Abstract
Autophagy eliminates dysfunctional mitochondria in an intricate process known as mitophagy. ULK1 is critical for the induction of autophagy, but its substrate(s) and mechanism of action in mitophagy remain unclear. Here, we show that ULK1 is upregulated and translocates to fragmented mitochondria upon mitophagy induction by either hypoxia or mitochondrial uncouplers. At mitochondria, ULK1 interacts with FUNDC1, phosphorylating it at serine 17, which enhances FUNDC1 binding to LC3. A ULK1-binding-deficient mutant of FUNDC1 prevents ULK1 translocation to mitochondria and inhibits mitophagy. Finally, kinase-active ULK1 and a phospho-mimicking mutant of FUNDC1 rescue mitophagy in ULK1-null cells. Thus, we conclude that FUNDC1 regulates ULK1 recruitment to damaged mitochondria, where FUNDC1 phosphorylation by ULK1 is crucial for mitophagy.
Keywords: FUNDC1, mitochondria, mitochondrial quality control, mitophagy, ULK1
Introduction
Maintenance of a healthy mitochondrial pool is essential for cellular homeostasis 1. Mitophagy is a major mechanism for mitochondrial quality control at the organelle level through two main mitophagy pathways, involving Parkin/FCCP or mitophagy receptors 2–7. Failure of mitophagy or autophagy results in a number of diseases, including neurodegeneration, diabetes and cancer 8–10.
ULK1 and ULK2 (the yeast ATG1 homologues) are Ser/Thr kinases required for early autophagosome formation 11–13. Mice deficient of either the ulk1 or the ulk2 gene displayed normal development but double knockout of both kinases in mice is lethal. The ability of ULK2 to compensate for the loss of ULK1 is further supported by evidence that ulk1-deficient cells displayed autophagy in response to glucose withdrawal, indicating that ULK1 and ULK2 are partially redundant protein kinases 14–16.
Recent studies suggest that ULK1 plays a more specific role in mitophagy. An ULK1-knockout mouse model shows defects in the autophagic clearance of mitochondria during erythroid maturation 17. ULK1 is associated with damaged mitochondria upon FCCP treatment 18. More recently, it has been found that phosphorylation of ULK1 by AMP-activated protein kinase (AMPK) connects cellular energy sensing to mitophagy 19. The ULK1 phosphorylation event was also reported by others, although the sites they identified are different 20–22. Despite the pivotal role of ULK1 in mitochondrial clearance, the molecular mechanisms by which ULK1 regulates mitophagy, the identity of the mitochondrial substrate of ULK1, and the subcellular localization of ULK1 upon mitophagy induction remain unknown. We previously identified that FUNDC1 is a novel mitophagy receptor 6. But how FUNDC1 is modulated by upstream signals to activate receptor-mediated mitophagy also remains poorly understood 23.
Here, we report that ULK1 is elevated and recruited to fragmented mitochondria in response to hypoxia or FCCP. The translocated ULK1 interacts with its substrate FUNDC1 and phosphorylates FUNDC1 at Ser-17. The ULK1-binding-deficient mutant of FUNDC1 or knockdown of FUNDC1 significantly impairs the translocation of ULK1 to fragmented mitochondria and inhibits mitophagy. The interaction between FUNDC1 and LC3 is enhanced by phosphorylation-mimetic mutant FUNDC1 (S17D) but is impaired by FUNDC1 (S17A). Hypoxia-, FCCP-, or FUNDC1-induced mitophagy is suppressed in ULK1−/− MEFs and can be restored by kinase-active form of ULK1 or FUNDC1 (S17D). Accordingly, both ULK1 and FUNDC1 are required for mitophagy and they collaboratively regulate mitophagy. Thus, our findings establish an unexpected link between ULK1 and FUNDC1 in response to different stresses.
Results
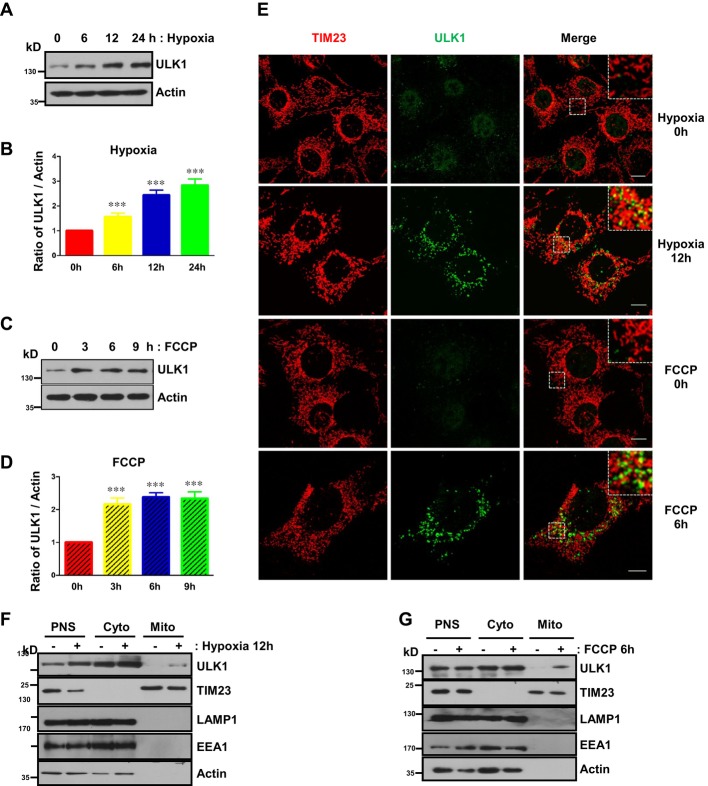
ULK1 is upregulated and translocates to fragmented mitochondria in response to mitophagy induced by hypoxia or FCCP
Hypoxia or the mitochondrial uncoupler FCCP induces extensive mitophagy 6, 24–26. We examined the expression of ULK1 under both conditions. The protein level of ULK1 is substantially elevated in response to hypoxia or FCCP treatment and has no significant changes upon short-term exposure to starvation but decreases after prolonged starvation (Fig 1A–D, and Supplementary Fig S1). Immunofluorescence results show that hypoxia or FCCP not only extensively induces mitochondrial fragmentation but also significantly enhances the number and size of ULK1-positive puncta (Fig 1E). A large number of ULK1-positive puncta translocate to fragmented mitochondria in both conditions compared with untreated control or with starvation condition (Fig 1E and Supplementary Fig S1). Subcellular fractionation assays further demonstrated that a proportion of ULK1 molecules are indeed co-fractionated with mitochondria in response to either hypoxia or FCCP (Fig 1F and G).
Figure 1. ULK1 is upregulated and translocates to fragmented mitochondria in response to mitophagy induced by hypoxia or FCCP.
A Levels of ULK1 in MEFs exposed to hypoxia (1% O2) for the indicated times.
B Quantification of the ratio of ULK1 to actin in (A).
C Levels of ULK1 in MEFs treated with FCCP (20 μM) for the indicated times.
D Quantification of the ratio of ULK1 to actin in (C).
E MEFs were exposed to hypoxia (1% O2) for 12 h or FCCP (20 μM) for 6 h and then fixed by 4% paraformaldehyde. Cells were stained with anti-TIM23 (mouse) and anti-ULK1 (rabbit) primary antibodies, then Alexa Fluor-488-labeled donkey anti-rabbit IgG and Alexa Fluor-555-labeled donkey anti-mouse IgG secondary antibodies before analysis by immunofluorescence microscopy. Scale bar, 10 μm.
F Immunoblots of subcellular fractions from control MEFs or MEFs that were exposed to hypoxia (1% O2) for 12 h. PNS, post-nuclear supernatant; Cyto, cytosol; Mito, mitochondria.
G Immunoblots of subcellular fractions from control MEFs or MEFs that were treated with FCCP (20 μM) for 6 h. PNS, post-nuclear supernatant; Cyto, cytosol; Mito, mitochondria.
Data information: All results are from three independent experiments. All quantitative data are presented as mean ± s.e.m. from 3 independent experiments; *** P < 0.001.
Source data are available online for this figure.
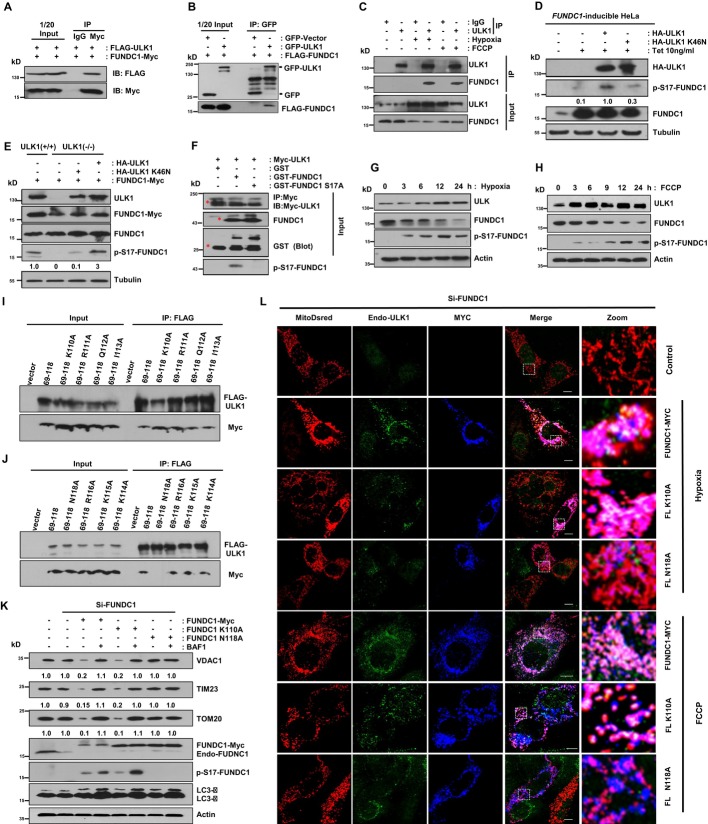
The mitophagy receptor FUNDC1 is a novel substrate of ULK1
The enrichment of ULK1 on fragmented mitochondria upon mitophagy induction suggests that there may be crosstalk between ULK1 and mitochondria. To find the mitochondrial substrate of ULK1, we used mass spectrometry to analyze the immunoprecipitants by the FLAG antibody from cell lysates that overexpressed FLAG-ULK1 and FUNDC1-MYC and found that FUNDC1-MYC is precipitated (Supplementary Fig S2).
Overexpression of FUNDC1 potentially induces mitophagy 6. Initially, we used transient transfection experiments to confirm that FUNDC1 is a binding partner of ULK1. Immunoprecipitation experiments revealed that FUNDC1-Myc interacts with FLAG-ULK1, but not with the IgG control (Fig 2A). Conversely, we also observed that FLAG-FUNDC1 coimmunoprecipitates with GFP-ULK1, but not with the GFP control (Fig 2B). We then confirmed the endogenous interaction between ULK1 and FUNDC1 upon mitophagy induction (Fig 2C).
Figure 2. The mitophagy receptor FUNDC1 is a novel substrate of ULK1 and FUNDC1 (N118A) that prevents its binding to ULK1, impairs ULK1 translocation to mitochondria, and inhibits mitophagy.
A HeLa cells were co-transfected with FUNDC1-Myc and FLAG-ULK1. Cells were lysed and immunoprecipitated with anti-Myc antibody 24 h after transfection.
B FLAG-FUNDC1 was co-transfected with GFP-vector or GFP-ULK1 for 24 h in HeLa cells, and then, cell lysates were immunoprecipitated with anti-GFP antibody.
C MEFs were exposed to hypoxia (1% O2) for 12 h or FCCP (20 μM) for 3 h, followed by immunoprecipitation using anti-ULK1 antibody.
D The expression of FUNDC1 is induced by 10 ng/ml tetracycline (Tet) in FUNDC1-inducible HeLa cells. Cells were transfected with HA-ULK1 or HA-ULK1 (K46N) for 24 h. Then, the cell lysates were prepared for immunoblotting using indicated antibodies.
E ULK1+/+ cells were transfected with FUNDC1-Myc, and ULK1 cells were transfected with FUNDC1-Myc alone, FUNDC1-Myc and HA-ULK1, or FUNDC1-Myc and HA-ULK1 (K46N). Cells were harvested and lysed for immunoblots 24 h after transfection.
F Purified GST-FUNDC1 and GST-FUNDC1 (S17A) were subjected to an in vitro kinase assay with Myc-ULK1 immunoprecipitated by anti-Myc antibody from Myc-ULK1-transfected cells about 24 h post-transfection. Phosphorylated FUNDC1 was detected by anti-p-S17-FUNDC1 antibody. The red asterisks mark the target bands.
G Western blot analysis of the kinetics of ULK1, FUNDC1, and the phosphorylated FUNDC1 in MEFs exposed to hypoxia (1% O2) for the indicated times.
H Western blot analysis of the kinetics of ULK1, FUNDC1, and the phosphorylated FUNDC1 in MEFs treated with FCCP (20 μM) for the indicated times.
I, J The single mutant which can abolish the ULK1 and FUNDC1 interaction was identified. HeLa cells were co-transfected with Flag-ULK1 and the indicated constructs. 24 h after transfection, cells were lysed for immunoprecipitation by anti-Flag antibody.
K MEFs were transfected with FUNDC1-Myc and its mutants after the endogenous FUNDC1 was knocked down by siRNA. 36 h after transfection, cells were harvested in the absence or presence of 50 nM bafilomycin A1 (BAF1) and then lysed for immunoblotting with the indicated antibodies.
L MEFs were co-transfected with FUNDC1-Myc or its mutants and MitoDsred after the endogenous FUNDC1 was knocked down by siRNA. 24 h post-transfection, cells were treated with hypoxic (1% O2) conditions for 12 h or FCCP (20 μM) for 6 h before fixed by 4% paraformaldehyde and stained with indicated antibodies. Scale bar, 10 μm.
Data information: All results are from three independent experiments.
Source data are available online for this figure.
Mass spectrometric analysis of cellular phosphoproteome revealed that FUNDC1 has a potential phosphorylation site at Ser-17 upon mitophagy induction (our unpublished observations), so we generated an anti-FUNDC1 (Ser-17)-specific antibody and verified its specificity (Supplementary Fig S3). We also generated a FUNDC1-inducible HeLa cell line using the Tetracycline-On system to see whether the phosphorylation state of FUNDC1 changes as mitophagy proceeds. Treatment with 10 ng/ml tetracycline elevated the expression of FUNDC1 and phosphorylated FUNDC1 (Fig 2D). Transfection with HA-ULK1 markedly promoted the phosphorylation of FUNDC1 at Ser-17, while transfection with the kinase-inactivated HA-ULK1 (K46N) showed much weaker phosphorylation since the mutant ULK1 (K46N) still has minor kinase activity (Fig 2E and Supplementary Fig S4) 27, 28. FUNDC1 (Ser-17) could not be phosphorylated in ULK1−/− MEFs, but when HA-ULK1 was reconstituted, the phosphorylation was significantly enhanced (Fig 2E). In vitro kinase assays further confirmed that Myc-ULK1 immunoprecipitated from cell lysates strongly phosphorylates GST-FUNDC1 at Ser-17 (Fig 2F). Time-course analysis of the phosphorylation of FUNDC1 at Ser-17 indicates that the level of ULK1 expression correlates with the phosphorylation of FUNDC1 upon mitophagy induction by hypoxia or FCCP (Fig 2G and H). However, starvation does not induce the phosphorylation of Ser-17 (Supplementary Fig S5).
FUNDC1 (N118A) prevents its binding to ULK1, impairs ULK1 translocation to mitochondria, and inhibits mitophagy
Next, we examined the role of FUNDC1 in the recruitment of ULK1 to mitochondria. Knockdown of Fundc1 markedly reduces mitochondria-associated ULK1 in response to hypoxia or FCCP but does not affect ULK1 puncta formation in starvation condition (Supplementary Figs S1 and S6). Next, we generated a series of FUNDC1 constructs to identify key elements required for the FUNDC1-ULK1 binding and for FUNDC1 mitochondrial localization. Immunoprecipitation reveals that N118 at FUNDC1 is essential for FUNDC1-ULK1 association (Fig 2I–N and Supplementary Fig S7). FUNDC1 loses its mitochondrial localization upon simultaneously deletion of the first and the second transmembrane domains (Fig 2I, Supplementary Figs S7 and S8). We found that mutation of N118 to Ala also reduced the translocation of ULK1 from cytosol to fragmented mitochondria and significantly inhibits mitophagy (Fig 2K and L).
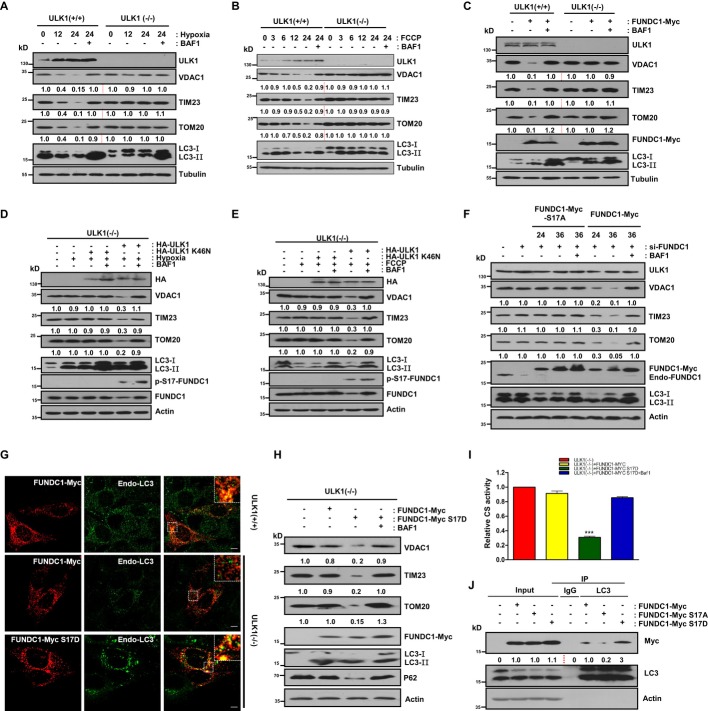
Kinase-active form of ULK1 is required for hypoxia-, FCCP-, or FUNDC1-induced mitophagy
We next investigated whether ULK1 is required for mitophagy. In ULK1+/+ MEFs, hypoxia, FCCP, or FUNDC1 overexpression promotes the autophagic degradation of the overall mitochondrial proteins (Fig 3A–C and Supplementary Fig S9). In ULK1−/− MEFs, however, the degradation is blocked (Fig 3A and C). We then assessed the role of the kinase-active form of ULK1 in hypoxia- or FCCP-induced mitophagy in ULK1-null MEFs. Wild-type ULK1 efficiently induces mitochondrial turnover by autophagy, whereas the kinase-inactivated form of ULK1 (K46N) does not, which suggests that the kinase activity of ULK1 is very important for mitophagy (Fig 3D and E). Mitophagy induced by hypoxia is reversed by the lysosomal ATPase inhibitor bafilomycin A1 (BAF1), but not by the proteasome inhibitor, MG132 (Fig 3A, Supplementary Figs S9,S10,S11). Mitophagy induced by FCCP, however, is reversed either by BAF1 or by MG132 but to a lesser extent by MG132 (Fig 3C, Supplementary Figs S9,S10,S11). These indicate that autophagy and proteasome are both involved in FCCP-induced mitochondrial protein turnover (Fig 3B, Supplementary Figs S9 and S11).
Figure 3. Mitophagy is restored in ULK1-null MEFs upon reconstitution of the kinase-active form of ULK1 or constitutively active FUNDC1 (S17D).
A ULK1+/+ and ULK1−/− cells were cultured under hypoxic (1% O2) condition for 12 h or 24 h in the absence or presence of 50 nM bafilomycin A1 (BAF1). Cell lysates were immunoblotted.
B ULK1+/+ and ULK1−/− cells were treated with FCCP (20 μM) for the indicated times in the absence or presence of 50 nM bafilomycin A1 (BAF1). Cell lysates were immunoblotted.
C ULK1+/+ and ULK1−/− cells were transfected with FUNDC1-Myc for 24 h and then harvested in the absence or presence of 50 nM bafilomycin A1 (BAF1). Cell lysates were immunoblotted.
D ULK1−/− cells were transfected with HA-ULK1 or HA-ULK1 (K46N), then cultured under hypoxic (1% O2) conditions for 24 h in the absence or presence of 50 nM bafilomycin A1 (BAF1). Cells lysates were immunoblotted.
E ULK1−/− cells were transfected with HA-ULK1 or HA-ULK1 (K46N) and then treated with FCCP (20 μM) for 24 h. Cells were harvested in the absence or presence of 50 nM bafilomycin A1 (BAF1). Cell lysates were immunoblotted.
F FUNDC1-knockdown MEFs were transfected with FUNDC1-Myc or FUNDC1-Myc (S17A) for the indicated times. Cell lysates were used for immunoblotting.
G ULK1+/+ cells were transfected with FUNDC1-Myc, and ULK1−/− cells were transfected with FUNDC1-Myc or FUNDC1-Myc (S17D). 24 h after transfection, cells were fixed with 4% paraformaldehyde and stained with anti-Myc (mouse) and anti-LC3 (rabbit) primary antibodies for immunofluorescence microscopy. Scale bar, 10 μm.
H ULK1−/− cells were transfected with FUNDC1-Myc or FUNDC1-Myc (S17D) for 24 h in the absence or presence of 50 nm bafilomycin A1 (BAF1) for an additional 6 h before harvesting. Cell lysates were immunoblotted.
I ULK1−/− cells were transfected with FUNDC1-Myc or FUNDC1-Myc (S17D) for 24 h in the absence or presence of 50 nm bafilomycin A1 (BAF1) for an additional 6 h before harvesting. Cell lysates were detected with the citrate synthase activity kit.
J HeLa cells were transfected with FUNDC1-Myc, FUNDC1-Myc (S17A), or FUNDC1-Myc (S17D). 24 h after transfection, cell lysates were immunoprecipitated with anti-LC3 antibody.
Data information: All results are from three independent experiments. All quantitative data are presented as mean ± s.e.m. from 3 independent experiments; *** P < 0.001.
Source data are available online for this figure.
Phosphorylation of FUNDC1 at Ser-17 restores mitophagy that is defective in ULK1-deficient cells by promoting FUNDC1-LC3 binding and colocalization
To elucidate the importance of the phosphorylation of FUNDC1 at Ser-17 by ULK1 in mitophagy, we compared the mitophagy-inducing ability of FUNDC1 (WT) with FUNDC1 (S17A) in Fundc1-KD MEFs. While FUNDC1 (WT) promotes effective mitophagy, FUNDC1 (S17A) cannot induce mitophagy (Fig 3F).
To further evaluate the role of phosphorylated Ser-17 in mitophagy, we constructed FUNDC1 (S17D), which mimics constitutive phosphorylation of FUNDC1, and compared its ability to mediate mitophagy with wild-type FUNDC1 in ULK1-deficient cells. Overexpression of FUNDC1 (WT) induces extensive colocalization of mitochondria with endogenous LC3 puncta in ULK1+/+ cells, which is largely suppressed in ULK1-deficient cells (Fig 3G). However, FUNDC1 (S17D) significantly restores the colocalization of mitochondria with LC3 dots in ULK1-deficient cells (Fig 3G). FUNDC1 (S17D) is able to rescue mitophagy in ULK1−/− MEFs, whereas FUNDC1 (WT) is not (Fig 3H and I). Immunoprecipitation demonstrated that compared to FUNDC1-Myc (WT), endogenous LC3 binds more strongly to FUNDC1-Myc (S17D) and much more weakly to FUNDC1-Myc (S17A) (Fig 3J). Since Ser-17 and Tyr-18 are two adjacent sites and Tyr-18 phosphorylation is inhibitory for mitophagy, we co-transfected SRC kinase and ULK1 to see whether two kinases have mutually inhibitory effects. As shown in Supplementary Fig S12, SRC inhibits the association of ULK1 puncta with mitochondria and suppresses phosphorylation of FUNDC1 at Ser-17 by ULK1.
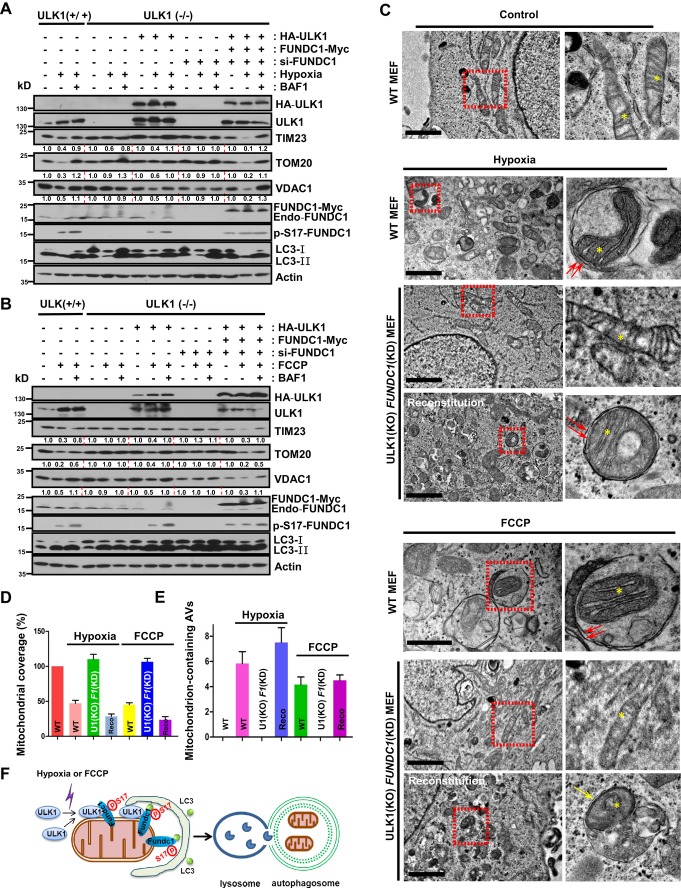
ULK1 and FUNDC1 cooperatively regulate mitophagy
Next, we determined the relationship between ULK1 and FUNDC1 in the regulation of mitophagy. Effective upregulation of ULK1 and autophagic degradation of mitochondrial proteins are readily observed in ULK1+/+ MEFs in exposure to hypoxia or FCCP treatment, while mitophagy is suppressed in ULK1−/− MEFs in response to both conditions (Fig 4A and B, and Supplementary Fig S13). When HA-ULK1 was reconstituted in ULK1−/− cells, mitophagy activity was recovered (Fig 4A and B, and Supplementary Fig S13). If FUNDC1 was knocked down in ULK1−/− cells, the autophagic removal of mitochondria was blocked compared to ULK1+/+ cells. However, mitophagy activity was much more strongly enhanced in ULK1- and FUNDC1-reconstituted ULK1−/− FUNDC1-KD cells than in ULK1 wild-type cells, or in ULK1−/− cells expressing HA-ULK1 alone (Fig 4A and B, and Supplementary Fig S13).
Figure 4. FUNDC1 and ULK1 cooperatively regulate mitophagy.
A ULK1+/+ or ULK1−/− cells were cultured under hypoxic (1% O2) or normoxic conditions in the absence or presence of 50 nM bafilomycin A1 (BAF1). ULK1−/− cells were transfected with HA-ULK1, and ULK1−/− FUNDC1-KD cells were transfected with HA-ULK1 and FUNDC1-Myc, then cultured under hypoxic (1% O2) conditions for 24 h in the absence or presence of 50 nM bafilomycin A1 (BAF1). Cells were harvested and lysed for immunoblotting assays.
B ULK1+/+ or ULK1−/− cells were treated with or without FCCP (20 μM) in the absence or presence of 50 nM bafilomycin A1 (BAF1). ULK1−/− cells were transfected with HA-ULK1 and ULK1−/− FUNDC1-KD cells were transfected with HA-ULK1 and FUNDC1-Myc and then treated with FCCP (20 μM) for 24 h in the absence or presence of 50 nM bafilomycin A1 (BAF1). Cells were harvested and lysed for immunoblotting assays.
C ULK1+/+, ULK1−/− FUNDC1-KD, or ULK1−/− FUNDC1-KD cells rescued by introduction of HA-ULK1 and FUNDC1-Myc were cultured under hypoxic (1% O2) conditions for 24 h or with FCCP (20 μM) for 24 h. Samples were analyzed by electron microscopy. Scale bar, 2 μm. The yellow asterisks mark mitochondria. The red arrows indicate double-membraned autophagic structures. The yellow arrow denotes autolysosomes.
D–E The mitochondrial coverage and mitochondrial-containing AVs shown in (C) were quantified. Recon, reconstitution.
F Provisional working model of the interplay between ULK1 and FUNDC1 in mitophagy regulation.
Data information: All results are from three independent experiments. All quantitative data are presented as mean ± s.e.m. from 3 independent experiments; ***P < 0.001.
Source data are available online for this figure.
Finally, we monitored mitophagy by electron microscopy. Normal-shaped mitochondria were abundant in control MEFs, while fewer mitochondria were observed in wild-type MEFs treated with hypoxia or FCCP (Fig 4C and E, and Supplementary Fig S14). Mitophagosomes were frequently observed in wild-type MEFs in response to hypoxia or FCCP (Fig 4C and E). In ULK1−/− FUNDC1-KD cells, however, the degradation of mitochondria was blocked and the formation of mitophagosomes was heavily suppressed under both conditions (Fig 4A and E, and Supplementary Fig S14). Mitophagy activity was restored when ULK1 and FUNDC1 expression was reconstituted in these cells (Fig 4A and E, and Supplementary Fig S14).
Taken together, our data show that in response to hypoxia, ULK1 is upregulated and associates with fragmented mitochondria, and this association in turn is required for interaction of ULK1 with the mitochondrial substrate FUNDC1 and for phosphorylation of FUNDC1 at Ser-17. The phosphorylation of FUNDC1 by ULK1 promotes the interaction between FUNDC1 and LC3, a critical step for bridging fragmented mitochondria and autophagosomes (Fig 4F).
Discussion
To date, several substrates of ULK1 in macroautophagy have been identified including Atg13, Atg9, and Beclin1 29–31. Here, our findings not only identified FUNDC1 as a novel substrate of ULK1 in selective mitophagy, but also revealed the specific role of ULK1 as well as how these two proteins coordinate to regulate this process.
It is well established that Parkin and PINK1 are required for mitophagy induced by the mitochondrial uncoupler FCCP 2, 32. PINK1 is stabilized at depolarized mitochondria and recruits Parkin to mediate the ubiquitination and degradation of a large number of substrates in the outer mitochondrial membrane, which is thought to be essential for subsequent mitophagy 33, 34. But the exact mechanism remains unknown. It has been proposed that in the initial steps of Parkin-mediated mitophagy, the structure containing the ULK1 complex is independently recruited to depolarized mitochondria and is required for further recruitment of the downstream Atg proteins, indicating that ULK1 has a more specific role in selective autophagy 18. The adaptor protein on mitochondria that recruits ULK1, however, has not been found.
We observed markedly increased expression and colocalization of ULK1 with mitochondria in cells exposed to both hypoxia and FCCP. Notably, ULK1 translocates to mitochondria even in the absence of Parkin overexpression in both HeLa cells and MEFs. We also discovered that after translocation, ULK1 binds endogenous FUNDC1 and phosphorylates it at Ser-17. Further, the ULK1-binding-deficient mutant of FUNDC1 or knockdown of Fundc1 substantially inhibits the translocation of ULK1 and mitophagy. Our data indicate that FUNDC1 is the mitochondrially localized substrate of ULK1, and probably acts as an adaptor for ULK1, thus allowing ULK1 to direct de novo synthesis of phagophores around dysfunctional mitochondria.
The function of mitophagy receptors is modified by phosphorylation but the kinases responsible for these events are less clear 23, 35, 36. In yeast, Atg32 is phosphorylated at two sites, Ser-114 and Ser-119, and phosphorylation of Ser-114 promotes Atg11–Atg32 interaction 36. Two mitogen-activated protein kinases (MAPKs), namely Slt2 and Hog1, may be indirectly responsible for triggering the mitophagy signaling pathway 37. NIX is involved in autophagic degradation of mitochondria during reticulocyte maturation by an unknown activation mechanism in mammals 23. At BNIP3, serine residues 17 and 24 are phosphorylated by unknown kinases, promoting its binding to LC3B and GATE-16.
Our result that the Ser-17-phosphorylated FUNDC1 binds to LC3 much stronger than FUNDC1 (WT) and FUNDC1 (S17A) suggests that, during mitophagy, FUNDC1 is activated through Ser-17 phosphorylation by ULK1 to build up a “mitophagy receptor-LC3” link between mitochondria and autophagosomes. The role of FUNDC1 Ser-17 phosphorylation contrasts with that of Tyr-18 phosphorylation since the phosphorylation of Tyr-18 in the LIR motif by SRC kinase inhibits FUNDC1-mediated mitophagy, while phosphorylation at Ser-17 promotes mitophagy (Supplementary Figs S3 and S12) 6.
Materials and Methods
Detailed experimental procedures for plasmids construction, siRNA sequences, antibodies, immunofluorescence, and biochemical assays are described in the Supplementary Materials and Methods.
Immunofluorescence microscopy
Cells were grown to 60% confluence on a coverslip. After treatment, cells were washed twice with PBS (Shanghai Sangon Biotech) and fixed with freshly prepared 4% paraformaldehyde at 37°C for 15 min. Antigen accessibility was increased by treatment with 0.1% Triton X-100 (Shanghai Sangon Biotech). After blocking with 1% BSA, cells were incubated with primary antibodies for 1 h at room temperature and, after washing with PBS, stained with a secondary antibody for further 50 min at room temperature. Cell images were captured with a TCS SPF5 II Leica confocal microscope.
Immunoprecipitation
Cells were transiently transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. About 24 h post-transfection, the cells were lysed in 1 ml of lysis buffer (50 mM Tris–Cl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 30% glycerin) containing 1 mM PMSF and protease inhibitor cocktail (Roche Applied Science) for 30 min on ice. After 10,000 g centrifugation for 10 min, the lysates were immunoprecipitated with specific antibody and protein A/G plus-agarose immunoprecipitation reagent (Santa Cruz Biotechnology) overnight at 4°C. Thereafter, the precipitants were washed five times with lysis buffer, and the immune complexes were eluted with sample buffer containing 1% SDS for 10 min at 100°C and analyzed by 12% SDS–PAGE.
Electron microscopy
Electron microscopy was described previously 38. Briefly, MEFs were fixed in 2.5% glutaraldehyde in 0.1 M sodium phosphate buffer, pH 7.4, at 37°C for 2 h, and then dehydrated in a graded ethanol series and embedded. Approximately 70-nm ultrathin sections were mounted on nickel grids. The samples were then stained and visualized using a 120-kV Jeol electron microscope (JEM-1400) at 80 kV. Images were captured using a Gatan-832 digital camera.
Statistical analysis
Assays for characterizing cell phenotypes were analyzed by Student’s t-test, and correlations between the groups were calculated using Pearson’s test. P-values < 0.01 were deemed statistically significant. Data were analyzed using GraphPad Prism 5 software (GraphPad Software, La Jolla, CA, USA).
Acknowledgments
We thank Dr. Isabel Hanson for editing the manuscript. We thank Drs. Hong Zhang and Li Yu for critically reading the manuscript. We are grateful to Dr. Quan Chen’s laboratory. This work was supported by grants from Guangdong Medical College (B2012043, B2012044, 701B01206), by the NSFC (No. 31301104, No. 81171244), by the NSF of Guangdong province (No. S2011010004095), by the Startup fund of the Ministry of Education for returning-back scholars of China ([2012]940), and by Foundation for Distinguished Young Talents in Higher Education of Guangdong, China (2013LYM_0035).
Author contributions
WXW, WLT, ZH, GC, LH, WL, XLZ, PX, LXL, CQZ, LL, YSZ, XLZ, and DF performed the research; WXW, WLT, LQZ, SFS, BZ, and DF designed the research; DF wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://embor.embopress.org
References
- Green DR, Kroemer G. The pathophysiology of mitochondrial cell death. Science. 2004;305:626–629. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Löhr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dötsch V, Ney PA, Dikic I. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, Huang L, Xue P, Li B, Wang X, Jin H, Wang J, Yang F, Liu P, Zhu Y, Sui S, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I, Johansen T, Kirkin V. Selective autophagy in cancer development and therapy. Cancer Res. 2010;70:3431–3434. doi: 10.1158/0008-5472.CAN-09-4027. [DOI] [PubMed] [Google Scholar]
- Meijer AJ, Codogno P. Autophagy: a sweet process in diabetes. Cell Metab. 2008;8:275–276. doi: 10.1016/j.cmet.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Tatsuta T, Langer T. Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J. 2008;27:306–314. doi: 10.1038/sj.emboj.7601972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EY, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem. 2007;282:25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Wirth M, Joachim J, Tooze SA. Autophagosome formation-The role of ULK1 and Beclin1-PI3KC3 complexes in setting the stage. Semin Cancer Biol. 2013;23:301–309. doi: 10.1016/j.semcancer.2013.05.007. [DOI] [PubMed] [Google Scholar]
- McAlpine F, Williamson LE, Tooze SA, Chan EY. Regulation of nutrient-sensitive autophagy by uncoordinated 51-like kinases 1 and 2. Autophagy. 2013;9:361–373. doi: 10.4161/auto.23066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci USA. 2011;108:11121–11126. doi: 10.1073/pnas.1107969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alers S, Loffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, Selak MA, Ney PA, Thompson CB. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Koyama-Honda I, Mizushima N. Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. J Cell Sci. 2012;125:1488–1499. doi: 10.1242/jcs.094110. [DOI] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler AS, Alers S, Dieterle AM, Keppeler H, Franz-Wachtel M, Kundu M, Campbell DG, Wesselborg S, Alessi DR, Stork B. Ulk1-mediated phosphorylation of AMPK constitutes a negative regulatory feedback loop. Autophagy. 2011;7:696–706. doi: 10.4161/auto.7.7.15451. [DOI] [PubMed] [Google Scholar]
- Shang L, Chen S, Du F, Li S, Zhao L, Wang X. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci USA. 2011;108:4788–4793. doi: 10.1073/pnas.1100844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Liu L, Zhu Y, Chen Q. Molecular signaling toward mitophagy and its physiological significance. Exp Cell Res. 2013;319:1697–1705. doi: 10.1016/j.yexcr.2013.03.034. [DOI] [PubMed] [Google Scholar]
- Band M, Joel A, Hernandez A, Avivi A. Hypoxia-induced BNIP3 expression and mitophagy: in vivo comparison of the rat and the hypoxia-tolerant mole rat, Spalax ehrenbergi. FASEB J. 2009;23:2327–2335. doi: 10.1096/fj.08-122978. [DOI] [PubMed] [Google Scholar]
- Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alers S, Loffler AS, Wesselborg S, Stork B. The incredible ULKs. Cell Commun Signal. 2012;10:7. doi: 10.1186/1478-811X-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, Hansmann I, Pfaffenwimmer T, Kijanska M, Stoffel I, Lee SS, Brezovich A, Lou JH, Turk BE, Aebersold R, Ammerer G, Peter M, Kraft C. Early steps in autophagy depend on direct phosphorylation of atg9 by the atg1 kinase. Mol Cell. 2014;53:471–483. doi: 10.1016/j.molcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo JH, Dorsey FC, Joshi A, Hennessy-Walters KM, Rose KL, McCastlain K, Zhang J, Iyengar R, Jung CH, Suen DF, Steeves MA, Yang CY, Prater SM, Kim DH, Thompson CB, Youle RJ, Ney PA, Cleveland JL, Kundu M. Hsp90-Cdc37 chaperone complex regulates Ulk1- and Atg13-mediated mitophagy. Mol Cell. 2011;43:572–585. doi: 10.1016/j.molcel.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii SR, Kishi C, Ishihara N, Mizushima N. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J Biol Chem. 2011;286:19630–19640. doi: 10.1074/jbc.M110.209338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, Hess S, Chan DC. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Massen S, Terenzio M, Lang V, Chen-Lindner S, Eils R, Novak I, Dikic I, Hamacher-Brady A, Brady NR. Modulation of serines 17 and 24 in the LC3-interacting region of Bnip3 determines pro-survival mitophagy versus apoptosis. J Biol Chem. 2013;288:1099–1113. doi: 10.1074/jbc.M112.399345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, Kanki T, Hirota Y, Kurihara Y, Saigusa T, Uchiumi T, Kang D. Phosphorylation of Serine 114 on Atg32 mediates mitophagy. Mol Biol Cell. 2011;22:3206–3217. doi: 10.1091/mbc.E11-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K, Wang K, Zhao M, Xu T, Klionsky DJ. Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J Cell Biol. 2011;193:755–767. doi: 10.1083/jcb.201102092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Zhao WL, Ye YY, Bai XC, Liu RQ, Chang LF, Zhou Q, Sui SF. Cellular internalization of exosomes occurs through phagocytosis. Traffic. 2010;11:675–687. doi: 10.1111/j.1600-0854.2010.01041.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.