T M. tus and the terminators: the arrest of replication in prokaryotes. Cell. 1989;59:581–583. doi: 10.1016/0092-8674(89)90001-9. [DOI] [PubMed] [Google Scholar]
- 2356.Kuempel P, Henson J M, Dircks L, Tecklenburg M L, Lim D F. dif, a recA-independent recombination site in the terminus region of the chromosome of Escherichia coli. New Biol. 1991;3:799–811. [PubMed] [Google Scholar]
- 2357.Kuhn J, Somerville R L. Mutant strains of Escherichia coli K12 that can use d-amino acids. Proc Natl Acad Sci USA. 1971;68:2484–2487. doi: 10.1073/pnas.68.10.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2358.Kullik I, Stevens J, Toledano M B, Storz G. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for DNA binding and multimerization. J Bacteriol. 1995;177:1285–1291. doi: 10.1128/jb.177.5.1285-1291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2359.Kullik I, Toledano M B, Tartaglia L A, Storz G. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J Bacteriol. 1995;177:1275–1284. doi: 10.1128/jb.177.5.1275-1284.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2360.Kumagai Y, Kato J I, Hoshino K, Akasaka T, Sato K, Ikeda H. Quinolone-resistant mutants of Escherichia coli DNA topoisomerase IV parC gene. Antimicrob Agents Chemother. 1996;40:710–714. doi: 10.1128/aac.40.3.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2361.Kumamoto C A. Molecular chaperone and protein translocation across the Escherichia coli inner membrane. Mol Microbiol. 1991;5:19–22. doi: 10.1111/j.1365-2958.1991.tb01821.x. [DOI] [PubMed] [Google Scholar]
- 2362.Kumamoto C A, Beckwith J R. Mutations in a new gene, secB, cause defective porin localization in Escherichia coli. J Bacteriol. 1983;154:253–260. doi: 10.1128/jb.154.1.253-260.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2363.Kumamoto C A, Beckwith J R. Evidence for specificity at an early step in protein export in Escherichia coli. J Bacteriol. 1985;163:267–274. doi: 10.1128/jb.163.1.267-274.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2364.Kumar A, Larsen C E, Preiss J. Biosynthesis of bacterial glycogen. Primary structure of Escherichia coli ADP-glucose:α-1,4-glucan, 4-glucosyltransferase as deduced from the nucleotide sequence of the glgA gene. J Biol Chem. 1986;261:16256–16259. [PubMed] [Google Scholar]
- 2365.Kumar A, Maples V F, Champney W S. Properties of adenyl cyclase and cyclic adenosine 3′,5′-monophosphate receptor protein-deficient mutants of Escherichia coli. J Bacteriol. 1976;125:545–555. doi: 10.1128/jb.125.2.545-555.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2366.Kumaresan K R, Jayaraman R. The sir locus of Escherichia coli: a gene involved in SOS-independent repair of mitomycin C-induced DNA damage. Mutat Res. 1990;235:85–92. doi: 10.1016/0921-8777(90)90061-9. [DOI] [PubMed] [Google Scholar]
- 2367.Kumari S, Tishel R, Eisenbach M, Wolfe A J. Cloning, characterization, and functional expression of acs, the gene which encodes acetyl coenzyme A synthetase in Escherichia coli. J Bacteriol. 1995;177:2878–2886. doi: 10.1128/jb.177.10.2878-2886.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2368.Kumura K, Sekiguchi M. Identification of the uvrD gene product of Escherichia coli as DNA helicase II and its induction by DNA-damaging agents. J Biol Chem. 1984;259:1560–1565. [PubMed] [Google Scholar]
- 2369.Kundu T K, Kusano S, Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase ςF holoenzyme involved in transcription of flagellar and chemotaxis genes. J Bacteriol. 1997;179:4264–4269. doi: 10.1128/jb.179.13.4264-4269.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2370.Kuo S C, Koshland D E. Sequence of the flaA (cheC) locus of Escherichia coli and discovery of a new gene. J Bacteriol. 1986;166:1007–1012. doi: 10.1128/jb.166.3.1007-1012.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2371.Kurahashi K. Enzyme formation in galactose-negative mutants of Escherichia coli. Science. 1957;125:114–116. doi: 10.1126/science.125.3238.114. [DOI] [PubMed] [Google Scholar]
- 2372.Kuramitsu S, Inoue K, Ogawa T, Ogawa H, Kagamiyama H. Aromatic amino acid aminotransferase of Escherichia coli: nucleotide sequence of the tyrB gene. Biochem Biophys Res Commun. 1985;133:134–139. doi: 10.1016/0006-291x(85)91851-0. [DOI] [PubMed] [Google Scholar]
- 2373.Kuramitsu S, Okuno S, Ogawa T, Ogawa H, Kagamiyama H. Aspartate aminotransferase of Escherichia coli: nucleotide sequence of the aspC gene. J Biochem (Tokyo) 1985;97:1259–1262. doi: 10.1093/oxfordjournals.jbchem.a135173. [DOI] [PubMed] [Google Scholar]
- 2374.Kuramitsu S, Ogawa T, Ogawa H, Kagamiyama H. Branched chain amino acid aminotransferase of Escherichia coli: nucleotide sequence of the ilvE gene and the deduced amino acid sequence. J Biochem (Tokyo) 1985;97:993–999. doi: 10.1093/oxfordjournals.jbchem.a135176. [DOI] [PubMed] [Google Scholar]
- 2375.Kurihara T, Nakamura H. Cloning of the nusA gene of Escherichia coli. Mol Gen Genet. 1983;190:189–195. doi: 10.1007/BF00330639. [DOI] [PubMed] [Google Scholar]
- 2376.Kurose N, Murata K, Kimura A. An Escherichia coli mutant having altered d-xylose uptake activity and cloning of a gene for d-xylose uptake. Agric Biol Chem. 1985;49:2597–2603. [Google Scholar]
- 2377.Kurose N, Murata K, Kimura A. Cloning of the d-xylose uptake gene linked to the xylA gene in Escherichia coli. Agric Biol Chem. 1987;51:2575–2578. [Google Scholar]
- 2378.Kusano K, Takahashi N, Yoshikura H, Kobayashi I. Involvement of RecE exonuclease and RecT annealing protein in DNA double-strand break repair by homologous recombination. Gene. 1994;138:17–25. doi: 10.1016/0378-1119(94)90778-1. [DOI] [PubMed] [Google Scholar]
- 2379.Kushiro M, Shimizu M, Tomita K. Molecular cloning and sequence determination of the tuf gene coding for the elongation factor Tu of Thermus thermophilus HB8. Eur J Biochem. 1987;170:93–98. doi: 10.1111/j.1432-1033.1987.tb13671.x. [DOI] [PubMed] [Google Scholar]
- 2380.Kusser W, Ishiguro E E. Suppression of mutations conferring penicillin tolerance by interference with the stringent control mechanism of Escherichia coli. J Bacteriol. 1987;169:4396–4398. doi: 10.1128/jb.169.9.4396-4398.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2381.Kusters R, Lentzen G, Eppens E, van Geel A, van der Weijden C C, Wintermeyer W, Luirink J. The functioning of the SRP receptor FtsY in protein-targeting in E. coli is correlated with its ability to bind and hydrolyse GTP. FEBS Lett. 1995;372:253–258. doi: 10.1016/0014-5793(95)00997-n. [DOI] [PubMed] [Google Scholar]
- 2382.Kuwajima G, Asaka J-I, Fujiwara T, Node K, Kondo E. Nucleotide sequence of the hag gene encoding flagellin of Escherichia coli. J Bacteriol. 1986;168:1479–1483. doi: 10.1128/jb.168.3.1479-1483.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2383.Kuwano M, Schlessinger D, Rinaldi G, Felicetti L, Tocchini-Valenti G P. G factor mutants of Escherichia coli: map location and properties. Biochem Biophys Res Commun. 1971;42:441–444. doi: 10.1016/0006-291x(71)90390-1. [DOI] [PubMed] [Google Scholar]
- 2384.Kwon O, Bhattacharyya D K, Meganathan R. Menaquinone (vitamin K2) biosynthesis: overexpression, purification, and properties of o-succinylbenzoyl-coenzyme A synthetase from Escherichia coli. J Bacteriol. 1996;178:6778–6781. doi: 10.1128/jb.178.23.6778-6781.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2385.La Teana A, Brandi A, Falconi M, Spurio R, Pon C L, Gualerzi C O. Identification of a cold shock transcriptional enhancer of the Escherichia coli gene encoding nucleoid protein H-NS. Proc Natl Acad Sci USA. 1991;88:10907–10911. doi: 10.1073/pnas.88.23.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2386.Labie C, Bouche F, Bouché J P. Minicell-forming mutants of Escherichia coli: suppression of both DicB- and MinD-dependent division inhibition by inactivation of the minC gene product. J Bacteriol. 1990;172:5852–5855. doi: 10.1128/jb.172.10.5852-5855.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2387.Lacroix J M, Loubens I, Tempete M, Menichi B, Bohin J P. The mdoA locus of Escherichia coli consists of an operon under osmotic control. Mol Microbiol. 1991;5:1745–1753. doi: 10.1111/j.1365-2958.1991.tb01924.x. [DOI] [PubMed] [Google Scholar]
- 2388.Lacroix J M, Tempete M, Menichi B, Bohin J P. Molecular cloning and expression of a locus (mdoA) implicated in the biosynthesis of membrane-derived oligosaccharides in Escherichia coli. Mol Microbiol. 1989;3:1173–1182. doi: 10.1111/j.1365-2958.1989.tb00267.x. [DOI] [PubMed] [Google Scholar]
- 2389.Lahti R, Perala M, Heikinheimo P, Pitkaranta T, Kukko-Kalske E, Heinonen J. Characterization of the 5′ flanking region of the Escherichia coli ppa gene encoding inorganic pyrophosphatase: mutations in the ribosome-binding site decrease the level of ppa mRNA. J Gen Microbiol. 1991;137:2517–2523. doi: 10.1099/00221287-137-11-2517. [DOI] [PubMed] [Google Scholar]
- 2390.Lahti R, Pitkaranta T, Valve E, Ilta I, Kukko-Kalske E, Heinonen J. Cloning and characterization of the gene encoding inorganic pyrophosphatase of Escherichia coli K-12. J Bacteriol. 1988;170:5901–5907. doi: 10.1128/jb.170.12.5901-5907.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2391.Laine B, Kmiecik D, Sautiere P, Biserte G, Cohen-Solal M. Complete amino-acid sequences of DNA-binding proteins HU-1 and HU-2 from Escherichia coli. Eur J Biochem. 1980;103:447–461. doi: 10.1111/j.1432-1033.1980.tb05968.x. [DOI] [PubMed] [Google Scholar]
- 2392.Laine B, Sautiere P, Spassky A, Rimsky S. A DNA-binding protein from E. coli: isolation, characterization and its relationship with proteins H1 and B1. Biochem Biophys Res Commun. 1984;119:1147–1153. doi: 10.1016/0006-291x(84)90895-7. [DOI] [PubMed] [Google Scholar]
- 2393.Laine P S, Meyer R R. Interaction of the heat shock protein GroEL of Escherichia coli with single-stranded DNA-binding protein: suppression of ssb-113 by groEL46. J Bacteriol. 1992;174:3204–3211. doi: 10.1128/jb.174.10.3204-3211.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2394.Laird A J, Ribbons D W, Woodrow G C, Young I G. Bacteriophage Mu-mediated gene transposition and in vitro cloning of the enterochelin gene cluster of Escherichia coli. Gene. 1980;11:347–357. doi: 10.1016/0378-1119(80)90074-8. [DOI] [PubMed] [Google Scholar]
- 2395.Laird A J, Young I G. Tn5 mutagenesis of the enterochelin gene cluster of Escherichia coli. Gene. 1980;11:359–366. doi: 10.1016/0378-1119(80)90075-x. [DOI] [PubMed] [Google Scholar]
- 2396.Lakshmi T M, Helling R B. Selection for citrate synthease deficiency in icd mutants of Escherichia coli. J Bacteriol. 1976;127:76–83. doi: 10.1128/jb.127.1.76-83.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2397.Lam H L, Tancula E, Dempsey W B, Winkler M E. Suppression of insertions in the complex pdxJ operon of Escherichia coli K-12 by lon and other mutations. J Bacteriol. 1992;174:1554–1567. doi: 10.1128/jb.174.5.1554-1567.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2398.Lam H M, Winkler M E. Characterization of the complex pdxH-tyrS operon of Escherichia coli K-12 and pleiotropic phenotypes caused by pdxH insertion mutations. J Bacteriol. 1992;174:6033–6045. doi: 10.1128/jb.174.19.6033-6045.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2399.Lamark T, Kaasen I, Eshoo M W, Falkenberg P, McDougall J, Strom A R. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol Microbiol. 1991;5:1049–1064. doi: 10.1111/j.1365-2958.1991.tb01877.x. [DOI] [PubMed] [Google Scholar]
- 2400.Lambalot R H, Gehring A M, Flugel R S, Zuber P, LaCelle M, Marahiel M A, Reid R, Khosla C, Walsh C T. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 2401.Lambalot R H, Walsh C T. Cloning, overproduction, and characterization of the Escherichia coli holo-acyl carrier protein synthase. J Biol Chem. 1995;270:24658–24661. doi: 10.1074/jbc.270.42.24658. [DOI] [PubMed] [Google Scholar]
- 2402.Lambden P R, Guest J R. A novel method for isolating chlorate-resistant mutants of Escherichia coli K12 by anaerobic selection on a lactate plus fumarate medium. J Gen Microbiol. 1976;93:173–176. doi: 10.1099/00221287-93-1-173. [DOI] [PubMed] [Google Scholar]
- 2403.Lamblin A F, Fuchs J A. Expression and purification of the cynR regulatory gene product: CynR is a DNA-binding protein. J Bacteriol. 1993;175:7990–7999. doi: 10.1128/jb.175.24.7990-7999.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2404.Lamblin A F, Fuchs J A. Functional analysis of the Escherichia coli K-12 cyn operon transcriptional regulation. J Bacteriol. 1994;176:6613–6622. doi: 10.1128/jb.176.21.6613-6622.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2405.Lander M, Pitt A R, Alefounder P R, Abell C, Battersby A R. Studies on the mechanism of hydroxymethylbilane synthase concerning the role of arginine residues in substrate binding. Biochem J. 1991;275:447–452. doi: 10.1042/bj2750447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2406.Landick R, Oxender D L. The complete nucleotide sequences of the Escherichia coli LIV-BP and LS-BP genes. J Biol Chem. 1985;260:8257–8261. [PubMed] [Google Scholar]
- 2407.Landick R, Vaughn V, Lau E T, VanBogelen R A, Erickson J, Neidhardt F C. Nucleotide sequence of the heat shock regulatory gene of E. coli suggests its protein products may be a transcription factor. Cell. 1984;38:175–182. doi: 10.1016/0092-8674(84)90538-5. [DOI] [PubMed] [Google Scholar]
- 2408.Landini P, Hajec L I, Nguyen L H, Burgess R R, Volkert M R. The leucine-responsive regulatory protein (Lrp) acts as a specific repressor for sigma s-dependent transcription of the Escherichia coli aidB gene. Mol Microbiol. 1996;20:947–955. doi: 10.1111/j.1365-2958.1996.tb02536.x. [DOI] [PubMed] [Google Scholar]
- 2409.Landini P, Hajec L I, Volkert M R. Structure and transcriptional regulation of the Escherichia coli adaptive response gene aidB. J Bacteriol. 1994;176:6583–6589. doi: 10.1128/jb.176.21.6583-6589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2410.Landini P, Volkert M R. RNA polymerase alpha subunit binding site in positively controlled promoters: a new model for RNA polymerase-promoter interaction and transcriptional activation in the Escherichia coli ada and aidB genes. EMBO J. 1995;14:4329–4335. doi: 10.1002/j.1460-2075.1995.tb00107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2411.Lange R, Fischer D, Hengge-Aronis R. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the ςS subunit of RNA polymerase in Escherichia coli. J Bacteriol. 1995;177:4676–4680. doi: 10.1128/jb.177.16.4676-4680.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2412.Lange R, Barth M, Hengge-Aronis R. Complex transcriptional control of the ςS-dependent stationary-phase-induced and osmotically regulated osmY (csi-5) gene suggests novel roles for Lrp, cyclic AMP (cAMP) receptor protein-cAMP complex, and integration host factor. J Bacteriol. 1993;175:7910–7917. doi: 10.1128/jb.175.24.7910-7917.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2413.Lange R, Hengge-Aronis R. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel ς factor ςS. J Bacteriol. 1991;173:4474–4481. doi: 10.1128/jb.173.14.4474-4481.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2413a.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 2414.Lange R, Hengge-Aronis R. The nlpD gene is located in an operon with rpoS on the Escherichia coli chromosome and encodes a novel lipoprotein with a potential function in cell wall formation. Mol Microbiol. 1994;13:733–743. doi: 10.1111/j.1365-2958.1994.tb00466.x. [DOI] [PubMed] [Google Scholar]
- 2415.Langley D, Guest J R. Biochemical studies of the alpha-keto acid dehydrogenase complexes of Escherichia coli K12: isolation and biochemical properties of deletion mutants. J Gen Microbiol. 1977;99:263–276. doi: 10.1099/00221287-99-2-263. [DOI] [PubMed] [Google Scholar]
- 2416.Langley D, Guest J R. Biochemical genetics of the alpha-keto acid dehydrogenase complexes of Escherichia coli K12: genetic characterization and regulatory properties of deletion mutants. J Gen Microbiol. 1978;106:103–117. doi: 10.1099/00221287-106-1-103. [DOI] [PubMed] [Google Scholar]
- 2417.Langridge J. Mutations conferring quantitative and qualitative increases in β-galactosidase activity in Escherichia coli. Mol Gen Genet. 1969;105:74–83. doi: 10.1007/BF00750315. [DOI] [PubMed] [Google Scholar]
- 2418.LaPorte D C, Thorsness P E, Koshland D E. Compensatory phosphorylation of isocitrate dehydrogenase. A mechanism for adaption to the intracellular environment. J Biol Chem. 1985;260:10563–10568. [PubMed] [Google Scholar]
- 2419.LaPorte D C, Chung T. A single gene codes for the kinase and phosphatase which regulate isocitrate dehydrogenase. J Biol Chem. 1985;260:15291–15297. [PubMed] [Google Scholar]
- 2420.LaRossa R, Vogeli G, Low K B, Soll D. Regulation of biosynthesis of aminoacyl-tRNA synthetases and of tRNA in Escherichia coli. II. Isolation of regulatory mutants affecting leucyl-tRNA synthetase levels. J Mol Biol. 1977;117:1033–1048. doi: 10.1016/s0022-2836(77)80011-9. [DOI] [PubMed] [Google Scholar]
- 2421.Larsen B, Wills N M, Gesteland R F, Atkins J F. rRNA-mRNA base pairing stimulates a programmed −1 ribosomal frameshift. J Bacteriol. 1994;176:6842–6851. doi: 10.1128/jb.176.22.6842-6851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2422.Larsen J E, Albrechtsen B, Valentin-Hansen P. Analysis of the terminator region after the deoCABD operon of Escherichia coli K-12 using a new class of single copy number operon-fusion vectors. Nucleic Acids Res. 1987;15:5125–5140. doi: 10.1093/nar/15.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2423.Larsen J N, Jensen K F. Nucleotide sequence of the pyrD gene of Escherichia coli and characterization of the flavoprotein dihydroorotate dehydrogenase. Eur J Biochem. 1985;151:59–65. doi: 10.1111/j.1432-1033.1985.tb09068.x. [DOI] [PubMed] [Google Scholar]
- 2424.Larson T J, Schumacher G, Boos W. Identification of the glpT-encoded sn-glycerol-3-phosphate permease of Escherichia coli, an oligomeric integral membrane protein. J Bacteriol. 1982;152:1008–1021. doi: 10.1128/jb.152.3.1008-1021.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2425.Larson T J, Ludtke D N, Bell R M. sn-Glycerol-3-phosphate auxotrophy of plsB strains of Escherichia coli: evidence that a second mutation, plsX, is required. J Bacteriol. 1984;160:711–717. doi: 10.1128/jb.160.2.711-717.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2426.Larson T J, Ehrmann M, Boos W. Periplasmic glycerophosphodiesterase of Escherichia coli, a new enzyme of the glp regulon. J Biol Chem. 1983;258:5424–5432. [PubMed] [Google Scholar]
- 2427.Laskowska E, Wawrzynow A, Taylor A. IbpA and IbpB, the new heat-shock proteins, bind to endogenous Escherichia coli proteins aggregated intracellularly by heat shock. Biochimie. 1996;78:117–122. doi: 10.1016/0300-9084(96)82643-5. [DOI] [PubMed] [Google Scholar]
- 2428.Lathe R, Bollen A, Herzog A. Revised location of the Escherichia coli gene coding for ribosomal protein S2. Mol Gen Genet. 1981;182:178–179. doi: 10.1007/BF00422787. [DOI] [PubMed] [Google Scholar]
- 2429.Lathe R, Buc H, Lecocq J-P, Bautz E K F. Prokaryotic histone-like protein interacting with RNA polymerase. Proc Natl Acad Sci USA. 1980;77:3548–3552. doi: 10.1073/pnas.77.6.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2430.Lathrop J T, Wei B Y, Touchie G A, Kadner R J. Sequences of the Escherichia coli BtuB protein essential for its insertion and function in the outer membrane. J Bacteriol. 1995;177:6810–6819. doi: 10.1128/jb.177.23.6810-6819.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2431.Latil M, Murgier M, Lazdunski A, Lazdunski C. Isolating and genetic mapping of Escherichia coli aminopeptidase mutants. Mol Gen Genet. 1976;148:43–47. doi: 10.1007/BF00268544. [DOI] [PubMed] [Google Scholar]
- 2432.Latil-Damotte M, Lares C. Relative order of glg mutations affecting glycogen biosynthesis in Escherichia coli K12. Mol Gen Genet. 1977;150:325–329. doi: 10.1007/BF00268132. [DOI] [PubMed] [Google Scholar]
- 2433.Laursen R A, L’Italien J J, Nagarkatti S, Miller D L. The amino acid sequence of elongation factor Tu of Escherichia coli. The complete sequence. J Biol Chem. 1981;256:8102–8109. [PubMed] [Google Scholar]
- 2434.Laval J. Role of DNA repair enzymes in the cellular resistance to oxidative stress. Pathol Biol (Paris) 1996;44:14–24. [PubMed] [Google Scholar]
- 2435.Lavina M, Pugsley A P, Moreno F. Identification, mapping, cloning, and characterization of a gene (sbmA) required for microcin B17 action on Escherichia coli K12. J Gen Microbiol. 1986;132:1685–1693. doi: 10.1099/00221287-132-6-1685. [DOI] [PubMed] [Google Scholar]
- 2436.Lawley B, Pittard J. Regulation of aroL expression by TyrR protein and Trp repressor in Escherichia coli K-12. J Bacteriol. 1994;176:6921–6930. doi: 10.1128/jb.176.22.6921-6930.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2437.Lawlis V B, Dennis M S, Chen E Y, Smith D H, Henner D J. Cloning and sequencing of the xylose isomerase and xylulose kinase genes of Escherichia coli. Appl Environ Microbiol. 1984;47:15–21. doi: 10.1128/aem.47.1.15-21.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2438.Lawrence J G, Ochman H, Hartl D L. Molecular and evolutionary relationships among enteric bacteria. J Gen Microbiol. 1991;137:1911–1921. doi: 10.1099/00221287-137-8-1911. [DOI] [PubMed] [Google Scholar]
- 2439.Lawrence J G, Roth J R. The cobalamin (coenzyme B12) biosynthetic genes of Escherichia coli. J Bacteriol. 1995;177:6371–6380. doi: 10.1128/jb.177.22.6371-6380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2440.Lawrence J G, Roth J R. Evolution of coenzyme B12 synthesis among enteric bacteria: evidence for loss and acquisition of a multigene complex. Genetics. 1996;142:11–24. doi: 10.1093/genetics/142.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2441.Lawther R P, Nichols B, Zurawski G, Hatfield G W. The nucleotide sequence preceding and including the beginning of the ilvE gene of the ilvGEDA operon of Escherichia coli. Nucleic Acids Res. 1979;7:2289–2301. doi: 10.1093/nar/7.8.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2442.Lawther R P, Calhoun D H, Adams C W, Hauser C A, Gray J E, Hatfield G W. Molecular basis of valine resistance in Escherichia coli K-12. Proc Natl Acad Sci USA. 1981;78:922–925. doi: 10.1073/pnas.78.2.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2443.Lawther R P, Calhoun D H, Gray J E, Adams C W, Hauser C A, Hatfield G W. DNA sequence fine-structure analysis of ilvG (IlvG+) mutations of Escherichia coli K-12. J Bacteriol. 1982;149:294–298. doi: 10.1128/jb.149.1.294-298.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2444.Lawther R P, Hatfield G W. Multivalent translational control of transcription termination at attenuator of ilvGEDA operon of Escherichia coli K-12. Proc Natl Acad Sci USA. 1980;77:1862–1866. doi: 10.1073/pnas.77.4.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2445.Lawther R P, Wek R C, Lopes J M, Pereira R, Taillon B E, Hatfield G W. The complete nucleotide sequence of the ilvGMEDA operon of Escherichia coli K-12. Nucleic Acids Res. 1987;15:2137–2155. doi: 10.1093/nar/15.5.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2446.Lazar S W, Kolter R. SurA assists the folding of Escherichia coli outer membrane proteins. J Bacteriol. 1996;178:1770–1773. doi: 10.1128/jb.178.6.1770-1773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2447.Lazzaroni J C, Fognini-Lefebvre N, Portalier R. Cloning of the lkyB (tolB) gene of Escherichia coli K12 and characterization of its product. Mol Gen Genet. 1986;204:285–288. doi: 10.1007/BF00425511. [DOI] [PubMed] [Google Scholar]
- 2448.Lazzaroni J C, Fognini-Lefebvre N, Portalier R. Cloning of the excC and excD genes involved in the release of periplasmic proteins by Escherichia coli K12. Mol Gen Genet. 1989;218:460–464. doi: 10.1007/BF00332410. [DOI] [PubMed] [Google Scholar]
- 2449.Lazzaroni J C, Portalier R. Genetic and biochemical characterization of periplasmic-leaky mutants of Escherichia coli K-12. J Bacteriol. 1981;145:1351–1358. doi: 10.1128/jb.145.3.1351-1358.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2450.Lazzaroni J C, Portalier R. The excC gene of Escherichia coli K-12 required for cell envelope integrity encodes the peptidoglycan-associated lipoprotein PAL. Mol Microbiol. 1992;6:735–742. doi: 10.1111/j.1365-2958.1992.tb01523.x. [DOI] [PubMed] [Google Scholar]
- 2451.Leach D R F, Lloyd R G, Coulson A F. The SbcCD protein of Escherichia coli is related to two putative nucleases in the UvrA superfamily of nucleotide-binding proteins. Genetica. 1992;87:95–100. doi: 10.1007/BF00120998. [DOI] [PubMed] [Google Scholar]
- 2452.Lebendiker M, Schuldiner S. Identification of residues in the translocation pathway of EmrE, a multidrug antiporter from Escherichia coli. J Biol Chem. 1996;271:21193–21199. doi: 10.1074/jbc.271.35.21193. [DOI] [PubMed] [Google Scholar]
- 2453.Lech K F, Lee C H, Isberg R R, Syvanen M. New gene in Escherichia coli K-12 (drpA): does its product play a role in RNA synthesis? J Bacteriol. 1985;162:117–123. doi: 10.1128/jb.162.1.117-123.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2454.Lecker S, Lill R, Ziegelhoffer T, Georgopoulos C, Bassford P J, Kumamoto C A, Wickner W. Three pure chaperone proteins of Escherichia coli—SecB, trigger factor and GroEL—form soluble complexes with precursor proteins in vitro. EMBO J. 1989;8:2703–2709. doi: 10.1002/j.1460-2075.1989.tb08411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2455.Leclerc G, Sirard C, Drapeau G R. The Escherichia coli cell division mutation ftsM1 is in serU. J Bacteriol. 1989;171:2090–2095. doi: 10.1128/jb.171.4.2090-2095.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2456.Lederberg E M. Genetic and functional aspects of galactose metabolism in Escherichia coli K-12. Symp Soc Gen Microbiol. 1960;10:115–131. [Google Scholar]
- 2457.Lederberg J. The selection of genetic recombinations with bacterial growth inhibitors. J Bacteriol. 1950;59:211–215. doi: 10.1128/jb.59.2.211-215.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2458.Lee C A, Saier M H. Mannitol-specific enzyme II of the bacterial phosphotransferase system. III. The nucleotide sequence of the permease gene. J Biol Chem. 1983;258:10761–10767. [PubMed] [Google Scholar]
- 2459.Lee C A, Fournier M J, Beckwith J R. Escherichia coli 6S RNA is not essential for growth or protein secretion. J Bacteriol. 1985;161:1156–1161. doi: 10.1128/jb.161.3.1156-1161.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2460.Lee C C, Kohara Y, Akiyama K, Smith C L, Craigen W J, Caskey C T. Rapid and precise mapping of the Escherichia coli release factor genes by two physical approaches. J Bacteriol. 1988;170:4537–4541. doi: 10.1128/jb.170.10.4537-4541.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2461.Lee E C, Hales L M, Gumport R I, Gardner J F. The isolation and characterization of mutants of the integration host factor (IHF) of Escherichia coli with altered, expanded DNA-binding specificities. EMBO J. 1992;11:305–313. doi: 10.1002/j.1460-2075.1992.tb05053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2462.Lee E H, Masai H, Allen G C, Jr, Kornberg A. The priA gene encoding the primosomal replicative n′ protein of Escherichia coli. Proc Natl Acad Sci USA. 1990;87:4620–4624. doi: 10.1073/pnas.87.12.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2463.Lee J, Kashlev M, Borukhov S, Goldfarb A. A beta subunit mutation disrupting the catalytic function of Escherichia coli RNA polymerase. Proc Natl Acad Sci USA. 1991;88:6018–6022. doi: 10.1073/pnas.88.14.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2464.Lee J H, Wendt J C, Shanmugam K T. Identification of a new gene, molR, essential for utilization of molybdate by Escherichia coli. J Bacteriol. 1990;172:2079–2087. doi: 10.1128/jb.172.4.2079-2087.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2465.Lee J H, Patel P, Sankar P, Shanmugam K T. Isolation and characterization of mutant strains of Escherichia coli altered in H2 metabolism. J Bacteriol. 1985;162:344–352. doi: 10.1128/jb.162.1.344-352.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2466.Lee J S, An G, Friesen J D, Isono K. Cloning and the nucleotide sequence of the genes for Escherichia coli ribosomal proteins L28 (rpmB) and L33 (rpmG) Mol Gen Genet. 1981;184:218–223. doi: 10.1007/BF00272908. [DOI] [PubMed] [Google Scholar]
- 2467.Lee J S, An G, Friesen J D, Fiil N. Location of the tufB promoter of E. coli: cotranscription of tufB with four transfer RNA genes. Cell. 1981;25:251–258. doi: 10.1016/0092-8674(81)90250-6. [DOI] [PubMed] [Google Scholar]
- 2468.Lee L G, Jacobson G R, Saier M H. Plasmid-directed synthesis of enzymes required for d-mannitol transport and utilization in Escherichia coli. Proc Natl Acad Sci USA. 1981;78:7336–7340. doi: 10.1073/pnas.78.12.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2469.Lee N, Gielow W, Martin R, Hamilton E, Fowler A. The organization of the araBAD operon of Escherichia coli. Gene. 1986;47:231–244. doi: 10.1016/0378-1119(86)90067-3. [DOI] [PubMed] [Google Scholar]
- 2470.Lee N, Gielow W, Wallace R G. Mechanism of araC autoregulation and the domains of two overlapping promoters, Pc and Pbad, in the l-arabinose regulatory region of Escherichia coli. Proc Natl Acad Sci USA. 1981;78:752–756. doi: 10.1073/pnas.78.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2471.Lee S H, Walker J R. Escherichia coli DnaX product, the tau subunit of DNA polymerase III, is a multifunctional protein with single-stranded DNA-dependent ATPase activity. Proc Natl Acad Sci USA. 1987;84:2713–2717. doi: 10.1073/pnas.84.9.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2472.Lee S H, Kanda P, Kennedy R C, Walker J R. Relation of the Escherichia coli dnaX gene to its two products, the τ and γ subunits of DNA polymerase III holoenzyme. Nucleic Acids Res. 1987;15:7663–7675. doi: 10.1093/nar/15.19.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2473.Lee S J, Xie A, Jiang W, Etchegaray J P, Jones P G, Inouye M. Family of the major cold-shock protein CspA (CS7.4) of Escherichia coli, whose members show a high sequence similarity with the eukaryotic Y-box binding proteins. Mol Microbiol. 1994;11:833–839. doi: 10.1111/j.1365-2958.1994.tb00361.x. [DOI] [PubMed] [Google Scholar]
- 2474.Lee Y S, Kim H, Hwang D S. Transcriptional activation of the dnaA gene encoding the initiator for oriC replication by IciA protein, an inhibitor of in vitro oriC replication in Escherichia coli. Mol Microbiol. 1996;19:389–396. doi: 10.1046/j.1365-2958.1996.485902.x. [DOI] [PubMed] [Google Scholar]
- 2475.Legname G, Buono P, Fossati G, Monzini N, Mascagni P, Modena D, Marcucci F. Evidence for GroES acting as a transcriptional regulator. Biochem Biophys Res Commun. 1996;229:412–418. doi: 10.1006/bbrc.1996.1818. [DOI] [PubMed] [Google Scholar]
- 2476.Legrain C, Halleux P, Stalon V, Glansdorff N. The dual genetic control of ornithine carbamoyltransferase in Escherichia coli. A case of bacterial hybrid enzymes. Eur J Biochem. 1972;27:93–102. doi: 10.1111/j.1432-1033.1972.tb01814.x. [DOI] [PubMed] [Google Scholar]
- 2477.Leifer Z, Engel R, Tropp B E. Transport of 3,4-dihydroxylbutyl-1-phosponate, an analogue of sn-glycerol 3-phosphate. J Bacteriol. 1977;130:968–971. doi: 10.1128/jb.130.2.968-971.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2478.Leinfelder W, Zehelein E, Mandrand-Berthelot M A, Bock A. Gene for a novel tRNA species that accepts l-serine and cotranslationally inserts selenocysteine. Nature. 1988;331:723–725. doi: 10.1038/331723a0. [DOI] [PubMed] [Google Scholar]
- 2479.Leinfelder W, Forchhammer K, Veprek B, Zehelein E, Bock A. In vitro synthesis of selenocysteinyl-tRNA(UCA) from seryl-tRNA(UCA): involvement and characterization of the selD gene product. Proc Natl Acad Sci USA. 1990;87:543–547. doi: 10.1073/pnas.87.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2480.Leinfelder W, Forchhammer K, Zinoni F, Sawers G, Mandrand-Berthelot M A, Bock A. Escherichia coli genes whose products are involved in selenium metabolism. J Bacteriol. 1988;170:540–546. doi: 10.1128/jb.170.2.540-546.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2481.Leisinger T, Haas D, Hegarty M P. Indospicine as an arginine antagonist in Escherichia coli and Pseudomonas aeruginosa. Biochim Biophys Acta. 1972;262:214–219. doi: 10.1016/0005-2787(72)90235-3. [DOI] [PubMed] [Google Scholar]
- 2482.Lejeune P, Danchin A. Mutations in the bglY gene increase the frequency of spontaneous deletions in Escherichia coli K12. Proc Natl Acad Sci USA. 1990;87:360–363. doi: 10.1073/pnas.87.1.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2483.Lejeune P, Bertin P, Walon C, Willemot K, Colson C, Danchin A. A locus involved in kanamycin, chloramphenicol and l-serine resistance is located in the bglY-galU region of the Escherichia coli K12 chromosome. Mol Gen Genet. 1989;218:361–363. doi: 10.1007/BF00331292. [DOI] [PubMed] [Google Scholar]
- 2484.Lelivelt M J, Kawula T H. Hsc66, an Hsp70 homolog in Escherichia coli, is induced by cold shock but not by heat shock. J Bacteriol. 1995;177:4900–4907. doi: 10.1128/jb.177.17.4900-4907.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2485.Lemaire H G, Muller-Hill B. Nucleotide sequences of the galE gene and the galT gene of E. coli. Nucleic Acids Res. 1986;14:7705–7711. doi: 10.1093/nar/14.19.7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2486.Lemire B D, Robinson J J, Weiner J H. Identification of the membrane anchor polypeptides of Escherichia coli fumarate reductase. J Bacteriol. 1982;152:1126–1131. doi: 10.1128/jb.152.3.1126-1131.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2487.Lemmon R D, Rowe J J, Tritz G. Isolation and characterization of mutants of Escherichia coli defective in pyridine nucleotide cycle enzymes. Curr Microbiol. 1980;4:31–35. [Google Scholar]
- 2488.Lemoine Y, Wach A, Jeltsch J M. To be free or not: the fate of pimelate in Bacillus sphaericus and in Escherichia coli. Mol Microbiol. 1996;19:645–647. doi: 10.1046/j.1365-2958.1996.t01-4-442924.x. [DOI] [PubMed] [Google Scholar]
- 2489.Lemotte P K, Walker G C. Induction and autoregulation of ada, a positively acting element regulating the response of Escherichia coli K-12 to methylating agents. J Bacteriol. 1985;161:888–895. doi: 10.1128/jb.161.3.888-895.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2490.Lengeler J W. Analysis of mutations affecting the dissimilation of galactitol (dulcitol) in Escherichia coli K12. Mol Gen Genet. 1977;152:83–91. doi: 10.1007/BF00264944. [DOI] [PubMed] [Google Scholar]
- 2491.Lengeler J W. Characterization of mutants of Escherichia coli K12, selected by resistance to streptozotocin. Mol Gen Genet. 1980;179:49–54. doi: 10.1007/BF00268445. [DOI] [PubMed] [Google Scholar]
- 2492.Lengeler J W, Steinberger H. Analysis of the regulatory mechanisms controlling the synthesis of the hexitol transport systems in Escherichia coli K12. Mol Gen Genet. 1978;164:163–169. doi: 10.1007/BF00267381. [DOI] [PubMed] [Google Scholar]
- 2493.Lennette E T, Apirion D. Genetic analysis of an Escherichia coli syndrome. J Bacteriol. 1971;108:1322–1328. doi: 10.1128/jb.108.3.1322-1328.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2494.Lenny A B, Margolin P. Locations of the opp and supX genes of Salmonella typhimurium and Escherichia coli. J Bacteriol. 1980;143:747–752. doi: 10.1128/jb.143.2.747-752.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2495.Leonardo M R, Clark D P. Locations of genes in the nar-adhE region of the Escherichia coli K-12 chromosome. J Bacteriol. 1991;173:1574–1575. doi: 10.1128/jb.173.5.1574-1575.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2496.Leonardo M R, Cunningham P R, Clark D P. Anaerobic regulation of the adhE gene, encoding the fermentative alcohol dehydrogenase of Escherichia coli. J Bacteriol. 1993;175:870–878. doi: 10.1128/jb.175.3.870-878.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2497.Leonardo M R, Dailly Y, Clark D P. Role of NAD in regulating the adhE gene of Escherichia coli. J Bacteriol. 1996;178:6013–6018. doi: 10.1128/jb.178.20.6013-6018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2498.Lerner C G, Inouye M. Pleiotropic changes resulting from depletion of Era, an essential GTP-binding protein in Escherichia coli. Mol Microbiol. 1991;5:951–957. doi: 10.1111/j.1365-2958.1991.tb00770.x. [DOI] [PubMed] [Google Scholar]
- 2499.Lerner T J, Zinder N. Another gene affecting sexual expression of Escherichia coli. J Bacteriol. 1982;150:156–160. doi: 10.1128/jb.150.1.156-160.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2500.Lesage P, Chiaruttini C, Graffe M, Dondon J, Milet M, Springer M. Messenger RNA secondary structure and translational coupling in the Escherichia coli operon encoding translation initiation factor IF3 and the ribosomal proteins, L35 and L20. J Mol Biol. 1992;228:366–386. doi: 10.1016/0022-2836(92)90827-7. [DOI] [PubMed] [Google Scholar]
- 2501.Lester B, Bonner D M. Genetic control of raffinose utilization in Escherichia coli. J Bacteriol. 1957;73:544–552. doi: 10.1128/jb.73.4.544-552.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2502.Letain T E, Postle K. TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in Escherichia coli. Mol Microbiol. 1997;24:271–283. doi: 10.1046/j.1365-2958.1997.3331703.x. [DOI] [PubMed] [Google Scholar]
- 2503.Leung H B, Kvalnes-Krick K L, Meyer S L, deRiel J K, Schramm V L. Structure and regulation of the AMP nucleosidase gene (amn) from Escherichia coli. Biochemistry. 1989;28:8726–8733. doi: 10.1021/bi00448a008. [DOI] [PubMed] [Google Scholar]
- 2504.Leung H B, Schramm V L. The structural gene for AMP nucleosidase. Mapping, cloning and overproduction of the enzyme. J Biol Chem. 1984;259:6972–6978. [PubMed] [Google Scholar]
- 2505.Levchenko I, Luo L, Baker T A. Disassembly of the Mu transposase tetramer by the ClpX chaperone. Genes Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- 2506.Levengood S K, Webster R E. Nucleotide sequences of the tolA and tolB genes and localization of their products, components of a multistep translocation system in Escherichia coli. J Bacteriol. 1989;171:6600–6609. doi: 10.1128/jb.171.12.6600-6609.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2507.Levengood S K, Beyer W F, Webster R E. TolA: a membrane protein involved in colicin uptake contains an extended helical region. Proc Natl Acad Sci USA. 1991;88:5939–5943. doi: 10.1073/pnas.88.14.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2508.Leveque F, Gazeau M, Blanquet S, Plateau P. Control of Escherichia coli lysyl-tRNA synthetase expression by anaerobiosis. J Bacteriol. 1991;173:7903–7910. doi: 10.1128/jb.173.24.7903-7910.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2509.Leveque F, Blanchin-Roland S, Fayat G, Plateau P, Blanquet S. Design and characterization of Escherichia coli mutants devoid of Ap4N-hydrolase activity. J Mol Biol. 1990;212:319–329. doi: 10.1016/0022-2836(90)90127-8. [DOI] [PubMed] [Google Scholar]
- 2510.Levin H L, Schachman H K. Regulation of aspartate transcarbamoylase synthesis in Escherichia coli: analysis of deletion mutations in the promoter region of the pyrBI operon. Proc Natl Acad Sci USA. 1985;82:4643–4647. doi: 10.1073/pnas.82.14.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2511.Levine C, Marians K J. Identification of dnaX as a high-copy suppressor of the conditional lethal and partition phenotypes of the parE10 allele. J Bacteriol. 1998;180:1232–1240. doi: 10.1128/jb.180.5.1232-1240.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2512.LeVine S M, Ardeshir F, Ames G F-L. Isolation and characterization of acetate kinase and phosphotransacetylase mutants of Escherichia coli and Salmonella typhimurium. J Bacteriol. 1980;143:1081–1085. doi: 10.1128/jb.143.2.1081-1085.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2513.Levitz R, Klar A, Sar N, Yagil E. A new locus in the phosphate specific transport (PST) region of Escherichia coli. Mol Gen Genet. 1984;197:98–103. doi: 10.1007/BF00327928. [DOI] [PubMed] [Google Scholar]
- 2514.Levitz R, Friedberg I, Brucker R, Fux A, Yagil E. The effect of the locus pstB on phosphate binding in the phosphate specific transport (PST) system of Escherichia coli. Mol Gen Genet. 1985;200:118–122. doi: 10.1007/BF00383323. [DOI] [PubMed] [Google Scholar]
- 2515.Levitz R, Bittan R, Yagil E. Complementation tests between alkaline phosphatase-constitutive mutants (phoS and phoT) of Escherichia coli. J Bacteriol. 1981;145:1432–1435. doi: 10.1128/jb.145.3.1432-1435.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2516.Lewis K. Multidrug resistance pumps in bacteria: variations on a theme. Trends Biochem Sci. 1994;19:119–123. doi: 10.1016/0968-0004(94)90204-6. [DOI] [PubMed] [Google Scholar]
- 2517.Lewis L A, Li K B, Gousse A, Pereira F, Pacheco N, Pierre S, Kodaman P, Lawson S. Genetic and molecular analysis of spontaneous respiratory deficient (res−) mutants of Escherichia coli K-12. Microbiol Immunol. 1991;35:289–301. doi: 10.1111/j.1348-0421.1991.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 2518.Lewis L K, Mount D W. Interaction of LexA repressor with the asymmetric dinG operator and complete nucleotide sequence of the gene. J Bacteriol. 1992;174:5110–5116. doi: 10.1128/jb.174.15.5110-5116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2519.Lewis L K, Jenkins M E, Mount D W. Isolation of DNA damage-inducible promoters in Escherichia coli: regulation of polB (dinA), dinG, and dinH by LexA repressor. J Bacteriol. 1992;174:3377–3385. doi: 10.1128/jb.174.10.3377-3385.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2520.Leyh T F, Taylor J C, Markham G D. The sulfate activation locus of Escherichia coli K12: cloning, genetic, and enzymatic characterization. J Biol Chem. 1988;263:2409–2416. [PubMed] [Google Scholar]
- 2521.Leyh T S, Vogt T F, Suo Y. The DNA sequence of the sulfate activation locus from Escherichia coli K-12. J Biol Chem. 1992;267:10405–10410. [PubMed] [Google Scholar]
- 2522.Li C, Peck D, Przybyla A E. Cloning of the 3′-phosphoadenylyl sulfate reductase and sulfite reductase genes from Escherichia coli K-12. Gene. 1987;53:227–234. doi: 10.1016/0378-1119(87)90011-4. [DOI] [PubMed] [Google Scholar]
- 2523.Li C, Ichikawa J K, Ravetto J J, Kuo H C, Fuchs J A, Fu J C, Clarke S. A new gene involved in stationary-phase survival located at 59 minutes on the Escherichia coli chromosome. J Bacteriol. 1994;176:6015–6022. doi: 10.1128/jb.176.19.6015-6022.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2524.Li C, Clarke S. A protein methyltransferase specific for altered aspartyl residues is important in Escherichia coli stationary-phase survival and heat-shock resistance. Proc Natl Acad Sci USA. 1992;89:9885–9889. doi: 10.1073/pnas.89.20.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2525.Li J, Horwitz R, McCracken S, Greenblatt J. NusG, a new Escherichia coli elongation factor involved in transcriptional antitermination by the N protein of phage lambda. J Biol Chem. 1992;267:6012–6019. [PubMed] [Google Scholar]
- 2526.Li J, Mason S W, Greenblatt J. Elongation factor NusG interacts with termination factor rho to regulate termination and antitermination of transcription. Genes Dev. 1993;7:161–172. doi: 10.1101/gad.7.1.161. [DOI] [PubMed] [Google Scholar]
- 2527.Li J, Stewart V J. Localization of upstream sequence elements required for nitrate and anaerobic induction of fdn (formate dehydrogenase-N) operon expression in Escherichia coli K-12. J Bacteriol. 1992;174:4935–4942. doi: 10.1128/jb.174.15.4935-4942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2528.Li J M, Russell C S, Cosloy S D. Cloning and structure of the hemA gene of Escherichia coli K-12. Gene. 1989;82:209–217. doi: 10.1016/0378-1119(89)90046-2. [DOI] [PubMed] [Google Scholar]
- 2529.Li J-M, Russell C S, Cosloy S D. The structure of the Escherichia coli hemB gene. Gene. 1989;75:177–184. doi: 10.1016/0378-1119(89)90394-6. [DOI] [PubMed] [Google Scholar]
- 2530.Li J-M, Umanoff H, Proenca R, Russell C S, Cosloy S D. Cloning of the Escherichia coli K-12 hemB gene. J Bacteriol. 1988;170:1021–1025. doi: 10.1128/jb.170.2.1021-1025.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2531.Li Q X, Dowhan W. Structural characterization of Escherichia coli phosphatidylserine decarboxylase. J Biol Chem. 1988;263:11516–11522. [PubMed] [Google Scholar]
- 2532.Li S, DeMoss J A. Promoter region of the nar operon of Escherichia coli: nucleotide sequence and transcription initiation signals. J Bacteriol. 1987;169:4614–4620. doi: 10.1128/jb.169.10.4614-4620.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2533.Li S, Rabi T, DeMoss J A. Delineation of two distinct regulatory domains in the 5′ region of the nar operon of Escherichia coli. J Bacteriol. 1985;164:25–32. doi: 10.1128/jb.164.1.25-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2534.Li S-J, Rock C O, Cronan J E., Jr The dedB (usg) open reading frame of Escherichia coli encodes a subunit of acetyl-coenzyme A carboxylase. J Bacteriol. 1992;174:5755–5757. doi: 10.1128/jb.174.17.5755-5757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2535.Li S J, Cronan J E., Jr The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-CoA carboxylase. J Biol Chem. 1992;267:855–863. [PubMed] [Google Scholar]
- 2536.Li S J, Cronan J E., Jr The genes encoding the two carboxyltransferase subunits of Escherichia coli acetyl-CoA carboxylase. J Biol Chem. 1992;267:16841–16847. [PubMed] [Google Scholar]
- 2537.Li S-J, Cronan J E., Jr Growth rate regulation of Escherichia coli acetyl coenzyme A carboxylase, which catalyzes the first committed step of lipid biosynthesis. J Bacteriol. 1993;175:332–340. doi: 10.1128/jb.175.2.332-340.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2538.Li X, Xlindahl L, Zengel J M. Ribosomal protein L4 from Escherichia coli utilizes nonidentical determinants for its structural and regulatory functions. RNA. 1996;2:24–37. [PMC free article] [PubMed] [Google Scholar]
- 2539.Li Z, Demple B. SoxS, an activator of superoxide stress genes in Escherichia coli. Purification and interaction with DNA. J Biol Chem. 1994;269:18371–18377. [PubMed] [Google Scholar]
- 2540.Lieb M. Spontaneous mutation at a 5-methylcytosine hotspot is prevented by very short patch (VSP) mismatch repair. Genetics. 1991;128:23–28. doi: 10.1093/genetics/128.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2541.Lieb M, Rehmat S. Very short patch repair of T:G mismatches in vivo: importance of context and accessory proteins. J Bacteriol. 1995;177:660–666. doi: 10.1128/jb.177.3.660-666.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2542.Liebke H, Gross C A, Walter W, Burgess R R. A new mutation, rpoD800, affecting the sigma subunit of E. coli RNA polymerase is allelic to two other sigma mutants. Mol Gen Genet. 1980;177:277–282. doi: 10.1007/BF00267439. [DOI] [PubMed] [Google Scholar]
- 2543.Liebke H, Hatfull G F. The sequence of the distal end of the E. coli ribosomal RNA rrnE operon indicates conserved features are shared by rrn operons. Nucleic Acids Res. 1985;13:5515–5525. doi: 10.1093/nar/13.15.5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2544.Lightner V A, Bell R M, Modrich P. The DNA sequences encoding plsB and dgk loci of Escherichia coli. J Biol Chem. 1983;258:10856–10861. [PubMed] [Google Scholar]
- 2545.Lightner V A, Larsen T J, Tailleur P, Kantor G D, Raetz C R H, Bell R M, Modrich P. Membrane phospholipid synthesis in Escherichia coli. Cloning of a structural gene (plsB) of the sn-glycerol-3-phosphate acyltransferase. J Biol Chem. 1980;255:9413–9420. [PubMed] [Google Scholar]
- 2546.Likhacheva N A, Samsonov V V, Samsonov V V, Sineoky S P. Genetic control of the resistance to phage C1 of Escherichia coli K-12. J Bacteriol. 1996;178:5309–5315. doi: 10.1128/jb.178.17.5309-5315.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2547.Liljestrand-Golden C A, Johnson J R. Physical organization of the metJB component of the Escherichia coli K-12 metJBLF gene cluster. J Bacteriol. 1984;157:413–419. doi: 10.1128/jb.157.2.413-419.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2548.Liljestrom P L, Liljestrom P. Nucleotide sequence of the melA gene, coding for alpha-galactosidase in Escherichia coli K-12. Nucleic Acids Res. 1987;15:2213–2220. doi: 10.1093/nar/15.5.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2549.Lilley P E, Stamford N P M, Vasudevan S G, Dixon N E. The 92-min region of the Escherichia coli chromosome: location and cloning of the ubiA and alr genes. Gene. 1993;129:9–16. doi: 10.1016/0378-1119(93)90690-5. [DOI] [PubMed] [Google Scholar]
- 2550.Lim C J, Haller B L, Fuchs J A. Thioredoxin is the bacterial protein encoded by fip that is required for filamentous bacteriophage f1 assembly. J Bacteriol. 1985;161:799–802. doi: 10.1128/jb.161.2.799-802.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2551.Lim C J, Geraghty D, Fuchs J A. Cloning and nucleotide sequence of the trxA gene of Escherichia coli K-12. J Bacteriol. 1985;163:311–316. doi: 10.1128/jb.163.1.311-316.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2552.Lim D, Oppenheim J D, Eckhardt T, Maas W K. Nucleotide sequence of the argR gene of Escherichia coli K-12 and isolation of its product, the arginine repressor. Proc Natl Acad Sci USA. 1987;84:6697–6701. doi: 10.1073/pnas.84.19.6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2553.Lima T M, Lim D. Isolation and characterization of host mutants defective in msDNA synthesis: role of ribonuclease H in msDNA synthesis. Plasmid. 1995;33:235–238. doi: 10.1006/plas.1995.1026. [DOI] [PubMed] [Google Scholar]
- 2554.Lin E C C, Iuchi S. Regulation of gene expression in fermentative and respiratory systems in Escherichia coli and related bacteria. Annu Rev Genet. 1991;25:361–387. doi: 10.1146/annurev.ge.25.120191.002045. [DOI] [PubMed] [Google Scholar]
- 2555.Lin R, Ernsting B, Hirshfield I N, Matthews R G, Neidhardt F C, Clark R L, Newman E B. The lrp gene product regulates expression of lysU in Escherichia coli K-12. J Bacteriol. 1992;174:2779–2784. doi: 10.1128/jb.174.9.2779-2784.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2556.Lin R-J, Hill C W. Mapping the xyl, mtl, and lct loci in Escherichia coli K-12. J Bacteriol. 1983;156:914–916. doi: 10.1128/jb.156.2.914-916.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2557.Lin R-J, Capage M, Hill C W. A repetitive DNA sequence, rhs, responsible for duplications within the Escherichia coli K-12 chromosome. J Mol Biol. 1984;177:1–18. doi: 10.1016/0022-2836(84)90054-8. [DOI] [PubMed] [Google Scholar]
- 2558.Lin R T, D’Ari R, Newman E B. The leucine regulon of Escherichia coli K-12: a mutation in rblA alters expression of l-leucine-dependent metabolic operons. J Bacteriol. 1990;172:4529–4535. doi: 10.1128/jb.172.8.4529-4535.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2559.Lindahl L, Zengel J M. Operon-specific regulation of ribosomal protein synthesis in Escherichia coli. Proc Natl Acad Sci USA. 1979;76:6542–6546. doi: 10.1073/pnas.76.12.6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2560.Lindquist S, Weston-Hafer K, Schmidt H, Pul C, Korfmann G, Erickson J, Sanders C, Martin H H, Normark S. AmpG, a signal transducer in chromosomal beta-lactamase induction. Mol Microbiol. 1993;9:703–715. doi: 10.1111/j.1365-2958.1993.tb01731.x. [DOI] [PubMed] [Google Scholar]
- 2561.Lindquist S, Galleni M, Lindberg F, Normark S. Signalling proteins in enterobacterial ampC beta-lactamase regulation. Mol Microbiol. 1989;3:1091–1102. doi: 10.1111/j.1365-2958.1989.tb00259.x. [DOI] [PubMed] [Google Scholar]
- 2562.Lindsey D F, Martinez C, Walker J R. Physical map location of the Escherichia coli attachment site for the P22 prophage (attP22) J Bacteriol. 1992;174:3834–3835. doi: 10.1128/jb.174.11.3834-3835.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2563.Lindsey D F, Mullin D A, Walker J R. Characterization of the cryptic lambdoid prophage DLP12 of Escherichia coli and overlap of the DLP12 integrase gene with the tRNA gene argU. J Bacteriol. 1989;171:6197–6205. doi: 10.1128/jb.171.11.6197-6205.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2564.Lindstrom P H R, Stuber D, Bjork G R. Genetic organization and transcription from the gene (trmA) responsible for synthesis of tRNA (uracil-5)-methyltransferase by Escherichia coli. J Bacteriol. 1985;164:1117–1123. doi: 10.1128/jb.164.3.1117-1123.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2565.Link C D, Reiner A M. Genotypic exclusion: a novel relationship between the ribitol-arabitol and galactitol genes of E. coli. Mol Gen Genet. 1983;189:337–339. doi: 10.1007/BF00337827. [DOI] [PubMed] [Google Scholar]
- 2566.Linn T, Greenblatt J. The NusA and NusG proteins of Escherichia coli increase the in vitro readthrough frequency of a transcriptional attenuator preceding the gene for the beta subunit of RNA polymerase. J Biol Chem. 1992;267:1449–1454. [PubMed] [Google Scholar]
- 2567.Linn T, Goman M, Scaife J G. Lambda transducing bacteriophage carrying deletions of the argCBH-rpoBC region of the Escherichia coli chromosome. J Bacteriol. 1979;140:479–489. doi: 10.1128/jb.140.2.479-489.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2568.Liochev S L, Fridovich I. Fumarase C, the stable fumarase of Escherichia coli, is controlled by the soxRS regulon. Proc Natl Acad Sci USA. 1992;89:5892–5896. doi: 10.1073/pnas.89.13.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2569.Lipinska B, King J, Ang D, Georgopoulos C. Sequence analysis and transcriptional regulation of the Escherichia coli grpE gene, encoding a heat shock protein. Nucleic Acids Res. 1988;16:7545–7562. doi: 10.1093/nar/16.15.7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2570.Lipinska B, Zylicz M, Georgopoulos C. The HtrA (DegP) protein, essential for Escherichia coli survival at high temperatures, is an endopeptidase. J Bacteriol. 1990;172:1791–1797. doi: 10.1128/jb.172.4.1791-1797.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2571.Lipinska B, Fayet O, Baird L, Georgopoulos C. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J Bacteriol. 1989;171:1574–1584. doi: 10.1128/jb.171.3.1574-1584.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2572.Lipinska B, Sharma S, Georgopoulos C. Sequence analysis and regulation of the htrA gene of Escherichia coli: a sigma 32-independent mechanism of heat-inducible transcription. Nucleic Acids Res. 1988;16:10053–10067. doi: 10.1093/nar/16.21.10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2573.Lipsett M N. Enzymes producing 4-thiouridine in Escherichia coli tRNA: approximate chromosomal locations of the genes and enzyme activities in a 4-thiouridine-deficient mutant. J Bacteriol. 1978;135:993–997. doi: 10.1128/jb.135.3.993-997.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2574.Little J W. Construction and characterization of a plasmid coding for a fragment of the Escherichia coli recA protein. Mol Gen Genet. 1979;177:13–22. doi: 10.1007/BF00267248. [DOI] [PubMed] [Google Scholar]
- 2575.Little J W, Mount D W, Yanisch-Perron C R. Purified lexA protein is a repressor of the recA and lexA genes. Proc Natl Acad Sci USA. 1981;78:4199–4203. doi: 10.1073/pnas.78.7.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2576.Little R. Isolation of recombinant plasmids and phage carrying the lexA gene of Escherichia coli K-12. Gene. 1980;10:237–247. doi: 10.1016/0378-1119(80)90053-0. [DOI] [PubMed] [Google Scholar]
- 2577.Little R, Fiil N, Dennis P P. Transcription and post-transcriptional control of ribosomal protein and ribonucleic acid polymerase genes. J Bacteriol. 1981;147:25–35. doi: 10.1128/jb.147.1.25-35.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2578.Liu J, Magasanik B. The glnB region of the Escherichia coli chromosome. J Bacteriol. 1993;175:7441–7449. doi: 10.1128/jb.175.22.7441-7449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2579.Liu J, Walsh C T. Peptidyl-prolyl cis-trans isomerase from Escherichia coli: a periplasmic homolog of cyclophilin that is not inhibited by cyclosporin A. Proc Natl Acad Sci USA. 1990;87:4028–4032. doi: 10.1073/pnas.87.11.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2580.Liu J, Burns D M, Beacham I R. Isolation and sequence analysis of the gene (cpdB) encoding periplasmic 2′3′-cyclic phosphodiesterase. J Bacteriol. 1986;165:1002–1010. doi: 10.1128/jb.165.3.1002-1010.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2581.Liu J, Duncan K, Walsh C T. Nucleotide sequence of a cluster of Escherichia coli enterobactin biosynthesis genes: identification of entA and purification of its product 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase. J Bacteriol. 1989;171:791–798. doi: 10.1128/jb.171.2.791-798.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2582.Liu J, Quinn N, Berchtold G A, Walsh C T. Overexpression, purification, and characterization of isochorismate synthase (EntC), the first enzyme involved in the biosynthesis of enterobactin from chorismate. Biochemistry. 1990;29:1417–1425. doi: 10.1021/bi00458a012. [DOI] [PubMed] [Google Scholar]
- 2583.Liu J D, Parkinson J S. Genetics and sequence analysis of the pcnB locus, an Escherichia coli gene involved in plasmid copy number control. J Bacteriol. 1989;171:1254–1261. doi: 10.1128/jb.171.3.1254-1261.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2584.Liu L, Yoshimura T, Endo K, Kishimoto K, Fuchikami Y, Manning J M, Esaki N, Soda K. Compensation for d-glutamate auxotrophy of Escherichia coli WM335 by d-amino acid aminotransferase gene and regulation of murI expression. Biosci Biotechnol Biochem. 1998;62:193–195. doi: 10.1271/bbb.62.193. [DOI] [PubMed] [Google Scholar]
- 2585.Liu L, Whalen W, Das A, Berg C M. Rapid sequencing of cloned DNA using a transposon for bidirectional priming: sequence of the Escherichia coli K-12 avtA gene. Nucleic Acids Res. 1987;15:9461–9469. doi: 10.1093/nar/15.22.9461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2585a.Liu M Y, Gui G, Wei B, Preston III J F, Oakford L, Yuksel U, Giedroc D P, Romeo T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- 2586.Liu M Y, Yang H, Romeo T. The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J Bacteriol. 1995;177:2663–2672. doi: 10.1128/jb.177.10.2663-2672.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2587.Liu S K, Tseng J N, Shiuan D, Hanawalt P C. Preferential mutagenesis of lacZ integrated at unique sites in the Escherichia coli chromosome. Mol Gen Genet. 1997;255:449–459. doi: 10.1007/s004380050517. [DOI] [PubMed] [Google Scholar]
- 2588.Liu X, DeMoss J A. Characterization of NarJ, a system-specific chaperone required for nitrate reductase biogenesis in Escherichia coli. J Biol Chem. 1997;272:24266–24271. doi: 10.1074/jbc.272.39.24266. [DOI] [PubMed] [Google Scholar]
- 2589.Liu X, Fujita N, Ishihama A, Matsumura P. The C-terminal region of the α subunit of Escherichia coli RNA polymerase is required for transcriptional activation of the flagellar level II operons by the FlhD/FlhC complex. J Bacteriol. 1995;177:5186–5188. doi: 10.1128/jb.177.17.5186-5188.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2590.Liu X, Matsumura P. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol. 1994;176:7345–7351. doi: 10.1128/jb.176.23.7345-7351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2591.Liu X, Matsumura P. An alternative sigma factor controls transcription of flagellar class-III operons in Escherichia coli: gene sequence, overproduction, purification and characterization. Gene. 1995;164:81–84. doi: 10.1016/0378-1119(95)00480-t. [DOI] [PubMed] [Google Scholar]
- 2592.Liveris D, Klotsky R A, Schwartz I. Growth rate regulation of translation initiation factor IF3 biosynthesis in Escherichia coli. J Bacteriol. 1991;173:3888–3893. doi: 10.1128/jb.173.12.3888-3893.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2593.Lloyd R G. Conjugational recombination in resolvase-deficient ruvC mutants of Escherichia coli K-12 depends on recG. J Bacteriol. 1991;173:5414–5418. doi: 10.1128/jb.173.17.5414-5418.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2594.Lloyd R G. Linkage distortion following conjugational transfer of sbcC+ to recBC sbcBC strains of Escherichia coli. J Bacteriol. 1991;173:5694–5698. doi: 10.1128/jb.173.18.5694-5698.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2595.Lloyd R G, Buckman C. Identification and genetic analysis of sbcC mutations in commonly used recBC sbcB strains of Escherichia coli K-12. J Bacteriol. 1985;164:836–844. doi: 10.1128/jb.164.2.836-844.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2596.Lloyd R G, Buckman C. Genetic analysis of the recG locus of Escherichia coli K-12 and of its role in recombination and DNA repair. J Bacteriol. 1991;173:1004–1011. doi: 10.1128/jb.173.3.1004-1011.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2597.Lloyd R G, Sharples G J. Molecular organization and nucleotide sequence of the recG locus of Escherichia coli K-12. J Bacteriol. 1991;173:6837–6843. doi: 10.1128/jb.173.21.6837-6843.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2598.Lloyd R G, Sharples G J. Dissociation of synthetic Holliday junctions by E. coli RecG protein. EMBO J. 1993;12:17–22. doi: 10.1002/j.1460-2075.1993.tb05627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2599.Lloyd R G, Sharples G J. Processing of recombination intermediates by the RecG and RuvAB proteins of Escherichia coli. Nucleic Acids Res. 1993;21:1719–1725. doi: 10.1093/nar/21.8.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2600.Lloyd R G, Picksley S M, Prescott C. Inducible expression of a gene specific to the recF pathway for recombination in Escherichia coli K12. Mol Gen Genet. 1983;190:162–167. doi: 10.1007/BF00330340. [DOI] [PubMed] [Google Scholar]
- 2601.Lloyd S A, Tang H, Wang X, Billings S, Blair D F. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J Bacteriol. 1996;178:223–231. doi: 10.1128/jb.178.1.223-231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2602.Lo T C Y, Sanwal B D. Genetic analysis of mutants of Escherichia coli defective in dicarboxylate transport. Mol Gen Genet. 1975;140:303–307. doi: 10.1007/BF00267321. [DOI] [PubMed] [Google Scholar]
- 2603.Lobner-Olesen A, Boye E. Different effects of mioC transcription on initiation of chromosomal and minichromosomal replication in Escherichia coli. Nucleic Acids Res. 1992;20:3029–3036. doi: 10.1093/nar/20.12.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2604.Lobner-Olesen A, Boye E, Marinus M G. Expression of the Escherichia coli dam gene. Mol Microbiol. 1992;6:1841–1851. doi: 10.1111/j.1365-2958.1992.tb01356.x. [DOI] [PubMed] [Google Scholar]
- 2605.Lobner-Olesen A, Marinus M G. Identification of the gene (aroK) encoding shikimic acid kinase I of Escherichia coli. J Bacteriol. 1992;174:525–529. doi: 10.1128/jb.174.2.525-529.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2606.Lobner-Olesen A, Atlung T, Rasmussen K V. Stability and replication control of Escherichia coli minichromosomes. J Bacteriol. 1987;169:2835–2842. doi: 10.1128/jb.169.6.2835-2842.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2607.Lobocka M, Hennig J, Wild J R, Klopotowski T. Organization and expression of the Escherichia coli K-12 dad operon encoding the smaller subunit of d-amino acid dehydrogenase and the catabolic alanine racemase. J Bacteriol. 1994;176:1500–1510. doi: 10.1128/jb.176.5.1500-1510.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2608.Loenen W A M, Daniel A S, Braymer H D, Murray N E. Organization and sequence of the hsd genes of Escherichia coli K-12. J Mol Biol. 1987;198:159–170. doi: 10.1016/0022-2836(87)90303-2. [DOI] [PubMed] [Google Scholar]
- 2609.Loewen P C. Isolation of catalase-deficient Escherichia coli mutants and genetic mapping of katE, a locus that affects catalase activity. J Bacteriol. 1984;157:622–626. doi: 10.1128/jb.157.2.622-626.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2610.Loewen P C, Triggs-Raine B L. Genetic mapping of katF, a locus that with katE affects the synthesis of a second catalase species in Escherichia coli. J Bacteriol. 1984;160:668–675. doi: 10.1128/jb.160.2.668-675.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2611.Loewen P C, Triggs-Raine B L, George C S, Hrabarchuk B E. Genetic mapping of katG, a locus that affects synthesis of the bifunctional catalase-peroxidase hydroperoxidase I in Escherichia coli. J Bacteriol. 1985;162:661–667. doi: 10.1128/jb.162.2.661-667.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2612.Loferer H, Hammar M, Normark S. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol Microbiol. 1997;26:11–23. doi: 10.1046/j.1365-2958.1997.5231883.x. [DOI] [PubMed] [Google Scholar]
- 2613.Lohman T M, Bjornson K P. Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 2614.Lohmeier E, Hagen D S, Dickie P, Weiner J H. Cloning and expression of the fumarate reductase gene of Escherichia coli. Can J Biochem. 1981;59:158–164. doi: 10.1139/o81-023. [DOI] [PubMed] [Google Scholar]
- 2615.Lois L M, Campos N, Putra S R, Danielsen K, Rohmer M, Boronat A. Cloning and characterization of a gene from Escherichia coli encoding a transketolase-like enzyme that catalyzes the synthesis of d-1-deoxyxylulose 5-phosphate, a common precursor for isoprenoid, thiamin, and pyridoxol biosynthesis. Proc Natl Acad Sci USA. 1998;95:2105–2110. doi: 10.1073/pnas.95.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2616.Lomax M S, Greenberg G R. Characteristics of the deo operon: role in thymine utilization and sensitivity to deoxyribonucleosides. J Bacteriol. 1968;96:501–514. doi: 10.1128/jb.96.2.501-514.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2617.Lombardo M J, Miller C G, Rudd K E. Physical mapping of the Escherichia coli pepT and potABCD genes. J Bacteriol. 1993;175:7745–7746. doi: 10.1128/jb.175.23.7745-7746.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2618.Lommatzsch J, Templin M F, Kraft A R, Vollmer W, Holtje J V. Outer membrane localization of murein hydrolases: MltA, a third lipoprotein lytic transglycosylase in Escherichia coli. J Bacteriol. 1997;179:5465–5470. doi: 10.1128/jb.179.17.5465-5470.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2619.Lomovskaya O, Kawai F, Matin A. Differential regulation of the mcb and emr operons of Escherichia coli: role of mcb in multidrug resistance. Antimicrob Agents Chemother. 1996;40:1050–1052. doi: 10.1128/aac.40.4.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2620.Lomovskaya O L, Kidwell J P, Matin A. Characterization of the ς38-dependent expression of a core Escherichia coli starvation gene, pexB. J Bacteriol. 1994;176:3928–3935. doi: 10.1128/jb.176.13.3928-3935.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2621.Lomovskaya O L, Lewis K. Emr, an Escherichia coli locus for multidrug resistance. Proc Natl Acad Sci USA. 1992;89:8938–8942. doi: 10.1073/pnas.89.19.8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2622.Lomovskaya O L, Lewis K, Matin A. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J Bacteriol. 1995;177:2328–2334. doi: 10.1128/jb.177.9.2328-2334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2623.Lonetto M, Gribskov M, Gross C A. The ς70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2624.Long W S, Slayman C L, Low K B. Production of giant cells of Escherichia coli. J Bacteriol. 1978;133:995–1007. doi: 10.1128/jb.133.2.995-1007.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2625.Loomis W F, Magasanik B. Genetic control of catabolite repression of the lac operon in Escherichia coli. Biochem Biophys Res Commun. 1965;20:230–234. doi: 10.1016/0006-291x(65)90351-7. [DOI] [PubMed] [Google Scholar]
- 2626.Loomis W F, Magasanik B. The catabolite repression gene of the lac operon in Escherichia coli. J Mol Biol. 1967;23:487–494. doi: 10.1016/s0022-2836(67)80120-7. [DOI] [PubMed] [Google Scholar]
- 2627.Lopata M, Schlieper D, von Wilcken-Bergmann B, Muller-Hill B. A lethal mutant of the catabolite gene activator protein CAP of Escherichia coli. Biol Chem. 1997;378:1153–1162. doi: 10.1515/bchm.1997.378.10.1153. [DOI] [PubMed] [Google Scholar]
- 2628.Lopes J M, Lawther R P. Analysis and comparison of the internal promoter, pE, of the ilvGMEDA operon from Escherichia coli K-12 and Salmonella typhimurium. Nucleic Acids Res. 1986;14:2779–2798. doi: 10.1093/nar/14.6.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2629.Lopez J, Webster R E. fipB and fipC: two bacterial loci required for morphogenesis of the filamentous bacteriophage f1. J Bacteriol. 1985;163:900–905. doi: 10.1128/jb.163.3.900-905.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2630.Lopilato J, Garwin J L, Emr S D, Silhavy T J, Beckwith J R. d-Ribose metabolism in Escherichia coli K-12: genetics, regulation, and transport. J Bacteriol. 1984;158:665–673. doi: 10.1128/jb.158.2.665-673.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2631.Lopilato J, Bortner S, Beckwith J R. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol Gen Genet. 1986;205:285–290. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- 2632.Lorence M C, Maika S D, Rupert C S. Physical analysis of phr gene transcription in Escherichia coli K-12. J Bacteriol. 1990;172:6551–6556. doi: 10.1128/jb.172.11.6551-6556.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2633.Lorenz E, Stauffer G V. MetR-mediated repression of the glyA gene in Escherichia coli. FEMS Microbiol Lett. 1996;144:229–233. doi: 10.1111/j.1574-6968.1996.tb08535.x. [DOI] [PubMed] [Google Scholar]
- 2634.Lorowitz W, Clark D P. Mutants of Escherichia coli with a temperature-sensitive alcohol dehydrogenase. J Bacteriol. 1982;152:935–938. doi: 10.1128/jb.152.2.935-938.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2635.Lother H, Messer W. Promoters in the E. coli replication origin. Nature. 1981;294:376–378. doi: 10.1038/294376a0. [DOI] [PubMed] [Google Scholar]
- 2636.Loubens I, Debarbieux L, Bohin A, Lacroix J M, Bohin J P. Homology between a genetic locus (mdoA) involved in the osmoregulated biosynthesis of periplasmic glucans in Escherichia coli and a genetic locus (hrpM) controlling pathogenicity of Pseudomonas syringae. Mol Microbiol. 1993;10:329–340. doi: 10.1111/j.1365-2958.1993.tb01959.x. [DOI] [PubMed] [Google Scholar]
- 2637.Loudon J A, Loughlin R E. Mutagenesis and regulation of the cysJ promoter of Escherichia coli K-12. Gene. 1992;122:17–25. doi: 10.1016/0378-1119(92)90027-m. [DOI] [PubMed] [Google Scholar]
- 2638.Lovett S T, Clark A J. Genetic analysis of the recJ gene of Escherichia coli K-12. J Bacteriol. 1984;157:190–196. doi: 10.1128/jb.157.1.190-196.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2639.Lovett S T, Clark A J. Cloning of the Escherichia coli recJ chromosomal region and identification of its encoded proteins. J Bacteriol. 1985;162:280–285. doi: 10.1128/jb.162.1.280-285.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2640.Lovett S T, Kolodner R D. Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc Natl Acad Sci USA. 1989;86:2627–2631. doi: 10.1073/pnas.86.8.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2641.Lovett S T, Kolodner R D. Nucleotide sequence of the Escherichia coli recJ chromosomal region and construction of RecJ-overexpression plasmids. J Bacteriol. 1991;173:353–364. doi: 10.1128/jb.173.1.353-364.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2642.Lovett S T, Sutera V A. Suppression of recJ exonuclease mutants of Escherichia coli by alterations in DNA helicases II (uvrD) and IV (helD) Genetics. 1995;140:27–45. doi: 10.1093/genetics/140.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2643.Low B. Restoration by the rac locus of recombinant forming ability in recB− and recC− merozygotes of Escherichia coli K-12. Mol Gen Genet. 1973;122:119–130. doi: 10.1007/BF00435185. [DOI] [PubMed] [Google Scholar]
- 2644.Lozoya E, Sanchez-Pescador R, Covarrubias A, Vichido I, Bolivar F. Tight linkage of genes that encode the two glutamate synthase subunits of Escherichia coli K-12. J Bacteriol. 1980;144:616–621. doi: 10.1128/jb.144.2.616-621.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2645.Lu A L, Yuen D S, Cillo J. Catalytic mechanism and DNA substrate recognition of Escherichia coli MutY protein. J Biol Chem. 1996;271:24138–24143. doi: 10.1074/jbc.271.39.24138. [DOI] [PubMed] [Google Scholar]
- 2646.Lu M, Campbell J L, Boye E, Kleckner N. SeqA: a negative modulator of replication initiation in E. coli. Cell. 1994;77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 2647.Lu M, Kleckner N. Molecular cloning and characterization of the pgm gene encoding phosphoglucomutase of Escherichia coli. J Bacteriol. 1994;176:5847–5851. doi: 10.1128/jb.176.18.5847-5851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2648.Lu M-F, Umbarger H E. Effects of deletion and insertion mutations in the ilvM gene of Escherichia coli. J Bacteriol. 1987;169:600–604. doi: 10.1128/jb.169.2.600-604.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2649.Lu Q, Zhang X, Almaula N, Mathews C K, Inouye M. The gene for nucleoside diphosphate kinase functions as a mutator gene in Escherichia coli. J Mol Biol. 1995;254:337–341. doi: 10.1006/jmbi.1995.0620. [DOI] [PubMed] [Google Scholar]
- 2650.Lu Z, Lin E C C. The nucleotide sequence of Escherichia coli genes for l-fucose dissimilation. Nucleic Acids Res. 1989;17:4883–4884. doi: 10.1093/nar/17.12.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2651.Lucht J M, Boos W, Bremer E. Alignment of genes from the 9-minute region (araJ to tsx) of the Escherichia coli K-12 linkage map to the physical map. J Bacteriol. 1992;174:1709–1710. doi: 10.1128/jb.174.5.1709-1710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2652.Luckey M, Neilands J B. Iron transport in Salmonella typhimurium LT-2: prevention, by ferrichrome, of adsorption of bacteriophages ES18 and ES18.h1 to a common cell envelope receptor. J Bacteriol. 1976;127:1036–1037. doi: 10.1128/jb.127.2.1036-1037.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2653.Luckey M, Pollack J R, Wayne R, Ames B N, Neilands J B. Iron uptake in Salmonella typhimurium: utilization of exogenous siderochromes as iron carriers. J Bacteriol. 1972;111:731–738. doi: 10.1128/jb.111.3.731-738.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2654.Ludtke D N, Bernstein H D, Hamilton E, Torriani A. Identification of the phoM gene product and its regulation in Escherichia coli K-12. J Bacteriol. 1984;159:19–25. doi: 10.1128/jb.159.1.19-25.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2655.Lugtenberg E J J, Wijsman H J W, van Zaane D. Properties of a d-glutamic acid-requiring mutant of Escherichia coli. J Bacteriol. 1973;114:499–506. doi: 10.1128/jb.114.2.499-506.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2656.Luirink J, ten Hagen-Jongman C M, van der Weijden C C, Oudega B, High S, Dobberstein B, Kusters R. An alternative protein targeting pathway in Escherichia coli: studies on the role of FtsY. EMBO J. 1994;13:2289–2296. doi: 10.1002/j.1460-2075.1994.tb06511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2657.Luisi-DeLuca C. Homologous pairing of single-stranded DNA and superhelical double-stranded DNA catalyzed by RecO protein from Escherichia coli. J Bacteriol. 1995;177:566–572. doi: 10.1128/jb.177.3.566-572.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2658.Lukomski S, Hull R, Hull S I. Identification of the O antigen polymerase (rfc) gene in Escherichia coli O4 by insertional mutagenesis using a nonpolar chloramphenicol resistance cassette. J Bacteriol. 1996;178:240–247. doi: 10.1128/jb.178.1.240-247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2659.Lum D, Wallace B J. Sequence and characterisation of three genes of a glutamate-aspartate binding protein-dependent transport system of Escherichia coli K12. 1995. GenBank submission U10981. [Google Scholar]
- 2660.Lum D, Lee C J, Wallace B J. Location of the gltP gene on the physical map of Escherichia coli K-12. J Bacteriol. 1993;175:5735. doi: 10.1128/jb.175.17.5735.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2661.Lundberg L G, Karlstrom H O, Nyman P O. Isolation and characterization of the dut gene of Escherichia coli. II. Restriction enzyme mapping and analysis of polypeptide products. Gene. 1983;22:127–131. doi: 10.1016/0378-1119(83)90071-9. [DOI] [PubMed] [Google Scholar]
- 2662.Lundberg L G, Thoresson H-O, Karlstrom H O, Nyman P O. Nucleotide sequence of the structural gene for dUTPase of Escherichia coli K-12. EMBO J. 1983;2:967–971. doi: 10.1002/j.1460-2075.1983.tb01529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2663.Lundberg U, Altman S. Processing of the precursor to the catalytic RNA subunit of RNase P from Escherichia coli. RNA. 1995;1:327–334. [PMC free article] [PubMed] [Google Scholar]
- 2664.Lundegaard C, Jensen K F. The DNA damage-inducible dinD gene of Escherichia coli is equivalent to orfY upstream of pyrE. J Bacteriol. 1994;176:3383–3385. doi: 10.1128/jb.176.11.3383-3385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2665.Lundrigan M, Earhart C F. Reduction in three iron-regulated outer membrane proteins and protein a by the Escherichia coli K-12 perA mutation. J Bacteriol. 1981;146:804–807. doi: 10.1128/jb.146.2.804-807.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2666.Lundrigan M D, Earhart C F. Gene envY of Escherichia coli K-12 affects thermoregulation of major porin expression. J Bacteriol. 1984;157:262–268. doi: 10.1128/jb.157.1.262-268.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2667.Lundrigan M D, DeVeaux L C, Mann B J, Kadner R J. Separate regulatory systems for the repression of metE and btuB by vitamin B12 in Escherichia coli. Mol Gen Genet. 1987;206:401–407. doi: 10.1007/BF00428878. [DOI] [PubMed] [Google Scholar]
- 2668.Lundrigan M D, Friedrich M J, Kadner R J. Nucleotide sequence of the Escherichia coli porin thermoregulatory gene envY. Nucleic Acids Res. 1989;17:800. doi: 10.1093/nar/17.2.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2669.Lundrigan M D, Kadner R J. Nucleotide sequence of the gene for the ferrienterochelin receptor fepA in Escherichia coli: homology among outer membrane receptors which interact with TonB. J Biol Chem. 1986;261:10797–10801. [PubMed] [Google Scholar]
- 2670.Lupo M, Halpern Y S. Gene controlling l-glutamic acid decarboxylase synthesis in Escherichia coli K-12. J Bacteriol. 1970;103:382–386. doi: 10.1128/jb.103.2.382-386.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2671.Lupski J R, Ruiz A A, Godson G N. Promotion, termination and anti-termination of the rpsU-dnaG-rpoD macromolecular synthesis operon of E. coli K-12. Mol Gen Genet. 1984;195:391–401. doi: 10.1007/BF00341439. [DOI] [PubMed] [Google Scholar]
- 2672.Lupski J R, Smiley B L, Blattner F R, Godson G N. Cloning and characterization of the Escherichia coli chromosomal region surrounding the dnaG gene, with a correlated physical and genetic map of dnaG generated via transposon Tn5 mutagenesis. Mol Gen Genet. 1982;185:120–128. doi: 10.1007/BF00333800. [DOI] [PubMed] [Google Scholar]
- 2673.Lupski J R, Smiley B L, Godson G N. Regulation of the rpsU-dnaG-rpoD macromolecular synthesis operon and the initiation of DNA replication in Escherichia coli K-12. Mol Gen Genet. 1983;189:48–57. doi: 10.1007/BF00326054. [DOI] [PubMed] [Google Scholar]
- 2674.Lutkenhaus J. Regulation of cell division in E. coli. Trends Genet. 1990;154:1339–1346. doi: 10.1016/0168-9525(90)90045-8. [DOI] [PubMed] [Google Scholar]
- 2675.Lutkenhaus J, Wolf-Watz H, Donachie W D. Organization of genes in the ftsA-envA region of the Escherichia coli genetic map and identification of a new fts locus (ftsA) J Bacteriol. 1980;142:615–620. doi: 10.1128/jb.142.2.615-620.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2676.Lutkenhaus J, Wu H C. Determination of transcriptional units and gene products from the ftsA region of Escherichia coli. J Bacteriol. 1980;143:1281–1288. doi: 10.1128/jb.143.3.1281-1288.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2677.Lutkenhaus J F. Coupling of DNA replication and cell division: sulB is an allele of ftsZ. J Bacteriol. 1983;154:1339–1346. doi: 10.1128/jb.154.3.1339-1346.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2678.Lutz S, Jacobi A, Schlensog V, Bohm R, Sawers G, Bock A. Molecular characterization of an operon (hyp) necessary for the activity of the three hydrogenase isoenzymes in Escherichia coli. Mol Microbiol. 1991;5:123–135. doi: 10.1111/j.1365-2958.1991.tb01833.x. [DOI] [PubMed] [Google Scholar]
- 2679.Lux R, Jahreis K, Bettenbrock K, Parkinson J S, Lengeler J W. Coupling the phosphotransferase system and the methyl-accepting chemotaxis protein-dependent chemotaxis signaling pathways of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:11583–11587. doi: 10.1073/pnas.92.25.11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2680.Lynch A S, Wang J C. Anchoring of DNA to the bacterial cytoplasmic membrane through cotranscriptional synthesis of polypeptides encoding membrane proteins or proteins for export: a mechanism of plasmid hypernegative supercoiling in mutants deficient in DNA topoisomerase I. J Bacteriol. 1993;175:1645–1655. doi: 10.1128/jb.175.6.1645-1655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2681.Lyngstadaas A, Lobner-Olesen A, Boye E. Characterization of three genes in the dam-containing operon of Escherichia coli. Mol Gen Genet. 1995;247:546–554. doi: 10.1007/BF00290345. [DOI] [PubMed] [Google Scholar]
- 2682.Lynn S P, Bauer C E, Chapman K E, Gardner J F. Identification and characterization of mutants affecting transcription termination at the threonine operon attenuator. J Mol Biol. 1985;183:529–541. doi: 10.1016/0022-2836(85)90169-x. [DOI] [PubMed] [Google Scholar]
- 2683.Ma C, Rupert C S. Promoters of the phr gene in Escherichia coli K-12. Mol Gen Genet. 1995;248:52–58. doi: 10.1007/BF02456613. [DOI] [PubMed] [Google Scholar]
- 2684.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol. 1993;175:6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2685.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 2686.Ma D, Alberti M, Lynch C, Nikaido H, Hearst J E. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol Microbiol. 1996;19:101–112. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- 2687.Ma J, Katsonouri A, Gennis R B. Subunit II of the cytochrome bo3 ubiquinol oxidase from Escherichia coli is a lipoprotein. Biochemistry. 1997;36:11298–11303. doi: 10.1021/bi9709710. [DOI] [PubMed] [Google Scholar]
- 2688.Ma J-C, Newman A J, Hayward R S. Internal promoters of the rpoBC operon of Escherichia coli. Mol Gen Genet. 1981;184:548–550. doi: 10.1007/BF00352538. [DOI] [PubMed] [Google Scholar]
- 2689.Ma X, Ehrhardt D W, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2690.Maas W K. Repression of arginine formation. Cold Spring Harbor Symp Quant Biol. 1961;26:183–190. doi: 10.1101/sqb.1961.026.01.023. [DOI] [PubMed] [Google Scholar]
- 2691.Maas W K. Genetic defect affecting an arginine permease and repression of arginine synthesis in E. coli. Fed Proc. 1965;24:1239–1242. [PubMed] [Google Scholar]
- 2691a.Maas W K. Mapping of genes involved in the synthesis of spermidine in Escherichia coli. Mol Gen Genet. 1972;119:1–9. doi: 10.1007/BF00270439. [DOI] [PubMed] [Google Scholar]
- 2692.Mabuchi K, Kanazawa H, Kayano T, Futai M. Nucleotide sequence of the gene coding for the δ subunit of proton-translocating ATPase of Escherichia coli. Biochem Biophys Res Commun. 1981;102:172–179. doi: 10.1016/0006-291x(81)91504-7. . (Erratum, 104:354, 1982.) [DOI] [PubMed] [Google Scholar]
- 2693.MacDonald H, Cole J A. Molecular cloning and functional analysis of the cysG and nirB genes of Escherichia coli K12, two closely-linked genes required for NADH-dependent nitrite reductase activity. Mol Gen Genet. 1985;200:328–334. doi: 10.1007/BF00425444. [DOI] [PubMed] [Google Scholar]
- 2694.MacDonald H, Pope N R, Cole J A. Isolation, characterization and complementation analysis of nirB mutants of Escherichia coli deficient only in NADH-dependent nitrite reductase activity. J Gen Microbiol. 1985;131:2771–2782. doi: 10.1099/00221287-131-10-2771. [DOI] [PubMed] [Google Scholar]
- 2695.Macfarlane J, Muller M. The functional integration of a polytopic membrane protein of Escherichia coli is dependent on the bacterial signal-recognition particle. Eur J Biochem. 1995;233:766–771. doi: 10.1111/j.1432-1033.1995.766_3.x. [DOI] [PubMed] [Google Scholar]
- 2696.Macinga D R, Cook G M, Poole R K, Rather P N. Identification and characterization of aarF, a locus required for production of ubiquinone in Providencia stuartii and Escherichia coli and for expression of 2′-N-acetyltransferase in P. stuartii. J Bacteriol. 1998;180:128–135. doi: 10.1128/jb.180.1.128-135.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2697.Macintyre G, Doiron K M, Cupples C G. The Vsr endonuclease of Escherichia coli: an efficient DNA repair enzyme and a potent mutagen. J Bacteriol. 1997;179:6048–6052. doi: 10.1128/jb.179.19.6048-6052.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2698.Mackay W J, Han S, Samson L D. DNA alkylation repair limits spontaneous base substitution mutations in Escherichia coli. J Bacteriol. 1994;176:3224–3230. doi: 10.1128/jb.176.11.3224-3230.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2699.Mackie G A. Cloning of fragments of λ dapB2 DNA and identification of the dapB gene product. J Biol Chem. 1980;255:8928–8935. [PubMed] [Google Scholar]
- 2700.Mackie G A. Nucleotide sequence of the gene for ribosomal protein S20 and its flanking regions. J Biol Chem. 1981;256:8177–8182. [PubMed] [Google Scholar]
- 2701.Mackie G A. Structure of the DNA distal to the gene for ribosomal protein S20 in Escherichia coli K12: presence of a strong terminator and an IS1 element. Nucleic Acids Res. 1986;14:6965–6981. doi: 10.1093/nar/14.17.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2702.Mackie G A. Specific endonucleolytic cleavage of the mRNA for ribosomal protein S20 of Escherichia coli requires the product of the ams gene in vivo and in vitro. J Bacteriol. 1991;173:2488–2497. doi: 10.1128/jb.173.8.2488-2497.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2703.MacLean M J, Ness L S, Ferguson G P, Booth I R. The role of glyoxalase I in detoxification of methylglyoxal and in the activation of the KefB K+ efflux system in Escherichia coli. Mol Microbiol. 1998;27:563–571. doi: 10.1046/j.1365-2958.1998.00701.x. [DOI] [PubMed] [Google Scholar]
- 2704.Macnab R M. Genetics and biogenesis of bacterial flagella. Annu Rev Genet. 1992;26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 2705.MacNeil D. General method, using Mu-Mudl dilysogens, to determine the direction of transcription of and generate deletions in the glnA region of Escherichia coli. J Bacteriol. 1981;146:260–268. doi: 10.1128/jb.146.1.260-268.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2706.MacNeil T, MacNeil D, Tyler B. Fine-structure deletion map and complementation analysis of the glnA-glnL-glnG region in Escherichia coli. J Bacteriol. 1982;150:1302–1313. doi: 10.1128/jb.150.3.1302-1313.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2707.MacNeil T, Roberts G P, MacNeil D, Tyler B. The products of glnL and glnG are bifunctional regulatory proteins. Mol Gen Genet. 1982;188:325–333. doi: 10.1007/BF00332696. [DOI] [PubMed] [Google Scholar]
- 2708.MacPherson A J S, Jones-Mortimer M C, Henderson P J F. Identification of the AraE transport protein of Escherichia coli. Biochem J. 1981;196:269–283. doi: 10.1042/bj1960269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2709.Madiraju M V, Templin A, Clark A J. Properties of a mutant recA-encoded protein reveal a possible role for Escherichia coli recF-encoded protein in genetic recombination. Proc Natl Acad Sci USA. 1988;85:6592–6596. doi: 10.1073/pnas.85.18.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2710.Magasanik B. Regulation of transcription of the glnALG operon of Escherichia coli by protein phosphorylation. Biochimie. 1989;71:1005–1012. doi: 10.1016/0300-9084(89)90104-1. [DOI] [PubMed] [Google Scholar]
- 2711.Magee T R, Kogoma T. Requirement of RecBC enzyme and an elevated level of activated RecA for induced stable DNA replication in Escherichia coli. J Bacteriol. 1990;172:1834–1839. doi: 10.1128/jb.172.4.1834-1839.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2712.Magnuson K, Jackowski S, Rock C O, Cronan J E., Jr Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol Rev. 1993;57:522–542. doi: 10.1128/mr.57.3.522-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2713.Magnuson K, Oh W, Larson T J, Cronan J E., Jr Cloning and nucleotide sequence of the fabD gene encoding malonyl coenzyme A-acyl carrier protein transacylase of Escherichia coli. FEBS Lett. 1992;299:262–266. doi: 10.1016/0014-5793(92)80128-4. [DOI] [PubMed] [Google Scholar]
- 2714.Magota K, Otsuji N, Miki T, Horiuchi T, Tsunasawa S, Kondo J, Sakiyama F, Amemura M, Shinagawa H, Nakata A. Nucleotide sequence of the phoS gene, the structural gene for the phosphate-binding protein of Escherichia coli. J Bacteriol. 1984;157:909–917. doi: 10.1128/jb.157.3.909-917.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2715.Maguin E, Brody H, Hill C W, D’Ari R. SOS-associated division inhibition gene sfiC is part of excisable element e14 in Escherichia coli. J Bacteriol. 1986;168:464–466. doi: 10.1128/jb.168.1.464-466.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2716.Maguire B A, Wild D G. The roles of proteins L28 and L33 in the assembly and function of Escherichia coli ribosomes in vivo. Mol Microbiol. 1997;23:237–245. doi: 10.1046/j.1365-2958.1997.2131578.x. [DOI] [PubMed] [Google Scholar]
- 2717.Magyar A, Zhang X, Kohn H, Widger W R. The antibiotic bicyclomycin affects the secondary RNA binding site of Escherichia coli transcription termination factor Rho. J Biol Chem. 1996;271:25369–25374. doi: 10.1074/jbc.271.41.25369. [DOI] [PubMed] [Google Scholar]
- 2718.Mahadevan S, Reynolds A E, Wright A. Positive and negative regulation of the bgl operon in Escherichia coli. J Bacteriol. 1987;169:2570–2578. doi: 10.1128/jb.169.6.2570-2578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2719.Mahajan S K, Chu C C, Willis D K, Templin A, Clark A J. Physical analysis of spontaneous and mutagen-induced mutants of Escherichia coli K-12 expressing DNA exonuclease VIII activity. Genetics. 1990;125:261–273. doi: 10.1093/genetics/125.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2720.Mahajan S K, Vartak N B, Datta A R. A new pleiotropic mutation causing defective carbohydrate uptake in Escherichia coli K-12: isolation, mapping, and preliminary characterization. J Bacteriol. 1988;170:2568–2574. doi: 10.1128/jb.170.6.2568-2574.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2721.Mahan M J, Csonka L N. Genetic analysis of the proBA genes of Salmonella typhimurium: physical and genetic analysis of the cloned proB+A+ genes of Escherichia coli and of a mutant allele that confers proline overproduction and enhanced osmotolerance. J Bacteriol. 1983;156:1249–1262. doi: 10.1128/jb.156.3.1249-1262.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2722.Mahdi A A, Sharples G J, Mandal T N, Lloyd R G. Holliday junction resolvases encoded by homologous rusA genes in Escherichia coli K-12 and phage 82. J Mol Biol. 1996;257:561–573. doi: 10.1006/jmbi.1996.0185. [DOI] [PubMed] [Google Scholar]
- 2723.Mahdi A A, Lloyd R G. Identification of the recR locus of Escherichia coli K-12 and analysis of its role in recombination and DNA repair. Mol Gen Genet. 1989;216:503–510. doi: 10.1007/BF00334397. [DOI] [PubMed] [Google Scholar]
- 2724.Maiden M C J, Jones-Mortimer M C, Henderson P J F. The cloning, DNA sequence, and overexpression of the gene araE coding for arabinose-proton symport in Escherichia coli K12. J Biol Chem. 1988;263:8003–8010. [PubMed] [Google Scholar]
- 2725.Maier D, Jacobi A, Sauter M, Bock A. The product of the hypB gene, which is required for nickel incorporation into hydrogenases, is a novel guanine nucleotide-binding protein. J Bacteriol. 1993;175:630–635. doi: 10.1128/jb.175.3.630-635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2726.Maier T, Binder U, Bock A. Analysis of the hydA locus of Escherichia coli: two genes (hydN and hypF) involved in formate and hydrogen metabolism. Arch Microbiol. 1996;165:333–341. doi: 10.1007/s002030050335. [DOI] [PubMed] [Google Scholar]
- 2727.Makaroff C A, Zalkin H. Regulation of Escherichia coli purF: analysis of the control region of a pur regulon gene. J Biol Chem. 1985;260:10378–10387. [PubMed] [Google Scholar]
- 2728.Makela P H, Stocker B. Genetics of polysaccharide biosynthesis. Annu Rev Genet. 1969;3:291–322. [Google Scholar]
- 2729.Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 2730.Maki H, Horiuchi T, Sekiguchi M. Structure and expression of the dnaQ mutator and the RNase H genes of Escherichia coli: overlap of the promoter regions. Proc Natl Acad Sci USA. 1983;80:7137–7141. doi: 10.1073/pnas.80.23.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2731.Maki S, Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. I. Purification and distinctive functions of subunits tau and gamma, the dnaZX gene products. J Biol Chem. 1988;263:6547–6554. [PubMed] [Google Scholar]
- 2732.Makino K, Shinagawa H, Nakata A. Cloning and characterization of the alkaline phosphatase positive regulator gene (phoB) of Escherichia coli. Mol Gen Genet. 1982;187:181–186. doi: 10.1007/BF00341438. [DOI] [PubMed] [Google Scholar]
- 2733.Makino K, Shinagawa H, Nakata A. Cloning and characterization of the alkaline phosphatase positive regulatory gene (phoM) of Escherichia coli. Mol Gen Genet. 1984;195:381–390. doi: 10.1007/BF00341438. [DOI] [PubMed] [Google Scholar]
- 2734.Makino K, Shinagawa H, Nakata A. Regulation of the phosphate regulon of Escherichia coli K-12: regulation and role of the regulatory gene phoR. J Mol Biol. 1985;184:231–240. doi: 10.1016/0022-2836(85)90376-6. [DOI] [PubMed] [Google Scholar]
- 2735.Makino K, Shinagawa H, Amemura M, Nakata A. Nucleotide sequence of the phoB gene, the positive regulatory gene for the phosphate regulon of Escherichia coli K-12. J Mol Biol. 1986;190:37–44. doi: 10.1016/0022-2836(86)90073-2. [DOI] [PubMed] [Google Scholar]
- 2736.Makino K, Shinagawa H, Amemura M, Nakata A. Nucleotide sequence of the phoR gene, a regulatory gene for the phosphate regulon of Escherichia coli. J Mol Biol. 1986;192:549–556. doi: 10.1016/0022-2836(86)90275-5. [DOI] [PubMed] [Google Scholar]
- 2737.Makino K, Kim S-K, Shinagawa H, Amemura M, Nakata A. Molecular analysis of the cryptic and functional phn operons for phosphonate use in Escherichia coli K-12. J Bacteriol. 1991;173:2665–2672. doi: 10.1128/jb.173.8.2665-2672.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2738.Malakooti J, Ely B, Matsumura P. Molecular characterization, nucleotide sequence, and expression of the fliO, fliP, fliQ, and fliR genes of Escherichia coli. J Bacteriol. 1994;176:189–197. doi: 10.1128/jb.176.1.189-197.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2739.Malakooti J, Yoshibumi K, Matsumara P. DNA sequence analysis, gene product identification, and localization of flagellar motor components of Escherichia coli. J Bacteriol. 1989;171:2728–2734. doi: 10.1128/jb.171.5.2728-2734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2740.Malcolm B A, Kirsch J F. Site-directed mutagenesis of aspartate aminotransferase from E. coli. Biochem Biophys Res Commun. 1985;132:915–921. doi: 10.1016/0006-291x(85)91894-7. [DOI] [PubMed] [Google Scholar]
- 2741.Maleszka R, Wang P Y, Schneider H. A ColE1 hybrid plasmid containing Escherichia coli genes complementing d-xylose negative mutants of Escherichia coli and Salmonella typhimurium. Can J Biochem. 1982;60:144–151. doi: 10.1139/o82-020. [DOI] [PubMed] [Google Scholar]
- 2742.Malo M S, Loughlin R E. Promoter elements and regulation of expression of the cysD gene of Escherichia coli K-12. Gene. 1990;87:127–131. doi: 10.1016/0378-1119(90)90504-k. [DOI] [PubMed] [Google Scholar]
- 2743.Maloy S R, Stewart V J. Autogenous regulation of gene expression. J Bacteriol. 1993;175:307–316. doi: 10.1128/jb.175.2.307-316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2744.Maloy S R, Nunn W D. Genetic regulation of the glyoxylate shunt in Escherichia coli K-12. J Bacteriol. 1982;149:173–180. doi: 10.1128/jb.149.1.173-180.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2745.Mandal T N, Mahdi A A, Sharples G J, Lloyd R G. Resolution of Holliday intermediates in recombination and DNA repair: indirect suppression of ruvA, ruvB, and ruvC mutations. J Bacteriol. 1993;175:4325–4334. doi: 10.1128/jb.175.14.4325-4334.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2746.Mandrand-Berthelot M A, Couchoux-Luthaud G, Santini C L, Giordano G. Mutants of Escherichia coli specifically deficient in respiratory formate dehydrogenase activity. J Gen Microbiol. 1988;134:3129–3139. doi: 10.1099/00221287-134-12-3129. [DOI] [PubMed] [Google Scholar]
- 2747.Mandrand-Berthelot M A, Ritzenthaler P, Mata-Gilsinger M. Construction and expression of hybrid plasmids containing the structural gene of the Escherichia coli K-12 3-deoxy-2-oxo-d-gluconate transport system. J Bacteriol. 1984;160:600–606. doi: 10.1128/jb.160.2.600-606.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2748.Mangroo D, Wu X Q, RajBhandary U L. Escherichia coli initiator tRNA: structure-function relationships and interactions with the translational machinery. Biochem Cell Biol. 1995;73:1023–1031. doi: 10.1139/o95-109. [DOI] [PubMed] [Google Scholar]
- 2749.Manna D, Gowrishankar J. Evidence for involvement of proteins HU and RpoS in transcription of the osmoresponsive proU operon in Escherichia coli. J Bacteriol. 1994;176:5378–5384. doi: 10.1128/jb.176.17.5378-5384.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2750.Maples V F, Kushner S R. DNA repair in Escherichia coli: identification of the uvrD gene product. Proc Natl Acad Sci USA. 1982;79:5616–5620. doi: 10.1073/pnas.79.18.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2751.Marceau M, McFall E, Lewis S D, Shafer J A. d-Serine dehydratase from Escherichia coli. DNA sequence and identification of catalytically inactive glycine to aspartic acid variants. J Biol Chem. 1988;263:16926–16933. [PubMed] [Google Scholar]
- 2752.March J B, Colloms M D, Hart-Davis D, Oliver I R, Masters M. Cloning and characterization of an Escherichia coli gene, pcnB, affecting plasmid copy number. Mol Microbiol. 1989;3:903–910. doi: 10.1111/j.1365-2958.1989.tb00239.x. [DOI] [PubMed] [Google Scholar]
- 2753.March P E, Ahnn J, Inouye M. The DNA sequence of the gene (rnc) encoding ribonuclease III of Escherichia coli. Nucleic Acids Res. 1985;13:4677–4685. doi: 10.1093/nar/13.13.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2754.March P E, Inoue M. Characterization of the lep operon of Escherichia coli. Identification of the promoter and the gene upstream of the signal peptidase I gene. J Biol Chem. 1985;260:7206–7213. [PubMed] [Google Scholar]
- 2755.March P E, Inouye M. GTP-binding membrane protein of Escherichia coli with sequence homology to initiation factor 2 and elongation factors Tu and G. Proc Natl Acad Sci USA. 1985;82:7500–7504. doi: 10.1073/pnas.82.22.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2756.Marconi R, Wigboldus J, Weissbach H, Brot N. Transcriptional start and MetR binding sites on the Escherichia coli metH gene. Biochem Biophys Res Commun. 1991;175:1057–1063. doi: 10.1016/0006-291x(91)91672-y. [DOI] [PubMed] [Google Scholar]
- 2757.Marcus M, Halpern Y S. Genetic analysis of glutamate transport in Escherichia coli. J Bacteriol. 1967;93:1409–1415. doi: 10.1128/jb.93.4.1409-1415.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2758.Marcus M, Halpern Y S. Genetic analysis of the glutamate permease in Escherichia coli K-12. J Bacteriol. 1969;97:1118–1128. doi: 10.1128/jb.97.3.1118-1128.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2759.Marek L E, Henson J M. Cloning and expression of the Escherichia coli K-12 sad gene. J Bacteriol. 1988;170:991–994. doi: 10.1128/jb.170.2.991-994.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2760.Margison G P, Cooper D P, Brennand J. Cloning of the E. coli O6-methylguanine and methylphosphotriester methyltransferase gene using a functional DNA repair assay. Nucleic Acids Res. 1985;13:1939–1952. doi: 10.1093/nar/13.6.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2761.Margison G P, Cooper D P, Potter P M. The E. coli ogt gene. Mutat Res. 1990;233:15–21. doi: 10.1016/0027-5107(90)90146-u. [DOI] [PubMed] [Google Scholar]
- 2762.Margolin P, Zumstein L, Sternglanz R, Wang J C. The Escherichia coli supX locus in topA, the structural gene for DNA topoisomerase I. Proc Natl Acad Sci USA. 1985;82:5437–5441. doi: 10.1073/pnas.82.16.5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2763.Marinus M G, Morris N R, Soll D, Kwong T C. Isolation and partial characterization of three Escherichia coli mutants with altered transfer ribonucleic acid methylases. J Bacteriol. 1975;122:257–265. doi: 10.1128/jb.122.1.257-265.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2764.Mark D F, Chang J W, Richardson C F. Genetic mapping of trxA, a gene affecting thioredoxin in Escherichia coli K12. Mol Gen Genet. 1977;155:145–152. doi: 10.1007/BF00393153. [DOI] [PubMed] [Google Scholar]
- 2765.Markham B E, Little J W, Mount D W. Nucleotide sequence of the lexA gene of Escherichia coli K-12. Nucleic Acids Res. 1981;9:4149–4161. doi: 10.1093/nar/9.16.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2766.Markham G D, DeParasis J, Gatmaitan J. The sequence of metK, the structural gene for S-adenosylmethionine synthetase in Escherichia coli. J Biol Chem. 1984;259:14505–14507. [PubMed] [Google Scholar]
- 2767.Markiewicz P, Kleina L G, Cruz C, Ehret S, Miller J H. Genetic studies of the lac repressor. XIV. Analysis of 4000 altered Escherichia coli lac repressors reveals essential and non-essential residues, as well as “spacers” which do not require a specific sequence. J Mol Biol. 1994;240:421–433. doi: 10.1006/jmbi.1994.1458. [DOI] [PubMed] [Google Scholar]
- 2768.Markovitz A. Regulatory mechanism for synthesis of capsular polysaccharide in mucoid mutants of Escherichia coli K-12. Proc Natl Acad Sci USA. 1964;51:239–240. doi: 10.1073/pnas.51.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2769.Markovitz A, Rosenbaum N. A regulator gene that is dominant on an episome and recessive on a chromosome. Proc Natl Acad Sci USA. 1965;54:1084–1091. doi: 10.1073/pnas.54.4.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2770.Marolda C L, Valvano M A. Genetic analysis of the dTDP-rhamnose biosynthesis region of the Escherichia coli VW187 (O7:K1) rfb gene cluster: identification of functional homologs of rfbB and rfbA in the rff cluster and correct location of the rffE gene. J Bacteriol. 1995;177:5539–5546. doi: 10.1128/jb.177.19.5539-5546.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2771.Marolda C L, Valvano M A. The GalF protein of Escherichia coli is not a UDP-glucose pyrophosphorylase but interacts with the GalU protein possibly to regulate cellular levels of UDP-glucose. Mol Microbiol. 1996;22:827–840. doi: 10.1046/j.1365-2958.1996.01531.x. [DOI] [PubMed] [Google Scholar]
- 2772.Marolewski A, Smith J M, Benkovic S J. Cloning and characterization of a new purine biosynthetic enzyme: a non-folate glycinamide ribonucleotide transformylase from E. coli. Biochemistry. 1994;33:2531–2537. doi: 10.1021/bi00175a023. [DOI] [PubMed] [Google Scholar]
- 2773.Marquardt J L, Siegele D A, Kolter R, Walsh C T. Cloning and sequencing of Escherichia coli murZ and purification of its product, a UDP-N-acetylglucosamine enolpyruvyl transferase. J Bacteriol. 1992;174:5748–5752. doi: 10.1128/jb.174.17.5748-5752.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2774.Marschall C, Hengge-Aronis R. Regulatory characteristics and promoter analysis of csiE, a stationary phase-inducible gene under the control of sigma S and the cAMP-CRP complex in Escherichia coli. Mol Microbiol. 1995;18:175–184. doi: 10.1111/j.1365-2958.1995.mmi_18010175.x. [DOI] [PubMed] [Google Scholar]
- 2775.Marschall C, Labrousse V, Kreimer M, Weichart D, Kolb A, Hengge-Aronis R. Molecular analysis of the regulation of csiD, a carbon starvation-inducible gene in Escherichia coli that is exclusively dependent on sigma s and requires activation by cAMP-CRP. J Mol Biol. 1998;276:339–353. doi: 10.1006/jmbi.1997.1533. [DOI] [PubMed] [Google Scholar]
- 2776.Marsh L, Walker G C. Cold sensitivity induced by overproduction of UmuDC in Escherichia coli. J Bacteriol. 1985;162:155–161. doi: 10.1128/jb.162.1.155-161.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2777.Martin F, Reinbolt J, Dirheimer G, Gangloff J, Eriani G. Selection of tRNA(Asp) amber suppressor mutants having alanine, arginine, glutamine, and lysine identity. RNA. 1996;2:919–927. [PMC free article] [PubMed] [Google Scholar]
- 2778.Martin J L, Bardwell J C, Kuriyan J. Crystal structure of the DsbA protein required for disulphide bond formation in vivo. Nature. 1993;365:464–468. doi: 10.1038/365464a0. [DOI] [PubMed] [Google Scholar]
- 2779.Martin R G, Rosner J L. Fis, an accessorial factor for transcriptional activation of the mar (multiple antibiotic resistance) promoter of Escherichia coli in the presence of the activator MarA, SoxS, or Rob. J Bacteriol. 1997;179:7410–7419. doi: 10.1128/jb.179.23.7410-7419.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2780.Martin R G, Jair K W, Wolf R E, Rosner J L. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J Bacteriol. 1996;178:2216–2223. doi: 10.1128/jb.178.8.2216-2223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2781.Martinez D, Whitehouse F. Selective autocytotoxicity in a model system of Escherichia coli recombinants. J Bacteriol. 1973;114:882–884. doi: 10.1128/jb.114.2.882-884.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2782.Martinez J, Steenbergen S, Vimr E. Derived structure of the putative sialic acid transporter from Escherichia coli predicts a novel sugar permease domain. J Bacteriol. 1995;177:6005–6010. doi: 10.1128/jb.177.20.6005-6010.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2783.Maruyama M, Horiuchi T, Maki H, Sekiguchi M. A dominant (mutD5) and a recessive (dnaQ49) mutator of Escherichia coli. J Mol Biol. 1983;167:757–771. doi: 10.1016/s0022-2836(83)80109-0. [DOI] [PubMed] [Google Scholar]
- 2784.Marvel C C, Arps P J, Rubin B C, Kammen H O, Penhoet E E, Winkler M E. hisT is part of a multigene operon in Escherichia coli K-12. J Bacteriol. 1985;161:60–71. doi: 10.1128/jb.161.1.60-71.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2785.Marykwas D L, Berg H C. A mutational analysis of the interaction between FliG and FliM, two components of the flagellar motor of Escherichia coli. J Bacteriol. 1996;178:1289–1294. doi: 10.1128/jb.178.5.1289-1294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2786.Masai H, Arai K. Operon structure of dnaT and dnaC genes essential for normal and stable DNA replication of Escherichia coli chromosome. J Biol Chem. 1988;263:15083–15093. [PubMed] [Google Scholar]
- 2787.Masai H, Bond M W, Arai K-I. Cloning of the Escherichia coli gene for primosomal protein i: the relationship to dnaT, essential for chromosomal DNA replication. Proc Natl Acad Sci USA. 1986;83:1256–1260. doi: 10.1073/pnas.83.5.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2788.Masters M. Location of purine genes on the physical map of Escherichia coli. J Bacteriol. 1990;172:1173. doi: 10.1128/jb.172.3.1173.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2789.Masters M, Paterson T, Popplewell A G, Owen-Hughes T, Pringle J H, Begg K J. The effect of DnaA protein levels and the rate of initiation at oriC on transcription originating in the ftsQ and ftsA genes: in vivo experiments. Mol Gen Genet. 1989;216:475–483. doi: 10.1007/BF00334393. [DOI] [PubMed] [Google Scholar]
- 2790.Masters M, March J B, Oliver I R, Collins J F. A possible role for the pcnB gene product of Escherichia coli in modulating RNA: RNA interactions. Mol Gen Genet. 1990;220:341–344. doi: 10.1007/BF00260507. [DOI] [PubMed] [Google Scholar]
- 2791.Masters M, Colloms M D, Oliver I R, He L, MacNaughton E J, Charters Y. The pcnB gene of Escherichia coli, which is required for ColE1 copy number maintenance, is dispensable. J Bacteriol. 1993;175:4405–4413. doi: 10.1128/jb.175.14.4405-4413.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2792.Masters P S, Hong J-S. Genetics of the glutamine transport system in Escherichia coli. J Bacteriol. 1981;147:805–819. doi: 10.1128/jb.147.3.805-819.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2793.Masuda Y, Ohtsubo E. Mapping and disruption of the chpB locus in Escherichia coli. J Bacteriol. 1994;176:5861–5863. doi: 10.1128/jb.176.18.5861-5863.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2794.Masuda Y, Miyakawa K, Nishimura Y, Ohtsubo E. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J Bacteriol. 1993;175:6850–6856. doi: 10.1128/jb.175.21.6850-6856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2795.Mata-Gilsinger M, Ritzenthaler P. Physical mapping of the exuT and uxaC operators by use of exu plasmids and generation of deletion mutants in vitro. J Bacteriol. 1983;155:973–982. doi: 10.1128/jb.155.3.973-982.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2796.Mata-Gilsinger M, Ritzenthaler P, Blanco C. Characterization of the operator sites of the exu regulon in Escherichia coli K-12 by operator-constitutive mutations and repressor titration. Genetics. 1983;105:829–842. doi: 10.1093/genetics/105.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2797.Mathew E, Zhi J, Freundlich M. Lrp is a direct repressor of the dad operon in Escherichia coli. J Bacteriol. 1996;178:7234–7240. doi: 10.1128/jb.178.24.7234-7240.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2798.Matin A. The molecular basis of carbon-starvation-induced general resistance in Escherichia coli. Mol Microbiol. 1991;5:3–10. doi: 10.1111/j.1365-2958.1991.tb01819.x. [DOI] [PubMed] [Google Scholar]
- 2799.Mat-Jan F, Williams C R, Clark D P. Mutations permitting the anaerobic growth of Escherichia coli on trehalose. FEMS Microbiol Lett. 1991;62:149–152. doi: 10.1016/0378-1097(91)90149-5. [DOI] [PubMed] [Google Scholar]
- 2800.Mat-Jan F, Alam K Y, Clark D P. Mutants of Escherichia coli deficient in the fermentative lactate dehydrogenase. J Bacteriol. 1989;171:342–348. doi: 10.1128/jb.171.1.342-348.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2801.Matsuhashi M, Maruyama I N, Takagaki Y, Tamaki S, Nishimura Y, Hirota Y. Isolation of a mutant of Escherichia coli lacking penicillin-sensitive d-alanine carboxypeptidase IA. Proc Natl Acad Sci USA. 1978;75:2631–2635. doi: 10.1073/pnas.75.6.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2802.Matsuhashi M, Tamaki S, Curtis S J, Strominger J L. Mutational evidence for identity of penicillin-binding protein 5 in Escherichia coli with the major d-alanine carboxypeptidase IA activity. J Bacteriol. 1979;137:644–647. doi: 10.1128/jb.137.1.644-647.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2803.Matsuhashi M, Takagaki Y, Maruyama I N, Tamaki S, Nishimura Y, Suzuki H, Ogino U, Hirota Y. Mutants of Escherichia coli lacking in highly penicillin-sensitive d-alanine carboxypeptidase activity. Proc Natl Acad Sci USA. 1977;74:2976–2979. doi: 10.1073/pnas.74.7.2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2804.Matsuhisa A, Suzuki N, Noda T, Shiba K. Inositol monophosphatase activity from the Escherichia coli suhB gene product. J Bacteriol. 1995;177:200–205. doi: 10.1128/jb.177.1.200-205.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2805.Matsui M, Oka A, Takanami M, Yasuda S, Hirota Y. Sites of DNA protein-binding in the replication origin of the Escherichia coli K-12 chromosome. J Mol Biol. 1985;184:529–533. doi: 10.1016/0022-2836(85)90299-2. [DOI] [PubMed] [Google Scholar]
- 2806.Matsumoto K. Phosphatidylserine synthase from bacteria. Biochim Biophys Acta. 1997;1348:214–227. doi: 10.1016/s0005-2760(97)00110-0. [DOI] [PubMed] [Google Scholar]
- 2807.Matsumoto Y, Shigesada K, Imai M. Autogenous regulation of the gene for transcription termination factor Rho in Escherichia coli: localization and function of its attenuators. J Bacteriol. 1986;166:945–958. doi: 10.1128/jb.166.3.945-958.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2808.Matsumura K, Ito K, Kawazu Y, Mikuni O, Nakamura Y. Suppression of temperature-sensitive defects of polypeptide release factors RF-1 and RF-2 by mutations or by an excess of RF-3 in Escherichia coli. J Mol Biol. 1996;258:588–599. doi: 10.1006/jmbi.1996.0271. [DOI] [PubMed] [Google Scholar]
- 2809.Matsumura P, Rydel J J, Linzmeier R, Vacante D. Overexpression and sequence of the Escherichia coli cheY gene and biochemical activities of the CheY protein. J Bacteriol. 1984;160:36–41. doi: 10.1128/jb.160.1.36-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2810.Matsunaga J, Simons E L, Simons R W. RNase III autoregulation: structure and function of rncO, the posttranscriptional “operator”. RNA. 1996;2:1228–1240. [PMC free article] [PubMed] [Google Scholar]
- 2811.Matsuoka M, McFadden B A. Isolation, hyperexpression, and sequencing of the aceA gene encoding isocitrate lyase in Escherichia coli. J Bacteriol. 1988;170:4528–4536. doi: 10.1128/jb.170.10.4528-4536.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2812.Matsuyama A, Yamamoto H, Nakano W. Cloning, expression, and nucleotide sequence of the Escherichia coli K-12 ackA gene. J Bacteriol. 1989;171:577–580. doi: 10.1128/jb.171.1.577-580.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2813.Matsuyama A, Yamamoto-Otake H, Hewitt J, MacGillivray R T, Nakano E. Nucleotide sequence of the phosphotransacetylase gene of Escherichia coli strain K12. Biochim Biophys Acta. 1994;1219:559–562. doi: 10.1016/0167-4781(94)90089-2. [DOI] [PubMed] [Google Scholar]
- 2814.Matsuyama S, Akimaru J, Mizushima S. SecE-dependent overproduction of SecY in Escherichia coli. Evidence for interaction between two components of the secretory machinery. FEBS Lett. 1990;269:96–100. doi: 10.1016/0014-5793(90)81128-b. [DOI] [PubMed] [Google Scholar]
- 2815.Matsuyama S, Yokota N, Tokuda H. A novel outer membrane lipoprotein, LolB (HemM), involved in the LolA (p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J. 1997;16:6947–6955. doi: 10.1093/emboj/16.23.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2816.Matsuyama S, Tajima T, Tokuda H. A novel periplasmic carrier protein involved in the sorting and transport of Escherichia coli lipoproteins destined for the outer membrane. EMBO J. 1995;14:3365–3372. doi: 10.1002/j.1460-2075.1995.tb07342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2817.Matsuyama S, Fujita Y, Sagara K, Mizushima S. Overproduction, purification and characterization of SecD and SecF, integral membrane components of the protein translocation machinery of Escherichia coli. Biochim Biophys Acta. 1992;1122:77–84. doi: 10.1016/0167-4838(92)90130-6. [DOI] [PubMed] [Google Scholar]
- 2818.Matsuzawa H, Asoh S, Kunai K, Muraiso K, Takasuga A, Ohta T. Nucleotide sequence of the rodA gene, responsible for the rod shape of Escherichia coli: rodA and the pbpA gene, encoding penicillin-binding protein 2, constitute the rodA operon. J Bacteriol. 1989;171:558–560. doi: 10.1128/jb.171.1.558-560.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2819.Mattern I E, Pittard J. Regulation of tyrosine biosynthesis in Escherichia coli K-12: isolation and characterization of operator mutants. J Bacteriol. 1971;107:8–15. doi: 10.1128/jb.107.1.8-15.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2820.Maupin-Furlow J A, Rosentel J K, Lee J H, Deppenmeier U, Gunsalus R P, Shanmugam K T. Genetic analysis of the modABCD (molybdate transport) operon of Escherichia coli. J Bacteriol. 1995;177:4851–4856. doi: 10.1128/jb.177.17.4851-4856.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2821.Maupin-Furlow J A, Shanmugam K T. Genetic regulation of formate hydrogenlyase of Escherichia coli: role of the fhlA gene product as a transcriptional activator for a new regulatory gene, fhlB. J Bacteriol. 1990;172:4798–4806. doi: 10.1128/jb.172.9.4798-4806.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2822.Maurizi M R. ATP-promoted interaction between ClpA and ClpP in activation of Clp protease from Escherichia coli. Biochem Soc Trans. 1991;19:719–723. doi: 10.1042/bst0190719. [DOI] [PubMed] [Google Scholar]
- 2823.Maurizi M R, Trisler P, Gottesman S. Insertional mutagenesis of the lon gene in Escherichia coli: lon is dispensable. J Bacteriol. 1985;164:1124–1135. doi: 10.1128/jb.164.3.1124-1135.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2824.Maurizi M R, Clark W P, Katayama Y, Rudikoff S, Pumphrey J, Bowers B, Gottesman S. Sequence and structure of ClpP, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J Biol Chem. 1990;265:12536–12545. [PubMed] [Google Scholar]
- 2825.Maxon M E, Redfield B, Cai X Y, Shoeman R, Fujita K, Fisher W, Stauffer, Weissbach H, Brot N. Regulation of methionine synthesis in Escherichia coli: effect of the MetR protein on the expression of the metE and metR genes. Proc Natl Acad Sci USA. 1989;86:85–89. doi: 10.1073/pnas.86.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2826.May G, Faatz E, Lucht J M, Haardt M, Bolliger M, Bremer E. Characterization of the osmoregulated Escherichia coli proU promoter and identification of ProV as a membrane-associated protein. Mol Microbiol. 1989;3:1521–1531. doi: 10.1111/j.1365-2958.1989.tb00138.x. [DOI] [PubMed] [Google Scholar]
- 2827.May G, Faatz E, Villarejo M, Bremer E. Binding protein dependent transport of glycine betaine and its osmotic regulation in Escherichia coli. Mol Gen Genet. 1986;205:225–233. doi: 10.1007/BF00430432. [DOI] [PubMed] [Google Scholar]
- 2828.May G, Dersch P, Haardt M, Middendorf A, Bremer E. The osmZ (bglY) gene encodes the DNA-binding protein H-NS (H1a), a component of the Escherichia coli K12 nucleoid. Mol Gen Genet. 1990;224:81–90. doi: 10.1007/BF00259454. [DOI] [PubMed] [Google Scholar]
- 2829.Mayaux J-F, Fayat G, Fromant M, Springer M, Grunberg-Manago M. Structural and transcriptional evidence for related thrS and infC expression. Proc Natl Acad Sci USA. 1983;80:6152–6156. doi: 10.1073/pnas.80.20.6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2830.Mazel D, Pochet S, Marliere P. Genetic characterization of polypeptide deformylase, a distinctive enzyme of eubacterial translation. EMBO J. 1994;13:914–923. doi: 10.1002/j.1460-2075.1994.tb06335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2831.McAlister-Henn L, Blaber M, Bradshaw R A, Nisco S J. Complete nucleotide sequence of the Escherichia coli gene encoding malate dehydrogenase. Nucleic Acids Res. 1987;15:4993. doi: 10.1093/nar/15.12.4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2832.McCalla D R, Kaiser C, Green M H L. Genetics of nitrofurazone resistance in Escherichia coli. J Bacteriol. 1978;133:10–16. doi: 10.1128/jb.133.1.10-16.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2833.McCaman M T, McPartland A, Villarejo M. Genetics and regulation of peptidase N in Escherichia coli. J Bacteriol. 1982;152:848–854. doi: 10.1128/jb.152.2.848-854.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2834.McCaman M T, Gabe J D. Sequence of the promoter and 5′ coding region of pepN, and the amino-terminus of peptidase N from Escherichia coli K-12. Mol Gen Genet. 1986;204:148–152. doi: 10.1007/BF00330202. [DOI] [PubMed] [Google Scholar]
- 2835.McCaman M T, Gabe J D. The nucleotide sequence of the pepN gene and its over-expression in Escherichia coli. Gene. 1986;48:145–153. doi: 10.1016/0378-1119(86)90360-4. [DOI] [PubMed] [Google Scholar]
- 2836.McCarthy J E G, Schairer H U, Sebald W. Translational initiation frequency of atp genes from Escherichia coli: identification of an intercistronic sequence that enhances translation. EMBO J. 1985;4:519–526. doi: 10.1002/j.1460-2075.1985.tb03659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2837.McCarty J S, Walker G C. DnaK mutants defective in ATPase activity are defective in negative regulation of the heat shock response: expression of mutant DnaK proteins results in filamentation. J Bacteriol. 1994;176:764–780. doi: 10.1128/jb.176.3.764-780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2838.McClain M S, Blomfield I C, Eberhardt K J, Eisenstein B I. Inversion-independent phase variation of type 1 fimbriae in Escherichia coli. J Bacteriol. 1993;175:4335–4344. doi: 10.1128/jb.175.14.4335-4344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2839.McConville M L, Charles H P. Isolation of haemin-requiring mutants of Escherichia coli K12. J Gen Microbiol. 1979;113:155–164. doi: 10.1099/00221287-113-1-155. [DOI] [PubMed] [Google Scholar]
- 2840.McConville M L, Charles H P. Mutants of Escherichia coli K12 accumulating porphobilinogen: a new locus, hemC. J Gen Microbiol. 1979;111:193–200. doi: 10.1099/00221287-111-1-193. [DOI] [PubMed] [Google Scholar]
- 2841.McCulloch R, Coggins L W, Colloms S D, Sherratt D J. Xer-mediated site-specific recombination at cer generates Holliday junctions in vivo. EMBO J. 1994;13:1844–1855. doi: 10.1002/j.1460-2075.1994.tb06453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2842.McCulloch R, Burke M E, Sherratt D J. Peptidase activity of Escherichia coli aminopeptidase A is not required for its role in Xer site-specific recombination. Mol Microbiol. 1994;12:241–251. doi: 10.1111/j.1365-2958.1994.tb01013.x. [DOI] [PubMed] [Google Scholar]
- 2843.McDowall K J, Hernandez R G, Lin-Chao S, Cohen S N. The ams-1 and rne-3071 temperature-sensitive mutations in the ams gene are in close proximity to each other and cause substitutions within a domain that resembles a product of the Escherichia coli mre locus. J Bacteriol. 1993;175:4245–4249. doi: 10.1128/jb.175.13.4245-4249.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2844.McEwen J, Silverman P. Chromosomal mutations of Escherichia coli that alter expression of conjugative plasmid functions. Proc Natl Acad Sci USA. 1980;77:513–517. doi: 10.1073/pnas.77.1.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2845.McEwen J, Silverman P. Genetic analysis of Escherichia coli K-12 chromosomal mutants defective in expression of F-plasmid functions: identification of genes cpxA and cpxB. J Bacteriol. 1980;144:60–67. doi: 10.1128/jb.144.1.60-67.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2846.McFall E, Runkel L. DNA sequences of the d-serine deaminase control region and the N-terminal portion of the gene. J Bacteriol. 1983;154:1508–1512. doi: 10.1128/jb.154.3.1508-1512.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2847.McFall E, Heincz M C. Identification and control of synthesis of the dsdC activator protein. J Bacteriol. 1983;153:872–877. doi: 10.1128/jb.153.2.872-877.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2848.McFarland N, McCarter L, Artz S, Kustu S. Nitrogen regulatory locus “glnR” of enteric bacteria is composed of cistrons ntrB and ntrC: identification of their protein products. Proc Natl Acad Sci USA. 1981;78:2135–2139. doi: 10.1073/pnas.78.4.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2849.McFarland N, McCarter L, Artz S, Kustu S. Characterization of λ glnA+ phages used as templates for in vitro synthesis of glutamine synthetase. Mol Gen Genet. 1982;185:152–157. [Google Scholar]
- 2850.McGovern K, Beckwith J. Membrane insertion of the Escherichia coli MalF protein in cells with impaired secretion machinery. J Biol Chem. 1991;266:20870–20876. [PubMed] [Google Scholar]
- 2851.McGraw B R, Marinus M G. Isolation and characterization of Dam+ revertants and suppressor mutations that modify secondary phenotypes of dam-3 strains of Escherichia coli K-12. Mol Gen Genet. 1980;178:309–315. doi: 10.1007/BF00270477. [DOI] [PubMed] [Google Scholar]
- 2852.McKown R L, Orle K A, Chen T, Craig N L. Sequence requirements of Escherichia coli att Tn7, a specific site of transposon Tn7 insertion. J Bacteriol. 1988;170:352–358. doi: 10.1128/jb.170.1.352-358.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2853.McLaggan D, Logan T M, Lynn D G, Epstein W. Involvement of gamma-glutamyl peptides in osmoadaptation of Escherichia coli. J Bacteriol. 1990;172:3631–3636. doi: 10.1128/jb.172.7.3631-3636.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2854.McMurry L M, Levy S B. Tn5 insertion in the polynucleotide phosphorylase (pnp) gene in Escherichia coli increases susceptibility to antibiotics. J Bacteriol. 1987;169:1321–1324. doi: 10.1128/jb.169.3.1321-1324.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2855.McNicholas P M, Chiang R C, Gunsalus R P. The Escherichia coli modE gene: effect of modE mutations on molybdate dependent modA expression. FEMS Microbiol Lett. 1996;145:117–123. doi: 10.1016/0378-1097(96)00398-9. [DOI] [PubMed] [Google Scholar]
- 2856.McPherson M J, Baron A J, Pappin D J C, Wooton J C. Respiratory nitrate reductase of Escherichia coli. Sequence identification of the large subunit gene. FEBS Lett. 1984;177:260–264. doi: 10.1016/0014-5793(84)81295-8. [DOI] [PubMed] [Google Scholar]
- 2857.McPherson M J, Wootton J C. A complete nucleotide sequence of the Escherichia coli gdhA gene. Nucleic Acids Res. 1983;11:5257–5266. doi: 10.1093/nar/11.15.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2858.Meador J, Kennell D. Cloning and sequencing the gene encoding Escherichia coli ribonuclease I: exact physical mapping using the genome library. Gene. 1990;95:1–7. doi: 10.1016/0378-1119(90)90406-h. [DOI] [PubMed] [Google Scholar]
- 2859.Meadow N D, Saffen D W, Dottin R P, Roseman S. Molecular cloning of the crr gene and evidence that it is the structural gene for IIIGlc, a phosphocarrier protein of the bacterial phosphotransferase system. Proc Natl Acad Sci USA. 1982;79:2528–2532. doi: 10.1073/pnas.79.8.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2860.Mechulam Y, Fayat G, Blanquet S. Sequence of the E. coli pheST operon and identification of the himA gene. J Bacteriol. 1985;163:787–791. doi: 10.1128/jb.163.2.787-791.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2861.Mechulam Y, Fromant M, Mellot P, Plateau P, Blanchin-Roland S, Fayat G, Blanquet S. Molecular cloning of the Escherichia coli gene for diadenosine 5′,5′′′-P1,P4-tetraphosphate pyrophosphohydrolase. J Bacteriol. 1985;164:63–69. doi: 10.1128/jb.164.1.63-69.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2862.Mechulam Y, Blanquet S, Fayat G. Dual level control of the Escherichia coli pheST-himA operon expression. tRNAPhe-dependent attenuation and transcriptional operator-repressor control by himA and the SOS network. J Mol Biol. 1987;197:453–470. doi: 10.1016/0022-2836(87)90558-4. [DOI] [PubMed] [Google Scholar]
- 2863.Mecsas J, Welch R, Erickson J, Gross C A. Identification and characterization of an outer membrane protein, OmpX, in Escherichia coli that is homologous to a family of outer membrane proteins including Ail of Yersinia enterocolitica. J Bacteriol. 1995;177:799–804. doi: 10.1128/jb.177.3.799-804.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2864.Medina V, Pontarollo R, Glaeske D, Tabel H, Goldie H. Sequence of the pckA gene of Escherichia coli K-12: relevance to genetic and allosteric regulation and homology of E. coli phosphoenolpyruvate carboxykinase with the enzymes from Trypanosoma brucei and Saccharomyces cerevisiae. J Bacteriol. 1990;172:7151–7156. doi: 10.1128/jb.172.12.7151-7156.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2865.Meek D W, Hayward R S. Nucleotide sequence of the rpoA-rplQ DNA of Escherichia coli: a second regulatory binding site for protein S4? Nucleic Acids Res. 1984;12:5813–5821. doi: 10.1093/nar/12.14.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2866.Meier U, Mayer H. Genetic location of genes encoding enterobacterial common antigen. J Bacteriol. 1985;163:756–762. doi: 10.1128/jb.163.2.756-762.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2867.Meier-Dieter U, Barr K, Starman R, Hatch L P, Rick P D. Nucleotide sequence of the Escherichia coli rfe gene involved in the synthesis of enterobacterial common antigen. Molecular cloning of the rfe-rff gene cluster. J Biol Chem. 1992;267:746–753. [PubMed] [Google Scholar]
- 2868.Meier-Dieter U, Starman R, Barr K, Mayer H, Rick P D. Biosynthesis of enterobacterial common antigen in Escherichia coli. Biochemical characterization of Tn10 insertion mutants defective in enterobacterial common antigen synthesis. J Biol Chem. 1990;265:13490–13497. [PubMed] [Google Scholar]
- 2869.Meinnel T, Lazennec C, Blanquet S. Mapping of the active site zinc ligands of peptide deformylase. J Mol Biol. 1995;254:175–183. doi: 10.1006/jmbi.1995.0609. [DOI] [PubMed] [Google Scholar]
- 2870.Meinnel T, Guillon J-M, Mechulam Y, Blanquet S. The Escherichia coli fmt gene, encoding methionyl-tRNAfMet formyltransferase, escapes metabolic control. J Bacteriol. 1993;175:993–1000. doi: 10.1128/jb.175.4.993-1000.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2871.Meinnel T, Blanquet S. Evidence that peptide deformylase and methionyl-tRNAfMet formyltransferase are encoded within the same operon in Escherichia coli. J Bacteriol. 1993;175:7737–7740. doi: 10.1128/jb.175.23.7737-7740.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2872.Meinnel T, Blanquet S. Enzymatic properties of Escherichia coli peptide deformylase. J Bacteriol. 1995;177:1883–1887. doi: 10.1128/jb.177.7.1883-1887.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2873.Meinnel T, Blanquet S, Dardel F. A new subclass of the zinc metalloproteases superfamily revealed by the solution structure of peptide deformylase. J Mol Biol. 1996;262:375–386. doi: 10.1006/jmbi.1996.0521. [DOI] [PubMed] [Google Scholar]
- 2874.Mejean V, Iobbi-Nivol C, Lepelletier M, Giordano G, Chippaux M, Pascal M C. TMAO anaerobic respiration in Escherichia coli: involvement of the tor operon. Mol Microbiol. 1994;11:1169–1179. doi: 10.1111/j.1365-2958.1994.tb00393.x. [DOI] [PubMed] [Google Scholar]
- 2875.Melamede R J, Hatahet Z, Kow Y W, Ide H, Wallace S S. Isolation and characterization of endonuclease VIII from Escherichia coli. Biochemistry. 1994;33:1255–1264. doi: 10.1021/bi00171a028. [DOI] [PubMed] [Google Scholar]
- 2876.Melefors O, von Gabain A. Genetic studies of cleavage-initiated mRNA decay and processing of ribosomal 9S RNA show that the Escherichia coli ams and rne loci are the same. Mol Microbiol. 1991;5:857–864. doi: 10.1111/j.1365-2958.1991.tb00759.x. [DOI] [PubMed] [Google Scholar]
- 2877.Mellies J, Wise A, Villarejo M. Two different Escherichia coli proP promoters respond to osmotic and growth phase signals. J Bacteriol. 1995;177:144–151. doi: 10.1128/jb.177.1.144-151.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2878.Melville S B, Gunsalus R P. Mutations in fnr that alter anaerobic regulation of electron transport-associated genes in Escherichia coli. J Biol Chem. 1990;265:18733–18736. [PubMed] [Google Scholar]
- 2879.Membrillo-Hernandez J, Cook G M, Poole R K. Roles of RpoS (ςS) IHF and ppGpp in the expression of the hmp gene encoding the flavohemoglobin (Hmp) of Escherichia coli K-12. Mol Gen Genet. 1997;254:599–603. doi: 10.1007/s004380050457. [DOI] [PubMed] [Google Scholar]
- 2880.Membrillo-Hernandez J, Ioannidis N, Poole R K. The flavohaemoglobin (HMP) of Escherichia coli generates superoxide in vitro and causes oxidative stress in vivo. FEBS Lett. 1996;382:141–144. doi: 10.1016/0014-5793(96)00154-8. [DOI] [PubMed] [Google Scholar]
- 2881.Membrillo-Hernandez J, Kim S O, Cook G M, Poole R K. Paraquat regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12 is SoxRS independent but modulated by ςS. J Bacteriol. 1997;179:3164–3170. doi: 10.1128/jb.179.10.3164-3170.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2882.Mendelson I, Gottesman M M, Oppenheim A B. HU and integration host factor function as auxiliary proteins in cleavage of phage lambda cohesive ends by terminase. J Bacteriol. 1991;173:1670–1676. doi: 10.1128/jb.173.5.1670-1676.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2883.Mendonca V M, Klepin H D, Matson S W. DNA helicases in recombination and repair: construction of a ΔuvrD ΔhelD ΔrecQ mutant deficient in recombination and repair. J Bacteriol. 1995;177:1326–1335. doi: 10.1128/jb.177.5.1326-1335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2884.Mendonca V M, Kaiser-Rogers K, Matson S W. Double helicase II (uvrD)-helicase IV(helD) deletion mutants are defective in the recombination pathways of Escherichia coli. J Bacteriol. 1993;175:4641–4651. doi: 10.1128/jb.175.15.4641-4651.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2885.Mendonca V M, Matson S W. Genetic analysis of delta helD and delta uvrD mutations in combination with other genes in the RecF recombination pathway in Escherichia coli: suppression of a ruvB mutation by a uvrD deletion. Genetics. 1995;141:443–452. doi: 10.1093/genetics/141.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2886.Meng S Y, Bennett G N. Nucleotide sequence of the Escherichia coli cad operon: a system for neutralization of low extracellular pH. J Bacteriol. 1992;174:2659–2669. doi: 10.1128/jb.174.8.2659-2669.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2887.Meng S Y, Bennett G N. Regulation of the Escherichia coli cad operon: location of a site required for acid induction. J Bacteriol. 1992;174:2670–2678. doi: 10.1128/jb.174.8.2670-2678.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2888.Meng W, Green J, Guest J R. FNR-dependent repression of ndh gene expression requires two upstream FNR-binding sites. Microbiology. 1997;143:1521–1532. doi: 10.1099/00221287-143-5-1521. [DOI] [PubMed] [Google Scholar]
- 2889.Mengin-Lecreulx D, Parquet C, Desviat L R, Pla J, Flouret B, Ayala J A. Organization of the murE-murG region of Escherichia coli: identification of the murD gene encoding the d-glutamic-acid-adding enzyme. J Bacteriol. 1989;171:6126–6134. doi: 10.1128/jb.171.11.6126-6134.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2890.Mengin-Lecreulx D, van Heijenoort J. Nucleotide sequence of the murD gene encoding the UDP-MurNAc-l-Ala-d-Glu synthetase of Escherichia coli. Nucleic Acids Res. 1990;18:183. doi: 10.1093/nar/18.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2891.Mengin-Lecreulx D, van Heijenoort J. Identification of the glmU gene encoding N-acetylglucosamine-1-phosphate uridyltransferase in Escherichia coli. J Bacteriol. 1993;175:6150–6157. doi: 10.1128/jb.175.19.6150-6157.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2892.Mengin-Lecreulx D, van Heijenoort J. Copurification of glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-phosphate uridyltransferase activities of Escherichia coli: characterization of the glmU gene product as a bifunctional enzyme catalyzing two consecutive steps in the pathway for UDP-N-acetylglucosamine synthesis. J Bacteriol. 1994;176:5788–5795. doi: 10.1128/jb.176.18.5788-5795.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2893.Mengin-Lecreulx D, van Heijenoort J. Characterization of the essential gene glmM encoding phosphoglucosamine mutase in Escherichia coli. J Biol Chem. 1996;271:32–39. doi: 10.1074/jbc.271.1.32. [DOI] [PubMed] [Google Scholar]
- 2894.Mengin-Lecreulx D, van Heijenoort J, Park J T. Identification of the mpl gene encoding UDP-N-acetylmuramate:l-alanyl-γ-d-glutamyl-meso-diaminopimelate ligase in Escherichia coli and its role in recycling of cell wall peptidoglycan. J Bacteriol. 1996;178:5347–5352. doi: 10.1128/jb.178.18.5347-5352.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2895.Mengin-Lecreulx D, Texier L, Rousseau M, van Heijenoort J. The murG gene of Escherichia coli codes for the UDP-N-acetylglucosamine:N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase involved in the membrane steps of peptidoglycan synthesis. J Bacteriol. 1991;173:4625–4636. doi: 10.1128/jb.173.15.4625-4636.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2896.Menon N K, Chatelus C Y, Dervartanian M, Wendt J C, Shanmugam K T, Peck H D, Przybyla A E. Cloning, sequencing, and mutational analysis of the hyb operon encoding Escherichia coli hydrogenase 2. J Bacteriol. 1994;176:4416–4423. doi: 10.1128/jb.176.14.4416-4423.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2897.Menon N K, Robbins J, Peck H D, Jr, Chatelus C Y, Choi E-S, Przybyla A E. Cloning and sequencing of a putative Escherichia coli [NiFe] hydrogenase-1 operon containing six open reading frames. J Bacteriol. 1990;172:1969–1977. doi: 10.1128/jb.172.4.1969-1977.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2898.Menon N K, Robbins J, Wendt J C, Shanmugam K T, Przybyla A E. Mutational analysis and characterization of the Escherichia coli hya operon, which encodes [NiFe] hydrogenase 1. J Bacteriol. 1991;173:4851–4861. doi: 10.1128/jb.173.15.4851-4861.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2899.Menzel R, Roth J R. Identification and mapping of a second proline permease in Salmonella typhimurium. J Bacteriol. 1980;141:1064–1070. doi: 10.1128/jb.141.3.1064-1070.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2900.Menzel R, Gellert M. Fusions of the Escherichia coli gyrA and gyrB control regions to the galactokinase gene are inducible by coumermycin treatment. J Bacteriol. 1987;169:1272–1278. doi: 10.1128/jb.169.3.1272-1278.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2901.Menzel R, Gellert M. Modulation of transcription by DNA supercoiling: a deletion analysis of the Escherichia coli gyrA and gyrB promoters. Proc Natl Acad Sci USA. 1987;84:4185–4189. doi: 10.1073/pnas.84.12.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2902.Merkel T J, Dahl J L, Ebright R H, Kadner R J. Transcription activation at the Escherichia coli uhpT promoter by the catabolite gene activator protein. J Bacteriol. 1995;177:1712–1718. doi: 10.1128/jb.177.7.1712-1718.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2903.Merkel W K, Nichols B P. Characterization and sequence of the Escherichia coli panBCD gene cluster. FEMS Microbiol Lett. 1996;143:247–252. doi: 10.1111/j.1574-6968.1996.tb08488.x. [DOI] [PubMed] [Google Scholar]
- 2904.Merrick M, Jones D H A, Thomas C M. Location of the rpoN gene on the physical map of Escherichia coli. J Bacteriol. 1993;175:1548–1549. doi: 10.1128/jb.175.5.1548-1549.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2905.Messer W, Meijer M, Bergmans H E N, Hansen F G, von Meyenburg K, Beck E, Schaller H. Origin of replication, oriC, of the Escherichia coli K12 chromosome: nucleotide sequence. Cold Spring Harbor Symp Quant Biol. 1978;43:139–145. doi: 10.1101/sqb.1979.043.01.020. [DOI] [PubMed] [Google Scholar]
- 2906.Metcalf W W, Wanner B L. Involvement of the Escherichia coli phn (psiD) gene cluster in assimilation of phosphorus in the form of phosphonates, phosphite, Pi esters, and Pi. J Bacteriol. 1991;173:587–600. doi: 10.1128/jb.173.2.587-600.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2907.Metcalf W W, Wanner B L. Evidence for a fourteen-gene, phnC to phnP locus for phosphonate metabolism in Escherichia coli. Gene. 1993;129:27–32. doi: 10.1016/0378-1119(93)90692-v. [DOI] [PubMed] [Google Scholar]
- 2908.Metcalf W W, Wanner B L. Mutational analysis of an Escherichia coli fourteen-gene operon for phosphonate degradation, using TnphoA′ elements. J Bacteriol. 1993;175:3430–3442. doi: 10.1128/jb.175.11.3430-3442.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2909.Metcalf W W, Steed P M, Wanner B L. Identification of phosphate starvation-inducible genes in Escherichia coli K-12 by DNA sequence analysis of psi::lacZ(Mu d1) transcriptional fusions. J Bacteriol. 1990;172:3191–3200. doi: 10.1128/jb.172.6.3191-3200.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2910.Metheringham R, Griffiths L, Crooke H, Forsythe S, Cole J A. An essential role for DsbA in cytochrome c synthesis and formate-dependent nitrite reduction by Escherichia coli K-12. Arch Microbiol. 1995;164:301–307. doi: 10.1007/BF02529965. [DOI] [PubMed] [Google Scholar]
- 2911.Metzer E, Halpern Y S. Mutations affecting the regulation of gamma-aminobutyrate utilization in Escherichia coli K-12. Curr Microbiol. 1980;4:51–55. [Google Scholar]
- 2912.Metzer E, Halpern Y S. In vivo cloning and characterization of the gabCTDP gene cluster of Escherichia coli K-12. J Bacteriol. 1990;172:3250–3256. doi: 10.1128/jb.172.6.3250-3256.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2913.Metzger S, Sarubbi E, Glaser G, Cashel M. Protein sequences encoded by the relA and the spoT genes of Escherichia coli are interrelated. J Biol Chem. 1989;264:9122–9125. [PubMed] [Google Scholar]
- 2914.Metzger S, Schreiber G, Aizenman E, Cashel M, Glaser G. Characterization of the relA1 mutation and a comparison of relA1 with new relA null alleles in Escherichia coli. J Biol Chem. 1989;264:21146–21152. [PubMed] [Google Scholar]
- 2915.Metzger S, Dror I B, Aizenman E, Schreiber G, Toone M, Friesen J D, Cashel M, Glaser G. The nucleotide sequence and characterization of the relA gene of Escherichia coli. J Biol Chem. 1988;263:15699–15704. [PubMed] [Google Scholar]
- 2916.Meyer R R, Rein D C, Glassberg J. The product of the lexC gene of Escherichia coli is single-stranded DNA-binding protein. J Bacteriol. 1982;150:433–435. doi: 10.1128/jb.150.1.433-435.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2917.Michael N P, Brehm J K, Anlezark G M, Minton N P. Physical characterisation of the Escherichia coli B gene encoding nitroreductase and its over-expression in Escherichia coli K12. FEMS Microbiol Lett. 1994;124:195–202. doi: 10.1111/j.1574-6968.1994.tb07284.x. [DOI] [PubMed] [Google Scholar]
- 2918.Michaeli S, Ron E Z. The metC gene in Escherichia coli K-12: isolation and studies of relatedness in Enterobacteriaceae. FEMS Microbiol Lett. 1984;22:31–35. [Google Scholar]
- 2919.Michaeli S, Ron E Z, Cohen G. Construction and physical mapping of plasmids containing the metA gene of Escherichia coli K-12. Mol Gen Genet. 1981;182:349–354. doi: 10.1007/BF00269682. [DOI] [PubMed] [Google Scholar]
- 2920.Michaeli S, Mevarech M, Ron E Z. Regulatory region of the metA gene of Escherichia coli K-12. J Bacteriol. 1984;160:1158–1162. doi: 10.1128/jb.160.3.1158-1162.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2921.Michaels M L, Cruz C, Miller J H. mutA and mutC: two mutator loci in Escherichia coli that stimulate transversions. Proc Natl Acad Sci USA. 1990;87:9211–9215. doi: 10.1073/pnas.87.23.9211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2922.Michaels M L, Pham L, Nghiem Y, Cruz C, Miller J H. MutY, an adenine glycosylase active on G-A mispairs, has homology to endonuclease III. Nucleic Acids Res. 1990;18:3841–3845. doi: 10.1093/nar/18.13.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2923.Michaud C, Parquet C, Flouret B, Blanot D, van Heijenoort J. Revised interpretation of the sequence containing the murE gene encoding the UDP-N-acetylmuramyl-tripeptide synthetase of Escherichia coli. Biochem J. 1990;269:277–278. doi: 10.1042/bj2690277. . (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2924.Miczak A, Chauhan A K, Apirion D. Two new genes located between 2758 and 2761 kilobase pairs on the Escherichia coli genome. J Bacteriol. 1991;173:3271–3272. doi: 10.1128/jb.173.11.3271-3272.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2925.Middendorf A, Schweizer H P, Vreemann J, Boos W. Mapping the markers in the his-gyrA region of Escherichia coli. Mol Gen Genet. 1984;197:175–181. doi: 10.1007/BF00327939. [DOI] [PubMed] [Google Scholar]
- 2926.Mihara H, Kurihara T, Yoshimura T, Soda K, Esaki N. Cysteine sulfinate desulfinase, a NIFS-like protein of Escherichia coli with selenocysteine lyase and cysteine desulfurase activities. Gene cloning, purification, and characterization of a novel pyridoxal enzyme. J Biol Chem. 1997;272:22417–22424. doi: 10.1074/jbc.272.36.22417. [DOI] [PubMed] [Google Scholar]
- 2927.Miki T, Kimura M, Hiraga S, Nagata T, Yura T. Cloning and physical mapping of the dnaA region of the Escherichia coli chromosome. J Bacteriol. 1979;140:817–824. doi: 10.1128/jb.140.3.817-824.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2928.Miki T, Ebina Y, Kishi F, Nakazawa A. Organization of the lexA gene of Escherichia coli and nucleotide sequence of the regulatory region. Nucleic Acids Res. 1981;9:529–543. doi: 10.1093/nar/9.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2929.Mikulskis A V, Cornelis G R. A new class of proteins regulating gene expression in enterobacteria. Mol Microbiol. 1994;11:77–86. doi: 10.1111/j.1365-2958.1994.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 2930.Mikuni O, Ito K, Moffat J, Matsumura K, McCaughan K, Nobukuni T, Tate W, Nakamura Y. Identification of the prfC gene, which encodes peptide-chain-release factor 3 of Escherichia coli. Proc Natl Acad Sci USA. 1994;91:5798–5802. doi: 10.1073/pnas.91.13.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2931.Mildener B, Fondy T P, Engel R, Tropp B E. Effects of halo analogs of glycerol 3-phosphate and dihydroxy-acetone phosphate upon Escherichia coli. Antimicrob Agents Chemother. 1981;19:678–681. doi: 10.1128/aac.19.4.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2932.Miles J S, Guest J R. Complete nucleotide sequence of the fumarase gene, fumA, of Escherichia coli. Nucleic Acids Res. 1984;12:3631–3642. doi: 10.1093/nar/12.8.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2933.Miles J S, Guest J R. Nucleotide sequence and transcriptional start point of the phosphomannose isomerase gene (manA) of Escherichia coli. Gene. 1984;32:41–48. doi: 10.1016/0378-1119(84)90030-1. [DOI] [PubMed] [Google Scholar]
- 2934.Miles J S, Guest J R. Subgenes expressing single lipoyl domains of the pyruvate dehydrogenase complex of Escherichia coli. Biochem J. 1987;245:869–874. doi: 10.1042/bj2450869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2935.Milkman R. An Escherichia coli homologue of eukaryotic potassium channel proteins. Proc Natl Acad Sci USA. 1994;91:3510–3514. doi: 10.1073/pnas.91.9.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2936.Millar G, Lewendon A, Hunter M G, Coggins J R. The cloning and expression of the aroL gene from Escherichia coli K12. Purification and complete amino acid sequence of shikimate kinase II, the aroL-gene product. Biochem J. 1986;237:427–437. doi: 10.1042/bj2370427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2937.Millar G, Coggins J R. The complete amino acid sequence of 3-dehydroquinate synthase of Escherichia coli K12. FEBS Lett. 1986;200:11–17. doi: 10.1016/0014-5793(86)80501-4. [DOI] [PubMed] [Google Scholar]
- 2938.Miller C G, Schwartz G. Peptidase-deficient mutants of Escherichia coli. J Bacteriol. 1978;135:603–611. doi: 10.1128/jb.135.2.603-611.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2939.Miller H I. Primary structure of the himA gene of Escherichia coli: homology with DNA-binding protein HU and association with the phenylalanyl-tRNA synthetase operon. Cold Spring Harbor Symp Quant Biol. 1984;49:691–698. doi: 10.1101/sqb.1984.049.01.078. [DOI] [PubMed] [Google Scholar]
- 2940.Miller H I, Friedman D I. Isolation of Escherichia coli mutants unable to support lambda integrative recombination. Cold Spring Harbor Symp Quant Biol. 1977;42:349–356. [Google Scholar]
- 2941.Miller H I, Friedman D I. An E. coli gene product required for λ site-specific recombination. Cell. 1980;20:711–719. doi: 10.1016/0092-8674(80)90317-7. [DOI] [PubMed] [Google Scholar]
- 2942.Miller H I, Nash H A. Direct role of the himA gene product in phage λ integration. Nature. 1981;290:523–526. doi: 10.1038/290523a0. [DOI] [PubMed] [Google Scholar]
- 2943.Miller H I, Kirk M, Echols H. SOS induction and autoregulation of the himA gene for site-specific recombination in Escherichia coli. Proc Natl Acad Sci USA. 1981;78:6754–6758. doi: 10.1073/pnas.78.11.6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2944.Miller H I, Kikuchi Y, Nash H A, Weisberg R, Friedman D I. Site-specific recombination of bacteriophage λ: the role of host gene products. Cold Spring Harbor Symp Quant Biol. 1978;43:1121–1126. doi: 10.1101/sqb.1979.043.01.125. [DOI] [PubMed] [Google Scholar]
- 2945.Miller K W, Wu H C. Cotranscription of the isoleucyl-tRNA synthetase (ileS) and prolipoprotein signal peptidase(lsp) genes: fine-structure mapping of the lsp internal promoter. J Biol Chem. 1987;262:389–393. [PubMed] [Google Scholar]
- 2946.Miller S, Douglas R M, Carter P, Booth I R. Mutations in the glutathione-gated KefC K+ efflux system of Escherichia coli that cause constitutive activation. J Biol Chem. 1997;272:24942–24947. doi: 10.1074/jbc.272.40.24942. [DOI] [PubMed] [Google Scholar]
- 2947.Milner J L, Wood J M. Insertion proQ220::Tn5 alters regulation of proline porter II, a transporter of proline and glycine betaine in Escherichia coli. J Bacteriol. 1989;171:947–951. doi: 10.1128/jb.171.2.947-951.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2948.Minagawa J, Nakamura H, Yamato I, Mogi T, Anraku Y. Transcriptional regulation of the cytochrome b562-o complex in Escherichia coli. Gene expression and molecular characterization of the promoter. J Biol Chem. 1990;265:11198–11203. [PubMed] [Google Scholar]
- 2949.Mineno J, Fukui H, Ishino Y, Kato I, Shinagawa H. Nucleotide sequence of the araD gene of Escherichia coli K12 encoding the l-ribulose 5-phosphate 4-epimerase. Nucleic Acids Res. 1990;18:6722. doi: 10.1093/nar/18.22.6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2950.Miner K M, Frank L. Sodium-stimulated glutamate transport in osmotically shocked cells and membrane vesicles of Escherichia coli. J Bacteriol. 1974;117:1093–1098. doi: 10.1128/jb.117.3.1093-1098.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2951.Minoshima S, Hayashi H. Studies on bacterial chemotaxis. VI. Effect of cheX mutation on the methylation of methyl-accepting chemotaxis protein of Escherichia coli. J Biochem (Tokyo) 1980;87:1371–1377. doi: 10.1093/oxfordjournals.jbchem.a132877. [DOI] [PubMed] [Google Scholar]
- 2952.Miranda-Rios J, Sanchez-Pescador R, Urdea M, Covarrubias A. The complete nucleotide sequence of the glnALG operon of Escherichia coli K12. Nucleic Acids Res. 1987;15:2757–2770. doi: 10.1093/nar/15.6.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2953.Miranda-Vizuete A, Damdimopoulos A E, Gustafsson J, Spyrou G. Cloning, expression, and characterization of a novel Escherichia coli thioredoxin. J Biol Chem. 1997;272:30841–30847. doi: 10.1074/jbc.272.49.30841. [DOI] [PubMed] [Google Scholar]
- 2954.Misra R. A novel ompC mutation of Escherichia coli K12 that reduces OmpC and OmpF levels in the outer membrane. Mol Microbiol. 1993;10:1029–1035. doi: 10.1111/j.1365-2958.1993.tb00973.x. [DOI] [PubMed] [Google Scholar]
- 2955.Misra R, Reeves P R. Molecular characterization of the Stc− mutation of Escherichia coli K-12. Gene. 1985;40:337–342. doi: 10.1016/0378-1119(85)90058-7. [DOI] [PubMed] [Google Scholar]
- 2956.Misra R, Benson S A. A novel mutation, cog, which results in production of a new porin protein (OmpG) of Escherichia coli K-12. J Bacteriol. 1989;171:4105–4111. doi: 10.1128/jb.171.8.4105-4111.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2957.Misra R, Miao Y. Molecular analysis of asmA, a locus identified as the suppressor of OmpF assembly mutants of Escherichia coli K-12. Mol Microbiol. 1995;16:779–788. doi: 10.1111/j.1365-2958.1995.tb02439.x. [DOI] [PubMed] [Google Scholar]
- 2958.Misra T K, Apirion D. Gene rne affects the structure of the ribonucleic acid-processing enzyme ribonuclease E of Escherichia coli. J Bacteriol. 1980;142:359–361. doi: 10.1128/jb.142.1.359-361.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2959.Missiakas D, Georgopoulos C, Raina S. The Escherichia coli rfaH gene, mutational analysis, cloning, sequencing, and transcriptional regulation: a positive transcriptional regulatory gene for htrM and htrL genes. 1992. GenBank submission M94888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2960.Missiakas D, Georgopoulos C, Raina S. Identification and characterization of the Escherichia coli gene dsbB, whose product is involved in the formation of disulfide bonds in vivo. Proc Natl Acad Sci USA. 1993;90:7084–7088. doi: 10.1073/pnas.90.15.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2961.Missiakas D, Georgopoulos C, Raina S. The Escherichia coli heat shock gene htpY: mutational analysis, cloning, sequencing, and transcriptional regulation. J Bacteriol. 1993;175:2613–2624. doi: 10.1128/jb.175.9.2613-2624.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2962.Missiakas D, Georgopoulos C, Raina S. The Escherichia coli dsbC (xprA) gene encodes a periplasmic protein involved in disulfide bond formation. EMBO J. 1994;13:2013–2020. doi: 10.1002/j.1460-2075.1994.tb06471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2963.Missiakas D, Schwager F, Betton J M, Georgopoulos C, Raina S. Identification and characterization of HsIV HsIU (ClpQ ClpY) proteins involved in overall proteolysis of misfolded proteins in Escherichia coli. EMBO J. 1996;15:6899–6909. [PMC free article] [PubMed] [Google Scholar]
- 2964.Missiakas D, Betton J M, Raina S. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol Microbiol. 1996;21:871–884. doi: 10.1046/j.1365-2958.1996.561412.x. [DOI] [PubMed] [Google Scholar]
- 2965.Missiakas D, Mayer M P, Lemaire M, Georgopoulos C, Raina S. Modulation of the Escherichia coli ςE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol Microbiol. 1997;24:355–371. doi: 10.1046/j.1365-2958.1997.3601713.x. [DOI] [PubMed] [Google Scholar]
- 2966.Missiakas D, Raina S. Signal transduction pathways in response to protein misfolding in the extracytoplasmic compartments of E. coli: role of two new phosphoprotein phosphatases PrpA and PrpB. EMBO J. 1997;16:1670–1685. doi: 10.1093/emboj/16.7.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2967.Mitra S, Pal B C, Foote R S. O6-methylguanine-DNA methyltransferase in wild-type and ada mutants of Escherichia coli. J Bacteriol. 1982;152:534–537. doi: 10.1128/jb.152.1.534-537.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2968.Miura K, Tomioka Y, Suzuki H, Yonezawa M, Hishinuma T, Mizugaki M. Molecular cloning of the nemA gene encoding N-ethylmaleimide reductase from Escherichia coli. Biol Pharm Bull. 1997;20:110–112. doi: 10.1248/bpb.20.110. [DOI] [PubMed] [Google Scholar]
- 2969.Miwa Y, Horiguchi T, Shigesada K. Structural and functional dissections of transcription termination factor rho by random mutagenesis. J Mol Biol. 1995;254:815–837. doi: 10.1006/jmbi.1995.0658. [DOI] [PubMed] [Google Scholar]
- 2970.Miyada C G, Soberon X, Itakura K, Wilcox G. The use of synthetic oligodeoxyribonucleotides to produce specific deletions in the araBAD promoter of Escherichia coli B/r. Gene. 1982;17:167–177. doi: 10.1016/0378-1119(82)90070-1. [DOI] [PubMed] [Google Scholar]
- 2971.Miyada C G, Horwitz A H, Cass L G, Timko J, Wilcox G. DNA sequence of the araC regulatory gene from Escherichia coli B/r. Nucleic Acids Res. 1980;8:5267–5274. doi: 10.1093/nar/8.22.5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2972.Miyajima A, Shibuya M, Kuchino Y, Kaziro Y. Transcription of the E. coli tufB gene: cotranscription with four tRNA genes and inhibition by guanosine-5′-diphosphate-3′-diphosphate. Mol Gen Genet. 1981;183:13–19. doi: 10.1007/BF00270131. [DOI] [PubMed] [Google Scholar]
- 2973.Miyake H, Yabu H, Satoh T, Yamamoto I. Characterization and transcriptional regulation of the modABCD genes for molybdenum transport in Escherichia coli. Nucleic Acids Symp Ser. 1995;34:91–92. [PubMed] [Google Scholar]
- 2974.Miyamoto K, Nakahigashi K, Nishimura K, Inokuchi H. Isolation and characterization of visible light-sensitive mutants of Escherichia coli K12. J Mol Biol. 1991;219:393–398. doi: 10.1016/0022-2836(91)90180-e. [DOI] [PubMed] [Google Scholar]
- 2975.Miyoshi Y, Yamagata H. Sucrose-dependent spectinomycin-resistant mutants of Escherichia coli. J Bacteriol. 1976;125:142–148. doi: 10.1128/jb.125.1.142-148.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2976.Mizote T, Nakayama H. The thiM locus and its relation to phosphorylation of hydroxyethylthiazole in Escherichia coli. J Bacteriol. 1989;171:3228–3232. doi: 10.1128/jb.171.6.3228-3232.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2977.Mizote T, Tsuda M, Nakazawa T, Nakayama H. The thiJ locus and its relation to phosphorylation of hydroxymethylpyrimidine in Escherichia coli. Microbiology. 1996;142:2969–2974. doi: 10.1099/13500872-142-10-2969. [DOI] [PubMed] [Google Scholar]
- 2978.Mizuno T, Wurtzel E, Inouye M. Cloning of the regulatory genes (ompR and envZ) for the matrix proteins of the Escherichia coli outer membrane. J Bacteriol. 1982;150:1462–1466. doi: 10.1128/jb.150.3.1462-1466.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2979.Mizuno T, Wurtzel E, Inouye M. Osmoregulation of gene expression. II. DNA sequence of the envZ gene of the ompB operon of Escherichia coli and characterization of its gene product. J Biol Chem. 1982;257:13692–13698. [PubMed] [Google Scholar]
- 2980.Mizuno T, Chou M-Y, Inouye M. A comparative study on the genes for three porins of the Escherichia coli outer membrane: DNA sequence of the osmoregulated ompC gene. J Biol Chem. 1983;258:6932–6940. [PubMed] [Google Scholar]
- 2981.Mizuno T, Chou M-Y, Inouye M. DNA sequence of the promoter region of the ompC gene and the amino acid sequence of the signal peptide of pro-OmpC protein of Escherichia coli. FEBS Lett. 1983;151:159–164. doi: 10.1016/0014-5793(83)80364-0. [DOI] [PubMed] [Google Scholar]
- 2982.Mizuno T, Chou M-Y, Inouye M. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA) Proc Natl Acad Sci USA. 1984;81:1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2983.Mizuno T, Mizushima S. Isolation and characterization of deletion mutants of ompR and envZ, regulatory genes for expression of the outer membrane proteins OmpC and OmpF in Escherichia coli. J Biochem (Tokyo) 1987;101:387–396. doi: 10.1093/oxfordjournals.jbchem.a121923. [DOI] [PubMed] [Google Scholar]
- 2984.Mizusawa S, Court D L, Gottesman S. Transcription of the sulA gene and expression by lexA. J Mol Biol. 1983;171:337–343. doi: 10.1016/0022-2836(83)90097-9. [DOI] [PubMed] [Google Scholar]
- 2985.Mizushima T, Sasaki S, Ohishi H, Kobayashi M, Katayama T, Miki T, Maeda M, Sekimizu K. Molecular design of inhibitors of in vitro oriC DNA replication based on the potential to block the ATP binding of DnaA protein. J Biol Chem. 1996;271:25178–25183. doi: 10.1074/jbc.271.41.25178. [DOI] [PubMed] [Google Scholar]
- 2986.Mizushima T, Katayama T, Sekimizu K. Effect on DNA topology by DnaA protein, the initiation factor of chromosomal DNA replication in Escherichia coli. Biochemistry. 1996;35:11512–11516. doi: 10.1021/bi953088f. [DOI] [PubMed] [Google Scholar]
- 2987.Model P, Jovanovic G, Dworkin J. The Escherichia coli phage-shock-protein (psp) operon. Mol Microbiol. 1997;24:255–261. doi: 10.1046/j.1365-2958.1997.3481712.x. [DOI] [PubMed] [Google Scholar]
- 2988.Moffat K G, Mackinnon A. Cloning of the Escherichia coli K-12 guaC gene following its transposition into the RP4:Mu cointegrate. Gene. 1985;40:141–143. doi: 10.1016/0378-1119(85)90034-4. [DOI] [PubMed] [Google Scholar]
- 2989.Mogi T, Yamamoto H, Nakao T, Yamato I, Anraku Y. Genetic and physical characterization of putP, the proline carrier gene of Escherichia coli K12. Mol Gen Genet. 1986;202:35–41. doi: 10.1007/BF00330513. [DOI] [PubMed] [Google Scholar]
- 2990.Mohan S, Kelly T M, Eveland S S, Raetz C R H, Anderson M S. An Escherichia coli gene (fabZ) encoding (3R)-hydroxymyristoyl acyl carrier protein dehydrase. J Biol Chem. 1994;269:32896–32903. [PubMed] [Google Scholar]
- 2991.Moir P D, Spiegelberg R, Oliver I R, Pringle J H, Masters M. Proteins encoded by the Escherichia coli replication terminus region. J Bacteriol. 1992;174:2102–2110. doi: 10.1128/jb.174.7.2102-2110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2992.Molnar J, Holland I B, Mandi Y. Selection of lon mutants in Escherichia coli by treatment with phenothiazines. Genet Res. 1977;30:13–20. doi: 10.1017/s0016672300017420. [DOI] [PubMed] [Google Scholar]
- 2993.Monticello R A, Angov E, Brusilow W S A. Effects of inducing expression of cloned genes for the F0 proton channel of the Escherichia coli F1F0 ATPase. J Bacteriol. 1992;174:3370–3376. doi: 10.1128/jb.174.10.3370-3376.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2994.Monticello R A, Brusilow W S A. Role of the δ subunit in enhancing proton conduction through the F0 of the Escherichia coli F1F0 ATPase. J Bacteriol. 1994;176:1383–1389. doi: 10.1128/jb.176.5.1383-1389.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2995.Moolenaar G F, Van Sluis C A, Backendorf C, Van de Putte P. Regulation of the Escherichia coli excision repair gene uvrC. Overlap between the uvrC structural gene and the region coding for a 24 kDa protein. Nucleic Acids Res. 1987;15:4273–4289. doi: 10.1093/nar/15.10.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2996.Moore R C, Boyle S M. Nucleotide sequence and analysis of the speA gene encoding biosynthetic arginine decarboxylase in Escherichia coli. J Bacteriol. 1990;172:4631–4640. doi: 10.1128/jb.172.8.4631-4640.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2997.Moore S, James E. Mapping of restriction sites in the argF gene of Escherichia coli by partial endonuclease digestion of end-labeled DNA. Gene. 1979;5:159–175. doi: 10.1016/0378-1119(79)90100-8. . (Erratum, 5:343–344, 1979.) [DOI] [PubMed] [Google Scholar]
- 2998.Moore S K, Garvin R T, James E. Nucleotide sequence of the argF regulatory region of Escherichia coli K-12. Gene. 1981;16:119–132. doi: 10.1016/0378-1119(81)90068-8. [DOI] [PubMed] [Google Scholar]
- 2999.Moralejo P, Egan S M, Hidalgo E, Aguilar J. Sequencing and characterization of a gene cluster encoding the enzymes for l-rhamnose metabolism in Escherichia coli. J Bacteriol. 1993;175:5585–5594. doi: 10.1128/jb.175.17.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3000.Moran M C, Mazaitis A J, Vogel R H, David A D. Clustered arg genes on a BamHI segment of the Escherichia coli chromosome. Gene. 1979;8:25–34. doi: 10.1016/0378-1119(79)90005-2. [DOI] [PubMed] [Google Scholar]
- 3001.Morgan S, Korner A, Low K B, Soll D. Regulation of biosynthesis of aminoacyl-tRNA synthetases and of tRNA in Escherichia coli. I. Isolation and characterization of a mutant with elevated levels of tRNA1Gln. J Mol Biol. 1977;117:1013–1031. doi: 10.1016/s0022-2836(77)80010-7. [DOI] [PubMed] [Google Scholar]
- 3002.Morimyo M. Isolation and characterization of methyl viologen-sensitive mutants of Escherichia coli K-12. J Bacteriol. 1988;170:2136–2142. doi: 10.1128/jb.170.5.2136-2142.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3003.Morimyo M, Hongo E, Hama-Inaba H, Machida I. Cloning and characterization of the mvrC gene of Escherichia coli K-12 which confers resistance against methyl viologen toxicity. Nucleic Acids Res. 1992;20:3159–3165. doi: 10.1093/nar/20.12.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3004.Morita T, Amemura M, Makino K, Shinagawa H, Magota K, Otsuji N, Nakata A. Hyperproduction of phosphate-binding protein, PhoS, and pre-PhoS proteins in Escherichia coli carrying a cloned phoS gene. Eur J Biochem. 1983;130:427–435. doi: 10.1111/j.1432-1033.1983.tb07169.x. [DOI] [PubMed] [Google Scholar]
- 3005.Moriya H, Kasai H, Isono K. Cloning and characterization of the hrpA gene in the terC region of Escherichia coli that is highly similar to the DEAH family RNA helicase genes of Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:595–598. doi: 10.1093/nar/23.4.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3006.Morona R, Klose M, Henning U. Escherichia coli K-12 outer membrane protein (OmpA) as a bacteriophage receptor: analysis of mutant genes expressing altered proteins. J Bacteriol. 1984;159:570–578. doi: 10.1128/jb.159.2.570-578.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3007.Morona R, Reeves P R. Molecular cloning of the tolC locus of Escherichia coli K-12 with the use of transposon Tn10. Mol Gen Genet. 1981;184:430–433. doi: 10.1007/BF00352517. [DOI] [PubMed] [Google Scholar]
- 3008.Morona R, Reeves P R. A new locus, stc, which affects the phenotype of tolC mutants of Escherichia coli K-12. Mol Gen Genet. 1982;187:335–341. [Google Scholar]
- 3009.Morona R, Reeves P R. The tolC locus of Escherichia coli affects the expression of three major outer membrane proteins. J Bacteriol. 1982;150:1016–1023. doi: 10.1128/jb.150.3.1016-1023.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3010.Morona R, Henning U. New locus (ttr) in Escherichia coli K-12 affecting sensitivity to bacteriophage T2 and growth on oleate as the sole carbon source. J Bacteriol. 1986;168:534–540. doi: 10.1128/jb.168.2.534-540.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3011.Morris J F, Newman E B. Map location of the ssd mutation in Escherichia coli. J Bacteriol. 1980;143:1504–1505. doi: 10.1128/jb.143.3.1504-1505.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3012.Morris P W, Binkley J P, Henson J M, Kuempel P. Cloning and location of the dgsA gene of Escherichia coli. J Bacteriol. 1985;163:785–786. doi: 10.1128/jb.163.2.785-786.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3013.Morris T W, Reed K E, Cronan J E., Jr Identification of the gene encoding lipoate-protein ligase A of Escherichia coli. Molecular cloning and characterization of the lplA gene and gene product. J Biol Chem. 1994;269:16091–16100. [PubMed] [Google Scholar]
- 3014.Morris T W, Reed K E, Cronan J E., Jr Lipoic acid metabolism in Escherichia coli: the lplA and lipB genes define redundant pathways for ligation of lipoyl groups to apoprotein. J Bacteriol. 1995;177:1–10. doi: 10.1128/jb.177.1.1-10.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3015.Mortarino M, Negri A, Tedeschi G, Simonic T, Duga S, Gassen H G, Ronchi S. l-Aspartate oxidase from Escherichia coli. I. Characterization of coenzyme binding and product inhibition. Eur J Biochem. 1996;239:418–426. doi: 10.1111/j.1432-1033.1996.0418u.x. [DOI] [PubMed] [Google Scholar]
- 3016.Mortensen K K, Hajnsdorf E, Regnier P, Sperling-Petersen H U. Improved recombinant tandem expression of translation initiation factor IF2 in RNASE E deficient E. coli cells. Biochem Biophys Res Commun. 1995;214:1254–1259. doi: 10.1006/bbrc.1995.2421. [DOI] [PubMed] [Google Scholar]
- 3017.Mortensen L, Dandanell G, Hammer K. Purification and characterization of the deoR repressor of Escherichia coli. EMBO J. 1989;8:325–331. doi: 10.1002/j.1460-2075.1989.tb03380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3018.Moskovitz J, Rahman M A, Strassman J, Yancey S D, Kushner S R, Brot N, Weissbach H. Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J Bacteriol. 1995;177:502–507. doi: 10.1128/jb.177.3.502-507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3019.Motamedi H, Lee Y, Schmidt F J. Tandem promoters preceding the gene for the M1 RNA component of Escherichia coli ribonuclease. Proc Natl Acad Sci USA. 1984;81:3959–3963. doi: 10.1073/pnas.81.13.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3020.Motojima K, Yamato I, Anraku Y, Nishimura A, Hirota Y. Amplification and characterization of the proline transport carrier of Escherichia coli K-12 by using proT+ hybrid plasmids. Proc Natl Acad Sci USA. 1979;76:6255–6259. doi: 10.1073/pnas.76.12.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3021.Mottl H, Nieland P, De Kort G, Wierenga J J, Keck W. Deletion of an additional domain located between SXXK and SXN active-site fingerprints in penicillin-binding protein 4 from Escherichia coli. J Bacteriol. 1992;174:3261–3269. doi: 10.1128/jb.174.10.3261-3269.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3022.Mottl H, Terpstra P, Keck W. Penicillin-binding protein 4 of Escherichia coli shows a novel type of primary structure among penicillin-interacting proteins. FEMS Microbiol Lett. 1991;62:213–220. doi: 10.1016/0378-1097(91)90160-c. [DOI] [PubMed] [Google Scholar]
- 3023.Movva N R, Nakamura K, Inouye M. Gene structure of the OmpA protein, a major surface protein of Escherichia coli required for cell-cell interaction. J Mol Biol. 1980;143:317–328. doi: 10.1016/0022-2836(80)90193-x. [DOI] [PubMed] [Google Scholar]
- 3024.Movva N R, Rao N N, Nakamura K, Inouye M. Regulatory region of the gene for the OmpA protein, a major outer membrane protein of Escherichia coli. Proc Natl Acad Sci USA. 1980;77:3845–3849. doi: 10.1073/pnas.77.7.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3025.Movva R N, Green P J, Nakamura K, Inouye M. Interaction of cAMP receptor protein with the ompA gene, a gene for a major outer membrane protein of Escherichia coli. FEBS Lett. 1981;128:186–190. doi: 10.1016/0014-5793(81)80077-4. [DOI] [PubMed] [Google Scholar]
- 3026.Moyed H S. False feedback inhibition: inhibition of tryptophan biosynthesis by 5-methyltryptophan. J Biol Chem. 1960;235:1098–1102. [PubMed] [Google Scholar]
- 3027.Moyed H S. Interference with the feed-back control of histidine biosynthesis. J Biol Chem. 1961;236:2261–2267. [PubMed] [Google Scholar]
- 3028.Moyed H S, Bertrand K. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol. 1983;155:768–775. doi: 10.1128/jb.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3029.Moyed H S, Broderick S H. Molecular cloning and expression of hipA, a gene of Escherichia coli K-12 that affects frequency of persistence after inhibition and murein synthesis. J Bacteriol. 1986;166:399–403. doi: 10.1128/jb.166.2.399-403.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3030.Muday G K, Johnson D I, Somerville R L, Herrmann K M. The tyrosine repressor negatively regulates aroH expression in Escherichia coli. J Bacteriol. 1991;173:3930–3932. doi: 10.1128/jb.173.12.3930-3932.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3031.Mudd E A, Krisch H M, Higgins C F. RNase E, an endoribonuclease, has a general role in the chemical decay of Escherichia coli mRNA: evidence that rne and ams are the same genetic locus. Mol Microbiol. 1990;4:2127–2135. doi: 10.1111/j.1365-2958.1990.tb00574.x. [DOI] [PubMed] [Google Scholar]
- 3032.Mueller E G, Buck C J, Palenchar P M, Barnhart L E, Paulson J L. Identification of a gene involved in the generation of 4-thiouridine in tRNA. Nucleic Acids Res. 1998;26:2606–2610. doi: 10.1093/nar/26.11.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3033.Mueller-Wriedt, K., B. F. Matzanke, V. Schoenemann, A. X. Trautwein, and K. Hantke. FhuF, an iron-regulated protein of Escherichia coli with a new type of [2FE-2S] center. Submitted for publication. [DOI] [PubMed]
- 3034.Muffler A, Fischer D, Altuvia S, Storz G, Hengge-Aronis R. The response regulator RssB controls stability of the ςS subunit of RNA polymerase in Escherichia coli. EMBO J. 1996;15:1333–1339. [PMC free article] [PubMed] [Google Scholar]
- 3035.Muffler A, Barth M, Marschall C, Hengge-Aronis R. Heat shock regulation of ςS turnover: a role for DnaK and relationship between stress responses mediated by ςS and ς32 in Escherichia coli. J Bacteriol. 1997;179:445–452. doi: 10.1128/jb.179.2.445-452.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3036.Mukhopadhyay B, Schellhorn H E. Induction of Escherichia coli hydroperoxidase I by acetate and other weak acids. J Bacteriol. 1994;176:2300–2307. doi: 10.1128/jb.176.8.2300-2307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3037.Mukhopadhyay S, Schellhorn H E. Identification and characterization of hydrogen peroxide-sensitive mutants of Escherichia coli: genes that require OxyR for expression. J Bacteriol. 1997;179:330–338. doi: 10.1128/jb.179.2.330-338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3038.Muller M M, Vianney A, Lazzaroni J C, Webster R E, Portalier R. Membrane topology of the Escherichia coli TolR protein required for cell envelope integrity. J Bacteriol. 1993;175:6059–6061. doi: 10.1128/jb.175.18.6059-6061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3039.Muller M M, Webster R E. Characterization of the tol-pal and cyd region of Escherichia coli K-12: transcript analysis and identification of two new proteins encoded by the cyd operon. J Bacteriol. 1997;179:2077–2080. doi: 10.1128/jb.179.6.2077-2080.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3040.Muller R, Dahm C, Schulte G, Leistner E. An isochorismate hydroxymutase isogene in Escherichia coli. FEBS Lett. 1996;378:131–134. doi: 10.1016/0014-5793(95)01436-5. [DOI] [PubMed] [Google Scholar]
- 3041.Muller V, Jones C J, Kawagishi I, Aizawa S, Macnab R M. Characterization of the fliE genes of Escherichia coli and Salmonella typhimurium and identification of the fliE protein as a component of the flagellar hook-basal body complex. J Bacteriol. 1992;174:2298–2304. doi: 10.1128/jb.174.7.2298-2304.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3042.Muller-Hill B, Crapo L, Gilbert W. Mutants that make more lac repressor. Proc Natl Acad Sci USA. 1968;59:1259–1264. doi: 10.1073/pnas.59.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3043.Mullin D A, Woldringh C L, Henson J M, Walker J R. Cloning of the Escherichia coli dnaZX region and identification of its product. Mol Gen Genet. 1983;192:73–79. doi: 10.1007/BF00327649. [DOI] [PubMed] [Google Scholar]
- 3044.Mulvey M R. Cloning and physical characterization of katE and katF required for catalase HPII expression of Escherichia coli. Gene. 1988;73:337–346. doi: 10.1016/0378-1119(88)90498-2. [DOI] [PubMed] [Google Scholar]
- 3045.Mulvey M R, Loewen P C. Nucleotide sequence of katF of Escherichia coli suggests KatF protein is a novel transcription factor. Nucleic Acids Res. 1989;17:9979–9991. doi: 10.1093/nar/17.23.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3046.Munch-Petersen A, Mygind B. Nucleoside transport systems in Escherichia coli K12: specificity and regulation. J Cell Physiol. 1976;89:551–560. doi: 10.1002/jcp.1040890410. [DOI] [PubMed] [Google Scholar]
- 3047.Munch-Petersen A, Mygind B, Nicolaisen A, Pihl N J. Nucleoside transport in cells and membrane vesicles from Escherichia coli K12. J Biol Chem. 1979;254:3730–3737. [PubMed] [Google Scholar]
- 3048.Munch-Petersen A, Nygaard P, Hammer-Jespersen K, Fiil N. Mutants constitutive for nucleoside-catabolizing enzymes in Escherichia coli K12. Eur J Biochem. 1972;27:208–215. doi: 10.1111/j.1432-1033.1972.tb01828.x. [DOI] [PubMed] [Google Scholar]
- 3049.Munekiyo R, Tsuzuki T, Sekiguchi M. A new locus of Escherichia coli that determines sensitivity to bacteriophage phiX174. J Bacteriol. 1979;138:1038–1040. doi: 10.1128/jb.138.3.1038-1040.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3050.Munro A W, Ritchie G Y, Lamb A J, Douglas R M, Booth I R. The cloning and DNA sequence of the gene for the glutathione-regulated potassium-efflux system KefC of Escherichia coli. Mol Microbiol. 1991;5:607–616. doi: 10.1111/j.1365-2958.1991.tb00731.x. [DOI] [PubMed] [Google Scholar]
- 3051.Murakami H, Inokuchi H, Hirota Y, Ozeki H, Yamagishi H. Characterization of the dnaA gene carried by lambda transducing phage. Mol Gen Genet. 1980;180:235–247. doi: 10.1007/BF00425835. [DOI] [PubMed] [Google Scholar]
- 3052.Murakami H, Kita K, Oya H, Anraku Y. Chromosomal location of the Escherichia coli cytochrome b556. Mol Gen Genet. 1984;196:1–5. doi: 10.1007/BF00334084. [DOI] [PubMed] [Google Scholar]
- 3053.Murakami H, Kita K, Oya H, Anraku Y. The Escherichia coli cytochrome b556 gene, cybA, is assignable as sdhC in the succinate dehydrogenase gene cluster. FEMS Microbiol Lett. 1985;30:307–311. [Google Scholar]
- 3054.Murakami H, Kita K, Anraku Y. Cloning of cybB, the gene for cytochrome b561 of Escherichia coli K12. Mol Gen Genet. 1984;198:1–6. doi: 10.1007/BF00328692. [DOI] [PubMed] [Google Scholar]
- 3055.Murakami H, Nagata T, Schwarz W, Wada C, Yura T. Novel dnaG mutation in a dnaP mutant of Escherichia coli. J Bacteriol. 1985;162:830–832. doi: 10.1128/jb.162.2.830-832.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3056.Muralikrishna P, Wickstrom E. Inducible high expression of the Escherichia coli infC gene subcloned behind a bacteriophage T7 promoter. Gene. 1989;80:369–374. doi: 10.1016/0378-1119(89)90301-6. [DOI] [PubMed] [Google Scholar]
- 3057.Muramatsu S, Mizuno T. Nucleotide sequence of the fabE gene and flanking regions containing a bent DNA sequence of Escherichia coli. Nucleic Acids Res. 1989;17:3982. doi: 10.1093/nar/17.10.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3058.Muramatsu T, Nishikawa K, Nemoto F, Kuchino Y, Nishimura S, Miyazawa T, Yokoyama K. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988;336:179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- 3059.Muramatsu T, Mizuno T. Nucleotide sequence of the region encompassing the int gene of a cryptic prophage and the dnaY gene flanked by a curved DNAQ sequence of Escherichia coli K12. Mol Gen Genet. 1990;220:325–328. doi: 10.1007/BF00260503. [DOI] [PubMed] [Google Scholar]
- 3060.Murata K, Tani K, Kato J-I, Chibata I. Isolation of Escherichia coli B mutant deficient in glutathione biosynthesis. Agric Biol Chem. 1981;45:2131–2132. [Google Scholar]
- 3061.Murayama N, Shimizu H, Takiguchi S, Baba Y, Amino H, Horiuchi T, Sekimizu K, Miki T. Evidence for involvement of Escherichia coli genes pmbA, csrA and a previously unrecognized gene tldD, in the control of DNA gyrase by letD (ccdB) of sex factor F. J Mol Biol. 1996;256:483–502. doi: 10.1006/jmbi.1996.0103. [DOI] [PubMed] [Google Scholar]
- 3062.Murgola E, Adelberg E A. Mutants of Escherichia coli K-12 with an altered glutamyl-transfer ribonucleic acid synthetase. J Bacteriol. 1970;103:178–183. doi: 10.1128/jb.103.1.178-183.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3063.Murgola E J, Pagel F T, Hijazi K A, Arkov A L, Xu W, Zhao S Q. Variety of nonsense suppressor phenotypes associated with mutational changes at conserved sites in Escherichia coli ribosomal RNA. Biochem Cell Biol. 1995;73:925–931. doi: 10.1139/o95-100. [DOI] [PubMed] [Google Scholar]
- 3064.Murgola E J, Guarneros G. Ribosomal RNA and peptidyl-tRNA hydrolase: a peptide chain termination model for lambda bar RNA inhibition. Biochimie. 1991;73:1573–1578. doi: 10.1016/0300-9084(91)90193-5. [DOI] [PubMed] [Google Scholar]
- 3065.Murgola E J, Xu W, Arkov A L. Mutations at three sites in the Escherichia coli 23S ribosomal RNA binding region for protein L11 cause UGA-specific suppression and conditional lethality. Nucleic Acids Symp Ser. 1995;114:70–72. [PubMed] [Google Scholar]
- 3066.Murooka Y, Higashiura T, Harada T. Genetic mapping of tyramine oxidase and arylsulfatase genes and their regulation in intergeneric hybrids of enteric bacteria. J Bacteriol. 1978;136:714–722. doi: 10.1128/jb.136.2.714-722.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3067.Murphy C K, Stewart E J, Beckwith J R. A double counter-selection system for the study of null alleles of essential genes in Escherichia coli. Gene. 1995;155:1–7. doi: 10.1016/0378-1119(94)00920-n. . (Erratum, 163:165, 1995.) [DOI] [PubMed] [Google Scholar]
- 3068.Murphy C K, Beckwith J R. Residues essential for the function of SecE, a membrane component of the Escherichia coli secretion apparatus, are located in a conserved cytoplasmic region. Proc Natl Acad Sci USA. 1994;91:2557–2561. doi: 10.1073/pnas.91.7.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3069.Murphy H, Kalman M, Cashel M. Identification of the gppB locus as two convergent arylsulfatase-like genes, aslA and aslB, capable of suppressing a guanosine pentaphosphate phosphatase missense mutation in Escherichia coli. 1992. GenBank submission. [Google Scholar]
- 3070.Murray K D, Bremer H. Control of spoT-dependent ppGpp synthesis and degradation in Escherichia coli. J Mol Biol. 1996;259:41–57. doi: 10.1006/jmbi.1996.0300. [DOI] [PubMed] [Google Scholar]
- 3071.Murray N E, Gough J A, Suri B, Bickle T A. Structural homologies among type I restriction-modification systems. EMBO J. 1982;1:535–539. doi: 10.1002/j.1460-2075.1982.tb01205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3072.Murray N E, Kelley W S. Characterization of λpolA transducing phages: effective expression of the E. coli polA gene. Mol Gen Genet. 1979;175:77–87. doi: 10.1007/BF00267858. [DOI] [PubMed] [Google Scholar]
- 3073.Muse W B, Bender R A. The nac (nitrogen assimilation control) gene from Escherichia coli. J Bacteriol. 1998;180:1166–1173. doi: 10.1128/jb.180.5.1166-1173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3074.Muto A, Sato M, Tadaki T, Fukushima M, Ushida C, Himeno H. Structure and function of 10Sa RNA: trans-translation system. Biochimie. 1996;78:985–991. doi: 10.1016/s0300-9084(97)86721-1. [DOI] [PubMed] [Google Scholar]
- 3075.Mutoh N, Inokuchi K, Mizushima S. Amino acid sequence of the signal peptides of OmpF, a major outer membrane protein of Escherichia coli. FEBS Lett. 1982;137:171–174. doi: 10.1016/0014-5793(82)80341-4. [DOI] [PubMed] [Google Scholar]
- 3076.Mutoh N, Simon M. Nucleotide sequence corresponding to five chemotaxis genes in Escherichia coli. J Bacteriol. 1986;165:161–166. doi: 10.1128/jb.165.1.161-166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3077.Mutoh N, Nagasawa T, Mizushima S. Specialized transducing bacteriophage lambda carrying the structural gene for a major outer membrane matrix protein of Escherichia coli K-12. J Bacteriol. 1981;145:1085–1090. doi: 10.1128/jb.145.2.1085-1090.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3078.Muzzin O, Campbell E A, Xia L, Severinova E, Darst S A, Severinov K. Disruption of Escherichia coli hepA, an RNA polymerase-associated protein, causes UV sensitivity. J Biol Chem. 1998;273:15157–15161. doi: 10.1074/jbc.273.24.15157. [DOI] [PubMed] [Google Scholar]
- 3079.Myler P J, Venkataraman G M, Lodes M J, Stuart K D. A frequently amplified region in Leishmania contains a gene conserved in prokaryotes and eukaryotes. Gene. 1994;148:187–193. doi: 10.1016/0378-1119(94)90688-2. [DOI] [PubMed] [Google Scholar]
- 3080.Mytelka D S, Chamberlin M J. Escherichia coli fliAZY operon. J Bacteriol. 1996;178:24–34. doi: 10.1128/jb.178.1.24-34.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3081.Nagano Y, Matsuno R, Sasaki Y. An essential gene of Escherichia coli that has sequence similarity to a chloroplast gene of unknown function. Mol Gen Genet. 1991;228:62–64. doi: 10.1007/BF00282448. [DOI] [PubMed] [Google Scholar]
- 3082.Nagasawa H, Sakagami Y, Suzuki A, Suzuki H, Hara H, Hirota Y. Determination of the cleavage site involved in C-terminal processing of penicillin-binding protein 3 of Escherichia coli. J Bacteriol. 1989;171:5890–5893. doi: 10.1128/jb.171.11.5890-5893.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3083.Nagasawa S, Ishige K, Mizuno T. Novel members of the two-component signal transduction genes in Escherichia coli. J Biochem (Tokyo) 1993;114:350–357. doi: 10.1093/oxfordjournals.jbchem.a124180. [DOI] [PubMed] [Google Scholar]
- 3084.Nagasawa S, Tokishita S, Aiba H, Mizuno T. A novel sensor-regulator protein that belongs to the homologous family of signal-transduction proteins involved in adaptive responses in Escherichia coli. Mol Microbiol. 1992;6:799–807. doi: 10.1111/j.1365-2958.1992.tb01530.x. [DOI] [PubMed] [Google Scholar]
- 3085.Nagase T, Ishii S, Imamoto F. Differential transcriptional control of the two tRNAfMet genes of Escherichia coli K-12. Gene. 1988;67:49–57. doi: 10.1016/0378-1119(88)90007-8. [DOI] [PubMed] [Google Scholar]
- 3086.Nagel R, Chan A, Rosen E. ruv and recG genes and the induced precise excision of Tn10 in Escherichia coli. Mutat Res. 1994;311:103–109. doi: 10.1016/0027-5107(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 3087.Naggert J, Narasimhan M L, DeVeaux L, Cho H, Randhawa Z I, Cronan J E, Green B N, Smith S. Cloning, sequencing, and characterization of Escherichia coli thioesterase II. J Biol Chem. 1991;266:11044–11050. [PubMed] [Google Scholar]
- 3088.Nagy P L, Marolewski A, Benkovic S J, Zalkin H. Formyltetrahydrofolate hydrolase, a regulatory enzyme that functions to balance pools of tetrahydrofolate and one-carbon tetrahydrofolate adducts in Escherichia coli. J Bacteriol. 1995;177:1292–1298. doi: 10.1128/jb.177.5.1292-1298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3089.Nagy P L, McCorkle G M, Zalkin H. purU, a source of formate for purT-dependent phosphoribosyl-N-formylglycinamide synthesis. J Bacteriol. 1993;175:7066–7073. doi: 10.1128/jb.175.21.7066-7073.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3090.Nahlik M S, Brickman T J, Ozenberger B A, McIntosh M A. Nucleotide sequence and transcriptional organization of the Escherichia coli enterobactin biosynthesis cistrons entB and entA. J Bacteriol. 1989;171:784–790. doi: 10.1128/jb.171.2.784-790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3091.Nahlik M S, Fleming T P, McIntosh M A. Cluster of genes controlling synthesis and activation of 2,3-dihydroxybenzoic acid in production of enterobactin in Escherichia coli. J Bacteriol. 1987;169:4163–4170. doi: 10.1128/jb.169.9.4163-4170.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3092.Naito S, Kitani T, Ogawa T, Okazaki T, Uchida H. Escherichia coli mutants suppressing replication-defective mutations of the ColE1 plasmid. Proc Natl Acad Sci USA. 1984;81:550–554. doi: 10.1073/pnas.81.2.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3093.Nakabeppu Y, Kondo H, Sekiguchi M. Cloning and characterization of the alkA gene of Escherichia coli that encodes 3-methyladenine DNA glycosylase II. J Biol Chem. 1984;259:13723–13729. [PubMed] [Google Scholar]
- 3094.Nakabeppu Y, Kondo H, Kawabata S I, Iwanaga S, Sekiguchi M. Purification and structure of the intact Ada regulatory protein of Escherichia coli K12, O6-methyl-guanine-DNA methyltransferase. J Biol Chem. 1985;260:7281–7288. [PubMed] [Google Scholar]
- 3095.Nakabeppu Y, Miyata T, Kondo H, Iwanaga S, Sekiguchi M. Structure and expression of the alkA gene of Escherichia coli involved in adaptive response to alkylating agents. J Biol Chem. 1984;259:13730–13736. [PubMed] [Google Scholar]
- 3096.Nakagawa J, Matsuhashi M. Molecular divergence of a major peptidoglycan synthetase with transglycosylase-transpeptidase activities in Escherichia coli—penicillin binding protein 1Bs. Biochem Biophys Res Commun. 1982;105:1546–1553. doi: 10.1016/0006-291x(82)90964-0. [DOI] [PubMed] [Google Scholar]
- 3097.Nakagawa J, Tamaki S, Matsuhashi M. Purified penicillin binding proteins 1Bs from Escherichia coli membrane showing activities of both peptidoglycan polymerase and peptidoglycan crosslinking enzyme. Agric Biol Chem. 1979;43:1379–1380. [Google Scholar]
- 3098.Nakahigashi K, Inokuchi H. Nucleotide sequence between the fadB and the rrnA operon from Escherichia coli. Nucleic Acids Res. 1990;18:6439. doi: 10.1093/nar/18.21.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3099.Nakahigashi K, Inokuchi H. Nucleotide sequence of the fadA and fadB genes from Escherichia coli. Nucleic Acids Res. 1990;18:4937. doi: 10.1093/nar/18.16.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3100.Nakahigashi K, Miyamoto K, Nishimura K, Inokuchi H. Isolation and characterization of a light-sensitive mutant of Escherichia coli K-12 with a mutation in a gene that is required for the biosynthesis of ubiquinone. J Bacteriol. 1992;174:7352–7359. doi: 10.1128/jb.174.22.7352-7359.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3101.Nakahigashi K, Nishimura K, Miyamoto K, Inokuchi H. Photosensitivity of a protoporphyrin-accumulating, light-sensitive mutant (visA) of Escherichia coli K-12. Proc Natl Acad Sci USA. 1991;88:10520–10524. doi: 10.1073/pnas.88.23.10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3102.Nakajima N, Ozeki H, Shimura Y. Organization and structure of an E. coli tRNA operon containing seven tRNA genes. Cell. 1981;23:239–249. doi: 10.1016/0092-8674(81)90288-9. [DOI] [PubMed] [Google Scholar]
- 3103.Nakajima N, Ozeki H, Shimura Y. In vitro transcription of the supB-E tRNA operon of Escherichia coli. J Biol Chem. 1982;257:11113–11120. [PubMed] [Google Scholar]
- 3104.Nakamura H. Novel acriflavine resistance genes, acrC and acrD, in Escherichia coli K-12. J Bacteriol. 1979;139:8–12. doi: 10.1128/jb.139.1.8-12.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3105.Nakamura H, Murakami H, Yamato I, Anraku Y. Nucleotide sequence of the cybB gene encoding cytochrome b561 in Escherichia coli K12. Mol Gen Genet. 1988;212:1–5. doi: 10.1007/BF00322437. [DOI] [PubMed] [Google Scholar]
- 3106.Nakamura H, Yamato I, Anraku Y, Lemieux L, Gennis R B. Expression of cyoA and cyoB demonstrates that the CO-binding heme component of the Escherichia coli cytochrome o complex is in subunit I. J Biol Chem. 1990;265:11193–11197. [PubMed] [Google Scholar]
- 3107.Nakamura H, Maruyama M, Soma M, Kato J-I, Suzuki H, Hirota Y. On the process of cellular division in Escherichia coli: nucleotide sequence of the gene for penicillin-binding protein. Mol Gen Genet. 1983;191:1–9. doi: 10.1007/BF00330881. [DOI] [PubMed] [Google Scholar]
- 3108.Nakamura H, Mizusawa S. In vivo evidence that the nusA and infB genes of E. coli are part of the same multi-gene operon which encodes at least four proteins. EMBO J. 1985;4:527–532. doi: 10.1002/j.1460-2075.1985.tb03660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3109.Nakamura K, Inouye M. DNA sequence of the gene for the outer membrane lipoprotein of E. coli: an extremely AT-rich promoter. Cell. 1979;18:1109–1117. doi: 10.1016/0092-8674(79)90224-1. [DOI] [PubMed] [Google Scholar]
- 3110.Nakamura K, Pirtle R M, Pirtle I L, Takeishi K, Inouye M. Messenger ribonucleic acid of the lipoprotein of the Escherichia coli outer membrane. II. The complete nucleotide sequence. J Biol Chem. 1980;255:210–216. [PubMed] [Google Scholar]
- 3111.Nakamura M, Yamada M, Hirota Y, Sugimoto K, Oka A, Takanami M. Nucleotide sequence of the asnA gene coding for asparagine synthetase of E. coli K-12. Nucleic Acids Res. 1981;9:4669–4676. doi: 10.1093/nar/9.18.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3112.Nakamura S, Nakamura M, Kojima T, Yoshida H. gyrA and gyrB mutations in quinolone-resistant strains of Escherichia coli. Antimicrob Agents Chemother. 1989;33:254–255. doi: 10.1128/aac.33.2.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3113.Nakamura Y. Hybrid plasmid carrying Escherichia coli genes for primase (dnaG) and RNA polymerase sigma factor (rpoD); gene organization and control of their expression. Mol Gen Genet. 1980;178:487–497. doi: 10.1007/BF00337853. [DOI] [PubMed] [Google Scholar]
- 3114.Nakamura Y, Kurihara T, Saito H, Uchida H. Sigma subunit of Escherichia coli RNA polymerase affects the function of λN gene. Proc Natl Acad Sci USA. 1979;76:4593–4597. doi: 10.1073/pnas.76.9.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3114a.Nakanishi A, Oshida T, Matsushita T, Imajoh-Ohmi S, Ohnuki T. Identification of DNA gyrase inhibitor (GyrI) in Escherichia coli. J Biol Chem. 1998;273:1933–1938. doi: 10.1074/jbc.273.4.1933. [DOI] [PubMed] [Google Scholar]
- 3115.Nakao T, Yamato I, Anraku Y. Nucleotide sequence of putC, the regulatory region for the put regulon of Escherichia coli K12. Mol Gen Genet. 1987;210:364–368. doi: 10.1007/BF00325707. [DOI] [PubMed] [Google Scholar]
- 3116.Nakao T, Yamato I, Anraku Y. Nucleotide sequence of putP, the proline carrier gene of Escherichia coli K12. Mol Gen Genet. 1987;208:70–75. doi: 10.1007/BF00330424. [DOI] [PubMed] [Google Scholar]
- 3117.Nakashima K, Sugiura A, Kanamaru K, Mizuno T. Signal transduction between the two regulatory components involved in the regulation of the kdpABC operon in Escherichia coli: phosphorylation-dependent functioning of the positive regulator KdpE. Mol Microbiol. 1993;7:109–116. doi: 10.1111/j.1365-2958.1993.tb01102.x. [DOI] [PubMed] [Google Scholar]
- 3118.Nakashima K, Sugiura A, Mizuno T. Functional reconstitution of the putative Escherichia coli osmosensor, KdpD, into liposomes. J Biochem (Tokyo) 1993;114:615–621. doi: 10.1093/oxfordjournals.jbchem.a124226. [DOI] [PubMed] [Google Scholar]
- 3119.Nakashima K, Kanamaru K, Mizuno T, Horikoshi K. A novel member of the cspA family of genes that is induced by cold shock in Escherichia coli. J Bacteriol. 1996;178:2994–2997. doi: 10.1128/jb.178.10.2994-2997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3120.Nakata A, Shinagawa H, Amemura M. Cloning of alkaline phosphatase isozyme gene (iap) of Escherichia coli. Gene. 1982;19:313–319. doi: 10.1016/0378-1119(82)90021-x. [DOI] [PubMed] [Google Scholar]
- 3121.Nakata A, Amemura M, Shinegawa H. Regulation of the phosphate regulon in Escherichia coli K-12: regulation of the negative regulatory gene phoU and identification of the gene product. J Bacteriol. 1984;159:979–985. doi: 10.1128/jb.159.3.979-985.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3122.Nakayama H. Genetic analysis of thiamin pyrophosphate biosynthesis in Escherichia coli. Vitamins. 1995;64:619–632. [Google Scholar]
- 3123.Nakayama H, Nakayama K, Nakayama R, Irino N, Nakayama Y, Hanawalt P C. Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K12: identification of a new mutation (recQ1) that blocks the RecF recombination pathway. Mol Gen Genet. 1984;195:474–480. doi: 10.1007/BF00341449. [DOI] [PubMed] [Google Scholar]
- 3124.Nakayama K, Irino N, Nakayama H. The recQ gene of Escherichia coli K-12: molecular cloning and isolation of insertion mutants. Mol Gen Genet. 1985;200:266–271. doi: 10.1007/BF00425434. [DOI] [PubMed] [Google Scholar]
- 3125.Nakayama N, Bond M W, Miyajima A, Kobori J, Arai K-I. Structure of Escherichia coli dnaC. Identification of a cysteine residue possibly involved in association with dnaB protein. J Biol Chem. 1987;262:10475–10480. [PubMed] [Google Scholar]
- 3126.Nakayama N, Arai N, Bond M W, Kaziro Y, Arai K-I. Nucleotide sequence of dnaB and the primary structure of the dnaB protein from Escherichia coli. J Biol Chem. 1984;259:97–101. [PubMed] [Google Scholar]
- 3127.Nakayashiki T, Nishimura K, Inokuchi H. Cloning and sequencing of a previously unidentified gene that is involved in the biosynthesis of heme in Escherichia coli. Gene. 1995;153:67–70. doi: 10.1016/0378-1119(94)00805-3. [DOI] [PubMed] [Google Scholar]
- 3128.Naom I S, Morton S J, Leach D R F, Lloyd R G. Molecular organization of sbcC, a gene that affects genetic recombination and the viability of DNA palindromes in Escherichia coli K-12. Nucleic Acids Res. 1989;17:8033–8044. doi: 10.1093/nar/17.20.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3129.Nara F, Matsuyama S, Mizuno T, Mizushima S. Molecular analysis of mutant ompR genes exhibiting different phenotypes as to osmoregulation of the ompF and ompC genes of Escherichia coli. Mol Gen Genet. 1986;202:194–199. doi: 10.1007/BF00331636. [DOI] [PubMed] [Google Scholar]
- 3130.Narasimhan M, Lampi J L, Cronan J E., Jr Genetic and biochemical characterization of an Escherichia coli K-12 mutant deficient in acyl-coenzyme A thioesterase II. J Bacteriol. 1986;165:911–917. doi: 10.1128/jb.165.3.911-917.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3131.Nargang F E, Subrahmanyam C S, Umbarger H E. Nucleotide sequence of ivlGEDA operon attenuator region of Escherichia coli. Proc Natl Acad Sci USA. 1980;77:1823–1827. doi: 10.1073/pnas.77.4.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3132.Naroditskaya V, Schlosser M J, Fang N Y, Lewis K. An E. coli gene emrD is involved in adaptation to low energy shock. Biochem Biophys Res Commun. 1993;196:803–809. doi: 10.1006/bbrc.1993.2320. [DOI] [PubMed] [Google Scholar]
- 3133.Nashimoto H, Uchida H. Late steps in the assembly of 30s ribosomal proteins in vivo in a spectinomycin-resistant mutant of Escherichia coli. J Mol Biol. 1975;96:443–453. doi: 10.1016/0022-2836(75)90171-0. [DOI] [PubMed] [Google Scholar]
- 3134.Nashimoto H, Uchida H. DNA sequencing of the Escherichia coli ribonuclease III gene and its mutations. Mol Gen Genet. 1985;201:25–29. doi: 10.1007/BF00397981. [DOI] [PubMed] [Google Scholar]
- 3135.Nasoff M S, Baker H V, Wolf R. DNA sequence of the Escherichia coli gene, gnd, for 6-phosphogluconate dehydrogenase. Gene. 1984;27:253–264. doi: 10.1016/0378-1119(84)90070-2. [DOI] [PubMed] [Google Scholar]
- 3136.Nass G, Thomale J. Alteration of structure or level of threonyl-tRNA synthetase in borrelidin-resistant mutants of Escherichia coli. FEBS Lett. 1974;39:182–186. doi: 10.1016/0014-5793(74)80046-3. [DOI] [PubMed] [Google Scholar]
- 3137.Nassau P M, Martin S L, Brown R E, Weston A, Monsey D, McNeil M R, Duncan K. Galactofuranose biosynthesis in Escherichia coli K-12: identification and cloning of UDP-galactopyranose mutase. J Bacteriol. 1996;178:1047–1052. doi: 10.1128/jb.178.4.1047-1052.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3138.Nassof M, Wolf R. Molecular cloning, correlation of genetic and restriction maps, and determination of the direction of transcription of gnd of Escherichia coli. J Bacteriol. 1980;143:731–741. doi: 10.1128/jb.143.2.731-741.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3139.Navarro C, Wu L F, Mandrand-Berthelot M A. The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol Microbiol. 1993;9:1181–1191. doi: 10.1111/j.1365-2958.1993.tb01247.x. [DOI] [PubMed] [Google Scholar]
- 3140.Navre M, Schachman H K. Synthesis of aspartate transcarbamoylase in Escherichia coli: transcriptional regulation of the pyrB-pyrI operon. Proc Natl Acad Sci USA. 1983;80:1207–1211. doi: 10.1073/pnas.80.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3141.Nazos P M, Mayo M M, Su T-Z, Anderson J J, Oxender D L. Identification of livG, a membrane-associated component of the branched-chain amino acid transport in Escherichia coli. J Bacteriol. 1985;163:1196–1202. doi: 10.1128/jb.163.3.1196-1202.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3142.Nazos P M, Antonucci T K, Landick R, Oxender D L. Cloning and characterization of livH, the structural gene encoding a component of the leucine transport system in Escherichia coli. J Bacteriol. 1986;166:565–573. doi: 10.1128/jb.166.2.565-573.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3143.Neely M N, Dell C L, Olson E R. Roles of LysP and CadC in mediating the lysine requirement for acid induction of the Escherichia coli cad operon. J Bacteriol. 1994;176:3278–3285. doi: 10.1128/jb.176.11.3278-3285.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3144.Neely M N, Olson E R. Kinetics of expression of the Escherichia coli cad operon as a function of pH and lysine. J Bacteriol. 1996;178:5522–5528. doi: 10.1128/jb.178.18.5522-5528.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3145.Neidhardt F C, VanBogelin R A, Lau E T. Molecular cloning and expression of a gene that controls the high-temperature regulon of Escherichia coli. J Bacteriol. 1983;153:597–603. doi: 10.1128/jb.153.2.597-603.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3146.Neidhardt F C, Phillips T A, VanBogelen R A, Smith M W, Georgalis Y, Subramanian A-R. Identity of the B56.5 protein, the A-protein, and the groE gene product of Escherichia coli. J Bacteriol. 1981;145:513–520. doi: 10.1128/jb.145.1.513-520.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3147.Neidhardt F C, Vaughn V, Phillips T A, Bloch P L. Gene-protein index of Escherichia coli K-12. Microbiol Rev. 1983;47:231–284. doi: 10.1128/mr.47.2.231-284.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3148.Nellemann L J, Holm F, Atlung T, Hansen F G. Cloning and characterization of the Escherichia coli phosphoglycerate kinase (pgk) gene. Gene. 1989;77:185–191. doi: 10.1016/0378-1119(89)90373-9. [DOI] [PubMed] [Google Scholar]
- 3149.Nelson K, Whittam T S, Selander R K. Nucleotide polymorphism and evolution in the glyceraldehyde-3-phosphate dehydrogenase gene (gapA) in natural populations of Salmonella and Escherichia coli. Proc Natl Acad Sci USA. 1991;88:6667–6671. doi: 10.1073/pnas.88.15.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3150.Nelson S O, Lengeler J W, Postman P W. Role of IIIGlc of the phosphoenol pyruvate-glucose phosphotransferase system in inducer exclusion in Escherichia coli. J Bacteriol. 1984;160:360–364. doi: 10.1128/jb.160.1.360-364.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3151.Ner S S, Bhayana V, Bell A W, Giles I G, Duckworth H W, Bloxham D P. Complete sequence of the gltA gene encoding citrate synthase in Escherichia coli. Biochemistry. 1983;22:5243–5249. [Google Scholar]
- 3152.Nersisyan A A, Mett I L, Badalyan Z R, Kochikyan A V, Mett I L, Kocharyan A M, Kocharyan S M, Sakanyan V A. Restriction mapping of recombinant plasmids carrying the genes of arginine biosynthesis from Escherichia coli K-12. Sov Genet (Engl Transl Genetika) 1986;22:805–809. [PubMed] [Google Scholar]
- 3153.Nersisyan A A, Fedorova Y A, Khurges E M. Cloning of Escherichia coli genes for proline biosynthesis. Sov Genet (Engl Transl Genetika) 1986;22:1493–1500. [PubMed] [Google Scholar]
- 3154.Nesin M, Lupski J R, Svec P, Godson G N. Possible new genes as revealed by molecular analysis of a 5-kb Escherichia coli chromosomal region 5′ to the rpsU-dnaG-rpoD macromolecular-synthesis operon. Gene. 1987;51:149–161. doi: 10.1016/0378-1119(87)90303-9. [DOI] [PubMed] [Google Scholar]
- 3155.Nettleton C J, Bull C, Baldwin T O, Fee J A. Isolation of the Escherichia coli iron superoxide dismutase gene: evidence that intracellular superoxide concentration does not regulate oxygen-dependent synthesis of the manganese superoxide dismutase. Proc Natl Acad Sci USA. 1984;81:4970–4973. doi: 10.1073/pnas.81.15.4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3156.Neuhard J, Thomassen K. Altered deoxyribonucleotide pools in P2 eductants of Escherichia coli K-12 due to deletion of the dcd gene. J Bacteriol. 1976;126:999–1001. doi: 10.1128/jb.126.2.999-1001.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3157.Neuhard J, Tarpo L. Location of the udk gene on the physical map of Escherichia coli. J Bacteriol. 1993;175:5742–5743. doi: 10.1128/jb.175.17.5742-5743.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3158.Neuhofen S, Theyssen H, Reinstein J, Trommer W E, Vogel P D. Nucleotide binding to the heat-shock protein DnaK as studied by ESR spectroscopy. Eur J Biochem. 1996;240:78–82. doi: 10.1111/j.1432-1033.1996.0078h.x. [DOI] [PubMed] [Google Scholar]
- 3159.Neuwald A F, Krishnan B R, Brikun I, Kulakauskas S T, Suziedelis K, Tomcsanyi T, Leyh T S, Berg D E. CysQ, a gene needed for cysteine synthesis in Escherichia coli K-12 only during aerobic growth. J Bacteriol. 1992;174:415–425. doi: 10.1128/jb.174.2.415-425.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3160.Neuwald A F, Berg D E, Stauffer G V. Mutational analysis of the Escherichia coli serB promoter region reveals transcriptional linkage to a downstream gene. Gene. 1992;120:1–9. doi: 10.1016/0378-1119(92)90002-7. [DOI] [PubMed] [Google Scholar]
- 3161.Neuwald A F, Stauffer G V. DNA sequence and characterization of the Escherichia coli serB gene. Nucleic Acids Res. 1985;13:7025–7039. doi: 10.1093/nar/13.19.7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3162.Neuwald A F, Stauffer G V. An Escherichia coli membrane protein with a unique signal sequence. Gene. 1989;82:219–228. doi: 10.1016/0378-1119(89)90047-4. [DOI] [PubMed] [Google Scholar]
- 3163.Neuwald A F, Stauffer G V. IS30 activation of an smp′-lacZ gene fusion in Escherichia coli. FEMS Microbiol Lett. 1990;56:13–17. doi: 10.1016/0378-1097(90)90116-8. [DOI] [PubMed] [Google Scholar]
- 3164.Nevers P, Saedler H. Mapping the characterization of an Escherichia coli mutant defective in ISl-mediated deletion formation. Mol Gen Genet. 1978;160:209–214. [Google Scholar]
- 3165.Newman A J, Hayward R S. Cloning of DNA of the rpoBC operon from the chromosome of Escherichia coli K12. Mol Gen Genet. 1980;177:527–533. doi: 10.1007/BF00271493. [DOI] [PubMed] [Google Scholar]
- 3166.Newman E B, Malik N, Walker C. l-Serine degradation in Escherichia coli K-12: directly isolated ssd mutants and their intragenic revertants. J Bacteriol. 1982;150:710–715. doi: 10.1128/jb.150.2.710-715.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3167.Newman E B, Lin R. Leucine-responsive regulatory protein: a global regulator of gene expression in E. coli. Annu Rev Microbiol. 1995;49:747–775. doi: 10.1146/annurev.mi.49.100195.003531. [DOI] [PubMed] [Google Scholar]
- 3168.Newman T, Levinthal M. A new map location for the ilvB locus of Escherichia coli. Genetics. 1980;96:59–77. doi: 10.1093/genetics/96.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3169.Newman T, Friden P, Sutton A, Freundlich M. Cloning and expression of the ilvB gene of Escherichia coli K-12. Mol Gen Genet. 1982;186:378–384. doi: 10.1007/BF00729457. [DOI] [PubMed] [Google Scholar]
- 3170.Newton W A, Snell E E. Formation and interrelationships of tryptophanase and tryptophan synthetases in Escherichia coli. J Bacteriol. 1965;89:355–364. doi: 10.1128/jb.89.2.355-364.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3171.Ng H, Gartner T K. Selection of mutants of Escherichia coli constitutive for tryptophanase. J Bacteriol. 1963;85:245–246. doi: 10.1128/jb.85.1.245-246.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3172.Nghiem Y, Cabrera M, Cupples C G, Miller J H. The mutY gene: a mutator locus in Escherichia coli that generates G · C→T · A transversions. Proc Natl Acad Sci USA. 1988;85:2709–2713. doi: 10.1073/pnas.85.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3173.Nichols B, Yanofsky C. Nucleotide sequences of trpA of Salmonella typhimurium and Escherichia coli: an evolutionary comparison. Proc Natl Acad Sci USA. 1979;76:5244–5248. doi: 10.1073/pnas.76.10.5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3174.Nichols B, Miozzari G F, van Cleemput M, Bennett G, Yanofsky C. Nucleotide sequences of the trpG regions of Escherichia coli, Shigella dysenteriae, Salmonella typhimurium and Serratia marcescens. J Mol Biol. 1980;142:503–517. doi: 10.1016/0022-2836(80)90260-0. [DOI] [PubMed] [Google Scholar]
- 3175.Nichols B, van Cleemput M, Yanofsky C. Nucleotide sequence of Escherichia coli trpE. Anthranilate synthetase component I contains no tryptophan residues. J Mol Biol. 1981;146:45–54. doi: 10.1016/0022-2836(81)90365-x. [DOI] [PubMed] [Google Scholar]
- 3176.Nichols B P, Green J M. Cloning and sequencing of Escherichia coli ubiC and purification of chorismate lyase. J Bacteriol. 1992;174:5309–5316. doi: 10.1128/jb.174.16.5309-5316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3177.Nicol S M, Fuller-Pace F V. The “DEAD box” protein DbpA interacts specifically with the peptidyltransferase center in 23S rRNA. Proc Natl Acad Sci USA. 1995;92:11681–11685. doi: 10.1073/pnas.92.25.11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3178.Niederhoffer E C, Naranjo C M, Bradley K L, Fee J A. Control of Escherichia coli superoxide dismutase (sodA and sodB) genes by the ferric uptake regulation (fur) locus. J Bacteriol. 1990;172:1930–1938. doi: 10.1128/jb.172.4.1930-1938.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3179.Niegemann E, Schulz A, Bartsch K. Molecular organization of the Escherichia coli gab cluster: nucleotide sequence of the structural genes gabD and gabP and expression of the GABA permease gene. Arch Microbiol. 1993;160:454–460. doi: 10.1007/BF00245306. [DOI] [PubMed] [Google Scholar]
- 3180.Nielson J, Jorgensen B B, von Meyenburg K, Hansen F G. The promoters of the atp operon of Escherichia coli K12. Mol Gen Genet. 1984;193:64–71. doi: 10.1007/BF00327415. [DOI] [PubMed] [Google Scholar]
- 3181.Niersbach M, Kreuzaler F, Geerse R H, Postma P W, Hirsch H J. Cloning and nucleotide sequence of the Escherichia coli K-12 ppsA gene, encoding PEP synthase. Mol Gen Genet. 1992;231:332–336. doi: 10.1007/BF00279808. [DOI] [PubMed] [Google Scholar]
- 3182.Nieto J M, Carmona M, Bolland S, Jubete Y, de la Cruz F, Juarez A. The hha gene modulates haemolysin expression in Escherichia coli. Mol Microbiol. 1991;5:1285–1293. doi: 10.1111/j.1365-2958.1991.tb01902.x. [DOI] [PubMed] [Google Scholar]
- 3183.Niki H, Jaffe A, Imamura N, Ogura T, Hiraga S. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 1991;10:183–193. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3184.Niki H, Ichinose C, Ogura T, Mori H, Morita M, Hasegawa M, Kusukawa N, Hiraga S. Chromosomal genes essential for stable maintenance of the mini-F plasmid in Escherichia coli. J Bacteriol. 1988;170:5272–5278. doi: 10.1128/jb.170.11.5272-5278.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3185.Niki H, Imamura R, Kitaoka M, Yamanaka K, Ogura T, Hiraga S. E. coli MukB protein involved in chromosome partition forms a homodimer with a rod-and-hinge structure having DNA binding and ATP/GTP binding activities. EMBO J. 1992;11:5101–5109. doi: 10.1002/j.1460-2075.1992.tb05617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3186.Niki H, Imamura R, Ogura T, Hiraga S. Nucleotide sequence of the tolC gene of Escherichia coli. Nucleic Acids Res. 1990;18:5547. doi: 10.1093/nar/18.18.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3187.Niki H, Ogura T, Hiraga S. Linear multimer formation of plasmid DNA in Escherichia coli hopE (recD) mutants. Mol Gen Genet. 1990;224:1–9. doi: 10.1007/BF00259444. [DOI] [PubMed] [Google Scholar]
- 3188.Nilles M L, Bertrand K. The Escherichia coli acrD gene encodes a homolog of the AcrB and EnvD multidrug efflux proteins. 1994. GenBank submission U12598. [Google Scholar]
- 3189.Nilsen I W, Bakke I, Vader A, Olsvik O, El-Gewely M R. Isolation of cmr, a novel Escherichia coli chloramphenicol resistance gene encoding a putative efflux pump. J Bacteriol. 1996;178:3188–3193. doi: 10.1128/jb.178.11.3188-3193.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3190.Nilsson O, Aberg A, Lundqvist T, Sjoberg B-M. Nucleotide sequence of the gene coding for the large subunit of ribonucleotide reductase of Escherichia coli. Nucleic Acids Res. 1988;16:4174. doi: 10.1093/nar/16.9.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3191.Nishi K, Morel-Deville F, Hershey J W B, Leighton T, Schnier J. An eIF-4A-like protein is a suppressor of an Escherichia coli mutant defective in 50S ribosomal subunit assembly. Nature. 1988;336:496–498. doi: 10.1038/336496a0. . (Erratum, 340:246.) [DOI] [PubMed] [Google Scholar]
- 3192.Nishida M, Kong K H, Inoue H, Takahashi K. Molecular cloning and site-directed mutagenesis of glutathione S-transferase from Escherichia coli. The conserved tyrosyl residue near the N terminus is not essential for catalysis. J Biol Chem. 1994;269:32536–32541. [PubMed] [Google Scholar]
- 3193.Nishijima M, Asami Y, Uetake N, Yamagoe S, Ohta A, Shibuya I. Disruption of the Escherichia coli cls gene responsible for cariolipin synthesis. J Bacteriol. 1988;170:775–780. doi: 10.1128/jb.170.2.775-780.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3194.Nishimura A. A new gene controlling the frequency of cell division per round of DNA replication in Escherichia coli. Mol Gen Genet. 1989;215:286–293. doi: 10.1007/BF00339730. [DOI] [PubMed] [Google Scholar]
- 3195.Nishimura K, Nakahigashi K, Inokuchi H. Location of the ubiA gene on the physical map of Escherichia coli. J Bacteriol. 1992;174:5762. doi: 10.1128/jb.174.17.5762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3196.Nishimura K, Nakayashiki T, Inokuchi H. Cloning and sequencing of the hemE gene encoding uroporphyrinogen III decarboxylase (UPD) from Escherichia coli K-12. Gene. 1993;133:109–113. doi: 10.1016/0378-1119(93)90233-s. [DOI] [PubMed] [Google Scholar]
- 3197.Nishimura K, Nakayashiki T, Inokuchi H. Cloning and identification of the hemG gene encoding protoporphyrinogen oxidase (PPO) of Escherichia coli. DNA Res. 1995;2:1–8. doi: 10.1093/dnares/2.1.1. [DOI] [PubMed] [Google Scholar]
- 3198.Nishimura Y, Suzuki H, Hirota Y, Park J T. A mutant of Escherichia coli defective in penicillin-binding protein 5 and lacking d-alanine carboxypeptidase IA. J Bacteriol. 1980;143:531–534. doi: 10.1128/jb.143.1.531-534.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3199.Nishiyama K, Hanada M, Tokuda H. Disruption of the gene encoding p12 (SecG) reveals the direct involvement and important function of SecG in the protein translocation of Escherichia coli at low temperature. EMBO J. 1994;13:3272–3277. doi: 10.1002/j.1460-2075.1994.tb06628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3200.Nishiyama K, Mizushima S, Tokuda H. The carboxyl-terminal region of SecE interacts with SecY and is functional in the reconstitution of protein translocation activity in Escherichia coli. J Biol Chem. 1992;267:7170–7176. [PubMed] [Google Scholar]
- 3201.Nishiyama K, Mizushima S, Tokuda H. A novel membrane protein involved in protein translocation across the cytoplasmic membrane of Escherichia coli. EMBO J. 1993;12:3409–3415. doi: 10.1002/j.1460-2075.1993.tb06015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3202.Nobelmann B, Lengeler J W. Molecular analysis of the gat genes from Escherichia coli and of their roles in galactitol transport and metabolism. J Bacteriol. 1996;178:6790–6795. doi: 10.1128/jb.178.23.6790-6795.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3203.Noble J A, Innis M A, Koonin E V, Rudd K E, Banuett F, Herskowitz I. The Escherichia coli hflA locus encodes a putative GTP-binding protein and two membrane proteins, one of which contains a protease-like domain. Proc Natl Acad Sci USA. 1993;90:10866–10870. doi: 10.1073/pnas.90.22.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3204.Noble M-A, Ishiguro E E. Temperature-sensitive mutation in lytF, a new gene involved in autolysis of Escherichia coli. J Gen Microbiol. 1993;139:3109–3113. doi: 10.1099/00221287-139-12-3109. [DOI] [PubMed] [Google Scholar]
- 3205.Noguchi S, Nishimura Y, Nishimura S. Isolation and characterization of an Escherichia coli mutant lacking tRNA-guanine transglycosylase. J Biol Chem. 1982;257:6544–6550. [PubMed] [Google Scholar]
- 3206.Nohno T, Saito T. Two transcriptional start sites found in the promoter region of Escherichia coli glutamine permease operon, glnHPQ. Nucleic Acids Res. 1987;15:2777. doi: 10.1093/nar/15.6.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3207.Nohno T, Saito T, Hong J-S. Cloning and complete nucleotide sequence of the Escherichia coli glutamine permease operon (glnHPQ) Mol Gen Genet. 1986;205:260–269. doi: 10.1007/BF00430437. [DOI] [PubMed] [Google Scholar]
- 3208.Nohno T, Kasai Y, Saito T. Cloning and sequencing of the Escherichia coli chlEN operon involved in molybdopterin biosynthesis. J Bacteriol. 1988;170:4097–4102. doi: 10.1128/jb.170.9.4097-4102.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3209.Nomura T, Aiba H, Ishihama A. Transcriptional organization of the convergent overlapping dnaQ-rnh genes of Escherichia coli. J Biol Chem. 1985;260:7122–7125. [PubMed] [Google Scholar]
- 3210.Nomura T, Fujita N, Ishihama A. Expression of the leuX gene in Escherichia coli: regulation at transcription and tRNA processing steps. J Mol Biol. 1987;197:659–670. doi: 10.1016/0022-2836(87)90472-4. [DOI] [PubMed] [Google Scholar]
- 3211.Nonet M L, Marvel C C, Tolan D R. The hisT-purF region of the Escherichia coli K-12 chromosome. Identification of additional genes of the hisT and purF operons. J Biol Chem. 1987;262:12209–12217. [PubMed] [Google Scholar]
- 3212.Normark S, Norlander L, Grundstrom T, Bloom G D, Boquel P, Frelat G. Septum formation-defective mutant of Escherichia coli. J Bacteriol. 1976;128:401–412. doi: 10.1128/jb.128.1.401-412.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3213.Normark S, Edlund T, Grundström T, Bergström S, Wolf-Watz H. Escherichia coli K-12 mutants hyperproducing chromosomal beta-lactamase by gene repetitions. J Bacteriol. 1977;132:912–922. doi: 10.1128/jb.132.3.912-922.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3214.Norregaard-Madsen M, Mygind B, Pedersen R, Valentin-Hansen P, Sogaard-Andersen L. The gene encoding the periplasmic cyclophilin homologue, PPIase A, in Escherichia coli, is expressed from four promoters, three of which are activated by the cAMP-CRP complex and negatively regulated by the CytR repressor. Mol Microbiol. 1994;14:989–997. doi: 10.1111/j.1365-2958.1994.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 3215.Norregaard-Madsen M, McFall E, Valentin-Hansen P. Organization and transcriptional regulation of the Escherichia coli K-12 d-serine tolerance locus. J Bacteriol. 1995;177:6456–6461. doi: 10.1128/jb.177.22.6456-6461.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3216.Norris V J, Alliotte T, Jaffe A, D’Ari R. DNA replication termination in Escherichia coli parB (a dnaG allele), parA, and gyrB mutants affected in DNA distribution. J Bacteriol. 1986;168:494–504. doi: 10.1128/jb.168.2.494-504.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3217.Norwood W I, Sadler J R. Pseudoreversion of lactose operator-constitutive mutants. J Bacteriol. 1977;130:100–106. doi: 10.1128/jb.130.1.100-106.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3218.Nouwen N, de Kruijff B, Tommassen J. prlA suppressors in Escherichia coli relieve the proton electrochemical gradient dependency of translocation of wild-type precursors. Proc Natl Acad Sci USA. 1996;93:5953–5957. doi: 10.1073/pnas.93.12.5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3219.Novel G, Novel M. Mutants d’Escherichia coli K12 affectés pour leur croissance sur methyl-β-d-glucuronide: localisation du gene de structure de la β-d-glucuronidase (uidA) Mol Gen Genet. 1973;120:319–335. [PubMed] [Google Scholar]
- 3220.Novel M, Novel G. Regulation of β-glucuronidase synthesis in Escherichia coli K-12: constitutive mutants specifically derepressed for uidA expression. J Bacteriol. 1976;127:406–417. doi: 10.1128/jb.127.1.406-417.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3221.Nuesch J, Schumperli D. Structural and functional organization of the gpt gene region of Escherichia coli. Gene. 1984;32:243–249. doi: 10.1016/0378-1119(84)90052-0. [DOI] [PubMed] [Google Scholar]
- 3222.Nunn W D, Simons R W. Transport of long-chain fatty acids by Escherichia coli: mapping and characterization of mutants in the fadL gene. Proc Natl Acad Sci USA. 1978;75:3377–3381. doi: 10.1073/pnas.75.7.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3223.Nunoshiba T, Hidalgo E, Amabile Cuevas C, Demple B. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J Bacteriol. 1992;174:6054–6060. doi: 10.1128/jb.174.19.6054-6060.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3224.Nunoshiba T, Hidalgo E, Li Z-W, Demple B. Negative autoregulation by the Escherichia coli SoxS protein: a dampening mechanism for the soxRS redox stress response. J Bacteriol. 1993;175:7492–7494. doi: 10.1128/jb.175.22.7492-7494.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3225.Nurminen M, Hirvas L, Vaara M. The outer membrane of lipid A-deficient Escherichia coli mutant LH530 has reduced levels of OmpF and leaks periplasmic enzymes. Microbiology. 1997;143:1533–1537. doi: 10.1099/00221287-143-5-1533. [DOI] [PubMed] [Google Scholar]
- 3226.Nurse K, Wrzesinski J, Bakin A, Lane B G, Ofengand J. Purification, cloning, and properties of the tRNA psi-55 synthase from Escherichia coli. RNA. 1995;1:102–112. [PMC free article] [PubMed] [Google Scholar]
- 3227.Nurse P, Zavitz K H, Marians K J. Inactivation of the Escherichia coli PriA DNA replication protein induces the SOS response. J Bacteriol. 1991;173:6686–6693. doi: 10.1128/jb.173.21.6686-6693.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3228.Nurse P, DiGate R J, Zavitz K H, Marians K J. Molecular cloning and DNA sequence analysis of Escherichia coli priA, the gene encoding the primosomal protein replication factor Y. Proc Natl Acad Sci USA. 1990;87:4615–4619. doi: 10.1073/pnas.87.12.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3229.Ny T, Bjork G R. Cloning and restriction mapping of the trmA gene coding for transfer ribonucleic acid (5-methyluridine)methyltransferase in Escherichia coli K-12. J Bacteriol. 1980;142:371–379. doi: 10.1128/jb.142.2.371-379.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3230.Nygaard P, Smith J M. Evidence for a novel glycinamide ribonucleotide transformylase in Escherichia coli. J Bacteriol. 1993;175:3591–3597. doi: 10.1128/jb.175.11.3591-3597.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3231.Nyström T. Glucose starvation stimulon of Escherichia coli: role of integration host factor in starvation survival and growth phase-dependent protein synthesis. J Bacteriol. 1995;177:5707–5710. doi: 10.1128/jb.177.19.5707-5710.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3232.Nystrom T, Neidhardt F C. Cloning, mapping, and nucleotide sequencing of a gene encoding a universal stress protein in Escherichia coli. Mol Microbiol. 1992;6:3187–3198. doi: 10.1111/j.1365-2958.1992.tb01774.x. [DOI] [PubMed] [Google Scholar]
- 3233.Nystrom T, Neidhardt F C. Isolation and properties of a mutant of Escherichia coli with an insertional activation of the uspA gene, which encodes a universal stress protein. J Bacteriol. 1993;175:3949–3956. doi: 10.1128/jb.175.13.3949-3956.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3234.Nyunoya H, Lusty C J. The carB gene of Escherichia coli: a duplicated gene coding for the large subunit of carbamoyl-phosphate synthetase. Proc Natl Acad Sci USA. 1983;80:4629–4633. doi: 10.1073/pnas.80.15.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3235.Oberto J, Bonnefoy E, Mouray E, Pellegrini O, Wikstrom P M, Rouviere-Yaniv J. The Escherichia coli ribosomal protein S16 is an endonuclease. Mol Microbiol. 1996;19:1319–1330. doi: 10.1111/j.1365-2958.1996.tb02476.x. [DOI] [PubMed] [Google Scholar]
- 3236.O’Connor M, Dahlberg A E. The involvement of two distinct regions of 23 S ribosomal RNA in tRNA selection. J Mol Biol. 1995;254:838–840. doi: 10.1006/jmbi.1995.0659. [DOI] [PubMed] [Google Scholar]
- 3237.O’Connor M, Willis N M, Bossi L, Gesteland R F, Atkins J F. Functional tRNAs with altered 3′ ends. EMBO J. 1993;12:2559–2566. doi: 10.1002/j.1460-2075.1993.tb05911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3238.O’Connor M, Gesteland R F, Atkins J F. Sequence of E. coli K12 valU operon which contains 3 genes for tRNA1Val and 1 gene for tRNALys. Nucleic Acids Res. 1990;18:672. doi: 10.1093/nar/18.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3239.O’Day K, Lopilato J, Wright A. Physical locations of bglA and serA on the Escherichia coli K-12 chromosome. J Bacteriol. 1991;173:1571. doi: 10.1128/jb.173.5.1571.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3240.Odoevskaya E R, Sineokii S P. Isolation and genetic study of the bacterial mutations gpr blocking the replication of certain lambdoid phages. Sov Genet (Engl Transl Genetika) 1987;23:432–440. [PubMed] [Google Scholar]
- 3241.O’Donovan G A, Edlin G, Fuchs J A, Neuhard J, Thomassen K. Deoxycytidine triphosphate deaminase: characterization of an Escherichia coli mutant deficient in the enzyme. J Bacteriol. 1971;105:666–672. doi: 10.1128/jb.105.2.666-672.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3242.Oeda K, Horiuchi T, Sekiguchi M. Molecular cloning of the uvrD gene of Escherichia coli that controls ultraviolet sensitivity and spontaneous mutation frequency. Mol Gen Genet. 1981;184:191–199. doi: 10.1007/BF00272904. [DOI] [PubMed] [Google Scholar]
- 3243.Oeda K, Horiuchi T, Sekiguchi M. The uvrD gene of E. coli encodes a DNA-dependent ATPase. Nature. 1982;298:98–100. doi: 10.1038/298098a0. [DOI] [PubMed] [Google Scholar]
- 3244.Ogawa T, Pickett G G, Kogoma T, Kornberg A. RNase H confers specificity in the dnaA-dependent initiation of replication at the unique origin of the Escherichia coli chromosome in vivo and in vitro. Proc Natl Acad Sci USA. 1984;81:1040–1044. doi: 10.1073/pnas.81.4.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3245.Ogawa T, Okazaki T. Cell cycle-dependent transcription from the gid and mioC promoters of Escherichia coli. J Bacteriol. 1994;176:1609–1615. doi: 10.1128/jb.176.6.1609-1615.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3246.Ogawa W, Kayahara T, Tsuda M, Mizushima T, Tsuchiya T. Isolation and characterization of an Escherichia coli mutant lacking the major serine transporter, and cloning of a serine transporter gene. J Biochem (Tokyo) 1997;122:1241–1245. doi: 10.1093/oxfordjournals.jbchem.a021887. [DOI] [PubMed] [Google Scholar]
- 3247.Ogden S, Haggerty D, Stoner C M, Kolodrubetz D, Schleif R. The Escherichia colil-arabinose operon: binding sites of the regulatory proteins and a mechanism of positive and negative regulation. Proc Natl Acad Sci USA. 1980;77:3346–3350. doi: 10.1073/pnas.77.6.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3248.Ogino T, Matsubara M, Kato N, Nakamura Y, Mizuno T. An Escherichia coli protein that exhibits phosphohistidine phosphatase activity towards the HPt domain of the ArcB sensor involved in the multistep His-Asp phosphorelay. Mol Microbiol. 1998;27:573–585. doi: 10.1046/j.1365-2958.1998.00703.x. [DOI] [PubMed] [Google Scholar]
- 3249.Ogura T, Miki T, Hiraga S. Copy-number mutants of the plasmid carrying the replication origin of the Escherichia coli chromosome: evidence for a control region of replication. Proc Natl Acad Sci USA. 1980;77:3993–3997. doi: 10.1073/pnas.77.7.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3250.Ogura T, Tomoyasu T, Yuki T, Morimura S, Begg K J, Donachie W D, Mori H, Niki H, Hiraga S. Structure and function of the ftsH gene in Escherichia coli. Res Microbiol. 1991;142:279–282. doi: 10.1016/0923-2508(91)90041-8. [DOI] [PubMed] [Google Scholar]
- 3251.Oh B K, Chauhan A K, Isono K, Apirion D. Location of a gene (ssrA) for a small, stable RNA (10Sa RNA) in the Escherichia coli chromosome. J Bacteriol. 1990;172:4708–4709. doi: 10.1128/jb.172.8.4708-4709.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3252.Oh B K, Apirion D. 10Sa RNA, a small stable RNA of Escherichia coli, is functional. Mol Gen Genet. 1991;229:52–56. doi: 10.1007/BF00264212. [DOI] [PubMed] [Google Scholar]
- 3253.Oh W, Larson T J. Physical locations of genes in the rne (ams)-rpmF-plsX-fab region of the Escherichia coli K-12 chromosome. J Bacteriol. 1992;174:7873–7874. doi: 10.1128/jb.174.23.7873-7874.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3254.O’Handley S F, Frick D N, Bullions L C, Mildvan A S, Bessman M J. Escherichia coli orf17 codes for a nucleoside triphosphate pyrophosphohydrolase member of the MutT family of proteins. Cloning, purification, and characterization of the enzyme. J Biol Chem. 1996;271:24649–24654. doi: 10.1074/jbc.271.40.24649. [DOI] [PubMed] [Google Scholar]
- 3255.Ohara O, Dorit R L, Gilbert W. Direct genomic sequencing of bacterial DNA: the pyruvate kinase I gene of Escherichia coli. Proc Natl Acad Sci USA. 1989;86:6883–6887. doi: 10.1073/pnas.86.18.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3256.Ohki M, Tamura F, Nishimura S, Uchida H. Nucleotide sequence of the Escherichia coli dnaJ gene and purification of the gene product. J Biol Chem. 1986;261:1778–1781. [PubMed] [Google Scholar]
- 3257.Ohki R, Morita R, Kawamata T, Uchida H, Ohki M. A complete deletion mutant of the Escherichia coli dnaKdnaJ operon. Biochim Biophys Acta. 1989;1009:94–98. doi: 10.1016/0167-4781(89)90085-7. [DOI] [PubMed] [Google Scholar]
- 3258.Ohki R, Kawamata T, Katoh Y, Hosoda F, Ohki M. Escherichia coli dnaJ deletion mutation results in loss of stability of a positive regulator, CRP. J Biol Chem. 1992;267:13180–13184. [PubMed] [Google Scholar]
- 3259.Ohmori, H. 1992. GenBank submission L02122.
- 3260.Ohmori H. Structural analysis of the rhlE gene of Escherichia coli. Jpn J Genet. 1994;69:1–12. doi: 10.1266/jjg.69.1. [DOI] [PubMed] [Google Scholar]
- 3261.Ohmori H, Kimura A, Nagata T, Sakakibara Y. Structural analysis of the dnaA and dnaN genes of Escherichia coli. Gene. 1984;28:159–170. doi: 10.1016/0378-1119(84)90253-1. [DOI] [PubMed] [Google Scholar]
- 3262.Ohmori H, Saito M, Yasuda T, Nagata T, Fujii T, Wachi M, Nagai K. The pcsA gene is identical to dinD in Escherichia coli. J Bacteriol. 1995;177:156–165. doi: 10.1128/jb.177.1.156-165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3263.Ohnishi Y. Genetic analysis of an Escherichia coli mutant with a lesion in stable RNA turnover. Genetics. 1974;76:185–194. doi: 10.1093/genetics/76.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3264.Ohsawa M, Mogi T, Yamamoto H, Yamato I, Anraku Y. Proline carrier mutant of Escherichia coli K-12 with altered cation sensitivity of substrate-binding activity: cloning, biochemical characterization and identification of the mutation. J Bacteriol. 1988;170:5185–5191. doi: 10.1128/jb.170.11.5185-5191.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3265.Ohsumi M, Sekiya T, Nishimura S, Ohki M. Nucleotide sequence of the regulatory region of malB operons in E. coli. J Biochem (Tokyo) 1983;94:243–247. doi: 10.1093/oxfordjournals.jbchem.a134335. [DOI] [PubMed] [Google Scholar]
- 3266.Ohta A, Waggoner K, Radominska-Pyrek A, Dowhan W. Cloning of genes involved in membrane lipid synthesis: effects of amplification of phosphatidylglycerophosphate synthase in Escherichia coli. J Bacteriol. 1981;147:552–562. doi: 10.1128/jb.147.2.552-562.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3267.Ohta A, Obara T, Asami Y, Shibuya I. Molecular cloning of the cls gene responsible for cardiolipin synthesis in Escherichia coli and phenotypic consequences of its amplification. J Bacteriol. 1985;163:506–514. doi: 10.1128/jb.163.2.506-514.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3268.Ohta M, Ina K, Kusuzaki K, Kido N, Arakawa Y, Kato N. Cloning and expression of the rfe-rff gene cluster of Escherichia coli. Mol Microbiol. 1991;5:1853–1862. doi: 10.1111/j.1365-2958.1991.tb00809.x. [DOI] [PubMed] [Google Scholar]
- 3269.Ohta T, Waggoner K, Louie K, Dowhan W. Cloning of genes involved in membrane lipid synthesis. Effects of amplification of phosphatidylserine synthetase in Escherichia coli. J Biol Chem. 1981;256:2219–2225. [PubMed] [Google Scholar]
- 3270.Ohta Y, Watanabe K, Kimura A. Complete nucleotide sequence of the E. coli N-acetylneuraminate lyase. Nucleic Acids Res. 1985;13:8843–8852. doi: 10.1093/nar/13.24.8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3271.Ohyama T, Igarashi K, Kobayashi H. Physiological role of the chaA gene in sodium and calcium circulations at a high pH in Escherichia coli. J Bacteriol. 1994;176:4311–4315. doi: 10.1128/jb.176.14.4311-4315.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3272.Ohyama T, Imaizumi R, Igarashi K, Kobayashi H. Escherichia coli is able to grow with negligible sodium ion extrusion activity at alkaline pH. J Bacteriol. 1992;174:7743–7749. doi: 10.1128/jb.174.23.7743-7749.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3273.Oka A, Sasaki H, Sugimoto K, Takami M. Sequence organization of replication origin of the Escherichia coli K-12 chromosome. J Mol Biol. 1984;176:443–458. doi: 10.1016/0022-2836(84)90171-2. [DOI] [PubMed] [Google Scholar]
- 3274.Oka A, Sugisaki H, Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981;147:217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- 3275.Oka A, Sugimoto K, Sasaki H, Takanami M. An in vitro method generating base substitutions in preselected regions of plasmid DNA: application to structural analysis of the replication origin of the Escherichia coli K-12 chromosome. Gene. 1982;19:59–69. doi: 10.1016/0378-1119(82)90189-5. [DOI] [PubMed] [Google Scholar]
- 3276.Oka A, Sugimoto K, Takanami M, Hirota Y. Replication origin of Escherichia coli K-12 chromosome: the size and structure of the minimum DNA segment carrying the information for autonomous replication. Mol Gen Genet. 1980;178:9–20. doi: 10.1007/BF00267207. [DOI] [PubMed] [Google Scholar]
- 3277.Okada K, Minehira M, Zhu X, Suzuki K, Nakagawa T, Matsuda H, Kawamukai M. The ispB gene encoding octaprenyl diphosphate synthase is essential for growth of Escherichia coli. J Bacteriol. 1997;179:3058–3060. doi: 10.1128/jb.179.9.3058-3060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3278.Okada Y, Wachi M, Hirata A, Suzuki K, Nagai K, Matsuhashi M. Cytoplasmic axial filaments in Escherichia coli cells: possible function in the mechanism of chromosome segregation and cell division. J Bacteriol. 1994;176:917–922. doi: 10.1128/jb.176.3.917-922.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3279.Okamoto K, Freundlich M. Mechanism for the autogenous control of the crp operon: transcriptional inhibition by a divergent RNA transcript. Proc Natl Acad Sci USA. 1986;83:5000–5004. doi: 10.1073/pnas.83.14.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3280.Okamura-Ikeda K, Ohmura Y, Fujiwara K, Motokawa Y. Cloning and nucleotide sequence of the gcv operon encoding Escherichia coli glycine cleavage system. Eur J Biochem. 1993;216:539–548. doi: 10.1111/j.1432-1033.1993.tb18172.x. [DOI] [PubMed] [Google Scholar]
- 3281.Okita T W, Rodriguez R L, Preiss J. Biosynthesis of bacterial glycogen. Cloning of the glycogen biosynthetic enzyme structural genes of Escherichia coli. J Biol Chem. 1981;256:6944–6952. [PubMed] [Google Scholar]
- 3282.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3283.Old I G, Margarita D, Glass R E, Saint Girons I. Nucleotide sequence of the metH gene of Escherichia coli K-12 and comparison with that of Salmonella typhimurium. Gene. 1990;87:15–21. doi: 10.1016/0378-1119(90)90490-i. [DOI] [PubMed] [Google Scholar]
- 3284.Old I G, Hunter M G, Wilson D T R, Knight S M, Weatherston C A, Glass R E. Cloning and characterization of the genes for the two homocysteine transmethylases of Escherichia coli. Mol Gen Genet. 1988;211:78–87. doi: 10.1007/BF00338396. [DOI] [PubMed] [Google Scholar]
- 3285.Oliver D B. Identification of five new essential genes involved in the synthesis of a secreted protein in Escherichia coli. J Bacteriol. 1985;161:285–291. doi: 10.1128/jb.161.1.285-291.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3286.Oliver D B, Beckwith J R. E. coli mutant pleiotropically defective in the export of secreted proteins. Cell. 1981;25:765–772. doi: 10.1016/0092-8674(81)90184-7. [DOI] [PubMed] [Google Scholar]
- 3287.Oliver D B, Beckwith J R. Identification of a new gene (secA) and gene product involved in the secretion of envelope proteins in Escherichia coli. J Bacteriol. 1982;150:686–691. doi: 10.1128/jb.150.2.686-691.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3288.Oliver G, Gosset G, Sanchez-Pescador R, Lozoya E, Ku L M, Flores N, Valle F, Becerril B, Bolivar F. Determination of the sequence nucleotide for the glutamate synthase structural genes of Escherichia coli. Gene. 1987;60:1–11. doi: 10.1016/0378-1119(87)90207-1. [DOI] [PubMed] [Google Scholar]
- 3289.Olsen A, Arnqvist A, Hammar M, Normark S. Environmental regulation of curli production in Escherichia coli. Infect Agents Dis. 1993;2:272–274. [PubMed] [Google Scholar]
- 3290.Olsen A, Arnqvist A, Sukupolvi S, Normark S. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol Microbiol. 1993;7:523–536. doi: 10.1111/j.1365-2958.1993.tb01143.x. [DOI] [PubMed] [Google Scholar]
- 3291.Olsen A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 3292.Olsen M K, Rosey E L, Tomich C-S C. Isolation and analysis of novel mutants of Escherichia coli prlA (secY) J Bacteriol. 1993;175:7092–7096. doi: 10.1128/jb.175.21.7092-7096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3293.Olsen S G, Greene K M, Brooker R J. Lactose permease mutants which transport malto-oligosaccharides. J Bacteriol. 1993;175:6269–6275. doi: 10.1128/jb.175.19.6269-6275.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3294.Olson E R, Dunyak D S, Jurss L M, Poorman R A. Identification and characterization of dppA, an Escherichia coli gene encoding a periplasmic dipeptide transport protein. J Bacteriol. 1991;173:234–244. doi: 10.1128/jb.173.1.234-244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3295.Olson M W, Dallmann H G, McHenry C S. DnaX complex of Escherichia coli DNA polymerase III holoenzyme. The χ · ψ complex functions by increasing the affinity of τ and γ for δ · δ′ to a physiologically relevant range. J Biol Chem. 1995;270:29570–29577. [PubMed] [Google Scholar]
- 3296.Omote H, Le N P, Park M Y, Maeda M, Futai M. Beta subunit Glu-185 of Escherichia coli H(+)-ATPase (ATP synthase) is an essential residue for cooperative catalysis. J Biol Chem. 1995;270:25656–25660. doi: 10.1074/jbc.270.43.25656. [DOI] [PubMed] [Google Scholar]
- 3297.O’Neill G P, Throbjarnardóttir S, Michelsen U, Pálsson S, Söll D, Eggertsson G. δ-Aminolevulinic acid dehydratase deficiency can cause δ-aminolevulinate auxotrophy in Escherichia coli. J Bacteriol. 1991;173:94–100. doi: 10.1128/jb.173.1.94-100.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3298.Ono M, Kuwano M. Mutation affecting the thermolability of the 50S ribosomal subunit in Escherichia coli. J Bacteriol. 1978;134:677–679. doi: 10.1128/jb.134.2.677-679.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3299.Ono M, Kuwano M. Chromosomal location of a gene for chemical longevity of messenger ribonucleic acid in a temperature-sensitive mutant of Escherichia coli. J Bacteriol. 1980;142:325–326. doi: 10.1128/jb.142.1.325-326.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3300.Onrust R, O’Donnell M. DNA polymerase III accessory proteins. II. Characterization of delta and delta′. J Biol Chem. 1993;268:11766–11772. [PubMed] [Google Scholar]
- 3301.Oram M, Fisher L M. An Escherichia coli DNA topoisomerase I mutant has a compensatory mutation that alters two residues between functional domains of the DNA gyrase A protein. J Bacteriol. 1992;174:4175–4178. doi: 10.1128/jb.174.12.4175-4178.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3302.Ordal G W, Adler J. Isolation and complementation of mutants in galactose taxis and transport. J Bacteriol. 1974;117:509–516. doi: 10.1128/jb.117.2.509-516.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3303.O’Regan M, Gloeckler R, Bernard S, Ledoux C, Ohsawa I, Lemoine Y. Nucleotide sequence of the bioH gene of Escherichia coli. Nucleic Acids Res. 1989;17:8004. doi: 10.1093/nar/17.19.8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3304.Orr G A, Fairweather N F, Holland I B, Pritchard R H. Isolation and characterization of a strain carrying a conditional lethal mutation in the cou gene of Escherichia coli K-12. Mol Gen Genet. 1979;177:103–112. doi: 10.1007/BF00267259. [DOI] [PubMed] [Google Scholar]
- 3305.Orth V, Chippaux M, Pascal M C. A mutant defective in electron transfer to nitrate in Escherichia coli K12. J Gen Microbiol. 1980;117:257–262. doi: 10.1099/00221287-117-1-257. [DOI] [PubMed] [Google Scholar]
- 3306.Osborne C, Chen L M, Matthews R G. Isolation, cloning, mapping, and nucleotide sequencing of the gene encoding flavodoxin in Escherichia coli. J Bacteriol. 1991;173:1729–1737. doi: 10.1128/jb.173.5.1729-1737.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3307.Osborne R S, Silhavy T J. PrlA suppressor mutations cluster in regions corresponding to three distinct topological domains. EMBO J. 1993;12:3391–3398. doi: 10.1002/j.1460-2075.1993.tb06013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3308.Oshima T, Aiba H, Baba T, Fujita K, Hayashi K, Honjo A, et al. A 718-kb DNA sequence of the Escherichia coli K-12 genome corresponding to the 12.7-28.0 min region on the linkage map. DNA Res. 1996;3:137–155. doi: 10.1093/dnares/3.3.137. [DOI] [PubMed] [Google Scholar]
- 3309.Ost K A, Deutscher M P. Escherichia coli orfE (upstream of pyrE) encodes RNase PH. J Bacteriol. 1991;173:5589–5591. doi: 10.1128/jb.173.17.5589-5591.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3310.Ostrowski J, Jagura-Burdzy G, Kredich N M. DNA sequences of the cysB regions of Salmonella typhimurium and Escherichia coli. J Biol Chem. 1987;262:5999–6005. [PubMed] [Google Scholar]
- 3311.Otsuji N, Soejima T, Maki S, Shinagawa H. Cloning of colicin E1 tolerant tolC (mtcB) gene of Escherichia coli K12 and identification of its gene product. Mol Gen Genet. 1982;187:30–36. doi: 10.1007/BF00384379. [DOI] [PubMed] [Google Scholar]
- 3312.Otsuka, A. J., M. R. Buoncristiani, P. K. Howard, J. Flamm, C. Johnson, R. Yamamoto, K. Uchida, C. C. Cook, J. Ruppert, and J. Matsuzaki. 1988. The Escherichia coli biotin biosynthetic enzyme sequences predicted from the nucleotide sequence of the bio operon. J. Biol. Chem. 19577–19585. [PubMed]
- 3313.Ou J T, Kuo L M. Suppression of the formation of polygenotypic recombinant colonies by a maf mutation in mating with HfrH. Genetics. 1979;93:345–351. doi: 10.1093/genetics/93.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3314.Ouzounis C A, Blencowe B J. Bacterial DNA replication initiation factor priA is related to proteins belonging to the ‘DEAD-box’ family. Nucleic Acids Res. 1991;19:6953. doi: 10.1093/nar/19.24.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3315.Ovchinnikov Y A, Monastyrskaya G S, Gubanov V V, Guryev S O, Salomatina I S, Shuvaeva T M, Lipkin V M, Sverdlov E D. The primary structure of E. coli RNA polymerase. Nucleotide sequence of the rpoC gene and amino acid sequence of the B′-subunit. Nucleic Acids Res. 1982;10:4035–4044. doi: 10.1093/nar/10.13.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3316.Ovchinnikov Y A, Monastyrskaya G S, Gubanov V V, Guryev S O, Chertov O Y, Modyanov N N, Grinkevich V A, Makarova I A, Marchenko T V, Polovnikova I N, Lipkin V M, Sverdlov E D. The primary structure of Escherichia coli RNA polymerase. Nucleotide sequence of the rpoB gene and amino-acid sequence of the B-subunit. Eur J Biochem. 1982;116:621–629. doi: 10.1111/j.1432-1033.1981.tb05381.x. [DOI] [PubMed] [Google Scholar]
- 3317.Ovchinnikov Y A, Aldanova N A, Grinkevich V A, Arzamazova N M, Moroz I N. The primary structure of a Leu, Ile and Val (LIV)-binding protein from Escherichia coli. FEBS Lett. 1977;78:313–316. doi: 10.1016/0014-5793(77)80331-1. [DOI] [PubMed] [Google Scholar]
- 3318.Ovchinnikov Y A, Lipkin V M, Modyanov N N, Chertov O Y, Smirnov Y V. Primary structure of alpha-subunit of DNA-dependent RNA polymerase from Escherichia coli. FEBS Lett. 1977;76:108–111. doi: 10.1016/0014-5793(77)80131-2. [DOI] [PubMed] [Google Scholar]
- 3319.Overbeeke N, Bergmans H, vanMansfield F, Lugtenberg B. Complete nucleotide sequence of phoE, the structural gene for the phosphate limitation inducible outer membrane pore protein of Escherichia coli K12. J Mol Biol. 1983;163:513–532. doi: 10.1016/0022-2836(83)90110-9. [DOI] [PubMed] [Google Scholar]
- 3320.Overduin P, Boos W, Tommassen J. Nucleotide sequence of the upg genes of Escherichia coli K-12: homology to the maltose system. Mol Microbiol. 1988;2:767–775. doi: 10.1111/j.1365-2958.1988.tb00088.x. [DOI] [PubMed] [Google Scholar]
- 3321.Oxender D L, Zurawski G, Yanofsky C. Attenuation in the Escherichia coli tryptophan operon: role of RNA secondary structure involving the tryptophan codon region. Proc Natl Acad Sci USA. 1979;76:5524–5528. doi: 10.1073/pnas.76.11.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3322.Oxender D L, Anderson J J, Daniels C J, Landick R, Gunsalus R P, Zurawski G, Yanofsky C. Amino-terminal sequence and processing of the precursor of the leucine-specific binding protein, and evidence for conformational differences between the precursor and the mature form. Proc Natl Acad Sci USA. 1980;77:2005–2009. doi: 10.1073/pnas.77.4.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3323.Oxender D L, Anderson J J, Daniels C J, Landick R, Gunsalus R P, Zurawski G, Selker E, Yanofsky C. Structural and functional analysis of cloned DNA containing genes responsible for branched-chain amino acid transport in Escherichia coli. Proc Natl Acad Sci USA. 1980;77:1412–1416. doi: 10.1073/pnas.77.3.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3324.Ozeki H, Inokuchi H, Yamao F, Kodaira M, Sakano H, Ikemura T, Shimura Y. Genetics of nonsense suppressor tRNAs in Escherichia coli. In: Soll D, Abelson J, Schimmel P, editors. Transfer RNA: biological aspects. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. pp. 341–362. [Google Scholar]
- 3325.Ozenberger B A, Nahlik M S, McIntosh M A. Genetic organization of multiple fep genes encoding ferric enterobactin transport functions in Escherichia coli. J Bacteriol. 1987;169:3638–3646. doi: 10.1128/jb.169.8.3638-3646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3326.Ozenberger B A, Brickman T J, McIntosh M A. Nucleotide sequence of Escherichia coli isochorismate synthetase gene entC and evolutionary relationship of isochorismate synthetase and other chorismate-utilizing enzymes. J Bacteriol. 1989;171:775–783. doi: 10.1128/jb.171.2.775-783.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3327.Ozer Z, Reardon J T, Hsu D S, Malhotra K, Sancar A. The other function of DNA photolyase: stimulation of excision repair of chemical damage to DNA. Biochemistry. 1995;34:15886–15889. doi: 10.1021/bi00049a002. [DOI] [PubMed] [Google Scholar]
- 3328.Pace N R, Smith D. Ribonuclease P: function and variation. J Biol Chem. 1990;265:3587–3590. [PubMed] [Google Scholar]
- 3329.Padan E, Maisler N, Taglicht D, Karpel R, Schuldiner S. Deletion of ant in Escherichia coli reveals its function in adaptation to high salinity and an alternative Na+/H+ antiporter systems(s) J Biol Chem. 1989;264:20297–20302. [PubMed] [Google Scholar]
- 3330.Padan E, Schuldiner S. Na+/H+ antiporters, molecular devices that couple the Na+ and H+ circulation in cells. J Bioenerg Biomembr. 1993;25:647–669. doi: 10.1007/BF00770252. [DOI] [PubMed] [Google Scholar]
- 3331.Padmanabha K P, Deutscher M P. RNase T affects Escherichia coli growth and recovery from metabolic stress. J Bacteriol. 1991;173:1376–1381. doi: 10.1128/jb.173.4.1376-1381.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3332.Paek K H, Walker G C. Escherichia coli dnaK null mutants are inviable at high temperature. J Bacteriol. 1987;169:283–290. doi: 10.1128/jb.169.1.283-290.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3333.Paetz W, Nass G. Biochemical and immunological characterization of threonyl-tRNA synthetase of two borrelidin-resistant mutants of Escherichia coli K-12. Eur J Biochem. 1973;35:331–337. doi: 10.1111/j.1432-1033.1973.tb02843.x. [DOI] [PubMed] [Google Scholar]
- 3334.Page L, Griffiths L, Cole J A. Different physiological roles of two independent pathways for nitrite reduction to ammonia by enteric bacteria. Arch Microbiol. 1990;154:349–354. doi: 10.1007/BF00276530. [DOI] [PubMed] [Google Scholar]
- 3335.Pahel G, Tyler B. A new glnA-linked regulatory gene for glutamine synthetase in Escherichia coli. Proc Natl Acad Sci USA. 1979;76:4544–4548. doi: 10.1073/pnas.76.9.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3336.Pahel G, Rothstein D M, Magasanik B. Complex glnA-glnL-glG operon of Escherichia coli. J Bacteriol. 1982;150:202–213. doi: 10.1128/jb.150.1.202-213.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3337.Pai C H. Biochemical and genetic characterization of dehydrobiotin resistant mutants of Escherichia coli. Mol Gen Genet. 1974;134:345–357. doi: 10.1007/BF00337469. [DOI] [PubMed] [Google Scholar]
- 3338.Palaniappan C, Sharma V, Hudspeth M E S, Meganathan R. Menaquinone (vitamin K2) biosynthesis: evidence that the Escherichia coli menD gene encodes both 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylic acid synthase and α-ketoglutarate decarboxylase activities. J Bacteriol. 1992;174:8111–8118. doi: 10.1128/jb.174.24.8111-8118.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3339.Palchaudhuri S, Patel V, McFall E. DNA sequence of the d-serine deaminase activator gene. J Bacteriol. 1988;170:330–334. doi: 10.1128/jb.170.1.330-334.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3340.Palm D, Goerl R, Weidinger G, Zeier R, Fischer B, Schinzel R, Ratcliffe A. E. coli maltodextrin phosphorylase: primary structure and deletion mapping of the C-terminal site. Z Naturforsch Sect C. 1987;42:394–400. doi: 10.1515/znc-1987-0411. [DOI] [PubMed] [Google Scholar]
- 3341.Palmer M L, Raker M A, Kennedy P J, Young J W, Barnes W, Rodriguez R L, Noller H F. Isolation and restriction mapping of plasmids containing ribosomal DNA sequences from the rrnB cistron. Mol Gen Genet. 1979;172:171–178. doi: 10.1007/BF00268279. [DOI] [PubMed] [Google Scholar]
- 3342.Palmer T, Vasishta A, Whitty P W, Boxer D H. Isolation of protein FA, a product of the mob locus required for molybdenum cofactor biosynthesis in Escherichia coli. Eur J Biochem. 1994;222:687–692. doi: 10.1111/j.1432-1033.1994.tb18913.x. [DOI] [PubMed] [Google Scholar]
- 3343.Palmer T, Santini C L, Iobbi-Nivol C, Eaves D J, Boxer D H, Giordano G. Involvement of the narJ and mob gene products in distinct steps in the biosynthesis of the molybdoenzyme nitrate reductase in Escherichia coli. Mol Microbiol. 1996;20:875–884. doi: 10.1111/j.1365-2958.1996.tb02525.x. [DOI] [PubMed] [Google Scholar]
- 3344.Palva E T, Saris P, Silhavy T J. Gene fusions to the ptsM/pel locus of Escherichia coli. Mol Gen Genet. 1985;199:427–433. doi: 10.1007/BF00330754. [DOI] [PubMed] [Google Scholar]
- 3345.Pannekoek H, Noordermeer I, Van de Putte P. Expression of the cloned uvrB gene of Escherichia coli: mode of transcription and orientation. J Bacteriol. 1979;139:54–63. doi: 10.1128/jb.139.1.54-63.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3346.Pao C C, Gallant J. A gene involved in the metabolic control of ppGpp synthesis. Mol Gen Genet. 1978;158:271–277. doi: 10.1007/BF00267198. [DOI] [PubMed] [Google Scholar]
- 3347.Pardee A B, Benz E J, St. Peter D A, Krieger J N, Meuth M, Triesmann H W. Hyperproduction and purification of nicotinamide deamidase, a microconstitutive enzyme of Escherichia coli. J Biol Chem. 1971;246:6792–6796. [PubMed] [Google Scholar]
- 3348.Parekh B S, Hatfield G W. Growth rate-related regulation of the ilvGMEDA operon of Escherichia coli K-12 is a consequence of the polar frameshift mutation in the ilvG gene of this strain. J Bacteriol. 1997;179:2086–2088. doi: 10.1128/jb.179.6.2086-2088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3349.Park H, Inouye M. Mutational analysis of the linker region of EnvZ, an osmosensor in Escherichia coli. J Bacteriol. 1997;179:4382–4390. doi: 10.1128/jb.179.13.4382-4390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3350.Park I S, Lin C H, Walsh C T. Gain of d-alanyl-d-lactate or d-lactyl-d-alanine synthetase activities in three active-site mutants of the Escherichia colid-alanyl-d-alanine ligase B. Biochemistry. 1996;35:10464–10471. doi: 10.1021/bi9603128. [DOI] [PubMed] [Google Scholar]
- 3351.Park J T. Turnover and recycling of the murein sacculus in oligopeptide permease-negative strains of Escherichia coli: indirect evidence for an alternative permease system and for a monolayered sacculus. J Bacteriol. 1993;175:7–11. doi: 10.1128/jb.175.1.7-11.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3352.Park J T, Raychaudhuri D, Li H, Normark S, Mengin-Lecreulx D. MppA, a periplasmic binding protein essential for import of the bacterial cell wall peptide l-alanyl-γ-d-glutamyl-meso-diaminopimelate. J Bacteriol. 1998;180:1215–1223. doi: 10.1128/jb.180.5.1215-1223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3353.Park M H, Wong B B, Lusk J E. Mutants in three genes affecting transport of magnesium in Escherichia coli: genetics and physiology. J Bacteriol. 1976;126:1096–1103. doi: 10.1128/jb.126.3.1096-1103.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3354.Park S J, Tseng C P, Gunsalus R P. Regulation of succinate dehydrogenase (sdhCDAB) operon expression in Escherichia coli in response to carbon supply and anaerobiosis: role of ArcA and Fnr. Mol Microbiol. 1995;15:473–482. doi: 10.1111/j.1365-2958.1995.tb02261.x. [DOI] [PubMed] [Google Scholar]
- 3355.Park S-J, Chao G, Gunsalus R P. Aerobic regulation of the sucABCD genes of Escherichia coli, which encode α-ketoglutarate dehydrogenase and succinyl coenzyme A synthetase: roles of ArcA, Fnr, and the upstream sdhCDAB promoter. J Bacteriol. 1997;179:4138–4142. doi: 10.1128/jb.179.13.4138-4142.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3356.Park S J, McCabe J, Turna J, Gunsalus R P. Regulation of the citrate synthase (gltA) gene of Escherichia coli in response to anaerobiosis and carbon supply: role of the arcA gene product. J Bacteriol. 1994;176:5086–5092. doi: 10.1128/jb.176.16.5086-5092.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3357.Park S-J, Gunsalus R P. Oxygen, iron, carbon, and superoxide control of the fumarase fumA and fumC genes of Escherichia coli: role of the arcA, fnr, and soxR gene products. J Bacteriol. 1995;177:6255–6262. doi: 10.1128/jb.177.21.6255-6262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3358.Park S K, Kim D W, Choe J, Kim H. RNA helicase activity of Escherichia coli SecA protein. Biochem Biophys Res Commun. 1997;235:593–597. doi: 10.1006/bbrc.1997.6834. [DOI] [PubMed] [Google Scholar]
- 3359.Park S K, Kim K I, Woo K M, Seol J H, Tanaka K, Ichihara A, Ha D B, Chung C H. Site-directed mutagenesis of the dual translational initiation sites of the clpB gene of Escherichia coli and characterization of its gene products. J Biol Chem. 1993;268:20170–20174. [PubMed] [Google Scholar]
- 3360.Park W, Takase I, Tamaki S, Jin I-J, Ishino F, Matsuhashi M. Cloning of the gene of Escherichia coli mutant penicillin-binding protein 5 that has no penicillin-releasing activity. Agric Biol Chem. 1985;49:881–883. [Google Scholar]
- 3361.Parker C T, Kloser A W, Schnaitman C A, Stein M A, Gottesman S, Gibson B W. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J Bacteriol. 1992;174:2525–2538. doi: 10.1128/jb.174.8.2525-2538.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3362.Parker C T, Pradel E, Schnaitman C A. Identification and sequences of the lipopolysaccharide core biosynthetic genes rfaQ, rfaP, and rfaG of Escherichia coli K-12. J Bacteriol. 1992;174:930–934. doi: 10.1128/jb.174.3.930-934.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3363.Parker J. Specific mistranslation in hisT mutants of Escherichia coli. Mol Gen Genet. 1982;187:405–409. doi: 10.1007/BF00332619. [DOI] [PubMed] [Google Scholar]
- 3364.Parker J. Identification of the purC gene product of Escherichia coli. J Bacteriol. 1984;157:712–717. doi: 10.1128/jb.157.3.712-717.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3365.Parker L L, Hall B G. A fourth Escherichia coli gene system with the potential to evolve beta-glucoside utilization. Genetics. 1988;119:485–490. doi: 10.1093/genetics/119.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3366.Parker L L, Hall B G. Characterization and nucleotide sequence of the cryptic cel operon of E. coli K12. Genetics. 1990;124:455–471. doi: 10.1093/genetics/124.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3367.Parkinson J S. Novel mutations affecting a signaling component for chemotaxis of Escherichia coli. J Bacteriol. 1980;142:953–961. doi: 10.1128/jb.142.3.953-961.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3368.Parquet C, Flouret B, Mengin-Lecreulx D, van Heijenoort J. Nucleotide sequence of the murF gene encoding the UDP-MurNAc-pentapeptide synthetase of Escherichia coli. Nucleic Acids Res. 1989;17:5379. doi: 10.1093/nar/17.13.5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3369.Parra F, Jones-Mortimer M C, Kornberg H L. Phosphotransferase-mediated regulation of carbohydrate utilization in Escherichia coli K12: the nature of the iex (crr) and gsr (tgs) mutations. J Gen Microbiol. 1983;129:337–348. doi: 10.1099/00221287-129-2-337. [DOI] [PubMed] [Google Scholar]
- 3370.Parra F, Britton P, Castle C, Jones-Mortimer M C, Kornberg H L. Two separate genes involved in sulphate transport in Escherichia coli K12. J Gen Microbiol. 1983;129:357–358. doi: 10.1099/00221287-129-2-357. [DOI] [PubMed] [Google Scholar]
- 3371.Parra-Lopez C, Baer M T, Groisman E A. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J. 1993;12:4053–4062. doi: 10.1002/j.1460-2075.1993.tb06089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3372.Parra-Lopez C, Lin R, Aspedon A, Groisman E A. A Salmonella protein that is required for resistance to antimicrobial peptides and transport of potassium. EMBO J. 1994;13:3964–3972. doi: 10.1002/j.1460-2075.1994.tb06712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3373.Parsons C A, Stasiak A, West S C. The E. coli RuvAB proteins branch migrate Holliday junctions through heterologous DNA sequences in a reaction facilitated by SSB. EMBO J. 1995;14:5736–5744. doi: 10.1002/j.1460-2075.1995.tb00260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3374.Parsot C, Boyen A, Cohen G N, Glansdorff N. Nucleotide sequence of Escherichia coli argB and argC genes: comparison of N-acetylglutamate kinase and N-acetylglutamate-gamma-semialdehyde dehydrogenase with homologous and analogous enzymes. Gene. 1988;68:275–283. doi: 10.1016/0378-1119(88)90030-3. [DOI] [PubMed] [Google Scholar]
- 3375.Parsot C, Cossart P, Saint-Girons I, Cohen G N. Nucleotide sequence of thrC and of the termination region of the threonine operon in Escherichia coli K12. Nucleic Acids Res. 1983;11:7331–7345. doi: 10.1093/nar/11.21.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3376.Pascal M C, Casse F. Genetic analysis of mutants of Escherichia coli K12 and Salmonella typhimurium LT2 deficient in hydrogenase activity. Mol Gen Genet. 1975;141:173–179. doi: 10.1007/BF00267682. [DOI] [PubMed] [Google Scholar]
- 3377.Pascal M C, Burini J-F, Chippaux M. Regulation of the trimethylamine N-oxide (TMAO) reductase in Escherichia coli: analysis of Tor:Mud1 operon fusion. Mol Gen Genet. 1984;195:351–355. doi: 10.1007/BF00332770. [DOI] [PubMed] [Google Scholar]
- 3378.Pascal M C, Chippaux M. Involvement of a gene of the chlE locus in the regulation of the nitrate reductase operon. Mol Gen Genet. 1982;185:334–338. doi: 10.1007/BF00330808. [DOI] [PubMed] [Google Scholar]
- 3379.Pascal M C, Chippaux M, Abou-Jaoude A, Blaschkowski H P, Knappe J. Mutants of Escherichia coli K12 with defects in anaerobic pyruvate metabolism. J Gen Microbiol. 1981;124:35–42. doi: 10.1099/00221287-124-1-35. [DOI] [PubMed] [Google Scholar]
- 3380.Pati S, Disilvestre D, Brusilow W S. Regulation of the Escherichia coli uncH gene by mRNA secondary structure and translational coupling. Mol Microbiol. 1992;6:3559–3566. doi: 10.1111/j.1365-2958.1992.tb01791.x. [DOI] [PubMed] [Google Scholar]
- 3381.Patzer S, Hantke K. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol. 1998;28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 3382.Pauli G, Overath P. ato operon: a highly inducible system for acetoacetate and butyrate degradation in Escherichia coli. Eur J Biochem. 1972;29:553–562. doi: 10.1111/j.1432-1033.1972.tb02021.x. [DOI] [PubMed] [Google Scholar]
- 3383.Pauza C D, Karels M J, Navre M, Schachman H K. Genes encoding Escherichia coli aspartate transcarbomoylase: the pyrB-pyrI operon. Proc Natl Acad Sci USA. 1982;79:4020–4024. doi: 10.1073/pnas.79.13.4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3384.Payne J W, Morley S, Armitage P, Payne G M. Transport and hydrolysis of antibacterial peptide analogues in Escherichia coli: backbone-modified aminoxy peptides. J Gen Microbiol. 1984;130:2253–2265. doi: 10.1099/00221287-130-9-2253. [DOI] [PubMed] [Google Scholar]
- 3385.Paz-Elizur T, Takeshita M, Goodman M, O’Donnell M, Livneh Z. Mechanism of translesion DNA synthesis by DNA polymerase II. Comparison to DNA polymerases I and III core. J Biol Chem. 1996;271:24662–24669. doi: 10.1074/jbc.271.40.24662. [DOI] [PubMed] [Google Scholar]
- 3386.Peakman T C, Crouzet J, Mayaux J-F, Busby S J, Mohan S, Harborne N, Wootton J C, Nicolson R, Cole J A. Nucleotide sequence, organisation and structural analysis of the products of genes in the nirB-cysG region of the Escherichia coli K-12 chromosome. Eur J Biochem. 1990;191:315–323. doi: 10.1111/j.1432-1033.1990.tb19125.x. [DOI] [PubMed] [Google Scholar]
- 3387.Peakman T C, Busby S J, Cole J A. Transcriptional control of the cysG gene of Escherichia coli K-12 during aerobic and anaerobic growth. Eur J Biochem. 1990;191:325–331. doi: 10.1111/j.1432-1033.1990.tb19126.x. [DOI] [PubMed] [Google Scholar]
- 3388.Pecher A, Zinoni F, Bock A. The seleno-peptide of formic dehydrogenase (formate hydrogen-lyase linked) from Escherichia coli: genetic analysis. Arch Microbiol. 1985;141:359–363. doi: 10.1007/BF00428850. [DOI] [PubMed] [Google Scholar]
- 3389.Pecher A, Blaschkowski H P, Knappe K, Bock A. Expression of pyruvate-formate lyase of Escherichia coli from the cloned structural gene. Arch Microbiol. 1982;132:365–371. doi: 10.1007/BF00413390. [DOI] [PubMed] [Google Scholar]
- 3390.Pedersen S, Skouv J, Kajitani M, Ishihama A. Transcriptional organization of the rpsA operon of Escherichia coli. Mol Gen Genet. 1984;196:135–140. doi: 10.1007/BF00334105. [DOI] [PubMed] [Google Scholar]
- 3391.Peekhaus N, Tong S, Reizer J, Saier M H, Murray E, Conway T. Characterization of a novel transporter family that includes multiple Escherichia coli gluconate transporters and their homologues. FEMS Microbiol Lett. 1997;147:233–238. doi: 10.1111/j.1574-6968.1997.tb10247.x. [DOI] [PubMed] [Google Scholar]
- 3392.Pegues J C, Chen L S, Gordon A W, Ding L, Coleman W G. Cloning, expression, and characterization of the Escherichia coli K-12 rfaD gene. J Bacteriol. 1990;172:4652–4660. doi: 10.1128/jb.172.8.4652-4660.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3393.Peist R, Koch A, Bolek P, Sewitz S, Kolbus T, Boos W. Characterization of the aes gene of Escherichia coli encoding an enzyme with esterase activity. J Bacteriol. 1997;179:7679–7686. doi: 10.1128/jb.179.24.7679-7686.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3394.Peist R, Schneider-Fresenius C, Boos W. The MalT-dependent and malZ-encoded maltodextrin glucosidase of Escherichia coli can be converted into a dextrinyltransferase by a single mutation. J Biol Chem. 1996;271:10681–10689. doi: 10.1074/jbc.271.18.10681. [DOI] [PubMed] [Google Scholar]
- 3395.Pelletier A J, Hill T M, Kuempel P. Location of sites that inhibit progression of replication forks in the terminus region in Escherichia coli. J Bacteriol. 1988;170:4293–4298. doi: 10.1128/jb.170.9.4293-4298.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3396.Pellicer M T, Badia J, Aguilar J, Baldoma L. glc locus of Escherichia coli: characterization of genes encoding the subunits of glycolate oxidase and the glc regulator protein. J Bacteriol. 1996;178:2051–2059. doi: 10.1128/jb.178.7.2051-2059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3397.Peng H, Marians K J. Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J Biol Chem. 1993;268:24481–24490. [PubMed] [Google Scholar]
- 3398.Peng H, Marians K J. The interaction of Escherichia coli topoisomerase IV with DNA. J Biol Chem. 1995;270:25286–25290. doi: 10.1074/jbc.270.42.25286. [DOI] [PubMed] [Google Scholar]
- 3399.Penninckx M, Gigot D. Synthesis of a peptide form of N-δ-(phosphonoacetyl)-l-ornithine. Its antibacterial effect through the specific inhibition of Escherichia colil-ornithine carbamoyltransferase. J Biol Chem. 1979;254:6392–6395. [PubMed] [Google Scholar]
- 3400.Perez-Roger I, Macian F, Armengod M-E. Transcription termination in the Escherichia coli dnaA gene is not mediated by the internal DnaA box. J Bacteriol. 1995;177:1896–1899. doi: 10.1128/jb.177.7.1896-1899.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3401.Peri K G, Waygood B. Sequence of cloned enzyme IIN-acetylglucosamine of the phosphoenolpyruvate:N-acetylglucosamine phosphotransferase system. Biochemistry. 1988;27:6054–6061. doi: 10.1021/bi00416a034. [DOI] [PubMed] [Google Scholar]
- 3402.Peri K G, Goldie H, Waygood B. Cloning and characterization of the N-acetylglucosamine operon of Escherichia coli. Biochem Cell Biol. 1990;68:123–137. doi: 10.1139/o90-017. [DOI] [PubMed] [Google Scholar]
- 3403.Perkins-Balding D, Prabha Dias D, Glasgow A C. Location, degree, and direction of DNA bending associated with the Hin recombinational enhancer sequence and Fis-enhancer complex. J Bacteriol. 1997;179:4747–4753. doi: 10.1128/jb.179.15.4747-4753.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3404.Perry K L, Elledge S J, Mitchell B B, Marsh L, Walker G C. umuDC and mucAB operons whose products are required for UV light- and chemical-induced mutagenesis: UmuD, MucA, and LexA proteins share homology. Proc Natl Acad Sci USA. 1985;82:4331–4335. doi: 10.1073/pnas.82.13.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3405.Persson B C, Gustafsson C, Berg D E, Bjork G R. The gene for a tRNA modifying enzyme, m5U54-methyltransferase, is essential for viability in Escherichia coli. Proc Natl Acad Sci USA. 1992;89:3995–3998. doi: 10.1073/pnas.89.9.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3406.Persson B C, Jager G, Gustafsson C. The spoU gene of Escherichia coli, the fourth gene of the spoT operon, is essential for tRNA (Gm18) 2′-O-methyltransferase activity. Nucleic Acids Res. 1997;25:4093–4097. doi: 10.1093/nar/25.20.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3407.Persson B C, Bylund G O, Berg D E, Wikstrom P M. Functional analysis of the ffh-trmD region of the Escherichia coli chromosome by using reverse genetics. J Bacteriol. 1995;177:5554–5560. doi: 10.1128/jb.177.19.5554-5560.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3408.Peruski L F., Jr Expression of heat shock protein D 48.5 of Escherichia coli is subject to modulation by catabolite repression. Microbiol Res. 1996;151:273–280. doi: 10.1016/s0944-5013(96)80024-3. [DOI] [PubMed] [Google Scholar]
- 3409.Petersen L, Downs D M. Mutations in apbC (mrp) prevent function of the alternative pyrimidine biosynthetic pathway in Salmonella typhimurium. J Bacteriol. 1996;178:5676–5682. doi: 10.1128/jb.178.19.5676-5682.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3410.Peterson J W, Chopra A K, Prasad R. Fine mapping of the rrnE, purHD, and hydGH operons on the Escherichia coli chromosome. J Bacteriol. 1991;173:3274–3275. doi: 10.1128/jb.173.11.3274-3275.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3411.Peterson K R, Mount D W. Analysis of the genetic requirements for viability of Escherichia coli K-12 DNA adenine methylase (dam) mutants. J Bacteriol. 1993;175:7505–7508. doi: 10.1128/jb.175.22.7505-7508.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3412.Petit C, Cayrol C, Lesca C, Kaiser P, Thompson C, Defais M. Characterization of dinY, a new Escherichia coli DNA repair gene whose products are damage inducible even in a lexA (Def) background. J Bacteriol. 1993;175:642–646. doi: 10.1128/jb.175.3.642-646.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3413.Pettigrew D W, Ma D-P, Conrad C A, Johnson J R. Escherichia coli glycerol kinase. Cloning and sequencing of the glpK gene and the primary structure of the enzyme. J Biol Chem. 1988;263:135–139. [PubMed] [Google Scholar]
- 3414.Pettigrew D W, Liu W Z, Holmes C, Meadow N D, Roseman S. A single amino acid change in Escherichia coli glycerol kinase abolishes glucose control of glycerol utilization in vivo. J Bacteriol. 1996;178:2846–2852. doi: 10.1128/jb.178.10.2846-2852.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3415.Pettis G S, McIntosh M A. Molecular characterization of the Escherichia coli enterobactin cistron entF and coupled expression of entF and the fes gene. J Bacteriol. 1987;169:4154–4162. doi: 10.1128/jb.169.9.4154-4162.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3416.Pettis G S, Brickman R J, McIntosh M A. Transcriptional mapping and nucleotide sequence of the Escherichia coli fepA-fes enterobactin region. Identification of a unique iron-regulated bidirectional promoter. J Biol Chem. 1988;263:18857–18863. [PubMed] [Google Scholar]
- 3417.Phillips G J, Prasher D, Kushner S R. Physical and biochemical characterization of cloned sbcB and xonA mutations from Escherichia coli K-12. J Bacteriol. 1988;170:2089–2094. doi: 10.1128/jb.170.5.2089-2094.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3418.Phillips G J, Kushner S R. Determination of the nucleotide sequence for the exonuclease I structural gene (sbcB) of Escherichia coli K-12. J Biol Chem. 1987;262:455–459. [PubMed] [Google Scholar]
- 3419.Phillips G J, Silhavy T J. The E. coli ffh gene is necessary for viability and efficient protein export. Nature. 1992;359:744–746. doi: 10.1038/359744a0. [DOI] [PubMed] [Google Scholar]
- 3420.Phillips T A, Vaughn V, Bloch P L, Neidhardt F C. Gene-protein index of Escherichia coli K-12, edition 2. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 919–966. [Google Scholar]
- 3421.Phoenix D A, Peters S E, Ramzan M A, Pratt J M. Analysis of the membrane-anchoring properties of the putative amphiphilic alpha-helical anchor at the C-terminus of Escherichia coli PBP 6. Microbiology. 1994;140:73–77. doi: 10.1099/13500872-140-1-73. [DOI] [PubMed] [Google Scholar]
- 3422.Pi J, Wookey P J, Pittard A J. Site-directed mutagenesis reveals the importance of conserved charged residues for the transport activity of the PheP permease of Escherichia coli. J Bacteriol. 1993;175:7500–7504. doi: 10.1128/jb.175.22.7500-7504.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3423.Pichersky E, Gottlieb D, Hess J F. Nucleotide sequence of the triose phosphate isomerase gene of Escherichia coli. Mol Gen Genet. 1984;195:314–320. doi: 10.1007/BF00332765. [DOI] [PubMed] [Google Scholar]
- 3423a.Pichoff S, Bouché J. Identification of a new Escherichia coli cell cycle gene. 1995. GenBank submission Z50870. [Google Scholar]
- 3424.Pickett C L, Hayes L, Earhart C F. Molecular cloning of the Escherichia coli K-12 entACGBE genes. FEMS Microbiol Lett. 1984;24:77–80. [Google Scholar]
- 3425.Picksley S M, Morton S J, Lloyd R G. The recN locus of Escherichia coli K12: molecular analysis and identification of the gene product. Mol Gen Genet. 1985;201:301–307. doi: 10.1007/BF00425675. [DOI] [PubMed] [Google Scholar]
- 3426.Pierard A, Glansdorff N, Gigot D, Crabeel M, Halleux P, Thiry L. Repression of Escherichia coli carbamoylphosphate synthase: relationships with enzyme synthesis in the arginine and pyrimidine pathways. J Bacteriol. 1976;127:291–301. doi: 10.1128/jb.127.1.291-301.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3427.Pierard A, Glansdorff N, Yashphe J. Mutations affecting uridine monophosphate pyrophosphorylase or the argR gene in Escherichia coli. Mol Gen Genet. 1972;118:235–245. doi: 10.1007/BF00333460. [DOI] [PubMed] [Google Scholar]
- 3428.Pierce J R, Earhart C F. Escherichia coli K-12 envelope proteins specifically required for ferrienterobactin uptake. J Bacteriol. 1986;166:930–936. doi: 10.1128/jb.166.3.930-936.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3429.Pierce J R, Pickett C L, Earhart C F. Two fep genes are required for ferrienterochelin uptake in Escherichia coli K-12. J Bacteriol. 1983;155:330–336. doi: 10.1128/jb.155.1.330-336.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3430.Pierson D E, Campbell A. Cloning and nucleotide sequence of bisC, the structural gene for biotin sulfoxide reductase in Escherichia coli. J Bacteriol. 1990;172:2194–2198. doi: 10.1128/jb.172.4.2194-2198.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3431.Pierson L S, III, Kahn M L. Integration of satellite bacteriophage P4 in Escherichia coli. DNA sequences of the phage and host regions involved in site-specific recombination. J Mol Biol. 1987;196:487–496. doi: 10.1016/0022-2836(87)90026-x. [DOI] [PubMed] [Google Scholar]
- 3432.Piette J, Nyunoya H, Lusty C J, Cunin R, Weyens G, Crabeel M, Charlier D, Glansdorff N, Pierard A. DNA sequence of the carA gene and the control region of carAB: tandem promoters, respectively controlled by arginine and the pyrimidines, regulate the synthesis of carbamoylphosphate. Proc Natl Acad Sci USA. 1984;81:4134–4138. doi: 10.1073/pnas.81.13.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3433.Piette J, Cunin R, Boyen A, Charlier D, Crabeel M, Van Vliet F, Glansdorff N, Squires C, Squires C L. The regulatory region of the divergent argECBH operon in Escherichia coli K-12. Nucleic Acids Res. 1982;10:8031–8048. doi: 10.1093/nar/10.24.8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3434.Piette J, Cunin R, Van Vliet F, Charlier D, Crabeel M, Ota Y, Glansdorff N. Homologous control sites and DNA transcription starts in the related argF and argI genes of Escherichia coli K12. EMBO J. 1982;1:853–857. doi: 10.1002/j.1460-2075.1982.tb01259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3435.Pilipcinec E, Huisman T T, Willemsen P T, Appelmelk B J, Graaf F K. Identification by Tn10 transposon mutagenesis of host factors involved in the biosynthesis of K99 fimbriae of Escherichia coli: effect of LPS core mutations. FEMS Microbiol Lett. 1994;123:201–206. doi: 10.1016/0378-1097(94)90295-x. [DOI] [PubMed] [Google Scholar]
- 3436.Pinkham J L, Platt T. The nucleotide sequence of the rho gene of E. coli K-12. Nucleic Acids Res. 1983;11:3531–3545. doi: 10.1093/nar/11.11.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3437.Pinn P A, Towner K J, O’Grady F W. Genetic analysis of chromosomal resistance to trimethoprim derived from clinical isolates of Escherichia coli. J Gen Microbiol. 1983;128:85–92. doi: 10.1099/00221287-129-1-85. [DOI] [PubMed] [Google Scholar]
- 3438.Pinner E, Padan E, Schuldiner S. Cloning, sequencing, and expression of the nhaB gene, encoding a Na+/H+ antiporter in Escherichia coli. J Biol Chem. 1992;267:11064–11068. [PubMed] [Google Scholar]
- 3439.Pinner E, Kotler Y, Padan E, Schuldiner S. Physiological role of nhaB, a specific Na+/H+ antiporter in Escherichia coli. J Biol Chem. 1993;268:1729–1734. [PubMed] [Google Scholar]
- 3440.Pistocchi R, Kashiwagi K, Miyamoto S, Nukui E, Kobayashi H, Igarashi K. Characteristics of the operon for a putrescine transport system that maps at 19 minutes on the Escherichia coli chromosome. J Biol Chem. 1993;268:146–152. [PubMed] [Google Scholar]
- 3441.Pittard A J, Davidson B E. TyrR protein of Escherichia coli and its role as repressor and activator. Mol Microbiol. 1991;5:1585–1592. doi: 10.1111/j.1365-2958.1991.tb01904.x. [DOI] [PubMed] [Google Scholar]
- 3442.Pittard J, Praszkier J, Certoma A, Eggertsson G, Gowrishankar J, Narasaiah G, Whipp M J. Evidence that there are only two tRNAPhe genes in Escherichia coli. J Bacteriol. 1990;172:6077–6083. doi: 10.1128/jb.172.10.6077-6083.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3443.Pla J, Sanchez M, Palacios P, Vicente M, Aldea M. Preferential cytoplasmic location of FtsZ, a protein essential for Escherichia coli septation. Mol Microbiol. 1991;5:1681–1686. doi: 10.1111/j.1365-2958.1991.tb01915.x. [DOI] [PubMed] [Google Scholar]
- 3444.Plamann M D, Stauffer G V. Characterization of the Escherichia coli gene for serine hydroxymethyltransferase. Gene. 1983;22:9–18. doi: 10.1016/0378-1119(83)90059-8. [DOI] [PubMed] [Google Scholar]
- 3445.Plamann M D, Stauffer L T, Urbanowski M L, Stauffer G V. Complete nucleotide sequence of the E. coli glyA gene. Nucleic Acids Res. 1983;11:2065–2075. doi: 10.1093/nar/11.7.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3446.Plamann M D, Rapp W D, Stauffer G V. Escherichia coli K12 mutants defective in the glycine cleavage enzyme system. Mol Gen Genet. 1983;192:15–20. doi: 10.1007/BF00327641. [DOI] [PubMed] [Google Scholar]
- 3447.Plasterk R H, Brinkman A A, Van de Putte P. DNA inversions in the chromosome of Escherichia coli and in bacteriophage Mu: relationship to other site-specific recombination systems. Proc Natl Acad Sci USA. 1983;80:5355–5358. doi: 10.1073/pnas.80.17.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3448.Plasterk R H, Van de Putte P. The invertible P-DNA segment in the chromosome of Escherichia coli. EMBO J. 1985;4:237–242. doi: 10.1002/j.1460-2075.1985.tb02341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3449.Plate C A S, Seely A, Laffler T G. Evidence for a protonmotive force related regulatory system in Escherichia coli and its effects on lactose transport. Biochemistry. 1986;25:6127–6132. doi: 10.1021/bi00368a044. [DOI] [PubMed] [Google Scholar]
- 3450.Platko J V, Willins D A, Calvo J M. The ilvIH operon of Escherichia coli is positively regulated. J Bacteriol. 1990;172:4563–4570. doi: 10.1128/jb.172.8.4563-4570.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3451.Platz A, Sjoberg B-M. Construction and characterization of hybrid plasmids containing the Escherichia coli region. J Bacteriol. 1980;143:561–568. doi: 10.1128/jb.143.2.561-568.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3452.Pledger W J, Umbarger H E. Isoleucine and valine metabolism in Escherichia coli. XXI. Mutations affecting derepression and valine resistance. J Bacteriol. 1973;113:183–194. doi: 10.1128/jb.114.1.183-194.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3453.Plumbridge J. Control of the expression of the manXYZ operon in Escherichia coli: Mlc is a negative regulator of the mannose PTS. Mol Microbiol. 1998;27:369–380. doi: 10.1046/j.1365-2958.1998.00685.x. [DOI] [PubMed] [Google Scholar]
- 3454.Plumbridge J A. Organization of the Escherichia coli chromosome between genes glnS and glnU, V. Mol Gen Genet. 1987;209:618–620. doi: 10.1007/BF00331173. [DOI] [PubMed] [Google Scholar]
- 3455.Plumbridge J A. Sequence of the nagBACD operon in Escherichia coli K12 and pattern of transcription within the nag regulon. Mol Microbiol. 1989;3:505–515. doi: 10.1111/j.1365-2958.1989.tb00197.x. [DOI] [PubMed] [Google Scholar]
- 3456.Plumbridge J A. Induction of the nag regulon of Escherichia coli by N-acetylglucosamine and glucosamine: role of the cyclic AMP-catabolite activator protein complex in expression of the regulon. J Bacteriol. 1990;172:2728–2735. doi: 10.1128/jb.172.5.2728-2735.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3457.Plumbridge J A. Repression and induction of the nag regulon of Escherichia coli K-12: the roles of nagC and nagA in the maintenance of the uninduced state. Mol Microbiol. 1991;5:2053–2062. doi: 10.1111/j.1365-2958.1991.tb00828.x. [DOI] [PubMed] [Google Scholar]
- 3458.Plumbridge J A. A dominant mutation in the gene for the Nag repressor of Escherichia coli that renders the nag regulon uninducible. J Gen Microbiol. 1992;138:1011–1017. doi: 10.1099/00221287-138-5-1011. [DOI] [PubMed] [Google Scholar]
- 3459.Plumbridge J A, Howe J G, Springer M, Touati-Schwartz D, Hershey J W B, Grunberg-Manago M. Cloning and mapping of a gene for translation initiation factor 1F2 in Escherichia coli. Proc Natl Acad Sci USA. 1982;79:5033–5037. doi: 10.1073/pnas.79.16.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3460.Plumbridge J A, Springer M. Genes for the two subunits of phenylalanyl-tRNA synthetase of Escherichia coli are transcribed from the same promoter. J Mol Biol. 1980;144:595–600. doi: 10.1016/0022-2836(80)90341-1. [DOI] [PubMed] [Google Scholar]
- 3461.Plumbridge J A, Springer M. Escherichia coli phenylalanyl-tRNA synthetase operon: transcription studies of wild-type mutated operons on multicopy plasmids. J Bacteriol. 1982;152:661–668. doi: 10.1128/jb.152.2.661-668.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3462.Plumbridge J A, Springer M. Organization of the Escherichia coli chromosome around the genes for translation initiation factor 1F2 (infB) and a transcription termination factor (nusA) J Mol Biol. 1983;167:227–243. doi: 10.1016/s0022-2836(83)80333-7. [DOI] [PubMed] [Google Scholar]
- 3463.Plumbridge J A, Springer M, Graffe M, Goursot R, Grunberg-Manago M. Physical localisation and cloning of the structural gene for E. coli initiation factor IF3 from a group of genes concerned with translation. Gene. 1980;11:33–42. doi: 10.1016/0378-1119(80)90084-0. [DOI] [PubMed] [Google Scholar]
- 3464.Plumbridge J A, Cochet O, Souza J M, Altamirano M M, Calcagno M L. Coordinated regulation of amino sugar-synthesizing and -degrading enzymes in Escherichia coli K-12. J Bacteriol. 1993;175:4951–4956. doi: 10.1128/jb.175.16.4951-4956.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3465.Plunkett G, Burland V D, Daniels D L, Blattner F R. Analysis of the Escherichia coli genome. III. DNA sequence of the region from 87.2 to 89.2 minutes. Nucleic Acids Res. 1993;21:3391–3398. doi: 10.1093/nar/21.15.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3466.Podkovyrov S M, Larson T J. Identification of promoter and stringent regulation of transcription of the fabH, fabD and fabG genes encoding fatty acid biosynthetic enzymes of Escherichia coli. Nucleic Acids Res. 1996;24:1747–1752. doi: 10.1093/nar/24.9.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3467.Pogliano J A, Beckwith J R. SecD and SecF facilitate protein export in Escherichia coli. EMBO J. 1994;13:554–561. doi: 10.1002/j.1460-2075.1994.tb06293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3468.Pogliano K J, Beckwith J R. The Cs sec mutants of Escherichia coli reflect the cold sensitivity of protein export itself. Genetics. 1993;133:763–773. doi: 10.1093/genetics/133.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3469.Pogliano K J, Beckwith J R. Genetic and molecular characterization of the Escherichia coli secD operon and its products. J Bacteriol. 1994;176:804–814. doi: 10.1128/jb.176.3.804-814.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3470.Pohlschroder M, Murphy C, Beckwith J R. In vivo analyses of interactions between SecE and SecY, core components of the Escherichia coli protein translocation machinery. J Biol Chem. 1996;271:19908–19914. doi: 10.1074/jbc.271.33.19908. [DOI] [PubMed] [Google Scholar]
- 3471.Polacco M, Cronan J E., Jr A mutant of Escherichia coli conditionally defective in the synthesis of holo-[acyl carrier protein] J Biol Chem. 1981;256:5750–5754. [PubMed] [Google Scholar]
- 3472.Polarek J W, Williams G, Epstein W. The products of the kdpDE operon are required for expression of the Kdp ATPase of Escherichia coli. J Bacteriol. 1992;174:2145–2151. doi: 10.1128/jb.174.7.2145-2151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3473.Polayes D A, Rice P W, Dahlberg A. DNA polymerase I activity in Escherichia coli is influenced by spot 42 RNA. J Bacteriol. 1988;170:2083–2088. doi: 10.1128/jb.170.5.2083-2088.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3474.Polissi A, Georgopoulos C. Mutational analysis and properties of the msbA gene of Escherichia coli, coding for an essential ABC family transporter. Mol Microbiol. 1996;20:1221–1233. doi: 10.1111/j.1365-2958.1996.tb02642.x. [DOI] [PubMed] [Google Scholar]
- 3475.Pon C L, Wittmann-Liebold B, Gualerzi C. Structure-function relationships in Escherichia coli initiation factors. II. Elucidation of the primary structure of initiation factor IF-1. FEBS Lett. 1979;101:157–160. doi: 10.1016/0014-5793(79)81316-2. [DOI] [PubMed] [Google Scholar]
- 3476.Pon C L, Calogero R A, Gualerzi C O. Identification, cloning, nucleotide sequence and chromosomal map location of hns, the structural gene of Escherichia coli DNA-binding protein H-NS. Mol Gen Genet. 1988;212:199–202. doi: 10.1007/BF00334684. [DOI] [PubMed] [Google Scholar]
- 3477.Ponce E, Flores N, Martinez A, Valle F, Bolivar F. Cloning of the two pyruvate kinase isoenzyme structural genes from Escherichia coli: the relative roles of these enzymes in pyruvate biosynthesis. J Bacteriol. 1995;177:5719–5722. doi: 10.1128/jb.177.19.5719-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3478.Pontis E, Sun X Y, Jornvall H, Krook M, Reichard P. ClpB proteins copurify with the anaerobic Escherichia coli reductase. Biochem Biophys Res Commun. 1991;180:1222–1226. doi: 10.1016/s0006-291x(05)81326-9. [DOI] [PubMed] [Google Scholar]
- 3479.Poole R K, Gibson F, Wu G. The cydD gene product, component of a heterodimeric ABC transporter, is required for assembly of periplasmic cytochrome c and of cytochrome bd in Escherichia coli. FEMS Microbiol Lett. 1994;117:217–223. doi: 10.1111/j.1574-6968.1994.tb06768.x. [DOI] [PubMed] [Google Scholar]
- 3480.Poole R K, Williams H D, Downie J A, Gibson F. Mutations affecting the cytochrome d-containing oxidase complex of Escherichia coli K12: identification and mapping of a fourth locus, cydD. J Gen Microbiol. 1989;135:1865–1874. doi: 10.1099/00221287-135-7-1865. [DOI] [PubMed] [Google Scholar]
- 3481.Poole R K, Hatch L P, Cleeter M W, Gibson F, Cox G B, Wu G. Cytochrome bd biosynthesis in Escherichia coli: the sequences of the cydC and cydD genes suggest that they encode the components of an ABC membrane transporter. Mol Microbiol. 1993;10:421–430. [PubMed] [Google Scholar]
- 3482.Poole R K, Anjum M F, Membrillo-Hernandez J, Kim S O, Hughes M N, Stewart V. Nitric oxide, nitrite, and Fnr regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12. J Bacteriol. 1996;178:5487–5492. doi: 10.1128/jb.178.18.5487-5492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3483.Poole R K, Rogers N J, D’Mello R A, Hughes M N, Orii Y. Escherichia coli flavohaemoglobin (Hmp) reduces cytochrome c and Fe(III)-hydroxamate K by electron transfer from NADH via FAD: sensitivity of oxidoreductase activity to haem-bound dioxygen. Microbiology. 1997;143:1557–1565. doi: 10.1099/00221287-143-5-1557. [DOI] [PubMed] [Google Scholar]
- 3484.Poon A P, Dhillon T S. Temperate coliphage HK253: attachment site and restricted transduction of proAB mutants of Escherichia coli K-12. J Virol. 1986;60:317–319. doi: 10.1128/jvi.60.1.317-319.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3485.Popkin P S, Maas W K. Escherichia coli regulatory mutation affecting lysine transport and lysine decarboxylase. J Bacteriol. 1980;141:485–492. doi: 10.1128/jb.141.2.485-492.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3486.Popp J L. Sequence and overexpression of the menD gene from Escherichia coli J. Bacteriol. 1989;171:4349–4354. doi: 10.1128/jb.171.8.4349-4354.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3487.Porco A, Isturiz T. Selection of lacZ operon fusions in genes of gluconate metabolism in E. coli. Characterization of a gntT::lacZ fusion. Acta Cient Venez. 1991;42:270–275. [PubMed] [Google Scholar]
- 3488.Poritz M A, Bernstein H D, Strub K, Zopf D, Wilhelm H, Walter P. An E. coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science. 1990;250:1111–1117. doi: 10.1126/science.1701272. [DOI] [PubMed] [Google Scholar]
- 3489.Portalier R, Robert-Baudouy J M, Stoeber F. Regulation of Escherichia coli K-12 hexuronate system genes: exu regulon. J Bacteriol. 1980;143:1095–1107. doi: 10.1128/jb.143.3.1095-1107.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3490.Porter A C, Kumamoto C, Aldape K, Simoni R D. Role of the b subunit of the Escherichia coli proton-translocating ATPase. A mutagenic analysis. J Biol Chem. 1985;260:8182–8187. [PubMed] [Google Scholar]
- 3491.Porter A C G, Brusilow W S, Simoni R D. Promoter for the unc operon of Escherichia coli. J Bacteriol. 1983;155:1271–1278. doi: 10.1128/jb.155.3.1271-1278.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3492.Portier C. Physical localization and direction of transcription of the structural gene for Escherichia coli ribosomal protein S15. Gene. 1982;18:261–266. doi: 10.1016/0378-1119(82)90164-0. [DOI] [PubMed] [Google Scholar]
- 3493.Portier C, Migot C, Grunberg-Manago M. Cloning of E. coli pnp gene from an episome. Mol Gen Genet. 1981;183:298–305. doi: 10.1007/BF00270632. [DOI] [PubMed] [Google Scholar]
- 3494.Portier C, Regnier P. Expression of the rspO and pnp genes: structural analysis of a DNA fragment carrying their control regions. Nucleic Acids Res. 1984;12:6091–6102. doi: 10.1093/nar/12.15.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3495.Post D A, Hove-Jensen B, Switzer R L. Characterization of the hemA-prs region of the Escherichia coli and Salmonella typhimurium chromosomes: identification of two open reading frames and implications for prs expression. J Gen Microbiol. 1993;139:259–266. doi: 10.1099/00221287-139-2-259. [DOI] [PubMed] [Google Scholar]
- 3496.Post L, Arfsten A E, Davis G R, Nomura M. DNA sequence of the promoter region for the a ribosomal protein operon in Escherichia coli. J Biol Chem. 1980;255:4653–4659. [PubMed] [Google Scholar]
- 3497.Post L, Nomura M. Nucleotide sequence of the intercistronic region preceding the gene for RNA polymerase subunit a in Escherichia coli. J Biol Chem. 1979;254:10604–10606. [PubMed] [Google Scholar]
- 3498.Post L, Nomura M. DNA sequences from the str operon of Escherichia coli. J Biol Chem. 1980;255:4660–4666. [PubMed] [Google Scholar]
- 3499.Post L E, Strycharz G D, Nomura M, Lewis H, Dennis P P. Nucleotide sequence of the ribosomal protein gene cluster adjacent to the gene for RNA polymerase subunit beta in Escherichia coli. Proc Natl Acad Sci USA. 1979;76:1697–1701. doi: 10.1073/pnas.76.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3500.Postle K, Good R F. DNA sequence of the Escherichia coli tonB gene. Proc Natl Acad Sci USA. 1983;80:5235–5239. doi: 10.1073/pnas.80.17.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3501.Postle K, Good R F. A bidirectional rho-independent transcription terminator between the E. coli tonB gene and an opposing gene. Cell. 1985;41:577–585. doi: 10.1016/s0092-8674(85)80030-1. [DOI] [PubMed] [Google Scholar]
- 3502.Postle K, Reznikoff W S. Identification of the Escherichia coli tonB gene product in minicells containing tonB hybrid plasmids. J Mol Biol. 1979;131:619–636. doi: 10.1016/0022-2836(79)90011-1. [DOI] [PubMed] [Google Scholar]
- 3503.Potter P M, Kleibl K, Cawkwell L, Margison G P. Expression of the ogt gene in wild-type and ada mutants of E. coli. Nucleic Acids Res. 1989;17:8047–8060. doi: 10.1093/nar/17.20.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3504.Potter P M, Harris L C, Margison G P. Mapping of OGT in the E. coli chromosome. Nucleic Acids Res. 1989;17:10505. doi: 10.1093/nar/17.24.10505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3505.Potter P M, Wilkinson M C, Fitton J, Carr F J, Brennand J, Cooper D P, Margison G P. Characterisation and nucleotide sequence of ogt, the O6-alkylguanine-DNA-alkyltransferase gene of E. coli. Nucleic Acids Res. 1987;15:9177–9193. doi: 10.1093/nar/15.22.9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3506.Potter S, Yang X, Boulanger M J, Ishiguro E E. Occurrence of homologs of the Escherichia coli lytB gene in gram-negative bacterial species. J Bacteriol. 1998;180:1959–1961. doi: 10.1128/jb.180.7.1959-1961.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3507.Poulsen L K, Refn A, Molin S, Andersson P. The gef gene from Escherichia coli is regulated at the level of translation. Mol Microbiol. 1991;5:1639–1648. doi: 10.1111/j.1365-2958.1991.tb01911.x. [DOI] [PubMed] [Google Scholar]
- 3508.Poulsen L K, Refn A, Molin S, Andersson P. Topographic analysis of the toxic Gef protein from Escherichia coli. Mol Microbiol. 1991;5:1627–1637. doi: 10.1111/j.1365-2958.1991.tb01910.x. [DOI] [PubMed] [Google Scholar]
- 3509.Poulsen L K, Larsen N W, Molin S, Andersson P. A family of genes encoding a cell-killing function may be conserved in all gram-negative bacteria. Mol Microbiol. 1989;3:1463–1472. doi: 10.1111/j.1365-2958.1989.tb00131.x. [DOI] [PubMed] [Google Scholar]
- 3510.Poulsen L K, Larsen N W, Molin S, Andersson P. Analysis of an Escherichia coli mutant strain resistant to the cell-killing function encoded by the gef gene family. Mol Microbiol. 1992;6:895–905. doi: 10.1111/j.1365-2958.1992.tb01540.x. [DOI] [PubMed] [Google Scholar]
- 3511.Poulsen P, Bonekamp F, Jensen K F. Structure of the Escherichia coli pyrE operon and control of pyrE expression by a UTP modulated intercistronic attenuation. EMBO J. 1984;3:1783–1790. doi: 10.1002/j.1460-2075.1984.tb02046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3512.Poulsen P, Andersen J T, Jensen K F. Molecular and mutational analysis of three genes preceding pyrE on the Escherichia coli chromosome. Mol Microbiol. 1989;3:393–404. doi: 10.1111/j.1365-2958.1989.tb00184.x. [DOI] [PubMed] [Google Scholar]
- 3513.Poulsen P, Jensen K F. Three genes preceding pyrE on the Escherichia coli chromosome are essential for survival and normal cell morphology in stationary culture and at high temperature. Res Microbiol. 1991;142:283–288. doi: 10.1016/0923-2508(91)90042-9. [DOI] [PubMed] [Google Scholar]
- 3514.Poulsen P, Jensen K F, Valentin-Hansen P, Carlsson P, Lundberg L G. Nucleotide sequence of the Escherichia coli pyrE gene and of the DNA in front of the protein-coding region. Eur J Biochem. 1983;135:223–229. doi: 10.1111/j.1432-1033.1983.tb07641.x. [DOI] [PubMed] [Google Scholar]
- 3515.Pouyssegur J, Stoeber F. Rameau degradatif commun des hexyronates chez Escherichia coli K12. Mecanisme d’induction des enzymes assurant le metabolisme du 2-ceto-3-desoxy-gluconate. Eur J Biochem. 1972;30:479–494. doi: 10.1111/j.1432-1033.1972.tb02120.x. [DOI] [PubMed] [Google Scholar]
- 3516.Powell B S, Court D L. Control of ftsZ expression, cell division, and glutamine metabolism in Luria-Bertani medium by the alarmone ppGpp in Escherichia coli. J Bacteriol. 1998;180:1053–1062. doi: 10.1128/jb.180.5.1053-1062.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3517.Powell B S, Court D L, Inada T, Nakamura Y, Michotey V, Cui X, Reizer A, Saier M H, Reizer J. Novel proteins of the phosphotransferase system encoded within the rpoN operon of Escherichia coli. Enzyme IIAntr affects growth on organic nitrogen and the conditional lethality of an era(ts) mutant. J Biol Chem. 1995;270:4822–4839. doi: 10.1074/jbc.270.9.4822. [DOI] [PubMed] [Google Scholar]
- 3518.Powell K A, Cox R, McConville M L, Charles H P. Mutations affecting porphyrin biosynthesis in Escherichia coli. Enzyme. 1973;16:65–73. doi: 10.1159/000459363. [DOI] [PubMed] [Google Scholar]
- 3519.Powers E L, Randall L L. Export of periplasmic galactose-binding protein in Escherichia coli depends on the chaperone SecB. J Bacteriol. 1995;177:1906–1907. doi: 10.1128/jb.177.7.1906-1907.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3520.Powers T, Walter P. Reciprocal stimulation of GTP hydrolysis by two directly interacting GTPases. Science. 1995;269:1422–1424. doi: 10.1126/science.7660124. [DOI] [PubMed] [Google Scholar]
- 3521.Pradel E, Marck C, Boquet P L. Nucleotide sequence and transcriptional analysis of the Escherichia coli agp gene encoding periplasmic acid glucose-1-phosphatase. J Bacteriol. 1990;172:802–807. doi: 10.1128/jb.172.2.802-807.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3522.Pradel E, Schnaitman C A. Effect of rfaH (sfrB) and temperature on expression of rfa genes of Escherichia coli K-12. J Bacteriol. 1991;173:6428–6431. doi: 10.1128/jb.173.20.6428-6431.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3523.Pradel E, Parker C T, Schnaitman C A. Structures of the rfaB, rfaI, rfaJ, and rfaS genes of Escherichia coli K-12 and their roles in assembly of the lipopolysaccharide core. J Bacteriol. 1992;174:4736–4745. doi: 10.1128/jb.174.14.4736-4745.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3524.Pradel E, Boquet P L. Acid phosphatases of Escherichia coli: molecular cloning and analysis of agp, the structural gene for a periplasmic acid glucose phosphatase. J Bacteriol. 1988;170:4916–4923. doi: 10.1128/jb.170.10.4916-4923.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3525.Pradel E, Boquet P L. Mapping of the Escherichia coli acid glucose-1-phosphatase gene agp and analysis of its expression in vivo by use of an agp-phoA protein fusion. J Bacteriol. 1989;171:3511–3517. doi: 10.1128/jb.171.6.3511-3517.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3526.Pradel E, Boquet P L. Utilization of exogenous glucose-1-phosphate as a source of carbon or phosphate by Escherichia coli K12: respective roles of acid glucose-1-phosphatase, hexose-phosphate permease, phosphoglucomutase and alkaline phosphatase. Res Microbiol. 1991;142:37–45. doi: 10.1016/0923-2508(91)90095-r. [DOI] [PubMed] [Google Scholar]
- 3527.Pramanik A, Wertheimer S J, Schwartz J J, Schwartz I. Expression of Escherichia coli infC: identification of a promoter in an upstream thrS coding sequence. J Bacteriol. 1986;168:746–751. doi: 10.1128/jb.168.2.746-751.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3528.Prasad I, Young B, Schaefler S. Genetic determination of the constitutive biosynthesis of phospho-β-glucosidase A in Escherichia coli K-12. J Bacteriol. 1973;114:909–915. doi: 10.1128/jb.114.3.909-915.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3529.Prasher D, Kasunic D A, Kushner S R. Physical and genetic characterization of the cloned sbcB (exonuclease I) region of the Escherichia coli genome. J Bacteriol. 1983;153:903–908. doi: 10.1128/jb.153.2.903-908.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3530.Prather N E, Mims B H, Murgola E. supG and supL in Escherichia coli code for mutant lysine tRNAs. Nucleic Acids Res. 1983;11:8283–8286. doi: 10.1093/nar/11.23.8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3531.Pratt D, Subramani S. Nucleotide sequence of the Escherichia coli xanthine-guanine phosphoribosyl transferase gene. Nucleic Acids Res. 1983;11:8817–8823. doi: 10.1093/nar/11.24.8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3532.Pratt L A, Silhavy T J. The response regulator SprE controls the stability of RpoS. Proc Natl Acad Sci USA. 1996;93:2488–2492. doi: 10.1073/pnas.93.6.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3533.Pratviel-Sosa F, Mengin-Lecreulx D, van Heijenoort J. Overproduction, purification and properties of the uridine diphsophate N-acetylmuramoyl-l-alanine:d-glutamate ligase from Escherichia coli. Eur J Biochem. 1991;202:1169–1176. doi: 10.1111/j.1432-1033.1991.tb16486.x. [DOI] [PubMed] [Google Scholar]
- 3534.Preiss J, Kleinhofs A, Klingmuller W. Cloning of seven differently complementing DNA fragments with chl functions from Escherichia coli K12. Mol Gen Genet. 1987;206:352–355. doi: 10.1007/BF00333594. [DOI] [PubMed] [Google Scholar]
- 3535.Prescott C D, Kornau H C. Mutations in E. coli 16s rRNA that enhance and decrease the activity of a suppressor tRNA. Nucleic Acids Res. 1992;20:1567–1571. doi: 10.1093/nar/20.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3536.Pressler U, Staudenmaier H, Zimmermann L, Braun V. Genetics of the iron dicitrate transport system of Escherichia coli. J Bacteriol. 1988;170:2716–2724. doi: 10.1128/jb.170.6.2716-2724.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3537.Prince W S, Villarejo M R. Osmotic control of proU transcription is mediated through direct action of potassium glutamate on the transcription complex. J Biol Chem. 1990;265:17673–17679. [PubMed] [Google Scholar]
- 3538.Prior T I, Kornberg H L. Nucleotide sequence of fruA, the gene specifying enzyme IIfru of the phosphoenolpyruvate-dependent sugar phosphotransferase system in Escherichia coli K12. J Gen Microbiol. 1988;134:2757–2768. doi: 10.1099/00221287-134-10-2757. [DOI] [PubMed] [Google Scholar]
- 3539.Pritchard R H, Ahmad S I. Fluorouracil and the isolation of mutants lacking uridine phosphorylase in E. coli: location of the gene. Mol Gen Genet. 1971;111:84–88. doi: 10.1007/BF00286557. [DOI] [PubMed] [Google Scholar]
- 3540.Privalle C T, Fridovich I. Induction of superoxide dismutase in Escherichia coli by heat shock. Proc Natl Acad Sci USA. 1987;84:2723–2726. doi: 10.1073/pnas.84.9.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3541.Privalle C T, Fridovich I. Transcriptional and maturational effects of manganese and iron on the biosynthesis of manganese-superoxide dismutase in Escherichia coli. J Biol Chem. 1992;267:9140–9145. [PubMed] [Google Scholar]
- 3542.Prodromou C, Haynes M J, Guest J R. The aconitase of Escherichia coli: purification of the enzyme and molecular cloning and map location of the gene (acn) J Gen Microbiol. 1991;137:2505–2515. doi: 10.1099/00221287-137-11-2505. [DOI] [PubMed] [Google Scholar]
- 3543.Prodromou C, Artymiuk P J, Guest J R. The aconitase of Escherichia coli: nucleotide sequence of the aconitase gene and amino acid sequence similarity with mitochondrial aconitases, the iron-responsive-element-binding protein and isopropylmalate isomerase. Eur J Biochem. 1992;204:599–609. doi: 10.1111/j.1432-1033.1992.tb16673.x. [DOI] [PubMed] [Google Scholar]
- 3544.Prody C A, Neilands J B. Genetic and biochemical characterization of the Escherichia coli K-12 fhuB mutation. J Bacteriol. 1984;157:874–880. doi: 10.1128/jb.157.3.874-880.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3545.Provence D L, Curtiss R. Role of crl in avian pathogenic Escherichia coli: a knockout mutation of crl does not affect hemagglutination activity, fibronectin binding, or curli production. Infect Immun. 1992;60:4460–4467. doi: 10.1128/iai.60.11.4460-4467.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3546.Pruss B M, Nelms J M, Park C, Wolfe A J. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J Bacteriol. 1994;176:2143–2150. doi: 10.1128/jb.176.8.2143-2150.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3547.Pruss B M, Matsumura P. A regulator of the flagellar regulon of Escherichia coli, flhD, also affects cell division. J Bacteriol. 1996;178:668–674. doi: 10.1128/jb.178.3.668-674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3548.Pucci M J, Discotto L F, Dougherty T J. Cloning and identification of the Escherichia coli murB DNA sequence, which encodes UDP-N-acetylenolpyruvoylglucosamine reductase. J Bacteriol. 1992;174:1690–1693. doi: 10.1128/jb.174.5.1690-1693.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3549.Pugsley A P. Translocation of a folded protein across the outer membrane in Escherichia coli. Proc Natl Acad Sci USA. 1992;89:12058–12062. doi: 10.1073/pnas.89.24.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3550.Pugsley A P. A mutation in the dsbA gene coding for periplasmic disulfide oxidoreductase reduces transcription of the Escherichia coli ompF gene. Mol Gen Genet. 1993;237:407–411. doi: 10.1007/BF00279445. [DOI] [PubMed] [Google Scholar]
- 3551.Pugsley A P, Dubreuil C. Molecular characterization of malQ, the structural gene for the Escherichia coli enzyme amylomaltase. Mol Microbiol. 1988;2:473–479. doi: 10.1111/j.1365-2958.1988.tb00053.x. [DOI] [PubMed] [Google Scholar]
- 3552.Pugsley A P, Moreno F, deLorenzo V. Microcin-E492-insensitive mutants of Escherichia coli K12. J Gen Microbiol. 1986;132:3253–3259. doi: 10.1099/00221287-132-12-3253. [DOI] [PubMed] [Google Scholar]
- 3553.Pugsley A P, Kornacker M G, Poquet I. The general protein-export pathway is directly required for extracellular pullulanase secretion in Escherichia coli K12. Mol Microbiol. 1991;5:343–352. doi: 10.1111/j.1365-2958.1991.tb02115.x. [DOI] [PubMed] [Google Scholar]
- 3554.Pugsley A P, Reeves P R. Iron uptake in colicin B-resistant mutants of Escherichia coli K-12. J Bacteriol. 1976;126:1052–1062. doi: 10.1128/jb.126.3.1052-1062.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3555.Puppe W, Zimmann P, Jung K, Lucassen M, Altendorf K. Characterization of truncated forms of the KdpD protein, the sensor kinase of the K+-translocating Kdp system of Escherichia coli. J Biol Chem. 1996;271:25027–25034. doi: 10.1074/jbc.271.40.25027. [DOI] [PubMed] [Google Scholar]
- 3556.Putney S D, Melendez D L, Schimmel P. Cloning, partial sequencing, and in vitro transcription of the gene for alanine tRNA synthetase. J Biol Chem. 1981;256:205–211. [PubMed] [Google Scholar]
- 3557.Putney S D, Royal N J, Neuman de Vegvar H, Herlihy W C, Biemann K, Schimmel P. Primary structure of a large aminoacyl-tRNA synthetase. Science. 1981;213:1497–1500. doi: 10.1126/science.7025207. [DOI] [PubMed] [Google Scholar]
- 3558.Putney S D, Schimmel P. An aminoacyl tRNA synthetase binds to a specific DNA sequence and regulates its gene transcription. Nature. 1981;291:632–635. doi: 10.1038/291632a0. [DOI] [PubMed] [Google Scholar]
- 3559.Puyo M F, Calsou P, Salles B. UV resistance of E. coli K-12 deficient in cAMP/CRP regulation. Mutat Res. 1992;282:247–252. doi: 10.1016/0165-7992(92)90130-a. [DOI] [PubMed] [Google Scholar]
- 3560.Qamar S, Marsh K, Berry A. Identification of arginine 331 as an important active site residue in the class II fructose-1,6-bisphosphate aldolase of Escherichia coli. Protein Sci. 1996;5:154–161. doi: 10.1002/pro.5560050119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3561.Qi F, Turnbough C L. Regulation of codBA operon expression in Escherichia coli by UTP-dependent reiterative transcription and UTP-sensitive transcriptional start site switching. J Mol Biol. 1995;254:552–565. doi: 10.1006/jmbi.1995.0638. [DOI] [PubMed] [Google Scholar]
- 3562.Qi H-Y, Sankaran K, Gan K, Wu H C. Structure-function relationship of bacterial prolipoprotein diacylglyceryl transferase: functionally significant conserved regions. J Bacteriol. 1995;177:6820–6824. doi: 10.1128/jb.177.23.6820-6824.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3563.Qi S-Y, Sukopolvi S, O’Connor C D. Outer membrane permeability of Escherichia coli K12: isolation, cloning and mapping of suppressors of a defined antibiotic hypersensitive mutant. Mol Gen Genet. 1991;229:421–427. doi: 10.1007/BF00267465. [DOI] [PubMed] [Google Scholar]
- 3564.Qian Q, Curran J F, Björk G R. The methyl group of the N6-methyl-N6-threonylcarbamoyladenosine in tRNA of Escherichia coli modestly improves the efficiency of the tRNA. J Bacteriol. 1998;180:1808–1813. doi: 10.1128/jb.180.7.1808-1813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3565.Qu J N, Makino S I, Adachi H, Koyama Y, Akiyama Y, Ito K, Tomoyasu T, Ogura T, Matsuzawa H. The tolZ gene of Escherichia coli is identified as the ftsH gene. J Bacteriol. 1996;178:3457–3461. doi: 10.1128/jb.178.12.3457-3461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3566.Quail M A, Dempsey C E, Guest J R. Identification of a fatty acyl responsive regulator (FarR) in Escherichia coli. FEBS Lett. 1994;356:183–187. doi: 10.1016/0014-5793(94)01264-4. [DOI] [PubMed] [Google Scholar]
- 3567.Quail M A, Haydon D J, Guest J R. The pdhR-aceEF-lpd operon of Escherichia coli expresses the pyruvate dehydrogenase complex. Mol Microbiol. 1994;12:95–104. doi: 10.1111/j.1365-2958.1994.tb00998.x. [DOI] [PubMed] [Google Scholar]
- 3568.Quinones A, Neumann S. The ssb-113 allele suppresses the dnaQ49 mutator and alters DNA supercoiling in Escherichia coli. Mol Microbiol. 1997;25:237–246. doi: 10.1046/j.1365-2958.1997.4531718.x. [DOI] [PubMed] [Google Scholar]
- 3569.Quinones A, Messer W. Discoordinate gene expression in the dnaA-dnaN operon of Escherichia coli. Mol Gen Genet. 1988;213:118–124. doi: 10.1007/BF00333407. [DOI] [PubMed] [Google Scholar]
- 3570.Quinones A, Juterbock W R, Messer W. Expression of the dnaA gene of Escherichia coli is inducible by DNA damage. Mol Gen Genet. 1991;227:9–16. doi: 10.1007/BF00260699. [DOI] [PubMed] [Google Scholar]
- 3571.Quintilla F X, Baldoma L, Badia J, Aguilar J. Aldehyde dehydrogenase induction by glutamate in Escherichia coli. Role of 2-oxoglutarate. Eur J Biochem. 1991;202:1321–1325. doi: 10.1111/j.1432-1033.1991.tb16506.x. [DOI] [PubMed] [Google Scholar]
- 3572.Quirk S, Seto D, Bhatnagar S K, Gauss P, Gold L, Bessman M J. Location and molecular cloning of the structural gene for the deoxyguanosine triphosphate triphosphohydrolase of Escherichia coli. Mol Microbiol. 1989;3:1391–1395. doi: 10.1111/j.1365-2958.1989.tb00121.x. [DOI] [PubMed] [Google Scholar]
- 3573.Quirk S, Bhatnagar S K, Bessman M J. Primary structure of the deoxyguanosine triphosphate triphosphohydrolase-encoding gene (dgt) of Escherichia coli. Gene. 1990;89:13–18. doi: 10.1016/0378-1119(90)90200-b. [DOI] [PubMed] [Google Scholar]
- 3574.Rabin R S, Stewart V J. Either of two functionally redundant sensor proteins, NarX and NarQ, is sufficient for nitrate regulation in Escherichia coli K-12. Proc Natl Acad Sci USA. 1992;89:8419–8423. doi: 10.1073/pnas.89.18.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3575.Rabin R S, Stewart V J. Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J Bacteriol. 1993;175:3259–3268. doi: 10.1128/jb.175.11.3259-3268.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3576.Radicella J P, Clark E A, Chen S, Fox M S. Patch length of localized repair events: role of DNA polymerase I in mutY-dependent mismatch repair. J Bacteriol. 1993;175:7732–7736. doi: 10.1128/jb.175.23.7732-7736.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3577.Radke K L, Siegel E C. Mutation preventing capsular polysaccharide synthesis in Escherichia coli K-12 and its effect on bacteriophage resistance. J Bacteriol. 1971;106:432–437. doi: 10.1128/jb.106.2.432-437.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3578.Raetz C R, Kantor G D, Nishijima M, Jones M L. Isolation of Escherichia coli mutants with elevated levels of membrane enzymes. J Biol Chem. 1981;256:2109–2112. [PubMed] [Google Scholar]
- 3579.Raftery L A, Yarus M. Site-specific mutagenesis of Escherichia coli gltT yields a weak, glutamic acid-inserting ochre suppressor. J Mol Biol. 1985;184:343–345. doi: 10.1016/0022-2836(85)90385-7. [DOI] [PubMed] [Google Scholar]
- 3580.Raha M, Sockett H, Macnab R M. Characterization of the fliL gene in the flagellar regulon of Escherichia coli and Salmonella typhimurium. J Bacteriol. 1994;176:2308–2311. doi: 10.1128/jb.176.8.2308-2311.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3581.Raha M, Kawagishi I, Muller V, Kihara M, Macnab R M. Escherichia coli produces a cytoplasmic alpha-amylase, AmyA. J Bacteriol. 1992;174:6644–6652. doi: 10.1128/jb.174.20.6644-6652.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3582.Raha M, Kihara M, Kawagishi I, Macnab R M. Organization of the Escherichia coli and Salmonella typhimurium chromosomes between flagellar regions IIIa and IIIb, including a large non-coding region. J Gen Microbiol. 1993;139:1401–1407. doi: 10.1099/00221287-139-7-1401. [DOI] [PubMed] [Google Scholar]
- 3583.Rahav-Manor O, Carmel O, Karpel R, Taglicht D, Glaser G, Schuldiner S, Padan E. NhaR, a protein homologous to a family of bacterial regulatory proteins (LysR) regulates nhaA, the sodium proton antiporter gene in Escherichia coli. J Biol Chem. 1992;267:10433–10438. [PubMed] [Google Scholar]
- 3584.Rahfeld J U, Rucknagel K P, Stoller G, Horne S M, Schierhorn A, Young K D, Fischer G. Isolation and amino acid sequence of a new 22-kDa FKBP-like peptidyl-prolyl cis/trans-isomerase of Escherichia coli. Similarity to Mip-like proteins of pathogenic bacteria. J Biol Chem. 1996;271:22130–22138. doi: 10.1074/jbc.271.36.22130. [DOI] [PubMed] [Google Scholar]
- 3585.Rahman M A, Nelson H, Weissbach H, Brot N. Cloning, sequencing, and expression of the Escherichia coli peptide methionine sulfoxide reductase gene. J Biol Chem. 1992;267:15549–15551. [PubMed] [Google Scholar]
- 3586.Rahman M A, Moskovitz J, Strassman J, Weissbach H, Brot N. Physical map location of the peptide methionine sulfoxide reductase gene on the Escherichia coli chromosome. J Bacteriol. 1994;176:1548–1549. doi: 10.1128/jb.176.5.1548-1549.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3587.Raibaud O, Vidal-Ingigliardi D, Kolb A. Genetic studies on the promoter of malT, the gene that encodes the activator of the Escherichia coli maltose regulon. Res Microbiol. 1991;142:937–942. doi: 10.1016/0923-2508(91)90003-s. [DOI] [PubMed] [Google Scholar]
- 3588.Raibaud O, Schwartz M. Restriction map of the Escherichia coli malA region and identification of the malT product. J Bacteriol. 1980;143:761–771. doi: 10.1128/jb.143.2.761-771.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3589.Raina S, Georgopoulos C. A new Escherichia coli heat shock gene, htrC, whose product is essential for viability only at high temperatures. J Bacteriol. 1990;172:3417–3426. doi: 10.1128/jb.172.6.3417-3426.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3590.Raina S, Georgopoulos C. The htrM gene, whose product is essential for Escherichia coli viability only at elevated temperatures, is identical to the rfaD gene. Nucleic Acids Res. 1991;19:3811–3819. doi: 10.1093/nar/19.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3591.Raina S, Missiakas D, Georgopoulos C. The Escherichia coli htrL gene, cloning, sequencing, and transcriptional regulation: evidence for its positive regulation by the rfaH gene product. 1992. GenBank submission M94888. [Google Scholar]
- 3592.Raina S, Missiakas D, Georgopoulos C. The rpoE gene encoding the sigma E (sigma 24) heat shock sigma factor of Escherichia coli. EMBO J. 1995;14:1043–1055. doi: 10.1002/j.1460-2075.1995.tb07085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3593.Raina S, Missiakas D, Baird L, Kumar S, Georgopoulos C. Identification and transcriptional analysis of the Escherichia coli htrE operon which is homologous to pap and related pilin operons. J Bacteriol. 1993;175:5009–5021. doi: 10.1128/jb.175.16.5009-5021.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3594.Raina S, Mabey L, Georgopoulos C. The Escherichia coli htrP gene product is essential for bacterial growth at high temperatures: mapping, cloning, sequencing, and transcriptional regulation of htrP. J Bacteriol. 1991;173:5999–6008. doi: 10.1128/jb.173.19.5999-6008.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3595.Rainwater S, Silverman P M. The Cpx proteins of Escherichia coli K-12: evidence that cpxA, ecfB, ssd, and eup mutations all identify the same gene. J Bacteriol. 1990;172:2456–2461. doi: 10.1128/jb.172.5.2456-2461.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3596.Raivio T L, Silhavy T J. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J Bacteriol. 1997;179:7724–7733. doi: 10.1128/jb.179.24.7724-7733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3597.Rajagopalan P T, Datta A, Pei D. Purification, characterization, and inhibition of peptide deformylase from Escherichia coli. Biochemistry. 1997;36:13910–13918. doi: 10.1021/bi971155v. [DOI] [PubMed] [Google Scholar]
- 3598.Rajapandi T, Oliver D. ssaD1, a suppressor of secA51(Ts) that renders growth of Escherichia coli cold sensitive, is an early amber mutation in the transcription factor gene nusB. J Bacteriol. 1994;176:4444–4447. doi: 10.1128/jb.176.14.4444-4447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3599.Rajkumari K, Kusano S, Ishihama A, Mizuno T, Gowrishankar J. Effects of H-NS and potassium glutamate on ςS- and ς70-directed transcription in vitro from osmotically regulated P1 and P2 promoters of proU in Escherichia coli. J Bacteriol. 1996;178:4176–4181. doi: 10.1128/jb.178.14.4176-4181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3600.Rakonjac J, Milic M, Ajdic-Predic D, Santos D, Ivanisevic R, Savic D J. nov: a new genetic locus that affects the response of Escherichia coli K-12 to novobiocin. Mol Microbiol. 1992;6:1547–1553. doi: 10.1111/j.1365-2958.1992.tb00876.x. [DOI] [PubMed] [Google Scholar]
- 3601.Raleigh E A. Organization and function of the mcrBC genes of Escherichia coli. Mol Microbiol. 1992;6:1079–1086. doi: 10.1111/j.1365-2958.1992.tb01546.x. [DOI] [PubMed] [Google Scholar]
- 3602.Raleigh E A, Wilson G. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc Natl Acad Sci USA. 1986;83:9070–9074. doi: 10.1073/pnas.83.23.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3603.Raleigh E A, Benner J, Bloom P, Braymer H D, DeCruz E, Dharmalingam K, Heitman J, Noyer-Weidner M, Piekarowicz A, Kretz P L, Short J M, Woodcock D. Nomenclature relating to restriction of modified DNA in Escherichia coli. J Bacteriol. 1991;173:2707–2709. doi: 10.1128/jb.173.8.2707-2709.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3604.Raleigh E A, Trimarchi R, Revel H. Genetic and physical mapping of the mcrA (rflA) and mcrB (rglB) loci of Escherichia coli K-12. Genetics. 1989;122:279–296. doi: 10.1093/genetics/122.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3605.Ralling G, Linn T. Relative activities of the transcriptional regulatory sites in the rplKAJLrpoBC gene cluster of Escherichia coli. J Bacteriol. 1984;158:279–285. doi: 10.1128/jb.158.1.279-285.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3606.Ramabhadram T V. Method for the isolation of Escherichia coli relaxed mutants, utilizing near-ultraviolet irradiation. J Bacteriol. 1976;127:1587–1589. doi: 10.1128/jb.127.3.1587-1589.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3607.Ramakrishnan T, Adelberg E A. Regulatory mechanisms in the biosynthesis of isoleucine and valine. II. Identification of two operator genes. J Bacteriol. 1965;89:654–660. doi: 10.1128/jb.89.3.654-660.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3608.Ramani N, Hedeshian M, Freundlich M. micF antisense RNA has a major role in osmoregulation of OmpF in Escherichia coli. J Bacteriol. 1994;176:5005–5010. doi: 10.1128/jb.176.16.5005-5010.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3609.Ramesh V, Gite S, Li Y, RajBhandary U L. Suppressor mutations in Escherichia coli methionyl-tRNA formyltransferase: role of a 16-amino acid insertion module in initiator tRNA recognition. Proc Natl Acad Sci USA. 1997;94:13524–13529. doi: 10.1073/pnas.94.25.13524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3610.Ramseier T M, Saier M H., Jr cAMP-cAMP receptor protein complex: five binding sites in the control region of the Escherichia coli mannitol operon. Microbiology. 1995;141:1901–1907. doi: 10.1099/13500872-141-8-1901. [DOI] [PubMed] [Google Scholar]
- 3611.Ramseier T M, Chien S Y, Saier M H., Jr Cooperative interaction between Cra and Fnr in the regulation of the cydAB operon of Escherichia coli. Curr Microbiol. 1996;33:270–274. doi: 10.1007/s002849900112. [DOI] [PubMed] [Google Scholar]
- 3612.Rancourt D E, Stephenson J T, Vickell G A, Wood J M. Proline excretion by Escherichia coli K12. Biotechnol Bioeng. 1984;26:74–80. doi: 10.1002/bit.260260114. [DOI] [PubMed] [Google Scholar]
- 3613.Randall K, Lever M, Peddie B A, Chambers S T. Competitive accumulation of betaines by Escherichia coli K-12 and derivative strains lacking betaine porters. Biochim Biophys Acta. 1995;1245:116–120. doi: 10.1016/0304-4165(95)00071-i. [DOI] [PubMed] [Google Scholar]
- 3614.Raskin D M, de Boer P A. The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell. 1997;91:685–694. doi: 10.1016/s0092-8674(00)80455-9. [DOI] [PubMed] [Google Scholar]
- 3615.Rasmussen L J, Samson L. The Escherichia coli MutS DNA mismatch binding protein specifically binds O(6)-methylguanine DNA lesions. Carcinogenesis. 1996;17:2085–2088. doi: 10.1093/carcin/17.9.2085. [DOI] [PubMed] [Google Scholar]
- 3616.Rasmussen L J, Marinus M G, Lobner-Olesen A. Novel growth rate control of dam gene expression in Escherichia coli. Mol Microbiol. 1994;12:631–638. doi: 10.1111/j.1365-2958.1994.tb01050.x. [DOI] [PubMed] [Google Scholar]
- 3617.Rasmussen P B, Holst B, Valentin-Hansen P. Dual-function regulators: the cAMP receptor protein and the CytR regulator can act either to repress or to activate transcription depending on the context. Proc Natl Acad Sci USA. 1996;93:10151–10155. doi: 10.1073/pnas.93.19.10151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3618.Ravi R S, Sozhamannan S, Dharmalingam K. Transposon mutagenesis and genetic mapping of the rglA and rglB loci of Escherichia coli. Mol Gen Genet. 1985;198:390–392. doi: 10.1007/BF00332928. [DOI] [PubMed] [Google Scholar]
- 3619.Ravnikar P D, Somerville R L. Localization of the structural gene for threonine dehydrogenase in Escherichia coli. J Bacteriol. 1986;168:434–436. doi: 10.1128/jb.168.1.434-436.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3620.Ravnikar P D, Somerville R L. Structural and functional analysis of a cloned segment of Escherichia coli DNA that specifies proteins of a C4 pathway of serine biosynthesis. J Bacteriol. 1987;169:4716–4721. doi: 10.1128/jb.169.10.4716-4721.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3621.Rawlings M, Cronan J E., Jr The gene encoding Escherichia coli acyl carrier protein lies within a cluster of fatty acid biosynthetic genes. J Biol Chem. 1992;267:5751–5754. [PubMed] [Google Scholar]
- 3622.Ray A, Apirion D. Characterization of DNA from the rne gene of Escherichia coli: uniqueness of the rne DNA. Biochem Biophys Res Commun. 1982;107:1361–1367. doi: 10.1016/s0006-291x(82)80148-4. [DOI] [PubMed] [Google Scholar]
- 3623.RayChaudhuri D, Park J T. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature. 1992;359:251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- 3624.Rayl E A, Green J M, Nichols B P. Escherichia coli aminodeoxychorismate synthase: analysis of pabB mutations affecting catalysis and subunit association. Biochim Biophys Acta. 1996;1295:81–88. doi: 10.1016/0167-4838(96)00029-5. [DOI] [PubMed] [Google Scholar]
- 3625.Raynal L C, Krisch H M, Carpousis A J. Bacterial poly(A) polymerase: an enzyme that modulates RNA stability. Biochimie. 1996;78:390–398. doi: 10.1016/0300-9084(96)84745-6. [DOI] [PubMed] [Google Scholar]
- 3626.Ream L W, Clark A J. Cloning and deletion mapping of the recF dnaN region of the Escherichia coli chromosome. Plasmid. 1983;10:101–110. doi: 10.1016/0147-619x(83)90062-8. [DOI] [PubMed] [Google Scholar]
- 3627.Ream L W, Margossian L J, Clark A J, Hansen F G, von Meyenburg K. Genetic and physical mapping of recF in Escherichia coli K-12. Mol Gen Genet. 1980;180:115–121. doi: 10.1007/BF00267359. [DOI] [PubMed] [Google Scholar]
- 3628.Reaney S K, Begg C, Bungard S J, Guest J R. Identification of the l-tartrate dehydratase genes (ttdA and ttdB) of Escherichia coli and evolutionary relationship with the class I fumarase genes. J Gen Microbiol. 1993;139:1523–1530. doi: 10.1099/00221287-139-7-1523. [DOI] [PubMed] [Google Scholar]
- 3629.Rebeck G W, Samson L D. Increased spontaneous mutation and alkylation sensitivity of Escherichia coli strains lacking the ogt O6-methylguanine DNA repair methyltransferase. J Bacteriol. 1991;173:2068–2076. doi: 10.1128/jb.173.6.2068-2076.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3630.Rech S, Deppenmeier U, Gunsalus R P. Regulation of the molybdate transport operon, modABCD, of Escherichia coli in response to molybdate availability. J Bacteriol. 1995;177:1023–1029. doi: 10.1128/jb.177.4.1023-1029.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3631.Reddy M, Gowrishankar J. Identification and characterization of ssb and uup mutants with increased frequency of precise excision of transposon Tn10 derivatives: nucleotide sequence of uup in Escherichia coli. J Bacteriol. 1997;179:2892–2899. doi: 10.1128/jb.179.9.2892-2899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3632.Reed K E, Cronan J E., Jr Lipoic acid metabolism in Escherichia coli: sequencing and functional characterization of the lipA and lipB genes. J Bacteriol. 1993;175:1325–1336. doi: 10.1128/jb.175.5.1325-1336.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3633.Reed K E, Morris T W, Cronan J E., Jr Mutants of Escherichia coli K-12 that are resistant to a selenium analog of lipoic acid identify unknown genes in lipoate metabolism. Proc Natl Acad Sci USA. 1994;91:3720–3724. doi: 10.1073/pnas.91.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3634.Reed R E, Baer M F, Guerrier-Takada C, Donis-Keller H, Altman S. Nucleotide sequence of the gene encoding the RNA subunit (M1 RNA) of ribonuclease P from Escherichia coli. Cell. 1982;30:627–636. doi: 10.1016/0092-8674(82)90259-8. [DOI] [PubMed] [Google Scholar]
- 3635.Reed R E, Altman S. Repeated sequences and open reading frames in the 3′ flanking region of the gene for the RNA subunit of Escherichia coli ribonuclease. Proc Natl Acad Sci USA. 1983;80:5359–5363. doi: 10.1073/pnas.80.17.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3636.Reeder T, Schleif R. Mapping, sequence, and apparent lack of function of araJ, a gene of the Escherichia coli arabinose regulon. J Bacteriol. 1991;173:7765–7771. doi: 10.1128/jb.173.24.7765-7771.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3637.Reeve E C R. Genetic analysis of some mutations causing resistance to tetracycline in Escherichia coli K12. Genet Res. 1968;11:303–309. doi: 10.1017/s0016672300011484. [DOI] [PubMed] [Google Scholar]
- 3638.Regnier P, Portier C. Initiation, attenuation and RNaseIII processing of transcripts from the Escherichia coli operon ribosomal protein S15 and polynucleotide phosphorylase. J Mol Biol. 1986;187:23–32. doi: 10.1016/0022-2836(86)90403-1. [DOI] [PubMed] [Google Scholar]
- 3639.Regnier P, Grunberg-Manago M, Portier C. Nucleotide sequence of the pnp gene of Escherichia coli encoding polynucleotide phosphorylase: homology of the primary structure of the protein with the RNA-binding domain of ribosomal protein S1. J Biol Chem. 1987;262:63–68. [PubMed] [Google Scholar]
- 3640.Regue M, Remenick J, Tokunaga M, Mackie G A, Wu H C. Mapping of the lipoprotein signal peptidase gene (lsp) J Bacteriol. 1984;158:632–635. doi: 10.1128/jb.158.2.632-635.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3641.Rehemtulla A, Kadam S K, Sanderson K E. Cloning and analysis of the sfrB (sex factor repression) gene of Escherichia coli K-12. J Bacteriol. 1986;166:651–657. doi: 10.1128/jb.166.2.651-657.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3642.Reidl J, Romisch K, Ehrmann M, Boos W. MalI, a novel protein involved in regulation of the maltose system of Escherichia coli, is highly homologous to the repressor proteins GalR, CytR, and LacI. J Bacteriol. 1989;171:4888–4899. doi: 10.1128/jb.171.9.4888-4899.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3643.Reidl J, Boos W. The malX malY operon of Escherichia coli encodes a novel enzyme II of the phosphotransferase system recognizing glucose and maltose and an enzyme abolishing the endogenous induction of the maltose system. J Bacteriol. 1991;173:4862–4876. doi: 10.1128/jb.173.15.4862-4876.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3644.Reiner A M. Xylitol and d-arabitol toxicities due to derepressed fructose, galactitol, and sorbitol phosphotransferases of Escherichia coli. J Bacteriol. 1977;132:166–173. doi: 10.1128/jb.132.1.166-173.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3645.Reitzer L J, Magasanik B. Expression of glnA in Escherichia coli is regulated at tandem promoters. Proc Natl Acad Sci USA. 1985;82:1979–1983. doi: 10.1073/pnas.82.7.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3646.Reizer J, Reizer A, Kornberg H L, Saier M H., Jr Sequence of the fruB gene of Escherichia coli encoding the diphosphoryl transfer protein (DTP) of the phosphoenolpyruvate:sugar phosphotransferase system. FEMS Microbiol Lett. 1994;118:159–162. doi: 10.1111/j.1574-6968.1994.tb06819.x. [DOI] [PubMed] [Google Scholar]
- 3647.Reizer J, Reizer A, Saier M H., Jr The cellobiose permease of Escherichia coli consists of three proteins and is homologous to the lactose permease of Staphylococcus aureus. Res Microbiol. 1990;141:1061–1067. doi: 10.1016/0923-2508(90)90079-6. [DOI] [PubMed] [Google Scholar]
- 3647a.Reizer J, Reizer A, Yamada M, Saier M H., Jr The glucitol permease of Escherichia coli: a tripartite permease of the phosphotransferase system. Microbiology. 1998;144:1–2. doi: 10.1099/00221287-144-6-1463. [DOI] [PubMed] [Google Scholar]
- 3648.Reizer J, Reizer A, Merrick M J, Plunkett G, Rose D J, Saier M H., Jr Novel phosphotransferase-encoding genes revealed by analysis of the Escherichia coli genome: a chimeric gene encoding an enzyme I homologue that possesses a putative sensory transduction domain. Gene. 1996;181:103–108. doi: 10.1016/s0378-1119(96)00481-7. [DOI] [PubMed] [Google Scholar]
- 3649.Reizer J, Ramseier T M, Reizer A, Charbit A, Saier M H., Jr Novel phosphotransferase genes revealed by bacterial genome sequencing: a gene cluster encoding a putative N-acetylgalactosamine metabolic pathway in Escherichia coli. Microbiology. 1996;142:231–250. doi: 10.1099/13500872-142-2-231. [DOI] [PubMed] [Google Scholar]
- 3650.Rensing C, Mitra B, Rosen B P. The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc Natl Acad Sci USA. 1997;94:14326–14331. doi: 10.1073/pnas.94.26.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3651.Repoila F, Gutierrez C. Osmotic induction of the periplasmic trehalase in Escherichia coli K12: characterization of the treA gene promoter. Mol Microbiol. 1991;5:747–755. doi: 10.1111/j.1365-2958.1991.tb00745.x. [DOI] [PubMed] [Google Scholar]
- 3652.Retallack D M, Friedman D I. A role for a small stable RNA in modulating the activity of DNA-binding proteins. Cell. 1995;83:227–235. doi: 10.1016/0092-8674(95)90164-7. [DOI] [PubMed] [Google Scholar]
- 3653.Retallack D M, Johnson L L, Friedman D I. Role for 10Sa RNA in the growth of lambda-P22 hybrid phage. J Bacteriol. 1994;176:2082–2089. doi: 10.1128/jb.176.7.2082-2089.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3654.Reuter K, Slany R, Ullrich F, Kersten H. Structure and organization of Escherichia coli genes involved in biosynthesis of the deazaguanine derivative queuine, a nutrient factor for eukaryotes. J Bacteriol. 1991;173:2256–2264. doi: 10.1128/jb.173.7.2256-2264.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3655.Reuter K, Chong S, Ullrich F, Kersten H, Garcia G A. Serine 90 is required for enzymic activity by tRNA-guanine transglycosylase from Escherichia coli. Biochemistry. 1994;33:7041–7046. doi: 10.1021/bi00189a004. [DOI] [PubMed] [Google Scholar]
- 3656.Reuven N B, Koonin E V, Rudd K E, Deutscher M P. The gene for the longest known Escherichia coli protein is a member of helicase superfamily II. J Bacteriol. 1995;177:5393–5400. doi: 10.1128/jb.177.19.5393-5400.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3657.Reynes J P, Tiraby M, Baron M, Drocourt F, Tiraby G. Escherichia coli thymidylate kinase: molecular cloning, nucleotide sequence, and genetic organization of the corresponding tmk locus. J Bacteriol. 1996;178:2804–2812. doi: 10.1128/jb.178.10.2804-2812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3658.Reynolds A E, Felton J, Wright A. Insertion of DNA activates the cryptic bgl operon in E. coli. K12. Nature. 1981;293:625–629. doi: 10.1038/293625a0. [DOI] [PubMed] [Google Scholar]
- 3659.Reynolds A E, Mahadevan S, LeGrice S F J, Wright A. Enhancement of bacterial gene expression by insertion elements or by mutation in a CAP-cAMP binding site. J Mol Biol. 1984;191:85–95. doi: 10.1016/0022-2836(86)90424-9. [DOI] [PubMed] [Google Scholar]
- 3660.Rhee K Y, Parekh B S, Hatfield G W. Leucine-responsive regulatory protein-DNA interactions in the leader region of the ilvGMEDA operon of Escherichia coli. J Biol Chem. 1996;271:26499–26507. doi: 10.1074/jbc.271.43.26499. [DOI] [PubMed] [Google Scholar]
- 3661.Rice P W, Dahlberg J E. A gene between polA and glnA retards growth of Escherichia coli when present in multiple copies: physiological effects of the gene for spot 42 RNA. J Bacteriol. 1982;152:1196–1210. doi: 10.1128/jb.152.3.1196-1210.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3662.Richardson K K, Fostel J, Skopek T R. Nucleotide sequence of the xanthine guanine phosphoribosyl transferase gene of E. coli. Nucleic Acids Res. 1983;11:8809–8816. doi: 10.1093/nar/11.24.8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3663.Richardson L V, Richardson J P. Rho-dependent termination of transcription is governed primarily by the upstream Rho utilization (rut) sequences of a terminator. J Biol Chem. 1996;271:21597–21603. doi: 10.1074/jbc.271.35.21597. [DOI] [PubMed] [Google Scholar]
- 3664.Richaud C, Printz C. Nucleotide sequence of the dapF gene and flanking regions from Escherichia coli K12. Nucleic Acids Res. 1988;16:10367. doi: 10.1093/nar/16.21.10367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3665.Richaud C, Richaud F, Martin C, Haziza C, Patte J C. Regulation of expression and nucleotide sequence of the Escherichia coli dapD gene. J Biol Chem. 1984;259:14824–14828. [PubMed] [Google Scholar]
- 3666.Richaud C, Higgins W, Mengin-Lecreulx D, Stragier P. Molecular cloning, characterization, and chromosomal localization of dapF, the Escherichia coli gene for diaminopimelate epimerase. J Bacteriol. 1987;169:1454–1459. doi: 10.1128/jb.169.4.1454-1459.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3667.Richaud F, Richaud C, Ratet P, Patte J C. Chromosomal location and nucleotide sequence of the Escherichia coli dapA gene. J Bacteriol. 1986;166:297–300. doi: 10.1128/jb.166.1.297-300.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3668.Richet E. On the role of the multiple regulatory elements involved in the activation of the Escherichia coli malEp promoter. J Mol Biol. 1996;264:852–862. doi: 10.1006/jmbi.1996.0682. [DOI] [PubMed] [Google Scholar]
- 3669.Richet E, Raibaud O. MalT, the regulatory protein of the Escherichia coli maltose system, is an ATP-dependent transcriptional activator. EMBO J. 1989;8:981–987. doi: 10.1002/j.1460-2075.1989.tb03461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3670.Richter G, Ritz H, Katzenmeier G, Volk R, Kohnle A, Lottspeich F, Allendorf D, Bacher A. Biosynthesis of riboflavin: cloning, sequencing, mapping, and expression of the gene coding for GTP cyclohydrolase II in Escherichia coli. J Bacteriol. 1993;175:4045–4051. doi: 10.1128/jb.175.13.4045-4051.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3671.Richter G, Fischer M, Krieger C, Eberhardt S, Luttgen H, Gerstenschlager I, Bacher A. Biosynthesis of riboflavin: characterization of the bifunctional deaminase-reductase of Escherichia coli and Bacillus subtilis. J Bacteriol. 1997;179:2022–2028. doi: 10.1128/jb.179.6.2022-2028.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3672.Richter G, Volk R, Krieger C, Lahm H-W, Rothlisberger U, Bacher A. Biosynthesis of riboflavin: cloning, sequencing, and expression of the gene coding for 3,4-dihydroxy-2-butanone 4-phosphate synthase of Escherichia coli. J Bacteriol. 1992;174:4050–4056. doi: 10.1128/jb.174.12.4050-4056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3673.Rida S, Caillet J, Alix J H. Amplification of a novel gene, sanA, abolishes a vancomycin-sensitive defect in Escherichia coli. J Bacteriol. 1996;178:94–102. doi: 10.1128/jb.178.1.94-102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3674.Rieul C, Bleicher F, Duclos B, Cortay J-C, Cozzone A J. Nucleotide sequence of the aceA gene coding for isocitrate lyase in Escherichia coli. Nucleic Acids Res. 1988;16:5689. doi: 10.1093/nar/16.12.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3675.Riggs P D, Derman A I, Beckwith J R. A mutation affecting the regulation of a secA-lacZ fusion defines a new sec gene. Genetics. 1988;118:571–579. doi: 10.1093/genetics/118.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3676.Riley M, Glansdorff N. Cloning the Escherichia coli K-12 argD gene specifying acetylornithine 8-transaminase. Gene. 1983;24:335–339. doi: 10.1016/0378-1119(83)90095-1. [DOI] [PubMed] [Google Scholar]
- 3677.Rimmele M, Boos W. Trehalose-6-phosphate hydrolase of Escherichia coli. J Bacteriol. 1994;176:5654–5664. doi: 10.1128/jb.176.18.5654-5664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3678.Ringquist S, Schneider D, Gibson T, Baron C, Bock A, Gold L. Recognition of the mRNA selenocysteine insertion sequence by the specialized translational elongation factor SELB. Genes Dev. 1994;8:376–385. doi: 10.1101/gad.8.3.376. [DOI] [PubMed] [Google Scholar]
- 3679.Rioux C R, Kadner R J. Vitamin B12 transport in Escherichia coli K12 does not require the btuE gene of the btuCED operon. Mol Gen Genet. 1989;217:301–308. doi: 10.1007/BF02464897. [DOI] [PubMed] [Google Scholar]
- 3680.Ritzenthaler P, Mata-Gilsinger M. Use of in vitro gene fusions to study the uxuR regulatory gene in Escherichia coli K-12; direction of transcription and regulation of its expression. J Bacteriol. 1982;150:1040–1047. doi: 10.1128/jb.150.3.1040-1047.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3681.Ritzenthaler P, Mata-Gilsinger M, Stoeber F. Construction and expression of hybrid plasmids containing Escherichia coli K-12 uxu genes. J Bacteriol. 1980;143:1116–1126. doi: 10.1128/jb.143.3.1116-1126.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3682.Ritzenthaler P, Mata-Gilsinger M, Stoeber F. Molecular cloning of Escherichia coli K-12 hexuronate system genes: exu region. J Bacteriol. 1981;145:181–190. doi: 10.1128/jb.145.1.181-190.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3683.Roa B B, Connolly D M, Winkler M E. Overlap between pdxA and ksgA in the complex pdxA-ksgA-apaG-apaH operon of Escherichia coli K-12. J Bacteriol. 1989;171:4767–4777. doi: 10.1128/jb.171.9.4767-4777.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3684.Robbins A R. Regulation of the Escherichia coli methylgalactoside transport system by gene mglD. J Bacteriol. 1975;123:69–74. doi: 10.1128/jb.123.1.69-74.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3685.Robbins J C, Oxender D L. Transport systems for alanine, serine, and glycine in Escherichia coli K-12. J Bacteriol. 1973;116:12–18. doi: 10.1128/jb.116.1.12-18.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3686.Robert-Baudouy J M, Portalier R C. Studies of mutations in glucoronate catabolism in Escherichia coli K-12. Mol Gen Genet. 1974;131:31–46. doi: 10.1007/BF00269385. [DOI] [PubMed] [Google Scholar]
- 3686a.Robert-Baudouy J M, Portalier R, Stoeber F. Regulation of hexuronate system genes in Escherichia coli K-12: multiple regulation of the uxu operon by exuR and uxuR gene products. J Bacteriol. 1981;145:211–220. doi: 10.1128/jb.145.1.211-220.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3687.Roberts J W, Carbon J. Nucleotide sequence studies of normal and genetically altered glycine transfer ribonucleic acids in Escherichia coli J. Biol Chem. 1975;250:5556–5572. [PubMed] [Google Scholar]
- 3688.Roberts L M, Reeve E C R. Two mutations giving low-level streptomycin resistance in Escherichia coli K-12. Genet Res. 1970;16:359–365. doi: 10.1017/s0016672300002640. [DOI] [PubMed] [Google Scholar]
- 3689.Roberts R E, Lienhard C I, Gaines C G, Smith J M, Guest J R. Genetic and molecular characterization of the guaC-nadC-aroP region of Escherichia coli K-12. J Bacteriol. 1988;170:463–467. doi: 10.1128/jb.170.1.463-467.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3690.Robertson A M, Sullivan P A, Jones-Mortimer M C, Kornberg H L. Two genes affecting glucarate utilization in Escherichia coli K12. J Gen Microbiol. 1980;117:377–382. doi: 10.1099/00221287-117-2-377. [DOI] [PubMed] [Google Scholar]
- 3691.Robinson A, Kenan D J, Hatfull G F, Sullivan N F, Spiegelberg R, Donachie W D. DNA sequence and transcriptional organization of essential cell division genes ftsQ and ftsA of Escherichia coli: evidence for overlapping transcriptional units. J Bacteriol. 1984;160:546–555. doi: 10.1128/jb.160.2.546-555.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3692.Robinson A, Kenan D J, Sweeney J, Donachie W D. Further evidence for overlapping transcriptional units in an Escherichia coli cell envelope-cell division gene cluster: DNA sequence and transcriptional organization of the ddl ftsQ region. J Bacteriol. 1986;167:809–817. doi: 10.1128/jb.167.3.809-817.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3693.Robinson A C, Begg K J, MacArthur E. Isolation and characterization of intragenic suppressors of an Escherichia coli ftsA mutation. Res Microbiol. 1991;142:623–631. doi: 10.1016/0923-2508(91)90075-l. [DOI] [PubMed] [Google Scholar]
- 3694.Robinson C L, Jackson J H. New acetohydroxy acid synthase activity from mutational activation of a cryptic gene in Escherichia coli K-12. Mol Gen Genet. 1982;186:240–246. doi: 10.1007/BF00331856. [DOI] [PubMed] [Google Scholar]
- 3695.Rocha M, Vazquez M, Garciarrubio A, Covarrubias A. Nucleotide sequence of the glnA-glnL intercistronic region of Escherichia coli. Gene. 1985;37:91–99. doi: 10.1016/0378-1119(85)90261-6. [DOI] [PubMed] [Google Scholar]
- 3696.Rock C O, Tsay J T, Heath R, Jackowski S. Increased unsaturated fatty acid production associated with a suppressor of the fabA6(Ts) mutation in Escherichia coli. J Bacteriol. 1996;178:5382–5387. doi: 10.1128/jb.178.18.5382-5387.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3697.Rockabrand D, Livers K, Austin T, Kaiser R, Jensen D, Burgess R, Blum P. Roles of DnaK and RpoS in starvation-induced thermotolerance of Escherichia coli. J Bacteriol. 1998;180:846–854. doi: 10.1128/jb.180.4.846-854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3698.Rockwell P, Gottesman M E. An Escherichia coli rpoB mutation that inhibits transcription of catabolite-sensitive operons. J Mol Biol. 1991;222:189–196. doi: 10.1016/0022-2836(91)90205-k. [DOI] [PubMed] [Google Scholar]
- 3699.Rodel W, Plaga W, Frank R, Knappe J. Primary structures of Escherichia coli pyruvate formate-lyase and pyruvate-formate-lyase-activating enzyme deduced from the DNA nucleotide sequences. Eur J Biochem. 1988;177:153–158. doi: 10.1111/j.1432-1033.1988.tb14356.x. [DOI] [PubMed] [Google Scholar]
- 3700.Rodionov D G, Pisabarro A G, de Pedro M A, Kusser W, Ishiguro E E. Beta-lactam-induced bacteriolysis of amino acid-deprived Escherichia coli is dependent on phospholipid synthesis. J Bacteriol. 1995;177:992–997. doi: 10.1128/jb.177.4.992-997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3701.Rodionov D G, Ishiguro E E. Dependence of peptidoglycan metabolism on phospholipid synthesis during growth of Escherichia coli. Microbiology. 1996;142:2871–2877. doi: 10.1099/13500872-142-10-2871. [DOI] [PubMed] [Google Scholar]
- 3702.Rodolakis A, Casse F, Starke J. Morphological mutants of Escherichia coli K-12. Mapping of the envC mutation. Mol Gen Genet. 1974;130:177–181. doi: 10.1007/BF00269088. [DOI] [PubMed] [Google Scholar]
- 3703.Rodriguez-Sainz M C, Hernandez-Chico C, Moreno F. Molecular characterization of pmbA, an Escherichia coli chromosomal gene required for the production of the antibiotic peptide MccB17. Mol Microbiol. 1990;4:1921–1932. doi: 10.1111/j.1365-2958.1990.tb02041.x. [DOI] [PubMed] [Google Scholar]
- 3704.Roecklein B A, Kuempel P L. In vivo characterization of tus gene expression in Escherichia coli. Mol Microbiol. 1992;6:1655–1661. doi: 10.1111/j.1365-2958.1992.tb00890.x. [DOI] [PubMed] [Google Scholar]
- 3705.Roeder W, Somerville R L. Cloning the trpR gene. Mol Gen Genet. 1979;176:361–368. doi: 10.1007/BF00333098. [DOI] [PubMed] [Google Scholar]
- 3706.Roehl R A, Vinopal R. New maltose Blu mutations in Escherichia coli K-12. J Bacteriol. 1979;139:683–685. doi: 10.1128/jb.139.2.683-685.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3707.Roehl R A, Vinopal R. Genetic locus, distant from ptsM, affecting enzyme IIA/IIB function in Escherichia coli K-12. J Bacteriol. 1980;142:120–130. doi: 10.1128/jb.142.1.120-130.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3708.Rogers K C, Crescenzo A T, Soll D. Aminoacylation of transfer RNAs with 2-thiouridine derivatives in the wobble position of the anticodon. Biochimie. 1995;77:66–74. doi: 10.1016/0300-9084(96)88106-5. [DOI] [PubMed] [Google Scholar]
- 3709.Rogers M J, Ohgi T, Plumbridge J A, Soll D. Nucleotide sequence of the Escherichia coli nagE and nagB genes: the structural genes for the N-acetylglucosamine transport protein of the bacterial phosphoenolpyruvate:sugar phosphotransferase system and for glucosamine-6-phosphate deaminase. Gene. 1988;62:197–207. doi: 10.1016/0378-1119(88)90558-6. [DOI] [PubMed] [Google Scholar]
- 3710.Rogers S D, Bhave M R, Mercer J R, Camakaris J, Lee B T. Cloning and characterization of cutE, a gene involved in copper transport in Escherichia coli. J Bacteriol. 1991;173:6742–6748. doi: 10.1128/jb.173.21.6742-6748.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3711.Rogers S G, Weiss R. Cloning of the exonuclease III gene of Escherichia coli. Gene. 1980;11:187–195. doi: 10.1016/0378-1119(80)90059-1. [DOI] [PubMed] [Google Scholar]
- 3712.Rohrbach M R, Braun V, Koster W. Ferrichrome transport in Escherichia coli K-12: altered substrate specificity of mutated periplasmic FhuD and interaction of FhuD with the integral membrane protein FhuB. J Bacteriol. 1995;177:7186–7193. doi: 10.1128/jb.177.24.7186-7193.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3713.Rohrwild M, Coux O, Huang H C, Moerschell R P, Yoo S J, Seol J H, Chung C H, Goldberg A L. HsIV-HsIU: a novel ATP-dependent protease complex in Escherichia coli related to the eukaryotic proteasome. Proc Natl Acad Sci USA. 1996;93:5808–5813. doi: 10.1073/pnas.93.12.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3714.Rojo F, Ayala J A, dePedro M A, Vazquez D. Analysis of the different molecular forms of penicillin-binding protein 1B in Escherichia coli ponB mutants lysogenized with special transducing λ (ponB+) bacteriophages. Eur J Biochem. 1984;144:571–576. doi: 10.1111/j.1432-1033.1984.tb08503.x. [DOI] [PubMed] [Google Scholar]
- 3715.Roland K L, Powell F E, Turnbough C L. Role of translation and attenuation in the control of pyrBI operon expression in Escherichia coli K-12. J Bacteriol. 1985;163:991–999. doi: 10.1128/jb.163.3.991-999.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3716.Roland K L, Martin L E, Esther C R, Spitznagel J K. Spontaneous pmrA mutants of Salmonella typhimurium LT2 define a new two-component regulatory system with a possible role in virulence. J Bacteriol. 1993;175:4154–4164. doi: 10.1128/jb.175.13.4154-4164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3717.Roldan-Arjona T, Sedgwick B. DNA base damage induced by ionizing radiation recognized by Escherichia coli UvrABC nuclease but not Nth or Fpg proteins. Mol Carcinog. 1996;16:188–196. doi: 10.1002/(SICI)1098-2744(199608)16:4<188::AID-MC2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 3718.Rolfes R J, Zalkin H. Escherichia coli gene purR encoding a repressor protein for purine nucleotide synthesis. J Biol Chem. 1988;263:19653–19661. [PubMed] [Google Scholar]
- 3719.Rolfes R J, Zalkin H. Regulation of Escherichia coli purF. Mutations that define the promoter, operator and purine repressor gene. J Biol Chem. 1988;263:19649–19661. [PubMed] [Google Scholar]
- 3720.Roman S J, Frantz B B, Matsumura P. Gene sequence, overproduction, purification and determination of the wild-type level of the Escherichia coli flagellar switch protein FliG. Gene. 1993;133:103–108. doi: 10.1016/0378-1119(93)90232-r. [DOI] [PubMed] [Google Scholar]
- 3721.Romeo T, Kumar A, Preiss J. Analysis of the Escherichia coli glycogen gene cluster suggests that catabolic enzymes are encoded among the biosynthetic genes. Gene. 1988;70:363–376. doi: 10.1016/0378-1119(88)90208-9. [DOI] [PubMed] [Google Scholar]
- 3722.Romeo T, Moore J, Smith J. A simple method for cloning genes involved in glucan biosynthesis: isolation of structural and regulatory genes for glycogen synthesis in Escherichia coli. Gene. 1991;108:23–29. doi: 10.1016/0378-1119(91)90483-r. [DOI] [PubMed] [Google Scholar]
- 3723.Romeo T, Gong M. Genetic and physical mapping of the regulatory gene csrA on the Escherichia coli K-12 chromosome. J Bacteriol. 1993;175:5740–5741. doi: 10.1128/jb.175.17.5740-5741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3724.Romeo T, Gong M, Liu M Y, Brun-Zinkernagel A M. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J Bacteriol. 1993;175:4744–4755. doi: 10.1128/jb.175.15.4744-4755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3725.Romling U, Bian Z, Hammar M, Sierralta W D, Normark S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol. 1998;180:722–731. doi: 10.1128/jb.180.3.722-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3726.Roncero C, Casadaban M J. Genetic analysis of the genes involved in synthesis of the lipopolysaccharide core in Escherichia coli K-12. J Bacteriol. 1992;174:3250–3260. doi: 10.1128/jb.174.10.3250-3260.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3727.Rondeaux S S, Hsu P-Y, DeMoss J A. Construction in vitro of a cloned nar operon from Escherichia coli. J Bacteriol. 1984;159:159–166. doi: 10.1128/jb.159.1.159-166.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3728.Rood J I, Laird A J, Williams J W. Cloning of the Escherichia coli K-12 dihydrofolate reductase gene following Mu-mediated transposition. Gene. 1980;8:255–265. doi: 10.1016/0378-1119(80)90003-7. [DOI] [PubMed] [Google Scholar]
- 3729.Roof W D, Foltermann F, Wild J R. The organization and regulation of the pyrBI operon in E. coli includes a Rho-independent attenuator sequence. Mol Gen Genet. 1982;187:391–400. doi: 10.1007/BF00332617. [DOI] [PubMed] [Google Scholar]
- 3730.Roof W D, Fang H Q, Young K D, Sun J, Young R. Mutational analysis of slyD, an Escherichia coli gene encoding a protein of the FKBP immunophilin family. Mol Microbiol. 1997;25:1031–1046. doi: 10.1046/j.1365-2958.1997.5201884.x. [DOI] [PubMed] [Google Scholar]
- 3731.Roof W D, Horne S M, Young K D, Young R. slyD, a host gene required for phi X174 lysis, is related to the FK506-binding protein family of peptidyl-prolyl cis-trans-isomerases. J Biol Chem. 1994;269:2902–2910. [PubMed] [Google Scholar]
- 3732.Roovers M, Charlier D, Feller A, Gigot D, Holemans F, Lissens W, Pierard A, Glansdorff N. carP, a novel gene regulating the transcription of the carbamoylphosphate synthetase operon of Escherichia coli. J Mol Biol. 1988;204:857–865. doi: 10.1016/0022-2836(88)90046-0. [DOI] [PubMed] [Google Scholar]
- 3733.Rosen B P. Basic amino acid transport in Escherichia coli: properties of canavanine-resistant mutants. J Bacteriol. 1973;116:627–635. doi: 10.1128/jb.116.2.627-635.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3734.Rosenberg H. Phosphate transport in prokaryotes. In: Rosen B P, Silver S, editors. Ion transport in prokaryotes. New York, N.Y: Academic Press, Inc.; 1987. pp. 205–246. [Google Scholar]
- 3735.Rosenfeld S A, Stevis P E, Ho N W Y. Cloning and characterization of the xyl gene from Escherichia coli. Mol Gen Genet. 1984;194:410–415. doi: 10.1007/BF00425552. [DOI] [PubMed] [Google Scholar]
- 3736.Rosentel J K, Healy F, Maupin-Furlow J A, Lee J H, Shanmugam K T. Molybdate and regulation of mod (molybdate transport), fdhF, and hyc (formate hydrogenlyase) operons in Escherichia coli. J Bacteriol. 1995;177:4857–4864. doi: 10.1128/jb.177.17.4857-4864.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3737.Rosner J L, Slonczewski J L. Dual regulation of inaA by the multiple antibiotic resistance (mar) and superoxide (soxRS) stress response systems of Escherichia coli. J Bacteriol. 1994;176:6262–6269. doi: 10.1128/jb.176.20.6262-6269.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3738.Rosner J L, Chai T J, Foulds J. Regulation of ompF porin expression by salicylate in Escherichia coli. J Bacteriol. 1991;173:5631–5638. doi: 10.1128/jb.173.18.5631-5638.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3739.Ross T K, Achberger C, Braymer H D. Characterization of the Escherichia coli modified cytosine restriction (mcrB) gene. Gene. 1987;61:277–289. doi: 10.1016/0378-1119(87)90191-0. [DOI] [PubMed] [Google Scholar]
- 3740.Ross T K, Achberger E C, Braymer H D. Nucleotide sequence of the McrB region of Escherichia coli K-12 and evidence for two independent translational initiation sites at the mcrB locus. J Bacteriol. 1989;171:1974–1981. doi: 10.1128/jb.171.4.1974-1981.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3741.Ross T K, Braymer H D. Localization of a genetic region involved in McrB restriction by Escherichia coli K-12. J Bacteriol. 1987;169:1757–1759. doi: 10.1128/jb.169.4.1757-1759.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3742.Ross W, Thompson J F, Newlands J T, Gourse R L. E. coli Fis protein activates ribosomal RNA transcription in vitro. EMBO J. 1990;9:3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3743.Rossi J J, Landy A. Structure and organization of the two tRNATyr gene clusters on the E. coli chromosome. Cell. 1979;16:523–534. doi: 10.1016/0092-8674(79)90027-8. [DOI] [PubMed] [Google Scholar]
- 3744.Rossi J J, Egan J, Hudson L, Landy A. The tyrT locus: termination and processing of a complex transcript. Cell. 1981;26:305–314. doi: 10.1016/0092-8674(81)90199-9. [DOI] [PubMed] [Google Scholar]
- 3745.Rossmann R, Sawers G, Bock A. Mechanism of regulation of the formate-hydrogenlyase pathway by oxygen, nitrate, and pH: definition of the formate regulon. Mol Microbiol. 1991;5:2807–2814. doi: 10.1111/j.1365-2958.1991.tb01989.x. [DOI] [PubMed] [Google Scholar]
- 3746.Rossmann R, Sauter M, Lottspeich F, Bock A. Maturation of the large subunit (HYCE) of Escherichia coli hydrogenase 3 requires nickel incorporation followed by C-terminal processing at Arg537. Eur J Biochem. 1994;220:377–384. doi: 10.1111/j.1432-1033.1994.tb18634.x. [DOI] [PubMed] [Google Scholar]
- 3747.Rossmann R, Maier T, Lottspeich F, Bock A. Characterisation of a protease from Escherichia coli involved in hydrogenase maturation. Eur J Biochem. 1995;227:545–550. doi: 10.1111/j.1432-1033.1995.tb20422.x. [DOI] [PubMed] [Google Scholar]
- 3748.Rostas K, Morton S J, Picksley S M, Lloyd R G. Nucleotide sequence and LexA regulation of the Escherichia coli recN gene. Nucleic Acids Res. 1987;15:5041–5049. doi: 10.1093/nar/15.13.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3748a.Rotering H, Braun V. Lipid deficiency on a lipoprotein mutant of Escherichia coli. FEBS Lett. 1977;83:41–44. doi: 10.1016/0014-5793(77)80637-6. [DOI] [PubMed] [Google Scholar]
- 3749.Rothery R A, Grant J L, Johnson J L, Rajagopalan K V, Weiner J H. Association of molybdopterin guanine dinucleotide with Escherichia coli dimethyl sulfoxide reductase: effect of tungstate and a mob mutation. J Bacteriol. 1995;177:2057–2063. doi: 10.1128/jb.177.8.2057-2063.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3750.Rothstein D M, Pahel G, Tyler B, Magasanik B. Regulation of expression from the glnA promoter of Escherichia coli in the absence of glutamine synthetase. Proc Natl Acad Sci USA. 1980;77:7372–7376. doi: 10.1073/pnas.77.12.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3751.Rotman B, Guzman R. Identification of the mglA gene product in the β-methylgalactoside transport system of Escherichia coli using plasmid DNA deletions generated in vitro. J Biol Chem. 1982;257:9030–9034. [PubMed] [Google Scholar]
- 3752.Rouviere P E, Gross C A. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 1996;10:3170–3182. doi: 10.1101/gad.10.24.3170. [DOI] [PubMed] [Google Scholar]
- 3753.Rowen L, Kobori J, Scherer S. Cloning of bacterial DNA replication genes in bacteriophage λ. Mol Gen Genet. 1982;187:501–509. doi: 10.1007/BF00332635. [DOI] [PubMed] [Google Scholar]
- 3754.Rowland G C, Giffard P M, Booth I R. Genetic studies of the phs locus of Escherichia coli, a mutation causing pleiotropic lesions in metabolism and pH homeostasis. FEBS Lett. 1984;173:295–300. doi: 10.1016/0014-5793(84)80794-2. [DOI] [PubMed] [Google Scholar]
- 3755.Rowland G C, Giffard P M, Booth I R. phs locus of Escherichia coli, a mutation causing pleiotropic lesions in metabolism, is an rpoA allele. J Bacteriol. 1985;164:972–975. doi: 10.1128/jb.164.2.972-975.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3756.Rowland G C, Lim P P, Glass R E. In vivo cloning of a carboxy-terminal rpoB allele which confers altered transcriptional properties. Folia Microbiol. 1995;40:588–594. doi: 10.1007/BF02818514. [DOI] [PubMed] [Google Scholar]
- 3756a.Rowley D L, Wolf R E., Jr Molecular characteristics of the Escherichia coli K-12 zwf gene encoding glucose-6-phosphate dehydrogenase. J Bacteriol. 1991;173:968–977. doi: 10.1128/jb.173.3.968-977.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3757.Rowsell E H, Smith J M, Wolfe A, Taylor B L. CheA, CheW, and CheY are required for chemotaxis to oxygen and sugars of the phosphotransferase system in Escherichia coli. J Bacteriol. 1995;177:6011–6014. doi: 10.1128/jb.177.20.6011-6014.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3758.Roy A M, Danchin A. Restriction map of the cya region of the Escherichia coli K12 chromosome. Biochimie. 1981;63:719–722. doi: 10.1016/s0300-9084(81)80220-9. [DOI] [PubMed] [Google Scholar]
- 3759.Roy A M, Danchin A. The cya locus of Escherichia coli K12: organization and gene products. Mol Gen Genet. 1982;188:465–471. doi: 10.1007/BF00330050. [DOI] [PubMed] [Google Scholar]
- 3760.Roy A M, Haziza C, Danchin A. Regulation of adenylate cyclase synthesis: nucleotide sequence of the control region. EMBO J. 1983;2:791–797. doi: 10.1002/j.1460-2075.1983.tb01502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3761.Roy A M, Coleman J. Mutations in firA, encoding the second acyltransferase in lipopolysaccharide biosynthesis, affect multiple steps in lipopolysaccharide biosynthesis. J Bacteriol. 1994;176:1639–1646. doi: 10.1128/jb.176.6.1639-1646.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3762.Rubin E M, Wilson G, Young F E. Expression of thymidylate synthetase activity in Bacillus subtilis upon integration of a cloned gene from E. coli. Gene. 1980;10:227–235. doi: 10.1016/0378-1119(80)90052-9. [DOI] [PubMed] [Google Scholar]
- 3763.Rudd K E. Escherichia coli K-12 on the internet. Trends Genet. 1996;12:156–157. doi: 10.1016/0168-9525(96)60012-2. . (News.) [DOI] [PubMed] [Google Scholar]
- 3763a.Rudd K E. Linkage map of Escherichia coli K-12, edition 10: the physical map. Microbiol Mol Biol Rev. 1998;62:985–1019. doi: 10.1128/mmbr.62.3.985-1019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3764.Rudd K E, Sofia H J, Koonin E V, Plunkett G, Lazar S, Rouviere P E. A new family of peptidyl-prolyl isomerases. Trends Biochem Sci. 1995;20:12–14. doi: 10.1016/s0968-0004(00)88940-9. [DOI] [PubMed] [Google Scholar]
- 3765.Ruijter G J, van Meurs G, Verwey M A, Postma P W, van Dam K. Analysis of mutations that uncouple transport from phosphorylation in enzyme IIGlc of the Escherichia coli phosphoenolpyruvate-dependent phosphotransferase system. J Bacteriol. 1992;174:2843–2850. doi: 10.1128/jb.174.9.2843-2850.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3766.Ruiz S M, Letourneau S, Cupples C G. Isolation and characterization of an Escherichia coli strain with a high frequency of C-to-T mutations at 5-methylcytosines. J Bacteriol. 1993;175:4985–4989. doi: 10.1128/jb.175.16.4985-4989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3767.Ruiz-Vazquez R, Cerda-Olmedo E. An Escherichia coli mutant refractory to nitrosoguanidine mutagenesis. Mol Gen Genet. 1980;178:625–631. doi: 10.1007/BF00337870. [DOI] [PubMed] [Google Scholar]
- 3768.Rule G S, Pratt E A, Chin C C Q, Wold F, Ho C. Overproduction and nucleotide sequence of the respiratory d-lactate dehydrogenase of Escherichia coli. J Bacteriol. 1985;161:1059–1068. doi: 10.1128/jb.161.3.1059-1068.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3769.Rupprecht K R, Gordon A W, Lundrigan M D, Gayda R C, Markovitz A, Earhart C F. ompT: Escherichia coli K-12 structural gene for protein a (3b) J Bacteriol. 1983;153:1104–1106. doi: 10.1128/jb.153.2.1104-1106.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3770.Rusnak F, Sakaitani M, Drueckhammer D, Reichert J, Walsh C T. Biosynthesis of the Escherichia coli siderophore enterobactin: sequence of the entF gene, expression and purification of EntF, and analysis of covalent phosphopantetheine. Biochemistry. 1991;30:2916–2927. doi: 10.1021/bi00225a027. [DOI] [PubMed] [Google Scholar]
- 3771.Russel M, Holmgren A. Construction and characterization of glutaredoxin-negative mutants of Escherichia coli. Proc Natl Acad Sci USA. 1988;85:990–994. doi: 10.1073/pnas.85.4.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3772.Russel M, Model P. A bacterial gene, fip, required for filamentous bacteriophage F1 assembly. J Bacteriol. 1983;154:1064–1076. doi: 10.1128/jb.154.3.1064-1076.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3773.Russel M, Model P. Characterization of the cloned fip gene and its product. J Bacteriol. 1984;157:526–532. doi: 10.1128/jb.157.2.526-532.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3774.Russel M, Model P. Direct cloning of the trxB gene that encodes thioredoxin reductase. J Bacteriol. 1985;163:238–242. doi: 10.1128/jb.163.1.238-242.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3775.Russel M, Model P. Thioredoxin is required for filamentous phage assembly. Proc Natl Acad Sci USA. 1985;82:29–33. doi: 10.1073/pnas.82.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3776.Russel M, Model P. Direct cloning of the trxB gene: role of thioredoxin reductase in filamentous phage assembly. In: Holmgren A, editor. Thioredoxin and glutaredoxin systems. New York, N.Y: Raven Press; 1986. p. 331. [Google Scholar]
- 3777.Russel M, Model P. Sequence of thioredoxin reductase from Escherichia coli. Relationship to other flavoprotein disulfide oxidoreductases. J Biol Chem. 1988;263:9015–9019. [PubMed] [Google Scholar]
- 3778.Russel M, Model P, Holmgren A. Thioredoxin or glutaredoxin in Escherichia coli is essential for sulfate reduction but not for deoxyribonucleotide synthesis. J Bacteriol. 1990;172:1923–1929. doi: 10.1128/jb.172.4.1923-1929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3779.Russell P W, Orndorff P E. Lesions in two Escherichia coli type 1 pilus genes alter pilus number and length without affecting receptor binding. J Bacteriol. 1992;174:5923–5935. doi: 10.1128/jb.174.18.5923-5935.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3780.Russell R R B, Pittard A J. New suppressor in Escherichia coli. J Bacteriol. 1971;107:736–740. doi: 10.1128/jb.107.3.736-740.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3781.Ryals J, Hsu R-Y, Lipsett M N, Bremer H. Isolation of single-site Escherichia coli mutants deficient in thiamine and 4-thiouridine syntheses: identification of a nuvC mutant. J Bacteriol. 1982;151:899–904. doi: 10.1128/jb.151.2.899-904.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3782.Ryden M, Murphy J B, Martin R, Isaksson L A, Gallant J. Mapping and complementation studies of the gene for release factor 1. J Bacteriol. 1986;168:1066–1069. doi: 10.1128/jb.168.3.1066-1069.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3783.Ryden S, Isaksson L A. A temperature-sensitive mutant of Escherichia coli that shows enhanced misreading of UAG/A and increased efficiency for some tRNA nonsense suppressors. Mol Gen Genet. 1984;193:38–45. doi: 10.1007/BF00327411. [DOI] [PubMed] [Google Scholar]
- 3784.Sa J H, Namgung M A, Lim C J, Fuchs J A. Expression of the Escherichia coli thioredoxin gene is negatively regulated by cyclic AMP. Biochem Biophys Res Commun. 1997;234:564–567. doi: 10.1006/bbrc.1997.6687. [DOI] [PubMed] [Google Scholar]
- 3785.Saarma U, Remme J. Novel mutants of 23S RNA: characterization of functional properties. Nucleic Acids Res. 1992;20:3147–3152. doi: 10.1093/nar/20.12.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3786.Sabe H, Miwa T, Kodaki T, Izui K, Hiraga S, Katsuki H. Molecular cloning of the phosphoenolpyruvate carboxylase gene, ppc, of Escherichia coli. Gene. 1984;31:279–283. doi: 10.1016/0378-1119(84)90222-1. [DOI] [PubMed] [Google Scholar]
- 3787.Sabnis N A, Yang H, Romeo T. Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. J Biol Chem. 1995;270:29096–29100. doi: 10.1074/jbc.270.49.29096. [DOI] [PubMed] [Google Scholar]
- 3788.Sacerdot C, Fayet G, Dessen P, Springer M, Plumbridge J A, Grunberg-Manago M, Blanquet S. Sequence of a 1.2 kb DNA fragment containing the structural gene for E. coli initiation factor IF3: presence of an AUU initiator codon. EMBO J. 1982;1:311–315. doi: 10.1002/j.1460-2075.1982.tb01166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3789.Sacerdot C, Vachon G, Laalami S, Morel-Deville F, Cenatiempo Y, Grunberg-Manago M. Both forms of translational initiation factor IF2 (alpha and beta) are required for maximal growth of Escherichia coli. Evidence for two translational initiation codons for IF2 beta. J Mol Biol. 1992;225:67–80. doi: 10.1016/0022-2836(92)91026-l. [DOI] [PubMed] [Google Scholar]
- 3790.Sadosky A B, Davidson A, Lin R-J, Hill C W. rhs gene family of Escherichia coli K-12. J Bacteriol. 1989;171:636–642. doi: 10.1128/jb.171.2.636-642.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3791.Sadosky A B, Gray J A, Hill C W. The RhsD-E subfamily of Escherichia coli K-12. Nucleic Acids Res. 1991;19:7177–7183. doi: 10.1093/nar/19.25.7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3792.Saedler H, Gullon A, Starlinger P. Negative control of the galactose operon in E. coli. Mol Gen Genet. 1968;102:79–88. doi: 10.1007/BF00341872. [DOI] [PubMed] [Google Scholar]
- 3793.Saffen D W, Presper K A, Doering T L, Roseman S. Sugar transport by the bacterial phosphotransferase system. Molecular cloning and structural analysis of the Escherichia coli ptsH, ptsI and crr genes. J Biol Chem. 1987;262:16241–16253. [PubMed] [Google Scholar]
- 3794.Sagara K, Matsuyama S, Mizushima S. SecF stabilizes SecD and SecY, components of the protein translocation machinery of the Escherichia coli cytoplasmic membrane. J Bacteriol. 1994;176:4111–4116. doi: 10.1128/jb.176.13.4111-4116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3795.Saget B M, Shevell D E, Walker G C. Alteration of lysine 178 in the hinge region of the Escherichia coli Ada protein interferes with activation of ada, but not alkA, transcription. J Bacteriol. 1995;177:1268–1274. doi: 10.1128/jb.177.5.1268-1274.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3796.Said B, Ghosn C R, Vu L, Nunn W D. Nucleotide sequencing and expression of the fadL gene involved in long-chain fatty acid transport in Escherichia coli. Mol Microbiol. 1988;2:363–370. doi: 10.1111/j.1365-2958.1988.tb00040.x. [DOI] [PubMed] [Google Scholar]
- 3797.Saier M H., Jr Cyclic AMP-independent catabolite repression in bacteria. FEMS Microbiol Lett. 1996;138:97–103. doi: 10.1111/j.1574-6968.1996.tb08141.x. [DOI] [PubMed] [Google Scholar]
- 3798.Saier M H, Jr, Straud H, Massman L S, Judice J J, Newman M J, Feucht B U. Permease-specific mutations in Salmonella typhimurium and Escherichia coli that release the glycerol, maltose, melibiose, and lactose transport systems from regulation by the phosphoenolpyruvate:sugar phosphotransferase system. J Bacteriol. 1978;133:1358–1367. doi: 10.1128/jb.133.3.1358-1367.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3799.Saiki K, Mogi T, Anraku Y. Heme O biosynthesis in Escherichia coli: the cyoE gene in the cytochrome bo operon encodes a protoheme IX farnesyltransferase. Biochem Biophys Res Commun. 1992;189:1491–1497. doi: 10.1016/0006-291x(92)90243-e. [DOI] [PubMed] [Google Scholar]
- 3800.Sain B, Murray N E. The hsd (host specificity) genes of E. coli K12. Mol Gen Genet. 1980;180:35–46. doi: 10.1007/BF00267350. [DOI] [PubMed] [Google Scholar]
- 3801.Saint-Girons I, Margarita D. Evidence for an internal promoter in the Escherichia coli threonine operon. J Bacteriol. 1985;161:461–462. doi: 10.1128/jb.161.1.461-462.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3802.Saint-Girons I, Duchange N, Cohen G N, Zakin M M. Structure and autoregulation of the metJ regulatory gene in Escherichia coli. J Biol Chem. 1984;259:14282–14285. [PubMed] [Google Scholar]
- 3803.Saint-Girons I, Duchange N, Zakin M M, Park I, Margarita D, Ferrara P, Cohen G N. Nucleotide sequence of metF, the E. coli structural gene for 5-10 methylene tetrahydrofolate reductase and of its control region. Nucleic Acids Res. 1983;11:6723–6732. doi: 10.1093/nar/11.19.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3804.Saito H, Uchida H. Initiation of the DNA replication of bacteriophage lambda in Escherichia coli K12. J Mol Biol. 1977;113:1–25. doi: 10.1016/0022-2836(77)90038-9. [DOI] [PubMed] [Google Scholar]
- 3804a.Saito M, Tsugawa A, Egawa K, Nakamua Y. Revised sequence of the nusA gene of Escherichia coli and identification of the nusA gene of Escherichia coli and identification of nusA11 (ts) and nusA1 mutations which cause changes in a hydrophobic amino acid cluster. Mol Gen Genet. 1986;205:380–382. doi: 10.1007/BF00430455. [DOI] [PubMed] [Google Scholar]
- 3805.Saito Y, Uraki F, Nakajima S, Asaeda A, Ono K, Kubo K, Yamamoto K. Characterization of endonuclease III (nth) and endonuclease VIII (nei) mutants of Escherichia coli K-12. J Bacteriol. 1997;179:3783–3785. doi: 10.1128/jb.179.11.3783-3785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3806.Sak B D, Eisenstark A, Touati D. Exonuclease III and the catalase hydroperoxidase II in Escherichia coli are both regulated by the katF gene product. Proc Natl Acad Sci USA. 1989;86:3271–3275. doi: 10.1073/pnas.86.9.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3807.Sakakibara Y. The dnaK gene of Escherichia coli functions in initiation of chromosome replication. J Bacteriol. 1988;170:972–979. doi: 10.1128/jb.170.2.972-979.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3808.Sakakibara Y. Suppression of thermosensitive initiation of DNA replication in a dnaR mutant of Escherichia coli by a rifampin resistance mutation in the rpoB gene. J Bacteriol. 1995;177:733–737. doi: 10.1128/jb.177.3.733-737.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3809.Sakakibara Y, Tsukano H, Sako T. Organization and transcription of the dnaA and dnaN genes of Escherichia coli. Gene. 1981;13:47–55. doi: 10.1016/0378-1119(81)90042-1. [DOI] [PubMed] [Google Scholar]
- 3810.Sakakibara Y, Mizukami T. A temperature-sensitive Escherichia coli mutant defective in DNA replication: dnaN, a new gene adjacent to the dnaA gene. Mol Gen Genet. 1980;178:541–553. doi: 10.1007/BF00337859. [DOI] [PubMed] [Google Scholar]
- 3811.Sakamoto H, Kimura A, Shimura Y. Processing of transcription products of the gene encoding the RNA component of RNase P. Proc Natl Acad Sci USA. 1983;80:6187–6191. doi: 10.1073/pnas.80.20.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3812.Sakashita H, Sakuma T, Akitomo Y, Ohkubo T, Kainosho M, Sekiguchi M, Morikawa K. Sequence-specific DNA recognition of the Escherichia coli Ada protein associated with the methylation-dependent functional switch for transcriptional regulation. J Biochem (Tokyo) 1995;118:1184–1191. doi: 10.1093/oxfordjournals.jbchem.a125005. [DOI] [PubMed] [Google Scholar]
- 3813.Sako T, Sakakibara Y. Coordination expression of Escherichia coli dnaA and dnaN genes. Mol Gen Genet. 1980;179:521–526. doi: 10.1007/BF00271741. [DOI] [PubMed] [Google Scholar]
- 3814.Saks M E, Sampson J R, Abelson J. Evolution of a transfer RNA gene through a point mutation in the anticodon. Science. 1998;279:1665–1670. doi: 10.1126/science.279.5357.1665. [DOI] [PubMed] [Google Scholar]
- 3815.Sakumi K, Igarashi K, Sekiguchi M, Ishihama A. The Ada protein is a class I transcription factor of Escherichia coli. J Bacteriol. 1993;175:2455–2457. doi: 10.1128/jb.175.8.2455-2457.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3816.Sakumi K, Nakabeppu Y, Yamamoto H, Kawabata S I, Iwanaga S, Sekiguchi M. Purification and structure of 3-methyladenine-DNA glycosylase I of Escherichia coli. J Biol Chem. 1986;261:15761–15766. [PubMed] [Google Scholar]
- 3817.Salanitro J P, Wegener W S. Growth of Escherichia coli on short-chain fatty acids: nature of the uptake system. J Bacteriol. 1971;108:893–900. doi: 10.1128/jb.108.2.893-901.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3818.Salavati R, Oliver D. Competition between ribosome and SecA binding promotes Escherichia coli secA translational regulation. RNA. 1995;1:745–753. [PMC free article] [PubMed] [Google Scholar]
- 3819.Saleh A Z, Yamanaka K, Niki H, Ogura T, Yamazoe M, Hiraga S. Carboxyl terminal region of the MukB protein in Escherichia coli is essential for DNA binding activity. FEMS Microbiol Lett. 1996;143:211–216. doi: 10.1111/j.1574-6968.1996.tb08482.x. [DOI] [PubMed] [Google Scholar]
- 3820.Salmond G P C, Lutkenhaus J, Donachie W D. Identification of new genes in a cell envelope-cell division gene cluster of Escherichia coli: cell envelope gene murG. J Bacteriol. 1980;144:438–440. doi: 10.1128/jb.144.1.438-440.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3821.Salmond G P C, Plakidou S. Genetic analysis of essential genes in the ftsE region of the Escherichia coli genetic map and identification of a new cell division gene ftsS. Mol Gen Genet. 1984;197:304–308. doi: 10.1007/BF00330978. [DOI] [PubMed] [Google Scholar]
- 3822.Salowe S P, Stubbe J. Cloning, overproduction, and purification of the B2 subunit of ribonucleoside-diphosphate reductase. J Bacteriol. 1986;165:363–366. doi: 10.1128/jb.165.2.363-366.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3823.Sambongi Y, Ferguson S J. Mutants of Escherichia coli lacking disulphide oxidoreductases DsbA and DsbB cannot synthesise an exogenous monohaem c-type cytochrome except in the presence of disulphide compounds. FEBS Lett. 1996;398:265–268. doi: 10.1016/s0014-5793(96)01256-2. [DOI] [PubMed] [Google Scholar]
- 3824.Sameshima J H, Wek R C, Hatfield G W. Overlapping transcription and termination of the convergent ilvA and ilvY genes of Escherichia coli. J Biol Chem. 1989;264:1224–1231. [PubMed] [Google Scholar]
- 3825.Sammartano L J, Tuveson R W, Davenport R. Control of sensitivity to inactivation by H2O2 and broad-spectrum near-UV radiation by the Escherichia coli katF locus. J Bacteriol. 1986;168:13–21. doi: 10.1128/jb.168.1.13-21.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3826.Sampei G, Mizobuchi K. Nucleotide sequence of the Escherichia coli purF gene encoding amidophosphoribosyltransferase for de novo purine nucleotide synthesis. Nucleic Acids Res. 1988;16:8717. doi: 10.1093/nar/16.17.8717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3827.Sampei G, Mizobuchi K. The organization of the purL gene encoding 5′-phosphoribosylformylglycinamide amidotransferase of Escherichia coli. J Biol Chem. 1989;264:21230–21238. [PubMed] [Google Scholar]
- 3828.Sampson B A, Misra R, Benson S A. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics. 1989;122:491–501. doi: 10.1093/genetics/122.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3829.Samsonov V V, Odoevskaya E R, Sineokii S P. Cloning and complementation analysis of Escherichia coli gpr locus affecting DNA replication of some lamdoid [sic] phages. Genetika. 1992;28:39–45. . (In Russian.) [PubMed] [Google Scholar]
- 3830.Sanatinia H, Kofoid E C, Morrison T B, Parkinson J S. The smaller of two overlapping cheA gene products is not essential for chemotaxis in Escherichia coli. J Bacteriol. 1995;177:2713–2720. doi: 10.1128/jb.177.10.2713-2720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3831.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 3832.Sancar A, Kacinski B M, Mott D L, Rupp W D. Identification of the uvrC gene product. Proc Natl Acad Sci USA. 1981;78:5450–5454. doi: 10.1073/pnas.78.9.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3833.Sancar A, Stachelek C, Konigsberg W, Rupp W D. Sequences of the recA gene and protein. Proc Natl Acad Sci USA. 1980;77:2611–2615. doi: 10.1073/pnas.77.5.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3834.Sancar A, Sancar G B, Rupp W D, Little J W, Mount D W. LexA protein inhibits transcription of the E. coli uvrA gene in vitro. Nature. 1982;298:96–98. doi: 10.1038/298096a0. [DOI] [PubMed] [Google Scholar]
- 3835.Sancar A, Williams K R, Chase J W, Rupp W D. Sequences of the ssb gene and protein. Proc Natl Acad Sci USA. 1981;78:4274–4278. doi: 10.1073/pnas.78.7.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3836.Sancar A, Clarke N D, Griswold J, Kennedy W J, Rupp W D. Identification of the uvrB gene product. J Mol Biol. 1981;148:63–76. doi: 10.1016/0022-2836(81)90235-7. [DOI] [PubMed] [Google Scholar]
- 3837.Sancar A, Wharton R P, Seltzer S, Kacinski B M, Clarke N D, Rupp W D. Identification of the uvrA gene product. J Mol Biol. 1981;148:45–62. doi: 10.1016/0022-2836(81)90234-5. [DOI] [PubMed] [Google Scholar]
- 3838.Sancar A, Rupp W D. A novel repair enzyme UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell. 1983;33:249–260. doi: 10.1016/0092-8674(83)90354-9. [DOI] [PubMed] [Google Scholar]
- 3839.Sancar G B, Sancar A, Little J W, Rupp W D. The uvrB gene of Escherichia coli has both lexA-repressed and lexA-independent promoter. Cell. 1982;28:523–530. doi: 10.1016/0092-8674(82)90207-0. [DOI] [PubMed] [Google Scholar]
- 3840.Sancar G B, Sancar A, Rupp W D. Sequences of the urrC gene and protein. Nucleic Acids Res. 1984;12:4593–4608. doi: 10.1093/nar/12.11.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3841.Sancar G B, Smith F W, Sancar A. Identification and amplification of the E. coli phr gene product. Nucleic Acids Res. 1983;11:6667–6678. doi: 10.1093/nar/11.19.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3842.Sancar G B, Smith F W, Lorence M C, Rupert C S, Sancar A. Sequences of the Escherichia coli photolyase gene and protein. J Biol Chem. 1984;259:6033–6038. [PubMed] [Google Scholar]
- 3843.Sanchez J C, Gimenez R, Schneider A, Fessner W D, Baldoma L, Aguilar J, Badia J. Activation of a cryptic gene encoding a kinase for l-xylulose opens a new pathway for the utilization of l-lyxose by Escherichia coli. J Biol Chem. 1994;269:29665–29669. [PubMed] [Google Scholar]
- 3844.Sanchez-Pescador R, Sanvicente E, Valle F, Bolivar F. Recombinant plasmids carrying the glutamate dehydrogenase structural gene from Escherichia coli. Gene. 1982;17:1–18. doi: 10.1016/0378-1119(82)90095-6. [DOI] [PubMed] [Google Scholar]
- 3845.Sandigursky M, Franklin W A. DNA deoxyribophosphodiesterase of Escherichia coli is associated with exonuclease I. Nucleic Acids Res. 1992;20:4699–4703. doi: 10.1093/nar/20.18.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3846.Sandler S J. Overlapping functions for recF and priA in cell viability and UV-inducible SOS expression are distinguished by dnaC809 in Escherichia coli K-12. Mol Microbiol. 1996;19:871–880. doi: 10.1046/j.1365-2958.1996.429959.x. [DOI] [PubMed] [Google Scholar]
- 3847.Sandler S J, Clark A J. Mutational analysis of sequences in the recF gene of Escherichia coli K-12 that affect expression. J Bacteriol. 1994;176:4011–4016. doi: 10.1128/jb.176.13.4011-4016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3848.Sandler S J, Chackerian B, Li J T, Clark A J. Sequence and complementation analysis of recF genes from Escherichia coli, Salmonella typhimurium, Pseudomonas putida and Bacillus subtilis: evidence for an essential phosphate binding loop. Nucleic Acids Res. 1992;20:839–845. doi: 10.1093/nar/20.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3849.Sands J F, Cummings H S, Sacerdot C, Dondon L, Grunberg-Manago M, Hershey J W B. Cloning and mapping of infA, the gene for protein synthesis initiation factor If1. Nucleic Acids Res. 1987;15:5157–5168. doi: 10.1093/nar/15.13.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3850.Sands J F, Regnier P, Cummings H S, Grunberg-Manago M, Hershey J W. The existence of two genes between infB and rpsO in the Escherichia coli genome: DNA sequencing and S1 nuclease mapping. Nucleic Acids Res. 1988;16:10803–10816. doi: 10.1093/nar/16.22.10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3851.Sanfacon H, Levasseur S, Roy P H, LaPointe J. Cloning of the gene for Escherichia coli glutamyl-tRNA synthetase. Gene. 1983;22:175–180. doi: 10.1016/0378-1119(83)90101-4. [DOI] [PubMed] [Google Scholar]
- 3852.Sankar P, Lee J H, Shanmugam K T. Cloning of hydrogenase genes and the fine structure of an operon essential for H2 metabolism in Escherichia coli. J Bacteriol. 1985;162:353–360. doi: 10.1128/jb.162.1.353-360.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3853.Sankar P, Lee J H, Shanmugam K T. Gene-product relationships of fhlA and fdv genes of Escherichia coli. J Bacteriol. 1988;170:5440–5445. doi: 10.1128/jb.170.12.5440-5445.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3854.Sankar P, Shanmugam K T. Biochemical and genetic analysis of hydrogen metabolism in Escherichia coli: the hydB gene. J Bacteriol. 1988;170:5433–5439. doi: 10.1128/jb.170.12.5433-5439.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3855.Sankar P, Shanmugam K T. Hydrogen metabolism in Escherichia coli: biochemical and genetic evidence for a hydF gene. J Bacteriol. 1988;170:5446–5451. doi: 10.1128/jb.170.12.5446-5451.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3856.Sanna M G, Simon M I. In vivo and in vitro characterization of Escherichia coli protein CheZ gain- and loss-of-function mutants. J Bacteriol. 1996;178:6275–6280. doi: 10.1128/jb.178.21.6275-6280.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3857.Sanna M G, Simon M I. Isolation and in vitro characterization of CheZ suppressors for the Escherichia coli chemotactic response regulator mutant CheYN23D. J Biol Chem. 1996;271:7357–7361. doi: 10.1074/jbc.271.13.7357. [DOI] [PubMed] [Google Scholar]
- 3858.Saporito S M, Smith-White B J, Cunningham R P. Nucleotide sequence of the xth gene of Escherichia coli K-12. J Bacteriol. 1988;170:4542–4547. doi: 10.1128/jb.170.10.4542-4547.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3859.Saporito S M, Cunningham R P. Nucleotide sequence of the nfo gene of Escherichia coli K-12. J Bacteriol. 1988;170:5141–5145. doi: 10.1128/jb.170.11.5141-5145.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3860.Saraste M, Gay N J, Eberle A N, Runswick M J, Walker J R. The atp operon: nucleotide sequence of the genes for the gamma, beta, and epsilon subunits of Escherichia coli ATP synthase. Nucleic Acids Res. 1981;9:5287–5296. doi: 10.1093/nar/9.20.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3861.Sargentini N J, Smith K C. Characterization of an Escherichia coli mutant (radB101) sensitive to gamma and uv radiation, and methyl methanesulfonate. Radiat Res. 1983;93:461–478. [PubMed] [Google Scholar]
- 3862.Sargentini N J, Smith K C. Genetic and phenotypic analyses indicating occurrence of the recN262 and radB101 mutations at the same locus in Escherichia coli. J Bacteriol. 1988;170:2392–2394. doi: 10.1128/jb.170.5.2392-2394.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3863.Saroja G N, Gowrishankar J. Role of SpoT and FNR in NH4+ assimilation and osmoregulation in GOGAT (glutamate synthase)-deficient mutants of Escherichia coli. J Bacteriol. 1996;178:4105–4114. doi: 10.1128/jb.178.14.4105-4114.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3864.Sarsero J P, Wookey P J, Pittard A J. Regulation of expression of the Escherichia coli K-12 mtr gene by TyrR protein and Trp repressor. J Bacteriol. 1991;173:4133–4143. doi: 10.1128/jb.173.13.4133-4143.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3865.Sarsero J P, Wookey P J, Gollnick P, Yanofsky C, Pittard A J. New family of integral membrane proteins involved in transport of aromatic amino acids in Escherichia coli. J Bacteriol. 1991;173:3231–3234. doi: 10.1128/jb.173.10.3231-3234.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3866.Sarthy A, Michaelis S, Beckwith J R. Deletion map of the Escherichia coli structural gene for alkaline phosphatase, phoA. J Bacteriol. 1981;145:288–292. doi: 10.1128/jb.145.1.288-292.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3867.Sarthy A, Michaelis S, Beckwith J R. Use of gene fusions to determine the orientation of gene phoA on the Escherichia coli chromosome. J Bacteriol. 1981;145:293–298. doi: 10.1128/jb.145.1.293-298.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3868.Sarubbi E, Rudd K E, Xiao H, Ikehara K, Kalman M, Cashel M. Characterization of the spoT gene of Escherichia coli. J Biol Chem. 1989;264:15074–15082. [PubMed] [Google Scholar]
- 3869.Sarubbi E, Rudd K E, Cashel M. Basal ppGpp level adjustments shown by new spoT mutants affect steady state growth rates and rrnA ribosomal promoter regulation in Escherichia coli. Mol Gen Genet. 1988;213:214–222. doi: 10.1007/BF00339584. [DOI] [PubMed] [Google Scholar]
- 3870.Sasaki M, Fujiyoshi T, Shimada K, Takagi Y. Fine structure of the recB and recC gene region of Escherichia coli. Biochem Biophys Res Commun. 1982;109:414–422. doi: 10.1016/0006-291x(82)91737-5. [DOI] [PubMed] [Google Scholar]
- 3871.Sasarman A, Nepveu A, Echelard Y, Dymetryszyn J, Drolet M, Goyer C. Molecular cloning and sequencing of the hemD gene of Escherichia coli K-12 and preliminary data on the Uro operon. J Bacteriol. 1987;169:4257–4262. doi: 10.1128/jb.169.9.4257-4262.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3872.Sasarman A, Letowski J, Czaika G, Ramirez V, Nead M S, Jacobs J M, Morais R. Nucleotide sequence of the hemG gene involved in the protoporphyrinogen oxidase activity of Escherichia coli K12. Can J Microbiol. 1993;39:1155–1161. doi: 10.1139/m93-174. [DOI] [PubMed] [Google Scholar]
- 3873.Sasarman A, Chartrand P, Lavoie M, Tardif D, Proschek R, Lapointe C. Mapping of a new hem gene in Escherichia coli K12. J Gen Microbiol. 1979;113:297–303. doi: 10.1099/00221287-113-2-297. [DOI] [PubMed] [Google Scholar]
- 3874.Sasarman A, Chartrand P, Proschek R, Desrochers M, Tardif D, Lapointe C. Uroporphyrin-accumulating mutant of Escherichia coli K-12. J Bacteriol. 1975;124:1205–1212. doi: 10.1128/jb.124.3.1205-1212.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3875.Sasarman A, Echelard Y, Letowski J, Tardif D, Drolet M. Nucleotide sequence of the hemX gene, the third member of the Uro operon of Escherichia coli K12. Nucleic Acids Res. 1988;16:11835. doi: 10.1093/nar/16.24.11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3876.Sastry S S, Jayaraman R. Nitrofurantoin-resistant mutants of Escherichia coli: isolation and mapping. Mol Gen Genet. 1984;196:379–380. doi: 10.1007/BF00328076. [DOI] [PubMed] [Google Scholar]
- 3877.Satishchandran C, Markham G D, Moore R C, Boyle S M. Locations of the speA, speB, speC, and metK genes on the physical map of Escherichia coli. J Bacteriol. 1990;172:4748. doi: 10.1128/jb.172.9.4748.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3878.Satishchandran C, Taylor J C, Markham G D. Novel Escherichia coli K-12 mutants impaired in S-adenosylmethionine synthesis. J Bacteriol. 1990;172:4489–4496. doi: 10.1128/jb.172.8.4489-4496.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3879.Satishchandran C, Hickman Y N, Markham G D. Characterization of the phosphorylated enzyme intermediate formed in the adenosine 5′-phosphosulfate kinase reaction. Biochemistry. 1992;31:11684–11688. doi: 10.1021/bi00162a003. [DOI] [PubMed] [Google Scholar]
- 3880.Sato T, Yura T. Regulatory mutations conferring constitutive synthesis of major outer membrane proteins (OmpC and OmpF) in Escherichia coli. J Bacteriol. 1981;145:88–96. doi: 10.1128/jb.145.1.88-96.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3881.Sauer M, Hantke K, Braun V. Ferric-coprogen receptor FhuE of Escherichia coli: processing and sequence common to all TonB-dependent outer membrane receptor proteins. J Bacteriol. 1987;169:2044–2049. doi: 10.1128/jb.169.5.2044-2049.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3882.Sauer M, Hantke K, Braun V. Sequence of the fhuE outer-membrane receptor gene of Escherichia coli K12 and properties of mutants. Mol Microbiol. 1990;4:427–437. doi: 10.1111/j.1365-2958.1990.tb00609.x. [DOI] [PubMed] [Google Scholar]
- 3883.Sauter M, Bohm R, Bock A. Mutational analysis of the operon (hyc) determining hydrogenase 3 formation in Escherichia coli. Mol Microbiol. 1992;6:1523–1532. doi: 10.1111/j.1365-2958.1992.tb00873.x. [DOI] [PubMed] [Google Scholar]
- 3884.Sawers G, Bock A. Anaerobic regulation of pyruvate formate-lyase from Escherichia coli K-12. J Bacteriol. 1988;170:5330–5336. doi: 10.1128/jb.170.11.5330-5336.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3885.Sawers G, Bock A. Novel transcriptional control of the pyruvate formate-lyase gene: upstream regulatory sequences and multiple promoters regulate anaerobic expression. J Bacteriol. 1989;171:2485–2498. doi: 10.1128/jb.171.5.2485-2498.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3886.Sawers G, Suppmann B. Anaerobic induction of pyruvate formate-lyase gene expression is mediated by the ArcA and FNR proteins. J Bacteriol. 1992;174:3474–3478. doi: 10.1128/jb.174.11.3474-3478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3887.Sawers G, Heider J, Zehelein E, Bock A. Expression and operon structure of the sel genes of Escherichia coli and identification of a third selenium-containing formate dehydrogenase isoenzyme. J Bacteriol. 1991;173:4983–4993. doi: 10.1128/jb.173.16.4983-4993.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3888.Saxena P, Walker J R. Expression of argU, the Escherichia coli gene coding for a rare arginine tRNA. J Bacteriol. 1992;174:1956–1964. doi: 10.1128/jb.174.6.1956-1964.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3889.Schaaper R M. Suppressors of Escherichia coli mutT: anitimutators for DNA replication errors. Mutat Res. 1996;350:17–23. doi: 10.1016/0027-5107(95)00086-0. [DOI] [PubMed] [Google Scholar]
- 3890.Schaefler S. Inducible system for the utilization of β-glucosides in Escherichia coli. I. Active transport and utilization of β-glucosides. J Bacteriol. 1967;93:254–263. doi: 10.1128/jb.93.1.254-263.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3891.Schaffer S, Hantke K, Braun V. Nucleotide sequence of the iron regulatory gene fur. Mol Gen Genet. 1985;200:110–113. doi: 10.1007/BF00383321. [DOI] [PubMed] [Google Scholar]
- 3892.Schaller K, Krauel A, Braun V. Temperature-sensitive, colicin M-tolerant mutant of Escherichia coli. J Bacteriol. 1981;147:135–139. doi: 10.1128/jb.147.1.135-139.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3893.Schatz P J, Bieker K L, Ottemann K M, Silhavy T J, Beckwith J R. One of three transmembrane stretches is sufficient for the functioning of the SecE protein, a membrane component of the E. coli secretion machinery. EMBO J. 1991;10:1749–1757. doi: 10.1002/j.1460-2075.1991.tb07699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3894.Schatz P J, Riggs P D, Jacq A, Fath M J, Beckwith J R. The secE gene encodes an integral membrane protein required for protein export in Escherichia coli. Genes Dev. 1989;3:1035–1044. doi: 10.1101/gad.3.7.1035. [DOI] [PubMed] [Google Scholar]
- 3895.Schaumberg T H, Kuempel P. Genetic mapping of the minB locus in Escherichia coli K-12. J Bacteriol. 1983;153:1063–1065. doi: 10.1128/jb.153.2.1063-1065.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3896.Schellenberg G D, Sarthy A, Larson A E, Backer M P, Crabb J W, Lidstrom M, Hall B D, Furlong C E. Xylose isomerase from Escherichia coli. Characterization of the protein and the structural gene. J Biol Chem. 1984;259:6826–6832. [PubMed] [Google Scholar]
- 3897.Schellenberg G D, Furlong C E. Resolution of the multiplicity of the glutamate and aspartate transport systems of Escherichia coli. J Biol Chem. 1977;252:9055–9064. [PubMed] [Google Scholar]
- 3898.Schellhorn H E. Regulation of hydroperoxidase (catalase) expression in Escherichia coli. FEMS Microbiol Lett. 1995;131:113–119. doi: 10.1111/j.1574-6968.1995.tb07764.x. [DOI] [PubMed] [Google Scholar]
- 3899.Schellhorn H E, Stones V L. Regulation of katF and katE in Escherichia coli K-12 by weak acids. J Bacteriol. 1992;174:4769–4776. doi: 10.1128/jb.174.14.4769-4776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3900.Schemidt R A, Hsu D K, Deckers-Hebestreit G, Altendorf K, Brusilow W S. The effects of an atpE ribosome-binding site mutation on the stoichiometry of the c subunit in the F1F0 ATPase of Escherichia coli. Arch Biochem Biophys. 1995;323:423–428. doi: 10.1006/abbi.1995.0063. [DOI] [PubMed] [Google Scholar]
- 3901.Schendel F J, Mueller E, Stubbe J, Shiau A, Smith J M. Formylglycinamide ribonucleotide synthetase from Escherichia coli: cloning, sequencing, overproduction, isolation, and characterization. Biochemistry. 1989;28:2459–2471. doi: 10.1021/bi00432a017. [DOI] [PubMed] [Google Scholar]
- 3902.Scherrer R, Moyed H S. Conditional impairment of cell division and altered lethality in hipA mutants of Escherichia coli K-12. J Bacteriol. 1988;170:3321–3326. doi: 10.1128/jb.170.8.3321-3326.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3903.Scheurmann R, Tam S, Burgers P M J, Lu C, Echols H. Identification of the E-subunit of Escherichia coli DNA polymerase III holoenzyme as the dnaQ gene product: a fidelity subunit for DNA replication. Proc Natl Acad Sci USA. 1983;80:7085–7089. doi: 10.1073/pnas.80.23.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3904.Schinina M E, Maffey L, Barra D, Bossa F, Puget K, Michelson A M. The primary structure of iron superoxide dismutase from Escherichia coli. FEBS Lett. 1987;221:87–90. doi: 10.1016/0014-5793(87)80357-5. [DOI] [PubMed] [Google Scholar]
- 3905.Schlaman H R, Risseeuw E, Franke-van Dijk M E, Hooykaas P J. Nucleotide sequence corrections of the uidA open reading frame encoding beta-glucuronidase. Gene. 1994;138:259–260. doi: 10.1016/0378-1119(94)90820-6. [DOI] [PubMed] [Google Scholar]
- 3906.Schleif R. Isolation and characterization of a streptolydigin resistant RNA polymerase. Nature. 1969;223:1068–1069. doi: 10.1038/2231068a0. [DOI] [PubMed] [Google Scholar]
- 3907.Schlensog V, Bock A. Identification and sequence analysis of the gene encoding the transcriptional activator of the formate hydrogenlyase system of Escherichia coli. Mol Microbiol. 1990;4:1319–1327. doi: 10.1111/j.1365-2958.1990.tb00711.x. [DOI] [PubMed] [Google Scholar]
- 3908.Schlensog V, Bock A. The Escherichia coli fdv gene probably encodes mutS and is located at minute 58.8 adjacent to the hyc-hyp gene cluster. J Bacteriol. 1991;173:7414–7415. doi: 10.1128/jb.173.23.7414-7415.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3909.Schlensog V, Lutz S, Bock A. Purification and DNA-binding properties of FHLA, the transcriptional activator of the formate hydrogenlyase system from Escherichia coli. J Biol Chem. 1994;269:19590–19596. [PubMed] [Google Scholar]
- 3910.Schleyer M, Bakker E P. Nucleotide sequence and 3′-end deletion studies indicate that the K+-uptake protein Kup from Escherichia coli is composed of a hydrophobic core linked to a large and partially essential hydrophilic C terminus. J Bacteriol. 1993;175:6925–6931. doi: 10.1128/jb.175.21.6925-6931.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3911.Schlictman D, Shankar S, Chakrabarty A M. The Escherichia coli genes sspA and rnk can functionally replace the Pseudomonas aeruginosa alginate regulatory gene algR2. Mol Microbiol. 1995;16:309–320. doi: 10.1111/j.1365-2958.1995.tb02303.x. [DOI] [PubMed] [Google Scholar]
- 3912.Schlindwein C, Giordano G, Santini C L, Mandrand-Berthelot M A. Identification and expression of the Escherichia coli fdhD and fdhE genes, which are involved in the formation of the respiratory formate dehydrogenase. J Bacteriol. 1990;172:6112–6121. doi: 10.1128/jb.172.10.6112-6121.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3913.Schlindwein C, Mandrand M A. Nucleotide sequence of the fdhE gene involved in respiratory formate dehydrogenase formation in Escherichia coli K-12. Gene. 1991;97:147–148. doi: 10.1016/0378-1119(91)90023-5. [DOI] [PubMed] [Google Scholar]
- 3914.Schlosser A, Hamann A, Bossemeyer D, Schneider H, Bakker E P. NAD+ binding to the Escherichia coli K+-uptake protein TrkA and sequence similarity between TrkA and domains of a family of dehydrogenases suggest a role for NAD+ in bacterial transport. Mol Microbiol. 1993;9:533–543. doi: 10.1111/j.1365-2958.1993.tb01714.x. [DOI] [PubMed] [Google Scholar]
- 3915.Schlosser A, Meldrof M, Stumpe S, Bakker E P, Epstein W. TrkH and its homolog, TrkG, determine the specificity and kinetics of cation transport by the Trk system of Escherichia coli. J Bacteriol. 1995;177:1908–1910. doi: 10.1128/jb.177.7.1908-1910.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3916.Schlosser A, Kluttig S, Hamann A, Bakker E P. Subcloning, nucleotide sequence, and expression of trkG, a gene that encodes an integral membrane protein involved in potassium uptake via the Trk system of Escherichia coli. J Bacteriol. 1991;173:3170–3176. doi: 10.1128/jb.173.10.3170-3176.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3917.Schmellik-Sandage C S, Tessman E S. Signal strains that can detect certain DNA replication and membrane mutants of Escherichia coli: isolation of a new ssb allele, ssb-3. J Bacteriol. 1990;172:4378–4385. doi: 10.1128/jb.172.8.4378-4385.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3918.Schmidt M, Delihas N. micF RNA is a substrate for RNase E. FEMS Microbiol Lett. 1995;133:209–213. doi: 10.1111/j.1574-6968.1995.tb07886.x. [DOI] [PubMed] [Google Scholar]
- 3919.Schmidt M G, Rollo E E, Grodberg J, Oliver D B. Nucleotide sequence of the secA gene and secA(Ts) mutations preventing protein export in Escherichia coli. J Bacteriol. 1988;170:3404–3414. doi: 10.1128/jb.170.8.3404-3414.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3920.Schnaitman C A, Parker C T, Klena J D, Pradel E, Pearson N B, Sanderson K E, MacLachlan P R. Physical maps of the rfa loci of Escherichia coli K-12 and Salmonella typhimurium. J Bacteriol. 1991;173:7410–7411. doi: 10.1128/jb.173.23.7410-7411.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3921.Schnaitman C A, Klena J D. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol Rev. 1993;57:655–682. doi: 10.1128/mr.57.3.655-682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3921a.Schneider B L, Kiupakis A K, Reitzer L J. Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli. J Bacteriol. 1998;180:1269–1297. doi: 10.1128/jb.180.16.4278-4286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3922.Schneider E, Freundlieb S, Tapio S, Boos W. Molecular characterization of the MalT-dependent periplasmic alpha-amylase of Escherichia coli encoded by MalS. J Biol Chem. 1992;267:5148–5154. [PubMed] [Google Scholar]
- 3923.Schneppe B, Deckers-Hebestreit G, McCarthy J E, Altendorf K. Translation of the first gene of the Escherichia coli unc operon. Selection of the start codon and control of initiation efficiency. J Biol Chem. 1991;266:21090–21098. [PubMed] [Google Scholar]
- 3924.Schnetz K, Rak B. Beta-glucoside permease represses the bgl operon of Escherichia coli by phosphorylation of the antiterminator protein and also interacts with glucose-specific enzyme III, the key element in catabolite control. Proc Natl Acad Sci USA. 1990;87:5074–5078. doi: 10.1073/pnas.87.13.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3925.Schnetz K, Toloczyki C, Rak B. β-Glucoside (bgl) operon of Escherichia coli K-12: nucleotide sequence, genetic organization, and possible evolutionary relationship to regulatory components of two Bacillus subtilis genes. J Bacteriol. 1987;169:2579–2590. doi: 10.1128/jb.169.6.2579-2590.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3926.Schnier J, Isono K. The DNA sequence of the gene rpsA of Escherichia coli coding for ribosomal protein S1. Nucleic Acids Res. 1982;10:1857–1865. doi: 10.1093/nar/10.6.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3927.Schnier J, Kimura M, Foulaki K, Subramanian A-R, Isono K, Wittmann-Liebold B. Primary structure of Escherichia coli ribosomal protein S1 and of its gene rpsA. Proc Natl Acad Sci USA. 1982;79:1008–1011. doi: 10.1073/pnas.79.4.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3928.Schnier J, Kitagawa M, Isono K. The nucleotide sequence of an Escherichia coli chromosomal region containing the genes of ribosomal proteins S6, S18, L9 and an open reading frame. Mol Gen Genet. 1986;204:126–132. doi: 10.1007/BF00330199. [DOI] [PubMed] [Google Scholar]
- 3929.Schoemaker J M, Markovitz A. Identification of the gene lon (capR) product as a 94-kilodalton polypeptide by cloning and deletion analysis. J Bacteriol. 1981;147:46–56. doi: 10.1128/jb.147.1.46-56.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3930.Schoenlein P V, Roa B B, Winkler M E. Divergent transcription of pdxB and homology between the pdxB and serA gene products in Escherichia coli K-12. J Bacteriol. 1989;171:6084–6092. doi: 10.1128/jb.171.11.6084-6092.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3931.Schofield M A, Agbunag R, Michaels M L, Miller J H. Cloning and sequencing of Escherichia coli mutR shows its identity to topB, encoding topoisomerase III. J Bacteriol. 1992;174:5168–5170. doi: 10.1128/jb.174.15.5168-5170.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3932.Scholle A, Vreemann J, Blank V, Nold A, Boos W, Manson M D. Sequence of the mglB gene from Escherichia coli K12: comparison of wild-type and mutant galactose chemoreceptors. Mol Gen Genet. 1987;208:247–253. doi: 10.1007/BF00330450. [DOI] [PubMed] [Google Scholar]
- 3933.Schonbrunner E R, Schmid F X. Peptidyl-prolyl cis-trans isomerase improves the efficiency of protein disulfide isomerase as a catalyst of protein folding. Proc Natl Acad Sci USA. 1992;89:4510–4513. doi: 10.1073/pnas.89.10.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3934.Schreiber G, Metzger S, Aizenman E, Roza S, Cashel M, Glaser G. Overexpression of the relA gene in Escherichia coli. J Biol Chem. 1991;266:3760–3767. [PubMed] [Google Scholar]
- 3935.Schroder I, Wolin C D, Cavicchioli R, Gunsalus R P. Phosphorylation and dephosphorylation of the NarQ, NarX, and NarL proteins of the nitrate-dependent two-component regulatory system of Escherichia coli. J Bacteriol. 1994;176:4985–4992. doi: 10.1128/jb.176.16.4985-4992.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3936.Schryvers A, Weiner J H. The anaerobic sn-glycerol-3-phosphate dehydrogenase: cloning and expression of the glpA gene of Escherichia coli and identification of the glpA products. Can J Biochem. 1982;60:224–231. doi: 10.1139/o82-027. [DOI] [PubMed] [Google Scholar]
- 3937.Schultz J E, Matin A. Molecular and functional characterization of a carbon starvation gene of Escherichia coli. J Mol Biol. 1991;218:129–140. doi: 10.1016/0022-2836(91)90879-b. [DOI] [PubMed] [Google Scholar]
- 3938.Schwartz I, Klotsky R A, Elseviers D, Gallagher P, Krauskopf M, Siddiqui M A Q, Wong J F H, Roe B A. Molecular cloning and sequencing of pheU, a gene for Escherichia coli tRNAPhe. Nucleic Acids Res. 1983;11:4379–4389. doi: 10.1093/nar/11.13.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3939.Schwartz J H, Mass W K, Simon E J. An impaired concentrating mechanism for amino acids in mutants of Escherichia coli resistant to l-canavanine and d-serine. Biochim Biophys Acta. 1959;32:582–583. doi: 10.1016/0006-3002(59)90650-x. [DOI] [PubMed] [Google Scholar]
- 3940.Schwartz M. Location of the maltose A and B loci on the genetic map of Escherichia coli. J Bacteriol. 1966;92:1083–1089. doi: 10.1128/jb.92.4.1083-1089.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3941.Schwartz M, Roa M, Debarbouille M. Mutations that affect lamB gene expression at a posttranscriptional level. Proc Natl Acad Sci USA. 1981;78:2937–2941. doi: 10.1073/pnas.78.5.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3942.Schweizer H P, Sweet G, Larson T J. Physical and genetic structure of the glpD-malT interval of the Escherichia coli chromosome. Mol Gen Genet. 1986;202:488–492. doi: 10.1007/BF00333282. [DOI] [PubMed] [Google Scholar]
- 3943.Schweizer H P, Datta P. Identification and DNA sequence of tdcR, a positive regulatory gene of the tdc operon of Escherichia coli. Mol Gen Genet. 1989;218:516–522. doi: 10.1007/BF00332418. [DOI] [PubMed] [Google Scholar]
- 3944.Schweizer H P, Datta P. The complete nucleotide sequence of the tdc region of Escherichia coli. Nucleic Acids Res. 1989;17:3994. doi: 10.1093/nar/17.10.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3944a.Schweizer H P, Datta P. Physical map location of the tdc operon of Escherichia coli. J Bacteriol. 1990;172:2825. doi: 10.1128/jb.172.6.2825.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3945.Schweizer H P, Datta P. Physical linkage and transcriptional orientation of the tdc operon on the Escherichia coli chromosome. Mol Gen Genet. 1991;228:125–128. doi: 10.1007/BF00282456. [DOI] [PubMed] [Google Scholar]
- 3946.Schweizer H P, Grussenmeyer T, Boos W. Mapping of two ugp genes coding for the pho regulon-dependent sn-glycerol-3-phosphate transport system of Escherichia coli. J Bacteriol. 1982;150:1164–1171. doi: 10.1128/jb.150.3.1164-1171.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3947.Schweizer H P, Larson T J. Cloning and characterization of the aerobic sn-glycerol-3-phosphate dehydrogenase structural gene glpD of Escherichia coli K-12. J Bacteriol. 1987;169:507–513. doi: 10.1128/jb.169.2.507-513.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3948.Schweizer H P, Boos W. Cloning of the ugp region containing the structural genes for the pho regulon-dependent sn-glycerol-3-phosphate transport system of Escherichia coli. Mol Gen Genet. 1983;192:177–186. doi: 10.1007/BF00327664. [DOI] [PubMed] [Google Scholar]
- 3949.Schweizer H P, Boos W. Characterization of the ugp region containing the genes for the phoB dependent sn-glycerol-3-phosphate transport system of Escherichia coli. Mol Gen Genet. 1984;197:161–168. doi: 10.1007/BF00327937. [DOI] [PubMed] [Google Scholar]
- 3950.Schweizer H P, Boos W, Larson T J. Repressor for the sn-glycerol-3-phosphate regulon of Escherichia coli K-12: cloning of the glpR gene and identification of its product. J Bacteriol. 1985;161:563–566. doi: 10.1128/jb.161.2.563-566.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3951.Sclafani R A, Wechsler J A. High frequency of genetic duplications in the dnaB region of the Escherichia coli K12 chromosome. Genetics. 1981;98:677–689. doi: 10.1093/genetics/98.4.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3952.Scofield M A, Lewis W S, Schuster S M. Nucleotide sequence of Escherichia coli asnB and deduced amino acid sequence of asparagine synthetase B. J Biol Chem. 1990;265:12895–12902. [PubMed] [Google Scholar]
- 3953.Scripture J B, Voelker C, Miller S, O’Donnell R T, Polgar L, Rade J, Horazdovsky B F, Hogg R H. High-affinity l-arabinose transport operon. Nucleotide sequence and analysis of gene products. J Mol Biol. 1987;197:37–46. doi: 10.1016/0022-2836(87)90607-3. [DOI] [PubMed] [Google Scholar]
- 3954.Scripture J B, Hogg R W. The nucleotide sequences defining the signal peptides of the galactose-binding protein and the arabinose-binding protein. J Biol Chem. 1983;258:10853–10855. [PubMed] [Google Scholar]
- 3955.Searles L L, Jones J W, Fournier M J, Grambow N, Tyler B, Calvo J M. Escherichia coli B/r leuK mutant lacking pseudouridine synthase I activity. J Bacteriol. 1986;166:341–345. doi: 10.1128/jb.166.1.341-345.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3956.Seaton B L, Vickery L E. A gene encoding a DnaK/hsp70 homolog in Escherichia coli. Proc Natl Acad Sci USA. 1994;91:2066–2070. doi: 10.1073/pnas.91.6.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3957.Sedgwick B. Genetic mapping of the ada and adc mutations affecting the adaptive response of Escherichia coli to alkylating agents. J Bacteriol. 1982;150:984–988. doi: 10.1128/jb.150.2.984-988.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3958.Sedgwick B. Molecular cloning of a gene which regulates the adaptive response to alkylating agents in Escherichia coli. Mol Gen Genet. 1983;191:466–472. doi: 10.1007/BF00425764. [DOI] [PubMed] [Google Scholar]
- 3959.Sedgwick B, Robins P. Isolation of mutants of Escherichia coli with increased resistance to alkylating agents: mutants deficient in thiols and mutants constitutive for the adaptive response. Mol Gen Genet. 1980;180:85–90. doi: 10.1007/BF00267355. [DOI] [PubMed] [Google Scholar]
- 3960.Sedgwick S G, Lodwick D, Doyle N, Crowne H, Strike P. Functional complementation between chromosomal and plasmid mutagenic DNA repair genes in bacteria. Mol Gen Genet. 1991;229:428–436. doi: 10.1007/BF00267466. [DOI] [PubMed] [Google Scholar]
- 3961.Sedivy J M, Daldal F, Fraenkel D. Fructose bisphosphatase of Escherichia coli: cloning of the structural gene and preparation of a chromosomal deletion. J Bacteriol. 1984;158:1048–1053. doi: 10.1128/jb.158.3.1048-1053.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3962.Seeger C, Poulsen C, Dandanell G. Identification and characterization of genes (xapA, xapB, and xapR) involved in xanthosine catabolism in Escherichia coli. J Bacteriol. 1995;177:5506–5516. doi: 10.1128/jb.177.19.5506-5516.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3963.Seifert J, Kunz N, Flachmann R, Laufer A, Jany K D, Gassen H G. Expression of the E. coli nadB gene and characterization of the gene product l-aspartate oxidase. Biol Chem Hoppe-Seyler. 1990;371:239–248. [PubMed] [Google Scholar]
- 3964.Selby C P, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 3965.Selby C P, Sancar A. Transcription-repair coupling and mutation frequency decline. J Bacteriol. 1993;175:7509–7514. doi: 10.1128/jb.175.23.7509-7514.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3966.Selby C P, Sancar A. Mechanisms of transcription-repair coupling and mutation frequency decline. Microbiol Rev. 1994;58:317–329. doi: 10.1128/mr.58.3.317-329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3967.Selby C P, Sancar A. Structure and function of transcription-repair coupling factor. I. Structural domains and binding properties. J Biol Chem. 1995;270:4882–4889. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- 3968.Selby C P, Sancar A. Structure and function of transcription-repair coupling factor. II. Catalytic properties. J Biol Chem. 1995;270:4890–4895. doi: 10.1074/jbc.270.9.4890. [DOI] [PubMed] [Google Scholar]
- 3969.Selby C P, Witkin E M, Sancar A. Escherichia coli mfd mutant deficient in “mutation frequency decline” lacks strand-specific repair: in vitro complementation with purified coupling factor. Proc Natl Acad Sci USA. 1991;88:11574–11578. doi: 10.1073/pnas.88.24.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3970.Seoane A S, Sabbaj A, McMurry L M, Levy S B. Multiple antibiotic susceptibility associated with inactivation of the prc gene. J Bacteriol. 1992;174:7844–7847. doi: 10.1128/jb.174.23.7844-7847.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3971.Seoane A S, Levy S B. Identification of new genes regulated by the marRAB operon in Escherichia coli. J Bacteriol. 1995;177:530–535. doi: 10.1128/jb.177.3.530-535.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3972.Seol J H, Kwon J A, Yoo S J, Kim H S, Kang M S, Chung C H. Site-directed mutagenesis of the Cys residues in ClpA, the ATPase component of protease Ti (ClpAP) in Escherichia coli. Biol Chem. 1997;378:1205–1209. [PubMed] [Google Scholar]
- 3973.Seol J H, Woo K M, Kang M S, Ha D B, Chung C H. Requirement of ATP hydrolysis for assembly of ClpA/ClpP complex, the ATP-dependent protease T1 in Escherichia coli. Biochem Biophys Res Commun. 1995;217:41–51. doi: 10.1006/bbrc.1995.2743. [DOI] [PubMed] [Google Scholar]
- 3974.Seol J H, Woo S K, Jung E M, Yoo S J, Lee C S, Kim K J, Tanaka K, Ichihara A, Ha D B, Chung C H. Protease Do is essential for survival of Escherichia coli at high temperatures: its identity with the htrA gene product. Biochem Biophys Res Commun. 1991;176:730–736. doi: 10.1016/s0006-291x(05)80245-1. [DOI] [PubMed] [Google Scholar]
- 3975.Seol W, Shatkin A J. A new gene located between pss and rrnG on the Escherichia coli chromosome. J Bacteriol. 1990;172:4745. doi: 10.1128/jb.172.9.4745.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3976.Seol W, Shatkin A J. Sequence of the distal end of E. coli ribosomal RNA rrnG operon. Nucleic Acids Res. 1990;18:3056. doi: 10.1093/nar/18.10.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3977.Seol W, Shatkin A J. Escherichia coli kgtP encodes an alpha-ketoglutarate transporter. Proc Natl Acad Sci USA. 1991;88:3802–3806. doi: 10.1073/pnas.88.9.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3978.Seol W, Shatkin A J. Membrane topology model of Escherichia coli αketoglutarate permease by PhoA fusion analysis. J Bacteriol. 1993;175:565–567. doi: 10.1128/jb.175.2.565-567.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3979.Serizawa H, Fukuda R. Structure of the gene for the stringent starvation protein of Escherichia coli. Nucleic Acids Res. 1987;15:1153–1163. doi: 10.1093/nar/15.3.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3980.Seta F D, Boschi-Muller S, Vignais M L, Branlant G. Characterization of Escherichia coli strains with gapA and gapB genes deleted. J Bacteriol. 1997;179:5218–5221. doi: 10.1128/jb.179.16.5218-5221.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3981.Seufert W, Messer W. Start sites for bidirectional in vitro DNA replication inside the replication origin, oriC, of Escherichia coli. EMBO J. 1987;6:2469–2472. doi: 10.1002/j.1460-2075.1987.tb02527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3982.Sevastopoulis C G, Wehr C T, Glaser D A. Large-scale automated isolation of Escherichia coli mutants with thermosensitive DNA replication. Proc Natl Acad Sci USA. 1977;74:3485–3489. doi: 10.1073/pnas.74.8.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3983.Sever I S, Kalyaeva E S, Danilevskaya O N. Mutations slowing down degradation of the BB′ subunits of Escherichia coli RNA polymerase. Sov Genet (Engl Transl Genetika) 1982;18:965–971. [Google Scholar]
- 3983a.Shafritz K M, Sandigursky M, Franklin W A. Exonuclease IX of Escherichia coli. Nucleic Acids Res. 1998;26:2593–2597. doi: 10.1093/nar/26.11.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3984.Shaibe E, Metzer E, Halpern Y S. Metabolic pathway for the utilization of l-arginine, l-ornithine, agmatine, and putrescine as nitrogen sources in Escherichia coli. J Bacteriol. 1985;163:933–937. doi: 10.1128/jb.163.3.933-937.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3985.Shankar S, Schlictman D, Chakrabarty A M. Regulation of nucleoside diphosphate kinase and an alternative kinase in Escherichia coli: role of the sspA and rnk genes in nucleoside triphosphate formation. Mol Microbiol. 1995;17:935–943. doi: 10.1111/j.1365-2958.1995.mmi_17050935.x. [DOI] [PubMed] [Google Scholar]
- 3985a.Shanley, M. S., et al. GenBank submission U17902.
- 3986.Shanmugam K T, Stewart V J, Gunsalus R P, Boxer D H, Cole J A, Chippaux M, DeMoss J A, Giordano G, Lin E C C, Rajagopalan K V. Proposed nomenclature for the genes involved in molybdenum metabolism in Escherichia coli and Salmonella typhimurium. Mol Microbiol. 1992;6:3452–3454. doi: 10.1111/j.1365-2958.1992.tb02215.x. [DOI] [PubMed] [Google Scholar]
- 3987.Shao Z, Newman E B. Sequencing and characterization of the sdaB gene from Escherichia coli K-12. Eur J Biochem. 1993;212:777–784. doi: 10.1111/j.1432-1033.1993.tb17718.x. [DOI] [PubMed] [Google Scholar]
- 3988.Shao Z, Lin R T, Newman E B. Sequencing and characterization of the sdaC gene and identification of the sdaCB operon in Escherichia coli K12. Eur J Biochem. 1994;222:901–907. doi: 10.1111/j.1432-1033.1994.tb18938.x. [DOI] [PubMed] [Google Scholar]
- 3989.Shapiro J A. Chromosomal location of the gene determining uridine diphosphoglucose formation in Escherichia coli K-12. J Bacteriol. 1966;92:518–520. doi: 10.1128/jb.92.2.518-520.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3990.Shapiro J A. A role for the Clp protease in activating Mu-mediated DNA rearrangements. J Bacteriol. 1993;175:2625–2626. doi: 10.1128/jb.175.9.2625-2631.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3991.Shapiro N, Keasling J D. The recA gene and cadmium toxicity in Escherichia coli K12. Microbios. 1996;86:23–26. [PubMed] [Google Scholar]
- 3992.Sharma B, Hill T M. TerF, the sixth identified replication arrest site in Escherichia coli, is located within the rcsC gene. J Bacteriol. 1992;174:7854–7858. doi: 10.1128/jb.174.23.7854-7858.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3993.Sharma R C, Sargentini N J, Smith K C. New mutation (mmrA1) in Escherichia coli K-12 that affects minimal medium recovery and postreplication repair after UV irradiation. J Bacteriol. 1983;154:743–747. doi: 10.1128/jb.154.2.743-747.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3994.Sharma R C, Dowhan W, Moses R E. Molecular structure of the uvrC gene of Escherichia coli: identification of DNA sequences required for transcription of the uvrC gene. Nucleic Acids Res. 1982;10:5209–5221. doi: 10.1093/nar/10.17.5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3995.Sharma S, Ohta A, Dowhan W, Moses R E. Cloning of the uvrC gene of Escherichia coli: expression of a DNA repair gene. Proc Natl Acad Sci USA. 1981;78:6033–6037. doi: 10.1073/pnas.78.10.6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3996.Sharma S, Stark T F, Beattie W G, Moses R E. Multiple control elements for the uvrC gene unit of Escherichia coli. Nucleic Acids Res. 1986;14:2301–2318. doi: 10.1093/nar/14.5.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3997.Sharma V, Suvarna K, Meganathan R, Hudspeth M E. Menaquinone (vitamin K2) biosynthesis: nucleotide sequence and expression of the menB gene from Escherichia coli. J Bacteriol. 1992;174:5057–5062. doi: 10.1128/jb.174.15.5057-5062.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3998.Sharma V, Hudspeth M E, Meganathan R. Menaquinone (vitamin K2) biosynthesis: localization and characterization of the menE gene from Escherichia coli. Gene. 1996;168:43–48. doi: 10.1016/0378-1119(95)00721-0. [DOI] [PubMed] [Google Scholar]
- 3999.Sharples G J, Benson F E, Illing G T, Lloyd R G. Molecular and functional analysis of the ruv region of Escherichia coli K-12 reveals three genes involved in DNA repair and recombination. Mol Gen Genet. 1990;221:219–226. doi: 10.1007/BF00261724. [DOI] [PubMed] [Google Scholar]
- 4000.Sharples G J, Lloyd R G. Location of a mutation in the aspartyl-tRNA synthetase gene of Escherichia coli K12. Mutat Res. 1991;264:93–96. doi: 10.1016/0165-7992(91)90122-k. [DOI] [PubMed] [Google Scholar]
- 4001.Sharples G J, Lloyd R G. Resolution of Holliday junctions in Escherichia coli: identification of the ruvC gene product as a 19-kilodalton protein. J Bacteriol. 1991;173:7711–7715. doi: 10.1128/jb.173.23.7711-7715.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4002.Sharples G J, Lloyd R G. An E. coli RuvC mutant defective in cleavage of synthetic Holliday junctions. Nucleic Acids Res. 1993;21:3359–3364. doi: 10.1093/nar/21.15.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4003.Sharples G J, Chan S N, Mahdi A A, Whitby M C, Lloyd R G. Processing of intermediates in recombination and DNA repair identification of a new endonuclease that specifically cleaves Holliday junctions. EMBO J. 1994;13:6133–6142. doi: 10.1002/j.1460-2075.1994.tb06960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4004.Sharrock R A, Gourse R L, Nomura M. Defective antitermination of rRNA transcription and derepression of rRNA and tRNA synthesis in the nusB5 mutant of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:5275–5279. doi: 10.1073/pnas.82.16.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4005.Shattuck-Eidens D M, Kadner R J. Exogenous induction of the Escherichia coli hexose phosphate transport systems defined by uhp-lac operon fusions. J Bacteriol. 1981;148:203–209. doi: 10.1128/jb.148.1.203-209.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4006.Shattuck-Eidens D M, Kadner R J. Molecular cloning of the uhp region and evidence for a positive activator for expression of the hexose phosphate transport system of Escherichia coli. J Bacteriol. 1983;155:1062–1070. doi: 10.1128/jb.155.3.1062-1070.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4007.Shaw D J, Robinson E C, Meganathan R, Bentley R, Guest J R. Recombinant plasmids containing menaquinone biosynthetic genes of Escherichia coli. FEMS Microbiol Lett. 1983;17:63–67. [Google Scholar]
- 4008.Shaw D J, Guest J R. Molecular cloning of the fnr gene of Escherichia coli K-12. Mol Gen Genet. 1981;181:95–100. doi: 10.1007/BF00339011. [DOI] [PubMed] [Google Scholar]
- 4009.Shaw D J, Guest J R. Amplification and product identification of the fnr gene of Escherichia coli. J Gen Microbiol. 1982;128:2221–2228. doi: 10.1099/00221287-128-10-2221. [DOI] [PubMed] [Google Scholar]
- 4010.Shaw D J, Guest J R. Nucleotide sequence of the fnr gene and primary structure of the Fnr region of Escherichia coli. Nucleic Acids Res. 1982;10:6119–6130. doi: 10.1093/nar/10.19.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4011.Shaw D J, Guest J R, Meganathan R, Bentley R. Characterization of Escherichia coli men mutants defective in conversion of o-succinylbenzoate to 1,4-dihydroxy-2-naphthoate. J Bacteriol. 1982;152:1132–1137. doi: 10.1128/jb.152.3.1132-1137.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4012.Shaw L, Grau F, Kaback H R, Hong J-S, Walsh C T. Vinylglycolate resistance in Escherichia coli. J Bacteriol. 1975;121:1047–1055. doi: 10.1128/jb.121.3.1047-1055.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4013.Shea C M, McIntosh M A. Nucleotide sequence and genetic organization of the ferric enterobactin transport system: homology to other periplasmic binding protein-dependent systems in Escherichia coli. Mol Microbiol. 1991;5:1415–1428. doi: 10.1111/j.1365-2958.1991.tb00788.x. [DOI] [PubMed] [Google Scholar]
- 4014.Sheldon R, Brenner S. Regulatory mutants of dihydrofolate reductase in Escherichia coli K12. Mol Gen Genet. 1976;147:91–97. doi: 10.1007/BF00337941. [DOI] [PubMed] [Google Scholar]
- 4015.Shen W-F, Squires C, Squires C L. Nucleotide sequence of the rrnG ribosomal RNA promoter region of Escherichia coli. Nucleic Acids Res. 1982;10:3303–3313. doi: 10.1093/nar/10.10.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4016.Shepard D, Oberfelder R W, Welch M M, McHenry C S. Determination of the precise location and orientation of the Escherichia coli dnaE gene. J Bacteriol. 1984;158:455–459. doi: 10.1128/jb.158.2.455-459.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4017.Shepley D P, Little J W. Mutant LexA proteins with specific defects in autodigestion. Proc Natl Acad Sci USA. 1996;93:11528–11533. doi: 10.1073/pnas.93.21.11528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4018.Sheppard D E. Dominance relationships among mutant alleles of regulatory gene araC in the Escherichia coli B/R l-arabinose operon. J Bacteriol. 1986;168:999–1001. doi: 10.1128/jb.168.2.999-1001.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4019.Shevchik V E, Condemine G, Robert-Baudouy J M. Characterization of DsbC, a periplasmic protein of Erwinia chrysanthemi and Escherichia coli with disulfide isomerase activity. EMBO J. 1994;13:2007–2012. doi: 10.1002/j.1460-2075.1994.tb06470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4020.Shi W, Bogdanov M, Dowhan W, Zusman D R. The pss and psd genes are required for motility and chemotaxis in Escherichia coli. J Bacteriol. 1993;175:7711–7714. doi: 10.1128/jb.175.23.7711-7714.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4021.Shi W, Zhou Y, Wild J, Adler J, Gross C A. DnaK, DnaJ, and GrpE are required for flagellum synthesis in Escherichia coli. J Bacteriol. 1992;174:6256–6263. doi: 10.1128/jb.174.19.6256-6263.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4022.Shi X, Waasdorp B C, Bennett G N. Modulation of acid-induced amino acid decarboxylase gene expression by hns in Escherichia coli. J Bacteriol. 1993;175:1182–1186. doi: 10.1128/jb.175.4.1182-1186.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4023.Shi X, Bennett G N. Effects of rpoA and cysB mutations on acid induction of biodegradative arginine decarboxylase in Escherichia coli. J Bacteriol. 1994;176:7017–7023. doi: 10.1128/jb.176.22.7017-7023.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4024.Shi X, Bennett G N. Plasmids bearing hfq and the hns-like gene stpA complement hns mutants in modulating arginine decarboxylase gene expression in Escherichia coli. J Bacteriol. 1994;176:6769–6775. doi: 10.1128/jb.176.21.6769-6775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4025.Shi X, Bennett G N. Effects of multicopy LeuO on the expression of the acid-inducible lysine decarboxylase gene in Escherichia coli. J Bacteriol. 1995;177:810–814. doi: 10.1128/jb.177.3.810-814.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4026.Shiba K, Ito K, Yura T. Mutation that suppresses the protein export defect of the secY mutation and causes cold-sensitive growth of Escherichia coli. J Bacteriol. 1984;160:696–701. doi: 10.1128/jb.160.2.696-701.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4027.Shiba K, Ito K, Yura T. Suppressors of the secY24 mutation: identification and characterization of additional ssy genes in Escherichia coli. J Bacteriol. 1986;166:849–856. doi: 10.1128/jb.166.3.849-856.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4028.Shiba K, Ito K, Yura T, Cerretti D P. A defined mutation in the protein export gene within the spc ribosomal protein operon of Escherichia coli: isolation and characterization of a new temperature-sensitive secY mutant. EMBO J. 1984;3:631–635. doi: 10.1002/j.1460-2075.1984.tb01859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4029.Shiba K, Ito K, Nakamura Y, Dondon J, Grunberg-Manago M. Altered translation initiation factor 2 in cold-sensitive ssyG mutant affects protein export in Escherichia coli. EMBO J. 1986;5:3001–3006. doi: 10.1002/j.1460-2075.1986.tb04598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4030.Shida T, Noda M, Sekiguchi J. The recognition of DNA containing an AP site by E. coli endonuclease VI (exonuclease III) Nucleic Acids Symp Ser. 1995;24:87–88. [PubMed] [Google Scholar]
- 4031.Shimamoto T, Yazyu H, Futai M, Tsuchiya T. Nucleotide sequence of the promoter region of the melibiose operon of Escherichia coli. Biochem Biophys Res Commun. 1984;121:41–46. doi: 10.1016/0006-291x(84)90685-5. [DOI] [PubMed] [Google Scholar]
- 4032.Shimizu H, Nishiyama K, Tokuda H. Expression of gpsA encoding biosynthetic sn-glycerol 3-phosphate dehydrogenase suppresses both the LB− phenotype of a secB null mutant and the cold-sensitive phenotype of a secG null mutant. Mol Microbiol. 1997;26:1013–1021. doi: 10.1046/j.1365-2958.1997.6392003.x. [DOI] [PubMed] [Google Scholar]
- 4033.Shimizu I, Kaji A. Identification of the promoter region of the ribosome-releasing factor cistron (ffr) J Bacteriol. 1991;173:5181–5187. doi: 10.1128/jb.173.16.5181-5187.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4034.Shimizu S, Dempsey W B. Genetic map position of the pdxlt gene in Escherichia coli. J Bacteriol. 1976;127:1593–1594. doi: 10.1128/jb.127.3.1593-1594.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4034a.Shimizu S, Dempsey W B. 3-Hydroxypyruvate substitutes for pyridoxine mutants of Escherichia coli K-12. J Bacteriol. 1978;134:944–949. doi: 10.1128/jb.134.3.944-949.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4035.Shimmin L C, Vanderwel D, Harkness R E, Currie B R, Galloway A, Ishiguro E E. Temperature-sensitive β-lactam-tolerant mutants of Escherichia coli. J Gen Microbiol. 1984;130:1315–1323. doi: 10.1099/00221287-130-6-1315. [DOI] [PubMed] [Google Scholar]
- 4036.Shimoike T, Taura T, Kihara A, Yoshihisa T, Akiyama Y, Cannon K, Ito K. Product of a new gene, syd, functionally interacts with SecY when overproduced in Escherichia coli. J Biol Chem. 1995;270:5519–5526. doi: 10.1074/jbc.270.10.5519. [DOI] [PubMed] [Google Scholar]
- 4037.Shimoike T, Akayama Y, Baba T, Taura T, Ito K. SecY variants that interfere with Escherichia coli protein export in the presence of normal secY. Mol Microbiol. 1992;6:1205–1210. doi: 10.1111/j.1365-2958.1992.tb01559.x. [DOI] [PubMed] [Google Scholar]
- 4038.Shin S, Park C. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J Bacteriol. 1995;177:4696–4702. doi: 10.1128/jb.177.16.4696-4702.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4039.Shinagawa H, Iwasaki H, Ishino Y, Nakata A. SOS-inducible DNA polymerase II of E. coli is homologous to replicative DNA polymerase of eukaryotes. Biochimie. 1991;73:433–435. doi: 10.1016/0300-9084(91)90110-m. [DOI] [PubMed] [Google Scholar]
- 4040.Shinagawa H, Makino K, Nakata A. Regulation of the pho regulon in Escherichia coli K-12. Genetic and physiological regulation of the positive regulatory gene phoB. J Mol Biol. 1983;168:477–488. doi: 10.1016/s0022-2836(83)80297-6. [DOI] [PubMed] [Google Scholar]
- 4041.Shinagawa H, Kato T, Ise T, Makino K, Nakata A. Cloning and characterization of the umu operon responsible for inducible mutagenesis in Escherichia coli. Gene. 1983;23:167–174. doi: 10.1016/0378-1119(83)90048-3. [DOI] [PubMed] [Google Scholar]
- 4041a.Shinozawa T. A mutant of Escherichia coli K-12 unable to support the multiplication of bacteriophage BF23. Virology. 1973;54:427–440. doi: 10.1016/0042-6822(73)90154-2. [DOI] [PubMed] [Google Scholar]
- 4042.Shoolingin-Jordan P M. Porphobilinogen deaminase and uroporphyrinogen III synthase: structure, molecular biology, and mechanism. J Bioenerg Biomembr. 1995;27:181–195. doi: 10.1007/BF02110033. [DOI] [PubMed] [Google Scholar]
- 4043.Short S A, Singer J T. Studies on deo operon regulation in Escherichia coli: cloning and expression of the deoB structural gene. Gene. 1984;31:205–211. doi: 10.1016/0378-1119(84)90211-7. [DOI] [PubMed] [Google Scholar]
- 4044.Shpanchenko O V, Zvereva M I, Dontsova O A, Nierhaus K H, Bogdanov A A. 5S rRNA sugar-phosphate backbone protection in complexes with specific ribosomal proteins. FEBS Lett. 1996;394:71–75. doi: 10.1016/0014-5793(96)00872-1. [DOI] [PubMed] [Google Scholar]
- 4045.Shrader T E, Tobias J W, Varshavsky A. The N-end rule in Escherichia coli: cloning and analysis of the leucyl, phenylalanyl-tRNA-protein transferase gene aat. J Bacteriol. 1993;175:4364–4374. doi: 10.1128/jb.175.14.4364-4374.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4046.Shukla D, Matsumura P. Mutations leading to altered CheA binding cluster on a face of CheY. J Biol Chem. 1995;270:24414–24419. doi: 10.1074/jbc.270.41.24414. [DOI] [PubMed] [Google Scholar]
- 4047.Shultz J, Hermodson M A, Garner C C, Herrmann K M. The nucleotide sequence of the aroF gene of Escherichia coli and the amino acid sequence of the encoded protein, the tyrosine-sensitive 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase. J Biol Chem. 1984;259:9655–9661. [PubMed] [Google Scholar]
- 4048.Shultz J, Silhavy T J, Berman M L, Fiil N, Emr S D. A previously unidentified gene in the spc operon of Escherichia coli K12 specifies a component of the protein export machinery. Cell. 1982;31:227–235. doi: 10.1016/0092-8674(82)90422-6. [DOI] [PubMed] [Google Scholar]
- 4049.Shurvinton C E, Lloyd R G, Benson F E, Attfield P V. Genetic analysis and molecular cloning of the Escherichia coli ruv gene. Mol Gen Genet. 1984;194:322–329. doi: 10.1007/BF00383535. [DOI] [PubMed] [Google Scholar]
- 4050.Shuttleworth H, Taylor J, Minton N P. Sequence of the gene for alkalinephosphatase from Escherichia coli JM83. Nucleic Acids Res. 1986;14:8689. doi: 10.1093/nar/14.21.8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4051.Siegel E. Complementation studies with the repair-deficient uvrD3, uvrE156, and recL152 mutations in Escherichia coli. Mol Gen Genet. 1981;184:526–530. doi: 10.1007/BF00352533. [DOI] [PubMed] [Google Scholar]
- 4052.Siegel E, Bryson V. Mutator gene of Escherichia coli. J Bacteriol. 1967;94:38–47. doi: 10.1128/jb.94.1.38-47.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4053.Siegele D A, Hu J C, Walter W A, Gross C A. Altered promoter recognition by mutant forms of the sigma 70 subunit of Escherichia coli RNA polymerase. J Mol Biol. 1989;206:591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- 4054.Siegele D A, Imlay K R, Imlay J A. The stationary-phase-exit defect of cydC (surB) mutants is due to the lack of a functional terminal cytochrome oxidase. J Bacteriol. 1996;178:6091–6096. doi: 10.1128/jb.178.21.6091-6096.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4055.Siegele D A, Kolter R. Isolation and characterization of an Escherichia coli mutant defective in resuming growth after starvation. Genes Dev. 1993;7:2629–2640. doi: 10.1101/gad.7.12b.2629. [DOI] [PubMed] [Google Scholar]
- 4056.Siggaard-Andersen M. Role of Escherichia coli beta-ketoacyl-ACP synthase I in unsaturated fatty acid synthesis. Carlsberg Res Commun. 1988;53:371–379. doi: 10.1007/BF02983312. [DOI] [PubMed] [Google Scholar]
- 4057.Siggard-Andersen M, Wissenbach M, Chuck J-A, Svendsen I, Olsen J G, von Wettstein-Knowles P. The fabJ-encoded beta-ketoacyl substrates. Proc Natl Acad Sci USA. 1994;91:11027–11031. doi: 10.1073/pnas.91.23.11027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4058.Sigmund C D, Ettayebi M, Morgan E A. Antibiotic resistance mutations in the 16s and 23s ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 1984;12:4653–4663. doi: 10.1093/nar/12.11.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4059.Signer E. Interaction of prophages at the att80 site with the chromosome of Escherichia coli. J Mol Biol. 1965;15:243–255. doi: 10.1016/s0022-2836(66)80224-3. [DOI] [PubMed] [Google Scholar]
- 4060.Silber K R, Keiler K C, Sauer R T. Tsp: a tail-specific protease that selectively degrades proteins with nonpolar C termini. Proc Natl Acad Sci USA. 1992;89:295–299. doi: 10.1073/pnas.89.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4061.Silber K R, Sauer R T. Deletion of the prc (tsp) gene provides evidence for additional tail-specific proteolytic activity in Escherichia coli K-12. Mol Gen Genet. 1994;242:237–240. doi: 10.1007/BF00391018. [DOI] [PubMed] [Google Scholar]
- 4062.Silver P, Wickner W. Genetic mapping of the Escherichia coli leader (signal) peptidase gene (lep): a new approach for determining the map position of a cloned gene. J Bacteriol. 1983;154:569–572. doi: 10.1128/jb.154.2.569-572.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4063.Silver S, Johnseine P, Whitney E, Clark D. Manganese-resistant mutants of Escherichia coli: physiological and genetic studies. J Bacteriol. 1972;110:186–195. doi: 10.1128/jb.110.1.186-195.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4064.Silverman M, Simon M. Genetic analysis of flagellar mutants in Escherichia coli. J Bacteriol. 1973;113:105–113. doi: 10.1128/jb.113.1.105-113.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4065.Silverman M, Simon M. Bacterial flagella. Annu Rev Biochem. 1977;31:397–419. doi: 10.1146/annurev.mi.31.100177.002145. [DOI] [PubMed] [Google Scholar]
- 4066.Silverman P, Nat K, McEwen J, Birchman R. Selection of Escherichia coli K-12 mutants that prevent expression of F-plasmid functions. J Bacteriol. 1980;143:1519–1523. doi: 10.1128/jb.143.3.1519-1523.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4067.Silverman P M. Gene cpxA is a new addition to the linkage map of Escherichia coli K-12. J Bacteriol. 1982;150:425–428. doi: 10.1128/jb.150.1.425-428.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4068.Silverman P M, Tran L, Harris R, Gaudin H M. Accumulation of the F plasmid TraJ protein in cpx mutants of Escherichia coli. J Bacteriol. 1993;175:921–925. doi: 10.1128/jb.175.4.921-925.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4069.Silverman P M, Rother S, Gaudin H. Arc and Sfr functions of the Escherichia coli K-12 arcA gene product are genetically and physiologically separable. J Bacteriol. 1991;173:5648–5652. doi: 10.1128/jb.173.18.5648-5652.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4070.Silverstone A E, Goman M, Scaife J G. ALT: a new factor involved in the synthesis of RNA by Escherichia coli. Mol Gen Genet. 1972;118:223–234. doi: 10.1007/BF00333459. [DOI] [PubMed] [Google Scholar]
- 4071.Simon G, Jourlin C, Ansaldi M, Pascal M C, Chippaux M, Mejean V. Binding of the TorR regulator to cis-acting direct repeats activates tor operon expression. Mol Microbiol. 1995;17:971–980. doi: 10.1111/j.1365-2958.1995.mmi_17050971.x. [DOI] [PubMed] [Google Scholar]
- 4072.Simon G, Méjean V, Jourlin C, Chippaux M, Pascal M-C. The torR gene of Escherichia coli encodes a response regulator protein involved in the expression of the trimethylamine N-oxide reductase genes. J Bacteriol. 1994;176:5601–5606. doi: 10.1128/jb.176.18.5601-5606.1994. . (Erratum, 177:275, 1995.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4073.Simons R W, Hughes K T, Nunn W D. Regulation of fatty acid degradation in Escherichia coli: dominance studies with strains merodiploid in gene fadR. J Bacteriol. 1980;143:726–730. doi: 10.1128/jb.143.2.726-730.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4074.Simons R W, Hughes K T, Nunn W D, Egan P A, Chute H T. Regulation of fatty acid degradation in Escherichia coli: isolation and characterization of strains bearing insertion and temperature-sensitive mutations in gene fadR. J Bacteriol. 1980;142:621–632. doi: 10.1128/jb.142.2.621-632.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4075.Simpson E B, Hancock T W, Buchanan C E. Transcriptional control of dacB, which encodes a major sporulation-specific penicillin-binding protein. J Bacteriol. 1994;176:7767–7769. doi: 10.1128/jb.176.24.7767-7769.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4076.Singer M, Walter W A, Cali B M, Rouviere P, Liebke H H, Gourse R L, Gross C A. Physiological effects of the fructose-1,6-diphosphate aldolase ts8 mutation on stable RNA synthesis in Escherichia coli. J Bacteriol. 1991;173:6249–6257. doi: 10.1128/jb.173.19.6249-6257.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4077.Singh J, Mukerji M, Mahadevan S. Transcriptional activation of the Escherichia coli bgl operon: negative regulation by DNA structural elements near the promoter. Mol Microbiol. 1995;17:1085–1092. doi: 10.1111/j.1365-2958.1995.mmi_17061085.x. [DOI] [PubMed] [Google Scholar]
- 4078.Singleton C K, Roeder W D, Bogosian G, Somerville R L, Weith H L. DNA sequence of the E. coli trpR gene and prediction of the amino acid sequence of Trp repressor. Nucleic Acids Res. 1980;8:1551–1560. doi: 10.1093/nar/8.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4079.Sirisena D M, MacLachlan P R, Liu S L, Hessel A, Sanderson K E. Molecular analysis of the rfaD gene, for heptose synthesis, and the rfaF gene, for heptose transfer, in lipopolysaccharide synthesis in Salmonella typhimurium. J Bacteriol. 1994;176:2379–2385. doi: 10.1128/jb.176.8.2379-2385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4080.Sirko A, Weglenska A, Hryniewicz M, Hulanicka D M. Characterization of the Escherichia coli gene encoding a new member of the short-chain dehydrogenase/reductase (SDR) family. Acta Biochim Pol. 1997;44:153–157. [PubMed] [Google Scholar]
- 4081.Sirko A E, Hryniewicz M, Hulanicka D, Bock A. Sulfate and thiosulfate transport in Escherichia coli K-12: nucleotide sequence and expression of the cysTWAM gene cluster. J Bacteriol. 1990;172:3351–3357. doi: 10.1128/jb.172.6.3351-3357.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4082.Sirko A E, Zatyka M, Hulanicka M D. Identification of the Escherichia coli cysM gene encoding O-acetylserine sulphydrylase B by cloning with mini-Mu-lac containing a plasmid replicon. J Gen Microbiol. 1987;133:2719–2725. doi: 10.1099/00221287-133-10-2719. [DOI] [PubMed] [Google Scholar]
- 4083.Sivasubramanian N, Jayaraman R. Mapping of two transcription mutations (tlnI and tlnII) conferring thiolutin resistance, adjacent to dnaZ and rho in Escherichia coli. Mol Gen Genet. 1980;180:609–615. doi: 10.1007/BF00268068. [DOI] [PubMed] [Google Scholar]
- 4084.Six S, Andrews S C, Unden G, Guest J R. Escherichia coli possesses two homologous anaerobic C4-dicarboxylate membrane transporters (DcuA and DcuB) distinct from the aerobic dicarboxylate transport system (Dct) J Bacteriol. 1994;176:6470–6478. doi: 10.1128/jb.176.21.6470-6478.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4085.Sjoberg B M, Eriksson S, Jornvall H, Carlquist M, Eklund H. Protein B1 of ribonucleotide reductase. Direct analytical data and comparisons with data indirectly deduced from the nucleotide sequence of the Escherichia coli nrdA gene. Eur J Biochem. 1985;150:423–427. doi: 10.1111/j.1432-1033.1985.tb09037.x. [DOI] [PubMed] [Google Scholar]
- 4086.Skarstad K, Thony B, Hwang D S, Kornberg A. A novel binding protein of the origin of the Escherichia coli chromosome. J Biol Chem. 1993;268:5365–5370. [PubMed] [Google Scholar]
- 4087.Skarstad K, von Meyenburg K, Hansen F G, Boye E. Coordination of chromosome replication initiation in Escherichia coli: effects of different dnaA alleles. J Bacteriol. 1988;170:852–858. doi: 10.1128/jb.170.2.852-858.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4088.Skinner A J, Cooper R A. An Escherichia coli mutant defective in the NAD-dependent succinate semialdehyde dehydrogenase. Arch Microbiol. 1982;132:270–275. doi: 10.1007/BF00407964. [DOI] [PubMed] [Google Scholar]
- 4089.Skjold A C, Ezekiel D H. Analysis of lambda insertions in the fucose utilization region of Escherichia coli K-12: use of λ fuc and λ argA transducing bacteriophages to partially order the fucose utilization genes. J Bacteriol. 1982;152:120–125. doi: 10.1128/jb.152.1.120-125.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4090.Skjold A C, Ezekiel D H. Regulation of d-arabinose utilization in Escherichia coli K-12. J Bacteriol. 1982;152:521–523. doi: 10.1128/jb.152.1.521-523.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4091.Skogman S G, Nilsson J. Molecular cloning and characterization of the gene for Escherichia coli valyl-tRNA synthetase. Gene. 1984;30:219–226. doi: 10.1016/0378-1119(84)90123-9. [DOI] [PubMed] [Google Scholar]
- 4092.Skorko-Glonek J, Wawrzynow A, Krzewski K, Kurpierz K, Lipinska B. Site-directed mutagenesis of the HtrA (DegP) serine protease, whose proteolytic activity is indispensable for Escherichia coli survival at elevated temperatures. Gene. 1995;163:47–52. doi: 10.1016/0378-1119(95)00406-v. [DOI] [PubMed] [Google Scholar]
- 4093.Skowyra D, Wickner S. GrpE alters the affinity of DnaK for ATP and Mg2+. Implications for the mechanism of nucleotide exchange. J Biol Chem. 1995;270:26282–26285. doi: 10.1074/jbc.270.44.26282. [DOI] [PubMed] [Google Scholar]
- 4094.Slany R, Bosl M, Crain P F, Kersten H. A new function of S-adenosylmethionine: the ribosyl moiety of AdoMet is the precursor of the cyclopentenediol moiety of the tRNA wobble base queuine. Biochemistry. 1993;32:7811–7817. doi: 10.1021/bi00081a028. [DOI] [PubMed] [Google Scholar]
- 4095.Slater A C, Jones-Mortimer M C, Kornberg H L. l-Sorbose phosphorylation in Escherichia coli K-12. Biochim Biophys Acta. 1981;646:365–367. doi: 10.1016/0005-2736(81)90346-1. [DOI] [PubMed] [Google Scholar]
- 4096.Slater S, Wold S, Lu M, Boye E, Skarstad K, Kleckner N. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell. 1995;82:927–936. doi: 10.1016/0092-8674(95)90272-4. [DOI] [PubMed] [Google Scholar]
- 4097.Slater S C, Lifsics M R, O’Donnell M E, Maurer R. holE, the gene coding for the theta subunit of DNA polymerase III of Escherichia coli: characterization of a holE mutant and comparison with a dnaQ (epsilon-subunit) mutant. J Bacteriol. 1994;176:815–821. doi: 10.1128/jb.176.3.815-821.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4098.Sledjeski D D, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 4099.Sledjeski D D, Gottesman S. Osmotic induction of capsule synthesis in Escherichia coli. J Bacteriol. 1996;178:1204–1206. doi: 10.1128/jb.178.4.1204-1206.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4100.Slettan A, Gebhardt K, Kristiansen E, Birkeland N-K, Lindqvist B H. Escherichia coli K-12 and B contain functional bacteriophage P2 ogr. J Bacteriol. 1992;174:4094–4100. doi: 10.1128/jb.174.12.4094-4100.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4101.Slocum M K, Parkinson J S. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: organization of the tar region. J Bacteriol. 1983;155:565–577. doi: 10.1128/jb.155.2.565-577.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4102.Slocum M K, Parkinson J S. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: null phenotypes of the tar and tap genes. J Bacteriol. 1985;163:586–594. doi: 10.1128/jb.163.2.586-594.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4103.Slupska M M, Baikalov C, Lloyd R, Miller J H. Mutator tRNAs are encoded by the Escherichia coli mutator genes mutA and mutC: a novel pathway for mutagenesis. Proc Natl Acad Sci USA. 1996;93:4380–4385. doi: 10.1073/pnas.93.9.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4104.Smallshaw J E, Kelln R A. Cloning, nucleotide sequence and expression of the Escherichia coli K-12 pyrH gene encoding UMP kinase. Genetics (Life Sci Adv) 1992;11:59–65. [Google Scholar]
- 4105.Smiley B L, Lupski J R, Svec P S, MacMacken R, Godson G N. Sequences of the Escherichia coli dnaG primase gene and regulation of its expression. Proc Natl Acad Sci USA. 1982;79:4550–4554. doi: 10.1073/pnas.79.15.4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4106.Smillie D A, Hayward R S, Suzuki T, Fujita N, Ishihama A. Locations of genes encoding alkyl hydroperoxide reductase on the physical map of the Escherichia coli K-12 genome. J Bacteriol. 1992;174:3826–3827. doi: 10.1128/jb.174.11.3826-3827.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4107.Smirnov G B, Ilyina T S, Romanova Y M, Markov A P, Nechaeva E V. Mutants of Escherichia coli affected in the process of transposition and genomic rearrangements. Cold Spring Harbor Symp Quant Biol. 1980;45:193–200. doi: 10.1101/sqb.1981.045.01.031. [DOI] [PubMed] [Google Scholar]
- 4108.Smirnov Y V, Lisenkov A F. Construction of the hybrid crp-lac operon and investigation of the role of the CRP-cAMP complex in its regulation in Escherichia coli. Sov Genet (Engl Transl Genetika) 1986;22:432–438. [PubMed] [Google Scholar]
- 4109.Smith A A, Greene R C. Cloning of the methionine regulatory gene, metJ, of Escherichia coli K12 and identification of its product. J Biol Chem. 1984;259:14279–14281. [PubMed] [Google Scholar]
- 4110.Smith B R, Schleif R. Nucleotide sequence of the l-arabinose regulatory region of Escherichia coli K12. J Biol Chem. 1978;253:6931–6933. [PubMed] [Google Scholar]
- 4111.Smith D K, Kassam T, Singh B, Elliott J F. Escherichia coli has two homologous glutamate decarboxylase genes that map to distinct loci. J Bacteriol. 1992;174:5820–5826. doi: 10.1128/jb.174.18.5820-5826.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4112.Smith D R, Calvo J M. Regulation of dihydrofolate reductase synthesis in Escherichia coli. Mol Gen Genet. 1979;175:31–38. doi: 10.1007/BF00267852. [DOI] [PubMed] [Google Scholar]
- 4113.Smith D R, Calvo J M. Nucleotide sequence of dihydrofolate reductase genes from trimethoprim-resistant mutants of Escherichia coli. Evidence that dihydrofolate reductase interacts with another essential gene product. Mol Gen Genet. 1982;187:72–78. doi: 10.1007/BF00384386. [DOI] [PubMed] [Google Scholar]
- 4114.Smith D R, Calvo J M. Nucleotide sequence of the E. coli gene coding for dihydrofolate reductase. Nucleic Acids Res. 1982;8:2255–2274. doi: 10.1093/nar/8.10.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4115.Smith J M, Daum H A., III Nucleotide sequence of the purM gene encoding 5′-phosphoribosyl-5-aminoimidazole synthetase of Escherichia coli K12. J Biol Chem. 1986;261:10632–10636. [PubMed] [Google Scholar]
- 4116.Smith J M, Daum H A., III Identification and nucleotide sequence of a gene encoding 5′-phosphoribosylglycinamide transformylase in Escherichia coli K12. J Biol Chem. 1987;262:10565–10569. [PubMed] [Google Scholar]
- 4117.Smith J M, Gots J S. purF-lac fusion and direction of purF transcription in Escherichia coli. J Bacteriol. 1980;143:1156–1164. doi: 10.1128/jb.143.3.1156-1164.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4118.Smith M W, Payne J W. Expression of periplasmic binding proteins for peptide transport is subject to negative regulation by phosphate limitation in Escherichia coli. FEMS Microbiol Lett. 1992;79:183–190. doi: 10.1111/j.1574-6968.1992.tb14038.x. [DOI] [PubMed] [Google Scholar]
- 4119.Smith R A, Parkinson J S. Overlapping genes at the cheA locus of Escherichia coli. Proc Natl Acad Sci USA. 1980;77:5370–5374. doi: 10.1073/pnas.77.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4120.Smith R L, Banks J L, Snavely M, Maguire M E. Sequence and topology of the corA magnesium transport systems of Salmonella typhimurium and Escherichia coli. Identification of a new class of transport protein. J Biol Chem. 1993;268:14071–14080. [PubMed] [Google Scholar]
- 4121.Smith T F, Sadler J R. The nature of lactose operator constitutive mutations. J Mol Biol. 1971;59:273–305. doi: 10.1016/0022-2836(71)90051-9. [DOI] [PubMed] [Google Scholar]
- 4122.Snyder W B, Davis L J, Danese P N, Cosma C L, Silhavy T J. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J Bacteriol. 1995;177:4216–4223. doi: 10.1128/jb.177.15.4216-4223.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4123.Snyder W B, Silhavy T J. Enhanced export of beta-galactosidase fusion proteins in prlF mutants is Lon dependent. J Bacteriol. 1992;174:5661–5668. doi: 10.1128/jb.174.17.5661-5668.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4124.Soballe B, Poole R K. Aerobic and anaerobic regulation of the ubiCA operon, encoding enzymes for the first two committed steps of ubiquinone biosynthesis in Escherichia coli. FEBS Lett. 1997;414:373–376. doi: 10.1016/s0014-5793(97)01041-7. [DOI] [PubMed] [Google Scholar]
- 4125.Sodergren E J, DeMoss J A. narI region of the Escherichia coli nitrate reductase (nar) operon contains two genes. J Bacteriol. 1988;170:1721–1729. doi: 10.1128/jb.170.4.1721-1729.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4126.Sodergren E J, Hsu P Y, DeMoss J A. Roles of the narJ and narI gene products in the expression of nitrate reductase in Escherichia coli. J Biol Chem. 1988;263:16156–16162. [PubMed] [Google Scholar]
- 4127.Sofia H J, Burland V D, Daniels D L, Plunkett G, Blattner F R. GenBank submission U00039. 1994. Analysis of the Escherichia coli genome. V. DNA sequence of the region from 76.0 to 81.5 minutes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4128.Sofia H J, Burland V D, Daniels D L, Plunkett G, Blattner F R. Analysis of the Escherichia coli genome. V. DNA sequence of the region from 76.0 to 81.5 minutes. Nucleic Acids Res. 1994;22:2576–2586. doi: 10.1093/nar/22.13.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4129.Sogaard-Andersen L, Pedersen H, Holst B, Valentin-Hansen P. A novel function of the cAMP-CRP complex in Escherichia coli: cAMP-CRP functions as an adaptor for the CytR repressor in the deo operon. Mol Microbiol. 1991;5:969–975. doi: 10.1111/j.1365-2958.1991.tb00772.x. [DOI] [PubMed] [Google Scholar]
- 4130.Sohail A, Lieb M, Dar M, Bhagwat A S. A gene required for very short patch repair in Escherichia coli is adjacent to the DNA cytosine methylase gene. J Bacteriol. 1990;172:4214–4221. doi: 10.1128/jb.172.8.4214-4221.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4131.Soll L. Pseudovirulent mutants of λb221 poriCasna resulting from mutations in or near oriC, the E. coli origin of DNA replication. Mol Gen Genet. 1980;178:391–396. doi: 10.1007/BF00270489. [DOI] [PubMed] [Google Scholar]
- 4132.Son H S, Rhee S G. Cascade control of Escherichia coli glutamine synthetase. Purification and properties of PII protein and nucleotide sequence of its structural gene. J Biol Chem. 1987;262:8690–8695. [PubMed] [Google Scholar]
- 4133.Sonden B, Uhlin B E. Coordinated and differential expression of histone-like proteins in Escherichia coli: regulation and function of the H-NS analog StpA. EMBO J. 1996;15:4970–4980. [PMC free article] [PubMed] [Google Scholar]
- 4134.Song W J, Jackowski S. Cloning, sequencing, and expression of the pantothenate kinase (coaA) gene of Escherichia coli. J Bacteriol. 1992;174:6411–6417. doi: 10.1128/jb.174.20.6411-6417.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4135.Song W-J, Jackowski S. coaA and rts are allelic and located at kilobase 3532 on the Escherichia coli physical map. J Bacteriol. 1992;174:1705–1706. doi: 10.1128/jb.174.5.1705-1706.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4136.Song Y, Sargentini N J. Escherichia coli DNA repair genes radA and sms are the same gene. J Bacteriol. 1996;178:5045–5048. doi: 10.1128/jb.178.16.5045-5048.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4137.Sood P, Lerner C G, Shimamoto T, Lu Q, Inouye M. Characterization of the autophosphorylation of Era, an essential Escherichia coli GTPase. Mol Microbiol. 1994;12:201–208. doi: 10.1111/j.1365-2958.1994.tb01009.x. [DOI] [PubMed] [Google Scholar]
- 4138.Sorensen K I, Hove-Jensen B. Ribose catabolism of Escherichia coli: characterization of the rpiB gene encoding ribose phosphate isomerase B and of the rpiR gene, which is involved in regulation of rpiB expression. J Bacteriol. 1996;178:1003–1011. doi: 10.1128/jb.178.4.1003-1011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4139.Sorensen P G, Lutkenhaus J, Young K, Eveland S S, Anderson M S, Raetz C R. Regulation of UDP-3-O-[R-3-hydroxymyristoyl]-N-acetylglucosamine deacetylase in Escherichia coli. The second enzymatic step of lipid a biosynthesis. J Biol Chem. 1996;271:258–263. doi: 10.1074/jbc.271.42.25898. [DOI] [PubMed] [Google Scholar]
- 4140.Spanjaard R A, Chen K S, Walker J R, van Duin J. Frameshift suppression at tandem AGA and AGG codons by cloned tRNA genes: assigning a codon to argU tRNA and T4 tRNA(Arg) Nucleic Acids Res. 1990;18:5031–5036. doi: 10.1093/nar/18.17.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4141.Sparkowski J, Das A. The nucleotide sequence of greA, a suppressor gene that restores growth of an Escherichia coli RNA polymerase mutant at high temperature. Nucleic Acids Res. 1990;18:6443. doi: 10.1093/nar/18.21.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4142.Sparkowski J, Das A. Location of a new gene, greA, on the Escherichia coli chromosome. J Bacteriol. 1991;173:5256–5257. doi: 10.1128/jb.173.17.5256-5257.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4143.Sparling P F, Blackman E. Mutation to erythromycin dependence in Escherichia coli K-12. J Bacteriol. 1973;116:74–83. doi: 10.1128/jb.116.1.74-83.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4144.Sparling P F, Ikeya Y, Elliot D. Two genetic loci for resistance to kasugamycin in Escherichia coli. J Bacteriol. 1973;113:704–710. doi: 10.1128/jb.113.2.704-710.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4145.Sparrow C P, Raetz C R. A trans-acting regulatory mutation that causes overproduction of phosphatidylserine synthase in Escherichia coli. J Biol Chem. 1983;258:9963–9967. [PubMed] [Google Scholar]
- 4146.Spears P A, Schauer D, Orndorff P E. Metastable regulation of type 1 piliation in Escherichia coli and isolation and characterization of a phenotypically stable mutant. J Bacteriol. 1986;168:179–185. doi: 10.1128/jb.168.1.179-185.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4147.Spence E L, Kawamukai M, Sanvoisin J, Braven H, Bugg T D. Catechol dioxygenases from Escherichia coli (MhpB) and Alcaligenes eutrophus (MpcI): sequence analysis and biochemical properties of a third family of extradiol dioxygenases. J Bacteriol. 1996;178:5249–5256. doi: 10.1128/jb.178.17.5249-5256.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4148.Spencer J B, Stolowich N J, Roessner C A, Scott A I. The Escherichia coli cysG gene encodes the multifunctional protein, siroheme synthase. FEBS Lett. 1993;335:57–60. doi: 10.1016/0014-5793(93)80438-z. [DOI] [PubMed] [Google Scholar]
- 4149.Spencer M E, Guest J R. Molecular cloning of four tricarboxylic acid cycle genes of Escherichia coli. J Bacteriol. 1982;151:542–552. doi: 10.1128/jb.151.2.542-552.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4150.Spencer M E, Guest J R. Transcription analysis of the sucAB, aceEF and lpd genes of Escherichia coli. Mol Gen Genet. 1985;200:145–154. doi: 10.1007/BF00383328. [DOI] [PubMed] [Google Scholar]
- 4151.Spencer M E, Darlison M G, Stephens P E, Guest J R. Nucleotide sequence of the sucB gene encoding the dihydrolipoamide succinyltransferase of Escherichia coli K12 and homology with the corresponding acetyltransferase. Eur J Biochem. 1984;141:361–374. doi: 10.1111/j.1432-1033.1984.tb08200.x. [DOI] [PubMed] [Google Scholar]
- 4152.Spira B, Yagil E. The relation between ppGpp and the PHO regulon in Escherichia coli. Mol Gen Genet. 1988;257:469–477. doi: 10.1007/s004380050671. [DOI] [PubMed] [Google Scholar]
- 4153.Spiro S, Guest J R. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol Rev. 1990;6:399–428. doi: 10.1111/j.1574-6968.1990.tb04109.x. [DOI] [PubMed] [Google Scholar]
- 4154.Spitzer E D, Weiss B. The dfp gene of Escherichia coli K-12, a locus affecting DNA synthesis, codes for a flavoprotein. J Bacteriol. 1985;164:994–1003. doi: 10.1128/jb.164.3.994-1003.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4155.Spitzer E D, Jimenez-Billini H E, Weiss B. β-Alanine auxotrophy associated with dfp, a locus affecting DNA synthesis in Escherichia coli. J Bacteriol. 1988;170:872–876. doi: 10.1128/jb.170.2.872-876.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4156.Sprague G F, Bell R M, Cronan J E., Jr A mutant of Escherichia coli auxotrophic for organic phosphates: evidence for two defects in inorganic phosphate transport. Mol Gen Genet. 1975;143:71–77. doi: 10.1007/BF00269422. [DOI] [PubMed] [Google Scholar]
- 4157.Spratt B G, Boyd A, Stoker N G. Defective and plaque-forming lambda transducing bacteriophage carrying penicillin-binding protein-cell shape genes: genetic and physical mapping and identification of gene products from the lip-dacA-rodA-pbpA-leuS region of the Escherichia coli chromosome. J Bacteriol. 1980;143:569–581. doi: 10.1128/jb.143.2.569-581.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4158.Spratt B G, Ginsburgh C L, Nunn W D. Isolation and genetic characterization of Escherichia coli mutants defective in propionate metabolism. J Bacteriol. 1981;146:1166–1169. doi: 10.1128/jb.146.3.1166-1169.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4159.Spratt B G, Zhou J, Taylor M, Merrick M J. Monofunctional biosynthetic peptidoglycan transglycosylases. Mol Microbiol. 1996;19:639–640. doi: 10.1046/j.1365-2958.1996.442924.x. [DOI] [PubMed] [Google Scholar]
- 4160.Spratt S K, Black P N, Ragozzino M M, Nunn W D. Cloning, mapping and expression of genes involved in the fatty acid-degradative multienzyme complex of Escherichia coli. J Bacteriol. 1984;158:535–542. doi: 10.1128/jb.158.2.535-542.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4161.Sprenger G A. Location of the transketolase (tkt) gene on the Escherichia coli physical map. J Bacteriol. 1992;174:1707–1708. doi: 10.1128/jb.174.5.1707-1708.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4162.Sprenger G A. Nucleotide sequence of the Escherichia coli K-12 transketolase (tkt) gene. Biochim Biophys Acta. 1993;1216:307–310. doi: 10.1016/0167-4781(93)90161-6. [DOI] [PubMed] [Google Scholar]
- 4163.Sprenger G A. Two open reading frames adjacent to the Escherichia coli K-12 transketolase (tkt) gene show high similarity to the mannitol phosphotransferase system enzymes from Escherichia coli and various gram-positive bacteria. Biochim Biophys Acta. 1993;1158:103–106. doi: 10.1016/0304-4165(93)90103-f. [DOI] [PubMed] [Google Scholar]
- 4164.Sprenger G A. Genetics of pentose-phosphate pathway enzymes of Escherichia coli K-12. Arch Microbiol. 1995;164:324–330. doi: 10.1007/BF02529978. [DOI] [PubMed] [Google Scholar]
- 4165.Sprenger G A, Schorken U, Sprenger G, Sahm H. Transaldolase B of Escherichia coli K-12: cloning of its gene, talB, and characterization of the enzyme from recombinant strains. J Bacteriol. 1995;177:5930–5936. doi: 10.1128/jb.177.20.5930-5936.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4166.Sprenger G A, Schorken U, Wiegert T, Grolle S, de Graaf A A, Taylor S V, Begley T P, Bringer-Meyer S, Sahm H. Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-d-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxal. Proc Natl Acad Sci USA. 1997;94:12857–12862. doi: 10.1073/pnas.94.24.12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4167.Spring K J, Jerlstrom P G, Burns D M, Beacham I R. l-Asparaginase genes in Escherichia coli: isolation of mutants and characterization of the ansA gene and its protein product. J Bacteriol. 1986;166:135–142. doi: 10.1128/jb.166.1.135-142.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4168.Springer M, Plumbridge J A, Trudel M, Graffe M, Grunberg-Manago M. Transcription units around the gene for E. coli translation initiation factor IF3 (infC) Mol Gen Genet. 1982;186:247–252. doi: 10.1007/BF00331857. [DOI] [PubMed] [Google Scholar]
- 4169.Springer M, Mayaux J-F, Fayat G, Plumbridge J A, Graffe M, Blanquet S, Grunberg-Manago M. Attenuation control of the Escherichia coli phenylalanyl-tRNA synthetase operon. J Mol Biol. 1985;181:467–478. doi: 10.1016/0022-2836(85)90420-6. [DOI] [PubMed] [Google Scholar]
- 4170.Springer M, Graffe M, Butler J S, Grunberg-Manago M. Genetic definition of the translational operator of the threonine-tRNA ligase gene in Escherichia coli. Proc Natl Acad Sci USA. 1986;83:4384–4388. doi: 10.1073/pnas.83.12.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4171.Springer M, Trudel M, Graffe M, Plumbridge J A, Fayat G, Mayaux J-F, Sacerdot C, Blanquet S, Grunberg-Manago M. Escherichia coli phenylalanyl-tRNA synthetase operon is controlled by attenuation in vivo. J Mol Biol. 1983;171:263–279. doi: 10.1016/0022-2836(83)90093-1. [DOI] [PubMed] [Google Scholar]
- 4172.Springer S E, Huber R E. Sulfate and selenate uptake and transport in wild and in two selenate-tolerant strains of Escherichia coli K-12. Arch Biochem. 1973;156:595–603. doi: 10.1016/0003-9861(73)90310-x. [DOI] [PubMed] [Google Scholar]
- 4173.Spurio R, Durrenberger M, Falconi M, La Teana A, Pon C L, Gualerzi C O. Lethal overproduction of the Escherichia coli nucleoid protein H-NS: ultramicroscopic and molecular autopsy. Mol Gen Genet. 1992;231:201–211. doi: 10.1007/BF00279792. [DOI] [PubMed] [Google Scholar]
- 4174.Spyrou G, Haggard-Ljungquist E, Krook M, Jornvall H, Nilsson E, Reichard P. Characterization of the flavin reductase gene (fre) of Escherichia coli and construction of a plasmid for overproduction of the enzyme. J Bacteriol. 1991;173:3673–3679. doi: 10.1128/jb.173.12.3673-3679.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4175.Squires C, Krainer A, Barry G, Shen W-F, Squires C L. Nucleotide sequence at the end of the gene for the RNA polymerase B′ subunit (rpoC) Nucleic Acids Res. 1981;9:6827–6840. doi: 10.1093/nar/9.24.6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4176.Squires C, DeFelice M, Wessler S R, Calvo J M. Physical characterization of the ilvH1 operon of Escherichia coli K-12. J Bacteriol. 1981;147:797–804. doi: 10.1128/jb.147.3.797-804.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4177.Squires C H, DeFelice M, Devereux J, Calvo J M. Molecular structure of ilvIH and its evolutionary relationship to ilvG in Escherichia coli K-12. Nucleic Acids Res. 1983;11:5299–5313. doi: 10.1093/nar/11.15.5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4178.Squires C L, Pedersen S, Ross B M, Squires C. ClpB is the Escherichia coli heat shock protein F84.1. J Bacteriol. 1991;173:4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4179.Srivastava B S. Radiation sensitivity of a mutant of the Escherichia coli K12 which affects transduction, transformation and rates of mutation. Mol Gen Genet. 1976;143:327–332. doi: 10.1007/BF00269411. [DOI] [PubMed] [Google Scholar]
- 4180.Staab J F, Elkins M F, Earhart C F. Nucleotide sequence of the Escherichia coli entE gene. FEMS Microbiol Lett. 1989;50:15–19. doi: 10.1016/0378-1097(89)90450-3. [DOI] [PubMed] [Google Scholar]
- 4181.Stacey K A, Oliver P. Novel pleiotropic mutation in Escherichia coli K12 which affects transduction, transformation and rates of mutation. J Gen Microbiol. 1977;98:569–578. doi: 10.1099/00221287-98-2-569. [DOI] [PubMed] [Google Scholar]
- 4182.Stader J, Gansheroff L J, Silhavy T J. New suppressors of signal-sequence mutations, prlG, are linked tightly to the secE gene of Escherichia coli. Genes Dev. 1989;3:1045–1052. doi: 10.1101/gad.3.7.1045. [DOI] [PubMed] [Google Scholar]
- 4183.Stader J, Matsumura P, Uacante D, Dean G E, Macnab R M. Nucleotide sequence of the Escherichia coli motB gene and site-limited incorporation of its product into the cytoplasmic membrane. J Bacteriol. 1986;166:244–252. doi: 10.1128/jb.166.1.244-252.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4184.Stadtman T C, Davis J N, Zehelein E, Bock A. Biochemical and genetic analysis of Salmonella typhimurium and Escherichia coli mutants defective in specific incorporation of selenium into formate dehydrogenase and tRNAs. Biofactors. 1989;2:35–44. [PubMed] [Google Scholar]
- 4185.Stahl F, Myers R. Old and new concepts for the role of chi in bacterial recombination. J Hered. 1995;86:327–329. doi: 10.1093/oxfordjournals.jhered.a111599. [DOI] [PubMed] [Google Scholar]
- 4186.Stalmach M E, Grothe S R, Wood J M. Two proline porters in Escherichia coli K-12. J Bacteriol. 1983;156:481–486. doi: 10.1128/jb.156.2.481-486.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4187.Stan-Lotter H, Clarke D M, Bragg P D. Isolation of a fourth cysteinyl-containing peptide of the alpha-subunit of the F1 ATPase from Escherichia coli necessitates revision of the DNA sequence. FEBS Lett. 1986;197:121–124. doi: 10.1016/0014-5793(86)80310-6. [DOI] [PubMed] [Google Scholar]
- 4188.Staskawicz B J, Panopoulos N J. Phaseolotoxin transport in Escherichia coli and Salmonella typhimurium via the oligopeptide permease. J Bacteriol. 1980;142:474–479. doi: 10.1128/jb.142.2.474-479.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4189.Staudenbauer W L. Replication of Escherichia coli DNA in vitro: inhibition by oxoline acid. J Biochem (Tokyo) 1976;62:491–497. doi: 10.1111/j.1432-1033.1976.tb10183.x. [DOI] [PubMed] [Google Scholar]
- 4190.Staudenmaier H, Van Hove B, Yaraghi Z, Braun V. Nucleotide sequences of the fecBCDE genes and locations of the proteins suggest a periplasmic-binding-protein-dependent transport mechanism for iron(III) dicitrate in Escherichia coli. J Bacteriol. 1989;171:2626–2633. doi: 10.1128/jb.171.5.2626-2633.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4191.Stauffer G, Plamann M D, Stauffer L T. Construction and expression of hybrid plasmids containing the Escherichia coli glyA gene. Gene. 1981;14:63–72. doi: 10.1016/0378-1119(81)90148-7. [DOI] [PubMed] [Google Scholar]
- 4192.Stauffer L T, Ghrist A C, Stauffer G V. The Escherichia coli gcvT gene encoding the T-protein of the glycine cleavage enzyme system. DNA Sequence. 1993;3:339–346. doi: 10.3109/10425179309020835. [DOI] [PubMed] [Google Scholar]
- 4193.Stauffer L T, Stauffer G V. Characterization of the gcv control region from Escherichia coli. J Bacteriol. 1994;176:6159–6164. doi: 10.1128/jb.176.20.6159-6164.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4194.Stauffer L T, Plamann M D, Stauffer G V. Cloning and characterization of the glycine-cleavage enzyme system of Escherichia coli. Gene. 1986;44:219–226. doi: 10.1016/0378-1119(86)90185-x. [DOI] [PubMed] [Google Scholar]
- 4195.Stebbins C E, Borukhov S, Orlova M, Polyakov A, Goldfarb A, Darst S A. Crystal structure of the GreA transcript cleavage factor from Escherichia coli. Nature. 1995;373:636–640. doi: 10.1038/373636a0. [DOI] [PubMed] [Google Scholar]
- 4196.Steed P M, Wanner B L. Use of the rep technique for allele replacement to construct mutants with deletions of the pstSCAB-phoU operon: evidence of a new role for the PhoU protein in the phosphate regulon. J Bacteriol. 1993;175:6797–6809. doi: 10.1128/jb.175.21.6797-6809.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4197.Steege D. A nucleotide change in the anticodon of an Escherichia coli serine transfer RNA results in supD− amber suppression. Nucleic Acids Res. 1983;11:3823–3832. doi: 10.1093/nar/11.11.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4198.Steffes C, Ellis J, Wu J, Rosen B P. The lysP gene encodes the lysine-specific permease. J Bacteriol. 1992;174:3242–3249. doi: 10.1128/jb.174.10.3242-3249.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4199.Steiert P S, Stauffer L T, Stauffer G V. The lpd gene product functions as the L protein in the Escherichia coli glycine cleavage enzyme system. J Bacteriol. 1990;172:6142–6144. doi: 10.1128/jb.172.10.6142-6144.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4200.Steinebach V, Benen J A, Bader R, Postma P W, de Vries S, Duine J A. Cloning of the maoA gene that encodes aromatic amine oxidase of Escherichia coli W3350 and characterization of the overexpressed enzyme. Eur J Biochem. 1996;237:584–591. doi: 10.1111/j.1432-1033.1996.0584p.x. [DOI] [PubMed] [Google Scholar]
- 4201.Steinum A L, Seeberg E. Nucleotide sequence of the tag gene from Escherichia coli. Nucleic Acids Res. 1986;14:3763–3772. doi: 10.1093/nar/14.9.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4202.Stephens D L, Choe M D, Earhart C F. Escherichia coli periplasmic protein FepB binds ferrienterobactin. Microbiology. 1995;141:1647–1654. doi: 10.1099/13500872-141-7-1647. [DOI] [PubMed] [Google Scholar]
- 4203.Stephens P E, Lewis H M, Darlison M G, Guest J R. Nucleotide sequence of the lipoamide dehydrogenase gene of Escherichia coli K12. Eur J Biochem. 1983;135:519–527. doi: 10.1111/j.1432-1033.1983.tb07683.x. [DOI] [PubMed] [Google Scholar]
- 4204.Stephens P E, Darlison M G, Lewis H M, Guest J R. The pyruvate dehydrogenase complex of Escherichia coli K-12. Nucleotide sequence encoding the pyruvate dehydrogenase component. Eur J Biochem. 1983;133:155–162. doi: 10.1111/j.1432-1033.1983.tb07441.x. [DOI] [PubMed] [Google Scholar]
- 4205.Sternberg N, Hamilton D, Austin S, Yarmolinsky M, Hoess R H. Site-specific recombination and its role in the life cycle of bacteriophage P1. Cold Spr Harb Symp Quant Biol. 1980;45:297–309. doi: 10.1101/sqb.1981.045.01.042. [DOI] [PubMed] [Google Scholar]
- 4206.Sternglanz R, DiNardo S, Voelkel K A, Nishimura Y, Hirota Y, Becherer K, Zumstein L, Wang J C. Mutations in the gene coding for Escherichia coli DNA topoisomerase I affect transcription and translation. Proc Natl Acad Sci USA. 1981;78:2747–2751. doi: 10.1073/pnas.78.5.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4207.Stevens F J, Wu T T. Growth on d-xylose of a mutant strain of Escherichia coli K12 using a novel isomerase and enzymes related to d-xylose metabolism. J Gen Microbiol. 1976;97:257–265. doi: 10.1099/00221287-97-2-257. [DOI] [PubMed] [Google Scholar]
- 4208.Stevenson G, Neal B, Liu D, Hobbs M, Packer N H, Batley M, Redmond J W, Lindquist L, Reeves P R. Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb gene cluster. J Bacteriol. 1994;176:4144–4156. doi: 10.1128/jb.176.13.4144-4156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4209.Stevenson G, Andrianopoulos K, Hobbs M, Reeves P R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4210.Steward K L, Linn T. In vivo analysis of overlapping transcription units in the rplKAJLrpoBC ribosomal protein-RNA polymerase gene cluster of Escherichia coli. J Mol Biol. 1991;218:23–31. doi: 10.1016/0022-2836(91)90870-c. [DOI] [PubMed] [Google Scholar]
- 4211.Steward K L, Linn T. Transcription frequency modulates the efficiency of an attenuator preceding the rpoBC RNA polymerase genes of Escherichia coli: possible autogenous control. Nucleic Acids Res. 1992;20:4773–4779. doi: 10.1093/nar/20.18.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4212.Stewart A. Genetic nomenclature guide, including information on genomic databases. Trends Genet (Suppl) 1995;11:4–42. [Google Scholar]
- 4213.Stewart P S, D’Ari R. Genetic and morphological characterization of an Escherichia coli chromosome segregation mutant. J Bacteriol. 1992;174:4513–4516. doi: 10.1128/jb.174.13.4513-4516.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4214.Stewart R C. Activating and inhibitory mutations in the regulatory domains of the methylesterase in bacterial chemotaxis. J Biol Chem. 1993;268:1921–1930. [PubMed] [Google Scholar]
- 4215.Stewart V, Parales J, Jr, Merkel S M. Structure of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J Bacteriol. 1989;171:2229–2234. doi: 10.1128/jb.171.4.2229-2234.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4216.Stewart V J. Requirement of Fnr and NarL functions for nitrate reductase expression in Escherichia coli K-12. J Bacteriol. 1982;151:1320–1325. doi: 10.1128/jb.151.3.1320-1325.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4217.Stewart V J. Nitrate regulation of anaerobic respiratory gene expression in Escherichia coli. Mol Microbiol. 1993;9:425–434. doi: 10.1111/j.1365-2958.1993.tb01704.x. [DOI] [PubMed] [Google Scholar]
- 4218.Stewart V J, Yanofsky C. Evidence for transcription antitermination control of tryptophanase operon expression in Escherichia coli K-12. J Bacteriol. 1985;164:731–740. doi: 10.1128/jb.164.2.731-740.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4219.Stewart V J, MacGregor C H. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J Bacteriol. 1982;151:788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4220.Stewart V J, Parales J., Jr Identification and expression of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J Bacteriol. 1988;170:1589–1597. doi: 10.1128/jb.170.4.1589-1597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4221.Stewart V J, Lin J T, Berg B L. Genetic evidence that genes fdhD and fdhE do not control synthesis of formate dehydrogenase-N in Escherichia coli K-12. J Bacteriol. 1991;173:4417–4423. doi: 10.1128/jb.173.14.4417-4423.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4222.Stewart V J, Landick R, Yanofsky C. Rho-dependent transcription termination in the tryptophanase operon leader region of Escherichia coli K-12. J Bacteriol. 1986;166:217–223. doi: 10.1128/jb.166.1.217-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4223.Stim K P, Bennett G N. Nucleotide sequence of the adi gene, which encodes the biodegradative acid-induced arginine decarboxylase of Escherichia coli. J Bacteriol. 1993;175:1221–1234. doi: 10.1128/jb.175.5.1221-1234.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4224.Stim-Herndon K P, Flores T M, Bennett G N. Molecular characterization of adiY, a regulatory gene which affects expression of the biodegradative acid-induced arginine decarboxylase gene (adiA) of Escherichia coli. Microbiology. 1996;142:1311–1320. doi: 10.1099/13500872-142-5-1311. [DOI] [PubMed] [Google Scholar]
- 4225.Stirling C J, Szatmari G, Stewart G J, Smith M C, Sherratt D J. The arginine repressor is essential for plasmid-stabilizing site-specific recombination at the ColE1 cer locus. EMBO J. 1988;7:4389–4395. doi: 10.1002/j.1460-2075.1988.tb03338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4226.Stirling C J, Stewart G J, Sherratt D J. Multicopy plasmid stability in Escherichia coli requires host-encoded functions that lead to plasmid site-specific recombination. Mol Gen Genet. 1988;214:80–84. doi: 10.1007/BF00340183. [DOI] [PubMed] [Google Scholar]
- 4227.Stirling C J, Colloms S D, Collins J F, Szatmari G, Sherratt D J. xerB, an Escherichia coli gene required for plasmid ColE1 site-specific recombination, is identical to pepA, encoding aminopeptidase A, a protein with substantial similarity to bovine lens leucine aminopeptidase. EMBO J. 1989;8:1623–1627. doi: 10.1002/j.1460-2075.1989.tb03547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4228.Stirling D A, Hulton C S J, Waddell L, Park S F, Stewart G S A B, Booth I R, Higgins C F. Molecular characterization of the proU loci of Salmonella typhimurium and Escherichia coli encoding osmoregulated glycine betaine transport systems. Mol Microbiol. 1989;3:1025–1038. doi: 10.1111/j.1365-2958.1989.tb00253.x. [DOI] [PubMed] [Google Scholar]
- 4229.Stitt B L, Revel H, Lielausis I, Wood W B. Role of the host cell in bacteriophage T4 development. II. Characterization of host mutants that have pleiotropic effects on T4 growth. J Virol. 1980;35:775–789. doi: 10.1128/jvi.35.3.775-789.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4230.Stoker K, Oltmann L F, Stouthamer A H. Randomly induced Escherichia coli K-12 Tn5 insertion mutants defective in hydrogenase activity. J Bacteriol. 1989;171:831–836. doi: 10.1128/jb.171.2.831-836.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4231.Stoker K, Reijnders W N, Oltmann L F, Stouthamer A H. Initial cloning and sequencing of hydHG, an operon homologous to ntrBC and regulating the labile hydrogenase activity in Escherichia coli K-12. J Bacteriol. 1989;171:4448–4456. doi: 10.1128/jb.171.8.4448-4456.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4232.Stoker N G, Broome-Smith J K, Edelman A, Spratt B G. Organization and subcloning of the dacA-rodA-pbpA cluster of cell shape genes in Escherichia coli. J Bacteriol. 1983;155:847–853. doi: 10.1128/jb.155.2.847-853.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4233.Stoker N G, Pratt J M, Spratt B G. Identification of the rodA gene product of Escherichia coli. J Bacteriol. 1983;155:854–859. doi: 10.1128/jb.155.2.854-859.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4234.Stokes H W, Hall B G. Sequence of the ebgR gene of Escherichia coli: evidence that the EBG and LAC operons are descended from a common ancestor. Mol Biol Evol. 1985;2:478–483. doi: 10.1093/oxfordjournals.molbev.a040373. [DOI] [PubMed] [Google Scholar]
- 4235.Stokes H W, Betts P W, Hall B G. Sequence of the ebgA gene of Escherichia coli: comparison with the lacZ gene. Mol Biol Evol. 1985;2:469–477. doi: 10.1093/oxfordjournals.molbev.a040372. [DOI] [PubMed] [Google Scholar]
- 4236.Stoller G, Rucknagel K P, Nierhaus K H, Schmid F X, Fischer G, Rahfeld J U. A ribosome-associated peptidyl-prolyl cis/trans isomerase identified as the trigger factor. EMBO J. 1995;14:4939–4949. doi: 10.1002/j.1460-2075.1995.tb00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4237.Stoner C M, Schleif R. Is the amino acid but not the nucleotide sequence of the Escherichia coli araC gene conserved? J Mol Biol. 1982;154:649–652. doi: 10.1016/s0022-2836(82)80020-x. [DOI] [PubMed] [Google Scholar]
- 4238.Stoner C M, Schleif R. The araE low affinity l-arabinose transport promoter. Cloning, sequence, transcription start site and DNA binding sites of regulatory proteins. J Mol Biol. 1983;171:369–381. doi: 10.1016/0022-2836(83)90035-9. [DOI] [PubMed] [Google Scholar]
- 4239.Storts D R X, Markovitz A. Construction and characterization of mutations in hupB, the gene encoding HU-β (HU-1) in Escherichia coli K-12. J Bacteriol. 1988;170:1541–1547. doi: 10.1128/jb.170.4.1541-1547.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4240.Storts D R X, Markovitz A. A novel rho promoter::Tn10 mutation suppresses an ftsQ1(Ts) missense mutation in an essential Escherichia coli cell division gene by a mechanism not involving polarity suppression. J Bacteriol. 1991;173:655–663. doi: 10.1128/jb.173.2.655-663.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4241.Storz G, Jacobson F S, Tartaglia L A, Morgan R W, Silveira L A, Ames B N. An alkyl hydroperoxide reductase induced by oxidative stress in Salmonella typhimurium and Escherichia coli: genetic characterization and cloning of ahp. J Bacteriol. 1989;171:2049–2055. doi: 10.1128/jb.171.4.2049-2055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4242.Storz G, Tartaglia L A, Ames B N. OxyR, a transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science. 1990;248:189–194. doi: 10.1126/science.2183352. [DOI] [PubMed] [Google Scholar]
- 4243.Stout V. Identification of the promoter region for the colanic acid polysaccharide biosynthetic genes in Escherichia coli K-12. J Bacteriol. 1996;178:4273–4280. doi: 10.1128/jb.178.14.4273-4280.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4244.Stout V, Torres-Cabassa A S, Maurizi M R, Gutnick D, Gottesman S. RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J Bacteriol. 1991;173:1738–1747. doi: 10.1128/jb.173.5.1738-1747.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4245.Stout V, Gottesman S. RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J Bacteriol. 1990;172:659–669. doi: 10.1128/jb.172.2.659-669.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4246.Stouthamer A H. A genetical and biochemical study of chlorate-resistant mutants of Salmonella typhimurium. Antonie Leeuwenhoek J Microbiol Serol. 1969;35:505–521. doi: 10.1007/BF02219168. [DOI] [PubMed] [Google Scholar]
- 4247.Stragier P, Richaud F, Borne F, Patte J C. Regulation of diaminopimelate decarboxylase synthesis in Escherichia coli. I. Identification of a lysR gene encoding an activator of the lysA gene. J Mol Biol. 1983;168:307–320. doi: 10.1016/s0022-2836(83)80020-5. [DOI] [PubMed] [Google Scholar]
- 4248.Stragier P, Patte J C. Regulation of diaminopimelate decarboxylase synthesis in Escherichia coli. III. Nucleotide sequence and regulation of the lysR gene. J Mol Biol. 1983;168:333–350. doi: 10.1016/s0022-2836(83)80022-9. [DOI] [PubMed] [Google Scholar]
- 4249.Stragier P, Danos O, Patte J C. Regulation of diaminopimelate decarboxylase synthesis in Escherichia coli. II. Nucleotide sequence of the lysA gene and its regulatory region. J Mol Biol. 1983;168:321–331. doi: 10.1016/s0022-2836(83)80021-7. [DOI] [PubMed] [Google Scholar]
- 4250.Straney R, Krah R, Menzel R. Mutations in the −10 TATAAT sequence of the gyrA promoter affect both promoter strength and sensitivity to DNA supercoiling. J Bacteriol. 1994;176:5999–6006. doi: 10.1128/jb.176.19.5999-6006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4251.Strauch K L, Johnson K R, Beckwith J R. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J Bacteriol. 1989;171:2689–2696. doi: 10.1128/jb.171.5.2689-2696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4252.Straus D, Walter W, Gross C A. DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of sigma 32. Genes Dev. 1990;4:2202–2209. doi: 10.1101/gad.4.12a.2202. [DOI] [PubMed] [Google Scholar]
- 4253.Strom A R, Kaasen I. Trehalose metabolism in Escherichia coli: stress protection and stress regulation of gene expression. Mol Microbiol. 1993;8:205–210. doi: 10.1111/j.1365-2958.1993.tb01564.x. [DOI] [PubMed] [Google Scholar]
- 4254.Studwell-Vaughan P S, O’Donnell M E. DNA polymerase III accessory proteins. V. Theta encoded by holE. J Biol Chem. 1993;268:11785–11791. [PubMed] [Google Scholar]
- 4255.Stuitje A R, deWind N, van der Spek J C, Pors T H, Meijer M. Dissection of promoter sequences involved in transcriptional activation of the Escherichia coli replication origin. Nucleic Acids Res. 1986;14:2333–2344. doi: 10.1093/nar/14.5.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4256.Sturla L, Bisso A, Zanardi D, Benatti U, De Flora A, Tonetti M. Expression, purification and characterization of GDP-d-mannose 4,6-dehydratase from Escherichia coli. FEBS Lett. 1997;412:126–130. doi: 10.1016/s0014-5793(97)00762-x. [DOI] [PubMed] [Google Scholar]
- 4257.Styrvold O B, Falkenberg P, Landfald B, Eshoo M W, Bjornsen T, Strom A R. Selection, mapping, and characterization of osmoregulatory mutants of Escherichia coli blocked in the choline-glycine betaine pathway. J Bacteriol. 1986;165:856–863. doi: 10.1128/jb.165.3.856-863.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4258.Su H S, Lang B F, Newman E B. l-Serine degradation in Escherichia coli K-12: cloning and sequencing of the sdaA gene. J Bacteriol. 1989;171:5095–5102. doi: 10.1128/jb.171.9.5095-5102.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4259.Su H S, Newman E B. A novel l-serine deaminase activity in Escherichia coli K-12. J Bacteriol. 1991;173:2473–2480. doi: 10.1128/jb.173.8.2473-2480.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4260.Su H S, Moniakis J, Newman E B. Use of gene fusions of the structural gene sdaA to purify l-serine deaminase 1 from Escherichia coli K-12. Eur J Biochem. 1993;211:521–527. doi: 10.1111/j.1432-1033.1993.tb17578.x. [DOI] [PubMed] [Google Scholar]
- 4261.Su T-Z, Schweizer H P, Oxender D L. Carbon-starvation induction of the ugp operon, encoding the binding protein-dependent sn-glycerol-3-phosphate transport system in Escherichia coli. Mol Gen Genet. 1991;230:28–32. doi: 10.1007/BF00290646. [DOI] [PubMed] [Google Scholar]
- 4262.Subrahmanyam C S, McCorkle G M, Umbarger H E. Physical location of the ilvO determinant in Escherichia coli K-12 deoxyribonucleic acid. J Bacteriol. 1980;142:547–555. doi: 10.1128/jb.142.2.547-555.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4263.Subrahmanyam C S, Noti J D, Umbarger H E. Regulation of ilvEDA expression occurs upstream of ilvG in Escherichia coli: additional evidence for an ilvGEDA operon. J Bacteriol. 1980;144:279–290. doi: 10.1128/jb.144.1.279-290.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4264.Sugai M, Wu H C. Export of the outer membrane lipoprotein is defective in secD, secE, and secF mutants of Escherichia coli. J Bacteriol. 1992;174:2511–2516. doi: 10.1128/jb.174.8.2511-2516.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4265.Sugimura K, Nishihara T. Purification, characterization, and primary structure of Escherichia coli protease VII with specificity for paired basic residues: identity of protease VII and OmpT. J Bacteriol. 1988;170:5625–5632. doi: 10.1128/jb.170.12.5625-5632.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4266.Sugiura A, Nakashima K, Tanaka K, Mizuno T. Clarification of the structural and functional features of the osmoregulated kdp operon of Escherichia coli. Mol Microbiol. 1992;6:1769–1776. doi: 10.1111/j.1365-2958.1992.tb01349.x. [DOI] [PubMed] [Google Scholar]
- 4267.Sukharev S I, Blount P, Martinac B, Kung C. Mechanosensitive channels of Escherichia coli: the MscL gene, protein, and activities. Annu Rev Physiol. 1997;59:633–657. doi: 10.1146/annurev.physiol.59.1.633. [DOI] [PubMed] [Google Scholar]
- 4268.Sukharev S I, Blount P, Martinac B, Blattner F R, Kung C. A large-conductance mechanosensitive channel in E. coli encoded by mscL alone. Nature. 1994;368:265–268. doi: 10.1038/368265a0. [DOI] [PubMed] [Google Scholar]
- 4269.Sukharev S I, Blount P, Martinac B, Guy H R, Kung C. MscL: a mechanosensitive channel in Escherichia coli. Soc Gen Physiol Ser. 1996;51:133–141. [PubMed] [Google Scholar]
- 4269a.Sukhodolets M V, Jin D J. RapA, a novel RNA polymerase-associated protein, is a bacterial homolog of SWI2/SNF2. J Biol Chem. 1998;273:7018–7023. doi: 10.1074/jbc.273.12.7018. [DOI] [PubMed] [Google Scholar]
- 4270.Sukhodolets V V, Dukhii D E. Unequal crossing over in Escherichia coli: genetic and physical mapping of duplications isolated in conjugational matings. Genetika. 1996;32:42–52. . (In Russian.) [PubMed] [Google Scholar]
- 4271.Sulavik M C, Gambino L F, Miller P F. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol Med. 1995;1:436–446. [PMC free article] [PubMed] [Google Scholar]
- 4272.Sullivan M A, Cannon J F, Webb F H, Bock A. Antisuppressor mutation in Escherichia coli defective in biosynthesis of 5-methylaminomethyl-2-thiouridine. J Bacteriol. 1985;161:368–376. doi: 10.1128/jb.161.1.368-376.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4273.Sullivan M A, Bock R M. Isolation and characterization of antisuppressor mutations in Escherichia coli. J Bacteriol. 1985;161:377–384. doi: 10.1128/jb.161.1.377-384.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4274.Sullivan N F, Donachie W D. Overlapping functional units in a cell division gene cluster in Escherichia coli. J Bacteriol. 1984;158:1198–1201. doi: 10.1128/jb.158.3.1198-1201.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4275.Sullivan N F, Donachie W D. Transcriptional organization within an Escherichia coli cell division gene cluster: direction of transcription of the cell separation gene envA. J Bacteriol. 1984;160:724–732. doi: 10.1128/jb.160.2.724-732.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4276.Sullivan S L, Gottesman M E. Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell. 1991;58:989–994. doi: 10.1016/0092-8674(92)90041-a. [DOI] [PubMed] [Google Scholar]
- 4277.Sumiya M, Davis E O, Packman L C, McDonald T P, Henderson P J. Molecular genetics of a receptor protein for d-xylose, encoded by the gene xylF, in Escherichia coli. Recept Chan. 1995;3:117–128. [PubMed] [Google Scholar]
- 4278.Sun L, Jacobson B A, Dien B S, Srienc F, Fuchs J A. Cell cycle regulation of the Escherichia coli nrd operon: requirement for a cis-acting upstream AT-rich sequence. J Bacteriol. 1994;176:2415–2426. doi: 10.1128/jb.176.8.2415-2426.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4279.Sun L, Fuchs J A. Escherichia coli ribonucleotide reductase expression in cell cycle regulated. Mol Biol Cell. 1992;3:1095–1105. doi: 10.1091/mbc.3.10.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4280.Sun L, Fuchs J A. Regulation of the Escherichia coli nrd operon: role of DNA supercoiling. J Bacteriol. 1994;176:4617–4626. doi: 10.1128/jb.176.15.4617-4626.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4281.Sun T-P, Webster R E. fii, a bacterial locus required for filamentous phage infection, and its relation to colicin-tolerant tolA and tolB. J Bacteriol. 1986;165:107–115. doi: 10.1128/jb.165.1.107-115.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4282.Sun T-P, Webster R E. Nucleotide sequence of a gene cluster involved in entry of E colicins and single-stranded DNA of infecting filamentous bacteriophages into Escherichia coli. J Bacteriol. 1987;169:2667–2674. doi: 10.1128/jb.169.6.2667-2674.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4283.Sun X, Eliasson R, Pontis E, Andersson J, Buist G, Sjoberg B M, Reichard P. Generation of the glycyl radical of the anaerobic Escherichia coli ribonucleotide reductase requires a specific activating enzyme. J Biol Chem. 1995;270:2443–2446. doi: 10.1074/jbc.270.6.2443. [DOI] [PubMed] [Google Scholar]
- 4284.Sun X Y, Harder J, Krook M, Jornvall H, Sjoberg B-M, Reichard P. A possible glycine radical in anaerobic ribonucleotide reductase from Escherichia coli: nucleotide sequence of the cloned nrdD gene. Proc Natl Acad Sci USA. 1993;90:577–581. doi: 10.1073/pnas.90.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4285.Sundararajan T A. Interference with glycerokinase induction in mutants of E. coli accumulating gal-1-P. Proc Natl Acad Sci USA. 1963;50:463–469. doi: 10.1073/pnas.50.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4286.Sung Y-C, Parsell D, Anderson P M, Fuchs J A. Identification, mapping, and cloning of the gene encoding cyanase in Escherichia coli K-12. J Bacteriol. 1987;169:2639–2642. doi: 10.1128/jb.169.6.2639-2642.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4287.Sung Y-C, Fuchs J A. Characterization of the cyn operon in Escherichia coli K12. J Biol Chem. 1988;263:14769–14775. [PubMed] [Google Scholar]
- 4288.Sung Y-C, Fuchs J A. The Escherichia coli K-12 cyn operon is positively regulated by a member of the lysR family. J Bacteriol. 1992;174:3645–3650. doi: 10.1128/jb.174.11.3645-3650.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4289.Sung Y-C, Anderson P M, Fuchs J A. Characterization of high-level expression and sequencing of the Escherichia coli K-12 cynS gene encoding cyanase. J Bacteriol. 1987;169:5224–5230. doi: 10.1128/jb.169.11.5224-5230.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4290.Sunnarborg A, Klumpp D J, Chung T, LaPorte D C. Regulation of the glyoxylate bypass operon: cloning and characterization of iclR. J Bacteriol. 1990;172:2642–2649. doi: 10.1128/jb.172.5.2642-2649.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4291.Sunshine M G, Kelly B. Extent of host deletions associated with bacteriophage P2-mediated education. J Bacteriol. 1971;108:695–704. doi: 10.1128/jb.108.2.695-704.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4292.Sunshine M, Sauer B. A bacterial mutation blocking P2 phage late gene expression. Proc Natl Acad Sci USA. 1975;72:2770–2774. doi: 10.1073/pnas.72.7.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4293.Sunshine M, Feiss M, Stuart J, Yochem J. A new host gene (groPC) necessary for lambda DNA replication. Mol Gen Genet. 1977;151:27–34. doi: 10.1007/BF00446909. [DOI] [PubMed] [Google Scholar]
- 4294.Suppmann B, Sawers G. Isolation and characterization of hypophosphite-resistant mutants of Escherichia coli: identification of the FocA protein, encoded by the pfl operon, as a putative formate transporter. Mol Microbiol. 1994;11:965–982. doi: 10.1111/j.1365-2958.1994.tb00375.x. [DOI] [PubMed] [Google Scholar]
- 4295.Surin B P, Jans D A, Fimmel A L, Shaw D C, Cox G B, Rosenberg H. Structural gene for the phosphate-repressible phosphate-binding protein of Escherichia coli has its own promoter: complete nucleotide sequence of the phoS gene. J Bacteriol. 1984;157:772–778. doi: 10.1128/jb.157.3.772-778.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4296.Surin B P, Rosenberg H, Cox G B. Phosphate-specific transport system of Escherichia coli: nucleotide sequence and gene-polypeptide relationships. J Bacteriol. 1985;161:189–198. doi: 10.1128/jb.161.1.189-198.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4297.Sutherland L, Cairney J, Elmore M J, Booth I R, Higgins C F. Osmotic regulation of transcription: induction of the proU betaine transport gene is dependent on accumulation of intracellular potassium. J Bacteriol. 1986;168:805–814. doi: 10.1128/jb.168.2.805-814.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4298.Sutherland P, McAlister-Henn L. Isolation and expression of the Escherichia coli gene encoding malate dehydrogenase. J Bacteriol. 1985;163:1074–1077. doi: 10.1128/jb.163.3.1074-1079.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4299.Sutton A, Newman T, Francis M, Freundlich M. Valine-resistant Escherichia coli K-12 strains with mutations in the ilvB operon. J Bacteriol. 1981;148:998–1001. doi: 10.1128/jb.148.3.998-1001.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4300.Sutton M D, Kaguni J M. Novel alleles of the Escherichia coli dnaA gene are defective in replication of pSC101 but not of oriC. J Bacteriol. 1995;177:6657–6665. doi: 10.1128/jb.177.22.6657-6665.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4301.Sutton M D, Kaguni J M. Novel alleles of the Escherichia coli dnaA gene. J Mol Biol. 1997;271:693–703. doi: 10.1006/jmbi.1997.1209. [DOI] [PubMed] [Google Scholar]
- 4302.Sutton M R, Fall R R, Nervi A M, Alberts A W, Vagelos P R, Bradshaw R A. Amino acid sequence of Escherichia coli biotin carboxyl carrier protein (9100) J Biol Chem. 1977;252:3934–3940. [PubMed] [Google Scholar]
- 4303.Suzuki H, Kumagai H, Echigo T, Tochikura T. Molecular cloning of Escherichia coli K-12 ggt and rapid isolation of γ-glutamyltranspeptidase. Biochem Biophys Res Commun. 1988;150:33–38. doi: 10.1016/0006-291x(88)90482-2. [DOI] [PubMed] [Google Scholar]
- 4304.Suzuki H, Kumagai H, Echigo T, Tochikura T. DNA sequence of the Escherichia coli K-12 γ-glutamyltranspeptidase gene, ggt. J Bacteriol. 1989;171:5169–5172. doi: 10.1128/jb.171.9.5169-5172.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4305.Suzuki H, Kumagai H, Tochikura T. Isolation, genetic mapping, and characterization of Escherichia coli K-12 mutants lacking γ-glutamyltranspeptidase. J Bacteriol. 1987;169:3926–3931. doi: 10.1128/jb.169.9.3926-3931.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4306.Suzuki H, Hashimoto W, Kumagai H. Escherichia coli K-12 can utilize an exogenous γ-glutamyl peptide as an amino acid source, for which γ-glutamyltranspeptidase is essential. J Bacteriol. 1993;175:6038–6040. doi: 10.1128/jb.175.18.6038-6040.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4307.Suzuki H, Van Heijenoort Y, Tamura T, Mizoguchi J, Hirota Y, Van Heijenoort J. In vitro peptidoglycan polymerization catalyzed by penicillin binding protein 1b of Escherichia coli K-12. FEBS Lett. 1980;110:245–249. doi: 10.1016/0014-5793(80)80083-4. [DOI] [PubMed] [Google Scholar]
- 4308.Suzuki T, Itoh A, Ichihara S, Mizushima S. Characterization of the sppA gene coding for protease IV, a signal peptide peptidase of Escherichia coli. J Bacteriol. 1987;169:2523–2528. doi: 10.1128/jb.169.6.2523-2528.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4309.Suzuki T, Ueguchi C, Mizuno T. H-NS regulates OmpF expression through micF antisense RNA in Escherichia coli. J Bacteriol. 1996;178:3650–3653. doi: 10.1128/jb.178.12.3650-3653.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4310.Svitil A L, Cashel M, Zyskind J W. Guanosine tetraphosphate inhibits protein synthesis in vivo. A possible protective mechanism for starvation stress in Escherichia coli. J Biol Chem. 1993;268:2307–2311. [PubMed] [Google Scholar]
- 4311.Swanberg S L, Wang J C. Cloning and sequencing of the Escherichia coli gyrA gene coding for the A subunit of DNA gyrase. J Mol Biol. 1987;197:729–736. doi: 10.1016/0022-2836(87)90479-7. [DOI] [PubMed] [Google Scholar]
- 4312.Swedberg G, Castensson S, Skold O. Characterization of mutationally altered dihydropteroate synthase and its ability to form a sulfonamide-containing dihydrofolate analog. J Bacteriol. 1979;137:129–136. doi: 10.1128/jb.137.1.129-136.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4313.Sweet G, Gandor C, Voegele R, Wittekindt N, Beuerle J, Truniger V, Lin E C C, Boos W. Glycerol facilitator of Escherichia coli: cloning of glpF and identification of the glpF product. J Bacteriol. 1990;172:424–430. doi: 10.1128/jb.172.1.424-430.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4314.Swift S, Kuhn J, Stewart G S. Selection and analysis of non-interactive mutants in the Escherichia coli tryptophan synthase alpha subunit. Mol Gen Genet. 1992;233:129–135. doi: 10.1007/BF00587570. [DOI] [PubMed] [Google Scholar]
- 4315.Swindle J, Ajioka J, Dawson D, Myers R, Carroll D, Georgopoulos C. The nucleotide sequence of the Escherichia coli K12 nusB (groNB) gene. Nucleic Acids Res. 1984;12:4977–4985. doi: 10.1093/nar/12.12.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4316.Szekely E, Simon M. DNA sequence adjacent to flagellar genes and evolution of flagellar-phase variation. J Bacteriol. 1983;155:74–81. doi: 10.1128/jb.155.1.74-81.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4317.Szentirmai A, Szentirmai M, Umbarger H E. Isoleucine and valine metabolism of Escherichia coli. XV. Biochemical properties of mutants resistant to thiaisoleucine. J Bacteriol. 1968;95:1672–1679. doi: 10.1128/jb.95.5.1672-1679.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4318.Szumanski M B, Boyle S M. Analysis and sequence of the speB gene encoding agmatine ureohydrolase, a putrescine biosynthetic enzyme in Escherichia coli. J Bacteriol. 1990;172:538–547. doi: 10.1128/jb.172.2.538-547.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4319.Szybalski E H, Szybalski W. A physical map of the Escherichia coli bio operon. Gene. 1982;19:93–103. doi: 10.1016/0378-1119(82)90193-7. [DOI] [PubMed] [Google Scholar]
- 4320.Ta D T, Seaton B L, Vickery L E. Localization of the ferredoxin (fdx) gene on the physical map of the Escherichia coli chromosome. J Bacteriol. 1992;174:5760–5761. doi: 10.1128/jb.174.17.5760-5761.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4321.Ta D T, Vickery L E. Cloning, sequencing, and overexpression of a [2FE-2S] ferredoxin gene from Escherichia coli. J Biol Chem. 1992;267:11120–11125. [PubMed] [Google Scholar]
- 4322.Tabata S, Oka A, Sugimoto K, Takanami M, Yasuda S, Hirota Y. The 245 base-pair oriC sequence of the E. coli chromosome directs bidirectional replication at an adjacent region. Nucleic Acids Res. 1983;11:2617–2626. doi: 10.1093/nar/11.9.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4323.Tabor C W, Tabor H. The speEspeD operon of Escherichia coli. Formation and processing of a proenzyme form of S adenosylmethionine decarboxylase. J Biol Chem. 1987;262:16037–16040. [PubMed] [Google Scholar]
- 4324.Tabor C W, Tabor H, Xie Q-W. Spermidine synthase of Escherichia coli: localization of the speE gene. Proc Natl Acad Sci USA. 1986;83:6040–6044. doi: 10.1073/pnas.83.16.6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4325.Tabor H, Hafner E W, Tabor C W. Construction of an Escherichia coli strain unable to synthesize putrescine, spermidine, or cadaverine: characterization of two genes controlling lysine decarboxylase. J Bacteriol. 1980;144:952–956. doi: 10.1128/jb.144.3.952-956.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4326.Taglicht D, Padan E, Schuldiner S. Overproduction and purification of a functional Na+/H+ antiporter coded by nhaA (ant) from Escherichia coli. J Biol Chem. 1991;266:11289–11294. [PubMed] [Google Scholar]
- 4327.Takagi J S, Ida N, Tokushige M, Sakamoto H, Shimura Y. Cloning and nucleotide sequence of the aspartase gene of Escherichia coli W. Nucleic Acids Res. 1985;13:2063–2074. doi: 10.1093/nar/13.6.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4328.Takahagi M, Iwasaki H, Nakata A, Shinagawa H. Molecular analysis of the Escherichia coli ruvC gene, which encodes a Holliday junction-specific endonuclease. J Bacteriol. 1991;173:5747–5753. doi: 10.1128/jb.173.18.5747-5753.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4329.Takahashi H. Genetic and physiological characterization of Escherichia coli K12 mutants (tabC) which induce the abortive infection of bacteriophage T4. Virology. 1978;87:256–265. doi: 10.1016/0042-6822(78)90131-9. [DOI] [PubMed] [Google Scholar]
- 4330.Takano K, Nakamura T, Sekiguchi M. Roles of two types of O6-methylguanine-DNA methyltransferases in DNA repair. Mutat Res. 1991;254:37–44. doi: 10.1016/0921-8777(91)90038-q. [DOI] [PubMed] [Google Scholar]
- 4331.Takano K, Nakabeppu Y, Maki H, Horiuchi T, Sekiguchi M. Structure and function of dnaQ and mutD mutators of Escherichia coli. Mol Gen Genet. 1986;205:9–13. doi: 10.1007/BF02428026. [DOI] [PubMed] [Google Scholar]
- 4332.Takase I, Ishino F, Wachi M, Kamada H, Doi M, Asoh S, Matsuzawa H, Ohta T, Matsuhashi M. Genes encoding two lipoproteins in the leuS-dacA region of the Escherichia coli chromosome. J Bacteriol. 1987;169:5692–5699. doi: 10.1128/jb.169.12.5692-5699.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4332a.Takata R, Isaksson L A. The temperature sensitive mutant 72C. II. Accumulation at high temperature of ppGpp and pppGpp in the presence of protein synthesis. Mol Gen Genet. 1978;161:15–21. doi: 10.1007/BF00266610. [DOI] [PubMed] [Google Scholar]
- 4333.Takata R, Mukai T, Hori K. Attenuation and processing of RNA from the rpsO-pnp transcription unit of Escherichia coli. Nucleic Acids Res. 1985;13:7289–7297. doi: 10.1093/nar/13.20.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4334.Takata R, Mukai T, Aoyagi M, Hori K. Nucleotide sequence of the gene for Escherichia coli ribosomal protein S15 (rpsO) Mol Gen Genet. 1984;197:225–229. doi: 10.1007/BF00330967. [DOI] [PubMed] [Google Scholar]
- 4335.Takayanagi K, Maeda S, Mizuno T. Expression of micF involved in porin synthesis in Escherichia coli: two distinct cis-acting elements respectively regulate micF expression positively and negatively. FEMS Microbiol Lett. 1991;67:39–44. doi: 10.1016/0378-1097(91)90440-l. [DOI] [PubMed] [Google Scholar]
- 4336.Takebe Y, Kaziro Y. In vitro construction of the tufB-lacZ fusion: analysis of the regulatory mechanism of tufB promoter. Mol Gen Genet. 1982;187:355–363. doi: 10.1007/BF00332612. [DOI] [PubMed] [Google Scholar]
- 4337.Takeda Y, Nishimura A, Nishimura Y, Yamada M, Yasuda S, Suzuki H, Hirota Y. Synthetic ColE1 plasmids carrying genes for penicillin-binding protein in Escherichia coli. Plasmid. 1981;6:86–98. doi: 10.1016/0147-619x(81)90056-1. [DOI] [PubMed] [Google Scholar]
- 4338.Takeda Y, Avila H. Structure and gene expression of the E. coli Mn-superoxide dismutase gene. Nucleic Acids Res. 1986;14:4577–4589. doi: 10.1093/nar/14.11.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4339.Takiff H E, Chen S M, Court D L. Genetic analysis of the rnc operon of Escherichia coli. J Bacteriol. 1989;171:2581–2590. doi: 10.1128/jb.171.5.2581-2590.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4340.Takiff H E, Baker T, Copeland T, Chen S M, Court D L. Locating essential Escherichia coli genes by using mini-Tn10 transposons: the pdxJ operon. J Bacteriol. 1992;174:1544–1553. doi: 10.1128/jb.174.5.1544-1553.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4341.Talarico T L, Ray P H, Dev I K, Merrill B M, Dallas W S. Cloning, sequence analysis, and overexpression of Escherichia coli folK, the gene coding for 7,8-dihydro-6-hydroxymethylpterin-pyrophosphokinase. J Bacteriol. 1992;174:5971–5977. doi: 10.1128/jb.174.18.5971-5977.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4342.Talens A, Boon K, Kraal B, Bosch L. Translational activities of EF-Tu [G222D] which cannot be reconciled with the classical scheme of the polypeptide chain elongation cycle. Biochem Biophys Res Commun. 1996;225:961–967. doi: 10.1006/bbrc.1996.1279. [DOI] [PubMed] [Google Scholar]
- 4343.Tamaki S, Matsuzawa H, Matsuhashi M. Cluster of mrdA and mrdB genes responsible for the rod shape and mecillinam sensitivity of Escherichia coli. J Bacteriol. 1980;141:52–57. doi: 10.1128/jb.141.1.52-57.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4344.Tamaki S, Sato T, Matsuhashi M. Role of lipopolysaccharides in antibiotic resistance and bacteriophage adsorption of Escherichia coli K-12. J Bacteriol. 1971;105:968–975. doi: 10.1128/jb.105.3.968-975.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4345.Tamura F, Nishimura S, Ohki M. The E. coli divE mutation, which differently inhibits synthesis of certain proteins, is in tRNA1Ser. EMBO J. 1984;3:1103–1107. doi: 10.1002/j.1460-2075.1984.tb01936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4346.Tamura T, Suzuki H, Nishimura Y, Mizoguchi J, Hirota Y. On the process of cellular division in Escherichia coli: isolation and characterization of penicillin-binding proteins 1a, 1b, and 3. Proc Natl Acad Sci USA. 1980;77:4499–4503. doi: 10.1073/pnas.77.8.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4347.Tanabe H, Goldstein J, Yang M, Inouye M. Identification of the promoter region of the Escherichia coli major cold shock gene, cspA. J Bacteriol. 1992;174:3867–3873. doi: 10.1128/jb.174.12.3867-3873.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4348.Tanaka S, Matsushita Y, Yoshikawa A, Isono K. Cloning and molecular characterization of the gene rimL which encodes an enzyme acetylating ribosomal protein L12 of Escherichia coli K12. Mol Gen Genet. 1989;217:289–293. doi: 10.1007/BF02464895. [DOI] [PubMed] [Google Scholar]
- 4349.Tao J S, Ishiguro E E. Nucleotide sequence of the murE gene of Escherichia coli. Can J Microbiol. 1989;35:1051–1054. doi: 10.1139/m89-175. [DOI] [PubMed] [Google Scholar]
- 4350.Tao K. oxyR-dependent induction of Escherichia coli grx gene expression by peroxide stress. J Bacteriol. 1997;179:5967–5970. doi: 10.1128/jb.179.18.5967-5970.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4351.Tao K, Makino K, Yonei S, Nakata A, Shinagawa H. Molecular cloning and nucleotide sequencing of oxyR, the positive regulatory gene of a regulon for an adaptive response to oxidative stress in Escherichia coli: homologies between OxyR protein and a family of bacterial activator proteins. Mol Gen Genet. 1989;218:371–376. doi: 10.1007/BF00332397. [DOI] [PubMed] [Google Scholar]
- 4352.Tapio S, Yeh F, Shuman H, Boos W. The malZ gene of Escherichia coli, a member of the maltose regulon, encodes a maltodextrin glucosidase. J Biol Chem. 1991;266:19450–19458. [PubMed] [Google Scholar]
- 4353.Taraseviciene L, Miczak A, Apirion D. The gene specifying RNase E (rne) and a gene affecting mRNA stability (ams) are the same gene. Mol Microbiol. 1991;5:851–855. doi: 10.1111/j.1365-2958.1991.tb00758.x. [DOI] [PubMed] [Google Scholar]
- 4354.Taraseviciene L, Bjork G R, Uhlin B E. Evidence for an RNA binding region in the Escherichia coli processing endoribonuclease RNase E. J Biol Chem. 1995;270:26391–26398. doi: 10.1074/jbc.270.44.26391. [DOI] [PubMed] [Google Scholar]
- 4355.Tardat B, Touati D. Two global regulators repress the anaerobic expression of MnSOD in Escherichia coli: Fur (ferric uptake regulation) and Arc (aerobic respiration control) Mol Microbiol. 1991;5:455–465. doi: 10.1111/j.1365-2958.1991.tb02129.x. [DOI] [PubMed] [Google Scholar]
- 4356.Tartaglia L A, Storz G, Ames B N. Identification and molecular analysis of oxyR-regulated promoters important for the bacterial adaptation to oxidative stress. J Mol Biol. 1989;210:709–719. doi: 10.1016/0022-2836(89)90104-6. [DOI] [PubMed] [Google Scholar]
- 4357.Taschner P E M, Verest J G J, Woldringh C L. Genetic and morphological characterization of ftsB and nrdB mutants of Escherichia coli. J Bacteriol. 1987;169:19–25. doi: 10.1128/jb.169.1.19-25.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4358.Tate C G, Muiry J A, Henderson P J F. Mapping, cloning, expression, and sequencing of the rhaT gene, which encodes a novel l-rhamnose-H+ transport protein in Salmonella typhimurium and Escherichia coli. J Biol Chem. 1992;267:6923–6932. [PubMed] [Google Scholar]
- 4359.Tate C G, Henderson P J F. Membrane topology of the l-rhamnose-H+ transport protein (RhaT) from enterobacteria. J Biol Chem. 1993;268:26850–26857. [PubMed] [Google Scholar]
- 4360.Taucher-Scholz G, Hoffmann-Berling H. Identification of the gene for DNA helicase II of Escherichia coli. Eur J Biochem. 1983;137:573–580. doi: 10.1111/j.1432-1033.1983.tb07864.x. [DOI] [PubMed] [Google Scholar]
- 4361.Taura T, Ueguchi C, Shiba K, Ito K. Insertional disruption of the nusB (ssyB) gene leads to cold-sensitive growth of Escherichia coli and suppression of the secY24 mutation. Mol Gen Genet. 1992;234:429–432. doi: 10.1007/BF00538702. [DOI] [PubMed] [Google Scholar]
- 4362.Taura T, Baba T, Akiyama Y, Ito K. Determinants of the quantity of the stable SecY complex in the Escherichia coli cell. J Bacteriol. 1993;175:7771–7775. doi: 10.1128/jb.175.24.7771-7775.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4363.Tawa P, Stewart R C. Mutational activation of CheA, the protein kinase in the chemotaxis system of Escherichia coli. J Bacteriol. 1994;176:4210–4218. doi: 10.1128/jb.176.14.4210-4218.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4364.Taylor A F. Movement and resolution of Holliday junctions by enzymes from E. coli. Cell. 1992;69:1063–1065. doi: 10.1016/0092-8674(92)90626-n. [DOI] [PubMed] [Google Scholar]
- 4365.Taylor A F, Smith G R. Monomeric RecBCD enzyme binds and unwinds DNA. J Biol Chem. 1995;270:24451–24458. doi: 10.1074/jbc.270.41.24451. [DOI] [PubMed] [Google Scholar]
- 4366.Taylor A F, Smith G R. Strand specificity of nicking of DNA at Chi sites by RecBCD enzyme. Modulation by ATP and magnesium levels. J Biol Chem. 1995;270:24459–24467. doi: 10.1074/jbc.270.41.24459. [DOI] [PubMed] [Google Scholar]
- 4367.Taylor A F, Siciliano P G, Weiss B. Cloning of the dut (deoxyuridine triphosphatase) gene of Escherichia coli. Gene. 1980;9:321–336. doi: 10.1016/0378-1119(90)90330-t. [DOI] [PubMed] [Google Scholar]
- 4368.Taylor A L. Current linkage map of Escherichia coli. Bacteriol Rev. 1970;34:155–175. doi: 10.1128/br.34.2.155-175.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4369.Taylor A L, Trotter C D. Revised linkage map of Escherichia coli. Bacteriol Rev. 1967;31:332–353. doi: 10.1128/br.31.4.332-353.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4370.Taylor A L, Adelberg E A. Linkage analysis with very high frequency males of Escherichia coli. Genetics. 1960;45:1233–1243. doi: 10.1093/genetics/45.9.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4371.Taylor A L, Thoman M S. The genetic map of Escherichia coli K-12. Genetics. 1964;50:659–663. doi: 10.1093/genetics/50.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4372.Taylor D E, Hou Y, Turner R J, Weiner J H. Location of a potassium tellurite resistance operon (tehA tehB) within the terminus of Escherichia coli K-12. J Bacteriol. 1994;176:2740–2742. doi: 10.1128/jb.176.9.2740-2742.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4373.Taylor J, Bedbrook J R, Grant F J, Kleinhofs A. Reconstitution of plant nitrate reductase by Escherichia coli extracts and the molecular cloning of chlA gene of Escherichia coli K-12. J Mol Appl Genet. 1983;2:261–271. [PubMed] [Google Scholar]
- 4374.Taylor R K, Hall M N, Enquist L, Silhavy T J. Identification of OmpR: a positive regulatory protein controlling expression of the major outer membrane matrix porin proteins of Escherichia coli K-12. J Bacteriol. 1981;147:255–258. doi: 10.1128/jb.147.1.255-258.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4375.Taylor R K, Garrett S, Sodergren E J, Silhavy T J. Mutations that define the promoter of ompF, a gene specifying a major outer membrane porin protein. J Bacteriol. 1985;162:1054–1060. doi: 10.1128/jb.162.3.1054-1060.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4376.Taylor W E, Straus D B, Grossman A D, Burton Z F, Gross C A, Burgess R R. Transcription from a heat-inducible promoter causes heat shock regulation of the sigma subunit of E. coli RNA polymerase. Cell. 1984;38:371–381. doi: 10.1016/0092-8674(84)90492-6. [DOI] [PubMed] [Google Scholar]
- 4377.Taylor W E, Burgess R R. Escherichia coli RNA polymerase binding and initiation of transcription on fragments of λ rifd 18 DNA containing promoters for λ genes and for rrnB, tufB, rplC, A, rplJ, L, and rpoB,C genes. Gene. 1979;6:331–365. doi: 10.1016/0378-1119(79)90073-8. [DOI] [PubMed] [Google Scholar]
- 4378.Tecklenburg M, Naumer A, Nagappan O, Kuempel P. The dif resolvase locus of the Escherichia coli chromosome can be replaced by a 33-bp sequence, but function depends on location. Proc Natl Acad Sci USA. 1995;92:1352–1356. doi: 10.1073/pnas.92.5.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4379.Tedeschi G, Negri A, Mortarino M, Ceciliani F, Simonic T, Faotto L, Ronchi S. l-Aspartate oxidase from Escherichia coli. II. Interaction with C4 dicarboxylic acids and identification of a novel l-aspartate: fumarate oxidoreductase activity. Eur J Biochem. 1996;239:427. doi: 10.1111/j.1432-1033.1996.0427u.x. [DOI] [PubMed] [Google Scholar]
- 4380.Tei H, Murata K, Kimura A. Molecular cloning of the cys genes (cysC, cysD, cysH, cysI, cysJ, and cysG) responsible for cysteine biosynthesis in Escherichia coli K-12. Biotechnol Appl Biochem. 1990;12:212–216. [PubMed] [Google Scholar]
- 4381.Tei H, Murata K, Kimura A. Structure and expression of cysX, the second gene in the Escherichia coli K-12 cysE locus. Biochem Biophys Res Commun. 1990;167:948–955. doi: 10.1016/0006-291x(90)90615-t. [DOI] [PubMed] [Google Scholar]
- 4382.Tei H, Watanabe K, Murata K, Kimura A. Analysis of the Escherichia coli K-12 cysB gene and its product using the method of gene fusion. Biochem Biophys Res Commun. 1990;167:962–969. doi: 10.1016/0006-291x(90)90617-v. [DOI] [PubMed] [Google Scholar]
- 4383.Tenson T, DeBlasio A, Mankin A. A functional peptide encoded in the Escherichia coli 23S rRNA. Proc Natl Acad Sci USA. 1996;93:5641–5646. doi: 10.1073/pnas.93.11.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4384.Tenson T, Mankin A. Comparison of functional peptide encoded in the Escherichia coli 23S rRNA with other peptides involved in cis-regulation of translation. Biochem Cell Biol. 1995;73:1061–1070. doi: 10.1139/o95-114. [DOI] [PubMed] [Google Scholar]
- 4385.Teo I, Sedwick B, Kilpatrick M W, McCarthy T V, Lindahl T. The intracellular signal for induction of resistance to alkylating agents in E. coli. Cell. 1986;45:315–324. doi: 10.1016/0092-8674(86)90396-x. [DOI] [PubMed] [Google Scholar]
- 4386.Tesfa-Selase F, Drabble W T. Regulation of the gua operon of Escherichia coli by the DnaA protein. Mol Gen Genet. 1992;231:256–264. doi: 10.1007/BF00279799. [DOI] [PubMed] [Google Scholar]
- 4387.Teslyar G E, Shavlovskii G M. Localization of genes coding cyclohexrolase [cyclohydrolase] II and riboflavin synthase on the Escherichia coli K-12 chromosome. Cytol Genet. 1983;17:57–59. [PubMed] [Google Scholar]
- 4388.Tessman I, Fassler J S, Bennett D C. Relative map location of the rep and rho genes of Escherichia coli. J Bacteriol. 1982;151:1637–1640. doi: 10.1128/jb.151.3.1637-1640.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4389.Tetart F, Bouché J P. Regulation of the expression of the cell-cycle gene ftsZ by DicF antisense RNA. Division does not require a fixed number of FtsZ molecules. Mol Microbiol. 1992;6:615–620. doi: 10.1111/j.1365-2958.1992.tb01508.x. [DOI] [PubMed] [Google Scholar]
- 4389a.Textor S, Wendisch V F, De Graaf A A, Muller U, Linder M I, Linder D. Propionate oxidation in Escherichia coli: evidence for operation of a methylcitrate cycle in bacteria. Arch Microbiol. 1997;168:428–436. doi: 10.1007/s002030050518. [DOI] [PubMed] [Google Scholar]
- 4390.Thaller M C, Schippa S, Bonci A, Cresti S, Rossolini G M. Identification of the gene (aphA) encoding the class B acid phosphatase/phosphotransferase of Escherichia coli MG1655 and characterization of its product. FEMS Microbiol Lett. 1997;146:191–198. doi: 10.1111/j.1574-6968.1997.tb10192.x. [DOI] [PubMed] [Google Scholar]
- 4391.Thapar R, Nicholson E M, Rajagopal P, Waygood E B, Scholtz J M, Klevit R E. Influence of N-cap mutations on the structure and stability of Escherichia coli HPr. Biochemistry. 1996;35:11268–11277. doi: 10.1021/bi960349s. [DOI] [PubMed] [Google Scholar]
- 4392.Theall G, Low K B, Soll D. Regulation of the biosynthesis of aminoacyl-tRNA synthetases and of tRNA in Escherichia coli. IV. Mutants with increased levels of leucyl- or seryl-tRNA synthetase. Mol Gen Genet. 1979;169:205–211. doi: 10.1007/BF00271672. [DOI] [PubMed] [Google Scholar]
- 4393.Thelen P, Tsuchiya T, Goldberg E B. Characterization and mapping of a major Na+/H+ antiporter gene of Escherichia coli. J Bacteriol. 1991;173:6553–6557. doi: 10.1128/jb.173.20.6553-6557.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4394.Thirion J P, Hofnung M. On some genetic aspects of phage λ resistance in E. coli K12. Genetics. 1972;71:207–216. doi: 10.1093/genetics/71.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4395.Thomas A, Lloyd R G. Altered regulation of the recA gene in Escherichia coli strains carrying a recA-linked suppressor of lexA. Mol Gen Genet. 1980;179:355–358. doi: 10.1007/BF00425464. [DOI] [PubMed] [Google Scholar]
- 4396.Thomas C D, Modha J, Razzaq T M, Cullis P M, Rivett A J. Controlled high-level expression of the lon gene of Escherichia coli allows overproduction of Lon protease. Gene. 1993;136:237–242. doi: 10.1016/0378-1119(93)90471-e. [DOI] [PubMed] [Google Scholar]
- 4397.Thomas J A, Valvano M A. Role of tol genes in cloacin DF13 susceptibility of Escherichia coli K-12 strains expressing the cloacin DF13-aerobactin receptor IutA. J Bacteriol. 1993;175:548–552. doi: 10.1128/jb.175.2.548-552.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4398.Thomas M S, Nomura M. Translational regulation of the L11 ribosomal protein operon of Escherichia coli: mutations that define the target site for repression by L1. Nucleic Acids Res. 1987;15:3085–3096. doi: 10.1093/nar/15.7.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4399.Thomas M S, Drabble W T. Molecular cloning and characterization of the gua regulatory region of Escherichia coli K12. Mol Gen Genet. 1984;195:238–245. doi: 10.1007/BF00332753. [DOI] [PubMed] [Google Scholar]
- 4400.Thomas M S, Drabble W T. Nucleotide sequence and organization of the gua promoter region of Escherichia coli. Gene. 1985;36:45–53. doi: 10.1016/0378-1119(85)90068-x. [DOI] [PubMed] [Google Scholar]
- 4401.Thomas S D, Jordan P M. Nucleotide sequence of the hemC locus encoding porphobilinogen deaminase of Escherichia coli K12. Nucleic Acids Res. 1986;14:6215–6226. doi: 10.1093/nar/14.15.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4402.Thome B M, Hoffschulte H K, Schiltz E, Muller M. A protein with sequence identity to Skp (FirA) supports protein translocation into plasma membrane vesicles of Escherichia coli. FEBS Lett. 1990;269:113–116. doi: 10.1016/0014-5793(90)81132-8. [DOI] [PubMed] [Google Scholar]
- 4403.Thome B M, Muller M. Skp is a periplasmic Escherichia coli protein requiring SecA and SecY for export. Mol Microbiol. 1991;5:2815–2821. doi: 10.1111/j.1365-2958.1991.tb01990.x. [DOI] [PubMed] [Google Scholar]
- 4404.Thomson G J, Howlett G J, Ashcroft A E, Berry A. The dhnA gene of Escherichia coli encodes a class I fructose bisphosphate aldolase. Biochem J. 1998;331:437–445. doi: 10.1042/bj3310437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4405.Thony B, Hwang D S, Fradkin L, Kornberg A. iciA, an Escherichia coli gene encoding a specific inhibitor of chromosomal initiation of replication in vitro. Proc Natl Acad Sci USA. 1991;88:4066–4070. doi: 10.1073/pnas.88.10.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4406.Thony-Meyer L, Fischer F, Kunzler P, Ritz D, Hennecke H. Escherichia coli genes required for cytochrome c maturation. J Bacteriol. 1995;177:4321–4326. doi: 10.1128/jb.177.15.4321-4326.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4407.Thorbjarnardottir S, Uemura H, Dingermann T, Rafner T, Thorsteinsdottir S, Soll D, Eggertsson G. Escherichia coli supH suppressor: temperature-sensitive missense suppression caused by an anticodon change in tRNA2Ser. J Bacteriol. 1985;161:207–211. doi: 10.1128/jb.161.1.207-211.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4408.Thorbjarnardottir S, Magnusdottir R A, Eggertsson G. Mutations determining generalized resistance to aminoglucoside antibiotics in Escherichia coli. Mol Gen Genet. 1978;161:89–98. doi: 10.1007/BF00266619. [DOI] [PubMed] [Google Scholar]
- 4409.Thorbjarnardottir S, Dingermann T, Rafnar T, Andresson O S, Soll D, Eggertsson G. Leucine tRNA family of Escherichia coli: nucleotide sequence of the supP(Am) suppressor gene. J Bacteriol. 1985;161:219–222. doi: 10.1128/jb.161.1.219-222.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4410.Thorne G M, Corwin L M. Mutations affecting aromatic amino acid transport in Escherichia coli and Salmonella typhimurium. J Gen Microbiol. 1975;90:203–216. doi: 10.1099/00221287-90-2-203. [DOI] [PubMed] [Google Scholar]
- 4411.Thorne-Holst M, Thony-Meyer L, Hederstedt L. Escherichia coli ccm in-frame deletion mutants can produce periplasmic cytochrome b but not cytochrome c. FEBS Lett. 1997;410:351–355. doi: 10.1016/s0014-5793(97)00656-x. [DOI] [PubMed] [Google Scholar]
- 4412.Tiedeman A, Keyhani J, Kamholz J, Daum III H A, Gots J S, Smith J M. Nucleotide sequence analysis of the purEK operon encoding 5′ phosphoribosyl-5-aminoimidazole carboxylase of Escherichia coli K-12. J Bacteriol. 1989;171:205–212. doi: 10.1128/jb.171.1.205-212.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4413.Tiedeman A, Smith J M. Nucleotide sequence of the guaB locus encoding IMP dehydrogenase of Escherichia coli K12. Nucleic Acids Res. 1985;13:1303–1316. doi: 10.1093/nar/13.4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4414.Tiedeman A, Smith J M, Zalkin H. Nucleotide sequence of the guaA gene encoding GMP synthetase of Escherichia coli K12. J Biol Chem. 1985;260:8676–8679. [PubMed] [Google Scholar]
- 4415.Tiedeman A A, DeMarini D J, Parker J, Smith J M. DNA sequence of the purC gene encoding 5′-phosphoribosyl-5-aminoimidazole-4-N-succinocarboxamide synthetase and organization of the dapA-purC region of Escherichia coli K-12. J Bacteriol. 1990;172:6035–6041. doi: 10.1128/jb.172.10.6035-6041.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4416.Tilly K, Murialdo H, Georgopoulos C. Identification of a second Escherichia coli groE gene whose product is necessary for bacteriophage morphogenesis. Proc Natl Acad Sci USA. 1981;78:1629–1633. doi: 10.1073/pnas.78.3.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4416a.Timms A R, Bridges B A. Reversion of the tyrosine ochre strain Escherichia coli WU3610 under starvation conditions depends on a new gene tas. Genetics. 1998;148:1627–1635. doi: 10.1093/genetics/148.4.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4417.Tisa L S, Adler J. Chemotactic properties of Escherichia coli mutants having abnormal Ca2+ content. J Bacteriol. 1995;177:7112–7118. doi: 10.1128/jb.177.24.7112-7118.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4417a.Tobe T, Sasakawa C, Okada N, Honma Y, Yoshikawa M. vacB, a novel chromosomal gene required for expression of virulence genes on the large plasmid of Shigella flexneri. J Bacteriol. 1992;174:6359–6367. doi: 10.1128/jb.174.20.6359-6367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4418.Tobe T, Ito K, Yura T. Isolation and physical mapping of temperature-sensitive mutants defective in heat shock induction of proteins in Escherichia coli. Mol Gen Genet. 1984;195:10–16. doi: 10.1007/BF00332716. [DOI] [PubMed] [Google Scholar]
- 4419.Tobe T, Kusukawa N, Yura T. Suppression of rpoH (htpR) mutations of Escherichia coli: heat shock response in suhA revertants. J Bacteriol. 1987;169:4128–4134. doi: 10.1128/jb.169.9.4128-4134.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4420.Tobey K L, Grant G A. The nucleotide sequence of the serA gene of Escherichia coli and the amino acid sequence of the encoded protein, D-3-phosphoglycerate dehydrogenase. J Biol Chem. 1986;261:12179–12183. [PubMed] [Google Scholar]
- 4421.Tobin J F, Schleif R. Positive regulation of the Escherichia coli rhamnose operon is mediated by the products of tandemly repeated regulatory genes. J Mol Biol. 1987;196:789–799. doi: 10.1016/0022-2836(87)90405-0. [DOI] [PubMed] [Google Scholar]
- 4422.Tokunaga M, Loranger J M, Chang S-Y, Regue M, Chang S, Wu H C. Identification of prolipoprotein signal peptidase and genomic organization of the lsp gene in Escherichia coli. J Biol Chem. 1985;260:5610–5615. [PubMed] [Google Scholar]
- 4423.Tolner J, Poolman B, Wallace B, Konings W N. Revised nucleotide sequence of the gltP gene, which encodes the proton-glutamate-aspartate transport protein of Escherichia coli K-12. J Bacteriol. 1992;174:2391–2393. doi: 10.1128/jb.174.7.2391-2393.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4424.Tomasiewicz H G, McHenry C S. Sequence analysis of the Escherichia coli dnaE gene. J Bacteriol. 1987;169:5735–5744. doi: 10.1128/jb.169.12.5735-5744.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4425.Tomioka S, Nikaido T, Miyakawa T, Matsuhashi M. Mutation of the N-acetylmuramyl-l-alanine amidase gene of Escherichia coli. J Bacteriol. 1983;156:463–465. doi: 10.1128/jb.156.1.463-465.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4426.Tommassen J, Lugtenberg B. Localization of phoE, the structural gene for outer membrane protein e in Escherichia coli K-12. J Bacteriol. 1981;147:118–123. doi: 10.1128/jb.147.1.118-123.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4427.Tommassen J, Lugtenberg B. Outer membrane protein e of Escherichia coli K-12 is co-regulated with alkaline phosphatase. J Bacteriol. 1980;143:151–157. doi: 10.1128/jb.143.1.151-157.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4428.Tommassen J, Eiglmeier K, Cole S T, Overduin P, Larson T J, Boos W. Characterization of two genes, glpQ and ugpQ, encoding glycerophosphoryl diester phosphodiesterases of Escherichia coli. Mol Gen Genet. 1991;226:321–327. doi: 10.1007/BF00273621. [DOI] [PubMed] [Google Scholar]
- 4429.Tommassen J, Koster M, Overduin P. Molecular analysis of the promoter region of the Escherichia coli K-12 phoE gene. Identification of an element, upstream from the promoter, required for the efficient expression of phoE protein. J Mol Biol. 1987;198:633–641. doi: 10.1016/0022-2836(87)90206-3. [DOI] [PubMed] [Google Scholar]
- 4430.Tommassen J, deGeus P, Lugtenberg B, Hackett J, Reeves P R. Regulation of the pho regulon of Escherichia coli K-12. Cloning of the regulator genes phoB and phoR and identification of their gene products. J Mol Biol. 1982;157:265–274. doi: 10.1016/0022-2836(82)90233-9. [DOI] [PubMed] [Google Scholar]
- 4431.Tommassen J, Hiemstra P, Overduin P, Lugtenberg B. Cloning of phoM, a gene involved in regulation of the synthesis of phosphate limitation inducible proteins in Escherichia coli K-12. Mol Gen Genet. 1984;195:190–194. doi: 10.1007/BF00332745. [DOI] [PubMed] [Google Scholar]
- 4432.Tommassen J, Overduin P, Lugtenberg B, Bergmans H. Cloning of phoE, the structural gene for the Escherichia coli phosphate limitation-inducible outer membrane pore protein. J Bacteriol. 1982;149:668–672. doi: 10.1128/jb.149.2.668-672.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4433.Tommassen J, van der Ley P, van der Ende A, Bergmans H, Lugtenberg B. Cloning of ompF, the structural gene for an outer membrane pore protein of Escherichia coli K12: physical localization and homology with the phoE gene. Mol Gen Genet. 1982;185:105–110. [Google Scholar]
- 4434.Tomochika K-I, Hong J-S. Transport-defective Escherichia coli ecf mutant permeable to protons and nucleotides. J Bacteriol. 1978;133:1008–1014. doi: 10.1128/jb.133.2.1008-1014.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4435.Tomoyasu T, Yuki T, Morimura S, Mori H, Yamanaka K, Niki H, Hiraga S, Ogura T. The Escherichia coli FtsH protein is a prokaryotic member of a protein family of putative ATPases involved in membrane functions, cell cycle control, and gene expression. J Bacteriol. 1993;175:1344–1351. doi: 10.1128/jb.175.5.1344-1351.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4436.Tong S, Porco A, Isturiz T, Conway T. Cloning and molecular genetic characterization of the Escherichia coli gntR, gntK, and gntU genes of GntI, the main system for gluconate metabolism. J Bacteriol. 1996;178:3260–3269. doi: 10.1128/jb.178.11.3260-3269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4437.Toone M, Rudd K E, Friesen J D. deaD, a new Escherichia coli gene encoding a presumed ATP-dependent RNA helicase, can suppress a mutation in rpsB, the gene encoding ribosomal protein S2. J Bacteriol. 1991;173:3291–3302. doi: 10.1128/jb.173.11.3291-3302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4438.Toone W M, Rudd K E, Friesen J D. Mutations causing aminotriazole resistance and temperature sensitivity reside in gyrB, which encodes the B subunit of DNA gyrase. J Bacteriol. 1992;174:5479–5481. doi: 10.1128/jb.174.16.5479-5481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4439.Toothman P. Restriction alleviation by bacteriophage lambda and lambda reverse. J Virol. 1981;38:621–631. doi: 10.1128/jvi.38.2.621-631.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4440.Tormo A, Almiron M, Kolter R. surA, an Escherichia coli gene essential for survival in stationary phase. J Bacteriol. 1990;172:4339–4347. doi: 10.1128/jb.172.8.4339-4347.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4441.Torres-Cabassa A S, Gottesman S. Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J Bacteriol. 1987;169:981–989. doi: 10.1128/jb.169.3.981-989.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4442.Torrey T A, Atlung T, Kogoma T. dnaA suppressor (dasF) mutants of Escherichia coli are stable DNA replication (sdrA/rnh) mutants. Mol Gen Genet. 1984;196:350–355. doi: 10.1007/BF00328070. [DOI] [PubMed] [Google Scholar]
- 4443.Torriani A, Rothman F. Mutants of Escherichia coli constitutive for alkaline phosphatase. J Bacteriol. 1961;81:835–836. doi: 10.1128/jb.81.5.835-836.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4444.Tosa T, Pizer L I. Biochemical bases for the antimetabolite action of l-serine hydroxamate. J Bacteriol. 1971;106:972–982. doi: 10.1128/jb.106.3.972-982.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4444a.Totemeyer S, Booth N A, Nichols W W, Dunbar B, Booth I R. From famine to feast: the role of methylglyoxal production in Escherichia coli. Mol Microbiol. 1998;27:553–562. doi: 10.1046/j.1365-2958.1998.00700.x. [DOI] [PubMed] [Google Scholar]
- 4445.Touati D. Cloning and mapping of the manganese superoxide dismutase gene (sodA) of Escherichia coli K-12. J Bacteriol. 1983;155:1078–1087. doi: 10.1128/jb.155.3.1078-1087.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4446.Touati E, Danchin A. The structure of the promoter and amino terminal region of the pH 2.5 acid phosphatase structural gene (appA) of E. coli: a negative control of transcription mediated by cyclic AMP. Biochimie. 1987;69:215–221. doi: 10.1016/0300-9084(87)90045-9. [DOI] [PubMed] [Google Scholar]
- 4447.Touati E, Dassa E, Dassa J, Boquet P L, Touati D. Are appR and katF the same Escherichia coli gene encoding a new sigma transcription initiation factor? Res Microbiol. 1991;142:29–36. doi: 10.1016/0923-2508(91)90094-q. [DOI] [PubMed] [Google Scholar]
- 4448.Touati E, Dassa E, Boquet P L. Pleiotropic mutations in appR reduce pH 2.5 acid phosphatase expression and restore succinate utilization in CRP-deficient strains of Escherichia coli. Mol Gen Genet. 1986;202:257–264. doi: 10.1007/BF00331647. [DOI] [PubMed] [Google Scholar]
- 4449.Tougu K, Marians K J. The interaction between helicase and primase sets the replication fork clock. J Biol Chem. 1996;271:21398–21405. doi: 10.1074/jbc.271.35.21398. [DOI] [PubMed] [Google Scholar]
- 4450.Tran P V, Bannor T A, Doktor S Z, Nichols B P. Chromosomal organization and expression of Escherichia coli pabA. J Bacteriol. 1990;172:397–410. doi: 10.1128/jb.172.1.397-410.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4451.Traub I, Gaisser S, Braun V. Activity domains of the TonB protein. Mol Microbiol. 1993;8:409–423. doi: 10.1111/j.1365-2958.1993.tb01584.x. [DOI] [PubMed] [Google Scholar]
- 4452.Traub I, Braun V. Energy-coupled colicin transport through the outer membrane of Escherichia coli K-12: mutated TonB proteins alter receptor activities and colicin uptake. FEMS Microbiol Lett. 1994;119:65–70. doi: 10.1111/j.1574-6968.1994.tb06868.x. [DOI] [PubMed] [Google Scholar]
- 4453.Trchunian A A, Vasilian A V. ATPase activity and K+ transport in membranes of anaerobically grown trk− mutants of Escherichia coli. Biokhimiya. 1993;58:1062–1070. . (In Russian with English summaries.) [PubMed] [Google Scholar]
- 4454.Treat M L, Weaver M L, Emmett M R, Johnson J R. Mutagenesis of the metJBLF gene cluster with transposon Tn5: localization of the metF transcription unit. Mol Gen Genet. 1984;193:370–375. doi: 10.1007/BF00330695. [DOI] [PubMed] [Google Scholar]
- 4455.Trempy J E, Kirby J E, Gottesman S. Alp suppression of Lon: dependence on the slpA gene. J Bacteriol. 1994;176:2061–2067. doi: 10.1128/jb.176.7.2061-2067.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4456.Triggs-Raine B L, Doble B W, Mulvey M R, Sorby P A, Loewen P C. Nucleotide sequence of katG, encoding catalase HPI of Escherichia coli. J Bacteriol. 1988;170:4415–4419. doi: 10.1128/jb.170.9.4415-4419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4457.Triggs-Raine B L, Loewen P C. Physical characterization of katG, encoding catalase HPI of Escherichia coli. Gene. 1987;52:121–128. doi: 10.1016/0378-1119(87)90038-2. [DOI] [PubMed] [Google Scholar]
- 4458.Triman K L. Mutational analysis of 16S ribosomal RNA structure and function in Escherichia coli. Adv Genet. 1995;33:1–39. doi: 10.1016/s0065-2660(08)60329-6. [DOI] [PubMed] [Google Scholar]
- 4459.Trimbur D E, Mortlock R P. Isolation and characterization of Escherichia coli mutants able to utilize the novel pentose l-ribose. J Bacteriol. 1991;173:2459–2464. doi: 10.1128/jb.173.8.2459-2464.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4460.Trisler P, Gottesman S. lon transcriptional regulation of genes necessary for capsular polysaccharide synthesis in Escherichia coli K-12. J Bacteriol. 1984;160:184–191. doi: 10.1128/jb.160.1.184-191.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4461.Tristram H, Neale S. The activity and specificity of the proline permease in wild-type and analogue-resistant strains of Escherichia coli. J Gen Microbiol. 1968;50:121–137. doi: 10.1099/00221287-50-1-121. [DOI] [PubMed] [Google Scholar]
- 4462.Tropp B E. Cardiolipin synthase from Escherichia coli. Biochim Biophys Acta. 1997;1348:192–200. doi: 10.1016/s0005-2760(97)00100-8. [DOI] [PubMed] [Google Scholar]
- 4463.Tropp B E, Ragolia L, Xia W, Dowhan W, Milkman R, Rudd K E, Ivanisevic R, Savic D J. Identity of the Escherichia coli cls and nov genes. J Bacteriol. 1995;177:5155–5157. doi: 10.1128/jb.177.17.5155-5157.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4464.Troup B, Jahn M, Hungerer C, Jahn D. Isolation of the hemF operon containing the gene for the Escherichia coli aerobic coproporphyrinogen III oxidase by in vivo complementation of a yeast HEM13 mutant. J Bacteriol. 1994;176:673–680. doi: 10.1128/jb.176.3.673-680.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4465.Trower M K. PCR cloning, sequence analysis and expression of the cybC genes encoding soluble cytochrome b-562 from Escherichia coli B strain OP7 and K strain NM522. Biochim Biophys Acta. 1993;1143:109–111. doi: 10.1016/0005-2728(93)90223-3. [DOI] [PubMed] [Google Scholar]
- 4466.Trucksis M, Zabel D J, Depew R E. Escherichia coli and Salmonella typhimurium supX genes specify deoxyribonucleic acid topoisomerase. J Bacteriol. 1981;147:679–681. doi: 10.1128/jb.147.2.679-681.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4467.Trucksis M, Depew R E. Identification and localization of a gene that specifies production of Escherichia coli DNA topoisomerase. Proc Natl Acad Sci USA. 1981;78:2164–2168. doi: 10.1073/pnas.78.4.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4468.Trun N J, Gottesman S. On the bacterial cell cycle: Escherichia coli mutants with altered ploidy. Genes Dev. 1990;4:2036–2047. doi: 10.1101/gad.4.12a.2036. [DOI] [PubMed] [Google Scholar]
- 4469.Trun N J, Gottesman S. Characterization of Escherichia coli mutants with altered ploidy. Res Microbiol. 1991;142:195–200. doi: 10.1016/0923-2508(91)90030-e. [DOI] [PubMed] [Google Scholar]
- 4470.Trun N J, Gottesman S, Lobner-Olesen A. Analysis of Escherichia coli mutants with altered DNA content. Cold Spring Harbor Symp Quant Biol. 1991;56:353–358. doi: 10.1101/sqb.1991.056.01.042. [DOI] [PubMed] [Google Scholar]
- 4471.Trun N J, Silhavy T J. Characterization and in vivo cloning of prlC, a suppressor of signal sequence mutations in Escherichia coli K12. Genetics. 1987;116:513–521. doi: 10.1093/genetics/116.4.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4472.Trun N J, Silhavy T J. PrlC, a suppressor of signal sequence mutations in Escherichia coli, can direct the insertion of the signal sequence into the membrane. J Mol Biol. 1989;205:665–676. doi: 10.1016/0022-2836(89)90312-4. [DOI] [PubMed] [Google Scholar]
- 4473.Truniger V, Boos W. Mapping and cloning of gldA, the structural gene of the Escherichia coli glycerol dehydrogenase. J Bacteriol. 1994;176:1796–1800. doi: 10.1128/jb.176.6.1796-1800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4474.Truniger V, Boos W, Sweet G. Molecular analysis of the glpFKX regions of Escherichia coli and Shigella flexneri. J Bacteriol. 1992;174:6981–6991. doi: 10.1128/jb.174.21.6981-6991.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4475.Truong Q C, Nguyen Van J C, Shlaes D, Gutmann L, Moreau N J. A novel, double mutation in DNA gyrase A of Escherichia coli conferring resistance to quinolone antibiotics. Antimicrob Agents Chemother. 1997;41:85–90. doi: 10.1128/aac.41.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4476.Tsai-Wu J J, Liu H F, Lu A L. Escherichia coli MutY protein has both N-glycosylase and apurinic/apyrimidinic endonuclease activities on A · C and A · G mispairs. Proc Natl Acad Sci USA. 1992;89:8779–8783. doi: 10.1073/pnas.89.18.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4477.Tsai-Wu J J, Radicella J P, Lu A L. Nucleotide sequence of the Escherichia coli micA gene required for A/G-specific mismatch repair: identity of micA and mutY. J Bacteriol. 1991;173:1902–1910. doi: 10.1128/jb.173.6.1902-1910.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4478.Tsaneva I R, Muller B, West S C. ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell. 1992;69:1171–1180. doi: 10.1016/0092-8674(92)90638-s. [DOI] [PubMed] [Google Scholar]
- 4479.Tsaneva I R, Muller B, West S C. RuvA and RuvB proteins of Escherichia coli exhibit DNA helicase activity in vitro. Proc Natl Acad Sci USA. 1993;90:1315–1319. doi: 10.1073/pnas.90.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4480.Tsaneva I R, Weiss B. soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J Bacteriol. 1990;172:4197–4205. doi: 10.1128/jb.172.8.4197-4205.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4481.Tsay J T, Oh W, Larson T J, Jackowski S, Rock C O. Isolation and characterization of the beta-ketoacyl-acyl carrier protein synthase III gene (fabH) from Escherichia coli K-12. J Biol Chem. 1992;267:6807–6814. [PubMed] [Google Scholar]
- 4482.Tse-Dinh Y-C, Wang J C. Complete nucleotide sequence of the topA gene encoding Escherichia coli DNA topoisomerase I. J Mol Biol. 1986;191:321–331. doi: 10.1016/0022-2836(86)90129-4. [DOI] [PubMed] [Google Scholar]
- 4483.Tso J Y, Zalkin H, van Cleemput M, Yanofsky C, Smith J M. Nucleotide sequence of Escherichia coli purF and deduced amino acid sequence of glutamine phosphoribosylpyrophosphate amidotransferase. J Biol Chem. 1982;257:3525–3531. [PubMed] [Google Scholar]
- 4484.Tsuchido T, VanBogelen R A, Neidhardt F C. Heat shock response in Escherichia coli influences cell division. Proc Natl Acad Sci USA. 1986;83:6959–6963. doi: 10.1073/pnas.83.18.6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4485.Tsuchihashi Z, Kornberg A. ATP interactions of the tau and gamma subunits of DNA polymerase III holoenzyme of Escherichia coli. J Biol Chem. 1989;264:17790–17795. [PubMed] [Google Scholar]
- 4486.Tsuchihashi Z, Kornberg A. Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc Natl Acad Sci USA. 1990;87:2516–2520. doi: 10.1073/pnas.87.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4487.Tsui H-C T, Feng G, Winkler M E. Transcription of the mutL repair, miaA tRNA modification, hfq pleiotropic regulator, and hflA region protease genes of Escherichia coli K-12 from clustered Eς32-specific promoters during heat shock. J Bacteriol. 1996;178:5719–5731. doi: 10.1128/jb.178.19.5719-5731.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4488.Tsui H C, Feng G, Winkler M E. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J Bacteriol. 1997;179:7476–7487. doi: 10.1128/jb.179.23.7476-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4489.Tsui H C, Zhao G, Feng G, Leung H C, Winkler M E. The mutL repair gene of Escherichia coli K-12 forms a superoperon with a gene encoding a new cell-wall amidase. Mol Microbiol. 1994;11:189–202. doi: 10.1111/j.1365-2958.1994.tb00300.x. [DOI] [PubMed] [Google Scholar]
- 4490.Tsui H C, Leung H C, Winkler M E. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 4491.Tsui P, Huang L, Freundlich M. Integration host factor binds specifically to multiple sites in the ompB promoter of Escherichia coli and inhibits transcription. J Bacteriol. 1991;173:5800–5807. doi: 10.1128/jb.173.18.5800-5807.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4492.Tsui P, Helu V, Freundlich M. Altered osmoregulation of ompF in integration host factor mutants of Escherichia coli. J Bacteriol. 1988;170:4950–4953. doi: 10.1128/jb.170.10.4950-4953.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4493.Tucker S D, Gopalakrishnan A S, Bollinger R, Dowhan W, Murgola E. Molecular mapping of glyW, a duplicate gene for tRNA3Gly of Escherichia coli. J Bacteriol. 1982;152:773–779. doi: 10.1128/jb.152.2.773-779.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4494.Tucker S D, Murgola E. Sequence analysis of the glyW region in Escherichia coli. Biochimie. 1985;67:1053–1057. doi: 10.1016/s0300-9084(85)80300-x. [DOI] [PubMed] [Google Scholar]
- 4495.Tuggle C K, Fuchs J A. Regulation of the operon encoding ribonucleotide reductase in Escherichia coli: evidence for both positive and negative control. EMBO J. 1986;5:1077–1085. doi: 10.1002/j.1460-2075.1986.tb04325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4496.Tuggle C K, Fuchs J A. Regulation of the operon encoding ribonucleotide reductase: role of the negative sites in nrd repression. J Bacteriol. 1990;172:1711–1718. doi: 10.1128/jb.172.4.1711-1718.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4497.Turlin E, Biville F, Gasser F. Complementation of Methylobacterium organophilum mutants affected in pyrroloquinoline quinone biosynthesis genes pqqE and pqqF by cloned Escherichia coli chromosomal DNA. FEMS Microbiol Lett. 1991;67:59–63. doi: 10.1016/0378-1097(91)90444-f. [DOI] [PubMed] [Google Scholar]
- 4498.Turnbough C L, Kerr K H, Funderburg W R, Donahue J P, Powell F E. Nucleotide sequence and characterization of the pyrF operon of Escherichia coli K-12. J Biol Chem. 1987;262:10239–10245. [PubMed] [Google Scholar]
- 4499.Turnbough C L, Hicks K L, Donahue J P. Attenuation control of pyrBI operon expression in Escherichia coli K-12. Proc Natl Acad Sci USA. 1983;80:368–372. doi: 10.1073/pnas.80.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4500.Turner R J, Taylor D E, Weiner J H. Expression of Escherichia coli TehA gives resistance to antiseptics and disinfectants similar to that conferred by multidrug resistance efflux pumps. Antimicrob Agents Chemother. 1997;41:440–444. doi: 10.1128/aac.41.2.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4501.Turner R J, Weiner J H, Taylor D E. Neither reduced uptake nor increased efflux is encoded by tellurite resistance determinants expressed in Escherichia coli. Can J Microbiol. 1995;41:92–98. doi: 10.1139/m95-012. . (Erratum, 41:656.) [DOI] [PubMed] [Google Scholar]
- 4502.Turnowsky F, Fuchs K, Jeschek C, Hogenauer G. envM genes of Salmonella typhimurium and Escherichia coli. J Bacteriol. 1989;171:6555–6565. doi: 10.1128/jb.171.12.6555-6565.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4503.Udaka S, Moyed H S. Inhibition of parental and mutant xanthosine 5-phosphate aminases by psicofuranine. J Biol Chem. 1963;238:2797–2803. [PubMed] [Google Scholar]
- 4504.Ueguchi C, Ito K. Multicopy suppression: an approach to understanding intracellular functioning of the protein export system. J Bacteriol. 1992;174:1454–1461. doi: 10.1128/jb.174.5.1454-1461.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4505.Ueguchi C, Kakeda M, Yamada H, Mizuno T. An analogue of the DnaJ molecular chaperone in Escherichia coli. Proc Natl Acad Sci USA. 1994;91:1054–1058. doi: 10.1073/pnas.91.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4506.Ueguchi C, Ohta T, Seto C, Suzuki T, Mizuno T. The leuO gene product has a latent ability to relieve bgl silencing in Escherichia coli. J Bacteriol. 1998;180:190–193. doi: 10.1128/jb.180.1.190-193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4507.Ueguchi C, Suzuki T, Yoshida T, Tanaka K, Mizuno T. Systematic mutational analysis revealing the functional domain organization of Escherichia coli nucleoid protein H-NS. J Mol Biol. 1996;263:149–162. doi: 10.1006/jmbi.1996.0566. [DOI] [PubMed] [Google Scholar]
- 4508.Ueki M, Wachi M, Jung H K, Ishino F, Matsuhashi M. Escherichia coli mraR gene involved in cell growth and division. J Bacteriol. 1992;174:7841–7843. doi: 10.1128/jb.174.23.7841-7843.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4509.Uemura H, Thorbjarnardottir S, Gamulin V, Yano J, Andresson O S, Soll D, Eggertsson G. supN ochre suppressor gene in Escherichia coli codes for tRNALys. J Bacteriol. 1985;163:1288–1289. doi: 10.1128/jb.163.3.1288-1289.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4510.Ueno-Nishio S, Mango S, Reitzer L J, Magasanik B. Identification and regulation of the glnL operator-promoter of the complex glnALG operon of Escherichia coli. J Bacteriol. 1984;160:379–384. doi: 10.1128/jb.160.1.379-384.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4511.Ukita T, Ikeda H. Role of the recJ gene product in UV-induced illegitimate recombination at the hotspot. J Bacteriol. 1996;178:2362–2367. doi: 10.1128/jb.178.8.2362-2367.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4512.Ulrich A K, de Mendoza D, Garwin J L, Cronan J E., Jr Genetic and biochemical analyses of Escherichia coli mutants altered in the temperature regulation of membrane lipid composition. J Bacteriol. 1983;154:221–230. doi: 10.1128/jb.154.1.221-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4513.Umbarger H E. Regulation of the biosynthesis of the branched-chain amino acids. Curr Top Microbiol Immunol. 1969;1:57–76. [Google Scholar]
- 4514.Umezu K, Nakayama K, Nakayama H. Escherichia coli RecQ protein is a DNA helicase. Proc Natl Acad Sci USA. 1990;87:5363–5377. doi: 10.1073/pnas.87.14.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4515.Urbanowski M L, Stauffer L T, Plamann L S, Stauffer G V. A new methionine locus, metR, that encodes a trans-acting protein required for activation of metE and metH in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1987;169:1391–1397. doi: 10.1128/jb.169.4.1391-1397.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4516.Ursini M V, Arcari P, De Felice M. Acetohydroxy acid synthetase isoenzymes of Escherichia coli K-12; a trans-acting regulatory locus for ilvH1 gene expression. Mol Gen Genet. 1981;181:491–496. doi: 10.1007/BF00428741. [DOI] [PubMed] [Google Scholar]
- 4517.Ursinus A, Holtje J V. Purification and properties of a membrane-bound lytic transglycosylase. J Bacteriol. 1994;176:338–343. doi: 10.1128/jb.176.2.338-343.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4518.Usui M, Sembongi H, Matsuzaki H, Matsumoto K, Shibuya I. Primary structures of the wild-type and mutant alleles encoding the phosphatidylglycerophosphate synthase of Escherichia coli. J Bacteriol. 1994;176:3389–3392. doi: 10.1128/jb.176.11.3389-3392.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4519.Utsumi R, Katayama S, Taniguchi M, Horie T, Ikeda M, Igaki S, Nakagawa H, Miwa A, Tanabe H, Noda M. Newly identified genes involved in the signal transduction of Escherichia coli K-12. Gene. 1994;140:73–77. doi: 10.1016/0378-1119(94)90733-1. [DOI] [PubMed] [Google Scholar]
- 4520.Utsumi R, Horie T, Katoh A, Kaino Y, Tanabe H, Noda M. Isolation and characterization of the heat-responsive genes in Escherichia coli. Biosci Biotechnol Biochem. 1996;60:309–315. doi: 10.1271/bbb.60.309. [DOI] [PubMed] [Google Scholar]
- 4521.Utsumi R, Nakamoto Y, Kawakumai M, Himeno M, Komano T. Involvement of cyclic AMP and its receptor protein in filamentation of an Escherichia coli fic mutant. J Bacteriol. 1982;151:807–812. doi: 10.1128/jb.151.2.807-812.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4522.Uzan M, Danchin A. A rapid test for the relA mutation in E. coli. Biochem Biophys Res Commun. 1976;69:751–758. doi: 10.1016/0006-291x(76)90939-6. [DOI] [PubMed] [Google Scholar]
- 4523.Uzan M, Danchin A. Correlation between the serine sensitivity and the derepressibility of the ilv genes in Escherichia coli relA− mutants. Mol Gen Genet. 1978;165:21–30. doi: 10.1007/BF00270372. [DOI] [PubMed] [Google Scholar]
- 4524.Uzan M, Favre R, Gallay E, Caro L. Genetical and structural analysis of a group of λilv and λrho transducing phages. Mol Gen Genet. 1981;182:462–470. doi: 10.1007/BF00293936. [DOI] [PubMed] [Google Scholar]
- 4525.Valent Q A, Kendall D A, High S, Kusters R, Oudega B, Luirink J. Early events in preprotein recognition in E. coli: interaction of SRP and trigger factor with nascent polypeptides. EMBO J. 1995;14:5494–5505. doi: 10.1002/j.1460-2075.1995.tb00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4526.Valentin-Hansen P. Tandem CRP binding sites in the deo operon of Escherichia coli K-12. EMBO J. 1982;1:1049–1054. doi: 10.1002/j.1460-2075.1982.tb01295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4527.Valentin-Hansen P, Boetius F, Hammer-Jespersen K, Svendsen I. The primary structure of Escherichia coli K12 2-deoxyribose 5-phosphate aldolase. Nucleotide sequence of the deoC gene and the amino acid sequence of the enzyme. Eur J Biochem. 1982;125:561–566. doi: 10.1111/j.1432-1033.1982.tb06719.x. [DOI] [PubMed] [Google Scholar]
- 4528.Valentin-Hansen P, Aiba H, Schumperli D. The structure of tandem regulatory regions in the deo operon of Escherichia coli K12. EMBO J. 1982;1:317–322. doi: 10.1002/j.1460-2075.1982.tb01167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4529.Valentin-Hansen P, Larson J E L, Hojrup P, Short S A, Barbier C S. Nucleotide sequence of the CytR regulatory gene of E. coli K-12. Nucleic Acids Res. 1986;14:2215–2228. doi: 10.1093/nar/14.5.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4530.Valentin-Hansen P, Hammer K, Larsen E L, Svendsen I. The internal regulated promoter of the deo operon of Escherichia coli K-12. Nucleic Acids Res. 1984;12:5211–5224. doi: 10.1093/nar/12.13.5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4531.Valentin-Hansen P, Hammer-Jespersen K, Boetius F, Svendsen I. Structure and function of the intercistronic regulatory deoC-deoA element of Escherichia coli K-12. EMBO J. 1984;3:179–183. doi: 10.1002/j.1460-2075.1984.tb01781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4532.Valentin-Hansen P, Hammer-Jespersen K, Buxton R S. Evidence for the existence of three promoters for the deo operon of Escherichia coli K12 in vitro. J Mol Biol. 1979;133:1–17. doi: 10.1016/0022-2836(79)90248-1. [DOI] [PubMed] [Google Scholar]
- 4533.Valentin-Hansen P, Hojrup P, Short S A. The primary structure of the deoR repressor from the Escherichia coli K-12. Nucleic Acids Res. 1985;13:5927–5936. doi: 10.1093/nar/13.16.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4534.Valentini G, Mattevi A, Barilla D, Galizzi A, Speranza M L. Recombinant pyruvate kinase type I from Escherichia coli: overproduction and revised C-terminus of the polypeptide. Biol Chem. 1997;378:719–721. [PubMed] [Google Scholar]
- 4535.Vales L D, Rabin B A, Chase J W. Subunit structure of Escherichia coli exonuclease VII. J Biol Chem. 1982;257:8799–8805. [PubMed] [Google Scholar]
- 4536.Vales L D, Rabin B A, Chase J W. Isolation and preliminary characterization of Escherichia coli mutants deficient in exonuclease VII. J Bacteriol. 1983;155:1116–1122. doi: 10.1128/jb.155.3.1116-1122.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4537.Vallari D S, Rock C O. Isolation and characterization of Escherichia coli pantothenate permease (panF) mutants. J Bacteriol. 1985;164:136–142. doi: 10.1128/jb.164.1.136-142.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4538.Vallari D S, Rock C O. Isolation and characterization of temperature-sensitive pantothenate kinase (coaA) mutants of Escherichia coli. J Bacteriol. 1987;169:5795–5800. doi: 10.1128/jb.169.12.5795-5800.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4539.Valle F, Becerril B, Chen E, Seeberg E, Heyneker H, Bolivar F. Complete nucleotide sequence of the glutamate dehydrogenase gene from Escherichia coli K-12. Gene. 1984;27:193–199. doi: 10.1016/0378-1119(84)90140-9. [DOI] [PubMed] [Google Scholar]
- 4540.Valle F, Sanvicente E, Seeberg P, Covarrubias A, Rodriguez R L, Bolivar F. Nucleotide sequence of the promoter and amino-terminal coding region of the glutamate dehydrogenase structural gene of Escherichia coli. Gene. 1983;23:199–209. doi: 10.1016/0378-1119(83)90052-5. [DOI] [PubMed] [Google Scholar]
- 4541.Van Bogelen R A, Vaughn V, Neidhardt F C. Gene for heat-inducible lysyl-tRNA synthetase (lysU) maps near cadA in Escherichia coli. J Bacteriol. 1983;153:1066–1068. doi: 10.1128/jb.153.2.1066-1068.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4542.van Buul C P J J, van Knippenberg P H. Nucleotide sequence of the ksgA gene of Escherichia coli: comparison of methyltransferases effecting dimethylation of adenosine in ribosomal RNA. Gene. 1985;38:65–72. doi: 10.1016/0378-1119(85)90204-5. [DOI] [PubMed] [Google Scholar]
- 4543.Van de Klundert J A, Van der Meide P H, Van de Putte P, Bosch L. Mutants of Escherichia coli altered in both genes coding for the elongation factor Tu. Proc Natl Acad Sci USA. 1978;75:4470–4473. doi: 10.1073/pnas.75.9.4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4544.van de Putte P, Plasterk R, Kuijpers A. A Mu gin complementing function and an invertible DNA region in Escherichia coli K-12 are situated on the genetic element e14. J Bacteriol. 1984;158:517–522. doi: 10.1128/jb.158.2.517-522.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4545.van de Zee J R, Postma P W, Hellingwerf K J. Non-physiological expression of UhpT does not lead to uncontrolled leakage of sugar phosphates out of Escherichia coli cells. FEMS Microbiol Lett. 1995;131:21–26. doi: 10.1111/j.1574-6968.1995.tb07748.x. [DOI] [PubMed] [Google Scholar]
- 4546.van Delft J H, Marinon B, Schmidt D S, Bosch L. Transcription of the tRNA-tufB operon of Escherichia coli: activation, termination and antitermination. Nucleic Acids Res. 1987;15:9515–9530. doi: 10.1093/nar/15.22.9515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4547.van den Berg E, Zwetsloot J, Noordermeer I, Pannekoeck H, Dekker B, Dijkema R, van Ormondt H. The structure and function of the regulatory element of the Escherichia coli uvrB gene. Nucleic Acids Res. 1981;9:5623–5643. doi: 10.1093/nar/9.21.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4548.Van der Meide P H, Vijgenboom E, Dicke M, Bosch L. Regulation of the expression of tufA and tufB, the two genes coding for elongation factor EF-Tu in Escherichia coli. FEBS Lett. 1982;139:325–330. doi: 10.1016/0014-5793(82)80881-8. [DOI] [PubMed] [Google Scholar]
- 4549.van der Ploeg J R, Weiss M A, Saller E, Nashimoto H, Saito N, Kertesz M X, Leisinger T. Identification of sulfate starvation-regulated genes in Escherichia coli: a gene cluster involved in the utilization of taurine as a sulfur source. J Bacteriol. 1996;178:5438–5446. doi: 10.1128/jb.178.18.5438-5446.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4550.van der Ploeg J R, Iwanicka-Nowicka R, Kertesz M A, Leisinger T, Hryniewicz M M. Involvement of CysB and Cbl regulatory proteins in expression of the tauABCD operon and other sulfate starvation-inducible genes in Escherichia coli. J Bacteriol. 1997;179:7671–7678. doi: 10.1128/jb.179.24.7671-7678.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4551.van der Woude M W, Kaltenbach L S, Low D A. Leucine-responsive regulatory protein plays dual roles as both an activator and a repressor of the Escherichia coli pap fimbrial operon. Mol Microbiol. 1995;17:303–312. doi: 10.1111/j.1365-2958.1995.mmi_17020303.x. [DOI] [PubMed] [Google Scholar]
- 4552.Van Dyk T K, Ayers B L, Morgan R W, Larossa R A. Constricted flux through the branched-chain amino acid biosynthetic enzyme acetolactate synthase triggers elevated expression of genes regulated by rpoS and internal acidification. J Bacteriol. 1998;180:785–792. doi: 10.1128/jb.180.4.785-792.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4553.Van Gelder P, Tommassen J. Demonstration of a folded monomeric form of porin PhoE of Escherichia coli in vivo. J Bacteriol. 1996;178:5320–5322. doi: 10.1128/jb.178.17.5320-5322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4554.van Heeswijk W C, Stegeman B, Hoving S, Molenaar D, Kahn D, Westerhoff H V. An additional PII in Escherichia coli: a new regulatory protein in the glutamine synthetase cascade. FEMS Microbiol Lett. 1995;132:153–157. doi: 10.1111/j.1574-6968.1995.tb07825.x. [DOI] [PubMed] [Google Scholar]
- 4555.van Heeswijk W C, Rabenberg M, Westerhoff H V, Kahn D. The genes of the glutamine synthetase adenylylation cascade are not regulated by nitrogen in Escherichia coli. Mol Microbiol. 1993;9:443–457. doi: 10.1111/j.1365-2958.1993.tb01706.x. [DOI] [PubMed] [Google Scholar]
- 4556.van Heeswijk W C, Kuppinger O, Merrick M, Kahn D. Localization of the glnD gene on a revised map of the 200-kilobase region of the Escherichia coli chromosome. J Bacteriol. 1992;174:1702–1703. doi: 10.1128/jb.174.5.1702-1703.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4557.van Heeswijk W C, Hoving S, Molenaar D, Stegeman B, Kahn D, Westerhoff H V. An alternative PII protein in the regulation of glutamine synthetase in Escherichia coli. Mol Microbiol. 1996;21:133–146. doi: 10.1046/j.1365-2958.1996.6281349.x. [DOI] [PubMed] [Google Scholar]
- 4558.Van Hove B, Staudenmaier H, Braun V. Novel two-component transmembrane transcription control: regulation of iron dicitrate transport in Escherichia coli K-12. J Bacteriol. 1990;172:6749–6758. doi: 10.1128/jb.172.12.6749-6758.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4559.Van Sluis C A, Moolenaar G F, Backendorf C. Regulation of the uvrC gene of Escherichia coli K12: localization and characterization of a damage-inducible promoter. EMBO J. 1983;2:2313–2318. doi: 10.1002/j.1460-2075.1983.tb01739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4560.Van Vliet F, Crabeel M, Boyen A, Tricot C, Stalon V, Falmagne P, Nakamura Y, Baumberg S, Glansdorff N. Sequences of the genes encoding argininosuccinate synthetase in Escherichia coli and Saccharomyces cerevisiae: comparison with methanogenic archaebacteria and mammals. Gene. 1990;95:99–104. doi: 10.1016/0378-1119(90)90419-r. [DOI] [PubMed] [Google Scholar]
- 4561.Van Vliet F, Cunin R, Jacobs A, Piette J, Gigot D, Lauwereys M, Pierard A, Glansdorff N. Evolutionary divergence of genes for ornithine and aspartate carbamoyl-transferase-complete sequence and mode of regulation of the Escherichia coli argF gene; comparison of argF with argI and pyrB. Nucleic Acids Res. 1984;12:6277–6289. doi: 10.1093/nar/12.15.6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4562.VanAlphen W, Lugtenberg B, Berendsen W. Heptose-deficient mutants of Escherichia coli K12 deficient in up to three major outer membrane proteins. Mol Gen Genet. 1976;147:263–269. doi: 10.1007/BF00582877. [DOI] [PubMed] [Google Scholar]
- 4563.Vanden Boom T, Reed K E, Cronan J E., Jr Lipoic acid metabolism in Escherichia coli: isolation of null mutants defective in lipoic acid biosynthesis, molecular cloning and characterization of the E. coli lip locus, and identification of the lipoylated protein of the glycine cleavage system. J Bacteriol. 1991;173:6411–6420. doi: 10.1128/jb.173.20.6411-6420.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4564.Vander Horn P B, Backstrom A D, Stewart V J, Begley T P. Structural genes for thiamine biosynthetic enzymes (thiCEFGH) in Escherichia coli. J Bacteriol. 1993;175:982–992. doi: 10.1128/jb.175.4.982-992.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4565.Vanet A, Plumbridge J A, Alix J-H. Cotranscription of two genes necessary for ribosomal protein L11 methylation (prmA) and pantothenate transport (panF) in Escherichia coli K-12. J Bacteriol. 1993;175:7178–7188. doi: 10.1128/jb.175.22.7178-7188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4566.Varga M E, Weiner J H. Physiological role of GlpB of anaerobic glycerol-3-phosphate dehydrogenase of Escherichia coli. Biochem Cell Biol. 1995;73:147–153. doi: 10.1139/o95-018. [DOI] [PubMed] [Google Scholar]
- 4567.Varshney U, Hutcheon T, van de Sande J H. Sequence analysis, expression and conservation of Escherichia coli uracil DNA glycosylase and its gene (ung) J Biol Chem. 1988;263:7776–7784. [PubMed] [Google Scholar]
- 4568.Vasudevan S G, Armarego W L, Shaw D C, Lilley P E, Dixon N E, Poole R K. Isolation and nucleotide sequence of the hmp gene that encodes a haemoglobin-like protein in Escherichia coli K-12. Mol Gen Genet. 1991;226:49–58. doi: 10.1007/BF00273586. [DOI] [PubMed] [Google Scholar]
- 4569.Veitinger S, Braun V. Localization of the entire fec region at 97.3 minutes on the Escherichia coli K-12 chromosome. J Bacteriol. 1992;174:3838–3839. doi: 10.1128/jb.174.11.3838-3839.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4570.Venables W A, Guest J R. Transduction of nitrate reductase loci of Escherichia coli by phages P1 and λ. Mol Gen Genet. 1968;103:127–140. doi: 10.1007/BF00427140. [DOI] [PubMed] [Google Scholar]
- 4571.Venkateswaran P S, Wu H C. Isolation and characterization of a phosphonomycin-resistant mutant of Escherichia coli K-12. J Bacteriol. 1972;110:935–944. doi: 10.1128/jb.110.3.935-944.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4572.Verde P, Frunzio R, di Nocera P P, Blasi F, Bruni C B. Identification, nucleotide sequence and expression of the regulatory region of the histidine operon of Escherichia coli K-12. Nucleic Acids Res. 1981;9:2075–2086. doi: 10.1093/nar/9.9.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4573.Verderber E, Lucast L J, Van Dehy J A, Cozart P, Etter J B, Best E A. Role of the hemA gene product and δ-aminolevulinic acid in regulation of Escherichia coli heme synthesis. J Bacteriol. 1997;179:4583–4590. doi: 10.1128/jb.179.14.4583-4590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4574.Verhof C, Lugtenberg B, van Boxtel R, de Graaff P, Verheij H M. Genetics and biochemistry of the peptidoglycan-associated proteins b and c of Escherichia coli K12. Mol Gen Genet. 1979;169:137–146. doi: 10.1007/BF00271664. [DOI] [PubMed] [Google Scholar]
- 4575.Verkamp E, Chelm B K. Isolation, nucleotide sequence, and preliminary characterization of the Escherichia coli K-12 hemA gene. J Bacteriol. 1989;171:4728–4735. doi: 10.1128/jb.171.9.4728-4735.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4576.Verkamp E, Backman V M, Bjornsson J M, Soll D, Eggertsson G. The periplasmic dipeptide permease system transports 5-aminolevulinic acid in Escherichia coli. J Bacteriol. 1993;175:1452–1456. doi: 10.1128/jb.175.5.1452-1456.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4577.Verkhovskaya M L, Verkhovsky M I, Wikstrom M. The respiration-driven active sodium transport system in E. coli does not function with lithium. FEBS Lett. 1996;388:217–218. doi: 10.1016/0014-5793(96)00531-5. [DOI] [PubMed] [Google Scholar]
- 4578.Versalovic J, Lupski J R. Missense mutations in the 3′ end of the Escherichia coli dnaG gene do not abolish primase activity but do confer the chromosome-segregation-defective (par) phenotype. Microbiology. 1997;143:585–594. doi: 10.1099/00221287-143-2-585. [DOI] [PubMed] [Google Scholar]
- 4579.Verwoert I, Verbree E C, van der Linden K H, Nijkamp H J, Stuitje A R. Cloning, nucleotide sequence, and expression of the Escherichia coli fabD gene, encoding malonyl coenzyme A-acyl carrier protein transacylase. J Bacteriol. 1992;174:2851–2857. doi: 10.1128/jb.174.9.2851-2857.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4580.Via P, Badia J, Baldoma L, Obradors N, Aguilar J. Transcriptional regulation of the Escherichia coli rhaT gene. Microbiology. 1996;142:1833–1840. doi: 10.1099/13500872-142-7-1833. [DOI] [PubMed] [Google Scholar]
- 4581.Vianney A, Muller M M, Clavel T, Lazzaroni J C, Portalier R, Webster R E. Characterization of the tol-pal region of Escherichia coli K-12: translational control of tolR expression by TolQ and identification of a new open reading frame downstream of pal encoding a periplasmic protein. J Bacteriol. 1996;178:4031–4038. doi: 10.1128/jb.178.14.4031-4038.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4582.Vianney A, Lewin T M, Beyer W F, Lazzaroni J C, Portalier R, Webster R E. Membrane topology and mutational analysis of the TolQ protein of Escherichia coli required for the uptake of macromolecules and cell envelope integrity. J Bacteriol. 1994;176:822–829. doi: 10.1128/jb.176.3.822-829.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4583.Vickery L E, Silberg J J, Ta D T. Hsc66 and Hsc20, a new heat shock cognate molecular chaperone system from Escherichia coli. Protein Sci. 1997;6:1047–1056. doi: 10.1002/pro.5560060511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4584.Vidal O, Longin R, Prigent-Combaret C, Dorel-Flamant C, Hooreman M, Lejeune P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol. 1998;180:2442–2449. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4585.Vimr E, Troy F A. Identification of an inducible catabolic system for sialic acids (nan) in Escherichia coli. J Bacteriol. 1985;164:845–853. doi: 10.1128/jb.164.2.845-853.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4586.Vinella D, Gagny B, Joseleau-Petit D, D’Ari R, Cashel M. Mecillinam resistance in Escherichia coli is conferred by loss of a second activity of the AroK protein. J Bacteriol. 1996;178:3818–3828. doi: 10.1128/jb.178.13.3818-3828.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4587.Vinella D, Joseleau-Petit D, Thevenet D, Bouloc P, D’Ari R. Penicillin-binding protein 2 inactivation in Escherichia coli results in cell division inhibition, which is relieved by FtsZ overexpression. J Bacteriol. 1993;175:6704–6710. doi: 10.1128/jb.175.20.6704-6710.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4588.Vinella D, D’Ari R. Thermoinducible filamentation in Escherichia coli due to an altered RNA polymerase beta subunit is suppressed by high levels of ppGpp. J Bacteriol. 1994;176:966–972. doi: 10.1128/jb.176.4.966-972.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4589.Vinella D, D’Ari R. Overview of controls in the Escherichia coli cell cycle. Bioessays. 1995;17:527–536. doi: 10.1002/bies.950170609. [DOI] [PubMed] [Google Scholar]
- 4590.Vinella D, D’Ari R, Bouloc P. Penicillin binding protein 2 is dispensable in Escherichia coli when ppGpp synthesis is induced. EMBO J. 1992;11:1493–1501. doi: 10.1002/j.1460-2075.1992.tb05194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4591.Visick J, Cai H, Clarke S. The l-isoaspartyl protein repair methyltransferase enhances survival of aging Escherichia coli subjected to secondary environmental stresses. J Bacteriol. 1998;180:2623–2629. doi: 10.1128/jb.180.10.2623-2629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4592.Visick J, Clarke S. RpoS- and OxyR-independent induction of HPI catalase at stationary phase in Escherichia coli and identification of rpoS mutations in common laboratory strains. J Bacteriol. 1997;179:4158–4163. doi: 10.1128/jb.179.13.4158-4163.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4593.Viswanathan V K, Green J M, Nichols B P. Kinetic characterization of 4-amino 4-deoxychorismate synthase from Escherichia coli. J Bacteriol. 1995;177:5918–5923. doi: 10.1128/jb.177.20.5918-5923.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4594.Vlahos C J, Dekker E E. The complete amino acid sequence and identification of the active-site arginine peptide of Escherichia coli 2-keto-4-hydroxyglutarate aldolase. J Biol Chem. 1988;263:11683–11691. [PubMed] [Google Scholar]
- 4595.Vlamis-Gardikas A, Aslund F, Spyrou G, Bergman T, Holmgren A. Cloning, overexpression, and characterization of glutaredoxin 2, an atypical glutaredoxin from Escherichia coli. J Biol Chem. 1997;272:11236–11243. doi: 10.1074/jbc.272.17.11236. [DOI] [PubMed] [Google Scholar]
- 4596.Vlazny D A, Hill C W. A stationary-phase-dependent viability block governed by two different polypeptides from the RhsA genetic element of Escherichia coli K-12. J Bacteriol. 1995;177:2209–2213. doi: 10.1128/jb.177.8.2209-2213.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4597.Voelkner P, Puppe W, Altendorf K. Characterization of the KdpD protein, the sensor kinase of the K+-translocating Kdp system of Escherichia coli. Eur J Biochem. 1993;217:1019–1026. doi: 10.1111/j.1432-1033.1993.tb18333.x. [DOI] [PubMed] [Google Scholar]
- 4598.Vogel R F, Entian K-D, Mecke D. Cloning and sequence of the mdh structural gene of Escherichia coli coding for malate dehydrogenase. Arch Microbiol. 1987;149:36–42. doi: 10.1007/BF00423133. [DOI] [PubMed] [Google Scholar]
- 4599.Vogler A P, Langeler J W. Analysis of the nag regulon from Escherichia coli K12 and Klebsiella pneumoniae and of its regulation. Mol Gen Genet. 1989;219:97–105. doi: 10.1007/BF00261163. [DOI] [PubMed] [Google Scholar]
- 4600.Vogler A P, Lengeler J W. Comparison of the sequences of the nagE operons from Klebsiella pneumoniae and Escherichia coli K12: enhanced variability of the enzyme IIN-acetylglucosamine in regions connecting functional domains. Mol Gen Genet. 1991;230:270–276. doi: 10.1007/BF00290677. [DOI] [PubMed] [Google Scholar]
- 4601.Vogler A P, Trentmann S, Lengeler J W. Alternative route for biosynthesis of amino sugars in Escherichia coli K-12 mutants by means of a catabolic isomerase. J Bacteriol. 1989;171:6586–6592. doi: 10.1128/jb.171.12.6586-6592.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4602.Volkert M R, Nguyen D C, Beard K C. Escherichia coli gene induction by alkylation treatment. Genetics. 1986;112:11–26. doi: 10.1093/genetics/112.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4603.Volkert M R, Hajec L I, Matijasevic Z, Fang F C, Prince R. Induction of the Escherichia coli aidB gene under oxygen-limiting conditions requires a functional rpoS (katF) gene. J Bacteriol. 1994;176:7638–7645. doi: 10.1128/jb.176.24.7638-7645.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4604.Volkert M R, Margossian L J, Clark A J. Evidence that rnmB is the operator of the Escherichia coli recA gene. Proc Natl Acad Sci USA. 1981;78:1786–1790. doi: 10.1073/pnas.78.3.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4605.Volkert M R, Loewen P C, Switala J, Crowley D, Conley M. The Δ(argF-lacZ)205(U169) deletion greatly enhances resistance to hydrogen peroxide in stationary-phase Escherichia coli. J Bacteriol. 1994;176:1297–1302. doi: 10.1128/jb.176.5.1297-1302.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4606.Voll M J, Leive L. Release of lipopolysaccharide in Escherichia coli resistant to the permeability increase induced by ethylenediaminetetraacetate. J Biol Chem. 1970;245:1108–1114. [PubMed] [Google Scholar]
- 4607.von Freiesleben U. DNA replication in Escherichia coli gyrB(Ts) mutants analysed by flow cytometry. Res Microbiol. 1991;142:223–227. doi: 10.1016/0923-2508(91)90034-8. [DOI] [PubMed] [Google Scholar]
- 4608.von Freiesleben U, Rasmussen K V, Schaechter M. SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol Microbiol. 1994;14:763–772. doi: 10.1111/j.1365-2958.1994.tb01313.x. [DOI] [PubMed] [Google Scholar]
- 4609.von Meyenburg K, Jorgensen B B, Nielsen J, Hansen F G. Promoters of the atp operon coding for the membrane-bound ATP synthase of Escherichia coli mapped by Tn10 insertion mutations. Mol Gen Genet. 1982;188:240–248. doi: 10.1007/BF00332682. [DOI] [PubMed] [Google Scholar]
- 4610.von Meyenburg K, Hansen F G, Riise E, Bergmans H E N, Meijer M, Messer W. Origin of replication, oriC, of the Escherichia coli K12 chromosome: genetic mapping and minichromosome replication. Cold Spring Harbor Symp Quant Biol. 1978;43:121–128. doi: 10.1101/sqb.1979.043.01.018. [DOI] [PubMed] [Google Scholar]
- 4611.von Meyenburg K, Hansen F G, Riise E, Bergmans H E N, Meijer M, Messer W. The origin of replication, oriC, of the Escherichia coli chromosome: genes near oriC and construction of oriC deletion mutations. ICN-UCLA Symp Mol Cell Biol. 1980;19:137–159. [Google Scholar]
- 4612.von Ossowski I, Mulvey M R, Leco P A, Borys A, Loewen P C. Nucleotide sequence of Escherichia coli katE which encodes catalase HPII. J Bacteriol. 1991;173:514–520. doi: 10.1128/jb.173.2.514-520.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4613.von Wilcken-Bergmann B, Muller-Hill B. Sequence of galR gene indicates a common evolutionary origin of lac and gal repressor in Escherichia coli. Proc Natl Acad Sci USA. 1982;79:2427–2431. doi: 10.1073/pnas.79.8.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4614.Vonder Haar R A, Umbarger H E. Isoleucine and valine metabolism in Escherichia coli. XIX. Inhibition of isoleucine biosynthesis by glycyl-leucine. J Bacteriol. 1972;112:142–147. doi: 10.1128/jb.112.1.142-147.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4615.Vuorio R, Vaara M. Mutants carrying conditionally lethal mutations in outer membrane genes omsA and firA (ssc) are phenotypically similar, and omsA is allelic to firA. J Bacteriol. 1992;174:7090–7097. doi: 10.1128/jb.174.22.7090-7097.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4616.Wachi M, Doi M, Tamaki S, Park W, Nakajima-Iijima S, Matsuhashi M. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J Bacteriol. 1987;169:4935–4940. doi: 10.1128/jb.169.11.4935-4940.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4617.Wachi M, Doi M, Ueda T, Ueki M, Tsuritani K, Nagai K, Matsuhashi M. Sequence of the downstream flanking region of the shape-determining genes mreBCD of Escherichia coli. Gene. 1991;106:135–136. doi: 10.1016/0378-1119(91)90578-y. [DOI] [PubMed] [Google Scholar]
- 4618.Wachi M, Doi M, Okada Y, Matsuhashi M. New mre genes mreC and mreD, responsible for formation of the rod shape of Escherichia coli cells. J Bacteriol. 1989;171:6511–6516. doi: 10.1128/jb.171.12.6511-6516.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4619.Wachi M, Matsuhashi M. Negative control of cell division by mreB, a gene that functions in determining the rod shape of Escherichia coli cells. J Bacteriol. 1989;171:3123–3127. doi: 10.1128/jb.171.6.3123-3127.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4620.Wackernagel W, Winkler U. A mutation in Escherichia coli enhancing the UV-mutability of phage lambda but not of its infectious DNA in a spheroplast assay. Mol Gen Genet. 1972;114:68–79. doi: 10.1007/BF00268748. [DOI] [PubMed] [Google Scholar]
- 4621.Wackett L P, Wanner B L, Venditti C P, Walsh C T. Involvement of the phosphate regulon and the psiD locus in carbon-phosphorus lyase activity of Escherichia coli K-12. J Bacteriol. 1987;169:1753–1756. doi: 10.1128/jb.169.4.1753-1756.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4622.Wada A, Igarashi K, Yoshimura S, Aimoto S, Ishihama A. Ribosome modulation factor: stationary growth phase-specific inhibitor of ribosome functions from Escherichia coli. Biochem Biophys Res Commun. 1995;214:410–417. doi: 10.1006/bbrc.1995.2302. [DOI] [PubMed] [Google Scholar]
- 4623.Wada A, Sako T. Primary structures of and genes for new ribosomal proteins A and B in Escherichia coli. J Biochem (Tokyo) 1987;101:817–820. doi: 10.1093/jb/101.3.817. [DOI] [PubMed] [Google Scholar]
- 4624.Wada A, Yamazaki Y, Fujita N, Ishihama A. Structure and probable genetic location of a “ribosome modulation factor” associated with 100S ribosomes in stationary-phase Escherichia coli cells. Proc Natl Acad Sci USA. 1990;87:2657–2661. doi: 10.1073/pnas.87.7.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4625.Wada C, Yura T. Mutants of Escherichia coli incapable of supporting replication of F-like plasmids at high temperature: isolation and characterization of mafA and mafB. J Bacteriol. 1979;140:864–873. doi: 10.1128/jb.140.3.864-873.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4626.Wada C, Yura T. Inhibition of initiation of mini-F plasmid replication in temperature-sensitive mafA mutants of Escherichia coli K-12. Plasmid. 1982;8:287–298. doi: 10.1016/0147-619x(82)90066-x. [DOI] [PubMed] [Google Scholar]
- 4627.Wada C, Yura T. Control of F plasmid replication by a host gene: evidence for interaction of the mafA gene product of Escherichia coli with the mini-F incC region. J Bacteriol. 1984;160:1130–1136. doi: 10.1128/jb.160.3.1130-1136.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4628.Wada C, Yura T, Hiraga S. Replication of Fpoh+ plasmid in mafA mutants of Escherichia coli defective in plasmid maintenance. Mol Gen Genet. 1977;152:211–217. doi: 10.1007/BF00268820. [DOI] [PubMed] [Google Scholar]
- 4629.Wagegg W, Braun V. Ferric citrate transport in Escherichia coli requires outer membrane receptor protein FecA. J Bacteriol. 1981;145:156–163. doi: 10.1128/jb.145.1.156-163.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4630.Wagner L A, Gesteland R F, Dayhuff T J, Weiss R B. An efficient Shine-Dalgarno sequence but not translation is necessary for lacZ mRNA stability in Escherichia coli. J Bacteriol. 1994;176:1683–1688. doi: 10.1128/jb.176.6.1683-1688.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4631.Wahab S Z, Elford R, Holmes W M. Nucleotide sequence of the Escherichia coli tRNA3Leu gene. Gene. 1989;81:193–194. doi: 10.1016/0378-1119(89)90351-x. [DOI] [PubMed] [Google Scholar]
- 4632.Wakayama Y, Takagi Y, Yano K. Gene responsible for protecting Escherichia coli from sodium dodecyl sulfate and toluidine blue plus light. J Bacteriol. 1984;159:527–532. doi: 10.1128/jb.159.2.527-532.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4633.Walderhaug M O, Polarek J W, Voelkner P, Daniel J, Hesse J E, Altendorf K, Epstein W. KdpD and KdpE, proteins that control expression of the kdpABC operon, are members of the two-component sensor-effector class of regulators. J Bacteriol. 1992;174:2152–2159. doi: 10.1128/jb.174.7.2152-2159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4634.Walkenhorst H M, Hemschemeier S K, Eichenlaub R. Molecular analysis of the molybdate uptake operon, modABCD, of Escherichia coli and modR, a regulatory gene. Microbiol Res. 1995;150:347–361. doi: 10.1016/S0944-5013(11)80016-9. [DOI] [PubMed] [Google Scholar]
- 4635.Walker J E, Gay N J, Saraste M, Eberle A N. DNA sequence around the Escherichia coli unc operon. Completion of the sequence of a 17 kilobase segment containing asnA, oriC, unc, glmS and phoS. Biochem J. 1984;224:799–815. doi: 10.1042/bj2240799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4636.Walker J R. Escherichia coli ras locus: its involvement in radiation repair. J Bacteriol. 1969;99:713–719. doi: 10.1128/jb.99.3.713-719.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4637.Walker J R. Defective excision repair of pyrimidine dimers in the ultraviolet-sensitive Escherichia coli ras− mutant. J Bacteriol. 1970;103:552–559. doi: 10.1128/jb.103.3.552-559.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4638.Wall D, Delaney J M, Fayet O, Lipinska B, Yamamoto T, Georgopoulos C. arc-dependent thermal regulation and extragenic suppression of the Escherichia coli cytochrome d operon. J Bacteriol. 1992;174:6554–6562. doi: 10.1128/jb.174.20.6554-6562.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4639.Wallace B, Yang Y J, Hong J-S, Lum D. Cloning and sequencing of a gene encoding a glutamate and aspartate carrier of Escherichia coli K-12. J Bacteriol. 1990;172:3214–3220. doi: 10.1128/jb.172.6.3214-3220.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4640.Wallace B J, Pittard J. Regulator gene controlling enzymes concerned in tyrosine biosynthesis in Escherichia coli. J Bacteriol. 1969;97:1234–1241. doi: 10.1128/jb.97.3.1234-1241.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4641.Wallace B J, Kushner S R. Genetic and physical analysis of the thioredoxin (trxA) gene of Escherichia coli K-12. Gene. 1984;32:399–408. doi: 10.1016/0378-1119(84)90015-5. [DOI] [PubMed] [Google Scholar]
- 4642.Wallace R G, Lee N, Fowler A V. The araC gene of Escherichia coli: transcriptional and translational start-points and complete nucleotide sequence. Gene. 1980;12:179–190. doi: 10.1016/0378-1119(80)90100-6. [DOI] [PubMed] [Google Scholar]
- 4643.Waller P R H, Sauer R T. Characterization of degQ and degS, Escherichia coli genes encoding homologs of the DegP protease. J Bacteriol. 1996;178:1146–1153. doi: 10.1128/jb.178.4.1146-1153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4644.Walsh C T, Liu J, Rusnak F, Sakaitani M. Molecular studies on enzymes in chorismate metabolism and the enterobactin biosynthetic pathway. Chem Rev. 1990;90:1105–1129. [Google Scholar]
- 4645.Walter E G, Weiner J H, Taylor D E. Nucleotide sequence and overexpression of the tellurite-resistance determinant from the IncHII plasmid pHH1508a. Gene. 1991;101:1–7. doi: 10.1016/0378-1119(91)90217-y. [DOI] [PubMed] [Google Scholar]
- 4646.Walton L, Richards C A, Elwell L P. Nucleotide sequence of the Escherichia coli uridine phosphorylase (udp) gene. Nucleic Acids Res. 1989;17:6741. doi: 10.1093/nar/17.16.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4647.Wandersman C, Moreno F, Schwartz M. Pleiotropic mutations rendering Escherichia coli K-12 resistant to bacteriophage TP1. J Bacteriol. 1980;143:1374–1383. doi: 10.1128/jb.143.3.1374-1383.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4648.Wandersman C, Delepelaire P. TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc Natl Acad Sci USA. 1990;87:4776–4780. doi: 10.1073/pnas.87.12.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4649.Wang A Y, Grogan D W, Cronan J E., Jr Cyclopropane fatty acid synthase of Escherichia coli: deduced amino acid sequence, purification, and studies of the enzyme active site. Biochemistry. 1992;31:11020–11028. doi: 10.1021/bi00160a011. [DOI] [PubMed] [Google Scholar]
- 4650.Wang E A, Koshland D E. Receptor structure in the bacterial sensing system. Proc Natl Acad Sci USA. 1980;77:7157–7161. doi: 10.1073/pnas.77.12.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4651.Wang E A, Mowry K L, Clegg D O, Koshland D E. Tandem duplication and multiple functions of a receptor gene in bacterial chemotaxis. J Biol Chem. 1982;257:4673–4676. [PubMed] [Google Scholar]
- 4652.Wang H, Matsumura P. Characterization of the CheAS/CheZ complex: a specific interaction resulting in enhanced dephosphorylating activity on CheY-phosphate. Mol Microbiol. 1996;19:695–703. doi: 10.1046/j.1365-2958.1996.393934.x. [DOI] [PubMed] [Google Scholar]
- 4653.Wang H, Di Gate R J, Seeman N C. An RNA topoisomerase. Proc Natl Acad Sci USA. 1996;93:9477–9482. doi: 10.1073/pnas.93.18.9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4654.Wang J C, Becherer K. Cloning of the gene topA encoding for DNA topoisomerase I and the physical mapping of the cysB-topA-trp region of Escherichia coli. Nucleic Acids Res. 1983;11:1773–1790. doi: 10.1093/nar/11.6.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4655.Wang L, Weiss B. dcd (dCTP deaminase) gene of Escherichia coli: mapping, cloning, sequencing, and identification as a locus of suppressors of lethal dut (dUTPase) mutations. J Bacteriol. 1992;174:5647–5653. doi: 10.1128/jb.174.17.5647-5653.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4656.Wang M-D, Liu J, Wang B, Berg C M. Cloning and characterization of the Escherichia coli K-12 alanine-valine transaminase (avtA) gene. J Bacteriol. 1987;169:4228–4234. doi: 10.1128/jb.169.9.4228-4234.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4657.Wang R F, O’Hara E B, Aldea M, Bargmann C I, Gromley H. Escherichia coli mrsC is an allele of hflB, encoding a membrane-associated ATPase and protease that is required for mRNA decay. J Bacteriol. 1998;180:1929–1938. doi: 10.1128/jb.180.7.1929-1938.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4658.Wang R F, Kushner S R. Identification and physical analysis of new genes in the argG region (69 min) of Escherichia coli chromosome. 1996. GenBank submission U01376. [Google Scholar]
- 4659.Wang X D, de Boer P A, Rothfield L I. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. EMBO J. 1991;10:3363–3372. doi: 10.1002/j.1460-2075.1991.tb04900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4660.Wanner B L. Control of phoR-dependent bacterial alkaline phosphatase clonal variation by the phoM region. J Bacteriol. 1987;169:900–903. doi: 10.1128/jb.169.2.900-903.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4661.Wanner B L. Gene regulation by phosphate in enteric bacteria. J Cell Biochem. 1993;51:47–54. doi: 10.1002/jcb.240510110. [DOI] [PubMed] [Google Scholar]
- 4662.Wanner B L, Sarthy A, Beckwith J R. Escherichia coli pleiotropic mutant that reduces amounts of several periplasmic and outer membrane proteins. J Bacteriol. 1979;140:229–239. doi: 10.1128/jb.140.1.229-239.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4663.Wanner B L, Chang B-D. The phoBR operon in Escherichia coli K-12. J Bacteriol. 1987;169:5569–5574. doi: 10.1128/jb.169.12.5569-5574.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4664.Wanner B L, Bernstein H D. Determining the phoM map location in Escherichia coli K-12 by using a nearby transposon Tn10 insertion. J Bacteriol. 1982;150:429–432. doi: 10.1128/jb.150.1.429-432.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4665.Wanner B L, Boline J A. Mapping and molecular cloning of the phn (psiD) locus for phosphonate utilization in Escherichia coli. J Bacteriol. 1990;172:1186–1196. doi: 10.1128/jb.172.3.1186-1196.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4666.Wanner B L, Wilmes M R, Hunter E. Molecular cloning of the wild-type phoM operon in Escherichia coli K-12. J Bacteriol. 1988;170:279–288. doi: 10.1128/jb.170.1.279-288.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4667.Wanner B L, Latterell P. Mutants affected in alkaline phosphatase expression: evidence for multiple positive regulators of the phosphate regulon in Escherichia coli. Genetics. 1980;96:353–366. doi: 10.1093/genetics/96.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4668.Wanner B L, Wieder S, McSharry R. Use of bacteriophage transposon Mu d1 to determine the orientation for three proC-linked phosphate-starvation-inducible (psi) genes in Escherichia coli K-12. J Bacteriol. 1981;146:93–101. doi: 10.1128/jb.146.1.93-101.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4669.Wanner B L, Metcalf W W. Molecular genetic studies of a 10.9-kb operon in Escherichia coli for phosphonate uptake and biodegradation. FEMS Microbiol Lett. 1992;79:133–139. doi: 10.1111/j.1574-6968.1992.tb14031.x. [DOI] [PubMed] [Google Scholar]
- 4670.Ward D F, Gottesman M E. The nus mutations affect transcription termination in Escherichia coli. Nature. 1981;292:212–215. doi: 10.1038/292212a0. [DOI] [PubMed] [Google Scholar]
- 4671.Wargel R J, Shadur C A, Neuhaus F C. Mechanism of d-cycloserine action: transport mutants for d-alanine, d-cycloserine, and glycine. J Bacteriol. 1971;105:1028–1035. doi: 10.1128/jb.105.3.1028-1035.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4672.Washburn B K, Kushner S R. Construction and analysis of deletions in the structural gene (uvrD) for DNA helicase II of Escherichia coli. J Bacteriol. 1991;173:2569–2575. doi: 10.1128/jb.173.8.2569-2575.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4673.Washburn B K, Kushner S R. Characterization of DNA helicase II from a uvrD252 mutant of Escherichia coli. J Bacteriol. 1993;175:341–350. doi: 10.1128/jb.175.2.341-350.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4674.Watanabe K, Yamano Y, Murata K, Kimura A. The nucleotide sequence of the gene for gamma-glutamylcysteine synthetase of Escherichia coli. Nucleic Acids Res. 1986;14:4393–4400. doi: 10.1093/nar/14.11.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4675.Watanabe M, Blobel G. SecB functions as a cytosolic signal recognition factor for protein export in E. coli. Cell. 1989;58:695–705. doi: 10.1016/0092-8674(89)90104-9. [DOI] [PubMed] [Google Scholar]
- 4676.Watanabe M, Blobel G. High-affinity binding of Escherichia coli SecB to the signal sequence region of a presecretory protein. Proc Natl Acad Sci USA. 1995;92:10133–10136. doi: 10.1073/pnas.92.22.10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4677.Watanabe T, Snell E E. Reversibility of the tryptophanase reaction: synthesis of tryptophan from indole, pyruvate, and ammonia. Proc Natl Acad Sci USA. 1972;69:1086–1090. doi: 10.1073/pnas.69.5.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4678.Watson G, Paigen K. Isolation and characterization of an Escherichia coli bacteriophage requiring cell wall galactose. J Virol. 1971;8:669–674. doi: 10.1128/jvi.8.5.669-674.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4679.Watson N, Apirion D. Molecular cloning of the gene for the RNA-processing enzyme RNase III of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:849–853. doi: 10.1073/pnas.82.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4680.Watson N, Dunyak D S, Rosey E L, Slonczewski J L, Olson E R. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J Bacteriol. 1992;174:530–540. doi: 10.1128/jb.174.2.530-540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4681.Waukau J, Forst S. Molecular analysis of the signaling pathway between EnvZ and OmpR in Escherichia coli. J Bacteriol. 1992;174:1522–1527. doi: 10.1128/jb.174.5.1522-1527.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4681a.Webb E, Downs D. Characterization of thiL, encoding thiamin-monophosphate kinase, in Salmonella typhimurium. J Biol Chem. 1997;272:15702–15707. doi: 10.1074/jbc.272.25.15702. [DOI] [PubMed] [Google Scholar]
- 4682.Webb G, Rohatgi K, Courtright J B. Location of gyrA on the physical map of the Escherichia coli chromosome. J Bacteriol. 1990;172:6617. doi: 10.1128/jb.172.12.6617.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4683.Webb M. The mechanism of acquired resistance to Co2+ and Ni2+ in gram-positive and gram-negative bacteria. Biochim Biophys Acta. 1970;222:440–446. doi: 10.1016/0304-4165(70)90134-0. [DOI] [PubMed] [Google Scholar]
- 4684.Weber R, Silverman P M. The cpx proteins of Escherichia coli K12. Structure of the cpxA polypeptide as an inner membrane component. J Mol Biol. 1988;203:467–478. doi: 10.1016/0022-2836(88)90013-7. [DOI] [PubMed] [Google Scholar]
- 4685.Webster C, Kempsell K, Booth I R, Busby S J. Organization of the regulatory region of the Escherichia coli melibiose operon. Gene. 1987;59:253–263. doi: 10.1016/0378-1119(87)90333-7. [DOI] [PubMed] [Google Scholar]
- 4686.Webster R E. The tol gene products and the import of macromolecules into Escherichia coli. Mol Microbiol. 1991;5:1005–1011. doi: 10.1111/j.1365-2958.1991.tb01873.x. [DOI] [PubMed] [Google Scholar]
- 4687.Weglenska A, Jacob B, Sirko A. Transcriptional pattern of Escherichia coli ihfB (himD) gene expression. Gene. 1996;181:85–88. doi: 10.1016/s0378-1119(96)00468-4. [DOI] [PubMed] [Google Scholar]
- 4688.Wei S-Q, Stader J. A new suppressor of a lamB signal sequence mutation, prlZ1, maps to 69 minutes on the Escherichia coli chromosome. J Bacteriol. 1994;176:5704–5710. doi: 10.1128/jb.176.18.5704-5710.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4689.Wei Y-F, Chen B J, Samson L D. Suppression of Escherichia coli alkB mutants by Saccharomyces cerevisiae genes. J Bacteriol. 1995;177:5009–5015. doi: 10.1128/jb.177.17.5009-5015.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4690.Weichart D, Lange R, Henneberg N, Hengge-Aronis R. Identification and characterization of stationary phase-inducible genes in Escherichia coli. Mol Microbiol. 1993;10:407–420. [PubMed] [Google Scholar]
- 4691.Weichert M J, Adhya S. A family of bacterial regulators homologous to Gal and Lac repressors. J Biol Chem. 1992;267:15869–15874. [PubMed] [Google Scholar]
- 4692.Weichert M J, Adhya S. Control of transcription of Gal repressor and isorepressor genes in Escherichia coli. J Bacteriol. 1993;175:251–258. doi: 10.1128/jb.175.1.251-258.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4693.Weichert M J, Adhya S. The galactose regulon of Escherichia coli. Mol Microbiol. 1993;10:245–251. doi: 10.1111/j.1365-2958.1993.tb01950.x. [DOI] [PubMed] [Google Scholar]
- 4694.Weidner U, Geier S, Ptock A, Friedrich T, Leif H, Weiss H. The gene locus of the proton-translocating NADH:ubiquinone oxidoreductase in Escherichia coli. Organization of the 14 genes and relationship between the derived proteins and subunits of mitochondrial complex I. J Mol Biol. 1993;233:109–122. doi: 10.1006/jmbi.1993.1488. [DOI] [PubMed] [Google Scholar]
- 4695.Weiner J H, Furlong C E, Heppel L A. A binding protein for l-glutamine and its relation to active transport in E. coli. Arch Biochem. 1971;142:715–717. doi: 10.1016/0003-9861(71)90538-8. [DOI] [PubMed] [Google Scholar]
- 4696.Weiner J H, Heppel L A. A binding protein for glutamine and its relation to active transport in E. coli. J Biol Chem. 1971;246:6933–6941. doi: 10.1016/0003-9861(71)90538-8. [DOI] [PubMed] [Google Scholar]
- 4697.Weiner L, Brissette J L, Model P. Stress-induced expression of the Escherichia coli phage shock protein operon is dependent on ς54 and modulated by positive and negative feedback mechanisms. Genes Dev. 1991;5:1912–1923. doi: 10.1101/gad.5.10.1912. [DOI] [PubMed] [Google Scholar]
- 4698.Weiner L, Model P. Role of an Escherichia coli stress-response operon in stationary-phase survival. Proc Natl Acad Sci USA. 1994;91:2191–2195. doi: 10.1073/pnas.91.6.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4699.Weinreich M D, Yigit H, Reznikoff W S. Overexpression of the Tn5 transposase in Escherichia coli results in filamentation, aberrant nucleoid segregation, and cell death: analysis of E. coli and transposase suppressor mutations. J Bacteriol. 1994;176:5494–5504. doi: 10.1128/jb.176.17.5494-5504.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4700.Weisberg R, Freundlich M, Friedman D I, Gardner J F, Goosen N, Nash H A, Oppenheim A B, Rouviere-Yaniv J. Nomenclature of the genes encoding IHF. Mol Microbiol. 1996;19:642. doi: 10.1046/j.1365-2958.1996.t01-2-442924.x. [DOI] [PubMed] [Google Scholar]
- 4701.Weiss B, Cunningham R P. Genetic mapping of nth, a gene affecting endonuclease III (thymine glycol-DNA glycosylase) in Escherichia coli K-12. J Bacteriol. 1985;162:607–610. doi: 10.1128/jb.162.2.607-610.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4702.Weiss D L, Johnson D I, Weith H L, Somerville R L. Structural analysis of the ileR locus of Escherichia coli K12. J Biol Chem. 1986;261:9966–9971. [PubMed] [Google Scholar]
- 4703.Weissbach H, Brot N. Regulation of methionine synthesis in Escherichia coli. Mol Microbiol. 1991;5:1593–1597. doi: 10.1111/j.1365-2958.1991.tb01905.x. [DOI] [PubMed] [Google Scholar]
- 4704.Weissborn A C, Liu Q, Rumley M K, Kennedy E P. UTP:α-d-glucose-1-phosphate uridylyltransferase of Escherichia coli: isolation and DNA sequence of the galU gene and purification of the enzyme. J Bacteriol. 1994;176:2611–2618. doi: 10.1128/jb.176.9.2611-2618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4705.Wek R C, Hauser C A, Hatfield G W. The nucleotide sequence of the ilvBN operon of Escherichia coli: sequence homologies of the acetohydroxy acid synthase isozyme. Nucleic Acids Res. 1985;13:3995–4010. doi: 10.1093/nar/13.11.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4706.Wek R C, Hatfield G W. Examination of the internal promoter, PE, in the ilvGMEDA operon of E. coli K-12. Nucleic Acids Res. 1986;14:2763–2777. doi: 10.1093/nar/14.6.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4707.Wek R C, Hatfield G W. Nucleotide sequence and in vivo expression of the ilvY and ilvC genes in Escherichia coli K12. Transcription from divergent overlapping promoters. J Biol Chem. 1986;261:2441–2450. [PubMed] [Google Scholar]
- 4708.Welch M M, McHenry C S. Cloning and identification of the product of the dnaE gene of Escherichia coli. J Bacteriol. 1982;152:351–356. doi: 10.1128/jb.152.1.351-356.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4709.Weng M, Makaroff C A, Zalkin H. Nucleotide sequence of Escherichia coli pyrG encoding CTP synthetase. J Biol Chem. 1986;261:5568–5574. [PubMed] [Google Scholar]
- 4710.Wertheimer S J, Klotsky R A, Schwartz I. Transcriptional patterns for the thrS-infC-rpLT operon of Escherichia coli. Gene. 1988;63:309–320. doi: 10.1016/0378-1119(88)90534-3. [DOI] [PubMed] [Google Scholar]
- 4711.Wessler S R, Calvo J M. Control of leu operon expression in Escherichia coli by a transcription attenuation mechanism. J Mol Biol. 1981;149:579–597. doi: 10.1016/0022-2836(81)90348-x. [DOI] [PubMed] [Google Scholar]
- 4712.West S C. The RuvABC proteins and Holliday junction processing in Escherichia coli. J Bacteriol. 1996;178:1237–1241. doi: 10.1128/jb.178.5.1237-1241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4713.West S C, Connolly B. Biological roles of the Escherichia coli RuvA, RuvB and RuvC proteins revealed. Mol Microbiol. 1992;6:2755–2759. doi: 10.1111/j.1365-2958.1992.tb01454.x. [DOI] [PubMed] [Google Scholar]
- 4714.West S C, Tsaneva I R, Hiom K, Benson F E. Late steps in genetic recombination: branch migration and Holliday junction resolution by RuvA, RuvB, and RuvC proteins. Cold Spring Harbor Symp Quant Biol. 1993;58:525–531. doi: 10.1101/sqb.1993.058.01.059. [DOI] [PubMed] [Google Scholar]
- 4715.Westh Hansen S V, Jensen N, Munch-Petersen A. Studies on the sequence and structure of the Escherichia coli K-12 nupG gene. Eur J Biochem. 1987;168:385–391. doi: 10.1111/j.1432-1033.1987.tb13431.x. [DOI] [PubMed] [Google Scholar]
- 4716.Westling-Haggström B, Normark S. Genetic and physiological analysis of an envB spherelike mutant of Escherichia coli K-12 and characterization of its transductants. J Bacteriol. 1975;123:75–82. doi: 10.1128/jb.123.1.75-82.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4717.Weston L A, Kadner R J. Identification of Uhp polypeptides and evidence for their role in exogenous induction of the sugar phosphate transport system of Escherichia coli. J Bacteriol. 1987;169:3546–3555. doi: 10.1128/jb.169.8.3546-3555.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4718.Whalen W A, Berg C M. Analysis of an avtA::Mu d1(Ap lac) mutant: metabolic role of transaminase C. J Bacteriol. 1982;150:739–746. doi: 10.1128/jb.150.2.739-746.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4719.Whipp M J, Pittard A J. A reassessment of the relationship between aroK- and aroL-encoded shikimate kinase enzymes of Escherichia coli. J Bacteriol. 1995;177:1627–1629. doi: 10.1128/jb.177.6.1627-1629.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4720.Whipp M J, Halsall D M, Pittard A J. Isolation and characterization of an Escherichia coli K-12 mutant defective in tyrosine- and phenylalanine-specific transport systems. J Bacteriol. 1980;143:1–7. doi: 10.1128/jb.143.1.1-7.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4721.Whitby M C, Vincent S D, Lloyd R G. Branch migration of Holliday junctions: identification of RecG protein as a junction specific DNA helicase. EMBO J. 1994;13:5220–5228. doi: 10.1002/j.1460-2075.1994.tb06853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4722.Whitchurch C B, Mattick J S. Escherichia coli contains a set of genes homologous to those involved in protein secretion, DNA uptake and the assembly of type-4 fimbriae in other bacteria. Gene. 1994;150:9–15. doi: 10.1016/0378-1119(94)90851-6. [DOI] [PubMed] [Google Scholar]
- 4723.White D G, Goldman J D, Demple B, Levy S B. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol. 1997;179:6122–6126. doi: 10.1128/jb.179.19.6122-6126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4724.White D G, Maneewannakul K, von Hofe E, Zillman M, Eisenberg W, Field A K, Levy S B. Inhibition of the multiple antibiotic resistance (mar) operon in Escherichia coli by antisense DNA analogs. Antimicrob Agents Chemother. 1997;41:2699–2704. doi: 10.1128/aac.41.12.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4725.White M K, Yudkin M. Complementation analysis of eleven tryptophanase mutations in Escherichia coli. J Gen Microbiol. 1979;114:471–475. doi: 10.1099/00221287-114-2-471. [DOI] [PubMed] [Google Scholar]
- 4726.White R J, West P W. An examination of the inhibitory effects of N-iodoacetylglucosamine on Escherichia coli and isolation of resistant mutants. Biochem J. 1970;118:81–87. doi: 10.1042/bj1180081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4727.White S, Tuttle F E, Blankenhorn D, Dosch D C, Slonczewski J L. pH dependence and gene structure of inaA in Escherichia coli. J Bacteriol. 1992;174:1537–1543. doi: 10.1128/jb.174.5.1537-1543.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4728.White-Ziegler C A, Low D A. Thermoregulation of the pap operon: evidence for the involvement of RimJ, the N-terminal acetylase of ribosomal protein S5. J Bacteriol. 1992;174:7003–7012. doi: 10.1128/jb.174.21.7003-7012.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4729.White-Ziegler C A, Blyn L B, Braaten B A, Low D A. Identification of an Escherichia coli genetic locus involved in thermoregulation of the pap operon. J Bacteriol. 1990;172:1775–1782. doi: 10.1128/jb.172.4.1775-1782.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4730.Whoriskey S K, Schofield M A, Miller J H. Isolation and characterization of Escherichia coli mutants with altered rates of deletion formation. Genetics. 1991;127:21–30. doi: 10.1093/genetics/127.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4731.Wiater A, Hulanicka M D. Properties of cysK mutants of Escherichia coli K12. Acta Biochim Pol. 1979;26:21–28. [PubMed] [Google Scholar]
- 4732.Wiborg O, Andersen C, Knudsen C R, Clark B F C, Nyborg J. Mapping Escherichia coli elongation factor Tu residues involved in binding of aminoacyl-tRNA. J Biol Chem. 1996;271:20406–20411. doi: 10.1074/jbc.271.34.20406. [DOI] [PubMed] [Google Scholar]
- 4733.Wickner W, Leonard M R. Escherichia coli preprotein translocase. J Biol Chem. 1996;271:29514–29516. doi: 10.1074/jbc.271.47.29514. [DOI] [PubMed] [Google Scholar]
- 4734.Wijsman H J W. Mutation affecting plasmolysis in Escherichia coli. J Bacteriol. 1972;110:789–790. doi: 10.1128/jb.110.2.789-790.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4735.Wijsman H J W, Pafort H C. Pleiotropic mutations in Escherichia coli conferring tolerance to glycine and sensitivity to penicillin. Mol Gen Genet. 1974;128:349–357. doi: 10.1007/BF00268522. [DOI] [PubMed] [Google Scholar]
- 4736.Wild J R, Obrepalska B. Regulation of expression of the dadA gene encoding d-amino acid dehydrogenase in Escherichia coli: analysis of dadA-lac fusions and direction of dadA transcription. Mol Gen Genet. 1982;186:405–410. doi: 10.1007/BF00729461. [DOI] [PubMed] [Google Scholar]
- 4737.Wild J R, Zakrzewska B, Walczak W, Klopotowski T. Two distinct types of mutations conferring to Escherichia coli K12 capability of d-tryptophan utilization. Acta Microbiol Pol. 1987;36:17–28. [PubMed] [Google Scholar]
- 4738.Wild J R, Foltermann F, Roof W D, O’Donovan G A. A mutation in the catalytic cistron of aspartate carbamoyltransferase affecting catalysis, regulatory response and holoenzyme assembly. Nature. 1981;292:373–375. doi: 10.1038/292373a0. [DOI] [PubMed] [Google Scholar]
- 4739.Wild J R, Henning J, Lobockz M, Walczak W, Klopotowski T. Identification of the dadX gene coding for the predominant isozyme of alanine racemase in Escherichia coli K12. Mol Gen Genet. 1985;198:315–322. doi: 10.1007/BF00383013. [DOI] [PubMed] [Google Scholar]
- 4740.Wild J R, Klopotowski T. d-Amino acid dehydrogenase of Escherichia coli K12: positive selection of mutants defective in enzyme activity and localization of the structural gene. Mol Gen Genet. 1981;181:373–378. doi: 10.1007/BF00425614. [DOI] [PubMed] [Google Scholar]
- 4741.Wilde R J, Guest J R. Transcript analysis of the citrate synthase and succinate dehydrogenase genes of Escherichia coli. J Gen Microbiol. 1986;132:3239–3251. doi: 10.1099/00221287-132-12-3239. [DOI] [PubMed] [Google Scholar]
- 4742.Williams M D, Ouyang T X, Flickinger M C. Starvation-induced expression of SspA and SspB: the effects of a null mutation in sspA on Escherichia coli protein synthesis and survival during growth and prolonged starvation. Mol Microbiol. 1994;11:1029–1043. doi: 10.1111/j.1365-2958.1994.tb00381.x. [DOI] [PubMed] [Google Scholar]
- 4743.Williams M G, Fortson M, Dykstra C C, Jensen P, Kushner S R. Identification and genetic mapping of the structural gene for an essential Escherichia coli membrane protein. J Bacteriol. 1989;171:565–568. doi: 10.1128/jb.171.1.565-568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4744.Williams M V, Kerr T J, Lemmon R D, Tritz G. Azaserine resistance in Escherichia coli: chromosomal location of multiple genes. J Bacteriol. 1980;143:383–388. doi: 10.1128/jb.143.1.383-388.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4745.Williams N, Fox D K, Shea C, Roseman S. Pel, the protein that permits λ DNA penetration of Escherichia coli, is encoded by a gene in ptsM and is required for mannose utilization by the phosphotransferase system. Proc Natl Acad Sci USA. 1986;83:8934–8938. doi: 10.1073/pnas.83.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4746.Williams S B, Stewart V J. Nitrate- and nitrite-sensing protein NarX of Escherichia coli K-12: mutational analysis of the amino-terminal tail and first transmembrane segment. J Bacteriol. 1997;179:721–729. doi: 10.1128/jb.179.3.721-729.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4747.Williamson J M, Anderson M S, Raetz C R. Acyl-acyl carrier protein specificity of UDP-GlcNAc acyltransferases from gram-negative bacteria: relationship to lipid A structure. J Bacteriol. 1991;173:3591–3596. doi: 10.1128/jb.173.11.3591-3596.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4748.Willins D A, Ryan C W, Calvo J M. Characterization of Lrp, an Escherichia coli regulatory protein that mediates a global response to leucine. J Biol Chem. 1991;266:10768–10774. [PubMed] [Google Scholar]
- 4749.Willis D K, Uhlin B E, Amini K S, Clark A J. Physical mapping of the srl recA region of Escherichia coli: analysis of Tn10 generated insertions and deletions. Mol Gen Genet. 1981;183:497–504. doi: 10.1007/BF00268771. [DOI] [PubMed] [Google Scholar]
- 4750.Willis D K, Fouts K E, Barbour S D, Clark A J. Restriction nuclease and enzymatic analysis of transposon-induced mutations of the Rac prophage which affect expression and function of recE in Escherichia coli. J Bacteriol. 1983;156:727–736. doi: 10.1128/jb.156.2.727-736.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4751.Willis D K, Satin L H, Clark A J. Mutation-dependent suppression of recB21 recC22 by a region cloned from the Rac prophage of Escherichia coli K-12. J Bacteriol. 1985;162:1166–1172. doi: 10.1128/jb.162.3.1166-1172.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4752.Willison J C. An essential gene (efg) located at 38.1 minutes on the Escherichia coli chromosome. J Bacteriol. 1992;174:5765–5766. doi: 10.1128/jb.174.17.5765-5766.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4753.Willsky G R, Bennett R L, Malamy M. Inorganic phosphate transport in Escherichia coli: involvement of two genes which play a role in alkaline phosphatase regulation. J Bacteriol. 1973;113:529–539. doi: 10.1128/jb.113.2.529-539.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4754.Wilson H R, Chan P T, Turnbough C L. Nucleotide sequence and expression of the pyrC gene of Escherichia coli K-12. J Bacteriol. 1987;169:3051–3058. doi: 10.1128/jb.169.7.3051-3058.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4755.Wilson R K, Brown T, Roe B A. Nucleotide sequence of pheW; a third gene for E. coli tRNAPhe. Nucleic Acids Res. 1986;14:5937. doi: 10.1093/nar/14.14.5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4756.Wilson R L, Stauffer G V. DNA sequence and characterization of GcvA, a LysR family regulatory protein for the Escherichia coli glycine cleavage enzyme system. J Bacteriol. 1994;176:2862–2868. doi: 10.1128/jb.176.10.2862-2868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4757.Wilson R L, Stauffer L T, Stauffer G V. Roles of the GcvA and PurR proteins in negative regulation of the Escherichia coli glycine cleavage enzyme system. J Bacteriol. 1993;175:5129–5134. doi: 10.1128/jb.175.16.5129-5134.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4758.Wilson R L, Urbanowski M L, Stauffer G V. DNA binding sites of the LysR-type regulator GcvA in the gcv and gcvA control regions of Escherichia coli. J Bacteriol. 1995;177:4940–4946. doi: 10.1128/jb.177.17.4940-4946.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4759.Wissenbach U, Keck B, Unden G. Physical map location of the new artPIQMJ genes of Escherichia coli, encoding a periplasmic arginine transport system. J Bacteriol. 1993;175:3687–3688. doi: 10.1128/jb.175.11.3687-3688.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4760.Wissenbach U, Six S, Bongaerts J, Ternes D, Steinwachs S, Unden G. A third periplasmic transport system for l-arginine in Escherichia coli: molecular characterization of the artPIQMJ genes, arginine binding and transport. Mol Microbiol. 1995;17:675–686. doi: 10.1111/j.1365-2958.1995.mmi_17040675.x. [DOI] [PubMed] [Google Scholar]
- 4761.Witkin E M, Roegner-Maniscalco V. Overproduction of DnaE protein (α subunit of DNA polymerase III) restores viability in a conditionally inviable Escherichia coli strain deficient in DNA polymerase I. J Bacteriol. 1992;174:4166–4168. doi: 10.1128/jb.174.12.4166-4168.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4762.Wittmann H G, Stoffler G, Apirion D, Rosen L, Tanaka K, Tamaki S, Takata R, Dekio S, Otaka E, Osawa S. Biochemical and genetic studies on two different types of erythromycin resistant mutants of Escherichia coli with altered ribosomal protein. Mol Gen Genet. 1973;127:175–189. doi: 10.1007/BF00333665. [DOI] [PubMed] [Google Scholar]
- 4763.Woisetschlager M, Hoedel-Neuhofer A, Hoegenauer G. Localization of the kdsA gene with the aid of the physical map of the Escherichia coli chromosome. J Bacteriol. 1988;170:5382–5384. doi: 10.1128/jb.170.11.5382-5384.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4764.Woisetschlager M, Hoegenauer G. The kdsA gene coding for 3-deoxy-d-manno-octulosonic acid 8-phosphate synthetase in part of an operon in Escherichia coli. Mol Gen Genet. 1987;207:369–373. doi: 10.1007/BF00331603. [DOI] [PubMed] [Google Scholar]
- 4765.Wojkowiak D, Georgopoulos C, Zylicz M. Isolation and characterization of ClpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. J Biol Chem. 1993;268:22609–22617. [PubMed] [Google Scholar]
- 4766.Wold M S, McMacken R. Regulation of expression of the Escherichia coli dnaG gene and amplification of the dnaG primase. Proc Natl Acad Sci USA. 1982;79:4907–4911. doi: 10.1073/pnas.79.16.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4767.Wold S, Crooke E, Skarstad K. The Escherichia coli Fis protein prevents initiation of DNA replication from oriC in vitro. Nucleic Acids Res. 1996;24:3527–3532. doi: 10.1093/nar/24.18.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4768.Wolf R, Cool J A. Mapping of insertion mutations in gnd of Escherichia coli with deletions defining the ends of the gene. J Bacteriol. 1980;141:1222–1229. doi: 10.1128/jb.141.3.1222-1229.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4769.Wolfe P B, Wickner W, Goodman J M. Sequence of the leader peptidase gene of Escherichia coli and the orientation of leader peptidase in the bacterial envelope. J Biol Chem. 1983;258:12073–12080. [PubMed] [Google Scholar]
- 4770.Wolfe S A, Smith J M. Nucleotide sequence and analysis of the purA gene encoding adenylosuccinate synthetase of Escherichia coli K12. J Biol Chem. 1988;1988:19147–19153. [PubMed] [Google Scholar]
- 4771.Wolf-Watz H, Masters M. Deoxyribonucleic acid and outer membrane: strains diploid for the oriC region show elevated levels of deoxyribonucleaic acid-binding protein and evidence for specific binding of the oriC region to outer membrane. J Bacteriol. 1979;140:50–58. doi: 10.1128/jb.140.1.50-58.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4772.Woo K M, Kim K I, Goldberg A L, Ha D B, Chung C H. The heat-shock protein ClpB in Escherichia coli is a protein-activated ATPase. J Biol Chem. 1992;267:20429–20434. [PubMed] [Google Scholar]
- 4773.Wood D, Darlison M G, Wilde R J, Guest J R. Nucleotide sequence encoding the flavoprotein and hydrophobic subunits of the succinate dehydrogenase of Escherichia coli. Biochem J. 1984;222:519–534. doi: 10.1042/bj2220519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4774.Wood E R, Matson S W. The molecular cloning of the gene encoding the Escherichia coli 75-kDa helicase and the determination of its nucleotide sequence and genetic map position. J Biol Chem. 1989;264:8297–8303. [PubMed] [Google Scholar]
- 4775.Wood J M. Genetics of l-proline utilization in Escherichia coli. J Bacteriol. 1981;146:895–901. doi: 10.1128/jb.146.3.895-901.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4776.Wood J M, Zadworny D. Characterization of an inducible porter required for l-proline catabolism by Escherichia coli K12. Can J Biochem. 1979;57:1191–1199. doi: 10.1139/o79-155. [DOI] [PubMed] [Google Scholar]
- 4777.Wood J M, Zadworny D. Amplification of the put genes and identification of the put gene products in Escherichia coli K12. Can J Biochem. 1980;58:787–796. doi: 10.1139/o80-110. [DOI] [PubMed] [Google Scholar]
- 4778.Woodgate R. Construction of a umuDC operon substitution mutation in Escherichia coli. Mutat Res. 1992;281:221–225. doi: 10.1016/0165-7992(92)90012-7. [DOI] [PubMed] [Google Scholar]
- 4779.Woodgate R, Rajagopalan M, Lu C, Echols H. UmuC mutagenesis protein of Escherichia coli: purification and interaction with UmuD and UmuD′. Proc Natl Acad Sci USA. 1989;86:7301–7305. doi: 10.1073/pnas.86.19.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4780.Wookey P, Rosenberg H. Involvement of inner and outer membrane components in the transport of iron and in colicin B action in Escherichia coli. J Bacteriol. 1978;133:661–666. doi: 10.1128/jb.133.2.661-666.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4781.Wookey P, Pittard J, Forrest S M, Davidson B E. Cloning of the tyrP gene and further characterization of the tyrosine-specific transport system in Escherichia coli K-12. J Bacteriol. 1984;160:169–174. doi: 10.1128/jb.160.1.169-174.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4782.Wookey P J, Pittard A J. DNA sequence of the gene (tyrP) encoding the tyrosine-specific transport system of Escherichia coli. J Bacteriol. 1988;170:4946–4949. doi: 10.1128/jb.170.10.4946-4949.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4783.Worsham P L, Konisky J. Use of cir-lac fusions to study transcriptional regulation of the colicin Ia receptor in Escherichia coli K-12. J Bacteriol. 1981;145:647–650. doi: 10.1128/jb.145.1.647-650.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4784.Wower I, Kowaleski M P, Sears L E, Zimmermann R A. Mutagenesis of ribosomal protein S8 from Escherichia coli: defects in regulation of the spc operon. J Bacteriol. 1991;124:1213–1221. doi: 10.1128/jb.174.4.1213-1221.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4785.Wright M. Mutants of Escherichia coli lacking endonuclease I, ribonuclease I, or ribonuclease II. J Bacteriol. 1971;107:87–94. doi: 10.1128/jb.107.1.87-94.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4785a.Wright M, Bultin G, Hurwitz J. The isolation and characterization of an adenosine triphosphate-dependent deoxyribonuclease directed by recB, C genes. J Biol Chem. 1971;246:6543–6555. [PubMed] [Google Scholar]
- 4786.Wrzesinski J, Bakin A, Nurse K, Lane B G, Ofengand J. Purification, cloning, and properties of the 16S RNA pseudouridine 516 synthase from Escherichia coli. Biochemistry. 1995;34:8904–8913. doi: 10.1021/bi00027a043. [DOI] [PubMed] [Google Scholar]
- 4787.Wrzesinski J, Nurse K, Bakin A, Lane B G, Ofengand J. A dual-specificity pseudouridine synthase: an Escherichia coli synthase purified and cloned on the basis of its specificity for ψ746 in 23S RNA is also specific for ψ32 tRNAPhe. RNA. 1995;1:437–448. [PMC free article] [PubMed] [Google Scholar]
- 4788.Wu A M, Chapman A B, Platt T, Guarente L P, Beckwith J R. Deletions of distal sequence affect termination of transcription at the end of the tryptophan operon in E. coli. Cell. 1980;19:829–836. doi: 10.1016/0092-8674(80)90073-2. [DOI] [PubMed] [Google Scholar]
- 4789.Wu A M, Christie G E, Platt T. Tandem termination sites in the tryptophan operon of Escherichia coli. Proc Natl Acad Sci USA. 1981;78:2913–2917. doi: 10.1073/pnas.78.5.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4790.Wu B, Wawrzynow A, Zylicz M, Georgopoulos C. Structure-function analysis of the Escherichia coli GrpE heat shock protein. EMBO J. 1996;15:4806–4816. [PMC free article] [PubMed] [Google Scholar]
- 4791.Wu B, Ang D, Snavely M, Georgopoulos C. Isolation and characterization of point mutations in the Escherichia coli grpE heat shock gene. J Bacteriol. 1994;176:6965–6973. doi: 10.1128/jb.176.22.6965-6973.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4792.Wu H C, Lai J S, Hayashi S, Giam C Z. Biogenesis of membrane lipoproteins in Escherichia coli. Biophys J. 1982;37:307–315. doi: 10.1016/S0006-3495(82)84679-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4792a.Wu, H. C., and K. Rudd. Personal communication.
- 4793.Wu J, Weiss B. Two divergently transcribed genes, soxR and soxS, control a superoxide response regulon of Escherichia coli. J Bacteriol. 1991;173:2864–2871. doi: 10.1128/jb.173.9.2864-2871.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4794.Wu J, Weiss B. Two-stage induction of the soxRS (superoxide response) regulon of Escherichia coli. J Bacteriol. 1992;174:3915–3920. doi: 10.1128/jb.174.12.3915-3920.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4795.Wu J Y, Siegel L M, Kredich N M. High-level expression of Escherichia coli NADPH-sulfite reductase: requirement for a cloned cysG plasmid to overcome limiting siroheme cofactor. J Bacteriol. 1991;173:325–333. doi: 10.1128/jb.173.1.325-333.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4796.Wu L F, Navarro C, Mandrand-Berthelot M A. The hydC region contains a multi-cistronic operon (nik) involved in nickel transport in Escherichia coli. Gene. 1991;107:37–42. doi: 10.1016/0378-1119(91)90294-l. [DOI] [PubMed] [Google Scholar]
- 4797.Wu L F, Mandrand-Berthelot M A. Genetic and physiological characterization of new Escherichia coli mutants impaired in hydrogenase activity. Biochimie. 1986;68:167–179. doi: 10.1016/s0300-9084(86)81081-1. [DOI] [PubMed] [Google Scholar]
- 4798.Wu L F, Mandrand-Berthelot M A. Characterization of the product of the cloned fdhF gene of Escherichia coli. J Gen Microbiol. 1987;133:2421–2426. doi: 10.1099/00221287-133-9-2421. [DOI] [PubMed] [Google Scholar]
- 4799.Wu L F, Mandrand-Berthelot M A. Regulation of the fdhF gene encoding the selenopolypeptide for benzyl viologen-linked formate dehydrogenase in Escherichia coli. Mol Gen Genet. 1987;209:129–134. doi: 10.1007/BF00329847. [DOI] [PubMed] [Google Scholar]
- 4800.Wu L F, Mandrand-Berthelot M A, Waugh R, Edmonds C J, Holt S E, Boxer D H. Nickel deficiency gives rise to the defective hydrogenase phenotype of hydC and fnr mutants in Escherichia coli. Mol Microbiol. 1989;3:1709–1718. doi: 10.1111/j.1365-2958.1989.tb00156.x. [DOI] [PubMed] [Google Scholar]
- 4801.Wu T-H, Wood D L, Stein P L, Comer M M. Transcription of a gene cluster coding for two aminoacyl-tRNA synthetases and an initiation factor in Escherichia coli. J Mol Biol. 1984;173:177–209. doi: 10.1016/0022-2836(84)90189-x. [DOI] [PubMed] [Google Scholar]
- 4802.Wu T-H, Marinus M G. Dominant negative mutator mutations in the mutS gene of Escherichia coli. J Bacteriol. 1994;176:5393–5400. doi: 10.1128/jb.176.17.5393-5400.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4803.Wu T T. Growth on d-arabitol of a mutant strain of Escherichia coli K12 using a novel dehydrogenase and enzymes related to l-1,2-propanediol and d-xylose metabolism. J Gen Microbiol. 1976;94:246–256. doi: 10.1099/00221287-94-2-246. [DOI] [PubMed] [Google Scholar]
- 4804.Wulfing C, Lombardero J, Pluckthun A. An Escherichia coli protein consisting of a domain homologous to FK506-binding proteins (FKBP) and a new metal binding motif. J Biol Chem. 1994;269:2895–2901. [PubMed] [Google Scholar]
- 4805.Wunderlich M, Glockshuber R. In vivo control of redox potential during protein folding catalyzed by bacterial protein disulfide-isomerase (DsbA) J Biol Chem. 1993;268:24547–24550. [PubMed] [Google Scholar]
- 4806.Wurgler S M, Richardson C C. Structure and regulation of the gene for dGTP triphosphohydrolase from Escherichia coli. Proc Natl Acad Sci USA. 1990;87:2740–2744. doi: 10.1073/pnas.87.7.2740. . (Erratum, 87:4022.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4807.Wurtzel E, Chou M-Y, Inouye M. Osmoregulation of gene expression. I. DNA sequence of the ompR gene of the ompB operon of Escherichia coli and characterization of its gene product. J Biol Chem. 1982;257:13685–13691. [PubMed] [Google Scholar]
- 4807a.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- 4808.Xiao H, Crombie R, Dong Z, Onrust R, O’Donnell M E. DNA polymerase III accessory proteins. III. holC and holD encoding chi and psi. J Biol Chem. 1993;268:11773–11778. [PubMed] [Google Scholar]
- 4809.Xiong H, Vik S B. Alanine-scanning mutagenesis of the epsilon subunit of the F1-F0 ATP synthase from Escherichia coli reveals two classes of mutants. J Biol Chem. 1995;270:23300–23304. doi: 10.1074/jbc.270.40.23300. [DOI] [PubMed] [Google Scholar]
- 4810.Xiong H, Vik S B. Construction and plasmid-borne complementation of strains lacking the epsilon subunit of the Escherichia coli F1F0 ATP synthase. J Bacteriol. 1995;177:851–853. doi: 10.1128/jb.177.3.851-853.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4811.Xiong X, Deeter J N, Misra R. Assembly-defective OmpC mutants of Escherichia coli K-12. J Bacteriol. 1996;178:1213–1215. doi: 10.1128/jb.178.4.1213-1215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4812.Xu C, Shi W, Rosen B P. The chromosomal arsR gene of Escherichia coli encodes a trans-acting metalloregulatory protein. J Biol Chem. 1996;271:2427–2432. doi: 10.1074/jbc.271.5.2427. [DOI] [PubMed] [Google Scholar]
- 4813.Xu J, Johnson R C. aldB, an RpoS-dependent gene in Escherichia coli encoding an aldehyde dehydrogenase that is repressed by Fis and activated by Crp. J Bacteriol. 1995;177:3166–3175. doi: 10.1128/jb.177.11.3166-3175.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4814.Xu J, Johnson R C. Fis activates the RpoS-dependent stationary-phase expression of proP in Escherichia coli. J Bacteriol. 1995;177:5222–5231. doi: 10.1128/jb.177.18.5222-5231.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4815.Xu J, Johnson R C. Identification of genes negatively regulated by Fis: Fis and RpoS comodulate growth-phase-dependent gene expression in Escherichia coli. J Bacteriol. 1995;177:938–947. doi: 10.1128/jb.177.4.938-947.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4816.Xu X, Kantrowitz E R. The importance of aspartate 327 for catalysis and zinc binding in Escherichia coli alkaline phosphatase. J Biol Chem. 1992;267:16244–16251. [PubMed] [Google Scholar]
- 4817.Xu Y, Beckett D. Evidence for interdomain interaction in the Escherichia coli repressor of biotin biosynthesis from studies of an N-terminal domain deletion mutant. Biochemistry. 1996;35:1783–1792. doi: 10.1021/bi952269e. [DOI] [PubMed] [Google Scholar]
- 4818.Yagil E, Be’eri H. Arsenate-resistant alkaline phosphatase-constitutive mutants of Escherichia coli. Mol Gen Genet. 1977;154:185–189. doi: 10.1007/BF00330835. [DOI] [PubMed] [Google Scholar]
- 4819.Yakushi T, Tajima T, Matsuyama S, Tokuda H. Lethality of the covalent linkage between mislocalized major outer membrane lipoprotein and the peptidoglycan of Escherichia coli. J Bacteriol. 1997;179:2857–2862. doi: 10.1128/jb.179.9.2857-2862.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4820.Yamada H, Kitagawa M, Kawakami K, Mizushima S. The gene coding for lipoprotein signal peptidase (lspA) and that for isoleucyl-tRNA synthetase (ileS) constitute a cotranscriptional unit in Escherichia coli. FEBS Lett. 1984;171:245–248. doi: 10.1016/0014-5793(84)80496-2. [DOI] [PubMed] [Google Scholar]
- 4821.Yamada H, Yoshida T, Tanaka K, Sasakawa C, Mizuno T. Molecular analysis of the Escherichia coli hns gene encoding a DNA-binding protein, which preferentially recognizes curved DNA sequences. Mol Gen Genet. 1991;230:332–336. doi: 10.1007/BF00290685. [DOI] [PubMed] [Google Scholar]
- 4822.Yamada M, Saier M H. Glucitol-specific enzymes of the phosphotransferase system in Escherichia coli. J Biol Chem. 1987;262:5455–5463. [PubMed] [Google Scholar]
- 4823.Yamada M, Saier M H. Physical and genetic characterization of the glucitol operon in Escherichia coli. J Bacteriol. 1987;169:2990–2994. doi: 10.1128/jb.169.7.2990-2994.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4824.Yamada M, Saier M H. Positive and negative regulators for glucitol (gut) operon expression in Escherichia coli. J Mol Biol. 1988;203:569–583. doi: 10.1016/0022-2836(88)90193-3. [DOI] [PubMed] [Google Scholar]
- 4825.Yamada M, Asaoka S, Saier M H, Yamada Y. Characterization of the gcd gene from Escherichia coli K-12 W3110 and regulation of its expression. J Bacteriol. 1993;175:568–571. doi: 10.1128/jb.175.2.568-571.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4826.Yamada M, Kawai T, Izu H. Analysis of the Escherichia coli gntT and gntU genes and comparison of the products with their homologues. Biosci Biotechnol Biochem. 1996;60:1548–1550. doi: 10.1271/bbb.60.1548. [DOI] [PubMed] [Google Scholar]
- 4827.Yamada M, Takeda Y, Okamoto K, Hirota Y. Physical map of the nrdA-nrdB-ftsB-glpT region of the chromosomal DNA of Escherichia coli. Gene. 1982;18:309–318. doi: 10.1016/0378-1119(82)90169-x. [DOI] [PubMed] [Google Scholar]
- 4828.Yamada M, Yamada Y, Saier M H. Nucleotide sequence and expression of the gutQ gene within the glucitol operon of Escherichia coli. DNA Sequence. 1990;1:141–145. doi: 10.3109/10425179009016042. [DOI] [PubMed] [Google Scholar]
- 4829.Yamagata H, Daishima K, Mizushima S. Cloning and expression of a gene coding for the prolipoprotein signal peptidase of Escherichia coli. FEBS Lett. 1983;158:301–304. doi: 10.1016/0014-5793(83)80600-0. [DOI] [PubMed] [Google Scholar]
- 4830.Yamagata H, Taguchi N, Daishima K, Mizushima S. Genetic characterization of a gene for prolipoprotein signal peptidase in Escherichia coli. Mol Gen Genet. 1983;192:10–14. doi: 10.1007/BF00327640. [DOI] [PubMed] [Google Scholar]
- 4831.Yamagishi J-I, Yoshida K, Yamayoshi M, Nakamura S. Nalidixic acid-resistant mutations of the gyrB gene of Escherichia coli. Mol Gen Genet. 1986;204:367–373. doi: 10.1007/BF00331012. [DOI] [PubMed] [Google Scholar]
- 4832.Yamagishi M, Matsushima H, Wada A, Sakagami M, Fujita N, Ishihama A. Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor: growth phase- and growth rate-dependent control. EMBO J. 1993;12:625–630. doi: 10.1002/j.1460-2075.1993.tb05695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4833.Yamaguchi K, Inouye M. Lipoprotein 28, an inner membrane protein of Escherichia coli encoded by nlpA, is not essential for growth. J Bacteriol. 1988;170:3747–3749. doi: 10.1128/jb.170.8.3747-3749.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4834.Yamamoto K, Takahashi N, Fujitani Y, Yoshikura H, Kobayashi I. Orientation dependence in homologous recombination. Genetics. 1996;143:27–36. doi: 10.1093/genetics/143.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4835.Yamamoto T, Tomiyama M, Mita H, Sode K, Karube I. Identification of proteins encoded in Escherichia coli hydA, hydB and analysis of the hydA locus. FEMS Microbiol Lett. 1990;54:187–192. doi: 10.1016/0378-1097(90)90280-4. [DOI] [PubMed] [Google Scholar]
- 4836.Yamamoto, Y. 1995. GenBank submission D49445.
- 4837.Yamamoto Y, Aiba H, Baba T, Hayashi K, Inada T, Isono K, Itoh T, Kimura S, Kitagawa M, Makino K, Miki T, Mitsuhashi N, Mizobuchi K, Mori H, Nakade S, Nakamura Y, Nashimoto H, Oshima T, Oyama S, Saito N, Sampei G, Satoh Y, Sivasundaram S, Tagami H, Takahashi H, Takeda J, Takemoto K, Uchara K, Wuda C, Yamagata S, Horiuchi T. Construction of a contiguous 874-kb sequence of the Escherichia coli K-12 genome corresponding to 50.0–68.8 min on the linkage map and analysis of its sequence features. DNA Res. 1997;4:91–113. doi: 10.1093/dnares/4.2.91. . (Suppl., 4:169–178.) [DOI] [PubMed] [Google Scholar]
- 4838.Yamamoto Y, Ogawa T, Shinagawa H, Nakayama T, Matsuo H, Ogawa H. Determination of the initiation sites of transcription and translation of the uvrD gene of Escherichia coli. J Biochem (Tokyo) 1986;99:1579–1590. doi: 10.1093/oxfordjournals.jbchem.a135631. [DOI] [PubMed] [Google Scholar]
- 4839.Yamamoto Y, Miwa Y, Miyoshi K, Furuyama J, Ohmori H. The Escherichia coli ldcC gene encodes another lysine decarboxylase, probably a constitutive enzyme. Genes Genet Syst. 1997;72:167–172. doi: 10.1266/ggs.72.167. [DOI] [PubMed] [Google Scholar]
- 4840.Yamanaka H, Kameyama M, Baba T, Fujii Y, Okamoto K. Maturation pathway of Escherichia coli heat-stable enterotoxin I: requirement of DsbA for disulfide bond formation. J Bacteriol. 1994;176:2906–2913. doi: 10.1128/jb.176.10.2906-2913.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4841.Yamanaka K, Fang L, Inouye M. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol Microbiol. 1998;27:247–255. doi: 10.1046/j.1365-2958.1998.00683.x. [DOI] [PubMed] [Google Scholar]
- 4842.Yamanaka K, Mitani T, Ogura T, Niki H, Hiraga S. Cloning, sequencing, and characterization of multicopy suppressors of a mukB mutation in Escherichia coli. Mol Microbiol. 1994;13:301–312. doi: 10.1111/j.1365-2958.1994.tb00424.x. [DOI] [PubMed] [Google Scholar]
- 4843.Yamanaka K, Ogura T, Koonin E V, Niki H, Hiraga S. Multicopy suppressors, mssA and mssB, of an smbA mutation of Escherichia coli. Mol Gen Genet. 1994;243:9–16. doi: 10.1007/BF00283870. [DOI] [PubMed] [Google Scholar]
- 4844.Yamanaka K, Ogura T, Niki H, Hiraga S. Characterization of the smtA gene encoding an S-adenosylmethionine-dependent methyltransferase of Escherichia coli. FEMS Microbiol Lett. 1995;133:59–63. doi: 10.1111/j.1574-6968.1995.tb07861.x. [DOI] [PubMed] [Google Scholar]
- 4845.Yamanaka K, Ogura T, Niki H, Hiraga S. Identification of two new genes, mukE and mukF, involved in chromosome partitioning in Escherichia coli. Mol Gen Genet. 1996;250:241–251. doi: 10.1007/BF02174381. [DOI] [PubMed] [Google Scholar]
- 4846.Yamao F, Inokuchi H, Cheng A, Ozeki H, Soll D. Escherichia coli glutaminyl-tRNA synthetase. I. Isolation and DNA sequence of the glnS gene. J Biol Chem. 1982;257:11639–11643. [PubMed] [Google Scholar]
- 4847.Yamashino T, Isomura M, Ueguchi C, Mizuno T. The yhhP gene encoding a small ubiquitous protein is fundamental for normal cell growth of Escherichia coli. J Bacteriol. 1998;180:2257–2261. doi: 10.1128/jb.180.8.2257-2261.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4848.Yamashita M, Azakami H, Yokoro N, Roh J H, Suzuki H, Kumagai H, Murooka Y. maoB, a gene that encodes a positive regulator of the monoamine oxidase gene (maoA) in Escherichia coli. J Bacteriol. 1996;178:2941–2947. doi: 10.1128/jb.178.10.2941-2947.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4849.Yamato I, Ohsawa M, Anraku Y. Defective cation-coupling mutants of Escherichia coli Na+/proline symport carrier. J Biol Chem. 1990;265:2450–2455. [PubMed] [Google Scholar]
- 4850.Yamato I, Anraku Y. Genetic and biochemical studies of transport systems for branched-chain amino acids in Escherichia coli K-12: isolation and properties of mutants defective in leucine-repressible transport activities. J Bacteriol. 1980;144:36–44. doi: 10.1128/jb.144.1.36-44.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4851.Yang C, Carlow D, Wolfenden R, Short S A. Cloning and nucleotide sequence of the Escherichia coli cytidine deaminase (cdd) gene. Biochemistry. 1992;31:4168–4174. doi: 10.1021/bi00132a003. [DOI] [PubMed] [Google Scholar]
- 4852.Yang G, Sandalova T, Lohman K, Lindqvist Y, Rendina A R. Active site mutants of Escherichia coli dethiobiotin synthetase: effects of mutations on enzyme catalytic and structural properties. Biochemistry. 1997;36:4751–4760. doi: 10.1021/bi9631677. [DOI] [PubMed] [Google Scholar]
- 4853.Yang H, Sasarman A, Inokuchi H, Adler J. Non-iron porphyrins cause tumbling to blue light by an Escherichia coli mutant defective in hemG. Proc Natl Acad Sci USA. 1996;93:2459–2463. doi: 10.1073/pnas.93.6.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4854.Yang H, Liu M Y, Romeo T. Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the CsrA gene product. J Bacteriol. 1996;178:1012–1017. doi: 10.1128/jb.178.4.1012-1017.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4855.Yang J, Camakaris H, Pittard A J. Mutations in the tyrR gene of Escherichia coli which affect TyrR-mediated activation but not TyrR-mediated repression. J Bacteriol. 1993;175:6372–6375. doi: 10.1128/jb.175.19.6372-6375.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4856.Yang J, Pittard J. Molecular analysis of the regulatory region of the Escherichia coli tyrB gene. J Bacteriol. 1987;169:4710–4715. doi: 10.1128/jb.169.10.4710-4715.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4857.Yang J, Ganesan S, Sarsero J P, Pittard A J. A genetic analysis of various functions of the TyrR protein of Escherichia coli. J Bacteriol. 1993;175:1767–1776. doi: 10.1128/jb.175.6.1767-1776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4858.Yang M, Luoh S M, Goddard A, Reilly D, Henzel W, Bass S. The bglX gene located at 47.8 min on the Escherichia coli chromosome encodes a periplasmic beta-glucosidase. Microbiology. 1996;142:1659–1665. doi: 10.1099/13500872-142-7-1659. [DOI] [PubMed] [Google Scholar]
- 4859.Yang M T, Scott H B N, Gardner J F. Transcription termination at the thr attenuator. Evidence that the adenine residues upstream of the stem and loop structure are not required for termination. J Biol Chem. 1995;270:23330–23336. doi: 10.1074/jbc.270.40.23330. [DOI] [PubMed] [Google Scholar]
- 4860.Yang S-Y, Schulz H. The large subunit of the fatty acid oxidation complex from Escherichia coli is a multifunctional polypeptide. Evidence for the existence of a fatty acid oxidation operon (fadAB) in Escherichia coli. J Biol Chem. 1983;258:9780–9785. [PubMed] [Google Scholar]
- 4861.Yang S-Y, Li J, He X-Y, Cosloy S D, Schulz H. Evidence that the fadB gene of the fadAB operon of Escherichia coli encodes 3-hydroxyacl-coenzyme A (CoA) epimerase, Δ3-cis-Δ2-trans-enoyl-CoA isomerase, and enoyl-CoA hydratase in addition to 3-hydroxyacyl-CoA dehydrogenase. J Bacteriol. 1988;170:2543–2548. doi: 10.1128/jb.170.6.2543-2548.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4862.Yang S-Y, Yang X-Y, Healy-Louie G, Schulz H, Elzinga M. Nucleotide sequence of the fadA gene. Primary structure of 3-ketoacyl-coenzyme A thiolase from Escherichia coli and the structural organization of the fadAB operon. J Biol Chem. 1990;265:10424–10429. . (Erratum, 166:16255, 1991.) [PubMed] [Google Scholar]
- 4863.Yang T P, Depew R E. Nucleotide sequence of a region duplicated in Escherichia coli toc mutants. Biochim Biophys Acta. 1992;1130:227–228. doi: 10.1016/0167-4781(92)90535-8. [DOI] [PubMed] [Google Scholar]
- 4864.Yang T P, Depew R E. Physical map of the tolC-htrP region of the Escherichia coli chromosome. J Bacteriol. 1992;174:1700–1701. doi: 10.1128/jb.174.5.1700-1701.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4865.Yang W, Ni L, Somerville R L. A stationary-phase protein of Escherichia coli that affects the mode of association between the trp repressor protein and operator-bearing DNA. Proc Natl Acad Sci USA. 1993;90:5796–5800. doi: 10.1073/pnas.90.12.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4866.Yang Y, Zhao G, Winkler M E. Identification of the pdxK gene that encodes pyridoxine (vitamin B6) kinase in Escherichia coli K-12. FEMS Microbiol Lett. 1996;141:89–95. doi: 10.1111/j.1574-6968.1996.tb08368.x. [DOI] [PubMed] [Google Scholar]
- 4867.Yang Y, Tsui H C, Man T, Winkler M E. Identification and function of the pdxY gene, which encodes a novel pyridoxal kinase involved in the salvage pathway of pyridoxal 5′-phosphate biosynthesis in Escherichia coli K-12. J Bacteriol. 1998;180:1814–1821. doi: 10.1128/jb.180.7.1814-1821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4868.Yang Y B, Lian J, Tai P C. Differential translocation of protein precursors across SecY-deficient membranes of Escherichia coli: SecY is not obligatorily required for translocation of certain secretory proteins in vitro. J Bacteriol. 1997;179:7386–7393. doi: 10.1128/jb.179.23.7386-7393.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4869.Yang Y-L, Polisky B. Suppression of ColE1 high-copy-number mutants by mutations in the polA gene of Escherichia coli. J Bacteriol. 1993;175:428–437. doi: 10.1128/jb.175.2.428-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4870.Yano R, Nagai H, Shiba K, Yura T. A mutation that enhances synthesis of ς32 and suppresses temperature-sensitive growth of the rpoH15 mutant of Escherichia coli. J Bacteriol. 1990;172:2124–2130. doi: 10.1128/jb.172.4.2124-2130.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4871.Yanofsky C, Platt T, Crawford I P, Nichols B, Christie G E, Horowitz H, VanCleemput M, Wu A M. The complete nucleotide sequence of the tryptophan operon of E. coli. Nucleic Acids Res. 1981;9:6647–6668. doi: 10.1093/nar/9.24.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4872.Yanofsky C, Horn V. Role of regulatory features of the trp operon of Escherichia coli in mediating a response to a nutritional shift. J Bacteriol. 1994;176:6245–6254. doi: 10.1128/jb.176.20.6245-6254.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4873.Yanofsky C, Horn V, Nakamura Y. Loss of overproduction of polypeptide release factor 3 influences expression of the tryptophanase operon of Escherichia coli. J Bacteriol. 1996;178:3755–3762. doi: 10.1128/jb.178.13.3755-3762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4874.Yao M, Kow Y W. Further characterization of Escherichia coli endonuclease V. Mechanism of recognition for deoxyinosine, deoxyuridine, and base mismatches in DNA. J Biol Chem. 1997;272:30774–30779. doi: 10.1074/jbc.272.49.30774. [DOI] [PubMed] [Google Scholar]
- 4875.Yao Z, Valvano M A. Genetic analysis of the O-specific lipopolysaccharide biosynthesis region (rfb) of Escherichia coli K-12 W3110: identification of genes that confer group 6 specificity to Shigella flexneri serotypes Y and 4a. J Bacteriol. 1994;176:4133–4143. doi: 10.1128/jb.176.13.4133-4143.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4876.Yarmolinsky M, Wiesmeyer H, Kalckar H M, Jordan E. Hereditary defects in galactose metabolism in Escherichia coli mutants. II. Galactose-induced sensitivity. Proc Natl Acad Sci USA. 1959;45:1786–1791. doi: 10.1073/pnas.45.12.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4877.Yasuda T, Nagata T, Ohmori H. Multicopy suppressors of the cold-sensitive phenotype of the pcsA68 (dinD68) mutation in Escherichia coli. J Bacteriol. 1996;178:3854–3859. doi: 10.1128/jb.178.13.3854-3859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4878.Yazyu H, Shiota-Niiya S, Shimamoto T, Kanazawa H, Futai M, Tsuchiya T. Nucleotide sequence of the melB gene and characteristics of deduced amino acid sequence of the melibiose carrier in Escherichia coli. J Biol Chem. 1985;259:4320–4326. [PubMed] [Google Scholar]
- 4879.Yerushalmi H, Lebendiker M, Schuldiner S. EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J Biol Chem. 1995;270:6856–6863. doi: 10.1074/jbc.270.12.6856. [DOI] [PubMed] [Google Scholar]
- 4880.Yerushalmi H, Lebendiker M, Schuldiner S. Negative dominance studies demonstrate the oligomeric structure of EmrE, a multidrug antiporter from Escherichia coli. J Biol Chem. 1996;271:31044–31048. doi: 10.1074/jbc.271.49.31044. [DOI] [PubMed] [Google Scholar]
- 4881.Yi Q-M, Lutkenhaus J. The nucleotide sequence of the essential cell-division gene ftsZ of Escherichia coli. Gene. 1985;36:241–247. doi: 10.1016/0378-1119(85)90179-9. [DOI] [PubMed] [Google Scholar]
- 4882.Yi Q-M, Rockenbach S, Ward J E, Lutkenhaus J. Structure and expression of the cell-division genes ftsQ, ftsA, and ftsZ. J Mol Biol. 1985;184:399–412. doi: 10.1016/0022-2836(85)90290-6. [DOI] [PubMed] [Google Scholar]
- 4883.Yim H H, Villarejo M. osmY, a new hyperosmotically inducible gene, encodes a periplasmic protein in Escherichia coli. J Bacteriol. 1992;174:3637–3644. doi: 10.1128/jb.174.11.3637-3644.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4884.Yim H H, Brems R L, Villarejo M. Molecular characterization of the promoter of osmY, an rpoS-dependent gene. J Bacteriol. 1994;176:100–107. doi: 10.1128/jb.176.1.100-107.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4885.Yin K-C, Blinkowa A, Walker J R. Nucleotide sequence of the Escherichia coli replication gene dnaZX. Nucleic Acids Res. 1986;14:6541–6550. doi: 10.1093/nar/14.16.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4886.Yoakum G H, Grossman L. Identification of E. coli uvrC protein. Nature. 1981;292:171–173. doi: 10.1038/292171a0. [DOI] [PubMed] [Google Scholar]
- 4887.Yokota T, Sugisaki H, Takanami M, Kaziro Y. The nucleotide sequence of the cloned tufA gene of Escherichia coli. Gene. 1980;12:25–31. doi: 10.1016/0378-1119(80)90012-8. [DOI] [PubMed] [Google Scholar]
- 4888.Yonezawa M, Takahata M, Banzawa N, Matsubara N, Watanabe Y, Narita H. Analysis of the NH2-terminal 83rd amino acid of Escherichia coli GyrA in quinolone-resistance. Microbiol Immunol. 1995;39:243–247. doi: 10.1111/j.1348-0421.1995.tb02196.x. [DOI] [PubMed] [Google Scholar]
- 4889.Yoo S J, Seol J H, Shin D H, Rohrwild M, Kang M S, Tanaka K, Goldberg A L, Chung C H. Purification and characterization of the heat shock proteins HslV and HslU that form a new ATP-dependent protease in Escherichia coli. J Biol Chem. 1996;271:14035–14040. doi: 10.1074/jbc.271.24.14035. [DOI] [PubMed] [Google Scholar]
- 4890.Yoo S J, Seol J H, Kang M S, Chung C H. Poly-l-lysine activates both peptide and ATP hydrolysis by the ATP-dependent HslVU protease in Escherichia coli. Biochem Biophys Res Commun. 1996;229:531–535. doi: 10.1006/bbrc.1996.1838. [DOI] [PubMed] [Google Scholar]
- 4891.Yoo S J, Seol J H, Kang M S, Ha D B, Chung C H. clpX encoding an alternative ATP-binding subunit of protease Ti (Clp) can be expressed independently from clpP in Escherichia coli. Biochem Biophys Res Commun. 1994;203:798–804. doi: 10.1006/bbrc.1994.2253. [DOI] [PubMed] [Google Scholar]
- 4892.York M K, Stodolsky M. Characterization of P1argF derivatives from Escherichia coli K12 transduction. Mol Gen Genet. 1981;181:230–240. doi: 10.1007/BF00268431. [DOI] [PubMed] [Google Scholar]
- 4893.Yoshida K, Kojima T, Yamagishi J-I, Nakamura S. Quinolone-resistant mutations of the gyrA gene of Escherichia coli. Mol Gen Genet. 1988;211:1–7. doi: 10.1007/BF00338386. [DOI] [PubMed] [Google Scholar]
- 4894.Yoshida T, Ueguchi C, Mizuno T. Physical map location of a set of Escherichia coli genes (hde) whose expression is affected by the nucleoid protein H-NS. J Bacteriol. 1993;175:7747–7748. doi: 10.1128/jb.175.23.7747-7748.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4895.Yoshikawa A, Isono S, Sheback A, Isono K. Cloning and nucleotide sequencing of the genes rimI and rimJ which encodes enzymes acetylating ribosomal proteins S18 and S5 of Escherichia coli K-12. Mol Gen Genet. 1987;209:481–488. doi: 10.1007/BF00331153. [DOI] [PubMed] [Google Scholar]
- 4896.Yoshikawa M, Okuyuma A, Tanaka N. A third kasugamycin resistance locus, ksgC, affecting ribosomal protein S2 in Escherichia coli K-12. J Bacteriol. 1975;122:796–797. doi: 10.1128/jb.122.2.796-797.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4897.Yoshimoto T, Nagai H, Ito K, Tsuru D. Location of the 7α-hydroxysteroid dehydrogenase gene (hdhA) on the physical map of the Escherichia coli chromosome. J Bacteriol. 1993;175:5730. doi: 10.1128/jb.175.17.5730.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4897a.Yoshimoto T, Tone H, Honda T, Osatomi K, Kobayashi R, Tsuru D. Sequencing and high expression of aminopeptidase P gene from Escherichia coli HB101. J Biochem. 1989;105:412–416. doi: 10.1093/oxfordjournals.jbchem.a122678. [DOI] [PubMed] [Google Scholar]
- 4898.Yoshimura M, Kimura A, Ohno M, Inokuchi H, Ozeki H. Identification of transfer RNA suppressors in Escherichia coli. III. Ochre suppressors of lysine tRNA. J Mol Biol. 1984;177:609–625. doi: 10.1016/0022-2836(84)90040-8. [DOI] [PubMed] [Google Scholar]
- 4899.Yoshimura M, Inokuchi H, Ozeki H. Identification of transfer RNA suppressors in Escherichia coli. IV. Amber suppressor Su+6, a double mutant of a new species of leucine tRNA. J Mol Biol. 1984;177:627–644. doi: 10.1016/0022-2836(84)90041-x. [DOI] [PubMed] [Google Scholar]
- 4900.Yoshimura M, Ashiuchi M, Esaki N, Kobatake C, Choi S-Y. Expression of glr (murI, dga) gene encoding glutamate racemase in Escherichia coli. J Biol Chem. 1993;268:24242–24246. [PubMed] [Google Scholar]
- 4901.Young I G, Rogers B L, Campbell H D, Jaworowski A, Shaw D C. Nucleotide sequence coding for the respiratory NADH dehydrogenase of Escherichia coli: UUG initiation codon. Eur J Biochem. 1981;116:165–170. doi: 10.1111/j.1432-1033.1981.tb05314.x. [DOI] [PubMed] [Google Scholar]
- 4902.Young I G, McCann L M, Stroobant P, Gibson F, Gibson F P. Characterization and genetic analysis of mutant strains of Escherichia coli K-12 accumulating the ubiquinone precursors 2-octaprenyl-6-methoxy-1,4-benzoquinone and 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinone. J Bacteriol. 1971;105:769–778. doi: 10.1128/jb.105.3.769-778.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4903.Young K, Silver L L, Bramhill D, Cameron P, Eveland S S, Raetz C R, Hyland S A, Anderson M S. The envA permeability/cell division gene of Escherichia coli encodes UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase. J Biol Chem. 1995;270:30384–30391. doi: 10.1074/jbc.270.51.30384. [DOI] [PubMed] [Google Scholar]
- 4904.Young R A. Transcription termination in the Escherichia coli ribosomal RNA operon rrnC. J Biol Chem. 1979;254:12725–12731. [PubMed] [Google Scholar]
- 4905.Yu F, Yamada H, Daishima K, Mizushima S. Nucleotide sequence of the lspA gene, the structural gene for lipoprotein signal peptidase of Escherichia coli. FEBS Lett. 1984;173:264–268. doi: 10.1016/0014-5793(84)81060-1. [DOI] [PubMed] [Google Scholar]
- 4906.Yu F, Inouye S, Inouye M. Lipoprotein-28, a cytoplasmic membrane lipoprotein from Escherichia coli. Cloning, DNA sequence, and expression of its gene. J Biol Chem. 1986;261:2284–2288. [PubMed] [Google Scholar]
- 4907.Yu F, Jen Y, Takeuchi E, Inouye M, Nakayama H, Tagaya M, Fukui T. Alpha-glucan phosphorylase from Escherichia coli. Cloning of the gene, and purification and characterization of the protein. J Biol Chem. 1988;263:13706–13711. [PubMed] [Google Scholar]
- 4908.Yu X C, Tran A H, Sun Q, Margolin W. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J Bacteriol. 1998;180:1296–1304. doi: 10.1128/jb.180.5.1296-1304.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4909.Yuan R, Hamilton D. Restriction and modification of DNA by a complex protein. Am Sci. 1982;70:61–69. [PubMed] [Google Scholar]
- 4910.Yuasa S, Sakakibara Y. Identification of dnaA and dnaN gene products of Escherichia coli. Mol Gen Genet. 1980;180:267–273. doi: 10.1007/BF00425838. [DOI] [PubMed] [Google Scholar]
- 4911.Yudkin M. Unstable mutations that relieve catabolite repression of tryptophanase synthesis by Escherichia coli. J Bacteriol. 1977;130:57–61. doi: 10.1128/jb.130.1.57-61.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4912.Yura T, Igarishi K. RNA polymerase mutants of Escherichia coli. I. Mutants resistant to streptovaricin. Proc Natl Acad Sci USA. 1968;61:1313–1319. doi: 10.1073/pnas.61.4.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4913.Yura T, Tobe T, Ito K, Osawa T. Heat shock regulatory gene (htpR) of Escherichia coli is required for growth at high temperature but is dispensable at low temperature. Proc Natl Acad Sci USA. 1984;81:6803–6807. doi: 10.1073/pnas.81.21.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4914.Zaitsev E, Alexseyev A, Lanzov V A, Satin L H, Clark A J. Nucleotide sequence between recA and alaSp in E. coli K12 and the sequence change in four recA mutations. Mutat Res. 1994;323:173–177. doi: 10.1016/0165-7992(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 4915.Zaitseva J, Zhang H, Binnie R A, Hermodson M. The proteins encoded by the rbs operon of Escherichia coli. II. Use of chimeric protein constructs to isolate and characterize RbsC. Protein Sci. 1996;5:1100–1107. doi: 10.1002/pro.5560050612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4916.Zakin M M, Duchange N, Ferrara P, Cohen G N. Nucleotide sequence of the metL gene of Escherichia coli. Its product, the bifunctional aspartokinase II-homoserine dehydrogenase II, and the bifunctional product of the thrA gene, aspartokinase I-homoserine dehydrogenase I, derive from a common ancestor. J Biol Chem. 1983;258:3028–2031. [PubMed] [Google Scholar]
- 4917.Zakin M M, Greene R C, Dautry-Varsat A, Duchange N, Ferrara P, Py M-C, Margarita D, Cohen G N. Construction and physical mapping of plasmids containing the metJBLF gene cluster of Escherichia coli K12. Mol Gen Genet. 1982;187:101–106. doi: 10.1007/BF00384390. [DOI] [PubMed] [Google Scholar]
- 4918.Zambrano M M, Kolter R. Escherichia coli mutants lacking NADH dehydrogenase I have a competitive disadvantage in stationary phase. J Bacteriol. 1993;175:5642–5647. doi: 10.1128/jb.175.17.5642-5647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4919.Zaniewski R, Deutscher M P. Genetic mapping of a mutation in Escherichia coli leading to a temperature-sensitive RNase D. Mol Gen Genet. 1982;185:142–147. doi: 10.1007/BF00333804. [DOI] [PubMed] [Google Scholar]
- 4920.Zavitz K H, DiGate R J, Marians K J. The PriB and PriC replication proteins of Escherichia coli: genes, DNA sequence, overexpression, and purification. J Biol Chem. 1991;266:13988–13995. [PubMed] [Google Scholar]
- 4921.Zawadzke L E, Bugg T D, Walsh C T. Existence of two d-alanine:d-alanine ligases in Escherichia coli: cloning and sequencing of the ddlA gene and purification and characterization of the DdlA and DdlB enzymes. Biochemistry. 1991;30:1673–1682. doi: 10.1021/bi00220a033. [DOI] [PubMed] [Google Scholar]
- 4922.Zdych E, Peist R, Reidl J, Boos W. MalY of Escherichia coli is an enzyme with the activity of a βC-S lyase (cystathionase) J Bacteriol. 1995;177:5035–5039. doi: 10.1128/jb.177.17.5035-5039.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4923.Zechiedrich E L, Cozzarelli N R. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes Dev. 1995;9:2859–2869. doi: 10.1101/gad.9.22.2859. [DOI] [PubMed] [Google Scholar]
- 4924.Zehnbauer B A, Markovitz A. Cloning of gene lon (capR) of Escherichia coli K-12 and identification of polypeptides specified by the cloned deoxyribonucleic acid fragment. J Bacteriol. 1980;143:852–863. doi: 10.1128/jb.143.2.852-863.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4925.Zehnbauer B A, Foley E C, Henderson G W, Markovitz A. Identification and purification of the Lon+ (capR+) gene product, a DNA-binding protein. Proc Natl Acad Sci USA. 1981;78:2043–2047. doi: 10.1073/pnas.78.4.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4926.Zeng G, Ye S, Larson T J. Repressor for the sn-glycerol 3-phosphate regulon of Escherichia coli K-12: primary structure and identification of the DNA-binding domain. J Bacteriol. 1996;178:7080–7089. doi: 10.1128/jb.178.24.7080-7089.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4927.Zengel J M, Mueckl D, Lindahl L. Protein L4 of the E. coli ribosome regulates an eleven gene r protein operon. Cell. 1980;21:523–535. doi: 10.1016/0092-8674(80)90490-0. [DOI] [PubMed] [Google Scholar]
- 4928.Zengel J M, Lindahl L. A secondary promoter for elongation factor Tu synthesis in the str ribosomal protein operon of Escherichia coli. Mol Gen Genet. 1982;185:487–492. doi: 10.1007/BF00334145. [DOI] [PubMed] [Google Scholar]
- 4929.Zengel J M, Archer R H, Lindahl L. The nucleotide sequence of the Escherichia coli fus gene, coding for elongation factor G. Nucleic Acids Res. 1984;12:2181–2192. doi: 10.1093/nar/12.4.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4930.Zenno S, Koike H, Kumar A N, Jayaraman R, Tanokura M, Saigo K. Biochemical characterization of NfsA, the Escherichia coli major nitroreductase exhibiting a high amino acid sequence homology to Frp, a Vibrio harveyi flavin oxidoreductase. J Bacteriol. 1996;178:4508–4514. doi: 10.1128/jb.178.15.4508-4514.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4931.Zenno S, Koike H, Tanokura M, Saigo K. Identification of the gene encoding the NfsB/NfnB nitroreductase associated with a low level of flavin reductase activity in Escherichia coli K12. 1994. GenBank submission D25414. [Google Scholar]
- 4932.Zenno S, Koike H, Tanokura M, Saigo K. Gene cloning, purification, and characterization of NfsB, a minor oxygen-insensitive nitroreductase from Escherichia coli, similar in biochemical properties to FRase I, the major flavin reductase in Vibrio fischeri. J Biochem (Tokyo) 1996;120:736–744. doi: 10.1093/oxfordjournals.jbchem.a021473. [DOI] [PubMed] [Google Scholar]
- 4933.Zhang A, Belfort M. Nucleotide sequence of a newly-identified Escherichia coli gene, stpA, encoding an H-NS-like protein. Nucleic Acids Res. 1992;20:6735. doi: 10.1093/nar/20.24.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4934.Zhang A, Rimsky S, Reaban M E, Buc H, Belfort M. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 1996;15:1340–1349. [PMC free article] [PubMed] [Google Scholar]
- 4935.Zhang A, Derbyshire V, Salvo J L, Belfort M. Escherichia coli protein StpA stimulates self-splicing by promoting RNA assembly in vitro. RNA. 1995;1:783–793. [PMC free article] [PubMed] [Google Scholar]
- 4936.Zhang G, Deng E, Baugh L R, Hamilton C M, Maples V F, Kushner S R. Conserved motifs II to VI of DNA helicase II from Escherichia coli are all required for biological activity. J Bacteriol. 1997;179:7544–7550. doi: 10.1128/jb.179.23.7544-7550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4937.Zhang H L, Malpure S, DiGate R J. Escherichia coli DNA topoisomerase III is a site-specific DNA binding protein that binds asymmetrically to its cleavage site. J Biol Chem. 1995;270:23700–23705. doi: 10.1074/jbc.270.40.23700. [DOI] [PubMed] [Google Scholar]
- 4938.Zhang J R, Deutscher M P. Escherichia coli RNase D: sequencing of the rnd structural gene and purification of the overexpressed protein. Nucleic Acids Res. 1988;16:6265–6278. doi: 10.1093/nar/16.14.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4939.Zhang M, Deutscher M P. Cloning, characterization and effects of overexpression of the Escherichia coli rnd gene encoding RNase D. J Bacteriol. 1988;170:522–527. doi: 10.1128/jb.170.2.522-527.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4940.Zhang P, Vigne J L, Mellon S H. Polyribonucleotide phosphorylase is a double-stranded DNA-binding protein. DNA Cell Biol. 1998;17:169–175. doi: 10.1089/dna.1998.17.169. [DOI] [PubMed] [Google Scholar]
- 4941.Zhang W, Evans D H. DNA strand transfer catalyzed by the 5′-3′ exonuclease domain of Escherichia coli DNA polymerase I. Nucleic Acids Res. 1995;23:4620–4627. doi: 10.1093/nar/23.22.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4942.Zhang X, Zhu L, Deutscher M P. Oligoribonuclease is encoded by a highly conserved gene in the 3′-5′ exonuclease superfamily. J Bacteriol. 1998;180:2779–2781. doi: 10.1128/jb.180.10.2779-2781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4943.Zhang X, Bremer H. Effects of Fis on ribosome synthesis and activity and on rRNA promoter activities in Escherichia coli. J Mol Biol. 1996;259:27–40. doi: 10.1006/jmbi.1996.0299. [DOI] [PubMed] [Google Scholar]
- 4944.Zhang Y, Craig J E, Gallagher M P. Location of the nupC gene on the physical map of the Escherichia coli K-12. J Bacteriol. 1992;174:5758–5759. doi: 10.1128/jb.174.17.5758-5759.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4945.Zhang Y, Hanna M M. NusA changes the conformation of Escherichia coli RNA polymerase at the binding site for the 3′ end of the nascent RNA. J Bacteriol. 1994;176:1787–1789. doi: 10.1128/jb.176.6.1787-1789.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4946.Zhao G, Pease A J, Bharani N, Winkler M E. Biochemical characterization of gapB-encoded erythrose 4-phosphate dehydrogenase of Escherichia coli K-12 and its possible role in pyridoxal 5′-phosphate biosynthesis. J Bacteriol. 1995;177:2804–2812. doi: 10.1128/jb.177.10.2804-2812.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4947.Zhao G, Winkler M E. An Escherichia coli K-12 tktA tktB mutant deficient in transketolase activity requires pyridoxine (vitamin B6) as well as the aromatic amino acids and vitamins for growth. J Bacteriol. 1994;176:6134–6138. doi: 10.1128/jb.176.19.6134-6138.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4948.Zhao G, Winkler M E. 4-Phospho-hydroxy-l-threonine is an obligatory intermediate in pyridoxal 5′-phosphate coenzyme biosynthesis in Escherichia coli K-12. FEMS Microbiol Lett. 1996;135:275–280. doi: 10.1111/j.1574-6968.1996.tb08001.x. [DOI] [PubMed] [Google Scholar]
- 4949.Zhao G, Winkler M E. A novel α-ketoglutarate reductase activity of the serA-encoded 3-phosphoglycerate dehydrogenase of Escherichia coli K-12 and its possible implications for human 2-hydroxyglutaric aciduria. J Bacteriol. 1996;178:232–239. doi: 10.1128/jb.178.1.232-239.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4950.Zhao S, Sandt C H, Feulner G, Vlazny D A, Gray J A, Hill C W. Rhs elements of Escherichia coli K-12: complex composites of shared and unique components that have different evolutionary histories. J Bacteriol. 1993;175:2799–2808. doi: 10.1128/jb.175.10.2799-2808.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4951.Zheng M, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. . (Comment.) [DOI] [PubMed] [Google Scholar]
- 4952.Zhou Y, Gottesman S. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J Bacteriol. 1998;180:1154–1158. doi: 10.1128/jb.180.5.1154-1158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4953.Zhou Y N, Chatterjee S, Roy S, Adhya S. The non-inducible nature of super-repressors of the gal operon in Escherichia coli. J Mol Biol. 1995;253:414–425. doi: 10.1006/jmbi.1995.0563. [DOI] [PubMed] [Google Scholar]
- 4954.Zhu N, Olivera B M, Roth J R. Genetic characterization of the pnuC gene, which encodes a component of the nicotinamide mononucleotide transport system in Salmonella typhimurium. J Bacteriol. 1989;171:4402–4409. doi: 10.1128/jb.171.8.4402-4409.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4955.Zhu Y, Lin E C C. A mutant crp allele that differentially activates the operons of the fuc regulon in Escherichia coli. J Bacteriol. 1988;170:2352–2358. doi: 10.1128/jb.170.5.2352-2358.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4956.Zientz E, Six S, Unden G. Identification of a third secondary carrier (DcuC) for anaerobic C4-dicarboxylate transport in Escherichia coli: roles of the three Dcu carriers in uptake and exchange. J Bacteriol. 1996;178:7241–7247. doi: 10.1128/jb.178.24.7241-7247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4957.Zijderveld C A L, Aarsman M E G, Nanninga N. Differences between inner membrane and peptidoglycan-associated PBP1B dimers of Escherichia coli. J Bacteriol. 1995;177:1860–1863. doi: 10.1128/jb.177.7.1860-1863.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4958.Zilhao R, Caillet J, Regnier P, Arraiano C M. Precise physical mapping of the Escherichia coli rnb gene, encoding ribonuclease II. Mol Gen Genet. 1995;248:242–246. doi: 10.1007/BF02190807. [DOI] [PubMed] [Google Scholar]
- 4959.Zilhao R, Plumbridge J A, Hajnsdorf E, Regnier P, Arraiano C M. Escherichia coli RNase II: characterization of the promoters involved in the transcription of rnb. Microbiology. 1996;142:367–375. doi: 10.1099/13500872-142-2-367. [DOI] [PubMed] [Google Scholar]
- 4960.Zilhao R, Camelo L, Arraiano C M. DNA sequencing and expression of the gene rnb encoding Escherichia coli ribonuclease II. Mol Microbiol. 1993;8:43–51. doi: 10.1111/j.1365-2958.1993.tb01201.x. [DOI] [PubMed] [Google Scholar]
- 4961.Zimmerman R A, Ikeya Y, Sparling P F. Alteration of ribosomal protein S4 by mutation linked to kasugamycin-resistance in Escherichia coli. Proc Natl Acad Sci USA. 1973;70:71–75. doi: 10.1073/pnas.70.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4962.Zimmermann L, Hantke K, Braun V. Exogenous induction of the iron dicitrate transport system of Escherichia coli K-12. J Bacteriol. 1984;159:271–277. doi: 10.1128/jb.159.1.271-277.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4963.Zinkewich-Peotti K, Fraser J M. New locus for exopolysaccharide overproduction in Escherichia coli K-12. J Bacteriol. 1988;170:1405–1407. doi: 10.1128/jb.170.3.1405-1407.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4964.Zinoni F, Birkmann A, Stadtman T C, Bock A. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc Natl Acad Sci USA. 1986;83:4650–4654. doi: 10.1073/pnas.83.13.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4965.Zinoni F, Birkmann A, Leinfelder W, Bock A. Cotranslational insertion of selenocysteine into formate dehydrogenase from Escherichia coli directed by a UGA codon. Proc Natl Acad Sci USA. 1987;84:3156–3160. doi: 10.1073/pnas.84.10.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4966.Zissler J, Signer E, Schaefer F. The role of recombination in growth of bacteriophage lambda. I. The gamma gene. In: Hershey A D, editor. The bacteriophage lambda. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1971. pp. 455–468. [Google Scholar]
- 4967.Zuckier G, Torriani A. Genetic and physiological tests of three phosphate-specific transport mutants of Escherichia coli. J Bacteriol. 1981;145:1249–1256. doi: 10.1128/jb.145.3.1249-1256.1981. . (Erratum, 148:397, 1981.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4968.Zurawski G, Gunsalus R P, Brown K D, Yanofsky C. Structure and regulation of aroH, the structural gene for the tryptophan-repressible 3-deoxy-d-arabino-heptulosonic acid-7-phosphate synthetase of Escherichia coli. J Mol Biol. 1981;145:47–73. doi: 10.1016/0022-2836(81)90334-x. [DOI] [PubMed] [Google Scholar]
- 4969.Zurawski G, Zurawski S M. Structure of the Escherichia coli S10 ribosomal protein operon. Nucleic Acids Res. 1985;13:4521–4526. doi: 10.1093/nar/13.12.4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4970.Zwaig N, de Zwaig R N, Isturiz T, Wecksler M. Regulatory mutations affecting the gluconate system in Escherichia coli. J Bacteriol. 1973;114:469–473. doi: 10.1128/jb.114.2.469-473.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4971.Zwiebel L, Inukai M, Nakamura K, Inouye M. Preferential selection of deletion mutations of the outer membrane lipoprotein gene of Escherichia coli by globomycin. J Bacteriol. 1981;145:654–656. doi: 10.1128/jb.145.1.654-656.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]