Abstract
Phospholipase D (PLD) hydrolyzes the phosphodiester bond of glycerophospholipids and produces phosphatidic acid (PA), which acts as a second messenger in many living organisms. A large number of PLDs have been identified in eukaryotes, and are viewed as promising targets for drug design because these enzymes are known to be tightly regulated and to function in the pathophysiology of many human diseases. However, the underlying molecular mechanisms of catalysis and regulation of eukaryotic PLD remain elusive. Here, we determined the crystal structure of full-length plant PLDα1 in the apo state and in complex with PA. The structure shows that the N-terminal C2 domain hydrophobically interacts with the C-terminal catalytic domain that features two HKD motifs. Our analysis reveals the catalytic site, substrate-binding mechanism, and a new Ca2+-binding site that is required for the activation of PLD. In addition, we tested several efficient small-molecule inhibitors against PLDα1, and suggested a possible competitive inhibition mechanism according to structure-based docking analysis. This study explains many long-standing questions about PLDs and provides structural insights into PLD-targeted inhibitor/drug design.
Subject terms: X-ray crystallography, Plant signalling
Introduction
Phospholipase D (PLD) belongs to the phospholipase superfamily and is widely distributed in prokaryotes and eukaryotes. It catalyzes the hydrolysis of phosphodiester bond of glycerophospholipid substrates to generate products including phosphatidic acid (PA) and hydrophilic headgroups such as choline, ethanolamine, serine, etc. Since the identification of PLD in plants,1 a large number of PLDs have been intensively studied in mammals and other eukaryotes.2–7 PLDs exert various biological functions mainly through its end product PA, and to a less extent through interaction proteins.5,6 In eukaryotic cells, PA serves as not only a structural intermediate lipid for membrane lipid synthesis but also a vital second messenger molecule binding with downstream targets to regulate various cellular events.8–10 PLD contributes to cell proliferation, inflammation, virus infection, and also plays roles in neurodegenerative diseases, human cancers, and plant stress responses.5,7,11–14
Eukaryotic PLDs typically contain two domains, a C-terminal catalytic domain comprising two HKD motifs and an N-terminal PX/PH or C2 domain. Based on their N-terminal domains, PLDs are classified as PX/PH-PLDs and C2-PLDs. Mammalian cells have two PLD isoenzymes, PLD1 and PLD2, and both are PX/PH-PLDs.3,15 Plants have a much larger number of PLD enzymes.16,17 For example, the model plant Arabidopsis encodes twelve PLD isoenzymes, classified into subtypes including PLDα (3), β (2), γ (3), δ, ε and ζ (2), among which two PLDζ isoenzymes are PX/PH-PLDs while all others are C2-PLDs. Although various PLDs differ significantly in their N-terminal domains, numerous lines of evidence suggest that most of these domains are closely related to the binding of membrane and lipids.5,6,18 In addition, it has been reported that the in vitro activity of C2-PLDs requires the C2 domain,19 whereas the activity of PX/PH-PLDs seems to be independent of its PX/PH domain.20,21 The sequences of the C-terminal catalytic domain among various eukaryotic PLDs show a high degree of similarity, suggesting a common catalytic mechanism. The activity of PLD is known to be modulated by small molecules and by interaction proteins.4–6 Specifically, calcium ions could activate C2-PLDs at various concentrations; in contrast, the influence of these ions on the activity of PX/PH-PLDs is apparently minor.15,17,22 Lipids have also been reported to affect the activities of PLDs, either as positive cofactors or as allosteric modulators.15,22,23 In addition, interaction proteins such as small G protein Rac1/Arf-1/Gα have also been demonstrated to influence the PLD activity directly in animals and plants.9,20,21,24 However, the underlying molecular mechanisms remain to be answered.
Since PLD plays important roles in the pathophysiology of many human diseases, it is considered as a promising drug target.7,25 Studies on screening lead compounds and inhibitors targeting PLDs have been increasing steadily in recent years. A number of small molecules have been found and their inhibitory effects on mammalian PLD1 and PLD2 have been tested.26–30 Several such compounds have been reported to have obvious cellular effects12,31–34; however, the lack of 3-D structure information of eukaryotic PLDs has hindered the pace of efficient drug design and optimization. We present here a structural and biochemical study of a plant C2-type PLDα, revealing the underlying molecular mechanisms of catalysis, activation, and regulation of the eukaryotic PLD superfamily. Our study may also inform drug design efforts targeting PLD.
Results and discussion
Activation and inhibition of PLDα1
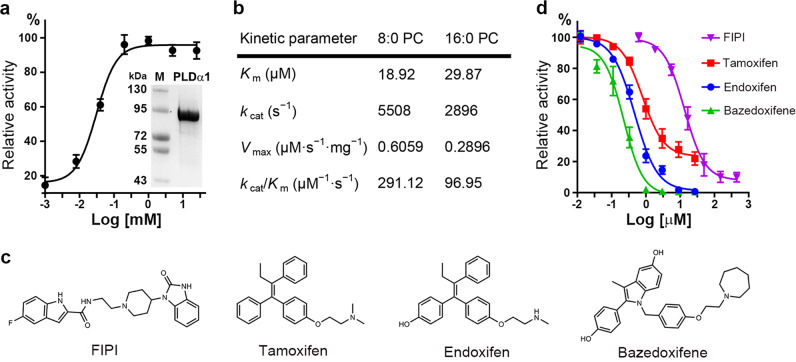
The gene encoding full-length Arabidopsis PLDα1 was cloned and recombinantly expressed in E. coli. Protein purification was performed through Ni2+ chelating, Source-Q, and gel filtration chromatography. The resulting PLDα1 protein was of high purity and homogeneity, suitable for use in biochemical analysis and crystallization (Fig. 1a). Consistent with the previous reports, we found that PLDα1 can be activated by Ca2+ in a concentration-dependent manner (Fig. 1a). The Ca2+ concentration required for optimal activity of PLDα1 was measured to be 1 mM; a Ca2+ concentration of 5 mM was chosen for the following activity assays. The kinetic parameters of PLDα1 were measured using both short- and long-chain phosphatidylcholine (PC, di8:0-PC and di16:0-PC) as substrates. The Km and kcat values of PLDα1 toward di8:0-PC were 18.9 μM and 5.5 × 103/s, respectively, comparable to those toward di16:0-PC (Km, 29.9 μM; kcat, 2.9 × 103/s) (Fig. 1b).
Fig. 1.
Characterization of plant PLDα1. a Activation of PLDα1 by Ca2+. Purified PLDα1 is shown on SDS-PAGE stained with Coomassie blue. Results are means ± SD, n = 3. b Kinetic parameters determined using di8:0-PC and di16:0-PC as substrates. c Chemical structures of the tested small-molecule inhibitors. d Inhibition curves of PLDα1 by the inhibitors in c (di8:0-PC was used as substrate). Results are means ± SD, n = 3
Small-molecule inhibitors have been reported for mammalian PLD1/PLD2, including FIPI, Raloxifene, Tamoxifen as well as their derivatives.7 We tested several small molecules with diverse basic structural skeletons for their inhibitory effect on PLDα1 (Fig. 1c). Among the tested small molecules, Bazedoxifene (a Raloxifene derivative) was the best inhibitor toward PLDα1 with an IC50 of 0.37 μM, followed by Endoxifen (a Tamoxifen derivative) with IC50 of 0.54 μM and Tamoxifen with IC50 of 0.89 μM (Fig. 1d). FIPI turned out to be the weakest inhibitor toward PLDα1 with an IC50 of 15.2 μM. Furthermore, we found that the Km increased and the Vmax remained unchanged with the addition of Bazedoxifene, inferring a substrate-competitive inhibition mode (Supplementary information, Fig. S1). These results suggest that C2-PLDs and PX/PH-PLDs share a common catalytic mechanism and can be inhibited by small molecules of similar structure.
Overall structure and domain constitution
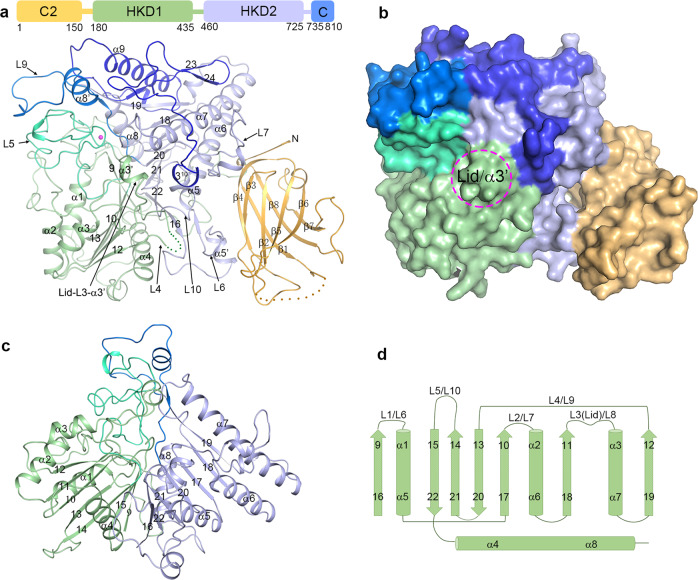
The crystal structure of Arabidopsis PLDα1 in apo form was determined at 1.8 Å using SAD method (Supplementary information, Table S1 and Fig. S2). The structure contains the full-length PLDα1, except that residues 22–46 and 313–326 are disordered (Fig. 2a). The overall structure can be divided into two domains, the N-terminal C2 domain (residues 1–150) and the C-terminal catalytic domain. The catalytic domain comprises three subdomains: the HKD1 subdomain (residues 180–435), the HKD2 subdomain (residues 460–725), and the C-terminus subdomain (residues 735–810) (Fig. 2a, b). The HKD1 and HKD2 subdomains are packed against each other, and together constitute the core of the catalytic domain (Fig. 2c), adopting a saddle-like conformation similar to the reported Streptomyces sp. PLDPMF structure.35
Fig. 2.
Overall structure of PLDα1. a Structure of PLDα1 is shown in ribbon cartoon with structure elements labeled. The C2 domain, HKD1 subdomain, HKD2 subdomain and C-terminus subdomain are colored in bright orange, pale green, light blue and blue, respectively. The L5 and L9 loops are colored in lime green and marine, respectively. The disordered regions (residues 22–46, and residues 313–326) are indicated with dotted lines. The Ca2+ ion is shown as a magenta sphere. A simplified domain partition is shown on the top, with starting/ending residue numbers. b Surface model shows the structure of PLDα1; color codes are the same as in a. Position of the substrate-binding pocket lid α3′ helix is circled. c, d Structure (c) and topology (d) of the HKD1 and HKD2 subdomains are shown. Structure elements are labeled
The C2 domain of PLDα1 comprises eight β-strands that collectively form two layers of antiparallel β-sheets, with β3/β2/β5/β6 strands as the top layer and β4/β1/β8/β7 strands as the bottom layer (Fig. 2a). The disordered residues 22–46 are located at the region connecting β1 and β2, which corresponds to the Ca2+-binding site in known C2 domain structures.36–38 However, no Ca2+ binding was found in our C2 domain structure, even though 200 mM Ca2+ was present during crystallization, indicating weak binding. The C2 domain is connected via a loop to the HKD1 subdomain, which is in turn connected to the HKD2 subdomain. The structures and topologies of HKD1 and HKD2 subdomains are very similar, and both adopt a three-layered sandwich conformation, with seven β-strands in the middle, and three or one α-helices positioned to each side (Fig. 2c, d). Loops of variable lengths connect the α-helices and β-strands of HKD1/2. The L1/L6 loops connect β9/β16 with α1/α5, the L2/L7 loops connect β10/β17 with α2/α6, the L3/L8 loops connect β11/β18 with α3/α7, the L4/L9 loops connect β12/β19 with β13/β20, and the L5/L10 loops connect β14/β21 with β15/β22. Small α-helix insertions (denoted with α′ in Fig. 2) are present in the L3 (α3′), L6 (α5′) and L9 (α8′) loops (Fig. 2a, d). Among these loops and insertions, α3′-L3 from the HKD1 subdomain covers the top of the substrate-binding pocket like a lid, indicating that the apo-form PLDα1 structure represents a “closed” conformation (Fig. 2a, b). The L5 loop of the HKD1 subdomain makes close interactions with the L9 loop of HKD2. Notably, a Ca2+-binding site was found in the L5/L9 loop interaction region near the substrate-binding pocket (Fig. 2a). The structure is ended by the C-terminus subdomain comprising 75 residues that form α9 helix, a long loop, and a C-terminus 310 helix. The architecture of PLDα1 is reminiscent of the previously reported human cytoplasmic PLA2 structure (cPLA2),39 which is also comprised of a C2 domain and a catalytic domain (Supplementary information, Fig. S3). However, different from the cPLA2 structure in which the C2 domain makes few interactions with the catalytic domain, the C2 domain of PLDα1 interacts extensively with the HKD2 subdomain (Fig. 2a, b); we later noted that these interactions are required for the enzyme activity of PLDα1 (below).
Substrate-binding pocket and conformational change
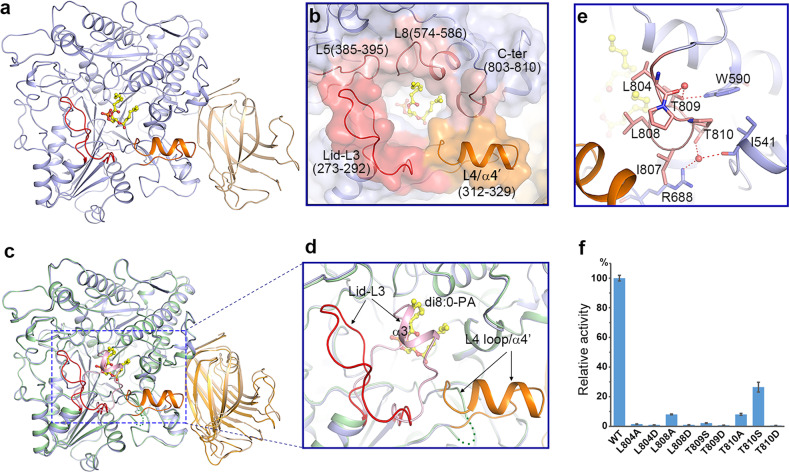
Attempting to study the mechanism of substrate binding and catalysis of PLDα1, we made numerous efforts aiming to determine the structure of a substrate-enzyme complex. In the end, we solved the crystal structure of PLDα1 in complex with the reaction product di8:0-PA (hereafter PLDα1-PA) (Fig. 3a and Supplementary information, Fig. S4). The di8:0-PA binds in a hydrophobic substrate-binding pocket positioned between the HKD1 and HKD2 subdomains (Fig. 3a and Supplementary information, Fig. S5b). The L3, L4 and L5 loops of the HKD1 subdomain, the L8 loop of the HKD2 subdomain, and the C-terminus 310 helix constitute the substrate-binding pocket, which is widely open to the solvent (Fig. 3b). Compared to the apo structure, there is a significant conformational change at the substrate-binding pocket induced by PA binding (Fig. 3c). Specifically, the α3′ helix sealing the substrate-binding pocket in the apo PLDα1 structure (Fig. 2a, b) unravels into a loop and flips away from the top in the PLDα1-PA structure (Fig. 3c, d); in addition, the disordered L4 loop adopts an α-helix structure (α4′ hereafter) upon the binding of PA (Fig. 3d). These conformational changes allow the substrate-binding pocket to convert from the aforementioned “closed” state to an “open” state (Fig. 3c and Supplementary information, Fig. S5). We also noted that the conformation of the C2 domain undergoes a slight positional twist away from the catalytic domain upon PA binding (Fig. 3c).
Fig. 3.
Substrate-binding pocket and conformational changes induced by substrate binding. a Ribbon structure of PLDα1 in complex with di8:0-PA. The catalytic domain and C2 domain are colored in slate blue and wheat, respectively. The Lid L3 loop, and pocket-forming L4 loop/α4′ are colored in red and orange, respectively. The di8:0-PA molecule is shown as a yellow stick model. b Zoom-in view shows the substrate-binding pocket. Among structure elements constituting the substrate-binding pocket, the L3 loop is colored in red, the L4 loop and α4′ are colored in orange, and the L5/L8 loops and the C-terminus 310 helix are colored in pink. c Structure superimposition of PLDα1 structure in apo form (the catalytic domain, C2 domain, and the substrate-binding pocket Lid-α3′ are colored in pale green, bright orange and light pink, respectively) and PA-binding form (colored codes are the same as in a) shows the conformational changes. The L4 loop (green dotted line) in the apo structure turns into α4′ helix (colored in orange) upon PA binding. d Zoom-in view of the substrate-binding pocket region showing conformational changes. e Interaction of the C-terminus with surrounding residues. The side chains of the interaction residues are shown with sticks, and water molecules are shown with red spheres. Red dotted lines indicate the hydrogen-bonding interactions. f Influence of C-terminal residue mutations on PLDα1 activity. Results are means ± SD, n = 3
Our structural data that the 310 helix at C-terminus contributes to the substrate-binding pocket in PLDα1 can well explain the previously reported results, which suggested that the C-terminal residues of PLDs, in both animals and plants, function in catalytic processes of these enzymes.19,40–42 This finding also motivated our speculation that perhaps the C-termini of all PLDs may function similarly. However, the amino acid sequences of the C-termini of human PLD1/2, plant PLDα1 and other C2-PLDs are variable (Supplementary information, Fig. S6). Zoom-in analysis of the PLDα1 structure reveals that two conserved hydrophobic residues Leu804 and Leu808, protrude from the C-terminus 310 helix into the substrate-binding pocket to bind with the acyl chains of PA (Fig. 3b, e). Mutation of these two residues to Ala or Asp dramatically reduced or eliminated the enzyme’s in vitro catalytic activity, experimentally supporting their functional roles in substrate binding (Fig. 3f). A strictly conserved Thr809 residue among the C2-PLD enzymes forms a hydrogen bond with residue Trp590 (again, invariable among C2-PLDs) to dictate the accurate position of the C-terminus (Fig. 3e and Supplementary information, Fig. S6), and mutation of Thr809 to Ser or Asp completely eliminated the activity. Similar results were obtained in T810D or T810A mutation variants (Fig. 3f). These results together reveal the functional mechanism of the C-terminus of PLDα1 and even other C2-PLDs, and infer that this may also be the case in PX/PH PLDs.
Active site
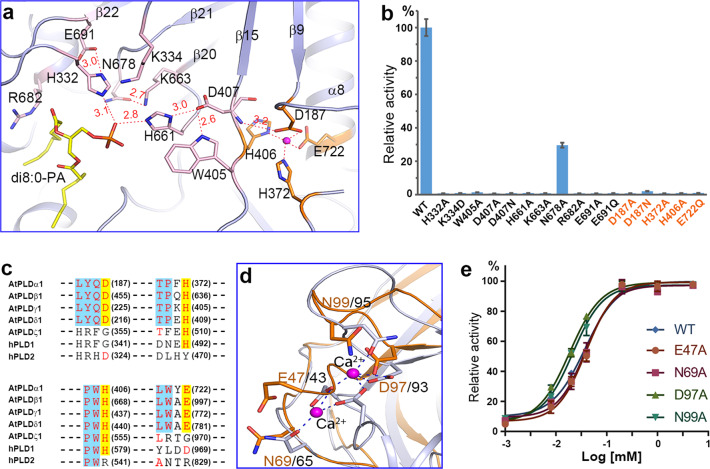
The active site of PLDα1 is located at the bottom of the substrate-binding pocket, and the residues forming the active site come from two HKD subdomains (Fig. 4a and Supplementary information, Fig. S6). Interestingly, the residues constituting the eukaryotic “HKD” motifs turn out to be spatial, but not sequential. Residues His332 and K334 from the L4 loop and Glu691 from β22 constitute the first HKD motif. Residues His661 from the L9 loop, K663 from β20, and Asp407 from the L5 loop constitute the second. The di8:0-PA binds to the active site, with the phosphate group forming hydrogen bonds with residues His332 and His661 (Fig. 4a), and the acyl-chains making hydrophobic interactions with residues Trp231, Tyr523, Val584, Ile587, Arg682, Leu804 and Leu808 (Fig. 4a and Supplementary information, Fig. S7). Other residues like Thr330, Asp361, Arg366, Trp405, Gln522, Asn678 and Asp689 stabilize the active site residues or substrate through hydrophilic or hydrophobic interactions (Supplementary information, Fig. S7). A hydrogen-bonding network can be clearly seen in the active site (Fig. 4a). Mutations of the HKD-motif residues could completely disrupt the enzyme activity, and other mutations could either eliminate or diminish the activity (Fig. 4b and Supplementary information, Fig. S8).
Fig. 4.
Active site and Ca2+-mediated activation of PLDα1. a Structure of the active site. The residues constituting the active site and the Ca2+-binding site are shown with sticks, and are colored in pink and orange, respectively. Ca2+ is shown as a magenta sphere. Dash lines indicate the interactions, and the interaction distances are labeled. b Enzyme activity of PLDα1 mutations at the active site residues and Ca2+-binding residues near the active site. Results are means ± SD, n = 3. c Amino acid sequence alignment of Ca2+-binding residues near the active site (colored in red and highlighted in yellow) among different PLDs. AtPLDα1, AtPLDβ1, AtPLDγ1, and AtPLDδ1 are C2-PLDs from Arabidopsis; AtPLDζ1 and hPLD1/2 are PX/PH-PLDs from Arabidopsis and human. The number of the Ca2+-binding residues and corresponding residues are shown in the parentheses. d Structure superimposition of C2 domains from PLDα1 (colored in bright orange) and cPLA2 (PDB ID: 1RLW, colored in gray) shows the residues involved in Ca2+ binding. e Influence of mutations of the C2 domain Ca2+-binding residues on Ca2+-mediated activation of PLDα1
Multiple sequence alignment reveals that most of the aforementioned residues are conserved among eukaryotic PLDs (Supplementary information, Fig. S6). Furthermore, comparison of the crystal structures indicated that most of the active site residues of PLDα1 are the same as those of the PLDPMF35 (Supplementary information, Fig. S9). These findings, viewed alongside our aforementioned results about the apparently common inhibitory interactions of small molecules (e.g., Tamoxifen), collectively suggest that the HKD family PLDs, although differing greatly in sequence and domain constitution, share a common catalytic mechanism. We propose that PLDα1 and even eukaryotic HKD family PLDs utilize a two-step catalytic mechanism as has been well studied in PLDPMF.43,44 The histidine residue (His332) from the first HKD motif acts as a nucleophile, which forms a phosphoenzyme intermediate and simultaneously releases the head group (choline, ethanolamine, etc.); whereas the histidine residue (H661) from the second HKD motif functions as a general acid in the cleavage of the phosphodiester bond to release PA.
Enzyme activation
We identified a Ca2+-binding site near the second HKD motif (Fig. 4a and Supplementary information, Fig. S10). Residues Asp187 from the L1 loop, His372 and His406 from the L5 loop, and Glu722 from α8 bind with the Ca2+ ion via four coordinates. In addition, residue Asp187 forms a hydrogen bond with the main chain nitrogen of residue Asp407 from the second HKD motif (Fig. 4a). Mutation of any one of these four residues results in complete loss of catalytic activity (Fig. 4b). The interaction network of Ca2+ probably defines the position of the loop preceding β15 and conformations of residues Asp407 and Trp405 that are critical for enzyme activity. Multiple sequence alignment shows that these Ca2+ coordination residues are strictly conserved among C2-PLDs, but not among PX/PH-PLDs (Fig. 4c). Accordingly, the C2-PLD enzymes are known to be activated by Ca2+, however, this is not the case with the PX/PH-PLDs.15,17,22 This distinction is informative, promoting reexamination of the mechanism of PLD activation by calcium ions. Previous studies have attributed the Ca2+-mediated activation of C2-PLDs to the calcium-binding motifs within the C2 domain,23,45 whereas our work establishes that Ca2+ binding at the catalytic domain near the second HKD motif contributes to the activation of PLDα1 and probably other C2-PLDs.
Most of the corresponding Ca2+-binding residues in the C2 domains of cPLA236,38 were found to be present in the PLDα1 structure (Fig. 4d), which are Glu47, Asn69, Asp97 and Asn99. We therefore mutated these residues to evaluate their functional impacts on Ca2+-mediated activation of PLDα1. Strikingly, the activation profiles of the tested mutant variants were not different from that of the wild type (Fig. 4e). Thus, our finding clearly suggests that Ca2+ binding in the C2 domain does not affect Ca2+-mediated activation of PLDα1. Instead, we show that the binding of Ca2+ near the active site in the catalytic domain can dictate the position of residues His407 and Trp405, and that both of these residues are essential for catalysis, thereby potentially explaining the nature of the mechanism underlying the Ca2+-mediated activation of C2-PLDs.
In addition to Ca2+, PLDs have been reported to be stimulated by phosphoinositide through direct binding. In mammalian and yeast PLD enzymes, a “RX3R” motif was proposed to be involved in PIP2 binding.46 While in plant PLDβ/γ, in addition to the phospholipid-binding C2 domain,45 a motif featured of “KX2K” (PI(4,5)P2 binding region 1, PBR1) was proposed to be involved in PIP2 binding.47 However, the “RX3R” and the “KX2K” motifs are replaced with “WDVMY” (residues 418–422) and “KQGG” (residues 430–433) in PLDα1, respectively. Seeing from the structure, these two motifs are both located on the α4 helix away from the PA-binding pocket (Supplementary information, Fig. S11), and no extra electron density was observed around this region. This infers that phosphoinositide might activate PLDs allosterically.
Function of C2 domain
Although the in vitro activity of PLDα1 seems to be independent of the Ca2+ binding to the C2 domain, it does require the C2 domain for catalysis. In our experiment, a PLDα1 mutant lacking the N-terminal C2 domain (PLDα1-ΔC2) completely lost the in vitro enzyme activity (Fig. 5a). This is in sharp contrast to the mammalian PLD1, wherein a mutant variant lacking its N-terminal PX/PH domain retained almost full catalytic activity.21
Fig. 5.
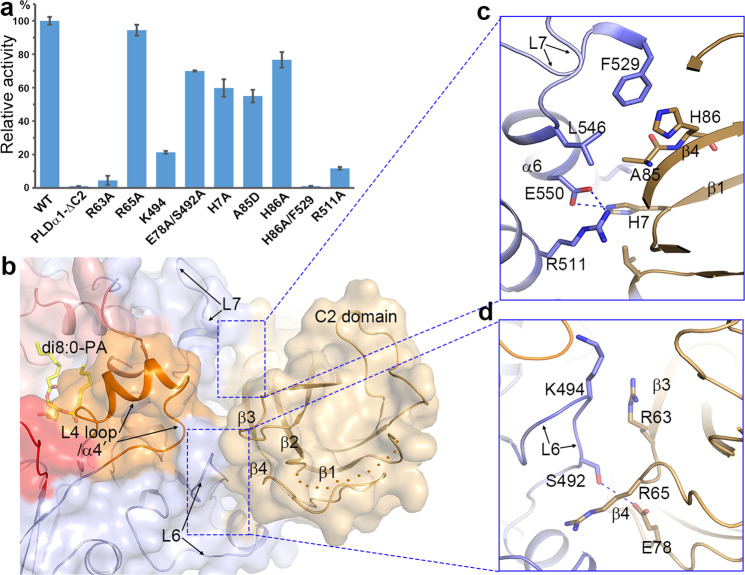
Interaction between the C2 domain and the catalytic domain and its influence on PLDα1 activity. a Influence of C2 domain deletion mutation (PLD-ΔC2) and interaction residue mutations on PLDα1 activity. Results are means ± SD, n = 3. b Surface view showing the interaction between C2 domain and catalytic domain. Color codes are the same as Fig. 3b. c, d Zoom-in views of the interactions. Residues involved in the interaction are shown with sticks
Seeing from the structure, the C2 domain binds to the HKD2 subdomain mainly through hydrophobic interactions (Fig. 5b and Supplementary information, Fig. S12). The interaction surface is constituted mainly of the L6/L7 loops and the α6 of the HKD2 subdomain, and β1/β3/β4 of the C2 domain, which buries 1770 Å2 of surface area (Fig. 5c, d). It is worth noting that many of the residues in the interaction surface, such as His7, Arg63, Glu78, Ala85, His86, Ser492, Lys494, Arg511, Phe529, Leu546, and Glu550, are conserved among C2-PLDs (Supplementary information, Fig. S6). Mutation of some of these residues, i.e., R63A, K494A, R511A and H86A/F529A, impaired the enzyme activity, while mutation of others such as H7A, A85D and E78A/S492A significantly reduced the activity. This suggests that the binding of C2 domain to the catalytic domain is essential for the activity of PLDα1. Since the L6 and L7 loops are in close proximity to the C-terminus and L4 loop/α’ helix that forms the substrate-binding pocket, we speculate that C2 domain may be required to regulate their conformations for substrate entry, binding and/or product release, which awaits further verification. Another important function of C2 domain could be targeting PLD to the membrane to access the lipid in vivo, which has been studied previously in PLDs and other C2 domain-containing proteins like cPLA2.37,38,47,48 Accordingly, positively charged surface patches are observed on the C2 domain and in between the C2 and catalytic domains, and these patches may be used by PLDα1 for lipid binding or membrane interaction (Supplementary information, Fig. S5). There are a number of studies suggesting that PLDα1 may undergo N-terminal cleavage for maturation,19,48–50 and a cleavage site has been proposed to exist in between residues 20 to 40. This region in the PLDα1 structure corresponds to β1 and to the disordered loop connecting β1 and β2 (Fig. 5b). Removal of this N-terminal fragment would destroy the C2 domain structure and therefore is unlikely to represent an in vivo maturation step. This also well explains why most N-terminal truncations of PLDα1 lost activity in vitro.19
In summary, a simplified functional model of C2-PLD was proposed based on our structure and literature (Supplementary information, Fig. S13). In the inactive state, the lid of the substrate-binding pocket of C2-PLD adopts a closed conformation to seal the pocket; when C2-PLD attaches to the membrane and the Ca2+ binds, the lid opens up to allow the membrane lipids to enter into the pocket; then the hydrolysis takes place and the products are released from the pocket. The C2 domain might be involved in the membrane attachment, substrate entry and/or product release of C2-PLDs.
Possible mechanism of small molecule inhibition
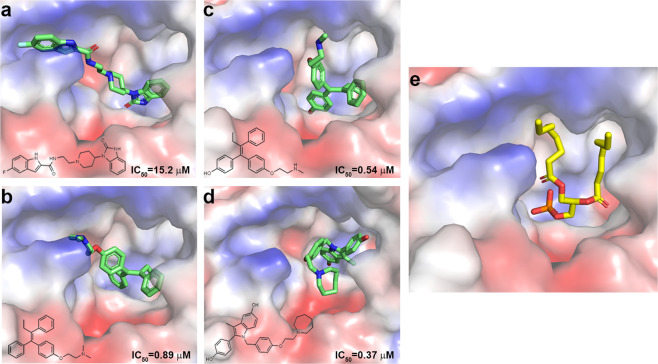
Given our finding that several small molecules efficiently inhibited PLDα1 (Fig. 1d), we next attempted to solve the structure of a PLDα1-inhibitor complex; however, these efforts have not been successful to date. We therefore used AutoDock to model inhibitors to the PLDα1-PA structure.51 Docking results indicated that all of the four tested inhibitors, FIPI, Tamoxifen, Endoxifen and Bazedoxifene, can be modeled into the substrate-binding pocket of PLDα1 (Fig. 6a–d). Furthermore, the binding positions of these four compounds overlap with the PA-binding site (Fig. 6e), inferring that the inhibitory effects of these compounds may result from occupation of the substrate-binding pocket, potentially preventing entry of the substrate. This is consistent with the competitive inhibition mode tested using Bazedoxifene (Supplementary information, Fig. S1).
Fig. 6.
Docking of small-molecule inhibitors. a–d Docking results show the binding of FIPI (a), Tamoxifen (b), Endoxifen (c), and Bazedoxifene (d) in the substrate-binding pocket of PLDα1 (electron static surface). Small-molecule inhibitors are shown with green sticks. The corresponding chemical structures and IC50 are shown. e Electron static surface view of the substrate-binding pocket of PLDα1-PA complex structure. di8:0-PA is shown with a yellow stick model
Comparison of the structures of these small molecules suggests that the extra hydroxyl group(s) of Endoxifen and Bazedoxifene, faces toward the bottom of the pocket. This may help explain our observations that they are apparently more efficient inhibitors of PLDα1 than the other examined compounds. Interestingly, both Endoxifen and Bazedoxifene are derivatives of selective oestrogen receptor modulators (SERM) that have been used in the treatment of breast cancers;52,53 these compounds are considered broad-spectrum PLD inhibitors with mild inhibitory activity.7 Zoom-in views of these modeled structure-inhibitor complexes indicate extra room within the pocket to be filled (Fig. 6a–d), thus providing structural insight into future strategies for the design and optimization of more efficient or specific PLD inhibitors.
Perspective
Eukaryotes encode three major classes of phospholipases, Phospholipase A, Phospholipase C, and Phospholipase D. Each class of phospholipase catalyzes a specific reaction to produce bioactive lipid molecules that regulate multiple cellular processes. These phospholipases are thus recognized as promising targets for drug design. Structure-based mechanistic study and inhibitor/drug design have been overwhelming in Phospholipase A and C, but have been greatly delayed for Phospholipase D due to the lack of three-dimensional structures. Our study thus provides a structural basis for further mechanistic understanding of PLD function, and opens the door for design of PLD-targeted inhibitors and drugs.
Materials and methods
Protein expression and purification
The gene encoding PLDα1 was amplified by PCR from the genomic DNA of Arabidopsis, and inserted into the pET-Duet vector with a 6× His tag at the N terminus. The constructs for point mutations were generated by one-step PCR or overlap PCR and verified by DNA sequencing. All the plasmids were transformed into the E. coli BL21 (DE3) strain. The transformed bacterial cells were grown in LB medium supplemented with ampicillin at 37 °C and induced by 0.25 mM isopropyl β-D-thiogalactopyranoside (IPTG) for 12 h at 16 °C. The cells were harvested and resuspended in buffer A (20 mM Tris-HCl, pH 8.0, 100 mM NaCl) supplemented with 1 mM PMSF. Cells were lysed by a high-pressure cell disruptor at 15,000 p.s.i. (pounds per square inch), and the lysate was centrifuged at 20,000 × g for 45 min. The supernatant was loaded onto a Ni2+-NTA affinity column (Qiagen) and washed with buffer A plus 20 mM imidazole. Proteins were eluted by buffer A plus 250 mM imidazole and purified using Source Q, followed by gel filtration using a Superdex 200 column (GE Healthcare) in buffer A. Peak fractions were collected and concentrated for subsequent structural and biochemical studies. For selenomethionine (SeMet)-derived protein expression, a previous protocol was adopted.54 In general, the constructs were transformed into E. coli B834 (DE3) cells, and the cells were cultured in M9 medium containing 50 mg/L SeMet.
Crystallization, data collection, and structure determination
To obtain the crystals of PLDα1 in complex with substrates, protein was incubated with 10-fold molar amounts of di8:0-PA on ice for 30 min before crystallization. Crystals for PLDα1 and PLDα1-PA complex were grown at 20 °C using the sitting-drop vapor-diffusion method by mixing 0.4 μL of protein with 0.4 μL of reservoir solution containing 20% (w/v) PEG 3000, 0.1 M Tris-HCl (pH 7.0), 0.2 M Ca(OAc)2. Crystals for data collection were directly flash-frozen in a nitrogen stream at 100 K. The data for the SeMet-derived PLDα1 and PLDα1 in apo form and di8:0-PA binding form were collected at Shanghai Synchrotron Radiation Facility (SSRF) beamline BL19U1/17U1 and were processed using the HKL3000 package.55 Structures of the PLDα1 was solved by the SAD method. The selenium sites were determined and initial phases were calculated using the HKL3000 package.55 Structures of PLDα1-PA complex was solved by molecular replacement using PHENIX56 with the apo form structure as the initial model. All the models were refined with PHENIX and manually built with Coot.57 The data collection and refinement statistics are summarized in Supplementary information, Table S1.
In vitro enzymatic assay
The commercial Amplex Red Phospholipase D Assay Kit (Invitrogen, A12219) was used to analyze the in vitro PLDα1 enzyme activity. Assay was performed in a volume of 200 μL containing 0.2 μg of protein, 125 μM di8:0PA and the supplied reagent of the kit. The reaction mixture was incubated at 37 °C for 10 min, and the fluorescence was measured (excitation at 530 nm and detection at 590 nm). For the assay of PLDα1 activation by Ca2+, the reaction buffer of the commercial kit was substituted with 50 mM Tris-HCl (pH 8.0). For the inhibition test and IC50 measurement, different concentrations of inhibitors of FIPI, Tamoxifen, Endoxifen, and Bazedoxifene (Sigma & Targetmol) were added to the assay system (0.03 μg protein was used alternatively), and the activity was measured using the same protocol. Data were analyzed with GraphPad Prism 5 software.
Data availability
The atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 6KZ8 and 6KZ9.
Supplementary information
Acknowledgements
We thank the staff members at BL19U1/BL17U1-SSRF for technical assistance with diffraction data collection. We also thank the staff members at the core facility center of Institute of Plant Physiology and Ecology for X-ray diffraction analysis. This work was supported by the National Key R&D Program of China (2018YFA0900600), the National Natural Science Foundation of China (31971120 and 31870720), the Chinese Academy of Sciences (XDB27020103 and QYZDB-SSW-SMC006), and Shanghai Science and Technology Commission (19XD1424500). J.L. is supported by the Foundation of Youth Innovation Promotion Association of the Chinese Academy of Sciences.
Author contributions
P.Z. conceived the project. J.L. carried out protein expression, purification and crystallization. J.L., F.Y. and P.Z. solved the crystal structures and designed mutations. F.Y. and J.L. carried out the enzymatic assay and data analysis. H.G., R.X. and W.Z. contributed to protein expression, purification and crystallization. M.Z. and F.H. contributed to diffraction data collection and analysis. P.Z. wrote the manuscript with inputs from all other authors.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Jianxu Li, Fang Yu
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-019-0244-6.
References
- 1.Wang X, Xu L, Zheng L. Cloning and expression of phosphatidylcholine-hydrolyzing phospholipase D from Ricinus communis L. J. Biol. Chem. 1994;269:20312–20317. doi: 10.1016/S0021-9258(17)31993-2. [DOI] [PubMed] [Google Scholar]
- 2.Hammond SM, et al. Human ADP-ribosylation factor-activated phosphatidylcholine-specific phospholipase D defines a new and highly conserved gene family. J. Biol. Chem. 1995;270:29640–29643. doi: 10.1074/jbc.270.50.29640. [DOI] [PubMed] [Google Scholar]
- 3.Rose K, Rudge SA, Frohman MA, Morris AJ, Engebrecht J. Phospholipase D signaling is essential for meiosis. Proc. Natl. Acad. Sci. USA. 1995;92:12151–12155. doi: 10.1073/pnas.92.26.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mansfeld J, Ulbrich-Hofmann R. Modulation of phospholipase D activity in vitro. Biochim. Biophys. Acta. 2009;1791:913–926. doi: 10.1016/j.bbalip.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Selvy PE, Lavieri RR, Lindsley CW, Brown HA. Phospholipase D: enzymology, functionality, and chemical modulation. Chem. Rev. 2011;111:6064–6119. doi: 10.1021/cr200296t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolesnikov YS, et al. Molecular structure of phospholipase D and regulatory mechanisms of its activity in plant and animal cells. Biochemistry. 2012;77:1–14. doi: 10.1134/S0006297912010014. [DOI] [PubMed] [Google Scholar]
- 7.Brown HA, Thomas PG, Lindsley CW. Targeting phospholipase D in cancer, infection and neurodegenerative disorders. Nat. Rev. Drug Disco. 2017;16:351–367. doi: 10.1038/nrd.2016.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao C, Du G, Skowronek K, Frohman MA, Bar-Sagi D. Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nat. Cell Biol. 2007;9:706–712. doi: 10.1038/ncb1594. [DOI] [PubMed] [Google Scholar]
- 9.Mishra G, Zhang W, Deng F, Zhao J, X. W. A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science. 2006;312:264–266. doi: 10.1126/science.1123769. [DOI] [PubMed] [Google Scholar]
- 10.Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- 11.Hong Y, et al. Plant phospholipases D and C and their diverse functions in stress responses. Prog. Lipid Res. 2016;62:55–74. doi: 10.1016/j.plipres.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Bruntz RC, Lindsley CW, Brown HA. Phospholipase D signaling pathways and phosphatidic acid as therapeutic targets in cancer. Pharm. Rev. 2014;66:1033–1079. doi: 10.1124/pr.114.009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jang JH, Lee CS, Hwang D, Ryu SH. Understanding of the roles of phospholipase D and phosphatidic acid through their binding partners. Prog. Lipid Res. 2012;51:71–81. doi: 10.1016/j.plipres.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Devaiah SP, Zhang W, Welti R. Signaling functions of phosphatidic acid. Prog. Lipid Res. 2006;45:250–278. doi: 10.1016/j.plipres.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Hammond SM, et al. Characterization of two alternately spliced forms of phospholipase D1. Activation of the purified enzymes by phosphatidylinositol 4,5-bisphosphate, ADP-ribosylation factor, and Rho family monomeric GTP-binding proteins and protein kinase C-alpha. J. Biol. Chem. 1997;272:3860–3868. doi: 10.1074/jbc.272.6.3860. [DOI] [PubMed] [Google Scholar]
- 16.Li G, Lin F, Xue HW. Genome-wide analysis of the phospholipase D family in Oryza sativa and functional characterization of PLD beta 1 in seed germination. Cell Res. 2007;17:881–894. doi: 10.1038/cr.2007.77. [DOI] [PubMed] [Google Scholar]
- 17.Qin C, Wang X. The Arabidopsis phospholipase D family. Characterization of a calcium-independent and phosphatidylcholine-selective PLDζ1 with distinct regulatory domains. Plant Physiol. 2002;128:1057–1068. doi: 10.1104/pp.010928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 19.Arhab Y, Abousalham A, Noiriel A. Plant phospholipase D mining unravels new conserved residues important for catalytic activity. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2019;1864:688–703. doi: 10.1016/j.bbalip.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Brown HA, Henage LG, Preininger AM, Xiang Y, Exton JH. Biochemical analysis of phospholipase D. Methods Enzymol. 2007;434:49–87. doi: 10.1016/S0076-6879(07)34004-4. [DOI] [PubMed] [Google Scholar]
- 21.Henage LG, Exton JH, Brown HA. Kinetic analysis of a mammalian phospholipase D: allosteric modulation by monomeric GTPases, protein kinase C, and polyphosphoinositides. J. Biol. Chem. 2006;281:3408–3417. doi: 10.1074/jbc.M508800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pappan K, Qin W, Dyer JH, Zheng L, Wang X. Molecular cloning and functional analysis of polyphosphoinositide-dependent phospholipase D, PLDbeta, from. Arabidopsis. J. Biol. Chem. 1997;272:7055–7061. doi: 10.1074/jbc.272.11.7055. [DOI] [PubMed] [Google Scholar]
- 23.Dressler L, et al. Metal ions and phosphatidylinositol 4,5-bisphosphate as interacting effectors of alpha-type plant phospholipase D. Phytochemistry. 2017;138:57–64. doi: 10.1016/j.phytochem.2017.02.024. [DOI] [PubMed] [Google Scholar]
- 24.Zhao J, Wang X. Arabidopsis phospholipase Dalpha1 interacts with the heterotrimeric G-protein alpha-subunit through a motif analogous to the DRY motif in G-protein-coupled receptors. J. Biol. Chem. 2004;279:1794–1800. doi: 10.1074/jbc.M309529200. [DOI] [PubMed] [Google Scholar]
- 25.Park JB, et al. Phospholipase signalling networks in cancer. Nat. Rev. Cancer. 2012;12:782–792. doi: 10.1038/nrc3379. [DOI] [PubMed] [Google Scholar]
- 26.Scott SA, et al. Discovery of desketoraloxifene analogues as inhibitors of mammalian, Pseudomonas aeruginosa, and NAPE phospholipase D enzymes. ACS Chem. Biol. 2015;10:421–432. doi: 10.1021/cb500828m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavieri RR, et al. Design, synthesis, and biological evaluation of halogenated N-(2-(4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-8-yl)ethyl)benzamides: discovery of an isoform-selective small molecule phospholipase D2 inhibitor. J. Med. Chem. 2010;53:6706–6719. doi: 10.1021/jm100814g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su W, et al. 5-Fluoro-2-indolyl des-chlorohalopemide (FIPI), a phospholipase D pharmacological inhibitor that alters cell spreading and inhibits chemotaxis. Mol. Pharm. 2009;75:437–446. doi: 10.1124/mol.108.053298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott SA, et al. Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat. Chem. Biol. 2009;5:108–117. doi: 10.1038/nchembio.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monovich L, et al. Optimization of halopemide for phospholipase D2 inhibition. Bioorg. Med. Chem. Lett. 2007;17:2310–2311. doi: 10.1016/j.bmcl.2007.01.059. [DOI] [PubMed] [Google Scholar]
- 31.Taylor HE, et al. Phospholipase D1 couples CD4+ T cell activation to c-Myc-dependent deoxyribonucleotide pool expansion and HIV-1 replication. PLoS Pathog. 2015;11:e1004864. doi: 10.1371/journal.ppat.1004864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Reilly MC, et al. Discovery of a highly selective PLD2 inhibitor (ML395): a new probe with improved physiochemical properties and broad-spectrum antiviral activity against influenza strains. ChemMedChem. 2014;9:2633–2637. doi: 10.1002/cmdc.201402333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oguin TH, et al. Phospholipase D facilitates efficient entry of influenza virus, allowing escape from innate immune inhibition. J. Biol. Chem. 2014;289:25405–25417. doi: 10.1074/jbc.M114.558817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheol Son J, et al. Phospholipase D inhibitor enhances radiosensitivity of breast cancer cells. Exp. Mol. Med. 2013;45:e38. doi: 10.1038/emm.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leiros I, Secundo F, Zambonelli C, Servi S, Hough E. The first crystal structure of a phospholipase D. Structure. 2000;8:655–667. doi: 10.1016/S0969-2126(00)00150-7. [DOI] [PubMed] [Google Scholar]
- 36.Sutton RB, Davletov BA, Berghuis AM, Südhof TC, Sprang SR. Structure of the first C2 domain of synaptotagmin I: a novel Ca2+/phospholipid-binding fold. Cell. 1995;80:929–938. doi: 10.1016/0092-8674(95)90296-1. [DOI] [PubMed] [Google Scholar]
- 37.Perisic O, Fong S, Lynch DE, Bycroft M, Williams RL. Crystal structure of a calcium-phospholipid binding domain from cytosolic phospholipase A2. J. Biol. Chem. 1998;273:1596–1604. doi: 10.1074/jbc.273.3.1596. [DOI] [PubMed] [Google Scholar]
- 38.Hirano Y, et al. Structural basis of phosphatidylcholine recognition by the C2-domain of cytosolic phospholipase A2α. Elife. 2019;8:e44760. doi: 10.7554/eLife.44760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dessen A, et al. Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism. Cell. 1999;97:349–360. doi: 10.1016/S0092-8674(00)80744-8. [DOI] [PubMed] [Google Scholar]
- 40.Xie Z, Ho WT, Exton JH. Conserved amino acids at the C-terminus of rat phospholipase D1 are essential for enzymatic activity. Eur. J. Biochem. 2000;267:7138–7146. doi: 10.1046/j.1432-1327.2000.01816.x. [DOI] [PubMed] [Google Scholar]
- 41.Liu MY, Gutowski S, Sternweis PC. The C terminus of mammalian phospholipase D is required for catalytic activity. J. Biol. Chem. 2001;276:5556–5562. doi: 10.1074/jbc.M006404200. [DOI] [PubMed] [Google Scholar]
- 42.Lerchner A, et al. Two highly homologous phospholipase D isoenzymes from Papaver somniferum L. with different transphosphatidylation potential. Biochim. Biophys. Acta. 2005;1737:94–101. doi: 10.1016/j.bbalip.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 43.Uesugi Y, Hatanaka T. Phospholipase D mechanism using Streptomyces PLD. Biochim. Biophys. Acta. 2009;1791:962–969. doi: 10.1016/j.bbalip.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 44.Leiros I, McSweeney S, Hough E. The reaction mechanism of phospholipase D from Streptomyces sp. strain PMF. Snapshots along the reaction pathway reveal a pentacoordinate reaction intermediate and an unexpected final product. J. Mol. Biol. 2004;339:805–820. doi: 10.1016/j.jmb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 45.Zheng L, Krishnamoorthi R, Zolkiewski M, Wang X. Distinct Ca2+ binding properties of novel C2 domains of plant phospholipase dalpha and beta. J. Biol. Chem. 2000;275:19700–19706. doi: 10.1074/jbc.M001945200. [DOI] [PubMed] [Google Scholar]
- 46.Sciorra VA, et al. Identification of a phosphoinositide binding motif that mediates activation of mammalian and yeast phospholipase D isoenzymes. EMBO J. 1999;18:5911–5921. doi: 10.1093/emboj/18.21.5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng L, Shan J, Krishnamoorthi R, Wang X. Activation of plant phospholipase Dbeta by phosphatidylinositol 4,5-bisphosphate: characterization of binding site and mode of action. Biochemistry. 2002;41:4546–4553. doi: 10.1021/bi0158775. [DOI] [PubMed] [Google Scholar]
- 48.Rahier R, Noiriel A, Abousalham A. Functional characterization of the N-terminal C2 domain from Arabidopsis thaliana phospholipase Dalpha and Dbeta. Biomed. Res. Int. 2016;2016:2721719. doi: 10.1155/2016/2721719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ben Ali Y, Carriere F, Abousalham A. High-level constitutive expression in Pichia pastoris and one-step purification of phospholipase D from cowpea (Vigna unguiculata L. Walp) Protein Expr. Purif. 2007;51:162–169. doi: 10.1016/j.pep.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 50.Ueki J, Morioka S, Komari T, Kumashiro T. Purification and characterization of phospholipase D (PLD) from rice (Oryza sativa L.) and cloning of cDNA for PLD from rice and maize (Zea mays L.) Plant Cell Physiol. 1995;36:903–914. doi: 10.1093/oxfordjournals.pcp.a078837. [DOI] [PubMed] [Google Scholar]
- 51.Morris GM, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cummings SR, et al. The effect of raloxifene on risk of breast cancer in postmenopausal women: results from the MORE randomized trial. Multiple outcomes of raloxifene evaluation. JAMA. 1999;281:2189–2197. doi: 10.1001/jama.281.23.2189. [DOI] [PubMed] [Google Scholar]
- 53.Plowman PN. Tamoxifen as adjuvant therapy in breast cancer. Current status. Drugs. 1993;46:819–833. doi: 10.2165/00003495-199346050-00003. [DOI] [PubMed] [Google Scholar]
- 54.Qi X, et al. Structural basis of rifampin inactivation by rifampin phosphotransferase. Proc. Natl. Acad. Sci. USA. 2016;113:3803–3808. doi: 10.1073/pnas.1523614113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D. Biol. Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 56.Adams PD, et al. Acta Crystallogr. D Biol. Crystallogr. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution; pp. 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 6KZ8 and 6KZ9.