Abstract
Purpose:
DNA-dependent protein kinase catalytic subunit (DNA-PKcs, herein referred as DNA-PK) is a multifunctional kinase of high cancer relevance. DNA-PK is deregulated in multiple tumor types, including prostate cancer, and is associated with poor outcomes. DNA-PK was previously nominated as a therapeutic target and DNA-PK inhibitors are currently undergoing clinical investigation. Although DNA-PK is well studied in DNA repair and transcriptional regulation, much remains to be understood about the way by which DNA-PK drives aggressive disease phenotypes.
Experimental Design:
Here, unbiased proteomic and metabolomic approaches in clinically relevant tumor models uncovered a novel role of DNA-PK in metabolic regulation of cancer progression. DNA-PK regulation of metabolism was interrogated using pharmacologic and genetic perturbation using in vitro cell models, in vivo xenografts, and ex vivo in patient-derived explants (PDE).
Results:
Key findings reveal: (i) the first-in-field DNA-PK protein interactome; (ii) numerous DNA-PK novel partners involved in glycolysis; (iii) DNA-PK interacts with, phosphorylates (in vitro), and increases the enzymatic activity of glycolytic enzymes ALDOA and PKM2; (iv) DNA-PK drives synthesis of glucose-derived pyruvate and lactate; (v) DNA-PK regulates glycolysis in vitro, in vivo, and ex vivo; and (vi) combination of DNA-PK inhibitor with glycolytic inhibitor 2-deoxyglucose leads to additive anti-proliferative effects in aggressive disease.
Conclusions:
Findings herein unveil novel DNA-PK partners, substrates, and function in prostate cancer. DNA-PK impacts glycolysis through direct interaction with glycolytic enzymes and modulation of enzymatic activity. These events support energy production that may contribute to generation and/or maintenance of DNA-PK–mediated aggressive disease phenotypes.
Translational Relevance.
DNA-dependent protein kinase catalytic subunit (DNA-PKcs, herein referred as DNA-PK) is a pleiotropic kinase, which is frequently overexpressed and hyperactivated in multiple cancer types. DNA-PK is associated with disease progression and metastasis, thus making DNA-PK an attractive therapeutic target. Targeting DNA-PK leads to anticancerous effects and DNA-PK inhibitors are currently being investigated in clinical trials with promising potential. However, further investigation into the mechanisms that drive DNA-PK protumorigenic functions is needed to aid in development of actionable, effective, and durable therapies for cancer treatment. This study, using unbiased proteomics and metabolomics, identified novel DNA-PK partners, substrates, and function in metabolic regulation of glycolysis, an energy-generating pathway that is also frequently deregulated in cancer. Findings herein identify a novel role for DNA-PK in driving metabolism and suggest new potential therapeutic strategies by co-targeting DNA-PK and glycolytic players in prostate cancer.
Introduction
DNA-dependent protein kinase catalytic subunit (DNA-PKcs, herein referred as DNA-PK) is a pleiotropic kinase of high cancer relevance (1). DNA-PK is dysregulated in multiple hematologic (2) and solid tumors including chronic lymphomas (3), colon (4), cervical (5), breast (6), lung (7), brain (8), and prostate cancer (9). DNA-PK levels increase as a function of disease progression where both high DNA-PK expression and activity are associated with aggressive disease (10, 11). Furthermore, high DNA-PK expression and activity in tumors are linked to poor response and resistance to therapy leading to decreased survival (12). Consequently, DNA-PK has been nominated as a therapeutic target in a subset of malignancies, including prostate cancer (1, 10, 11). In prostate cancer, DNA-PK was identified as the most deregulated kinase in tumors from patients that develop metastatic castration-resistant prostate cancer (CRPC; ref. 9), a stage of the disease that remains fatal. Moreover, in another cohort of patients with advanced prostate cancer, high levels of DNA-PK independently predicted for recurrence, metastasis, and overall survival (13). Downregulation of DNA-PK via pharmacologic and genetic perturbation in prostate cancer results in sensitization of tumors to therapy, decreased tumor size, decreased metastasis, and increased survival (9, 13–15). Current mechanistic understanding of DNA-PK protumorigenic functions have led to clinical trials in various cancers where DNA-PK inhibitors are being assessed as monotherapy and/or in combination with standard of care (10, 11), including metastatic prostate cancer (15).
Although the antitumorigenic effects of targeting DNA-PK are thought to be due to downregulation of canonical DNA-PK functions in (i) DNA damage repair through regulation of nonhomologous end joining (NHEJ; ref. 14) and (ii) transcriptional regulation of protumorigenic processes such as androgen receptor (AR) signaling, metastatic networks, Wnt signaling, epithelial–mesenchymal transition (EMT), and inflammatory response (9, 13, 15), much remains to be understood about DNA-PK functions in cancer. DNA-PK is a highly abundant protein (16) that localizes mainly to the nucleus. Importantly, DNA-PK is also found in various other compartments including the cytoplasm (17), lipid rafts (18), cytoskeleton (19), and plasma membrane (20), where it plays key roles through modulation of various substrates and signaling cascades of cancer relevance to promote tumor growth/progression. For instance, in the cytoplasm, DNA-PK is involved in the regulation of innate and adaptive immunity, inflammation, stress response, and cell death (1). DNA-PK exerts these functions through interaction with and/or phosphorylation of key proteins mediating critical oncogenic signaling pathways. These pathways have been shown to aid in tumor progression and resistance to therapy. Thus, identifying and further understanding the molecular mechanisms that DNA-PK utilizes to promote tumorigenesis, in the nucleus and beyond, is of utmost importance.
To uncover novel DNA-PK functions, partners, and substrates contributing to aggressive disease, various molecular approaches were employed using clinically relevant CRPC models. First, through unbiased proteomics, the first-in-field DNA-PK protein–protein interactome was identified in CRPC. Intriguingly, numerous novel DNA-PK partners engaged in various metabolic processes, including glycolysis. These findings are important because cancer cells are known to undergo metabolic adaptations, including upregulation of glycolysis, known as the Warburg effect, to meet higher energetic demands needed for proliferation and metastasis (21). Metabolomic studies performed in CRPC in the presence of DNA-PK inhibitor (DNA-PKi) demonstrated that DNA-PK modulates a multitude of cancer relevant metabolic pathways. Mechanistic studies showed that DNA-PK interacts with glycolytic enzymes aldolase A (ALDOA), previously described but not explored in metabolic regulation (22), and a novel partner pyruvate kinase M2 (PKM2) both in the nucleus and the cytoplasm. DNA-PK also phosphorylates these enzymes in vitro and promotes their activity in CRPC cells, xenograft, and patient with prostate cancer tissue models. Functional assays showed that pharmacologic inhibition and genetic perturbation of DNA-PK reduces glycolysis and TCA cycle activity, with DNA-PK downregulation decreasing synthesis of glucose-derived pyruvate and lactate production. The studies herein are the first to show that DNA-PK regulates glycolysis in vitro, in vivo, and ex vivo in patient-derived explant (PDE) models. Thus, these data have uncovered a novel role of DNA-PK as a metabolic regulator, through direct interaction and phosphorylation of key glycolytic enzymes that promotes glycolysis, a key pathway used to generate energy and intermediates needed for macromolecular synthesis to support cancer cell growth and metastasis. Collectively, these data expand the current knowledge of DNA-PK functions and inform new avenues for potential therapeutic targeting of DNA-PK in concert with glycolytic inhibitors in cancer.
Materials and Methods
Cell lines
CRPC cell lines, C4–2 and 22Rv1 were obtained from and authenticated by ATCC. C4–2 cells were grown in IMEM media supplemented with 5% FBS, and 22Rv1 cells were grown in DMEM media supplemented with 10% FBS, both supplemented with 1% l-glutamine (2 mmol/L) and 1% penicillin–streptomycin (100 units/mL). Cells were checked for mycoplasma (ATCC, 30–1012K) upon thawing.
Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME)
RIME was performed as described previously (23). Briefly, immunoprecipitation was performed using G magnetic beads (Dynabeads) linked to DNA-PK (Thermo Fisher Scientific, #MS-423-P) or IgG (Santa Cruz Biotechnology, sc-2025) antibodies, after cells were crosslinked with 1% formaldehyde for 8 minutes and neutralized with glycine for 5 minutes. Mass spectrometry (MS) was used to identify endogenous DNA-PK protein partners. Proteins (≥3 peptides) were considered interactors if they were identified in the DNA-PK pulldown but not the IgG control.
Steady-state metabolomics
Steady-state metabolomics were performed by Metabolon, Inc., and data visualization was performed using Metabolon's built in Cytoscape plugin. Briefly, C4–2 cells were plated in complete media overnight and treated with DNA-PK inhibitor, NU7441 (1 μmol/L), or DMSO for 24 hours in quadruplicate. Cell pellets were collected, flash frozen, and stored at −80°C, according to Metabolon sample guidelines. Metabolite identification was performed through ultra-high-performance liquid-phase chromatography and gas-chromatography separation, coupled with MS. Compounds were identified by comparison to libraries maintained by Metabolon.
Seahorse assay
Cellular oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured with a Seahorse XFe bioanalyzer (Seahorse Bioscience) using a Seahorse XF Cell Mito Stress Test Kit (Agilent Seahorse Bioscience, #103020–100) and Seahorse XF Glycolysis Stress Test Kit (Agilent Seahorse Bioscience, #103015–100), respectively, according to manufacturer's instructions. Briefly, cells were transfected with siDNAPK/siCtrl (C4–2, 1 × 104/well; 22Rv-1, 2 × 104/well) or pretreated with NU7441/DMSO (1 μmol/L; C4–2, 0.5 × 104/well; 22Rv-1, 1 × 104/well) and then seeded in a Seahorse XF 96-well assay plate. For the Mito Stress Test mitochondrial inhibitors oligomycin, FCCP, and rotenone/antimycin A were used at a final concentration of 1, 1, and 0.5 μmol/L, respectively. For the glycolysis stress test, glucose, oligomycin, and 2-deoxy-glucose (2-DG) were used at a final concentration of 10 mmol/L, 1 μmol/L, and 50 mmol/L, respectively.
In vitro [U-13C6]-d-glucose tracing
C4–2 cells were plated in complete media with dialyzed FBS overnight and treated with NU7441 (1 μmol/L) or DMSO for 24 hours prior to a 2-hour pulse with [U-13C6]-D-glucose (Sigma, 389374) as described previously (24). Metabolites were extracted with ice-cold buffer (methanol: acetonitrile: MilliQ water at 50:30:20, v/v), separated on a SeQuant ZIC-pHILIC column (Merck, 150460) and identified using a Q Exactive HF mass spectrometer. Compounds were identified using an in-house library, with isotopologues corrected against naturally occurring stable carbons.
Xenografts
C4–2 cells (5 × 106 cells per injection) were resuspended in a 1:1 mixture of 1X PBS and Matrigel (BD Biosciences 354234) and injected subcutaneously into the flank of 5- to 6-week-old SCID male mice following protocol approved by IACUC at Thomas Jefferson University. Upon tumor reaching 150 mm3, mice were treated with the DNA-PKi NU7441 (Selleck Chemicals, 25 mg/kg) or vehicle daily for 4 days via intraperitoneal injection. After 4 days, tumors were harvested and processed for pyruvate kinase activity and pyruvate levels as described in Supplemental Experimental Methods. Tissue was used for hematoxylin and eosin (H&E) staining, and immunohistochemistry staining for pDNA-PK (Abcam ab18192), DNA-PK (Abcam, ab168854), PKM2 (Cell Signaling, #4053S), GLUT-1 (Abcam, ab115730) and MCT1 (Proteintech, 20139–1-AP) performed by Thomas Jefferson Pathology Core.
PDE
PDE experiments were conducted as described previously (25). Briefly, tissues samples were treated with NU7441 (1 μmol/L) or DMSO for 6 days. PDEs were used for H&E staining and IHC staining for pDNA-PK (Abcam, ab18192) and DNA-PK (Cell Signaling Technology, #12311) performed by Thomas Jefferson Pathology Core, and to measure pyruvate kinase activity and pyruvate levels. The Thomas Jefferson University Institutional Review Board has reviewed this protocol and deemed this research in compliance with federal regulations [45 CFR 46.102(f)]. Prostate tissues were also collected with written informed consent from previously-untreated patients undergoing radical prostatectomy at St Andrew's Hospital, Adelaide, Australia, through the Australian Prostate Cancer BioResource. A longitudinal section of each tissue was removed prior to ex vivo culture, fixed in formalin, and paraffin embedded for assessment for histology and IHC. Ethical approval for tissue collection and experimentation was obtained from St Andrew's and the University of Adelaide Human Research Ethics committees and all experiments with patient material were performed in accordance with the National Health and Medical Research Council of Australia guidelines. Prostate specimens were dissected and cultured ex vivo as PDEs in RPMI medium containing 10% FCS on gelatin sponges for 24 hours in the presence of DMSO (vehicle) or NU7441 (1 or 2 μmol/L). After culture, explants were transferred into a new well containing treatment medium with U-13C-D-glucose (including DMSO, 1 or 2 μmol/L NU7441) for a further 2 hours. PDEs were then harvested and snap frozen for metabolomics analysis.
Statistical analysis
All experiments were performed in technical triplicate with at least three biological replicates per condition. In vitro, in vivo, and ex vivo data are displayed as mean ± SEM. Statistical significance was determined using Student t test and two-way ANOVA on GraphPad Prism Software (version 9.0.2).
Data availability
The data generated and/or analyzed during this study are available upon reasonable request from the corresponding author.
Results
DNA-PK interacts and promotes activity of glycolytic enzymes
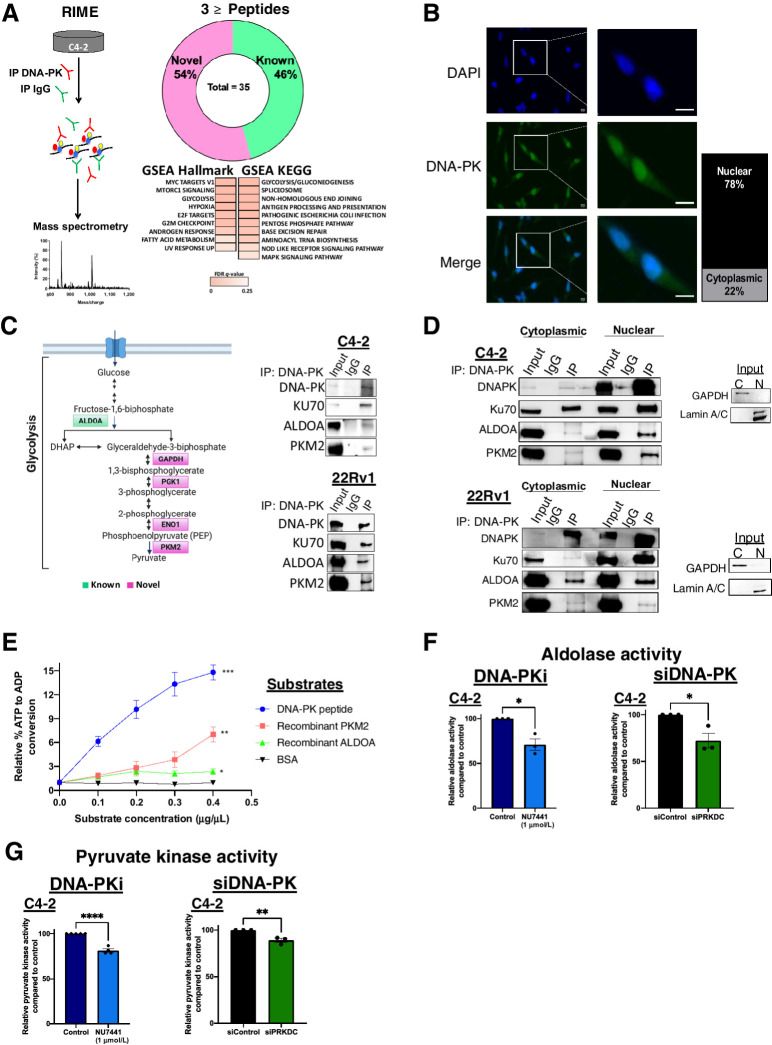
Abundant expression of DNA-PK in tumors is strongly associated with poor outcome, and far exceeds the levels required for its established roles including DNA repair, transcriptional regulation, and genome stability (26). To address the significant gaps in understanding the pleiotropic functions of DNA-PK in human malignancies, an unbiased proteomic approach was utilized. Given the role of DNA-PK in promoting prostate cancer aggressive features and metastatic potential (13), DNA-PK function was examined in CRPC models. RIME (ref. 23; Fig. 1A, left) was performed to map the DNA-PK protein–protein interactome. As shown, these studies identified 144 unique proteins, consisting of 17% known and 83% novel partners, with every distinct peptide found specifically in DNA-PK immunoprecipitation, but not in IgG control, given designation of a protein (Supplementary Fig. S1A). Using an increased stringency filter of three or more distinct peptides per protein, 35 DNA-PK–interacting proteins were identified, of which 54% were novel DNA-PK partners (Fig. 1A, right; Supplementary Table S1). Gene set enrichment analysis (GSEA) Hallmark and KEGG analyses (FDR <0.25; Fig. 1A, right) were performed, and as expected known DNA-PK modulated pathways including DNA repair pathways (NHEJ and base excision repair), androgen response, hypoxia, and G2M checkpoint. Specifically, DNA-PK interacted with peptides derived from known partners in NHEJ, Ku70, and Ku80, as well as PARP1, HSP90, and TRIM28 (KAP1; refs. 27–30), which lends further confidence in the protein–protein interactome mapping as these proteins and associated pathways have been previously identified as regulated by DNA-PK (1, 10, 31). Strikingly, novel DNA-PK partners were highly enriched for proteins involved in metabolic pathways including the pentose phosphate pathway, fatty acid metabolism, and glycolysis/gluconeogenesis (Fig. 1A; Supplementary Fig. S1A). These findings suggest a potential role for DNA-PK in cancer metabolism. Increased metabolic activity is an important characteristics of cancer cells and has been to linked advanced disease and resistance to therapy (32). Thus, understanding the potential role of DNA-PK in metabolism, through interaction with metabolic enzymes, could uncover novel mechanisms by which DNA-PK promotes aggressive disease phenotypes in CRPC.
Figure 1.
DNA-PK interacts with glycolytic enzymes and promotes enzymatic activity. A, Schematic of RIME proteins in C4–2 cell line. Donut plot of known and novel DNA-PK partners, GSEA hallmark and KEGG gene set analysis. B, DNA-PK localization in C4–2 cells: DAPI (blue), DNA-PK (green). DNA-PK levels are quantified (right). C, Simplified schematic of glycolysis pathway highlighting known (green) and novel (magenta) DNA-PK partners identified by RIME. ALDOA and PKM2 interaction with DNA-PK in CRPC whole cell lysates. D, Cytoplasmic and nuclear Interaction of DNA-PK with ALDOA and PKM2 in CRPC cell lines (C4–2 top, 22Rv1 bottom). E,In vitro DNA-PK kinase assay using ALDOA and PKM2 as substrates, DNA-PK peptide (positive control), and BSA (negative control). Aldolase activity assay (F) and pyruvate kinase activity assay (G) after DNA-PK inhibition (NU7441, 1 μmol/L, 3 hours) and DNA-PK knockdown (96 hours posttransfection). Data collected in at least three biological replicates, represented as mean ± SEM (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Although DNA-PK functions have predominantly been studied in the nucleus, the kinase also localizes to the cytoplasm (33, 34), where several metabolic processes primarily take place. Consistent with previous reports, DNA-PK localized to both compartments with 78% of DNA-PK identified as nuclear and 22% as cytoplasmic in CRPC cells (Fig. 1B; Supplementary Fig. S1B), indicating that DNA-PK has the potential to interact with cytoplasmic metabolic factors. To further explore the link between DNA-PK and metabolic regulation, glycolysis was prioritized as the most enriched pathway identified in the interactome studies through RIME (Fig. 1A). Specifically, glycolysis is an energy generating pathway, often upregulated in cancer cells due to high energetic demands for establishing and maintaining aggressive phenotypes (35). DNA-PK was found to interact with multiple members of the glycolysis/gluconeogenesis pathway by RIME (Fig. 1C, left): ALDOA, GAPDH, phosphoglycerate kinase 1 (PGK1), enolase alpha (ENO1), and pyruvate kinase M2 (PKM2). To interrogate the regulation of glycolysis by DNA-PK in a systematic manner, ALDOA and PKM2 were prioritized for further investigation. ALDOA was previously reported to interact with DNA-PK (22), although functionally not investigated in metabolism, and thus identification of this interaction with DNA-PK in CRPC cells supports the validity of the novel DNA-PK RIME analyses performed. In addition, PKM2 is the downstream glycolytic factor that catalyzes the last and irreversible step in glycolysis generating pyruvate and energy, thus, potentially linking DNA-PK to regulation of energy production through glycolysis. To validate RIME-nominated partners, co-immunoprecipitation (Co-IP) analyses were performed in CRPC cells in whole cell lysates, with Ku70 serving as a positive control (Fig. 1C, left). The interactions were confirmed using reverse Co-IPs, further validating these partners (Supplementary Fig. S1C). Co-IPs on nuclear and cytoplasmic fractionated lysates revealed that DNA-PK interacts with ALDOA and PKM2 in both cellular compartments (Fig. 1D), supporting the potential role of DNA-PK in the regulation of ALDOA and PKM2 functions in glycolysis. Combined, these data are the first to identify DNA-PK as a putative effector of glycolytic regulation.
Despite the wealth of knowledge surrounding DNA-PK function in DNA repair (36), substrates outside of DNA repair remain largely understudied. Therefore, a putative functional interaction was explored next, to uncover whether ALDOA and PKM2 are DNA-PK substrates for phosphorylation. DNA-PK has been shown to phosphorylate the majority of its substrates utilizing a consensus sequence previously delineated as either a serine or threonine followed by a glutamine (S/T[Q]; ref. 37), or sometimes a leucine or tyrosine residue (non-S/T[Q]; ref. 37). However, DNA-PK also has been shown to phosphorylate substrates that do not adhere to these sequences (38), therefore making substrate identification complex. On the basis of the S/T[Q] and non-S/T[Q] preferred sequences, seven potential sites were predicted on ALDOA and PKM2 (Supplementary Fig. S1D). Here, in vitro DNA-PK kinase assays were utilized to capture all the possible phosphorylation events despite adherence or not to the consensus sequences. These analyses used recombinant human ALDOA and PKM2 as substrates, DNA-PK peptide substrate as a positive control, and BSA as a negative control. Resulting data shows that DNA-PK phosphorylates both ALDOA and PKM2 in a dose-dependent manner as compared with BSA (Fig. 1E). Thus, this investigation identified ALDOA and PKM2 as novel DNA-PK substrates in vitro.
This functional interaction suggests that DNA-PK may affect downstream pathway activities, which was further assessed by DNA-PK activity suppression or depletion. Suppressing DNA-PK activity using a specific DNA-PKi (1 μmol/L NU7441) reduced aldolase activity by 29% and 27% after 3 hours in C4–2 and 22Rv1 cells, respectively (Fig. 1F; Supplementary Fig. S1E). Similarly, pyruvate kinase activity was assessed upon DNA-PKi and was reduced by 19% and 4% in C4–2 and 22Rv1 cells, respectively (Fig. 1G; Supplementary Fig. S1F). To account for any off-target effects, siRNA-mediated DNA-PK knockdown (siPRKDC, referred herein as siDNA-PK) was utilized to validate results in C4–2 cells, resulting in a 28% reduction in aldolase activity (Fig. 1F) and an 11% decrease in pyruvate kinase activity (Fig. 1G). Thus, these results indicate that DNA-PK promotes ALDOA and PKM2 enzymatic activity to a degree that could impact downstream metabolism. Importantly, the modulation of ALDOA and PKM2 activity was not due to changes in protein levels of glycolytic enzymes upon DNA-PKi and siDNA-PK, or off-target effect on mTOR/PI3K or AMPK pathways (Supplementary Figs. S2A–S2C). In summary, these data show for the first time that DNA-PK interacts, phosphorylates, and induces activity of key glycolytic enzymes in CRPC, revealing a potential role of DNA-PK in metabolic regulation.
DNA-PK activity modulates metabolic pathways and results in significant changes in carbohydrate metabolism
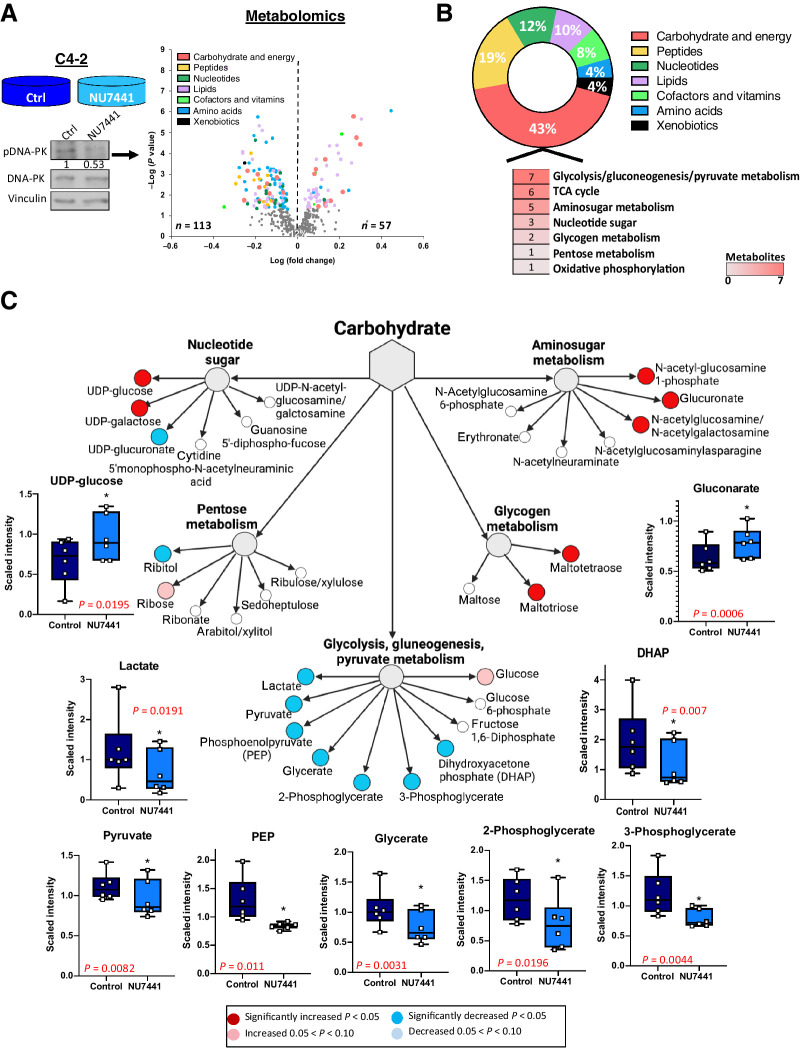
Cancer cells alter their metabolism to support the acquisition and maintenance of malignant phenotypes, including by increase of glycolysis and lactic acid production in aerobic conditions, also known as the Warburg effect (32, 39). Because DNA-PK interacts and affects enzymatic activity of glycolytic enzymes, it is important to discern the role of DNA-PK in metabolism and assess biological outcome in CRPC. To this end, untargeted metabolic profiling using steady-state metabolomics was performed in CRPC cells treated with DNA-PKi (NU7441, 1 μmol/L, 24 hours). This analysis captured the DNA-PK–dependent alterations that occur at this particular time in CRPC cells and identified 113 decreased and 57 increased metabolites after DNA-PKi as compared with control (P < 0.05; Fig. 2A, left). Metabolite hits significantly changed compared with total metabolites detected in each pathway were enriched for carbohydrate and energy metabolism (43%), peptide (19%), nucleotide (12%), and lipid pathways (10%) (Fig. 2B; Supplementary Fig. S3A). Within the carbohydrate and energy super-pathway, glycolysis/gluconeogenesis/pyruvate was the top altered subpathway after DNA-PKi with seven decreased metabolites. Dihydroxyacetone phosphate (DHAP), 3-phosphoglycerate, 2-phosphoglycerate, glycerate, PEP, pyruvate, and lactate were significantly reduced after DNA-PKi, suggesting a potential role for DNA-PK in catabolism in cancer cells (Fig. 2C, blue = significantly decreased, red = significantly increased). Strikingly, four of the decreased metabolites (DHAP, 3-phosphoglycerate, PEP, and pyruvate) are downstream of the RIME-identified DNA-PK partners (ALDOA, PGK1, ENO1, and PKM2, respectively; Supplementary Fig. S3B). Considering that DNA-PK inhibition decreased the enzymatic activity of ALDOA and PKM2, identification of downregulated DHAP and pyruvate metabolites provide additional evidence that DNA-PK directly alters glycolysis. Together, these data demonstrate that DNA-PK activity modulates cancer metabolism, specifically glycolysis via interaction with glycolytic enzymes, which was postulated to support the high energetic demands of advanced cancer, including CRPC.
Figure 2.
DNA-PK activity modulates metabolic pathways and results in significant changes in carbohydrate metabolism. A, Steady-state metabolomics performed in C4–2 cells in DMSO vs. NU7441 (1 μmol/L, 24 hours). Increased and decreased metabolites shown in the scatter plot for each super-pathway. B, Relative percent of metabolites changed (up and down) compared with total number of metabolites identified in each super-pathway. C, Metabolites changed in each carbohydrate subpathway with glycolysis having the most changed metabolites shown by box and whisker plots.
DNA-PK impacts glycolytic activity in CRPC
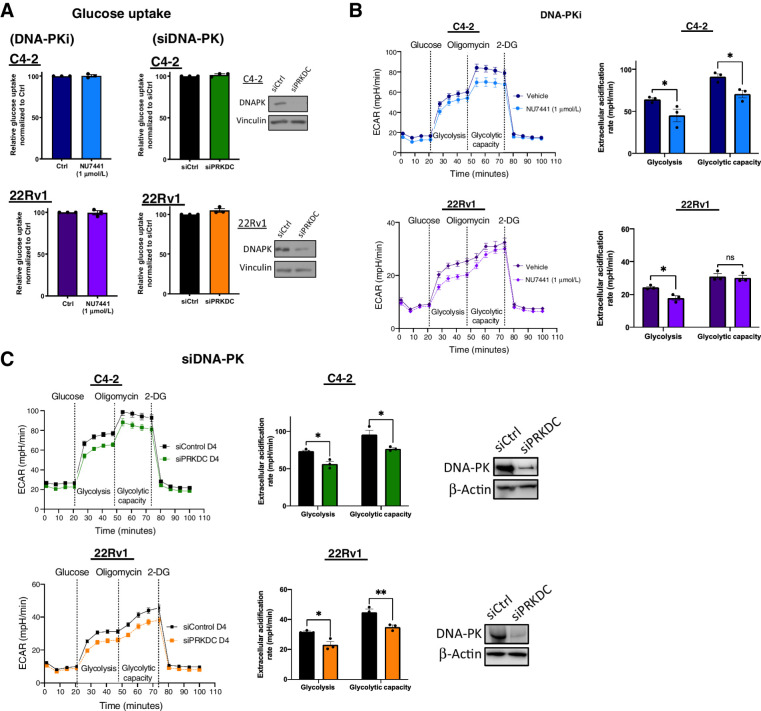
To assess whether changes in glucose availability led to decreased levels of downstream glycolysis metabolites, glucose uptake was interrogated. Although often upregulated in cancer (32), glucose uptake was unchanged in CRPC cells after suppression of DNA-PK activity (DNA-PKi) and DNA-PK depletion (siDNA-PK; Fig. 3A). These data indicate that DNA-PK does not alter glucose availability for glycolysis and alteration in glucose uptake is not responsible for the observed decrease in downstream metabolites. Therefore, to understand the potential reasons for the observed downregulation of glycolytic intermediates, glycolytic activity was investigated by monitoring the extracellular acidification rate (ECAR). ECAR was measured after stimulation with glucose to evaluate the rate of glycolysis under basal conditions (labeled as Glycolysis in the graph) and was decreased by 22% and 19% in C4–2 and 22Rv1 cells after inhibition of DNA-PK activity with DNA-PKi, respectively (Fig. 3B). Similarly, upon DNA-PK depletion (siDNA-PK), glycolytic rate was decreased by 23% and 26% for C4–2 and 22Rv1, respectively (Fig. 3C). These data suggest that DNA-PK promotes glycolysis in CRPC. However, under stress cells have the ability to adapt and may utilize glycolysis at maximum capacity (labeled as Glycolytic Capacity in the graph) to meet energetic demands without help from the TCA cycle. This can be modeled by using an inhibitor of oxidative phosphorylation (oligomycin) that halts mitochondrial ATP generation, forcing cancer cells to increase glycolytic flux to reach maximum capacity as shown by an increase in ECAR. After treatment with DNA-PKi, glycolytic capacity was reduced by 17% in C4–2 cells, whereas there was no significant difference in 22Rv1 cells (Fig. 3B). Upon DNA-PK knockdown, glycolytic capacity was reduced by 20% and 15% for C4–2 and 22Rv1 cells, respectively (Fig. 3C). These findings show that under stress, DNA-PK downregulation reduces the ability of CRPC cells to perform at maximum glycolytic capacity, likely resulting in decreased energy production. Together these data suggest that DNA-PK sustains glycolysis through regulation of glycolytic activity in CRPC.
Figure 3.
DNA-PK impacts overall glycolytic activity. A, Glucose uptake was measured upon DNA-PK inhibition (NU7441, 1 μmol/L, 24 hours) and DNA-PK knockdown (96 hours posttransfection) using a fluorescence glucose analog, 2NBDG. B and C, Glycolysis stress test seahorse performed upon DNA-PK inhibition (B; NU7441, 1 μmol/L, 24 hours) and DNA-PK knockdown (C; 96 hours posttransfection). ECAR was quantified and shown (right). Glycolysis was measured after glucose stimulation and prior to oligomycin treatment. Glycolytic capacity was measured after oligomycin stimulation and prior to 2-DG. Data collected in at least three biological replicates, represented as mean ± SEM (ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
DNA-PK regulates synthesis of pyruvate and lactate in CRPC
To further confirm the role of DNA-PK in impacting aerobic glycolysis, pyruvate and lactate were assessed as the end products of glycolysis. As expected from steady-state metabolomics (Fig. 2C) and reduced glycolytic activity (Fig. 3B and C), a 42% decrease in pyruvate levels and 10% decrease in lactate production were observed in C4–2 cells after DNA-PKi (Fig. 4A; Supplementary Fig. S4A). Similarly, DNA-PK knockdown resulted in a 20% and 23% decrease in pyruvate levels and lactate production, respectively (Fig. 4B; Supplementary Fig. S4B). Furthermore, ectopic overexpression of ALDOA and PKM2 rescue the reduced levels of pyruvate after DNA-PK inhibition (Fig. 4C; Supplementary Fig. S4C). These findings further confirm that DNA-PK modulates glycolysis in CRPC through ALDOA and PKM2.
Figure 4.
DNA-PK regulates glycolysis. A, Pyruvate levels and lactate production was measured upon DNA-PK inhibition (NU7441, 1 μmol/L, 24 hours). B, DNA-PK knockdown (96 hours posttransfection). C, Pyruvate levels upon overexpression of ALDOA and PKM2 as validated via protein levels (left). D, Glucose tracing through glycolysis pathway to GADP, 3PG, pyruvate, and lactate generation through metabolic flux analysis. C4–2 cells were treated with NU7441 (1 μmol/L, 24 hours) or vehicle control, followed by 2 hours pulse with [U-13C6]-D-glucose. Data collected in at least three biological replicates, represented as mean ± SEM (ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
In addition to being converted to lactate through aerobic glycolysis, pyruvate can serve as a link between glycolysis and other metabolic processes such as TCA cycle (40). The impact of DNA-PK on the TCA cycle was investigated via oxygen consumption rate (OCR), as a measure of mitochondrial function. In line with decreased levels of pyruvate observed, thus potentially less glycolytic intermediates entering the TCA cycle, the basal OCR levels (the ability to meet energetic demands under baseline conditions) were significantly decreased after DNA-PKi (Supplementary Fig. S4D) and DNA-PK depletion (Supplementary Fig. S4E). Furthermore, maximal OCR (the maximum electron chain capacity) was also significantly decreased (Supplementary Figs. S4C and S4D). Together, these findings show that DNA-PK downregulation reduces TCA cycle activity.
Nevertheless, the lower pyruvate/lactate levels and TCA activity may be due to changes in utilization and/or synthesis of glucose-derived metabolites. To explore these possibilities, [U-13C6]-D-glucose tracing metabolomics using a stable isotope to label all six carbons in glucose, was performed to follow glucose carbons through glycolysis after DNA-PKi treatment (Fig. 4C, left). As depicted in the schematic (Fig. 4C) uniformly labeled glucose going through glycolysis would generate fully labeled pyruvate and lactate represented as m+3 (all carbons labeled). After 2 hours of [U-13C6]-D-glucose labeling, intracellular glucose (m+6) was unaffected by DNA-PK inhibition, whereas m+3 glucose-derived metabolites downstream of ALDOA were significantly reduced including GADP, glycerate 3-phosphate (3PG), pyruvate, and lactate (Fig. 4D). Because no changes were observed in glucose uptake and both pyruvate and lactate levels were also decreased in steady state metabolomics, these data show decreased synthesis of these glucose-derived metabolites downstream of ALDOA, rather than increased utilization. Combined, findings herein suggest that DNA-PK is a critical player in cancer metabolism as a positive regulator of aerobic glycolysis by promoting synthesis of glucose-derived pyruvate and lactate.
DNA-PK regulates glycolysis in vivo and in human prostate cancer tissues
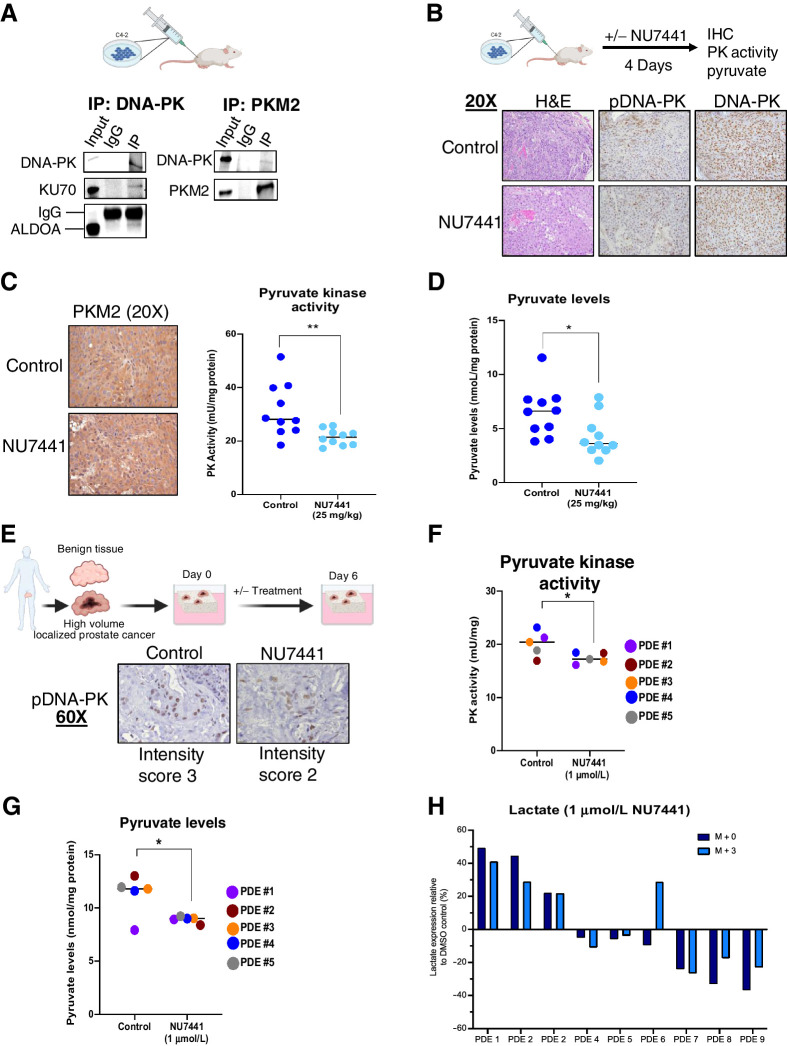
Considering the ability of DNA-PK to interact with ALDOA and PKM2, as well as promote glycolysis in vitro, these findings were further investigated in vivo via a xenograft model of CRPC. Importantly, DNA-PK interacts with ALDOA and PKM2 in vivo as shown by Co-IPs (Fig. 5A), supporting the in vitro data (Fig. 1C; Supplementary S1C). Mice were also treated with DNA-PKi for 4 days and tissue was used for histology analysis to assess histoarchitecture via H&E, pDNA-PK, and total DNA-PK levels (Fig. 5B). Furthermore, glycolytic markers GLUT-1, PKM2, and MCT1 levels were interrogated (Fig. 5C; Supplementary Fig. S5). Similar to the in vitro studies, glycolytic marker levels, including PKM2, were unaffected by DNA-PKi (Fig. 5C, left), however in vivo pyruvate kinase activity was significantly reduced (22.6%). Thus, regulation of pyruvate kinase activity by DNA-PK is maintained in vivo. Because pyruvate levels were decreased in vitro after DNA-PKi and siDNA-PK (Fig. 4A and B), this endpoint was assessed as a measure of glycolytic activity. After treatment with DNA-PKi in vivo, pyruvate levels were also significantly decreased by 34.3% (Fig. 5D). These findings show that DNA-PK activity drives the enzymatic activity of glycolytic enzyme PKM2 and the generation of glycolytic pathway product, pyruvate, in vivo.
Figure 5.
DNA-PK regulates glycolysis in in vivo and ex vivo models. A, DNA-PK interacts with ALDOA and PKM2 in C4–2 xenografts. B, Representative IHC images for H&E, pDNA-PK, and DNA-PK after DNA-PK inhibition (25 mg/kg, 4 continuous days) effect. C and D, DNA-PK inhibition does not impact PKM2 expression but reduces pyruvate kinase activity (C) and pyruvate levels (D) in vivo. E–G, DNA-PK inhibition (1 μmol/L, 6 days) reduces pDNA-PK levels (E), pyruvate kinase activity (F), and pyruvate levels (G) in PDE. H, DNA-PK inhibition reduces lactate synthesis ex vivo (*, P < 0.05; **, P < 0.01).
To challenge this concept in human prostate cancer tissues, PDEs were interrogated, utilizing human tumor samples obtained from high volume disease post-radical prostatectomy, as previously described and summarized in Fig. 5E, left (25, 41). Notably, these tumor tissues retain histoarchitecture, AR expression, proliferation rate, and tumor microenvironment features of the original tumor (25, 41). Upon resection, these tissues were treated with DNA-PKi or vehicle control before undergoing IHC (Supplementary Fig. S6A) and metabolic assay interrogation. Treatment of five PDEs with DNA-PKi led to a slight decrease in pDNA-PK staining intensity compared with control (Fig. 5E, bottom). Pyruvate kinase activity and pyruvate levels served as endpoints for the effect of DNA-PK downregulation on PKM2 and glycolysis in PDEs. A 13.5% decrease of pyruvate kinase activity was observed upon treatment with DNA-PKi (Fig. 5F), which shows that DNA-PK directly affects the activity of PKM2 in PDEs, the last and irreversible step in glycolysis to generate energy and pyruvate. As expected, downstream pyruvate levels were also decreased by 20.7% upon DNA-PKi, further highlighting the effect of DNA-PK on glycolysis, this energy producing pathway to support cancer cell growth and metastasis (Fig. 5G). Furthermore, [U-13C6]-D-glucose tracing metabolomics performed in additional nine PDEs (Supplementary Fig. S6B; Supplementary Table S2) showed heterogeneity in response to DNA-PKi, measured by the flux of glucose-derived carbons to lactate (m+3 labeling was 51% ± 9% of total lactate levels across all nine explants). Flux analysis also showed a decrease in lactate synthesis (m+3) after DNA-PKi (NU7441, 1 μmol/L; Fig. 5H), which is further pronounced at a higher dose (NU7441, 2 μmol/L; Supplementary Fig. S6C). Only highly abundant lactate showed efficient 13C-labelling, with other metabolites such as pyruvate not shown as they were below detection limits in approximately two thirds of explant samples, similar to our C4–2 labeling. In summary, these data show for the first time that DNA-PK regulates glycolysis in vitro, in vivo, and ex vivo via interaction, regulation of glycolytic enzyme activity, and glycolytic pathway activity as observed by pyruvate and lactate production levels. Together, these data show that DNA-PK–dependent regulation of glycolysis can contribute to proliferation and promoting aggressive disease which can be leveraged for therapeutic targeting.
Combination of DNA-PK and glycolysis inhibitor decreases proliferation in CRPC
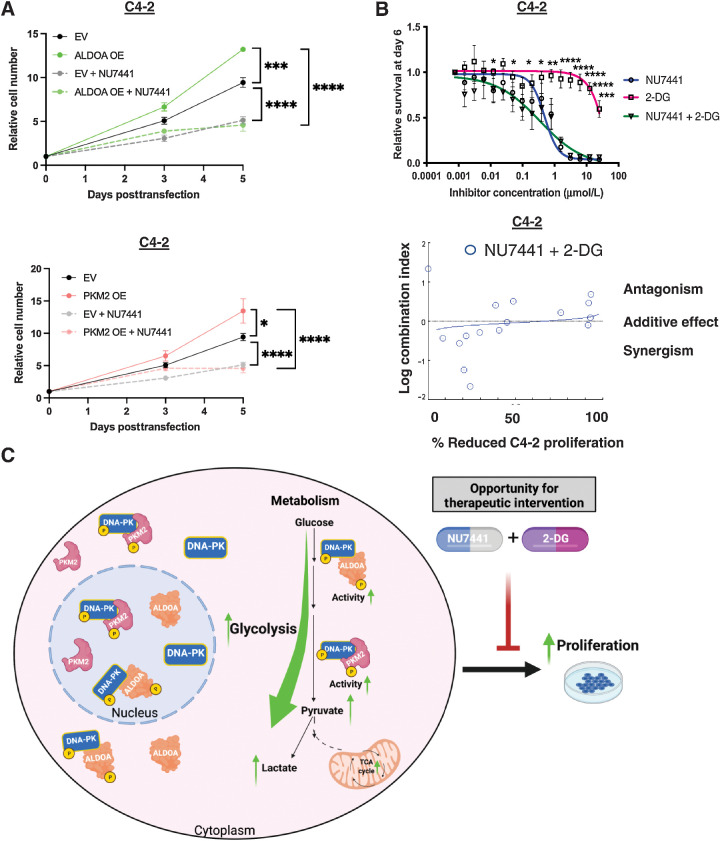
Next, knowing the impact of other DNA-PK functions in promoting proliferation and metastasis, the potential contribution of the role of DNA-PK in glycolysis through regulation of ALDOA and PKM2 in CRPC proliferation was investigated. As expected, CRPC cells expressing empty vector (EV) treated with DNA-PKi (NU7441, 1 μmol/L) showed significantly reduced proliferation compared with EV alone (Fig. 6A). Importantly, cells expressing ectopic ALDOA and PKM2, showed a significant increase in proliferation when compared with EV and EV + NU7441 cells (Fig. 6A). However, treatment with DNA-PKi led to significant decrease of proliferation despite ALDOA and PKM2 overexpression on Day 5, potentially due to inhibition of other DNA-PK functions that modulate proliferation. These data suggest that upregulation of glycolytic metabolite pyruvate via ALDOA and PKM2 overexpression alone is not enough to rescue proliferation. Nevertheless, the findings herein show that DNA-PK plays a novel role in regulation of glycolysis through ALDOA and PKM2. Regulation of glycolysis by DNA-PK is important for the production of glucose-derived metabolites and glycolytic activity that may contribute to energy generation, biosynthesis, and various other functions that promote aggressive disease.
Figure 6.
DNA-PK regulation of glycolysis contributes to aggressive disease and can be mitigated by co-targeting DNA-PK and glycolysis. A, ALDOA and PKM2 overexpression rescue blunted proliferation by DNA-PK inhibition. B, Co-targeting DNA-PK and glycolysis has additive anti-proliferative effect when using titrating DNA-PKi and 2-DG concentration (0–25 μmol/L). C, Summary schematic of the role of DNA-PK in glycolysis (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Considering that glycolysis is upregulated in most cancers and DNA-PK's regulation of glycolytic activity, co-targeting DNA-PK and glycolysis was investigated. CRPC cells were treated with glycolysis inhibitor 2-deoxyglucose (2-DG), DNA-PKi (NU7441), and 2-DG + NU7441 combination (Fig. 6B; Supplementary Fig. S7A). 2-DG showed limited activity as a single agent but in combination with DNA-PK inhibition showed a more potent anti-proliferative effect (Fig. 6B, top). To calculate combination indices (CI) CompuSyn software was used, where CI < 1, CI = 0, and CI > 1 indicate synergistic, additive, and antagonistic effects of the two drugs in combination, respectively. These data revealed that co-targeting DNA-PK and glycolysis together leads to combinatorial anti-proliferative effects in CRPC (additive effect at higher doses of 2-DG; Fig. 6B, bottom; Supplementary S7B). Together, these data show that DNA-PK contributes to aggressive disease by modulating glycolysis and this can be targeted via a combination of DNA-PK and glycolysis inhibition (Fig. 6C).
Discussion
DNA-PK expression and activity are elevated in multiple cancers including prostate cancer and have been associated with aggressive disease and poor outcome (9, 10, 13). DNA-PK is a multifunctional protein, but mechanisms by which DNA-PK promotes aggressive cancer phenotypes beyond DNA damage repair remain understudied. Therefore, understanding the mechanisms by which DNA-PK drives aggressive disease is critical for development of effective therapeutic strategies in advanced cancers. This study identifies a novel role for DNA-PK as a key contributor to CRPC by regulating cancer metabolism via glycolysis. Findings herein uncovered the first DNA-PK protein–protein interactome in CRPC, where novel DNA-PK partners are involved in various cancer relevant pathways, including glycolysis. This study revealed that DNA-PK interacts and phosphorylates (in vitro) glycolytic proteins ALDOA and PKM2. However, the exact phosphorylation sites and whether these events occur in vivo remains to be elucidated. Furthermore, DNA-PK was found to promote the enzymatic activity of glycolytic proteins ALDOA and PKM2. DNA-PK also increases synthesis of glycolytic products pyruvate and lactate, and regulates glycolysis in in vitro, in vivo, and ex vivo CRPC models. Moreover, regulation glycolysis by DNA-PK may contribute to aggressive disease, beyond proliferation, and combination of DNA-PK and glycolysis inhibitors such as 2-DG leads to larger anti-proliferative effects than 2-DG alone. In sum, this study identified novel partners, substrates, and function of DNA-PK as a regulator of cancer metabolism through glycolysis, potentially impacting energy and macromolecule generation needed for proliferation and metastasis.
RIME analyses identified the first-in-field DNA-PK interactome with multiple novel DNA-PK partners involved in regulation of glycolysis. In addition to ALDOA and PKM2, RIME-identified DNA-PK partners included GAPDH, PGK1, and ENO1 (Fig. 1A). GAPDH, PGK1, and ENO1 catalyze the 6th, 7th, and 9th steps in glycolysis, respectively. GAPDH, largely known as a housekeeping gene, is upregulated in various cancer types and often correlates with reduced survival (42). Although much remains to be understood about the role of GAPDH in cancer beyond glycolysis, GAPDH has been shown to modulate DNA repair, replication, cell death, and cytoskeletal organization (42, 43). Therefore, further analysis of the DNA-PK/GAPDH interaction will provide insights into the mechanisms and biological outcomes on glycolysis and other cancer relevant pathways. Further, PGK1 is the glycolytic enzyme that catalyzes the first ATP producing step in glycolysis and plays a key role in coordinating energy production from glycolysis with one carbon metabolism, redox homeostasis, and serine synthesis (42, 44). Interrogation of the DNA-PK/PGK1 interaction will help further delineate the role of DNA-PK in metabolic modulation through potential regulation of biosynthesis and redox metabolism in cancer. High levels of PGK1 expression correlate with poor prognosis and therapy response, however PGK1 can also play a tumor suppressor role in cancers such as lung, colon, hepatocellular, and prostate cancer (44). Moreover, PGK1 is a kinase involved in oncogenic signaling, thus understanding the processes driven by DNA-PK/PGK1 interaction may identify novel mechanisms that drive proliferation and metastasis. Finally, ENO1 is a glycolytic enzyme that has been shown to contribute to proliferation, metastasis, and therapy resistance through its role in glycolysis (45). ENO1 is overexpressed and promotes tumor growth in head and neck, hepatocellular, and gastric cancers (45). Further interrogation of the DNA-PK/ENO1 interaction could expand our understanding of the role of DNA-PK in glycolysis and how these proteins together may impact response to therapy. It is also important to not only understand the impact of the DNA-PK interactions with these glycolytic partners, but also determine whether DNA-PK regulates these proteins via phosphorylation, changes in enzymatic activity, and/or via allosteric regulation. Furthermore, although ALDOA and PKM2 overexpression alone did not significantly rescue proliferation after DNA-PKi, further interrogation should focus on identifying which protein (GAPDH, PGK1, and ENO1 alone; combination of 2 or more; or all DNA-PK glycolysis partner proteins together) may drive DNA-PK–dependent proliferation via regulation of glycolysis. Together, the potential interaction of DNA-PK with glycolytic proteins could impact their role in glycolysis, overall energy metabolism, and potentially other oncogenic pathways.
As seen with GAPDH, PGK1, and ENO1, glycolytic proteins have functions beyond glycolysis both in the cytoplasm and the nucleus. Also observed herein (Fig. 1D), DNA-PK interacts with ALDOA and PKM2 in both cellular compartments. Although the functional consequences of these DNA-PK interactions were studied in the cytoplasm herein, these interactions may affect various nuclear processes. Both ALDOA and PKM2 are known to play roles in cell proliferation, motility, epithelial–mesenchymal transition, p53, Wnt signaling, and DNA repair (46, 47). ALDOA has been shown to induce DNA-PK activity after dietary restriction (30% decrease in caloric intake) in liver and ovarian cancer (22), thus suggesting a potential function for the DNA-PK–ALDOA interaction in regulation of DNA repair in the nucleus. Furthermore, the regulation of DNA-PK activity by ALDOA and vice versa, supports the possibility of a feedback loop leading to crosstalk between regulation of DNA repair and metabolism, which merits further investigation. Importantly, various metabolic factors including fumarase, glutamine synthase, ATP-citrate lyase, and PKM2 have been shown to play a critical role in DNA repair pathways including NHEJ and homologous recombination (HR; ref. 48). Specifically, PKM2 has been shown to colocalize and phosphorylate H2AX (48), a DNA-PK substrate during NHEJ, at sites of DNA damage. These findings suggest that DNA-PK/ALDOA and DNA-PK/PKM2 interactions may play a role in nuclear functions, including DNA repair. The DNA damage repair process and the phosphorylation of DNA repair proteins require high energy phosphate such as ATP which is generated during glycolysis or oxidative phosphorylation (49). Therefore, DNA-PK interaction with metabolic partners and promotion of glycolysis/TCA activity may be important in supporting the energy demands of cells undergoing DNA repair. In addition, these interactions may generate intermediates that serve as building blocks for macromolecule synthesis or generation of acetyl and methyl moieties to modify chromatin and drive protumorigenic transcriptional networks, which would support cancer cell proliferation, metastasis, and resistance to therapies (42). Although these functions are studied separately, there may be coordinated crosstalk between the DNA-PK–mediated pathways in the nucleus and cytoplasm. Studying these functions in tandem, in the presence and absence of genotoxic or metabolic stress, may provide additional insights into how DNA-PK regulates and coordinates between its protumorigenic functions in cancer. This may lead to identification of novel approaches to target these pathways alone or in various combinations maximize antitumorigenic benefit.
Considering the ability of DNA-PK to promote multiple protumorigenic pathways, DNA-PK has been nominated as therapeutic target and DNA-PK inhibitors are currently being investigated in clinical trials as monotherapy or in combination with standard of care in various cancers (11, 31). The data herein and overexpression of various glycolytic proteins including ALDOA and PKM2 in cancer, suggest that patients with tumors overexpressing DNA-PK and/or ALDOA/PKM2 may benefit from combination of DNA-PK and ALDOA/PKM2 inhibitors. In that prostate cancer targeting PKM2 via siRNA leads to decrease proliferation and metastasis in prostate cancer (50, 51), thus combination with DNA-PKi may show greater anticancerous activity. Moreover, because upregulated glycolysis is a hallmark of cancer (32), patients may benefit from combination therapy of DNA-PKi and glycolytic inhibitors to target different nodes of the pathway, as suggested by data in Fig. 6B. Various glycolytic inhibitors have been identified and are being investigated in clinical trials including glucose transporter inhibitors, PFKFB3 inhibitors, 2-DG, and lonidamine (LND; ref. 52). 2-DG has been shown to possess single-agent activity in vitro but its interrogation was halted in phase I/II clinical trials for treatment of solid tumors, including hormone refractory prostate cancer, due to limited efficacy on inhibiting tumor growth and high toxicities (52). Similarly, LND showed promise but clinical investigation was terminated due to liver toxicities and modest effects in suppressing tumor growth (52). Combination of these drugs with DNA-PKi at lower doses may mitigate the observed high toxicities and have greater anti-proliferative effects on prostate cancer tumors. Data herein (Fig. 6B), shows that using DNA-PK inhibitor at IC25 and IC50 and combining it with titrating doses of 2-DG leads to combinatorial anti-proliferative effects, supporting the use of this combination for future studies. Although NU7441 is not clinically actionable, investigation of the new generation of DNA-PK inhibitors in combination with low doses of 2-DG is of high clinical relevance in CRPC. Furthermore, the genetic tumor landscape should be considered when using DNA-PK inhibitors to enhance response. Tumors with alterations in drivers of disease such as AR amplification/mutations, amplification of c-Myc, PI3K mutations, loss/mutation in tumor suppressor p53 and PTEN are associated with metabolic rewiring that augment energy producing pathways (53, 54), including glycolysis. These oncogenic events positively regulate glycolytic enzyme expression and activity, thus targeting these tumors with DNA-PK inhibitors alone or in combination with glycolytic inhibitors could lead to more robust anti-proliferative effects in aggressive disease.
In addition, targeting other metabolic pathways regulated by DNA-PK could provide therapeutic benefit. DNA-PK was found to interact with de novo lipid synthesis protein FASN through RIME. FASN is overexpressed and considered a hallmark of prostate cancer progression (55). Recent studies have suggested FASN is a therapeutic target in CRPC, therefore presenting an opportunity for co-targeting FASN and DNA-PK in CRPC. Interrogation of the DNA-PK/FASN complex and its consequences in lipid synthesis and beyond are essential to better understand the impact of this interaction in cancer proliferation and metastasis. Lipid metabolism is important in cancer because it provides lipid species needed for structural components of cellular membranes and is involved in redox homeostasis and energy generation (56). Lipids can also act as signaling molecules for cancer relevant pathways promoting proliferation and metastasis (56). Because lipid metabolism is upregulated in many cancers, targeting lipogenic enzymes (FASN, ACLY, ACC) and intracellular cellular lipid homeostasis (CPT1a, PPARγ, lipin2) are being investigated preclinically and in clinical trials (56, 57). Clinical trials focusing on lipid metabolism include ABT510 targeting CD36 in melanoma (phase I), GPR119 agonist targeting AMPK in various cancers (phase I/II), and perhexiline targeting CPT1, which is approved in Australia and New Zealand (57). In addition, multiple FASN inhibitors (C93, IPI-9119, TVB-2640) have shown promising results in preclinical models and TVB-2640 has entered phase I and II trials in colon cancer and breast cancer, respectively. Combined, targeting lipid metabolism in cancer is a promising area and combination with DNA-PK could enhance anticancerous benefits.
In summary, DNA-PK is a frequently overexpressed and hyperactive multifunctional protein that drives aggressive phenotypes in prostate cancer (10, 13). Although the role of DNA-PK is well studied in DNA repair, much remains to be uncovered about the underpinning mechanisms of DNA-PK protumorigenic functions. The study herein utilized unbiased proteomic and metabolomic approaches to identify novel DNA-PK partners, substrates and function that promote energy producing pathway, glycolysis to contribute in generation of aggressive disease. Furthermore, data herein nominate a combinatorial treatment of DNA-PK and glycolysis inhibitors for further investigation in CRPC. Findings presented here expand the knowledge of the field, encourage further interrogation of DNA-PK function in metabolic rewiring and inform potential novel combinatorial strategies that can result in more effective, durable therapies for management and treatment of CRPC.
Supplementary Material
Acknowledgments
We gratefully thank all the members of the Knudsen laboratory for their intellectual and technical support. We also want to thank the Grabocka Laboratory at Thomas Jefferson University, for their assistance with immunofluorescence experiments and Kayla Bremert for tissue collection through the Australian Prostate Cancer BioResource. Moreover, we want to thank the Translational Pathology core facility at SKCC, and the Bioanalytical Mass Spectrometry Facility (BMSF) at the Mark Wainwright Analytical Centre (MWAC) and UNSW Sydney. In addition, we thank the following institutions that supported this work: the NIH/NCI grants to K.E. Knudsen (5R01CA17640105, 5R01CA18256905) and the Sidney Kimmel Cancer Center (5P30CA056036), the Prostate Cancer Foundation Young Investigator Award to A.A. Shafi, and NCI F99 grant to J.J. McCann (F99CA212225). J. Holst and L.M. Butler are supported by the Tour de Cure Senior Research Grant (RSP-171–18/19). L.M. Butler was supported by a Principal Cancer Research Fellowship produced with the financial and other support of Cancer Council SA's Beat Cancer Project on behalf of its donors and the State Government of South Australia through the Department of Health.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
L.G. Gomella reports personal fees from Merck and Pfizer/Astellas during the conduct of the study, as well as personal fees from Lantheus outside the submitted work; L.G. Gomella also has a patent for Shed Cell Technology licensed to NuView. L.M. Butler reports grants from Cancer Council South Australia and Tour de Cure during the conduct of the study, as well as nonfinancial support from Novartis outside the submitted work. J. Holst reports grants from Tour de Cure during the conduct of the study, as well as personal fees from MetabloQ Pharmaceuticals outside the submitted work. K.E. Knudsen reports other support from CellCentric, Genentech, Janssen, Celgene, and Sanofi outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
E. Dylgjeri: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. V. Kothari: Validation, investigation, methodology, writing–review and editing. A.A. Shafi: Conceptualization, methodology, writing–review and editing. G. Semenova: Investigation, methodology, writing–review and editing. P.T. Gallagher: Methodology. Y.F. Guan: Investigation, methodology. A. Pang: Investigation, methodology. J.F. Goodwin: Conceptualization. S. Irani: Investigation, methodology. J.J. McCann: Methodology, writing–review and editing. A.C. Mandigo: Writing–review and editing. S. Chand: Writing–review and editing. C.M. McNair: Writing–review and editing. I. Vasilevskaya: Writing–review and editing. M.J. Schiewer: Writing–review and editing. C.D. Lallas: Resources. P.A. McCue: Data curation. L.G. Gomella: Resources. E.L. Seifert: Methodology, writing–review and editing. J.S. Carroll: Methodology. L.M. Butler: Resources, investigation, methodology, writing–review and editing. J. Holst: Resources, methodology, writing–review and editing. W.K. Kelly: Conceptualization, resources, supervision, funding acquisition, writing–review and editing. K.E. Knudsen: Conceptualization, resources, supervision, funding acquisition, writing–review and editing.
References
- 1. Goodwin JF, Knudsen KE. Beyond DNA repair: DNA-PK function in cancer. Cancer Discov 2014;4:1126–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cornell L, Munck JM, Alsinet C, Villanueva A, Ogle L, Willoughby CE, et al. DNA-PK-A candidate driver of hepatocarcinogenesis and tissue biomarker that predicts response to treatment and survival. Clin Cancer Res 2015;21:925–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Munster P, Mita M, Mahipal A, Nemunaitis J, Massard C, Mikkelsen T, et al. First-in-human phase I study of a dual mTOR kinase and DNA-PK inhibitor (CC-115) in advanced malignancy. Cancer Manag Res 2019;11:10463–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hosoi Y, Watanabe T, Nakagawa K, Matsumoto Y, Enomoto A, Morita A, et al. Up-regulation of DNA-dependent protein kinase activity and Sp1 in colorectal cancer. Int J Oncol 2004;25:461–8. [PubMed] [Google Scholar]
- 5. Beskow C, Skikuniene J, Holgersson Å, Nilsson B, Lewensohn R, Kanter L, et al. Radioresistant cervical cancer shows upregulation of the NHEJ proteins DNA-PKcs, Ku70 and Ku86. Br J Cancer 2009;101:816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Soderlund Leifler K, Queseth S, Fornander T, Stenmark Askmalm M. Low expression of Ku70/80, but high expression of DNA-PKcs, predict good response to radiotherapy in early breast cancer. Int J Oncol 2010;37:1547–54. [DOI] [PubMed] [Google Scholar]
- 7. Xing J, Wu X, Vaporciyan AA, Spitz MR, Gu J. Prognostic significance of ataxia-telangiectasia mutated, DNA-dependent protein kinase catalytic subunit, and Ku heterodimeric regulatory complex 86-kD subunit expression in patients with non–small cell lung cancer. Cancer 2008;112:2756–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shao C-J, Fu J, Shi H-L, Mu Y-G, Chen Z-P. Activities of DNA-PK and Ku86, but not Ku70, may predict sensitivity to cisplatin in human gliomas. J Neurooncol 2008;89:27–35. [DOI] [PubMed] [Google Scholar]
- 9. Kothari V, Goodwin JF, Zhao SG, Drake JM, Yin Y, Chang SL, et al. DNA-dependent protein kinase drives prostate cancer progression through transcriptional regulation of the Wnt signaling pathway. Clin Cancer Res 2019;25:5608–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mohiuddin IS, Kang MH. DNA-PK as an emerging therapeutic target in cancer. Front Oncol 2019;9:635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Medová M, Medo M, Hovhannisyan L, Muñoz-Maldonado C, Aebersold DM, Zimmer Y. DNA-PK in human malignant disorders: mechanisms and implications for pharmacological interventions. Pharmacol Ther 2020;215:107617. [DOI] [PubMed] [Google Scholar]
- 12. Hsu FM, Zhang S, Chen BP. Role of DNA-dependent protein kinase catalytic subunit in cancer development and treatment. Transl Cancer Res 2012;1:22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goodwin JF, Kothari V, Drake JM, Zhao S, Dylgjeri E, Dean JL, et al. DNA-PKcs-mediated transcriptional regulation drives prostate cancer progression and metastasis. Cancer Cell 2015;28:97–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goodwin JF, Schiewer MJ, Dean JL, Schrecengost RS, de Leeuw R, Han S, et al. A hormone-DNA repair circuit governs the response to genotoxic insult. Cancer Discov 2013;3:1254–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dylgjeri E, McNair C, Goodwin JF, Raymon HK, McCue PA, Shafi AA, et al. Pleiotropic impact of DNA-PK in cancer and implications for therapeutic strategies. Clin Cancer Res 2019;25:5623–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jackson SP. DNA-dependent protein kinase. Int J Biochem Cell Biol 1997;29:935–8. [DOI] [PubMed] [Google Scholar]
- 17. Ferguson BJ, Mansur DS, Peters NE, Ren H, Smith GL. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife 2012;1:e00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lucero H, Gae D, Taccioli GE. Novel localization of the DNA-PK complex in lipid rafts: a putative role in the signal transduction pathway of the ionizing radiation response. J Biol Chem 2003;278:22136–43. [DOI] [PubMed] [Google Scholar]
- 19. Kotula E, Faigle W, Berthault N, Dingli F, Loew D, Sun J-S, et al. DNA-PK target identification reveals novel links between DNA repair signaling and cytoskeletal regulation. PLoS One 2013;8:e80313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Feng J, Park J, Cron P, Hess D, Hemmings BA. Identification of a PKB/Akt hydrophobic motif Ser-473 kinase as DNA-dependent protein kinase. J Biol Chem 2004;279:41189–96. [DOI] [PubMed] [Google Scholar]
- 21. Wei Q, Qian Y, Yu J, Wong CC. Metabolic rewiring in the promotion of cancer metastasis: mechanisms and therapeutic implications. Oncogene 2020;39:6139–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma D, Chen X, Zhang P-Y, Zhang H, Wei L-J, Hu S, et al. Upregulation of the ALDOA/DNA-PK/p53 pathway by dietary restriction suppresses tumor growth. Oncogene 2018;37:1041–8. [DOI] [PubMed] [Google Scholar]
- 23. Mohammed H, D'Santos C, Serandour AA, Ali HR, Brown GD, Atkins A, et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep 2013;3:342–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mackay GM, Zheng L, van den Broek NJ, Gottlieb E. Analysis of cell metabolism using LC-MS and isotope tracers. Methods Enzymol 2015;561:171–96. [DOI] [PubMed] [Google Scholar]
- 25. Shafi AA, Schiewer MJ, de Leeuw R, Dylgjeri E, McCue PA, Shah N, et al. Patient-derived models reveal impact of the tumor microenvironment on therapeutic response. Eur Urol Oncol 2018;1:325–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Davis AJ, Chen DJ. DNA double strand break repair via non-homologous end-joining. Transl Cancer Res 2013;2:130–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol Cell 2006;22:511–9. [DOI] [PubMed] [Google Scholar]
- 28. Solier S, Kohn KW, Scroggins B, Xu W, Trepel J, Neckers L, et al. Heat shock protein 90alpha (HSP90alpha), a substrate and chaperone of DNA-PK necessary for the apoptotic response. Proc Natl Acad Sci U S A 2012;109:12866–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bunch H, Zheng X, Burkholder A, Dillon ST, Motola S, Birrane G, et al. TRIM28 regulates RNA polymerase II promoter-proximal pausing and pause release. Nat Struct Mol Biol 2014;21:876–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lees-Miller SP, Chen YR, Anderson CW. Human cells contain a DNA-activated protein kinase that phosphorylates simian virus 40 T antigen, mouse p53, and the human Ku autoantigen. Mol Cell Biol 1990;10:6472–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Damia G. Targeting DNA-PK in cancer. Mutat Res 2020;821:111692. [DOI] [PubMed] [Google Scholar]
- 32. DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv 2016;2:e1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liccardi G, Hartley JA, Hochhauser D. EGFR nuclear translocation modulates DNA repair following cisplatin and ionizing radiation treatment. Cancer Res 2011;71:1103–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huston E, Lynch MJ, Mohamed A, Collins DM, Hill EV, MacLeod R, et al. EPAC and PKA allow cAMP dual control over DNA-PK nuclear translocation. Proc Natl Acad Sci U S A 2008;105:12791–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ganapathy-Kanniappan S, Geschwind JF. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer 2013;12:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Davis AJ, Chen BP, Chen DJ. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amst) 2014;17:21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dobbs TA, Tainer JA, Lees-Miller SP. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst) 2010;9:1307–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hartley KO, Gell D, Smith GCM, Zhang H, Divecha N, Connelly MA, et al. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell 1995;82:849–56. [DOI] [PubMed] [Google Scholar]
- 39. Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab 2016;23:27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Choi SYC, Collins CC, Gout PW, Wang Y. Cancer-generated lactic acid: a regulatory, immunosuppressive metabolite? J Pathol 2013;230:350–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Centenera MM, Raj GV, Knudsen KE, Tilley WD, Butler LM. Ex vivo culture of human prostate tissue and drug development. Nat Rev Urol 2013;10:483–7. [DOI] [PubMed] [Google Scholar]
- 42. Yu X, Li S. Non-metabolic functions of glycolytic enzymes in tumorigenesis. Oncogene 2017;36:2629–36. [DOI] [PubMed] [Google Scholar]
- 43. Zhang JY, Zhang F, Hong CQ, Giuliano AE, Cui XJ, Zhou GJ, et al. Critical protein GAPDH and its regulatory mechanisms in cancer cells. Cancer Biol Med 2015;12:10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. He Y, Luo Y, Zhang D, Wang X, Zhang P, Li H, et al. PGK1-mediated cancer progression and drug resistance. Am J Cancer Res 2019;9:2280–302. [PMC free article] [PubMed] [Google Scholar]
- 45. Yang T, Shu X, Zhang H-W, Sun L-X, Yu L, Liu J, et al. Enolase 1 regulates stem cell-like properties in gastric cancer cells by stimulating glycolysis. Cell Death Dis 2020;11:870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zahra K, Dey T, Ashish , Mishra SP, Pandey U. Pyruvate kinase M2 and cancer: the role of PKM2 in promoting tumorigenesis. Front Oncol 2020;10:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gizak A, Wiśniewski J, Heron P, Mamczur P, Sygusch J, Rakus D. Targeting a moonlighting function of aldolase induces apoptosis in cancer cells. Cell Death Dis 2019;10:712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sobanski T, Rose M, Suraweera A, O'Byrne K, Richard DJ, Bolderson E. Cell metabolism and DNA repair pathways: implications for cancer therapy. Front Cell Dev Biol 2021;9:633305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Turgeon MO, Perry NJS, Poulogiannis G. DNA damage, repair, and cancer metabolism. Front Oncol 2018;8:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dey P, Kundu A, Sachan R, Park JH, Ahn MY, Yoon K, et al. PKM2 knockdown induces autophagic cell death via AKT/mTOR pathway in human prostate cancer cells. Cell Physiol Biochem 2019;52:1535–52. [DOI] [PubMed] [Google Scholar]
- 51. Guo W, Zhang Z, Li G, Lai X, Gu R, Xu W, et al. Pyruvate kinase M2 promotes prostate cancer metastasis through regulating ERK1/2-COX-2 signaling. Front Oncol 2020;10:544288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li J, Eu JQ, Kong LR, Wang L, Lim YC, Goh BC, et al. Targeting metabolism in cancer cells and the tumour microenvironment for cancer therapy. Molecules 2020;25:4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zadra G, Loda M. Metabolic vulnerabilities of prostate cancer: diagnostic and therapeutic opportunities. Cold Spring Harb Perspect Med 2018;8:a030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chetta P, Zadra G. Metabolic reprogramming as an emerging mechanism of resistance to endocrine therapies in prostate cancer. Cancer Drug Resistance 2021;4:143–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zadra G, Ribeiro CF, Chetta P, Ho Y, Cacciatore S, Gao X, et al. Inhibition of de novo lipogenesis targets androgen receptor signaling in castration-resistant prostate cancer. Proc Natl Acad Sci U S A 2019;116:631–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fernandez LP, Gomez de Cedron M, Ramirez de Molina A. Alterations of lipid metabolism in cancer: implications in prognosis and treatment. Front Oncol 2020;10:577420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Butler LM, Perone Y, Dehairs J, Lupien LE, de Laat V, Talebi A, et al. Lipids and cancer: emerging roles in pathogenesis, diagnosis and therapeutic intervention. Adv Drug Deliv Rev 2020;159:245–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated and/or analyzed during this study are available upon reasonable request from the corresponding author.



![Figure 4. DNA-PK regulates glycolysis. A, Pyruvate levels and lactate production was measured upon DNA-PK inhibition (NU7441, 1 μmol/L, 24 hours). B, DNA-PK knockdown (96 hours posttransfection). C, Pyruvate levels upon overexpression of ALDOA and PKM2 as validated via protein levels (left). D, Glucose tracing through glycolysis pathway to GADP, 3PG, pyruvate, and lactate generation through metabolic flux analysis. C4–2 cells were treated with NU7441 (1 μmol/L, 24 hours) or vehicle control, followed by 2 hours pulse with [U-13C6]-D-glucose. Data collected in at least three biological replicates, represented as mean ± SEM (ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/5020/9365345/aad635c26740/1446fig4.jpg)