Neuroendocrine carcinomas (NEC) of the gastrointestinal system show similar histopathologic features and share some genomic features, but considerable differences exist between pancreatic NECs and nonpancreatic NECs.
Abstract
The neuroendocrine carcinoma of the gastrointestinal system (GIS-NEC) is a rare but highly malignant neoplasm. We analyzed 115 cases using whole-genome/exome sequencing, transcriptome sequencing, DNA methylation assays, and/or ATAC-seq and found GIS-NECs to be genetically distinct from neuroendocrine tumors (GIS-NET) in the same location. Clear genomic differences were also evident between pancreatic NECs (Panc-NEC) and nonpancreatic GIS-NECs (Nonpanc-NEC). Panc-NECs could be classified into two subgroups (i.e., “ductal-type” and “acinar-type”) based on genomic features. Alterations in TP53 and RB1 proved common in GIS-NECs, and most Nonpanc-NECs with intact RB1 demonstrated mutually exclusive amplification of CCNE1 or MYC. Alterations of the Notch gene family were characteristic of Nonpanc-NECs. Transcription factors for neuroendocrine differentiation, especially the SOX2 gene, appeared overexpressed in most GIS-NECs due to hypermethylation of the promoter region. This first comprehensive study of genomic alterations in GIS-NECs uncovered several key biological processes underlying genesis of this very lethal form of cancer.
Significance:
GIS-NECs are genetically distinct from GIS-NETs. GIS-NECs arising in different organs show similar histopathologic features and share some genomic features, but considerable differences exist between Panc-NECs and Nonpanc-NECs. In addition, Panc-NECs could be classified into two subgroups (i.e., “ductal-type” and “acinar-type”) based on genomic and epigenomic features.
This article is highlighted in the In This Issue feature, p. 587
Introduction
Neuroendocrine neoplasms (NEN), characterized by neuroendocrine differentiation, can arise in most epithelial organs of the body. The digestive system is the most common location, accounting for two thirds of NENs, with the pancreas as a major primary site. These neoplasms include a number of distinct entities with widely differing etiologies, clinical features, and morphologic and genomic findings.
The World Health Organization (WHO) Classification of Digestive System Tumours was recently updated (1). The highlight of this updated classification is a new distinction between well-differentiated neuroendocrine tumors (NET; Supplementary Fig. S1A) and poorly differentiated neuroendocrine carcinomas (NEC; Supplementary Fig. S1B and S1C). NETs have histologically low-grade nuclear features, and are graded as G1, G2, or G3 on the basis of proliferation activity as assessed by the mitotic rate and the Ki-67 proliferation index. NECs have high-grade, carcinoma-like nuclear features and characteristically exhibit aggressive clinical behavior, frequent metastases, and poor survival (2). NECs may be pure or mixed with variable amounts of adenocarcinoma, squamous cell carcinoma, or other components (ref. 3; Supplementary Fig. S1D).
Somatic mutations in pancreatic NETs (Panc-NET) have been well characterized (4, 5). In contrast, little is known about molecular drivers of the neuroendocrine carcinoma of the gastrointestinal system (GIS-NEC), for which few specimens have been available for analysis. We previously reported abnormal immunolabeling of p53 and RB1 protein to be frequent in 19 pancreatic NECs (Panc-NEC; ref. 6). In the present study, taking advantage of international collaboration, we conducted a comprehensive genomic analysis of a relatively large series of cases, the results implicating several previously unknown biological processes in the pathogenesis of neuroendocrine neoplasms of the gastrointestinal system (GIS-NEN).
Results
Clinicopathologic Features of GIS-NENs
An international cohort of 114 clinically and pathologically well-characterized GIS-NEN cases (115 independent lesions), comprising 79 from Japan, 23 from the United States, and 12 from the Netherlands, underwent broad analyses. Included were 60 GIS-NECs (18 pancreatic, 14 gastric, 10 biliary, 9 colorectal, 6 ampullary, and 3 esophageal) and 55 GIS-NETs (48 pancreatic, 6 colorectal, and 1 nonampullary duodenal). Of note, two (cases NE067 and NE115) of nine patients with colorectal NEC suffered from inflammatory bowel disease (i.e., ulcerative colitis), one for 20 years and the other for 18 years (Supplementary Table S1).
Pathologic diagnosis was based on criteria of the 2019 WHO classification of digestive system tumors. A flow chart for the entire analysis is provided in Supplementary Fig. S2. Clinicopathologic features are detailed in Supplementary Tables S1–S3. The age of patients with Panc-NECs was significantly greater than the age of patients with Panc-NETs (Supplementary Table S3).
Kaplan–Meier analyses and log-rank tests were performed to determine relationships between clinicopathologic features and five-year disease-specific survival rates (Supplementary Fig. S3A–S3D). Patients with GIS-NECs had significantly worse outcomes than did patients with GIS-NETs (P = 4.32 × 10−15). In addition, patients with Panc-NECs had poorer five-year disease-specific survival than patients with nonpancreatic GIS-NECs (Nonpanc-NEC; P = 0.0382). There was also a significant difference in survival between patients with GIS-NETG3 and patients with GIS-NETG1/G2 (P = 0.00309).
Significantly Mutated Genes in GIS-NECs
We performed whole-genome sequencing (WGS), transcrip-tome sequencing [RNA sequencing (RNA-seq)], DNA methyla-tion analysis, and/or Assay for Transposase Accessible Chromatin sequencing (ATAC-seq) on 76 frozen samples including 4 organoids (Supplementary Tables S4 and S5). Whole-exome sequencing (WES) was also conducted on formalin-fixed and paraffin-embedded (FFPE) material from 56 cases (Supplementary Table S6). We further examined the integration of virus genomes previously reported to be associated with neuroendocrine carcinomas (Supplementary Table S7). To accurately evaluate the frequency, distribution, and clonality of somatic mutations, targeted gene sequencing was performed on 107 cases. This targeted sequencing provided deeper read coverage for 78 genes with recurrent mutations identified in WGS/WES, or for examples selected as being identified as driver genes in a previous report (ref. 5; Supplementary Tables S8–S10). An oncoplot based on targeted gene sequencing is shown in Supplementary Fig. S4. Significantly mutated genes (SMG) in GIS-NECs (Supplementary Table S11) and GIS-NETs (Supplementary Table S12) were investigated by WGS and/or WES and validated by targeted gene sequencing. The median numbers of nonsynonymous mutations in GIS-NECs and GIS-NETs were 60 and 10.5, respectively. The SMGs in GIS-NECs are TP53, KRAS, RB1, CCNE1, CDKN2A, and MYC. KRAS gene alterations were mainly detected in Panc-NECs, APC in colorectal NECs, and ELF3 in ampullary (7) and biliary tract NECs (8). Thus, driver mutations initially detected in conventional carcinomas in each body site were found to be to a large extent organ-specific. Recurrent mutations in noncoding DNA regions were not observed.
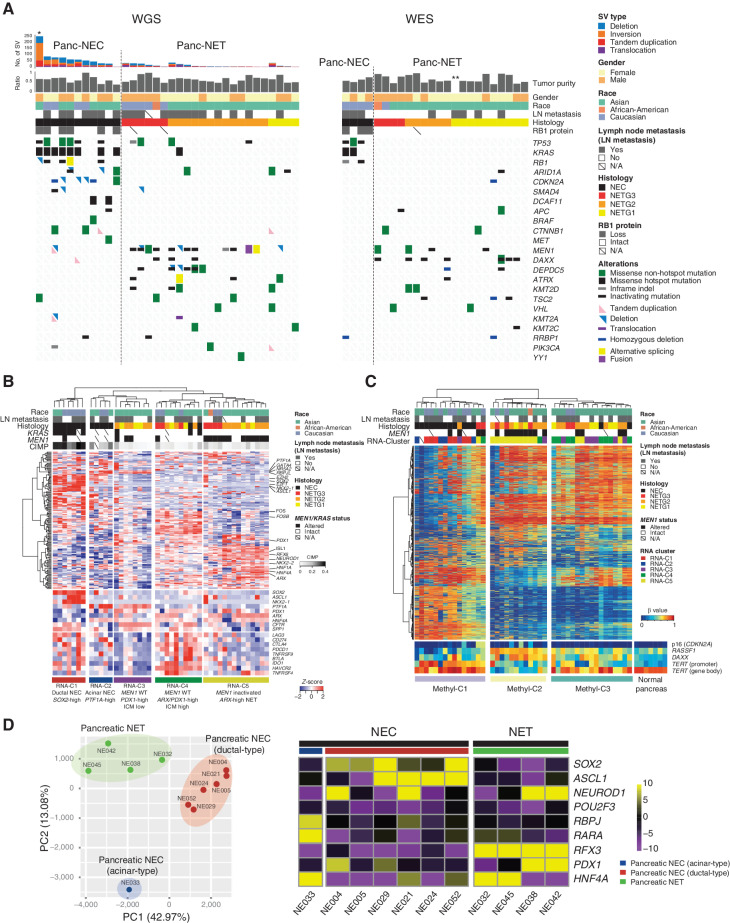
Structural Variation in GIS-NECs
Differences were noted at the genomic level between Panc-NECs and Nonpanc-NECs (Fig. 1), the number of structural variations (SV) being significantly larger in the latter (P = 3.82 × 10−9; Fig. 1; Supplementary Table S13). We identified loci frequently affected by SVs by counting breakpoints within 1 Mb windows genome-wide (Supplementary Fig. S5A and S5B). Loci characteristically affected contained the PTPRD gene in Panc-NECs as with pancreatic ductal adenocarcinoma (PDAC; ref. 9) and Panc-NET (5). SMAD4 and CDKN2A, which are frequently targeted for SV in PDAC, were not found to be targeted in Panc-NECs. In Nonpanc-NECs, recurrent SV-affected genes listed in the COSMIC Cancer Gene Census were CLTC, TCF, NOTCH1, CREBBP, and ZNFR3, in descending order of prevalence. We identified whole-genome duplication (WGD) in 17 (33.4%) of 51 GIS-NECs (Supplementary Table S14). To determine chromothripsis presence, we applied ShatterSeek (10) to 57 GIS-NENs (35 GIS-NECs and 22 GIS-NETs), with positive results in 14 of the 35 (40.0%) GIS-NECs (Supplementary Table S15). Of these 14, 12 (85.8%) harbored TP53 mutations statistically significantly (P = 0.0320) as compared with the prevalence of only 10 of 21 GIS-NECs without chromothripsis. This result is in agreement with previous findings for other types of neoplasm (11, 12). Recurrent chromothripsis involving chromosome 11 was detected in four GIS-NECs (Supplementary Fig. S5C). In contrast, GIS-NETs were not found to harbor regions of chromothripsis. Although chromothripsis has been associated with a poor prognosis, fusion genes created by chromosome shattering may have therapeutic potential for GIS-NECs with this feature (13).
Figure 1.
Genomic alterations of GIS-NECs. A, Landscape of genomic alterations in GIS-NEC cases. The left oncoplot indicates WGS data, and representative gene expression data are obtained from frozen samples. The cases are arranged from left to right according to descending order of the number of SVs in each primary organ. Asterisks, organoid samples. The right oncoplot shows WES data in patients differing from patients available on the WGS data. B, Hematoxylin and eosin (H&E) staining and synaptophysin immunolabeling of TP53 and RB1 double knockout (TR-KO) organoids before and after blocking Notch signaling with a γ-secretase inhibitor (DAPT). The synaptophysin-positive cells were increased by the Notch inhibitor. Scale bar, 100 μm. C, Unsupervised hierarchical cluster analysis with 2,000 high variant probes for DNA methylation in GIS-NECs. D, Integration of RNA-seq and DNA methylation array data comparing GIS-NECs with normal tissues. RNA-seq data were filtered using significant differentially expressed gene (DEG; abs [log2FC] ≥ 1) with significant FDR values (<0.05). DNA methylation assay data were filtered using differentially methylated regions (DMR; abs [Δβ value] ≥ 0.1) with significant adjusted P values (<0.05). In the area of the figure showing high levels of gene expression and hypermethylation, 199 DMRs are situated, of which 39 (19.6%) are transcription factors (TF; red dots) including SOX2 and ASCL1. In contrast, in the area of the figure showing high levels of gene expression and hypomethylation, 424 DMRs are situated, of which 28 (6.6%) are TFs. CHGA, chromogranin A. E, Schematic of NET–AKR fusion genes detected in two gastric NECs. The neuroepithelioma transforming gene1 (NET1) is a specific guanine nucleotide exchange factor for RhoA. Both aldo-keto reductase family 1 members C3 (AKR1C3) and C4 (AKR1C4) are reductase enzymes that play critical roles in the biotransformation of endogenous substrates such as steroids. The chimeric genes demonstrate in-frame fusion of the NET1 amino terminus (exons 1–3) and the AKR1C3 carboxyl terminus (exons 2–9) or the AKR1C4 carboxyl terminus (exons 6–9). NLS, nuclear localization signal; DH, Dbl homology; PH, pleckstrin homology; PDZ, post-synaptic density 95; aa, amino acids. F, Gastric NEC with the Merkel cell polyomavirus (MCPyV; case NE002). The read depth along the polyomavirus genome is shown in blue, and read pairs bridging the polyomavirus genome and the integration site on chromosome 8 are indicated by red lines. Polyomavirus genes are indicated by large T antigen, small T Antigen, VP1, VP2, and VP3.
Genomic Distinction between Panc-NECs and Nonpanc-NECs
The number of nonsynonymous mutations was significantly larger in Nonpanc-NECs than in Panc-NECs (P = 0.00238). Loss of RB1 protein and TP53 mutations proved prevalent in both. In Nonpanc-NECs with intact RB1, CCNE1 and MYC amplification was a mutually exclusive event. In addition, alterations of Notch family genes were frequently detected in Nonpanc-NECs (12/23, 52.1%; Fig. 1A; Supplementary Fig. S5A), in contrast to their relative paucity in conventional gastrointestinal adenocarcinomas (14). Notch family genes are known to act as tumor suppressors and master regulators of neuroendocrine differentiation in pulmonary NEC, i.e., small-cell lung cancer (SCLC; ref. 15). In contrast to Nonpanc-NECs, Panc-NECs lacked any SV of Notch family genes except for one organoid, which demonstrated an unstable SV phenotype (>200 SVs; Fig. 1A). No significant differences in lymph node metastasis, tumor purity, age, and race were evident between Panc-NECs and Nonpanc-NECs.
To determine the effect of dysregulation of Notch signaling in TP53- and RB1-deficient cells, we tested whether blockage with a γ-secretase inhibitor would alter the expression of synaptophysin in TP53 and RB1 double knockout (TR-KO) organoids, generated from normal colon epithelium using CRISPR/Cas9 (16). These TR-KO cells had increased synaptophysin levels after administration of the γ-secretase inhibitor (Fig. 1B and Supplementary Text). In TP53 KO gastric organoids, the same phenomenon was also observed (Supplementary Fig. S6).
Genomic Distinction between Small-Cell Type GIS-NECs, Large-Cell Type GIS-NECs, and Mixed Neuroendocrine–Nonneuroendocrine Neoplasms
Genetic differences based on morphologic subclassification of GIS-NECs were investigated. Regarding frequently altered genes, targeted deep sequence data, derived from both frozen and FFPE samples, showed RB1 gene mutations to be significantly more prevalent in small-cell than in large-cell type GIS-NECs (P = 0.00513) and mixed neuroendocrine–nonneuroendocrine neoplasms (MiNEN; P = 0.0239; Supplementary Fig. S7A). However, WGS data, derived solely from frozen samples, demonstrated SVs in the RB1 gene in large-cell type GIS-NECs (Supplementary Fig. S7B). These findings indicate that inactivation mechanisms of RB1 may be different between small-cell and large-cell GIS-NECs.
DNA Methylation Status and Transcriptome Profiling of GIS-NECs
Unsupervised hierarchical cluster analysis of methylation status divided GIS-NECs into three groups (Fig. 1C). The third group (Methyl-C3) consisted mainly of Panc-NECs and the second group (Methyl-C2) mostly of Nonpanc-NECs. The remaining cluster (Methyl-C1) featured a CpG island methylator phenotype (CIMP) status due to alterations of DNA methylation–related genes and/or MLH1 promoter hypermethylation. The microsatellite instability (MSI) scores using MSIsensor-pro (17) were significantly higher (P = 0.00794) in Methyl-C1 (median, 0.91) than in Methyl-C2 and Methyl-C3 (median, 0.09) cases.
Transcriptome analysis confirmed overexpression of transcription factors for neuroendocrine differentiation (NE-TF), especially SOX2, in GIS-NECs (Fig. 1A; Supplementary Fig. S8A–S8C). Interestingly, overexpression of SOX2 and other NE-TFs (e.g., ASCL1) was regulated by hypermethylation of the promoter regions of these genes, rather than by hypomethylation of the promoter (Fig. 1D; Supplementary Fig. S9A). Gene expression of SOX2 was strongly correlated (ρ = 0.620, P = 2.40 × 10−9) with DNA methylation status of that gene's promoter (Supplementary Fig. S9B and S9C). Furthermore, in GIS-NECs with increased expression of SOX2, ATAC-seq demonstrated an open chromatin status in the region surrounding the SOX2 gene (Supplementary Fig. S9D).
NET1–AKR1C3/4 Fusion
A recurrent novel fusion gene, NET1–AKR1C3/4, was found in two gastric NECs [2/60 (3.3%) GIS-NECs; Fig. 1E]. The neuroepithelioma transforming gene 1 (NET1) is a specific guanine nucleotide exchange factor (GEF) for RhoA. Aldo-keto reductase family 1 members C3 (AKR1C3) and C4 (AKR1C4) are both reductase enzymes that play critical roles in biotransformation of substrates such as steroids. The chimeric genes featured in-frame fusion of the NET1 amino terminus (exons 1–3) and the AKR1C3 carboxyl terminus (exons 2–9) or the AKR1C4 carboxyl terminus (exons 8–11; Fig. 1E; Supplementary Fig. S10A). The same exons of the NET1 gene were fused with AKR1C, which harbors nuclear localization signals without enzymatic activity of Rho GEFs. To assess the function of this fusion, an immortalized normal epithelial cell line of gastric fundus origin (HFundEC4N) expressing NET1–AKR fusion proteins was established, and RNA-seq of NET1–AKR1C3 and NET1–AKR1C4 fusion gene–expressing cells was performed (see Supplementary Text and Supplementary Fig. S10B–S10D). Analysis of upregulated genes showed that pathways related to cell-cycle regulation were top ranked in both NET1–AKR1C3 and NET1–AKR1C4– expressing cells, but neuroendocrine markers [e.g., SYP (synaptophysin), CHGA (chromogranin A), and NCAM1] were not increased (Supplementary Fig. S10B). Recently, the Pan-Cancer Analysis of Whole Genomes (PCAWG) Consortium of the International Cancer Genome Consortium (ICGC) and The Cancer Genome Atlas (TCGA) identified AKR1C genes as novel structural-variant driver candidates (18).
Nonpanc-NECs Caused by Virus Infection
Case NE002, a gastric NEC with Merkel cell polyomavirus (MCPyV), appeared to be caused by monoclonal integration of MCPyV (Supplementary Text and Supplementary Fig. S11A–S11H). Merkel cell carcinoma is a highly aggressive neuroendocrine carcinoma of the skin whose main etiologic agent is MCPyV, detected in 80% of cases (19). A complete and accurate examination of the skin of the patient revealed no suspicious lesions. The MCPyV large T antigen directly binds to and inactivates RB1 (20), and MCPyV integration was found in an intron of the CNGB3 gene on chromosome 8q21 (Fig. 1F). The patient had regularly taken steroids for polymyalgia rheumatica, suggesting a state of chronic drug-induced immune suppression.
Recently, a significant subset of rectal and anal NEC was concluded to be driven by high-risk human papillomavirus infection, without genomic alteration in RB1, TP53, or CCNE1 (21). In the present study, case NE066 with a rectal NEC showing intact RB1 and p53 and no amplification of CCNE1/MYC proved positive for high-risk human papillomavirus (HPV-18). The patient had regularly taken Famciclovir for herpes simplex virus infection, which suggests an immunocompromised state.
Nonpanc-NECs Expressing Tuft Cell Lineage Markers
Three GIS-NECs (colon, NE030; esophagus, NE055; stomach, NE023) exhibited high expression of POU2F3 (Fig. 1A; Supplementary Fig. S8A), recently recognized as one of four key transcriptional regulators (i.e., ASCL1, NEUROD1, POU2F3, and YAP1) distinguishing biological subtypes of SCLC and a marker of the tuft cell lineage (22). Tuft cells have been identified in not only the lung (23) but also the gastrointestinal tract (24), suggesting that they might act as a cell of origin for a proportion of Nonpanc-NECs.
LINE1-MET Chimeric Transcript
In most GIS-NECs (74.3%, 26/35) but not present in GIS-NETs (0%, 0/30), LINE1-MET chimeric transcripts could be identified, presumably due to demethylation of the long interspersed nuclear element (LINE1) antisense promoter (ref. 25; Supplementary Fig. S12A–S12C). LINE1-MET chimeric transcripts have been reported in some cancers, especially aggressive tumor subtypes (26).
“Ductal-type Panc-NEC” and “Acinar-type Panc-NEC”
A focus on genomic distinction between Panc-NECs and Panc-NETs (Fig. 2) revealed significantly higher SV numbers in the former (P = 3.82 × 10−9; Fig. 2A). Panc-NECs were classified into two groups based on multiomics data (Fig. 2A and B), the first group featuring high expression of pancreatic ductal lineage markers (e.g., SPP1 and CFTR) and harboring KRAS mutations. In this “ductal-type Panc-NEC,” transcription factors, such as SOX2, ASCL1, NKX2-1, EZH2, and E2F1, were characteristically overexpressed (Fig. 2B; Supplementary Fig. S8A). Unlike PDAC (27), loss of p16 and Smad4 protein immunolabeling was rare (Supplementary Table S16), indicating a pathway to progression different from PDAC development. Most “ductal-type Panc-NEC” showed RB1 protein loss and TP53 mutations, and generally CIMP (Fig. 2B). In the second Panc-NEC group and one gastric NEC (NE020) lacking KRAS mutation, the PTF1A gene, a marker of acinar cell differentiation (28), was mostly overexpressed. However, IHC staining for acinar cell markers (trypsin, chymotrypsin, and BCL10) was negative except for the case of one mixed acinar–neuroendocrine carcinoma (NE051), in which trypsin, chymotrypsin, and BCL10 were found to be partially positive (Supplementary Table S16). The “acinar-type Panc-NEC” featured altered WNT sig-naling (e.g., mutations of APC and CTNNB1 genes) and characteristically alterations of CDKN2A (i.e., homozygous deletion, large deletion, or promoter hypermethylation; Fig. 2A) and overexpression of transcription factors, such as PTF1A, GATA4, NR5A2, and RBPJL (Fig. 2B). Of note, hotspot mutations of DCAF11 [c.714G>T: NE016, tumor variant allele frequency (TVAF) = 0.511; NE051, TVAF = 0.400] and hemizygous loss of chromosomes 3 and 4 were observed in all cases in this group (Supplementary Tables S4 and S14). It was found that Panc-NEC patients whose tumors were KRAS wild-type had a significantly (P = 0.00279) better prognosis than their counterparts with tumors that harbored KRAS mutations (Supplementary Fig. S3D).
Figure 2.
Genomic alterations of Panc-NECs and Panc-NETs. A, Landscape of genomic alterations in Panc-NECs and Panc-NETs. The left oncoplot indicates WGS data obtained from frozen samples. The cases are arranged from left to right according to descending order in the number of SV in each pancreatic neuroendocrine neoplasm based on WHO classification 2019. The right oncoplot shows WES data differing from patients available on the WGS data. Asterisk, organoid sample; double asterisk, a case for which tumor purity was not calculated due to the limited number of mutations. B, Unsupervised clustering analysis using gene expression of high variant 160 TFs. The expression of representative TFs specific to each cluster and immune checkpoint molecules is shown at the bottom. C, Unsupervised clustering of the methylation levels with 2,000 high variant CpG probes in Panc-NECs and Panc-NETs. D, Principal component analysis, based on reads of distal elements by ATAC-seq, could clearly discriminate between Panc-NETs (green), acinar-type Panc-NECs (blue, NE033), and ductal-type Panc-NECs (red). Motif enrichment analysis could also clearly discriminate between Panc-NETs (subgroups: PDX1-high and HNF4A-high), acinar-type Panc-NECs, and ductal-type Panc-NECs.
Deleterious germline variants in the PALB2 gene have been reported in 3% to 4% of familial PDAC cases (29). Here we identified a deleterious germline mutation (frameshift deletion) of PALB2 in one patient (NE103) with Panc-NEC (small-cell type) at 30 years of age harboring somatic loss of heterozygosity (LOH) of the PALB2 wild-type allele (Supplementary Table S17). In addition, a pathogenic germline variant of MAD1L1 was identified in one Panc-NEC (NE071). We also detected pathogenic germline variants in VHL (n = 2), APC (n = 1), FANCG (n = 1), and EPHB2 (n = 1) in patients with GIS-NETs (Supplementary Table S17).
Genomic Features of Panc-NETs Based on RNA-seq
Two driver genes (i.e., MEN1 and DAXX) were significantly (FDR-corrected P < 0.1) mutated in the 55 GIS-NETs (Supplementary Table S12). The MEN1 gene was frequently included in SV in Panc-NETs (Supplementary Fig. S5A). One pathogenic germline large deletion (NE042) resulted in a novel ATG2A–MEN1 fusion transcript (Supplementary Fig. S13A and S13B). This patient (23-year-old female) with the germ-line large deletion had a family history (father, parathyroid tumor; brother, pituitary tumor) consistent with MEN1 syndrome (Supplementary Table S1). Most Panc-NETG3s harbored alterations of driver genes (e.g., TP53 mutation, CDKN2A hypermethylation) in addition to MEN1 mutations (Fig. 2A and C).
The Panc-NETs were roughly divided into three clusters based on RNA-seq data. In RNA cluster 5 (C5 in Fig. 2B), ARX was overexpressed and the MEN1 gene was characteristically inactivated (30). RNA cluster 3 (C3 in Fig. 2B) showed high expression of PDX1. The remaining cluster (C4 in Fig. 2B) featured high expression of both ARX and PDX1. Of note, increased expression of immune checkpoint molecules was found in C4. All Panc-NETG3s belonged to the ARX-high groups (C4 or C5).
Clinically, the Panc-NETs analyzed in this study were diagnosed as nonfunctional tumors. We nevertheless comprehensively investigated the expression of 94 neuropeptides listed in the neuropeptide database (http://www.neuropeptides.nl/) and TPH1 and selected 27 high variant neuropeptides and TPH1 [standard deviation of transcripts per million (TPM) > 3.0 among Panc-NETs] for further analysis (Supplementary Fig. S14). Unexpectedly, expressed neuropeptides included neuropeptides not normally produced in the pancreas. In addition, we found that Panc-NETs with high-level expression of gamma cell–specific, delta cell–specific, and epsilon cell–specific hormones in addition to alpha and beta cell–specific were present.
DNA Methylation Status in Panc-NECs and Panc-NETs
Cluster analysis of DNA methylation results stratified pancreatic NENs into three subgroups (Fig. 2C). First, Panc-NECs and Panc-NETs branched. Second, Panc-NETs were classified into two clusters. The classification of Panc-NETs based on epigenomes generally resembled that of transcriptomes (Fig. 2B and C). DAXX hypermethylation was found in almost all of Panc-NETs (Fig. 2C). Hypermethylation of CDKN2A was evident in 3 of 7 Panc-NETG3s, compared with 2 of 23 Panc-NETG1/G2s (P = 0.0332; Fig. 2C). The two clusters of Panc-NETs were strongly linked with MEN1 findings. Methylation cluster 2 (Methyl-C2) harboring MEN1 alterations showed greater hypermethylation of RASSF1, PDX1, and CDX2 and hypomethylation of HNF4A and 18 genes listed in the COSMIC Cancer Gene Census (e.g., MGMT and TERT), as compared with methylation cluster 3 (Methyl-C3; Supplementary Fig. S15A). A focus on DNA methylation modifiers from RNA-seq data showed that the expression of IDH2 and TET3, methylation erasers, and UHRF1 and DNMT1, methylation maintainers, was significantly higher in Methyl-C1 mostly consisting of NECs than that in Methyl-C2 and/or C3 (Supplementary Fig. S15B).
ATAC-seq in Panc-NECs and Panc-NETs
We performed ATAC-seq for seven Panc-NECs and four Panc-NETs (Fig. 2D; Supplementary Fig. S16). Principal component analysis (PCA) of chromatin accessibility at distal elements (n = 191,422) revealed distinct clusters (Fig. 2D). Motif enrichment analysis of transcription factors showed that the SOX2 and/or ASCL1 motif was enriched in “ductal-type Panc-NECs” and the RBPJ and RARA were enriched in the “acinar-type Panc-NEC.” Regarding four Panc-NETs, the RFX3 motif was enriched in all, but the enrichment of two motifs, PDX1 (NE042 and NE038, Methyl-C3 in Fig. 2C) and HNF4A (NE032 and NE045, Methyl-C2 in Fig. 2C), were mutually exclusive.
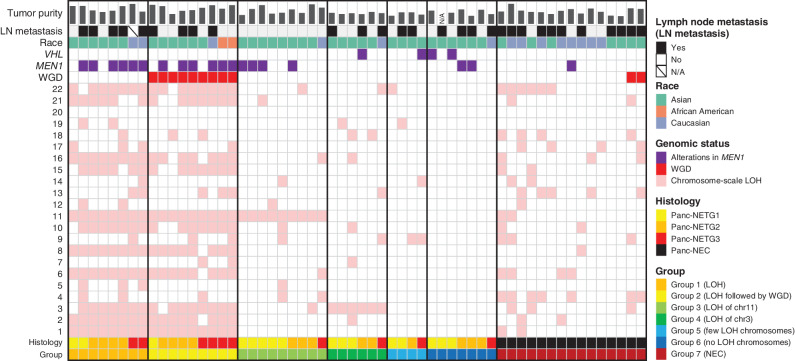
Genome-wide LOH and Subsequent WGD in Panc-NETs
Striking genome-wide LOH that affected nearly half their chromosomes was observed in 17 of 43 (39.6%) Panc-NETs (13 NETG1s, 20 NETG2s, and 10 NETG3s) that underwent WGS and/or WES (Fig. 3). The most common targets of haploidy were chromosomes 1, 2, 3, 6, 8, 10, 11, 15, 16, 21, and 22, generally in accord with the previous report (5, 31). In these patients, MEN1 alterations (mutations and/or SVs) were frequently observed (11/17, 64.7%). Of interest, in nearly half of Panc-NETs (9/17, 53.0%) with genome-wide LOH, WGD occurred after the LOH (Fig. 3; Supplementary Table S14).
Figure 3.
Landscape of DNA copy-number alterations in Panc-NETs and Panc-NECs. Examination of the chromosome-level copy number allowed stratification into six groups in Panc-NETs. Group 1: recurrent pattern of whole chromosomal loss, affecting specific chromosomes (1, 2, 3, 6, 8, 10, 11, 15, 16, 21, and 22); group 2: recurrent pattern of whole chromosomal loss, affecting specific chromosomes and subsequent WGD; group 3, chromosome-scale LOH to chromosome 11; group 4, chromosome-scale LOH to chromosome 3; group 5, chromosome-scale LOH to nonspecific one or two chromosome(s); group 6, no chromosome-scale LOH. Chromosomes 1–22 are depicted from bottom to top, and individual samples are shown from left to right. Pink indicates chromosome-scale LOH.
Sawicki and colleagues (32) recently reported that menin, which is encoded by the MEN1 gene, during early mitosis is localized to the mitotic spindle poles and to the mitotic spindle, and during cytokinesis to the intercellular bridge microtubules. Menin depletion has the following effects: during early mitosis, defects in spindle assembly and chromosome congression; during anaphase, lagging chromosomes; and defective cytokinesis. We discovered a novel group of Panc-NETs with loss of one copy in entire chromosome 3, in addition to a group with loss of one copy in entire chromosome 11 as previously reported (5), which were mutually exclusive in nearly one third of the Panc-NETs (15/43, 34.9%). Presumably, MEN1 on chromosome 11 and VHL on chromosome 3 could be targeted during tumorigenesis. In addition, the frequency of lymph node metastasis in Panc-NETs in group 3 (loss of one copy in entire chromosome 11) was significantly lower than Panc-NETs in other groups (P = 0.00293).
Novel Splicing Machinery by Large Deletion of SNRNP70
Among two neoplasms exhibiting RB1 exon skipping, case NE005, with exon 15 skipping, had a large deletion of SNRNP70, an RNA splicing factor (Supplementary Fig. S17A–S17D). This carcinoma had the most alternative splicing events among all neoplasms in the present study using the rMATS software (33). To the best of our knowledge, there has been no documentation of any relationship between aberrant SNRNP70 and cancer, but it has been reported that knockdown of the SNRNP70 gene dramatically induced significant alternative splicing events in the HepG2 cell line (ref. 34; Supplementary Fig. S17E), suggesting that inactivation of RB1 in NE005 might be caused by such a novel splicing event.
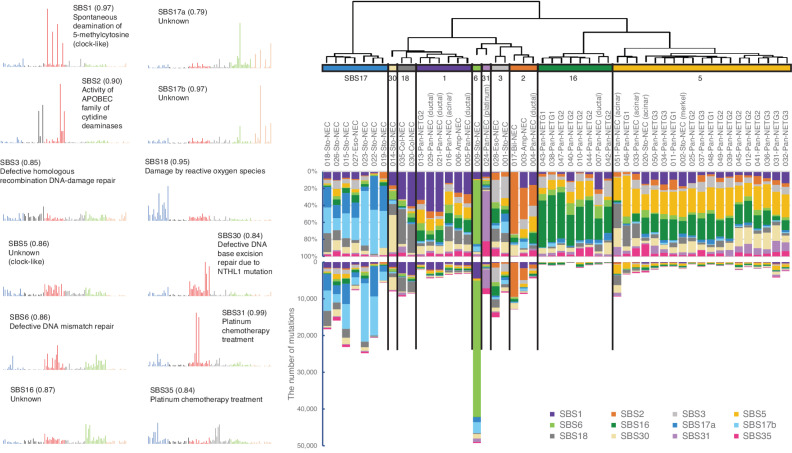
Mutational Signatures in GIS-NENs
De novo extraction of mutational signatures by nonnegative matrix factorization (NMF) analysis using WGS data defined 12 robust mutational signatures (Supplementary Fig. S18), in accord with any of the Cosmic Single Base Substitution (SBS) Signatures (v3.1; Fig. 4). SCLC is a representative smoking-related disease, with a typical mutational signature (SBS4; ref. 35). However, smoking-related signatures were not detected in GIS-NECs in this series, suggesting distinct mutational processes. Clustering analysis showed that GIS-NECs derived from the same organ were generally grouped. Panc-NETs were divided into two clusters, namely dominant contribution of SBS5 or SBS16. “Ductal-type Panc-NECs” were clustered into the group that was characterized by dominant contribution of SBS1, whereas “acinar-type Panc-NECs” were clustered in the group consisting of Panc-NETs (dominant contribution of SBS5). Most gastric NECs were clustered in the group exhibiting dominant contributions of SBS17a and SBS17b. The number of gastric NECs with a dominant contribution of SBS17 is likely high (cases with SBS17 accounted for >50% of total signatures: 6/10, 60%), compared with the Chinese WGS cohort of 168 conventional gastric cancers (14). We also identified a rare signature [i.e., SBS31 in a Panc-NEC (NE024)] in the updated COSMIC mutational signatures (v3.1). Although the number of cases with this mutational signature is relatively few (SBS31, six cases reported in the ICGC/TCGA PCAWG; ref. 36), it should be borne in mind that the signature may be due to platinum drug treatment (37). Indeed, our case received platinum-based chemotherapy before the carcinoma was harvested. Findings of a mutational signature analysis of GIS-NECs by deconstructSigs (38) using WGS and WES data are provided in Supplementary Fig. S19.
Figure 4.
Mutational signature analysis of GIS-NENs. Hierarchical clustering by de novo extraction of mutational signatures with nonnegative matrix factorization of available WGS data. Stability plotting indicated 12 mutational signatures (>0.85; Supplementary Fig. S18). Their profiles and functions were assigned based on COSMIC SBS Signatures (v3.1). Sto, stomach; Eso, esophagus, Col, colorectum; Pan, pancreas; Amp, ampullary; Bil, bile duct; Platinum, platinum chemotherapy treatment; Merkel, gastric NEC with Merkel cell polyomavirus. The parentheses in the left figure indicate cosine similarity to COSMIC signatures.
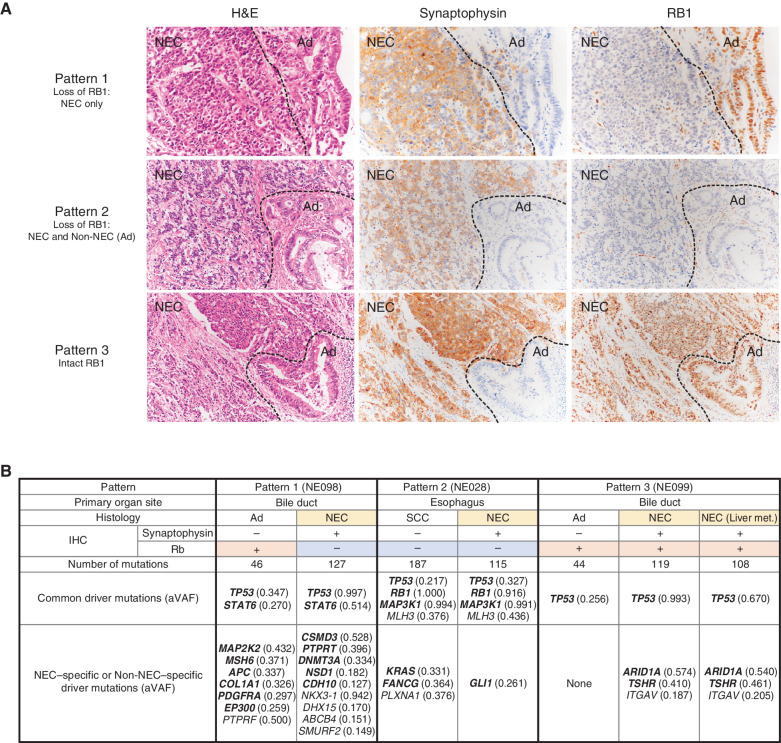
Divergent Genetic Alterations in NEC and Non-NEC Components
Several of the neuroendocrine carcinomas had a nonneuroendocrine component (MiNENs). The nonneuroendocrine components included adenocarcinoma or squamous cell carcinoma. We observed three patterns of RB1 immunolabeling in these MiNENs: pattern 1, loss of RB1 only in the NEC component; pattern 2, loss of RB1 in both NEC and Non-NEC components; and pattern 3, intact RB1 in both components (Fig. 5A). Most cases belonged to pattern 1 or 3, but given the existence of NECs showing pattern 2, we conducted WES using macrodissected FFPE materials of NEC and Non-NEC components independently in the same patients (n = 3; Fig. 5B; Supplementary Table S18). In all three cases, the phenotypically different cells had a common origin because they shared specific genomic aberrations. In case NE028 (esophageal NEC), RB1 protein was lost in both the NEC and Non-NEC components. The number of mutations was larger in the Non-NEC component than in the NEC components. Four driver mutations detected by OncodriveMUT (39) were common in both components, and driver mutations of oncogenic genes including KRAS (p.G12A) had accumulated in the Non-NEC component (Fig. 5B).
Figure 5.
Bidirectional differentiation in GIS-NECs. A, Representative microscopic features of GIS-NECs with nonneuroendocrine carcinoma elements (Non-NEC; adenocarcinoma or squamous cell carcinoma). Based on RB1 immunolabeling, these GIS-NECs are classified into three patterns. Pattern 1, loss of RB1 only in the NEC component; pattern 2, loss of RB1 in both the NEC and Non-NEC components; pattern 3, intact RB1 in both the NEC and Non-NEC components. The NEC components are positive for synaptophysin in all cases. NEC, NEC component; Ad, adenocarcinoma component. B, In three cases of GIS-NECs with Non-NEC elements (adenocarcinoma or squamous cell carcinoma), NEC and Non-NEC components were separately macrodissected from FFPE materials. WES was performed for each component. The table shows driver mutations (TVAF ≥ 0.1) identified by OncodriveMUT (39) in the NEC and Non-NEC components in these cases. The genes in bold font are listed in the COSMIC Cancer Gene Census. SCC, squamous cell carcinoma component; aVAF, variant allele frequency adjusted to tumor purity; Liver met., liver metastasis.
Multiregion Analysis Using Multiomics Data in an Autopsied Patient with Panc-NEC
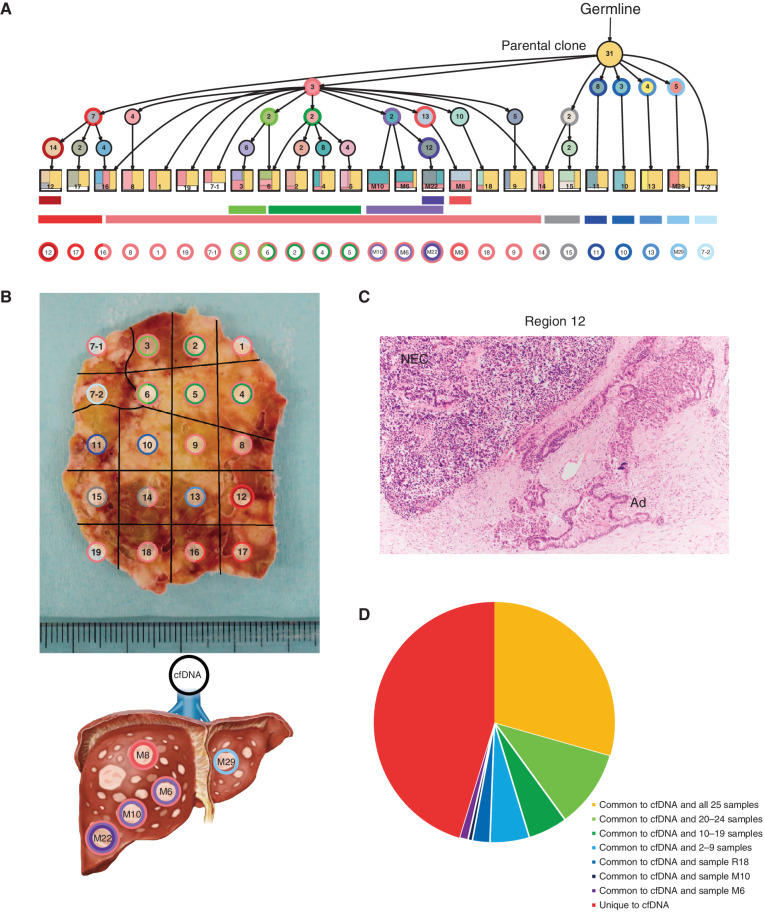
We also performed multiregion WES, whole-transcriptome sequencing, and methylation assay on 20 regions of the primary and five separate metastases from an autopsied patient with Panc-NEC (case NE004). The number of nonsynonymous somatic mutations ranged from 30 to 47 with an average of 38.5 (Supplementary Table S19). Because somatic phylogenetics is complicated by sample heterogeneity, we inferred the evolutional lineage tree based on variant allele frequency (VAF)–based clustering of each sample to detect subclones, in this case using the method that automates the phylogenetic inference of cancer progression from multiple somatic samples (LICHeE; ref. 40; Fig. 6A). We also inferred a phylogenic tree based on mutations and LOH patterns using randomized axelerated maximum likelihood (RAxML; ref. 41), which showed widely spreading branches (Supplementary Fig. S20A and S20B). All 20 regions and 5 liver metastases shared mutations in major driver genes (i.e., KRAS, RB1, and TP53). In Fig. 6B, sections are colored corresponding to the colors of the evolutional lineage tree in Fig. 6A. Interestingly, ploidy levels and copy-number analysis demonstrated that WGD occurred in only adjacent regions 12, 16, and 17 in the primary Panc-NEC (Supplementary Table S20). In this case, WGD occurred in the same subclone as a late event. Of note, histologic examination showed adenocarcinoma elements only in region 12 (WGD region; Fig. 6C), where synaptophysin was positive in only NEC components, but RB1 protein was negative in both immunohistochemically.
Figure 6.
Geographic mapping of subclones based on multiregion WES and proposed clonal evolution of the Panc-NEC in the autopsied patient. A, Proposal clonal evolution model drawn according to the evolutional lineage tree based on VAF of the mutations (LICHeE; ref. 40) in 20 primary regions and 5 liver metastases. The numbers inside the circles are for mutations used by LICHeE to infer the subclonal structure. The colors in each subdivision describe the mutation groups characterizing cells in this subpopulation. The numbers and colors inside the squares indicate the region numbers shown in B and the composition of subpopulations, respectively. B, Macroscopic picture of the maximum section through the primary Panc-NEC. Sections are marked, corresponding to the colors of the predicted subclones based on the evolutional lineage tree. C, Microscopic picture of region 12 in the primary Panc-NEC (H&E staining). Copy-number analysis demonstrated that WGD occurred in only the adjacent regions 12, 16, and 17 (Supplementary Table S20). Adenocarcinoma component observed together with NEC only in region 12. NEC, NEC component; Ad, adenocarcinoma component. D, Pie chart showing the relationship between mutations detected in plasma cfDNA and in tissue samples (20 primary regions and 5 liver metastases).
One liver metastasis (M29) was located in different branches from four other liver metastases in both lineage and phylogenic trees and diverged in the early stage of the RAxML phylogenetic tree, indicating that this metastasis (M29) occurred at an earlier stage of disease compared with the others. We next performed hierarchical clustering based on the methylation data (Supplementary Fig. S20C–S20E). The map based on methylation assay was similar to that based on somatic mutations and LOH status, regions 12, 16, and 17 (WGD regions) being clearly separated.
At autopsy, we obtained blood from inferior vena cava from this patient and performed WES using circulating cell-free DNA (cfDNA; Supplementary Table S21). The cfDNA concentration was extremely high (5,000 ng/mL plasma). We compared somatic mutations in cfDNA with the mutations in tissue samples: 29.4% (50/170) of cfDNA mutations were shared with all 25 samples, while 45.3% (77/170) of the cfDNA mutations were unique to cfDNA (Fig. 6D). We also observed that common mutations shared with many samples (e.g., KRAS, TP53, and RB1) tend to have higher TVAF in cfDNA. Comparison of TVAFs in cfDNA to median aVAFs (variant allele frequencies adjusted to tumor purity) among the 25 samples showed a strong correlation (Spearman rank correlation: ρ = 0.850, P < 2.2 × 10−16), as shown in Supplementary Fig. S20F and S20G. The mutational signature of cfDNA belonged to the different cluster than that for tissue samples (Supplementary Fig. S19).
Discussion
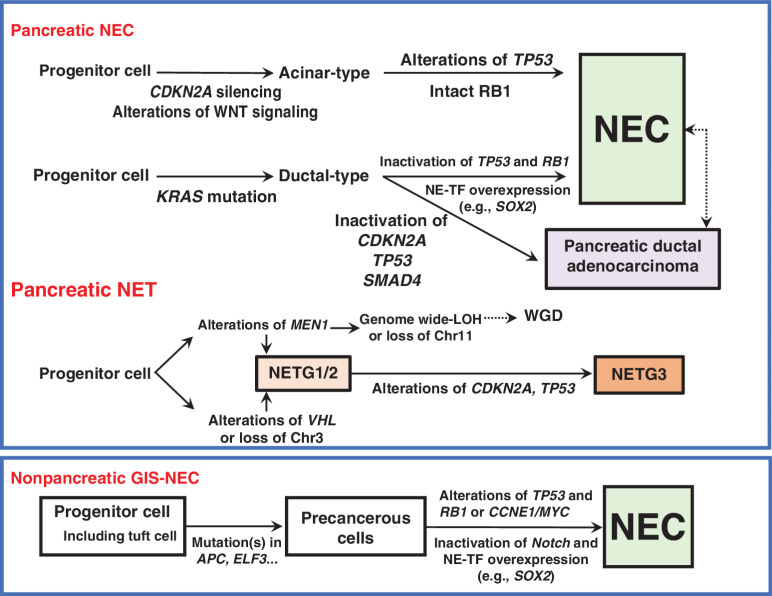
In the present study, we first verified that Panc-NECs are genetically distinct from Panc-NETs. Most Panc-NETs harbor MEN1, DAXX gene mutations, and hemizygous loss of specific chromosome(s). GIS-NECs harbor TP53 mutations and RB1 alterations and/or CCNE1/MYC amplification. Of interest, Panc-NECs and Nonpanc-NECs have moderately different genomic characteristics, the latter exhibiting more SVs and nonsynonymous mutations. Panc-NECs could be further divided into “ductal” or “acinar” types. A schematic representation of genomic alterations of GIS-NENs during tumor progression is shown in Fig. 7.
Figure 7.
Schematic diagram of genomic alterations involved in the genesis of pancreatic NEC, pancreatic NET, and nonpancreatic NEC of the gastrointestinal system (GIS). NE-TF, transcription factor for neuroendocrine differentiation.
In addition to prevalent TP53 mutations and RB1 alterations and/or CCNE1/MYC amplification in GIS-NECs, we found that inactivation of Notch family genes was characteristic in Nonpanc-NECs, and a Notch inhibitor induced neuroendocrine features in genome-engineered organoids. The Notch pathway is known to be involved in the clinical behavior of SCLC through its action on a number of biological processes such as neuroendocrine differentiation (42).
Our findings suggest that NE-TFs, especially the SOX2 gene, have crucial roles in GIS-NECs. Akiyama and colleagues reported five of seven Panc-NECs to be positive for SOX2 immunohistochemically (43). SOX2 is an embryonic stem cell transcription factor that is essential both for the pluripotency of embryonic stem cells and for the maintenance of physiologically migratory neural progenitor cells (44). Mu and colleagues (45) reported that lineage plasticity is promoted by SOX2, in particular in neuroendocrine transdifferentiation in TP53- and RB1-deficient prostate cancers. Harold and colleagues (46) reported the SOX2-dependent conversion of an undifferentiated aggressive cancer cell to a differentiated neuron-like phenotype in Merkel cell carcinomas. Unexpectedly, the high expression of SOX2 in GIS-NECs observed here appeared to be regulated by hypermethylation of the promoter region of SOX2, so-called paradoxical gene activation. Recently, growing evidence has been presented that this association may not always hold true, and promoter hypermethylation also appears to be associated with high transcriptional activity (47). In addition, ATAC-seq showed open chromatin peaks in the region surrounding the SOX2 gene. Thus, our comprehensive genomic profiling demonstrated that epigenetic aberrations play a significant role in GIS-NECs.
Most GIS-NECs presumably arise from precursor lesions that typically give rise to nonneuroendocrine carcinomas of the respective organ, since organ-specific initial genetic events were detected (e.g., KRAS in ductal-type Panc-NECs and APC in colorectal NECs). In addition, the observation that tubulovillous adenomas were located adjacent to the colorectal NEC in two cases (NE035 and NE084) supports this hypothesis.
There is a possibility that NEC and Non-NEC components develop bidirectionally. In case NE028, RB1 protein was lost in both the NEC and Non-NEC components. Four driver mutations were common in both components, and driver mutations of oncogenic genes including KRAS (p.G12A) had accumulated in the Non-NEC component (Fig. 5B). In addition, the evolutional lineage tree of our autopsied case indicated that the Non-NEC component (adenocarcinoma) was linked to a unique WGD-positive subclone. Although spontaneous transformation from conventional adenocarcinoma or squamous cell carcinoma to NEC has been reported (48), the possibility of the reverse occurring was recently postulated from a case study of SCLC (49). In addition, Calbo and colleagues (50) reported that RasV12 promoted transition from a neuroendocrine to a nonneuroendocrine phenotype in Rb1- and Trp53-deficient mouse SCLC cells.
We here observed that two GIS-NECs were etiologically linked to viruses (MCPyV for a gastric NEC and HPV for a colorectal NEC), indicating that more attention should be devoted to this possibility in future analyses in clinical practice. It has been reported that the combination of dinaciclib, a CDK1/2/5/9 inhibitor, with other drugs (e.g., AKT inhibitors) showed selective and potent activity in CCNE1-amplified carcinomas (51). In addition, early-phase trials of Chk1 and Wee1 inhibitors identified high frequencies of CCNE1 amplification among responders, though the sample sizes were limited (52). These drugs might offer promising therapeutic strategies for CCNE1-amplified GIS-NECs.
In conclusion, Panc-NECs are genetically distinct from Panc-NETs and may not be etiologically related. GIS-NECs arising in different organs show similar histopathologic features and share some genomic features, but considerable differences exist between Panc-NECs and Nonpanc-NECs.
Methods
Patients and Tissue Samples
This study included 115 GIS-NENs from patients evaluated and treated at the National Cancer Center Hospital (Tokyo, Japan), the National Cancer Center Hospital East (Chiba, Japan), Tokyo Women's Medical University Hospital (Tokyo, Japan), Osaka University Hospital (Osaka, Japan), Wakayama Medical University Hospital (Wakayama, Japan), Hiroshima University Hospital (Hiroshima, Japan), Kagawa University Hospital (Kagawa, Japan), the Johns Hopkins Hospital (Baltimore, MD), and the University Medical Center Utrecht (Utrecht, the Netherlands). Tumors and corresponding nontumor tissues were frozen or FFPE after surgical resection (Supplementary Table S1). In addition, two frozen samples were taken at autopsy, four organoid samples were derived from biopsied materials following upper or lower gastrointestinal endoscopy, and three specimens of liver metastasis were taken via biopsy. We performed macrodissection in order to enrich the tumor content relative to the surrounding normal cells in most frozen and FFPE tumor tissues. The study was conducted in accordance with the ethical guidelines of each country. This study was approved by the institutional review boards of each contributing institute. Written informed consent was obtained from all patients.
The histologic features of all GIS-NENs were reviewed, and each case was also assessed for the mitotic index and the Ki-67 proliferation index according to the WHO Classification of Tumours (5th Edition, Digestive System Tumours; ref. 1). Mitotic rates were expressed as the number of mitoses/2 mm2 as determined by examining 50 fields of 0.2 mm2. The Ki-67 value was determined by counting at least 500 cells in the regions of highest labeling (hotspots). In cases in which the grade differed for mitotic count compared with the Ki-67 proliferation index, the higher grade was assumed. As for the biopsy samples, the Ki-67 proliferation index value was assigned. NECs were divided into small-cell type NECs and large-cell type NECs. Specifically, carcinomas with a diffuse, infiltrative growth pattern, small-sized to medium-sized cells with minimal cytoplasm, and fusiform nuclei with finely granular chromatin pattern, inconspicuous nucleoli, and nuclear molding by adjacent nuclei were categorized as small-cell type NEC (Supplementary Fig. S1B). In contrast, carcinomas with a more pronounced nesting pattern and abundant necrosis, and containing neoplastic cells with a moderate amount of amphophilic cytoplasm, large nuclei with coarsely clumped chromatin, and prominent nucleoli were categorized as large-cell type NEC (Supplementary Fig. S1C). MiNENs are classified as having a neuroendocrine component that is combined with a nonneuroendocrine component, each morphologically and immunohistochemically recognizable as discrete and constituting ≥30% of the neoplasm (Supplementary Fig. S1D).
Due to the lack of easily recognizable morphologic criteria, the distinction of a NETG3 from an NEC is challenging (53). Therefore, all the evaluations were first performed by a pathologist at each institution according to WHO Classification of Tumours, and then we selected a single representative hematoxylin and eosin (H&E) slide to represent the high-grade region of the tumor, which was blinded and reviewed by a single central pathologist specialized in gastrointestinal and hepatopancreatobiliary pathology (T. Furukawa). Cases for which a consensus diagnosis was not achieved (ambiguous morphologic cases) were removed.
IHC
Paraffin-embedded samples for each tumor were immunolabeled using the antibodies listed in Supplementary Table S22. To discriminate between pure Panc-NECs, MiNEN, and pure acinar cell carcinoma (54), IHC for acinar cell markers (trypsin, chymotrypsin, and BCL10) was performed for all of the Panc-NECs. IHC labeling was carried out using a Bond Max instrument (Leica Microsystems) or VENTANA BenchMark ULTRA (Roche Diagnostics), as previously described (27).
DNA-Sequencing Analysis
Genomic DNA was extracted from frozen tissues using a QIAamp DNA Mini Kit (QIAGEN) for WGS or from FFPE tissues using a QIAamp DNA FFPE Tissue Kit (QIAGEN) for WES. For WGS, short-fragment libraries (insert size 350 bp) were prepared with 1–2 μg aliquots of genomic DNA from tumor and matched nonneoplastic tissues using a TruSeq DNA PCR-Free Library Prep Kit (Illumina) according to the manufacturer's instructions. The libraries were made from DNA fragmented with an average size of 150–200 bp using a Covaris S2 or E220ev system (Covaris). Paired-end sequencing of the libraries was performed using a HiSeq 2000/2500 sequencer (Illumina). WGS depths were 44.7 on average (range, 27.3–69.0). Whole-exome capture libraries were prepared from tumor and matched normal DNA using a TruSeq Exome Library Prep Kit (Illumina), according to the manufacturer's instructions. DNA libraries were sequenced in a paired-end mode with 100-bp reads, using a HiSeq 2000/2500 sequencer (Illumina). WES depths were 360.9 on average (range, 58.9–1876.2).
With the autopsied patient, blood was collected from the inferior vena cava, and WES was performed using circulating cfDNA. Before DNA extraction, plasma samples were centrifuged at 16,000 × g for 10 minutes at 4°C in order to remove cell debris. Circulating cfDNA was extracted from 6 mL of plasma using a QIAamp DNA Circulating Nucleic Acid kit (QIAGEN). DNA libraries were sequenced in a paired-end mode with 100-bp reads, using a HiSeq 2500 sequencer (Illumina). Paired-end sequencing of the libraries was performed in the same way as exome sequencing on DNA from tissue specimens. WES depths were 579.3 on average.
Targeted deep resequencing was performed to validate mutations and accurately evaluate the VAF using the SureSelect Target Capture System (Agilent Technologies). In addition to the SMGs in WGS/WES, cancer-related genes and previously identified SMGs in a Panc-NET cohort (5) were included in target regions (total of 78 genes; Supplementary Table S8). Preparation of sequence libraries and target capture were carried out using SureSelect XT Low Input Kit (Agilent Technologies). All samples were sequenced on a HiSeq 2500 (Illumina) platform with paired read of 100 bp. The depths of targeted deep resequencing were 2,306.0 on average (range, 217.6–5131.8).
Mutation Calling
Paired-end reads were aligned to the human reference genome (GRCh37) using BWA-MEM for both tumor and nontumor samples. Potential PCR duplications, for which paired-end reads aligned to the same genomic position, were removed, and pileup files were generated using SAMtools (55) and programs developed in-house. To find somatic point mutations [single-nucleotide variations (SNV)] and short indels, the following cutoff values for the base selection were used: (i) a mapping quality score of at least 20; (ii) a base quality score of at least 10. Then somatic mutations were selected using the following two filtering conditions; (iii) the numbers of reads supporting a mutation in each tumor sample were at least 4 or 8 when TVAF ≥ 0.15 or 0.15 > TVAF ≥ 0.05, respectively, and at least a quality score of one base of these reads must be greater than 30; (iv) the VAF of the matched nontumor sample was less than 0.03 with a read depth of at least 8. To further exclude sequence-context–dependent errors, the sequence reads of all nontumor samples were grouped and used to discriminate true positives from false positives. The VAF of grouped nontumor samples (NVAF) with a sequence depth more than or equal to 10 and a VAF less than 0.2 was calculated at each mutated genomic position. Then the following filter was applied: (v) NVAF was less than 0.03 or 0.01 when TVAF ≥ 0.15 or 0.15 > TVAF ≥ 0.05, respectively; (vi) the ratio of TVAF to NVAF had to be more than or equal to 20, at each mutated genomic position; (vii) the ratio of nontumor samples with a VAF more than or equal to 0.1 must be less than 0.002; and finally (viii) mutations with a strand bias (between forward and reverse reads) greater than 95% were removed.
Detection of Germline Variants
We performed germline analysis on WGS and WES data using DNA derived from nonneoplastic tissues. Sequence reads were mapped onto the Human Genome Reference, GRCh37/hg19, using default settings by BWA-MEM. The following reads were removed using an in-house program: (i) low mapping quality reads (mapping quality score < 10); (ii) PCR duplication reads; (iii) reads with a low alignment score [alignment score (AS tag in SAM file) < 60] and a mismatch rate ≥ 0.05; and (iv) reads with a high suboptimal-alignment score [the suboptimal-alignment score (XS tag in SAM file) ≥ 90] and a mismatch rate ≥ 0.05. The germline SNVs/indel call analysis pipeline was based on the Genome Analysis Tool Kit (GATK; v3.5: https://software.broadinstitute.org/gatk/) best practices (56). After running indel realignment and base quality score recalibration using IndelRealigner and BaseRecalibrator in GATK, samples were subjected to HaplotypeCaller runs in genomic VCF mode. The genomic VCF files for each sample were merged by GenotypeGVCF, and calls were filtered by VariantRecalibrator. Additionally, the following calls were filtered using an in-house program: (i) variants with average read depths <8 and (ii) variants with minor allele frequencies of all cases <0.2. Pathogenic gene variants were defined as annotated as “pathogenic” on the ClinVar (57) database. Rare variants were defined with an allele frequency less than 1% in the International Genome Sample Resource (IGSR; ref. 58), the Exome Aggregation Consortium database (ExAC; ref. 59), the NHLBI Grand Opportunity Exome Sequencing Project (GO-ESP; ref. 60), and the Integrative Japanese Genome Variation Database (iJGVD; Tohoku Medical Megabank; ref. 61). In addition, nonsense or frameshift variants were expected to be pathogenic. Detected variants were visually reviewed using Integrated Genomics Viewer (Broad Institute, Cambridge, MA).
Copy-Number Analysis
The initial copy-number ratio was estimated by comparing the read depths of tumor and nontumor samples. The copy-number ratio was adjusted using the tumor purity of each sample. Our calculation formula was as follows: R′(x) = 1 − (1 − R(x))/tumor purity, where R′(x) is the adjusted copy-number ratio at probe x, and R(x) is the ratio of tumor read depth to nontumor read depth. Amplifications were selected if the adjusted copy-number ratio >8, and homozygous deletions were selected if the adjusted copy-number ratio was ≤0.
Processing SMGs
SMGs were determined by three separate tests (the aggregated somatic alteration method, ref. 62; the activation bias method; and the inactivation bias method). The P values from individual significance tests were combined as geometric means, and multiple testing adjustments (FDR-corrected P < 0.1) were performed using the method described by Benjamini and Hochberg (63).
Estimation of SMGs by the aggregated somatic alteration method.
SMGs were estimated by aggregating somatic substitutions, short indels, focal amplifications, and homozygous deletions, as reported previously (62). The expected number of each alteration was estimated from the background alteration rate, and tests of significance were performed by assuming a Poisson distribution (P value).
Estimation of SMGs by the inactivation bias method.
The number of samples with inactivating alterations (nonsense, read-through, splice-site and frameshift mutations, and homozygous deletions) was compared with the number of samples with other alterations using the Fisher exact test.
Estimation of SMGs by the activation bias method.
The number of activating alterations (hotspot missense mutations and focal amplifications) was compared with the number of other alterations using the Fisher exact test. With at least two identical mutations in the same genomic position, the position was defined as a hotspot. With at least two identical mutations within 5 bp of the hotspot, these were all counted as hotspot mutations. Hotspot missense mutations often show activating functions but sometimes inactivating or unknown functions. Therefore, if the P value with the inactivation bias method was <0.05, the gene was considered a tumor suppressor gene, and the P value of the activation bias method was set to 1.
Mutational Signature Analysis
De novo mutational signature analysis using WGS data was performed using NMF software (64), and factorized signatures were compared with COSMIC mutational signatures v2 and v3.1 (https://cancer.sanger.ac.uk/cosmic/signatures). The contribution of COSMIC mutational signatures in WGS and WES data was calculated using the deconstructSigs (38) R package. Cases with a small number of mutations (<50) were excluded from deconstructSigs analysis. Clustering by contribution of mutational signatures was performed using unsupervised hierarchical clustering with cosine distance and Ward linkage.
Detection of Somatic Structural Variants
An in-house pipeline was developed to detect all types of SVs. As inputs into the pipeline, 100-bp paired-end reads of approximately 600-bp fragments from paired tumor and nontumor samples were used for cases with WGS data available. All paired-end reads were aligned to the human reference genome (hg19) using BWA-MEM with a −T 0 option, and then PCR duplication and reads with low alignment quality were filtered out. After filtering, the sequence data sets were processed using two independent algorithms. The first one used paired-end reads mapped discordantly, for which both ends align to the reference genome uniquely with improper spacing, orientation, or both (65). The second one utilized single reads split and mapped apart (so-called soft-clipped reads) to identify SV breakpoints. All soft-clipped reads with enough mapping quality were collected and the genomic positions of split–alignment junctions were extracted as SV breakpoint candidates. For each SV candidate, sequences of upstream and downstream of the breakpoint were obtained from the reference sequence, and they were concatenated to reconstruct a local rearrangement sequence. All input reads were realigned to the reconstructed sequences and divided into two groups: SV-support reads or reference-support reads. The number of reads of the two groups, from tumor and nontumor samples, were evaluated using the Fisher exact test. Finally, outputs from the two algorithms using paired-end and soft-clipped reads were integrated. False-positive SVs were filtered out based on the following cutoff values: (i) for translocation SVs, the number of support reads must be eight or more with four or more paired-end reads; (ii) for other SV types, the number of support reads must be four or more with two or more paired-end reads; (iii) read depth at SV breakpoint must be 10 or more; (iv) SV allele frequency must be 0.07 or more; (v) the total length of alignment region of soft-clipped reads supporting an SV must be 1.6 times the read length or more.
Inference of WGD and Chromosome-Scale LOH Patterns
WGD and chromosome-scale LOH patterns were inferred from somatic copy numbers estimated by FACETS (66). First, copy-number analysis was performed using cnv_facets (version 0.16.0, https://github.com/dariober/cnv_facets), and genomes were annotated as WGD genomes if the estimated ploidy was larger than three. Second, we considered the possibility that chromosome-scale LOH occurred extensively before WGD, making the ploidy values almost indistinguishable between diploid (non-LOH) and tetraploid (extensive LOH followed by WGD) genomes. Even in such cases, WGD should be detectable, as most chromosomes in a doubled genome have even copy numbers. Therefore, for each autosome (chr1–22), we classified chromosomal segments into three groups based on their allelic copy numbers: even–even, even–odd, or odd–odd. If the total segment length of even–even allelic copy numbers was the longest, the chromosome was annotated as doubled chromosomes. If ≥10 chromosomes were doubled, we inferred that the entire genome was doubled by WGD even if the ploidy was not larger than three. Chromosome-scale LOH was inferred if at least 90% of the genes on the chromosome had a lesser (minor) allele copy number zero.
Detection of Virus-Derived Sequence Reads in the WGS Data Set
Paired-end reads were mapped to the human genome (hg19) using BWA (version 0.7.15), and unmapped reads were mapped to the virus genomes as shown in Supplementary Table S7.
Whole-Transcriptome Sequencing (RNA-seq)
Total RNA was extracted from frozen tissues using the illustra RNAspin Mini RNA Isolation Kit (GE Healthcare). A whole-transcriptome sequencing library was prepared using the TruSeq RNA Access Library Prep Kit (Illumina). Sequencing was performed on a HiSeq 2500 with paired read of 100 bp. Detection of gene fusions was performed using our in-house pipeline (8) and fusionfusion (https://github.com/Genomon-Project/fusionfusion), as previously reported (67). For calculating gene expression, the paired-end reads of RNA-seq were mapped and aligned to the coding-regions of known RNA sequences in the RefSeq databases using Bowtie (68) version 0.12.7, and TPMs were calculated for each gene. Gene expression of 1,672 transcriptional factors (TF) was extracted according to the FANTOM5 TF database (https://fantom.gsc.riken.jp/5/sstar/) and 100 to 200 high variant TFs were used for clustering. Differential gene expression was performed by the Wilcoxon–Mann–Whitney test.
DNA Methylation Analysis
Comprehensive DNA methylation analysis was performed using the Infinium MethylationEPIC BeadChip Kit (Illumina). The methylation level of CpG sites was represented by β values ranging from 0 (completely unmethylated) to 1 (completely methylated). The R-package ChAMP pipeline (version 2.18.2) was used for data quality control, preprocessing, normalization, and detection of differentially methylated regions (DMR; ref. 69). Probes with detection P > 0.01, probes with <3 beads in at least 5% of samples per probe, non-CpG probes, SNP-related probes, multihit probes, and probes located in chromosome X and Y were filtered out. When the ratio of probes with detection P > 0.01 was over 0.1, samples were removed. BMIQ was used for normalization and ProbeLasso was applied for defining DMRs with the following parameters: minProbes = 3, meanLassoRadius = 1,200, minDmrSep = 1,000, minDmrSize = 50, adjPvalProbe = 0.05.
ATAC-seq
ATAC-seq was performed with a modified protocol based on Omni-ATAC for frozen tissue (70). Approximately 20 mg or 50-μm-thick section(s) of frozen tissue was prepared to extract nuclei. Aliquots of 50,000 nuclei for each sample were incubated with 50 μL of Transposition mix [2.5 μL Tagment DNA Enzyme1 (Illumina), 25 μL 2 × Tagment DNA Buffer (Illumina), 0.5 μL 10% Tween-20, 0.5 μL 1% digitonin, 16.5 μL PBS and 5 μL water] in a Thermomixer. Transposed fragments were purified using a MinElute Reaction Cleanup Kit (QIAGEN), and PCR was performed according to the original method. Sequence library was pooled at 5 nmol/L and sequenced on HiSeq X (Illumina) with 150-bp pair-end reads to obtain at least 20 million leads per sample.
Quality control and adapter removal of raw sequencing reads were performed using Trim Galore! (version 0.2.2) and reads were aligned against hg38 by Bowtie2 (version 2.3.5.1). Peak calling was performed with Genrich (version 0.6, available at https://github.com/jsh58/Genrich) with default parameters. Transcription start site enrichment, quantification of read counts, and quantile normalization between samples were performed using R-package ChrAccR (version 0.9.11). Distal element accessibility was used for PCA in line with a previous report (71). The R-package chromVARmotifs pipeline (version 0.2.2) was applied for motif enrichment analysis (72), and deviation scores for motif enrichment were calculated by chromVAR function. The motif data set was downloaded from the JASPAR 2020 website (available at http://jaspar.genereg.net/downloads/).
HPV Genotyping
HPV genotyping was performed by HPV 14 Screening and 16, 18, 45 Typing Real-TM Quant (Sacace Biotechnologies), a real-time PCR kit for quantitative detection and genotyping of HPV types 16, 18, and 45 and quantitative detection of HPV types 31, 33, 35, 39, 51, 58, 59, 66, and 68. In brief, a total of 25 μL of PCR reaction containing 10 μL of DNA per sample was run using the CFX 96 Real-Time PCR Detection System (Bio-Rad Laboratories) and the results were automatically analyzed using the Microsoft Excel format supplied with the kit.
Generation, Gene Manipulation, and IHC of Colon and Gastric Organoids
For organoid experiments, normal colon organoids with genetic engineering of TP53 KO and RB1 KO, and gastric organoids with TP53 KO were used in this experiment as previously reported (16). Culture conditions were as previously described, with Advanced Dulbecco's Modified Eagle Medium/F12 supplemented with penicillin/streptomycin, 10 mmol/L HEPES, 2 mmol/L Gluta-MAX, 1 × B27 (Thermo Fisher Scientific), 10 nmol/L gastrin I (MilliporeSigma), and 1 mmol/L N-acetylcysteine (FUJIFILM Wako Pure Chemical Corporation). Complete medium was prepared by supplementing the basal culture medium with the following niche factors: 50 ng/mL mouse recombinant EGF (Thermo Fisher Scientific), 50 ng/mL human recombinant FGF2 (PeproTech), 100 ng/mL human recombinant IGF1 (BioLegend), 100 ng/mL mouse recombinant noggin (PeproTech), 1 μg/mL recombinant human R-spondin-1 (Bio-Techne), 25% Afamin-Wnt-3A serum-free conditioned medium, and 500 nmol/L A83-01 (Tocris Bioscience). For organoid experiments, N-[N-(3,5-difluorophenacetyl)-1-alanyl]-S-phenylglycine t-butyl ester (DAPT; MilliporeSigma, 10 μmol/L) was added on day 1. EGF/FGF2/IGF1 were removed from the culture media on day 5, and the organoids were collected on day 10, fixed with 4% paraformaldehyde, and immunostained using mouse antisynaptophysin.
Comparison of Mutation Patterns between NEC and Non-NEC Components in the Same Patients
The numbers of mutations with TVAF ≥ 0.1 for NE028 and NE099 samples, and with TVAF ≥ 0.2 for NE098 due to a higher frequency of false positives caused by DNA damages characteristic to FFPE samples are shown in Fig. 5B. All mutations with TVAF ≥ 0.1 were then annotated by Annovar (73) and Cancer Genome Interpreter (https://www.cancergenomeinterpreter.org; ref. 39), known or predicted driver mutations being shown. We calculated aVAF from TVAF and purity as aVAF = TVAF/purity if TVAF < 0.75, and aVAF = min (1, (2/purity × TVAF/(1 + TVAF)) otherwise, where the two formulae account for copy number–neutral cases and LOH cases, respectively.
Multiregion Analysis in an Autopsied Patient with Pancreatic NEC
To cast light on tumor heterogeneity and genesis of the disease, multiregion WES, transcriptome sequencing, and DNA methylation assays were conducted using samples obtained from an autopsied patient with a Panc-NEC (case NE004). At autopsy (two hours after death), diffuse liver metastases, retention of pleural fluid (600 mL), and abdominal dropsy (1,500 mL) were grossly identified. At the beginning of the autopsy, blood was collected from the inferior vena cava, and WES was performed using circulating cfDNA. The primary tumor and multiple liver metastases were sampled for snap-freezing in liquid nitrogen. We sectioned a maximum surface of the primary tumor, which presented a heterogeneous appearance, and divided it into 20 pieces in a grid shape (Fig. 6B). Genomic DNA and RNA were extracted from each unit and five liver metastases. We performed WES and transcriptome sequencing and DNA methylation assay on each sample obtained from 20 regions, 5 liver metastases and matched normal spleen tissue.
Inference of the Branching Pattern and Subclonal Structure from the Multiregion Sequencing Data in the Autopsied Patient
WES reads were mapped to the reference genome (hg19) using BWA (version 0.7.15, https://github.com/lh3/bwa) and duplicate reads were removed using Picard MarkDuplicates (version 2.9.0, Broad Institute, Picard Toolkit; GitHub Repository; 2019; https://github.com/broadinstitute/picard). Copy-number analysis was performed using cnv_facets (version 0.16.0, https://github.com/dariober/cnv_facets), and chromosomal regions were annotated as LOH regions if CNLR_MEDIAN < −1.5. To infer phylogenetic relationships among samples, a multiple sequence alignment was constructed from somatic mutation patterns in individual samples (i.e., each column represents a locus where somatic mutations were observed in one or more samples, and each sample had a reference base if no somatic mutation was detected or an alternative base if a somatic mutation was detected). Nonmutated loci in LOH regions were treated as alignment gaps. The phylogenetic tree (Supplementary Fig. S20A) was inferred using RAxML (version 8.2.12, https://github.com/stamatak/standard-RAxML; ref. 41) with the GTRGAMMA substitution model. Subclones were inferred from somatic mutation patterns with TVAFs, using LICHeE (version 1.0, https://github.com/viq854/lichee; ref. 40) with the following options: –tree 1 –absent 0.05 –present 0.1. Distinct colors were assigned to (i) all subclonal lineages that diverged directly from the clonal lineage, (ii) subclonal lineages spanning multiple regions, and (iii) subclonal lineages having more than 10 lineage-specific mutations (Fig. 6A). We calculated aVAF from TVAF and tumor purity as aVAF = TVAF/purity if TVAF < 0.75, and aVAF = min (1, (2/purity × TVAF/(1 + TVAF)) otherwise, where the two formulae account for copy number–neutral cases and LOH cases, respectively.
Functional Analysis of NET1–AKR Fusion Genes
An immortalized epithelial cell line of gastric fundus origin (HFundEC4N) was established and maintained as described previously with minor modification (74). NET1–AKR1C3 and NET1–AKR1C4 cDNAs were chemically synthesized. The NET1–AKR fusion-expressing HFundEC4N cell lines were generated by infection with the retroviral vectors pQCXIP-Puro-NET1-AKR1C3, pQCXIP-Puro-NET1-AKR1C3-FLAG, pQCXIP-Puro-NET1-AKR1C4, or pQCXIP-Puro-NET1-AKR1C4-FLAG. Total RNA was extracted from NET1–AKR1C3 or NET1–AKR1C4 fusion-expressing cells using an RNeasy Mini kit (QIAGEN). Libraries were prepared using TruSeq stranded mRNA Sample Preparation Kit (Illumina). RNA-seq was performed with NovaSeq 6000 (Illumina). Cells were used for no more than five passages after infection and tested for Mycoplasma contamination by PCR (e-Myco Mycoplasma PCR Detection Kit; the latest test date: February 18, 2019).
Data Availability
The raw sequencing data reported in this paper has been deposited in the National Bioscience Database Center with the accession number JGAS000359.
Supplementary Material
Acknowledgments
We thank all patients and their families who participated in this study, Y. Kawahara (Osaka University, Osaka, Japan), Y. Shimada (Kyoto University, Kyoto, Japan), J. Shibahara (Kyorin University, Tokyo, Japan), T. Sakatani (Nippon Medical School Hospital, Tokyo, Japan), T. Ushiku (The University of Tokyo, Tokyo, Japan), A. Fukagawa (National Cancer Center Research Institute, Tokyo, Japan) for expert advice, and E. Arakawa, K. Igarashi (National Cancer Center Research Institute, Tokyo, Japan), and M. Oda (Hiroshima University Hospital, Hiroshima, Japan) for expert technical assistance. This work was supported by grants from the National Cancer Center Research and Development Fund (29-A-6 to T. Shibata); Practical Research for Innovative Cancer Control from the Japan Agency for Medical Research and Development (AMED; JP21ck0106558 to S. Yachida; JP21ck0106693 to S. Yachida; JP21ck0106690 to S. Yachida, Y. Totoki, and T. Shibata; JP21ck0106547 to S. Yachida and T. Shibata; JP15ck0106138 and JP16ck0106138 to C. Morizane); Project for Cancer Research and Therapeutic Evolution (P-CREATE) from AMED (JP17cm0106612 to S. Yachida, Y. Totoki, and E. Takai); United States–Japan Cooperative Medical Science Program from AMED (JP20jk0210009 to S. Yachida, T. Kiyono, and T. Shibata); Integrated Frontier Research for Medical Science Division, Institute for Open and Transdisciplinary Research Initiatives, Osaka University (to S. Yachida); Joint Research Project of the Institute Medical Science, the University of Tokyo (to S. Yachida and T. Shibata); the Takeda Science Foundation (to S. Yachida); the Yasuda Medical Foundation (to S. Yachida); the Mitsubishi Foundation (to S. Yachida); and the Princess Takamatsu Cancer Research Fund (to S. Yachida). The National Cancer Center Research Biobank is supported by the National Cancer Center Research and Development Fund, Japan.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
S. Mitsunaga reports personal fees and other support from Chugai and Ono, and other support from Sumitomo Dainippon Pharma, Astellas, Ajinomoto, and TORAY outside the submitted work. T. Okumura reports grants from Grant-in-Aid for Scientific Research outside the submitted work. C. Morizane reports grants from Japan Agency for Medical Research and Development during the conduct of the study, as well as personal fees from Novartis, Teijin Pharma, and Servier, grants and personal fees from Yakult, Taiho Pharmaceutical, Eisai, and MSD K.K., and grants from ONO Pharmaceutical, J-Pharma, AstraZeneca, Merck biopharma, Daiichi Sankyo, and Daiichi Sankyo RD Novare outside the submitted work. T. Furukawa reports grants from Tokyo Central Pathology outside the submitted work. T. Kiyono reports personal fees and nonfinancial support from the National Cancer Center during the conduct of the study, as well as grants from AMED and JSPS KAKENHI outside the submitted work. L. Brosens reports personal fees from Bristol Myers Squibb B.V. outside the submitted work. R.H. Hruban reports a patent for GNAS in cysts licensed to Exact Sciences. No disclosures were reported by the other authors.
Authors’ Contributions
S. Yachida: Conceptualization, data curation, funding acquisition, validation, investigation, writing–original draft, project administration, writing–review and editing. Y. Totoki: Data curation, software, formal analysis, writing–original draft. M. Noë: Resources and data curation. Y. Nakatani: Data curation, software, formal analysis, visualization, methodology, writing–original draft. M. Horie: Data curation, formal analysis, investigation, visualization, methodology, writing–original draft. K. Kawasaki: Resources, investigation, methodology, writing–original draft. H. Nakamura: Data curation, software, formal analysis, and methodology. M. Saito-Adachi: Data curation, software, formal analysis, methodology, writing–original draft. M. Suzuki: Investigation, visualization, methodology, writing–original draft. E. Takai: Conceptualization, investigation, writing–original draft. N. Hama: Data curation, formal analysis, and methodology. R. Higuchi: Resources and data curation. S. Hirono: Resources and data curation. S. Shiba: Resources and data curation. M. Kato: Data curation and formal analysis. E. Furukawa: Data curation and formal analysis. Y. Arai: Investigation and methodology. H. Rokutan: Investigation and methodology. T. Hashimoto: Resources and investigation. S. Mitsunaga: Resources and data curation. M. Kanda: Resources and data curation. H. Tanaka: Investigation, methodology, writing–original draft. S. Takata: Validation and investigation. A. Shimomura: Resources and data curation. M. Oshima: Resources and data curation. W.M. Hackeng: Resources and data curation. T. Okumura: Resources and data curation. K. Okano: Resources and data curation. M. Yamamoto: Resources and data curation. H. Yamaue: Resources and data curation. C. Morizane: Conceptualization, resources, data curation, supervision, and funding acquisition. K. Arihiro: Resources, data curation, and supervision. T. Furukawa: Conceptualization, resources, data curation, supervision, methodology, writing–review and editing. T. Sato: Resources, data curation, supervision, and methodology. T. Kiyono: Conceptualization, resources, data curation, supervision, investigation, visualization, and methodology. L. Brosens: Conceptualization, resources, data curation, and supervision. L.D. Wood: Conceptualization, resources, data curation, supervision, and project administration. R.H. Hruban: Conceptualization, resources, data curation, supervision, investigation, visualization, methodology, writing–original draft, writing–review and editing. T. Shibata: Conceptualization, data curation, supervision, funding acquisition, methodology, project administration, writing–review and editing.
References
- 1. WHO Classification of Tumors Editorial Board. WHO classification of tumors: digestive system tumours. Lyon, France: International Agency of Research on Cancer; 2019. [Google Scholar]
- 2. Strosberg JR, Coppola D, Klimstra DS, Phan AT, Kulke MH, Wiseman GAet al. The NANETS consensus guidelines for the diagnosis and management of poorly differentiated (high-grade) extrapulmonary neuroendocrine carcinomas. Pancreas 2010;39:799–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Milione M, Maisonneuve P, Pellegrinelli A, Grillo F, Albarello L, Spaggiari Pet al. Ki67 proliferative index of the neuroendocrine component drives MANEC prognosis. Endocr Relat Cancer 2018;25:583–93. [DOI] [PubMed] [Google Scholar]
- 4. Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra Aet al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 2011;331:1199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Scarpa A, Chang DK, Nones K, Corbo V, Patch AM, Bailey Pet al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature 2017;543:65–71. [DOI] [PubMed] [Google Scholar]
- 6. Yachida S, Vakiani E, White CM, Zhong Y, Saunders T, Morgan Ret al. Small cell and large cell neuroendocrine carcinomas of the pancreas are genetically similar and distinct from well-differentiated pancreatic neuroendocrine tumors. Am J Surg Pathol 2012;36:173–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yachida S, Wood LD, Suzuki M, Takai E, Totoki Y, Kato Met al. Genomic sequencing identifies ELF3 as a driver of ampullary carcinoma. Cancer Cell 2016;29:229–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato Met al. Genomic spectra of biliary tract cancer. Nat Genet 2015;47:1003–10. [DOI] [PubMed] [Google Scholar]
- 9. Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey Pet al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015;518:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cortes-Ciriano I, Lee JJ, Xi R, Jain D, Jung YL, Yang Let al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat Genet 2020;52:331–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Quigley DA, Dang HX, Zhao SG, Lloyd P, Aggarwal R, Alumkal JJet al. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell 2018;174:758–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rucker FG, Dolnik A, Blatte TJ, Teleanu V, Ernst A, Thol Fet al. Chromothripsis is linked to TP53 alteration, cell cycle impairment, and dismal outcome in acute myeloid leukemia with complex karyotype. Haematologica 2018;103:e17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Voronina N, Wong JKL, Hubschmann D, Hlevnjak M, Uhrig S, Heilig CEet al. The landscape of chromothripsis across adult cancer types. Nat Commun 2020;11:2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xing R, Zhou Y, Yu J, Yu Y, Nie Y, Luo Wet al. Whole-genome sequencing reveals novel tandem-duplication hotspots and a prognostic mutational signature in gastric cancer. Nat Commun 2019;10:2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong Get al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kawasaki K, Toshimitsu K, Matano M, Fujita M, Fujii M, Togasaki Ket al. An organoid biobank of neuroendocrine neoplasms enables genotype–phenotype mapping. Cell 2020;183:1420–35. [DOI] [PubMed] [Google Scholar]
- 17. Jia P, Yang X, Guo L, Liu B, Lin J, Liang Het al. MSIsensor-pro: fast, accurate, and matched-normal-sample-free detection of microsatellite instability. Genomics Proteomics Bioinformatics 2020;18:65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rheinbay E, Nielsen MM, Abascal F, Wala JA, Shapira O, Tiao Get al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature 2020;578:102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kervarrec T, Samimi M, Gaboriaud P, Gheit T, Beby-Defaux A, Houben Ret al. Detection of the Merkel cell polyomavirus in the neuroendocrine component of combined Merkel cell carcinoma. Virchows Arch 2018;472:825–37. [DOI] [PubMed] [Google Scholar]
- 20. Harms PW, Harms KL, Moore PS, DeCaprio JA, Nghiem P, Wong MKKet al. The biology and treatment of Merkel cell carcinoma: current understanding and research priorities. Nat Rev Clin Oncol 2018;15:763–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shamir ER, Devine WP, Pekmezci M, Umetsu SE, Krings G, Federman Set al. Identification of high-risk human papillomavirus and Rb/E2F pathway genomic alterations in mutually exclusive subsets of colorectal neuroendocrine carcinoma. Mod Pathol 2019;32:290–305. [DOI] [PubMed] [Google Scholar]
- 22. Poirier JT, George J, Owonikoko TK, Berns A, Brambilla E, Byers LAet al. New approaches to SCLC therapy: from the laboratory to the clinic. J Thorac Oncol 2020;15:520–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Plasschaert LW, Zilionis R, Choo-Wing R, Savova V, Knehr J, Roma Get al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 2018;560:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schutz B, Ruppert AL, Strobel O, Lazarus M, Urade Y, Buchler MWet al. Distribution pattern and molecular signature of cholinergic tuft cells in human gastro-intestinal and pancreatic-biliary tract. Sci Rep 2019;9:17466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cervantes-Ayalc A, Ruiz Esparza-Garrido R, Velazquez-Flores MA. Long interspersed nuclear elements 1 (LINE1): the chimeric transcript L1-MET and its involvement in cancer. Cancer Genet 2020;241:1–11. [DOI] [PubMed] [Google Scholar]
- 26. Miglio U, Berrino E, Panero M, Ferrero G, Coscujuela Tarrero L, Miano Vet al. The expression of LINE1-MET chimeric transcript identifies a subgroup of aggressive breast cancers. Int J Cancer 2018;143:2838–48. [DOI] [PubMed] [Google Scholar]
- 27. Oshima M, Okano K, Muraki S, Haba R, Maeba T, Suzuki Yet al. Immunohistochemically detected expression of 3 major genes (CDKN2A/p16, TP53, and SMAD4/DPC4) strongly predicts survival in patients with resectable pancreatic cancer. Ann Surg 2013;258:336–46. [DOI] [PubMed] [Google Scholar]
- 28. Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet 2002;32:128–34. [DOI] [PubMed] [Google Scholar]
- 29. Hofstatter EW, Domchek SM, Miron A, Garber J, Wang M, Componeschi Ket al. PALB2 mutations in familial breast and pancreatic cancer. Fam Cancer 2011;10:225–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cejas P, Drier Y, Dreijerink KMA, Brosens LAA, Deshpande V, Epstein CBet al. Enhancer signatures stratify and predict outcomes of non-functional pancreatic neuroendocrine tumors. Nat Med 2019;25:1260–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lawrence B, Blenkiron C, Parker K, Tsai P, Fitzgerald S, Shields Pet al. Recurrent loss of heterozygosity correlates with clinical outcome in pancreatic neuroendocrine cancer. NPJ Genom Med 2018;3:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sawicki MP, Gholkar AA, Torres JZ. Menin associates with the mitotic spindle and is important for cell division. Endocrinology 2019;160:1926–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YNet al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc Natl Acad Sci U S A 2014;111:E5593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Van Nostrand EL, Freese P, Pratt GA, Wang X, Wei X, Xiao Ret al. A large-scale binding and functional map of human RNA-binding proteins. Nature 2020;583:711–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alexandrov LB, Ju YS, Haase K, Van Loo P, Martincorena I, Nik-Zainal Set al. Mutational signatures associated with tobacco smoking in human cancer. Science 2016;354:618–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Yet al. The repertoire of mutational signatures in human cancer. Nature 2020;578:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pich O, Muinos F, Lolkema MP, Steeghs N, Gonzalez-Perez A, Lopez-Bigas N. The mutational footprints of cancer therapies. Nat Genet 2019;51:1732–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 2016;17:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tamborero D, Rubio-Perez C, Deu-Pons J, Schroeder MP, Vivancos A, Rovira Aet al. Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med 2018;10:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Popic V, Salari R, Hajirasouliha I, Kashef-Haghighi D, West RB, Batzoglou S. Fast and scalable inference of multi-sample cancer lineages. Genome Biol 2015;16:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014;30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ito T, Kudoh S, Ichimura T, Fujino K, Hassan WA, Udaka N. Small cell lung cancer, an epithelial to mesenchymal transition (EMT)-like cancer: significance of inactive Notch signaling and expression of achaete-scute complex homologue 1. Hum Cell 2017;30:1–10. [DOI] [PubMed] [Google Scholar]
- 43. Akiyama T, Shida T, Yoshitomi H, Takano S, Kagawa S, Shimizu Het al. Expression of sex determining region Y-box 2 and pancreatic and duodenal homeobox 1 in pancreatic neuroendocrine tumors. Pancreas 2016;45:522–7. [DOI] [PubMed] [Google Scholar]
- 44. Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat Neurosci 2003;6:1162–8. [DOI] [PubMed] [Google Scholar]
- 45. Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen CCet al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 2017;355:84–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harold A, Amako Y, Hachisuka J, Bai Y, Li MY, Kubat Let al. Conversion of Sox2-dependent Merkel cell carcinoma to a differentiated neuron-like phenotype by T antigen inhibition. Proc Natl Acad Sci U S A 2019;116:20104–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Smith J, Sen S, Weeks RJ, Eccles MR, Chatterjee A. Promoter DNA hypermethylation and paradoxical gene activation. Trends Cancer 2020;6:392–406. [DOI] [PubMed] [Google Scholar]
- 48. Scardoni M, Vittoria E, Volante M, Rusev B, Bersani S, Mafficini Aet al. Mixed adenoneuroendocrine carcinomas of the gastrointestinal tract: targeted next-generation sequencing suggests a monoclonal origin of the two components. Neuroendocrinology 2014;100:310–6. [DOI] [PubMed] [Google Scholar]
- 49. Watanabe K, Kage H, Shinozaki-Ushiku A, Kohsaka S, Takai D, Nakajima Jet al. Spontaneous transdifferentiation from small cell lung carcinoma to squamous cell carcinoma. J Thorac Oncol 2019;14:e31–e4. [DOI] [PubMed] [Google Scholar]
- 50. Calbo J, van Montfort E, Proost N, van Drunen E, Beverloo HB, Meuwissen Ret al. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell 2011;19:244–56. [DOI] [PubMed] [Google Scholar]
- 51. Au-Yeung G, Lang F, Azar WJ, Mitchell C, Jarman KE, Lackovic Ket al. Selective targeting of cyclin E1-amplified high-grade serous ovarian cancer by cyclin-dependent kinase 2 and AKT inhibition. Clin Cancer Res 2017;23:1862–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. da Costa A, Baiocchi G. Genomic profiling of platinum-resistant ovarian cancer: the road into druggable targets. Semin Cancer Biol 2021;77:29–41. [DOI] [PubMed] [Google Scholar]
- 53. Tang LH, Basturk O, Sue JJ, Klimstra DS. A practical approach to the classification of WHO Grade 3 (G3) well-differentiated neuroendocrine tumor (WD-NET) and poorly differentiated neuroendocrine carcinoma (PD-NEC) of the pancreas. Am J Surg Pathol 2016;40:1192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. La Rosa S, Adsay V, Albarello L, Asioli S, Casnedi S, Franzi Fet al. Clinicopathologic study of 62 acinar cell carcinomas of the pancreas: insights into the morphology and immunophenotype and search for prognostic markers. Am J Surg Pathol 2012;36:1782–95. [DOI] [PubMed] [Google Scholar]
- 55. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer Net al. The sequence alignment/map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine Aet al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;43:11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Landrum MJ, Chitipiralla S, Brown GR, Chen C, Gu B, Hart Jet al. ClinVar: improvements to accessing data. Nucleic Acids Res 2020;48:D835–D44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HMet al. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell Tet al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet 2013;9:e1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nagasaki M, Yasuda J, Katsuoka F, Nariai N, Kojima K, Kawai Yet al. Rare variant discovery by deep whole-genome sequencing of 1,070 Japanese individuals. Nat Commun 2015;6:8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Totoki Y, Tatsuno K, Covington KR, Ueda H, Creighton CJ, Kato Met al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet 2014;46:1267–73. [DOI] [PubMed] [Google Scholar]
- 63. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 1995;57:289–300. [Google Scholar]
- 64. Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 2013;3:246–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Totoki Y, Yoshida A, Hosoda F, Nakamura H, Hama N, Ogura Ket al. Unique mutation portraits and frequent COL2A1 gene alteration in chondrosarcoma. Genome Res 2014;24:1411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016;44:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha Set al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tian Y, Morris TJ, Webster AP, Yang Z, Beck S, Feber Aet al. ChAMP: updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics 2017;33:3982–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna Set al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 2017;14:959–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Corces MR, Granja JM, Shams S, Louie BH, Seoane JA, Zhou Wet al. The chromatin accessibility landscape of primary human cancers. Science 2018;362:eaav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods 2017;14:975–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Inagawa Y, Yamada K, Yugawa T, Ohno S, Hiraoka N, Esaki Met al. A human cancer xenograft model utilizing normal pancreatic duct epithelial cells conditionally transformed with defined oncogenes. Carcinogenesis 2014;35:1840–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data reported in this paper has been deposited in the National Bioscience Database Center with the accession number JGAS000359.



![Figure 1. Genomic alterations of GIS-NECs. A, Landscape of genomic alterations in GIS-NEC cases. The left oncoplot indicates WGS data, and representative gene expression data are obtained from frozen samples. The cases are arranged from left to right according to descending order of the number of SVs in each primary organ. Asterisks, organoid samples. The right oncoplot shows WES data in patients differing from patients available on the WGS data. B, Hematoxylin and eosin (H&E) staining and synaptophysin immunolabeling of TP53 and RB1 double knockout (TR-KO) organoids before and after blocking Notch signaling with a γ-secretase inhibitor (DAPT). The synaptophysin-positive cells were increased by the Notch inhibitor. Scale bar, 100 μm. C, Unsupervised hierarchical cluster analysis with 2,000 high variant probes for DNA methylation in GIS-NECs. D, Integration of RNA-seq and DNA methylation array data comparing GIS-NECs with normal tissues. RNA-seq data were filtered using significant differentially expressed gene (DEG; abs [log2FC] ≥ 1) with significant FDR values (<0.05). DNA methylation assay data were filtered using differentially methylated regions (DMR; abs [Δβ value] ≥ 0.1) with significant adjusted P values (<0.05). In the area of the figure showing high levels of gene expression and hypermethylation, 199 DMRs are situated, of which 39 (19.6%) are transcription factors (TF; red dots) including SOX2 and ASCL1. In contrast, in the area of the figure showing high levels of gene expression and hypomethylation, 424 DMRs are situated, of which 28 (6.6%) are TFs. CHGA, chromogranin A. E, Schematic of NET–AKR fusion genes detected in two gastric NECs. The neuroepithelioma transforming gene1 (NET1) is a specific guanine nucleotide exchange factor for RhoA. Both aldo-keto reductase family 1 members C3 (AKR1C3) and C4 (AKR1C4) are reductase enzymes that play critical roles in the biotransformation of endogenous substrates such as steroids. The chimeric genes demonstrate in-frame fusion of the NET1 amino terminus (exons 1–3) and the AKR1C3 carboxyl terminus (exons 2–9) or the AKR1C4 carboxyl terminus (exons 6–9). NLS, nuclear localization signal; DH, Dbl homology; PH, pleckstrin homology; PDZ, post-synaptic density 95; aa, amino acids. F, Gastric NEC with the Merkel cell polyomavirus (MCPyV; case NE002). The read depth along the polyomavirus genome is shown in blue, and read pairs bridging the polyomavirus genome and the integration site on chromosome 8 are indicated by red lines. Polyomavirus genes are indicated by large T antigen, small T Antigen, VP1, VP2, and VP3.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/c657/9394397/8e0be143173e/692fig1.jpg)