Abstract
Arbuscular mycorrhizal (AM) fungi - Glomeromycota and Endogonomycetes - comprise multiple species and higher-level taxa that have remained undescribed. We propose a mixed morphology- and DNA-based classification framework to promote taxonomic communication and shed light into the phylogenetic structure of these ecologically essential fungi. Based on eDNA samples and long reads as type materials, we describe 15 new species and corresponding genera (Pseudoentrophosporakesseensis, Hoforsarebekkae, Kahvenarebeccae, Kelottijaerviashannonae, Kungsaengenashadiae, Langduoadianae, Lehetuaindrekii, Lokrumastenii, Moosteastephanieae, Nikkaluoktamahdiehiae, Parniguacraigii, Riederbergasylviae, Ruuacoralieae, Tammsaareavivikae and Unemaeeanathalieae), the genus Parvocarpum as well as 19 families (Pseudoentrophosporaceae, Hoforsaceae, Kahvenaceae, Kelottijaerviaceae, Kungsaengenaceae, Langduoaceae, Lehetuaceae, Lokrumaceae, Moosteaceae, Nikkaluoktaceae, Parniguaceae, Riederbergaceae, Ruuaceae, Tammsaareaceae, Unemaeeaceae, Bifigurataceae, Planticonsortiaceae, Jimgerdemanniaceae and Vinositunicaceae) and 17 orders (Hoforsales, Kahvenales, Kelottijaerviales, Kungsaengenales, Langduoales, Lehetuales, Lokrumales, Moosteales, Nikkaluoktales, Parniguales, Riederbergales, Ruuales, Tammsaareales, Unemaeeales, Bifiguratales and Densosporales), and propose six combinations (Diversisporabareae, Diversisporanevadensis, Fuscutatacerradensis, Fuscutatareticulata, Viscosporadeserticola and Parvocarpumbadium) based on phylogenetic evidence. We highlight further knowledge gaps in the phylogenetic structure of AM fungi and propose an alphanumeric coding system for preliminary communication and reference-based eDNA quality-filtering of the remaining undescribed genus- and family-level groups. Using AM fungi as examples, we hope to offer a sound, mixed framework for classification to boost research in the alpha taxonomy of fungi, especially the “dark matter fungi”.
Key words: Dark taxa, DNA-based classification, holotype, molecular phylogeny, species description
Introduction
Arbuscular mycorrhizal (AM) fungi play a crucial role in mineral nutrition and stress alleviation of a vast majority of vascular plants (Brundrett and Tedersoo 2018), especially in the grassland and tropical forest ecosystems (Soudzilovskaia et al. 2019). Besides higher plants, AM fungi also associate with certain liverworts (Marchantiophyta) and hornworts (Anthocerotophyta), forming arbuscule-like structures in their thalli and improving their access to nutrients in soil (Ligrone et al. 2007; Bidartondo and Duckett 2010).
Traditionally, only members of the phylum Glomeromycota (occasionally considered as subphylum Densosporales within Mucoromycota, sensu Spatafora et al. (2016)) have been recognised as AM mycobionts (Smith and Read 2008; Varma et al. 2017). However, there is strong morphological and molecular evidence for AM associations between Endogonomycetes (subphylum Mucoromycotina within Mucoromycota) and various plant groups, including hornworts, liverworts and herbaceous vascular plant species (Bidartondo et al. 2011; Desiro et al. 2013; Bonfante and Venice 2020). Further molecular evidence for endogonomycete associations in plant roots (Orchard et al. 2017b) suggests that these fungi may be as important as Glomeromycota in AM associations in evolutionary (Hoysted et al. 2018) and ecological (Orchard et al. 2017b) terms. Except for Geosiphon, taxa of Glomeromycota are recognised as obligate root symbionts, but such information is lacking for Endogonomycetes. Certain small groups of Endogonomycetes are known as ectomycorrhizal symbionts (Tedersoo and Smith 2017) or saprotrophs (Berch and Fortin 1983; Hirose et al. 2014). Members of Endogonomycetes may form macroscopic (semi)hypogeous fruiting bodies containing sexual zygospores or asexual chlamydospores. A few species of Glomeromycota form such fruiting bodies containing chlamydospores (also termed glomerospores), but most species of Glomeromycota produce single glomerospores on hyphal tips in soil. The few in vitro culturable species of Glomeromycota can be grown exclusively in co-culture (except Geosiphon) with plant roots (but see Tanaka et al. (2022)). Some saprotrophic, ectomycorrhizal and AM species of Endogonomycetes can be grown in pure culture (e.g. Field et al. (2015)).
Given the high attention on Glomeromycota as the primary AM root symbionts, their taxonomy is relatively well established, with three classes, six orders, 17 families and 52 genera accepted (Wijayawardene et al. 2022; Błaszkowski et al. 2022, 2023; da Silva et al. 2023). Conversely, the Endogonomycetes comprise a single order (Endogonales), two families and seven genera (Wijayawardene et al. 2022). The DNA samples of two genera of Endogonomycetes (Peridiospora and Sclerogone) have never been sequenced due to difficulties accessing old collections.
Much of the Glomeromycota DNA barcoding and phylogenetics research relies on the rRNA internal transcribed spacer (ITS) region and 5’ quarter of the 28S gene (LSU). However, all nuclear LSU, ITS region and 18S rRNA gene (SSU) are nearly equally used for molecular identification from soil and plant roots. The geographically most inclusive studies have focused on either the SSU marker (Davison et al. 2015; Vasar et al. 2022) or the ITS region (Tedersoo et al. 2014, 2021; Kivlin 2020; Mikryukov et al. 2023). Due to the paucity of species-level reference data, the SSU-based surveys suffer relatively more from poor species- and genus-level identification (Tedersoo et al. 2024). In Endogonomycetes, taxonomic studies have used all SSU, ITS and LSU markers. Short-read endogonomycete ITS1 and ITS2 sequences derived from general soil fungal surveys are common in the International Nucleotide Sequence Databases Consortium (INSDC), but the few endogonomycete AM-focused studies have used a long marker fragment of SSU (e.g. Bidartondo et al. (2011); Albornoz et al. (2022)). For the identification from soil or roots, sufficient coverage of both groups requires the use of specific primers. Therefore, focusing on one of the AM groups reduces the amplification of the other (Seeliger et al. 2023). Molecular identification of AM fungi has been heavily biased towards the Glomeromycota, whereas Endogonomycetes have been virtually ignored in 99% of molecular surveys of AM fungi in the last 15 years. For both groups, several undescribed family- or order-level taxa have been revealed based on eDNA, suggesting that much of the taxonomic and phylogenetic diversity remains yet to be described (Desiro et al. 2013; Öpik et al. 2014).
Historically, species of both Endogonomycetes and Glomeromycota have been described in the genus Endogone Link (erected by Link (1809)) that was used in a cross-phylum sense until 1980s, although the genus Glomus Tul. & C. Tul. was erected for G.macrocarpum nearly 150 years earlier (Tulasne and Tulasne 1844). Most of the glomeromycotan species were later transferred to Glomus under the family Glomeraceae (Pirozynski and Dalpé 1989), order Glomerales (Morton and Benny 1990), class Glomeromycetes (Cavalier-Smith 1998) and phylum Glomeromycota (Schüssler et al. 2001). In the last two decades, the initial large genera Glomus and Endogone were split into multiple smaller genera based on combined morphological and molecular analyses. Additional families and orders of Glomeromycota were described by Schüssler et al. (2001), Walker and Schüssler (2004) and Błaszkowski et al. (2021). Gigasporales and Entrophosporales were erected from Glomerales more recently (Gautam and Patel 2007; Błaszkowski et al. 2022). Paraglomerales and Archaeosporales were assigned class rank (Oehl et al. 2011). Recently, the class Endogonomycetes was erected (Doweld 2014) to include the Endogonales (Jaczewski and Jaczewski 1931), covering the mucoromycotan families Endogonaceae (Saccardo 1889) and Densosporaceae (Desiro et al. 2017) as well as two order-level clades “GS21” and “GS22” (Tedersoo et al. 2017) recognised based on soil eDNA samples.
The main purpose of this article is to develop a mixed phylogenetic classification framework that integrates environmental DNA (eDNA) sequences into a specimen-based classification system, which is particularly relevant for high-diversity and cryptic taxonomic groups, such as AM fungi with predicted richness of thousands of species. Already three decades ago, it was stated: “It is unavoidable that DNA will serve as character source for contemporary taxonomic descriptions” (cf. Reynolds and Taylor (1991:311)). Such a mixed morphology- and eDNA-based classification framework is expected to facilitate species discovery and promote work on alpha taxonomy. “Leaving this diversity unnamed or unclassified is not an option, as it would continue to be an enormous and increasing impediment to communication and research in the field” (cf. Lücking and Hawksworth (2018:146)). Fungal species with names improve our capacity to refer to particular organisms and facilitate biodiversity surveys, conservation planning and assessment of toxic, pathogenic and mutualistic organisms in a direct way (Ryberg and Nilsson 2018; Lücking et al. 2021). Furthermore, a well-structured taxonomic hierarchy would offer additional possibilities for using phylodiversity and evolutionary methods without performing phylogenetic analyses (Tedersoo et al. 2018), and it would improve taxonomy-aware chimera filtering in reference-based methods for metabarcoding analyses (Nilsson et al. 2010). The main shortfalls of sequence-based classification include eroding the concept of physical type material and parallel classifications based on specimens and sequences or using different DNA markers (Hongsanan et al. 2018; Lücking and Hawksworth 2018; Thines et al. 2018). Therefore, many leading fungal taxonomists do not approve use of DNA sequences (Thines et al. 2018; Zamora et al. 2018) or eDNA sample (Hongsanan et al. 2018) as holotypes.
Here, we use the mixed specimen-eDNA phylogenetic classification framework to shed light into the phylogenetic diversity of the two groups of AM fungi - Glomeromycota and Endogonomycetes. By using eDNA samples as holotypes (Reynolds and Taylor 1991; Renner 2016), DNA sequences as lectotypes and diagnoses based on molecular differences in ITS and LSU marker genes (Renner 2016), we first describe novel species for the highly divergent groups of AM fungi following the International Code of Nomenclature for Algae, Fungi and Plants (Turland et al. 2018; Lücking et al. 2021). Building on these species, we then introduce novel families and orders. Finally, we provide a large number of taxonomically re-annotated and novel SSU, ITS and LSU sequences, equipped with preliminary alphanumeric taxonomic identifiers, where relevant, to the scientific community.
Materials and methods
We downloaded the sequence data identified as Glomeromycota, Mucoromycota and uncultured fungi from three nucleotide sequence databases - NCBI (Sayers et al. 2024; https://www.ncbi.nlm.nih.gov/), UNITE v.9.1 (Abarenkov et al. 2024; https://unite.ut.ee/) and EUKARYOME v.1.7 (Tedersoo et al. 2024; https://eukaryome.org/). We also added rRNA gene sequences from scaffolds in the Joint Genome Institute data portal (https://genome.jgi.doe.gov/portal/). The unidentified fungi were first assigned to rough taxonomic groups based on BLASTn queries against identified sequences in EUKARYOME v.1.7. For sequences affiliated with Glomeromycota or Endogonomycetes, we conducted phylogenetic analyses separately for the SSU, LSU and a longer fragment spanning much of SSU, ITS and LSU. A large part of the ITS region was not used for the phylogeny reconstruction because of alignment unreliability. The sequences of Glomeromycota and Endogonomycetes were aligned using MAFFT v.7 (Katoh and Standley 2013), followed by manual trimming of overarching and misaligned ends and manual correction in case of obvious misalignments using AliView v.1.26 (Larsson 2014). The alignments were further trimmed to exclude unalignable regions and subjected to ClipKIT v.1.4.0 (Steenwyk et al. 2020) to remove phylogenetically uninformative positions, including rare introns and insertions. Based on the alignments, we visually evaluated mismatches to commonly used primers targeting SSU, ITS and LSU regions.
Phylogenetic analyses were performed using IQ-TREE v.2.2.5 (Minh et al. 2020), with standard options including 1000 trees and 1000 ultrafast bootstrap replicates. The trees were visualised and used for taxonomic re-annotation in FigTree v.1.4.4 (Rambaut 2018). Various taxa of Mucoromycota with relatively short branches were tested as potential outgroups. The first three rounds of alignments and analyses were primarily used to detect and remove low-quality reads and chimeric sequences. From the fourth round onwards, the reads were assigned to clades corresponding to putative genera, families and orders, following the monophyly criterion and accounting for the level of sequence divergence in previously described groups. We included at least one read from each described species to delimit clades and assign taxonomy. For both Glomeromycota and Endogonomycetes, we focused mainly on the long fragment covering the ITS and LSU regions because of: 1) the greatest taxonomic resolution in the ITS2 and D2 subregion of LSU, 2) the occurrence of the largest number of described species and 3) the presence of most abundant and diverse set of eDNA reads from soil and roots falling into these groups (Tedersoo et al. 2024).
Diagnoses of species were prepared based on molecular characters in the ITS and LSU regions by selecting the most characteristic short barcodes (20–30 bases) for the target species using multiple sequence alignments. The barcodes typically had no ambiguous position for the target species and had at least two differences from closely-related species. We also estimated the number of mutations (i.e. alignment mismatches) allowed for the target species to be separable from related species (typically set to 0 or 1). For the entire alignment length of ITS and LSU, we estimated the maximum proportion of differences amongst sequences corresponding to the target species (i.e. within-species variability).
For establishing higher-ranking taxa such as genera, families and orders, we used the following criteria: i) monophyly; ii) bootstrap support >95; iii) phylogenetic breadth and divergence roughly comparable to previously described taxa; and iv) minimising the number of novel taxa (i.e. preferably retaining larger groups if there were multiple alternative splitting possibilities). Based on a visual assessment of the ITS and LSU alignments and phylograms, we predicted the approximate number of (potential) species for each newly-described genus (but extrapolation to unobserved taxa was not attempted).
The eDNA samples with the highest proportions of target reads were selected as holotypes, except in the cases where long reads spanning SSU, ITS and LSU were available along with the stored DNA samples. Lectotypes were identified amongst the highest-quality sequences derived from these holotype DNA samples. Most of the type materials and additional samples were derived from composite topsoil samples (40 subsamples of 5 cm diam. to 5 cm depth from 2500-m2 area) of the Global Soil Mycobiome consortium (GSMc) project (Tedersoo et al. 2021), FunAqua sediment samples (V. Prins et al., unpublished) or from various soil samples sequenced by Jamy et al. (2022). Both eDNA and corresponding substrate samples are maintained as vouchered collections in the repository of the University of Tartu (acronym TUE, with 6-digit accession numbers). The sequences were first deposited in the EUKARYOME database (denoted by “EUK” with 7-digit accession numbers) and subsequently submitted to the INSDC and UNITE databases. EUKARYOME v.1.9.2 includes 55,648 and 10,081 annotated reads of Glomeromycota and Endogonomycetes, respectively.
Results
Phylogeny of Glomeromycota
The SSU-ITS-LSU phylogram supported the separation of all described Glomeromycota orders and families, and placed these into expected positions (Fig. 1, Suppl. material 1) as in previous analyses based on rRNA gene and partial genomes (Stockinger et al. 2012; Montoliu-Nerin et al. 2021; Rosling et al. 2024). A vast majority of valid genera were separated from each other with high statistical support. As exceptions, the genera Otospora Oehl, Palenzuela & N.Ferrol (O.bareae) and Tricispora Oehl, Sieverd., G.A.Silva & Palenz. (T.nevadensis) were nested within Diversispora C.Walker & A.Schüssler, whereas species of Dentiscutata Sieverd., F.A.Souza & Oehl (D.cerradensis and D.reticulata) were placed within Fuscutata Oehl, F.A.Souza & Sieverd. The type species of Dentiscutata (D.nigra) was not sequenced for the ITS region, but an analysis of the LSU region indicated that D.nigra is placed separately from other species of Dentiscutata that clustered with Fuscutata (Suppl. material 2). Corymbiglomuscorymbiforme Błaszk. & Chwat – the type species of this genus – was nested within the genus Redeckera C.Walker & A.Schüssler, whereas C.globiferum (Koske & C.Walker) Błaszk. & Chwat served as a sister group to species of Redeckera. Furthermore, the recently described genus Blaszkowskia G.A.Silva & Oehl was nested within Viscospora. Where relevant, we propose new combinations (see below).
Figure 1.
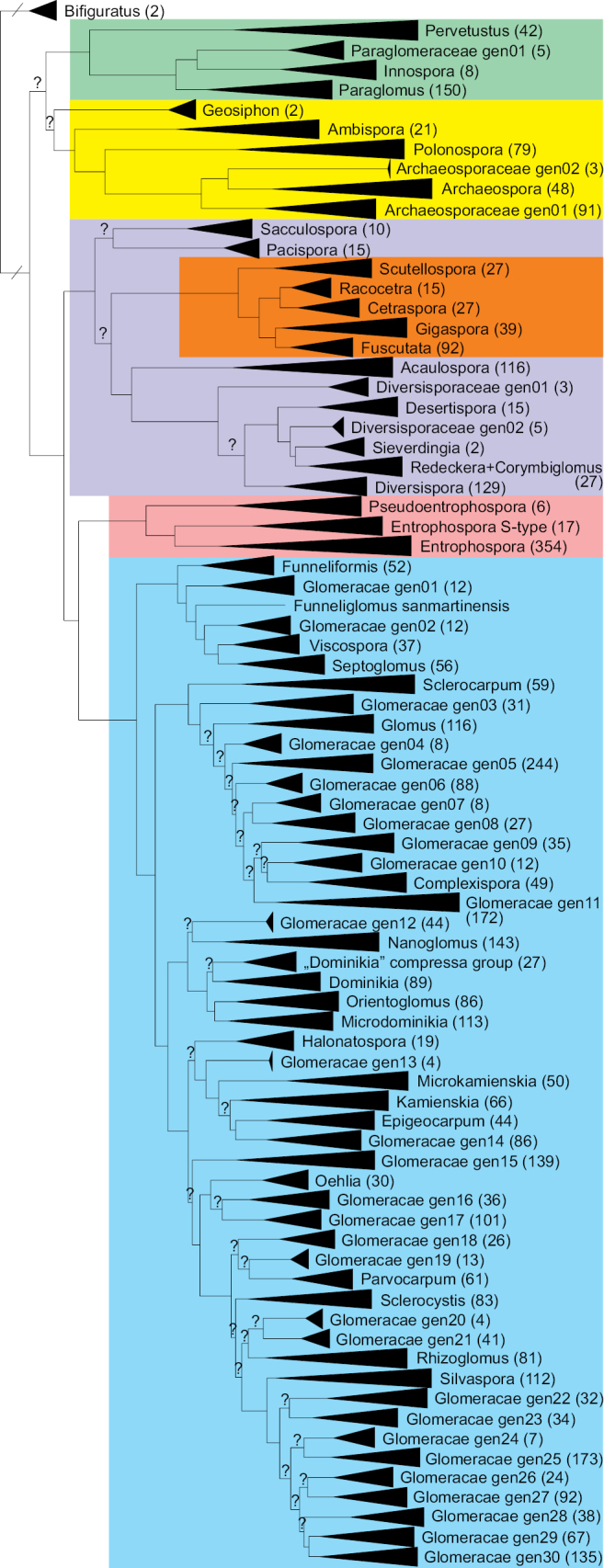
Phylogenetic position of genera and genus-level groups of Glomeromycota based on Maximum Likelihood analysis of SSU-ITS-LSU sequences. The clades are collapsed, with the number of sequences included in parentheses. Question marks above branches indicate low ultra-rapid bootstrap support (< 95). Orders are highlighted in different colours. For the genus Entrophospora, the so-called S-type reads (VanKuren et al. 2013) were included for comparison. The uncollapsed phylogram is given in Suppl. material 1.
Of previously described species, Dominikiacompressa (Sieverd., Oehl, Palenz., Sánchez-Castro & G.A.Silva) Oehl, Palenz., Sánchez-Castro & G.A.Silva (basionym Glomuscompressum Sieverd., Oehl, Palenz., Sánchez-Castro & G.A.Silva) formed a well-supported group in a sister position to the rest of Dominikia Błaszk., Chwat & Kovács, but their close relationship was poorly supported and inconsistent amongst various phylograms prepared. Thus, D.compressa is currently being transferred to a new genus (J. Błaszkowski et al., in prep.). Similarly, Glomusbadium Oehl, D.Redecker & Sieverd. was placed outside the genus Glomus as a well-supported small clade, but its sister relationships with other genera remain unresolved. Based on both phylogenetic and morphological characters, we propose to treat G.badium as a new genus, herein designated as Parvocarpum (see below).
Our phylogenetic analysis revealed a large number of previously undescribed or unsequenced taxa. One of these taxa was located as a deep clade in the Entrophosporales, which warrants consideration as a new family outside the Entrophosporaceae. We describe the new species, genera and families based on eDNA samples and sequences. The Archaeosporaceae and Diversisporaceae families each revealed two novel genus-level taxa, whereas the Paraglomeraceae harboured one new genus-level taxon. The most prominent family – Glomeraceae – was found to include 30 novel genus-level taxa clearly distinct from current delimitations of known genera based on our criteria. We propose informal alphanumeric labels for these genera to enable their communication (see below). For the Glomeraceae, it is most likely that, upon DNA sequencing of materials belonging to unsequenced species, many will fall into these unnamed groups (like the cases of D.compressa and G.badium).
Phylogeny of Endogonomycetes
The SSU-5.8S-LSU phylogram resolved the internal structure of Endogonomycetes reasonably well, except the order of divergence for most of the 17 main, deep-branching groups (Fig. 2, Suppl. material 3). The hitherto described and sequenced species of Endogone and Jimgerdemannia Trappe, Desirò, M.E.Sm., Bonito & Bidartondo, as well as Densospora McGee and Sphaerocreas Sacc. & Ellis, formed two relatively large order-level groups that were distantly related to each other and surrounded by eDNA sequences derived from soil. Species of Bifiguratus Torres-Cruz & Porras-Alfaro formed a small, deep-branching, order-level group, with no clear sister group. For each of the additional 14 order-level groups, we described new species based on eDNA sample and sequence information. These 14 species were further assigned to genera and families based on the internal branching structure of these orders, with other unnamed groups labelled alphanumerically. Two potentially order-level groups remain undescribed and unlabelled, because these were found from a single locality and their phylogenetic position may change with extra sequences and more precise alignments.
Figure 2.
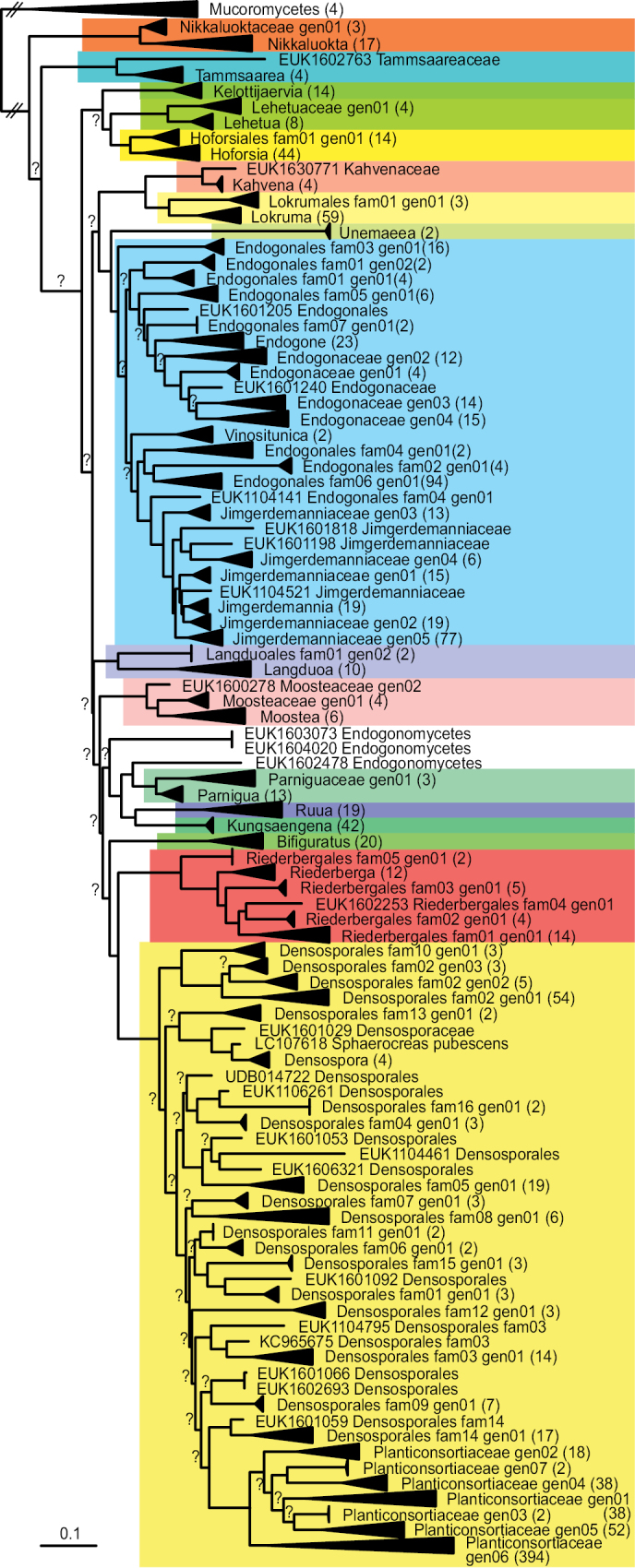
Phylogenetic position of genera and genus-level groups of Endogonomycetes based on Maximum Likelihood analysis of SSU-5.8S-LSU sequences. The clades are collapsed, with the number of sequences included in parentheses. Question marks above branches indicate low ultra-rapid bootstrap support (< 95). Orders are highlighted in different colours. The uncollapsed phylogram is given in Suppl. material 3.
The additional SSU phylogram confirmed the separation of the main orders, although nearly half of them lacked SSU sequence data or were represented by a single read (Suppl. material 4). Furthermore, the SSU phylogram indicated that the vast majority of putative AM fungi were widely dispersed in the groups corresponding to Densosporales (mostly Densosporaceae and Planticonsortiaceae), Endogonales (including Endogonaceae, Jimgerdemanniaceae and other family-level taxa) and Hoforsales (formerly clade GS22), and to some extent in Bifiguratales. However, a few root-derived sequences fell outside these groups, suggesting that certain other orders lacking the SSU sequence data may also host AMF.
Taxonomic combinations, emendations and descriptions in Glomeromycota
. Diversispora bareae
(Palenz., N.Ferrol & Oehl) Tedersoo & Magurno comb. nov.
DA57AE55-27B2-5141-BD8F-039113A0E4D3
853545
Otospora bareae Palenz., N.Ferrol & Oehl, in Palenzuela, Ferrol, Boller, Azcón-Aguilar & Oehl, Mycologia 100(2): 298 (2008). Basionym.
Description.
As presented originally in Palenzuela et al. (2008).
Diagnosis.
Diversisporabareae differs from other species of the Diversispora by producing acaulosporoid (otosporoid) spores compared with diversisporoid and entrophosporoid (tricisporoid) spores in other described species. Glomerospores with inner flexible hyaline layer and pigmented sporiferous saccule. Phylogenetically belongs to Diversispora based on the SSU-ITS-LSU phylogram (Fig. 1, Suppl. material 1).
Notes.
The new combination invites an amendment of the genus Diversispora to accommodate species with otosporoid spores.
. Diversispora nevadensis
(Palenz., N.Ferrol, Azcón-Aguilar & Oehl) Tedersoo & Magurno comb. nov.
64AF15C7-631D-5EF7-B44C-D219A627DB40
853546
Entrophospora nevadensis Palenz., N.Ferrol, Azcón-Aguilar & Oehl, in Palenzuela, Barea, Ferrol & Azcón-Aguilar, Mycologia 102(3): 627 (2010). Basionym.
Description.
Diagnosis.
Diversisporanevadensis differs from other species of the Diversispora by producing entrophosporoid (tricisporoid) spores compared with diversisporoid and acaulosporoid (otosporoid) spores in other species. Glomerospores with inner flexible hyaline wall layers without granular beaded surface and no Melzer reaction. Phylogenetically nested in Diversispora based on the SSU-ITS-LSU phylogram (Fig. 1, Suppl. material 1).
Notes.
The new combination invites an amendment of the genus Diversispora to accommodate species with entrophosporoid (tricisporoid) spores.
. Diversispora
C.Walker & A.Schüssler emend. Tedersoo & Magurno
1613A7B6-E4AE-55A3-A34A-9797979835A7
28884
Type species.
Diversisporaspurca (C.M.Pfeiffer, C.Walker & Bloss) C.Walker & Schüssler.
Description.
Spores diversisporoid, rarely otosporoid or tricisporoid. Diversisporoid spores formed singly, in clusters or in large disorganised fruiting bodies with high spore numbers. Spores with 1–4 wall layers; pores often closed with a septum. Subtending hyphal pores rarely open. Otosporoid spores formed laterally on the persistent neck of a sporiferous saccule. Tricisporoid spores with inner flexible hyaline wall layers (formed de novo) without granular beaded surface and no Melzer reaction. Spore pores generally closed by a septum at the spore base, arising from the innermost wall lamina or inner layer or from both. Forms a monophyletic group within Diversisporaceae based on the SSU-ITS-LSU phylogram (Fig. 1, Suppl. material 1).
. Fuscutata cerradensis
(Spain & J. Miranda) Tedersoo & Magurno comb. nov.
C73638C5-35FF-5D57-8DD7-7EAC3C32C667
853547
Scutellospora cerradensis Spain & J. Miranda, Mycotaxon 60: 130 (1996). Basionym.
Dentiscutata cerradensis Sieverd., F.A.Souza & Oehl, Mycotaxon 106: 342 (2009). Synonym.
Description.
Diagnosis.
Fuscutatacerradensis differs from other species of the Fuscutata by spore wall ornamentation, three-walled spores and dark-pigmented multilobed germinal shield produced in the inner wall. Phylogenetically forms a monophyletic clade with F.heterogama - the type species of genus - based on the SSU-ITS-LSU phylogram (Fig. 1, Suppl. material 1).
Notes.
The new combination invites an amendment of genus Fuscutata to accommodate species with dark, multilobed germinal shields. However, we decided not to prepare an amendment for Fuscutata because the genus Dentiscutata, their close relative, requires additional information to confirm their status, supported only in the LSU sequence of D.nigra.
. Fuscutata reticulata
(Koske, D.D.Mill. & C.Walker) Tedersoo & Magurno comb. nov.
BBD6E7DB-B766-5D21-BBAE-BB0CC7E23B0A
853548
Gigaspora reticulata Koske, D.D.Mill. & C.Walker, Mycotaxon 16(2): 429 (1983). Basionym.
Dentiscutata reticulata (Koske, D.D.Mill. & C.Walker) Sieverd., F.A.Souza & Oehl, Mycotaxon 106: 342 (2009). Synonym.
Description.
See Koske et al. (1983).
Diagnosis.
Fuscutatareticulata differs from other species of the Fuscutata by spore wall ornamentation, three-walled spores and dark-pigmented, multilobed germinal shield produced in the inner wall. Phylogenetically forms a monophyletic clade with F.heterogama - type species of genus - based on the SSU-ITS-LSU phylogram (Fig. 1, Suppl. material 1).
Notes.
See note of F.cerradensis.
. Viscospora deserticola
(Trappe, Bloss & J.A.Menge) Tedersoo & Magurno comb. nov.
40E147F5-E4AD-5218-9307-DE6B082407FB
853549
Glomus deserticola Trappe, Bloss & J.A.Menge, Mycotaxon 20 (1): 123 (1984). Basionym.
Blaszkowskia deserticola (Trappe, Bloss & J.A.Menge) Oehl & G.A.Silva, Mycol. Progr. 22 (11, no. 74): 5 (2023). Synonym.
Description.
See Trappe et al. (1984).
Diagnosis.
Subtending hyphae pigmented over long distances (>100 μm) unlike in other species of Viscospora and Septoglomus. Differs from other species of Viscospora by spore colour (da Silva et al. 2023).
Notes.
Transferred from Blaszkowskia to Viscospora because of phylogenetic nestedness within Viscospora and recognition as a separate genus would render Viscospora paraphyletic and leave many orphan taxa in the Septoglomus-Viscospora clade (Suppl. material 1; da Silva et al. (2023)).
. Parvocarpum
Magurno gen. nov.
EAB06364-E6B2-57A0-BADE-E28C34CA886E
853558
Type species.
Parvocarpumbadium (Oehl, Redecker & Sieverd.) Magurno.
Description.
Producing glomoid-like spores surrounding a central plexus of interwoven hyphae in small organised fruiting bodies, lacking a peridium. Spores with inner flexible hyaline layer and short subtending hyphae. Forms a monophyletic group within Glomeraceae based on SSU-ITS-LSU phylogram (Fig. 1, Suppl. material 1).
Notes.
Based on ITS and LSU sequences, Parvocarpum includes 10–20 species.
. Parvocarpum badium
(Oehl, Redecker & Sieverd.) Magurno comb. nov.
35B241B1-E9A2-51F5-B40C-EE7943B4FEDF
853560
Glomus badium Oehl, D.Redecker & Sieverd., Angew. Botan. 79: 39 (2005). Basionym.
Funneliformis badius (Oehl, Redecker & Sieverd.) C.Walker & A.Schüssler. Synonymy.\
Description.
See Oehl et al. (2005).
Etymology.
parvus (Latin) = small; and carpum (Latin) = body, referring to the small size of fruiting bodies produced.
Diagnosis.
P.badium differs from other genera of the Glomeraceae by producing glomoid-like spores surrounding a central plexus of interwoven hyphae in small organised fruiting bodies, lacking a peridium. Spores with inner flexible hyaline layer and short subtending hyphae. Phylogenetically distinct from G.macrocarpum and other Glomussens. str. species based on the SSU-ITS-LSU phylogram (Fig. 1, Suppl. material 1).
Notes.
Phylogenetic position of P.badium within the genus Parvocarpum is unresolved because of a single available short read.
. Pseudoentrophosporaceae
Tedersoo & Magurno fam. nov.
439F61CA-62E7-587A-925F-F81FC46C1DBE
853564
Type genus.
Pseudoentrophospora Tedersoo & Magurno.
Description.
Covers the monophyletic group in Entrophosporales (Fig. 1). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1631429, EUK1105140 and EUK0135500 (Suppl. material 1).
Notes.
Recognised based on eDNA sequences only. Currently monogeneric.
. Pseudoentrophospora
Tedersoo & Magurno gen. nov.
799AECC1-5536-50BB-8D37-1A618B2DA4F8
853565
Type species.
Pseudoentrophosporakesseensis Tedersoo & Magurno.
Description.
Covers the monophyletic group in Pseudoentrophosporaceae (Fig. 1). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1631429, EUK1105140 and EUK0135500 (Suppl. material 1).
Notes.
Recognised based on eDNA sequences only. There are potentially 3–6 species in Pseudoentrophospora based on ITS sequences, some of which are represented by sequences EUK1105140 (tropical rainforest soil in El Yunque, Puerto Rico, 18.29°N, -65.78°E); EUK1010525 (GSMc plot S056, tropical rainforest soil in Pegaima Mountains, Guyana, 5.43567°N, -60.08825°E); and EUK0133825 (flooded grassland soil in Dijle, Belgium, 5.83°N, 4.65°E).
. Pseudoentrophospora kesseensis
Tedersoo & Magurno sp. nov.
3EF89F07-3237-56C3-AB4B-37AEDC4F9360
853566
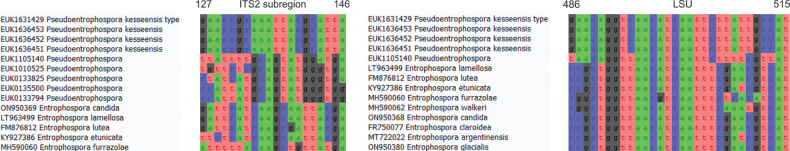
Diagnosis.
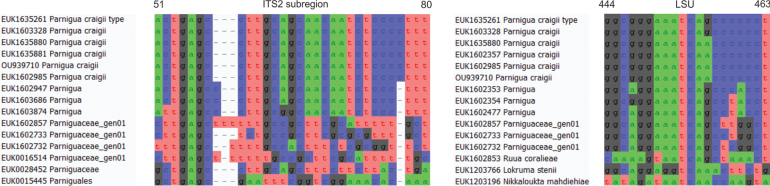
Differs from other species of Pseudoentrophospora and Entrophospora based on the ITS region (ITS2 positions 127–146 gaaccgcaaattacgcatta, one mismatch allowed) and LSU (positions 486–515 gaacaggtcaacatcaattcttattgccat, one mismatch allowed) as indicated in Fig. 3.
Figure 3.
Diagnostic barcodes for Pseudoentrophosporakesseensis relative to closely-related taxa in ITS2 and LSU.
Type.
Soil eDNA sample TUE101916 (holotype); eDNA sequence EUK1631429 (lectotype); GSMc plot G4940, coppiced Juniperus-Acer woodland (soil sample TUE001916) in Kesse Island, Estonia, 58.63443°N, 23.43938°E.
Description.
Other eDNA sequences EUK1636430–EUK1636432 from the type locality.
Etymology.
pseudo (Greek) = false; Entrophospora (Latin) refers to a related fungal genus; and kesseensis (Latin) indicates locality of the type species. The name depicts phylogenetic relatedness to Entrophosphora and the only locality where the type species has been recorded.
Notes.
Found from a single site, with ITS and LSU sequences differing up to 0.5% and 1%, respectively. The ITS1 subregion harbours only 58 bases, being amongst the shortest across fungi (excl. microsporidians).
Taxonomic descriptions of Endogonomycetes
. Endogonomycetes
Doweld emend. Tedersoo
2E5A33C3-7C77-5BC3-8FE8-3A22AA0ADEB6
550357
Type order.
Endogonales Jacz. & P.A.Jacz.
Description.
Fruiting body absent, rarely present - hypogeous or on debris, globose, irregular, sometimes resupinate, 1–20 mm in diam., may be composed of aggregated zygosporangial clusters. Reproductive structures as zygosporangia (in Endogone, Jimgerdemannia) or chlamydospores (in Vinositunica, Densospora), aggregated in the fruiting body or as chlamydospores on extraradical hyphae (in Planticonsortium). Chlamydospore wall continuous, multilayered, with dense subtending hyphae, lacking septa. Hyphae filamentous, coenocytic, sometimes with secondary septa, rarely yeast-like (in Bifiguratus). Forms a monophyletic group in Mucoromycota, as the least inclusive clade covering accessions UDB025468, UDB28692, EUK1201418, EUK1203196, EUK1602762, EUK1202520, EUK1203766, EUK1107335 and EUK1602357 (Suppl. material 3).
Notes.
Endogonomycetes harbours currently 17 orders and two unassigned, potentially order-level groups represented by sequences EUK1604020 and EUK1603073 (GSMc plot G3308, Juniperuscommunis coppiced grassland soil in Atla, Estonia, 58.30122°N, 21.93600°E); and EUK1602478 (GSMc plot G4627, mixed forest soil in Tudusoo, Estonia, 59.11368°N, 26.75944°E).
. Hoforsales
Tedersoo ord. nov.
BE12D337-D710-57AD-9C3D-BA3EBCFB80C5
853567
Type family.
Hoforsaceae Tedersoo.
Description.
Covers the monophyletic group in Endogonomycetes (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1100001, EUK1602331 and EUK1602346 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Hoforsaceae and another potentially family-level group, which is represented by sequence EUK1631675 (GSMc plot G4124, Populustremula forest soil in Mäla, Estonia, 58.58693°N, 23.28597°E). Hoforsales corresponds to clade GS22 (sensu Tedersoo et al. (2017)).
. Hoforsaceae
Tedersoo fam. nov.
F545EB4C-FA9F-5CAC-A79E-85C9BAF058D5
853569
Type genus.
Hoforsa Tedersoo.
Description.
Covers the monophyletic group in Hoforsales (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1100001, EUK1107311 and EUK1602325 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently monogeneric.
. Hoforsa
Tedersoo gen. nov.
80B9FA9A-DD5D-56D0-87AE-A603667661BB
853570
Type species.
Hoforsarebekkae Tedersoo.
Description.
Covers the monophyletic group in Hoforsaceae (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1100001, EUK1107311 and EUK1602325 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. There are potentially about 20 species in Hoforsa based on ITS and LSU sequences, with examples including taxa represented by sequences EUK1107311 (bog peat in Svartberget, Sweden, 64.24°N, 19.76°E) and AM260926 (bog peat, Scotland) first isolated by Rebekka Artz (Artz et al. 2007). Most taxa are found from various soils, but the LSU sequence AB982123 originates from an ectomycorrhizal root of Dipterocarpaceae (Lambir, Malaysia). The most common taxon at 99% LSU sequence similarity (EUK1602281) has been recorded from 31 localities in Estonia and Latvia. The genus has a global distribution and it occurs commonly in soil samples but rarely in roots.
. Hoforsa rebekkae
Tedersoo sp. nov.
2025C719-6EB2-5F88-82D3-678705158197
853571
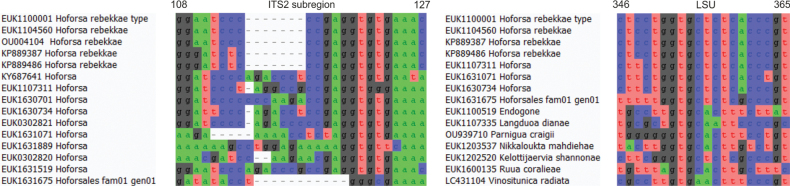
Diagnosis.
Separation from other species of Hoforsa based on the ITS region (ITS2 positions 108–127 ggratcycccgaggtgtgaaac; one mismatch allowed) and LSU (positions 546–565 ctcctggtgctctcacccgt; no mismatch allowed) as indicated in Fig. 4.
Figure 4.
Diagnostic barcodes for Hoforsarebekkae relative to closely-related taxa in ITS2 and LSU.
Type.
Soil eDNA sample: TUE128830 (holotype); eDNA sequence EUK1100001 (lectotype); Pinussylvestris forest near Hofors, Sweden (60.49°N, 16.30°E).
Description.
Other sequences: EUK1104560 (type locality); OU004104 (San Francisco, Ecuador, root sample); and KP889387 and KP889486 (both coniferous forest soil in British Columbia, Canada).
Etymology.
Hofors (Swedish) refers to type locality; and Rebekka (Scotch) refers to the first name Rebekka Artz who was the first to collect materials from this genus.
Notes.
Found from three sites across three continents, with ITS sequences differing up to 3.5% and LSU sequences up to 0.5%.
. Kahvenales
Tedersoo ord. nov.
00199A53-C7D8-5E01-9356-0081EBCF84FA
853572
Type family.
Kahvenaceae Tedersoo.
Description.
Covers the monophyletic group in Endogonomycetes (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1634339 and EUK1630771 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Kahvenaceae.
. Kahvenaceae
Tedersoo fam. nov.
8DEBCDA3-A185-54F9-BBA8-F5B2295633EC
853573
Type genus.
Kahvena Tedersoo.
Description.
Covers the monophyletic group in Kahvenales (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1634339 and EUK1630771 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Kahvena.
. Kahvena
Tedersoo gen. nov.
F24FD4C7-2094-5DC7-A432-F2D97A5626EB
853574
Type species.
Kahvenarebeccae Tedersoo.
Description.
Covers the monophyletic group in Kahvenaceae (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1634339 and EUK1630771 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Based on ITS sequences, Kahvena is comprised of two species; the other represented by sequences EUK1630771 (GSMc plot G4185, Picea-Pinus forest soil in Ristipalo, Estonia, 58.10241°N, 27.47874°E) and ON963629 (Pinussylvestris forest soil, Lithuania).
. Kahvena rebeccae
Tedersoo sp. nov.
EB20E4EE-572F-53DB-849A-4606F20139EB
853575
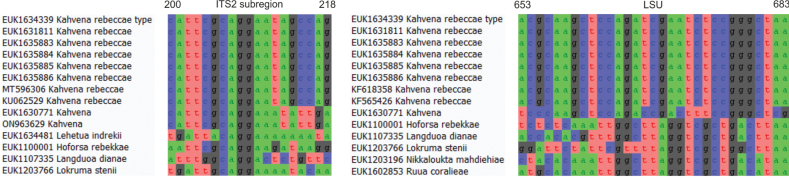
Diagnosis.
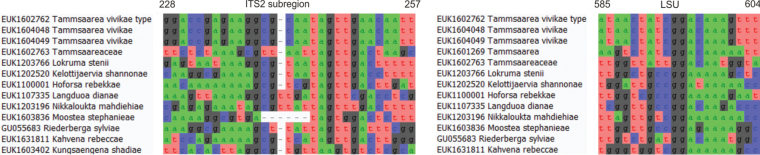
Separation from other species of Kahvena based on the ITS region (ITS2 positions 200–218 cattcgcaggaatagccag; one mismatch allowed) and from other species of Endogonomycetes based on LSU (positions 653–683 acgcaagctccagatcgaatctccgggctaa; one mismatch allowed) as indicated in Fig. 5.
Figure 5.
Diagnostic barcodes for Kahvenarebeccae relative to closely-related taxa in ITS2 and LSU.
Type.
Soil eDNA sample TUE100738 (holotype); eDNA sequence EUK1634339 (lectotype); GSMc plot G4196, Populus-Picea-Pinus forest (soil sample TUE000738) in Kahvena, Estonia (58.27991°N, 25.23165°E).
Description.
Other sequences: EUK1635883–EUK1635886 (type locality); EUK1631811 (GSMc plot G2767, mixed woodland soil at Mäebe, Estonia, 58.30937°N, 22.07618°E); KF618358 (Piceamariana forest soil, AK, USA); MT596306 (Tobiotsuka Kofun, Japan, 34.6355°N, 133.6814°E); KU062529 (unknown source); and KF565426 (Duke Forest, NC, USA, 35.97°N, -79.09°E), isolated by Rebecca C. Mueller (Mueller et al. 2014).
Etymology.
Kahvena (Estonian) refers to type locality; and Rebecca (English) refers to the first name of Rebecca C. Mueller, who collected the first materials belonging to this genus and the type species.
Notes.
Found from temperate and subarctic forests in Europe, Asia and North America, with ITS and LSU sequences differing up to 4% (excluding a 29-base deletion in EUK1631811 and KU062529) and 1.5%, respectively. Considered as a single species because of high intraspecific variation amongst common sequence variants in the type locality (2% in ITS and 1% in LSU, representing both indels and substitutions).
. Kelottijaerviales
Tedersoo ord. nov.
D66B982B-4882-54A1-B6F9-D49D9073B6CD
853576
Type family.
Kelottijaerviaceae Tedersoo.
Description.
Covers the monophyletic group in Endogonomycetes (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1202520 and EUK1633699 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Kelottijaerviaceae.
. Kelottijaerviaceae
Tedersoo fam. nov.
BBC931E5-D7C2-51AF-8556-A9DB1B4080D9
853577
Type genus.
Kelottijaervia Tedersoo.
Description.
Covers the monophyletic group in Kelottijaerviales (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1202520 and EUK1633699 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Kelottijaervia.
. Kelottijaervia
Tedersoo gen. nov.
B94EFDDD-356D-5FE4-A92E-A2C0ACB93E7A
853578
Type species.
Kelottijaerviashannonae Tedersoo.
Description.
Covers the monophyletic group in Kelottijaerviaceae (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1202520 and EUK1633699 (Suppl. material 3).
Notes.
Based on ITS and LSU sequences, Kelottijaervia is comprised of about five species that are represented by sequences EUK1603128 (GSMc plot G2755X, Pinussylvestris forest soil, Liiva-Putla, Estonia, 58.38859°N, 22.65545°E); EUK0302816 (plot G5403, mixed coniferous forest in Kõrveküla, Estonia, 58.43789°N, 26.75099°E); EUK1104755 (Pinussylvestris forest soil near Hofors, Sweden, 60.49°N, 16.30°E); and KP889573 (coniferous forest soil in British Columbia, Canada). The genus seems to prefer acidic coniferous forest habitats.
. Kelottijaervia shannonae
Tedersoo sp. nov.
E3587C65-1087-50AE-BCB0-CA7ADC90CAE1
853579
Diagnosis.
Separation from other species of Kelottijaervia based on the ITS region (positions 212–239 taatgtgagtgcaggaaatattatgact; one mismatch allowed) and LSU (positions 600–619 ctttggggtggcggtcgctg; one mismatch allowed) as indicated in Fig. 6.
Figure 6.
Diagnostic barcodes for Kelottijaerviashannonae relative to closely-related taxa in ITS2 and LSU.
Type.
eDNA sample TUE100189 (holotype); eDNA sequence EUK1202520 (lectotype); GSMc plot G2836 Finland, subpolar Betulapubescens forest (soil sample TUE000189) in Kelottijärvi, Finland, 68.60353°N, 21.74517°E.
Description.
Other sequences: EUK1603540, (GSMc plot G4196, Populus-Picea-Pinus forest soil in Kahvena, Estonia, 58.27991°N, 25.23165°E); EUK1603663 (GSMc plot G4406, mixed coniferous forest soil in Tarumaa, Estonia, 59.20745°N, 27.15333°E); EUK1602832 (GSMc plot G5828, Malusdomestica orchard soil in Mooste, Estonia, 58.15335°N, 27.19642°E); and KP889965 (coniferous forest soil in British Columbia, Canada) that was first isolated by Shannon H.A. Guichon (Guichon 2015).
Etymology.
Kelottijärvi (Finnish) refers to type locality; and Shannon (English) refers to the first name of Shannon H.A. Guichon who collected the first materials belonging to this genus.
Notes.
Found in Estonia, Finland and Canada, with ITS and LSU sequences displaying up to 2% and 1% of differences, respectively.
. Kungsaengenales
Tedersoo ord. nov.
180EF327-4C34-574C-B09B-21F423A4C97D
853580
Type family.
Kungsaengenaceae Tedersoo.
Description.
Covers the monophyletic group in Endogonomycetes (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1603402 and EUK1602136 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Kungsaengenaceae.
. Kungsaengenaceae
Tedersoo fam. nov.
87BBC899-4CD0-53E2-8551-701053D6C46F
853581
Type genus.
Kungsaengena Tedersoo.
Description.
Covers the monophyletic group in Kungsaengenales (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1603402 and EUK1602136 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Kungsaengena and a genus-level unassigned species represented by sequence EUK0013897 (GSMc plot G2907, subtropical forest soil in Cuc Phuong, Viet Nam, 20.34902°N, 105.59649°E).
. Kungsaengena
Tedersoo gen. nov.
A71ACC98-9F38-590A-B2E6-17C3DCF2E964
853582
Type species.
Kungsaengenashadiae Tedersoo.
Description.
Covers the monophyletic group in Kungsaengenaceae (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1603402 and EUK1602136 (Suppl. material 3).
Notes.
Based on ITS and LSU sequences, Kungsaengena comprises 5–6 species. Other putative species in this genus are represented by sequences EUK1603803 (GSMc plot G5906, stadium soil in Karksi-Nuia, Estonia, 58.10088°N, 25.55959°E); EUK1603124 (GSMc plot G5003, Pinussylvestris forest soil in Naissaar, Estonia, 59.5634°N, 24.5451°E); EUK1217319 (FunAqua sample W0279s, lake sediment near Bezdan, Serbia, 45.82031°N, 18.9599°E); and MW215857 (forest nursery soil in Lithuania).
. Kungsaengena shadiae
Tedersoo sp. nov.
4E91B05A-4FD2-5067-B51F-B76F2A2DE7F7
853583
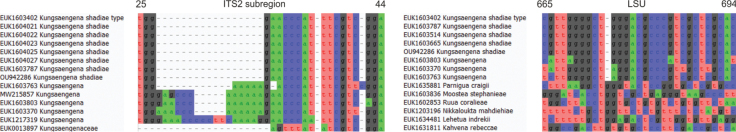
Diagnosis.
separation from other species of Kungsaengena based on the ITS region (ITS2 positions 25–44 tgggaacccatttcgtcgga; one mismatch allowed) and LSU (positions 665–694 cgttggggctgggacgcccgtcgctcgcac; one mismatch allowed) as indicated in Fig. 7.
Figure 7.
Diagnostic barcodes for Kungsaengenashadiae relative to closely-related taxa in ITS2 and LSU.
Type.
eDNA sample TUE128324 (holotype); eDNA sequence EUK1603402 (lectotype); GSMc plot G5763, wet grassland (soil sample TUE028324) in Haage, Estonia, 58.35555°N, 26.61277°E).
Description.
other sequences: EUK1604022 (GSMc plot G5906, football field soil in Karksi-Nuia, Estonia, 58.10088°N, 25.55959°E); EUK1604023 (GSMc plot G5844, wet pasture soil in Tuhala, Estonia, 59.23003°N, 25.00283°E); EUK1604025 (GSMc plot G4444, Estonia, mixed forest soil in Altnurga, Estonia, 58.53676°N, 26.28321°E); and OU942286 (grassland soil in Kungsängen, Sweden, 59.837°N, 17.661°E), isolated by Shadi Eshghi Sahraei (Eshghi Sahraei et al. 2022).
Etymology.
Kungsängen (Swedish) refers to type locality; and Shadi (Persian) refers to the first name of Shadi Eshghi Sahraei who analysed materials collected from the type locality.
Notes.
Found from the Baltic States and Sweden, with ITS and LSU sequences differing up to 15% and 1%, respectively. The ITS region is infested with microsatellite-like regions and homopolymers, and many sequence variants have long deletions in multiple positions. K.shadiae seems to be generalist in terms of habitat type.
. Langduoales
Tedersoo ord. nov.
D17889EB-3E73-5C99-A9AB-5605ED77DBE2
853584
Type family.
Langduoaceae Tedersoo.
Description.
Covers the monophyletic group in Endogonomycetes (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1107335, EUK1103607 and EUK1632831 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Langduoaceae and another potentially family-level group, which is represented by sequences EUK1632831 (GSMc plot G4104, Salixalba wetland forest soil in Koiva, Estonia, 57.68283°N, 26.20146°E); EUK1603795 (GSMc plot G5906, football field in Karksi-Nuia, Estonia, 58.10088°N, 25.55959°E); and EUK1602996 (GSMc plot G4171, mixed coniferous forest soil in Nõmmeotsa, Estonia, 58.48765°N, 26.22523°E).
. Langduoaceae
Tedersoo fam. nov.
DC725A98-B736-55A5-8E83-D039AA3741FE
853585
Type genus.
Langduoa Tedersoo.
Description.
Covers the monophyletic group in Langduoales (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1107335, EUK1103607 and EUK1632829 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently represented by Langduoa.
. Langduoa
Tedersoo gen. nov.
E64A7959-C72F-5134-BD05-FAC855682359
853586
Type species.
Langduoadianae Tedersoo.
Description.
Covers the monophyletic group in Langduoaceae (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1107335, EUK1103607 and EUK1632829 (Suppl. material 3).
Notes.
Based on ITS sequences, Langduoa is comprised of 40–50 species. The genus is distributed globally in multiple habitat types, but not found from roots so far. Most Langduoa species are poorly separable based on the LSU marker. Other putative species in Langduoa are represented by sequences EUK1103607 (tropical rainforest soil in El Yunque, Puerto Rico, 18.29°N, -65.78°E); EUK1631446 (GSMc plot G4189, Populustremula forest soil in Tammsaare, Estonia, 57.84444°N, 27.20141°E); and MW215048 (tree nursery soil in Lithuania), which was recorded by Diana Marčiulynienė (Marčiulynienė et al. 2021).
. Langduoa dianae
Tedersoo sp. nov.
540EE31A-230A-5EA2-B48A-7CBFA29997CF
853587
Diagnosis.
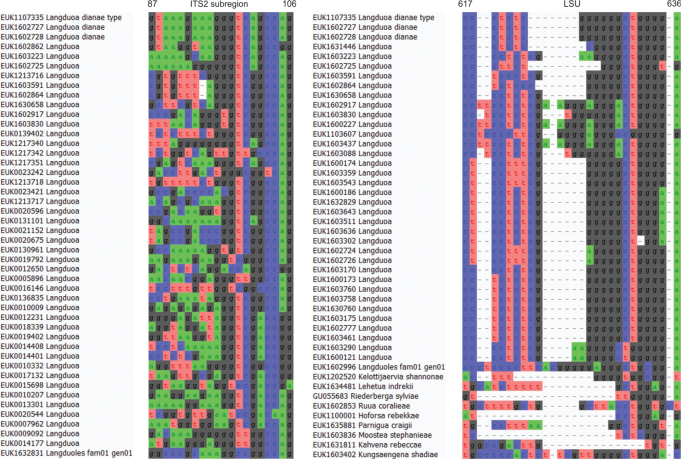
Separation from other species of Langduoa based on the ITS region (positions 87–106 actgagccttgcagcaacaatctccccttt; no mismatch allowed) and LSU (positions 617–636 ccctctcggggggctgggga; no mismatch allowed) as indicated in Fig. 8.
Figure 8.
Diagnostic barcodes for Langduoadianae relative to closely-related taxa in ITS2 and LSU.
Type.
Soil eDNA sample TUE128827 (holotype); eDNA sequence: EUK1107335 (lectotype); montane grassland in Langduo, Tibet, 29.4°N, 94.4°E.
Description.
Other sequences: EUK1602727 and EUK1602728 (both from GSMc plot G5906, stadium grassland soil in Karksi-Nuia, Estonia, 58.10088°N, 25.55959°E); EUK1604031 (GSMc plot G4185, Picea-Pinus forest soil in Ristipalo, Estonia, 58.10241°N, 27.47874°E); and EUK1604032 (GSMc plot G4766, soil of coppiced garden dominated by Fraxinus and Ulmus in Ruudiküla, Estonia, 58.33630°N, 25.78084°E).
Etymology.
Langduo (Tibetan) refers to type locality; and Diana (Lithuanian) refers to the first name of Diana Marčiulynienė who was the first to record this genus.
Notes.
Found from grassland soils in Estonia and Tibet, with ITS and LSU sequences differing up to 0.2%. So far, not found from the roots.
. Lehetuales
Tedersoo ord. nov.
E66B1F7F-6F8C-5AEC-9358-ADF9EF53D43F
853588
Type family.
Lehetuaceae Tedersoo.
Description.
Covers the monophyletic group in Endogonomycetes (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1603180, EUK1602375 and EUK1602377 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Lehetuaceae.
. Lehetuaceae
Tedersoo fam. nov.
CAD10076-055E-5EC8-B4C1-E73F5316175F
853589
Type genus.
Lehetua Tedersoo.
Description.
Covers the monophyletic group in Lehetuales (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1603180, EUK1602375 and EUK1602377 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Lehetua and another potentially genus-level group that is represented by sequences EUK1602869 (GSMc plot G4531, Piceaabies forest soil in Selisoo, Estonia, 57.621658°N, 27.179296°E) and EUK1603296 (GSMc plot S590, Populustremula forest soil in Lehetu, Estonia, 59.01857°N, 24.28041°E); and unassigned sequences EUK0025664 (GSMc plot G5536, tropical rainforest soil in Bamboesi, Suriname, 5.54086°N, -54.03131°E) and EUK0030289 (GSMc plot AV120, tropical rainforest soil in El Zafire, Colombia, -3.9997°N, 69.8947°E).
. Lehetua
Tedersoo gen. nov.
6B88749C-F6F5-588C-A1D9-7124A2204C64
853590
Type species.
Lehetuaindrekii Tedersoo.
Description.
Covers the monophyletic group in Lehetuaceae (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1603180, EUK1602366 and EUK1602374 (Suppl. material 3).
Notes.
Based on ITS and LSU sequences, Lehetua is comprised of 8–10 species. Other putative ITS-based species in Lehetua are represented by sequences EUK1602811 (GSMc plot G4105, Piceaabies forest soil in Lepa, Estonia, 57.70158°N, 26.23993°E); EUK1603124 (GSMc plot G5003, Pinussylvestris forest soil in Naissaar, Estonia; 59.5634°N, 24.5451°E); and EUK0022184 (GSMc plot AV106, Pseudomonotestropenbosii rainforest soil in El Zafire, Colombia, -3.995°N, -69.898°E).
. Lehetua indrekii
Tedersoo sp. nov.
EB330B0E-F11F-5A3C-AA19-7758D63BE4E1
853591
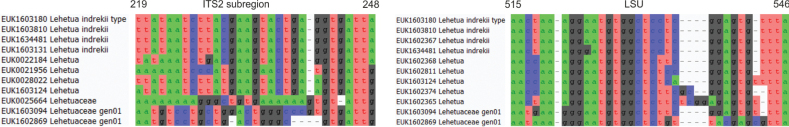
Diagnosis.
Separation from other species of Lehetua based on the ITS region (positions 219–248 ttataatcttacgaagtactgaggtgatta; one mismatch allowed) and LSU (positions 515–546 aactaaaggratgtggctcctcggagtgttta; one mismatch allowed) as indicated in Fig. 9.
Figure 9.
Diagnostic barcodes for Lehetuaindrekii relative to closely-related taxa in ITS2 and LSU.
Type.
Soil eDNA sample TUE103095 (holotype); type sequence EUK1603180 (lectotype); GSMc plot S590, Populustremula forest (soil sample TUE003095) in Lehetu, Estonia, 59.01857°N, 24.28041°E.
Description.
Other sequences: EUK1603180 (type locality); EUK1602367 (LSU only; type locality; also found in 50 other sites in Estonia); EUK1634481 (GSMc plot G4195, Quercusrobur woodland soil in Lustivere, Estonia, 58.66293°N, 26.08465°E); EUK1603818 (GSMc plot G5824, managed grassland soil in Kuremaa, Estonia, 58.74138°N, 26.52727°E); EUK1603131 (GSMc plot G4105, Piceaabies forest soil in Lepa, Estonia, 57.70158°N, 26.23993°E); EUK0021956 (GSMc plot G5150, subarctic grassland soil in Kokelv, Norway, 70.61116°N, 24.62483°E); and EUK0023592 (GSMc plot S035, mixed deciduous forest soil in Kedrovaya Pad, Russia, 43.10834°N, 131.55447°E).
Etymology.
Lehetu (Estonian) refers to type locality (also meaning “leafless”); and Indrek (Estonian) refers to the first name of Indrek Hiiesalu who collected materials from the type locality.
Notes.
Found in Baltic States, Scandinavia and Russia, with ITS and LSU sequences differing up to 3.5% and 0.2%, respectively. Seems to be a generalist in terms of habitat type and soil pH; so far, not found from roots.
. Lokrumales
Tedersoo ord. nov.
21ACC375-A4D8-5CC7-BA4A-8CED06A36780
853594
Type family.
Lokrumaceae Tedersoo.
Description.
Covers the monophyletic group in Endogonomycetes (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1203766, EUK1600125 and EUK1600268 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Lokrumaceae and another potentially family-level taxon, represented by sequences EUK1602809 (GSMc plot G4499, rich, calcareous Piceaabies forest soil in Kurisoo, Estonia; 59.12808°N, 25.76395°E); EUK1603041 and EUK1603145 (both GSMc plot G4185, Picea-Pinus forest soil in Ristipalo, Estonia, 58.10241°N, 27.47874°E).
. Lokrumaceae
Tedersoo fam. nov.
EC7E1DA2-F922-56B7-BB63-8064778B6AB6
853595
Type genus.
Lokruma Tedersoo.
Description.
Covers the monophyletic group in Lokrumales (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1203766, EUK1600125 and EUK1600078 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Lokruma and a few sequences not assigned to any genus; these include EUK0014543 and EUK0006923 (both GSMc plot G5106, subtropical forest soil in Brejo da Lapa, Brazil, -22.3582°N, -44.7383°E) and EUK1602939 (GSMc plot G4464, Quercusrobur forest soil in Ruu, Estonia, 59.45059°N, 25.22166°E).
. Lokruma
Tedersoo gen. nov.
118E7589-2DE2-5053-8FC7-469A0F7D1B6C
853596
Type species.
Lokrumastenii Tedersoo.
Description.
Covers the monophyletic group in Lokrumaceae (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1203766, EUK1600125 and EUK1600078 (Suppl. material 3).
Notes.
Based on ITS sequences, Lokruma is comprised of 35–40 species, some of which are represented by sequences EUK1200048 (GSMc plot G5130, grassland soil in Angera, Italy, 45.77336°N, 8.59657°E); EUK1602967 (GSMc plot G4626, Picea-Populus forest soil in Kõrve, Estonia, 59.07754°N, 26.76144°E); and EUK1603058 (Piceaabies forest soil in Serga, Estonia, 57.76052°N, 27.47502°E). Given the relatively high intraspecific differences and low interspecific differences, the LSU region is not optimal for distinguishing species of Lokruma.
. Lokruma stenii
Tedersoo sp. nov.
76664418-0381-5937-993C-E8A1D5B82667
853597
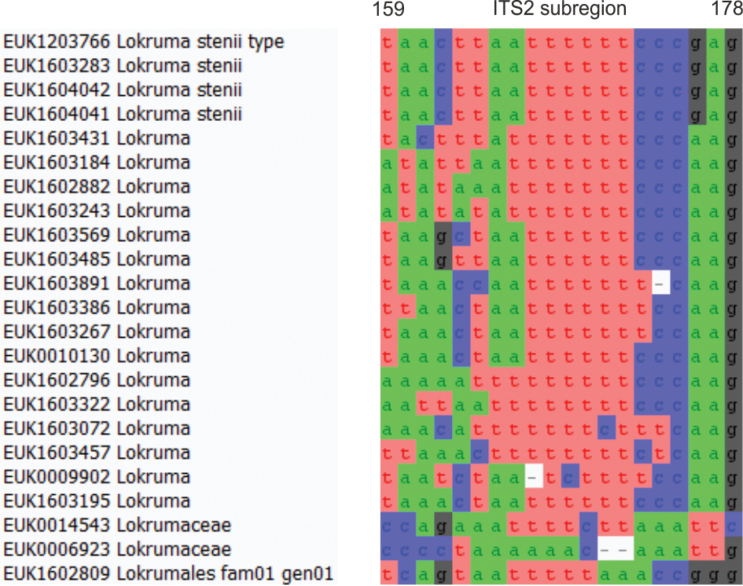
Diagnosis.
Separation from other species of Lokruma based on the ITS region (positions 159–178 taacttaattttttcccgag; one mismatch allowed) as shown in Fig. 10. There are no short barcodes in the first 700 bp of LSU that allow distinguishing L.stenii all from other congeners.
Figure 10.
Diagnostic barcodes for Lokrumastenii relative to closely-related taxa in ITS2.
Type.
Soil eDNA sample TUE103193 (holotype); type sequence EUK1203766 (lectotype); GSMc plot S689, Pinushalepensis forest (soil sample TUE003193) in Lokrum, Croatia, 42.6223°N, 18.1241°E.
Description.
Other sequences: EUK1603283 (GSMc plot G4301, Betulapendula forest soil in Männamaa, Estonia, 58.83258°N, 22.63346°E); EUK1604041 (GSMc plot S480, Populus-Picea forest soil in Käru, Estonia, 58.80407°N, 25.22249°E); EUK1604042 (GSMc plot G4734, Populus-Alnus forest soil in Urissaare, Estonia, 58.02673°N, 24.65739°E); and EUK1600039 (LSU: GSMc plot HB19, Populusxwettsteinii forest plantation soil, Oja, Estonia, 58.82747°N, 26.37799°E).
Etymology.
Lokrum (Serbo-Croatian) refers to type locality; and Sten (Estonian) refers to the first name of Sten Anslan who collected the materials from the type locality.
Notes.
Found in Croatia and Estonia, with ITS and LSU sequences displaying up to 1% of differences.
. Moosteales
Tedersoo ord. nov.
253AD908-CEEC-516A-9648-8AF7F4409EF3
853598
Type family.
Moosteaceae Tedersoo.
Description.
Covers the monophyletic group in Endogonomycetes (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1604044, JQ311412 and EUK1600278 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Moosteaceae.
. Moosteaceae
Tedersoo fam. nov.
6C93DE73-BECB-5482-8FDE-AC38CD9FDEE3
853600
Type genus.
Moostea Tedersoo.
Description.
Covers the monophyletic group in Moosteales (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1604044, JQ311412 and EUK1600278 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Moostea and two other potential genera. One of these is represented by sequences EUK0030179 (GSMc plot G4146, mixed forest soil in High Point Reserve Park, NJ, USA, 41.31569°N, -74.66485°E); EUK1600279 (GSMc plot G5826, Malusdomestica orchard soil in Tabivere, Estonia, 58.54286°N, 26.61575°E); and JQ311412 (microcosm soil in Los Alamos, NM, USA), isolated by Stephanie A. Eichorst (Eichorst and Kuske 2012). The other genus is represented by sequences EUK1600278 (GSMc plot S570, Betulapubescens wetland forest soil in Nõmme, Estonia, 58.47962°N, 22.94584°E); EUK0029679 (GSMc plot G2749, Eucalyptus spp. woodland soil near Lake Copperfield, Australia, -13.84191°N, 131.81858°E); and EUK0028885 (GSMc plot G5081, Coccoloba sp. woodland soil near Lagoa Grande, Brazil, -10.6342°N, -36.7579°E).
. Moostea
Tedersoo gen. nov.
72EA30F4-A8C0-596C-A6E9-85F1EAE8E3B5
853601
Type species.
Moosteastephanieae Tedersoo.
Description.
Covers the monophyletic group in Moosteaceae (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1604044, EUK1103239 and EUK1600287 (Suppl. material 3).
Notes.
The ITS sequences are poorly alignable because of long deletions and inserts in certain species. Based on ITS sequences, Moostea is comprised of 25–30 species, some of which are represented by sequences EUK1103239 (tropical rainforest soil in El Yunque, Puerto Rico, 18.29°N, -65.78°E); EUK1603515 (GSMc plot G5835, airfield soil in Ridali, Estonia, 57.93692°N, 26.98099°E); and EUK0014332 (GSMc plot S1225, grassland soil in Ayapel, Colombia, 8.27825°N, -75.1257°E).
. Moostea stephanieae
Tedersoo sp. nov.
0EF302DD-BD47-544E-A9A2-605C87529DC0
853603
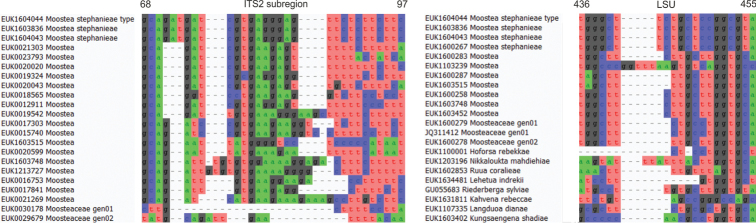
Diagnosis.
Separation from other species of Moostea based on the ITS region (positions 68–97 gcagatgatcgtgagggagttctcttcttc; one mismatch allowed) and LSU (positions 436–455 tgggcttctgctccggcgta; one mismatch allowed) as indicated in Fig. 11.
Figure 11.
Diagnostic barcodes for Moosteastephanieae relative to closely-related taxa in ITS2 and LSU.
Type.
Soil eDNA sample TUE128417 (holotype); eDNA sequence EUK1604044 (lectotype); GSMc plot G5828, Malusdomestica orchard (soil sample TUE028417) in Mooste, Estonia, 58.15335°N, 27.19642°E.
Description.
Other sequences: EUK1600287 (LSU: type locality); EUK1604043 and EUK1603823 (both GSMc plot G5835, airfield soil in Ridali, Estonia, 57.93692°N, 26.98099°E).
Etymology.
Mooste (Estonian) refers to type locality; and Stephanie (English) refers to the first name of Stephanie A. Eichorst who collected the first materials from the respective family.
Notes.
Found in two sites in Estonia, with ITS and LSU sequences displaying up to 1% and 0.3% differences, respectively.
. Nikkaluoktales
Tedersoo ord. nov.
57DD4DAD-2A4B-500C-8AB0-FBBC3019945B
853604
Type family.
Nikkaluoktaceae Tedersoo.
Description.
Covers the monophyletic group in Endogonomycetes (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1203196, EUK1600291 and EUK1600248 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Nikkaluoktaceae.
. Nikkaluoktaceae
Tedersoo fam. nov.
D9F0E42A-593A-5E2A-AD53-C7C9823073F6
853605
Type genus.
Nikkaluokta Tedersoo.
Description.
Covers the monophyletic group in Nikkaluoktales (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1203196, EUK1600291 and EUK1600248 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Nikkaluokta and another potentially genus-level group that is represented by sequences EUK1602730 (GSMc plot S554, Betula-Quercus woodland soil in Mädapea, Estonia, 59.32169°N, 26.2621°E); EUK1602729 (GSMc plot FF14, Piceaabies forest soil in Kõdesi, Estonia, 58.61484°N, 27.12781°E); and EUK1600257 (GSMc plot G4464, Quercusrobur forest soil in Ruu, Estonia, 59.45059°N, 25.22166°E).
. Nikkaluokta
Tedersoo gen. nov.
5DE29B8F-AEEF-59BB-8DB1-3345828F9AE4
853606
Type species.
Nikkaluoktamahdiehiae Tedersoo.
Description.
Covers the monophyletic group in Nikkaluoktales (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1203196, EUK1600291, EUK1600289, EUK1600235, EUK1600225, EUK1600250 and EUK1600248 (Suppl. material 3).
Notes.
Based on ITS and LSU sequences, Nikkaluokta is comprised of 15–20 species, some of which are represented by sequences EUK1603884 (GSMc plot G4406, mixed coniferous forest soil in Tarumaa, Estonia, 59.20745°N, 27.15333°E); EUK1603411 (GSMc plot G4462, Salixviminalis energy plantation soil in Kambja, Estonia, 58.25166°N, 26.71276°E); and EUK0006485 (GSMc plot MX23, Pinushartwegii montane forest soil in Iztaccihuatl, Mexico, 19.12622°N, -98.65972°E).
. Nikkaluokta mahdiehiae
Tedersoo sp. nov.
A2004B52-C41B-510F-93D8-14C7C8D1AD94
853607
Diagnosis.
Separation from other species of Nikkaluokta based on the ITS region (positions 97–116 cctgggcaaatttttttttc; one mismatch allowed) and LSU (positions 687–717 cttggatataagaagtggaatctacacaaat; one mismatch allowed) as indicated in Fig. 12.
Figure 12.
Diagnostic barcodes for Nikkaluoktamahdiehiae relative to closely-related taxa in ITS2 and LSU.
Type.
Soil eDNA sample TUE100497 (holotype); eDNA sequence EUK1203196 (lectotype); subarctic Pinussylvestris forest (soil sample TUE000497) in Nikkaluokta, Sweden, 67.85596°N, 19.47575°E.
Description.
Other sequences: EUK1203537 (type locality) and EUK1603797 (GSMc plot G5003, Pinussylvestris forest soil in Naissaare, Estonia, 59.56340°N, 24.54510°E).
Etymology.
Nikkaluokta (Sami) refers to type locality; and Mahdieh (Persian) refers to the first name of Mahdieh Hosseyni Moghaddam who sequenced the type materials using target capture protocols.
Notes.
Found in Sweden and Estonia, with ITS and LSU sequences displaying up to 1% and 0.2% differences, respectively.
. Parniguales
Tedersoo ord. nov.
84530192-F9FD-5536-B614-A92CE860FCCC
853608
Type family.
Parniguaceae Tedersoo.
Description.
Covers the monophyletic group in Endogonomycetes (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1635261, EUK1602353, EUK1602857 and EUK1602732 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently represented by Parniguaceae.
. Parniguaceae
Tedersoo fam. nov.
6227D941-6F9F-5139-8DEE-A97F9BE5A65B
853609
Type genus.
Parnigua Tedersoo.
Description.
Covers the monophyletic group in Parniguales (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1635261, EUK1602353, EUK1602857 and EUK1602732 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently represented by Parnigua and another potentially genus-level group, which is characterised by sequences EUK0016514 (GSMc plot S1218, urban park soil in Qujing, China, 25.52619°N, 103.74497°E), EUK0028452 (GSMc plot G3060, Vateriaindica forest in Hebri, India, 13.45437°N, 75.02213°E), EUK1602857 (GSMc plot G5771, grassland soil in Hino, Estonia, 57.57566°N, 27.22649°E), EUK1602732 (GSMc plot G5777, grassland soil in Eoste, Estonia, 58.11427°N, 27.08404°E) and EUK1602733 (GSMc plot G5816, Trifoliumpratense cropland soil in Hermani, Estonia, 58.80705°N, 25.75639°E).
. Parnigua
Tedersoo gen. nov.
DCC9362C-CDEF-56DD-BFCC-23F4F2B5B9C2
853610
Type species.
Parniguacraigii Tedersoo.
Description.
Covers the monophyletic group in Parniguaceae (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1635261 and EUK1602353 (Suppl. material 3).
Notes.
Based on stringent criteria, there are around five species in this genus, but all these may represent a single variable biological species. In this genus, across and within species, the ITS region has very low variability when compared with LSU (up to 3% differences across species). Other putative species in Parnigua are represented by sequences EUK1602947 (GSMc plot G4444, mixed forest soil in Altnurga, Estonia, 58.53676°N, 26.28321°E); EUK1603686 (GSMc plot G5844, wet pasture land soil in Tuhala, Estonia, 59.23003°N, 25.00283°E); EUK1633696 (GSMc plot G4207 Tiliacordata forest soil in Ubari, Estonia, 59.492609°N, 25.285663°E); EUK1603848 (GSMc plot G5883, flooded grassland soil in Kasari, Estonia, 58.73608°N, 23.98599°E); EUK1602353 (GSMc plot G4389, Quercus-Tilia forest soil in Naha, Estonia, 57.520914°N, 26.601199°E); MF484762 (agricultural soil in England); and MW163928 (Crocussativus cropland soil in Aosta Valley, Italy). The genus can be found from various soils but not from roots. However, SSU sequences are lacking, and links to AM fungi in SSU-based studies cannot be tested.
. Parnigua craigii
Tedersoo sp. nov.
EB911EB9-126A-5830-815B-5352D47ACEA6
853611
Diagnosis.
Separation from other species of Parnigua based on the ITS region (positions 51–80 actgagccttgcagcaacaatctccccttt; no mismatch allowed) and LSU (positions 444–463 ggcgggaaatcagcccccct; no mismatch allowed) as indicated in Fig. 13.
Figure 13.
Diagnostic barcodes for Parniguacraigii relative to closely-related taxa in ITS2 and LSU.
Type.
Soil eDNA sample TUE102228 (holotype); type sequence: EUK1635261 (lectotype); GSMc plot G5251, Quercusrobur woodland (soil sample TUE002228) in Parnigu, Estonia, 58.64096°N, 26.38468°E.
Description.
Other sequences: EUK1635874 (GSMc plot G4499, calcareous Piceaabies forest soil in Kurisoo, Estonia; 59.12808°N, 25.76395°E); EUK1635875 (GSMc plot G4746, Betulapendula forest soil in Karjamõisa, Estonia, 57.59761°N, 26.35493°E); EUK1635878 (GSMc plot G4794, Ulmus-Fraxinus forest soil in Lõhtsuu, Estonia, 57.91781°N, 26.52069°E); EUK1603328 (GSMc plot G4167, Salixpentandra peat soil in Tammispää, Estonia, 58.92051°N, 27.01118°E); EUK1602985 (GSMc plot G5923, Malusdomestica orchard soil in Kalnabeites, Latvia, 57.1333°N, 24.8566°E); OU939710 (grassland soil in Kungsängen, Sweden, 59.837°N, 17.661°E); and MH625006 (grassland soil in Wakanui, New Zealand, -43.668°N, 172.470°E), first isolated by Craig R. Anderson (Anderson et al. 2018).
Etymology.
Parnigu (Estonian) refers to type locality; and Craig (English) refers to the first name of Craig R. Anderson who was the first to record this species.
Notes.
Found from Estonia, Sweden and New Zealand, with ITS and LSU sequences differing up to 0.5%. Found in all croplands, grasslands, deciduous and coniferous forests.
. Riederbergales
Tedersoo ord. nov.
4E815FED-5175-5ABF-BDE7-D2E8BCEEDB04
853612
Type family.
Riederbergaceae Tedersoo.
Description.
Covers the monophyletic group in Endogonomycetes (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1602903, EUK1603115, EUK1602258, EUK1602253, EUK1602251 and EUK1104709 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Riederbergaceae and four additional potentially family-level taxa represented by sequences EUK1100540 (bog peat soil in Svartberget, Sweden, 64.24°N, 19.76°E); EUK1602254 (GSMc plot G5826, Malusdomestica orchard in Tabivere, Estonia, 58.54286°N, 26.61575°E); EUK1602251, EUK1602253 and EUK1602257 (all GSMc plot G5828, Estonia, Malusdomestica orchard soil in Mooste, Estonia, 58.15335°N, 27.19642°E). Sequences EUK0031975 (GSMc plot S1082, Araucariaaraucana forest, Nahuelbuta, Chile, -37.78985°N, -73.0038°E) and EUK1217433 (GSMc plot G4777, maritime grassland (saltmarsh) soil in Härs-hämani, Estonia, 59.33103°N, 23.92720°E) represent additional, monospecific, potentially family-level groups not included in the phylograms due to the lack of LSU sequences.
. Riederbergaceae
Tedersoo fam. nov.
45351A61-6079-5AB7-84BC-A513AA72CD3E
853613
Type genus.
Riederberga Tedersoo.
Description.
Covers the monophyletic group in Riederbergales (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1602903, EUK1602242 and EUK1602243 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Riederberga.
. Riederberga
Tedersoo gen. nov.
2EEC473B-405C-52F3-AFFE-F114FBBC1D31
853614
Type species.
Riederbergasylviae Tedersoo.
Description.
Covers the monophyletic group in Riederbergaceae (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1602903, EUK1602242 and EUK1602243 (Suppl. material 3).
Notes.
Based on ITS and LSU sequences, Riederberga is comprised of 5–6 species, some of which are represented by sequences EUK1602859 (GSMc plot G4770, Populusberolinensis dominated coppiced garden in Ubasalu, Estonia, 59.06755°N, 24.47842°E); EUK1602912 (GSMc plot G4772, Juniperuscommunis calcareous woodland soil in Kohatu, Estonia, 58.95934°N, 24.30017°E); EUK1602761 (GSMc plot G4434, mixed woodland soil in Kalli, Estonia, 58.53770°N, 24.06659°E); and EUK1603687 (GSMc plot G4229, Quercusrobur woodland soil in Niidiaia, Estonia, 58.88603°N, 24.47280°E).
. Riederberga sylviae
Tedersoo sp. nov.
9E7F08B8-4B5F-5D4A-A6FD-E53BEF71D3D8
853615
Diagnosis.
Separation from other species of Riederberga based on the ITS region (ITS2 positions 186–215 gctttggacggcatgcgaatctgcatcaca; one mismatch allowed) and LSU (positions 656–685 tcaccaatcgacgtcaatcggcatgcgtct; one mismatch allowed) as indicated in Fig. 14.
Figure 14.
Diagnostic barcodes for Riederbergasylviae relative to closely-related taxa in ITS2 and LSU.
Type.
Soil eDNA sample TUE128372 (holotype); eDNA sequence: EUK1602903 (lectotype); GSMc plot G5783, wet grassland (soil sample TUE028372) in Altnurga, Estonia, 58.55682°N, 26.29259°E.
Description.
Other sequences: EUK1604046 and EUK1604047 (both type locality); and GU055683 (ITS part considered; managed grassland soil in Riederberg, Austria, 48.25°N, 16.07°E), collected by Sylvia Klaubauf (Klaubauf et al. 2010).
Etymology.
Riederberg (German) refers to type locality; and Sylvia (German) refers to the first name of Sylvia Klaubauf, who first collected the materials of type species and the entire order from the type habitat.
Notes.
Found in Austria and Estonia, with ITS and LSU sequences displaying up to 1% differences.
. Ruuales
Tedersoo ord. nov.
F93E6A4C-D77E-56AB-BB21-164B965DB770
853616
Type family.
Ruuaceae Tedersoo.
Description.
Covers the monophyletic group in Endogonomycetes (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1603424, EUK1600239, EUK1600169 and EUK1600180 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Ruuaceae.
. Ruuaceae
Tedersoo fam. nov.
A5F825F3-3F11-54D4-BC46-D16EE26D497C
853617
Type genus.
Ruua Tedersoo.
Description.
Covers the monophyletic group in Ruuales (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1603424, EUK1600239, EUK1600169 and EUK1600180 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Ruua and another genus-level taxon represented by sequence EUK1602764 (GSMc plot G4189, Populustremula forest soil in Tammsaare, Estonia, 57.84444°N, 27.20141°E).
. Ruua
Tedersoo gen. nov.
12C8B808-14E0-5604-BAF2-30B66BF99003
853618
Type species.
Ruuacoralieae Tedersoo.
Description.
Covers the monophyletic group in Ruuaceae (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1603424, EUK1600239, EUK1600169 and EUK1600180 (Suppl. material 3).
Notes.
Based on ITS and LSU sequences, Ruua is comprised of 3–4 potential species that are represented by sequences EUK1632165 (GSMc plot S510, village habitat soil in Kihnu, Estonia, 58.1282°N, 23.9815°E); EUK1603289 (GSMc plot G4450, Fraxinus-Tilia forest soil in Nigula, Estonia, 58.0190°N, 24.6803°E); EUK1103406 (freshwater in Skogaryd, Sweden, 58.37°N, 12.16°E); and FN610984 (Fagussylvatica forest soil in Breuil-Chenue, France, 47.301°N, 4.076°E), isolated by Coralie Damon (Damon et al. 2010).
. Ruua coralieae
Tedersoo sp. nov.
857DC242-41B5-5BCD-9ACA-B042FC3E784D
853619
Diagnosis.
Separation from other species of Ruua based on the ITS region (positions 217–243 gaaaaaaaaagaaaggaaagaaaaggt; one mismatch allowed) and LSU (positions 470–489 tagtgcacttgctttcgcac; no mismatch allowed) as indicated in Fig. 15.
Figure 15.
Diagnostic barcodes for Ruuacoralieae relative to closely-related taxa in ITS2 and LSU.
Type.
eDNA sample TUE101598 (holotype); eDNA sequence EUK1603424; GSMc plot G4464, Quercusrobur forest (soil sample TUE101598) in Ruu, Estonia, 59.45059°N, 25.22166°E.
Description.
Other sequences: EUK1602853 and EUK1600135 (type locality); EUK1604050 (GSMc plot G5002, Tilia-Quercus forest soil in Naissaar, Estonia, 59.57530°N, 24.53590°E); and EUK1604051 (GSMc plot S480, Populus-Picea forest soil in Käru, Estonia, 58.80407°N, 25.22249°E).
Etymology.
Ruu (Estonian) refers to type locality; and Coralie (French) refers to the first name of Coralie Damon, who collected the first materials belonging to this genus.
Notes.
Found from three sites in Estonia, with ITS and LSU sequences displaying up to 0.3% differences.
. Tammsaareales
Tedersoo ord. nov.
95DA88D2-FCCD-598F-A206-6AC87C816BAB
853620
Type family.
Tammsaareaceae Tedersoo.
Description.
Covers the monophyletic group in Endogonomycetes (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1602762, EUK1635767 and EUK1602763 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Tammsaareaceae.
. Tammsaareaceae
Tedersoo fam. nov.
F9F1308F-B1A7-54BA-9336-11364D537E08
853621
Type genus.
Tammsaarea Tedersoo.
Description.
Covers the monophyletic group in Tammsaareales (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1602762, EUK1635767 and EUK1602763 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Tammsaarea and the sequence EUK1602763 (GSMc plot G5835, airfield soil in Ridali, Estonia, 57.93692°N, 26.98099°E).
. Tammsaarea
Tedersoo gen. nov.
04D6E4EC-698B-5CA4-9FE1-217FEDFA3169
853622
Type species.
Tammsaareavivikae Tedersoo.
Description.
Covers the monophyletic group in Tammsaareaceae (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1602762 and EUK1635767 (Suppl. material 3).
Notes.
Based on ITS sequences, Tammsaarea is comprised of two species; the other being represented by LSU sequences EUK1601269, EUK1635767 and EUK1635768 (all GSMc plot G4185, Picea-Pinus forest soil in Ristipalo, Estonia, 58.10241°N, 27.47874°E).
. Tammsaarea vivikae
Tedersoo sp. nov.
5A06EDBE-08CB-5D3E-BC1C-68891B139752
853683
Diagnosis.
Separation from other species of Tammsaarea and other species of Endogonomycetes based on ITS (positions 228–257 ggaccgagaaggcgcaatagttgaacaatt; one mismatch allowed) and LSU (positions 585–604 ataactatcggacaaagttt; one mismatch allowed) as indicated in Fig. 16.
Figure 16.
Diagnostic barcodes for Tammsaareavivikae relative to closely-related taxa in ITS2 and LSU.
Type.
eDNA sample TUE100731 (holotype); eDNA sequence EUK1602762 (lectotype); GSMc plot G4189, Populustremula forest (soil sample TUE000731) in Tammsaare, Estonia, 57.84444°N, 27.20141°E.
Description.
Other sequences EUK1604048 and EUK1604049 (type locality).
Etymology.
Tammsaare (Estonian) refers to the type locality and one of the most famous Estonian writers, Anton Hansen Tammsaare; and Vivika (Estonian) refers to the first name of Vivika Adamson who provided access to the type locality.
Notes.
Found from a single locality in Estonia, with ITS and LSU sequences differing up to 0.5% and 0.3%, respectively.
. Unemaeeales
Tedersoo ord. nov.
826EEC8F-A6E8-55EB-AC2E-0EEBCAE0171C
853684
Type family.
Unemaeeaceae Tedersoo.
Description.
Covers the monophyletic group in Endogonomycetes (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1630871 and EUK1635889 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Unemaeeaceae.
. Unemaeeaceae
Tedersoo fam. nov.
97D136DC-C849-579B-AA7C-D1763568E03E
853685
Type genus.
Unemaeea Tedersoo.
Description.
Covers the monophyletic group in Unemaeeales (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1630871 and EUK1635889 (Suppl. material 3).
Notes.
Recognised based on eDNA sequences only. Currently includes Unemaeea and multiple poorly alignable ITS sequences with no LSU, for example EUK1217297 (FunAqua sample W0006s, lake sediment in Petrolandia, Brazil, -8.9908°N, -38.2251°E) and FJ528738 (Araucaria spp. plantation soil, Gadgarra, Australia, -17.1641°N, 145.6469°E) that was isolated by Nathalie J.A. Curlevski (Curlevski et al. 2010). It seems that several Unemaeeaceae spp. have preferential habitat in anoxic soils and sediments.
. Unemaeea
Tedersoo gen. nov.
66AC7F74-232A-5273-99D5-FF5E6CA159D3
853686
Type species.
Unemaeeanathalieae Tedersoo.
Description.
Covers the monophyletic group in Unemaeeales (Fig. 2). Phylogenetically delimited as the least inclusive clade covering sequence accessions EUK1630871 and EUK1635889 (Suppl. material 3).
Notes.
Based on ITS and LSU sequences, Unemaeea is comprised of three species; others represented by sequences EUK1217289 (freshwater lake sediment near Bezdan, Serbia, 45.82031°N, 18.9599°E) and KX196132 (deciduous forest soil in Champaign County, IL, USA).
. Unemaeea nathalieae
Tedersoo sp. nov.
89C10250-C799-54E9-97E2-E2AAFE6E3EF9
853687
Diagnosis.
Separation from other species of Unemaeea based on the ITS region (5.8S positions 122–151 gtcagtgtttgccacggagtatgccggctt; no mismatch allowed) and from other species of Endogonomycetes based on LSU (positions 694–723 gggcttgtcatggcagagggacacgtcgta; no mismatch allowed) as indicated in Fig. 17.
Figure 17.
Diagnostic barcodes for Unemaeeanathalieae relative to closely-related taxa in ITS2 and LSU.
Type.
Soil eDNA sample TUE100213 (holotype); eDNA sequence EUK1630871 (lectotype); GSMc plot G3318, marshland (soil sample TUE000213) in Unemäe, Estonia, 58.28253°N, 22.46296°E.
Description.
Other sequences: EUK1635887–EUK1635890 (type locality) and EUK1213720 (FunAqua sample W0581s, river sediment in Floresti, Romania, 46.75472°N, 23.49923°E).
Etymology.
Unemäe (Estonian) refers to the type locality; and Nathalie (English) refers to the first name of Nathalie J.A. Curlevski who collected the first materials belonging to this genus.
Notes.
The end of 5.8S and start of LSU are strongly diverged compared with other species of Unemaeea and Endogonomycetes. As no other confamilial LSU sequences are available, the diagnostic positions are compared against the most divergent, unalignable part across Endogonomycetes. Found in anoxic soil in Estonia and Romania, with ITS sequences displaying up to 4% differences.
. Bifiguratales
Tedersoo ord. nov.
EEE1045F-7B8B-50D9-BA60-97274AFC4591
853688
Type family.
Bifigurataceae Tedersoo.
Description.
Covers the monophyletic group in Endogonomycetes (Fig. 2). Cultured mycelium filamentous, aseptate, coenocytic, 2 μm diam., mucose in appearance, commonly producing budding yeast-like cells; chlamydospores intercalary, 5–10 μm diam., forming on hyphal tips. Phylogenetically delimited by the least inclusive clade covering sequence accessions HM123225, EUK1104879, KF568171 and KF567389.
Notes.
Comprised of a single family Bifigurataceae. Order description is adapted from Torres-Cruz et al. (2017).
. Bifigurataceae
Tedersoo fam. nov.
42100643-7102-5F0F-871D-8A0DB31BB616
853689
Type genus.
Bifiguratus T.J.Torres-Cruz & A.Porras-Alfaro.
Description.
Cultured mycelium filamentous, aseptate, coenocytic, 2 μm diam., mucose in appearance, commonly producing budding yeast-like cells; chlamydospores intercalary, 5–10 μm diam., forming on hyphal tips. Phylogenetically delimited by the least inclusive clade covering sequence accessions HM123225, EUK1104879, KF568171 and KF567389.
Notes.
Comprised of a single genus Bifiguratus that is commonly found in soil and occasionally in roots of non-AM plants. No sexual structures have been revealed. Family description is adapted from Torres-Cruz et al. (2017).
. Densosporales
Tedersoo ord. nov.
861CE3AB-89AA-5874-BB2A-133E6C3004A1
853690
Type family.
Densosporaceae Desirò, M.E.Sm., Bidartondo, Trappe & Bonito.
Description.
Densosporales is defined as a monophyletic group in Endogonomycetes (Fig. 2, Suppl. material 3) that corresponds to Densosporaceae sensu Desiro et al. (2017). Covers Densosporacae and Planticonsortiaceae and the least inclusive clade with sequence accessions UDB028692, EUK1104889, EUK1104816 and EUK1601509 (Suppl. material 3).
Notes.
Densosporales harbours roughly one half of the Endogonomycetes based on LSU data. It comprises Densosporacae, Planticonsortiaceae and 16 additional family-level groups collectively covering >200 species. LSU has much greater phylogenetic resolution compared with SSU (Suppl. materials 3, 4), and the potential utility of the ITS region seems to vary greatly by family. Many more ITS-LSU sequences are needed to understand family- and genus-level composition of Densosporales.
. Densosporaceae
Desirò, M.E.Sm., Bidartondo, Trappe & Bonito, emend. Tedersoo
01C2B117-0490-54D9-9444-92E16980CE17
821851
Type genus.
Densospora McGee.
Description.
Phylogenetic diagnosis as in Desiro et al. (2017), but includes a more limited phylogenetic group - the least inclusive clade comprised of Densospora spp. (accessions JF414167 and UDB28692), Sphaerocreaspubescens (accession LC107618) and accession EUK1601029 (Suppl. material 3).
Notes.
Based on SSU phylogeny (Suppl. material 4), one or both of the genera Densospora and Sphaerocreas are paraphyletic, and their relationships require further research. Most species in both genera remain to be sequenced, including D.tubiformis (P.A.Tandy) McGee - the type species of Densospora.
. Planticonsortiaceae
Tedersoo fam. nov.
77431B6A-8B3B-5CCC-A716-2450B6002A98
853691
Type genus.
Planticonsortium C.Walker & D.Redecker.
Description.
Emanating hyphae 0.5–4 μm diam., forming colourless to brown chlamydospores (10–12 μm, up to 35 μm diam.), sometimes rope-like strands; appressoria swollen, frequently with several thin hyphae giving an insect-like appearance. Intraradical mycelium 0.5–4 μm diam., smooth to angular, with (sub-)globose swellings, forming comb-like (ctenoid), fan-shaped, palmate, antler-like, digitate or feather-like structures appearing clasped around epidermal and cortical cells; forming finely branched arbuscules. All hyphae stain darkly in acidic blue stains, more strongly for extraradical hyphae. Monophyletic group in Densosporales (Fig. 2, Suppl. materials 3, 4).
Notes.
Planticonsortiaceae covers roughly one third of Endogonomycetes reads based on LSU (Suppl. material 3) and SSU (Suppl. material 4), but is poorly represented in the ITS dataset. This may be due to the highly divergent and relatively long ITS region (800–1200 bases). Based on the LSU phylogram, Planticonsortiaceae harbours seven genus-level groups with >100 putative species. The description is adapted from Walker et al. (2018).
. Endogonales
Jacz. & P.A.Jacz., emend. Tedersoo
794A3239-8DBB-56F1-832D-0144835F524F
90720
Type family.
Endogonaceae Paol.
Description.
Fruiting body hypogeous or on debris, globose, irregular, sometimes resupinate, 1–10 mm in diam., may be composed of aggregated zygosporangial clusters, with zygospores formed on apposed suspensors. Hyphae of fruiting body tissue coenocytic, aseptate, sometimes with secondary septa that form micropores. Reproductive structures as zygosporangia, rarely azygosporangia (co-existing with zygosporangia in Endogonepisiformis) or chlamydospores (in Vinositunica), distributed randomly or radially in fruiting bodies, 100–700 μm diam., with yellow granular contents. Zygosporangial wall comprises outer sporangiothecium with 1–4 openings and inner eusporium with no openings. Azygosporangia rare, with a single-layered wall and separated from the single suspensor by a gametangial septum. Chlamydospore wall continuous, multilayered, with dense subtending hyphae, lacking septa. Forms a monophyletic group in Endogonomycetes as the least inclusive clade covering accessions EUK1601498, EUK1100757, LC002628, LC431107, EUK1104693 and UDB025468.
Notes.
Includes taxa with or without fruiting bodies and with ectomycorrhizal, arbuscular mycorrhizal and saprotrophic lifestyles. Endogonales harbours Endogonaceae, Jimgerdemanniaceae and Vinositunicaceae families, as well as seven potentially family-level taxa, collectively comprising >200 species based on ITS and LSU sequences. Order description is adapted from Morton and Benny (1990) and Yamamoto et al. (2020).
. Endogonaceae
Paol., emend. Tedersoo
B44E017E-4A61-5D53-B312-17EBAC90BB9D
81877
Type genus.
Endogone Link.
Description.
Fruiting body hypogeous or on debris, globose, irregular, sometimes resupinate, 1–10 mm diam., may be composed of aggregated zygosporangial clusters, with zygospores formed on apposed suspensors. Hyphae of fruiting body tissue coenocytic, aseptate, sometimes with secondary septa that form micropores. Reproductive structures as zygosporangia, rarely azygosporangia (co-existing with zygosporangia in Endogonepisiformis) distributed randomly or radially in fruiting bodies, 100–700 μm diam., with yellow granular contents. Zygosporangial wall comprises outer sporangiothecium with 1–4 openings and inner eusporium with no openings. Azygosporangia rare, with a single-layered wall, and separated from the single suspensor by a gametangial septum. Forms a monophyletic group in Endogonales as the least inclusive clade covering accessions LC002628, EUK1601764 and EUK1601442.
Notes.
Covers species of Endogone that are saprotrophic or potentially ectomycorrhizal (/endogone2 and /endogone3 lineages, sensuTedersoo and Smith (2017)) and four closely-related genus-level taxa.
. Jimgerdemanniaceae
Tedersoo fam. nov.
9FA53440-1DC9-54F3-94C5-49B14EA58F10
853692
Type genus.
Jimgerdemannia Trappe, Desirò, M.E.Sm., Bonito & Bidartondo.
Description.
Includes Jimgerdemannia and closely-related genera that form a monophyletic group in Endogonales, with the least inclusive clade covering accessions KC568319, EUK1631035, JN890102, UDB025468 and OU942919 (Suppl. material 3).
Notes.
Jimgerdemanniaceae covers an ectomycorrhizal genus Jimgerdemannia and six genus-level taxa that are soil-inhabiting, potentially arbuscular mycorrhizal and probably not producing macroscopic fruiting bodies.
. Vinositunicaceae
Tedersoo fam. nov.
B3DEA742-7B7D-587B-839E-1117637F08A0
853693
Type genus.
Vinositunica Koh.Yamam., Degawa & A.Yamada.
Description.
Fruiting bodies epigeous or semi-hypogeous, reniform or irregular, often with a short stipe-like sterile base, 2–20 mm in diam. Peridium white, partly purple, in a single layer, composed of coenocytic aseptate hyphae. Gleba pale yellow to purplish-grey, composed of numerous radially or randomly distributed chlamydospores. Chlamydospores granular, with yellow contents, broadly ellipsoid, 50–700 μm diam, terminal on single subtending hypha. Cell wall composed of purplish to vinaceous outer layer and colourless inner layer.
Notes.
Vinositunicaceae includes the genus Vinositunica. This group has not been found from root or soil eDNA samples thus far, and ITS sequences are not available. Probably humus saprotrophs. Family description is adapted from Yamamoto et al. (2020).
Primer bias
To evaluate whether some part of the dark diversity of putative AM fungi can be accounted for by primer bias as suggested for Glomeromycota (Kohout et al. 2014; van Geel et al. 2014; Seeliger et al. 2023), we tested the commonly used SSU, ITS and LSU primers for critical mismatches based on multiple sequence alignments. The AMV4.5NF (Sato et al. 2005) and AM-Sal-F (Seeliger et al. 2023) primers, proposed to cover both AM fungal groups, exhibited several (near-)terminal mismatches to many groups of Glomeromycota and one central and one near-terminal mismatch to many groups of Endogonomycetes. The FRE-F (Seeliger et al. 2023) primer had multiple mismatches to most target Endogonomycetes groups including a terminal mismatch to some groups. The reverse SSU primers AMDGR (Sato et al. 2005) and FRE-R (Seeliger et al. 2023) matched well with Glomeromycota, but had one or more (near)-terminal mismatches to several groups of Endogonomycetes. Regarding the ITS-LSU primers, ITS1F (Gardes and Bruns 1993), ITS1 (White et al. 1990), gITS7ngs and ITS4ngsUni (Tedersoo and Lindahl 2016) had single central mismatches to a few Glomeromycota and Endogonomycetes lineages, whereas ITS9munngs (Tedersoo and Lindahl 2016) had no mismatches. The fungi-specific primer ITS1catta (Tedersoo and Anslan 2019) had (near)-terminal mismatches to several minor lineages of both AM groups.
Of Glomeromycota-specific primers, wSSUmcf (Krüger et al. 2009) matched well to all target lineages. The primer wLSUmbr (Krüger et al. 2009) had one central mismatch to Pervetustus and Archaeosporales, suggesting a negligible bias.
Of Endogonomycetes-specific primers designed and tested initially, ITS3-End displayed mismatches to multiple groups, while LR3-End had 1–2 central mismatches to Jimgerdemanniaceae and terminal mismatches to Unemaeeaceae. For Endogonomycetes, we thus recommend use of universal forward primers gITS7ngs or LROR or the newly-designed LF350End (ccgatagcgaacaagtac; also amplifies many other fungi) in combination with the combination of reverse primers LR3-End2 (aycattahgycagcgacc; >99% of Endogonomycetes) and LR3-End2a (aycattahgycagccgtta; Unemaeeaceae). These primer pairs yield amplicons of 900–1200 bases, ca. 700 bases and ca. 400 bases, respectively. For simultaneous amplification of Glomeromycota and Endogonomycetes, only universal or fungal primers can be recommended (e.g., forward primers gITS7ngs, LROR and LF350 combined with a reverse primer TW13; White et al. (1990)) along with deep sequencing to 105 reads.
Discussion
In this paper, we describe 15 new species of potentially AM fungi belonging to Glomeromycota and Endogonomycetes from soil eDNA samples. These new species and six re-combinations lead to 16 new genera, 19 new families and 17 new orders that are well delimited by phylogenetic analyses of rRNA genes. The high taxonomic and phylogenetic resolution at the levels of species to class render long-read rRNA gene sequences highly useful for both species delimitation and phylogeny reconstruction. Future studies using protein-encoding genes or whole-genome analyses will be useful for solving phylogenetic uncertainties related to rapid rRNA gene evolution in certain groups (e.g. Entrophosporales) and unsettled branching order (e.g. endogonomycete orders). For this study, the genomes that were available for only 13 described genera of Glomeromycota and two genera of Endogonomycetes (Rosling et al. 2024) would have added no extra value. Our phylogenies indicate that eDNA from soil and sediment habitats may substantially add to novel phylogenetic diversity in these groups, especially in Endogonomycetes. Studies combining fine root staining and DNA sequencing should improve our understanding of the symbiotic potential of these newly-described groups and the evolution of AM associations in general.
We rely on public long-read rRNA gene sequences to describe new species in previously unrecognised family- and order-level taxa, using eDNA samples as holotypes and sequences as lectotypes. Previous DNA-based taxonomic studies on fungi have described new species in well-known genera (Bridge and Hughes 2012; Kirk 2012; Kalsoom Khan et al. 2020) or families (de Beer et al. 2016; Lücking and Moncada 2017) based on typifying sequences of the ITS region. The species described here are usually represented by both ITS and LSU regions from multiple eDNA samples. This allows us to estimate rough intraspecific variation and interspecific distances, and develop continuous, 20–30-base diagnostic barcodes (see also Kalsoom Khan et al. (2020)) for ITS and LSU regions separately. This contrasts with other studies that point to single diagnostic differences scattered across the entire marker length (de Beer et al. 2016; Lücking and Moncada 2017), or provide no sequence-diagnostic features. The continuous barcodes are better findable for the human eye and software, such as custom BLAST algorithms (Camacho et al. 2009), Cutadapt (Martin 2011), CAOS-R (Bergmann 2024) and SeqKit2 (Shen et al. 2024). For nearly all species (except Parniguacraigii), these diagnostic barcodes are more informative for the ITS region than LSU due to greater variability and taxonomic resolution. The species of Langduoa and Lokruma have relatively lower LSU short barcode resolution compared with other taxa. Nonetheless, species from all groups can be distinguished well based on ITS1 or ITS2 sequences and usually by LSU sequences.
The newly-described species, genera, families and orders are represented exclusively by eDNA sequences supplied with metadata ranging from none to ample background information about location and environmental properties, depending on the source of reads and success in contacting the data producers or material collectors. Besides fragmented information about habitat and distribution, soil eDNA provides no information about biotic interactions or functioning. Given the paucity of data from non-soil habitats outside northern Europe, we refrain from speculating about the distribution and functional role of the described species and higher-level taxa.
The Glomeromycota SSU-ITS-LSU phylogram is congruent with previous studies at the level of families and orders (Oehl et al. 2011; Redecker et al. 2013; Blaszkowski et al. 2021, 2022; Montoliu-Nerin et al. 2021; Rosling et al. 2024). The newly-described entrophosporalean family Pseudoentrophosporaceae does not affect the overall phylogenetic structure of Glomeromycota, but expands the phylogenetic breadth of Entrophosporales. Besides this new family, we also recorded multiple potentially new genera, most of which have also been revealed in previous analyses of root and soil materials. In this paper, we refrain from formally describing these for two main reasons. First, most of these genera are relatively common, and there are high chances that the corresponding species have been described but yet to be sequenced; here, we sincerely hope that the current study motivates the publishing of these materials, kept in several research teams’ drawers. Second, we are surveying hundreds of global soil and sediment samples using the ITS- and LSU-orientated Glomeromycota- and Endogonomycetes-specific primers, which will likely reveal novel diversity and improve delimiting the new putative taxa. For their short-term communication, we propose alphanumeric labels that facilitate quality-filtering, especially chimaera control, for forthcoming eDNA studies. The currently accepted genera and proposed genus-level groups for Glomeromycota and Endogonomycetes are provided in Suppl. materials 5, 6, respectively.
The Endogonomycetes SSU-5.8S-LSU phylogram only marginally reflects the SSU-focused multigene phylograms of Desiro et al. (2017) and Yamamoto et al. (2020) and the SSU-based phylogram of Albornoz et al. (2022). Here, we distinguish 17 well-supported orders in Endogonomycetes, including Densosporales and Bifiguratales (ord. nov.) and Endogonales (sens. str.), as well as entirely new groups represented by no sequenced culture, spore or fruiting body specimen. For anchoring names to these orders, we describe well-chosen representative species, genera and families based on eDNA and long-read sequences. To communicate these groups’ internal structure, we propose alphanumeric codes for putative families and genera as for the Glomeromycota. Furthermore, our analyses indicate much greater phylogenetic and species-level resolution for the LSU marker than the SSU (Suppl. materials 3, 4). The sequence data accumulated thus far also reveal much more information available for LSU compared with ITS and SSU at the level of species and orders (Desiro et al. 2017; Suppl. material 3). However, these datasets are biased for soil (ITS and LSU) or plant samples (SSU). As a downside of the ITS-LSU approach, the genus Planticonsortium lacks sequence data for these markers and cannot be reliably assigned to any of the multiple Planticonsortiaceae genus-level groups. However, ultra-long reads should be able to bridge the SSU and LSU and provide insights into Planticonsortiaceae soon.
We have encountered several conflicting situations by focusing on the mixed morphology- and eDNA-based classification. Undoubtedly, there is a potential risk of parallel morphological and DNA-based descriptions, especially given that nearly half of the accepted species of AM fungi are represented by no sequence data. However, the high-throughput sequencing methods have been available for >15 years, making it increasingly less likely that old spore collections from microscope slides will be successfully sequenced soon.
In addition to the parallel morphology-based and DNA-based descriptions, the focus on different morphological characters may also hamper the taxonomy of AM fungi. We find that a pair of glomeromycete genera, Redeckera and Corymbiglomus, that can be seemingly well delimited by morphological characters, are not clearly separated in phylogenetic analysis. Importantly, Redeckera spp. are described based on the small glomerospores clustering in large fruiting bodies, whereas species in Corymbiglomus are distinguished based on glomerospores on hyphal tips. The presence of spore dimorphism, as recently revealed for Entrophospora-Claroideoglomus (Blaszkowski et al. 2022), might be behind the inconsistency between phylogenetic and morphological data. Since Diversisporaceae harbours multiple described and undescribed genera, we leave the taxonomy of the Redeckera-Corymbiglomus group to be settled in further studies. Furthermore, species of Endogonomycetes have been described based on chlamydospores on hyphae (Planticonsortium), chlamydosporic fruiting bodies (Vinositunica and Densospora), zygosporangial fruiting bodies (Endogone and Jimgerdemannia) and features of pure cultures (Bifiguratus), potentially resulting in parallel classification based on different characters. Here, the massive amount of available eDNA sequences enables us to bridge a vast majority of these taxa (except Vinositunica and Planticonsortium) and translate the various types of morphological descriptions into a common DNA-based language.
Conclusions
This study offers the first example of a mixed morphological and eDNA-based classification from species to order level in the fungal kingdom. Our approach of typifying both eDNA samples and sequences and preparing diagnoses based on DNA barcodes will likely boost alpha and higher-level taxonomic research in fungi and potentially in non-fungal organisms. Such a mixed classification would help provide human-readable names to many of the “dark matter” fungi (Nilsson et al. 2016) and tremendously reduce the number of entirely unidentified fungi. To avoid parallel DNA-based classifications, we propose that the description of new species should primarily focus on the universal fungal barcode - the ITS region, preferably supplemented with at least one additional taxonomically or phylogenetically informative marker - LSU or SSU in the case of amplicons - or a protein-encoding gene for genomes derived from tissues. Accordingly, we propose using both the ITS and LSU markers for the two groups of AM fungi, considering taxonomic resolution, availability of specific primers and the large number of previously described reference species.
Our research also points out that, in addition to registering newly-described fungal taxa, we urgently need a linked system (related to, for example, INSDC, MycoBank and/or UNITE - the leading platforms that cross-communicate fungal species and molecular sequence data) for mandatory registering of taxonomic emendations and taxonomic updates of sequences, especially when new taxa are erected based on already published sequences. Such sequence registration would minimise the risk that taxon names of the sequences in databases evolve in different directions and that new species are described several times based on the same or related sequences.
Supplementary Material
Acknowledgements
We are indebted to Bruno T. Goto for taxonomic discussions about Glomeromycota taxonomy and comments on an earlier version of the manuscript. We thank David Bass, Fabien Burki and Mahwash Jamy for donating eDNA samples to TUE.
Additional information
Conflict of interest
The authors have declared that no competing interests exist.
Ethical statement
No ethical statement was reported.
Funding
The EUKARYOME project led by the Mycology and Microbiology Center is funded by the Es-tonian Science Foundation (grant MOBERC106) and the King Saud University Distinguished Scientist Fellowship Programme (DSFP-2024).
Author contributions
Conceptualization: LT. Data curation: VM. Funding acquisition: SA. Investigation: FM.
Author ORCIDs
Franco Magurno https://orcid.org/0000-0002-3117-8149
Saad Alkahtani https://orcid.org/0000-0001-7381-5110
Vladimir Mikryukov https://orcid.org/0000-0003-2786-2690
Data availability
All of the data that support the findings of this study are available in the main text or Supplementary Information.
Supplementary materials
Maximum Likelihood phylogram indicating phylogenetic relationships amongst Glomeromycota based on SSU-ITS-LSU sequences
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Leho Tedersoo, Franco Magurno, Saad Alkahtani, Vladimir Mikryukov
Data type
Maximum Likelihood phylogram indicating phylogenetic relationships amongst Gigasporales based on LSU sequences
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Leho Tedersoo, Franco Magurno, Saad Alkahtani, Vladimir Mikryukov
Data type
Maximum Likelihood phylogram indicating phylogenetic relationships amongst Endogonomycetes based on SSU-5.8S-LSU sequences
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Leho Tedersoo, Franco Magurno, Saad Alkahtani, Vladimir Mikryukov
Data type
Maximum Likelihood phylogram indicating phylogenetic relationships amongst Endogonomycetes based on SSU sequences
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Leho Tedersoo, Franco Magurno, Saad Alkahtani, Vladimir Mikryukov
Data type
Currently recognised orders, families and genera and proposed taxonomic groups in Glomeromycota
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Leho Tedersoo, Franco Magurno, Saad Alkahtani, Vladimir Mikryukov
Data type
Currently recognised orders, families and genera and proposed taxonomic groups in Endogonomycetes
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Leho Tedersoo, Franco Magurno, Saad Alkahtani, Vladimir Mikryukov
Data type
References
- Abarenkov K, Nilsson RH, Larsson KH, Taylor AF, May TW, Frøslev TG, Pawlowska J, Lindahl B, Põldmaa K, Truong C, Vu D, Hosoya T, Niskanen T, Piirmann T, Ivanov F, Zirk A, Peterson M, Cheeke TE, Ishigami Y, Jansson AT, Jeppesen TS, Kristiansson E, Mikryukov V, Miller JT, Oono R, Ossandon FJ, Paupério J, Saar I, Schigel D, Suija A, Tedersoo L, Kõljalg U. (2024) The UNITE database for molecular identification and taxonomic communication of fungi and other eukaryotes: Sequences, taxa and classifications reconsidered. Nucleic Acids Research 52(D1): D791–D797. 10.1093/nar/gkad1039 [DOI] [PMC free article] [PubMed]
- Albornoz FE, Ryan MH, Bending GD, Hilton S, Dickie IA, Gleeson DB, Standish RJ. (2022) Agricultural land‐use favours Mucoromycotinian, but not Glomeromycotinian, arbuscular mycorrhizal fungi across ten biomes. The New Phytologist 233(3): 1369–1382. 10.1111/nph.17780 [DOI] [PubMed] [Google Scholar]
- Anderson CR, Peterson ME, Frampton RA, Bulman SR, Keenan S, Curtin D. (2018) Rapid increases in soil pH solubilise organic matter, dramatically increase denitrification potential and strongly stimulate microorganisms from the Firmicutes phylum. PeerJ 6: e6090. 10.7717/peerj.6090 [DOI] [PMC free article] [PubMed]
- Artz RRE, Anderson IC, Chapman SJ, Hagn A, Schloter M, Potts JM, Campbell CD. (2007) Changes in fungal community composition in response to vegetational succession during the natural regeneration of cutover peatlands. Microbial Ecology 54(3): 508–522. 10.1007/s00248-007-9220-7 [DOI] [PubMed] [Google Scholar]
- Berch SM, Fortin JA. (1983) Endogonepisiformis: Axenic culture and associations with Sphagnum, Pinussylvestris, Alliumcepa and Alliumporrum. Canadian Journal of Botany 61(3): 899–905. 10.1139/b83-100 [DOI]
- Bergmann T. (2024) CAOS-R: Character-based barcoding. Methods in Molecular Biology (Clifton, N.J. ) 2744: 347–357. 10.1007/978-1-0716-3581-0_22 [DOI] [PubMed] [Google Scholar]
- Bidartondo MI, Duckett JG. (2010) Conservative ecological and evolutionary patterns in liverwort-fungal symbioses. Proceedings. Biological Sciences 277(1680): 485–492. 10.1098/rspb.2009.1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidartondo MI, Read DJ, Trappe JM, Merckx V, Ligrone R, Duckett JG. (2011) The dawn of symbiosis between plants and fungi. Biology Letters 7(4): 574–577. 10.1098/rsbl.2010.1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaszkowski J, Niezgoda P, Meller E, Milczarski P, Zubek S, Malicka M, Uszok S, Casieri L, Goto BT, Magurno F. (2021) New taxa in Glomeromycota: Polonosporaceae fam. nov., Polonospora gen. nov., and P.polonica comb. nov. Mycological Progress 20(8): 941–951. 10.1007/s11557-021-01726-4 [DOI] [Google Scholar]
- Blaszkowski J, Sanchez-García M, Niezgoda P, Zubek S, Fernández F, Vila A, Al-Yahya’ei MN, Symanczik S, Milczarski P, Malinowski R, Cabello M, Goto BT, Casieri L, Malicka M, Bierza W, Magurno F. (2022) A new order, Entrophosporales, and three new Entrophospora species in Glomeromycota. Frontiers in Microbiology 13: 4615. 10.3389/fmicb.2022.962856 [DOI] [PMC free article] [PubMed]
- Bonfante P, Venice F. (2020) Mucoromycota: Going to the roots of plant-interacting fungi. Fungal Biology Reviews 34(2): 100–113. 10.1016/j.fbr.2019.12.003 [DOI] [Google Scholar]
- Bridge P, Hughes K. (2012) Mortierellasignyensis K. Voigt, P.M. Kirk & Bridge. Index Fungorum: Published Numbers 7: 1.
- Brundrett MC, Tedersoo L. (2018) Evolutionary history of mycorrhizal symbioses and global host plant diversity. The New Phytologist 220(4): 1108–1115. 10.1111/nph.14976 [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. (2009) BLAST+: Architecture and applications. BMC Bioinformatics 10(1): 421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. (1998) A revised six-kingdom system of life. Biological Reviews of the Cambridge Philosophical Society 73: 203–266. 10.1017/S0006323198005167 [DOI] [PubMed] [Google Scholar]
- Curlevski NJ, Xu Z, Anderson IC, Cairney JW. (2010) Converting Australian tropical rainforest to native Araucariaceae plantations alters soil fungal communities. Soil Biology & Biochemistry 42(1): 14–20. 10.1016/j.soilbio.2009.08.001 [DOI] [Google Scholar]
- da Silva GA, Corazon-Guivin MA, de Assis DM, Oehl F. (2023) Blaszkowskia, a new genus in Glomeraceae. Mycological Progress 22(11): 74. 10.1007/s11557-023-01919-z [DOI]
- Damon C, Barroso G, Férandon C, Ranger J, Fraissinet-Tachet L, Marmeisse R. (2010) Performance of the COX1 gene as a marker for the study of metabolically active Pezizomycotina and Agaricomycetes fungal communities from the analysis of soil RNA. FEMS Microbiology Ecology 74(3): 693–705. 10.1111/j.1574-6941.2010.00983.x [DOI] [PubMed] [Google Scholar]
- Davison J, Moora M, Öpik M, Adholeya A, Ainsaar L, Ba A, Burla S, Diedhiou AG, Hiiesalu I, Jairus T, Johnson NC, Kane A, Koorem K, Kochar M, Ndiaye C, Pärtel M, Reier Ü, Saks Ü, Singh R, Vasar M, Zobel M. (2015) Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism. Science 349(6251): 970–973. 10.1126/science.aab1161 [DOI] [PubMed] [Google Scholar]
- de Beer ZW, Marincowitz S, Duong TA, Kim JJ, Rodrigues A, Wingfield MJ. (2016) Hawksworthiomyces gen. nov. (Ophiostomatales), illustrates the urgency for a decision on how to name novel taxa known only from environmental nucleic acid sequences (ENAS). Fungal Biology 120(11): 1323–1340. 10.1016/j.funbio.2016.07.004 [DOI] [PubMed] [Google Scholar]
- Desiro A, Duckett JG, Pressel S, Villarreal JC, Bidartondo MI. (2013) Fungal symbioses in hornworts: A chequered history. Proceedings. Biological Sciences 280(1759): 20130207. 10.1098/rspb.2013.0207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiro A, Rimington WR, Jacob A, Pol NV, Smith ME, Trappe JM, Bidartondo MI, Bonito G. (2017) Multigene phylogeny of Endogonales, an early diverging lineage of fungi associated with plants. IMA Fungus 8(2): 245–264. 10.5598/imafungus.2017.08.02.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doweld AB. (2014) Nomenclatural novelties. Index Fungorum: Published Numbers 57: 1.
- Eichorst SA, Kuske CR. (2012) Identification of cellulose-responsive bacterial and fungal communities in geographically and edaphically different soils by using stable isotope probing. Applied and Environmental Microbiology 78(7): 2316–2327. 10.1128/AEM.07313-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshghi Sahraei S, Furneaux B, Kluting K, Zakieh M, Rydin H, Hytteborn H, Rosling A. (2022) Effects of operational taxonomic unit inference methods on soil microeukaryote community analysis using long‐read metabarcoding. Ecology and Evolution 12(3): e8676. 10.1002/ece3.8676 [DOI] [PMC free article] [PubMed]
- Field KJ, Rimington WR, Bidartondo MI, Allinson KE, Beerling DJ, Cameron DD, Duckett JG, Leake JR, Pressel S. (2015) First evidence of mutualism between ancient plant lineages (Haplomitropsida liverworts) and Mucoromycotina fungi and its response to simulated Paleozoic changes in atmospheric CO2). The New Phytologist 205(2): 743–756. 10.1111/nph.13024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardes M, Bruns TD. (1993) ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizas and rusts. Molecular Ecology 2(2): 113–118. 10.1111/j.1365-294X.1993.tb00005.x [DOI] [PubMed] [Google Scholar]
- Gautam SP, Patel US. (2007) Rhizoendomutualmycota (REMM): A new phylum for the farmer’s friend number one. The Mycorrhizae, Diversity, Ecology and Applications. Hindu Publishing, Delhi.
- Guichon SHA. (2015) Mycorrhizal fungi: unlocking their ecology and role in the establishment and growth performance of different conifer species in nutrient-poor coastal forests. PhD thesis. University of British Columbia, Vancouver.
- Hirose D, Degawa Y, Yamamoto K, Yamada A. (2014) Sphaerocreaspubescens is a member of the Mucoromycotina closely related to fungi associated with liverworts and hornworts. Mycoscience 55(3): 221–226. 10.1016/j.myc.2013.09.002 [DOI] [Google Scholar]
- Hongsanan S, Jeewon R, Purahong W, Xie N, Liu JK, Jayawardena RS, Ekanayaka AH, Dissanayake A, Raspé O, Hyde KD, Stadler M, Peršoh D. (2018) Can we use environmental DNA as holotypes? Fungal Diversity 92(1): 1–30. 10.1007/s13225-018-0404-x [DOI]
- Hoysted GA, Kowal J, Jacob A, Rimington WR, Duckett JG, Pressel S, Orchard S, Ryan MH, Field KJ, Bidartondo MI. (2018) A mycorrhizal revolution. Current Opinion in Plant Biology 44: 1–6. 10.1016/j.pbi.2017.12.004 [DOI] [PubMed] [Google Scholar]
- Jaczewski AA, Jaczewski PA. (1931) Opredelitel Gribov. Sovershennye Griby (diploidnye stadii). I. Fikomicety. Moscow: Nauk Press.
- Jamy M, Biwer C, Vaulot D, Obiol A, Jing H, Peura S, Massana R, Burki F. (2022) Global patterns and rates of habitat transitions across the eukaryotic tree of life. Nature Ecology and Evolution 6(10): 1458–1470. 10.1038/s41559-022-01838-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsoom Khan F, Kluting K, Tångrot J, Urbina H, Ammunet T, Eshghi Sahraei S, Rydén M, Ryberg M, Rosling A. (2020) Naming the untouchable–environmental sequences and niche partitioning as taxonomical evidence in fungi. IMA Fungus 11(1): 23. 10.1186/s43008-020-00045-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution 30(4): 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk PM. (2012) Nomenclatural novelties: Piromycescryptodigmaticus Fliegerová, K. Voigt & P.M. Kirk. Index Fungorum: Published Numbers 1: 1.
- Kivlin SN. (2020) Global mycorrhizal fungal range sizes vary within and among mycorrhizal guilds but are not correlated with dispersal traits. Journal of Biogeography 47(9): 1994–2001. 10.1111/jbi.13866 [DOI] [Google Scholar]
- Klaubauf S, Inselbacher E, Zechmeister-Boltenstern S, Wanck W, Gottsberger R, Strauss J, Gorfer M. (2010) Molecular diversity of fungal communities in agricultural soils from lower Austria. Fungal Diversity 44(1): 65–75. 10.1007/s13225-010-0053-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohout P, Sudova R, Janouskova M, Ctvrtlikova M, Hejda M, Pankova H, Slavikova R, Stajerova K, Vosatka M, Sykorova Z. (2014) Comparison of commonly used primer sets for evaluating arbuscular mycorrhizal fungal communities: Is there a universal solution? Soil Biology & Biochemistry 68: 482–493. 10.1016/j.soilbio.2013.08.027 [DOI]
- Koske RE, Miller DD, Walker C. (1983) Gigasporareticulata: A newly described endomycorrhizal fungus from New England. Mycotaxon 16: 429–435. [Google Scholar]
- Krüger M, Stockinger H, Krüger C, Schüssler A. (2009) DNA-based species level detection of Glomeromycota: One PCR primer set for all arbuscular mycorrhizal fungi. The New Phytologist 183(1): 212–223. 10.1111/j.1469-8137.2009.02835.x [DOI] [PubMed] [Google Scholar]
- Larsson A. (2014) AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30(22): 3276–3278. 10.1093/bioinformatics/btu531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligrone R, Carafa A, Lumini E, Bianciotto V, Bonfante P, Duckett JG. (2007) Glomeromycotean associations in liverworts: A molecular, cellular, and taxonomic analysis. American Journal of Botany 94(11): 1756–1777. 10.3732/ajb.94.11.1756 [DOI] [PubMed] [Google Scholar]
- Link HF. (1809) Observationes in ordines plantarum naturales. Dissertatio Ima. Magazin für die Neuesten Entdeckungen in der Gesammten Naturkunde. Gesellschaft Naturforschender Freunde zu Berlin 3: 3–42. [Google Scholar]
- Lücking R, Hawksworth DL. (2018) Formal description of sequence-based voucherless Fungi: Promises and pitfalls, and how to resolve them. IMA Fungus 9(1): 143–165. 10.5598/imafungus.2018.09.01.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lücking R, Moncada B. (2017) Dismantling Marchandiomphalina into Agonimia (Verrucariaceae) and Lawreymyces gen. nov. (Corticiaceae): Setting a precedent to the formal recognition of thousands of voucherless fungi based on type sequences. Fungal Diversity 84(1): 119–138. 10.1007/s13225-017-0382-4 [DOI] [Google Scholar]
- Lücking R, Aime MC, Robbertse B, Miller AN, Aoki T, Ariyawansa HA, Cardinali G, Crous PW, Druzhinina IS, Geiser DM, Hawksworth DL, Hyde KD, Irinyi L, Jeewon R, Johnston PR, Kirk PM, Malosso E, May TW, Meyer W, Nilsson HR, Öpik M, Robert V, Stadler M, Thines M, Vu D, Yurkov AM, Zhang N, Schoch CL. (2021) Fungal taxonomy and sequence-based nomenclature. Nature Microbiology 6(5): 540–548. 10.1038/s41564-021-00888-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 17(1): 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Marčiulynienė D, Marčiulynas A, Lynikienė J, Vaičiukynė M, Gedminas A, Menkis A. (2021) DNA-metabarcoding of belowground fungal communities in bare-root forest nurseries: Focus on different tree species. Microorganisms 9(1): 150. 10.3390/microorganisms9010150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikryukov V, Dulya O, Zizka A, Bahram M, Hagh-Doust N, Anslan S, Prylutskyi O, Delgado-Baquerizo M, Maestre FT, Nilsson H, Pärn J, Tedersoo L. (2023) Connecting the multiple dimensions of global soil fungal diversity. Science Advances 9(48): eadj8016. 10.1126/sciadv.adj8016 [DOI] [PMC free article] [PubMed]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R. (2020) IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37(5): 1530–1534. 10.1093/molbev/msaa015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoliu-Nerin M, Sánchez-García M, Bergin C, Kutschera VE, Johannesson H, Bever JD, Rosling A. (2021) In-depth phylogenomic analysis of arbuscular mycorrhizal fungi based on a comprehensive set of de novo genome assemblies. Frontiers in Fungal Biology 2: 716385. 10.3389/ffunb.2021.716385 [DOI] [PMC free article] [PubMed]
- Morton JB, Benny GL. (1990) Revised classification of arbuscular mycorrhizal fungi (Zygomycetes): A new order, Glomales, two new suborders, Glomineae and Gigasporineae, and two new families, Acaulosporaceae and Gigasporaceae, with an emendation of Glomaceae. Mycotaxon 37: 471–491.
- Mueller RC, Balasch MM, Kuske CL. (2014) Contrasting soil fungal community responses to experimental nitrogen addition using the large subunit rRNA taxonomic marker and cellobiohydrolase I functional marker. Molecular Ecology 23(17): 4406–4417. 10.1111/mec.12858 [DOI] [PubMed] [Google Scholar]
- Nilsson RH, Abarenkov K, Veldre V, Nylinder S, de Wit P, Brosche S, Alfredsson JF, Ryberg M, Kristiansson E. (2010) An open source chimera checker for the fungal ITS region. Molecular Ecology Resources 10(6): 1076–1081. 10.1111/j.1755-0998.2010.02850.x [DOI] [PubMed] [Google Scholar]
- Nilsson RH, Wurzbacher C, Bahram M, Coimbra VRM, Larsson E, Tedersoo L, Eriksson J, Duarte Ritter C, Svantesson S, Sánchez-García M, Ryberg M, Kristiansson E, Abarenkov K. (2016) Top 50 most wanted fungi. MycoKeys 12: 29–40. 10.3897/mycokeys.12.7553 [DOI] [Google Scholar]
- Oehl F, Redecker D, Sieverding E. (2005) Glomusbadium, a new sporocarpic mycorrhizal fungal species from European grasslands with higher soil pH. Journal of Applied Botany and Food Quality 79: 38–43. [Google Scholar]
- Oehl F, Sieverding E, Palenzuela J, Ineichen K, da Silva GA. (2011) Advances in Glomeromycota taxonomy and classification. IMA Fungus 2(2): 191–199. 10.5598/imafungus.2011.02.02.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öpik M, Davison J, Moora M, Zobel M. (2014) DNA-based detection and identification of Glomeromycota: The virtual taxonomy of environmental sequences. Botany 92(2): 135–147. 10.1139/cjb-2013-0110 [DOI] [Google Scholar]
- Orchard S, Hilton S, Bending GD, Dickie IA, Standish RJ, Gleeson DB, Jeffery RP, Powell JR, Walker C, Bass D, Monk J, Simonin A, Ryan MH. (2017a) Fine endophytes (Glomustenue) are related to Mucoromycotina, not Glomeromycota. The New Phytologist 213(2): 481–486. 10.1111/nph.14268 [DOI] [PubMed]
- Orchard S, Standish RJ, Dickie IA, Renton M, Walker C, Moot D, Ryan MH. (2017b) Fine root endophytes under scrutiny: A review of the literature on arbuscule-producing fungi recently suggested to belong to the Mucoromycotina. Mycorrhiza 27(7): 619–638. 10.1007/s00572-017-0782-z [DOI] [PubMed]
- Palenzuela J, Ferrol N, Boller T, Azcón-Aguilar C, Oehl F. (2008) Otosporabareai, a new fungal species in the Glomeromycetes from a dolomitic shrub land in Sierra de Baza National Park (Granada, Spain). Mycologia 100(2): 296–305. 10.1080/15572536.2008.11832484 [DOI] [PubMed] [Google Scholar]
- Palenzuela J, Barea JM, Ferrol N, Azcón-Aguilar C, Oehl F. (2010) Entrophosporanevadensis, a new arbuscular mycorrhizal fungus from Sierra Nevada National Park (southeastern Spain). Mycologia 102(3): 624–632. 10.3852/09-145 [DOI] [PubMed] [Google Scholar]
- Pirozynski KA, Dalpé Y. (1989) Geological history of the Glomaceae with particular reference to mycorrhizal symbiosis. Symbiosis 7: 1–36. [Google Scholar]
- Rambaut A. (2018) FigTree v1.4.4. https://github.com/rambaut/figtree/releases
- Redecker D, Schüssler A, Stockinger H, Stürmer SL, Morton JB, Walker C. (2013) An evidence-based consensus for the classification of arbuscular mycorrhizal fungi (Glomeromycota). Mycorrhiza 23(7): 515–531. 10.1007/s00572-013-0486-y [DOI] [PubMed] [Google Scholar]
- Renner SS. (2016) A return to Linnaeus’s focus on diagnosis, not description: The use of DNA characters in the formal naming of species. Systematic Biology 65(6): 1085–1095. 10.1093/sysbio/syw032 [DOI] [PubMed] [Google Scholar]
- Reynolds DR, Taylor JW. (1991) DNA specimens and the ‘International code of botanical nomenclature’. Taxon 40(2): 311–315. 10.2307/1222985 [DOI] [Google Scholar]
- Rosling A, Eshghi Sahraei S, Kalsoom Khan F, Desirò A, Bryson AE, Mondo SJ, Grigoriev IV, Bonito G, Sánchez-García M. (2024) Evolutionary history of arbuscular mycorrhizal fungi and genomic signatures of obligate symbiosis. BMC Genomics 25(1): 529. 10.1186/s12864-024-10391-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryberg M, Nilsson RH. (2018) New light on names and naming of dark taxa. MycoKeys 30: 31–39. 10.3897/mycokeys.30.24376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccardo PA. (1889) Discomyceteae et Phymatosphaeriaceae. Sylloge Fungorum 8: 1–1143.
- Sato K, Suyama Y, Saito M, Sugawara K. (2005) A new primer for discrimination of arbuscular mycorrhizal fungi with polymerase chain reaction‐denature gradient gel electrophoresis. Grassland Science 51(2): 179–181. 10.1111/j.1744-697X.2005.00023.x [DOI] [Google Scholar]
- Sayers EW, Cavanaugh M, Clark K, Pruitt KD, Sherry ST, Yankie L, Karsch-Mizrachi I. (2024) GenBank 2024 update. Nucleic Acids Research 52(D1): D134–D137. 10.1093/nar/gkad903 [DOI] [PMC free article] [PubMed]
- Schüssler A, Schwarzott D, Walker C. (2001) A new fungal phylum, the Glomeromycota: Phylogeny and evolution. Mycological Research 105(12): 1413–1421. 10.1017/S0953756201005196 [DOI] [Google Scholar]
- Seeliger M, Hilton S, Muscatt G, Walker C, Bass D, Albornoz F, Standish RJ, Gray ND, Mercy L, Rempelos L, Schneider C. (2023) New fungal primers reveal the diversity of Mucoromycotinian arbuscular mycorrhizal fungi and their response to nitrogen application. Research Square Preprints 3: 3463087. 10.21203/rs.3.rs-3463087/v1 [DOI]
- Shen W, Sipos B, Zhao L. (2024) SeqKit2: A Swiss army knife for sequence and alignment processing. iMeta 5(3): e191. 10.1002/imt2.191 [DOI] [PMC free article] [PubMed]
- Smith SE, Read DJ. (2008) Mycorrhizal Symbiosis, 3rd edn. 787 pp. Academic Press, London, UK.
- Soudzilovskaia NA, van Bodegom PM, Terrer C, van’t Zelfde M, McCallum I, McCormack ML, Fisher JB, Brundrett MC, de Sá NC, Tedersoo L. (2019) Global mycorrhizal plant distribution linked to terrestrial carbon stocks. Nature Communications 10(1): 5077. 10.1038/s41467-019-13019-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain JL, Miranda JD. (1996) Glomusbrasilianum: An ornamented species in the Glomaceae. Mycotaxon 60: 137–142.
- Spatafora JW, Chang Y, Benny GL, Lazarus K, Smith ME, Berbee ML, Bonito G, Corradi N, Grigoriev I, Gryganskyi A, James TY, O’Donnell K, Roberson RW, Taylor TN, Uehling J, Vilgalys R, White MM, Stajich JE. (2016) A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108(5): 1028–1046. 10.3852/16-042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwyk JL, Buida TJ III, Li Y, Shen XX, Rokas A. (2020) ClipKIT: A multiple sequence alignment trimming software for accurate phylogenomic inference. PLoS Biology 18(12): e3001007. 10.1371/journal.pbio.3001007 [DOI] [PMC free article] [PubMed]
- Stockinger H, Peyret-Guzzon M, Koegel S, Bouffaud M-L, Redecker D. (2014) The largest subunit of RNA polymerase II as a new marker gene to study assemblages of arbuscular mycorrhizal fungi in the field. PLoS ONE 9: e107783. 10.1371/journal.pone.0107783 [DOI] [PMC free article] [PubMed]
- Tanaka S, Hashimoto K, Kobayashi Y, Yano K, Maeda T, Kameoka H, Ezawa T, Saito K, Akiyama K, Kawaguchi M. (2022) Asymbiotic mass production of the arbuscular mycorrhizal fungus Rhizophagusclarus. Communications Biology 5(1): 43. 10.1038/s42003-021-02967-5 [DOI] [PMC free article] [PubMed]
- Tedersoo L, Anslan S. (2019) Towards PacBio-based pan-eukaryote metabarcoding using full-length ITS sequences. Environmental Microbiology Reports 11(5): 659–668. 10.1111/1758-2229.12776 [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Lindahl B. (2016) Fungal identification biases in microbiome projects. Environmental Microbiology Reports 8(5): 774–779. 10.1111/1758-2229.12438 [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Smith ME. (2017) Ectomycorrhizal fungal lineages: Detection of four new groups and notes on consistent recognition of ectomycorrhizal taxa in high-throughput sequencing studies. Ecological Studies 230: 125–142. 10.1007/978-3-319-56363-3_6 [DOI] [Google Scholar]
- Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R, Villarreal-Ruiz L, Vasco-Palacios A, Quang Thu P, Suija A, Smith ME, Sharp C, Saluveer E, Saitta A, Ratkowsky D, Pritsch K, Riit T, Põldmaa K, Piepenbring M, Phosri C, Peterson M, Parts K, Pärtel K, Otsing E, Nouhra E, Njouonkou AL, Nilsson RH, Morgado LN, Mayor J, May TW, Kohout P, Hosaka K, Hiiesalu I, Henkel TW, Harend H, Guo L, Greslebin A, Grelet G, Geml J, Gates G, Dunstan W, Dunk C, Drenkhan R, Dearnaley J, De Kesel A, Dang T, Chen X, Buegger F, Brearley FQ, Bonito G, Anslan S, Abell S, Abarenkov K. (2014) Global diversity and geography of soil fungi. Science 346(6213): 1078. 10.1126/science.1256688 [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Bahram M, Puusepp R, Nilsson RH, James TY. (2017) Novel soil-inhabiting clades fill gaps in the fungal tree of life. Microbiome 5(1): 42. 10.1186/s40168-017-0259-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedersoo L, Sánchez-Ramírez S, Kõljalg U, Bahram M, Döring M, Schigel D, May T, Ryberg M, Abarenkov K. (2018) High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Diversity 90(1): 135–159. 10.1007/s13225-018-0401-0 [DOI] [Google Scholar]
- Tedersoo L, Mikryukov V, Anslan S, Bahram M, Khalid AN, Corrales A, Agan A, Vasco-Palacios AM, Saitta A, Antonelli A, Rinaldi AC, Verbeken A, Sulistyo BP, Tamgnoue B, Furneaux B, Ritter CD, Nyamukondiwa C, Sharp C, Marín C, Dai DQ, Gohar D, Sharmah D, Biersma EM, Cameron EK, De Crop E, Otsing E, Davydov EA, Albornoz FE, Brearley FQ, Buegger F, Gates G, Zahn G, Bonito G, Hiiesalu I, Hiiesalu I, Zettur I, Barrio IC, Pärn J, Heilmann-Clausen J, Ankuda J, Kupagme JY, Sarapuu J, Maciá-Vicente JG, Fovo JD, Geml J, Alatalo JM, Alvarez-Manjarrez J, Monkai J, Põldmaa K, Runnel K, Adamson K, Bråthen KA, Pritsch K, Tchan KI, Armolaitis K, Hyde KD, Newsham KK, Panksep K, Adebola LA, Lamit LJ, Saba M, da Silva Cáceres ME, Tuomi M, Gryzenhout M, Bauters M, Bálint M, Wijayawardene N, Hagh-Doust N, Yorou NS, Kurina O, Mortimer PE, Meidl P, Nilsson RH, Puusepp R, Casique-Valdés R, Drenkhan R, Garibay-Orijel R, Godoy R, Alfarraj S, Rahimlou S, Põlme S, Dudov SV, Mundra S, Ahmed T, Netherway T, Henkel TW, Roslin T, Fedosov VE, Onipchenko VG, Yasanthika WAE, Lim YW, Piepenbring M, Klavina D, Kõljalg U, Abarenkov K. (2021) The Global Soil Mycobiome consortium dataset for boosting fungal diversity research. Fungal Diversity 111(1): 573–588. 10.1007/s13225-021-00493-7 [DOI] [Google Scholar]
- Tedersoo L, Hosseyni Moghaddam MS, Mikryukov V, Hakimzadeh A, Bahram M, Nilsson RH, Yatsiuk I, Geisen S, Schwelm A, Piwosz K, Prous M, Chmolowska D, Rueckert S, Skaloud P, Laas P, Thines M, Jung J-H, Alkahtani S, Anslan S. (2024) EUKARYOME: The rRNA gene reference database for identification of all eukaryotes. Database 2024: baae043. 10.1093/database/baae043 [DOI] [PMC free article] [PubMed]
- Thines M, Crous PW, Aime MC, Aoki T, Cai L, Hyde KD, Miller AN, Zhang N, Stadler M. (2018) Ten reasons why a sequence-based nomenclature is not useful for fungi anytime soon. IMA Fungus 9(1): 177–183. 10.5598/imafungus.2018.09.01.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Cruz TJ, Billingsley Tobias TL, Almatruk M, Hesse CN, Kuske CR, Desirò A, Niccolò Benucci GR, Bonito G, Stajich JE, Dunlap C, Arnold AE, Porras-Alfaro A. (2017) Bifiguratusadelaidae gen. et sp. nov., a new member of Mucoromycotina in endophytic and soil-dwelling habitats. Mycologia 109(3): 363–378. 10.1080/00275514.2017.1364958 [DOI] [PubMed] [Google Scholar]
- Trappe JM, Bloss HE, Menge JA. (1984) Glomusdeserticola sp.nov. Mycotaxon 20: 123–127. [Google Scholar]
- Tulasne L-R, Tulasne C. (1844) Fungi nonnulli hypogaei, novi v. minus cogniti. Giornale Botanico Italiano 1: 55–63. [Google Scholar]
- Turland NJ, Wiersema JH, Barrie FR, Greuter W, Hawksworth DL. (2018) International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code). Koeltz Botanical Books, Glashütten. 10.12705/Code.2018 [DOI]
- van Geel M, Busschaert P, Honnay O, Lievens B. (2014) Evaluation of six primer pairs targeting the nuclear rRNA operon for characterization of arbuscular mycorrhizal fungal (AMF) communities using 454 pyrosequencing. Journal of Microbiological Methods 106: 93–100. 10.1016/j.mimet.2014.08.006 [DOI] [PubMed] [Google Scholar]
- VanKuren NW, den Bakker HC, Morton JB, Pawlowska TE. (2013) Ribosomal RNA gene diversity, effective population size, and evolutionary longevity in asexual Glomeromycota. Evolution 67(1): 207–224. 10.1111/j.1558-5646.2012.01747.x [DOI] [PubMed] [Google Scholar]
- Varma A, Prasad R, Tuteja N [Eds] (2017) Mycorrhiza-function, diversity, state of the art. Springer, Berlin. 10.1007/978-3-319-53064-2 [DOI]
- Vasar M, Davison J, Sepp SK, Oja J, Al-Quraishy S, Bueno CG, Cantero JJ, Fabiano EC, Decocq G, Fraser L, Hiiesalu I, Hozzein WN, Koorem K, Moora M, Mucina L, Onipchenko V, Öpik M, Pärtel M, Phosri C, Vahter T, Tedersoo L, Zobel M. (2022) Global taxonomic and phylogenetic assembly of AM fungi. Mycorrhiza 32(2): 135–144. 10.1007/s00572-022-01072-7 [DOI] [PubMed] [Google Scholar]
- Walker C, Schüssler A. (2004) Nomenclatural clarifications and new taxa in the Glomeromycota. Mycological Research 108(9): 981–982. 10.1017/S0953756204231173 [DOI]
- Walker C, Gollotte A, Redecker D. (2018) A new genus, Planticonsortium (Mucoromycotina), and new combination (P.tenue), for the fine root endophyte, Glomustenue (basionym Rhizophagustenuis). Mycorrhiza 28(3): 213–219. 10.1007/s00572-017-0815-7 [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns TD, Lee SB, Taylor JW. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (Eds) PCR Protocols: A Guide to Methods and Applications, 315–322. Academic Press, New York. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Wijayawardene NN, Hyde KD, Dai DQ, Sánchez-García M, Goto BT, Saxena RK, Erdogdu M, Selçuk F, Rajeshkumar KC, Aptroot A, Błaszkowski J, Boonyuen N, da Silva GA, de Souza FA, Dong W, Ertz D, Haelewaters D, Jones EBG, Karunarathna SC, Kirk PM, Kukwa M, Kumla J, Leontyev DV, Lumbsch HT, Maharachchikumbura SSN, Marguno F, Martínez-Rodríguez P, Mešić A, Monteiro JS, Oehl F, Pawłowska J, Pem D, Pfliegler WP, Phillips AJL, Pošta A, He MQ, Li JX, Raza M, Sruthi OP, Suetrong S, Suwannarach N, Tedersoo L, Thiyagaraja V, Tibpromma S, Tkalčec Z, Tokarev YS, Wanasinghe DN, Wijesundara DSA, Wimalaseana SDMK, Madrid H, Zhang GQ, Gao Y, Sánchez-Castro I, Tang LZ, Stadler M, Yurkov A, Thines M. (2022) Outline of Fungi and fungus-like taxa – 2021. Mycosphere 13(1): 53–453. 10.5943/mycosphere/13/1/2 [DOI] [Google Scholar]
- Yamamoto K, Degawa Y, Yamada A. (2020) Taxonomic study of Endogonaceae in the Japanese islands: New species of Endogone, Jimgerdemannia, and Vinositunica, gen. nov. Mycologia 112(2): 309–328. 10.1080/00275514.2019.1689092 [DOI] [PubMed] [Google Scholar]
- Zamora JC, Svensson M, Kirschner R, Olariaga I, Ryman S, Parra LA, Geml J, Rosling A, Adamčík S, Ahti T, Aime MC, Ainsworth AM, Albert L, Albertó E, García AA, Ageev D, Agerer R, Aguirre-Hudson B, Ammirati J, Andersson H, Angelini C, Antonín V, Aoki T, Aptroot A, Argaud D, Sosa BIA, Aronsen A, Arup U, Asgari B, Assyov B, Atienza V, Bandini D, Baptista-Ferreira JL, Baral H-O, Baroni T, Barreto RW, Beker H, Bell A, Bellanger J-M, Bellù F, Bemmann M, Bendiksby M, Bendiksen E, Bendiksen K, Benedek L, Bérešová-Guttová A, Berger F, Berndt R, Bernicchia A, Biketova AY, Bizio E, Bjork C, Boekhout T, Boertmann D, Böhning T, Boittin F, Boluda CG, Boomsluiter MW, Borovička J, Brandrud TE, Braun U, Brodo I, et al. (2018) Considerations and consequences of allowing DNA sequence data as types of fungal taxa. IMA Fungus 9(1): 167–175. 10.5598/imafungus.2018.09.01.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maximum Likelihood phylogram indicating phylogenetic relationships amongst Glomeromycota based on SSU-ITS-LSU sequences
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Leho Tedersoo, Franco Magurno, Saad Alkahtani, Vladimir Mikryukov
Data type
Maximum Likelihood phylogram indicating phylogenetic relationships amongst Gigasporales based on LSU sequences
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Leho Tedersoo, Franco Magurno, Saad Alkahtani, Vladimir Mikryukov
Data type
Maximum Likelihood phylogram indicating phylogenetic relationships amongst Endogonomycetes based on SSU-5.8S-LSU sequences
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Leho Tedersoo, Franco Magurno, Saad Alkahtani, Vladimir Mikryukov
Data type
Maximum Likelihood phylogram indicating phylogenetic relationships amongst Endogonomycetes based on SSU sequences
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Leho Tedersoo, Franco Magurno, Saad Alkahtani, Vladimir Mikryukov
Data type
Currently recognised orders, families and genera and proposed taxonomic groups in Glomeromycota
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Leho Tedersoo, Franco Magurno, Saad Alkahtani, Vladimir Mikryukov
Data type
Currently recognised orders, families and genera and proposed taxonomic groups in Endogonomycetes
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Leho Tedersoo, Franco Magurno, Saad Alkahtani, Vladimir Mikryukov
Data type
Data Availability Statement
All of the data that support the findings of this study are available in the main text or Supplementary Information.