Abstract
The single-celled cotton (Gossypium hirsutum) fiber provides an excellent model to investigate how human selection affects phenotypic evolution. To gain insight into the evolutionary genomics of cotton domestication, we conducted comparative transcriptome profiling of developing cotton fibers using RNA-Seq. Analysis of single-celled fiber transcriptomes from four wild and five domesticated accessions from two developmental time points revealed that at least one-third and likely one-half of the genes in the genome are expressed at any one stage during cotton fiber development. Among these, ∼5,000 genes are differentially expressed during primary and secondary cell wall synthesis between wild and domesticated cottons, with a biased distribution among chromosomes. Transcriptome data implicate a number of biological processes affected by human selection, and suggest that the domestication process has prolonged the duration of fiber elongation in modern cultivated forms. Functional analysis suggested that wild cottons allocate greater resources to stress response pathways, while domestication led to reprogrammed resource allocation toward increased fiber growth, possibly through modulating stress-response networks. This first global transcriptomic analysis using multiple accessions of wild and domesticated cottons is an important step toward a more comprehensive systems perspective on cotton fiber evolution. The understanding that human selection over the past 5,000+ years has dramatically re-wired the cotton fiber transcriptome sets the stage for a deeper understanding of the genetic architecture underlying cotton fiber synthesis and phenotypic evolution.
Author Summary
Ever since Darwin biologists have recognized that comparative study of crop plants and their wild relatives offers a powerful framework for generating insights into the mechanisms that underlie evolutionary change. Here, we study the domestication process in cotton, Gossypium hirsutum, an allopolyploid species (containing two different genomes) which initially was domesticated approximately 5000 years ago, and which primarily is grown for its single-celled seed fibers. Strong directional selection over the millennia was accompanied by transformation of the short, coarse, and brown fibers of wild plants into the long, strong, and fine white fibers of the modern cotton crop plant. To explore the evolutionary genetics of cotton domestication, we conducted transcriptome profiling of developing cotton fibers from multiple accessions of wild and domesticated cottons. Comparative analysis revealed that the domestication process dramatically rewired the transcriptome, affecting more than 5,000 genes, and with a more evenly balanced usage of the duplicated copies arising from genome doubling. We identify many different biological processes that were involved in this transformation, including those leading to a prolongation of fiber elongation and a reallocation of resources toward increased fiber growth in modern forms. The data provide a rich resource for future functional analyses targeting crop improvement and evolutionary objectives.
Introduction
Ever since Darwin's time, biologists have recognized that human domestication of wild plants and animals offers promising opportunities for enhancing our understanding of the evolutionary process. As highlighted in recent reviews [1], [2], comparisons among wild and domesticated forms of crop plants often lead to insights into the genetic architecture and developmental mechanisms that underlie traits subjected to strong directional human selection. The power of this approach is magnified by the recent advent of high-throughput “omics” technologies, which hold promise for leading us to an eventual systems-level understanding of phenotypic change. Domesticated forms of cultivated species differ from their wild counterparts in numerous traits, particularly those subjected to intentional directional selection, e.g., loss of seed dormancy, larger and/or more fruits, determinate growth, annualized habit, and earlier flowering. Insights into the evolution of this “domestication syndrome” [3] are made possible by comparative studies of wild and domesticated representatives of individual cultivated species [1], [2].
Upland cotton (Gossypium hirsutum L.) is the most important domesticated fiber plant in the world, accounting for more than 90% of global cotton production. Originally native to the northern coast of the Yucatan peninsula in Mexico, upland cotton is widely cultivated in over 50 countries in both hemispheres [4]. The trait for which cotton was initially domesticated is the remarkably elongated, single-celled epidermal trichomes, or hairs, that cover the cottonseed surface (colloquially termed “fibers”). These seed hairs vary greatly in length, color, strength, and density among the myriad wild, semi-domesticated, feral and modern annualized forms that collectively comprise the species G. hirsutum. In truly wild G. hirsutum trichomes are short (typically <1.5 cm), coarse, and are various shades of tan to brown. Gossypium hirsutum was initially domesticated at least 5000 years ago, and following millennia of directional selection, domesticated forms now produce long, strong, and fine white fibers along with a dramatically enhanced fiber yield. In addition to this increase in fiber length, strength, and quality, the domestication process brought about other morphological transformations, including decreased plant stature, earlier flowering, and loss of seed dormancy.
Gossypium hirsutum is an allotetraploid containing two diverged sets of chromosomes, “A” and “D”, which became reunited in a common nucleus as a result of a hybridization event approximately 1–2 million years ago (mya). This merger of an African/Asian, A genome (similar to modern G. arboreum) and an American, D genome (much like modern G. raimondii) gave rise to a new allopolyploid lineage that diversified into five species (AD1 to AD5) [4], [5]. Considering the importance of polyploidy as a major evolutionary process in plants and its prevalence in all flowering plants [6], comparative analyses of wild and domesticated cottons may provide new perspectives about how human selection affects duplicated genes in allopolyploids. In addition, many important crops, such as alfalfa, potato, wheat, soybean, and cabbage, are obvious polyploids, so studying gene expression in allopolyploid cotton has the potential to offer novel insights on the role of polyploidy in crop evolution (e.g., Bao et al.[7]).
To date, and despite its importance to understanding molecular mechanisms governing fiber development, there have been only a handful studies of global gene expression in G. hirsutum, using expressed sequence tags (ESTs) [8], [9] and microarrays [10]–[14]. In addition, most have focused on comparisons of modern, annualized G. hirsutum and its fiberless/lintless mutants. The single notable exception is the study of Rapp et al., who used microarrays to compare truly wild and domesticated G. hirsutum [13]. Notably, Rapp et al. explored global gene expression patterns in wild and domesticated G. hirsutum cotton fibers across five temporal/developmental time points, and found that about one quarter of all genes examined exhibited expression changes during domestication, indicating massive alteration of the cotton fiber transcriptome by domestication and crop improvement [13]. However, a limitation of the study of Rapp et al. is that they employed only one accession representing each of the wild and domesticated gene pools, raising the possibility that some of the observed differential expression might simply reflect expression variation that is unconnected to the evolutionary transformation of interest [13]. Also, the microarray methodology relies on less precise probe/target hybridization, is subject to high background noise, and has a narrower range of gene expression quantification, in comparison to profiling using RNA-Seq data [15]. Finally, the genome sequence for G. raimondii only recently became available [16], providing deeper annotation and better discrimination among homologs (and homoeologs), and hence enhanced power to decipher gene expression level changes across the whole genome.
Here, to gain insight into the evolutionary genomics of cotton domestication, we conducted comparative transcriptome profiling of developing cotton fibers from multiple accessions of wild and domesticated G. hirsutum using RNA-Seq data. Two developmental stages were studied, 10 and 20 days post anthesis (dpa), representing key stages of primary cell wall growth and the transition to secondary cell wall growth, respectively [17], [18]. By examining gene expression levels digitally, we found that approximately one-third of the genes in the genome are expressed in cotton fiber regardless of lineage, accession, and developmental stages. Notably, nearly 5,000 genes are diagnosed as being differentially expressed as a consequence of cotton fiber domestication. These data suggest that human selection has reprogrammed the transcriptome on a massive scale, and that part of this rewiring entails a reallocation from stress response pathways toward fiber growth.
Results
We performed global transcriptome profiling of developing cotton fibers from wild and domesticated G. hirsutum using RNA-Seq. A total of 310 million (M) reads was generated from 20 libraries, and on average 70% of these uniquely mapped to the reference genome (Table 1). To determine how many genes were expressed in fibers and whether there was variation among accessions, we first evaluated the number of expressed genes. Since we did not include external controls, such as the External RNA Controls Consortium (ERCC) controls, we used arbitrary measures for “expression”, such as RPKM = 2 or 5 ( = 32 or 80 short reads on average for a 1.6 kb-gene; RPKM = Reads Per Kilobase of gene model per Million mapped reads) [19]. Based on the criterion of RPKM≧5, approximately 12,700 (33.9%) and 12,000 (33.0%) genes were expressed at 10 and 20 dpa, respectively, in most accessions (Table 2); three domesticated accessions (Cascot L-7, Coker 315, and CRB250) showed lower numbers of expressed genes at 20 dpa compared to other accessions, which we attribute to the higher proportions of redundantly mapped reads in these accessions (data not shown). This is consistent with the fiber transcriptome diversity obtained from domesticated G. hirsutum cv. TM-1 [20] and from diploid cotton G. arboreum [21]. In general, more genes were expressed in wild than domesticated cottons at both developmental stages. More genes were expressed at 20 dpa than 10 dpa in wild cottons, while the opposite was observed for domesticated cottons (Table 2). Before identifying differentially expressed genes during domestication and development, variation among samples was evaluated using Multidimensional scaling (MDS). Because the transcriptome profile from Maxxa 10 dpa exhibited a large distance from other domesticated cottons and was embedded in wild cottons (Figure S1), it was excluded from further analyses. However, in general, samples clustered as expected, indicating that the variation among samples is largely explained by developmental stage and domestication.
Table 1. Accession information used in this study and the number and percentage of short reads mapped onto the Cotton D genome reference assembly.
| Species | Origin | Stage | Total Reads | Mapped Reads (%) |
| G. hirsutum var. yucatanense (TX2090) | Yucatan, Mexico | 10 dpa 20 dpa | 9,598,358 10,517,180 | 7,554,107 (78.7) 8,175,498 (77.7) |
| G. hirsutum var. yucatanense (TX2094) | Yucatan, Mexico | 10 dpa 20 dpa | 11,139,818 14,110,893 | 8,326,479 (74.7) 10,627,792 (75.3) |
| G. hirsutum var. yucatanense (TX2094 = YUC*) | Yucatan, Mexico | 10 dpa 20 dpa | 10,444,338 10,991,115 | 8,908,976 (85.3) 7,851,707 (71.4) |
| G. hirsutum var. yucatanense (TX2095) | Yucatan, Mexico | 10 dpa 20 dpa | 42,989,086 15,142,099 | 33,550,251 (78.0) 11,646,705 (76.9) |
| G. hirsutum var. palmeri (TX665) | Yucatan, Mexico | 10 dpa 20 dpa | 14,559,567 29,461,705 | 11,627,366 (79.9) 23,229,997 (78.8) |
| G. hirsutum cv. Cascot L-7 | Plains in U.S.A | 10 dpa 20 dpa | 10,688,995 31,345,469 | 8,488,820 (79.4) 17,785,760 (56.7) |
| G. hirsutum cv. Coker 315 | Eastern U.S.A | 10 dpa 20 dpa | 14,201,314 10,615,133 | 11,065,757 (77.9) 5,881,452 (55.4) |
| G. hirsutum cv. CRB252† | Eastern U.S.A | 10 dpa 20 dpa | 9,986,108 8,344,787 | 7,583,439 (75.9) 4,720,388 (56.6) |
| G. hirsutum cv. Maxxa* | Western U.S.A | 10 dpa 20 dpa | 12,266,868 15,484,446 | 9,410,233 (76.7) 12,353,194 (79.8) |
| G. hirsutum cv. Texas Marker 1 (TM1) | Delta in U.S.A. | 10 dpa 20 dpa | 14,706,919 13,078,240 | 11,766,434 (80.0) 9,003,797 (68.8) |
CRB252-Derives from a double cross, SG 248/PHY 72//ST 474/Maxxa.
data from the study number SRP001603 at NCBI SRA. Fiber samples from TX2094 and Maxxa were collected in different growing seasons; thus, this TX2094 was noted as “YUC” to be distinguished from TX2094, which was collected in the same year as the other materials.
Table 2. The number of genes expressed in Gossypium hirsutum and G. arboretum.
| Accession | Tissue* | domestication | RPKM≧2 (%) | RPKM≧5 (%) | N1† | Reference |
| TX665 | 10 | wild | 17,724 (47.3) | 12,501 (33.3) | This study | |
| TX2090 | 10 | wild | 18,846 (50.2) | 13,674 (36.5) | ||
| TX2095 | 10 | wild | 17,741 (47.3) | 12,383 (33.0) | ||
| TX2094 | 10 | wild | 19,057 (50.8) | 13,849 (36.9) | ||
| YUC | 10 | wild | 20,396 (54.4) | 15,096 (40.3) | ||
| Cascot L-7 | 10 | domesticated | 15,977 (42.6) | 11,150 (29.7) | ||
| Coker 315 | 10 | domesticated | 16,885 (45.5) | 11,852 (31.6) | ||
| CRB 252 | 10 | domesticated | 17,264 (46.0) | 12,260 (32.7) | ||
| TM1 | 10 | domesticated | 16,544 (44.1) | 11,884 (31.7) | ||
| Maxxa | 10 | domesticated | 17,465 (46.6) | 12,055 (32.1) | ||
| TX665 | 20 | wild | 18,487 (49.3) | 13,068 (34.8) | This study | |
| TX2090 | 20 | wild | 19,850 (52.9) | 15,033 (40.1) | ||
| TX2095 | 20 | wild | 19,393 (51.7) | 14,094 (37.6) | ||
| TX2094 | 20 | wild | 20,396 (54.4) | 15,026 (40.1) | ||
| YUC | 20 | wild | 19,877 (53.0) | 14,660 (39.1) | ||
| Cascot L-7 | 20 | domesticated | 13,154 (35.1) | 7,900 (21.1) | ||
| Coker 315 | 20 | domesticated | 10,804 (28.8) | 6,640 (17.7) | ||
| CRB 252 | 20 | domesticated | 14,223 (37.9) | 8,674 (23.1) | ||
| TM-1 | 20 | domesticated | 17,212 (45.9) | 11,730 (31.3) | ||
| Maxxa | 20 | domesticated | 16,652 (44.4) | 11,506 (30.7) | ||
| TM-1† | 2 | domesticated | 11,094 (29.6) | [20] | ||
| TM-1† | 7 | domesticated | 10,278 (27.4) | |||
| TM-1† | 10 | domesticated | 10,814 (28.8) | |||
| TM-1† | 20 | domesticated | 11,078 (29.5) | |||
| TM-1† | 25 | domesticated | 10,716 (28.6) | |||
| TM-1† | various§ | domesticated | 11,913 (31.8) | |||
| TX2094 | young leaf | wild | 18,794 (50.1) | 11,350 (32.5) | [55] | |
| Maxxa | young leaf | domesticated | 19,618 (52.3) | 12,183 (32.5) | ||
| G. arboreum | 10 to 24 | domesticated diploid | 12,227 (32.6) | [21] |
numbers indicate day post anthesis (dpa) fibers.
denotes the number of genes expressed at levels significantly different from zero (P<0.01) [20].
leaves, stems, petals, anthers, calyx, and bracts.
Transcriptomic change during development in wild and domesticated G. hirsutum
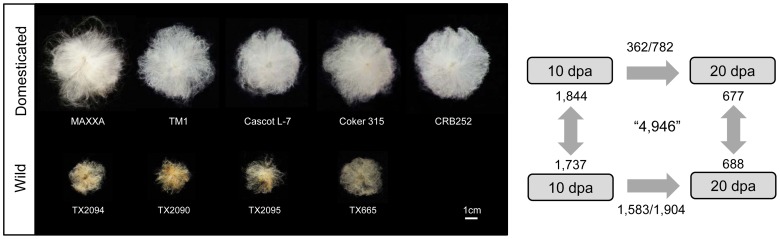
We profiled the transcriptome during development in wild and domesticated cottons using two developmental stages, 10 and 20 dpa. Both wild and domesticated cottons showed more gene up-regulation than down-regulation during the transition from 10 to 20 dpa, e.g., 782 vs. 362 in domesticated cottons (Figure 1). However, three times as many genes (3,487 vs. 1,144) were differentially expressed during development in wild cottons compared to domesticated cottons (Figure 1). This pattern is also supported by the MDS plot, which showed less variation between the two developmental stages of domesticated cottons compared to wild cottons (Figure S1). Our results with respect to developmental variation differ from those of Rapp et al. [13], where differential expression was observed at 2.6-times as many genes in domesticated cotton relative to a wild accession (5,851 vs. 2,207 with 1.5-fold change; Table S1). However, our results parallel those from a second domesticated allopolyploid, G. barbadense, using either microarrays [22] or RNA-Seq data (M.J. Yoo et al., unpublished data). To clarity the difference between the two studies, we reanalyzed our data using the same accessions used in Rapp et al. [13]. The inclusion of TX2094 and YUC as two biological replicates resulted in 10 times as many DE genes compared to a single accession analysis (TX2094 vs. “TX2094+YUC” = 436 vs. 4,520; Table S1) and a 30% increase in DE genes relative to the “All accession” analysis (4,520 vs. 3,487; Table S1). As for domesticated cottons, since we have only one replicate for TM1, we included a second domesticated cotton for comparison, which resulted in a 17∼21% increase compared to the “All accession” analysis (Table S1).These results suggest that the observed conflict between the two studies likely is explained by technical differences among platforms and the reference genome used.
Figure 1. Number of genes differentially expressed during fiber development within and between wild and domesticated cottons.
Left: Representative images of individual seeds with attached fiber are presented for domesticated (top) and wild (bottom) accessions. Right: Number of differentially expressed genes in developing cotton fiber within and between wild and domesticated cottons (RPKM≧1, FDR<0.05, fold-change≧1.5). For example, between two developmental stages within domesticated cottons, 362 genes were up-regulated at 10 dpa, whereas 762 genes were more highly expressed at 20 dpa. Similarly, between wild and domesticated cottons at 10 dpa, 1,844 genes were up-regulated in domesticated cottons, while 1,737 genes were more highly expressed in wild cottons.
As expected based on our understanding of primary and secondary cell wall biosynthesis in cotton [17], [18], the two developmental stages were clearly differentiated by the expression patterns of genes involved in cell wall biogenesis. Cellulose synthase (CesA) genes, such as CesA4, CesA6, CesA7, and CesA8 were more up-regulated at 20 than 10 dpa in wild cottons, while they exhibited less differential expression during fiber development in domesticated cottons (Figure S2A, B). Among them, four CesA genes were highly expressed at 20 dpa in domesticated G. hirsutum, consistent with a previous report [23], but only CesA8, a homologue of GhCesA1, was up-regulated at 20 dpa compared to 10 dpa in domesticated cottons (Figure S2A, B). Cellulose synthase-like (Csl) genes, particularly CslA and CslC, responsible for glucomannan and xyloglucan synthesis, respectively [24], were up-regulated at 10 dpa in both wild and domesticated accessions (Figure S2A, C). Additional differentially expressed genes related to cell wall biogenesis exhibited various patterns during fiber development. For example, β-galactosidase was up-regulated at 10 dpa, while β-1,3-glucosidase and β-xylosidase were up-regulated at 20 dpa; these two enzymes are thought to function in hydrolyzing galactan, glucan, and xylogucan, respectively, into monosaccharides, such as glucose, which can be further processed to either cellulose or pectin [25]–[27]. Xyloglucan endotransglycosylase (XTH) genes, which encode proteins involved in xyloglucan breakdown and subsequent rejoining with different acceptor chains, showed variable expression patterns during development (Figure S3). For example, XTH5 and XTH28 were up-regulated at 10 and 20 dpa, respectively, in both wild and domesticated cottons. Genes related to pectin synthesis, for example, UDP-D-glucuronate-4-epimerase, β-galactosidase, and pectate lyase, were also up-regulated at 10 dpa relative to 20 dpa in both wild and domesticated accessions. However, UDP-glucose-6-dehydrogenases, which oxidize UDP-glucose into UDP-glucuronate, were up-regulated at 10 dpa of domesticated cottons, but down-regulated in wild cottons. Since the foregoing genes represent only a small portion of the total number of differentially expressed genes, we further investigated the difference in development between wild and domesticated cottons using functional analyses (see below).
Transcriptomic changes accompanying domestication
To investigate transcriptomic changes in cotton fiber that distinguish wild from domesticated cottons, and hence reflect the presumptive effects of human selection, we compared the gene expression patterns of wild and domesticated cottons from multiple accessions. A total of 4,946 (13.2%) genes were differentially expressed between wild and domesticated cottons (Figure 1), approximately evenly split between genes that were differentially up- and down-regulated between these two pools. However, nearly three times as many genes were differentially expressed at 10 relative to 20 dpa, a result that at least partially mirrors the data in Rapp et al. [13] (1.7-fold more genes differentially expressed at 10 dpa relative to 20 dpa); the two studies differ in that there was a greater bias toward up-regulation in domesticated than in wild cotton in the earlier study. However, if strict criteria for differential expression are applied, such as RPKM≧5 and more than 2-fold change (All accessions (RPKM> = 5) and Rapp et al. in Table S2), the two studies yield similar results; for example, about 60% of differentially expressed genes at both developmental time points were up-regulated in domesticated cottons relative to wild cottons, although it looks like there are more differentially expressed genes at 20 dpa in domesticated cottons than in wild cottons in Rapp et al. [13] compared to this study (not statistically significant; P = 0.1106) (Table S1). A single accession analysis resulted in an extremely small number of DE genes (<1% of the genes in the reference genome), perhaps due to the lack of biological replicates, while inclusion of two TX2094 samples in RNA-Seq data showed more DE genes compared to multiple accession analysis ((TX2094+YUC)−TM1 vs. All accession = 3,609 vs. 2,910 at 10 dpa, 3,299 vs. 1,339 at 20 dpa; Table S2). These results suggest that variation among biological replicates plays an important role in analyzing RNA-Seq data; that is, more biological replicates from one accession increase the power of DE gene detection (see the previous section). However, at the same time, including multiple accessions facilitates discovery of DE genes across multiple accessions (e.g., (TX2094+TX665+TX2095)−(TM1+CRB250+cascot7) vs. All accession = 1,254 vs. 3,581 at 10 dpa, 876 vs. 1,365 at 20 dpa; Table S2).
We evaluated whether the effects of human selection were biased with respect to the genomic distribution of the effected loci. To do this we tabulated differentially expressed genes by chromosome, and then calculated an expectation based on a null hypothesis of equal distribution, calibrated by the number of genes in each scaffold. This analysis revealed that chromosomes 8 and 1 were differentially targeted during domestication at 10 and 20 dpa, respectively (red text in Table S3). With respect to the latter observation about chromosome 1, the results reflect a putative nuclear mitochondrial DNA (NUMT) sequence block (Figure S4) [16] that contained an unexpectedly high number of up-regulated genes at 20 dpa in both “domestication” (wild vs. domesticated) and “development” (10 vs. 20 dpa) contrasts. For example, during domestication 36 of 84 differentially expressed genes on chromosome 1 were included in this NUMT block and 12 genes are found to be mitochondrial genes, including eight NADH dehydrogenase and four cytochrome-c-related genes.
The comparison of wild and domesticated cottons highlights the fact that the transcriptome of developing cotton fibers was highly altered by five thousand years or more of domestication and crop improvement. To explore this complexity, we first investigated genes previously inferred to be involved in fiber initiation, elongation, and secondary wall biosynthesis (reviewed in [28]). Interestingly, most of the genes involved in the first (initiation) and third (secondary wall biosynthesis) of these stages were up-regulated in wild cottons compared to domesticated cottons at 10 and 20 dpa, respectively (Table S4). In contrast, many genes involved in fiber elongation were highly up-regulated in domesticated cotton compared to wild cottons at 10 dpa, while several genes from this same developmental stage were up-regulated in wild cottons at 20 dpa, encoding annexin, actin depolymerizing factor, FASCICLIN-like arabinogalactan-protein, and tubulin alpha-2 chain (Table S4). Considering expression levels of these genes, differentiation between wild and domesticated cottons was greater during fiber elongation, which is also true at the global transcriptome level, as reported here.
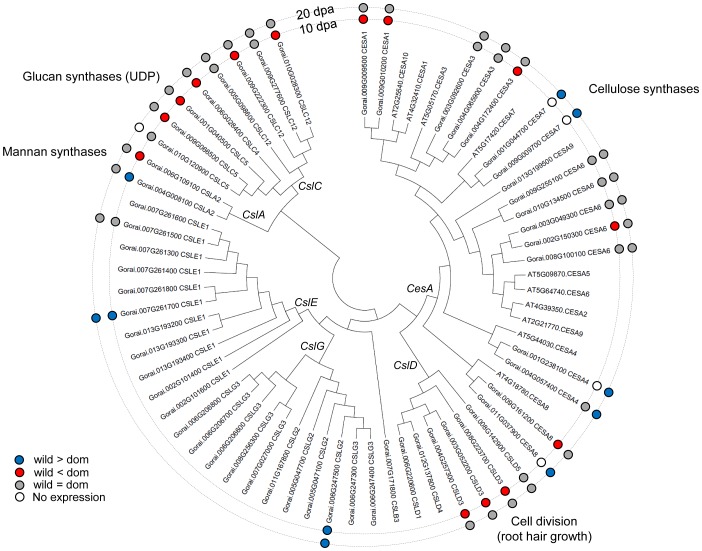
Among the 3,581 genes differentially expressed between wild and domesticated cotton at 10 dpa, we tabulated the most highly up-regulated genes in wild or domesticated cottons, relative to their counterparts, with respect to fold change (with RPKM≧50). This analysis reveals that many genes involved in fiber elongation were over-expressed in domesticated cottons, including profilin 1, HXXXD-type acyl-transferase family protein, expansin A8, beta-6 tubulin, FASCICLIN-like arabinogalactan 9 (FLA9), and 3-ketoacyl-CoA synthase 2 (KCS2) (Table 3). KCS genes are involved in fatty acid elongation and are known to be highly expressed during fiber elongation [29]–[31]. In this study, nine and three of 27 KCSs were up-regulated in domesticated cotton compared to wild types at 10 and 20 dpa, respectively (Figure S5). Notably, five of nine differentially expressed KCSs showed A-homoeolog expression bias, while the other four exhibited no bias (Table S5). CesA and Csl play critical roles in cell wall biosynthesis [17], [18], [23] and were differentially expressed between wild and domesticated cottons. Five of 18 CesA, five of seven CslC and three of six CslD genes in the reference genome were up-regulated in domesticated cotton at 10 dpa (Figure 2).
Table 3. The most abundantly up-regulated genes in domesticated cottons (log2FC>0) or wild cottons (log2FC<0) relative to their counterparts at 10 dpa.
| GoraiID | Sequence description | Expression level (RPKM) | log2FC* | |
| wild | dom | |||
| Gorai.009G028500 | Profilin 1 | 7 | 755 | 7.117 |
| Gorai.012G006600 | HXXXD-type acyl-transferase family protein | 17 | 292 | 4.390 |
| Gorai.010G045500 | glycerol-3-phosphate acyltransferase 3 | 4 | 56 | 4.014 |
| Gorai.006G240600 | Lipid-transfer protein | 38 | 229 | 2.923 |
| Gorai.002G193500 | proline-rich protein 2 | 383 | 2260 | 2.880 |
| Gorai.012G014400 | expansin A8 | 10 | 52 | 2.706 |
| Gorai.004G211800 | beta-6 tubulin | 103 | 431 | 2.378 |
| Gorai.006G150100 | NDR1/HIN1-like 1 | 26 | 102 | 2.311 |
| Gorai.006G000200 | Chalcone and stilbene synthase family protein | 29 | 112 | 2.264 |
| Gorai.011G035400 | GDSL-like Lipase/Acylhydrolase protein | 354 | 1346 | 2.245 |
| Gorai.002G245900 | gamma tonoplast intrinsic protein | 246 | 924 | 2.230 |
| Gorai.013G177600 | glutathione S-transferase TAU 19 | 14 | 52 | 2.174 |
| Gorai.013G270600 | heat shock protein 70B | 25 | 89 | 2.125 |
| Gorai.006G150600 | Eukaryotic aspartyl protease family protein | 383 | 1293 | 2.075 |
| Gorai.008G155400 | FASCICLIN-like arabinoogalactan 9 | 569 | 1901 | 2.058 |
| Gorai.011G165200 | 3-ketoacyl-CoA synthase 2 | 63 | 203 | 2.018 |
| Gorai.002G161400 | polygalacturonase 2 | 18 | 59 | 2.009 |
| Gorai.006G148600 | syntaxin of plants 121 | 31 | 100 | 2.009 |
| Gorai.010G069000 | alpha/beta-Hydrolases superfamily protein | 30 | 97 | 2.005 |
| Gorai.002G248400.2 | plasma membrane intrinsic protein 2 | 27 | 88 | 2.001 |
| Gorai.008G131700 | O-methyltransferase 1 (OMT1) | 365 | 3 | −6.422 |
| Gorai.007G336600 | Plant invertase/pectin methylesterase inhibitor | 134 | 2 | −5.544 |
| Gorai.007G029700 | HXXXD-type acyl-transferase family protein | 213 | 4 | −5.341 |
| Gorai.008G198200 | Cytochrome P450 superfamily protein (CYP75B1) | 239 | 8 | −4.642 |
| Gorai.001G136100 | Lipid-transfer protein | 57 | 3 | −4.155 |
| Gorai.013G023400 | Chalcone-flavanone isomerase family protein | 709 | 36 | −3.988 |
| Gorai.010G255300 | Clathrin light chain protein | 62 | 3 | −3.947 |
| Gorai.011G161300 | Chalcone and stilbene synthase family protein | 1541 | 88 | −3.811 |
| Gorai.004G205900 | leucoanthocyanidin dioxygenase | 1744 | 101 | −3.785 |
| Gorai.008G062900 | flavanone 3-hydroxylase | 5324 | 349 | −3.611 |
| Gorai.011G161200 | Chalcone and stilbene synthase family protein | 1120 | 74 | −3.604 |
| Gorai.004G105500 | NAD(P)-binding Rossmann-fold superfamily protein | 443 | 33 | −3.447 |
| Gorai.009G182300 | ethylene-forming enzyme | 112 | 9 | −3.360 |
| Gorai.005G035100 | Chalcone and stilbene synthase family protein | 1752 | 143 | −3.297 |
| Gorai.004G184100 | NAD(P)-binding Rossmann-fold superfamily protein | 329 | 29 | −3.209 |
| Gorai.001G134900 | Cytochrome P450 superfamily protein (CYP75B1) | 2157 | 193 | −3.166 |
| Gorai.007G322600 | metallothionein 3 | 82 | 8 | −3.133 |
| Gorai.004G277300 | Histone superfamily protein | 84 | 8 | −3.064 |
Genes were filtered by RPKM≧50 in either wild or domesticated cottons. Bold indicates genes involved in phenylpropanoid metabolism. Eleven and four genes up-regulated in domesticated and wild cottons, respectively, were removed because of no annotation.
Log2 Fold Change calculated from raw mapped read numbers using DESeq software.
Figure 2. The phylogeny of cellulose synthase (CesA) and cellulose synthase-like (CSL) genes.
Inside and outside circles show differential expression between wild and domesticated cottons at 10 and 20-regulation at domesticated cottons, while blue one indicates up-regulation at wild cottons during domestication. Grey circle shows no differential expression, while white circle designates no expression (zero read count).
FLAs have been classified into four groups [32], but the function of only a few FLAs are known. For example, in Arabidopsis, FLA4 or SOS5 (At3g36550) plays a role in cell expansion [33], and several FLA homologues of G. hirsutum were highly expressed during fiber elongation [34]. We also observed several FLA homologues that were up-regulated in domesticated cottons compared to wild cottons at 10 dpa, including two SOS5 homologues and three of four AtFLA7 homologues (Figure S6).
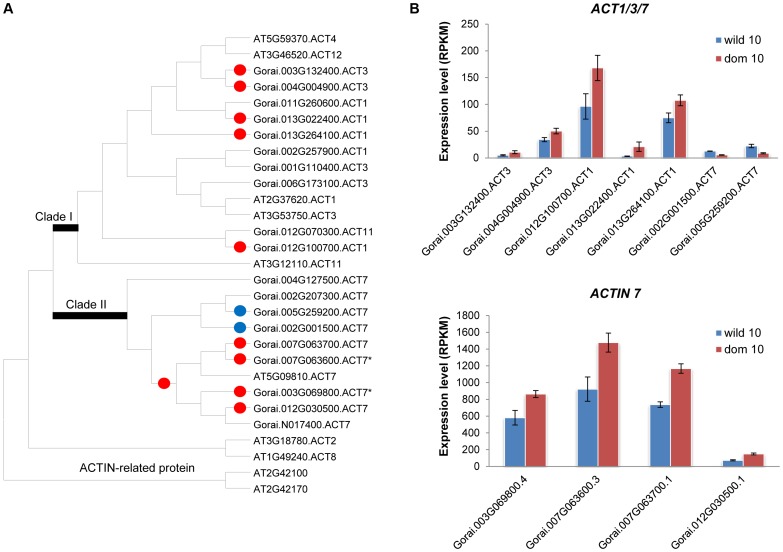
Profilin (PRF) and its partners (e.g., actin, tubulin, and villin), which play an important role in actin polymerization [7], [35], were also up-regulated in domesticated cotton, and their expression levels were high, except for villins (Figure S7). Consistent with Bao et al. [7], PRF1 exhibited the highest expression differences between wild and domesticated cottons, and PRF3 and PRF4 were up-regulated in domesticated cottons relative to wild cottons (Figure S7). However, the other two PRF genes were not differentially expressed (cf. ref [7]), a conflict perhaps explained by the larger number of accessions studied here. As for ACTIN (ACT), there were two main clades of Gossypium ACTs, ACT1/3/4/11/12 (clade I) and ACT7 (clade II) (Figure 3). These two clades are distinct in their gene expression patterns; members of ACT1/3/4/11/12 generally are expressed in reproductive organs, such as pollen, pollen tubes, and ovules of Arabidopsis, while ACT7 is expressed in vegetative tissues, including root hairs and trichomes along with ACT2 and ACT8 [36]–[39]. In the present study, at 10 dpa nine and two ACT genes were up-regulated in domesticated and wild cottons, respectively (Figure 3). Interestingly, all of the genes closely related to Arabidopsis ACT7 were commonly and highly up-regulated in domesticated cottons, except Gorai.N017400. Previously identified GhACT1, which was shown to participate in fiber elongation [40], showed the highest similarity to Gorai.007G063600 (probably GhACT2; see Table S4) included in the ACT7 clade. Notably, Gorai.007G063600 seems to be duplicated; its duplicate, Gorai.007G063700, was also up-regulated in domesticated cottons with expression levels similar to that of Gorai.007G063600 (Figure 3). Other ACT genes, which are members of clade I or which are up-regulated in wild cottons, exhibited relatively low expression levels compared to ACT7 homologues.
Figure 3. Phylogenetic relationship of ACTIN and their gene expression patterns at 10 dpa.
(A) Phylogenetic relationship of G. raimondii ACTIN genes. Red and blue dots on the node indicate up- or down regulation at domesticated cottons, respectively. Asterisk (*) shows ACTIN homologues previously studied. (B) Expression patterns of differentially expressed ACT genes. Bar denotes standard error.
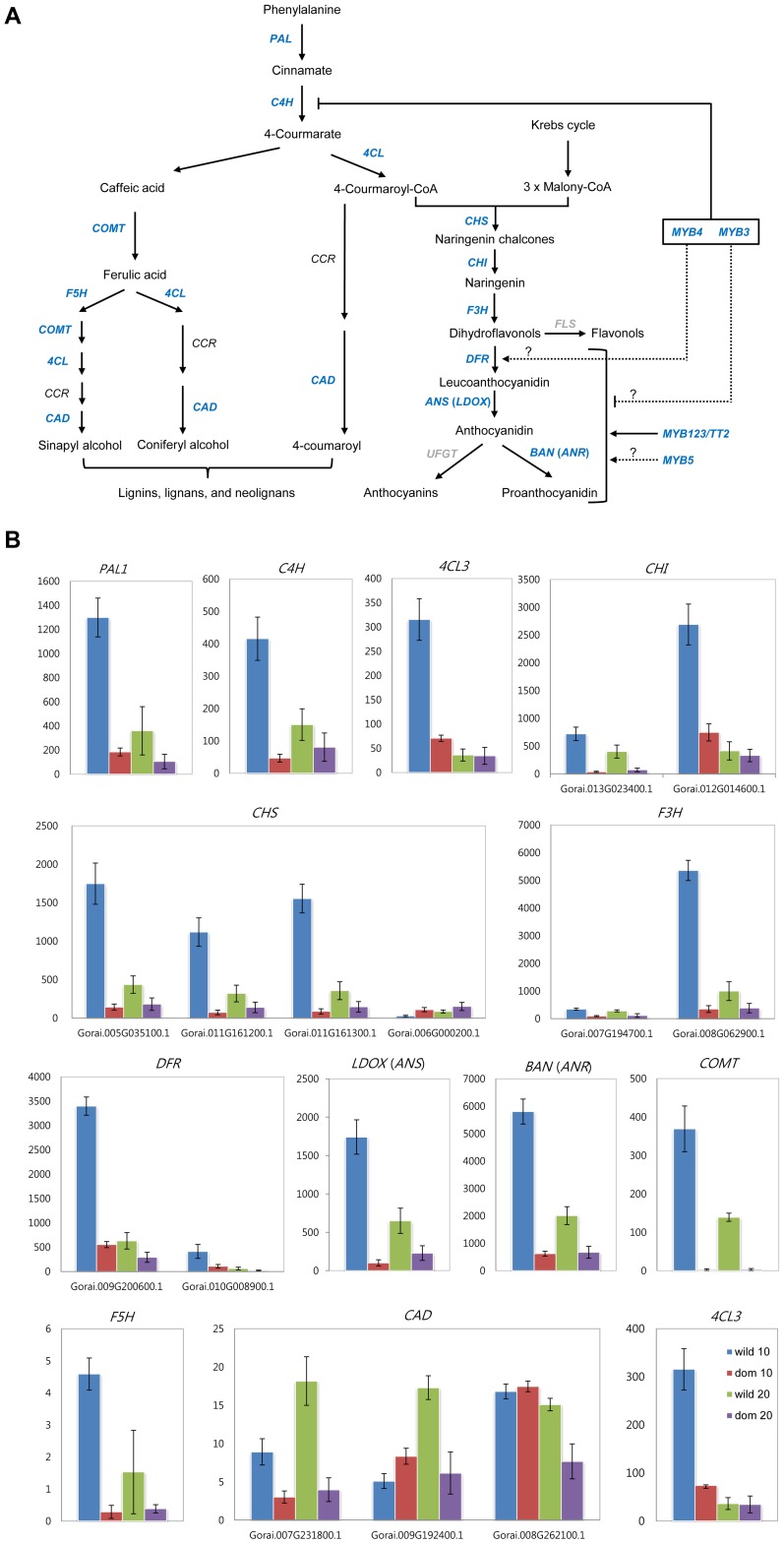
In contrast to some key genes observed to be up-regulated under domestication, in wild cottons some genes involved in phenylpropanoid metabolism, such as flavonoid biosynthesis and anthocyanin biosynthesis, were highly up-regulated at both developmental time points compared to their counterparts in domesticated cottons (Figure 4). For example, PHENYLALANINE AMMONIA LYASE 1 (PAL1) exhibited 5.6 times higher expression than in domesticated cottons at 10 dpa, and other genes involved in this pathway showed similar patterns (Figure 4B). In addition, some MYB transcription factors (TFs) were also up-regulated in wild cotton compared to domesticated cotton (Figure 5, Figure S8). In particular, half of the 23 differentially expressed MYB TFs in wild cottons were related to the phenylpropanoid pathway, as noted earlier [16], and their up-regulation was observed at both developmental stages (Figure S8).
Figure 4. Expression patterns of genes related to phenylpropanoid pathway (A) and their actual expression levels (B) in wild and domesticated cottons.
Blue text in (A) indicates up-regulation in wild cottons relative to domesticated cottons at 10 dpa, and bar in (B) denotes standard error. PAL, phenylalanine ammonium lyase; C4H, cinnamate-4-hydroxylase; 4CL, 4-coumaroyl-CoA synthase; CHI, chalcone isomerase; F3H, flavonol 3-hydroxylase; FLS, flavonol synthase; DFR, dihydroflavonol-4-reductase; ANS, anthocyanin synthase, LDOX, leucoanthocyanidin dioxygenase; ANR, anthocyanidin reductase; COMT, F5H, CCR, CCD.
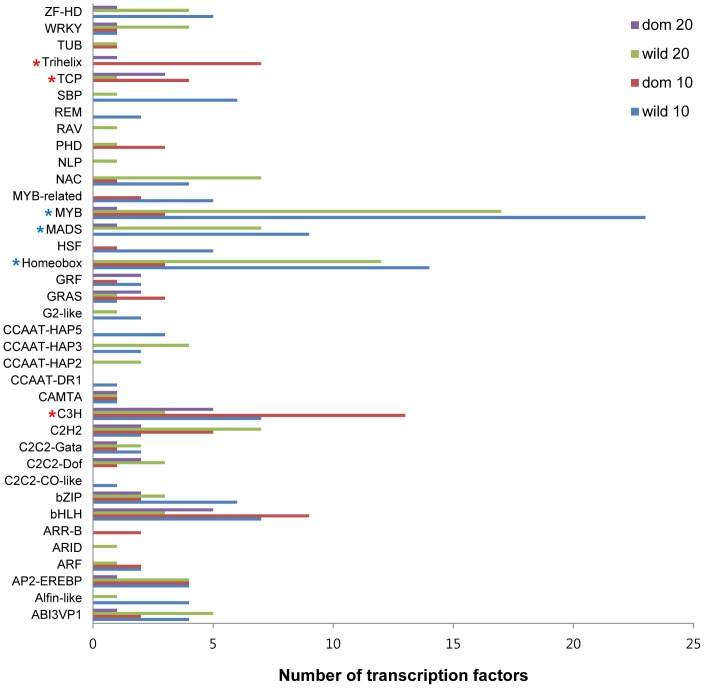
Figure 5. Number of TFs differentially expressed between wild and domesticated cotton.
Blue and red asterisk (*) represent over-represented TFs in wild and domesticated cottons relative to their counterparts at both developmental time points, respectively (proportion test; P<0.05).
At 20 dpa, similar sets of genes were differentially expressed, but their expression levels were relatively lower compared to those observed in 10 dpa (e.g., Figure S3, S4, S5, S6, S7, S8). Many of the genes up-regulated in domesticated cottons were related to protein synthesis (see below) or were found in a putative nuclear mitochondrial DNA (NUMT) sequence block (Table S3; see above). In addition and importantly, 14–19% of the differentially expressed genes encode unknown proteins, in agreement with previous reports [13], [21]; these genes become obvious targets for future functional analysis, to discover their roles in cellular development and in evolution.
Among the 2,830 TFs that are annotated in the cotton genome, fewer than 10% were differentially expressed between wild and domesticated cottons. Specifically, 266 (184 vs. 82 up-regulated in wild vs. domesticated, or the reverse) and 132 (100 vs. 32 up-regulated in wild vs. domesticated, or the reverse) were differentially expressed at 10 and 20 dpa, respectively. Among these, only 48 TFs were expressed at the level of RPKM≧50, indicating that the majority of TFs are not highly expressed in fibers. Of these 48 highly expressed TFs, 27 and 16 were up-regulated in wild and domesticated cottons relative to their counterparts, respectively. Five genes were not differentially expressed including GLABRA2 (GL2) and MYB60 which have been functionally studied. GL2 regulates cell wall-related gene expression (CeSA5 and XTH17) during root development in Arabidopsis [41], while MYB60 is involved in stomatal regulation and root growth under drought stress in grapevine [42] and Arabidopsis [43], or repressing anthocyanin biosynthesis in lettuce [44]. As for TFs up-regulated in wild cottons relative to domesticated cottons, three TFs families were the most commonly represented, including homeobox, MADS and MYB TFs (Figure 5, Table S6). This is consistent with previous studies on the importance of MYBs in fiber development [45]–[48], but over-representation of MADS genes has not previously been reported for cotton fibers. MADS genes that were differentially expressed were related to carpel (e.g., AGAMOUS, SHATTERPROOF1, SEPALLATA) and seed development (SEEDSTICK); results here suggest the possibility that these genes have found a new role in fiber development. In domesticated cottons, three TFs, C3H, TCP, and trihelix, were the most commonly represented classes among the 48 TFs (Figure 5, Table S6); this includes a TF that recently has been identified as important for fiber development in both G. barbadense [49] and G. hirsutum [50]. Gorai.007G036800, a homologue of GhTCP14 [50], was up-regulated in domesticated cottons relative to wild cottons, supporting its relatedness to fiber elongation.
Functional analyses of differential expression
To evaluate whether specific biological processes were enriched in representation by either development or domestication, two different functional analyses were performed, the Singular Enrichment Analysis (SEA) and the Parametric Analysis of Gene set Enrichment (PAGE). Although SEA and PAGE deploy different strategies, both methods yielded similar results. Thus, we present only SEA results here (Table S7), to highlight some of the differences between wild and domesticated cottons. In general, during development more biological processes were differentially regulated in wild cottons than domesticated cottons (wild vs. domesticated = 71 vs. 1 biological processes (P) in Table S7A), as expected based on the degree of differential expression found in comparison of two developmental time points in wild and domesticated cottons (Figure 1). For example, at 10 dpa in wild cottons, genes related to lipid metabolism were enriched, including fatty acid biosynthetic process, very-long-chain fatty acid (VLCFA) metabolic process, sterol biosynthetic process, and steroid biosynthetic process, and secondary metabolites biosynthesis process was also up-regulated, including phenylpropanoid, coumarin, flavonoid, and anthocyanin biosynthesis processes (Table S7A). In addition, gibberellic acid (GA) mediated signaling pathway was also over-represented at 10 dpa in wild cottons, noting that GA is required for fiber initiation and elongation [51], [52]. At 20 dpa in wild cottons, in addition to in cell wall organization or biogenesis, genes involved in response to abiotic and biotic stimuli, such as water deprivation, organic substance, chemical and hormone stimuli were over-represented relative to 10 dpa (Table S7A). In domesticated cottons, there was no biological process enriched between 10 and 20 dpa based on PAGE analysis (data not shown), while SEA results indicated that genes related to lipid metabolic process were up-regulated at 10 dpa compared to 20 dpa (Table S7A).
In the domestication contrast, SEA results showed that more biological processes were up-regulated in domesticated cottons than in wild cottons (123 in domesticated cottons vs. 49 in wild cottons; Table S7B). Many up-regulated genes in wild cottons relative to domesticated cottons at 10 dpa were related to protein-DNA complex assembly, nucleosome assembly, response to disaccharide stimulus, and secondary metabolite synthetic processes, such as anthocyanin, flavonoid, and phenylpropanoids (Table S7B). Consistent with this result, cellular components, such as chromosome and nucleosome, and molecular function of transcription factor activity were highly enriched in wild cottons (Table S7B). At 20 dpa of wild cottons, genes involved in cell wall macromolecule metabolism and amine catabolism were up-regulated relative to domesticated cottons, suggesting that secondary cell wall synthesis is active in wild cottons. Amine catabolism involves protein degradation, which may generate nitrogen-containing compounds for secondary metabolite synthesis. In fact, most genes related to amine catabolism were associated with phenylpropanoid biosynthesis, for example, 4-coumarate:CoA ligase 1 (4CL1), PAL1, cinnamyl alcohol dehydrogenase (CAD), and cinnamoyl CoA reductase 1 (CCR1). On the other hand, domesticated cottons were defined by fiber elongation-related processes (e.g., vesicle-mediated transport, actin cytoskeleton organization, and cellulose metabolism) at 10 dpa and energy generation and protein synthesis at 20 dpa (Table S7B). For example, many genes differentially expressed at 20 dpa of domesticated cottons compared to wild cottons were involved in RNA elongation, cellular respiration along with oxidative phosphorylation, and protein synthesis (translation). Also, some genes involved in fatty acid biosynthesis were up-regulated, perhaps to facilitate membrane growth and turnover during fiber elongation and maturation [53].
Homoeolog-specific biases and change during development and domestication
To explore whether there is a bias in usage of parental gene copies (homoeologs) during development and domestication, a phenomenon termed homoeolog expression bias [54], [55], the relative contribution of homoeologs to total gene expression was investigated. An average of 17,800 genes had homoeolog-specific reads, of which 17.5 to 53.5% showed unequal (biased) expression in any one case (Table 4). Notably, by far the highest percentage of genes showing biased homoeolog contributions to the transcriptome was at 10 dpa for domesticated cottons, a rate nearly twice that observed at 20 dpa. In addition, more genes at 10 dpa than at 20 dpa exhibit homoeolog bias in all comparisons (Table 4). Considering the entire data sets, which include all genes having a minimum number of homoeolog-specific reads (RPKM≧1), there is no global bias in homoeolog expression in either wild or domesticated cottons; that is, despite appreciable gene-level bias, the number of genes that exhibit either At or Dt bias (where the lower case t designates homoeolog in the allopolyploid) are approximately equal (balanced homoeolog bias, sensu Grover et al. [54]). This same result also characterizes most other comparisons.
Table 4. Homoeolog-specific bias in developing cotton fiber.
| Total # genesa | At = Dt | At>Dt | At<Dt | Total biased genes (%) | |
| Entire data | |||||
| wild 10 dpa | 18,315 | 13,815 | 2,258 | 2,242 | 4,500 (24.6%) |
| wild 20 dpa | 19,163 | 14,941 | 2,141 | 2,081 | 4,222 (22.0%) |
| dom 10 dpa | 17,551 | 11,641 | 2,966 | 2,944 | 5,910 (33.7%) |
| dom 20 dpa | 15,901 | 13,126 | 1,382 | 1,393 | 2,775 (17.5%) |
| DE genes during development | |||||
| wild 10 dpa | 2,315 | 1,335 | 468 | 512 | 980 (42.3%) |
| wild 20 dpa | 2,612 | 1,632 | 490 | 490 | 980 (37.5%) |
| dom 10 dpa | 626 | 291 | 163 | 172 | 335 (53.5%) |
| dom 20 dpa | 693 | 422 | 131 | 140 | 271 (39.1%) |
| DE genes during domestication | |||||
| wild 10 dpa | 1,691 | 1,256 | 223 | 212 | 435 (25.7%) |
| dom 10 dpa | 1,605 | 1,066 | 289 | 250 | 539 (33.6%) |
| wild 20 dpa | 951 | 567 | 183 | 201 | 384 (40.4%) |
| dom 20 dpa | 631 | 360 | 131 | 140 | 271 (42.9%) |
At = A-homoeolog; Dt = D-homoeolog; At = Dt, equal expression of homoeologs; At>Dt, biased expression of the At homoeolog; At<Dt, biased expression of the Dt homoeolog; DE = differential expression from the contrasts of development (10 vs. 20 dpa) or domestication (wild vs. dom).
a includes genes having at least RPKM≧1 in either A or D-homoeolog specific reads across all biological replicates.
To assess how homoeolog usage is affected during development and by domestication, we compared the same homoeolog from two different developmental stages or from the two pools of wild vs. domestication cottons. During cotton fiber development from 10 dpa to 20 dpa, we observed more homoeolog expression change in wild cottons than in domesticated cottons (4,358 of 22,012 (19.8%) in wild vs. 2,110 of 19,974 (10.6%) in domesticated; Table 5) although there is no difference in DE genes between wild and domesticated cottons (Table 5). There were more homoeolog changes at 10 dpa than at 20 dpa (3,350 of 20,994 (16.0%) at 10 dpa vs. 2,433 of 21,230 (11.5%) at 20 dpa; Table 5) as a result of domestication. However, we observed balance in most comparisons; for example, there are similar numbers of At and Dt down- or up-regulation during fiber development in wild cottons (down-regulation of At vs. Dt = 290 vs. 321, up-regulation of At vs. Dt = 320 vs. 336; Table 5). When combined with the results from Table 4 (described above), we infer that homoeolog modulations (both expression and change) were balanced in cotton fiber regardless of development and domestication.
Table 5. Homoeolog expression changes across two developmental stages and during domestication.
| Comparison | Homoeolog changes from 10 dpa to 20 dpa | Gene pairs in entire data | Gene pairs in DE genes | ||||||
| wild cotton | % | dom. cotton | % | wild cotton | % | dom. cotton | % | ||
| Development | No changes | 17,654 | 80.2 | 17,864 | 89.4 | 306 | 9.5 | 86 | 9.1 |
| Total changed | 4,358 | 19.8 | 2,110 | 10.6 | 2,929 | 90.5 | 861 | 90.9 | |
| both down-regulated | 766 | 3.5 | 246 | 1.2 | 692 | 21.4 | 142 | 15.0 | |
| At down-regulated | 615 | 2.8 | 302 | 1.5 | 290 | 9.0 | 76 | 8.0 | |
| Dt down-regulated | 603 | 2.7 | 283 | 1.4 | 321 | 9.9 | 79 | 8.3 | |
| both up-regulated | 1,045 | 4.7 | 389 | 1.9 | 966 | 29.9 | 273 | 28.8 | |
| At up-regulated | 694 | 3.2 | 478* | 2.4 | 320 | 9.9 | 150 | 15.8 | |
| Dt up-regulated | 622 | 2.8 | 411 | 2.1 | 336 | 10.4 | 141 | 14.9 | |
| At up, Dt down | 7 | 0.0 | 0 | 0.0 | 4 | 0.1 | 0 | 0.0 | |
| At down, Dt up | 6 | 0.0 | 1 | 0.0 | 0 | 0.0 | 0 | 0.0 | |
indicates that the number of genes belonging to “At up-regulated” and “Dt down-regulated” is statistically different from “Dt up-regulated” and “At down-regulated”, respectively (P<0.05).
Discussion
The complex cotton fiber transcriptome
Cotton fiber development involves an extraordinarily complex biology regulated by multiple and diverse pathways and transcriptional regulatory networks. In this study, we generated global transcriptome profiles of developing cotton fibers from multiple accessions of wild and domesticated G. hirsutum. Using RNA-Seq, we determined that at least one-third and likely about half of the genes (depending on the RPKM threshold) in the cotton genome are expressed in developing fibers. This number is consistent with previous estimates of the fiber transcriptome diversity generated for G. hirsutum cv. TM1 and G. arboreum, notwithstanding the technical differences among studies [20], [21]. It is striking that the genic diversity in the transcriptome of fibers, which are single cells, is comparable to that of entire young leaves of G. hirsutum (Table 2) [55], which are far more complex organs comprising multiple different cell types and with varying cellular specializations and diverse metabolic roles. This comparison justifies the perspective that the cotton fiber transcriptome is extraordinarily rich and that it is subject to complex transcriptional regulation during fiber development.
One of the justifications for the experimental design used in the present study was to attempt to account for expression variation that might occur within wild and within domesticated G. hirsutum and hence account for this variation to strengthen inferences about the differences between these groups. Accordingly, we selected multiple accessions within each pool. For the two developmental stages studied here, 10 and 20 dpa, we estimate that, respectively, 3.1% (1,144) and 9.3% (3,487) of the duplicate gene pairs in the tetraploid cotton genome were differentially expressed between 10 and 20 dpa in domesticated and wild cottons, respectively. Importantly, when we reanalyze our RNA-Seq data, restricting our attention to the same two accessions as used in Rapp et al. [13], TM1 (domesticated) and TX2094 (wild), we observe about a 40% increase in the number of differentially expressed genes (6,908 vs. 4,946) (Table S2). These data indicate that inclusion of multiple accessions narrowed the differences between the two pools “wild” and “domesticated”, boosting confidence in inferences regarding the effects of human selection, and in the process identifying expression variation arising from other causes.
In addition to bolstering the notion that the fiber transcriptome is highly diverse and dynamic, the results presented here include deep and rich data sets that can be mined for clues regarding processes of cellular development and those that have been most strongly affected by human-mediated directional selection under domestication. The data also provide new information on homoeolog usage and biases in a polyploid cell type. Each of these topics is discussed in more detail in the following.
Domestication prolonged fiber elongation during development
A body of work has established that cotton fiber development consists of stages that are well-defined temporally, i.e., fiber initiation (0–3 dpa), primary cell wall synthesis and elongation (3–15 dpa), transition to secondary cell wall growth (15–20 dpa), secondary wall biosynthesis (20–40 dpa), and maturation (40–50 dpa) [18]. Our observation of relatively little differentiation between 10 and 20 dpa in domesticated cottons suggests that the fiber primary elongation developmental program had continued to 20 dpa, consistent with a previous study based on fiber growth curves [56]. In domesticated cotton, the rates of fiber growth and maturation were the highest between 10 and 15 dpa, but extended up to 30 dpa (20 days of fiber elongation), while fibers of wild cottons elongated fastest between 15 and 20 dpa (5 days of elongation) [56], [57]. In particular, cotton fiber from wild G. hirsutum already reached >90% of maturation around 20 dpa, indicating early termination of fiber elongation, and likely entry into the transition phase leading to secondary wall synthesis. Our transcriptome profiling results showed that gene expression patterns were significantly more differentiated between 10 and 20 dpa in wild cottons, perhaps reflecting this subtle temporal shift in the fiber developmental program. Based on fiber growth curve analyses, fiber elongation appears modest until 15 dpa in wild cottons, yet is almost complete by 20 dpa [56]. Thus, 10 dpa from wild cottons represents an early stage of fiber elongation in wild, relative to domesticated, G. hirsutum, while by 20 dpa fibers from wild cotton likely have completed primary cell wall synthesis and have entered the transition to secondary wall synthesis. This inference is also supported by expression patterns of genes previously reported from gene-by-gene surveys; wild cottons showed more up-regulations of genes related to fiber initiation and secondary wall biosynthesis at 10 and 20 dpa, respectively, compared to domesticated cottons (Table S4). This difference in developmental timing might account for the three-fold increase in the number of differential expressed genes between the two developmental stages of wild cottons relative to domesticated cottons (Figure 1).
Previous studies support this interpretation of a period of prolonged fiber elongation under domestication, and in parallel in different domesticated cotton species. Notably, similar conclusions have been reached for diploid domesticated cotton, G. arboreum [58], and in a second domesticated allopolyploid cotton, G. barbadense [59]. In addition, Hu et al. showed, in a recent, high-throughput iTRAQ proteomic analysis, that the proteome of domesticated cotton during early fiber elongation (5–10 dpa) was similar to that of later developmental stage of wild cottons (10–20 dpa) [59]. Thus, it seems that human domestication may have induced parallel prolongations and developmental shifts on the fiber elongation period in both diploid and allopolyploid species, as evidenced by growth curve analysis [56], and both transcriptomic and proteomic analyses [58], [59]. These studies, as well as light microscopy observations [13] which demonstrate that wild and domesticated G. hirsutum share similar timing and morphology of early wall thickening, point to the need to develop a deeper understanding of the underlying developmental programs and architecture of fiber growth and evolution. In a recent metabolic profiling study [12] of a lintless mutant and its wild type (WT) G. hirsutum relative, 487 metabolites identified from nine developmental time points clearly differentiated the metabolomic profiles of the lintless mutant from that of WT cotton during elongation, but there was no clear differentiation between the two forms during fiber initiation (−3 to 3 dpa). This suggests that the short period of fiber elongation in the lintless mutant, where fiber cells become arrested at about 6 mm of linear growth, resembles, to a certain extent, the wild representatives of the domesticated species. Considering the evolutionary, morphological transformation of fiber from lintless in wild to linted in domesticated cottons [4], prolonged fiber elongation was a key innovation for longer fiber, which is apparent at the transcript, protein, and metabolite levels. Additional insight into the nature of this developmental shift will probably arise from further integrated studies of various “omics”, combined with a denser sampling of developmental time points.
Analysis of differential expression showed that wild cottons deployed a higher number of biological processes compared to domesticated cottons, for example SEA results showed there were 77 vs. 1 in wild vs. domesticated cottons, respectively (Table S7A). In particular, as consistent with little differentiation between 10 and 20 dpa in domesticated cottons, only one biological pathway, lipid metabolic process, was over-represented at 10 dpa relative to 20 dpa. However, in wild cottons, different metabolic pathways were over-represented in the DE gene sets that characterize development, including fatty acid biosynthesis and secondary metabolite biosynthesis at 10 dpa and cell wall organization and biogenesis at 20 dpa (Table S7A). Interestingly, the GA mediated signaling pathway was enriched at 10 dpa in wild cottons. Considering that GA is required for fiber growth [52] and shows the highest level in 10 dpa fibers [51], up-regulation of this pathway indicates active fiber elongation at 10 dpa compared to 20 dpa in wild cottons. For example, GAST1 PROTEIN HOMOLOG 4 (GASA4) was known to promote GA response and regulate redox status in Arabidopsis [60], and two cotton homologues (Gorai.006G017000, Gorai.012G054200) were highly expressed and up-regulated at 10 dpa compared to 20 dpa in wild cottons. On the other hand, “response to stress” pathways were enriched in 20 dpa wild cottons, thus, genes related to stress were up-regulated, including those in the dehydrin family protein (EARLY RESPONSIVE TO DEHYDRATION 10; Gorai.002G119600), senescence-associated genes (Gorai.012G124700), late embryogenesis abundant like 5 (Gorai.002G119600), and cold-regulated 47 (Gorai.009G189500). These genes are known to be expressed in response to abiotic stress, such as high salinity, drought, and cold in Arabidopsis [61]–[63], implying that wild cottons are utilizing stress-response pathways at 20 dpa. This inference was also supported by up-regulation of many ROS genes (see below), suggesting that up-regulation of stress-related gene expression in wild cottons could have resulted in halting fiber elongation and promoting the transition to secondary wall biosynthesis.
Many genes related to phenylpropanoid biosynthesis were up-regulated during fiber development in wild cottons (Figure 4A) with expression levels that were significantly higher at 10 than at 20 dpa (Figure 4B), in agreement with previous studies [64], [65]. The same differential regulation characterized wild vs. domesticated cottons at both developmental time points (discussed further below).
Wild and domesticated cottons exhibited no differences in homoeolog utilization at both expression (Table 4) and change (Table 5). This is consistent with Yoo et al. [55] and Rambani et al. [66] which showed an overall equal usage of both homoeologs in young leaf and petal tissues of both TX2094 and Maxxa, respectively. These results suggest that domestication has not affected the utilization of homoeologs from the two co-resident genomes of allopolyploid cotton. However, further study is required to evaluate whether this equal usage of homoeologs was derived from vertical inheritance of progenitor A and D genome conditions, of if instead trans-acting regulatory factors overwhelmed pre-existing, evolved cis- and trans- differences that accumulated during evolutionary divergence of the two progenitor diploids
Domestication may have reallocated resources from stress-response pathways to fiber growth
Over the course of several thousands of years of domestication and selection, the short, coarse, and brown fibers of wild G. hirsutum were transformed into the long, strong, and fine white fibers that characterize modern upland cultivars. Recent large-scale transcriptomic and proteomics analyses have begun to reveal some of the molecular underpinnings of this remarkable morphological modification [12], [13], [20]–[22], [58], [59], [67]. Here we tried to build on this initial insight into the effects of the domestication and plant improvement process by generating transcriptomes from multiple accessions, thus permitting gene expression changes resulting from domestication to be isolated from those arising from other causes. The wild and domesticated cottons selected exhibit the typical fiber characteristics of their respective pools in fiber color and fiber length (Figure 1), and they also were highly differentiated with respect to their transcriptomes (Figure S1). One key result is the observation of greater gene expression differentiation between wild and domesticated cottons at 10 than 20 dpa (Figure 1), consistent with previous studies [11], [13]. These were partitioned almost equally toward either wild or domesticated cottons (Figure 1; dom vs. wild = 1,839 vs. 1,736). However, if we consider only the more highly expressed genes, i.e., those with a RPKM≧5, twice as many genes were up-regulated in domesticated cottons compared to wild cottons at 10 dpa (dom vs. wild = 1,476 vs. 733), implicating a ramping up of cellular machinery involved in primary wall synthesis and perhaps down regulation of other pathways (see below). A corollary implication is that the majority of up-regulated genes in the wild cottons compared to domesticated cottons at 10 dpa were expressed at a lower level (RPKM<5). Similar trends were observed in the 20 dpa comparison, but with less imbalance and smaller absolute numbers (dom vs. wild = 349 vs. 302 with RPKM≧5).
Gene enrichment analyses indicate that specific biological processes were enriched as a consequence of domestication. In particular, the combined significance of functional suggestions become apparent when one considers carbohydrate and fatty acid metabolism with respect to glycolysis, cell wall component biosynthesis, the pentose phosphate pathway, and phenylpropanoid biosynthesis [68]–[70]. In domesticated cottons, carbon resources appear to be more heavily invested in cell wall component biosynthesis, such as cellulose and matrix polysaccharides, as well as energy production through glycolysis (Figure 6). In addition, acetyl-CoA, a product of glycolysis, is linked to synthesis of VLCFAs that are precursors for phospholipids and sphingolipids, essential components of plasma membranes [70]. VLCFAs accumulate preferentially in elongating fibers and KCSs, the rate-limiting enzyme in biosynthesis of VCLFAs [71], are also up-regulated during fiber elongation [29]–[31]. In this study, we also observed several KCSs that were highly expressed in domesticated cottons at 10 dpa (Figure S5). Notably, five of nine KCSs differentially expressed exhibited A-homoeolog expression bias (Table S5), implying that domestication process could have selected maternal parental copy only. Further study on the genome scale is required to elucidate whether this phenomenon is stochastic or linked to specific pathway(s). Other genes related to this pathway were also up-regulated in domesticated cotton compared to wild cottons, including beta-ketoacyl reductase (KCR), fatty acid hydroxylase (CER3/WAX2), fatty acid reductase (CER4/FAR3), lipid transport protein (LTP), and ATP-binding cassette transporter (WBC1) (Figure 6). Noteworthy, CesA, CslC and CslD genes were up-regulated in domesticated cottons at 10 dpa only, while CesA genes were up-regulated in wild cottons at 20 dpa (Figure 2). This indicates that CslC and CslD genes have become up-regulated by domestication early in fiber development. Other fiber elongation-related genes, e.g., profilin and its partners, were also up-regulated in domesticated cottons at 10 dpa (Figure 3, Figure S7), including members of one sub-clade of ACT7 (Figure 3). In Arabidopsis, ACT7 is a vegetative actin, along with ACT2 and ACT8, (the latter two have no obvious homologs in cotton), and are involved in root growth and epidermal cell specification [39]. Here, we observe two sub-clades of ACT7 in cotton, one up-regulated in wild cottons, while the other is up-regulated in domesticated cottons (Figure 3). Thus, the domestication process may have recruited enhanced utilization of one sub-clade of ACT7 for greater fiber elongation.
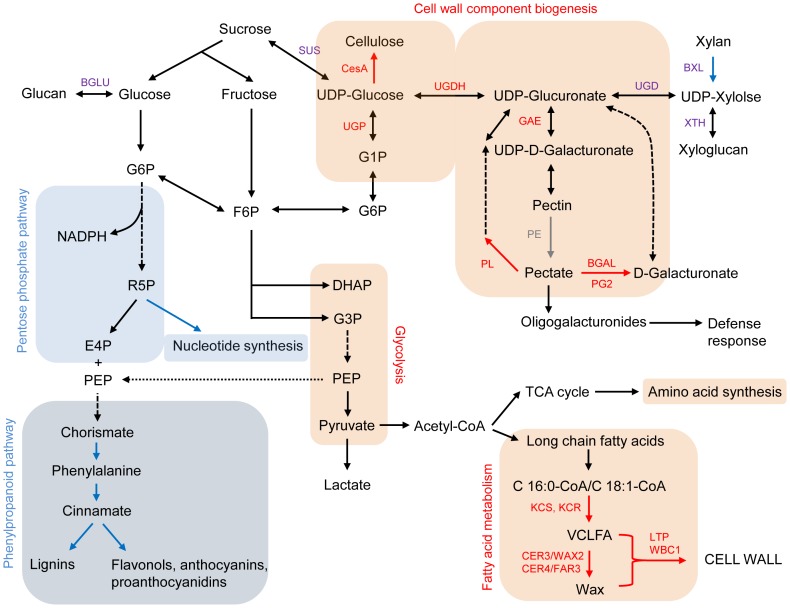
Figure 6. Carbohydrate and fatty acid metabolisms, focusing on cell wall biosynthesis [68]–[70].
Red text and line indicate up-regulated genes or pathways in domesticated cottons relative to wild cottons at 10 dpa, while blue text and lines show up-regulated genes or pathways in wild cottons compared to domesticated cottons at 10 dpa. Purple text indicates that the genes were up- or down-regulated during domestication. Blue and pink shaded boxes show enriched pathways in wild and domesticated cottons compared to their counterparts, respectively. ADPG, ADP-Glucose (ADPG); BGAL, β-galactosidase; BGLU, β-1,3-glucosidase; BXL, β-xylosidase; CER3/WAX2, fatty acid hydroxylase superfamily; CER4/FAR3, fatty acid reductase 3; DAHP, 3-deoxy-D-arabino-heptulosonate-7-phosphate; DHAP, dihydroxyacetone phosphate; E4P, erythrose-4-phosphate; F6P, Fructose-6-Phosphate; GAE, UDP-D-glucuronate-4-epimerase; G1P, Glucose-1-Phosphate; G3P, Glyceraldehyde-3-phosphate; G6P, Glucose-6-Phosphate; KCR, beta-ketoacyl reductase; KCS, 3-ketoacyl-CoA synthase; LTP, lipid transfer protein; PE, pectinesterase; PEP, phosphoenolpyruvate; PG2, polygalacturonase 2; PHS2, alpha-glucan phosphorylase 2; PL, Pectate lyase; R5P, Ribose-5-phosphate; SUS, sucrose synthase; UGD, UDP-glucuronate decarboxylase; UGDH, UDP-glucose-6-dehydrogenase; UGP, UDP-glucose pyrophosphorylase; VLCFA, very long chain fatty acids; WBC1, ATP-binding cassette transporter white-brown complex homolog protein 1; XTH, Xyloglucan endotransglycosylase.
In wild cottons, nucleotide biosynthesis and phenylpropanoid biosynthesis were enriched, based on differential expression. In particular, many genes related to phenylpropanoid biosynthesis were up-regulated during fiber development and domestication in wild cottons (Figure 4A), and their expression levels were much higher at 10 than 20 dpa (Figure 4B), in agreement with previous studies [64], [65]. Notably, phenylpropanoids, particularly flavonoids, are known to inhibit fiber elongation [64], but protect cells from abiotic and biotic stresses [72]. The involvement of flavonoids in fiber processes has been shown in many studies at the transcript, protein, and metabolic levels [12], [13], [20], [64], [73]. Tan et al. [64], in particular, showed that the flavonoid naringenin negatively regulates fiber development and that higher levels of naringenin accumulate in short, brown fibers.
This up-regulation of genes related to phenylpropanoid biosynthesis as well as nucleotide biosynthesis is illustrated in the model suggested in Figure 6, presented within the conceptual framework of carbon/nitrogen balance. For optimal growth and development, carbon and nitrogen metabolism need to be tightly coordinated [74]. Based on the presumed function of the differentially expressed genes, more C compound related pathways are enriched in domesticated cottons relative to wild cottons, as reflected in the greater allocation to cell wall component synthesis (e.g., cellulose, VCLFA), energy generation through glycolysis, and amino acid synthesis (Figure 6). In turn, these biological processes might lower C/N, giving rise to less accumulation of anthocyanin. In contrast, nitrogen related pathways were enriched in wild cottons relative to domesticated cotton, as represented by nucleotide biosynthesis and phenylpropanoid biosynthesis. These two pathways can redirect carbon flow to nitrogen metabolism by diverting glucose-6-phosphate (G6P) into the pentose phosphate pathway or phosphoenolpyruvate (PEP) to the phenylpropanoid pathway (Figure 6). It may be, therefore, that the domestication process reallocated carbon resources toward carbohydrate and fatty acid metabolism. This speculation is also supported by a comparative metabolomics survey [12] of a lintless mutant and its wild type progenitor; the lintless mutant exhibited up-regulation of genes related to nitrogen compound metabolism along with accumulation of nitrogen compounds, compared to its WT. This phenomenon remains to be demonstrated at the metabolic level in wild and domesticated cottons.
Perhaps related to the above are differences in the deployment of stress-response pathways. For example, GAST1 protein homolog 1 (GASA1; Gorai.010G004400), involved in diverse developmental programs and stress responses [75], was highly up-regulated in wild cottons compared to domesticated cottons at 20 dpa. GASA genes have been reported to promote cell elongation in petunia flower [76], [77] or arrest cell elongation in gerbera [78] and strawberry [79]. Possibly, up-regulation of GASA1 in wild cottons at 20 dpa implies a negative regulation of cotton fiber elongation and/or modulation of stress response. Analyses of 176 genes related to the reactive oxygen species (ROS)-scavenging network [80] support the possibility of greater ROS sensitivity of wild cottons at 20 dpa; for example, during development 27 ROS genes were differentially expressed in wild cottons (7 vs. 20 genes = 10 vs. 20 dpa), while there were only 8 ROS genes identified in domesticated cottons, and also more ROS genes were up-regulated in wild than in domesticated cottons. ROS plays different roles depending on concentrations and context; ROS at low concentrations are involved as secondary messengers in several plant hormone responses, including seed germination, lignin biosynthesis, programmed cell death, and osmotic stress, while at high concentrations ROS are known to cause oxidative damage to proteins, lipids, and DNA [81]. It has been suggested that proper regulation of ROS homeostasis is necessary for cotton fiber elongation [28], [58]. For example, many ROS genes were up-regulated in parallel in domesticated diploid and polyploid cottons during early fiber elongation (2 dpa) [82], but only a few genes were investigated during fiber elongation, including ascorbate peroxidase (APX) [83], copper/zinc superoxide dismutase (CSD) [17], and peroxidase (POX) [84], which were all up-regulated in domesticated cottons relative to wild cottons at 10 dpa (Table S4). H2O2 accumulated at low levels during early elongation and peaks at 20 dpa in domesticated cottons [83], [85], but its levels have not been examined in wild accessions. Interestingly, recent analysis of transcriptomic profiles of a lintless mutant compared to its wild type G. hirsutum at 8 and 12 dpa showed higher expression levels of genes related to stress-response processes [12]. Up-regulation of stress-related genes in the lintless mutant and wild accessions of G. hirsutum relative to domesticated cottons suggests elevated levels of ROS in mutant and wild cotton fibers. It would be interesting to carefully evaluate the levels of different ROS molecules during development in wild vs. cultivated cotton under controlled conditions.
Concluding remarks
Although the transcriptomic data presented here are extraordinarily complex, as is usually the case in comparative profiling experiments, the data allow a speculative scenario to emerge from a consideration of the different classes of genes and pathways that are enriched under domestication. Specifically, we raise the suggestion that initial domestication of G. hirsutum, followed by several millennia of improvement and breeding, resulted in a shift or reallocation of resources from stress-related pathways in wild cottons to greater growth in domesticated forms. We envision that the reallocation and accompanying divergence in multiple pathways led to a prolonged period of fiber elongation, which at maturity are recognized now as the long, white, and fine fibers of modern cotton commerce. This scenario should become testable using a combination of forward genetic tools combined with advanced segregating populations (e.g., isogenic introgression lines), in conjunction with genomic, transcriptomic, proteomic, and metabolomic profiling. This systems approach holds the promise of improving our understanding of the evolutionary modification of a remarkable single-celled structure, while simultaneously providing clues to advance cotton breeding objectives.
Materials and Methods
Plant materials and library construction
Four wild and five domesticated G. hirsutum were selected for fiber transcriptome profiling based on their geographic origins and cotton fiber traits (Table 1; Figure 1). Wild cottons were originally from Yucatan, Mexico [86], while domesticated cottons were from four major cotton cultivation areas, i.e., Plains, Delta, and eastern and western U.S.A. Between three and twenty ovaries were collected for domesticated and wild cottons, respectively, from two developmental stages, 10 and 20 dpa, and were immediately dissected to harvest ovules, which were snap-frozen in liquid nitrogen until extraction. RNA was extracted using either a hot borate/lithium chloride procedure [87] or a CTAB extraction protocol [88], then purified by the RNeasy Plant Mini Kit (Qiagen, Stanford, CA, USA). Purified RNAs were quantified and qualified with Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). After mRNA purification using the MicroPoly(A) Purist kit (Ambion, Austin, TX, USA), RNA-Seq libraries were constructed with NEBNext mRNA Sample Prep Master Mix Set 1 following the manufacturer's suggestion (New England Biolabs, MA, USA). The constructed libraries, indexed with six nucleotide sequences, were pooled together with equimolar amounts and were sequenced on the Illumina HiSeq 2000 sequencer with 100 base reads at the Genomics Core Facility at the University of Oregon. Short read sequences were deposited in the NCBI Sequence Read Archive (SRA) with a study number SRP017061.
Analysis of RNA-seq data: Mapping and differential expression
Raw reads were sorted into the correct accession according to their indexed nucleotides. After trimming the indexed sequences, reads were filtered based on the quality scores (Q = 20) and read length (≧17 bp) with a fastx tool kit (http://hannonlab.cshl.edu/fastx_toolkit/index.html). Fastq formatted reads were mapped to the reference genome (Cotton D V2.0; 37,505 genes) [16] using GSNAP [89]. Reads with SNP information between A and D genome progenitors were parsed into A or D homoeolog-specific bins (At or Dt) using PolyCat (http://bioinfo3.pgml.uga.edu/polyCat/upload.html) [90].
Before identifying differentially expressed genes in each comparison of domestication (wild vs. domesticated G. hirsutum) and development (10 vs. 20 dpa), we examined the sample relations based on a multidimensional scale (or principal coordinate) using the edgeR package (ver. 2.0.5) in R software (ver. 2.16.0) [91]. If one sample shows a large distance from the others, that sample was removed for computing differential expression. The DESeq package (ver. 2.1.0) was used to detect differentially expressed genes in each contrast of domestication and development [92], and differential expression was defined when a gene showed at least 1.5-fold change with RPKM≧1 (RPKM: Reads Per Kilobase of gene model per Million mapped reads) [19] in all biological replicates of either wild or domesticated G. hirsutum. Also, to evaluate whether specific chromosomes or chromosome regions were selected during domestication and development, we investigated the distribution of differentially expressed genes on the 13 chromosomes in the haploid diploid cotton genome. For homoeolog-specific read counts, expression bias was evaluated using Fisher's exact test of the edgeR package. The distribution of p-values was controlled for a false discovery rate (FDR) by the BH method [93] at α = 0.05. Homoeolog-specific reads were analyzed as described in Yoo et al. [55], and differential expression was delimited by 1.5-fold expression changes with RPKM≧1 in either At or Dt reads across all biological replicates. In addition, we traced homoeolog expression changes during development and by domestication via comparing homoeolog expression patterns in each contrast. For example, At reads at 10 dpa were more down-regulated or highly expressed in domesticated cottons than in wild cottons, this expression change was tabulated as reflecting down- or up-regulation of At, respectively, during domestication.
For several gene families where some members are known to be involved in fiber development, we examined expression patterns of individual paralogs and (homoeologs) based on their phylogenetic relationships. Sequences were annotated by homology search against Arabidopsis thaliana and aligned via Clustal W [94]. Phylogenetic trees were constructed using MEGA 5.05 with a default option of Maximum Parsimony [95], and majority rule consensus trees were constructed.
Functional analysis of differentially expressed genes
To explore the nature of the biological pathways that were altered by domestication or that change during development, differentially expressed genes in each contrast were analyzed by SEA tool of agriGO (http://bioinfo.cau.edu.cn/agriGO/index.php) which performs GO term enrichment in one set of genes by comparing it to a reference list using fold changes [96]. For SEA, we used genes identified as differentially expressed (RPKM≧5 in either wild or domesticated cottons) in each contrast, with multi-test adjustment of the Benjamini-Yekutieli method (FDR<0.05) [97], and a minimum 5 mapping entries.
Supporting Information
Multidimensional scaling (MDS) plot showing relationships among samples in two dimensions. Because Maxxa at 10 dpa exhibited a large distance from other domesticated cottons and was embedded in wild cottons from 10 dpa, it was removed from further analysis. Red and blue circles indicate domesticated cottons from 10 and 20 dpa, respectively, while red and blue triangles represent wild cottons from 10 and 20 dpa, respectively.
(PPTX)
(A) CesA and Csl gene expression patterns during development. Inside and outside circles show the expression variation in domesticated and wild cottons, respectively. Blue and red circles indicate up-regulation at 10 and 20 dpa, respectively. Grey circles denote lack of differential expression, while white circles indicate lack of expression (zero read count). (B) Expression levels of CesA genes. (C) Expression levels of Csl genes. Bars in the charts show standard errors.
(PPTX)
Expression patterns of XTH genes in developing cotton fibers. Bar in the chart shows standard error.
(PPTX)
A putative nuclear-mitochondrial DNA sequence block (NUMT) (red-circled area) showing the fold changes across development of domesticated cottons (red cross: log2 dom20/dom10) or between wild and domesticated cottons at 20 dpa (blue diamond: log2 dom20/wild20) on chromosome 1.
(PPTX)
3-ketoacyl-CoA synthase (KCS) gene expression patterns. (A) Phylogenetic relationships of 27 KCS genes in G. raimondii. Genes in red and bold were up-regulated at domesticated cottons at 10 and 20 dpa, respectively. Asterisk (*) indicates A homoeolog expression bias in domesticated cottons. (B) Gene expression levels of differentially expressed KCSs. The expression level of Gorai.002G218500 is shown as half of its original value to allow comparison with other genes. Bar denotes standard error.
(PPTX)
Phylogenetic relationship of FLA homologues from Arabidopsis thaliana and Gossypium raimondii. Several FLA homologues from G. hirsutum were included (e.g., GhFLA; Huang et al. 2013) and group information is based on MacMillan et al. (2010). Blue and red arrows indicate genes up-regulated in wild and domesticated cottons at 10 dpa, respectively.
(PPTX)
The expression patterns of PROFILIN genes and several of its partners in wild and domesticated cottons.
(PPTX)
Expression patterns of MYB transcription factor related to phenylpropanoid biosynthesis.
(PPTX)
Number of differentially expressed genes in developing cotton fibers within wild and domesticated cottons using subsets of RNA-Seq data and two different techniques.
(XLSX)
Number of differentially expressed genes in developing cotton fibers between wild and domesticated cottons using subsets of RNA-Seq data and two different techniques. Grey shaded data are from the entire data set analysis, and red text indicates the use of biological replicates from the same accession. As for RNA-Seq data, RPKM≧1 was considered for differential expression if not specified.
(XLSX)
Chromosomal distribution of differentially expressed genes on Cotton D genome. The first table is from comparison of development, and the second one is from domestication comparison within wild and domesticated cotton. Numbers in red indicates statistically over-represented chromosome based on Chi-square test (P<0.05).
(XLSX)
Genes related to fiber development and their expression patterns in wild and domesticated cottons. Red and bold text indicates up-regulated in wild or domesticated cottons relative to their counterparts (RPKM≧5, P<0.05). dpa = days post anthesis. RPKM = Reads Per Kilobase of gene model per Million mapped reads.
(DOCX)
Homoeolog-specific regulation of 3-ketoacyl-CoA synthase (KCS) genes in developing cotton fiber. Expression levels were normalized with RPKM.
(XLSX)
Transcription factors that are the most highly expressed (RPKM≧100) in developing fibers and/or which are differentially expressed between wild and domesticated cottons (bold text, P<0.05). Underlined indicates differential expression when one domesticated accession was excluded.
(XLSX)
Comparison of results from Single Enrichment Analysis (SEA) of differentially expressed genes across two developmental stages and between wild and domesticated cottons. In ontology, P, M, and C indicate Biological Process, Molecular Function, and Cellular Components, respectively. The yellow-to-red colored blocks in CM (colorful mode) represent the level of up-regulation of each term; gray blocks indicate that the term is not significant. Numbers under CM correspond to the each comparison set, e.g., (1) shows SEA results for up-regulated at 10 dpa compared to 20 dpa in domesticated cottons.
(XLSX)
Acknowledgments
We thank Justin Page and Joshua Udall for providing access to PolyCat prior to its release. We also express our gratitude to Corrinne Grover and Jin Koh for bioinformatics assistance.
Funding Statement
This work was supported by the National Science Foundation Plant Genome Program award # 0817707. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Olsen KM, Wendel JF (2013) A bountiful harvest: genomic insights into crop domestication phenotypes. Annu Rev Plant Biol 64: 47–70. [DOI] [PubMed] [Google Scholar]
- 2. Olsen KM, Wendel JF (2013) Crop plants as models for understanding plant adaptation and diversification. Front Plant Sci 4: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication. Cell 127: 1309–1321. [DOI] [PubMed] [Google Scholar]
- 4.Wendel JF, Flagel L, Adams KL (2012) Jeans, genes, and genomes: cotton as a model for studying polyploidy. In: Soltis PS, Soltis DE, editors. Polyploidy and Genome Evolution. Berlin: Springer. pp. 181–207. [Google Scholar]
- 5. Wendel JF, Cronn RC (2003) Polyploidy and the evolutionary history of cotton. Adv Agron 78: 139–186. [Google Scholar]
- 6. Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, et al. (2011) Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100. [DOI] [PubMed] [Google Scholar]
- 7. Bao Y, Hu G, Flagel LE, Salmon A, Bezanilla M, et al. (2011) Parallel up-regulation of the profilin gene family following independent domestication of diploid and allopolyploid cotton (Gossypium). Proc Natl Acad Sci U S A 108: 21152–21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Udall JA, Swanson JM, Haller K, Rapp RA, Sparks ME, et al. (2006) A global assembly of cotton ESTs. Genome Res 16: 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lacape JM, Claverie M, Vidal RO, Carazzolle MF, Guimaraes Pereira GA, et al. (2012) Deep sequencing reveals differences in the transcriptional landscapes of fibers from two cultivated species of cotton. PLoS One 7: e48855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hinchliffe DJ, Turley RB, Naoumkina M, Kim HJ, Tang Y, et al. (2011) A combined functional and structural genomics approach identified an EST-SSR marker with complete linkage to the Ligon lintless-2 genetic locus in cotton (Gossypium hirsutum L.). BMC Genomics 12: 445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu K, Sun J, Yao L, Yuan Y (2012) Transcriptome analysis reveals critical genes and key pathways for early cotton fiber elongation in Ligon lintless-1 mutant. Genomics 100: 42–50. [DOI] [PubMed] [Google Scholar]
- 12. Naoumkina M, Hinchliffe DJ, Turley RB, Bland JM, Fang DD (2013) Integrated metabolomics and genomics analysis provides new insights into the fiber elongation process in Ligon lintless-2 mutant cotton (Gossypium hirsutum L.). BMC Genomics 14: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rapp RA, Haigler CH, Flagel L, Hovav RH, Udall JA, et al. (2010) Gene expression in developing fibres of Upland cotton (Gossypium hirsutum L.) was massively altered by domestication. BMC Biol 8: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu Y, Machado AC, White RG, Llewellyn DJ, Dennis ES (2006) Expression profiling identifies genes expressed early during lint fibre initiation in cotton. Plant Cell Physiol 47: 107–127. [DOI] [PubMed] [Google Scholar]
- 15. Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, et al. (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492: 423–427. [DOI] [PubMed] [Google Scholar]
- 17. Kim HJ, Triplett BA (2001) Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol 127: 1361–1366. [PMC free article] [PubMed] [Google Scholar]
- 18. Haigler CH, Betancur L, Stiff MR, Tuttle JR (2012) Cotton fiber: a powerful single-cell model for cell wall and cellulose research. Front Plant Sci 3: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y (2008) RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18: 1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hovav R, Udall JA, Hovav E, Rapp R, Flagel L, et al. (2008) A majority of cotton genes are expressed in single-celled fiber. Planta 227: 319–329. [DOI] [PubMed] [Google Scholar]
- 21. Arpat AB, Waugh M, Sullivan JP, Gonzales M, Frisch D, et al. (2004) Functional genomics of cell elongation in developing cotton fibers. Plant Mol Biol 54: 911–929. [DOI] [PubMed] [Google Scholar]
- 22. Chaudhary B, Hovav R, Rapp R, Verma N, Udall JA, et al. (2008) Global analysis of gene expression in cotton fibers from wild and domesticated Gossypium barbadense . Evol Dev 10: 567–582. [DOI] [PubMed] [Google Scholar]
- 23. Betancur L, Singh B, Rapp RA, Wendel JF, Marks MD, et al. (2010) Phylogenetically distinct cellulose synthase genes support secondary wall thickening in arabidopsis shoot trichomes and cotton fiber. J Integr Plant Biol 52: 205–220. [DOI] [PubMed] [Google Scholar]
- 24. Liepman AH, Cavalier DM (2012) The CELLULOSE SYNTHASE-LIKE A and CELLULOSE SYNTHASE-LIKE C families: recent advances and future perspectives. Front Plant Sci 3: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carpita NC (2011) Update on mechanisms of plant cell wall biosynthesis: how plants make cellulose and other (1->4)-beta-D-glycans. Plant Physiol 155: 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Farrokhi N, Burton RA, Brownfield L, Hrmova M, Wilson SM, et al. (2006) Plant cell wall biosynthesis: genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnol J 4: 145–167. [DOI] [PubMed] [Google Scholar]
- 27. Lerouxel O, Cavalier DM, Liepman AH, Keegstra K (2006) Biosynthesis of plant cell wall polysaccharides - a complex process. Curr Opin Plant Biol 9: 621–630. [DOI] [PubMed] [Google Scholar]
- 28.Stiff MR, Haigler CH (2012) Recent advances in cotton fiber development. In: Oosterhuis DM, Cothren JT, editors. Flowering and fruiting in cotton. Tennessee: The Cotton Foundation. pp. 163–192. [Google Scholar]
- 29. Ji SJ, Lu YC, Feng JX, Wei G, Li J, et al. (2003) Isolation and analyses of genes preferentially expressed during early cotton fiber development by subtractive PCR and cDNA array. Nucleic acids research 31: 2534–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qin YM, Pujol FM, Hu CY, Feng JX, Kastaniotis AJ, et al. (2007) Genetic and biochemical studies in yeast reveal that the cotton fibre-specific GhCER6 gene functions in fatty acid elongation. J Exp Bot 58: 473–481. [DOI] [PubMed] [Google Scholar]
- 31. Qin YM, Hu CY, Pang Y, Kastaniotis AJ, Hiltunen JK, et al. (2007) Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. Plant cell 19: 3692–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. MacMillan CP, Mansfield SD, Stachurski ZH, Evans R, Southerton SG (2010) Fasciclin-like arabinogalactan proteins: specialization for stem biomechanics and cell wall architecture in Arabidopsis and Eucalyptus. Plant J 62: 689–703. [DOI] [PubMed] [Google Scholar]
- 33. Shi H, Kim Y, Guo Y, Stevenson B, Zhu JK (2003) The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant cell 15: 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang GQ, Gong SY, Xu WL, Li W, Li P, et al. (2013) A fasciclin-like arabinogalactan protein, GhFLA1, is involved in fiber initiation and elongation of cotton. Plant Physiol 161: 1278–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Argiriou A, Kalivas A, Michailidis G, Tsaftaris A (2012) Characterization of PROFILIN genes from allotetraploid (Gossypium hirsutum) cotton and its diploid progenitors and expression analysis in cotton genotypes differing in fiber characteristics. Mol Biol Rep 39: 3523–3532. [DOI] [PubMed] [Google Scholar]
- 36. McKinney EC, Meagher RB (1998) Members of the Arabidopsis actin gene family are widely dispersed in the genome. Genetics 149: 663–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gilliland LU, Pawloski LC, Kandasamy MK, Meagher RB (2003) Arabidopsis actin gene ACT7 plays an essential role in germination and root growth. Plant J 33: 319–328. [DOI] [PubMed] [Google Scholar]
- 38. McDowell JM, An YQ, Huang S, McKinney EC, Meagher RB (1996) The arabidopsis ACT7 actin gene is expressed in rapidly developing tissues and responds to several external stimuli. Plant physiology 111: 699–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kandasamy MK, McKinney EC, Meagher RB (2009) A single vegetative actin isovariant overexpressed under the control of multiple regulatory sequences is sufficient for normal Arabidopsis development. Plant cell 21: 701–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li XB, Fan XP, Wang XL, Cai L, Yang WC (2005) The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant cell 17: 859–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tominaga-Wada R, Iwata M, Sugiyama J, Kotake T, Ishida T, et al. (2009) The GLABRA2 homeodomain protein directly regulates CESA5 and XTH17 gene expression in Arabidopsis roots. Plant J 60: 564–574. [DOI] [PubMed] [Google Scholar]
- 42. Galbiati M, Matus JT, Francia P, Rusconi F, Canon P, et al. (2011) The grapevine guard cell-related VvMYB60 transcription factor is involved in the regulation of stomatal activity and is differentially expressed in response to ABA and osmotic stress. BMC Plant Biol 11: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oh JE, Kwon Y, Kim JH, Noh H, Hong SW, et al. (2011) A dual role for MYB60 in stomatal regulation and root growth of Arabidopsis thaliana under drought stress. Plant Mol Biol 77: 91–103. [DOI] [PubMed] [Google Scholar]
- 44. Park JS, Kim JB, Cho KJ, Cheon CI, Sung MK, et al. (2008) Arabidopsis R2R3-MYB transcription factor AtMYB60 functions as a transcriptional repressor of anthocyanin biosynthesis in lettuce (Lactuca sativa). Plant Cell Rep 27: 985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Machado A, Wu Y, Yang Y, Llewellyn DJ, Dennis ES (2009) The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J 59: 52–62. [DOI] [PubMed] [Google Scholar]
- 46. Walford SA, Wu Y, Llewellyn DJ, Dennis ES (2011) GhMYB25-like: a key factor in early cotton fibre development. Plant J 65: 785–797. [DOI] [PubMed] [Google Scholar]
- 47. Pu L, Li Q, Fan X, Yang W, Xue Y (2008) The R2R3 MYB transcription factor GhMYB109 is required for cotton fiber development. Genetics 180: 811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Suo J, Liang X, Pu L, Zhang Y, Xue Y (2003) Identification of GhMYB109 encoding a R2R3 MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton (Gossypium hirsutum L.). Biochim Biophys Acta 1630: 25–34. [DOI] [PubMed] [Google Scholar]
- 49. Hao J, Tu L, Hu H, Tan J, Deng F, et al. (2012) GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J Exp Bot 63: 6267–6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang MY, Zhao PM, Cheng HQ, Han LB, Wu XM, et al. (2013) The Cotton transcription factor TCP14 functions in auxin-mediated epidermal cell differentiation and elongation. Plant Physiol 162: 1669–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xiao YH, Li DM, Yin MH, Li XB, Zhang M, et al. (2010) Gibberellin 20-oxidase promotes initiation and elongation of cotton fibers by regulating gibberellin synthesis. J Plant Physiol 167: 829–837. [DOI] [PubMed] [Google Scholar]
- 52. Beasley CA, Ting IP (1974) The effects of plant growth substances on in vitro fiber development from unfertilized cotton ovules. Am J Bot 61: 188–194. [Google Scholar]
- 53. Wanjie SW, Welti R, Moreau RA, Chapman KD (2005) Identification and quantification of glycerolipids in cotton fibers: reconciliation with metabolic pathway predictions from DNA databases. Lipids 40: 773–785. [DOI] [PubMed] [Google Scholar]
- 54. Grover CE, Gallagher JP, Szadkowski EP, Yoo MJ, Flagel LE, et al. (2012) Homoeolog expression bias and expression level dominance in allopolyploids. New Phytol 196: 966–971. [DOI] [PubMed] [Google Scholar]
- 55. Yoo MJ, Szadkowski E, Wendel JF (2013) Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 110: 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Applequist WL, Cronn R, Wendel JF (2001) Comparative development of fiber in wild and cultivated cotton. Evol Dev 3: 3–17. [DOI] [PubMed] [Google Scholar]
- 57. Seagull RW, Oliveri V, Murphy K, Binder A, Kothari S (2000) Cotton fiber growth and development 2. Changes in cell diameter and wall birefringence. J Cotton Sci 4: 97–104. [Google Scholar]
- 58. Hovav R, Udall JA, Chaudhary B, Hovav E, Flagel L, et al. (2008) The evolution of spinnable cotton fiber entailed prolonged development and a novel metabolism. PLoS Genet 4: e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hu G, Koh J, Yoo MJ, Grupp K, Chen S, et al. (2013) Proteomic profiling of developing cotton fibers from wild and domesticated Gossypium barbadense . New Phytol 200: 570–582. [DOI] [PubMed] [Google Scholar]
- 60. Rubinovich L, Weiss D (2010) The Arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. Plant J 64: 1018–1027. [DOI] [PubMed] [Google Scholar]
- 61. Hundertmark M, Hincha DK (2008) LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana . BMC Genomics 9: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Seo PJ, Park JM, Kang SK, Kim SG, Park CM (2011) An Arabidopsis senescence-associated protein SAG29 regulates cell viability under high salinity. Planta 233: 189–200. [DOI] [PubMed] [Google Scholar]
- 63. Hsieh TH, Li CW, Su RC, Cheng CP, Sanjaya, et al. (2010) A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta 231: 1459–1473. [DOI] [PubMed] [Google Scholar]
- 64. Tan J, Tu L, Deng F, Hu H, Nie Y, et al. (2013) A genetic and metabolic analysis revealed that cotton fiber cell development was retarded by flavonoid naringenin. Plant Physiol 162: 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Feng H, Tian X, Liu Y, Li Y, Zhang X, et al. (2013) Analysis of flavonoids and the flavonoid structural genes in brown fiber of upland cotton. PLoS One 8: e58820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rambani A, Page JT, Udall JA (2013) Polyploidy and the petal transcriptome of Gossypium . BMC Plant Biol (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Padmalatha KV, Dhandapani G, Kanakachari M, Kumar S, Dass A, et al. (2012) Genome-wide transcriptomic analysis of cotton under drought stress reveal significant down-regulation of genes and pathways involved in fibre elongation and up-regulation of defense responsive genes. Plant Mol Biol 78: 223–246. [DOI] [PubMed] [Google Scholar]
- 68. Wang CY, Chiou CY, Wang HL, Krishnamurthy R, Venkatagiri S, et al. (2008) Carbohydrate mobilization and gene regulatory profile in the pseudobulb of Oncidium orchid during the flowering process. Planta 227: 1063–1077. [DOI] [PubMed] [Google Scholar]
- 69. Babb VM, Haigler CH (2001) Sucrose phosphate synthase activity rises in correlation with high-rate cellulose synthesis in three heterotrophic systems. Plant Physiol 127: 1234–1242. [PMC free article] [PubMed] [Google Scholar]
- 70. Padmalatha KV, Patil DP, Kumar K, Dhandapani G, Kanakachari M, et al. (2012) Functional genomics of fuzzless-lintless mutant of Gossypium hirsutum L. cv. MCU5 reveal key genes and pathways involved in cotton fibre initiation and elongation. BMC Genomics 13: 624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lassner MW, Lardizabal K, Metz JG (1996) A jojoba beta-Ketoacyl-CoA synthase cDNA complements the canola fatty acid elongation mutation in transgenic plants. Plant cell 8: 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pourcel L, Routaboul JM, Cheynier V, Lepiniec L, Debeaujon I (2007) Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends Plant Sci 12: 29–36. [DOI] [PubMed] [Google Scholar]
- 73. Al-Ghazi Y, Bourot S, Arioli T, Dennis ES, Llewellyn DJ (2009) Transcript profiling during fiber development identifies pathways in secondary metabolism and cell wall structure that may contribute to cotton fiber quality. Plant Cell Physiol 50: 1364–1381. [DOI] [PubMed] [Google Scholar]
- 74. Zheng ZL (2009) Carbon and nitrogen nutrient balance signaling in plants. Plant Signal Behav 4: 584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nahirnak V, Almasia NI, Hopp HE, Vazquez-Rovere C (2012) Snakin/GASA proteins: involvement in hormone crosstalk and redox homeostasis. Plant Signal Behav 7: 1004–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ben-Nissan G, Weiss D (1996) The petunia homologue of tomato gast1: transcript accumulation coincides with gibberellin-induced corolla cell elongation. Plant Mol Biol 32: 1067–1074. [DOI] [PubMed] [Google Scholar]
- 77. Ben-Nissan G, Lee JY, Borohov A, Weiss D (2004) GIP, a Petunia hybrida GA-induced cysteine-rich protein: a possible role in shoot elongation and transition to flowering. Plant J 37: 229–238. [DOI] [PubMed] [Google Scholar]
- 78. Kotilainen M, Helariutta Y, Mehto M, Pollanen E, Albert VA, et al. (1999) GEG participates in the regulation of cell and organ shape during corolla and carpel development in Gerbera hybrida . Plant cell 11: 1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. de la Fuente JI, Amaya I, Castillejo C, Sanchez-Sevilla JF, Quesada MA, et al. (2006) The strawberry gene FaGAST affects plant growth through inhibition of cell elongation. J Exp Bot 57: 2401–2411. [DOI] [PubMed] [Google Scholar]
- 80. Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9: 490–498. [DOI] [PubMed] [Google Scholar]
- 81. Sharma P, Jha AB, Dubey RS, Pessarakli M (2012) Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J Bot 10.1155/2012/217037. [Google Scholar]
- 82. Chaudhary B, Hovav R, Flagel L, Mittler R, Wendel JF (2009) Parallel expression evolution of oxidative stress-related genes in fiber from wild and domesticated diploid and polyploid cotton (Gossypium). BMC Genomics 10: 378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Yang YW, Bian SM, Yao Y, Liu JY (2008) Comparative proteomic analysis provides new insights into the fiber elongating process in cotton. J Proteome Res 7: 4623–4637. [DOI] [PubMed] [Google Scholar]
- 84. Mei W, Qin Y, Song W, Li J, Zhu Y (2009) Cotton GhPOX1 encoding plant class III peroxidase may be responsible for the high level of reactive oxygen species production that is related to cotton fiber elongation. J Genet Genomics 36: 141–150. [DOI] [PubMed] [Google Scholar]
- 85. Potikha TS, Collins CC, Johnson DI, Delmer DP, Levine A (1999) The involvement of hydrogen peroxide in the differentiation of secondary walls in cotton fibers. Plant Physiol 119: 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Brubaker CL, Wendel JF (1994) Reevaluating the origin of domesticated cotton (Gossypium hirsutum: Malvaceae) using nuclear restriction fragment length polymorphisms (RFLPs). Am J Bot 81: 1309–1326. [Google Scholar]
- 87.Wilkins TA, Smart LB (1996) Isolation of RNA from plant tissue. In: Krieg PA, editor. A Laboratory Guide to RNA: solation, Analysis and Synthesis. New York: Wiley-Liss. pp. 21–41. [Google Scholar]
- 88. Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19: 11–15. [Google Scholar]
- 89. Wu TD, Nacu S (2010) Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26: 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Page JT, Gingle AR, Udall JA (2013) PolyCat: a resource for genome categorization of sequencing reads from allopolyploid organisms. G3 3: 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 57: 289–300. [Google Scholar]
- 94. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. Ann Stat 29: 1165–1188. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multidimensional scaling (MDS) plot showing relationships among samples in two dimensions. Because Maxxa at 10 dpa exhibited a large distance from other domesticated cottons and was embedded in wild cottons from 10 dpa, it was removed from further analysis. Red and blue circles indicate domesticated cottons from 10 and 20 dpa, respectively, while red and blue triangles represent wild cottons from 10 and 20 dpa, respectively.
(PPTX)
(A) CesA and Csl gene expression patterns during development. Inside and outside circles show the expression variation in domesticated and wild cottons, respectively. Blue and red circles indicate up-regulation at 10 and 20 dpa, respectively. Grey circles denote lack of differential expression, while white circles indicate lack of expression (zero read count). (B) Expression levels of CesA genes. (C) Expression levels of Csl genes. Bars in the charts show standard errors.
(PPTX)
Expression patterns of XTH genes in developing cotton fibers. Bar in the chart shows standard error.
(PPTX)
A putative nuclear-mitochondrial DNA sequence block (NUMT) (red-circled area) showing the fold changes across development of domesticated cottons (red cross: log2 dom20/dom10) or between wild and domesticated cottons at 20 dpa (blue diamond: log2 dom20/wild20) on chromosome 1.
(PPTX)
3-ketoacyl-CoA synthase (KCS) gene expression patterns. (A) Phylogenetic relationships of 27 KCS genes in G. raimondii. Genes in red and bold were up-regulated at domesticated cottons at 10 and 20 dpa, respectively. Asterisk (*) indicates A homoeolog expression bias in domesticated cottons. (B) Gene expression levels of differentially expressed KCSs. The expression level of Gorai.002G218500 is shown as half of its original value to allow comparison with other genes. Bar denotes standard error.
(PPTX)
Phylogenetic relationship of FLA homologues from Arabidopsis thaliana and Gossypium raimondii. Several FLA homologues from G. hirsutum were included (e.g., GhFLA; Huang et al. 2013) and group information is based on MacMillan et al. (2010). Blue and red arrows indicate genes up-regulated in wild and domesticated cottons at 10 dpa, respectively.
(PPTX)
The expression patterns of PROFILIN genes and several of its partners in wild and domesticated cottons.
(PPTX)
Expression patterns of MYB transcription factor related to phenylpropanoid biosynthesis.
(PPTX)
Number of differentially expressed genes in developing cotton fibers within wild and domesticated cottons using subsets of RNA-Seq data and two different techniques.
(XLSX)
Number of differentially expressed genes in developing cotton fibers between wild and domesticated cottons using subsets of RNA-Seq data and two different techniques. Grey shaded data are from the entire data set analysis, and red text indicates the use of biological replicates from the same accession. As for RNA-Seq data, RPKM≧1 was considered for differential expression if not specified.
(XLSX)
Chromosomal distribution of differentially expressed genes on Cotton D genome. The first table is from comparison of development, and the second one is from domestication comparison within wild and domesticated cotton. Numbers in red indicates statistically over-represented chromosome based on Chi-square test (P<0.05).
(XLSX)
Genes related to fiber development and their expression patterns in wild and domesticated cottons. Red and bold text indicates up-regulated in wild or domesticated cottons relative to their counterparts (RPKM≧5, P<0.05). dpa = days post anthesis. RPKM = Reads Per Kilobase of gene model per Million mapped reads.
(DOCX)
Homoeolog-specific regulation of 3-ketoacyl-CoA synthase (KCS) genes in developing cotton fiber. Expression levels were normalized with RPKM.
(XLSX)
Transcription factors that are the most highly expressed (RPKM≧100) in developing fibers and/or which are differentially expressed between wild and domesticated cottons (bold text, P<0.05). Underlined indicates differential expression when one domesticated accession was excluded.
(XLSX)
Comparison of results from Single Enrichment Analysis (SEA) of differentially expressed genes across two developmental stages and between wild and domesticated cottons. In ontology, P, M, and C indicate Biological Process, Molecular Function, and Cellular Components, respectively. The yellow-to-red colored blocks in CM (colorful mode) represent the level of up-regulation of each term; gray blocks indicate that the term is not significant. Numbers under CM correspond to the each comparison set, e.g., (1) shows SEA results for up-regulated at 10 dpa compared to 20 dpa in domesticated cottons.
(XLSX)