Activating MET tyrosine kinase domain mutations were detected across diverse cancer types and showed sensitivity to several MET inhibitors, suggesting that this newly characterized molecular cancer subgroup can be a targetable oncogenic driver.
Abstract
Activating point mutations in the MET tyrosine kinase domain (TKD) are oncogenic in a subset of papillary renal cell carcinomas. Here, using comprehensive genomic profiling among >600,000 patients, we identify activating MET TKD point mutations as putative oncogenic driver across diverse cancers, with a frequency of ∼0.5%. The most common mutations in the MET TKD defined as oncogenic or likely oncogenic according to OncoKB resulted in amino acid substitutions at positions H1094, L1195, F1200, D1228, Y1230, M1250, and others. Preclinical modeling of these alterations confirmed their oncogenic potential and also demonstrated differential patterns of sensitivity to type I and type II MET inhibitors. Two patients with metastatic lung adenocarcinoma harboring MET TKD mutations (H1094Y, F1200I) and no other known oncogenic drivers achieved confirmed partial responses to a type I MET inhibitor. Activating MET TKD mutations occur in multiple malignancies and may confer clinical sensitivity to currently available MET inhibitors.
Significance: The identification of targetable genomic subsets of cancer has revolutionized precision oncology and offers patients treatments with more selective and effective agents. Here, we demonstrate that activating, oncogenic MET tyrosine kinase domain mutations are found across a diversity of cancer types and are responsive to MET tyrosine kinase inhibitors.
Introduction
The discovery of new targetable genomic alterations continues to advance the field of precision oncology across cancer types. Dysregulation of MET gene signaling can occur through a variety of molecular mechanisms, which can be targeted using novel therapeutic strategies (1–3). MET exon 14 skipping (METex14) alterations, which occur in ∼3% of non–small cell lung cancers (NSCLC) and lead to impaired degradation of the MET receptor, are targetable with MET tyrosine kinase inhibitors (TKI) such as capmatinib and tepotinib, and may also be sensitive to the EGFR:MET bispecific antibody amivantamab (1, 4). Variants in the extracellular SEMA domain of MET have also been reported to have tumorigenic potential in lung cancers (5, 6). MET gene amplification has been found in NSCLC, gastric cancer, esophageal cancer, and other malignancies and is responsive to treatment with MET TKIs. MET gene fusions have been described in NSCLC, glioblastomas, and other cancers and can be successfully targeted with MET TKIs (7–9). Furthermore, MET protein overexpression is also a therapeutic target for which MET antibody–drug conjugates are being developed, such as telisotuzumab vedotin (1, 4, 10).
In addition to these molecular alterations, activating point mutations in the MET tyrosine kinase domain (TKD) have been reported in 13% to 20% of type 1 papillary renal cell carcinoma (PRCC), as either sporadic or germline mutations, and may confer sensitivity to multikinase inhibitors with activity against MET (11, 12). Somatic MET mutations Y1230C and Y1235H have been also identified in head and neck squamous cell carcinomas (13). We sought to perform a comprehensive pan-cancer analysis of malignancies with MET TKD mutations, characterize their oncogenic potential, identify their sensitivity pattern to MET TKIs, and determine whether these alterations were clinically actionable.
Results
Type and Frequency of MET TKD Mutations Across Cancer Types
The prevalence of MET TKD mutations, located between amino acids p.V1078 and p.I1345 (NM_000245.2, variant 2), across cancer types was established in two independent cohorts: cohort #1, totaling 145,949 unique patients with cancer (Supplementary Table S1), and cohort #2, totaling 459,339 unique patients with cancer. Because NSCLC with METex14 alterations can develop MET TKD mutations at the time of acquired resistance to MET TKIs (14), we excluded cases with MET TKD mutations and concurrent METex14 alterations in order to focus on MET TKD mutations as a de novo oncogenic driver event. The prevalence of MET TKD mutations without concurrent METex14 alterations in each cohort was approximately 0.5% (Supplementary Fig. S1).
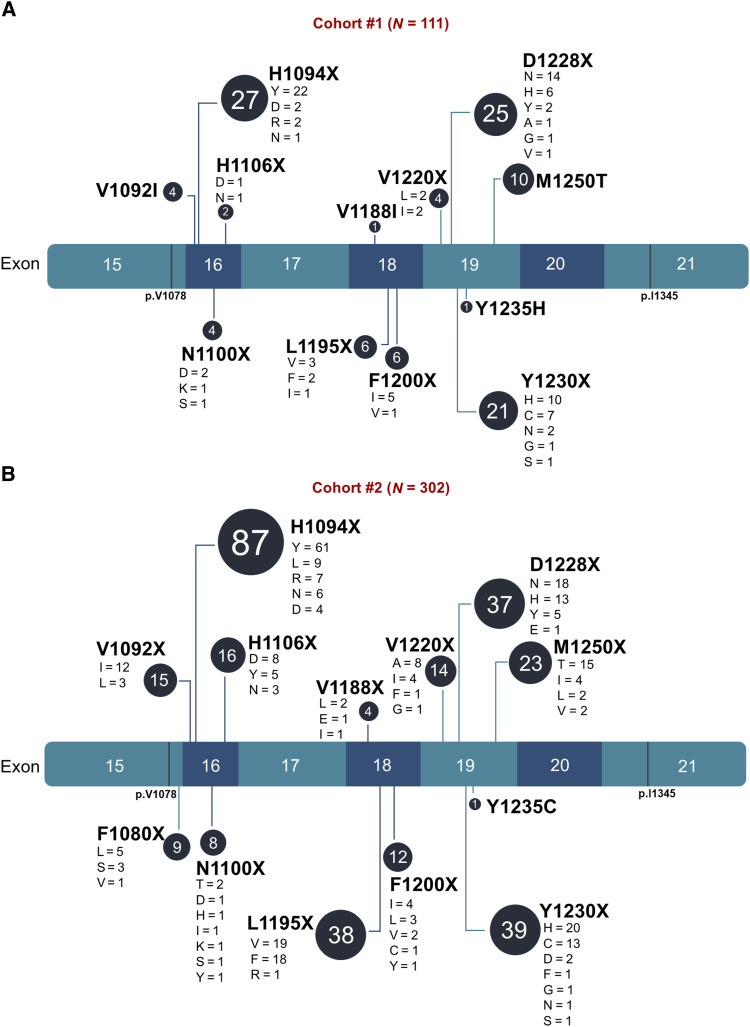
In cohort #1, a total of 711 MET TKD mutations were detected in the absence of METex14 alterations in 664 (0.45%) of 145,949 unique patients with cancer; 83% of these mutations (N = 590/711) were missense, and 16% (N = 111/711) were classified as oncogenic/likely oncogenic according to OncoKB annotation (Supplementary Fig. S2A and S2B). The distribution and frequency of mutations along the MET kinase domain in cohort #1 are shown in Supplementary Fig. S2C and detailed in Supplementary Table S2. The most commonly mutated residue was R1170 (representing 6.9% of the total mutation count), and the oncogenic/likely oncogenic MET TKD mutations included H1094X, D1228X, Y1230X, M1250T, L1195X, F1200X, V1092I, N1100X, V1220X, H1106X, V1188I, and Y1235H, listed in order of decreasing frequency (Fig. 1A).
Figure 1.
Oncogenic/likely oncogenic MET TKD mutations detected in pan-cancer cohorts. A, Lollipop plot of the oncogenic/likely oncogenic MET TKD mutations according to OncoKB in cohort #1 (N = 111 MET TKD mutations in 105 unique patients). B, Lollipop plot of the oncogenic/likely oncogenic MET TKD mutations according to OncoKB in cohort #2 (N = 302 MET TKD mutations in 300 unique patients).
In cohort #2, 2,559 MET TKD mutations without concurrent METex14 alterations were detected in 2,477 (0.54%) of 459,339 unique patients with cancer; 85% (N = 2,157/2,559) were missense, and 12% (N = 302/2,559) were classified as oncogenic/likely oncogenic (Supplementary Fig. S2D and S2E). The distribution and frequency of mutations along the MET kinase domain in cohort #2 are shown in Supplementary Fig. S2F and detailed in Supplementary Table S3. As in cohort #1, the most commonly mutated residue was R1170 (representing 4.9% of the total mutation count), and the oncogenic/likely oncogenic MET TKD mutations detected included H1094X, Y1230X, L1195X, D1228X, M1250X, H1106X, V1092X, V1220X, F1200X, F1080X, N1100X, V1188X, and Y1235C, listed in order of decreasing frequency (Fig. 1B).
Characterization of MET TKD Mutations with Unknown Biological Function
To investigate the pathogenicity of missense MET TKD mutations with unknown significance in OncoKB, we used three in silico tools for scoring the effect of a specific amino acid substitution: PolyPhen-2, SIFT, and pathogenic 3D feature index for kinase (P3DFikinase). In cohort #1, among unique MET TKD mutations with unknown significance in OncoKB (N = 275), 43% (N = 119) showed concordance for likely pathogenicity across all three scores (PolyPhen value > 0.908, SIFT < 0.05, and P3DFikinase ≥1: categorized as “probably pathogenic”). A total of 7% (N = 18) were considered to be benign variants across all three scores (PolyPhen value ≤ 0.446, SIFT ≥ 0.05, and P3DFikinase ≤ 0: categorized as “probably benign”). The remaining 50% (N = 137) had discordant scoring for pathogenicity (categorized as “uncertain”). Similarly, in cohort #2, among unique MET TKD mutations with unknown significance in OncoKB (N = 675), 45% (N = 307) were “probably pathogenic,” 7% (N = 46) were “probably benign,” and 48% (N = 322) were “uncertain.” The list of missense mutations with unknown biologic function and their pathogenicity scores using all three tools for cohorts #1 and #2 are summarized in Supplementary Table S4 and Supplementary Table S5, respectively. Merging the two cohorts and considering unique missense mutations with unknown significance in OncoKB (N = 765), 344 were categorized as “probably pathogenic” and 52 as “probably benign”; 369 were categorized as “uncertain”. Next, to provide an overview of the three-dimensional amino acid positions characterizing “probably pathogenic” mutations that could help in understanding their impact on protein structure, we investigated whether “probably pathogenic” and “probably benign” MET TKD mutations were characterized by different structural features. “Probably pathogenic” mutations were enriched in α-helices [odds ratio (OR) = 10.45; P < 0.0001; Supplementary Fig. S3A], had a lower relative solvent accessible surface area (RSA) [median: 0.03 (0–0.74) vs. 0.53 (0.20–0.91); P < 0.001; Supplementary Fig. S3B], and were more associated with substitutions of hydrophobic and aliphatic (OR = 2.44; P = 0.01) and special (OR = 4.58; P = 0.001) amino acids (Supplementary Fig. S3C).
Distribution and Characterization of MET TKD Mutations by Cancer Type
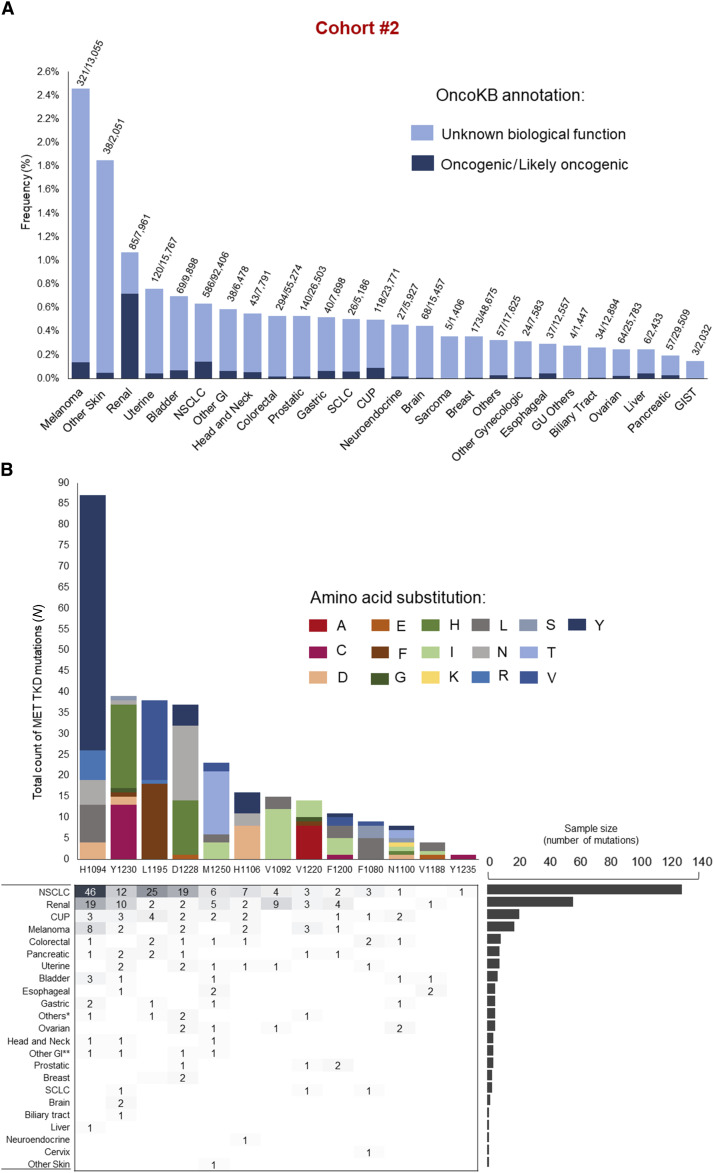
The distribution of MET TKD mutations (without concurrent METex14 alterations) varied across cancer types (Supplementary Fig. S4A and S4B). In cohort #1, a total of 105 cancer cases harbored MET TKD mutations classified as oncogenic/likely oncogenic; the most frequent cancer types were renal cell carcinoma (N = 34/3,226; 1.05%), followed by NSCLC (N = 44/23,195; 0.2%), cancer of unknown primary (CUP; N = 2/1,184; 0.2%) and melanoma (N = 7/5,916; 0.12%), as shown in Supplementary Tables S6 and S7; the frequency of cases with MET TKD mutations in each individual dataset from cohort #1 is indicated in Supplementary Fig. S5. Cohort #2 showed a similar incidence of mutations across cancer types (Fig. 2A and B), with a total of 300 cases harboring oncogenic/likely oncogenic MET TKD mutations; similarly to cohort #1, the most frequent cancer types were renal cell carcinoma (N = 57/7,961; 0.72%), followed by NSCLC (N = 129/92,406; 0.14%) and melanoma (N = 18/13,055; 0.14%), as shown in Supplementary Tables S8 and S9.
Figure 2.
Type and frequency of MET TKD mutations across cancer types in cohort #2. A, Frequency of MET TKD mutations in various cancer types in cohort #2 according to OncoKB annotation; the numbers above each bar indicate the number of cases with MET TKD mutations (regardless of OncoKB annotation) out of the total of cases of each cancer type. B, Detailed oncogenic/likely oncogenic MET TKD mutations (N = 302 MET TKD mutations in 300 unique patients) according to cancer type in cohort #2. GI, gastrointestinal cancers; SCLC, small cell lung cancer; CUP, cancer of unknown primary; GU, genitourinary; GIST, gastrointestinal stromal tumors. *Others: endocrine tumor (N = 1), lung underspecified (N = 2), salivary gland carcinoma (N = 1), adenoid cystic carcinoma (N = 1); **Other GI: small intestine (N = 3), anus (N = 1).
MET TKD Mutations in NSCLC
Among 23,195 patients with NSCLC in cohort #1, 123 MET TKD mutations without concurrent METex14 alterations were identified in 120 (0.52%) patients (Supplementary Fig. S6A). Of these, 47 MET TKD mutations in 44 patients (0.19%) were considered to be oncogenic/likely oncogenic, including D1228N/H/G/V/Y (N = 16), H1094Y/D/R (N = 13), Y1230H/C/N (N = 7), and others (Supplementary Fig. S6B and S6C). In cohort #2, among 92,406 patients with NSCLC, 590 MET TKD mutations without concurrent METex14 alterations were identified in 586 (0.63%) patients (Supplementary Fig. S6D). Of these, 129 patients (0.14%) had NSCLC with an oncogenic/likely oncogenic MET TKD mutation, including H1094Y/N/D/L/R (N = 46), L1195V/F (N = 25), and D1228N/H/Y (N = 19), and others (Supplementary Fig. S6E and S6F).
Among the 44 patients with NSCLC harboring oncogenic/likely oncogenic MET TKD mutations in cohort #1, the median age was 63 (range: 30–89), 61% (N = 27) were female, 66% (N = 27) were white, and 90% (N = 39) had adenocarcinoma histology (Supplementary Table S10). MET TKD mutation was the sole oncogenic driver detected in 46% of cases (N = 20/44) and the remaining 55% (N = 24/44) of cases had a concurrent driver alteration in another gene, including EGFR (23%), KRAS (18%), and others (Supplementary Fig. S7A). Among cases with available MET copy number status, concurrent MET amplification was present in 30% (N = 12/40) of cases (Supplementary Fig. S7B). Detailed genomic characteristics for each case are shown in Supplementary Table S11. The median variant allele frequency (VAF) of oncogenic/likely oncogenic MET TKD mutations for NSCLC cases was 26% (range: 1.6%–79%) for Dana-Farber Cancer Institute (DFCI) cases and 21% (range: 0.02%–75%) for Memorial Sloan–Kettering Cancer Center (MSKCC) cases.
Twenty-nine cases from DFCI and MSKCC in cohort #1 had clinicopathological and treatment history annotations. Among these, 13 cases showed oncogenic/likely oncogenic MET TKD mutations as a de novo genomic event in NSCLC in treatment-naïve patients: in seven cases MET TKD mutations were detected in the absence of other driver alterations; one case harbored a MET TKD mutation with concurrent MET amplification; four cases showed a MET TKD mutation along with an oncogenic KRAS mutation (two with KRAS G12C, one with G12A, and one with Q61H), as shown in Table 1.
Table 1.
Clinicopathologic characteristics of patients with NSCLC harboring oncogenic/likely oncogenic MET tyrosine kinase domain mutations as a de novo genomic event from DFCI and MSKCC in cohort #1.
| Pt ID | MET TKD mutation | Concurrent MET amplification | Sex | Race | Age at diagnosis | Smoking status | Pack-years | Stage at diagnosis | Stage at MET TKD mutation detection | Histology | PD-L1 TPS | TMB (mut/Mb) | Concurrent driver | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MET TKD mutation as a sole driver | #21 | H1106D | No | F | W | 34 | Fo | 10 | IIB | IIB | Poorly Differentiated | NA | 23.0 | No |
| #22 | V1188I | No | F | W | 86 | Fo | 7 | IIIA | IIIA | LUAD | NA | 5.3 | No | |
| #23 | V1220I | No | M | W | 70 | Fo | 30 | IV | IV | LUAD | 10 | 9.0 | No | |
| #35 | H1094Y | No | F | W | 57 | Fo | 20 | IIA | IIA | LUAD | NA | 55.0 | No | |
| #39 | H1094Y | No | M | W | 72 | Fo | NA | IV | IV | Poorly Differentiated | 90% | 16.5 | No | |
| #42 | N1100S | No | M | W | 81 | Fo | 15 | IIB | IV | LUAD | NA | 7.0 | No | |
| #41 | L1195V | No | M | W | 88 | Fo | 80 | IV | IV | LUAD | 0% | 13.0 | No | |
| #16 | D1228H | Yes | F | W | 44 | Fo | 60 | IB | IV | SCC | 1 | 17.5 | No | |
| #20 | F1200I | No | M | W | 78 | Fo | 25 | IV | IV | LUAD | 90% | 14.0 | No | |
|
MET TKD mutation with a co-occurring driver |
#24 | V1220L | No | F | W | 70 | Fo | 10 | IB | IV | LUAD | 0 | 13.7 | KRAS G12C |
| #27 | D1228H | No | F | W | 74 | Fo | 30 | IV | IV | LUAD | 50% | 8.8 | KRAS G12C | |
| #38 | H1094Y | No | F | W | 78 | Fo | 35 | NA | IV | LUAD | 100% | 3.5 | KRAS G12A | |
| #33 | H1094R | No | F | W | 80 | Fo | 23 | IIIB | IIIB | LUAD | 70 | 19.8 | KRAS Q61H |
Abbreviations: F, female; Fo, former; LUAD, lung adenocarcinoma; M, male; NA, not available.; patient, identification; Pt, ID; SCC, squamous cell carcinoma; TKD, tyrosine kinase domain; TMB, tumor mutational burden; TPS, tumor proportion score; W, white.
In the remaining 16 cases, MET TKD mutations were acquired as a putative resistance mechanism to other targeted therapies: eight cases as an on-target resistance mechanism after treatment with MET TKIs; seven cases as a bypass mechanism of resistance in EGFR-mutant (N = 5) or ROS1-rearranged (N = 2) NSCLC after treatment with EGFR or ROS1 TKIs, respectively; one MET TKD mutation co-occurred with a KLC4-ALK fusion but next-generation sequencing (NGS) analysis before treatment with an ALK TKI was not available.
Among 129 patients with MET TKD-mutant NSCLC from cohort #2, 57% (N = 73) were male, 74% (N = 96) had European ancestry, and 67% (N = 86) had adenocarcinoma histology (Supplementary Table S12). Oncogenic/likely oncogenic MET TKD mutations were found as a sole oncogenic driver in 60% (N = 78) of cases; the two most common mutations were at residues H1094X (N = 25) and L1195X (N = 18) (Supplementary Table S13). In the other 40% (N = 51) of cases, concurrent KRAS- (N = 33), EGFR- (N = 11), ROS1- (N = 3), BRAF- (N = 2), and ALK- (N = 2) activating alterations were detected (Supplementary Fig. S7C). Concurrent MET amplification was found in 10% (N = 13) of cases (Supplementary Fig. S7D); among them, a concurrent oncogenic alteration was found in five cases (Supplementary Table S14). In 25 cases (19%, N = 25/129), point mutations at residue H1094 were detected without concurrent oncogenic drivers (most commonly H1094Y, N = 17), followed by L1195 [14%, N = 18, most commonly L1195F (N = 9) and L1195V (N = 9)], as shown in Supplementary Fig. S8 and listed in Supplementary Table S13. In this cohort, the median VAF of oncogenic/likely oncogenic MET TKD mutations was 16% (range: 1.3%–82%) in tissue samples (N = 119) and 0.87% (range: 0.15%–10%) in liquid biopsies (N = 10).
Comparing NSCLC with oncogenic/likely oncogenic de novo MET TKD mutations (N = 13) to those with METex14 alterations available from the DFCI dataset (N = 123) in cohort #1 (Supplementary Table S15), there was no difference in age, sex, race, PD-L1 tumor proportion score (TPS), or concurrent MET amplification status. NSCLCs harboring oncogenic/likely oncogenic MET TKD mutations had a higher tumor mutational burden (TMB) z score compared to METex14-altered cases (P < 0.001). In cohort #2, compared to METex14-altered NSCLC, patients with MET TKD-mutant NSCLC without other oncogenic drivers were younger (P < 0.0001), more often male (P < 0.0001), and had a higher TMB (median 9.8 vs. 3.8 mut/Mb, P < 0.0001). Differences in ancestry were also observed (P = 0.005). There was no difference in sex, PD-L1 TPS, or concurrent MET amplification status (Supplementary Table S16).
Considering the genomic landscape of NSCLC harboring oncogenic/likely oncogenic MET TKD mutations as a sole oncogenic driver in cohort #2, the most commonly mutated genes included TP53 (77%), CDKN2A (35%), CDKN2B (21%) (Supplementary Fig. S9A); 50% (N = 39/78) of cases had a TMB <10 mut/Mb (Supplementary Fig. S9B). When comparing the genomic profile of MET TKD-mutant cases (N = 78) to METex14-altered cases (N = 2,036), NSCLC with MET TKD mutations was significantly enriched in alterations in TP53, KEAP1, TERT, and NFE2L2 (q-value < 0.05; Supplementary Fig. S9C).
Last, we queried a separate third cohort of NSCLC from Caris Life Sciences, where MET TKD mutations were detected in 0.66% (N = 280/42,328) of cases (Supplementary Fig. S10A) and 0.08% (N = 35/42,328) of patients had a NSCLC harboring an oncogenic/likely oncogenic MET TKD mutation (Supplementary Fig. S10B). Median VAF for oncogenic/likely oncogenic MET TKD mutations in tumor tissue was 16% (range: 7%–68%). The most common oncogenic/likely oncogenic MET TKD mutations were H1094Y (N = 11) and D1228N (N = 11; Supplementary Fig. S10C). Among cases tested for concurrent actionable genomic drivers (N = 24/35), 62.5% (N = 15/24) had MET TKD mutations in the absence of additional driver alterations (Supplementary Fig. S10D); concurrent MET amplification was detected in 6% of cases (2 out of 33 cases with available data; Supplementary Fig. S10E). Demographic features, TMB, and PD-L1 of NSCLCs with MET TKD mutations as a sole oncogenic driver compared to those with METex14 alterations are summarized in Supplementary Table S17: similar to cohort #2, patients with MET TKD-mutant NSCLC were younger (P = 0.006), with tumors characterized by higher TMB (P < 0.0001) compared to those with METex14-altered NSCLC.
MET TKD Mutations in Malignancies Other Than NSCLC
In cohort #1, 61 patients had a malignancy other than NSCLC harboring an oncogenic/likely oncogenic MET TKD mutation. The median age was 63 years old (range: <18–84), 66% (N = 40) of patients were male, and 85% (N = 44) were white; concurrent MET amplification was detected in 4% (N = 2) of cases. MET TKD mutations were most common in renal cell carcinoma (N = 34, 56%), with a high prevalence in PRCC subtype (N = 29 PRCCs among 34 renal cell carcinomas), followed by melanoma (N = 7, 12%). Clinicopathological and genomic characteristics of these cases are summarized in Table 2 with details of each case described in Supplementary Table S18. Additional detailed clinicopathologic and genomic characteristics of the RCC cases in cohort #1 are summarized in Supplementary Table S19.
Table 2.
Clinicopathologic and genomic characteristics of 61 cases of cancers other than NSCLC harboring oncogenic/likely oncogenic MET tyrosine kinase domain mutations in cohort #1.
| Characteristics | Total (N = 61) |
|---|---|
| Age | |
| Median (range) | 63 (<18–84) |
| Sex | |
| Female | 21 (34%) |
| Male | 40 (66%) |
| Race | |
| White | 44 (85%) |
| Asian | 2 (4%) |
| Black | 6 (11%) |
| Other/unknown | 9 |
| Tumor type | |
| Renal cell carcinomaa | 34 (56%) |
| Melanoma | 7 (12%) |
| Uterine endometrioid carcinoma | 4 (6%) |
| Cholangiocarcinoma | 2 (3.3%) |
| Colon adenocarcinoma | 1 (1.6%) |
| CUP | 2 (3.3%) |
| Esophageal adenocarcinoma | 2 (3.3%) |
| Ovarian cancer | 2 (3.3%) |
| Bladder urothelial carcinoma | 1 (1.6%) |
| Stomach adenocarcinoma | 1 (1.6%) |
| GIST | 1 (1.6%) |
| Hepatocellular carcinoma | 1 (1.6%) |
| Adenocarcinoma NOS | 1 (1.6%) |
| Pancreatic adenocarcinoma | 1 (1.6%) |
| Sarcoma | 1 (1.6%) |
| Stage of disease reported at MET TKD mutation detection | |
| I | 16 (45%) |
| II | 2 (6.5%) |
| III | 5 (14.5%) |
| IV | 12 (34%) |
| NA | 25 |
| MSI genotype | |
| Deficient | 3 (9%) |
| Proficient | 30 (91%) |
| NA | 28 |
| Concurrent MET amplification | |
| Yes | 2 (4%) |
| No | 49 (96%) |
| NA | 10 |
Abbreviations: CUP, cancer of unknown primary; GIST, gastrointestinal stromal tumors; MSI, microsatellite instability; NA, not available; NOS, not otherwise specified.
85% (N = 29/34) of renal cancer were papillary renal cell carcinoma (Others: three renal cell carcinoma, two renal clear cell carcinoma).
In cohort #2, MET TKD mutations were identified in 171 cases of malignancies other than NSCLC, as summarized in Supplementary Table S20. The median age of patients was 63 years old (range: 35–89+), 71% (N = 121) were male. The most common tumor type was renal cancer (33%, N = 57), notably PRCC (N = 33/57), followed by CUP (12%, N = 21), and melanoma (11%, N = 18). The median TMB was 5 mut/Mb, and 65% (N = 111/171) of cases had a TMB < 10 mut/Mb. Concurrent MET amplification was detected in 4.1% (N = 7) of cases. Additional detailed clinicopathologic and genomic characteristics of the RCC cases in cohort #2 are summarized in Supplementary Table S21.
Examining the frequency of oncogenic/likely oncogenic MET TKD mutations in RCC compared to NSCLC, we observed an enrichment of D1228X (cohorts #1 and #2) and L1195X (cohort #2) mutations in NSCLC and an enrichment of M1250X (cohort #1) and V1092X (cohort #2) mutations in RCC. The frequencies of H1094X mutations were similar in RCC and NSCLC (Supplementary Tables S22 and S23).
Antitumor Activity of the MET Inhibitor Elzovantinib (TPX-0022) in Two Patients with MET TKD-Mutant Lung Adenocarcinoma
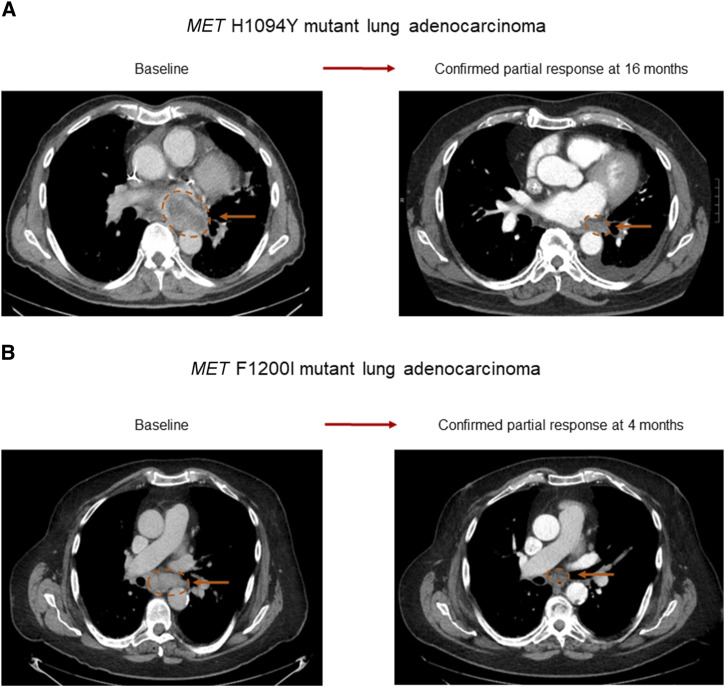
Hypothesizing that lung cancers with de novo MET TKD mutations lacking other oncogenic driver alterations could be clinically targeted with MET TKIs, we enrolled two patients with MET TKD-mutant lung adenocarcinoma in a clinical trial (NCT03993873) with the MET inhibitor elzovantinib (TPX-0022). The first patient was a 64-year-old man with a 5 pack-year history of former tobacco use who presented with metastatic lung adenocarcinoma with a PD-L1 TPS of 60%. Genomic analysis of a tissue biopsy of the left lung mass at diagnosis showed a MET H1094Y mutation and no other driver alterations in KRAS, EGFR, ALK, ROS1, BRAF, HER2, RET, or NTRK; there were no MET amplification and no METex14 alteration. The patient was initially treated with first-line carboplatin, pemetrexed, and pembrolizumab, and after an initial partial response, his cancer progressed within 9 months. A repeat biopsy of a mesenteric lymph node, obtained for clinical trial eligibility purposes, underwent NGS (FoundationOne CDx) and confirmed the presence of the MET H1094Y mutation, and the lack of other oncogenic drivers; the cancer was microsatellite stable and had a TMB of 3 mut/Mb. The patient experienced a confirmed partial response to elzovantinib, with the sum of diameters of target lesions decreasing by −67.7% at maximal response (Fig. 3A) and a duration of response of 21 months. At the time of disease progression, a repeat lung biopsy showed the MET H1094Y mutation and high-level EGFR gene amplification (52 copies), which was not present on his pre-elzovantinib tumor sample.
Figure 3.
Antitumor activity of MET inhibitor in two patients with NSCLC harboring MET TKD mutations. Confirmed partial response achieved with the type Ib MET inhibitor elzovantinib (TPX-0022) in A a 64-year-old man with MET H1094Y mutant lung adenocarcinoma and in B an 80-year-old man with MET F1200I mutant lung adenocarcinoma.
The second patient was an 80-year-old man with a 25 pack-year history of prior tobacco use, who was diagnosed with metastatic lung adenocarcinoma with a PD-L1 TPS of 90%, and no known targetable genomic alterations at initial diagnosis; liquid biopsy by Guardant360 CDx showed a MET F1200I mutation and no other oncogenic driver alterations. He initially responded to first-line carboplatin, pemetrexed, and pembrolizumab but experienced disease progression after 6 months. A repeat biopsy of the right upper lung lobe sequenced via the Dana-Farber OncoPanel platform showed a MET F1200I mutation and low copy number gain of MET (five copies). He experienced a confirmed partial response to elzovantinib, with a maximal reduction in target lesions of −51.4% (Fig. 3B). After 6 months of treatment, the patient was hospitalized with a bacterial pneumonia and due to clinical deterioration, elzovantinib was discontinued.
MET TKD Mutations Activate the MET Signaling Pathway and Induce Transformation of Ba/F3 Cells
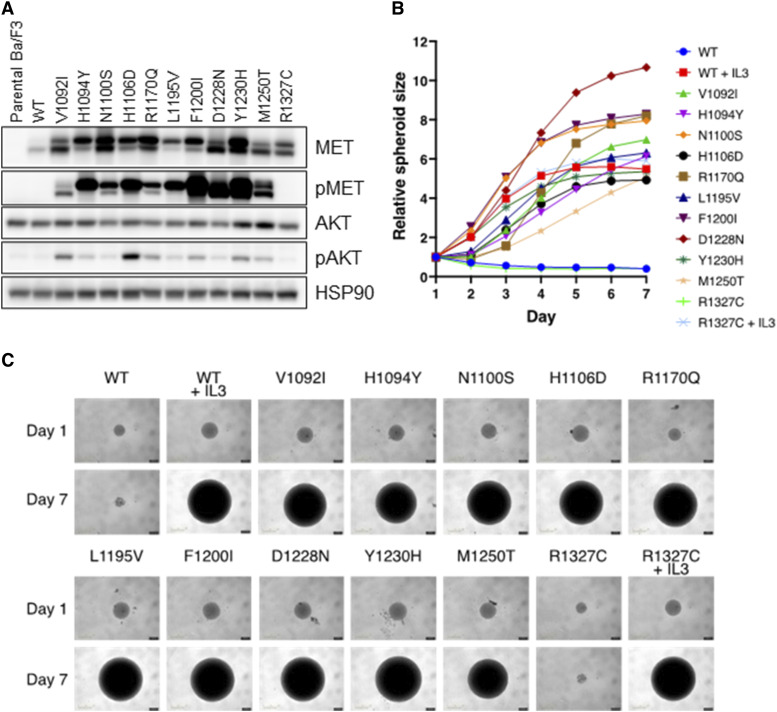
We further investigated in vitro the oncogenicity of the MET TKD mutations that were most commonly detected in our cohorts. We not only focused on the oncogenic/likely oncogenic MET TKD mutations but also included those of unknown significance, categorized as “probably pathogenic,” “uncertain,” and “probably benign” in our in silico scoring. For the oncogenic/likely oncogenic mutations, we included V1092I, H1094Y, N1100S, H1106D, L1195V, F1200I, D1228N, Y1230H, and M1250T. H1094Y and F1200I were also the mutations detected in patients with NSCLC who responded to the MET TKI elzovantinib. R1170Q was the most frequent mutation of unknown significance in cohort #1 and categorized as “probably pathogenic” in silico; whereas R1327C was the second most frequent mutation and categorized as “uncertain” in silico. We also analyzed the four most frequent mutations categorized as “probably benign” in cohort #1, specifically N1081S, G1102D, M1229V, and I1345T. MET TKD mutations were transduced in murine pro-B Ba/F3 cells; Ba/F3 cells require interleukin-3 (IL-3) for growth but can become IL-3 independent when transformed by oncogenes. All transduced MET mutant constructs resulted in constitutive phosphorylation of MET except for MET wild-type (WT), R1327C, and mutations which were classified as “probably benign” in silico (Fig. 4A; Supplementary Fig. S11A), suggesting that these MET TKD mutations are activating mutations. In addition, MET receptor phosphorylation status was coupled with the downstream activation of AKT, further corroborating oncogene dependence in cellular signaling. Phenotypically, Ba/F3 cells displaying activation of MET and AKT showed IL-3-independent cell proliferation as shown by spheroid assay (Fig. 4B and C; Supplementary Fig. S11B and S11C). Together, these signaling and functional studies support the conclusion that all tested MET TKD mutations, except R1327C and the “probably benign” mutations, are oncogenically activating in the MET WT genetic background, that is, in the absence of METex14 alteration.
Figure 4.
In vitro analysis of oncogenic potential of MET TKD mutations in Ba/F3 cells. A, Phosphorylation of MET and AKT was analyzed in Ba/F3 cells stably transduced with MET TKD mutants. B, IL-3-independent cell proliferation of Ba/F3 cells expressing MET TKD mutants, shown by relative spheroid size measured daily from day 1 to day 7. C, Spheroid images of Ba/F3 cells expressing MET TKD mutants taken on day 1 and day 7. Ba/F3 cells expressing MET WT and R1327C were also cultured in media containing IL-3.
MET TKD Activating Mutations Have Differential Sensitivity to MET TKIs
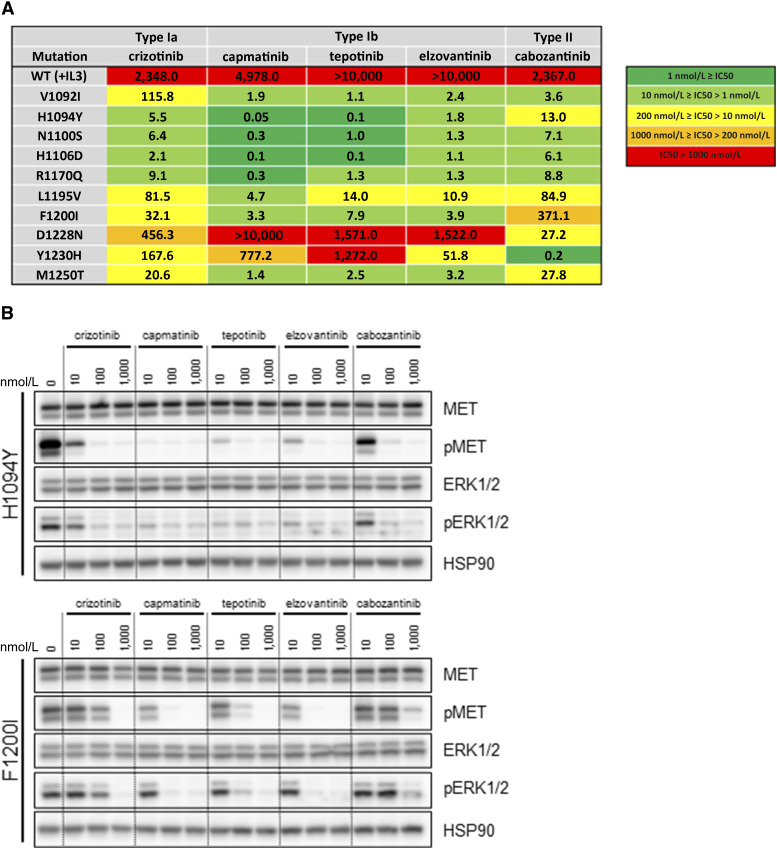
To understand the clinical relevance of these MET TKD mutations as potentially actionable oncogenic driver, we further assessed the sensitivity pattern of the MET TKD mutations to currently available type Ia, Ib, and II MET TKIs. When transiently expressed in 293T cells, all MET TKD mutants with the exception of MET R1327C showed phosphorylation of MET and activation of ERK-mediated transcription (Supplementary Fig. S12A). The type Ia MET TKI crizotinib inhibited phosphorylation of MET and the downstream effector ERK in MET WT, H1094Y, N1100S, H1106D, and R1170Q, whereas the other MET TKD mutants conferred mild to strong resistance to crizotinib (Supplementary Fig. S12B). We next analyzed sensitivity of Ba/F3 cells transformed by the MET TKD mutations in response to MET TKIs. Growth inhibition assays under drug treatment showed that Ba/F3 cells expressing MET TKD mutants showed a differential response to MET TKIs as demonstrated by IC50 values (Fig. 5A) and cell growth curves (Supplementary Fig. S13). Especially, F1200I was resistant to type II MET TKI, D1228N was resistant to both type Ia and Ib MET TKIs, and Y1230H was resistant to type Ib TKIs, except for elzovantinib.
Figure 5.
Sensitivity of MET TKD mutants to MET TKIs. A, IC50 values of Ba/F3 cells expressing MET TKD mutants in response to MET TKIs. Ba/F3 cells expressing MET WT and cultured in media containing IL-3 are shown as control (first row). B, Western blot analysis of Ba/F3 cells expressing MET H1094Y and F1200I in response to MET TKIs.
We next performed a signaling analysis for MET H1094Y and F1200I, detected in patients with NSCLC who responded to the MET TKI elzovantinib (Fig. 5B); and MET R1170Q, a mutation most commonly detected in cohort #1 but with unknown significance according to OncoKB and categorized as “probably pathogenic” by in silico scoring (Supplementary Fig. S14). In concordance with what was observed with the growth inhibition assays, Ba/F3 MET H1094Y and F1200I models showed sensitivity to type Ib MET TKIs (capmatinib, tepotinib, and elzovantinib) as demonstrated by the loss of MET and ERK phosphorylation. Conversely, Ba/F3 MET F1200I conferred mild resistance to the type Ia MET TKI crizotinib and particularly strong resistance to the type II MET TKI cabozantinib, as evidenced by the retained phosphorylation of MET and ERK. MET R1170Q showed sensitivity to all MET TKIs both in the Ba/F3 model (Supplementary Fig. S14A) and when transiently overexpressed in 293T cells (Supplementary Fig. S14B).
To validate the oncogenic function of MET TKD mutants in a more physiologically relevant context, we transduced MET WT as a control, as well as the MET TKD mutants H1094Y and F1200I, in the PC9 human lung cancer cell line (Supplementary Fig. S15A). PC9 cells harbor an EGFR exon 19 deletion alteration and are typically highly sensitivity to treatment with the EGFR inhibitor osimertinib. When treated with osimertinib, PC9 cells expressing MET TKD mutants showed increased viability (Supplementary Fig. S15B), increased proliferation, and decreased apoptosis (Supplementary Fig. S15C), compared to both parental PC9 cells and those transduced with MET WT. Similarly, osimertinib led to the suppression of ERK and AKT phosphorylation in both parental and cells transduced with MET WT, whereas phosphorylation was retained in PC9 cells transduced with MET TKD mutants. This phosphorylation was inhibited with combination of EGFR and MET TKI treatment (Supplementary Fig. S15D). Last, PC9 cells transduced with MET H1094Y and F1200I showed a very similar sensitivity pattern to MET TKIs compared to that observed in Ba/F3 cells (Supplementary Fig. S15E).
Structural Analysis of MET H1094Y, MET F1200I, and MET R1170Q
To understand the activation mechanism associated with MET TKD mutations, we conducted 1,000 ns long molecular dynamics (MD) simulations to probe change to the active ATP bound MET kinase domain (15, 16) with focus on H1094Y, F1200I, and R1170Q. The equilibrated trajectories were used to calculate the free energy of ATP binding via a single trajectory method molecular mechanics/generalized born surface area (MM/GBSA) approach (Supplementary Fig. S16A–S16D; ref. 17). All the three mutations enhance ATP binding relative to the WT (Supplementary Table S24). We then sought to better characterize how each of these mutations (H1094Y, F1200I, and R1170Q) led to MET activation.
The H1094Y mutation caused the stabilization of π–π stacking interactions with Y1158 by more than ∼85% of total simulation trajectory as measured by distance between the geometric centers of phenolic rings within 5.0 ångström (Å) (Supplementary Figs. S17A–S17C and S18A–S18C), stabilizing the ATP hinge hydrogen bonding network. This drives the increase in affinity to ATP that was reported via the single trajectory MM/GBSA calculations.
In the MET WT, F1200 is involved in a complex π–π stacking network that bridges F1223 of the Asp-Phe-Gly (DFG) motif and an additional phenyl ring of F1134. The inter-ring distances as measured by distance between the geometric centers of phenyl rings are relatively tightly distributed for π–π stacking interactions of F1134 to F1200 and F1223 to F1200 (Supplementary Fig. S19A–S19C). However, F1134 and F1223 are too distal for π–π stacking interactions. This suggests that F1200 intercalates between these residues and facilitates a larger electronically coupled system. F1200I provides steric bulk that can direct the conformations of F1134 and F1223 to facilitate the formation of new π–π stacking interactions between these residues. The loss of aromatic side chain in position 1,200 locks the π–π stacking interactions that bridge the alpha C helix and DFG motifs. Therefore, the F1200I mutation led to a conformation bias that stabilize the DFG in the activation loop, as shown by the root mean square fluctuation (RMSF) for residues between 1213 and 1235. The stabilization of the activation loop leads to the increase of the ATP binding that was demonstrated through the single trajectory MM/GBSA calculations.
Molecular dynamic simulation for R1170Q showed that this mutation led to the formation of a water hydrogen bonding network between ATP-D1164-N1167, boosting the ATP binding to the kinase domain (Supplementary Fig. S20A–SC).
Discussion
MET alterations, including METex14 alterations, gene amplification, rearrangement, and overexpression, promote carcinogenesis in a wide spectrum of cancer types (1). Detailed detection of these oncogenic alterations using comprehensive molecular profiling widens the range of patients who could benefit from specific targeted therapies (4, 18). Here, we provide clinical, preclinical, and structural evidence that activating de novo MET TKD mutations are a currently undercharacterized subset of oncogenic drivers that can be actionable in NSCLC, and potentially in other malignancies.
Our comprehensive analysis of pan-cancer cohorts revealed that MET TKD mutations without concurrent METex14 alterations were detected in various cancer types, including renal cell carcinoma, NSCLC, and melanoma. Several recurrent mutations that were detected across pan-cancer cohorts had previously been demonstrated as being de novo oncogenic drivers in papillary renal cell carcinoma, such as H1094Y, F1200I, V1092I, L1195V, and M1250T (11). IL-3-independent Ba/F3 cell proliferation was also observed for several MET TKD mutations other than H1094Y and F1200I, such as V1092I, L1195V, M1250T, D1228N, Y1230H, and others, some with differential responses to MET TKIs; this characterization should enable more precise treatment selection for this rare subgroup of patients with MET TKD-mutant cancers. The detection and recognition of these targetable oncogenic/likely oncogenic MET TKD mutations more broadly across various malignancies will hopefully increase the number of patients who can benefit from MET-directed therapies, further highlighting the clinical need to implement comprehensive genomic profiling into daily practice (18).
Focusing on NSCLC, oncogenic/likely oncogenic MET TKD mutations were detected in 0.14% to 0.19% of patients, either as a sole oncogenic driver or with concurrent oncogenic drivers such as KRAS or EGFR. Importantly, two patients with NSCLC harboring activating MET TKD mutations (H1094Y and F1200I) but no other concurrent oncogenic drivers, enrolled in a phase 1 study of type Ib MET TKI elzovantinib (TPX-0022) showed partial response. Taken together, these two clinical cases, along with preclinical and structural analyses, strongly suggest that de novo MET TKD mutations are oncogenic drivers in NSCLC and more importantly, can be clinically actionable with MET-targeted therapies.
Considering clinicopathologic features of NSCLC with MET TKD mutations, patients with MET TKD-mutant NSCLC were significantly younger than patients with NSCLC harboring METex14 alterations, but older than the typical age for EGFR-, ALK-, or ROS1-positive lung cancers (19, 20). Though the numbers of patients with detailed smoking history were limited, all patients with MET TKD-mutant NSCLC had a history of tobacco use, in contrast to certain other oncogenic subsets of NSCLC, such as EGFR-, ALK-, or ROS1-mutant lung cancers, where percentage of never smokers is relevantly lower compared to former or current smokers (20, 21). Higher rates of tobacco use have also been reported in METex14-altered NSCLC compared to EGFR, ALK, and ROS1, suggesting a possible link between smoking and MET signaling, as demonstrated in the preclinical setting (22, 23). The higher percentage of tobacco use could also explain the higher median TMB found in this subset of NSCLC (24). There was no difference in PD-L1 expression levels between MET TKD-mutant and METex14-altered NSCLC cases. Whether MET TKD-mutant NSCLC is sensitive to treatment with immune checkpoint inhibitors will be of interest in further studies.
In MET TKD-mutant NSCLC, the most common concurrently altered gene was TP53, with an enrichment of TP53 and KEAP1/NFE2L2 alterations when compared to METex14-altered NSCLC. In EGFR-mutant NSCLC, mutations in KEAP1 and NFE2L2 are associated with shorter median time to treatment failure on EGFR TKIs, and TP53 mutations also represent a poor prognostic factor in patients receiving EGFR TKI (25, 26). Whether these concurrent mutations in MET TKD mutant NSCLC impair sensitivity to MET TKI treatment remains an unanswered question.
In addition, our analysis of NSCLCs with concurrent oncogenic alterations revealed that MET TKD mutations can emerge after treatment with other targeted therapies, such as EGFR or ROS1 TKIs. This suggests that acquired MET TKD mutations can mediate resistance not only in METex14-altered NSCLC but also in other oncogene-driven tumors. Furthermore, a fraction of NSCLCs with MET TKD mutations had concurrent KRAS mutations. This co-occurrence of KRAS and MET alterations was observed in a previous study reporting that 4.1% of KRAS-mutant NSCLC had concurrent MET mutations, specifically METex14 alterations, and 15% had concurrent MET amplification (27). Whether MET TKD mutations may contribute to increased oncogenicity or resistance to KRAS inhibitors in KRAS-driven NSCLC will require further investigation.
Until now, MET TKD mutations in NSCLC have been largely characterized as an on-target mechanism of resistance to MET TKIs in METex14-altered tumors (14, 28). Surprisingly, many of the de novo MET TKD mutations that we detected across pan-cancer cohorts were the same mutations that were previously identified as being on-target mechanism of resistance to MET TKIs, such as H1094Y, D1228H/N, Y1230C/H, L1195V, and F1200I/L (14, 28). As would be expected, MET TKD mutations reported to confer resistance to MET TKIs in NSCLCs with METex14 alterations also showed the same pattern of resistance when seen as de novo MET TKD mutations. For example, the D1228X mutation is known as an on-target resistance mechanism in METex14-altered NSCLC after MET TKI treatment, conferring resistance to type Ia/Ib MET TKIs but retaining sensitivity to type II MET TKIs (14, 28). Our preclinical modeling also demonstrates the same pattern of sensitivity/resistance for MET D1228N as a de novo oncogenic driver. These findings suggest that de novo activating MET TKD mutations not only have an activating and oncogenic potential but can also intrinsically interfere with TKI binding, showing that the mechanism of action for MET TKD mutations is multifaceted, affecting both ATP and drug binding.
Regarding missense MET TKD mutations with “unknown” significance in OncoKB, three in silico scores were used to categorize mutations; among them, Ba/F3 cell models were used to investigate the two top mutated residues in cohort #1, R1170, specifically R1170Q missense mutation and R1327, specifically R1327C missense mutation. Contrary to R1327C which was categorized as “uncertain” by in silico scores, R1170Q was categorized as “probably pathogenic” and showed IL-3-independent cell proliferation, as well as increased ATP binding through structural simulation, suggesting a potential oncogenic role for this mutation. We also modeled the most frequent four mutations categorized as “probably benign” according to in silico scores in cohort #1, and they did not show IL-3-independent cell proliferation in Ba/F3 cells. Taken together, in silico tools may be used as an initial reference to select uncharacterized but potentially pathogenic mutations which would be candidates for further in vitro and structural modeling.
Limitations to this study include incomplete clinical, therapeutic, and genomic data (including TMB and gene copy number) in several cohorts. Cohort #2 from Foundation Medicine (FMI) included tissue samples but also a small percentage of liquid samples, in which certain co-alterations (such as MET amplification) might be underreported for cases with low tumor fraction, though this should have minimal impact since the majority of the cohort was tissue sample. Furthermore, due to the wide variety of MET TKD mutations detected, only a selected and representative subset were included in our preclinical and structural models. Last, a percentage of MET TKD mutations could be passenger mutations in cases with high TMB. Nonetheless, MET TKD mutations were also identified in tumors with low TMB (<10 mut/Mb): looking at pan-cancer cases (other than NSCLC) in cohort #2, 65% had a low TMB, considering NSCLCs with MET TKD mutations as a sole oncogenic driver in cohort #2, 50% had a low TMB, suggesting a role as oncogenic driver rather than passenger mutation in these cases.
Our study showed that MET TKD mutations can be a de novo oncogenic driver in different cancer types, and that these MET TKD mutations have differential sensitivity to MET TKIs. Although the overall incidence of oncogenic MET TKD mutations is <1%, identification of MET TKD mutations across various tumor types will be critical for patient care given their sensitivity to clinically available MET inhibitors. MET TKD mutations are detectable both in tissue and liquid biopsies through many currently available NGS platforms (29). However, as some sequencing assays only cover a portion of the MET gene to detect actionable METex14 alterations, more comprehensive genomic sequencing which also covers the MET kinase domain (exons 15–21) should be considered in order to detect actionable activating MET TKD mutations. Therefore, our study highlights the importance of performing comprehensive genomic tumor profiling to detect rare but targetable driver alterations. Finally, our clinical and in vitro findings may pave the way for future clinical trial design in this newly characterized molecular cancer subgroup.
Methods
Patient Population and Tumor Genomic Profiling
To determine the type and frequency of MET TKD mutations, two pan-cancer cohorts with available genomic profiling were queried: cohort #1, which merged publicly available data from GENIE v.11.0 (https://www.aacr.org/wp-content/uploads/2022/03/GENIE_data_guide_11.0-public.pdf), China Pan-cancer (OrigiMed2020, Nature 2022; ref. 30) and The Cancer Genome Atlas (TCGA) PanCancer Atlas studies from cBioPortal, pan-cancer data from DFCI, and NSCLCs from MSKCC; cohort #2 was from the FMI genomic dataset. Patients from DFCI were included if they had consented to institutional review board (IRB)-approved protocol #19-201 for translational and clinical pan-cancer studies (data cutoff: March 2022). Patients from MSKCC with advanced NSCLC with NGS data were included if they had consented to IRB-approved institutional protocols (data cutoff: June 2022). Duplicate patients from either the DFCI or the MSKCC datasets who were also in the GENIE v.11.0 dataset were identified and only considered once. For the FMI genomic analysis, approval for this study, including a waiver of informed consent and a Health Insurance Portability and Accountability Act waiver of authorization, was obtained from the Western Institutional Review Board Copernicus Group (Protocol No. 20152817; data cutoff: June 2022). This study was conducted in accordance with the Declaration of Helsinki.
For cohort #1, genomic profiling for cases from GENIE v.11.0, China Pan-cancer, and 32 TCGA studies (listed in Supplementary Table S25) was performed according to each sequence pipeline and downloaded (https://www.aacr.org/wp-content/uploads/2022/03/GENIE_data_guide_11.0-public.pdf; https://www.cbioportal.org/study/summary?id=laml_tcga_pan_can_atlas_2018%2Cacc_tcga_pan_can_atlas_2018%; https://www.cbioportal.org/study/summary?id=pan_origimed_2020). For cases from DFCI, targeted exome NGS was performed using the validated OncoPanel assay in the Center for Cancer Genome Discovery at DFCI for 277 (v1, April 2013–July 2014), 302 (v2, July 2014–September 2016), or 447 (v3, September 2016–ongoing) cancer-associated genes, as previously described (31, 32). For NSCLC from MSKCC NGS was performed using the MSK-IMPACT NGS platform for 341 (v1), 410 (v2), and 468 (v3) unique cancer-associated genes, as previously published (33).
For cohort #2, pan-cancer cases submitted for comprehensive genomic profiling (CGP) during routine clinical care were interrogated in the FMI dataset (data cut off: June 2022). For tissue samples (N = 426,427), CGP of formalin-fixed, paraffin-embedded sections was performed using FoundationOne or FoundationOne CDx, as previously described (34–38). Liquid biopsy samples (N = 32,912) were profiled using a validated, FDA-approved NGS panel assay (FoundationOne Liquid CDx). Circulating cell-free DNA was extracted from peripheral whole blood and CGP was performed using hybridization-captured, adaptor ligation-based libraries as previously described (39). Additional detailed methods for cohort #1 and cohort #2 are reported in Supplementary Methods.
A separate cohort of NSCLC molecularly profiled at Caris Life Sciences (Phoenix, AZ) between 2017 and 2022 was interrogated for MET TKD mutations. For this cohort, the study was conducted in accordance with the guidelines of the Declaration of Helsinki, Belmont Report, and U.S. Common Rule. In keeping with 45 CFR 46.101(b)(4), this study was performed using retrospective and de-identified clinical data. Additional detailed methods for Caris Life Sciences cohort are reported in Supplementary Methods.
Classification of MET Tyrosine Kinase Domain Mutations
Each dataset was first queried for any type of MET alteration, and alterations within the MET tyrosine kinase domain were identified if they were located between amino acids p.V1078 and p.I1345 (NM_000245.2, variant 2; ref. 1). Cases with concurrent METex14 alterations were removed from the final analysis since MET TKD mutations may represent acquired resistance mutations after treatment with MET TKIs (14). The frequency of MET TKD mutations in each cohort was determined according to cancer type. MET TKD mutations were classified as nonsense, insertions/deletions, splice site, or missense mutations. Nonsense, insertions/deletions, and splice site mutations were classified as unknown biological function. Missense mutations were categorized for biological function according to OncoKB annotation (https://www.oncokb.org/; version 4.1), as either “oncogenic,” “likely oncogenic,” or “unknown.” For cohorts #1 and #2, “unknown” missense mutations were further classified according to PolyPhen-2 score, SIFT score, and P3DFikinase (40–42). Missense mutations were classified into four categories according to PolyPhen-2 values: “Probably Damaging” (value >0.908), “Possibly Damaging” (value greater than 0.446 and less than or equal to 0.908), “Benign” (value less than or equal to 0.446), and “Unknown” (https://www.ensembl.org/info/genome/variation/prediction/protein_function.html). The SIFT score classified the missense mutations as “Deleterious” (SIFT value <0.05) and “Tolerated” (SIFT value greater than or equal to 0.05; https://www.ensembl.org/info/genome/variation/prediction/protein_function.html). For P3DFikinase, missense mutations were defined pathogenic with a P3DFikinase value ≥1 (42).
Definition of Concurrent Driver Alterations
For cohort #1, considering tumors other than NSCLC, concurrent driver alterations were annotated for cases with oncogenic/likely oncogenic MET TKD mutations as reported below: (i) relevant concurrent driver alterations defined for each tumor type according to the latest NCCN guidelines 2023 (https://www.nccn.org/guidelines/category_1); (ii) other concurrent driver alterations with an OncoKB level of evidence (OncoKB Therapeutic Level of Evidence V2; ref. 43) NTRK- and RET-rearrangement, due to recent FDA approval in solid tumors (44, 45). For NSCLCs with oncogenic/likely oncogenic MET TKD mutations, activating alterations in KRAS, EGFR, BRAF, HER2, RET, ALK, ROS1, NTRK1/2/3, were classified as concurrent driver alterations, if present (46).
Regarding MET gene copy number, for cases in GENIE v.11.0, China Pan-cancer, and TCGA, MET amplification was defined according to each platform, when assessed. For OncoPanel, MET was considered amplified when ≥6 copies of the gene were detected, for MSK-IMPACT, MET amplification was defined in case of fold change ≥2. In cohort #2, MET amplification includes amplification ≥4 copies above the overall ploidy of the specimen.
Cell Lines
The 293T and PC9 cells were purchased from ATCC (in 2009) and Sigma Aldrich (in 2014), respectively. The 293T cells were cultured in DMEM, supplemented with 10% FBS and 1% streptomycin/penicillin. The 293T and PC9 cell line were authenticated in August 2016 using the Promega GenePrint 10 System at the RTSF Genomics Core at Michigan State University. Ba/F3 cells were a generous gift from the laboratory of Dr. David Weinstock in 2014 and were not authenticated, because their short tandem repeat profile has not been publicly available. Transformed Ba/F3 cells and PC9 cells were maintained in RPMI1640 supplemented with 10% FBS, 1% streptomycin/penicillin, and IL-3 was added for parental Ba/F3 cells. All cell lines used in the study tested negative for Mycoplasma as determined by the Universal Mycoplasma Detection Kit (30-1012K; ATCC) in November 2023. Antibodies and compounds are listed in Supplementary Methods.
Expression Vectors
The full-length human MET, transcript variant 2, cDNA (NM_000245.2) was amplified from a banked tumor specimen with an unrelated genetic alteration and cloned into the pCDH-EF1-FHC vector (Addgene plasmid # 64874) using the In-Fusion HD Seamless Cloning Kit (Takara Bio; 638916) following the manufacturer’s protocols. The cDNA is cloned in frame with a single FLAG tag on the C-terminus. Individual MET TKD mutations were introduced using the QuikChange Lightning or QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies; 210519 and 200521), with the exception of the Y1230H mutation, which was introduced using the Q5 Site-Directed Mutagenesis Kit (NEB; E0552S). PCR conditions and primer sequences were designed using the manufacturers’ online tools. The mutagenic primers used for mutagenesis are listed in Supplementary Table S26. All constructs were validated by DNA sequencing.
Viral Transduction
To establish stable MET TKD mutant expression, Ba/F3 cells were transduced with lentivirus according to standard procedures (47). To produce lentiviral particles, 293T cells were first transfected with the lentivirus construct in combination with the packaging plasmids psPAX2 and pMD2.G (Addgene) using FuGENE HD Transfection Reagent (Promega) as per the manufacturer’s protocol (Supplementary Methods). Viral supernatants produced in 293T cells were harvested at 48 hours after transfection, filtered, and then centrifuged with Ba/F3 or PC9 cells supplemented with 10 μg/mL polybrene for 90 minutes at 2,100 rpm. Ba/F3 or PC9 cells with successful lentiviral integration were selected with 1 μg/mL puromycin. Transformed Ba/F3 cells were maintained in RPMI1640 supplemented with 10% FBS and 1% streptomycin/penicillin.
Cell Growth Inhibition Assay and Incucyte Assays
For cell growth inhibition assay in Ba/F3 cells, stable MET TKD mutant-driven and IL-3-independent Ba/F3 cells were treated with dose-escalated inhibitor over the course of 72 hours. For PC9 cells, cells transduced with MET WT or MET TKD mutants were treated with dose-escalated MET TKIs in combination with 100 nmol/L osimertinib. Endpoint cell viability was assessed using Cell Titer Glo (Promega) and read by FLUOstar Omega. All experimental points were set up in five to six wells. The represented data are representative of several biological replicates.
For Incucyte growth assay, individual MET TKD mutant Ba/F3 cells were seeded in 96-well round bottom plate at 3,000 cells/well and allowed to pool into the center of the well before imaging. For confluency assay, PC9 cells were plated into 96-well plate at 2,500 cells/well and drug added the next day. For apoptosis assay, CellEvent Caspase 3/7 Green ReadyProbes reagent (Thermo Fisher Scientific) was also added to the media. Growth rate, confluency, and relative apoptosis were monitored and analyzed using the IncucyteS3 Live-Cell Imaging Analysis System (Essen Bioscience). IC50 value for cell growth inhibition assay and cell growth, confluency, relative apoptosis were analyzed using GraphPad Prism software (v. 9.5.1).
Western Blotting
Cell lysis, SDS-PAGE, and immunoblotting were done as described previously (48). Briefly, cells were lysed with RIPA lysis buffer supplemented with protease and phosphatase inhibitors. Total cell lysate was subjected to SDS-PAGE and transferred to PVDF membranes. Blots were developed on Amersham Imager 600 (GE Healthcare Life Sciences).
Structural MET Models Analysis
The cocrystal structure of MET with ATP (PDBID:3DKC) was used to build the simulation system. The protein preparation wizard in Schrodinger 2022-37 was used for initial modeling, and water molecules near ATP and Mg+2 were retained. The initial structure contained three engineered residues and served as a template for generating active state ATP-bound MET kinase homology models for mutants. The canonical sequence was used to model the WT structure, and appropriate substitutions were made to generate the H1094Y, F1200I, and R1170Q mutants. Co-factors (ATP, Mg+2 ion, and a water molecule) were included in building the homology models. ATP parameterization was done based on the initial ATP bound in the WT MET crystal. The complex underwent global unrestrained minimization using the OPLS4 force field with implicit solvent model. Quantum mechanical calculations were performed afterward for charge generation, utilizing the B3LYP-D3/6-311G(d,p)++ basis set.
Model systems were generated using CHARMM-GUI enhanced sampler for the three ATP-containing MET, i.e., WT, H1094Y, F1200I, and R1170Q. A rectangular water box was fitted to the protein structure and neutralizing ions and salt (KCl) concentration of 150 mmol/L were added. The FF19SB force field was used for parameterizing protein residues, and hydrogen mass repartitioning was employed to enable a 4-fs time step. All-atom MD simulations were conducted using the PMEMD in AMBER22. The production phase simulations after equilibration consisted of conventional MD and Gaussian accelerated MD steps.
Analysis of the MD simulations was performed on the last 500 ns of the production trajectories. Water and ions were removed except for Mg+2, and distance measurements, root mean square fluctuation (RMSF), and root-mean-square deviation (RMSD) values were calculated. The MMPBSA.py utility in AmberTools was used to calculate the free energy of binding ΔG based on a single trajectory MM/GBSA protocol.
Additional methods are explained in Supplementary Methods section.
Statistical Analysis
Categorical and continuous variables were summarized descriptively using percentages and medians. The Wilcoxon-Rank Sum test and Kruskal–Wallis test were used to examine differences between continuous variables, and Fisher’s exact test or χ2 test were used to compare associations between categorical variables. P-values were based on a two-sided hypothesis with confidence intervals at the 95% level, and P < 0.05 was considered statistically significant. These analyses were performed using R v3.6.1. Analysis of co-occurring and mutually exclusive gene alterations between groups was limited to genes altered in at least five cases and targeted across all the assay versions. For each gene, substitutions, short insertions/deletions, rearrangements, and copy number changes of known or likely functional significance detected using FMI assay were included. A Fisher’s exact test with false discovery rate-based correction for multiple testing was applied for co-mutation analysis. Regarding structural MET models analysis, statistical analysis was performed using a Mann–Whitney–Wilcoxon test in GraphPad Prism Software to assess interaction distances.
Data Availability
The data that support the finding of our study are available upon request from the corresponding author.
Supplementary Material
Additional information regarding the methods used in this study
Supplementary Table 1. Prevalence of MET tyrosine kinase domain (TKD) mutations in each dataset of the cohort #1.
Frequency of MET tyrosine kinase domain (TKD) mutations in cohort #1 according to amino acid residue, out of a total of 711 MET TKD mutations.
Frequency of MET tyrosine kinase domain (TKD) mutations in cohort #2 according to amino acid residue, out of a total of 2,559 MET TKD mutations.
List of missense mutations with unknown biologic function and their pathogenicity scores using all three in silico tools in cohort #1
List of missense mutations with unknown biologic function and their pathogenicity scores using all three in silico tools in cohorts #2
Prevalence of MET tyrosine kinase domain (TKD) mutations across cancer types in cohort #1.
Prevalence of MET tyrosine kinase domain (TKD) mutations among “other cancers” from Supplementary Table 6.
Prevalence of MET tyrosine kinase domain (TKD) mutations across cancer types in cohort #2.
Prevalence of MET tyrosine kinase domain (TKD) mutations across tumor in “other cancers” from Supplementary Table 8.
Characteristics of 44 NSCLC cases harboring oncogenic / likely oncogenic MET TKD mutations in cohort #1.
Detailed genomic characteristics for each case of NSCLC with oncogenic / likely oncogenic MET tyrosine kinase domain (TKD) mutations in cohort #1.
Characteristics of 129 NSCLC cases harboring oncogenic / likely oncogenic MET TKD mutations from cohort #2.
Concurrent driver mutations according to oncogenic / likely oncogenic MET TKD mutations detected in cohort #2.
Concurrent genomic drivers in cases with oncogenic / likely oncogenic MET tyrosine kinase domain (TKD) mutations and co-occurring MET gene amplification in cohort #2.
Comparison between clinicopathologic characteristic of NSCLCs with oncogenic / likely oncogenic MET tyrosine kinase domain (TKD) mutations as a de novo genomic event to NSCLCs with MET exon 14 alterations in cohort #1.
Comparison between clinicopathologic characteristic of NSCLCs with oncogenic / likely oncogenic MET tyrosine kinase domain (TKD) mutations without concurrent other drivers to NSCLCs with MET exon 14 alterations in cohort #2.
Comparison of clinicopathologic characteristics between NSCLCs with oncogenic / likely oncogenic MET tyrosine kinase domain (TKD) mutations without other concurrent drivers and NSCLCs with MET exon 14 alterations in the Caris Life Sciences cohort.
Detailed clinicopathologic and genomic characteristics of the 61 cases with a malignancy other than NSCLC harboring an oncogenic / likely oncogenic MET TKD mutation in cohort #1.
Detailed clinicopathologic and genomic characteristics related to renal cell cancer cases in cohort #1.
Clinicopathologic and genomic characteristics of 171 cases of cancers other than NSCLC harboring oncogenic / likely oncogenic MET TKD mutations in cohort #2.
Detailed clinicopathologic and genomic characteristics related to renal cell cancer cases in cohort #2.
Oncogenic / likely oncogenic MET TKD mutations in RCC compared to those detected in NSCLC in cohort #1.
Oncogenic / likely oncogenic MET TKD mutations in RCC compared to those detected in NSCLC in cohort #2.
Single trajectory generalized Born and surface area solvation (MM/GBSA) calculated free energy of ATP binding into the orthosteric pocket.
TCGA PanCancer Atlas Studies included in cohort #1 downloaded from cBioPortal.
Mutagenic primers used for mutagenesis.
Study flowchart. Cohort #1 (multi-institutional cohort): GENIE V.11.0, China Pan-cancer (OrigiMed2020, Nature 2022), The Cancer Genome Atlas (TCGA) Pan-Cancer Atlas studies, Dana-Farber Cancer Institute (DFCI) and NSCLC from Memorial Sloan Kettering Cancer Center (MSKCC); cohort #2: Foundation Medicine Pan-Cancer cohort.
Overview of MET TKD mutations detected in pan-cancer cohorts. (A) Type (missense, truncating, splice site, in-frame) of MET TKD mutations detected in the cohort #1 (711 MET TKD mutations among 664 unique patients with a tumor harboring MET TKD mutations without concurrent METex14 alterations). (B) Type of MET TKD mutations according to OncoKB annotation (biological function: oncogenic / likely oncogenic, unknown) detected in cohort #1. (C) Distribution of MET TKD mutations across MET kinase domain for cohort #1. (D) Type (missense, truncating, splice site, in-frame) of MET TKD mutations detected in cohort #2 (2,559 MET TKD mutations among 2,477 unique patients with a tumor harboring MET TKD mutations without concurrent METex14 alterations). (E) Type of MET TKD mutations according to OncoKB annotation (biological function: oncogenic / likely oncogenic, unknown) detected in cohort #2. (F) Distribution of MET TKD mutation across MET kinase domain for cohort #2.
Comparison of missense MET TKD defined as “probably pathogenic” and “probably benign” according to three in silico tools. Comparison of (A) secondary structure features, (B) relative solvent accessible surface area (RSA), and (C) mutant amino acid properties of missense MET TKD mutations of unknown biological function according to OncoKB. Mutations were defined as “probably pathogenic” for likely pathogenicity across all three tools and as “probably benign” for likely benign across all three scores. OR: odds ratio; 95% CI: 95% confidence interval.
Type and frequency of MET TKD mutations across cancer types in cohort #1. (A) Frequency of MET TKD mutations in various cancer types in cohort #1 according to OncoKB status; the numbers above each bar indicate the number of cases with MET TKD mutations (regardless OncoKB annotations) out of the total of cases of each cancer type. (B) Detailed oncogenic / likely oncogenic MET TKD mutations according to cancer type in cohort #1.
Prevalence of MET TKD mutation in various cancer types according to OncoKB status in each dataset in cohort #1.
MET TKD mutations in NSCLC in cohort #1 and #2. (A) Flowchart of the NSCLC subgroup in cohort #1 (123 MET TKD mutations in 120 unique patients); (B) Prevalence of MET TKD mutations in NSCLC cases according to each dataset in cohort #1; (C) Lollipop plot of the oncogenic / likely oncogenic MET TKD mutations detected in NSCLC cases of cohort #1 (47 oncogenic / likely oncogenic MET TKD mutations in 44 unique patients); (D) Flowchart of the NSCLC subgroup in cohort #2 (590 MET TKD mutations in 586 unique patients); (E) Prevalence of MET TKD mutations in NSCLC cases according to cohort #2 (590 MET TKD mutations in 586 unique patients); (F) Lollipop plot of the oncogenic / likely oncogenic MET TKD mutations detected in NSCLC cases of cohort #2 (129 oncogenic / likely oncogenic MET TKD mutations in 129 unique patients).
Genomic characterization of NSCLC harboring oncogenic / likely oncogenic MET TKD mutations in cohort #1 and #2. Genomic characteristics of 44 NSCLC cases harboring oncogenic / likely oncogenic MET TKD mutations from cohort #1: (A) concurrent driver alterations and (B) concurrent MET amplification status (information unavailable for 4 cases). Genomic characteristics of 129 NSCLC cases harboring oncogenic / likely oncogenic MET TKD mutations from cohort #2: (C) concurrent driver alterations and (D) concurrent MET amplification status.
Concurrent oncogenic driver alterations according to specific oncogenic / likely oncogenic MET TKD mutations detected in NSCLC cases (N=129) of cohort #2.
Genomic characterization of NSCLC cases harboring oncogenic / likely oncogenic MET TKD mutations in cohort #2. (A) Oncoprint summarizing the genomic profile and (B) the TMB value of the 78 NSCLC cases with oncogenic / likely oncogenic MET TKD mutations without other concurrent driver alterations according to NCCN guidelines. (C) Comparison of the genomic profile of NSCLC cases with oncogenic / likely oncogenic MET TKD mutations (N=78) and METex14-altered NSCLC (N=2036)
MET TKD mutations in NSCLC cohort of Caris Life Sciences. (A) Flowchart of the NSCLC subgroup in the Caris Life Sciences dataset. (B) Prevalence of MET TKD mutations in NSCLC cases in the Caris Life Sciences dataset (MET TKD mutations in 280 unique patients). (C) Lollipop plot of the oncogenic / likely oncogenic MET TKD mutations detected in NSCLC cases in the Caris Life Sciences dataset (35 oncogenic / likely oncogenic MET TKD mutations in 35 unique patients). (D) concurrent driver alterations (information available for 24 cases, unavailable for 11 cases) and (E) concurrent MET amplification status (information available for 33 cases, unavailable for 2 cases) of NSCLC cases harboring oncogenic / likely oncogenic MET TKD mutations in the Caris Life Sciences dataset.
In vitro analysis of “probably benign” MET TKD mutations in Ba/F3 cells. (A) Phosphorylation of MET was analyzed in Ba/F3 cells stably transduced with MET TKD mutants. MET H1094Y and F1200I were included as positive controls for oncogenic mutations (OncoKB, also detected in the two patient cases) and MET WT was included as negative control. (B) IL-3-independent cell proliferation of Ba/F3 cells expressing “probably benign” MET TKD mutants, shown by relative spheroid size measured daily from day 1 to day 7. (C) Spheroid images of Ba/F3 cells expressing MET TKD mutants taken on day 1 and day 7.
Sensitivity pattern of MET TKD mutations in 293T cells. (A) ERK-mediated transcriptional activation assessed by luciferase assay in 293T cells co-transfected with MET WT or TKD mutant constructs, an ERK activation reporter vector (pGL4.33), and a control co-reporter vector (pRL-TK). The lower panel shows western blot analysis of transfected samples. (B) Western blot analysis of MET and ERK1/2 phosphorylation in response to 1 μM crizotinib. MET TKD mutants showing sensitivity and resistance to crizotinib are colored in green and orange, respectively.
Growth inhibition assays of Ba/F3 cells expressing MET TKD mutants under treatment with MET TKIs.
Western blot analysis of MET and ERK1/2 phosphorylation in MET R1170Q cell models. MET and ERK1/2 phosphorylation in (A) Ba/F3 cells stably transduced with MET R1170Q and (B) 293T cells transiently overexpressing MET R1170Q. Cells were treated with MET TKIs at the indicated dose for Ba/F3 cells or at 1 μM for 293T cells.
In vitro analysis of MET H1094Y and F1200I mutations in PC9 cells. (A) Establishment of NSCLC cancer cell lines expressing MET TKD mutations. PC9 cells were transduced with wild-type (WT) MET (as control) and MET H1094Y and F1200I. (B) Growth inhibition assay of PC9 cells in response to EGFR inhibition with osimertinib. (C) Cell proliferation and relative apoptosis were monitored under EGFR inhibition with osimertinib (300 nM). (D) Phosphorylation of MET, EGFR, AKT, and ERK was analyzed in PC9 cells expressing MET TKD mutants. PC9 cells were treated with osimertinib (1 μM) with or without capmatinib (1 μM) for 48 hrs. (E) IC50 values of PC9 cells (upper table) and Ba/F3 cells (lower table) expressing MET TKD mutants in response to MET TKIs. PC9 cells were treated with gradient dose of MET TKI combined with a fixed dose of osimertinib (100 nM) for 72 hrs.
Heavy atom root-mean-square deviation. Heavy atom root-mean-square deviation (RMSD) of each simulation: (A) MET wild-type, (B) MET H1094Y, (C) MET F1200I, and (D) R1170Q. Each frame is taken in a 0.1 ns interval.
Equilibration of interactions that drives ATP affinity in the presence MET H1094Y mutant. (A) Representative structure of the ATP bound cMET H1094Y with the regions of interest highlighted [subsection of the 5 pleated beta sheet bundle containing H1094Y and direct interactions (red), hinge binding region (green)]. (B) H1094Y forms novel π- π stacking interactions (T stacking with an upper cut-off of 5.0 angstroms) with Y1158. (C) π- π stacking from Y1158 stabilizes the ATP hydrogen bonding network.
Time resolved interaction distances of ATP in the presence of MET WT and MET H1094Y mutant. (A) Hydrogen bonding between P1157 backbone carbonyl and ATP, (B) Inter ring center distances for Y1158-ATP π–π stacking interactions, and (C) Hydrogen bonding between M1159 backbone amine and ATP distances for ATP interactions between MET wild-type (orange) and MET H1094Y (blue) over the last 500 ns of each trajectory.
Equilibration of interactions that drives ATP affinity in the presence MET F1200I mutant. (A) Representative structure of ATP bound MET F1200I with the regions of interest highlighted [alpha C helix (red), DFG site (green), mutation site (blue)]. (B) π - π stacking interactions that bridge the alpha C helix and DFG site are conformationally controlled through residues position 1200. π - π stacking interactions were calculated between ring centers. Distances between F1200I and neighboring residues were measured from the F1200I side chain center of mass to the ring centers of the aromatic rings. (C) root-mean-square-fluctuation (RMSF) calculations show a stabilization of the activation loop in the F1200I system.
Equilibration of interactions that drives ATP affinity in the presence MET R1170Q mutant. (A) Representative structure of ATP bound MET R1170Q with the regions of interest highlighted [hinge site (green), and mutation site (blue)]. (B) Mutation at position 1170 facilitates the formation of a hydrogen bonding network. (C) Interatomic distances for non solvent mediated hydrogen bonding.
Acknowledgments
F. Pecci acknowledges funding support from Turning Point Therapeutics, a wholly owned subsidiary of Bristol Myers Squibb Company. P.C. Ma acknowledges funding support from Frank and Franco Cancer Research Endowment, Vemon M. and Jolene E. Chinchilli Family Endowment, and GLY Foundation Cancer Research Endowment (PSCI). P.A. Jänne acknowledges funding support from R01 CA222823, Mock Family Fund for Lung Cancer Research, Kaifer Family Fund for Lung Cancer Research and Goldstein Family Research Fund. M.F. Chen acknowledges funding support from T32-CA009207 and a ASCO Young Investigator Award. S. Iqbal is supported by the Merkin Institute of Transformative Technologies in Healthcare. S. Baldacci thanks the Ligue Contre le Cancer (grant NAAPMRC2.2020.LCC/SB) and the Philippe Foundation for their financial support. S. Nakazawa received funding from Daiichi Sankyo Foundation of Life Science. M.M. Awad acknowledges funding support from V Foundation for Cancer Research, Elva J. and Clayton L. McLaughlin Fund for Lung Cancer Research, Team Stuie, Team LUNGStrong, John Hallick Family Fund, Stephen Wald Family Fund, and the Angela Marotta Inho Foundation.
Footnotes
Note Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
S. Nakazawa reports a Research Fellowship from Daiichi Sankyo Foundation of Life Science. B. Ricciuti reports personal fees from AstraZeneca and Regeneron; and personal fees from Amgen outside the submitted work. G. Harada reports personal fees from BMS, Merck, and J&J; personal fees and other support from MSD, AstraZeneca, Lilly, and Takeda; and personal fees from Daiichi Sankyo outside the submitted work. J.K. Lee reports personal fees from Foundation Medicine and Roche during the conduct of the study. J.V. Alessi reports other support from AstraZeneca outside the submitted work. A. Di Federico reports personal fees from Hanson-Wade outside the submitted work. S. Baldacci reports non-financial support from Lilly, GSK, Roche, Pfizer, and Janssen; grants from Intergroupe Francophone de Cancérologie Thoracique and Lille University; personal fees from Boehringer Ingelheim; personal fees and non-financial support from MSD; and personal fees from AstraZeneca outside the submitted work. M.F. Chen reports grants from NCI and ASCO; other support from Nordisk, Quest, and DOCS; and other support from FIGS outside the submitted work. Z. Zimmerman reports other support from Turning Point Therapeutics during the conduct of the study. R.S. Heist reports other support from AbbVie, AstraZeneca, Daichii Sankyo, Claim, Lilly, Novartis, Merck, Sanofi, and Regeneron; grants from Daichii Sankyo, Lilly, Mirati, Novartis, Erasca, Mythic, AbbVie, Agios, and Corvus; and grants from Turning Point outside the submitted work. A. Elliott reports personal fees from Caris Life Sciences outside the submitted work. A.M. Vanderwalde reports other support from Caris Life Sciences during the conduct of the study; other support from George Clinical outside the submitted work. B. Halmos reports grants and personal fees from AstraZeneca, Merck, Boehringer Ingelheim, Pfizer, Janssen, Daiichi, AbbVie, Amgen, Takeda, and BMS; grants from GSK, Advaxis, Black Diamond, Forward Pharma, and Arrivent; personal fees from Genentech; and personal fees from Precede outside the submitted work. S.V. Liu reports grants and personal fees from AbbVie, AstraZeneca, Bristol-Myers Squibb, Elevation Oncology, Genentech, Gilead, Merck, Merus, RAPT, and Turning Point Therapeutics; personal fees from Amgen, Boehringer Ingelheim, Catalyst, Daiichi Sankyo, Guardant Health, Janssen, Jazz Pharmaceuticals, Mirati, Novartis, Pfizer, Regeneron Revolution Medicines, Sanofi, Takeda, grants from Alkermes, Ellipses, Nuvalent, OSE Immunotherapeutics, Puma; and personal fees from Candel Therapeutics outside the submitted work. J. Che reports a scientific cofounder for Matchpoint therapeutics and M3 bioinformatics & Technology Inc. He is an equity holder and scientific consultant for Matchpoint, Allorion, and Soltego. A.B. Schrock reports personal fees from Foundation Medicine and Roche during the conduct of the study. A. Drilon reports personal fees from 14ner/Elevation Oncology, Amgen, AbbVie, ArcherDX, AstraZeneca, Beigene, BergenBio, Blueprint Medicines, Chugai Pharmaceutical, EcoR1, EMD Serono, Entos, Exelixis, Helsinn, Hengrui Therapeutics, Ignyta/Genentech/Roche, Janssen, Loxo/Bayer/Lilly, Merus, Monopteros, MonteRosa, Novartis, Nuvalent, Pfizer, Prelude, Repare RX, Takeda/Ariad/Millenium, Treeline Bio, TP Therapeutics, Tyra Biosciences, Verastem during the conduct of the study; other support from Foundation Medicine, Teva, Taiho, GlaxSmithKlein, other support from mBrace, Treeline, other support from Boehringer Ingelheim, Merck, Puma, and personal fees from Wolters Kluwer, UpToDate outside the submitted work; in addition, A. Drilon has a patent for Selpercatinib-Osimertinib (filed/pending) pending; and CME Honoraria: Answers in CME, Applied Pharmaceutical Science, Inc., AXIS, Clinical Care Options, EPG Health, Harborside Nexus, I3 Health, Imedex, Liberum, Medendi, Medscape, Med Learning, MJH Life Sciences, MORE Health, Ology, OncLive, Paradigm, Peerview Institute, PeerVoice, Physicians Education Resources, Remedica Ltd., Research to Practice, RV More, Targeted Oncology, TouchIME, WebMD. P.A. Jänne reports grants and personal fees from AstraZeneca, Boehringher Ingelheim, Eli Lilly, and Daiichi Sankyo; personal fees from Pfizer, Roche/Genentech, Chugai Pharmaceuticals, SFJ Pharmaceuticals, Voronoi, Biocartis, Novartis, Sanofi Oncology, Takeda Oncology, Mirati Therapeutics, Transcenta, Silicon Therapeutics, Syndax, Nuvalent, Bayer, Eisai, Allorion Therapeutics, Accutar Biotech, AbbVie, Mone Rosa, Scorpion Therapeutics, Merus, Frontier Medicines, Hongyun Biotechnology, Duality Biologics, and Blueprint Medicines; grants from PUMA and REvolution Medicines outside the submitted work; in addition, P.A. Jänne has a patent for EGFR mutations issued and licensed to Lab Corp. M.M. Awad reports personal fees from Genmab, Janssen, Genentech, Merck, Novartis, Mirati, and Blueprint; grants and personal fees from Bristol-Myers Squibb and AstraZeneca; grants from Amgen; and grants and personal fees from Lilly during the conduct of the study. No disclosures were reported by the other authors.
Authors’ Contributions
F. Pecci: Conceptualization, data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft. S. Nakazawa: Conceptualization, data curation, software, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft. B. Ricciuti: Conceptualization, data curation, formal analysis, supervision, methodology, writing–original draft. G. Harada: Data curation, formal analysis, investigation, methodology, writing–original draft. J.K. Lee: Conceptualization, data curation, formal analysis, methodology, writing–original draft. J.V. Alessi: Conceptualization, data curation, formal analysis, methodology, writing–original draft. A. Barrichello: Data curation, writing–original draft. V.R. Vaz: Data curation, writing–original draft. G. Lamberti: Data curation, visualization, methodology, writing–original draft. A. Di Federico: Data curation, formal analysis, methodology, writing–original draft. M.M. Gandhi: Data curation, writing–original draft. D. Gazgalis: Conceptualization, software, formal analysis, investigation, methodology, writing–original draft. W.W. Feng: Conceptualization, data curation, methodology, writing–original draft. J. Jiang: Data curation, methodology, writing–original draft. S. Baldacci: Data curation, methodology, writing–original draft. M. Locquet: Data curation, writing–original draft. F.H. Gottlieb: Data curation, methodology, writing–original draft. M.F. Chen: Data curation, writing–original draft. E. Lee: Data curation. D. Haradon: Data curation. A. Smokovich: Data curation. E. Voligny: Data curation. T. Nguyen: Data curation, investigation, writing–original draft. V.K. Goel: Data curation, investigation, writing–original draft. Z. Zimmerman: Conceptualization, data curation, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft. S. Atwal: Conceptualization, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft. X. Wang: Conceptualization, data curation, formal analysis, supervision, investigation, methodology, writing–original draft. M. Bahcall: Conceptualization, data curation, formal analysis, supervision, methodology, writing–original draft. R.S. Heist: Conceptualization, data curation, formal analysis, supervision, methodology, writing–original draft. S. Iqbal: Conceptualization, data curation, formal analysis, supervision, methodology, writing–original draft. N. Gandhi: Conceptualization, data curation, formal analysis, supervision, methodology, writing–original draft. A. Elliott: Data curation, formal analysis, methodology, writing–original draft. A.M. Vanderwalde: Data curation, writing–original draft. P.C. Ma: Data curation, supervision, writing–original draft. B. Halmos: Data curation, supervision, writing–original draft. S.V. Liu: Conceptualization, data curation, formal analysis, supervision, methodology, writing–original draft. J. Che: Conceptualization, data curation, software, formal analysis, supervision, methodology, writing–original draft. A.B. Schrock: Conceptualization, data curation, software, formal analysis, supervision, methodology, writing–original draft. A. Drilon: Conceptualization, data curation, software, formal analysis, supervision, funding acquisition, visualization, methodology, writing–original draft. P.A. Jänne: Conceptualization, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft. M.M. Awad: Conceptualization, data curation, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft.
References
- 1. Recondo G, Che J, Jänne PA, Awad MM. Targeting MET dysregulation in cancer. Cancer Discov 2020;10:922–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wolf J, Seto T, Han J-Y, Reguart N, Garon EB, Groen HJM, et al. Capmatinib in MET exon 14–mutated or MET-amplified non–small-cell lung cancer. N Engl J Med 2020;383:944–57. [DOI] [PubMed] [Google Scholar]
- 3. Paik PK, Felip E, Veillon R, Sakai H, Cortot AB, Garassino MC, et al. Tepotinib in non–small-cell lung cancer with MET exon 14 skipping mutations. N Engl J Med 2020;383:931–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Remon J, Hendriks LEL, Mountzios G, García-Campelo R, Saw SPL, Uprety D, et al. MET alterations in NSCLC—current perspectives and future challenges. J Thorac Oncol 2023;18:419–35. [DOI] [PubMed] [Google Scholar]
- 5. Stella G, Senetta R, Balderacchi A, Cassoni P, Comoglio P, Benvenuti S. Oncogenic potential of MET SEMA mutations affecting brain metastases from NSCLC is sustained by their microrheological features. Eur Respir J 2017;50:PA3306. [Google Scholar]
- 6. Ma PC, Kijima T, Maulik G, Fox EA, Sattler M, Griffin JD, et al. c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res 2003;63:6272–81. [PubMed] [Google Scholar]
- 7. Camidge DR, Otterson GA, Clark JW, Ignatius Ou SH, Weiss J, Ades S, et al. Crizotinib in patients with MET-amplified NSCLC. J Thorac Oncol 2021;16:1017–29. [DOI] [PubMed] [Google Scholar]
- 8. Cheng F, Guo D. MET in glioma: signaling pathways and targeted therapies. J Exp Clin Cancer Res 2019;38:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang W, Zhao X, Zheng A, Liu Z, Ma J, Zhang X, et al. Identification of MET fusions in solid tumors: a multicenter, large scale study in China. Int J Cancer 2023;152:1259–68. [DOI] [PubMed] [Google Scholar]
- 10. Camidge DR, Barlesi F, Goldman JW, Morgensztern D, Heist R, Vokes E, et al. Phase Ib study of telisotuzumab vedotin in combination with erlotinib in patients with c-met protein–expressing non–small-cell lung cancer. J Clin Oncol 2023;41:1105–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cancer Genome Atlas Research Network; Linehan WM, Spellman PT, Ricketts CJ, Creighton CJ, Fei SS, et al. Comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med 2016;374:135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tovar EA, Graveel CR. MET in human cancer: germline and somatic mutations. Ann Transl Med 2017;5:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hunt JL, Barnes L, Lewis JS, Mahfouz ME, Slootweg PJ, Thompson LDR, et al. Molecular diagnostic alterations in squamous cell carcinoma of the head and neck and potential diagnostic applications. Eur Arch Otorhinolaryngol 2014;271:211–23. [DOI] [PubMed] [Google Scholar]
- 14. Recondo G, Bahcall M, Spurr LF, Che J, Ricciuti B, Leonardi GC, et al. Molecular mechanisms of acquired resistance to MET tyrosine kinase inhibitors in patients with MET exon 14–mutant NSCLC. Clin Cancer Res 2020;26:2615–25. [DOI] [PubMed] [Google Scholar]
- 15. Miao Y, Feher VA, McCammon JA. Gaussian accelerated molecular dynamics: unconstrained enhanced sampling and free energy calculation. J Chem Theor Comput 2015;11:3584–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miao Y, Bhattarai A, Wang J. Ligand Gaussian accelerated molecular dynamics (LiGaMD): characterization of ligand binding thermodynamics and kinetics. J Chem Theor Comput 2020;16:5526–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miller BR, McGee TD, Swails JM, Homeyer N, Gohlke H, Roitberg AE. MMPBSA.py: an efficient program for end-state free energy calculations. J Chem Theor Comput 2012;8:3314–21. [DOI] [PubMed] [Google Scholar]
- 18. Matsubara J, Mukai K, Kondo T, Yoshioka M, Kage H, Oda K, et al. First-line genomic profiling in previously untreated advanced solid tumors for identification of targeted therapy opportunities. JAMA Netw Open 2023;6:e2323336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sacher AG, Dahlberg SE, Heng J, Mach S, Jänne PA, Oxnard GR. Association between younger age and targetable genomic alterations and prognosis in non–small-cell lung cancer. JAMA Oncol 2016;2:313–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hendriks LE, Kerr KM, Menis J, Mok TS, Nestle U, Passaro A, et al. Oncogene-addicted metastatic non-small-cell lung cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol 2023;34:339–57. [DOI] [PubMed] [Google Scholar]
- 21. Couraud S, Zalcman G, Milleron B, Morin F, Souquet PJ. Lung cancer in never smokers—a review. Eur J Cancer 2012;48:1299–311. [DOI] [PubMed] [Google Scholar]
- 22. Tu C-Y, Cheng F-J, Chen C-M, Wang S-L, Hsiao Y-C, Chen C-H, et al. Cigarette smoke enhances oncogene addiction to c-MET and desensitizes EGFR-expressing non-small cell lung cancer to EGFR TKIs. Mol Oncol 2018;12:705–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim SY, Yin J, Bohlman S, Walker P, Dacic S, Kim C, et al. Characterization of MET exon 14 skipping alterations (in NSCLC) and identification of potential therapeutic targets using whole transcriptome sequencing. JTO Clin Res Rep 2022;3:100381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang X, Ricciuti B, Nguyen T, Li X, Rabin MS, Awad MM, et al. Association between smoking history and tumor mutation burden in advanced non-small cell lung cancer. Cancer Res 2021;81:2566–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hellyer JA, Stehr H, Das M, Padda SK, Ramchandran K, Neal JW, et al. Impact of KEAP1/NFE2L2/CUL3 mutations on duration of response to EGFR tyrosine kinase inhibitors in EGFR mutated non-small cell lung cancer. Lung Cancer 2019;134:42–5. [DOI] [PubMed] [Google Scholar]
- 26. Frank R, Scheffler M, Merkelbach-Bruse S, Ihle MA, Kron A, Rauer M, et al. Clinical and pathological characteristics of KEAP1- and NFE2L2-mutated non–small cell lung carcinoma (NSCLC). Clin Cancer Res 2018;24:3087–96. [DOI] [PubMed] [Google Scholar]
- 27. Scheffler M, Ihle MA, Hein R, Merkelbach-Bruse S, Scheel AH, Siemanowski J, et al. K-Ras mutation subtypes in NSCLC and associated Co-occuring mutations in other oncogenic pathways. J Thorac Oncol 2019;14:606–16. [DOI] [PubMed] [Google Scholar]
- 28. Fujino T, Kobayashi Y, Suda K, Koga T, Nishino M, Ohara S, et al. Sensitivity and resistance of MET exon 14 mutations in lung cancer to eight MET tyrosine kinase inhibitors in vitro. J Thorac Oncol 2019;14:1753–65. [DOI] [PubMed] [Google Scholar]
- 29. Husain H, Pavlick DC, Fendler BJ, Madison RW, Decker B, Gjoerup O, et al. Tumor fraction correlates with detection of actionable variants across > 23,000 circulating tumor DNA samples. JCO Precis Oncol 2022;6:e2200261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu L, Yao H, Chen H, Wang A, Guo K, Gou W, et al. Landscape of somatic alterations in large-scale solid tumors from an Asian population. Nat Commun 2022;13:4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ricciuti B, Wang X, Alessi JV, Rizvi H, Mahadevan NR, Li YY, et al. Association of high tumor mutation burden in non–small cell lung cancers with increased immune infiltration and improved clinical outcomes of PD-L1 blockade across PD-L1 expression levels. JAMA Oncol 2022;8:1160–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garcia EP, Minkovsky A, Jia Y, Ducar MD, Shivdasani P, Gong X, et al. Validation of OncoPanel: a targeted next-generation sequencing assay for the detection of somatic variants in cancer. Arch Pathol Lab Med 2017;141:751–8. [DOI] [PubMed] [Google Scholar]
- 33. Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015;17:251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee JK, Sivakumar S, Schrock AB, Madison R, Fabrizio D, Gjoerup O, et al. Comprehensive pan-cancer genomic landscape of KRAS altered cancers and real-world outcomes in solid tumors. NPJ Precis Oncol 2022;6:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Milbury CA, Creeden J, Yip WK, Smith DL, Pattani V, Maxwell K, et al. Clinical and analytical validation of FoundationOne®CDx, a comprehensive genomic profiling assay for solid tumors. PLoS One 2022;17:e0264138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Clark TA, Chung JH, Kennedy M, Hughes JD, Chennagiri N, Lieber DS, et al. Analytical validation of a hybrid capture-based next-generation sequencing clinical assay for genomic profiling of cell-free circulating tumor DNA. J Mol Diagn 2018;20:686–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee JK, Madison R, Classon A, Gjoerup O, Rosenzweig M, Frampton GM, et al. Characterization of non–small-cell lung cancers with MET exon 14 skipping alterations detected in tissue or liquid: clinicogenomics and real-world treatment patterns. JCO Precis Oncol 2021;5:PO.21.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Woodhouse R, Li M, Hughes J, Delfosse D, Skoletsky J, Ma P, et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One 2020;15:e0237802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet 2013;76:7.20.1–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 2012;40:W452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Iqbal S, Pérez-Palma E, Jespersen JB, May P, Hoksza D, Heyne HO, et al. Comprehensive characterization of amino acid positions in protein structures reveals molecular effect of missense variants. Proc Natl Acad Sci U S A 2020;117:28201–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chakravarty D, Gao J, Phillips S, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol 2017;2017:PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hong DS, Bauer TM, Lee JJ, Dowlati A, Brose MS, Farago AF, et al. Larotrectinib in adult patients with solid tumours: a multi-centre, open-label, phase I dose-escalation study. Ann Oncol 2019;30:325–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Subbiah V, Cassier PA, Siena S, Garralda E, Paz-Ares L, Garrido P, et al. Pan-cancer efficacy of pralsetinib in patients with RET fusion–positive solid tumors from the phase 1/2 ARROW trial. Nat Med 2022;28:1640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Owen DH, Singh N, Ismaila N, Masters G, Riely GJ, Robinson AG, et al. Therapy for stage IV non-small-cell lung cancer with driver alterations: ASCO living guideline, version 2023.2. J Clin Oncol 2023;41:e63–72. [DOI] [PubMed] [Google Scholar]
- 47. Ercan D, Zejnullahu K, Yonesaka K, Xiao Y, Capelletti M, Rogers A, et al. Amplification of EGFR T790M causes resistance to an irreversible EGFR inhibitor. Oncogene 2010;29:2346–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature 2009;462:1070–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional information regarding the methods used in this study
Supplementary Table 1. Prevalence of MET tyrosine kinase domain (TKD) mutations in each dataset of the cohort #1.
Frequency of MET tyrosine kinase domain (TKD) mutations in cohort #1 according to amino acid residue, out of a total of 711 MET TKD mutations.
Frequency of MET tyrosine kinase domain (TKD) mutations in cohort #2 according to amino acid residue, out of a total of 2,559 MET TKD mutations.
List of missense mutations with unknown biologic function and their pathogenicity scores using all three in silico tools in cohort #1
List of missense mutations with unknown biologic function and their pathogenicity scores using all three in silico tools in cohorts #2
Prevalence of MET tyrosine kinase domain (TKD) mutations across cancer types in cohort #1.
Prevalence of MET tyrosine kinase domain (TKD) mutations among “other cancers” from Supplementary Table 6.
Prevalence of MET tyrosine kinase domain (TKD) mutations across cancer types in cohort #2.
Prevalence of MET tyrosine kinase domain (TKD) mutations across tumor in “other cancers” from Supplementary Table 8.
Characteristics of 44 NSCLC cases harboring oncogenic / likely oncogenic MET TKD mutations in cohort #1.
Detailed genomic characteristics for each case of NSCLC with oncogenic / likely oncogenic MET tyrosine kinase domain (TKD) mutations in cohort #1.
Characteristics of 129 NSCLC cases harboring oncogenic / likely oncogenic MET TKD mutations from cohort #2.
Concurrent driver mutations according to oncogenic / likely oncogenic MET TKD mutations detected in cohort #2.
Concurrent genomic drivers in cases with oncogenic / likely oncogenic MET tyrosine kinase domain (TKD) mutations and co-occurring MET gene amplification in cohort #2.
Comparison between clinicopathologic characteristic of NSCLCs with oncogenic / likely oncogenic MET tyrosine kinase domain (TKD) mutations as a de novo genomic event to NSCLCs with MET exon 14 alterations in cohort #1.
Comparison between clinicopathologic characteristic of NSCLCs with oncogenic / likely oncogenic MET tyrosine kinase domain (TKD) mutations without concurrent other drivers to NSCLCs with MET exon 14 alterations in cohort #2.
Comparison of clinicopathologic characteristics between NSCLCs with oncogenic / likely oncogenic MET tyrosine kinase domain (TKD) mutations without other concurrent drivers and NSCLCs with MET exon 14 alterations in the Caris Life Sciences cohort.
Detailed clinicopathologic and genomic characteristics of the 61 cases with a malignancy other than NSCLC harboring an oncogenic / likely oncogenic MET TKD mutation in cohort #1.
Detailed clinicopathologic and genomic characteristics related to renal cell cancer cases in cohort #1.
Clinicopathologic and genomic characteristics of 171 cases of cancers other than NSCLC harboring oncogenic / likely oncogenic MET TKD mutations in cohort #2.
Detailed clinicopathologic and genomic characteristics related to renal cell cancer cases in cohort #2.
Oncogenic / likely oncogenic MET TKD mutations in RCC compared to those detected in NSCLC in cohort #1.
Oncogenic / likely oncogenic MET TKD mutations in RCC compared to those detected in NSCLC in cohort #2.
Single trajectory generalized Born and surface area solvation (MM/GBSA) calculated free energy of ATP binding into the orthosteric pocket.
TCGA PanCancer Atlas Studies included in cohort #1 downloaded from cBioPortal.
Mutagenic primers used for mutagenesis.
Study flowchart. Cohort #1 (multi-institutional cohort): GENIE V.11.0, China Pan-cancer (OrigiMed2020, Nature 2022), The Cancer Genome Atlas (TCGA) Pan-Cancer Atlas studies, Dana-Farber Cancer Institute (DFCI) and NSCLC from Memorial Sloan Kettering Cancer Center (MSKCC); cohort #2: Foundation Medicine Pan-Cancer cohort.
Overview of MET TKD mutations detected in pan-cancer cohorts. (A) Type (missense, truncating, splice site, in-frame) of MET TKD mutations detected in the cohort #1 (711 MET TKD mutations among 664 unique patients with a tumor harboring MET TKD mutations without concurrent METex14 alterations). (B) Type of MET TKD mutations according to OncoKB annotation (biological function: oncogenic / likely oncogenic, unknown) detected in cohort #1. (C) Distribution of MET TKD mutations across MET kinase domain for cohort #1. (D) Type (missense, truncating, splice site, in-frame) of MET TKD mutations detected in cohort #2 (2,559 MET TKD mutations among 2,477 unique patients with a tumor harboring MET TKD mutations without concurrent METex14 alterations). (E) Type of MET TKD mutations according to OncoKB annotation (biological function: oncogenic / likely oncogenic, unknown) detected in cohort #2. (F) Distribution of MET TKD mutation across MET kinase domain for cohort #2.
Comparison of missense MET TKD defined as “probably pathogenic” and “probably benign” according to three in silico tools. Comparison of (A) secondary structure features, (B) relative solvent accessible surface area (RSA), and (C) mutant amino acid properties of missense MET TKD mutations of unknown biological function according to OncoKB. Mutations were defined as “probably pathogenic” for likely pathogenicity across all three tools and as “probably benign” for likely benign across all three scores. OR: odds ratio; 95% CI: 95% confidence interval.
Type and frequency of MET TKD mutations across cancer types in cohort #1. (A) Frequency of MET TKD mutations in various cancer types in cohort #1 according to OncoKB status; the numbers above each bar indicate the number of cases with MET TKD mutations (regardless OncoKB annotations) out of the total of cases of each cancer type. (B) Detailed oncogenic / likely oncogenic MET TKD mutations according to cancer type in cohort #1.
Prevalence of MET TKD mutation in various cancer types according to OncoKB status in each dataset in cohort #1.
MET TKD mutations in NSCLC in cohort #1 and #2. (A) Flowchart of the NSCLC subgroup in cohort #1 (123 MET TKD mutations in 120 unique patients); (B) Prevalence of MET TKD mutations in NSCLC cases according to each dataset in cohort #1; (C) Lollipop plot of the oncogenic / likely oncogenic MET TKD mutations detected in NSCLC cases of cohort #1 (47 oncogenic / likely oncogenic MET TKD mutations in 44 unique patients); (D) Flowchart of the NSCLC subgroup in cohort #2 (590 MET TKD mutations in 586 unique patients); (E) Prevalence of MET TKD mutations in NSCLC cases according to cohort #2 (590 MET TKD mutations in 586 unique patients); (F) Lollipop plot of the oncogenic / likely oncogenic MET TKD mutations detected in NSCLC cases of cohort #2 (129 oncogenic / likely oncogenic MET TKD mutations in 129 unique patients).
Genomic characterization of NSCLC harboring oncogenic / likely oncogenic MET TKD mutations in cohort #1 and #2. Genomic characteristics of 44 NSCLC cases harboring oncogenic / likely oncogenic MET TKD mutations from cohort #1: (A) concurrent driver alterations and (B) concurrent MET amplification status (information unavailable for 4 cases). Genomic characteristics of 129 NSCLC cases harboring oncogenic / likely oncogenic MET TKD mutations from cohort #2: (C) concurrent driver alterations and (D) concurrent MET amplification status.
Concurrent oncogenic driver alterations according to specific oncogenic / likely oncogenic MET TKD mutations detected in NSCLC cases (N=129) of cohort #2.
Genomic characterization of NSCLC cases harboring oncogenic / likely oncogenic MET TKD mutations in cohort #2. (A) Oncoprint summarizing the genomic profile and (B) the TMB value of the 78 NSCLC cases with oncogenic / likely oncogenic MET TKD mutations without other concurrent driver alterations according to NCCN guidelines. (C) Comparison of the genomic profile of NSCLC cases with oncogenic / likely oncogenic MET TKD mutations (N=78) and METex14-altered NSCLC (N=2036)
MET TKD mutations in NSCLC cohort of Caris Life Sciences. (A) Flowchart of the NSCLC subgroup in the Caris Life Sciences dataset. (B) Prevalence of MET TKD mutations in NSCLC cases in the Caris Life Sciences dataset (MET TKD mutations in 280 unique patients). (C) Lollipop plot of the oncogenic / likely oncogenic MET TKD mutations detected in NSCLC cases in the Caris Life Sciences dataset (35 oncogenic / likely oncogenic MET TKD mutations in 35 unique patients). (D) concurrent driver alterations (information available for 24 cases, unavailable for 11 cases) and (E) concurrent MET amplification status (information available for 33 cases, unavailable for 2 cases) of NSCLC cases harboring oncogenic / likely oncogenic MET TKD mutations in the Caris Life Sciences dataset.
In vitro analysis of “probably benign” MET TKD mutations in Ba/F3 cells. (A) Phosphorylation of MET was analyzed in Ba/F3 cells stably transduced with MET TKD mutants. MET H1094Y and F1200I were included as positive controls for oncogenic mutations (OncoKB, also detected in the two patient cases) and MET WT was included as negative control. (B) IL-3-independent cell proliferation of Ba/F3 cells expressing “probably benign” MET TKD mutants, shown by relative spheroid size measured daily from day 1 to day 7. (C) Spheroid images of Ba/F3 cells expressing MET TKD mutants taken on day 1 and day 7.
Sensitivity pattern of MET TKD mutations in 293T cells. (A) ERK-mediated transcriptional activation assessed by luciferase assay in 293T cells co-transfected with MET WT or TKD mutant constructs, an ERK activation reporter vector (pGL4.33), and a control co-reporter vector (pRL-TK). The lower panel shows western blot analysis of transfected samples. (B) Western blot analysis of MET and ERK1/2 phosphorylation in response to 1 μM crizotinib. MET TKD mutants showing sensitivity and resistance to crizotinib are colored in green and orange, respectively.
Growth inhibition assays of Ba/F3 cells expressing MET TKD mutants under treatment with MET TKIs.
Western blot analysis of MET and ERK1/2 phosphorylation in MET R1170Q cell models. MET and ERK1/2 phosphorylation in (A) Ba/F3 cells stably transduced with MET R1170Q and (B) 293T cells transiently overexpressing MET R1170Q. Cells were treated with MET TKIs at the indicated dose for Ba/F3 cells or at 1 μM for 293T cells.
In vitro analysis of MET H1094Y and F1200I mutations in PC9 cells. (A) Establishment of NSCLC cancer cell lines expressing MET TKD mutations. PC9 cells were transduced with wild-type (WT) MET (as control) and MET H1094Y and F1200I. (B) Growth inhibition assay of PC9 cells in response to EGFR inhibition with osimertinib. (C) Cell proliferation and relative apoptosis were monitored under EGFR inhibition with osimertinib (300 nM). (D) Phosphorylation of MET, EGFR, AKT, and ERK was analyzed in PC9 cells expressing MET TKD mutants. PC9 cells were treated with osimertinib (1 μM) with or without capmatinib (1 μM) for 48 hrs. (E) IC50 values of PC9 cells (upper table) and Ba/F3 cells (lower table) expressing MET TKD mutants in response to MET TKIs. PC9 cells were treated with gradient dose of MET TKI combined with a fixed dose of osimertinib (100 nM) for 72 hrs.
Heavy atom root-mean-square deviation. Heavy atom root-mean-square deviation (RMSD) of each simulation: (A) MET wild-type, (B) MET H1094Y, (C) MET F1200I, and (D) R1170Q. Each frame is taken in a 0.1 ns interval.
Equilibration of interactions that drives ATP affinity in the presence MET H1094Y mutant. (A) Representative structure of the ATP bound cMET H1094Y with the regions of interest highlighted [subsection of the 5 pleated beta sheet bundle containing H1094Y and direct interactions (red), hinge binding region (green)]. (B) H1094Y forms novel π- π stacking interactions (T stacking with an upper cut-off of 5.0 angstroms) with Y1158. (C) π- π stacking from Y1158 stabilizes the ATP hydrogen bonding network.
Time resolved interaction distances of ATP in the presence of MET WT and MET H1094Y mutant. (A) Hydrogen bonding between P1157 backbone carbonyl and ATP, (B) Inter ring center distances for Y1158-ATP π–π stacking interactions, and (C) Hydrogen bonding between M1159 backbone amine and ATP distances for ATP interactions between MET wild-type (orange) and MET H1094Y (blue) over the last 500 ns of each trajectory.
Equilibration of interactions that drives ATP affinity in the presence MET F1200I mutant. (A) Representative structure of ATP bound MET F1200I with the regions of interest highlighted [alpha C helix (red), DFG site (green), mutation site (blue)]. (B) π - π stacking interactions that bridge the alpha C helix and DFG site are conformationally controlled through residues position 1200. π - π stacking interactions were calculated between ring centers. Distances between F1200I and neighboring residues were measured from the F1200I side chain center of mass to the ring centers of the aromatic rings. (C) root-mean-square-fluctuation (RMSF) calculations show a stabilization of the activation loop in the F1200I system.
Equilibration of interactions that drives ATP affinity in the presence MET R1170Q mutant. (A) Representative structure of ATP bound MET R1170Q with the regions of interest highlighted [hinge site (green), and mutation site (blue)]. (B) Mutation at position 1170 facilitates the formation of a hydrogen bonding network. (C) Interatomic distances for non solvent mediated hydrogen bonding.
Data Availability Statement
The data that support the finding of our study are available upon request from the corresponding author.