This phase I trial with an alternative and biologically informed dose-escalation design demonstrated targeted therapy resensitization with SHP2 inhibitor combinations in diverse oncogene-driven cancers.
Abstract
Rationally targeted therapies have transformed cancer treatment, but many patients develop resistance through bypass signaling pathway activation. PF-07284892 (ARRY-558) is an allosteric SHP2 inhibitor designed to overcome bypass-signaling-mediated resistance when combined with inhibitors of various oncogenic drivers. Activity in this setting was confirmed in diverse tumor models. Patients with ALK fusion–positive lung cancer, BRAFV600E-mutant colorectal cancer, KRASG12D-mutant ovarian cancer, and ROS1 fusion–positive pancreatic cancer who previously developed targeted therapy resistance were treated with PF-07284892 on the first dose level of a first-in-human clinical trial. After progression on PF-07284892 monotherapy, a novel study design allowed the addition of oncogene-directed targeted therapy that had previously failed. Combination therapy led to rapid tumor and circulating tumor DNA (ctDNA) responses and extended the duration of overall clinical benefit.
Significance:
PF-07284892–targeted therapy combinations overcame bypass-signaling-mediated resistance in a clinical setting in which neither component was active on its own. This provides proof of concept of the utility of SHP2 inhibitors in overcoming resistance to diverse targeted therapies and provides a paradigm for accelerated testing of novel drug combinations early in clinical development.
See related commentary by Hernando-Calvo and Garralda, p. 1762.
This article is highlighted in the In This Issue feature, p. 1749
INTRODUCTION
Although single-agent targeted therapies have transformed cancer therapy for patients with oncogene-driven tumors, primary and secondary resistance can limit efficacy. Given that rationally selected combinations of targeted therapy are poised to overcome resistance, an increase in the number of combination therapy trials is an inevitable next step in precision medicine evolution. Unfortunately, traditional first-in-human (FIH) phase I clinical trials are inefficient at investigating drug combinations. FIH dose escalations are typically designed to treat patients with a single drug at a time. Drugs are only later combined after monotherapy exploration, a process that delays study completion. Furthermore, the utility of this strategy is questionable when monotherapy is unlikely to be effective. Designing combination trials that circumvent these limitations is a challenge.
This problem applies to two important circumstances. First, combination therapy is required for oncogene-driven cancers that acquire off-target resistance to targeted therapy. For example, reactivation of mitogen-associated protein kinase (MAPK) pathway signaling may be induced shortly after starting targeted therapy treatment due to bypass receptor tyrosine kinase (RTK) activation (e.g., EGFR, KIT; refs. 1–3) or after initial tumor response due to acquired genetic alterations (e.g., RAS mutations, MET amplification; refs. 4–6). Second, targeted therapy combinations may be required for prevalent oncogenic drivers deemed intractable to single-agent targeted therapy, like select de novo KRAS and PIK3CA mutations (7–10).
Both situations may be addressed by combination targeted therapies that include an inhibitor of SHP2, a protein tyrosine phosphatase. SHP2 reinforces activation of the MAPK and potentially other pathways through multiple mechanisms: dephosphorylation of inhibitory tyrosine phosphorylation on positive regulators (e.g., RTKs, GAB1, and RAS; refs. 11–13); dephosphorylation of activating tyrosine phosphorylation on negative regulators (e.g., Sprouty; ref. 14); direct recruitment of Grb and SOS to RTKs (15, 16); and stimulation of RAS GDP–GTP exchange (17). Bypass resistance can be overcome in preclinical models by combining a SHP2 inhibitor with inhibitors of diverse oncogenic drivers (8, 17, 18). A SHP2 inhibitor is unlikely to work on its own, however. FIH trials of the first SHP2 inhibitors to enter the clinic (e.g., RMC-4630 and TNO155) clearly showed limited monotherapy activity and were not optimally designed to explore rational combinations (19, 20).
In this study, we describe a bench-to-bedside approach to the characterization of combinations with a novel SHP2 inhibitor (PF-07284892/ARRY-558) that distinguishes itself from prior drug development programs by leveraging early introduction of combination therapy in a phase I trial. As opposed to treating patients with a SHP2 inhibitor alone in dose escalation, each patient with an oncogene-driven cancer was permitted the addition of matched targeted therapy after a lead-in period to characterize monotherapy safety, dose-limiting toxicity (DLT), pharmacokinetics (PK), pharmacodynamics (PD), and efficacy prior to introduction of one of three rational combinations. This strategy resulted in the rapid rescue of progressive disease on single-agent PF-07284892 and the establishment of proof-of-concept combination therapy activity.
RESULTS
PF-07284892 Design and Preclinical Pharmacology and Monotherapy Activity
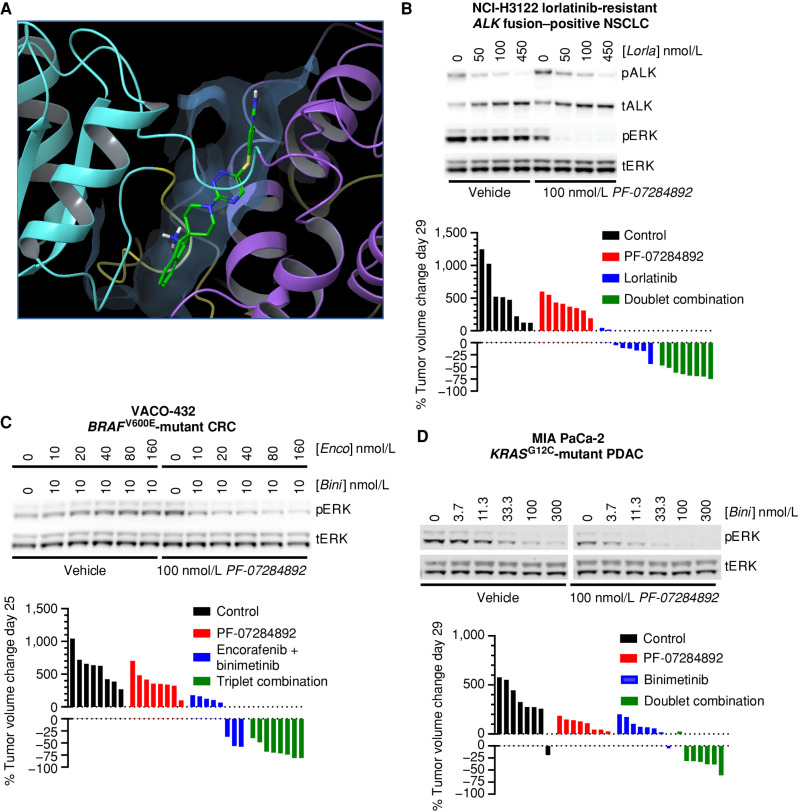
PF-07284892 (ARRY-558), an allosteric SHP2 inhibitor, was discovered by Array BioPharma (Methods). The X-ray crystal structure of PF-07284892 bound to SHP2 shows that PF-07284892 is encased in a tunnel at an interface between the N- and C-proximal SH2 and phosphatase domains (Fig. 1A; Supplementary Table S1). Whereas the overall binding mode is like other SHP2 inhibitors, one difference is the fused dihydroindene phenyl of PF-07284892, which fits deeper into the tunnel and forms enhanced contacts with several residues (Supplementary Fig. S1A and S1B).
Figure 1.
PF-07284892 promotes antitumor efficacy in multiple oncogene-addicted models with up-front or acquired resistance to targeted therapies. A, X-ray crystal structure of PF-07284892–bound SHP2. N-SH2, C-SH2, and PTP domains are yellow, cyan, and violet; inhibitor is green/blue. See Supplementary Table S1 for data collection and refinement statistics. B–D, Top, the indicated human cancer cell lines were treated in vitro with each agent at the indicated concentrations for 4 (H3122 lorR-06), 18 (VACO-432), or 24 (MIA PaCa-2) hours followed by preparation of cell lysates and analysis of the indicated protein by immunoblot. Quantitation of each band is shown in Supplementary Fig. S3. Bottom, immunodeficient mice (8 per group) were xenografted subcutaneously with the same human tumor cells used for in vitro signaling analysis. When tumors reached ∼200 mm3, animals were treated orally with vehicle, PF-07284892 30 mg/kg q.o.d., lorlatinib 3 mg/kg qd, encorafenib 20 mg/kg qd + binimetinib 3.5 mg/kg b.i.d., binimetinib 3.5 mg/kg b.i.d., or with the indicated combinations (at the monotherapy doses). Tumor sizes on days 25 to 29 were normalized to day 1 prior to treatment. b.i.d., twice daily; Bini, binimetinib; C, carboxy-proximal; CRC, colorectal cancer; Enco, encorafenib; Lorla, lorlatinib; N, amino-proximal; NSCLC, non–small cell lung cancer; p, phosphorylated; PDAC, pancreatic ductal adenocarcinoma; PTP, phosphatase; qd, daily; q.o.d., every other day; t, total.
In cell-free systems, PF-07284892 inhibited SHP2 biochemical activity with an IC50 of 21 nmol/L (±5 nmol/L, n = 20) and demonstrated >∼1,000-fold selectivity for SHP2 over 21 other phosphatases, including the closely related SHP1 protein (Supplementary Table S2). In cell-based assays, PF-07284892 potently inhibited phosphorylated ERK (pERK) with low nanomolar IC50 values (Supplementary Table S3). PF-07284892 demonstrated favorable PK properties in animals (Supplementary Table S4). The potential for improved brain penetration compared with other SHP2 inhibitors was also observed (Supplementary Table S5). Intermittent dosing (e.g., every 2–3 days) maintained efficacy and was better tolerated (as determined by peripheral blood counts and hematocrit) than continuous daily dosing while maintaining efficacy (Supplementary Fig. S2A–S2D).
Combination Therapy Preclinical Activity
To determine the effect of PF-07284892 treatment in combination with appropriate targeted therapies, three human tumor cell lines were investigated: NCI-H3122 lorR-06, an EML4–ALK fusion–positive lung cancer model with acquired resistance to next-generation ALK inhibitors (see Methods); VACO-432, a BRAFV600E-mutant colorectal cancer model; and MIA PaCa-2, a KRASG12C-mutant pancreatic cancer model. Preclinically, SHP2 had been shown to contribute to primary or secondary targeted therapy resistance for each of the three driver-tumor contexts (8, 17, 18, 21).
The following targeted therapies were chosen to combine with PF-07284892: lorlatinib (for ALK fusions), encorafenib + cetuximab or binimetinib (for BRAFV600E mutants), and binimetinib (for KRASG12C mutants). These drugs were chosen based on clinical availability (e.g., we did not have access to a KRASG12C inhibitor for clinical studies) and thus the potential to translate findings to the clinic. Agents were used at concentrations (in vitro) and doses (in vivo) informed by nonclinical studies (PF-07284892) or that approximated human exposures at clinically approved doses (all other combination agents).
For each cell line, treatment in vitro with PF-07284892 in combination with oncogene-matched targeted therapy led to maximum inhibition of pERK levels (60% to >90%) compared with PF-07284892 or each targeted therapy regimen alone (Fig. 1B–D, top; quantitation in Supplementary Fig. S3A–S3C, respectively).
Consistent with pERK suppression, oral treatment of mouse xenografts of each cell line with oncogene-matched targeted therapy in combination with PF-07284892 resulted in maximal tumor regression compared with either component alone (Fig. 1B–D, bottom; additional in vivo models in Supplementary Fig. S4A–S4C). PF-07284892 at the same single oral dose (30 mg/kg) used for efficacy studies caused significant suppression of pERK in MIA PaCa-2 tumors (Supplementary Fig. S4D).
Proof-of-Concept Clinical Activity
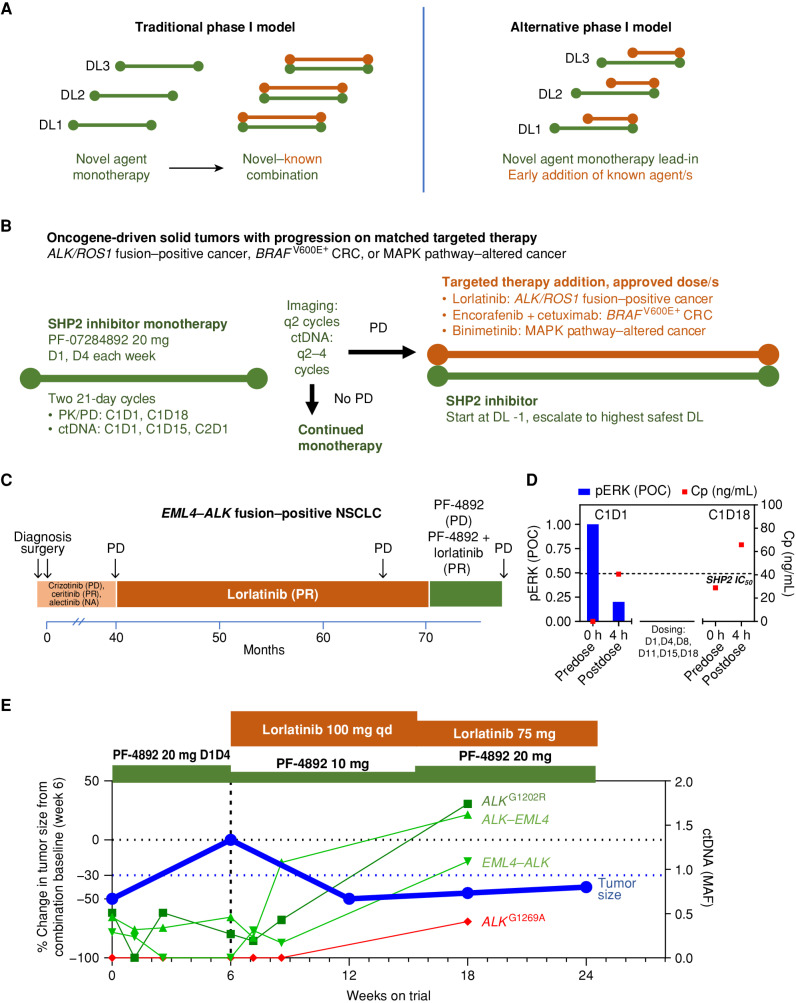
We designed an FIH phase I clinical trial of PF-07284892 in patients whose solid tumors demonstrated acquired or intrinsic resistance to oncogene-matched targeted therapy (NCT04800822). Compared with traditional phase I designs (Fig. 2A), a novel dose-escalation study design was used that allowed the addition of (the same or similar) previously used oncogene-directed targeted therapy after a lead-in period of SHP2 inhibitor monotherapy (Fig. 2B; Supplementary Fig. S5). Twice weekly dosing of PF-07284892 was planned from the start of the trial based on nonclinical studies demonstrating improved therapeutic index (TI; Supplementary Fig. S2A–S2D). The sequential approach of giving monotherapy followed by combination therapy enabled assessment of safety, PK, preliminary efficacy, and contribution of effect for both PF-07284892 monotherapy and the added targeted therapies. We highlight below the first four patients enrolled to the first dose level and transitioned from monotherapy to combination. The disposition of all eight patients treated at this dose level and adverse events (AE) experienced by these patients are summarized in Supplementary Tables S6 and S7, respectively.
Figure 2.
Proof-of-concept clinical activity in an ALK fusion–positive NSCLC patient with resistance to multiple ALK inhibitors. A, Overview of traditional vs. alternative phase I combination trial design. Left, traditional phase I first-in-human trials require a new anticancer agent to be investigated as monotherapy, with the maximum tolerated dose/recommended dose for expansion identified, prior to allowing a combination with a second anticancer drug. If the investigational agent is ineffective on its own, treated patients do not have the opportunity to benefit from a potentially efficacious combination. Right, an alternative design allows patients to receive treatment with a potentially effective combination after an initial period of treatment with the study drug as monotherapy. B, Implementation of early combination testing strategy with the investigational SHP2 inhibitor PF-07284892. Prior to trial enrollment, eligible patients had experienced PD with appropriate targeted therapy. Patients begin treatment with PF-07284892 monotherapy on study. Early combination with appropriate targeted therapy (lorlatinib, encorafenib + cetuximab, or binimetinib, each at the approved dose) may be initiated after a minimum of 6 weeks of PF-07284892 monotherapy, in the absence of ongoing grade ≥3 toxicity or DLT, and after PD (symptoms of PD without tumor growth ≥20% was allowed). At the start of the combination, the PF-07284892 dose must be lowered if the monotherapy dose level the patient was enrolled to has not yet been cleared from a safety perspective. The dose may subsequently be escalated to the highest monotherapy dose that has been cleared. C, The patient's previous systemic therapies included four approved ALK inhibitors. Parentheses show the best overall response to each treatment. D, Peripheral blood was isolated from the patient prior to and 4 hours after dosing with PF-07284892 on C1D1 and C1D18, and levels of PF-07284892 in plasma and of pERK in ex vivo CSF1-stimulated peripheral blood monocytes were analyzed (C1D18 samples for pERK were not available). The last dose of PF-07284892 prior to the C1D18 predose sampling was C1D15. The horizontal dashed line indicates PF-07284892 concentration required to inhibit 50% of pERK in cells in vitro. E, Changes in the sum of the longest tumor diameters of target lesions (blue, normalized to the start of combination) and in EML4–ALK, ALK–EML4, ALKG1202R (shades of green), and ALKG1269A (red) in ctDNA. C, cycle; Cp, plasma concentration; CRC, colorectal cancer; ctDNA, circulating tumor DNA; D, day; DL, dose level; MAF, mean allele frequency; NA, not available; PD, progressive disease; pERK, phosphorylated ERK; PF-4892, PF-07284892; PK/PD, pharmacokinetics/pharmacodynamics; POC, percent of control; PR, partial response; qd, every day.
EML4–ALK Fusion–Positive Lung Cancer.
A patient with EML4–ALK fusion–positive non–small cell lung cancer (NSCLC) was previously treated with the ALK inhibitors crizotinib, ceritinib, alectinib, and lorlatinib (Fig. 2C). Two weeks after discontinuing lorlatinib for progression, the patient started treatment with PF-07284892 monotherapy [20 mg orally twice weekly (b.i.w.) on days 1 and 4] at dose level 1. PK analysis indicated sustained plasma levels of PF-07284892 despite intermittent dosing [29 ng/mL predose cycle 1 day 18 (C1D18, 3 days after prior dose) vs. 41 ng/mL 4 hours after C1D1 dose (Fig. 2D, red squares); complete PK curves shown in Supplementary Fig. S6]. PD analysis indicated an 80% decrease in the levels of pERK in peripheral blood monocytes 4 hours after dosing on C1D1 (Fig. 2D, blue bars). Circulating tumor DNA (ctDNA) showed clear decrease in EML4–ALK but less in ALK–EML4 or ALKG1202R (Fig. 2E).
Despite this, CT imaging after 6 weeks of PF-07284892 monotherapy demonstrated progressive disease (+50%; Fig. 2E). In a “typical” phase I clinical trial, this patient would have discontinued treatment. Instead, the patient continued PF-07284892 (at an initial lower dose of 10 mg b.i.w.; Fig. 2E and Methods), and lorlatinib was added [100 mg daily (qd), the dose the patient previously progressed on].
A partial response (PR; −50%) was achieved after 6 weeks of combination therapy (study week 12); precombination therapy imaging was used as a baseline for this comparison (Fig. 2E). ALK–EML4 and ALKG1202R in ctDNA showed early decreases after 1 week of combination and then all three ctDNA species increased after 3 weeks of combination (study week 9; Fig. 2E). After 9 weeks of combination (study week 15), the PF-07284892 dose was increased to 20 mg b.i.w., whereas the dose of lorlatinib was reduced to 75 mg qd (for grade 2 depression related to lorlatinib); no AEs required PF-07284892 dose modification, and there were no DLTs. The PR was confirmed after 12 weeks of combination therapy (study week 18), with a slight increase in target lesions, further increase in ALK fusions and ALKG1202R, and new appearance of ALKG1269A, in ctDNA. The patient remained on the combination for 4.5 months (6 months on trial) until progressive disease (Fig. 2E).
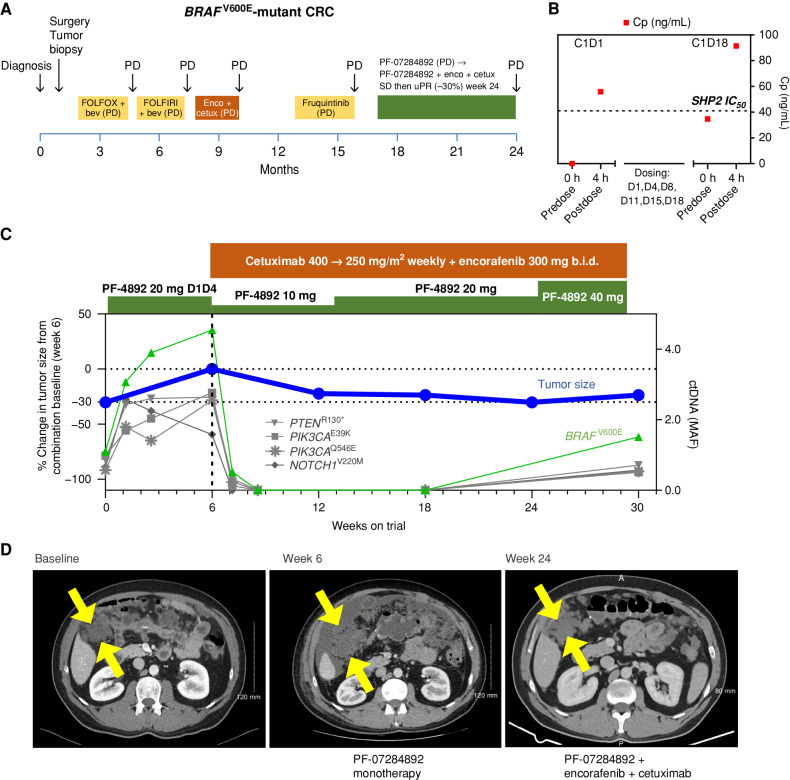
BRAF V600E-Mutant Colorectal Cancer.
A patient with metastatic BRAFV600E colorectal cancer was treated with chemotherapy, encorafenib + cetuximab, and fruquintinib (VEGFR inhibitor; Fig. 3A). PF-07284892 monotherapy (same dose level as the prior patient) was initiated. PK analysis indicated sustained plasma levels of PF-07284892 (35 ng/mL C1D18 vs. 56 ng/mL C1D1; Fig. 3B). Blood samples were not available for pERK analysis. Progressive disease (+43% and new lesions) was observed after 6 weeks, together with increased BRAFV600E (and additional mutations) in ctDNA (Fig. 3C and D).
Figure 3.
PF-07284892 overcomes intrinsic resistance to encorafenib + cetuximab in a BRAFV600E-mutant CRC patient. A, The patient's previous systemic therapies were chemotherapy + bevacizumab, encorafenib + cetuximab, and fruquintinib, with the best overall response PD, indicating primary progression/intrinsic resistance to each therapy. B, Levels of PF-07284892 (red squares) in plasma, as in Fig. 2D; blood samples for pERK were not available. C, Change in the sum of the longest tumor diameters of target lesions and in BRAFV600E (and other mutations) in ctDNA over time, as in Fig. 2E. D, Imaging of a right-sided intra-abdominal target lesion mass during study treatment. The patient experienced one AE with monotherapy (grade 2 ascites not related to study treatment) and three grade 1 AEs with combination treatment (headache, fatigue, and acneiform rash, the latter a known toxicity of cetuximab). bev, bevacizumab; b.i.d., twice daily; C, cycle; cetux, cetuximab; Cp, plasma concentration; CRC, colorectal cancer; D, day; enco, encorafenib; FOLFIRI, folinic acid, fluorouracil, irinotecan; FOLFOX, folinic acid, fluorouracil, oxaliplatin; MAF, mean allele frequency; PD, progressive disease; PF-4892, PF-07284892; SD, stable disease; uPR, unconfirmed partial response.
PF-07284892 (initially dose decreased like the prior patient) was continued, and encorafenib + cetuximab were added at their approved doses. After 6 weeks of the combination (study week 12), rapid and complete disappearance of BRAFV600E (and other mutations) in ctDNA occurred together with a −22% decrease in the peritoneal target lesion and resolution of malignant ascites (Fig. 3C). After 7 weeks of combination (study week 13), the PF-07284892 dose was increased to 20 mg b.i.w. like the prior patient. BRAFV600E in ctDNA remained undetectable through 12 weeks of combination (study week 18). At 18 weeks of combination (study week 24), tumor reduction reached −30% (Fig. 3C and D). Despite increasing the dose of PF-07284892 to 40 mg b.i.w., BRAFV600E was again detected in ctDNA together with a +20% increase in target lesion after 24 weeks of combination (study week 30), indicating progressive disease (Fig. 3C and D). The patient remained on combination therapy without disease progression for 6 months, three times longer than prestudy encorafenib + cetuximab. There were no DLTs and no dose modifications for AEs.
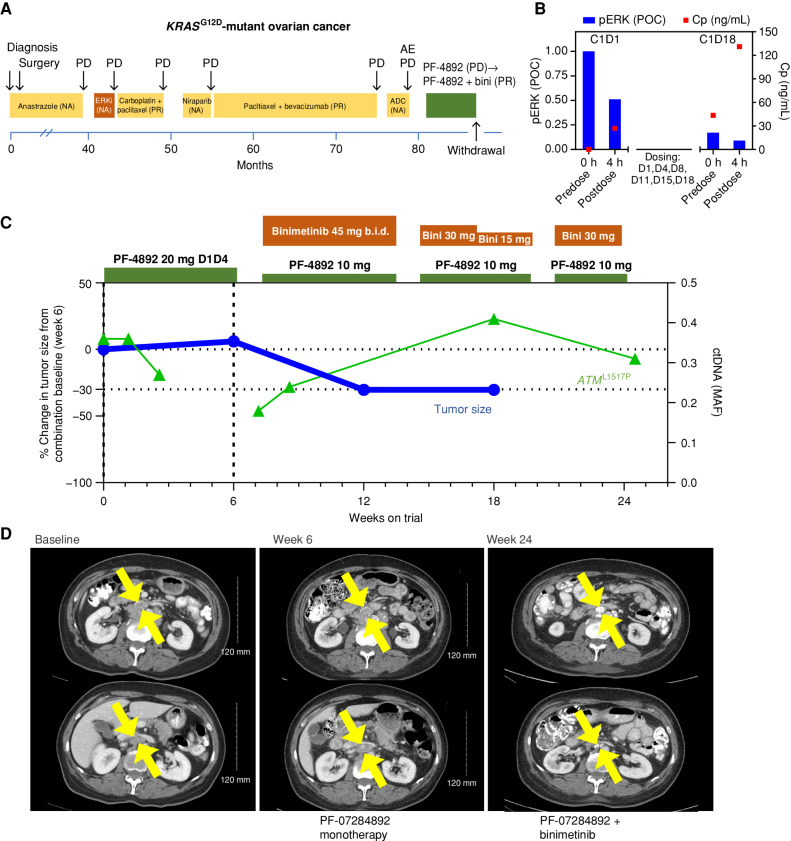
KRAS G12D-Mutant Ovarian Cancer.
A patient with KRASG12D-mutant low-grade serous ovarian cancer underwent surgical resection and adjuvant anastrozole. For recurrent/metastatic disease, she received ASN-007 (investigational ERK inhibitor), two lines of chemotherapy, niraparib, and SGN-STNV (antibody–drug conjugate; Fig. 4A). PF-07284892 monotherapy was initiated at the same dose as prior patients. PK/PD analysis demonstrated sustained PF-07284892 and inhibition of pERK in peripheral blood monocytes, including on C1D18, 3 days after the last prior dose (Fig. 4B). Nevertheless, after 6 weeks of monotherapy, imaging demonstrated +6% tumor growth in two target lesion mediastinal lymph nodes, together with worsening disease-related abdominal pain and distension (Fig. 4C and D).
Figure 4.
PF-07284892 sensitizes a patient with KRASG12D-mutant ovarian cancer to the MAPK pathway inhibitor binimetinib. A, Prior treatments for metastatic disease included ASN-007 (investigational ERK inhibitor), chemotherapy, niraparib, and SGN-STNV (investigational antibody–drug conjugate). B, Levels of PF-07284892 in plasma (red squares) and pERK in ex vivo CSF1-stimulated peripheral blood monocytes (blue bars), as in Fig. 2D. C, Change in the sum of the longest tumor diameters of target lesions and of ATML1517P in ctDNA over time, as in Fig. 2E. D, Imaging of two abdominal target lesions during study treatment. All treatment-related AEs were grade 1 except for edema (grades 1–2, starting on monotherapy, worsening on the combination, and leading to dose modification), fatigue (grades 2–3), weight gain, diarrhea, and eczema (each grade 2 and resolved by the end of treatment). ADC, antibody–drug conjugate; b.i.d., twice daily; bini, binimetinib; C, cycle; Cp, plasma concentration; D, day; ERKi, ERK inhibitor; MAF, mean allele frequency; NA, not available; PD, progressive disease; PF-4892, PF-07284892; POC, percentage of control.
The patient continued PF-07284892 (dose decreased like the prior patients), and binimetinib was added at the approved dose. A PR (−34%) was achieved after 6 weeks of combination therapy (study week 12; Fig. 4C and D). Despite binimetinib dose reduction for edema, the patient continued combination treatment with PR for 5 months, at which time treatment was discontinued for persistent edema. There were no PF-07284892 dose modifications and no DLTs.
Although the founder KRASG12D mutation was not detected in ctDNA, ATML1517P appeared to decrease early during monotherapy and combination (study week 6 sample was not available) and increased at later time points (Fig. 4C).
GOPC–ROS1 Fusion–Positive Pancreatic Cancer.
A patient with GOPC–ROS1 fusion–positive pancreatic adenocarcinoma was previously treated with gemcitabine + abraxane + nivolumab followed by repotrectinib (progressed after ∼10 months; Supplementary Fig. S7A). PF-07284892 monotherapy was initiated at the same dose as prior patients. PK analysis indicated sustained plasma levels of PF-07284892 despite intermittent dosing (Supplementary Fig. S7B). After 6 weeks of monotherapy, imaging demonstrated progressive disease in two target lesion liver metastases (+26% overall), together with an increase in the GOPC–ROS1 and CTNNB1S45F in ctDNA (Supplementary Fig. S7C and S7D).
The patient continued PF-07284892, and lorlatinib was added at 100 mg qd. After 2 weeks of combination (study week 8), both mutations in ctDNA decreased by 95% compared with baseline before starting any study treatment (Supplementary Fig. S7C). After 6 weeks of combination (study week 12), target liver lesions decreased by −35% compared with precombination imaging consistent with PR, confirmed after 12 weeks of combination (study week 18; Supplementary Fig. S7C and S7D). She remained on the combination for a total of 7.5 months until progressive disease.
DISCUSSION
This study underscores the utility of early combination therapy exploration in drug development programs that include a novel agent. PF-07284892 is a highly potent, selective, allosteric SHP2 inhibitor. Compared with other SHP2 inhibitors, PF-07284892 has a long half-life, causing sustained pERK inhibition in preclinical models and peripheral blood monocytes from patients despite intermittent dosing, enhanced target binding, and potential intracranial coverage. Most importantly, significant preclinical antitumor activity was observed only in oncogene-driven cancers when administered with matched targeted therapy.
Traditional phase I trials would have explored PF-07284892 with a dose-escalation design that only featured monotherapy or only allowed combination after full monotherapy dose exploration. This strategy raises ethical concerns, as patients risk exposure not only to subtherapeutic doses in early dose levels but also ineffective treatment even at later doses. All patients in this series unsurprisingly had primary progression on PF-07284892 monotherapy. These findings are consistent with trials of other SHP2 inhibitors in which little single-agent activity was observed (20, 22). Furthermore, patients with florid disease progression may clinically deteriorate and miss the opportunity to receive subsequent cancer-directed therapies altogether.
Proof-of-concept data presented here demonstrate that the early addition of combination therapy in dose escalation is safe and feasible. Primary progression on SHP2 inhibitor monotherapy was rescued with the addition of matched targeted therapy in patients with cancers driven by BRAFV600E, KRASG12D, and ALK/ROS1 fusions, each of whom had progressed on the same/similar targeted therapy prior to study entry. Benefit correlated with molecular response in founder or companion alterations in ctDNA after combination therapy initiation. These outcomes were enabled by the trial's contemporary study design approved by regulatory authorities. This design is scientifically informed and patient-centric and should be considered by other drug development stakeholders for future trials.
The preclinical and clinical combination therapy activity presented here supports the role of SHP2 as an “Achilles’ heel” whose inhibition may sensitize or resensitize diverse tumors to targeted therapy. Of note, although responses to the up-front combination of KRASG12C and SHP2 inhibitors (e.g., JDQ443 and TNO155 or adagrasib and RMC-4630) have previously been described in KRASG12C-mutant cancers, the patients had not first received either agent as monotherapy, challenging ascertaining the need for combination treatment (20, 23). Our data support combination therapy in other oncogene-driven cancers. We are not aware of previous reports of responses to SHP2 + fusion kinase inhibitor therapy in ALK/ROS1 fusion–positive cancer patients.
Early combination therapy exploration likewise enables early safety data readouts. Other SHP2 inhibitors had narrow TIs, requiring intermittent dosing to mitigate on-target toxicity. PF-07284892 preclinical data demonstrated consistent activity with a wider TI with intermittent dosing. This led us to pursue intermittent dosing in patients from the start (Supplementary Fig. S2A–S2D). Although two of four patients required dose decreases of the combination agent due to drug-related depression (lorlatinib) or edema (binimetinib), none required dose reduction of PF-07284892 for AEs, and mostly low-grade toxicities were observed with combination therapy in the rest. Although earlier in development, PF-07284892's overall safety profile appears like other investigational SHP2 inhibitors (compared in Supplementary Table S8). We note the potential for consistent target inhibition by PF-07284892 (as measured by inhibition of pERK in patient monocytes) despite intermittent dosing.
There are limitations to this study. As summarized in Supplementary Table S6, some patients were not able to start or receive sufficient combination treatment due to clinical deterioration, disease progression, and/or toxicity during monotherapy. The number of patients treated with combination therapy should be increased to firmly establish the contribution of individual components to observed clinical effects. Higher doses of PF-07284892 should be explored to determine if even greater pERK inhibition and efficacy in patients are achieved.
Two patients had previously been treated with targeted therapy that differed from the actual combination agent provided on study: an investigational ERK inhibitor for the patient with ovarian cancer and repotrectinib for the patient with pancreatic cancer. Although it is possible that the efficacy observed in these two patients was driven primarily by the study combination agent patients were not previously exposed to (binimetinib or lorlatinib, respectively), this seems less likely. The ERK inhibitor inhibits the MAPK pathway downstream of binimetinib, and the absence of acquired ROS1 resistance mutations in GOPC–ROS1 may be consistent with occult bypass resistance to repotrectinib that is not generally sensitive to single-agent targeted therapy. Furthermore, lorlatinib is not clearly active after repotrectinib (24).
Regarding long-term combination therapy toxicities, clearly attributing side effects to individual drug components or their combination may be challenging, although in a traditional phase I design, if monotherapy is ineffective, most patients will discontinue treatment early and long-term side effects cannot be interrogated. Finally, although knowing the clinical AE profile of similar drugs is helpful, it is not necessary for this type of trial design if robust nonclinical data are available on potential efficacy, toxicities, and drug–drug interactions that support drug combination.
In summary, we describe the discovery and early clinical development of the investigational SHP2 inhibitor PF-07284892 using a novel FIH phase I clinical trial design that allowed each patient to receive one of three rational combinations of targeted therapy and PF-07284892. This approach led to tumor and ctDNA reductions and extended clinical benefit for four patients treated at the first dose level. We are not aware of another trial in which the first patients in dose escalation had access to rational combination treatments instituted immediately upon monotherapy progression. This phase I study is ongoing and will more fully evaluate up-front combination therapy safety and efficacy at higher doses in patients with prior progression on appropriate targeted treatment.
METHODS
PF-07282892 Structure and Design
See US Patent 11,634,417, Example No. 6.
SHP2 Protein Expression and Purification
N-terminal poly-His tagged SHP2(2–257) for enzyme assay and SHP2(1–525) for crystallography were expressed in Escherichia coli BL21(DE3) and purified by standard methods, including metal affinity and anionic ion exchange, followed by size-exclusion column chromatography. The poly-His tag was removed from SHP2(1–525) by the TEV protease that recognizes the TEV cleavage sequence engineered between the poly-His tag and the SHP2(1–525) amino sequence. The SH2(1–525) was concentrated to 8 mg/mL in a buffer of 25 mmol/L Tris, pH 8.0, 150 mmol/L NaCl, and 1 mmol/L tris(2-carboxyethyl)phosphine (TCEP) for crystallization.
Cocrystallization, X-ray Data Collection, Data Processing, and Structure Solution of SHP2 with PF-07282892
The inhibitor-bound SHP2 protein complex was prepared by mixing purified SHP2(1-525) with a 5-fold molar excess of PF-07282892 (in DMSO) and incubated on ice for at least 30 minutes. An equal volume of SHP2:PF-07282892 complex and the reservoir solution, which consists of 9% PEG3350, 0.1M Bicine, pH 9.2, 30 mmol/L ammonium acetate, and 5% Tacsimate, were mixed with additional microcrystal seeds to promote the crystallization process. Crystallization was performed by the hanging-drop vapor diffusion method at 14°C for ∼7 days before harvesting. Rectangular plate crystals with sizes of 200 to 300 μm in two dimensions were collected and cryoprotected in the reservoir solution containing 25% (v/v) glycerol and flash-frozen in a 100K nitrogen gas stream. X-ray diffraction data were collected on a Rigaku FRE SuperBright X-ray generator equipped with Confocal VariMax optics and EIGER 1M detector. Diffraction data were processed using Mosflm in the CCP4 software package (25). Structures were solved by molecular replacement using a published SHP structure (PDB ID: 2SHP) as a search model. Itinerate rounds of structure refinement (26) and manual model rebuilding were performed with Refmac5 (27) and COOT (28). Data collection and refinement statistics are shown in Supplementary Table S1. The image in Fig. 1A was generated using the Maestro graphical interface (version 13.3.121; SchrÖdinger).
SHP2 Enzyme Assays
C-terminal, Histidinex6 (His6) tagged full-length SHP2 (amino acids 2–527) was recombinantly expressed in and purified from E. coli using standard methods. Fluorescence intensity kinetic assays were configured for full-length SHP2 to monitor the amount of 6,8-difluoro-7-hydroxy-4-methylcoumarin (DiFMU) formed upon hydrolysis of 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP). The assay mixture consisted of 25 mmol/L K+HEPES, pH 7.4, 0.01% Triton X-100, 1 mmol/L DTT, 50 mmol/L KCl, 100 μg/mL bovine γ-globulin, 50 μmol/L DiFMUP, 1 μmol/L SHP2 activating peptide [LN(pY)IDLDLV(dPEG8)LST(pY)-ASINFQK-amide], 1 nmol/L full-length SHP2 [His6-tagged SHP2(2–527)], and 2% (v/v) DMSO (from compound). Compounds were typically diluted in DMSO across a 10-point dosing range created using a 3-fold serial dilution protocol at a top dose of 20 μmol/L. The assays were run in 384-well, polystyrene, low-volume, nontreated, black microtiter plates (Costar 4511) in a final volume of 20 μL. Low control wells lacked enzymes. The assays were initiated by the addition of a mixture of SHP2 and the activating peptide and, following a 15-second mix on an orbital shaker, were read in kinetic mode for 15 minutes (30 seconds/cycle) at ambient temperature on a PerkinElmer EnVision microplate reader (λEx = 355 nm, λEm = 460 nm). Initial velocities (slopes of the tangents at t = 0) were estimated from exponential fits to the slightly nonlinear progress curves and then were converted to percent of control (POC) using the following equation:
where  is the average of the uninhibited controls and
is the average of the uninhibited controls and  is the average of the background samples. A 4-parameter logistic model was fit to the POC data for each compound. From that fit, the IC50 was estimated and is defined as the concentration of compound at which the curve crosses 50 POC.
is the average of the background samples. A 4-parameter logistic model was fit to the POC data for each compound. From that fit, the IC50 was estimated and is defined as the concentration of compound at which the curve crosses 50 POC.
Phosphatase Profiling
PF-07284892 was evaluated by Eurofins, Inc. with Phosphatase Profiler for 22 phosphatases [20 human phosphatases, YoPH (bacterial) and lambda (phage)]. Compounds were run at 10,000, 1,000, and 100 nmol/L final compound concentration according to Eurofin's specifications. IC50 values were estimated from the POC of the three concentrations evaluated.
Cell Lines and Xenografts
NCI-H3122, HT29, MIA PaCa-2, and NCI-H1975 cells were obtained from ATCC, KYSE-520 from DSMZ, RT 112/84 from the European Collection of Animal Cell Cultures, and EBC-1 from the Japanese Collection of Research Bioresources Cell Bank. All cell lines were authenticated by STR profiling and regularly evaluated for Mycoplasma (MycoAlert, Lonza, Inc.). The CR5087 patient-derived xenograft (PDX) was obtained from Crown Biosciences. The MGH915-4Y10 PDX was provided by Dr. Aaron Hata, MGH Cancer Center. LorR-06 (EML4–ALK) fusion-positive NSCLC cell lines were derived from parental NCI-H3122 cells by long-term in vitro culture in the presence of lorlatinib. See Supplementary Methods for additional molecular validation of cell lines and PDXs.
Quantitative pERK Cell Analysis
Cells were seeded at 5 × 104/well into clear, black-bottom, 96-well plates, incubated overnight at 37°C, and incubated for 1 hour with a 9-point dilution series of each inhibitor, followed by 3.7% formaldehyde fixation, costaining with pERK and GAPDH antibodies, costaining with goat secondary antibodies conjugated to IrDye 800CW (for pERK) or IRDye 680RD (for GAPDH), and analysis of staining intensity by InCell Western (LI-COR, Inc.). Signal intensity for pERK was normalized to GAPDH and the DMSO-treated control samples to generate POC data, which were then plotted versus compound concentration using GraphPad Prism 5 software to generate IC50 data using a 3-parameter curve fit. See Supplementary Table S9 for sources of primary antibodies.
Immunoblotting
Each cell line was seeded in 12-well plates at 2.5 × 105 cells/well (1 mL total volume), incubated overnight at 37°C, and incubated for the indicated times with the indicated concentrations of each inhibitor dissolved in DMSO (DMSO used for vehicle control). Cells were lysed with 100 μL cell lysis buffer (Cell Signaling Technology), processed for SDS-PAGE, and transferred to nitrocellulose membrane for immunoblot analyses with the indicated primary and appropriate antibodies, followed by imaging analysis (Bio-Rad ChemiDoc XR/ECL for Fig. 1B; LI-COR Odyssey Scanner/fluorescent imaging for Fig. 1C and D). See Supplementary Table S9 for sources of antibodies.
Animal Care, Xenograft Preparation, and Treatment
All procedures performed on animals were in accordance with regulations and established guidelines and were reviewed and approved by Pfizer's Institutional Animal Care and Use Committee. Tissue samples were collected in accordance with regulations and established guidelines for the humane treatment of research animals. All animals were obtained at 6 to 8 weeks of age (Envigo), housed in groups of five, and allowed a 1-week acclimation period before cancer cell injection. Food, water, temperature, and humidity were prepared per Pharmacology Testing Facility performance standards (standard operating procedures), which are in accordance with the 2011 Guide for the Care and Use of Laboratory Animals (National Research Council) and AAALAC International.
Each cell line (5 × 106 cells) or PDX (cell suspension prepared with Miltenyi gentleMACS) was injected subcutaneously into the right flank of female Foxn1nu mice and allowed to grow to approximately 200 mm3 (efficacy) or approximately 500 mm3 (PK/PD) prior to randomization by tumor size into dosing groups of eight (for efficacy) or three (for PK/PD) analysis. Animals were dosed by oral gavage with vehicle (1% carboxymethylcellulose/0.5% Tween-80), PF-07284892 (20% hydroxypropyl-β-cyclodextrin in 50 mmol/L citric acid, pH 4), lorlatinib (water for injection, pH 3), encorafenib + binimetinib (1% carboxymethylcellulose/0.5% Tween-80), binimetinib (1% carboxymethylcellulose/0.5% Tween-80), or their combination with PF-07284892.
Animal Efficacy Studies
Animals were dosed with vehicle, PF-07284892 (30 mg/kg every other day), appropriate targeted therapy [lorlatinib, 3 mg/kg qd, encorafenib 20 mg/kg qd + binimetinib 3.5 mg/kg twice daily (b.i.d.), binimetinib 3.5 mg/kg b.i.d.], or the combination of each targeted therapy with PF-07284892 at the doses used for monotherapy. Tumor size was determined by the formula (length × width × width)/2. For each animal, tumor size on days 22 to 29 of treatment was normalized to tumor size on day 0 immediately prior to first treatment and displayed as a percentage of tumor volume change.
Animal PK Assessment
Concentrations of PF-07284892 in mouse, rat, dog, and monkey plasma and mouse brains were determined by liquid chromatography–tandem mass spectrometry (LC-MS/MS) following protein precipitation. Oral and intravenous mouse, rat, dog, and monkey pharmacokinetic parameters were calculated using the noncompartmental analysis of total PF-07284892 concentration in plasma, and mouse brain-to-plasma ratios were calculated following adjustment for the unbound fraction of PF-07284892 in both brain homogenate and plasma.
For the quantitation of PF-07284892 concentrations in plasma, blood samples were collected into tubes containing K2EDTA as an anticoagulant and processed to the plasma component via centrifugation. A 12-point calibration curve, ranging from 0.282 to 50,000 ng/mL, was prepared in duplicate. A solution of 400 μg/mL PF-07284892 in DMSO was serially diluted 3-fold, and 2.5 μL of each standard solution was added to 20 μL of naive plasma. To mimic extraction of the standard curve, 2.5 μL of DMSO was added to 20 μL aliquots of test plasma. Both calibration and test plasma samples were spiked with 2.5 μL of internal standard in DMSO.
For the quantitation of PF-07284892 in brain, whole brains were harvested from mice administered an oral dose of PF-07284892, weighed, and homogenized in matrix D lysing tubes (MP Biomedicals, Inc.) following the addition of 500 μL of 4:1 water:methanol. A 10-point calibration curve, ranging from 0.508 to 10,000 ng/mL, was prepared in duplicate. A solution of 400 μg/mL PF-07284892 in DMSO was serially diluted 3-fold in DMSO, and then 2.5 μL of each standard solution was added to 100 μL of naive mouse brain homogenate. To mimic extraction of the standard curve, 2.5 μL of DMSO was added to 100 μL aliquots of test brain homogenate. Both calibration and test brain homogenate samples were spiked with 2.5 μL of internal standard in DMSO.
Proteins in plasma and brain preparations were precipitated by the addition of 300 μL of acetonitrile. Samples were vortex-mixed, and precipitated proteins were removed via centrifugation. A 75 μL aliquot of the supernatant was transferred to a 96-well plate and diluted 1:1 with water for analysis.
Animal PK/PD Studies
Each animal was administered a single dose of 30 mg/kg PF-07284892 by oral gavage. The mice were euthanized at 1, 8, 24, and 48 hours after dose by CO2 inhalation, and tumors were excised, flash-frozen in liquid nitrogen, and stored at −80°C. Blood was collected into tubes containing 10% v/v EDTA, processed to the plasma component via centrifugation, and stored at −80°C for bioanalysis. To determine the levels of pERK in tumors, tumor tissue was homogenized in lysis buffer (1% NP-40, 20 mmol/L Tris, pH 8.0, 137 mmol/L NaCl, 10% glycerol, 2 mmol/L EDTA, protease and phosphatase inhibitors) and centrifuged twice to separate insoluble material. Protein levels in each lysate were determined using Coomassie protein reagent (Pierce, Inc.). pERK levels were determined in 250 μg of each sample by immunoblot and normalized to GAPDH in the same lysate and vehicle-treated controls, and results were expressed as POC pERK levels in tumors. To determine the levels of PF-07284892 in plasma, see “Animal PK Assessment” above.
Clinical Studies
Trial Design.
Patients were treated on the FIH phase I clinical trial of PF-07284892 (NCT04800822). The FDA and Institutional Review Boards from each site approved the trial, the studies were conducted in accordance with the Declaration of Helsinki and in compliance with all International Council for Harmonisation Good Clinical Practice guidelines, and each patient (or legal guardians/representatives) provided written informed consent. Early combination was initiated after 2 cycles/6 weeks of PF-07284892 monotherapy, absent ongoing grade ≥3 toxicity or DLT, after radiographic evidence of disease progression (symptoms of progressive disease without tumor growth ≥20% was allowed), and starting at one dose level lower than the highest safe monotherapy dose of PF-07284892 based on an acceptable rate of first treatment cycle DLTs. Intrapatient dose escalation of PF-07284892 to previously determined safe dose(s) was permitted after 1 cycle and absent grade ≥3 toxicity.
Treatment and Response Assessment.
PF-07284892 was administered to patients as a powder in capsule. Lorlatinib, encorafenib, cetuximab, and binimetinib were administered at their labeled doses. Dose modifications and interruptions followed a prescribed algorithm. AEs were graded using Common Terminology Criteria for Adverse Events (CTCAE) version 5.0. Response was evaluated by CT imaging using RECIST version 1.1 every 6 weeks.
PD Assessment.
Plasma samples were collected prior to treatment and at defined intervals after dosing on days 1 and 18 of cycle 1 at the starting PF-07284892 monotherapy dose. PF-07284892 plasma concentrations were determined using a validated LC-MS/MS assay.
Patient PD Assessment.
Whole blood was collected from patients in 4 mL collection tubes and shipped on the same day with refrigerated gel packs to the Pfizer Early Clinical Development Precision Medicine Flow Cytometry Lab in Groton, CT. Samples received within the 24-hour stability window were processed and analyzed. Whole blood samples were stimulated with 9 ng/mL recombinant human M-CSF (R&D Systems) for 12 minutes at room temperature. Unstimulated samples were used as a control sample. Whole blood was stained by incubating with a mixture of antibodies to anti-CD45-APC-H7 (clone 2D1, Becton Dickinson), anti-CD14-BV421 (clone M5E2, Becton Dickinson), ERK1/2-AF-647 (clone 137F5, Cell Signaling Technology), and pERK1/2 T202/Y204-PE (clone 20A, Becton Dickinson) for 30 minutes at room temperature in the dark. Samples were analyzed on a Becton Dickinson FACSCanto Flow Cytometer using BD FACSDiva software v8.0.1, FlowJo V10. Whole blood samples were gated to exclude doublets by using FSC-A versus FSC-H. Cells were gated for CD45+ and CD14+ cells. Median fluorescent intensity (MFI) of ERK1/2 in CD14+ monocytes and MFI of pERK1/2 in CD14+ monocytes are reported. MFI of pERK1/2 in CD14+ monocytes at 9 ng/mL is normalized to the unstimulated sample to account for a change in cell numbers.
Patient Plasma Cell-Free DNA Extraction and Analyses.
Whole blood was collected from patients in 2 × 10 mL collection tubes containing K2EDTA as anticoagulant. Within 30 minutes, plasma was processed by centrifugation at approximately 1,600 × g for 10 minutes using a prechilled centrifuge set to 4°C. Plasma was then transferred to a 15-mL centrifuge tube, centrifuged as above at approximately 3,000 × g for 25 minutes, aliquoted, and frozen at −80°C until shipment to Guardant Health. Extraction of cell-free DNA and next-generation sequencing were performed at Guardant Health using the G360 73-gene panel (Panel v 2.10, bioinformatics pipeline v 3.5.2). Mean allele frequency of each mutation was plotted.
Data Availability
Upon request, and subject to review, Pfizer, Inc. will provide the data that support the findings of this study. Subject to certain criteria, conditions, and exceptions, Pfizer, Inc. may also provide access to the related individual deidentified participant data. See https://www.pfizer.com/science/clinical-trials/trial-data-and-results for more information.
Supplementary Material
Supplementary Data Title Page, Methods, References
Supplementary Table 1: X-ray crystallography data collection and refinement statistics
Supplementary Table 2: Phosphatase Profiling of PF-07284892
Supplementary Table 3: IC50 values for PF-07284892 in cell-based assays
Supplementary Table 4: Pharmacokinetics of PF-07284892 in preclinical species
Supplementary Table 5: Plasma and brain tissue concentrations, brain-to-plasma ratios, and brain concentration over IC50 values for PF-07284892 and TNO155 at dose levels preferred for concomitant administration
Supplementary Table 6: Disposition of all patients enrolled to Dose Level 1
Supplementary Table 7: Adverse events for Dose Level 1 patients
Supplementary Table 8: Comparison of clinical safety data for SHP2 inhibitors
Supplementary Table 9: Sources of antibodies
Supplementary Figure 1: Comparison of the structures of PF-07284892, RMC445507, TNO155 bound to SHP2
Supplementary Figure 2: Intermittent dosing of PF-07284892 preserves efficacy and improves tolerability preclinically
Supplementary Figure 3: Preclinical development of PF-07284892
Supplementary Figure 4: PF-07284892 combination efficacy in additional oncogenic driver-positive mouse cancer models and monotherapy PK/PD
Supplementary Figure 5: Study schema
Supplementary Figure 6: PK profiles for patients
Supplementary Figure 7: PF-07284892 sensitizes a patient with GOPC-ROS1 fusion-positive pancreatic cancer to lorlatinib
Acknowledgments
We thank Pfizer Boulder Research & Development and Array BioPharma colleagues, including Shannon L. Winski, James F. Blake, Mark Chicarelli, and Lauren Hanson. The study was funded by Pfizer, Inc. A. Drilon was supported in part by the NCI of the NIH under award numbers P30 CA008748 and 1R01CA273224-01.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
A. Drilon reports personal fees from Ignyta, Genentech, Roche, Loxo, Bayer, Lilly, Takeda, Ariad, Millenium, TP Therapeutics, AstraZeneca, Pfizer, Blueprint Medicines, Helsinn, BeiGene, BerGenBio, Hengrui Therapeutics, Exelixis, Tyra Biosciences, Verastem, MORE Health, AbbVie, 14ner/Elevation Oncology, ArcherDX, Monopteros, Novartis, EMD Serono, Medendi, Repare RX, Nuvalent, Merus, Chugai Pharmaceutical, Remedica Ltd, mBrace, AXIS, EPG Health, Harborside, Nexus, Liberum RV, More Ology, Amgen, TouchIME, Janssen, Entos, Treeline Bio, Prelude, Applied Pharmaceutical Science, Inc., AiCME I3 Health, MonteRosa, EcoR1, and Treeline during the conduct of the study; other support from Pfizer, Exelixis, GSK, Teva, Taiho, and PharmaMar outside the submitted work; a patent for selpercatinib–osimertinib pending; and equity in mBrace, royalties from Wolters Kluwer, other (food/beverage) from Merck, Puma, Merus, and Boehringer Ingelheim, and CME honoraria from Medscape, OncLive, PeerVoice, Physicians Education Resources, Targeted Oncology, Research to Practice, Axis, Peerview Institute, Paradigm Medical Communications, WebMD, MJH Life Sciences, AXIS, EPG Health, JNCC/Harborside, and I3 Health. M.R. Sharma reports other support from Pfizer, Inc. during the conduct of the study; personal fees from Pliant Therapeutics outside the submitted work; and Pfizer, Inc. stock ownership as part of a diverse investment portfolio. M.L. Johnson reports grants and other support from Daiichi Sankyo during the conduct of the study; grants and other support from AbbVie, Amgen, Arcus Biosciences, AstraZeneca, Black Diamond, Calithera Biosciences, Daiichi Sankyo, Genentech/Roche, Genmab, Genocea Biosciences, GSK, Gritstone Oncology, Ideaya Biosciences, Immunocore, Novartis, Janssen, Incyte, Merck, Mirati Therapeutics, Regeneron Pharmaceuticals, Revolution Medicines, Sanofi, Takeda Pharmaceuticals, and Turning Point Therapeutics, grants from Acerta, Adaptimmune, Apexigen, Array BioPharma, Artios Pharma, Atreca, Beigene, BerGenBio, BioAtla, Boehringer Ingelheim, Bristol Myers Squibb, Carisma Therapeutics, Checkpoint Therapeutics, City of Hope National Medical Center, Corvus Pharmaceuticals, Curis, CytomX, Dracen Pharmaceuticals, Dynavax, Lilly, Elicio Therapeutics, EMD Serono, EQRx, Erasca, Exelixis, Fate Therapeutics, Guardant Health, Harpoon, Helsinn Healthcare SA, Hengrui Therapeutics, Hutchinson MediPharma, IGM Biosciences, Immunitas Therapeutics, Jounce Therapeutics, Kadmon Pharmaceuticals, Kartos Therapeutics, Loxo Oncology, Lycera, Memorial Sloan Kettering, Merus, Mythic Therapeutics, NeoImmune Tech, Neovia Oncology, Numab Therapeutics, Nuvalent, OncoMed Pharmaceuticals, Palleon Pharmaceuticals, Pfizer, PMV Pharmaceuticals, Rain Therapeutics, RasCal Therapeutics, Relay Therapeutics, Ribon Therapeutics, Rubius Therapeutics, Seven and Eight Biopharmaceuticals/Birdie Biopharmaceuticals, Shattuck Labs, Silicon Therapeutics, StemCentRx, Syndax Pharmaceuticals, Tarveda, TCR2 Therapeutics, Tempest Therapeutics, Tizona Therapeutics, TMUNITY Therapeutics, University of Michigan, Vyriad, WindMIL Therapeutics, and Y-mAbs Therapeutics, and other support from Arrivent, Astellas, Axelia Oncology, EcoR1, iTeos, Jazz Pharmaceuticals, Molecular Axiom, Oncorus, Pyramid Biosciences, Seagen, Synthekine, and VBL Therapeutics outside the submitted work. T.A. Yap reports grants and personal fees from Acrivon, Artios, AstraZeneca, Bayer, BeiGene, Pfizer, Merck, Clovis, EMD Serono, Genentech, ImmuneSensor, F-Star, and Blueprint, grants from Bristol Myers Squibb, Boundless Bio, BioNTech, Repare, Sanofi, Constellation, Cyteir, Eli Lilly, Forbius, GSK, Haihe, Ideaya, Ionis, Ipsen, Jounce, Karyopharm, KSQ, Kyowa, Mirati, Novartis, Ribon Therapeutics, Regeneron, Rubius, Scholar Rock, Seattle Genetics, Tesaro, Vivace, and Zenith, personal fees from AbbVie, Adagene, Almac, Aduro, Amphista, Athena, Atrin, Ayoro, Axiom, Baptist Health System, Boxer, Bristol Myers Squibb, C4 Therapeutics, Calithera, Cancer Research UK, Circle Pharma, CUHK Committee, Cybrexa, Dark Blue Therapeutics, Diffusion, Ellipses Life, Genmab, Gerson and Lehrman Group, Glenmark, GLG, Globe Life Sciences, GSK, Guidepoint, Idience, Ignyta, I-Mab, Institut Gustave Roussy, Intellisphere, Janssen, Kyn, LRG1, MEI Pharma, Mereo, Natera, Nexys, Novocure, OHSU, OncoSec, Ono Pharma, Panangium, Pegascy, PER, Piper-Sandler, Pliant Therapeutics, Prolynx, Radiopharm Theranostics, resTORbio, Roche, SchrÖdinger, Synthis Therapeutics, Terremoto Biosciences, Tessellate Bio, TD2 Theragnostics, Tome Biosciences, Varian, Versant, Vibliome, Xinthera, Zai Labs, Zentalis, and ZielBio, and personal fees and other support from Seagen during the conduct of the study. S. Gadgeel reports personal fees from Genentech/Roche, Pfizer, Bristol Myers Squibb, AstraZeneca, Blueprint, Novartis, Daichii Sankyo, Merck, Eisai, Gilead, Mirati, GSK, Takeda, and Arcus and nonfinancial support from Mirati outside the submitted work. G. Feng is an employee of and a shareholder in Pfizer, Inc. C.E. Wong reports a patent for WO2020/201991 A1 issued. R.J. Hinklin reports a patent for WO2020201991 issued and a patent for WO2020081848 issued. Y. Jiang reports a patent for WO2020201991 pending. P. Wei reports other support from Pfizer, Inc. outside the submitted work. K.A. Ching reports personal fees from Pfizer, Inc. during the conduct of the study; personal fees from Pfizer, Inc. outside the submitted work; and is an employee of and shareholder in Pfizer, Inc. and may own shares and/or options of Pfizer, Inc. stock. P.A. Lee reports personal fees from Pfizer, Inc. and other support from Pfizer, Inc. outside the submitted work. S.M. Rothenberg reports personal fees and other support from Pfizer, Inc. outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
A. Drilon: Conceptualization, investigation, writing–original draft, writing–review and editing. M.R. Sharma: Investigation, writing–review and editing. M.L. Johnson: Investigation, writing–review and editing. T.A. Yap: Investigation, writing–review and editing. S. Gadgeel: Investigation, writing–review and editing. D. Nepert: Conceptualization, investigation, methodology, project administration, writing–review and editing. G. Feng: Investigation, visualization, project administration, writing–review and editing. M.B. Reddy: Conceptualization, investigation, writing–review and editing. A.S. Harney: Investigation, visualization, methodology. M. Elsayed: Investigation. A.W. Cook: Investigation. C.E. Wong: Investigation. R.J. Hinklin: Investigation. Y. Jiang: Investigation. E.N. Brown: Investigation, visualization. N.A. Neitzel: Investigation. E.R. Laird: Visualization, writing–review and editing. W. Wu: Supervision, investigation. A. Singh: Investigation, visualization, methodology, writing–review and editing. P. Wei: Resources, investigation, visualization, methodology, writing–review and editing. K.A. Ching: Resources, methodology. J.J. Gaudino: Resources, supervision, investigation, methodology. P.A. Lee: Supervision, investigation. D.P. Hartley: Supervision, writing–review and editing. S.M. Rothenberg: Conceptualization, formal analysis, supervision, visualization, methodology, writing–original draft, writing–review and editing.
References
- 1. Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov 2012;2:227–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Katayama R, Shaw AT, Khan TM, Mino-Kenudson M, Solomon BJ, Halmos B, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung cancers. Sci Transl Med 2012;4:120ra17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012;483:100–3. [DOI] [PubMed] [Google Scholar]
- 4. Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039–43. [DOI] [PubMed] [Google Scholar]
- 5. Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov 2014;4:80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dagogo-Jack I, Yoda S, Lennerz JK, Langenbucher A, Lin JJ, Rooney MM, et al. MET alterations are a recurring and actionable resistance mechanism in ALK-positive lung cancer. Clin Cancer Res 2020;26:2535–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bosch A, Li Z, Bergamaschi A, Ellis H, Toska E, Prat A, et al. PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor-positive breast cancer. Sci Transl Med 2015;7:283ra51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nichols RJ, Haderk F, Stahlhut C, Schulze CJ, Hemmati G, Wildes D, et al. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat Cell Biol 2018;20:1064–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med 2019;380:1929–40. [DOI] [PubMed] [Google Scholar]
- 10. Lu H, Liu C, Velazquez R, Wang H, Dunkl LM, Kazic-Legueux M, et al. SHP2 Inhibition overcomes RTK-mediated pathway reactivation in KRAS-mutant tumors treated with MEK inhibitors. Mol Cancer Ther 2019;18:1323–34. [DOI] [PubMed] [Google Scholar]
- 11. Agazie YM, Hayman MJ. Molecular mechanism for a role of SHP2 in epidermal growth factor receptor signaling. Mol Cell Biol 2003;23:7875–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Montagner A, Yart A, Dance M, Perret B, Salles JP, Raynal P. A novel role for Gab1 and SHP2 in epidermal growth factor-induced ras activation. J Biol Chem 2005;280:5350–60. [DOI] [PubMed] [Google Scholar]
- 13. Liotti F, Kumar N, Prevete N, Marotta M, Sorriento D, Ieranò C, et al. PD-1 blockade delays tumor growth by inhibiting an intrinsic SHP2/Ras/MAPK signalling in thyroid cancer cells. J Exp Clin Cancer Res 2021;40:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hanafusa H, Torii S, Yasunaga T, Matsumoto K, Nishida E. Shp2, an SH2-containing protein-tyrosine phosphatase, positively regulates receptor tyrosine kinase signaling by dephosphorylating and inactivating the inhibitor sprouty. J Biol Chem 2004;279:22992–5. [DOI] [PubMed] [Google Scholar]
- 15. Bennett AM, Tang TL, Sugimoto S, Walsh CT, Neel BG. Protein-tyrosine-phosphatase SHPTP2 couples platelet-derived growth factor receptor beta to Ras. Proc Natl Acad Sci U S A 1994;91:7335–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li W, Nishimura R, Kashishian A, Batzer AG, Kim WJ, Cooper JA, et al. A new function for a phosphotyrosine phosphatase: linking GRB2-Sos to a receptor tyrosine kinase. Mol Cell Biol 1994;14:509–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fedele C, Ran H, Diskin B, Wei W, Jen J, Geer MJ, et al. SHP2 Inhibition prevents adaptive resistance to MEK inhibitors in multiple cancer models. Cancer Discov 2018;8:1237–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dardaei L, Wang HQ, Singh M, Fordjour P, Shaw KX, Yoda S, et al. SHP2 inhibition restores sensitivity in ALK-rearranged non-small-cell lung cancer resistant to ALK inhibitors. Nat Med 2018;24:512–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ou SI, Koczywas M, Ulahannan S, Janne PA, Pacheco JM, Burris HA, et al. The SHP2 inhibitor RMC-4630 in patients with KRAS-mutant non-small cell lung cancer: preliminary evaluation of a first-in-man phase 1 clinical trial. Sixth AACR-IASLC International Joint Conference: Lung Cancer Translational Science from the Bench to the Clinic; 2020 Jan 11–14; San Diego, CA. Plenary Session 1. Available from: https://www.revmed.com/media/shp2-inhibitor-rmc-4630-patients-kras-mutant-non-small-cell-lung-cancer-preliminary-0.
- 20. Brana I, Shapiro G, Johnson ML, Yu HA, Robbrecht D, Tan DSW, et al. Initial results from a dose finding study of TNO155, a SHP2 inhibitor, in adults with advanced solid tumors. J Clin Oncol 39, 2021. (suppl 15; abstr 3005). [Google Scholar]
- 21. Ahmed TA, Adamopoulos C, Karoulia Z, Wu X, Sachidanandam R, Aaronson SA, et al. SHP2 drives adaptive resistance to ERK signaling inhibition in molecularly defined subsets of ERK-dependent tumors. Cell Rep 2019;26:65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koczywas M, Haura EB, Janne PA, Pacheco JM, Ulahannan S, Wang JS, et al. Anti-tumor activity and tolerability of the SHP2 inhibitor RMC-4630 as a single agent in patients with RAS-addicted solid cancers. American Association for Cancer Research Annual Meeting 2021, Virtual Meeting I; 2021 Apr 10–15. Presentation LB001. Available from: https://www.revmed.com/media/anti-tumor-activity-and-tolerability-shp2-inhibitor-rmc-4630-single-agent-patients-ras.
- 23. Falchook G, Li BT, Marrone KA, Bestvina CM, Langer CJ, Krauss JC, et al. Sotorasib in combination with RMC-4630, a SHP2 inhibitor, in KRAS p.G12C-mutated NSCLC and other solid tumors [abstract]. J Thorac Oncol 2022;17(9 Suppl S8);OA03.03. Available from: 10.1016/j.jtho.2022.07.022. [DOI] [Google Scholar]
- 24. Drilon A, Ou SI, Cho BC, Kim D-W, Lee J, Lin JJ, et al. Repotrectinib (TPX-0005) is a next-generation ROS1/TRK/ALK inhibitor that potently inhibits ROS1/TRK/ALK solvent-front mutations. Cancer Discov 2018;8:1227–36. [DOI] [PubMed] [Google Scholar]
- 25. Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 2011;67:235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pannu NS, Murshudov GN, Dodson EJ, Read RJ. Incorporation of prior phase information strengthens maximum-likelihood structure refinement. Acta Crystallogr D Biol Crystallogr 1998;54:1285–94. [DOI] [PubMed] [Google Scholar]
- 27. Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 2011;67:355–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004;60:2126–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data Title Page, Methods, References
Supplementary Table 1: X-ray crystallography data collection and refinement statistics
Supplementary Table 2: Phosphatase Profiling of PF-07284892
Supplementary Table 3: IC50 values for PF-07284892 in cell-based assays
Supplementary Table 4: Pharmacokinetics of PF-07284892 in preclinical species
Supplementary Table 5: Plasma and brain tissue concentrations, brain-to-plasma ratios, and brain concentration over IC50 values for PF-07284892 and TNO155 at dose levels preferred for concomitant administration
Supplementary Table 6: Disposition of all patients enrolled to Dose Level 1
Supplementary Table 7: Adverse events for Dose Level 1 patients
Supplementary Table 8: Comparison of clinical safety data for SHP2 inhibitors
Supplementary Table 9: Sources of antibodies
Supplementary Figure 1: Comparison of the structures of PF-07284892, RMC445507, TNO155 bound to SHP2
Supplementary Figure 2: Intermittent dosing of PF-07284892 preserves efficacy and improves tolerability preclinically
Supplementary Figure 3: Preclinical development of PF-07284892
Supplementary Figure 4: PF-07284892 combination efficacy in additional oncogenic driver-positive mouse cancer models and monotherapy PK/PD
Supplementary Figure 5: Study schema
Supplementary Figure 6: PK profiles for patients
Supplementary Figure 7: PF-07284892 sensitizes a patient with GOPC-ROS1 fusion-positive pancreatic cancer to lorlatinib
Data Availability Statement
Upon request, and subject to review, Pfizer, Inc. will provide the data that support the findings of this study. Subject to certain criteria, conditions, and exceptions, Pfizer, Inc. may also provide access to the related individual deidentified participant data. See https://www.pfizer.com/science/clinical-trials/trial-data-and-results for more information.