Abstract
We study invasion and survival of weakly beneficial mutations arising in linkage to an established migration–selection polymorphism. Our focus is on a continent–island model of migration, with selection at two biallelic loci for adaptation to the island environment. Combining branching and diffusion processes, we provide the theoretical basis for understanding the evolution of islands of divergence, the genetic architecture of locally adaptive traits, and the importance of so-called “divergence hitchhiking” relative to other mechanisms, such as “genomic hitchhiking”, chromosomal inversions, or translocations. We derive approximations to the invasion probability and the extinction time of a de novo mutation. Interestingly, the invasion probability is maximized at a nonzero recombination rate if the focal mutation is sufficiently beneficial. If a proportion of migrants carries a beneficial background allele, the mutation is less likely to become established. Linked selection may increase the survival time by several orders of magnitude. By altering the timescale of stochastic loss, it can therefore affect the dynamics at the focal site to an extent that is of evolutionary importance, especially in small populations. We derive an effective migration rate experienced by the weakly beneficial mutation, which accounts for the reduction in gene flow imposed by linked selection. Using the concept of the effective migration rate, we also quantify the long-term effects on neutral variation embedded in a genome with arbitrarily many sites under selection. Patterns of neutral diversity change qualitatively and quantitatively as the position of the neutral locus is moved along the chromosome. This will be useful for population-genomic inference. Our results strengthen the emerging view that physically linked selection is biologically relevant if linkage is tight or if selection at the background locus is strong.
Keywords: local adaptation, gene flow, linked selection, establishment, adaptive divergence
ADAPTATION to local environments may generate a selective response at several loci, either because the fitness-related traits are polygenic or because multiple traits are under selection. However, populations adapting to spatially variable environments often experience gene flow that counteracts adaptive divergence. The dynamics of polygenic adaptation is affected by physical linkage among selected genes, and hence by recombination (Barton 1995). Recombination allows contending beneficial mutations to form optimal haplotypes, but it also breaks up existing beneficial associations (Fisher 1930; Muller 1932; Hill and Robertson 1966; Lenormand and Otto 2000). On top of that, finite population size causes random fluctuations of allele frequencies that may lead to fixation or loss. Migration and selection create statistical associations even among physically unlinked loci.
The availability of genome-wide marker and DNA-sequence data has spurred both empirical and theoretical work on the interaction of selection, gene flow, recombination, and genetic drift. Here, we study the stochastic fate of a locally beneficial mutation that arises in linkage to an established migration–selection polymorphism. We also investigate the long-term effect on linked neutral variation of adaptive divergence with gene flow.
Empirical insight on local adaptation with gene flow emerges from studies of genome-wide patterns of genetic differentiation between populations or species. Of particular interest are studies that have either related such patterns to function and fitness (e.g., Nadeau et al. 2012, 2013) or detected significant deviations from neutral expectations (e.g., Karlsen et al. 2013), thus implying that some of this divergence is adaptive. One main observation is that in some organisms putatively adaptive differentiation (e.g., measured by elevated FST) is clustered at certain positions in the genome (Nosil and Feder 2012 and references therein). This has led to the metaphor of genomic islands of divergence or speciation (Turner et al. 2005). Other studies did not identify such islands, however (see Strasburg et al. 2012, for a review of plant studies).
These findings have stimulated theoretical interest in mechanistic explanations for the presence or absence of genomic islands. Polygenic local adaptation depends crucially on the genetic architecture of the selected traits, but, in the long run, local adaptation may also lead to the evolution of this architecture. Here, we define genetic architecture as the number of, and physical distances between, loci contributing to local adaptation, and the distribution of selection coefficients of established mutations.
Using simulations, Yeaman and Whitlock (2011) have shown that mutations contributing to adaptive divergence in a quantitative trait may physically aggregate in the presence of gene flow. In addition, these authors reported cases where the distribution of mutational effects changed from many divergent loci with mutations of small effect to few loci with mutations of large effect. Such clustered architectures reduce the likelihood of recombination breaking up locally beneficial haplotypes and incorporating maladaptive immigrant alleles. This provides a potential explanation for genomic islands of divergence. However, it is difficult to explain the variability in the size of empirically observed islands of divergence, especially the existence of very long ones. Complementary mechanisms have been proposed, such as the accumulation of adaptive mutations in regions of strongly reduced recombination (e.g., at chromosomal inversions; Guerrero et al. 2012; McGaugh and Noor 2012) or the assembly of adaptive mutations by large-scale chromosomal rearrangements (e.g., transpositions of loci under selection; Yeaman 2013).
It is well established that spatially divergent selection can cause a reduction in the effective migration rate (Charlesworth et al. 1997; Kobayashi et al. 2008; Feder and Nosil 2010). This is because migrants tend to carry combinations of alleles that are maladapted, such that selection against a locally deleterious allele at one locus also eliminates incoming alleles at other loci. The effective migration rate can be reduced either by physical linkage to a gene under selection or by statistical associations among physically unlinked loci. Depending on whether physical or statistical linkage is involved, the process of linkage-mediated differentiation with gene flow has, by some authors, been called “divergence hitchhiking” or “genomic hitchhiking,” respectively (Nosil and Feder 2012; Feder et al. 2012; Via 2012). These two processes are not mutually exclusive, and, recently, interest in assessing their relative importance in view of explaining observed patterns of divergence has been growing. If not by inversions or translocations, detectable islands of divergence are expected as a consequence of so-called divergence hitchhiking, but not of genomic hitchhiking. This is because physical linkage reduces the effective migration rate only locally (i.e., in the neighborhood of selected sites), whereas statistical linkage may reduce it across the whole genome. Yet, if many loci are under selection, it is unlikely that all of them are physically unlinked (Barton 1983), and so the two sources of linkage disequilibrium may be confounded.
A number of recent studies have focused on the invasion probability of neutral or locally beneficial de novo mutations in the presence of divergently selected loci in the background (Feder and Nosil 2010; Yeaman and Otto 2011; Feder et al. 2012; Flaxman et al. 2013; Yeaman 2013). They showed that linkage elevates invasion probabilities only over very short map distances, implying that physical linkage provides an insufficient explanation for both the abundance and size of islands of divergence. Such conclusions hinge on assumptions about the distribution of effects of beneficial mutations, the distribution of recombination rates along the genome, and the actual level of gene flow. These studies were based on time-consuming simulations (Feder and Nosil 2010; Feder et al. 2012; Flaxman et al. 2013; Yeaman 2013) or heuristic ad hoc aproximations (Yeaman and Otto 2011; Yeaman 2013) that provide limited understanding. Although crucial, invasion probabilities on their own might not suffice to gauge the importance of physical linkage in creating observed patterns of divergence. In finite populations, the time to extinction of adaptive mutations is also relevant. It codetermines the potential of synergistic interactions among segregating adaptive alleles.
Here, we fill a gap in existing theory to understand the role of physical linkage in creating observed patterns of divergence with gene flow. First, we provide numerical and analytical approximations to the invasion probability of locally beneficial mutations arising in linkage to an existing migration–selection polymorphism. This sheds light on the ambiguous role of recombination and allows for an approximation to the distribution of fitness effects of successfully invading mutations. Second, we obtain a diffusion approximation to the proportion of time the beneficial mutation segregates in various frequency ranges (the sojourn-time density) and the expected time to its extinction (the mean absorption time). From these, we derive an invasion-effective migration rate experienced by the focal mutation. Third, we extend existing approximations of the effective migration rate at a neutral site linked to two migration–selection polymorphisms (Bürger and Akerman 2011) to an arbitrary number of such polymorphisms. These formulae are used to predict the long-term footprint of polygenic local adaptation on linked neutral variation. We extend some of our analysis to the case of standing, rather than de novo, adaptive variation at the background locus.
Methods
Model
We consider a discrete-time version of a model with migration and selection at two biallelic loci (Bürger and Akerman 2011). Individuals are monoecious diploids and reproduce sexually. Soft selection occurs at the diploid stage and then a proportion m (0 < m < 1) of the island population is replaced by immigrants from the continent (Haldane 1930). Migration is followed by gametogenesis, recombination with probability r (0 ≤ r ≤ 0.5), and random union of gametes including population regulation. Generations do not overlap.
We denote the two loci by A and B and their alleles by A1 and A2, and B1 and B2, respectively. Locus A is taken as the focal locus and locus B as background locus. The four haplotypes 1, 2, 3, and 4 are A1B1, A1B2, A2B1, and A2B2. On the island, the frequencies of A1 and B1 are p and q, and the linkage disequilibrium is denoted by D (see Supporting Information, File S1, sect.1, for details).
Biological scenario
We assume that the population on the continent is fixed for alleles A2 and B2. The island population is of size N and initially fixed for A2 at locus A. At locus B, the locally beneficial allele B1 has arisen some time ago and is segregating at migration–selection balance. Then, a weakly beneficial mutation occurs at locus A, resulting in a single copy of A1 on the island. Its fate is jointly determined by direct selection on locus A, linkage to the selected locus B, migration, and random genetic drift. If A1 occurs on the beneficial background (B1), the fittest haplotype is formed and invasion is likely unless recombination transfers A1 to the deleterious background (B2). If A1 initially occurs on the B2 background, a suboptimal haplotype is formed (A1B2; Equation 1 below) and A1 is doomed to extinction unless it recombines onto the B1 background early on. These two scenarios occur proportionally to the marginal equilibrium frequency of B1. Overall, recombination is therefore expected to play an ambiguous role.
Two aspects of genetic drift are of interest: random fluctuations when A1 is initially rare and random sampling of alleles between successive generations. In the first part of the article, we focus exclusively on the random fluctuations when A1 is rare, assuming that N is so large that the dynamics is almost deterministic after an initial stochastic phase. In the second part, we allow for small to moderate population size N on the island. The long-term invasion properties of A1 are expected to differ in the two cases (Ewens 2004, pp. 167–171). With N sufficiently large and parameter combinations for which a fully polymorphic internal equilibrium exists under deterministic dynamics, the fate of A1 is decided very early on. If it survives the initial phase of stochastic loss, it will reach the (quasi-) deterministic equilibrium frequency and stay in the population for a very long time (Petry 1983). This is what we call invasion, or establishment. Extinction will finally occur, because migration introduces A2, but not A1. Yet, extinction occurs on a timescale much longer than is of interest for this article. For small or moderate N, however, genetic drift will cause extinction of A1 on a much shorter timescale, even for moderately strong selection. In this case, stochasticity must be taken into account throughout, and interest shifts to the expected time A1 spends in a certain range of allele frequencies (sojourn time) and the expected time to extinction (absorption time).
As an extension of this basic scenario, we allow the background locus to be polymorphic on the continent. Allele B1 is assumed to segregate at a constant frequency qc. This reflects, for instance, a polymorphism maintained at drift–mutation or mutation–selection balance. It could also apply to the case where the continent is a metapopulation or receives migrants from other populations. A proportion qc of haplotypes carried by immigrants to the focal island will then be A2B1, and a proportion 1 − qc will be A2B2.
Fitness and evolutionary dynamics
We define the relative fitness of a genotype as its expected relative contribution to the gamete pool from which the next generation of zygotes is formed. We use wij for the relative fitness of the genotype composed of haplotypes i and j (i, j ∈ {1, 2, 3, 4}). Ignoring parental and position effects in heterozygotes, we distinguish nine genotypes. We then have wij = wji for all i ≠ j and w23 = w14.
The extent to which analytical results can be obtained for general fitnesses is limited (Ewens 1967; Karlin and McGregor 1968). Unless otherwise stated, we therefore assume absence of dominance and epistasis, i.e., allelic effects combine additively within and between loci. The matrix of relative genotype fitnesses wij (Equation 27 in File S1) may then be written as
| (1) |
where a and b are the selective advantages on the island of alleles A1 and B1 relative to A2 and B2, respectively. To enforce positive fitnesses, we require that 0 < a, b < 1, and a + b < 1. We assume that selection in favor of A1 is weaker than selection in favor of B1 (a < b). Otherwise, A1 could be maintained in a sufficiently large island population independently of B1, whenever B1 is not swamped by gene flow (Haldane 1930). As our focus is on the effect of linkage on establishment of A1, this case is not of interest.
The deterministic dynamics of the haplotype frequencies are given by the recursion equations in File S1, Equation 28 (see also File S2). A crucial property of these dynamics is the following. Whenever a marginal one-locus migration–selection equilibrium EB exists such that the background locus B is polymorphic and locus A is fixed for allele A2, this equilibrium is asymptotically stable. After occurrence of A1, EB may become unstable, in which case a fully polymorphic (internal) equilibrium emerges and is asymptotically stable, independently of whether the continent is monomorphic (qc = 0) or polymorphic (0 < qc < 1) at the background locus. Therefore, in the deterministic model, invasion of A1 via EB is always followed by an asymptotic approach toward an internal equilibrium (see File S1, sect. 3 and 6).
Casting our model into a stochastic framework is difficult in general. By focusing on the initial phase after occurrence of A1, the four-dimensional system in Equation 28 can be simplified to a two-dimensional system (Equation 29 in File S1). This allows for a branching-process approach as described in the following.
Two-type branching process
As shown in File S1, sect. 2, for rare A1, we need to follow only the frequencies of haplotypes A1B1 and A1B2. This corresponds to A1 initially occurring on the B1 or B2 background, respectively, and holds as long as A1 is present in heterozygotes only. Moreover, it is assumed that allele B1 is maintained constant at the marginal one-locus migration–selection equilibrium EB of the dynamics in Equation 28. At this equilibrium, the frequency of B1 is
| (2) |
for a monomorphic continent (see File S1, sect. 3, for details, and Equation 39 for a polymorphic continent).
To model the initial stochastic phase after occurrence of A1 for large N, we employed a two-type branching process in discrete time (Harris 1963). We refer to haplotypes A1B1 and A1B2 as types 1 and 2, respectively. They are assumed to propagate independently and contribute offspring to the next generation according to type-specific distributions. We assume that the number of j-type offspring produced by an i-type parent is Poisson-distributed with parameter λij (i ∈ {1, 2}). Because of independent offspring distributions, the probability-generating function (pgf) for the number of offspring of any type produced by an i-type parent is , where for i, j ∈ {1, 2} (File S1, sect. 4). The λij depend on fitness, migration, and recombination and are derived from the deterministic model (Equation 33 in File S1). The matrix L = (λij), i, j ∈ {1, 2}, is called the mean matrix. Allele A1 has a strictly positive invasion probability if ν > 1, where ν is the leading eigenvalue of L. The branching process is called supercritical in this case.
We denote the probability of invasion of A1 conditional on initial occurrence on background B1 (B2) by π1 (π2), and the corresponding probability of extinction by Q1 (Q2). The latter are found as the smallest positive solution of
| (3a) |
| (3b) |
such that si < 1 (i ∈ {1, 2}). Then, π1 = 1 − Q1 and π2 = 1 − Q2 (Haccou et al. 2005). The overall invasion probability of A1 is given as the weighted average of the two conditional probabilities,
| (4) |
(cf. Ewens 1967, 1968; Kojima and Schaffer 1967). File S1, sect. 4, gives further details and explicit expressions for additive fitnesses.
Diffusion approximation
The branching process described above models the initial phase of stochastic loss and applies as long as the focal mutant A1 is rare. To study long-term survival of A1, we employ a diffusion approximation. We start from a continuous-time version of the deterministic dynamics in Equation 28, assuming additive fitnesses as in Equation 1. For our purpose, it is convenient to express the dynamics in terms of the allele frequencies (p, q) and the linkage disequilibrium (D), as given in Equation 87 in File S1. Changing to the diffusion scale, we measure time in units of 2Ne generations, where Ne is the effective population size.
We introduce the scaled selection coefficients α = 2Ne a and β = 2Neb, the scaled recombination rate ρ = 2Ner, and the scaled migration rate μ = 2Nem. As it is difficult to obtain analytical results for the general two-locus diffusion problem (Ethier and Nagylaki 1980, 1988, 1989; Ewens 2004), we assume that recombination is much stronger than selection and migration. Then, linkage disequilibrium decays on a faster timescale, whereas allele frequencies evolve on a slower one under quasi-linkage equilibrium (QLE) (Kimura 1965; Nagylaki et al. 1999; Kirkpatrick et al. 2002). In addition, we assume that the frequency of the beneficial background allele B1 is not affected by establishment of A1 and stays constant at Here, is the frequency of B1 at the one-locus migration–selection equilibrium when time is continuous, (Equations 88 and 89 in File S1). As further shown in File S1, sect. 6, these assumptions lead to a one-dimensional diffusion process. The expected change in p per unit time is
| (5) |
if the continent is monomorphic. The first term is due to direct selection on the focal locus, the second reflects migration, and the third represents the interaction of all forces.
For a polymorphic continent, M(p) is given by Equation 116 in File S1, and the interaction term includes the continental frequency qc of B1. In both cases, assuming random genetic drift according to the Wright–Fisher model, the expected squared change in p per unit time is V(p) = p(1 − p) (Ewens 2004). We call M(p) the infinitesimal mean and V(p) the infinitesimal variance (Karlin and Taylor 1981, p. 159).
Let the initial frequency of A1 be p0. We introduce the sojourn-time density (STD) t(p; p0) such that the integral approximates the expected time A1 segregates at a frequency between p1 and p2 before extinction, conditional on p0. Following Ewens (2004, Equations 4.38 and 4.39), we define
| (6) |
with subscript QLE for the assumption of quasi-linkage equilibrium. The densities ti,QLE(p; p0) are
| (7a) |
| (7b) |
where Integration over p yields the expected time to extinction,
| (8) |
or the mean absorption time, in units of 2Ne generations. A detailed exposition is given in File S1, sect. 7. See File S10 for Mathematica Notebooks.
Simulations
We conducted two types of simulation, one for the branching-process regime and another for a finite island population with Wright–Fisher random drift. In the branching-process regime, we simulated the absolute frequency of the two types of interest (A1B1 and A1B2) over time. Each run was initiated with a single individuum and its type determined according to Equation 2. Every generation, each individual produced a Poisson-distributed number of offspring of either type (see above). We performed n = 106 runs. Each run was terminated if either the mutant population went extinct (no invasion), reached a size of 500/(2a) (invasion), or survived for more than 5 × 104 generations (invasion). We estimated the invasion probability from the proportion of runs that resulted in invasion, and its standard error as
In the Wright–Fisher-type simulations, each generation was initiated by zygotes built from gametes of the previous generation. Viability selection, migration, and gamete production including recombination (meiosis) were implemented according to the deterministic recursions for the haplotype frequencies in Equation 28. Genetic drift was simulated through the formation of Ne (rather, the nearest integer) zygotes for the next generation by random union of pairs of gametes. Gametes were sampled with replacement from the gamete pool in which haplotypes were represented according to the deterministic recursions. Replicates were terminated if either allele A1 went extinct or a maximum of 109 generations was reached. Unless otherwise stated, for each parameter combination we performed 1000 runs, each with 1000 replicates. Replicates within a given run provided one estimate of the mean absorption time, and runs provided a distribution of these estimates. Java source code and JAR files are available in File S11.
Results
Establishment in a large island population
We first describe the invasion properties of the beneficial mutation A1, which arises in linkage to a migration–selection polymorphism at the background locus B. Because we assume that the island population is large, random genetic drift is ignored after A1 has overcome the initial phase during which stochastic loss is likely. Numerical and analytical results were obtained from the two-type branching process and confirmed by simulations (see Methods). We turn to the case of small to moderate population size further below. (See lines 511, 515, and 516.)
Conditions for the invasion of A1:
Mutation A1 has a strictly positive invasion probability whenever
| (9) |
(File S1, sect. 4, and File S3). Here, wi is the marginal fitness of type i and the mean fitness of the resident population (see Equations 30 and 31 in File S1). Setting m = 0, we recover the invasion condition obtained by Ewens (1967) for a panmictic population in which allele B1 is maintained at frequency by overdominant selection. All remaining results in this subsection assume additive fitnesses as in Equation 1.
For a monomorphic continent (qc = 0), it follows from Equation 9 that A1 can invade only if m < m*, where
| (10) |
In terms of the recombination rate, A1 can invade only if r < r*, where
| (11) |
(see File S1, sects. 3 and 4, File S2, and Figure S1 for details).
For a polymorphic continent (0 < qc < 1), A1 has a strictly positive invasion probability whenever r and qc are below the critical values r* and derived in File S1, sect. 3 (cf. File S4, Figure S2). In this case, we could not determine the critical migration rate m* explicitly. For an analysis in continuous time, see File S1, sect. 6, and Figure S7, Figure S8, and Figure S9.
Invasion probability:
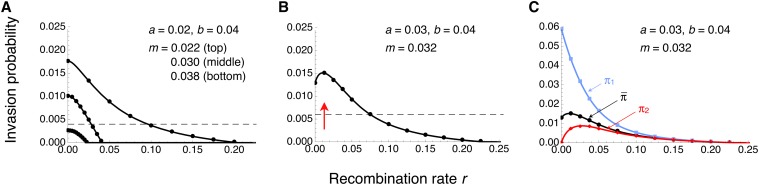
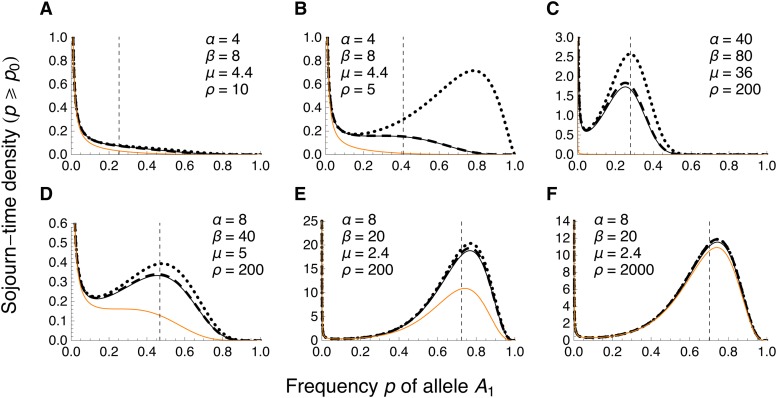
We obtained exact conditional invasion probabilities, π1 and π2, of A1 by numerical solution of the pair of transcendental equations in Equation 3. From these, we calculated the average invasion probability according to Equation 4, with as in Equation 2 (Figure 1 and Figure S3 for a monomorphic continent). Haldane (1927) approximated the invasion probability without migration and linked selection by 2a, i.e., twice the selective advantage of A1 in a heterozygote. With linked selection, the map distance over which is above, say, 10% of 2a can be large despite gene flow (Figure 1, A and B).
Figure 1.
Invasion probability of A1 as a function of the recombination rate for a monomorphic continent. (A and B) Weighted average invasion probabilitiy across the two genetic backgrounds B1 and B2 (Equations 2 and 4). For comparison, horizontal dashed lines give 10% of Haldane’s (1927) approximation 2a, valid for m = 0 and r = 0.5. (B) The optimal recombination rate ropt, defined as the recombination rate at which is maximized (red arrow), is nonzero. (C) Same as in B, but in addition to the weighted average, the invasion probabilities of A1 conditional on initial occurrence on the B1 or B2 background are shown in blue or red, respectively. Note the difference in the scale of the vertical axis between B and C. In A–C, curves show exact numerical solutions to the branching process. Dots represent the point estimates across 106 simulations under the branching-process assumptions (see Methods). Error bars span twice the standard error on each side of the point estimates, but are too short to be visible.
Analytical approximations were obtained by assuming that the branching process is slightly supercritical, i.e., that the leading eigenvalue of the mean matrix L is of the form ν = 1 + ξ, with ξ > 0 small. We denote these approximations by π1(ξ) and π2(ξ). The expressions are long (File S5) and not shown here. For weak evolutionary forces (a, b, m, r ≪ 1), π1(ξ) and π2(ξ) can be approximated by
| (12a) |
| (12b) |
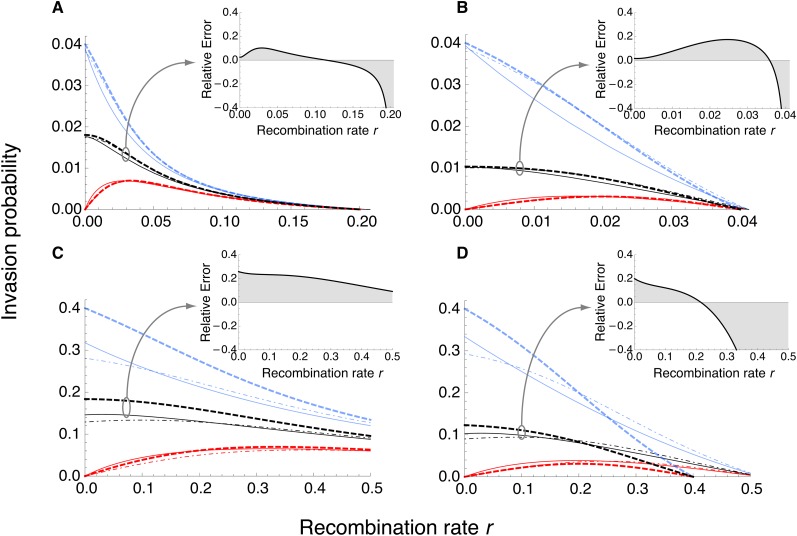
where R2 = b2 + 2br − 4mr + r2 and The approximate average invasion probability is obtained according to Equation 4, with as in Equation 2. Formally, these approximations are justified if ξ ≪ 1 (File S1, sect. 4). Figure 2 suggests that the assumption of weak evolutionary forces is more crucial than ξ small and that if it is fulfilled, the approximations are very good (compare Figure 2, A–D).
Figure 2.
Approximation to the invasion probability of A1 for a monomorphic continent. Invasion probabilities are shown for A1 initially occurring on the beneficial background B1 (blue), on the deleterious background B2 (red), and as a weighted average across backgrounds (black). Analytical approximations assuming a slightly supercritical branching process (dot-dashed curves) and, in addition, weak evolutionary forces (Equation 12; thick dashed curves) are compared to the exact numerical branching-process solution (solid curves). Inset figures show the error of the analytical approximation (thick dashed black curve) relative to (solid black curve), (A) a = 0.02, b = 0.04, m = 0.022. (B) a = 0.02, b = 0.04, m = 0.03. (C) a = 0.2, b = 0.4, m = 0.22. (D) a = 0.2, b = 0.4, m = 0.3. As expected, the analytical approximations are very good for weak evolutionary forces (top row), but less so for strong forces (bottom row).
For a polymorphic continent, exact and approximate invasion probabilities are derived in File S3 and File S5 (see also sect. 4 in File S1). The most important, and perhaps surprising, effect is that the average invasion probability decreases with increasing continental frequency qc of the beneficial background allele B1 (Figure S4). As a consequence, invasion requires tighter linkage if qc > 0. This is because the resident island population has a higher mean fitness when a proportion qc > 0 of immigrating haplotypes carry the B1 allele, which makes it harder for A1 to become established. Competition against fitter residents therefore compromises the increased probability of recombining onto a beneficial background (B1) when A1 initially occurs on the deleterious background (B2). However, a closer look suggests that if A1 is sufficiently beneficial and recombination sufficiently weak (r ≪ a), there are cases where the critical migration rate below which A1 can invade is maximized at an intermediate qc (Figure S5, right column). In other words, for certain combinations of m and r, the average invasion probability as a function of qc is maximized at an intermediate (nonzero) value of qc (Figure S6).
For every combination of selection coefficients (a, b) and recombination rate (r), the mean invasion probability decreases as a function of the migration rate m. This holds for a monomorphic and a polymorphic continent (Figure S3 and Figure S5, respectively). In both cases, migrants carry only allele A2 and, averaged across genetic backgrounds, higher levels of migration make it harder for A1 to invade (cf. Bürger and Akerman 2011).
Optimal recombination rate:
Deterministic analysis showed that A1 can invade if and only if recombination is sufficiently weak; without epistasis, large r is always detrimental to establishment of A1 (Bürger and Akerman 2011; File S1, sect. 3). In this respect, stochastic theory is in line with deterministic predictions. However, considering the average invasion probability as a function of r, we could distinguish two qualitatively different regimes. In the first one, decreases monotonically with increasing r (Figure 1A). In the second one, is maximized at an intermediate recombination rate ropt (Figure 1B). A similar dichotomy was previously found for a panmictic population in which the background locus is maintained polymorphic by heterozygote superiority (Ewens 1967) and has recently been reported in the context of migration and selection in simulation studies (Feder and Nosil 2010; Feder et al. 2012). As shown in File S1, sect. 5, ropt > 0 holds in our model whenever
| (13) |
where w1 (w2) is the marginal fitness of type 1 (2) and the mean fitness of the resident population (defined in Equations 30 and 31 in File S1). Here, is the invasion probability of A1 conditional on background B1 and complete linkage (r = 0). Setting m = 0, we recover Equation 36 of Ewens (1967) for a panmictic population with overdominance at the background locus.
Inequality (13) is very general. In particular, it also holds with epistasis or dominance. However, explicit conclusions require calculation of , , and wi, which themselves depend on and hence on m (cf. Equation 2). For mathematical convenience, we resorted to the assumption of additive fitnesses (Equation 1). For a monomorphic continent, to first order in a. Moreover, we found that
| (14) |
is a necessary condition for ropt > 0, where
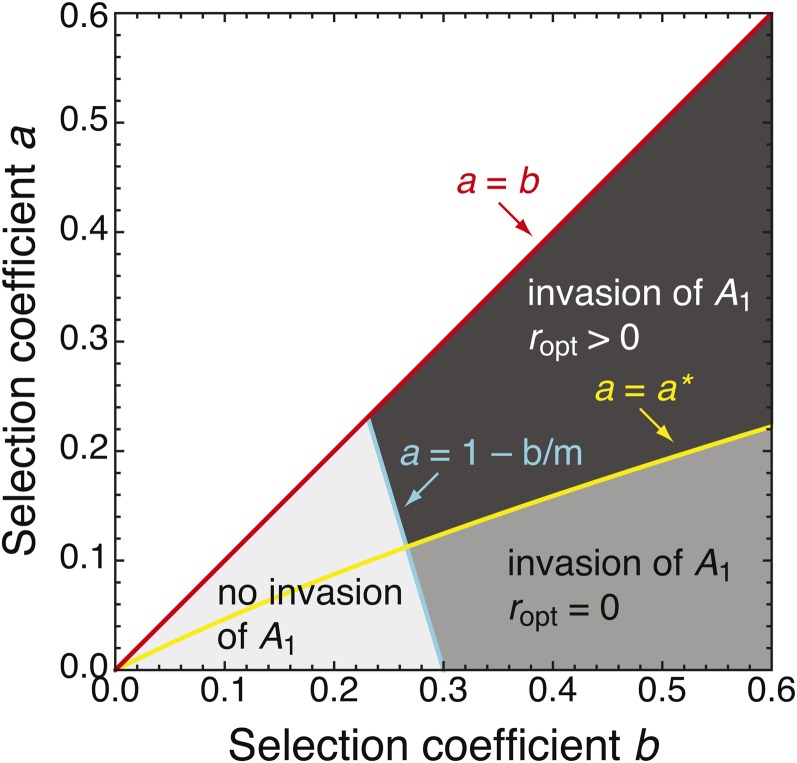
(File S1, sect. 5). Thus, A1 must be sufficiently beneficial for ropt > 0 to hold. Figure 3 shows the division of the parameter space where A1 can invade into two areas where ropt = 0 or ropt > 0 holds.
Figure 3.
Optimal recombination rate and regions of invasion. The dark shaded area indicates where the optimal recombination rate r is positive (ropt > 0; cf. Figure 1B). The medium shaded area shows the parameter range for which ropt = 0 (cf. Figure 1A). Together, these two areas indicate where A1 can invade via the marginal one-locus migration–selection equilibrium EB if r is sufficiently small. The light shaded area shows where EB does not exist and A1 cannot invade via EB. The area above a = b is not of interest, as we focus on mutations that are weakly beneficial compared to selection at the background locus (a < b). The critical selection coefficient a∗ is given in Equation 14 and the migration rate is m = 0.3 (other values of m yield qualitatively similar diagrams). The continent is monomorphic (qc = 0).
The two regimes ropt = 0 and ropt > 0 arise from the ambiguous role of recombination. On the one hand, when A1 initially occurs on the deleterious background (B2), some recombination is needed to transfer A1 onto the beneficial background (B1) and rescue it from extinction. This is reminiscent of Hill and Robertson’s (1966) result that recombination improves the efficacy of selection in favor of alleles that are partially linked to other selected sites (Barton 2010). On the other hand, when A1 initially occurs on the beneficial background, recombination is always deleterious, as it breaks up the fittest haplotype on the island (A1B1). This interpretation is confirmed by considering π1 and π2 separately as functions of r (Figure 1C). Whereas π1(r) always decreases monotonically with increasing r, π2(r) is always 0 at r = 0 (File S1, sect. 5) and then increases to a maximum at an intermediate recombination rate (compare blue to red curve in Figure 1C). As r increases further, π1(r) and π2(r) both approach 0. We recall from Equation 4 that the average invasion probability is given by Depending on , either π1 or π2 makes a stronger conbribution to , which then leads to either ropt > 0 or ropt = 0.
A more intuitive interpretation of Equation 14 is as follows. If A1 conveys a weak advantage on the island (a < a*), it will almost immediately go extinct when it initially arises on background B2. Recombination has essentially no opportunity of rescuing A1, even if r is large. Therefore, π2 contributes little to If A1 is sufficiently beneficial on the island (a > a*), however, it will survive for some time even when arising on the deleterious background. Recombination now has time to rescue A1 if r is sufficiently different from 0 (but not too large). In this case, π2 makes an important contribution to and leads to ropt > 0. For a polymorphic continent, ropt > 0 may also hold ( File S1, sect. 5). However, in such cases, ropt approaches zero quickly with increasing qc (File S6 and Figure S4).
Distribution of fitness effects of successful mutations:
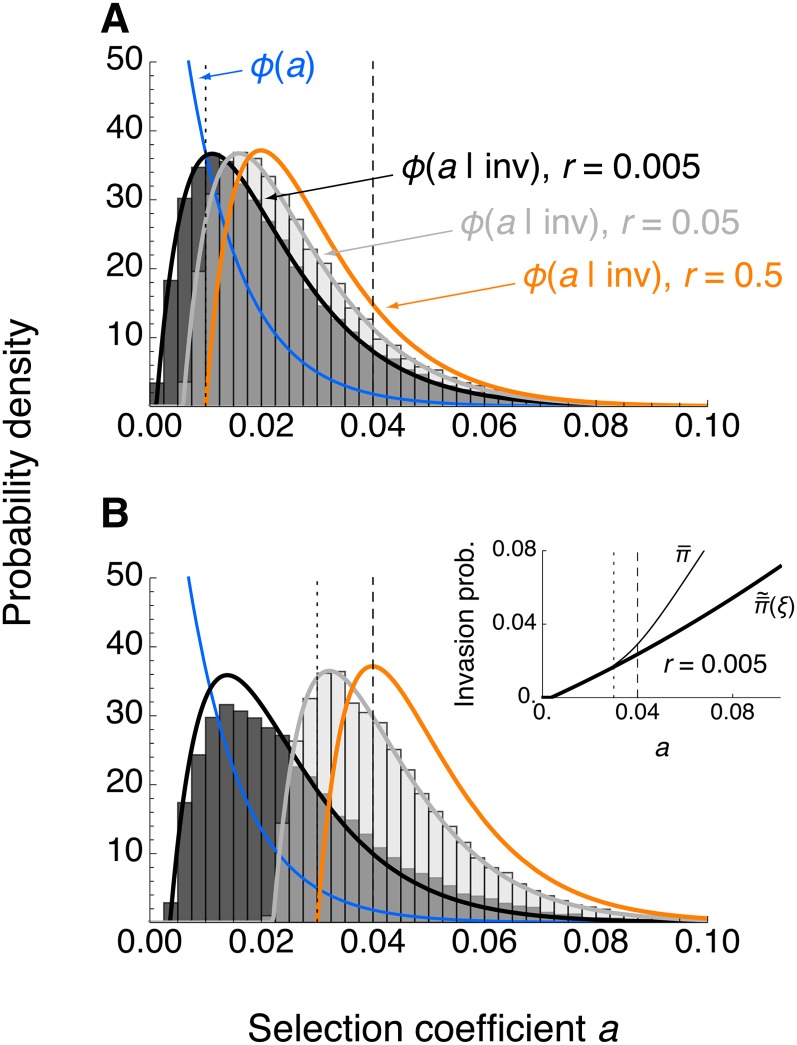
Using Equation 12 we can address the distribution of fitness effects (DFE) of successfully invading mutations. This distribution depends on the distribution of selection coefficients a of novel mutations (Kimura 1979), which in general is unknown (Orr 1998). In our scenario, the island population is at the marginal one-locus migration–selection equilibrium EB before the mutation A1 arises. Unless linkage is very tight, the selection coefficient a must be above a threshold for A1 to effectively withstand gene flow (this threshold is implicitly defined by Equation 10). Therefore, we assumed that a is drawn from the tail of the underlying distribution, which we took to be exponential (Gillespie 1983, 1984; Orr 2002, 2003; Barrett et al. 2006; Eyre-Walker and Keightley 2007) (for alternatives, see Cowperthwaite et al. 2005; Barrett et al. 2006; Martin and Lenormand 2008). We further assumed that selection is directional with a constant fitness gradient (Equation 1). We restricted the analysis to the case of a monomorphic continent. As expected, linkage to a migration–selection polymorphism shifts the DFE of successfully invading mutations toward smaller effect sizes (Figure 4). Comparison to simulated histograms in Figure 4 suggests that the approximation based on Equation 12 is very accurate.
Figure 4.
DFE of successfully invading mutations for a monomorphic continent. The DFE of successfully invading mutations was obtained as , where with and as in Equations 2 and 12, respectively. The mutational input distribution was assumed to be exponential, φ(a) = λe−aλ (blue). Vertical lines denote a = m (dotted) and a = b (dashed). Histograms were obtained from simulations under the branching-process assumptions (intermediate shading indicates where histograms overlap). Each represents 2.5 × 104 realizations in which A1 successfully invaded (see Methods). As a reference, the one-locus model (no linkage) is shown in orange. (A) Relatively weak migration: b = 0.04, m = 0.01. (B) Migration three times stronger: b = 0.04, m = 0.03. In A and B, λ = 100 and is shown for a recombination rate of r = 0.005 (black) and r = 0.05 (gray). The inset in B shows why the fit is worse for r = 0.005: in this case, underestimates the exact invasion probability (Equation 4) for large a.
Survival in a finite island population
We now turn to island populations of small to moderate size N. In this case, genetic drift is strong enough to cause extinction on a relevant timescale even after successful initial establishment. Our focus is on the sojourn-time density and the mean absorption time of the locally beneficial mutation A1 (see Methods). We also derive an approximation to the effective migration rate experienced by A1.
Sojourn-time density:
A general expression for the STD was given in Equation 7. Here, we describe some properties of the exact numerical solution and then discuss analytical approximations (see also File S7). Because A1 is a de novo mutation, it has an initial frequency of p0 = 1/(2N). For simplicity, we assumed that the effective population size on the island is equal to the actual population size, i.e., Ne = N (this assumption is relaxed later). As p0 = 1/(2N) is very close to zero in most applications, we used t2,QLE(p; p0) as a proxy for tQLE(p; p0) (cf. Equation 6).
The STD always has a peak at p = 0, because most mutations go extinct after a very short time (Figure 5). However, for parameter combinations favorable to invasion of A1 (migration weak relative to selection, or selection strong relative to genetic drift), the STD has a second mode at an intermediate allele frequency p. Then, allele A1 may spend a long time segregating in the island population before extinction. The second mode is usually close to—but slightly greater than—the corresponding deterministic equilibrium frequency (solid black curves in Figure 5, C–F for a monomorphic continent). The peak at this mode becomes shallower as the continental frequency qc of B1 increases (Figure S10).
Figure 5.
Diffusion approximation to the sojourn-time density of A1 under quasi-linkage equilibrium for a monomorphic continent. Comparison of the STD t2,QLE(p; p0) (thin black; Equation 7b) to the approximation valid for small p0, (dashed black; Equation 109 in File S1), and the one based on the additional assumption of ρ ≫ max(α, β, μ), (dotted; Equation 17b). The STD for the one-locus model, , is shown in orange as a reference. Vertical lines give the deterministic frequency of A1 at the fully polymorphic equilibrium (computed in File S7). (A) Weak evolutionary forces relative to genetic drift. (B) As in A, but with half the scaled recombination rate ρ. The assumption of ρ ≫ max(α, β, μ) is violated and hence is a poor approximation of t2,QLE(p; p0). (C) Strong evolutionary forces relative to genetic drift. The STD has a pronounced mode different from p = 0, but overestimates t2,QLE(p; p0) considerably. (D) Strong assymmetry in selection coefficients and moderate migration. As in C, the STD has a pronounced mode different from p = 0, but now approximates t2,QLE(p; p0) better. (E) Recombination 10 times stronger than selection at locus B. (F) As in E, but with recombination 100 times stronger than selection at locus B. In A–F, p0 = 0.005, which corresponds to an island population of size N = 100 and a single initial copy of A1.
The effect on the STD of linkage is best seen from a comparison to the one-locus model (OLM), for which the STD is given by Ewens (2004) as
| (15) |
If invasion of A1 is unlikely without linkage, but selection at the background locus is strong, even loose linkage has a large effect and causes a pronounced second mode in the STD (compare orange to black curves in Figure 5C). In cases where A1 can be established without linkage, the STD of the one-locus model also shows a second mode at an intermediate allele frequency p. Yet, linkage to a background polymorphism leads to a much higher peak, provided that selection at the background locus is strong and the recombination rate not too high (Figure 5, D–F). Specifically, comparison of Figure 5E with Figure 5F suggests that the effect of linkage becomes weak if the ratio of the (scaled) recombination rate to the (scaled) selection coefficient at the background locus, ρ/β, becomes much larger than ∼10. In other words, for a given selective advantage b of the beneficial background allele, a weakly beneficial mutation will profit from linkage if it occurs within ∼b × 103 map units (centimorgans) from the background locus. This assumes that one map unit corresponds to r = 0.01.
An analytical approximation of the STD can be obtained under two simplifying assumptions. The first is that the initial frequency p0 of A1 is small (p0 on the order of 1/(2Ne) ≪ 1). The second concerns the infinitesimal mean M(p) of the change in the frequency of A1: assuming that recombination is much stronger than selection and migration, we may approximate Equation 5 by
| (16) |
for a monomorphic continent. The STDs in Equation 7 can then be approximated by
| (17a) |
| (17b) |
Here, we use ∼ to denote the assumption of p0 small, and a subscript ρ ≫ 0 for the assumption of ρ ≫ max(α, β, μ). For a polymorphic continent, expressions analogous to Equations 16 and 17 are given in Equations 117 and 119 in File S1.
Better approximations than those in Equations 17 and 119 are obtained by making only one of the two assumptions above. We denote by and the approximations of the STD in Equation 7 based on the assumption p0 ≪ 1 (Equations 108 and 109 in File S1). Alternatively, the approximations obtained from the assumption ρ ≫ max(α, β, μ) in M(p) are called t1,QLE,ρ≫0(p; p0) and t2,QLE,ρ≫0(p; p0) (Equation 113).
In the following, we compare the different approximations to each other and to stochastic simulations. Conditional on p0 = 1/(2N), the approximation (Equations 109) is indeed very close to the exact numerical value t2,QLE(p; p0) from Equation 7b. This holds across a wide range of parameter values, as seen from comparing solid to dashed curves in Figure 5 (monomorphic continent) and Figure S10 (polymorphic continent). The accuracy of the approximation from Equation 17b is rather sensitive to violation of the assumption ρ ≫ max(α, β, μ), however (dotted curves deviate from other black curves in Figure 5, B and C). The same applies to a polymorphic continent, but the deviation becomes smaller as qc increases from zero (Figure S10A).
Comparison of the diffusion approximation to sojourn-time distributions obtained from stochastic simulations shows a very good agreement, except at the boundary p = 0. There, the continuous solution of the diffusion approximation is known to provide a suboptimal fit to the discrete distribution (Figure S11 and Figure S12).
Based on the analytical approximations above, we may summarize the effect of weak linkage relative to the one-locus model as follows. For a monomorphic continent, the ratio of to t2,OLM(p; p0) is where γ = 2μ(β − μ)/ρ. The exponent γ is a quadratic function of μ and linear in β. For weak migration, suggesting the following rule of thumb. For the focal allele to spend at least the -fold amount of time at frequency p compared to the case without linkage, we require
| (18) |
For example, allele A1 will spend at least twice as much time at frequency P = 0.5 (0.8) if βμ ≳ 0.72ρ (0.31ρ). Because we assumed weak migration and QLE, we conducted numerical explorations to check when this rule is conservative, meaning that it does not predict a larger effect of linked selection than is observed in simulations. We found that, first, genetic drift must not dominate, i.e., 1 < α, β, μ, ρ holds. Second, migration, selection at the background locus and recombination should roughly satisfy μ < β/4 < 0.1ρ. This condition applies only to the validity of Equation 18, which is based on in Equation 17b. It does not apply to , which fits simulations very well if ρ is as low as 1.25β (Figure S11D). For related observations in different models, see Slatkin (1975) and Barton (1983).
Mean absorption time:
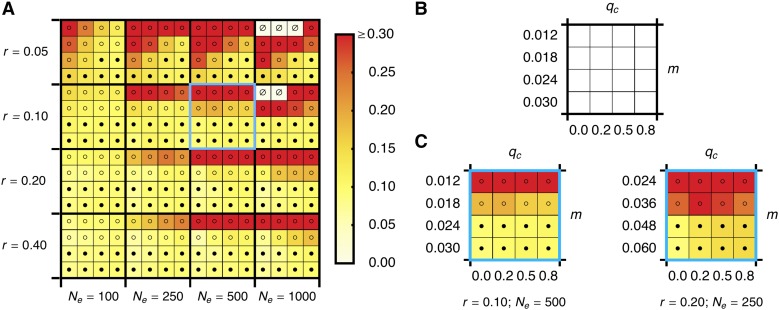
The mean absorption time is obtained by numerical integration of the STD as outlined in Methods. Comparison to stochastic simulations shows that the diffusion approximation from Equation 8 is fairly accurate: the absolute relative error is <15%, provided that the QLE assumption is not violated and migration is not too weak (Figure 6).
Figure 6.
Relative error of the diffusion approximation to the mean absorption time of A1. (A) The error of from Equation 8 relative to simulations for various parameter combinations. Squares bounded by thick lines delimit combinations of values of the recombination rate r and the effective population size Ne. Within each of them, values of the migration rate m and the continental frequency qc of B1 are as shown in B. No negative relative errors were observed. For better resolution, we truncated values >0.30 (the maximum was 3.396 for Ne = 1000, r = 0.05, m = 0.018, qc = 0.0). Open (solid) circles indicate that the marginal one-locus equilibrium is unstable (stable) and A1 can (not) be established under deterministic dynamics. Parameter combinations for which simulations were too time consuming are indicated by ∅. Selection coefficients are a = 0.02 and b = 0.04. (C) The left plot corresponds to the square in A that is framed in blue. The right plot shows the fit of the diffusion approximation to simulations conducted with unscaled parameters twice as large and Ne half as large, as on the left side. Scaled parameters are equal on both sides. As expected, the diffusion approximation is worse on the right side. Simulations were as described in Methods. See Table S1 for numerical values.
Given the approximations to the STD derived above, various degrees of approximation are available for the mean absorption time, too. Their computation is less prone to numerical issues than that of the exact expressions. Extensive numerical computations showed that if p0 = 1/(2N) and Ne = N, the approximations based on the assumption of p0 small ( and as given in Equations 110 and 115) provide an excellent fit to their more exact counterparts ( and in Equations 8 and 114, respectively). See also Table S2 and S4. Across a wide range of parameter values, the absolute relative error never exceeds 1.8% (Figure S13, A and C). In contrast, the approximation based on the assumption of ρ ≫ 0, , is very sensitive to violations of this assumption. For large effective population sizes and weak migration, the relative error becomes very high if recombination is not strong enough (Figure S13B; Table S3).
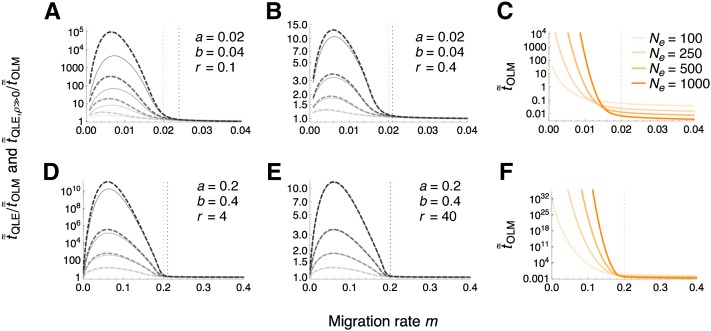
The effect of linkage is again demonstrated by a comparison to the one-locus model. If selection is strong relative to recombination, the mean absorption time with linkage, , is increased by several orders of magnitude compared to the one-locus case, (Figure 7, A and D; Table S5). The effect is reduced, but still notable, when the recombination rate becomes substantially higher than 10 times the strength of selection in favor of the beneficial background allele, i.e., ρ/β ≫ 10 (Figure 7, B and E). Importantly, large ratios of are not an artifact of being very small, as Figure 7, C and F confirm. Moreover, is maximized at intermediate migration rates: for very weak migration, A1 has a fair chance of surviving for a long time even without linkage ( is large); for very strong migration, and both tend to zero and approaches unity.
Figure 7.
Mean absorption time of A1 under quasi-linkage equilibrium relative to the one-locus model (OLM). In panels (A), (B), (D) and (E), thin solid curves show the ratio and thick dashed curves , as a function of the migration rate m. The effective population size Ne increases from light to dark gray, taking values of 100, 250, 500, and 1000. Vertical lines denote the migration rate below which A1 can invade in the deterministic one-locus (orange) and two-locus (black) model. (A) Recombination is too weak for the assumption ρ ≫ max(α, β, μ) to hold. (B) As in (A), but with recombination four times stronger. (D) Evolutionary forces – other than drift – are ten times stronger than in (B). (E) As in (D), but with recombination ten times stronger. Panels (C) and (F) show the mean absorption time (in multiples of 2Ne) under the one-locus model for the respective row. For m close to 0, numerical procedures are unstable and we truncated the curves. As m converges to 0, and are expected to approach unity, however.
As expected from deterministic theory (Bürger and Akerman 2011; see also File S1, sect. 3) and invasion probabilities calculated above, the mean absorption time decreases as a function of the migration rate m (Figure S14). A noteworthy interaction exists between m and the effective population size Ne. For small m, the mean absorption time increases with Ne, whereas for large m, it decreases with Ne. Interestingly, the transition occurs at a value of m lower than the respective critical migration rate below which A1 can invade in the deterministic model (Figure S14). Hence, there exists a small range of intermediate values of m for which deterministic theory suggests that A1 will invade, but the stochastic model suggests that survival of A1 lasts longer in island populations of small rather than large effective size. Similar, but inverted, relations hold for the dependence of the mean absorption time on the selective advantage a of allele A1 and Ne (Figure S15).
For the parameter ranges we explored, the mean absorption time decreases with increasing continental frequency qc of B1. As for the invasion probabilities, competition against a fitter resident population has a negative effect on maintenance of the focal mutation A1. For a given recombination rate, the effect depends on the relative strength of migration and selection, though: increasing qc from 0 to 0.8 decreases the mean absorption time by a considerable amount only if m is low or a is large enough; otherwise, genetic drift dominates (Figure S16). This effect is more pronounced for weak than for strong recombination (Figure S17).
So far, we assumed that the initial frequency of A1 is small, i.e., p0 = 1/(2N), and that Ne = N. In many applications, Ne < N holds and hence 1/(2N) < 1/(2Ne). Approximations based on the assumption of p0 being small, i.e., on the order of 1/(2Ne) or smaller, then cause no problem. However, Ne > N may hold in certain models, e.g., with spatial structure (Whitlock and Barton 1997), and p0 = 1/(2N) may be much greater than 1/(2Ne). We therefore investigated the effect of violating the assumption of p0 ≤ 1/(2Ne). For this purpose, we fixed the initial frequency at p0 = 0.005 (e.g., a single copy of A1 in a population of actual size N = 100) and then assessed the relative error of our approximations for various Ne ≥ 100. As expected, the approximate mean absorption times based on the assumption of p0 small ( and ) deviate further from their exact conterparts ( and , respectively) as Ne increases from 100 to 104 (Figure S13, D and F). See also Table S6 and S7. For strong migration, the relative error tends to be negative, while it is positive for weak migration (blue vs. red boxes in Figure S13, D and F). The assumption of ρ ≫ max(α, β, μ) in M(p) does not lead to any further increase of the relative error, though (Figure S13E; Table S7). Moreover, violation of p0 ≤ 1/(2Ne) has almost no effect on the ratio of the two-locus to the one-locus absorption time, (compare Table S9 to Table S5).
Invasion-effective migration rate:
Comparison of the sojourn-time densities given in Equations 15 and 17 suggests that if μ in the one-locus model is replaced by μe = μ(μ − β + ρ)/ρ, one obtains the STD for the two-locus model. Hence, μe denotes the scaled migration rate in a one-locus model such that allele A1 has the same sojourn properties as it would have if it arose in linkage (decaying at rate ρ) to a background polymorphism maintained by selection against migration at rate μ. In other words, if the assumptions stated above hold, we may use single-locus migration–selection theory, with μ replaced by μe, to describe two-locus dynamics. Transforming from the diffusion to the natural scale, we therefore define an invasion-effective migration rate as
| (19) |
which, for small m, is approximately
| (20) |
(Figure S18A). Note that me and are nonnegative only if r ≥ b − m and r ≥ b, respectively. As we assumed quasi-linkage equilibrium in the derivation, these conditions do not impose any further restriction.
Petry (1983) previously derived an effective migration rate for a neutral site linked to a selected site. In our notation, it is given by
| (21) |
(see Bengtsson 1985 and Barton and Bengtsson 1986 for an extension of the concept). Petry (1983) obtained this approximation by comparing the moments of the stationary allele-frequency distribution for the two-locus model to those for the one-locus model. He assumed that selection and recombination are strong relative to migration and genetic drift. To first order in r−1, i.e., for loose linkage, Petry’s is equal to our in Equation 20. As we derived under the assumption of QLE, convergence of to is reassuring. Effective gene flow decreases with the strength of background selection b, but increases with the recombination rate r (Figure S18, B and C).
Long-term effect on linked neutral variation
Selection maintaining genetic differences across space impedes the homogenizing effect of gene flow at closely linked sites (Bengtsson 1985; Barton and Bengtsson 1986). This has consequences for the analysis of sequence or marker data, as patterns of neutral diversity may reveal the action of recent or past selection at nearby sites (Maynard Smith and Haigh 1974; Kaplan et al. 1989; Takahata 1990; Barton 1998). We investigated the impact of a two-locus polymorphism contributing to local adaptation on long-term patterns of linked genetic variation. For this purpose, we included a neutral locus C with alleles C1 and C2. Allele C1 segregates on the continent at a constant frequency nc (0 ≤ nc < 1), for example at drift–mutation equilibrium. This may require that the continental population is very large, such that extinction or fixation of C1 occurs over sufficiently long periods of time compared to the events of interest on the island. The neutral locus is on the same chromosome as A and B, to the left (C–A–B), in the middle (A–C–B), or to the right (A–B–C) of the two selected loci (without loss of generality, A is to the left of B). We denote the recombination rate between locus X and Y by rXY, where rXY = rYX, and assume that the recombination rate is additive. For example, if the configuration is A–C–B, we set rAB = rAC + rCB.
Unless linkage to one of the selected loci is complete, under deterministic dynamics, allele C1 will reach the equilibrium frequency on the island, independently of its initial frequency on the island. Recombination affects only the rate of approach to this equilibrium, not its value. We focus on the case where the continent is monomorphic at locus B (qc = 0). Selection for local adaptation acts on loci A and B, and migration–selection equilibrium will be reached at each of them (File S1, sect. 6). Gene flow from the continent will be effectively reduced in their neighborhood on the chromosome. Although the expected frequency of C1 remains nc throughout, drift will cause variation around this mean to an extent that depends on the position of C on the chromosome. It may take a long time for this drift–migration equilibrium to be established, but the resulting signal should be informative for inference.
To investigate the effect of selection at two linked loci, we employed the concept of an effective migration rate according to Bengtsson (1985), Barton and Bengtsson (1986), and Kobayashi et al. (2008). As derived in File S1, sect. 8, and File S8, for continuous time and weak migration, the effective migration rates for the three configurations are
| (22a) |
| (22b) |
| (22c) |
We note that has been previously derived (Bürger and Akerman 2011, Equation 4.30). From Equation 22, we define the effective migration rate experienced at a neutral site as
| (23) |
Equation 23 subsumes the effect on locus C of selection at loci A and B. It can be generalized to an arbitrary number of selected loci. Let Ai (i = 1,… ,I) and Bj (j = 1,… ,J) be the ith and jth locus to the left and right of the neutral locus, respectively. We find that the effective migration rate at the neutral locus is
| (24) |
where ai (bj) is the selection coefficient at locus Ai (Bj), and () the recombination rate between the neutral locus and Ai (Bj). Each of the terms in the round brackets in Equation 24 is reminiscent of Petry’s (1983) effective migration rate for a neutral linked site (Equation 21). For weak linkage, these terms are also similar to the invasion-effective migration rate experienced by a weakly beneficial mutation (Equation 20). This suggests that the effective migration rate experienced by a linked neutral site is approximately the same as that experienced by a linked weakly beneficial mutation, which corroborates the usefulness of Equation 24. In the following, we study different long-term properties of the one-locus drift-migration model by substituting effective for actual migration rates.
Mean absorption time:
Suppose that C1 is absent from the continent (nc = 0), but present on the island as a de novo mutation. Although any such mutant allele is doomed to extinction, recurrent mutation may lead to a permanent influx and, at mutation–migration equilibrium, to a certain level of neutral differentiation between the continent and the island. Here, we ignore recurrent mutation and focus on the fate of a mutant population descending from a single copy of C1. We ask how long it will survive on the island, given that a migration–selection polymorphism is maintained at equilibrium at both selected loci in the background (A, B). Standard diffusion theory predicts that the mean absorption (extinction) time of C1 is approximately (Ewens 2004, pp. 171–175). We replace the scaled actual migration rate μ by , with from Equation 23. This assumes that the initial frequency of C1 on the island is n0 = 1/(2N) and that Ne = N. For moderately strong migration , is of order log(2Ne), meaning that C1 will on average remain in the island population for a short time. However, if locus C is tightly linked to one of the selected loci, or if configuration A–C–B applies and A and B are sufficiently close, the mean absorption time of C1 is strongly elevated (Figure S19).
Stationary distribution of allele frequencies:
In contrast to above, assume that C1 is maintained at a constant frequency nc ∈ (0, 1) on the continent. Migrants may therefore carry both alleles, and genetic drift and migration will lead to a stationary distribution of allele frequencies given by
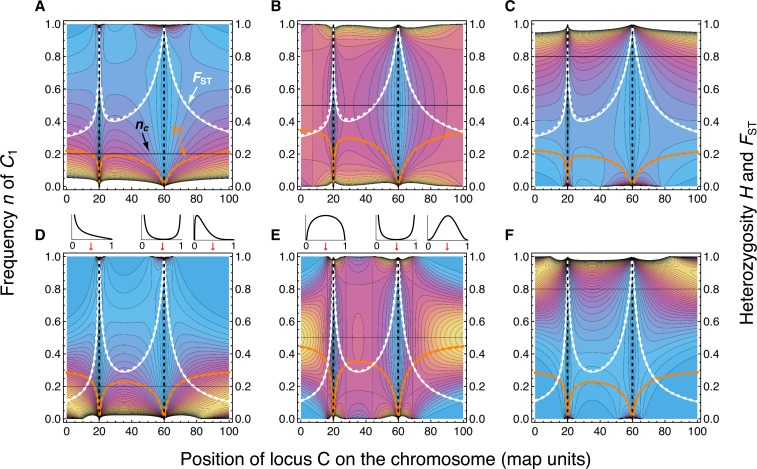
where Γ(x) is the Gamma function (Wright 1940, pp. 239–241). As above, we replace μ by to account for the effect of linked selection. The mean of the distribution φ(n) is nc, independently of μe, whereas the stationary variance is (Wright 1940). The expected heterozygosity is , and the divergence from the continental population is (see File S9 for details). Depending on the position of the neutral locus, φ(n) may change considerably in shape, for example, from L- to U- to bell-shaped (Figure 8). The pattern of φ(n), H and FST along the chromosome reveals the positions of the selected loci, and their rate of change per base pair contains information about the strength of selection if the actual migration rate is known.
Figure 8.
The effect of linked selection on neutral diversity and population divergence. Shown are top views of the stationary allele frequency distribution on the island for a neutral biallelic locus C linked to two selected sites at (locus A) 20 and (locus B) 60 map units from the left end of the chromosome. Density increases from light blue to yellow (high peaks were truncated for better resolution). Orange and white curves show the expected diversity (heterozygosity H) and population divergence (FST) as a function of the position of the neutral site. Solid curves use exact, numerically computed values of the effective migration rate and dashed curves use the approximations given in Equation 23. One map unit (cM) corresponds to r = 0.01 and the effective size of the island population is Ne = 100. The continental frequency nc of allele C1 is indicated by a horizontal black line and, from left to right, equal to 0.2, 0.5 and 0.8. (A–C) Relatively strong drift and weak migration compared to selection: α = 4, β = 80, μ = 2. (D–F) Relatively weak drift and migration on the same order of magnitude as selection at locus A: α = 40, β = 800, μ = 48. Note that H is sensitive to nc, whereas FST is not. On top of D and E, allele frequency distributions that result from taking vertical slices at positions indicated by red arrows are shown (15, 59 and 90 cM).
Rate of coalescence:
As a third application, we study the rate of coalescence for a sample of size two taken from the neutral locus C, assuming that migration–selection equilibrium has been reached a long time ago at the selected loci A and B. We restrict the analysis to the case of strong migration compared to genetic drift, for which results by Nagylaki (1980) (forward in time) and Notohara (1993) (backward in time) apply (see Wakeley 2009, for a detailed review). The strong-migration limit follows from a separation of timescales: going back in time, migration spreads the lineages on a faster timescale, whereas genetic drift causes lineages to coalesce on a slower one.
For a moment, let us assume that there are two demes of size N1 and N2, and denote the total number of diploids by . We define the relative deme size and let the backward migration rates m1 and m2 denote the fractions of individuals in deme 1 and 2 in the current generation that were in deme 2 and 1 in the previous generation, respectively. The strong-migration limit then requires that is large (Wakeley 2009). Importantly, the relative deme sizes ci are constant in the limit of . Under these assumptions, it can be shown that the rate of coalescence for a sample of two is independent of whether the two lineages were sampled from the same or different demes. The rate of coalescence is given by
| (25) |
(Wakeley 2009, p. 193). The coalescent-effective population size is defined as the actual total population size times the inverse of the rate of coalescence, (Sjödin et al. 2005).
In our context, we substitute from Equation 23 for m1 in G. To be consistent with the assumption of continent–island migration—under which we studied the migration–selection dynamics at A and B—we require N2 ≫ N1 and m2 ≪ m1. This way, the assumptions of N1m1 and N2m2 being large can still be fulfilled. However, note that m2 ≪ m1 does not automatically imply ; depending on the strength of selection and recombination, may become very small. Hence, in applying the theory outlined here, one should bear in mind that the approximation may be misleading if is small (for instance, if locus C is tightly linked to either A or B). The neutral coalescent rate G is strongly increased in the neighborhood of selected sites; accordingly, is increased (Figure S20). Reassuringly, this pattern parallels those for linked neutral diversity and divergence in Figure 8.
Discussion
We have provided a comprehensive analysis of the fate of a locally beneficial mutation that arises in linkage to a previously established migration–selection polymorphism. In particular, we obtained explicit approximations to the invasion probability. These reveal the functional dependence on the key parameters and substitute for time-consuming simulations. Further, we found accurate approximations to the mean extinction time, showing that a unilateral focus on invasion probabilities yields an incomplete understanding of the effects of migration and linkage. Finally, we derived the effective migration rate experienced by a neutral or weakly beneficial mutation that is linked to arbitrarily many migration–selection polymorphisms. This opens up a genome-wide perspective of local adaptation and establishes a link to inferential frameworks.
Insight from stochastic modeling
Previous theoretical studies accounting for genetic drift in the context of polygenic local adaptation with gene flow were mainly simulation based (Yeaman and Whitlock 2011; Feder et al. 2012; Flaxman et al. 2013) or did not model recombination explicitly (Lande 1984, 1985; Barton 1987; Rouhani and Barton 1987; Barton and Rouhani 1991; but see Barton and Bengtsson 1986). Here, we used stochastic processes to model genetic drift and to derive explicit expressions that provide an alternative to simulations. We distinguished between the stochastic effects due to initial rareness of a de novo mutation on the one hand and the long-term effect of finite population size on the other.
For a two-locus model with a steady influx of maladapted genes, we found an implicit condition for invasion of a single locally beneficial mutation linked to the background locus (Equation 9). This condition is valid for arbitrary fitnesses, i.e., any regime of dominance or epistasis. It also represents an extension to the case of a panmictic population in which the background polymorphism is maintained by overdominance, rather than migration–selection balance (Ewens 1967). Assuming additive fitnesses, we derived simple explicit conditions for invasion in terms of a critical migration or recombination rate (Equations 10 or 11, respectively). Whereas these results align with deterministic theory (Bürger and Akerman 2011), additional quantitative and qualitative insight emerged from studying invasion probabilities and extinction times. Specifically, invasion probabilities derived from a two-type branching process (Equations 3 and 12) capture the ambiguous role of recombination breaking up optimal haplotypes on the one hand and creating them on the other. Diffusion approximations to the sojourn and mean absorption time shed light on the long-term effect of finite population size. A comparison between the dependence of invasion probabilities and extinction times on migration and recombination rate revealed important differences (discussed further below). Deterministic theory fails to represent such aspects, and simulations provide only limited understanding of functional relationships.
Recently, Yeaman (2013) derived an ad hoc approximation of the invasion probability, using the so-called “splicing approach” (Yeaman and Otto 2011). There, the leading eigenvalue of the appropriate Jacobian (Bürger and Akerman 2011) is taken as a proxy for the selection coefficient and inserted into Kimura’s (1962) formula for the one-locus invasion probability in a panmictic population. Yeaman’s (2013) method provides a fairly accurate approximation to the invasion probability if A1 initially occurs on the beneficial background B1 (at least for tight linkage). However, it does not describe the invasion probability of an average mutation (Figure S22) and hence does not predict the existence of a nonzero optimal recombination rate. As a consequence, Yeaman’s (2013) conclusion that physically linked selection alone is of limited importance for the evolution of clustered architectures is likely conservative, because it is based on an approximation that inflates the effect of linked selection.
Nonzero optimal recombination rate
We have shown that the average invasion probability of a linked beneficial mutation can be maximized at a nonzero recombination rate (ropt > 0). Equation 13 provides a general condition for when this occurs. With additive fitnesses, the local advantage of the focal mutation must be above a critical value (Equation 14). Otherwise, the invasion probability is maximized at ropt = 0.
Existence of a nonzero optimal recombination rate in the absence of epistasis and dominance is noteworthy. For a panmictic population in which the polymorphism at the background locus is maintained by overdominance, Ewens (1967) has shown that the optimal recombination rate may be nonzero, but this requires epistasis. In the context of migration, the existence of ropt > 0 has been noted and discussed in a simulation study (Feder et al. 2012, Figure 5), but no analytical approximation or explanation that captures this feature has been available. In principle, ropt > 0 suggests that the genetic architecture of polygenic adaptation may evolve such as to optimize the recombination rates between loci harboring adaptive mutations. Testing this prediction requires modifier-of-recombination theory (e.g., Otto and Barton 1997; Martin et al. 2006; Roze and Barton 2006; Kermany and Lessard 2012). While we expect evolution toward the optimal recombination rate in a deterministic model (Lenormand and Otto 2000), it is important to determine if and under which conditions this occurs in a stochastic model and what the consequences for polygenic adaptation are.
For instance, in a model with two demes and a quantitative trait for which the fitness optima are different in the two demes, Yeaman and Whitlock (2011) have shown that mutations contributing to adaptive divergence in the presence of gene flow may cluster with respect to their position on the chromosome. Moreover, architectures with many weakly adaptive mutations tended to become replaced by architectures with fewer mutations of larger effect. Although our migration model is different, existence of a nonzero optimal recombination rate suggests that there might be a limit to the degree of clustering of locally adaptive mutations. It is worth recalling that our result of ropt > 0 applies to the average invading mutation (black curves in Figure 1 and Figure 2). For any particular mutation that arises on the beneficial background, r = 0 is (almost) always optimal (blue curves in Figure 1 and Figure 2; see Figure S3D for an exception).
Long-term dynamics of adaptive divergence
Finite population size on the island eventually leads to extinction of a locally beneficial mutation even after successful initial establishment. This is accounted for neither by deterministic nor branching-process theory. Employing a diffusion approximation, we have shown that linkage of the focal mutation to a migration–selection polymorphism can greatly increase the time to extinction and thus alter the long-term evolutionary dynamics. In such cases, the timescale of extinction may become similar to that on which mutations occur. This affects the rate at which an equilibrium between evolutionary forces is reached. We provided a rule of thumb for when the time spent by the focal allele at a certain frequency exceeds a given multiple of the respective time without linkage. Essentially, the product of the background selection coefficient times the migration rate must be larger than a multiple of the recombination rate (Equation 18).
The effect of linked selection can also be expressed in terms of an invasion-effective migration rate (Equations 19 and 20). Both our rule of thumb and the formula for the effective migration rate provide a means of quantifying the importance of linkage to selected genes in the context of local adaptation. In practice, however, their application requires accurate estimates of the recombination map, the selective advantage of the beneficial background allele, and the actual migration rate.
A nontrivial effect of gene flow
Our stochastic modeling allows for a more differentiated understanding of the role of gene flow in opposing adaptive divergence. Whereas deterministic theory specifies a critical migration rate beyond which a focal mutation of a given advantage cannot be established (Bürger and Akerman 2011; see also Figure S7 and Figure S8), the potential of invasion is far from uniform if migration is below this critical value (Figure S3, Figure S5, and Figure S14). For instance, we may define the relative advantage of linkage to a migration–selection polymorphism as the ratio of the quantity of interest with a given degree of linkage to that without linkage.
A comparison of the two quantities of interest in our case—invasion probability and mean extinction time—with respect to migration is instructive (Figure 7 and Figure S21). Starting from zero migration, the relative advantage of linkage in terms of the invasion probability initially increases with the migration rate very slowly, but then much faster as the migration rate approaches the critical value beyond which an unlinked focal mutation cannot invade (Figure S21A). Beyond this critical value, the relative advantage is infinite until migration is so high that even a fully linked mutation cannot be established. In contrast, we have shown that the relative advantage of linkage in terms of the mean extinction time is maximized at an intermediate migration rate (Figure 7).
In conclusion, for very weak migration, the benefit of being linked to a background polymorphism is almost negligible. For intermediate migration rates, the potential of invasion is elevated by linked selection; this is mainly due to a substantially increased mean extinction time of those still rather few mutations that successfully survive the initial phase of stochastic loss. This argument is based on the increase of the mean extinction time relative to unlinked selection. Because, for large populations, absolute extinction times become very large as the migration rate decreases (Figure 7, C and F), the biological relevance of this comparison may be confined to cases in which the mean extinction time of an unlinked mutation is not extremely high. For migration rates close to the critical migration rate, however, any relative advantage of linkage seems to arise via an increased invasion probability, not via an increased mean extinction time. This is because, in this case, the latter is close to that for no linkage (compare Figure S21A to Figure 7, A and B). A final statement about the relative importance of invasion probability vs. mean extinction time is not appropriate at this point. This would require extensive numerical work, along with a derivation of a diffusion approximation to the mean extinction time for tight linkage. However, for small populations, our results show that linked selection can increase the mean extinction time to an extent that is biologically relevant, while, at the same time, not affecting the invasion probability much. This suggests that invasion probabilities may not be a sufficient measure for the importance of physical linkage in adaptive divergence.
Standing variation at the background locus
We have extended some of our analyses to the case where the background locus is polymorphic on the continent and immigrants may therefore carry both the locally beneficial or deleterious allele. This represents a compromise between the extremes of adaptation from standing vs. de novo genetic variation. We have shown that the presence of the beneficial background allele on the continent, and hence among immigrants, leads to a lower invasion probability and a shorter extinction time for the focal de novo mutation. This effect is due to increased competition against a fitter resident population. While this result is of interest as such, it should not be abused to gauge the relative importance of standing vs. de novo variation in the context of local adaptation. For this purpose, invasion probabilities and extinction times of single mutations do matter, but are not sufficient metrics on their own. Factors such as the mutation rate, the mutational target size, and the distribution of selection coefficients must be taken into account (Hermisson and Pennings 2005).
Footprint of polygenic local adaptation
A number of previous studies have quantified the effect of divergent selection or genetic conflicts on linked neutral variation in discrete (Bengtsson 1985; Charlesworth et al. 1997) and continuous space (Barton 1979; Petry 1983). They all concluded that a single locus under selection leads to a pronounced reduction in effective gene flow only if selection is strong or if linkage to the neutral site is tight. Whereas Bengtsson (1985) found that additional, physically unlinked, loci under selection had no substantial effect on neutral differentiation, Feder and Nosil (2010) recently suggested that such loci may have an appreciable effect as long as they are not too numerous. When these authors added a large number of unlinked loci under selection, this resulted in a genome-wide reduction of the effective migration rate, such that the baseline level of neutral divergence was elevated and any effect of linkage to a single selected locus unlikely to be detected. However, for large numbers of selected loci, it is no longer justified to assume that all of them are physically unlinked. This was noted much earlier by Barton and Bengtsson (1986), who therefore considered a linear genome with an arbitrary number of selected loci linked to a focal neutral site. They showed that a large number of linked selected loci is needed to cause a strong reduction in effective migration rate. In such cases, the majority of other genes must be linked to some locus under selection.
The concept of an effective migration rate has played a key role in most of the studies mentioned above (see Barton and Bengtsson 1986, and Charlesworth et al. 1997 for a more comprehensive review). However, for models with more than one linked locus under selection, previous studies relied on numerical solutions or simulations to compute the effective migration rate. Recently, Bürger and Akerman (2011) derived an analytical approximation for a neutral site that is flanked by two selected loci. We have generalized their result to alternative genetic architectures and an arbitrary number of selected loci (Equation 24). From this, we predicted the long-term footprint of polygenic local adaptation in terms of the distribution of allele frequencies, population divergence, and coalescent rate at the neutral site. When considered as a function of the position of the neutral site on the chromosome, these quantities reveal patterns that can hopefully be used for inference about the selective process (Figure 8 and Figure S20).
We have considered only the case where migration–selection equilibrium has been reached at the selected loci. It would be interesting, although more demanding, to study the transient phase during which locally beneficial mutations (such as A1 in our case) rise in frequency from p0 = 1/(2N) to the (pseudo-)equilibrium frequency. We expect this to create a temporary footprint similar to that of a partial sweep (Pennings and Hermisson 2006a,b; Pritchard et al. 2010; Ralph and Coop 2010). Theoretical progress hinges on a description of the trajectory of the linked sweeping alleles, accounting in particular for the stochastic “lag phase” at the beginning. It will then be of interest to study recurrent local sweeps and extend previous theory for panmictic populations (Coop and Ralph 2012; Lessard and Kermany 2012) to include population structure, migration, and spatially heterogeneous selection. The hitchhiking effect of a beneficial mutation in a subdivided population has been described in previous studies (e.g., Slatkin and Wiehe 1998; Kim and Maruki 2011), but these did not account for additional linked loci under selection.
One limitation to our prediction of the coalescence rate at linked neutral sites is the assumption of strong migration relative to genetic drift (Nagylaki 1980; Notohara 1993). As the effective migration rate decays to zero if the neutral site is very closely linked to a selected site (Figure S18C), this assumption will be violated. Therefore, our predictions should be interpreted carefully when linkage is tight. Moreover, and even though this seems widely accepted, we are not aware of a rigorous proof showing that an effective migration rate can sufficiently well describe the effect of local selection on linked neutral genealogies (cf. Barton and Etheridge 2004).
Another limitation is that our prediction of linked neutral diversity and divergence (Figure 8) holds only for drift–migration equilibrium. For closely linked neutral sites, which experience very low rates of effective migration, it may take a long time for this equilibrium to be reached. By that time, other evolutionary processes such as background selection and mutation will have interfered with the dynamics at the focal site.
Further limitations and future extensions
We assumed no dominance and no epistasis. Both are known to affect the rate of adaptation and the maintenance of genetic variation (e.g., Charlesworth et al. 1987; Bank et al. 2012). Empirical results on dominance effects of beneficial mutations are ambiguous (Vicoso and Charlesworth 2006). Some studies showed no evidence for a deviation from additivity, whereas others suggested weak recessivity (reviewed in Orr 2010 and Presgraves 2008). Empirical evidence for epistasis comes from studies reporting genetic incompatibilities between hybridizing populations (Lowry et al. 2008; Presgraves 2010). In the classical Dobzhansky–Muller model (Bateson 1909; Dobzhansky 1936; Muller 1942), such incompatibilities may become expressed during secondary contact after allopatry, even if divergence is neutral. With gene flow, genetic incompatibilities can be maintained only if the involved alles are locally beneficial (Bank et al. 2012). Bank et al. (2012) derived respective conditions using deterministic theory. An extension to a stochastic model focusing on invasion probabilities and extinction times would be desirable.
Our model assumed one-way migration. While this is an important limiting case and applies to a number of natural systems (e.g., King and Lawson 1995), an extension to two-way migration is of interest, because natural populations or incipient species often exchange migrants mutually (e.g., Janssen and Mundy 2013; Nadeau et al. 2013). Such theory will allow for a direct comparison to recent simulation studies (Feder and Nosil 2010; Yeaman and Whitlock 2011; Feder et al. 2012; Flaxman et al. 2013; Yeaman 2013). It will also have a bearing on the evolution of suppressed recombination in sex chromosomes (e.g., Rice 1984, 1987; Fry 2010; Jordan and Charlesworth 2012; Charlesworth 2013). Deterministic theory suggests that linkage becomes less crucial for the maintenance of locally beneficial alleles the more symmetric gene flow is (Akerman and Bürger 2014).
When describing the distribution of fitness effects of successful beneficial mutations, we considered only a single mutation. Future studies should investigate a complete adaptive walk, allowing for mutations at multiple loci to interact via dominance, epistasis, and linkage. Moreover, it would then seem justified to relax the assumption of a constant fitness gradient, especially in the proximity of an optimum, and to account for the fact that the input DFE is not necessarily exponential (Martin and Lenormand 2008).
In our derivations of sojourn and mean absorption times, we assumed QLE. As expected, the approximations break down if recombination is weak (e.g., Figure 6). For tight linkage, when linked selection is most beneficial, an alternative diffusion process needs to be developed. However, to determine how weak physical linkage may be such that an invading mutation still has an advantage, an approximation that is accurate for moderate and loose linkage is required. Therefore, the assumption of QLE does not restrict the scope of our results that address the limits to the importance of linked selection.
Conclusion
This study advances our understanding of the effects of physical linkage and maladaptive gene flow on local adaptation. We derived explicit approximations to the invasion probability and extinction time of benefical de novo mutations that arise in linkage to an established migration–selection polymorphsim. In addition, we obtained an analytical formula for the effective migration rate experienced by a neutral or weakly beneficial site that is linked to an arbitrary number of selected loci. These approximations provide an efficient alternative to simulations (e.g., Feder and Nosil 2010; Feder et al. 2012). Our results strengthen the emerging view that physically linked selection (and hence so-called divergence hitchhiking) is biologically relevant only if linkage is tight or if selection at the background locus is strong (Petry 1983; Barton and Bengtsson 1986; Feder et al. 2012; Flaxman et al. 2013). When these conditions are met, however, the effect of linkage can be substantial. A definite statement about the importance of “divergence hitchhiking” vs. “genome hitchhiking” and complementary processes (cf. Yeaman 2013) seems premature, though; it will require further empirical and theoretical work. We suggest that future theoretical studies (i) obtain analogous approximations for bi- rather than unidirectional gene flow, (ii) account for epistasis and dominance, (iii) incorporate the distribution of fitness effects of beneficial mutations, and (iv) employ a stochastic modifier-of-recombination model to assess the importance of nonzero optimal recombination rates. Extensions of this kind will further enhance our understanding of polygenic local adaptation and its genetic footprint.
Supplementary Material
Acknowledgments
We thank Ada Akerman for sharing an unpublished manuscript and Mathematica code, Josef Hofbauer for help with finding the derivative of the invasion probability as a function of the recombination rate, the Biomathematics Group at the University of Vienna for stimulating discussions, and Samuel Flaxman and Samuel Yeaman for useful comments on the manuscript. S.A. and R.B. acknowledge financial support by the Austrian Science Fund (FWF), projects P21305 and P25188. The computational results presented have been achieved in parts using the Vienna Scientific Cluster (VSC).
Footnotes
Communicating editor: L. M. Wahl
Literature cited
- Akerman A., Bürger R., 2014. The consequences of gene flow for local adaptation and differentiation: a two-locus two-deme model. J. Math. Biol. 68: 1135–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank C., Bürger R., Hermisson J., 2012. The limits to parapatric speciation: Dobzhansky–Muller incompatibilities in a continent–island model. Genetics 191: 845–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett R. D. H., M’Gonigle L. K., Otto S. P., 2006. The distribution of beneficial mutant effects under strong selection. Genetics 174: 2071–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N., 1979. Gene flow past a cline. Heredity 43: 333–339. [Google Scholar]
- Barton N., Bengtsson B. O., 1986. The barrier to genetic exchange between hybridising populations. Heredity 57: 357–376. [DOI] [PubMed] [Google Scholar]
- Barton N. H., 1983. Multilocus clines. Evolution 37: 454–471. [DOI] [PubMed] [Google Scholar]
- Barton N. H., 1987. The probability of establishment of an advantageous mutant in a subdivided population. Genet. Res. 50: 35–40. [DOI] [PubMed] [Google Scholar]
- Barton N. H., 1995. Linkage and the limits to natural selection. Genetics 140: 821–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. H., 1998. The effect of hitchhiking on neutral genealogies. Genet. Res. 72: 123–133. [Google Scholar]
- Barton N. H., 2010. Genetic linkage and natural selection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 2559–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. H., Etheridge A. M., 2004. The effect of selection on genealogies. Genetics 166: 1115–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. H., Rouhani S., 1991. The probability of fixation of a new karyotype in a continuous population. Evolution 45: 499–517. [DOI] [PubMed] [Google Scholar]
- Bateson W., 1909. Heredity and variation in modern lights, pp. 85–101 in Darwin and Modern Science, edited by Seward A. C. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Bengtsson B. O., 1985. The flow of genes through a genetic barrier, pp. 31–42 in Evolution: Essays in Honour of John Maynard Smith, Vol. 1 Chap. 3, edited by Greenwood P. J., Harvey P., Slatkin M. Cambridge University Press, New York. [Google Scholar]
- Bürger R., Akerman A., 2011. The effects of linkage and gene flow on local adaptation: a two-locus continent–island model. Theor. Popul. Biol. 80: 272–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Coyne J. A., Barton N. H., 1987. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 130: 113–146. [Google Scholar]
- Charlesworth B., Nordborg M., Charlesworth D., 1997. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet. Res. 70: 155–174. [DOI] [PubMed] [Google Scholar]
- Charlesworth D., 2013. Plant sex chromosome evolution. J. Exp. Bot. 64: 405–420. [DOI] [PubMed] [Google Scholar]
- Coop G., Ralph P., 2012. Patterns of neutral diversity under general models of selective sweeps. Genetics 192: 205–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowperthwaite M. C., Bull J. J., Meyers L. A., 2005. Distributions of beneficial fitness effects in RNA. Genetics 170: 1449–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., 1936. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21: 113–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethier S. N., Nagylaki T., 1980. Diffusion approximations of Markov-chains with 2 time scales with applications to population-genetics. Adv. Appl. Probab. 12: 14–49. [Google Scholar]
- Ethier S. N., Nagylaki T., 1988. Diffusion approximations of Markov-chains with 2 time scales with applications to population-genetics 2. Adv. Appl. Probab. 20: 525–545. [Google Scholar]
- Ethier S. N., Nagylaki T., 1989. Diffusion approximations of the two-locus Wright–Fisher model. J. Math. Biol. 27: 17–28. [DOI] [PubMed] [Google Scholar]
- Ewens W. J., 1967. The probability of fixation of a mutant: the two-locus case. Evolution 21: 532–540. [DOI] [PubMed] [Google Scholar]
- Ewens W. J., 1968. Some applications of multiple-type branching processes in population genetics. J. R. Stat. Soc. Ser. B. Stat. Methodol. 30: 164–175. [Google Scholar]
- Ewens W. J., 2004. Mathematical Population Genetics, Ed. 2 Springer-Verlag, New York. [Google Scholar]
- Eyre-Walker A., Keightley P. D., 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8: 610–618. [DOI] [PubMed] [Google Scholar]
- Feder J. L., Nosil P., 2010. The efficacy of divergence hitchhiking in generating genomic islands during ecological speciation. Evolution 64: 1729–1747. [DOI] [PubMed] [Google Scholar]
- Feder J. L., Gejji R., Yeaman S., Nosil P., 2012. Establishment of new mutations under divergence and genome hitchhiking. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection. Oxford University Press, New York. [Google Scholar]
- Flaxman S. M., Feder J. L., Nosil P., 2013. Genetic hitchhiking and the dynamic buildup of genomic divergence during speciation with gene flow. Evolution 67: 2577–2591. [DOI] [PubMed] [Google Scholar]
- Fry J. D., 2010. The genomic location of sexually antagonistic variation: some cautionary comments. Evolution 64: 1510–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J. H., 1983. A simple stochastic gene substitution model. Theor. Popul. Biol. 23: 202–215. [DOI] [PubMed] [Google Scholar]
- Gillespie J. H., 1984. Molecular evolution over the mutational landscape. Evolution 38: 1116–1129. [DOI] [PubMed] [Google Scholar]
- Guerrero R. F., Rousset F., Kirkpatrick M., 2012. Coalescent patterns for chromosomal inversions in divergent populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haccou, P., J. Peter, and V. A. Vatutin, 2005 Branching Processes: Variation, Growth, and Extinction of Populations, Vol. 5, Cambridge Studies in Adaptive Dynamics. Cambridge University Press, New York. [Google Scholar]
- Haldane J. B. S., 1927. A mathematical theory of natural and artificial selection. v. Selection and mutation. Math. Proc. Camb. Philos. Soc. 23: 838–844. [Google Scholar]
- Haldane J. B. S., 1930. A mathematical theory of natural and artificial selection. VI. Isolation. Math. Proc. Camb. Philos. Soc. 26: 220–230. [Google Scholar]
- Harris, T. E., 1963 The Theory of Branching Processes, Vol. 119, Die Grundlehren der Mathematischen Wissenschaften, Ed. 1. Springer-Verlag, Berlin. [Google Scholar]
- Hermisson J., Pennings P. S., 2005. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics 169: 2335–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., Robertson A., 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269–294. [PubMed] [Google Scholar]
- Janssen K., Mundy N. I., 2013. Molecular population genetics of the melanic plumage polymorphism in Arctic skuas (Stercorarius parasiticus): evidence for divergent selection on plumage colour. Mol. Ecol. 22: 4634–4643. [DOI] [PubMed] [Google Scholar]
- Jordan C. Y., Charlesworth D., 2012. The potential for sexually antagonistic polymorphism in different genome regions. Evolution 66: 505–516. [DOI] [PubMed] [Google Scholar]
- Kaplan N. L., Hudson R. R., Langley C. H., 1989. The “hitchhiking effect” revisited. Genetics 123: 887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., McGregor J., 1968. Rates and probabilities of fixation for two locus random mating finite populations without selection. Genetics 58: 141–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Taylor H. M., 1981. A Second Course in Stochastic Processes, Vol. 2 Academic Press, San Diego, CA. [Google Scholar]
- Karlsen B. O., Klingan K., Emblem Å., Jørgensen T. E., Jueterbock A., et al. , 2013. Genomic divergence between the migratory and stationary ecotypes of Atlantic cod. Mol. Ecol. 22: 5098–5111. [DOI] [PubMed] [Google Scholar]
- Kermany A. R., Lessard S., 2012. Effect of epistasis and linkage on fixation probability in three-locus models: an ancestral recombination-selection graph approach. Theor. Popul. Biol. 82: 131–145. [DOI] [PubMed] [Google Scholar]
- Kim Y., Maruki T., 2011. Hitchhiking effect of a beneficial mutation spreading in a subdivided population. Genetics 189: 213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1962. On the probability of fixation of mutant genes in a population. Genetics 47: 713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1965. Attainment of quasi linkage equilibrium when gene frequencies are changing by natural selection. Genetics 52: 875–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1979. Model of effectively neutral mutations in which selective constraint is incorporated. Proc. Natl. Acad. Sci. USA 76: 3440–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R. B., Lawson R., 1995. Color-pattern variation in Lake Erie water snakes: the role of gene flow. Evolution 49: 885–896. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M., Johnson T., Barton N., 2002. General models of multilocus evolution. Genetics 161: 1727–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Hammerstein P., Telschow A., 2008. The neutral effective migration rate in a mainland–island context. Theor. Popul. Biol. 74: 84–92. [DOI] [PubMed] [Google Scholar]
- Kojima K., Schaffer H. E., 1967. Survival process of linked mutant genes. Evolution 21: 518–531. [DOI] [PubMed] [Google Scholar]
- Lande R., 1984. The expected fixation rate of chromosomal inversions. Evolution 38: 743–752. [DOI] [PubMed] [Google Scholar]
- Lande R., 1985. The fixation of chromosomal rearrangements in a subdivided population with local extinction and colonization. Heredity 54: 323–332. [DOI] [PubMed] [Google Scholar]
- Lenormand T., Otto S. P., 2000. The evolution of recombination in a heterogeneous environment. Genetics 156: 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard S., Kermany A. R., 2012. Fixation probability in a two-locus model by the ancestral recombination–selection graph. Genetics 190: 691–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry D. B., Modliszewski J. L., Wright K. M., Wu C. A., Willis J. H., 2008. The strength and genetic basis of reproductive isolating barriers in flowering plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363: 3009–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Lenormand T., 2008. The distribution of beneficial and fixed mutation fitness effects close to an optimum. Genetics 179: 907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Otto S. P., Lenormand T., 2006. Selection for recombination in structured populations. Genetics 172: 593–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J., Haigh J., 1974. Hitch-hiking effect of a favorable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- McGaugh S. E., Noor M. A. F., 2012. Genomic impacts of chromosomal inversions in parapatric drosophila species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1932. Some genetic aspects of sex. Am. Nat. 66: 118–138. [Google Scholar]
- Muller H. J., 1942. Isolating mechanisms, evolution and temperature. Biol. Symp. 6: 71–125. [Google Scholar]
- Nadeau N. J., Whibley A., Jones R. T., Davey J. W., Dasmahapatra K. K., et al. , 2012. Genomic islands of divergence in hybridizing Heliconius butterflies identified by large-scale targeted sequencing. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau N. J., Martin S. H., Kozak K. M., Salazar C., Dasmahapatra K. K., et al. , 2013. Genome-wide patterns of divergence and gene flow across a butterfly radiation. Mol. Ecol. 22: 814–826. [DOI] [PubMed] [Google Scholar]
- Nagylaki T., 1980. The strong-migration limit in geographically structured populations. J. Math. Biol. 9: 101–114. [DOI] [PubMed] [Google Scholar]
- Nagylaki T., Hofbauer J., Brunovský P., 1999. Convergence of multilocus systems under weak epistasis or weak selection. J. Math. Biol. 38: 103–133. [DOI] [PubMed] [Google Scholar]
- Nosil P., Feder J. L., 2012. Genomic divergence during speciation: causes and consequences. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notohara M., 1993. The strong-migration limit for the genealogical process in geographically structured populations. J. Math. Biol. 31: 115–122. [DOI] [PubMed] [Google Scholar]
- Orr H. A., 1998. The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution 52: 935–949. [DOI] [PubMed] [Google Scholar]
- Orr H. A., 2002. The population genetics of adaptation: the adaptation of DNA sequences. Evolution 56: 1317–1330. [DOI] [PubMed] [Google Scholar]
- Orr H. A., 2003. The distribution of fitness effects among beneficial mutations. Genetics 163: 1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 2010. The population genetics of beneficial mutations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S. P., Barton N. H., 1997. The evolution of recombination: removing the limits to natural selection. Genetics 147: 879–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings P. S., Hermisson J., 2006a Soft sweeps II: molecular population genetics of adaptation from recurrent mutation or migration. Mol. Biol. Evol. 23: 1076–1084. [DOI] [PubMed] [Google Scholar]
- Pennings P. S., Hermisson J., 2006b Soft sweeps III: the signature of positive selection from recurrent mutation. PLoS Genet. 2: e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry D., 1983. The effect on neutral gene flow of selection at a linked locus. Theor. Popul. Biol. 23: 300–313. [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., 2008. Sex chromosomes and speciation in Drosophila. Trends Genet. 24: 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves D. C., 2010. The molecular evolutionary basis of species formation. Nat. Rev. Genet. 11: 175–180. [DOI] [PubMed] [Google Scholar]
- Pritchard J. K., Pickrell J. K., Coop G., 2010. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20: R208–R215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph P., Coop G., 2010. Parallel adaptation: One or many waves of advance of an advantageous allele? Genetics 186: 647–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice W. R., 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution 38: 735–742. [DOI] [PubMed] [Google Scholar]
- Rice W. R., 1987. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution 41: 911–914. [DOI] [PubMed] [Google Scholar]
- Rouhani S., Barton N., 1987. Speciation and the “shifting balance” in a continuous population. Theor. Popul. Biol. 31: 465–492. [Google Scholar]
- Roze D., Barton N. H., 2006. The Hill–Robertson effect and the evolution of recombination. Genetics 173: 1793–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjödin P., Kaj I., Krone S., Lascoux M., Nordborg M., 2005. On the meaning and existence of an effective population size. Genetics 169: 1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M., 1975. Gene flow and selection in a two-locus system. Genetics 81: 787–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M., Wiehe T., 1998. Genetic hitch-hiking in a subdivided population. Genet. Res. 71: 155–160. [DOI] [PubMed] [Google Scholar]
- Strasburg J. L., Sherman N. A., Wright K. M., Moyle L. C., Willis J. H., et al. , 2012. What can patterns of differentiation across plant genomes tell us about adaptation and speciation? Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 364–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N., 1990. A simple genealogical structure of strongly balanced allelic lines and transspecies evolution of polymorphism. Proc. Natl. Acad. Sci. USA 87: 2419–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Hahn M. W., Nuzhdin S. V., 2005. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 3: e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via S., 2012. Divergence hitchhiking and the spread of genomic isolation during ecological speciation-with-gene-flow. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B., Charlesworth B., 2006. Evolution on the X chromosome: unusual patterns and processes. Nat. Rev. Genet. 7: 645–653. [DOI] [PubMed] [Google Scholar]
- Wakeley J., 2009. Coalescent Theory: An Introduction, Roberts & Company, Greenwood Village, CO. [Google Scholar]
- Whitlock M. C., Barton N. H., 1997. The effective size of a subdivided population. Genetics 146: 427–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1940. Breeding structure of populations in relation to speciation. Am. Nat. 74: 232–248. [Google Scholar]
- Yeaman S., 2013. Genomic rearrangements and the evolution of clusters of locally adaptive loci. Proc. Natl. Acad. Sci. USA 110: 1743–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman S., Otto S. P., 2011. Establishment and maintenance of adaptive genetic divergence under migration, selection, and drift. Evolution 65: 2123–2129. [DOI] [PubMed] [Google Scholar]
- Yeaman S., Whitlock M. C., 2011. The genetic architecture of adaptation under migration–selection balance. Evolution 65: 1897–1911. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.