Abstract
Close to 50 % of the human genome harbors repetitive sequences originally derived from mobile DNA elements, and in normal cells this sequence compartment is tightly regulated by epigenetic silencing mechanisms involving chromatin-mediated repression. In cancer cells, repetitive DNA elements suffer abnormal demethylation, with potential loss of silencing. We used a genome-wide microarray approach to measure DNA methylation changes in cancers of the head and neck, and to compare these changes to alterations found in adjacent non-tumor tissues. We observed specific alterations at thousands of small clusters of CpG dinucletides associated with DNA repeats. Among the 257,599 repetitive elements probed, 5 to 8% showed disease-related DNA methylation alterations. In dysplasia, a large number of local events of loss of methylation appear in apparently stochastic fashion. Loss of DNA methylation is most pronounced for certain members of the SVA, HERV, LINE-1P, AluY, and MaLR families. The methylation levels of retrotransposons are discretely stratified, with younger elements being highly methylated in healthy tissues, while in tumors these young elements suffer the most dramatic loss of methylation. Wilcoxon test statistics reveal that a subset of primate LINE-1 elements is demethylated preferentially in tumors, as compared to non-tumoral adjacent tissue. Sequence analysis of these strongly demethylated elements reveals genomic loci harboring full-length, as opposed to truncated elements, while possible enrichment for functional LINE-1 ORFs is weaker. Our analysis suggests that in non-tumor adjacent tissues there is generalized and highly variable disruption of epigenetic control across the repetitive DNA compartment, while in tumor cells a specific subset of LINE-1 retrotransposons that arose during primate evolution suffers the most dramatic DNA methylation alterations.
Keywords: mobile DNA, natural selection, methylation, epigenetic control, disease progression, DNA repair
1. Introduction
The DNA of most tumors has a reduced content of methylated cytosine residues. This so-called global “hypomethylation” affects primarily DNA sequences that belong to interspersed DNA repeats. In normal human tissues, DNA repeats are predominantly methylated, consistent with the requirement to maintain genomic stability by transcriptional silencing of retroelements whose potential deleterious functions include DNA mobilization as well as the facilitation of recombination events in somatic cells. There have been a considerable number of reports of transcriptional activation of retrotransposons in the context of loss of DNA methylation. Expression of human endogenous retroviruses (HERVs) has been detected in breast cancer (Wang-Johanning et al., 2001), ovarian cancer (Menendez et al., 2004, Wang-Johanning et al., 2007), leukemia cell lines, (Patzke et al., 2002), urothelial and renal cell carcinomas (Florl et al., 1999). Increased transcriptional expression of HERV-K has been reported in teratocarcinoma (Löwer et al., 1984; Herbst et al., 1998), breast cancer cells and adjacent tissues (Wang-Johanning et al., 2003, Golan et al., 2008), and in melanoma (Muster et al., 2003; Büscher et al., 2006, Serafino et al., 2009). Stauffer et al. (2004) used massively parallel signature sequencing (MPSS) to define the number and type of transcripts of endogenous retroviruses of the LTR family in various cancers. This study reported that HERV-H, a relatively young retrotransposon, was expressed in cancers of the intestine, bone marrow, bladder and cervix, and was more highly expressed than the other families in cancers of the stomach, colon and prostate. Recently Alves et al. (2008) have reported that a specific HERVH element present in the X chromosome is selectively transcribed in 60% of colon cancers, and in a high proportion of metastatic colon cancers. There is evidence for context-specific induction of LINE-1 transcription during oxidative stress (Teneng et al., 2007). In a relatively large study of squamous head and neck carcinomas, Smith et al. (2007) reported that the DNA methylation level of LINE-1 elements was significantly reduced, and correlated with environmental insults such as alcohol use and smoking, as well as tumor stage.
Here we report a systematic study of DNA methylation changes occurring in the repetitive DNA compartment of squamous carcinomas of the head and neck. In contrast to previous studies, we use a novel microarray-based approach to obtain discrete DNA methylation data at hundreds of thousands of individual repetitive DNA loci in the human genome. We then use extensive annotation resources for different subfamilies of repeats to evaluate possible relationships between loss of epigenetic silencing in the context of natural history of cancer, and the evolutionary history of repetitive element sub-compartments in the human genome.
2. Materials and Methods
A microarray analysis method developed in our laboratory permits genome-wide assessment of DNA methylation status using restriction endonucleases (see Supplementary Materials section 1). Among the 339,314 probes in the microarray, 257,599 are dedicated to the measurement of the methylation levels of individual members of interspersed DNA repeat families.
2.1 Principle of the DNA methylation analysis method
Multiple displacement amplification (MDA, Dean et al., 2002; Lage et al., 2003; Lage et al., 2005) is an isothermal amplification method based on random priming and DNA hyper-branching, catalyzed by a strand-displacing DNA polymerase. The yield of the MDA reaction is strongly influenced by the size of the DNA used as template (Lage et al., 2005). We have systematically studied the dependence of amplification yield using DNA templates of different size, and also built a computational model of the reaction that fits the experimental data. The results of this analysis (data not shown) indicate that the yield of DNA derived from any sequence segment depends on template size, and additionally on the distance of the sequence segment from the nearest DNA terminus on the template molecule. We reasoned that a specific cleavage event in a genomic DNA molecule could be detected by measuring DNA amplification yield using a DNA microarray, and that a probe in the microarray would be able to measure a local reduction in sequence representation due to cleavage, even if that cleavage event occurred as far as 1200 bases upstream or downstream from the location of the probe. This property enables the use of probe designs that measure cleavage events not only in unique DNA sequences overlapping a probe, but also cleavage events within repetitive DNA sequences that contain CpG dinucleotides, located in the vicinity of a probe of unique sequence, within a window of approximately 2400 bases surrounding the probe. Below we will present experimental data that help to define the approximate size of the window that enables probing-at-a-distance.
2.2 Microarray probe design
DNA probes of unique sequence (uniqueness assessed using merEngine, Healy et al., 2003) were designed to map as closely as possible to every CpG island in the human genome. We examined the DNA sequences located within a window of plus or minus 4 kb from loci coding for microRNAs, and noticed that many of these regions contain small clusters of CpG residues. We then created a relatively lax “CpG islet” specification, requiring that a region in the genome contain a minimum of 7 CpG residues, that the ratio of the CG count to the GC content be larger than 0.53, and that the region be no shorter than 200 bases to be nominated as a CpG islet. Using this specification, 453 out of the 532 microRNA loci in the Sanger database (Griffiths-Jones, 2006) are associated with at least one CpG islet within a window of +/− 4 kb. By contrast, based on the more restrictive Takai and Jones definition (2002), the equivalent count of CpG islands in the vicinity of microRNA loci is 141. The total count of CpG islets in the human genome using our relaxed specification is approximately 500,000. We designed a custom microarray containing probes for all CpG islands and CpG islets, in order not to miss DNA methylation changes that may occur in tumors in CpG-rich regions that would not fit the standard CpG island definition. Five broad classes of CpG islands and CpG islets were probed: promoter associated, unique, non-promoter associated, interspersed repeat associated (Jurka, 1998; Smit, 1996–2004), tandem repeat associated (Benson, 1999), and microRNA locus associated (Griffiths-Jones, 2006). A subset of the probes were replicated on the array surface, bringing the total number of probes in the microarray to 377,000.
2.3 Experimental work flow for microarray analysis
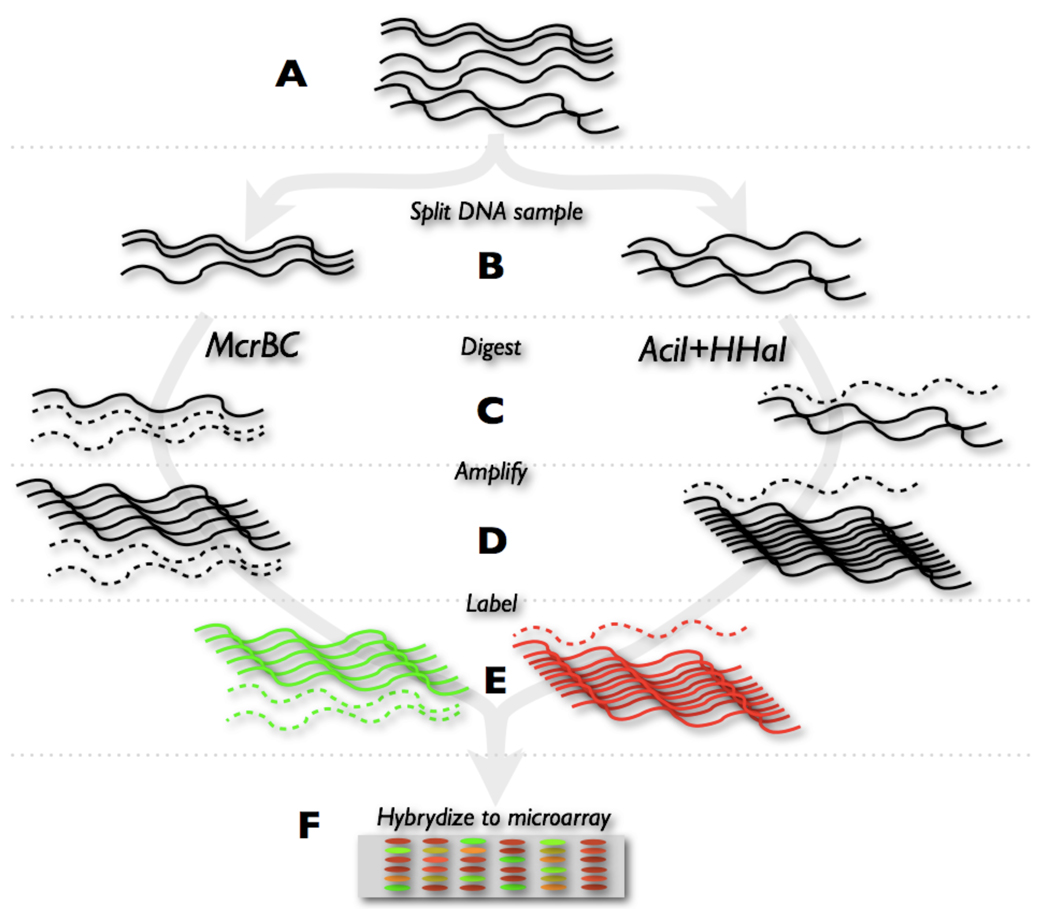
Relative methylation was measured by splitting the DNA sample in two equal aliquots, and digesting each aliquot with either methylation-sensitive or methylation-dependent restriction endonucleases, respectively, as shown diagrammatically in Figure 1. Each of the two digests was amplified by MDA, and then labeled with a different dye, followed by mixing after labeling, and processed for DNA microarray analysis as described in Section 1 of Supplementary Materials. The enzymes we used to sample DNA methylation have fairly high sampling efficiency when used individually, as ascertained using sequence analysis. Supplementary Table S1 documents the theoretical sampling efficiency of the mixture of the methylation-sensitive endonucleases AciI (recognition site CCGC) and HhaI (recognition site GCGC). The table also documents the sampling efficiency of the methylation-dependent endonuclease McrBC (recognition site PumC[N40-3000]PumC). Of course, the enzymes do not sample all CpG residues in the genome, but this limitation is alleviated by the fact that most neighboring CpG residues in a CpG island tend to have similar methylation status at any given time. By theoretical sampling efficiency we mean that known cleavage sites exist in the sequence within or near a CpG island, which may or may not be methylated in any given DNA sample, but would cause a fluorescence intensity change in either channel of the microarray whenever a methylation change occurred. Since we sample DNA using two separate digestion reactions, probed loci should be capable of reporting DNA methylation changes based on the presence or absence of cleavage in a single color channel, as well as detecting signal alterations in both color channels, reflecting changes in the combined cleavage susceptibility to the two classes of endonuclease used. The last column on Supplementary Table S1 indicates that if one considers theoretical cleavage sites for both sets of enzymes in combination, the potential sampling efficiency increases to 99.9% of all probed CpG islands. The labeled DNA is hybridized to the custom microarray, and subsequently the ratio of intensities is generated for locus-specific methylation levels associated with each probe. In order to illustrate the relationship between the probe location and the location of restriction endonuclease sites in CpG rich domains associated with specific repetitive elements, we show two detailed maps of interspersed repetitive elements that were probed in the microarray. Figure 2A shows a map of a LINE-1PA3 element that was probed using a unique sequence located within 150 bases of the 5’-terminus of the retroelement. The Figure shows the location of the CpG islet in the retroelement, as well as the location of all possible restriction endonuclease sites within and around the element. Figure 2B shows a similar map, in this case corresponding to a THE1C element. More examples of probed MaLR subclass of elements can be found in Supplementary Figure S22 which shows the locations of the probes and the restriction endonuclease cutting sites in the CpG islands associated with these elements.
Figure 1.
A simplified diagram summarizing the steps discussed in the methods section A) DNA is first acquired from a tissue material B) the DNA is split into two equal aliquots C) each of them is then digested with methylation sensitive or dependent enzymes D) the DNA is then amplified E) labeled and F) hybridized to a microarray.
Figure 2.
Examples of probe design and microarray response for two probes near repetitive elements. The figure shows the genomic context of a repetitive element, the locations of probes, CpG islands, other repeats, potential enzyme cuts as well as outcomes from 6 methylation experiments.
The top part was generated using the UCSC genome browser. Each of the 6 tracks labeled on the right with a green (normal sample), yellow (non-tumor adjacent sample) or red box (tumor sample) corresponds to a single methylation experiment. The text underneath provides a summary of a region using ASCII characters (generated using a tool ASCIIMap). The 6 ASCIIMap tracks show the location of the probe (o and highlighted with an arrow) and ~700 bases up- and downstream (:) which together form a region where the probe’s signal is coming from.The location of a CpG island is marked underneath (*) as are the locations of repetitive elements in the area (1-Line1, 2-Line2, T-LTR(MaLR), S- SINE, =-other, A-Alu, etc.). The vertical bars (|) indicate the presence of an enzyme recognition site for AciI, HHaI and McrBC enzymes respectively.
The resolution of 1 character is about 100 nucleotides.
A) L1PA3, the total region shown is approximately 16kb wide.
B) THE1C, the total region shown is approximately 11kb wide.
More ASCIIMAPs are shown in Supplementary figure S22.
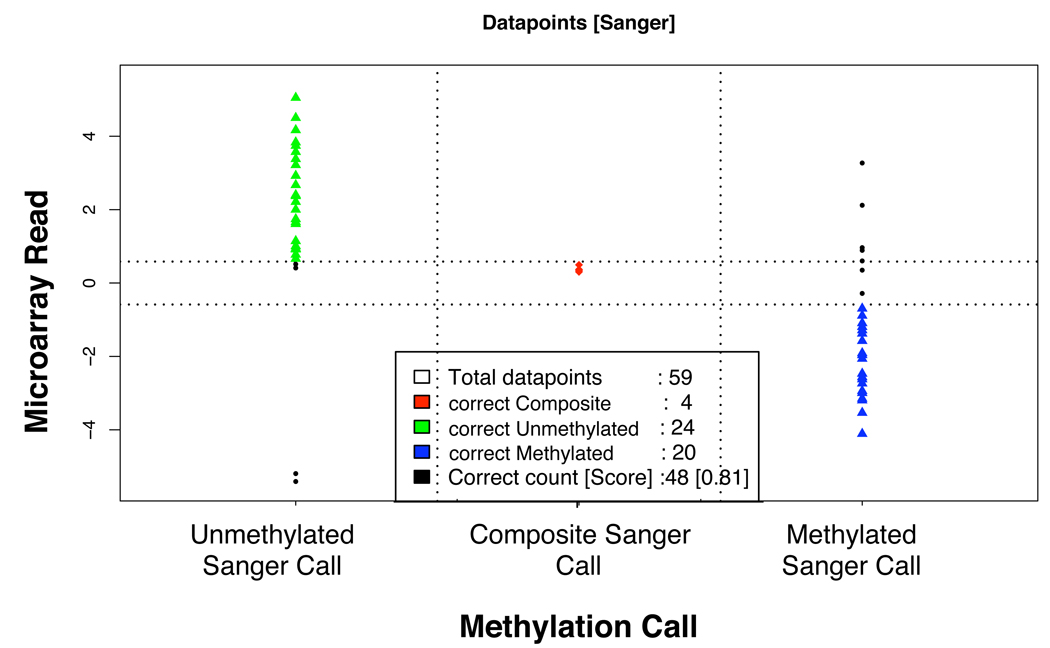
The experimental data obtained from 74 different probe loci in the microarray was independently validated by bisulfite sequencing using either Sanger sequencing of individual clones of PCR products, or using the Sequenom EpiTyper platform, which is based on sequencing of transcribed RNA by mass spectrometry. In Figure 3, we show the results of the Sanger-based analysis, which was performed for a total of 59 different microarray probes. For 48 of the 59 probes, there was agreement between the microarray methylation result and the bisulfite Sanger sequencing result, for a concordance of 81.4%. We show exemplary data from the bisulfite sequencing validation analysis in Supplementary Figures S2A, S2B, and S2C. These figures present the methylation results from the microarray analysis, the map position of the probes, and the bisulfite sequencing result for one gene promoter region, one AluSq element and one AluY element. In these three validation experiments there is agreement between the microarray result and the bisulfite sequencing data. It should be noted, however, that in the case of the probe that samples the AluSq element (Supplementary Figure S2B) there are neighboring sequences belonging to an MLT1C LTR element, whose methylation status will influence our measurements. We made an effort, through extensive probe annotation data, to keep track of these complex cases. The results we will present below are generated by calculating the average methylation of hundreds, or even thousands of repetitive elements belonging to specific families of repeats. One would expect that this averaging process will minimize the influence of the surrounding sequence context.
Figure 3.
Correlation between the microarray read-out and the results of Sanger sequencing. Based on the count of CpGs methylated or demethylated in all the clones of the sequencing result of a locus, the sequences were classified as un-methylated (green), composite (red) or methylated (dark blue). The black dots represent disagreement between the microarray call and the sequencing result.
2.4 Specimen Sample Acquisition and DNA preparation
Tumor samples and adjacent non-tumor tissue were obtained through the Tissue Procurement Program of the Surgical Pathology Laboratory at Yale New Haven Hospital. All patients provided informed consent (IRB/HIC # 14414). Representative histological sections of all specimens were reviewed to confirm the nature of the sample. After informed consent, oral epithelial cells from subjects with no known risk for oral cancer were obtained by scraping. Available clinical data for each of the samples is presented in Supplementary Table S4. DNA from all tissues was obtained using MasterPure DNA Purification Kit (EPICENTRE). The protocol follows: for every reaction a mix of 150 µL of Tissue and Cell Lysis solution and 1.5 µL of proteinase K from the kit was created. Lysate from about 8mm3 of specimen was collected. The lysate was vortexed every 5 min until the tissue was completely dissolved. The incubation at 65 degrees followed for 30–60 min. Subsequently 0.5 µL of RNase was added to each tube and incubated for 30 min at 37 degrees. 75 µL of MPC protein precipitation agent was added to the lysed sample. After centrifugation for 10 min at 15,000 rpm the supernatant was transferred to a labeled 1.5mL tube. With 250 µL of isopropanol added to the supernatant the tube was inverted multiple times. The DNA was then transferred using Pasteur pipet and resuspended in 100 µL of TE (0.1 mM EDTA). The DNA was then stored for 2 days at 4 degrees. Subsequent quantitation was done using PicoGreen fluorescence.
200 ng genomic DNA extracted from the head and neck tumor or the corresponding non-tumor adjacent tissues were digested by two sets of restriction enzymes respectively. One genomic sample was digested by McrBC (New England Biolabs), the other was digested by AciI and HhaI (New England Biolabs). 20 units of each enzyme were used to set up 45 µl reaction in the recommended buffer (McrBC: 50 mM NaCl, 10 mM Tris-HCl, 10 mM MgCl2, 1 mM DTT supplemented with 100 µg/mL BSA, and 5 mM GTP; AciI and Hha: 50 mM Tris-HCl, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT supplemented with 100 µg/mLBSA). Reactions were incubated at 37 C for 6 hours and then boosted with an additional 10 units of the corresponding enzyme for another 12 hrs, and finally inactivated at 65C for 20 minutes. One aliquot of each digested genomic DNA (20ng) was subjected to whole genome amplification respectively using REPLI-G kit (Qiagen) with 8 hours incubation at 30C. The amplified DNA sample was then purified by QIAEX II kit (Qiagen) with slightly modified protocol (3 instead of 2 washes with PE buffer and finally eluted in water rather than EB buffer). 4 µg of the purified genomic DNA sample was submitted to Nimblegen for labeling and hybridization.
3. Results And Discussion
3.1 Methylation patterns of major classes and families of DNA repeats
We analyzed the DNA methylation profiles of 33 tumors and 17 non-tumor adjacent tissue samples obtained from patients with head and neck squamous carcinoma (HNSCC). We also generated DNA methylation profiles from the buccal epithelia of 10 normal individuals, which served as controls. In addition, an analysis of sperm DNA was performed in technical triplicate to assess the reproducibility of the microarray results. We calculated an average methylation value for selected subsets of “genomic probe compartments”. An exemplary profile of average methylation for two extremely different genomic probe compartments can be found in Supplementary Figure S4A, which shows DNA methylation values for all SVA elements as well as methylation values for all genes for each of the tissue sample experiments. We then combine all average metrics per tissue class, generating a distribution of averages, shown in Supplementary Figure S4B. The figures below (4 through 9) all use a box and whiskers distribution plot to display DNA methylation trends for different classes of repetitive elements.
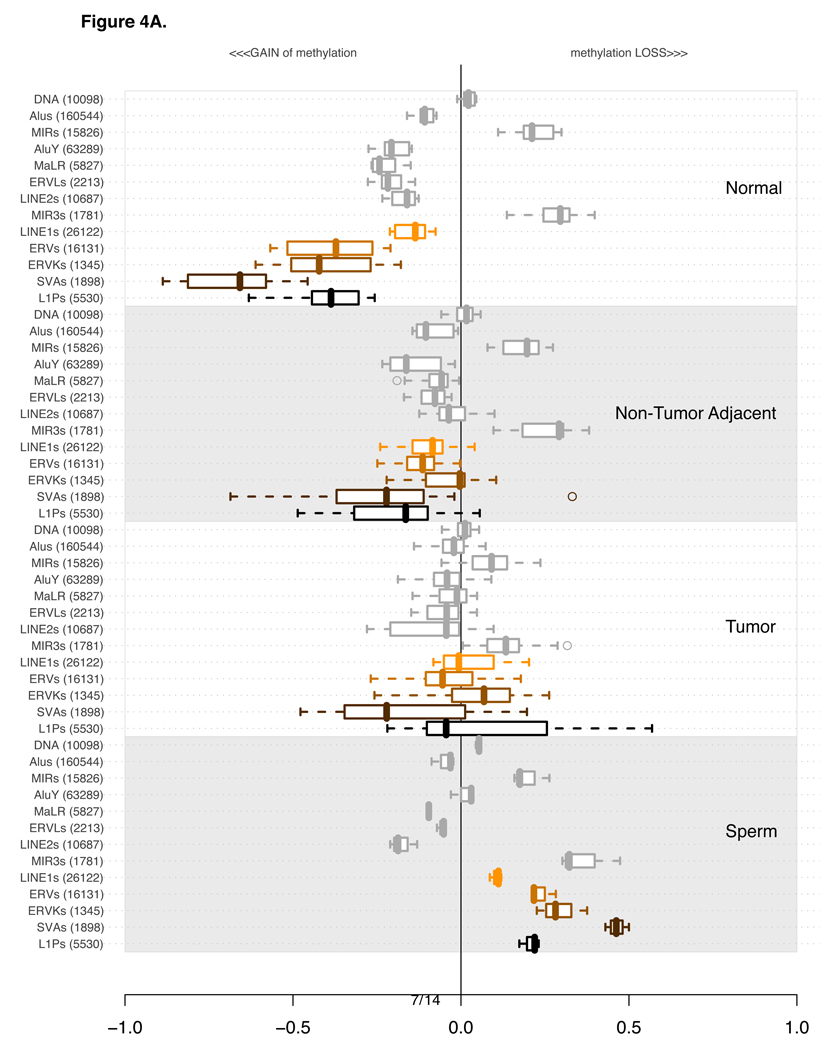
Figure 4A depicts the distribution of all averages of DNA methylation values across all experiments for each of the major repetitive element families (as summarized by Mandal and Kazazian, 2008).Two primate-specific families of repeats, AluY (Alu) and L1P (LINE-1), were also included and will be discussed at length in sections 3.3 and 3.6, respectively. Gain of methylation is represented by values on the negative scale of the x-axis, and loss of methylation by values on the positive scale, towards the right side of the plots. Each subsection of the plot features the same families of repetitive elements in the same order for normal, non-tumoral adjacent, tumor and replicated sperm experiments. The order of repetitive families was established based on the information content (Shannon entropy metrics) of the methylation values among normal and tumor experiments (Supplementary Materials Section 3). The 5 most “informative” families of repeats are plotted towards the bottom of the plot’s subsection and highlighted in colors. The most informative is a primate-specific subset of probes for a subcategory of LINE-1. Annotation provided by RepBase (Jurka, 1998) allowed us to investigate subclasses of repetitive elements in greater detail. Section 3.6 expands on the analysis of L1P elements delving into finer subcategories of these repetitive elements.
Figure 4.
The four sections of the plot indicate 4 distinct classes of tissue types used for methylation profiling: Normals (10 experiments), non-tumor adjacent (17 experiments), Tumors (33 Experiments) and Sperm (3 replicate experiments). Each of the four sections contains the methylation levels of the same 13 categories of repetitive elements.
Per category, the values are summarized using a box-and-whisker plot. A line within each box indicates the median value. Box boundaries are drawn based on 1st and 3rd quartiles. The dashed lines extending from the box indicate the extreme values of the distribution. Outliers, if any, are indicated by a circle.
The classes and families of repetitive elements are indicated on the left of the box-and-whisker segment. The number in parenthesis next to the category description indicates the number of probes corresponding to the number of repetitive elements uniquely probed in the genome.
The order of categories is constant in all four of the subsections. It was established based on the extent of variation in the plotted distributions using Shannon entropy information content metric. Only Normal and Tumor experiments were used to calculate the Shannon’s Information metric. For a more detailed explanation see Supplementary Section 3.
A) Distribution of average methylation levels per category. In each of the 4 subsections of the plot the pertinent experiments contributed an average methylation level for all probes in proximity of a specific class of repetitive element.
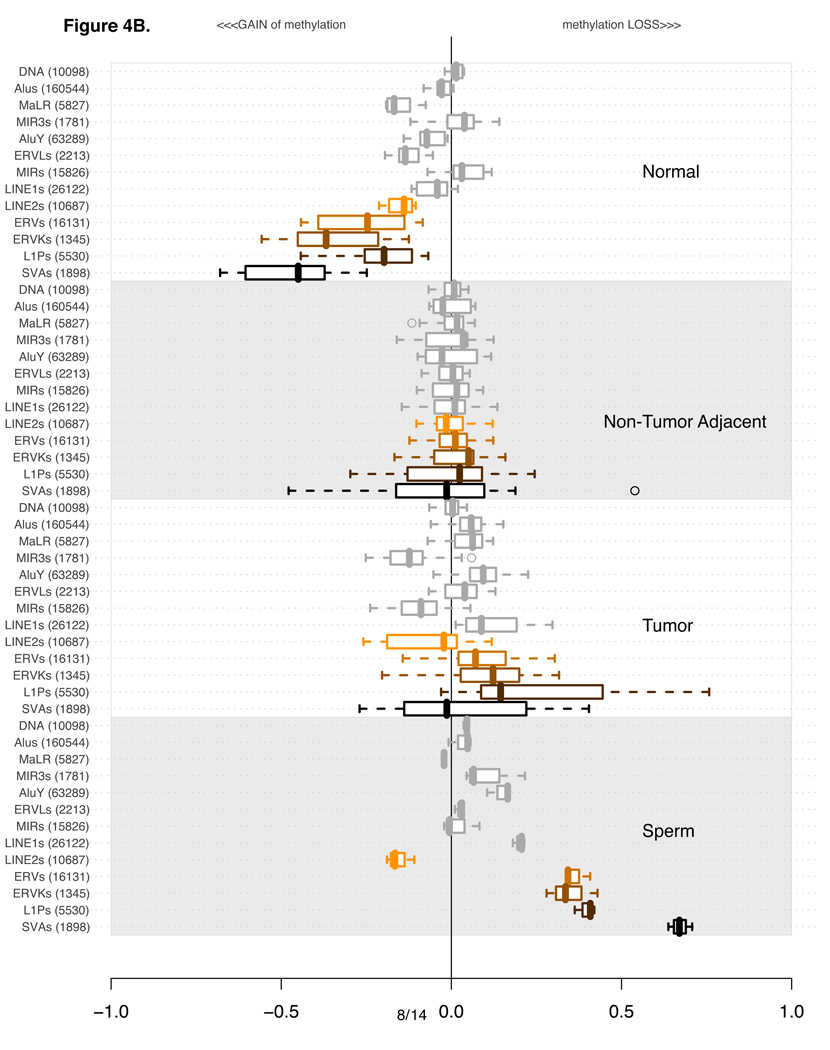
B) As in part A, except this time every experiment is normalized using an average of all tumor-adjacent experiments.
An alternative way of examining the DNA methylation data is to compare methylation levels that have been normalized with respect to the values of the non-tumor adjacent tissue, as we show in Figure 4B. This adjacent-tissue-normalized plot makes it clear that the average methylation levels of several classes of repetitive elements in tumors are not dramatically different from those in the non-tumor adjacent material, while the normal buccal mucosa shows much higher methylation, especially for SVA, ERV, ERVK, and L1P elements. Remarkably, it appears that the cumulative differences of loss of methylation for certain classes of elements such as SVA are as large between the normal and the non-tumoral adjacent tissue as they are between tumors and normal tissue samples. Previous reports in the head and neck cancer literature (Smith et al., 2007) indicate that normal buccal epithelium of individuals exposed to cigarette smoke have abnormally reduced levels of global methylation of LINE-1 elements, as determined by bisulfite sequencing of LINE-1 PCR amplicons. Even lower methylation levels were reported by these authors in cancer tissues, with advanced tumors (stages II and IV) showing the lowest methylation levels. It has also been reported that in colon cancer patients, non-tumor adjacent regions of colonic mucosa show significant loss of methylation of LINE-1 retrotransposons as compared to colonic mucosa of normal individuals (Suter et al., 2004), while colon cancer tissue shows even more pronounced loss of methylation. In our microarray study, Figures 4A and 4B suggest that various subcategories of larger repetitive element families contribute disproportionately to the DNA methylation changes of their parent category. In the following subsections we will adhere to the plot style in Figure 4A, which most accurately represents the raw data generated by the microarray analysis, and also shows the best fit to the DNA methylation values obtained independently by bisulfite sequencing of PCR products of specific probed loci. For example, in Figure 4A it is clear that MIR elements (representing DNA transposons), are relatively unmethylated in normal mucosa, in non-tumor adjacent tissue, and in sperm. Notably, only in tumor tissue do we observe gain of methylation of these DNA transposons. These interesting relative methylation relationships are more difficult to observe, and less indicative of the actual methylation levels, as displayed in Figure 4B due to the effects of normalization.
The following sections will focus on exploring in greater detail the dynamics of methylation patterns among various sequence sub-compartments of a given class of repetitive elements. To facilitate navigation through the perhaps unfamiliar nomenclature that identifies the various subclasses of elements, we have included four tables that list the different subclasses within a class, in chronological order of estimated evolutionary age. These tables (Supplementary Tables S2A through S2D) can be found in the supplementary materials. We are not including a table for the ERV retroelements, because the evolutionary ages and phylogenetic relationships of these elements are still a subject of investigation and revised annotation. To facilitate additional exploration of the data sets that follow, the Supplementary Figures section includes a set of Figures (S5 through S10, matching Figure 4 through Figure 9) where the methylation levels are grouped by subclass of repetitive element, rather than by tissue type, as we have done in this manuscript.
Figure 9.
Distributions of average methylation levels per lineages of L1P in subsets of experiments. In each of the 4 subsections of the plot, the pertinent experiments contributed an average methylation level for all probes in proximity of a specific class of repetitive element indicated on the left. The values are summarized using a box-and-whisker plot. A single star indicates a family only present in primates (< 25 MYA, younger than the origin of baboon species). A double star indicates a human specific lineage (< 5 MYA, not present in the chimpanzee genome). Supplementary Table S2 contains more detailed information about the ages of each of the subfamilies.
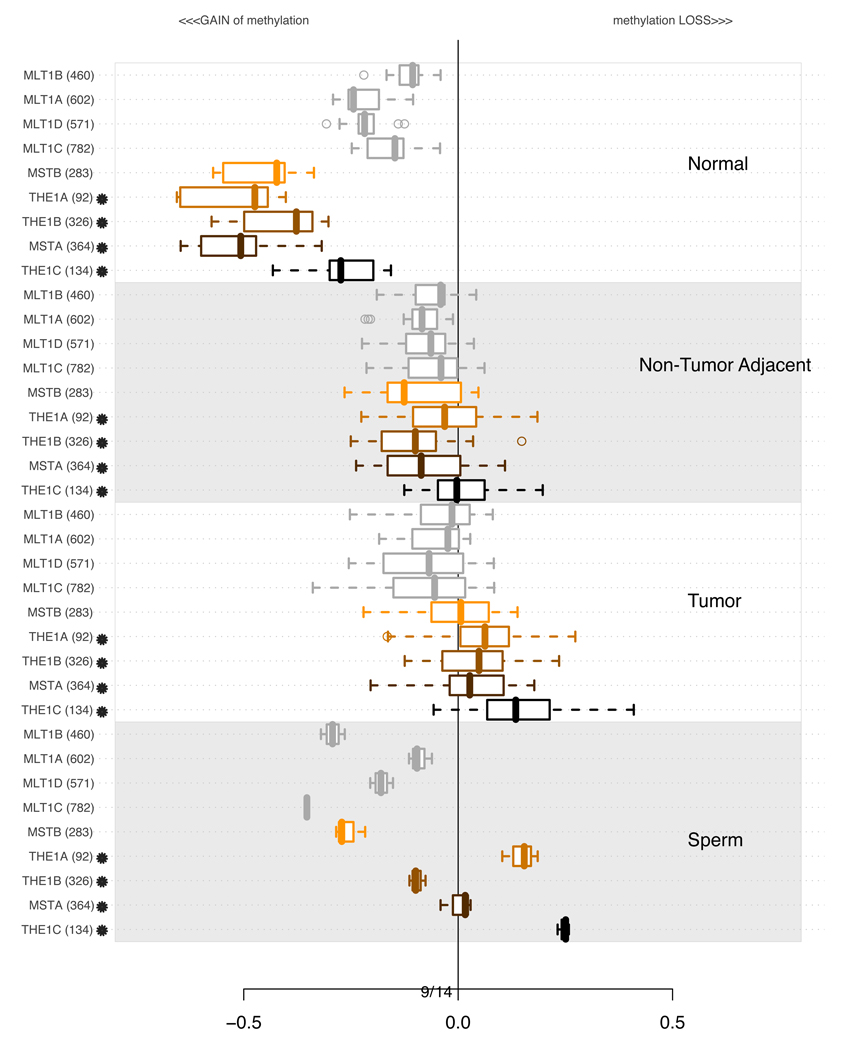
3.2 Methylation of MaLR elements
Since we observed that AluY and L1P, two primate-specific subfamilies of repeats scored higher using the information content statistic than their respective (and all-inclusive) parents, we investigated a relatively well-annotated family tree of “mammalian apparent LTR retrotransposons” (MaLR) (Smit, 1993). An analysis involving specific subsets of MaLR is shown in Figure 5. Here, again, the youngest members of the MaLR elements, THE1A, THE1B, THE1C, MSTA, and MSTB which only exist in simians and humans, are methylated in normal tissue but show marked loss of methylation in most tumors. Older MaLR subfamilies are less methylated in normal tissue, and show less striking loss of methylation in tumors. It is intriguing that among the younger families, THE1A and THE1B retroelements, which we observe to be demethylated in most non-tumoral adjacent tissue as well as tumors, have been identified as key sequence attributes of human recombination hotspots (Myers et al., 2005, 2008). The possible contribution of these subfamilies of demethylated MaLR elements to recombinational events in tumors is an interesting subject for future investigation.
Figure 5.
Distributions of average methylation levels per lineages of MaLR (Smit, 1993) in subsets of experiments. In each of the 4 subsections of the plot, the pertinent experiments contributed an average methylation level for all probes in proximity of a specific class of repetitive element indicated on the left. The values are summarized using a box-and-whisker plot. A star next to a family name indicates that the family is primate specific, and the estimated time of its origin in the genome is less than 60 Millions years ago (MYA). Supplementary Table S2 contains more detailed information about the ages of each of the subfamilies.
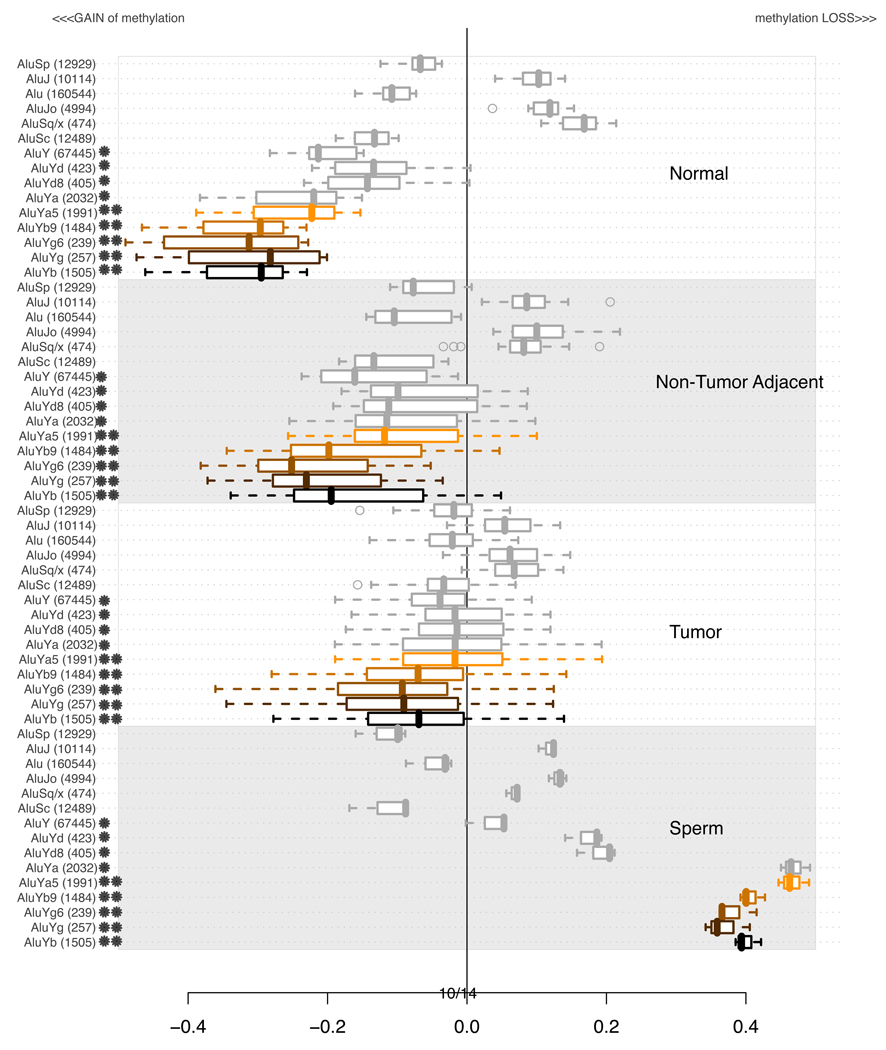
3.3 Methylation of Alu elements
Alu elements are the most abundant class of repetitive elements in the human genome with over one million copies and spanning over 30 lineages. The most detailed published analysis of Alu DNA methylation in normal cells and cancer cells was reported by Rodriguez et al. (2008). These authors targeted unmethylated SmaI sites within Alu sequences, and found that normal colon epithelial cells contain a subpopulation of undermethylated Alus, while in tumor cells the number of unmethylated Alu sequences is doubled. They also reported an increased methylation of the younger Alu subfamilies. Our microarray-based analysis, shown in Figure 6, includes only those Alu lineages for which we were able to probe more than 200 unique locations. As observed for other classes of elements, the younger elements (AluY) are more highly methylated in normal adult tissues, yet are suffering a greater loss of DNA methylation in many tumors. Interestingly, the oldest Alu elements remain methylated in sperm, while the younger ones show loss of methylation in this tissue. The most informative among normal and tumor tissues lineage of Alu’s is AluYb. Coincidentally, it is also the most active of all Alu lineages and found primarily in human genomes (Jurka, 1993; Carter et al., 2004). AluYg, the next most informative lineage remains relatively unknown. Among other, less informative lineages, the middle-age AluS families lose methylation in tumor tissue, while the members of the oldest, AluJ lineages remain methylated at an intermediate level, and constant in all 4 tissue types.
Figure 6.
Distributions of average methylation levels per lineages of Alu in subsets of experiments. In each of the 4 subsections of the plot, the pertinent experiments contributed an average methylation level for all probes in proximity of a specific class of repetitive element indicated on the left. The values are summarized using a box-and-whisker plot. A star next to a family’s name indicates that the age of that family is estimated to be less than 25 MYA (Xing et al., 2004) or the time of origin of the baboon species. A double star indicates a human specific family (i.e. less than 4 MYO). Supplementary Table S2 contains detailed information about the ages of each of the subfamilies.
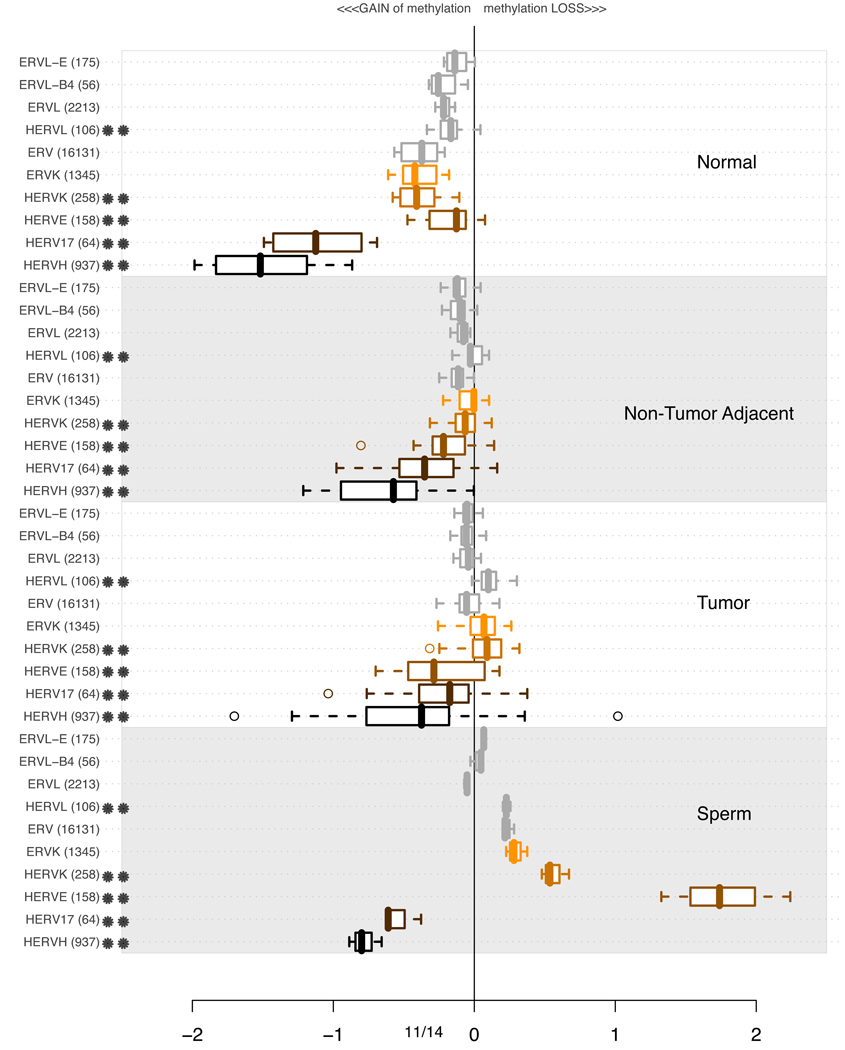
3.4 Methylation of ERV elements
Endogenous Retrovirus (ERV) Families are a heterogeneous group of sequences with over 60 lineages according to RepBase (Jurka, 1998). There are reports of ERV sequences being involved in extensive chromosomal rearrangement during the last 30 million years in primate evolution (Romano et al., 2006). Per-lineage analysis pertaining to the methylation pattern of ERV is presented in Figure 7. Similarly to MaLR and Alu discussed in 3.2 and 3.3, Human Endogeneous Retrovirus (HERV) families appear heavily methylated in the normal tissues. The gradual loss of methylation is apparent for HERVH and HERV17 families. To an extent, the methylation levels of HERVE and KERVK also vary among normal, tumors and non-tumoral adjacent tissues. So far, for MaLR, Alu and ERV families of ancient repetitive elements, predating the mammalian radiation, the microarray DNA methylation analysis suggests that young, primate specific lineages appear more susceptible to de-methylation in disease than other, older lineages.
Figure 7.
Distributions of average methylation levels per lineages of ERV in subsets of experiments. In each of the 4 subsections of the plot, the pertinent experiments contributed an average methylation level for all probes in proximity of a specific class of repetitive element indicated on the left. The values are summarized using a box-and-whisker plot. A double star indicates a primarily human lineage (Carter et al., 2004).
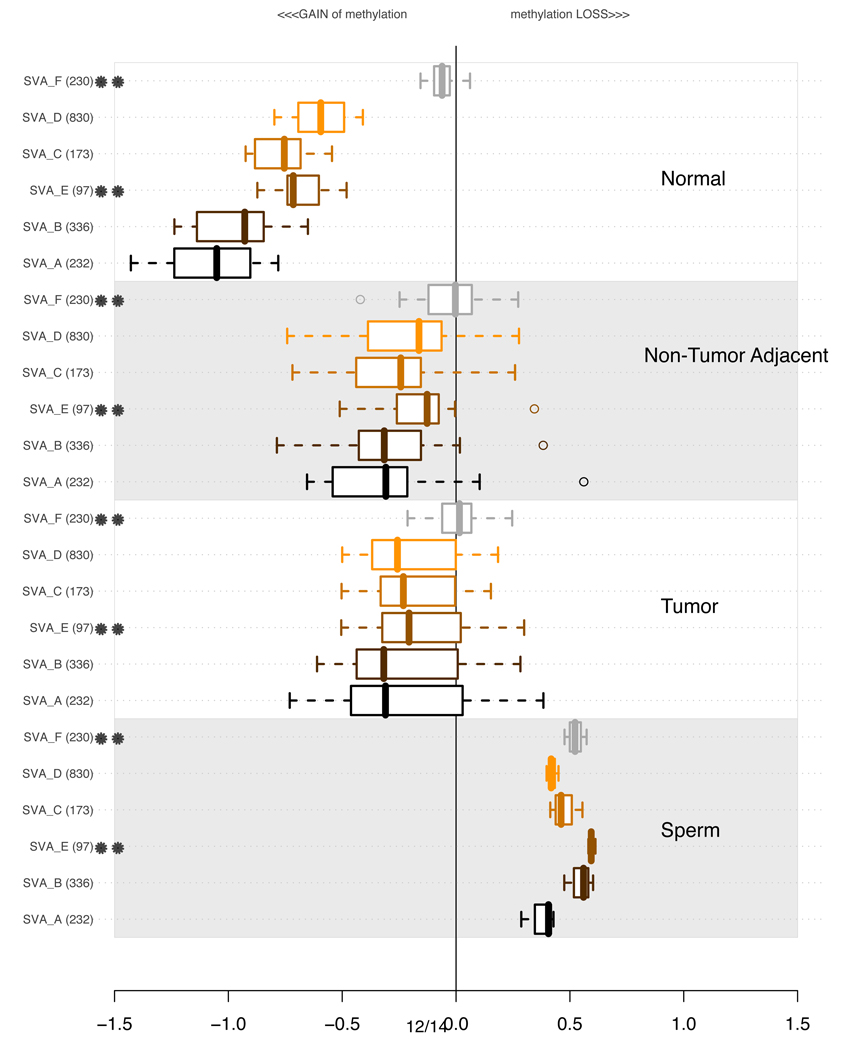
3.5 Methylation of SVA elements
We performed a similar analysis for SVA elements, which have been extensively mobilized in the human genome after the divergence of hominids from chimpanzees (Xing et al., 2007; Wang et al., 2007; Macfarlane and Simmonds, 2004). SVA elements consist of a combination of sequences derived from other retroelements (Babushok and Kazazian, 2007) and are known to be non-autonomous, depending on LINE-1 elements for mobilization. Wang et al. (2005) have estimated the evolutionary age of different subfamilies of SVA elements, named SVA-A through SVA-F. Our analysis reveals that the youngest SVA subfamilies show an unusual relationship between evolutionary age and the level of dysregulation, as shown in Figure 8. SVA-F elements, which are human specific, and only 3 MY old, are significantly less methylated than other, older subfamilies, and their methylation level does not change much in different samples, with the exception of sperm, where these elements show loss of methylation. On the other hand, the SVA-A elements, which are the oldest SVA subfamily (16.81 MY), are strongly methylated in normal oral tissues, but their loss of methylation is strikingly variable among different samples, with tumors showing the greatest level of variation. Thus, the magnitude and trends of DNA methylation changes for the youngest SVA elements seems to diverge from the patterns observed for AluY, MaLR, and ERV elements. The dramatic DNA methylation dysregulation affecting most SVA subfamilies in non-tumoral adjacent tissue is particularly striking.
Figure 8.
Distributions of average methylation levels per lineages of hominid specific SVA (Wang et al., 2005) in subsets of experiments. In each of the 4 subsections of the plot, the pertinent experiments contributed an average methylation level for all probes in proximity of a specific class of repetitive element indicated on the left. The values are summarized using a box-and-whisker plot. A double star indicates a human specific lineage (i.e. with age estimated at less than 4MY). Supplementary Table S2 contains more detailed information about the ages of each of the subfamilies.
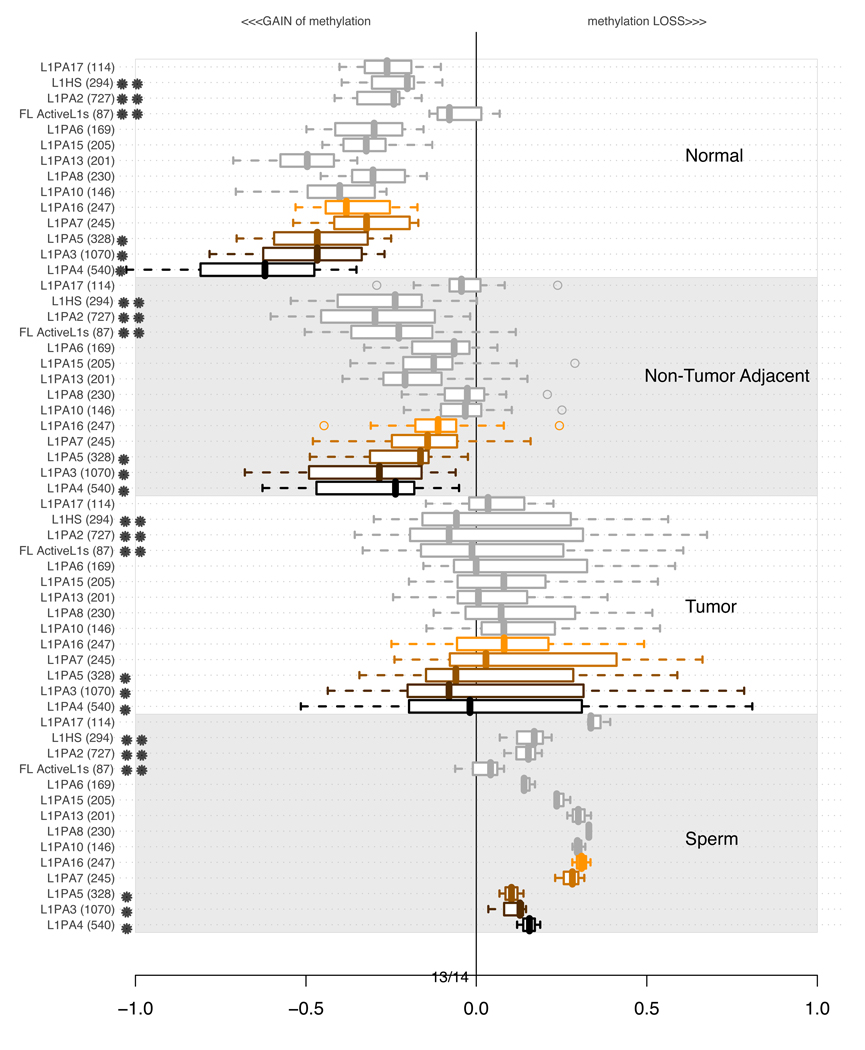
3.6 Methylation of LINE-1 elements
To expand our understanding of the LINE-1 family, we investigated lineages of this category. Figure 9 shows the categories which we could probe in at least 100 unique genomic loci. Comparing the values across the four classes of experiments, it is apparent (Figure 9) that younger, primate specific classes of LINE-1 elements (LINE-1PA3 (L1PA3) and LINE-1PA4 (L1PA4) and LINE-1PA5 (L1PA5), none of which exist in the baboon or marmoset) are more strongly methylated in normal tissue, and suffer more dramatic losses in DNA methylation in tumors and sperm. However, similarly to our observations for SVA subfamilies, the newest LINE-1 families that are strictly human specific (L1PA2, L1HS, Full Length active LINE-1 (Penzkofer et al., 2005)) are not as highly methylated in normal tissue, and not as dramatically demethylated in tumorigenesis as the longer-established lineages, the primate-specific LINE-1PA subfamilies.
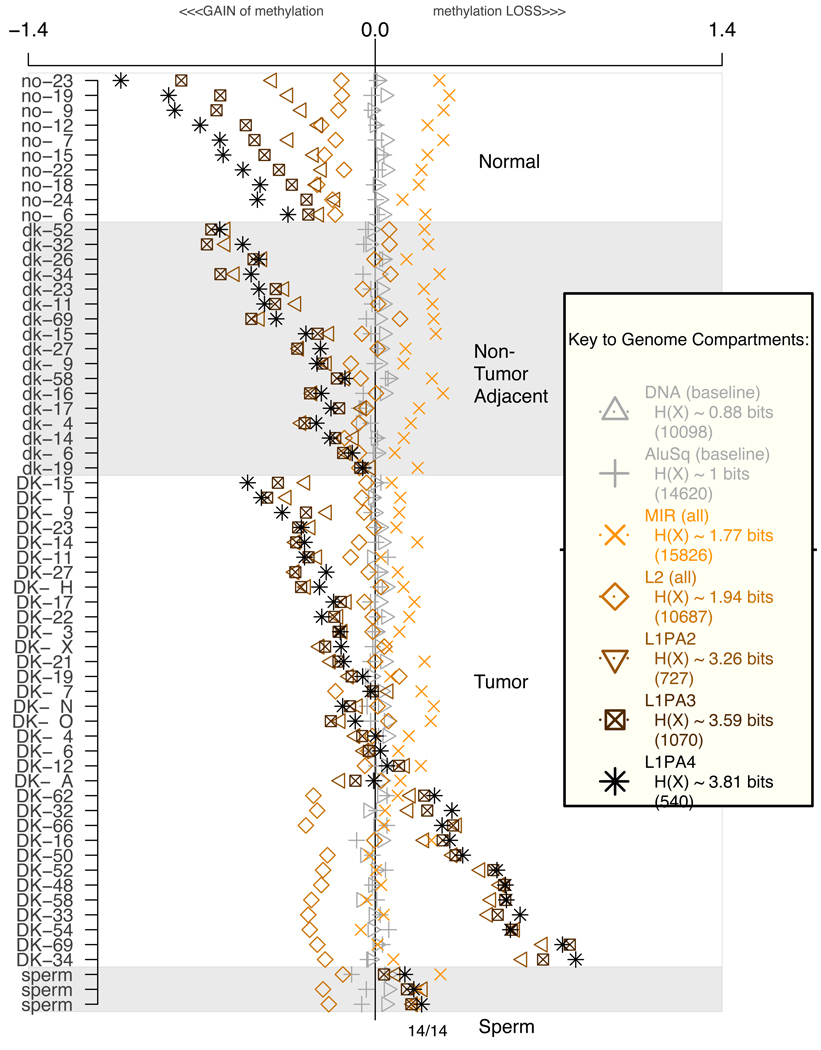
It is relevant to explore potential correlations or anti-correlations among the DNA methylation metrics within individual experiments. With this question in mind, Figure 10 reports methylation levels in individual experiments for the family of MIR repeats, as well as the family of L2 repeats, as compared to L1PA3 and L1PA4 methylation levels. The data illustrates completely distinct and sometime opposing trends in their levels of methylation, demonstrating that the observed metrics for the L1PA methylation levels are not due to normalization artifacts. In most cases the youngest members of each retrotransposon family are strongly methylated in normal buccal tissues, as shown by their negative values for all 10 samples from healthy adults. In tumors, the corresponding retroelement families shift dramatically to a relatively unmethylated state, as shown by the predominantly positive values. In the adjacent non-tumor tissue, the methylation level is variable, reflecting different degrees of epigenetic dysregulation in these tissues among different patients. These results underscore the fact that the methylation level of different families can be regulated (and dysregulated) independently in different tissues. Interestingly, our data also shows that for any member of a retroelement subfamily, the methylation level in tissues from different patients can vary within certain bounds depending on the genomic sequence context, while in the sperm experiment, which represents a single individual, the methylation levels for any given family converge to a distinct and strikingly narrow range of values, characteristic of each repeat family.
Figure 10.
Average methylation levels of repetitive element categories per experiment. Numbers in parenthesis indicate how many probes were averaged per experiment (See also Supplementary Section 3)
3.7 Analysis of relative CpG content among different repetitive element subfamilies
We explored the formal possibility that some of the differences in DNA methylation levels could be influenced by the CpG content of the DNA sequences being probed. This analysis involves analyzing the count of CpG residues in the repeats and the immediately surrounding sequences, as shown for a single repetitive element family in Supplementary Figure S11, and for all repetitive element families in Supplementary Figures S12 through S16. For example, an analysis of the CpG content of all classes of MaLR elements (see Supplementary Figure S12) shows that for those elements that we probed, the CpG content, as well as the frequency of endonuclease recognition sites is not noticeably different. MLT1C elements, which show much lower methylation changes relative to MSTA elements, have almost identical metrics of CpG count and endonuclease sites (see Supplementary Figure S17). In the case of SVA elements, the analysis in Supplementary Figure S18 shows that the CpG content, as well as the frequency of endonuclease recognition sites is noticeably higher for the SVA-F elements than for the SVA-B elements that we probed. Yet, the SVA-B elements, in spite of their somewhat lower frequency of potential endonuclease cutting sites, show more dramatic differences in methylation between normal samples and tumor samples relative to the SVA-F elements. A comparison of the CpG content of L1PA17 elements shows a higher content of CpGs and endonuclease sites within a +/− 400 base window of the probes, as compared to L1PA4 elements (Supplementary Figure S19), which show lower values for both metrics. Yet, it is the L1PA4 elements that show the greater changes in DNA methylation. The L1HS (human-specific) subfamily shows a somewhat higher frequency of endonuclease sites compared to the L1PA3 subfamily (Supplementary Figure S20), and yet the L1HS methylation levels change to a lower degree in different tissues. Analysis of the Alu elements show that the AluY subfamilies have a higher content of CpG residues and endonuclease sites compared to the relatively older AluJ and AluS subfamilies (Supplementary Figure S13). While these differences could partially contribute to the observed smaller changes in DNA methylation observed for the older elements, the differences cannot account for all observations in DNA methylation changes. It is important to note that, using different methodology, Rodriguez et al. (2008) obtained evidence suggesting that in normal colonic epithelia the older members of the Alu family are less methylated than the younger members. In our own analysis, the related AluYd8 and AluYb9 subfamilies have almost identical metrics for CpG content and endonuclease sites (Supplementary Figure S21), and yet show marked differences in their DNA methylation changes among normal tissues, tumors, and sperm. Taken together, these observations argue against differences in the content of CpG dinucleotides as a trivial explanation for the observed differences in DNA methylation levels among different families of repetitive elements.
3.8 Properties of probes capable of best distinguishing non-tumor adjacent tissue from tumor tissue
The foregoing analysis does not help us to identify events that could be tumor-specific. To address this issue, we ranked each individual probe associated with a repetitive element on the basis of its ability to differentiate tumors from non-tumoral adjacent tissue using a Wilcoxon test. We performed a statistical analysis involving those probes that displayed altered methylation, by calculating the probe values (ratios) in tumor samples, and the likelihood of random methylation changes as a function of the total number of probes belonging to any one family of repeats. The probes were ranked based on the P-values generated by a hypergeometric t-test, as shown in Supplementary Table S3. The entries with the most significant P-values include members of the LINE-1P, AluY, LTR, and SVA families of interspersed repeats. Among the primate-specific L1 elements, the L1PA3, L1PA2, and L1PA4 are among the most highly enriched. Among the LTR elements, the LTR7, LTR33, and HERV elements are high on the list. AluY represents the youngest family of Alu elements, and they rank much higher than older Alu elements. The HERV and SVA elements are among the few retrotransposon families known to have been extensively mobilized in the human genome after the divergence of hominids from chimpanzees (Xing et al., 2007; Wang et al., 2005; Macfarlane and Simmonds, 2004).
The data in Tables 1A and 1B summarizes salient properties of the subset of LINE-1 elements that we identify, using the Wilcoxon test, as the best DNA methylation probe variables for distinguishing tumors from non-paired non-tumoral adjacent tissue. In Table 1A, the column corresponding to relative enrichment of a set of elements shows that the highest value (4.757) corresponds to a subset of the L1PA4 subfamily. Members of the L1PA3 subfamily are also highly enriched among the most significant probes. The column specifying the median length of the elements shows that for L1PA5 and L1PA6 there is a noticeable increase in the length of the elements corresponding to the most significant probes (almost a 2-fold increase relative to all probed elements, in the case of L1PA6). A longer length could be associated with a higher likelihood of having an intact L1 promoter, as well as a higher probability of generating a full-length LINE-1 RNA transcriptional product. The table also shows enrichment of probes mapping to full-length L1 elements (FLI-L1) and ORF2-competent L1 elements (ORF2-L1, Jurka, 1998; Penzkofer, et al., 2005). L1PA4 elements, which are the most highly enriched among the significant probes, are unlikely to code for functional ORF2 proteins, and thus unlikely to generate reverse transcriptase. This observation suggests that possible positive selection in tumors for long L1 elements among the most significant probes is not operating at the level of conservation of ORF2 protein-coding function.
Table 1.
Table 1A.
Enrichment of significant probes in all probes associated with young L1P lineages. Highlighted in bold are the primate L1P lineages that appear in post-baboon species. The 15,587 probes are the most significant probes characterized in the Supplementary Table S3. Enrichment is calculated based on all 339,314 probes in the microarray. Hypergeometric test score is recorded as well. The two highest enrichment values and two highest p-values are highlighted in bold.
Table 1B.
A continuation of the table from A. For selected families of L1PAs discussed in Table 1A we tried to address the issue whether the statistically significant members (Supplementary table S3) of L1 are more homologous to a promoter region of an active i.e. intact and relatively young, full length Line1 element.
A consensus promoter region was obtained from one of the L1HS characterized by L1Base as full lentgth and active. To generate this table, the alignment of the 700 bases long promoter region was performed against all members of each lineages and against subset of significant members of each of the lineages. Software BLAT and parameters designated to result in 80% homology (-minIdentity=80 -tileSize=10) were used. The count of alignments per lineage was recorded in the table. The “%” columns show the percentage of all elements in the group (either all repeats or subset of significant repeats only) for which a BLAT alignment was found. Note that the percentage does not reflect on sequence conservation, but is a mere statement that an alignment using BLAT and the specified parameters is possible for a fraction of sequences in a group. Subsequently, a hypergeometric test was used to provide statistical significance between the count of alignments in an entire group and only significant members of the group.
| Table 1A. Enrichment of significant probes in all probes associated with young L1P lineages. | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Family | TOTAL GENOMIC | ALL PROBED | 15,587 Most Significant Probes | Length Selection |
||||||
| N | median length |
N | median length |
exp | obs | obs/ exp |
hyper-geo pval |
median length |
||
| FLI-L1 | 145 | 8,047 | 87 | 6,029 | 4 | 9 | 2.252 | 6.61E-03 | 6,028 | (1) |
| ORF2-L1 | 103 | 8,047 | 91 | 6,026 | 4 | 13 | 3.110 | 7.00E-05 | 6,026 | 0 |
| L1HS | 1696 | 987 | 294 | 6,025 | 14 | 43 | 3.184 | 7.25E-12 | 6,026 | 2 |
| L1PA2 | 4666 | 771 | 727 | 6,024 | 33 | 115 | 3.444 | 3.56E-31 | 6,023 | (1) |
| L1PA3 | 10281 | 952 | 1070 | 6,031 | 49 | 226 | 4.598 | 2.19E-83 | 6,031 | 1 |
| L1PA4 | 11462 | 713 | 540 | 6,130 | 25 | 118 | 4.757 | 2.80E-46 | 6,132 | 2 |
| L1PA5 | 10904 | 623 | 328 | 5,659 | 15 | 46 | 3.053 | 6.91E-12 | 6,115 | 457 |
| L1PA6 | 5617 | 888 | 169 | 2,600 | 8 | 13 | 1.675 | 2.46E-02 | 4,076 | 1,476 |
| L1PA7 | 12334 | 768 | 245 | 1,018 | 11 | 23 | 2.044 | 4.46E-04 | 469 | (549) |
| L1PA8 | 7715 | 394 | 159 | 328 | 7 | 20 | 2.738 | 1.44E-05 | 275 | (54) |
| L1PA8A | 2210 | 707 | 72 | 677 | 3 | 8 | 2.419 | 5.65E-03 | 499 | (178) |
| L1PA10 | 6962 | 474 | 146 | 676 | 7 | 8 | 1.193 | 2.30E-01 | 1,217 | 542 |
| L1PA11 | 3886 | 511 | 76 | 601 | 3 | 5 | 1.432 | 1.36E-01 | 1,278 | 678 |
| L1PA12 | 2320 | 679 | 59 | 644 | 3 | 3 | 1.107 | 2.87E-01 | 372 | (272) |
| L1PA13 | 10722 | 457 | 201 | 503 | 9 | 13 | 1.408 | 8.12E-02 | 532 | 29 |
| L1PA14 | 3561 | 558 | 66 | 557 | 3 | 7 | 2.309 | 1.06E-02 | 337 | (220) |
| L1PA15 | 8233 | 547 | 173 | 502 | 8 | 16 | 2.013 | 2.73E-03 | 450 | (52) |
| L1PA15-16 | 1201 | 727 | 32 | 331 | 1 | 3 | 2.041 | 5.75E-02 | 224 | (107) |
| L1PA16 | 13098 | 482 | 247 | 489 | 11 | 22 | 1.939 | 1.15E-03 | 488 | (1) |
| L1PA17 | 4722 | 368 | 114 | 368 | 5 | 4 | 0.764 | 6.05E-01 | 560 | 192 |
| Table 1B. Increase of L1HS promoter homology in L1P members with significantly altered methylation patterns. | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Family | TOTAL GENOMIC |
ALL PROBED | 15,587 Most Significant Probes |
Improvement in count of alignments |
Hyper- geo pval |
||||
| N | N | promoter alignments |
% | N | promoter alignments |
% | |||
| FLI-L1 | 145 | 87 | 86 | 98.85% | 9 | 9 | 100.00% | 1.15% | 0.00E+00 |
| L1HS | 1696 | 294 | 280 | 95.24% | 43 | 43 | 100.00% | 4.76% | 0.00E+00 |
| L1PA2 | 4666 | 727 | 672 | 92.43% | 115 | 111 | 96.52% | 4.09% | 1.53E-02 |
| L1PA3 | 10281 | 1070 | 972 | 90.84% | 226 | 218 | 96.46% | 5.62% | 8.39E-05 |
| L1PA4 | 11462 | 540 | 429 | 79.44% | 118 | 107 | 90.68% | 11.23% | 7.57E-05 |
| L1PA5 | 10904 | 328 | 178 | 54.27% | 46 | 37 | 80.43% | 26.17% | 1.62E-05 |
| L1PA6 | 5617 | 169 | 62 | 36.69% | 13 | 8 | 61.54% | 24.85% | 1.41E-02 |
| L1PA7 | 12334 | 245 | 32 | 13.06% | 23 | 3 | 13.04% | (0.02%) | 3.25E-01 |
| L1PA8 | 7715 | 159 | 8 | 5.03% | 20 | 0 | 0.00% | (5.03%) | 6.68E-01 |
An additional level of analysis, shown in the Table 1B involved measurement of the level of homology of sequences near the 5’ end of each L1 sequence with an exemplar sequence represented by the first 700 bases of an active LINE-1 element of the class FLI-L1, which contain an active promoter. Using BLAT (Kent, 2002), we selected those 5’-end sequences of different subclasses of L1PA elements scoring with a homology of 80% or better. The table shows that among the L1PA elements present in the subset of the 15,587 most significant probes, there is a much higher percentage of sequences with good homology matches to the active L1 exemplar. This suggests that possible selection in tumors for demethylated L1 elements could involve specific features of the sequence at the 5’-end of the element, which harbor potential forward promoter as well as antisense promoter activity (discussed in section 3.9, below). If, for any given class of elements (i.e. L1PA5), a potentially active promoter exists, it may be more likely to be associated with a full-length L1PA5 elements. Along this line of thought, the apparent length-selection could be a by-product of functional promoter selection.
3.9 Possible functional significance of the enrichment of demethylated LINE-1 elements in tumors
The simplest interpretation of the age-stratified dysregulation DNA methylation of repetitive DNA observed among normal tissue, non-tumoral adjacent tissue and tumors is that the younger members of repetitive DNA families are the most likely to be transcribed, and that these RNA transcripts are best able in normal cells to trigger RNA-directed chromatin silencing. Silencing efficiency would be additionally enhanced in the younger elements due to reduced sequence divergence, as recently proposed by Reiss and Mager (2007). Paradoxically, for the very youngest members of retrotransposon families, exemplified in our data set by SVA-F and full-length, active LINE-1s, the emergence of optimal silencing may still remain incomplete, for lack of sufficient evolutionary time for RNA-mediated silencing traits to be selected and fixed. Such a hypothesis could explain why the very youngest, human specific retotransposon families are relatively under-methylated in normal tissue, as compared to their relatively older and more “mature” primate siblings. It has been reported that heterochromatic piRNA loci interact with potentially active transposons in Drosophila resulting in transposon control (Brennecke et al., 2007). Normal transcriptional events involving retrotransposon sequences occur in human oocytes (Georgiu et al, 2009) an are well documented in the murine germ-line, where DNA is transiently demethylated, and where piRNAs have been implicated in reestablishing silencing (Aravin et al., 2007, 2008, Kuramochi-Miyagawa, 2008). Unfortunately, our understanding of the evolutionary history of piRNAs remains extremely limited, particularly with regards to the mechanism responsible for generation of new functional piRNA sequences, as novel subclasses of retrotransposons enter the genome.
In another plausible scenario, the most recently evolved repetitive elements will have accumulated fewer mutations or truncations deleterious to their function, and their selective loss of epigenetic silencing could be associated with functions that increase the fitness of tumors, therefore subjecting to positive selection. An example of such a function would be the transcriptional activation of genes with oncogenic potential as a result of loss of methylation of cryptic promoter or enhancer sequences within a full-length retrotransposon. For example, Roman-Gomez et al (2005) reported that L1 hypomethylation led to activation of c-MET gene transcription driven by an L1 antisense promoter (Speek, 2001, Nigumann et al, 2002) located within intron one of the c-MET gene in patients with blast crisis chronic myeloid leukemia (BC-CML), where these transcriptional events may contribute to disease progression. More recently, Lin et al. (2006) reported the induction of an abnormal chimeric transcript in esophageal adenocarcinomas, initiated from the antisense promoter located in the 5’-UTR of a full-length LINE-1 element. Another function that could be subject to positive selection in tumor cell lineages is the transcriptional activation of a retrotransposon ORF coding for a reverse transcriptase. It has been reported that the reverse transcriptase inhibitor efavirenz antagonizes the growth of H69 human small-cell lung carcinomas in nude mice (Sinibaldi-Vallebona et al., 2005). The same group has recently reported that inhibition of the reverse transcriptase messenger RNA of LINE-1 elements or HERV-K elements leads to loss of tumorigenic potential in cell lines (Oriccio et al., 2007). Of course, an important caveat is that the reported occurrence in cancer cells of transcripts or proteins derived from retrotransposons could be merely coincidental, not causal.
An interesting functional hypothesis regarding L1 retrotransposon sequences is the possible unselfish participation of expressed and reverse-transcribed LINE-1 elements in nonstandard DNA double strand break repair in the context of oncogenesis, where normal repair mechanisms are disrupted (Helleday et al., 2007). Repair of double-strand breaks by gene conversion involving different endogenous LINE-1 elements has been reported in the mouse (Tremblay et al., 2000). DNA repair by endonuclease-independent LINE-1 retrotransposition was first reported by Morrish et al. (2002, see commentary by Eickbush, 2002) using a model reporter vector transfected into CHO cells. This pathway was found to be dependent on reverse transcriptase activity, and resulted in integration of a truncated LINE-1 sequence lacking target site duplications. Recently Sen et al. (2007) characterized sites in the human genome where L1 elements have integrated without signs of endonuclease-related activity, and found that the structural features of these loci suggested that they arose by double-strand break repair, resulting in translocations or deletions. Also relevant are the findings of Srikanta et al (2009), who scanned the human, chimpanzee, and rhesus macaque genomes, and reported 23 instances of Alu integration events most likely mediated by endonuclease-independent DNA repair (EIDR). Observations of truncated LINE-1 insertions in the context of physiological stress have been reported in two mouse models, lambda-MYC lymphomas and endogenous oxidative stress caused by deficient G6PD expression. In these two models (Rockwood et al, 2004), the LINE-1 insertions, plausibly generated by the EIDR mechanism, have been captured within a chromosomally integrated lac-Z reporter vector. The observed insertions represent predominantly incomplete elements, and their frequency (25% of all events) is higher than the frequency of LINE-1 sequences in the mouse genome (10%).
Additional experimental work will be required to assess unambiguously whether or not EIDR involving LINE-1 and Alu elements is ubiquitous in human cancer cells, and whether or not it has adaptative value, possibly enhancing the viability of DNA repair-deficient tumor cells. More experimental work will also be required to discover additional examples of activation of L1 antisense promoters capable of driving abnormal expression of neighboring proto-oncogenes or regulatory transcripts such as microRNAs or large noncoding RNAs (Guttman et al., 2009). The rapid rate of progress in high-throughput, low cost DNA sequencing will soon make it possible to sequence a large number of human tumor genomes to elucidate the sequences found at sites of genomic rearrangements, insertions, and deletions (CGP, 2009). Emerging genome analysis tools will also facilitate the design of experiments to assess the potential adaptative value of EIDR mediated by retroelements. A fruitful line of inquiry for the future will be the study of relationships between retrotransposon sequences and specific RNA molecules involved in localized epigenetic silencing. We await the discovery of detailed mechanisms underlying the machinery that controls retrotransposon-silencing functions in somatic cells of mammals, which would provide an experimental handle to elucidate the mechanisms leading to differential methylation or repeats during the micro-evolution of tumor progenitor cell lineages.
An important question not addressed in this study is the potential contribution of disruption of maintenance DNA methylation in cancer tissues. This question has not been systematically studied for most human tumor tissues, yet it should be pointed out that the abnormally hypermethylated status of many tumor suppressor genes is propagated faithfully in human cancers as well as cancer cell lines. Whether or not the maintenance of DNA methylation of retrotransposons is governed by independent mechanisms, and whether our observations are due to failure of maintenance of retrotransposon methylation during the multiple divisions undergone by cancer cells, remains a subject for future studies.
4. Conclusions
A novel microarray method for analysis of DNA methylation, based on the use of methylation sensitive as well as methylation dependent endonucleases, enables the interrogation of methylation levels in all compartments of the genome, including repetitive elements. Analysis of a substantial set of samples of squamous carcinomas of the head and neck, as well as non-tumor adjacent tissue and normal controls, reveals a complex framework of epigenetic dysregulation, where loss of methylation differentially affect distinct families of repetitive elements. Predominantly the younger, primate-specific members of retroelement families suffer the most dramatic loss of methylation, with the exception of some extremely young, human-specific retroelements. These complex patterns of differential susceptibility to disruption of silencing are probably a result of the natural history of evolutionary domestication of retroelements in genomes, in interplay with a minimal time requirement for strong silencing to be established. Primate-specific subfamilies of LINE-1 elements appear to suffer a particularly pronounced loss of methylation in tumors, with the most dramatic changes apparently observed for those primate retroelements with conserved promoter regions and longer sequences. Whether these special epigenetic alterations of primate-specific LINE-1 elements could implicate potential functions of these elements in cancerogenesis, or merely reflect the abnormal dynamics of establishment and maintenance of silencing of this class of elements in tumors remains a fascinating subject for future work.
Supplementary Material
Acknowledgments
This work was supported by a pilot grant from the Yale Cancer Center and by NIH grant No. R21 CA116079-01, awarded to PML.
Abbreviations
- DSB
double strand break
- EIDR
endonuclease-independent DNA repair
- ERV
endogenous retrovirus
- HERV
human endogenous retrovirus
- HNSCC
head and neck squamous cell carcinoma
- LINE
long interspersed nuclear element
- LTR
long terminal repeat
- MaLR
mammalian apparent LTR retrotransposons
- MDA
multiple displacement amplification
- MPSS
massively parallel sequencing
- ORF
open reading frame
- SINE
short interspersed nuclear element
- SVA
SINE-VNTR-Alu
- VNTR
variable-number tandem repeat
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alves PM, Lévy N, Stevenson BJ, Bouzourene H, Theiler G, Bricard G, Viatte S, Ayyoub M, Vuilleumier H, Givel JC, Rimoldi D, Speiser DE, Jongeneel CV, Romero PJ, Lévy F. Identification of tumor-associated antigens by large-scale analysis of genes expressed in human colorectal cancer. Cancer Immun. 2008;8:11. [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babushok DV, Kazazian HH. Progress in understanding the biology of the human mutagen LINE-1. Hum Mutat. 2007;28:527–539. doi: 10.1002/humu.20486. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Research. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Büscher K, Hahn S, Hofmann M, Trefzer U, Ozel M, Sterry W, Löwer J, Löwer R, Kurth R, Denner J. Expression of the human endogenous retrovirus-K transmembrane envelope, Rec and Np9 proteins in melanomas and melanoma cell lines. Melanoma Res. 2006;16:223–234. doi: 10.1097/01.cmr.0000215031.07941.ca. [DOI] [PubMed] [Google Scholar]
- Carter AB, Salem AH, Hedges DJ, Keegan CN, Kimball B, Walker JA, Watkins WS, Jorde LB, Batzer MA. Genome-wide analysis of the human Alu Yb-lineage. Hum Genomics. 2004;1:167–178. doi: 10.1186/1479-7364-1-3-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CGP. Cancer Genome Project. 2009 http://www.sanger.ac.uk/genetics/CGP/
- Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, Driscoll M, Song W, Kingsmore SF, Egholm M, Lasken RS. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci U S A. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush TH. Repair by retrotransposition. Nature Genet. 2002;31:126–127. doi: 10.1038/ng897. [DOI] [PubMed] [Google Scholar]
- Florl AR, Löwer R, Schmitz-Dräger BJ, Schulz WA. DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Br J Cancer. 1999;80:1312–1321. doi: 10.1038/sj.bjc.6690524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiou I, Noutsopoulos D, Dimitriadou E, Markopoulos G, Apergi A, Lazaros L, Vaxevanoglou T, Pantos K, Syrrou M, Tzavaras T. Retrotransposon RNA expression and evidence for retrotransposition events in human oocytes. Hum Mol Genet. 2009;18:1221–1228. doi: 10.1093/hmg/ddp022. [DOI] [PubMed] [Google Scholar]
- Golan M, Hizi A, Resau JH, Yaal-Hahoshen N, Reichman H, Keydar I, Tsarfaty I. Human endogenous retrovirus (HERV-K) reverse transcriptase as a breast cancer prognostic marker. Neoplasia. 2008;10:521–533. doi: 10.1593/neo.07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006 doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy J, Thomas E, Schwartz J, Wigler M. Annotating large genomes with exact word matches. Genome Res. 2003;13:2306–2315. doi: 10.1101/gr.1350803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst) 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Herbst H, Sauter M, Kühler-Obbarius C, Löning T, Mueller-Lantzsch N. Human endogenous retrovirus (HERV)-K transcripts in germ cell and trophoblastic tumours. APMIS. 1998;106:216–220. doi: 10.1111/j.1699-0463.1998.tb01338.x. [DOI] [PubMed] [Google Scholar]
- Jurka J. A new subfamily of recently retroposed human Alu repeats. Nucleic Acids Research. 1993;21:2252. doi: 10.1093/nar/21.9.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. Repeats in genomic DNA: mining and meaning. Curr Opin Struct Biol. 1998;8:333–337. doi: 10.1016/s0959-440x(98)80067-5. [DOI] [PubMed] [Google Scholar]
- Kapitonov V, Jurka J. The age of Alu subfamilies. J Mol Evol. 1996;42:59–65. doi: 10.1007/BF00163212. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lage JM, Lizardi PM. Introduction to Whole genome Amplification. In: Hughes S, Lasken R, editors. Whole Genome Amplification: Methods Express. Scion Publishing Limited; 2005. [Google Scholar]
- Lage JM, Leamon JH, Pejovic T, Hamann S, Lacey M, Dillon D, Segraves R, Vossbrinck B, González A, Pinkel D, Albertson DG, Costa J, Lizardi PM. Whole genome analysis of genetic alterations in small DNA samples using hyperbranched strand displacement amplification and array-CGH. Genome Res. 2003;13:294–307. doi: 10.1101/gr.377203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Ying S. Gene silencing in vitro and in vivo using intronic microRNAs. Methods Mol Biol. 2006;342:295–312. doi: 10.1385/1-59745-123-1:295. [DOI] [PubMed] [Google Scholar]
- Löwer R, Löwer J, Frank H, Harzmann R, Kurth R. Human teratocarcinomas cultured in vitro produce unique retrovirus-like viruses. J Gen Virol. 1984;65:887–898. doi: 10.1099/0022-1317-65-5-887. [DOI] [PubMed] [Google Scholar]
- Macfarlane C, Simmonds P. Allelic variation of HERV-K(HML-2) endogenous retroviral elements in human populations. J Mol Evol. 2004;59:642–656. doi: 10.1007/s00239-004-2656-1. [DOI] [PubMed] [Google Scholar]
- Mandal PK, Kazazian HH. SnapShot: Vertebrate transposons. Cell. 2008;135:192–192. doi: 10.1016/j.cell.2008.09.028. e1. [DOI] [PubMed] [Google Scholar]
- Menendez L, Benigno BB, McDonald JF. L1 and HERV-W retrotransposons are hypomethylated in human ovarian carcinomas. Molecular Cancer. 2004;3:12. doi: 10.1186/1476-4598-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- Muster T, Waltenberger A, Grassauer A, Hirschl S, Caucig P, Romirer I, Födinger D, Seppele H, Schanab O, Magin-Lachmann C, Löwer R, Jansen B, Pehamberger H, Wolff K. An endogenous retrovirus derived from human melanoma cells. Cancer Res. 2003;63:8735–8741. [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- Nigumann P, Redik K, Mätlik K, Speek M. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics. 2002;79:628–634. doi: 10.1006/geno.2002.6758. [DOI] [PubMed] [Google Scholar]
- Oricchio E, Sciamanna I, Beraldi R, Tolstonog G, Schumann G, Spadafora C. Distinct roles for LINE-1 and HERV-K retroelements in cell proliferation, differentiation and tumor progression. Oncogene. 2007;26:4226–4233. doi: 10.1038/sj.onc.1210214. [DOI] [PubMed] [Google Scholar]
- Pace JK, Feschotte C. The evolutionary history of human DNA transposons: evidence for intense activity in the primate lineage. Genome Res. 2007;17:422–432. doi: 10.1101/gr.5826307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzke S, Lindeskog M, Munthe E, Aasheim HC. Characterization of a novel human endogenous retrovirus, HERV-H/F, expressed in human leukemia cell lines. Virology. 2002;303:164–173. doi: 10.1006/viro.2002.1615. [DOI] [PubMed] [Google Scholar]
- Penzkofer T, Dandekar T, Zemojtel T. L1Base: from functional annotation to prediction of active LINE-1 elements. Nucleic Acids Research. 2005;33:D498–D500. doi: 10.1093/nar/gki044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss D, Mager DL. Stochastic epigenetic silencing of retrotransposons: does stability come with age? Gene. 2007;390:130–135. doi: 10.1016/j.gene.2006.07.032. [DOI] [PubMed] [Google Scholar]
- Rockwood LD, Felix K, Janz S. Elevated presence of retrotransposons at sites of DNA double strand break repair in mouse models of metabolic oxidative stress and MYC-induced lymphoma. Mutat Res. 2004;548:117–125. doi: 10.1016/j.mrfmmm.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Vives L, Jordà M, Morales C, Muñoz M, Vendrell E, Peinado MA. Genome-wide tracking of unmethylated DNA Alu repeats in normal and cancer cells. Nucleic Acids Res. 2008;36:770–784. doi: 10.1093/nar/gkm1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman-Gomez J, Jimenez-Velasco A, Agirre X, Cervantes F, Sanchez J, Garate L, Barrios M, Castillejo JA, Navarro G, Colomer D, Prosper F, Heiniger A, Torres A. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24:7213–7223. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- Romano CM, Ramalho RF, Zanotto PM. Tempo and mode of ERV-K evolution in human and chimpanzee genomes. Arch Virol. 2006;151:2215–2228. doi: 10.1007/s00705-006-0792-1. [DOI] [PubMed] [Google Scholar]
- Sen SK, Huang CT, Han K, Batzer MA. Endonuclease-independent insertion provides an alternative pathway for L1 retrotransposition in the human genome. Nucleic Acids Research. 2007;35:3741–3751. doi: 10.1093/nar/gkm317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafino A, Balestrieri E, Pierimarchi P, Matteucci C, Moroni G, Oricchio E, Rasi G, Mastino A, Spadafora C, Garaci E, Vallebona PS. The activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp Cell Res. 2009;315:849–862. doi: 10.1016/j.yexcr.2008.12.023. [DOI] [PubMed] [Google Scholar]
- Sinibaldi-Vallebona P, Lavia P, Garaci E, Spadafora C. A role for endogenous reverse transcriptase in tumorigenesis and as a target in differentiating cancer therapy. Genes Chromosom. Cancer. 2005;45:1–10. doi: 10.1002/gcc.20266. [DOI] [PubMed] [Google Scholar]
- Smit AF. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Research. 1993;21:1863–1872. doi: 10.1093/nar/21.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF, Hubley R, Green P. RepeatMasker Open-3.0. 1996–2004 < http://www.repeatmasker.org>.
- Smith IM, Mydlarz WK, Mithani SK, Califano JA. DNA global hypomethylation in squamous cell head and neck cancer associated with smoking, alcohol consumption and stage. Int J Cancer. 2007;121:1724–1728. doi: 10.1002/ijc.22889. [DOI] [PubMed] [Google Scholar]
- Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol. 2001;21:1973–1985. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanta D, Sen SK, Huang CT, Conlin EM, Rhodes RM, Batzer MA. An alternative pathway for Alu retrotransposition suggests a role in DNA double-strand break repair. Genomics. 2009;93:205–212. doi: 10.1016/j.ygeno.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer Y, Theiler G, Sperisen P, Lebedev Y, Jongeneel CV. Digital expression profiles of human endogenous retroviral families in normal and cancerous tissues. Cancer Immun. 2004;4:2. [PubMed] [Google Scholar]
- Suter CM, Martin DI, Ward RL. Hypomethylation of L1 retrotransposons in colorectal cancer and adjacent normal tissue. Int J Colorectal Dis. 2004;19:95–101. doi: 10.1007/s00384-003-0539-3. [DOI] [PubMed] [Google Scholar]
- Takai D, Jones P. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teneng I, Stribinskis V, Ramos KS. Context-specific regulation of LINE-1. Genes Cells. 2007;12:1101–1110. doi: 10.1111/j.1365-2443.2007.01117.x. [DOI] [PubMed] [Google Scholar]
- Tremblay A, Jasin M, Chartrand P. A double-strand break in a chromosomal LINE element can be repaired by gene conversion with various endogenous LINE elements in mouse cells. Mol Cell Biol. 2000;20:54–60. doi: 10.1128/mcb.20.1.54-60.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Xing J, Grover D, Hedges D, Han K, Walker J, Batzer M. SVA elements: a hominid-specific retroposon family. J Mol Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- Wang-Johanning F, Frost AR, Jian B, Azerou R, Lu DW, Chen DT, Johanning GL. Detecting the expression of human endogenous retrovirus E envelope transcripts in human prostate adenocarcinoma. Cancer. 2003;98:187–197. doi: 10.1002/cncr.11451. [DOI] [PubMed] [Google Scholar]
- Wang-Johanning F, Frost AR, Johanning GL, Khazaeli MB, LoBuglio AF, Shaw DR, Strong TV. Expression of human endogenous retrovirus k envelope transcripts in human breast cancer. Clin Cancer Res. 2001;7:1553–1560. [PubMed] [Google Scholar]
- Wang-Johanning F, Liu J, Rycaj K, Huang M, Tsai K, Rosen DG, Chen DT, Lu DW, Barnhart KF, Johanning GL. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int J Cancer. 2007;120:81–90. doi: 10.1002/ijc.22256. [DOI] [PubMed] [Google Scholar]
- Xing J, Hedges D, Han K, Wang H, Cordaux R, Batzer M. Alu element mutation spectra: molecular clocks and the effect of DNA methylation. J Mol Biol. 2004;344:675–682. doi: 10.1016/j.jmb.2004.09.058. [DOI] [PubMed] [Google Scholar]
- Xing J, Witherspoon DJ, Ray DA, Batzer MA, Jorde LB. Mobile DNA elements in primate and human evolution. Am J Phys Anthropol. 2007 Suppl 45:2–19. doi: 10.1002/ajpa.20722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.