In addition to de novo F-actin assembly at the division site, directed transport of F-actin cables assembled elsewhere can contribute to actomyosin ring assembly during cytokinesis.
Abstract
In many eukaryotes, cytokinesis requires the assembly and constriction of an actomyosin-based contractile ring. Despite the central role of this ring in cytokinesis, the mechanism of F-actin assembly and accumulation in the ring is not fully understood. In this paper, we investigate the mechanism of F-actin assembly during cytokinesis in Schizosaccharomyces pombe using lifeact as a probe to monitor actin dynamics. Previous work has shown that F-actin in the actomyosin ring is assembled de novo at the division site. Surprisingly, we find that a significant fraction of F-actin in the ring was recruited from formin-Cdc12p nucleated long actin cables that were generated at multiple nonmedial locations and incorporated into the ring by a combination of myosin II and myosin V activities. Our results, together with findings in animal cells, suggest that de novo F-actin assembly at the division site and directed transport of F-actin cables assembled elsewhere can contribute to ring assembly.
Introduction
Cytokinesis is the final step in the process of cell division, during which two daughter cells are generated. In many eukaryotic cells, ranging from yeasts to human, cytokinesis requires the function of an actomyosin-based contractile ring. This ring consists of overlapping actin filaments (Satterwhite and Pollard, 1992; Kamasaki et al., 2007) that interact with the molecular motor myosin II (Schroeder, 1973). Force generation by myosin II then leads to ring closure and cell division (Balasubramanian et al., 2004; Wolfe and Gould, 2005; Pollard and Wu, 2010). Both assembly and constriction of the actomyosin ring are influenced by a large array of actin-modulating proteins, which regulate actin polymerization, cross-linking, and disassembly (Balasubramanian et al., 1992, 1994; Chang et al., 1997; Kitayama et al., 1997; Wu et al., 2001, 2003; Nakano and Mabuchi, 2006).
Although we have gained a detailed understanding of many aspects of cytokinesis, the mechanisms by which actin filaments assemble at the division site are still not fully understood. Studies of actin dynamics have suffered from lack of fully functional and fluorescently tagged versions of actin (Doyle and Botstein, 1996; Kovar et al., 2005; Wu and Pollard, 2005). Several studies, using probes that serve as a surrogate for the actin cytoskeleton, have shown that actin filaments assemble de novo at the division site (Noguchi and Mabuchi, 2001; Wu et al., 2006). In addition, in some other cell types, actin filaments can be transported to the division site through a process known as cortical flow (White and Borisy, 1983; Bray and White, 1988; Cao and Wang, 1990; Lee et al., 1998). More recently, increasing evidence suggests that de novo assembly and cortical flow of actin filaments can jointly contribute to actomyosin ring assembly (Chen et al., 2008; Zhou and Wang, 2008; Alsop et al., 2009).
The fission yeast Schizosaccharomyces pombe is one of the most tractable cell types that divides using an actomyosin-based contractile ring (Balasubramanian et al., 2004; Wolfe and Gould, 2005; Pollard and Wu, 2010). Genetic analyses have uncovered a large number of proteins, including formins, myosins, and other actin-binding proteins, as key elements of the cytokinetic machinery (Balasubramanian et al., 1992, 1994, 1998; Fankhauser et al., 1995; McCollum et al., 1995; Chang et al., 1996, 1997; Kitayama et al., 1997; May et al., 1997). The actomyosin ring is assembled at the cell middle in early mitosis through a pathway initiated by the anillin-related protein Mid1p (Wu et al., 2003). Ring maturation and maintenance during later stages of mitosis depend on a pathway requiring the F-BAR protein Cdc15p (Wu et al., 2006; Hachet and Simanis, 2008; Huang et al., 2008; Mishra and Oliferenko, 2008; Vavylonis et al., 2008). In a set of elegant studies, actin filaments for cytokinesis have been shown to be assembled de novo at the division site by the formin Cdc12p either from a series of cortical nodes (Wu et al., 2006; Vavylonis et al., 2008) or from a single dominant spot (Chang, 1999; Arai and Mabuchi, 2002). Actin cables that run along the long axis of the cell have also been identified. However, with the exception of one study (Arai and Mabuchi, 2002), actin cables have been thought to solely contribute to cell polarization in interphase (Feierbach and Chang, 2001; Nakano et al., 2002).
In this study, we use lifeact (LA), a recently developed marker for labeling F-actin in living cells (Riedl et al., 2008), as a probe to monitor actin dynamics in S. pombe cells. We find that actin filaments for the cytokinetic ring are organized from actin cables that are assembled throughout the cell, including the cell middle. Nonmedially assembled actin filaments are transported to the division site and compacted through a combination of myosin II and myosin V activities. Our study suggests that similar mechanisms operate for F-actin assembly at the division sites of fission yeast and animal cells.
Results
LA decorates all actin structures stained by phalloidin
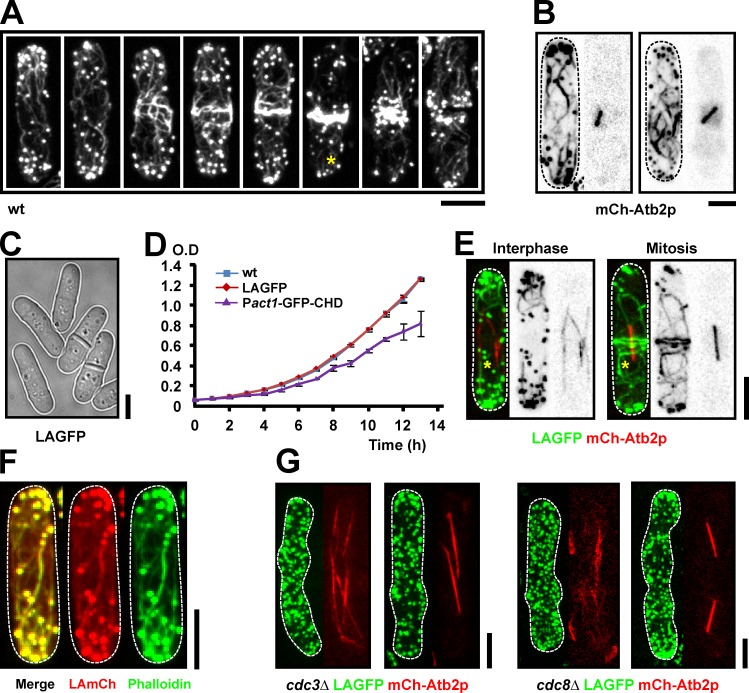
Studies in fission yeast have shown that during cytokinesis, actin filaments are assembled at the division site, either from an array of cortical nodes (Wu et al., 2006; Vavylonis et al., 2008) or from spotlike precursors (Chang, 1999; Arai and Mabuchi, 2002). However, consistent with a previous study on fixed cells (Arai and Mabuchi, 2002), we found that Alexa Fluor 488 phalloidin stains prominent actin cables distributed throughout the cell cortex of both interphase and mitotic wild-type (wt) S. pombe cells (Fig. 1 A). Furthermore, actin cables were less confined to the medial region of the cell during cytokinesis than had been appreciated previously. Using the mCherry (mCh)-Atb2p fusion protein as a microtubule marker for mitotic progression, we found that in early mitotic cells (cells with short spindles) actin cables covered the whole-cell cortex and were oriented along the cell axis (Fig. 1 B). In addition, short actin cables were also located at the cell ends in cells with mature actomyosin rings (Fig. 1 A, asterisk).
Figure 1.
LA is a reliable tool to visualize F-actin in living fission yeast cells. (A) wt cells were fixed with paraformaldehyde and stained with Alexa Fluor 488 phalloidin. The asterisk shows a short cable located at the end of a mitotic cell. (B) Cells carrying mCh-Atb2p were stained as in A. (C) Bright-field image of LAGFP cells. (D) Growth curves of wt, LAGFP, and Pact1-GFP-CHD cells grown in YES at 30°C. Error bars show means ± SD of three independent experiments. (E) LAGFP mCh-Atb2p cells in interphase and mitosis. Asterisks indicate actin cables. (F) LAmCh cells were stained as in A. (G) Images of interphase and mitotic cells of cdc3Δ LAGFP mCh-Atb2p and cdc8Δ LAGFP mCh-Atb2p germinated from spores. Dashed lines indicate the cell boundary. Bars, 5 µm.
Fluorescently labeled LA has been shown to faithfully label F-actin in vivo in various organisms (Vidali et al., 2009; Berepiki et al., 2010; Delgado-Alvarez et al., 2010; Riedl et al., 2010). To gain an understanding of the dynamics of actin cables during cytokinesis, we generated yeast strains expressing LA fused to GFP (LAGFP), mGFP (LAmGFP), or mCh (LAmCh), under control of the fission yeast act1 promoter. Cells expressing these fusion proteins resembled wt cells in morphology (Fig. 1, C and F; and Fig. S1, A and B), growth rate (Fig. 1 D), and cell division (Video 1, middle). LAGFP-expressing cells did not show any genetic interactions with temperature-sensitive mutants defective in cdc3-profilin, cdc8-tropomyosin, or cdc12-formin (unpublished data). LAGFP expression lowered the restrictive temperature of adf1-1, defective in the actin-severing protein cofilin (Nakano and Mabuchi, 2006), suggesting an actin-stabilizing effect of LA, which might be caused by the slight dimerization tendency of GFP (Zacharias et al., 2002). Consistent with this possibility, LAGFP, but not LAmGFP, led to weak phenotypic effects when expressed in the cytokinesis checkpoint mutant clp1Δ (∼10% of cells with more than two nuclei; Mishra et al., 2004). We tested actin stability in cells expressing LAGFP by treating them with the actin polymerization inhibitor Latrunculin A (LatA). In this assay, almost no difference was observed between wt cells and LAGFP-expressing cells, and most F-actin was lost within 4 min of addition of LatA (Fig. S2). In contrast, a similar assay performed on cells expressing GFP–calponin homology domain (CHD; a routinely used marker for the study of actin dynamics) showed stable actin structures persisting even up to 12 min (Fig. S2). These experiments suggested that cells expressing LA fusion proteins phenotypically resembled wt cells, and although minor actin cytoskeletal defects were observed, these were not as severe as in cells expressing GFP-CHD, under our assay conditions.
Fluorescence from all three fusion proteins (LAGFP, LAmGFP, and LAmCh) was detected in actin patches, cables, and rings (Fig. 1, E and F; and Fig. S1). Importantly, cables were labeled in interphase as well as during mitosis (Fig. 1 E). Staining of LAmCh-expressing cells with Alexa Fluor 488 phalloidin revealed a nearly complete overlap of the labeled structures (Fig. 1 F). Actin cables depend on Cdc3p-profilin for their assembly and Cdc8p-tropomyosin for their stability (Balasubramanian et al., 1992, 1994; Pelham and Chang, 2001; Arai and Mabuchi, 2002). Consequently, cables were either absent or very short in length in germinated cdc3Δ and cdc8Δ spores expressing LAGFP (Fig. 1 G). In summary, we established LA as a reliable marker to study actin dynamics in fission yeast without any significant interference to cell physiology.
Nonmedially assembled actin cables incorporate into the actomyosin ring
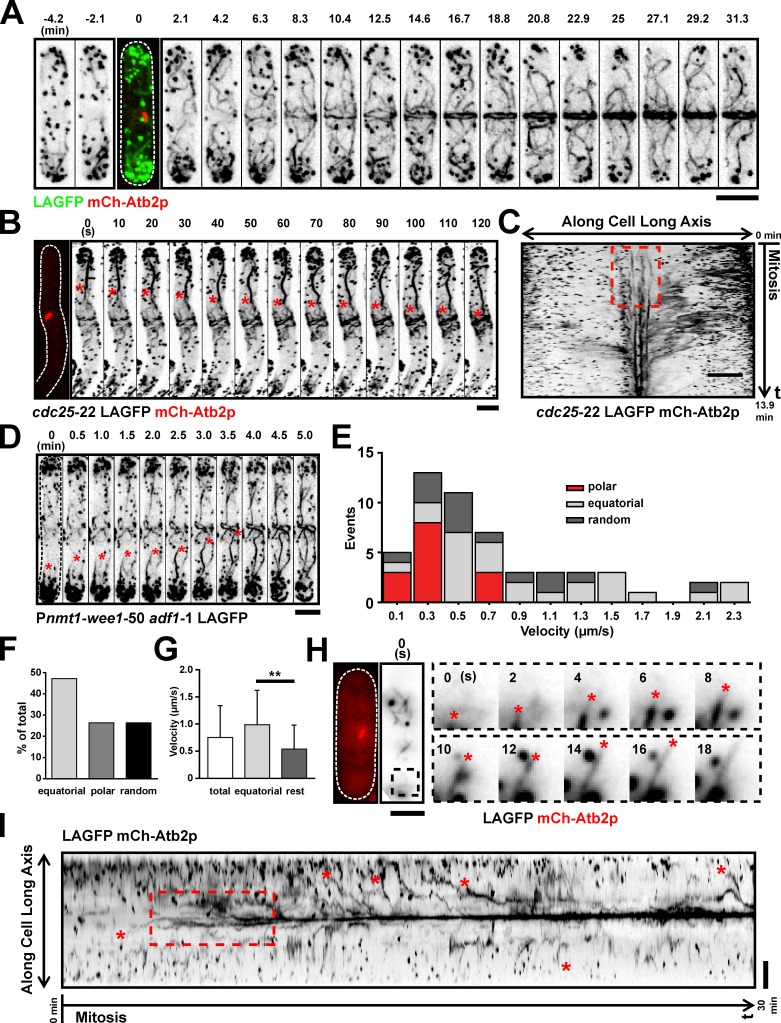
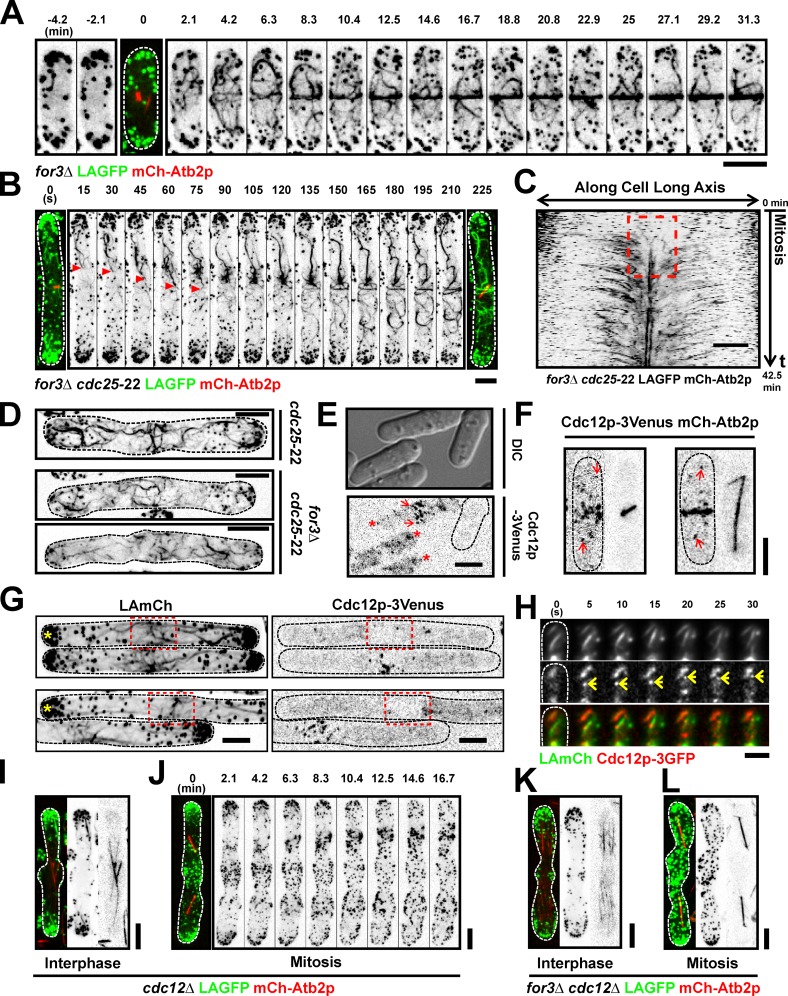
To address the mechanisms responsible for F-actin accumulation at the division site, we monitored actin dynamics during mitosis of wt cells coexpressing LAGFP and mCh-Atb2p. Time-lapse imaging of >30 cells revealed that actin cables were present in interphase (Fig. 2 A, −4.2 and −2.1 min; and Video 1, left) and throughout mitosis (Fig. 2 A, time point 0 min shows a short mitotic spindle marked with mCh-Atb2p; and Video 1, left). These cables were initially distributed throughout the cell and then appeared to incorporate into the maturing ring (Fig. 2 A and Video 1, left). Actin dynamics during ring assembly was nearly identical in cells expressing LAGFP and LAmGFP (Fig. S1 and Video 1, middle and right). Because ring assembly dynamics was similar for both fusion proteins and because LAGFP gave the strongest signal, we used LAGFP as the actin cable marker for the rest of this study.
Figure 2.
Nonmedially assembled actin cables migrate toward the middle during ring formation. (A) Time-lapse images of a LAGFP mCh-Atb2p cell. The third image (time 0 min) is shown as a merged image to indicate the cell cycle stage. Video 1. (B) Time-lapse images of a cdc25-22 LAGFP mCh-Atb2p cell after release from 36 to 24°C. The first micrograph on the left was taken before the video was started. Asterisks show a nonmedially assembled actin cable migrating to the medial region during ring assembly. Time interval of this video was 5 s. Video 2 (left cell). (C) Kymograph of ring assembly in a cdc25-22 LAGFP mCh-Atb2p cell revealing a flow of actin fluorescence signal to the cell middle. The broken box illustrates de novo nucleation of actin filaments at the division site. Numbers indicate the duration of the video. (D) Time-lapse images of a mitotic nmt1-wee1-50 adf1-1 LAGFP cell at 36°C. Asterisks show a cable migrating from the nonmedial region to the cell middle. Video 3 (left cell). (E) Stacked histogram showing distributions of cable extension velocities for polar, equatorial, and randomly oriented cables in mitotic LAGFP mCh-Atb2p cells. 59 cables were counted from one single experiment. (F) Graph showing the fraction of cables extending toward the cell middle (equatorial), cell poles (polar), or in other directions (random) in mitotic cells expressing LAGFP and mCh-Atb2p. 59 cables were counted from one single experiment. (G) Graph showing extension rates of cables grouped by their orientations. Error bars show means ± SD. Left to right, n = 59, 28, and 31 cables; **, P < 0.01. Data were counted from TIRFM videos in E, F, and G. (H) TIRFM images showing typical extension of an actin cable in a mitotic cell expressing LAGFP and mCh-Atb2p. The images on the left indicate the mitotic spindle and cable at the start of the video. The montage shows an extending equatorial cable (end marked with asterisks) from the black dashed box. (I) Kymograph of LAGFP signal in a wt cell passing through mitosis (Video 4). Asterisks indicate nonmedial actin cables that migrate to the middle during ring assembly. The broken box illustrates de novo nucleation of actin filaments at the division site. Numbers indicate the duration of the video. Time interval of this video was 2 s. Dashed lines indicate the cell boundary. Bars, 5 µm.
Next, we followed actin ring assembly in cdc25-22 cells, in which the increased cell length (>25 µm at division), after heat arrest and release (Russell and Nurse, 1986), allowed better visualization of the incorporation of nonmedially assembled actin cables into the actomyosin ring (Fig. 2 B, a nonmedial cable growing toward the division site is marked with asterisks; and Video 2, left). Full incorporation of the cable into the ring can be seen in time points >400 s in Video 2. Kymographs clearly showed movement of actin cables (from nonmedial regions) toward the medial region of the cell (Fig. 2 C). Analysis of actin filaments in adf1-1 mutants (Nakano and Mabuchi, 2006) also showed striking examples of incorporation of nonmedially nucleated actin cables into the forming actomyosin ring (Fig. 2 D and Video 3), suggesting that cables might be partially severed before incorporation into the ring.
To confirm whether actin filaments could indeed be nucleated at nonmedial locations, we used a total internal reflection microscopy (TIRFM) setup to follow actin cables in mitotic cells with increased contrast and sensitivity (Yu et al., 2011). Because actin cables in S. pombe are not entirely associated with the cell cortex (Pelham and Chang, 2001), we used highly inclined laser illumination. With this method, the signal to noise ratio can be drastically improved, and photobleaching can be reduced compared with wide-field imaging. At the same time, illumination depth is much higher than with conventional TIRFM (Tokunaga et al., 2008). To better characterize extension rates, we separated actin cables in mitotic cells according to their orientation (Fig. 2, E and F). We found that the majority of cables extended toward the cell equator (Fig. 2 F, equatorial: 47.2%), and the remaining cables either grew toward the poles or in other random orientations (Fig. 2 F, polar and random orientations: each 26.4%). We measured the velocities of cable extensions by tracking the leading ends over time (Fig. 2, E and G). On average, cables extended with 0.75 ± 0.08 µm/s (mean ± SEM, n = 53). However, equatorial cables grew at significantly faster rates than polar or randomly oriented cables (Fig. 2 G, equatorial: 0.99 ± 0.13 µm/s, mean ± SEM, n = 25, and rest: 0.54 ± 0.08 µm/s, mean ± SEM, n = 28; P < 0.01, unpaired t test with Welch correction). In TIRFM images, we were able to find cables assembling throughout the cell that grew in all directions, including toward the cell middle (Fig. 2 H). The preferred extension toward the cell middle could also be clearly detected in kymographs (Fig. 2 I, extensions toward the equator marked with red asterisks; and cell used to make the kymograph is shown in Video 4). The kymographs also showed that actin fluorescence was present in a less focused manner at the division site early in mitosis (Fig. 2, C and I, marked broken boxes). This observation is consistent with the nucleation of actin filaments from multiple locations, including at the division site, as proposed by other investigators (Wu et al., 2006; Vavylonis et al., 2008).
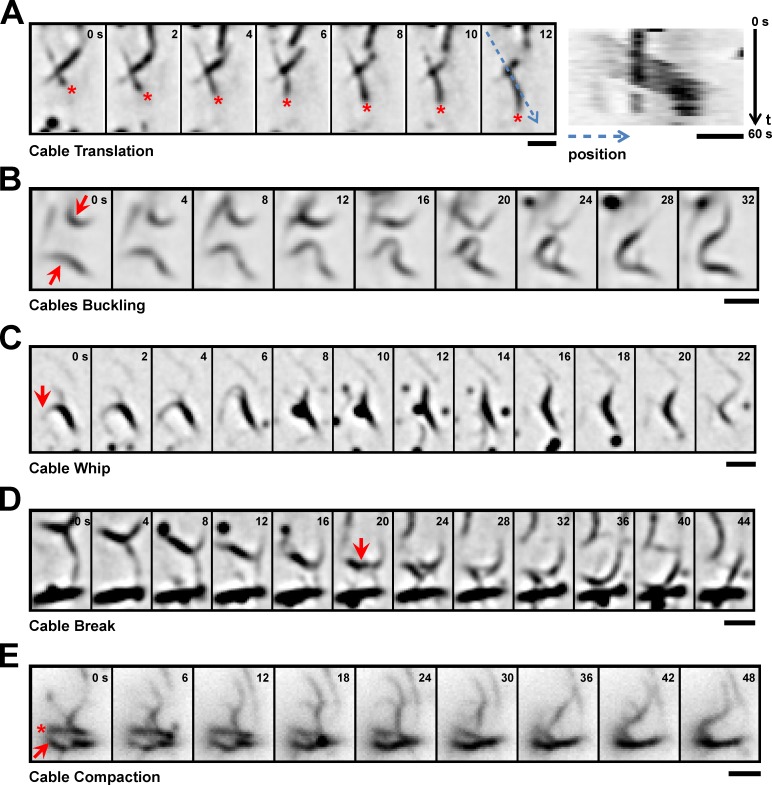
Besides the presumably polymerization-driven extension of cables, we also observed a variety of behaviors that suggest additional factors may be involved in cable reorganization. First, some cables moved rapidly through the cell without an apparent change in length (Fig. 3 A). This was reminiscent of cable translation by myosins in budding yeast (Yu et al., 2011). Second, we observed extensive buckling of individual cables (Fig. 3 B) or cable bundles (Fig. 3 C, cable whip). This lateral motion sometimes was accompanied by breaking or even fragmentation of longer cables (Fig. 3 D). Finally, we observed cables being directly pulled and incorporated into the medial ring (Fig. 3 E). Interestingly, this compaction often occurred perpendicular to the cable axis and was coupled to transient bundling and zipping up of neighboring cables (Fig. 3 E, asterisk). In summary, actin cables in early mitosis can originate at sites distant from the medial ring, frequently grow toward the ring, and undergo extensive lateral reorganization—ultimately leading to compaction into a medial ring.
Figure 3.
Examples of cable dynamics. (A) Translation of a single actin cable. The leading end is marked by an asterisk. Kymograph on the right was made by reslicing a one-pixel-wide line along the path (indicated by the blue dotted line in the 12-s time point) of the moving cable. Numbers on the right corners of the kymograph indicate the start time and end time. (B) Buckling of cables (marked by arrows). The two cables fused at the end of the sequence. (C) Whiplike bending of a cable bundle. Note the rotation of the marked (arrow) cable by nearly 180°. (D) Break in an apparently continuous cable (break position indicated by arrow). (E) Lateral translation of a cable bundle (asterisk) and incorporation into the medial ring (arrow). All images in this figure were obtained from TIRFM videos. Time points are given in seconds. Bars, 2 µm.
Actin cable dynamics can also be observed when utrophin (Utr)-CH is used as a probe
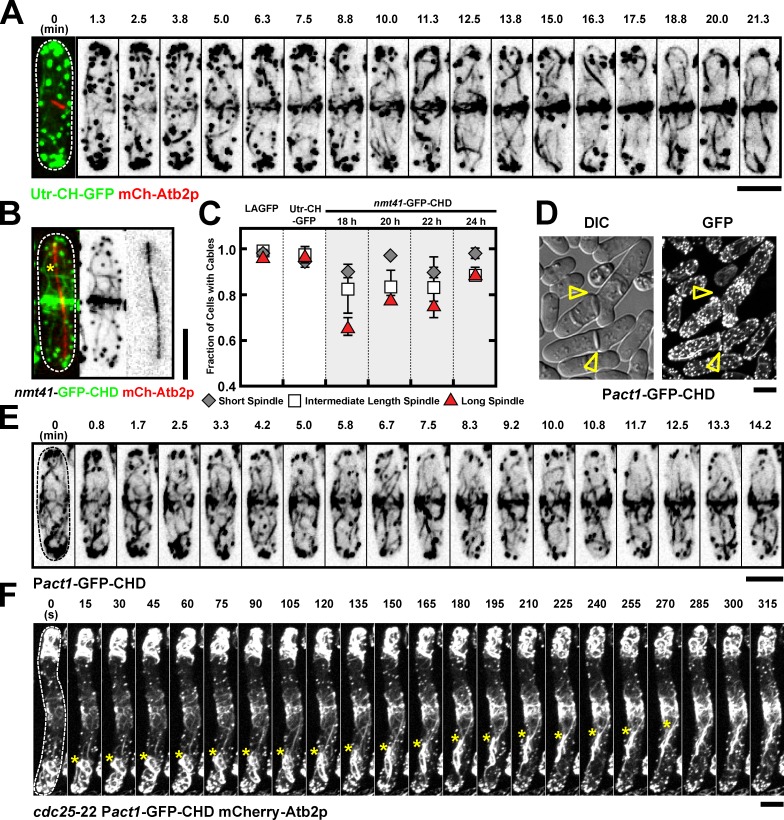
Previous work, using the CHD of Rng2p, has suggested that actin filaments in the ring arise predominantly from de novo nucleation (Wu et al., 2006; Vavylonis et al., 2008). In contrast, our study has shown that actin cables nucleated/assembled over the entire cortex incorporate into the actomyosin ring. In light of this disagreement, we investigated actin assembly using a third probe. Work in other organisms has shown that the CHD of mammalian Utr (Utr-CH) decorates actin very efficiently (Burkel et al., 2007). We therefore generated S. pombe cells in which a Utr-CH-GFP fusion was expressed under control of the fission yeast act1 promoter. Time-lapse imaging of cells undergoing mitosis and cytokinesis again provided evidence for migration and incorporation of nonmedial actin cables into the actomyosin ring (Fig. 4 A and Video 5, left). We therefore concluded that the observed incorporation of actin cables into the assembling actomyosin ring was not an artifact of the LA probe used.
Figure 4.
Incorporation of actin cables into the actomyosin ring can also be observed by using Utr-CH and Pact1-GFP-CHD. (A) Time-lapse images of a Utr-CH-GFP mCh-Atb2p cell. Time 0 (cell with short spindle) is shown as a merged image to indicate the cell cycle stage. Video 5 (left cell). (B) Images of an nmt41-GFP-CHD mCh-Atb2p after 20-h induction. Asterisks show cables in the nonmedial region. (C) Graph showing percentage of cells with cables in LAGFP, Utr-CH-GFP, and nmt41-GFP-CHD (18-, 20-, 22-, and 24-h induction). mCh-Atb2p was used in all these cells to measure spindle length. Cells with spindles were grouped into three categories: short, intermediate, and long spindles. Error bars show means ± SD of two independent experiments (n > 30 cells/experiment). (D) Images of Pact1-GFP-CHD cells cultured in YES. Arrowheads identify cells with septation defects. (E) Time-lapse images of a Pact1-GFP-CHD cell. Video 5 (right cells). (F) Time-lapse images of a cdc25-22 Pact1-GFP-CHD mCh-Atb2p cell after release from 36 to 24°C. Asterisks show cables migrating toward the middle. Video 6 (left cell). DIC, differential interference contrast. Dashed lines indicate the cell boundary. Bars, 5 µm.
We next tested whether actin cables could also be found throughout mitosis using the previously used marker GFP-CHD (Rng2), expressed under control of the inducible nmt41 promoter. We found that the proportion of early mitotic, midmitotic, and late mitotic cells with actin cables organized along the long axis was slightly reduced in cells expressing GFP-CHD (Rng2; Fig. 4, B and C) compared with control cells expressing LAGFP and Utr-CH-GFP (Fig. 4 C). The number of visible cables was particularly reduced for shorter induction times of GFP-CHD, in which fluorescence signals were weaker (Fig. 4 C). In addition, in time-lapse microscopy, the GFP-CHD signal was rapidly bleached. To enable unambiguous evaluation of the CHD probe, we generated a strain in which GFP-CHD was expressed under control of the constitutive act1 promoter. Cells expressing Pact1-GFP-CHD exhibited slightly slower growth and a weak cytokinesis defect (Fig. 1 D and Fig. 4 D). In addition, actin cables in Pact1-GFP-CHD cells were even more stabilized than in Pnmt41-GFP-CHD cells (Fig. S2 D). Time-lapse imaging of actin cable dynamics in Pact1-GFP-CHD cells confirmed that during actomyosin ring assembly, nonmedial actin cables migrate toward the cell division site and are incorporated into the actomyosin ring (Fig. 4, E and F; Video 5, right; and Video 6). Notably, GFP fluorescence in the cell middle of elongated cdc25-22 cells was more pronounced for Pact1-GFP-CHD than for the other markers (Fig. 4 F). In summary, our results indicate that nonmedial actomyosin cables are incorporated into the actomyosin ring, and this can be observed using LAGFP, GFP-CHD, or Utr-CH-GFP as probes for F-actin.
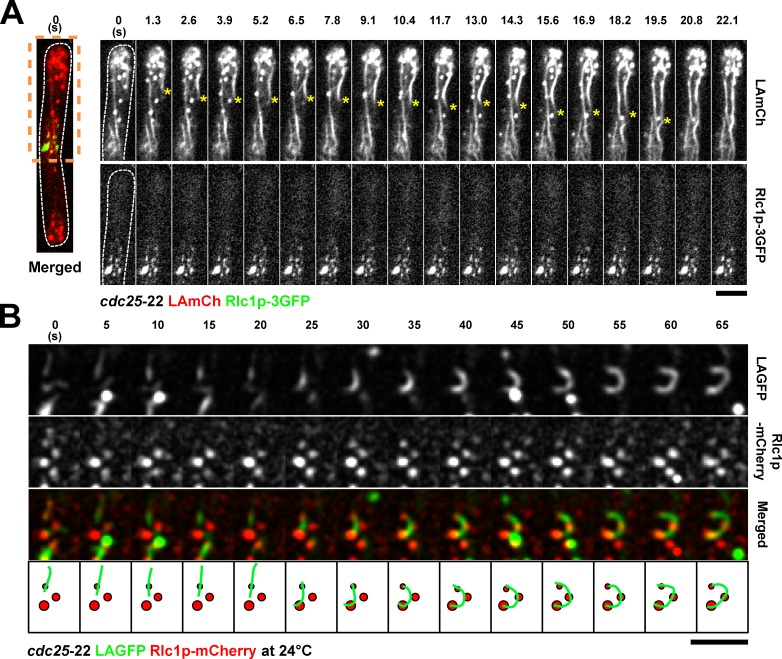
We next addressed the relationship between medially placed nodes (containing myosin II, formins, and Cdc15p) and actin cables. We therefore performed fast time-lapse microscopy of cells coexpressing LAmCh and the myosin II marker Rlc1p-3GFP. In this experiment, single-plane images were acquired every 1.3 s to provide a high time resolution of the process. In these cells, it was clear that the actin migrated toward the Rlc1p-3GFP node–containing medial region of the cell (Fig. 5 A and Video 7). TIRFM performed with a similar strain provided examples of capture and buckling of LAGFP-labeled actin cables by Rlc1p-mCh–containing nodes (Fig. 5 B).
Figure 5.
Double-color imaging of nodes and LA-labeled actin cables. (A) Spinning-disk single-plane images of a cdc25-22 LAmCh Rlc1p-3GFP cell after release from 36 to 24°C. Image on the left is shown as a merged image to indicate the spatial relationship between nodes and nonmedial actin cables. Time-lapse images of the region of the cell within the orange dashed box are shown on the right. Time interval is 1.3 s. Asterisks show the migration of a nonmedial cable toward the middle. Video 7. (B) TIRFM time-lapse double-color images of a cdc25-22 LAGFP Rlc1p-mCh cell at 24°C, showing an actin cable that was captured and bent by three static Rlc1p nodes. The bottom is a schematic representation of the merged montage. Green, actin cable; red, cortical nodes. Dashed lines indicate the cell boundary. Bars, 5 µm.
Cdc12p is essential for assembly of nonmedial actin cables
Previous work has shown that the formin For3p plays an important role in the nucleation of actin cables in interphase, the formin Cdc12p nucleates actin filaments at the division site, and the formin Fus1p participates in cell fusion upon nutrient starvation (Chang et al., 1997; Petersen et al., 1998; Feierbach and Chang, 2001; Nakano et al., 2002). We tested whether For3p was also responsible for the generation of actin cables in mitotic cells. As expected, unlike in wt cells, we could not detect prominent actin cables in interphase for3Δ cells expressing LAGFP (Fig. 6 A, −4.2 and −2.1 min; and Video 8, left). In contrast, a large number of cables connected to the actomyosin ring were visible in mitotic for3Δ cells, as in wt cells. To better follow the origin of cables, we imaged cdc25-22 for3Δ cells expressing LAGFP and mCh-Atb2p. This experiment provided further evidence that For3p-independent actin cables assembled at nonmedial locations incorporated into the assembling actomyosin ring (Fig. 6 B, growing end of a nonmedial actin cable is shown with arrowheads; and Video 8, right). Again, kymographs established that nonmedially assembled actin cables incorporated into the actomyosin ring (Fig. 6 C). Analysis of cdc25-22 for3Δ cells also provided clear evidence for the assembly of actin filaments at the cell division site (Fig. 6 C, marked with a broken box). Furthermore, TIRFM of for3Δ cells revealed that actin filaments were assembled both at the cell division site, as reported by other investigators (Fig. S3, arrows; Wu et al., 2006; Vavylonis et al., 2008) as well as away from the cell middle that grew toward the division site (Fig. S3, red asterisks). To establish that the nonmedial actin cables detected in for3Δ cells were not an artifact of the presence of LA, we fixed and stained cdc25-22 and cdc25-22 for3Δ with phalloidin. This experiment clearly demonstrated the presence of nonmedial actin cables in mitotic cdc25-22 and cdc25-22 for3Δ cells (Fig. 6 D).
Figure 6.
Nonmedially assembled actin cables are nucleated by forminlike protein Cdc12p. (A) Time-lapse images of a for3Δ LAGFP mCh-Atb2p cell. The third panel (time 0 with short spindle) is shown as a merged image to indicate the cell cycle stage. Video 8 (left cell). (B) Time-lapse images of a for3Δ cdc25-22 LAGFP mCh-Atb2p cell after release from 36 to 24°C. Arrowheads point to an actin cable migrating to the cell middle during ring assembly. Video 8 (right cell). (C) Kymograph of a for3Δ cdc25-22 LAGFP mCh-Atb2p cell after release from 36 to 24°C, revealing a flow of F-actin signals from the nonmedial region to the cell middle. The broken box illustrates de novo nucleation of actin filaments at the division site. Numbers indicate duration of the video. Time interval, 15 s. (D) cdc25-22 and for3Δ cdc25-22 cells were fixed with paraformaldehyde and stained with Alexa Fluor 488 phalloidin. (E) Cdc12p-3Venus nodes (arrows) and speckles (asterisks) could be clearly distinguished from the background at 515 nm. Cdc12p-3Venus LAmCh cells and wt cells were grown in YES at 24°C and mixed before imaging. Dashed line shows the boundary of the wt cell. (F) Images of two mitotic Cdc12p-3Venus mCh-Atb2p cells. Arrows show nonmedially located Cdc12p-3Venus speckles during mitosis. (G) Maximum intensity projected images of cdc25-22 LAmCh Cdc12p-3Venus cells after release from 36 to 24°C. Asterisks point out the cells with medial accumulation of actin cables but without accumulation of Cdc12p-3Venus nodes. Red broken boxes show the medial region of the indicated cells. (H) TIRFM time-lapse double-color images of a LAmCh Cdc12p-3GFP cell, showing a Cdc12p-3GFP speckle moving rapidly along a LAmCh-labeled actin cable. Arrows point to the Cdc12p-3GFP speckle. (I) Image of an interphase cdc12Δ LAGFP mCh-Atb2p cell germinated from spore. (J) Time-lapse images of a mitotic cdc12Δ LAGFP mCh-Atb2p cell germinated from spore. Video 9 (left cell). (K and L) Images of interphase and mitotic for3Δ cdc12Δ LAGFP mCh-Atb2p cells germinated from spores. Video 9 (right cell). DIC, differential interference contrast. Dashed lines indicate the cell boundary. Bars, 5 µm.
Because For3p did not play an obvious role in the assembly of nonmedial actin cables in mitosis, we tested whether Cdc12p played a role in the organization of these actin cables. This was possible because Cdc12p has recently been shown to localize to speckles scattered throughout the cell in addition to its localization to the division site (Coffman et al., 2009). We confirmed this result using a Cdc12p-3Venus strain, in which, but not in cells not carrying this fusion, fluorescent speckles were observed in the medial region and elsewhere in the cell throughout mitosis (Fig. 6, E and F, cell not expressing Cdc12p-3Venus marked with a dotted line in E). In the course of the experiments of localization of Cdc12p-3Venus, we also found that Cdc12p-3Venus was not clearly detected in 23.8% of elongated cdc25-22 cells (i.e., less than four nodes) with medial actin filaments (Fig. 6 G). This observation suggested that actin cables transported to the medial region might initiate ring assembly, and further accumulation of Cdc12p nodes might enhance actin assembly at the division site.
We set out to image nucleation of actin filaments by Cdc12p speckles. Because of the low concentration of Cdc12p speckles, we were unable to find cases of nucleation of actin filaments. This observation is similar to work in budding yeast, in which nucleation of actin filaments (that make up actin cables) by Bni1p has only been rarely observed. However, we were able to see examples of a processive movement of Cdc12p speckles on actin cables (Fig. 6 H), suggesting that the observed Cdc12p speckles were likely functional moieties.
We then set out to genetically assess the role of Cdc12p in nonmedial actin cable assembly. Toward this goal, germinated cdc12Δ spores expressing LAGFP and mCh-Atb2p were imaged by time-lapse microscopy. Actin cables were detected in interphase cdc12Δ cells (Fig. 6 I) but not prominently in mitotic cdc12Δ cells (Fig. 6 J and Video 9, left), suggesting that Cdc12p was essential for actin cable nucleation at nonmedial locations as well. Interphase and mitotic cables were fully abolished in germinated for3Δ cdc12Δ spores (Fig. 6, K and L; and Video 9, right), establishing that the formin Fus1p played no obvious role in actin cable assembly in vegetative cells. Collectively, these experiments established that, during mitosis, the formin Cdc12p played a key role in the assembly of actin filaments in the medial and nonmedial regions of the cell.
Mid1p-dependent and septation initiation network (SIN)–dependent ring assembly pathways are not required for actin filament assembly during mitosis
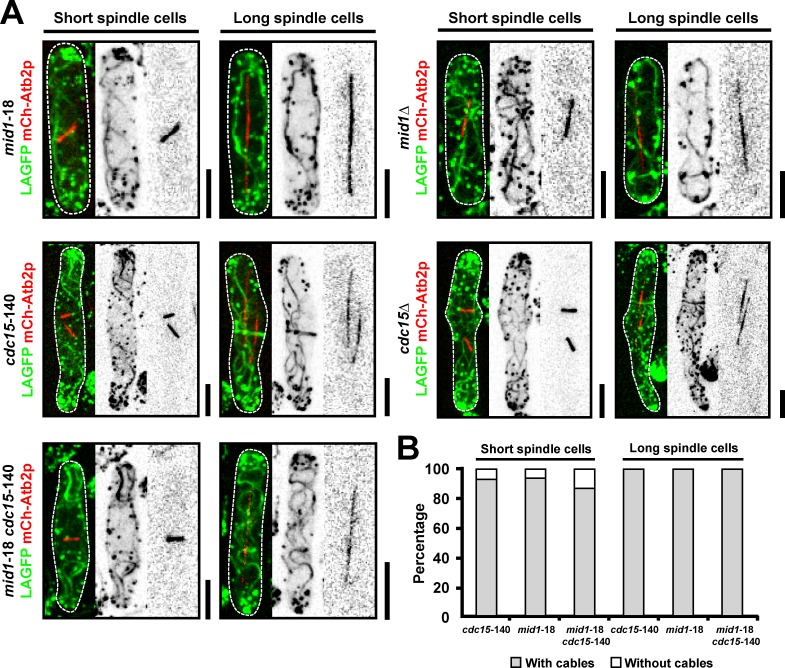
Previous studies have shown that actomyosin ring assembly in fission yeast can occur through one of two pathways (Hachet and Simanis, 2008; Huang et al., 2008). The first of these, the Mid1p nodes–dependent pathway, operates in early mitosis (Wu et al., 2006; Vavylonis et al., 2008), whereas the SIN via the F-BAR protein Cdc15p operates during late stages of mitosis (Hachet and Simanis, 2008; Huang et al., 2008). However, whether assembly of actin filaments during mitosis depends on Mid1p or Cdc15p has not been studied. To this end, we investigated the assembly of actin cables in mitotic mid1-18, mid1Δ, cdc15-140, cdc15Δ, and mid1-18 cdc15-140 cells expressing LAGFP and mCh-Atb2p as probes (Fig. 7, A and B; and Fig. S4, A–C). Interestingly, actin cable assembly was not compromised in mid1-18 and mid1Δ cells even during early stages of mitosis. As shown previously (Wachtler et al., 2006), actin rings were clearly visualized in cdc15-140 and cdc15Δ cells (Fig. 7 A), and nonmedial actin cables were readily observed and found to incorporate into actomyosin rings in these cells (Fig. 7, A and B). Moreover, actin cable assembly was not compromised in mid1-18 cdc15-140 double mutant cells, which are simultaneously defective in both pathways (Fig. 7, A and B; and Fig. S4 C). We also established that the SIN pathway, which is known to be required for Cdc15p function, was dispensable for actin cable assembly during mitosis (Fig. S4 D). Collectively, these experiments established that assembly of actin filaments for cytokinesis could proceed in the simultaneous absence of Mid1p and Cdc15p.
Figure 7.
Mid1p-dependent and Cdc15p-dependent pathways are dispensable for actin filament assembly. (A) mid1-18, cdc15-140, and mid1-18 cdc15-140 cells expressing LAGFP and mCh-Atb2p were shifted to 36°C and imaged by spinning-disk microscopy at 36°C. mid1Δ and cdc15Δ cells expressing LAGFP and mCh-Atb2p were grown at 24°C and imaged by spinning-disk microscopy at 24°C. The cdc15Δ LAGFP mCh-Atb2p cell was germinated from spores. (B) Quantitation of the mutant cells in A with actin cables. Cells with spindles were grouped into two categories: short and long spindles. A single representative image from three independent experiments is shown. In each experiment, 100 cells were counted for each bar. Dashed line indicates the cell boundary. Bars, 5 µm.
Myo2p and Myo51p participate in organizing nonmedially assembled actin cables into the actomyosin ring
Next, we tested the mechanisms by which actin cables migrate from the nonmedial regions of the cell to the cell equator. We considered the possibility that the migration of nuclei toward the cell ends might generate a counter flow of actin cables to the cell middle. However, nonmedial actin cables clearly coalesced into rings even when uninucleate cells were forced to assemble rings upon expression of an activated form of Cdc12p (Yonetani and Chang, 2010) or upon SIN activation (Fig. S5, A and B; Minet et al., 1979).
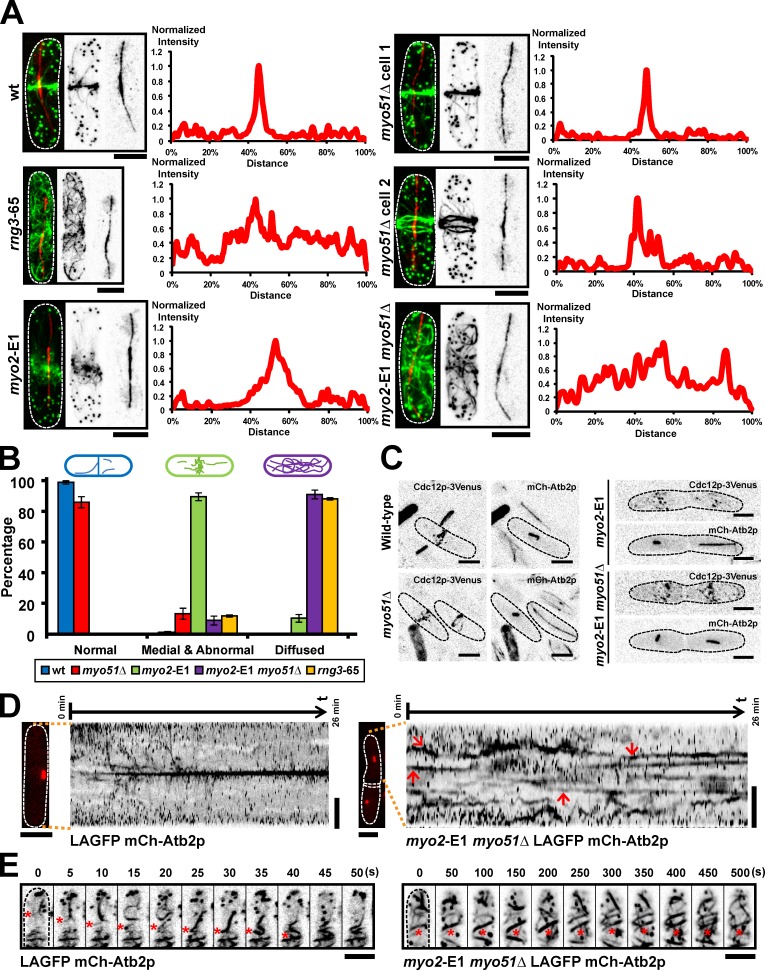
We then investigated the behavior of actin cables in various mutants defective in actomyosin ring assembly. Of all the mutants tested, rng3-65, a mutant defective in the UCS (UNC-45/CRO1/She4) domain–containing myosin assembly factor and activator (Wong et al., 2000; Lord and Pollard, 2004; Mishra et al., 2005), failed to compact actin cables to the cell division site (Fig. 8, A and B). Notably, whereas fluorescence intensity plots over the cell length showed a sharp compaction of actin fluorescence in wt cells, such fluorescence was scattered throughout the entire cell length of rng3-65 cells (Fig. 8, A and B). Because fission yeast Rng3p functions as a type II myosin activator, we tested the distribution of actin cables in the myo2-E1 mutant (Fig. 8, A and B). Although actin cables were not properly compacted into a ring, actin cables concentrated at the medial region of the cell, suggesting that an additional molecule also participates in actin cable migration. UCS proteins have been shown to function together with type V myosins in budding yeast (Toi et al., 2003; Wesche et al., 2003), leading us to consider a role for type V myosin participates in actin cable migration. Two myosin Vs have been identified in fission yeast, of which Myo51p localizes to the actomyosin ring (Motegi et al., 2001; Win et al., 2001). We found that ∼10% of myo51Δ cells failed to compact actin cables into a ring (Fig. 8, A and B, see cell 2 in myo51Δ). This experiment suggested that Myo51p participated in, but was not essential for, actin cable migration to the division site. Given the phenotypes of myo2-E1 and myo51Δ mutants, we considered the possibility that these proteins might perform an overlapping role in this process. To this end, we tested the distribution of actin cables in myo2-E1 myo51Δ cells. Strikingly, in these cells, we found that actin cables, as in rng3-65 cells, were scattered throughout the cell (Fig. 8, A and B). Time-lapse imaging was performed on wt, myo2-E1, and myo2-E1 myo51Δ cells expressing LAGFP and mCh-Atb2p to assess the behavior of actin cables in these cells. Kymographs generated from such time-lapse sequences showed a rapid migration of actin cables to the division site in wt cells (Fig. 8, D and E, left; Fig. 2 I; and Video 10, left). Actin cables were slower to organize into a ring in myo2-E1 (Video 10, middle). Importantly, actin cables were very stable and less mobile (Fig. 8, D and E, right; and Video 10, right) and appeared to be more bundled (Fig. 8 E and Video 10, right) in myo2-E1 myo51Δ cells.
Figure 8.
Myo2p and Myo51p participate in actin cable reorganization during actomyosin ring assembly. (A) Cells of indicated genotype (without any fluorescent marker) were treated with HU for 4 h at 24°C and then shifted to 36°C for 2.5 h. HU was washed out, and cells continued to grow at 36°C for an additional 2 h. Cells were fixed and stained with Alexa Fluor 488 phalloidin and the TAT1 antibody to visualize F-actin and tubulin, respectively. Fluorescence intensities of F-actin in various mutants were measured along the cell length. The data shown are from a single representative experiment out of more than three repeats (n ≥ 10 cells/experiment). (B) Quantitation of different F-actin localization patterns in various mutants. Error bars show means ± SD of three independent experiments (n = 100 cells/experiment). Cells were grouped into three categories based on the actin patterns. Normal: cells with normal sharp actin ring. Medial and abnormal: cells with actin cables concentrated at the medial region of the cell but not properly compacted into a ring. Diffused: cells with actin cables scattered throughout the cell. (C) Images of wt, myo2-E1, myo51Δ, and myo2-E1 myo51Δ cells expressing both Cdc12p-3Venus and mCh-Atb2p. All cells were shifted to 36°C for 4 h before imaging. (D) Kymographs of LAGFP mCh-Atb2p and myo2-E1 myo51Δ LAGFP mCh-Atb2p cells. Spinning-disk single-plane images were taken every 5 s. Maximum intensity projected mCh-Atb2p images on the left of the kymographs were taken just before the start of the videos. Arrows in the kymograph of myo2-E1 myo51Δ LAGFP mCh-Atb2p cell indicate the stable actin structures observed in mitosis. Numbers on top of each kymograph indicate the start time and end time of the video. Video 10. (E) Stable actin structures were observed in myo2-E1 myo51Δ LAGFP mCh-Atb2p cells. Cell culturing and imaging conditions were the same as in D. Asterisks in the left montage show the migration of an actin cable to the middle in LAGFP mCh-Atb2p cell. Asterisks in the right montage show the stable actin structures in myo2-E1 myo51Δ LAGFP mCh-Atb2p cell. Video 10. Dashed lines indicate the cell boundary. Bars, 5 µm.
Because medial actin cables were detected in the division site even before accumulation of a large number of Cdc12p nodes (Fig. 6 G), it was possible that Cdc12p was itself transported along actin cables. We therefore tested whether Myo2p and Myo51p played a role in the accumulation of Cdc12p speckles and nodes to the division site. To this end, we followed the localization of Cdc12p-3Venus in wt, myo51Δ, myo2-E1, and myo2-E1 myo51Δ mutants (Fig. 8 C). In all cases, Cdc12p was detected in the medial region of the cell in nodes, suggesting that Myo2p, Myo51p, and the accumulation of actin cables in the middle of the cell did not play a role in the accumulation of Cdc12p at the division site (Fig. 8 C).
Discussion
In this study, we have addressed the mechanism of actin filament assembly at the division site in the fission yeast S. pombe. We have generated yeast strains expressing LA fused to fluorescent proteins as a probe to monitor actin dynamics (Riedl et al., 2008). LAGFP, LAmGFP, and LAmCh decorate actin patches, actin cables, and the actomyosin ring. The structures decorated by LA qualitatively were nearly identical to F-actin patterns generated upon staining with phalloidin. In particular, among the probes available (LAGFP, GFP-CHD, and actin-GFP), LAGFP allowed the most convincing visualization of actin cables. Because the cables decorated by LA were lost upon treatment with LatA or upon loss of function of Cdc3p-profilin, formins, and Cdc8p-tropomyosin, we believe LA is an ideal probe to study actin cable dynamics.
Using a variety of probes, we have shown that actin filaments and cables for cytokinesis are assembled throughout the cell, including the cell division site. The fact that a significant number of actin filaments for cytokinesis are assembled at nonmedial locations and are incorporated into the actomyosin ring was deduced from several lines of investigations. First, phalloidin staining of wt cells showed that actin cables connected to the ring are detected throughout mitosis and cytokinesis. Second, in time-lapse experiments of wt cells and elongated cdc25-22 cells, actin cables, unconnected to the cell middle, were found to grow toward the cell middle and incorporate into the actomyosin ring. Such incorporation was particularly clearer in cells defective for Adf1p cofilin function, suggesting that Adf1p might also participate in the reorganization of long actin cables into short filaments that incorporate into the actomyosin ring. Third, the generation of kymographs from such wt and cdc25-22 cells showed that nonmedially assembled actin cables incorporate into the actomyosin ring. Fourth, TIRFM showed possible actin nucleation events, in some instances, even from the cell ends and the growth of these actin cables toward the cell middle. Finally, under our experimental conditions, we have found that Cdc12p-formin is not detected at the division site in 23.8% of cells containing medial F-actin. Based on these observations, and through similar observations with Utr-CH-GFP, we conclude that actin filaments for cytokinesis were assembled throughout the cell including at the division site.
Previous studies have used the CHD of Rng2p (expressed under nmt1 promoter) as a probe to monitor actin dynamics during cytokinesis (Wu et al., 2006; Vavylonis et al., 2008) and arrived at the conclusion that actin filaments for cytokinesis are predominantly assembled at the division site. Our conclusions are thus at odds with previous work. We have recently found that the GFP-CHD, but not LAGFP, interaction with actin filaments leads to stabilization of actin structures in LatA sensitivity assays (Fig. S2). This observation is consistent with the in vitro ability of the N-terminal region of Rng2p (containing the CHD) to cause bundling of actin filaments (Takaine et al., 2009). In addition, CHD is a fragment of Rng2p, a bona fide component of the cortical nodes. It is therefore possible that CHD might associate with cortical nodes by interacting with other components of the actomyosin ring. We believe that the fact that CHD is fragment of a bona fide component of the cortical nodes and that it is able to bundle and generate more stable actin filaments might lead to a more robust detection of actin filaments assembled at the medial cortical nodes. Interestingly, we have found that expression of GFP-CHD under control of the act1 promoter, despite leading actin-stabilizing effects in LatA sensitivity experiments allowed better visualization of actin cable dynamics. In such a strain, we were able to detect both incorporation of nonmedial actin cables into the forming actomyosin ring and de novo generation of actin filaments in the ring. Thus, actin filaments do indeed assemble at the division site, but those assembled elsewhere also contribute toward the formation of the actomyosin ring.
Our observations are similar to studies of actin filament assembly during cytokinesis in animal cells. In animal cells, two major mechanisms contribute to actin filament assembly and accumulation at the division site. In some mammalian cell types, actin filaments for cytokinesis are transported from the polar regions of the cell to the cleavage furrow through a process that depends on myosin II (White and Borisy, 1983; Bray and White, 1988; Cao and Wang, 1990; Lee et al., 1998). In other cell types (such as in Xenopus laevis embryos), actin filaments for cytokinesis are assembled de novo at the division site (Noguchi and Mabuchi, 2001). Two recent studies have shown that both mechanisms coexist in some other cell types (Chen et al., 2008; Zhou and Wang, 2008; Alsop et al., 2009). Our work with fission yeast is consistent with the idea that two overlapping mechanisms contribute to actin filament assembly at the division site.
Previous work has shown that the formin For3p assembles actin cables, whereas the formin Cdc12p regulates actomyosin ring assembly (Chang et al., 1997; Feierbach and Chang, 2001). We have observed assembly of actin cables at nonmedial locations in mitotic cells devoid of For3p, suggesting that For3p is not essential for the assembly of such actin cables. Recent work has shown that although Cdc12p concentrates in the actomyosin ring, a significant fraction of Cdc12p also localized to speckles throughout the cell (Coffman et al., 2009), raising the possibility that these Cdc12p speckles might contribute to the assembly of such nonmedial actin cables. Consistently, loss of Cdc12p function leads to a failure in assembly of both medial and nonmedial actin cables. These results established that Cdc12p could generate actin cables for cytokinesis both from the division site and from other cellular locations. Currently the fractions of actin filaments in the actomyosin ring that are nucleated by medial and nonmedial Cdc12p are unknown. It is also unknown whether assembly at one or the other locations alone might suffice for actomyosin ring assembly. Future studies with mutant alleles of Cdc12p that show distinct localization patterns will be required to answer these questions.
Previous work has shown that actomyosin ring assembly in fission yeast proceeds through one of two pathways (Mishra and Oliferenko, 2008). The Mid1p nodes–dependent pathway operates in early mitosis (Wu et al., 2006; Vavylonis et al., 2008), whereas the SIN- and Cdc15p-dependent pathway operates in a node-independent manner during later stages of mitosis (Hachet and Simanis, 2008; Huang et al., 2008). We have found that actin cables are assembled in early and late mitotic cells lacking both of these pathways. These observations suggest that the two previously described mechanisms participate in the organization of actin cables into a ring but not in the nucleation of actin filaments during cytokinesis.
How do the nonmedially assembled actin cables migrate to and incorporate into the medial actomyosin ring? We have shown that actin cables exhibit three types of motions: toward the cell ends, toward the cell middle, and random. We have found that cable movement to the cell middle dominates over the other two. The velocity of cable movement to the cell middle is also higher than the velocity of cables directed elsewhere. These observations suggested that actin-based motors contribute to the directional migration of actin cables to the medial division site. The behavior of actin cables, such as buckling, breakage, whipping, and translation, are likely a consequence of directional force impacting these actin cables. Our analysis of cells defective in myosin II (Myo2p) and myosin V (Myo51p) and the myosin activator/chaperone Rng3p has shown that a combination of Myo2p and Myo51p functions might contribute to actin cable accumulation at the division site (Wong et al., 2000; Lord and Pollard, 2004; Mishra et al., 2005). Because Cdc12p localization was not affected in myo2-E1 myo51Δ cells, it is unlikely that Cdc12p transport along actin cables contributes to the dispersed actin cable phenotype. Rather, the dispersed localization of actin cables in cells defective in Myo2p and Myo51p should reflect a role for these proteins in actin cable movement to the cell division site.
In summary, our study has provided evidence for a novel mechanism involved in actin filament assembly during cytokinesis in fission yeast. The mechanisms we describe bear similarity to those that operate in animal cells. Given the conservation of mechanisms involved in actin filament assembly between yeast and animals, future experiments in fission yeast should provide a more complete understanding of eukaryotic cytokinesis mechanisms.
Materials and methods
Yeast strains, medium, and culture conditions
The fission yeast strains used in this study are listed in Table S1. Conventional fission yeast genetic and molecular biology techniques, such as strain construction, DNA-mediated transformation, and fluorescent protein expression, were performed as previously described (Moreno et al., 1991). For yeast transformation, mid–log phase cells were spun down and washed with equal volume of water. After washing with 1 ml LiAc/TE (Tris-EDTA) buffer, cells were incubated with both sonicated salmon sperm DNA and plasmid (or DNA fragment) at room temperature for 15 min, 240 µl polyethylene glycol/LiAc/TE solution was added, and cells were incubated at 30°C for 45 min. Then, 43 µl DMSO was added, and cells were given a heat shock at 42°C for 5 min. Cells were spun down, resuspended in water, and spread onto appropriate plate.
Plasmid construction
To construct the pJK148-Pact1-LAGFP plasmid (pCDL1484), oligonucleotides MOH4378 (with restriction site for EcoRI underlined in the following sequence: 5′-CCGGAATTCCAAAACCTTCTCTGCC-3′) and MOH4379 (contains the N-terminal sequence of LA: 5′-TTCTTGATAAGGTCAGCGACACCCATGGTCTTGTCTTTTGAGGGTT-3′) were used in a PCR to obtain the 1-kb actin promoter from yeast genomic DNA. The purified fragment was then used as a template, together with MOH4380 (with restriction site BamHI underlined in the following sequence and containing the C-terminal sequence of LA: 5′-CGCGGATCCTTCCTTAGAAATAGACTCGAACTTCTTGATAAGGTCAGCGAC-3′) and MOH4378, in a PCR to generate an actin promoter-LA fragment with EcoRI and BamHI sites at two ends. This actin promoter-LA fragment and pJK148-linker-GFP plasmid (pCDL1485; a gift from S. Oliferenko, Temasek Life Sciences Laboratory, Singapore; EGFP sequence was inserted between NdeI and NotI) were double digested by EcoRI and BamHI. The digested actin promoter-LA fragment was ligated into the pJK148-linker-GFP plasmid by T4 DNA ligase and transformed into Escherichia coli. The plasmid was confirmed by sequencing (a point mutation was found at −408 in the actin promoter region; T was changed to A). The final codon-optimized LA sequence is 5′-ATGGGTGTCGCTGACCTTATCAAGAAGTTCGAGTCTATTTCTAAGGAA-3′ (this sequence only encodes the first 16 amino acids of the original LA peptide).
To construct the pJK148-Pact1-LAmGFP plasmid (pCDL1546), a mutation L221K (Zacharias et al., 2002) was introduced into EGFP in the pJK148-Pact1-LAGFP plasmid (pCDL1484) by site-directed mutagenesis using high fidelity Pfu DNA polymerase (Agilent Technologies). Oligonucleotides used are as follows: MOH4936, 5′-CCACATGGTCCTTAAGGAGTTTGTAACAGC-3′, and MOH4937, 5′-GCTGTTACAAACTCCTTAAGGACCATGTGG-3′.
To construct the pJK148-Pact1-LAmCh plasmid (pCDL1564), the EGFP sequence of pCDL1484 was replaced by the mCh sequence by restriction digestion and ligation method (NdeI and NotI restriction sites were used). To construct the pJK148-Pact1-Utr-CH-GFP plasmid (pCDL1561), the LA sequence of pCDL1484 was replaced by the first 783 base pairs (cloned from the pCDL1560 mCh-Utr plasmid; a gift from A. Bershadsky, Mechanobiology Institute, Singapore; Burkel et al., 2007) of the human Utr gene by invert PCR (van den Ent and Löwe, 2006) using high fidelity Pfu DNA polymerase. Oligonucleotides used are as follows: MOH5182, 5′-CGAACCAAAAAACCCTCAAAAGACAAGACCATGGCCAAGTATGGAGAAC-3′, and MOH5183, 5′-CATATGACCACCGGGACCACCGGATCCGTCTATGGTGACTTGCTGAGGT-3′. Two point mutations were found within the Utr gene, a silent mutation T339C and a missense mutation A707G (Gln236Arg).
To construct the pJK148-Pact1-GFP-CHD plasmid (pCDL1581), LA sequence in the pJK148-Pact1-LAGFP plasmid (pCDL1484) was first deleted by invert PCR to generate the pJK148-Pact1-GFP plasmid (pCDL1577). Oligonucleotides used are as follows: MOH5262, 5′-AACCCTCAAAAGACAAGACCATGAGTAAAGGAGAAGAACTTTTCACTG-3′, and MOH5263, 5′-AGTTCTTCTCCTTTACTCATGGTCTTGTCTTTTGAGGGTTTTTTGGTTCG-3′. Then, the linker sequence and the sequence encoding the first 189 amino acids of fission yeast protein Rng2p (these are exactly the same linker-CHD sequences used in the previous nmt41-GFP-CHD strain) were cloned from pCDL270 (pREP42-GFP-CHD plasmid) by PCR. The oligonucleotides used are as follows: MOH5264, 5′-CACATGGCATGGATGAACTATACAAAGGGCATATGTCGACAATGGACGT-3′, and MOH5265, 5′-GAGCTCCACCGCGGTGGCGGCCGCTTAAGCTTTGAAGTTAGGAAGGAT-3′. This PCR fragment was used as a primer for invert PCR to insert the linker-CHD sequence into pJK148-Pact1-GFP plasmid (pCDL1577), generating the pJK148-Pact1-GFP-CHD plasmid (pCDL1581).
Construction of yeast strains carrying the pJK148 constructs
The integrative pJK148 plasmids (pJK148-Pact1-LAGFP, pJK148-Pact1-LAmGFP, pJK148-Pact1-LAmCh, pJK148-Pact1-Utr-CH-GFP, and pJK148-Pact1-GFP-CHD) were digested by NruI within the leu1+ gene. The digested products were purified and transformed into the yeast cells. The transformants were selected on minimal medium lacking leucine (leu−), and the colonies were further screened visually for fluorescence. The pJK148 constructs carrying strains were then used to cross with the other strains of interest.
Diploid strains were transformed by NruI-digested pJK148-Pact1-LAGFP plasmid. Positive strains were grown on ade− leu− ura− or ade− leu− his− (depending on the marker used to delete the essential gene in the diploid). The diploids carrying LAGFP were patched out every 4 d until nonsporulating strains were obtained. The LAGFP-expressing nonsporulating strains were then used to cross with cells expressing both LAGFP and mCh-Atb2p (MBY6659 or MBY7114) to get the sporulating diploid strains carrying both LAGFP and mCh-Atb2p.
Construction of the Pnmt1-wee1-50 strain
To make the Pnmt1-wee1-50 strain (with KanMX6 marker), a DNA fragment containing the KanMX6 cassette and nmt1 promoter was cloned from plasmid pFA6a-KanMX6-P3nmt1-3HA (pCDL953; a gift from J.-q. Wu, Ohio State University, Columbus, OH) and then transformed into the wee1-50 strain (MBY169). The oligonucleotides used are as follows: MOH5304, 5′-GCATTCCAATTCAATTTAATTAAATCAAAAATTTCATATCTATTTTTTTGTTAAATTGCCACATTTTCCATACAGAAAACGAATTCGAGCTCGTTTAAAC-3′, and MOH5305, 5′-GTAGCACGATTTAGATTCATGGAGCGTTGGGACCGCCGTAAGCCATAAGATCTATGACTGCTGGTATTAGAAGAAGAGCTCATCATGATTTAACAAAGCGACTATA-3′. The transformants were selected on G418 plates and replicate to complete minimal medium for screening the positive clones, which showed a G2-arrested phenotype.
LatA treatment
LatA (obtained from Enzo Life Sciences) was used at a final concentration of 50 µM in all experiments. After adding LatA, cells were shook manually every minute to make sure all the cells were evenly treated with LatA.
Hydroxyurea (HU) treatment
HU (obtained from Sigma-Aldrich) was used at a stock concentration of 1.2 M in all the experiments. In Fig. 8 (A and B), cells were cultured in YES (yeast extract with supplements) medium at 24°C overnight till mid–log phase. 1-ml cells were treated with 10 µl HU at 24°C for 4 h. After adding another 10 µl HU, cells were shifted to 36°C for an additional 2.5 h. Then, HU was washed out (with 36°C YES medium), and cells were grown at 36°C for another 2 h (cells were at 36°C for 4.5 h before fixation). Then, cells were fixed with formaldehyde (final concentration of 3.7%) and stained with Alexa Fluor 488 phalloidin and antitubulin (TAT1, anti–α-tubulin mouse monoclonal antibody; a gift from K. Gull, University of Oxford, Oxford, England, UK) antibody to visualize F-actin and tubulin, respectively (Moreno et al., 1991).
In Fig. S5 B, cdc16-116 LAGFP cells were cultured in YES medium at 24°C overnight till mid–log phase. 1-ml cells were treated with 10 µl HU at 24°C for 4 h. After adding another 10 µl HU, cells were cultured at 24°C for an additional 1 h. Then, cells were shifted up to 36°C for 25 min and imaged on a YES agarose pad (cells were still immersed in HU) at 36°C.
Induction of the nmt1 promoter
For the expression of GFP-CHD under the nmt41 promoter, nmt41-GFP-CHD–containing cells (MBY2309 and MBY7132) were first grown in YES at 24°C overnight till mid–log phase and washed with complete minimal medium three times. Then, cells were resuspended in complete minimal medium for 18–24 h to induce the expression of GFP-CHD.
For overexpression of Wee1-50p under the nmt1 promoter to arrest cells in G2 phase, nmt1-wee1-50 adf1-1 LAGFP cells (MBY6866) were first grown in leu− medium with thiamine at 24°C overnight till mid–log phase. After washing with leu− medium without thiamine three times, cells were resuspended in leu− medium without thiamine and cultured at 24°C for 16–20 h for the induction of Wee1-50p. After being shifted up to 36°C in a water bath shaker for 20 min, cells were put on a YES agarose pad and imaged in 36°C spinning-disk chamber. Cells would enter into mitosis after around 30 min from the time they were shifted up in the water bath shaker.
Microscope image acquisition and data analysis
For time-lapse videos, mid–log phase cells were concentrated by centrifugation at 4,200 rpm for 10 s to 1 min. 1 µl cells was placed on a glass slide with either YES + 2% agarose pad or selection medium + 2% agarose pad and then sealed under a coverslip using VALAP (vaseline, lanolin, and paraffin) and imaged at the respective temperature. For time-lapse videos at 36°C, temperature was controlled and maintained by a full enclosure incubation chamber.
Bright-field images were captured using a microscope (IX71; Olympus; Plan Apochromat 100×/1.45 NA oil objective lens) equipped with a charge-coupled device camera (CoolSNAP HQ; Photometrics) and MetaMorph (v6.2r6) software (Molecular Devices). Spinning-disk images were captured either by spinning disk using a microscope (Axiovert 200M; Carl Zeiss) or spinning disk (microLAMBDA). Spinning disk using a microscope (Axiovert 200M; Plan Apochromat 100×/1.4 NA oil objective lens) was equipped with a spinning-disk system (CSU-21; Yokogawa Corporation of America), camera (ORCA-ER; Hamamatsu Photonics), and MetaMorph (v7.6.5.0) software. A 491-nm solid-state laser (Calypso; Cobolt) and 561-nm solid-state laser (Jive; Cobolt) were used for excitation. microLAMBDA spinning disk using a microscope (Eclipse Ti; Nikon; Plan Apochromat VC 100×/1.40 NA oil objective lens) was equipped with a spinning-disk system (CSUX1FW; Yokogawa Corporation of America), camera (CoolSNAP HQ2), and MetaMorph (v7.7.7.0) software. A 491-nm diode-pumped solid-state (DPSS) laser (Calypso), 515-nm DPSS laser (Fandango; Cobolt), and 561-nm DPSS laser (Jive) were used for excitation. 3D (0.5-µm step size) spinning-disk images were collected with an interval time of 5, 10, 15, 25, or 30 s. Unless specified otherwise, all these 3D images were shown by 2D maximum intensity projection. Double-color, single-plane spinning-disk images were collected with an interval time of 1.3 s.
TIRFM images were acquired on a custom setup obtained from TILL Photonics based on an automated iMic stand and a 1.45 NA 100× objective (Olympus). Lasers at 488 nm (Sapphire; Coherent; 75 mW) and 561 nm (Jive; 75 mW) were coupled through an acousto-optic tunable filter and a galvanometer-driven scan head to adjust TIRFM angles. Images were collected with an electron multiplying charge-coupled device camera (iXon DU-897; Andor Technology) at maximum gain after a 2× magnifying lens (Andor Technology). Acquisition was controlled by the Live Acquisition software package (TILL Photonics). Cable extension rates were quantified from TIRFM series with a 200-ms frame rate. Cable ends were tracked for at least four frames. Rates are given as means ± SD. Unpaired t tests with Welch correction were performed to test significance of variations. For TIRFM double-color time-lapse images, the interval times were 5 s. Switching between two filters took ∼1 s. TIRFM images were notified in the figure legends. If not specified, the fluorescent images were taken by spinning disk.
Images were analyzed using ImageJ (National Institutes of Health) or MetaMorph. All maximum intensity projected spinning-disk images were background subtracted using the Process/Subtract Background function in ImageJ. After background subtraction, some time-lapse videos were bleach corrected using the Plugins/Stacks–T-functions/Bleach Correction plugin in ImageJ. For making kymographs in Fig. 2 (C and I) and Fig. 6 C, a line was first drawn from one pole to another along the cell long axis of the cell of interest and then the Image/Stacks/Reslice function in ImageJ was used to make the kymograph. Many lines were drawn in one cell, but only one representative kymograph of each cell was shown. For the graphs in Fig. 8 A, a square was first drawn outside each cell and then the Analyze/Plot Profile function in ImageJ was used to measure the average intensities along the cell long axis. In some images or videos, different channels of the same cell taken at the same time point or different videos were combined together using the Plugins/Stacks-Building/Stack Combiner plugin or Plugins/Stacks-Building/Stack Inserter in ImageJ. All time-lapse videos were edited by ImageJ or MetaMorph software and saved in MP4 format with H.264 compression. Graphs in this paper were either made by Excel (Microsoft) or Illustrator (Adobe).
Online supplemental material
Fig. S1 shows dynamics of LA-labeled actin structures during ring assembly in LAGFP and LAmGFP cells. Fig. S2 shows that CHD stabilizes actin structures in fission yeast cells. Fig. S3 shows the assembly of mitotic actin cables from both the medial region and nonmedial region in a for3Δ LAGFP mCh-Atb2p cell. Fig. S4 shows that the assembly of mitotic actin cables is independent of Cdc15p, Mid1p, or Cdc7p. Fig. S5 shows that the movement of nuclei is not required for the migration of actin cables to the cell division site. Video 1 shows actin dynamics in LAGFP mCh-Atb2p, LAGFP, and LAmGFP cells at 24°C. Video 2 shows nonmedially assembled actin cables migrated toward the middle in four blocked and released cdc25-22 LAGFP mCh-Atb2p cells. Video 3 shows actin cables migrated from the nonmedial region toward the middle during ring assembly in two nmt1-wee1-50 adf1-1 LAGFP cells at 36°C. Video 4 is a TIRFM video that shows actin dynamics in a mitotic LAGFP mCh-Atb2p cell at 24°C. Video 5 shows that nonmedial actin cables can also be observed in wt cells expressing Utr-CH-GFP mCh-Atb2p or Pact1-GFP-CHD. Video 6 shows nonmedial actin cables migrated toward the middle in two blocked and released cdc25-22 Pact1-GFP-CHD mCh-Atb2p cells. Video 7 is a single-plane, 1.3-s, spinning-disk double-color video that shows nonmedially assembled actin cables migrated toward the cell middle in a blocked and released the cdc25-22 LAmCh Rlc1p-3GFP cell. Video 8 shows actin dynamics in a for3Δ LAGFP mCh-Atb2p cell at 24°C and an actin cable migrated from the nonmedial region toward the middle in a for3Δ cdc25-22 LAGFP mCh-Atb2p cell after release from 36 to 24°C. Video 9 shows that mitotic actin cables are either not prominent or not detectable during mitosis in a germinated cdc12Δ LAGFP mCh-Atb2p cell or for3Δ cdc12Δ LAGFP mCh-Atb2p cell at 24°C. Video 10 is a single-plane spinning-disk video of LAGFP mCh-Atb2p, myo2-E1 LAGFP mCh-Atb2p, and myo2-E1 myo51Δ LAGFP mCh-Atb2p cells at 36°C. Table S1 shows S. pombe strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201209044/DC1.
Supplementary Material
Acknowledgments
We wish to especially thank Issei Mabuchi, Kathleen L. Gould (Vanderbilt University, Nashville, TN), Daniel P. Mulvihill, Jian-qiu Wu, Snezhana Oliferenko, Fred Chang, Alexander Bershadsky, Keith Gull, and Kentaro Nakano for the gifts of fission yeast strains, plasmids, and antibodies. Special thanks are given to all members of the Cell Division Laboratory for discussion, Meredith Calvert, Mithilesh Mishra, and Ramanujam Srinivasan for critical reading of the manuscript, and Snezhana Oliferenko for supporting R. Thadani. We also express our sincere gratitude to Meredith Calvert and Fiona Chia from the Temasek Life Sciences Laboratory microscopy unit for expert assistance.
This work was supported by research funds from the Temasek Life Sciences Laboratory, Mechanobiology Institute, National University of Singapore, Singapore Millennium Foundation, and Max Planck Society.
Footnotes
Abbreviations used in this paper:
- CHD
- calponin homology domain
- DPSS
- diode pumped solid state
- HU
- hydroxyurea
- LA
- lifeact
- LatA
- Latrunculin A
- mCh
- mCherry
- SIN
- septation initiation network
- TIRFM
- total internal reflection microscopy
- Utr
- utrophin
- wt
- wild type
References
- Alsop G.B., Chen W., Foss M., Tseng K.F., Zhang D. 2009. Redistribution of actin during assembly and reassembly of the contractile ring in grasshopper spermatocytes. PLoS ONE. 4:e4892 10.1371/journal.pone.0004892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai R., Mabuchi I. 2002. F-actin ring formation and the role of F-actin cables in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 115:887–898 [DOI] [PubMed] [Google Scholar]
- Balasubramanian M.K., Helfman D.M., Hemmingsen S.M. 1992. A new tropomyosin essential for cytokinesis in the fission yeast S. pombe. Nature. 360:84–87 10.1038/360084a0 [DOI] [PubMed] [Google Scholar]
- Balasubramanian M.K., Hirani B.R., Burke J.D., Gould K.L. 1994. The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. J. Cell Biol. 125:1289–1301 10.1083/jcb.125.6.1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian M.K., McCollum D., Chang L., Wong K.C., Naqvi N.I., He X., Sazer S., Gould K.L. 1998. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics. 149:1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian M.K., Bi E., Glotzer M. 2004. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr. Biol. 14:R806–R818 10.1016/j.cub.2004.09.022 [DOI] [PubMed] [Google Scholar]
- Berepiki A., Lichius A., Shoji J.Y., Tilsner J., Read N.D. 2010. F-actin dynamics in Neurospora crassa. Eukaryot. Cell. 9:547–557 10.1128/EC.00253-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray D., White J.G. 1988. Cortical flow in animal cells. Science. 239:883–888 10.1126/science.3277283 [DOI] [PubMed] [Google Scholar]
- Burkel B.M., von Dassow G., Bement W.M. 2007. Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell Motil. Cytoskeleton. 64:822–832 10.1002/cm.20226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L.G., Wang Y.L. 1990. Mechanism of the formation of contractile ring in dividing cultured animal cells. II. Cortical movement of microinjected actin filaments. J. Cell Biol. 111:1905–1911 10.1083/jcb.111.5.1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F. 1999. Movement of a cytokinesis factor cdc12p to the site of cell division. Curr. Biol. 9:849–852 10.1016/S0960-9822(99)80372-8 [DOI] [PubMed] [Google Scholar]
- Chang F., Woollard A., Nurse P. 1996. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J. Cell Sci. 109:131–142 [DOI] [PubMed] [Google Scholar]
- Chang F., Drubin D., Nurse P. 1997. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J. Cell Biol. 137:169–182 10.1083/jcb.137.1.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Foss M., Tseng K.F., Zhang D. 2008. Redundant mechanisms recruit actin into the contractile ring in silkworm spermatocytes. PLoS Biol. 6:e209 10.1371/journal.pbio.0060209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman V.C., Nile A.H., Lee I.J., Liu H., Wu J.Q. 2009. Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis. Mol. Biol. Cell. 20:5195–5210 10.1091/mbc.E09-05-0428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Alvarez D.L., Callejas-Negrete O.A., Gómez N., Freitag M., Roberson R.W., Smith L.G., Mouriño-Pérez R.R. 2010. Visualization of F-actin localization and dynamics with live cell markers in Neurospora crassa. Fungal Genet. Biol. 47:573–586 10.1016/j.fgb.2010.03.004 [DOI] [PubMed] [Google Scholar]
- Doyle T., Botstein D. 1996. Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc. Natl. Acad. Sci. USA. 93:3886–3891 10.1073/pnas.93.9.3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Reymond A., Cerutti L., Utzig S., Hofmann K., Simanis V. 1995. The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell. 82:435–444 10.1016/0092-8674(95)90432-8 [DOI] [PubMed] [Google Scholar]
- Feierbach B., Chang F. 2001. Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr. Biol. 11:1656–1665 10.1016/S0960-9822(01)00525-5 [DOI] [PubMed] [Google Scholar]
- Hachet O., Simanis V. 2008. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev. 22:3205–3216 10.1101/gad.1697208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Yan H., Balasubramanian M.K. 2008. Assembly of normal actomyosin rings in the absence of Mid1p and cortical nodes in fission yeast. J. Cell Biol. 183:979–988 10.1083/jcb.200806151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamasaki T., Osumi M., Mabuchi I. 2007. Three-dimensional arrangement of F-actin in the contractile ring of fission yeast. J. Cell Biol. 178:765–771 10.1083/jcb.200612018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama C., Sugimoto A., Yamamoto M. 1997. Type II myosin heavy chain encoded by the myo2 gene composes the contractile ring during cytokinesis in Schizosaccharomyces pombe. J. Cell Biol. 137:1309–1319 10.1083/jcb.137.6.1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D.R., Wu J.Q., Pollard T.D. 2005. Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast. Mol. Biol. Cell. 16:2313–2324 10.1091/mbc.E04-09-0781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E., Shelden E.A., Knecht D.A. 1998. Formation of F-actin aggregates in cells treated with actin stabilizing drugs. Cell Motil. Cytoskeleton. 39:122–133 [DOI] [PubMed] [Google Scholar]
- Lord M., Pollard T.D. 2004. UCS protein Rng3p activates actin filament gliding by fission yeast myosin-II. J. Cell Biol. 167:315–325 10.1083/jcb.200404045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May K.M., Watts F.Z., Jones N., Hyams J.S. 1997. Type II myosin involved in cytokinesis in the fission yeast, Schizosaccharomyces pombe. Cell Motil. Cytoskeleton. 38:385–396 [DOI] [PubMed] [Google Scholar]
- McCollum D., Balasubramanian M.K., Pelcher L.E., Hemmingsen S.M., Gould K.L. 1995. Schizosaccharomyces pombe cdc4+ gene encodes a novel EF-hand protein essential for cytokinesis. J. Cell Biol. 130:651–660 10.1083/jcb.130.3.651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet M., Nurse P., Thuriaux P., Mitchison J.M. 1979. Uncontrolled septation in a cell division cycle mutant of the fission yeast Schizosaccharomyces pombe. J. Bacteriol. 137:440–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M., Oliferenko S. 2008. Cytokinesis: catch and drag. Curr. Biol. 18:R247–R250 10.1016/j.cub.2008.01.029 [DOI] [PubMed] [Google Scholar]
- Mishra M., Karagiannis J., Trautmann S., Wang H., McCollum D., Balasubramanian M.K. 2004. The Clp1p/Flp1p phosphatase ensures completion of cytokinesis in response to minor perturbation of the cell division machinery in Schizosaccharomyces pombe. J. Cell Sci. 117:3897–3910 10.1242/jcs.01244 [DOI] [PubMed] [Google Scholar]
- Mishra M., D’souza V.M., Chang K.C., Huang Y., Balasubramanian M.K. 2005. Hsp90 protein in fission yeast Swo1p and UCS protein Rng3p facilitate myosin II assembly and function. Eukaryot. Cell. 4:567–576 10.1128/EC.4.3.567-576.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- Motegi F., Arai R., Mabuchi I. 2001. Identification of two type V myosins in fission yeast, one of which functions in polarized cell growth and moves rapidly in the cell. Mol. Biol. Cell. 12:1367–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K., Mabuchi I. 2006. Actin-depolymerizing protein Adf1 is required for formation and maintenance of the contractile ring during cytokinesis in fission yeast. Mol. Biol. Cell. 17:1933–1945 10.1091/mbc.E05-09-0900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K., Imai J., Arai R., Toh-E A., Matsui Y., Mabuchi I. 2002. The small GTPase Rho3 and the diaphanous/formin For3 function in polarized cell growth in fission yeast. J. Cell Sci. 115:4629–4639 10.1242/jcs.00150 [DOI] [PubMed] [Google Scholar]
- Noguchi T., Mabuchi I. 2001. Reorganization of actin cytoskeleton at the growing end of the cleavage furrow of Xenopus egg during cytokinesis. J. Cell Sci. 114:401–412 [DOI] [PubMed] [Google Scholar]
- Pelham R.J., Jr, Chang F. 2001. Role of actin polymerization and actin cables in actin-patch movement in Schizosaccharomyces pombe. Nat. Cell Biol. 3:235–244 10.1038/35060020 [DOI] [PubMed] [Google Scholar]
- Petersen J., Nielsen O., Egel R., Hagan I.M. 1998. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J. Cell Biol. 141:1217–1228 10.1083/jcb.141.5.1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., Wu J.Q. 2010. Understanding cytokinesis: lessons from fission yeast. Nat. Rev. Mol. Cell Biol. 11:149–155 10.1038/nrm2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J., Flynn K.C., Raducanu A., Gärtner F., Beck G., Bösl M., Bradke F., Massberg S., Aszodi A., Sixt M., Wedlich-Söldner R. 2010. Lifeact mice for studying F-actin dynamics. Nat. Methods. 7:168–169 10.1038/nmeth0310-168 [DOI] [PubMed] [Google Scholar]
- Russell P., Nurse P. 1986. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell. 45:145–153 10.1016/0092-8674(86)90546-5 [DOI] [PubMed] [Google Scholar]
- Satterwhite L.L., Pollard T.D. 1992. Cytokinesis. Curr. Opin. Cell Biol. 4:43–52 10.1016/0955-0674(92)90057-J [DOI] [PubMed] [Google Scholar]
- Schroeder T.E. 1973. Actin in dividing cells: contractile ring filaments bind heavy meromyosin. Proc. Natl. Acad. Sci. USA. 70:1688–1692 10.1073/pnas.70.6.1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaine M., Numata O., Nakano K. 2009. Fission yeast IQGAP arranges actin filaments into the cytokinetic contractile ring. EMBO J. 28:3117–3131 10.1038/emboj.2009.252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toi H., Fujimura-Kamada K., Irie K., Takai Y., Todo S., Tanaka K. 2003. She4p/Dim1p interacts with the motor domain of unconventional myosins in the budding yeast, Saccharomyces cerevisiae. Mol. Biol. Cell. 14:2237–2249 10.1091/mbc.E02-09-0616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M., Imamoto N., Sakata-Sogawa K. 2008. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods. 5:159–161 10.1038/nmeth1171 [DOI] [PubMed] [Google Scholar]
- van den Ent F., Löwe J. 2006. RF cloning: a restriction-free method for inserting target genes into plasmids. J. Biochem. Biophys. Methods. 67:67–74 10.1016/j.jbbm.2005.12.008 [DOI] [PubMed] [Google Scholar]
- Vavylonis D., Wu J.Q., Hao S., O’Shaughnessy B., Pollard T.D. 2008. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 319:97–100 10.1126/science.1151086 [DOI] [PubMed] [Google Scholar]
- Vidali L., Rounds C.M., Hepler P.K., Bezanilla M. 2009. Lifeact-mEGFP reveals a dynamic apical F-actin network in tip growing plant cells. PLoS ONE. 4:e5744 10.1371/journal.pone.0005744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachtler V., Huang Y., Karagiannis J., Balasubramanian M.K. 2006. Cell cycle-dependent roles for the FCH-domain protein Cdc15p in formation of the actomyosin ring in Schizosaccharomyces pombe. Mol. Biol. Cell. 17:3254–3266 10.1091/mbc.E05-11-1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesche S., Arnold M., Jansen R.P. 2003. The UCS domain protein She4p binds to myosin motor domains and is essential for class I and class V myosin function. Curr. Biol. 13:715–724 10.1016/S0960-9822(03)00264-1 [DOI] [PubMed] [Google Scholar]
- White J.G., Borisy G.G. 1983. On the mechanisms of cytokinesis in animal cells. J. Theor. Biol. 101:289–316 10.1016/0022-5193(83)90342-9 [DOI] [PubMed] [Google Scholar]
- Win T.Z., Gachet Y., Mulvihill D.P., May K.M., Hyams J.S. 2001. Two type V myosins with non-overlapping functions in the fission yeast Schizosaccharomyces pombe: Myo52 is concerned with growth polarity and cytokinesis, Myo51 is a component of the cytokinetic actin ring. J. Cell Sci. 114:69–79 [DOI] [PubMed] [Google Scholar]
- Wolfe B.A., Gould K.L. 2005. Split decisions: coordinating cytokinesis in yeast. Trends Cell Biol. 15:10–18 10.1016/j.tcb.2004.11.006 [DOI] [PubMed] [Google Scholar]
- Wong K.C., Naqvi N.I., Iino Y., Yamamoto M., Balasubramanian M.K. 2000. Fission yeast Rng3p: an UCS-domain protein that mediates myosin II assembly during cytokinesis. J. Cell Sci. 113:2421–2432 [DOI] [PubMed] [Google Scholar]
- Wu J.Q., Pollard T.D. 2005. Counting cytokinesis proteins globally and locally in fission yeast. Science. 310:310–314 10.1126/science.1113230 [DOI] [PubMed] [Google Scholar]
- Wu J.Q., Bähler J., Pringle J.R. 2001. Roles of a fimbrin and an alpha-actinin-like protein in fission yeast cell polarization and cytokinesis. Mol. Biol. Cell. 12:1061–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.Q., Kuhn J.R., Kovar D.R., Pollard T.D. 2003. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev. Cell. 5:723–734 10.1016/S1534-5807(03)00324-1 [DOI] [PubMed] [Google Scholar]
- Wu J.Q., Sirotkin V., Kovar D.R., Lord M., Beltzner C.C., Kuhn J.R., Pollard T.D. 2006. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J. Cell Biol. 174:391–402 10.1083/jcb.200602032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonetani A., Chang F. 2010. Regulation of cytokinesis by the formin cdc12p. Curr. Biol. 20:561–566 10.1016/j.cub.2010.01.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.H., Crevenna A.H., Bettenbühl M., Freisinger T., Wedlich-Söldner R. 2011. Cortical actin dynamics driven by formins and myosin V. J. Cell Sci. 124:1533–1541 10.1242/jcs.079038 [DOI] [PubMed] [Google Scholar]
- Zacharias D.A., Violin J.D., Newton A.C., Tsien R.Y. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 296:913–916 10.1126/science.1068539 [DOI] [PubMed] [Google Scholar]
- Zhou M., Wang Y.L. 2008. Distinct pathways for the early recruitment of myosin II and actin to the cytokinetic furrow. Mol. Biol. Cell. 19:318–326 10.1091/mbc.E07-08-0783 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.